- 1VIB-KU Leuven Center for Microbiology, Leuven, Belgium
- 2Laboratory of Molecular Cell Biology, Institute of Botany and Microbiology, KU Leuven, Leuven, Belgium
Despite being one of the most important human fungal pathogens, Candida albicans has not been studied extensively at the level of protein-protein interactions (PPIs) and data on PPIs are not readily available in online databases. In January 2018, the database called “Biological General Repository for Interaction Datasets (BioGRID)” that contains the most PPIs for C. albicans, only documented 188 physical or direct PPIs (release 3.4.156) while several more can be found in the literature. Other databases such as the String database, the Molecular INTeraction Database (MINT), and the Database for Interacting Proteins (DIP) database contain even fewer interactions or do not even include C. albicans as a searchable term. Because of the non-canonical codon usage of C. albicans where CUG is translated as serine rather than leucine, it is often problematic to use the yeast two-hybrid system in Saccharomyces cerevisiae to study C. albicans PPIs. However, studying PPIs is crucial to gain a thorough understanding of the function of proteins, biological processes and pathways. PPIs can also be potential drug targets. To aid in creating PPI networks and updating the BioGRID, we performed an exhaustive literature search in order to provide, in an accessible format, a more extensive list of known PPIs in C. albicans.
Introduction
Fungal Infections and Candida albicans
One to five million fungal species are estimated to exist of which only 400–600 (< 0.1%) are documented to be pathogenic to humans and of those only about 100 are commonly found as human pathogens (Taylor et al., 2001; Blackwell, 2011; de Pauw, 2011; Köhler et al., 2014; Kastora et al., 2017). However, these fungal pathogens/infections are still often overlooked and underestimated even though they have evolved from uncommon to a major global health problem, paradoxically due to the introduction of new medical therapies (Brown et al., 2012; Köhler et al., 2014; Editorial, 2017). Recent outbreaks have drawn more attention to fungal infections (Benedict et al., 2017). It is estimated that over a billion people are affected by superficial fungal infections and about 1.5 million people die due to invasive fungal diseases worldwide each year (Brown et al., 2012; Bongomin et al., 2017). Only a few classes of antifungals are available for the treatment of invasive infections and resistance is on the rise (Fairlamb et al., 2016; Editorial, 2017; Perlin, 2017). These invasive fungal infections are associated with high mortality rates (up to 50%) and occur mainly in patients who had major surgery, are immunocompromised or on heavy antibiotic treatments. However, major infections in healthy individuals are increasing. The majority of those fungal-related deaths are attributed to only four genera of fungi: Cryptococcus, Aspergillus, Pneumocystis, and Candida (Spellberg, 2008; Brown et al., 2012; Editorial, 2017). Specifically Candida infections are the 4th most common cause of hospital-acquired infections (Brown et al., 2012; Noble et al., 2017) and in the United States alone candidiasis is estimated to cost up to $2–4 billion yearly (Uppuluri et al., 2017). Within the genus Candida, Candida albicans is currently the most medically important species (Brown et al., 2012) but Candida auris is gaining a lot of recent publicity (see below).
Candida albicans is a pleiomorphic diploid fungus generally only found in humans. Up to 70% of humans are hosts of this fungus (Noble et al., 2017). It is generally considered as a commensal, but it can turn pathogenic in certain circumstances. These circumstances are often regarded as “caused” or provided by the host (e.g., being immunocompromised or taking antibiotic treatments) and not so much as actively generated by C. albicans itself (Köhler et al., 2014; Mitchell, 2016; Noble et al., 2017). C. albicans can cause superficial mucosal infections such as oral thrush or vulvovaginal candidiasis but also life-threatening invasive infections (Spellberg, 2008; Brown et al., 2012; Köhler et al., 2014; Noble et al., 2017). This switch from a commensal to a potentially lethal pathogen is still not fully understood (Noble et al., 2017). Recently, the 1st fungal cytolytic peptide toxin in C. albicans, candidalysin, was described and is thought to play a pivotal role in the pathogenicity of C. albicans (Mitchell, 2016; Moyes et al., 2016). C. albicans can thrive in the human body. It is able to evade the immune system, colonize every organ and form biofilms on implanted medical devices (Brown et al., 2012; Mathé and Van Dijck, 2013; Noble et al., 2017; Sherrington et al., 2017). It is not surprising that it is used as a model organism to study fungal pathogenesis (Kabir et al., 2012). C. albicans has a few remarkable characteristics that make it challenging to study. It is a diploid organism without a complete sexual cycle (no meiosis found so far) and has a non-canonical codon usage (CUG translated as serine rather than leucine), in addition, it lacks stable episomal plasmids and has a low transformation efficiency (Datta et al., 1990; Murad et al., 2000; Noble and Johnson, 2007; Stynen et al., 2010; Hickman et al., 2015). In recent years, as novel research tools have been developed, the whole genome has been sequenced and the ORFs have been made available for the community in GatewayTM-adapted vectors (Jones et al., 2004; Kaplanek et al., 2006; Muzzey et al., 2013; Legrand et al., 2018).
One important feature to study in order to understand an organism is its protein-protein interactions (PPIs) (Auerbach et al., 2002; De Las Rivas and Fontanillo, 2010; Khan et al., 2011). Advances in high-throughput detection techniques mean that mapping large physical PPI networks has become a possibility for several organisms (Auerbach et al., 2002; Schächter, 2002; Uetz, 2002; De Las Rivas and Fontanillo, 2010), which will help in compiling the interactome of the studied organisms (Bonetta, 2010; De Las Rivas and Fontanillo, 2010). Several techniques are available to study PPIs (von Mering et al., 2002; Khan et al., 2011; Rao et al., 2014; Podobnik et al., 2016) but the most prominent techniques to study PPIs on a high-throughput scale are the tandem-affinity purification (TAP) followed by mass spectrometry (Gingras et al., 2005; Krogan et al., 2006; De Las Rivas and Fontanillo, 2010) and the yeast two-hybrid (Y2H) assay (Uetz, 2002; De Las Rivas and Fontanillo, 2010; Xing et al., 2016). There are, however, about 25,000 CUG codons in C. albicans, complicating the use of Saccharomyces cerevisiae as a host organism to study C. albicans PPIs using the Y2H technique (Skrzypek et al., 2017). A possible solution is to change the CUG codons (Hoppen et al., 2007; Badrane et al., 2008) or work with only parts of the protein (Xu and Mitchell, 2001) to minimize translation problems, yet most researchers did not change the CUG codons when performing experiments in S. cerevisiae (Oughtred et al., 2016, 2018; Chatr-Aryamontri et al., 2017). In one study, it was observed that changing the CUG codons led to the discovery of a PPI not found when using non-altered CUG codons (Feng et al., 2017). An adapted Candida two-hybrid (C2H) system was developed to overcome the problem with the non-canonical codon usage and the first small-scale high-throughput screen was performed with this system (Stynen et al., 2010; Schoeters et al., 2018). The TAP-tag has also been used to study PPIs in C. albicans, but only a limited number of PPIs or complexes have been studied with this technique (see below).
Results
Databases to Store PPI Data and Curation of the Literature Describing PPIs Demonstrated in C. albicans
Several large-scale chromatin immunoprecipitation (ChiP) studies have already been done on C. albicans to study protein-DNA interactions, leading to several networks (Zordan et al., 2007; Tuch et al., 2008; Nobile et al., 2012; Hernday et al., 2013; Znaidi et al., 2013) and even leading to a few PPIs in C. albicans (Znaidi et al., 2013). However, limited by the difficulties encountered in C. albicans research (Noble and Johnson, 2007; Palzer et al., 2013) only a small number of PPIs have been detected in C. albicans. In contrast to the ChiP studies, no large scale high-throughput PPI screens have been performed for C. albicans and only a limited number of papers have described PPIs in C. albicans (Wang et al., 2014; Chatr-Aryamontri et al., 2017; Márkus et al., 2017; Oughtred et al., 2018). Several (public) databases are available to screen for PPIs, but they are extremely limited when it comes to C. albicans PPIs. The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database does not even mention C. albicans (Szklarczyk et al., 2017) and the Database for Interacting Proteins (DIP) only holds a handful interactions (Salwinski et al., 2004). Another database, The Molecular INTeraction Database (MINT), only mentions 24 PPIs for C. albicans (Licata et al., 2012). The IntAct MINT mentions 25 interaction protein pairs (Kerrien et al., 2012). The Biological General Repository for Interaction Datasets (BioGRID) contained 188 PPIs extracted from 42 publications, release 3.4.156 (January 2018). These 188 interactions (using 139 unique genes) can further be divided into 128 unique protein interacting pairs (non-redundant interactions) (Chatr-Aryamontri et al., 2017; Oughtred et al., 2018). Because the BioGRID contains the largest number of PPIs for C. albicans, we decided to further continue our work with the BioGRID. A literature search revealed many PPIs that were not yet mentioned in this database. The absence of many interactions from the literature in the BioGRID (and other databases) also shows that PPI data are not generally sent to databases such as the BioGRID. A remarkable example is that the BioGRID database, release 3.4.156, mentioned only one interaction that was found with crystallography (van den Berg et al., 2016) while several structures of C. albicans proteins or enzymes showing PPIs have already been studied with crystallography (Whitlow et al., 1997; Senay et al., 2003; Echt et al., 2004; Morgunova et al., 2007; Raczynska et al., 2007; Hast and Beese, 2008; Santini et al., 2008; Arachea et al., 2010; Rocha et al., 2011; Nakamura et al., 2013; Sheng et al., 2013; Yan et al., 2015; Nasser et al., 2016; Tonthat et al., 2016; Hong et al., 2017; Sinha and Rule, 2017; Dostal et al., 2018; Garcia et al., 2018; Kiliszek et al., 2019) and mentioned in the protein data bank1. A second observation during our literature search for PPIs in C. albicans is that certain “keywords” commonly used in papers studying PPIs are not often used by C. albicans researchers in their manuscripts.
To update the BioGRID with regard to C. albicans PPI data we performed an exhaustive literature search using several keywords (Table 1) in combination with “C. albicans” using Google search and PubMed. In addition to a literature search for novel PPIs we also checked the already available data for potential mistakes in order to correct them. All the novel interactions and/or mistakes in the already available data found during our literature search were sent to the BioGRID in order to update their data.
Putting PPIs in C. albicans in Perspective
If one compares the data for C. albicans with the data available for S. cerevisiae (up to 171,000 non-unique interactions) (von Mering et al., 2002; Oughtred et al., 2018) or certain bacteria (Parrish et al., 2007; Wang et al., 2010a; Oughtred et al., 2018), then it is easy to see that C. albicans PPI data are lagging behind significantly even though it is a highly studied organism. Another observation is that the golden standard reference website for the Candida community2 (Skrzypek et al., 2017, 2018) does not have a direct link or a file to PPIs in C. albicans in contrast to the yeast genome database3 that integrated, in a separate “tab”, all the interactions for a protein of interest mentioned on the BioGRID (Cherry et al., 2012). The combination “PPI” and “C. albicans” is also rarely used in the published literature (Table 2). This difference in found papers using the search term “PPI” and the name of the organism also shows that there is less work done regarding PPIs for C. albicans compared with the other four organisms (Table 2) and/or that C. albicans researchers are less inclined to use the term “PPIs” in their manuscripts. Four organisms were compared with C. albicans. S. cerevisiae and Schizosaccharomyces pombe were used since they are also yeast models. Escherichia coli is the bacterial “counterpart,” serving as a model organism for bacteria while Arabidopsis thaliana is a plant model.
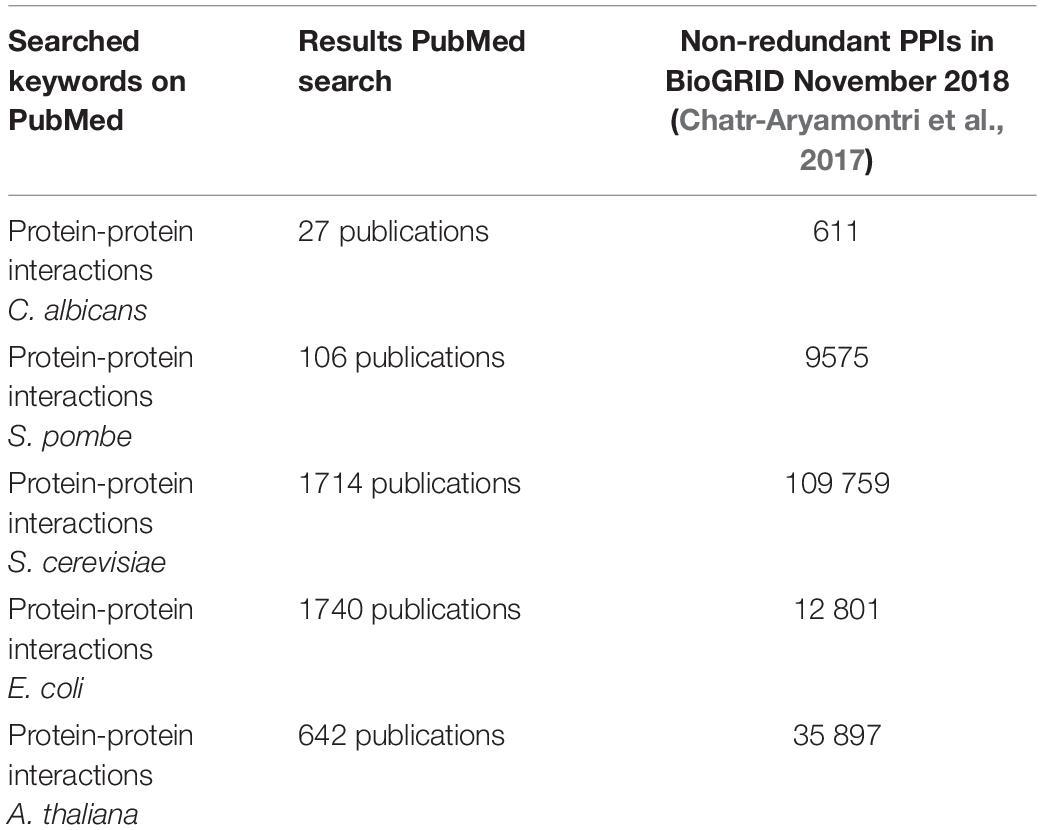
Table 2. A search on pubmed (https://www.ncbi.nlm.nih.gov/pubmed/) using “PPIs” and the name of the organism resulted in X publications, while a search on the Biological General Repository for Interaction Datasets (https://wiki.thebiogrid.org/doku.php/statistics) shows how many PPIs are curated.
