- 1Division of Bacteriology, National Institute of Cholera and Enteric Diseases, Kolkata, India
- 2Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama University, Okayama, Japan
- 3Collaborative Research Center of Okayama University for Infectious Diseases in India, National Institute of Cholera and Enteric Diseases, Kolkata, India
- 4Center for Human Microbial Ecology, Translational Health Science and Technology Institute, Faridabad, India
The self-transferring integrative and conjugative elements (ICEs) are large genomic segments carrying several bacterial adaptive functions including antimicrobial resistance (AMR). SXT/R391 family is one of the ICEs extensively studied in cholera-causing pathogen Vibrio cholerae. The genetic characteristics of ICE-SXT/R391 in V. cholerae are dynamic and region-specific. These ICEs in V. cholerae are strongly correlated with resistance to several antibiotics such as tetracycline, streptomycin and trimethoprim-sulfamethoxazole. We screened V. cholerae O1 strains isolated from cholera patients in Kolkata, India from 2008 to 2015 for antibiotic susceptibility and the presence of ICEs, and subsequently sequenced their conserved genes. Resistance to tetracycline, streptomycin and trimethoprim-sulfamethoxazole was detected in strains isolated during 2008–2010 and 2014–2015. The genes encoding resistance to tetracycline (tetA), trimethoprim-sulfamethoxazole (dfrA1 and sul2), streptomycin (strAB), and chloramphenicol (floR) were detected in the ICEs of these strains. There was a decrease in overall drug resistance in V. cholerae associated with the ICEs in 2011. DNA sequence analysis also showed that AMR in these strains was conferred mainly by two types of ICEs, i.e., ICETET (comprising tetA, strAB, sul2, and dfrA1) and ICEGEN (floR, strAB, sul2, and dfrA1). Based on the genetic structure, Kolkata strains of V. cholerae O1 had distinct genetic traits different from the ICEs reported in other cholera endemic regions. Transfer of AMR was confirmed by conjugation with sodium azide resistant Escherichia coli J53. In addition to the acquired resistance to streptomycin and trimethoprim-sulfamethoxazole, the conjugally transferred (CT) E. coli J53 with ICE showed higher resistance to chloramphenicol and tetracycline than the donor V. cholerae. Pulsed-field gel electrophoresis (PFGE) based clonal analysis revealed that the V. cholerae strains could be grouped based on their ICEs and AMR patterns. Our findings demonstrate the epidemiological importance of ICEs and their role in the emergence of multidrug resistance (MDR) in El Tor vibrios.
Introduction
The Gram-negative pathogen Vibiro cholerae O1 has caused seven pandemics in the history of cholera and tends to cause several epidemics in developing countries (Lekshmi et al., 2018). This pathogen has more than 200 serogroups, but only the serogroups O1 and O139 are associated with epidemic cholera (Lekshmi et al., 2018). The ongoing seventh pandemic is linked with the El Tor biotype of serogroup O1 that has spread in the cholera endemic regions of the world (Lekshmi et al., 2018). The emergence and spread of antimicrobial resistant (AMR) V. cholerae, especially those resistant to nalidixic acid, tetracycline, and trimethoprim-sulfamethoxazole, has been reported since the 1980s (Ghosh and Ramamurthy, 2011). Resistance to these antimicrobials has been strongly associated with the presence of integrative and conjugative elements (ICEs) of the SXT/R391 family and its discovery has greatly changed the understanding of AMR in V. cholerae.
SXT/R391 ICEs have been characterized/classified based on the conserved core genes, and their integration into the 5′-end of the prfC gene that encodes peptide chain release factor 3 (Hochhut and Waldor, 1999). More than 1000 ICEs have been updated in the ICEberg database1. Mobility of SXT/R391 ICEs occurs between bacteria by conjugation, resulting in the transfer of several functions including AMR, resistance to heavy metals, regulation of motility and biofilm formation (Waldor et al., 1996; Bordeleau et al., 2010). Five insertion hotspots (H1 to H5) and four variable regions (VRI to VRIV) are also carried by the ICEs (Wozniak et al., 2009). The structure of ICEs changes periodically contributing to the differences in AMR profiles of V. cholerae. More than 50 ICEs have been grouped within the SXT/R391 family, of which 30 are reported in clinical and environmental V. cholerae strains (Pande et al., 2012). Between 1992 and 2001, 15 ICEs were identified in India and Bangladesh, of which six (SXTMO10, ICEVchInd4, ICEVchBan5, ICEVchBan10, ICEVchBan9, and ICEVchInd5) were completely sequenced and annotated (Ceccarelli et al., 2011).
Tetracycline has been the drug of choice in treating cholera cases for a long time (World Health Organization [WHO], 2005). A sudden upsurge in the tetracycline resistance (TetR), from 1% in 2004 to 76% in 2007, was reported among V. cholerae in Kolkata and it decreased to about 50% in 2009 (Bhattacharya et al., 2011). Similar trends have been observed previously in large cholera epidemics in Tanzania and Madagascar due to extensive prophylactic use of tetracycline (Mhalu et al., 1979; Dromigny et al., 2002). Only a few studies have been carried out to understand the mechanisms of AMR due to ICEs in India (Roychowdhury et al., 2008; Bhattacharya et al., 2011; Kutar et al., 2013). In this study, we screened the AMR patterns of V. cholerae O1 Ogawa strains isolated from cholera patients in Kolkata, India from 2008 to 2015 and examined the type of ICEs present by analyzing their backbone genes. Our study revealed the differences between the sequence types of ICEs and recent changes in AMR patterns of V. cholerae.
Materials and Methods
Clinical Specimens and Bacterial Strains
Stool specimens were collected from the Infectious Diseases Hospital (IDH) and B. C. Roy Children Hospital (BCH), Kolkata, before the patients were treated with antibiotics. Clinical symptoms of diarrheal patients included loose/watery stools with or without dehydration, abdominal cramps, vomiting and fever. Dysentery patients had frequent passage of stool with blood/mucus and mild to severe abdominal pain. For the isolation of V. cholerae, all the stool specimens/rectal swabs were enriched in alkaline peptone water (pH 8.0) (Difco, Sparks, MD, United States) for 6 h, followed by inoculation and overnight incubation in thiosulphate citrate bile-salts sucrose agar (TCBS, Eiken, Tokyo, Japan) plates. Sucrose-positive strains were confirmed serologically using commercially available V. cholerae O1 poly and monovalent antisera (Denka-Seiken, Tokyo, Japan). To obtain the AMR pattern from 2008 to 2015, 546 out of 1591 strains were randomly selected covering each month of the study period. Sodium azide resistant (AzR) Escherichia coli J53 (Martínez-Martínez et al., 1998) was used for the conjugation experiments. All the strains were preserved in Luria Bertani (LB) broth (Difco) containing 15% glycerol at −80°C. E. coli ATCC 25922 (Clinical and Laboratory Standards Institute [CLSI], 2014) was used as a control strain in antimicrobial susceptibility testing.
Antibiotic Susceptibility Testing
Susceptibilities of V. cholerae strains to ampicillin (AMP, 10 μg), ceftriaxone (CRO, 30 μg), chloramphenicol (CHL, 30 μg), nalidixic acid (NA, 30 μg), ciprofloxacin (CIP, 5 μg), ofloxacin (OFX, 5 μg), norfloxacin (NOR, 10 μg), imipenem (IPM, 10 μg), streptomycin (STR, 10 μg), azithromycin (AZM, 15 μg), tetracycline (TET, 30 μg), trimethoprim-sulfamethoxazole (SXT, 1.25 and 23.75 μg) and gentamicin (GEN, 10 μg), were determined by Kirby-Bauer disk diffusion technique using commercial disks (BD, Sparks, MD, United States) as per the Clinical and Laboratory Standards Institute guidelines (Clinical and Laboratory Standards Institute [CLSI], 2014, 2015).
Detection of Antibiotic Resistance Encoding Genes
Total nucleic acid of V. cholerae strains was extracted using a QIAamp DNA mini kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The integrase gene (intSXT) present in ICE was amplified by PCR using previously described primer pair int1-F and int1-B (Dalsgaard et al., 2001). Beside intSXT, PCR was also performed to detect the presence of resistance encoding genes for chloramphenicol (floR and cat), streptomycin (strA and strB), and sulfonamide (sul1 and sul2) (Sarkar et al., 2015a). Primer pairs VCtetA.F-(5′- ACGGTATCCTGCTGGCACTGTATG-3′) and VCtetA.R-(5′- CATCCATATCCAGCCATCCCAACT-3′) and VctetR.F-(5′-GA AGTGGGAATGGAAGGGCTGAC-3′) and VctetR.R-(5′-AG CCTCTGTGCCATCATCTTG-3′) were designed to detect the TetR encoding gene (tetA), and the repressor protein (tetR) for a regulatory portion of resistance cassettes, respectively. Representative amplicons were purified using a PCR product purification kit (Qiagen) and sequenced using the ABI Big Dye terminator cycle sequencing ready reaction kit, version 3.1 (Applied Biosystems, Foster City, CA, United States) in an automated DNA sequencer (ABI 3730, Applied Biosystems). The sequences were assembled and analyzed using DNASTAR software (DNASTAR Inc., Madison, WI, United States).
Conjugation
To test the mobility of the ICEs, conjugation assay was carried out using a representative ICE-positive V. cholerae O1 strain as donor with E. coli J53 (AzR, Martínez-Martínez et al., 1998). In brief, overnight cultures of the bacteria were mixed at 1:2 donor-to-recipient ratios in 1 ml of LB broth and allowed to grow overnight at 37°C. The donor and recipient suspensions were diluted serially in phosphate buffer saline (PBS) and plated on TCBS and MacConkey agar plates, respectively, to confirm the purity and count the number of colonies. To detect the conjugally transferred E. coli J53 (CT-E. coli J53), MacConkey agar supplemented with streptomycin (100 μg/ml) and sodium azide (AZD, 100 μg/ml) was used. Transconjugants were confirmed as ICE-positive by PCR analysis, followed by PCR amplicon sequencing. To confirm the resistance phenotype, antibiotic susceptibility patterns of the donor, recipient and transconjugants were determined after their growth on Mueller-Hinton (MH, Difco) agar by disk diffusion method. An increase in resistance of transconjugants was quantified by determining the MICs of CHL, STR, TET, and SXT using E-test strips (AB bioMérieux, Solna, Sweden).
Pulsed-Field Gel Electrophoresis (PFGE)
Clonal analysis of representative V. cholerae O1 strains isolated between 2008 and 2015 was made following the PulseNet protocol (Cooper et al., 2006). V. cholerae O1 strains were used after digesting the DNA with NotI [New England Biolabs (NEB), Ipswich, MA, United States]. XbaI (NEB) digested Salmonella Braendruff H-9812 was used as a DNA size marker. The PFGE run conditions were generated by the auto-algorithm mode of the CHEF Mapper system (Bio-Rad, Hercules, CA, United States). PFGE profiles were analyzed by the BioNumerics version 4.0 software (Applied Maths, Sint-Martens-Latem, Belgium) using the Dice coefficient and unweighted pair group method using arithmetic averages (UPGMA).
