- 1International Institute of Tropical Agriculture, Ibadan, Nigeria
- 2Department of Crop and Soil Sciences, Kwame Nkrumah University of Science and Technology, Kumasi, Ghana
- 3Agricultural Research Service, United States Department of Agriculture, Tucson, AZ, United States
Increasing knowledge of the deleterious health and economic impacts of aflatoxin in crop commodities has stimulated global interest in aflatoxin mitigation. Current evidence of the incidence of Aspergillus flavus isolates belonging to vegetative compatibility groups (VCGs) lacking the ability to produce aflatoxins (i.e., atoxigenic) in Ghana may lead to the development of an aflatoxin biocontrol strategy to mitigate crop aflatoxin content. In this study, 12 genetically diverse atoxigenic African A. flavus VCGs (AAVs) were identified from fungal communities associated with maize and groundnut grown in Ghana. Representative isolates of the 12 AAVs were assessed for their ability to inhibit aflatoxin contamination by an aflatoxin-producing isolate in laboratory assays. Then, the 12 isolates were evaluated for their potential as biocontrol agents for aflatoxin mitigation when included in three experimental products (each containing four atoxigenic isolates). The three experimental products were evaluated in 50 maize and 50 groundnut farmers’ fields across three agroecological zones (AEZs) in Ghana during the 2014 cropping season. In laboratory assays, the atoxigenic isolates reduced aflatoxin biosynthesis by 87–98% compared to grains inoculated with the aflatoxin-producing isolate alone. In field trials, the applied isolates moved to the crops and had higher (P < 0.05) frequencies than other A. flavus genotypes. In addition, although at lower frequencies, most atoxigenic genotypes were repeatedly found in untreated crops. Aflatoxin levels in treated crops were lower by 70–100% in groundnut and by 50–100% in maize (P < 0.05) than in untreated crops. Results from the current study indicate that combined use of appropriate, well-adapted isolates of atoxigenic AAVs as active ingredients of biocontrol products effectively displace aflatoxin producers and in so doing limit aflatoxin contamination. A member each of eight atoxigenic AAVs with superior competitive potential and wide adaptation across AEZs were selected for further field efficacy trials in Ghana. A major criterion for selection was the atoxigenic isolate’s ability to colonize soils and grains after release in crop field soils. Use of isolates belonging to atoxigenic AAVs in biocontrol management strategies has the potential to improve food safety, productivity, and income opportunities for smallholder farmers in Ghana.
Introduction
Following its discovery nearly 60 years ago, aflatoxin contamination of key staple, economically important crops has attracted global attention (Wu, 2015). Developed nations have stringent aflatoxin standards for food/feed crops, milk, and their derived products (Cheli et al., 2014). This allows protecting consumers from health risks associated with aflatoxin exposure (JECFA, 2018). Aflatoxin contamination not only threatens public health but also curtails trade and economic opportunities from farm enterprises when crops exceed tolerance thresholds (Dzirasah, 2015; Kraemer et al., 2016). In contrast, although aflatoxin standards exist in many developing countries such as Ghana (GSA, 2001, 2013), these are poorly enforced. Maize and groundnut in Ghana are prone to aflatoxin contamination. A recent study mirrored the high prevalence of aflatoxin contamination reported frequently over 50 years with concentrations, in most cases, far exceeding the 15 and 10 ppb acceptable threshold for maize and groundnut, respectively, set by the Ghana Standards Authority (Agbetiameh et al., 2018). The two crops constitute major staple and cash crops for millions with per-capita consumption of 44 (US$ 15) and 12 kg (US$ 25) per annum for maize and groundnut, respectively (MoFA, 2011). Consequently, aflatoxin exposure is common and widespread across Ghana. Exposure begins in the unborn child in the uterus and throughout life (Lamplugh et al., 1988; Kumi et al., 2015). Several studies have documented the myriad of health problems associated with aflatoxins in Ghanaians (Shuaib et al., 2010; Jolly et al., 2013; Afum et al., 2016; UNICEF, 2017).
Aflatoxins are produced by fungi belonging to Aspergillus section Flavi (Frisvad et al., 2019). A. flavus, the most common aflatoxin-producing species worldwide (Klich, 2007), can be subdivided into two distinct morphotypes, the L and S morphotypes (Cotty, 1989). The S morphotype produces numerous small sclerotia (avg. dia <400 μm), few conidia, and consistently high B aflatoxin levels (Cotty, 1989). The L morphotype produces fewer, larger sclerotia (avg. dia >400 μm), numerous conidia, and variable levels of B aflatoxins. There are L morphotype genotypes that lack the ability to produce aflatoxins (i.e., atoxigenic) due to deletions, inversions, or defects in one or more of the aflatoxin biosynthesis genes (Adhikari et al., 2016). Aspergillus fungi can be further subdivided into vegetative compatibility groups (VCGs). Members of a VCG descend from the same clonal lineage and therefore are isolated subpopulations (Leslie, 1993; Grubisha and Cotty, 2010, 2015). Diversity among VCGs can be assessed using simple sequence repeat (SSR) markers. Closely related SSR haplotypes in most cases belong to the same VCG (Grubisha and Cotty, 2010, 2015).
Across the globe, several lineages resembling the A. flavus S morphotype have been detected with some of them producing copious amounts of both B and G aflatoxins (Probst et al., 2014; Singh and Cotty, 2019). In West Africa, fungi with S morphotype producing both B and G aflatoxins were known as unnamed taxon SBG (Cardwell and Cotty, 2002; Atehnkeng et al., 2008; Donner et al., 2009; Probst et al., 2014). Unknown taxon SBG fungi may be any of the recently described species A. aflatoxiformans, A. austwickii, A. cerealis, or A. minisclerotigenes (Pildain et al., 2008; Frisvad et al., 2019). Here we refer as SBG strains to all fungi with S morphotype producing both B and G aflatoxins.
Interactions between atoxigenic and aflatoxin-producing fungi are complex and coupled with other factors determine the extent of crop aflatoxin content (Cotty and Jaime-Garcia, 2007; Mehl et al., 2012; Atehnkeng et al., 2016). In regions where atoxigenic A. flavus have been detected, such genotypes have become valuable active ingredients in biocontrol formulations to mitigate crop contamination (Cotty et al., 2007; Atehnkeng et al., 2008; Abbas et al., 2011; Probst et al., 2011; Tran-Dinh et al., 2014; Mauro et al., 2015; Zhou et al., 2015; Alanis Zanon et al., 2016; Bandyopadhyay et al., 2016). Displacement of toxigenic fungi from the crop environment by the deployment of carefully selected atoxigenic A. flavus genotypes results in drastic aflatoxin reductions. This has been demonstrated in various crops grown commercially in the United States, Nigeria, Kenya, Senegal, The Gambia, and Italy (Cotty et al., 2007; Dorner, 2010; Doster et al., 2014; Bandyopadhyay et al., 2016; Mauro et al., 2018). This intervention is highly cost-effective in reducing aflatoxin contamination, curtailing aflatoxin-related diseases, and increasing access to local and international premium markets (Wu and Khlangwiset, 2010; Mehl et al., 2012).
In Ghana, aflatoxin management techniques have focused largely on traditional postharvest interventions (Florkowski and Kolavalli, 2013) and more recently on hermetically sealed bags (Paudyal et al., 2017; Danso et al., 2019). In many cases, postharvest technologies are insufficient in curtailing aflatoxin content to safe levels because crop infection and contamination often begins in the field (Mahuku et al., 2019). Once crops become contaminated, aflatoxins cannot be completely removed (Grenier et al., 2014). The aflatoxin biocontrol strategy that targets the source of infection and contamination, the aflatoxin-producing fungi, has not been developed for the farming system in Ghana. However, several atoxigenic A. flavus isolates are associated with both maize and groundnut grown across diverse agroecological zones (AEZs) in Ghana (Agbetiameh et al., 2018). The potential of atoxigenic isolates native to Ghana to competitively displace aflatoxin producers and limit crop aflatoxin content has not been investigated.
Atoxigenic biocontrol products are applied during crop development in a formulation (e.g., sterile wheat, sorghum, barley) that gives the active ingredient fungi reproductive advantages over the fungi naturally residing in the treated soils (Mehl et al., 2012). Spores of the beneficial fungi reproduce on the grain, colonize other organic matter substrates in the field, and then become associated with the treated crop during its development (Mehl et al., 2012; Bandyopadhyay et al., 2016). Criteria to select atoxigenic biocontrol agents include wide distribution of the atoxigenic AAV to which they belong over the target nation and superior ability to limit aflatoxin contamination when challenged with highly toxigenic genotypes (Probst et al., 2011; Atehnkeng et al., 2016). It is also necessary to select genotypes with superior abilities to both out-compete other fungi while in the soil and to efficiently move to the crop to provide the intended protection.
The objectives of this study were to: (i) evaluate 12 native atoxigenic A. flavus isolates belonging to genetically diverse atoxigenic AAVs for their abilities to reduce aflatoxin production in laboratory assays; (ii) assess comparative abilities of the 12 isolates to establish in soil and crop (maize and groundnut) niches across three AEZs; (iii) determine the extent of aflatoxin reduction by experimental biocontrol products constituted with the candidate isolates; and (iv) select isolates of superior atoxigenic AAVs for use as active ingredients in biocontrol formulations for crop aflatoxin mitigation in Ghana. Native, ecologically adapted atoxigenic AAVs with wide distribution across several AEZs, and with potential as biocontrol agents were detected. Ability to disperse from soil and establish in grains in the field as an ecological criterion for selection of biocontrol active ingredients is a novelty of this study. The identified atoxigenic AAVs are biological resources that can be used to formulate biocontrol products for aflatoxin mitigation. Use of the representative isolates of the selected AAVs may allow for enhanced crop value and food safety and reduce aflatoxin exposure in humans and livestock.
Materials and Methods
Microsatellite Genotyping
In a previous study, 4,736 A. flavus L morphotype isolates were examined for their aflatoxin-production potential and it was found that 847 isolates lacked aflatoxin-producing abilities (Agbetiameh et al., 2018). We characterized the 847 atoxigenic isolates using SSR markers developed for A. flavus (Grubisha and Cotty, 2009). DNA extraction, multiplex-PCR, and microsatellite genotyping were conducted following previously described protocols (Grubisha and Cotty, 2009, 2010; Callicott and Cotty, 2015; Islam et al., 2018). Over 20% of isolates were subjected to at least three independent PCR and genotyping assays for all loci. This allowed to assess consistency of the data.
Population Genetic Analyses
After genotyping, isolates were manually assigned to haplotypes defined by identity across 17 SSR markers (Grubisha and Cotty, 2009). Haplotype frequency was calculated following sample correction, such that a haplotype was only counted once per individual sample. Frequencies were then calculated on a per sample basis (data not shown). Twelve atoxigenic isolates were chosen (Table 1) for testing based on a combination of per sample haplotype frequency, presence in other West African countries, and similarity to atoxigenic biocontrol active ingredients already in use in other West African countries (Figure 1). Frequently encountered haplotypes were assumed to be already well adapted to Ghana. Isolates belonging to AAVs already selected as active ingredients of biocontrol products have a known ability to reduce aflatoxins when properly applied to crops.
