
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 09 August 2019
Sec. Extreme Microbiology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.01841
This article is part of the Research Topic Microbial Life Under Stress: Biochemical, Genomic, Transcriptomic, Proteomic, Bioinformatics, Evolutionary Aspects and Biotechnological Applications of Poly-Extremophilic Bacteria View all 27 articles
Forced aeration is one of the major energy consumption factors of the bioleaching process of run-of-mine ore. The effect of aeration in the microbial community has scarcely been studied at industrial level. Leptospirillum ferriphilum is one of the most representative species of the Fe3+ producing population in this kind of systems. We analyzed the effect of oxygen availability on L. ferriphilum by growth activity and transcriptional dynamics of its two terminal oxidases (cbb3 and bd complexes) under different experimental test: culture reactor, bioleaching column, and industrial heap tests. Relatively low O2 availability triggered important changes in the microbial community composition, cell growth, microbial activity and cydAB genes transcription in all cases of study. We assessed the potential role of the terminal oxidases on the adaptation to variable aeration conditions in different lifestyles of L. ferriphilum and identified transcriptional markers associated to oxygen metabolism in an industrial system. An interesting hypothesis about the possible role of the cbb3 complex in the response to oxidative stress as well as their role as a high oxygen-affinity oxidase in L. ferriphilum is proposed and discussed. This study successfully proves the function of the cydAB genes as valid genetic markers for low-grade copper industrial bioleaching systems.
It is known that forced aeration is the main O2 and CO2 source in bioleaching systems, both sustaining microbial activity and subsequent metal recovery (Ghorbani et al., 2016; Lakshmanan et al., 2016). Nowadays it is acknowledged that forced aeration is estimated to account for 30% of the cost of energy in a copper bioleaching process. However, it is not possible to measure the gas availability through the entire ore-heap as it implies a huge economic cost. Actually, the operation practices regarding aeration includes only switching on or off the blowers systems in the process and it is not possible to know the real effect of these changes on the microbial activity and metal recovery. For this reason, a current biotechnological and operational challenge is to identify an effective and reliable method that allows monitoring O2 and CO2 availability in industrial bioleaching process as well as their effect on the microbial consortia.
Dissolved oxygen (DO) is the main final electron acceptor in most bioleaching microorganisms (aerobic; chemolithoautotrophic) for energy generation by iron (II) and/or reduced sulfur oxidization pathway (Levicán et al., 2012). Prokaryotic cells have the ability to induce branched-respiratory chains with multiple oxidases that, depending on species, can have different affinities for oxygen (Bueno et al., 2012). This ability provides them with a flexible respiratory system able to adapt to variable oxygen availability by modulating gene expression as a function of the oxygen levels (Levicán et al., 2012). This trait confers an extraordinary adaptive ability to microorganisms to both colonize different environments and to opportunely respond to oxygen variations (Bueno et al., 2012).
Several works have described the diversity of species inhabiting bioleaching systems and acid mine drainage environments and their relevance in the metal recovery process (Bond and Banfield, 2001; Demergasso et al., 2005; Xie et al., 2007; Remonsellez et al., 2009; Bobadilla-Fazzini et al., 2017). Leptospirillum ferriphilum is a Gram-negative and extreme acidophilic bacterium that can derive energy only from ferrous iron oxidization and can only use oxygen as final electron acceptor (Coram and Rawlings, 2002). This species is recognized as one of the most stable and abundant microbial bacterium of bioleaching systems (Demergasso et al., 2005; Galleguillos et al., 2009; Remonsellez et al., 2009) because of its wide temperature tolerance range (37–48°C) (Acosta et al., 2014), its high Fe2+ affinity and its high Fe3+ tolerance (Coram and Rawlings, 2002). These traits make it also one of the key player in the industrial bioleaching processes (Galleguillos et al., 2013).
It has been proposed that species of the Leptospirillum genus probably have the ability to flexibilize their cellular respiration in response to changes in oxygen-tension (Levicán et al., 2012), but the mechanisms underlying this ability are still unknown. Direct effect of DO availability in the performance of biomining microorganisms has scarcely been studied and the available knowledge is more related to the effect of oxygen on metal recovery (Cautivo and Gentina, 2009) than to microbial growth and activity.
It is known that under O2-limited conditions, some other bacteria induce high-oxygen-affinity cytochrome oxidases to respire the traces of gas (Bueno et al., 2012). The cbb3-type (Hemme-Copper Oxidase family, Type A) and bd-type (Quinol Oxidase Family only found in bacteria and archaea) oxidases have been described as cytochromes with high-oxygen-affinity in several species, such as Escherichia coli, Rhodobacter capsulatus, and Bradyrhizobium japonicum (Borisov et al., 2011; Bueno et al., 2012). Additionally, in E. coli, the bd-type cytochrome has been associated to protection against oxidative stress (Lindqvist et al., 2000). In bioleaching species, there is one study related to this topic and was performed on the most studied acidophilic species, Acidithiobacillus ferrooxidans, that has terminal cytochrome c oxidase aa3-type and terminal cytochrome bd and bo-type ubiquinol oxidases (Brasseur et al., 2004). The authors observed that A. ferrooxidans synthesizes cytochrome bd oxidase in highly aerated cultures and suggested that this oxidase probably plays an active role in the rapid consumption of oxygen for maintaining a low intracellular oxygen concentrations compatible with nitrogenase function. Analysis performed on Leptospirillum genomes showed that all species from this genus presented both cbb3 and bd-type terminal oxidases (Levicán et al., 2012) and, unlike A. ferrooxidans, Leptospirillum did not have cytochromes with recognized low-oxygen-affinity. Levicán et al. (2012) inferred, through a genomic analysis, that Leptospirillum sp. would be able to sustain a high iron oxidization rate even at low oxygen levels due to the presence of the high-oxygen-affinity cbb3 cytochrome, which represents an adaptive advantage for the genus in the system.
It has been reported that expression of genes associated to cytochromes with high-oxygen-affinity like cbb3 and bd-type oxidases is strongly induced in cells grown under microaerobic or anaerobic conditions (Pitcher and Watmough, 2004), while those associated to cytochromes with low-oxygen-affinity, as cytochrome bo3 ubiquinol oxidase, are expressed in cells grown under aerobic conditions in E. coli (Pitcher and Watmough, 2004). Parro et al. (2007) mentioned an induction of the operon cydAB in Leptospirillum ferrooxidans from one site (S1) of the Tinto River having a relatively lower oxygen availability (13.1% sat O2 vs. 64.8% sat O2), suggesting that bd-type oxidase would have a high-affinity for oxygen in this species. However, the transcriptomic data of cydAB was not properly described in that study so results are inconclusive.
Omics information of chemolithoautotrophic and acidophilic species has been increasing through time (Orellana and Jerez, 2011; Bonnefoy and Holmes, 2012; Aliaga et al., 2013, 2015; Quatrini and Johnson, 2018). However, comprehension of the metabolic response of these microorganisms to the environmental variability inherent to bioleaching processes is still scarce. The aim of this research was to study the transcriptomic response of the terminal cytochrome cbb3 and bd oxidases in L. ferriphilum growing under different laboratory and industrial environments and different oxygen availabilities with the biotechnological purpose to identify and validate molecular markers to monitor the aeration parameter in industrial bioleaching processes.
Leptospirillum ferriphilum type-strain DSM 14647 was grown in 3 L reactors with 1 L of ABS medium (g/L), (NH4)2SO4 (0.15), Na2SO4 (0.066), KCl (0.05), MgSO4∗7H2O (0.5), KH2PO4 (0.05), Ca(NO3)2∗4H2O (0.014), trace element (Bryan et al., 2012), and with FeSO4 7H2O (50 mM) at 37°C, pH 1.5, agitation 200 rpm and three different conditions: 3, 10, and 23% (control) of O2 in the inlet air, respectively. Oxygen concentration was regulated by gas mixture (atmospheric compressed air and N2) and feeding flow rate at 400 mL/min in the inlet flow. Experiment was reproduced to obtain an experimental repetition. The input and output air O2 concentration (%) in the reactors and the DO concentration of the cultures were monitored using a Micro-Oxymax respirometer (Columbus Instruments) and a portable DO meter, respectively. Cell growth curves were built with data of cell count in Neubauer camera every 3 h for 3 days. All cultures were sampled at the late exponential growth phase when ferrous consumption reached ∼90% and cells were in abundance and metabolically active. Cell concentrations of reactors at the sampling time were in the range of 9.0 × 107 to 1.5 × 108 cell/mL in both experiments. Cells were recovered by vacuum filtration through nitrocellulose membrane (0.2 μm), and were immediately preserved at −80°C in RNAlater® solution (Invitrogen TM AM7021) until nucleic acid isolation.