Whole Genome Sequence Analysis
The whole genome sequences submitted from our previous study (Imamura et al., 2017) were used in the analysis. The open reading frames (ORFs) from the contigs were generated by contig integrator for sequence assembly (CISA) using Glimmer-MG program2. Nucleotide sequences and amino acid sequences were obtained from these ORFs and translated in the appropriate frame. The predicted ORFs were annotated using CANoPI (Contig Annotator Pipeline) that also includes BlastX search for each ORF sequence against the “nr” database of NCBI3. From the whole genome sequence data of representative strains (TetR IDH 1986 and TetS IDH 4268), we have used part of the ICE region in the analysis. The contigs were aligned, assembled and compared with SEQMAN, assembly module of DNASTAR’s LASERGENE with published sequences like ICEVchInd5 (GQ463142), ICEVchBan5 (GQ463140), MO10 (AY055428), etc. For confirmation, PCR was performed targeting important short regions of the ICEs (rumAB, traI, traC, setR, traA-traC, and traG) with previously described primers (Bani et al., 2007). Published ICE sequences were used for homology search. ORF search and gene prediction were performed for the complete ICE region with EditSEQ, Lasergene software (DNASTAR), and pairwise alignment was analyzed by blastN and blastP homology search using the NCBI database.
Nucleotide Sequence Submission
The AMR encoding gene cassettes and their flanking sequences of representative ICE of TetR and TetS V. cholerae O1 have been submitted in GenBank (Accession numbers MK165649 and MK165650, respectively).
Ethics and Biosafety Statements
The Ethics and Biosafety Committees of National Institute of Cholera and Enteric Diseases, Kolkata approved this study (A:1/2015-IEC). Each participant/parent in the case of children gave written informed consent. All the experiments were performed following Biosafety Level-2 standards.
Results
Prevalence of Cholera
During 8 years of surveillance from 2008 to 2015, the isolation rate of V. cholerae O1 Ogawa was about 11% (1591 of 14237 tested samples) (Figure 1). The incidence of this pathogen in BCH samples was very low (∼2%) but was found to be much higher (∼18%) in IDH samples. As shown in Figure 1, the mean incidence of cholera in IDH/BCH fluctuated between 4.9% (2014) and 27.2% (2009). Except for children ≤5 years, V. cholerae O1 remained one of the important bacterial pathogens. The incidence of V. cholerae O1 varies in certain extent from year to year (Figure 1).
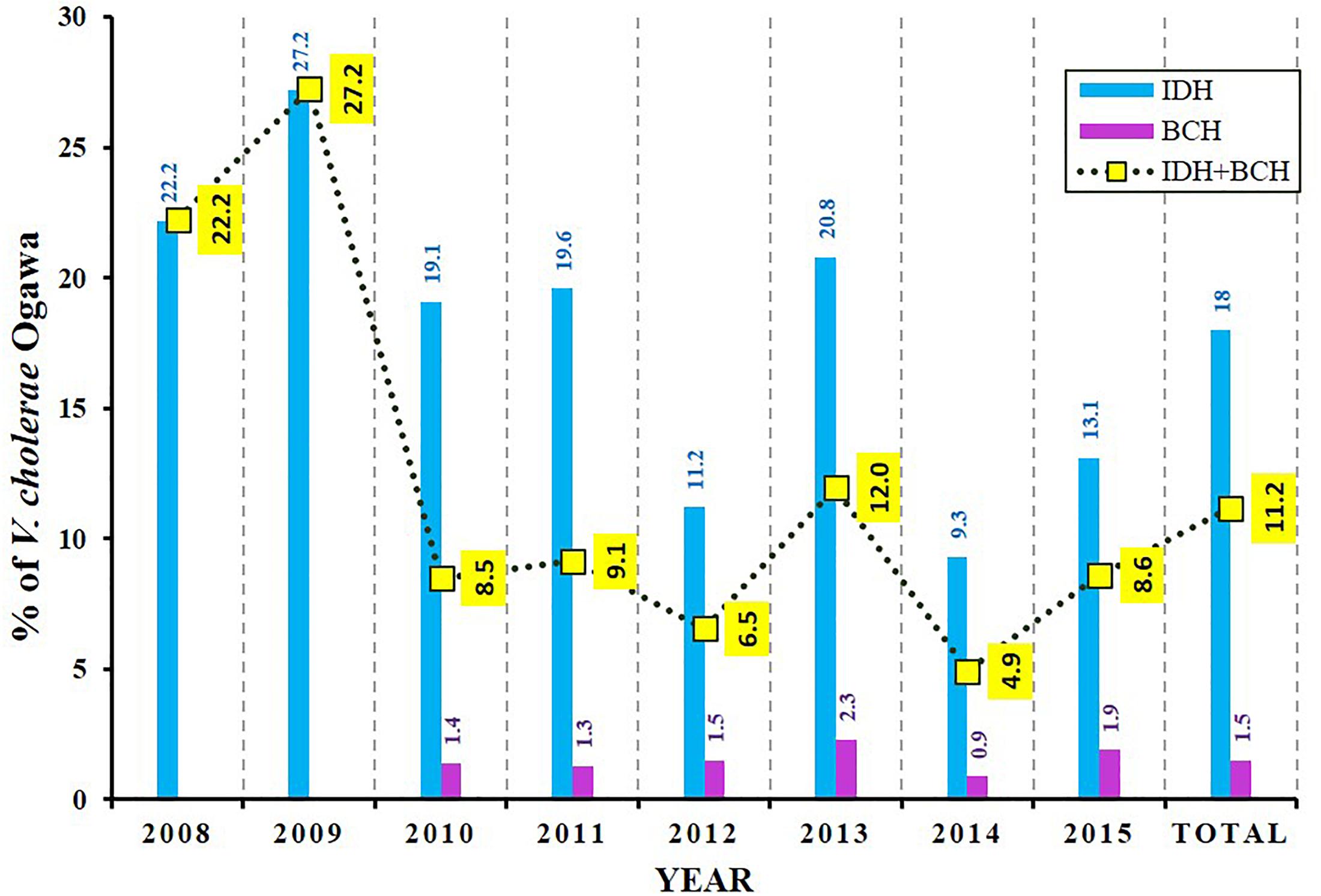
Figure 1. Isolation rate of V. cholerae O1 Ogawa among diarrheal patients. BCH sample collection was started from 2010 onward. The dotted line with yellow boxes represent the mean incidence of cholera in IDH and BCH.
Antimicrobial Resistance
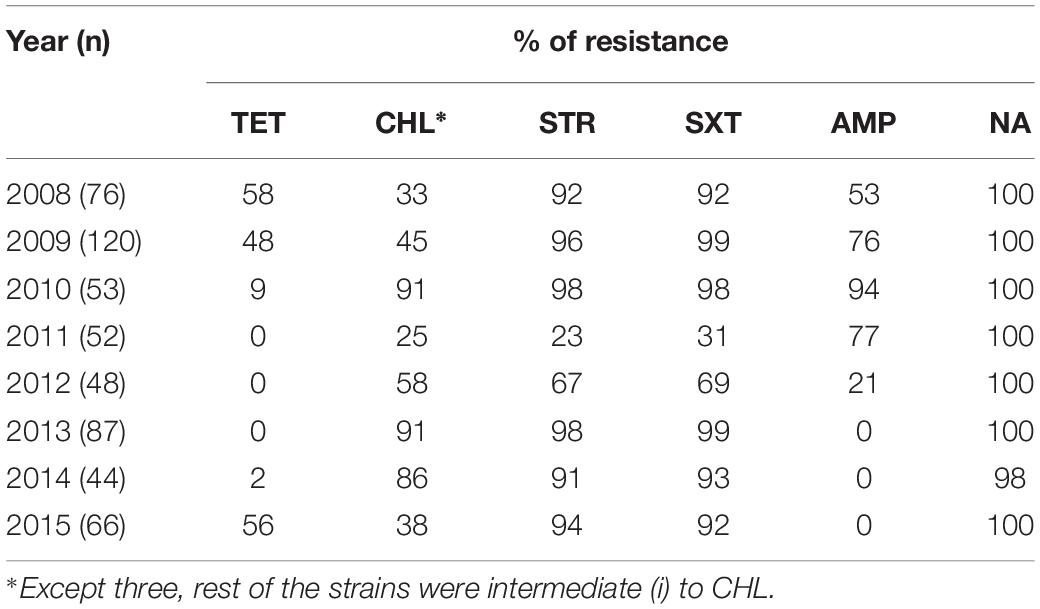
All the V. cholerae O1 strains isolated were consistently resistant to NA. TetR gradually decreased from 58% in 2008 to 48% in 2009, followed by a further drop in 2010 (9%). Thereafter, all the strains isolated between 2011 and 2013 were found to be susceptible to TET (Table 1). Remarkably, TetR trait increased again in 2015 (56%). There was a marked change in AMP resistance each year with highest in 2010 (94%) and lowest in 2012 (21%) (Table 1). About three fourth of the strains were resistant to AMP in 2009 and 2011 (>76%). Thereafter, most of the V. cholerae isolated from 2013 to 2015 were found to be susceptible to AMP.
Throughout the study period, only three V. cholerae strains were found to be fully resistant to CHL and the rest of the floR containing strains showed intermediate resistance [CHL(i)] to this antibiotic. Interestingly, resistance to TET was found to be inversely proportional to CHL(i), i.e., strains showing TetR had intermediate resistance to CHL. The CHL(i) trait increased in 2010 (91%) when TetR was very low (9%) but dropped to 38% with the re-emergence of TetR in 2015 (56%). Resistance to STR and SXT were detected in most of the V. cholerae O1 strains. Resistance to these antimicrobials was >90% from 2008 to 2010 and 2013 to 2015. Interestingly, there was a sudden decrease in STR and SXT resistance (23 and 31%, respectively, in 2011) followed by an increase in 2012 (67 and 69%, respectively) (Table 1).
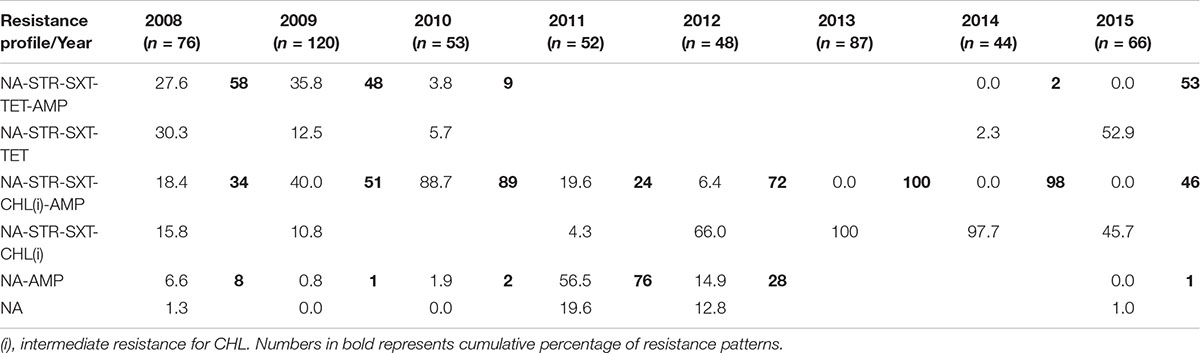
This study shows the changing profile of MDR in V. cholerae from Kolkata; MDR profiles NA-STR-SXT-TET-AMP and NA-STR-SXT-TET were predominant during 2008, 2009 and 2015 (Table 2), while from 2009 to 2010 and 2012 to 2014 the MDR profiles NA-STR-SXT-CHL(i), and NA-STR-SXT-CHL(i)-AMP were found in more than 50% of the V. cholerae O1 strains.