The knowledge of PPIs is important for fully unraveling the complexity of organisms (De Las Rivas and Fontanillo, 2010). Besides the importance of PPIs for the fundamental understanding of an organism, they also form potential targets for specific drugs (Khan et al., 2011; Bakail and Ochsenbein, 2016; Nishikawa et al., 2016). The latter is very important since finding novel drug targets is hard due to the similarities between the eukaryotic pathogen and the eukaryotic host (Ismail et al., 2018). The currently limited availability of antifungals, rising resistance, and the increase of fungal infections underscore the need to identify new drug targets (Brown et al., 2012; Fairlamb et al., 2016; Perlin, 2017). PPIs might thus play an important role in the development of novel, very specific, antifungal drugs (Kingwell, 2016; Liu et al., 2018). An interesting example is blocking the interaction of Cdc42 with its CRIB-domain binding effectors and thereby inhibiting hyphal growth (Su et al., 2007).
Several techniques have been used to detect the interactions described for C. albicans. The majority of them were found using affinity-capture techniques or by using the traditional Y2H technique as described below. The most important techniques used to study PPIs in C. albicans are described below.
Approaches to Study PPIs in C. albicans
Several techniques are available to detect PPIs (Rao et al., 2014; Podobnik et al., 2016), but only a select few have been adapted for use in C. albicans (Boysen et al., 2009; Stynen et al., 2010; Palzer et al., 2013; Subotić et al., 2017). For a complete overview of all the used techniques and detected protein interactions in C. albicans, we refer to the BioGRID website4 (Chatr-Aryamontri et al., 2017). So far, no large-scale or genome-wide screens have been performed, however, Prof. Whiteway’s lab has performed two tandem affinity purifications (TAP) and detected more than 200 PPIs (Tebbji et al., 2014, 2017). Prof. Liu’s lab has also used a TAP approach to identify 103 interacting proteins for the Wor1 protein (Alkafeef et al., 2018) while the labs of Profs. Emili, Gingras, and Cowen found more than 250 PPIs when studying Hsp90 (O’Meara et al., 2019). The labs of Profs. Dickman and Sudbery used the Stable Isotope Labeling with Amino Acids in Cell Culture (SILAC) technique combined with mass-spectrometry (MS) and identified 126 interacting proteins for Cdc14 (Kaneva et al., 2019).
Co-immunoprecipitation (Co-IP) is a type of affinity purification technique that is referred to as “affinity-western” in the BioGRID database (Chatr-Aryamontri et al., 2017; Oughtred et al., 2018). Many of the studied interactions with the classic Co-IP technique were experiments to validate interactions found with other techniques such as the Y2H technique (Ni et al., 2004; Fang and Wang, 2006; Kaneko et al., 2006; Berggård et al., 2007; Hoppen et al., 2007; Badrane et al., 2008; Lu et al., 2008; Lavoie et al., 2010; Sun et al., 2013; Lee H.J. et al., 2015). An expert with this technology is Prof. Wang who has used Co-IP a lot in his lab and showed several interactions (Zheng et al., 2004, 2007; Li et al., 2007, 2008, 2012; Sinha et al., 2007; Bai et al., 2011; Zeng et al., 2012; Wang et al., 2013, 2016; Gao et al., 2014; Huang et al., 2014a, b; Guan et al., 2015; Au Yong et al., 2016; Liu et al., 2016; Yao et al., 2016, 2017; Yang et al., 2018). Several tags have been used, such as a Flag-tag (Umeyama et al., 2002; Singh et al., 2011), Myc-tag (Cheetham et al., 2007, 2011; Sinha et al., 2007; Kos et al., 2016), GFP or derived tags (Bishop et al., 2010; Greig et al., 2015), HA-tag (Ni et al., 2004; Askew et al., 2011; Sellam et al., 2019), TAP-tag (see below) or the protein A tag with a TEV protease site (Blackwell et al., 2003). The Co-IP technique is rather limited and is not suited for high-throughput systems (Sun et al., 2013).
Tandem-affinity purification is another affinity purification technique that uses a TAP tag to perform a two-step specific affinity purification process. The original tag incorporated two protein A domains and the calmodulin binding peptide separated by a tobacco etch virus (TEV) protease site to provide the two-step affinity purification (Rigaut et al., 1999). Different versions of the TAP-tag were later designed without the TEV protease site (Xu et al., 2010). The TAP technique has the advantage that it can be used in a large-scale high-throughput setup where protein complexes are purified in two steps followed by protein identification with MS (Xu et al., 2010; Rao et al., 2014; Podobnik et al., 2016; O’Meara et al., 2019). It has, for example, been proven to be very efficient to perform large-scale screenings to identify PPIs and protein complexes in S. cerevisiae (Gavin et al., 2002; Krogan et al., 2006). A TAP tag was first successfully utilized in C. albicans by Kaneko et al. (2004) to purify the C. albicans septin protein complex (Kaneko et al., 2004), which was later confirmed by another group (Sinha et al., 2007). Both groups used a TAP-tagged Cdc11 protein and showed the same interactions except for two extra interactions found in the study from Sinha et al. (2007). One of these interactions was the Gin4 protein shown to interact with Cdc11. The Cdc11-Gin4 interaction was interestingly reported to be detected only after a 150 min hyphal induction of the cells, but not after a 10 min induction (Sinha et al., 2007). Kaneko et al. (2004) did not find this interaction, but they only induced hyphal growth for 90 min (Kaneko et al., 2004), showing the need to induce hyphal growth long enough to have Gin4 interact with the septin complex. Later studies not using a TAP tag approach also demonstrated the interaction of Gin4 with members of the septin complex (Li et al., 2012; Au Yong et al., 2016). The TAP-tag has also been applied to study several other complexes (Corvey et al., 2005; Lavoie et al., 2008; Blackwell and Brown, 2009; Ryan et al., 2012; Zhang et al., 2012; Tebbji et al., 2014, 2017; Guan et al., 2015; Lee J.E. et al., 2015; Rao et al., 2016; Alkafeef et al., 2018). The mediator complex was studied twice (Zhang et al., 2012; Tebbji et al., 2014) and both studies showed an overlap in found proteins (subunits) for the mediator complex, However, Tebbji et al. (2014) found a total of 179 proteins interacting with Med7 while Zhang et al. (2012) tagged Med8 and used a pre-purification with heparin sepharose to bind the intact mediator complex followed by a TAP and only purified the 25 subunits of the mediator complex itself (Zhang et al., 2012; Tebbji et al., 2014). Whether this difference is caused by the use of the pre-purification step is unclear. More recently O’Meara et al. (2019) applied the TAP-tag to identify well over 250 PPIs.
The TAP-tags can also be used in a type of Co-IP experiment, tagging both the bait and prey constructs and not using an MS approach to identify the preys (Kaneko et al., 2006; Singh et al., 2009; Chen and Noble, 2012; Shapiro et al., 2012; Li et al., 2017).
Using TAP-tag approaches with a two-step purification process has the advantage to produce cleaner protein samples for MS (Kaneko et al., 2004; Blackwell and Brown, 2009). However, single-step purification protocols followed by MS have also led to the discovery of several PPIs (Tseng et al., 2010; Li et al., 2012; Guan et al., 2015; Xie et al., 2017). Two of those studies used a GFP-tagged protein to purify the complexes (Li et al., 2012; Guan et al., 2015) while Xie et al. (2017) used a Flag-tagged Ydj1 protein (Xie et al., 2017). Tseng et al. (2010) used E. coli-expressed, His-tagged Cdc14 protein to pull down interacting proteins from a Cdc14 deletion mutant cell lysate (Tseng et al., 2010). Proteins found in a single-step purification approach should be confirmed with another technique (Tseng et al., 2010; Guan et al., 2015) as it often leads to false positives.
SILAC is a technique that takes advantage of the in vivo incorporation of non-radioactive isotope-labeled amino acids. It can be used to detect the up- or down-regulation of proteins. For this, growth medium is supplemented with a labeled amino acid that is then incorporated into the proteins of the cells grown on this medium (Ong et al., 2002). By subsequently mixing cells grown in this medium with cells grown in regular medium, lysing the cells, purifying the protein(s), and then digesting the purified proteins, the relative abundance of isotope-labeled and unlabeled proteins in the mixture can be determined by MS (Ong et al., 2002). In C. albicans the technique was first used in 2018 to perform a quantitative proteomic analysis of Cdc14 (Ong et al., 2002; Kaneva et al., 2018). Later the technique was used to study the proteome changes while transcriptionally repressing or pharmacologically inhibiting Hsp90 (O’Meara et al., 2019).
However, the technique can also be combined with an affinity purification step to identify PPIs. It was used to identify interacting proteins for Cdc14. This was achieved by growing a C. albicans strain with a phosphatase-dead, substrate-trapping Cdc14 protein with a Myc-tag (Cdc14PD-Myc, the “bait”) on medium supplemented with heavy isotope-labeled arginine and lysine (heavy medium). In parallel, a wild-type strain was grown on light medium. After mixing the bait and wild-type strain in a 1:1 ratio, the cells are lysed and the bait was pulled down followed by SDS-page and trypsin digestion. In the subsequent MS analysis, Cdc14-specific interacting proteins showed a heavy-to-light (H:L) ratio greater than 1:1 because specific interacting proteins will originate from the heavy medium, whereas non-specifically bound proteins will have a 1:1 ratio. A total of 126 interacting proteins for Cdc14 could be detected. Remarkably, only a few of the found interactions have also been demonstrated for the orthologous proteins in S. cerevisiae (Kaneva et al., 2019). See also Supplementary Tables S1, S2.
The SILAC approach (combined with affinity-purification) has several advantages: (1) it can be used in a high-throughput setup, (2) there is no forced cell localization (e.g., two-hybrid techniques force proteins into the nucleus), and (3) post-translational modifications can be preserved. The SILAC technique is also quantitative and not qualitative as is the traditional TAP-tag approach. The samples in SILAC are also analyzed as a whole single sample, thus minimizing the bias in sample handling (Ong et al., 2003; Emmott and Goodfellow, 2014).
Bimolecular Fluorescence Complementation (BiFC) is a technique that can be used to study PPIs in vivo in their native environment and location (Kerppola, 2008) and is one of many protein complementation techniques (PCA) (Zhou et al., 2011). Since its discovery, this technique has been applied in several organisms in a high-throughput setup (Miller et al., 2015). However, it proved difficult to apply this system in C. albicans and only recently it was used to detect several binary interactions (Mamouei et al., 2017; Subotić et al., 2017). Subotić et al. (2017) worked with an overexpression plasmid system with the genes under the control of the MET3 promoter rather than endogenously tagged proteins. Mamouei et al. (2017) also used the MET3 promoter, but were also able to use the native promoter for the Ftr1 and Fet34 BiFC constructs. Besides the need to codon-optimize the fluorophores (CUG codons), another potential explanation for the difficulties with adapting this system is the autofluorescence of C. albicans (Diaz et al., 2005; Graus et al., 2017). In addition to BiFC, several other PCAs are used for the detection of PPIs e.g., the split luciferase system (Stynen et al., 2012). However, so far, no other PCAs have been applied for PPI detection in C. albicans (Chatr-Aryamontri et al., 2017), despite the fact that several luciferases have been optimized for and used in C. albicans, for example, for the study of biofilm formation and drug susceptibility (Jacobsen et al., 2014; Kucharíková et al., 2015; Dorsaz et al., 2017). A split luciferase system can thus potentially be developed for use in C. albicans. It is also possible to use fluorophores and luciferases in a Förster Resonance Energy Transfer (FRET) or Bioluminescence Resonance Energy Transfer (BRET) system, however, BRET has so far not been reported in C. albicans (Chatr-Aryamontri et al., 2017). A FRET biosensor has been shown to work in C. albicans (Jain et al., 2018) and Candida glabrata (Demuyser et al., 2018).
The Vesicle Capture Interaction (VCI) assay was developed for use in C. albicans to circumvent the codon usage problem when using the model organism S. cerevisiae. VCI can be used to detect binary interactions. The technique is based on the targeting to endocytic vesicles by the Endosomal Sorting Complex Required for Transport (ESCRT) of which Snf7 (Vps32) is a subunit. The technique uses a vps4Δ mutant strain that promotes vesicular accumulation of Snf7. A bait protein is fused to the ESCRT subunit Snf7 while the prey protein is fused to a GFP protein. When bait and prey interact, a punctate GFP signal can be detected compared with a diffuse signal if no interaction occurs. The technique uses the native promoters so overexpression is avoided and real-time imaging facilitates the detection of transient interactions. The system was, so far as we know, only used in two studies from the lab that developed it (Boysen et al., 2009; Argimón et al., 2011). It remains an open question how applicable it is on a high-throughput setup.
The expanded genetic code system is a technique adapted for use in C. albicans. It relies on the incorporation of a synthetic photo-cross-linker amino acid, p-azido-L-phenylalanine (AzF), in a “bait” protein to covalently capture the binding partner (prey) after UV-activation. To incorporate AzF into the protein of interest, an amber stop codon needs to be introduced into this protein in a C. albicans strain that expresses the optimized orthogonal tRNA and tRNA synthetase for AzF. It is at the amber stop codon where AzF, provided in the medium, is incorporated (rather than terminating the translation). So far, only two interactions, TUP1 and TSA1 have been studied using this technique (Palzer et al., 2013). The technique has been proven to be valuable but has some limitations such as high dependence on the incorporation efficiency of AzF and the lack of site selectivity (Wang et al., 2009). The amounts of mutant protein is also reduced significantly compared with the wild-type (Palzer et al., 2013). The technique is still relatively new, and given its complexity and high cost, it seems unlikely that it will be used in a high-throughput setup soon. So far, no other studies applying this technique have been reported in C. albicans.
The Yeast two-hybrid (Y2H) system has become, since the first publication, one of the most used systems to detect PPIs in vivo (Auerbach et al., 2002; Silva et al., 2015). Several adaptations have been created and the Y2H system has been used to perform large-scale or even genome-scale PPI assays for several organisms (Legrain and Selig, 2000; Auerbach et al., 2002; Schächter, 2002; Hart et al., 2006). Compared to another technique often used in high-throughput screenings, mass spectrometry (MS) of purified complexes, the Y2H technique is easier and cheaper to use (von Mering et al., 2002; Brückner et al., 2009; Silva et al., 2015) and multiple commercial plasmids and yeast strains are available. It however, suffers from large amounts of false positives and negatives. The system also forces proteins into the nucleus, making it hard to use for certain proteins (e.g., cell membrane components) (Uetz et al., 2000; Silva et al., 2015). A potential solution for the problematic forced nuclear movement is the removal of parts of the protein of interest (Weber et al., 2002; Miwa et al., 2004). Despite these problems, it is still one of the most used and best techniques for high-throughput screening of PPIs (Silva et al., 2015).
