
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 25 July 2019
Sec. Food Microbiology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.01710
This article is part of the Research Topic Food Safety and Foodborne Pathogen – A Global Perspective on the Diversity, Combating Multidrug Resistance and Management View all 39 articles
 Yunyi Zhang1†
Yunyi Zhang1† Shilei Dong2†
Shilei Dong2† Honghu Chen1
Honghu Chen1 Jiancai Chen1
Jiancai Chen1 Junyan Zhang1
Junyan Zhang1 Zhen Zhang1
Zhen Zhang1 Yong Yang1
Yong Yang1 Ziyan Xu3
Ziyan Xu3 Li Zhan1*
Li Zhan1* Lingling Mei1*
Lingling Mei1*Listeria monocytogenes is an important foodborne pathogen causing public concern. A total of 3354 retail foods in bulk were sampled and screened for L. monocytogenes. Seventy-three (2.2%) samples including 21 ready-to-eat (RTE) foods and 52 raw foods were confirmed positive for L. monocytogenes. Sushi and salmon sashimi occupied the top two slots in RTE foods with relatively high presence rate of 12.9 and 6.9%, respectively. Meanwhile, L. monocytogenes was found to be distributed unequally in raw foods; the presence rates in raw meat (3.5%) and poultry (3.8%) were significantly higher than that in raw seafood (1.3%). Notably, L. monocytogenes was not detected in raw freshwater food. The L. monocytogenes isolates belonged to four serotypes, 1/2a, 1/2b, 1/2c, and 4b, with the most prevalent serotype being 1/2a (47.9%). Eighteen sequence types (STs) and eighteen virulence types (VTs) containing four newly assigned VTs (VT180, VT181, VT182, and VT183) were determined via multilocus sequence typing (MLST) and multi-virulence-locus sequence typing (MVLST). Among the 73 L. monocytogenes isolates, 23 (31.5%) belonged to epidemic clones (ECs) including ECI, ECIV, ECV, ECVI, ECVIII and ECXI among which ECV was predominant. Antibiotic susceptibility tests revealed a high resistance rate (11.0%) to tetracycline. Moreover, we identified the distribution patterns of virulence genes of four Listeria pathogenicity islands (LIPI) in L. monocytogenes isolates. prfA, hly, plcA, plcB, mpl, actA genes in LIPI-1 and inlA, inlB, inlC, inlJ genes in LIPI-2 were detected in approximately all L. monocytogenes isolates. The distribution of both LIPI-3 genes and LIPI-4 genes exhibited association with lineage and ST. LIPI-4 genes were present exclusively in ST87 isolates. Relatedness analysis revealed the absence of distinct association between STs, ECs, LIPI-3 and LIPI-4 distribution and specific food groups. This study provided fundamental data for Chinese food safety authorities to grasp the contamination status of L. monocytogenes in foods, assess the potential risk of this pathogen and further address the safety issue of retail foods in bulk in China.
Listeria monocytogenes is an important foodborne pathogen which can cause severe human listeriosis, particularly in older adults, newborns, pregnant women and immune-compromised individuals, resulting in septicemia, abortion, preterm delivery, stillbirth, meningitis, encephalomyelitis, or even death (Lomonaco et al., 2009; Lamont et al., 2011). In some cases, this bacterium’s capability to infect the central nervous system (CNS) of immunocompetent adults was also revealed (Guo and Liang, 2014; Giménez-Muñoz et al., 2015). The case-fatality rate of listeriosis attains 20–30% in several regions around the world (Centers for Disease Control and Prevention, 2017; European Food Safety Authority [EFSA] and European Centre for Disease Prevention and Control [ECDC], 2017; Food Standards Australia New Zealand, 2017; Li et al., 2018; Salama et al., 2018). Although not common, listeriosis shares a large proportion of the public health and economic burden resulting from foodborne pathogens. Almost 1500 listeriosis cases were reported annually in the United States, only accounting for 0.02% among major foodborne pathogens infection cases. However, 4% of hospitalization and 19% of deaths were caused by L. monocytogenes infection (Scallan et al., 2011).
Listeria monocytogenes extensively inhabits environments including surface soil, rivers, decaying vegetation, effluents from sewage treatments, wild animal intestines and domestic animal feces, due to its robust survival ability under various unfavorable conditions such as high salinity, low pH and low temperature (Donnelly, 2001; Vermeulen et al., 2007; Soni et al., 2013; Ribeiro and Destro, 2014; Ribeiro et al., 2014; Haley et al., 2015; Wang et al., 2017b). Wide distribution of L. monocytogenes offers opportunities to contaminate diverse foods, raw material and the environments of processing plants for food production (Heisick et al., 1989; Bula et al., 1995; Zhu et al., 2012; Wang et al., 2017b; Bergholz et al., 2018). Intake of contaminated food by L. monocytogenes has been widely considered to be the entry point of infection. In China, retail foods in bulk account for a large percentage of food consumption. Foods in bulk are defined as foods that are not divided into parts and/or packaged in separate units when sold in various sales locations. Compared with prepackaged foods, foods in bulk are prone to be directly exposed to the environment during the sales process, which makes it easier for them to be contaminated by pathogens. Surveying the prevalence, phenotypic and genotypic characteristics of L. monocytogenes in food products in bulk can provide valuable information on the distribution profiles of this pathogen, allowing Chinese food safety authorities to assess the potential risk of L. monocytogenes in bulk foods, and take appropriate hygiene measures to improve microbial safety. Epidemic clones (ECs) of L. monocytogenes are defined as genetically similar isolates descended from a common ancestor, involved in either a single large outbreak or geographically and temporally unrelated outbreaks (Kathariou, 2002; Cantinelli et al., 2013). Eleven known ECs including ECI - ECXI have been identified thus far (Chen et al., 2007, 2016; Lomonaco et al., 2013; Filipello et al., 2017). EC isolates were usually closely associated with listeriosis massive outbreaks (Haley et al., 2015). Since the first determined ECI outbreak in 1981, outbreaks of ECI were reported in western Switzerland during the time period 1983–1987, in California, United States in 1985, and in France in 1992 (Kathariou, 2002; Cantinelli et al., 2013). ECII caused an United States multistate hotdog outbreak and another turkey deli meat outbreak (Chen et al., 2011). In 2000, ECIII caused an United States outbreak attributed to intake of turkey deli meat. ECIV caused the United Kingdom and Ireland outbreak in 1987–1988, and an outbreak in Italy in 1997. ECV caused several major outbreaks occurring in the United Kingdom, Italy, Canada, and United States (Chen et al., 2011). ECVI and ECVII were newly defined in 2011, and these two epidemic clones led to 147 cases in 28 U.S. states, causing 33 deaths and one miscarriage (Lomonaco et al., 2013). ECVIII caused an United States milk chocolate outbreak in 1994, and an outbreak in Pennsylvania in 1987 (Chen et al., 2016). ECIX caused an United States multistate caramel apple outbreak in 2014–2015 (Chen et al., 2016). ECX caused a France pork rillettes outbreak in 1999–2000 (Chen et al., 2016). In 2012, ECXI caused an United States ricotta salata outbreak (Filipello et al., 2017). Clearly, ECs concealed in food products have the potential risk to cause L. monocytogenes infection, and even listeriosis outbreaks. Initially, ECs were confirmed based on the pulsed-field gel electrophoresis (PFGE) typing, ribotyping, and multilocus enzyme electrophoresis (MEE) (Kathariou, 2002; Kathariou et al., 2006). In subsequent studies, multi-virulence-locus sequence typing (MVLST) was developed and verified to discriminate ECs accurately (Chen et al., 2007; Lomonaco et al., 2013). Additionally, in some previous studies, the PCR assay was used to rapidly screen specific ECs markers to determine epidemic clones. However, these results were presumptive and should be confirmed using MVLST (Chen and Knabel, 2007; Knabel et al., 2012; Lomonaco et al., 2012). To date, ECs among L. monocytogenes strains isolated from foods in China were screened by means of PCR, and limited from ECI to ECIII (Wu et al., 2015, 2016). Accurate and comprehensive data for ECs distribution in foodborne L. monocytogenes isolates in China were still sparse. The aims of this study were (i) to reveal the prevalence of L. monocytogenes in retail foods in bulk sampled in Zhejiang Province, and (ii) to gain information on the phenotypic and genotypic characteristics, especially the EC profiles of L. monocytogenes isolates. Zhejiang Province is located on the southeast coast of China with a population of 57 million people. In 2018, the annual GDP (Gross Domestic Product) in Zhejiang Province was 81 billion USD. According to 2017 statistical data, the production of raw meat including pork, beef, and mutton was 869.900 tons, the total fishery production was 594.45 million tons and the number of poultry raised was 25,139,100 (Department of Agriculture of Zhejiang Province, 2017). The annual per capita raw meat, poultry and seafood consumption was 27.3, 11.5, and 26.4 kg respectively in 2017 (China Statistical Yearbook Committee, 2017).
