- School of Agriculture and Biology/State Key Laboratory of Microbial Metabolism, Shanghai Jiao Tong University, Shanghai, China
Xanthomonas translucens pv. cerealis (Xtc) causes bacterial leaf streak (BLS) of important cereal crops, including wheat (Triticum aestivum) and barley (Hordeum vulgare). Transcription activator-like effectors (TALEs) play vital roles in many plant diseases caused by Xanthomonas spp., however, TALEs have not been previously characterized in Xtc. In this study, the whole genome of NXtc01, a virulent strain of Xtc from Xinjiang, China, was sequenced and compared with genomes of other Xanthomonas spp. Xtc NXtc01 consists of a single 4,622,298 bp chromosome that encodes 4,004 genes. Alignment of the NXtc01 sequence with the draft genome of Xtc strain CFBP 2541 (United States) revealed a single giant inversion and differences in the location of two tal genes, which were designated tal1 and tal2. In NXtc01, both tal genes are located on the chromosome, whereas tal2 is plasmid-encoded in CFBP 2541. The repeat variable diresidues (RVDs) at the 12th and 13th sites within Tal2 repeat units were identical in both strains, whereas Tal1 showed differences in the third RVD. Xtc NXtc01 and CFBP 2541 encoded 35 and 33 non-TALE type III effectors (T3Es), respectively. tal1, tal2, and tal-free deletion mutants of Xtc NXtc01 were constructed and evaluated for virulence. The tal1 and tal-free deletion mutants were impaired with respect to symptom development and growth in wheat, suggesting that tal1 is a virulence factor in NXtc01. This was confirmed in gain-of-function experiments that showed the introduction of tal1, but not tal2, restored virulence to the tal-free mutant. Furthermore, we generated a hrcC deletion mutant of NXtc01; the hrcC mutant was non-pathogenic on wheat and unable to elicit a hypersensitive response in the non-host Nicotiana benthamiana. Our data provide a platform for exploring the roles of both TALEs and non-TALEs in promoting BLS on wheat.
Introduction
Xanthomonas is a large genus of plant-associated, Gram-negative bacteria and contains 27 species that cause disease on approximately 124 monocots and 268 dicots (Chan and Goodwin, 1999). Xanthomonas spp. are subdivided into pathovars on the basis of host specificity and can also be divided into two main phylogenetic groups based on sequence analysis of 16S rDNA, gyrB, dnaK, rpoD, and fyuA (Hauben et al., 1997; Young et al., 2008). The smaller Group 1 includes diverse Xanthomonas spp. including X. translucens, X. albilineans, X. hyacinthi, X. theicola and X. sacchari, whereas the larger Group 2 contains X. oryzae, X. campestris, X. vasicola, X. axonopodis, X. euvesicatoria, and X. citri. Some species are comprised of numerous pathovars (Parkinson et al., 2007); for example, X. translucens includes X. translucens pv. cerealis (Xtc), X. translucens pv. translucens (Xtt), and X. translucens pv. undulosa (Xtu).
X. translucens causes disease in a wide variety of economically-important crops. An emerging disease of global importance is bacterial leaf streak (BLS), which is caused by Xtc, Xtt and Xtu (Pesce et al., 2015; Jaenicke et al., 2016; Peng et al., 2016; Falahi Charkhabi et al., 2017). Xtc has a broad host range with respect to cereals and infects wheat (Triticum spp.), triticale (Triticosecale), barley (Hordeum vulgare), rye (Secale cereale), and oats (Avena spp.) (Cunfer and Scolari, 1982; Bragard et al., 1997; Adhikari et al., 2012); symptoms are particularly severe on wheat (Adhikari et al., 2012). Reported yield losses due to BLS in cereals range from 8 to 34% depending on host susceptibility and the amount of inoculum, while the highest documented yield loss is 40% (Shane et al., 1987; Forster and Schaad, 1988; Duveiller and Maraite, 1993). BLS can also impact grain quality when it occurs on spikes (Mehta, 1990).
Xanthomonas pathogens use similar strategies to release virulence factors into the host including the production of exopolysaccharides (EPS), cell wall-degrading enzymes and protein secretion systems (Büttner and Bonas, 2010). Xanthomonas spp. produce type III effectors (T3Es) that can modulate virulence in host plants; T3Es are delivered into plant cells via the type III secretion system (T3SS), which is encoded by the hrp [hypersensitive response (HR) and pathogenicity] regulon (White et al., 2009). This hrp regulon is positioned on approximately 20 kb genomic island that encode more than 20 hrp genes in many operons. Individual genes have been classified as hrp, hrc (hrp conserved) and hpa (hrp associated). Originally, the “hrp” and “hrc” designations indicated loci that are required for non-host hypersensitive reaction and pathogenicity, but not all of them have this phenotype. hrc genes sequences are clearly conserved among animal and plant pathogens, while the sequences of hrp genes are unique among Xanthomonas and some other genera with some exceptions (Bogdanove et al., 1996). The hpa genes have their role in pathogenicity, depending on the gene. Some function in targeting the type-III secreted proteins to the secretion apparatus, some are themselves translocated into host cells, and some have unknown roles (Kim et al., 2003; Lorenz et al., 2008).
In phytopathogenic bacteria hrp gene clusters are classified into two groups, based on the repertoire genes organization and regulation. Group I includes Pseudomonas syringae, Erwinia spp., and Pantoea stewartii subsp. stewartii while group II comprises Ralstonia solanacearum, Acidovorax avenae, and Xanthomonas spp. (Alfano and Collmer, 1997). The expression of group II hrp genes are controlled by two key regulatory genes, hrpG and hrpX (syn. hrpB) (Büttner and Bonas, 2010). In Xanthomonas the hrp gene cluster was previously known as a pathogenicity island (Noël et al., 2002) but it is reported missing from some Xanthomonas spp. The largest and well-studied Xanthomonas group, Group 2 contain a canonical hrp gene clusters but some studies demonstrated the absence of Hrp T3SS from the group 2 strains that are pathogenic on cannabis and barley (Ignatov et al., 2015; Jacobs et al., 2015). The diverse and smaller group of Xanthomonas, Group 1, contains Hrp T3SS with the exceptions of X. albilneans and X. sacchari (Pieretti et al., 2009; Studholme et al., 2011), all others revealed Hrp T3SS but differing degree of genetic organization to those from group 2 Xanthomonas (Pesce et al., 2017). In X. translucens the mutants of hrp structural genes hrcC of Xtu strain XT4699 pathogenic on various members of Poaceae were impaired in causing symptoms (Peng et al., 2016) while mutants of hrcR, hrpE, and hrpG of Xtg29 pathogenic on forage grass reduced disease symptoms but multiplication of mutants was not affected relative to wild type (Wichmann et al., 2013).
Hrp T3SS is involved in the translocation of type III secreted effector proteins that play a key role in complex process of pathogenesis and host adaptation (Büttner and Bonas, 2010). These T3Es are categorized into transcription activator-like effectors (TALEs) and non-TALEs, which are commonly known as Xanthomonas outer proteins or Xops. The TALE proteins localize to the host plant nucleus, where they recognize and activate effector-binding elements (EBEs) that reside in the promoter regions of host susceptibility (S) or resistance (R) genes. Common structural features of TALEs include an N-terminal domain that harbors the type III secretion signal, a central repeat region (CRR) that interacts with host EBEs, a C-terminal domain that contains nuclear localization signals, and an acidic activation domain. The CRR encodes tandem repeats of 33–35 amino acids and residues at positions 12 and 13 are known as repeat-variable diresidues (RVDs); residue 13 is responsible for base-specific DNA targeting (Boch and Bonas, 2010; Mak et al., 2013; Ji et al., 2016). Evolutionary modifications occur in TALEs by substitution, recombination and deletion events in the nucleotide sequence, but individual RVDs are highly conserved (Erkes et al., 2017). TALE-like proteins can occur in other organisms; examples include the RipTALEs in Ralstonia solanacearum, the MOrTL proteins in marine microorganisms and the Burkholderia rhizoxinica Bat proteins (de Lange et al., 2015).
The number of tal genes in Xanthomonas spp. is highly variable; for example, strains of X. translucens pv. translucens, pv. undulosa, and pv. cerealis were shown to harbor eight, seven and two tal genes, respectively (Pesce et al., 2015; Jaenicke et al., 2016; Peng et al., 2016; Falahi Charkhabi et al., 2017). Some TALEs function to induce plant host S genes, and this fosters pathogen growth and disease development. For example, X. oryzae pv. oryzae (Xoo), which causes bacterial leaf blight (BLB) in rice, targets the host S genes OsSWEET11 via TALE PthXo1, OsSWEET13 via PthXo2 and OsSWEET14 by TALEs AvrXa7, Tal5, TalC and PthXo3 (Yang et al., 2006; Streubel et al., 2013; Zhou et al., 2015). In the BLS pathogen X. oryzae pv. oryzicola (Xoc), the Tal7 effector activates the rice gene Os09g29100, which suppresses the host immune response (Cai et al., 2017). TALEs can also induce the expression of plant transcription factors; for example, PthXo6 and PthXo7 induce OsTFX1 and OsTFIIAγ1, respectively (Sugio et al., 2007; Ma et al., 2018). Similarly, AvrHah1 of X. gardneri activates the expression of two bHLH (basic helix-loop-helix) transcription factors (Schwartz et al., 2017).
Some TALEs activate host resistance (R) genes that restrict bacterial growth and disease development through rapid cell death. For example, the pepper R gene, Bs3, is activated by AvrBs3 in X. euvesicatoria, and rice R genes Xa10, Xa23, and Xa27 are activated by AvrXa10, AvrXa23, and AvrXa27 in Xoo (Gu et al., 2005; Römer et al., 2007; Ji et al., 2014; Tian et al., 2014; Wang et al., 2015). Individual TALEs can also deploy disease resistance through direct protein-protein interaction rather than targeting host R or S genes (Read et al., 2016). Overall, it is well-accepted that TALE proteins in pathogens and their targets in host plants are engaged in an ongoing, co-evolutionary arms race (Ji et al., 2016).
Although several X. translucens strains genome sequences are available and revealed many shared virulence factors (Wichmann et al., 2013; Pesce et al., 2015; Hersemann et al., 2016; Jaenicke et al., 2016; Peng et al., 2016; Falahi Charkhabi et al., 2017; Langlois et al., 2017). Among them, TALEs play important role in plant diseases. Recently, a complete genome sequence of highly virulent Iranian strain Xtu ICMP11055 revealing seven TALE genes. Indel mutagenesis and complementation analysis revealed that two TALE genes, Tal2 and Tal4b contribute to the disease development (Falahi Charkhabi et al., 2017), no other X. translucens pathovars TALEs are reported that contribute to virulence up-to-date.
Bacterial leaf streak of wheat, which is caused by Xtc, is an important, emerging disease that has led to significant yield losses in Xinjiang Province, China. Xtc NXtc01 is a virulent strain isolated from wheat cultivated in Xinjiang. To better understand the mechanistic basis of pathogenesis in the Xtc/wheat interaction, we sequenced the whole genome of Xtc using SMRT technology and compared it with the genomes of X. translucens, X. campestris, and X. oryzae. We also compared NXtc01 genes encoding T3SS gene cluster and T3Es, particularly TALEs, with homologs in other Xanthomonas spp. Two tal genes were deleted in NXtc01, and their role in Xtc virulence was investigated.
Materials and Methods
Bacterial Strains, Growth Conditions, Plasmids and Primers
The bacterial strains, plasmids and primers used in this study are provided in Supplementary Tables S1, S2. X. translucens pv. cerealis strains were cultured in NB medium at 28∘C, and Escherichia coli was grown in LB medium at 37∘C (Miller, 1972). Plasmids were transferred into Xtc NXtc01 and E. coli DH5α by electroporation and heat shock, respectively (Hanahan et al., 1991). Antibiotics used for screening are as follows (μg ml–1): kanamycin (Km), 20; ampicillin (Ap), 100; and spectinomycin (Sp), 25.
