- 1School of Life Science and Technology, Tongji University, Shanghai, China
- 2CAS Key Laboratory of Insect Developmental and Evolutionary Biology, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China
- 3University of Chinese Academy of Sciences, Beijing, China
- 4Laboratory of Malaria and Vector Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Rockville, MD, United States
The malaria development in the mosquito midgut is a complex process that results in considerable parasite losses. The mosquito gut microbiota influences the outcome of pathogen infection in mosquitoes, but the underlying mechanisms through which gut symbiotic bacteria affect vector competence remain elusive. Here, we identified two Serratia strains (Y1 and J1) isolated from field-caught female Anopheles sinensis from China and assessed their effect on Plasmodium development in An. stephensi. Colonization of An. stephensi midgut by Serratia Y1 significantly renders the mosquito resistant to Plasmodium berghei infection, while Serratia J1 has no impact on parasite development. Parasite inhibition by Serratia Y1 is induced by the activation of the mosquito immune system. Genome-wide transcriptomic analysis by RNA-seq shows a similar pattern of midgut gene expression in response to Serratia Y1 and J1 in sugar-fed mosquitoes. However, 24 h after blood ingestion, Serratia Y1 modulates more midgut genes than Serratia J1 including the c-type lectins (CTLs), CLIP serine proteases and other immune effectors. Furthermore, silencing of several Serratia Y1-induced anti-Plasmodium factors like the thioester-containing protein 1 (TEP1), fibrinogen immunolectin 9 (FBN9) or leucine-rich repeat protein LRRD7 can rescue parasite oocyst development in the presence of Serratia Y1, suggesting that these factors modulate the Serratia Y1-mediated anti-Plasmodium effect. This study enhances our understanding of how gut bacteria influence mosquito-Plasmodium interactions.
Introduction
Malaria continues to be one of the most devastating infectious diseases and a major public health problem in tropical and subtropical regions. The increase in mosquito insecticide resistance and parasite drug resistance, combined with the lack of an effective vaccine, have stalled the steady reduction in global malaria cases (World Health Organization, 2018), making the development of new malaria interventions a global priority.
Malaria is caused by Plasmodium parasites and is transmitted to humans by Anopheles mosquitoes. The most severe bottleneck during Plasmodium development occurs in the mosquito midgut, where the majority of ingested parasites are killed (Vaughan et al., 1992; Ghosh et al., 2000; Pradel, 2007; Simon et al., 2009). The malaria parasite encounters a hostile environment in the mosquito midgut and undergoes a chain of complex developmental transitions that are required for successful transmission (Smith et al., 2014). Several reports have shown that the mosquito midgut microbiota affects mosquito susceptibility to parasite infection (Pumpuni et al., 1993, 1996; Dong et al., 2009; Cirimotich et al., 2011; Boissiere et al., 2012; Gendrin and Christophides, 2013; Bahia et al., 2014; Smith et al., 2014; Stathopoulos et al., 2014). Diverse species of bacteria colonize the midgut of both laboratory-reared and field caught mosquitoes, and some can inhibit Plasmodium development (Pumpuni et al., 1996; Straif et al., 1998; Gonzalez-Ceron et al., 2003). Likewise, elimination of the gut bacteria with antibiotics renders the mosquito more susceptible to Plasmodium infection, which can be reverted by reintroduction of bacteria in the midgut (Dong et al., 2009; Gendrin et al., 2015). However, the mechanisms by which the specific gut bacteria negatively impact malaria parasite development in the mosquito midgut are not completely understood.
The number of bacteria in the mosquito midgut increases exponentially within 24 h of a blood meal ingestion (Pumpuni et al., 1996; Wang et al., 2012), resulting in induction of the midgut immune responses (Dong et al., 2009; Meister et al., 2009). This immune activation can also modulate the mosquito defense against the malaria parasite (Meister et al., 2005, 2009; Bahia et al., 2014). RNA transcription profiling of microbe-free aseptic and septic mosquitoes identified many genes up-regulated by gut bacteria, including several anti-Plasmodium factors (Dong et al., 2009). Although, there is overlap between the mosquito antibacterial and anti-malarial immune responses, some antibacterial immune genes have no impact on Plasmodium development (Dimopoulos et al., 1997; Oduol et al., 2000; Dong et al., 2006, 2009). In addition, the effect of the gut microbiota on Plasmodium infection may be exerted through direct interactions with bacteria-produced anti-Plasmodium factors or by the formation of a physical barrier that blocks the parasite’s access to the midgut epithelium (Cirimotich et al., 2011; Bando et al., 2013; Bahia et al., 2014; Song et al., 2018). Conversely, a positive correlation was reported between the presence of Enterobacteriaceae bacteria in the midgut of field-caught mosquitoes and the Plasmodium falciparum infection status (Boissiere et al., 2013). These indicate that the effect of mosquito gut bacteria on parasite infection is complex and may depend on species-specific or strain-specific interactions.
In this study, we examined the influence of two Serratia strains isolated from field-caught Anopheles sinensis, the main malaria vector in Asia, on Plasmodium infection. Serratia strain Y1 inhibited Plasmodium berghei infection in An. stephensi mosquitoes, whereas Serratia strain J1 had no impact on parasite infection. Gene expression and RNA interference analysis show that the inhibition of Plasmodium by Serratia Y1 is mediated by the bacterial activation of the mosquito immune system. Understanding the interactions and mechanisms through which gut commensal bacteria actively shape mosquito immunity may yield new insights into vector-pathogen interactions and may help in the development of new vector-based disease interventions.
Materials and Methods
Ethics Statement
This study was carried out in strict accordance with the guidelines of the Shanghai Institutes for Biological Sciences Animal Care and Use Committee, and all animal work was approved by the committee.
Mosquito Rearing, Oral Bacterial Introduction and Infection with Plasmodium berghei
Anopheles stephensi (Nijmegen strain) and An. sinensis (Jiangsu strain) mosquitoes were maintained at 27°C with 70 ± 5% relative humidity under 12 h/12 h day-night cycle. Adult mosquitoes were maintained on 10% (w/v) sucrose. The larvae were reared on cat food pellets and ground fish food supplement. Axenic female mosquitoes were generated via treatment with oral antibiotics as previously described (Wei et al., 2017). Female mosquitoes were reared with 10% sucrose solution containing penicillin (10 unit/ml), streptomycin (10 μg/ml) and gentamicin (15 μg/ml). Three days after antibiotic treatment, the antibiotic solution was replaced by sterile water, and mosquitoes were starved for 8 h and fed for 24–48 h on a cotton pad soaked with a 5% sucrose solution containing Serratia bacteria at a final concentration of 107 CFU/ml. The control group was only given 5% sucrose solution without any bacteria. The mosquitoes were allowed to feed on wild-type P. berghei-mCherry (Graewe et al., 2009) infected mouse. On each experiment, control and experimental groups were fed on the same infected mouse. Fully engorged mosquitoes were separated within 24 h and provided with a cotton pad soaked with 5% (w/v) sterile sucrose solution that was replaced twice a day. Oocyst numbers in midguts were determined on day 10 after infection, stained with 0.1% (w/v) mercurochrome.
Direct PCR Amplification of the 16S rRNA Gene was Performed from the Isolated Bacteria
16S rRNA Gene Cloning and Sequencing
A fragment of approximately 1.5 kb of the 16S rRNA gene was PCR amplified from isolated bacteria from the mosquito midgut using the following primers: 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′). PCR mixtures used to amplify bacterial sequences contained about 50 ng DNA; 0.4 mM each dNTP; 2.5 U Taq DNA polymerase; 3 mM MgCl2 (50 mM magnesium chloride); 1X PCR buffer (200 mM Tris, pH 8.4, 500 mM KCl); 25 pmol of each primer and sterile water to a final volume of 50 μl. PCR amplification was carried out with the following conditions: 95°C for 3 min; 35 cycles at 95°C for 15 s, 58°C for 30 s and 72°C for 2 min, and a final extension at 72°C for 5 min. Reactions were carried out in an ProFlex PCR System (Applied Biosystems). Amplification products were analyzed by electrophoresis on a 1% agarose gel and visualized under UV light after staining with ethidium bromide. PCR products were purified using the Wizard® SV Gel and PCR Clean-Up System (Promega, Madison, WI, USA) according to the manufacturer recommendations. Purified PCR products were cloned into a pGEM-T easy vector according to manufacturer instructions and transformed into competent DH5α Escherichia coli cells by heat-shock for 90 s. Recombinant colonies were identified using blue and white screening on LB agar medium containing 100 μg/ml ampicillin, 2 mM IPTG, and 0.004% X-gal. The plates were incubated at 37°C overnight. The white colonies were selected and cultured in Luria-Bertani (LB) broth complemented with 100 μg/ml ampicillin. Plasmid extraction were done using an alkaline lysis method. The 16S rRNA gene was sequenced by Sangon Biotech (Shanghai, China) Co., Ltd. The sequences obtained were compared with GenBank database for bacterial species identification.
