- 1College of Food Biological Engineering, Xuzhou University of Technology, Xuzhou, China
- 2Key Construction Laboratory of Food Resources Development and the Quality Safety in Jiangsu, Xuzhou University of Technology, Xuzhou, China
- 3College of Environmental Engineering, Xuzhou University of Technology, Xuzhou, China
- 4Logistics & Security Department, Shanghai Civil Aviation College, Shanghai, China
- 5Environment Monitoring Station, Zaozhuang Municipal Bureau of Ecology and Environment, Zaozhuang, China
- 6Henan Key Laboratory of Cold Chain Food Quality and Safety Control, Zhengzhou University of Light Industry, Zhengzhou, China
The development of multidrug- and toxin-resistant bacteria as a result of increasing industrialization and sustained and intense antimicrobial use in aquaculture results in human health problems through increased incidence of food-borne illnesses. Integrative and conjugative elements (ICEs) are self-transmissible mobile genetic elements that allow bacteria to acquire complex new traits through horizontal gene transfer and encode a wide variety of genetic information, including resistance to antibiotics and heavy metals; however, there is a lack of studies of ICEs of environmental origin in Asia. Here, we determined the prevalence, genotypes, heavy metal resistance and antimicrobial susceptibility of 997 presumptive strains of Vibrio parahaemolyticus (tlh+, tdh–), a Gram-negative bacterium that causes gastrointestinal illness in humans, isolated from four species of aquaculture shrimp in Jiangsu, China. We found that 59 of the 997 isolates (5.9%) were ICE-positive, and of these, 9 isolates tested positive for all resistance genes. BLAST analysis showed that similarity for the eight strains to V. parahaemolyticus was 99%. Tracing the V. parahaemolyticus genotypes, showed no significant relevance of genotype among the antimicrobial resistance strains bearing the ICEs or not. Thus, in aquaculture, ICEs are not the major transmission mediators of resistance to antibiotics or heavy metals. We suggest future research to elucidate mechanisms that drive transmission of resistance determinants in V. parahaemolyticus.
Introduction
Microbes rapidly acquire or donate new genes and phenotypes through the process of horizontal gene transfer between organisms that is a key driver of microbial evolution (Pan et al., 2019). Integrative and conjugative elements (ICEs) are self-transmissible modular mobile genetic elements (MGEs) integrated into a host genome that are passively propagated during chromosomal replication and cell division, and mediate the acquisition of complex new traits in bacteria (Johnson and Grossman, 2015). Recent studies have shown these MGEs contain cargo genes encoding traits including resistance, virulence, novel carbon source metabolism, and degradation of aromatic compounds, that may benefit the recipient bacteria (Rubio-Cosials et al., 2018; Xu et al., 2018). ICEs tend to be mosaic and modular, ranging from 20 to >500 kb. ICEs are excised from the host chromosome and then transfer to recipients via conserved conjugation machinery in the type IV secretion system (Flores-Ríos et al., 2019), prior to reintegration into the host chromosome.
Vibrio parahaemolyticus is a Gram-negative, halophilic, mesophilic, aerobic bacterium common in warm climate marine and estuarine environments. Pathogenic strains in food cause serious health issues to humans, including gastroenteritis, septicemia, and wound infection (He et al., 2016). Shrimp represents an important reservoir of V. parahaemolyticus, particularly in fresh and refrigerated stock, but it is also recorded from frozen stock. China is the world’s largest producer of aquatic products (He et al., 2016). However, industrial development and use of antimicrobials in aquaculture have led to increased heavy metal pollution and development of multidrug resistant (MDR) bacteria that are problematic in many aquatic systems as they drive incidence of food-borne illnesses (Lopatek et al., 2018). Previous studies have revealed that bacteria can acquire resistance via conjugation or transformation to induce a wide variety of disease and adapt to the harsh environment (Matyar, 2012). The World Health Organization (WHO) produced a global map of antimicrobial resistance, warning that a “post-antibiotic” world could soon become a reality in April 2014 (WHO, 2014). Recent studies indicated that drugs which were once lifesavers are now worthless, for instance, chloramphenicol, once a physician’s first choice against typhoid, is no longer effective in many parts of the world and resistance has spread around the world (Woolhouse and Farrar, 2014). Antimicrobial resistance is a global problem that requires global solutions, better surveillance is essential. Nevertheless, to date, no global approaches were conducted on further demonstrating the characterization of the V. parahaemolyticus isolates present in shrimp-production industry, despite their great significance in economy and human health.
The discovery and early studies of ICEs were stimulated by interest in bacterial resistance to antibiotics and heavy metals, and how that resistance was spread. MGEs with ICE-like properties have been described in several species of Gammaproteobacteria, particularly Vibrio (Liu et al., 2019). However, few studies report on ICEs in V. parahaemolyticus isolates from Asia. Hence, in this study, we focused on analyzing the V. parahaemolyticus strains from different shrimp samples in Jiangsu, China to determine the antibiotic and heavy metal resistance of these bacteria and to investigate the relationship between antibiotic and heavy metal. Molecular characterization and phenotypes of antibiotic resistance and heavy metals have been characterized. The information will facilitate the better understanding of this bacterium and facilitate related risk assessment and health management for consuming seafood.
Materials and Methods
Sample Collection
Freshwater shrimp (Procambarus clarkii, Macrobrachium nipponense, Penaeus vannamei, and Macrobrachium rosenbergii), which are commonly shrimp breeds in China were collected once a month from Xuzhou Kaiming Fish Market, Jiangsu, China from May to October 2016–2018. P. clarkii and M. nipponense are key economic species in Jiangsu; P. vannamei and M. rosenbergii are cultured widely in the littoral provinces of southeastern China. We randomly collected samples of each species following a modified version of a standard protocol (Kaysner and DePaola, 2004). Samples were placed in sterile plastic bags (Shanghai Sangon Biological Engineering Technology and Services Co., Ltd., Shanghai, China) and taken to the laboratory on ice for homogenization within 2 h of collection.
V. parahaemolyticus Isolation and Identification
Vibrio parahaemolyticus was isolated and identified as described by the Chinese Government Standard (GB17378-2007) and He et al. (2016). Shrimp (25 g) were rinsed with sterile saline solution then homogenized for 60 s in sterile bags (Stomacher) containing 225 mL of sterile saline. Serial 10-fold dilutions (to 1:104) were prepared, and 100 μl of each dilution were spread on thiosulfate citrate-bile salts-sucrose plates (TCBS; Beijing Land Bridge Technology Company Ltd., Beijing, China) which were incubated for 16 h at 37°C. Putative V. parahaemolyticus colonies (which are green on TCBS) were placed separately in wells of 96-well microtiter plates containing 200 μl of sterile alkaline peptone water plus 3% NaCl (pH 8.5 ± 0.2).
