- 1State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agriculture Sciences, Beijing, China
- 2Biointeractions and Plant Health, Wageningen Plant Research, Wageningen, Netherlands
Peroxisomes are involved in a wide range of important cellular functions. Here, the role of the peroxisomal membrane protein PEX3 in the plant-pathogen and mycotoxin producer Fusarium graminearum was studied using knock-out and complemented strains. To fluorescently label peroxisomes’ punctate structures, GFP and RFP fusions with the PTS1 and PTS2 localization signal were transformed into the wild type PH-1 and ΔFgPex3 knock-out strains. The GFP and RFP transformants in the ΔFgPex3 background showed a diffuse fluorescence pattern across the cytoplasm suggesting the absence of mature peroxisomes. The ΔFgPex3 strain showed a minor, non-significant reduction in growth on various sugar carbon sources. In contrast, deletion of FgPex3 affected fatty acid β-oxidation in F. graminearum and significantly reduced the utilization of fatty acids. Furthermore, the ΔFgPex3 mutant was sensitive to osmotic stressors as well as to cell wall-damaging agents. Reactive oxygen species (ROS) levels in the mutant had increased significantly, which may be linked to the reduced longevity of cultured strains. The mutant also showed reduced production of conidiospores, while sexual reproduction was completely impaired. The pathogenicity of ΔFgPex3, especially during the process of systemic infection, was strongly reduced on both tomato and on wheat, while to production of deoxynivalenol (DON), an important factor for virulence, appeared to be unaffected.
Introduction
Peroxisomes are a class of dynamic single-membrane-bound organelles, found in almost all eukaryotic cells. In recent years, it has become increasingly clear that peroxisomes are involved in a wide range of important cellular functions, including long-chain fatty acid β-oxidation, scavenging of reactive oxygen species (ROS), glyoxylate detoxification, and generation of reactive nitrogen species (RNS) signal molecules (Wanders and Waterham, 2006; Brown and Baker, 2008; Sandalio and Romero-Puertas, 2015; Tripathi and Walker, 2016). In addition, peroxisomes display multiple functions in filamentous fungi, such as the formation of Woronin bodies, which can seal septal pores after cellular wounding (Wanner and Theimer, 1982), metabolism of specific carbon sources (Hynes et al., 2006), and the biosynthesis of secondary metabolites (Brakhage et al., 2004; Meijer et al., 2010; Reverberi et al., 2012). Moreover, they are involved in virulence (Min et al., 2012) and sexual development (Peraza-Reyes and Berteaux-Lecellier, 2013).
The biogenesis of peroxisomes requires multiple peroxisomal proteins encoded by PEX genes (Distel et al., 1996). Currently, more than 30 PEX genes have been implicated in for instance the import of peroxisomal matrix proteins, assembly of the peroxisomal membrane, peroxisome proliferation and maintenance (Wanders and Waterham, 2005; Sibirny, 2016). All peroxisome matrix proteins are synthesized in the cytosol and transported to the peroxisome by targeting. There are two distinct peroxisomal targeting signals: PTS1 or PTS2 (Lazarow and Fujiki, 1985). PTS1 is located at the C-terminal end of matrix proteins and consists of a highly conserved tripeptide of -[S/C/A]-[K/R/H]-[L/M] (Notzel et al., 2016). PTS2 contains a conserved sequence [R/K]-[L/V/I]-X5-[H/Q]-[L/A], located at the N-terminal of matrix protein (Gould et al., 1989; Petriv et al., 2004). PEX5 and PEX7 are receptors for PTS1 and PTS2, respectively, that escort the cargo to the peroxisome membrane (Leon et al., 2006). In addition, PEX3, PEX16 and PEX19 are essential for the proper localization of peroxisome membrane proteins (PMPs). PEX3 has been proposed to act as a docking factor for PEX19 and to be involved in peroxisomal membrane biogenesis. In the early stages of peroxisome formation, PEX19 binds PMPs in the cytosol through a peroxisomal membrane-targeting signal (mPTS), composed of a PMP-binding domain and a membrane-anchoring domain (Rottensteiner et al., 2004; Matsuzono et al., 2006; Schueller et al., 2010). PEX3 can form a ternary complex with PEX19 and PMPs. The binding of PEX3 and PEX19 is an essential step in the targeting of PMPs to the peroxisomal membrane and absence of either PEX3 or PEX19 results in peroxisome deficiency (Pieuchot and Jedd, 2012).
Fusarium head blight, caused by the filamentous ascomycete F. graminearum species complex (FGSC), is one of the most devastating diseases affecting global wheat and barley production (Goswami and Kistler, 2004). F. graminearum is the predominant species and occurs in most FHB areas around the world (Bai and Shaner, 2004; Zhang et al., 2012; van der Lee et al., 2015). In recent years, FHB has attracted attention not only because of yield losses of economic importance, but also because of the resulting mycotoxin contamination, e.g., deoxynivalenol and zearalenone, which present a threat to the safety of human and livestock (Windels, 2000; Goswami and Kistler, 2004). F. graminearum produces both sexual (ascospores) and asexual (macroconidia) propagules, but ascospores are generally considered to be the primary inoculum of the disease (Bai and Shaner, 1996). These ascospores are formed in sexual fruiting bodies called perithecia and are forcibly discharged into the air to facilitate dispersal. The period of flowering is the most susceptible stage of wheat for head blight, and the presence of ascospores during this period is a critical epidemiological factor.
Few reports on the role of peroxisomes on growth, development and toxin production are available for F. graminearum. Deletion of FgPex5 and FgPex6 genes resulted in decreased metabolism of ROS, retarded vegetative growth on media with long-chain fatty acids, reduced sexual development and diminished pathogenicity (Min et al., 2012). A recent study (Chen et al., 2018) showed that peroxisomal proliferation was highly stimulated upon induction of DON biosynthesis and during plant infection by F. graminearum. Furthermore, deletion of FgPex13, FgPex14 or FgPex33 exhibited an increased accumulation of endogenous ROS, reduced phosphorylation of MAP kinase FgMgv1 and reduced virulence (Chen et al., 2018). FgPex4, FgPex1 and FgPex10 are involved in cell wall integrity (Zhang et al., 2019a, b). These results demonstrate that peroxisomes play pivotal roles in development, cell wall integrity, DON biosynthesis and virulence in F. graminearum.
In this study, we investigated the function of PEX3 in F. graminearum, as there are no reports on the role of PEX3 on the peroxisome assembly in F. graminearum or any other filamentous fungi. Our results demonstrate that PEX3 is crucial for the formation of mature peroxisomes, hyphal development, conidiation, sexual reproduction, the utilization of long-chain carbon sources, ROS metabolism, stress responses, and pathogenicity in F. graminearum.
Materials and Methods
Strains and Cultural Conditions
Fusarium graminearum wild-type PH-1 and transformants were cultured on PDA (200 g potato, 20 g dextrose, 20 g agar per liter) agar plates at 25oC. For growth assays, the growth rates on PDA, complete medium (CM) (1 g KH2PO4, 0.5 g MgSO4⋅7H2O, 0.5 g KCl, 2 g NaNO3, 200 μL of trace element solution (5 g citric acid, 5 g ZnSO4⋅7H2O, 0.25 g CuSO4⋅5H2O, 50 mg MnSO4⋅H2O, 50 mg H3BO3 and 50 mg NaMoO4⋅2H2O per 100 mL), 10 mL of vitamin stock solution (100 mg thiamine, 75 mg nicotinamide, 50 mg ascorbic acid, 5 mg folic acid, 5 mg biotin, 30 mg riboflavin, 75 mg pyridoxine, 200 mg Choline chloride, 5 mg p-aminobenzoic acid, 200 mg Ca-pantothenate, 4 g Inositol, absolute ethanol and ddH2O are volumetric to 1 L in a ratio of 1:1), 2 g yeast extract, 2 g peptone, 1 g casamino acids, 30 g sucrose and 20 g agar per liter) and minimal medium (MM) (1 g KH2PO4, 0.5 g MgSO4⋅7H2O, 0.5 g KCl, 2 g NaNO3, 200 μL of trace element solution, 30 g sucrose and 20 g agar per liter) were measured during 3 days after inoculation. Sensitivity to various stress agents was tested by growing the strains on MM supplemented with either 0.08 M LiCl, 1 M NaCl, 1 M Sorbitol, 1.2 M KCl, 1 M glucose. Cell wall integrity was evaluated by inoculating the strains on MM amended with 0.01% SDS, 5 mM caffeine, 0.3 g/L congo red (CR) or 0.3 mg/L calcofluor white. Determination of utilization of different carbon sources was conducted on MM supplemented with either 2% sucrose, 40 mM ethanol (C2), 40 mM sodium butyrate (C4), 40 mM sodium valerate (C5), 40 mM sodium caproate (C6), 3 mM sodium laurate (C12), 3 mM sodium tridecanoate (C13), 3 mM sodium myristate (C14), 3 mM sodium laurate (C16) or sodium oleate (C18) as the sole carbon source. Five replicates were used for each strain and three independent repeats of the experiment were performed.
Strain Construction
For deletion of FgPex3 gene, we firstly constructed a FgPex3 gene knockout vector as described below. Genomic DNA of wild-type strain PH-1 was used as template to amplify upstream and downstream fragments of FgPex3 using primer pairs R1/R2 and R3/R4, respectively followed by ligation into vector pKH-KO, carrying the hygromycin resistance gene hph. For the construction of the gene complementation vector, the fragment containing promoter, gene, and terminator of FgPex3 was amplified using primers R5/R6 and ligated into vector pKN (carrying the G418 resistance gene neo). Furthermore, we constructed a vector named as pPTS (carries the G418 resistance gene neo), which contains a GFP-PTS1 and a PTS2-DsRed fusion gene. PTS1 (SKL) was fused to the C-terminus of GFP and PTS2 was added to the N-terminus of DsRed. The GFP ORF with PTS1 signal was amplified with the forward primer GFP-Nco1 and the reverse fusion primer GFP-SKL that contains a tripeptide SKL encoding serial. The DsRed ORF with PTS2 signal was amplified using the forward fusion primer DsRed-F, which contains a PTS2 encoding region derived from N-terminal of Saccharomyces cerevisiae thiolase gene (GeneID: 854646), and the reverse primer DsRed-Kpn1. This vector was used as peroxisomal marker. The gene deletion, complementation and PTS-containing constructs were transformed into corresponding strains using the protocols described previously (Turgeon et al., 1987). PDA plates containing 100 μg/mL hygromycin B (AMRESCO, United States) or 200 μg/mL G418 (AMRESCO, United States) were used to screen the corresponding transformants. Putative positive transformants were identified by PCR assays, R7/R8 and H850/H852 primers were used for detecting the present of FgPex3 and hph gene respectively. Transformants were further confirmed by Southern hybridization, FgPex3 and hph probe were amplified by R11/R12 and R13/R14. The primers used are listed in Supplementary Table S1.
