- 1Key Laboratory of Edible Fungal Resources and Utilization (South), National Engineering Research Center of Edible Fungi, Shanghai Academy of Agricultural Sciences, Institute of Edible Fungi, Shanghai, China
- 2Key Laboratory of Agricultural Genetics and Breeding of Shanghai, National Engineering Research Center of Edible Fungi, Shanghai Academy of Agricultural Sciences, Institute of Edible Fungi, Shanghai, China
- 3Hongzhen Agricultural Science and Technology Co. Ltd., Jinghong, China
Phlebopusportentosus (Berk. and Broome) Boedijin is a popular edible mushroom found in China and Thailand. To date, P. portentosus is the only species in the order Boletales that can be successfully cultivated worldwide. The use of a casing layer or casing soil overlaying the substrate is a crucial step in the production of this mushroom. In this study, bacterial profiling and dynamic succession analyses of casing soil during the cultivation of P. portentosus were performed. One hundred and fifty samples were collected, and MiSeq sequencing of the V3-V4 region of the 16S rRNA gene was conducted. After performing a decontamination procedure, only 38 samples were retained, including 6 casing soil-originated samples (OS), 6 casing soil samples (FHCS) and 5 upper substrate samples (FHCU) from the period of complete colonization by mycelia; 6 casing soil samples (PCS) and 5 upper substrate samples (PCU) from the primordium period; and 6 casing soil samples (FCS) and 4 upper substrate samples (FCU) from fruit body period. The results revealed that bacterial diversity increased sharply from the hyphal to the primordium stage and then decreased during harvesting. The non-metric multidimensional scaling (NMDS) ordination and analysis of similarities (ANOSIM) analysis suggested that the community composition during different stages was significantly different in casing soil. The most abundant phyla in all of the samples were Proteobacteria, Chloroflexi, Acidobacteria, Actinobacteria, Saccharibacteria, and Bacteroidetes. Burkholderia was the most abundant genus in all the samples except the OS samples. The relative abundance of Burkholderia in the FHCS samples (55.79%) decreased to 35.14% in the PCS samples and then increased to 45.60% in the FCS samples. The abundances of Acidobacterium, Rhizobium, Acidisphaera, Bradyrhizobium, and Bacillus increased from the FHCS to PCS samples. The linear discriminant analysis (LDA) effect size (LEfSe) suggested that Acidobacterium and Acidisphaera are micromarkers for PCS, whereas Bradyrhizobium, Roseiarcus, and Pseudolabrys were associated with fruit body stages. The network analyses resulted in 23 edges, including 4 negative and 19 positive edges. Extensive mutualistic interactions may occur among casing soil bacteria. Furthermore, these bacteria play important roles in mycelial elongation, primordium formations, and the production of increased yields.
Introduction
Phlebopus portentosus (Berk. and Broome) Boedijin (also known as tropical black bolete) belongs to the order Boletales and is a popular edible mushroom in both China and Thailand (Zhang et al., 2017). P. portentosus is widely distributed in Asia, America, Oceania, China, Thailand, Indonesia, Sri Lanka, Vietnam, Australia, New Zealand, Brazil, Mexico, and other tropical regions (Heinemann and Rammeloo, 1982; Segedin, 1987; Watling, 2001; Sanmee et al., 2003; Bandala et al., 2004; Ji, 2007). P. portentosus was previously reported to form a mutualistic relationship with the roots of some plants, characterizing it as an ectomycorrhizal (ECM) fungus (Kumla et al., 2016). However, unlike its close relatives in the family Boletaceae, such as Boletus edulis, P. portentosus can be artificially cultivated and produce sporocarps in artificial substrate in vitro (Ji, 2007; Ji et al., 2011; Kumla et al., 2015). Up to now, the cultivation of P. portentosus has been highly industrialized, as in the Agaricus mushroom industry, which has resulted in daily production of 2 tons of P. portentosus. To date, this is the only species in the Boletales which can be successfully cultivated worldwide.
For the commercial cultivation of P. portentosus, a crucial step in the production of this mushroom is the addition of a casing layer or casing soil overlaying the substrate (Kumla et al., 2015). Many mushrooms need a casing layer to induce the formation of fruiting bodies or increase yields, e.g., Agaricus bisporus, Agaricus blazei, Copyinds comatus, Grifola frondosa, Ganoderma lucidum, Stropharia rugosoannulata, and so on (Martos et al., 2017; Kertesz and Thai, 2018; Carrasco et al., 2019b). Casing soil-associated bacteria and fungi are known to influence the growth and development of Agaricus bisporus and other commercial mushrooms (Kertesz and Thai, 2018). Kertesz and Thai (2018) summarized the roles that these bacteria and fungi play, including the conversion of the lignocellulose substrate into nutrient-rich compost for mushroom growth; interactions with the fungal mycelia during hyphal elongation and proliferation through the substrate; and the induction of primordial fruiting body formation (Segedin, 1987). For example, Pseudomonas, Bacillus and other bacteria can increase mycelial growth, primordial formation, and yield in some species of the genera Agaricus and Pleurotus (Park and Agnihotri, 1969; Rainey, 1991; Cho et al., 2003; Cai et al., 2009; Young et al., 2012; Zarenejad et al., 2012; Chen et al., 2013; Rossouw and Korsten, 2017). Furthermore, the bioactive factors in the casing soil may promote fruiting body formation in P. portentosus.
Several studies have revealed that some bacteria isolated from the casing soil of P. portentosus have a significant effect on mycelial growth and fruiting body formation (Ji et al., 2011; Liu et al., 2011, 2012; Wang et al., 2013; Wu et al., 2013). Bacillus species from the phylum Firmicutes and Brevibacterium species from the phylum Actinobacteria, as well as other Actinobacteria strains, have been shown to play important roles in promoting mycelial elongation, fruiting body formation, and increased individual weight (Ji et al., 2011; Liu et al., 2011, 2012; Wang et al., 2013; Wu et al., 2013). The results also suggested that there was a complicated bacterial network influencing the development of P. portentosus. To characterize the bacterial structure and dynamics in compost and casing soil in other mushrooms, molecular methods and next-generation sequencing have been widely used at an unprecedented scale, e.g., PCR-DGGE, PLFA, 454 pyrosequencing and MiSeq methodologies (Szekely et al., 2009; De Gannes et al., 2013; Kertesz et al., 2016). However, only cultivation methods have been used to assess the bacterial community present in the casing soil used for P. portentosus (Ji et al., 2011; Liu et al., 2011, 2012; Wang et al., 2013; Wu et al., 2013; Kertesz and Thai, 2018). The use of the MiSeq next-generation sequencing platform to detect the profiles and dynamic succession bacteria in casing soil for P. portentosus may provide insights into the interactions between casing soil-associated bacteria and P. portentosus.
In this study, an examination of the bacterial structures and dynamics associated with casing soil at different stages during the cultivation of P. portentosus was conducted using MiSeq sequencing of 16S ribosomal DNA (rDNA). As a unique cultivated species of the order Boletales, P. portentosus may be another useful model to study the mechanisms of casing soil to influence the growth of mushrooms.
Materials and Methods
Artificial Cultivation of Phlebopus portentosus
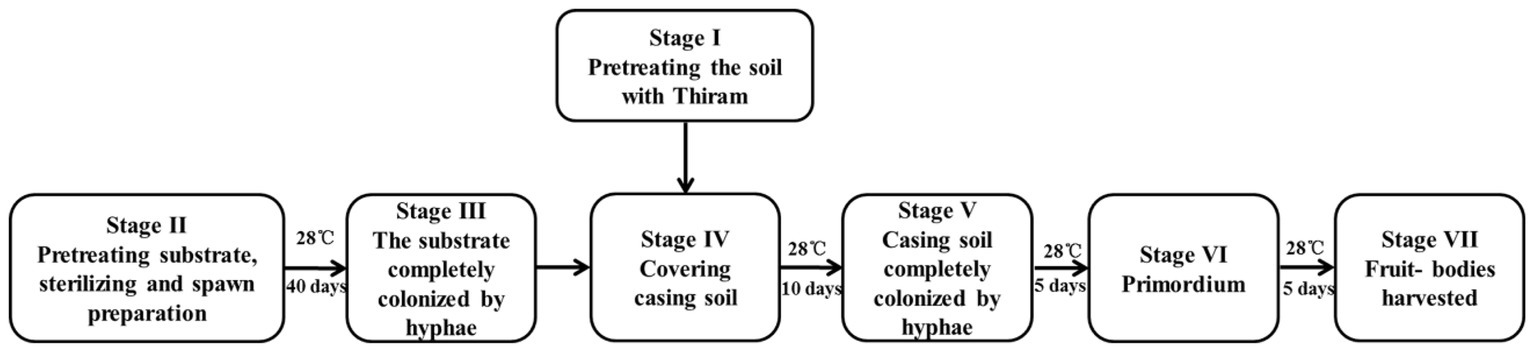
The P. portentosus strain 17026, which was isolated from a natural mature fruiting body, was provided by Hongzhen Agricultural Science and Technology Co. Ltd. The strain was maintained on modified PDA medium at 30°C in the dark. All the primary and secondary spawn preparation and culturing of the mushrooms was conducted at Hongzhen Agricultural Science and Technology Co. Ltd. The substrate was primarily composed of sawdust, starch, and soil. The substrate was placed into 1,100-ml culture bottles (8.5 cm diameter and 14.0 cm height) and sterilized at 121°C for 2 h (S, stage II). The sterilized bottles were inoculated with the spawn of P. portentosus strain 17026 and incubated at 28°C for approximately 40 days to obtain completely colonized hyphae (H, stage III), after which the substrate-hyphae in culture bottles were covered with 2 cm of casing soil (stage IV). After continuous incubation for 10 days, the casing soil was full of fungal hyphae (FH, stage V). The primordia of the fruit bodies appeared after 5 days (P, stage VI), at which time the fruit bodies were harvested (F, stage VII). The entire cultivation process was shown in Figure 1.
