
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 13 September 2019
Sec. Microbial Physiology and Metabolism
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.02140
This article is part of the Research Topic Fundamentals of, and Applications Based on, Quorum Sensing and Quorum Sensing Interference View all 21 articles
Streptococcus mutans, a bacterium with high cariogenic potential, coordinates competence for natural transformation and bacteriocin production via the XIP and CSP pheromones. CSP is effective in inducing bacteriocin responses but not competence in chemically defined media (CDM). This is in contrast to XIP, which is a strong inducer of competence in CDM but can also stimulate bacteriocin genes as a late response. Interconnections between the pathways activated by the two pheromones have been characterized in certain detail in S. mutans UA159, but it is mostly unknown whether such findings are representative for the species. In this study, we used bioassays based on luciferase reporters for the bacteriocin gene cipB and the alternative sigma factor sigX to investigate various S. mutans isolates for production and response to CSP and XIP pheromones in CDM. Similar to S. mutans UA159, endogenous CSP was undetectable in the culture supernatants of all tested strains. During optimization of the bioassay using the cipB reporter, we discovered that the activity of exogenous CSP used as a standard was reduced over time during S. mutans growth. Using a FRET-CSP reporter peptide, we found that S. mutans UA159 was able to degrade CSP, and that such activity was not significantly different in isogenic mutants with deletion of the protease gene htrA or the competence genes sigX, oppD, and comR. CSP cleavage was also detected in all the wild type strains, indicating that this is a conserved feature in S. mutans. For the XIP pheromone, endogenous production was observed in the supernatants of all 34 tested strains at peak concentrations in culture supernatants that varied between 200 and 26000 nM. Transformation in the presence of exogenous XIP was detected in all but one of the isolates. The efficiency of transformation varied, however, among the different strains, and for those with the highest transformation rates, endogenous XIP peak concentrations in the supernatants were above 2000 nM XIP. We conclude that XIP production and inducing effect on transformation, as well as the ability to degrade CSP, are conserved functions among different S. mutans isolates. Understanding the functionality and conservation of pheromone systems in S. mutans may lead to novel strategies to prevent or treat unbalances in oral microbiomes that may favor diseases.
Natural genetic transformation is widely distributed in bacteria. In streptococci it occurs during a genetically programmed differentiated state called competence. During this state the bacteria become capable of taking up DNA from the environment and incorporate it into their genomes. The capacity for natural transformation has been reported for more than 80 bacterial species (Johnsborg et al., 2007; Johnston et al., 2014). Among streptococci, competence for natural transformation includes most species of the mitis, salivarius, bovis, anginosus, and mutans groups (Johnsborg et al., 2007; Fontaine et al., 2010; Desai et al., 2012; Morrison et al., 2013). The core of the machinery necessary for streptococcal transformation relies on the transcription of the conserved alternative sigma factor SigX, also known as ComX. SigX orchestrates a core response in streptococcal species characterized by the induction of 27 to 30 genes (Khan et al., 2016). The functions of the core genes are predominantly related to transformation, most of them coding for competence effector proteins for DNA binding, uptake and recombination (Li et al., 2001; Mashburn-Warren et al., 2010; Khan et al., 2016).
Streptococcus mutans is a member of the mutans group and part of the human oral microbiota. Environmental factors disturbing the ecological balance in the oral cavity, such as increased sugar intake, may favor S. mutans growth. The S. mutans acidogenic and aciduric properties may then contribute to tooth demineralization and dental caries (Marsh et al., 2011). Natural transformation was first reported in S. mutans in 1981 (Perry and Kuramitsu, 1981). This and later studies showed that transformation in S. mutans, at least in the laboratory setting, is restricted to a limited range of strains and depends on environmental conditions not yet fully understood (Perry and Kuramitsu, 1981). Studies investigating the S. mutans pan-genome have recently revealed the ubiquity of sigX and competence effector genes in S. mutans (Cornejo et al., 2013; Palmer et al., 2013; Song et al., 2013), indicating that competence may be a conserved feature in S. mutans. Moreover, the extensive horizontal gene transfer observed in the genomes of S. mutans clinical isolates (Cornejo et al., 2013) indicates that transformation occurs in their natural habitat and may be a widespread feature in the species.
In S. mutans the competent state is triggered by two linear peptides, the CSP (competence stimulating peptide) (Li et al., 2001) and the XIP (sigX-inducing peptide) (Figure 1; Mashburn-Warren et al., 2010). CSP was the first described pheromone and, until recently, the only one known to activate the competence system for genetic transformation in S. mutans. The comC gene, which encodes CSP, is downstream of the comDE operon, encoding the ComD histidine kinase and the ComE response regulator. At least in the synthetic form, CSP is thought to bind to the ComD histidine kinase of the ComDE two-component system, leading to phosphorylation of the ComE response regulator (Peterson et al., 2004). Phosphorylated ComE directly up-regulates the transcription of clusters of bacteriocin-related genes (Petersen et al., 2006; Mashburn-Warren et al., 2010; Federle and Morrison, 2012; Khan et al., 2016). These include among others the SMU_1914 gene (also known as nlmC and cipB), encoding mutacin V, and the putative immunity protein SMU_1913, found in a comE downstream region. Mutacin V has been proposed to link the early CSP bacteriocin-inducing response to the competence response (Ween et al., 1999; van der Ploeg, 2005; Kreth et al., 2007; Dufour et al., 2011; Hung et al., 2011) by mechanisms that remain elusive. Essential for competence development is the activation of the XIP pheromone encoding gene, comS, followed by the upregulation of the alternative sigma factor SigX, the master core regulator of competence. In rich media, ComS seems to be processed to XIP inside the cells. Intracellular XIP and/or ComS then binds to ComR to activate the competence pathway (Underhill et al., 2018). The predicted CSP is in general conserved among S. mutans strains (Li et al., 2001; Petersen et al., 2006), and there are indications that S. mutans may form a single CSP pherotype (Petersen et al., 2006; Hossain and Biswas, 2012). The C-terminal of the CSP precursor (ComC) is exported outside of the cells, where it is further processed into the active 18 amino acid peptide (18-CSP) (Petersen et al., 2006; Hossain and Biswas, 2012).
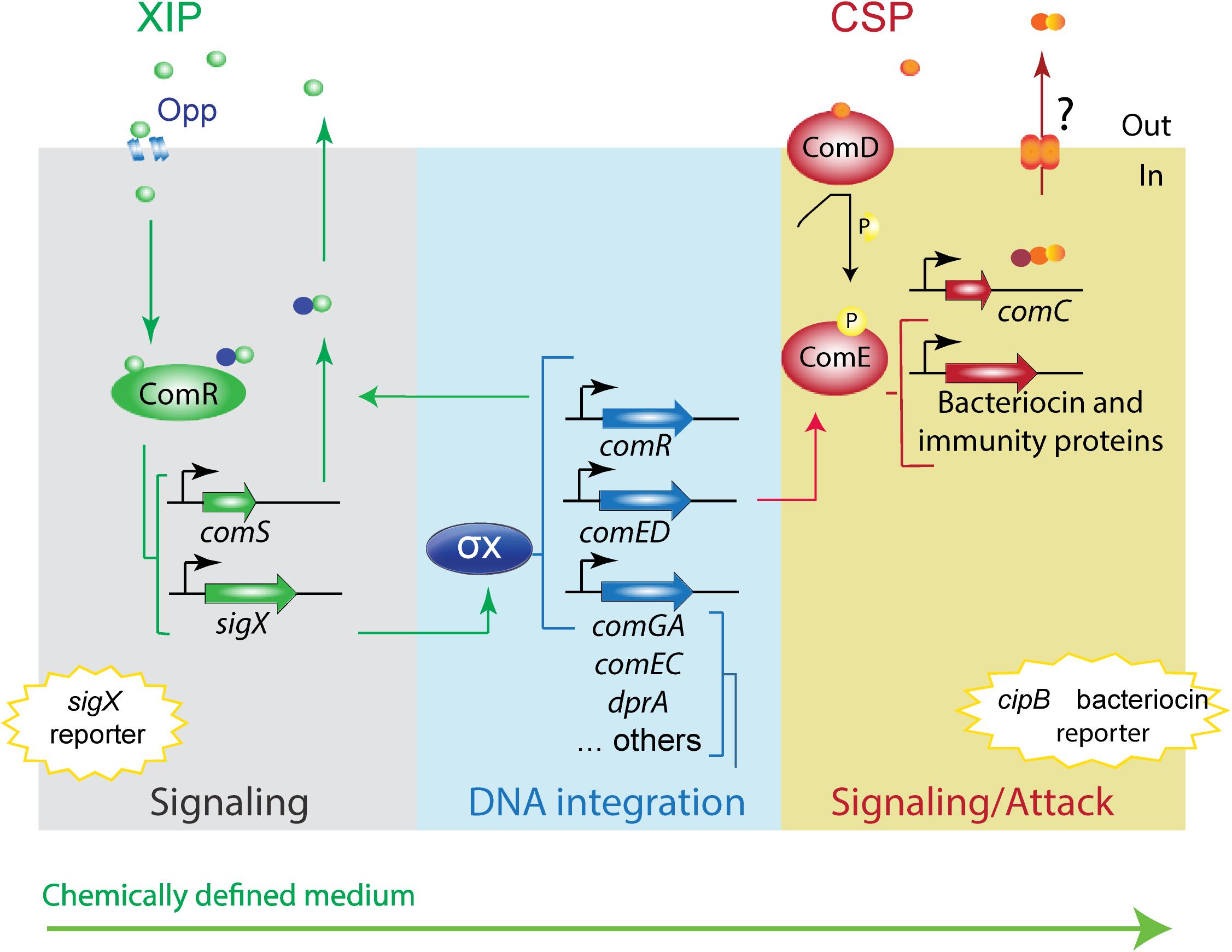
Figure 1. Model of S. mutans XIP pheromone signaling pathway in peptide-free chemically defined medium. The model is based on the UA159 S. mutans strain. The S. mutans XIP pheromone is exported outside of the cells. Upon reaching a certain threshold, XIP is internalized via the Opp permease, and binds to the ComR regulator to activate comS, thus creating a positive feedback loop. ComS is the precursor of XIP but can also function in its unprocessed form as an internal signal to activate the ComR-pheromone signaling pathway. The ComR-pheromone complex activates sigX. This codes for the alternative sigma factor X (σX). σX is the master regulator of competence for natural transformation that regulates the expression of the streptococcal core set of genes involved in the competence response, most of them with functions that enable DNA acquisition and integration into its genome. σX induces also comR (Khan et al., 2017) and comED (Reck et al., 2015; Son et al., 2015; Khan et al., 2017). ComED is a two-component system comprising the histidine kinase ComD and the cognate response regulator ComE. ComD autophosphorylates upon CSP pheromone binding and transfers the phosphoryl group to ComE, which in turn induces the expression of bacteriocin and immunity proteins, including the gene for the non-lantibiotic bacteriocin cipB. In rich media, cipB expression is required for induction of sigX by an as yet unknown mechanism. In CDM, CSP induces the ComED pathway, but it is unknown whether the CSP pheromone system is endogenously activated. It is also unknown why in CDM cipB is stimulated by CSP, but not the competence response. The conservation of the CSP and XIP signaling pathways in CDM were examined in this study using sigX and cipB luciferase reporter bioassays.
