- 1State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, Fujian Agriculture and Forestry University, Fuzhou, China
- 2Key Laboratory for Biopesticide and Chemical Biology, Ministry of Education, Fujian Agriculture and Forestry University, Fuzhou, China
- 3Fujian Key Laboratory of Plant Virology, Institute of Plant Virology, Fujian Agriculture and Forestry University, Fuzhou, China
- 4Department of Forest Mycology and Plant Pathology, Swedish University of Agricultural Sciences, Uppsala, Sweden
Evolution of fungicide resistance in plant pathogens is one of major concerns in sustainable plant disease management. In this study, the genetics and potential of developing resistance to a demethylation inhibitor (DMI) fungicide, difenoconazole, in the fungal pathogen Alternaria alternata was investigated using a comparative analysis of genetic variation in molecular (Single Sequence Repeats, SSR) and phenotypic (fungicide tolerance) markers. No difenoconazole resistance was found in the 215 A. alternata isolates sampled from seven different ecological zones in China despite the widespread use of the fungicide for more than 20 years. This result suggests that the risk of developing resistance to difenoconazole in A. alternata is low and we hypothesize that the low risk is likely caused by fitness penalties incurred by resistant mutants and the multiple mechanisms involving in developing resistance. Heritability and plasticity account for ∼24 and 3% of phenotypic variation, respectively, indicating that genetic adaptation by sequence variation plays a more important role in the evolution of difenoconazole resistance than physiological adaptation by altering gene expression. Constraining selection in the evolution of A. alternata resistance to difenoconazole was documented by different patterns of population differentiation and isolate-by-distance between SSR markers and difenoconazole tolerance. Though the risk of developing resistance is low, the findings of significant differences in difenoconazole tolerance among isolates and populations, and a skewing distribution toward higher tolerance suggests that a stepwise accumulation of tolerance to the fungicide might be occurring in the pathogen populations. As a consequence, dynamic management programs guided by evolutionary principles such as spatiotemporal rotations of fungicides with different modes of action are critical to prevent the continued accumulation of tolerance or the evolution of resistance to difenoconazole and other DMI fungicides.
Introduction
Plant diseases caused by pathogenic fungi have been and continue to be one of the major factors threatening global food security and social stability and development (Brent and Hollomon, 2007; Savary et al., 2012; Chourasiya et al., 2013; Hahn, 2014). Outbreaks of several plant diseases in history such as the Irish Potato Famine, Bengal Rice Famine and the southern corn leaf blight pandemic in United States not only caused severe economic losses, but in the case of the first two diseases, also the death of millions of people from starvation (Strange and Scott, 2005). To counter such losses and human casualty, fungicides have been an essential weapon in the disease control armory especially when genetic controls fail. The application of fungicide not only benefits society directly and immediately by reducing food losses but also indirectly and in the long term, by improving the quality and longevity of human life and supporting economic development (Cooper and Dobson, 2007).
Due to their short generation time and large population size (Angelini et al., 2015; Delmas et al., 2017), fungal pathogens can rapidly evolve and adapt to meet changing environments including those imposed by the introduction of new fungicides in agricultural systems. Indeed, many fungicides have been rendered ineffective by the development of resistance in target pathogens (Avenot et al., 2016; Rupp et al., 2016; Delmas et al., 2017; Jørgensen et al., 2018). Resistant pathotypes usually originate from mutation and/or recombination (sexual and asexual) in a single population, increase frequency through natural selection and migrate to other populations directly or via intermediary stepping-stone populations (Gisi et al., 2002; Angelini et al., 2015).
Development of fungicide resistance is a complex and multifactorial process. It depends on many factors including: (i) properties of the fungicides such as their chemistry and mode of action, (ii) characteristics of the pathogens such as their reproductive rate and genetic adaptability, and (iii) ecological features such as host traits and environmental conditions (Brent and Hollomon, 2007; Damicone and Smith, 2009; Hahn, 2014; Muneret et al., 2018). In terms of their mode of action, fungicides can be either site-specific or site non-specific. Resistance to site-specific fungicides is a qualitative trait and can be developed by single point mutations occurring at the target sites. In this class of fungicides, resistant pathotypes can rapidly emerge in pathogen populations shortly after the fungicides are commercialized. For example, resistance to Quinone outside inhibitors (QoI) emerged in Blumeria graminis f. sp. tritici only 2 years after the fungicides were commercialized (Sierotzki et al., 2000; Villa et al., 2017). On the other hand, resistance to site non-specific fungicides is quantitatively inherited and involves a series of changes in genes governing the uptake, transport, storage and metabolism of the fungicide (de Waard et al., 2006). Therefore, the risk of developing resistance to this class of fungicides is expected to be lower than the risk involved with the use of site-specific fungicides.
Human activities also play an important role in the evolution of fungicide resistance, mainly through their influences on the direction and intensity of natural selection acting upon pathogens. Continuous and widespread application of fungicides with the same mode of action creates strong, directional selection for resistant mutants, thereby facilitating the development of fungicide resistance in pathogen populations. On the other hand, mixture or rotation of fungicides with different modes of action generates disruptive selection that reduces the probability of emergence of resistant mutants – a strategy that enhances the durability of fungicide efficacy (Brent and Hollomon, 2007).
Difenoconazole, approved in the 1988 in Europe (Bowyer and Denning, 2014), is synthesized as a novel demethylation inhibitor (DMI) fungicide, targeting sterol 14α-demethylase (CYP51), an important regulatory enzyme in the ergosterol biosynthetic pathway (Zarn et al., 2003; Price et al., 2015). The fungicide has been used worldwide in agricultural system due to its excellent, fast-acting and prominent systemic activity (Dong et al., 2013; Ge et al., 2017). It has both protective and curative efficacy and is typically used to control a broad spectrum of foliar, seed- and soil-borne diseases caused by Ascomycetes, Basidiomycetes. Deuteromycetes and oomycetes (Horsfield et al., 2010; Gveroska, 2013; Elansky et al., 2016). Resistance to difenoconazole is a polygenic trait with multiple mechanisms contributing to the final phenotype (Wyand and Brown, 2005; Leroux and Walker, 2011; Cools and Fraaije, 2013; Villani et al., 2015; Pereira et al., 2017). Possibly due to the intensive application of difenoconazole, isolates with reduced sensitivity have been detected in Phoma ligulicola (Jones et al., 2007) and Venturia inaequalis (Stevic′ et al., 2010). Cross-resistance was also found between difenoconazole and other DMI fungicides such as myclobutanil, fenbuconazole, and flusilazole in the pathogen V. inaequalis (Stevic′ et al., 2010; Pfeufer and Ngugi, 2012).
Early blight is a destructive foliar disease reducing the photosynthetic ability of potato, tomato and other important Solanaceae crops in the world (Gent and Schwartz, 2003; MacDonald et al., 2007; Meng et al., 2015b). It can cause significant economic loss to farmers when environmental conditions (e.g., nitrogen and water shortages’) favor for epidemics. The causal agents of early blight are haploid filamentous fungi from the genus Alternaria (Parvez, 2003; Leiminger and Hausladen, 2012). In China, Alternaria alternata has replaced A. solani as the main pathogen inducing potato early blight (Meng et al., 2015b; Zheng et al., 2015). It is a global pathogen dispersed by rain-splash, wind or infected plant materials (Reis et al., 2006). Although no sexual fruiting bodies have been documented yet, population genetic and phylogenetic analyses suggest that cryptic sex may occur frequently in the life-cycle of the pathogen (Meng et al., 2015a). Besides potato early blight, A. alternata can also cause catastrophic diseases on other numerous food and ornamental crops (Wier et al., 1998; Woudenberg et al., 2013) although host specificity has been reported in the pathogen (Elena, 2006; Woudenberg et al., 2015). In potato industry, cultivars with major-gene resistance to A. alternata are rare and management of early blight is mainly relied on the application of fungicides. Difenoconazole has been used to battle against A. alternata for many years around the world including China (Zheng et al., 2013; Avenot et al., 2016; Fonseka and Gudmestad, 2016; Wang et al., 2016).
Knowledge of genetic and evolutionary mechanisms responsible for the development of fungicide resistance in pathogen populations is important as a basis for formulating effective approaches to plant disease management. This knowledge can be acquired through statistical analyses of genetic variation and its spatial dynamics in fungicide sensitivity (Qin et al., 2016). Therefore, the main objectives of the present study were to: (i) evaluate the potential for developing difenoconazole resistance in A. alternata, by quantifying genetic and environmental factors contributing to phenotypic variation of difenoconazole tolerance using a common garden experiment and (ii) determine the role of natural selection in the evolution of difenoconazole resistance by comparing the spatial distribution of genetic variation in SSR markers and difenoconazole tolerance.