In C. albicans, in spite of the codon usage problem, the Y2H technique is still responsible for the discovery of a large fraction of PPIs (Chatr-Aryamontri et al., 2017). Most researchers used the traditional Y2H system to detect PPIs (Gkourtsa et al., 2016; Chatr-Aryamontri et al., 2017; Oughtred et al., 2018) but it is also possible to use an adapted Y2H system: the SRYTH (Ste11p/Ste50p Related Yeast Two-Hybrid) system, which allows cytoplasmic PPI analysis (Mallick et al., 2016). This system makes use of the essential interaction of Ste11 and Ste50 to activate the high osmolarity glycerol (HOG) pathway in S. cerevisiae, in the absence of the SLN1–SSK1–SSK2/SSK22 pathway, in order to survive under osmotic stress (Wu et al., 2006; Mallick et al., 2016). Ste11 and Ste50 interact with each other through their sterile alpha motif (SAM) domain. These SAM domains can, however, be replaced by two proteins of interest (bait and prey). If those proteins interact, then Ste11 and Ste50 are brought together, the HOG pathway is activated and the cells survive under osmotic stress (Wu et al., 2006; Mallick et al., 2016). This system was used to study the mating pheromone pathway of C. albicans (Côte et al., 2011) and several transcription factors (Mallick and Whiteway, 2013).
The Candida two-hybrid (C2H) system is a special adaptation of the Y2H system (see Figure 1). It was designed in 2010 to address the codon usage problem in S. cerevisiae. Compared with the traditional Y2H system, it uses an integrative approach because plasmids are not very stable in C. albicans. Expression of the bait and prey constructs is driven by the MET3 promoter and can be up-regulated by omitting methionine or both methionine and cysteine from the medium for a higher expression of the bait and prey constructs. With this system, several interactions were detected in a low-throughput setup (Stynen et al., 2010). The system was later used to confirm the interaction between the MAP kinases Cek1 and Cek2 (Correia et al., 2016), an interaction that is remarkably not found with the SRYTH system (Côte et al., 2011). The C2H system was also used to validate the PPIs found with the BiFC assay (Subotić et al., 2017). In the most recent paper using the C2H system, Wangsanut et al. (2018) tried to demonstrate the interaction between transcription factors Grf10 and Bas1 but without success (Wangsanut et al., 2018, 2019). The C2H system had been limited to small-scale studies until recently when it was adapted to a high-throughput setup (Legrand et al., 2018; Schoeters et al., 2018).
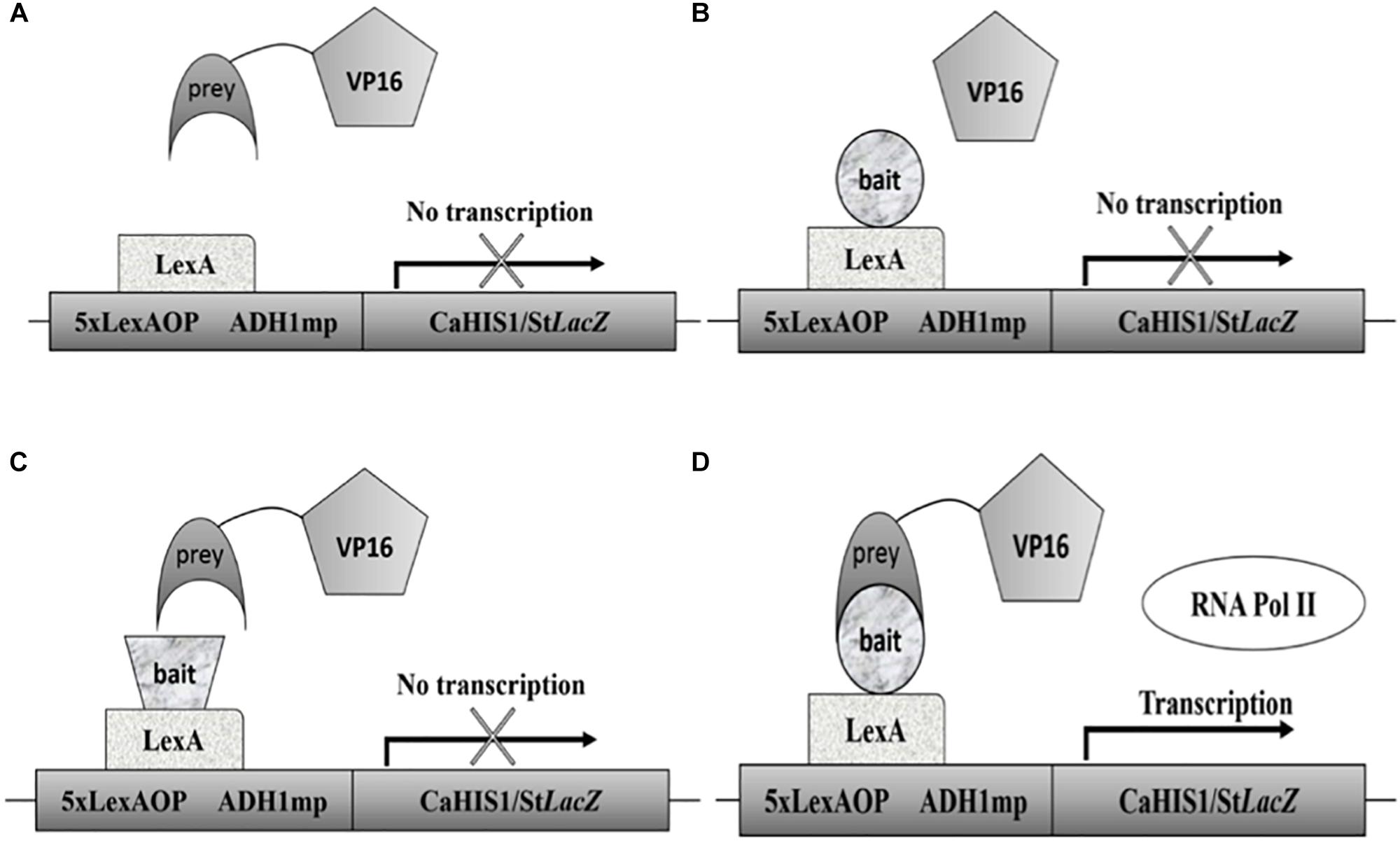
Figure 1. Schematic representation of the C2H system. (A) When no bait protein is attached to the DBD no interaction is possible and no transcription will take place. (B) Represents a situation in which no prey is attached to the AD leading to no transcription. In (C) both a prey and bait are present, but they do not interact so no transcription happens. Situation (D) depicts the interaction between a prey and bait protein thus leading to recruitment of RNA polymerase II and subsequent transcription. A common problem is the auto-activation by bait proteins. It is thus always essential to test auto-activation using the setup seen in part B.
The CTG clade of Candida includes nine potential pathogens (Gabaldón et al., 2016). All of these can benefit from using the C2H system to study PPIs. One emerging pathogen in particular, C. auris, raises concerns due to its resistance against antifungals and sudden emergence (Lu et al., 2018). Table 3 lists the CTG clade species, all of which can be studied with the C2H system.
Comparing Data Between C. albicans and S. cerevisiae
Saccharomyces cerevisiae has been studied extensively regarding PPIs. Release 3.5.174, July 2019 from the BioGRID contains over 114,693 PPIs for S. cerevisiae while C. albicans only has 876 non-redundant interactions mentioned (Chatr-Aryamontri et al., 2017). While S. cerevisiae often functions as a model organism for fungi (in general) it is not always a good idea to extrapolate data from S. cerevisiae to C. albicans. While many pathways perform similar cellular functions, differences are also present, making it harder to simply extrapolate data from S. cerevisiae to construct pathways in C. albicans (Kobayashi and Cutler, 1998; Román et al., 2005, 2009; Bahn et al., 2007; Biswas et al., 2007; Cheetham et al., 2007; de Dios et al., 2010; Smith et al., 2010). Interestingly, of the 1,208 non-redundant PPIs in C. albicans (see Supplementary Tables S1, S2, based on release 3.5.174 from BioGRID and our literature search), we were able to find only 249 PPIs that were also demonstrated in S. cerevisiae. Several of the PPIs identified using Mcu1 and Snf6 as bait in C. albicans were not found in S. cerevisiae as it does not have orthologs of the two proteins according to the CGD database (based on BLAST analysis: query coverage of 13% between CaSnf6 and ScSnf6). Snf6 was however, identified as a member of the SWI/SNF complex and it was shown that the N-terminal domain, which interacts with Snf2, is conserved in S. cerevisiae (Tebbji et al., 2017); in addition, although Wor1, Hsp90, Med7, and Cdc14 together have over 500 interacting proteins in C. albicans, only a few of these interactions can be found with S. cerevisiae orthologs (Supplementary Tables S1, S2). The low overlap of PPIs between orthologs in C. albicans and S. cerevisiae might indicate big differences in PPIs and protein functions between the two organisms. This is no surprise given that C. albicans co-evolved with its host, being a commensal and potential pathogen, whereas S. cerevisiae is a saprophyte and only occasionally becomes pathogenic to humans. The two fungi are evolutionarily separated for 140–850 millions years (Kobayashi and Cutler, 1998; Biswas et al., 2007; Skrzypek et al., 2017) and only approximately 55% of the genes in both organisms have orthologs (we only looked at orthologous genes and not “best hits”) (Skrzypek et al., 2017). An important remark is that while in S. cerevisiae many interactions are studied with multiple techniques, this is not the case for C. albicans. Using different techniques is crucial to confirm PPIs (von Mering et al., 2002; Yu et al., 2008) so perhaps the low overlap is also partly due to the limited number of techniques used for detection of PPIs in C. albicans (see Supplementary Tables S1, S2).
An interesting, but often overlooked, dataset comprises the data of PPIs that were tested but could not be demonstrated as they are often not mentioned in the published literature. To further look into the differences between PPIs in C. albicans and S. cerevisiae, we also looked into the literature for PPIs that were investigated but not demonstrated for C. albicans. We then also compared this with known data in S. cerevisiae. With this information, we were able to construct Table 4. Our lab currently also hosts a more extensive and up-to-date list of interactions tested but not detected in C. albicans at: https://docs.google.com/spreadsheets/d/1nZDAPyyfCaqqtvAkU_Xt8wZcOPK6-17jJxQxKSen58g/edit#gid=93661881.
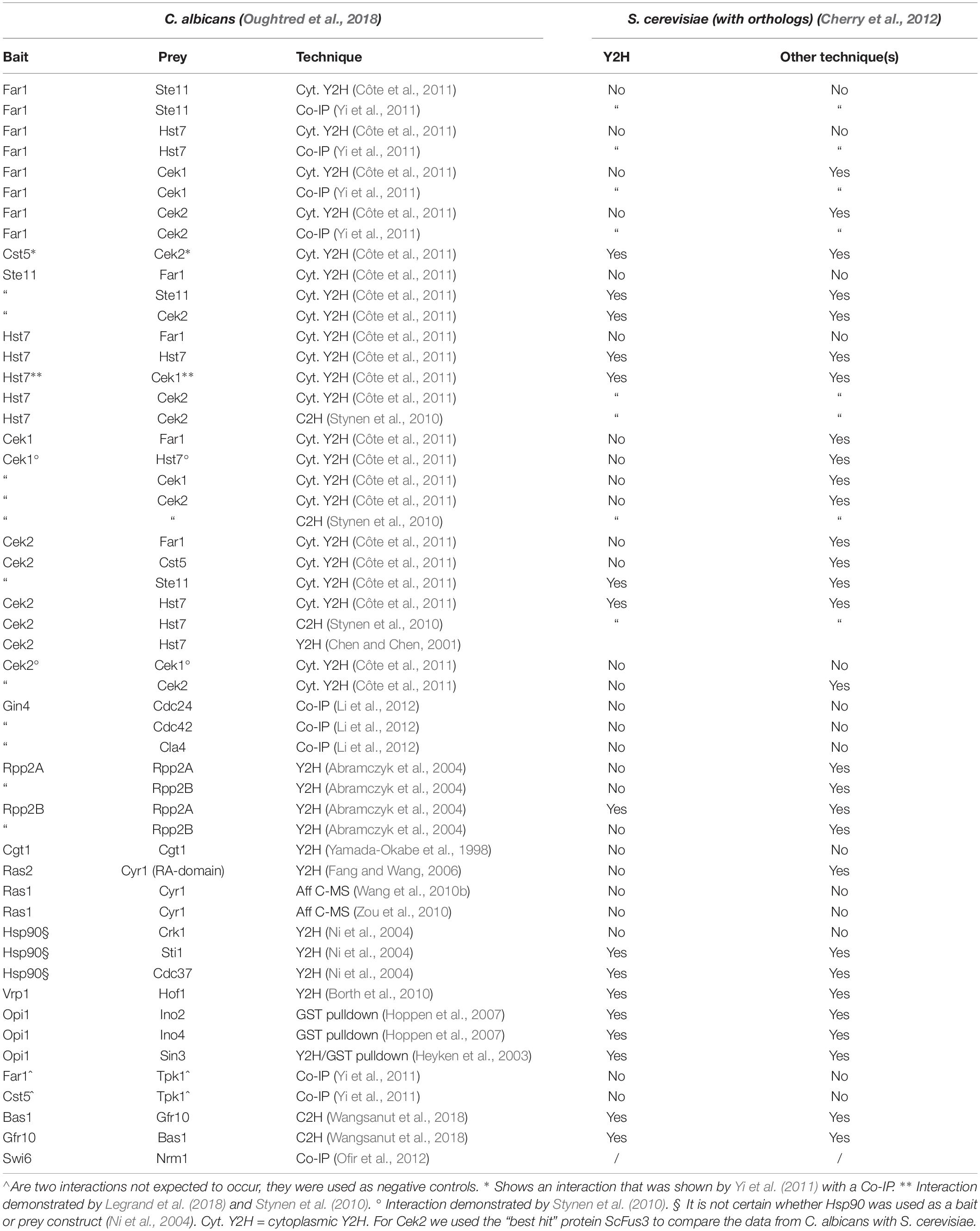
Table 4. A (limited) set of PPIs mentioned in several papers that could not be detected for C. albicans proteins.
Apart from helping elucidate differences between two organisms, the “non-interacting” protein pairs might also indicate which technique could be used to study certain proteins or interactions.
