
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 04 September 2019
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.02030
This article is part of the Research Topic "One Health" Approach For Revealing Reservoirs And Transmission Of Antimicrobial Resistance View all 27 articles
 Dandan Dong1,2,3†
Dandan Dong1,2,3† Manli Li3,4†‡
Manli Li3,4†‡ Zhenzhen Liu1,2
Zhenzhen Liu1,2 Jiantao Feng1,2
Jiantao Feng1,2 Nan Jia1
Nan Jia1 Hui Zhao1
Hui Zhao1 Baohua Zhao4
Baohua Zhao4 Tingting Zhou1
Tingting Zhou1 Xianglilan Zhang3*
Xianglilan Zhang3* Yigang Tong3,5*
Yigang Tong3,5* Yuanqi Zhu1,2*
Yuanqi Zhu1,2*Acquisition of the blaNDM–1 gene by Proteus mirabilis is a concern because it already has intrinsic resistance to polymyxin E and tigecycline antibiotics. Here, we describe a P. mirabilis isolate that carries a pPrY2001-like plasmid (pHFK418-NDM) containing a blaNDM–1 gene. The pPrY2001-like plasmid, pHFK418-NDM, was first reported in China. The pHFK418-NDM plasmid was sequenced using a hybrid approach based on Illumina and MinION platforms. The sequence of pHFK418-NDM was compared with those of the six other pPrY2001-like plasmids deposited in GenBank. We found that the multidrug-resistance encoding region of pHFK418-NDM contains ΔTn10 and a novel transposon Tn6625. Tn6625 consists of ΔTn1696, Tn6260, In251, ΔTn125 (carrying blaNDM–1), ΔTn2670, and a novel mph(E)-harboring transposon Tn6624. In251 was first identified in a clinical isolate, suggesting that it has been transferred efficiently from environmental organisms to clinical isolates. Genomic comparisons of all these pPrY2001-like plasmids showed that their relatively conserved backbones could integrate the numerous and various accessory modules carrying multifarious antibiotic resistance genes. Our results provide a greater depth of insight into the horizontal transfer of resistance genes and add interpretive value to the genomic diversity and evolution of pPrY2001-like plasmids.
Urinary tract infections (UTIs) are the most common bacterial infections (Gastmeier et al., 1998). Cases of UTIs can be classified as uncomplicated or complicated (Beahm et al., 2017). Clinically, Proteus mirabilis is most frequently a pathogen of UTIs, particularly in patients suffering from complicated cUTIs (Schaffer and Pearson, 2015). Although Escherichia coli is the primary urinary tract pathogen, P. mirabilis ranks third as the cause of UTIs and accounts for 4.1% of urinary tract infection isolates in CANWARD surveillance study, 4.6% in southern China, respectively (Karlowsky et al., 2011; Li et al., 2017). Because this pathogen is intrinsically resistant to nitrofurantoin, polymyxin, and tigecycline antibiotics (Ramos et al., 2018), acquiring additional carbapenemase antibiotic resistance is worrisome (Reffert and Smith, 2014). Currently, fosfomycin, which is previously used mainly as oral treatment for UTIs, has gained clinicians’ attention worldwide because of its activity against multidrug-resistant bacteria (Reffert and Smith, 2014; Giske, 2015). In additionally, fosfomycin resistance rates are generally low but substantially higher when carbapenemase producers are considered (Giske, 2015). One such resistance gene is blaNDM–1 (New Delhi metallo-β-lactamase), which was initially identified in a Klebsiella pneumoniae strain (Yong et al., 2009). Isolates of this species that harbor the blaNDM–1 gene can hydrolyze nearly all β-lactam antibiotics except aztreonam. Therefore, the acquisition of blaNDM–1 by P. mirabilis would be problematic, as it would greatly reduce the therapeutic options for treating infections caused by it.
The blaNDM–1 gene is mainly and widely spread by an ISAba125-bounded composite transposon Tn125 (Poirel et al., 2012; Ranjan et al., 2016), and blaNDM–1-carrying plasmids are commonly found in IncA (Solgi et al., 2017), IncC (Harmer and Hall, 2017), IncT (Mataseje et al., 2016), IncR (Gamal et al., 2016), IncFII (Lin et al., 2016), IncX (Wang et al., 2018), and IncN (Wang et al., 2018) incompatible groups. However, blaNDM–1-carrying plasmids have gradually appeared in some unknown incompatibility groups. The blaNDM–1-harboring pHFK418-NDM plasmid and six other plasmids have been assigned into the same unknown incompatibility group based on their replicons. The six plasmids are pPrY2001 (Accession no. KF295828) (Mataseje et al., 2014), p06-1619-1 (Accession no. KX832929) (Marquez-Ortiz et al., 2017), p16Pre36-NDM (Accession no. KX832927) (Marquez-Ortiz et al., 2017), pPp47 (Accession no. MG516912) (Dolejska et al., 2018), pPm60 (Accession no. MG516911) (Dolejska et al., 2018), and pC131 (Accession no. KX774387). The earliest reported plasmid, pPrY2001, is considered to be the reference plasmid, so the above-named plasmids are called pPrY2001-like plasmids (Marquez-Ortiz et al., 2017; Dolejska et al., 2018). Up to now, no studies in the published scientific literature have thoroughly analyzed and compared in detail the structures and genomes of this unknown incompatibility group.
Here, we studied the blaNDM–1-harboring plasmid, pHFK418-NDM, a known pPrY2001-like plasmid according to its replicon, which was first isolated from a clinical P. mirabilis HFK418 strain in China. We elucidated the complete sequence of pHFK418-NDM (which carries two novel transposons, Tn6624 and Tn6625) and compared it with six other pPrY2001-like plasmids to obtain insight into the horizontal transfer of resistance genes and the diversity and evolution of pPrY2001-like plasmids.
The study was approved by the Medical Ethics Committee at the Affiliated Hospital of Qingdao University, China, and written informed consent was received from the patient. The P. mirabilis HFK418 strain was isolated from the urine specimen of a patient with epidemic encephalitis at the Affiliated Hospital of Qingdao University, China, in 2017. Referring to the method described in Ranjan et al. (2016), this strain was multiple tested for purity by routine laboratory methods, then the pure strain was cryopreserved at −80°C in 50% glycerol. The pure isolate was revived in Luria-Bertani (LB) broth (BD Biosciences, United States) with 4 μg/ml meropenem to experiments. The P. mirabilis HFK418 isolate was identified and subjected to antimicrobial susceptibility testing using the VITEK compact-2 automated system (bioMérieux, France). In addition, fosfomycin MICs were further determined by fosfomycin E-tests (bioMérieux). CLSI (Clinical and Laboratory Standards Institute) 2018 breakpoints were used (M100-S28) (CLSI, 2018).
The major acquired extended-spectrum β-lactamase (Dallenne et al., 2010; Hussain et al., 2014; Ranjan et al., 2017), fosfomycin (Dantas Palmeira et al., 2018), chloramphenicol (White et al., 1999), lincosamide (Garcia-Martin et al., 2018), and carbapenemase genes (Chen et al., 2015; Ranjan et al., 2016, 2017) were detected by PCR, after which all the PCR amplicons were sequenced on the ABI 3730 platform (Applied Biosystems, United States). The sodium azide-resistant E. coli J53AziR strain was used as the recipient and the P. mirabilis HFK418 isolate as the donor for the conjugative transfer of the plasmids. The conjugal transfer tests were performed as described previously (Srijan et al., 2018), and the conjugation frequency was calculated as transconjugants divided by number of donors.
To determine whether the blaNDM–1 gene was expressed in both P. mirabilis HFK418 and the E. coli J53AziR transconjugant HFK418-NDM-J53 strain, we performed an imipenem–EDTA E-test (AB-BioMérieux, Sweden) to assess the class B carbapenemase activity.