Conidiation Assays
For conidiation assays, three mycelial plugs (9 mm in diameter) of each strain were inoculated into mung bean liquid medium (MBL) (30 g mung bean, 1 L ddH2O, boiled for 10 min, filtered and sterilized) and incubated at 25°C on a shaking table (180 rpm). Each strain was set up in three technical replicates. After 4 days of cultivation, the number of conidia was counted for each strain using a hemocytometer. Conidial sizes were observed with a Leica TCS SP5 imaging system.
Sexual Reproduction Assay
For sexual reproduction, mycelial plugs of each strain were placed onto carrot agar (400 g carrot, 20 g agar per liter) medium and cultured at 25°C. After 4 days, aerial hyphae were removed and the plates were incubated under a cycle of 12 h of UV-light and 12 h of darkness for more than 1 week, during which period aerial hyphae were pressed down with 0.2% Tween-60 whenever they appeared. Formation of the perithecia was observed under a digital microscope VHX-2000 (KEYENCE, United States).
Fluorescence Microscopy
Localization of GFP- and DsRed proteins in spores and mycelium was detected using a Zeiss LSM800 Confocal System (Germany), with excitation at 488 nm and emission at 520 nm for GFP, or excitation at 543 nm and emission at 607 nm for DsRed.
Measurement of ROS Accumulation and Viability
The wild-type strain PH-1 and the FgPex3 gene deletion mutant were inoculated into CM medium or CM medium containing 0.6 M NaCl for 3 days. Then the strains were stained with 20 mL of 0.2% NBT and cultured at 25°C for 30–45 min. Finally, excess of staining agent was removed by rinsing with ethanol and plates were placed in the incubator for 30–45 min and then used for photo recording. Mycelial plugs from the original cultures were transferred to fresh CM agar plates were cultured for 7 days, 30 days, 60 days, 90 days, respectively. The growth was observed and photographed.
Pathogenicity Determination
Conidia produced in the MBL medium were harvested and resuspended to a final concentration of 106 conidia/mL in sterile water as previously described (Kong et al., 2018). For infection on flowering wheat (cv. Yangmai 158) grown in the greenhouse, a spikelet at the lower-middle part of a wheat head was inoculated with 10 μL of conidial suspension, with 30 replicates per strain. The inoculated ear was sprinkled with water and incubated in a humidity chamber for 3 days. Symptomatic spikelets in each inoculated wheat head were recorded and photographed. Infection assays on tomato were performed as previous described (Di Pietro et al., 2001). In brief, a 10 μL aliquot of a conidial suspension (106 conidia/mL) was injected into wounded tomato after surface disinfection. Inoculated tomatoes were incubated in 25°C, 80–100% humidity with 12 h of daylight and were investigated 3 days after inoculation.
DON Production Assays
One milliliter spore suspension with a concentration of 105 spores/mL was added to a flask with 30 g of sterilized rice kernels. Each strain was cultured for 7 days with 3 replicates at 25°C, shaking the flasks on a daily basis. The infected rice medium was dried in an oven at 60°C and subsequently ground to a fine powder. 5 g samples were mixed with 25 mL distilled water for 3 min, centrifuged and filtered, and 50 μL filtrate was taken for analysis using the DON Plate Kit ELISA from Beacon Analytical Systems (Portland, ME, United States) according to the manufacturer’s instructions.
Results
Identification of FgPex3 in F. graminearum
The F. graminearum PEX3 ortholog (FGSG_06942, assigned as FgPex3 in the present work) was identified from the genome data-base (King et al., 2015) through BLASTP searches using the S. cerevisiae PEX3 protein (GenBank accession number: NP_010616) as a query. The FgPex3 gene consists of three exons and two introns, and encodes a polypeptide of 523 amino acid residues. The predicted protein sequences of FgPex3 exhibited moderate identity (25%) with that of S. cerevisiae. The PEX3 amino acid sequences from several pathogenic fungi (Magnaporthe oryzae, Neurospora crassa, Aspergillus flavus, A. nidulans and Fusarium oxysporum) were downloaded from the NCBI database, and the phylogenetic tree based on the amino acid sequences revealed that FgPex3 is most closely related to FoPex3 and MoPex3 (Figure 1).
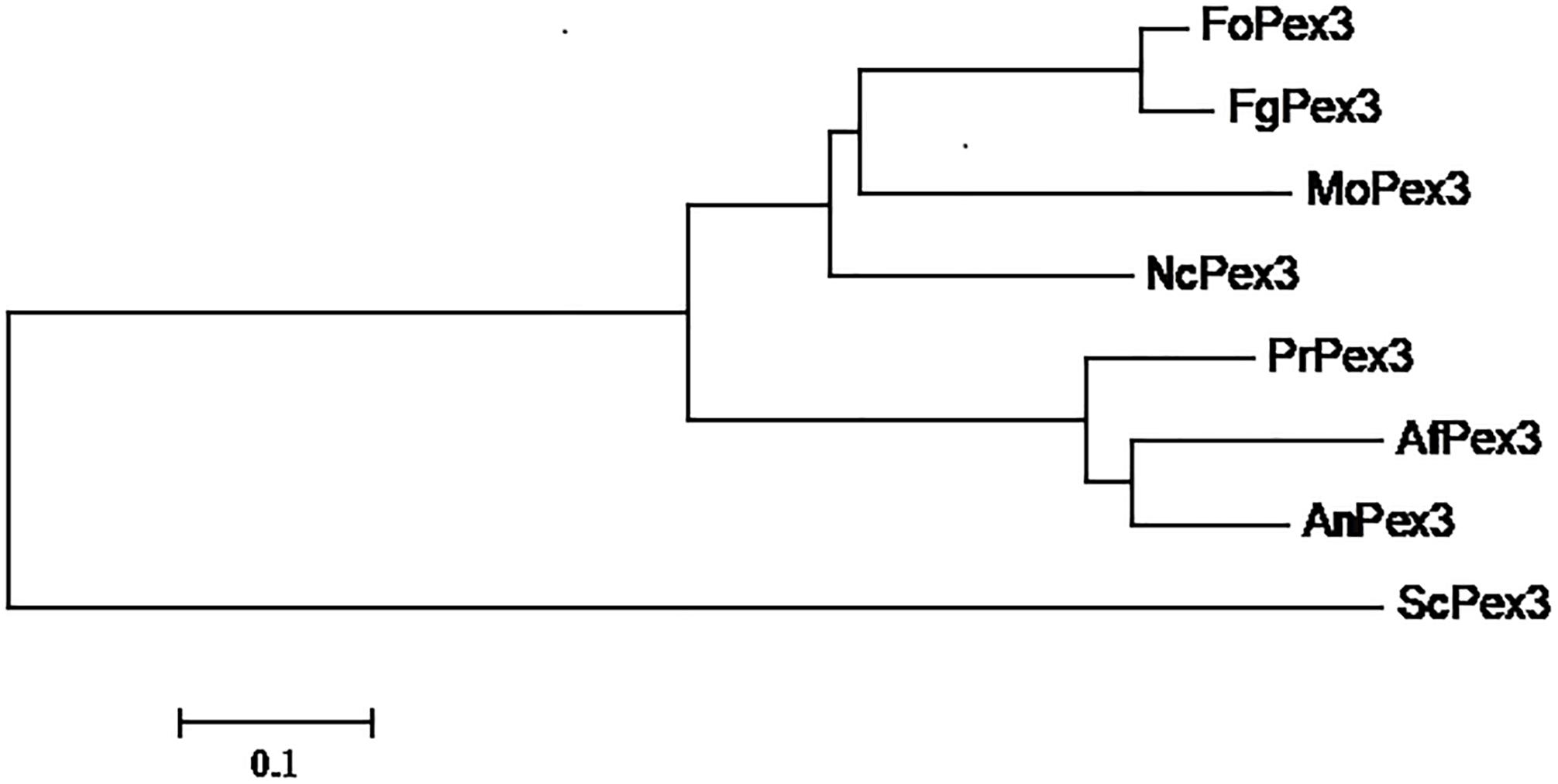
Figure 1. Phylogenetic relationship of Pex3 protein homologs. Phylogenetic relationship of Pex3 homologs calculated using the neighbor-joining method with the MEGA 6.0 program. Abbreviations and accession numbers of the sequences are as follows: FoPex3 (EWZ44752.1) from Fusarium oxysporum Fo47; FgPex3 (XP_011326612.1) from Fusarium graminearum PH-1; MoPex3 (XP_003717143.1) from Magnaporthe oryzae 70–15; NcPex3 (XP_962987.1) from Neurospora crassa OR74A; PrPex3 (XP_002567187.1) from Penicillium rubens Wisconsin 54–1255; AfPex3 (XP_002383001.1) from Aspergillus flavus NRRL3357; AnPex3 (XP_659885.1) from Aspergillus nidulans FGSC A4; ScPex3 (NP_010616.3) from Saccharomyces cerevisiae S288C.
Construction of the ΔFgPex3 Mutant and FgPex3 Complemented Strain
To determine the functions of FgPex3, we performed gene replacement through homologous recombination (Figure 2A), and the resulting hygromycin-resistant strains were tested by PCR. As expected, the fragment within the FgPex3 nucleotide sequence could be detected in the wild-type, but not in the ΔFgPex3 mutant (Figure 2B). Replacement of the FgPex3 gene in these mutants was further confirmed by Southern hybridization (Figure 2C). Complementation of this mutant was obtained by ectopic insertion of the FgPex3 gene into the genome and identification by PCR (primers R7/R8) (data not shown) as well as by Southern hybridization (Figure 2D).