Generation of Fluorescent Strains of Serratia Y1 and J1
To integrate an enhanced fluorescent protein gene (egfp) into the chromosome of Serratia Y1 and J1, the transposon plasmid pBAM2-GFP (Wang et al., 2017) was transformed into a donor strain E. coli S17-1λpir. Freshly cultured transformed donor strain and the recipient strains (Serratia Y1 and J1) cells were washed and resuspended with 10 mM MgSO4 solution to a final OD600 of 0.1 of each strain, mixed (1:1) and co-cultured on Luria-Bertani (LB) agar plates at 37°C for conjugation mating. After 5 h incubation, the fluorescence gene was mobilized and integrated into the genome of the recipient strain Serratia Y1 and J1. The co-culture was seriously diluted and plated on LB agar plates containing 100 μg/ml of kanamycin. The plates were incubated overnight at 30°C and fluorescent colonies were identified under fluorescent microscopy.
Colonization of Serratia Y1 and J1 in the Mosquito Midgut
To test colonization of Serratia Y1 and J1 in adult An. sinensis and An. Stephensi mosquitoes, GFP-labeled Serratia strains (Y1-GFP and J1-GFP) were used in this assay. The bacteria were cultured overnight in LB broth medium at 30°C, washed twice in sterile 1 × PBS and resuspended in 5% sterile sucrose solution to obtain 1 × 107 cells/ml. The bacteria were introduced into 2 day-old female mosquitoes by feeding on a cotton pad moistened with bacterial suspension for 24–48 h. Two days later, the mosquitoes were allowed to feed on a non-infected mouse. Before and post-blood meal, 10 mosquitoes were collected at different time points, and were surface-sterilized by washing in 75% ethanol for 3 min and then rinsing them in sterile PBS three times. The midguts were dissected under sterile conditions and homogenized in 100 μl sterile PBS. The bacterial load was determined by plating 10-fold serial dilutions of the homogenates on LB agar plates containing 100 μg/ml of kanamycin and incubating the plates overnight at 30°C. The fluorescent colonies were counted by fluorescent microscopy.
RNA Extraction and Transcriptome Sequencing
Serratia Y1 and J1 were separately fed to 3-day-old aseptic mosquitoes in a sugar meal for 24–48 h, then mosquitoes were allowed to feed on non-infected blood. About 50 female mosquitoes were collected from two biological replicates before (0 h) and 24 h after blood meal. The aseptic females that did not feed on Y1 and J1bacteria were used as the controls. Mosquito midguts were dissected in ice-cold PBS and total RNA was extracted using Direct-zol RNA Miniprep Kit (The Epigenetics Company, USA) followed by RNase-free DNase I treatment. Messenger RNA (mRNA) was purified, and reverse-transcribed into cDNA libraries using the NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, Boston, Massachusetts, USA). The cDNA libraries were sequenced on an Illumina HiSeq 2000 platform.
Assembly and Annotation of Transcriptomes
Before assembling the clean reads, the raw reads were preprocessed using filter-fq software. For the transcriptome analysis, the clean reads were aligned in paired-end mode against the Anopheles stephensi SDA-500 genome1 using HISAT v0.1.6-beta and assembled with Cufflinks with the default settings2. The gene annotation was performed by FunCat3. Secreted proteins were predicted by SignalP 3.04.
Differential Expression Genes, Clustering and Functional Enrichment Analysis
Differential expression genes (DEGs) were identified for each time point, which were compared to the data of the control group using Cuffdiff. Differential expression was detected using log2 (fold change) ≥ 1.0 and adjusted p < 0.05. The combined transcriptomes were used as the background to search for GO terms enriched within the DEGs using http://bioinfo.cau.edu.cn/agriGO/ and a p < 0.01 as the parameters for determining significantly enriched terms. Similarly, pathways significantly enriched with the DEGs were identified by mapping all DEGs to terms in the KEGG database using KOBAS2.0 with a p < 0.05.
Validation of DEGs by qRT-PCR Analysis
First-strand cDNA was synthesized from total RNA using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara) according to the manufacturer’s instructions. Quantitative real-time PCR (qRT-PCR) analysis was performed with the PikoReal 96 (Thermo, USA) using the AceQ qPCR SYBR Green Master Mix (Vazyme). The housekeeping As S7 gene was used as an endogenous control. The primers are shown in Supplementary Table S2.
Double-Stranded RNA Synthesis and Gene Silencing in Adult Mosquitoes
Forward and reverse primers with T7 promoter sequence (5′-TAATACGACTCACTATAGGG-3′) were used to amplify the fragment of the enhanced green fluorescent protein (eGFP) gene from plasmid pBacRMCE-Ac5-EGFP (Supplementary Table S3). Fragments of the Rel1, Rel2, TEP1, FBN9, and LRRD7 genes were amplified from An. stephensi cDNA. Purified PCR products were used as template for in vitro double-strand RNA (dsRNA) transcription using the MEGAscript RNAi Kit (Thermo Fisher Scientific). Pure dsRNAs were diluted to approximately 3 μg/μl in DEPC-water. Two-day-old female mosquitoes were intrathoracically injected with 138 nl of either dsRel1, dsRel2, dsTEP1, dsFBN9, dsLRRD7, or dsGFP as a control. The silencing efficiency was evaluated by qPCR at selected time points after injection.
Statistical Analysis
Significant difference in oocyst intensity between two samples was analyzed using the Mann-Whitney test. The statistics were performed using GraphPad Prism version 5.00 for Windows (GraphPad Software). p < 0.05 was considered to be statistically significant.
Results
Effect of Serratia Strains on Plasmodium Berghei Infection in the Mosquito
We isolated two dominant Serratia strains, termed Y1 and J1, from the midguts of field-collected adult female An. sinensis mosquitoes in China. The 16S rRNA gene sequence showed 99% similarity to Serratia marcescens. The two Serratia strains, Y1 and J1, could stably colonize the midgut of both An. sinensis and An. stephensi mosquitoes, and rapidly proliferate by more than 200-fold 24 h after a blood meal (Figure 1A). These two strains did not significantly affect mosquito survival whether the mosquitoes fed on sugar or blood (Supplementary Figure S1).