Screening and Identification of Virulence and ICE Genes
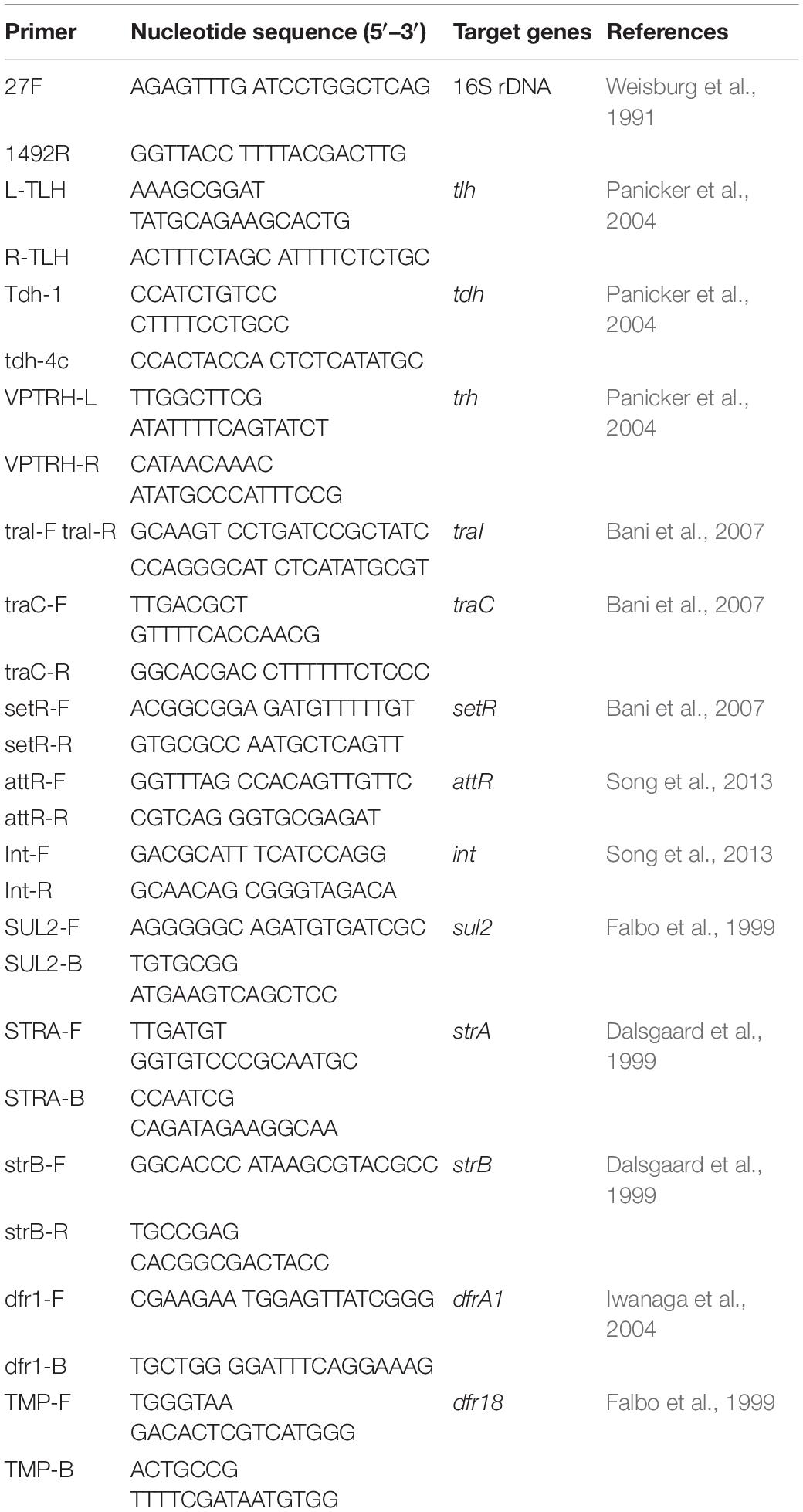
Presumptive V. parahaemolyticus colonies (N = 25 per species of shrimp) were selected, screened, and identified using PCR-based screening of the species-specific marker gene thermolabile hemolysin (tlh). Table 1 lists primers used in this study. In tlh-positive V. parahaemolyticus strains, the virulence-associated genes thermostable direct hemolysin (tdh) and TDH-related hemolysin (trh), SXT/R391-like ICE conserved genes (int, attR, traC, setR, and traI), and the typical resistance genes for streptomycin (strA/strB), trimethoprim (dfrA1/dfr18), and sulfamethoxazole/trimethoprim (sul2) were identified using primers as described by He et al. (2015a, 2016) and Beker et al. (2018). Strain taxonomy was determined from 16S rRNA gene sequences, obtained using primer pair 27F and 1492R; sequencing was performed by Shanghai Sangon Biological Engineering Technology and Services Co., Ltd., (Shanghai, China) (He et al., 2016). V. parahaemolyticus ATCC33846 (tdh+, trh–) and ATCC17802 (tdh–, trh+) were used as positive controls. Genomic DNA was prepared using a MiniBest bacterial genomic DNA extraction kit (v. 2.0; Japan TaKaRa BIO, Dalian, China). DNA was amplified using a Mastercycler pro PCR thermal cycler (Eppendorf, Hamburg, Germany). DNA sequences were assembled into contigs using ContigExpress software1. Protein functions were analyzed using BLAST2.
Susceptibility to Antimicrobials and Heavy Metals
In vitro susceptibility of isolates to antimicrobial agents according to the guidance of the Performance Standards for Antimicrobial Disk Susceptibility Tests of the Clinical and Laboratory Standards Institute (CLSI) (2006, Approved Standard-Ninth Edition, M2-A9, Vol. 26 No. 1) following the approach described by Song et al. (2013) and He et al. (2015a). Mueller-Hinton agar medium (Oxoid, United Kingdom), and the discs (Oxoid, United Kingdom) were used in this study. Examined antimicrobial agents included: 30 μg of chloramphenicol (CHL); 10 μg of gentamicin (CN); 25 μg of sulfamethoxazole/trimethoprim 19:1 (SXT); 5 μg of rifampin (RIF); 30 μg of tetracycline (TET); 10 μg of ampicillin (AMP); 100 μg of spectinomycin (SPT); 30 μg of kanamycin (KAN); 5 μg of trimethoprim (TM); and 10 μg of streptomycin (STR). The assays were performed in triplicate experiments, and reference strain Escherichia coli ATCC25922 was purchased from the Institute of Industrial Microbiology (Shanghai, China) and used for quality control. Broth Dilution Testing (microdilution) was used to measure quantitatively the minimal inhibitory concentration (MIC) in vitro of the tested heavy metals against the strains, according to the Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically (2006, CLSI, Approved Standard-Seventh Edition, M7-A7, Vol. 26 No. 2). The heavy metals tested were: NiCl2, CrCl3, CdCl2, PbCl2, CuCl2, ZnCl2, BaCl2, and HgCl2 (Sinopharm Chemical Reagent Co., Ltd., Shanghai, China); E. coli K12 MG1655 strain was used as the control.
Molecular Typing of V. parahaemolyticus Strains
Vibrio parahaemolyticus was cultured in Luria-Bertani broth (Beijing Land Bridge Technology Co.) following methods described by He et al. (2015b). Genomic DNA was extracted using a CHEF Bacterial DNA Plug Kit (Bio-Rad Laboratories, Hercules, CA, United States). Agarose plugs were prepared by mixing equal volumes of cell suspension, and each plug was digested using NotI (Japan TaKaRa BIO). Electrophoresis was performed at 6 V/cm, 14°C, at a field angle of 120°, using 1% SeaKem Gold agarose (Lonza, Basel, Switzerland). Pulsed-field gel electrophoresis (PFGE) patterns were visualized under 260-nm light; images were recorded using the UVP EC3 Imaging system (UVP LLC), and data were analyzed using NTSYSpc 2.10e software.