ICE Comprising Antimicrobial Resistance Genes
While analyzing the sequences of the resistance gene clusters, two types of ICEs could be detected, i.e., ICETET (Acc No. MK165649; TetR IDH 1986) and ICEGEN (Acc No. MK165650; TetS IDH 4268). The superscript “GEN” stands for “general.” Although the ICEGEN was very similar to the ICEVchInd5 with 99% identity at 100% query coverage, the ICETET had only 99% identity at 70% query coverage. The structure of these two ICEs with ORFs is shown in Figure 2. The ICEGEN was found to be larger (96.7 kb) than ICETET (91.5 kb). SXT and STR resistant V. cholerae O1 strains were positive for intSXT. Detection of ICEs was >90% in 2008 and 2009, with highest in 2010 (98%), followed by an abrupt decrease in 2011 (23%). However, in 2012, 68% of the V. cholerae O1 strains harbored the ICEs. Interestingly, except for NA, the intSXT negative strains were susceptible to most of the antimicrobials tested in this study. In the 1st type, ICETET carried a TET efflux pump encoding gene (tetAR; tetA is a gene encoding TET efflux pump and tetR is a repressor protein regulating the tetA expression) and in the 2nd type, ICEGEN harbored CHL efflux pump encoding gene (floR). ICEGEN has high similarity (99%) with the ICEVchInd5, the most common ICE detected among seventh-pandemic El Tor vibrios (Spagnoletti et al., 2014; Bioteau et al., 2018). This ICE also has very high similarity to the ICEVchHai1 from the Haitian V. cholerae lineage (Sjölund-Karlsson et al., 2011).
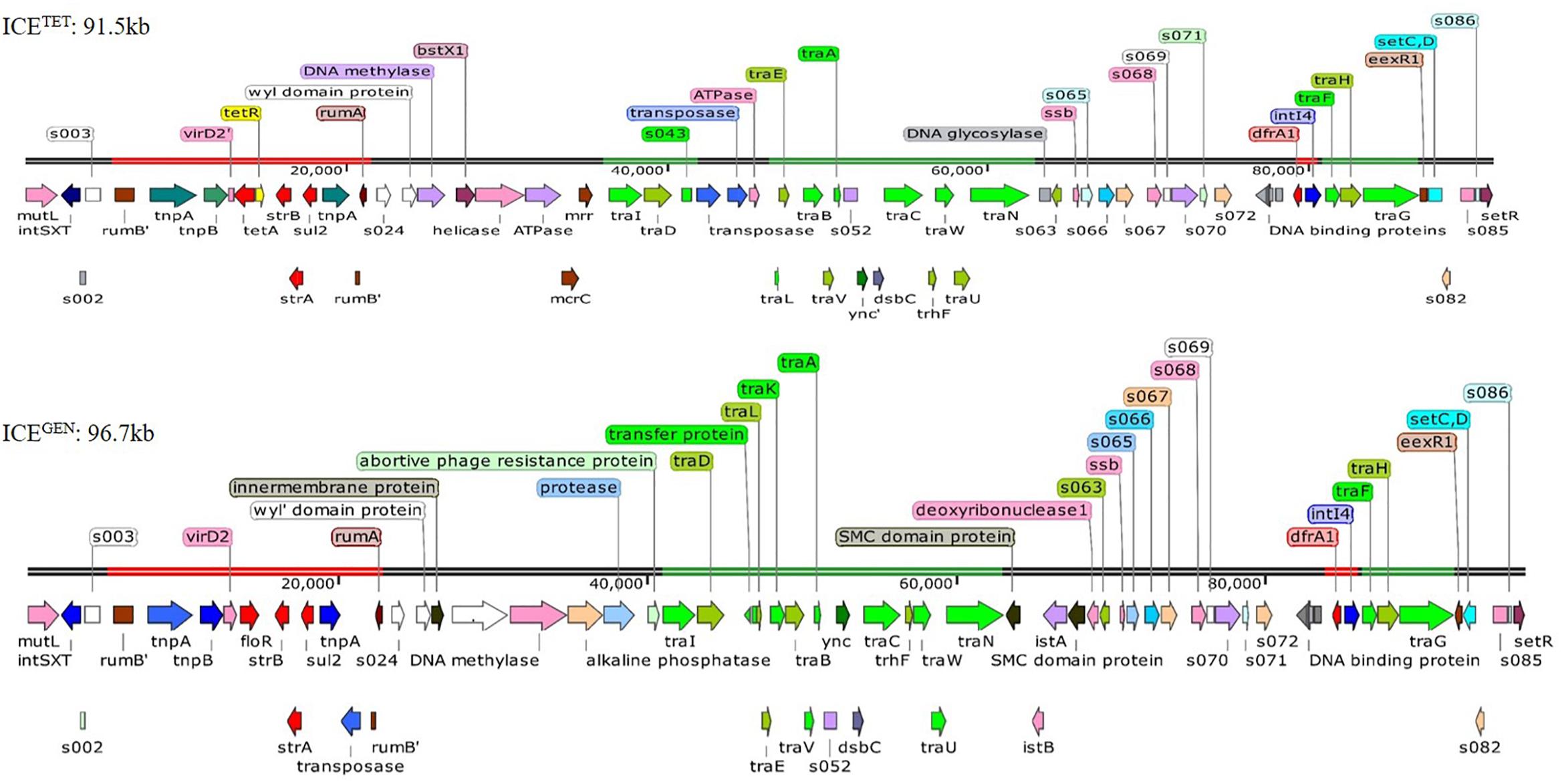
Figure 2. Structure of the two ICEs found in MDR V. cholerae O1 Ogawa strains. The AMR genes are shown in red, the genes responsible for the transfer are presented in green, and transposases and integrases are shown in blue. The other shades represented miscellaneous features.
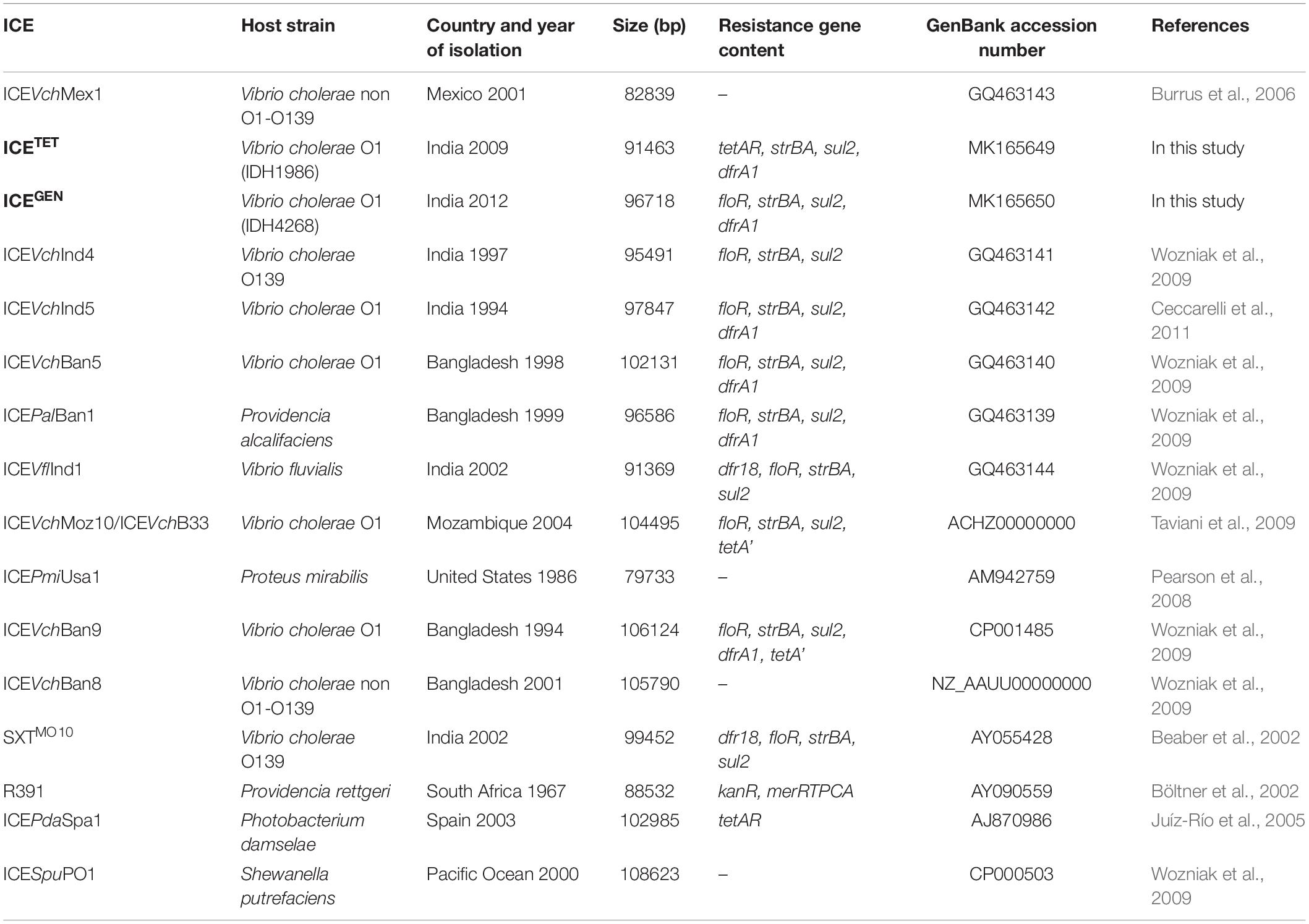
The ICEGEN and ICETET had sul2, strBA in the AMR gene cluster conferring resistance to SXT and STR, respectively. Generally, in V. cholerae, the presence of tet alleles within the ICE gene clusters is uncommon. In the prototype SXTMO10, resistance gene cluster comprised dfr18, floR, strBA, sul2 encoding resistance to trimethoprim, CHL, STR, and sulfamethoxazole, respectively (Table 3). In ICEVchInd4, there was a major deletion of dfr18 gene in the cluster. In IDH1986 and IDH14268 strains, a class 4 integron carrying the trimethoprim resistance encoding dfrA1 was identified in H3 located within the s073-traF locus. Such arrangement exists in ICEVchInd5 backbone (Figure 2) and ICEVchInd1. But, tetA gene was absent in these ICEs.