The XIP pheromone system is found in several streptococci (Gardan et al., 2009; Fontaine et al., 2010; Mashburn-Warren et al., 2010; Fleuchot et al., 2011; Morrison et al., 2013). In S. mutans UA159 the XIP pheromone encoded by comS has been identified in culture supernatants grown in chemically defined medium (CDM) lacking peptides (Mashburn-Warren et al., 2010; Khan et al., 2012). XIP is produced as a pro-peptide (ComS) that is possibly exported and processed into the active XIP (N-GLDWWSL) (Mashburn-Warren et al., 2010; Desai et al., 2012; Khan et al., 2012). The presence of an exporter has, however, never been demonstrated, and recent studies indicate that release of XIP or ComS by autolysis is sufficient to promote intercellular communication in CDM (Kaspar et al., 2017). Under such conditions, response to the XIP depends on the Opp oligopeptide permease system, indicating that the peptide is internalized (Mashburn-Warren et al., 2010). Once inside the cells XIP is thought to bind to the Rgg-like regulator ComR to activate transcription of sigX and comS. In contrast with XIP, CSP shows no activity or low potency in triggering competence in CDM (Desai et al., 2012; Wenderska et al., 2012; Reck et al., 2015) but can still induce the expression of bacteriocin-related genes of the ComE regulon (Reck et al., 2015). When it comes to transformation, the use of synthetic CSP has led to higher levels of S. mutans transformation (Gaustad and Morrison, 1998; Petersen and Scheie, 2010) but has not extended the range of strains transformed in the absence of the synthetic pheromone. As for the synthetic XIP, evidence for competence induction and endogenous pheromone production has so far been restricted to the reference strain UA159, with few exceptions (Palmer et al., 2013).
In this study we investigated the functional conservation of the S. mutans CSP and XIP pheromone signaling systems in CDM. Extracellular CSP activity was not detected in any of the tested strains. Exposure to synthetic CSP revealed that S. mutans can indeed degrade CSP, a behavior that was conserved in the examined strains, and that at least in strain UA159 it was not abolished by deletion of sigX, comR, oppD, or the htrA protease gene. For the XIP pheromone, endogenous production was found in all tested strains, and transformation was found in all but one of the isolates grown in CDM. The results thus indicate the presence of a single S. mutans XIP pherogroup and suggest that S. mutans has a conserved ability to suppress CSP activity in CDM.
The Streptococcus mutans strains used in this study and their relevant characteristics are listed in Table 1. The strains used in the study were selected from the collection of strains in our laboratory, representing strains known to be transformed (UA159, V403, OMZ175, LML-2, LML-4, GS5, NG8, BM71, LT11) and others in which genetic competence has not yet been characterized. Todd-Hewitt broth (THB; Becton Dickinson) was used to grow all the strains used for DNA extraction and DNA sequencing. Chemically Defined Medium (CDM) (Mashburn-Warren et al., 2010) was used to perform the genetic transformation assays and bioassay to measure the activity of exogenously added CSP in the supernatants of S. mutans and also to verify and quantify the concentration of extracellular native XIP in culture supernatants (Desai et al., 2012; Khan et al., 2012). The carbohydrate source was glucose at 1% final concentration, unless specified. The antibiotics erythromycin, kanamycin and spectinomycin were used at final concentrations of 10, 500, and 500 μg mL–1, respectively. THB agar and THB agar supplemented with erythromycin were used to enumerate the total and transformed number of S. mutans.
The deletion mutants were constructed using the PCR-ligation mutagenesis strategy (Lau et al., 2002). Sequence information was obtained from the S. mutans UA159 genome. Mutants with deletion of sigX and htrA were constructed. AscI or FseI sites were incorporated into the 5′-ends of the oligonucleotide primers and both ends of the resistance cassette. The kanamycin resistance cassette was amplified using primer pair FP001-FP068. The comX flanking regions were amplified with the primers pairs FP462 (CTTGGTAGCAGGAGAGCAC), FP463 (AAAG CACAGCCTGCTTCAAT) and FP714 (TGCCGAACA CAGCAGTTAAG), FP715 (CATTCCCTCTTGTTGCCAAT). The htrA flanking regions were amplified with the primers pairs FP807 (TCCCTCCAATAACGAAGGTCA), FP808 (GGTAAGT GTTGA TATGACCCCT) and FP809 (GAAGGTAGCGTCTA TCAGCGA), FP810 (GCAGTCGAGGTTGATAGGGA). The resultant amplicons were digested with AscI or FseI whereas the kanamycin resistance cassette was digested with both enzymes. The upstream and downstream amplicons of target genes were ligated to the kanamycin cassette using T4 DNA ligase. The two ligated products were mixed and PCR amplified with distal primers. The resultant amplicons were used to transform S. mutans. Gene deletion was confirmed by PCR amplification and gel electrophoresis.
The chromosomal DNA of S. mutans strains were isolated as previously described (Petersen and Scheie, 2000). The primers FP678 (5′-ATGCGGAAGCTAAAAAGAGC-3′) and FP679 (5′-TCCAGTCTTCCTATCTGAGCAA-3′) were used to amplify a region of 431 bp, which contains the tRNA transcriptional terminator of comR (9 bp stem, a 4 nt loop and a T-rich region at the 3′ side of the stem-loop), the comS promoter and the comS sequence. First, a PCR was performed (20 μL reaction volume containing 2.5 mM MgCl2, 2 pmol of each primer, 0.2 mM of each dNTP, 2 μL of 10× buffer, 50–100 ng of genomic DNA and 2 units of TaqDNA polymerase using a thermocycling profile of initial denaturing period of 3 min at 94°C; followed by 30 cycles of 30 s at 94°C, 30 s at 55°C and 2.5 min at 72°C; with a final extension period of 3 min at 72°C and the products were visualized on a 1% agarose gel to check the amplified fragments by the primers (Petersen and Scheie, 2000). The sequencing was performed bi-directionally in a single experiment for all the samples using BigDye® Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, United States) on an ABI 3730 DNA analyzer (Applied Biosystems, Foster City, CA, United States). All the sequence analysis was done using the Sequencher 5.0 software (Gene Codes, Ann Arbor, MI, United States). The electropherograms were checked and comparative analysis of the bidirectional sequences was performed using the S. mutans UA159 sequence as a reference.