Two copper bioleaching columns (col1/control and col2/air-test, 1 m high and 0.6 m in diameter) were simultaneously run and controlled. Samples were collected at different operational days (OD) (Table 1) for population and transcriptomic analysis between February and March 2015. Columns were operated at an average temperature of 21–25°C in an open circuit system fed with raffinate and inoculated with a microbial consortium from the Escondida Mine (MEL) industrial bioleaching heap, located 170 km south-east of Antofagasta, Chile (Figure 1). All physicochemical parameters (pH, temperature, nutrient-NH4) were maintained stable during the operation except the inlet air of col2 that was intentionally interrupted two times by air displacement with N2 (Table 1). The percentage of inlet-oxygen, when it was turned on, was constantly 21%. Input (-in) and output (-out) O2 concentration (percentage of O2 in the air) were continually monitored in both columns by a Micro-Oxymax respirometer (Columbus Instruments). Cell collection was performed before and after the input-airflow interventions of col2 by filtration of 40 L of PLS (Pregnant Leach Solution) through a 0.2 μm nitrocellulose membranes. Cells were preserved as described in Section “Cultures of Leptospirillum ferriphilum.”
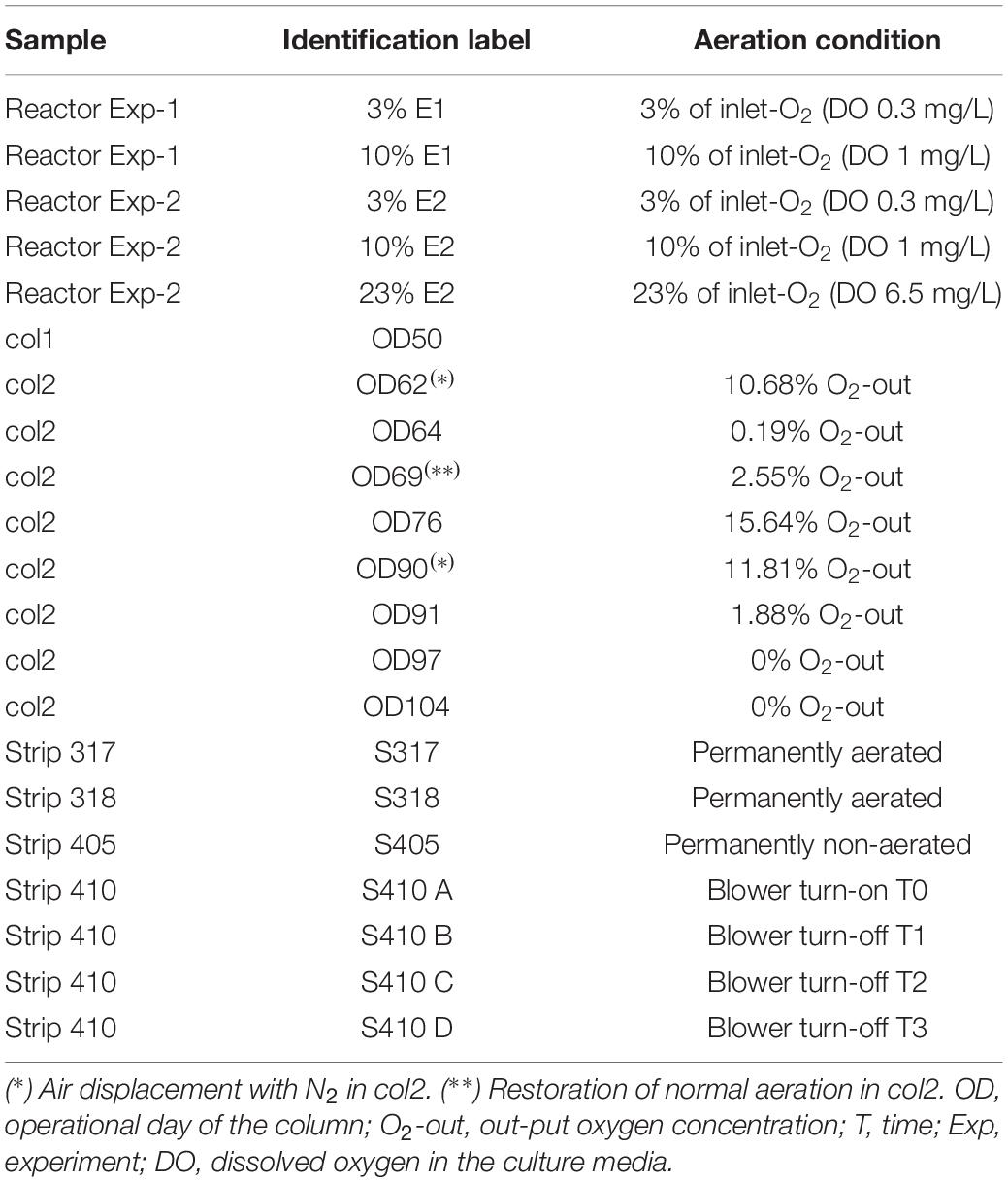
Table 1. Samples from L. ferriphilum cultures, bioleaching columns and industrial heap with details of their respective air (O2) feeding situation.
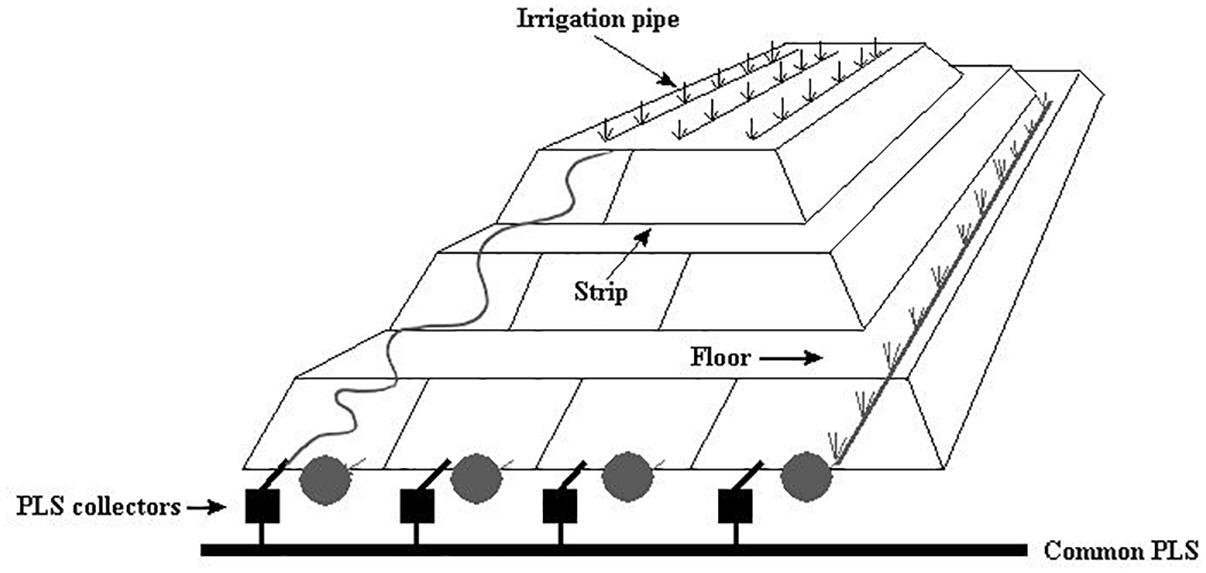
Figure 1. Schematic view of heap bioleaching at the Escondida mine adapted from Acosta et al. (2014).
The industrial bioleaching heap from Escondida Mine was built with ROM (run of mine) material ore characterized as low-grade sulfide minerals (Soto et al., 2013; Acosta et al., 2014). This heap is divided into lifts and strips. The strips are irrigated with raffinate which percolates vertically up to the bottom, finally obtaining the PLS (Figure 1). Six PLS samples from four strips (317, 318, 405, and 410), each with different aeration conditions, were sampled (Table 1). Briefly, PLS (40 L) of strips 317 and 318 (permanently aerated, with both blowers in turn-on) and 405 strip (permanently non-aerated, with blowers in turn-off) were obtained in a first sampling. In addition, PLS (40 L) from the aerated strip 410 was obtained before (one sample) and after (three samples) an aeration interruption. Aeration suspension was carried out by turning off the blowers (forced aeration) located in the base of the MEL heap. All cell samples were collected and preserved as described above.
