- 1Key Laboratory of Plant Protection Resources and Pest Management of Ministry of Education, Research and Development Center of Biorational Pesticides, College of Plant Protection, Northwest A&F University, Yangling, China
- 2Plant Quarantine and Protection Bureau of Zhumadian, Zhumadian, China
This study was conducted to purify and identify metabolites of antimicrobial activity against phytopathogens from Xenorhabdus nematophila YL001. Three dipeptide compounds were purified from its cell-free cultural broth and identified as (±)-nematophin, cyclo (L-Pro-Gly), and N, N′-dimethyl-cyclo (L-Phe-L-Leu). Nematophin demonstrated a wider antifungal spectrum than the other two compounds. It also exhibited strong inhibitory effects on mycelial growth of Rhizoctonia solani and Phytophthora infestans with EC50 values of 40.00 and 51.25 μg/ml, respectively. Its (S)-configuration structure [(+)-nematophin] was also synthesized and exhibited higher antimicrobial activity than the enantiomeric mixture. The detached leaf assay revealed that nematophin possessed significant preventive and curative efficacy against R. solani on broad bean leaves showing corresponding control efficacies of 93.01 and 94.93% at 1,000 μg/ml, comparable to those of a chemical fungicide (carbendazim) at 500 μg/ml. Additionally, the pot experiments indicated that nematophin could effectively inhibit the disease extension on rice and broad bean plants caused by R. solani. Nematophin also exerted some adverse influences on the sclerotial development of R. solani by dramatically suppressing their formation and maturation at 40.00 μg/ml, as well as their germination at 15.00 μg/ml. Morphological and ultrastructural observations showed that the hyphae of R. solani became twisted, shriveled, and deformed at the growing points after exposure to nematophin at 40.00 μg/ml, and that the subcellular fractions also became abnormal concurrently, especially the mitochondrial structure. These results indicate that nematophin has great potential to be used as a bio-pesticide in agricultural production.
Introduction
Xenorhabdus nematophila, a well-studied genus of entomopathogenic bacteria, lives in symbiosis with Steinernema nematodes. Previous research has revealed that X. nematophila is a potent producer of natural compounds with versatile biological activities including antifungal, antibacterial, and antimalarial activities (Crawford et al., 2012; Challinor and Bode, 2015; Engel et al., 2017; Dreyer et al., 2018). Many of these metabolites belong to peptides such as xenocoumacins, nematophin, xenorhabdins, rhabduscins, and xenorxides (Li et al., 1997a,b; Ji et al., 2004; Böszörményi et al., 2009; Helge, 2011; Yang et al., 2011). Such peptides are of diverse structures and resultant multiple bioactivities including antimicrobial activity against plant pathogens (Li et al., 1997a; Yang et al., 2011; Dreyer et al., 2018). Besides, whole genome programs have shown that approximately 7.5% of genomic genes encode proteins involving the secondary metabolism biosynthesis in the strain X. nematophila ATCC19061, most of which encoded molecules are cryptic (Chaston et al., 2011). Given that the culture supernatants of many Xenorhabdus species exhibited potent antimicrobial activity against many plant pathogens (Fang et al., 2014; Hazir et al., 2016; Sharma et al., 2016), it is feasible to discover novel secondary metabolites of antifungal activity from X. nematophila as bio-pesticides for agricultural production.
Plant diseases caused by fungi are destructive attacks on crop production, generating enormous economic losses worldwide. Rhizoctonia solani is a necrotrophic fungal pathogen that infects more than 200 plant species worldwide, resulting in severe losses in crop yield (Lewis and Lumsden, 2001; Zheng et al., 2013; Mayo et al., 2015; Zhou et al., 2016). It can also produce sclerotia, a special resting structure surviving in soil for many years, which is the main source of infection in the disease cycle (Townsend and Willetts, 1954; Feng et al., 2016; Moni et al., 2016). Currently, the most common strategy for disease management is the application of chemical fungicides. Their overuse, however, has posed serious threats to human health, environmental safety and ecological balance (Nicolopoulou-Stamati et al., 2016; Rohr et al., 2017). Bio-pesticides are effective alternatives to the chemical fungicides to overcome their adverse impacts (Kumar, 2018; Lengai and Muthomi, 2018). Moreover, searching for bioactive compounds from microbes for control of the difficult-to-control plant pathogens is becoming a promising lead in the development of novel antimicrobial agents for agriculture production (Kim and Hwang, 2007; Mnif and Ghribi, 2015; Kanagaraj Muthu-Pandian et al., 2018).
Previous studies revealed that the cell-free culture of X. nematophila YL001 exhibited potent antimicrobial activity against some plant pathogens in vitro and in vivo, including P. infestans and B. cinerea (Guo et al., 2017; Zhang et al., 2018). However, no purified bioactive compounds were obtained from this bacterial strain, although many metabolites of novel structures and strong antimicrobial activity were purified and identified from strains of genus Xenorhabdus (Dreyer et al., 2018) including odilorhabdins (Pantel et al., 2018), cabanillasin (Houard et al., 2013), and bicornutin-A (Böszörményi et al., 2009). These studies mainly focused on their medical applications as antimicrobial agents. Currently, there is still limited information available about the antimicrobial potency of the metabolites of X. nematophila against phytopathogens (Li et al., 1997a; Ji et al., 2004; Lang et al., 2008; Gualtieri et al., 2009; Dreyer et al., 2018), especially the in vivo antimicrobial data (Yang et al., 2011; Zhou et al., 2016). In this paper, we tried to isolate and identify metabolites of antimicrobial potency against agricultural pathogenic fungi and oomycetes from X. nematophila YL001 and determine their antimicrobial activity in vitro. Subsequently, we evaluated the in vivo control efficacy of (±)-nematophin against R. solani. The effects of nematophin on the sclerotial development of R. solani, as well as on the hyphal morphology and ultrastructure, were also observed in this paper. The results revealed that N,N′-dimethyl-cyclo (L-Phe-L-Leu) possessed efficient antifungal activity against Exserohilum turcicum, and that nematophin could be used as a potential candidate for bio-pesticides.
Materials and Methods
Strains and Growth Conditions
X. nematophila YL001 was isolated from its nematode symbiont, Steinernema sp. YL001, which was obtained from the soybean rhizosphere soil of Yangling, China (E: 107°59′—108°08′; N: 34°14′—34°20′; Soil type: Lou soil; average annual temperature: 12.9°C; average annual sunshine hours: 2163.8 h; average annual rainfall: 635.1 mm). Its morphological and biochemical characterization was performed in our laboratory with results listed in Supplementary Tables S1, S2. Additionally, its species was also identified as X. nematophila by 16S rDNA amplification (GenBank number: EU124381; Supplementary Figure S1A) and the genome sequence (GenBank number: NZ_CP032329; Supplementary Figure S1B).
The tested strains of eight phytopathogenic fungi (R. solani, E. turcicum, Fusarium graminearum, Verticillium dahlia, Botrytis cinerea, Sclerotinia sclerotiorum, Alternaria alternata, and Gaeumannomyces graminis) and two fungal-like oomycetes (Phytophthora infestans and Phytophthora capsici) were collected from fields in different regions of China which were identified and preserved in the laboratory of the Research and Development Center of Biorational Pesticides, Northwest A&F University, Yangling, Shaanxi, China. The information of their collection places and host plants can be seen in Supplementary Table S3. Each fungal or oomycete strain was activated by growing on the petri dishes containing potato-dextrose agar (PDA) culture medium for 2 or 3 days at 26°C before use.
Microbial Fermentation
A single colony of cells of X. nematophila YL001 was cultured in a 500-ml flask containing 200 ml of fresh Luria-Bertani medium (LB: 1.0% Bacto tryptone, 0.5% yeast extract, and 1% NaCl in water; pH 7.2) for 12 h at 28°C with shaking at 180 rev/min. Then, 3.5 L of the culture broth (OD600 = 0.8) was transferred as a seed into a 70-L fermenter (Eastbio, China) containing 35 L of TSB medium (per liter of distilled water: peptone from soymeal 3 g, peptone from casein 17 g, NaCl 5 g, glucose 2.5 g, and K2HPO4 2.5 g; pH 7.2) and the system was incubated at 28°C with continuous agitation of 160 rpm and aeration of 0.25 v/v/min. After 48-h culture, the broth of X. nematophila YL001 was centrifuged (10,000 g, 20 min, 4°C) to remove the bacterial cells. Then, the cell-free broth was concentrated to 7 L on a rotary evaporator at 35°C and stored at 4°C for use.
Isolation and Characterization of Dipeptide Compounds
The concentrated cell-free broth (7 L) was extracted with petroleum ether (4 × 7 L), chloroform (4 × 7 L), and ethyl acetate (4 × 7 L), consecutively. The petroleum ether and ethyl acetate layers were dried over anhydrous sodium sulfate, filtered, and evaporated by a rotary evaporator to give their respective residues that were subjected to further purification.
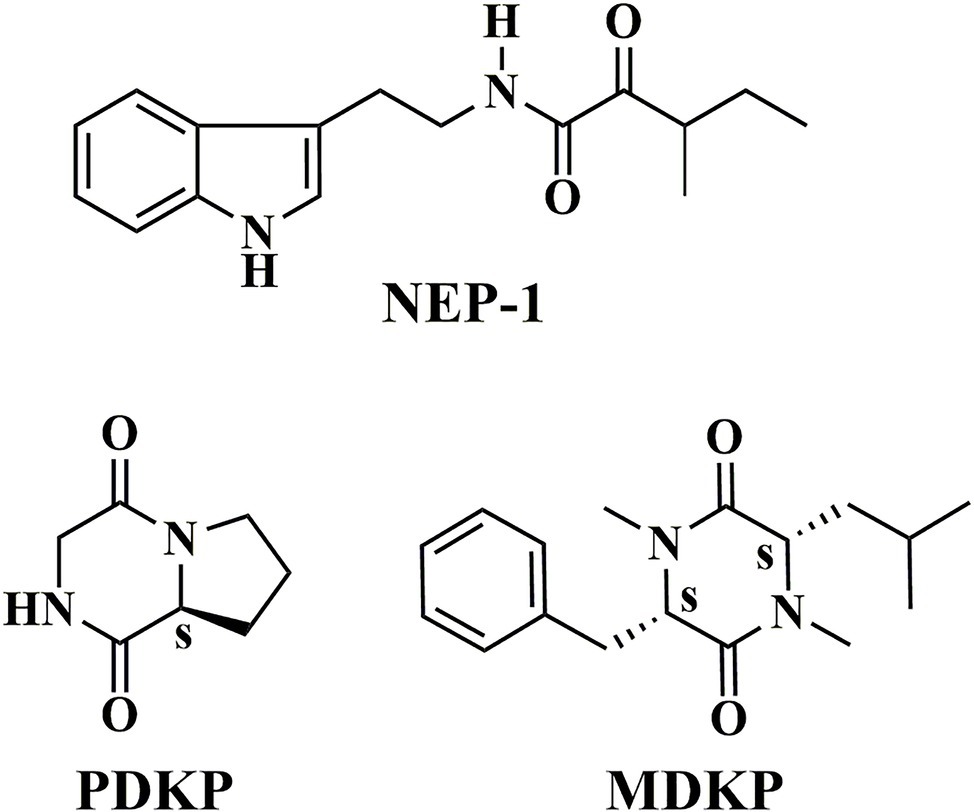
The extract of petroleum ether (2.8 g) was then loaded onto a silica gel (90 g, 200–300 mesh, Qingdao Haiyang Chemical Co., Ltd., China) and eluted with a series of petroleum ether/ethyl acetate mixtures (1 L, v/v, 100:0, 75:1, 50:1, 30:1, 25:1, 15:1, 10:1, 5:1, 3:1, 2:1, 1:1, 0:1), sequentially. Each fraction was monitored by thin-layer chromatography (TLC). NEP-1 was obtained from the eluent solutions of 25:1, 15:1, and 10:1, while PDKP was obtained from the eluent solutions of 5:1 and 3:1 (Figure 1).