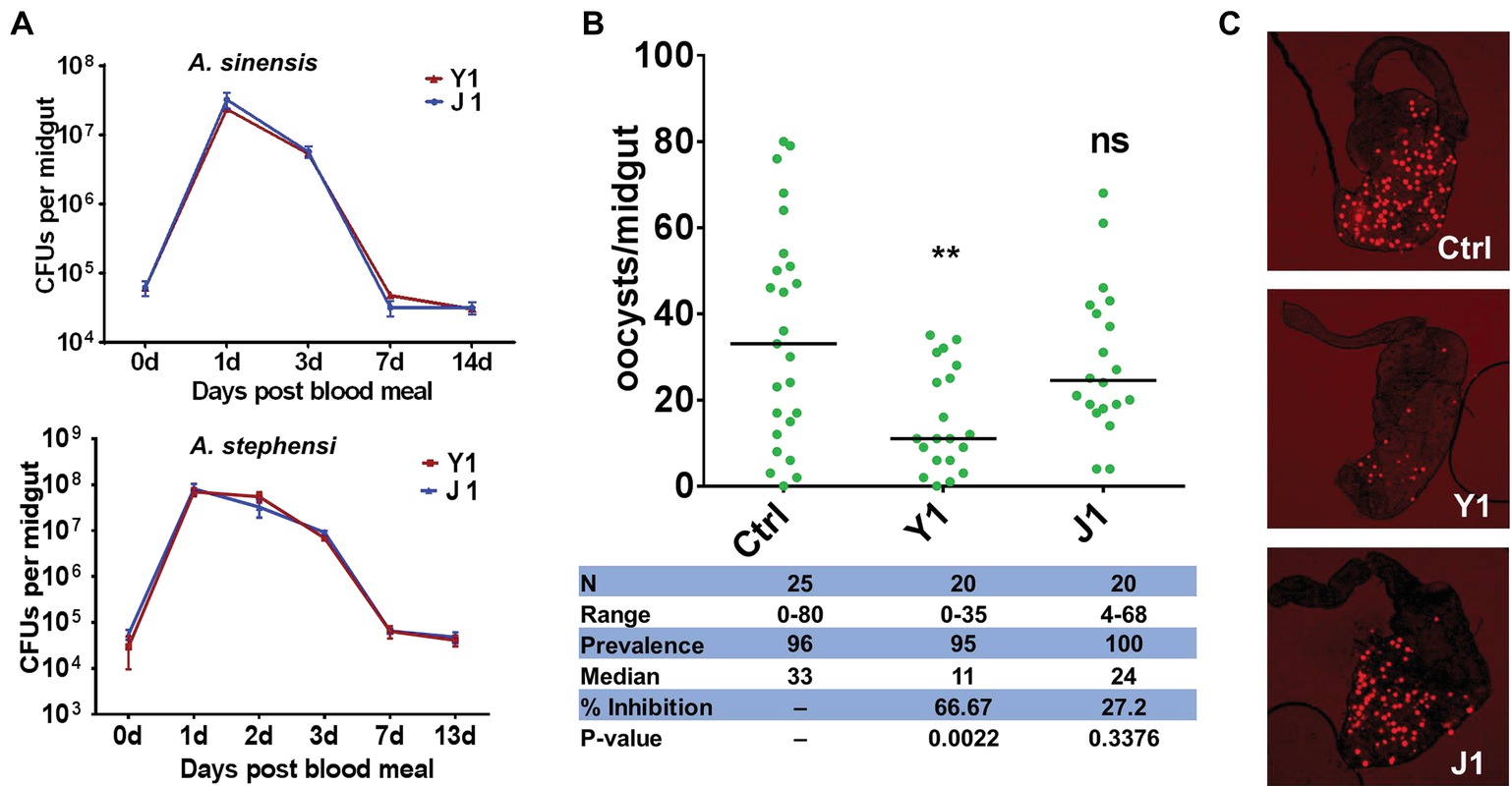
Figure 1. Effect of different Serratia strains on Plasmodium berghei oocyst development. (A) Serratia strains Y1 and J1 stably colonize the midgut of female An. sinensis and An. stephensi mosquitoes and rapidly proliferate after a blood meal. The eGFP-tagged strains Y1-GFP and J1-GFP were fed to 3-day-old female An. sinensis and An. stephensi mosquitoes in a 5% sugar meal for 24 h, then mosquitoes were allowed to feed on a blood meal. Fluorescent bacteria colony-forming units (CFUs) were determined by plating serially diluted homogenates of midguts on LB agar plates containing 100 mg/ml of kanamycin. Data were pooled from three biological replicates (shown are means ± SEM). (B) Oocyst infection intensity in An. stephensi mosquitoes colonized with Serratia Y1 or J1 after feeding on P. berghei mCherry infected mice. Each dot represents the oocysts number from individual midguts, and the horizontal lines indicate the median number of oocysts. Inhibition = [(# of oocyst in control group – # of oocyst in experimental group)/# of mosquitoes in control group] × 100; Median, median oocyst number per midgut; N, number of mosquitoes analyzed; Prevalence, percentage of mosquitoes carrying at least one oocyst; Range, range of oocyst numbers per midgut. The experiments were repeated three times with similar results (Supplementary Figure S2). The Mann-Whitney test was used to determine significance in oocysts numbers, **p < 0.01. (C) mCherry oocysts in the midgut epithelium of mosquitoes that had been fed with only sugar solution (top panel), Serratia Y1 (center panel) or J1 (bottom panel).
The downstream analyses were performed in An. stephensi because it is difficult to maintain An. sinensis under laboratory conditions. Investigation of the effect of the two Serratia strains on Plasmodium development in the midgut of An. stephensi show that Serratia Y1 significantly reduced oocyst formation (p < 0.001), while Serratia J1 did not affect P. berghei oocyst formation (Figures 1B,C; Supplementary Figure S2). Thus, we identified two mosquito S. marcescens strains, one of them inhibiting Plasmodium infection in the mosquito midgut.
Serratia Y1 Inhibits Plasmodium Development Through Activation of the Mosquito Immune System
To test whether Serratia Y1 exerts the parasite inhibitory effect through stimulation of the mosquito immune system, we used RNA interference (RNAi) to silence the expression of Rel1 and Rel2, the NF-κB transcription factors of Toll and IMD signaling pathways that regulate mosquito innate immunity, respectively. We compared the oocyst numbers in the midguts of An. stephensi mosquito cohorts that had been injected with double-stranded RNA either for GFP (dsGFP), dsRel1 or dsRel2. Inhibition of the Toll pathway by Rel1 knockdown significantly increased P. berghei infection in mosquitoes fed with Y1 (Figure 2; Supplementary Figure S3). Silencing of Rel2 did not rescue P. berghei oocyst development in the presence of Y1 (Figure 2; Supplementary Figure S3). These results suggested that Serratia Y1 exerts its anti-Plasmodium activity through the activation of the Toll immune pathway.
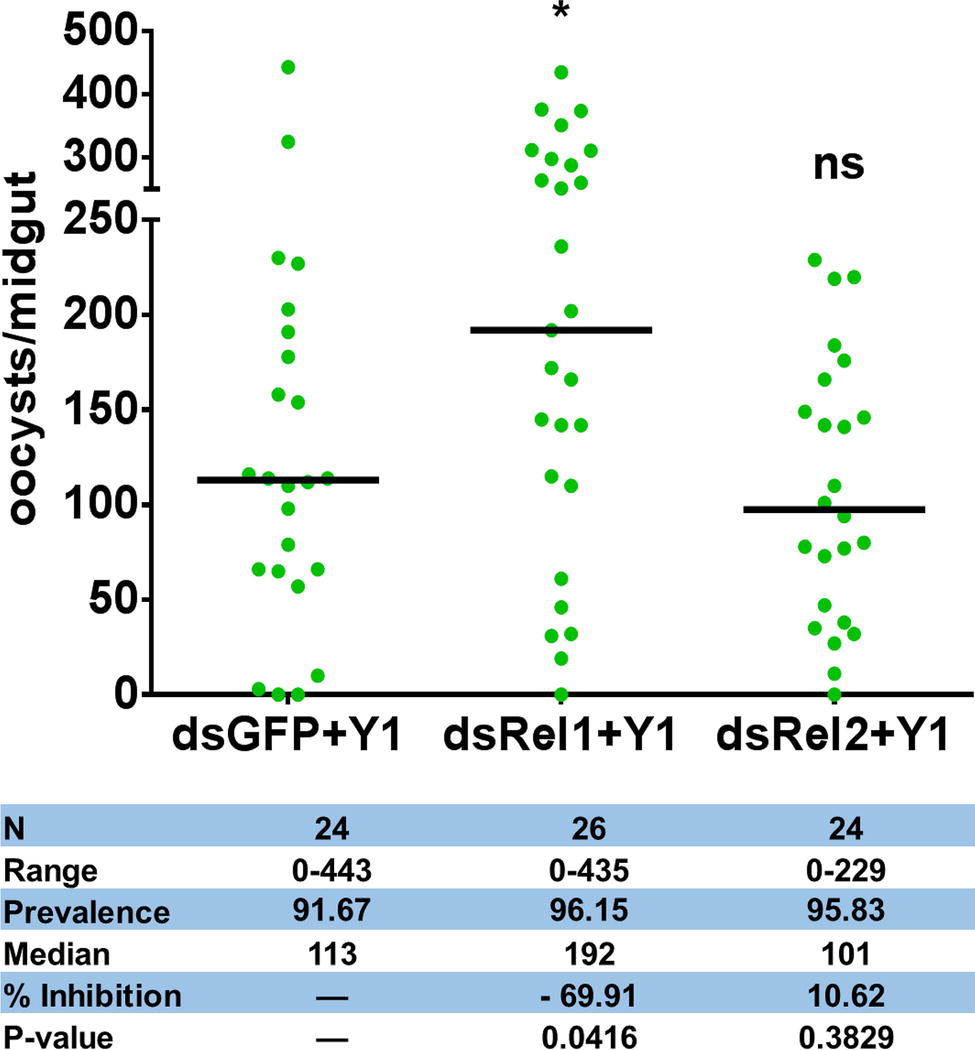
Figure 2. Effect of Rel1 and Rel2 silencing on the Serratia Y1-mediated anti-Plasmodium activity. Rel1 and Rel2 were silenced in An. stephensi mosquitoes by systemic injection of dsGFP, dsRel1 or dsRel2 RNA. The injected mosquitoes were fed on a sugar meal containing Serratia Y1. Three days later, all mosquito groups were allowed to feed on the same P. berghei infected mouse. The injected double-stranded RNA (ds) and presence (Y1) of Serratia Y1 are indicated below each column. Each dot represents the number of oocysts from an individual midgut, and the horizontal lines indicate the median number of oocysts. The experiments were repeated three times with similar results (Supplementary Figure S3). The Mann-Whitney test was used to determine significance in oocysts numbers, *p < 0.05.