Genomic DNA from cells was isolated from eight column samples (OD 62, 64, 69, 76, 90, 91, 97, and 104) and seven industrial samples (S317, 318, 405, 410A, 410B, 410C, and 410D) (Table 1) as previously described (Demergasso et al., 2005). Effect of aeration on bioleaching community and population dynamics were determined by absolute quantification of 16S rRNA genes associated to nine acidophilic species using quantitative Real-Time PCR (q-PCR) as previously described (Marín et al., 2017).
RNA of all samples (Supplementary Table 1) was isolated and purified as previously described (Marín et al., 2017). Reverse transcription was performed from digested (RQ1 RNase- Free DNase, Promega M 6101) total RNA using the First Strand cDNA Synthesis Kit (Thermo Scientific, cat. No. K1612) in presence of ribonuclease inhibitor RNasin® (Promega, N2511) and random primers, following directions provided by the manufacturer. Retrotranscripts were stored at −20°C for 1 week or −80°C for longer time.
The metagenomic analysis of a mix of industrial bioleaching samples from MEL was previously performed (Acosta et al., 2017). L. ferriphilum related sequences were separated trough mapping with CLC Genomic Workbench 8.1 (QIAGEN Bioinformatics) against Leptospirillum genomes. A representative reconstructed genome was chosen based on the coverage and the depth of the sequencing. The sequences of the reconstructed genome were then annotated using the RAST Server (Aziz et al., 2008).
Primers were designed for genes coding proteins related to terminal oxidases cbb3 and bd from L. ferriphilum (Table 2). In addition, primers were designed for 16S rRNA and alaS genes from L. ferriphilum. These housekeeping genes were used as endogenous controls for the gene expression calculation. The chosen genes of L. ferriphilum strains ML04 (genome paper), SpCl (genome paper) and the reconstructed MEL genome were aligned with ClustalW (Larkin et al., 2007) and conserved regions were used as base for the primer design. Finally, the promissory candidate primers were evaluated with Primer-BLAST (Ye et al., 2012).
All primers were validated in their specificity, repeatability and sensibility (Table 2, Supplementary Figures 1–5, and Supplementary Table 1). Pearsons (R) coefficients of 0.99, optimal efficiencies (E) (100–110%) and dynamic ranges of 1 × 102–1 × 107 were obtained for all selected primers (Table 2).
Relative gene expression analysis was performed by RT-qPCR (two steps) using a real-time PCR machine Rotor-Gen Q (Qiagen). Amplification reaction was composed by: 1X SensiMixTM SYBR No-ROX (BIOLINE, cat. No. QT650-05), 0.1 μM of each specific primer designed for selected genes (Table 2), 1 μL of previously generated cDNA as template and water to complete a final volume of 10 μL. The thermal cycling protocol was as follows: initial denaturation at 95°C for 10 min followed by 40 cycles at 95°C for 30 s, 60°C for 30 s, and 70°C for 30 s. Each qPCR run was performed using technical triplicates. Relative quantification of transcripts was calculated by Pfaffl mathematical model (Pfaffl, 2001) using Excel and REST© 2009 software (Pfaffl et al., 2002). Pfaffl model has advantage to consider the real Efficiency (E) value of qPCR reaction in the calculation unlike other available models such as 2–ΔΔ CQ (Livak and Schmittgen, 2001). Pfaffl model has been successfully used in previous gene expression analysis for acidophilic microorganisms (Galleguillos et al., 2013; Acosta et al., 2014; Marín et al., 2017).
It is known that to use one or more validated reference genes is crucial to obtain reliable and repeatable relative expression data (Vandesompele et al., 2002). In this work, all relative gene expression analyses were normalized using two reference genes previously validated by geNorm (Vandesompele et al., 2002) and NormFinder (Andersen et al., 2004) algorithms: alaS and 16S rRNA. The relative expression data was represented as Log2 (ratio). All RT-qPCR analyses performed in this work presented negative controls without amplification. Significant differences in relative expression were adjusted at 1 < Log2-ratio < −1. In other words, all data (Log2-ratio) > −1 and < 1 was considered as a non-significant difference of relative gene expression.
RNA concentrations obtained from samples of the pure cultures, experimental column and MEL strips ranged between 26.4 and 173.0 ng/μL (Supplementary Table 1). Efficiency obtained for each primer set was between 1.80 and 2.17 (Table 2), and high specificity (Supplementary Figures 1–5) and sensitivity (results not shown) for designed primers were achieved in all cases.
Growth of L. ferriphilum DSM 14647 changed depending on the analyzed O2 conditions. Less cell growth was observed in reactor with 3% (0.3 mg/L of DO in the culture medium) and 10% (1 mg/L of DO in the culture medium) of inlet-O2 with respect to the control condition with 23% of inlet-O2 (6.5 mg/L of DO in the culture medium) (Figure 2A). In addition, differences in the time needed to reach the late exponential growth phase and to complete ferrous oxidization between the three O2 conditions analyzed was observed (Figures 2A,B). Although the growth and ferrous oxidizing activity of L. ferriphilum were not identical in both experimental repetitions, a similar tendency in the repetitions growing at 3 and 10% of inlet-O2 was observed (Figures 2A,B). L. ferriphilum reached the higher cell growth and complete ferrous oxidization at 40 h of cultivation in the control (23% O2), while it was unable to completely oxidize the substrate at O2 restricted incubation (3 and 10% of inlet-O2) up to 45 and 51 h of cultivation, respectively (Figures 2A,B).
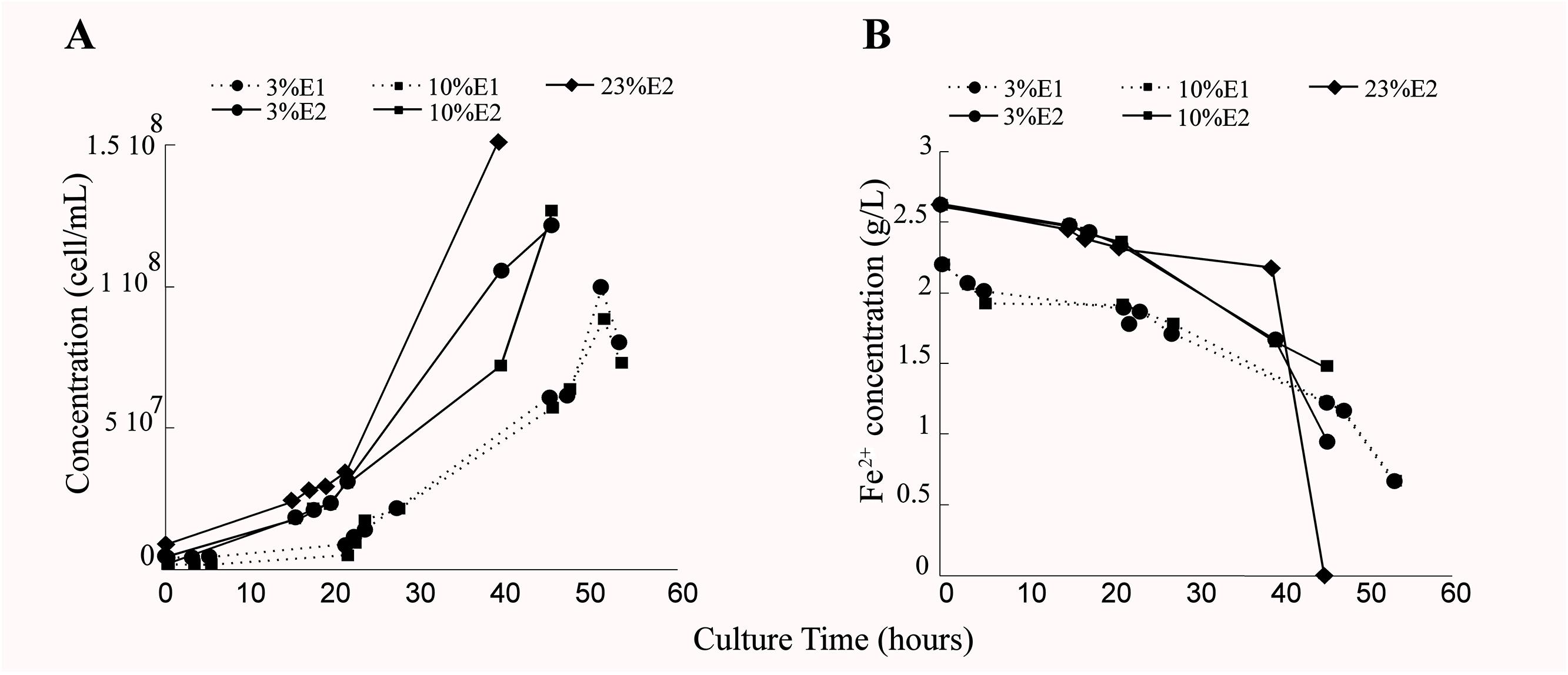
Figure 2. Cell growth (cell/mL) (A) and ferrous consumption (g/L) (B) of Leptospirillum ferriphilum DSM 14647 under the analyzed conditions (3, 10, and 23% of inlet-O2). The dotted and continuous lines are showing the results of the experimental repetitions E1 and E2, respectively.