The extract of ethyl acetate (4.1 g) was also purified by column chromatography on a silica gel (60 g, 200–300 mesh) using a series of chloroform/methanol mixtures (1 L, v/v, 50:1, 40:1, 20:1, 10:1, 5:1, 0:1) as the eluate. Each fraction was monitored by thin-layer chromatography (TLC). MDKP was obtained from the eluent solution of 100% methanol (Figure 1).
Structural identification of the three metabolites was performed on the basis of spectroscopic analysis. The mass spectra (MS) of the three compounds were obtained by a Mariner Mass 5,304 instrument (California, USA) or an AB Sciex TripleTOF 5,600 + System (Framingham, MA). 1H and 13C nuclear magnetic resonance (NMR) data were acquired on a Bruker AVANCE III 500 spectrometer (Rheinstetten, Germany) with tetramethylsilane as the internal standard. The optical rotation data were recorded on an Anton Paar MCP 300 polarimeter (Graz, Austria). The circular dichroism (CD) spectra were collected on an Applied Photophysics Chirascan spectropolarimeter (Leatherhead, UK).
NEP-1: 1H NMR (500 MHz, CDCl3) δ 8.08 (bs, 1H), 7.61 (dd, J = 7.8, 1.0 Hz, 1H), 7.38 (d, J = 8.1 Hz, 1H), 7.14 (td, J = 8.0, 7.0, 1.0 Hz, 1H), 7.14 (td, J = 8.0, 7.0, 1.0 Hz, 1H), 7.06 (bs, 1H), 7.04 (d, J = 2.3 Hz, 1H), 3.65 (q, J = 6.7 Hz, 2H), 3.50 (h, J = 6.9 Hz, 1H), 3.03 (t, J = 6.9 Hz, 2H), 1.80–1.67 (m, 1H), 1.50–1.34 (m, 1H), 1.09 (d, J = 7.0 Hz, 3H), 0.89 (t, J = 7.4 Hz, 3H); 13C NMR (125 MHz, CDCl3) δ 202.48, 160.18, 136.58, 127.29, 122.45, 122.14, 119.72, 118.78, 112.68, 111.40, 40.51, 39.66, 25.58, 25.31, 15.29, 11.62; ESI-MS (m/z): [M − H]− calculated for C16H19N2O2 271.14, found 271.10; HRESI-MS (m/z): [M + H]+ calculated for C16H21N2O2 273.1603, found 273.1608; [M + Na]+ calculated for C16H20N2NaO2 295.1422, found 295.1429.
PDKP: : −185.95 (c 0.83, EtOH); 1H NMR (500 MHz, D2O) δ 4.36 (t, J = 6.6 Hz, 1H), 4.21 (d, J = 17.3 Hz, 1H), 3.92 (d, J = 17.3 Hz, 1H), 3.65–3.48 (m, 2H), 2.47–2.29 (m, 1H), 2.47–2.29 (m, 1H), 2.05–1.86 (m, 2H); 13C NMR (125 MHz, D2O) δ 171.88, 165.96, 58.66, 45.52, 45.40, 27.84, 21.76; ESI-MS (m/z): [M + H]+ calculated for C7H11N2O2 155.08, found 154.97; [M + Na]+ calculated for C7H10N2O2Na 177.06, found 176.97; HRESI-MS (m/z): [M + Na]+ calculated for C7H10N2O2Na 177.0640, found 177.0642.
MDKP: : −7.1 (c 0.38, CHCl3); 1H NMR (500 MHz, CDCl3) δ 7.38–7.25 (m, 1H), 7.17–7.12 (m, 1H), 4.21 (t, J = 4.7 Hz, 1H), 3.65 (dd, J = 9.2, 4.2 Hz, 1H), 3.33 (dd, J = 14.0, 4.9 Hz, 1H), 3.20 (dd, J = 14.0, 4.5 Hz, 1H), 2.98 (s, 3H), 2.90 (s, 3H), 1.75–1.68 (m, 1H), 0.87 (d, J = 6.5 Hz, 3H), 0.75 (d, J = 6.7 Hz, 3H), 0.70 (ddd, J = 13.7, 9.3, 4.2 Hz, 1H), 0.32 (ddd, J = 14.2, 9.2, 5.2 Hz, 1H); 13C NMR (125 MHz, CDCl3) δ 166.57, 165.19, 135.73, 130.02, 128.88, 127.60, 64.02, 60.06, 42.46, 37.72, 32.81, 32.55, 25.14, 22.71, 21.48; HRESI-MS (m/z): [M + H]+ calculated for C17H25N2O2 289.1916, found 289.1907.
Synthesis of an Enantiomeric Mixture of Nematophin (NEP-2) and Its (S)-Configuration Structure (NEP-3)
(±)-Nematophin (NEP-2) was readily synthesized by one-pot amidation of (±)-2-keto-3-methylvaleric acid [(±)-KMVA] and tryptamine using ethyl-dimethylaminopropyl-carbodiimide hydrochloride (EDCl) as the condensation agent and N, N-dimethyl-4-aminopyridine (DMAP) as the catalyzer (Supplementary Scheme S1).
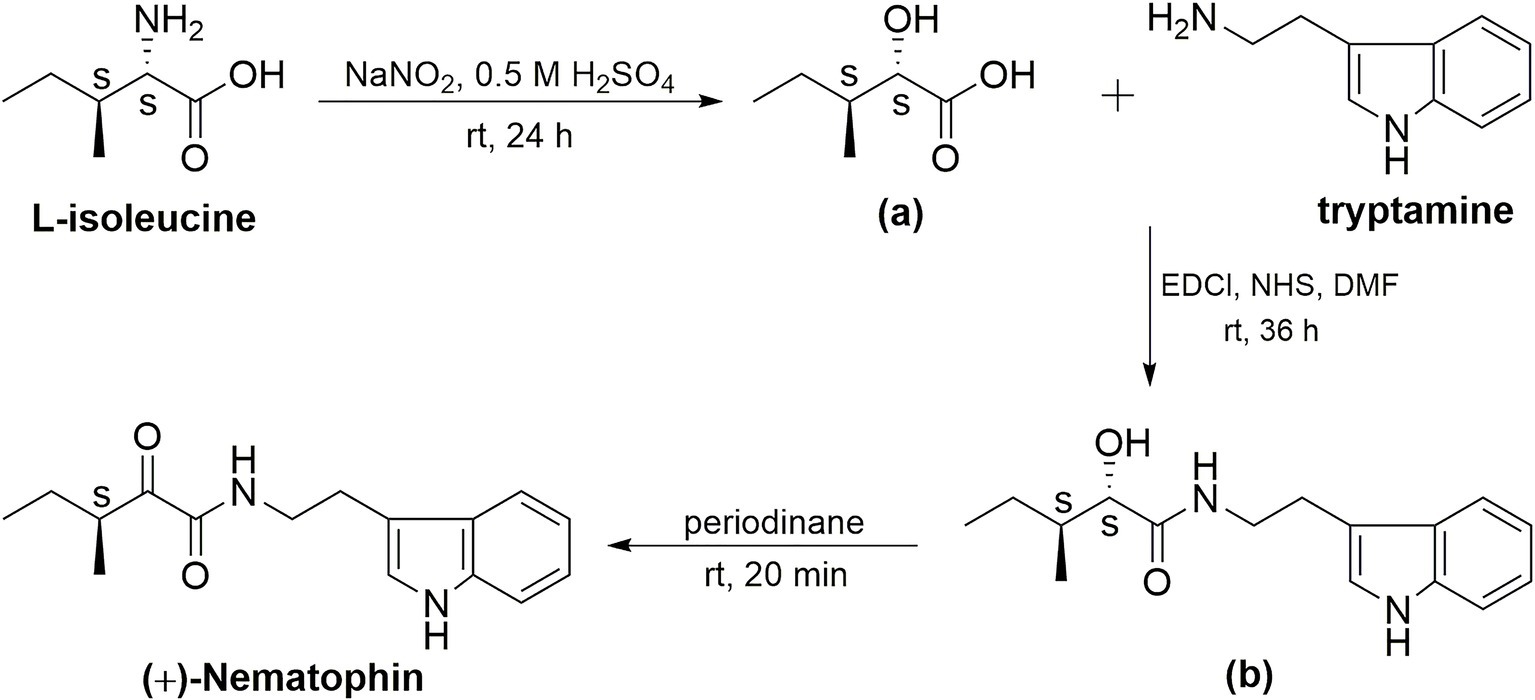
(+)-Nematophin (NEP-3) was also prepared by three reaction steps (Figure 2) according to a previously reported method with some modifications (Paik et al., 2003). L-Isoleucine [(2S, 3S)-isoleucine] was used as the chiral template material to prepare (2S, 3S)-hydroxypentanoic acid (a) by diazotization and hydrolysis in 0.5 M H2SO4 solution. Then, coupling of compound a with tryptamine was performed by a one-pot amidation method (EDCl/NHS) to give (2′S, 3′S)-N-(indol-3-ylethyl)-2′-hydroxy-3′-methylpentanamide (b). Finally, the title compound (3′S)-N-[2-(1H-Indol-3-yl) ethyl]-3-methyl-2-oxopentanamide [(+)-nematophin] was obtained by oxidation of b with Dess-Martin reagent (periodinane) in dichloromethane.
All the chemicals used were purchased from Aladdin Co. Ltd. (Beijing, China). The detailed synthetic procedures can be seen in Supplementary Material. The synthetic intermediates and target compounds were characterized identically to the reported structures by 1H NMR, 13C NMR, and ESI-MS (Supplementary Figures S16–S28).
Effects of Three Isolated Compounds on Mycelial Growth of the Phytopathogenic Fungi and Oomycetes in vitro
The antimicrobial activity of three isolated compounds against the mycelial growth of 10 plant pathogens was performed as described in previous research with slight modifications (Zhang et al., 2018). Briefly, the three compounds were dissolved in dimethyl sulfoxide (DMSO) to prepare their respective stock solutions with a concentration of 10,000 μg/ml. A stock solution (150 μl) was mixed with 15 ml of molten PDA medium at a low temperature (about 40°C) to make a final drug concentration of 100 μg/ml (1% DMSO), and then the mixture was immediately poured into a petri dish (60-mm diameter) to form a plate of 2–3 mm thickness. PDA supplemented with the same volume of DMSO (1%) served as the blank control. Carbendazim (MBC) and mancozeb (MZ) at a concentration of 0.5 μg/ml were used as positive controls. After cooling, a 5-mm diameter mycelial disc was placed in the center of each plate with the inoculum side down. The plates were incubated at 28°C in dark until the mycelia reached the edges of the control dishes. Each experiment was repeated three times. The mycelia growth inhibition rate was calculated using the following formula:
where Dc and Dt represent the mycelial growth diameter of the control and treatment group, respectively. The diameter of the plug is 0.5 cm.
The antimicrobial activity of NEP-2 and -3 against five plant pathogens (R. solani, S. sclerotiorum, P. infestans, F. graminearum, and P. capsici) was also evaluated using the method as described above. The final concentration of NEP-2 or -3 in each test medium was 100 μg/ml and PDA plates supplemented with 1% DMSO were used as the controls.
The half-maximal effective concentration (effective dose for 50% inhibition, EC50) was derived from analysis of the concentration-dependent inhibition rates. PDA plates with serial concentrations of NEP-1 (2.5, 5, 10, 20, 40, 60, 80, and 100 μg/ml) were used for test by the method described above. All experiments were independently performed three times under the same conditions.