Bacterial genomic DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega, United States), followed by the MiSeq (Illumina, United States) and MinION (Oxford Nanopore, United Kingdom) sequencing. The short Illumina reads were trimmed to remove the poor quality sequences, and the resultant contigs were assembled using Newbler3.0 (Nederbragt, 2014). The longest single read obtained by the MinION sequencer was 98 kb, thereby crossing the repetitive shufflon regions in the plasmid (Laver et al., 2015). The long reads from the MinION combined with the short Illumina reads were hybrid assembled using SPAdesv3.11.1 (Bankevich et al., 2012). Hybrid assembly produced several scaffolds and BLASTN analysis confirmed that the scaffold in our study has the highest similarity to the plasmid p16Pre36-NDM (Accession no. KX832927) with coverage of 69% and identity of 96%. As most of the published plasmids are in a circle form, further bioinformatics analysis confirmed that this scaffold can be successfully cyclized using our in-house script. The correctness was then proved by mapping the high-throughput sequencing reads to the cyclized scaffold using CLC Genomics Workbench 9.0, with a mean reads mapping coverage of 111x. The consensus sequence acquired from CLC Genomics Workbench 9.0 was finally treated as the complete sequence of our plasmid pHFK418-NDM.
Open reading frames (ORFs) and pseudogenes that were predicted by RAST2.0 (Brettin et al., 2015) were further annotated using BLASTP/BLASTN (Boratyn et al., 2013) against the RefSeq databases (O’Leary et al., 2016) and UniProtKB/Swiss-Prot (Boutet et al., 2016). Mobile elements, resistance genes, and other features were annotated by INTEGRALL (Moura et al., 2009), ISfinder (Siguier et al., 2006), ResFinder (Kleinheinz et al., 2014), PlasmidFinder (Carattoli et al., 2014), and the Tn Number Registry (Roberts et al., 2008) online databases. Comparisons of the multiple and paired sequences were conducted using MUSCLE 3.8.31 and BLASTN, respectively. Gene organization diagrams were drawn in Inkscape0.48.11.
The complete nucleotide sequence of plasmid pHFK418-NDM has been deposited in the National Center for Biotechnology Information nucleotide database2 under accession number MH491967.
Plasmid pHFK418-NDM from P. mirabilis HFK418 was transferable to E. coli J53AziR in the conjugation experiments, thereby generating the blaNDM–1-positive E. coli J53AziR transconjugant HFK418-NDM-J53 strain. The conjugation frequency was 1.5 × 10–2.
Imipenem-EDTA E-tests were positive in both P. mirabilis HFK418 and HFK418-NDM-J53. These two strains were highly resistant to ampicillin, cefazolin, cefuroxime, ceftazidime, ceftriaxone, imipenem, and meropenem, but not to aztreonam, revealing that pHFK418-NDM is a conjugative NDM-encoding plasmid with carbapenemase activity (Table 1 and Supplementary Figure S1).
PCR screening for antimicrobial resistance genes showed that P. mirabilis HFK418 carries blaNDM–1, blaOXA–1, blaCTX–M–65, fosA3, catB5, lnu(G), and blaOXA–10 genes. The complete sequence of pHFK418-NDM is 145,619 bp with a mean G + C content of 42.8%, and 157 ORFs (Table 2 and Supplementary Figure S2). Based on the replicon, pHFK418-NDM was assigned into the unknown incompatibility group of pPrY2001-like plasmids. The linear genomic comparison conducted between pHFK418-NDM and six other pPrY2001-like plasmids [pPrY2001 (Mataseje et al., 2014), p06-1619-1 (Marquez-Ortiz et al., 2017), pC131, pPp47 (Dolejska et al., 2018), pPm60 (Dolejska et al., 2018), and p16Pre36-NDM (Marquez-Ortiz et al., 2017)] showed that the highest sequence homology belonged to pHFK418-NDM with >69% query coverage and >99% identity (Supplementary Figure S3 and Supplementary Data Sheet S2).
The genomic structures of the pPrY2001-like plasmids comprised two major regions: the backbone and accessory module. The backbone could be further divided into three parts: the replication genes (repA and its iterons), the conjugal transfer genes (tiv, rlx, and cpl), and the plasmid maintenance genes (parFG, MazFE, stbB, ssb, and flhC). Each plasmid’s backbone was able to integrate two or more accessory modules by transposition or recombination events. pHFK418-NDM contains two accessory modules, the Tn6901 related region and the multidrug-resistant (MDR) region, while the MDR region contains Tn6625 and ΔTn10 (Supplementary Figures S2, S3).
Our pairwise comparison analysis of the pPrY2001-like plasmids backbones showed that they shared >96% nucleotide identity across >42%, indicating that their backbones were relatively conserved. However, there were three major differences among all their backbones. (I) the parC gene (centromere, binding sites for parG) did not exist in pPrY2001, and the copy numbers of the 8-bp tandem repeat (TGTGTata) within the parC gene varied among the other plasmids (4 for p06-1619-1, pC131, and pPm60; 5 for pPp47, pHFK418-NDM, and p16Pre36-NDM). (II) Compared with the conjugal transfer region in the other plasmids, the rlx gene from pPrY2001 is disrupted into Δrlx-3′ and Δrlx-5′ by insertion of ISPrre5 (named in this study). (III) The hybrid backbone of plasmid p16Pre36-NDM was acquired from a pPrY2001-like plasmid and the IncC2 plasmid (the orf1847 and rhs2 marked genes) (Supplementary Figure S3).
We found that the blaNDM–1-carrying ΔTn125 transposon is present in the MDR region of pHFK418-NDM, p16Pre36-NDM (the MDR region-2), and pPrY2001. Tn125, an ISAba125-bounded composite transposon in plasmid pNDM-BJ01, was acquired from Acinetobacter lwoffii (Poirel et al., 2012). It is made up of ISAba125, blaNDM–1, bleMBL (bleomycin resistance), trpF, dsbD, cutA, groES, groEL and ISCR21, and is bordered by 3-bp direct repeats (DRs: target site duplication signals for transposition). In the MDR region of these three plasmids, ΔTn125 has undergone the deletion of ISAba125 downstream of ISCR27. In addition, ΔTn125 from pHFK418-NDM and p16Pre36-NDM contain the following differences: ΔTn125 in pHFK418-NDM has a ΔdsbD–trpF–bleMBL–blaNDM–1–ISAba125 structure, while the ISCR21–groEL–groES–cutA–dsbD fragment, which occurs upstream of blaNDM–1 in p16Pre36-NDM, was generated by complex recombination events (Figure 1A).