Materials and Methods
Origin of Alternaria alternata Isolates
Seven A. alternata populations with a total of 215 genetically distinct isolates from potato host were included in the current analysis of difenoconazole tolerance. The isolates were sampled during the 2011 and 2012 potato growing seasons from seven fields distributed across various ecological niches and agro-ecosystems of China including Fujian (FJN), Heilongjiang (HLJ), Henan (HNN), Hubei (HBI), Inner Mongolia (IMG), Shandong (SDG), and Yunnan (YNN). They were collected from plants separated by >100 cm and The A. alternata isolates were molecularly assayed with eight microsatellite markers (PAS1, PAS2, PAS3, PAS4, PAS5, PAS6, PAS7, and Ad8) previously and genotypes of the isolates were determined by GenClone 2.0 (Arnaud-Haond et al., 2007) using the allele information at each of the eight SSR loci. Detailed information on pathogen collection, isolation, DNA extraction and microsatellite genotyping as well as primer sequences can be found in these previous publications (Benichou et al., 2009; Meng et al., 2015a,b). Because potato early blight can be induced by several species in the Alternaria genus, the identity of these isolates were checked morphologically by spore characterization under a light microscope (Meng et al., 2015a, 2015b) and molecularly by PCR amplifications of ITS (Internal transcribed spacer) regions and histone 3 gene (Meng et al., 2015a; Zheng et al., 2015) to ensure they are all A. alternata.
Measurement of Difenoconazole Tolerance
A total of 215 isolates were tested for difenoconazole tolerance by a growth rate assay using a common garden design (Moloney et al., 2009; Yang et al., 2016; He et al., 2018). The pathogen isolates taken out from long-term storage were activated on PDA plates for 6 days. Mycelial plugs (ϕ = 5 mm) taken from the edge of the colony were transferred to fresh PDA plates with or without amendment of 0.02, 0.06, or 0.12 μg/mL of difenoconazole (technical grade). Preliminary experiments indicated these concentrations provided the best resolution with the least experimental error. Many isolates did not grow at a higher difenoconazole concentration while growth rates of these isolates were unchanged at a lower concentration. The PDA plates inoculated with different A. alternata isolates were divided into three groups each corresponding to one difenoconazole concentration and arranged according to a completely randomized design with three replicates in a single incubator set to 24°C. Pathogen inoculated on the PDA plates without difenoconazole ingredient was set as a control for each isolate and controls were included in each fungicide concentration. To minimize experimental errors, plate preparation, pathogen inoculation and colony measurement for the entire fungicide tolerance assay were completed by the same student with all experimental activities for a single fungicide concentration being assessed on the same day. Pathogen colonies were digitalized daily between 2nd and 6th post-inoculation and Assess (Lamari, 2002) was used to estimate the size (area) of the colonies. In total, 12900 [215 isolates × 3 replicates × 4 treatment (3 fungicide concentrations + 1 control) × 5 measurements] colonies were measured, generating a large number of data points to evaluate the spatial pattern, adaptive mechanism and evolutionary history of difenoconazole resistance in the A. alternata populations.
Data Analysis
Intrinsic growth rate of the pathogen isolates was estimated as described previously (Qin et al., 2016; Yang et al., 2016; He et al., 2018). A logistic model was applied for the estimate (Aguayo et al., 2014) using the daily colony sizes of isolates measured over the 6 days of inoculation and the intrinsic growth rate was estimated separately for each of the three difenoconazole concentrations. The colony size of the pathogen isolates at the first day of inoculation was set to 0.2 cm2 (πr2 = 3.14 × 0.252). the size of the mycelial plug initiated the colonies and the capacity of colony growth (K, the maximum colony size) for the logistic model was set to 63.6 cm2 (πr2 = 3.14 × 4.52), the area of a Petri dish with a diameter of 9 cm. Difenoconazole tolerance was measured by the relative intrinsic growth rate (RGR) of the pathogen isolates in the presence to the absence of difenoconazole (Zhan et al., 2006; Brunner et al., 2016; He et al., 2018). The percentile of difenoconazole tolerance in the combined population of the 215 A. alternata isolates was tabulated using a bin system as described previously (Qin et al., 2016; Wu et al., 2019). General linear model procedure was used to evaluate the contribution of population, fungicide concentration, pathogen genotype and genotype-concentration interaction to the phenotypic variance of difenoconazole tolerance and least significant difference (Kokalisburelle et al., 2013) was used to determine the spatial variation of difenoconazole tolerance in the A. alternata populations from different locations.
Single Sequence Repeats data of the 215 isolates were taken from a previous study (Meng et al., 2015a) and used to estimate Nei’s gene diversity, population differentiation GST and the effective number of migrants (Nem) in neutral markers (Nei, 1973) using Popgene 3.249 (Yeh et al., 2000). Phenotypic variance in difenoconazole tolerance was partitioned into sources attributable to isolate (I, random effect), population (P, random effect), and fungicide concentration (C, fixed effect) using SAS GLM and VARCOMP programs (SAS 9.4, SAS Institute) according to the model:
Where Yripc refers to the mean RGR of replicate r for isolate i in population p at concentration c; M, P, I(P), I(P) × C, P × C and Eripc refer to the overall population mean, genetic variance among populations, genetic variance within populations, variance due to genotype × concentration interaction, variance responses of populations to dose effect and the variance among replicates, respectively (Zhan and McDonald, 2011). In common garden experiments with asexual species, any among-replicate variation in the phenotypic value of an isolate can be treated as environmental effect. Therefore, variance among replicates in this case is equivalent to the environmental variance of RGR (Zhan et al., 2005). Population differentiation (QST) in difenoconazole tolerance as measured by RGR was estimated in a similar way to the estimation of population differentiation of SSR marker loci (GST) using the formula described in previous publications (Qin et al., 2016; Yang et al., 2016).
Heritability of RGR in a population was estimated by dividing genetic variance within populations by total phenotypic variance and phenotypic plasticity was calculated by dividing the variance of isolate-concentration interaction by the total phenotypic variance (Tonsor et al., 2013). Statistical difference between the overall GST in SSR loci and overall QST in fungicide sensitivity was tested using the standard deviation of QST constructed from 100 resamplings of the original data (Zhan and McDonald, 2011). Difenoconazole tolerance measured by RGR among A. alternata populations from collection sites were compared by Least significant difference (LSD) test (Ott and Longnecker, 2008). Physical distance between collection sites was estimated by Google Earth. Isolation-by-distance in A. alternata was inferred by association analysis between Napierian logarithm of the pair-wise physical distance and pair-wise gene flow among the pathogen populations, and association of fungicide resistance (RGR) among difenoconazole doses was evaluated by Pearson’s correlation (Lawrence and Lin, 1989).
Results
Frequency Distribution of Alternaria alternata Tolerance (RGR) to Difenoconazole
Between 28 and 32 (total 215) genetically distinct A. alternata isolates originating from seven field populations were assayed for growth rate in the absence and presence of difenoconazole. The growth rate of the 215 A. alternata isolates displayed a continuous, unimodal distribution both in the absence and presence of difenoconazole (Figure 1A). The average growth rate of colonies declined as the concentration of the fungicide increased (r = −0.96, P = 0.04; Figure 1B). In the absence of the fungicide, the average growth rate was 0.765 cm2/day. This value declined linearly to 0.640 cm2/day at a fungicide concentration of 0.12 μg/mL.

Figure 1. Growth rate of Alternaria alternata collected from seven potato fields across China: (A) distribution of growth rate at in the presence and absence of difenoconazole, (B) correlation between difenoconazole concentrations and average growth rate of A. alternata populations collected from seven potato fields across China.
Distribution of Difenoconazole Tolerance Measured by RGR Between Presence and Absence of the Fungicide
Relative intrinsic growth rate of the 215 isolates also displayed a continuous, unimodal distribution in all three concentrations (Figure 2), ranging from 0.74 to 1.07 with an average of 0.92 at the 0.02 μg/mL, 0.63 to 1.04 with an average of 0.88 at the 0.06 μg/mL and 0.53 to 1.02 with an average of 0.85 at the 0.12 μg/mL, respectively. As fungicide concentration increased, the mean RGR of A. alternata populations decreased but the ratio of RGR in the fastest and slowest growth isolates increased. In the 0.02 μg/ml treatment, the ratio of RGR between the fastest and slowest growth isolates in the population was 1.44. This value increased to 1.66 and 1.92 for the 0.06 and 0.12 μg/mL treatments, respectively. Association analysis showed that A. alternata tolerance in different concentrations was positively and significantly correlated (P < 0.0001, Figure 3).