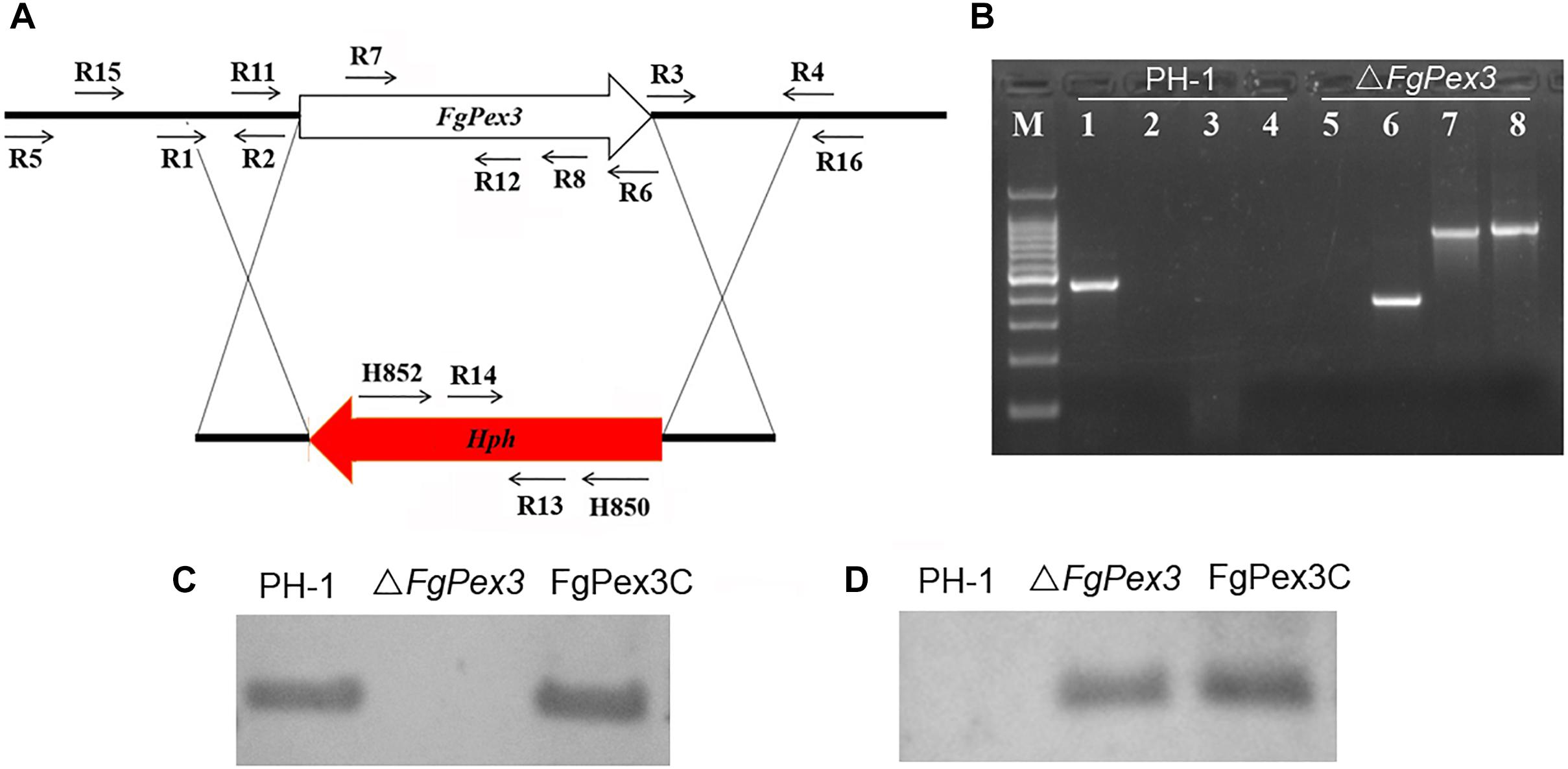
Figure 2. The FgPex3 gene replacement construct. (A) Schematic representation gene disruption strategy. FgPex3 is denoted by an open arrow; hygromycin resistance cassette (hph) is represented by a red arrow. The annealing sites of primers are indicated by small black arrows. (B) PCR analysis of PH-1 and FgPex3 deletion mutant using primers R7/R8, H850/H852, H852/R16, and R15/H850, respectively. 1–4: PCR products amplified with the above primers using PH-1 as template; 5–8: PCR products amplified with the above primers using ΔFgPex3 as template. (C,D) Southern blot hybridization analyses of transformants with hybridization probe R11/R12 (FgPex3) and R13/R14 (hph).
FgPex3 Is Indispensable for the Mature Peroxisome Biogenesis
It is reported that yeast pex3 atg1 mutant cells contain abundant reticular and vesicular structures — preperoxisomal vesicles (PPVs) that harbor key proteins of the peroxisomal receptor docking complex Pex13 and Pex14 as well as the matrix proteins Pex8 and alcohol oxidase. Besides, Pex3 first appeared on the PPVs upon its reintroduction in pex3 cells, after which these structures mature into normal peroxisomes (Aksit and van der Klei, 2018). That is to say, Pex3 is essential for the maturation of peroxisomes. To test whether the deletion of FgPex3 had any effect on the peroxisome biogenesis, we analyzed the localization of the fluorescent hybrid proteins GFP-PTS1 and PTS2-DsRed, which represent the distribution of peroxisomes. In conidia and in mycelium of wild-type strain PH-1, the two marker proteins were found to be well distributed in peroxisomes (visible as discrete entities in Figure 3), while in spores and mycelia of ΔFgPex3, the two marker proteins exhibited a diffuse fluorescence pattern across the cytoplasm (Figure 3). This indicated that FgPex3 is required for the biogenesis and formation of mature peroxisomes.
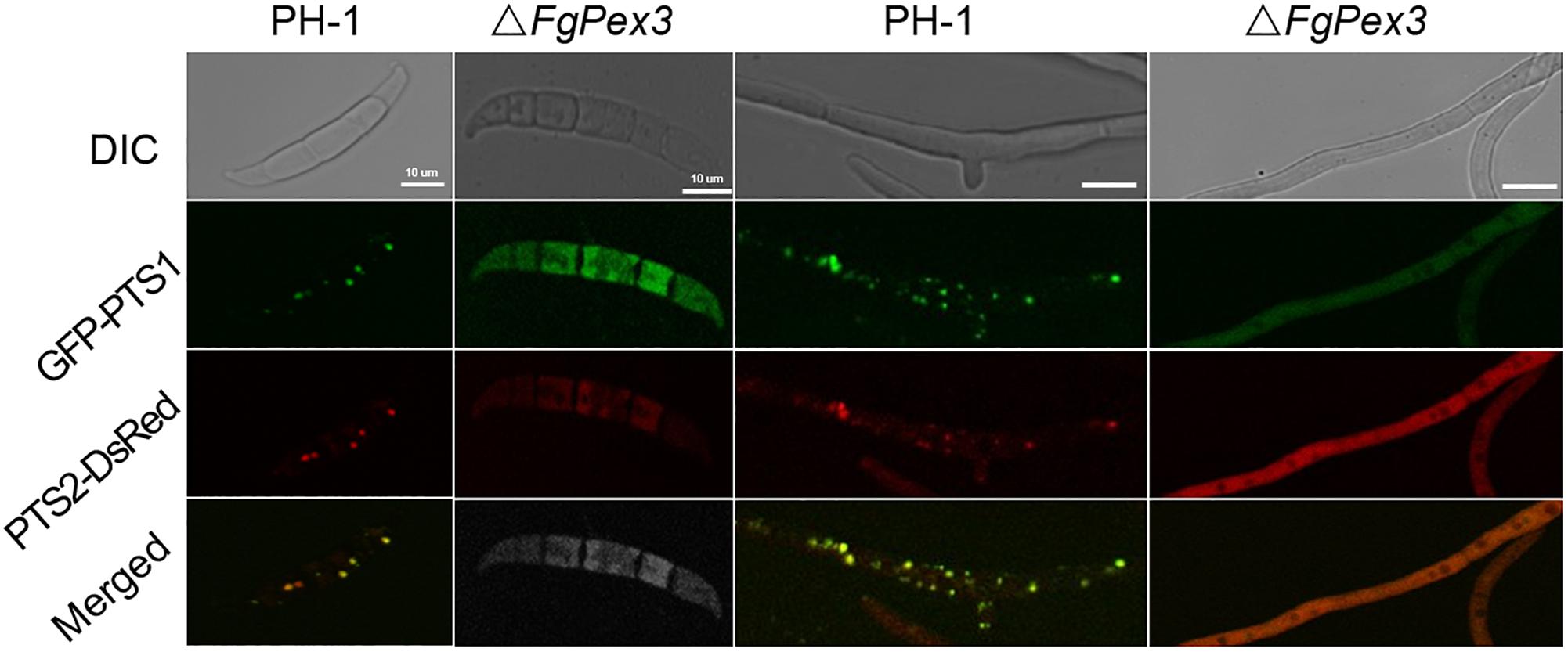
Figure 3. Peroxisomal targeting fusion proteins in spores (left) and mycelia (right) of PH-1 and ΔFgPex3. The fluorescence of GFP-PTS1 and PTS2-DsRed localized to peroxisomes as discernable structures in the wild type strain PH-1, whereas both types of fluorescence exhibited a diffuse pattern in the cytoplasm of the mutant ΔFgPex3. DIC, differential interference contrast; GFP-PTS1, green fluorescent protein contained in the peroxisomal targeting signal 1; DsRed-PTS2, red fluorescent protein contained in the peroxisomal targeting signal 2; Merge, overlays of green and red fluorescence proteins images. Scale bar = 10 μm.
