
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 31 July 2019
Sec. Microbial Physiology and Metabolism
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.01725
Bacillus methanolicus is a Gram-positive, thermophilic, methanol-utilizing bacterium. As a facultative methylotroph, B. methanolicus is also known to utilize D-mannitol, D-glucose and, as recently discovered, sugar alcohol D-arabitol. While metabolic pathways for utilization of methanol, mannitol and glucose are known, catabolism of arabitol has not yet been characterized in B. methanolicus. In this work we present the elucidation of this hitherto uncharted pathway. In order to confirm our predictions regarding genes coding for arabitol utilization, we performed differential gene expression analysis of B. methanolicus MGA3 cells grown on arabitol as compared to mannitol via transcriptome sequencing (RNA-seq). We identified a gene cluster comprising eight genes that was up-regulated during growth with arabitol as a sole carbon source. The RNA-seq results were subsequently confirmed via qRT-PCR experiments. The transcriptional organization of the gene cluster identified via RNA-seq was analyzed and it was shown that the arabitol utilization genes are co-transcribed in an operon that spans from BMMGA3_RS07325 to BMMGA3_RS07365. Since gene deletion studies are currently not possible in B. methanolicus, two complementation experiments were performed in an arabitol negative Corynebacterium glutamicum strain using the four genes discovered via RNA-seq analysis as coding for a putative PTS for arabitol uptake (BMMGA3_RS07330, BMMGA3_RS07335, and BMMGA3_RS07340 renamed to atlABC) and a putative arabitol phosphate dehydrogenase (BMMGA3_RS07345 renamed to atlD). C. glutamicum is a natural D-arabitol utilizer that requires arabitol dehydrogenase MtlD for arabitol catabolism. The C. glutamicum mtlD deletion mutant was chosen for complementation experiments. Heterologous expression of atlABCD as well as the arabitol phosphate dehydrogenase gene atlD from B. methanolicus alone restored growth of the C. glutamicum ΔmtlD mutant with arabitol. Furthermore, D-arabitol phosphate dehydrogenase activities could be detected in crude extracts of B. methanolicus and these were higher in arabitol-grown cells than in methanol- or mannitol-grown cells. Thus, B. methanolicus possesses an arabitol inducible operon encoding, amongst others, a putative PTS system and an arabitol phosphate dehydrogenase for uptake and activation of arabitol as growth substrate.
Bacillus methanolicus is an aerobic, Gram-positive, thermophilic, methanol-utilizing bacterium originally isolated from freshwater marsh soil (Schendel et al., 1990; Arfman et al., 1992). Methylotrophs, such as B. methanolicus, utilize carbon sources without C-C bonds also called C1 substrates. The key intermediate for biological C1 fixation is formaldehyde, and B. methanolicus belongs to the group of facultative methylotrophs that fix formaldehyde via the ribulose monophosphate (RuMP) cycle (Anthony, 1982; Arfman et al., 1992). What makes methanol an attractive feedstock is the fact that it is abundant and cheap, and that addition of methanol to fermentation broth reduces the risk of microbial contamination in fermentative processes due to toxicity of its derivative – formaldehyde (Irla et al., 2015; Müller et al., 2015a). Furthermore, methanol presents a non-food alternative to conventional feedstock generally used in biotechnological processes. The ability to utilize methanol as carbon source, in addition to its high growth temperature, makes B. methanolicus MGA3 a promising candidate for biotechnological amino acid production. It has been successfully used for methanol-based production of the amino acids L-lysine and L-glutamate (Brautaset et al., 2007), and has been engineered for production of the compounds cadaverine (Naerdal et al., 2015; Irla et al., 2016) and γ-aminobutyric acid (GABA) (Irla et al., 2017). Furthermore, tools for gene expression have been recently developed, comprising gene co-expression from two different plasmids and controlled inducible gene expression systems with both rolling circle and theta replicating plasmids (Irla et al., 2016).
Bacillus methanolicus methylotrophy has been extensively studied and characterized in recent years. This contributed to a broader understanding of methanol metabolism and its regulation by fully sequencing the MGA3 genome (Heggeset et al., 2012; Irla et al., 2014), achieving a comprehensive analysis of the transcriptional landscape using RNA-seq (Irla et al., 2015) and accomplishing proteome (Müller et al., 2014) and metabolome (Müller et al., 2015b; Carnicer et al., 2016) studies. These studies not only increased our understanding of methanol metabolism in B. methanolicus, but also yielded insight into catabolic pathways of alternative carbon sources. B. methanolicus is known to utilize D-mannitol and D-glucose as sole carbon and energy sources and metabolic pathways for the utilization of these substrates have already been described (Heggeset et al., 2012). Both mannitol and glucose enter the cells via a phosphotransferase system (PTS) as mannitol 1-phosphate and glucose 6-phosphate, respectively, and are converted to fructose 6-phosphate. Here, we describe and characterize utilization of the pentose sugar alcohol D-arabitol for growth of B. methanolicus MGA3. Two alternative pathways for arabitol utilization have been described in bacteria (Figure 1): uptake of arabitol via a permease followed by intracellular oxidation and phosphorylation to yield xylulose 5-phosphate, route described for proteobacteria and actinobacteria as in e.g., Corynebacterium glutamicum (Laslo et al., 2012), Enterobacter aerogenes (Charnetzky and Mortlock, 1974), Klebsiella pneumoniae (Heuel et al., 1997), Rhizobium trifolii (Primrose and Ronson, 1980), and Pseudomonas fluorescens (Brünker et al., 1998), or PTS-mediated uptake and phosphorylation followed by oxidation to pentose phosphates as described for the firmicutes Listeria moncytogenes (Kentache et al., 2016) and Enterococcus avium (Povelainen et al., 2003).
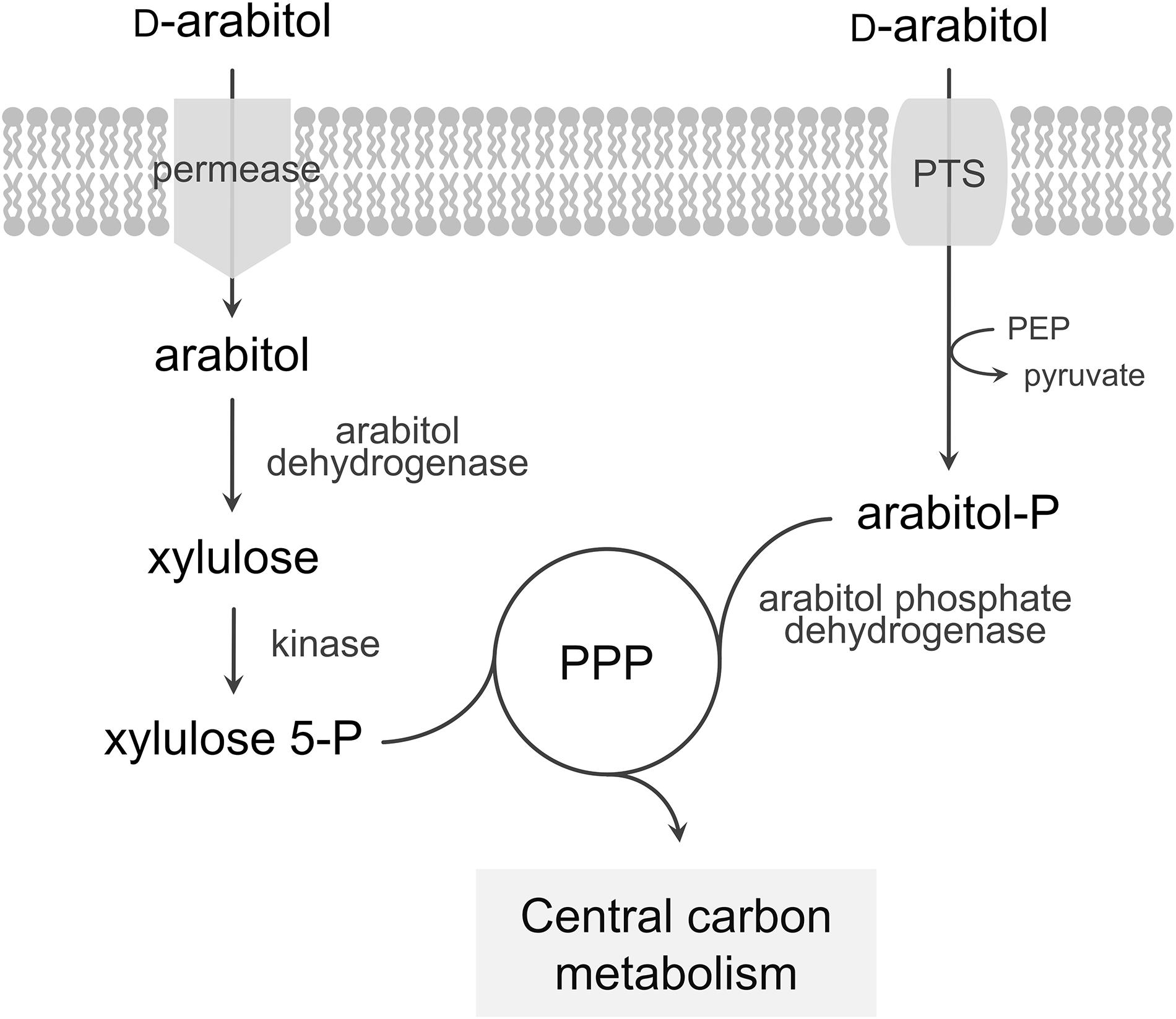
Figure 1. Model of the pathways for arabitol catabolism in bacteria. The first route represents uptake of arabitol via a permease, a dehydrogenase and a kinase while the second route represents uptake via a PTS and a phosphate dehydrogenase. PTS, phosphotransferase system; PEP, phosphoenolpyruvate; PPP, pentose phosphate pathway.
Arabitol is ubiquitous in nature and has been found in plants and fungi, often alongside mannitol, being involved in osmoprotection and carbohydrate storage (Daly et al., 1967; Plemenitaš et al., 2008; Wang et al., 2019). It has additionally been reported that arabitol conferred drought tolerance when provided by a lichenous fungi to the green algae Trebouxia sp. (Kosugi et al., 2013). Yeast or yeast-like fungi produce extracellular glycolipids, some of which have been reported to exceptionally contain mannitol and arabitol residues (Kulakovskaya and Kulakovskaya, 2014b). Their role in metabolism includes promotion of solubilization and absorption of hydrophobic substrates, extracellular reserve of carbon sources and antibiotic activity (Kulakovskaya and Kulakovskaya, 2014a). Roselipins, consisting of C20-fatty acids with three hydroxyl groups, mannose and arabitol residues, are extracellular glycolipids synthesized by Clonostachys rosea (Tabata et al., 1999). Mannosylerythritol lipids are major extracellular glycolipids of the Pseudozyma genera. In Pseudozyma parantarctica the rarely occurring mannosylarabitol lipids and mannosylmannitol lipids have been described (Morita et al., 2009, 2012). The natural ecological niches of these extracellular glycolipid yeast producers include soil, nectaries and leaves of plants (Kulakovskaya and Kulakovskaya, 2014a), coexisting together with B. methanolicus in similar habitats. Additionally, some yeasts possess the ability of transforming glucose into arabitol (Kordowska-Wiater, 2015). It has previously been reported that methanol is a by-product of pectin metabolism during cell wall synthesis and pathogen attack in plants, which in turn assists in plant immunity (Fall and Benson, 1996; Komarova et al., 2014). Bacterial isolates from leaf surfaces showed the presence of B. methanolicus on Citrus paradisi plants (Izhaki et al., 2013). These findings indicate that methanol, mannitol, glucose and arabitol might be present in the natural habitat of B. methanolicus.
In the present study, we characterized growth of B. methanolicus MGA3 on arabitol. Based on our differential transcriptome analysis, the genes coding for proteins involved in arabitol utilization were identified, and their functionality confirmed by genetic complementation and enzyme assays.