Antimicrobial Activity Assay of NEP-1 Against R. solani and P. infestans by a Detached Leaf Method
The efficacies of NEP-1 on broad bean leaves infected with R. solani and potato leaves infected with P. infestans were evaluated according to the studies (Arfaoui et al., 2018) with some modifications. Broad bean (Vicia faba L.) was grown in a growth chamber (25 ± 1°C, 75 ± 10% RH, and a 12:12 LD photoperiod) for 21 days. Potato tuber was grown for 30 days under the same conditions. NEP-1 solutions of 500 and 1,000 μg/ml were prepared by dissolving it in an aqueous solution (sterile water composed of 0.5% tween 20, v/v). MBC solution of 500 μg/ml in the same mixture was used as the positive control against R. solani and MZ solution of 500 μg/ml against P. infestans. Sterile water containing 0.5% tween 20 was used as the blank control. All experiments were arranged in a plant growth chamber (25 ± 1°C, 75 ± 10% RH, and a 12:12 LD photoperiod) with nine leaves per treatment and were repeated three times.
For the protective activity assay, nine leaves of broad bean or potato were as a group and sprayed with a pre-prepared sample solution (1 ml for each leaf). After incubation for 12 h, a 5-mm diameter mycelial disc was inoculated onto the center of each leaf that was wounded at the inoculation site using a sterilized needle (avoiding the main vein of the leaves) in advance. After incubation for another 2 days, the lesion area was quantified from the captured images with Image J 1.38x software1, and the control efficacy was also calculated according to the following formula:
where Ac and At represent the disease area of the blank control and treatment group, respectively. The area of the mycelial disc is 0.25 cm2.
For the curative activity assay, nine leaves of broad bean or potato were as a group and each leaf was inoculated with a diameter mycelial disc as described above. After incubation for 12 h (~3 mm lesion around each disc), nine leaves of a group were sprayed with a pre-prepared sample solution of NEP-1 (500 or 1,000 μg/ml), MBC (500 μg/ml) or MZ (500 μg/ml) (1 ml for each leaf). The subsequent operations were as described above.
Biocontrol Efficacies of NEP-1 Against R. solani on Rice and Broad Bean Plants Grown in a Greenhouse
A pot experiment was performed to assess the in vivo biocontrol efficiency of NEP-1 against R. solani. Potted rice plants (35 days after seeding) and broad bean plants (30 days after seeding) grown in a greenhouse were used as the host plants. The temperature of the greenhouse ranged from 28°C (±2°C) in the daytime to 22°C (±2°C) during nighttime, and the relative humidity was controlled at 85% (±5%). The procedures of fungal inoculum, pathogen inoculation, and disease evaluation were according to the reported literature (Park et al., 2008; Wu et al., 2019) with some modifications. Briefly, 10 agar blocks (0.5-cm diameter) from the outer edge of a 3-day-old inoculated PDA plate were inoculated in a 250-ml Erlenmeyer flask with 100 ml of PDB (potato dextrose broth) medium. After incubation on a shaker (160 rpm) under darkness for 7 days, liquid cultured mycelia were harvested and cut into small mycelial balls (0.1 g in wet weight). A mycelial ball was placed beneath the leaf sheath and covered with aluminum foil immediately. When typical lesions appeared after 3 days, the aluminum foil was removed. Then the plants of a group were sprayed with a sample solution of NEP-1 (500 or 1,000 μg/ml), MBC (500 μg/ml), or sterile water. Three pots with more than five rice plants of each and nine pots with one broad bean plant of each were used as a group, respectively. Each experiment was performed with three replications. After 4-day incubation, the lesion length in each stem of the inoculated plants was recorded.
Effects of NEP-1 on the Sclerotial Development of R. solani
The effects of NEP-1 on sclerotial formation were performed according to a previously published procedure (Soltani et al., 2017). Briefly, serial concentrations of NEP-1 in DMSO were added to PDA medium at final concentrations of 15, 20, 30, and 40 μg/ml, respectively. PDA supplemented with DMSO (1%) served as the control. A 5-mm diameter plug of R. solani was inoculated onto the center of each plate maintained at 28°C for 6 days until sclerotia manifested in the control plates. Additionally, the dynamic developmental process of sclerotia treated with 40 μg/ml of NEP-1 was also observed at fixed time intervals of 3, 4, 6, and 12 days. All plates were photographed with a Nikon D500 camera (Tokyo, Japan), and the sclerotia phenotypes were observed by a LECIA M165 FC stereoscopic microscope (Leica-ULTRACUT, Wetzlar, Germany).
The effects of NEP-1 on the sclerotial germination were measured according to a previously described method (Kazempour, 2004; Soltani et al., 2017). Uniform and healthy sclerotia were collected from PDA plates and soaked in solutions of various concentrations of NEP-1 (15, 20, and 30 μg/ml) with 1% DMSO for 5 min. Then, the sclerotia treated with NEP-1 at a certain concentration were placed onto PDA plates (one sclerotia per plate) and maintained at 28°C to observe their germination. The sclerotia treated with 1% DMSO were used as the controls. Each concentration was tested with 12 sclerotia and each test was repeated three times.
Effects of NEP-1 on Hyphal Morphology and Ultrastructure of R. solani
Determination of the effects of NEP-1 on the hyphal morphology and ultrastructure was conducted according to the method of Soner Soylu (Soylu et al., 2010). A prepared mycelial agar disc from a 2-day-old culture was inoculated in the center of the PDA plate with 40.00 μg/ml (EC50) of NEP-1, and the plate was incubated at 28°C for 2 days in the dark. PDA plates without NEP-1 treatment were used as the controls.
For the scanning electron microscopy (SEM) observations, mycelial discs (2 mm × 4 mm × 4 mm) were fixed with 2.5% glutaraldehyde in a phosphate buffer (0.1 M, pH = 7.2) overnight at 4°C. Each sample was washed three times (20 min each) with 0.1 M phosphate buffer (pH = 7.2) to remove the excess glutaraldehyde. After fixation, the samples were dehydrated in a graded ethanol series (twice at 30, 50, 70, 80, and 90% and three times at 100%, v/v) for 20 min in each solution. After dehydration, the samples were dipped into isoamylacetate three times (20 min each) for replacement of ethanol. Finally, each sample was dried at the critical point using supercritical carbon dioxide. All samples were fixed on a holder using double-sided tape and coated with gold using an E1010 sputter coating machine (Hitachi, Tokyo, Japan) for 90 s at 9 mA. Samples were then imaged using a JSM-6360LV SEM instrument (JEOL, Tokyo, Japan).
For the transmission electron microscopy (TEM) observations, the pre-fixed mycelia were the same as described in the SEM method. After washing with a phosphate buffer (0.1 M, pH = 7.2) three times, the samples were post-fixed with 1% osmic acid for 2 h. Then, the samples were washed again with the phosphate buffer (0.1 M, pH 7.2) immediately followed by dehydration as described in the SEM method. Samples were dipped into ethanol twice (30 min each), after which the specimens were passed through a solution of epoxy resin/epoxy propane (1:1, v/v) for 1 h and embedded in epoxy media at 55°C for 48 h. Blocks were sectioned using a diamond knife with an Ultramicrotome (Leica-ULTRACUT, Wetzlar, Germany) into ultrathin sections of approximately 70 nm. The ultrathin sections were contrasted with 2% uranyl acetate and 2% lead citrate for 30 min prior to examination on a JEM-1230 TEM (Hitachi, Tokyo, Japan). At least three samples from each of the treated and control groups were examined by SEM and TEM.
Data Processing and Analysis
All datasets were analyzed by ANOVA using Statistica Software (Statsoft, Tulsa, OK, USA). The EC50 was calculated by linear regression of the log of the colony diameter versus the various NEP-1 concentrations. When the ANOVA was significant, means were separated with the least significant difference test (LSD, p < 0.05).
Results
Structure Identification
In the present study, three metabolites (Figure 1) were isolated from the fermented supernatant of X. nematophila YL001. Their structures were characterized by 1H NMR, 13C NMR, and MS spectra. According to the relevant spectral data reported, the three compounds were identified as nematophin (NEP-1, Supplementary Figures S5–S8) (Li et al., 1997a), hexahydropyrrolo[1, 2-α]pyrazine-1,4-dione (PDKP, Supplementary Figures S9–S12) (Bishay et al., 2018) and 1,4-dimethyl-3-(2-methylpropyl)-6-(phenylmethyl)-2,5-piperazinedione (MDKP, Supplementary Figures S13–S15) (Nakao et al., 2016), respectively. They are dipeptide compounds in structure. Referring to the existing optical rotation data, the absolute stereostructures of PDKP and MDKP were determined as (8αS)-hexahydropyrrolo[1, 2-α]pyrazine-1,4-dione [cyclo(L-Pro-Gly)] (Campbell et al., 2009) and (3S, 6S)-1,4-dimethyl-3-(2-methylpropyl)-6-(phenylmethyl)-2,5-piperazinedione [N, N′-dimethyl-cyclo(L-Phe-L-Leu)] (Nakao et al., 2016), respectively.
To confirm the stereo structure of NEP-1, an enantiomeric mixture of nematophin (NEP-2, Supplementary Figures S16–S19) and its (S)-configuration structure (NEP-3, Supplementary Figures S25–S28) were synthesized and characterized in this study. Their circular dichroism (CD) spectra were recorded with the results shown in Supplementary Figure S2. An obvious positive absorption band was observed from 300 to 400 nm for the NEP-3 solution. Interestingly, no absorption signals were detected in this wavelength range for both NEP-1 and NEP-2 solutions. Moreover, different from NEP-1 and NEP-2 of racemic property, NEP-3 also exhibited a specific rotation value of 31.66 (c 0.58) in CHCl3. These findings indicate that NEP-1 is an enantiomeric mixture.
Examination of Inhibitory Effects of Three Isolated Compounds on Mycelia Growth of Agricultural Pathogenic Fungi and Oomycetes in vitro
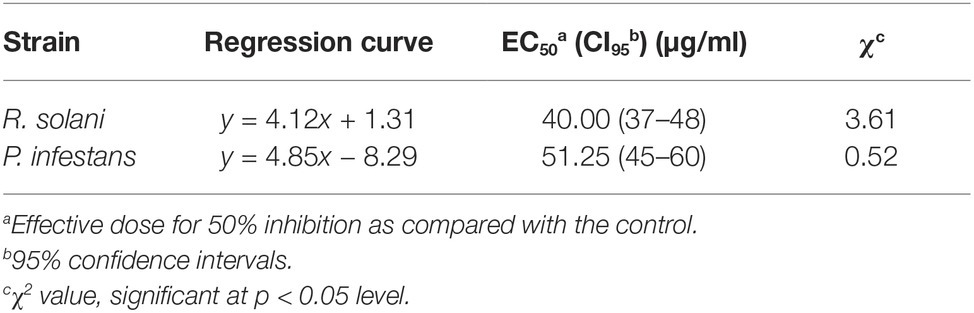
A previous study revealed that NEP-1 had strong in vitro bioactivity against a series of fungal and bacterial species (Li et al., 1997a). Piperazine-2,5-diones including PDKP and MDKP are an important class of cyclodipeptide compounds of versatile bioactivities (Borthwick, 2012). However, scarce work described their antimicrobial activity against plant pathogens. Thus, to further explore their potential as bio-pesticides, we evaluated their inhibitory effects on mycelial growth of 10 common phytopathogens in vitro with results listed in Table 1. NEP-1 showed a broad antimicrobial spectrum and exhibited high inhibitory effects against R. solani, P. infestans, and F. graminearum, with inhibition rates of 82.74, 80.41, and 80.93%, respectively. R. solani was most sensitive to NEP-1. Moreover, the EC50 values of NEP-1 against R. solani and P. infestans were determined to be 40 and 51.25 μg/ml, respectively (Table 2). Noteworthily, NEP-1 also exhibited higher inhibition rates at 100 μg/ml than MBC at 0.5 μg/ml against the selected pathogens except F. graminearum, P. capsica, and S. sclerotiorum. Similarly, compared to the antimicrobial activity of MZ at 0.5 μg/ml, higher inhibition rates were observed for NEP-1 at 100 μg/ml against the tested pathogens except E. turcicum and G. gramini. PDKP displayed a moderate inhibitory effect against G. graminis (inhibition rate of 35%) and weak inhibitory effects against R. solani, P. infestans, S. sclerotiorum, and A. alternate (inhibition rate < 40%) at 100 μg/ml. MDKP, however, remarkably inhibited the mycelial growth of E. turcicum with an inhibition rate of 67.60% at 100 μg/ml, higher than that of NEP-1. These results indicate that NEP-1 is an excellent antifungal agent with a broad spectrum and that MDKP has the potential to control the maize leaf spot disease caused by E. turcicum.