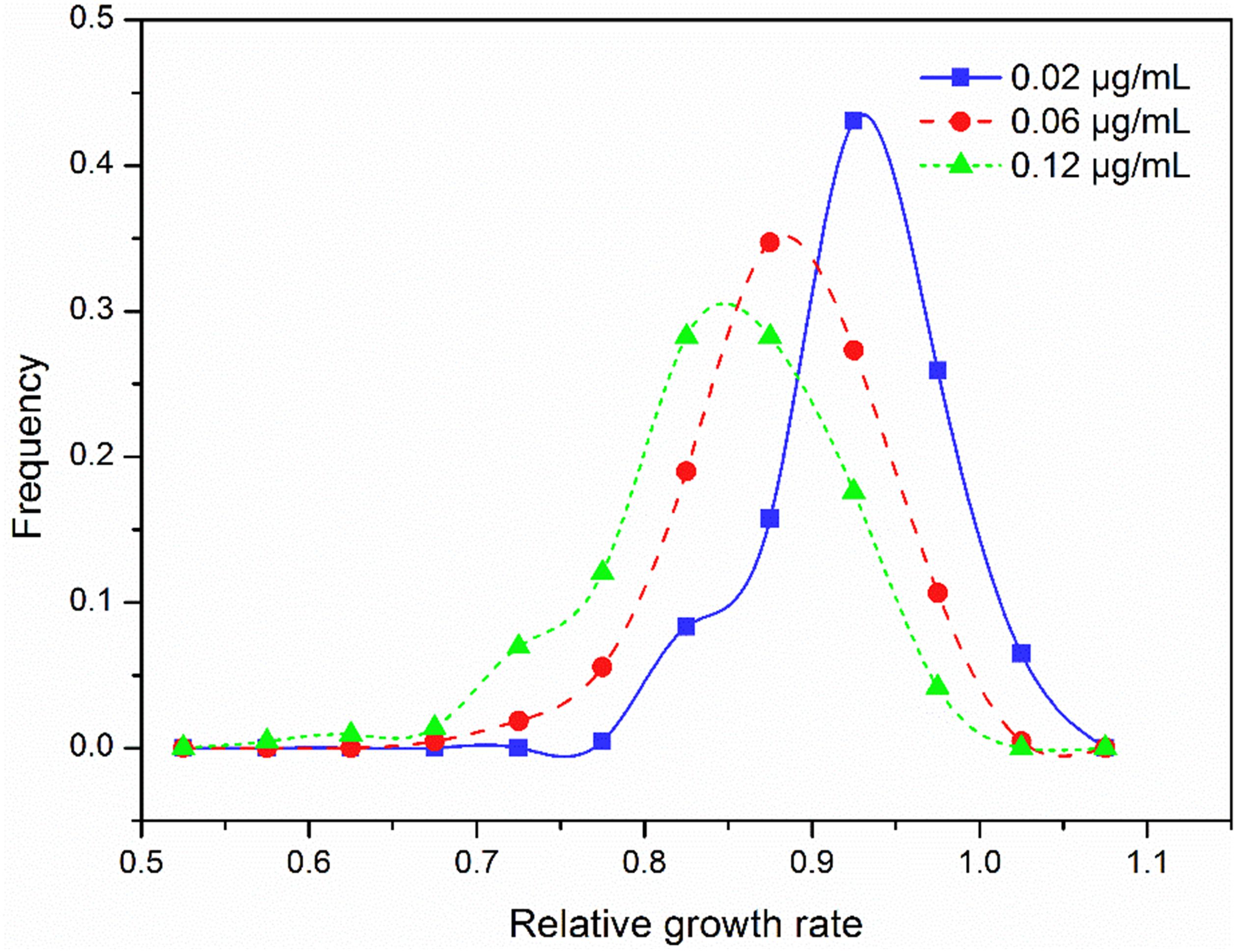
Figure 2. Frequency distribution of difenoconazole tolerance measured by the relative growth rate in the presence to the absence of difenoconazole at three concentrations in the 215 isolates of Alternaria alternata collected from seven potato fields across China.
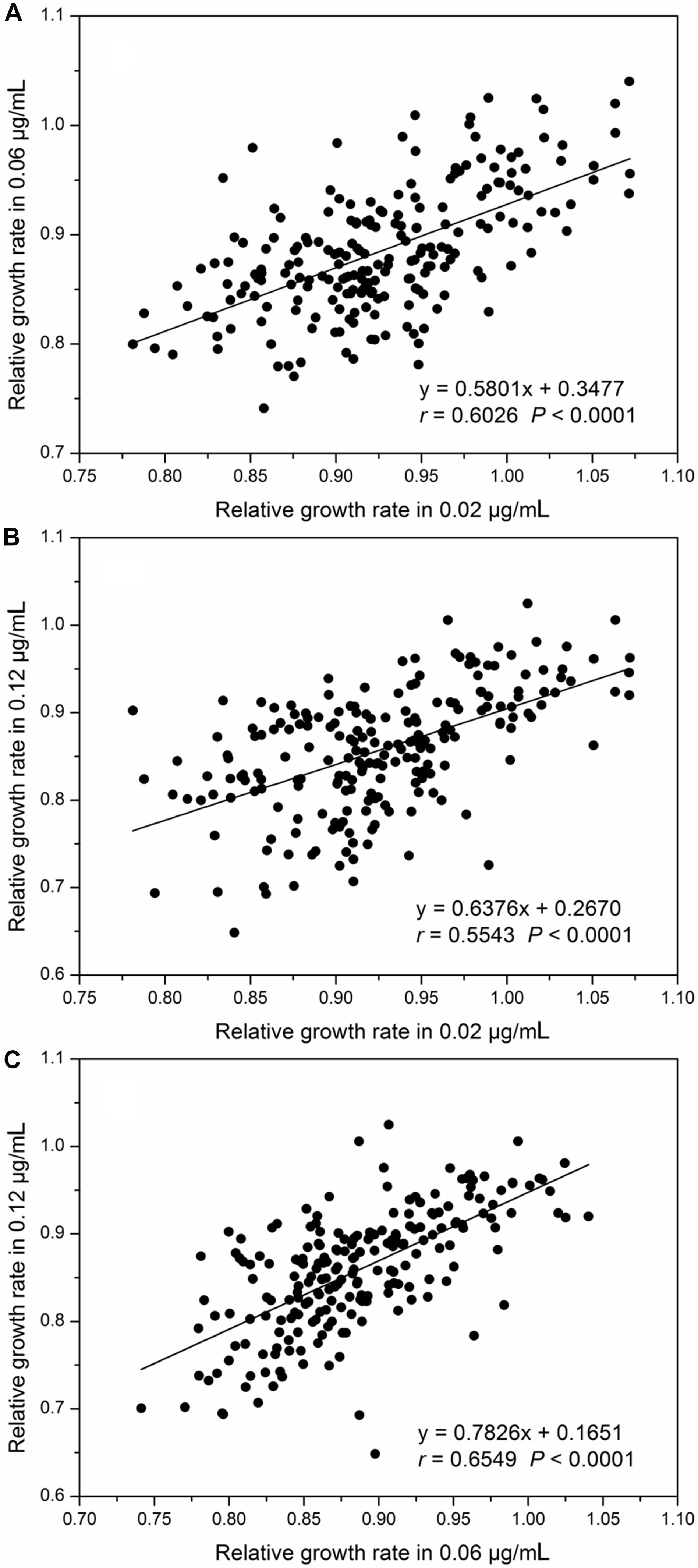
Figure 3. Correlation between relative growth rates (difenoconazole tolerance) in different concentrations: (A) difenoconazole concentrations at 0.02 and 0.06 μg/mL, (B) difenoconazole concentrations at 0.02 and 0.12 μg/mL, and (C) difenoconazole concentrations at 0.06 and 0.12 μg/mL.
Spatial Variation in Difenoconazole Tolerance (RGR) Among Alternaria alternata Populations
Population, isolate and fungicide dose all contributed significantly (P < 0.0001) to the performance of difenoconazole tolerance in A. alternata as measured by RGR (Table 1). The 215 A. alternata isolates also responded differentially to different concentrations. Least significant difference (LSD) analysis indicated that the pathogen population sampled from Shandong showed the least tolerance to difenoconazole while the pathogen population sampled from Heilongjiang was most tolerant to difenoconazole (Table 2). The populations sampled from Hubei and Inner Mongolia showed an intermediate level of tolerance (Table 2).
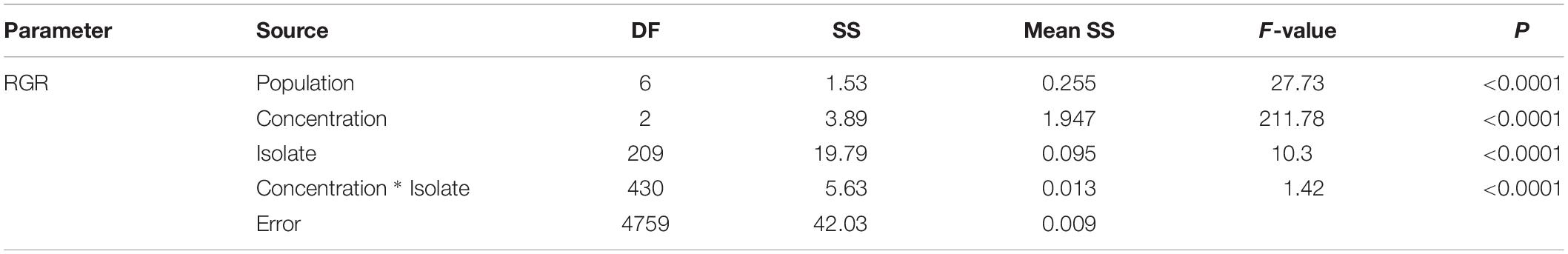
Table 1. Analysis of variance (ANOVA) of difenoconazole tolerance in the 215 isolates of Alternaria alternata sampled from seven potato fields in China.