Mosquito Global Transcriptome Responses to Serratia Challenge
To gain a better understanding of the differences in the mosquito immune response to Serratia Y1 and J1, we compared the midgut gene transcriptional profile of mosquitoes colonized by Serratia Y1 or J1 before (0 h) and 24 h after a blood meal. RNA-seq data from six RNA libraries are summarized in Supplementary Table S1. Genome-wide transcriptomic profiles showed a similar pattern of midgut gene expression in response to Serratia Y1 and J1 in sugar-fed mosquitoes (Figure 3A). A total of 375 genes were differentially regulated in the Serratia Y1 challenged group (p < 0.05), of which 225 genes were up-regulated and 150 genes were down-regulated (Figure 3C). In the Serratia J1 challenged group, 357 genes were differentially regulated including 223 up-regulated and 134 down-regulated genes (Figures 3A,C). The majority of the DEGs (257 genes) had similar expression pattern between the two groups (Figure 3C). At 24 h post-blood meal, Serratia Y1 modulated 370 DGEs while Serratia J1 regulated only 252 DGEs (Figures 3B,C). Interestingly, a total of 146 upregulated and 3 downregulated genes were specific for the Serratia challenged Y1 group. However, the Serratia J1 challenged group only showed 7 upregulated and 24 downregulated genes specific to this group (Figures 3B,C). This indicates that Serratia Y1 regulates more genes in the midgut after the mosquito takes a blood meal than Serratia J1.
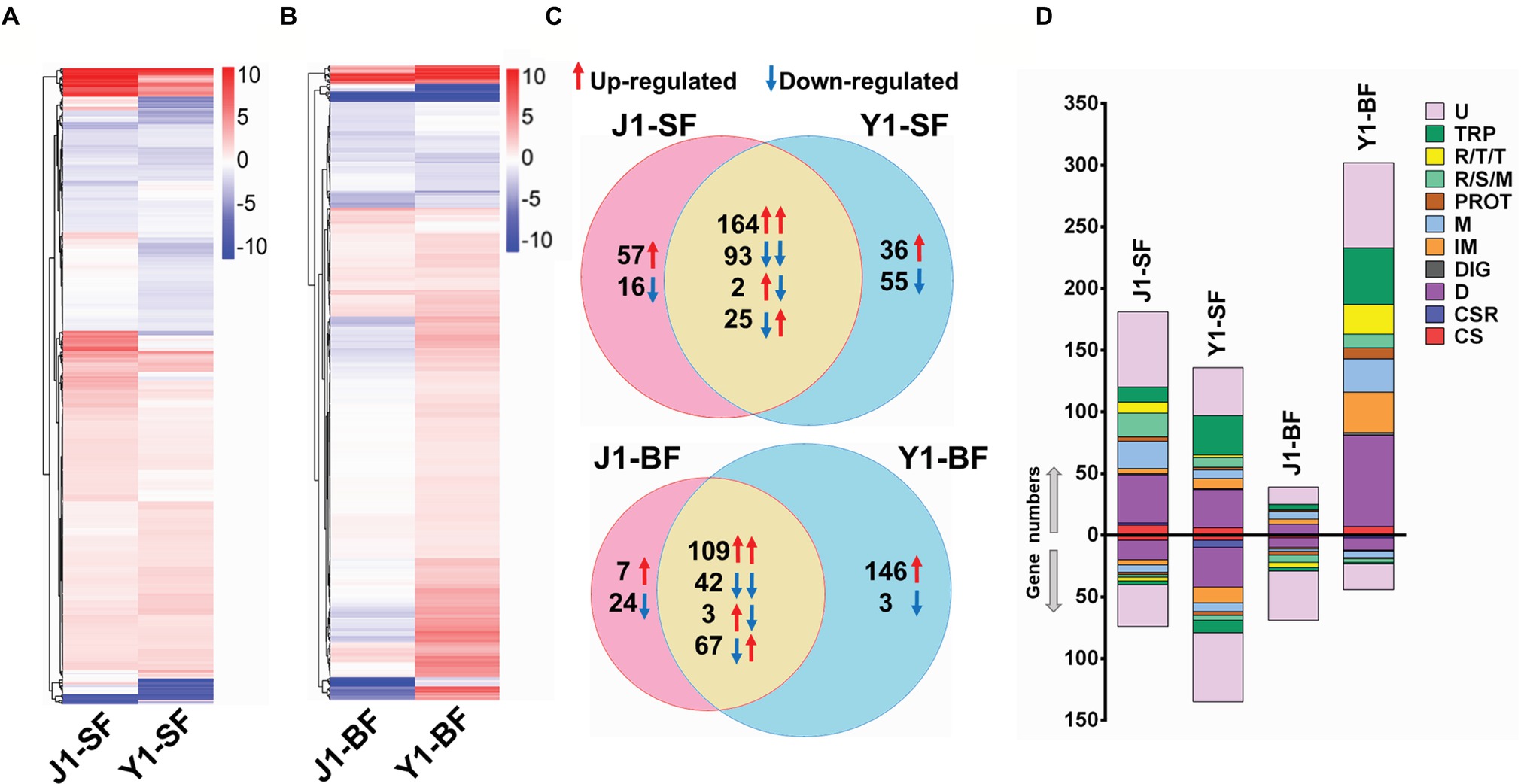
Figure 3. Comparative analysis of transcriptional profiles in mosquito midguts challenged with Serratia Y1 or J1. (A,B) Heat maps and hierarchical clustering analysis of differentially expressed genes (DEGs) in the midgut of An. stephensi mosquitoes fed with Serratia Y1 or J1 before (SF, sugar fed) and 24 h after blood meal (BF, blood fed). DEGs in each group were identified with log 2 (fold change) of ≥1 (p < 0.05) by comparison of transcript abundance between bacteria challenged and aseptic mosquitoes. Colors from white to red indicate up-regulation; colors from white to blue indicate down-regulation. (C) Venn diagram showing the number of shared and specific DEGs in the midgut of mosquitoes fed with Serratia Y1 or J1 before and 24 h after blood meal. Red and blue arrows indicate the mosquitoes challenged with Serratia Y1 and J1, respectively. (D) Global gene regulation in the different groups. Proportions and numbers of genes belonging to distinct functional groups were up- (red arrow) or down-regulated (blue arrow) in the corresponding groups. U, unknown functions; TRP, transport; R/T/T, replication, transcription, translation; R/S/M, oxidoreductive, stress-related and mitochondrial; PROT, proteolysis; M, metabolism; IM, immunity; DIG, digestion; D, diverse; CSR, chemosensory protein; CS, cytoskeletal, structural.
Gene Ontology (GO) enrichment analysis was performed to show the potential biological function of the differentially expressed genes. For 0 h before blood meal, we found that, 7 of 48 gene functional groups (14.6%) belonging to extracellular matrix, extracellular region part, extracellular space and transcription regulator were only enriched in the Serratia J1 challenged group, while 37 gene functional groups (77%) were enriched in both Y1 and J1 groups (Supplementary Figure S4). At 24 h post-blood meal, 19 of 50 GO terms (38%) were only presented in the Serratia Y1 challenged group (Supplementary Figure S5). KEGG classification was also used to analyze the corresponding metabolic pathways involved in the regulation of mosquito genes induced by Serratia challenge. Before blood meal, 39 and 42 pathways of level 2 were enriched in the Serratia Y1 challenged group and the Serratia J1 challenged group, respectively (Supplementary Figure S6A). At 24 h post-blood meal, 42 and 34 pathways of level 2 were enriched in the Serratia Y1 challenged group and the Serratia J1 challenged group, respectively (Supplementary Figure S6B). Pathways including global and overview maps, signal transduction, immune system, transport and catabolism, signaling molecules and interaction, amino acid metabolism, digestive system and endocrine system were overrepresented in the Serratia Y1 challenged group (Supplementary Figure S6B).
Serratia Y1 Modulates Mosquito Immune Genes
Given that Serratia Y1 exerts anti-Plasmodium activity by stimulating the mosquito immune pathway (Figure 2), the expression profiles of genes involved in mosquito immunity were further analyzed. In sugar-fed mosquitoes, the number of immune regulated genes was similar between Serratia Y1 and Serratia J1 colonized mosquitoes (Figure 3D). At 24 h post-blood meal, there were marked differences in immune gene regulation between Serratia Y1 and J1 challenged groups. A total of 33 immunity genes were up-regulated in mosquitoes fed with Serratia Y1. However, in mosquitoes fed with Serratia J1 only 4 immune genes were up-regulated and 1 down-regulated (Figure 3D). In mosquitoes challenged by Serratia Y1, the antimicrobial immune genes with the highest upregulation were Defensin 1(DEF1), Cecropins 1 (CEC1), Cecropins 2 (CEC2) and Gambicin 1 (GAM1). In addition, except for CLIPD 2 and CLIPA 3, all other genes encoding the CLIP family were up-regulated by Serratia Y1 (Figure 4A). The expression profiles of CTL family genes were also differently regulated in the midgut of mosquitoes fed with Serratia Y1 and J1 (Figure 4B).