The strains used in this study are listed in Table 1. C. glutamicum ATCC 13032 was used as the expression host and Escherichia coli DH5α was used as the general cloning host. E. coli strains were routinely cultivated at 37°C and 180 rpm in Lysogeny Broth (LB) media or on LB plates [1% (w/v) agar] supplemented with 25 μg mL-1 kanamycin if relevant. B. methanolicus strains were cultivated at 50°C and 200 rpm in minimal MVcMY media for pre-cultures or MVcM for main cultures as previously described (Brautaset et al., 2003) with 200 mM methanol, 5, 10, 15, and 50 mM mannitol or 5, 10, 15, 30, and 60 mM arabitol. For co-consumption experiments, 15 mM mannitol and 15 mM arabitol were added to the media. Main cultures of all B. methanolicus experiments were inoculated at a start optical density (OD600) of 0.2. C. glutamicum strains were routinely cultivated at 30°C and 120 rpm in LB media with 30 mM glucose for pre-cultures and in minimal CGXII media (Eggeling and Bott, 2005) for main cultures with 30 mM glucose or 30 mM arabitol. Media were supplemented with 25 μg mL-1 kanamycin when necessary and 1 mM IPTG was added for induction of gene expression at inoculation of the main cultures, which was done at an initial OD600 of 0.5. Cultivations were performed in 500 mL baffled shake flasks with 50 mL media volume and in biological triplicates in all cases.
The description of all plasmids constructed in this study is presented in Table 1. Molecular cloning was performed as described by Sambrook and Russell (2001). Primer sequences used in this study were obtained from Metabion (Planegg/Steinkirchen, Germany) and are listed in Supplementary Table 1. Total DNA isolation from B. methanolicus MGA3 was performed as previously described (Eikmanns et al., 1994). Inserts were amplified by polymerase chain reactions (PCRs) using ALLinTM HiFi DNA Polymerase (HighQu, Kraichtal, Germany) and purified with the NucleoSpin® Gel and PCR Clean-up kit (Macherey-Nagel, Düren, Germany). For plasmid isolation, the GeneJET Plasmid Miniprep Kit (Thermo Fisher Scientific, Waltham, MA, United States) was used. Ends of DNA fragments (PCR-amplified fragments and plasmid pVWEx1 cut with restriction enzymes) were joined by means of the isothermal DNA assembly method (Gibson et al., 2009). Transformation of chemically competent E. coli cells was done following the procedure of Mandel and Higa (1970). Colony PCRs were performed using the Taq polymerase (New England Biolabs, Ipswich, England) with primers acF1, acR1 and acR2 (Supplementary Table 1). All cloned DNA fragments were verified by sequencing (Sequencing Core Facility, Bielefeld University). C. glutamicum competent cells and electroporation were prepared as previously described (Eggeling and Bott, 2005).
Plasmids for inducible gene expression in C. glutamicum were constructed on the basis of pVWEx1 (Peters-Wendisch et al., 2001). The atlABCD and atlABCDEF genes were PCR-amplified from B. methanolicus MGA3 genomic DNA using the primers acF1 and acR1 or acF1 and acR2, respectively, and the atlD gene was amplified using primers aPDF and aPDR (Supplementary Table 1). The resulting PCR product was joined with BamHI digested pVWEx1 via Gibson assembly.
In order to perform total RNA extraction from B. methanolicus MGA3 cells, the NucleoSpin RNA isolation Kit (Machery-Nagel, Düren, Germany) and the RNase-free DNase set (Qiagen, Hilden, Germany) were used according to the manufacturer’s instructions. B. methanolicus cultures were grown in minimal MVcM media containing 200 mM methanol, 15 mM mannitol or 15 mM arabitol. Cells were harvested in the middle of the exponential growth phase at an OD600 of 1.0 followed by total RNA isolation individually for each cultivation condition. The RNA material was tested for contaminating DNA using primers PRIF and PRIR for the amplification of the proI gene and primers MRF1 and MRR1 for the amplification of the mtlR gene (Supplementary Table 1). No product was obtained for any of the tested samples (data not shown). The quality of the samples was subsequently verified by capillary gel electrophoresis (Agilent Bioanalyzer 2100 system using the Agilent RNA 6000 Pico kit; Agilent Technologies, Böblingen, Germany) and the concentration checked by DropSenseTM 16 (Trinean, Ghent, Belgium). The RNA material was subsequently used either for RNA-seq analysis, qRT-PCR or RT-PCR analysis of operon structure.
Isolated RNA samples from B. methanolicus MGA3 were pooled in equal parts and the total RNA was subsequently used for the cDNA library preparation. The library was prepared and sequenced on a single flow cell of a MiSeq Desktop Sequencer system (Illumina, San Diego, CA, United States) in paired-end mode following a protocol that allowed for the analysis of the whole transcriptome (Mentz et al., 2013). Previous to mapping of the generated reads onto the reference genome, the sequences were trimmed using the tool Trimmomatic version 0.33 (Bolger et al., 2014) to a minimal length of 35 base pairs. The trimmed reads were mapped to the B. methanolicus MGA3 reference sequences of the chromosome as well as the two plasmids pBM19 and pBM69 (GenBank accession numbers CP007739, CP007741, and CP007740, respectively) using the software for short read alignment Bowtie (Langmead et al., 2009). For the visualization of the mapped reads the ReadXplorer software was used (Hilker et al., 2014). The differential gene expression analysis was performed with the statistical method DESeq (Anders and Huber, 2010) using the same software. In order to designate a gene as differentially expressed, the cut-off values were set to a change in expression level higher than 30, for which the P-value was adjusted to be equal to or less than 0.01. Sequences of differentially expressed genes that coded for proteins of unknown function were subjected to BLASTx analysis for identification of protein family conservations (Altschul et al., 1990).
qRT-PCR was performed in order to validate the generated data in RNA-seq analysis. Isolated RNA samples from B. methanolicus MGA3 grown on either mannitol, arabitol or a combination of both were used as template. The optimization of a qRT-PCR protocol included a series of standard PCRs using different primer concentrations (250, 400, and 600 mM) and different annealing temperatures (48 to 65°C). For each gene to be analyzed, a pair of primers were designed for the amplification of about 200 bp using the primer design tool of Clone Manager 9 (Scientific & Educational Software, Denver, CO, United States) (Supplementary Table 1). The experiments were performed with the LightCycler® 96 System (Roche Diagnostics, Penzberg, Germany) using the SensiFASTTM SYBR® No-ROX One-Step Kit (Bioline, Luckenwalde, Germany) according to the manufacturer’s instructions. After the optimization process, each reaction mixture contained 400 mM of each primer and 50 ng of RNA in a final volume of 20 μL. The qRT-PCR profile was chosen to be performed as follows: the reverse transcription was performed at 45°C for 10 min, the polymerase activation at 95°C for 2 min followed by 40 cycles of a three-step amplification composed of a denaturation step at 95°C for 5 s, annealing at 55°C for 10 s and extension at 72°C for 5 s and, lastly, dissociation curve analyses were done from 65°C up to 95°C in 0.5°C increments for 5 s each step. Amplification of repB, the pBM19 replication initiator gene, was used for sample normalization following the recommendations of Jakobsen et al. (2006). They could confirm by qRT-PCR that the repB transcript levels were similar in cells utilizing mannitol and methanol and, additionally, our RNA-seq data showed similar repB expression between mannitol and arabitol, too (data not shown). Relative quantification was done by means of the comparative threshold cycle method: the 2-ΔΔCT method (Livak and Schmittgen, 2001). All measurements were performed in technical replicates.
Analysis of the transcriptional organization of the arabitol gene cluster was done via RT-PCR. Isolated RNA from B. methanolicus MGA3 grown on arabitol was used as template for cDNA synthesis using the BioScriptTM Reverse Transcriptase kit (Bioline, Luckenwalde, Germany) according to the manufacturer’s instructions with gene specific primer RT03, which hybridizes in BMMGA3_RS07365 (Supplementary Table 1). In order to detect if the referred gene cluster was transcribed as a single mRNA molecule, the resulting cDNA was then used as template for PCR using primers spanning gene borders of all genes putatively present in the arabitol gene cluster (Supplementary Table 1).
Bacillus methanolicus MGA3 cells were grown in minimal media with methanol, mannitol or arabitol and collected by centrifugation at 4°C and 4,000 rpm for 10 min in the middle of the exponential growth phase at an OD600 of around 1.0. The pelleted cells were re-suspended in Tris-HCl buffer (pH 7.2) and disrupted by sonication (UP 200 S, Dr. Hielscher GmbH, Teltow, Germany) on ice at an amplitude of 55% and a duty cycle of 0.5 for 9 min with a 30 s pause in between. To obtain lysates, the samples were centrifuged for 60–90 min at 4°C and 14,000 rpm to remove cell debris, the supernatants were then collected and protein concentration was determined by means of the Bradford method (Bradford, 1976) using bovine serum albumin as reference. Determination of the arabitol phosphate dehydrogenase activity in the reductive reaction using xylulose 5-phosphate as substrate was performed following the indications of Povelainen et al. (2003). The assay mixture contained 20 mM Tris-HCl buffer (pH 7.2), 1 mM DTT, 0.3 mM NADH, 50–200 μL of crude extract and 0.2–2 mM xylulose 5-phosphate in a total volume of 1 mL. The oxidation rate of NADH was monitored at 340 nm and 30°C for 3 min on a Shimadzu UV-1202 spectrophotometer (Shimadzu, Duisburg, Germany).
For the quantification of growth substrates mannitol and arabitol in a cultivation broth a high-pressure liquid chromatography (HPLC) system was used (1200 series, Agilent Technologies Deutschland GmbH, Böblingen, Germany) as in Pérez-García et al. (2016). The supernatants of the cell cultures were obtained by centrifugation at 14,000 rpm and at room temperature for 15 min. The analysis was carried out using a column for organic acids (300 mm × 8 mm, 10 μm particle size, 25 Å pore diameter, CS Chromatographie Service GmbH, Langerwehe, Germany) with mobile phase of 5 mM sulphuric acid at a flow rate of 0.8 mL min-1, at 60°C and for 17 min. The detection was executed with a refractive index detector (RID G1362A, 1200 series, Agilent Technologies).
In search of gratuitous inducer of an mtlR promoter described by Irla et al. (2016), arabitol was tested as one of the potentially feasible compounds. While it was shown that addition of arabitol to cultivation broth does not lead to induction of expression of genes controlled by PmtlR, the authors have discovered that arabitol serves as sole carbon and energy source for B. methanolicus (Irla et al., 2016). Due to the fact that at that time only two alternative carbon sources had been described for this facultative methylotroph, it was interesting to investigate the physiology and genetic background of arabitol utilization in B. methanolicus. For that purpose, B. methanolicus MGA3 was cultivated in MVcM minimal media containing 10, 15, 30, or 60 mM arabitol. Arabitol was completely consumed in all tested conditions except when 60 mM was used, since about 20 mM remained in the supernatant when growth stopped (Supplementary Figure 1). Although arabitol supported growth of MGA3 as a sole source of carbon and energy, the strain grew at a lower growth rate than with its preferred sugar alcohol substrate mannitol: the growth rate using arabitol was 0.20 ± 0.01 h-1 as compared to 0.37 ± 0.01 h-1 when mannitol was used (Table 2). Accordingly, the substrate consumption rate was higher for mannitol than for arabitol (7.4 ± 0.5 vs. 5.7 ± 0.1 mmol g CDW-1 h-1), which was also the case for the biomass yield (0.28 ± 0.01 vs. 0.24 ± 0.01 g CDW g-1 or carbon normalized biomass yield 0.70 ± 0.02 vs. 0.60 ± 0.02 g CDW g carbon-1) (Table 2). As shown in Figure 2A, a relation between growth rate and substrate concentration according to Monod kinetics indicated that 2.9 ± 0.9 mM arabitol supported growth with a half-maximal growth rate.
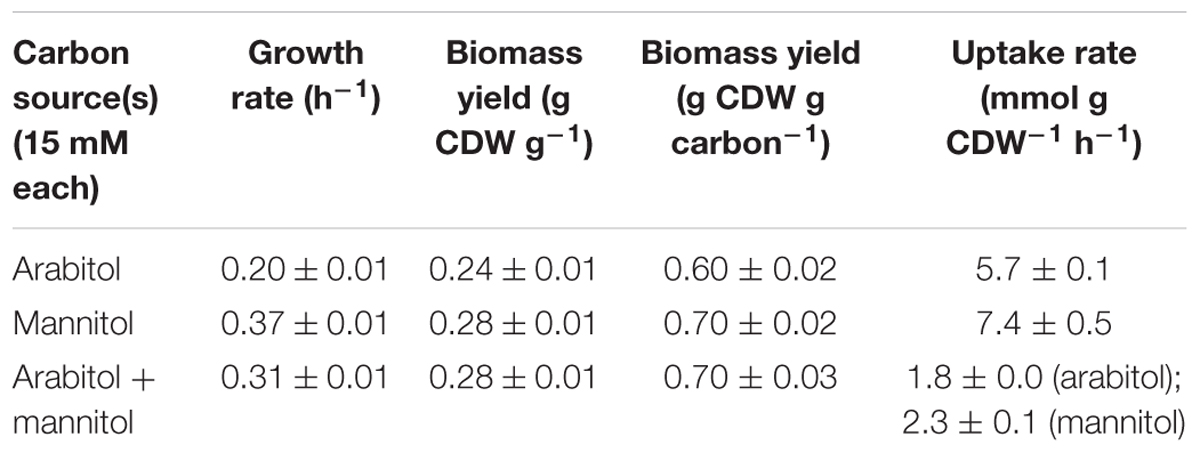
Table 2. Growth rates, uptake rates, biomass yields, and other characteristics of B. methanolicus MGA3 grown on arabitol, mannitol, or a combination of both.