The CSP bioassay was performed to measure the production of native CSP and activity of exogenously added CSP in the supernatants of S. mutans during growth, and the XIP bioassay was to verify and quantify the concentration of extracellular native XIP in culture supernatants. A PcipB-luc reporter (SM059) was used in the CSP bioassay as previously described (Khan et al., 2012), and a PsigX-luc reporter in a comS deletion background (SM091) was used to perform the XIP bioassay as described by Desai et al. (2012), with slight modifications (Khan et al., 2012).
To estimate CSP production and response, the supernatants were collected by 10× dilution of precultures at OD600 of 0.6 in CDM containing 10% bovine serum albumin (BSA). When added, 250 nM CSP concentration was used. Supernatants were collected for 6 h during growth. For XIP production and response, overnight cultures of S. mutans strains were diluted in fresh CDM to an optical density at 600 nm (OD600) of 0.05 and incubated in air at 37°C. At each hour, from the second until the 10 h of bacterial growth, the OD600 was measured and 1 mL of the culture was centrifuged (10,000 g for 10 min at 4°C) to collect the supernatant. The indicator strains SM059 and SM091 were grown in CDM to an OD600 of 0.05 and stored in microcentrifuge tubes with 10% of glycerol at −80°C. The entire assay was performed with stock cultures from the same batch. The bioassay was performed adding 10 μL of each culture supernatant to 50 μL of the indicator strain, 40 μL fresh CDM and 20 μL of 1.0 mM D-luciferin (Synchem, Felsberg-Altenburg, Germany) in a flat-bottom 96-well plate (Nunc, Rochester, NY, United States). Luminescence was measured by reading the plates in a multidetection microplate reader (Synergy HT; BioTek Instruments, Winooski, VT, United States). To quantify the XIP concentrations in the culture supernatants, standard curves were performed at the same time, replacing the culture supernatant with 10 μL of CDM-standard solutions with different XIP concentrations. CDM without synthetic peptides was used to obtain background values that were subtracted from the sample values. For strains exhibiting no XIP activity in the supernatants, the experiment was repeated at least twice. The software SigmaPlot (version 12.0, Systat Software, Inc.) was used to calculate the XIP concentrations using the relative light unit (RLU) values.
Overnight cultures of all S. mutans strains were diluted 1:10 in fresh CDM and incubated in 5% CO2 at 37°C. The OD600 was followed until the cultures reached an absorbance value of 0.6, then 10% of glycerol was added and they were stored at −80°C. To perform the assay the stored cultures were diluted in fresh CDM 1:10 to an OD600 of 0.05 and incubated in air for 2 h at 37°C. The OD was measured and aliquots of 150 μL of the bacterial suspensions were transferred to 1.5 mL microcentrifuge tubes. The plasmid pVA838 (Table 1) was added to all tubes at a final concentration of 1 μg mL–1 and XIP was added in the experimental group at a final concentration of 1,000 nM. The samples were then incubated in air for an additional 4 h at 37°C, at which time the OD was measured again and the bacterial suspensions were serially diluted (up to 1 to 106) in PBS. The suspensions were plated in duplicate on THB agar and THB agar supplemented with erythromycin 10 μg mL–1. The plates were incubated in 5% CO2 for 48 h at 37°C before counting the colony-forming units (CFU). The number of genetically transformed cells was divided per total CFU to obtain the values for transformation frequency. Three independent experiments were performed to evaluate transformation in CDM.
In order to evaluate genetic transformation in the non-responsive strains, an independent experiment extending the incubation time to 10 h before plating was performed. In addition, at 6 h an extra load of pVA838 was added, which increased the final concentration to 2 μg mL–1. For the remaining non-responsive strains, transformation was attempted using the 6.3 kb aRJ02 chromosomal PCR amplicon with a kanamycin marker, which is expected to have a higher sensitivity for detection of transformability (Table 1; Morrison et al., 2015; Salvadori et al., 2017).
The synthetic XIP (ComS11–17; GLDWWSL) (Mashburn-Warren et al., 2010; Khan et al., 2012) and the 18-CSP (ComC26–43; SGSLSTFFRLFNRSFTQA) (Petersen et al., 2006) were synthesized by GenScript (GenScript Biotech Corporation, Piscataway, NJ, United States), both with an estimated purity of 98% (Khan et al., 2012). The stock solutions of both peptides were stored in small aliquots at −20°C. The final concentration used in the genetic transformation assays for XIP was 1,000 nM. For FRET-assays to investigate protease activity, the 18-CSP was synthesized using methoxycoumarin-acetic-acidyl (MCA) on the N-terminal and Lys-Dinitrophenyl on the C-terminal (Dnp) (MCA-SGSLSTFFRLFNRSFTQA-Dnp; GenScript). On cleavage of the peptide by potential proteases, the Dnp quencher separates from the MCA fluorophore. The fluorophore is then activated giving an increase in fluorescence.
Pre-cultures of S. mutans were diluted in 2 mL CDM supplemented with 2% BSA to an OD600 of 0.04 to 0.05, and incubated at 37°C in air atmosphere to an OD600 of 0.06. The cultures were then distributed into the wells of a 96-well plate (115 μL in each well), and 5 μL of FRET-18CSP peptide was added (4 μM final concentration). The plate was incubated at 37°C in air, and fluorescence and optical density at 600 nm were measured at different time points during growth in a multi-detection microplate reader (Synergy, Cytation 3; excitation 325 nm, emission 392 nm).
Supernatants from S. mutans UA159 PcipB-luc reporter (SM059) and ΔhtrA (SM165) were collected by centrifugation (6000 g for 10 min at 4°C) at different time points during growth, corresponding to early, mid- and late exponential phases. The supernatants were then distributed into the wells of a 96-well plate (115 μL in each well), and 5 μL of FRET-18CSP peptide was added (4 μM final concentration). The plate was incubated at 37°C in air atmosphere for 1 h, and fluorescence was measured in a multi-detection microplate reader (Synergy, Cytation 3; excitation 325 nm, emission 392 nm). Supernatants without FRET-18CSP were used for detection of background fluorescence, which were then subtracted from the values in the tested samples exposed to FRET-18CSP.
In this study we used a PcipBluc reporter for detection of extracellular CSP activity, in CDM as previously described (Khan et al., 2012), except that high sensitivity was only achieved in the presence of BSA (Figure 2A). BSA was chosen because it is known to enhance competence of S. mutans in rich media (Ahn et al., 2006), and in S. pneumoniae, BSA prevents CSP proteolysis (Cassone et al., 2012).