The population dynamics of the microbial consortia in the experimental columns and the industrial strips showed that L. ferriphilum was the predominant species in these systems (Figure 3). However, the cell concentration changed depending on the aeration. In the bioleaching column, the cell concentration decreased when the aeration was suspended (OD62, 69, 97, and 104) and increased when the aeration was restored (OD76, 90, and 91) (Figure 3A). Moreover, a significant decrease of L. ferriphilum concentration after the second aeration suspension in OD90 was noticed. Even lower concentrations of L. ferriphilum in the bioleaching column were observed at OD 69 and 104, after 7 and 14 days of aeration suspension respectively (Figure 3A). The same tendency was observed in samples of the industrial bioleaching heap where most of non-aerated strips (S405, S410B, S410C, S410D) presented lower cellular concentration compared to aerated strips (S317, S410A) (Figure 3B and Table 1).
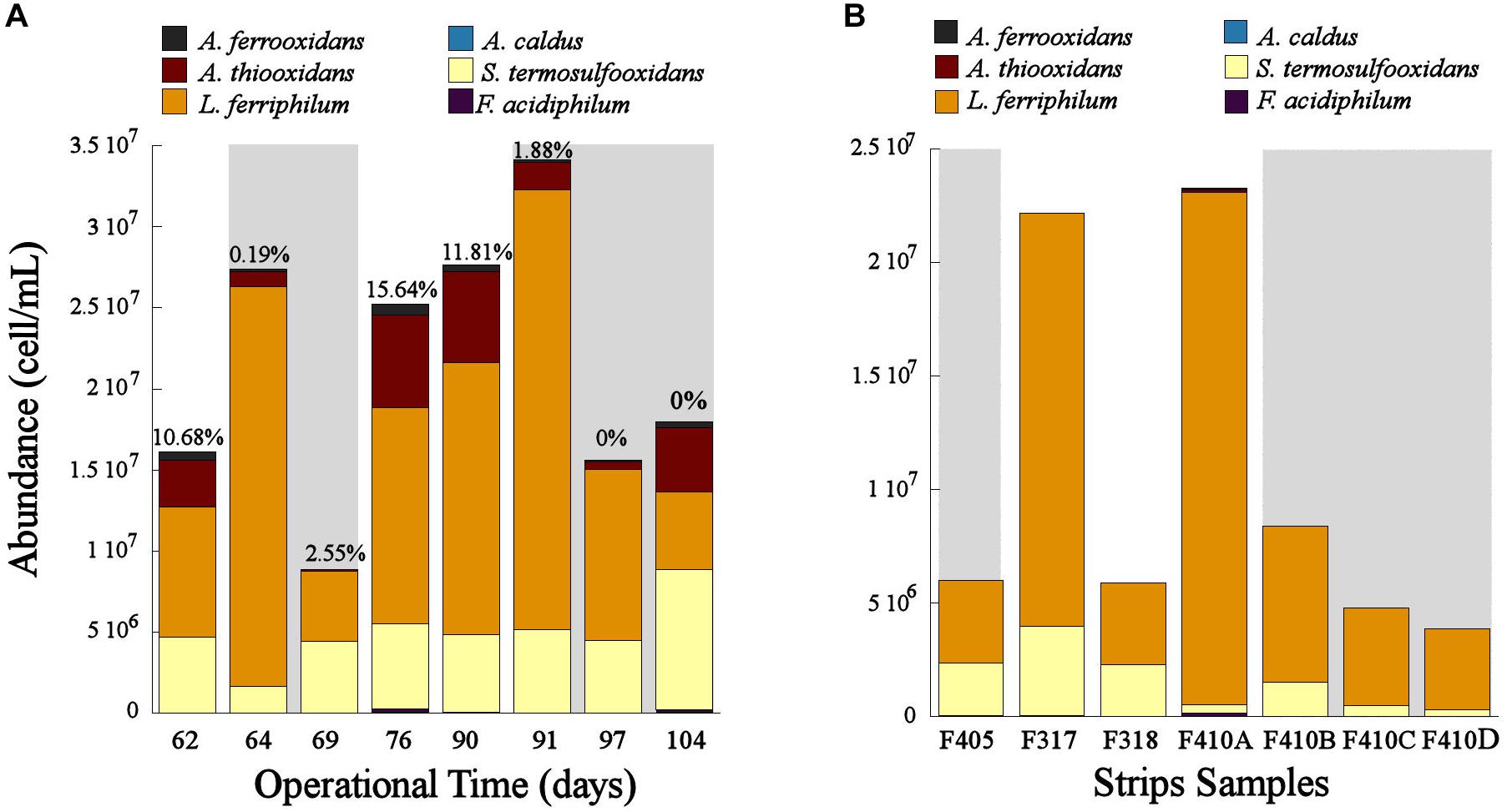
Figure 3. Population dynamics of the microbial community in the bioleaching column (A) and in the industrial bioleaching strips (B) in response to variable O2 availabilities. Values above each bar shows the O2-out level in the different operational days of the bioleaching column. Gray area shows the samples with low O2 availability (A) and the strips with interrupted aeration (B).
No significant differences (1 > Log2 ratio > −1) in transcription levels of cbb3 and cydAB genes were observed between cultures at 3 and 10% of O2 in both experimental replicates of pure cultures, while both cytochrome complexes were overexpressed 4.5 and 3.3 folds, respectively, in the control reactor with 23% O2 in the feed (Figure 4).
