
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 06 August 2019
Sec. Microbiotechnology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.01750
This article is part of the Research Topic Microorganisms Degrading Organic Pollutants and Their Potential for the Bioremediation of Contaminated Environments View all 48 articles
 Juanping Wang1,2,3
Juanping Wang1,2,3 Chang Wang1,2
Chang Wang1,2 Qi Li1
Qi Li1 Mengyuan Shen1,2
Mengyuan Shen1,2 Peng Bai1,2
Peng Bai1,2 Jionghui Li1,2
Jionghui Li1,2 Yan Lin1
Yan Lin1 Nanqin Gan1
Nanqin Gan1 Tao Li1*
Tao Li1* Jindong Zhao1,4
Jindong Zhao1,4
The bacterium Novosphingobium sp. THN1 (THN1) is capable of degrading microcystin-LR (MC-LR). To study the ability of THN1 to degrade MC-LR and its possible mechanism(s) of regulation, we analyzed the effect of carbon concentrations on the degradation process. The MC-LR degradation rate peaked early and then declined during MC-LR biodegradation. Decreased levels of carbon in the medium caused the degradation peak to occur earlier. The expression of the functional gene mlrA, encoding a microcystinase, showed a similar trend to the MC-LR degradation rate at various carbon concentrations (r2 = 0.717, p < 0.05), suggesting that regulation of mlrA expression may play an important role in MC-LR degradation by THN1. The total bacterial biomass decreased when the carbon source was limited and did not correlate with the MC-LR degradation rate. Transcriptomic analysis showed that MC-LR degradation differentially regulated 62.16% (2597/4178) of THN1 genes. A considerable number of differentially expressed genes (DEGs) during MC-LR degradation encoded proteins related to carbon-, nitrogen-, and amino acid-related pathways. At 2 h of MC-LR degradation, most DEGs (29/33) involved in carbon and nitrogen metabolism were downregulated. This indicated that MC-LR may regulate carbon and nitrogen pathways of Novosphingobium sp. THN1. KEGG pathway analysis indicated that the upregulated DEGs during MC-LR degradation were mainly related to amino acid degradation and substrate metabolism pathways. Particularly, we detected increased expression of glutathione metabolism-related genes from transcriptomic data at 2 h of MC-LR degradation compared with the gene expression of 0 h, such as GST family protein, glutathione peroxidase, S-(hydroxymethyl) glutathione dehydrogenase, and glutathione-dependent disulfide-bond oxidoreductase that have been reported to be involved in microcystin degradation.
Microcystins (MCs) are a family of monocyclic heptapeptides having a general structure composed of five D-amino acids and two variable L-amino acids. They are synthesized by multienzyme complexes encoded by the mcy gene cluster and produced by some genera of cyanobacteria, including Microcysits, Anabaena, Planktothrix, and Nostoc (Carmichael, 1992; Watanabe et al., 1992; Tillett et al., 2000; Keil et al., 2002). More than 200 different structural analogs of MCs have been identified from cyanobacterial blooms and cultures (Zastepa et al., 2015). Microcystin-LR is the most common and potent analog, followed by microcystin-RR and microcystin-YR (Chen et al., 2016). Many of the threats that cyanobacterial blooms pose to humans and animals are from MCs (Carmichael et al., 2001; Azevedo et al., 2002). These compounds are active in inhibition of protein phosphatase 1 and 2A and tumor promotion (Mackintosh et al., 1990; Campos and Vasconcelos, 2010), causing severe health risks to plants, animals, and humans. MCs are also structurally stable and resistant to many physical and chemical processes such as high temperature, sunlight, chemical hydrolysis, and oxidation (Wormer et al., 2010; Rastogi et al., 2014). However, some bacterial populations occurring in natural waters have been reported to degrade MCs effectively (Edwards et al., 2008; Dziga et al., 2013; Kormas and Lymperopoulou, 2013), providing a promising approach to elimination of MCs from natural waters. Many indigenous bacteria from lakes and reservoirs that can degrade MCs have been isolated to investigate MC degradation (Eleuterio and Batista, 2010; Jiang et al., 2011; Yang et al., 2014; Zhang et al., 2015; Zhu et al., 2016). Sphingomonadaceae bacteria have been focused on because many are effective in MC degradation (Valeria et al., 2006; Zhang et al., 2010; Xiao et al., 2011; Maghsoudi et al., 2016; Ding et al., 2018).
Typically, a cluster of four genes (mlrA, mlrB, mlrC, and mlrD) has been characterized as being responsible for MC degradation (Bourne et al., 1996, 2001). The mlrA gene encodes a microcystinase (MlrA), which can cleave the cyclic MC structure into a 2100-times less toxic linearized molecule by breaking it at the Arg–Adda peptide bond and is considered to be an important marker for the detection of MC-degrading bacteria (Bourne et al., 1996; Dziga et al., 2012). MlrB hydrolyzes the linearized MC-LR at Leu–Ala to form a tetrapeptide (Bourne et al., 1996). MlrC can decompose both linearized MC-LR and the tetrapeptide into Adda through the fracture of the Adda–Glu bond (Hashimoto et al., 2009; Shimizu et al., 2012; Dziga et al., 2016). MlrD was predicted to be an oligopeptide transporter because of its potential transmembrane spanning regions (Bourne et al., 2001). Apart from the mlr gene cluster, some alternative MC degradation pathways have been reported. Edwards et al. (2008) found demethylation, hydrolysis, decarboxylation, and condensation of microcystin LF (MC-LF), with nodularin as a novel intermediate degradation product. Dziga et al. (2017) detected four products of dmMC-LR in temperate freshwater bodies, including cyclic dmMC-LR, two cyclic dmMC-LRs with different modifications in the Arg–Asp–Leu region, and a tetrapeptide. Zhang et al. (2010) found that Sphingopyxis sp. USTB-05 can degrade MC-RR through hydrolysis and dehydration reactions to form a mostly linear MC-RR with two small peptide rings. A recent report identified some new MC-LR metabolites, such as Mdha–Ala, MeAsp–Arg, and Leu, indicating that there may be other novel types of hydrolases (Ding et al., 2018). More recently, a novel degradation pathway in which MC-LR is hydrolyzed by cleaving the Ala–Mdha peptide to a new linear MC-LR intermediate in anaerobic conditions was proposed (Zhu et al., 2019). Previous studies revealed that the glutathione-S-transferase (GST) and the CAAX-type II amino-terminal protease present in MC-degrading bacteria are involved in MC degradation (Ding et al., 2000; Saito et al., 2003; Gehringer et al., 2004; Maatouk et al., 2004; Dziga et al., 2013; Kansole and Lin, 2016). Analysis of whole genome gene expression may help identify further enzymes that are activated during MC degradation.
Novosphingobium sp. THN1, isolated from a water sample from Lake Taihu, China, belongs to the family Sphingomonadaceae (Jiang et al., 2011). This bacterial strain can degrade MC-LR effectively, eliminating 91.2% of MC-LR in THN1 culture during the first 12 h and completely removing MC-LR after 60 h. Like other microcystin-degrading Sphingomonadaceae, THN1 contains a mlr gene cluster that was confirmed to be involved in MC degradation (Bourne et al., 2001). The mlrA gene expression of THN1 during MC degradation was investigated (Jiang et al., 2011), and the results revealed that MC-LR induced upregulation of mlrA, with the highest transcription level occurring at 45 min.
Biodegradation of MCs in the environment has been widely studied. Environmental factors such as nutrient conditions (Li et al., 2011), oxygen status (Holst et al., 2003), temperature (Park et al., 2001; Chen et al., 2010), and MC-LR and chlorophyll a concentration (Li et al., 2015) in water are reported to affect the biodegradation rate of MCs. Glucose was used to investigate the effect of organic carbon on MC-LR degradation and contradictory effects have been noted in previous studies. Some studies showed that glucose added as exogenous carbon sources increased the removal percentage of MC-LR by Lactobacillus plantarum IS-10506, Lactobacillus plantarum IS-20506, and Bacillus nanhaiencis JZ-2013 in medium (Surono et al., 2008; Zhang et al., 2015), while others drew the opposite conclusion that addition of glucose to cultures repressed MC-LR degradation (Eleuterio and Batista, 2010; Li et al., 2011, 2012). Furthermore, mlrA gene expression and gene abundance were assessed during MC-LR biodegradation. Some previous studies of MC degradation by the microbial communities in environmental samples confirmed that the abundance of the mlrA gene in the community increased with MC-LR removal (Eleuterio and Batista, 2010; Ho et al., 2010; Li et al., 2012; Lezcano et al., 2016). A recent study showed that the variety and the number of MC-degrading microbes decreased dramatically on the addition of high concentrations of glucose (Ma et al., 2016). Divergent responses of functional gene expression to various nutrient conditions were revealed during MC-LR biodegradation by Novosphingobium sp. THN1. However, to the best of our knowledge, no previous work has assessed both mlrA gene expression and bacterial biomass during MC degradation. It is unclear whether bacterial populations, functional gene expression, or both, regulate MC degradation. The reported contradictory nutrient effects on MC-LR biodegradation may be due to differences in cell growth state or regulation of gene expression.
The aim of this study was to analyze the effect of carbon availability on the degradation rate of MC-LR by strain THN1 and explore the possible regulatory mechanism(s). We first assessed the MC-LR biodegradation rate at different carbon concentrations. Then, we linked the MC-LR biodegradation rate with mlrA gene expression and bacterial growth at different carbon concentrations. Significantly, we applied transcriptomics to identify the gene regulation mechanisms of MC-LR biodegradation at different carbon concentrations. Results from our transcriptomic data may help to clarify the whole-genome gene regulation of MC-LR biodegradation, and identify novel genes that are involved in MC-LR biodegradation.
Novosphingobium sp. strain THN1 was obtained from the Laboratory of Harmful Algae Biology, Institute of Hydrobiology, Chinese Academy of Sciences. The strain was incubated in R2A medium (Table 1) at 37°C in the dark while shaking at 200 rpm.
MC-LR (≥95% purity, Lot No.: L1101003, CAS No.: 101043-37-2) was purchased from Taiwan Algal Science Incorporation and stored at -20°C. Upon use, MC-LR was dissolved in methanol to prepare a stock solution.
The ingredients of R2A medium are listed in Table 1. Glucose, starch, and sodium pyruvate are the main carbon sources. To investigate the effect of carbon availability on MC-LR biodegradation, we decreased the three main carbon ingredients of R2A medium to 70% and 40% (70%C_R2A and 40%C_R2A medium, respectively). The ingredients of the modified media are also listed in Table 1.
Exponentially growing cultures of THN1 were harvested and centrifuged at 6000× g for 10 min. Cell pellets were washed and resuspended at a final absorbance of 0.3 measured at 600 nm (OD600) in media with different organic carbon concentrations (100%C_R2A, 70%C_R2A, and 40%C_R2A). MC-LR was then spiked into the culture at a final concentration of 1.5 or 3 mg L-1. Autoclaved cultures were set as controls to account for any abiotic loss of MCs. All the groups were shaken at 37°C in the dark. In addition, R2A medium containing 1 or 5 mg L-1 MC-LR was prepared to examine the growth of THN1 in different concentrations of MC-LR; bacterial cultures spiked with an equivalent amount of sterile ultrapure water were used as controls.