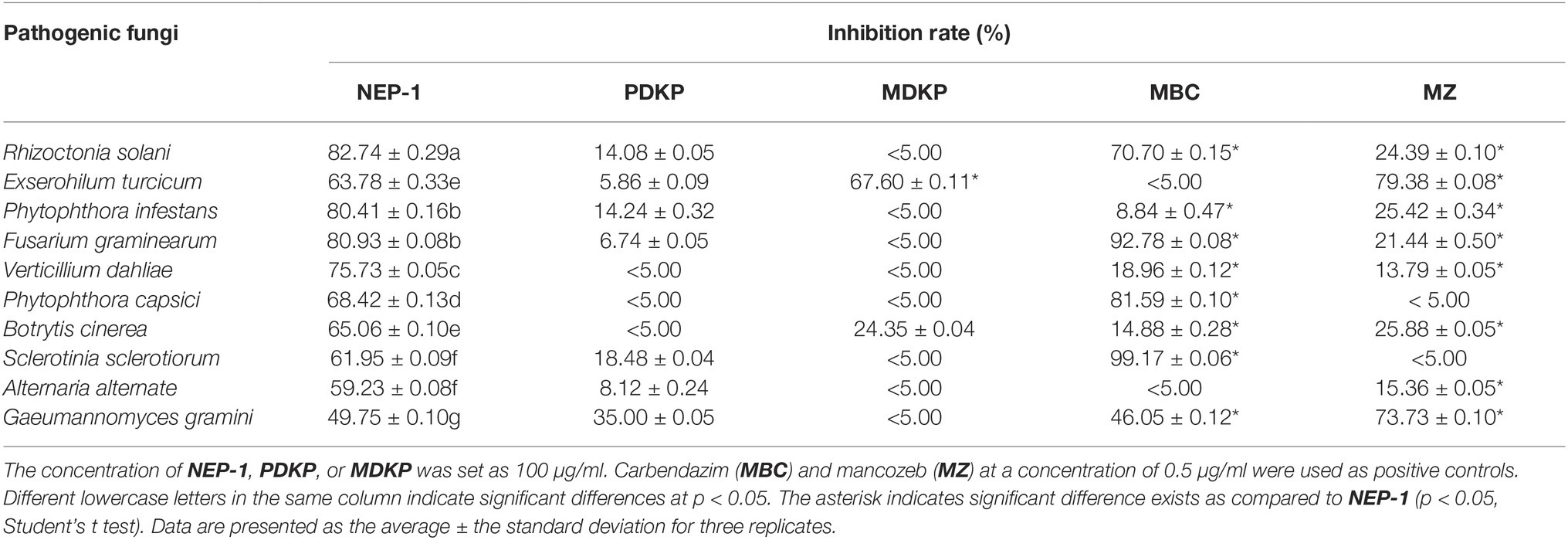
Table 1. Inhibitory effects of the three isolated compounds on the mycelial growth of 10 plant pathogens.
To explore whether the configuration of nematophin affects its antimicrobial activity, we tested the inhibition of three nematophin compounds (NEP-1, -2, and -3) on mycelia growth of five plant pathogens. As revealed in Table 3, compared to NEP-1 and -2, NEP-3 exhibited stronger inhibition against the five selected pathogens and significantly higher inhibition rates against S. sclerotiorum and P. capsica. Besides, no obvious differences were observed between NEP-1 and -2 in the inhibition rates. These results reveal that the (S)-configuration structure may possess higher antimicrobial activity than the (R)-enantiomer of nematophin and that NEP-1 is an enantiomeric mixture. Given that NEP-1 can be easily obtained through the bacterial fermentation of low cost, it was selected as the subject for subsequent experiments.
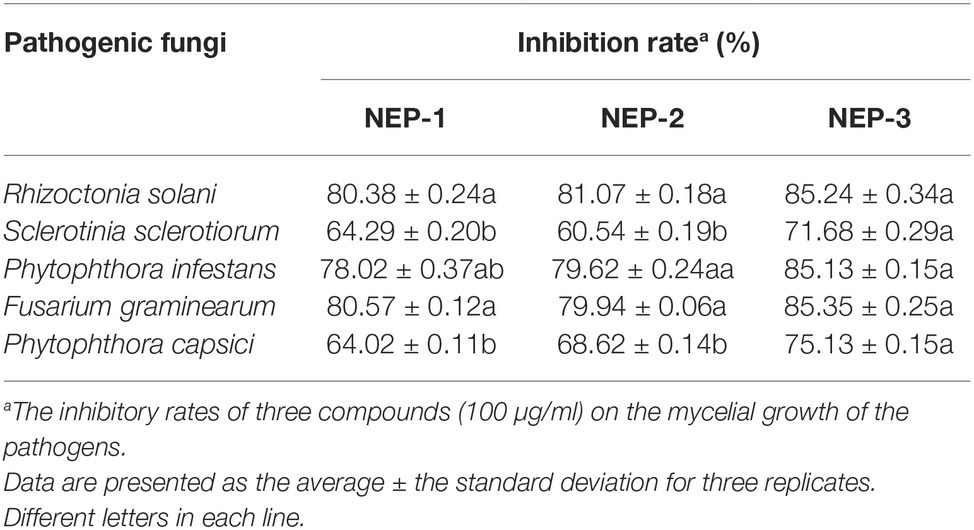
Table 3. Inhibitory effects of three nematophin compounds of decided configuration on the mycelial growth of five plant pathogens.
Examination the Control Efficacies of NEP-1 Against R. solani and P. infestans on Their Respective Host Plant Leaves
As described above, NEP-1 exhibited strong in vitro antimicrobial activity against R. solani and P. infestans. However, the antimicrobial activity in vivo was scarcely reported in previous studies. Thus, we explored the control efficacy of NEP-1 against R. solani on broad bean leaves and against P. infestans on potato leaves. As shown in Figures 3A,C, NEP-1 could protect the bean leaves from the infection of R. solani effectively, with preventive efficacies of 53.05% at 500 μg/ml and 93.01% at 1,000 μg/ml, which is comparable to that of the MBC treatment at 500 μg/ml (98.01%). Moreover, NEP-1 also exhibited high curative activity for the diseased leaves infected by R. solani, with control efficacies of 83.59 and 94.93% at 500 and 1,000 μg/ml, respectively (Figures 3B,D).
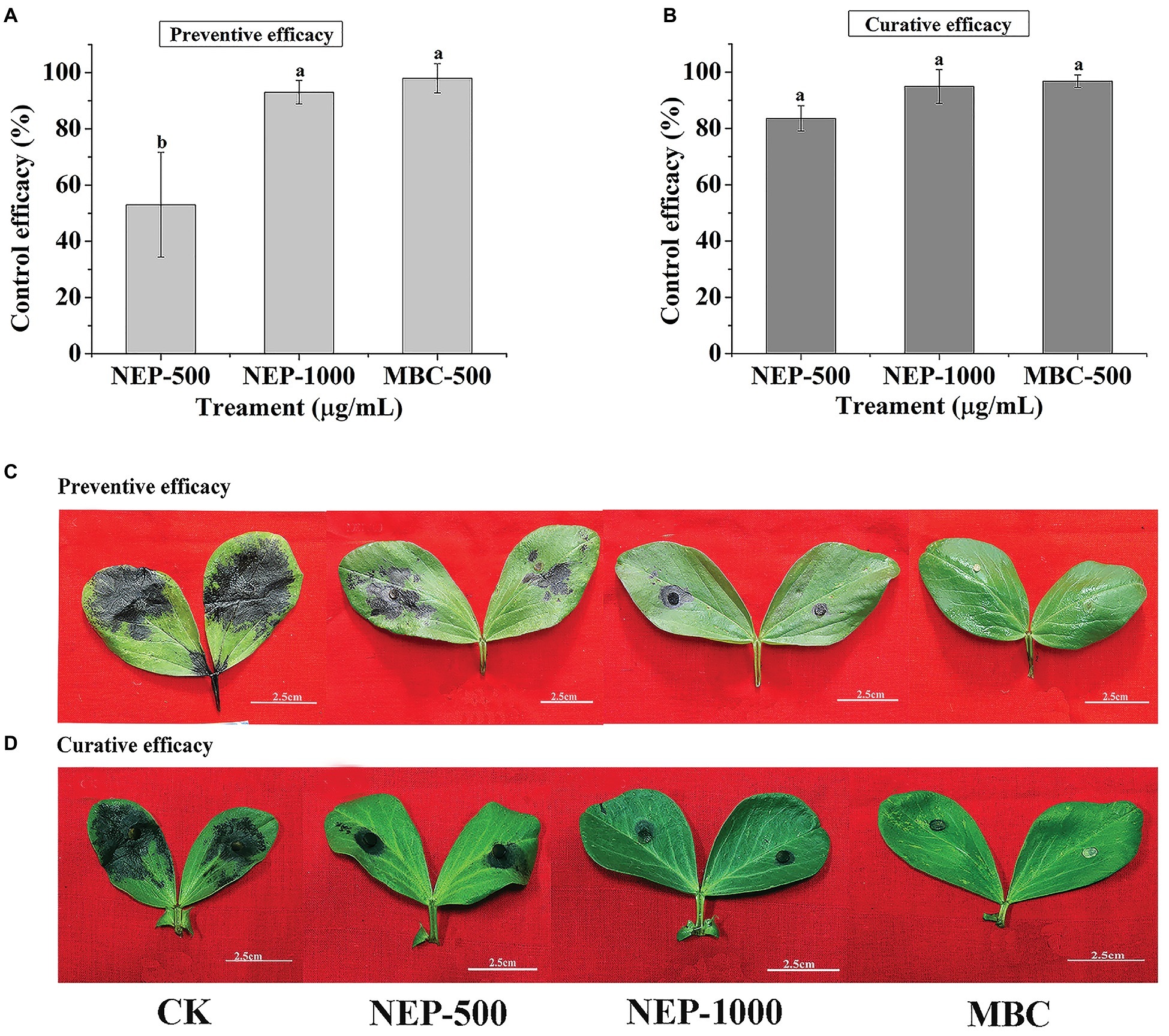
Figure 3. In vivo antifungal activity of NEP-1 and carbendazim against R. solani. (A,C) Protective activity of NEP-1 and (B,D) curative activity of NEP-1. Preventive/curative efficacy: the control efficacy of a compound that was sprayed on broad bean leaves 12 h before/after inoculation with R. solani. NEP500, 500 μg/ml of NEP-1; NEP1000, 1,000 μg/ml of NEP-1; MBC, 500 μg/ml of carbendazim.
The control efficacy of NEP-1 against P. infestans on potato leaves is shown in Supplementary Figure S3. The results showed that NEP-1 possessed weak preventive efficacy, with control efficacies of 15.58% at 500 μg/ml and 36.56% at 1,000 μg/ml (Supplementary Figures S3A,C), lower than that of MZ treatment at 500 μg/ml. Additionally, NEP-1 exhibited moderate curative activity with control efficacies of 39.63% at 500 μg/ml and 56.41% and 1,000 μg/ml, lower than that of referenced MZ at 500 μg/ml (Supplementary Figures S3B,D). Overall, the disease development control assay indicated that NEP-1 exhibited high control efficacy against R. solani in vivo. Therefore, R. solani was selected as the target pathogen for further experiments.