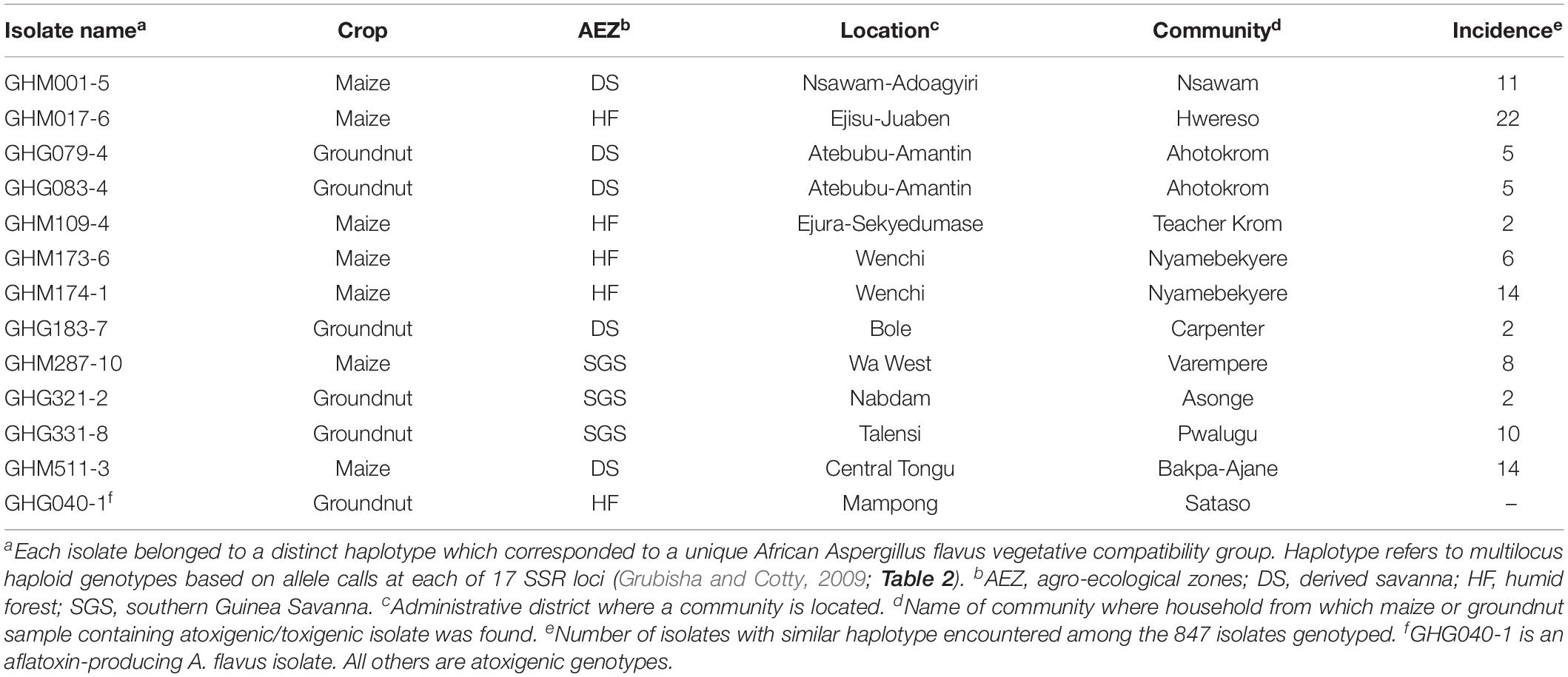
Table 1. Origin of a toxigenic isolate and one atoxigenic isolate each of 12 haplotypes of Aspergillus flavus used in the current study.
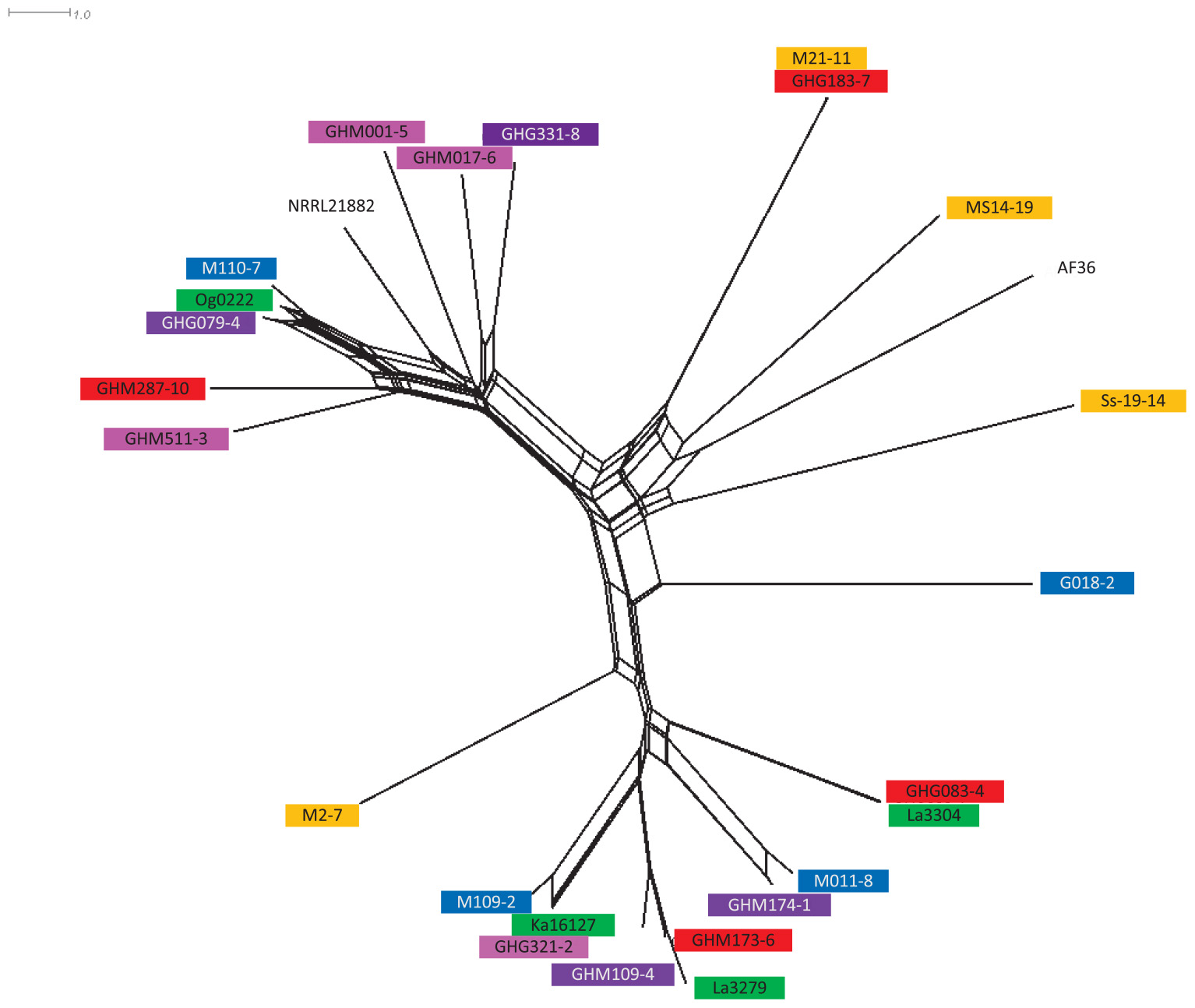
Figure 1. NeighborNet splitstree of 12 selected atoxigenic Aspergillus flavus haplotypes from Ghana with other active ingredients of registered aflatoxin biological control products in West Africa. La3279, La3304, Ka16127, and Og0222 are the active ingredients of AflasafeTM (used in Nigeria, in green); G018-2, M011-8, M109-2, and M110-7 are the active ingredients of Aflasafe BF01 (used in Burkina Faso, in blue); M2-7, M21-11, MS14-19, and Ss19-14 are the active ingredients of Aflasafe SN01 (used in Senegal and The Gambia, in orange); AF36 is the active ingredient of AF36 PrevailTM; and NRRL21882 is the active ingredient of Afla-GuardTM (both registered for use in the United States) (Ortega-Beltran and Bandyopadhyay, 2019). Isolates composing experimental product A are in purple, isolates composing experimental product B are in red, and isolates composing experimental product C are in pink. Length of branches are proportional to distances between isolates.
Simple sequence repeat data were re-coded from amplicon size to the number of repeats prior to assessing genetic relationships among all haplotypes. Phylogenetic relationships among the 12 selected genotypes and other registered biocontrol genotypes were assessed with Genodive (Meirmans and Van Tienderen, 2004) after which SplitsTree 4.14.6 (Huson and Bryant, 2005) was used to create a NeighborNet tree (Figure 1).
Atoxigenic Aspergillus flavus L Morphotype Isolates
The population genetic analyses revealed 12 dominant atoxigenic SSR haplotypes widely distributed across different locations of Ghana (Table 1). The origin and distribution of atoxigenic and aflatoxin-producing genotypes is summarized in Table 1. Tester pairs of VCGs were developed for 11 of the 12 SSR haplotype groups following previously described protocols (Cove, 1976; Bayman and Cotty, 1991). It was not possible to obtain a complementary pair of nit auxotrophs for isolate GHG183-7. The concordance between SSR haplotype and VCG for 11 of the 12 groups was then tested using vegetative compatibility analyses. These VCGs were termed as AAVs.
Laboratory Competition Assays
Representative isolates of the 12 SSR haplotypes were evaluated for their ability to limit aflatoxin accumulation when challenged with A. flavus isolate GHG040-1, a potent aflatoxin producer native to Ghana, in laboratory competition assays as described by Probst et al. (2011).
To prepare inocula, single-spored isolates, maintained for long-term storage on silica grains, were grown on 5–2 agar [(5% V-8 juice (Campbell Soup Company, Camden, NJ, United States), 2% Bacto-agar (Difco Laboratories Inc., Detroit, MI, United States), pH 6.0)] at 31°C for 7 days (Cotty, 1989). Spore suspensions of each isolate were prepared in 0.1% TWEEN 80® and adjusted to 106 spores ml–1 using a turbidimeter (Atehnkeng et al., 2014). A 1-ml spore suspension of the individual atoxigenic isolates and the aflatoxin producer, and mixtures of each atoxigenic/aflatoxin-producing isolate (ratio = 1:1) were separately inoculated on 10 g of autoclaved maize grains. Maize inoculated with 1-ml sterile distilled water served as negative control. Inoculated grains, five replications per treatment, were incubated for 7 days (31°C, dark). The experiment was conducted twice (test 1 and test 2). In test 1, all except atoxigenic isolate GHG083-4 was evaluated.
Following incubation, aflatoxins were extracted from maize fermentations as previously described (Agbetiameh et al., 2018). Briefly, fermentations were combined with 50 ml 70% methanol. Suspensions were shaken on a Roto-Shake Genie (Scientific Industries, Bohemia, NY, United States) for 30 min at 400 rpm and filtered through Whatman No. 1 filter paper (Whatman International Ltd., Maidstone, United Kingdom). Filtrates were collected in 250 ml separatory funnels, combined with 5 ml distilled water, and extracted with 15 ml methylene chloride. The methylene chloride phase was filtered through a bed of 25 g anhydrous sodium sulfate contained in fluted Whatman No. 4 filter paper, combined, and evaporated to dryness in a fume hood (Cotty and Cardwell, 1999). Residues were dissolved in 1 ml methylene chloride, spotted (4 μl) alongside aflatoxin standards (Supelco, Bellefonte, PA, United States) on thin layer chromatography (TLC) Aluminum (20 cm × 10 cm) Silica gel 60 F254 plates (Merck, Darmstadt, Germany) and developed with diethyl ether–methanol–water (96:3:1) (Probst and Cotty, 2012). Aflatoxins were quantified directly on TLC plates with a scanning densitometer (CAMAG TLC Scanner 3) and quantification software (winCATS 1.4.2, Camag, AG, Muttenz, Switzerland) (Agbetiameh et al., 2018).
Formulation of Experimental Biocontrol Products
Three experimental biocontrol products (named A, B, and C) were composed each with four representative atoxigenic isolates of different haplotypes and manufactured in Ibadan, Nigeria (Table 2). To prepare each product, spores of the four atoxigenic isolates were obtained from 5-day-old cultures grown on 5–2 agar to prepare inoculum in bulk. Spores were dislodged and suspended in 0.1% TWEEN 80® and adjusted to 106 spores ml–1 as above. Spores of each atoxigenic isolate were independently reproduced in glass bottles containing sterilized sorghum grain as follows. Prior to inoculation, sorghum grain was pre-conditioned in sterile 1-L plastic bottles. Moisture content of sorghum grain was increased to 30% by adding sterile distilled water and bottles were rolled for 4 h on a 240 Vac Benchtop Roller (Wheaton, Millville, NJ, United States). Thirty grams of pre-conditioned grain were added to 250-ml glass bottles along with two Teflon balls (1/2″ diameter) and autoclaved (20 min, 121°C). Each cooled bottle containing sorghum was independently inoculated with 4 ml of spore suspension of each atoxigenic isolate. After incubation (7 days, 31°C), 125 ml sterile 0.1% TWEEN® 20 was added to each bottle to harvest spores. Bottles were placed on a Roto-Shake Genie reciprocal shaker (Scientific Industries, Bohemia, NY, United States) at 200 rpm for 20 min. The Teflon balls facilitated dislodging spores from sorghum grains. For each atoxigenic strain, a suspension was adjusted to 4 × 107 spores ml–1 as above. To prepare 100 kg of each experimental product, a spore suspension (1 l, 4 × 107 spores ml–1) of the constituent atoxigenic genotypes was individually combined with 150 ml of a polymer (SentryTM, Precision Laboratories, Waukegan, IL, United States) and 200 ml of a blue non-toxic dye (PrismTM, Milliken and Company, Spartanburg, SC, United States) and coated on roasted, sterile sorghum grain with a seed treater (Bandyopadhyay et al., 2016). Following phytosanitary certification by the Nigeria Plant Quarantine Service and the issuance of import permit by the Plant Protection and Regulatory Services Directorate (PPRSD) of Ghana’s Ministry of Food and Agriculture (MoFA), the three experimental products were transported to Ghana for evaluation in farmer field trials.