Case Study Regarding PPIs Shown in C. albicans
The cell wall is a dynamic structure that offers a first line of defense against external influences. As C. albicans can be present in any host niche, it must have a huge range of possible adaptations to external stresses (Ene et al., 2015; Román et al., 2016). Several pathways have been documented that play a role in these adaptation processes. Four of the most studied and important pathways are depicted in a simplified version in Figure 2. The cAMP-PKA pathway acts through a Ras1-independent or -dependent mechanism as a response to several external influences such as the quorum-sensing molecules homoserine lactone (HSL) and farnesol, amino acids, CO2, serum, N-acetylglucosamine (GlcNac) or glucose. The HOG pathway is important to react upon osmotic and oxidative stress while the cell wall integrity (PKC) and Cek1-mediated (or SVG) pathways play a role in the response toward cell wall damage and stress. All four pathways play a crucial role in morphogenesis and the survival of C. albicans under stress conditions (Biswas et al., 2007; de Dios et al., 2010; Noble et al., 2017). While there is considerable knowledge regarding how the pathways function in C. albicans, there is still a lot to demonstrate in terms of PPIs. Figure 2, for example, shows that only 8 out of the 26 depicted potential PPIs have been demonstrated in C. albicans (Chatr-Aryamontri et al., 2017). A ninth PPI that could be shown in Figure 2 is the direct interaction between Msb2 and Cst20 (van Wijlick et al., 2016). In S. cerevisiae all interactions for the depicted pathways in Figure 2 have already been demonstrated (Cherry et al., 2012).
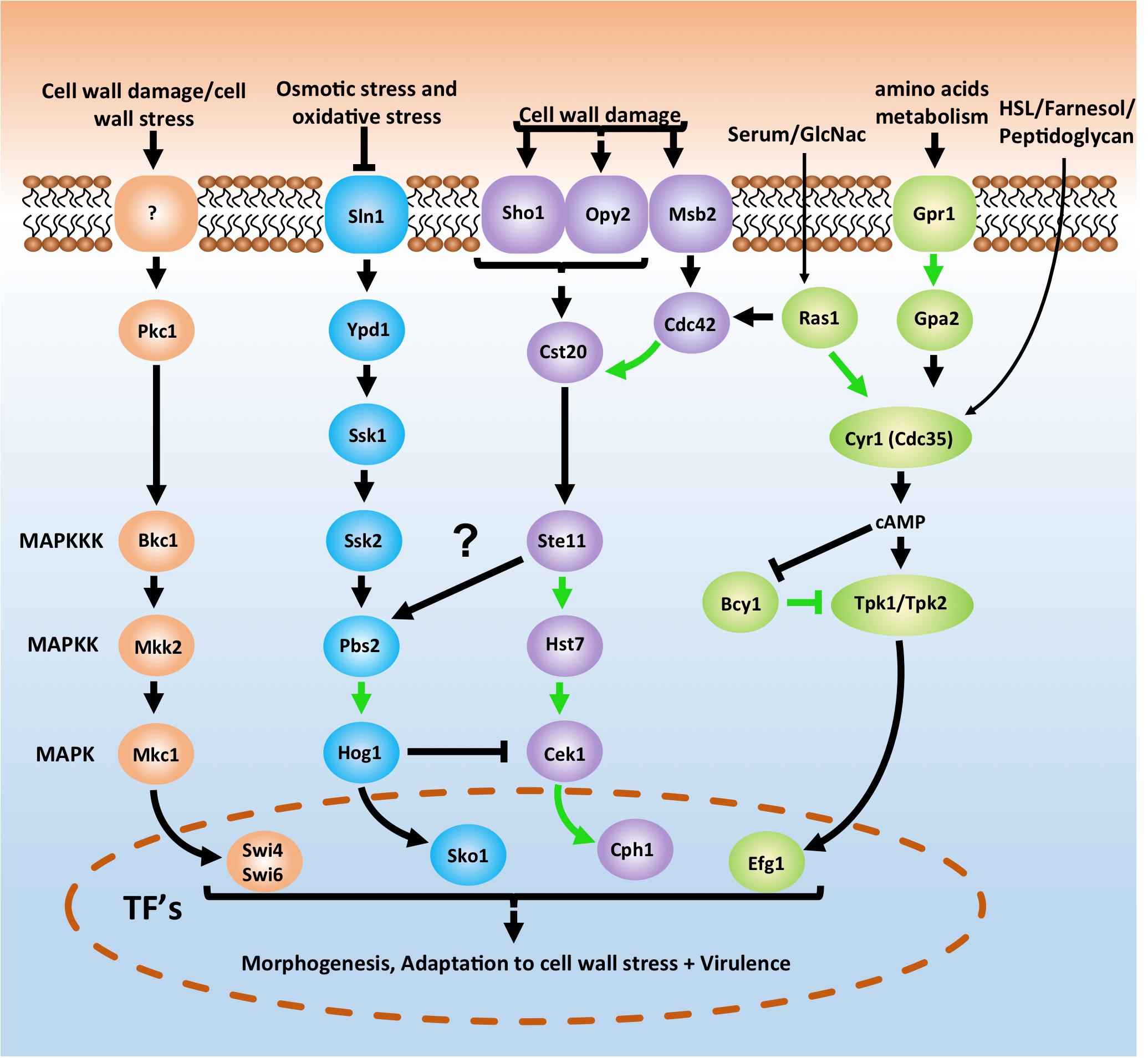
Figure 2. Basic schematic representation of the cAMP-PKA pathway (green) and three MAPK pathways in Candida albicans which are important for morphogenesis, adaptation to stress and survival. The cell integrity pathway (also known as PKC pathway) is depicted in orange, the HOG pathway is shown in blue and the CEK1 mediated pathway (also known as SVG pathway) is represented in purple. Pathways depicted here are an oversimplified version. A direct interaction between Msb2 and Cst20 (van Wijlick et al., 2016) is for example not depicted here. For a more in depth overview see Refs. (Biswas et al., 2007; de Dios et al., 2010; Sudbery, 2011; Huang, 2012; Noble et al., 2017; Burch et al., 2018). Notice also the arrow between Ste11 and Pbs2. In yeast the Sho1 branch plays a role in osmotic stress signaling to Hog1 (Hohmann, 2002). However, in C. albicans this does not seem to be the case as Ssk2 is the only MAPKKK signaling to Hog1 (Cheetham et al., 2007; Román et al., 2009). Arrows in green depict physical interactions between two proteins that have already been demonstrated in C. albicans. Arrows in black are Protein-protein interactions not yet demonstrated in C. albicans. See text for more details.
Conclusion
Of the plethora of available techniques to study PPIs, only a select few have been used intensively for large-scale high-throughput screenings. Two of the most important techniques are the TAP-MS and Y2H system (De Las Rivas and Fontanillo, 2010; Podobnik et al., 2016). Both techniques have been used to examine a large number of PPIs for several organisms, leading to a profound knowledge on the studied organisms and even the start of the construction of the interactome of the studied organisms (Ito et al., 2001; von Mering et al., 2002). PPI studies in C. albicans are, however, lagging behind. C. albicans is comparable with S. cerevisiae in terms of ORFs, respectively, 6,198 vs. 6,572. Both organisms serve as model organisms and are fully sequenced. However, a staggering 70% of the ORFs are still not characterized in C. albicans versus 12% in S. cerevisiae (Cherry et al., 2012; Skrzypek et al., 2017). Looking at PPIs the difference is even more extreme (based on release 3.5.174, July 2019); 1,080 interactions for C. albicans are mentioned in the BioGRID vs. 171,959 for S. cerevisiae (Chatr-Aryamontri et al., 2017; Oughtred et al., 2018). Supplementary Tables S1, S2 give an overview of interactions found in C. albicans that also have been detected in S. cerevisiae using orthologs. Supplementary Table S2 also shows that the majority of PPIs shown in C. albicans have been demonstrated with only one technique while in S. cerevisiae the majority of PPI have been demonstrated using multiple techniques. Interactions should, ideally, always be validated with two or more techniques (Auerbach et al., 2002; Gavin et al., 2002; von Mering et al., 2002). Despite being one of the first fully sequenced fungal pathogens (Jones et al., 2004), the difficulties encountered when working with C. albicans have slowed down the progress (Noble and Johnson, 2007). This is mainly due to the non-canonical codon usage in C. albicans and the long-held misconception that the gene function of C. albicans is very similar to that of S. cerevisiae. The latter has, however, been shown to be more complicated (Boysen et al., 2009). Until now, only 249 out of a total of 1,208 non-redundant PPIs in C. albicans have also been demonstrated in S. cerevisiae (see also Supplementary Tables S1, S2). A critical note here is that our tables also contain PPIs demonstrated in other C. albicans strains than the wild type-strain SC5314 (or its derivative strains) and PPIs deposited at the rcsb protein structure database, but not yet published.
Knowing that a similar organism, S. cerevisiae, has an estimated total of 30,000 to 40,000 interactions (Grigoriev, 2003; Sambourg and Thierry-Mieg, 2010) or even more (Hart et al., 2006), there is still a lot to discover for C. albicans. Besides giving fundamental knowledge, PPIs can also be used as very specific drug targets (Khan et al., 2011). Previously thought undruggable, PPIs have become increasingly interesting targets for drug development (Modell et al., 2016). Given the limited availability of antifungals, rising resistance, lack of antifungal vaccines, difficulties in antifungal drug development, and the increase of fungal infections worldwide (Brown et al., 2012; Fairlamb et al., 2016, Fairlamb et al., 2017; Patin et al., 2018) PPIs might become crucial in the future development of novel, specific antifungals.
In the “omics” era, the enormous amount of information generated by a wide range of large-scale, high-throughput assays creates severe problems for data storage and sorting, which emphasizes the importance of data collection and curation (Costanzo et al., 2006; Reguly et al., 2006). Open databases such as the Candida genome database (CGD) (Skrzypek et al., 2017) and the BioGRID (Oughtred et al., 2016, 2018; Chatr-Aryamontri et al., 2017) are crucial tools for Candida researchers. Integration of the BioGRID PPI dataset into the CGD would be a substantial improvement of the CGD. Currently the CGD only mentions the BioGRID as an external link on the summary page of the genes.
While using PPI data from S. cerevisiae to aid in constructing C. albicans PPI networks, caution is advised considering the low overlap between the PPI data (Supplementary Tables S1, S2). Das et al. (2019), for example, constructed a PPI network with a focus on proteins important for hyphae formation in C. albicans using data from S. cerevisiae, a species generally regarded as only forming pseudohyphae, as a control for the validation of interactions (Arkowitz and Bassilana, 2011; Das et al., 2019). Comprehensive, high-quality databases of C. albcians genome sequences and PPIs will make it possible to resolve the C. albicans interactome based on C. albicans data rather than inferences from data obtained in S. cerevisiae (Fraser et al., 2003; Tsai et al., 2014; Wang et al., 2014; Márkus et al., 2017; Das et al., 2019). PPI databases, such as the BioGRID, play a crucial role in elucidating the protein interaction networks but also rely on external help to grow and keep up with the most recent research (the BioGRID relies on researchers to send in their data as it does not actively track PPI data from C. albicans). The latter was noticeable by the absence of a huge number of interactions in the BioGRID at the start of this review (compare releases 3.4.156 and 3.5.174), yet it is of the utmost importance for researchers to send in their data as the whole research community depends on such databases to perform their experiments with a high-quality dataset (Costanzo et al., 2006; Cusick et al., 2009; Oughtred et al., 2016).
With an exhaustive literature search, we tried to include most of the PPIs known in C. albicans in the BioGRID dataset but are aware that some might still be missing. We therefore hope that future studies on PPIs in C. albicans will be sent to databases such as the BioGRID (an excel template for submitting PPI data can be found here5) so that a more complete overview regarding PPIs can be achieved for C. albicans.
Author Contributions
FS initiated this review, compiled all the data, provided all the input to BioGRID, and wrote the draft version of the manuscript. FS and PV wrote the final version of the manuscript.