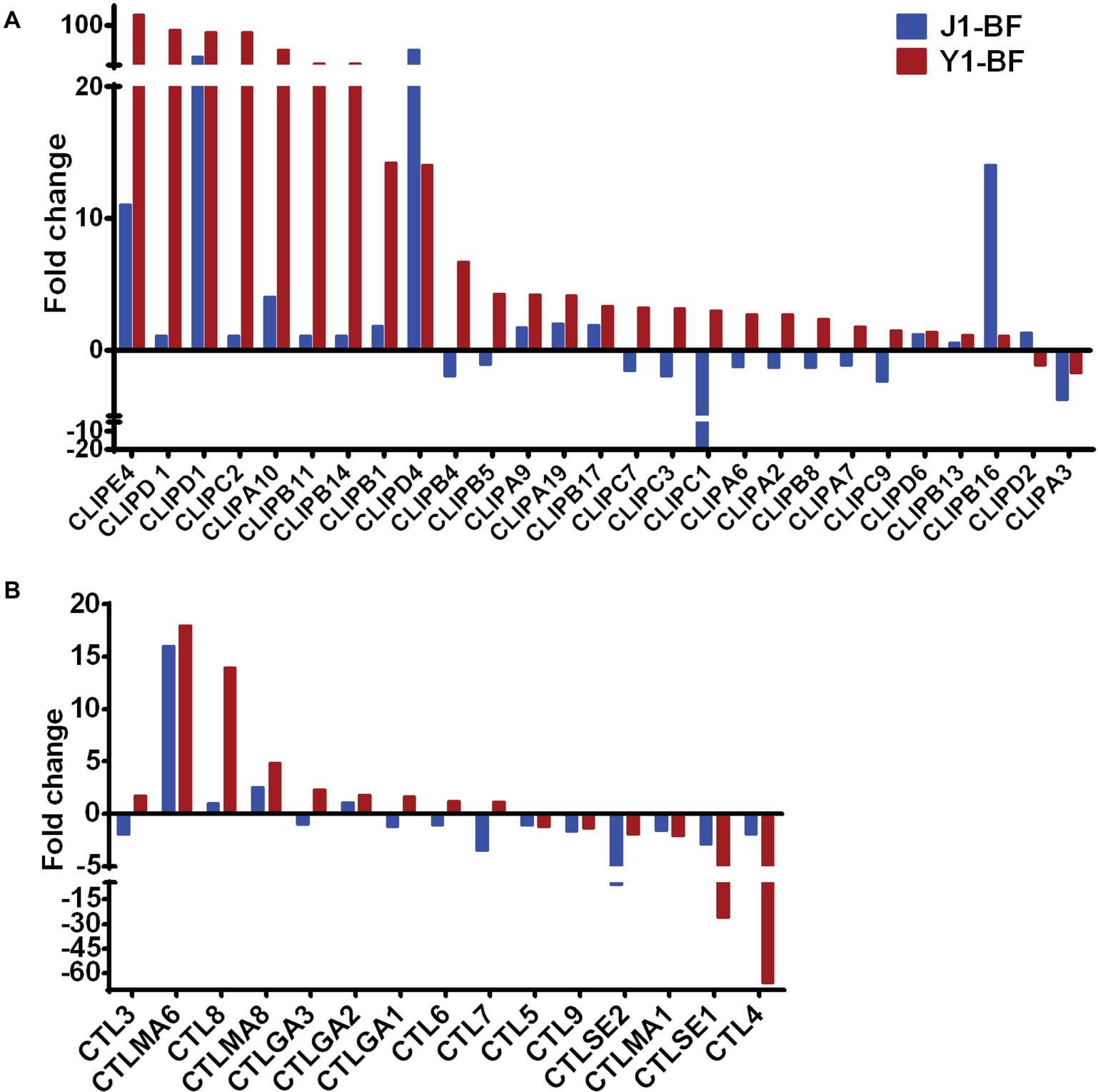
Figure 4. Transcript abundance of CLIP and CTL family genes. (A) Fold change in transcript levels of the CLIP family genes in the midgut of mosquitoes fed with Serratia Y1 or J1, as compared with the aseptic control mosquitoes. (B) Fold change in transcript levels of the CTL family in the midgut of mosquitoes fed with Serratia Y1 or J1, relative to the aseptic mosquitoes.
Serratia Y1 Modulates the Anti-Plasmodium Effector Genes
To validate the RNA-seq results, we focused on the transcriptional regulation of several potent anti-Plasmodium and anti-bacterial effector genes encoding the thioester-containing protein 1 (TEP1), Anopheles Plasmodium responsive leucine-rich repeat protein (APL1A), leucine-rich repeat protein LRRD7, fibrinogen immunolectin 9 (FBN9), Plasmodium protective c-type lectin 4 (CTL4) and Gambicin (GAM1). All these genes were differentially regulated in the midgut of mosquitoes fed with Serratia Y1 at 24 h post-blood meal (Figure 5A). Real-time quantitative PCR (qPCR) confirmed that TEP1, APL1A, LRRD7, FBN9, and GAM1 were significantly up-regulated, while the protective agonist CTL4 gene was significantly down-regulated in the mosquitoes fed with Y1 at 24 h post-blood meal (Figures 5A–C). The other two antimicrobial peptides (DEF1 and CEC1) were also significantly up-regulated (Figure 5D).
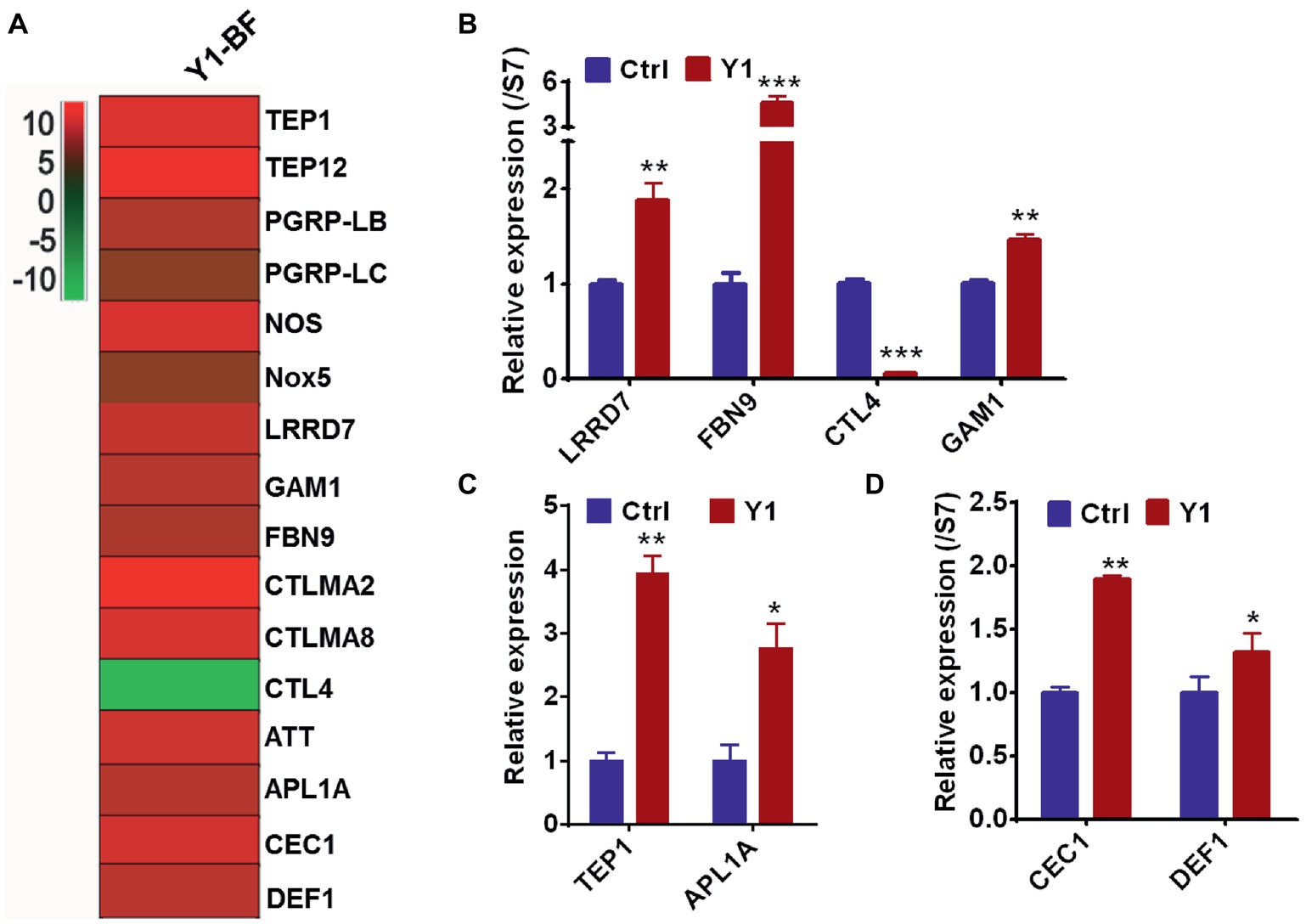
Figure 5. Serratia Y1 elicits expression of immune genes in the midgut of mosquitoes 24 h post-blood meal. (A) Fold change in transcript levels of immune marker genes in the midgut of Y1-containing mosquitoes 24 h post-blood meal obtained from RNA-seq data. (B) Relative expression of immune factor genes (LRRD7, FBN9, CTL4, and GAM1) in the midgut of mosquitoes 24 h post-blood meal detected by qPCR. (C) Relative expression of TEP1 and APL1A in the midgut of mosquitoes 24 h post-blood meal detected by qPCR. (D) Relative expression of DEF1 and CEC1 in the midgut of mosquitoes 24 h post-blood meal detected by qPCR. mRNA levels of the tested genes were normalized to that of the housekeeping gene S7. Error bars indicate SD of three technical replicates. Experiments were repeated three times with similar results, *p < 0.05; **p < 0.01; ***p < 0.001.
