- 1National Research Institute of Fisheries and Environment of Inland Sea, Fisheries Research and Education Agency, Hatsukaiti, Japan
- 2Department of Biotechnology, College of Life Sciences, Ritsumeikan University, Kusatsu, Japan
- 3Japan Science and Technology Agency, Precursory Research for Embryonic Science and Technology, Kawaguchi, Japan
- 4Laboratory of Genome Informatics, National Institute for Basic Biology, National Institutes of Natural Sciences, Okazaki, Japan
- 5Core Research Facilities, National Institute for Basic Biology, Okazaki, Japan
- 6Department of Biochemistry and Molecular Biology, Graduate School of Science and Engineering, Saitama University, Sakura-ku, Japan
- 7Laboratory of Environmental Biology, Department of Applied Aquabiology, National Fisheries University, Fisheries Research and Education Agency, Yamaguchi, Japan
Aquaculture industries are under threat from noxious red tides, but harm can be mitigated by precautions such as early harvesting and restricting fish feeding to just before the outbreak of a red tide. Therefore, accurate techniques for forecasting red-tide outbreaks are strongly needed. Omics analyses have the potential to expand our understanding of the eco-physiology of these organisms at the molecular level, and to facilitate identification of molecular markers for forecasting their population dynamics and occurrence of damages to fisheries. Red tides of marine raphidophytes, especially Chattonella species, often extensively harm aquaculture industries in regions with a temperate climate around the world. A red tide of Chattonella tends to develop just after an input of nutrients along the coast. Chattonella displays diurnal vertical migration regulated by a weak blue light, so it photosynthesizes in the surface layer during the daytime and takes up nutrients in the bottom layer during the nighttime. Superoxide produced by Chattonella cells is a strong candidate for the cause of its toxicity to bacteria and fishes. Here we conducted mRNA-seq of Chattonella antiqua to identify genes with functions closely related to the dynamics of the noxious red tide, such as photosynthesis, photoreception, nutrient uptake, and superoxide production. The genes related to photosynthetic pigment biosynthesis and nutrient uptake had high similarity with those of model organisms of plants and algae and other red-tide microalgae. We identified orthologous genes of photoreceptors such as aureochrome (newly five genes), the cryptochrome/photolyase (CRY/PHR) family (6-4PHR, plant CRY or cyclobutane pyrimidine dimer [CPD] Class III, CPD Class II, and CRY-DASH), and phytochrome (four genes), which regulate various physiological processes such as flagellar motion and cell cycle in model organisms. Six orthologous genes of NADPH oxidase, which produces superoxide on the cell membrane, were found and divided into two types: one with 5–6 transmembrane domains and another with 11 transmembrane domains. The present study should open the way for analyzing the eco-physiological features of marine raphidophytes at the molecular level.
Introduction
Red tides sometimes impact fish and shellfish aquaculture industries and tourism in coastal waters around the world. Long-term studies have provided information on taxonomy, life cycle, and eco-physiology of organisms that cause harmful red tides, but no practical method to control the causative microorganisms has been established (Anderson, 1997). On the other hand, precautionary field interventions such as early harvesting, restricting fish feeding, and transport of fish cages into zones free of red tides have shown efficacy in mitigating the damage (Kim, 2006). These interventions require monitoring of the distribution of noxious flagellates and forecasting the occurrence of a red tide; however, development of forecasting techniques based on environmental factors by using numerical and statistical models has been difficult because the mechanism underlying the development of red tides remains unknown (Glibert et al., 2010).
Understanding the molecular mechanism of red-tide formation should lead to development of molecular markers that forecast the impact and its toxicity. However, molecular analyses require genetic information. Recently, whole transcriptome analysis by total mRNA-seq has been used to rapidly accumulate genetic information and data on physiological molecular processes in noxious red-tide organisms (Ryan et al., 2014; Kimura et al., 2015; Guo et al., 2016). For non-model organisms with large genome sizes, the large cost and effort required to sequence the whole genome may make total mRNA-seq analysis a better alternative for obtaining genetic information.
Marine raphidophytes are stramenopiles, and most of them cause noxious red tides in temperate coastal waters around the world. Heterosigma akashiwo sometimes forms dense red tides that cause harm to fish aquaculture industries around the world (Smayda, 1998). In Japan, the genus Chattonella, including C.antiqua, C. marina, and C. ovata, has caused the most harm to the aquaculture industry (Imai and Yamaguchi, 2012). Chattonella subsalsa forms red tides in the United States and Europe (Zhang et al., 2006; Satta et al., 2017) but not in Japan.
Red tides of H. akashiwo and Chattonella spp. tend to occur frequently in eutrophic areas (Imai et al., 2006) or just after an influx of nutrients (Nakamura and Watanabe, 1983; Nakamura et al., 1989; Shikata et al., 2008), although the nutrient requirement of C. antiqua is not higher than that of other red-tide flagellates (Nakamura, 1985). Information on pathways and enzymes related to uptake and metabolisms of nutrients may resolve this apparent discrepancy. Heterosigma akashiwo and C. antiqua undergo diurnal vertical migrations that enable them to photosynthesize in the surface layers during the daytime and take up nutrients in bottom layers during the nighttime (Watanabe et al., 1991). The rhythm of diurnal vertical migration is reset by light within a specific range of wavelengths (Shikata et al., 2013). Moreover, negative phototaxis and positive geotaxis are also induced by light with different wavelengths (Shikata et al., 2016). Therefore, the motility can be regulated by photoreceptors that have unique spectra for absorbance and action. The gene encoding a blue light receptor aureochrome has been isolated in H. akashiwo and C. antiqua (Ishikawa et al., 2009; Ji et al., 2017), but there is little information on the genes encoding other photoreceptors such as a UV/blue light receptor, cryptochrome (CRY) and photolyase (PHR), and a red/far-red light receptor, phytochrome, which function in physiological control in various organisms (Ahmad and Cashmore, 1993; Furuya, 1993; Smith, 2000; Lin and Todo, 2005).
Ichthyotoxicity of marine raphidophytes, especially Chattonella, has also been energetically studied. The main hypothesis is that the fish die from lack of oxygen due to gill lesions and induction of excessive mucus production from gill cells by Chattonella cells (Ishimatsu et al., 1996). Because Chattonella cells produce large quantities of superoxide known as one of reactive oxygen species (ROS) compared to other phytoplankters (Oda et al., 1997), superoxide is suspected as a factor contributing to gill damage. Moreover, C. marina strongly inhibits marine bacteria proliferation, but the toxic effect is completely suppressed by the addition of catalase and superoxide dismutase (Oda et al., 1992). Therefore, the ability to produce superoxide may enable Chattonella to outcompete other microorganisms. However, there is little information on molecules and enzymes related to superoxide generation in Chattonella, although it is well known that NADPH oxidase (NOX) produces superoxide in membranes of cells and phagosome in plants and animals (Canton and Grinstein, 2014).
Here, we sequenced mRNA of C. antiqua, which frequently blooms and causes tremendous damage to the aquaculture industry in Japan, to seek genes closely related to development and toxicity of a red tide: e.g., those involved in photosynthesis, photoreception, nutrient uptake, and superoxide production. We then compared the data with sequences of model organisms, such as Arabidopsis and Chlamydomonas, noxious red-tide dinoflagellates in Japan (Karenia mikimotoi, Heterocapsa circularisquama) and other raphidophyte species (H. akashiwo, C. subsalsa).
Materials and Methods
Culture Conditions
We used a clonal axenic strain of C. antiqua (3KGY, Shikata et al., 2015, 2016) isolated from the Yatsushiro Sea, Japan. Cultures were maintained in 50-mL Erlenmeyer flasks containing 25 mL of modified SWM-3 medium (Chen et al., 1969; Shikata et al., 2011) with salinity of 30 at 25°C under 200 μmol photons m–2 s–1 of white fluorescent light illumination (FL20SW, Toshiba Lighting and Technology Corporation, Kanagawa, Japan) on a 12 h:12 h light:dark cycle (light period: local time [LT] 0600–1800). The photon flux density was measured with a Quantum Scalar Laboratory Irradiance Sensor (QSL-2101, Biospherical Instruments Inc., San Diego, CA, United States). During exponential growth (10,000 cells mL–1), C. antiqua cells were harvested during the daytime (LT 1100–1200, n = 3 flasks) or nighttime (LT 1900–2000, n = 3 flasks).
We also used a clonal axenic strain of H. circularisquama (N-30) isolated from Asoura, Japan; H. circularisquama is a dinoflagellate that forms a red tide that kills shellfish, specifically. The cultures were maintained under the same conditions as C. antiqua, and cell harvesting was conducted during the daytime (LT 1100–1200, n = 1 flask).
RNA Extraction and cDNA Library
First, 25 mL of C. antiqua culture or 50 mL of H. circularisquama culture were concentrated to 5 mL by using a Nuclepore filter (pore diameter, 3 μm; Whatman, Kent, United Kingdom). Shortly thereafter, 5 mL of RNase inhibitor (47.5 mL of ethanol plus 2.68 g phenol) were added to the cell suspension and vigorously mixed. Cell pellets were obtained by centrifugation at 2,500 g for 5 min at room temperature and stored at −80°C until RNA extraction. Total RNA was isolated from the cell pellets and purified using RNeasy Plant Mini kits (QIAGEN, Valencia, CA, United States). The quantity and quality of total RNA was determined using an automated electrophoresis system, ExperionTM (Bio-Rad Laboratories, Hercules, CA, United States). cDNA libraries for C. antiqua and H. circularisquama were constructed by using an Illumina Gene Expression Sample Prep Kit (Illumina Inc., San Diego, CA, United States). Sequencing was conducted using an Illumina HiSeq 2000 system (Illumina Inc., San Diego, CA, United States).
Assembly and Expression Analysis
Adapters and low-quality bases were removed from the raw data by using Cutadapt (Martin, 2011). The combined reads of all six samples of C. antiqua as well as the reads of H. circularisquama were assembled using Trinity software (Grabherr et al., 2011). The original reads of C. antiqua samples were mapped onto the contigs by using Bowtie2 (Langmead and Salzberg, 2012), and then the abundance of each contig was estimated using RSEM (Li and Dewey, 2011). The degree of differential gene expression between day and night samples was evaluated using EdgeR (Robinson et al., 2010). The raw read transcriptome sequences were submitted to the DDBJ Sequence Read Archive database under BioProject IDs PRJDB7469 and PRJDB7513. The assembled and annotated sequence data can be obtained from the web site http://hab.nibb.ac.jp.
Gene Annotation
For each contig of C. antiqua, coding sequences (CDS) were extracted using TransDecoder1. Similarity searches were conducted using BLASTX (criterion, E -value < 0.001) against the NCBI non-redundant protein (NR) database to extract top hits. Motifs and domains were searched using InterProScan (Jones et al., 2014). Gene ontologies (GOs) were assigned to each contig with the default parameter setting (E -value < 10–6), and the Fisher’s exact test implemented in Blast2go (Conesa et al., 2005) was used to identify significantly enriched GO terms in each differentially expressed gene set.
Comparative Genome/Transcriptome Analysis
The translated gene sets of C. antiqua and H. circularisquama were compared with those from three existing RNA-seq datasets of harmful algae and 10 existing genome sequences of various algae and a plant model organism, Arabidopsis thaliana (Supplementary Table S1). RNA sequence data of C. subsalsa and H. akashiwo were obtained from the NCBI Sequence Read Archive database (accession numbers, SRR1300240 and SRR1296916, respectively) and assembled using Trinity software. Contig sequences constructed from the RNA sequence of K. mikimotoi were obtained from the authors (Kimura et al., 2015). Coding sequences were extracted from the above K. mikimotoi contig data, and the contig data for C. antiqua and H. circularisquama (obtained in this study) by using TransDecoder software. For comparative analysis, draft genome sequences of the following microorganisms and algae—Emiliania huxleyi, Cyanidioschyzon merolae, Phaeodactylum tricornutum, Thalassiosira pseudonana, Nannochloropsis gaditana, Phytophthora sojae, Saprolegnia diclina, and Ectocarpus siliculosus—and complete genome sequences of A. thaliana and Chlamydomonas reinhardtii, were obtained from the NCBI. The DomClust program (Uchiyama, 2006) implemented in the RECOG system (Uchiyama, 2017)2 was used to identify orthologous relationships among the proteins identified in the above genomes and transcriptomes. RECOG was also used to manage and visualize the data obtained from comparative analysis among C. antiqua and the other organisms in the search for orthologous genes related to photoreception, nutrient uptake, and superoxide production.