A total of 3354 retail foods in bulk including 1196 RTE foods and 2158 raw foods were sampled from supermarkets, farm fairs, fish markets, and restaurants in eleven geographic regions of Zhejiang Province, China from March 2016 to December 2016. The detailed information for food samples is shown in Table 1. Raw foods in this study were defined as not-ready-to-eat raw foods. During the sampling process, sterile containers were used to transfer representative units or portions of foods to the laboratory by aseptic practices. Frozen or chilled foods were transported in thermal insulated containers. Frozen foods including ice cream, frozen meat, frozen poultry and frozen seafood were stored at about −18°C and kept frozen prior to sampling. Chilled foods included chilled meat and chilled poultry stored at 0 – 5°C, as well as chilled seafood and salmon sashimi, were stored in crushed ice prior to sampling. Other categories of foods were stored at ambient temperature prior to sampling. Sample analysis was initiated as soon as the sample was received. All RTE foods except ice cream were sampled on the day of production. Ice cream was sampled during the shelf life period. Isolation of L. monocytogenes was carried out following the recommendation of the National Food Safety Standard of China – Food microbiological examination: L. monocytogenes (GB4789.30-2010; National Standard of the People’s Republic of China, 2010). Twenty-five grams of each sample was suspended in 225 mL Listeria enrichment broth LB1 (Huankai, Guangzhou, China) and homogenized for 2 min. The homogenate was incubated at 30°C for 24 h. 0.1 mL of each culture was transferred into 10 mL of listeria enrichment broth LB2 (Huankai, Guangzhou, China) and incubated at 30°C for 24 h, followed by spreading on CHROMagar Listeria plates (CHROMAgar, Paris, France). After incubation at 36°C for 48 h, five suspect colonies on each plate were selected for identification. If less than five colonies grew on any one plate, all suspect colonies were selected for identification. VITEK 2 compact system (Bio Mérieux, Lyon, France) and the following PCR method (Huang et al., 2007) were performed to identify each suspect colony. PCR was applied to discriminate L. monocytogenes from other Listeria species. L. monocytogenes strains EGDe (ATCC BAA-679) and 10403S were included as positive control strains for PCR. Serotyping was conducted adopting serum agglutination according to the manufacture’s instruction (Denka Seiken, Tokyo, Japan). All confirmed strains were stored at −70°C for further analysis.
The total DNA of L. monocytogenes isolates was extracted using Bacterial DNA Kit (Omega, United States) according to the manufacturer’s protocol. DNA quality was tested by electrophoresis on 0.8% agarose gel and the concentration was determined by using Spectrophotometer NanoDrop 1000 (Thermo, United States). Prepared DNA was stored at −20°C for MLST, MVLST and virulence genes analysis. Seven housekeeping genes abcZ, bglA, cat, dapE, dat, ldh, lhkA were amplified according to the scheme advised in Institute Pasteur MLST database1 (Moura et al., 2016). Each amplicon was sequenced bidirectionally using 3730XL Genetic Analyser (Applied Biosystems, United States). Assembled nucleotide sequences were queried against Institute Pasteur MLST database. Sequence type (ST), clonal complex (CC) and lineage were then designated to each isolate. The phylogenic tree was constructed harnessing MEGA 5.1 (Tamura et al., 2011). Based on the concatenated sequence (3288 bp) of seven housekeeping genes, a neighbor-joining statistical method was employed. The number of Bootstrap replications was 1,000. The Genbank database accession numbers for abcZ, bglA, cat, dapE, dat, ldh, lhkA genes sequences of L. monocytogenes isolates in this study were MK368885 – MK368957, MK368812 – MK368884, MK368958 – MK369030, MK369031 – MK369103, MK369104 – MK369176, MK369177 – MK369249 and MK369250 – MK369322, respectively.
Multi-virulence-locus sequence typing analysis was carried out following the method developed by Zhang et al. (2004). Six virulence genes including prfA, inlB, inlC, dal, clpP, and lisR were amplified and then sequenced. The nucleotide sequences were trimmed to the correct length and concatenated in a specific order as mentioned in the MVLST database2. Alignment was performed between the concatenated sequence of each isolate and the reference sequence of each virulence type (VT) available in the MVLST database by using BLAST 2.2.28+. VT and EC of each isolate were determined according to the alignment results. New VTs were designated by the MVLST database. The Genbank database accession numbers for prfA, inlB, inlC, dal, clpP, and lisR gene sequences of L. monocytogenes isolates in the current study were MK392393–MK392465, MK369469–MK369541, MK369542–MK369614, MK369396–MK369468, MK369323–MK369395, and MK369615–MK369687. The polymorphism of both MLST and MVLST genes was analyzed using DnaSP 5.10 software (Librado and Rozas, 2009). Analysis parameters included number of polymorphic sites, G + C content, Ks (the number of synonymous substitutions per synonymous site), Ka (the number of non-synonymous substitutions per non-synonymous site), π (average pairwise nucleotide difference per site) and Tajima’s D test for neutrality. Tajima’s D is a population genetic test statistic to distinguish between neutral evolution and non-neutral evolution of DNA sequences (Tajima, 1989).
Antibiotic susceptibility of L. monocytogenes isolates was assayed using the broth micro-dilution minimum inhibitory concentrations (MICs) method according to the Clinical and Laboratory Standards Institute guidelines (Clinical and Laboratory Standards Institute, 2015). Breakpoints for Penicillin and Ampicillin were found in CLSI documents M45-A3 (Clinical and Laboratory Standards Institute, 2015). Since there is no relevant criteria for Erythromycin, Clindamycin, Quinupristin/dalfopristin, Vancomycin, Tetracycline, Gentamicin, Rifampin, Levofloxacin, Ciprofloxacin, Gatifloxacin, and Oxacillin, the susceptibility results for these antibiotics were interpreted based on the breakpoints of Staphylococcus spp (Clinical and Laboratory Standards Institute, 2017) as reported previously (Conter et al., 2009; Lungu et al., 2011; Chen et al., 2015; Khen et al., 2015; Tahoun et al., 2017). L. monocytogenes isolates resistant to three or more types of antibiotics belonging to different antibiotic classes were defined as multi-drug resistant (Magiorakos et al., 2012). Escherichia coli ATCC29522 and S. aureus ATCC29213 were used as quality control strains.
The detailed information of primers used for virulence genes detection is shown in Table 2. Primers for llsA, llsG, llsH, llsB, llsY, llsD, llsP in LIPI-3 and all LIPI-4 genes were designed using PrimerPremier 5.0. The genome sequences of LIPI-3 positive L. monocytogenes strains including FSL N1-017, FSL R2-503, H7858 and NCTC 11994 were obtained from the NCBI Refseq genome database. Multiple alignment of nucleotide sequences of genes llsA, llsG, llsH, llsB, llsY, llsD, llsP mining from the above genomes were performed using clustal v1.83. The conserved region of each gene was used as a template for the primer’s design. Primers for LIPI-4 genes were designed according to the LIPI-4 sequence of L. monocytogenes strain LM09-00558 (Maury et al., 2016). PCR was conducted employing the performance system and conditions described previously (Zhang et al., 2017). The amplicons were analyzed with electrophoresis on 1% agarose gel. To validate the new primers for LIPI-3 and LIPI-4 genes, all associated positive PCR products were sequenced. The DNA sequences were analyzed using the BLAST algorithm on the website http://www.ncbi.nlm.nih.gov/BLAST.