Genomic and Plasmid DNA Extraction
All bacterial strains were stored at −80∘C in 50% glycerol. The NXtc01 genomic DNA used for PacBio sequencing was extracted from a 24 h culture using the Hipure bacterial DNA kit (Magen, Guangzhou, Guangdong, China) as recommended by the manufacturer. Genomic DNA was isolated from the Xtc mutant strains using the same method and prepared for Southern blotting (see below). DNA quality and quantity were checked with a NanoDrop spectrophotometer and screened for plasmids by agarose gel electrophoresis (Chakrabarty et al., 2010). Plasmids were isolated from E. coli DH5α using the Plasmid DNA Mini Kit (GBS Biotechnology, China).
PacBio Sequencing, Assembly and Annotation
The NXtc01 genome was sequenced on a PacBio RSII platform (Pacific Biosciences) using Single Molecule, Real Time (SMRT) DNA sequencing. Totally 304,280 reads (2,107,977,050 total reads base) were obtained with average length 6,927 bp and 287.72x coverage after filtering data through SMRT analysis software v2.3. The genome assembly was done using de novo assembly, HGAP protocol available with SMRT analysis packages and can be accessed at SMRT Analysis portal v2.1. The verification of assembly was performed on CheckM with default parameters (Parks et al., 2015). The genome was annotated using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) and deposited in GenBank as accession number PRJNA528834. The Circos was used to generate the circular genome map of NXtc01 including all the predicted ORFs (Krzywinski et al., 2009) as shown in Figure 1.
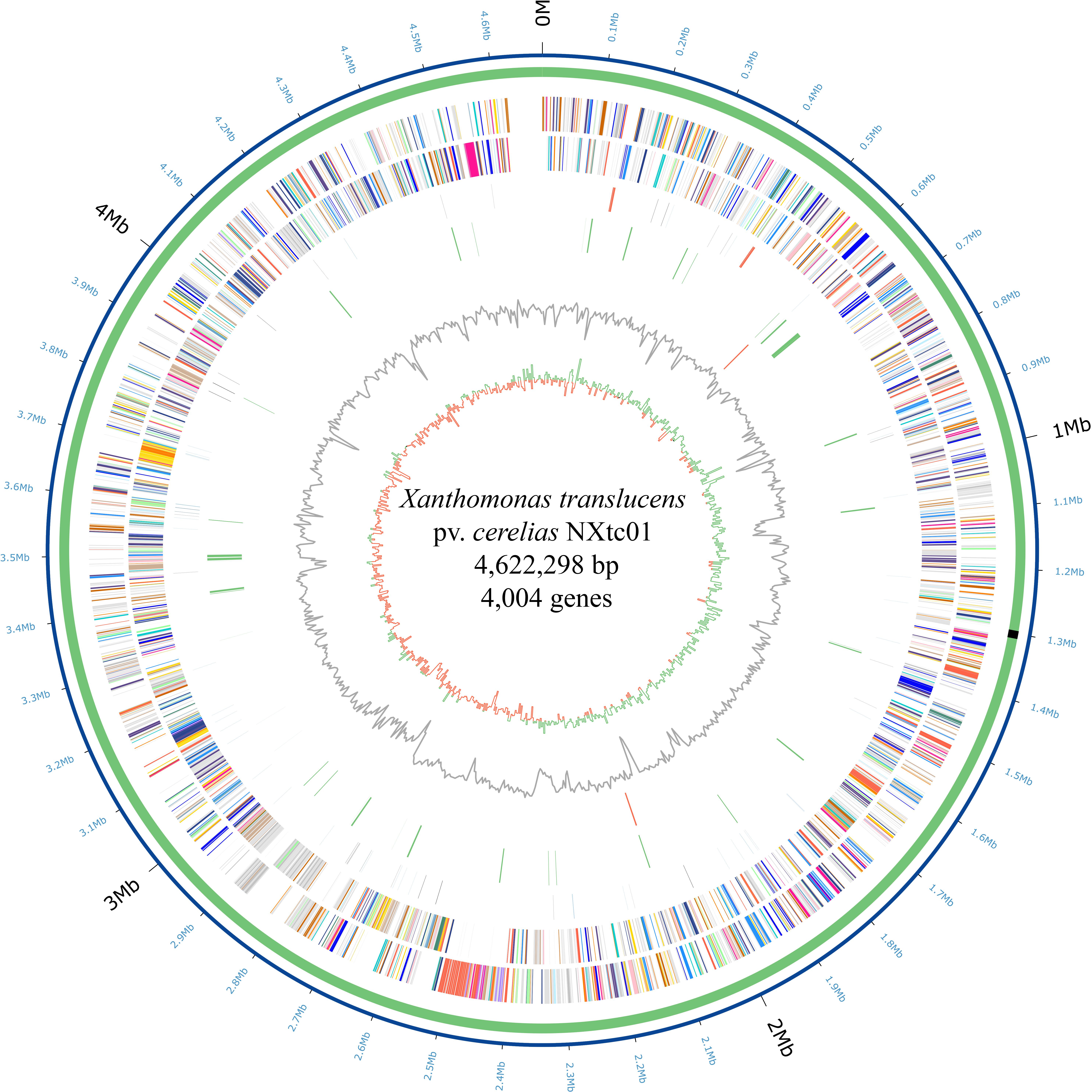
Figure 1. Circular representation of the X. translucens pv. cerealis strain NXtc01 genome starting from gyrB. Inner to outermost rings illustrate GC skew (G − C)/(G + C), GC contents, tal genes (orange), IS elements (green), tRNA (blue), rRNA (orange) and ncRNA (black) genes, reverse COGs (cluster of orthologous groups), forward COGs (Supplementary Table S4), chromosome (green), and coordinates (Mb). The T3SS genes cluster is indicated as black rectangle on chromosome.
Construction of Phylogenetic Trees and Tal RVDs Comparison
A total of 12 strains were selected for homology searches. Ortho MCL v. 2.0 was used to generate groups of orthologous proteins with default parameters1. The resulting set of 1,270 single copy genes was used selected to construct a phylogenetic tree as described previously by Zhu et al. (2011).
In addition to the Xtc NXtc01, all available X. translucens genome sequences were obtained from the NCBI and used for TALEs phylogenetic analysis. TALE genes were predicted and analyzed in each genome using AnnoTALE v1.2 (Grau et al., 2016). DisTAL v1.1 was used to align and phylogenetically classify TAL effectors based on their repeat arrangement (Pérez-Quintero et al., 2015).
For the analysis of TALEs RVDs, we used AnnoTALE version 1.2 that provides 516 TALE genes of 33 Xanthomonas strains. First we analyzed all the X. translucens genomes and merged the TALEs RVDs output file into available 516 TALEs RVDs. Finally, we grouped the TALEs into classes on the basis of RVDs that indicates possible functional and evolutionary relationship (Grau et al., 2016; Erkes et al., 2017).
Identification of Genes Encoding the T3SS and T3Es
Genes encoding the T3SS and effectors (T3Es) were identified in the NXtc01 genome using BLASTn and BLASTp as described by Peng et al. (2016) and Falahi Charkhabi et al. (2017).
Cloning and Expression of NXtc01 tal Genes
NXtc01 BamHI-digested genomic DNA (50 μg) was subjected to 1.2% agarose gel electrophoresis as described by Cernadas et al. (2014). Regions containing tal genes (tal1, 2,745 bp; tal2, 3,872 bp) were gel-purified and ligated into BamHI-linearized and CIP (calf intestinal phosphatase)-treated pBluescript II. Ligated products were transformed into DH5α by heat shock. Tal-containing clones were confirmed by colony blots, restriction digestion and Sanger DNA sequencing.
For expression in NXtc01, pBluescript II clones containing tal genes were digested at SphI sites to release the CRR and ligated into SphI-linearized, CIP-treated pZW vector. This vector contains a FLAG-tagged derivative of avrXa10 with the N- and C-terminal domains of the effector (without the CRR), a strong lac promoter, and transcription termination signals (Yang et al., 2000). Ligated products were used to transform DH5α; the orientation of tal genes was then analyzed by PCR using primers TALN18S-F/TalN-R (Supplementary Table S2) and confirmed by sequence analysis. For expression in Xanthomonas, pZW vectors harboring tal genes were ligated into pHMI at the HindIII site and transferred into the tal-free strain; these were designated as PHZW-tal1 and PHZW-tal2. To confirm of tal gene expression, bacterial cells containing the C-terminally flag-tagged tal genes (PHZW-tal) were harvested and washed in phosphate-buffered saline (PBS). Cells were harvested by centrifugation, diluted in PBS and boiled for 8 min. Proteins were separated on 8% SDS-PAGE gels and transferred to polyvinylidene difluoride membranes for immunoblotting using mouse anti-flag as the primary antibody (Transgene, Beijing, China). Primary antibodies were detected and visualized using goat anti-mouse IgG (H + L) and the EasySee Western Kit (Transgene) (Supplementary Figure S1C).
Mutagenesis of NXtc01 tal and hrcC Genes
The tal1, tal2, tal-free, and hrcC mutants were generated by homologous recombination using suicide vector pKMS1 (Zou et al., 2011). For tal1, unique 640 bp upstream and 425 bp downstream sequences flanking tal1 gene were amplified using primer sets T1Nt-F/T1Nt-R and T1Ct-F/T1Ct-R (Supplementary Table S2), respectively. The primers contained XbaI (T1Nt-R and T1Ct-F), SmaI (T1Ct-R), and PstI (T1Nt-F) sites. The two PCR products were gel-purified, digested with XbaI and ligated at 22∘C for 2 h. The ligation of upstream and downstream fragments was confirmed by PCR using primer set T1Nt-F/T1Ct-R. The desired fragment was gel-purified, digested with SmaI and PstI and cloned into pKMS1. The ligation mixture was introduced into DH5α cells and cultured on LB containing Km. The resulting construct, pKMSTal1 (Supplementary Table S1), was confirmed by colony PCR using a M13 universal primer set and sequenced. pKMSTal1 was introduced into Xtc NXtc01, plated on NAN containing Km (NANKm), and incubated for 4 days. NANKm colonies were then cultured overnight on NAS (NA containing 10% sucrose) and NAN/NANKm, individually. Colonies that grew only on NAN/NANKm were then transferred to NAN broth for 10 h at 28∘C (∼OD600 ≤ 0.1). The culture was plated on NAS for 2 days and colonies were transferred to NA and NAKm. Colonies that did not grow on NAKm were analyzed by PCR with primer set T1Nt-F/T1Ct-R (Supplementary Table S2 and Supplementary Figure S1A) and were further confirmed by Southern blot analysis (Supplementary Figure S1B).
Similar to the strategy mentioned above, pKMSTal2 was constructed by cloning fragments flanking the CRR in tal2 (upstream fragment, 698 bp; downstream, 413 bp) into pKMS1 using primer sets T2Ct-F/T2Ct-R and T2Nt-F/T2Nt-R (Supplementary Tables S1, S2 and Supplementary Figure S1A). Construct pKMShrcC was constructed by cloning fragments that flank the hrcC gene (upstream fragment, 610 bp; downstream, 694 bp) using primer sets hrcC-up-F/hrcC-up-R and hrcC-do-F/hrcC-do-R (Supplementary Tables S1, S2 and Supplementary Figure S1A). A tal-free mutant was generated by transferring pKMSTal2 into the Xtc NXtc01 tal1 mutant (Δtal1) following the same strategy described above (Supplementary Tables S1, S2 and Supplementary Figure S1A). The tal2, hrcC, and tal-free mutants were generated and screened using the protocol described for the tal1 mutant.
Southern Blot Analysis
Genomic DNA was extracted from NXtc01 and tal mutants and digested with BamHI for 5 h. Digested DNA fragments were separated in 1.3% agarose gels in TAE buffer at 4∘C, 80 V for 14–16 h and then transferred to Hybond N+ nylon membranes (Pall Corporation, NY, United States). The SphI fragment of the NXtc01 tal1 gene was used as a probe and labeled with digoxigenin (DIG) using the Dig-High Prime Labeling and Detection kit (Roche, Germany). DIG labeling, hybridization, washing, blocking, and detection were conducted as recommended by the manufacturer.