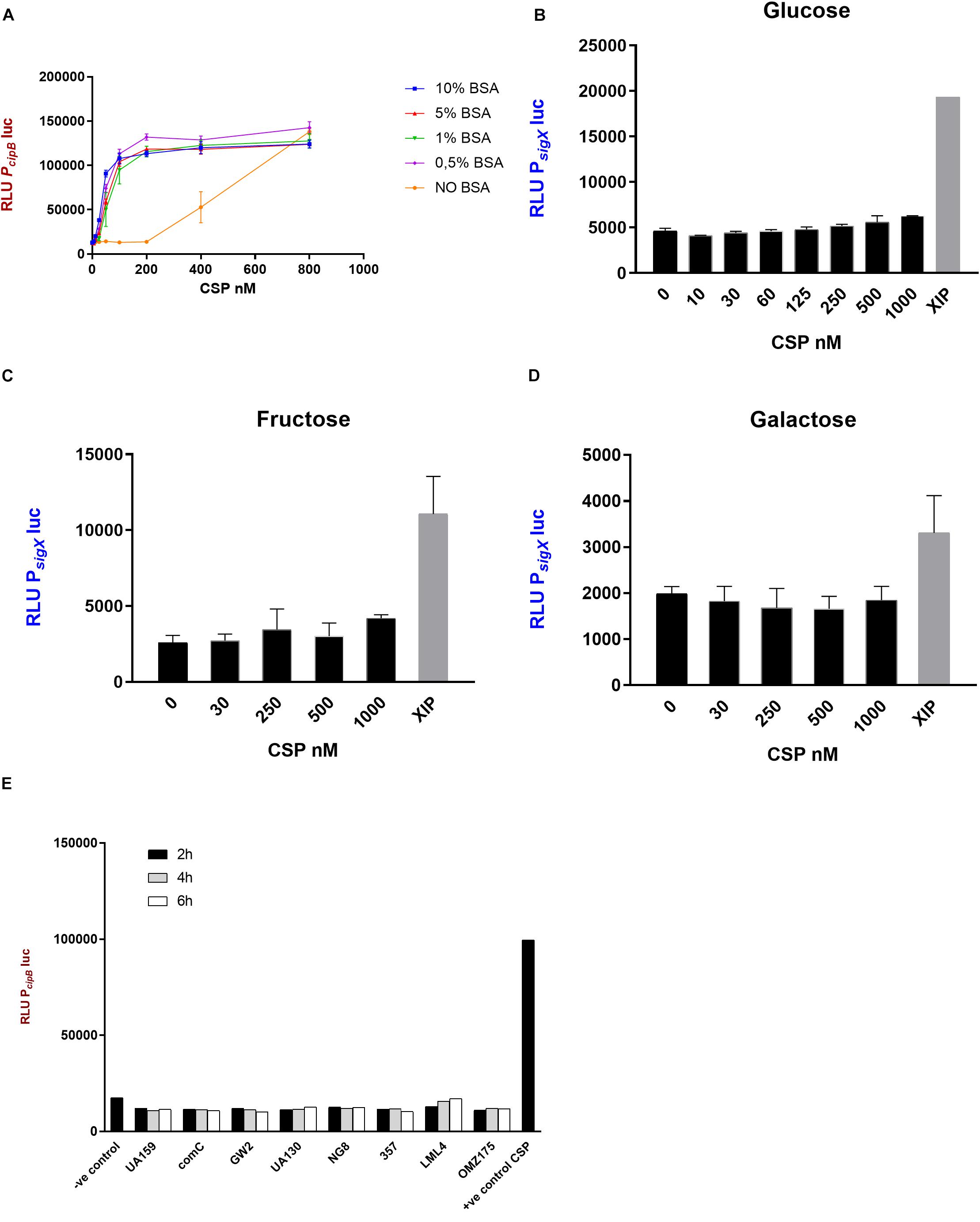
Figure 2. CSP activity in CDM. (A) The indicator strain SM059 (PcipBluc) was exposed to serial dilutions of synthetic CSP in the presence and absence of BSA. Different concentrations of BSA were tested. 10% BSA (blue), 5% BSA (red), 1% BSA (green), 0.5% BSA (pink), No BSA (orange). Bars correspond to standard errors of two to three independent experiments; RLU values were measured in a 96-well plate using a multi-detection microplate reader. (B–D) Activity of PsigXluc was determined after addition of various concentrations of CSP and 1 μM XIP in CDM supplemented with (B) glucose, (C) fructose and (D) galactose. Mean and standard deviation for two independent experiments. (E) Supernatants were collected from different S. mutans strains at 2 h (black bars), 4 h (gray bars) and 6 h (white bars) growth in CDM and examined for CSP activity using SM059 (PcipBluc) indicator strain. Medium alone was used as a negative control, and CSP (250 nM) was used as a positive control.
The potency of CSP in inducing the activity of the cipB promoter was increased by approximately 33-fold in CDM supplemented with BSA compared with CDM alone (Figure 2A). BSA concentrations from 0.5 to 10% gave similar results (Figure 2A).
Since the induction of cipB was significantly enhanced in the presence of BSA, we decided to further examine the intriguing fact that CSP fails to stimulate sigX in CDM (Desai et al., 2012; Wenderska et al., 2012; Reck et al., 2015). Our hypothesis was that increased stimulation of the early response by CSP, as observed with BSA supplementation, would enable the activation of sigX, which is a late response in complex media. We used a similar bioassay as described above, but using the promoter of sigX linked to the luciferase gene instead. The results showed that CSP concentrations up to 1000 nM failed to induce the PsigXluc reporter in CDM supplemented with BSA and glucose (Figure 2B).
Since the carbohydrate source, at least in complex media, has a large influence on the induction of sigX expression by CSP (Moye et al., 2016), we also investigated sigX expression in CDM supplemented with fructose and galactose (Figures 2C,D). CSP still failed in demonstrating sigX induction. Thus, although CSP induces strong expression of cipB in CDM supplemented with BSA and different sources of carbohydrate, it fails to induce the late response characterized by the induction of sigX expression at concentrations as high as 1000 nM.
Lack of endogenous CSP activity in the supernatants of S. mutans UA159 grown in CDM has been previously reported by our group (Khan et al., 2012). It is, however, unknown whether this is a conserved feature in the species. We investigated 6 other strains of S. mutans, and the comC deletion mutant SM004, for the CSP activity in their supernatants by growing them in CDM in the presence of BSA (Figure 2E). No cipB-inducing activity was detected in culture supernatants collected at early-, mid- or late- exponential phases of growth, thus indicating that the lack of extracellular CSP activity under such growth conditions may represent a conserved feature in S. mutans.
Supernatants of S. mutans cultures exposed to 250 nM synthetic CSP were collected at different time points during growth in CDM. CSP activity in the supernatants of S. mutans UA159 was measured by using the cipB luciferase reporter described above. Within the first 3 h, CSP had almost completely disappeared from the culture supernatants (Figure 3A).
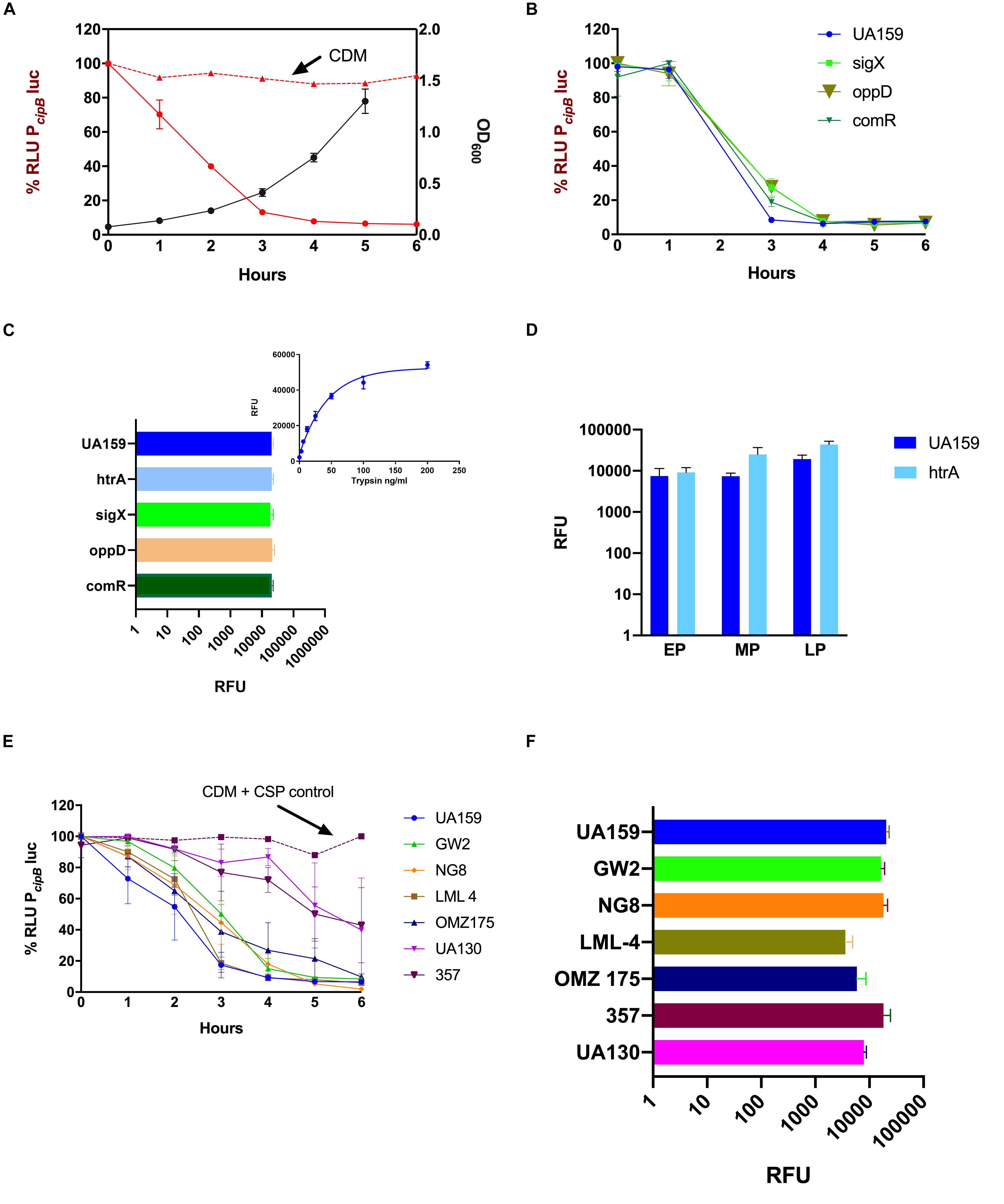
Figure 3. Suppression of CSP activity during growth in CDM. Frozen stock cultures at OD600 0.6 were diluted 10× in CDM containing 10% BSA, and 250 nM synthetic CSP (A,B,E) or 4 μM FRET-CSP (C,D,F). (A,B,E) The PcipBluc reporter was used to measure CSP concentration in the supernatants of different strains (A) S. mutans UA159 growth curve was measured at OD600 (black line) and indicates the time points when supernatants were collected. CSP activity in CDM alone (positive control) is represented by the red dashed line and in the supernatants of S. mutans UA159 by the red solid line. (B) Loss of CSP activity in the supernatants of UA159, ΔsigX, ΔoppD and ΔcomR. (C,F) CSP cleavage measured at 240 min growth in the presence of the FRET-18CSP peptide, and recorded as relative fluorescence units (RFU) for panel (C) UA159, ΔhtrA, ΔsigX, ΔoppD, and ΔcomR, and (F) UA159, GW2, NG, LML-4, OMZ175, 357, and UA130. In panel (C), right upper corner, a standard curve showing degradation of FRET-18CSP by trypsin is included as a reference. (D) FRET-18CSP proteolytic activity in supernatants of UA159 and the ΔhtrA mutant collected at early (EP), mid- (MP) and late (LP) exponential phase of growth. RFU background values of the corresponding strains without FRET-CSP were subtracted. Error bars show standard error of mean from two to three independent experiments, with three parallels each.