Chi-square analysis or Fisher’s exact test was performed to determine if a significant difference in the prevalence and serotype distribution of L. monocytogenes among different food groups could be found. The significance level was set at a p-value of <0.05. All analyses were performed using the SPSS v 21.0 software package. Association between food groups and genotyping data, antibiotic resistance profiles, and Listeria pathogenicity islands (LIPI) distribution was calculated based on the Gini coefficient (Henri et al., 2016) using the Stata software. A coefficient value of smaller than 0.4 reveals no association between genotypes, antibiotic resistance profiles or LIPI distribution and food types, a value between 0.4 and 0.6 indicates moderate association, and a value greater than 0.6 indicates unequal dispersion of genotypes, antibiotic resistance profiles or LIPI within food groups.
The prevalence of L. monocytogenes in various analyzed food samples was shown in Table 1. Seventy-three out of 3354 samples were confirmed to be L. monocytogenes positive with the total rate of 2.2%. Among them, 21 strains (1.8%) were isolated from 1196 ready-to-eat (RTE) foods and 52 strains (2.4%) were isolated from 2158 raw foods. No statistically significant difference in the prevalence of L. monocytogenes was present between RTE foods and raw foods (χ2 = 1.545, p > 0.05). RTE foods contaminated with pathogenic microorganisms have been widely considered to be the major source of foodborne pathogen infections due to the absence of further cooking, baking or pasteurizing processes prior to consumption (European Food Safety Authority [EFSA], 2013; Pouillot et al., 2015; Luchansky et al., 2017). Here L. monocytogenes was isolated in five RTE food categories including sushi, salmon sashimi, salad, vegetables in sauce and cooked meat. The occurrence of L. monocytogenes in sushi and salmon sashimi exhibited the top two highest rates (12.9 and 6.9%) among RTE foods and significantly higher than that in all screened RTE foods (1.8%) (χ2 = 27.762, p = 0; χ2 = 5.082, p = 0.024) and cooked meat (1.1%) (χ2 = 14.076, p = 0; χ2 = 4.123, p = 0.042). Relatively high presence rates of L. monocytogenes in sushi and salmon sashimi poses a potential risk of causing human L. monocytogenes infection. RTE meat products are considered one of the important food sources of human L. monocytogenes infection throughout the world (European Food Safety Authority [EFSA], 2013; Iannetti et al., 2016; Raheem, 2016). According to our data, the prevalence rate of L. monocytogenes in cooked meat was close to that in prepackaged heated meat at the end of its shelf-life in the European Union as well as meat products in Italy (2.07 and 1.66% respectively) (European Food Safety Authority [EFSA], 2013; Iannetti et al., 2016).
Listeria monocytogenes was distributed unequally in raw foods. Presence rates in raw meat (3.5%) and raw poultry (3.8%) were significantly higher than that in raw seafood (1.3%) (χ2 = 5.663, p = 0.017; χ2 = 6.415, p = 0.011). It is worthy to note that L. monocytogenes was not detected in 396 samples of raw freshwater food. We further analyzed if rates of L. monocytogenes in raw foods under different storage conditions were different. The results did not show a significant difference among fresh, chilled and frozen foods. Due to the common eating habit in China of cooking foods completely, raw foods were not predicted to have a high risk of causing L. monocytogenes infection in China. However, transfer of L. monocytogenes might occur from contaminated raw foods to RTE foods during the process of storage after purchase. In particular, cross contamination may be more likely to occur due to lack of tight packaging for foods in bulk.
Four different serotypes, 1/2a, 1/2b, 1/2c, and 4b were identified in the 73 L. monocytogenes isolates (Figure 1). 1/2a was the most prevalent serotype, accounting for 47.9% of the identified isolates. Serotype 1/2a strains have exhibited extensive distribution in various foods around the world, which might be due to superior adaptability in different environments compared to other serotype strains (Korsak et al., 2012; Schmitz-Esser et al., 2015; Wu et al., 2015). Serotypes 1/2b and 1/2c had similar presence rates at 21.9 and 23.3%, respectively. Among the 73 isolated strains, only five (6.8%) were confirmed as serotype 4b. Serotype distribution difference among different food groups was analyzed based on Chi-square or Fisher analysis, as appropriate. The prevalence of serotype 1/2a in raw poultry was significantly higher than that in raw seafood (χ2 = 8.478, p = 0.004) and RTE food (χ2 = 6.025, p = 0.014). Meanwhile, there was no obvious distribution difference of serotypes 1/2b, 1/2c, and 4b among different food groups (p > 0.05). The current study showed that a total of 52 (71.2%) isolates were grouped into 1/2a, 1/2b, and 4b serotypes, which were the main causative agents of clinical cases worldwide and were responsible for 98% of listeriosis outbreaks (Buchrieser et al., 1993; Raheem, 2016). Among all the confirmed serotypes, no serotypes 3a, 3b, 3c, 4a, 4c, 4d, 4e, or 7 were detected, which was consistent with a previous report (Wu et al., 2015) in China indicating that those serotypes were rarely identified from food and clinical samples.
Eighteen different sequence types (STs) were classified among all 73 L. monocytogenes isolates, which were further assigned to sixteen clonal complexes (CCs) and one singleton by using MLST analysis, with the Simpson’s index of diversity (DI) at 0.892 (Table 3). Among the 18 STs, ST9 (23.3%) was predominant, followed by ST155 (16.4%), ST8 (12.3%), and ST121 (9.6%). Many studies have shown that hypervirulence exists in L. monocytogenes with different STs. Although ST9, ST155, ST8, and ST121 were all revealed to be involved in L. monocytogenes infection, they tended to be isolated from food products and food processing environments. Only a small number of clinical cases were induced by these ST types (Ebner et al., 2015; Schmitz-Esser et al., 2015; Wu et al., 2016; Maury et al., 2016; Wang et al., 2018). For instance, while ST9 and ST121 were two major STs among L. monocytogenes foodborne isolates during 1999–2014 in France, ST121 included the fewest clinical isolates (Henri et al., 2016). ST1 and ST2 clones have been verified to be strongly associated with clinical origin particularly in human CNS or maternal-neonatal (MN) listeriosis cases (Maury et al., 2016). According to our data, one isolate from fresh pork and three isolates from sushi, chilled beef and chilled seafood were genotyped into ST1 and ST2, respectively. Although the presence rates (1.4 and 4.1%) were relatively low, the existence of ST1 and ST2 in foods, particularly in RTE foods, poses a potential risk on food safety due to their high virulence. ST87 clones were detected in four samples including two RTE foods. ST87 strains were scarcely isolated from food, environmental or clinical samples in North America, South America, Europe, and Australia (Maury et al., 2016). In contrast, ST87 was the predominant ST in clinical L. monocytogenes isolates and closely related to CNS and MN infection in China (Wang et al., 2015, 2018; Wu et al., 2016).
A total of eighteen VTs including four newly assigned VTs (VT180, VT181, VT182, and VT183) were determined in all L. monocytogenes isolates (Figure 2). Among 73 strains, 23 (31.5%) of them were epidemic clones including ECI, ECIV, ECV, ECVI, ECVIII, and ECXI, which is different from previous reports indicating that ECI and ECIII are the only detected ECs in China (Wu et al., 2015, 2016). ECV was most common; 10 out of 23 (43.5%) EC isolates were classified into this group.