The soil used for the casing layers was collected from banana fields (banana was the most dominant plant species) near the company (21.68 N 100.72 E). The total nitrogen (TN) concentrations of soil samples were measured using a Kjeltec TM2300 azotometer (FOSS, Sweden); total phosphorus (TP) was estimated using the colorimetric method with ammonium molybdate in hydrochloric acid; and total potassium (TK) was determined using a flame photometer (Gao et al., 2019). Humic acids (HAs) were extracted using the classic alkali/acid fractionation procedure (Valdrighi et al., 1996). Subsequently, the soil was pretreated with Thiram (stage I) to avoid fungal contamination and then used as the casing soil. There were three treatments performed for this study for stage IV, (1) normal casing soil (90 samples); (2) no covering casing soil (24 samples); and (3) sterilized casing soil (36 samples).
Sampling
Each culture bottle was divided into two sections, an upper section and a lower section. During each stage, samples were collected from the upper and lower sections of each bottle, with six repetitions conducted. In stage I, only the casing soil was collected; in stages II and III, six upper section samples and six lower section samples were collected; in stages V and VI, six casing soil samples, six upper and six lower section samples were collected; and in stage VII, six casing soil samples, six upper section samples, six lower section samples, and six fruit bodies were collected. For treatments 2 and 3, samples were collected used the same strategies from the stages V and VI. Thus, a total of 150 samples were collected in this study.
DNA Extraction, Polymerase Chain Reaction Amplification, and MiSeq Sequencing
DNA extractions from the substrate and casing soil were conducted using a commercial PowerSoil DNA Isolation Kit (MoBio Laboratories, Carlsbad, CA, USA) according to the manufacturer’s protocol. The primer pair 338F/806R was used to amplify the V3-V4 hypervariable region of the bacterial 16S rRNA gene (Zhang et al., 2018). A 6 bp-barcode was added to the 5′ end of the forward and reverse primers for discriminating different samples. Polymerase chain reaction (PCR) amplification was performed in a 50 μl volume containing 25 μl of 2 × PCR MIX, 2 μl of template DNA, 1 μl of barcoded primers, and 21 μl of H2O. The following thermocycling conditions were used for PCR amplification: 94°C for 2 min, followed by 30 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 45 s, with a final extension at 72°C for 10 min (Langenheder and Szekely, 2011). Three PCR repetitions for each sample were performed to offset PCR bias. The mixed PCR products were purified using a QIAquick Gel Extraction kits (QIAGEN, Germany) and quantified via Real Time PCR. Illumina MiSeq sequencing was performed at Personalbio Company (Shanghai, China).
Data Processing
The paired-end reads were assembled into one scaffold using Mothur v.1.40.0 (Schloss et al., 2009) with a minimum overlap of 10 bp. The assembled reads were filtered and removed according to the criterions: length < 200 bp; average quality score ≤ 30; contained ambiguous base calls and did not exactly align with primer sequences and barcode tags. The qualifying reads were sorted into different samples based on their unique barcodes. Importantly, a non-standard pipeline suggested by Łukasik et al., 2017 was to filter out suspected contaminant genotypes or noise from the dataset. The samples from stage II were used as the negative controls. The procedure consisted of four steps. In the first step, the unique sequences (results from Mothur using the script unique.seqs), unless having a relative abundance in at least one experimental library that was at least 10 times its maximum relative abundance in any of the negative controls, were excluded from the dataset. In the second step, to avoid the retention of low-abundance sequences in samples, the unique sequences that did not account for at least 0.1% of at least one experimental library were removed. In the third step, samples where the number of the reads filtered over 75% of the starting number of reads were discarded, as these samples were deemed to be too highly contaminated. In the fourth step, the steps above were repeated. Finally, only high-quality, uncontaminated reads were retained.
Operational Taxonomic Unit Clustering, Taxonomy, Alpha Diversity, Beta Diversity, and Statistics
The reads in the dataset were clustered into operational taxonomic units (OTUs) using QIIME at a 97% sequence similarity (Caporaso et al., 2010). The microbial diversity and richness were evaluated using the Shannon and ACE indices, both of which were tested by t-tests. The distance matrices of community composition (Hellinger-transformation of the OTU data) of casing soil were generated by calculating dissimilarities using the Bray-Curtis method. To further visualize the community composition dissimilarities in each stage, non-metric multidimensional scaling (NMDS) together with analysis of similarities (ANOSIM) was conducted using the metaMDS and anosim command in the vegan package (version 2.4.1). Taxonomic classifications of the effective sequences were carried out using the RDP Classifier (Wang et al., 2007). As the relative abundance of bacterial compositions at the phylum, class, and genus levels did not satisfy the normality of distribution and homogeneity of variance after the square root and log transformation, a nonparametric Kruskal-Wallis test was used. Linear discriminant analysis effect size (LEfSe, LDA = 4.0) was used to identify bacterial markers from the different stages. To construct the co-occurrence network of the core bacteria from the different stages, bivariate correlation analysis was performed, and the network was generated using Cytoscape (version 3.2.1). Networks were visualized using a prefuse force directed layout in which the nodes represent the bacterial genera and the edges represent the correlation (negative-blue; positive-red). All of the reads were submitted to the NCBI Sequence Read Archive under the accession numbers SRR9018053-SRR9018090.
Results
Phlebopus portentosus Cultivation and the Physicochemical Properties of Casing Soil
After a 60-day incubation period, different results were obtained from the three treatments. The samples with no casing soil and sterilized casing soil did not form fruiting bodies (Figures 2A,B), whereas the formation of healthy fruit bodies was induced by the normal casing soil (Figure 2C). The average weight of the individual fruiting body was 126 g.
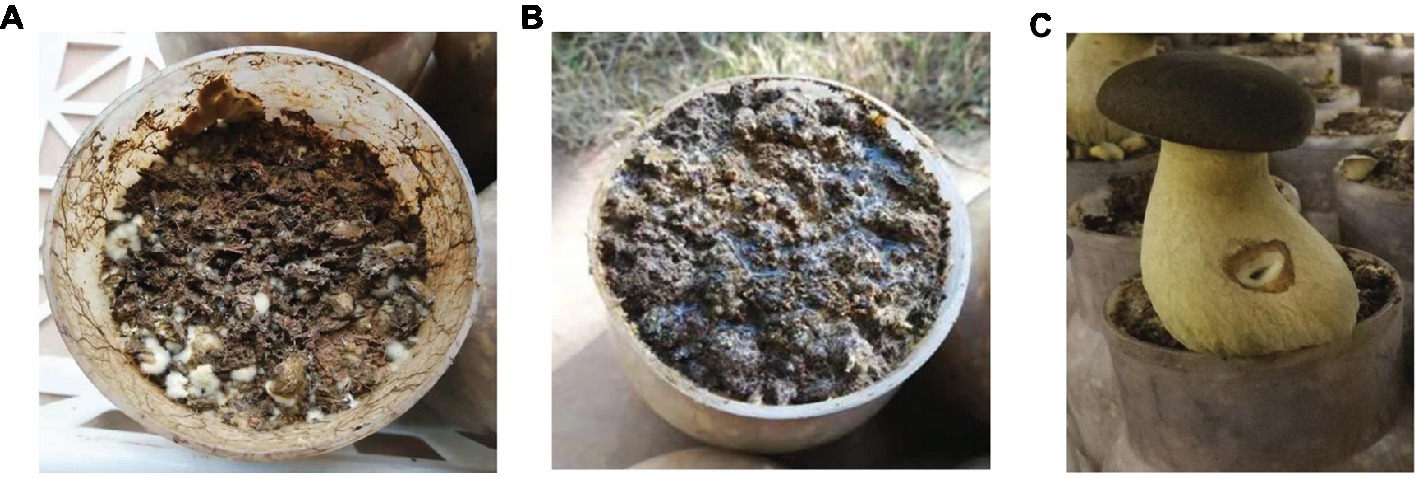
Figure 2. The cultivation of P. portentosus in the three treatments. (A) No covering casing soil; (B) sterilized casing soil; (C) normal covering casing soil.
The total N, P, K, and organic matter contents in the normal and sterilized casing soil were not different significantly (Supplementary Table S1).
Sequencing and Filtering Information
The DNA extractions and V3-V4 16S rRNA gene region amplifications were successful in all the samples except one from stage VII. Finally, 5,094,718 assembled high-quality reads were generated from the 149 samples. The number of reads across all the samples ranged from 24,308 to 72,653 (Supplementary Table S2), and the average number of reads in each sample was 34,192 (Supplementary Table S2). After the first decontamination step, only 682,922 reads were retained, accounting for 13.40% of the total reads (Supplementary Table S2), and the number of reads across all of the samples ranged from 47 to 17,858 (Supplementary Table S1). One hundred and eleven samples retained less than 25% of the starting number of reads after the filtering steps and were removed from subsequent analyses. Finally, 424,200 high-quality, contamination-free reads were retained in 38 samples (Supplementary Table S1). The 38 samples included 24 casing soil samples from stages I (OS), V (FHCS), VI (PCS), and VII (FCS), 5 upper substrate samples from stage V (FHCU), 5 upper substrate samples from stage VI (PCU), and 4 upper substrate samples from stage VII (FCU) (Supplementary Table S1). The results also showed that samples from the lower substrate, samples taken before covering the soil, and fruit bodies were all filtered out. Because the number of reads in each sample was highly uneven, 7,096 reads per sample were randomly selected to balance the reads in each sample for subsequent analyses.