Phylogenetic Analysis
We inferred species phylogeny among the above organisms based on the concatenated protein sequence of the conserved and nearly one-to-one orthologs. For this purpose, we extracted orthologous groups (OGs) containing at least one or two genes for each genome and at least one gene for each transcriptome (we considered no upper limit for transcriptome data because they generally contain multiple isoforms from a single gene, which can generate more apparent “paralogs” than reality). We selected the longest sequence if multiple genes existed in the organism. Multiple sequence alignments were then constructed using MAFFT (Katoh and Standley, 2013), conserved blocks were extracted using Gblocks (Castresana, 2000), and a phylogenetic tree was drawn based on the concatenated conserved block by using FastTree (Price et al., 2010).
We also conducted phylogenetic analyses on some orthologous genes related to photoreception, nutrient uptake, and superoxide production. The amino acid sequences of typical domains and motifs were aligned using ClustalX 2.1 with the default options (Thompson et al., 1997). Neighbor-Joining (NJ) trees were constructed with ClustalX 2.1 and MEGA software (Kumar et al., 2016). Maximum-likelihood (ML, Guindon and Gascuel, 2003) and most parsimony (MP) trees were determined using MEGA, based on various models; bootstrap analysis for NJ trees (1,000 replications), ML and NP trees (100 replications) was performed.
Results
Sequencing, de novo Assembly, and Annotation
RNA sequencing generated 186 million paired reads (2 × 101 bp) from daytime and nighttime C. antiqua cDNA libraries, yielding assembled sequences consisting of 67,823 contigs with a mean length of 1,091 bp and N50 of 1,838 bp. A total of 45.6 million paired reads (2 × 101 bp) were also generated from the H. circularisquama library, yielding assembled sequences consisting of 195,802 contigs with a mean length of 698 bp and N50 of 997 bp. The average GC contents of C. antiqua and H. circularisquama samples were 41.9 and 63.1%, respectively (Table 1).
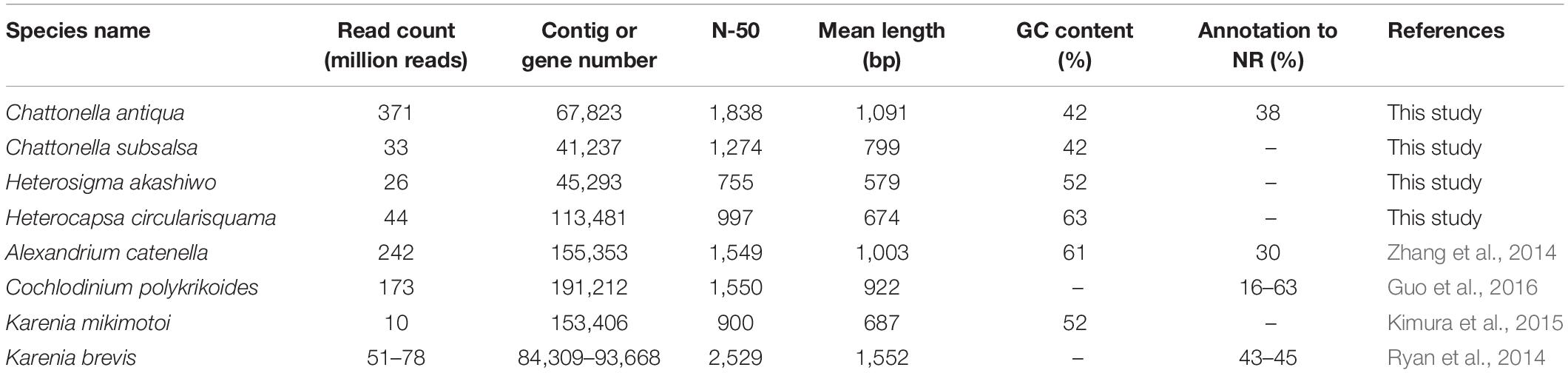
Table 1. Comparison of parameters in mRNA-seq among Chattonella antiqua and the other harmful flagellates.
TransDecoder identified 39,031 CDSs among the contigs of C. antiqua samples. Each CDS was then searched against the NCBI NR protein sequence database by using BLASTP with an e -value cutoff of 10–3, resulting in annotation of 25,808 CDSs (top-hit entries), of which 55% belonged to stramenopiles (Supplementary Figure S1).
GO analysis using GO slim (Gene Ontology Consortium, 2004) was performed to classify the predicted functions of C. antiqua transcripts. Of the 39,031 CDSs identified, 4,966 (13%) were assigned with one or more GO terms in the biological process category, 3,854 (10%) in the cellular component category, and 6,006 (15%) in the molecular function category.
To evaluate the completeness of the gene set obtained from C. antiqua transcriptome analysis, we conducted Benchmarking Universal Single-Copy Orthologs (BUSCO) analysis (Simao et al., 2015). Of the 303 BUSCOs in the “eukaryota_odb9” dataset, 93.4% were complete (either single copy or duplicated) in the C. antiqua gene set (Supplementary Figure S2). This percentage was much larger than those for other gene sets obtained from mRNA-seq analyses of raphidophytes and dinoflagellates causing noxious red tides (Supplementary Table S1) and was comparable or even better those for gene sets obtained from genome sequence analyses of other kinds of algae, protists, and a plant (Supplementary Table S1), supporting the better coverage of the present RNA-seq study than previous ones.
Comparative Genome and Transcriptome Analysis Among Various Algae
To identify orthologous relationships among the proteins, we performed DomClust orthologous clustering analysis of the 15 proteome sequence sets listed in Supplementary Table S1. We identified 148,421 OGs (including singletons) among which 18,039 contained C. antiqua genes, and we examined the presence/absence patterns of orthologs among various algae (Supplementary Figure S3). The number of shared OGs between two genomes is likely to reflect the closeness between them. As expected, C. subsalsa and H. akashiwo belonging to Raphidophyceae had the largest numbers of shared OGs with C. antiqua (Supplementary Figure S4). The next organism that had the largest shared OGs with C. antiqua was Ectocarpus siliculosus.
We then constructed a phylogenetic tree based on concatenated alignment of universally conserved and nearly one-to-one OGs (Supplementary Figure S5). There were 97 OGs satisfying this condition, yielding a concatenated alignment of length 8487 amino acids. The phylogenetic position of C. antiqua was consistent with the above observation from the shared OG analysis.
Genes Differentially Expressed Between Daytime and Nighttime
We evaluated the expression level of each gene (isoforms of the same gene were merged) in each sample to identify genes that were differentially expressed between daytime and nighttime. As a result, 1,777 and 658 genes were identified as significantly upregulated during the daytime and nighttime, respectively (false discovery rate [FDR] ≤ 0.001). These gene sets were functionally characterized on the basis of GOslim term assignment in terms of biological process (Supplementary Figure S6), cellular component (Supplementary Figure S7), and molecular function (Supplementary Figure S8). We compared four gene sets: all genes, highly expressed genes (the top 1,000 most highly expressed genes in the sample combined with gene sets of daytime and nighttime), genes significantly upregulated during the daytime, and genes significantly upregulated during the nighttime. Some GO terms uniquely appeared in the most frequent categories in either the daytime-specific gene set (highlighted red in Supplementary Figures S6c, S7c, S8c) or nighttime-specific gene set (highlighted blue in Supplementary Figures S6d, S7d, S8d). We also performed Fisher’s exact tests to identify significantly enriched GO terms in either of the differentially expressed gene sets (Supplementary Table S2). GO terms of biological processes enriched in the daytime-specific gene set included “photosynthesis” (GO:0015979; FDR = 2.6 × 10–6) and “carbohydrate metabolic process” (GO:0005975; FDR = 2.2 × 10–9), and those enriched in the nighttime-specific gene set included “cofactor metabolic process” (GO:0051186; FDR = 4.9 × 10–4) and “sulfur compound metabolic process” (GO:0006790; FDR = 7.8 × 10–3).
Genes Related to Photoreception
Using BLASTP and the RECOG system, we searched for genes related to the photosynthetic pigment pathway and light signal transduction in C. antiqua and other red-tide flagellates. First, we searched for genes related to photosynthetic pigment biosynthesis with Arabidopsis amino acid sequences as the reference (Castelfranco and Beale, 1983). Almost all genes encoding enzymes required for biosynthesis of chlorophyll a were found in C. antiqua, as well as the other red-tide flagellates (Supplementary Figure S9); percent identities and e -values for C. antiqua were 36–67% and ≤2.00E−42, respectively (Supplementary Data S1). Most of the genes encoding enzymes required for biosynthesis of carotenoids were also found in C. antiqua and the other red-tide flagellates (Supplementary Figure S10); percent identities and e -values for C. antiqua were 25–63% and ≤3.00E−12, respectively (Supplementary Data S1). Among genes encoding hydroxylases that convert β-carotene to zeaxanthin, genes for a cytochrome P450 monooxygenase (CYP97 type) was found in C. antiqua and the other red-tide flagellates, but those encoding a nonheme di-iron hydroxylase (BCH type) were not found in any red-tide flagellates. Phylogenetic trees of CYP97-type hydroxylase genes are shown in Figure 1 (ML) and Supplementary Figure S11 (NJ, MP). In these trees, the CYP97-type hydroxylase genes seemed to be divided into two groups: algae and plants. In the algae group, the orthologs of the C. antiqua CYP97-type hydroxylase gene formed a clade together with the other raphidophytes with strong bootstrap support and were located near clades containing other stramenopiles such as diatoms and brown algae. Genes encoding neoxanthin synthase (NSY) were not found in any red-tide flagellate.
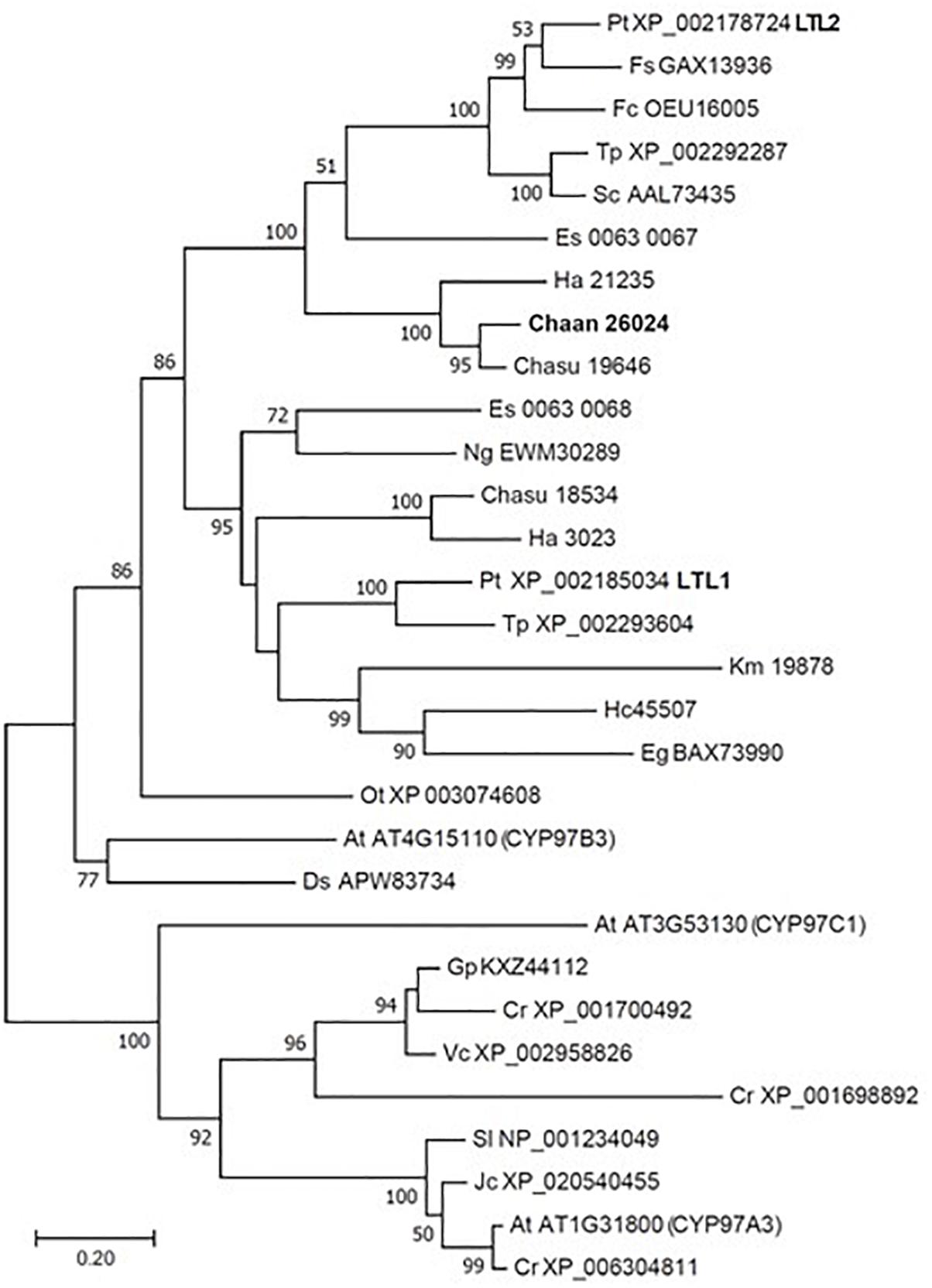
Figure 1. Maximum-likelihood phylogenetic tree of cytochrome P450 monooxygenase (CYP97 type) genes. The tree was inferred using the LG+G model. Numbers represent support values (>50%) obtained with 100 bootstrap replicates. Each gene is labeled with the abbreviated scientific name and accession ID. Abbreviations of scientific names are as follows: (Raphidophyceae, stramenopiles) Chaan, Chattonella antiqua; Chasu, Chattonella subsalsa; Ha, Heterosigma akashiwo; (Bacillariophyceae, stramenopiles); Fs, Fistulifera solaris; Fc, Fragilariopsis cylindrus; Pt, Phaeodactylum tricornutum; Sc, Skeletonema costatum; Tp, Thalassiosira pseudonana; (Eustigmataceae, stramenopiles) Ng, Nannochloropsis gaditana; (Phaeophyceae, stramenopiles) Es, Ectocarpus siliculosus; (Brassicaceae) At, Arabidopsis thaliana; (Bathycoccaceae) Ot, Ostreococcus tauri; (Chlorophyceae) Ds, Dunaliella salina; Gp, Gonium pectoral; Vc, Volvox carteri; Cr, Chlamydomonas reinhardtii; (Dinophyceae) Hc, Heterocapsa circularisquama; Km, Karenia mikimotoi; (Euglenaceae) Eg, Euglena gracilis; (Euphorbiaceae) Jc, Jatropha curcas; (Solanaceae) Sl, Solanum lycopersicum.