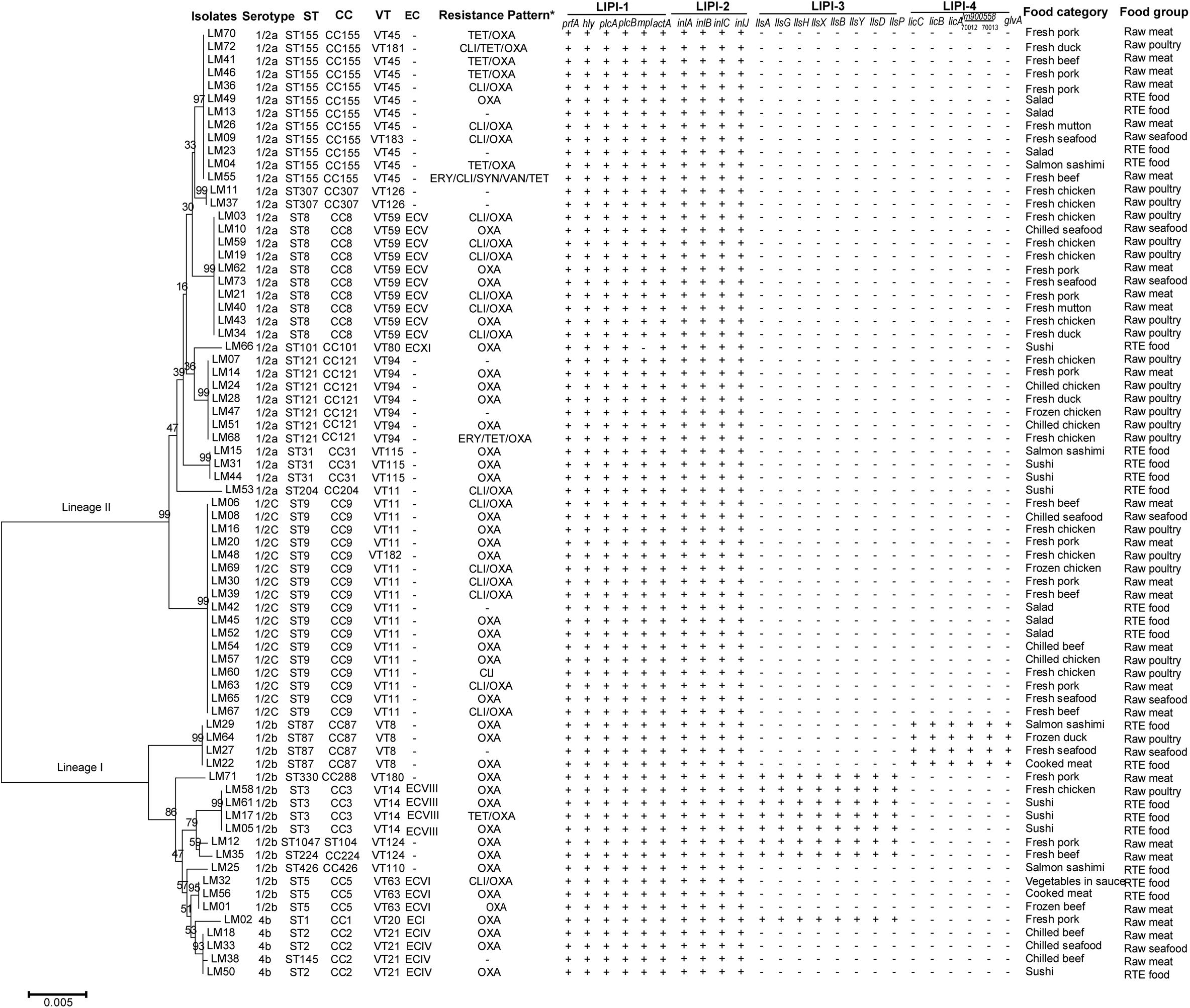
Figure 2. Characteristics of Listeria monocytogenes strains isolated from retail foods in bulk in Zhejiang Province, China. The neighbor-joining phylogeny tree was computed by MEGA 5.1 with Bootstrap replications number set 1,000. *: TET, tetracycline; OXA, oxacillin; CLI, clindamycin; ERY, erythromycin; SYN, quinupristin/dalfopristin; VAN, vancomycin.
To explore the polymorphisms of MLST and MVLST genes in L. monocytogenes population investigated in this study, the number of polymorphic sites, Ka/Ks ratio, π and Tajima’s D of associated genes in 73 L. monocytogenes isolates were calculated (Table 4). Seven MLST genes contained a total of 151 polymorphic sites (4.6%, range 4.2–12.1% per gene). Six MVLST virulence genes contained a total of 150 polymorphic sites (5.7%, range 1.1–14.2% per gene). Dat and dal had the two highest percentages of polymorphic sites, respectively. Ka/Ks of all MLST genes as well as MVLST genes dal, inlB, inlC, prfA were less than one, indicating these genes evolved under purifying (negative) selection. Meanwhile, Ka/Ks of clpP was 3.551, which indicated clpP was under positive selection during the course of genetic evolution (Zhang and Yu, 2006). Through comparison analysis of Ka/Ks between our data and previous reports (Cantinelli et al., 2013; Wu et al., 2016), MLST genes in different L. monocytogenes populations demonstrated similar selective strength. However, the selective strength for MVLST genes presented to be variable in L. monocytogenes populations from different ecological niches. For instance, inlB in L. monocytogenes outbreak strains in France was under neutral selection. Meanwhile, clpP was under purifying (negative) selection (Cantinelli et al., 2013). The average nucleotide diversity was close between MLST genes and MVLST genes, with π equaling 1.8% (range 1.0–5.6% per gene) and 2.1% (range 0.3–6.1% per gene) respectively. Tajima’s D test illustrated that abcZ, bglA, ldh, lhkA, inlB, inlC, lisR, and prfA evolved neutrally, whereas cat, dapE, dat, clpP, dal evolved under balancing selection.
The susceptibilities of L. monocytogenes isolates to thirteen antibiotics were shown in Table 5. All 73 strains were susceptible to penicillin, ampicillin, gentamicin and rifampin. More than 95% of strains were susceptible to erythromycin (97.3%), quinupristin/dalfopristin (98.6%), vancomycin (98.6%), levofloxacin (98.6%) and ciprofloxacin (95.9%). The results were similar to previous reports from China, the United States, Ireland, and Poland (Wieczorek et al., 2012; Shen et al., 2013; Chen et al., 2015; Khen et al., 2015; Wieczorek and Osek, 2017) and consistent with the general thought that the Listeria genus is naturally susceptible to ampicillin, penicillin, gentamicin and erythromycin, which are usually active against Gram-positive bacteria (Wang et al., 2013; Chen et al., 2015; Wu et al., 2015, 2016). On the other hand, L. monocytogenes strains isolated from raw milk, milk equipment and farm workers in Egypt showed lower susceptibility rates to gentamicin (19.0%), rifampin (0%) and ciprofloxacin (42.9%) (Tahoun et al., 2017). The most prevalent detected antibiotic resistance type was resistance to oxacillin. 86.3% of L. monocytogenes isolates were resistant to this antibiotic. Compared with previously reported resistance rates (1.0–4.5%) to tetracycline of L. monocytogenes isolated from various types of raw foods in China and beef chain in Ireland (Wieczorek et al., 2012; Shen et al., 2013; Wang et al., 2013; Chen et al., 2015; Khen et al., 2015), a much higher resistance rate of 11.0% to tetracycline was observed among our isolates. In the reference listeriosis therapy scheme, ampicillin or penicillin G combined with gentamicin was recommended as the treatment of choice. Meanwhile, vancomycin, trimethoprim-sulfamethoxazole and erythromycin were usually used as alternatives, especially for pregnant women (Hof, 2004). Both our results and previous reports (Rodas-Suarez et al., 2006; Morvan et al., 2010; Lungu et al., 2011; Wang et al., 2013; Chen et al., 2015; Tahoun et al., 2017) showed relatively low resistance rates to ampicillin, penicillin, gentamicin, vancomycin, trimethoprim-sulfamethoxazole and erythromycin, which revealed that the antibiotic treatment might be efficient for most of the L. monocytogenes strains. Including three (4.1%) multidrug-resistant strains, resistant to Erythromycin/Clindamycin/Quinupristin/dalfopristin/ Vancomycin/Tetracycline, Clindamycin/Tetracycline/Oxacillin and Erythromycin/Tetracycline/Oxacillin, respectively, all 73 isolates were grouped into seven antibiotic resistance patterns. It is worthy to note that one strain was identified as resistant to six types of antibiotics (Figure 2).