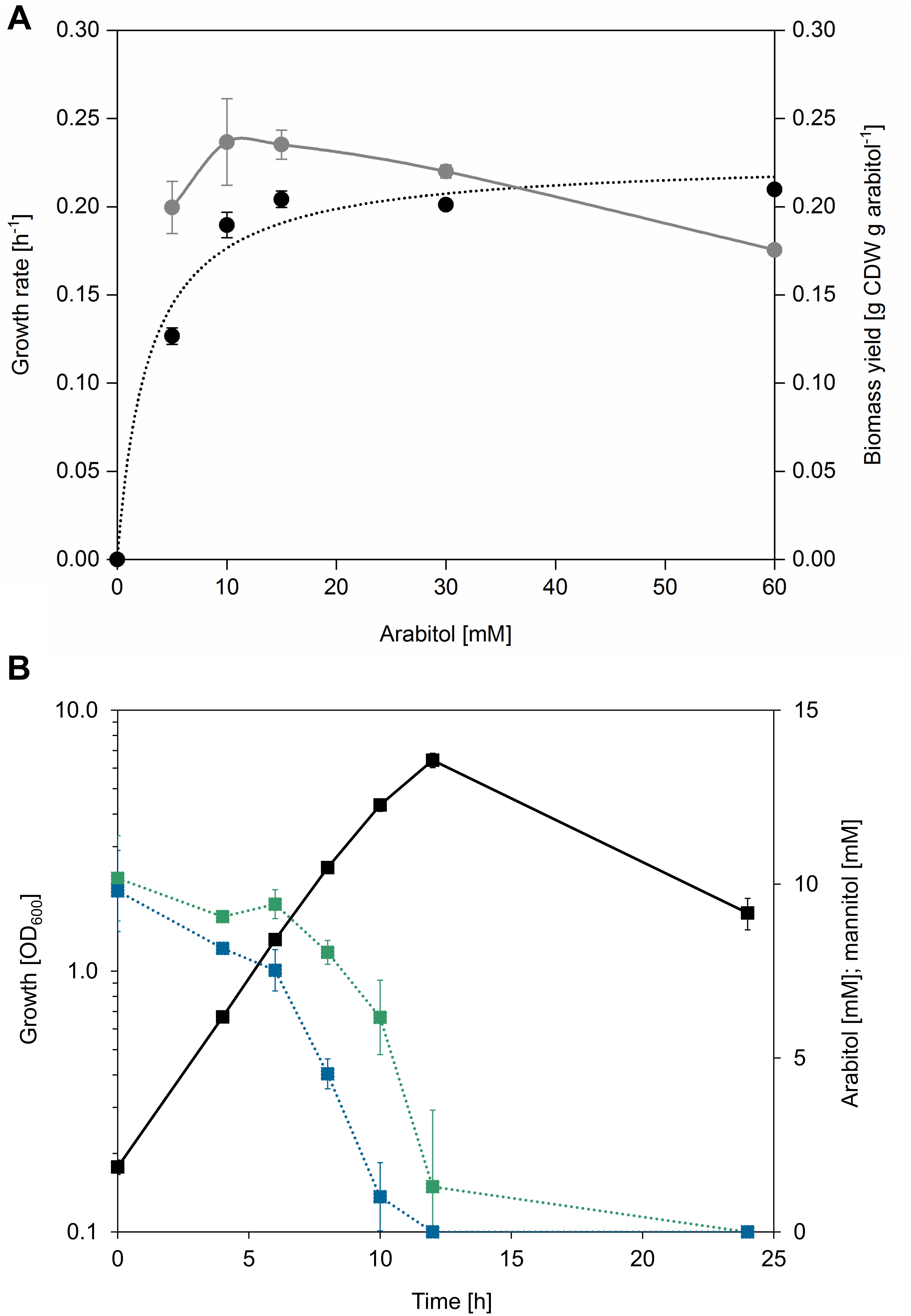
Figure 2. (A) Growth rates (black dots) and biomass yields (gray dots) of B. methanolicus MGA3 grown in minimal media containing 5, 10, 15, 30, and 60 mM arabitol. Full arabitol consumption was observed for all conditions except when 60 mM was used, in which case 19.1 ± 0.78 mM residual arabitol was detected after growth stopped. A relation between growth rate and substrate concentration was generated with the Michaelis Menten model using the OriginPro software version 2018 (OriginLab Corporation, Northampton, MA, United States). (B) Growth (black squares, solid lines) and residual substrate concentration (mannitol: blue squares, dotted lines; arabitol: green squares, dotted lines) of B. methanolicus MGA3 in minimal media containing a mixture of 15 mM mannitol and 15 mM arabitol. Mean values and standard deviations of triplicate shake flask cultures are given.
In order to test if arabitol and mannitol are utilized sequentially or simultaneously, growth of MGA3 in minimal media containing a mixture of 15 mM arabitol and 15 mM mannitol was analyzed (Figure 2B). MGA3 did not show biphasic growth in that experiment, and the maximum OD600 of 6.34 ± 0.41 was reached. Mannitol was utilized faster than arabitol and co-consumption of both sugar alcohols was observed between 6 and 12 h, at which point the growth stopped and both substrates were fully consumed (Figure 2B). As expected, the uptake rates for mannitol and arabitol during co-consumption (2.3 ± 0.1 and 1.8 ± 0.0 mmol g CDW-1 h-1, respectively) were lower than when either carbon source was provided as sole substrate (7.4 ± 0.5 and 5.7 ± 0.1 mmol g CDW-1 h-1, respectively) and the biomass yield using both mannitol and arabitol was the same as when only mannitol was provided to the media (0.28 ± 0.01 g CDW g-1) (Table 2).
In order to elucidate the genetic background of arabitol utilization, a differential gene expression analysis of B. methanolicus MGA3 cultivated with either 15 mM mannitol or 15 mM arabitol as sole carbon source was performed by RNA-seq. Sequencing of the prepared cDNA libraries from RNA isolated under the two chosen conditions resulted in 3,200,444 raw reads for cDNA library of mannitol grown cells and 2,728,707 raw reads for cDNA library of arabitol grown cells. Of these reads, 3,163,610 were mapped onto the genome of B. methanolicus MGA3 for the mannitol condition, leading to a coverage of 89.50% for the chromosome and 9.16 and 0.53% for the natural plasmids pBM19 and pBM69, respectively. For the arabitol condition, 2,684,972 reads were mapped onto the genome with an 88.16% coverage for the chromosome, 10.09% for pBM19 and 0.71% for pBM69 (Supplementary Table 2). During growth on arabitol, 48 genes showed significantly higher and 24 genes significantly lower RNA levels than during growth with mannitol (Supplementary Table 3). Of those, four genes involved in mannitol uptake and catabolism (Heggeset et al., 2012; Irla et al., 2016) and eight genes putatively involved in arabitol metabolism were selected and are shown in Table 3. As expected, genes coding for proteins related to mannitol metabolism showed higher RNA levels during growth on mannitol as compared to growth on arabitol. On the other hand, a gene cluster including genes BMMGA3_RS07325 to BMMGA3_RS07360 showed higher RNA levels during growth on arabitol as compared to mannitol-based growth (Table 3). The cluster comprises four genes annotated as coding for a PTS for galactitol uptake (BMMGA3_RS07330, BMMGA3_RS07335 and BMMGA3_RS07340) and a sorbitol dehydrogenase gene (BMMGA3_RS07345). However, neither galactitol nor sorbitol supported growth of B. methanolicus MGA3 (data not shown). Therefore, it was concluded that these genes may, in fact, be involved in arabitol metabolism.
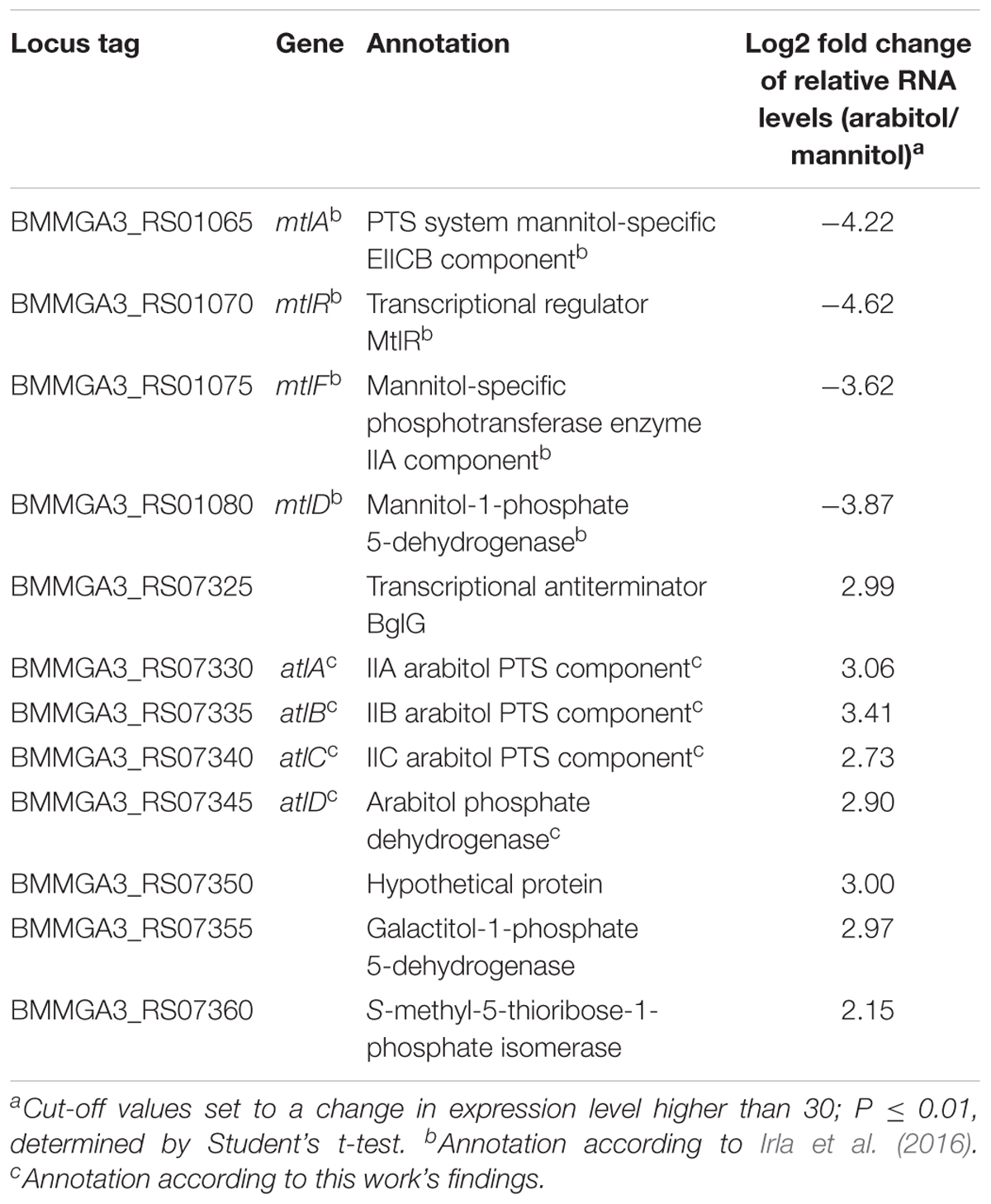
Table 3. Key genes of mannitol and arabitol metabolism with altered expression in B. methanolicus MGA3 cultivated with arabitol in comparison to mannitol as sole carbon source.