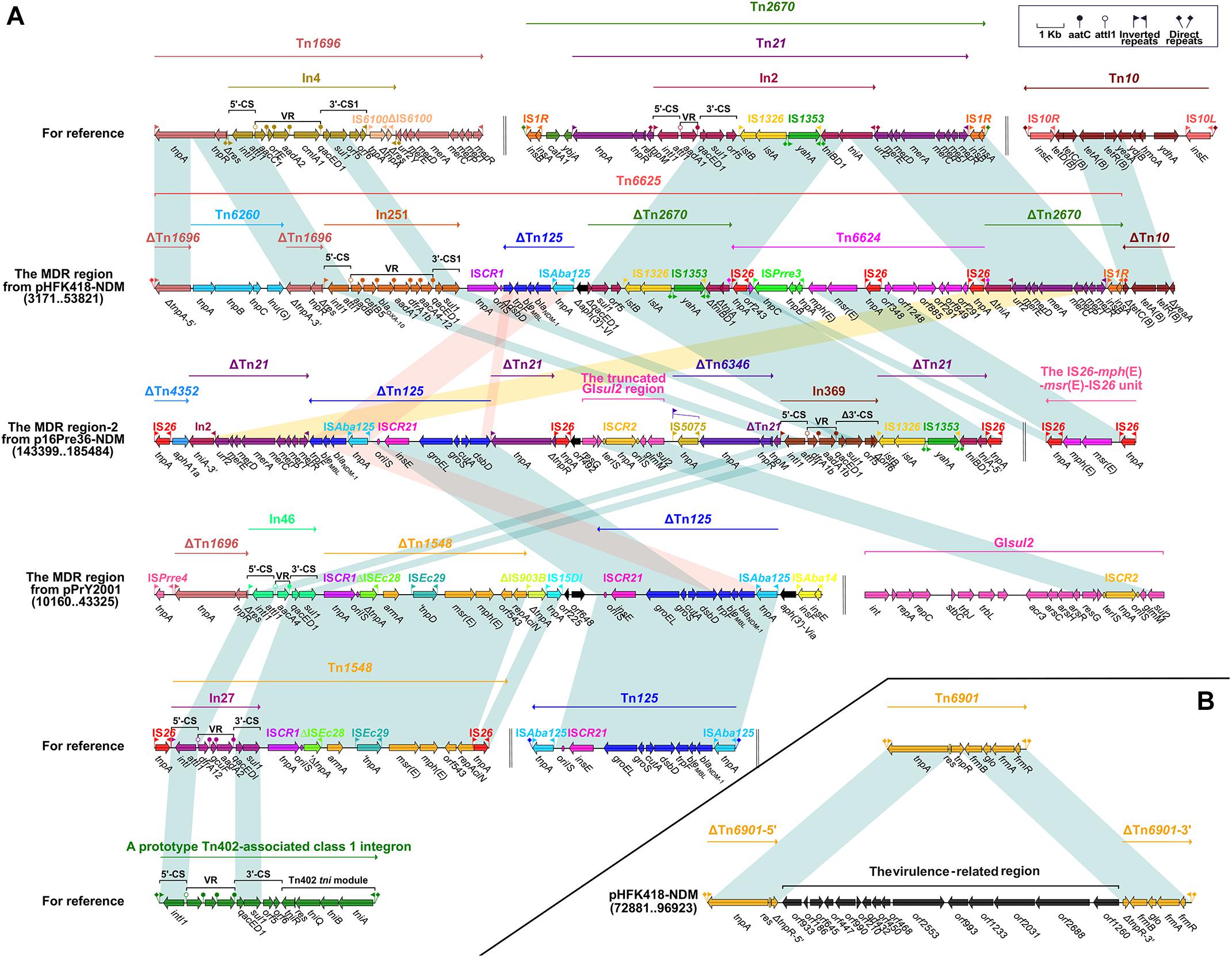
Figure 1. The MDR region harboring blaNDM–1 of pPrY2001-like plasmids and comparison with related regions (A). Accessory module outside the MDR region of pHFK418-NDM (B). Genes, mobile elements and other features are colored based on function classification. Numbers in parentheses show the nucleotide positions within the corresponding plasmids. Shading regions denote shared DNA regions of homology (>95% nucleotide identity). MDR: multidrug-resistant.
Integron In251, which is located upstream of ΔTn125 in pHFK418-NDM, belongs to the prototypic Tn402-associated class 1 integron. This class 1 integron can be divided sequentially into an IRi (inverted repeat at the integrase end), a 5′-conserved segment (5′-CS: intI1–attI1), a variable region (VR: containing one or more resistant genes), a 3′-conserved segment (3’-CS: qacED1–sul1–orf5–orf6), the Tn402 tni module (tniA–tniB–tniQ–res–tniR) and IRt (inverted repeat at the tni end), and is surrounded by 5-bp DRs. Furthermore, In369 (in MDR region-2 from p16Pre36-NDM), In46 (in the MDR region from pPrY2001), In809 (in the MDR region-1 from pPm60), and In1129 (in the MDR region-1 from p16Pre36-NDM) are also different derivatives from the prototypical Tn402-associated class 1 integron. The structures of In251, In369, and In46 are arranged as IRi, 5′-CS, VR (aadB–catB5–blaOXA–10–aadA1–dfrA1–aacA4-12 in In251, dfrA1b–aadA1b in In369, and aacA4 in In46), and Δ3′-CS (qacED1–sulI in In251and In46, qacED1–sulI–orf5–Δorf6 in In369), without the Tn402 tni module and IRt. The Tn402 tni module and IRt have been replaced downstream by other mobile elements. In809 and In1129 each have the following common structure: IRi, 5′-CS, VR, 3′-CS, and IRt, and their Tn402 tni module has been lost during the evolutionary process. A difference between In809 and In1129 is apparent in the variable region (dfrA1–aadA27c in In1129, blaIMP–4–qacG2–aacA4′–catB3 in In809). ΔTn1696 is embedded upstream of the class 1 integrons In251, In46, In809, and In1460 (in the MDR region-1 from pPp47). The Tn1696 prototype comprises an IRL (inverted repeat left)–tnpA (transposase)–tnpR (resolvase)–res (resolution site)–mer (mercury resistance)–IRR (inverted repeat right) structure, and a res site is interrupted by insertion of In4 into 75-bp Δres-5′ and 45-bp Δres-3′. Compared with the structure of Tn1696, ΔTn1696 has the same IRL–tnpA–tnpR–Δres-5′ module in the MDR region of pHFK418-NDM, pPrY2001, pPm60, and pPp47. The ΔTn1696 tnpA from pHFK418-NDM and pPm60 is segmented into two fragments, ΔtnpA-5′ and ΔtnpA-3′, by insertion of Tn6260. Belonging to the Tn554 family, Tn6260 consists of tnpA, tnpB, tnpC, and lnu(G) (lincosamide resistance), as identified in Enterococcus thailandicus a523 (Ybazeta et al., 2017), Virgibacillus halodenitrificans PDB-F2 (Tao et al., 2016), and E. faecalis E531 (Zhu et al., 2017). Up until now, Tn6260 only appeared in pPrY2001-like plasmids when pHFK418-NDM and pPm60 were present. Moreover, ISPmi3 split tnpB of Tn6260 from pPm60 into two parts, ΔtnpB-5′ and ΔtnpB-3′, which are surrounded by 8-bp DRs (Figures 1A, 2).