FgPex3 Influences Vegetative Growth
As shown in Figure 4A, the colony morphology of ΔFgPex3 mutant strains had great differences on PDA medium compared with that of wild-type PH-1 and the complemented strain, FgPex3C. On PDA medium, ΔFgPex3 showed short and spare aerial hyphae, while on MM and CM media, no morphological differences could be observed between the three strains, with the exception that ΔFgPex3 seemed to grow slightly slower. The colony diameter of ΔFgPex3 (43.36 + 2.21 mm) was significantly smaller than that of PH-1 (49.68 + 2.12 mm) and FgPex3C (49.95 + 3.21 mm) after 3 days at 25°C (P < 0.05).
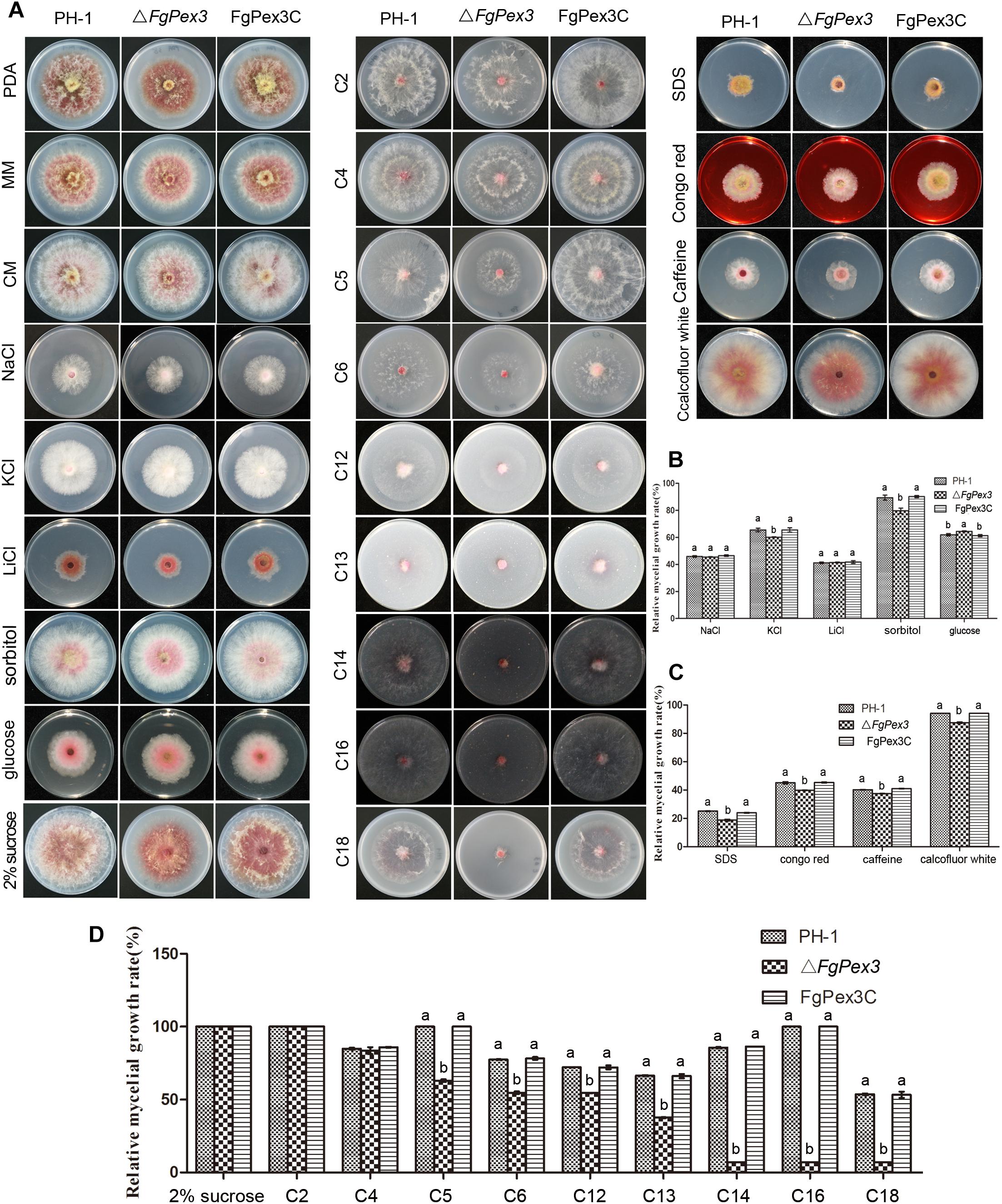
Figure 4. Growth analysis of F. graminearum strains under different culture conditions. (A) The wild-type PH-1, deletion mutant and the complemented strain were cultured on three basic media (PDA, CM, and MM) as well as MM plates supplemented with osmotic stress agents (NaCl, KCl, LiCl, sorbitol or glucose), MM plates amended with either cell wall damaging agents (SDS, Congo red, caffeine or calcofluor white) or with various carbon sources (2% sucrose, 40 mM sodium acetate (C2), 40 mM sodium butyrate (C4), 40 mM sodium valerate (C5), 40 mM sodium hexanoate (C6), 3 mM laurate acid (C12), 3 mM tridecanoic acid (C13), 3 mM myristic acid (C14), 3 mM palmitic acid (C16) or 3 mM oleic acid (C18) as the sole carbon source) at 25°C for 3 days. (B–D) Colony diameters of the tested strains were measured to calculate the inhibition rate and statistically analyzed. Error bars represent the deviation from five replicates, and the same letter on the bars for each treatment indicates no significant difference at P = 0.05.
FgPex3 Is Essential for Stress Responses and Cell Wall Integrity of F. graminearum
To determine whether ΔFgPex3 was defective in various forms of stress response, we assayed its growth on MM plates with different stress agents compared to growth on MM plates without any stress agent. In osmotic sensitivity tests, ΔFgPex3 proved more sensitive to potassium chloride (1.2 M) and sorbitol (1 M), and its relative growth rates decreased significantly (P < 0.05). When growing on a medium containing glucose (1 M), the sensitivity of ΔFgPex3 decreased and the relative growth rates increased (P < 0.05). ΔFgPex3 did not show any obvious response to sodium chloride (1.2 M) or lithium chloride (0.1 M) (Figures 4A,B). To investigate whether FgPex3 plays a role in maintaining cell wall integrity in F. graminearum, we examined how the gene deletion mutant responded to a range of cell wall-damaging agents. As shown in Figure 4A, the ΔFgPex3 mutant was more sensitive toward SDS (0.01%), Congo red (0.3 mg/L), caffeine (5 mM) and calcofluor white (0.3 mg/L) than either the wild-type or the complemented strain and its growth rate was significantly decreased (Figure 4C, P < 0.05).
Deletion of FgPex3 Affects the Capacity to Utilize of Long-Chain Carbon Sources
For the determination of the capacity to use carbon sources, PH-1, ΔFgPex3 mutant and FgPex3C was cultured on MM plates with 2% (58.4 mM) sucrose, 40 mM sodium acetate (C2), 40 mM sodium butyrate (C4), sodium valerate (C5), 40 mM sodium caproate (C6), 3 mM sodium laurate (C12), 3 mM sodium tridecanoate (C13), 3 mM sodium myristate (C14), 3 mM sodium palmitate (C16) or 3 mM sodium oleate (C18) as the sole carbon source.
Compared with wild-type strain PH-1 and the complemented strain FgPex3C, ΔFgPex3 mutant showed no significant difference in relative growth rates on MM medium with sucrose, C2 and C4 as the sole carbon sources. In contrast, relative growth rates of ΔFgPex3 mutant on MM medium with C5, C6, C12, C13, C14, C16 and C18 as sole carbon sources were significantly decreased (P < 0.05). With the increase of fatty acid carbon chain length, the growth of the ΔFgPex3 mutant was reduced to very tiny colonies on C14, C16 or C18 media (Figures 4A,D).
FgPex3 Is Involved in the Conidial Production and Sexual Development
The ΔFgPex3 mutant produced significantly less conidia compared to the wild-type and complemented strain FgPex3C in MBL medium (P < 0.05) (Figure 5A). Furthermore, microscopic examination showed that the conidia of ΔFgPex3 mutant were shorter than those of wild-type and complemented strains (Figure 5B). These results indicate that FgPex3 is important for conidiogenesis.