Diversity and Richness in Different Samples
Two hundred and thirty-one OTUs were clustered in all the samples. The microbial diversity and richness were calculated by assessing the OTU composition in each sample. In the casing soil samples, the OTU diversity varied significantly during the growth of P. portentosus, where the Shannon index was 3.30 in the original casing soil samples (OS, stage I) and decreased to 2.79 in the FHCS samples (stage V) (Figure 3A). A sharp increasing in the Shannon index value (3.71) was observed in the PCS stage (stage VII) (Figure 3A), and then decreased to 3.31 before the fruit bodies were harvested (stage FCS). The OTU richness continually increased from 172 in the OS (stage I) to 182 in stage the FHCS (stage V). For the samples from upper substrate, the Shannon index increased and became stable in stage V (Figure 3B), whereas the ACE index values in the different upper substrate samples were nearly identical (Figure 3B).
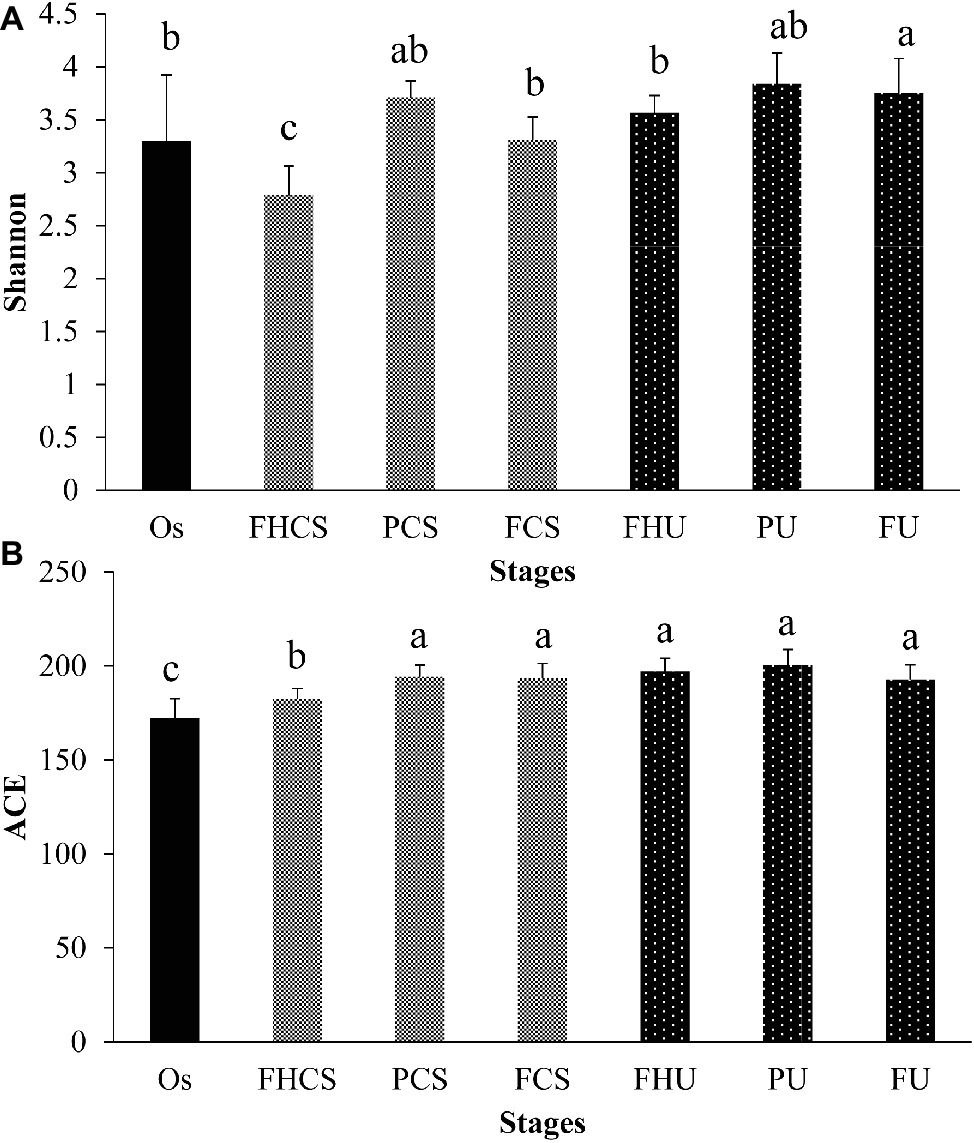
Figure 3. Microbial diversity and richness. The Shannon and ACE indices were calculated at a depth of 7,096 reads in each sample. The bars indicate the mean; the error bars denote standard deviation; and the lowercase letters above the bars indicate significant differences. FHCS, casing soil from stage V (casing soil completely colonized by hyphae); PCS, casing soil from stage VI (primordial stage); FCS, casing soil from stage VII (fruiting body stage); FHU, upper portion of the substrates from stage V; PU, upper portion of the substrates from stage VI; FU, upper portion of the substrates from stage VII. (A) Shannon, (B) ACE.
Comparisons of Community Composition in Different Stages
The results of the non-metric multidimensional scaling (NMDS) ordination analysis suggested that the community composition at different stages was significantly different. The bacterial community was different between the OS and CS and CU samples (Figure 4A). The fungus influenced the bacterial structures in the casing soil samples from the different stages (Figure 4B), demonstrating that a folded linearity fluctuate of bacterial structure occurred during P. portentosus development. There were no differences between samples from the upper substrate (p > 0.05, Figure 4C). The results of the ANOSIM analysis were in agreement with those of the NMDS analysis.
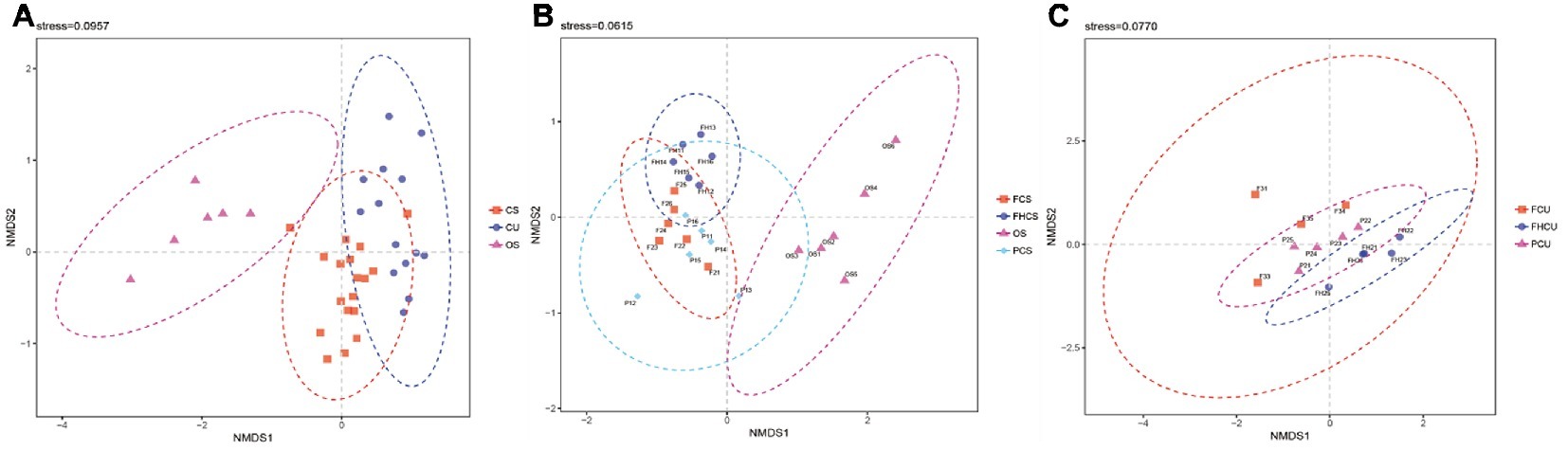
Figure 4. Non-metric multidimensional scaling (NMDS) ordination of the bacterial community composition at different stages of P. portentosus cultivation. Ellipses in the plots denote 95% confidence intervals for the centroids of the CS, CU, and OS samples. FHCS: casing soil from stage V (casing soil completely colonized by hyphae); PCS, casing soil from stage VI (primordial stage); FCS, casing soil from stage VII (fruiting body stage); FHU, upper portion of the substrates from stage V; PU, upper portion of the substrates from stage VI; FU, upper portion of the substrates from stage VII. (A) NMDS for CS, CU and OS, (B) NMDS for FCS, FHCS, OS and PCS, (C) NMDS for FCU, FHCU and PCU.