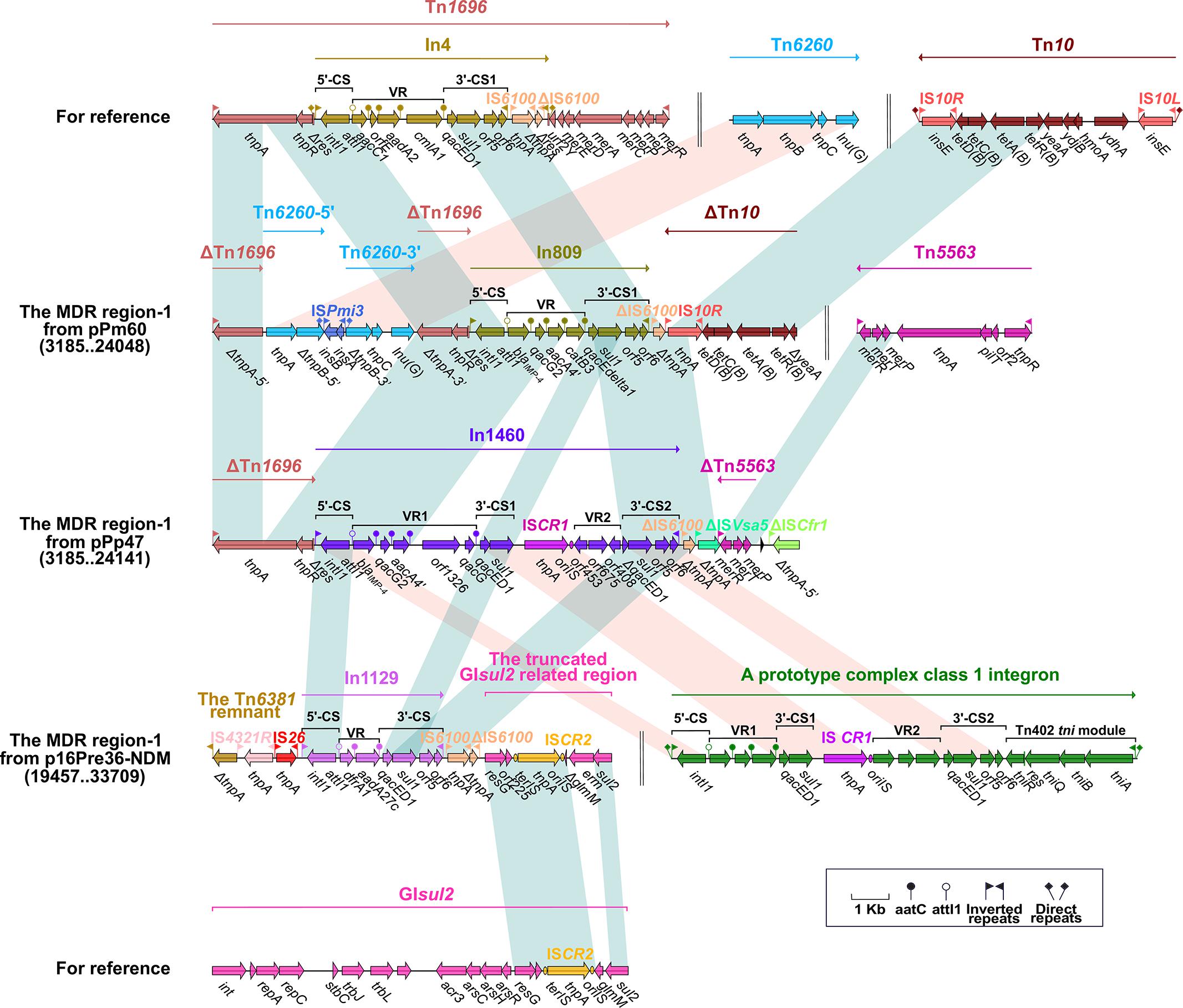
Figure 2. The MDR region carrying blaIMP–4 and comparison with related regions. Genes are denoted by arrows. Mobile elements, genes, and other features are colored based on function classification. Numbers in parentheses denote the nucleotide positions within the corresponding plasmids. Shading regions show shared DNA regions of homology (>95% nucleotide identity). MDR: multidrug-resistant.
ΔTn2670 from pHFK418-NDM is integrated downstream of ΔTn125. Flanked by 9-bp DRs, Tn2670 is organized as IS1R, catA1 (chloramphenicol resistance), ybjA (acetyl transferase), Tn21, and IS1R, and was initially discovered in plasmid R100 from Shigella flexneri (Partridge and Hall, 2004). Tn21, a Tn3-family transposon unit, contains an IRL–tnpA–tnpR–res–tnpM (modulator protein)–In2–urf2–the mer operon–IRR module, and a presumed ancestral urf2M gene is interrupted by insertion of In2 to generate tnpM and urf2 (Liebert et al., 1999). In2 comprises IRi, 5′-CS, VR (aadA1), 3′-CS, IS1326, IS1353, the tni module, and IRt, and is delimited by 5-bp DRs. In terms of the structure of Tn2670, ΔTn21 can be divided into four segments in the MDR region from p16Pre36-NDM; namely, (I) IRL, tnpA, and ΔtnpR, (II) tnpM, (III), In2 (IS1326, IS1353, the disrupted tni module), and (IV), In2 (the disrupted tni module and IRt), urf2, the mer operon, and IRR. These four segments fall within different positions by virtue of transposition or recombination events. In pHFK418-NDM, ΔTn2670 reserves a fragment from the 3′-CS of In2 to IS1R, but its tniA gene is segmented into two fragments (ΔtniAIn2-5′ and ΔtniAIn2-3′) by insertion of Tn6624 (Figure 1A).
Tn6624, a novel IS26-based transposon unit, has been inserted into the pHFK418-NDM plasmid from P. mirabilis HFK418. Delimited by 8-bp DRs (CATCGGCG), it has the following mosaic structure: IS26, a novel IS66-family ISPrre3, mph(E) (macrolide resistance), msr(E) (macrolide efflux protein), IS26, a fragment with an unknown function, and IS26. The mph(E)–msr(E)–IS26 fragment originated from the IS26–mph(E)–msr(E)–IS26 transposon unit and was initially identified in the chromosomal integrative conjugative element from Pasteurella multocida (Michael et al., 2012). Three copies of IS26 are present in Tn6624, which promotes the formation and transposition of Tn6624. Another novel 48,068 bp multidrug resistance transposon, Tn6625, was found in the pHFK418-NDM plasmid from P. mirabilis HFK418. The ΔTn1696, Tn6260, In251, ΔTn125, Tn6624, and ΔTn2670 mobile elements have been described in detail above, and all of them are included in the large composite Tn6625 transposon. Tn6625 carries twelve resistance genes, bounded by 3-bp DRs (TTG). Tn6625 contains integron In25, which has so far only been found in wastewater-isolated Providencia VIGAT3 (Guo et al., 2011). Thus, In251 was first isolated from clinical P. mirabilis HFK418, suggesting that it has been efficiently transferred from environmental micro-organisms to clinical isolates (Figure 1A).
The MDR region of pHFK418-NDM includes Tn6625 and ΔTn10. Delimited by 9-bp DRs, Tn10 is arranged sequentially as IS10L, ydhA, hmoA, ydjB, yeaA, tetR, tetA (tetracycline resistance), tetC, tetD, and IS10R, as identified in the conjugative R27 plasmid from Salmonella typhi (Lawley et al., 2000). ΔTn10 was found in the MDR region of pHFK418-NDM, pPp47, and pPm60, and comprises a common fragment (tetD–tetC–tetA–tetR–ΔyeaA). But IS10R is absent in pHFK418-NDM, truncated in pPp47, and intact in pPm60. Tn10 is also integrated between orf153 and orf489 in the backbone of p16Pre36-NDM, bracketed by 9-bp DRs. Tn10 is an integral transposon in p16Pre36-NDM, but its IS10R has two segments (ΔIS10R-5′ and ΔIS10R-3′) and is disrupted by insertion of ISKpn26 with 4-bp DRs (Figures 1A, 2, 3).