To establish further evidence that Serratia Y1 impacts P. berghei infection through activation of anti-Plasmodium effector genes, we chose three induced potent anti-Plasmodium factors, TEP1, FBN9, and LRRD7 for validation in mosquitoes colonized with Serratia Y1. Midgut mRNA levels for TEP1, FBN9, and LRRD7 in the dsRNAs-injected mosquitoes were markedly reduced compared to the dsGFP treated control (Supplementary Figure S7). Silencing of the three effector genes resulted in a significant increase of oocyst numbers when compared to the dsGFP -injected control in the Serratia Y1 colonized mosquitoes (Figure 6; Supplementary Figure S8). This result indicates that the depletion of dsTEP1, dsFBN9, or dsLRRD7 significantly reverses the refractoriness conferred by Serratia Y1 infection.
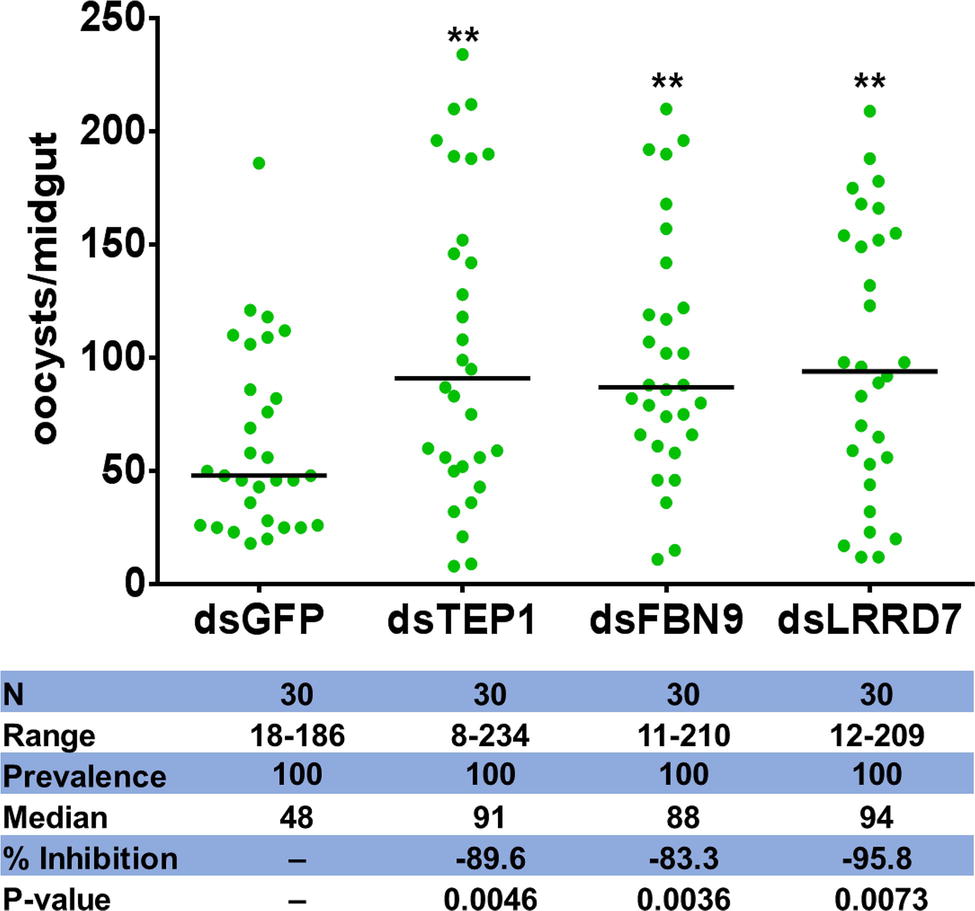
Figure 6. Oocyst loads following silencing of TEP1, FBN9 and LRRD7 genes. An. stephensi female mosquitoes were injected with dsRNA solution for TEP1, FBN9, LRRD7 or GFP as a control. Injected mosquitoes were colonized with Serratia Y1 and then fed on a P. berghei infected mouse 3 days after dsRNA injection. Mosquito midguts were dissected 10 days post-blood feeding to determine the number of oocysts per mosquito. Each dot represents the number of oocysts from individual midguts, and the horizontal lines indicate the median number of oocysts. The experiments were repeated three times with similar results (Supplementary Figure S8). The Mann-Whitney test was used to determine significance in oocysts numbers, **p < 0.01.
Discussion
Mosquito gut bacteria form a resident community that has co-evolved with the insect host. In addition to playing important roles in digestion and harvesting energy, commensal bacteria are important factors determining the outcome of pathogen infection. Other mechanisms such as induction of mosquito innate immune responses have also been proposed, although the exact effect and mode of action by individual commensal bacteria are largely unknown. Interestingly, the presence, outgrowth, or loss of certain bacterial components of gut microbiota correlates with increased or decreased susceptibility to Plasmodium and dengue virus infection (Apte-Deshpande et al., 2012, 2014; Boissiere et al., 2012; Bahia et al., 2014). A previous study also showed that different strains of the same bacterial species can induce different outcomes on Plasmodium infections (Bando et al., 2013), indicating that the effect of gut bacteria on Plasmodium infection is complex and may involve species-specific or strain-specific interactions. In this study, we identified two mosquito symbiotic Serratia strains, from field caught An. sinensis mosquitoes, with different effect on Plasmodium development. Serratia Y1 shows anti-Plasmodium activity, but Serratia J1 does not influence parasite development in the midgut of mosquitoes. In mosquitoes, anti-Plasmodium and antibacterial immune defenses are largely controlled by the Toll and Imd immune signaling pathways, and the Toll pathway seems to be most efficient against the rodent parasite P. berghei (Meister et al., 2005, 2009; Garver et al., 2009). Some immune factors and AMP genes are regulated by both Toll and Imd pathways (Luna et al., 2006). This dual activation may be modulated by independent stimulation or by cross-regulation of the two signaling pathways (De Gregorio et al., 2002). Previous study also showed that the Toll and Imd pathways can interact synergistically and activate innate immune responses in Drosophila, demonstrating that cross-regulation of the two pathways occur (Tanji et al., 2007). Our findings also show that Serratia Y1 and J1 induce some genes that are regulated by both Toll and Imd pathways. We further showed that Serratia Y1 indirectly antagonizes P. berghei infection through activation of the mosquito Toll immune pathway.
Since the midgut is the primary site of response to the invading Plasmodium, identification of factors regulated by gut bacteria would provide insight into the mechanisms of how commensal bacteria activate immune-mediated anti-Plasmodium activity. Toward this we investigated the influence of Serratia Y1- or J1 on the mosquito midgut transcriptome. Before blood meal, the number of regulated immune genes in the Serratia Y1 and J1 groups was very similar, which may indicate that the mosquitoes modulate similar basal immune responses to limit the over-proliferation of the symbiotic bacteria and maintain gut homeostasis. Normally, when mosquitoes ingest sugar, the symbiotic bacteria in the midgut are kept at a relatively low level (Pumpuni et al., 1996; Wang et al., 2012). This healthy status requires mosquitoes to deploy basal immunity to maintain a symbiotic relationship with the bacteria (Wei et al., 2017). The gut microbiota and the host mosquito have adapted to coexist during long-term adaptive coevolution, which may be the reason why the gene expression patterns of the Serratia Y1 and J1 mosquitoes are similar when they feed on sugar.