Due to the recognizable role of CSP in the induction of competence, it was crucial to determine if the loss of CSP activity was dependent on the development of competence. For this, we investigated the CSP activity in the culture supernatants of UA159 deletion mutants for sigX, oppD, and comR (Table 1). In all of them, a dramatic reduction in activity of exogenously added CSP was observed (Figure 3B).
To determine whether such an inhibitory effect on CSP activity could be due to proteolytic cleavage, we supplemented the CDM medium with a FRET-18CSP reporter peptide (Figure 3C). We found that all three isogenic mutants (sigX, oppD, and comR) grown under such conditions promoted increase in fluorescence activity, as measured after 240 min incubation at 37°C (Figure 3C), thus suggesting that the ability of S. mutans to degrade CSP does not require expression of key elements of the competence regulon. We also examined the effect of inactivating the htrA gene coding for the S. mutans HtrA protease, which in S. pneumoniae inactivates the CSP and cleaves misfolded proteins. No reduction in CSP degradation was observed for the htrA mutant (Figure 3C). CSP activity was indeed slightly higher for the culture supernatants of the htrA mutant at mid- and late exponential phases of growth (Figure 3D).
Finally, we tested whether suppression of CSP activity is extended to other S. mutans strains. In all the strains tested, reduction in CSP activity was observed, though at different levels (Figure 3E). When grown in the presence of the FRET-18CSP reporter peptide, an increase in fluorescence was observed for all strains (Figure 3F). We conclude that S. mutans suppresses CSP activity during growth in CDM, and that the mechanisms involved are active in the absence of htrA, sigX, comR or oppD, indicating the involvement of competence-regulated independent factors. The potential proteolytic activity leading to CSP inactivation seems to be conserved in S. mutans.
S. mutans UA159 produces and responds to exogenous XIP in CDM (Mashburn-Warren et al., 2010; Desai et al., 2012). We investigated whether XIP production is conserved in S. mutans. We used the PsigXluc ΔcomS indicator (SM091) to estimate the XIP concentration in the supernatants of the different strains during growth. The minimum detection level was set at 10 nM, as determined by standard curve measurements using synthetic XIP. Our results showed the presence of extracellular XIP activity in all strains tested. The highest XIP concentrations in the supernatants of growing cultures ranged from approximately 200 to 26,000 nM (Figure 4A), with most showing XIP accumulation starting at the mid-exponential growth phase and reaching maximal values at early stationary phase (data not shown). We conclude that the growth conditions that support extracellular XIP pheromone accumulation by UA159 may also sustain XIP production by a majority of the strains.
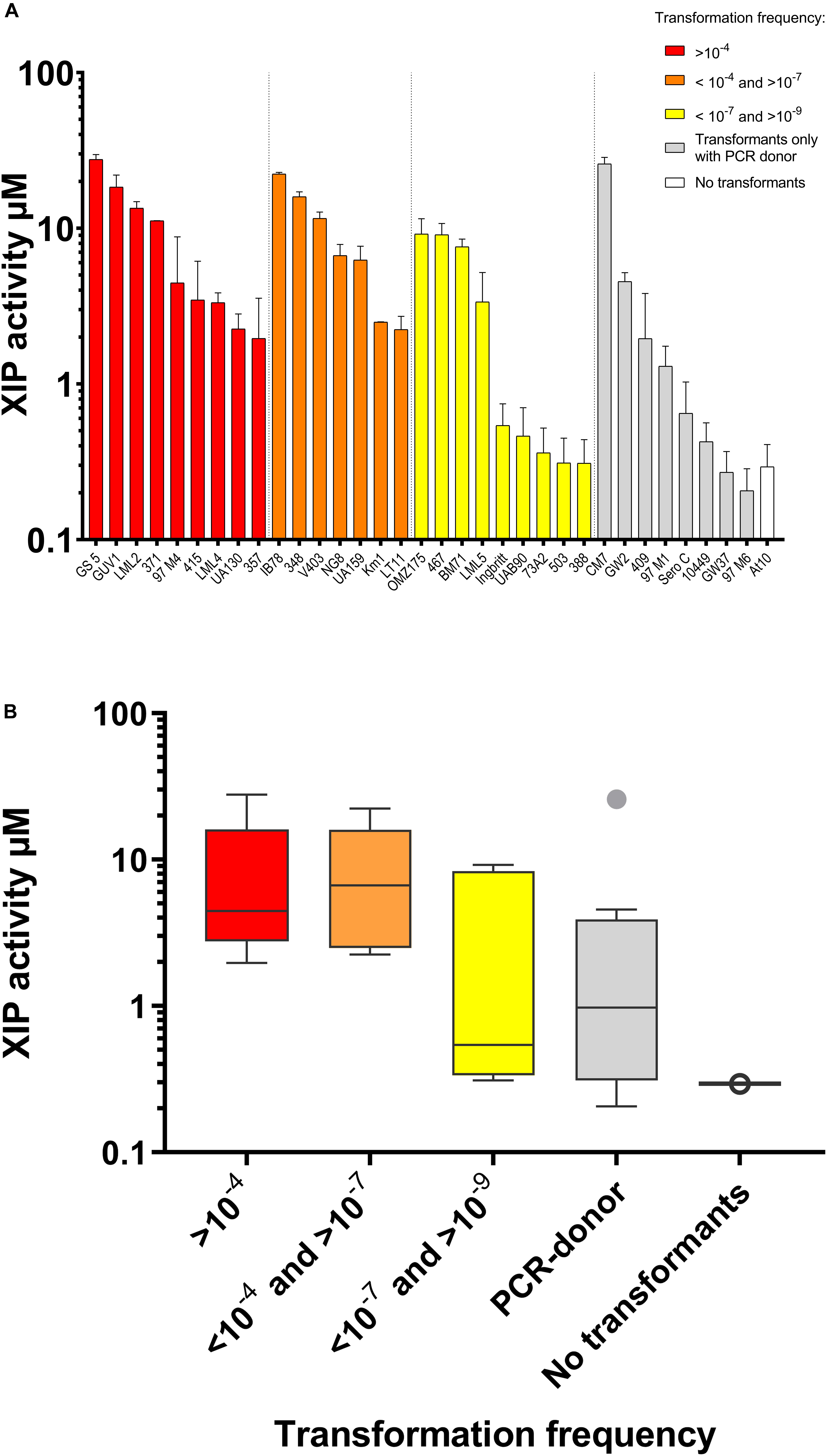
Figure 4. Conservation of XIP production and transformation in CDM. Extracellular XIP concentration was measured in the supernatants collected from S. mutans strains grown overnight at 37°C in air. (A) Columns show average of XIP-equivalent concentration from three independent experiments. Bars correspond to standard error. The indicator strain used was a ΔcomS PsigX-luc reporter (SM091). Bars in different colors indicate transformation frequency in CDM supplemented with synthetic XIP by a plasmid donor (pVA838): (>10–4 in red, between 10–4 and 10–7 in orange, and between 10–7 and 10–8 in yellow). For those that were not transformed by the plasmid donor, transformation frequency with a 6.3 Kb PCR donor designed to be integrated into the chromosome by homologous recombination are shown (all frequencies below 10–7; gray). Only At10 yielded no transformants (white). (B) Whisker plots showing the average of XIP activity of the strains shown in panel (A), grouped according to transformation frequency.