Detection of ICETET in V. cholerae O1 decreased from 2008 (58% TetR) to 2010 (9% TetR). All the V. cholerae O1 strains isolated during 2011–2013 lacked ICETET. In 2015, however, the tetAR was again detected in a higher number of strains (56% TetR). In contrast, ICEGEN was detected throughout the study period. AMR gene cassettes located within the rumB locus are also different. From 2011 to 2013, the tetAR locus in ICETET was replaced by floR gene of ICEGEN. This feature marked the difference of ICETET from ICEVchLao1, where floR and tetA were concurrently present.
Based on the presence of the AMR encoding genes harbored by these elements, the genetic background of ICETET appears to be very different from the other ICEs carrying the tet. The ICEPdaSpa1 was found to have only the TET resistance determinant located within rumBA operon (Table 3). Whereas, in the ICEVchLao1, resistance genes of CHL (floR), STR (strBA) and sulfamethoxazole (sul2) were present along with tetA. But, the ICEVchLao1 did not carry dfrA1 or dfr18 that confer resistance to trimethoprim in SXTET and SXTMO10, respectively. Within the resistance gene cluster of 2008–2010 strains of V. cholerae in Kolkata, a deletion of floR gene, which was present upstream of the tetA gene in ICEVchLao1 and ICEVchBan9 was detected.
Genetic Structure of the ICEs
Generally, the genetic organization of ICETET and ICEGEN was similar to that of the other members of this family. Many ORFs were commonly shared by these ICEs; most of them being in the conserved core genes (Beaber et al., 2002). Five conserved insertion hotspots are located between s043 (traJ) and traL (H1), traA and s054 (H2), s073 and traF (H3), traN and s063 (H4), and s025 and traID (H5) (Wozniak et al., 2009).
Five ORFs were found in the H1 of ICETET that include tbp (integrase catalytic subunit), a hypothetical protein (HP), transposase, ISPsy4 transposition helper protein and DNA helicase family protein. These ORFs present in H1 are unique compared to other reported ICEs. Instead of mosA, mosT that encode toxin-antitoxin reported in the H2 of other ICEs, the ICEGEN and ICETET have 3 ORFs with ynd (transcriptional regulator with AbiEi antitoxin N-terminal domain), ync (nucleotidyl transferase AbiEii/AbiGii toxin family protein) and dsbC (disulfide isomerase DsbC). H3 of ICEGEN and ICETET contains 7 ORFs with bleR (glyoxalase/bleomycin resistance), araC (AraC family transcriptional regulator; helix-turn-helix domain protein), a hypothetical protein, XRE family transcriptional regulators, a putative membrane protein, dfrA1 (trimethoprim-resistance) and intI4 (site-specific recombinase IntI4). Of these, AraC, XRE, and DFRA1 were reported in ICEVchMoz10. H3 in ICEGEN and ICETET is varied from ICEVchInd4, SXTMO10, ICER391 ICEVchMex1, ICEVflInd1, ICEPmiUSA1, ICESpuPO1 (Wozniak et al., 2009). H4 of ICETET was small with 2 ORFs, whereas the ICEGEN had 5 ORFs with two SMC (structural maintenance of chromosome) domain proteins, istB (ATP binding domain), istA (integrase catalytic subunit) and deoxyribonuclease I. The ORF content of H4 in these ICEs is different from the others. In ICEGEN and ICETET, the H5 has 10–11 gene combinations with the new ORFs of WYL domain protein, N-6 DNA methylase, restriction endonuclease subunit S, BstXI (restriction endonuclease protein), ATPases associated with diverse cellular activities (AAA) family protein, McrC (putative protein) in ICETET and WYL domain-containing protein with three conserved amino acids, BrxC (BREX system P-loop protein), PglX (BREX-1 system adenine-specific DNA-methyltransferase) and abortive phage resistance protein in ICEGEN. These changes in the hotspot regions may not have an obvious effect on the ICE, as they did not influence its transfer. VR-II has an insertion of single ORF, mutL similar to the ICE contigs circulating in India and Bangladesh. In the VRIII of ICETET, 12 ORFs [Tn3 (transposase), tnpA (transposase), tnpB (InsA transposase), truncated virD2, tetA, tetR, IS91 transposase, strB, strA, sul2, tnpA tn3 transposase, s021] were identified within the two rumB portions. In the case of ICEGEN, 14 ORFs [Tn3 (trnansposase), tnpA (transposase), tnpB (InsA transposase), virD2 (relaxase), floR, LysR family protein, truncated transpoase, strB, strA, sul2, tnpA tn3 transposase, truncated s021, putative transpoase, truncated mutL] have been detected.
The restriction-modification system is composed of genes encoding the functions of DNA modification, recombination, and repair (Wozniak and Waldor, 2009). ICEGEN and ICETET were found to have a type I restriction-modification system in the H5. In the ICE backbones, there were sequences in the ORFs located between s024 and traI in Kolkata strains (Figure 2). In ICEGEN carrying strains, after the traN locus, there was an insertion of istBA gene flanked by gene encoding SMC domain protein. This arrangement was not observed in V. cholerae strains with ICETET. Though these two types of ICEs had same traFHG locus, ORFs encoding transposases and ATPase were found incorporated between the traD and traE locus only in ICETET. In contrast, the ICEGEN possessed an intact transfer region (Figure 2). In ICEVchInd4, there was a major deletion of dfr18 gene in the cluster. In strains with ICEGEN or ICETET, a class 4 integron carrying the trimethoprim resistance encoding dfrA1 was identified in the H3 region located within the s073-traF locus. Similar gene configuration exists in the ICEVchInd1 and ICEVchInd5 backbones. In the 2008–2010 strains of V. cholerae in Kolkata, TetR in ICE was primarily due to tetA, whose presence was previously reported in ICEPdaSpa1 of Photobacterium damselae, ICEVchLao1 and ICEVchBan9 of V. cholerae O1 from Laos and Bangladesh, respectively (Table 3).
The tra loci appeared to be derived from a common ancestor and were mostly present in ICEs of V. cholerae strains. These loci are crucial for the transfer of ICEs and generating the conjugation machinery (Wozniak et al., 2009). Similar to the other ICEs backbone, the tra genes are arranged in four clusters in IDH1986 and IDH4268 strains, spanning more than 25 kb. Cluster 1 contains the genes and sequences necessary for transfer initiation, the nickase (encoded by traI), and the coupling protein (encoded in the traD). The mating pair formation function is controlled by three gene clusters: (i) traLEKBVA, (ii) traC/trhF/traWUN, and (iii) traFHG (Figure 2).
Comparison of Conserved Genes in the ICEs
ICETET and ICEGEN shared the same exclusion group (EexR). This EexR system might have been transferred from R391 type ICEs (Marrero and Waldor, 2007). The site-specific integration of the ICE is mediated through integrase enzyme encoded in the int. The int of ICETET and ICEGEN harboring V. cholerae O1 is identical to those present in the strains that have ICEPalBan1 of P. alcalifaciens, ICEVfInd1 of V. fluvialis and ICEVchBan5, ICEVchBan9 and ICEVchInd5 of V. cholerae (Figure 3). These ICEs are distinct from those reported in Proteus mirabilis, Providencia rettgeri, Shewanella putrefaciens, P. damselae as well as in other V. cholerae with ICEVchMex1, ICEVchInd4, and SXTMO10. SetR and SetC/D are the key regulators of ICEs, which are closely followed by the genes encoding for inner membrane proteins (Eex and TraG) of the donor and recipient cells. Eex and TraG facilitate entry-exclusion in the SXT/R391 family of ICEs. In the cluster tree, eex genes of the ICETET and ICEGEN showed high homology with ICE identified in ICEVchBan5, ICEVchBan9, ICEVchInd5, but was distantly related to other ICEs of V. cholerae and other species (Figure 4). setR in the ICETET and ICEGEN are identical with that in ICEVchInd4, ICEVchInd5, ICEVchBan5, ICEVchBan9, SXTMO10, ICEVfInd1, ICEPalBan1 but different from ICEVchMex1 and ICEs of other species (Figure 5).
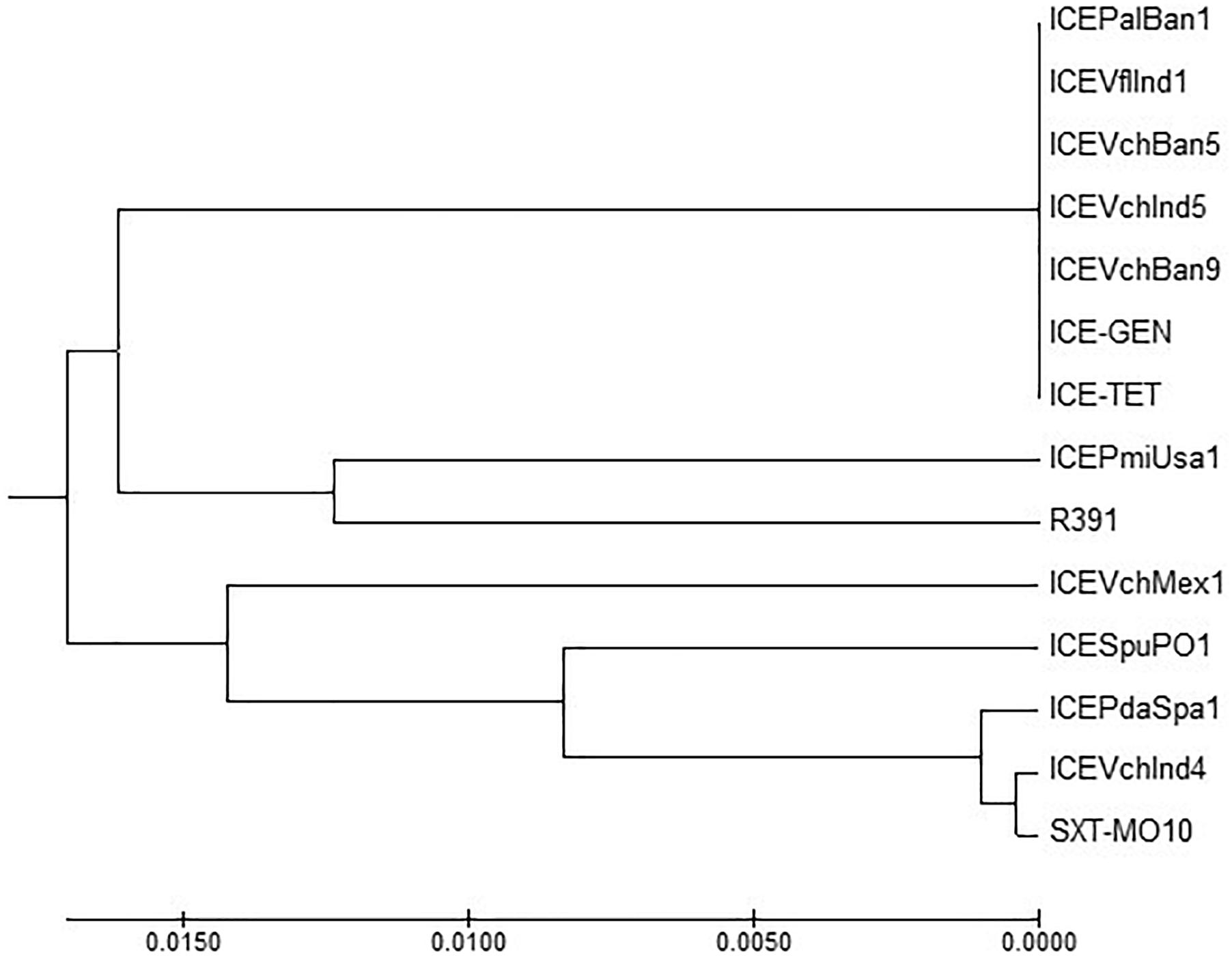
Figure 3. MEGA7 analysis based (Kumar et al., 2016) evolutionary relationships of taxa of int of V. cholerae O1 strains.