Biocontrol Efficacies of NEP-1 Against R. solani on Rice and Broad Bean Plants
To further explore the potency of NEP-1 as a new biopesticide, we evaluated the biocontrol potential of NEP-1 against the infestation of R. solani on rice and broad bean plants using a pot experiment. The morphological variations as well as the average lesion length of the inoculated plants in different treatment groups are illustrated in Figure 4. In the blank control groups, both of the rice and broad bean plants were severely infected by R. solani, with an apparent symptom of sheath blight disease (Figures 4C,D). However, the disease severity was effectively alleviated in the NEP-1 treatment groups (Figures 4C,D). Correspondingly, the lesion length on the inoculated plants in each NEP-1 treatment group was much smaller than that in the corresponding blank control group (Figures 4A,B). Moreover, no obvious difference was observed in the lesion length on the rice plants between the NEP-1 treatment group (1,000 μg/ml) and the MBC treatment group (500 μg/ml) (Figure 4B). These results indicated that NEP-1 had a favorable biocontrol efficacy against the disease extension on rice and broad bean plants caused by R. solani.
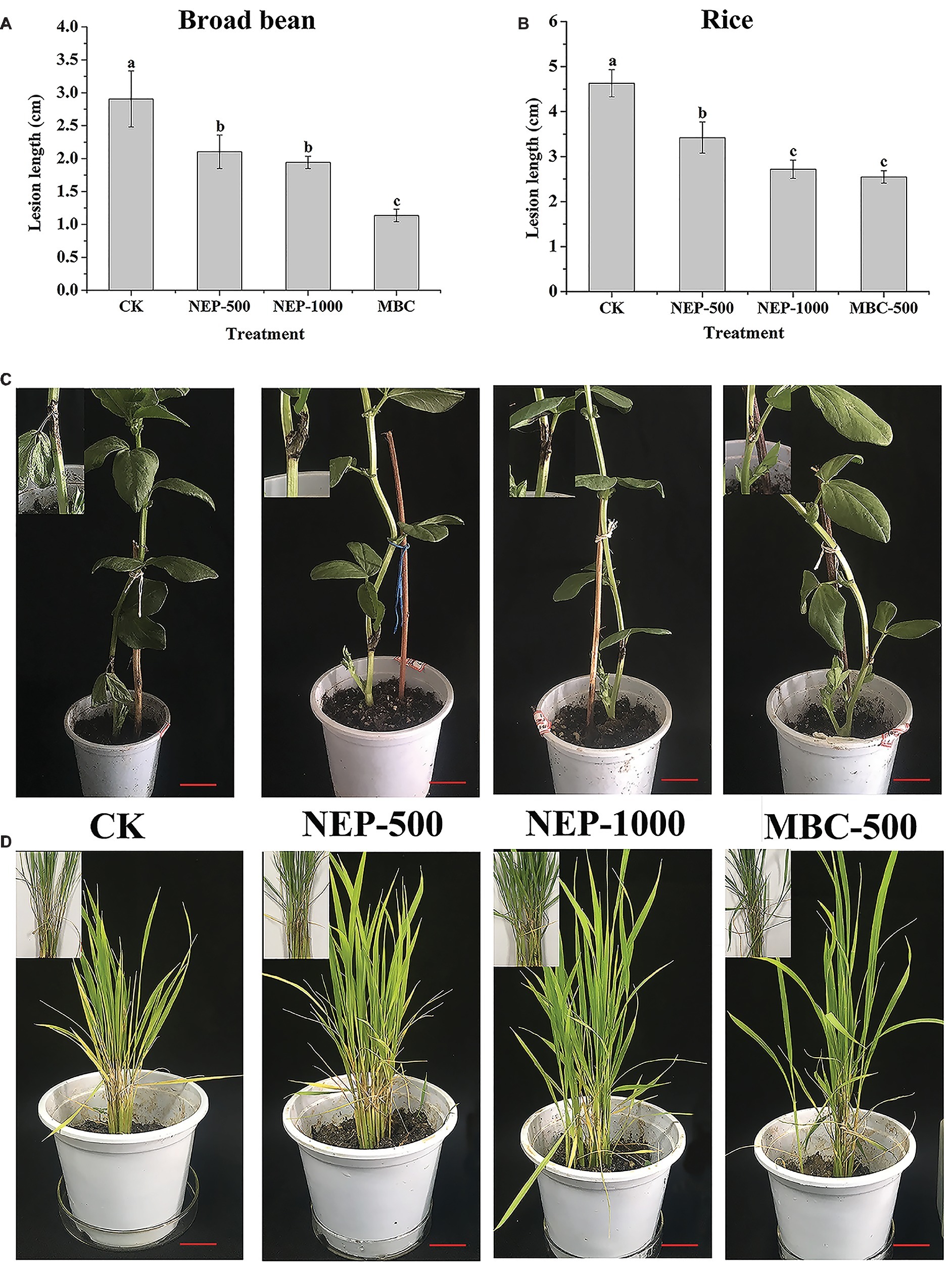
Figure 4. The lesion length and morphology of the plants infected by R. solani in pot experiments. (A) The lesion length of the broad bean plants; (B) the lesion length of the rice plants; (C) the morphology of the broad bean plants; and (D) the morphology of the rice plants. Insert: the disease area of the host plants. NEP500, 500 μg/ml of NEP-1; NEP1000, 1,000 μg/ml of NEP-1; MBC, 500 μg/ml of carbendazim; and CK was the blank control. Scale bar: 2.5 cm.
Effects of NEP-1 on the Sclerotial Development of R. solani
Suppressing sclerotial formation and germination is a crucial strategy for control of R. solani (Soltani et al., 2017). Thus, we tested the capacity of NEP-1 to suppress the sclerotial development of R. solani. As illustrated in Figure 5, on the plates with 40 μg/ml of NEP-1, few sclerotia were observed at the 6th day after inoculation and only a few abnormal sclerotium associated with large droplet secretion appeared after 12-day incubation. We further tested the effects of NEP-1 on the sclerotial development of R. solani at low concentrations (15, 20, and 30 μg/ml) with results shown in Supplementary Figure S4. Compared with the control plates, sclerotial formation was considerably suppressed with an increase of the NEP-1 concentration after 6 days of incubation. There were a very limited number of sclerotia occurring in the plates with 15.00 μg/ml of NEP-1 (Supplementary Figure S4B). Meanwhile, the sclerotial maturation was also prevented by NEP-1 and no mature sclerotia were observed when the concentration of NEP-1 was above 20.00 μg/ml (Supplementary Figure S4). Moreover, NEP-1 could also suppress the sclerotial germination significantly even at a very low concentration (15.00 μg/ml) (Figure 6). The above results revealed that NEP-1 could inhibit the sclerotial development of R. solani including the formation, maturation, and germination, and that NEP-1 might be an excellent antifungal agent for control of such sclerotia-forming fungal pathogens as R. solani.
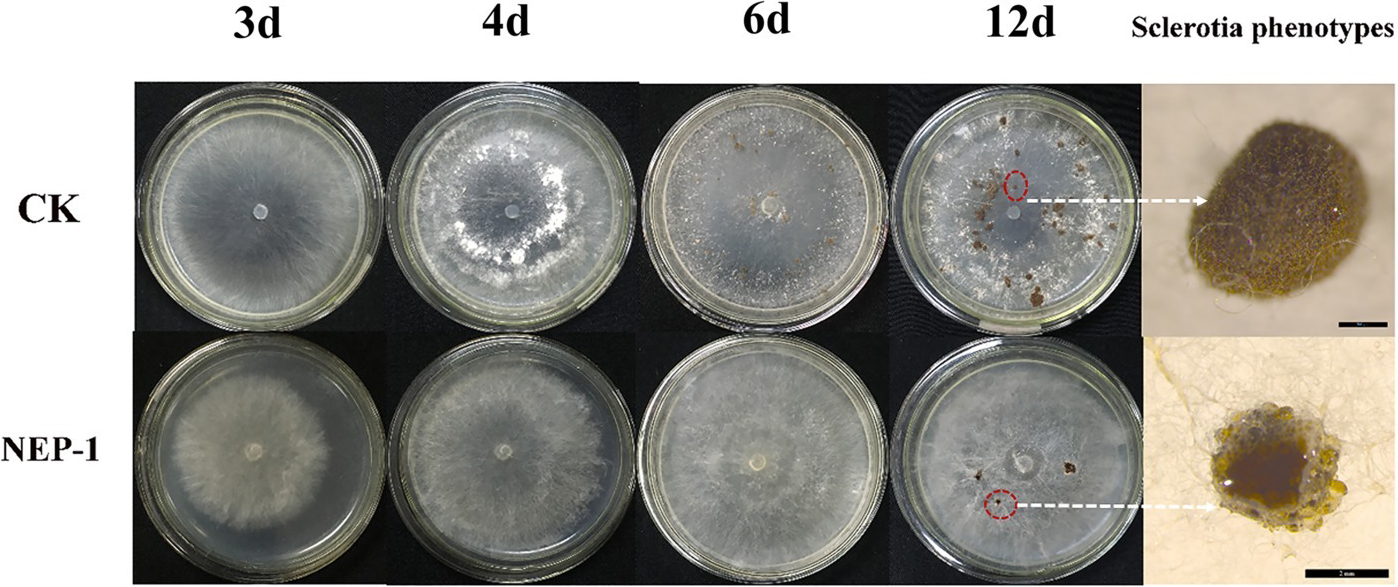
Figure 5. Effects of NEP-1 on the sclerotial development of R. solani. CK, Control plate; NEP-1, plate treated with NEP-1 at 40.00 μg/ml; plates were photographed at different incubating times (3, 4, 6, and 12 days). Sclerotia phenotypes were observed by a stereoscopic microscope after 12-day incubation. Scale bar: CK, 500 μm; NEP-1, 2 mm.
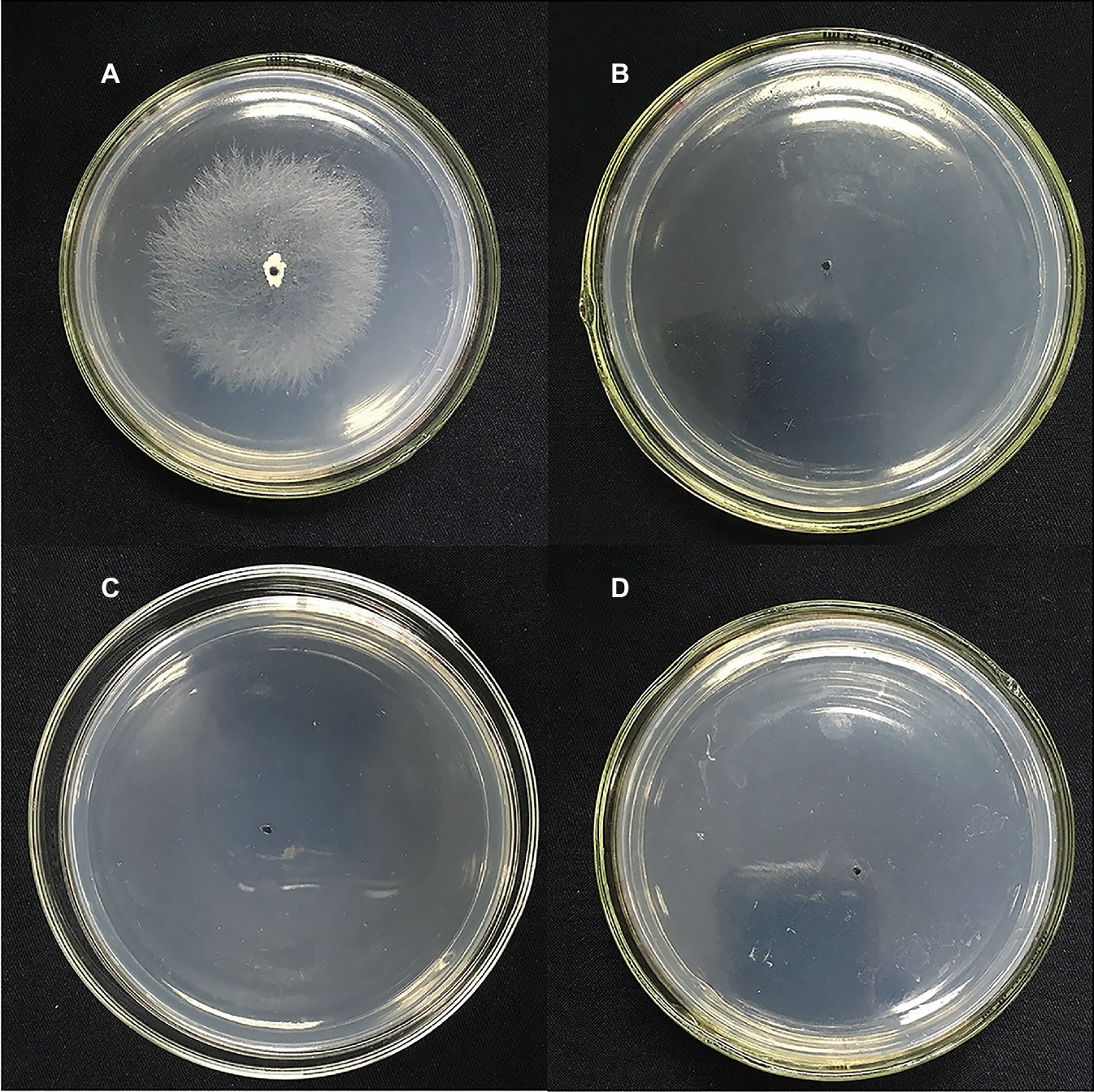
Figure 6. Effects of NEP-1 on the sclerotial germination of R. solani. (A) Control plate; (B) plate treated with NEP-1 at 15.00 μg/ml; (C) plate treated with NEP-1 at 20.00 μg/ml; and (D) plate treated with NEP-1 at 30.00 μg/ml.