Natural transformation without the addition of synthetic pheromone was detected in 23 of the 34 strains (data not shown). Because optimal levels of XIP may not be present at the time of competence under in vitro conditions, it was of interest to learn whether addition of synthetic XIP would result in higher transformation levels. Our results showed that synthetic XIP extended the range of transformed strains. Three of the strains with detectable XIP levels in the supernatants, including OMZ175, UAB90, and KM1 were transformed only upon addition of synthetic XIP. In the strains that were not transformed with the initial protocol, we increased the time during which they grew in the presence of synthetic XIP and donor DNA to 10 h, but even then no transformants were obtained (Murchison et al., 1986). We next used a more sensitive protocol based on the use of PCR large fragments as donor DNA (Morrison et al., 2015; Salvadori et al., 2017). With addition of the synthetic pheromone, 33 out of 34 strains were naturally transformable. Only strain At10 was not transformed, thus indicating that a majority of S. mutans are amenable to transformation. Overall, strains that exhibited lower transformation frequencies showed lower levels of XIP activity in their culture supernatants (Figure 4B).
The S. mutans comS gene encoding the XIP precursor is essential for competence development. Given the variability in transformation efficiency observed above, and that some strains were not amenable to transformation, we investigated whether the comS gene and its putative promoter sequences were conserved in the S. mutans strains included in the study. The results showed that comS was identical in all 34 strains (Figure 5). Highly conserved sequences were also found in the comS putative promoter. In this region the only difference in relation to UA159 was the presence or absence of an additional adenine between the −10 element and the comS translation initiation site. The additional adenine was present in 22 out of the 34 strains evaluated. In all the strains, comS was located 57–58 nt downstream of the tRNA transcriptional terminator of comR (SMU_61) and the putative −10 element was 32–33 nt upstream of the comS translation initiation site. While this study was being performed, new genomic sequences for 78 more strains of S. mutans were made available on public databases (Maruyama et al., 2009). In order to compare our results with the new genomic sequences we ran BLAST using the sequence shown in Figure 5 against the CoGe database1 (Lyons et al., 2011). The comS sequences in these strains were identical to the S. mutans UA159. In the comS promoter the sequences varied in only a single base pair at the same position as that observed in 22 of the strains sequenced in our study. Taken together the results indicate that the comS gene and promoter regions are highly conserved in the S. mutans strains analyzed, suggesting that variations in transformation levels and XIP production are not correlated with differences in this region.
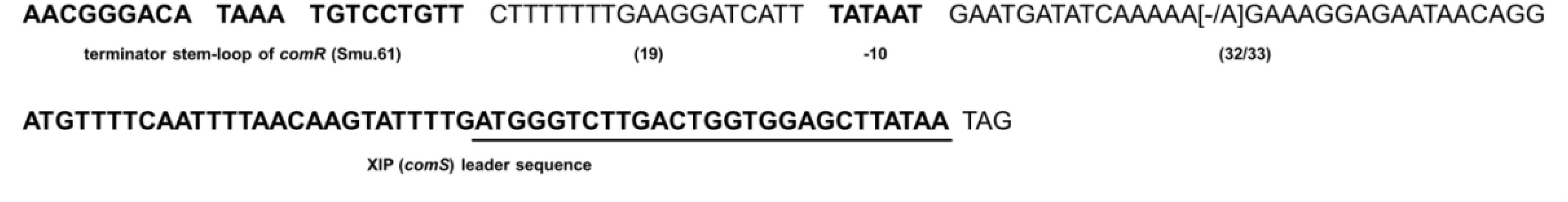
Figure 5. Conservation of comS promoter and comS sequences in S. mutans strains. The first sequence in bold shows the terminator stem-loop of comR (Smu.61) thought to function as part of the comS promoter. Downstream the stem-loop is a 19 bp sequence, followed by the putative –10 element of the comS promoter distant 32 to 33 bases from the comS gene sequence shown in bold. Underlined is the sequence corresponding to the XIP functional heptapeptide (ComS11–17 – GLDWWSL). Variable bases compared with S. mutans UA159 are in brackets.
In complex media, the CSP pheromone triggers the expression of bacteriocin-related genes, followed by a late response characterized by increased expression of the XIP-encoding gene comS and competence development (Kreth et al., 2005; Lemme et al., 2011; Khan et al., 2016). The mechanisms leading to activation of comS expression and competence remain unknown. It is also still unknown why CSP fails in stimulating competence in peptide-free defined medium. It has been suggested that S. mutans may perhaps produce a protease that, similar to the HtrA protease in S. pneumoniae, could inactivate the CSP (Desai et al., 2012). This possible explanation did not seem to match with the later finding that CSP is actually active in CDM, in that it can stimulate cipB and all other genes regulated by ComED (Reck et al., 2015). Our results show that S. mutans can indeed inactivate CSP by a mechanism that most probably involves proteolysis, given the results obtained with the FRET-18CSP reporter peptide. We confirmed previous results that CSP induces cipB (Reck et al., 2015) and found that a CSP concentration as low as 1 nM in the presence of BSA was sufficient to activate the early bacteriocin response. This is opposite to the CSP concentration to activate sigX, which requires values that exceed the maximum concentration used in this study (1000 nM) (data not shown). A higher threshold for sigX activation by CSP has also been observed in complex media (Son et al., 2012). A question that remains is why the threshold of CSP concentration to activate sigX, and therefore competence, is higher than for cipB activation, when CipB is thought to be the main link of CSP to sigX activation. One possibility is that at high concentrations CSP may activate or repress other two-component systems than ComED, which would then trigger a different link to competence. It is a well known phenomenon that although pheromones are usually highly specific to their cognate receptors, at high concentrations they may bind to other non-specific receptors (Hawver et al., 2016). However, several other mechanisms are possible, since the competence system is part of a signaling network of high complexity, affected by a variety of other systems not directly regulated by ComED, as recently reviewed (Kaspar and Walker, 2019). Irrespective of the mechanism, the higher threshold for CSP to induce competence indicates that CSP degradation may affect the competence response to this pheromone.
Inactivation of the S. mutans CSP by proteases produced by other streptococcal species has been known for more than 10 years (Wang and Kuramitsu, 2005). However, this is the first indication that S. mutans can also inhibit the activity of its own CSP, most probably via proteolytic activity. The genomes of S. mutans have more than 65 known or putative proteases, according to the MEROPS database2, but only HtrA has shown a role in autologous CSP cleavage. However, unlike for S. pneumoniae (Cassone et al., 2012), the HtrA protease in S. mutans was not required for CSP inactivation.
In S. pneumoniae, luciferase and gfp reporter strains for CSP activity have been successfully used to identify pneumococcal CSP pheromones in culture supernatants (Moreno-Gamez et al., 2017). Also in the seminal study that identified the S. pneumoniae competence factor, the CSP was isolated and purified from the supernatant (Havarstein et al., 1995). In S. mutans UA159 grown in complex medium, endogenous CSP has been identified by mass spectrometry in the supernatant fraction (Hossain and Biswas, 2012). Based on these findings, we hypothesized that if S. mutans produces CSP in CDM, we would probably be able to detect it in its culture supernatant. However, despite the high sensitivity of the protocol used in the present study, we could not detect any CSP-inducing activity in the supernatants of the different strains tested. Thus, if CSP is produced, it is either rapidly degraded outside of the cells or it remains associated with the cells.
In contrast to CSP, endogenous XIP activity was present in the supernatants of most strains. Moreover, activation of the XIP system in CDM enabled transformation of all but one of the 34 strains tested. The comS gene encoding the pre-processed form of XIP was indeed found in all the strains, which is in line with the results of a recent study reporting comS as part of the S. mutans core genome (Cornejo et al., 2013). In general, higher levels of XIP detection in the supernatants correlated with increased transformability. Of note, some strains that produced high concentrations of endogenous XIP needed to be stimulated by synthetic XIP to transform. This was, however, not surprising, given the fact that maximum concentrations were in most cases observed only close to the stationary phase, which is a period during which competence under laboratory conditions is already shut off. For some of the strains that produced low levels of XIP, transformation was only detected by using a large PCR amplicon as DNA donor, which is known to result in increased recovery of transformants (Morrison et al., 2015; Junges et al., 2017).
Taken together, our results indicate that the XIP system is conserved in S. mutans, and that all S. mutans strains may belong to a single pherotype. Moreover, the conserved ability of S. mutans to cleave CSP under conditions that favor accumulation of XIP may provide an adaptive advantage to S. mutans, by allowing them to fine-tune competence and bacteriocin responses in response to environmental changes. While the knowledge on pheromone signaling for any species is mostly based on models derived from limited selected strains, understanding how the system works in different strains of a species is of high relevance for the development of strategies aiming at interfering with their communication systems.
All datasets generated for this study are included in the manuscript and or supplementary files.
All authors conceptualized the manuscript, drafted and critically revised the manuscript and approved the final version of the manuscript for publication.