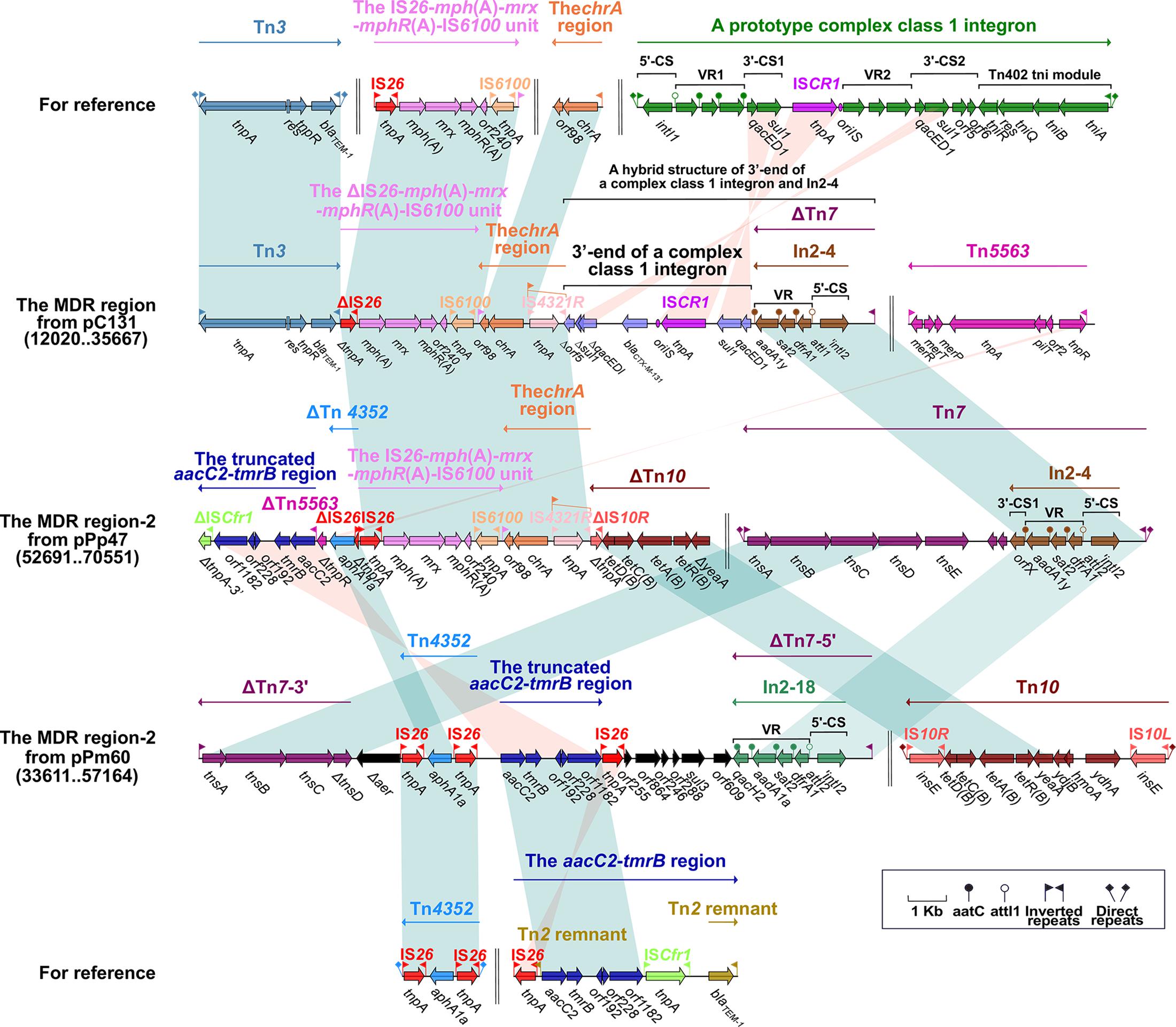
Figure 3. The MDR region harboring class 2 integron and comparison with related regions. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on function classification. Numbers in parentheses denote the nucleotide positions within the corresponding plasmids. Shading regions indicate shared DNA regions of homology (>95% nucleotide identity). MDR: multidrug-resistant.
There are other transposon units also (ΔTn6346, the truncated GIsul2 region, and ΔTn1548) in the MDR region of p16Pre36-NDM and pPrY2001, except as described above. ΔTn6346 and the truncated GIsul2 region are embedded in the MDR region of p16Pre36-NDM. Tn6346, a Tn3-family transposon, was discovered in heavy metal-tolerant Achromobacter AO22 (Ng et al., 2009). In the MDR region-2 of p16Pre36-NDM, ΔTn6346 is arranged in turn as the IS5075 interrupted-IRL, tnpA, tnpR, and 121-bp Δres, and the lost mer operon and IRR were replaced by tnpM from ΔTn21. GIsul2 is arranged sequentially as int (integrase), several conjugation transfer genes, resG (resolvase), ISCR2, glmM (phosphoglucosamine mutase) and sul2, which are found in various bacterial species (Nigro and Hall, 2011). In the MDR region-2 from p16Pre36-NDM, ΔGIsul2 comprises resG, orf225, ISCR2, glmM, and sul2. In the MDR region-1 of p16Pre36-NDM, the truncated GIsul2 related region has a resG–orf225–ISCR2–ΔglmM–erm (rRNA adenine N-6-methyltransferase)–sul2 structure. The erm resistance gene is present 100-bp downstream of ISCR2. The truncation of glmM and the appearance of erm are correlated with ISCR2-mediated transposition. ΔTn1548 is present in the MDR region of pPrY2001, and Tn1548 was initially discovered in plasmid pCTX-M3 from Citrobacter freundii (Dolejska et al., 2013). Compared with the structure of Tn1548, ΔTn1548 comprises ISCR1, ΔISEc28, armA (aminoglycoside resistance), ISEc29, msr(E), mph(E), orf543, and ΔrepAci (Figure 1A).
The blaIMP–4 gene is integrated into the integron (In809 and In1460) of the MDR region-1 from pPm60 and pPp47. In809 is a prototype Tn402-associated class 1 integron, and its VR region includes blaIMP–4, qacG2, aacA4′, and catB3. In1460 is a complex class 1 integron made up of IRi, 5′-CS, VR1 (variable region 1), 3′-CS1 (qacED1–sul1), ISCR1 (common region), VR2 (variable region 2), 3′-CS2 (qacED1–sul1–orf5–orf6), the Tn402 tni module and IRt, bounded by 5-bp DRs. In1460 comprises IRi, 5′-CS, VR1 (blaIMP–4–qacG2–aacA4′–orf1326–qacG), 3′-CS1 (qacED1–sul1), ISCR1, VR2 (Δorf453–orf675–orf408), 3′-CS2 (ΔqacED1–sul1–orf5–orf6), and IRt. The insertion of VR2 between ISCR1 and 3′-CS2 truncates qacED1 at the 5′ terminal of the 3′-CS2 (Figure 2).
In2-4 (in the MDR region from pC131) and In2-18 (in the MDR region-2 from pPm60) can be classified as class 2 integrons, embedded in ΔTn7. The class 2 integron is found in transposon Tn7 and its derivatives (Hansson et al., 2002). Bracketed by 5-bp DRs, Tn7 contains IRL–In2-4–the tns module (tnsE–tnsD–tnsC–tnsB–tnsA)–IRR (Peters, 2014). Integron In2-4 contains 5′-CS (intI2–attI2), VR (dfrA [dihydrofolate reductase]–sat2 [streptothricin acetyltransferase]–aadA1y [aminoglycoside adenylyltransferase]), and 3′-CS (ybeA, also known as orfX) (Hansson et al., 2002). ΔTn7 from pC131 has a core module (from IRL to the VR of In2-4), and its missing portion has been replaced by the 3’-end of a complex class 1 integron. The 3′ end of the complex class 1 integron includes 3′-CS1, ISCR1, VR2 (blaCTX–M–131), and Δ3′-CS2 (ΔqacED1–Δsul1–Δorf5); the truncated 3’-CS2 results from its positioning between VR2 and the chrA region. It is apparent that ΔTn7 from pPm60 is arranged as an IRL, In2-18 (5′-CS and VR [dfrA1–sat2–aadA1a–qacH2]), a truncated tns module (ΔtnsD–tnsC–tnsB–tnsA), and an IRR. Insertion of the accessory region (from Δaer to orf609) means that ΔTn7 is split into two separate portions; namely, ΔTn7-5′ and ΔTn7-3′ (Figure 3).
The transposon unit IS26–mph(A)–mrx–mphR(A)–IS6100 and the chrA region were found to be inserted into the upstream region of the 3’-end of a complex class 1 integron in pC131. The macrolide resistance unit IS26–mph(A)–mrx–mphR(A)–IS6100 is considered to be a mobile element, and the mph(A)–mrx–mphR(A) operon encodes a phosphotransferase, a positive regulator factor, and a negative transcription factor (Partridge, 2011). This transposon unit is truncated in the MDR region of pC131, but it is present as an intact structure in the MDR region-2 of pPp47, while in pC131, the transposon unit (ΔIS26–mph(A)–mrx–mphR(A)–IS6100) is situated between Tn3 and the chrA region. Tn3 carries the class A beta-lactamase-encoding blaTEM–1 gene, which was initially observed as an R1 plasmid in E. coli (Bailey et al., 2011). Here, Tn3 is an unabridged transposon in pC131, but its tnpA is a pseudogene. The chrA region (IRLchrA–chrA [chromate resistance]–orf98) is derived from a Tn21-like transposon in plasmid pCNB1 from Comamonas, and is often closely linked to IRt–IS6100 (Partridge, 2011). The chrA region, which is connected with the IS26–mph(A)–mrx–mphR(A)–IS6100 unit in pC131 and pPp47, has arisen through IS6100-mediated recombination. The chrA region includes the IS4321R interrupted-IRLchrA, chrA and orf98 in pC131 and pPp47. Insertion of IS4321R, IRLchrA is disrupted and forms two parts, ΔIRLchrA-5′ and ΔIRLchrA-3′ (Figure 3).