The molecular determinants of L. monocytogenes virulence has been investigated for many years (Tilney and Portnoy, 1989; Cossart, 2011). Four Listeria pathogenicity islands (LIPI) have been verified thus far, which are involved in invasion, survival and colonization of L. monocytogenes in host tissues. LIPI-1 contains six genes including hly, prfA, plcA, plcB, mpl, and actA (Hamon and Cossart, 2011; Travier and Lecuit, 2014; Mitchell et al., 2015; Rupp et al., 2015; Wang et al., 2017a; Hadjilouka et al., 2018; Poimenidou et al., 2018). LIPI-2 encodes a series of internalin family proteins which interact with the molecular cell surface and are essential for host cell adherence and virulence (McGann et al., 2007). LIPI-3 contributes to the expression of listeriolysin S (LLS), which is a post-translationally modified hemolytic peptide acting as a bacteriocin to alter the host intestinal microbiota, and plays an important role in the survival of L. monocytogenes in polymorphonucleocytes (PMNs) and virulence in the murine model (Cotter et al., 2008; Quereda et al., 2016). LIPI-4 encodes a cellobiose-family phosphotransfer system (PTS) and is involved in neural and placental infection (Maury et al., 2016). The distribution of virulence genes of these LIPIs in L. monocytogenes isolates were tested in this study (Figure 2). LIPI-1 genes were detected in approximately all L. monocytogenes isolates except one ST101 strain isolated from sushi, in which mpl was absent. inlA, inlB, inlC and inlJ of LIPI-2 existed in all 73 strains. 11.0% of isolates harbored all LIPI-3 genes and four isolates (5.5%) were determined LIPI-4 genes positive.
Remarkably, the distribution of both LIPI-3 and LIPI-4 exhibited apparent association with L. monocytogenes lineage and ST. LIPI-3 positive strains belonged to ST1, ST3, ST224, ST330, and ST1047; these STs were grouped into lineage I, in accordance with the finding that LIPI-3 was identified exclusively in a subset of lineage I (Clayton et al., 2014; Quereda et al., 2016). LIPI-4 was verified to exist uniquely in CC4 L. monocytogenes and closely linked to high virulence in CNS and MN listeriosis (Maury et al., 2016). A subsequent study pointed out that ST619, CC87 strains carried LIPI-4 fragment ptsA (Wang et al., 2018). In this study, we detected six genes encoding PTS sugar transporter subunit EIIC (licC), EIIB (licB), EIIA (licA), PTS systems associated protein (gene locus in Genbank, lm900558-70012), transcriptional antiterminator (gene locus in Genbank, lm900558-70013) and maltose-6′-phosphate-glucosidase (glvA) (Maury et al., 2016) of LIPI-4 in all ST87 isolates, which confirmed that this L. monocytogenes group carried all LIPI -4 genes. Meanwhile, the test of virulence genes was based on the PCR method employing target gene specific primers. There are some disadvantages to this method, including (i) polymorphism might exist in the primer annealing regions in the genomes of certain L. monocytogenes strains, leading to invalid or inefficient binding to primers and then false negative results, (ii) specificity of primers is not sufficient to avoid amplification of non-target regions. In recent years, whole genome sequencing (WGS) was utilized in many studies on L. monocytogenes, including the determination of virulence genes profiles (Maury et al., 2016; Fox et al., 2017; Rychli et al., 2017; Pightling et al., 2018), which can avoid false negative or positive results significantly. Genetic diversity of virulence genes can be further analyzed based on the nucleotide sequence.
Additionally, Gini coefficient analysis did not find an association between STs, ECs, LIPI-3 and LIPI-4 distribution of L. monocytogenes isolates and food groups. Values of the coefficient for eighteen STs ranged from 0.0 to 0.38. Seven ECs ranged from 0.0 to 0.25: 0.25 for LIPI-3 and 0.17 for LIPI-4, which indicated that STs, ECs, LIPI-3, and LIPI-4 were distributed uniformly within the four food groups: RTE food, raw meat, raw poultry and raw seafood. Meanwhile, moderate association was demonstrated between antibiotic resistance to oxacillin and raw poultry (coefficient value of 0.46), indicating that L. monocytogenes isolated from RTE food and raw poultry tend to be resistant to oxacillin.
In summary, a comprehensive study of prevalence and characteristics of L. monocytogenes isolated from retail foods in bulk in Zhejiang Province, China was performed. Both RTE foods and raw foods were included, showing a wide range of food categories. The potential risk of causing human L. monocytogenes infection by certain foods with relatively high contamination rates, including sushi and salmon sashimi, should arouse public concern. Distribution differences of serotype 1/2a among different food groups revealed that this serotype of L. monocytogenes might have specific ecological niches. To the best of our knowledge, this is the first time the distribution of ECs (ECI-ECXI) has been investigated in foods sampled in China. Furthermore, the discovery of multidrug-resistant strains and the particularly high resistance rate (11.0%) to tetracycline among L. monocytogenes isolates indicates a potential public health problem. According to our relatedness analysis, ECs and LIPI-3 or LIPI-4 positive isolates were distributed equally among various food groups. The present study provides initial data for Chinese food safety authorities to address the issue of microbial safety of retail bulk foods in China. One recommendation is to strengthen the monitoring of retail foods in bulk with relatively high detection rates of L. monocytogenes, including sushi and salmon sashimi. In addition, the public must recognize the potential risks of certain foods with high contamination rate of L. monocytogenes, and a national standard should be developed for the detection limit of L. monocytogenes for bulk foods with high risk of causing L. monocytogenes infection.
LZ, LM, and YZ designed the experiments. YZ, LZ, HC, JC, JZ, ZZ, YY, and ZX carried out the experiments. YZ and SD analyzed the experimental results and wrote the manuscript.
This work was supported by the National Key Research and Development Program of China (2017YFC1601503), the National Research Project of the 13th Five-Year Plan (2018ZX10714002), and the Medical Scientific Research Foundation of Zhejiang Province (2018KY034 and 2016KYB065).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank Dr. Hongliang Yang of Houston Methodist Research Institute and Kaity Mussio of University of California, Berkeley, for their help to revise and polish the language throughout the manuscript, and Drs. Mingbin Liang and Rong Zhang at Zhejiang Provincial Center for Disease Control and Prevention for their contribution on the statistical analysis.
Bergholz, T. M., Shah, M. K., Burall, L. S., Rakic-Martinez, M., and Datta, A. R. (2018). Genomic and phenotypic diversity of Listeria monocytogenes clonal complexes associated with human listeriosis. Appl. Microbiol. Biotechnol. 102, 3475–3485. doi: 10.1007/s00253-018-8852-5
Buchrieser, C., Brosch, R., Catimel, B., and Rocourt, J. (1993). Pulsed-field gel electrophoresis applied for comparing Listeria monocytogenes strains involved in outbreaks. Can. J. Microbiol. 39, 395–401.
Bula, C. J., Bille, J., and Glauser, M. P. (1995). An epidemic of food-borne listeriosis in western Switzerland: description of 57 cases involving adults. Clin. Infect. Dis. 20, 66–72.
Cantinelli, T., Chenal-Francisque, V., Diancourt, L., Frezal, L., Leclercq, A., Wirth, T., et al. (2013). “Epidemic clones” of Listeria monocytogenes are widespread and ancient clonal groups. J. Clin. Microbiol. 51, 3770–3779. doi: 10.1128/JCM.01874-13
Centers for Disease Control and Prevention (2017). Listeria (Listeriosis): People at Risk. Available at: https://www.cdc.gov/listeria/risk.htmlU3T (accessed December 9, 2017).