Plant Inoculation and Quantification of Bacterial Populations
Wheat cv. Yangmai 158 was planted in a greenhouse with a 12 h photoperiod and cultivated at 28∘C with 75% RH. The bacterial suspentions were prepared by inoculating cells into 5 ml NA broth containing the appropriate antibiotics. Bacterial cells were grown for 12 h, harvested, washed and suspended in 10 mM MgCl2 to OD600 = 0.2 (1.6 × 108CFU/ml). To access the tal contribution, wheat plants (3 weeks old) were spray-inoculated with the wild-type (WT), tal mutants (Δtal1, Δtal2, and tal-free) and tal complemented mutants (PHZW-tal1 and PHZW-tal2) with a hand atomizer, which simulates natural entry of the pathogen into stomates, hydathodes and lenticels. Symptoms were compared at 11 days post-inoculation (dpi). For the T3SS virulence assays, the WT, ΔhrcC mutant, and ΔhrcC/hrcC were spray-inoculated into young leaves of 3 weeks old wheat plants and needless syringe infiltrated into 3 weeks old N. benthamiana plants; symptoms were evaluated at 11 dpi (wheat) and 24 hpi (N. benthamiana).
For quantification of bacterial growth, three replicate leaf samples (2 × 0.5 cm) were excised from five plants and surface-sterilized in 70% ethanol and double-distilled water (ddw). Samples were macerated in 1 ml ddw and incubated at 4∘C for 1 h; serial dilutions were then plated for enumeration of bacteria. The experiment was repeated at least three times, and the Student’s t-test was used to determine significant difference.
Results
X. translucens pv. cerealis NXtc01 Genome Sequence
The NXtc01 genome is encoded by a 4,622,298 bp chromosome that lacks autonomous plasmid DNA (Figure 1). The genome, as annotated by the NCBI Prokaryotic Genome Annotation Pipeline (PGAP), contains overall G + C content is 67.23%, and 4,004 coding DNA sequences (CDS) with an average length of 1,011 bp. NXtc01 has two rRNA operons, 54 tRNA genes and other non-coding RNA genes, which are characteristics similar to other Xanthomonas genomes (Table 1). NXtc01 genome consists of 0.13% repetitive regions of 6,269 bp. A total of 43 simple repeat sequences and one satellite sequence were predicted via Repbase. In genome 80 complete and 56 partial insertion sequence (IS) elements were predicted using the ISfinder database (Table 1). To access the completeness and contamination of this genome CheckM was used, which is 99.86 and 0.05%. These results revealed the high quality assembled genome.
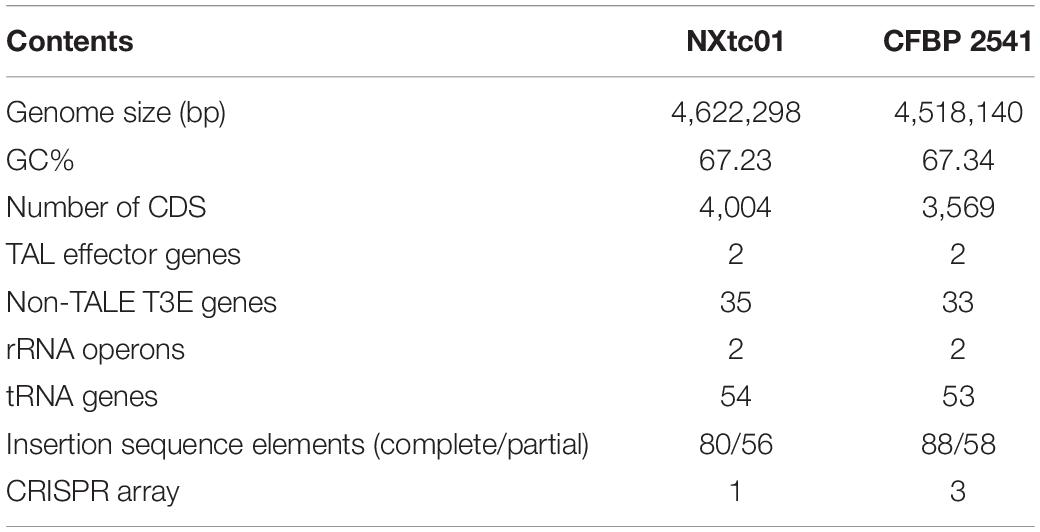
Table 1. Comparison of X. translucens pv. cerealis genomes from strains NXtc01 and CFBP 2541, which were isolated from Triticum aestivum in Xinjiang, China and Bromus inermis in the United States, respectively.
Genome-wide, single-copy gene comparisons were made between NXtc01, the draft genome of Xtc CFBP 2541 (accession no. PRJNA268946) and other Xanthomonas genome sequences [X. albilineans GPE PC73, PRJEA16687 (Pieretti et al., 2009); X. translucens pv. undulosa 4699, PRJNA248137 (Peng et al., 2016); X. translucens pv. translucens DSM 18974T, PRJEB647 (Jaenicke et al., 2016); X. campestris pv. campestris 8004, PRJNA15 (Qian et al., 2005); X. euvesicatoria 85-10 (previously known as X. campestris pv. vesicatoria) PRJNA341901 (Thieme et al., 2005); X. citri pv. vignicola CFBP7111, PRJNA390891 (Ruh et al., 2017a); X. axonopodis pv. citri 306, PRJNA297 (da Silva et al., 2002); X. campestris pv. musacearum NCPPB 4384, PRJNA73881 (Wasukira et al., 2012); X. oryzae pv. oryzae PXO99A, PRJNA131967 (Salzberg et al., 2008); X. oryzae pv. oryzicola RS105, PRJNA280380 (Wilkins et al., 2015)]. This analysis revealed that NXtc01 groups with Xtc, Xtu and Xtt and resides on the same branch as Xtc CFBP 2541 (Figure 2). Thus, Xtc NXtc01 and CFBP 2541 are closely related, even though they were isolated from different continents.
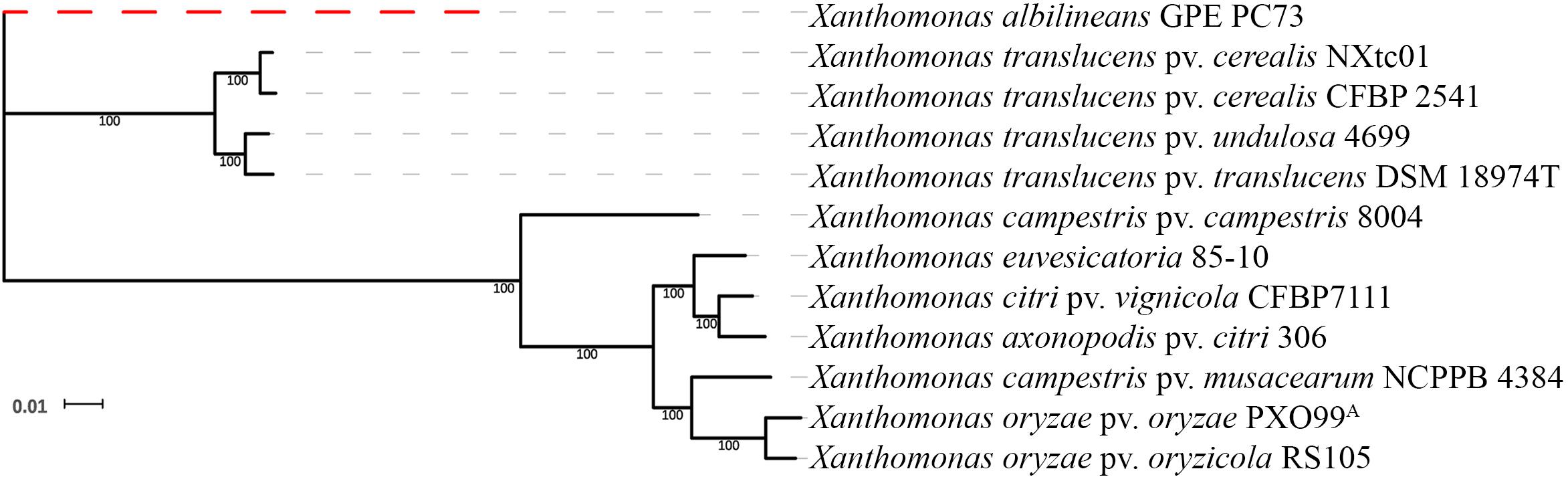
Figure 2. Phylogenetic relationships based on single copy genes. The tree was constructed from a concatenated alignment of 1,270 single copy genes and shows the relationship of NXtc01 to the following Xanthomonas spp. (accession numbers in parenthesis): Xtc CFBP 2541 (PRJNA268946); X. translucens pv. undulosa 4699 (PRJNA248137); X. translucens pv. translucens DSM 18974T (PRJEB647); X. campestris pv. campestris 8004 (PRJNA15); X. euvesicatoria 85-10 (PRJNA341901); X. citri pv. vignicola CFBP7111 (PRJNA390891); X. axonopodis pv. citri 306 (PRJNA297); X. campestris pv. musacearum NCPPB 4384 (PRJNA73881); X. oryzae pv. oryzae PXO99A (PRJNA131967); and X. oryzae pv. oryzicola RS105 (PRJNA280380). X. albilineans GPE PC73 (PRJEA16687) was used as an outgroup.
Comparative Analysis of NXtc01 and CFBP 2541 Genomes
A comparison of the NXtc01 genome with the draft genome of CFBP 2541 is illustrated in Figure 3 starting from same gene, gyrB. The structure of both genomes is quite similar but with a single giant inversion that is bordered on one side with putative IS elements and/or integrases. The NXtc01 genome is 104 kb larger than CFBP 2541, however, this could be due to incomplete sequencing in the draft genome of CFBP 2541. Both strains harbor two tal genes that are identical in size but located in different genomic positions. CFBP 2541 contains a plasmid-borne TALE gene, named tal2 (Pesce et al., 2015), whereas the gene is chromosomally encoded in NXtc01, possibly due to horizontal transfer (Figure 3). IS elements are frequently found in both genomes (Table 1); implicating possible involvement in the gain or loss of some regions. Likely, IS elements found at the CFBP 2541 plasmid and NXtc01 chromosome regions containing tal2 (Figure 3) indicating the introgression that might be transposon mediated. The genomes of Xtc NXtc01 and CFBP 2541 also contained one and three CRISPR array2, respectively (Table 1).