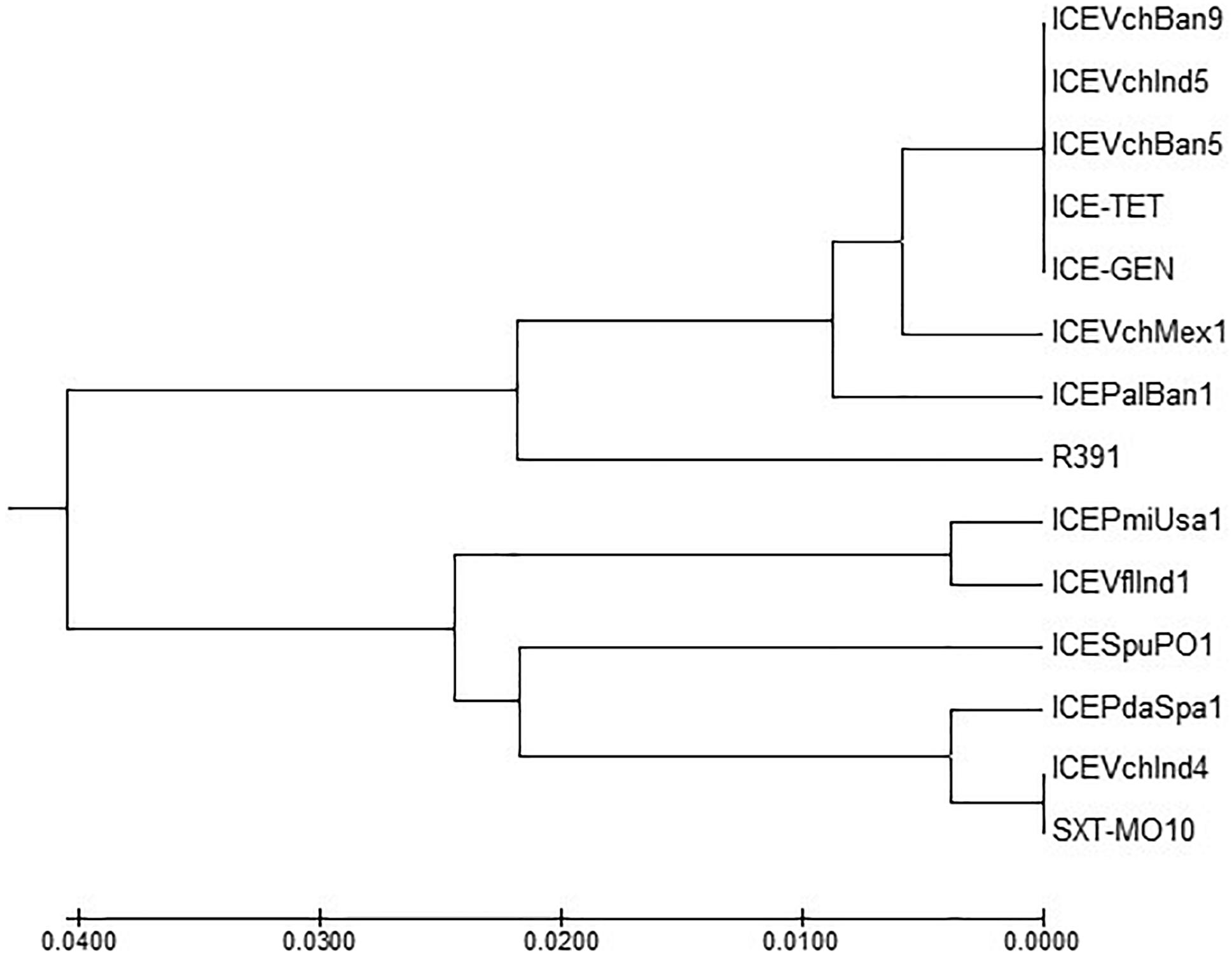
Figure 4. MEGA7 analysis based (Kumar et al., 2016) evolutionary relationships of taxa of eex of V. cholerae O1 strains.
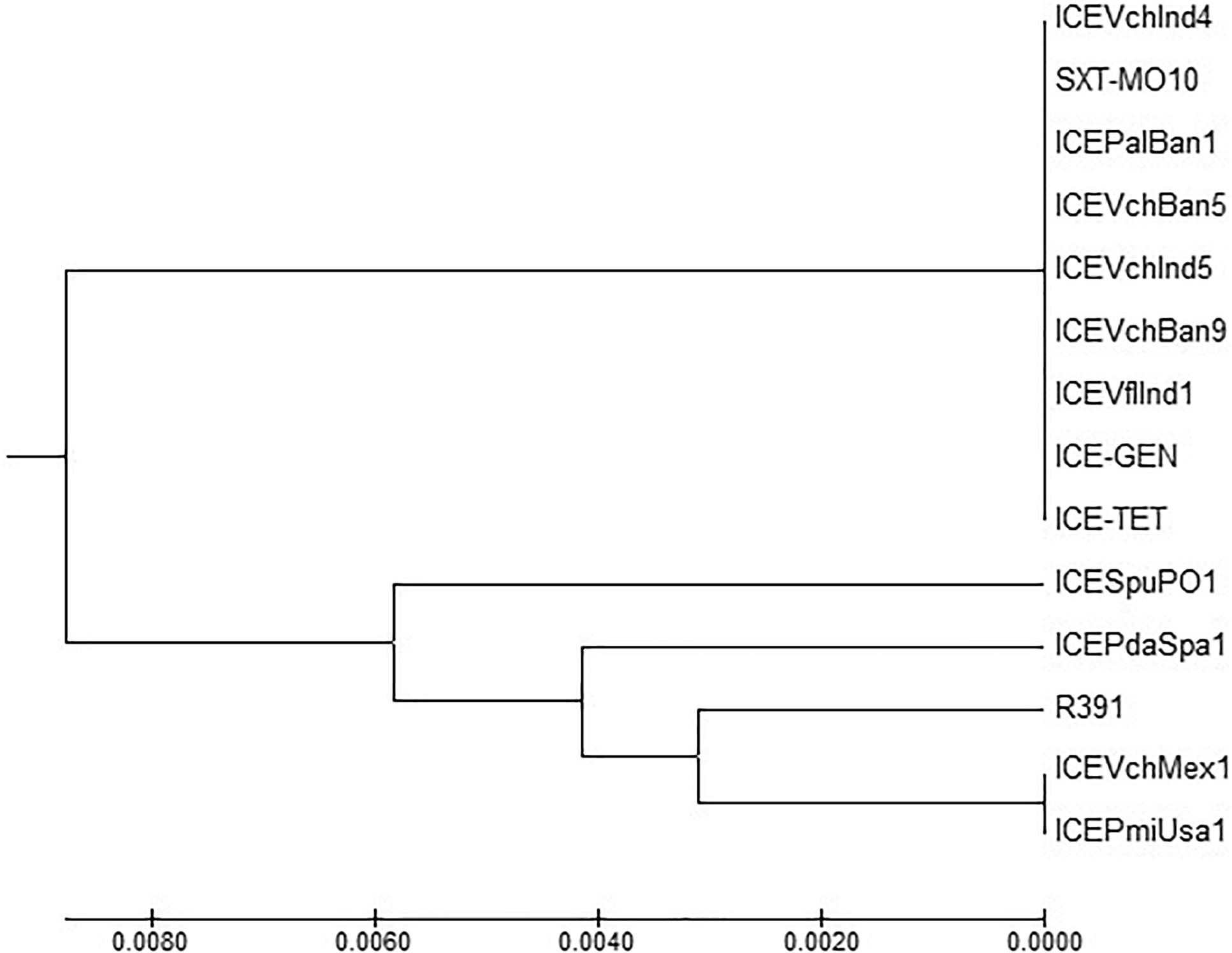
Figure 5. MEGA7 analysis based (Kumar et al., 2016) evolutionary relationships of taxa of setR of V. cholerae O1 strains.
Transfer of ICEs
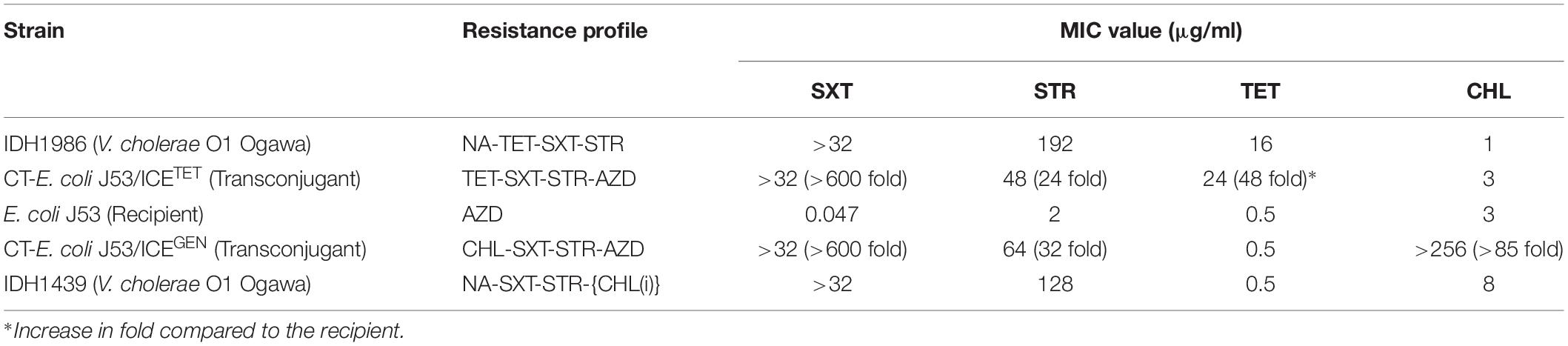
To test the transferability of the V. cholerae ICEs, we selected ICETET and ICEGEN carrying strains (IDH1986 and IDH1439, respectively). Both the types of ICEs could be transferred to E. coli J53 by conjugation. The transconjugants acquired additional resistance against SXT and STR (Table 4). Remarkably, CT-E. coli J53 from ICEGEN was highly resistant to CHL compared to the donor V. cholerae O1 strain, which showed reduced susceptibility to this antibiotic. Similarly, CT-E. coli J53 from ICETET expressed more resistance against TET than the donor Vibrio (Table 4). The frequency of transfer ranged from 3 × 10–5 to 5 × 10–6 transconjugants/recipient.