Bacterial Compositions in Different Samples
At the phylum level, 97.6% of the total sequences were assigned to 16 known phyla. The most abundant phyla in all the samples were Proteobacteria, Chloroflexi, Acidobacteria, Actinobacteria, Saccharibacteria, and Bacteroidetes, accounting for 60.00, 15.18, 7.62, 6.40, 3.48, and 2.48% of the total phyla, respectively (Supplementary Figure S1). The relative abundance of Proteobacteria sharply increased from the casing soil OS samples (23.55%) to the FHCS samples (68.39%). Subsequently, the percentage of Proteobacteria decreased to 60.49% in the PCS samples and increased to 65.84% in the FCS samples (Figure 5A). The observed changes in the abundances of Acidobacteria, Actinobacteria, and Bacteroidetes were the same, with an initial decrease from the OS to FHCS samples followed by an increase from the FHCS to PCS samples and a subsequent further decrease from the PCS to the FCS samples (Figures 5C,F,D). The relative abundance of Chloroflexi sharply decreased from 46.61% in the OS samples to 13.15% in the FHCS samples (Figure 5B), whereas that of Saccharibacteria exhibited a slow decrease from the OS to the FHCS samples before increasing to a plateau in stage VI (PCS) (Figure 5E).
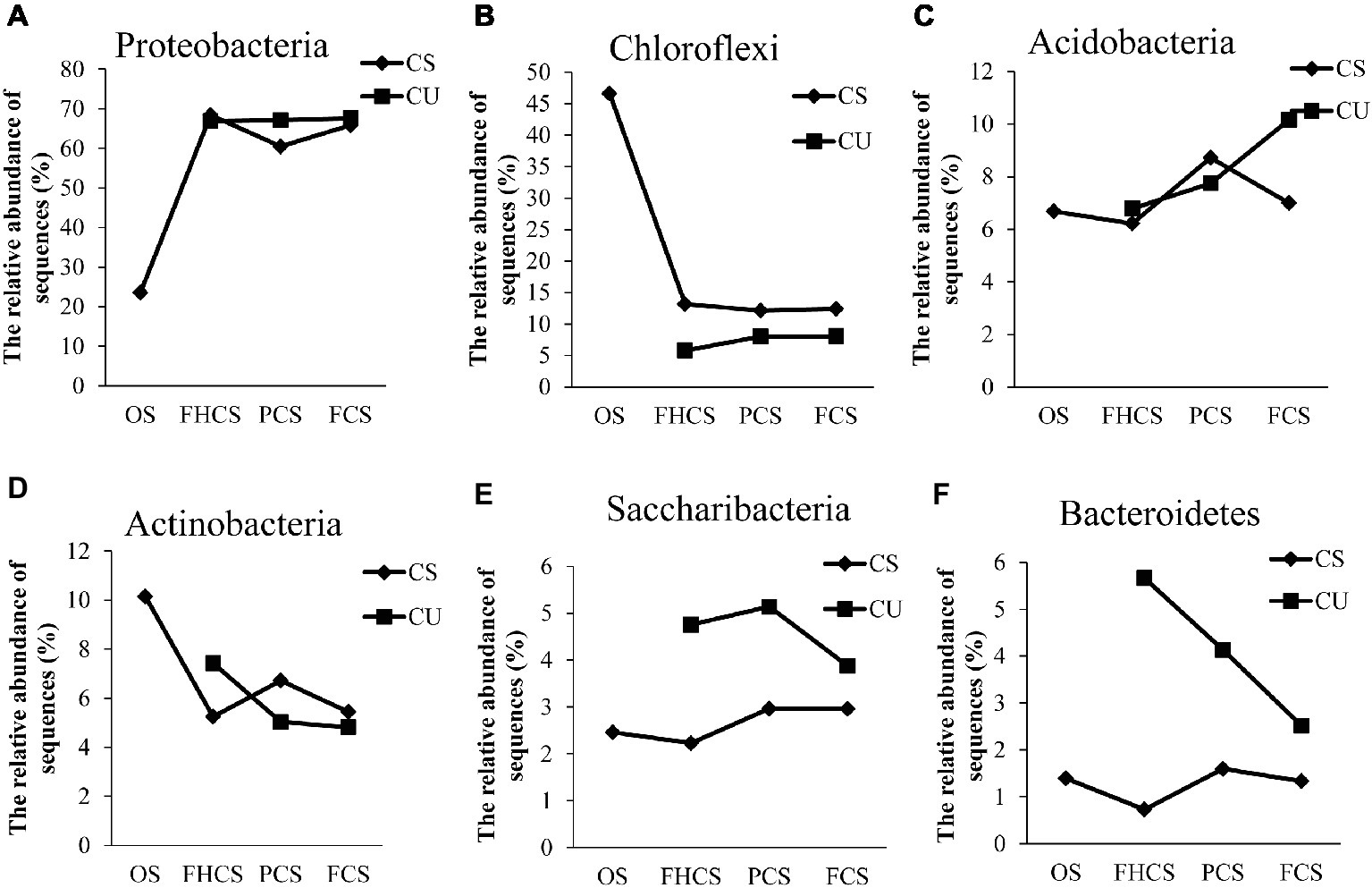
Figure 5. The dynamics of some phyla in the casing soil and upper substrate samples during the development of P. portentosus. (A) Proteobacteria; (B) Chloroflexi; (C) Acidobacteria; (D) Actinobacteria; (E) Saccharibacteria; (F) Bacteroidetes. CS, samples from casing soil; CU, samples from the upper substrate. FHCS, casing soil from stage V (casing soil completely colonized by hyphae); PCS, casing soil from stage VI (primordial stage); FCS, casing soil from stage VII (fruiting body stage); FHU, upper portion of the substrates from stage V; PU, upper portion of the substrates stage VI; FU, upper part of the substrates stage VII.
The relative abundances of Proteobacteria and Chloroflexi in upper substrate nearly did not vary throughout the process (Figures 5A,B). In contrast, a continued increase in the abundance of Acidobacteria was observed, from 6.78 to 10.16% in the FHCU and FCU samples, respectively (Figures 5A,B). The relative abundance of Actinobacteria and Bacteroidetes continued decreasing (Figures 5D,F). The percentage of Saccharibacteria increased from the OS samples (4.75%) to the FHCS samples (5.14%) and then decreased to 3.88% in the stage VI (PCS) samples (Figure 5E).
At the genus level, 76 genera were detected and annotated in all of the samples, 53.10% of which belonged to Burkholderia (36.26%), Thermogemmatispora (5.70%), Acidobacterium (4.17%), Bryobacter (2.43%), Sphingomonas (2.27%), and Rhizomicrobium (2.27%) (Supplementary Figure S2). The relative abundance of Burkholderia was higher in the FH samples (55.79%) than in the OS samples (11.95%) but decreased to 35.14% in the PCS samples before increasing to 45.60% in the FCS samples (Figure 6A). The same increasing trends of Acidobacterium, Rhizobium, and Acidisphaera were observed from the FHCS to the PCS samples, while the same decreasing tendencies were observed from the PCS to the FCS samples (Figures 6B–D). The relative abundance of Thermogemmatispora exhibited a continual decrease from 26.68 to 1.10% in the OS and FCS samples, respectively (Figure 6E), as did that of Actinoallomurus, which decreased from 5.13% in the OS samples to 1.41 and 1.88% in the PCS and FCS samples, respectively (Figure 6F). The abundance of Bradyrhizobium increased to 0.47% in the PCS samples and 0.88% in FCS samples, which was 100 times higher than that observed in the OS and FHCS samples (Figure 6G). A sharp increase in the abundance of Bacillus was also observed, from 0.049% in PCS samples to 0.37% in FCS (Figure 6H). A higher abundance of Sphingomonas was detected in the FHCS (1.67%) and PCS (1.71%) samples than in the OS (1.19%) and FCS (0.89%) samples (Figure 6I).