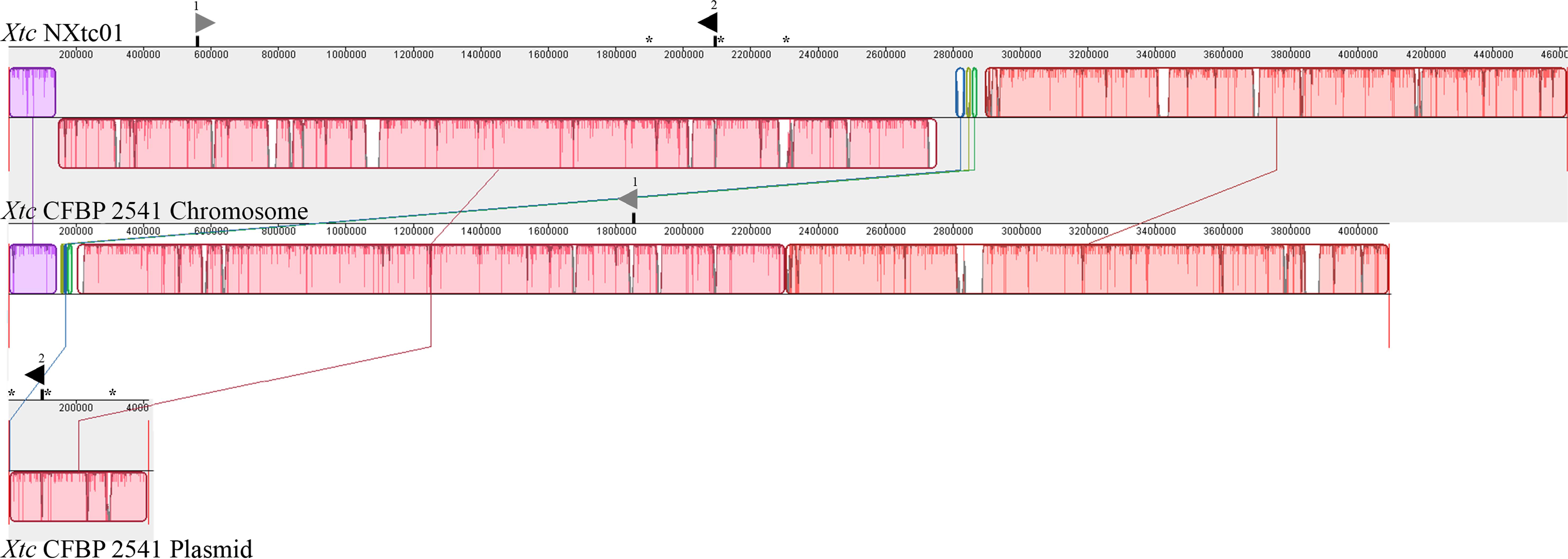
Figure 3. Organization of the whole genome and tal genes of Xtc strains NXtc01 and CFBP 2541. Genomes starting from gyrB gene were aligned using progressive MAUVE with default parameters (http://darlinglab.org/mauve). Horizontal axes show sequence coordinates in base pairs. The colored, locally collinear blocks (LCB) shows conserved and highly similar genomic regions; white areas inside colored regions indicates sequence elements specific to one genome that were not aligned. Height of similarity profile is present inside each block. The colored lines that connect LCBs represent translocations of homologous regions. Blocks above or below the horizontal bar indicate regions that aligned in the forward or reverse orientation, respectively (same-colored LCBs on opposite side of the center line indicates an inversion). Asterisks indicates IS elements that bordered tal2. The two tal genes are represented as triangles above the sequence coordinates, pointing toward transcriptional orientation on chromosomal or plasmid DNA. Black filled triangles indicate tal2 on chromosome in NXtc01 and on plasmid in CFBP 2541, encode identical RVDs. Gray filled triangles indicate tal1 (see Table 2 for RVD sequences).
TALE phylogenetic tree of X. translucens strains were created by aligning TALE-CRR using DisTAL. All 27 TALEs (Xtc NXtc01 = 2, Xtc CFBP 2541 = 2, Xtu ICMP11055 = 7, Xtu 4699 = 8 and Xtt DSM18974T = 8) were classified into 17 groups. Both Tal1 and Tal2 of Xtc stains NXtc01 and CFBP 2541 grouped separately from pv. translucens and pv. undulosa TALEs (Figure 4). Together, two of these groups contained nearly identical TALEs from all X. translucens strains, while all others were exclusive to Xtu or Xtt strains (Figure 4).
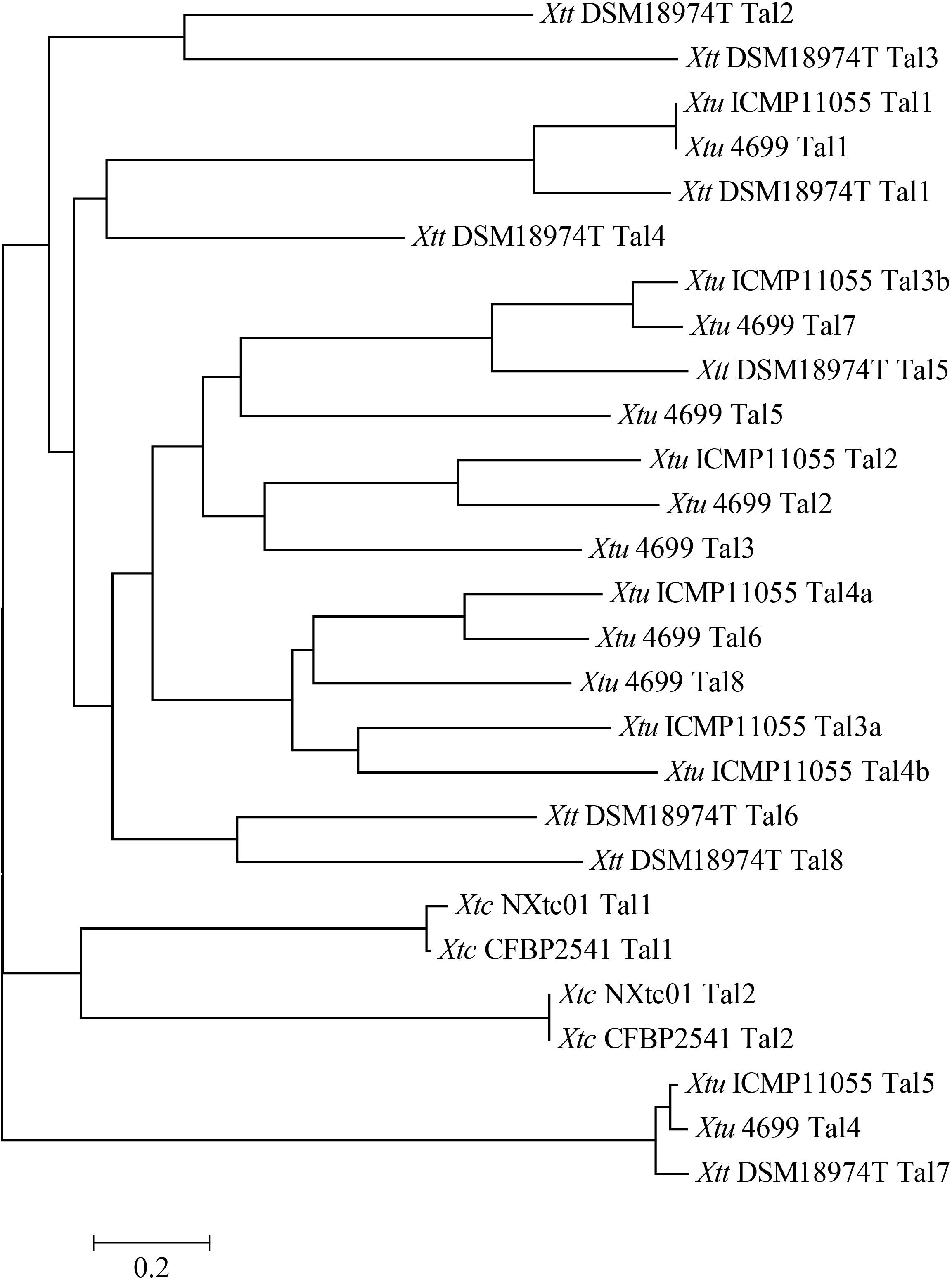
Figure 4. Phylogenetic tree based on TALEs repeat arrangement alignments. DisTAL was used to build tree for a set of 27 TAL effector sequences from 5 different X. translucens strains. TALEs were classified into 17 groups showing the relationship of Xtc NXtc01 to several other X. translucens strains published previously (Pesce et al., 2015; Jaenicke et al., 2016; Peng et al., 2016; Falahi Charkhabi et al., 2017). Scale is shown below the tree.
In Nxtc01, Tal1, and Tal2 contained 11.5 and 15.5 RVDs, respectively (Table 2). Tal1 in NXtc01 and CFBP 2541 contained 11.5 RVDs that were nearly identical except for variation in the RVD of the third repeat (Table 2). Tal1 in NXtc01 and CFBP 2541 RVDs did not show relatedness to TALEs annotated in Xtu 4699, Xtt DSM 18974T, or Xtu ICMP11055. Tal2 consisted of 15.5 RVDs that were identical in Xtc NXtc01 and CFBP 2541; Tal2 was also closely related to Tal4, Tal5, and Tal7 in Xtu 4699, Xtu ICMP11055, and Xtt DSM 18974T, respectively (Table 2).
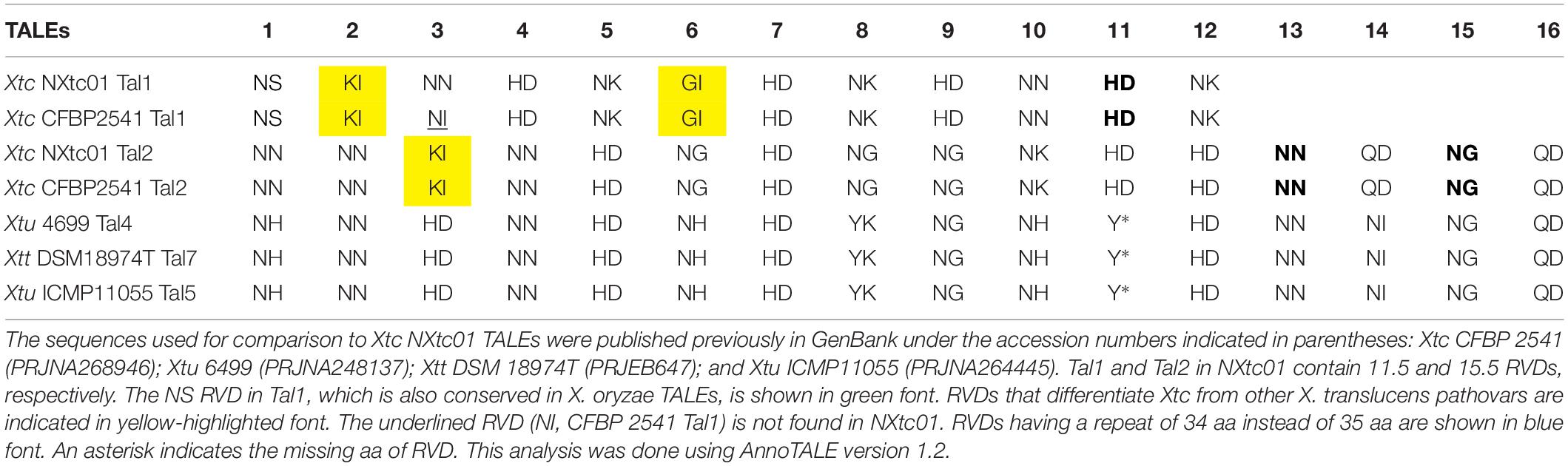
Table 2. Comparison of RVDs in Tal1 and Tal2 with RVDs in other X. translucens strains and pathovars.
In addition to Tal1 and Tal2, the Xtc NXtc01 and CFBP 2541 genomes encode 35 and 33 non-TALE T3Es, respectively. Many of these non-TALE genes are Xop-encoding genes that are also conserved in Xtu ICMP11055 (Falahi Charkhabi et al., 2017), Xtt DSM 18974T (Jaenicke et al., 2016), X. campestris pv. campestris 8004 (Qian et al., 2005), Xoo PX099A (Salzberg et al., 2008), and Xoc BLS256 (Bogdanove et al., 2011; Supplementary Table S3).
Type III Secretion System (T3SS) in X. translucens pv. cerealis
The organization of the T3SS in Xtc NXtc01 was identical to Xtc CFBP 2541, Xtu ICMP11055 and Xtt DSM 18974T (Supplementary Figure S2). Interestingly, we identify putatively encoding an hrp gene cluster covering 20,486 bp region (from hpaD to hrcC) in Xtc NXtc01. This region consisted of eight hrp, 11 hrc and four hpa genes (Supplementary Figure S2). The 23 genes that encode this cluster are conserved among the three X. translucens pathovars (Supplementary Figure S2), but have less similarity based on aa identity and genetic organization at the species level. The two hrp regulatory genes, hrpG and hrpX, flank the hrp cluster of NXtc01 (Supplementary Figure S2), and this organization is conserved in other X. translucens pathovars (Supplementary Figure S2; Falahi Charkhabi et al., 2017; Pesce et al., 2017).