We found that Tn4352 and the truncated aacC2–tmrB region are integrated between ΔTn7-3′ and ΔTn7-5′ in pPm60. Flanked by 8-bp DRs at both ends, Tn4352 is an IS26-bounded structure (IS26–aphA1a–IS26), and the aphA1a resistance gene confers resistance to kanamycin and neomycin (Wrighton and Strike, 1987). Although Tn4352 is complete in the MDR region-2 from pPm60, it is truncated in the MDR region-2 from p16Pre36-NDM and pPp47. Furthermore, the structure of ΔTn4352 is IS26–aphA1a in p16Pre36-NDM and ΔIS26–aphA1a in pPp47. The orientation of Tn4352 in p16Pre36-NDM is direct, but reversed in pPp47and pPm60. The aacC2–tmrB region is present in plasmids pCTX-M3 and pU302L, is derived from transposon Tn2 from the Tn3-family, and contains a IS26 mobile element at its right-hand end (Partridge, 2011). The aacC2 and tmrB genes account for aminoglycoside and tunicamycin resistance, respectively. The truncated aacC2–tmrB region in pPm60 is composed of an aacC2–tmrB–orf192–orf228–orf1182 segment. The direction of the truncated aacC2–tmrB region is direct in pPm60, but reversed in pPp47. Owing to the insertion of a 28,064 bp exogenous region (with an unknown function), the truncated aacC2–tmrB region from pPp47 is segmented into two parts: ΔISCfr1-3′ exists in the MDR region-1, while a fragment from ΔISCfr1-5′ to the aacC2 gene is embedded in the MDR region-2. Similarly, ΔTn5563 is also located in the two MDR regions of pPp47. Tn5563 was originally discovered in plasmid pRA2 from Pseudomonas aeruginosa (Yeo et al., 1998), and two segments of ΔTn5563 in pPp47 are arranged as follows: the reverse segment (the mer operon and IRR) is present in MDR region-1 and the direct fragment (IRL and ΔtnpR) is present in MDR region-2 (Figures 1A, 2, 3).
We found that Tn6901 has a complete structure in pHFK418-NDM, but it is interrupted by insertion of the virulence-related region to generate two segments, ΔTn6901-5′ and ΔTn6901-3′. Tn6901 is made up of an IRL–tnpA–res–tnpR–frmB (S-formylglutathione hydrolase)–glo (glyoxalase resistance)–frmA (S-glutathione dehydrogenase)–frmR (negative transcriptional regulator)–IRR structure in plasmid Rts1 from Proteus vulgaris, flanked by 5-bp DRs (Murata et al., 2002). Tn6901 is inserted between orf1528 and orf942 in the backbone of pHFK418-NDM, bracketed by 5-bp DRs. pHFK418-NDM, a pPrY2001-like plasmid, is the only virulence gene-carrying plasmid, indicating that this plasmid can not only carry a large number of drug resistance genes, but also integrate virulence genes within it (Figure 1B).
It is known that xerC and xerD genes are site-specific recombinases in the lambda integrase family, where it was found that xer-mediated recombination events resulted in the transmission of resistance gene between plasmids and chromosomal locations (Merino et al., 2010). The dfrA6-ereA region is located downstream of the conjugal transfer region in p16Pre36-NDM, and has undergone xer-mediated recombination. The dfrA6-ereA region consists of xerC, recD, xerD, dfrA6 (trimethoprim resistance), ereA (erythromycin resistance), and dinB (Supplementary Figure S2).
The blaNDM–1-harboring pHFK418-NDM plasmid, a pPrY2001-like plasmid group member, was first recovered from a clinical multidrug resistant P. mirabilis HFK418 isolate in China. Our data have revealed that the pHFK418-NDM plasmid contains two novel transpositions, Tn6624 and Tn6625. Tn6625, a large composite transposon, has integrated a variety of mobile elements, such as the blaNDM–1-carrying ΔTn125, mph(E)-harboring Tn6624, and In251. In251 was first identified from the above-mentioned clinical isolate, suggesting that it had been efficiently transferred from environmental organisms to clinical isolates. The pHFK418-NDM plasmid was found to have the ability for conjugal transfer, and to harbor a large numbers of resistance and virulence genes.
The pPrY2001-like plasmids described above harbor a wide variety of antimicrobial resistance genes, with the exception of p06-1619-1. Their relatively conserved backbones have integrated a great variety of accessory modules in the form of resistance genes, gene clusters, insertion sequences, transposons, and integrons, all of which enhance the diversification and evolution of the pPrY2001-like plasmids. Our findings augment our current understanding on the horizontal transfer of resistance genes and the genetic diversity and evolution of pPrY2001-like plasmids.
YZ, YT, and XZ conceived the study and designed the experimental procedures. DD, ZL, JF, NJ, and HZ performed the experiments. DD and ML analyzed the data. YZ, YT, XZ, BZ, and TZ contributed to reagents and materials. YZ, YT, and DD wrote the manuscript.
This work was supported in part by the National Natural Science Foundation of China (Grant No. 81802107), the National Science and Technology Major Project (Grant No. 2018ZX10201001), the Science and Technology Plan of Yantai City (2019MSGY132), and the Science and Technology Development Plan of Yeda Hospital of Yantai (201801).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We gratefully acknowledge Prof. Dujun Li for his helpful discussion and continuous encouragement.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02030/full#supplementary-material
Bailey, J. K., Pinyon, J. L., Anantham, S., and Hall, R. M. (2011). Distribution of the blaTEM gene and blaTEM-containing transposons in commensal Escherichia coli. J. Antimicrob. Chemother. 66, 745–751. doi: 10.1093/jac/dkq529
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Beahm, N. P., Nicolle, L. E., Bursey, A., Smyth, D. J., and Tsuyuki, R. T. (2017). The assessment and management of urinary tract infections in adults: guidelines for pharmacists. Can. Pharm. J. 150, 298–305. doi: 10.1177/1715163517723036
Boratyn, G. M., Camacho, C., Cooper, P. S., Coulouris, G., Fong, A., Ma, N., et al. (2013). BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41, W29–W33. doi: 10.1093/nar/gkt282
Boutet, E., Lieberherr, D., Tognolli, M., Schneider, M., Bansal, P., Bridge, A. J., et al. (2016). UniProtKB/swiss-prot, the manually annotated section of the uniprot knowledgebase: how to use the entry view. Methods Mol. Biol. 1374, 23–54. doi: 10.1007/978-1-4939-3167-5_2
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Carattoli, A., Zankari, E., Garcia-Fernandez, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Chen, Z., Li, H., Feng, J., Li, Y., Chen, X., Guo, X., et al. (2015). NDM-1 encoded by a pNDM-BJ01-like plasmid p3SP-NDM in clinical Enterobacter aerogenes. Front. Microbiol. 6:294. doi: 10.3389/fmicb.2015.00294
CLSI (2018). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fifth Informational Supplement M100-S28. Wayne, PA: CLSI.