Effects of NEP-1 on Hyphal Morphology and Ultrastructure of R. solani
To better understand the mode of anti-fungal action of NEP-1, the effects of NEP-1 on the hyphal morphology of R. solani were observed by SEM. As shown in Figure 7A, in the absence of NEP-1, the mycelia of R. solani exhibited a normal morphology of smooth, uniform, and robust hyphae with plump growing points. However, the NEP-1 treated sample displayed an altered hyphal morphology characterized by irregular hyphae of the twisted, shriveled and distorted morphology, or with deformities at the growing points (Figures 7B–D).
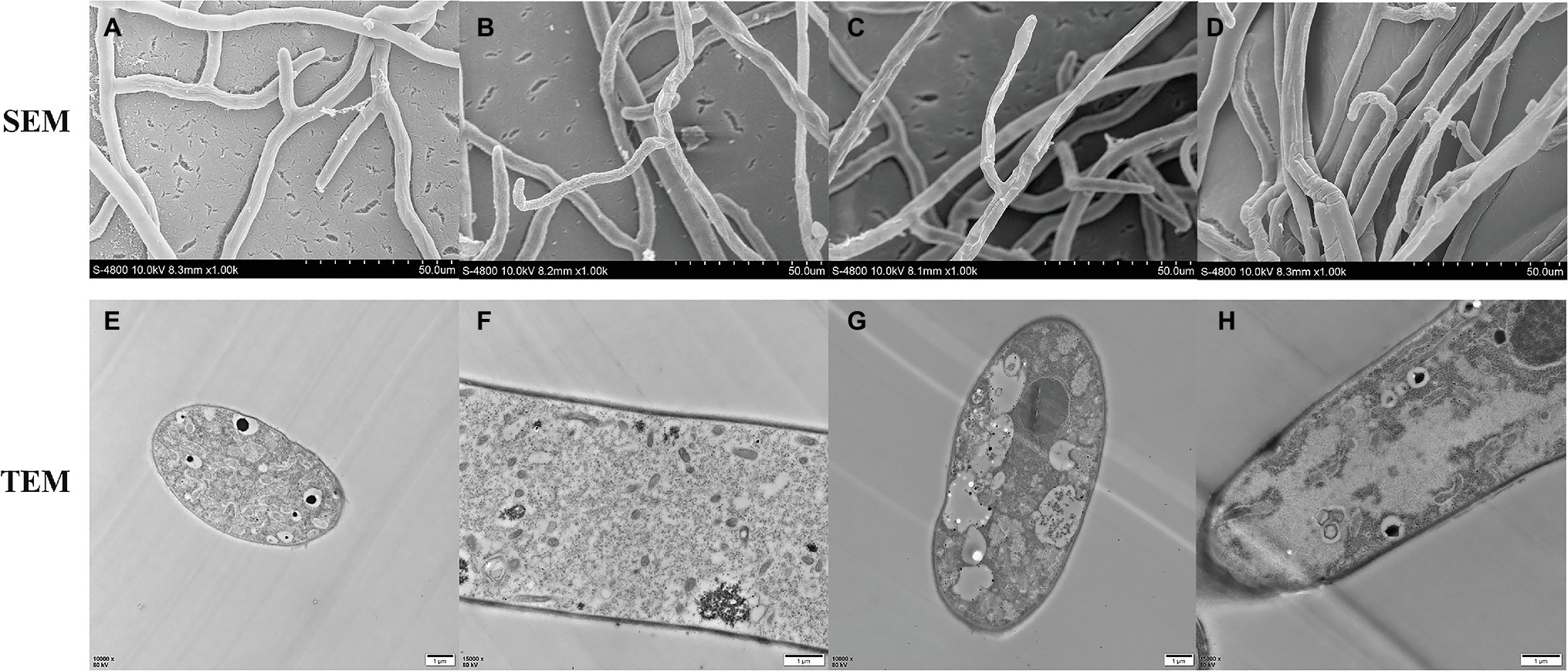
Figure 7. Effects of NEP-1 on the hyphal morphology and ultrastructure of R. solani. (A,E,F) Healthy hyphae in control petri plates; (B–D,G,H) hyphae treated with NEP-1 (40.00 μg/ml). Scale bar: 50.0 μm for SEM and 1 μm for TEM.
TEM was employed to evaluate the ultrastructural alterations of R. solani with the results illustrated in Figure 7. The control sample represented a typical fungal ultrastructure of intact cell walls of normal thicknesses, evenly distributed cellular cytoplasm, and regularly shaped organelles in the mycelial cells (Figures 7E,F). For the sample treated with NEP-1 (EC50 of 40.00 μg/ml), the mitochondrial abnormalities were clearly observed of hazy outlines and vacuolar degeneration, as well as the reduction in number (Figures 7G,H). Furthermore, the vacuolization and disorganization of the cytoplasm were also found in the mycelial cells exposed to NEP-1 (Figures 7G,H).
Discussion
X. nematophila YL001 is a valuable producer of natural compounds with pesticidal properties. Its fermentation broth was indicated to possess strong antimicrobial activity against some plant pathogens in vitro and in vivo, including P. infestans and B. cinerea (Guo et al., 2017; Zhang et al., 2018). Additionally, several pesticidal compounds have been identified from other X. nematophila stains, such as xenocoumacins (McInerney et al., 1991; Yang et al., 2011; Zhou et al., 2016), nematophin (Li et al., 1997a,b), PAX (peptide-antimicrobial-Xenorhabdus) peptides (Gualtieri et al., 2009; Fuchs et al., 2011), benzylideneacetone (Ji et al., 2004), xenortides, and xenematides (Lang et al., 2008). Xenocoumacins and nematophin were also present in the culture supernatant of X. nematophila YL001, which was confirmed by the HPLC-MS analysis in our previous study (Guo et al., 2017). Based on this, we try to isolate and identify new metabolites from the strain of X. nematophila YL001. In the present study, three dipeptide compounds were isolated and identified from the cell-free culture. NEP-1 is an antibiotic which was first isolated from strain X. nematophila BC1 (Li et al., 1997a). PDKP, a cyclodipeptide of proline and glycine, has been isolated previously from a sea marine sponge of undescribed species of Callyspongia (Chen et al., 2014), as well as many microorganisms such as Quambalaria cyanescens (Bishay et al., 2018) and Bacillus amyloliquefaciens (Li et al., 2018). MDKP, a dimethylated cyclodipeptide of phenylalanine and leucine, was first synthesized by Nakao et al. (2016) and its core structure piperazine-2,5-dione was found in a variety of natural products from fungi, bacteria, plants, and mammals (Borthwick, 2012). Despite this, to the best of our knowledge, it was first identified as a natural compound in this study. Moreover, this is also the first report of the presence of PDKP and MDKP in the X. nematophila fermentation broth. To gain more information about their pesticidal properties, we also evaluated their antimicrobial activity against 10 common phytopathogens. NEP-1 was demonstrated to possess a broad-spectrum antimicrobial activity in vitro (Table 1) and high control efficacy against R. solani in vivo (Figures 3, 4). Despite the weak antimicrobial activity against the selected plant pathogens, PDKP was determined to be a potent acaricide against Tetranychus urticae with a LC50 value of 95.96 μg/ml in a recent study (Li et al., 2018). MDKP was proved to have moderate antifungal activity against E. turcicum (Table 1). These results reveal the potential value of the three compounds in the crop protection.
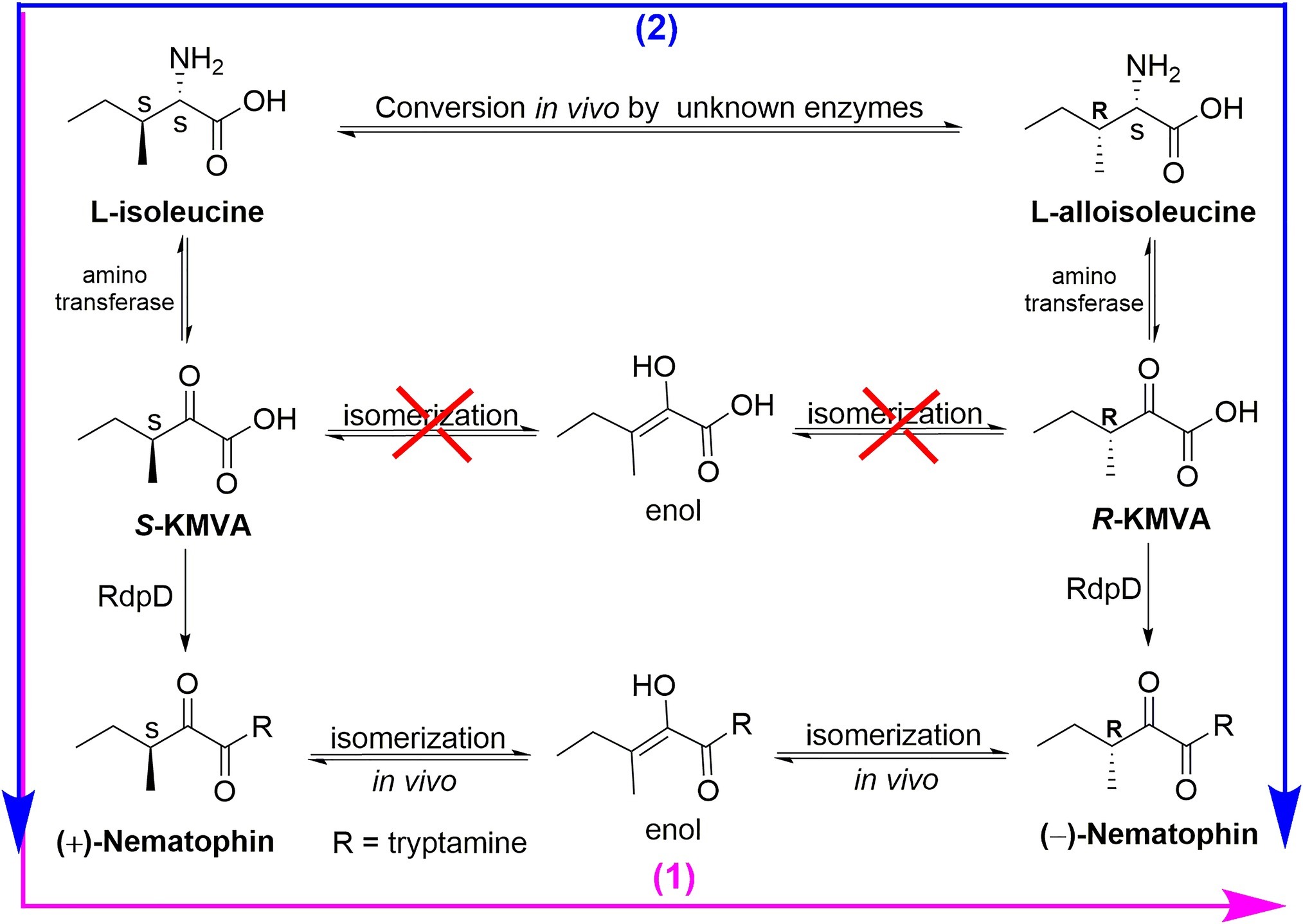
The biosynthetic origin of (±)-nematophin (NEP-1) may be involved in two pathways in X. nematophila (Figure 8). The CD spectra (Supplementary Figure S2) and specific rotation of NEP-1, -2, and -3 reveal that NEP-1 is an enantiomeric mixture of nematophin, denying the previous inference that the natural nematophin is an enantiomer of (S)-configuration (Paik et al., 2003). Based on the biosynthetic process of nematophin, we deduced two possible pathways relevant to the formation of NEP-1. One is that (+)-nematophin (NEP-3) is derived from L-isoleucine (L-Ile) via the aminotransferase to generate 2-keto-3S-methylvaleric acid (S-KMVA) (Mamer, 2001), followed by enolization-induced racemization to form the nematophin stereoisomers in vivo. Previous studies revealed that the racemization of S-KMVA did not occur in vivo, and that L-alloisoleucine (L-allo-Ile) was derived from L-Ile rather than R-KMVA (Mamer and Reimer, 1992; Mamer and Lépine, 1996). Thus, the other pathway may be that NEP-1 is derived from the authigenic L-Ile and L-allo-Ile, which can be converted to S-KMVA and R-KMVA, respectively. Conversion of L-Ile to L-allo-Ile was verified in bacteria, fungi, plants and mammalian systems and L-allo-Ile is also present in many natural cyclic peptide antibiotics (Mamer, 2001; Li et al., 2016), providing evidence for the second pathway. However, the actual mechanism of the racemization needs further clarification.