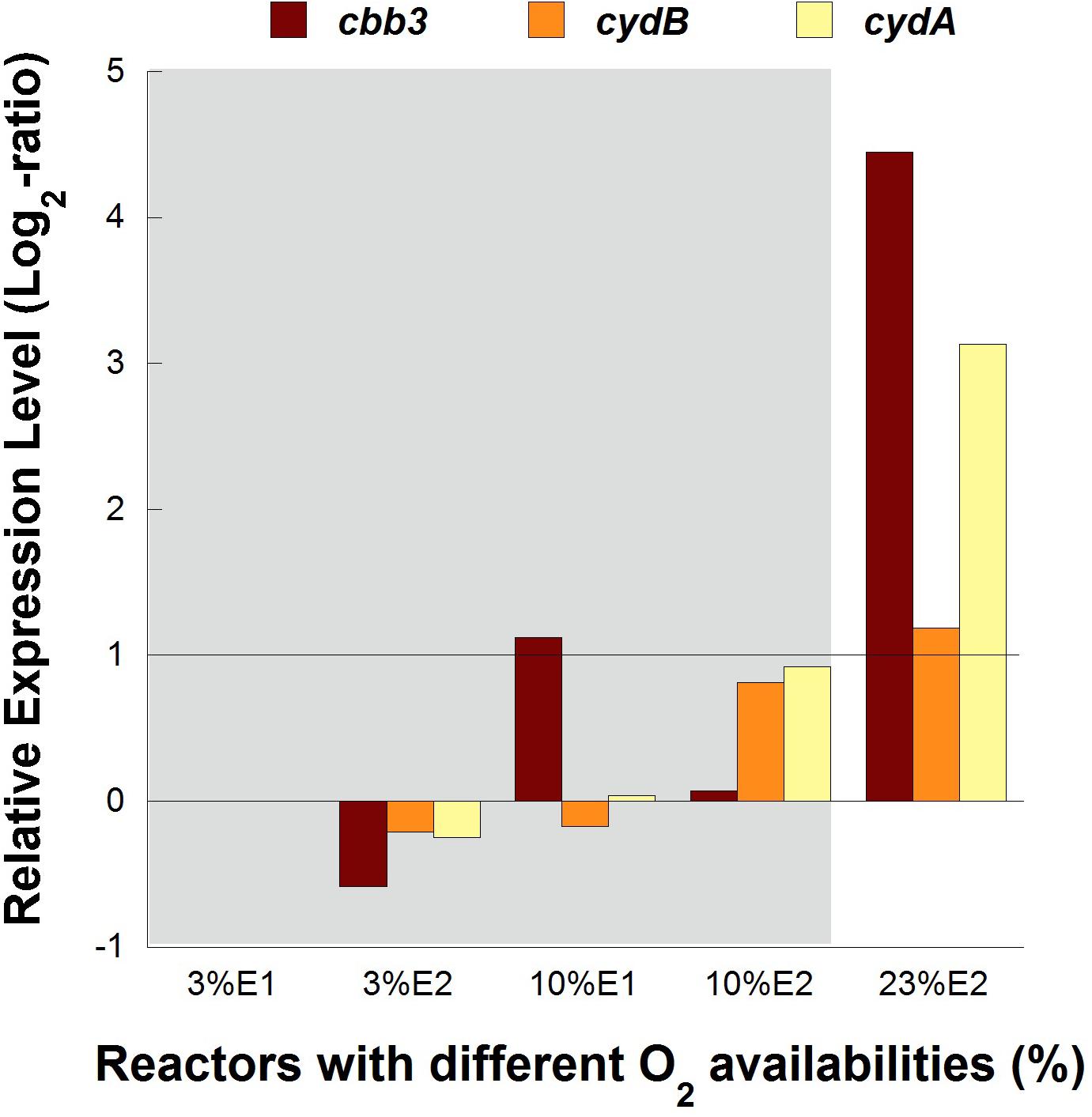
Figure 4. Transcription dynamics of cbb3, cydA, and cydB genes in L. ferriphilum DSM 14647 growing in pure cultures with different O2 concentrations in the air. Gray area shows the reactors with lower O2 levels.
A drop in the percentage of O2-out (from approximately 11 to 0%) in the bioleaching column test (col2) was recorded after both aeration interruption events (days 62–66 and 90–104, respectively) (Figure 5). Also, similar transcription levels (1 > Log2 ratio > −1) were observed for the control sample and the samples taken before the aeration interruption (OD62) in col2 for all analyzed genes, and then an abrupt decrease in their transcription (Log2 ratio < −1) was evidenced after O2 displacement with N2 (OD64 and OD91) (Figure 5). The cbb3 gene showed a greater transcription level (observed as a minor underexpression with respect to the control) than cydAB genes under limited O2 conditions (OD64, 69, 91, 97, and 104) (Figure 5), while all three genes tended to restore their transcription levels after the aeration was restarted in the column (Figure 5). Although the cbb3 gene did not show transcriptional dynamics clearly related to oxygen availability, an evident increase of their transcription (perceived as a gradual decrease in their underexpression through time) after 7 and 13 days of exposition to low O2 availability (OD69 and OD104, respectively) was observed (Figure 5). This result differs from that observed in L. ferriphilum DSM 14647 pure cultures where the cbb3 gene was overexpressed in the highest O2 condition (23%) (Figure 4). This increment in transcription apparently induced by low oxygen levels in col2 was not observed in cydAB genes, which maintained their underexpression (with respect to the control) along all the period time when aeration was suspended in the col2 (OD64, 0.19% O2; OD69, 2.55% O2; OD91, 1.88% O2; OD97, 0% O2; and OD104, 0% O2) (Figure 5).
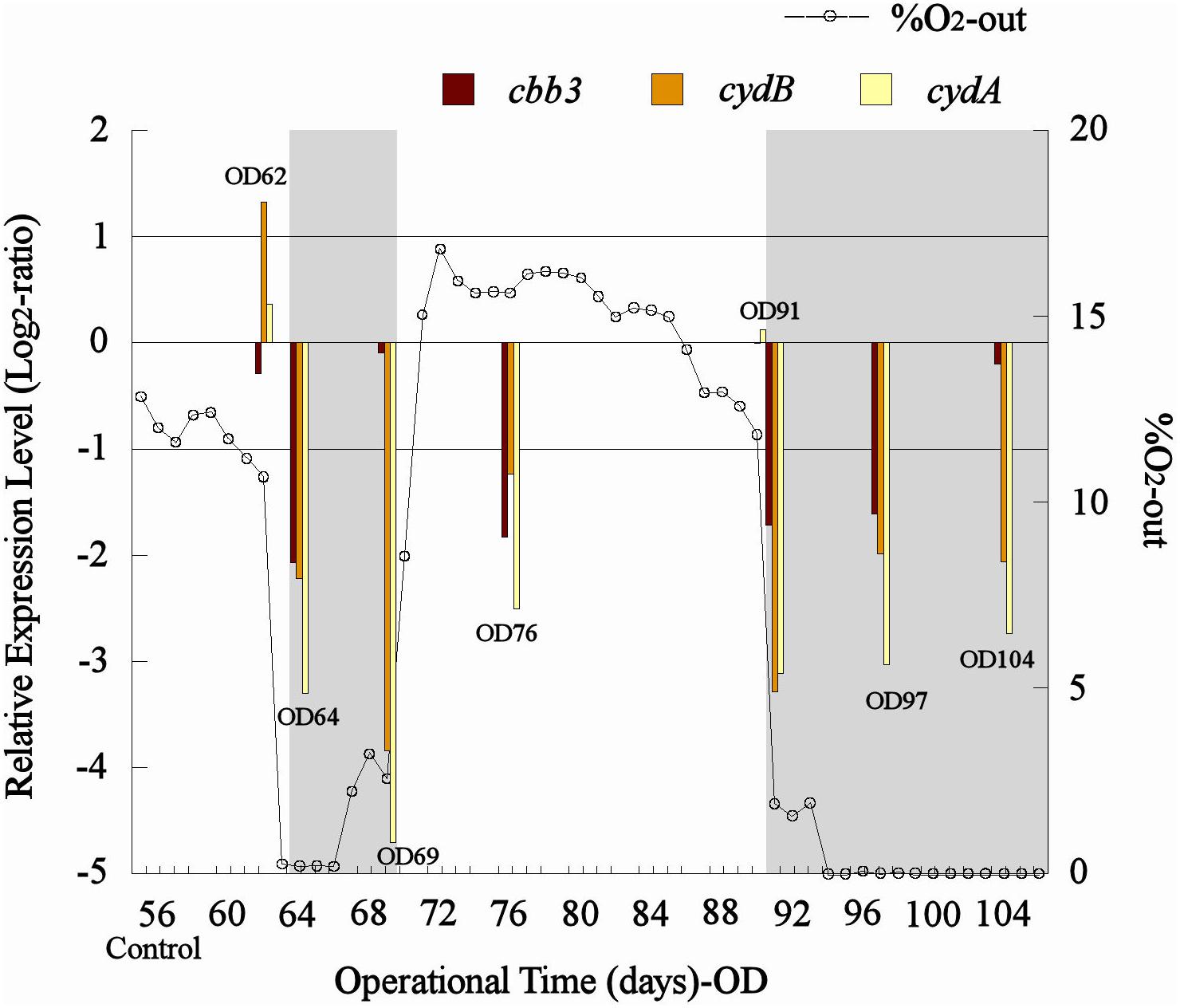
Figure 5. Transcriptional dynamics of cbb3, cydA, and cydB genes of L. ferriphilum during aeration interventions in the bioleaching column. OD, operational day of the column. The dotted line shows the O2-out (%) from col2. Gray area shows periods when aeration was interrupted in the column. When underexpressed bars are approaching zero, that means that gene transcription is increasing.
The cbb3 gene presented very different transcriptional dynamics in both studied industrial scenarios. In the industrial scenario of uninterrupted aeration condition, the cbb3 gene did not show significant transcriptional differences between aerated (S317 and S318) and non-aerated strips (S405) (Figure 6A). However, an overexpression of the cbb3 gene was observed at 14 (S410B) and 28 (S410D) days after aeration interruption of the industrial strip S410 (Figure 6B). On the other hand, cydAB genes maintained a congruent transcriptional dynamic in both industrial scenarios because they were significantly overexpressed in the aerated strips with respect to the non-aerated ones (Figure 6A) and underexpressed in all samples without aeration (S410B and S410D) (Figure 6B).