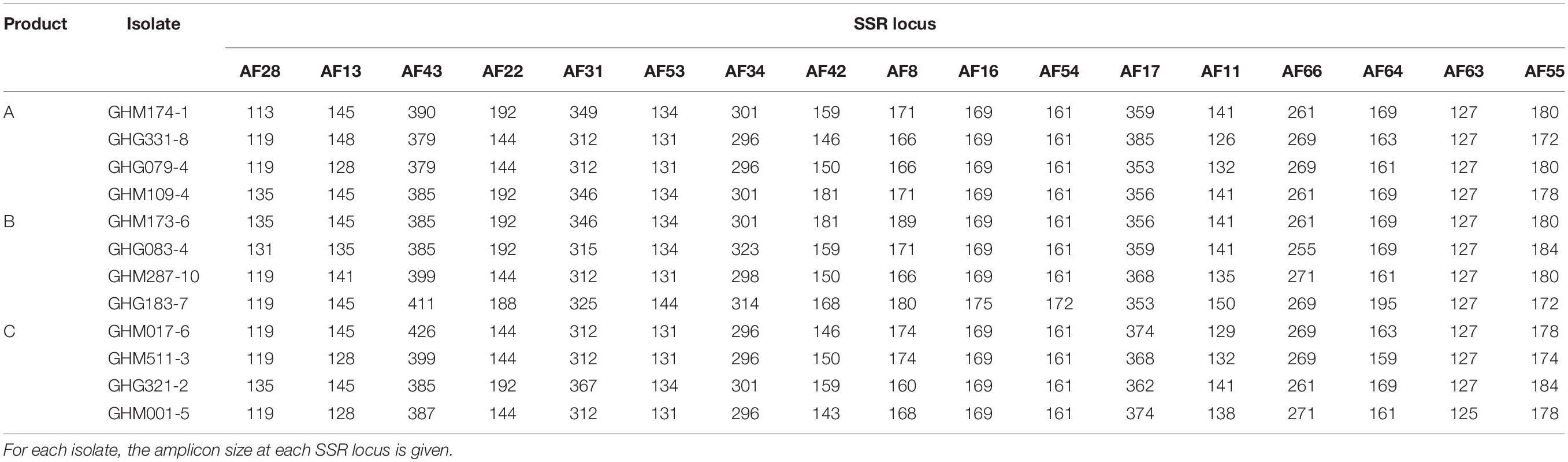
Table 2. Composition of experimental aflatoxin biocontrol products, each containing a mixture of four atoxigenic Aspergillus flavus vegetative compatibility groups represented by their type isolates.
Field Sites, Plots, and Trial Establishment
Field trials were conducted in 2014 during the major cropping season in Northern Ghana and minor season in the Middle Belt. The trials were conducted in five regions located in three AEZs. In each region, the fields were distributed in two districts. The two cropping seasons and the AEZs’ characteristics have been described previously (Agbetiameh et al., 2018). Farmers and their field selection was done in collaboration with Agricultural Extension Agents from the Department of Agriculture of MoFA in the respective districts following stakeholder sensitization and training workshops. In each district, five maize and five groundnut fields (size ≥ 2 ha) were selected. Farmers grew their crops according to their own agronomic practices. Each field was divided into four equal-sized plots separated by 5 m from each other. Assignment of plots to treatments across field locations was done using a randomized complete block design (RCBD). Three plots within a block were assigned treatment to one of the three experimental products. The remaining plot was left untreated and served as control. In each district, treatments were replicated five times. When field sizes were <2 ha (mostly groundnut fields), individual fields in a group of four nearby fields were considered as plots. Experimental products were broadcasted by hand (10 kg ha–1) to field soils 2 weeks before flowering and following weeding and/or fertilizer application by farmers. From each plot, before product application and also at harvest, soil samples (up to 2.5 cm depth) were taken randomly from at least 15 different spots resulting in a composite sample of about 150 g (Atehnkeng et al., 2014). Grain samples comprising 25 maize ears and approximately 1-kg groundnut (in-shell) were collected at harvest.
Analysis of Aspergillus Section Flavi in Soils and Grains
Soil samples were dried in a forced-air oven (50°C, 48 h). Samples with clods were pulverized and sieved through 2 mm wire mesh to remove gravel and large particles. Grains were manually shelled, and 500 g were milled using a laboratory blender (Waring Commercial, Springfield, MO, United States) for 1 min in a 250 ml stainless steel blending jar (MC-2). Milled samples were stored at 4°C before aflatoxin and microbial analyses. The blending jar was washed between samples with 80% ethanol to prevent microbial and aflatoxin cross contamination. Aspergillus section Flavi fungi in soil and grains were isolated using dilution plate technique on modified rose Bengal Agar as described previously (Atehnkeng et al., 2014). Plates were incubated for 3 days (31°C, dark). From each sample, 12 discrete Aspergillus species colonies were sub-cultured on 5–2 agar (31°C, 7 days) and then assigned to their corresponding species based on macroscopic and microscopic characteristics (Pitt and Hocking, 2009). Sporulating cultures of each isolate were saved as agar plugs in 4 ml vials containing 2 ml sterile distilled water until further characterization.
Aflatoxin Determination in Grain Samples
Aflatoxin levels in maize and groundnut sampled at harvest were examined to determine the extent of contamination in grains from treated and control plots. Aflatoxins were extracted from maize by combining 20 g ground sample with 100 ml of 70% methanol (Atehnkeng et al., 2008). For groundnut, 20 g of ground sample was combined with 100 ml of 80% methanol (Cole and Dorner, 1993). Aflatoxins were extracted, combined, separated on TLC plates, and quantified as described above.
Incidence of Atoxigenic Genotypes
Frequencies of A. flavus belonging to the applied AAVs of the three experimental products were examined in soils and grains. Nitrate non-utilizing (nit) auxotrophs were generated for all recovered A. flavus L morphotype isolates (Grubisha and Cotty, 2010). Briefly, a spore suspension of each isolate (approximately 1,000 spores in 15 μl) was seeded into a well at the center of a plate containing mutant selection medium (Czapek-dox broth, 25 g l–1 KClO3, 10 ml l–1 rose Bengal, 2% Bacto-agar, pH 7.0). Seeded plates were incubated at 31°C for 7–30 days. Spontaneous auxotrophic sectors were transferred to a purification medium (Czapek-dox broth, 15 g l–1 KClO3, 2% Bacto-agar, pH 6.5) for 3 days to clean up and stabilize nit mutants. A mutant sector was subsequently transferred onto 5–2 agar, and incubated for 5 days at 31°C. Plugs of sporulating mutants were stored in 4 ml glass vials containing 2 ml sterile distilled water for use in complementation assays. Assignment of mutants of isolates to an AAV was based on pairing the isolate auxotroph with complementary tester auxotrophs of each applied AAV (Grubisha and Cotty, 2010). A single complementation test was performed on starch agar (36 g l–1 dextrose, 3 g l–1 NaNO3, 2% Bacto-agar, 2% soluble starch, pH 6.0) (Cotty and Taylor, 2003) where three wells (3 mm dia, 1 cm apart) were made in a triangular pattern at the center of the plate. Two wells were each seeded with 15 μl of either of the tester pair while the third well was seeded with the isolate auxotroph being characterized. Plates were incubated for 5–10 days at 31°C. Auxotrophs forming a stable heterokaryon with one or both tester auxotrophs of an applied AAV were assigned to that AAV and were considered to be the applied genotype. In all, a total of 47,520 vegetative compatibility tests were conducted.
Data Analysis
All statistical tests were performed with SAS (version 9.4, SAS Institute Inc., Cary, NC, United States). Prior to data analysis, all response variables were log-transformed to stabilize variances. Means of the response variables were subjected to analysis of variance (ANOVA) and separated with Fisher’s protected least significant difference (LSD) test (α = 0.05). Pairwise comparison means of response variables from treated and control plots were conducted using Student’s t-test (α = 0.05). Applied AAVs were ranked separately by their incidence in soil and grain samples across different geographical locations. To calculate the rank, the proportion of the number of (i) AEZ (n = 3), (ii) regions (n = 5), (iii) districts (n = 10), and (iv) samples (n = 30) where the AAV was detected and (v) the proportion of isolates of the AAV detected (n = 360) was summed. Higher the sum, higher (1 = highest, 11 = lowest) the rank. For example, AAV GHM287-10 in maize was detected in the 3 AEZ (3/3 = 1.0), 5 regions (5/5 = 1.0), 8 districts (8/10 = 0.8), 17 samples (17/30 = 0.57), and 75 isolates were detected (75/360 = 0.21) for a total of 3.58.
Results
Identification of Dominant Atoxigenic Genotypes
Out of the 847 atoxigenic A. flavus L morphotype isolates identified previously (Agbetiameh et al., 2018), there were 454 unique and diverse haplotypes. Among those haplotypes, 12 were widely distributed across Ghana (Table 1) but not closely related (Figure 1). AAV grouping of 11 of the 12 groups concurred with the grouping revealed by SSRs (data not shown). Mutants of isolate GHG183-7 did not complement with tester pairs of any of the 11 AAVs. Therefore, GHG183-7 was considered another AAV.
The SSR signatures for identifying the representative isolates of AAVs constituting the experimental products are reported in Table 2. None of the locus was monomorphic among the examined isolates. The number of alleles per locus ranged from 2 to 7 (Table 2).
Aflatoxin Inhibition Potential of Atoxigenic Genotypes in Competition Tests
When inoculated individually, none of the 12 atoxigenic isolates produced aflatoxins on maize grains (LOD = 0.1 μg kg–1), as in the previous study (Agbetiameh et al., 2018). The aflatoxin-producing isolate GHG040-1 produced high aflatoxin B1 levels (>51.0 mg kg–1) on maize grains in both tests, as expected. Marked variations (P < 0.01) were detected in the aflatoxin inhibition potential of atoxigenic isolates when co-inoculated with the aflatoxin producer. Aflatoxin reductions ranged from 92.8 to 98.7% (Table 3). In test 1, atoxigenic isolates GHM173-6 and GHM511-3 significantly (P < 0.0001) reduced aflatoxin accumulation by the aflatoxin producer to <1.0 mg kg–1, the lowest level among all combinations. GHG183-7 had the least aflatoxin inhibition potential (5.59 mg kg–1). However, that level was also significantly (P < 0.0001) lower than in grains inoculated solely with the aflatoxin producer. GHG083-4 was not selected when test 1 was conducted, hence no aflatoxin inhibition data were generated in test 1 (Table 3).