Funding
Work in the PVD’s laboratory was supported by grants from the Interuniversity Attraction Poles Programme initiated by the Belgian Science Policy Office (IAP P7/28) and the Fund for Scientific Research Flanders (FWO, Grant G062616N).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the people from BioGRID and in specific Dr. Rose Oughtred for their help and interaction. We would also like to thank both reviewers for their constructive comments which very much improved this review.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01792/full#supplementary-material
Footnotes
- ^ https://www.rcsb.org
- ^ http://www.candidagenome.org
- ^ https://www.yeastgenome.org
- ^ https://thebiogrid.org
- ^ https://wiki.thebiogrid.org/lib/exe/fetch.php/biogrid_data_submission_form.xls
References
Abramczyk, D., Tchorzewski, M., Krokowski, D., Boguszewska, A., and Grankowski, N. (2004). Overexpression, purification and characterization of the acidic ribosomal P-proteins from Candida albicans. Biochim. Biophys. Acta 1672, 214–223. doi: 10.1016/j.bbagen.2004.04.005
Alkafeef, S. S., Yu, C., Huang, L., and Liu, H. (2018). Wor1 establishes opaque cell fate through inhibition of the general co-repressor Tup1 in Candida albicans. PLoS Genet. 14:e1007176. doi: 10.1371/journal.pgen.1007176
Arachea, B. T., Liu, X., Pavlovsky, A. G., and Viola, R. E. (2010). Expansion of the aspartate beta-semialdehyde dehydrogenase family: the first structure of a fungal ortholog. Acta Crystallogr. D Biol. Crystallogr. 66(Pt 2), 205–212. doi: 10.1107/S0907444909052834
Argimón, S., Fanning, S., Blankenship, J. R., and Mitchell, A. P. (2011). Interaction between the Candida albicans high-osmolarity glycerol (HOG) pathway and the response to human beta-defensins 2 and 3. Eukaryot. Cell 10, 272–275. doi: 10.1128/EC.00133-10
Arkowitz, R. A., and Bassilana, M. (2011). Polarized growth in fungi: symmetry breaking and hyphal formation. Semin. Cell Dev. Biol. 22, 806–815. doi: 10.1016/j.semcdb.2011.08.010
Askew, C., Sellam, A., Epp, E., Mallick, J., Hogues, H., Mullick, A., et al. (2011). The zinc cluster transcription factor Ahr1p directs Mcm1p regulation of Candida albicans adhesion. Mol. Microbiol. 79, 940–953. doi: 10.1111/j.1365-2958.2010.07504.x
Au Yong, J. Y., Wang, Y. M., and Wang, Y. (2016). The Nim1 kinase Gin4 has distinct domains crucial for septin assembly, phospholipid binding and mitotic exit. J. Cell Sci. 129, 2744–2756. doi: 10.1242/jcs.183160
Auerbach, D., Thaminy, S., Hottiger Michael, O., and Stagljar, I. (2002). The post-genomic era of interactive proteomics: facts and perspectives. Proteomics 2, 611–623. doi: 10.1002/1615-9861(200206)2:6<611:AID-PROT611<3.0.CO;2-Y
Badrane, H., Nguyen, M. H., Cheng, S., Kumar, V., Derendorf, H., Iczkowski, K. A., et al. (2008). The Candida albicans phosphatase Inp51p interacts with the EH domain protein Irs4p, regulates phosphatidylinositol-4,5-bisphosphate levels and influences hyphal formation, the cell integrity pathway and virulence. Microbiology 154(Pt 11), 3296–3308. doi: 10.1099/mic.0.2008/018002-0
Bahn, Y.-S., Xue, C., Idnurm, A., Rutherford, J. C., Heitman, J., and Cardenas, M. E. (2007). Sensing the environment: lessons from fungi. Nat. Rev. Microbiol. 5, 57–69. doi: 10.1038/nrmicro1578
Bai, C., Xu, X. L., Wang, H. S., Wang, Y. M., Chan, F. Y., and Wang, Y. (2011). Characterization of a hyperactive Cyr1 mutant reveals new regulatory mechanisms for cellular cAMP levels in Candida albicans. Mol. Microbiol. 82, 879–893. doi: 10.1111/j.1365-2958.2011.07859.x
Bakail, M., and Ochsenbein, F. (2016). Targeting protein–protein interactions, a wide open field for drug design. C. R. Chim. 19, 19–27. doi: 10.1016/j.crci.2015.12.004
Benedict, K., Richardson, M., Vallabhaneni, S., Jackson, B. R., and Chiller, T. (2017). Emerging issues, challenges, and changing epidemiology of fungal disease outbreaks. Lancet Infect. Dis. 17, e403–e411. doi: 10.1016/s1473-3099(17)30443-7
Berggård, T., Linse, S., and James, P. (2007). Methods for the detection and analysis of protein-protein interactions. Proteomics 7, 2833–2842. doi: 10.1002/pmic.200700131
Bishop, A., Lane, R., Beniston, R., Chapa-y-Lazo, B., Smythe, C., and Sudbery, P. (2010). Hyphal growth in Candida albicans requires the phosphorylation of Sec2 by the Cdc28-Ccn1/Hgc1 kinase. EMBO J. 29, 2930–2942. doi: 10.1038/emboj.2010.158
Biswas, S., Van Dijck, P., and Datta, A. (2007). Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol. Mol. Biol. Rev. 71, 348–376. doi: 10.1128/MMBR.00009-06
Blackwell, C., and Brown, J. D. (2009). “The application of tandem-affinity purification to Candida albicans,” in Candida albicans: Methods and Protocols, eds R. L. Cihlar, and R. A. Calderone, (Totowa, NJ: Humana Press), 133–148. doi: 10.1007/978-1-60327-151-6_13
Blackwell, C., Russell, C. L., Argimon, S., Brown, A. J., and Brown, J. D. (2003). Protein A-tagging for purification of native macromolecular complexes from Candida albicans. Yeast 20, 1235–1241. doi: 10.1002/yea.1036
Blackwell, M. (2011). The fungi: 1, 2, 3. 5.1 million species? Am. J. Bot. 98, 426–438. doi: 10.3732/ajb.1000298
Bongomin, F., Gago, S., Oladele, R. O., and Denning, D. W. (2017). Global and multi-national prevalence of fungal diseases-estimate precision. J. Fungi 3:E57. doi: 10.3390/jof3040057
Borth, N., Walther, A., Reijnst, P., Jorde, S., Schaub, Y., and Wendland, J. (2010). Candida albicans Vrp1 is required for polarized morphogenesis and interacts with Wal1 and Myo5. Microbiology 156(Pt 10), 2962–2969. doi: 10.1099/mic.0.041707-0
Boysen, J. H., Fanning, S., Newberg, J., Murphy, R. F., and Mitchell, A. P. (2009). Detection of protein-protein interactions through vesicle targeting. Genetics 182, 33–39. doi: 10.1534/genetics.109.101162
Brown, G. D., Denning, D. W., Gow, N. A., Levitz, S. M., Netea, M. G., and White, T. C. (2012). Hidden killers: human fungal infections. Sci. Transl. Med. 4:165rv113. doi: 10.1126/scitranslmed.3004404
Brückner, A., Polge, C., Lentze, N., Auerbach, D., and Schlattner, U. (2009). Yeast two-hybrid, a powerful tool for systems biology. Int. J. Mol. Sci. 10, 2763–2788. doi: 10.3390/ijms10062763
Burch, J. M., Mashayekh, S., Wykoff, D. D., and Grimes, C. L. (2018). Bacterial derived carbohydrates bind Cyr1 and trigger hyphal growth in Candida albicans. ACS Infect. Dis. 4, 53–58. doi: 10.1021/acsinfecdis.7b00154
Chatr-Aryamontri, A., Oughtred, R., Boucher, L., Rust, J., Chang, C., Kolas, N. K., et al. (2017). The BioGRID interaction database: 2017 update. Nucleic Acids Res. 45, D369–D379. doi: 10.1093/nar/gkw1102
Cheetham, J., MacCallum, D. M., Doris, K. S., da Silva Dantas, A., Scorfield, S., Odds, F., et al. (2011). MAPKKK-independent regulation of the Hog1 stress-activated protein kinase in Candida albicans. J. Biol. Chem. 286, 42002–42016. doi: 10.1074/jbc.M111.265231
Cheetham, J., Smith, D. A., da Silva, Dantas, A., Doris, K. S., Patterson, M. J., et al. (2007). A Single MAPKKK regulates the Hog1 MAPK pathway in the pathogenic fungus Candida albicans. Mol. Biol. Cell 18, 4603–4614. doi: 10.1091/mbc.E07-06-0581
Chen, C., and Noble, S. M. (2012). Post-transcriptional regulation of the sef1 transcription factor controls the virulence of Candida albicans in its mammalian host. PLoS Pathog. 8:e1002956. doi: 10.1371/journal.ppat.1002956
Chen, J., and Chen, J. (2001). Knocking out of Cek2 and Cks1 affect the phenotype of Candida albicans. Shi Yan Sheng Wu Xue Bao 34, 25–33.
Cherry, J. M., Hong, E. L., Amundsen, C., Balakrishnan, R., Binkley, G., Chan, E. T., et al. (2012). Saccharomyces genome database: the genomics resource of budding yeast. Nucleic Acids Res. 40, D700–D705. doi: 10.1093/nar/gkr1029
Correia, I., Román, E., Prieto, D., Eisman, B., and Pla, J. (2016). Complementary roles of the Cek1 and Cek2 MAP kinases in Candida albicans cell-wall biogenesis. Future Microbiol. 11, 51–67. doi: 10.2217/fmb.15.127
Corvey, C., Koetter, P., Beckhaus, T., Hack, J., Hofmann, S., Hampel, M., et al. (2005). Carbon Source-dependent assembly of the Snf1p kinase complex in Candida albicans. J. Biol. Chem. 280, 25323–25330. doi: 10.1074/jbc.M503719200
Costanzo, M. C., Arnaud, M. B., Skrzypek, M. S., Binkley, G., Lane, C., Miyasato, S. R., et al. (2006). The candida genome database: facilitating research on Candida albicans molecular biology. FEMS Yeast Res. 6, 671–684. doi: 10.1111/j.1567-1364.2006.00074.x
Côte, P., Sulea, T., Dignard, D., Wu, C., and Whiteway, M. (2011). Evolutionary reshaping of fungal mating pathway scaffold proteins. mBio 2:e00230-10. doi: 10.1128/mBio.00230-10
Cusick, M. E., Yu, H., Smolyar, A., Venkatesan, K., Carvunis, A.-R., Simonis, N., et al. (2009). Literature-curated protein interaction datasets. Nat. Methods 6, 39–46. doi: 10.1038/nmeth.1284
Das, S., Bhuyan, R., Bagchi, A., and Saha, T. (2019). Network analysis of hyphae forming proteins in Candida albicans identifies important proteins responsible for pathovirulence in the organism. Heliyon 5:e01916. doi: 10.1016/j.heliyon.2019.e01916
Datta, A., Ganesan, K., and Natarajan, K. (1990). “Current trends in Candida albicans research,” in Advances in Microbial Physiology, eds A. H. Rose, and D. W. Tempest, (Cambridge, MA: Academic Press), 53–88. doi: 10.1016/s0065-2911(08)60110-1
de Dios, C. H., Roman, E., Monge, R. A., and Pla, J. (2010). The role of MAPK signal transduction pathways in the response to oxidative stress in the fungal pathogen Candida albicans: implications in virulence. Curr. Protein Peptide Sci. 11, 693–703. doi: 10.2174/138920310794557655
De Las Rivas, J., and Fontanillo, C. (2010). Protein-protein interactions essentials: key concepts to building and analyzing interactome networks. PLoS Comput. Biol. 6:e1000807. doi: 10.1371/journal.pcbi.1000807
de Pauw, B. E. (2011). What are fungal infections? Mediterr. J. Hematol. Infect. Dis. 3:e2011001. doi: 10.4084/MJHID.2011.001
Demuyser, L., Van Genechten, W., Mizuno, H., Colombo, S., and Van Dijck, P. (2018). Introducing fluorescence resonance energy transfer-based biosensors for the analysis of cAMP-PKA signalling in the fungal pathogen Candida glabrata. Cell Microbiol. 20:e12863. doi: 10.1111/cmi.12863
Diaz, G., Polonelli, L., Conti, S., Messana, I., Cabras, T., Putzolu, M., et al. (2005). Mitochondrial alterations and autofluorescent conversion of Candida albicans induced by histatins. Microsc. Res. Tech. 66, 219–228. doi: 10.1002/jemt.20161
Dorsaz, S., Coste, A. T., and Sanglard, D. (2017). Red-shifted firefly luciferase optimized for Candida albicans in vivo bioluminescence imaging. Front. Microbiol. 8:1478. doi: 10.3389/fmicb.2017.01478
Dostal, J., Brynda, J., Blaha, J., Machacek, S., Heidingsfeld, O., and Pichova, I. (2018). Crystal structure of carbonic anhydrase CaNce103p from the pathogenic yeast Candida albicans. BMC Struct. Biol. 18:14. doi: 10.1186/s12900-018-0093-4
Echt, S., Bauer, S., Steinbacher, S., Huber, R., Bacher, A., and Fischer, M. (2004). Potential anti-infective targets in pathogenic yeasts: structure and properties of 3,4-dihydroxy-2-butanone 4-phosphate synthase of Candida albicans. J. Mol. Biol. 341, 1085–1096. doi: 10.1016/j.jmb.2004.06.053
Emmott, E., and Goodfellow, I. (2014). Identification of protein interaction partners in mammalian cells using SILAC-immunoprecipitation quantitative proteomics. J. Vis. Exp. 89:e51656. doi: 10.3791/51656
Ene, I. V., Walker, L. A., Schiavone, M., Lee, K. K., Martin-Yken, H., Dague, E., et al. (2015). Cell wall remodeling enzymes modulate fungal cell wall elasticity and osmotic stress resistance. mBio 6:e00986. doi: 10.1128/mBio.00986-15
Fairlamb, A. H., Gow, N. A. R., Matthews, K. R., and Waters, A. P. (2016). Drug resistance in eukaryotic microorganisms. Nat. Microbiol. 1, 16092–16092. doi: 10.1038/nmicrobiol.2016.92
Fang, H. M., and Wang, Y. (2006). RA domain-mediated interaction of Cdc35 with Ras1 is essential for increasing cellular cAMP level for Candida albicans hyphal development. Mol. Microbiol. 61, 484–496. doi: 10.1111/j.1365-2958.2006.05248.x