Dallenne, C., Da Costa, A., Decre, D., Favier, C., and Arlet, G. (2010). Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65, 490–495. doi: 10.1093/jac/dkp498
Dantas Palmeira, J., Ferreira, H., Madec, J. Y., and Haenni, M. (2018). Pandemic Escherichia coli ST648 isolate harbouring fosA3 and blaCTX-M-8 on an IncI1/ST113 plasmid: a new successful combination for the spread of fosfomycin resistance? J. Glob. Antimicrob. Resist. 15, 254–255. doi: 10.1016/j.jgar.2018.10.025
Dolejska, M., Papagiannitsis, C. C., Medvecky, M., Davidova-Gerzova, L., and Valcek, A. (2018). Characterization of the complete nucleotide sequences of IMP-4-encoding plasmids, belonging to diverse inc families, recovered from Enterobacteriaceae isolates of wildlife origin. Antimicrob Agents Chemother 62:e02434-17. doi: 10.1128/AAC.02434-17
Dolejska, M., Villa, L., Poirel, L., Nordmann, P., and Carattoli, A. (2013). Complete sequencing of an IncHI1 plasmid encoding the carbapenemase NDM-1, the ArmA 16S RNA methylase and a resistance-nodulation-cell division/multidrug efflux pump. J. Antimicrob. Chemother. 68, 34–39. doi: 10.1093/jac/dks357
Gamal, D., Fernandez-Martinez, M., Salem, D., El-Defrawy, I., Montes, L. A., Ocampo-Sosa, A. A., et al. (2016). Carbapenem-resistant Klebsiella pneumoniae isolates from Egypt containing blaNDM–1 on IncR plasmids and its association with rmtF. Int. J. Infect. Dis. 43, 17–20. doi: 10.1016/j.ijid.2015.12.003
Garcia-Martin, A. B., Perreten, V., Rossano, A., Schmitt, S., Nathues, H., and Zeeh, F. (2018). Predominance of a macrolide-lincosamide-resistant Brachyspira hyodysenteriae of sequence type 196 in Swiss pig herds. Vet. Microbiol. 226, 97–102. doi: 10.1016/j.vetmic.2018.10.007
Gastmeier, P., Kampf, G., Wischnewski, N., Hauer, T., Schulgen, G., Schumacher, M., et al. (1998). Prevalence of nosocomial infections in representative German hospitals. J. Hosp. Infect. 38, 37–49. doi: 10.1016/s0195-6701(98)90173-6
Giske, C. G. (2015). Contemporary resistance trends and mechanisms for the old antibiotics colistin, temocillin, fosfomycin, mecillinam and nitrofurantoin. Clin. Microbiol. Infect. 21, 899–905. doi: 10.1016/j.cmi.2015.05.022
Guo, X., Xia, R., Han, N., and Xu, H. (2011). Genetic diversity analyses of class 1 integrons and their associated antimicrobial resistance genes in Enterobacteriaceae strains recovered from aquatic habitats in China. Lett. Appl. Microbiol. 52, 667–675. doi: 10.1111/j.1472-765X.2011.03059.x
Hansson, K., Sundstrom, L., Pelletier, A., and Roy, P. H. (2002). IntI2 integron integrase in Tn7. J. Bacteriol. 184, 1712–1721. doi: 10.1128/jb.184.6.1712-1721.2002
Harmer, C. J., and Hall, R. M. (2017). Evolution in situ of ARI-A in pB2-1, a type 1 IncC plasmid recovered from Klebsiella pneumoniae, and stability of Tn4352B. Plasmid 94, 7–14. doi: 10.1016/j.plasmid.2017.10.001
Hussain, A., Ranjan, A., Nandanwar, N., Babbar, A., Jadhav, S., and Ahmed, N. (2014). Genotypic and phenotypic profiles of Escherichia coli isolates belonging to clinical sequence type 131 (ST131), clinical non-ST131, and fecal non-ST131 lineages from India. Antimicrob. Agents Chemother. 58, 7240–7249. doi: 10.1128/AAC.03320-14
Karlowsky, J. A., Lagace-Wiens, P. R., Simner, P. J., DeCorby, M. R., Adam, H. J., Walkty, A., et al. (2011). Antimicrobial resistance in urinary tract pathogens in Canada from 2007 to 2009: CANWARD surveillance study. Antimicrob. Agents Chemother. 55, 3169–3175. doi: 10.1128/AAC.00066-11
Kleinheinz, K. A., Joensen, K. G., and Larsen, M. V. (2014). Applying the ResFinder and VirulenceFinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage 4:e27943. doi: 10.4161/bact.27943
Laver, T., Harrison, J., O’Neill, P. A., Moore, K., Farbos, A., Paszkiewicz, K., et al. (2015). Assessing the performance of the oxford nanopore technologies MinION. Biomol. Detect. Quantif. 3, 1–8. doi: 10.1016/j.bdq.2015.02.001
Lawley, T. D., Burland, V., and Taylor, D. E. (2000). Analysis of the complete nucleotide sequence of the tetracycline-resistance transposon Tn10. Plasmid 43, 235–239. doi: 10.1006/plas.1999.1458
Li, X., Chen, Y., Gao, W., Ye, H., Shen, Z., Wen, Z., et al. (2017). A 6-year study of complicated urinary tract infections in southern China: prevalence, antibiotic resistance, clinical and economic outcomes. Ther. Clin. Risk Manag. 13, 1479–1487. doi: 10.2147/TCRM.S143358
Liebert, C. A., Hall, R. M., and Summers, A. O. (1999). Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63, 507–522.
Lin, D., Xie, M., Li, R., Chen, K., Chan, E. W., and Chen, S. (2016). IncFII conjugative plasmid-mediated transmission of blaNDM–1 elements among animal-borne Escherichia coli strains. Antimicrob. Agents Chemother. 61:e02285-16. doi: 10.1128/AAC.02285-16
Marquez-Ortiz, R. A., Haggerty, L., Olarte, N., Duarte, C., Garza-Ramos, U., Silva-Sanchez, J., et al. (2017). Genomic epidemiology of NDM-1-encoding plasmids in latin american clinical isolates reveals insights into the evolution of multidrug resistance. Genome Biol. Evol. 9, 1725–1741. doi: 10.1093/gbe/evx115
Mataseje, L. F., Boyd, D. A., Lefebvre, B., Bryce, E., Embree, J., Gravel, D., et al. (2014). Complete sequences of a novel blaNDM–1-harbouring plasmid from Providencia rettgeri and an FII-type plasmid from Klebsiella pneumoniae identified in Canada. J Antimicrob. Chemother. 69, 637–642. doi: 10.1093/jac/dkt445
Mataseje, L. F., Peirano, G., Church, D. L., Conly, J., Mulvey, M., and Pitout, J. D. (2016). Colistin-Nonsusceptible Pseudomonas aeruginosa sequence Type 654 with blaNDM–1 arrives in north america. Antimicrob. Agents Chemother. 60, 1794–1800. doi: 10.1128/AAC.02591-15
Merino, M., Acosta, J., Poza, M., Sanz, F., Beceiro, A., Chaves, F., et al. (2010). OXA-24 carbapenemase gene flanked by XerC/XerD-like recombination sites in different plasmids from different Acinetobacter species isolated during a nosocomial outbreak. Antimicrob. Agents Chemother. 54, 2724–2727. doi: 10.1128/AAC.01674-09
Michael, G. B., Kadlec, K., Sweeney, M. T., Brzuszkiewicz, E., Liesegang, H., Daniel, R., et al. (2012). ICEPmu1, an integrative conjugative element (ICE) of Pasteurella multocida: structure and transfer. J. Antimicrob. Chemother. 67, 91–100. doi: 10.1093/jac/dkr411
Moura, A., Soares, M., Pereira, C., Leitao, N., Henriques, I., and Correia, A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25, 1096–1098. doi: 10.1093/bioinformatics/btp105
Murata, T., Ohnishi, M., Ara, T., Kaneko, J., Han, C. G., Li, Y. F., et al. (2002). Complete nucleotide sequence of plasmid Rts1: implications for evolution of large plasmid genomes. J. Bacteriol. 184, 3194–3202. doi: 10.1128/jb.184.12.3194-3202.2002