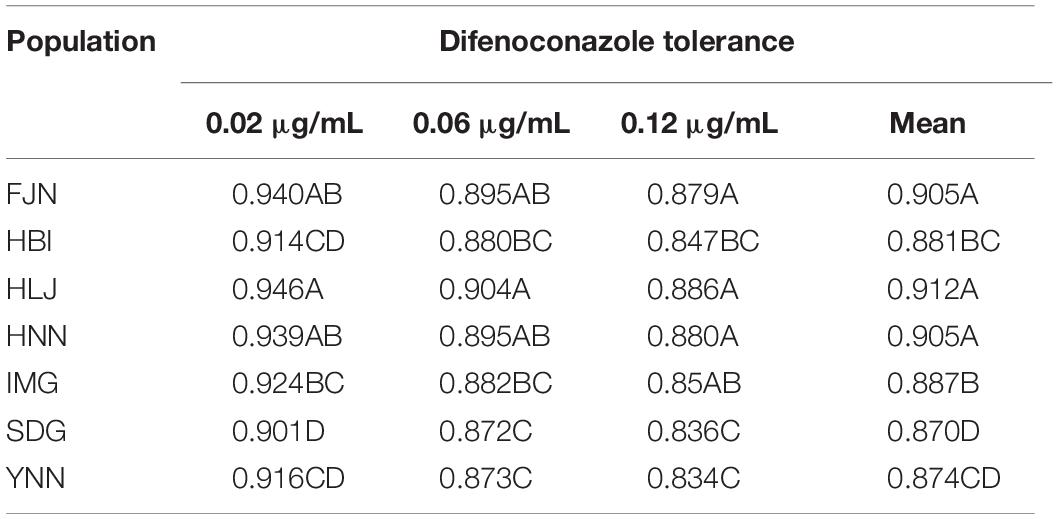
Table 2. Difenoconazole tolerance at three concentrations and mean of three concentrations in the 215 isolates of Alternaria alternata collected from seven potato fields across China.
Genetic Variations in RGR and SSR Marker Loci
The contribution of genetic architecture and gene expression to difenoconazole tolerance in the pathogen populations was measured by heritability and phenotypic plasticity. Heritability in the seven populations ranged from 0.17 to 0.47 with an average of 0.24; while phenotypic plasticity in the seven populations ranged from 0 to 0.17 with an average of 0.03 (Table 3). Heritability was higher than phenotypic plasticity in all seven populations. The A. alternata population collected from Yunnan displayed the highest heritability in difenoconazole tolerance while that collected from Hubei displayed the lowest heritability (Table 3). The average SSR diversity in the seven A. alternata field populations ranged from 0.31 to 0.62 with an overall diversity of 0.44 when the isolates from the seven populations were pooled (Table 3). The highest SSR variation was found in the A. alternata population sampled from Fujian while that the lowest SSR variation was found in the A. alternata population collected from Yunnan. No association (r = −0.49, P = 0.26) was detected between heritability of difenoconazole tolerance and genetic variation in SSR markers.
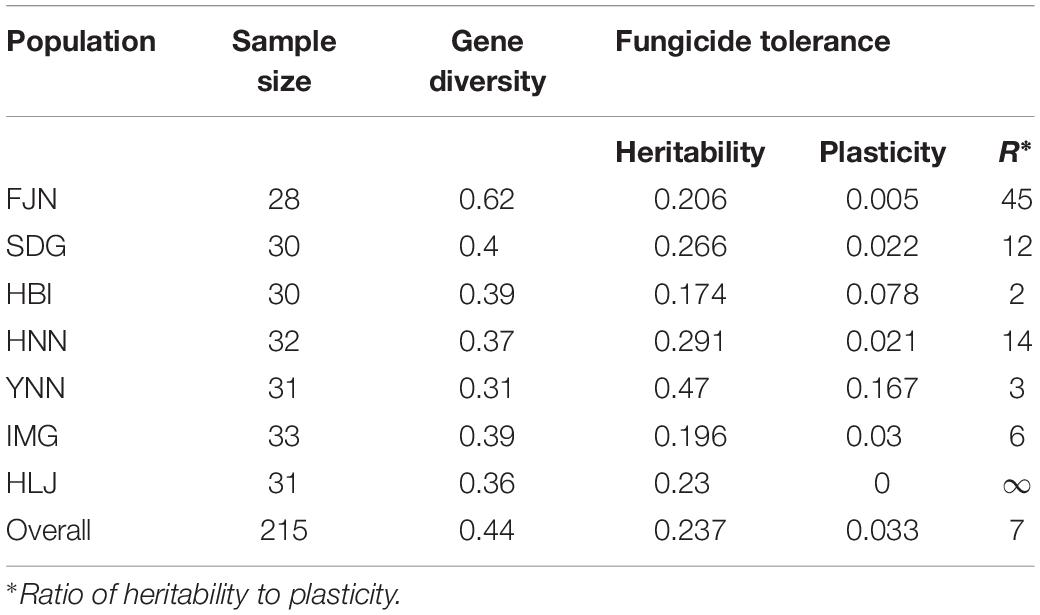
Table 3. Sample size, gene diversity in SSR marker loci, quantitative genetic parameters related to the level of tolerance to difenoconazole in the seven Alternaria alternata populations from potato.
Comparison of QST in Difenoconazole Tolerance (RGR) and GST SSR Marker Loci
Pair-wise QST in difenoconazole tolerance ranged from 0.00 to 0.12 and pair-wise GST in SSR markers ranged from 0.01 to 0.15. Thirteen out of twenty-one pairs of GST were higher than QST (Table 4). The overall GST (0.12) in the SSR markers across the seven pathogen populations was significantly higher than overall QST (0.028) in difenoconazole tolerance.
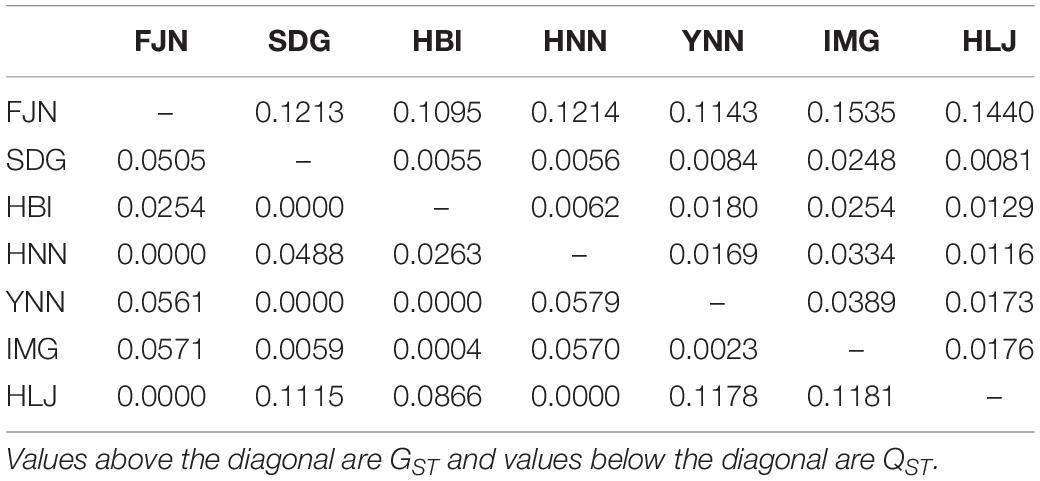
Table 4. Pair-population differentiation of SSR marker loci (GST) and difenoconazole tolerance (QST) among the seven populations of Alternaria alternata sampled from potato.
Isolation-by-Distance in Difenoconazole Tolerance (RGR) and SSR Marker Loci
The geographic distances between pairs of collection sites ranged from 341 to 3146 kilometers, with a mean of 1386 kilometers. The pair-wise gene flow in SSR markers ranged from 3.73 to 99.5 with an average of 45.14 and the pair-wise gene flow in SSR markers ranged from 2.76 to 90.41 with an average of 31.14, There was no correlation between the Napierian logarithm of pair-wise geographic distance and gene flow in difenoconazole tolerance but a significant negative correlation occurred between the Napierian logarithm of pair-wise geographic distances and gene flow in SSR marker loci (Figure 4).