Feng, J., Duan, Y., Qin, Y., Sun, W., Zhuang, Z., Zhu, D., et al. (2017). The N-terminal pY33XL motif of CaPsy2 is critical for the function of protein phosphatase 4 in CaRad53 deactivation, DNA damage-induced filamentation and virulence in Candida albicans. Int. J. Med. Microbiol. 307, 471–480. doi: 10.1016/j.ijmm.2017.09.017
Fraser, H. B., Wall, D. P., and Hirsh, A. E. (2003). A simple dependence between protein evolution rate and the number of protein-protein interactions. BMC Evol. Biol. 3:11. doi: 10.1186/1471-2148-3-11
Gabaldón, T., Naranjo-Ortiz, M. A., and Marcet-Houben, M. (2016). Evolutionary genomics of yeast pathogens in the Saccharomycotina. FEMS Yeast Res. 16:fow064. doi: 10.1093/femsyr/fow064
Gao, J., Wang, H., Wong, A. H., Zeng, G., Huang, Z., Wang, Y., et al. (2014). Regulation of Rfa2 phosphorylation in response to genotoxic stress in Candida albicans. Mol. Microbiol. 94, 141–155. doi: 10.1111/mmi.12749
Garcia, M. D., Chua, S. M. H., Low, Y. S., Lee, Y. T., Agnew-Francis, K., Wang, J. G., et al. (2018). Commercial AHAS-inhibiting herbicides are promising drug leads for the treatment of human fungal pathogenic infections. Proc. Natl. Acad. Sci. U.S.A. 115, E9649–E9658. doi: 10.1073/pnas.1809422115
Gavin, A.-C., Bösche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., et al. (2002). Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147. doi: 10.1038/415141a
Gingras, A.-C., Aebersold, R., and Raught, B. (2005). Advances in protein complex analysis using mass spectrometry. J. Physiol. 563(Pt 1) 11–21. doi: 10.1113/jphysiol.2004.080440
Gkourtsa, A., van den Burg, J., Avula, T., Hochstenbach, F., and Distel, B. (2016). Binding of a proline-independent hydrophobic motif by the Candida albicans Rvs167-3 SH3 domain. Microbiol. Res. 190, 27–36. doi: 10.1016/j.micres.2016.04.018
Graus, M. S., Neumann, A. K., and Timlin, J. A. (2017). Hyperspectral fluorescence microscopy detects autofluorescent factors that can be exploited as a diagnostic method for Candida species differentiation. J. Biomed. Opt. 22:16002. doi: 10.1117/1.JBO.22.1.016002
Greig, J. A., Sudbery, I. M., Richardson, J. P., Naglik, J. R., Wang, Y., and Sudbery, P. E. (2015). Cell cycle-independent phospho-regulation of Fkh2 during hyphal growth regulates Candida albicans pathogenesis. PLoS Pathog. 11:e1004630. doi: 10.1371/journal.ppat.1004630
Grigoriev, A. (2003). On the number of protein–protein interactions in the yeast proteome. Nucleic Acids Res. 31, 4157–4161. doi: 10.1093/nar/gkg466
Guan, G., Wang, H., Liang, W., Cao, C., Tao, L., Naseem, S., et al. (2015). The mitochondrial protein Mcu1 plays important roles in carbon source utilization, filamentation, and virulence in Candida albicans. Fungal Genet. Biol. 81, 150–159. doi: 10.1016/j.fgb.2015.01.006
Hart, G. T., Ramani, A. K., and Marcotte, E. M. (2006). How complete are current yeast and human protein-interaction networks? Genome Biol. 7:120. doi: 10.1186/gb-2006-7-11-120
Hast, M. A., and Beese, L. S. (2008). Structure of protein geranylgeranyltransferase-I from the human pathogen Candida albicans complexed with a lipid substrate. J. Biol. Chem. 283, 31933–31940. doi: 10.1074/jbc.M805330200
Hernday, A. D., Lohse, M. B., Fordyce, P. M., Nobile, C. J., DeRisi, J. L., and Johnson, A. D. (2013). Structure of the transcriptional network controlling white-opaque switching in Candida albicans. Mol. Microbiol. 90, 22–35. doi: 10.1111/mmi.12329
Heyken, W.-T., Wagner, C., Wittmann, J., Albrecht, A., and Schüller, H.-J. (2003). Negative regulation of phospholipid biosynthesis in Saccharomyces cerevisiae by a Candida albicans orthologue of OPI1. Yeast 20, 1177–1188. doi: 10.1002/yea.1031
Hickman, M. A., Paulson, C., Dudley, A., and Berman, J. (2015). Parasexual ploidy reduction drives population heterogeneity through random and transient aneuploidy in Candida albicans. Genetics 200, 781–794. doi: 10.1534/genetics.115.178020
Hohmann, S. (2002). Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66, 300–372. doi: 10.1128/MMBR.66.2.300-372.2002
Hong, H., Cai, Y., Zhang, S., Ding, H., Wang, H., and Han, A. (2017). Molecular basis of substrate specific acetylation by N-Terminal acetyltransferase NatB. Structure 25, 641.e3–649.e3. doi: 10.1016/j.str.2017.03.003
Hoppen, J., Dietz, M., Warsow, G., Rohde, R., and Schüller, H.-J. (2007). Ribosomal protein genes in the yeast Candida albicans may be activated by a heterodimeric transcription factor related to Ino2 and Ino4 from S. cerevisiae. Mol. Genet. Genomics 278, 317–330. doi: 10.1007/s00438-007-0253-x
Huang, G. (2012). Regulation of phenotypic transitions in the fungal pathogen Candida albicans. Virulence 3, 251–261. doi: 10.4161/viru.20010
Huang, Z.-X., Wang, H., Wang, Y.-M., and Wang, Y. (2014a). Novel mechanism coupling cyclic AMP-protein kinase a signaling and golgi trafficking via Gyp1 phosphorylation in polarized growth. Eukaryot. Cell 13, 1548–1556. doi: 10.1128/EC.00231-14
Huang, Z. X., Zhao, P., Zeng, G. S., Wang, Y. M., Sudbery, I., and Wang, Y. (2014b). Phosphoregulation of Nap1 plays a role in septin ring dynamics and morphogenesis in Candida albicans. mBio 5:e00915-13. doi: 10.1128/mBio.00915-13
Ismail, T., Fatima, N., Muhammad, S. A., Zaidi, S. S., Rehman, N., Hussain, I., et al. (2018). Prioritizing and modelling of putative drug target proteins of Candida albicans by systems biology approach. Acta Biochim. Pol. 65, 209–218. doi: 10.18388/abp.2017_2327
Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M., and Sakaki, Y. (2001). A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. U.S.A. 98, 4569–4574. doi: 10.1073/pnas.061034498
Jacobsen, I. D., Lüttich, A., Kurzai, O., Hube, B., and Brock, M. (2014). In vivo imaging of disseminated murine Candida albicans infection reveals unexpected host sites of fungal persistence during antifungal therapy. J. Antimicrob. Chemother. 69, 2785–2796. doi: 10.1093/jac/dku198
Jain, P., Sethi, S. C., Pratyusha, V. A., Garai, P., Naqvi, N., Singh, S., et al. (2018). Ras signaling activates glycosylphosphatidylinositol (GPI) anchor biosynthesis via the GPI-N-acetylglucosaminyltransferase (GPI-GnT) in Candida albicans. J. Biol. Chem. 293, 12222–12238. doi: 10.1074/jbc.RA117.001225
Jones, T., Federspiel, N. A., Chibana, H., Dungan, J., Kalman, S., Magee, B. B., et al. (2004). The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. U.S.A. 101, 7329–7334. doi: 10.1073/pnas.0401648101
Kabir, M. A., Hussain, M. A., and Ahmad, Z. (2012). Candida albicans: a model organism for studying fungal pathogens. ISRN Microbiol. 2012:538694. doi: 10.5402/2012/538694
Kaneko, A., Umeyama, T., Hanaoka, N., Monk, B. C., Uehara, Y., and Niimi, M. (2004). Tandem affinity purification of the Candida albicans septin protein complex. Yeast 21, 1025–1033. doi: 10.1002/yea.1147
Kaneko, A., Umeyama, T., Utena-Abe, Y., Yamagoe, S., Niimi, M., and Uehara, Y. (2006). Tcc1p, a novel protein containing the tetratricopeptide repeat motif, interacts with Tup1p to regulate morphological transition and virulence in Candida albicans. Eukaryot. Cell 5, 1894–1905. doi: 10.1128/EC.00151-06
Kaneva, I. N., Longworth, J., Sudbery, P. E., and Dickman, M. J. (2018). Quantitative proteomic analysis in Candida albicans using SILAC-based mass spectrometry. Proteomics 18:1700278. doi: 10.1002/pmic.201700278
Kaneva, I. N., Sudbery, I. M., Dickman, M. J., and Sudbery, P. E. (2019). Proteins that physically interact with the phosphatase Cdc14 in Candida albicans have diverse roles in the cell cycle. Sci. Rep. 9:6258. doi: 10.1038/s41598-019-42530-1
Kaplanek, P., de Boer, A., Gross, U., de Groot, P., Hube, B., and Weig, M. (2006). Candida and candidosis today: where are we, and where to go? The interdisciplinary forum on candidosis (IFOCAN) 2005, Gottingen (Germany), 23-25 September 2005. FEMS Yeast Res. 6, 1290–1294. doi: 10.1111/j.1567-1364.2006.00184.x
Kastora, S. L., Herrero-de-Dios, C., Avelar, G. M., Munro, C. A., and Brown, A. J. P. (2017). Sfp1 and Rtg3 reciprocally modulate carbon source-conditional stress adaptation in the pathogenic yeast Candida albicans. Mol. Microbiol. 105, 620–636. doi: 10.1111/mmi.13722
Kerppola, T. K. (2008). Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 37, 465–487. doi: 10.1146/annurev.biophys.37.032807.125842
Kerrien, S., Aranda, B., Breuza, L., Bridge, A., Broackes-Carter, F., Chen, C., et al. (2012). The IntAct molecular interaction database in 2012. Nucleic Acids Res. 40, D841–D846. doi: 10.1093/nar/gkr1088
Khan, S. H., Ahmad, F., Ahmad, N., Flynn, D. C., and Kumar, R. (2011). Protein-protein interactions: principles, techniques, and their potential role in new drug development. J. Biomol. Struct. Dyn. 28, 929–938. doi: 10.1080/07391102.2011.10508619
Kiliszek, A., Rypniewski, W., Rzad, K., Milewski, S., and Gabriel, I. (2019). Crystal structures of aminotransferases Aro8 and Aro9 from Candida albicans and structural insights into their properties. J. Struct. Biol. 205, 26–33. doi: 10.1016/j.jsb.2019.02.001
Kingwell, K. (2016). Antifungals: protein-protein interaction inhibitor tackles drug resistance. Nat. Rev. Drug Discov. 15, 232–233. doi: 10.1038/nrd.2016.50
Kobayashi, S. D., and Cutler, J. E. (1998). Candida albicans hyphal formation and virulence: is there a clearly defined role? Trends Microbiol. 6, 92–94. doi: 10.1016/S0966-842X(98)01218-9
Köhler, J. R., Casadevall, A., and Perfect, J. (2014). The spectrum of fungi that infects humans. Cold Spring Harb. Perspect. Med. 5:a019273. doi: 10.1101/cshperspect.a019273
Kos, I., Patterson, M. J., Znaidi, S., Kaloriti, D., da Silva Dantas, A., Herrero-de-Dios, C. M., et al. (2016). Mechanisms underlying the delayed activation of the Cap1 transcription factor in Candida albicans following combinatorial oxidative and cationic stress important for phagocytic potency. mBio 7:e00331. doi: 10.1128/mBio.00331-16
Krogan, N. J., Cagney, G., Yu, H., Zhong, G., Guo, X., Ignatchenko, A., et al. (2006). Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643. doi: 10.1038/nature04670
Kucharíková, S., Vande Velde, G., Himmelreich, U., and Van Dijck, P. (2015). Candida albicans biofilm development on medically-relevant foreign bodies in a mouse subcutaneous model followed by bioluminescence imaging. J. Vis. Exp. 95:e52239. doi: 10.3791/52239
Lavoie, H., Hogues, H., Mallick, J., Sellam, A., Nantel, A., and Whiteway, M. (2010). Evolutionary tinkering with conserved components of a transcriptional regulatory network. PLoS Biol. 8:e1000329. doi: 10.1371/journal.pbio.1000329
Lavoie, H., Sellam, A., Askew, C., Nantel, A., and Whiteway, M. (2008). A toolbox for epitope-tagging and genome-wide location analysis in Candida albicans. BMC Genomics 9:578. doi: 10.1186/1471-2164-9-578
Lee, H. J., Kim, J. M., Kang, W. K., Yang, H., and Kim, J. Y. (2015). The NDR kinase Cbk1 downregulates the transcriptional repressor Nrg1 through the mRNA-binding protein Ssd1 in Candida albicans. Eukaryot. Cell 14, 671–683. doi: 10.1128/EC.00016-15
Lee, J. E., Oh, J. H., Ku, M., Kim, J., Lee, J. S., and Kang, S. O. (2015). Ssn6 has dual roles in Candida albicans filament development through the interaction with Rpd31. FEBS Lett. 589, 513–520. doi: 10.1016/j.febslet.2015.01.011
Legrain, P., and Selig, L. (2000). Genome-wide protein interaction maps using two-hybrid systems. FEBS Lett. 480, 32–36. doi: 10.1016/S0014-5793(00)01774-9
Legrand, M., Bachellier-Bassi, S., Lee, K. K., Chaudhari, Y., Tournu, H., Arbogast, L., et al. (2018). Generating genomic platforms to study Candida albicans pathogenesis. Nucleic Acids Res. 46, 6935–6949. doi: 10.1093/nar/gky594
Li, C. R., Au Yong, J. Y., Wang, Y. M., and Wang, Y. (2012). CDK regulates septin organization through cell-cycle-dependent phosphorylation of the Nim1-related kinase Gin4. J. Cell Sci. 125(Pt 10), 2533–2543. doi: 10.1242/jcs.104497