NEP-1 has great potential to be developed as a new biopesticide for agricultural production. NEP-1 not only represented broad-spectrum antimicrobial activity against 10 agricultural pathogenic fungi and oomycetes in vitro (Table 1) but also exerted a comparable control efficacy against R. solani in vivo at 1,000 μg/ml to MBC at 500 μg/ml on the rice plants (Figure 4). Moreover, NEP-1 could also inhibit the sclerotial formation, maturation and germination of R. solani substantially even at a low concentration (15 μg/ml) (Supplementary Figures S4, S5), beneficial to control the disease extension during its epidemic period and its outbreak in the next year effectively. Besides high bioactivity (in vitro and in vivo), easy production at an industrial scale is also another key factor affecting the development of a microbial pesticide (Montesinos, 2003). NEP-1 and its analogs can be synthesized through chemical methods due to its simple structure (Li et al., 1997b). In addition, researchers have also illuminated its biosynthesis pathway and achieved its heterologous production in Escherichia coli (Cai et al., 2017). In this paper, we also synthesized an enantiomeric mixture of nematophin (NEP-2) and its (S)-configuration structure (NEP-3), with high yields and easy operating conditions. The ultrastructure data suggest that NEP-1 may diffuse into the exposed cells and then disrupt the cells by altering the subcellular structures, especially the mitochondrial structure, which are similar to those induced by camptothecin (CPT) against R. solani, indicating that they may possess a similar antifungal mechanism (Sirikantaramas et al., 2008; Dai et al., 2017). However, more work is needed to explore the effects of NEP-1 on the mitochondria-related pathways of R. solani to further clarify its mechanism of action as a potent antifungal agent.
Conclusions
Three dipeptide compounds were purified from the cultural broth and identified as (±)-nematophin (NEP-1), cyclo (L-Pro-Gly) and N,N′-dimethyl-cyclo (L-Phe-L-Leu). NEP-1 demonstrated a wider antimicrobial spectrum than the other two compounds. It exhibited strong inhibitory effects on mycelial growth of R. solani and P. infestans. Its (S)-configuration structure (NEP-3, (+)-nematophin) was also synthesized and exhibited higher antifungal activity than the enantiomeric mixture. Besides, NEP-1 not only possessed significant preventive and curative efficacy against R. solani on broad bean leaves, but also could effectively inhibit the disease extension on rice and broad bean plants. The sclerotial formation, maturation and germination of R. solani was also significantly inhibited by NEP-1 at even a low concentration (15.00 μg/ml). The electron microscopic observations showed that the mycelium morphology of R. solani was adversely affected by nematophin at 40.00 μg/ml, as well as the subcellular structures. These results indicate that NEP-1 has great potential to be used as a bio-pesticide in agricultural production.
Data Availability
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
SZ, QL, YW, and XZ conceived and designed the experiments. SZ, QL, YH, JH, and ZY performed the experiments. QL and YH analyzed the data. JH and ZY contributed to reagents, materials, and analysis tools. SZ wrote the paper.
Funding
This work was supported by the Key Research and Development Program of Shaanxi Province of China (No. 2019NY-189), the Natural Science Foundation of China (No. 31171910), and the Natural Science Foundation of Shaanxi Province (No. 2014JZ004).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Hongli Zhang and Zhanfeng Zhang for technical assistance with NMR and MS spectra who are working in State Key Laboratory of Crop Stress Biology for Arid Areas, Northwest A&F University. We thank Yanqing Wang for technical assistance with stereoscopic microscope who is working in Life Science Research Core Services (LSRCS), Northwest A&F University. We also thank Yunfei Zhang for technical assistance with synthesized the compound and for critical reading of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01765/full#supplementary-material
Footnotes
References
Arfaoui, A., El, A. H., and Daayf, F. (2018). Pre-treatment of soybean plants with calcium stimulates ROS responses and mitigates infection by Sclerotinia sclerotiorum. Plant Physiol. Biochem. 122, 121–128. doi: 10.1016/j.plaphy.2017.11.014
Bishay, D. W., Abdel-Baky, A. M., Moharram, A. M., Malak, L. G., Srivedavyasasri, R., and Ross, S. A. (2018). Secondary metabolites from the fungus Quambalaria cyanescens. Chem. Nat. Compd. 54, 274–277. doi: 10.1007/s10600-018-2322-2
Borthwick, A. D. (2012). 2,5-Diketopiperazines: synthesis, reactions, medicinal chemistry, and bioactive natural products. Chem. Rev. 112, 3641–3716. doi: 10.1021/cr200398y
Böszörményi, E., Ersek, T., Fodor, A., Fodor, A., Foeldes, L. S., Hevesi, M., et al. (2009). Isolation and activity of Xenorhabdus antimicrobial compounds against the plant pathogens Erwinia amylovora and Phytophthora nicotianae. J. Appl. Microbiol. 107, 746–759. doi: 10.1111/j.1365-2672.2009.04249.x
Cai, X., Challinor, V. L., Zhao, L., Reimer, D., Adihou, H., Grün, P., et al. (2017). Biosynthesis of the antibiotic nematophin and its elongated derivatives in entomopathogenic bacteria. Org. Lett. 19, 806–809. doi: 10.1021/acs.orglett.6b03796
Campbell, J., Lin, Q., Geske, G. D., and Blackwell, H. E. (2009). New and unexpected insights into the modulation of LuxR-type quorum sensing by cyclic dipeptides. ACS Chem. Biol. 4, 1051–1059. doi: 10.1021/cb900165y
Challinor, V. L., and Bode, H. B. (2015). Bioactive natural products from novel microbial sources. Ann. N. Y. Acad. Sci. 1354, 82–97. doi: 10.1111/nyas.12954
Chaston, J. M., Suen, G., Tucker, S. L., Andersen, A. W., Bhasin, A., Bode, E., et al. (2011). The entomopathogenic bacterial endosymbionts Xenorhabdus and Photorhabdus: convergent lifestyles from divergent genomes. PLoS One 6:e27909. doi: 10.1371/journal.pone.0027909
Chen, Y., Peng, Y., Gao, C., and Huang, R. (2014). A new diketopiperazine from South China Sea marine sponge Callyspongia sp. Nat. Prod. Res. 28, 1010–1014. doi: 10.1080/14786419.2014.903397
Crawford, J. M., Portmann, C., Zhang, X., Roeffaers, M. B., and Clardy, J. (2012). Small molecule perimeter defense in entomopathogenic bacteria. Proc. Natl. Acad. Sci. USA 109, 10821–10826. doi: 10.1073/pnas.1201160109
Dai, T. T., Xu, Z., Zhou, X., Li, B., and Mao, S. F. (2017). The inhibitory effect of the plant alkaloid camptothecin on the rice sheath blight pathogen Rhizoctonia solani. Int. J. Agric. Biol. 19, 558–562. doi: 10.13227/j.hjkx.201704135
Dreyer, J., Malan, A. P., and Dicks, L. M. T. (2018). Bacteria of the genus Xenorhabdus, a novel source of bioactive compounds. Front. Microbiol. 9:3177. doi: 10.3389/fmicb.2018.03177
Engel, Y., Windhorst, C., Lu, X., Goodrichblair, H., and Bode, H. B. (2017). The global regulators Lrp, LeuO, and HexA control secondary metabolism in entomopathogenic bacteria. Front. Microbiol. 8:209. doi: 10.3389/fmicb.2017.00209
Fang, X., Zhang, M., Tang, Q., Wang, Y., and Zhang, X. (2014). Inhibitory effect of Xenorhabdus nematophila TB on plant pathogens Phytophthora capsici and Botrytis cinerea in vitro and in planta. Sci. Rep. 4, 1–7. doi: 10.1038/srep04300
Feng, S., Shu, C., Wang, C., Jiang, S., and Zhou, E. (2016). Survival of Rhizoctonia solani AG-1 IA, the causal agent of rice sheath blight, under different environmental conditions. J. Phytopathol. 165, 44–52. doi: 10.1111/jph.12535
Fuchs, S. W., Proschak, A., Jaskolla, T. W., Karas, M., and Bode, H. B. (2011). Structure elucidation and biosynthesis of lysine-rich cyclic peptides in Xenorhabdus nematophila. Org. Biomol. Chem. 9, 3130–3132. doi: 10.1039/c1ob05097d
Gualtieri, M., Aumelas, A., and Thaler, J.-O. (2009). Identification of a new antimicrobial lysine-rich cyclolipopeptide family from Xenorhabdus nematophila. J. Antibiot. 62, 295–302. doi: 10.1038/ja.2009.31
Guo, S., Zhang, S., Fang, X., Liu, Q., Gao, J., Bilal, M., et al. (2017). Regulation of antimicrobial activity and xenocoumacins biosynthesis by pH in Xenorhabdus nematophila. Microb. Cell Factories 16:203. doi: 10.1186/s12934-017-0813-7
Hazir, S., Shapiro-Ilan, D. I., Bock, C. H., Hazir, C., Leite, L. G., and Hotchkiss, M. W. (2016). Relative potency of culture supernatants of Xenorhabdus and Photorhabdus spp. on growth of some fungal phytopathogens. Eur. J. Plant Pathol. 146, 1–13. doi: 10.1007/s10658-016-0923-9
Helge, B. (2011). Structure elucidation and biosynthesis of lysine-rich cyclic peptides in Xenorhabdus nematophila. Org. Biomol. Chem. 9, 3130–3132. doi: 10.1039/c1ob05097d
Houard, J., Aumelas, A., Noël, T., Pages, S., and Gualtieri, M. (2013). Cabanillasin, a new antifungal metabolite, produced by entomopathogenic Xenorhabdus cabanillasii JM26. J. Antibiot. 66, 617–620. doi: 10.1038/ja.2013.58
Ji, D., Yi, Y., Kang, G. H., Choi, Y. H., Kim, P., Baek, N. I., et al. (2004). Identification of an antibacterial compound, benzylideneacetone, from Xenorhabdus nematophila against major plant-pathogenic bacteria. FEMS Microbiol. Lett. 239, 241–248. doi: 10.1016/j.femsle.2004.08.041
Kanagaraj Muthu-Pandian, C., Sengottayan, S.-N., Vel, T., Karthi, S., Sreenath Kumar, C., Sreenath Kumar, C., et al. (2018). Bacterial compounds, as biocontrol agent against early blight (Alternaria solani) and tobacco cut worm (Spodoptera litura Fab.) of tomato (Lycopersicon esculentum Mill.) Arch. Phytopathol. Plant Protect. 51, 729–753. doi: 10.1080/03235408.2018.1496525
Kazempour, M. N. (2004). Biological control of Rhizoctonia solani, the causal agent of rice sheath blight by antagonistics bacteria in greenhouse and field conditions. Plant Pathol. J. 3, 88–96. doi: 10.3923/ppj.2004.88.96
Kim, B. S., and Hwang, B. K. (2007). Microbial fungicides in the control of plant diseases. J. Phytopathol. 155, 641–653. doi: 10.1111/j.1439-0434.2007.01314.x
Kumar, V. V. (2018). “Biofertilizers and biopesticides in sustainable agriculture” in Role of rhizospheric microbes in soil: Volume 1: Stress management and agricultural sustainability. ed. V. S. Meena (Singapore: Springer Singapore), 377–398.