Results and Discussion
V. parahaemolyticus Isolation and Identification
Procambarus clarkii, M. nipponense, P. vannamei, and M. rosenbergii are common species of shrimp consumed in Jiangsu, China, and PCR analysis showed that 997 of 1800 (55.4%) bacterial isolates obtained from them tested positive for the V. parahaemolyticus-specific tlh gene. There were distinct temporal patterns of tlh-positive prevalence (Figure 1); over the 3-year study, tlh gene abundance was greater during the summer months (July and August) when temperature is highest.
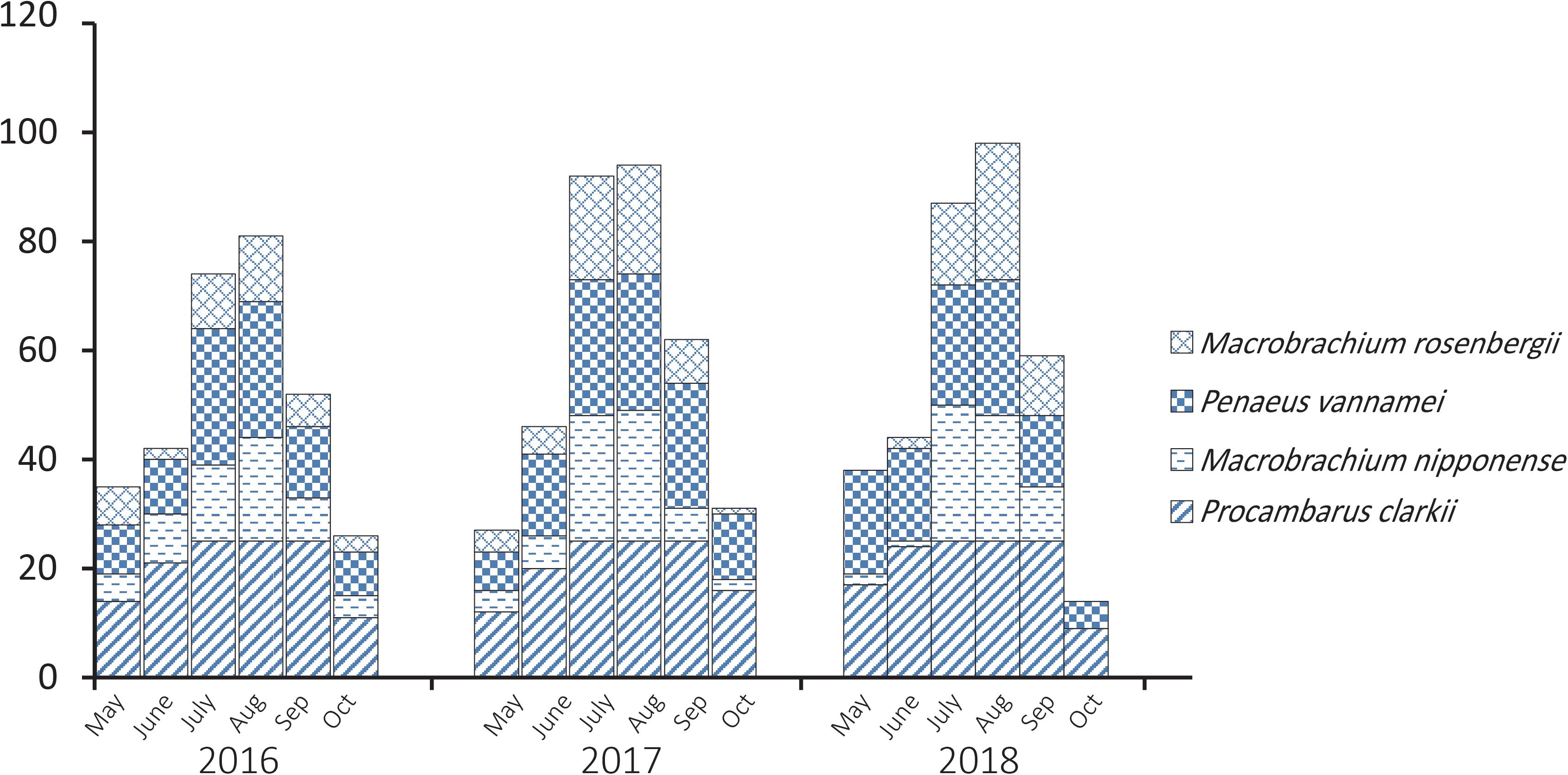
Figure 1. Temporal patterns of prevalence of Vibrio parahaemolyticus isolated from aquaculture shrimp.
Prevalence of Virulence Associated-Genes and Conserved ICE Genes
Pathogenic V. parahaemolyticus produces two major toxic proteins, TDH and TRH, that are important in the diarrheal diseases caused by this species (Boyd et al., 2008). We tested the 997 tlh-positive strains for the presence of virulence-associated toxin genes tdh and trh, and found that most isolates were considered not virulent; we did not amplify tdh from any isolate, and amplified trh from only two isolates (obtained from P. clarkii in August 2018). Similar very low incidence of pathogenic V. parahaemolyticus has been reported in many non-clinical samples (He et al., 2016; Hu and Chen, 2016; Martinez-Urtaza et al., 2016; Lopatek et al., 2018; Zhao et al., 2018; Jiang et al., 2019).