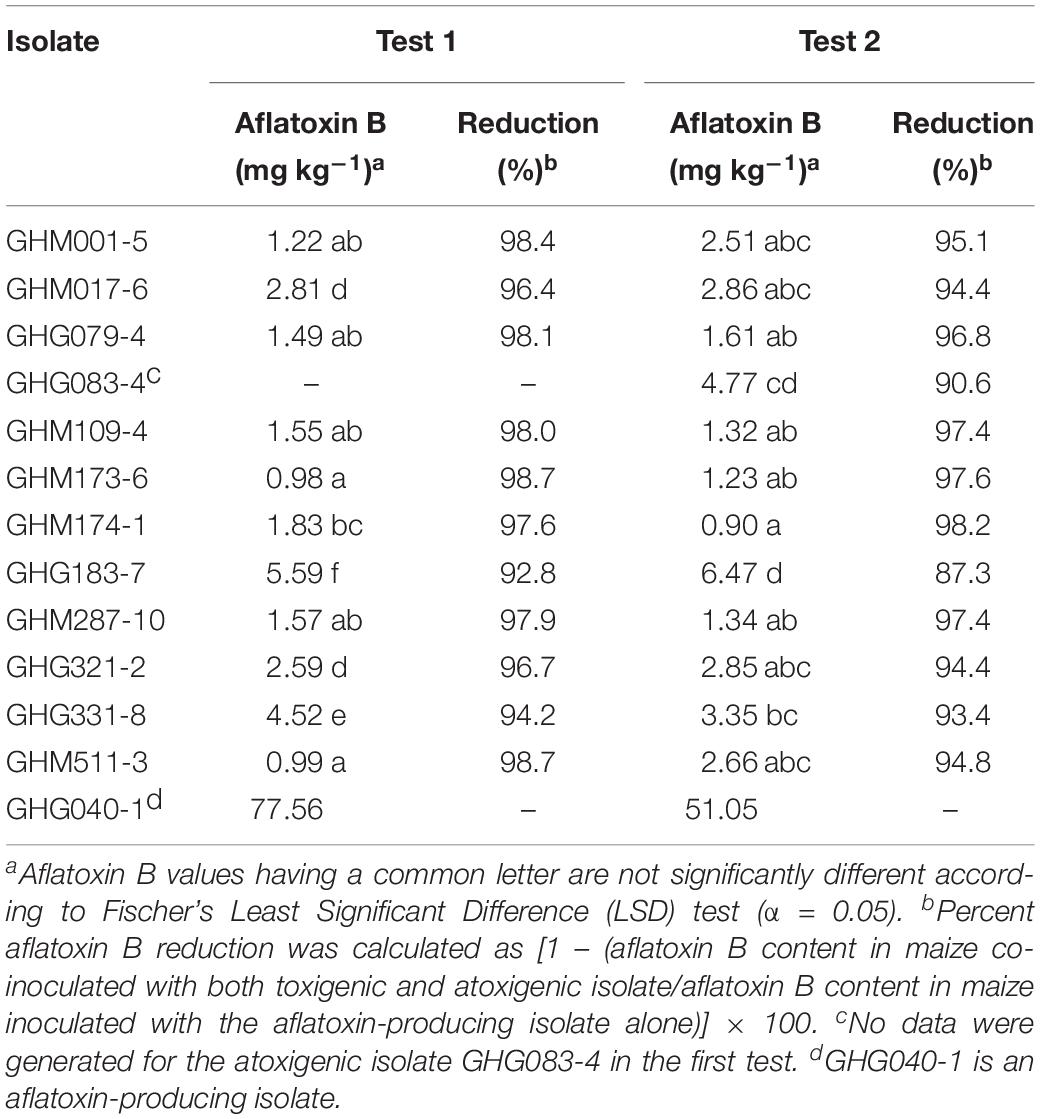
Table 3. Aflatoxin B (B1 + B2) content of maize in μg/kg during co-inoculation of atoxigenic isolates and an aflatoxin-producer.
Similar results were observed in test 2. Aflatoxin reductions ranged from 87.3 to 98.2% (Table 3). The lowest toxin inhibition (6.47 mg kg–1) was by GHG183-7, as in test 1. GHM174-1 reduced aflatoxin the most (0.90 mg kg–1).
Quality Control of the Experimental Products
All carrier grains of all batches of the experimental products were colonized only by A. flavus. Other microorganisms were not recovered in any of the grains. The recovered A. flavus fungi were solely composed of the active ingredient AAVs composing the experimental products. Other AAVs of A. flavus were not detected in any of the batches. In each experimental product, each of the four active ingredient AAVs was found on 25 ± 3% carrier grains of the examined batches. Each gram of product contained, on average, 3500 ± 300 colony forming units (CFUs) of the active ingredient fungi.
Aflatoxin Concentration in Crop Samples
Field trials were conducted in 2014 in 10 districts from five regions located in three AEZs in Ghana (Figure 2). Across all AEZs, substantially (P < 0.05) less aflatoxins accumulated in grains from plots treated with the experimental products, compared to untreated grains. Treated groundnut contained 70.5–99.7% less aflatoxins than those untreated. Across AEZs, aflatoxin levels in treated groundnut ranged from 1 to 61 μg kg–1 with those from humid forest (HF) containing safe levels. Aflatoxin content in untreated groundnut ranged from 58 to 302 μg kg–1 (Table 4). In maize, up to 100% reduction was detected in treated crops. Aflatoxin concentration was below 0.1 μg kg–1 in treated maize while it ranged from 0.8 to 7.8 μg kg–1 in control plots (Table 4).
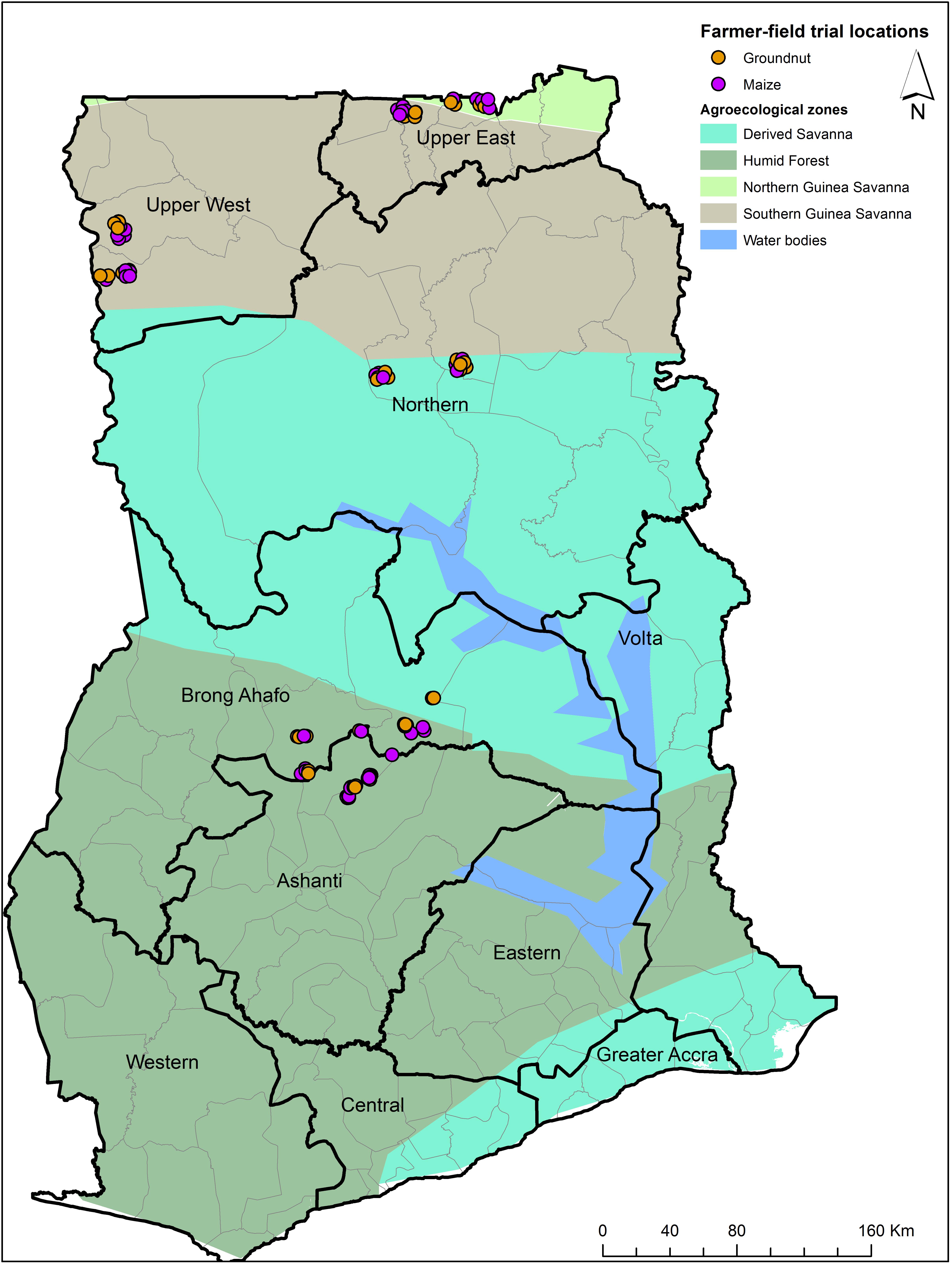
Figure 2. Map of Ghana indicating locations where field trials were conducted in maize and groundnut during 2014.
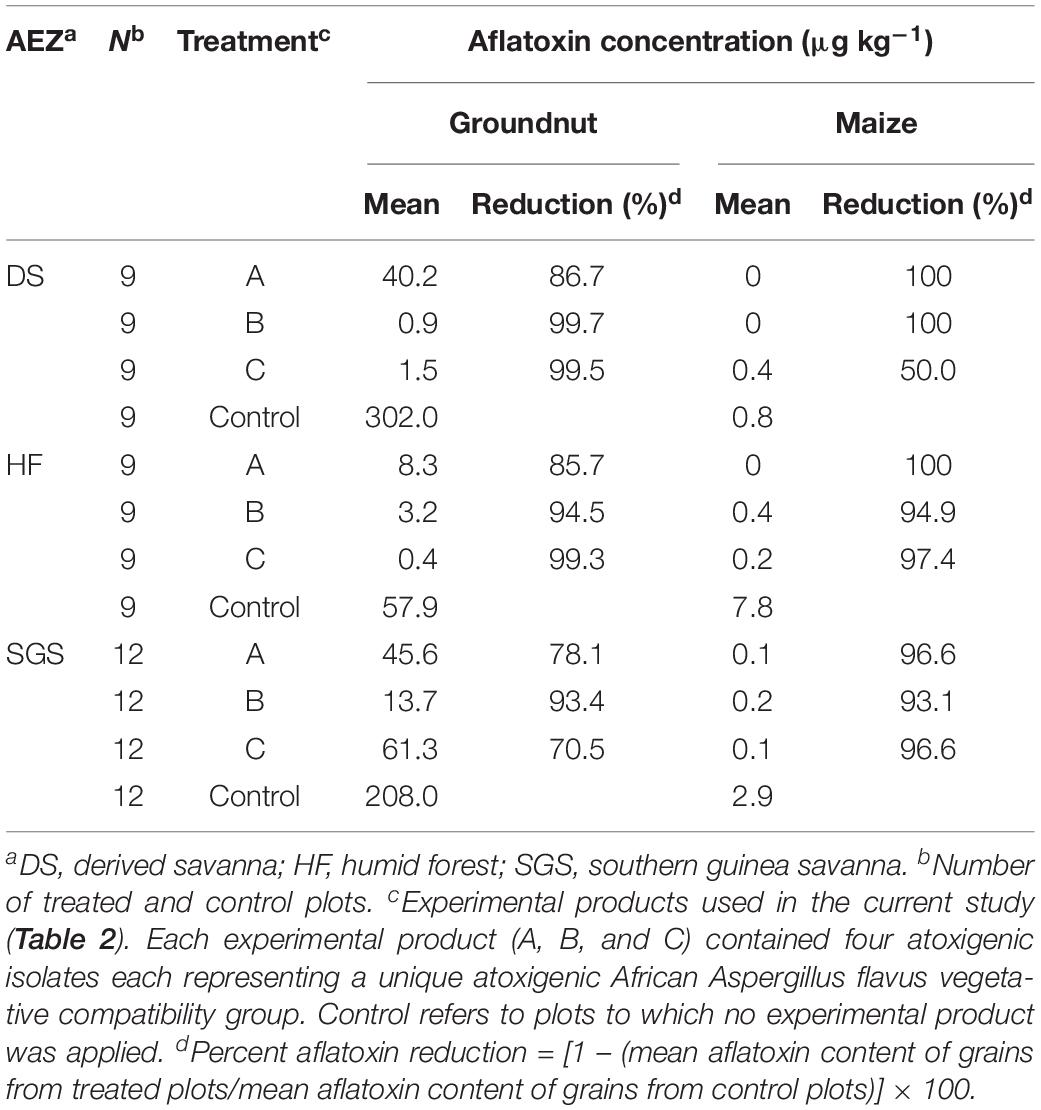
Table 4. Aflatoxin content (μg kg–1) in groundnut and maize kernels at harvest from treated and control fields across three agroecological zones (AEZs) in Ghana during 2014 cropping season.
Aspergillus Fungal Communities in Soils and Grains
Four main members within Aspergillus section Flavi (A. flavus L morphotype, SBG strains, A. parasiticus, and A. tamarii) were recovered from soil before application and at harvest, and on grain collected at harvest. In all substrates, A. flavus L morphotype dominated the communities with frequencies greater than 83% (Table 5). Prior to application of experimental products, incidence of L morphotype in field soils ranged from 87.7% in HF to 99.1% in derived savannah (DS). Frequencies of SBG strains, A. parasiticus, and A. tamarii were low (range = 0–9.9%; Table 5).