Next, we searched for genes encoding photoreceptors, such as blue-absorbing light–oxygen–voltage (LOV) protein, the CRY/PHR family (CPF), and red-absorbing phytochrome (Table 2). By using reference sequences of the xanthophycean algae Vaucheria frigida, from which aureochrome was first isolated (Takahashi et al., 2007), orthologous aureochrome genes with a LOV domain and a basic region/leucine zipper (bZIP) domain were identified in raphidophytes only; percent identities and e -values for C. antiqua were 41–63% and ≤3.00E-52, respectively (Supplementary Data S2). Several genes with LOV domains were found in red-tide dinoflagellates, but no gene for aureochrome was found. Phylogenetic trees of aureochrome genes were constructed using orthologous genes in red-tide flagellates and other stramenopiles (Figure 2 [ML] and Supplementary Figure S12 [NJ, MP]). In the ML tree, four groups were formed, and the aureochrome genes of raphidophytes were divided into each group. Within each group, aureochrome genes of the three raphidophytes formed a clade, with strong bootstrap support.
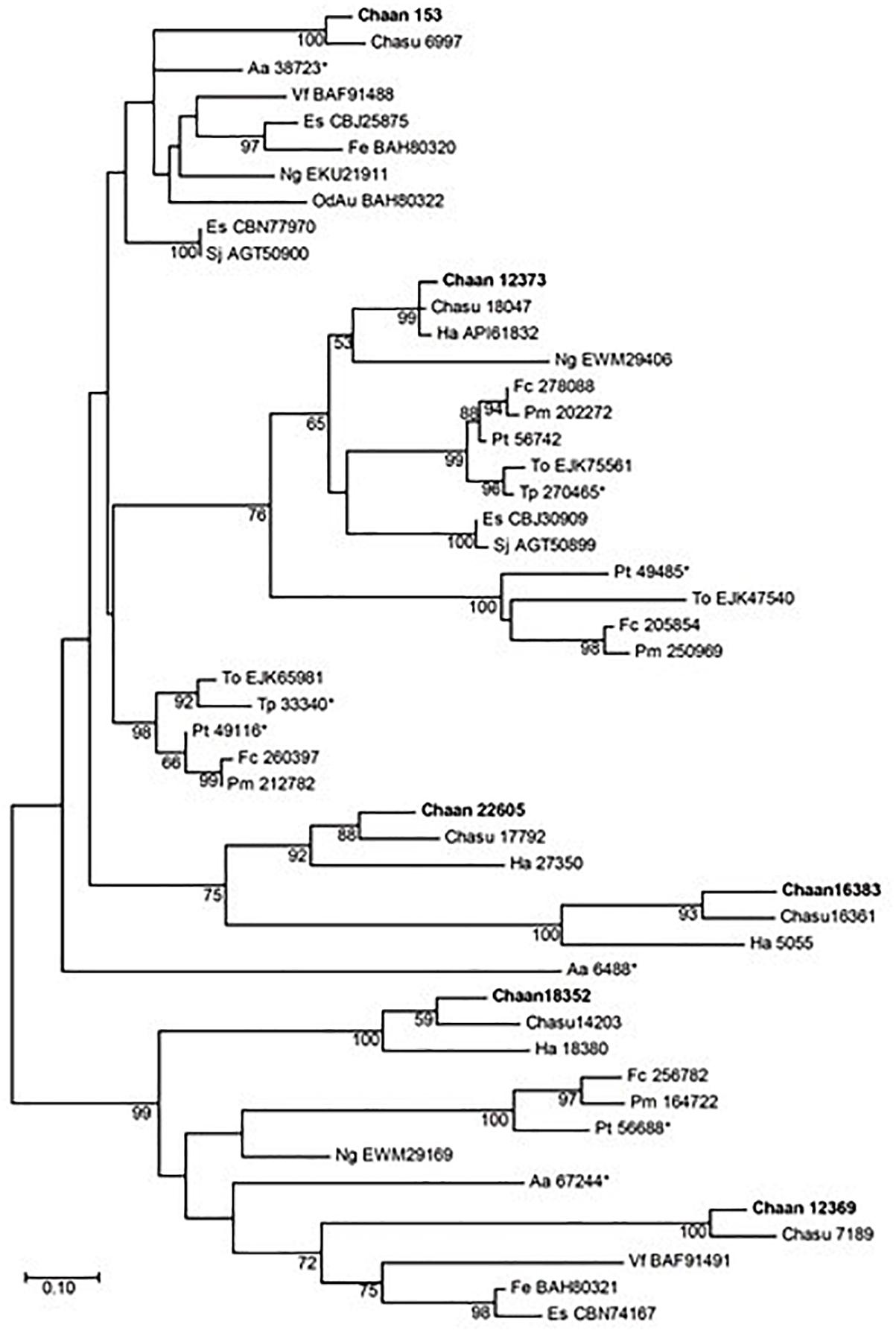
Figure 2. The maximum-likelihood phylogenetic tree of aureochrome genes. The tree was inferred using the Le_Gascuel_2008+gamma model. Numbers represent support values (>50%) obtained with 100 bootstrap replicates. Each gene is labeled with the abbreviated scientific name and accession ID. Sequences with asterisks were obtained from Costa et al. (2013). Abbreviations of scientific names are as follows: (Raphidophyceae) Chaan, Chattonella antiqua; Chasu, Chattonella subsalsa; Ha, Heterosigma akashiwo; (Bacillariophyceae) Fc, Fragilariopsis cylindrus; Pt, Phaeodactylum tricornutum; Pm, Pseudo-nitzschia multiseries; To, Thalassiosira oceanica; (Chrysophyceae) Od, Ochromonas danica; (Eustigmataceae) Ng, Nannochloropsis gaditana; (Pelagomonadaceae) Aa, Aureococcus anophagefferens; (Phaeophyceae) Es, Ectocarpus siliculosus; Fe, Fucus evanescens; Sj, Saccharina japonica; (Xanthophyceae) Vf, Vaucheria frigida.
Comparison with Arabidopsis references revealed orthologous genes of CPF in all five red-tide flagellates examined. In the phylogenic tree, CPF contains four superclasses: 6-4pyrimidine pyrimidone dimer (6-4PHR), the superclass including plant CRY, plant CRY-like, and Class I and III CPDs, CRY-DASH, and the Class II cyclobutane pyrimidine dimer (CPD) photolyases (Fortunato et al., 2015). We constructed phylogenetic trees of CPF genes using orthologous genes in red-tide flagellates and other stramenopiles (Figure 3 and Supplementary Figures S13-1 [ML], S13-2 [NJ, MP]). In these trees, the seven CPF genes of C. antiqua seemed to be divided into 6-4PHR, plant CRY or CPD Class III, CPD II, and CRY-DASH (Figure 3); percent identities and e -values were 21–50% and ≤1E+00, respectively, in C. antiqua and other raphidophytes (Supplementary Data S2). The photolyase homology region (PHR) was found in all CPF orthologous genes of C. antiqua. Methylenetetrahydrofolate reductase (MHFR) domain was found in N-terminal site of Chaan_17231, which was nested in the superclass including plant CRY, plant CRY-like, and Class I and III CPDs. transcription elongation factor S-II (TFIIS) domain was found in N-terminal site of Chaan_9990, which was nested in the CPD Class II.
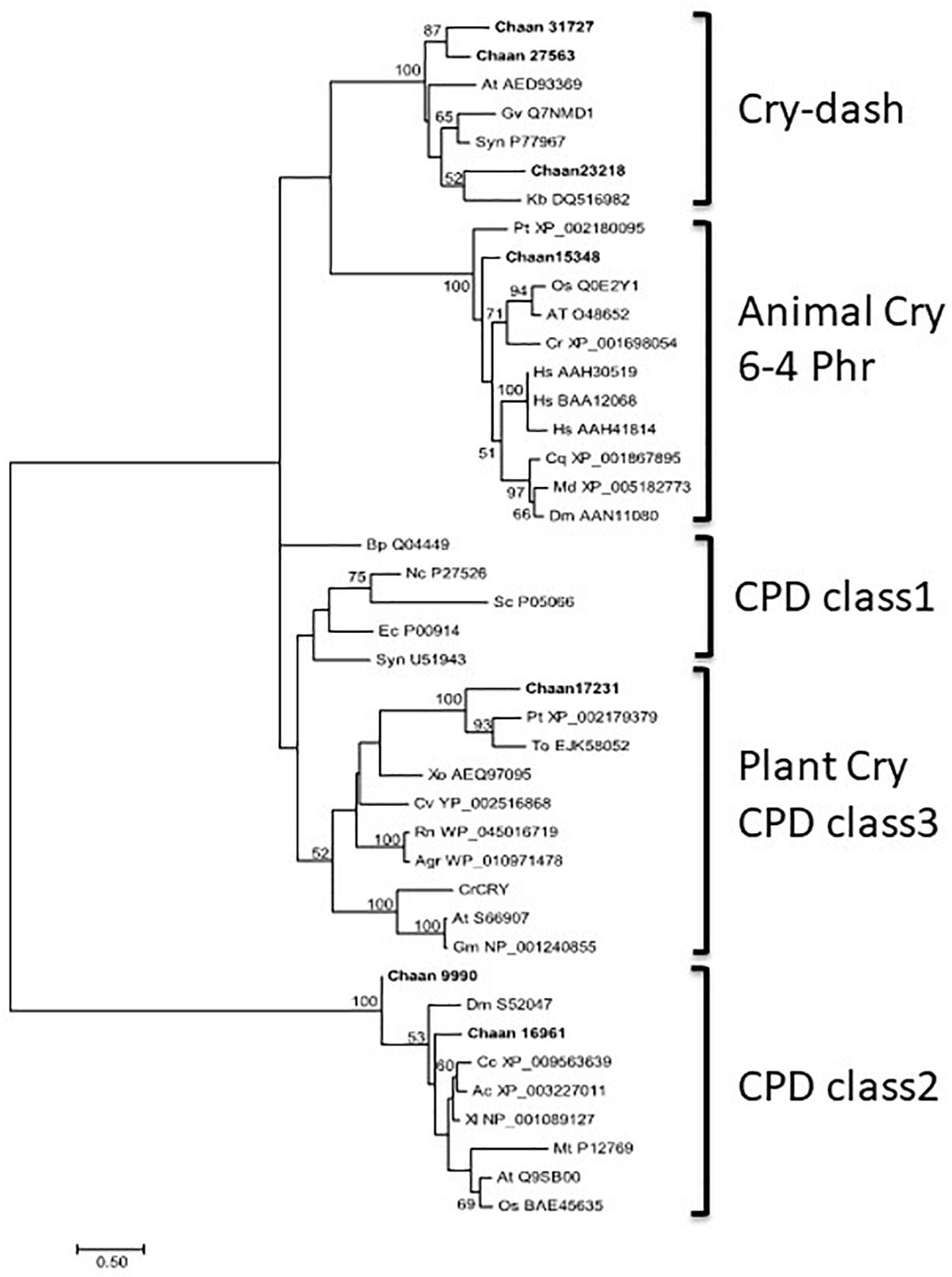
Figure 3. Maximum-likelihood phylogenetic tree of the photolyase/cryptochrome family. The tree was inferred using the LG+G model. Numbers represent support values (>50%) obtained with 100 bootstrap replicates. Each gene is labeled with the abbreviated scientific name and accession ID. Abbreviations of species names are as follows: (raphidophyte) Chaan, Chattonella antiqua; (diatom) Pt, Phaeodactylum tricornutum; To, Thalassiosira oceanica; (dinoflagellate) Kb, Karenia brevis; (green algae) Cr, Chlamydomonas reinhardtii; (plant) At, Arabidopsis thaliana; Gm, Glycine max; Os, Oryza sativa; (mammal) Hs, Homo sapiens; (bird) Cc, Cuculus canorus; (amphibia) Xl, Xenopus laevis; (reptile) Ac, Anolis carolinensis; (insect) Cq, Culex quinquefasciatus; Dm, Drosophila melanogaster; Md, Musca domestica; (fungi) Nc, Neurospora crassa; (yeast) Sc, Saccharomyces cerevisiae; (cyanobacteria) Gv, Gloeobacter violaceus; Syn, Synechocystis sp.; (bacteria) Agr, Agrobacterium tumefaciens; Bp, Bacillus pseudofirmus; Ec, Escherichia coli; (proteobacteria) Cv, Caulobacter vibrioides; Rn, Rhizobium nepotum; Xo, Xanthomonas oryzae; (Archaea) Mt, Methanothermobacter thermautotrophicus.