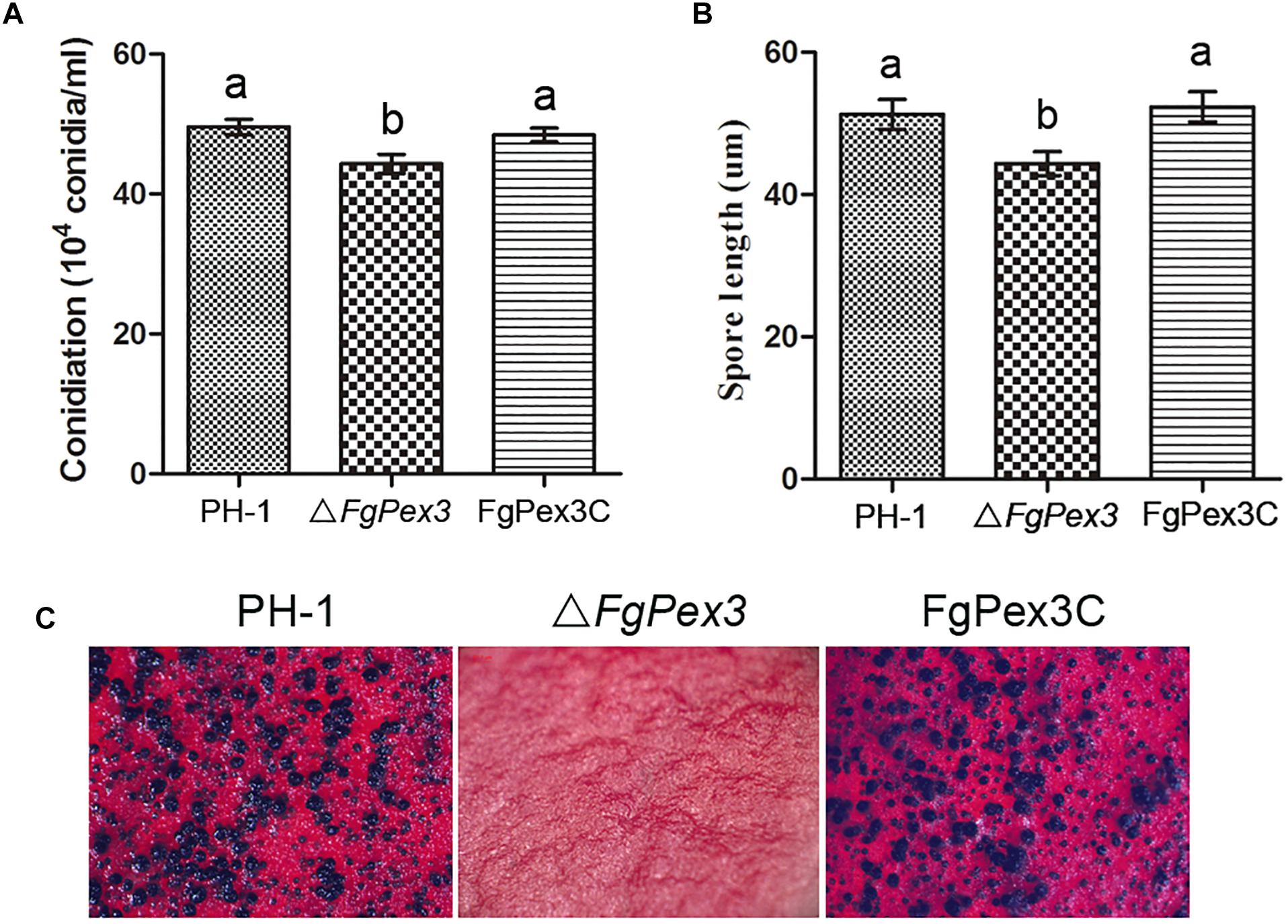
Figure 5. Conidiation and sexual reproduction of PH-1, ΔFgPex3 and FgPex3C. (A) Number of conidia produced in MBL medium during 6 days at 25°C. Means and standard errors were calculated from three independent replicates. Error bars represent standard deviation. Values on the bars followed by the same letter indicate no significant difference at P = 0.05. (B) Size of conidia produced by PH-1, ΔFgPex3 and FgPex3C strains. Error bars indicate the standard deviations of three repeated experiments. Values on the bars followed by the same letter are not significantly different at P = 0.05. (C) Perithecia formation of PH-1, ΔFgPex3 and FgPex3C on carrot agar plates were photographed at 2 weeks post-fertilization. Mutant ΔFgPex3 failed to produce any perithecia.
Because ascospores play a vital role in the primary infection of F. graminearum, the production of perithecia on carrot agar plates was determined as described (Kong et al., 2018). At 14 days post fertilization, the wild type strain PH-1 and the complemented strain FgPex3C produced mature perithecia, while the ΔFgPex3 mutant failed to produce any perithecia under the same conditions (Figure 5C). The absence of perithecia indicates that FgPex3 is essential for sexual reproduction.
FgPex3 Affects the Virulence of F. graminearum
Infection experiments were performed to see if the ΔFgPex3 mutant was affected in the pathogenicity on tomato and wheat. Inoculations were performed on tomatoes and wheat spikelets. The infected area of the ΔFgPex3 mutant on tomato was substantially smaller and did not expand beyond the inoculation site (Figure 6). Most of the wheat heads inoculated with the ΔFgPex3 mutant showed rarely symptoms. Under the same experimental conditions, PH-1 and FgPex3C strains caused extensive lesions on tomato fruits and typical scab symptoms on wheat heads (Figure 6). These results revealed that FgPex3 plays an important role in the infection of (host) plants. The displayed reduced virulence and failure to spread to neighboring spikelets is reminiscent of mutants in the DON pathway (Proctor et al., 1995; Seong et al., 2009). However, when we examined the DON accumulation in the FgPex3 deletion mutant, no significant difference in DON production among the wild type, ΔFgPex3 mutant and the complemented strain FgPex3C was observed (Figure 7). This indicates that DON production is still possible and does not require FgPex3.

Figure 6. Virulence of PH-1, ΔFgPex3 and FgPex3C on infected plants. Virulence of the three strains was determined by point inoculations of conidial suspensions on five tomato fruits and 30 wheat heads. Infected tomato fruits and wheat heads were photographed at 3 days and 21 days post inoculation, respectively.
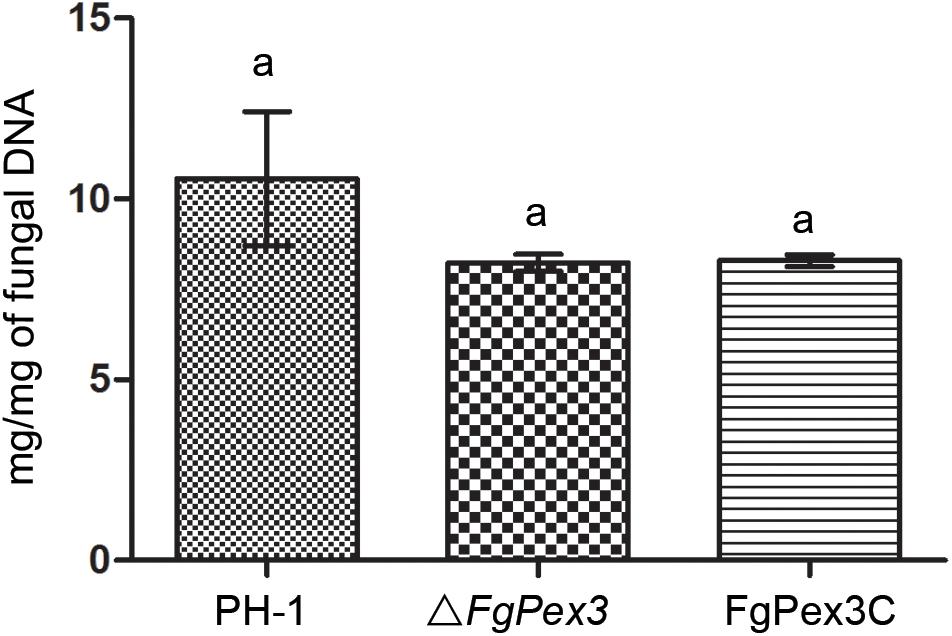
Figure 7. DON production of the wild-type, mutant and complemented strain on infected rice kernels. Values on the bars followed by the same letter indicate no significant difference at P = 0.05.
ROS Accumulation and Viability of ΔFgPex3
Under different stress conditions, most eukaryotic organisms will produce large amounts of ROS, including hydrogen peroxide (H2O2), superoxide radicals and singlet oxygen (Baxter et al., 2014; Mor et al., 2014), to resist environmental changes. Nitroblue tetrazolium (NBT) can react with superoxide radicals to form blue formazan. When NBT was added to the growth plate, the color of the plate on which ΔFgPex3 mutant was growing on, was significantly deeper than that of wild-type strain PH-1 (Figure 8A), indicating increased accumulation of ROS in ΔFgPex3.
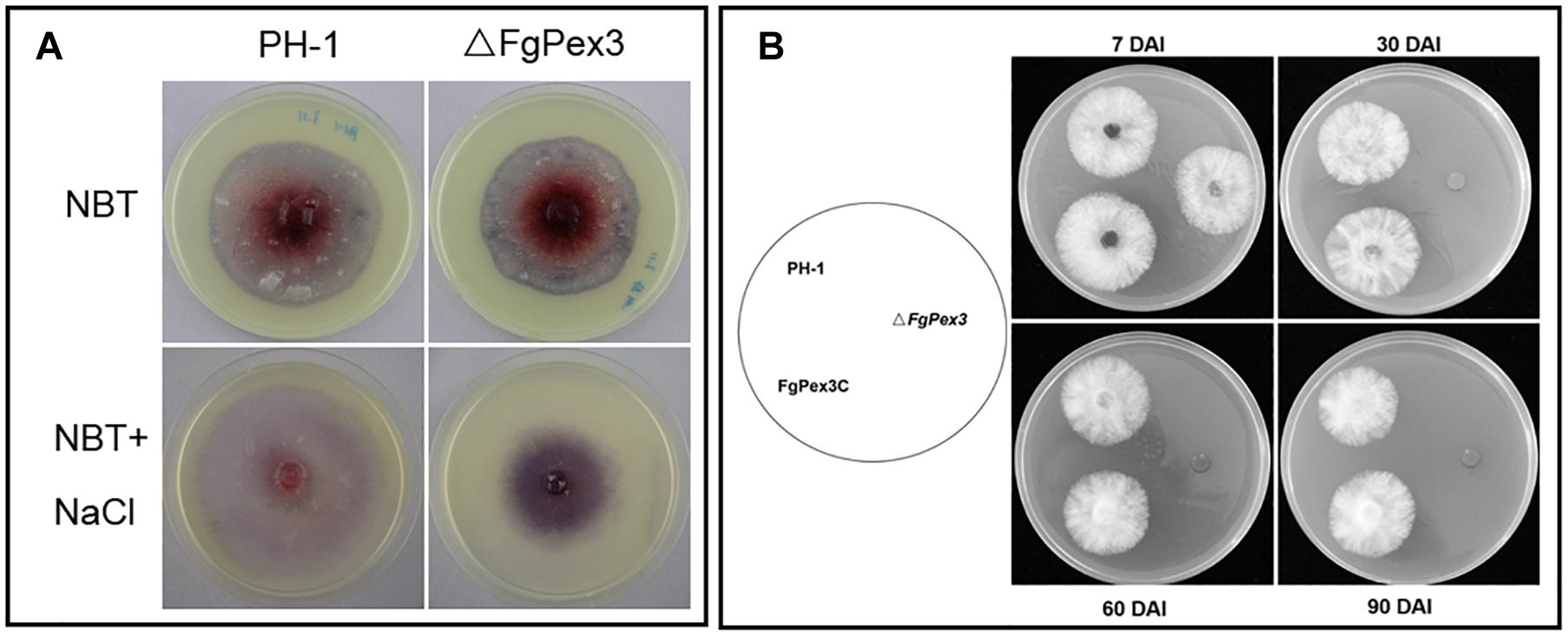
Figure 8. Reduced ability of ΔFgPex3 to scavenge reactive oxygen species (ROS) survival assay. (A) Tetranitroblue tetrazolium chloride (NBT) staining for ROS accumulation in cultures of ΔFgPex3 and the wild-type. In plates at the bottom 0.6 M NaCl was added to promote ROS production. The dark blue color of the colony indicated ROS accumulation. (B) Strains PH-1,ΔFgPex3, FgPex3C were inoculated onto CM plates and incubated for 7–90 days. Mycelial plugs from the cultures were transferred to fresh CM agar plates, and the viability of the culture was observed. DAI, days after incubation.