At reaction times of 0, 1, 2, and 3 h, samples were taken and centrifuged at 6000× g for 10 min. The supernatants were collected for MC-LR analysis while the precipitates were used for RNA extraction. Moreover, 3-mL samples were collected and monitored for bacterial growth by measuring OD600 values. The experiments were conducted in triplicate and the average values were used for analysis.
Cell cultures collected at 0, 1, 2, and 3 h were centrifuged at 6000× g for 10 min. The supernatant was collected and filtered through 0.22-μm syringe filters (PTEE Hydrophilic, Millipore, United States) before analysis. The concentration of MC-LR (standard) for preparing the calibration curve ranged between 0.1 and 5 μg L-1.
The MC-LR concentration in the supernatant was determined using an enzyme-linked immunosorbent assay kit (IHB, CAS, China) according to Lei’s method (Lei et al., 2004). Briefly, samples were diluted based on predicted MC-LR concentration, so that the final MC-LR concentration in each sample was theoretically within the range of standards supplied. Anti-MC-LR monoclonal antibodies were mixed with MC-LR standards or diluted samples in microtiter plates (Nunc, Denmark). After incubation at 37°C for 1 h, the plates were washed, and europium-labeled antimouse IgG conjugate (Perkin-Elmer), diluted 1:500 in assay buffer (Perkin-Elmer), was added at 100 μL/well. After a further incubation for 1 h at 37°C, the plates were washed six times, and enhancement solution (Perkin-Elmer) was added at 100 μL/well. The plates underwent rotation incubation for 5 min and then the concentration of MC-LR was measured at 450 nm using a Microplate Spectrophotometer Reader (Thermo ScientificTM MultiskanTM GO Microplate Spectrophotometer type 357, Thermo Scientific, Helsinki, Finland).
The average biodegradation percentage was calculated by dividing the concentration of MCs initially spiked into the cultures by the remaining MC concentration in the sample. The biodegradation rate was the average biodegradation percentage per h.
Bacterial RNA from Novosphingobium sp. THN1 was extracted using an E.Z.N.A. Bacterial RNA kit (Omega) according to the manufacturer’s protocol. The amount and purity of the extracted RNA were determined using comparison of the optical density at 260 and 280 nm by spectrophotometry (NanoDrop 8000, Thermo Fisher Inc., United States) and agarose gel electrophoresis to evaluate integrity (Rastogi et al., 2014). Samples were then stored at -80°C. After digestion with DNase I (Promega), 2 μg of total RNA was reverse transcribed using a RevertAid first-strand cDNA synthesis kit (Thermo Fisher Scientific, Waltham, MA, United States) according to the kit manual.
Real-time qPCR reactions were performed on the Roche LightCycler 480 Real-Time PCR system (Roche, United States) using cDNA (see section “RNA Extraction and Reverse Transcription”). Two pairs of specific primers, qmlrAF/qmlrAR, and q16SF/q16SR, were used to quantify the number of copies of the mlrA and 16S ribosomal ribonucleic acid (rRNA) genes, respectively (Table 2). The 16S rRNA gene acted as the housekeeping gene in qPCR assays. Real-time qPCR assay and analysis were performed as described previously (Li et al., 2014). All reactions were completed in a total volume of 20 μL, containing 10 μL Master Mix (SYBR Green, Toyobo, Japan), 1 μL (10 μmol L-1) of each primer, 1 μL cDNA template, and ddH2O. The qRT-PCR program was: 95°C for 4 min, followed by 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 45 s. Gene expression results were assessed through the Ct value. The relative expression ratio (Rastogi et al., 2014) was calculated by formula 2-ΔΔCt based on the equation: ΔΔCt = (Ct, target gene – Ct,16SrRNA)stress – (Ct, target gene – Ct,16SrRNA)control. All assays were performed in triplicate and the results are reported as means [±standard deviation (SD)].
RNA samples extracted from cells spiked with MC-LR at 0 and 2 h in 100%C_R2A and 40%C_R2A media were used for transcriptome sequencing. The mRNA was sheared into fragments in fragmentation buffer, and used for first-strand cDNA synthesis using reverse transcriptase and random primers. This was followed by second strand cDNA synthesis using DNA polymerase I and RNase H. These cDNA fragments were purified with a QiaQuick PCR extraction kit, and underwent end repair and ligation of adapters. The products were purified, and fragments with an approximate size of 350 bp were selected by agarose gel-electrophoresis. Sequencing libraries were constructed by amplifying the selected fragments by PCR. The libraries were sequenced using the Illumina HiSeq 2000 platform, and raw reads were generated. Qualified sequences were mapped to the Novosphingobium sp. THN1 genome (Wang et al., 2018) using Bowtie2 (Coronado et al., 2012) with no more than five mismatched bases. The datasets generated for this study can be found in NCBI Gene Expression Omnibus, GSE125827.
To compare the differences in gene expression between the control (0 h) and MC-LR-treated samples, mRNA abundance was estimated by the number of uniquely mapped reads per kilobase per million reads (RPKM) method (Bullard et al., 2010). The calculated gene expression was used to compare the differentially expressed genes (DEGs) between samples. A false discovery rate (FDR) control was used to obtain the true number of DEGs (Audic and Claverie, 1997). Differential expression between the treatment and reference conditions was computed with DESeq (Anders and Huber, 2010). The DEGs were defined as those with an FDR < 0.001 and an RPKM ratio of the two samples |log2 ratio|≥ 1. The fold-changes of DEGs were calculated as the log2 ratios of the gene abundance comparing MC-LR treated-samples and the control sample.
The biological functions of the DEGs were identified to investigate the pattern of transcriptome regulation that occurred during MC-LR biodegradation at different carbon concentrations. Clusters of Orthologous Groups (COG) annotations of the DEGs were used for functional classification. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotations were performed1 to determine the cellular pathways involving the DEGs.
Figure 1 shows degradation kinetics of 1.5 mg L-1 MC-LR by Novosphingobium sp. strain THN1 at different concentrations of carbon sources. No obvious decrease in the initial MC-LR concentration was observed in controls, indicating that any loss of MC-LR in the experimental groups was attributable to biodegradation.
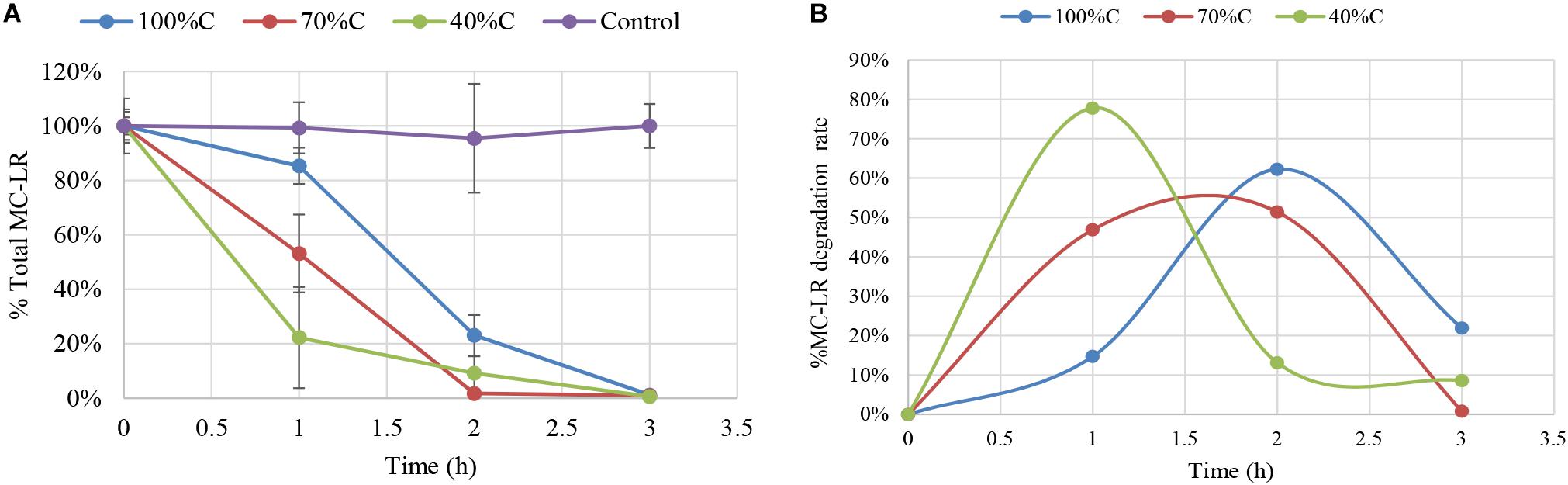
Figure 1. Biodegradation of total microcystin (MC)-LR by Novosphingobium sp. strain THN1 incubated at different carbon concentrations. (A) The remaining MC-LR percentage after 3 h; (B) The biodegradation percentage of MC-LR over 3 h. The initial MC-LR concentration was 1.5 mg L-1.
At all carbon concentrations, MC-LR concentration declined dramatically in the first 3 h and >95% of MC-LR was degraded. The MC-LR degradation rate peaked early and then declined. MC-LR biodegradation by THN1 was different at different carbon concentrations. In comparison with the MC-LR biodegradation in R2A medium, decreased carbon concentration caused significant stimulation of MC-LR biodegradation at 1 h: the remaining MC-LR percentage under sequentially decreased carbon concentrations (100%C_R2A, 70%C_R2A, and 40%C_R2A) was 85.31 ± 6.65%, 53.17 ± 14.32%, and 22.26 ± 18.58%, respectively (Figure 1A). Correspondingly, the MC-LR degradation rate increased as the carbon level decreased. The biodegradation percentage of MC-LR at 2 and 3 h were different from that of 1 h. At 2 h of MC-LR degradation, the biodegradation rate decreased as the carbon concentration decreased. The biodegradation rate at 3 h in 40%C_R2A medium was lower than in 100%C_R2A medium and higher than in 70%C_R2A medium. As Figure 1B shows, the degradation peaks occurred at 1, 1.5, and 2 h for 40%C_R2A, 70%C_R2A, and 100%C_R2A, respectively. Decreased carbon stimulates the advance of the degradation peak.
The MC-degrading bacterium THN1 did not exhibit significant differences in growth at any carbon concentration when 1.5 mg L-1 MC-LR was added. The treatment cultures and controls reached similar OD600 values in the first 5 h (Figure 2A). As expected, the growth of THN1 in decreased carbon concentrations was slower than that in normal R2A medium. This result was consistent with previous observations on growth of MC-degrading bacteria that the presence of exogenous C and/or N stimulate the growth of such bacteria during MC-degradation (Zhang et al., 2015; Lezcano et al., 2016). The lower the carbon concentration, the slower the growth and thus the lower the cell population. Meanwhile, the OD600 of THN1 declined during the monitoring period in control medium without carbon sources, indicating that THN1 cannot grow in inorganic medium lacking a carbon source.