We deleted the hrcC gene, which encodes an outer ring protein of the T3SS (Li et al., 2017), and evaluated the phenotypes of the ΔhrcC mutant on wheat and the non-host N. benthamiana. In contrast to the virulent NXtc01 strain, the ΔhrcC mutant was non-pathogenic on wheat cv. Yangmai 158 and did not elicit an HR on N. benthamiana (Figures 5A,C). For complementation analysis, a recombinant plasmid containing the entire coding region of hrcC (1,848 bp) was constructed and designated pH1-Flag(-)hrcC (Supplementary Table S1). This plasmid was introduced into ΔhrcC, resulting in strain ΔhrcC/hrcC; production of HrcC in trans was confirmed by western blot analysis (Supplementary Figure S1C). The pathogenicity of ΔhrcC, ΔhrcC/hrcC and wild-type NXtc01 were compared by assessing disease symptoms in wheat leaves at 11 dpi, and an HR on N. benthamiana at 24 hpi. Both the wild-type and the complementing strain ΔhrcC/hrcC produced similar symptoms on wheat and the HR on N. benthamiana, indicating that the hrp phenotype was restored to ΔhrcC when the hrcC gene was supplied in trans (Figures 5A,C).
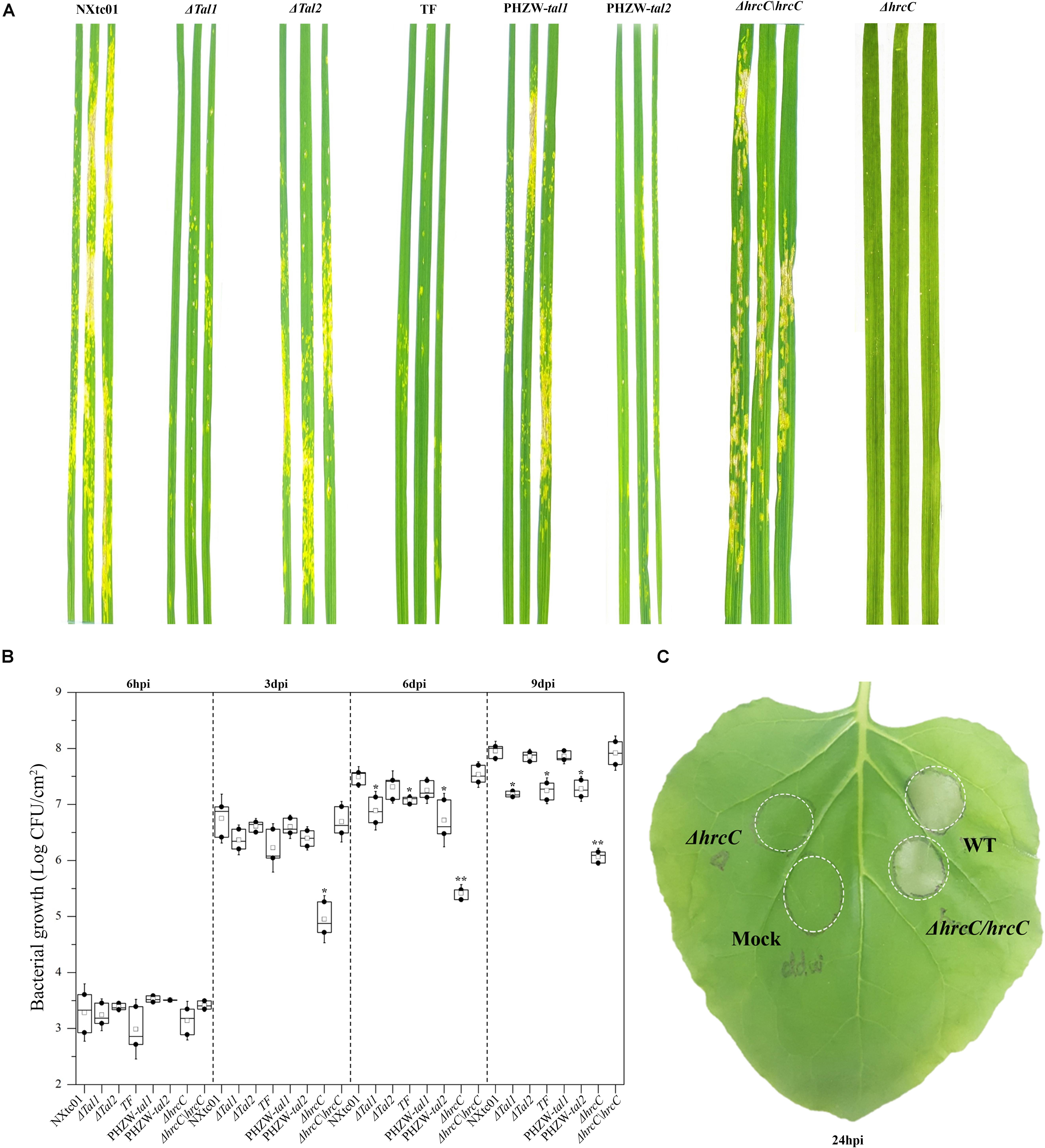
Figure 5. Aggressiveness and bacterial growth of wild type Xtc NXtc01, deletion mutants and complemented strains. NXtc01 (WT), Δtal1, Δtal2, tal-free (TF), ΔhrcC, PHZW-tal1, PHZW-tal2, and ΔhrcC/hrcC were sprayed-inoculated (OD600 = 0.2) onto 3 weeks old wheat plants. (A) Symptoms were compared and photographed at 11 dpi. (B) Bacterial populations were quantified at 6 hpi and 3, 6, and 9 dpi. The box plot displays replicates (n = 3) and mean value (middle box); error bars show mean ± SD. ∗p < 0.05, ∗∗p < 0.01 (Student’s t-test); indicates a significant difference compared to WT and ΔhrcC/hrcC. (C) The inocula (OD600 = 0.2) of WT, ΔhrcC, ΔhrcC/hrcC, and mock (ddw) were infiltrated into three to four leaves of 20 days old N. benthamiana plants using a needless syringe, and infiltrated areas were marked. Symptoms were observed and compared at 24 hpi. (A–C) Experiments were repeated at least three times with similar results. WT, Xtc NXtc01 (wild-type); Δtal1, NXtc01 tal1 deletion mutant; Δtal2, NXtc01 tal2 deletion mutant; TF, NXtc01 tal-free mutant; ΔhrcC, NXtc01 hrcC deletion mutant; PHZW-tal1, NXtc01 TF with tal1 in trans; PHZW-tal2, NXtc01 TF with tal2 in trans; ΔhrcC/hrcC, NXtc01 ΔhrcC containing hrcC in trans.
In addition, we also test the bacterial proliferation of ΔhrcC and ΔhrcC/hrcC in planta. The bacterial multiplication in mutant ΔhrcC was reduced as compared to virulent NXtc01 strain. While ΔhrcC/hrcC have restored bacterial growth similar like wild type strain NXtc01 (Figure 5B).
Non-TALEs of NXtc01
The NXtc01 genome contained 35 non-TALEs; 10 are conserved T3Es, including AvrBs2, XopF, XopK, XopL, XopN, XopP, XopQ, XopR (frameshift mutation), XopX, and XopZ (Supplementary Table S3). X. translucens pathovars contained multiple copies of AvrBs2 (n = 2), XopF (n = 2), XopL (n = 2–4), XopP (n = 2–4) and XopX (n = 3) (Supplementary Table S3). Other T3Es conserved in X. translucens pathovars include XopAA, XopAD, XopAF1, XopAM, XopAP, XopB, XopC2, XopG1, XopV, and XopY. The T3Es varied among Xanthomonas species, pathovars and strains. For example, AvrBs1 is present in Xtc CFBP 2541 and Xcc 8004 but was not present in NXtc01 (Supplementary Table S3); whereas AvrXccA1, XopAZ, and XopJ were detected in NXtc01 but not in CFBP 2541. AvrXccA1 and XopAZ were present in NXtc01 but not the other X. translucens pathovars, and XopJ was only present in NXtc01 and DSM 18974T.
TALEs of NXtc01
The number (n = 2) of TALEs in Xtc Nxtc01 were confirmed by Southern blot analysis (Supplementary Figure S1B). Furthermore, we also sequenced the tal genes after cloning hybridizing BamHI fragments; the corresponding sequences were identical to the tal genes in the genome. The sizes of TALE genes are; tal1 2,898 bp and tal2 3,342 bp but tal2 N-terminus lack classically conserved BamHI site (Supplementary Figure S1B). The two TALEs encode 11.5 and 15.5 RVDs, respectively. Some unusual RVDs found in Tal1 and/or Tal2 include GI and KI, which help differentiate Xtc from other Xanthomonas spp. (Table 2), and QD, which is occurring once in TALEs of X. oryzae pv. oryzicola CFBP 7331 and CFBP 7341 (Wilkins et al., 2015). Tal1 contains the NS RVD, which is common in X. oryzae, and Tal2 contains QD at the distal end, which is conserved in other X. translucens pathovars (Table 2). The Tal1 and Tal2 RVDs in NXtc01 are identical to those in Xtc CFBP 2541 strain except for the third RVD in Tal1 (Table 2).
Tal1NXtc01 Contribution to Virulence
To examine whether the NXtc01 TALEs contribute to virulence, tal deletion mutants were generated by homologous recombination using pKMSTal1 and pKMSTal2. Putative mutants were initially screened via colony PCR using pKMSTal1 and pKMSTal2 as positive controls (Supplementary Figure S1A). The screened mutants were further verified by Southern blot hybridization with a DIG-labeled SphI fragment of tal1. The deletion mutants were designated Δtal1,Δtal2 and TF (tal-free mutant) (Supplementary Figure S1B). Suspensions of the wild-type NXtc01, Δtal1, Δtal2, and the tal-free mutant were sprayed on leaves of wheat cv. Yangmai 158, and symptoms were evaluated at 11 dpi. The Δtal1 and tal-free mutants showed a significant reduction in disease symptoms as compared with the wild-type strain (Figure 5A), and the population growth of these two mutants was significantly lower than the wild-type (Figure 5B). In contrast, the Δtal2 mutant was not impaired in symptom development, and its growth in planta was not significantly different from the wild-type NXtc01 (Figures 5A,B).
To further investigate the contribution of tal genes in virulence, we expressed individual tal genes in the tal-free mutant. Recombinant plasmids containing tal1 and tal2 were constructed and designated pHZW-tal1 and pHZW-tal2 (Supplementary Table S1), respectively, and introduced into the NXtc01 tal-free strain. The expression of individual tal genes in the tal-free mutant was confirmed by immunoblotting (Supplementary Figure S1C). The introduction of tal1, but not tal2, restored full virulence to the tal-free mutant (Figure 5A) and its multiplication in planta (Figure 5B); indicating that tal1 contributes to NXtc01 virulence in wheat.
Discussion
In this study, we sequenced the genome of Xtc NXtc01, a virulent strain isolated from wheat grown in Xinjiang, China. The NXtc01 genome was compared with the draft genome of Xtc CFBP 2541 and other strains of X. translucens, X. campestris, and X. oryzae. The Xtc NXtc01 and CFBP 2541 genomes are slightly different in sequence and size, and show one giant rearrangement relative to each other. Interestingly, tal2 is chromosomally-encoded in NXtc01 but plasmid-borne in CFBP 2541, which is likely due to transposon mediated horizontal gene transfer (Siguier et al., 2006; Ruh et al., 2017b; Chen et al., 2018). Comparative analysis based on single-copy genes revealed homogeneity among X. translucens strains; in other words, X. translucens are divided into two subgroups apart from X. campestris and X. oryzae. Xtc grouped together uniquely, beside a group of Xtu and Xtt. On the basis of genetic similarity, this result suggests that Xtc is a robust biological entity with a common genetic basis for host specificity apart from their geographical distribution.