Li, C. R., Lee, R. T., Wang, Y. M., Zheng, X. D., and Wang, Y. (2007). Candida albicans hyphal morphogenesis occurs in Sec3p-independent and Sec3p-dependent phases separated by septin ring formation. J. Cell Sci. 120(Pt 11), 1898–1907. doi: 10.1242/jcs.002931
Li, C.-R., Wang, Y.-M., and Wang, Y. (2008). The IQGAP Iqg1 is a regulatory target of CDK for cytokinesis in Candida albicans. EMBO J. 27, 2998–3010. doi: 10.1038/emboj.2008.219
Li, X., Robbins, N., O’Meara, T. R., and Cowen, L. E. (2017). Extensive functional redundancy in the regulation of Candida albicans drug resistance and morphogenesis by lysine deacetylases Hos2, Hda1, Rpd3 and Rpd31. Mol. Microbiol. 103, 635–656. doi: 10.1111/mmi.13578
Licata, L., Briganti, L., Peluso, D., Perfetto, L., Iannuccelli, M., Galeota, E., et al. (2012). MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 40, D857–D861. doi: 10.1093/nar/gkr930
Liu, N., Tu, J., Dong, G., Wang, Y., and Sheng, C. (2018). Emerging new targets for the treatment of resistant fungal infections. J. Med. Chem. 61, 5484–5511. doi: 10.1021/acs.jmedchem.7b01413
Liu, Q., Han, Q., Wang, N., Yao, G., Zeng, G., Wang, Y., et al. (2016). Tpd3-Pph21 phosphatase plays a direct role in Sep7 dephosphorylation in Candida albicans. Mol. Microbiol. 101, 109–121. doi: 10.1111/mmi.13376
Lu, P.-L., Liu, W.-L., Lo, H.-J., Wang, F.-D., Ko, W.-C., Hsueh, P.-R., et al. (2018). Are we ready for the global emergence of multidrug-resistant Candida auris in Taiwan? J. Formos. Med. Assoc. 117, 462–470. doi: 10.1016/j.jfma.2017.10.005
Lu, Y., Su, C., Mao, X., Raniga, P. P., Liu, H., and Chen, J. (2008). Efg1-mediated recruitment of NuA4 to promoters is required for hypha-specific Swi/Snf binding and activation in Candida albicans. Mol. Biol. Cell 19, 4260–4272. doi: 10.1091/mbc.E08-02-0173
Mallick, J., Jansen, G., Wu, C., and Whiteway, M. (2016). “SRYTH: a new yeast two-hybrid method,” in Candida Species: Methods and Protocols, eds R. Calderone, and R. Cihlar, (New York, NY: Springer), 31–41. doi: 10.1007/978-1-4939-3052-4_3
Mallick, J., and Whiteway, M. (2013). The evolutionary rewiring of the ribosomal protein transcription pathway modifies the interaction of transcription factor heteromer Ifh1-Fhl1 (interacts with forkhead 1-forkhead-like 1) with the DNA-binding specificity element. J. Biol. Chem. 288, 17508–17519. doi: 10.1074/jbc.M112.436683
Mamouei, Z., Zeng, G., Wang, Y. M., and Wang, Y. (2017). Candida albicans possess a highly versatile and dynamic high-affinity iron transport system important for its commensal-pathogenic lifestyle. Mol. Microbiol. 106, 986–998. doi: 10.1111/mmi.13864
Márkus, B., Szabó, K., Pfliegler, W. P., Petrényi, K., Boros, E., Pócsi, I., et al. (2017). Proteomic analysis of protein phosphatase Z1 from Candida albicans. PLoS One 12:e0183176. doi: 10.1371/journal.pone.0183176
Mathé, L., and Van Dijck, P. (2013). Recent insights into Candida albicans biofilm resistance mechanisms. Curr. Genet. 59, 251–264. doi: 10.1007/s00294-013-0400-3
Miller, K. E., Kim, Y., Huh, W.-K., and Park, H.-O. (2015). Bimolecular fluorescence complementation (BiFC) analysis: advances and recent applications for genome-wide interaction studies. J. Mol. Biol. 427, 2039–2055. doi: 10.1016/j.jmb.2015.03.005
Mitchell, A. P. (2016). Fungus produces a toxic surprise. Nature 532, 41–42. doi: 10.1038/nature17319
Miwa, T., Takagi, Y., Shinozaki, M., Yun, C. W., Schell, W. A., Perfect, J. R., et al. (2004). Gpr1, a putative G-protein-coupled receptor, regulates morphogenesis and hypha formation in the pathogenic fungus Candida albicans. Eukaryot. Cell 3, 919–931. doi: 10.1128/EC.3.4.919-931.2004
Modell, A. E., Blosser, S. L., and Arora, P. S. (2016). Systematic targeting of protein-protein interactions. Trends Pharmacol. Sci. 37, 702–713. doi: 10.1016/j.tips.2016.05.008
Morgunova, E., Saller, S., Haase, I., Cushman, M., Bacher, A., Fischer, M., et al. (2007). Lumazine synthase from Candida albicans as an anti-fungal target enzyme: structural and biochemical basis for drug design. J. Biol. Chem. 282, 17231–17241. doi: 10.1074/jbc.M701724200
Moyes, D. L., Wilson, D., Richardson, J. P., Mogavero, S., Tang, S. X., Wernecke, J., et al. (2016). Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature 532, 64–68. doi: 10.1038/nature17625
Murad, A. M., Lee, P. R., Broadbent, I. D., Barelle, C. J., and Brown, A. J. (2000). CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16, 325–327. doi: 10.1002/1097-0061(20000315)16:4<325::aid-yea538>3.0.co;2-#
Muzzey, D., Schwartz, K., Weissman, J. S., and Sherlock, G. (2013). Assembly of a phased diploid Candida albicans genome facilitates allele-specific measurements and provides a simple model for repeat and indel structure. Genome Biol. 14:R97. doi: 10.1186/gb-2013-14-9-r97
Nakamura, A., Nemoto, T., Heinemann, I. U., Yamashita, K., Sonoda, T., Komoda, K., et al. (2013). Structural basis of reverse nucleotide polymerization. Proc. Natl. Acad. Sci. U.S.A. 110, 20970–20975. doi: 10.1073/pnas.1321312111
Nasser, L., Weissman, Z., Pinsky, M., Amartely, H., Dvir, H., and Kornitzer, D. (2016). Structural basis of haem-iron acquisition by fungal pathogens. Nat. Microbiol. 1:16156. doi: 10.1038/nmicrobiol.2016.156
Ni, J., Gao, Y., Liu, H., and Chen, J. (2004). Candida albicans Cdc37 interacts with the Crk1 kinase and is required for Crk1 production. FEBS Lett. 561, 223–230. doi: 10.1016/s0014-5793(04)00172-3
Nishikawa, J. L., Boeszoermenyi, A., Vale-Silva, L. A., Torelli, R., Posteraro, B., Sohn, Y. J., et al. (2016). Inhibiting fungal multidrug resistance by disrupting an activator-mediator interaction. Nature 530, 485–489. doi: 10.1038/nature16963
Nobile, C. J., Fox, E. P., Nett, J. E., Sorrells, T. R., Mitrovich, Q. M., Hernday, A. D., et al. (2012). A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148, 126–138. doi: 10.1016/j.cell.2011.10.048
Noble, S. M., Gianetti, B. A., and Witchley, J. N. (2017). Candida albicans cell-type switching and functional plasticity in the mammalian host. Nat. Rev. Microbiol. 15, 96–108. doi: 10.1038/nrmicro.2016.157
Noble, S. M., and Johnson, A. D. (2007). Genetics of Candida albicans, a diploid human fungal pathogen. Annu. Rev. Genet. 41, 193–211. doi: 10.1146/annurev.genet.41.042007.170146
Ofir, A., Hofmann, K., Weindling, E., Gildor, T., Barker, K. S., Rogers, P. D., et al. (2012). Role of a Candida albicans Nrm1/Whi5 homologue in cell cycle gene expression and DNA replication stress response. Mol. Microbiol. 84, 778–794. doi: 10.1111/j.1365-2958.2012.08056.x
O’Meara, T. R., O’Meara, M. J., Polvi, E. J., Pourhaghighi, M. R., Liston, S. D., Lin, Z.-Y., et al. (2019). Global proteomic analyses define an environmentally contingent Hsp90 interactome and reveal chaperone-dependent regulation of stress granule proteins and the R2TP complex in a fungal pathogen. PLoS Biol. 17:e3000358. doi: 10.1371/journal.pbio.3000358
Ong, S.-E., Blagoev, B., Kratchmarova, I., Kristensen, D. B., Steen, H., Pandey, A., et al. (2002). Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteom. 1, 376–386. doi: 10.1074/mcp.M200025-MCP200
Ong, S.-E., Kratchmarova, I., and Mann, M. (2003). Properties of 13C-substituted arginine in stable isotope labeling by amino acids in cell culture (SILAC). J. Proteome Res. 2, 173–181. doi: 10.1021/pr0255708
Oughtred, R., Chatr-aryamontri, A., Breitkreutz, B. J., Chang, C. S., Rust, J. M., Theesfeld, C. L., et al. (2016). Use of the BioGRID database for analysis of yeast protein and genetic interactions. Cold Spring Harb. Protoc. 2016:rot088880. doi: 10.1101/pdb.prot088880
Oughtred, R., Stark, C., Breitkreutz, B. J., Rust, J., Boucher, L., Chang, C., et al. (2018). The BioGRID interaction database: 2019 update. Nucleic Acids Res. 47, D529–D541. doi: 10.1093/nar/gky1079
Palzer, S., Bantel, Y., Kazenwadel, F., Berg, M., Rupp, S., and Sohn, K. (2013). An expanded genetic code in Candida albicans to study protein-protein interactions in vivo. Eukaryot. Cell 12, 816–827. doi: 10.1128/EC.00075-13
Parrish, J. R., Yu, J., Liu, G., Hines, J. A., Chan, J. E., Mangiola, B. A., et al. (2007). A proteome-wide protein interaction map for Campylobacter jejuni. Genome Biol. 8:R130. doi: 10.1186/gb-2007-8-7-r130
Patin, E. C., Thompson, A., and Orr, S. J. (2018). Pattern recognition receptors in fungal immunity. Semin. Cell Dev. Biol. 89, 24–33. doi: 10.1016/j.semcdb.2018.03.003
Perlin, D. S. (2017). “Antifungals,” in Candida albicans: Cellular and Molecular Biology, ed. R. Prasad, (Cham: Springer International Publishing), 471–489.
Podobnik, M., Kraševec, N., Bedina Zavec, A., Naneh, O., Flašker, A., Caserman, S., et al. (2016). How to study protein-protein interactions. Acta Chim. Slov. 63, 424–439. doi: 10.17344/acsi.2016.2419
Priest, S. J., and Lorenz, M. C. (2015). Characterization of virulence-related phenotypes in Candida species of the CUG clade. Eukaryot. cell 14, 931–940. doi: 10.1128/EC.00062-15
Raczynska, J., Olchowy, J., Konariev, P. V., Svergun, D. I., Milewski, S., and Rypniewski, W. (2007). The crystal and solution studies of glucosamine-6-phosphate synthase from Candida albicans. J. Mol. Biol. 372, 672–688. doi: 10.1016/j.jmb.2007.07.002
Rao, K. H., Ghosh, S., and Datta, A. (2016). Env7p Associates with the golgin protein Imh1 at the trans-golgi network in Candida albicans. mSphere 1:e00080-16. doi: 10.1128/mSphere.00080-16
Rao, V. S., Srinivas, K., Sujini, G. N., and Kumar, G. N. S. (2014). Protein-protein interaction detection: methods and analysis. Int. J. Proteom. 2014:147648. doi: 10.1155/2014/147648
Reguly, T., Breitkreutz, A., Boucher, L., Breitkreutz, B.-J., Hon, G. C., Myers, C. L., et al. (2006). Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J. Biol. 5:11. doi: 10.1186/jbiol36
Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Séraphin, B. (1999). A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030. doi: 10.1038/13732
Rocha, R., Pereira, P. J., Santos, M. A., and Macedo-Ribeiro, S. (2011). Unveiling the structural basis for translational ambiguity tolerance in a human fungal pathogen. Proc. Natl. Acad. Sci. U.S.A. 108, 14091–14096. doi: 10.1073/pnas.1102835108
Román, E., Correia, I., Salazin, A., Fradin, C., Jouault, T., Poulain, D., et al. (2016). The Cek1mediated MAP kinase pathway regulates exposure of alpha1,2 and beta1,2mannosides in the cell wall of Candida albicans modulating immune recognition. Virulence 7, 558–577. doi: 10.1080/21505594.2016.1163458
Román, E., Cottier, F., Ernst, J. F., and Pla, J. (2009). Msb2 signaling mucin controls activation of Cek1 mitogen-activated protein kinase in Candida albicans. Eukaryot. Cell 8, 1235–1249. doi: 10.1128/EC.00081-09
Román, E., Nombela, C., and Pla, J. (2005). The Sho1 adaptor protein links oxidative stress to morphogenesis and cell wall biosynthesis in the fungal pathogen Candida albicans. Mol. Cell. Biol. 25, 10611–10627. doi: 10.1128/mcb.25.23.10611-10627.2005
Ryan, O., Shapiro, R. S., Kurat, C. F., Mayhew, D., Baryshnikova, A., Chin, B., et al. (2012). Global gene deletion analysis exploring yeast filamentous growth. Science 337, 1353–1356. doi: 10.1126/science.1224339
Salwinski, L., Miller, C. S., Smith, A. J., Pettit, F. K., Bowie, J. U., and Eisenberg, D. (2004). The database of interacting proteins: 2004 update. Nucleic Acids Res. 32, D449–D451. doi: 10.1093/nar/gkh086
Sambourg, L., and Thierry-Mieg, N. (2010). New insights into protein-protein interaction data lead to increased estimates of the S. cerevisiae interactome size. BMC Bioinform. 11:605. doi: 10.1186/1471-2105-11-605
Santini, S., Monchois, V., Mouz, N., Sigoillot, C., Rousselle, T., Claverie, J. M., et al. (2008). Structural characterization of CA1462, the Candida albicans thiamine pyrophosphokinase. BMC Struct. Biol. 8:33. doi: 10.1186/1472-6807-8-33
Schächter, V. (2002). Bioinformatics of large-scale protein interaction networks. Biotechniques 32(3 Suppl.), S16–S27.