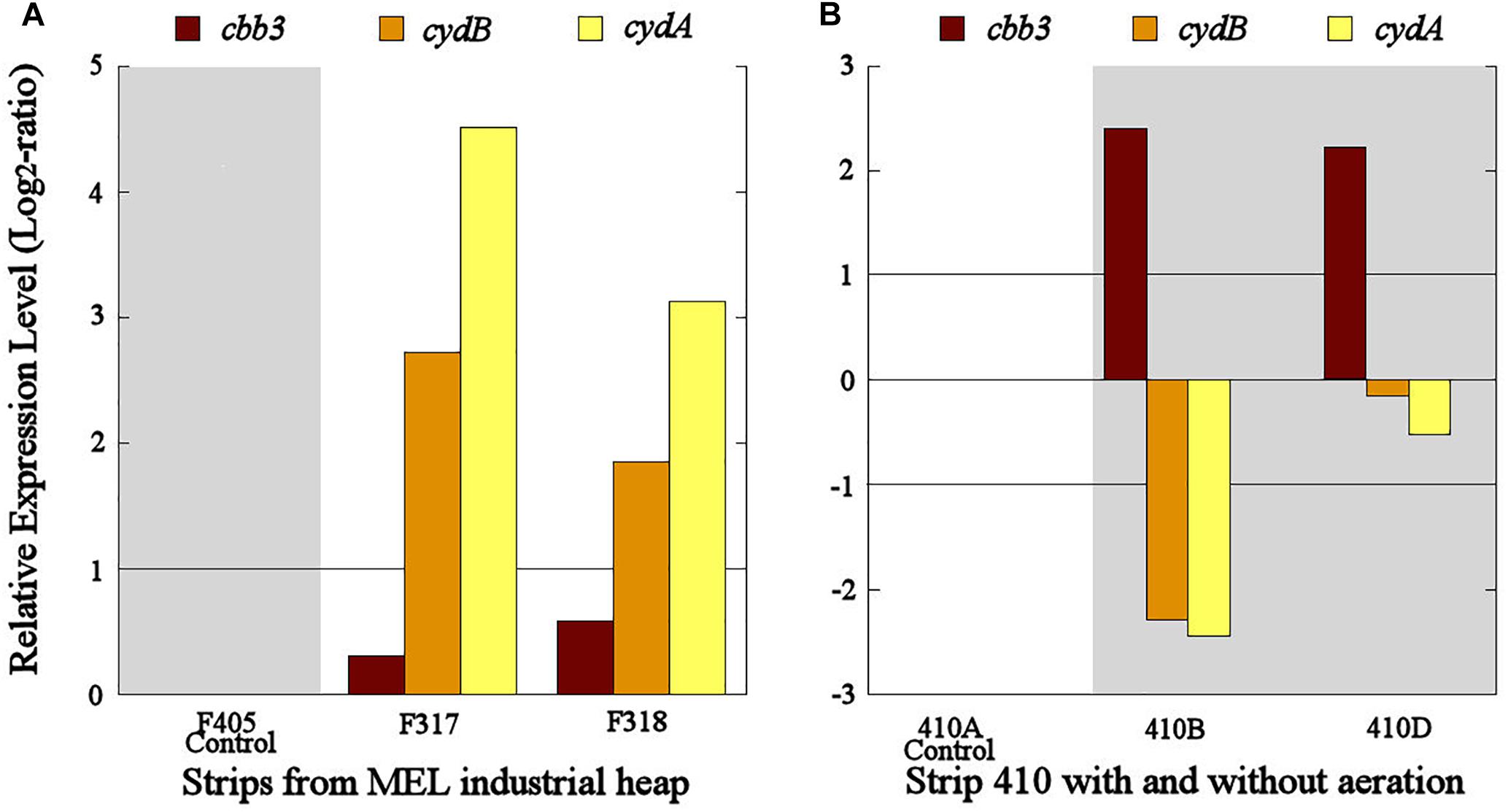
Figure 6. Transcriptional dynamics of cbb3, cydA, and cydB genes in L. ferriphilum from strips S317 (with aeration), S318 (with aeration), S405 (without aeration) (A) and S410 (before and after the aeration suspension) (B) of the industrial copper bioleaching heap. Gray area shows absence or low O2 availability.
A very similar performance was observed in the cell growth and the ferrous oxidation rate at both repetitions in bioreactors tests with 3 and 10% of inlet-O2 (Figures 2A,B), suggesting that this specific difference in the O2 availability did not influence the growth and the oxidizing activity of this species. However, the differences in the cell growth curve and also the incomplete ferrous oxidation observed in cultures with 3–10% (45 and 51 h of culture) with respect to 23% of input-O2 strongly suggest a scarcity of final electron acceptor (O2) availability during the Fe2+ oxidation under those decreased O2 conditions (Figure 2B).
Leptospirillum ferriphilum was the predominant species in the bioleaching column and in the industrial strips (Figure 3). This result agrees with previous studies describing this species as one of the principal ferrous oxidizers in bioleaching systems (Demergasso et al., 2005; Galleguillos et al., 2009, 2013). The decrease in cell concentration of L. ferriphillum during the suspension of the aeration (OD62, 69, 97, and 104) and its subsequent increase observed after the oxygen resumption in the bioleaching column (OD76, 90, and 91) (Figure 3A) are both evidence of an inhibited growth of this species triggered by an oxygen starvation. Similar results of inhibited cell growth in response to low O2 availability has been also observed in L. ferriphilum from other bioleaching systems (Xingyu et al., 2010). Moreover, our results suggest that the growth of L. ferriphilum is probably affected after a sustained absence of oxygen in the system and not in an immediate short time as observed in Figure 3A that shows a drop in L. ferriphilum planktonic cell concentration only after 7 and 6 days after the first and second restriction of aeration, respectively.
The results of this study evidenced that L. ferriphilum as well as other species of the bioleaching community such as Sulfobacillus sp. and A. thiooxidans were negatively affected by the relatively low oxygen concentration when ranged from 2.5 to 0% of O2-out in the bioleaching column (Figure 3A). Similar results were also observed in the industrial strips without forced aeration (Figure 3B).
The overexpression of cbb3 and cydAB evidenced in the pure culture reactor with 23% O2 (Figure 4) would suggest that both terminal oxidases (cbb3 and bd-type) were performing as cytochromes with low-oxygen-affinity in L. ferriphilum DSM 14647 because they were overexpressed only at the higher O2 concentration (23% O2) (Bueno et al., 2012). However, only results of cydAB genes agree with those observed in the bioleaching column and in the industrial bioleaching strips. A pronounced and sustained under-expression with respect to the control was observed along all the time that aeration was suspended in the col2 (OD64, 0.19% O2; OD69, 2.55% O2; OD91, 1.88% O2; OD97, 0% O2; and OD104, 0% O2) (Figure 5), suggesting that the cydAB genes consistently presented transcriptional dynamics related to a terminal oxidase with low-oxygen-affinity in L. ferriphilum. These results differ from reported for this cytochrome complex in species such as E. coli (Borisov et al., 2011), Rho. capsulatus (Bueno et al., 2012) and L. ferrooxidans (Parro et al., 2007) where bd-type oxidase is described as a cytochrome with high-oxygen-affinity. Since none of the L. ferriphilum sequenced genomes have any described low-oxygen-affinity cytochromes it is possible that the bd-type complex could be fulfilling this function. In a number of organisms, the bd oxygen reductase is induced under O2-limited conditions as well as under other growth conditions that can be considered stressful, such as Fe deficiency (Borisov et al., 2011). However, in this study the bd complex was overexpressed at the same time that L. ferriphilum presented the best growth and ferrous oxidation rate so the observed transcriptional dynamics is most rather associated with an oxygen availability than a stress condition. On the other hand, Parro et al. (2007) reported that cydAB transcription rose in sectors of Rio Tinto with low oxygen saturation conditions (13 vs. 65% sat. O2 in water) but the scarcity of details in the transcriptomic data presented in that study did not allow us to perform a robust comparative analysis.