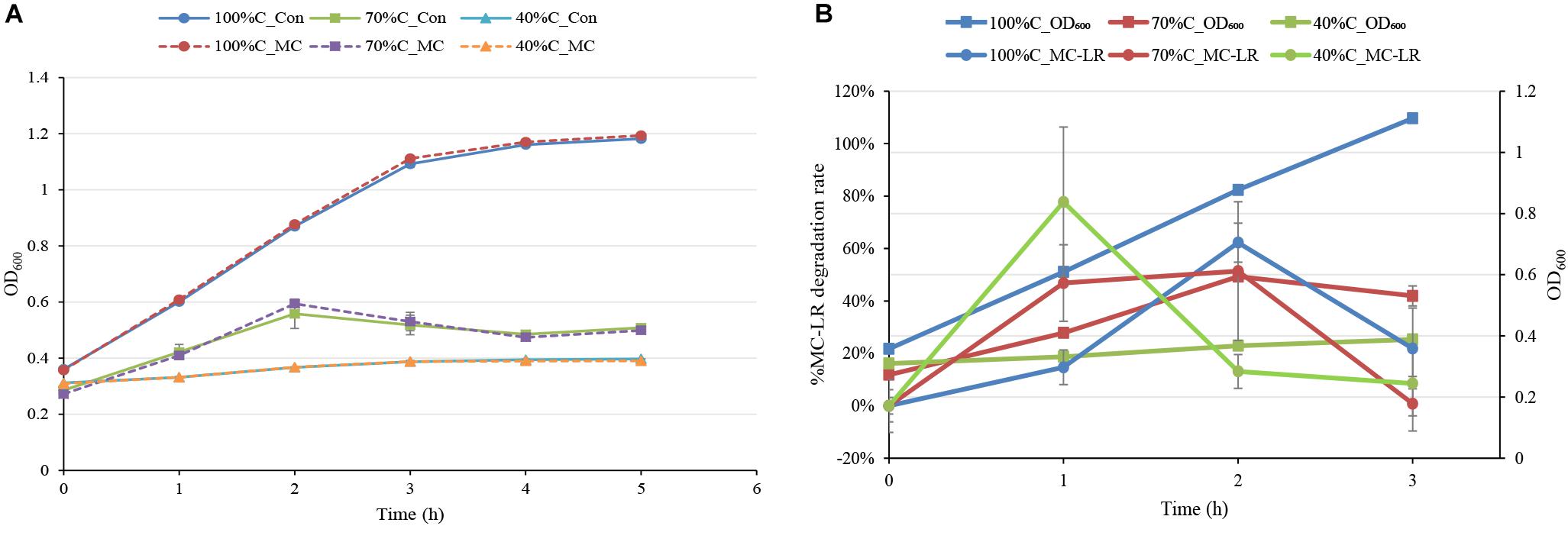
Figure 2. (A) The absorbance of THN1 cultures at 600 nm at different carbon concentrations during MC-LR biodegradation. (B) Kinetic relationship between MC-LR removal and THN1 growth at different carbon concentrations.
As previous observation showed that the abundance of the mlrA gene responded positively to the release of MCs in waters (Zhu et al., 2014; Li et al., 2015; Lezcano et al., 2016), we assessed the growth of THN1 at different concentrations of MC-LR (1, 1.5, and 5 mg L-1). There was no significant difference in the cell growth of cultures treated with MC-LR and controls (data not shown). These findings suggest that MC-LR cannot promote THN1 growth. Figure 2B showed that THN1 growth was not correlated with MC-LR degradation rate. The increased mlrA gene abundance in response to MCs in waters remained to be explored.
Figure 3 shows mlrA gene expression in THN1 cells in the first 3 h of MC-LR biodegradation at different carbon concentrations. After addition of MC-LR, expression of mlrA increased immediately and subsequently declined at all carbon concentrations. Exposure to MC-LR induced upregulation of mlrA expression in the first hour at all carbon concentrations. This indicates that MC-LR promotes the activity of MC-degrading enzymes. Consistently, Shimizu et al. (2011) revealed that the addition of MCs increased the MC degradation rate and mlrA gene expression of Sphingopyxis sp. C-1 in 30 min.
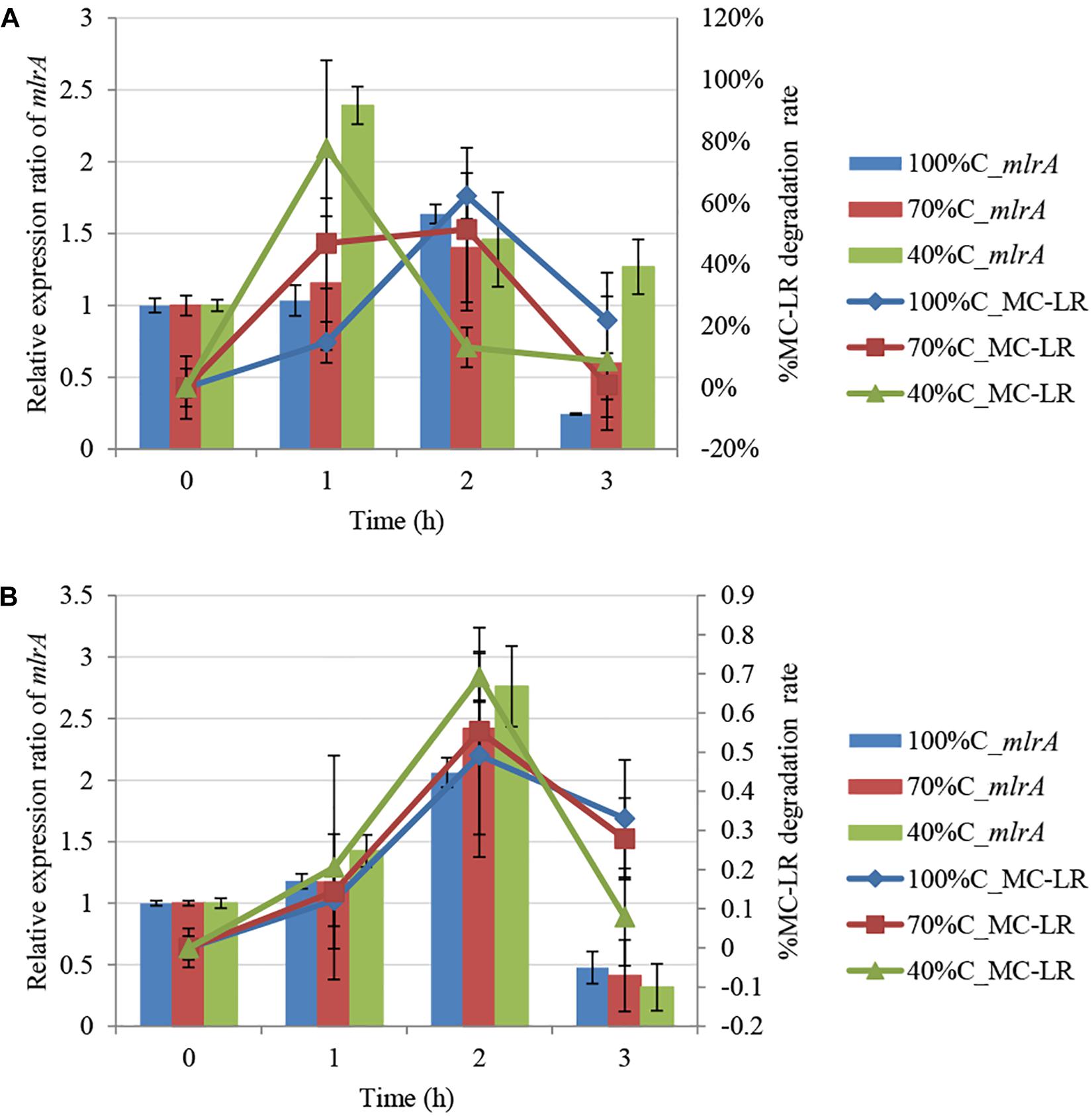
Figure 3. Kinetic relationship between MC-LR removal and mlrA gene expression at different carbon concentrations: (A) 1.5 mg L-1 MC-LR; (B) 3 mg L-1 MC-LR.
Furthermore, we found that as the carbon concentration declined, the relative expression ratio of mlrA increased at 1 h and decreased at 3 h. At 1 h, the ratio of mlrA gene expression compared with the level of mlrA in the absence of MC-LR at sequentially decreased carbon concentrations (100%C_R2A, 70%C_R2A, and 40%C_R2A) was 1.034 ± 0.107, 1.156 ± 0.465, and 2.392 ± 0.131, respectively, i.e., lower carbon concentrations induced more mlrA gene expression. The relative expression ratio of mlrA gene at 3 h at sequentially decreased carbon concentrations (100%C_R2A, 70%C_R2A, and 40%C_R2A) was 0.2445 ± 0.007, 0.598 ± 0.466, and 1.2685 ± 0.19, respectively. We suspect that a decrease of carbon concentration induced sensitive mlrA gene expression; the upregulation peak of mlrA was earlier and the subsequent decrease slower at lower carbon concentrations.
During MC-LR biodegradation by THN1, there was no positive correlation between the bacterial biomass and MC-LR degradation at different carbon concentrations. However, mlrA gene expression showed a similar trend to the degradation of MC-LR. At 1 h, at decreased carbon concentration, mlrA gene expression was upregulated relative to that of 0 h and the MC-LR degradation percentage increased. At 2 h, the expression ratio of mlrA gene in 70%C_R2A medium was a little less than that in 40%C_R2A medium, which mirrored exactly the MC-LR degradation percentage at the two carbon concentrations. The removal trend of MC-LR followed closely the trend of mlrA gene expression during 1–3 h at each carbon concentration (Figure 3). There was a positive correlation between the MC-LR degradation percentage and the expression ratios of mlrA at different organic carbon concentrations (100%C_R2A, 70%C_R2A, and 40%C_R2A) (r2 = 0.717, p < 0.05), while the biomass was not related to the degradation rate. This correlation was also found in the degradation of 3 mg L-1 MC-LR (Figure 3B). Thus, we suspect that MC-LR biodegradation may be gene regulated rather than biomass regulated; MC-LR use may lead to an increase in the expression of bacterial MC-LR biodegradation-related genes and the upregulation of mlr gene expression helps bacteria degrade more MC-LR.
To further explore gene expression during MC-LR degradation, we used RNA-Seq analyses to examine genome-wide gene expression in strain THN1 at 2 h of MC-LR degradation in 100%C_R2A and 40%C_R2A carbon conditions. At 2 h, DEGs shared similar gene expression patterns in 100%C_R2A and 40%C_R2A carbon conditions (Figure 4A). In comparison with 0 h, 62.16% (2597/4178) and 63.50% (2653/4178) of genes were differentially expressed at 2 h of MC-LR degradation at 100%C_R2A and 40%C_R2A, respectively. A total of 2377 genes were commonly differentially expressed at both carbon concentrations, indicating a large proportion of DEGs induced by MC-LR (Figure 4B). Among the common DEGs, 47.50% were upregulated and 52.2% were downregulated after 2 h of MC-LR degradation compared to 0 h. Globally speaking, the COG distribution of DEGs at 100%C_R2A and 40%C_R2A during MC-LR degradation was similar (Figure 5). During MC-LR degradation, a large proportion of DEGs were grouped in categories COG-C (Energy production and conversion), COG-E (Amino acid metabolism), COG-G (Carbohydrate transport and metabolism), COG-I (Lipid metabolism), COG-J (Translation, ribosomal structure and biogenesis), and COG-K (Transcription). At 2 h, in comparison to 100%C_R2A carbon conditions, many upregulated genes at 40%C_R2A grouped in COG-C (Energy production and conversion), COG-I (Lipid metabolism), and COG-Q (Secondary metabolites biosynthesis, transport and catabolism).