Using Arabidopsis reference sequences, four orthologous genes of the phytochrome family were identified in C. antiqua; percent identities and e -values were 23–27% and ≤1.00E−30, respectively (Supplementary Data S2). Using domain search by NCBI, we found no phy-GAF domain, but a domain COG4251 (Bacteriophytochrome: light-regulated signal transduction histidine kinase) which diatom phytochromes with photoreception ability (Fortunato et al., 2016) contain in Chattonella phytochromes. Moreover, some cysteine residues were found in the N-terminal photosensory module, indicating that chromophore can bind there. No phytochrome gene was found in other red-tide flagellates than C. antiqua and C. subsalsa. Phylogenetic trees of phytochrome genes were constructed using orthologous genes in C. antiqua and other organisms (Figure 4 [ML] and Supplementary Figure S14 [NJ, MP]). In the ML tree, all phytochrome genes of C. antiqua formed a clade located next to the clade of diatom phytochrome genes.
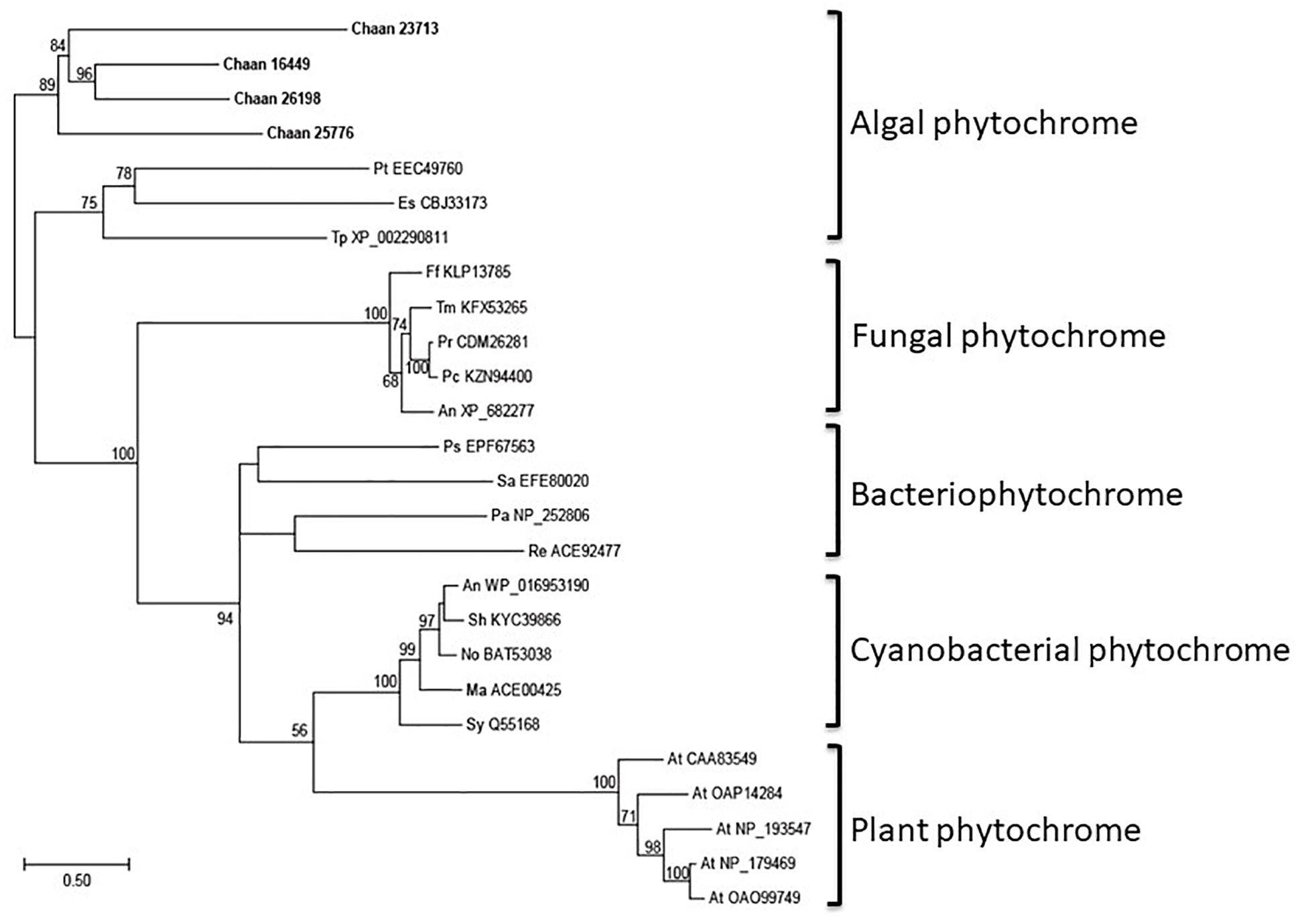
Figure 4. The maximum-likelihood phylogenetic tree of phytochrome family genes. The tree was inferred using the LG+G+I model. Numbers represent support values (>50%) obtained with 100 bootstrap replicates. Each gene is labeled with the abbreviated scientific name and accession ID. Abbreviations of species names are as follows: (raphidophyte) Chaan, Chattonella antiqua; (brown algae) Es, Ectocarpus siliculosus; (diatom) Pt, Phaeodactylum tricornutum; Tp, Thalassiosira pseudonana; (plant) At, Arabidopsis thaliana; (fungi) An, Aspergillus nidulans; Ff, Fusarium fujikuroi; Pc, Penicillium chrysogenum; Pr, Penicillium roqueforti; Tm, Talaromyces marneffei; (cyanobacteria) Ma, Microcystis aeruginosa; No, Nostoc sp.; Sh, Scytonema hofmannii; Sy, Synechocystis sp.; (bacteria) An, Anabaena sp.; Ps, Pseudomonas syringae; Sa, Streptomyces albus; (proteobacteria) Pa, Pseudomonas aeruginosa; Re, Rhizobium etli.
Genes Related to Nutrient Uptake and Initial Metabolism
We then searched for genes related to the uptake of and initial steps in the metabolism of nitrogen and phosphorus in C. antiqua and the other red-tide flagellates by using reference sequences of a model microalga, Chlamydomonas reinhardtii (chlorophyte) (Sanz-Luque et al., 2015; Dyhrman, 2016; Supplementary Data S3). As observed for the other red-tide flagellates, orthologous genes encoding most of the enzymes required for uptake and initial metabolism of nitrogen and phosphorus were found in C. antiqua (Figures 5, 6); percent identities and e -values were 28–57% and ≤9E-13 for nitrogen-related genes and 24–54% and ≤9E-10 for phosphorus-related genes (Supplementary Data S3). Orthologous genes of alkaline phosphatase (APA), which is required for utilization of organic phosphorus, were found in C. antiqua, K. mikimotoi, and H. circularisquama. Multi-sequence alignment showed that four motifs (Lin et al., 2015) are conserved in the APA genes of these red-tide flagellates (Supplementary Figure S15). Phylogenetic trees of APA genes were constructed using orthologous genes in C. antiqua and other red-tide flagellates (Figure 7 [ML] and Supplementary Figure S16 [NJ and MP]). In the phylogenetic trees, the APA genes of red-tide flagellates formed a clade with the same or a closely related taxonomic group: for instance, the APA gene of C. antiqua was nested in the same clade as APA genes of diatoms.
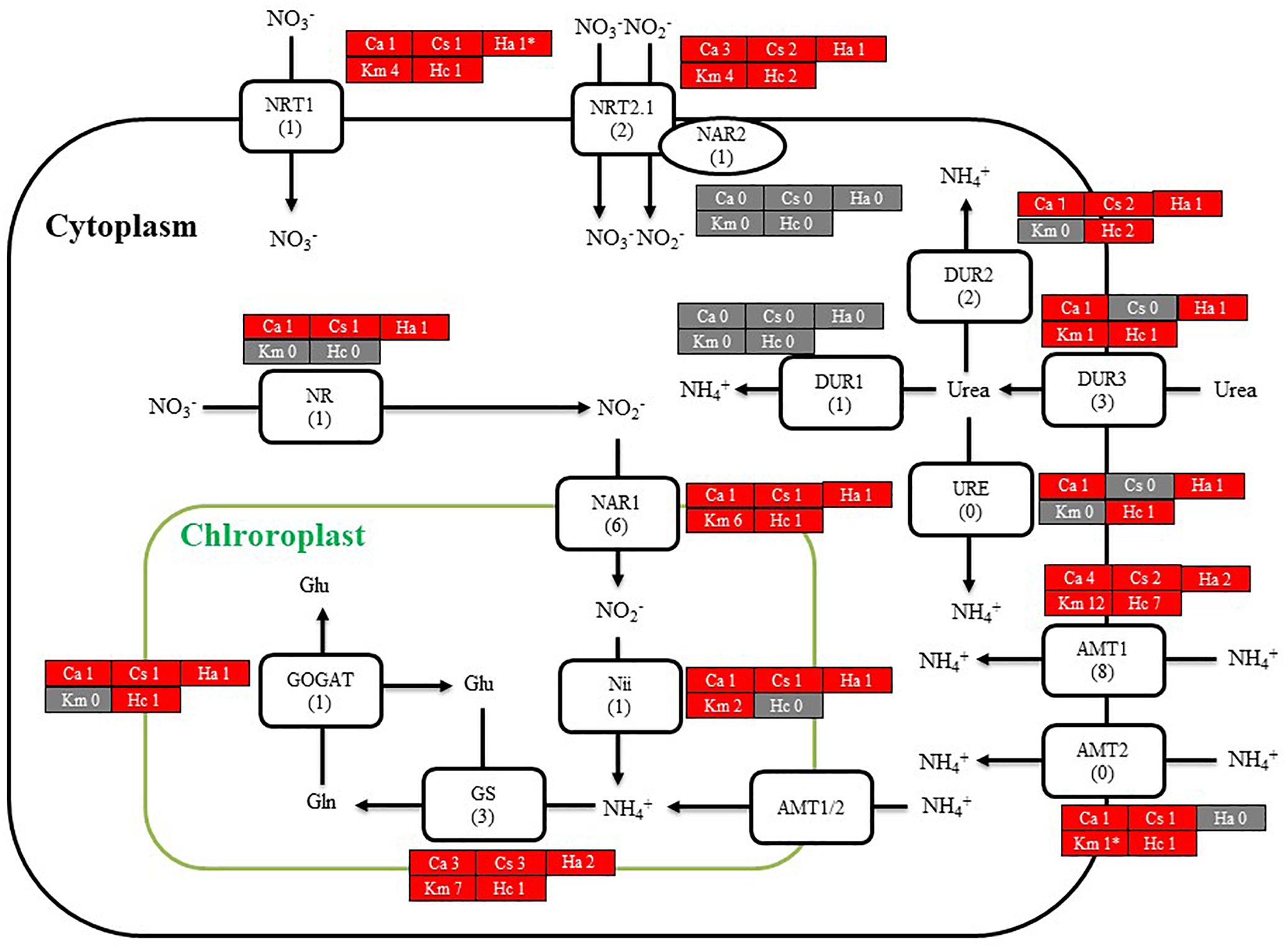
Figure 5. A cell model illustrating the uptake and initial metabolism of nitrogen in Chattonella antiqua (Ca), Chattonella subsalsa (Cs), Heterosigma akashiwo (Ha), Karenia mikimotoi (Km), and Heterocapsa circularisquama (Hc). Red and gray boxes indicate that orthologous genes were found and not found, respectively, in the RNA-seq data. The numbers next to a species name and below a protein name represent the gene numbers of each species and a model alga, Chlamydomonasreinhardtii. Abbreviations of protein names are as follows: AMT, ammonium transporter; DUR, urea transporter; GOGAT, glutamine oxoglutarate amino transferase; GS, glutamine synthetase; NAR, nitrate assimilation-related component; Nii, nitrite reductase; NR, nitrate reductase; NRT, nitrate transporter; and URE, urease.