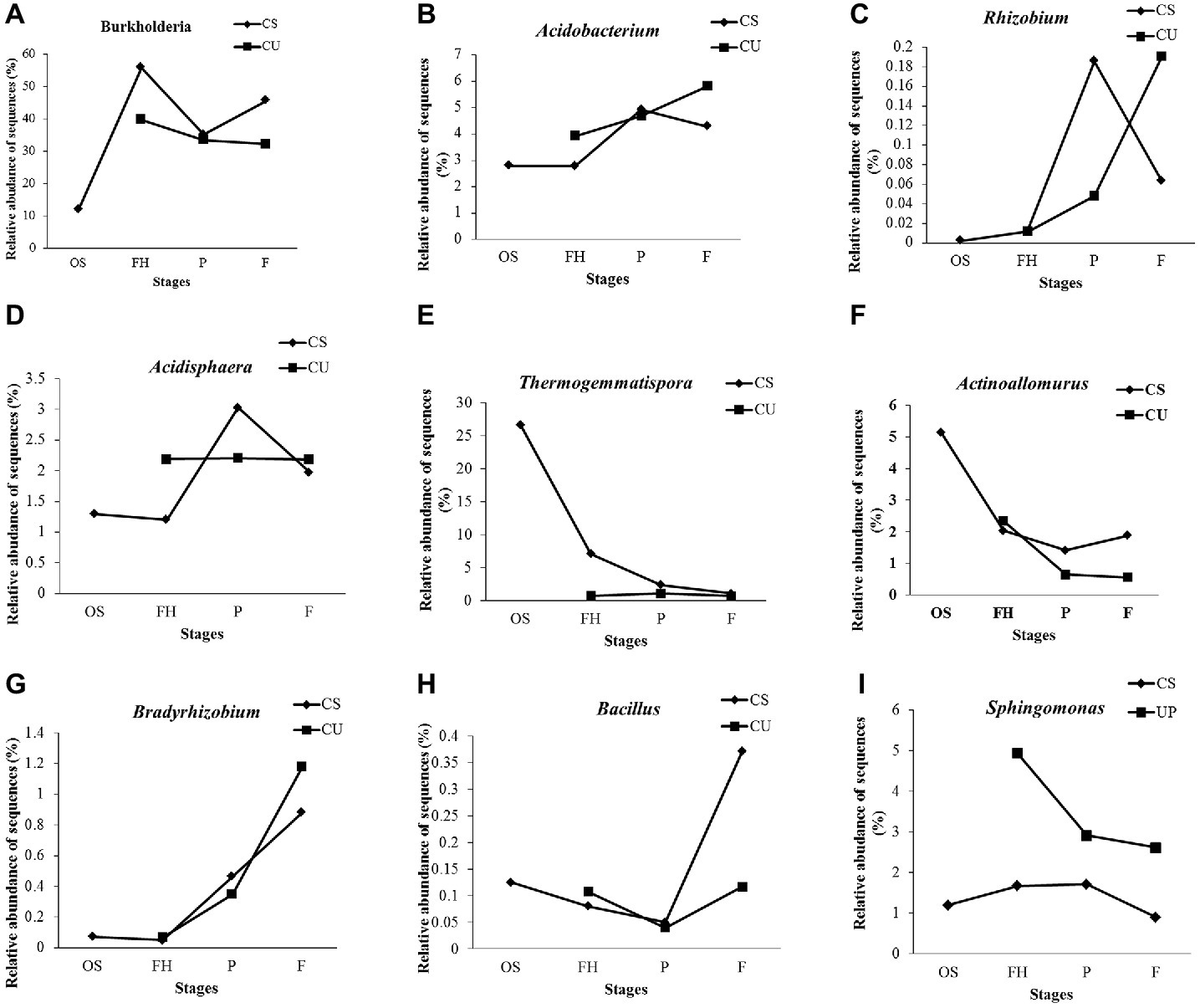
Figure 6. The dynamics of some genera in the casing soil and upper substrate samples during the development of P. portentosus. (A) Burkholderia; (B) Acidobacterium; (C) Acidobacteria; (D) Acidisphaera; (E) Thermogemmatispora; (F) Actinoallomurus; (G) Bradyrhizobium; (H) Bacillus; (I) Sphingomonas. CS, samples from the casing soil; CU, samples from the upper substrate.
Except for Bacillus (Figure 6H), the relative abundances of the eight other detected genera continually increased, decreased, or remained stable in the upper substrate (CU) sample throughout the entire process of P. portentosus cultivation. The abundances of Acidisphaera and Thermogemmatispora remained stable during the growth process (Figures 6D,E).
Microbiota Biomarkers in the Different Stages or Samples
The linear discriminant analysis (LDA) effect size method was used to identify the greatest differences in taxa among the different casing soil samples. The differential features were identified based on the OTU compositions. LEfSe identified 35, 20, 13, and 13 clades or taxa in the PCS, OS, FHCS, and FCS samples, respectively (Figure 7). At the genus level, Thermogemmatispora, Crenotalea, Acidothermus, Nevskia, Rhizomicrobium, and Thermobispora were significantly enriched in OS samples; Burkholderia, Massilia, and Sinomonas were significantly enriched in the FHCS samples; Sorangium, Acidobacterium, Burkholderia, Bordetella, Leifsonia, Reyranella, Hyphomicrobium, Aquicella, Legionella, Dyella, Devosia, and Acidisphaera were significantly enriched in the PCS samples; and Bradyrhizobium, Roseiarcus, and Pseudolabrys were the most significantly enriched in the fruiting body stages (FCS).
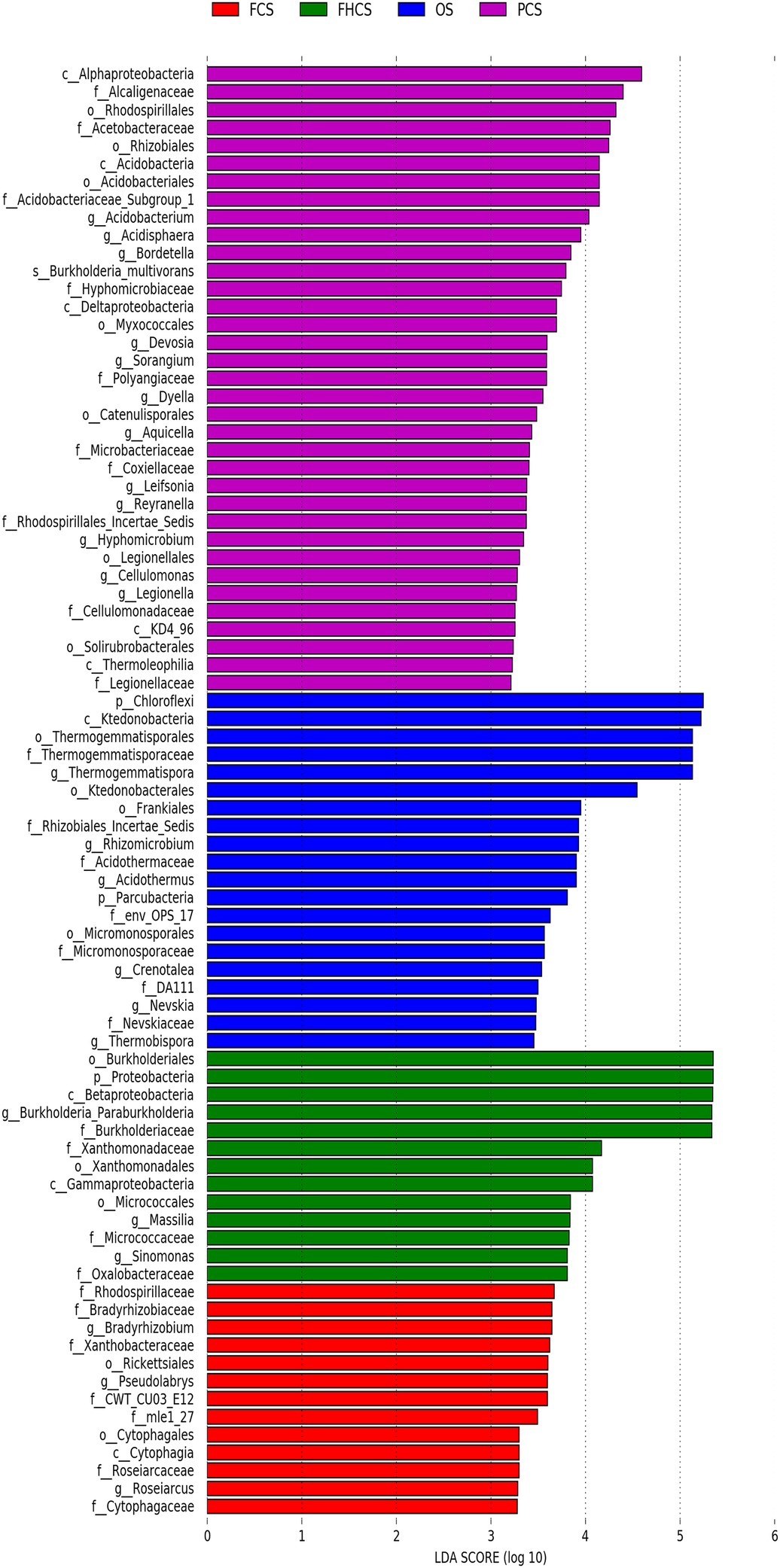
Figure 7. Identification of crucial bacteria associated with different stages. LEfSe reports the taxonomic representation of statistically and biologically consistent differences between the OS, FHCS, PCS, and FCS bacterial communities. OS, original casing soil; FHCS, casing soil completed with colonized hyphae; PCS, casing soil during the primordial period; FCS, casing soil during the fruiting body period.
Topological Consistency Between Operational Taxonomic Units Across Networks
The top 20 OTUs were present at least in 2 networks that resulted in 23 edges, including 4 negative and 19 positive edges (Figure 8), revealing that 19 pairs of OTUs had consistently positive correlations and 4 pairs of OTUs had consistently negative correlations. The negative correlations were only observed between OTU123_Acidothermus and OTU9_Xanthomonadaceae, OTU10_Burkholderia and OTU161_Thermogemmatispora, OTU188_Rhizomicrobium and OTU161_Thermogemmatispora, OTU8_Alcaligenaceae and OTU161_Thermogemmatispora, and OTU8_Alcaligenaceae and OTU122_Actinoallomurus.