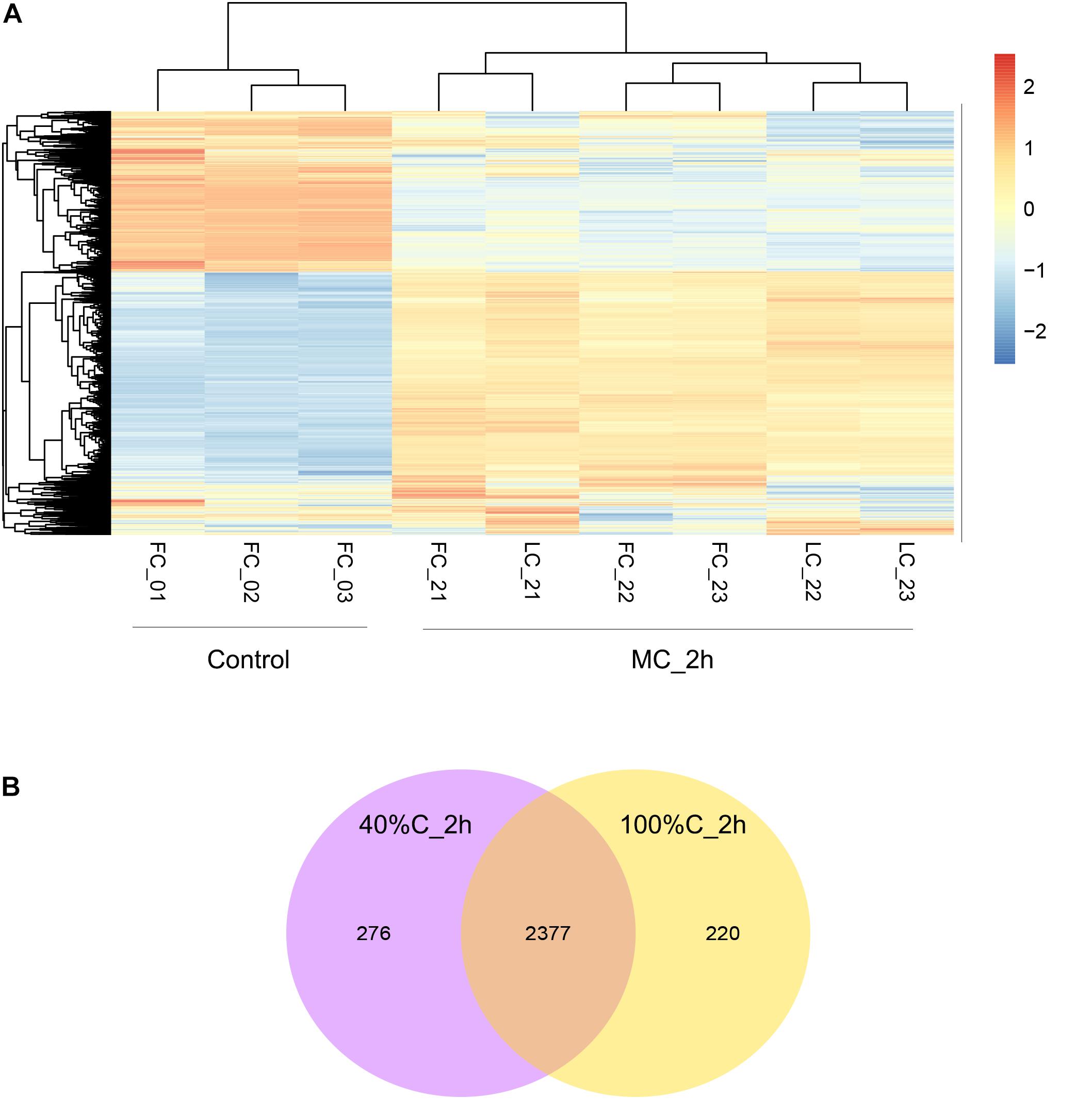
Figure 4. (A) Hierarchical clustering of THN1 gene expression on exposure to MC-LR at 100%C_R2A and 40%C_R2A at 0 and 2 h. FC_0∗ refers to transcriptional profiles at 0 h in 100%C_R2A medium, FC_2∗ to that at 2 h in 100%C_R2A medium, and LC_2∗ refers to transcriptional profiles at 2 h in 40%C_R2A medium. (B) Venn diagram grouping the genes differentially expressed on exposure to MC-LR at 2 h compared to 0 h.
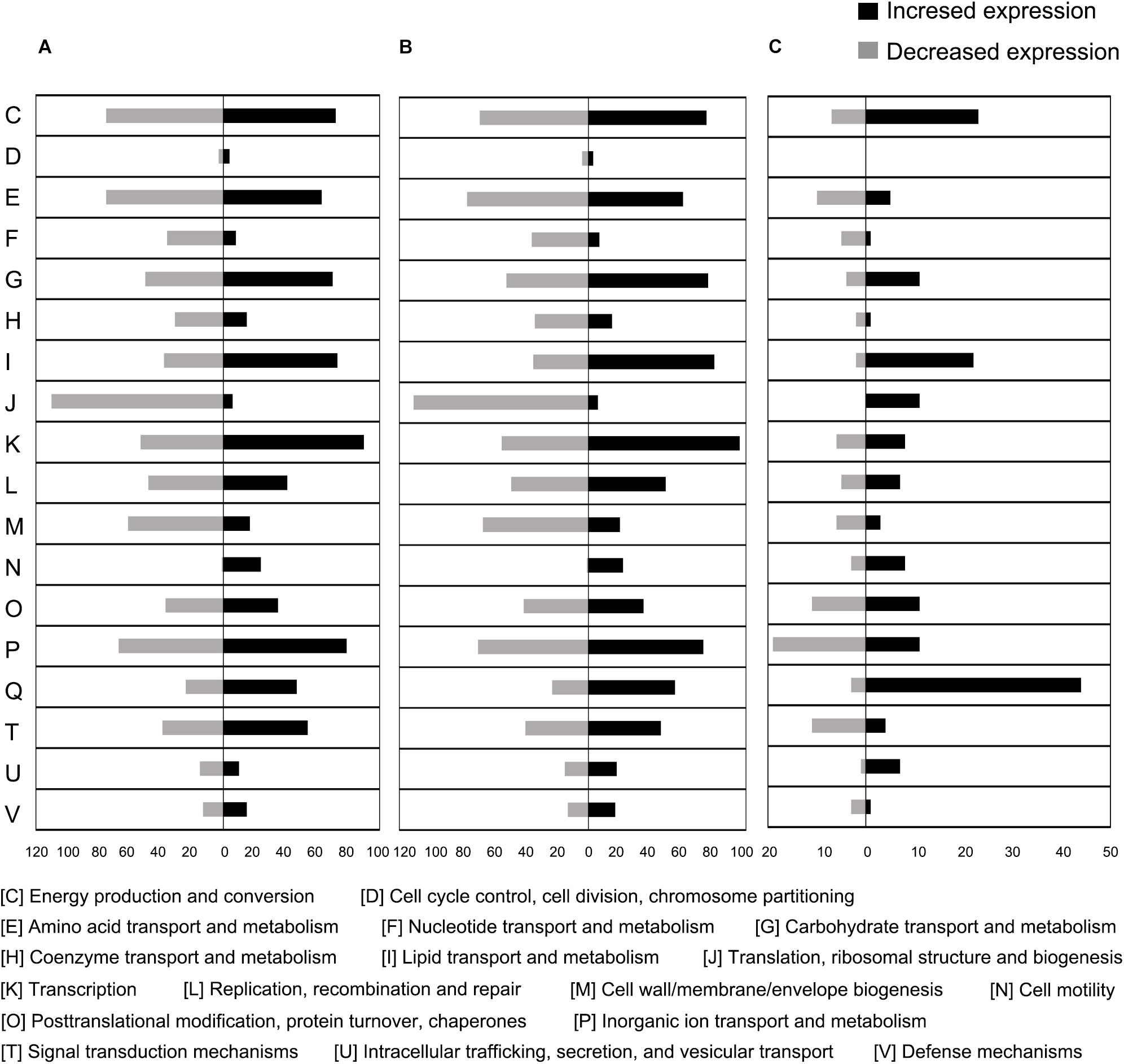
Figure 5. Proportional abundance of differentially expressed genes in conditions of MC-LR exposure, categorized by Clusters of Orthologous Genes (COG). (A) 100%C_R2A at 2 h compared to 0 h, (B) 40%C_R2A at 2 h compared to 0 h, (C) 100%C_R2A at 2 h compared to 40%C_R2A at 2 h.
Kyoto Encyclopedia of Genes and Genomes pathway analysis indicated that the upregulated DEGs during MC-LR degradation were mainly related to amino acid degradation and substrate metabolism pathways, such as valine, leucine and isoleucine degradation, tryptophan metabolism, butanoate metabolism, arginine and proline metabolism, and aminobenzoate degradation. Downregulated DEGs during MC-LR degradation were involved in the ribosome, biosynthesis of amino acids, oxidative phosphorylation, and biosynthesis of secondary metabolites (Figure 6). The expression of DEGs increased at 40%C_R2A compared to 100%C_R2A (Figure 7) including genes for valine, leucine and isoleucine degradation, the ribosome, benzoate degradation, and aminobenzoate degradation, while those that decreased included genes for pyrimidine metabolism, purine metabolism, the pentose phosphate pathway, nitrogen metabolism, alanine, aspartate, and glutamate metabolism. This suggests that MC-LR degradation differentially regulated a large number of genes involved in carbon, nitrogen, and amino acid metabolism. During MC-LR degradation, decreased carbon concentration also upregulated carbon- and nitrogen-related pathways.