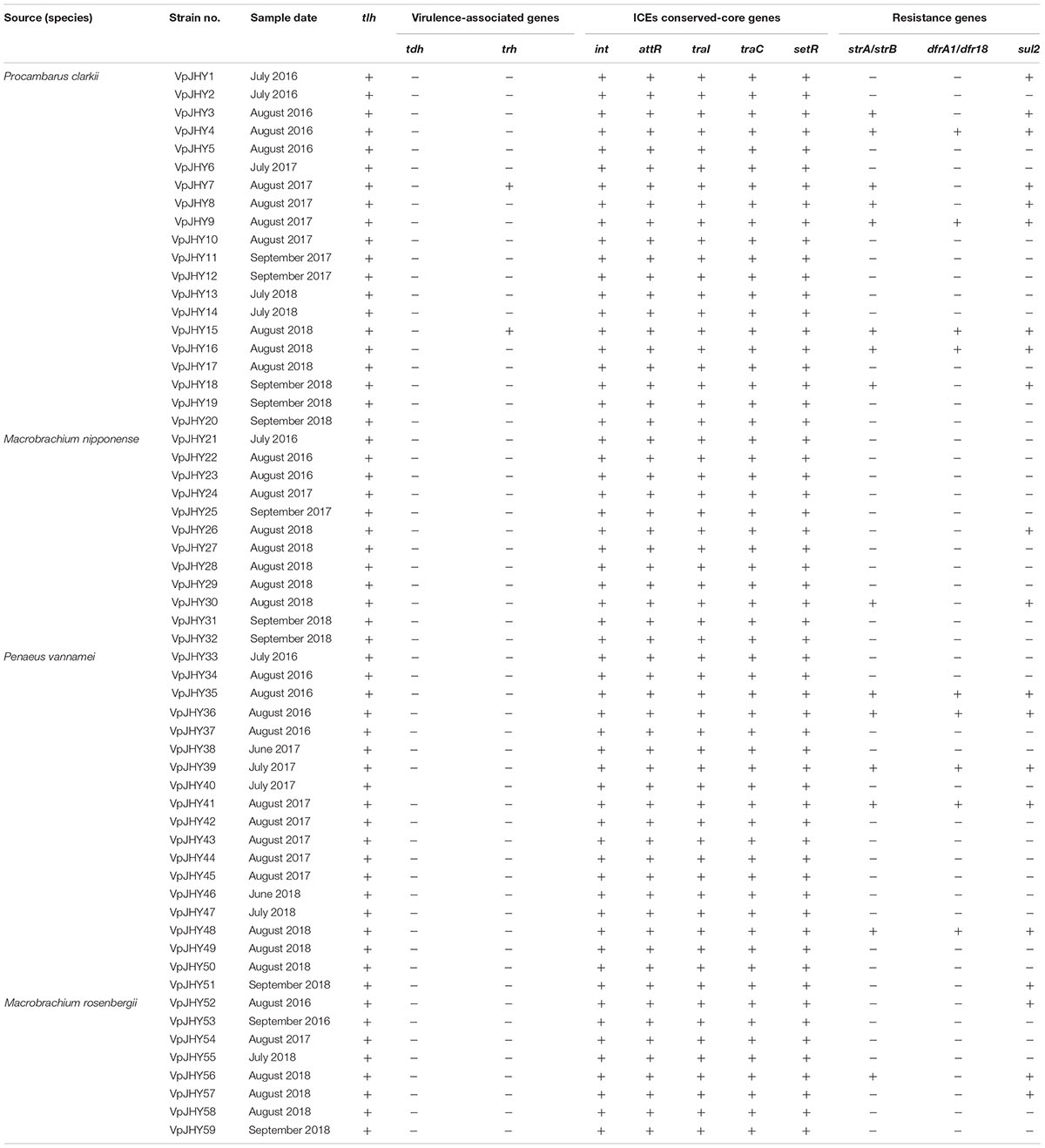
Analysis of the highly conserved core genes of SXT/R391-like ICEs revealed that 59 (5.9%) of the isolates tested positive for all five genes (int, attR, traC, setR, and traI). Occurrence was highest in isolates recovered from P. clarkii (33.9%), followed by P. vannamei (32.2%), M. nipponense (20.3%), and M. rosenbergii (13.6%), and resistance genes strA/strB, dfrA1/dfr18, and sul2 were recorded in 25.4, 15.3, and 33.9% of the 59 isolates, respectively, with nine strains testing positive for all three resistance genes (Table 2).
Antimicrobial Susceptibility and Heavy Metal Tolerance
Our results revealed distinct antibiotic-resistance patterns for the 59 isolates that were positive for the highly conserved ICE genes. All isolates were AMP resistant, and resistance to STR, TM, RIF, and SXT was 25.4, 22.0, 18.6, and 16.9%, respectively; resistance to CHL, SPT, and KAN was 15.3% (Figure 2). We found that approximately 84.7, 81.3, 74.6, and 72.9% of the isolates exhibited intermediate susceptibility to SPT, KAN, STR, and TM, and while 15.3% of isolates showed intermediate patterns of susceptibility to CN and TET, none of the isolates was resistant to these two antibiotics. We found nine isolates tested positive for the three resistance genes strA/strB, dfrA1/dfr18, sul2, and these strains were resistant to CHL, SXT, RIF, AMP, SPT, KAN, TM, and STR, with intermediate susceptibility to CN and TET. BLAST analysis showed that the 16S rRNA gene sequence of isolate VpJHY15 shared 99% similarity with that from Proteus vulgaris3, and similarity for the other eight strains (VpJHY4, VpJHY9, VpJHY16, VpJHY35, VpJHY36, VpJHY39, VpJHY41, and VpJHY48) to V. parahaemolyticus was 99%.
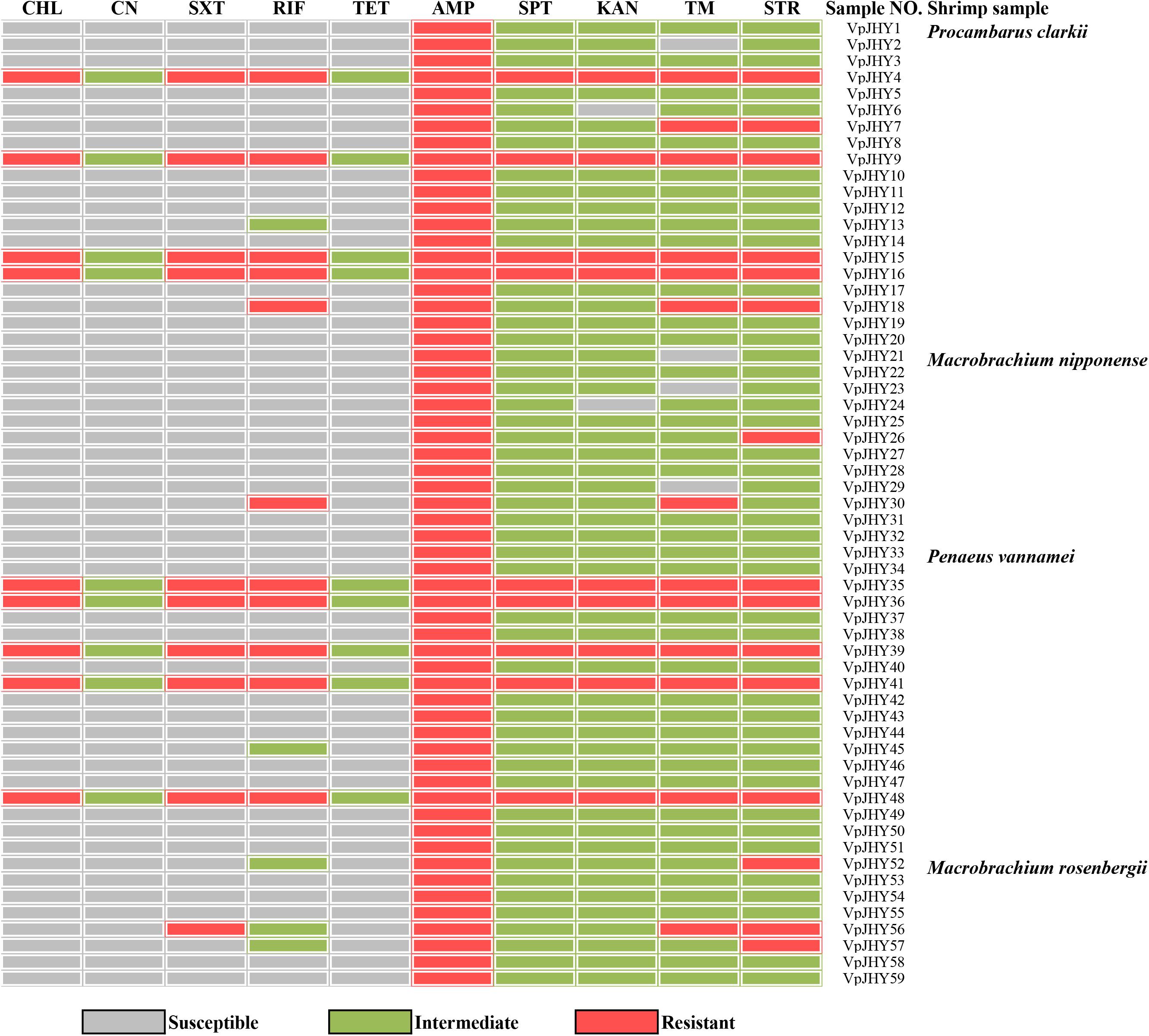
Figure 2. Drug resistance of V. parahaemolyticus isolates. CHL, chloramphenicol; CN, gentamicin; SXT, sulfamethoxazole-trimethoprim; RIF, rifampicin; TET, tetracycline; AMP, ampicillin; SPT, spectinomycin; KAN, kanamycin; TM, trimethoprim, and STR, streptomycin.