In many phytopathogenic bacteria, the T3SS is critical for virulence and modulates the delivery of effector proteins into plant cells. The T3SS of group 1 and group 2 Xanthomonas are very different; for example, X. albilineans belongs to group 1 and has a T3SS different from that encoded by the hrp gene cluster. Xanthomonas spp. in group 2 encode a typical Hrp T3SS (Marguerettaz et al., 2011), which is comprised of seven hrp operons, 11 conserved hrc (hrp-conserved) genes and other hpa (hrp-associated) genes (Tampakaki et al., 2010; Li et al., 2017). The products of the nine hrc genes are structural components of the T3SS for secretion of T3Es into plants (Li et al., 2017). A comparison of genes encoding the T3SS in seven different Xanthomonas strains revealed that the organization of the cluster in NXtc01 and other X. translucens strains is conserved (Supplementary Figure S2). Notably, the organization of regulatory genes hrpG and hrpX in NXtc01 is different from the position of hrpG/X in Xcc 8004, Xoo PXO99A, and Xoc BLS256 (Supplementary Figure S2). Interestingly, a genetic organization similar to NXtc01 has been reported for the T3SS in R. solanacearum for hrpB (homologous to hrpX) and hrpG (Salanoubat et al., 2002; Pesce et al., 2017).
In addition, we also demonstrated that a mutant of the conserved hrp gene hrcC of wheat pathogen Xtc NXtc01 resulted completely loss of symptoms and hardly colonize the leaf upon inoculation (Figures 5A,B), similar as barley pathogens Xt pv. hordei and pv. translucens hrcT mutant and wheat pathogen Xtu XT4699 hrcC mutant, respectively, a typical phenotype like group 2 Xanthomonas (Peng et al., 2016; Pesce et al., 2017). In contrast grass pathogen Xtg29 hrcR, hrcE, and hrpG mutants can’t eliminate the symptoms completely and colonization is also unaffected (Wichmann et al., 2013). The hrc gene products are structural components of the T3SS, which functions as a conduit for secretion of T3Es into plants (Kim et al., 2003; Li et al., 2017). Possibly the T3SS import more T3Es into small grain cereals than the grasses for colonization and symptoms development. Therefore, it will be worthwhile to compare the hrcC or hrcT mutants in all other X. translucens pathovars.
We extended our analysis of mutant hrcC in non-host N. benthamiana. Infiltration of Xtc NXtc01 T3SS-deficient mutant, a ΔhrcC does not show any visible HR. while the complementation showed non-host HR, similar like wild type (Figure 5C). Thus non-host HR largely depends on translocation of T3Es, likely XopQ (Adlung et al., 2016).
This study also reveals a repertoire of non-TALEs in NXtc01 that have been identified in other Xanthomonas spp. Xtc NXtc01 and X. translucens strains contain two copies of AvrBs2 (Supplementary Table S3), which contains a putative glycerophosphoryl-diester phosphodiesterase domain thought to be involved in plant host signaling and osmotic adaptation (Swords et al., 1996; Li et al., 2015). X. translucens strains also contain a single copy of XopK and two copies of XopF (Supplementary Table S3); these effectors presumably suppress Xoo-mediated PAMP-triggered immunity in rice (Mondal et al., 2016; Qin et al., 2018). Xtc NXtc01 and CFBP 2541 both contain copies of XopL and XopN, which were essential for X. axonopodis pv. phaseoli virulence (Kumar et al., 2016; Soni and Mondal, 2018). XopL encodes an E3 ubiquitin ligase that interacts with plant-specific E2 enzymes and ubiquitinates unidentified target proteins in X. euvesicatoria 85-10 (Singer et al., 2013). Several other non-TALE core T3Es of Xanthomonas that play important roles, likely XopR in Xoo strain 13571 is required for full virulence but the mechanism is yet not understood (Zhao et al., 2013). Another core effector, the XopZ that potentially interferes the bacterial proliferation and host innate immunity in Xoo PXO99A (Song and Yang, 2010). XopQ of Xoo strain BXO43 suppressing the rice immune responses by interacting Gf14f and Gf14g, the two rice 14-3-3 proteins (Deb et al., 2019). XopP hijacked the proteasomal pathway by interacting with E3 ubiquitin ligase PUB44 in Xoo (Ishikawa et al., 2014). Less conserved T3Es, such as XopB, were shown to be important in the multiplication of X. euvesicatoria 85-10 in planta and inhibited the immune responses trigged by other T3Es (Schulze et al., 2012). Other T3Es, notably XopY, XopAA, and XopAP (Yamaguchi et al., 2013a, b; Teper et al., 2016), also provide valuable avenues of research for probing the roles of these effectors in NXtc01. Furthermore, the conserved sets of X. translucens non-TALEs may ultimately help us identify pyramided R genes for BLS resistance.
Plant diseases are driven by molecular interactions between pathogen effectors and host defense genes, and there is evolutionary pressure for pathogens to develop new virulence strategies to counteract host defense mechanisms (Jones and Dangl, 2006). The pathogenesis of Xanthomonas spp. largely depends on T3Es that are translocated into plant cells by the T3SS (Gürlebeck et al., 2006). In order to understand how X. translucens TALome (TALEs per genome) differ from each other within and between strains, different tools were applied (Pérez-Quintero et al., 2015; Grau et al., 2016). X. translucens encodes very diverse TAL effectors that were classified exclusively into 17 groups. TALE phylogenetic tree showed that all X. translucens pathovars form distinct groups, despite the overall genomic similarities (Figure 4).
In this study, we investigated the role of two TALEs, tal1 and tal2, in the virulence of Xtc NXtc01; tal1, but not tal2, was required for a full level of virulence. Although the TALE repertoire of X. translucens pathovars is somewhat variable, tal2NXtc01 was highly similar to tal2 in CFBP 2541, tal4 in Xtu 4699, tal5 in Xtu ICMP11055, and tal7 in Xtt DSM 18974T with respect to the number and sequence of RVDs and repeat length. Interestingly, the TALEs in Xtc NXtc01 share some features (e.g., repeat length, RVD composition) with the RipTALs of R. solanacearum, which may be the result of convergent evolution and/or recombination events (Schandry et al., 2016).
Numerous reports show that tal genes contribute to virulence in Xtu, Xoc, Xoo, X. euvesicatoria, X. campestris pv. malvacearum, X. citri subspecies citri, X. axonopodis pv. vesicatoria, and X. axonopodis pv. manihotis (Yang, 1994; Wichmann and Bergelson, 2004; Antony et al., 2010; Cernadas et al., 2014; Cohn et al., 2014; Hu et al., 2014; Cox et al., 2017; Falahi Charkhabi et al., 2017). The NXtc01 genome provides an excellent platform for future comparative genomic studies aimed at understanding Xtc pathogenicity in wheat. Deletion mutagenesis of NXtc01 and complementation analysis with the tal-free strain clearly demonstrated that tal1NXtc01 contributes to virulence on wheat. The deletion mutants and genetic constructs described in this study provide valuable new tools to identify the wheat gene(s) that is potentially activated by Tal1NXtc01.
Data Availability
The datasets generated for this study can be found in NCBI, PRJNA528834.
Author Contributions
SS and GC designed the study, with assistance from FH, WM, XX, and ZX. SS conducted the experiments, with assistance from FH, XX, WM, and LZ. SW and BZ did phylogenetic analysis and also helped in other computational analysis. SS and GC prepared the manuscript. All authors read and approved the final version of the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China Research Grant No. 31830072 and Chinese Scholarship Council (2017GXZ018098).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Baishi Hu, Nanjing Agricultural University and Miss Yangyang Yang (a master degree student in this lab) who isolated Xtc strain from wheat fields in Xinjiang Province of China, Dr. Lulu Cai (a previous Ph.D. student in this lab) for helpful discussions in the experiments and Mr. Liu Longyu (a senior Ph.D. student in this lab) for helping in Southern blot experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02040/full#supplementary-material
Footnotes
References
Adhikari, T. B., Gurung, S., Hansen, J. M., and Bonman, J. M. (2012). Pathogenic and genetic diversity of Xanthomonas translucens pv. undulosa in North Dakota. Phytopathology 102, 390–402. doi: 10.1094/PHYTO-07-11-0201
Adlung, N., Prochaska, H., Thieme, S., Banik, A., Blüher, D., John, P., et al. (2016). Non-host resistance induced by the Xanthomonas effector XopQ is widespread within the genus Nicotiana and functionally depends on EDS1. Front. Plant Sci. 7:1796. doi: 10.3389/fpls.2016.01796
Alfano, J. R., and Collmer, A. (1997). The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J. Bacteriol. 179, 5655–5662. doi: 10.1128/jb.179.18.5655-5662.1997
Antony, G., Zhou, J., Huang, S., Li, T., Liu, B., White, F., et al. (2010). Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3. Plant Cell 22, 3864–3876. doi: 10.1105/tpc.110.078964
Boch, J., and Bonas, U. (2010). Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu. Rev. Phytopathol. 48, 419–436. doi: 10.1146/annurev-phyto-080508-081936
Bogdanove, A. J., Beer, S. V., Bonas, U., Boucher, C. A., Collmer, A., Coplin, D. L., et al. (1996). Unified nomenclature for broadly conserved hrp genes of phytopathogenic bacteria. Mol. Microbiol. 20, 681–683. doi: 10.1046/j.1365-2958.1996.5731077.x
Bogdanove, A. J., Koebnik, R., Lu, H., Furutani, A., Angiuoli, S. V., Patil, P. B., et al. (2011). Two new complete genome sequences offer insight into host and tissue specificity of plant pathogenic Xanthomonas spp. J. Bacteriol. 193, 5450–5064. doi: 10.1128/JB.05262-11
Bragard, C., Singer, E., Alizadeh, A., Vauterin, L., Maraite, H., and Swings, J. (1997). Xanthomonas translucens from small grains: diversity and phytopathological relevance. Phytopathology 87, 1111–1117. doi: 10.1094/PHYTO.1997.87.11.1111
Büttner, D., and Bonas, U. (2010). Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol. Rev. 34, 107–133. doi: 10.1111/j.1574-6976.2009.00192.x
Cai, L., Cao, Y., Xu, Z., Ma, W., Zakria, M., Zou, L., et al. (2017). A transcription activator-like effector Tal7 of Xanthomonas oryzae pv. oryzicola activates rice gene Os09g29100 to suppress rice immunity. Sci. Rep. 7:5089. doi: 10.1038/s41598-017-04800-8
Cernadas, R. A., Doyle, E. L., Niño-Liu, D. O., Wilkins, K. E., Bancroft, T., Wang, L., et al. (2014). Code-assisted discovery of TAL effector targets in bacterial leaf streak of rice reveals contrast with bacterial blight and a novel susceptibility gene. PLoS Pathog. 10:e1003972. doi: 10.1371/journal.ppat.1003972
Chakrabarty, P., Chavhan, R., Ghosh, A., and Gabriel, D. (2010). Rapid and efficient protocols for throughput extraction of high quality plasmid DNA from strains of Xanthomonas axonopodis pv malvacearum and Escherichia coli. J. Plant Biochem. Biotechnol. 19, 99–102. doi: 10.1007/bf03323444
Chan, J. W., and Goodwin, P. H. (1999). The molecular genetics of virulence of Xanthomonas campestris. Biotechnol. Adv. 17, 489–508. doi: 10.1016/s0734-9750(99)00025-7
Chen, N. W., Serres-Giardi, L., Ruh, M., Briand, M., Bonneau, S., Darrasse, A., et al. (2018). Horizontal gene transfer plays a major role in the pathological convergence of Xanthomonas lineages on common bean. BMC Genomics 19:606. doi: 10.1186/s12864-018-4975-4
Cohn, M., Bart, R. S., Shybut, M., Dahlbeck, D., Gomez, M., Morbitzer, R., et al. (2014). Xanthomonas axonopodis virulence is promoted by a transcription activator-like effector–mediated induction of a SWEET sugar transporter in cassava. Mol. Plant Microbe Interact. 27, 1186–1198. doi: 10.1094/MPMI-06-14-0161-R
Cox, K. L., Meng, F., Wilkins, K. E., Li, F., Wang, P., Booher, N. J., et al. (2017). TAL effector driven induction of a SWEET gene confers susceptibility to bacterial blight of cotton. Nat. Commun. 8:15588. doi: 10.1038/ncomms15588
Cunfer, B., and Scolari, B. (1982). Xanthomonas campestris pv. translucens on tritcale and other small grains. Phytopathology 72, 683–686.
da Silva, A. R., Ferro, J. A., Reinach, F. D. C., Farah, C. S., Furlan, L. R., Quaggio, R. B., et al. (2002). Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417, 459–463.
de Lange, O., Wolf, C., Thiel, P., Krüger, J., Kleusch, C., Kohlbacher, O., et al. (2015). DNA-binding proteins from marine bacteria expand the known sequence diversity of TALE-like repeats. Nucleic Acids Res. 43, 10065–10080. doi: 10.1093/nar/gkv1053
Deb, S., Gupta, M. K., Patel, H. K., and Sonti, R. V. (2019). Xanthomonas oryzae pv. oryzae XopQ protein suppresses rice immune responses through interaction with two 14-3-3 proteins but its phospho-null mutant induces rice immune responses and interacts with another 14-3-3 protein. Mol. Plant Pathol. 20, 976–989. doi: 10.1111/mpp.12807
Duveiller, E., and Maraite, H. (1993). Study on yield loss due to Xanthomonas campestris pv. undulosa in wheat under high rainfall temperate conditions/Untersuchungen zur Ertragsreduktion durch Xanthomonas campestris pv. undulosa in Brotweizen unter gemäßigten Klimabedingungen mit hoher Niederschlagsmenge. Z. Pflanzenkr. Pflanzenschutz 100, 453–459.