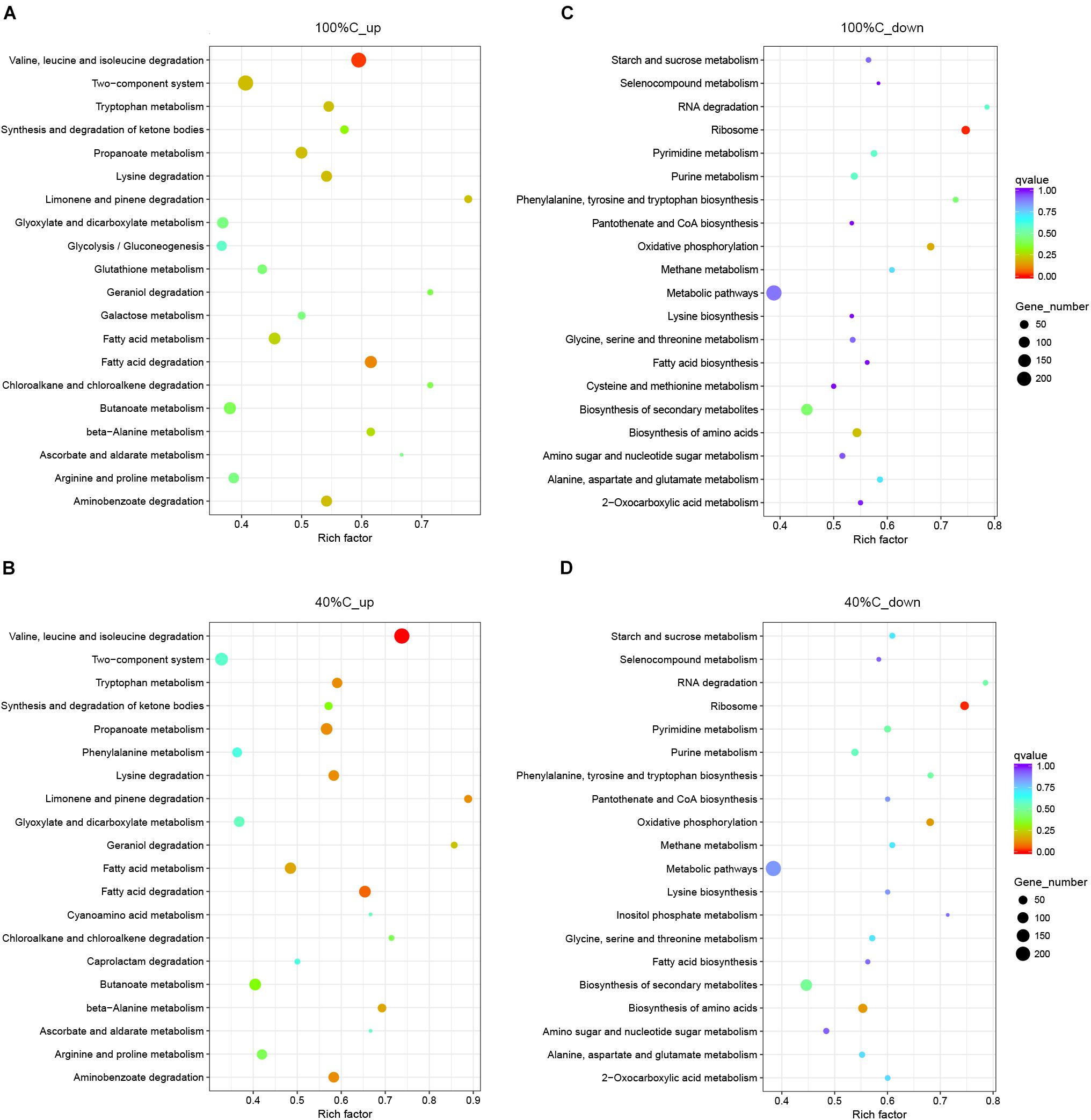
Figure 6. Functional categories of upregulated (A,B) and downregulated genes (C,D) on exposure to MC-LR at 100%C_R2A and 40%C_R2A, respectively. The size of the circles represent the total number of genes that are assigned to each functional category. The rich factor represents the ratio between enriched gene numbers and annotated gene numbers. The Q-value represents the corrected p-value. If the rich factor is larger and the Q-value is closer to 0, the enrichment is more significant. Functional categories with the top 20 enrichments are shown.
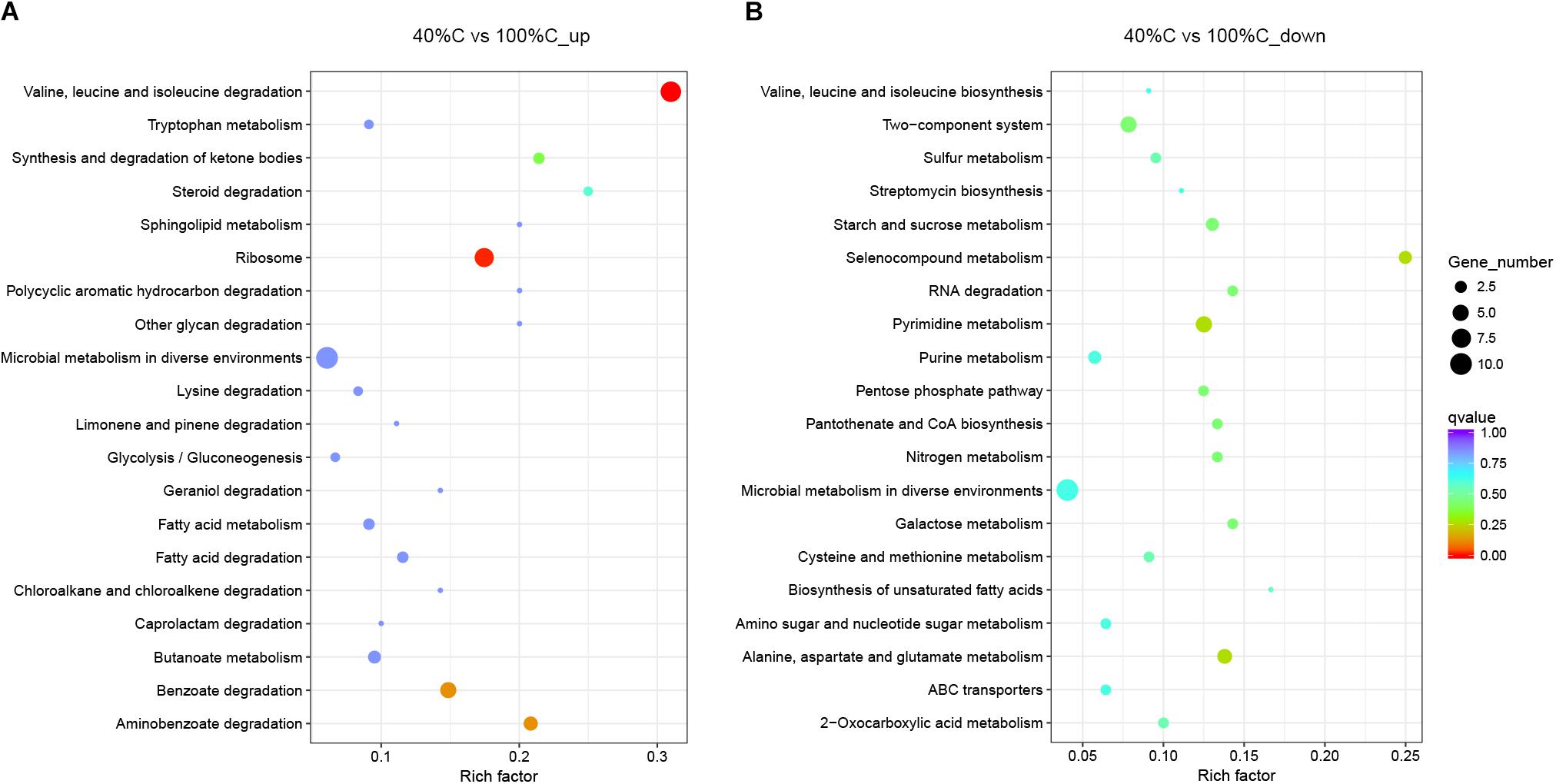
Figure 7. Functional categories of upregulated (A) and downregulated genes (B) after 2 h of exposure to MC-LR in 40%C_R2A compared to that in 100%C_R2A. The size of the circles represent the total number of genes that are assigned to each functional category. The rich factor represents the ratio between enriched gene numbers and annotated gene numbers. The Q-value represents the corrected p-value. If the rich factor is larger and the Q-value is closer to 0, the enrichment is more significant. Functional categories with the top 20 enrichments are shown.
A considerable number of DEGs during MC-LR degradation encoded proteins related to carbon-, nitrogen- and amino acid-related pathways. At 2 h of MC-LR degradation, most DEGs (29/33) involved in carbon metabolism were downregulated (Table 3). Decreased carbon concentration increased the fold-downregulation. MC-LR degradation upregulated ncd2/pcd, which encode nitronate monooxygenase, and gdhA, which encodes glutamate dehydrogenase (NADP+), while downregulating other genes involved in nitrogen metabolism (Table 4). Decreased carbon concentration also increased the fold-downregulation of nitrogen metabolism-related genes. Furthermore, at 2 h of MC-LR degradation compared with the gene expression of 0 h, we detected increased expression of glutathione (GSH) metabolism-related genes from transcriptomic data, such as GST family protein, glutathione peroxidase, S-(hydroxymethyl) glutathione dehydrogenase, and glutathione-dependent disulfide-bond oxidoreductase. Previous studies revealed that the glutathione pathway was involved in MC degradation (Gehringer et al., 2004; Maatouk et al., 2004; Dziga et al., 2013). We thus suspect that these degradative enzymes may act here in MC degradation. More studies are expected to reveal the functions of these upregulated genes.