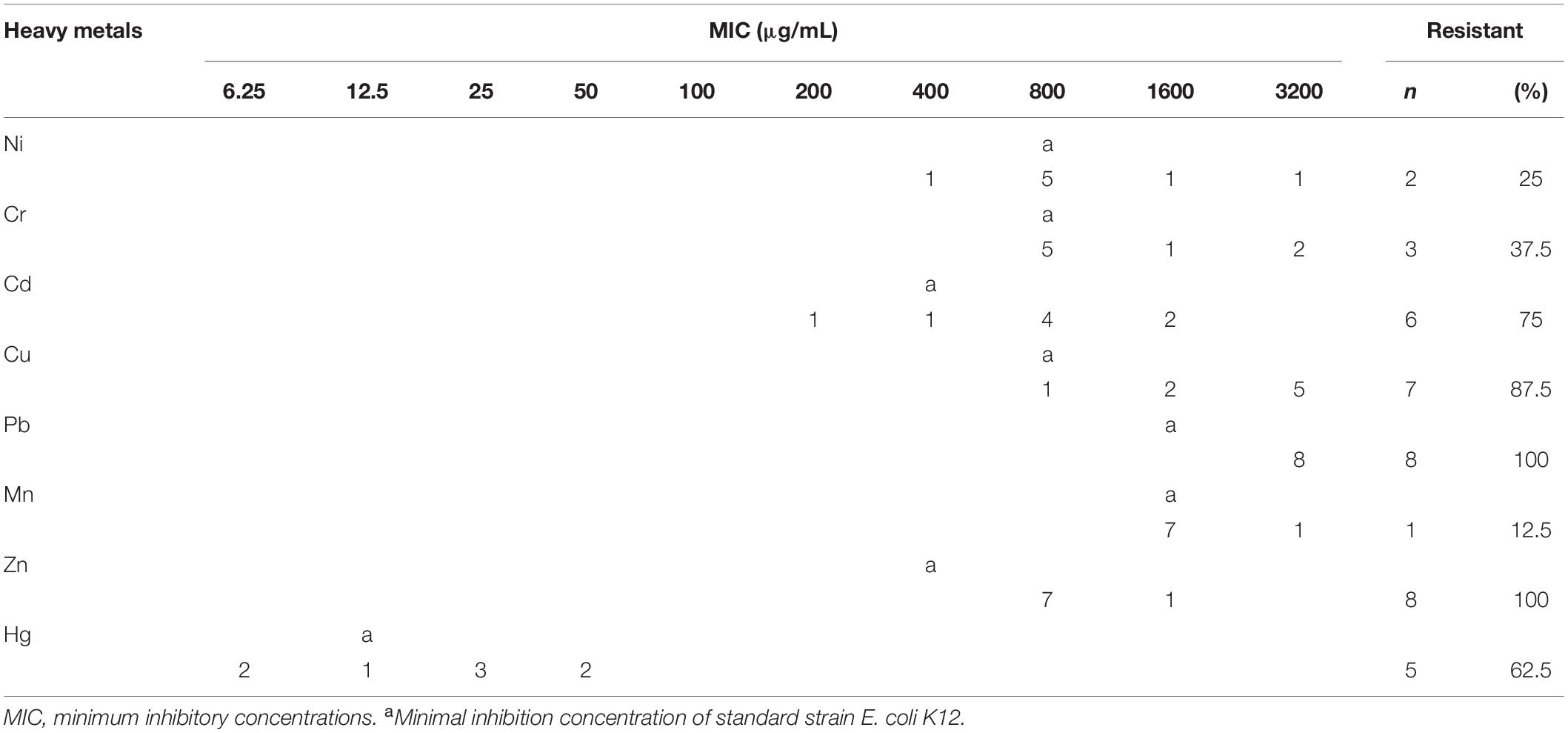
Therefore, in this study, eight V. parahaemolyticus isolates tested positive for SXT/R391-like ICE conserved genes (int, attR, traC, setR, traI) and associated typical resistance genes (strA/strB, dfrA1/dfr18, sul2). These strains may be described as having MDR phenotypes because they were resistant to at least one agent in ≥3 categories of antimicrobial (Thapa Shrestha et al., 2015). These isolates were also examined for susceptibility to heavy metals (Malik and Aleem, 2011), we found minimal inhibitory concentrations (MICs) of 3200 μg/mL for Ni2+, Cr3+, Cd2+, Cu2+, Pb2+, and Mn2+; 1600 μg/ml for Zn2+; and 50 μg/ml for Hg2+ (Table 3). All strains were resistant to Zn2+ and Pb2+, and most also displayed tolerance to Cu2+ (87.5%), Cd2+ (75%), and Hg2+ (62.5%), while a few were resistant to Cr3+ (37.5%), Ni2+ (25%), and Mn2+ (12.5%). V. parahaemolyticus isolates derived from the four shrimp species showed tolerance to at least four heavy-metal agents (Table 4). While we found that resistance to heavy metals in V. parahaemolyticus did not vary with shrimp species, tolerance was shown to be very prevalent in ICE-positive strains with more than eight antibiotic resistance phenotypes. These contrasting results may be a result of inappropriate, variable releases of industrial wastes to aquaculture environments (Jiang et al., 2019), because industrial pollutants enhance selection for antibiotic resistance and vice versa. Abundant double-resistant bacteria threaten human health if contaminated products are consumed (Silva et al., 2018).
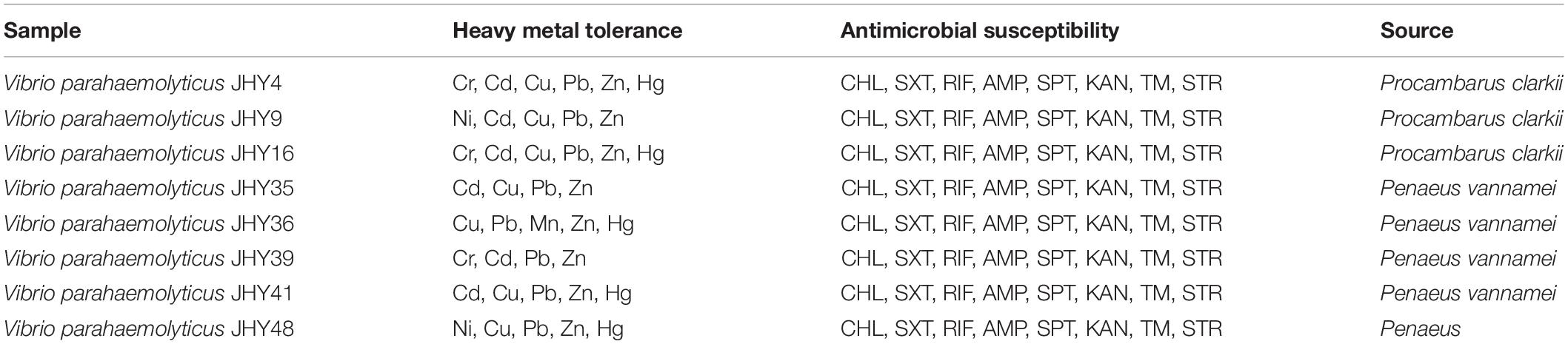
Table 4. Susceptibility of V. parahaemolyticus harboring the SXT/R391 family of integrative and conjugative elements to heavy metals and antibiotics.