To investigate a possible function of FgPex3 on the viability of F. graminearum, we performed survival assays of mycelia. ΔFgPex3 mutant, wild-type strain PH-1 and FgPex3C were cultured for 7, 30, 60, and 90 days in CM medium, and mycelial plugs were inoculated onto fresh CM plates. There was no significant difference in viability among 7-day-old mycelia. However, 30-, 60- and 90-day-old mycelia of ΔFgPex3 were no longer viable. Both PH-1 and FgPex3C strains remained viable (Figure 8B). In conclusion, the deletion of FgPex3 may affect the metabolism of peroxisomes in F. graminearum, and the accumulation of harmful substances such as ROS causes cell death, resulting in a decline of viability of ΔFgPex3 beyond seven days of cultivation.
Discussion
Peroxisomes are involved in a variety of metabolic reactions in almost all eukaryotic cells. Previously, several peroxisomal proteins were shown to be associated with biosynthesis of secondary metabolites and virulence in filamentous fungi (van der Klei and Veenhuis, 2013). However, the role of PEX3, an integral peroxisomal membrane protein that interacts with the cytosolic shuttling receptor PEX19 was not studied. This is surprising, as PEX3 was suggested to be responsible for the early steps of peroxisome biogenesis as well as assembly of peroxisomal membranes and/or the correct translocation of PMPs (Ghaedi et al., 2000; Schliebs and Kunau, 2004). Here, we functionally analyzed PEX3 from F. graminearum using targeted gene deletion methods, and found that the loss of FgPex3 led to a plethora of developmental defects in hyphal growth, asexual and sexual reproduction, stress responses as well as virulence in F. graminearum. This is the first demonstration that PEX3 plays an important role in pathogenicity of filamentous fungus.
Eukaryotic cells are highly organized and specific functions are performed by specialized cellular compartments such as Golgi apparatus, ER, vacuole, mitochondria nucleolus and peroxisomes. Peroxisomes are devoid of genome or transcription machinery, therefore, all peroxisomal membrane and matrix proteins have to be imported from the cytosol (Purdue and Lazarow, 2001). It has been suggested that all PMPs accumulate in the endoplasmic reticulum (ER) in pex3 cells, and that upon reintroduction of Pex3 these PMPs are incorporated in two types of vesicles that fuse to form mature peroxisomes (van der Zand et al., 2010, 2012). According to this model, Pex3 is important for the exit of PMPs from the ER into PPVs. However, recent findings showed that pex3 atg1 cells contain PPVs, which are the targets for the reintroduced Pex3, which then mature into normal peroxisomes (Knoops et al., 2014; Wroblewska et al., 2017). Based on this, Pex3 is required for the formation of mature peroxisomes. In our study, ΔFgPex3 matrix proteins (PTS1 or PTS2 containing proteins) were mislocated, demonstrating that FgPex3 is also required for the formation of mature peroxisomes in F. graminearum.
Fatty acid β-oxidation is the main pathway for fatty acid degradation in eukaryotes. In plants, β-oxidation pathway plays a central role in plant reproduction, seed development and germination and post-germinative seedling establishment (Pan et al., 2018). Earlier studies in yeast suggest that fungi lack the mitochondrial fatty acid β-oxidation pathway (Hiltunen et al., 1992; Kurihara et al., 1992). However, related studies have shown that A. nidulans has both mitochondrial and peroxisomal fatty acid β-oxidation pathways (Maggio-Hall and Keller, 2004). Bioinformatic analyses indicate that both these pathways exist in most filamentous fungi, including F. graminearum (Shen and Burger, 2009). It has been reported that deletion of pex genes in various fungi can cause defects in fatty acid utilization. For example, deletion of the pex7 gene in M. oryzae and in F. graminearum leads to short-chain fatty acid utilization disorder, while gene deletion of pex5 or pex6 in F. graminearum gave rise to reduced growth rate on media containing either long-chain or short-chain fatty acids (Goh et al., 2011; Min et al., 2012). Our study showed that the deletion of FgPex3 in F. graminearum significantly reduced the utilization of both long-chain (C > = 12)and short-chain (C = 5 or 6) fatty acids. However, unlike the ΔFgPex5 and ΔFgPex6 deletion mutants, deletion of FgPex3 gradually decreased the utilization of fatty acids with increasing of carbon chain length and it was almost impossible to grow and expand on medium with carbon chains longer than 14 (myristic acid, palmitic acid, oleic acid). In studies with animal cells it was also observed that the substrate for peroxisomal fatty acid β-oxidation is primarily long-chain to very-long-chain fatty acids (Reddy and Hashimoto, 2001). We hypothesize that similarly to these animal systems the deletion of the FgPex3 affects the fatty acid β-oxidation process in F. graminearum, and significantly reduces the utilization of long-chain fatty acids.
Triacylglycerol produced by fatty acid β-oxidation is important for the production of ascospores by Ascomycetes (Lee et al., 2011). During the process of sexual reproduction of ascomycetes, the expression of genes associated with peroxisomal proteins and fatty acids biosynthesis is up-regulated and lipid content is gradually reduced (Guenther et al., 2009). Effective use of lipids can provide both energy and metabolites required for formation female sexual reproductive structures. In Podospora anserina and A. nidulans, peroxisomal proteins play important roles in sexual reproduction (Bonnet et al., 2006; Hynes et al., 2008). The FgPex3 gene-deficient F. graminearum mutant was unable to produce perithecia on carrot agar medium, which could be linked to fatty acid β-oxidation in peroxisomes as these play an important role in sexual reproduction of F. graminearum (Peraza-Reyes and Berteaux-Lecellier, 2013).
Oxidative reactions are subject of the peroxisome metabolism, which generate massive amounts of ROS, mainly H2O2 (Chen et al., 2016). Besides H2O2, it has been demonstrated that peroxisomes also produce superoxide radicals as a consequence of their normal metabolism (Del Rio and Lopez-Huertas, 2016). ROS can be scavenged by catalases and peroxidases, which are abundant in peroxisomes. In Hansenula polymorpha the degradation of the peroxisomes caused by the exposure to high methanol conditions also resulted in an increase in formation of ROS (van Zutphen et al., 2011). In this study, the disruption of FgPex3 in F. graminearum blocked peroxisome biogenesis and resulted in a significant increase in the accumulation of superoxide radicals, which reflects a higher ROS level. Like FgPex5 and FgPex6 deletion mutant, the survival of the FgPex3 gene deletion mutant was significantly reduced. These results indicate that FgPex3 is involved in mature peroxisomes formation, causing abnormal ROS metabolism, accumulation of ROS, and ultimately cell death.
The filamentous fungi, as an important group of plant pathogenic fungi, have attracted extensive attention for their pathogenicity. In the rice blast fungus, M. oryzae, the loss of MoPex5, MoPex7 or MoPex14/17 led to decreased conidia production, reduction of fatty acid utilization, ROS degradation and cell wall integrity as well as diminished virulence (Goh et al., 2011; Wang et al., 2013; Li et al., 2017). In F. graminearum, PEX5 and PEX6 deletion mutants showed greatly reduced pathogenicity (Min et al., 2012). In this study, we observed that the pathogenicity of the ΔFgPex3 mutant is also strongly reduced both on its natural host wheat as well as on the alternative host tomato. To our knowledge, this is the first time that the fungal PEX3 has been found to be involved in pathogenicity.
Peroxisomal fatty acid β-oxidation plays an important role in the synthesis of glycerol and melanin, thereby affecting the infection of plant-pathogenic fungi like Colletotrichum lagenarium and M. oryzae in host plants (Kimura et al., 2001; Ramos-Pamplona and Naqvi, 2006; Wang et al., 2013). The FgPex3 gene deletion mutant strain can produce lesions on tomato and infect inoculated wheat spikelets, which indicates that peroxisomes have little effects on the initial stage of pathogen infection. However, PEX3 plays a pivotal role during the process of systemic infection. Loss of peroxisomal function may result in reduced utilization of carbon sources from both the pathogen itself and from host plants, resulting in a slower growth rates. This slower growth rate could provide host plants time needed to prepare to ward off the fungal invasion, for instance by thickening the cell walls of the wheat head (Jansen et al., 2005), resulting in decreased pathogenicity. DON is an important factor for the virulence of F. graminearum on wheat. Although the disruption of FgPex3 in F. graminearum still allowed DON production, pathogenicity is markedly reduced, indicating the importance of additional factors for virulence in Δ FgPex3.
In summary, we have functionally characterized FgPex3, an integral peroxisomal membrane protein involved in the mature peroxisomes biogenesis, fungal development and pathogenicity in F. graminearum. We discovered that FgPex3 plays an essential role in the peroxisome biogenesis, fungal development, lipid degradation, scavenging of ROS, cell wall integrity and virulence. We also observed that the development of sexual fruiting bodies is blocked in the mutant. As the sexual ascospores are a critical driver for the epidemiology, fungicides that target the PEX3 or other proteins that are essential for peroxisome formation could be good targets for fungicide development.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
Author Contributions
JF, WC, and HZ conceived and designed the study. XK and XW carried out the experiments, analyzed the data, prepared the figures and tables, and wrote the manuscript. JinX and JingX provided assistance for data analysis. HZ, BB, AD, TL, and CW wrote and revised the drafts of the manuscript. All authors read and approved the final manuscript.