Erkes, A., Reschke, M., Boch, J., and Grau, J. (2017). Evolution of transcription activator-like effectors in Xanthomonas oryzae. Genome Biol. Evol. 9, 1599–1615. doi: 10.1093/gbe/evx108
Falahi Charkhabi, N., Booher, N. J., Peng, Z., Wang, L., Rahimian, H., Shams-Bakhsh, M., et al. (2017). Complete genome sequencing and targeted mutagenesis reveal virulence contributions of Tal2 and Tal4b of Xanthomonas translucens pv. undulosa ICMP11055 in bacterial leaf streak of wheat. Front. Microbiol. 8:1488. doi: 10.3389/fmicb.2017.01488
Forster, R., and Schaad, N. (1988). Control of black chaff of wheat with seed treatment and a foundation seed health program. Plant Dis. 72, 935–938.
Grau, J., Reschke, M., Erkes, A., Streubel, J., Morgan, R. D., Wilson, G. G., et al. (2016). AnnoTALE: bioinformatics tools for identification, annotation, and nomenclature of TALEs from Xanthomonas genomic sequences. Sci. Rep. 6:21077. doi: 10.1038/srep21077
Gu, K., Yang, B., Tian, D., Wu, L., Wang, D., Sreekala, C., et al. (2005). R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435, 1122–1125. doi: 10.1038/nature03630
Gürlebeck, D., Thieme, F., and Bonas, U. (2006). Type III effector proteins from the plant pathogen Xanthomonas and their role in the interaction with the host plant. J. Plant Physiol. 163, 233–255. doi: 10.1016/j.jplph.2005.11.011
Hanahan, D., Jessee, J., and Bloom, F. R. (1991). Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 204, 63–113. doi: 10.1016/0076-6879(91)04006-a
Hauben, L., Vauterin, L., Swings, J., and Moore, E. (1997). Comparison of 16S ribosomal DNA sequences of all Xanthomonas species. Int. J. Syst. Evol. Microbiol. 47, 328–335. doi: 10.1099/00207713-47-2-328
Hersemann, L., Wibberg, D., Widmer, F., Vorhölter, F.-J., and Kölliker, R. (2016). Draft genome sequences of three Xanthomonas translucens pathovar reference strains (pv. arrhenatheri, pv. poae and pv. phlei) with different specificities for forage grasses. Stand. Genomic Sci. 11:50. doi: 10.1186/s40793-016-0170-x
Hu, Y., Zhang, J., Jia, H., Sosso, D., Li, T., Frommer, W. B., et al. (2014). Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. U.S.A. 111, E521–E529.
Ignatov, A. N., Kyrova, E. I., Vinogradova, S. V., Kamionskaya, A. M., Schaad, N. W., and Luster, D. G. (2015). Draft genome sequence of Xanthomonas arboricola strain 3004, a causal agent of bacterial disease on barley. Genome Announc. 3:e01572-14. doi: 10.1128/genomeA.01572-14
Ishikawa, K., Yamaguchi, K., Sakamoto, K., Yoshimura, S., Inoue, K., Tsuge, S., et al. (2014). Bacterial effector modulation of host E3 ligase activity suppresses PAMP-triggered immunity in rice. Nat. Commun. 5:5430. doi: 10.1038/ncomms6430
Jacobs, J. M., Pesce, C., Lefeuvre, P., and Koebnik, R. (2015). Comparative genomics of a cannabis pathogen reveals insight into the evolution of pathogenicity in Xanthomonas. Front. Plant Sci. 6:431. doi: 10.3389/fpls.2015.00431
Jaenicke, S., Bunk, B., Wibberg, D., Spröer, C., Hersemann, L., Blom, J., et al. (2016). Complete genome sequence of the barley pathogen Xanthomonas translucens pv. translucens DSM 18974T (ATCC 19319T). Genome Announc. 4:e01334-16. doi: 10.1128/genomeA.01334-16
Ji, Z., Ji, C., Liu, B., Zou, L., Chen, G., and Yang, B. (2016). Interfering TAL effectors of Xanthomonas oryzae neutralize R-gene-mediated plant disease resistance. Nat. Commun. 7:13435. doi: 10.1038/ncomms13435
Ji, Z.-Y., Xiong, L., Zou, L.-F., Li, Y.-R., Ma, W.-X., Liu, L., et al. (2014). AvrXa7-Xa7 mediated defense in rice can be suppressed by transcriptional activator-like effectors TAL6 and TAL11a from Xanthomonas oryzae pv. oryzicola. Mol. Plant Microbe Interact. 27, 983–995. doi: 10.1094/MPMI-09-13-0279-R
Kim, J.-G., Park, B. K., Yoo, C.-H., Jeon, E., Oh, J., and Hwang, I. (2003). Characterization of the Xanthomonas axonopodis pv. glycines Hrp pathogenicity island. J. Bacteriol. 185, 3155–3166. doi: 10.1128/jb.185.10.3155-3166.2003
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kumar, R., Soni, M., and Mondal, K. K. (2016). XopN-T3SS effector of Xanthomonas axonopodis pv. punicae localizes to the plasma membrane and modulates ROS accumulation events during blight pathogenesis in pomegranate. Microbiol. Res. 193, 111–120. doi: 10.1016/j.micres.2016.10.001
Langlois, P. A., Snelling, J., Hamilton, J. P., Bragard, C., Koebnik, R., Verdier, V., et al. (2017). Characterization of the Xanthomonas translucens complex using draft genomes, comparative genomics, phylogenetic analysis, and diagnostic LAMP assays. Phytopathology 107, 519–527. doi: 10.1094/PHYTO-08-16-0286-R
Li, L., Li, R.-F., Ming, Z.-H., Lu, G.-T., and Tang, J.-L. (2017). Identification of a novel type III secretion-associated outer membrane-bound protein from Xanthomonas campestris pv. campestris. Sci. Rep. 7:42724. doi: 10.1038/srep42724
Li, S., Wang, Y., Wang, S., Fang, A., Wang, J., Liu, L., et al. (2015). The type III effector AvrBs2 in Xanthomonas oryzae pv. oryzicola suppresses rice immunity and promotes disease development. Mol. Plant Microbe Interact. 28, 869–880. doi: 10.1094/MPMI-10-14-0314-R
Lorenz, C., Kirchner, O., Egler, M., Stuttmann, J., Bonas, U., and Büttner, D. (2008). HpaA from Xanthomonas is a regulator of type III secretion. Mol. Microbiol. 69, 344–360. doi: 10.1111/j.1365-2958.2008.06280.x
Ma, W., Zou, L., Ji, Z., Xu, X., Xu, Z., Yang, Y., et al. (2018). Xanthomonas oryzae pv. oryzae TALE proteins recruit OsTFIIAγ1 to compensate for the absence of OsTFIIAγ5 in bacterial blight in rice. Mol. Plant Pathol. 19, 2248–2262. doi: 10.1111/mpp.12696
Mak, A. N.-S., Bradley, P., Bogdanove, A. J., and Stoddard, B. L. (2013). TAL effectors: function, structure, engineering and applications. Curr. Opin. Struct. Biol. 23, 93–99. doi: 10.1016/j.sbi.2012.11.001
Marguerettaz, M., Pieretti, I., Gayral, P., Puig, J., Brin, C., Cociancich, S., et al. (2011). Genomic and evolutionary features of the SPI-1 type III secretion system that is present in Xanthomonas albilineans but is not essential for xylem colonization and symptom development of sugarcane leaf scald. Mol. Plant Microbe Interact. 24, 246–259. doi: 10.1094/MPMI-08-10-0188
Mehta, Y. (1990). Management of Xanthomonas campestris pv. undulosa and hordei through cereal seed testing. Seed Sci. Technol. 18, 467–476.
Miller, J. H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
Mondal, K. K., Verma, G., Junaid, A., and Mani, C. (2016). Rice pathogen Xanthomonas oryzae pv. oryzae employs inducible hrp-dependent XopF type III effector protein for its growth, pathogenicity and for suppression of PTI response to induce blight disease. Eur. J. Plant Pathol. 144, 311–323. doi: 10.1007/s10658-015-0768-7
Noël, L., Thieme, F., Nennstiel, D., and Bonas, U. (2002). Two novel type III-secreted proteins of Xanthomonas campestris pv. vesicatoria are encoded within the hrp pathogenicity island. J. Bacteriol. 184, 1340–1348. doi: 10.1128/jb.184.5.1340-1348.2002
Parkinson, N., Aritua, V., Heeney, J., Cowie, C., Bew, J., and Stead, D. (2007). Phylogenetic analysis of Xanthomonas species by comparison of partial gyrase B gene sequences. Int. J. Syst. Evol. Microbiol. 57, 2881–2887. doi: 10.1099/ijs.0.65220-0
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Peng, Z., Hu, Y., Xie, J., Potnis, N., Akhunova, A., Jones, J., et al. (2016). Long read and single molecule DNA sequencing simplifies genome assembly and TAL effector gene analysis of Xanthomonas translucens. BMC Genomics 17:21. doi: 10.1186/s12864-015-2348-9
Pérez-Quintero, A. L., Lamy, L., Gordon, J., Escalon, A., Cunnac, S., Szurek, B., et al. (2015). QueTAL: a suite of tools to classify and compare TAL effectors functionally and phylogenetically. Front. Plant Sci. 6:545. doi: 10.3389/fpls.2015.00545
Pesce, C., Bolot, S., Cunnac, S., Portier, P., Fischer-Le Saux, M., Jacques, M.-A., et al. (2015). High-quality draft genome sequence of the Xanthomonas translucens pv. cerealis pathotype strain CFBP 2541. Genome Announc. 3:e01574-14. doi: 10.1128/genomeA.01574-14
Pesce, C., Jacobs, J. M., Berthelot, E., Perret, M., Vancheva, T., Bragard, C., et al. (2017). Comparative genomics identifies a novel conserved protein, HpaT, in proteobacterial type III secretion systems that do not possess the putative translocon protein HrpF. Front. Microbiol. 8:1177. doi: 10.3389/fmicb.2017.01177
Pieretti, I., Royer, M., Barbe, V., Carrere, S., Koebnik, R., Cociancich, S., et al. (2009). The complete genome sequence of Xanthomonas albilineans provides new insights into the reductive genome evolution of the xylem-limited Xanthomonadaceae. BMC Genomics 10:616. doi: 10.1186/1471-2164-10-616
Qian, W., Jia, Y., Ren, S.-X., He, Y.-Q., Feng, J.-X., Lu, L.-F., et al. (2005). Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv. campestris. Genome Res. 15, 757–767. doi: 10.1101/gr.3378705
Qin, J., Zhou, X., Sun, L., Wang, K., Yang, F., Liao, H., et al. (2018). The Xanthomonas effector XopK harbours E3 ubiquitin-ligase activity that is required for virulence. New Phytol. 220, 219–231. doi: 10.1111/nph.15287
Read, A. C., Rinaldi, F. C., Hutin, M., He, Y.-Q., Triplett, L. R., and Bogdanove, A. J. (2016). Suppression of Xo1-mediated disease resistance in rice by a truncated, non-DNA-binding TAL effector of Xanthomonas oryzae. Front. Plant Sci. 7:1516. doi: 10.3389/fpls.2016.01516
Römer, P., Hahn, S., Jordan, T., Strauß, T., Bonas, U., and Lahaye, T. (2007). Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science 318, 645–648. doi: 10.1126/science.1144958
Ruh, M., Briand, M., Bonneau, S., Jacques, M.-A., and Chen, N. W. (2017b). Xanthomonas adaptation to common bean is associated with horizontal transfers of genes encoding TAL effectors. BMC Genomics 18:670. doi: 10.1186/s12864-017-4087-6
Ruh, M., Briand, M., Bonneau, S., Jacques, M.-A., and Chen, N. W. (2017a). First complete genome sequences of Xanthomonas citri pv. vignicola strains CFBP7111, CFBP7112, and CFBP7113 obtained using long-read technology. Genome Announc. 5:e00813-17. doi: 10.1128/genomeA.00813-17
Salanoubat, M., Genin, S., Artiguenave, F., Gouzy, J., Mangenot, S., Arlat, M., et al. (2002). Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415, 497–502.