Phylogenetic Relationships of Resistant V. parahaemolyticus Isolates
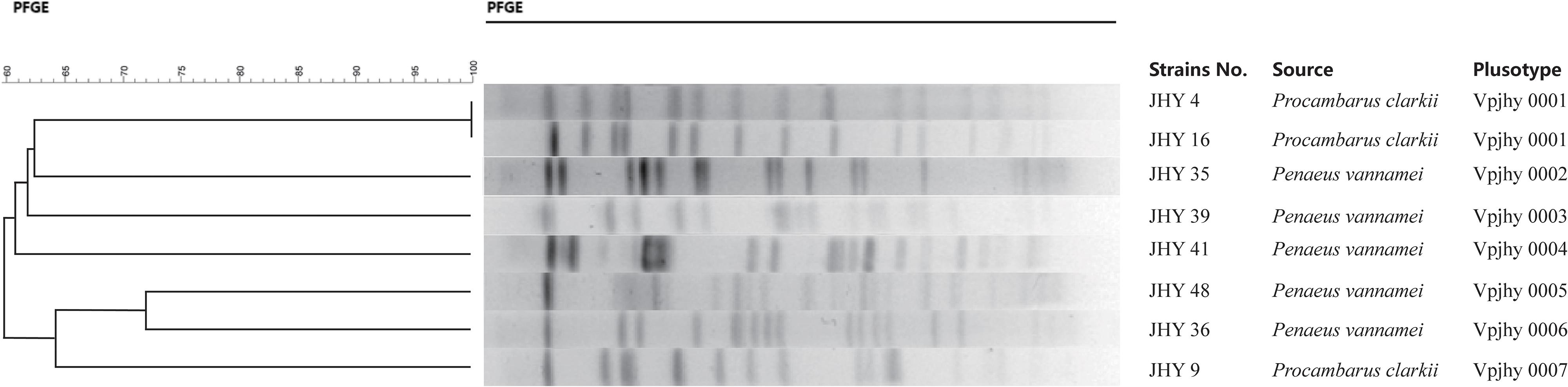
Genomic DNA of the eight V. parahaemolyticus strains was individually digested with the restriction endonuclease NotI, and the resulting DNA fragments were resolved by PFGE. This analysis revealed different genomic finger prints of the strains tested (Figure 3). Fingerprinting analysis of the relatedness of the eight ICE-positive isolates produced 14 to 19 restriction bands that ranged from 20.5 to 1135 kb. Cluster analysis of the PFGE profiles revealed seven pulsotypes with ≥87% similarity, which indicates isolates belonging to the same epidemic strain (Seifert et al., 2005). The isolates were assigned to four distinct clusters, with 62.5% assigned to Clusters A to C and one that was more distantly related assigned to Cluster D. Simpson’s diversity index (0.9872) indicated high diversity among these isolates. Clustering analysis of the genomic fingerprints revealed eight distinguishable NotI-PFGE types, demonstrating that the MDR of ICEs-positive strains isolated for the prevalence analysis exhibited various genotypes. According to our previous studies, the presence or absence of ICEs has no significant relevance among these strains in terms of antimicrobial resistance (He et al., 2015a), it indicating that resistance determinants may spread among genetic lineages within the V. parahaemolyticus population. MGEs that carry resistance genes (Song et al., 2013) may be responsible for the high variation among the genotypes and resistance phenotypes of isolates. Therefore, we suggest future research to elucidate the precise mechanisms of resistance determinant transmission in V. parahaemolyticus populations.
Conclusion
Bacteria secrete toxin proteins or effectors into external media or directly into eukaryotic target cells to facilitate adaption to environmental stress conditions; this response is key during the process of infection (Park et al., 2004; Cascales, 2008; Tseng et al., 2009). Previous studies have revealed that clinical isolates of V. parahaemolyticus produce beta-hemolysis in Wagatsuma agar, in a process known as the Kanagawa phenomenon that is linked to TDH-secreted proteins. These proteins have been recognized as primary virulence factors and effectors (Naim et al., 2001; Ono et al., 2006; Igbinosa and Okoh, 2008). Studies from different regions of the world show that tdh and/or trh genes are found in 90–99.8% of clinical strains, whereas only 0.2–10% of environmental V. parahaemolyticus isolates are potentially pathogenic, based on the presence of tdh and/or trh (Letchumanan et al., 2015b; Hu and Chen, 2016; Taiwo et al., 2017; Rortana et al., 2018; Jiang et al., 2019).
Previous studies disclosed that V. parahaemolyticus is a very diverse species and is an opportunistic pathogen in aquatic environments that is highly successful in adapting to changing environmental conditions (Song et al., 2013; Letchumanan et al., 2015a, b). Increasing aquaculture production and industrialization has led to large amounts of antibiotics being used to prevent or treat disease outbreaks in shrimp farming; consequently, multidrug resistant (MDR) and heavy metals resistant pathogens have emerged and posed serious problems in many aquatic systems. It revealed that bacteria could acquire resistance via conjugation or transformation, allowing them to adapt to the harsh environment and to cause a wide variety of diseases (Matyar, 2012). Incidents of human food poisoning from aquaculture products pose a becoming a serious clinical issue.
In the present study, we evaluated the prevalence, antimicrobial susceptibility, heavy metal resistance and genotypes of V. parahaemolyticus from four species of shrimp obtained from fish markets in Jiangsu (in 2016–2018), China. The results showed that ICEs-positive isolates have been given more antibiotic resistance, and the MDR strains also showed more heavy metal resistance. Consistent with previous reports (Song et al., 2013; Hu and Chen, 2016; Kang et al., 2018), we found that AMP resistance dominated among the isolates and was present in all samples tested. Although TET, sulfonamides, and quinolones are used widely in aquaculture (Holmström et al., 2003), we did not find evidence of resistance to TET in the V. parahaemolyticus isolates. Nine isolates that tested positive for all resistance genes (strA/strB, dfrA1/dfr18, sul2) exhibited intermediate susceptibility to CN and TET, indicating potential resistance to these two antibiotics. Our results for susceptibility of the V. parahaemolyticus isolates to CHL contrast with previous work (He et al., 2016), where we found all ICE-positive strains were resistant to CHL, possibly as a result of the ban, since 2002, of the use of CHL and derivatives (including chloramphenicol succinate) in the breeding industry in China (China Department of Agriculture, Bulletin No. 193). We suggest that ICE-positive V. parahaemolyticus isolates may indicate tolerance to heavy metal agents.