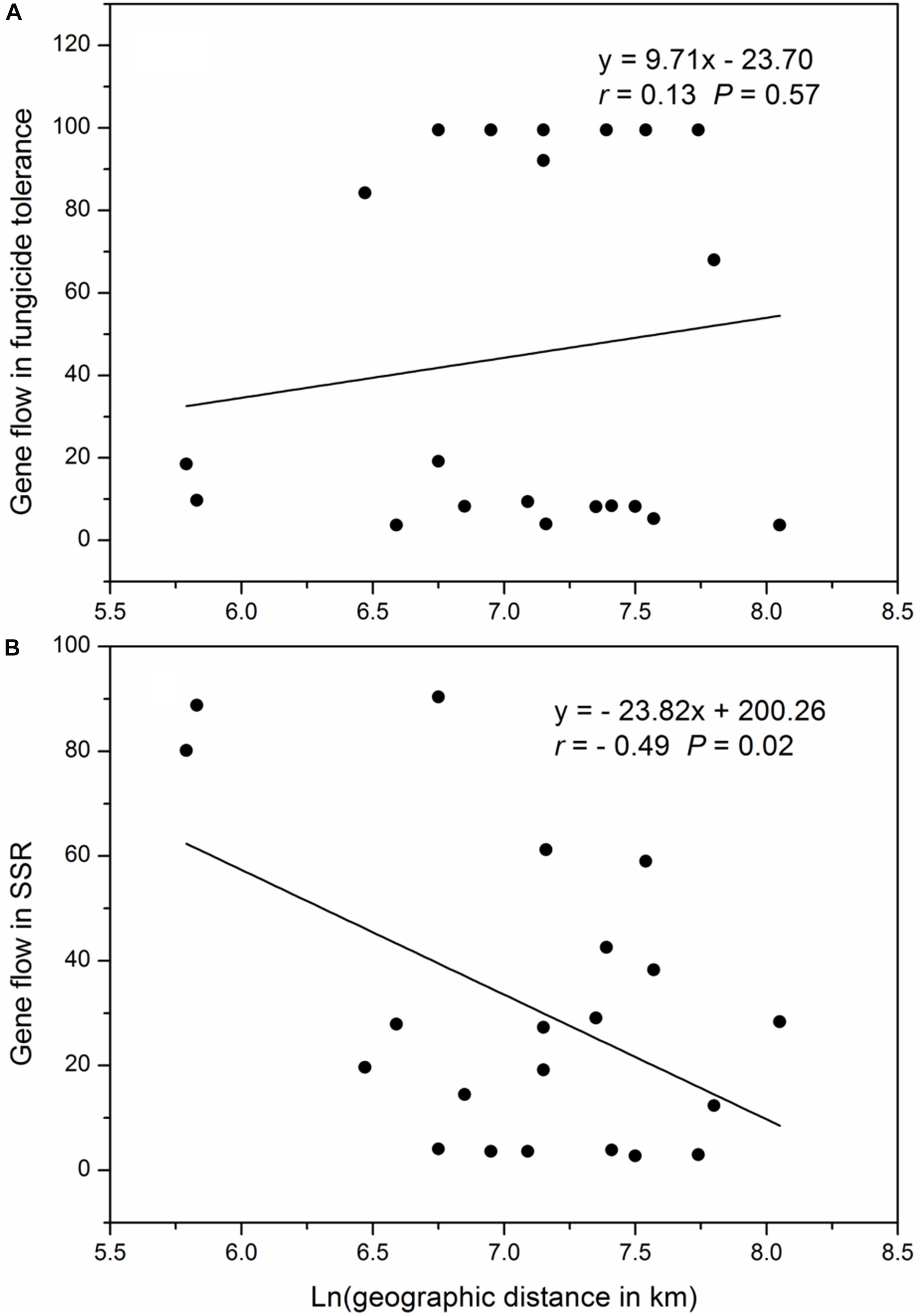
Figure 4. Correlation between Napierian logarithm of the pair-wise geographic distance and pair-wise gene flow among the seven populations of Alternaria alternata sampled from potato: (A) relative growth rate (difenoconazole tolerance) and (B) SSR marker loci.
Discussion
We used a common garden design to investigate the genetics and evolutionary trajectory of fungicide resistance in a large number of pathogen isolates originating from seven distinct regions of China. Our results indicated that the mycelial growth of A. alternata isolates was significantly inhibited by small doses of difenoconazole (Figure 2). In addition, no resistant isolates were detected in the widely dispersed pathogen populations sampled despite the continuous and extensive use of the fungicide for several decades in the region (Liang et al., 2009; Chen et al., 2013). Difenoconazole has been used to control many cereal, vegetable and fruit diseases including potato early blight in China since 1998 and ∼10% of its global production is sold in Chinese markets recent years1 ,2 (Zheng et al., 2013). However, this observation is consistent with previous results showing that difenoconazole is still effective in controlling many plant pathogens such as A. solani, Rhizoctonia cerealis, Colletotrichum capsici, Didymella applanata, and Ceratocystis fimbriata (Reuveni and Sheglov, 2002; Gopinath et al., 2006; Mirkovic et al., 2015; Wang et al., 2016; Scruggs et al., 2017) and suggests that there is a low risk of developing difenoconazole resistance in many plant pathogens (Hamada et al., 2011; Fonseka and Gudmestad, 2016).
Several factors may contribute to the low risk of developing difenoconazole resistance in plant pathogens. Unlike other site-specific fungicides such as QoI and benzimidazole in which resistance is a qualitative trait and can be quickly developed in pathogen populations (Mcgrath, 2001; Villani et al., 2015; Blake et al., 2018), resistance to DMI fungicides such as difenoconazole is a quantitative trait resulted from the combined effect of: (i) point mutations in the CYP51 (Cools and Fraaije, 2013; Mair et al., 2016); (ii) overexpression of the CYP51 enzyme (Ma et al., 2006); (iii) overexpression of genes encoding efflux pump proteins (Price et al., 2015); (iv) alteration in sterol biosynthesis pathways (Karaoglanidis et al., 2003); and (v) changes in pathogen cell wall composition and reduced positive influx (Leroux and Walker, 2011). Evolution of resistance to DMIs, such as difenoconazole, involves sequential accumulation of multiple amino acid substitutions in many independent genes of the pathogen genome. Indeed, using a standard experimental protocol involving a repeated series of infection passaging, mutants that were resistant to some other fungicides but not difenoconazole were detected (Hamada et al., 2011). The finding of a continuous and unimodal distribution in difenoconazole tolerance (Figure 2) accords with the multiple mechanisms involved in the development of difenoconazole resistance.
The low risk of developing difenoconazole resistance in A. alternata may also be attributed to the fitness penalty associated with resistance. Previous studies have shown that DMI-resistant mutants exhibited significantly lower fitness compared to their wild-type parents (Karaoglanidis et al., 2001; Reimann and Deising, 2005; Cox et al., 2007; Zhang et al., 2017). In the current study, comparative analysis of spatial distribution in genetic variation indicates the evolution of difenoconazole resistance in A. alternata is under constraining selection as demonstrated by a significantly lower population differentiation in difenoconazole tolerance (QST) than in SSR marker loci (GST).
In the evolution of fungicide resistance, selection for resistant mutants due to their ability to reduce the efficacy of fungicides or selection against resistant mutants due to severe fitness costs can lead to constraining evolution (Qin et al., 2016; He et al., 2018). We hypothesize that the observed constraining evolution is caused by fitness costs of mutations to difenoconazole resistance because difenoconazole tolerance in A. alternata isolates was negatively associated with their aggressiveness though not significantly (data not shown). Constraining selection in the evolution of difenoconazole resistance is also supported by the different patterns of gene flow detected between difenoconazole tolerance measurements and SSR marker loci. Isolation-by-distance was observed in neutral (SSR) loci but not in difenoconazole tolerance (Figure 4), suggesting selection against mutants harmonizes the genetic difference of geographically distant populations accumulated by random drift.
Alternaria alternata can infect a wide range of plants including many wild species (Bashan et al., 1991; Thomma, 2003) which are usually not exposed to synthetic fungicides. Previously, we hypothesized that the wild species may serve as a reservoir of pathotypes with high fungicide sensitivity, and that continuous influx of these sensitive pathotypes from wild species prevents or substantially reduces the risk of developing resistance to the non-specific fungicide mancozeb in agriculture (He et al., 2018). This dilution effect through immigrant populations (Andersson and Hughes, 2011) may also explain the low risk of developing difenoconazole resistance in A. alternata.
Genetic effects (heritability) account for ∼25% of the phenotypic variation in the tolerance of A. alternata to difenoconazole compared to less than 5% attributable to isolate–concentration interactions (plasticity) (Table 3). This result indicates that genetic variation plays a more important role in the evolution of difenoconazole resistance in A. alternata than epigenetic variation. This observation is consistent with the positive correlation of difenoconazole tolerance among different concentrations (Figure 3). Heritability leads to lasting adaptation of organisms to environments through changes in gene composition. Plasticity, on the other hand, is a phenomenon whereby a genotype produces different phenotypes through methylation (Angers et al., 2010; Bossdorf et al., 2010), or changes gene expression in response to environmental fluctuations (Winkler and Breaker, 2005; Chen et al., 2015). Unlike other site-specific fungicides that select for strong pathogen–fungicide interactions (i.e., polarized pathogen genotypes that are either highly sensitive or resistant), DMI, as well as non-specific fungicides, selects for pathogen genotypes differing quantitatively in fungicide tolerance. This group of fungicides thus maximizes the accumulation of genetic effect but reduces the effect of particular genotype–environment (concentration) interactions (i.e., plasticity) (He et al., 2018).
Because of their unique mode of action, DMIs are an excellent alternative chemistry and an effective partner to use together with other fungicides to manage plant diseases (Avenot et al., 2016). Although the risk of developing resistance to DMI fungicides is generally low, these fungicides also face resistance problems and cases associated with increased tolerance resulting in reduced or loss of efficacy have been reported for many pathogens (Thomas et al., 2012). For example, it has been reported that Alternaria species in California pistachio orchards have become less sensitive to DMIs as a result of regular sprays (Brent and Hollomon, 2007). The finding of significant differences in difenoconazole tolerance among isolates and populations (Table 2) and a skewing of the tolerance distribution toward the right (Figure 2) suggests that a stepwise accumulation of tolerance to difenoconazole might be occurring in the early blight pathogen populations in China. In the study, growth inhibition was only tested in three concentrations due to a large number of isolates. This restriction in dose treatments does not allow us to calculate the half maximum effective concentration robustly (Sebaugh, 2011). A rough estimate found that approximate 1% of A. alternata isolates had an EC50 value 10-fold greater than the baseline. These isolates have a high potential of developing resistance to difenoconazole in future. Therefore, dynamic management programs formulated by evolutionary principles (Zhan et al., 2014, 2015) such as spatiotemporal rotations of fungicides used alone or in combination with fungicides with different modes of action modes are critical to generate diversifying selection to prevent or mitigate the evolution of resistance to difenoconazole and other DMI fungicides (Valencia-Botín et al., 2013).