Chen, M., Wu, Q., Zhang, J., Wu, S., and Guo, W. (2015). Prevalence, enumeration, and pheno- and genotypic characteristics of Listeria monocytogenes isolated from raw foods in South China. Front. Microbiol. 6:1026. doi: 10.3389/fmicb.2015.01026
Chen, Y., Gonzalez-Escalona, N., Hammack, T. S., Allard, M. W., Strain, E. A., and Brown, E. W. (2016). Core genome multilocus sequence typing for identification of globally distributed clonal groups and differentiation of outbreak strains of Listeria monocytogenes. Appl. Environ. Microbiol. 82, 6258–6272. doi: 10.1128/AEM.01532-16
Chen, Y., and Knabel, S. J. (2007). Multiplex PCR for simultaneous detection of bacteria of the genus Listeria, Listeria monocytogenes, and major serotypes and epidemic clones of L. monocytogenes. Appl. Environ. Microbiol. 73, 6299–6304. doi: 10.1128/AEM.00961-07
Chen, Y., Strain, E. A., Allard, M., and Brown, E. W. (2011). Genome sequence of listeria monocytogenes strains J1816 and J1-220, associated with human outbreaks. J. Bacteriol. 193, 3424–3425. doi: 10.1128/JB.05048-11
Chen, Y., Zhang, W., and Knabel, S. J. (2007). Multi-virulence-locus sequence typing identifies single nucleotide polymorphisms which differentiate epidemic clones and outbreak strains of Listeria monocytogenes. J. Clin. Microbiol. 45, 835–846. doi: 10.1128/JCM.01575-06
China Statistical Yearbook Committee (2017). Per Capita Major Food Consumption of Urban Households in Different Regions in 2017. Available at: http://www.yearbookchina.com/navipage-n3018103001000692.html (accessed June 16, 2019).
Clayton, E. M., Daly, K. M., Guinane, C. M., Hill, C., Cotter, P. D., and Ross, P. R. (2014). Atypical Listeria innocua strains possess an intact LIPI-3. BMC Microbiol. 14:58. doi: 10.1186/1471-2180-14-58
Clinical and Laboratory Standards Institute (2015). Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria, 3rd Edn. Wayne, PA: Clinical and Laboratory Standard Institute.
Clinical and Laboratory Standards Institute (2017). Performance Standards for Antimicrobial Susceptibility Testing. Twenty-Seven Informational Supplement. Wayne, PA: Clinical and Laboratory Standard Institute.
Conter, M., Paludi, D., Zanardi, E., Ghidini, S., Vergara, A., and Ianieri, A. (2009). Characterization of antimicrobial resistance of foodborne Listeria monocytogenes. Int. J. Food Microbiol. 128, 497–500. doi: 10.1016/j.ijfoodmicro.2008.10.018
Cossart, P. (2011). Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc. Natl. Acad. Sci. U.S.A. 108, 19484–19491. doi: 10.1073/pnas.1112371108
Cotter, P. D., Draper, L. A., Lawton, E. M., Daly, K. M., Groeger, D. S., and Casey, P. G. (2008). Listeriolysin S, a novel peptide haemolysin associated with a subset of lineage I Listeria monocytogenes. PLoS Pathog. 4:e1000144. doi: 10.1371/journal.ppat.1000144
Department of Agriculture of Zhejiang Province (2017). Animal Husbandry Production of Zhejiang Province, 2016 -2017. Available at: http://www.zjagri.gov.cn/art/2018/11/29/art_1591988_30653695.html (accessed June 18 2019).
Donnelly, C. W. (2001). Listeria monocytogenes: a continuing challenge. Nutr. Rev. 59, 183–194. doi: 10.1111/j.1753-4887.2001.tb07011.x
Ebner, R., Stephan, R., Althaus, D., Brisse, S., Maury, M., and Tasara, T. (2015). Phenotypic and genotypic characteristics of Listeria monocytogenes strains isolated during 2011–2014 from different food matrices in Switzerland. Food Control 57, 321–326. doi: 10.1016/j.foodcont.2015.04.030
European Food Safety Authority [EFSA] (2013). Analysis of the baseline survey on the prevalence of Listeria monocytogenes in certain ready-to-eat foods in the EU, 2010-2011 Part A: Listeria monocytogenes prevalence estimates. EFSA J. 11:3241. doi: 10.2903/j.efsa.2013.3241
European Food Safety Authority [EFSA], and European Centre for Disease Prevention and Control [ECDC]. (2017). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2016. EFSA J. 15:e5077. doi: 10.2903/j.efsa.2017.5077
Filipello, V., Gallina, S., Amato, E., Losio, M. N., Pontello, M., Decastelli, L., et al. (2017). Diversity and persistence of Listeria monocytogenes within the Gorgonzola PDO production chain and comparison with clinical isolates from the same area. Int. J. Food Microbiol. 245, 73–78. doi: 10.1016/j.ijfoodmicro.2017.01.012
Food Standards Australia New Zealand (2017). Listeria: Food Safety and Recalls. Available at: http://www.foodstandards.gov.au/consumer/safety/listeria/Pages/default.aspx (accessed December 9, 2017).
Fox, E., Casey, A., Jordan, K., Coffey, A., Gahan, C., and Mcauliffe, O. (2017). Whole genome sequence analysis; an improved technology that identifies underlying genotypic differences between closely related Listeria monocytogenes strains. Innov. Food Sci. Emerg. 44, 89–96. doi: 10.1016/j.ifset.2017.07.010
Giménez-Muñoz, Á, Campello, I., Pérez Trullén, J. M., Alfaro, J., Sánchez Valiente, S., and Sanz Moncasi, P. (2015). Rhombencephalitis due to Listeria monocytogenes: a clinicopathologic study of a case. Neurologist 20, 97–100. doi: 10.1097/NRL.0000000000000060
Guo, Y. Z., and Liang, W. F. (2014). A case of meningitis caused by Listeria monocytogenes infection in immunocompetent adult. Chin. J. Infect. Dis. 32, 177–178.
Hadjilouka, A., Paramithiotis, S., and Drosinos, E. H. (2018). Genetic analysis of the listeria pathogenicity Island 1 of Listeria monocytogenes 1/2a and 4b isolates. Curr. Microbiol. 75, 857–865. doi: 10.1007/s00284-018-1458-4
Haley, B. J., Sonnier, J., Schukken, Y. H., Karns, J. S., and Van Kessel, J. A. (2015). Diversity of Listeria monocytogenes within a U.S. dairy herd, 2004-2010. Foodborne Pathog. Dis. 12, 844–850. doi: 10.1089/fpd.2014.1886
Hamon, M. A., and Cossart, P. (2011). K+ efflux is required for histone H3 dephosphorylation by Listeria monocytogenes listeriolysin O and other pore-forming toxins. Infect. Immun. 79, 2839–2846. doi: 10.1128/IAI.01243-10
Heisick, J. E., Wagner, D. E., Nierman, M. L., and Peeler, J. T. (1989). Listeria spp. found on fresh market produce. Appl. Environ. Microbiol. 55, 1925–1927.
Henri, C., Félix, B., Guillier, L., Leekitcharoenphon, P., Michelon, D., Mariet, J. F., et al. (2016). Population genetic structure of Listeria monocytogenes strains as determined by pulsed-field gel electrophoresis and multilocus sequence typing. Appl. Environ. Microbiol. 82, 5720–5728. doi: 10.1128/AEM.00583-16
Hof, H. (2004). An update on the medical management of listeriosis. Expert Opin. Pharmacother. 5, 1727–1735. doi: 10.1517/14656566.5.8.1727
Huang, B., Eglezos, S., Heron, B. A., Smith, H., Graham, T., Bates, J., et al. (2007). Comparison of multiplex PCR with conventional biochemical methods for the identification of Listeria spp. Isolates from food and clinical samples in Queensland, Australia. J. Food Prot. 70, 1874–1880.