This work was supported in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001 (AR postdoctoral grant), and by the program International Partnership for Outstanding Education, Research, and Innovation (INTPART), RCN grant number 274867.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Andreas Podbielski for the kind gift of pFW5-luc and Roy Russel for providing the S. mutans strains LML-2, LML-4, LML-5, and At10. We are grateful to Anne Karin Kristoffersen for helpful assistance in DNA sequence analysis.
Ahn, S. J., Wen, Z. T., and Burne, R. A. (2006). Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect. Immun. 74, 1631–1642. doi: 10.1128/iai.74.3.1631-1642.2006
Ajdic, D., Mcshan, W. M., Mclaughlin, R. E., Savic, G., Chang, J., Carson, M. B., et al. (2002). Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. U.S.A. 99, 14434–14439. doi: 10.1073/pnas.172501299
Biswas, S., and Biswas, I. (2012). Complete genome sequence of Streptococcus mutans GS-5, a serotype c strain. J. Bacteriol. 194, 4787–4788. doi: 10.1128/JB.01106-12
Cassone, M., Gagne, A. L., Spruce, L. A., Seeholzer, S. H., and Sebert, M. E. (2012). The HtrA protease from Streptococcus pneumoniae digests both denatured proteins and the competence-stimulating peptide. J. Biol. Chem. 287, 38449–38459. doi: 10.1074/jbc.M112.391482
Cornejo, O. E., Lefebure, T., Bitar, P. D., Lang, P., Richards, V. P., Eilertson, K., et al. (2013). Evolutionary and population genomics of the cavity causing bacteria Streptococcus mutans. Mol. Biol. Evol. 30, 881–893. doi: 10.1093/molbev/mss278
Coykendall, A. L. (1971). Genetic heterogeneity in Streptococcus mutans. J. Bacteriol. 106, 192–196.
Cvitkovitch, D. G., Boyd, D. A., Thevenot, T., and Hamilton, I. R. (1995). Glucose transport by a mutant of Streptococcus mutans unable to accumulate sugars via the phosphoenolpyruvate phosphotransferase system. J. Bacteriol. 177, 2251–2258. doi: 10.1128/jb.177.9.2251-2258.1995
Desai, K., Mashburn-Warren, L., Federle, M. J., and Morrison, D. A. (2012). Development of competence for genetic transformation by Streptococcus mutans in a chemically defined medium. J. Bacteriol. 194, 3774–3780. doi: 10.1128/JB.00337-12
Dufour, D., Cordova, M., Cvitkovitch, D. G., and Levesque, C. M. (2011). Regulation of the competence pathway as a novel role associated with a streptococcal bacteriocin. J. Bacteriol. 193, 6552–6559. doi: 10.1128/JB.05968-11
Federle, M. J., and Morrison, D. A. (2012). One if by land, two if by sea: signalling to the ranks with CSP and XIP. Mol. Microbiol. 86, 241–245. doi: 10.1111/mmi.12029
Fleuchot, B., Gitton, C., Guillot, A., Vidic, J., Nicolas, P., Besset, C., et al. (2011). Rgg proteins associated with internalized small hydrophobic peptides: a new quorum-sensing mechanism in streptococci. Mol. Microbiol. 80, 1102–1119. doi: 10.1111/j.1365-2958.2011.07633.x
Fontaine, L., Boutry, C., De Frahan, M. H., Delplace, B., Fremaux, C., Horvath, P., et al. (2010). A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius. J. Bacteriol. 192, 1444–1454. doi: 10.1128/JB.01251-09
Gardan, R., Besset, C., Guillot, A., Gitton, C., and Monnet, V. (2009). The oligopeptide transport system is essential for the development of natural competence in Streptococcus thermophilus strain LMD-9. J. Bacteriol. 191, 4647–4655. doi: 10.1128/JB.00257-09
Gaustad, P., and Morrison, D. A. (1998). Induction of transformation in streptococci by synthetic competence stimulating peptides. Methods Cell Sci. 20, 65–70. doi: 10.1007/978-94-017-2258-2_7
Havarstein, L. S., Coomaraswamy, G., and Morrison, D. A. (1995). An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U.S.A. 92, 11140–11144. doi: 10.1073/pnas.92.24.11140
Hawver, L. A., Jung, S. A., and Ng, W. L. (2016). Specificity and complexity in bacterial quorum-sensing systems. FEMS Microbiol. Rev. 40, 738–752. doi: 10.1093/femsre/fuw014
Hossain, M. S., and Biswas, I. (2012). An extracelluar protease, sepm, generates functional competence-stimulating peptide in Streptococcus mutans UA159. J. Bacteriol. 194, 5886–5896. doi: 10.1128/JB.01381-12
Hung, D. C., Downey, J. S., Ayala, E. A., Kreth, J., Mair, R., Senadheera, D. B., et al. (2011). Characterization of DNA binding sites of the ComE response regulator from Streptococcus mutans. J. Bacteriol. 193, 3642–3652. doi: 10.1128/JB.00155-11
Johnsborg, O., Eldholm, V., and Havarstein, L. S. (2007). Natural genetic transformation: prevalence, mechanisms and function. Res. Microbiol. 158, 767–778. doi: 10.1016/j.resmic.2007.09.004
Johnston, C., Martin, B., Fichant, G., Polard, P., and Claverys, J. P. (2014). Bacterial transformation: distribution, shared mechanisms and divergent control. Nat. Rev. Microbiol. 12, 181–196. doi: 10.1038/nrmicro3199
Junges, R., Khan, R., Tovpeko, Y., Amdal, H. A., Petersen, F. C., and Morrison, D. A. (2017). Markerless genome editing in competent streptococci. Methods Mol. Biol. 1537, 233–247. doi: 10.1007/978-1-4939-6685-1_14
Kaspar, J., Underhill, S. A. M., Shields, R. C., Reyes, A., Rosenzweig, S., Hagen, S. J., et al. (2017). Intercellular communication via the comX-inducing peptide (XIP) of Streptococcus mutans. J. Bacteriol. 199:e00404-17. doi: 10.1128/JB.00404-17
Kaspar, J. R., and Walker, A. R. (2019). Expanding the vocabulary of peptide signals in Streptococcus mutans. Front. Cell Infect. Microbiol. 9:194. doi: 10.3389/fcimb.2019.00194
Khan, R., Junges, R., Amdal, H. A., Chen, T., Morrison, D. A., and Petersen, F. C. (2017). A positive feedback loop mediated by sigma X enhances expression of the streptococcal regulator ComR. Sci. Rep. 7:5984. doi: 10.1038/s41598-017-04768-5
Khan, R., Rukke, H. V., Hovik, H., Amdal, H. A., Chen, T., Morrison, D. A., et al. (2016). Comprehensive transcriptome profiles of Streptococcus mutans UA159 map core streptococcal competence genes. mSystems 1:e00038-15.