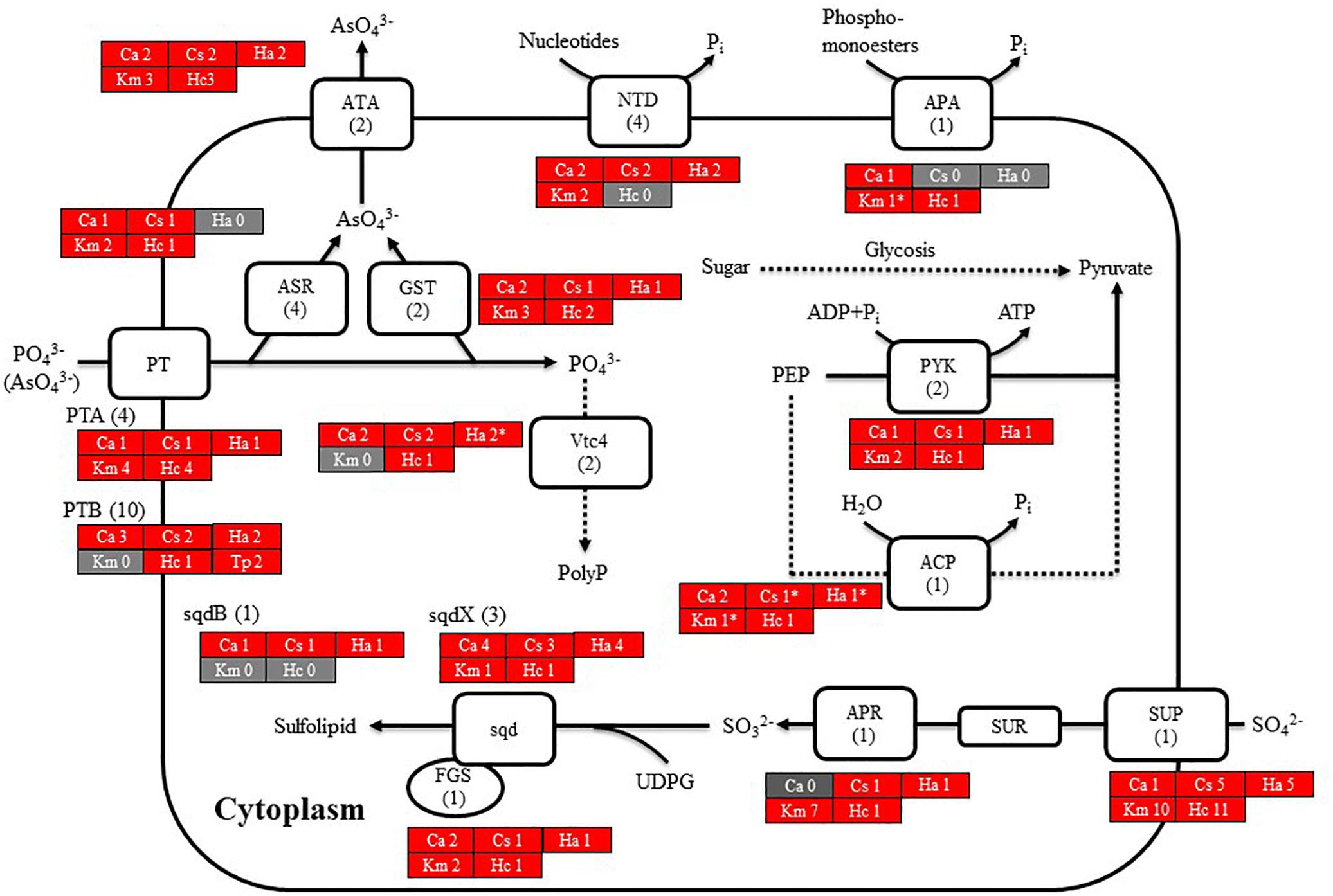
Figure 6. Cell model illustrating the uptake and initial metabolism of phosphate and sulfur compound in Chattonella antiqua (Ca), Chattonella subsalsa (Cs), Heterosigma akashiwo (Ha), Karenia mikimotoi (Km), and Heterocapsa circularisquama (Hc). Red and gray boxes indicate that orthologous genes were found and not found, respectively, in the RNA-seq data. The numbers next to a species name and below a protein name represent the gene numbers of each species and a model alga, Chlamydomonasreinhardtii. Abbreviations of protein names are as follows: ACP, acid phosphatase; APA, alkaline phosphatase; APR, adenosine-5′-phosphosulfate reductase; ASR, arsenate reductase; ATA, arsenite translocating ATPase; FGS, ferredoxin-dependent glutamate synthase; GST, glutathione S-transferase; NTD, 5′-nucleotidase; sqd, sulfolipid SQDG biosynthesis protein; PT, proton/phosphate symporter; PYK, pyruvate kinase; and Vtc4, vacuolar transporter chaperone 4.
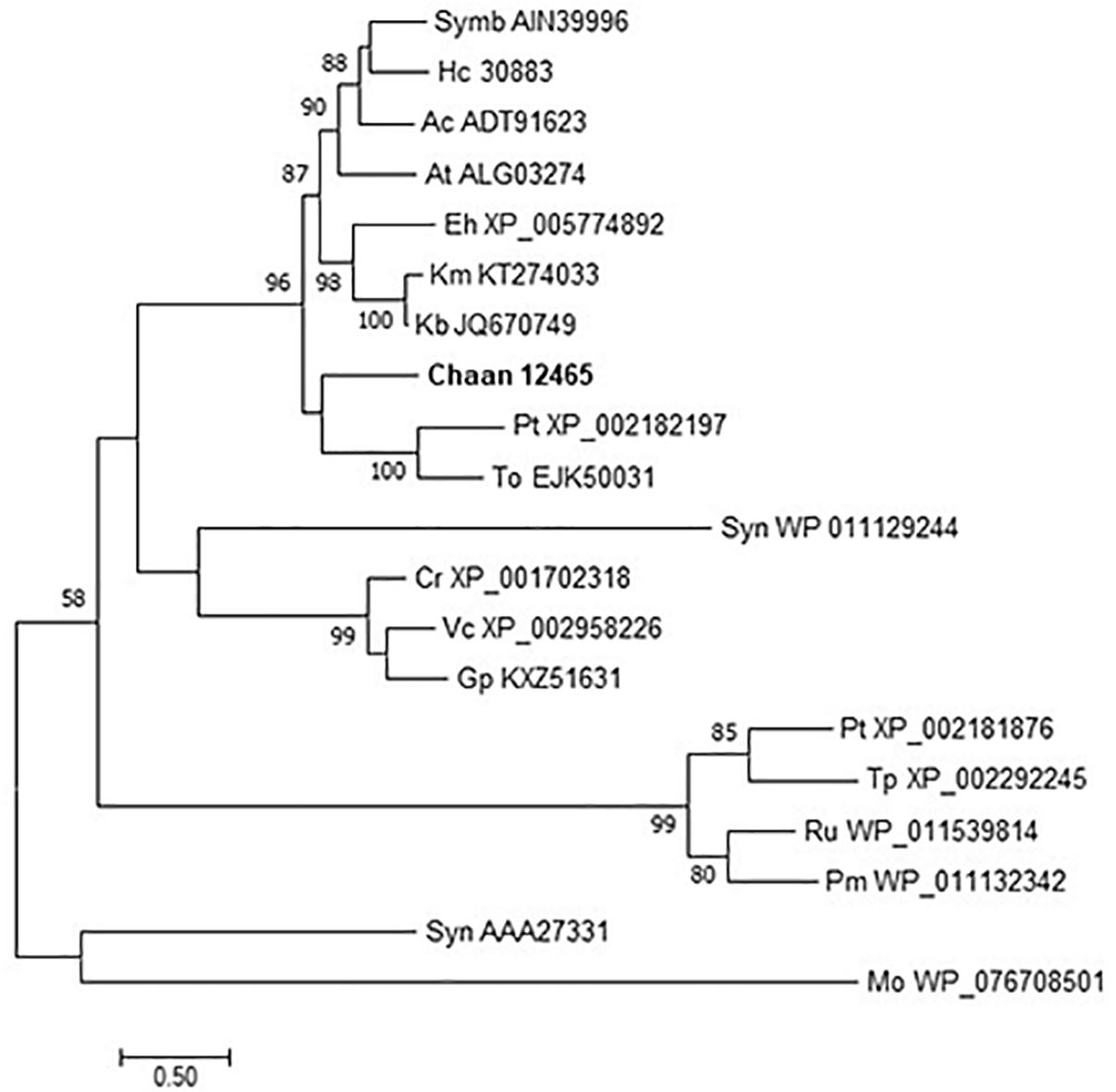
Figure 7. The maximum-likelihood phylogenetic tree of alkaline phosphatase genes. The tree was inferred using the WAG+G model. Numbers represent support values (>50%) obtained with 100 bootstrap replicates. Each gene is labeled with the abbreviated scientific name and accession ID. Abbreviations of species names are as follows: (Raphidophyceae) Chaan, Chattonella antiqua; (Bacillariophyceae) Pt, Phaeodactylum tricornutum; To, Thalassiosira oceanica; Tp, Thalassiosira pseudonana; (Chlorophyceae) Cr, Chlamydomonas reinhardtii; Gp, Gonium pectoral; Vc, Volvox carteri; (Dinophyceae) At, Alexandrium tamarense; Ac, Amphidinium carterae; Hc, Heterocapsa circularisquama; Kb, Karenia brevis; Km, Karenia mikimotoi; Symb, Symbiodinium sp.; (Prymnesiophyceae) Eh, Emiliania huxleyi; (cyanobacteria) Syn, Synechocystis sp.; Pm, Prochlorococcus marinus; Syn, Synechococcus sp.; (bacteria) Mo, Microbacterium oleivorans; and (proteobacteria) Ru, Ruegeria sp.
Genes Related to Ichthyotoxicity
To identify NOX genes in C. antiqua and the other red-tide flagellates we conducted searches using the sequence of Arabidopsis respiratory burst oxidase homolog protein A gene as the reference (Supplementary Data S3). Six NOX genes were found in C. antiqua; percent identities and e -values were 22–29% and ≤1.00E−14, respectively. In each of the other red-tide flagellates, two to four NOX genes were found. Multi-sequence alignment showed that four motifs, i.e., FAD-isoalloxazine binding site, motif 2, NADPH-ribulose binding site, and NADPH binding site (Finegold et al., 1996; Torres et al., 1998; Hervé et al., 2006) were conserved with high identities in the NOX genes of C. antiqua (Supplementary Figure S17). However, an EF-hand domain was not found in any NOX genes in the red-tide flagellates. Analyses using the TMHMM Server v. 2.0 to predict transmembrane helices in proteins indicated that two of the six C. antiqua NOX genes had five or six transmembrane domains (TMDs) (Chaan 29325, 9302), and the others had 11 TMDs (Chaan 9627, 13055, 16934, 26008) (Figure 8 and Supplementary Figure S18). These two types of C. antiqua NOX genes fell into different clades in the ML phylogenic tree (Figure 8). Similarly, some NOX genes of the other red-tide flagellates, i.e., C. subsalsa and K. mikimotoi, contained several TMDs (Chasu 21390, 5581, Km 24021, 27871, 29037) and others contained more TMDs (Chasu 14907, 16972, Km 12137, 11538), which were phylogenetically divided into clades in the same manner as the C. antiqua genes. All NOX genes of H. circularisquama were the type with several TMDs.