Funding
This project received funding from the National Key R&D Program of China (2017YFE0126700, 2018YFD0200500, and 2016YFE0112900), the EU Horizon 2020 Research and Innovation Programme under grant agreement no. 678781 (MycoKey), the National Natural Science Foundation of China (No. 31201477), and the Fundamental Research Funds for Central Non-profit Scientific Institution (Y2017XM01).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02088/full#supplementary-material
References
Aksit, A., and van der Klei, I. J. (2018). Yeast peroxisomes: how are they formed and how do they grow? Int. J. Biochem. Cell Biol. 105, 24–34. doi: 10.1016/j.biocel.2018.09.019
Bai, G. H., and Shaner, G. (1996). Variation in Fusarium graminearum and cultivar resistance to wheat scab. Plant Dis. 80, 975–979. doi: 10.1094/Pd-80-0975
Bai, G. H., and Shaner, G. (2004). Management and resistance in wheat and barley to Fusarium head blight. Annu. Rev. Phytopathol. 42, 135–161. doi: 10.1146/annurev.phyto.42.040803.140340
Baxter, A., Mittler, R., and Suzuki, N. (2014). ROS as key players in plant stress signalling. J. Exp. Bot. 65, 1229–1240. doi: 10.1093/jxb/ert375
Bonnet, C., Espagne, E., Zickler, D., Boisnard, S., Bourdais, A., and Berteaux-Lecellier, V. (2006). The peroxisomal import proteins PEX2, PEX5 and PEX7 are differently involved in Podospora anserina sexual cycle. Mol. Microbiol. 62, 157–169. doi: 10.1111/j.1365-2958.2006.05353.x
Brakhage, A. A., Sprote, P., Al-Abdallah, Q., Gehrke, A., Plattner, H., and Tuncher, A. (2004). Regulation of penicillin biosynthesis in filamentous fungi. Adv. Biochem. Eng. Biotechnol. 88, 45–90. doi: 10.1007/b99257
Brown, L. A., and Baker, A. (2008). Shuttles and cycles: transport of proteins into the peroxisome matrix. Mol. Membr. Biol. 25, 363–375. doi: 10.1080/09687680802130583
Chen, X. L., Wang, Z., and Liu, C. Y. (2016). Roles of peroxisomes in the rice blast fungus. Biomed Res. Int. 2016, 1–10. doi: 10.1155/2016/9343417
Chen, Y., Zheng, S. Y., Ju, Z. Z., Zhang, C. Q., Tang, G. F., Wang, J., et al. (2018). Contribution of peroxisomal docking machinery to mycotoxin biosynthesis, pathogenicity and pexophagy in the plant pathogenic fungus Fusarium graminearum. Environ. Microbiol. 20, 3224–3245. doi: 10.1111/1462-2920.14291
Del Rio, L. A., and Lopez-Huertas, E. (2016). ROS generation in peroxisomes and its role in cell signaling. Plant Cell Physiol. 57, 1364–1376. doi: 10.1093/pcp/pcw076
Di Pietro, A., Garcia-Maceira, F. I., Meglecz, E., and Roncero, M. I. G. (2001). A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 39, 1140–1152. doi: 10.1046/j.1365-2958.2001.02307.x
Distel, B., Erdmann, R., Gould, S. J., Blobel, G., Crane, D. I., Cregg, J. M., et al. (1996). A unified nomenclature for peroxisome biogenesis factors. J. Cell Biol. 135, 1–3. doi: 10.1083/jcb.135.1.1
Ghaedi, K., Tamura, S., Okumoto, K., Matsuzono, Y., and Fujiki, Y. (2000). The peroxin Pex3p initiates membrane assembly in peroxisome biogenesis. Mol. Biol. Cell 11, 2085–2102. doi: 10.1091/mbc.11.6.2085
Goh, J., Jeon, J., Kim, K. S., Park, J., Park, S. Y., and Lee, Y. H. (2011). The PEX7-mediated peroxisomal import system is required for fungal development and pathogenicity in Magnaporthe oryzae. PLoS One 6:e28220. doi: 10.1371/journal.pone.0028220
Goswami, R. S., and Kistler, H. C. (2004). Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 5, 515–525. doi: 10.1111/J.1364-3703.2004.00252.X
Gould, S. J., Keller, G. A., Hosken, N., Wilkinson, J., and Subramani, S. (1989). A conserved tripeptide sorts proteins to peroxisomes. J. Cell Biol. 108, 1657–1664. doi: 10.1083/jcb.108.5.1657
Guenther, J. C., Hallen-Adams, H. E., Bucking, H., Shachar-Hill, Y., and Trail, F. (2009). Triacylglyceride metabolism by Fusarium graminearum during colonization and sexual development on wheat. Mol. Plant Microbe Interact. 22, 1492–1503. doi: 10.1094/Mpmi-22-12-1492
Hiltunen, J. K., Wenzel, B., Beyer, A., Erdmann, R., Fossa, A., and Kunau, W. H. (1992). Peroxisomal multifunctional beta-oxidation protein of Saccharomyces cerevisiae. Molecular analysis of the FOX2 gene and gene product. J. Biol. Chem. 267, 6646–6653. doi: 10.1016/0014-5793(92)80867-G
Hynes, M. J., Murray, S. L., Duncan, A., Khew, G. S., and Davis, M. A. (2006). Regulatory genes controlling fatty acid catabolism and peroxisomal functions in the filamentous fungus Aspergillus nidulans. Eukaryot. Cell 5, 794–805. doi: 10.1128/EC.5.5.794-805.2006
Hynes, M. J., Murray, S. L., Khew, G. S., and Davis, M. A. (2008). Genetic analysis of the role of peroxisomes in the utilization of acetate and fatty acids in Aspergillus nidulans. Genetics 178, 1355–1369. doi: 10.1534/genetics.107.085795
Jansen, C., von Wettstein, D., Schafer, W., Kogel, K. H., Felk, A., and Maier, F. J. (2005). Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. Proc. Natl. Acad. Sci. U.S.A. 102, 16892–16897. doi: 10.1073/pnas.0508467102
Kimura, A., Takano, Y., Furusawa, I., and Okuno, T. (2001). Peroxisomal metabolic function is required for appressorium-mediated plant infection by Colletotrichum lagenarium. Plant Cell 13, 1945–1957. doi: 10.1105/tpc.13.8.1945
King, R., Urban, M., Hammond-Kosack, M. C. U., Hassani-Pak, K., and Hammond-Kosack, K. E. (2015). The completed genome sequence of the pathogenic ascomycete fungus Fusarium graminearum. BMC Genomics 16:544. doi: 10.1186/s12864-015-1756-1
Knoops, K., Manivannan, S., Cepinska, M. N., Krikken, A. M., Kram, A. M., Veenhuis, M., et al. (2014). Preperoxisomal vesicles can form in the absence of Pex3. J. Cell Biol. 204, 659–668. doi: 10.1083/jcb.201310148
Kong, X., van Diepeningen, A. D., van der Lee, T. A. J., Waalwijk, C., Xu, J., Xu, J., et al. (2018). The Fusarium graminearum histone acetyltransferases are important for morphogenesis, DON biosynthesis, and pathogenicity. Front. Microbiol. 9:654. doi: 10.3389/fmicb.2018.00654
Kurihara, T., Ueda, M., Okada, H., Kamasawa, N., Naito, N., Osumi, M., et al. (1992). Beta-oxidation of butyrate, the short-chain-length fatty acid, occurs in peroxisomes in the yeast Candida tropicalis. J. Biochem. 111, 783–787. doi: 10.1093/oxfordjournals.jbchem.a123836
Lazarow, P. B., and Fujiki, Y. (1985). Biogenesis of peroxisomes. Annu. Rev. Cell Biol. 1, 489–530. doi: 10.1146/annurev.cb.01.110185.002421
Lee, S., Son, H., Lee, J., Min, K., Choi, G. J., Kim, J. C., et al. (2011). Functional analyses of two acetyl coenzyme A synthetases in the ascomycete Gibberella zeae. Eukaryot. Cell 10, 1043–1052. doi: 10.1128/EC.05071-1
Leon, S., Goodman, J. M., and Subramani, S. (2006). Uniqueness of the mechanism of protein import into the peroxisome matrix: transport of folded, co-factor-bound and oligomeric proteins by shuttling receptors. BBA-Mol. Cell Res. 1763, 1552–1564. doi: 10.1016/j.bbamcr.2006.08.037
Li, L., Wang, J. Y., Chen, H. L., Chai, R. Y., Zhang, Z., Mao, X. Q., et al. (2017). Pex14/17, a filamentous fungus-specific peroxin, is required for the import of peroxisomal matrix proteins and full virulence of Magnaporthe oryzae. Mol. Plant Pathol. 18, 1238–1252. doi: 10.1111/mpp.12487
Maggio-Hall, L. A., and Keller, N. P. (2004). Mitochondrial beta-oxidation in Aspergillus nidulans. Mol. Microbiol. 54, 1173–1185. doi: 10.1111/j.1365-2958.2004.04340.x
Matsuzono, Y., Matsuzaki, T., and Fujiki, Y. (2006). Functional domain mapping of peroxin Pex19p: interaction with Pex3p is essential for function and translocation. J. Cell Sci. 119, 3539–3550. doi: 10.1242/jcs.03100
Meijer, W. H., Gidijala, L., Fekken, S., Kiel, J. A. K. W., van den Berg, M. A., Lascaris, R., et al. (2010). Peroxisomes are required for efficient penicillin biosynthesis in Penicillium chrysogenum. Appl. Environ. Microbiol. 76, 5702–5709. doi: 10.1128/Aem.02327-9
Min, K., Son, H., Lee, J., Choi, G. J., Kim, J. C., and Lee, Y. W. (2012). Peroxisome function is required for virulence and survival of Fusarium graminearum. Mol. Plant Microbe Interact. 25, 1617–1627. doi: 10.1094/Mpmi-06-12-0149-R