After ingestion of a Plasmodium infected blood meal by the mosquito, the parasite undergoes sexual development in the midgut lumen to form a motile ookinete that invades the midgut epithelium around 24–26 h after blood feeding (Sinden and Billingsley, 2001; Baton and Ranford-Cartwright, 2005). The bacterial numbers in the mosquito midgut also increase by 100- to 1,000-fold 24 h after a blood meal (Cirimotich et al., 2011), activating the mosquito immune system to limit the over-proliferation of gut bacteria (Dong et al., 2009). We found that the c-type lectins (CTLs) and CLIP serine proteases were were up-regulated in the Serratia Y1 group. Many CLIP family genes have been showed to affect Plasmodium development. CLIPA2, A5 and A7 suppress parasite melanization, and CLIPA2 and CLIPA5 interact synergistically to block ookinete invasion (Barillas-Mury, 2007), while CLIPB3, B4, B8 and B17 promote ookinete invasion (Volz et al., 2006). CLIPB14 and CLIPB15 are also involved in killing Plasmodium ookinetes and participate in the response against bacteria (Volz et al., 2005). Interestingly, expression of CTL4 was severely down-regulated in the Serratia Y1 challenged mosquitoes at 24 h post-blood meal. CTL 4 and CTLMA 2 are present in the hemolymph and up-regulated 24 h after blood ingestion and can protect the rodent Plasmodium ookinetes from destruction (Osta et al., 2004). FBN9, a member of the fibrinogen domain immunolectin family (FBN), was up-regulated in the Serratia Y1 fed mosquitoes at 24 h post-blood meal. A previous study suggested that FBN9 interacts with gram-positive and gram-negative bacteria and also affects Plasmodium development (Dong and Dimopoulos, 2009). The complement-like protein TEP1 and the leucine-rich repeat (LRR) protein APL1, which mediate lysis of Plasmodium parasites in the mosquito midgut (Blandin et al., 2008; Garver et al., 2009; Dong et al., 2011), were also up-regulated in the Serratia Y1 group at 24 h post-blood meal. Other factors including LRRD7, TEP12, NOX5, PGRP-LB, NOS and PGRP-LC, which had been shown to affect Plasmodium infection (Luckhart et al., 1998; Dong et al., 2006; Meister et al., 2009; Oliveira Gde et al., 2012), were also up-regulated in the Serratia Y1 group at 24 h post-blood meal. We further demonstrated that silencing of TEP1, FBN9, or LRRD7 markedly reverses the parasite inhibition conferred by Serratia Y1 infection. Previous studies reported that these three genes can strongly influence both P. falciparum and P. berghei development (Blandin et al., 2004; Dong et al., 2006). Taken together, Serratia Y1 elicited a strong immune response in the mosquito midgut after taking a blood meal and knock-down of several highly elicited immune genes induced a significant increase of P. berghei oocysts, further confirming that Serratia Y1 interferes with P. berghei development by eliciting the mosquito’s immune response.
In summary, our data shows that Serratia Y1 inhibits Plasmodium development through stimulation of the mosquito immunity. Our work establishes an important framework of knowledge for further investigations into the molecular interactions between gut bacteria, Anopheles mosquitoes and pathogens, which would enhance our understanding of how gut bacteria inhibit Plasmodium development in the midgut of mosquitoes.
Data Availability
RNA sequencing datasets from this work were deposited in the National Center for Biotechnology Information Sequence Read Archive (accession no. PRJNA520745).
Author Contributions
SW conceived the study. SW and LB designed the experiments. LB performed the majority of experiments and generated fluorescent strains and performed transcriptome analysis. LB and GW performed RNAi analysis. LW and JV-R discussed results and provided advice. LB and SW analyzed the data. JV-R edited the manuscript. LB, LW, and SW wrote the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (grants 31830086, 31772534, 31472044, 31501703 to SW), the Key Research Program of the Chinese Academy of Sciences (grant KFZD-SW-219 to SW), the Strategic Priority Research Program of Chinese Academy of Sciences (grant XDB11010500 to SW), and One Hundred Talents Program of the Chinese Academy of Sciences (grant 2013OHTP01 to SW). The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01580/full#supplementary-material
Footnotes
1. https://www.vectorbase.org/organisms/anopheles-stephensi
References
Apte-Deshpande, A., Paingankar, M., Gokhale, M. D., and Deobagkar, D. N. (2012). Serratia odorifera a midgut inhabitant of Aedes aegypti mosquito enhances its susceptibility to dengue-2 virus. PLoS One 7:e40401. doi: 10.1371/journal.pone.0040401
Apte-Deshpande, A. D., Paingankar, M. S., Gokhale, M. D., and Deobagkar, D. N. (2014). Serratia odorifera mediated enhancement in susceptibility of Aedes aegypti for chikungunya virus. Indian J. Med. Res. 139, 762–768. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4140042/.
Bahia, A. C., Dong, Y., Blumberg, B. J., Mlambo, G., Tripathi, A., BenMarzouk-Hidalgo, O. J., et al. (2014). Exploring Anopheles gut bacteria for Plasmodium blocking activity. Environ. Microbiol. 16, 2980–2994. doi: 10.1111/1462-2920.12381
Bando, H., Okado, K., Guelbeogo, W. M., Badolo, A., Aonuma, H., Nelson, B., et al. (2013). Intra-specific diversity of Serratia marcescens in Anopheles mosquito midgut defines Plasmodium transmission capacity. Sci. Rep. 3:1641. doi: 10.1038/srep01641
Barillas-Mury, C. (2007). CLIP proteases and Plasmodium melanization in Anopheles gambiae. Trends Parasitol. 23, 297–299. doi: 10.1016/j.pt.2007.05.001
Baton, L. A., and Ranford-Cartwright, L. C. (2005). How do malaria ookinetes cross the mosquito midgut wall? Trends Parasitol. 21, 22–28. doi: 10.1016/j.pt.2004.11.001
Blandin, S. A., Marois, E., and Levashina, E. A. (2008). Antimalarial responses in Anopheles gambiae: from a complement-like protein to a complement-like pathway. Cell Host Microbe 3, 364–374. doi: 10.1016/j.chom.2008.05.007
Blandin, S., Shiao, S. H., Moita, L. F., Janse, C. J., Waters, A. P., Kafatos, F. C., et al. (2004). Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell 116, 661–670. doi: 10.1016/s0092-8674(04)00173-4
Boissiere, A., Gimonneau, G., Tchioffo, M. T., Abate, L., Bayibeki, A., Awono-Ambene, P. H., et al. (2013). Application of a qPCR assay in the investigation of susceptibility to malaria infection of the M and S molecular forms of An. gambiae s.s. in Cameroon. PLoS One 8:e54820. doi: 10.1371/journal.pone.0054820
Boissiere, A., Tchioffo, M. T., Bachar, D., Abate, L., Marie, A., Nsango, S. E., et al. (2012). Midgut microbiota of the malaria mosquito vector Anopheles gambiae and interactions with Plasmodium falciparum infection. PLoS Pathog. 8:e1002742. doi: 10.1371/journal.ppat.1002742
Cirimotich, C. M., Dong, Y., Clayton, A. M., Sandiford, S. L., Souza-Neto, J. A., Mulenga, M., et al. (2011). Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science 332, 855–858. doi: 10.1126/science.1201618
De Gregorio, E., Spellman, P. T., Tzou, P., Rubin, G. M., and Lemaitre, B. (2002). The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 21, 2568–2579. doi: 10.1093/emboj/21.11.2568
Dimopoulos, G., Richman, A., Muller, H. M., and Kafatos, F. C. (1997). Molecular immune responses of the mosquito Anopheles gambiae to bacteria and malaria parasites. Proc. Natl. Acad. Sci. USA 94, 11508–11513. doi: 10.1073/pnas.94.21.11508
Dong, Y., Aguilar, R., Xi, Z., Warr, E., Mongin, E., and Dimopoulos, G. (2006). Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PLoS Pathog. 2:e52. doi: 10.1371/journal.ppat.0020052
Dong, Y., Das, S., Cirimotich, C., Souza-Neto, J. A., McLean, K. J., and Dimopoulos, G. (2011). Engineered Anopheles immunity to Plasmodium infection. PLoS Pathog. 7:e1002458. doi: 10.1371/journal.ppat.1002458
Dong, Y., and Dimopoulos, G. (2009). Anopheles fibrinogen-related proteins provide expanded pattern recognition capacity against bacteria and malaria parasites. J. Biol. Chem. 284, 9835–9844. doi: 10.1074/jbc.M807084200