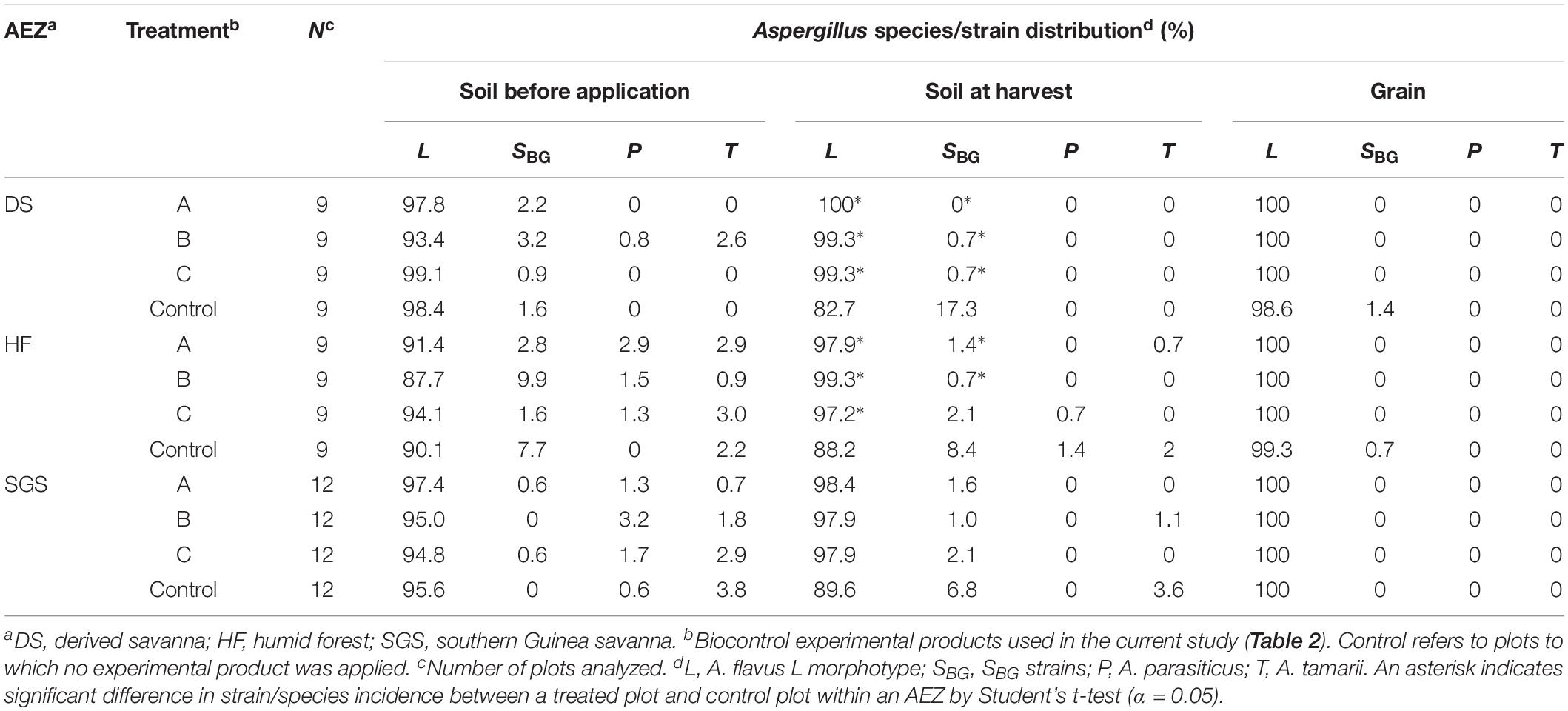
Table 5. Community structure of Aspergillus section Flavi in soils and maize samples from control and treated plots across three agroecological zones (AEZs) in Ghana.
Frequencies of A. flavus L morphotype in treated soils at harvest ranged from 97.2% in HF to 100% in DS. Across AEZs, in control plots, relatively lower L morphotype frequencies were detected in soil at harvest compared to soil before application. Significantly (P < 0.05) higher L morphotype frequencies were observed across treated plots in both DS and HF. Generally, incidences of SBG strains, A. parasiticus, and A. tamarii were lower in treated soils at harvest, compared to soil before application of experimental products. At harvest in DS, the proportions of SBG strains were significantly (P < 0.05) higher in untreated soils than in treated soils (Table 5). Aspergillus communities in treated maize kernels across all three AEZs were entirely composed of the L morphotype. In control maize, the L morphotype dominated and minor frequencies of SBG strains (up to 1.4%) were found (Table 5). A similar trend in frequencies of L morphotype, SBG strains, A. parasiticus, and A. tamarii was observed in soils from groundnut fields and groundnut kernels, except that communities in treated groundnut, in addition to the L morphotype, harbored minor proportions of SBG strains (up to 0.7%) (Table 6).
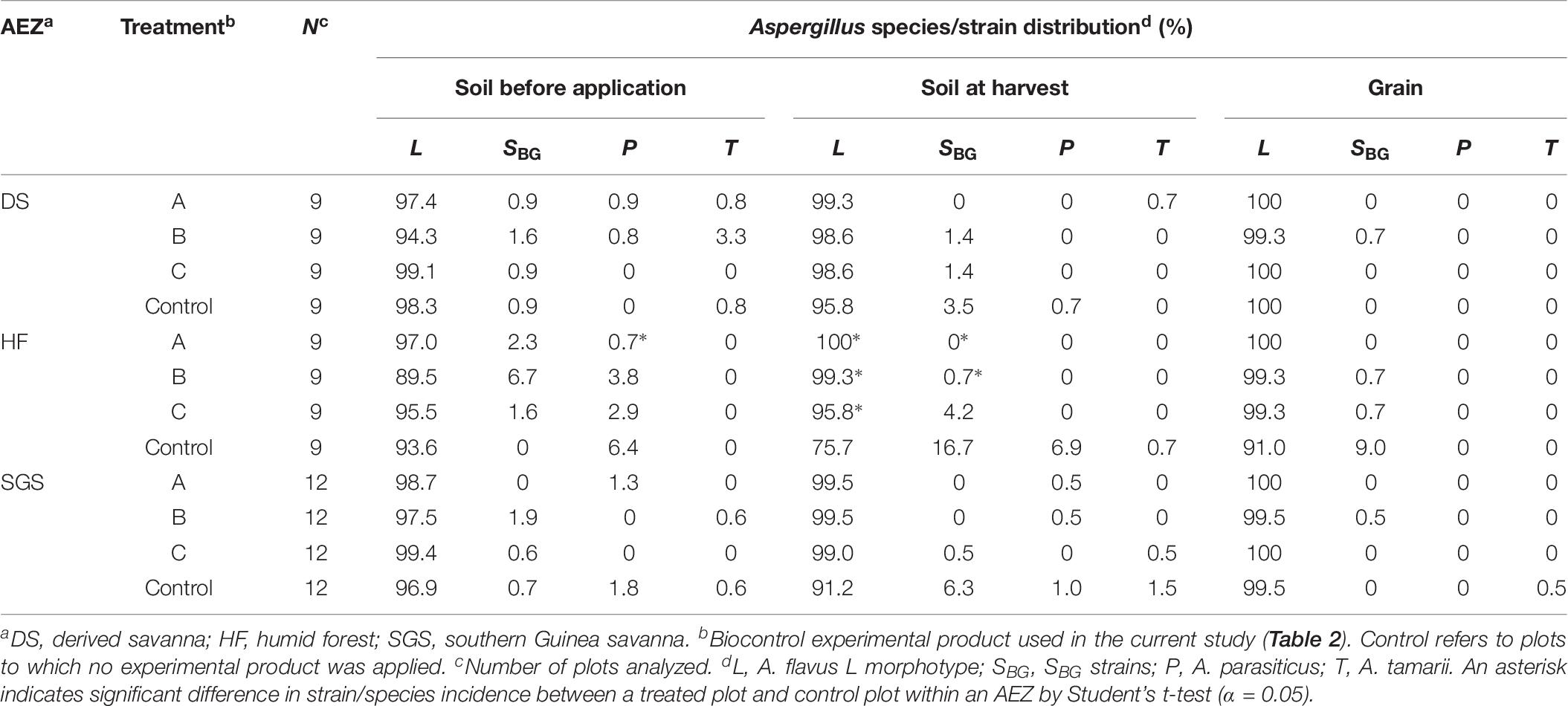
Table 6. Community structure of Aspergillus section Flavi in soils and groundnut samples from treated and control plots across three agroecological zones (AEZs) in Ghana.
Incidence of Applied Atoxigenic AAVs in Grains After Treatment
The individual atoxigenic AAVs composing the applied experimental products showed varying abilities to disperse from treated soils and establish in the grain of treated and control plots. Each AAV was assigned a rank based on their incidence across AEZ, regions, districts, samples, and number of AAV individuals detected. For instance, 75 isolates belonging to AAV GHM287-10 were recovered from 17 maize samples from 8 out of 10 districts in all five regions across all three AEZs, thus being the most dominant applied AAV in treated grains (rank = 1, Table 7). The same AAV was also frequently isolated from control grains (rank = 2, in control grains). Barring a few exceptions (e.g., AAV GHM511-3), most AAVs with high post-release incidence in grains from treated plots also had relatively high incidence in control plots. In contrast, AAV GHM173-6 was the least frequently isolated from grains of both treated and control plots (Table 7).
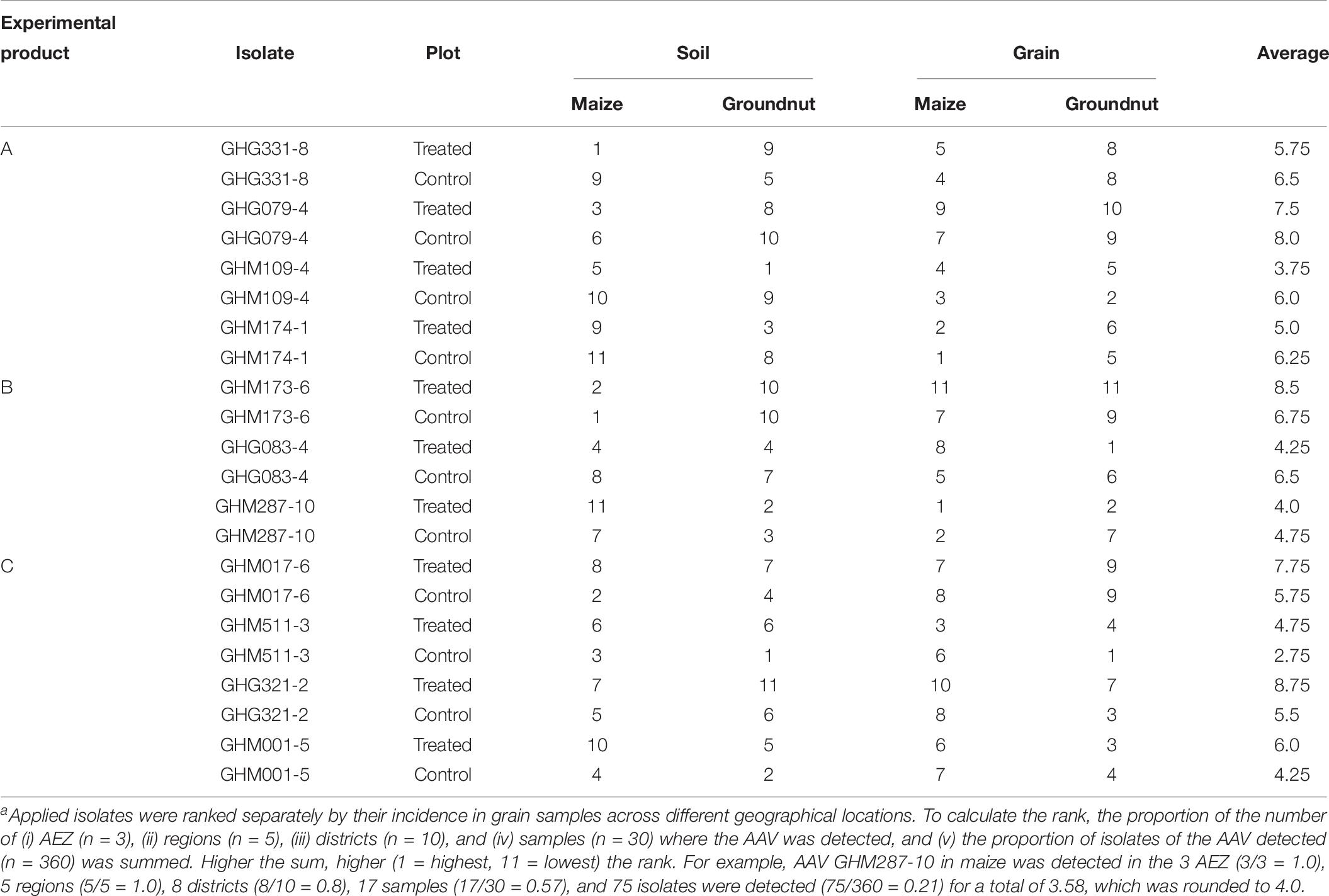
Table 7. Rankingsa of isolates belonging to atoxigenic African Aspergillus flavus vegetative compatibility groups (AAVs) in soils and grain from both maize and groundnut plots treated with three experimental products and their corresponding controls in three agroecological zones (AEZs) in Ghana.