Iannetti, L., Acciari, V. A., Antoci, S., Addante, N., Bardasi, L., Bilei, S., et al. (2016). Listeria monocytogenes in ready-to-eat foods in Italy: prevalence of contamination at retail and characterisation of strains from meat products and cheese. Food Control 68, 55–61. doi: 10.1016/j.foodcont.2016.03.036
Kathariou, S. (2002). Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 65, 1811–1829.
Kathariou, S., Graves, L., Buchrieser, C., Glaser, P., Siletzky, R. M., and Swaminathan, B. (2006). Involvement of closely related strains of a new clonal group of Listeria monocytogenes in the 1998-99 and 2002 multistate outbreaks of foodborne listeriosis in the United States. Foodborne Pathog. Dis. 3, 292–302. doi: 10.1089/fpd.2006.3.292
Khen, B. K., Lynch, O. A., Carroll, J., McDowell, D. A., and Duffy, G. (2015). Occurrence, antibiotic resistance and molecular characterization of Listeria monocytogenes in the beef chain in the Republic of Ireland. Zoonoses Public Health 62, 11–17. doi: 10.1111/zph.12106
Knabel, S. J., Reimer, A., Verghese, B., Lok, M., Ziegler, J., Farber, J., et al. (2012). Sequence typing confirms that a predominant Listeria monocytogenes clone caused human listeriosis cases and outbreaks in Canada from 1988 to 2010. J. Clin. Microbiol. 50, 1748–1751. doi: 10.1128/JCM.06185-11
Korsak, D., Borek, A., Daniluk, S., Grabowska, A., and Pappelbaum, K. (2012). Antimicrobial susceptibilities of Listeria monocytogenes strains isolated from food and food processing environment in Poland. Int. J. Food Microbiol. 158, 203–208. doi: 10.1016/j.ijfoodmicro.2012.07.016
Lamont, R. F., Sobel, J., Mazaki-Tovi, S., Kusanovic, J. P., Vaisbuch, E., and Kim, S. K. (2011). Listeriosis in human pregnancy: a systematic review. J. Perinat. Med. 39, 227–236. doi: 10.1515/JPM.2011.035
Li, W., Bai, L., Fu, P., Han, H., Liu, J., and Guo, Y. (2018). The epidemiology of Listeria monocytogenes in China. Foodborne Pathog. Dis. 15, 459–466. doi: 10.1089/fpd.2017.2409
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Lomonaco, S., Decastelli, L., Nucera, D., Gallina, S., Manila Bianchi, D., and Civera, T. (2009). Listeria monocytogenes in Gorgonzola: subtypes, diversity and persistence over time. Int. J. Food Microbiol. 128, 516–520. doi: 10.1016/j.ijfoodmicro.2008.10.009
Lomonaco, S., Patti, R., Knabel, S. J., and Civera, T. (2012). Detection of virulence-associated genes and epidemic clone markers in Listeria monocytogenes isolates from PDO Gorgonzola cheese. Int. J. Food Microbiol. 160, 76–79. doi: 10.1128/JCM.06185-11
Lomonaco, S., Verghese, B., Gerner-Smidt, P., Tarr, C., Gladney, L., and Joseph, L. (2013). Novel epidemic clones of Listeria monocytogenes, United States, 2011. Emerg. Infect. Dis. 19, 147–150. doi: 10.3201/eid1901.121167
Luchansky, J. B., Chen, Y., Porto-Fett, A. C. S., Pouillot, R., Shoyer, B. A., Johnson-DeRycke, R., et al. (2017). Survey for Listeria monocytogenes in and on ready-to-eat foods from retail establishments in the United States (2010 through 2013): assessing potential changes of pathogen prevalence and levels in a decade. J Food Prot. 80, 903–921. doi: 10.4315/0362-028X.JFP-16-420
Lungu, B., O’Bryan, C. A., Muthaiyan, A., Milillo, S. R., Johnson, M. G., Crandall, P. G., et al. (2011). Listeria monocytogenes: antibiotic resistance in food production. Foodborne Pathog. Dis. 8, 569–578. doi: 10.1089/fpd.2010.0718
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multi-drug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Maury, M. M., Tsai, Y. H., Charlier, C., Touchon, M., Chenal-Francisque, V., and Leclercq, A. (2016). Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 48, 308–313. doi: 10.1038/ng.3501
McGann, P., Ivanek, R., Wiedmann, M., and Boor, K. J. (2007). Temperature-dependent expression of Listeria monocytogenes internalin and internalin-like genes suggests functional diversity of these proteins among the listeriae. Appl. Environ. Microbiol. 73, 2806–2814. doi: 10.1128/AEM.02923-06
Mitchell, G., Ge, L., Huang, Q., Chen, C., Kianian, S., and Roberts, M. F. (2015). Avoidance of autophagy mediated by PlcA or ActA is required for Listeria monocytogenes growth in macrophages. Infect. Immun. 83, 2175–2184. doi: 10.1128/IAI.00110-15
Montero, D., Bodero, M., Riveros, G., Lapierre, L., Gaggero, A., Vidal, R. M., et al. (2015). Molecular epidemiology and genetic diversity of Listeria monocytogenes isolates from a wide variety of ready-to-eat foods and their relationship to clinical strains from listeriosis outbreaks in Chile. Front. Microbiol. 6:384. doi: 10.3389/fmicb.2015.00384
Morvan, A., Moubareck, C., Leclercq, A., Herve-Bazin, M., Bremont, S., and Lecuit, M. (2010). Antimicrobial resistance of Listeria monocytogenes strains isolated from humans in France. Antimicrob. Agents Chemother. 54, 2728–2731. doi: 10.1128/AAC.01557-09
Moura, A., Criscuolo, A., Pouseele, H., Maury, M. M., Leclercq, A., and Tarr, C. (2016). Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2:16185. doi: 10.1038/nmicrobiol.2016.185
National Standard of the People’s Republic of China (2010). National Food Safety Standard Food Microbiological Examination: Listeria monocytogenes (GB4789.30-2010). Beijing: Ministry of Health of People’s Republic of China.