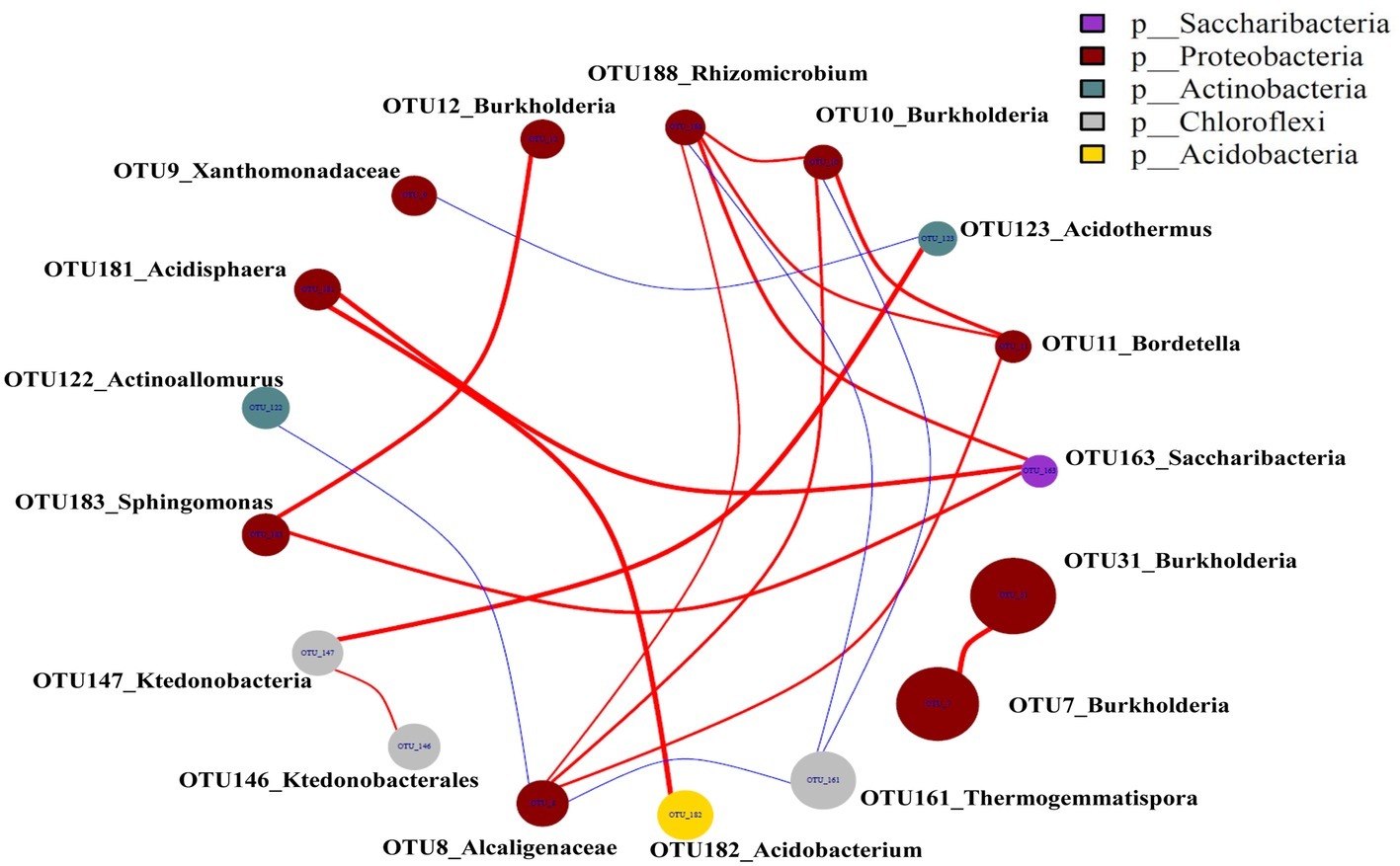
Figure 8. Microbial networks in the casing soil and upper substrate samples based on the top 20 OTUs. The red color edges were positive associations, and the blue edges were negative associations (p < 0.05). A connection indicates a strong positive Spearman’s correlation coefficient if p > 0.75 and a negative Spearman’s correlation coefficient occurred if p < −0.75.
Discussion
In this study, an investigation of the dynamics of microbiota in casing soil during the development of P. portentosus was conducted based on 16S rRNA sequencing using the MiSeq platform. The results revealed that the bacterial community varied across the P. portentosus cultivation process. Interestingly, there was also a dynamic change in the relative abundance of some specific taxa that were either positively or negatively associated with mycelial elongation, primordial stimulation, and fruiting body formation.
An important decontamination procedure was used in this study to filter suspected contaminating sequences generated from the sequencing, PCR, or negative samples. It was previously reported that bacterial DNA is present in reagents and in the laboratory environment that can alter the microbial communities of samples, particularly those with a relatively low template concentration (Salter et al., 2014). These suspicious artifacts derived from one sample may affect the amplification process for other samples (Kircher et al., 2012). Thus, Łukasik et al. (2017) recommended a non-standard analysis pipeline based on the use of some negative controls to filter out the suspected contaminant genotypes or sequences from the MiSeq-generated dataset. In our study, although the sterilized samples used as negative controls were considered to be DNA- or bacteria-free, PCR amplification and MiSeq sequencing were successful. These results suggested that bacterial DNA was still present in these samples, although no active bacteria survived. These DNA templates used for PCR may have influenced the estimations of bacterial communities. In addition, the identified rare sequences were also excluded in this study. This step is crucial since these sequences may exhibit an increased influence under some reweighting schemes, and it may be beneficial to delineate their true presence from simple noise effects (Pylro et al., 2014). We also recommended that a decontaminating procedure was conducted in other investigations of bacterial communities.
Like Agaricus, the presence of a diverse bacterial population in casing layers has been shown to be essential for primordial formation, as fruiting did not occur with sterilized casing layers (Eger, 1963; Hayes et al., 1969; Park and Agnihotri, 1969; Eger, 1972). The production of the mushroom fruiting bodies were induced when isolated bacteria from casing soils returned to the sterile casing layer over axenically grown cultures of A. bisporus in compost, which revealed that bacteria was the essential factor for fruiting body formation (Eger, 1972). In this study, the primordia or fruit bodies of P. portentosus could not form when no casing soil and sterilized casing soil were used (Figure 2A). Some physicochemical properties in the normal and sterilized soil were similar, indicating that biotic factors may play important roles in the formation of fruiting bodies, e.g., bacteria and fungi. The Shannon index values of bacteria increased sharply from the hyphal stage to the primordium stage and then decreased during the harvesting stage in the process of P. portentosus cultivation. These results were in agreement with those from Agaricus, where the bacterial diversity in casing soil increased dramatically after the initiation of the first primordia but decreased during later flushes (Cai et al., 2009; Partanen et al., 2010). The formation of primordia and fruit bodies in Agaricus was previously shown to be reinitiated by the addition of bacterial inoculums or adsorptive carbon-based materials. These results revealed that the stimulatory role of the bacteria involved the removal of an inhibitor of primordial formation, including 1-octen-3-ol, which controls the early differentiation of vegetative hyphae to multicellular knots (Hayes et al., 1969; Rainey, 1991; Noble et al., 2003, 2009; Eastwood et al., 2013; Colauto et al., 2016). And some ACC deaminase-producing bacteria lower the levels of ethylene produced by Agaricus to stimulate primordial formation (Wood and Hammond, 1977; Chen et al., 2013; Zhang et al., 2016). However, these results were not confirmed in P. portentosus, for which the stimulatory mechanisms need to be further studied.
Members of the genus Burkholderia were the major bacteria observed to be associated with P. portentosus in this study. This finding corroborated previous studies that also detected this taxon in association with ECM root tips and surrounding soil of Suillus luteus, Tricholoma matsutake, Leccinum extremiorientale, and so on (Ryota et al., 2012; Li et al., 2016; Oh et al., 2016; Qin et al., 2016; Nishino et al., 2018). Burkholderia species have emerged common and potentially important bacteria associated with plants, animals, and especially fungi (Santos et al., 2001; Stoyanova et al., 2007), with some of these bacteria having been shown to be useful in promoting plant growth (Stoyanova et al., 2007). No Burkholderia living within the hyphae of Rhizopus resulted in losing the abilities to produce reproductive sporangia or spores (Partida-Martinez et al., 2007), which revealed that Burkholderia can affect fungal development and spore production. Members of the genus Burkholderia have been shown to have the ability to fix nitrogen associated with root tips. Whether in vitro nitrogen is an important interaction between Burkholderia and P. portentosus requires further study. However, some Burkholderia are the most common pathogens detected in other mushroom and can inhibit mushroom formation, including mycelial elongation and primordial formation in Agaricus and Pleurotus (Mignucci et al., 2000; Largeteau and Savoie, 2010). Another surprising finding was that no Pseudomonas species were detected, as these bacteria are accepted to play influential roles in the development of Pleurotus and Agaricus mushroom fruiting bodies (Noble et al., 2003; Kertesz and Thai, 2018). Meanwhile, the most abundant genus detected in the casing soil of Agaricus bisporus was Flavobacterium (Carrasco et al., 2019a). The differences in the interactions between fungi and bacteria may be associated with specific ecotypes (ECM and saprophytic fungi) and genetic backgrounds (secretions: enzymes or product of metabolisms).
The relative abundances of Acidobacterium, Bradyrhizobium, Rhizobium, and Acidisphaera increased along with the development of P. portentosus, which may be associated with the formation of P. portentosus primordia. Rhizobium in the casing soil of A. bisporus was previously shown to increase pinning (Park and Agnihotri, 1969), and Acidobacterium and Bradyrhizobium were enriched in the active shiro of Suillus granulatus and Tricholoma matsutake during the fruiting process in pine (Ryota et al., 2012; Zhu et al., 2013; Zhang et al., 2015). Acidobacterium and Acidisphaera were identified as micro-markers for primordial formation, while Bradyrhizobium was the signature genus for fruiting body formation in P. portentosus. The relative abundance of Bacillus only increased after primordial formation, suggesting that Bacillus plays important roles in the process of P. portentosus fruiting body maturation. It was previously demonstrated that Brevibacterium and Bacillus could promote mycelial elongation, fruiting body formation, and increased individual weight in P. portentosus (Liu et al., 2012) and other mushrooms (Zhu et al., 2013; Ekinci and Dursun, 2014; Kertesz and Thai, 2018). However, some other bacterial genera may also be associated with the development of P. portentosus, e.g., Sorangium, Bordetella, Reyranella, Hyphomicrobium, Aquicella, Legionella, Dyella, Devosia, Roseiarcus, and Pseudolabrys, as extensive mutualistic interactions may occur among casing soil bacteria (Figure 8; Shi et al., 2016).