Data Availability
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
MH-H collected the pathogen isolates, generated and analyzed the data, and wrote the manuscript. Y-PW, E-JW, L-LS, L-NY, TW, L-PS, and WZ generated the data and wrote the manuscript. JZ conceived and designed the experiments, analyzed the data, and wrote the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31761143010 and U1405213), the Ministry of Science and Technology (Grant No. 2018YFD0200802), and the China’s Agricultural Research System (Grant No. CARS-10-P20).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Jeremy Burdon to proofread the manuscript.
Footnotes
- ^ https://www.marketresearchandstatistics.com/ad/difenoconazole-global-product-intelligence/
- ^ http://www.agrichem.cn/news/2016/3/8/201638135612812.shtml
References
Aguayo, J., Elegbede, F., Husson, C., Saintonge, F. X., and Marcais, B. (2014). Modeling climate impact on an emerging disease, the Phytophthora alni-induced alder decline. Glob. Change Biol. 20, 3209–3221. doi: 10.1111/gcb.12601
Andersson, D. I., and Hughes, D. (2011). Persistence of antibiotic resistance in bacterial populations. FEMS Micro Biol. Rev. 35, 901–911. doi: 10.1111/j.1574-6976.2011.00289.x
Angelini, R. M. D. M., Pollastro, S., and Faretra, F. (2015). Genetics of Fungicide Resistance. Japan: Springer.
Angers, B., Castonguay, E., and Massicotte, R. (2010). Environmentally induced phenotypes and DNA methylation: how to deal with unpredictable conditions until the next generation and after. Mol. Ecol. 19, 1283–1295. doi: 10.1111/j.1365-294X.2010.04580.x
Arnaud-Haond, S., Duarte, C. M., Alberto, F., and Serrao, E. A. (2007). Standardizing methods to address clonality in population studies. Mol. Ecol. 16, 5115–5139. doi: 10.1111/j.1365-294x.2007.03535.x
Avenot, H. F., Solorio, C., Morgan, D. P., and Michailides, T. J. (2016). Sensitivity and cross-resistance patterns to demethylation-inhibiting fungicides in California populations of Alternaria alternata pathogenic on pistachio. Crop Prot. 88, 72–78. doi: 10.1016/j.cropro.2016.05.012
Bashan, Y., Levanony, H., and Or, R. (1991). Wild beets as an important inoculum source of Alternaria alternata, a cause of leaf blight of cotton in Israel. Can. J. Bot. 69, 2608–2615. doi: 10.1139/b91-325
Benichou, S., Dongo, A., Henni, D. E., Peltier, D., and Simoneau, P. (2009). Isolation and characterization of microsatellite markers from the phytopathogenic fungus Alternaria dauci. Mol. Ecol. Resour. 9, 390–392. doi: 10.1111/j.1755-0998.2008.02237.x
Blake, J. J., Gosling, P., Fraaije, B. A., Burnett, F. J., Knight, S. M., Kildea, S., et al. (2018). Changes in field dose-response curves for demethylation inhibitor (DMI) and quinone outside inhibitor (QoI) fungicides against Zymoseptoria tritici, related to laboratory sensitivity phenotyping and genotyping assays. Pest Manag. Sci. 74, 302–313. doi: 10.1002/ps.4725
Bossdorf, O., Arcuri, D., Richards, C. L., and Pigliucci, M. (2010). Experimental alteration of DNA methylation affects the phenotypic plasticity of ecologically relevant traits in Arabidopsis thaliana. Evol. Ecol. 24, 541–553. doi: 10.1007/s10682-010-9372-7
Bowyer, P., and Denning, D. W. (2014). Environmental fungicides and triazole resistance in Aspergillus. Pest Manag. Sci. 70, 173–178. doi: 10.1002/ps.3567
Brent, K. J., and Hollomon, D. W. (2007). Fungicide Resistance: the Assessment of Risk. London: Fungicide Resistance Action Committee.
Brunner, P. C., Stefansson, T. S., Fountaine, J., Richina, V., and McDonald, B. A. (2016). A global analysis of CYP51 diversity and azole sensitivity in Rhynchosporium commune. Phytopathology 106, 355–361. doi: 10.1094/PHYTO-07-15-0158-R
Chen, J., Nolte, V., and Schlotterer, C. (2015). Temperature-related reaction norms of gene expression: regulatory architecture and functional implications. Mol. Biol. Evol. 32, 2393–2402. doi: 10.1093/molbev/msv120
Chen, Y., Zhang, Y., Yao, J., Li, Y. F., Yang, X., Wang, W. X., et al. (2013). Frequency distribution of sensitivity of Ustilaginoidea virens to four EBI fungicides, prochloraz, difenoconazole, propiconazole and tebuconazole, and their efficacy in controlling rice false smut in Anhui Province of China. Phytoparasitica 41, 277–284. doi: 10.1007/s12600-013-0288-y
Chourasiya, P. K., Lal, A. A., and Simon, S. (2013). Effect of certain fungicides and botanicals against early blight of tomato caused by Alternaria solani(Ellis and Martin) under Allahabad Uttar Pradesh, India conditions. Int. J. Agric. Sci. Res. 3, 151–155.
Cools, H. J., and Fraaije, B. A. (2013). Update on mechanisms of azole resistance in Mycosphaerella graminicola and implications for future control. Pest Manag. Sci. 69, 150–155. doi: 10.1002/ps.3348
Cooper, J., and Dobson, H. (2007). The benefits of pesticides to mankind and the environment. Crop Prot. 26, 1337–1348. doi: 10.1016/j.cropro.2007.03.022
Cox, K. D., Bryson, P. K., and Schnabel, G. (2007). Instability of propiconazole resistance and fitness in Monilinia fructicola. Phytopathology 97, 448–453. doi: 10.1094/PHYTO-97-4-0448
Damicone, J. P., and Smith, D. L. (2009). Fungicide Resistance Management. Division of Agricultural Sciences and Natural Resources. Oklahoma: Oklahoma State University.
de Waard, M. A., Andrade, A. C., Hayashi, K., Schoonbeek, H. J., Stergiopoulos, I., and Zwiers, L. H. (2006). Impact of fungal drug transporters on fungicide sensitivity, multidrug resistance and virulence. Pest Manag. Sci. 62, 195–207. doi: 10.1002/ps.1150
Delmas, C. E., Dussert, Y., Deliere, L., Couture, C., Mazet, I. D., Richart Cervera, S., et al. (2017). Soft selective sweeps in fungicide resistance evolution: recurrent mutations without fitness costs in grapevine downy mildew. Mol. Ecol. 26, 1936–1951. doi: 10.1111/mec.14006
Dong, F., Li, J., Chankvetadze, B., Cheng, Y., Xu, J., Liu, X., et al. (2013). Chiral triazole fungicide difenoconazole: absolute stereochemistry, stereoselective bioactivity, aquatic toxicity, and environmental behavior in vegetables and soil. Environ. Sci. Technol. 47, 3386–3394. doi: 10.1021/es304982m
Elansky, S. N., Mita, E. D., Skolotneva, E. S., Pobedinskaya, M. A., and Kokaeva, L. Yu (2016). Effect of difenoconazole on the formation of oospores by Phytophthora infestans (Mont) de Bary. J. Plant Pathol. 98, 123–127.
Elena, K. (2006). Alternaria brown spot of Minneola in Greece; evaluation of citrus species susceptibility. Eur. J. Plant Pathol. 115, 259–262. doi: 10.1007/s10658-006-9005-8
Fonseka, D. L., and Gudmestad, N. C. (2016). Spatial and temporal sensitivity of Alternaria species associated with potato foliar diseases to demethylation inhibiting and anilino-pyrimidine fungicides. Plant Dis. 100, 1848–1857. doi: 10.1094/PDIS-01-16-0116-RE
Ge, J., Cui, K., Yan, H., Li, Y., Chai, Y., Liu, X., et al. (2017). Uptake and translocation of imidacloprid, thiamethoxam and difenoconazole in rice plants. Environ. Pollut. 226, 479–485. doi: 10.1016/j.envpol.2017.04.043
Gent, D. H., and Schwartz, H. F. (2003). Validation of potato early blight disease forecast models for Colorado using various sources of meteorological data. Plant Dis. 87, 78–84. doi: 10.1094/PDIS.2003.87.1.78
Gisi, U., Sierotzki, H., Cook, A., and McCaffery, A. (2002). Mechanisms influencing the evolution of resistance to Qo inhibitor fungicides. Pest Manag. Sci. 58, 859–867. doi: 10.1002/ps.565
Gopinath, K., Radhakrishnan, N. V., and Jayaraj, J. (2006). Effect of propiconazole and difenoconazole on the control of anthracnose of chilli fruits caused by Colletotrichum capsici. Crop Prot. 25, 1024–1031. doi: 10.1016/j.cropro.2006.02.001
Gveroska, B. (2013). The effectiveness of fungicides in the control of Alternaria alternata depending on their impact on pathogen biology. Tobacco 63, 36–46.