Abilities of the applied AAVs to move into groundnut kernels also varied. Generally, incidence of applied AAVs was relatively lower in groundnut than in maize (Table 8). The most prevalent applied AAV was GHG083-4 with 52 member isolates found in 15 samples from all 10 districts in all the regions of the three AEZs. On the other hand, no isolate of AAV GHM173-6 was recovered in groundnut from any field (Table 7).
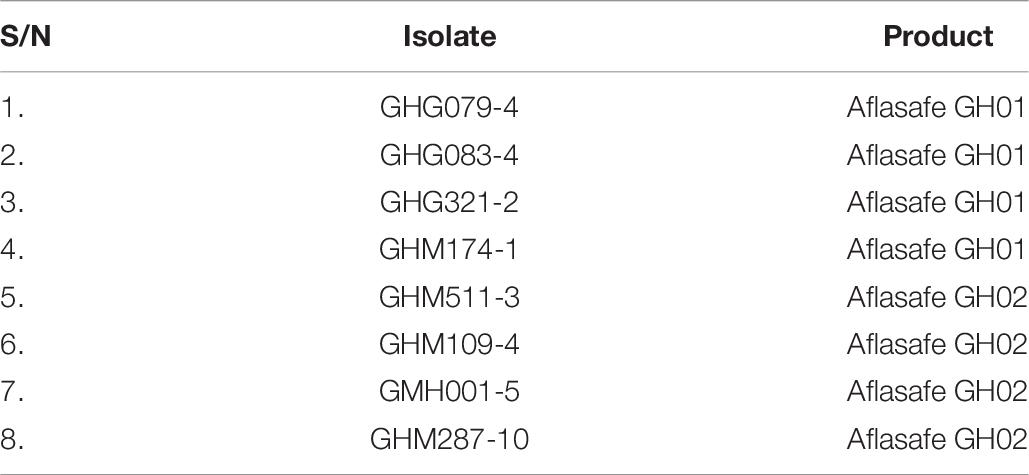
Table 8. Selected isolates belonging to atoxigenic African Aspergillus flavus vegetative compatibility groups composing two biocontrol products for further evaluation in Ghana.
There were some AAVs with high ranking positions in both crops. For example GHM287-10 was the 1st and 2nd ranked AAV in treated maize and groundnut, respectively (Table 7). However, also in treated grains, GHG083-4 was the 1st and 8th ranked AAV in groundnut and maize, respectively. Success of establishment of an AAV in one crop was not always associated with success in the other crop.
Selected Isolates of Atoxigenic AAVs for Aflatoxin Biocontrol in Ghana
Based on incidence of the candidate AAVs in maize and groundnut grains following their release across locations, regions, and AEZs, and SSR data (Table 2), one representative atoxigenic isolate of eight AAVs with widest distribution and with superior ability to reduce aflatoxin contamination in grains were selected as active ingredients of two biocontrol products (Table 8).
Discussion
In the current study, 12 atoxigenic AAVs native to Ghana were identified and a representative isolate of each AAV were evaluated for their potential as biocontrol agents for aflatoxin mitigation of both maize and groundnut grown across various AEZs. The 12 evaluated isolates successfully inhibited aflatoxin production (range = 87.3–98.7% less) when co-inoculated with a potent aflatoxin-producing A. flavus isolate native to Ghana in laboratory tests. Aflatoxin reduction levels were comparable to those detected in elite atoxigenic A. flavus isolates endemic to the United States (Cotty and Bayman, 1993; Ortega-Beltran et al., 2019), Nigeria (Atehnkeng et al., 2008), Kenya (Probst et al., 2011), Italy (Mauro et al., 2015), and China (Zhou et al., 2015). In sub-Saharan Africa, specifically Nigeria, Kenya, Senegal, The Gambia, and Burkina Faso, similar evaluations resulted in identification and selection of unique AAVs for the development of atoxigenic products tailored to each country (Atehnkeng et al., 2008; Probst et al., 2011; Bandyopadhyay et al., 2016). To our knowledge, the current work is the first published study of selection of active ingredients of an aflatoxin biocontrol product supported by information on their ability to disperse to crops from a formulated product applied on soil.
For over two decades, atoxigenic aflatoxin biocontrol has been demonstrated as the most effective and sustainable strategy to reduce crop aflatoxin content (Cotty, 1994; Dorner, 2004, 2010; Cotty et al., 2007; Mehl et al., 2012; Atehnkeng et al., 2014; Doster et al., 2014; Grubisha and Cotty, 2015). This strategy is based on the deployment of native atoxigenic isolates of VCGs that both competitively displace aflatoxin-producers and inhibit aflatoxin biosynthesis. Isolates belonging to atoxigenic VCGs locally adapted to specific AEZs and cropping systems, and with superior competitive ability to exclude aflatoxin producers from the target crop or environment are used in aflatoxin management programs (Cotty et al., 2007; Dorner, 2010; Abbas et al., 2011; Mehl et al., 2012; Doster et al., 2014; Bandyopadhyay et al., 2016). In keeping with this paradigm, 12 isolates belonging to genetically diverse SSR haplotypes/AAVs with wide distribution across Ghana (Islam et al., 2015) were identified from 847 atoxigenic isolates described previously (Agbetiameh et al., 2018) using 17 SSR loci (Grubisha and Cotty, 2009).
Mehl et al. (2012) emphasized that VCG analyses provide insights into the diversity of fungal communities including aflatoxin production and inhibition potentials. Indeed, variation in aflatoxin inhibition among representative isolates of the 12 atoxigenic AAVs was expected. GHG183-7 was least effective at inhibiting aflatoxin contamination in laboratory assays. This suggests that GHG183-7 is a poor competitor during host colonization (Mehl and Cotty, 2010) compared to the other evaluated isolates. Atehnkeng et al. (2008) emphasized that reduced competitiveness in laboratory conditions may provide an early signal of low competitiveness during crop development and, subsequently, less efficacy in practice. Furthermore, Atehnkeng et al. (2008) suggested exclusion of atoxigenic isolates with considerably less competitive abilities prior to expensive, time consuming field studies. Apart from being the least competitive isolate, we were unable to obtain a complementary pair of nit auxotrophs for this isolate. Whether this isolate is self-incompatible as reported in studies of Aspergillus and other genera (Correll et al., 1987; Krnjaja et al., 2013) needs to be clarified. Consequently, frequencies of AAV GHG183-7 were not evaluated even though an isolate of that AAV was a constituent of experimental product B.
Use of native AAVs in biocontrol programs offers better adaptation to target agroecosystems (Probst et al., 2011) and long-term establishment of A. flavus communities with low aflatoxin-producing potential (Mehl et al., 2012). Genetic variability among A. flavus individuals results in differential adaptation to various agroecological niches (Cotty and Mellon, 2006; Mehl and Cotty, 2013). Indeed, this phenomenon was expected among the 12 atoxigenic isolates evaluated in the current study. Studies of adaptive potentials of these isolates across three AEZs suggest extents of adaptation of their corresponding AAVs to the conditions of the three evaluated AEZs. For instance, the atoxigenic isolate GHM173-6 was the most effective at reducing aflatoxin concentrations in laboratory assays (Table 3) and was also one of the isolates most commonly found in treated and untreated maize soil (Table 7 and Supplementary Table 1). This notwithstanding, GHM173-6 was the least encountered in maize grain from all field locations across regions and was never recovered from groundnut (Table 7 and Supplementary Tables 2–4). On the contrary, GHM511-3 exhibited both high aflatoxin inhibition potential and high recovery on both maize and groundnut across regions and all three AEZs (Tables 3, 7). These observations support both competitiveness and crop adaptation as important criteria for selection of active ingredient AAVs for biocontrol formulations.
A major objective of the field evaluations of multiple isolates was to detect atoxigenic isolates belonging to AAVs with superior ability to establish in the crop after introduction in formulated product on the soil (Table 7). Apart from aflatoxin reduction of the experimental products, this portion of the research allowed identification of AAVs with greatest abilities to compete in the presence of both other atoxigenic isolates and aflatoxin producers under field conditions. Similarly in Nigeria, one of the four constituent AAVs of the initial experimental product established poorly in field evaluations (Atehnkeng et al., 2014) and hence was not included as an active ingredient of the final multi-AAV biocontrol product Aflasafe®.
Bandyopadhyay et al. (2016) underscored the importance of distribution and incidence of AAVs with potential as aflatoxin biocontrol agents as proxies for adaptation, competitiveness, and fitness in target environments. However, superior adaptation should also reflect increased efficacy in the target crop (Mauro et al., 2015). We report substantial reductions in aflatoxin concentrations in both groundnut (70–100% less) and maize (50–100% less) from plots treated with mixtures of atoxigenic isolates belonging to genetically diverse AAVs across all three AEZs. Lower than expected aflatoxin levels were also detected in maize from control plots across AEZs and may reflect the effect of drift of atoxigenic fungi from treated plots to adjacent control plots due to the relatively short separation distance (5 m). Indeed, most AAVs of the applied isolates were detected in control crops (Table 7). Conidia of A. flavus are common constituents of air currents dispersed over short and long distances (Bennett, 2010). Thus, a distance of at least 500 m between a treated and a control plot is necessary to avoid inter-plot interference (Bock et al., 2004; Atehnkeng et al., 2014).
Atehnkeng et al. (2014) demonstrated that mixtures of atoxigenic isolates are effective at reducing aflatoxin contamination in maize. Our results suggest that atoxigenic isolates mixtures belonging to distinct AAVs can be strategically designed for aflatoxin reduction in both maize and groundnut cropping systems in Ghana. Eight atoxigenic isolates belonging to atoxigenic AAVs were selected as active ingredients of two biocontrol products for aflatoxin mitigation and subsequently registered with Ghana’s Environmental Protection Agency (Table 8). Six of the eight selected isolates had total or partial deletions in the cyclopiazonic acid (CPA) gene cluster while two produced undetectable amount of CPA (unpublished data). For the selection of the active ingredient AAVs, we considered their frequency of occurrence (Table 1), the competitive potential against aflatoxin producers (Table 3) and the relative adaptation in the evaluated maize and groundnut treated and control soils and crops (Table 7). This systematic evaluation protocol offered the opportunity to select the best possible combinations of active ingredients among the evaluated AAVs. However, all experimental products evaluated in the current study were efficient in reducing aflatoxin contamination of both maize and groundnut and each of the 12 AAVs were able to disperse to and increase frequency on the target crops. The selection strategy provides a basis for use of the most detected AAVs. However, even use of the most poorly adapted isolates examined here would provide better crop protection and increased food safety than failure to use atoxigenic strain-based biocontrol.
Application of atoxigenic A. flavus isolates on a target crop is a deliberate action to reshape fungal community composition in favor of the applied atoxigenic isolates due to founder events and competitive exclusion resulting in displacement of aflatoxin producers (Cotty and Bayman, 1993; Cotty et al., 2007; Mehl et al., 2012). Effective displacement of resident aflatoxin producers is achieved through proper timing of biocontrol applications during critical crop developmental stages (2–3 weeks before crop flowering) prior to the natural increase of the local Aspergillus population (Bandyopadhyay et al., 2016). Timed applications offer atoxigenic genotypes the advantage of becoming the founding population (Cotty and Mellon, 2006; Cotty et al., 2007) to quickly multiply and disperse to other nutrient sources and the target crop so that aflatoxin producers become less frequent (Bandyopadhyay et al., 2016).