PFGE Analysis
Pulsed-field gel electrophoresis was performed to identify the clonal relationship between ICETET and ICEGEN carrying V. choleare strains. It was found that the V. cholerae O1 strains displayed clonal clusters reflecting their MDR profile, which indirectly revealed the composition of AMR encoding genes in the ICEs (Figure 6). Cluster A represented Vibrio strains devoid of the ICEs. These strains were only resistant to NA. Strains with ICEGEN were present in cluster B. These strains are resistant to NA, SXT and exhibited intermediate susceptibility to CHL. Cluster C contained the ICETET harboring strains that showed resistance to NA, SXT, and TET (Figure 6).
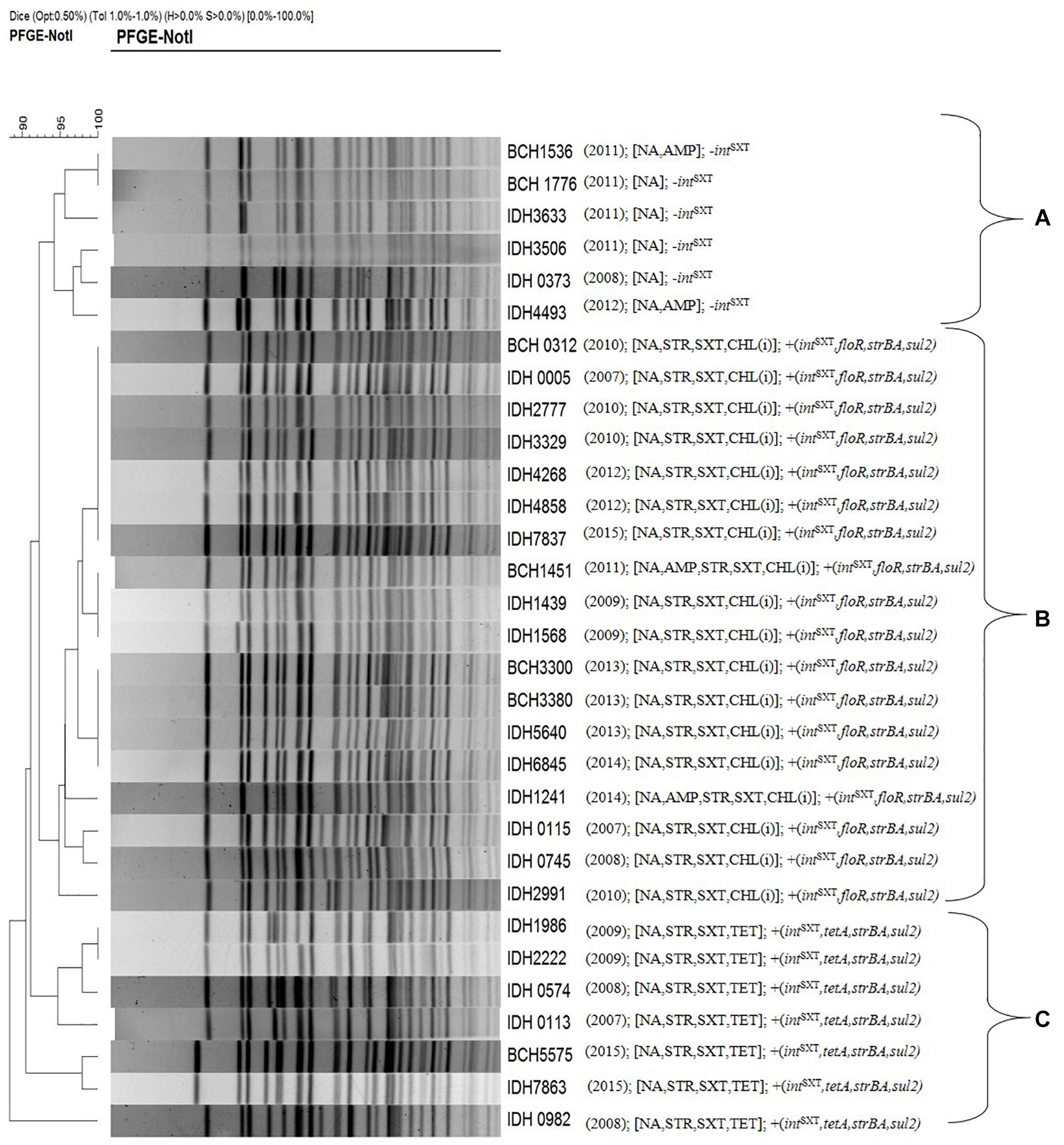
Figure 6. PFGE profile of the representative V. cholerae O1 strains with their antimicrobial resistance. Cluster A, strains devoid of intSXT or ICE; Cluster B, strains carrying ICEGEN; Cluster C, strains having ICETET. Number in the parenthesis represents the year of isolation.
Discussion
Cholera is endemic in the Indian subcontinent and it has spread to several other parts of the world (Mutreja et al., 2011). In Kolkata, MDR V. cholerae is associated with sporadic cholera for many years (Garg et al., 2000; Nair et al., 2010). V. cholerae O1 was susceptible to several antibiotics before 1980s, but developed resistance to SXT in the following years (Ghosh and Ramamurthy, 2011). V. cholerae O1 El Tor biotype that re-emerged in 1994 may have acquired SXT resistance phenotype from the O139 serogroup (Ramamurthy et al., 2003). Investigations conducted almost during the same period in several cholera endemic regions in India showed that the isolation rate of V. cholerae O1 was lesser than Kolkata, but the AMR pattern followed nearly the same trend, especially to tetracycline (Taneja et al., 2010; Das et al., 2011; Bhattacharya et al., 2012; Borkakoty et al., 2012; Mandal et al., 2012; Roy et al., 2012; Palewar et al., 2015; Bhuyan et al., 2016; Jain et al., 2016; Torane et al., 2016; Pal et al., 2018).
From 2010 to 2012, V. cholerae strains with AMR profiles of NA-STR-SXT-TET-AMP and NA-STR-SXT-TET were completely replaced with NA-STR-SXT-CHL(i)-AMP and NA-STR-SXT-CHL(i) along with NA-AMP and NA. Strains with the AMR profile of NA-STR-SXT-TET appeared again in 2015 (53%). Though the number of V. cholerae strains with the NA-SXT-STR-CHL(i) profile was highest from 2013 to 2014 (98–100%), it has reached to 46% with the re-emergence of TetR in 2015. The appearance of TetR in V. cholerae O1 Ogawa in 2008 has been reported from northern parts of India (Taneja et al., 2010). TetR has been previously reported mostly in Inaba serotype (Jesudason, 2006; Roychowdhury et al., 2008). Presence of tetA, floR, strBA, sul2, dfrA1 within the AMR gene cassettes has positive correlation with the phenotypic expression of drug resistance against TET, CHL, STR, and SXT (Dalsgaard et al., 2001; Hochhut et al., 2001; Wang et al., 2016). It is interesting to note that although dfrA18 conferring resistance to trimethoprim was reported in MO10, later it was replaced by the dfrA1 allele in a class IV integron located in the H3 (Wozniak et al., 2009).
In our study, floR and tetA genes were not found to coexist within the VRIII present in the rumB locus. Previous reports, however, had shown the presence of both floR and tetA in the V. cholerae ICEVchLao1 isolated from the Laos, ICEVchB33 from Beira, Mozambique (Iwanaga et al., 2004; Taviani et al., 2009). Depending upon the presence of resistance cassettes in the ICEs, we found two types of ICEs in our study namely ICEGEN and ICETET. Though the ICE backbone of ICEGEN was similar to those of SXTMO10 and SXTET, it had 99% structural similarity to ICEVchInd5. Lineages of ICEVchInd5 of V. cholerae O1 strains causing epidemics in the Indian subcontinent might have spread to Africa (Valia et al., 2013).
ICEGEN circulating in V. cholerae strains from Kolkata belonged to the group 1 ICE, which comprised ICEVchInd5 (India, 1994–2005), ICEVchBan5 (Bangladesh, 1998), ICEVchHai1 (Haiti, 2010), ICEVchNig1 (Nigeria, 2010), and ICEVchNep1 (Nepal, 1994) (Marin et al., 2014). Type I restriction-modification system systems of ICEGEN and ICETET were also reported in the other ICEs families, such as ICEVchMex1 and ICESpuPO1 (Burrus et al., 2006; Pembroke and Piterina, 2006). ICEs are constantly spreading in different geographical areas. ICEVchB33, which is different from other ICEs of SXT/R391 was first identified in V. cholerae O1 strains from India in 1994 and then Mozambique in 2004 (Taviani et al., 2009). Similar to V. cholerae O1 from India with ICEVchInd1, the other ICEs identified in Vietnam, Laos, and Mozambique (ICEVchVie1, ICEVchLao1, and ICEVchB33, respectively) lack the trimethoprim resistance encoding dfr18, but carried virD2 and floR, conferring resistance to CHL (Taviani et al., 2009). Majority of the V. cholerae O1 isolated in Kolkata from 1989 to 1990 had STXMO10/ICEVchInd4. This ICE was replaced by ICEVchInd5/ICEVchBan5 in the subsequent years (Weill et al., 2017, 2019).
In this study, the ICETET detected in V. cholerae O1 strains had significant structural dissimilarities with ICEVchBan9 (Bangladesh, 1994), ICEVchMoz10 (Mozambique, 2004), ICEVchB33 (Beira, 2004), and ICEVchLao1 (Iwanaga et al., 2004; Taviani et al., 2009; Marin et al., 2014). Nevertheless, structural variations, unstable core region, and the transfer region of both the ICEs found in our study were very much similar and shared a common ancestral backbone. In many ICEs, the core genes such as int, bet, exo, and setR are usually associated with phages, and genes such as tra are associated with plasmids (Wozniak et al., 2009; Armshaw and Pembroke, 2013). Having the same exclusion group (eexR1), ICEGEN and ICETET were mutually exclusive and therefore did not co-exist in a strain. ICE sequences reconfirmed that there were two ICE types that kept emerging in different years. The key modifications between them indicated that they may have diverse origins or be derived from a common ancestor and could have later evolved independently.
We could transfer the ICEGEN and ICETET from V. cholerae O1 to E. coli J53 by conjugation. The frequency of transfer observed was high (10–5 to 10–6), indicating that the ICEs were promiscuous due to the presence of an active tra region (Kiiru et al., 2009; Pande et al., 2012). Our study showed that only the resistances conferred by genes present in ICE were transferable and that the level of expression was different, being more in the transconjugants with respect to the donor vibrios. This could be due to “gene dosage” effect or absence of repressor in the new genetic environment of the recipient E. coli. Transconjugants showing higher drug resistance have been described in the previous reports as well (Petroni et al., 2002; Sarkar et al., 2015b). The co-existence of ICEs with plasmids and class 1 integrons in clinical as well as environmental V. cholerae has been reported (Thungapathra et al., 2002; Pande et al., 2012). The involvement of plasmids carrying the ICEs was not tested in this study. We also observed that resistance to NA and AMP were not transferable, indicating that the resistance to these antimicrobials could be contributed by the chromosomal factors such as mutations and efflux pumps (Ghosh and Ramamurthy, 2011).