Transcriptional dynamics of cydAB genes in all industrial strips were also congruent with that observed in pure cultures and bioleaching column experiments (Figures 4–6). Both genes were always overexpressed and underexpressed when O2 concentrations were apparently sufficient or limiting, respectively. Based on these results, the bd-type cytochrome oxidase of L. ferriphilum consistently fulfills the function of a low-oxygen-affinity oxidase and validate cydAB genes as genetic markers associated to O2 availability for copper bioleaching systems. This finding differs from reported in L. ferrooxidans by Parro et al. (2007) who observed that transcription dynamics of bd-type cytochrome oxidase (cydAB genes) were induced at relatively low oxygen concentrations just like a cytochrome with high-oxygen-affinity. This could be another example of metabolic differences among related species triggered by adaptation processes, as it was exemplified above.
In contrast with the transcriptional dynamics of cydAB genes associated to the high O2-availability that was observed in all the experiments, the cbb3 gene showed different transcriptional dynamics: (i) a gradual increase in the transcription level when forced aeration (O2) was abruptly cut off from the system, as in the case of the bioleaching column (Figure 5) and strip S410 (Figure 6B) and (ii) an increase in the transcription level in the reactor test with relatively high O2 concentration (23% O2) (Figure 4). The increased transcription of the cbb3 gene observed at 7 and 14 days (OD97 and OD104) after the aeration suspension in the bioleaching column (Figure 5) and in the industrial strip 410 (Figure 6B), respectively, suggests that this terminal oxidase could be fulfilling the function of a high oxygen-affinity cytochrome when O2 conditions change abruptly in the system and it remains over time. This cbb3 function has been previously proposed for L. ferriphilum (Levicán et al., 2012) and for other species such as B. japonicum and Rhodobacter sphaeroides (Bueno et al., 2012). However, the cbb3 gene does not seem to fulfill that function in L. ferriphilum DSM 14647, as it showed increased transcription levels at elevated oxygen concentrations in the reactor tests (Figure 4).
These results did not allow us to confirm the cbb3 gene as a validated genetic marker associated solely to oxygen availability. Thus, it is possible to infer that the cbb3 gene in L. ferriphilum is probably repressed or induced depending on oxygen availability and/or oxidative stress level. Aerobic respiration not only provides energy to the cell but, consequently, it also produces reactive oxygen species (ROS) such as hydrogen peroxide and hydroxyl radical that in excess trigger cellular damage and oxidative stress (Sigler et al., 1999). The role of terminal oxidases in preventing ROS production (Lindqvist et al., 2000; Poole and Cook, 2000; Giuffre et al., 2014) or in response to oxidative stress induced by excessive air sparging in continuous culture and by the growth on minerals (Christel et al., 2018) has been described. It is necessary to perform a deeper and more specific investigation to unravel the real function of this gene in the energy metabolism of L. ferriphilum. It is important to note that the L. ferriphilum type-strain (DSM 14647) is a laboratory- adapted strain and not one adapted to bioleaching industrial system as the case of L. ferriphilum from the bioleaching column (col2) and from the industrial strips from MEL heap. Adaptive differences among closely related strains has been widely reported (Brem et al., 2002). As an example, it has been reported that there are strains with (Llinás et al., 2006) and without (Gunasekera et al., 2017) nitrogen fixation ability within the L. ferriphilum species.
As far as we know, this is the first report of a transcriptomic approach related to the oxygen affinity of cytochromes oxidases in L. ferriphilum from which it was also possible to identify validated genetic markers for an industrial and operative copper bioleaching system.
The results of this study allowed us to confirm that the oxygen availability is a key factor influencing the growth and gene transcription dynamics in L. ferriphilum. The bd-type cytochrome has a performance associated to a terminal oxidase with low-oxygen-affinity while cbb3-type cytochrome could be associated to different metabolic functions depending on the oxygen availability and oxidative stress level in this species. The consistent behavior of cydAB genes at different experimental scales provided the evidence to consider it as valid genetic markers associated to oxygen availability in this industrial heap bioleaching system.
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
SM and CD conceived the presented idea, verified the analytical methods, and involved in the planning and supervision of the work. MC and SM developed the theory, performed the experiments, and wrote the manuscript with support from CD. SM performed both the identification of the candidate genes and the design of all primers for the gene-expression analysis by qPCR. MC carried out the gene expression experiments. DC involved in the design and supervision of the columns and reactor experiments. MC, SM, CD, and PG discussed the results and contributed to the final manuscript.
This work was funded by the FONDEF projects IT13I20042 and IT16M100045 of the Comisión Chilena de Ciencia y Tecnología (CONICYT).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was performed with the industrial samples provided by Minera Escondida Ltda. (MEL).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01841/full#supplementary-material
Acosta, M., Galleguillos, P., Ghorbani, Y., Tapia, P., Contador, Y., Velásquez, A., et al. (2014). Variation in microbial community from predominantly mesophilic to thermotolerant and moderately thermophilic species in an industrial copper heap bioleaching operation. Hydrometallurgy 150, 281–289. doi: 10.1016/j.hydromet.2014.09.010
Acosta, M., Galleguillos, P., Guajardo, M., and Demergasso, C. (2017). Microbial survey on industrial bioleaching heap by high-throughput 16S sequencing and metagenomics analysis. Solid State Phenomena 262, 219–223. doi: 10.4028/www.scientific.net/ssp.262.219
Aliaga, D., Comolli, L., Thomas, B., and Banfield, J. (2015). Community transcriptomics reveals unexpected high microbial diversity in acidophilic biofilm communities. ISME J. 9, 1014–1023. doi: 10.1038/ismej.2014.200
Aliaga, D., Dasari, M., Thomas, B., Shah, M., VerBerkmoes, N., Hettich, R., et al. (2013). New group in the leptospirillum clade: cultivation-independent community genomics, proteomics, and transcriptomics of the new species “Leptospirillum group IV UBA BS.”. Appl. Environ. Microbiol. 79, 5384–5393. doi: 10.1128/AEM.00202-13
Andersen, C., Jensen, J., and Ørntoft, T. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. doi: 10.1158/0008-5472.CAN-04-0496
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bobadilla-Fazzini, R., Piña, P., Gautier, V., Brunel, K., and Parada, P. (2017). Mesophilic inoculation enhances primary and secondary copper sulfide bioleaching altering the microbial & mineralogical ore dynamics. Hydrometallurgy 168, 7–12. doi: 10.1016/j.hydromet.2016.07.016
Bond, P., and Banfield, J. (2001). Design and performance of rRNA targeted oligonucleotide probes for in situ detection and phylogenetic identification of microorganisms inhabiting acid mine drainage environments. Microb. Ecol. 41, 149–161. doi: 10.1007/s002480000063
Bonnefoy, V., and Holmes, D. (2012). Genomic insights into microbial iron oxidation and iron uptake strategies in extremely acidic environments. Environ. Microbiol. 14, 1597–1611. doi: 10.1111/j.1462-2920.2011.02626.x
Borisov, V. B., Gennis, R. B., Hemp, J., and Verkhovsky, M. I. (2011). The cytochrome bd respiratory oxygen reductases. Biochim. Biophys. Acta 1807, 1398–1413. doi: 10.1016/j.bbabio.2011.06.016
Brasseur, G., Levican, G., Bonnefoy, V., Holmes, D., Jedlicki, E., and Lemesle-Meunier, D. (2004). Apparent redundancy of electron transfer pathways via bc1complexes and terminal oxidases in the extremely acidophilic chemoautotrophic Acidithiobacillus ferrooxidans. Biochim. Biophys. Acta 1656, 114–126. doi: 10.1016/j.bbabio.2004.02.008
Brem, R., Yvert, G., Clinton, R., and Kruglyak, L. (2002). Genetic dissection of transcriptional regulation in budding yeast. Science 296, 752–755. doi: 10.1126/science.1069516
Bryan, C., Davis-Belmar, C., Wyk, N., Fraser, M., Dew, D., Rautenbach, G., et al. (2012). The effect of CO2 availability on the growth, iron oxidation and CO2-fixation rates of pure cultures of Leptospirillum ferriphilum and Acidithiobacillus ferrooxidans. Biotechnol. Bioeng. 109, 1693–1703. doi: 10.1002/bit.24453
Bueno, E., Mesa, S., Bedmar, E., Richardson, D., and Delgado, M. (2012). Bacterial adaptation of respiration from oxic to microoxic and anoxic conditions: redox control. Antioxid. Redox Signal. 16, 819–852. doi: 10.1089/ars.2011.4051
Cautivo, D., and Gentina, J. C. (2009). Influence of CO2 and O2 feeding rates on the continuous bioleaching of a chalcopyrite concentrate using Sulfolobus metallicus. Adv. Mater. Res. 4028, 71–73.