Hahn, M. (2014). The rising threat of fungicide resistance in plant pathogenic fungi: botrytis as a case study. J. Biol. Chem. 7, 133–141. doi: 10.1007/s12154-014-0113-1
Hamada, M. S., Yin, Y., and Ma, Z. (2011). Sensitivity to iprodione, difenoconazole and fludioxonil of Rhizoctonia cerealis isolates collected from wheat in China. Crop Prot. 30, 1028–1033. doi: 10.1016/j.cropro.2011.04.004
He, M. H., Li, D. L., Zhu, W., Wu, E. J., Yang, L. N., Wang, Y. P., et al. (2018). Slow and temperature-mediated pathogen adaptation to a nonspecific fungicide in agricultural ecosystem. Evol. Appl. 11, 182–192. doi: 10.1111/eva.12526
Horsfield, A., Wicks, T., Davies, K., Wilson, D., and Paton, S. (2010). Effect of fungicide use strategies on the control of early blight (Alternaria solani) and potato yield. Aust. Plant Pathol. 39, 368–375.
Jones, S., Pethybridge, S., Hay, F., Groom, T., and Wilson, C. (2007). Baseline sensitivity of australian phoma ligulicola isolates from pyrethrum to azoxystrobin and difenoconazole. J. Phytopathol. 155, 377–380. doi: 10.1111/j.1439-0434.2007.01241.x
Jørgensen, P., Aktipis, A., Brown, Z., Downes, S., Dunn, R., Carrière, Y., et al. (2018). Antibiotic and pesticide susceptibility and the Anthropocene operating space. Nat. Sustain. 1, 632–641. doi: 10.1038/s41893-018-0164-3
Karaoglanidis, G. S., Menkissoglu-Spiroudi, U., and Thanassoulopoulos, C. C. (2003). Sterol composition of DMI-resistant and -sensitive field isolates of Cercospora beticola. J. Phytopathol. 151, 431–435. doi: 10.1046/j.1439-0434.2003.00746.x
Karaoglanidis, G. S., Thanassoulopoulos, C. C., and Ioannidis, P. M. (2001). Fitness of cercospora beticola field isolates - resistant and - sensitive to demethylation inhibitor fungicides. Eur. J. Plant Pathol. 107, 337–347.
Kokalisburelle, N., Butler, D. M., and Rosskopf, E. N. (2013). Evaluation of cover crops with potential for use in anaerobic soil disinfestation (ASD) for susceptibility to three species of Meloidogyne. J. Nematol. 45, 272–278.
Lamari, L. (2002). Assess: Image Analysis Software Forplant Disease Quantification. Saint Paul, MN: APS Press.
Lawrence, I., and Lin, K. (1989). A concordance correlation coefficient to evaluate reproducibility. Biometrics 45, 255–268.
Leiminger, J. H., and Hausladen, H. (2012). Early blight control in potato using disease-orientated threshold values. Plant Dis. 96, 124–130. doi: 10.1094/PDIS-05-11-0431
Leroux, P., and Walker, A. S. (2011). Multiple mechanisms account for resistance to sterol 14alpha-demethylation inhibitors in field isolates of Mycosphaerella graminicola. Pest Manag. Sci. 67, 44–59. doi: 10.1002/ps.2028
Liang, W., Tai, L., Jin, X., Zuo, Y., Wang, H., and Sun, M. (2009). Laboratory screening and proportioning tests on fungicides against the potato early blight caused by Alternaria solani. Plant Prot. 35, 168–171.
Ma, Z., Proffer, T. J., Jacobs, J. L., and Sundin, G. W. (2006). Overexpression of the 14alpha-demethylase target gene (CYP51) mediates fungicide resistance in Blumeriella jaapii. Appl. Environ. Microbiol. 72, 2581–2585. doi: 10.1128/aem.72.4.2581-2585.2006
MacDonald, W., Peters, R. D., Coffin, R. H., and Lacroix, C. (2007). Effect of strobilurin fungicides on control of early blight (Alternaria solani) and yield of potatoes grown under two N fertility regimes. Phytoprotection 88, 9–15.
Mair, W. J., Deng, W., Mullins, J. G., West, S., Wang, P., Besharat, N., et al. (2016). Demethylase inhibitor fungicide resistance in Pyrenophora teres f. sp. teres associated with target site modification and inducible overexpression of Cyp51. Front. Microbiol. 7:1279. doi: 10.3389/fmicb.2016.01279
Mcgrath, M. T. (2001). Fungicide resistance in cucurbit powdery mildew: experiences and challenges. Plant Dis. 85, 236–245. doi: 10.1094/pdis.2001.85.3.236
Meng, J. W., Zhu, W., He, M. H., Wu, E. J., Duan, G. H., Xie, Y. K., et al. (2015a). Population genetic analysis reveals cryptic sex in the phytopathogenic fungus Alternaria alternata. Sci. Rep. 5:18250. doi: 10.1038/srep18250
Meng, J. W., Zhu, W., He, M. H., Wu, E. J., Yang, L. N., Shang, L. P., et al. (2015b). High genotype diversity and lack of isolation by distance in the Alternaria solani populations from China. Plant Pathol. 64, 434–441. doi: 10.1111/ppa.12275
Mirkovic, B., Tanovic, B., Stevic, M., Hrustic, J., Mihajlovic, M., Delibasic, G., et al. (2015). Toxicity of mancozeb, chlorothalonil, captan, fluopyram, boscalid, and difenoconazole to Didymella applanata isolates from Serbia. J. Environ. Sci. Health Part B 50, 845–850. doi: 10.1080/03601234.2015.1062648
Moloney, K. A., Holzapfel, C., Tielbörger, K., Jeltsch, F., and Schurr, F. M. (2009). Rethinking the common garden in invasion research. Perspect. Plant Ecol. Evol. Syst. 11, 311–320. doi: 10.1016/j.ppees.2009.05.002
Muneret, L., Mitchell, M., Seufert, V., Aviron, S., Djoudi, E. Z., Pétillon, J., et al. (2018). Evidence that organic farming promotes pest control. Nat. Sustain. 1, 361–368. doi: 10.1038/s41893-018-0102-4
Nei, M. (1973). Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. U.S.A. 70, 3321–3323. doi: 10.1073/pnas.70.12.3321
Ott, R. L., and Longnecker, M. T. (2008). An Introduction to Statistical Methods and Data Analysis. Boston, MA: Cengage Learning.
Parvez, E. (2003). Evaluation of different protectant and eradicant fungicides against early and late blight of potato caused by Alternaria solani(Ellis and Mart) Jones and Grout and Phytophthora infestans (Mont.) de Bary under field conditions. Pak. J. Biol. Sci. 21, 347–359.
Pereira, D. A., McDonald, B. A., and Brunner, P. C. (2017). Mutations in the CYP51 gene reduce DMI sensitivity in Parastagonospora nodorum populations in Europe and China. Pest Manag. Sci. 73, 1503–1510. doi: 10.1002/ps.4486
Pfeufer, E. E., and Ngugi, H. K. (2012). Orchard factors associated with resistance and cross resistance to sterol demethylation inhibitor fungicides in populations of Venturia inaequalis from Pennsylvania. Phytopathology 102, 272–282. doi: 10.1094/PHYTO-04-11-0117
Price, C. L., Parker, J. E., Warrilow, A. G., Kelly, D. E., and Kelly, S. L. (2015). Azole fungicides - understanding resistance mechanisms in agricultural fungal pathogens. Pest Manag. Sci. 71, 1054–1058. doi: 10.1002/ps.4029
Qin, C. F., He, M. H., Chen, F. P., Zhu, W., Yang, L. N., and Wu, E. J. (2016). Comparative analyses of fungicide sensitivity and SSR marker variations indicate a low risk of developing azoxystrobin resistance in Phytophthora infestans. Sci. Rep. 6:20483. doi: 10.1038/srep20483
Reimann, S., and Deising, H. B. (2005). Inhibition of efflux transporter-mediated fungicide resistance in Pyrenophora tritici-repentis by a derivative of 4’-hydroxyflavone and enhancement of fungicide activity. Appl. Environ. Microbiol. 71, 3269–3275. doi: 10.1128/aem.71.6.3269-3275.2005
Reis, R. F., de Goes, A., Mondal, S. N., Shilts, T., Brentu, F. C., and Timmer, L. W. (2006). Effect of lesion age, humidity, and fungicide application on sporulation of Alternaria alternata, the cause of brown spot of tangerine. Plant Dis. 90, 1051–1054. doi: 10.1094/PD-90-1051
Reuveni, M., and Sheglov, D. (2002). Effects of azoxystrobin, difenoconazole, polyoxin B (polar) and trifloxystrobin on germination and growth of Alternaria alternata and decay in red delicious apple fruit. Crop Prot. 21, 951–955. doi: 10.1016/s0261-2194(02)00073-x
Rupp, S., Weber, R. W., Rieger, D., Detzel, P., and Hahn, M. (2016). Spread of Botrytis cinerea strains with multiple fungicide resistance in German horticulture. Front. Microbiol. 7:2075. doi: 10.3389/fmicb.2016.02075
Savary, S., Ficke, A., Aubertot, J. N., and Hollier, C. (2012). Crop losses due to diseases and their implications for global food production losses and food security. Food Secur. 4, 519–537. doi: 10.1007/s00203-017-1426-6
Scruggs, A. C., Basaiah, T., Adams, M. L., and Quesada-Ocampo, L. M. (2017). Genetic diversity, fungicide sensitivity, and host resistance to Ceratocystis fimbriata infecting sweetpotato in North Carolina. Plant Dis. 101, 994–1001. doi: 10.1094/PDIS-11-16-1583-RE
Sebaugh, J. L. (2011). Guidelines for accurate EC50/IC50 estimation. Pharm. Stat. 10, 128–134. doi: 10.1002/pst.426
Sierotzki, H., Wullschleger, J., and Gisi, U. (2000). Point mutation in cytochrome b gene conferring resistance to strobilurin fungicides in Erysiphe graminis f. sp. Tritici field isolates. Pestic. Biochem. Physiol. 68, 107–112. doi: 10.1006/pest.2000.2506
Stevic′, M., Vukša, P., and Elezovic′, I. (2010). Resistance of Venturia inaequalis to demethylation inhibiting (DMI) fungicides. Z?emdirbyste? 97, 65–72.