To validate the RNA-seq results, qRT-PCR experiments were performed. As shown in Figure 3A, the expression levels of the targeted genes detected by qRT-PCR were in accordance with the gene expression patterns obtained by RNA-seq analysis (Table 3). Additionally, RNA levels of mtlD, mtlR, atlC, and atlD were determined by qRT-PCR during growth on the mixture of mannitol and arabitol as combined carbon sources. Differential expression was observed for genes mtlD and atlD, which code for catabolic enzymes, whereas RNA levels of the regulatory gene mtlR and the transport gene atlC did not change significantly (Figure 3B).
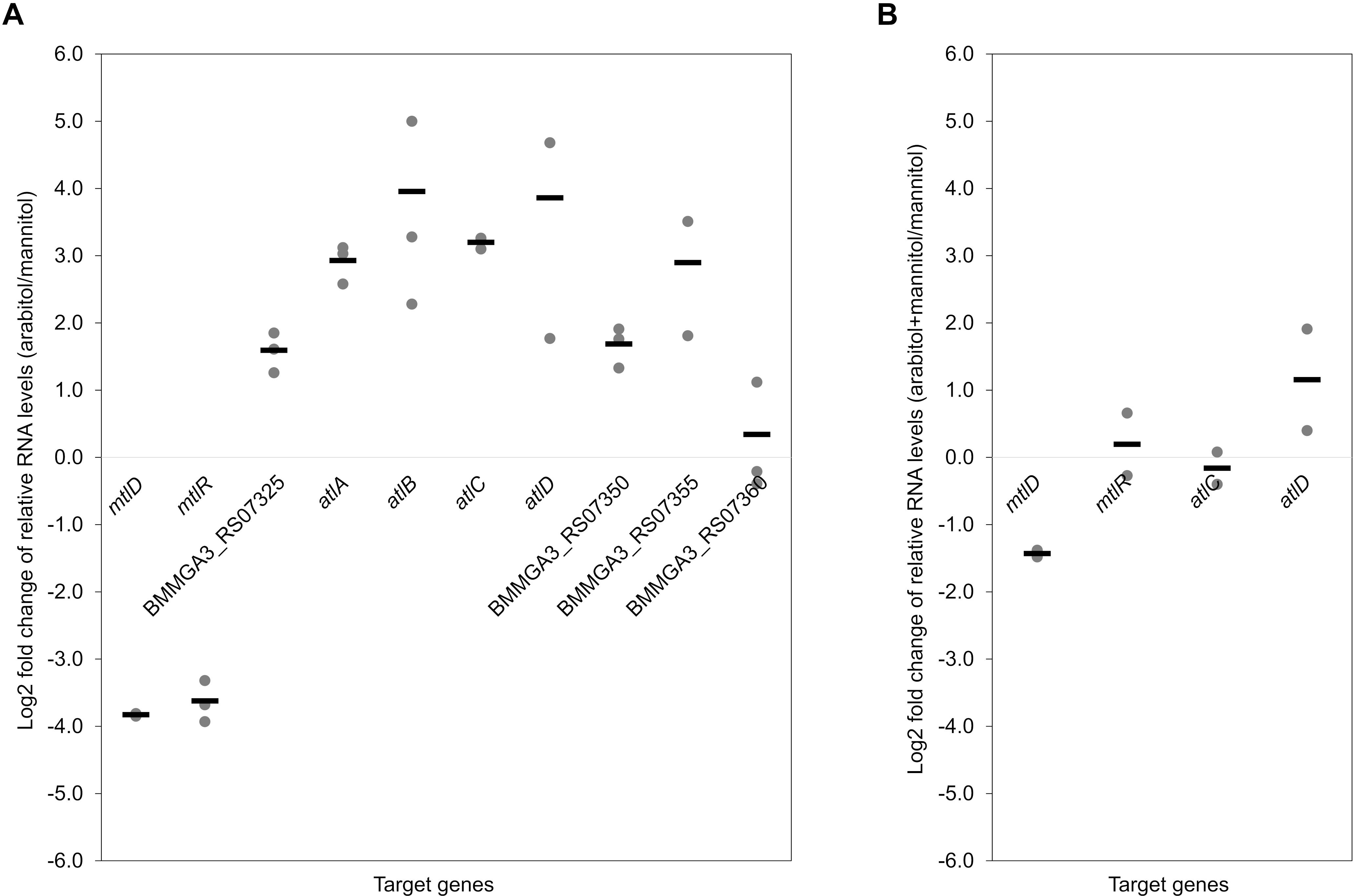
Figure 3. Dot plot graph depicts relative gene expression levels (shown in Log2 fold change) obtained in qRT-PCR of B. methanolicus MGA3 grown on arabitol in comparison to mannitol (A) and on a mixture of arabitol and mannitol in comparison to mannitol (B). Graph represents individual data points and means as a bar.
In order to verify the hypothesis that the atlABCD genes code for a PTS and an arabitol phosphate dehydrogenase and support arabitol catabolism, the arabitol-negative C. glutamicum ΔmtlD mutant (Laslo et al., 2012) was used for genetic complementation experiments. This experiment could not have been performed with an arabitol-negative B. methanolicus because gene deletion studies are currently not possible in this bacterium. C. glutamicum ΔmtlD is unable to grow with arabitol as sole carbon source (Laslo et al., 2012). Therefore, atlABCD genes from MGA3 were cloned into C. glutamicum expression vector pVWEx1, and the resulting vector pVWEx1-atlABCD was used to transform C. glutamicum ΔmtlD with the aim of restoring growth on arabitol. While the ΔmtlD mutant transformed with pVWEx1 empty vector was unable to grow on arabitol, heterologous expression of B. methanolicus MGA3-derived atlABCD genes from vector pVWEx1 allowed for growth with arabitol as sole carbon source and led to complete arabitol consumption (Figure 4A). However, since C. glutamicum ΔmtlD still possesses the arabitol transporter rbtT (Laslo et al., 2012), the possibility that the strain could still import arabitol via the native permease and that AtlD would subsequently take over its oxidation to xylulose could not be excluded. Therefore, we performed a complementation experiment in C. glutamicum ΔmtlD heterologously expressing the B. methanolicus atlD gene alone. Indeed, atlD could complement the deficiency of the ΔmtlD strain as efficiently as when the four atlABCD genes were heterologously expressed (Figure 4B and Supplementary Table 4). As expected, growth of C. glutamicum wild type (WT) with pVWEx1 empty vector on arabitol (Figure 4) and growth of C. glutamicum ΔmtlD on glucose was unaffected (Supplementary Figure 2). Thus, the atlD gene was shown to be sufficient to restore growth on arabitol of the arabitol-negative C. glutamicum ΔmtlD strain and provided evidence that atlD functions in arabitol utilization in B. methanolicus MGA3. Although the functionality of the atlABC genes could not be shown, their genetic organization and proximity to atlD in addition to the results obtained from the differential gene expression analysis strongly support their involvement in arabitol uptake, hence the genes were re-annotated to atlABCD (Table 3).
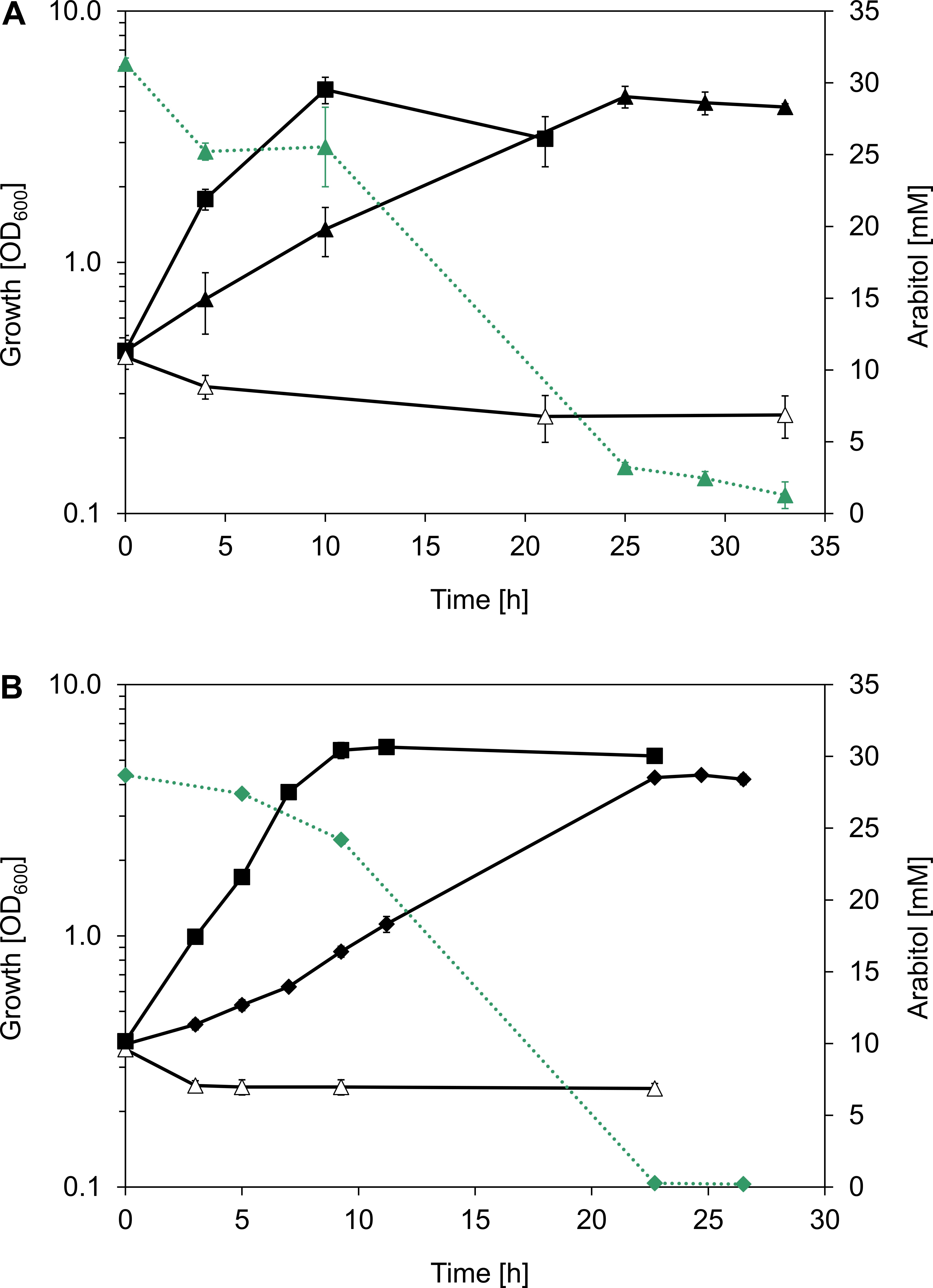
Figure 4. B. methanolicus MGA3 arabitol pathway genes complement an arabitol-negative C. glutamicum strain. (A) Growth (black, solid lines) of C. glutamicum strains WT(pVWEx1) (full squares), ΔmtlD(pVWEx1) (empty triangles), and ΔmtlD(pVWEx1-atlABCD) expressing atlABCD genes from B. methanolicus MGA3 under IPTG induction (full triangles) in minimal media containing 30 mM arabitol. (B) Growth (black, solid lines) of C. glutamicum strains WT(pVWEx1) (full squares), ΔmtlD(pVWEx1) (empty triangles) and ΔmtlD(pVWEx1-atlD) expressing the atlD gene from B. methanolicus MGA3 under IPTG induction (full diamonds) in minimal media containing 30 mM arabitol. Arabitol consumption (green, dotted lines) is depicted for C. glutamicum ΔmtlD(pVWEx1-atlABCD) and C. glutamicum ΔmtlD(pVWEx1-atlD). Mean values and standard deviations of triplicate shake flask cultures are given.
In an additional experiment, the C. glutamicum ΔmtlD mutant strain was complemented with the atlABCD and the two consecutive BMMGA3_RS07350 and BMMGA3_RS07355 genes. BMMGA3_RS07350 codes for a hypothetical protein and BMMGA3_RS07355 is annotated as coding for a galactitol-1-phosphate 5-dehydrogenase. However, growth and uptake rates weren’t significantly different than when only atlD or atlABCD from B. methanolicus MGA3 were used for complementation (Supplementary Table 4).