As shown in the PFGE analysis, the clonal relatedness of V. cholerae strains isolated during different years corresponded with the MDR profiles. ICE integrase-negative strains isolated in 2008, 2011, and 2012 were found to cluster together (cluster A). V. cholerae O1 strains harboring either ICEGEN or ICETET were also grouped in different clusters (B and C, respectively). A similar observation was made with the outbreak strains of V. cholerae O1 in Kenya (Kiiru et al., 2009).
In conclusion, our findings revealed the existence of two types of ICEs in V. cholerae O1 strains from Kolkata. The ICEGEN that contained conserved backbone genes was most commonly detected in V. cholerae O1 circulating around Kolkata. Features of the Kolkata V. cholerae O1 strains with ICE carrying the TetR encoding genes are unique and the sequence of the ICETET had several variations from other sequenced ICEs. Also the ICETET harboring V. cholerae O1 strains reappeared after 4 years of disappearance in Kolkata. Unique PFGE clusters of V. cholerae O1 harboring different ICEs are linked with the AMR patterns. The primer pair designed in this study may be useful in the detection of ICEs carrying the tet. The transmission potential of ICEs identified in this study was very high, as evidenced from the conjugation assay. Therefore, the impact of ICE regulation and interactions between bacteria prevailing in the same ecological niches should be explored in detail. Emergence of new types of ICEs may pose challenges in the existing cholera management strategies.
Author Contributions
AG, TR, and KO conceived and designed the experiments. AS, DM, and GC performed the experiments. KO contributed reagents, materials, and analysis tools. TR and AM analyzed the data. AS and TR wrote the manuscript. All authors discussed the results, and reviewed and commented on the manuscript.
Funding
This work was supported in part by the Department of Biotechnology, New Delhi, India (Grant No. BT/MB/THSTI/HMC-SFC/2011), the Japan Initiative for Global Research Network on Infectious Diseases (J-GRID), the Ministry of Education, Culture, Sports, Science and Technology in Japan, the Japan Agency for Medical Research and Development (AMED; Grant No. JP18fm0108002), and the Indian Council of Medical Research. AG is J. C. Bose Chair Professor of the National Academy of Sciences, India.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer AB declared a past co-authorship with several of the authors, GC, AM, TR, and AG, to the handling Editor.
Footnotes
- ^ http://db-mml.sjtu.edu.cn/ICEberg
- ^ http://www.cbcb.umd.edu/software/glimmer-mg
- ^ www.scigenom.com/CANoPI
References
Armshaw, P., and Pembroke, J. (2013). “Integrative conjugative elements (ICEs) of the SXT/R391 group as vehicles for acquisition of resistance determinants, stable maintenance and transfer to a wide range of enterobacterial pathogens,” in Microbial Pathogens and Strategies for Combating Them: Science, Technology and Education, ed. A. Méndez-Vilas (Badajoz: Formatex Research Center), 439–446.
Bani, S., Mastromarino, P. N., Ceccarelli, D., Le Van, A., Salvia, A. M., Ngo Viet, Q. T., et al. (2007). Molecular characterization of ICEVchVie0 and its disappearance in Vibrio cholerae O1 strains isolated in 2003 in Vietnam. FEMS Microbiol. Lett. 266, 42–48. doi: 10.1111/j.1574-6968.2006.00518.x
Beaber, J. W., Hochhut, B., and Waldor, M. K. (2002). Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J. Bacteriol. 184, 4259–4269. doi: 10.1128/JB.184.15.4259-4269.2002
Bhattacharya, D., Sayi, D. S., Thamizhmani, R., Bhattacharjee, H., Bharadwaj, A. P., Roy, A., et al. (2012). Emergence of multidrug-resistant Vibrio cholerae O1 biotype El Tor in Port Blair, India. Am. J. Trop. Med. Hyg. 86, 1015–1017. doi: 10.4269/ajtmh.2012.11-0327
Bhattacharya, K., Kanungo, S., Sur, D., Lal Sarkar, B., Manna, B., Lopez, A. L., et al. (2011). Tetracycline-resistant Vibrio cholerae O1, Kolkata, India. Emerg. Infect. Dis. 17, 568–569. doi: 10.3201/eid1703.101176
Bhuyan, S. K., Vairale, M. G., Arya, N., Yadav, P., Veer, V., Singh, L., et al. (2016). Molecular epidemiology of Vibrio cholerae associated with flood in Brahamputra River valley, Assam, India. Infect. Genet. Evol. 40, 352–356. doi: 10.1016/j.meegid.2015.11.029
Bioteau, A., Durand, R., and Burrus, V. (2018). Redefinition and unification of the SXT/R391 family of integrative and conjugative elements. Appl. Environ. Microbiol. 84:e0485-18. doi: 10.1128/AEM.00485-18
Böltner, D., MacMahon, C., Pembroke, J. T., Strike, P., and Osborn, A. M. (2002). R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J. Bacteriol. 184, 5158–5169. doi: 10.1128/jb.184.18.5158-5169.2002
Bordeleau, E., Brouillette, E., Robichaud, N., and Burrus, V. (2010). Beyond antibiotic resistance: integrating conjugative elements of the SXT/R391 family that encode novel diguanylate cyclases participate to c-di-GMP signalling in Vibrio cholerae. Environ. Microbiol. 12, 510–523. doi: 10.1111/j.1462-2920.2009.02094.x
Borkakoty, B., Biswas, D., Devi, U., Yadav, K., and Mahanta, J. (2012). Emergence of classical ctxB genotype 1 and tetracycline resistant strains of Vibrio cholerae O1 El Tor in Assam, India. Trans. R. Soc. Trop. Med. Hyg. 106, 382–386. doi: 10.1016/j.trstmh.2012.03.005
Burrus, V., Quezada-Calvillo, R., Marrero, J., and Waldor, M. K. (2006). SXT-related integrating conjugative element in New World Vibrio cholerae. Appl. Environ. Microbiol. 72, 3054–3057. doi: 10.1128/AEM.72.4.3054-3057.2006
Ceccarelli, D., Spagnoletti, M., Bacciu, D., Danin-Poleg, Y., Mendiratta, D. K., Koshi, Y., et al. (2011). ICEVchInd5 is prevalent in epidemic Vibrio cholerae O1 El Tor strains isolated in India. Int. J. Med. Microbiol. 301, 318–324. doi: 10.1016/j.ijmm.2010.11.005
Clinical and Laboratory Standards Institute [CLSI] (2014). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fourth Informational Supplement. CLSI Document M100-S24. Wayne, PA: Clinical and Laboratory Standards Institute.
Clinical and Laboratory Standards Institute [CLSI] (2015). Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria: 3rd Edition, CLSI Document M45. Wayne, PA: Clinical and Laboratory Standards Institute.
Cooper, K. L., Luey, C. K., Bird, M., Terajima, J., Nair, G. B., Kam, K. M., et al. (2006). Development and validation of a PulseNet standardized pulsed-field gel electrophoresis protocol for subtyping of Vibrio cholerae. Foodborne Pathog. Dis. 3, 51–58. doi: 10.1089/fpd.2006.3.51
Dalsgaard, A., Forslund, A., Sandvang, D., Arntzen, L., and Keddy, K. (2001). Vibrio cholerae O1 outbreak isolates in Mozambique and South Africa in 1998 are multiple-drug resistant, contain the SXT element and the aadA2 gene located on class 1 integrons. J. Antimicrob. Chemother. 48, 827–838. doi: 10.1093/jac/48.6.827
Das, S., Choudhry, S., Saha, R., Ramachandran, V. G., Kaur, K., and Sarkar, B. L. (2011). Emergence of multiple drug resistance Vibrio cholerae O1 in East Delhi. J. Infect. Dev. Ctries 5, 294–298.
Dromigny, J. A., Rakoto-Alson, O., Rajaonatahina, D., Migliani, R., Ranjalahy, J., and Mauclére, P. (2002). Emergence and rapid spread of tetracycline-resistant Vibrio cholerae strains, Madagascar. Emerg. Infect. Dis. 8, 336–338. doi: 10.3201/eid0803.010258
Garg, P., Chakraborty, S., Basu, I., Datta, S., Rajendran, K., Bhattacharya, T., et al. (2000). Expanding multiple antibiotic resistance among clinical strains of Vibrio cholerae isolated from 1992-7 in Calcutta, India. Epidemiol. Infect. 124, 393–399. doi: 10.1017/S0950268899003957
Ghosh, A., and Ramamurthy, T. (2011). Antimicrobials & cholera: are we stranded. Indian J. Med. Res. 133, 225–231.
Hochhut, B., Lotfi, Y., Mazel, D., Faruque, S. M., Woodgate, R., and Waldor, M. K. (2001). Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob. Agents Chemother. 45, 2991–3000. doi: 10.1128/AAC.45.11.2991-3000.2001
Hochhut, B., and Waldor, M. K. (1999). Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol. Microbiol. 32, 99–110. doi: 10.1046/j.1365-2958.1999.01330.x
Imamura, D., Morita, M., Sekizuka, T., Mizuno, T., Takemura, T., Yamashiro, T., et al. (2017). Comparative genome analysis of VSP-II and SNPs reveals heterogenic variation in contemporary strains of Vibrio cholerae O1 isolated from cholera patients in Kolkata, India. PLoS Negl. Trop. Dis. 11:e0005386. doi: 10.1371/journal.pntd.0005386
Iwanaga, M., Toma, C., Miyazato, T., Insisiengmay, S., Nakasone, N., and Ehara, M. (2004). Antibiotic resistance conferred by a class I integron and SXT constin in Vibrio cholerae O1 strains isolated in Laos. Antimicrob. Agents Chemother. 48, 2364–2369. doi: 10.1128/AAC.48.7.2364-2369.2004
Jain, M., Kumar, P., and Goel, A. K. (2016). Emergence of tetracycline resistant Vibrio cholerae O1 biotype El Tor serotype Ogawa with classical ctxB gene from a cholera outbreak in Odisha, Eastern India. J. Pathog. 2016:1695410. doi: 10.1155/2016/1695410
Jesudason, M. V. (2006). Change in serotype and appearance of tetracycline resistance in V. cholerae O1 in Vellore, South India. Indian J. Med. Microbiol. 24, 152–153. doi: 10.4103/0255-0857.25224
Juíz-Río, S., Osorio, C. R., de Lorenzo, V., and Lemos, M. L. (2005). Subtractive hybridization reveals a high genetic diversity in the fish pathogen Photobacterium damselae subsp. piscicida: evidence of a SXT-like element. Microbiology 151, 2659–2669. doi: 10.1099/mic.0.27891-0
Kiiru, J. N., Saidi, S. M., Goddeeris, B. M., Wamae, N. C., Butaye, P., and Kariuki, S. M. (2009). Molecular characterisation of Vibrio cholerae O1 strains carrying an SXT/R391-like element from cholera outbreaks in Kenya: 1994-2007. BMC Microbiol. 9:275. doi: 10.1186/1471-2180-9-275
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kutar, B. M., Rajpara, N., Upadhyay, H., Ramamurthy, T., and Bhardwaj, A. K. (2013). Clinical isolates of Vibrio cholerae O1 El Tor Ogawa of 2009 from Kolkata, India: preponderance of SXT element and presence of Haitian ctxB variant. PLoS One 8:e56477. doi: 10.1371/journal.pone.0056477
Lekshmi, N., Joseph, I., Ramamurthy, T., and Thomas, S. (2018). Changing facades of Vibrio cholerae: an enigma in the epidemiology of cholera. Indian J. Med. Res. 147, 133–141. doi: 10.4103/ijmr.IJMR_280_17
Mandal, J., Dinoop, K. P., and Parija, S. C. (2012). Increasing antimicrobial resistance of Vibrio cholerae O1 biotype E1 tor strains isolated in a tertiary-care centre in India. J. Health Popul. Nutr. 30, 12–16.