Dong, Y., Manfredini, F., and Dimopoulos, G. (2009). Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 5:e1000423. doi: 10.1371/journal.ppat.1000423
Garver, L. S., Dong, Y., and Dimopoulos, G. (2009). Caspar controls resistance to Plasmodium falciparum in diverse Anopheline species. PLoS Pathog. 5:e1000335. doi: 10.1371/journal.ppat.1000335
Gendrin, M., and Christophides, G. (2013). The Anopheles mosquito microbiota and their impact on pathogen transmission. 525–539. INTECH. doi: 10.5772/55107
Gendrin, M., Rodgers, F. H., Yerbanga, R. S., Ouédraogo, J. B., Basáñez, M. G., Cohuet, A., et al. (2015). Antibiotics in ingested human blood affect the mosquito microbiota and capacity to transmit malaria. Nat. Commun. 6, 1–7. doi: 10.1038/ncomms6921
Ghosh, A., Edwards, M. J., and Jacobs-Lorena, M. (2000). The journey of the malaria parasite in the mosquito: hopes for the new century. Parasitol. Today 16, 196–201. doi: 10.1016/S0169-4758(99)01626-9
Gonzalez-Ceron, L., Santillan, F., Rodriguez, M. H., Mendez, D., and Hernandez-Avila, J. E. (2003). Bacteria in midguts of field-collected Anopheles albimanus block Plasmodium vivax sporogonic development. J. Med. Entomol. 40, 371–374. doi: 10.1603/0022-2585-40.3.371
Graewe, S., Retzlaff, S., Struck, N., Janse, C. J., and Heussler, V. T. (2009). Going live: a comparative analysis of the suitability of the RFP derivatives RedStar, mCherry and tdTomato for intravital and in vitro live imaging of Plasmodium parasites. Biotechnol. J. 4, 895–902. doi: 10.1002/biot.200900035
Luckhart, S., Vodovotz, Y., Cui, L., and Rosenberg, R. (1998). The mosquito Anopheles stephensi limits malaria parasite development with inducible synthesis of nitric oxide. Proc. Natl. Acad. Sci. USA 95, 5700–5705. doi: 10.1073/pnas.95.10.5700
Luna, C., Hoa, N. T., Lin, H., Zhang, L., Nguyen, H. L., Kanzok, S. M., et al. (2006). Expression of immune responsive genes in cell lines from two different Anopheline species. Insect Mol. Biol. 15, 721–729. doi: 10.1111/j.1365-2583.2006.00661.x
Meister, S., Agianian, B., Turlure, F., Relogio, A., Morlais, I., Kafatos, F. C., et al. (2009). Anopheles gambiae PGRPLC-mediated defense against bacteria modulates infections with malaria parasites. PLoS Pathog. 5:e1000542. doi: 10.1371/journal.ppat.1000542
Meister, S., Kanzok, S. M., Zheng, X. L., Luna, C., Li, T. R., Hoa, N. T., et al. (2005). Immune signaling pathways regulating bacterial and malaria parasite infection of the mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. USA 102, 11420–11425. doi: 10.1073/pnas.0504950102
Oduol, F., Xu, J. N., Niare, O., Natarajan, R., and Vernick, K. D. (2000). Genes identified by an expression screen of the vector mosquito Anopheles gambiae display differential molecular immune response to malaria parasites and bacteria. Proc. Natl. Acad. Sci. USA 97, 11397–11402. doi: 10.1073/pnas.180060997
Oliveira Gde, A., Lieberman, J., and Barillas-Mury, C. (2012). Epithelial nitration by a peroxidase/NOX5 system mediates mosquito antiplasmodial immunity. Science 335, 856–859. doi: 10.1126/science.1209678
Osta, M. A., Christophides, G. K., and Kafatos, F. C. (2004). Effects of mosquito genes on Plasmodium development. Science 303, 2030–2032. doi: 10.1126/science.1091789
Pradel, G. (2007). Proteins of the malaria parasite sexual stages: expression, function and potential for transmission blocking strategies. Parasitology 134, 1911–1929. doi: 10.1017/S0031182007003381
Pumpuni, C. B., Beier, M. S., Nataro, J. P., Guers, L. D., and Davis, J. R. (1993). Plasmodium falciparum: inhibition of sporogonic development in Anopheles stephensi by gram-negative bacteria. Exp. Parasitol. 77, 195–199. doi: 10.1006/expr.1993.1076
Pumpuni, C. B., Demaio, J., Kent, M., Davis, J. R., and Beier, J. C. (1996). Bacterial population dynamics in three anopheline species: the impact on Plasmodium sporogonic development. Am. J. Trop. Med. Hyg. 54, 214–218. doi: 10.4269/ajtmh.1996.54.214
Simon, N., Scholz, S. M., Moreira, C. K., Templeton, T. J., Kuehn, A., Dude, M. A., et al. (2009). Sexual stage adhesion proteins form multi-protein complexes in the malaria parasite Plasmodium falciparum. J. Biol. Chem. 284, 14537–14546. doi: 10.1074/jbc.M808472200
Sinden, R. E., and Billingsley, P. F. (2001). Plasmodium invasion of mosquito cells: hawk or dove? Trends Parasitol. 17, 209–211. doi: 10.1016/S1471-4922(01)01928-6
Smith, R. C., Vega-Rodriguez, J., and Jacobs-Lorena, M. (2014). The Plasmodium bottleneck: malaria parasite losses in the mosquito vector. Mem. Inst. Oswaldo Cruz 109, 644–661. doi: 10.1590/0074-0276130597
Song, X., Wang, M., Dong, L., Zhu, H., and Wang, J. (2018). PGRP-LD mediates A. stephensi vector competency by regulating homeostasis of microbiota-induced peritrophic matrix synthesis. PLoS Pathog. 14:e1006899. doi: 10.1371/journal.ppat.1006899
Stathopoulos, S., Neafsey, D. E., Lawniczak, M. K., Muskavitch, M. A., and Christophides, G. K. (2014). Genetic dissection of Anopheles gambiae gut epithelial responses to Serratia marcescens. PLoS Pathog. 10:e1003897. doi: 10.1371/journal.ppat.1003897
Straif, S. C., Mbogo, C. N., Toure, A. M., Walker, E. D., Kaufman, M., Toure, Y. T., et al. (1998). Midgut bacteria in Anopheles gambiae and An. funestus (Diptera: Culicidae) from Kenya and Mali. J. Med. Entomol. 35, 222–226. doi: 10.1093/jmedent/35.3.222
Tanji, T., Hu, X., Weber, A. N., and Ip, Y. T. (2007). Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster. Mol. Cell. Biol. 27, 4578–4588. doi: 10.1128/MCB.01814-06
Vaughan, J. A., Noden, B. H., and Beier, J. C. (1992). Population dynamics of Plasmodium falciparum sporogony in laboratory-infected Anopheles gambiae. J. Parasitol. 78, 716–724. doi: 10.2307/3283550
Volz, J., Muller, H. M., Zdanowicz, A., Kafatos, F. C., and Osta, M. A. (2006). A genetic module regulates the melanization response of Anopheles to Plasmodium. Cell. Microbiol. 8, 1392–1405. doi: 10.1111/j.1462-5822.2006.00718.x
Volz, J., Osta, M. A., Kafatos, F. C., and Muller, H. M. (2005). The roles of two clip domain serine proteases in innate immune responses of the malaria vector Anopheles gambiae. J. Biol. Chem. 280, 40161–40168. doi: 10.1074/jbc.M506191200
Wang, S., Dos-Santos, A. L. A., Huang, W., Liu, K. C., Oshaghi, M. A., Wei, G., et al. (2017). Driving mosquito refractoriness to Plasmodium falciparum with engineered symbiotic bacteria. Science 357, 1399–1402. doi: 10.1126/science.aan5478
Wang, S., Ghosh, A. K., Bongio, N., Stebbings, K. A., Lampe, D. J., and Jacobs-Lorena, M. (2012). Fighting malaria with engineered symbiotic bacteria from vector mosquitoes. Proc. Natl. Acad. Sci. USA 109, 12734–12739. doi: 10.1073/pnas.1204158109
Wei, G., Lai, Y., Wang, G., Chen, H., Li, F., and Wang, S. (2017). Insect pathogenic fungus interacts with the gut microbiota to accelerate mosquito mortality. Proc. Natl. Acad. Sci. USA 114, 5994–5999. doi: 10.1073/pnas.1703546114
World Health Organization (2018). “World malaria report 2018” in Geneva Switzerland WHO. Available at: www.who.int/malaria/publications/worldmalariareport-2018/en/
Keywords: Anopheles mosquito, gut symbiont, antiparasitic defense, malaria transmission, immune activation
Citation: Bai L, Wang L, Vega-Rodríguez J, Wang G and Wang S (2019) A Gut Symbiotic Bacterium Serratia marcescens Renders Mosquito Resistance to Plasmodium Infection Through Activation of Mosquito Immune Responses. Front. Microbiol. 10:1580. doi: 10.3389/fmicb.2019.01580
Edited by:
Rhoel Dinglasan, University of Florida, United StatesReviewed by:
Isabelle Morlais, Institut de Recherche pour le Développement (IRD), FranceSandrine Eveline Nsango, University of Douala, Cameroon
Mathilde Gendrin, Institut Pasteur de la Guyane, French Guiana
Copyright © 2019 Bai, Wang, Vega-Rodríguez, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sibao Wang, c2J3YW5nQHNpYnMuYWMuY24=
 Liang Bai
Liang Bai Lili Wang
Lili Wang Joel Vega-Rodríguez
Joel Vega-Rodríguez Guandong Wang2,3
Guandong Wang2,3 Sibao Wang
Sibao Wang