Wide usage of teracyclines, sulfonamides, and (fluoro) quinolones in aquaculture has been reported (Holmström et al., 2003). In addition, animal fecal used as fertilizer of aquaculture ponds is a common practice in integrated aquaculture-agriculture system herein. Manure and urine from field-herding cattles and goats were continuously discharged directly into ponds, which could change the bacterial community composition and bring more antimicrobials and even heavy metals in the aquaculture environment. In the Vibrionaceae, ICEs have been demonstrated to bestow resistance to multiple antibiotics and some complex new traits through horizontal gene transfer, which could be beneficial under certain environmental conditions (Makino et al., 2003; Balado et al., 2013). However, the present study revealed that ICEs are not the major transmission mediators of resistance to antibiotics or heavy metals. Thus, we speculate that many of the antibiotic resistance genes are intrinsic, and their expressions are activated when the environmental conditions became hostile. Furthermore, few of them are commonly transferred via conjugation or transformation. These data will aid future research in controlling of aquaculture diseases, forecasting food safety incidence and improving our understanding of V. parahaemolyticus prevalence and behavior in the aquaculture.
Data Availability
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
YH, SW, XZ, and JZ participated in the design and/or discussion of the study. FS, BH, and XL carried out the major experiments. YH and SW analyzed the data. YH wrote the manuscript. SW, XZ, and JZ revised the manuscript. All authors read and approved the final version of the manuscript.
Funding
This work was supported by grants from the National Science Foundation for Young Scientists of China (No. 31701566), the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (No. 18KJB550011), the Subsidized Project of Brand Major Construction in Colleges and Universities of Jiangsu Province (PPZY2015B153 Food Science and Engineering), the Research Projects of Xuzhou University of Technology (No. XCX2019136), and the Henan Key Laboratory of Cold Chain Food Quality and Safety Control (No. CCFQ2018-YB-03).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Balado, M., Lemos, M. L., and Osorio, C. R. (2013). Integrating conjugative elements of the SXT/R391 family from fish-isolated Vibrios encode restriction-modification systems that confer resistance to bacteriophages. FEMS Microbiol. Ecol. 83, 457–467. doi: 10.1111/1574-6941.12007
Bani, S., Mastromarino, P. N., Ceccarelli, D., Le Van, A., Salvia, A. M., Viet, Q. T. N., et al. (2007). Molecular characterization of ICEVchVie0 and its disappearance in Vibrio cholerae O1 strains isolated in 2003 in Vietnam. FEMS Microbiol. Lett. 266, 42–48.
Beker, M., Rose, S., Lykkebo, C. A., and Douthwaite, S. (2018). Integrative and conjugative elements (ICEs) in Pasteurellaceae species and their detection by multiplex PCR. Front. Microbiol. 9:1329. doi: 10.3389/fmicb.2018.01329
Boyd, E. F., Cohen, A. L., Naughton, L. M., Ussery, D. W., Binnewies, T. T., Stine, O. C., et al. (2008). Molecular analysis of the emergence of pandemic Vibrio parahaemolyticus. BMC Microbiol. 8:110. doi: 10.1186/1471-2180-8-110
Cascales, E. (2008). The type VI secretion toolkit. EMBO Rep. 9, 735–741. doi: 10.1038/embor.2008.131
Dalsgaard, A., Forslund, A., Tam, N. V., Vinh, D. X., and Cam, P. D. (1999). Cholera in Vietnam: changes in genotypes and emergence of class I integrons containing aminoglycoside resistance gene cassettes in Vibrio cholerae O1 strains isolated from 1979 to 1996. J. Clin. Microbiol. 37, 734–741.
Falbo, V., Carattoli, A., Tosini, F., Pezzella, C., Dionisi, A. M., and Luzzi, I. (1999). Antibiotic resistance conferred by a conjugative plasmid and a class I integron in Vibrio cholerae O1 El Tor strains isolated in Albania and Italy. Antimicrob. Agents Chemother. 43, 693–696.
Flores-Ríos, R., Moya-Beltrán, A., Pareja-Barrueto, C., Arenas-Salinas, M., Valenzuela, S., Orellana, O., et al. (2019). The type IV secretion system of ICEAfe1: formation of a conjugative pilus in Acidithiobacillus ferrooxidans. Front. Microbiol. 10:30. doi: 10.3389/fmicb.2019.00030
He, Y., Jin, L., Sun, F., Hu, Q., and Chen, L. (2016). Antibiotic and heavy-metal resistance of Vibrio parahaemolyticus isolated from fresh shrimps in Shanghai fish markets, China. Environ. Sci. Pollut. Res. Int. 23, 15033–15040. doi: 10.1007/s11356-016-6614-4
He, Y., Tang, Y., Sun, F., and Chen, L. (2015a). Detection and characterization of integrative and conjugative elements (ICEs)-positive Vibrio cholerae isolates from aquacultured shrimp and the environment in Shanghai, China. Mar. Pollut. Bull. 101, 526–532. doi: 10.1016/j.marpolbul.2015.10.062
He, Y., Wang, H., and Chen, L. (2015b). Comparative secretomics reveals novel virulence-associated factors of Vibrio parahaemolyticus. Front. Microbiol. 6:707. doi: 10.3389/fmicb.2015.00707
Holmström, K., Gräslund, S., Wahlström, A., Poungshompoo, S., Bengtsson, B. E., and Kautsky, N. (2003). Antibiotic use in shrimp farming and implications for environmental impacts and human health. Int. J. Food Sci. Technol. 38, 255–266.
Hu, Q., and Chen, L. (2016). Virulence and antibiotic and heavy metal resistance of Vibrio parahaemolyticus isolated from crustaceans and shellfish in shanghai, China. J. Food Prot. 79, 1371–1377. doi: 10.4315/0362-028X.JFP-16-031
Igbinosa, E. O., and Okoh, A. I. (2008). Emerging Vibrio species: an unending threat to public health in developing countries. Res. Microbiol. 159, 495–506. doi: 10.1016/j.resmic.2008.07.001
Iwanaga, M., Toma, C., Miyazato, T., Insisiengmay, S., Nakasone, N., and Ehara, M. (2004). Antibiotic resistance conferred by a class I integron and SXT constin in Vibrio cholerae O1 strains isolated in Laos. Antimicrob. Agents Chemother. 48, 2364–2369.
Jiang, Y., Chu, Y., Xie, G., Li, F., Wang, L., Huang, J., et al. (2019). Antimicrobial resistance, virulence and genetic relationship of Vibrio parahaemolyticus in seafood from coasts of Bohai Sea and Yellow Sea, China. Int. J. Food Microbiol. 290, 116–124. doi: 10.1016/j.ijfoodmicro.2018.10.005
Johnson, C. M., and Grossman, A. D. (2015). Integrative and conjugative elements (ICEs): what they do and how they work. Annu. Rev. Genet. 49, 577–601. doi: 10.1146/annurev-genet-112414-055018
Kang, C. H., Shin, Y., Yu, H., Kim, S., and So, J. S. (2018). Antibiotic and heavy-metal resistance of Vibrio parahaemolyticus isolated from oysters in Korea. Mar. Pollut. Bull. 135, 69–74. doi: 10.1016/j.marpolbul.2018.07.007
Kaysner, C., and DePaola, A. (2004). Bacteriological Analytical Manual Chapter 9: Vibrio. Silver Spring, MD: Food U.S. and Drug Administration.