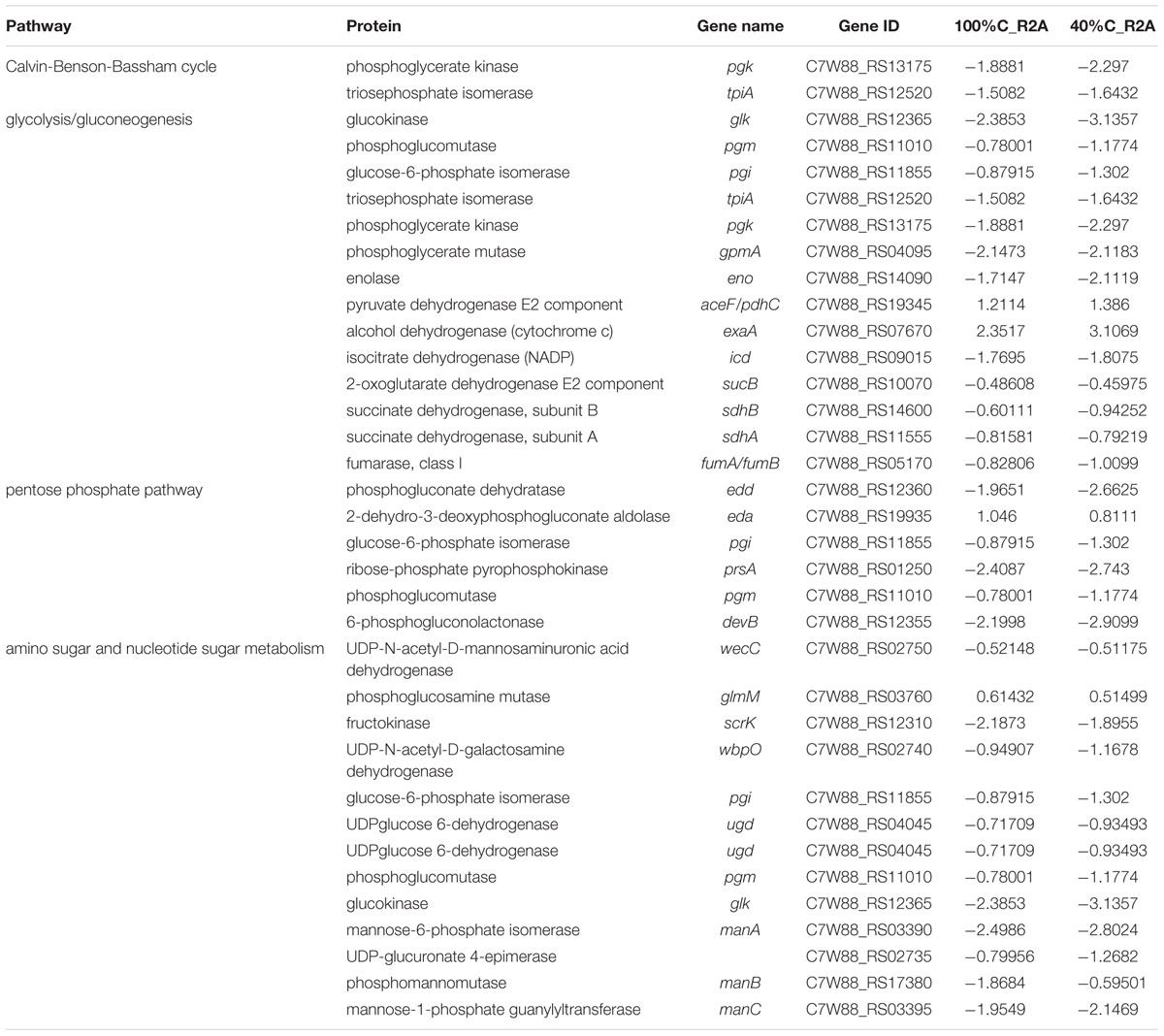
Table 3. Genes involved in carbon metabolism and their differential expression in 100%C_R2A and 40%C_R2A after 2 h of exposure to MC-LR relative to that at 0 h.
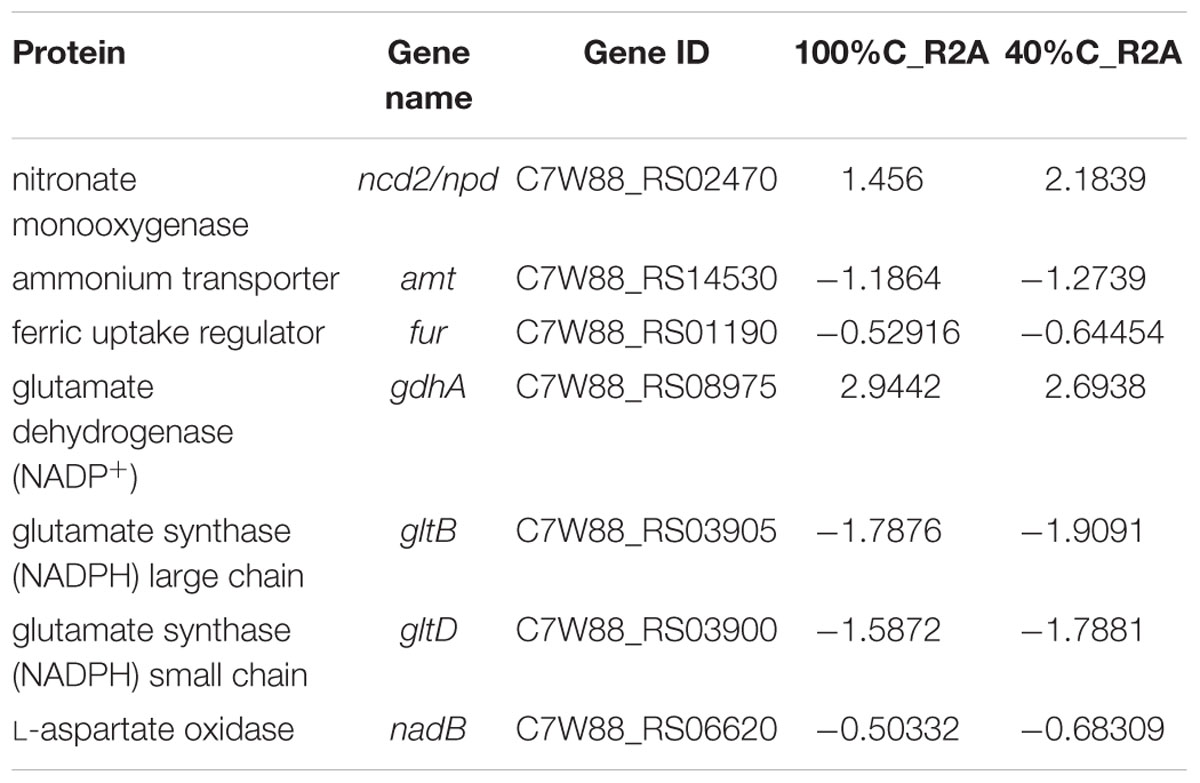
Table 4. Genes involved in nitrogen metabolism and their differential expression in 100%C_R2A and 40%C_R2A after 2 h of exposure to MC-LR relative to that at 0 h.
Biodegradation is an ecofriendly and effective strategy for MC removal, and has been applied practically in water purification processes (Li et al., 2011, 2015; Huisman et al., 2018). However, in natural environments, MC-LR biodegradation can be inhibited by high variability of environmental factors such as temperature, bacterial composition, pH, and the presence of exogenous nutrients (Li et al., 2017; Lezcano et al., 2018). Carbon concentration is a main factor affecting the MC-LR biodegradation rate. Contradictory effects of glucose have been noted in previous studies. Park et al. (2001) reported that Sphingomonas sp. Y2 could use MCs as a carbon source. In the presence of alternative organic carbon sources, MC degradation was much slower than that in organic-free medium. Holst et al. (2003) observed a stimulatory effect on MC-LR degradation by glucose addition in anoxic conditions, and stimulation of MC degradation activity by glucose. Some other studies have also shown that MC-degrading bacteria may preferentially use glucose over MCs as a carbon source. We speculate that in our study, a preference of Novosphingobium sp. strain THN1 for the carbon sources in the medium over MC-LR resulted in the inhibition of MC-LR biodegradation at high carbon concentrations. However, this explanation does not apply to the results of Li et al. (2011), who reported a stimulatory effect of the addition of glucose on MC-LR biodegradation by biofilm. In their study, enhanced inhibition of MC-LR degradation was correlated with increased proliferation of MC-degrading bacteria as the concentration of nutrients increased. Varied strain characteristics were probably attributable to the presence of different species or unique functional genes.
Cao et al. (2018) investigated the effect of different types of organic matter on MC-LR degradation in soils. They observed that MC-degradation was stimulated by the addition of humic acid but inhibited by the addition of glucose and glycine. Our study may provide advice for regulating optimal carbon concentrations to biodegrade MC-LR efficiently. In the field scale, we may decrease the carbon concentrations by throwing heterotrophic bacteria into the lake or pond in order to reduce the inhibition of high carbon concentrations on MC-LR degradation.
The kinetics of MC-biodegradation and mlrA gene abundance have been evaluated in various nutrient conditions. Expression profiles of the mlrA gene were detected during MC-LR degradation by MC-degrading Sphingopyxis sp. m6 isolated from Lake Taihu (Ding et al., 2018). In that study, mlrA gene expression showed a rapid increase in the first hour and then a gradual decline to the control level from 2 to 6 h. That result was consistent with our observations. Previously, a correlation between proteolytic activity and MC removal was found for Lactobacillus rhamnosus GG, Lactobacillus rhamnosus LC-705, and Bifidobacteriu longum 46 and both these parameters were dependent on glucose as an energy source (Nybom et al., 2012). These reports combined with our results may suggest that MC degradation mainly results from the expression of functional mlrA genes. The question of why different concentrations of organic carbon affected mlrA gene expression the way they did remains to be explored.
Microcystin biodegradation kinetics of several isolated bacterial populations have been quantified. Zhang et al. (2015) revealed that when 10 mg/L of glucose and ammonium chloride were added as exogenous carbon and nitrogen sources, the degradation percentage of MC-LR by Bacillus nanhaiencis JZ-2013 increased from about 80% to about 90%. At the same time, the bacterial growth also improved. When combined with our results, one may conclude that the improved bacterial growth resulted from the added carbon or nitrogen sources, but not from MCs. However, Valeria et al. (2006) observed that MC-RR biodegradation by Sphingomonas sp. CBA4 was accompanied by a slight increase in bacterial density. Also, Sphingopyxis sp. m6 maintained moderate growth in the first 3 h of MC-LR degradation and then rapidly increased after MC-LR was decomposed (Ding et al., 2018). However, in our study, the presence or absence of MC-LR did not result in a significant difference in bacterial growth. MC-LR cannot be assimilated as a carbon source for THN1 growth. Recently, a comparison of several unstructured kinetic models to describe MC biodegradation by isolated degrading populations was reported (Manheim et al., 2019). Model predictions suggest that MC concentrations in the environment are well below saturating levels for optimal bacterial growth. Thus, MC degradation may fail in promoting bacterial growth. Previously, two studies reported that during MC-LR degradation, the degrading bacteria did not use MC-LR as a carbon source and MC-LR might be removed via a xenobiotic mechanism. As a result, the total bacterial concentration was not increased by MC-LR (Mou et al., 2013; Kansole and Lin, 2016). Xenobiotic MC-LR degradation may also explain why MC-LR degradation did not lead to an increase in bacterial growth in the present study.
Alternative microcystin enzymatic degradation pathways apart from the mlr gene cluster have been reported (Dziga et al., 2013). CAAX type II amino-terminal protease belonging to the CAAX Proteases and Bacteriocin Processing Enzymes family might encode a microcystinase function (Read et al., 2002; Pei et al., 2011) and be involved in MC-LR degradation (Kansole and Lin, 2016). Mou et al. (2013) reported over-representation of GST and cytochrome P450 oxidase during MC-LR degradation, which are proposed to catalyze the synthetic metabolism of MC-LR to cysteine and GSH conjugates in animals (Campos and Vasconcelos, 2010). Several studies also found some MC-degrading bacteria that did not contain mlr genes (Manage et al., 2009; Yang et al., 2014; Zhu et al., 2019). Also, here, we detected increased expression of GSH metabolism-related genes in our transcriptomic data (GST family protein, glutathione peroxidase, S-[hydroxymethyl] glutathione dehydrogenase, and glutathione-dependent disulfide-bond oxidoreductase). As the transcriptomics analysis in this study showed that MC-LR degradation upregulated degradative genes aside from mlr, these degradative genes may be involved in MC-LR degradation by bacteria and account for the production of novel metabolites. More studies should thus be performed to explore the functions of these degradative genes during MC-LR degradation in the natural environment.
Microcystin removal efficiency was also dependent on temperature, pH, and cell density when glucose was added to the medium (Nybom et al., 2008). Determination of the regulatory mechanisms of different environment factors will better clarify MC-LR degradation regulation in practice.