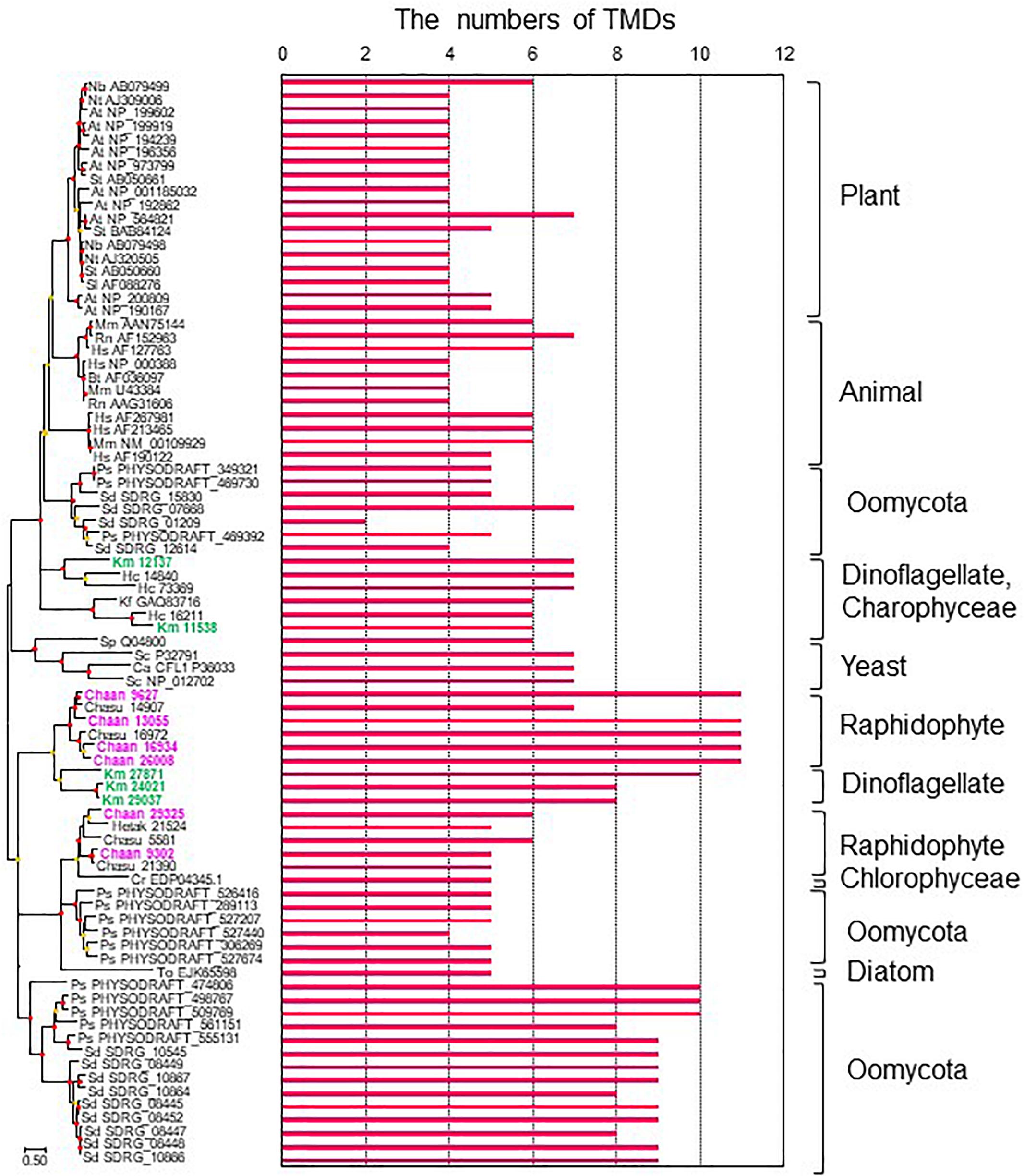
Figure 8. The maximum-likelihood phylogenetic tree and the numbers of transmembrane domains (TMDs) of NADPH oxidase genes. The tree was inferred using the LG+G+I model. Red and orange circles on nodes represent that the bootstrap value was ≥80% and 50–79%, respectively. Each gene is labeled with the abbreviated scientific name and accession ID. Abbreviations of species names are as follows: (Raphidophyceae) Chaan, Chattonella antiqua; Chasu, Chattonella subsalsa; Ha, Heterosigma akashiwo; (Bacillariophyceae) To, Thalassiosira oceanica; (Chlorophyceae) Cr, Chlamydomonas reinhardtii; (Dinophyceae) Km, Karenia mikimotoi; Hc, Heterocapsa circularisquama; (Klebsormidiophyceae) Kf, Klebsormidium flaccidum; (Oomycota) Ps, Phytophthora sojae; Sd, Saprolegnia diclina; (plant) At, Arabidopsis thaliana; Nb, Nicotiana benthamiana; Nt, Nicotiana tabacum; Sl, Solanum lycopersicum; St, Solanum tuberosum; (mammal) Bt, Bos Taurus; Hs, Homo sapiens; Mm, Mus musculus; Rn, Rattus norvegicus; (yeast) Sp, Schizosaccharomyces pombe; Sc, Saccharomyces cerevisiae; and (imperfect yeast) Ca, Candida albicans. The numbers of TMDs were predicted by TMHMM Server v. 2.0.
Discussion
In the present study, we conducted RNA-seq of Chattonella antiqua to identify genes with functions closely related to the dynamics of a noxious red tide. The read counts (371 million reads) were higher than those in previous studies of red-tide flagellates (10–242 million reads; Table 1). As a result, the contig number (84,309–191,212 contigs) was lower, and N50 and mean length of the contigs were slightly larger than those in the previous studies, with the exception of Karenia brevis (N50, ∼1,500 nt; mean length, ∼1,000, Ryan et al., 2014). This taken together with the results of the BUSCO analysis, where more than 90% of the eukaryotic BUSCOs were identified as complete in the C. antiqua gene set (Supplementary Figure S2), suggests that the larger number of reads used in the present study allowed us to obtain improved assembly of mRNA sequences of C. antiqua. However, by searching the NR database, we were able to assign functional annotations to only a small fraction of contigs (<40%). This was in part because no CDS was predicted in many contigs, suggesting the existence of non-coding RNAs, and in part because information on the genomes and functions of raphidophytes is poor.
In GO analysis, transcripts assigned to terms such as “photosynthesis” and “carbohydrate metabolic process” were significantly enriched in the genes expressed predominantly in the daytime (Supplementary Table S2). This was as expected because cellular carbon content in C. antiqua increases due to photosynthesis during the light period (Kohata and Watanabe, 1988). GO terms such as “DNA metabolic process” and “chromosome organization” uniquely appeared in the most frequent categories in the nighttime-specific gene set, although these terms were not significantly enriched (Supplementary Figure S6). Again, this was as expected because the nuclear DNA content of C. antiqua (G1/S transition) begins to increase ∼10 h after the onset of light irradiation, and cell division occurs during the nighttime (Nemoto and Furuya, 1985; Nemoto et al., 1987). The gene ontology data thus indicate that the sequence data roughly covered representative transcripts expressed during daytime and nighttime.
Using the reference sequences of model organisms and the RECOG system, orthologous genes related to processes important for red-tide development and toxicity—photosynthesis, photoreception, nutrient uptake, and superoxide production—were identified in the RNA-seq data of C. antiqua. As in other harmful red-tide species, orthologous genes of almost all enzymes required for biosynthesis of chlorophyll a and carotenoids were found in C. antiqua (Supplementary Figures S9, S10). In the carotenoid biosynthesis system, the NSY gene was not found in C. antiqua or other red-tide flagellates; this was as predicted because neoxanthin has not been detected in raphidophytes or dinoflagellates (Takaichi, 2011). Hydroxylation is required for the biosynthesis of xanthophylls from carotenes (Tian and DellaPenna, 2004). In plants, there are two types of carotenoid hydroxylase genes, i.e., CYP97 type and BCH type, which are reported to play a role in hydroxylation of α-carotene and β-carotene, respectively (Ruiz-Sola and Rodríguez-Concepción, 2012). Although raphidophytes and dinoflagellates do not produce α-carotene (Takaichi, 2011), only CYP97-type genes were found in the five red-tide algae. According to the phylogenetic tree (Figure 1), the C. antiqua CYP97-type gene belongs to the same clade as genes of lutein deficient like proteins (LUT) in diatoms (LTL1, LTL2), which are putative genes encoding CYP97-type enzymes involved in the hydroxylation of β-carotene (Coesel et al., 2008; Bertrand, 2010). Similarly, the CYP97-type genes in the other five red-tide species also belonged to the same clades as LTL1 and LTL2. These findings indicate that coastal phytoplankters such as raphidophytes, diatoms and dinoflagellates may universally utilize CYP97-type genes for hydroxylation of β-carotene.
Recently, a plethora of photoreceptor-like sequences from marine microalgae have been identified by omics approaches, and their functions and structures have been thoroughly analyzed (Jaubert et al., 2017). Some swimming behaviors, such as diurnal vertical migration and negative phototaxis, are regulated by specific spectral bands of light in C. antiqua and other red-tide flagellates (Shikata et al., 2013, 2015, 2016). The present study identified genes for blue light receptors such as aureochrome and CRY and a red/far-red light receptor phytochrome. In addition to the aureochrome gene reported previously (Ishikawa et al., 2009), the present study newly identified five aureochrome genes in C. antiqua (Figure 2). In H. akashiwo, one of the four aureochrome genes (Haaureo1) exhibits a clear diel rhythm, with the highest and the lowest transcript abundances occurring at dawn and dusk under a light:dark cycle, although the rhythm disappears under continuous dark conditions (Ji et al., 2017). Here, one of the aureochrome genes in C. antiqua (Chaan22605) was shown to belong to the same clade as Haaureo1. We cannot strictly compare the expression patterns between Chaan22605 and Haaureo1 because the light:dark cycle used by Ji et al. (2017) differed from that used here. Nevertheless, we observed that the expression level of Chaan22605 was significantly higher in the daytime (roughly corresponding to T4 in Ji et al., 2017) than in the nighttime (corresponding to T12 in Ji et al., 2017) (Supplementary Data S2), which corresponds with the expression rhythm of Haaureo1.
CPF contains photolyase and CRY. Under blue light irradiation, the photolyase repairs DNA damage caused by ultraviolet exposure. The photolyase has two types, i.e., CPD and 6-4PHR (Lin and Todo, 2005), of which CPD is phylogenetically divided into Class I (in prokaryotes) and Class II (in eukaryotes). CRY contributes to photomorphogenesis as a blue light receptor in plants and controls circadian rhythm in animals. According to sequence similarities and phylogenetical analyses, CRYs are clustered into three subfamilies: plant CRY, animal CRY, and CRY-DASH. CPF genes have also been reported in diatoms and dinoflagellates and are suspected of contributing to light-dependent regulation of the cell cycle and circadian rhythm (Brunelle et al., 2007; Oliveri et al., 2014). Here, C. antiqua genes encoding 6-4PHR, plant CRY or CPD Class III, CPD Class II, and CRY-DASH were identified (Figure 3).
All phytochrome genes of C. antiqua seem to be nested within a clade of algal phytochrome genes (Figure 4). As in plants, physiological phenomena of microalgae are regulated by red light. Precision of geotaxis in the green alga C. reinhardtii, and alterations of cell speed during phototaxis of benthic diatoms, are regulated by red light (Sineshchekov et al., 2000; McLachlan et al., 2009). It is suspected that phytochromes may contribute to these physiological phenomena, because C. reinhardtii and diatoms have phytochrome genes (Bowler et al., 2008). In C. antiqua, the negative geotaxis during the daytime disappears under red light, regardless of irradiation direction (Shikata et al., 2016). These findings suggest that photoreceptors, such as aureochrome, CPF, and phytochrome, may be involved in the regulation of swimming behaviors in C. antiqua; however, biochemical and functional analyses such as transformation techniques and dsRNA transfection (RNAi) are required to verify the contribution of photoreceptors to each swimming trait.
Chattonella antiqua can utilize dissolved inorganic nitrogen (nitrate, nitrite, and ammonium) and urea as a nitrogen source for growth (Nakamura and Watanabe, 1983; Fukao et al., 2007), and dissolved inorganic phosphorus as a phosphorus source (Nakamura and Watanabe, 1983). The present study revealed orthologous genes of transporters and enzymes required for uptake and primary metabolism of nitrogen and phosphorus (Figures 5, 6). Orthologs of transporters for uptakes of sulfate ion and enzymes for synthesis of sulfolipid (sulfoquinovosyldiacylglycerol) which act as a substitute for phospholipids under phosphorus deficient conditions (Dyhrman, 2016) were also identified (Figure 6).