The atlABCD genes are clustered on the B. methanolicus genome and are arranged in the same transcriptional orientation as several neighboring genes (Figure 5A). Previous transcription analyses indicated that genes BMMGA3_RS07325 to BMMGA3_RS07355 might be organized in an operon (Irla et al., 2015). In addition to the previously described genes showing higher RNA levels during growth on arabitol in comparison to mannitol (Table 3), our RNA-seq data suggested that BMMGA3_RS07365 might also be co-transcribed with the preceding genes based on reads spanning two genes. For independent confirmation of that assumption, RT-PCR experiments were performed. RNA obtained during growth on arabitol was reverse transcribed with primer RT03 annealing downstream of BMMGA3_RS07365 and used as template for PCRs with primer pairs spanning the gene borders (Figure 5B). The observed amplification products indicated that indeed genes BMMGA3_RS07325 to BMMGA3_RS07365 are co-transcribed as an operon (Figure 5C). As positive control, B. methanolicus MGA3 genomic DNA was used as template (Figure 5D). The absence of contaminations of the RNA preparation with genomic DNA was confirmed in PCRs using RNA as template (Figure 5E). Thus, genes atlA, atlB, atlC, and atlD are part of a larger operon that ranges from BMMGA3_RS07325 to BMMGA3_RS07365.
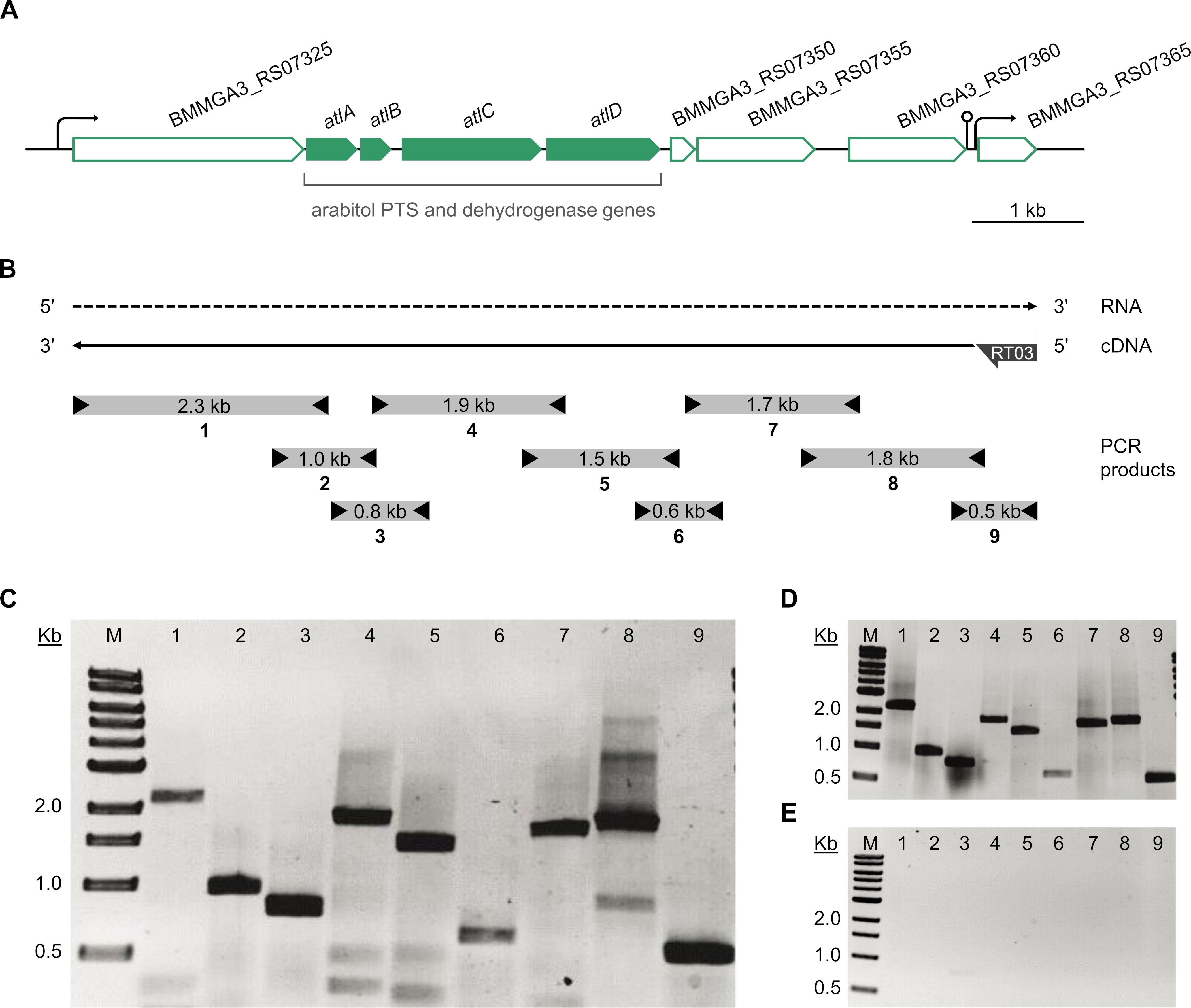
Figure 5. Transcriptional organization of the arabitol operon. (A) Genetic organization of the arabitol operon in B. methanolicus MGA3. Arrows represent the length and direction of the coding regions. (B) Representation of the RT-PCR analysis of the operon structure. The dashed arrow depicts the RNA transcript, the solid arrow depicts the cDNA generated by reverse transcription and subsequently used as template for PCR. Boxes with arrowheads depict the location and size of amplified PCR products (1–9) using primer pairs that spanned gene borders and the arrow at the cDNA 5′ end depicts the location of the gene specific primer RT03 hybridizing with BMMGA3_RS07365. (C) Agarose gel electrophoresis of the amplified PCR products using the RT-generated cDNA as template. (D) Agarose gel electrophoresis of the amplified PCR products using B. methanolicus MGA3 genomic DNA template as positive control. (E) Agarose gel electrophoresis of the amplified PCR products using no-RT (RNA) template as negative control. Lane M: 0.5–10 kb DNA ladder (NEB). Lanes 1 to 9: PCR products (1: 2,273 bp; 2: 958 bp; 3: 792 bp; 4: 1,858 bp; 5: 1,466 bp; 6: 629 bp; 7: 1,708 bp; 8: 1,794 bp; 9: 554 bp).
In order to confirm whether B. methanolicus MGA3 possesses an arabitol phosphate dehydrogenase and to assay if its activity is increased during growth with arabitol, crude extracts of MGA3 cells grown on arabitol, mannitol or methanol were prepared and arabitol phosphate dehydrogenase activities were determined. The enzyme assays were carried as described by Povelainen et al. (2003) with xylulose 5-phosphate as substrate. Since arabitol 1-phosphate and arabitol 5-phosphate were not available, arabitol phosphate oxidation could not be assayed. Instead, reduction of xylulose 5-phosphate was assayed and arabitol phosphate dehydrogenase activity was shown in B. methanolicus crude extracts. As expected, the highest enzyme activity (0.05 ± 0.01 U mg-1) was detected in extracts of cells grown on arabitol (Table 4). Surprisingly, mannitol grown cells showed, albeit reduced, arabitol phosphate dehydrogenase activity (0.02 ± 0.00 U mg-1), while methanol grown cells lacked detectable arabitol phosphate dehydrogenase activity (Table 4). Using the crude extracts prepared from arabitol grown cells, the KM value for the substrate xylulose 5-phosphate was determined to be 0.03 ± 0.02 mM. Although this is a rough estimate obtained with crude extracts rather than with the purified enzyme, the sub-millimolar KM value indicates high affinity of the arabitol phosphate dehydrogenase from B. methanolicus for the substrate xylulose 5-phosphate and is in line with the arabitol concentration supporting half-maximal growth (KS of 1.2 ± 0.3 mM).

Table 4. Specific activities of arabitol phosphate dehydrogenase in cell extracts of B. methanolicus MGA3 grown in minimal medium containing different carbon sources.
Growth of B. methanolicus MGA3 on the sugar alcohol D-arabitol is characterized for the first time. Based on an RNA-seq analysis of global gene expression during growth on this sugar alcohol, genetic and biochemical investigation of a role of atlABCD encoding a PTS and arabitol phosphate dehydrogenase in uptake and activation of arabitol is demonstrated. Figure 6 depicts the proposed pathway for arabitol uptake and catabolism next to the known pathways for methanol, mannitol and glucose utilization operating in B. methanolicus MGA3.
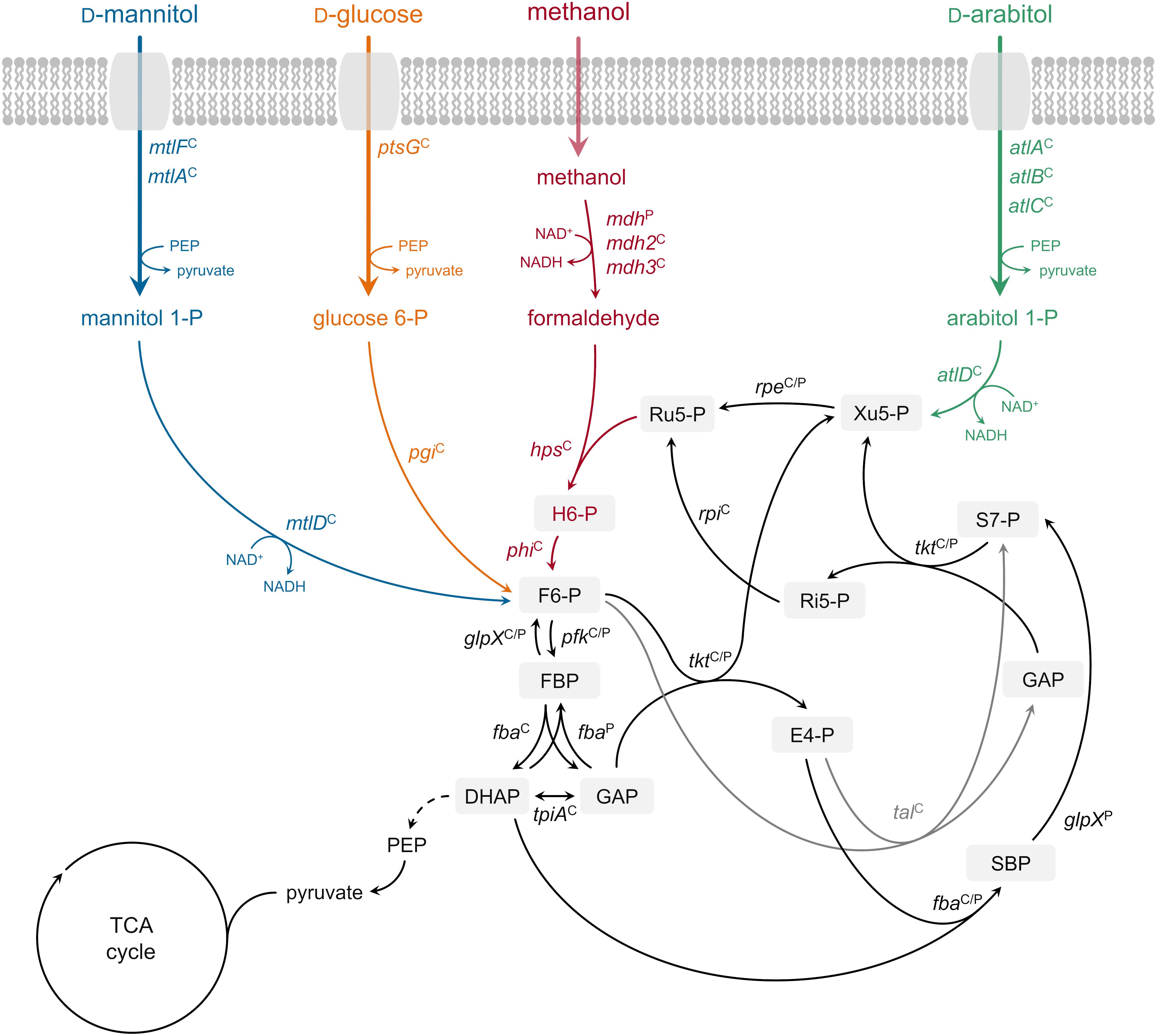
Figure 6. Ribulose monophosphate cycle and proposed methylotrophic and carbohydrate assimilation pathways in B. methanolicus MGA3. Genes: mdh, methanol dehydrogenase (EC 1.1.1.244); hps, 3-hexulose-6-phosphate synthase (EC 4.1.2.43); phi, 6-phospho-3-hexuloisomerase (EC 5.3.1.27); pfk, 6-phosphofructokinase, (EC 2.7.1.11); fba, fructose-bisphosphate aldolase (EC 4.1.2.13); tkt, transketolase (EC 2.2.1.1); glpX, fructose-bisphosphatase (EC 3.1.3.1); tal, transaldolase (EC 2.2.1.2); rpe, ribulosephosphate 3-epimerase (EC 5.1.3.1); rpi, ribose-5-phosphate isomerase (EC 5.3.1.6); tpiA, triosephosphate isomerase (EC 5.3.1.1); ptsG, PTS-glucose-specific transporter subunit IICBA (EC 2.7.1.69) pgi, glucose-6-phosphate isomerase (EC 5.3.1.9); mtlF, mannitol-specific phosphotransferase enzyme IIA component (EC 2.7.1.69); mtlA, PTS system mannitol-specific enzyme IIBC component (EC 2.7.1.69); mtlD, mannitol-1-phosphate 5-dehydrogenase (EC 1.1.1.17); atlA, IIA arabitol PTS component; atlB, IIB arabitol PTS component; atlC, IIC arabitol PTS component; atlD, arabitol phosphate dehydrogenase. Metabolites: H6-P, 3-hexulose 6-phosphate; F6-P, fructose 6-phosphate; FBP, fructose 1,6-bisphosphate; GAP, glyceraldehyde 3-phosphate; DHAP, dihydroxyacetone phosphate; E4-P, erythrose 4-phosphate; SBP, sedoheptulose 1,7-bisphosphate; S7-P, sedoheptulose 7-phosphate; Ri5-P, ribose 5-phosphate; Xu5-P, xylulose 5-phosphate; Ru5-P, ribulose 5-phosphate; PEP, phosphoenolpyruvate; TCA tricarboxylic acid. Superscript “C”: chromosomally encoded; superscript “P”: natural plasmid pBM19 encoded. Phosphoenolpyruvate–protein phosphotransferase [enzyme EI (EC 2.7.3.9)] and phosphocarrier protein (HPr) are not depicted.