Strange, R. N., and Scott, P. R. (2005). Plant disease: a threat to global food security. Annu. Rev. Phytopathol. 43, 83–116. doi: 10.1146/annurev.phyto.43.113004.133839
Thomas, A., Langston, D. B., and Stevenson, K. L. (2012). Baseline sensitivity and cross-resistance to succinate-dehydrogenase-inhibiting and demethylation-inhibiting fungicides in Didymella bryoniae. Plant Dis. 96, 979–984. doi: 10.1094/PDIS-09-11-0744-RE
Thomma, B. P. (2003). Alternaria spp.: from general saprophyte to specific parasite. Mol. Plant Pathol. 4, 225–236. doi: 10.1046/j.1364-3703.2003.00173.x
Tonsor, S. J., Elnaccash, T. W., and Scheiner, S. M. (2013). Developmental instability is genetically correlated with phenotypic plasticity, constraining heritability, and fitness. Evolution 67, 2923–2935. doi: 10.1111/evo.12175
Valencia-Botín, A. J., Jeffers, S. N., Palmer, C. L., and Buck, J. W. (2013). Fungicides used alone, in combinations, and in rotations for managing gladiolus rust in Mexico. Plant Dis. 97, 1491–1496. doi: 10.1094/PDIS-03-13-0272-RE
Villa, F., Cappitelli, F., Cortesi, P., and Kunova, A. (2017). Fungal biofilms: targets for the development of novel strategies in plant disease management. Front. Microbiol. 8:654. doi: 10.3389/fmicb.2017.00654
Villani, S. M., Biggs, A. R., Cooley, D. R., Raes, J. J., and Cox, K. D. (2015). Prevalence of myclobutanil resistance and difenoconazole insensitivity in populations of Venturia inaequalis. Plant Dis. 99, 1526–1536. doi: 10.1094/PDIS-01-15-0002-RE
Wang, H., Huang, Y., Wang, J., Chen, X., Wei, K., Wang, M., et al. (2016). Activities of azoxystrobin and difenoconazole against Alternaria alternata and their control efficacy. Crop Prot. 90, 54–58. doi: 10.1016/j.cropro.2016.08.022
Wier, T. L., Huff, D. R., Christ, B. J., and Romaine, C. P. (1998). RAPD-PCR analysis of genetic variation among isolates of Alternaria solani and Alternaria alternata from potato and tomato. Mycologia 90, 813–821. doi: 10.1080/00275514.1998.12026975
Winkler, W. C., and Breaker, R. R. (2005). Regulation of bacterial gene expression by riboswitches. Microbiology 59, 487–517. doi: 10.1146/annurev.micro.59.030804.121336
Woudenberg, J. H., Groenewald, J. Z., Binder, M., and Crous, P. W. (2013). Alternaria redefined. Stud. Mycol. 75, 171–212. doi: 10.3114/sim0015
Woudenberg, J. H., Seidl, M. F., Groenewald, J. Z., de Vries, M., Stielow, J. B., Thomma, B. P., et al. (2015). Alternaria section Alternaria: species, formae speciales or pathotypes? Stud. Mycol. 82, 1–21. doi: 10.1016/j.simyco.2015.07.001
Wu, E. J., Wang, Y. P., Shen, L. L., Yahuza, L., Tian, J. C., Yang, L. N., et al. (2019). Strategies of Phytophthora infestans adaptation to local UV radiation conditions. Evol. Appl. 12, 415–424. doi: 10.1111/eva.12722
Wyand, R. A., and Brown, J. K. (2005). Sequence variation in the CYP51 gene of Blumeria graminis associated with resistance to sterol demethylase inhibiting fungicides. Fungal Genet. Biol. 42, 726–735. doi: 10.1016/j.fgb.2005.04.007
Yang, L. N., Zhu, W., Wu, E. J., Yang, C., Thrall, P. H., Burdon, J. J., et al. (2016). Trade-offs and evolution of thermal adaptation in the Irish potato famine pathogen Phytophthora infestans. Mol. Ecol. 25, 4047–4058. doi: 10.1111/mec.13727
Yeh, F. C., Yang, R., Boyle, T. J., Ye, Z., and Xiyan, J. M. (2000). PopGene32, Microsoft Windows-Based Freeware for Population Genetic Analysis, version 1.32. Molecular Biology and Biotechnology Centre. Edmonton, AB: University of Alberta.
Zarn, J. A., Brüschweiler, B. J., and Schlatter, J. R. (2003). Azole fungicides affect mammalian steroidogenesis by inhibiting sterol 14 alpha-demethylase and aromatase. Environ. Health Perspect. 111, 255–261. doi: 10.1289/ehp.5785
Zhan, J., Linde, C. C., Jurgens, T., Merz, U., Steinebrunner, F., and McDonald, B. A. (2005). Variation for neutral markers is correlated with variation for quantitative traits in the plant pathogenic fungus Mycosphaerella graminicola. Mol. Ecol. 14, 2683–2693. doi: 10.1111/j.1365-294x.2005.02638.x
Zhan, J., and McDonald, B. A. (2011). Thermal adaptation in the fungal pathogen Mycosphaerella graminicola. Mol. Ecol. 20, 1689–1701. doi: 10.1111/j.1365-294X.2011.05023.x
Zhan, J., Stefanato, F. L., and McDonald, B. A. (2006). Selection for increased cyproconazole tolerance in Mycosphaerella graminicola through local adaptation and in response to host resistance. Mol. Plant Pathol. 7, 259–268. doi: 10.1111/j.1364-3703.2006.00336.x
Zhan, J., Thrall, P. H., and Burdon, J. J. (2014). Achieving sustainable plant disease management through evolutionary principles. Trends Plant Sci. 19, 570–575. doi: 10.1016/j.tplants.2014.04.010
Zhan, J., Thrall, P. H., Papaix, J., Xie, L., and Burdon, J. J. (2015). Playing on a pathogen’s weakness: using evolution to guide sustainable plant disease control strategies. Annu. Rev. Phytopathol. 53, 19–43. doi: 10.1146/annurev-phyto-080614-120040
Zhang, C., Diao, Y., Wang, W., Hao, J., Imran, M., Duan, H., et al. (2017). Assessing the risk for resistance and elucidating the genetics of Colletotrichum truncatum that is only sensitive to some DMI fungicides. Front. Microbiol. 8:1779. doi: 10.3389/fmicb.2017.01779
Zheng, H. H., Wang, T. Y., Zhao, J., and Wu, X. H. (2013). Research progress on potato early blight and its integrated control. Chin. Plant Prot. 33, 18–22.
Keywords: Alternaria alternata, population genetics, difenoconazole tolerance, fitness penalties, genetic variation
Citation: He M-H, Wang Y-P, Wu E-J, Shen L-L, Yang L-N, Wang T, Shang L-P, Zhu W and Zhan J (2019) Constraining Evolution of Alternaria alternata Resistance to a Demethylation Inhibitor (DMI) Fungicide Difenoconazole. Front. Microbiol. 10:1609. doi: 10.3389/fmicb.2019.01609
Received: 25 March 2019; Accepted: 27 June 2019;
Published: 10 July 2019.
Edited by:
Jack Wong, The Chinese University of Hong Kong, ChinaReviewed by:
Mingqiang Wang, The University of Texas Health Science Center at Houston, United StatesSlawomir Milewski, Gdańsk University of Technology, Poland
Copyright © 2019 He, Wang, Wu, Shen, Yang, Wang, Shang, Zhu and Zhan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiasui Zhan, amlhc3VpLnpoYW5AZmFmdS5lZHUuY24=
 Meng-Han He1,2
Meng-Han He1,2 Jiasui Zhan
Jiasui Zhan