Salzberg, S. L., Sommer, D. D., Schatz, M. C., Phillippy, A. M., Rabinowicz, P. D., Tsuge, S., et al. (2008). Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics 9:204. doi: 10.1186/1471-2164-9-204
Schandry, N., de Lange, O., Prior, P., and Lahaye, T. (2016). TALE-like effectors are an ancestral feature of the Ralstonia solanacearum species complex and converge in DNA targeting specificity. Front. Plant Sci. 7:1225. doi: 10.3389/fpls.2016.01225
Schulze, S., Kay, S., Büttner, D., Egler, M., Eschen-Lippold, L., Hause, G., et al. (2012). Analysis of new type III effectors from Xanthomonas uncovers XopB and XopS as suppressors of plant immunity. New Phytol. 195, 894–911. doi: 10.1111/j.1469-8137.2012.04210.x
Schwartz, A. R., Morbitzer, R., Lahaye, T., and Staskawicz, B. J. (2017). TALE-induced bHLH transcription factors that activate a pectate lyase contribute to water soaking in bacterial spot of tomato. Proc. Natl. Acad. Sci. U.S.A. 114, E897–E903. doi: 10.1073/pnas.1620407114
Shane, W., Baumer, J., and Teng, P. (1987). Crop losses caused by Xanthomonas streak on spring wheat and barley. Plant Dis. 71, 927–930.
Siguier, P., Pérochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, (Suppl. 1), D32–D36.
Singer, A. U., Schulze, S., Skarina, T., Xu, X., Cui, H., Eschen-Lippold, L., et al. (2013). A pathogen type III effector with a novel E3 ubiquitin ligase architecture. PLoS Pathog. 9:e1003121. doi: 10.1371/journal.ppat.1003121
Song, C., and Yang, B. (2010). Mutagenesis of 18 type III effectors reveals virulence function of XopZPXO99 in Xanthomonas oryzae pv. oryzae. Mol. Plant Microbe Interact. 23, 893–902. doi: 10.1094/MPMI-23-7-0893
Soni, M., and Mondal, K. K. (2018). Xanthomonas axonopodis pv. punicae uses XopL effector to suppress pomegranate immunity. J. Integr. Plant Biol. 60, 341–357. doi: 10.1111/jipb.12615
Streubel, J., Pesce, C., Hutin, M., Koebnik, R., Boch, J., and Szurek, B. (2013). Five phylogenetically close rice SWEET genes confer TAL effector-mediated susceptibility to Xanthomonas oryzae pv. oryzae. New Phytol. 200, 808–819. doi: 10.1111/nph.12411
Studholme, D. J., Wasukira, A., Paszkiewicz, K., Aritua, V., Thwaites, R., Smith, J., et al. (2011). Draft genome sequences of Xanthomonas sacchari and two banana-associated xanthomonads reveal insights into the Xanthomonas group 1 clade. Genes 2, 1050–1065. doi: 10.3390/genes2041050
Sugio, A., Yang, B., Zhu, T., and White, F. F. (2007). Two type III effector genes of Xanthomonas oryzae pv. oryzae control the induction of the host genes OsTFIIAγ1 and OsTFX1 during bacterial blight of rice. Proc. Natl. Acad. Sci. U.S.A. 104, 10720–10725. doi: 10.1073/pnas.0701742104
Swords, K., Dahlbeck, D., Kearney, B., Roy, M., and Staskawicz, B. J. (1996). Spontaneous and induced mutations in a single open reading frame alter both virulence and avirulence in Xanthomonas campestris pv. vesicatoria avrBs2. J. Bacteriol. 178, 4661–4669. doi: 10.1128/jb.178.15.4661-4669.1996
Tampakaki, A. P., Skandalis, N., Gazi, A. D., Bastaki, M. N., Panagiotis, F. S., Charova, S. N., et al. (2010). Playing the “Harp”: evolution of our understanding of hrp/hrc genes. Annu. Rev. Phytopathol. 48, 347–370. doi: 10.1146/annurev-phyto-073009-114407
Teper, D., Burstein, D., Salomon, D., Gershovitz, M., Pupko, T., and Sessa, G. (2016). Identification of novel Xanthomonas euvesicatoria type III effector proteins by a machine-learning approach. Mol. Plant Pathol. 17, 398–411. doi: 10.1111/mpp.12288
Thieme, F., Koebnik, R., Bekel, T., Berger, C., Boch, J., Büttner, D., et al. (2005). Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J. Bacteriol. 187, 7254–7266. doi: 10.1128/jb.187.21.7254-7266.2005
Tian, D., Wang, J., Zeng, X., Gu, K., Qiu, C., Yang, X., et al. (2014). The rice TAL effector–dependent resistance protein XA10 triggers cell death and calcium depletion in the endoplasmic reticulum. Plant Cell 26, 497–515. doi: 10.1105/tpc.113.119255
Wang, C., Zhang, X., Fan, Y., Gao, Y., Zhu, Q., Zheng, C., et al. (2015). XA23 is an executor R protein and confers broad-spectrum disease resistance in rice. Mol. Plant 8, 290–302. doi: 10.1016/j.molp.2014.10.010
Wasukira, A., Tayebwa, J., Thwaites, R., Paszkiewicz, K., Aritua, V., Kubiriba, J., et al. (2012). Genome-wide sequencing reveals two major sub-lineages in the genetically monomorphic pathogen Xanthomonas campestris pathovar musacearum. Genes 3, 361–377. doi: 10.3390/genes3030361
White, F. F., Potnis, N., Jones, J. B., and Koebnik, R. (2009). The type III effectors of Xanthomonas. Mol. Plant Pathol. 10, 749–766. doi: 10.1111/j.1364-3703.2009.00590.x
Wichmann, F., Vorhölter, F. J., Hersemann, L., Widmer, F., Blom, J., Niehaus, K., et al. (2013). The noncanonical type III secretion system of Xanthomonas translucens pv. graminis is essential for forage grass infection. Mol. Plant Pathol. 14, 576–588. doi: 10.1111/mpp.12030
Wichmann, G., and Bergelson, J. (2004). Effector genes of Xanthamonas axonopodis pv. vesicatoria promote transmission and enhance other fitness traits in the field. Genetics 166, 693–706. doi: 10.1534/genetics.166.2.693
Wilkins, K. E., Booher, N. J., Wang, L., and Bogdanove, A. J. (2015). TAL effectors and activation of predicted host targets distinguish Asian from African strains of the rice pathogen Xanthomonas oryzae pv. oryzicola while strict conservation suggests universal importance of five TAL effectors. Front. Plant Sci. 6:536. doi: 10.3389/fpls.2015.00536
Yamaguchi, K., Nakamura, Y., Ishikawa, K., Yoshimura, Y., Tsuge, S., and Kawasaki, T. (2013a). Suppression of rice immunity by Xanthomonas oryzae type III effector Xoo 2875. Biosci. Biotechnol. Biochem. 77, 796–801.
Yamaguchi, K., Yamada, K., Ishikawa, K., Yoshimura, S., Hayashi, N., Uchihashi, K., et al. (2013b). A receptor-like cytoplasmic kinase targeted by a plant pathogen effector is directly phosphorylated by the chitin receptor and mediates rice immunity. Cell Host Microbe 13, 347–357. doi: 10.1016/j.chom.2013.02.007
Yang, B., Sugio, A., and White, F. F. (2006). Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. U.S.A. 103, 10503–10508. doi: 10.1073/pnas.0604088103
Yang, B., Zhu, W., Johnson, L. B., and White, F. F. (2000). The virulence factor AvrXa7 of Xanthomonas oryzae pv. oryzae is a type III secretion pathway-dependent nuclear-localized double-stranded DNA-binding protein. Proc. Natl. Acad. Sci. U.S.A. 97, 9807–9812. doi: 10.1073/pnas.170286897
Yang, Y. (1994). Host-specific symptoms and increassed release of Xanthomonas citri and X. campestris pv. malvacearum from leaves are determined by the 102-bp tandem repeats of pthA and avrb6, respectively. Mol. Plant Microbe Interact. 7, 345–355.
Young, J., Park, D.-C., Shearman, H., and Fargier, E. (2008). A multilocus sequence analysis of the genus Xanthomonas. Syst. Appl. Microbiol. 31, 366–377. doi: 10.1016/j.syapm.2008.06.004
Zhao, S., Mo, W.-L., Wu, F., Tang, W., Tang, J.-L., Szurek, B., et al. (2013). Identification of non-TAL effectors in Xanthomonas oryzae pv. oryzae Chinese strain 13751 and analysis of their role in the bacterial virulence. World J. Microbiol. Biotechnol. 29, 733–744. doi: 10.1007/s11274-012-1229-5
Zhou, J., Peng, Z., Long, J., Sosso, D., Liu, B., Eom, J. S., et al. (2015). Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 82, 632–643. doi: 10.1111/tpj.12838
Zhu, B., Zhou, S., Lou, M., Zhu, J., Li, B., Xie, G., et al. (2011). Characterization and inference of gene gain/loss along burkholderia evolutionary history. Evol. Bioinform. Online 7, 191–200. doi: 10.4137/EBO.S7510
Keywords: bacterial leaf streak, Xanthomonas translucens pv. cerealis, SMRT sequencing, TALE, T3SS, virulence
Citation: Shah SMA, Haq F, Ma W, Xu X, Wang S, Xu Z, Zou L, Zhu B and Chen G (2019) Tal1NXtc01 in Xanthomonas translucens pv. cerealis Contributes to Virulence in Bacterial Leaf Streak of Wheat. Front. Microbiol. 10:2040. doi: 10.3389/fmicb.2019.02040
Received: 18 April 2019; Accepted: 19 August 2019;
Published: 04 September 2019.
Edited by:
Jens Staal, Flanders Institute for Biotechnology, BelgiumReviewed by:
Boris Szurek, Institut de Recherche pour le Développement, FranceLaurent D. Noël, Centre National de la Recherche Scientifique (CNRS), France
Copyright © 2019 Shah, Haq, Ma, Xu, Wang, Xu, Zou, Zhu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gongyou Chen, Z3lvdWNoZW5Ac2p0dS5lZHUuY24=
 Syed Mashab Ali Shah
Syed Mashab Ali Shah Fazal Haq
Fazal Haq Wenxiu Ma
Wenxiu Ma Xiameng Xu
Xiameng Xu Sai Wang
Sai Wang Zhengyin Xu
Zhengyin Xu Lifang Zou
Lifang Zou Bo Zhu
Bo Zhu Gongyou Chen
Gongyou Chen