The cultivation of P. portentosus is similar to that of Lentinula grown in fully sterilized substrate, as the enzymes secreted by mycelia are crucial to the growth and crop performance of Lentinula (Philippoussis et al., 2003). Although the addition of specific bacteria could potentially stimulate either mushroom growth or fruiting body formation, this possibility has not been well investigated in P. portentosus. The high bacterial diversity present in casing soil plays important roles in mushroom mycelial elongation, primordial formation, and increased yields. Thus, the recovery of the bacteria from casing soil used to grow P. portentosus as well as non-casing soil used in the cultivated process should be the focus of future investigations.
Data Availability
All the sequences generated for this study have been submitted to NCBI under the accession numbers of SRR9018053-SRR9018090.
Author Contributions
R-HY, D-PB, K-PJ, and QT contributed in the conception and design of the study. TG, YL, and G-YJ performed the sampling. R-HY performed the analysis and wrote the manuscript draft. All authors contributed to manuscript revision and approved the final version.
Funding
This work was supported by the Shanghai Action Plan for Scientific and Technological Innovation (17391900400) and the Shanghai Agriculture Applied Technology Development Program (G2014070106).
Conflict of Interest Statement
K-PJ and G-YJ are employed by the Hongzhen Agricultural Science and Technology Co. Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The supplementary material for this article is available online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01927/full#supplementary-material
References
Bandala, V. M., Montoya, L., and Jarvio, D. (2004). Two interesting records of Boletes found in coffee plantations in eastern Mexico. Persoonia 18, 365–380.
Cai, W., Yao, H., Feng, W., Jin, Q., Liu, Y., Li, N., et al. (2009). Microbial community structure of casing soil during mushroom growth. Pedosphere 19, 446–452. doi: 10.1016/S1002-0160(09)60137-5
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Carrasco, J., Tello, M. L., de Toro, M., Tkacz, A., Poole, P., Pérez-Clavijo, M., et al. (2019a). Casing microbiome dynamics during button mushroom cultivation: implications for dry and wet bubble diseases. Microbiology 1–14. doi: 10.1099/mic.0.000792
Carrasco, J., Tello, M. L., Perez, M., and Preston, G. (2019b). Biotechnological requirements for the commercial cultivation of macrofungi: substrate and casing layer. Biol. Macrofungi 159–175. doi: 10.1007/978-3-030-02622-6_7
Chen, S., Qiu, C., Huang, T., Zhou, W., Qi, Y., Gao, Y., et al. (2013). Effect of 1-aminocyclopropane-1-carboxylic acid deaminase producing bacteria on the hyphal growth and primordium initiation of Agaricus bisporus. Fungal Ecol. 6, 110–118. doi: 10.1016/j.funeco.2012.08.003
Cho, Y., Kim, J., Crowley, D. E., and Cho, B. (2003). Growth promotion of the edible fungus Pleurotus ostreatus by fluorescent pseudomonads. FEMS Microbiol. Lett. 218, 271–276. doi: 10.1016/s0378-1097(02)01144-8
Colauto, N. B., Fermor, T. R., Eira, A. F. D., and Linde, G. A. (2016). Pseudomonas putida stimulates primordia on Agaricus bitorquis. Curr. Microbiol. 72, 482–488. doi: 10.1007/s00284-015-0982-8
De Gannes, V., Eudoxie, G., and Hickey, W. J. (2013). Prokaryotic successions and diversity in composts as revealed by 454-pyrosequencing. Bioresour. Technol. 133, 573–580. doi: 10.1016/j.biortech.2013.01.138
Eastwood, D. C., Herman, B., Noble, R., Dobrovinpennington, A., Sreenivasaprasad, S., and Burton, K. S. (2013). Environmental regulation of reproductive phase change in Agaricus bisporus by 1-octen-3-ol, temperature and CO2. Fungal Genet. Biol. 55, 54–66. doi: 10.1016/j.fgb.2013.01.001
Eger, G. (1963). Untersuchungen zur fruchtkörperbildung des kulturchampignons. Mushroom Sci. 5, 314–320.
Eger, G. (1972). Experiments and comments on the action of bacteria on sporophore initiation in Agaricus bisporus. Mushroom Sci. 8, 719–725.
Ekinci, M., and Dursun, A. (2014). The effects of compost added bacteria, organic fertilizer and their mixtures on yield and quality of mushroom (Agaricus bisporus). C. R. Acad. Bulg. Sci. 67, 1441–1450.
Gao, X. S., Yi, X., Deng, L. J., Li, Q. Q., Wang, C. Q., Li, B., et al. (2019). Spatial variability of soil total nitrogen, phosphorus and potassium in Renshou County of Sichuan Basin, China. J. Integr. Agric. 18, 279–289. doi: 10.1016/S2095-3119(18)62069-6
Hayes, W., Randle, P. E., and Last, F. (1969). The nature of the microbial stimulus affecting sporophore formation in Agaricus bisporus (Lange) Sing. Ann. Appl. Biol. 64, 177–187. doi: 10.1111/j.1744-7348.1969.tb02867.x
Heinemann, P., and Rammeloo, J. (1982). Observations sur le genre Phlebopus (Boletineae). Mycotaxon 15, 384–404.
Ji, K. (2007). Artificial fungal colony and its fruiting of Phlebopus portentosus (Boletaceae) in pot. Acta Bot. Yunnanica 19, 554–558. doi: 10.1360/yc-007-1324
Ji, K., Cao, Y., Zhang, C., He, M., Liu, J., Wang, W., et al. (2011). Cultivation of Phlebopus portentosus in southern China. Mycol. Prog. 10, 293–300. doi: 10.1007/s11557-010-0700-7
Kertesz, M., Safianowicz, K., and Bell, T. (2016). New insights into the microbial communities and biological activities that define mushroom compost. Sci. Cultiv. Edible Fungi 19, 161–165.
Kertesz, M. A., and Thai, M. (2018). Compost bacteria and fungi that influence growth and development of Agaricus bisporus and other commercial mushrooms. Appl. Microbiol. Biotechnol. 102, 1639–1650. doi: 10.1007/s00253-018-8777-z
Kircher, M., Sawyer, S., and Meyer, M. (2012). Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 40:e3. doi: 10.1093/nar/gkr771
Kumla, J., Danell, E., and Lumyong, S. (2015). Improvement of yield for a tropical black bolete, Phlebopus portentosus, cultivation in northern Thailand. Mycoscience 56, 114–117. doi: 10.1016/j.myc.2014.04.005
Kumla, J., Hobbie, E. A., Suwannarach, N., and Lumyong, S. (2016). The ectomycorrhizal status of a tropical black bolete, Phlebopus portentosus, assessed using mycorrhizal synthesis and isotopic analysis. Mycorrhiza 26, 333–343. doi: 10.1007/s00572-015-0672-1
Langenheder, S., and Szekely, A. J. (2011). Species sorting and neutral processes are both important during the initial assembly of bacterial communities. ISME J. 5, 1086–1094. doi: 10.1038/ismej.2010.207
Largeteau, M. L., and Savoie, J.-M. (2010). Microbially induced diseases of Agaricus bisporus: biochemical mechanisms and impact on commercial mushroom production. Appl. Microbiol. Biotechnol. 86, 63–73. doi: 10.1007/s00253-010-2445-2
Li, Q., Li, X., Chen, C., Li, S., Huang, W., Xiong, C., et al. (2016). Analysis of bacterial diversity and communities associated with Tricholoma matsutake fruiting bodies by barcoded pyrosequencing in Sichuan province, Southwest China. J. Microbiol. Biotechnol. 26, 89–98. doi: 10.4014/jmb.1505.05008
Liu, J., Ji, K., Cao, Y., Zhang, C., and He, M. (2011). Selection of bacteria beneficial to the mycelial growth of Phlebopus portentosus. Acta Edulis Fungi 18, 36–38. doi: 10.16488/j.cnki.1005-9873.2011.02.014
Liu, J., Wang, W. B., Ji, K. P., Zhang, C. X., and He, M. X. (2012). Effect of bacteria in the casing soil for fruit of Phlebopus portentosus. Microbiology/Weishengwuxue Tongbao 39, 1622–1628. doi: 10.13344/j.microbiol.china.2012.11.012
Łukasik, P., Newton, J. A., Sanders, J. G., Hu, Y., Moreau, C. S., Kronauer, D. J. C., et al. (2017). The structured diversity of specialized gut symbionts of the new world army ants. Mol. Ecol. 26, 3808–3825. doi: 10.1111/mec.14140
Martos, E. T., Zied, D. C., Junqueira, P. P. G., Rinker, D. L., Silva, R. D., Toledo, R. C. C., et al. (2017). Casing layer and effect of primordia induction in the production of Agaricus subrufescens mushroom. Agric. Nat. Resour. 51, 231–234. doi: 10.1016/j.anres.2017.04.003
Mignucci, J. S., Hernández-Bacó, C., Rivera-Vargas, L., Betancourt, C., and Alameda, M. (2000). Diseases and pests research on oyster mushrooms (Pleurotus spp.) in Puerto Rico. Int. J. Mushroom Sci. 3, 21–26.