Nederbragt, A. J. (2014). On the middle ground between open source and commercial software - the case of the Newbler program. Genome Biol. 15:113. doi: 10.1186/gb4173
Ng, S. P., Davis, B., Palombo, E. A., and Bhave, M. (2009). A Tn5051-like mer-containing transposon identified in a heavy metal tolerant strain Achromobacter sp. AO22. BMC Res. Notes 2:38. doi: 10.1186/1756-0500-2-38
Nigro, S. J., and Hall, R. M. (2011). GIsul2, a genomic island carrying the sul2 sulphonamide resistance gene and the small mobile element CR2 found in the Enterobacter cloacae subspecies cloacae type strain ATCC 13047 from 1890, Shigella flexneri ATCC 700930 from 1954 and Acinetobacter baumannii ATCC 17978 from 1951. J. Antimicrob. Chemother. 66, 2175–2176. doi: 10.1093/jac/dkr230
O’Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745. doi: 10.1093/nar/gkv1189
Partridge, S. R. (2011). Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol. Rev. 35, 820–855. doi: 10.1111/j.1574-6976.2011.00277.x
Partridge, S. R., and Hall, R. M. (2004). Complex multiple antibiotic and mercury resistance region derived from the r-det of NR1 (R100). Antimicrob. Agents Chemother. 48, 4250–4255. doi: 10.1128/aac.48.11.4250-4255.2004
Peters, J. E. (2014). Tn7. Microbiol. Spectr. 2:MDNA3-0010-2014. doi: 10.1128/microbiolspec.MDNA3-0010-2014
Poirel, L., Bonnin, R. A., Boulanger, A., Schrenzel, J., Kaase, M., and Nordmann, P. (2012). Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56, 1087–1089. doi: 10.1128/AAC.05620-11
Ramos, A. C., Cayo, R., Carvalhaes, C. G., Jove, T., da Silva, G. P., Sancho, F. M. P., et al. (2018). Dissemination of multidrug-resistant Proteus mirabilis clones carrying a novel integron-borne blaIMP-1 in a Tertiary Hospital. Antimicrob. Agents Chemother. 62:e01321-17. doi: 10.1128/AAC.01321-17
Ranjan, A., Shaik, S., Mondal, A., Nandanwar, N., Hussain, A., Semmler, T., et al. (2016). Molecular epidemiology and genome dynamics of new delhi metallo-beta-lactamase-producing extraintestinal pathogenic Escherichia coli strains from india. Antimicrob. Agents Chemother. 60, 6795–6805. doi: 10.1128/AAC.01345-16
Ranjan, A., Shaik, S., Nandanwar, N., Hussain, A., Tiwari, S. K., Semmler, T., et al. (2017). Comparative genomics of Escherichia coli isolated from skin and soft tissue and other extraintestinal infections. mBio 8:e01070-17. doi: 10.1128/mBio.01070-17
Reffert, J. L., and Smith, W. J. (2014). Fosfomycin for the treatment of resistant gram-negative bacterial infections. insights from the society of infectious diseases pharmacists. Pharmacotherapy 34, 845–857. doi: 10.1002/phar.1434
Roberts, A. P., Chandler, M., Courvalin, P., Guedon, G., Mullany, P., Pembroke, T., et al. (2008). Revised nomenclature for transposable genetic elements. Plasmid 60, 167–173. doi: 10.1016/j.plasmid.2008.08.001
Schaffer, J. N., and Pearson, M. M. (2015). Proteus mirabilis and urinary tract infections. Microbiol. Spectr. 3:UTI-0017-2013. doi: 10.1128/microbiolspec.UTI-0017-2013
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36.
Solgi, H., Giske, C. G., Badmasti, F., Aghamohammad, S., Havaei, S. A., Sabeti, S., et al. (2017). Emergence of carbapenem resistant Escherichia coli isolates producing blaNDM and blaOXA-48-like carried on IncA/C and IncL/M plasmids at two Iranian university hospitals. Infect. Genet. Evol. 55, 318–323. doi: 10.1016/j.meegid.2017.10.003
Srijan, A., Margulieux, K. R., Ruekit, S., Snesrud, E., Maybank, R., Serichantalergs, O., et al. (2018). Genomic characterization of nonclonal mcr-1-Positive multidrug-resistant Klebsiella pneumoniae from clinical samples in Thailand. Microb. Drug Resist. 24, 403–410. doi: 10.1089/mdr.2017.0400
Tao, P., Li, H., Yu, Y., Gu, J., and Liu, Y. (2016). Ectoine and 5-hydroxyectoine accumulation in the halophile Virgibacillus halodenitrificans PDB-F2 in response to salt stress. Appl. Microbiol. Biotechnol. 100, 6779–6789. doi: 10.1007/s00253-016-7549-x
Wang, J., Zeng, Z. L., Huang, X. Y., Ma, Z. B., Guo, Z. W., Lv, L. C., et al. (2018). Evolution and Comparative Genomics of F33:A-:B- Plasmids Carrying blaCTX-M-55 or blaCTX-M-65 in Escherichia coli and Klebsiella pneumoniae Isolated from Animals, Food Products, and Humans in China. mSphere 3:e00137-18. doi: 10.1128/mSphere.00137-18
White, P. A., Stokes, H. W., Bunny, K. L., and Hall, R. M. (1999). Characterisation of a chloramphenicol acetyltransferase determinant found in the chromosome of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 175, 27–35. doi: 10.1016/s0378-1097(99)00171-8
Wrighton, C. J., and Strike, P. (1987). A pathway for the evolution of the plasmid NTP16 involving the novel kanamycin resistance transposon Tn4352. Plasmid 17, 37–45. doi: 10.1016/0147-619x(87)90006-0
Ybazeta, G., Douglas, L., Graham, J., Fraleigh, N. L., Murad, Y., Perez, J., et al. (2017). Complete Genome Sequence of Enterococcus thailandicus strain a523 Isolated from Urban Raw Sewage. Genome Announc. 5:e01298-17. doi: 10.1128/genomeA.01298-17
Yeo, C. C., Tham, J. M., Kwong, S. M., Yiin, S., and Poh, C. L. (1998). Tn5563, a transposon encoding putative mercuric ion transport proteins located on plasmid pRA2 of Pseudomonas alcaligenes. FEMS Microbiol. Lett. 165, 253–260. doi: 10.1016/s0378-1097(98)00286-9
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. doi: 10.1128/AAC.00774-09
Keywords: Proteus mirabilis, blaNDM–1, transposons, plasmids, multidrug-resistant
Citation: Dong D, Li M, Liu Z, Feng J, Jia N, Zhao H, Zhao B, Zhou T, Zhang X, Tong Y and Zhu Y (2019) Characterization of a NDM-1- Encoding Plasmid pHFK418-NDM From a Clinical Proteus mirabilis Isolate Harboring Two Novel Transposons, Tn6624 and Tn6625. Front. Microbiol. 10:2030. doi: 10.3389/fmicb.2019.02030
Received: 15 December 2018; Accepted: 19 August 2019;
Published: 04 September 2019.
Edited by:
Ziad Daoud, University of Balamand, LebanonReviewed by:
Arif Hussain, International Centre for Diarrhoeal Disease Research, BangladeshCopyright © 2019 Dong, Li, Liu, Feng, Jia, Zhao, Zhao, Zhou, Zhang, Tong and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianglilan Zhang, emhhbmd4aWFuZ2xpbGFuQGdtYWlsLmNvbQ==; Yigang Tong, dG9uZy55aWdhbmdAZ21haWwuY29t; Yuanqi Zhu, enl1ZG90YWxpeXVuQDE2My5jb20=
†These authors have contributed equally to this work
‡Present address: Manli Li, Center for Advanced Measurement Science, National Institute of Metrology, Beijing, China
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.