Nakamura (1985) reported that C. antiqua cannot utilize dissolved organic phosphorus and has no APA activity; however, an ortholog gene of APA was found in the present study. It has been reported that C. marina and C. ovata can grow not only with inorganic phosphorus but also with some kinds of dissolved organic phosphorus (Yamaguchi et al., 2008; Wang et al., 2011). Opinions about the ability to utilize dissolved organic phosphorus and to produce APA in H. akashiwo differ (Yamaguchi et al., 2004; Wang et al., 2011; Wang and Liang, 2015), but no ortholog gene of APA was detected in the RNA-seq data of H. akashiwo in the present study. On the other hand, Haley et al. (2017) reported that expression level of the APA gene was significantly increased under low phosphorus conditions in H. akashiwo. Differences in culture conditions may change a level of the gene expression of APA, and the ability to produce APA may be different among each culture strains.
To our knowledge, the present study is the first report on putative sequences of NOX genes of raphidophytes, including C. antiqua, although a previous report of a Southern blot analysis has suggested the presence of a gene encoding a gp91phox homolog in C. marina (Kim et al., 2000). Moreover, we also found multiple NOX genes in the other four red-tide flagellates examined. We observed that none of the NOX genes of the five red-tide flagellates had an EF-hand domain; absence of such a domain appears to be a common feature in algal NOX genes (Anderson et al., 2011). Ca2+ signaling through the EF-hand domain contributes to regulation of ROS production in plants (Sagi and Fluhr, 2006; Kadota et al., 2015), but algae may have another mechanism. Two types of NOX genes were found in C. antiqua : one with several TMDs and another with numerous TMDs (Figure 8). Interestingly, the red-tide dinoflagellate K. mikimotoi (Alveolata), which is not phylogenically close to C. antiqua (Stramenopiles) but kills fishes and produces relatively large amounts of ROS in the similar way (Yamasaki et al., 2004), also has NOX genes with numerous TMDs; these genes belong to the clade of C. antiqua NOX genes with numerous TMDs in the phylogenetic tree (Figure 8). Most animals and plants have multiple NOX genes with different induction patterns and functions (Canton and Grinstein, 2014). It is reported that the superoxide production level is higher during the light period than during the dark period in C. antiqua (Kim et al., 2005). Two genes of NOX with numerous TMDs (Chaan26008, Chaan16934) expressed significantly higher during the daytime than during the night time (Supplementary Data S4, FDR > 0.001). Chattonella cells stimulated with fish mucus and galacturonic acid increased the generation of superoxide (Nakamura et al., 1998; Kim et al., 2000). It is interesting how two types of Chattonella NOX work to produce superoxide.
In the present study, RNA-seq and comparative analyses with other organisms identified genes related to biosynthesis of pigments for photosynthesis, light signal transduction, nutrient uptake, and ichthyotoxicity of C. antiqua. The next step is functional analysis of each gene to enrich our understanding of the eco-physiological features of marine raphidophytes at the molecular level.
Data Availability
The datasets generated for this study can be found in DDBJ, BioProject IDs PRJDB7469 and PRJDB7513.
Author Contributions
TS designed the research projects, carried out the sampling from algal cultures, performed the phylogenic analyses, and wrote the manuscript. FT performed the phylogenic analyses and wrote a part of the manuscript. HN performed the bioinformatic analyses. ShS sequenced the mRNA of C. antiqua. YK designed the project and promoted acquirement of the research grants. SeS prepared the algal culture samples. KY and YN provided the information on photosynthesis and wrote a part of the section “Discussion.” YY provided the information on toxicity of red-tide flagellates and wrote a part of the section “Discussion.” IU performed the bioinformatic analyses and wrote a part of the manuscript. All authors read and approved the final manuscript.
Funding
This work was partially supported by the Japanese Fisheries Agency-aided project for addressing red tides and oxygen-depleted water masses, PRESTO Japan Science and Technology Corporation, a grant-in-aid for scientific research (17K07891), from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and NIBB Collaborative Research Program (#11-203, #12-204, #13-367, #14-380, #15-356, #16-441, #17-440, #18-460, #19-452) to TS.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Mineo Yamaguchi (Kitasato University), Prof. Masahiro Kasahara (Ritsumeikan University), and Dr. Rei Narikawa (Shizuoka University) for thoughtful comments and valuable information on eco-physiology of raphidophytes, photoreceptors, and NADPH oxidase. We are grateful to Dr. Yukihiko Matsuyama (Fisheries Research and Education Agency) for providing the culture strain of H. circularisquama. Computational resources were provided by the Data Integration and Analysis Facility, National Institute for Basic Biology.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01764/full#supplementary-material
Footnotes
References
Ahmad, M., and Cashmore, A. R. (1993). HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366, 162–166. doi: 10.1038/366162a0
Anderson, A., Bothwell, J. H., Laohavisit, A., Smith, A. G., and Davies, J. M. (2011). NOX or not? evidence for algal NADPH oxidases. Trends Plant Sci. 16, 579–581. doi: 10.1016/j.tplants.2011.09.003
Bertrand, M. (2010). Carotenoid biosynthesis in diatoms. Photosynth. Res. 106, 89–102. doi: 10.1007/s11120-010-9589-x
Bowler, C., Allen, A. E., Badger, J. H., Grimwood, J., Jabbari, K., Kuo, A., et al. (2008). The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244. doi: 10.1038/nature07410
Brunelle, S. A., Hazard, E. S., Sotka, E. E., and Dolah, F. M. V. (2007). Characterization of a dinoflagellate cryptochrome blue-light receptor with a possible role in circadian control of the cell cycle 1. J. Phycol. 43, 509–518. doi: 10.1111/j.1529-8817.2007.00339.x
Canton, J., and Grinstein, S. (2014). Priming and activation of NADPH oxidases in plants and animals. Trends Immunol. 35, 405–407. doi: 10.1016/j.it.2014.07.007
Castelfranco, P. A., and Beale, S. I. (1983). Chlorophyll biosynthesis: recent advances and areas of current interest. Annu. Rev. Plant Physiol. 34, 241–276. doi: 10.1146/annurev.pp.34.060183.001325
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Chen, L. C. M., Edelstein, T., and McLachlan, J. (1969). Bonnemaisonia hamifera harlot in nature and in culture. J. Phycol. 5, 211–220. doi: 10.1111/j.1529-8817.1969.tb02605.x
Coesel, S., Oborník, M., Varela, J., Falciatore, A., and Bowler, C. (2008). Evolutionary origins and functions of the carotenoid biosynthetic pathway in marine diatoms. PLoS One 3:e2896. doi: 10.1371/journal.pone.0002896
Conesa, A., Gotz, S., Garcia-Gomez, J. M., Terol, J., Talon, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Costa, B. S., Sachse, M., Jungandreas, A., Bartulos, C. R., Gruber, A., Jakob, T., et al. (2013). Aureochrome 1a is involved in the photoacclimation of the diatom Phaeodactylum tricornutum. PLoS One 8:e74451. doi: 10.1371/journal.pone.0074451
Dyhrman, S. T. (2016). “Nutrients and their acquisition: phosphorus physiology in microalgae,” in The physiology of Microalgae, eds M. A. Borowitzka, J. Beardall, and J. A. Raven (New York, NY: Springer), 155–183. doi: 10.1007/978-3-319-24945-2_8
Finegold, A. A., Shatwell, K. P., Segal, A. W., Klausner, R. D., and Dancis, A. (1996). Intramembrane bis-heme motif for transmembrane electron transport conserved in a yeast iron reductase and the human NADPH oxidase. J. Biol. Chem. 271, 31021–31024. doi: 10.1074/jbc.271.49.31021
Fortunato, A. E., Annunziata, R., Jaubert, M., Bouly, J. P., and Falciatore, A. (2015). Dealing with light: the widespread and multitasking cryptochrome/photolyase family in photosynthetic organisms. J. Plant Physiol. 172, 42–54. doi: 10.1016/j.jplph.2014.06.011
Fortunato, A. E., Jaubert, M., Enomoto, G., Bouly, J. P., Raniello, R., Thaler, M. J., et al. (2016). Diatom phytochromes reveal the existence of far-red-light-based sensing in the ocean. Plant Cell 28, 616–628. doi: 10.1105/tpc.15.00928
Fukao, T., Nishijima, T., Yamaguchi, H., and Adachi, M. (2007). Utilization of urea by the 6 species of red tide phytoplankton. Bull. Plankton Soc. JPN. 54, 1–8.
Furuya, M. (1993). Phytochromes: their molecular species, gene families, and functions. Annu. Rev. Plant Biol. 44, 617–645. doi: 10.1146/annurev.arplant.44.1.617
Gene Ontology Consortium (2004). The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32, D258–D261.
Glibert, P. M., Allen, J. I., Bouwman, A. F., Brown, C. W., Flynn, K. J., Lewitus, A. J., et al. (2010). Modeling of HABs and eutrophication: status, advances, challenges. J. Mar. Syst. 83, 262–275. doi: 10.1016/j.jmarsys.2010.05.004
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 29:644. doi: 10.1186/1471-2105-12-S14-S2
Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. doi: 10.1080/10635150390235520
Guo, R., Wang, H., Suh, Y. S., and Ki, J. S. (2016). Transcriptomic profiles reveal the genome-wide responses of the harmful dinoflagellate Cochlodinium polykrikoides when exposed to the algicide copper sulfate. BMC Genomics 17:29. doi: 10.1186/s12864-015-2341-3
Haley, S. T., Alexander, H., Juhl, A. R., and Dyhrman, S. T. (2017). Transcriptional response of the harmful raphidophyte Heterosigma akashiwo to nitrate and phosphate stress. Harmful Algae 68, 258–270. doi: 10.1016/j.hal.2017.07.001
Hervé, C., Tonon, T., Collén, J., Corre, E., and Boyen, C. (2006). NADPH oxidases in eukaryotes: red algae provide new hints!Curr. Genet. 49, 190–204. doi: 10.1007/s00294-005-0044-z
Imai, I., and Yamaguchi, M. (2012). Life cycle, physiology, ecology and red tide occurrences of the fish-killing Raphidophyte chattonella. Harmful Algae 14, 46–70. doi: 10.1016/j.hal.2011.10.014
Imai, I., Yamaguchi, M., and Hori, Y. (2006). Eutrophication and occurrences of harmful algal blooms in the seto inland sea Japan. Plankton Benthos Res. 1, 71–84. doi: 10.3800/pbr.1.71
Ishikawa, M., Takahashi, F., Nozaki, H., Nagasato, C., Motomura, T., and Kataoka, H. (2009). Distribution and phylogeny of the blue light receptors aureochromes in eukaryotes. Planta 230, 543–552. doi: 10.1007/s00425-009-0967-6
Ishimatsu, A., Sameshima, M., Tamura, A., and Oda, T. (1996). Histological analysis of the mechanisms of chattonella-induced hypoxemia in yellowtail. Fish. Sci. 62, 50–58. doi: 10.2331/fishsci.62.50
Jaubert, M., Bouly, J. P., d’Alcalà, M. R., and Falciatore, A. (2017). Light sensing and responses in marine microalgae. Curr. Opin. Plant Biol. 37, 70–77. doi: 10.1016/j.pbi.2017.03.005
Ji, N., Li, L., Lin, L., and Lin, S. (2017). Identification and expression analysis of blue light receptor aureochrome in the harmful alga Heterosigma akashiwo (raphidophyceae). Harmful Algae 61, 71–79. doi: 10.1016/j.hal.2016.11.016
Jones, P., Binns, D., Chang, H.-Y., Fraser, M., Li, W., McAnulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Kadota, Y., Shirasu, K., and Zipfel, C. (2015). Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol. 56, 1472–1480. doi: 10.1093/pcp/pcv063
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kim, D., Oda, T., Ishimatsu, A., and Muramatsu, T. (2000). Galacturonic-acid-induced increase of superoxide production in red tide phytoplankton chattonella marina and Heterosigma akashiwo. Biosci. Biotechnol. Biochem. 64, 911–914. doi: 10.1271/bbb.64.911
Kim, D., Watanabe, M., Nakayasu, Y., and Kohata, K. (2005). Changes in O2–and H2O2 production by chattonella antiqua during diel vertical migration under nutrient stratification. Aquat. Microb. Ecol. 39, 183–191. doi: 10.3354/ame039183
Kim, H. G. (2006). “Mitigation and controls of HABs,” in Ecology of Harmful Algae, Ecological Studies, eds E. Granéli and J. T. Turner (New York, NY: Springer), 327–338. doi: 10.1007/978-3-540-32210-8_25
Kimura, K., Okuda, S., Nakayama, K., Shikata, T., Takahashi, F., Yamaguchi, H., et al. (2015). RNA sequencing revealed numerous polyketide synthase genes in the harmful dinoflagellate Karenia mikimotoi. PLoS One 10:e0142731. doi: 10.1371/journal.pone.0142731
Kohata, K., and Watanabe, M. (1988). Diel changes in the composition of photosynthetic pigments and cellular carbon and nitrogen in Chattonella antiqua (raphidophyceae). J. Phycol. 24, 58–66. doi: 10.1111/j.1529-8817.1988.tb04456.x
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12:323. doi: 10.1186/1471-2105-12-323
Lin, X., Wang, L., Shi, X., and Lin, S. (2015). Rapidly diverging evolution of an atypical alkaline phosphatase (PhoAaty) in marine phytoplankton: insights from dinoflagellate alkaline phosphatases. Front. Microbiol. 6:868. doi: 10.3389/fmicb.2015.00868
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12.