The finding that B. methanolicus MGA3 is able to catabolize low arabitol concentrations effectively (KS value of about 3 mM) is in line with our prediction that arabitol is taken up by the cells via arabitol PTS encoded by arabitol inducible genes atlABC. High substrate affinity has been reported in several organisms in relation to PTS-mediated uptake (Nothaft et al., 2003; Lindner et al., 2011; Opačić et al., 2012), as is also the case for mannitol in B. methanolicus: the KS value for mannitol was determined to be 0.2 ± 0.1 mM with genes mtlF, mtlA, and mtlD known to code for a mannitol-specific PTS enzyme IIA component, a mannitol-specific PTS enzyme IIBC component and a mannitol-1-phosphate 5-dehydrogenase, respectively (Heggeset et al., 2012; Irla et al., 2016). However, the observed growth rates and substrate uptake rates were higher for mannitol than for arabitol, and the biomass yield for the C5 sugar alcohol arabitol was lower than for the C6 sugar alcohol mannitol (Table 2). Methanol allows growth rate and biomass yield similar to those observed for mannitol-based growth (Jakobsen et al., 2006; Nilasari et al., 2012). Thus, if a mixture of methanol, mannitol and arabitol is present in the natural habitat, arabitol is expected to contribute to growth of B. methanolicus albeit slower and less efficient as compared to methanol and mannitol. This is likely even more pronounced if high arabitol concentrations are encountered in its ecological niche, since B. methanolicus cannot utilize arabitol efficiently at concentrations exceeding 30 mM, as biomass yields with 60 mM arabitol, for example, were only moderately higher than with 30 mM and residual arabitol was observed when growth stopped (Supplementary Figure 1).
Co-consumption of arabitol with mannitol and monophasic growth were observed with an equimolar mixture of both sugar alcohols as combined carbon source (Table 2). Monophasic growth with simultaneous substrate consumption has been previously reported for e.g., C. glutamicum in a mixture of glucose and pyruvate (Cocaign et al., 1993), glucose and fructose (Dominguez et al., 1997) or glucose and acetate (Wendisch et al., 2000), Mycobacterium tuberculosis in a mixture of glucose, acetate, and/or glycerol (de Carvalho et al., 2010) and Bacillus subtilis in a mixture of glucose and malate (Kleijn et al., 2010), although consumption of single carbon substrates in a preferred order displaying diauxic growth, consequence of catabolite repression, is a more widespread mechanism in most bacteria (Kovárová-Kovar and Egli, 1998). The fact that both arabitol and mannitol uptake rates were more than three-fold lower during growth with the mixture than with either sugar alcohol alone (Table 2) indicated regulation of carbon source utilization. This assumption is confirmed by our qRT-PCR analysis (Figure 3B) where mtlD transcript levels are lower in the cells cultivated on mixture of arabitol and mannitol in comparison to mannitol only. Inhibition of glucose uptake during growth on glucose in the presence of arabitol was previously reported by Laslo et al. (2012) both for the wild type and the C. glutamicum ΔmtlD mutant strain. Co-consumption of glucose and xylose led, too, to lower consumption rates for glucose in the E. coli ΔptsG mutant (Matsuoka and Shimizu, 2013), while glucose was shown to inhibit pentose uptake in Saccharomyces cerevisiae (Subtil and Boles, 2012).
Differential gene expression analysis of B. methanolicus MGA3 grown on arabitol as compared to mannitol detected higher RNA levels for mtlA, mtlR, mtlF, and mtlD in mannitol-grown cells (Table 3) as expected (Heggeset et al., 2012; Müller et al., 2014; Irla et al., 2016). The higher RNA levels observed for the putative arabitol PTS genes atlA, atlB, and atlC as well as arabitol phosphate dehydrogenase gene atlD during growth on arabitol compared to growth with mannitol might be due to arabitol induction or lack of mannitol repression. In C. glutamicum, arabitol induces expression of rbtT, mtlD, sixA, xylB and atlR and glucose represses expression of mtlD (Laslo et al., 2012). In B. methanolicus, RNA levels for atlD were higher while lower for mtlD during growth with both carbon sources as compared to growth with mannitol alone (Figure 3B). By contrast, the RNA levels of both mtlR and atlC were comparable under both conditions. This may indicate arabitol repression of mtlR and mannitol repression of atlC in the arabitol/mannitol mixture condition. Moreover, this observation is in line with the finding that both carbon sources are utilized faster when present alone as compared to growth with a mixture of mannitol and arabitol (Table 2). Contrarily, in Pseudomonas fluorescens DSM 50106 mannitol, arabitol and glucitol are inducers for transcription of mtl operon coding for proteins involved in transport and utilization of those sugar alcohols and the gene expression is regulated by the transcriptional regulator MtlR (Hoffmann and Altenbuchner, 2015).
Here, the physiological role of the altABCD genes in the utilization of arabitol by B. methanolicus was analyzed via complementation studies. Growth with arabitol of the arabitol growth-deficient C. glutamicum ΔmtlD strain was restored by heterologous expression of atlABCD as well as atlD from B. methanolicus. However, involvement of atlABC in arabitol uptake in B. methanolicus could not be confirmed via complementation experiments: the phenotypes of C. glutamicum ΔmtlD(pVWEx1-atlABCD) and C. glutamicum ΔmtlD(pVWEx1-atlD) did not show significant differences (Figure 4 and Supplementary Table 4). The fact that the atlABC genes were not required for arabitol uptake in the ΔmtlD mutant was additionally supported by the displayed substrate affinity: the arabitol affinity of the PTS in B. methanolicus was estimated at the level of 2.9 ± 0.9 mM, whereas the arabitol KS for C. glutamicum ΔmtlD(pVWEx1-atlABCD) was almost ten-fold higher, namely 9.4 ± 0.3 mM (Supplementary Figure 3A). This result is similar to arabitol KS in C. glutamicum WT(pVWEx1) of 8.3 ± 2.0 mM (Supplementary Figure 3B) or wild type C. glutamicum 7.91 ± 0.52 mM (Laslo et al., 2012). The difference of KS of B. methanolicus-derived PTS system in the genetic background of B. methanolicus and C. glutamicum ΔmtlD might be due to the presence of the native arabitol permease rbtT in the latter. The observation that only atlD was necessary to complement the C. glutamicum ΔmtlD deficiency implies that either AtlD has arabitol dehydrogenase activity besides the here determined arabitol phosphate dehydrogenase activity (Table 4) or that C. glutamicum can import arabitol via an additional uptake route, which would be supported by the fact that C. glutamicum ΔrbtT, although poorly, can still grow on arabitol (Laslo et al., 2012). Despite the fact that the atlABC genes were not found necessary to complement C. glutamicum ΔmtlD, their participation in arabitol uptake in B. methanolicus cannot be excluded. As seen from the differential gene expression analysis, atlABCD were clearly up-regulated under arabitol conditions (Table 3). Moreover, transcriptional organization experiments revealed that said genes are part of the same operon and the genetic organization is in accordance to previously reported arabitol PTS and dehydrogenase genes (Povelainen et al., 2003; Kentache et al., 2016). BLASTp analyses recognized AtlABC as part of the multienzyme PTS complex involved in the transport and phosphorylation of carbohydrates. The PTS phosphorylation cascade involves the general PTS components phosphoenolpyruvate–protein phosphotransferase enzyme I (EI) and phosphocarrier protein (HPr), and the carbohydrate-specific permease enzyme II, consisting of two cytoplasmic domains (IIA and IIB) and a transmembrane channel domain (IIC, with or without IID depending on the system) (Saier, 2015; Kentache et al., 2016). Homology comparisons identified AtlA as PTS sugar transporter subunit IIA inside the family of fructose/mannitol specific IIA subunit enzymes (cd00211), AtlB as subunit IIB of enzyme II of the galactitol-specific PTS (cd05566) and AtlC as PTS galactitol-specific IIC component (COG3775). EI autophosphorylates using phosphoenolpyruvate (PEP) as phosphoryl donor, which in turn transfers the phosphoryl group to HPr (Saier, 2015; Kentache et al., 2016). Following this, we propose that HPr phosphorylates arabitol-specific EIIA AtlA, which subsequently transfers the phosphoryl group to EIIB component AtlB and, in the last step, donates its phosphoryl group to arabitol bound to EIIC transmembrane domain AtlC, releasing arabitol-phosphate into the cytoplasm. Sequence comparison of characterized transmembrane permease IIC for the arabitol AltC from L. monocytogenes (Saklani-Jusforgues et al., 2001) with B. methanolicus AtlC showed identity at the level of 56%.
Furthermore, the comparison of the sequence of the putative arabitol phosphate dehydrogenase encoded by altD gene with an amino acid sequence of characterized arabitol 1-phosphate dehydrogenase from E. avium (Povelainen et al., 2003) showed 51% identity between these proteins. Arabitol phosphate dehydrogenase (AtlD) activities were tested in crude extracts of MGA3 cells grown on arabitol, mannitol or methanol (Table 4). The results confirm that B. methanolicus MGA3 indeed possesses an arabitol phosphate dehydrogenase with increased activity during growth with arabitol, which is in accordance with the differential gene expression and the qRT-PCR analyses comparing arabitol- to mannitol-based growth (Table 3 and Figure 3). Additionally, the KM value for xylulose 5-phosphate was determined to be 0.03 ± 0.02 mM using crude extracts from cells grown on arabitol, which shows high substrate affinity (Supplementary Figure 4). Despite the fact that the reaction was not assayed in the physiological direction (i.e., oxidation of arabitol 1-phosphate or arabitol 5-phosphate to xylulose 5-phosphate), the enzyme kinetics are in accordance with the KS value of 2.9 ± 0.9 mM determined for arabitol. Although AtlD affinity for the substrate xylulose 5-phosphate was not determined using the purified enzyme, our results are in the range of previously characterized arabitol phosphate dehydrogenase in E. avium (Povelainen et al., 2003). Taken all together, these results strongly indicate the functionality of the altABCD encoded proteins in the arabitol utilization in B. methanolicus MGA3.