Khan, R., Rukke, H. V., Ricomini Filho, A. P., Fimland, G., Arntzen, M. O., Thiede, B., et al. (2012). Extracellular identification of a processed type II ComR/ComS pheromone of Streptococcus mutans. J. Bacteriol. 194, 3781–3788. doi: 10.1128/JB.00624-12
Knox, K. W., Hardy, L. N., and Wicken, A. J. (1986). Comparative studies on the protein profiles and hydrophobicity of strains of Streptococcus mutans serotype c. J. Gen. Microbiol. 132, 2541–2548. doi: 10.1099/00221287-132-9-2541
Kreth, J., Hung, D. C., Merritt, J., Perry, J., Zhu, L., Goodman, S. D., et al. (2007). The response regulator ComE in Streptococcus mutans functions both as a transcription activator of mutacin production and repressor of CSP biosynthesis. Microbiology 153, 1799–1807. doi: 10.1099/mic.0.2007/005975-0
Kreth, J., Merritt, J., Shi, W., and Qi, F. (2005). Co-ordinated bacteriocin production and competence development: a possible mechanism for taking up DNA from neighbouring species. Mol. Microbiol. 57, 392–404. doi: 10.1111/j.1365-2958.2005.04695.x
Lau, P. C., Sung, C. K., Lee, J. H., Morrison, D. A., and Cvitkovitch, D. G. (2002). PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49, 193–205. doi: 10.1016/s0167-7012(01)00369-4
Lemme, A., Grobe, L., Reck, M., Tomasch, J., and Wagner-Dobler, I. (2011). Subpopulation-specific transcriptome analysis of competence-stimulating-peptide-induced Streptococcus mutans. J. Bacteriol. 193, 1863–1877. doi: 10.1128/JB.01363-10
Li, Y. H., Lau, P. C., Lee, J. H., Ellen, R. P., and Cvitkovitch, D. G. (2001). Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183, 897–908. doi: 10.1128/jb.183.3.897-908.2001
Lindler, L. E., and Macrina, F. L. (1986). Characterization of genetic transformation in Streptococcus mutans by using a novel high-efficiency plasmid marker rescue system. J. Bacteriol. 166, 658–665. doi: 10.1128/jb.166.2.658-665.1986
Lyons, E., Freeling, M., Kustu, S., and Inwood, W. (2011). Using genomic sequencing for classical genetics in E. coli K12. PLoS One 6:e16717. doi: 10.1371/journal.pone.0016717
Macrina, F. L., Tobian, J. A., Jones, K. R., Evans, R. P., and Clewell, D. B. (1982). A cloning vector able to replicate in Escherichia coli and Streptococcus sanguis. Gene 19, 345–353. doi: 10.1016/0378-1119(82)90025-7
Marsh, P. D., Moter, A., and Devine, D. A. (2011). Dental plaque biofilms: communities, conflict and control. Periodontol 2000, 16–35. doi: 10.1111/j.1600-0757.2009.00339.x
Maruyama, F., Kobata, M., Kurokawa, K., Nishida, K., Sakurai, A., Nakano, K., et al. (2009). Comparative genomic analyses of Streptococcus mutans provide insights into chromosomal shuffling and species-specific content. BMC Genomics 10:358. doi: 10.1186/1471-2164-10-358
Mashburn-Warren, L., Morrison, D. A., and Federle, M. J. (2010). A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol. Microbiol. 78, 589–606. doi: 10.1111/j.1365-2958.2010.07361.x
Moreno-Gamez, S., Sorg, R. A., Domenech, A., Kjos, M., Weissing, F. J., Van Doorn, G. S., et al. (2017). Quorum sensing integrates environmental cues, cell density and cell history to control bacterial competence. Nat. Commun. 8:854. doi: 10.1038/s41467-017-00903-y
Morrison, D. A., Guedon, E., and Renault, P. (2013). Competence for natural genetic transformation in the Streptococcus bovis group streptococci S. infantarius and S. macedonicus. J. Bacteriol. 195, 2612–2620. doi: 10.1128/JB.00230-13
Morrison, D. A., Khan, R., Junges, R., Amdal, H. A., and Petersen, F. C. (2015). Genome editing by natural genetic transformation in Streptococcus mutans. J. Microbiol. Methods 119, 134–141. doi: 10.1016/j.mimet.2015.09.023
Moye, Z. D., Son, M., Rosa-Alberty, A. E., Zeng, L., Ahn, S. J., Hagen, S. J., et al. (2016). Effects of carbohydrate source on genetic competence in Streptococcus mutans. Appl. Environ. Microbiol. 82, 4821–4834. doi: 10.1128/AEM.01205-16
Murchison, H. H., Barrett, J. F., Cardineau, G. A., and Curtiss, R. III. (1986). Transformation of Streptococcus mutans with chromosomal and shuttle plasmid (pYA629) DNAs. Infect. Immun. 54, 273–282.
Palmer, S. R., Miller, J. H., Abranches, J., Zeng, L., Lefebure, T., Richards, V. P., et al. (2013). Phenotypic heterogeneity of genomically-diverse isolates of Streptococcus mutans. PLoS One 8:e61358. doi: 10.1371/journal.pone.0061358
Perch, B., Kjems, E., and Ravn, T. (1974). Biochemical and serological properties of Streptococcus mutans from various human and animal sources. Acta Pathol. Microbiol. Scand. B Microbiol. Immunol. 82, 357–370. doi: 10.1111/j.1699-0463.1974.tb02338.x
Perry, D., and Kuramitsu, H. K. (1981). Genetic transformation of Streptococcus mutans. Infect. Immun. 32, 1295–1297.
Petersen, F. C., Fimland, G., and Scheie, A. A. (2006). Purification and functional studies of a potent modified quorum-sensing peptide and a two-peptide bacteriocin in Streptococcus mutans. Mol. Microbiol. 61, 1322–1334. doi: 10.1111/j.1365-2958.2006.05312.x
Petersen, F. C., and Scheie, A. A. (2000). Genetic transformation in Streptococcus mutans requires a peptide secretion-like apparatus. Oral. Microbiol. Immunol. 15, 329–334. doi: 10.1034/j.1399-302x.2000.150511.x
Petersen, F. C., and Scheie, A. A. (2010). Natural transformation of oral streptococci. Methods Mol. Biol. 666, 167–180. doi: 10.1007/978-1-60761-820-1_12
Petersen, F. C., Tao, L., and Scheie, A. A. (2005). DNA binding-uptake system: a link between cell-to-cell communication and biofilm formation. J. Bacteriol. 187, 4392–4400. doi: 10.1128/jb.187.13.4392-4400.2005
Peterson, S. N., Sung, C. K., Cline, R., Desai, B. V., Snesrud, E. C., Luo, P., et al. (2004). Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 51, 1051–1070. doi: 10.1046/j.1365-2958.2003.03907.x
Reck, M., Tomasch, J., and Wagner-Dobler, I. (2015). The alternative sigma factor sigX controls bacteriocin synthesis and competence, the two quorum sensing regulated traits in Streptococcus mutans. PLoS Genet. 11:e1005353. doi: 10.1371/journal.pgen.1005353
Salvadori, G., Junges, R., Khan, R., Amdal, H. A., Morrison, D. A., and Petersen, F. C. (2017). Natural transformation of oral streptococci by use of synthetic pheromones. Methods Mol. Biol. 1537, 219–232. doi: 10.1007/978-1-4939-6685-1_13
Son, M., Ahn, S. J., Guo, Q., Burne, R. A., and Hagen, S. J. (2012). Microfluidic study of competence regulation in Streptococcus mutans: environmental inputs modulate bimodal and unimodal expression of comX. Mol. Microbiol. 86, 258–272. doi: 10.1111/j.1365-2958.2012.08187.x
Son, M., Shields, R. C., Ahn, S. J., Burne, R. A., and Hagen, S. J. (2015). Bidirectional signaling in the competence regulatory pathway of Streptococcus mutans. FEMS Microbiol. Lett. 362:fnv159. doi: 10.1093/femsle/fnv159
Song, L., Wang, W., Conrads, G., Rheinberg, A., Sztajer, H., Reck, M., et al. (2013). Genetic variability of mutans streptococci revealed by wide whole-genome sequencing. BMC Genomics 14:430. doi: 10.1186/1471-2164-14-430
Tao, L., Macalister, T. J., and Tanzer, J. M. (1993). Transformation efficiency of EMS-induced mutants of Streptococcus mutans of altered cell shape. J. Dent. Res. 72, 1032–1039. doi: 10.1177/00220345930720060701
Underhill, S. A. M., Shields, R. C., Kaspar, J. R., Haider, M., Burne, R. A., and Hagen, S. J. (2018). Intracellular signaling by the comRS system in Streptococcus mutans genetic competence. mSphere 4:e00042-19.
van der Ploeg, J. R. (2005). Regulation of bacteriocin production in Streptococcus mutans by the quorum-sensing system required for development of genetic competence. J. Bacteriol. 187, 3980–3989. doi: 10.1128/jb.187.12.3980-3989.2005
van Loveren, C., Buijs, J. F., and Ten Cate, J. M. (1993). Protective effect of topically applied fluoride in relation to fluoride sensitivity of mutans Streptococci. J Dent Res 72, 1184–1190. doi: 10.1177/00220345930720080401
Wang, B. Y., and Kuramitsu, H. K. (2005). Interactions between oral bacteria: inhibition of Streptococcus mutans bacteriocin production by Streptococcus gordonii. Appl. Environ. Microbiol. 71, 354–362. doi: 10.1128/aem.71.1.354-362.2005
Waterhouse, J. C., and Russell, R. R. (2006). Dispensable genes and foreign DNA in Streptococcus mutans. Microbiology 152, 1777–1788. doi: 10.1099/mic.0.28647-0
Ween, O., Gaustad, P., and Havarstein, L. S. (1999). Identification of DNA binding sites for ComE, a key regulator of natural competence in Streptococcus pneumoniae. Mol. Microbiol. 33, 817–827. doi: 10.1046/j.1365-2958.1999.01528.x
Keywords: pheromone, streptococcus, competence, natural transformation, quorum-sensing, CSP, XIP, ComS
Citation: Ricomini Filho AP, Khan R, Åmdal HA and Petersen FC (2019) Conserved Pheromone Production, Response and Degradation by Streptococcus mutans. Front. Microbiol. 10:2140. doi: 10.3389/fmicb.2019.02140
Received: 14 May 2019; Accepted: 30 August 2019;
Published: 13 September 2019.
Edited by:
Cristina García-Aljaro, University of Barcelona, SpainReviewed by:
Indranil Biswas, The University of Kansas, United StatesCopyright © 2019 Ricomini Filho, Khan, Åmdal and Petersen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fernanda C. Petersen, Zi5jLnBldGVyc2VuQG9kb250LnVpby5ubw==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.