In the current study, substantial displacement of aflatoxin producers from soils and crops occurred in treated plots across all three AEZs. The displacement was observed also in the non-treated crops. The A. flavus L morphotype largely dominated communities of Aspergillus section Flavi in soils collected before treatment, soils at harvest, and grains from both treated and control plots. The L morphotype is recognized as the most successful colonizer of soil and other substrates including grains in similar studies (Alanis Zanon et al., 2013; Atehnkeng et al., 2014; Doster et al., 2014). Frequencies of A. parasiticus were low (<1%), as reported previously in Ghana (Agbetiameh et al., 2018). Factors leading to low frequencies of this species in groundnut in West Africa remain unknown. Similarly, A. parasiticus is not common in portions of the Middle East (Lisker et al., 1993). In other regions of Southern Africa and North America, A. parasiticus is an important causal agent of groundnut aflatoxin contamination (Horn and Dorner, 1998; Kachapulula et al., 2017).
Conclusion
Twelve atoxigenic African A. flavus vegetative compatibility groups (AAVs) commonly occurring across Ghana were characterized. The potential of a representative member of each AAV to inhibit aflatoxin contamination of maize grains was assessed in laboratory assays. AAV adaptation in maize and groundnut cropping systems in three AEZs in Ghana was assessed. The results formed the basis for selection of eight superior atoxigenic A. flavus isolates, each belonging to an unique AAV, as active ingredients of two biocontrol products, Aflasafe GH01 and Aflasafe GH02, for use on maize and groundnut in Ghana (Table 8). The unique SSR patterns of the eight atoxigenic isolates (Table 2) can serve as a resource for identification of the active ingredients of each of Aflasafe GH01 and Aflasafe GH02 after field application. Use of the identified atoxigenic AAVs offers a sustainable management option for aflatoxin mitigation in maize and groundnut for smallholder farmers in Ghana providing an inexpensive opportunity for improved food safety, productivity, and income.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
Author Contributions
DA, JA, PC, and RB contributed to the conception and design of the experiments. DA and JA conducted the experiments and field studies, and collected and analyzed the data. AO-B, RA, PC, and RB provided the guidance. KC, M-SI, and PC conducted the molecular studies. DA, AO-B, KC, PC, and RB drafted the manuscript. All authors read, reviewed, and approved the final version of the manuscript. RB and AO-B secured funds for the study.
Funding
This study was funded by the Bill & Melinda Gates Foundation (OPP1007117), the USAID-Africa RISING, CGIAR Research Program on Agriculture for Nutrition and Health (A4NH) and the European Union MycoKey program in partnership with the International Institute of Tropical Agriculture (IITA), the Kwame Nkrumah University of Science and Technology (KNUST), and the United States Department of Agriculture – Agricultural Research Service (USDA-ARS).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We specially thank the maize and groundnut farmers in Ghana for willingly allowing the trials to be conducted in their fields and for participating in this study. We also thank the staff of Ghana’s MoFA for assisting in field selection, experimental product application and monitoring, and sample collection. The use of trade, firm, or corporation names in these methods is for the information and convenience of the reader. Such use does not constitute an official endorsement or approval by the USDA-ARS, of any product or service to the exclusion of others that may be suitable. In addition, USDA-ARS makes no warranties as to the merchantability or fitness of the methodologies described on these pages for any particular purpose, or any other warranties expressed or implied. These methodologies provide a guide and do not replace published work. USDA-ARS is not liable for any damages resulting from the use or misuse of these methodologies.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02069/full#supplementary-material
References
Abbas, H. K., Weaver, M. A., Horn, B. W., Carbone, I., Monacell, J. T., and Shier, W. T. (2011). Selection of Aspergillus flavus isolates for biological control of aflatoxins in corn. Toxin Rev. 30, 59–70.
Adhikari, B. N., Bandyopadhyay, R., and Cotty, P. J. (2016). Degeneration of aflatoxin gene clusters in Aspergillus flavus from Africa and North America. AMB Express 6:62. doi: 10.1186/s13568-016-0228-6
Afum, C., Cudjoe, L., Hills, J., Hunt, R., Padilla, L. A., Elmore, S., et al. (2016). Association between aflatoxin M1 and liver disease in HBV/HCV infected persons in Ghana. Int. J. Environ. Res. Public Health 13:377. doi: 10.3390/ijerph13040377
Agbetiameh, D., Ortega-Beltran, A., Awuah, R. T., Atehnkeng, J., Cotty, P. J., and Bandyopadhyay, R. (2018). Prevalence of aflatoxin contamination in maize and groundnut in Ghana: population structure, distribution, and toxigenicity of the causal agents. Plant Dis. 102, 764–772. doi: 10.1094/PDIS-05-17-0749-RE
Alanis Zanon, M. S., Barros, G. G., and Chulze, S. N. (2016). Non-aflatoxigenic Aspergillus flavus as potential biocontrol agents to reduce aflatoxin contamination in peanuts harvested in Northern Argentina. Int. J. Food Microbiol. 231, 63–68. doi: 10.1016/j.ijfoodmicro.2016.05.016
Alanis Zanon, M. S., Chiotta, M., Giaj-Merlera, G., Barros, G. G., and Chulze, S. N. (2013). Evaluation of potential biocontrol agent for aflatoxin in Argentinean peanuts. Int. J. Food Microbiol. 162, 220–225. doi: 10.1016/j.ijfoodmicro.2013.01.017
Atehnkeng, J., Donner, M., Ojiambo, P. S., Ikotun, B., Augusto, J., Cotty, P. J., et al. (2016). Environmental distribution and genetic diversity of vegetative compatibility groups determine biocontrol strategies to mitigate aflatoxin contamination of maize by Aspergillus flavus. Microb. Biotechnol. 9, 75–88. doi: 10.1111/1751-7915.12324
Atehnkeng, J., Ojiambo, P. S., Cotty, P. J., and Bandyopadhyay, R. (2014). Field efficacy of a mixture of atoxigenic Aspergillus flavus Link:Fr vegetative compatibility groups in preventing aflatoxin contamination in maize (Zea mays L.). Biol. Control 72, 62–70. doi: 10.1016/j.biocontrol.2014.02.009
Atehnkeng, J., Ojiambo, P. S., Donner, M., Ikotun, T., Sikora, R. A., Cotty, P. J., et al. (2008). Distribution and toxigenicity of Aspergillus species isolated from maize kernels from three agro-ecological zones in Nigeria. Int. J. Food Microbiol. 122, 74–84. doi: 10.1016/j.ijfoodmicro.2007.11.062
Bandyopadhyay, R., Ortega-Beltran, A., Akande, A., Mutegi, C., Atehnkeng, J., Kaptoge, L., et al. (2016). Biological control of aflatoxins in Africa: current status and potential challenges in the face of climate change. World Mycotoxin J. 9, 771–789. doi: 10.3920/wmj2016.2130
Bayman, P., and Cotty, P. J. (1991). Vegetative compatibility and genetic diversity in the Aspergillus flavus population of a single field. Can. J. Bot. 69, 1707–1711.
Bennett, J. W. (2010). “An overview of the genus Aspergillus,” in Aspergillus molecular biology and genomics, eds M. Machida, and K. Gomi, (Norfolk: Caister Academic Press), 1–17.
Bock, C. H., Mackey, B., and Cotty, P. J. (2004). Population dynamics of Aspergillus flavus in the air of an intensively cultivated region of south-west Arizona. Plant Pathol. 53, 422–433. doi: 10.1111/j.0032-0862.2004.01015.x
Callicott, K. A., and Cotty, P. J. (2015). Method for monitoring deletions in the aflatoxin biosynthesis gene cluster of Aspergillus flavus with multiplex PCR. Lett. Appl. Microbiol. 60, 60–65. doi: 10.1111/lam.12337
Cardwell, K. F., and Cotty, P. J. (2002). Distribution of Aspergillus section Flavi among field soils from four agroecological zones of the Republic of Bénin, West Africa. Plant Dis. 86, 434–439. doi: 10.1094/pdis.2002.86.4.434
Cheli, F., Battaglia, D., Gallo, R., and Dell’Orto, V. (2014). EU legislation on cereal safety: an update with a focus on mycotoxins. Food Control 37, 315–325. doi: 10.1016/j.foodcont.2013.09.059
Cole, R. J., and Dorner, J. W. (1993). Extraction of aflatoxins from naturally contaminated peanuts with different solvents and solvent/peanut ratios. J. AOAC Int. 77, 1509–1511.
Correll, J. C., Klittich, C. J. R., and Leslie, J. F. (1987). Nitrate nonutilizing mutants of Fusarium oxysporum and their use in vegetative compatibility tests. Phytopathology 77, 1640–1646.
Cotty, P. J. (1989). Virulence and cultural characteristics of two Aspergillus flavus strains pathogenic on cotton. Phytopathology 79, 808–814.
Cotty, P. J. (1994). Influence of field application of an atoxigenic strain of Aspergillus flavus on the populations of A. flavus infecting cotton bolls and on the aflatoxin content of cottonseed. Phytopathology 84, 1270–1277.
Cotty, P. J., Antilla, L., and Wakelyn, P. J. (2007). “Competitive exclusion of aflatoxin producers: Farmer-driven research and development,” in Biological Control: A Global Perspective, eds C. Vincent, M. S. Goettel, and G. Lazarovits, (Wallingford: CAB International), 241–253. doi: 10.1079/9781845932657.0241
Cotty, P. J., and Bayman, P. (1993). Competitive exclusion of a toxigenic strain of Aspergillus flavus by an atoxigenic strain. Phytopathology 83, 1283–1287.
Cotty, P. J., and Cardwell, K. F. (1999). Divergence of West African and North American communities of Aspergillus section Flavi. Appl. Environ. Microbiol. 65, 2264–2266.
Cotty, P. J., and Jaime-Garcia, R. (2007). Influences of climate on aflatoxin producing fungi and aflatoxin contamination. Int. J. Food Microbiol. 119, 109–115. doi: 10.1016/j.ijfoodmicro.2007.07.060
Cotty, P. J., and Mellon, J. E. (2006). Ecology of aflatoxin producing fungi and biocontrol of aflatoxin contamination. Mycotoxin Res. 22, 110–117. doi: 10.1007/BF02956774
Cotty, P. J., and Taylor, D. R. (2003). Influence of complementation medium composition on vegetative compatibility analyses of Aspergillus flavus. Phytopathology 93:S18.
Cove, D. J. (1976). Chlorate toxicity in Aspergillus nidulans: the selection and characterisation of chlorate resistant mutants. Heredity 36, 191–203. doi: 10.1038/hdy.1976.24
Danso, J. K., Osekre, E. A., Opit, G. P., Arthur, F. H., Campbell, J. F., Mbata, G., et al. (2019). Impact of storage structures on moisture content, insect pests and mycotoxin levels of maize in Ghana. J. Stored Prod. Res. 81, 114–120. doi: 10.1016/j.jspr.2018.11.012
Donner, M., Atehnkeng, J., Sikora, R. A., Bandyopadhyay, R., and Cotty, P. J. (2009). Distribution of Aspergillus section Flavi in soils of maize fields in three agroecological zones of Nigeria. Soil Biol. Biochem. 41, 37–44. doi: 10.1016/j.soilbio.2008.09.013
Dorner, J. W. (2004). Biological control of aflatoxin contamination of crops. J. Toxicol. Toxin Rev. 23, 425–450. doi: 10.1081/txr-200027877
Dorner, J. W. (2010). Efficacy of a biopesticide for control of aflatoxins in corn. J. Food Prot. 73, 495–499. doi: 10.4315/0362-028x-73.3.495
Doster, M. A., Cotty, P. J., and Michailides, T. J. (2014). Evaluation of the atoxigenic Aspergillus flavus strain AF36 in pistachio orchards. Plant Dis. 98, 948–956. doi: 10.1094/PDIS-10-13-1053-RE
Dzirasah, D. (2015). EU threatens ban over quality of Ghana’s cereals. The Ghanaian Times, Thursday, May 14, 2015.
Florkowski, W. J., and Kolavalli, S. (2013). Aflatoxin Control Strategies in the Groundnut Value Chain in Ghana. Ghana Strategy Support Program. Working Paper 33. Washington, DC: International Food Policy Research Institute.