Addition of arabitol to cultivation broth not only induced expression of atlABCD, but also of adjacent genes, i.e., eight genes from BMMGA3_RS07325 to BMMGA3_RS07360 and all part of the arabitol operon as established via RT-PCR analysis (Figure 5). BMMGA3_RS07325 codes for a putative transcriptional antiterminator BglG. The bgl-sac family of antiterminator proteins are effectors of substrate-induced antitermination of catabolic operons and include, e.g., SacT and SacY of B. subtilis (Debarbouille et al., 1990; Manival, 1997) and BglG of E. coli (Nussbaum-Shochat and Amster-Choder, 1999; Raveh et al., 2009). These antiterminator proteins have been extensively characterized and act as RNA-binding proteins abrogating termination of transcription and allowing transcription elongation (Rutberg, 1997; Van Assche et al., 2015). A BLASTp analysis showed that the protein encoded by BMMGA3_RS07325 shared 30% identity to BglG of E. coli, 35% identity to SacY and 37% identity to SacT of B. subtilis, and 40% identity to BglG of L. monocytogenes (Gorski et al., 2003). Additionally, both E. coli bglG and B. subtilis sacT are located upstream of genes coding for PTS components involved in the utilization of ß-glucosides and sucrose, respectively (Rutberg, 1997). Similar gene order, where gene coding for antiterminator is upstream of genes encoding arabitol PTS, was observed in an arabitol gene cluster in L. monocytogenes and E. avium (Gorski et al., 2003; Povelainen et al., 2003). Interestingly, the other B. methanolicus wild type strain PB1 (NCIMB13113) is unable to grow on arabitol (data not shown). Both strains have been previously reported to show physiological differences (Heggeset et al., 2012). A BLAST analysis comparing the arabitol operon sequences of MGA3 and PB1 showed an incomplete BMMGA3_RS07325 gene sequence in the genome of the PB1 strain. These findings may indicate that BMMGA3_RS07325 functions as regulator of arabitol catabolism in B. methanolicus MGA3, and that its truncated form in B. methanolicus PB1 leads to absence of growth on arabitol.
RT-PCR and RNA-seq analysis revealed that genes BMMGA3_RS07325 to BMMGA3_RS07365 are co-transcribed as an operon (Figure 5). Accordingly, conserved promoter motifs were present upstream of BMMGA3_RS07325 (Irla et al., 2015). Using the ARNold tool for identification of transcriptional terminators (Naville et al., 2011), additional promoter motifs were found between BMMGA3_RS07360 and BMMGA3_RS07365 overlapping with a putative terminator structure (data not shown). These findings suggest the presence of a sub-operon starting at BMMGA3_RS07365 that would most likely not be involved in arabitol metabolism since it is not induced by arabitol. It is therefore remarkable that BMMGA3_RS07365 is co-transcribed alongside the arabitol inducible genes.
Bacillus methanolicus MGA3 is a facultative methyloptroph with a narrow substrate spectrum. Here, we have identified and characterized growth with D-arabitol. RNA-seq analysis revealed evidence for arabitol inducible catabolism of this sugar alcohol via a PTS AtlABC and an arabitol phosphate dehydrogenase AtlD, and genetic complementation studies confirmed functionality of the latter during arabitol metabolism. The role of a second putative arabitol phosphate dehydrogenase co-transcribed with atlABCD and metabolic fluxes during growth with arabitol remain to be studied. Once established for B. methanolicus, gene deletion experiments combined with biochemical characterization of the enzymes and 13C labeling experiments will help to further our understanding on how this sugar alcohol is catabolized as sole or combined carbon source.
All datasets generated for this study are included in the manuscript and/or the Supplementary Files. The data sets supporting the results of this article are available in the NCBI Gene Expression Omnibus database under the accession number GSE133849, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE133849.
ML carried out the experimental procedures. ML, MI, and LB analyzed the data. ML prepared a draft of the manuscript. ML, MI, LB, and VW finalized the manuscript. VW coordinated the study. All authors read and approved the final version of the manuscript.
This work was supported by the ERASysAPP project MetAPP (No. 031A603) and the ERA CoBioTech project C1Pro (No. 722361). Support for the Article Processing Charge by the Deutsche Forschungsgemeinschaft and the Open Access Publication Fund of Bielefeld University is acknowledged. The funding bodies had no role in the design of the study or collection, analysis, or interpretation of data or in writing of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Anika Winkler, Dr. Tobias Busche, and Prof. Dr. Jörn Kalinowski from the technology platform Genomics of CeBiTec for the kind assistance with the preparation and sequencing of the cDNA libraries and for the bioinformatics advice. We additionally thank Lara Paparić, Maximilian Schöne, and Lucas Riedel for the technical assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01725/full#supplementary-material
Abe, S., Takayama, K.-I., and Kinoshita, S. (1967). Taxonomical studies on glutamic acid-producing bacteria. J. Gen. Appl. Microbiol. 13, 279–301. doi: 10.2323/jgam.13.279
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Arfman, N., Dijkhuizen, L., Kirchhof, G., Ludwig, W., Schleifer, K.-H., Bulygina, E. S., et al. (1992). Bacillus methanolicus sp. nov., a new species of thermotolerant, methanol-utilizing, endospore-forming bacteria. Int. J. Syst. Bacteriol. 42, 439–445. doi: 10.1099/00207713-42-3-439
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Brautaset, T., Jakobsen,ØM., Josefsen, K. D., Flickinger, M. C., and Ellingsen, T. E. (2007). Bacillus methanolicus: a candidate for industrial production of amino acids from methanol at 50°C. Appl. Microbiol. Biotechnol. 74, 22–34. doi: 10.1007/s00253-006-0757-z
Brautaset, T., Williams, M. D., Dillingham, R. D., Kaufmann, C., Bennaars, A., Crabbe, E., et al. (2003). Role of the Bacillus methanolicus citrate synthase II gene, citY, in regulating the secretion of glutamate in l-Lysine-Secreting mutants. Appl. Environ. Microbiol. 69, 3986–3995. doi: 10.1128/AEM.69.7.3986-3995.2003
Brünker, P., Altenbuchner, J., and Mattes, R. (1998). Structure and function of the genes involved in mannitol, arabitol and glucitol utilization from Pseudomonas fluorescens DSM50106. Gene 206, 117–126. doi: 10.1016/S0378-1119(97)00574-X
Carnicer, M., Vieira, G., Brautaset, T., Portais, J.-C., and Heux, S. (2016). Quantitative metabolomics of the thermophilic methylotroph Bacillus methanolicus. Microb. Cell Factories 15:92. doi: 10.1186/s12934-016-0483-x
Charnetzky, W. T., and Mortlock, R. P. (1974). D-Arabitol catabolic pathway in Klebsiella aerogenes. J. Bacteriol. 119, 170–175.
Cocaign, M., Monnet, C., and Lindley, N. D. (1993). Batch kinetics of Corynebacterium glutamicum during growth on various carbon substrates: use of substrate mixtures to localise metabolic bottlenecks. Appl. Microbiol. Biotechnol. 40, 526–530. doi: 10.1007/BF00175743
Daly, J. M., Knoche, H. W., and Wiese, M. V. (1967). Carbohydrate and lipid metabolism during germination of uredospores of Puccinia graminis tritici 1. Plant Physiol. 42, 1633–1642. doi: 10.1104/pp.42.11.1633
de Carvalho, L. P. S., Fischer, S. M., Marrero, J., Nathan, C., Ehrt, S., and Rhee, K. Y. (2010). Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chem. Biol. 17, 1122–1131. doi: 10.1016/j.chembiol.2010.08.009
Debarbouille, M., Arnaud, M., Fouet, A., Klier, A., and Rapoport, G. (1990). The sacT gene regulating the sacPA operon in Bacillus subtilis shares strong homology with transcriptional antiterminators. J. Bacteriol. 172, 3966–3973. doi: 10.1128/jb.172.7.3966-3973.1990
Dominguez, H., Cocaign-Bousquet, M., and Lindley, N. D. (1997). Simultaneous consumption of glucose and fructose from sugar mixtures during batch growth of Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 47, 600–603. doi: 10.1007/s002530050980
Eggeling, L., and Bott, M. (2005). Handbook of Corynebacterium Glutamicum. Boca Raton, FL: CRC Press.
Eikmanns, B. J., Thum-Schmitz, N., Eggeling, L., Lüdtke, K. U., and Sahm, H. (1994). Nucleotide sequence, expression and transcriptional analysis of the Corynebacterium glutamicum gltA gene encoding citrate synthase. Microbiology 140(Pt 8), 1817–1828. doi: 10.1099/13500872-140-8-1817
Fall, R., and Benson, A. A. (1996). Leaf methanol — the simplest natural product from plants. Trends Plant Sci. 1, 296–301. doi: 10.1016/S1360-1385(96)88175-0
Gibson, D. G., Young, L., Chuang, R.-Y., Venter, J. C., Hutchison, C. A., and Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. doi: 10.1038/nmeth.1318
Gorski, L., Palumbo, J. D., and Mandrell, R. E. (2003). Attachment of Listeria monocytogenes to radish tissue is dependent upon temperature and flagellar motility. Appl. Environ. Microbiol. 69, 258–266. doi: 10.1128/AEM.69.1.258-266.2003
Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580. doi: 10.1016/s0022-2836(83)80284-8
Heggeset, T. M. B., Krog, A., Balzer, S., Wentzel, A., Ellingsen, T. E., and Brautaset, T. (2012). Genome Sequence of Thermotolerant Bacillus methanolicus: features and regulation related to methylotrophy and production of L-Lysine and L-Glutamate from methanol. Appl. Environ. Microbiol. 78, 5170–5181. doi: 10.1128/AEM.00703-12
Heuel, H., Turgut, S., Schmid, K., and Lengeler, J. W. (1997). Substrate recognition domains as revealed by active hybrids between the D-arabinitol and ribitol transporters from Klebsiella pneumoniae. J. Bacteriol. 179, 6014–6019. doi: 10.1128/jb.179.19.6014-6019.1997
Hilker, R., Stadermann, K. B., Doppmeier, D., Kalinowski, J., Stoye, J., Straube, J., et al. (2014). ReadXplorer–visualization and analysis of mapped sequences. Bioinformatics 30, 2247–2254. doi: 10.1093/bioinformatics/btu205
Hoffmann, J., and Altenbuchner, J. (2015). Functional characterization of the mannitol promoter of Pseudomonas fluorescens DSM 50106 and its application for a mannitol-inducible expression system for Pseudomonas putida KT2440. PLoS One 10:e0133248. doi: 10.1371/journal.pone.0133248
Irla, M., Heggeset, T. M. B., Nærdal, I., Paul, L., Haugen, T., Le, S. B., et al. (2016). Genome-based genetic tool development for Bacillus methanolicus: theta- and rolling circle-replicating plasmids for inducible gene expression and application to methanol-based cadaverine production. Front. Microbiol. 7:1481. doi: 10.3389/fmicb.2016.01481
Irla, M., Naerdal, I., Brautaset, T., and Wendisch, V. F. (2017). Methanol-based γ-aminobutyric acid (GABA) production by genetically engineered Bacillus methanolicus strains. Ind. Crops Prod. 106, 12–20. doi: 10.1016/j.indcrop.2016.11.050
Irla, M., Neshat, A., Brautaset, T., Rückert, C., Kalinowski, J., and Wendisch, V. F. (2015). Transcriptome analysis of thermophilic methylotrophic Bacillus methanolicus MGA3 using RNA-sequencing provides detailed insights into its previously uncharted transcriptional landscape. BMC Genomics 16:73. doi: 10.1186/s12864-015-1239-4
Irla, M., Neshat, A., Winkler, A., Albersmeier, A., Heggeset, T. M. B., Brautaset, T., et al. (2014). Complete genome sequence of Bacillus methanolicus MGA3, a thermotolerant amino acid producing methylotroph. J. Biotechnol. 188, 110–111. doi: 10.1016/j.jbiotec.2014.08.013
Izhaki, I., Fridman, S., Gerchman, Y., and Halpern, M. (2013). Variability of bacterial community composition on leaves between and within plant species. Curr. Microbiol. 66, 227–235. doi: 10.1007/s00284-012-0261-x
Jakobsen, O. M., Benichou, A., Flickinger, M. C., Valla, S., Ellingsen, T. E., and Brautaset, T. (2006). Upregulated transcription of plasmid and chromosomal ribulose monophosphate pathway genes is critical for methanol assimilation rate and methanol tolerance in the methylotrophic bacterium Bacillus methanolicus. J. Bacteriol. 188, 3063–3072. doi: 10.1128/JB.188.8.3063-3072.2006
Kentache, T., Milohanic, E., Cao, T. N., Mokhtari, A., Aké, F. M., Pham, Q. M. M., et al. (2016). Transport and catabolism of pentitols by Listeria monocytogenes. MMB 26, 369–380. doi: 10.1159/000447774
Kleijn, R. J., Buescher, J. M., Le Chat, L., Jules, M., Aymerich, S., and Sauer, U. (2010). Metabolic fluxes during strong carbon catabolite repression by malate in Bacillus subtilis. J. Biol. Chem. 285, 1587–1596. doi: 10.1074/jbc.M109.061747
Komarova, T. V., Sheshukova, E. V., and Dorokhov, Y. L. (2014). Cell wall methanol as a signal in plant immunity. Front. Plant Sci. 5:101. doi: 10.3389/fpls.2014.00101
Kordowska-Wiater, M. (2015). Production of arabitol by yeasts: current status and future prospects. J. Appl. Microbiol. 119, 303–314. doi: 10.1111/jam.12807
Kosugi, M., Miyake, H., Yamakawa, H., Shibata, Y., Miyazawa, A., Sugimura, T., et al. (2013). Arabitol provided by lichenous fungi enhances ability to dissipate excess light energy in a symbiotic green alga under desiccation. Plant Cell Physiol. 54, 1316–1325. doi: 10.1093/pcp/pct079
Kovárová-Kovar, K., and Egli, T. (1998). Growth kinetics of suspended microbial cells: from single-substrate-controlled growth to mixed-substrate kinetics. Microbiol. Mol. Biol. Rev. 62, 646–666.