Pightling, A. W., Pettengill, J. B., Luo, Y., Baugher, J. D., Rand, H., and Strain, E. (2018). Interpreting whole-genome sequence analyses of foodborne bacteria for regulatory applications and outbreak investigations. Front. Microbiol. 10:1482. doi: 10.3389/fmicb.2018.01482
Poimenidou, S. V., Dalmasso, M., Papadimitriou, K., Fox, E. M., Skandamis, P. N., and Jordan, K. (2018). Virulence gene sequencing highlights similarities and differences in sequences in Listeria monocytogenes serotype 1/2a and 4b strains of clinical and food origin from 3 different geographic locations. Front. Microbiol. 9:1103. doi: 10.3389/fmicb.2018.01103
Pouillot, R. D., Gallagher, J., Hoelzer, T. K., Kause, J., and Dennis, S. B. (2015). Listeria monocytogenes in retail delicatessens: an interagency risk assessment—model and baseline results. J. Food Prot. 78, 134–145. doi: 10.4315/0362-028X.JFP-14-235
Quereda, J. J., Dussurget, O., Nahori, M. A., Ghozlane, A., Volant, S., Dillies, M. A., et al. (2016). Bacteriocin from epidemic Listeria strains alters the host intestinal microbiota to favor infection. Proc. Natl. Acad. Sci. U.S.A. 113, 5706–5711. doi: 10.1073/pnas.1523899113
Raheem, D. (2016). Outbreaks of listeriosis associated with deli meats and cheese: an overview. AIMS Microbiol. 2, 230–250. doi: 10.3934/microbiol.2016.3.230
Ribeiro, V. B., and Destro, M. T. (2014). Listeria monocytogenes serotype 1/2b and 4b isolates from human clinical cases and foods show differences in tolerance to refrigeration and salt stress. J. Food Prot. 77, 1519–1526. doi: 10.4315/0362-028X.JFP-13-548
Ribeiro, V. B., Mujahid, S., Orsi, R. H., Bergholz, T. M., Wiedmann, M., and Boor, K. J. (2014). Contributions of sigma(B) and PrfA to Listeria monocytogenes salt stress under food relevant conditions. Int. J. Food Microbiol. 177, 98–108. doi: 10.1016/j.ijfoodmicro.2014.02.018
Rodas-Suarez, O. R., Flores-Pedroche, J. F., Betancourt-Rule, J. M., Quinones-Ramirez, E. I., and Vazquez-Salinas, C. (2006). Occurrence and antibiotic sensitivity of Listeria monocytogenes strains isolated from oysters, fish, and estuarine water. Appl. Environ. Microbiol. 72, 7410–7412. doi: 10.1128/AEM.00956-06
Rupp, S., Aguilar-Bultet, L., Jagannathan, V., Guldimann, C., Drogemuller, C., and Pfarrer, C. (2015). A naturally occurring prfA truncation in a Listeria monocytogenes field strain contributes to reduced replication and cell-to-cell spread. Vet. Microbiol. 179, 91–101. doi: 10.1016/j.vetmic.2015.03.002
Rychli, K., Wagner, E. M., Ciolacu, L., Zaiser, A. Z., Tasara, T., Wagner, M., et al. (2017). Comparative genomics of human and nonhuman Listeria monocytogenes sequence type 121 strains. PLoS One 12:e0176857. doi: 10.1371/journal.pone.0176857
Salama, P. J., Embarek, P. K. B., Bagaria, J., and Fall, I. S. (2018). Learning from listeria: safer food for all. Lancet 391, 2305–2306. doi: 10.1016/S0140-6736(18)31206-6
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M. A., and Roy, S. L. (2011). Foodborne illness acquired in the United States–major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.P11101
Schmitz-Esser, S., Müller, A., Stessl, B., and Wagner, M. (2015). Genomes of sequence type 121 Listeria monocytogenes strains harbor highly conserved plasmids and prophages. Front. Microbiol. 6:380. doi: 10.3389/fmicb.2015.00380
Shen, J., Rump, L., Zhang, Y., Chen, Y., Wang, X., and Meng, J. (2013). Molecular subtyping and virulence gene analysis of Listeria monocytogenes isolates from food. Food Microbiol. 35, 58–64. doi: 10.1016/j.fm.2013.02.014
Soni, D. K., Singh, R. K., Singh, D. V., and Dubey, S. K. (2013). Characterization of Listeria monocytogenes isolated from Ganges water, human clinical and milk samples at Varanasi, India. Infect. Genet. Evol. 14, 83–91. doi: 10.1016/j.meegid.2012.09.019
Tahoun, A., Abou Elez, R. M. M., Abdelfatah, E. N., Elsohaby, I., El-Gedawy, A. A., and Elmoslemany, A. M. (2017). Listeria monocytogenes in raw milk, milking equipment and dairy workers: molecular characterization and antimicrobial resistance patterns. J. Glob. Antimicrob. Resist. 10, 264–270. doi: 10.1016/j.jgar.2017.07.008
Tajima, F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tilney, L. G., and Portnoy, D. A. (1989). Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J. Cell Biol. 109, 1597–1608.
Travier, L., and Lecuit, M. (2014). Listeria monocytogenes ActA: a new function for a ’classic’ virulence factor. Curr. Opin. Microbiol. 17, 53–60. doi: 10.1016/j.mib.2013.11.007
Vermeulen, A., Gysemans, K. P., Bernaerts, K., Geeraerd, A. H., Van Impe, J. F., and Debevere, J. (2007). Influence of pH, water activity and acetic acid concentration on Listeria monocytogenes at 7 degrees C: data collection for the development of a growth/no growth model. Int. J. Food Microbiol. 114, 332–341. doi: 10.1016/j.ijfoodmicro.2006.09.023
Wang, H., Luo, L., Zhang, Z., Deng, J., Wang, Y., and Miao, Y. (2018). Prevalence and molecular characteristics of Listeria monocytogenes in cooked products and its comparison with isolates from listeriosis cases. Front. Med. 12:104–112. doi: 10.1007/s11684-017-0593-9
Wang, X. M., Lü, X. F., Yin, L., Liu, H. F., Zhang, W. J., and Si, W. (2013). Occurrence and antimicrobial susceptibility of Listeria monocytogenes isolates from retail raw foods. Food Control 32, 153–158. doi: 10.1016/j.foodcont.2012.11.032
Wang, Y., Feng, H., Zhu, Y., and Gao, P. (2017a). Structural insights into glutathione-mediated activation of the master regulator PrfA in Listeria monocytogenes. Protein Cell. 8, 308–312. doi: 10.1007/s13238-017-0390-x
Wang, Y., Lu, L., Lan, R., Salazar, J. K., Liu, J., and Xu, J. (2017b). Isolation and characterization of Listeria species from rodents in natural environments in China. Emerg. Microbes Infect. 6:e44. doi: 10.1038/emi.2017.28
Wang, Y., Jiao, Y., Lan, R., Xu, X., Liu, G., and Wang, X. (2015). Characterization of Listeria monocytogenes isolated from human Listeriosis cases in China. Emerg. Microbes Infect. 4:e50. doi: 10.1038/emi.2015.50
Wieczorek, K., Dmowska, K., and Osek, J. (2012). Prevalence, characterization, and antimicrobial resistance of Listeria monocytogenes isolates from bovine hides and carcasses. Appl. Environ. Microbiol. 78, 2043–2045. doi: 10.1128/AEM.07156-11
Wieczorek, K., and Osek, J. (2017). Prevalence, genetic diversity and antimicrobial resistance of Listeria monocytogenes isolated from fresh and smoked fish in Poland. Food Microbiol. 64, 164–171. doi: 10.1016/j.fm.2016.12.022
Wu, S., Wu, Q., Zhang, J., Chen, M., and Guo, W. (2016). Analysis of multilocus sequence typing and virulence characterization of Listeria monocytogenes isolates from Chinese retail ready-to-eat food. Front. Microbiol. 7:168. doi: 10.3389/fmicb.2016.00168
Wu, S., Wu, Q., Zhang, J., Chen, M., Yan, Z., and Hu, H. (2015). Listeria monocytogenes prevalence and characteristics in retail raw foods in China. PLoS One 10:e0136682. doi: 10.1371/journal.pone.0136682
Zhang, W., Jayarao, B. M., and Knabel, S. J. (2004). Multi-virulence-locus sequence typing of Listeria monocytogenes. Appl. Environ. Microbiol. 70, 913–920.
Zhang, Y., Chen, J., Feng, C., Zhan, L., Zhang, J., Li, Y., et al. (2017). Quantitative prevalence, phenotypic and genotypic characteristics of Bacillus cereus isolated from retail infant foods in China. Foodborne Pathog. Dis. 14, 564–572. doi: 10.1089/fpd.2017.2287
Zhang, Z., and Yu, J. (2006). Evaluation of six methods for estimating synonymous and nonsynonymous substitution rates. Genomics Proteomics Bioinformatics 3, 173–181. doi: 10.1016/S1672-0229(06)60030-2
Keywords: Listeria monocytogenes, prevalence, retail foods in bulk, antibiotic resistance, multilocus sequence typing (MLST), multi-virulence-locus sequence typing (MVLST), epidemic clone (EC)
Citation: Zhang Y, Dong S, Chen H, Chen J, Zhang J, Zhang Z, Yang Y, Xu Z, Zhan L and Mei L (2019) Prevalence, Genotypic Characteristics and Antibiotic Resistance of Listeria monocytogenes From Retail Foods in Bulk in Zhejiang Province, China. Front. Microbiol. 10:1710. doi: 10.3389/fmicb.2019.01710
Received: 09 October 2018; Accepted: 11 July 2019;
Published: 25 July 2019.
Edited by:
Learn-Han Lee, Monash University Malaysia, MalaysiaReviewed by:
Marc Heyndrickx, Institute for Agricultural and Fisheries Research (ILVO), BelgiumCopyright © 2019 Zhang, Dong, Chen, Chen, Zhang, Zhang, Yang, Xu, Zhan and Mei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Zhan, bHpoYW5AY2RjLnpqLmNu; Lingling Mei, bGxtZWlAY2RjLnpqLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.