Lang, G., Kalvelage, T., Peters, A., Wiese, J., and Imhoff, J. F. (2008). Linear and cyclic peptides from the entomopathogenic bacterium Xenorhabdus nematophilus. J. Nat. Prod. 71, 1074–1077. doi: 10.1021/np800053n
Lengai, G. M. W., and Muthomi, J. W. (2018). Biopesticides and their role in sustainable agricultural production. J. Biosci. Med. 6, 7–41. doi: 10.4236/jbm.2018.66002
Lewis, J. A., and Lumsden, R. D. (2001). Biocontrol of damping-off of greenhouse-grown crops caused by Rhizoctonia solani with a formulation of Trichoderma spp. Crop Prot. 20, 49–56. doi: 10.1016/S0261-2194(00)00052-1
Li, J., Chen, G., and Webster, J. M. (1997a). Nematophin, a novel antimicrobial substance produced by Xenorhabdus nematophilus (Enterobactereaceae). Can. J. Microbiol. 43, 770–773. doi: 10.1139/m97-110
Li, J., Chen, G., and Webster, J. M. (1997b). Synthesis and antistaphylococcal activity of nematophin and its analogues. Bioorg. Med. Chem. Lett. 7, 1349–1352. doi: 10.1016/s0960-894x(97)00223-0
Li, Q., Qin, X., Liu, J., Gui, C., Wang, B., Li, J., et al. (2016). Deciphering the biosynthetic origin of l-allo-isoleucine. J. Am. Chem. Soc. 138, 408–415. doi: 10.1021/jacs.5b11380
Li, X. Y., Wang, Y. H., Yang, J., Cui, W. Y., and He, Y. Q. (2018). Acaricidal activity of cyclodipeptides from Bacillus amyloliquefaciens W1 against Tetranychus urticae. J. Agric. Food Chem. 66, 10163–10168. doi: 10.1021/acs.jafc.8b03806
Mamer, O. A. (2001). Initial catabolic steps of isoleucine, the R-pathway and the origin of alloisoleucine. J. Chromatogr. B Biomed. Sci. Appl. 758, 49–55. doi: 10.1016/S0378-4347(01)00111-6
Mamer, O. A., and Lépine, F. L. (1996). 15N conservation in the metabolic conversion of isoleucine to alloisoleucine in the rat. J. Mass Spectrom. 31, 1382–1388. doi: 10.1002/(SICI)1096-9888(199612)31:12<1382::AID-JMS435>3.0.CO;2-V
Mamer, O. A., and Reimer, M. L. (1992). On the mechanisms of the formation of L-alloisoleucine and the 2-hydroxy-3-methylvaleric acid stereoisomers from L-isoleucine in maple syrup urine disease patients and in normal humans. J. Biol. Chem. 267, 22141–22147. doi: 10.1016/S0022-5193(05)80771-4
Mayo, S., Gutiérrez, S., Malmierca, M. G., Lorenzana, A., Campelo, M. P., Hermosa, R., et al. (2015). Influence of Rhizoctonia solani and Trichoderma spp. in growth of bean (Phaseolus vulgaris L.) and in the induction of plant defense-related genes. Front. Plant Sci. 6:685. doi: 10.3389/fpls.2015.00685
McInerney, B. V., Taylor, W. C., Lacey, M. J., Akhurst, R. J., and Gregson, R. P. (1991). Biologically active metabolites from Xenorhabdus spp., part 2. Benzopyran-1-one derivatives with gastroprotective activity. J. Nat. Prod. 54, 785–795. doi: 10.1021/np50075a006
Mnif, I., and Ghribi, D. (2015). Potential of bacterial derived biopesticides in pest management. Crop Prot. 77, 52–64. doi: 10.1016/j.cropro.2015.07.017
Moni, Z. R., Ali, M. A., Alam, M. S., Rahman, M. A., Bhuiyan, M. R., Mian, M. S., et al. (2016). Morphological and genetical variability among Rhizoctonia solani isolates causing sheath blight disease of rice. Rice Sci. 23, 42–50. doi: 10.1016/j.rsci.2016.01.005
Montesinos, E. (2003). Development, registration and commercialization of microbial pesticides for plant protection. Int. Microbiol. 6, 245–252. doi: 10.1007/s10123-003-0144-x
Nakao, M., Hiroyama, Y., Fukayama, S., and Sano, S. (2016). N4-methylation changes the conformation of (3S,6S)-3-alkyl-6-benzylpiperazine-2,5-diones from folded to extended. J. Mol. Struct. 1116, 37–44. doi: 10.1016/j.molstruc.2016.03.019
Nicolopoulou-Stamati, P., Maipas, S., Kotampasi, C., Stamatis, P., and Hens, L. (2016). Chemical pesticides and human health: the urgent need for a new concept in agriculture. Front. Public Health 4:148. doi: 10.3389/fpubh.2016.00148
Paik, S., Park, M. K., Jhun, S. H., Park, H. K., and Suh, S. I. (2003). Isolation and synthesis of tryptamine derivatives from a symbiotic bacterium Xenorhabdus nematophilus PC. ChemInform 34, 2101–2118. doi: 10.1002/chin.200345209
Pantel, L., Florin, T., Dobosz-Bartoszek, M., Racine, E., Sarciaux, M., Serri, M., et al. (2018). Odilorhabdins, antibacterial agents that cause miscoding by binding at a new ribosomal site. Mol. Cell 70:83. doi: 10.1016/j.molcel.2018.03.001
Park, D. S., Sayler, R., Hong, Y. G., Nam, M. H., and Yang, Y. (2008). A method for inoculation and evaluation of rice sheath blight disease. Plant Dis. 92, 25–29. doi: 10.1094/PDIS-92-1-0025
Rohr, J. R., Brown, J., Battaglin, W. A., McMahon, T. A., and Relyea, R. A. (2017). A pesticide paradox: fungicides indirectly increase fungal infections. Ecol. Appl. 27, 2290–2302. doi: 10.1002/eap.1607
Sharma, K., Walia, S., Ganguli, S., and Kundu, A. (2016). Analytical characterization of secondary metabolites from Indian Xenorhabdus species the symbiotic bacteria of entomopatathogenic nematode (Steinernema spp.) as antifungal agent. Natl. Acad. Sci. Lett. 39, 175–180. doi: 10.1007/s40009-016-0453-1
Sirikantaramas, S., Yamazaki, M., and Saito, K. (2008). Mutations in topoisomerase I as a self-resistance mechanism coevolved with the production of the anticancer alkaloid camptothecin in plants. Proc. Natl. Acad. Sci. USA 105, 6782–6786. doi: 10.1073/pnas.0801038105
Soltani, N. M., Ghs, B., Khatami, M., Amini, A., and Aghighi, S. (2017). In vitro and in vivo antifungal properties of silver nanoparticles against Rhizoctonia solani, a common agent of rice sheath blight disease. IET Nanobiotechnol. 11, 236–240. doi: 10.1049/iet-nbt.2015.0121
Soylu, E. M., Kurt, S., and Soylu, S. (2010). In vitro and in vivo antifungal activities of the essential oils of various plants against tomato grey mould disease agent Botrytis cinerea. Int. J. Food Microbiol. 143, 183–189. doi: 10.1016/j.ijfoodmicro.2010.08.015
Townsend, B. B., and Willetts, H. J. (1954). The development of sclerotia of certain fungi. Trans. Br. Mycol. Soc. 37, 213–221. doi: 10.1016/S0007-1536(54)80003-9
Wu, Z. M., Yang, Y., and Li, K. T. (2019). Antagonistic activity of a novel antifungalmycin N2 from Streptomyces sp. N2 and its biocontrol efficacy against Rhizoctonia solani. FEMS Microbiol. Lett. 366, 1–8. doi: 10.1093/femsle/fnz018
Yang, X., Qiu, D., Yang, H., Liu, Z., Zeng, H., and Yuan, J. (2011). Antifungal activity of xenocoumacin 1 from Xenorhabdus nematophilus var. pekingensis against Phytophthora infestans. World J. Microbiol. Biotechnol. 27, 523–528. doi: 10.1007/s11274-010-0485-5
Zhang, S., Fang, X., Tang, Q., Ge, J., Wang, Y., and Zhang, X. (2018). CpxR negatively regulates the production of xenocoumacin 1, a dihydroisocoumarin derivative produced by Xenorhabdus nematophila. Microbiology 8:e00674. doi: 10.1002/mbo3.674
Zheng, A., Lin, R., Zhang, D., Qin, P., Xu, L., Peng, A., et al. (2013). The evolution and pathogenic mechanisms of the rice sheath blight pathogen. Nat. Commun. 4:1424. doi: 10.1038/ncomms2427
Zhou, S., Liu, Y., Zhang, M., Li, B., Chen, X., and Liang, W. (2016). Comparison of the virulence and cognate virulence factors of multinucleate, binucleate and uninucleate Rhizoctonia isolates, causing sheath blight on maize plants. Eur. J. Plant Pathol. 145, 501–506. doi: 10.1007/s10658-015-0855-9.aa
Keywords: Xenorhabdus nematophila YL001, dipeptides, nematophin, enantiomeric mixture, antimicrobial activity, sclerotial development
Citation: Zhang S, Liu Q, Han Y, Han J, Yan Z, Wang Y and Zhang X (2019) Nematophin, an Antimicrobial Dipeptide Compound From Xenorhabdus nematophila YL001 as a Potent Biopesticide for Rhizoctonia solani Control. Front. Microbiol. 10:1765. doi: 10.3389/fmicb.2019.01765
Edited by:
Víctor Flors, University of Jaume I, SpainReviewed by:
M. Y. Sreenivasa, University of Mysore, IndiaFrédérique Reverchon, Instituto de Ecología (INECOL), Mexico
Copyright © 2019 Zhang, Liu, Han, Han, Yan, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yonghong Wang, yhwang@nwafu.edu.cn
 Shujing Zhang1
Shujing Zhang1 Yonghong Wang
Yonghong Wang