Mor, A., Koh, E., Weiner, L., Rosenwasser, S., Sibony-Benyamini, H., and Fluhr, R. (2014). Singlet oxygen signatures are detected independent of light or chloroplasts in response to multiple stresses. Plant Physiol. 165, 249–261. doi: 10.1104/pp.114.236380
Notzel, C., Lingner, T., Klingenberg, H., and Thoms, S. (2016). Identification of new fungal peroxisomal matrix proteins and revision of the PTS1 consensus. Traffic 17, 1110–1124. doi: 10.1111/tra.12426
Pan, R., Liu, J., and Hu, J. (2018). Peroxisomes in plant reproduction and seed-related development. J. Integr. Plant Biol. 67, 784–802. doi: 10.1111/jipb.12765
Peraza-Reyes, L., and Berteaux-Lecellier, V. (2013). Peroxisomes and sexual development in fungi. Front. Physiol. 4:244. doi: 10.3389/fphys.2013.00244
Petriv, O. I., Tang, L., Titorenko, V. I., and Rachubinski, R. A. (2004). A new definition for the consensus sequence of the peroxisome targeting signal type 2. J. Mol. Biol. 341, 119–134. doi: 10.1016/j.jmb.2004.05.064
Pieuchot, L., and Jedd, G. (2012). Peroxisome assembly and functional diversity in eukaryotic microorganisms. Annu. Rev. Microbiol. 66, 237–263. doi: 10.1146/annurev-micro-092611-150126
Proctor, R. H., Hohn, T. M., and Mccormick, S. P. (1995). Reduced virulence of Gibberella-zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol. Plant Microbe Interact. 8, 593–601. doi: 10.1094/Mpmi-8-0593
Purdue, P. E., and Lazarow, P. B. (2001). Peroxisome biogenesis. Annu. Rev. Cell Dev. Bi. 17, 701–752. doi: 10.1146/annurev.cellbio.17.1.701
Ramos-Pamplona, M., and Naqvi, N. I. (2006). Host invasion during rice-blast disease requires carnitine-dependent transport of peroxisomal acetyl-CoA. Mol. Microbiol. 61, 61–75. doi: 10.1111/j.1365-2958.2006.05194.x
Reddy, J. K., and Hashimoto, T. (2001). Peroxisomal beta-oxidation and peroxisome proliferator-activated receptor alpha: an adaptive metabolic system. Annu. Rev. Nutr. 21, 193–230. doi: 10.1146/annurev.nutr.21.1.193
Reverberi, M., Punelli, M., Smith, C. A., Zjalic, S., Scarpari, M., Scala, V., et al. (2012). How peroxisomes affect aflatoxin biosynthesis in Aspergillus flavus. PLoS One 7:e48097. doi: 10.1371/journal.pone.0048097
Rottensteiner, H., Kramer, A., Lorenzen, S., Stein, K., Christiane, L. F., Volkmer-Engert, R., et al. (2004). Peroxisomal membrane proteins contain common Pex19p-binding sites that are an integral part of their targeting signals. Mol. Biol. Cell 15, 3406–3417. doi: 10.1091/mbc-E04-03-0188
Sandalio, L. M., and Romero-Puertas, M. C. (2015). Peroxisomes sense and respond to environmental cues by regulating ROS and RNS signalling networks. Ann. Bot. 116, 475–485. doi: 10.1093/aob/mcv074
Schliebs, W., and Kunau, W. H. (2004). Peroxisome membrane biogenesis: the stage is set. Curr. Biol. 14, R397–R399. doi: 10.1016/j.cub.2004.05.017
Schueller, N., Holton, S. J., Fodor, K., Milewski, M., Konarev, P., Stanley, W. A., et al. (2010). The peroxisomal receptor Pex19p forms a helical mPTS recognition domain. EMBO J. 29, 2491–2500. doi: 10.1038/emboj.2010.115
Seong, K. Y., Pasquali, M., Zhou, X. Y., Song, J., Hilburn, K., McCormick, S., et al. (2009). Global gene regulation by Fusarium transcription factors Tri6 and Tri10 reveals adaptations for toxin biosynthesis. Mol. Microbiol. 72, 354–367. doi: 10.1111/j.1365-2958.2009.06649.x
Shen, Y. Q., and Burger, G. (2009). Plasticity of a key metabolic pathway in fungi. Funct. Integr. Genomics 9, 145–151. doi: 10.1007/s10142-008-0095-6
Sibirny, A. A. (2016). Yeast peroxisomes: structure, functions and biotechnological opportunities. Fems Yeast Res. 16, fow038. doi: 10.1093/femsyr/fow038
Tripathi, D. N., and Walker, C. L. (2016). The peroxisome as a cell signaling organelle. Curr. Opin. Cell Biol. 39, 109–112. doi: 10.1016/j.ceb.2016.02.017
Turgeon, B. G., Garber, R. C., and Yoder, O. C. (1987). Development of a fungal transformation system based on selection of sequences with promoter activity. Mol. Cell. Biol. 7, 3297–3305. doi: 10.1128/MCB.7.9.3297
van der Klei, I. J., and Veenhuis, M. (2013). The versatility of peroxisome function in filamentous fungi. Subcell Biochem. 69, 135–152. doi: 10.1007/978-94-007-6889-5_8
van der Lee, T., Zhang, H., van Diepeningen, A., and Waalwijk, C. (2015). Biogeography of Fusarium graminearum species complex and chemotypes: a review. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 32, 453–460. doi: 10.1080/19440049.2014.984244
van der Zand, A., Braakman, I., and Tabak, H. F. (2010). Peroxisomal membrane proteins insert into the endoplasmic reticulum. Mol. Biol. Cell 21, 2057–2065. doi: 10.1091/mbc.E10-02-0082
van der Zand, A., Gent, J., Braakman, I., and Tabak, H. F. (2012). Biochemically distinct vesicles from the endoplasmic reticulum fuse to form peroxisomes. Cell 149, 397–409. doi: 10.1016/j.cell.2012.01.054
van Zutphen, T., Veenhuis, M., and van der Klei, I. J. (2011). Damaged peroxisomes are subject to rapid autophagic degradation in the yeast Hansenula polymorpha. Autophagy 7, 863–872. doi: 10.4161/auto.7.8.15697
Wanders, R. J., and Waterham, H. R. (2006). Peroxisomal disorders: the single peroxisomal enzyme deficiencies. Biochim. Biophys. Acta 1763, 1707–1720. doi: 10.1016/j.bbamcr.2006.08.010
Wanders, R. J. A., and Waterham, H. R. (2005). Peroxisomal disorders I: biochemistry and genetics of peroxisome biogenesis disorders. Clin. Genet. 67, 107–133. doi: 10.1111/j.1399-0004.2004.00329.x
Wang, J., Zhang, Z., Wang, Y., Li, L., Chai, R., Mao, X., et al. (2013). PTS1 peroxisomal import pathway plays shared and distinct roles to PTS2 pathway in development and pathogenicity of Magnaporthe oryzae. PLoS One 8:e55554. doi: 10.1371/journal.pone.0055554
Wanner, G., and Theimer, R. R. (1982). Two types of microbodies in Neurospora crassa. Ann. N. Y. Acad. Sci. 386, 269–284. doi: 10.1111/j.1749-6632.1982.tb21422.x
Windels, C. E. (2000). Economic and social impacts of Fusarium head blight: changing farms and rural communities in the Northern Great Plains. Phytopathology 90, 17–21. doi: 10.1094/Phyto.2000.90.1.17
Wroblewska, J. P., Cruz-Zaragoza, L. D., Yuan, W., Schummer, A., Chuartzman, S. G., de Boer, R., et al. (2017). Saccharomyces cerevisiae cells lacking Pex3 contain membrane vesicles that harbor a subset of peroxisomal membrane proteins. BBA-Mol. Cell Res. 1864, 1656–1667. doi: 10.1016/j.bbamcr.2017.05.021
Zhang, H., Van der Lee, T., Waalwijk, C., Chen, W. Q., Xu, J., Xu, J. S., et al. (2012). Population analysis of the Fusarium graminearum species complex from wheat in China show a shift to more aggressive isolates. PLoS One 7:e31722. doi: 10.1371/journal.pone.0031722
Zhang, L., Liu, C., Wang, L., Sun, S., Liu, A., Liang, Y., et al. (2019a). FgPEX1 and FgPEX10 are required for the maintenance of Woronin bodies and full virulence of Fusarium graminearum. Curr. Genet. [Epub ahead of print],
Keywords: Fusarium graminearum, peroxisome, FgPex3, fungal growth, virulence
Citation: Kong X, Zhang H, Wang X, van der Lee T, Waalwijk C, van Diepeningen A, Brankovics B, Xu J, Xu J, Chen W and Feng J (2019) FgPex3, a Peroxisome Biogenesis Factor, Is Involved in Regulating Vegetative Growth, Conidiation, Sexual Development, and Virulence in Fusarium graminearum. Front. Microbiol. 10:2088. doi: 10.3389/fmicb.2019.02088
Received: 11 April 2019; Accepted: 23 August 2019;
Published: 20 September 2019.
Edited by:
Jens Staal, Flanders Institute for Biotechnology, BelgiumReviewed by:
Richard Rachubinski, University of Alberta, CanadaLuisa M. Sandalio, Spanish National Research Council (CSIC), Spain
Copyright © 2019 Kong, Zhang, Wang, van der Lee, Waalwijk, van Diepeningen, Brankovics, Xu, Xu, Chen and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wanquan Chen, d3FjaGVuQGlwcGNhYXMuY24=; Jie Feng, amZlbmdAaXBwY2Fhcy5jbg==
†These authors have contributed equally to this work
‡Present address: Xiangjiu Kong, School of Life Sciences, Shandong University, Qingdao, China
 Xiangjiu Kong
Xiangjiu Kong Hao Zhang
Hao Zhang Xiaoliang Wang1
Xiaoliang Wang1 Theo van der Lee
Theo van der Lee Cees Waalwijk
Cees Waalwijk Anne van Diepeningen
Anne van Diepeningen Balazs Brankovics
Balazs Brankovics Wanquan Chen
Wanquan Chen