Christel, S., Herold, M., Bellenberg, S., El Hajjami, M, Buetti-Dinh, A., Pivkin, I. V., et al. (2018). Multi-omics reveals the lifestyle of the acidophilic, mineraloxidizing model species Leptospirillum ferriphilum. Appl. Environ. Microbiol. 84:e2091-17. doi: 10.1128/AEM.02091-17
Coram, N., and Rawlings, D. (2002). Molecular relationship between two groups of the genus Leptospirillum and the finding that Leptospirillum ferriphilum sp., nov. dominates south african commercial biooxidation tanks that operate at 40°C. Appl. Environ. Microbiol. 68, 838–845. doi: 10.1128/AEM.68.2.838
Demergasso, C., Galleguillos, P., Escudero, L., Zepeda, V., Castillo, D., and Casamayor, E. (2005). Molecular characterization of microbial populations in a low-grade copper ore bioleaching test heap. Hydrometallurgy 80, 241–253. doi: 10.1016/j.hydromet.2005.07.013
Galleguillos, P., Hallberg, K., and Johnson, D. (2009). Microbial diversity and genetic response to stress conditions of extremophilic bacteria isolated from the escondida copper mine. Adv. Mater. Res. 71–73, 55–58. doi: 10.4028/www.scientific.net/amr.71-73.55
Galleguillos, P., Music, V., Acosta, M., Salazar, C. N., Quatrini, R., Shmaryahu, A., et al. (2013). Temporal dynamics of genes involved in metabolic pathways of C and N of L. ferriphilum, in the industrial bioleaching process of Escondida mine, chile. Adv. Mater. Res. 825, 162–165. doi: 10.4028/www.scientific.net/amr.825.162
Ghorbani, Y., Franzidis, J., and Petersen, J. (2016). Heap leaching technology - current state, innovations, and future directions: a review. Miner. Process. Extr. Metall. Rev. 37, 73–119. doi: 10.1080/08827508.2015.1115990
Giuffre, A., Borisov, V. B., Arese, M., Sarti, P., and Forte, E. (2014). Cytochrome bd oxidase and bacterial tolerance to oxidative stress. Biochim. Biophys. Acta 1837, 1178–1187. doi: 10.1016/j.bbabio.2014.01.016
Gunasekera, T., Bowen, L., Zhou, C., Howard-Byerly, S., Foley, W., Striebich, R., et al. (2017). Transcriptomic analyses elucidate adaptive differences of closely related strains of Pseudomonas aeruginosa in fuel. Appl. Environ. Microbiol. 83, 1–24. doi: 10.1128/AEM.03249-16
Lakshmanan, V., Roy, R., and Ramachandran, V. (2016). Innovative Process Development in Metallurgical Industry. New York, NY: Springer, doi: 10.1007/978-3-319-21599-0
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Levicán, G., Gómez, M. J., Chávez, R., Orellana, O., Moreno-Paz, M., and Parro, V. (2012). Comparative genomic analysis reveals novel facts about Leptospirillum spp. cytochromes. J. Mol. Microbiol. Biotechnol. 22, 94–104. doi: 10.1159/000338105
Lindqvist, A., Membrillo-Hernández, J., Poole, R., and Cook, G. (2000). Roles of respiratory oxidases in protecting Escherichia coli K12 from oxidative stress. Antonie van Leeuwenhoek 78, 23–31. doi: 10.1023/A:1002779201379
Livak, K., and Schmittgen, T. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2^(−ΔΔCT) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Llinás, M., Bozdech, Z., Wong, E., Adai, A., and DeRisi, J. (2006). Comparative whole genome transcriptome analysis of three Plasmodium falciparum strains. Nucleic Acids Res. 34, 1166–1173. doi: 10.1093/nar/gkj517
Marín, S., Acosta, M., Galleguillos, P., Chibwana, C., Strauss, H., and Demergasso, C. (2017). Is the growth of microorganisms limited by carbon availability during chalcopyrite bioleaching? Hydrometallurgy 168, 13–20. doi: 10.1016/j.hydromet.2016.10.003
Orellana, L., and Jerez, C. (2011). A genomic island provides Acidithiobacillus ferrooxidans ATCC 53993 additional copper resistance: a possible competitive advantage. Appl. Microbiol. Biotechnol. 92, 761–767. doi: 10.1007/s00253-011-3494-x
Parro, V., Moreno-Paz, M., and González-Toril, E. (2007). Analysis of environmental transcriptomes by DNA microarrays. Environ. Microbiol. 9, 453–464. doi: 10.1111/j.1462-2920.2006.01162.x
Pfaffl, M. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Pfaffl, M., Horgan, G., and Dempfle, L. (2002). Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. doi: 10.1093/nar/30.9.e36
Pitcher, R., and Watmough, N. (2004). The bacterial cytochrome cbb3 oxidases. Biochim. Biophys. Acta 1655, 388–399. doi: 10.1016/j.bbabio.2003.09.017
Poole, R. K., and Cook, G. M. (2000). Redundancy of aerobic respiratory chains in bacteria? routes, reasons and regulation. Adv. Microb. Physiol. 43, 165–224. doi: 10.1016/s0065-2911(00)43005-5
Quatrini, R., and Johnson, D. B. (2018). Microbiomes in extremely acidic environments: functionalities and interactions that allow survival and growth of prokaryotes at low pH. Curr. Opin. Microbiol. 43, 139–147. doi: 10.1016/j.mib.2018.01.011
Remonsellez, F., Galleguillos, F., Moreno-Paz, M., Parro, V., Acosta, M., and Demergasso, C. (2009). Dynamic of active microorganisms inhabiting a bioleaching industrial heap of low-grade copper sulfide ore monitored by real-time PCR and oligonucleotide prokaryotic acidophile microarray. Microb. Biotechnol. 2, 613–624. doi: 10.1111/j.1751-7915.2009.00112.x
Sigler, K., Chaloupka, J., Brozmanová, J., Stadler, N., and Höfer, M. (1999). Oxidative stress in microorganisms—I. Microbial vs. higher cells—damage and defenses in relation to cell aging and death. Folia Microbiol. 44, 587–624. doi: 10.1007/bf02825650
Soto, P., Galleguillos, P., Seron, M., Zepeda, V., Demergasso, C., and Pinilla, C. (2013). Parameters influencing the microbial oxidation activity in the industrial bioleaching heap at Escondida mine, Chile. Hydrometallurgy 133, 51–57. doi: 10.1016/j.hydromet.2012.11.011
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:research0034. doi: 10.1186/gb-2002-3-7-research0034
Xie, X., Xiao, S., He, Z., Liu, J., and Qiu, G. (2007). Microbial populations in acid mineral bioleaching systems of tong shankou copper mine, China. J. Appl. Microbiol. 103, 1227–1238. doi: 10.1111/j.1365-2672.2007.03382.x
Xingyu, L., Bowei, C., Jiankang, W., and Renman, R. (2010). Leptospirillum forms a minor portion of the population in zijinshan commercial non-aeration copper bioleaching heap identified by 16S rRNA clone libraries and real-time PCR. Hydrometallurgy 104, 399–403. doi: 10.1016/j.hydromet.2010.03.024
Keywords: Leptospirillum ferriphilum, heap bioleaching, oxygen availability, electron transport chain, genetic markers, industrial monitoring
Citation: Cortés M, Marín S, Galleguillos P, Cautivo D and Demergasso C (2019) Validation of Genetic Markers Associated to Oxygen Availability in Low-Grade Copper Bioleaching Systems: An Industrial Application. Front. Microbiol. 10:1841. doi: 10.3389/fmicb.2019.01841
Received: 22 May 2019; Accepted: 26 July 2019;
Published: 09 August 2019.
Edited by:
Davide Zannoni, University of Bologna, ItalyReviewed by:
Sabrina Hedrich, Federal Institute for Geosciences and Natural Resources, GermanyCopyright © 2019 Cortés, Marín, Galleguillos, Cautivo and Demergasso. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cecilia Demergasso, Y2RlbWVyZ2FAdWNuLmNs
†These authors share first co-authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.