We decreased the carbon concentration of R2A medium to assess MC-LR degradation by the MC-degrading bacterium Novosphingobium sp. THN1 and explored the possible regulatory mechanisms. MC-LR degradation peaked early during MC-LR biodegradation and then declined. Decreases in the carbon level stimulated the advance of the degradation peak. The mlrA expression level correlated with the MC-LR degradation rate by strain THN1. During MC-LR degradation, most genes involved in the ribosome, biosynthesis of amino acids, nitrogen biosynthesis, starch and sucrose metabolism, and biosynthesis of secondary metabolites were downregulated. This indicated that MC-LR regulates carbon and nitrogen pathways of Novosphingobium sp. THN1. Genes involved in amino acid degradation and substrate metabolism pathways were upregulated during MC-LR degradation.
TL and JZ conceived the study and designed the experiments. JW performed the experiments, carried out the data analysis, and prepared the first draft of the manuscript. CW and JL assisted in the formatting of the figures. QL and MS assisted in the data analysis. PB and YL helped to revise the manuscript. NG assisted in determining the MC-LR concentrations. All authors participated in the discussion of the manuscript, agreed to the final content, and read and approved the final manuscript.
This research was supported by the National Natural Science Foundation of China (Grant No. 91851118), the Science and Technology Basic Resources Investigation Program of China (Grant No. 2017FY100300), the Key Research Program of the Chinese Academy of Sciences (Grant No. KFZD-SW-219), the State Key Laboratory of Freshwater Ecology and Biotechnology (Grant No. 2019FBZ01), and the National Key R&D Program of China (Grant No. 2018YFA0903100).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the Laboratory of Biology of Harmful Algae, Institute of Hydrobiology, Chinese Academy of Sciences for providing Novosphingobium sp. THN1. We thank Professor Nanqin Gan from the Laboratory of Algal Resources and Toxicology, Institute of Hydrobiology, Chinese Academy of Sciences for providing the enzyme-linked immunosorbent assay kit to determine MC-LR concentrations. We also thank Liwen Bianji, Edanz Group China (http://www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106
Audic, S., and Claverie, J. M. (1997). The significance of digital gene expression profiles. Genome Res. 7, 986–995. doi: 10.1101/Gr.7.10.986
Azevedo, S. M. F. O., Carmichael, W. W., Jochimsen, E. M., Rinehart, K. L., Lau, S., Shaw, G. R., et al. (2002). Human intoxication by microcystins during renal dialysis treatment in Caruaru-Brazil. Toxicology 181, 441–446. doi: 10.1016/S0300-483x(02)00491-2
Bourne, D. G., Jones, G. J., Blakeley, R. L., Jones, A., Negri, A. P., and Riddles, P. (1996). Enzymatic pathway for the bacterial degradation of the cyanobacterial cyclic peptide toxin microcystin LR. Appl. Environ. Microbiol. 62, 4086–4094.
Bourne, D. G., Riddles, P., Jones, G. J., Smith, W., and Blakeley, R. L. (2001). Characterisation of a gene cluster involved in bacterial degradation of the cyanobacterial toxin microcystin LR. Environ. Toxicol. 16, 523–534. doi: 10.1002/tox.10013.abs
Bullard, J. H., Purdom, E., Hansen, K. D., and Dudoit, S. (2010). Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. Bmc Bioinformatics 11:94. doi: 10.1186/1471-2105-11-94
Campos, A., and Vasconcelos, V. (2010). Molecular mechanisms of microcystin toxicity in animal cells. Int. J. Mol. Sci. 11, 268–287. doi: 10.3390/ijms11010268
Cao, Q., Steinman, A. D., Yao, L., and Xie, L. (2018). Effects of light, microorganisms, farming chemicals and water content on the degradation of microcystin-LR in agricultural soils. Ecotoxicol. Environ. Saf. 156, 141–147. doi: 10.1016/j.ecoenv.2018.03.030
Carmichael, W. W. (1992). Cyanobacteria secondary metabolites - the cyanotoxins. J. Appl. Bacteriol. 72, 445–459. doi: 10.1111/j.1365-2672.1992.tb01858.x
Carmichael, W. W., Azevedo, S. M. F. O., An, J. S., Molica, R. J. R., Jochimsen, E. M., Lau, S., et al. (2001). Human fatalities from cyanobacteria: chemical and biological evidence for cyanotoxins. Environ. Health Perspect. 109, 663–668. doi: 10.2307/3454781
Chen, L., Chen, J., Zhang, X. Z., and Xie, P. (2016). A review of reproductive toxicity of microcystins. J. Hazard. Mater. 301, 381–399. doi: 10.1016/j.jhazmat.2015.08.041
Chen, X., Yang, X., Yang, L., Xiao, B., Wu, X., Wang, J., et al. (2010). An effective pathway for the removal of microcystin LR via anoxic biodegradation in lake sediments. Water Res. 44, 1884–1892. doi: 10.1016/j.watres.2009.11.025
Coronado, E., Roggo, C., Johnson, D. R., and van der Meer, J. R. (2012). Genome-wide analysis of salicylate and dibenzofuran metabolism in Sphingomonas wittichii RW1. Front. Microbiol. 3:300. doi: 10.3389/Fmicb.2012.00300
Ding, Q., Liu, K., Xu, K., Sun, R., Zhang, J., Yin, L., et al. (2018). Further Understanding of degradation pathways of microcystin-LR by an indigenous Sphingopyxis sp. in environmentally relevant pollution concentrations. Toxins 10, E536. doi: 10.3390/toxins10120536
Ding, W. X., Shen, H. M., and Ong, C. N. (2000). Critical role of reactive oxygen species and mitochondrial permeability transition in microcystin-induced rapid apoptosis in rat hepatocytes. Hepatology 32, 547–555. doi: 10.1053/jhep.2000.16183
Dziga, D., Maksylewicz, A., Maroszek, M., Budzynska, A., Napiorkowska-Krzebietke, A., Toporowska, M., et al. (2017). The biodegradation of microcystins in temperate freshwater bodies with previous cyanobacterial history. Ecotoxicol. Environ. Saf. 145, 420–430. doi: 10.1016/j.ecoenv.2017.07.046
Dziga, D., Wasylewski, M., Wladyka, B., Nybom, S., and Meriluoto, J. (2013). Microbial degradation of microcystins. Chem. Res. Toxicol. 26, 841–852. doi: 10.1021/tx4000045
Dziga, D., Wladyka, B., Zielinska, G., Meriluoto, J., and Wasylewski, M. (2012). Heterologous expression and characterisation of microcystinase. Toxicon 59, 578–586. doi: 10.1016/j.toxicon.2012.01.001
Dziga, D., Zielinska, G., Wladyka, B., Bochenska, O., Maksylewicz, A., Strzalka, W., et al. (2016). Characterization of enzymatic activity of MlrB and MlrC proteins involved in bacterial degradation of cyanotoxins microcystins. Toxins 8, E76. doi: 10.3390/toxins8030076
Edwards, C., Graham, D., Fowler, N., and Lawton, L. A. (2008). Biodegradation of microcystins and nodularin in freshwaters. Chemosphere 73, 1315–1321. doi: 10.1016/j.chemosphere.2008.07.015
Eleuterio, L., and Batista, J. R. (2010). Biodegradation studies and sequencing of microcystin-LR degrading bacteria isolated from a drinking water biofilter and a fresh water lake. Toxicon 55, 1434–1442. doi: 10.1016/j.toxicon.2010.02.020
Gehringer, M. M., Shephard, E. G., Downing, T. G., Wiegand, C., and Neilan, B. A. (2004). An investigation into the detoxification of microcystin-LR by the glutathione pathway in Balb/c mice. Int. J. Biochem. Cell Biol. 36, 931–941. doi: 10.1016/j.biocel.2003.10.012
Hashimoto, E., Kato, H. Y., Nozawa, Y., Tsuji, K., Hirooka, E., and Harada, K. (2009). Further investigation of microbial degradation of microcystin using the advanced marfey method. Chem. Res. Toxicol. 22, 391–398. doi: 10.1021/tx8003517
Ho, L., Hoefel, D., Palazot, S., Sawade, E., Newcombe, G., Saint, C. P., et al. (2010). Investigations into the biodegradation of microcystin-LR in wastewaters. J. Hazard. Mater. 180, 628–633. doi: 10.1016/j.hazmat.2010.04.081
Holst, T., Jørgensen, N. O., Jørgensen, C., and Johansen, A. (2003). Degradation of microcystin in sediments at oxic and anoxic, denitrifying conditions. Water Res. 37, 4748–4760. doi: 10.1016/s0043-1354(03)00413-5
Huisman, J., Codd, G. A., Paerl, H. W., Ibelings, B. W., Verspagen, J. M. H., and Visser, P. M. (2018). Cyanobacterial blooms. Nat. Rev. Microbiol. 16, 471–483. doi: 10.1038/s41579-018-0040-1
Jiang, Y., Shao, J., Wu, X., Xu, Y., and Li, R. (2011). Active and silent members in the mlr gene cluster of a microcystin-degrading bacterium isolated from lake taihu. China. FEMS Microbiol. Lett. 322, 108–114. doi: 10.1111/j.1574-6968.2011.02337.x
Kansole, M., and Lin, T.-F. (2016). Microcystin-LR biodegradation by Bacillus sp.: reaction rates and possible genes involved in the degradation. Water 8:508. doi: 10.3390/w8110508
Keil, C., Forchert, A., Fastner, J., Szewzyk, U., Rotard, W., Chorus, I., et al. (2002). Toxicity and microcystin content of extracts from a Planktothrix bloom and two laboratory strains. Water Res. 36, 2133–2139. doi: 10.1016/S0043-1354(01)00417-1
Kormas, K. A., and Lymperopoulou, D. S. (2013). Cyanobacterial toxin degrading bacteria: who are they? Biomed Res. Int. 2013:463894. doi: 10.1155/2013/463894
Lei, L. M., Wu, Y. S., Gan, N. Q., and Song, L. R. (2004). An ELISA-like time-resolved fluorescence immunoassay for microcystin detection. Clin. Chim. Acta 348, 177–180. doi: 10.1016/j.cccn.2004.05.019
Lezcano, M. A., Moron-Lopez, J., Agha, R., Lopez-Heras, I., Nozal, L., Quesada, A., et al. (2016). Presence or absence of mlr genes and nutrient concentrations co-determine the microcystin biodegradation efficiency of a natural bacterial community. Toxins 8:E318. doi: 10.3390/toxins8110318
Lezcano, M. A., Quesada, A., and El-Shehawy, R. (2018). Seasonal dynamics of microcystin-degrading bacteria and toxic cyanobacterial blooms: interaction and influence of abiotic factors. Harmful Algae 71, 19–28. doi: 10.1016/j.hal.2017.11.002