Marin, M. A., Fonseca, E. L., Andrade, B. N., Cabral, A. C., and Vicente, A. C. (2014). Worldwide occurrence of integrative conjugative element encoding multidrug resistance determinants in epidemic Vibrio cholerae O1. PLoS One 9:e108728. doi: 10.1371/journal.pone.0108728
Marrero, J., and Waldor, M. K. (2007). The SXT/R391 family of integrative conjugative elements is composed of two exclusion groups. J. Bacteriol. 189, 3302–3305. doi: 10.1128/JB.01902-06
Martínez-Martínez, L., Pascual, A., and Jacoby, G. A. (1998). Quinolone resistance from a transferable plasmid. Lancet 351, 797–799.
Mhalu, F. S., Mmari, P. W., and Ijumba, J. (1979). Rapid emergence of El Tor Vibrio cholerae resistant to antimicrobial agents during first six months of fourth cholera epidemic in Tanzania. Lancet 1, 345–347. doi: 10.1016/S0140-6736(79)92889-7
Mutreja, A., Kim, D. W., Thomson, N. R., Connor, T. R., Lee, J. H., Kariuki, S., et al. (2011). Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477, 462–465. doi: 10.1038/nature10392
Nair, G. B., Ramamurthy, T., Bhattacharya, M. K., Krishnan, T., Ganguly, S., Saha, D. R., et al. (2010). Emerging trends in the etiology of enteric pathogens as evidenced from an active surveillance of hospitalized diarrhoeal patients in Kolkata. India. Gut Pathog. 2:4. doi: 10.1186/1757-4749-2-4
Pal, B. B., Nayak, S. R., and Khuntia, H. K. (2018). Epidemiology and antibiogram profile of Vibrio cholerae isolates between 2004-2013 from Odisha. India. Jpn. J. Infect. Dis. 71, 99–103. doi: 10.7883/yoken.JJID.2017.193
Palewar, M. S., Choure, A. C., Mudshingkar, S., Dohe, V., Kagal, A., Bhardwaj, R., et al. (2015). Typing and antibiogram of Vibrio cholerae isolates from a tertiary care hospital in Pune: a 3 year study. J. Glob. Infect. Dis. 7, 35–36. doi: 10.4103/0974-777X.146375
Pande, K., Mendiratta, D. K., Vijayashri, D., Thamke, D. C., and Narang, P. (2012). SXT constin among Vibrio cholerae isolates from a tertiary care hospital. Indian J. Med. Res. 135, 346–350.
Pearson, M. M., Sebaihia, M., Churcher, C., Quail, M. A., Seshasayee, A. S., Luscombe, N. M., et al. (2008). Complete genome sequence of uropathogenic Proteus mirabilis, a master of both adherence and motility. J. Bacteriol. 190, 4027–4037. doi: 10.1128/JB.01981-07
Pembroke, J. T., and Piterina, A. V. (2006). A novel ICE in the genome of Shewanella putrefaciens W3-18-1: comparison with the SXT/R391 ICE-like elements. FEMS Microbiol. Lett. 264, 80–88. doi: 10.1111/j.1574-6968.2006.00452.x
Petroni, A., Corso, A., Melano, R., Cacace, M. L., Bru, A. M., Rossi, A., et al. (2002). Plasmidic extended-spectrum beta-lactamases in Vibrio cholerae O1 El Tor isolates in Argentina. Antimicrob. Agents Chemother. 46, 1462–1468. doi: 10.1128/AAC.46.5.1462-1468.2002
Ramamurthy, T., Yamasaki, S., Takeda, Y., and Nair, G. B. (2003). Vibrio cholerae O139 Bengal: odyssey of a fortuitous variant. Microbes Infect. 5, 329–344. doi: 10.1016/S1286-4579(03)00035-2
Roy, S., Parande, M. V., Mantur, B. G., Bhat, S., Shinde, R., Parande, A. M., et al. (2012). Multidrug-resistant Vibrio cholerae O1 in Belgaum, South India. J. Med. Microbiol. 61, 1574–1579. doi: 10.1099/jmm.0.049692-0
Roychowdhury, A., Pan, A., Dutta, D., Mukhopadhyay, A. K., Ramamurthy, T., Nandy, R. K., et al. (2008). Emergence of tetracycline-resistant Vibrio cholerae O1 serotype Inaba, in Kolkata, India. Jpn. J. Infect. Dis. 61, 128–129.
Sarkar, A., Pazhani, G. P., Chowdhury, G., Ghosh, A., and Ramamurthy, T. (2015b). Attributes of carbapenemase encoding conjugative plasmid pNDM-SAL from an extensively drug-resistant Salmonella enterica serovar Senftenberg. Front. Microbiol. 6:969. doi: 10.3389/fmicb.2015.00969
Sarkar, A., Pazhani, G. P., Dharanidharan, R., Ghosh, A., and Ramamurthy, T. (2015a). Detection of integron-associated gene cassettes and other antimicrobial resistance genes in enterotoxigenic Bacteroides fragilis. Anaerobe 33, 18–24. doi: 10.1016/j.anaerobe.2015.01.008
Sjölund-Karlsson, M., Reimer, A., Folster, J. P., Walker, M., Dahourou, G. A., Batra, D. G., et al. (2011). Drug-resistance mechanisms in Vibrio cholerae O1 outbreak strain, Haiti, 2010. Emerg. Infect. Dis. 17, 2151–2154. doi: 10.3201/eid1711.110720
Spagnoletti, M., Ceccarelli, D., Rieux, A., Fondi, M., Taviani, E., Fani, R., et al. (2014). Acquisition and evolution of SXT-R391 integrativeconjugative elements in the seventh-pandemic Vibrio cholerae lineage. mBio 5:e01356-14. doi: 10.1128/mBio.01356-14
Taneja, N., Samanta, P., Mishra, A., and Sharma, M. (2010). Emergence of tetracycline resistance in Vibrio cholerae O1 biotype El Tor serotype Ogawa from north India. Indian J. Pathol. Microbiol. 53, 865–866. doi: 10.4103/0377-4929.72014
Taviani, E., Grim, C. J., Chun, J., Huq, A., and Colwell, R. R. (2009). Genomic analysis of a novel integrative conjugative element in Vibrio cholerae. FEBS Lett. 583, 3630–3636. doi: 10.1016/j.febslet.2009.10.041
Thungapathra, M., Amita, M., Sinha, K. K., Chaudhuri, S. R., Garg, P., Ramamurthy, T., et al. (2002). Occurrence of antibiotic resistance gene cassettes aac(6′)-Ib, dfrA5,dfrA12, and ereA2 in class I integrons in non-O1, non-O139 Vibrio cholerae strains in India. Antimicrob. Agents Chemother. 46, 2948–2955. doi: 10.1128/AAC.46.9.2948-2955.2002
Torane, V., Kuyare, S., Nataraj, G., Mehta, P., Dutta, S., and Sarkar, B. (2016). Phenotypic and antibiogram pattern of V. cholerae isolates from a tertiary care hospital in Mumbai during 2004-2013: a retrospective cross-sectional study. BMJ Open 6:e012638. doi: 10.1136/bmjopen-2016-012638
Valia, R., Taviani, E., Spagnoletti, M., Ceccarelli, D., Cappuccinelli, P., and Colombo, M. M. (2013). Vibrio cholerae O1 epidemic variants in Angola: a retrospective study between 1992 and 2006. Front. Microbiol. 4:354. doi: 10.3389/fmicb.2013.00354
Waldor, M. K., Tschape, H., and Mekalanos, J. J. (1996). A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J. Bacteriol. 178, 4157–4165. doi: 10.1128/jb.178.14.4157-4165.1996
Wang, R., Yu, D., Yue, J., and Kan, B. (2016). Variations in SXT elements in epidemic Vibrio cholerae O1 El Tor strains in China. Sci. Rep. 6:22733. doi: 10.1038/srep22733
Weill, F. X., Domman, D., Njamkepo, E., Almesbahi, A. A., Naji, M., Nasher, S. S., et al. (2019). Genomic insights into the 2016-2017 cholera epidemic in Yemen. Nature 7738, 230–233. doi: 10.1038/s41586-018-0818-3
Weill, F. X., Domman, D., Njamkepo, E., Tarr, C., Rauzier, J., Fawal, N., et al. (2017). Genomic history of the seventh pandemic of cholera in Africa. Science 358, 785–789. doi: 10.1126/science.aad5901
World Health Organization [WHO] (2005). The Treatment of Diarrhoea: A Manual for Physicians and Other Senior Health Workers, 4th rev. WHO/CDD/SER/80.2. Geneva: World Health Organization.
Wozniak, R. A., Fouts, D. E., Spagnoletti, M., Colombo, M. M., Ceccarelli, D., Garriss, G., et al. (2009). Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet. 5:e1000786. doi: 10.1371/journal.pgen.1000786
Keywords: cholera, V. cholerae O1, tetracycline, antimicrobial resistance, multidrug resistance, integrative conjugative element
Citation: Sarkar A, Morita D, Ghosh A, Chowdhury G, Mukhopadhyay AK, Okamoto K and Ramamurthy T (2019) Altered Integrative and Conjugative Elements (ICEs) in Recent Vibrio cholerae O1 Isolated From Cholera Cases, Kolkata, India. Front. Microbiol. 10:2072. doi: 10.3389/fmicb.2019.02072
Received: 16 November 2018; Accepted: 22 August 2019;
Published: 06 September 2019.
Edited by:
Rustam Aminov, University of Aberdeen, United KingdomReviewed by:
Pramod Kumar, All India Institutes of Medical Sciences, New Delhi, IndiaAshima Kushwaha Bhardwaj, Independent Researcher, Gurugram, India
Copyright © 2019 Sarkar, Morita, Ghosh, Chowdhury, Mukhopadhyay, Okamoto and Ramamurthy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thandavarayan Ramamurthy, dHJhbXVAdGhzdGkucmVzLmlu; cmFtYTFtdXJ0aHlAeWFob28uY29t
†These authors have contributed equally to this work
 Anirban Sarkar
Anirban Sarkar Daichi Morita2†
Daichi Morita2† Amit Ghosh
Amit Ghosh Goutam Chowdhury
Goutam Chowdhury Keinosuke Okamoto
Keinosuke Okamoto Thandavarayan Ramamurthy
Thandavarayan Ramamurthy