Schoeters, F., Munro, C. A., d’Enfert, C., and Van Dijck, P. (2018). A high-throughput Candida albicans two-hybrid system. mSphere 3:e00391-18. doi: 10.1128/mSphere.00391-18
Sellam, A., Chaillot, J., Mallick, J., Tebbji, F., Richard Albert, J., Cook, M. A., et al. (2019). The p38/HOG stress-activated protein kinase network couples growth to division in Candida albicans. PLoS Genet. 15:e1008052. doi: 10.1371/journal.pgen.1008052
Senay, C., Ferrari, P., Rocher, C., Rieger, K. J., Winter, J., Platel, D., et al. (2003). The Mtr2-Mex67 NTF2-like domain complex. Structural insights into a dual role of Mtr2 for yeast nuclear export. J. Biol. Chem. 278, 48395–48403. doi: 10.1074/jbc.M308275200
Shapiro, R. S., Zaas, A. K., Betancourt-Quiroz, M., Perfect, J. R., and Cowen, L. E. (2012). The Hsp90 co-chaperone Sgt1 governs Candida albicans morphogenesis and drug resistance. PLoS One 7:e44734. doi: 10.1371/journal.pone.0044734
Sheng, Y., Durazo, A., Schumacher, M., Gralla, E. B., Cascio, D., Cabelli, D. E., et al. (2013). Tetramerization reinforces the dimer interface of MnSOD. PLoS One 8:e62446. doi: 10.1371/journal.pone.0062446
Sherrington, S. L., Sorsby, E., Mahtey, N., Kumwenda, P., Lenardon, M. D., Brown, I., et al. (2017). Adaptation of Candida albicans to environmental pH induces cell wall remodelling and enhances innate immune recognition. PLoS Pathog. 13:e1006403. doi: 10.1371/journal.ppat.1006403
Silva, J. V., Freitas, M. J., Felgueiras, J., and Fardilha, M. (2015). The power of the yeast two-hybrid system in the identification of novel drug targets: building and modulating PPP1 interactomes. Expert Rev. Proteom. 12, 147–158. doi: 10.1586/14789450.2015.1024226
Singh, R. P., Prasad, H. K., Sinha, I., Agarwal, N., and Natarajan, K. (2011). Cap2-HAP complex is a critical transcriptional regulator that has dual but contrasting roles in regulation of iron homeostasis in Candida albicans. J. Biol. Chem. 286, 25154–25170. doi: 10.1074/jbc.M111.233569
Singh, S. D., Robbins, N., Zaas, A. K., Schell, W. A., Perfect, J. R., and Cowen, L. E. (2009). Hsp90 governs echinocandin resistance in the pathogenic yeast Candida albicans via calcineurin. PLoS Pathog. 5:e1000532. doi: 10.1371/journal.ppat.1000532
Sinha, I., Wang, Y. M., Philp, R., Li, C. R., Yap, W. H., and Wang, Y. (2007). Cyclin-dependent kinases control septin phosphorylation in Candida albicans hyphal development. Dev. Cell 13, 421–432. doi: 10.1016/j.devcel.2007.06.011
Sinha, K., and Rule, G. S. (2017). The structure of thymidylate kinase from Candida albicans reveals a unique structural element. Biochemistry 56, 4360–4370. doi: 10.1021/acs.biochem.7b00498
Skrzypek, M. S., Binkley, J., Binkley, G., Miyasato, S. R., Simison, M., and Sherlock, G. (2017). The Candida genome database (CGD): incorporation of Assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res. 45, D592–D596. doi: 10.1093/nar/gkw924
Skrzypek, M. S., Binkley, J., and Sherlock, G. (2018). Using the Candida genome database. Methods Mol. Biol. 1757, 31–47. doi: 10.1007/978-1-4939-7737-6
Smith, D. A., Morgan, B. A., and Quinn, J. (2010). Stress signalling to fungal stress-activated protein kinase pathways. FEMS Microbiol. Lett. 306, 1–8. doi: 10.1111/j.1574-6968.2010.01937.x
Spellberg, B. (2008). Novel insights into disseminated candidiasis: pathogenesis research and clinical experience converge. PLoS Pathog. 4:e38. doi: 10.1371/journal.ppat.0040038
Stynen, B., Tournu, H., Tavernier, J., and Van Dijck, P. (2012). Diversity in genetic in vivo methods for protein-protein interaction studies: from the yeast two-hybrid system to the mammalian split-luciferase system. Microbiol. Mol. Biol. Rev. 76, 331–382. doi: 10.1128/MMBR.05021-11
Stynen, B., Van Dijck, P., and Tournu, H. (2010). A CUG codon adapted two-hybrid system for the pathogenic fungus Candida albicans. Nucleic Acids Res. 38:e184. doi: 10.1093/nar/gkq725
Su, Z., Li, H., Li, Y., and Ni, F. (2007). Inhibition of the pathogenically related morphologic transition in Candida albicans by disrupting Cdc42 binding to its effectors. Chem. Biol. 14, 1273–1282. doi: 10.1016/j.chembiol.2007.10.008
Subotić, A., Swinnen, E., Demuyser, L., De Keersmaecker, H., Mizuno, H., Tournu, H., et al. (2017). A bimolecular fluorescence complementation tool for identification of protein-protein interactions in Candida albicans. G3 7, 3509–3520. doi: 10.1534/g3.117.300149
Sudbery, P. E. (2011). Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 9, 737–748. doi: 10.1038/nrmicro2636
Sun, X., Hong, P., Kulkarni, M., Kwon, Y., and Perrimon, N. (2013). PPIRank - an advanced method for ranking protein-protein interations in TAP/MS data. Proteome Sci. 11:S16. doi: 10.1186/1477-5956-11-S1-S16
Szklarczyk, D., Morris, J. H., Cook, H., Kuhn, M., Wyder, S., Simonovic, M., et al. (2017). The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368. doi: 10.1093/nar/gkw937
Taylor, L. H., Latham, S. M., and Woolhouse, M. E. (2001). Risk factors for human disease emergence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356, 983–989. doi: 10.1098/rstb.2001.0888
Tebbji, F., Chen, Y., Richard Albert, J., Gunsalus, K. T., Kumamoto, C. A., Nantel, A., et al. (2014). A functional portrait of Med7 and the mediator complex in Candida albicans. PLoS Genet. 10:e1004770. doi: 10.1371/journal.pgen.1004770
Tebbji, F., Chen, Y., Sellam, A., and Whiteway, M. (2017). The genomic landscape of the fungus-specific SWI/SNF complex subunit, Snf6, in Candida albicans. mSphere 2:e00497-17. doi: 10.1128/mSphere.00497-17
Tonthat, N. K., Juvvadi, P. R., Zhang, H., Lee, S. C., Venters, R., Spicer, L., et al. (2016). Structures of pathogenic fungal FKBP12s reveal possible self-catalysis function. mBio 7:e00492-16. doi: 10.1128/mBio.00492-16
Tsai, P.-W., Chen, Y.-T., Yang, C.-Y., Chen, H.-F., Tan, T.-S., Lin, T.-W., et al. (2014). The role of Mss11 in Candida albicans biofilm formation. Mol. Genet. Genomics 289, 807–819. doi: 10.1007/s00438-014-0846-0
Tseng, T. L., Lai, W. C., Jian, T., Li, C., Sun, H. F., Way, T. D., et al. (2010). Affinity purification of Candida albicans CaCdc4-associated proteins reveals the presence of novel proteins involved in morphogenesis. Biochem. Biophys. Res. Commun. 395, 152–157. doi: 10.1016/j.bbrc.2010.03.162
Tuch, B. B., Galgoczy, D. J., Hernday, A. D., Li, H., and Johnson, A. D. (2008). The evolution of combinatorial gene regulation in fungi. PLoS Biol. 6:e38. doi: 10.1371/journal.pbio.0060038
Uetz, P. (2002). Two-hybrid arrays. Curr. Opin. Chem. Biol. 6, 57–62. doi: 10.1016/s1367-5931(01)00288-5
Uetz, P., Giot, L., Cagney, G., Mansfield, T. A., Judson, R. S., Knight, J. R., et al. (2000). A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627. doi: 10.1038/35001009
Umeyama, T., Nagai, Y., Niimi, M., and Uehara, Y. (2002). Construction of FLAG tagging vectors for Candida albicans. Yeast 19, 611–618. doi: 10.1002/yea.863
Uppuluri, P., Khan, A., and Edwards, J. E. (2017). “Current trends in Candidiasis,” in Candida albicans: Cellular and Molecular Biology, ed. R. Prasad, (Cham: Springer International Publishing), 5–23. doi: 10.1007/978-3-319-50409-4_2
van den Berg, B., Chembath, A., Jefferies, D., Basle, A., Khalid, S., and Rutherford, J. C. (2016). Structural basis for Mep2 ammonium transceptor activation by phosphorylation. Nat. Commun. 7:11337. doi: 10.1038/ncomms11337
van Wijlick, L., Swidergall, M., Brandt, P., and Ernst, J. F. (2016). Candida albicans responds to glycostructure damage by Ace2-mediated feedback regulation of Cek1 signaling. Mol. Microbiol. 102, 827–849. doi: 10.1111/mmi.13494
von Mering, C., Krause, R., Snel, B., Cornell, M., Oliver, S. G., Fields, S., et al. (2002). Comparative assessment of large-scale data sets of protein–protein interactions. Nature 417, 399–403. doi: 10.1038/nature750
Wang, H., Gao, J., Wong, A. H., Hu, K., Li, W., Wang, Y., et al. (2013). Rfa2 is specifically dephosphorylated by Pph3 in Candida albicans. Biochem. J. 449, 673–681. doi: 10.1042/BJ20120952
Wang, H., Huang, Z. X., Au Yong, J. Y., Zou, H., Zeng, G., Gao, J., et al. (2016). CDK phosphorylates the polarisome scaffold Spa2 to maintain its localization at the site of cell growth. Mol. Microbiol. 101, 250–264. doi: 10.1111/mmi.13386
Wang, Q., Parrish, A. R., and Wang, L. (2009). Expanding the genetic code for biological studies. Chem. Biol. 16, 323–336. doi: 10.1016/j.chembiol.2009.03.001
Wang, Y., Cui, T., Zhang, C., Yang, M., Huang, Y., Li, W., et al. (2010a). Global protein-protein interaction network in the human pathogen Mycobacterium tuberculosis H37Rv. J. Proteome Res. 9, 6665–6677. doi: 10.1021/pr100808n
Wang, Y., Zou, H., Fang, H. M., and Zhu, Y. (2010b). Linking cellular actin status with cAMP signaling in Candida albicans. Virulence 1, 202–205. doi: 10.4161/viru.1.3.11836
Wang, Y.-C., Tsai, I. C., Lin, C., Hsieh, W.-P., Lan, C.-Y., Chuang, Y.-J., et al. (2014). Essential functional modules for pathogenic and defensive mechanisms in Candida albicans infections. BioMed. Res. Int. 2014:136130. doi: 10.1155/2014/136130
Wangsanut, T., Tobin, J. M., and Rolfes, R. J. (2018). Functional mapping of transcription factor Grf10 that regulates adenine-responsive and filamentation genes in Candida albicans. mSphere 3:e00467-18. doi: 10.1128/mSphere.00467-18
Wangsanut, T., Tobin, J. M., and Rolfes, R. J. (2019). Functional mapping of transcription factor Grf10 that regulates adenine-responsive and filamentation genes in Candida albicans. mSphere 4:e00094-19. doi: 10.1128/mSphere.00094-19
Weber, Y., Swoboda, R. K., and Ernst, J. F. (2002). Sec20p-interacting proteins (Tip20p, Ufe1p) in the retrograde secretory pathway of the fungal pathogen Candida albicans. Mol. Genet. Genomics 268, 468–476. doi: 10.1007/s00438-002-0777-z
Whitlow, M., Howard, A. J., Stewart, D., Hardman, K. D., Kuyper, L. F., Baccanari, D. P., et al. (1997). X-ray crystallographic studies of Candida albicans dihydrofolate reductase: high resolution structures of the holoenzyme and an inhibited ternary complex. J. Biol. Chem. 272:30289. doi: 10.1074/jbc.272.48.30289
Wu, C., Jansen, G., Zhang, J., Thomas, D. Y., and Whiteway, M. (2006). Adaptor protein Ste50p links the Ste11p MEKK to the HOG pathway through plasma membrane association. Genes Dev. 20, 734–746. doi: 10.1101/gad.1375706
Xie, J. L., Bohovych, I., Wong, E. O. Y., Lambert, J. P., Gingras, A. C., Khalimonchuk, O., et al. (2017). Ydj1 governs fungal morphogenesis and stress response, and facilitates mitochondrial protein import via Mas1 and Mas2. Microb. Cell 4, 342–361. doi: 10.15698/mic2017.10.594
Xing, S., Wallmeroth, N., Berendzen, K. W., and Grefen, C. (2016). Techniques for the analysis of protein-protein interactions in vivo. Plant Physiol. 171, 727–758. doi: 10.1104/pp.16.00470
Xu, W., and Mitchell, A. P. (2001). Yeast PalA/AIP1/Alix homolog Rim20p associates with a PEST-like region and is required for its proteolytic cleavage. J. Bacteriol. 183, 6917–6923. doi: 10.1128/JB.183.23.6917-6923.2001
Xu, X., Song, Y., Li, Y., Chang, J., Zhang, H., and An, L. (2010). The tandem affinity purification method: an efficient system for protein complex purification and protein interaction identification. Protein Expr. Purif. 72, 149–156. doi: 10.1016/j.pep.2010.04.009
Yamada-Okabe, T., Mio, T., Matsui, M., Kashima, Y., Arisawa, M., and Yamada-Okabe, H. (1998). Isolation and characterization of the Candida albicans gene for mRNA 5′-triphosphatase: association of mRNA 5′-triphosphatase and mRNA 5′-guanylyltransferase activities is essential for the function of mRNA 5′-capping enzyme in vivo. FEBS Lett. 435, 49–54. doi: 10.1016/S0014-5793(98)01037-0
Yan, L., Sun, S., Wang, W., Shi, J., Hu, X., Wang, S., et al. (2015). Structures of the yeast dynamin-like GTPase Sey1p provide insight into homotypic ER fusion. J. Cell Biol. 210, 961–972. doi: 10.1083/jcb.201502078
Yang, S. L., Zeng, G., Chan, F. Y., Wang, Y. M., Yang, D., and Wang, Y. (2018). Sac7 and Rho1 regulate the white-to-opaque switching in Candida albicans. Sci. Rep. 8:875. doi: 10.1038/s41598-018-19246-9
Yao, G., Wan, J., Liu, Q., Mu, C., Wang, Y., and Sang, J. (2017). Characterization of Pph3-mediated dephosphorylation of Rad53 during methyl methanesulfonate-induced DNA damage repair in Candida albicans. Biochem. J. 474, 1293–1306. doi: 10.1042/BCJ20160889
Yao, G., Wan, J., Mu, C., Liu, Q., Wang, Y., and Sang, J. (2016). Sds22 participates in Glc7 mediated Rad53 dephosphorylation in MMS-induced DNA damage in Candida albicans. Fungal Genet. Biol. 93, 50–61. doi: 10.1016/j.fgb.2016.06.003
Yi, S., Sahni, N., Daniels, K. J., Lu, K. L., Huang, G., Garnaas, A. M., et al. (2011). Utilization of the mating scaffold protein in the evolution of a new signal transduction pathway for biofilm development. mBio 2:e00237-10. doi: 10.1128/mBio.00237-10
Yu, H., Braun, P., Yildirim, M. A., Lemmens, I., Venkatesan, K., Sahalie, J., et al. (2008). High Quality binary protein interaction map of the yeast interactome network. Science 322, 104–110. doi: 10.1126/science.1158684
Zeng, G., Wang, Y.-M., and Wang, Y. (2012). Cdc28–Cln3 phosphorylation of Sla1 regulates actin patch dynamics in different modes of fungal growth. Mol. Biol. Cell 23, 3485–3497. doi: 10.1091/mbc.E12-03-0231
Zhang, A., Petrov, K. O., Hyun, E. R., Liu, Z., Gerber, S. A., and Myers, L. C. (2012). The Tlo proteins are stoichiometric components of Candida albicans mediator anchored via the Med3 subunit. Eukaryot. Cell 11, 874–884. doi: 10.1128/EC.00095-12
Zheng, X., Wang, Y., and Wang, Y. (2004). Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 23, 1845–1856. doi: 10.1038/sj.emboj.7600195
Zheng, X.-D., Lee, R. T. H., Wang, Y.-M., Lin, Q.-S., and Wang, Y. (2007). Phosphorylation of Rga2, a Cdc42 GAP, by CDK/Hgc1 is crucial for Candida albicans hyphal growth. EMBO J. 26, 3760–3769. doi: 10.1038/sj.emboj.7601814
Zhou, J., Lin, J., Zhou, C., Deng, X., and Xia, B. (2011). An improved bimolecular fluorescence complementation tool based on superfolder green fluorescent protein. Acta Biochim. Biophys. Sin. 43, 239–244. doi: 10.1093/abbs/gmq128
Znaidi, S., Nesseir, A., Chauvel, M., Rossignol, T., and d’Enfert, C. (2013). A comprehensive functional portrait of two heat shock factor-type transcriptional regulators involved in Candida albicans morphogenesis and virulence. PLoS Pathog. 9:e1003519. doi: 10.1371/journal.ppat.1003519
Zordan, R. E., Miller, M. G., Galgoczy, D. J., Tuch, B. B., and Johnson, A. D. (2007). Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 5:e256. doi: 10.1371/journal.pbio.0050256
Keywords: Candida albicans, protein-protein interactions, Candida two-hybrid system, yeast two-hybrid system, BioGRID, S. cerevisiae
Citation: Schoeters F and Van Dijck P (2019) Protein-Protein Interactions in Candida albicans. Front. Microbiol. 10:1792. doi: 10.3389/fmicb.2019.01792
Received: 24 June 2019; Accepted: 19 July 2019;
Published: 07 August 2019.
Edited by:
James Bernard Konopka, Stony Brook University, United StatesReviewed by:
Yue Wang, Institute of Molecular and Cell Biology (A*STAR), SingaporeMalcolm Whiteway, Concordia University, Canada
Copyright © 2019 Schoeters and Van Dijck. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patrick Van Dijck, UGF0cmljay52YW5kaWpja0BrdWxldXZlbi52aWIuYmU=
 Floris Schoeters
Floris Schoeters Patrick Van Dijck
Patrick Van Dijck