McLachlan, D. H., Brownlee, C., Taylor, A. R., Geider, R. J., and Underwood, G. J. (2009). Light-induced motile responses of the estuarine benthic diatoms navicula perminuta and Cylindrotheca closterium (bacillariophyceae). J. Phycol. 45, 592–599. doi: 10.1111/j.1529-8817.2009.00681.x
Nakamura, A., Okamoto, T., Komatsu, N., Ooka, S., Oda, T., Ishimatsu, A., et al. (1998). Fish mucus stimurates the generation of superoxide anion by Chattonella marina and Heterosigma akashiwo. Fish. Sci. 64, 866–869. doi: 10.2331/fishsci.64.866
Nakamura, Y. (1985). Alkaline phosphatase activity of Chattonella antiqua and Heteroslgma akashiwo. Res. Rep. Natl. Inst. Environ. Stud. 80, 67–72.
Nakamura, Y., Umemori, T., and Watanabe, M. (1989). Chemical environment for red tides due to Chattonella antiqua Part 2. daily monitoring of the marine environment throughout the outbreak period. J. Oceanogr. Soc. JPN. 45, 116–128. doi: 10.1007/bf02108885
Nakamura, Y., and Watanabe, M. M. (1983). Growth characteristics of Chattonella antiqua Part 2, effects of nutrients on growth. J. Oceanogr. Soc. JPN. 39, 151–155. doi: 10.1007/bf02070258
Nemoto, Y., and Furuya, M. (1985). Inductive and inhibitory effects of light on cell division in Chattonella antiqua. Plant Cell Physiol. 26, 669–674. doi: 10.1093/oxfordjournals.pcp.a076956
Nemoto, Y., Kuroiwa, T., and Furuya, M. (1987). Photocontrol of nuclear DNA replication in Chattonella antiqua (raphidophyceae). Plant Cell Physiol. 28, 1043–1049.
Oda, T., Ishimatsu, A., Shimada, M., Takeshita, S., and Muramatsu, T. (1992). Oxygen-radical-mediated toxic effects of the red tide flagellate Chattonella marina on vibrio alginolyticus. Mar. Biol. 112, 505–509. doi: 10.1007/bf00356297
Oda, T., Nakamura, A., Shikayama, M., Kawano, I., Ishimatsu, A., and Muramatsu, T. (1997). Generation of reactive oxygen species by raphidophycean phytoplankton. Biosci. Biotech. Biochem. 61, 1658–1662. doi: 10.1271/bbb.61.1658
Oliveri, P., Fortunato, A. E., Petrone, L., Ishikawa-Fujiwara, T., Kobayashi, Y., Todo, T., et al. (2014). The cryptochrome/photolyase family in aquatic organisms. Mar. Genomics 14, 23–37. doi: 10.1016/j.margen.2014.02.001
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). Edger: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Ruiz-Sola, M. Á, and Rodríguez-Concepción, M. (2012). Carotenoid biosynthesis in arabidopsis: a colorful pathway. Arabidopsis Book 10:e0158. doi: 10.1199/tab.0158
Ryan, D. E., Pepper, A. E., and Campbell, L. (2014). De novo assembly and characterization of the transcriptome of the toxic dinoflagellate karenia brevis. BMC Genomics 15:888. doi: 10.1186/1471-2164-15-888
Sagi, M., and Fluhr, R. (2006). Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol. 141, 336–340. doi: 10.1104/pp.106.078089
Sanz-Luque, E., Chamizo-Ampudia, A., Llamas, A., Galvan, A., and Fernandez, E. (2015). Understanding nitrate assimilation and its regulation in microalgae. Front. Plant Sci. 6:899. doi: 10.3389/fpls.2015.00899
Satta, C. T., Padedda, B. M., Sechi, N., Pulina, S., Loria, A., and Lugliè, A. (2017). Multiannual chattonella subsalsa biecheler (raphidophyceae) blooms in a mediterranean lagoon (santa giusta lagoon, sardinia island, italy). Harmful Algae 67, 61–73. doi: 10.1016/j.hal.2017.06.002
Shikata, T., Matsunaga, S., Iseki, M., Nishide, H., Higashi, S., Kamei, Y., et al. (2013). Blue light regulates the rhythm of diurnal vertical migration in the raphidophyte red-tide alga Chattonella antiqua. J. Plankton Res. 35, 542–552. doi: 10.1093/plankt/fbt006
Shikata, T., Matsunaga, S., Kuwahara, Y., Iwahori, S., and Nishiyama, Y. (2016). Light spectrum regulates cell accumulation during daytime in the raphidophyte Chattonella antiqua causing noxious red tides. J. Photochem. Photobiol. B Biol. 160, 128–133. doi: 10.1016/j.jphotobiol.2016.03.046
Shikata, T., Matsunaga, S., Nishide, H., Sakamoto, S., Onistuka, G., and Yamaguchi, M. (2015). Diurnal vertical migration rhythms and their photoresponse in four phytoflagellates causing harmful algal blooms. Limnol. Oceanogr. 60, 1251–1264. doi: 10.1002/lno.10095
Shikata, T., Nagasoe, S., Matsubara, M., Yoshikawa, S., Yamasaki, Y., Shimasaki, Y., et al. (2008). Factors influencing the initiation of blooms of the raphidophyte Heterosigma akashiwo and the diatom skeletonema costatum in a port in japan. Limnol. Oceanogr. 53, 2503–2518. doi: 10.4319/lo.2008.53.6.2503
Shikata, T., Sakurada, K., Jomoto, Y., Oyama, N., Onji, M., Yoshida, M., et al. (2011). Growth dynamics of Chattonella antiqua in relation to nutrients in the yatsushiro sea. Nippon Suisan Gakk. 77, 40–52. doi: 10.2331/suisan.77.40
Simao, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Sineshchekov, O., Lebert, M., and Häder, D. P. (2000). Effects of light on gravitaxis and velocity in Chlamydomonas reinhardtii. J. Plant Physiol. 157, 247–254. doi: 10.1016/s0176-1617(00)80045-0
Smayda, T. J. (1998). “Ecophysiology and bloom dynamics of Heterosigma akashiwo (raphidophyceae),” in Physiological Ecology of Harmful Algal Blooms, eds D. M. Anderson, A. D. Cembella, and G. M. Hallegraeff (Berlin: Springer-Verlag), 113–131.
Smith, H. (2000). Phytochromes and light signal perception by plants—an emerging synthesis. Nature 407:585. doi: 10.1038/35036500
Takahashi, F., Yamagata, D., Ishikawa, M., Fukamatsu, Y., Ogura, Y., Kasahara, M., et al. (2007). Aureochrome, a photoreceptor required for photomorphogenesis in stramenopiles. Proc. Natl. Acad. Sci. U.S.A. 104, 19625–19630. doi: 10.1073/pnas.0707692104
Takaichi, S. (2011). Carotenoids in algae: distributions, biosyntheses and functions. Mar. Drugs 9, 1101–1118. doi: 10.3390/md9061101
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Tian, L., and DellaPenna, D. (2004). Progress in understanding the origin and functions of carotenoid hydroxylases in plants. Arch. Biochem. Biophys. 430, 22–29. doi: 10.1016/j.abb.2004.02.003
Torres, M. A., Onouchi, H., Hamada, S., Machida, C., Hammond-Kosack, K. E., and Jones, J. D. (1998). Six Arabidopsis thaliana homologues of the human respiratory burst oxidase (gp91phox). Plant J. 14, 365–370. doi: 10.1046/j.1365-313x.1998.00136.x
Uchiyama, I. (2006). Hierarchical clustering algorithm for comprehensive orthologous-domain classification in multiple genomes. Nucleic Acids Res. 34, 647–658. doi: 10.1093/nar/gkj448
Uchiyama, I. (2017). “Ortholog identification and comparative analysis of microbial genome using MBGD and RECOG,” in Protein Function Prediction. Methods in Molecular Biology, ed. D. Kihara (New York, NY: Humana press).
Wang, Z. H., and Liang, Y. (2015). Growth and alkaline phosphatase activity of Chattonella marina and Heterosigma akashiwo in response to phosphorus limitation. J. Environ. Sci. 28, 1–7. doi: 10.1016/j.jes.2014.04.015
Wang, Z. H., Liang, Y., and Kang, W. (2011). Utilization of dissolved organic phosphorus by different groups of phytoplankton taxa. Harmful Algae 12, 113–118. doi: 10.1016/j.hal.2011.09.005
Watanabe, M., Kohata, K., and Kimura, T. (1991). Diel vertical migration and nocturnal uptake of nutrients by Chattonella antiqua under stable stratification. Limnol. Oceanogr. 36, 593–602. doi: 10.4319/lo.1991.36.3.0593
Yamaguchi, H., Nishijima, T., Nishitani, H., Fukami, K., and Adachi, M. (2004). Organic phosphorus utilization and alkaline phosphatase production of 3 red tide phytoplankton. Nippon Suisan Gakk. 70, 123–130. doi: 10.2331/suisan.70.123
Yamaguchi, H., Sakamoto, S., and Yamaguchi, M. (2008). Nutrition and growth kinetics in nitrogen-and phosphorus-limited cultures of the novel red tide flagellate chattonella ovata (raphidophyceae). Harmful Algae 7, 26–32. doi: 10.1016/j.hal.2007.05.011
Yamasaki, Y., Kim, D. I., Matsuyama, Y., Oda, T., and Honjo, T. (2004). Production of superoxide anion and hydrogen peroxide by the red tide dinoflagellate Karenia mikimotoi. J. Biosci. Bioeng. 97, 212–215. doi: 10.1263/jbb.97.212
Zhang, Y., Fu, F. X., Whereat, E., Coyne, K. J., and Hutchins, D. A. (2006). Bottom-up controls on a mixed-species HAB assemblage: a comparison of sympatric Chattonella subsalsa and Heterosigma akashiwo (raphidophyceae) isolates from the delaware inland bays, USA. Harmful Algae 5, 310–320. doi: 10.1016/j.hal.2005.09.001
Keywords: aureochrome, cryptochrome, harmful algae, NADPH-oxidase, phytochrome, pigment biosynthesis, transcriptome, transporter
Citation: Shikata T, Takahashi F, Nishide H, Shigenobu S, Kamei Y, Sakamoto S, Yuasa K, Nishiyama Y, Yamasaki Y and Uchiyama I (2019) RNA-Seq Analysis Reveals Genes Related to Photoreception, Nutrient Uptake, and Toxicity in a Noxious Red-Tide Raphidophyte Chattonella antiqua. Front. Microbiol. 10:1764. doi: 10.3389/fmicb.2019.01764
Received: 22 March 2019; Accepted: 16 July 2019;
Published: 31 July 2019.
Edited by:
Anne Michelle Wood, University of Oregon, United StatesReviewed by:
Fei Gan, University of California, Berkeley, United StatesMohammad Moniruzzaman, Virginia Tech, United States
Copyright © 2019 Shikata, Takahashi, Nishide, Shigenobu, Kamei, Sakamoto, Yuasa, Nishiyama, Yamasaki and Uchiyama. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tomoyuki Shikata, c2hpa2F0YXRAYWZmcmMuZ28uanA=; Ikuo Uchiyama, dWNoaXlhbWFAbmliYi5hYy5qcA==
 Tomoyuki Shikata
Tomoyuki Shikata Fumio Takahashi2,3
Fumio Takahashi2,3 Hiroyo Nishide
Hiroyo Nishide Yasuhiro Kamei
Yasuhiro Kamei