Letchumanan, V., Pusparajah, P., Tan, L. T. H., Yin, W. F., Lee, L. H., and Chan, K. G. (2015a). Occurrence and antibiotic resistance of Vibrio parahaemolyticus from shellfish in Selangor, Malaysia. Front. Microbiol. 6:1417. doi: 10.3389/fmicb.2015.01417
Letchumanan, V., Yin, W. F., Lee, L. H., and Chan, K. G. (2015b). Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front. Microbiol. 6:33. doi: 10.3389/fmicb.2015.00033
Liu, M., Li, X., Xie, Y., Bi, D., Sun, J., Li, J., et al. (2019). ICEberg 2.0: an updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 47, D660–D665. doi: 10.1093/nar/gky1123
Lopatek, M., Wieczorek, K., and Osek, J. (2018). Antimicrobial resistance, virulence factors, and genetic profiles of Vibrio parahaemolyticus from seafood. Appl. Environ. Microbiol. 84:e00537-18. doi: 10.1128/AEM.00537-18
Makino, K., Oshima, K., Kurokawa, K., Yokoyama, K., Uda, T., Tagomori, K., et al. (2003). Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361, 743–749.
Malik, A., and Aleem, A. (2011). Incidence of metal and antibiotic resistance in Pseudomonas spp. From the river water, agricultural soil irrigated with wastewater and groundwater. Environ. Monit. Assess. 178, 293–308. doi: 10.1007/s10661-010-1690-2
Martinez-Urtaza, J., Powell, A., Jansa, J., Rey, J. L., Montero, O. P., Campello, M. G., et al. (2016). Epidemiological investigation of a foodborne outbreak in Spain associated with U.S. West Coast genotypes of Vibrio parahaemolyticus. Springerplus 5:87. doi: 10.1186/s40064-016-1728-1
Matyar, F. (2012). Antibiotic and heavy metal resistance in bacteria isolated from the Eastern Mediterranean Sea coast. B. Environ. Contam. Tox. 89, 551–556. doi: 10.1007/s00128-012-0726-4
Naim, R., Yanagihara, I., Iida, T., and Honda, T. (2001). Vibrio parahaemolyticus thermostable direct hemolysin can induce an apoptotic cell death in Rat-1 cells from inside and ouside of the cells. FEMS Microbiol. Lett. 195, 237–244.
Ono, T., Park, K. S., Ueta, M., Iida, T., and Honda, T. (2006). Identification of proteins secreted via Vibrio parahaemolyticus type III secretion system 1. Infect. Immun. 74, 1032–1042.
Pan, Z., Liu, J., Zhang, Y., Chen, S., Ma, J., Dong, W., et al. (2019). A novel integrative conjugative element mediates transfer of multi-drug resistance between Streptococcus suis strains of different serotypes. Vet. Microbiol. 229, 110–116. doi: 10.1016/j.vetmic.2018.11.028
Panicker, G., Call, D. R., Krug, M. J., and Bej, A. K. (2004). Detection of pathogenic Vibrio spp. In shellfish by using multiplex PCR and DNA microarrays. Appl. Environ. Microbiol. 70, 7436–7444.
Park, K. S., Ono, T., Rokuda, M., Jang, M. H., Okada, K., Iida, T., et al. (2004). Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect. Immun. 72, 6659–6665.
Rortana, C., Wajjwalku, W., Boonyawiwat, V., Hrianpreecha, C., Thongratsakul, S., and Amavisit, P. (2018). Antimicrobial resistance and pirAB-like profiles of Vibrio parahaemolyticus in Pacific white shrimp. Agric. Nat. Resour. 52, 377–381.
Rubio-Cosials, A., Schulz, E. C., Lambertsen, L., Smyshlyaev, G., Rojas-Cordova, C., Forslund, K., et al. (2018). Transposase-DNA complex structures reveal mechanisms for conjugative transposition of antibiotic resistance. Cell 173, 208–220.e20. doi: 10.1016/j.cell.2018.02.032
Seifert, H., Dolzani, L., Bressan, R., Van Der Reijden, T., Van Strijen, B., Stefanik, D., et al. (2005). Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J. Clin. Microbiol. 43, 4328–4335.
Silva, I. P., Carneiro, C. S., Saraiva, M. A. F., Oliveira, T. A. S., Sousa, O. V., and Evangelista-Barreto, N. S. (2018). Antimicrobial resistance and potential virulence of Vibrio parahaemolyticus isolated from water and bivalve mollusks from Bahia, Brazil. Mar. Pollut. Bull. 131, 757–762. doi: 10.1016/j.marpolbul.2018.05.007
Song, Y., Yu, P., Li, B., Pan, Y., Zhang, X., Cong, J., et al. (2013). The mosaic accessory gene structures of the SXT/R391-like integrative and conjugative elements derived from Vibrio spp. Isolated from aquatic products and environment in the Yangtze River Estuary, China. BMC Microbiol. 13:214. doi: 10.1186/1471-2180-13-214
Taiwo, M., Baker-Austin, C., Powell, A., Hodgson, E., Natås, O. B., and Walker, D. I. (2017). Comparison of toxR and tlh based PCR assays for Vibrio parahaemolyticus. Food Control 77, 116–120.
Thapa Shrestha, U., Adhikari, N., Maharjan, R., Banjara, M. R., Rijal, K. R., Basnyat, S. R., et al. (2015). Multidrug resistant Vibrio cholerae O1 from clinical and environmental samples in Kathmandu city. BMC Infect. Dis. 15:104. doi: 10.1186/s12879-015-0844-9
Tseng, T. T., Tyler, B. M., and Setubal, J. C. (2009). Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol. 9(Suppl. 1):S2. doi: 10.1186/1471-2180-9-S1-S2
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703.
Woolhouse, M., and Farrar, J. (2014). Policy: an intergovernmental panel on antimicrobial resistance. Nature 509, 555–557.
Xu, J., Jia, H., Cui, G., Tong, H., Wei, J., Shao, D., et al. (2018). ICEAplChn1, a novel SXT/R391 integrative conjugative element (ICE), carrying multiple antibiotic resistance genes in Actinobacillus pleuropneumoniae. Vet. Microbiol. 220, 18–23. doi: 10.1016/j.vetmic.2018.05.002
Zhao, S., Ma, L., Wang, Y., Fu, G., Zhou, J., Li, X., et al. (2018). Antimicrobial resistance and pulsed-field gel electrophoresis typing of Vibrio parahaemolyticus isolated from shrimp mariculture environment along the east coast of China. Mar. Pollut. Bull. 136, 164–170. doi: 10.1016/j.marpolbul.2018.09.017
Keywords: Vibrio parahaemolyticus, antimicrobial susceptibility, integrative and conjugative elements, heavy metal resistance, genotypes
Citation: He Y, Wang S, Zhang J, Zhang X, Sun F, He B and Liu X (2019) Integrative and Conjugative Elements-Positive Vibrio parahaemolyticus Isolated From Aquaculture Shrimp in Jiangsu, China. Front. Microbiol. 10:1574. doi: 10.3389/fmicb.2019.01574
Received: 25 March 2019; Accepted: 24 June 2019;
Published: 18 July 2019.
Edited by:
Daniela Ceccarelli, Research Executive Agency, European Commission, BelgiumReviewed by:
Maria M. Lleo, University of Verona, ItalyLearn-Han Lee, Monash University Malaysia, Malaysia
Biao Kan, National Institute for Communicable Disease Control and Prevention (China CDC), China
Copyright © 2019 He, Wang, Zhang, Zhang, Sun, He and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuai Wang, aGFuZHNvbWV3QGZveG1haWwuY29t; Jianping Zhang, NDIxMzI1MDUxQHFxLmNvbQ==; Xueyang Zhang, amp5ODYxMTIwQDE2My5jb20=
†These authors have contributed equally to this work
 Yu He
Yu He Shuai Wang1,2*†
Shuai Wang1,2*†