Frisvad, J. C., Hubka, V., Ezekiel, C. N., Hong, S. B., Nováková, A., Chen, A. J., et al. (2019). Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Stud. Mycol. 93, 1–63. doi: 10.1016/j.simyco.2018.06.001
Grenier, B., Loureiro-Bracarense, A.-P., Leslie, J. F., and Oswald, I. P. (2014). “Physical and chemical methods for mycotoxin decontamination in maize,” in Mycotoxin Reduction in Grain Chains, eds J. F. Leslie, and A. F. Logrieco, (Oxford: John Wiley & Sons, Inc.), 116–129. doi: 10.1002/9781118832790.ch9
Grubisha, L. C., and Cotty, P. J. (2009). Twenty-four microsatellite markers for the aflatoxin-producing fungus Aspergillus flavus. Mol. Ecol. Resour. 9, 264–267. doi: 10.1111/j.1755-0998.2008.02378.x
Grubisha, L. C., and Cotty, P. J. (2010). Genetic isolation among sympatric vegetative compatibility groups of the aflatoxin-producing fungus Aspergillus flavus. Mol. Ecol. 19, 269–280. doi: 10.1111/j.1365-294X.2009.04467.x
Grubisha, L. C., and Cotty, P. J. (2015). Genetic analysis of the Aspergillus flavus vegetative compatibility group to which a biological control agent that limits aflatoxin contamination in US crops belongs. Appl. Environ. Microbiol. 81, 5889–5899. doi: 10.1128/aem.00738-15
Horn, B. W., and Dorner, J. W. (1998). Soil populations of Aspergillus species from section Flavi along a transect through peanut-growing regions of the United States. Mycologia 90, 767–776. doi: 10.1080/00275514.1998.12026969
Huson, D. H., and Bryant, D. (2005). Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267. doi: 10.1093/molbev/msj030
Islam, M.-S., Callicott, K. A., Atehnkeng, J., Augusto, J., Bonkoungou, S., Agbetiameh, D., et al. (2015). Genetic diversity of L-strain isolates of Aspergillus flavus of potential use for aflatoxin biocontrol in sub-Saharan Africa. Phytopathology 105(Suppl. 4), S4.63.
Islam, M. S., Callicott, K. A., Mutegi, C., Bandyopadhyay, R., and Cotty, P. J. (2018). Aspergillus flavus resident in Kenya: high genetic diversity in an ancient population primarily shaped by clonal reproduction and mutation-driven evolution. Fungal Ecol. 35, 20–33. doi: 10.1016/j.funeco.2018.05.012
JECFA, (2018). Safety Evaluation of Certain Contaminants in Food: Prepared by the Eighty-Third Meeting of the Joint FAO/WHO Expert Committee on Food Additives (JECFA). Geneva: World Health Organization and Food and Agriculture Organization of the United Nations.
Jolly, P., Inusah, S., Lu, B., Ellis, W., Nyarko, A., Phillips, T., et al. (2013). Association between high aflatoxin B1 levels and high viral load in HIV-positive people. World Mycotoxin J. 6, 255–261. doi: 10.1080/19440049.2011.581698
Kachapulula, P. W., Akello, J., Bandyopadhyay, R., and Cotty, P. J. (2017). Aspergillus section Flavi community structure in Zambia influences aflatoxin contamination of maize and groundnut. Int. J. Food Microbiol. 261, 49–56. doi: 10.1016/j.ijfoodmicro.2017.08.014
Klich, M. A. (2007). Aspergillus flavus: the major producer of aflatoxin. Mol. Plant Pathol. 8, 713–722. doi: 10.1111/j.1364-3703.2007.00436.x
Kraemer, K., Cordaro, J., Fanzo, J., Gibney, M., Kennedy, E., Labrique, A., et al. (2016). “The critical role of food safety in ensuring food security,” in Good Nutrition: Perspectives for the 21st Century, eds M. Eggersdorfer, K. Kraemer, J. Cordaro, J. Fanzo, M. Gibney, E. Kennedy, et al. (Basel: Karger Publishers), 312–325.
Krnjaja, V., Lević, J., Stanković, S., and Vasić, T. (2013). The use of vegetative compatibility tests for identification of biodiversity of phytopathogenic fungi. Pestic. Phytomed. 28, 157–165. doi: 10.2298/pif1303157k
Kumi, J., Dotse, E., Asare, G. A., and Ankrah, N.-A. (2015). Urinary aflatoxin M1 exposure in Ghanaian children weaned on locally prepared nutritional food. Afr. J. Sci. Res. 4, 28–32.
Lamplugh, S. M., Hendrickse, R. G., Apeagyei, F., and Mwanmut, D. D. (1988). Aflatoxins in breast milk, neonatal cord blood, and serum of pregnant women. Bri. Med. J. 296:968. doi: 10.1136/bmj.296.6627.968
Leslie, J. F. (1993). Fungal vegetative compatibility. Annu. Rev. Phytopathol. 31, 127–150. doi: 10.1146/annurev.py.31.090193.001015
Lisker, N., Michaeli, R., and Frank, Z. R. (1993). Mycotoxigenic potential of Aspergillus flavus strains isolated from groundnuts growing in Israel. Mycopathologia 122, 177–183. doi: 10.1007/bf01103479
Mahuku, G., Nzioki, H. S., Mutegi, C., Kanampiu, F., Narrod, C., and Makumbi, D. (2019). Pre-harvest management is a critical practice for minimizing aflatoxin contamination of maize. Food Cont. 96, 219–226. doi: 10.1016/j.foodcont.2018.08.032
Mauro, A., Battilani, P., and Cotty, P. J. (2015). Atoxigenic Aspergillus flavus endemic to Italy for biocontrol of aflatoxins in maize. BioControl 60, 125–134. doi: 10.1007/s10526-014-9624-5
Mauro, A., Garcia-Cela, E., Pietri, A., Cotty, P. J., and Battilani, P. (2018). Biological control products for aflatoxin prevention in Italy: commercial field evaluation of atoxigenic Aspergillus flavus active ingredients. Toxins 10, 1–14.
Mehl, H. L., and Cotty, P. J. (2010). Variation in competitive ability among isolates of Aspergillus flavus from different vegetative compatibility groups during maize infection. Phytopathology 100, 150–159. doi: 10.1094/PHYTO-100-2-0150
Mehl, H. L., and Cotty, P. J. (2013). Influence of plant host species on intraspecific competition during infection by Aspergillus flavus. Plant Pathol. 62, 1310–1318. doi: 10.1111/ppa.12038
Mehl, H. L., Jaime, R., Callicott, K. A., Probst, C., Garber, N. P., Ortega-Beltran, A., et al. (2012). Aspergillus flavus diversity on crops and in the environment can be exploited to reduce aflatoxin exposure and improve health. Ann. N.Y. Acad. Sci. 1273, 7–17. doi: 10.1111/j.1749-6632.2012.06800.x
Meirmans, P. G., and Van Tienderen, P. H. (2004). GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Mol. Ecol. Notes 4, 792–794. doi: 10.1111/j.1471-8286.2004.00770.x
MoFA. (2011). Agriculture in Ghana. Facts and Figures (2010). Accra: Statistics, Research and Information Directorate (SRID).
Ortega-Beltran, A., and Bandyopadhyay, R. (2019). Comments on “Trial summary on the comparison of various non-aflatoxigenic strains of Aspergillus flavus on mycotoxin levels and yield in maize” by M.S. Molo, et al. Agron. J. 111, 942–946. doi: 10.2134/agronj2019.04.0281
Ortega-Beltran, A., Moral, J., Picot, A., Puckett, R. D., Cotty, P. J., and Michailides, T. J. (2019). Atoxigenic Aspergillus flavus isolates endemic to almond, fig, and pistachio orchards in California with potential to reduce aflatoxin contamination in these crops. Plant Dis. 103, 905–912. doi: 10.1094/PDIS-08-18-1333-RE
Paudyal, S., Opit, G. P., Osekre, E. A., Arthur, F. H., Bingham, G. V., Payton, M. E., et al. (2017). Field evaluation of the long-lasting treated storage bag, deltamethrin incorporated, (ZeroFly® Storage Bag) as a barrier to insect pest infestation. J. Stored Prod. Res. 70, 44–52. doi: 10.1016/j.jspr.2016.11.003
Pildain, M. B., Frisvad, J. C., Vaamonde, G., Cabral, D., Varga, J., and Samson, R. A. (2008). Two novel aflatoxin-producing Aspergillus species from Argentinean peanuts. Int. J. Syst. Evol. Microbiol. 58, 725–735. doi: 10.1099/ijs.0.65123-0
Probst, C., Bandyopadhyay, R., and Cotty, P. J. (2014). Diversity of aflatoxin-producing fungi and their impact on food safety in sub-Saharan Africa. Int. J. Food Microbiol. 174, 113–122. doi: 10.1016/j.ijfoodmicro.2013.12.010
Probst, C., Bandyopadhyay, R., Price, L. E., and Cotty, P. J. (2011). Identification of atoxigenic Aspergillus flavus isolates to reduce aflatoxin contamination of maize in Kenya. Plant Dis. 95, 212–218. doi: 10.1094/PDIS-06-10-0438
Probst, C., and Cotty, P. J. (2012). Relationships between in vivo and in vitro aflatoxin production: reliable prediction of fungal ability to contaminate maize with aflatoxins. Fungal Biol. 116, 503–510. doi: 10.1016/j.funbio.2012.02.001
Shuaib, F., Jolly, P. E., Ehiri, J. E., Yatich, N., Jiang, Y., Funkhouser, E., et al. (2010). Association between birth outcomes and aflatoxin B1 biomarker blood levels in pregnant women in Kumasi, Ghana. Trop. Med. Int. Health 15, 160–167. doi: 10.1111/j.1365-3156.2009.02435.x
Singh, P., and Cotty, P. J. (2019). Characterization of Aspergilli from dried red chilies (Capsicum spp.): insights into the etiology of aflatoxin contamination. Int. J. Food Microbiol. 289, 145–153. doi: 10.1016/j.ijfoodmicro.2018.08.025
Tran-Dinh, N., Pitt, J. I., and Markwell, P. (2014). Selection of non-toxigenic strains of Aspergillus flavus for biocontrol of aflatoxins in maize in Thailand. Biocontrol Sci. Tech. 24, 652–661. doi: 10.1080/09583157.2014.888398
UNICEF, (2017). Levels and Trends in Child Malnutrition: UNICEF/WHO/World Bank Group Joint Child Malnutrition Estimates. Key Findings of the 2017 edition. Global Database on Child Growth and Malnutrition. Available: http://www.who.int/nutgrowthdb/estimates2016/en/ (accessed April 4, 2017).
Wu, F. (2015). Global impacts of aflatoxin in maize: trade and human health. World Mycotoxin J. 8, 137–142. doi: 10.3920/wmj2014.1737
Wu, F., and Khlangwiset, P. (2010). Health economic impacts and cost-effectiveness of aflatoxin-reduction strategies in Africa: case studies in biocontrol and post-harvest interventions. Food Addit. Contam. 27, 496–509. doi: 10.1080/19440040903437865
Keywords: aflatoxin, biocontrol, strain selection, efficacy trials, safer food
Citation: Agbetiameh D, Ortega-Beltran A, Awuah RT, Atehnkeng J, Islam M-S, Callicott KA, Cotty PJ and Bandyopadhyay R (2019) Potential of Atoxigenic Aspergillus flavus Vegetative Compatibility Groups Associated With Maize and Groundnut in Ghana as Biocontrol Agents for Aflatoxin Management. Front. Microbiol. 10:2069. doi: 10.3389/fmicb.2019.02069
Received: 03 July 2019; Accepted: 22 August 2019;
Published: 06 September 2019.
Edited by:
Federica Giacometti, University of Bologna, ItalyReviewed by:
Paolo Bonilauri, Experimental Zooprophylactic Institute of Lombardy and Emilia Romagna (IZSLER), ItalyCarol Verheecke-Vaessen, Cranfield University, United Kingdom
Copyright © 2019 Agbetiameh, Ortega-Beltran, Awuah, Atehnkeng, Islam, Callicott, Cotty and Bandyopadhyay. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ranajit Bandyopadhyay, ci5iYW5keW9wYWRoeWF5QGNnaWFyLm9yZw==
†Present address: Md-Sajedul Islam, United States Department of Agriculture – Animal and Plant Health Inspection Service, Plant Epidemiology and Risk Analysis Laboratory, Raleigh, NC, United States; Peter J. Cotty, Independent Researcher, Tucson, AZ, United States
 Daniel Agbetiameh
Daniel Agbetiameh Alejandro Ortega-Beltran
Alejandro Ortega-Beltran Richard T. Awuah2
Richard T. Awuah2 Kenneth A. Callicott
Kenneth A. Callicott Peter J. Cotty
Peter J. Cotty Ranajit Bandyopadhyay
Ranajit Bandyopadhyay