Nishino, K., Matsubara, K., Tanaka, C., Yamaguchi, M., Fujita, T., Yamada, A., et al. (2018). Seasonal changes in the content of the (oxalato) aluminate complex, an antimicrobial substance of the “shiro” of Tricholoma matsutake, and the bacterial community structure in the shiro area. Mushroom Sci. Biotechnol. 25, 9–16. doi: 10.24465/msb.25.1_9
Noble, R., Dobrovinpennington, A., Hobbs, P. J., Pederby, J., and Rodger, A. (2009). Volatile C8 compounds and pseudomonads influence primordium formation of Agaricus bisporus. Mycologia 101, 583–591. doi: 10.2307/20619219
Noble, R., Fermor, T. R., Lincoln, S., Dobrovinpennington, A., Evered, C., Mead, A., et al. (2003). Primordia initiation of mushroom (Agaricus bisporus) strains on axenic casing materials. Mycologia 95, 620–629. doi: 10.1099/00221287-95-2-313
Oh, S.-Y., Fong, J. J., Park, M. S., and Lim, Y. W. (2016). Distinctive feature of microbial communities and bacterial functional profiles in Tricholoma matsutake dominant soil. PLoS One 11:e0168573. doi: 10.1371/journal.pone.0168573
Park, J. Y., and Agnihotri, V. P. (1969). Sporophore production of Agaricus bisporus in aseptic environments. Antonie Van Leeuwenhoek 35, 523–528. doi: 10.1007/BF02219169
Partanen, P., Hultman, J., Paulin, L., Auvinen, P., and Romantschuk, M. (2010). Bacterial diversity at different stages of the composting process. BMC Microbiol. 10:94. doi: 10.1186/1471-2180-10-94
Partida-Martinez, L. P., Monajembashi, S., Greulich, K. O., and Hertweck, C. (2007). Endosymbiont-dependent host reproduction maintains bacterial-fungal mutualism. Curr. Biol. 17, 773–777. doi: 10.1016/j.cub.2007.03.039
Philippoussis, A., Diamantopoulou, P., and Zervakis, G. (2003). Correlation of the properties of several lignocellulosic substrate to the crop performance of the Shiitake mushroom Lentinula edodes. World J. Microbiol. Biotechnol. 19, 551–557. doi: 10.1023/a:1025100731410
Pylro, V. S., Roesch, L. F. W., Morais, D. K., Clark, I., Hirsch, P. R., and Totola, M. R. (2014). Data analysis for 16S microbial profiling from different benchtop sequencing platforms. J. Microbiol. Methods 107, 30–37. doi: 10.1016/j.mimet.2014.08.018
Qin, Y., Li, Q.-Q., Zheng, Y.-H., Ding, G.-L., and Guo, X. (2016). Bacteria diversity of shiroes soil growing three species of wild Boletus. Microbiol. China 43, 2148–2153. doi: 10.13344/j.microbiol.china.150823
Rainey, P. B. (1991). Effect of Pseudomonas putida on hyphal growth of Agaricus bisporus. Fungal Biol. 95, 699–704. doi: 10.1016/S0953-7562(09)80817-4
Rossouw, W., and Korsten, L. (2017). Cultivable microbiome of fresh white button mushrooms. Lett. Appl. Microbiol. 64, 164–170. doi: 10.1111/lam.12698
Ryota, K., Fau, S. Z., Fau, K. J., Fau, A. M., Fau, S. R., Fau, N. A., et al. (2012). Detecting nonculturable bacteria in the active mycorrhizal zone of the pine mushroom Tricholoma matsutake. J. Microbiol. 50, 199–206. doi: 10.1007/s12275-012-1371-7
Salter, S. J., Cox, M. J., Turek, E. M., Calus, S. T., Cookson, W. O. C. M., Moffatt, M. F., et al. (2014). Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12:87. doi: 10.1186/s12915-014-0087-z
Sanmee, R., Dell, B., Lumyong, P., Izumori, K., and Lumyong, S. (2003). Nutritive value of popular wild edible mushrooms from northern Thailand. Food Chem. 82, 527–532. doi: 10.1016/S0308-8146(02)00595-2
Santos, P. E. L., Bustilloscristales, R., and Caballeromellado, J. (2001). Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl. Environ. Microbiol. 67, 2790–2798. doi: 10.1128/AEM.67.6.2790-2798.2001
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Segedin, B. P. (1987). An annotated checklist of Agarics and Boleti recorded from New Zealand. New Zeal. J. Bot. 25, 185–215. doi: 10.1080/0028825X.1987.10410067
Shi, S., Nuccio, E. E., Shi, Z. J., He, Z., Zhou, J., and Firestone, M. K. (2016). The interconnected rhizosphere: high network complexity dominates rhizosphere assemblages. Ecol. Lett. 19, 926–936. doi: 10.1111/ele.12630
Stoyanova, M., Pavlina, I., Moncheva, P., and Bogatzevska, N. (2007). Biodiversity and incidence of Burkholderia species. Biotechnol. Biotechnol. Equip. 21, 306–310. doi: 10.1080/13102818.2007.10817465
Szekely, A., Sipos, R., Berta, B., Vajna, B., Hajdu, C., and Marialigeti, K. (2009). DGGE and T-RFLP analysis of bacterial succession during mushroom compost production and sequence-aided T-RFLP profile of mature compost. Microb. Ecol. 57, 522–533. doi: 10.1007/s00248-008-9424-5
Valdrighi, M. M., Pera, A., Agnolucci, M., Frassinetti, S., Lunardi, D., and Vallini, G. (1996). Effects of compost-derived humic acids on vegetable biomass production and microbial growth within a plant (Cichorium intybus)-soil system: a comparative study. Agric. Ecosyst. Environ. 58, 133–144. doi: 10.1016/0167-8809(96)01031-6
Wang, W.-B., Cao, Y., Ji, K.-P., Liu, J., Zhang, C.-X., and He, M.-X. (2013). Effect of Actinomycetes of casing on fruiting of Phlebopus portentosus. Edible Fungi of China 32, 36–38. doi: 10.13629/j.cnki.53-1054.2013.02.016
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Watling, R. (2001). The relationships and possible distributional patterns of boletes in south-east Asia. Fungal Biol. 105, 1440–1448. doi: 10.1017/S0953756201004877
Wood, D. A., and Hammond, J. B. W. (1977). Ethylene production by axenic fruiting cultures of Agaricus bisporus. Appl. Environ. Microbiol. 34, 228–229. doi: 10.1002/jobm.3630181007
Wu, Y., Liu, J., Cao, Y., He, M.-X., Zhang, Ch.-X., Wang, W.-B., et al. (2013). Effect of adding compound strains of bacteria and Actinomycetes on fruit body growth of Phlebopus portentosus. Southwest China J. Agric. Sci. 26, 1984–1987. doi: 10.16213/j.cnki.scjas.2013.05.064
Young, L., Chu, J., and Young, C. (2012). Beneficial bacterial strains on Agaricus blazei cultivation. Pesq. Agrop. Brasileira 47, 815–821. doi: 10.1590/S0100-204X2012000600012
Zarenejad, F., Yakhchali, B., and Rasooli, I. (2012). Evaluation of indigenous potent mushroom growth promoting bacteria (MGPB) on Agaricus bisporus production. World J. Microbiol. Biotechnol. 28, 99–104. doi: 10.1007/s11274-011-0796-1
Zhang, J., Chen, Q., Qin, L., Wang, S., and Zhang, G. (2015). Dynamics of bacterial community of Suillus granulatus mushroom shiro in Pine (Pinus tabuliformis) forests. Chin. J. App. Environ. Biol. 21, 974–980. doi: 10.3724/SP.J.1145.2015.03001
Zhang, C., He, M., Liu, J., Xu, X., Cao, Y., Gao, F., et al. (2017). Brief introduction to a unique edible Bolete-Phlebopus portentosus in Southern China. J. Agric. Sci. Technol. B 7, 386–394. doi: 10.17265/2161-6264/2017.06.003
Zhang, C., Huang, T., Shen, C., Wang, X., Qi, Y., Shen, J., et al. (2016). Downregulation of ethylene production increases mycelial growth and primordia formation in the button culinary-medicinal mushroom, Agaricus bisporus (Agaricomycetes). Int. J. Med. Mushrooms 18, 1131–1140. doi: 10.1615/IntJMedMushrooms.v18.i12.80
Zhang, B., Yan, L., Li, Q., Zou, J., Tan, H., Tan, W., et al. (2018). Dynamic succession of substrate-associated bacterial composition and function during Ganoderma lucidum growth. PeerJ 6:e4975. doi: 10.7717/peerj.4975
Keywords: casing soil, decontamination, bacterial diversity, dynamics, MiSeq
Citation: Yang R-H, Bao D-P, Guo T, Li Y, Ji G-Y, Ji K-P and Tan Q (2019) Bacterial Profiling and Dynamic Succession Analysis of Phlebopus portentosus Casing Soil Using MiSeq Sequencing. Front. Microbiol. 10:1927. doi: 10.3389/fmicb.2019.01927
Edited by:
Baokai Cui, Beijing Forestry University, ChinaReviewed by:
Liang-Dong Guo, Institute of Microbiology (CAS), ChinaZheng Wang, Yale University, United States
Copyright © 2019 Yang, Bao, Guo, Li, Ji, Ji and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kai-Ping Ji, amtwY25sY0AxMjYuY29t; Qi Tan, c3lqMEBzYWFzLnNoLmNu; dHFzYWFzQDE2My5jb20=
†These authors have contributed equally to this work
 Rui-Heng Yang
Rui-Heng Yang Da-Peng Bao
Da-Peng Bao Ting Guo1,2
Ting Guo1,2 Qi Tan
Qi Tan