Kulakovskaya, E., and Kulakovskaya, T. (2014a). “Biological activities of extracellular yeast glycolipids,” in Extracellular Glycolipids of Yeasts, eds E. Kulakovskaya and T. Kulakovskaya (Amsterdam: Elsevier), 35–64. doi: 10.1016/B978-0-12-420069-2.00004-2
Kulakovskaya, E., and Kulakovskaya, T. (2014b). “Structure and occurrence of yeast extracellular glycolipids,” in Extracellular Glycolipids of Yeasts, eds E. Kulakovskaya and T. Kulakovskaya (Amsterdam: Elsevier), 1–13. doi: 10.1016/B978-0-12-420069-2.00001-7
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. doi: 10.1186/gb-2009-10-3-r25
Laslo, T., von Zaluskowski, P., Gabris, C., Lodd, E., Ruckert, C., Dangel, P., et al. (2012). Arabitol metabolism of Corynebacterium glutamicum and Its Regulation by AtlR. J. Bacteriol. 194, 941–955. doi: 10.1128/JB.06064-11
Lindner, S. N., Seibold, G. M., Henrich, A., Krämer, R., and Wendisch, V. F. (2011). Phosphotransferase system-independent glucose utilization in Corynebacterium glutamicum by inositol permeases and glucokinases▽. Appl. Environ. Microbiol. 77, 3571–3581. doi: 10.1128/AEM.02713-10
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mandel, M., and Higa, A. (1970). Calcium-dependent bacteriophage DNA infection. J. Mol. Biol. 53, 159–162. doi: 10.1016/0022-2836(70)90051-3
Manival, X. (1997). From genetic to structural characterization of a new class of RNA-binding domain within the SacY/BglG family of antiterminator proteins. EMBO J. 16, 5019–5029. doi: 10.1093/emboj/16.16.5019
Matsuoka, Y., and Shimizu, K. (2013). Catabolite regulation analysis of Escherichia coli for acetate overflow mechanism and co-consumption of multiple sugars based on systems biology approach using computer simulation. J. Biotechnol. 168, 155–173. doi: 10.1016/j.jbiotec.2013.06.023
Mentz, A., Neshat, A., Pfeifer-Sancar, K., Pühler, A., Rückert, C., and Kalinowski, J. (2013). Comprehensive discovery and characterization of small RNAs in Corynebacterium glutamicum ATCC 13032. BMC Genomics 14:714. doi: 10.1186/1471-2164-14-714
Morita, T., Fukuoka, T., Imura, T., and Kitamoto, D. (2012). Formation of the two novel glycolipid biosurfactants, mannosylribitol lipid and mannosylarabitol lipid, by Pseudozyma parantarctica JCM 11752T. Appl. Microbiol. Biotechnol. 96, 931–938. doi: 10.1007/s00253-012-4230-x
Morita, T., Fukuoka, T., Konishi, M., Imura, T., Yamamoto, S., Kitagawa, M., et al. (2009). Production of a novel glycolipid biosurfactant, mannosylmannitol lipid, by Pseudozyma parantarctica and its interfacial properties. Appl. Microbiol. Biotechnol. 83, 1017–1025. doi: 10.1007/s00253-009-1945-4
Müller, J. E. N., Heggeset, T. M. B., Wendisch, V. F., Vorholt, J. A., and Brautaset, T. (2015a). Methylotrophy in the thermophilic Bacillus methanolicus, basic insights and application for commodity production from methanol. Appl. Microbiol. Biotechnol. 99, 535–551. doi: 10.1007/s00253-014-6224-3
Müller, J. E. N., Meyer, F., Litsanov, B., Kiefer, P., and Vorholt, J. A. (2015b). Core pathways operating during methylotrophy of Bacillus methanolicus MGA3 and induction of a bacillithiol-dependent detoxification pathway upon formaldehyde stress. Mol. Microbiol. 98, 1089–1100. doi: 10.1111/mmi.13200
Müller, J. E. N., Litsanov, B., Bortfeld-Miller, M., Trachsel, C., Grossmann, J., Brautaset, T., et al. (2014). Proteomic analysis of the thermophilic methylotroph Bacillus methanolicus MGA3. Proteomics 14, 725–737. doi: 10.1002/pmic.201300515
Naerdal, I., Pfeifenschneider, J., Brautaset, T., and Wendisch, V. F. (2015). Methanol-based cadaverine production by genetically engineered Bacillus methanolicus strains. Microb. Biotechnol. 8, 342–350. doi: 10.1111/1751-7915.12257
Naville, M., Ghuillot-Gaudeffroy, A., Marchais, A., and Gautheret, D. (2011). ARNold: a web tool for the prediction of Rho-independent transcription terminators. RNA Biol. 8, 11–13. doi: 10.4161/rna.8.1.13346
Nilasari, D., Dover, N., Rech, S., and Komives, C. (2012). Expression of recombinant green fluorescent protein in Bacillus methanolicus. Biotechnol. Prog. 28, 662–668. doi: 10.1002/btpr.1522
Nothaft, H., Parche, S., Kamionka, A., and Titgemeyer, F. (2003). In Vivo analysis of HPr reveals a fructose-specific phosphotransferase system that confers high-affinity uptake in Streptomyces coelicolor. J. Bacteriol. 185, 929–937. doi: 10.1128/JB.185.3.929-937.2003
Nussbaum-Shochat, A., and Amster-Choder, O. (1999). BglG, the transcriptional antiterminator of the bgl system, interacts with the ’ subunit of the Escherichia coli RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 96, 4336–4341. doi: 10.1073/pnas.96.8.4336
Opačić, M., Hesp, B. H., Fusetti, F., Dijkstra, B. W., and Broos, J. (2012). Structural investigation of the transmembrane C domain of the mannitol permease from Escherichia coli using 5-FTrp fluorescence spectroscopy. Biochim. Biophys. Acta 1818, 861–868. doi: 10.1016/j.bbamem.2011.11.001
Pérez-García, F., Peters-Wendisch, P., and Wendisch, V. F. (2016). Engineering Corynebacterium glutamicum for fast production of L-lysine and L-pipecolic acid. Appl. Microbiol. Biotechnol. 100, 8075–8090. doi: 10.1007/s00253-016-7682-6
Peters-Wendisch, P. G., Schiel, B., Wendisch, V. F., Katsoulidis, E., Möckel, B., Sahm, H., et al. (2001). Pyruvate carboxylase is a major bottleneck for glutamate and lysine production by Corynebacterium glutamicum. J. Mol. Microbiol. Biotechnol. 3, 295–300.
Plemenitaš, A., Vaupotiè, T., Lenassi, M., Kogej, T., and Gunde-Cimerman, N. (2008). Adaptation of extremely halotolerant black yeast Hortaea werneckii to increased osmolarity: a molecular perspective at a glance. Stud. Mycol. 61, 67–75. doi: 10.3114/sim.2008.61.06
Povelainen, M., Eneyskaya, E. V., Kulminskaya, A. A., Ivanen, D. R., Kalkkinen, N., Neustroev, K. N., et al. (2003). Biochemical and genetic characterization of a novel enzyme of pentitol metabolism: D-arabitol-phosphate dehydrogenase. Biochem. J. 371, 191–197. doi: 10.1042/bj20021096
Primrose, S. B., and Ronson, C. W. (1980). Polyol metabolism by Rhizobium trifolii. J. Bacteriol. 141, 1109–1114.
Raveh, H., Lopian, L., Nussbaum-Shochat, A., Wright, A., and Amster-Choder, O. (2009). Modulation of transcription antitermination in the bgl operon of Escherichia coli by the PTS. Proc. Natl. Acad. Sci. 106, 13523–13528. doi: 10.1073/pnas.0902559106
Rutberg, B. (1997). Antitermination of transcription of catabolic operons. Mol. Microbiol. 23, 413–421. doi: 10.1046/j.1365-2958.1997.d01-1867.x
Saier, M. H. (2015). The Bacterial phosphotransferase system: new frontiers 50 years after its discovery. J. Mol. Microbiol. Biotechnol. 25, 73–78. doi: 10.1159/000381215
Saklani-Jusforgues, H., Fontan, E., and Goossens, P. L. (2001). Characterisation of a Listeria monocytogenes mutant deficient in D-arabitol fermentation. Res. Microbiol. 152, 175–177. doi: 10.1016/S0923-2508(01)01189-5
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning: A Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory Press.
Schendel, F. J., Bremmon, C. E., Flickinger, M. C., and Hanson, R. S. (1990). L-Lysine Production at 50°C by mutants of a newly isolated and characterized methylotrophic Bacillus sp. Appl. Environ. Microbiol. 56, 963–970.
Subtil, T., and Boles, E. (2012). Competition between pentoses and glucose during uptake and catabolism in recombinant Saccharomyces cerevisiae. Biotechnol. Biofuels 5:14. doi: 10.1186/1754-6834-5-14
Tabata, N., Ohyama, Y., Tomoda, H., Abe, T., Namikoshi, M., and Omura, S. (1999). Structure elucidation of roselipins, inhibitors of diacylglycerol acyltransferase produced by Gliocladium roseum KF-1040. J. Antibiot. 52, 815–826. doi: 10.7164/antibiotics.52.815
Van Assche, E., Van Puyvelde, S., Vanderleyden, J., and Steenackers, H. P. (2015). RNA-binding proteins involved in post-transcriptional regulation in bacteria. Front. Microbiol. 6:141. doi: 10.3389/fmicb.2015.00141
Wang, X., Bai, J., Wang, W., and Zhang, G. (2019). Leaf metabolites profiling between red and green phenotypes of Suaeda salsa by widely targeted metabolomics. Funct. Plant Biol. doi: 10.1071/FP18182 [Epub ahead of print].
Wendisch, V. F., de Graaf, A. A., Sahm, H., and Eikmanns, B. J. (2000). Quantitative determination of metabolic fluxes during coutilization of two carbon sources: comparative analyses with Corynebacterium glutamicum during growth on acetate and/or glucose. J. Bacteriol. 182, 3088–3096. doi: 10.1128/jb.182.11.3088-3096.2000
Keywords: Bacillus methanolicus, differential transcriptome analysis, mannitol metabolism, arabitol metabolism, monophasic growth, operon organization
Citation: López MG, Irla M, Brito LF and Wendisch VF (2019) Characterization of D-Arabitol as Newly Discovered Carbon Source of Bacillus methanolicus. Front. Microbiol. 10:1725. doi: 10.3389/fmicb.2019.01725
Received: 04 April 2019; Accepted: 12 July 2019;
Published: 31 July 2019.
Edited by:
Ilana Kolodkin-Gal, Weizmann Institute of Science, IsraelReviewed by:
Fabian M. Commichau, University of Göttingen, GermanyCopyright © 2019 López, Irla, Brito and Wendisch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Volker F. Wendisch, dm9sa2VyLndlbmRpc2NoQHVuaS1iaWVsZWZlbGQuZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.