Li, J., Li, R., and Li, J. (2017). Current research scenario for microcystins biodegradation - A review on fundamental knowledge, application prospects and challenges. Sci. Total Environ. 595, 615–632. doi: 10.1016/j.scitotenv.2017.03.285
Li, J., Peng, L., Li, J., and Qiao, Y. (2014). Divergent responses of functional gene expression to various nutrient conditions during microcystin-LR biodegradation by Novosphingobium sp. THN1. Strain. Bioresour. Technol. 156, 335–341. doi: 10.1016/j.biortech.2013.12.118
Li, J., Shimizu, K., Akasako, H., Lu, Z., Akiyama, S., Goto, M., et al. (2015). Assessment of the factors contributing to the variations in microcystins biodegradability of the biofilms on a practical biological treatment facility. Bioresour. Technol. 175, 463–472. doi: 10.1016/j.biortech.2014.10.047
Li, J., Shimizu, K., Sakharkar, M. K., Utsumi, M., Zhang, Z., and Sugiura, N. (2011). Comparative study for the effects of variable nutrient conditions on the biodegradation of microcystin-LR and concurrent dynamics in microcystin-degrading gene abundance. Bioresour. Technol. 102, 9509–9517. doi: 10.1016/j.biortech.2011.07.112
Li, J. M., Shimizu, K., Maseda, H., Lu, Z. J., Utsumi, M., Zhang, Z. Y., et al. (2012). Investigations into the biodegradation of microcystin-LR mediated by the biofilm in wintertime from a biological treatment facility in a drinking-water treatment plant. Bioresour. Technol. 106, 27–35. doi: 10.1016/j.biortech.2011.11.099
Ma, G. X., Pei, H. Y., Hu, W. R., Xu, X. C., Ma, C. X., and Pei, R. T. (2016). Effects of glucose on microcystin-LR removal and the bacterial community composition through anoxic biodegradation in drinking water sludge. Environ. Technol. 37, 64–73. doi: 10.1080/09593330.2015.1063705
Maatouk, I., Bouaïcha, N., Plessis, M. J., and Périn, F. (2004). Detection by 32P-postlabelling of 8-oxo-7,8-dihydro-2’-deoxyguanosine in DNA as biomarker of microcystin-LR- and nodularin-induced DNA damage in vitro in primary cultured rat hepatocytes and in vivo in rat liver. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 564, 9–20. doi: 10.1016/j.mrgentox.2004.06.010
Mackintosh, C., Beattie, K. A., Klumpp, S., Cohen, P., and Codd, G. A. (1990). Cyanobacterial microcystin-LR is a potent and specific inhibitor of protein phosphatase-1 and phosphatase-2a from both mammals and higher-plants. FEBS Lett. 264, 187–192. doi: 10.1016/0014-5793(90)80245-E
Maghsoudi, E., Fortin, N., Greer, C., Maynard, C., Page, A., Duy, S. V., et al. (2016). Cyanotoxin degradation activity and mlr gene expression profiles of a Sphingopyxis sp. isolated from lake champlain, Canada. Environ. Sci. Process. Impacts 18, 1417–1426. doi: 10.1039/c6em00001k
Manage, P. M., Edwards, C., Singh, B. K., and Lawton, L. A. (2009). Isolation and identification of novel microcystin-degrading bacteria. Appl. Environ. Microbiol. 75, 6924–6928. doi: 10.1128/AEM.01928
Manheim, D. C., Detwiler, R. L., and Jiang, S. C. (2019). Application of unstructured kinetic models to predict microcystin biodegradation: towards a practical approach for drinking water treatment. Water Res. 149, 617–631. doi: 10.1016/j.watres.2018.11.014
Mou, X., Lu, X., Jacob, J., Sun, S., and Heath, R. (2013). Metagenomic identification of bacterioplankton taxa and pathways involved in microcystin degradation in lake erie. PLoS One 8:e61890. doi: 10.1371/journal.pone.0061890
Nybom, S. M., Collado, M. C., Surono, I. S., Salminen, S. J., and Meriluoto, J. A. (2008). Effect of glucose in removal of microcystin-LR by viable commercial probiotic strains and strains isolated from dadih fermented milk. J. Agric. Food Chem. 56, 3714–3720. doi: 10.1021/jf071835x
Nybom, S. M. K., Dziga, D., Heikkila, J. E., Kull, T. P. J., Salminen, S. J., and Meriluoto, J. A. O. (2012). Characterization of microcystin-LR removal process in the presence of probiotic bacteria. Toxicon 59, 171–181. doi: 10.1016/j.toxicon.2011.11.008
Park, H. D., Sasaki, Y., Maruyama, T., Yanagisawa, E., Hiraishi, A., and Kato, K. (2001). Degradation of the cyanobacterial hepatotoxin microcystin by a new bacterium isolated from a hypertrophic lake. Environ. Toxicol. 16, 337–343. doi: 10.1002/tox.1041
Pei, J., Mitchell, D. A., Dixon, J. E., and Grishin, N. V. (2011). Expansion of Type II CAAX proteases reveals evolutionary origin of γ-secretase subunit APH-1. J. Mol. Biol. 410, 18–26. doi: 10.1016/j.jmb.2011.04.066
Rastogi, R. P., Sinha, R. P., and Incharoensakdi, A. (2014). The cyanotoxin-microcystins: current overview. Rev. Environ. Sci. Bio Technol. 13, 215–249. doi: 10.1007/s11157-014-9334-6
Read, T. D., Salzberg, S. L., Pop, M., Shumway, M., Umayam, L., Jiang, L., et al. (2002). Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science 296, 2028–2033. doi: 10.1126/science.1071837
Saito, T., Okano, K., Park, H.-D., Itayama, T., Inamori, Y., Neilan, B. A., et al. (2003). Detection and sequencing of the microcystin LR-degrading gene, mlrA, from new bacteria isolated from Japanese lakes. FEMS Microbiol. Lett. 229, 271–276. doi: 10.1016/s0378-1097(03)00847-4
Shimizu, K., Maseda, H., Okano, K., Itayama, T., Kawauchi, Y., Chen, R., et al. (2011). How microcystin-degrading bacteria express microcystin degradation activity. Lakes Res. Res. Manag. 16, 169–178. doi: 10.1111/j.1440-1770.2011.00480.x
Shimizu, K., Maseda, H., Okano, K., Kurashima, T., Kawauchi, Y., Xue, Q., et al. (2012). Enzymatic pathway for biodegrading microcystin LR in Sphingopyxis sp. C-1. J. Biosci. Bioeng. 114, 630–634. doi: 10.1016/j.jbiosc.2012.07.004
Surono, I. S., Collado, M. C., Salminen, S., and Meriluoto, J. (2008). Effect of glucose and incubation temperature on metabolically active Lactobacillus plantarum from dadih in removing microcystin-LR. Food Chem. Toxicol. 46, 502–507. doi: 10.1016/j.fct.2007.08.017
Tillett, D., Dittmann, E., Erhard, M., von Dohren, H., Borner, T., and Neilan, B. A. (2000). Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: an integrated peptide-polyketide synthetase system. Chem. Biol. 7, 753–764. doi: 10.1016/S1074-5521(00)00021-1
Valeria, A. M., Ricardo, E. J., Stephan, P., and Alberto, W. D. (2006). Degradation of microcystin-RR by Sphingomonas sp. CBA4 isolated from San Roque reservoir (Cordoba - Argentina). Biodegradation 17, 447–455. doi: 10.1007/s10532-005-9015-9
Wang, J. P., Wang, C., Li, J. H., Bai, P., Li, Q., Shen, M. Y., et al. (2018). Comparative genomics of degradative Novosphingobium strains with special reference to microcystin-degrading Novosphingobium sp. THN1. Front. Microbiol. 9:2238. doi: 10.3389/Fmicb.2018.02238
Watanabe, M. F., Tsuji, K., Watanabe, Y., Harada, K., and Suzuki, M. (1992). Release of heptapeptide toxin (microcystin) during the decomposition process of Microcystis aeruginosa. Natural Toxins 1, 48–53. doi: 10.1002/nt.2620010110
Wormer, L., Huerta-Fontela, M., Cires, S., Carrasco, D., and Quesada, A. (2010). Natural photodegradation of the cyanobacterial toxins microcystin and cylindrospermopsin. Environ. Sci. Technol. 44, 3002–3007. doi: 10.1021/es9036012
Xiao, C. B., Yan, H., Wang, J. F., Wei, W., Ning, J., and Pan, G. (2011). Microcystin-LR biodegradation by Sphingopyxis sp. USTB-05. Front. Environ. Sci. Eng. China 5:526–532. doi: 10.1007/s11783-010-0261-7
Yang, F., Zhou, Y. L., Sun, R. L., Wei, H. Y., Li, Y. H., Yin, L. H., et al. (2014). Biodegradation of microcystin-LR and-RR by a novel microcystin-degrading bacterium isolated from Lake Taihu. Biodegradation 25, 447–457. doi: 10.1007/s10532-013-9673-y
Zastepa, A., Pick, F. R., Blais, J. M., and Saleem, A. (2015). Analysis of intracellular and extracellular microcystin variants in sediments and pore waters by accelerated solvent extraction and high performance liquid chromatography-tandem mass spectrometry. Anal. Chim. Acta 872, 26–34. doi: 10.1016/j.aca.2015.02.056
Zhang, J., Shi, H., Liu, A. M., Cao, Z. Y., Hao, J. S., and Gong, R. M. (2015). Identification of a new microcystin-degrading bacterium isolated from Lake Chaohu, China. Bull. Environ. Contam. Toxicol. 94, 661–666. doi: 10.1007/s00128-015-1531-7
Zhang, M., Pan, G., and Yan, H. (2010). Microbial biodegradation of microcystin-RR by bacterium Sphingopyxis sp. USTB-05. J. Environ. Sci. 22, 168–175. doi: 10.1016/s1001-0742(09)60089-9
Zhu, F. P., Han, Z. L., Duan, J. L., Shi, X. S., Wang, T. T., Sheng, G. P., et al. (2019). A novel pathway for the anaerobic biotransformation of microcystin-LR using enrichment cultures. Environ. Pollut. 247, 1064–1070. doi: 10.1016/j.envpol.2019.02.013
Zhu, L., Wu, Y. L., Song, L. R., and Gan, N. Q. (2014). Ecological dynamics of toxic Microcystis spp. and microcystin-degrading bacteria in Dianchi Lake, China. Appl. Environ. Microbiol. 80, 1874–1881. doi: 10.1128/Aem.02972-13
Keywords: microcystin-LR, biodegradation, Novosphingobium sp. THN1, carbon availability, gene expression, transcriptome
Citation: Wang J, Wang C, Li Q, Shen M, Bai P, Li J, Lin Y, Gan N, Li T and Zhao J (2019) Microcystin-LR Degradation and Gene Regulation of Microcystin-Degrading Novosphingobium sp. THN1 at Different Carbon Concentrations. Front. Microbiol. 10:1750. doi: 10.3389/fmicb.2019.01750
Received: 31 January 2019; Accepted: 15 July 2019;
Published: 06 August 2019.
Edited by:
Mariusz Cycoń, Medical University of Silesia, PolandReviewed by:
Naresh Singhal, The University of Auckland, New ZealandCopyright © 2019 Wang, Wang, Li, Shen, Bai, Li, Lin, Gan, Li and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tao Li, bGl0YW9AaWhiLmFjLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.