- Research Group of Microbiology, Department of Bioengineering Sciences, Vrije Universiteit Brussel, Brussels, Belgium
Two out of the three major uptake systems for arginine in Escherichia coli are encoded by the artJ-artPIQM gene cluster. ArtJ is the high-affinity periplasmic arginine-specific binding protein (ArgBP-I), whereas artI encodes the arginine and ornithine periplasmic binding protein (AO). Both ArtJ and ArtI are supposed to combine with the inner membrane-associated ArtQMP2 transport complex of the ATP-binding cassette-type (ABC). Transcription of artJ is repressed by arginine repressor (ArgR) and the artPIQM operon is regulated by the transcriptional regulators ArgR and Leucine-responsive regulatory protein (Lrp). Whereas repression by ArgR requires arginine as corepressor, repression of PartP by Lrp is partially counteracted by leucine, its major effector molecule. We demonstrate that binding of dimeric Lrp to the artP control region generates four complexes with a distinct migration velocity, and that leucine has an effect on both global binding affinity and cooperativity in the binding. We identify the binding sites for Lrp in the artP control region, reveal interferences in the binding of ArgR and Lrp in vitro and demonstrate that the two transcription factors act as competitive repressors in vivo, each one being a more potent regulator in the absence of the other. This competitive behavior may be explained by the partial steric overlap of their respective binding sites. Furthermore, we demonstrate ArgR binding to an unusual position in the control region of the lrp gene, downstream of the transcription initiation site. From this unusual position for an ArgR-specific operator, ArgR has little direct effect on lrp expression, but interferes with the negative leucine-sensitive autoregulation exerted by Lrp. Direct arginine and ArgR-dependent repression of lrp could be observed with a 25-bp deletion mutant, in which the ArgR binding site was artificially moved to a position immediately downstream of the lrp transcription initiation site. This finding is reminiscent of a previous observation made for the carAB operon encoding carbamoylphosphate synthase, where ArgR bound in overlap with the downstream promoter P2 does not block transcription initiated 67 bp upstream at the P1 promoter, and further supports the hypothesis that ArgR does not act as an efficient roadblock.
Introduction
Arginine is a particularly important amino acid for bacterial cell growth, physiology, and survival in stress conditions. It is not only essential for protein synthesis, but plays an equally important role in extreme acid resistance and pH homeostasis, is a precursor for the biosynthesis of the polyamines spermidine and spermine, and may serve as a source of nitrogen, carbon, and energy upon degradation via distinct catabolic routes (Charlier and Glansdorff, 2004; Reitzer, 2005; Leroy and Charlier, 2017). Hence, the biosynthesis, catabolism, uptake, and export of arginine are tightly regulated. Here, we focus on regulation of arginine uptake in Escherichia coli K-12 that possesses several transport systems of the ATP-binding cassette-type (ABC) for the import of arginine (Figure 1A), which is energetically more favorable than its biosynthesis (Saier, 2000; Hosie and Poole, 2001; Burkovski and Krämer, 2002; Davidson and Chen, 2004; Biemans-Oldehinkel et al., 2006). These various import systems are encoded by two gene clusters, artPIQM-artJ and argT-hisJQMP (Figure 1B). They differ in substrate specificity and affinity for arginine, and exhibit differences in the regulation of synthesis and activity (for an overview, see Caldara et al., 2007). These differences indicate that distinct transport mechanisms may be active in different growth conditions and fulfill different physiological needs. Each gene cluster encodes two periplasmic binding proteins, ArtJ and ArtI and HisJ and ArgT, respectively. ArtJ (alias ArgBP-I) is the arginine-specific binding protein, and has the highest affinity for arginine (Kd 0.4 μM) (Rosen, 1971; Celis, 1981). ArtI binds arginine and its precursor ornithine (Wissenbach et al., 1993, 1995), ArgT (LAO) binds the basic amino acids lysine, arginine (Kd 1.5 μM), and ornithine (Celis et al., 1973; Rosen, 1973), whereas HisJ binds histidine (Kd 0.11 μM) and with low affinity also arginine (Kd 10 μM) (Kustu and Ames, 1973). HisJ and ArgT, which share 70% amino acid sequence identity, associate with the HisQMP2 complex consisting of the integral membrane proteins, HisQ and HisM, and two membrane-associated HisP subunits that bind ATP and provide the energy for active transport (Kerppola et al., 1991; Ames et al., 2001). In analogy, ArtJ and ArtI are supposed to associate with the ArtQMP2 complex that is similar to HisQMP2.
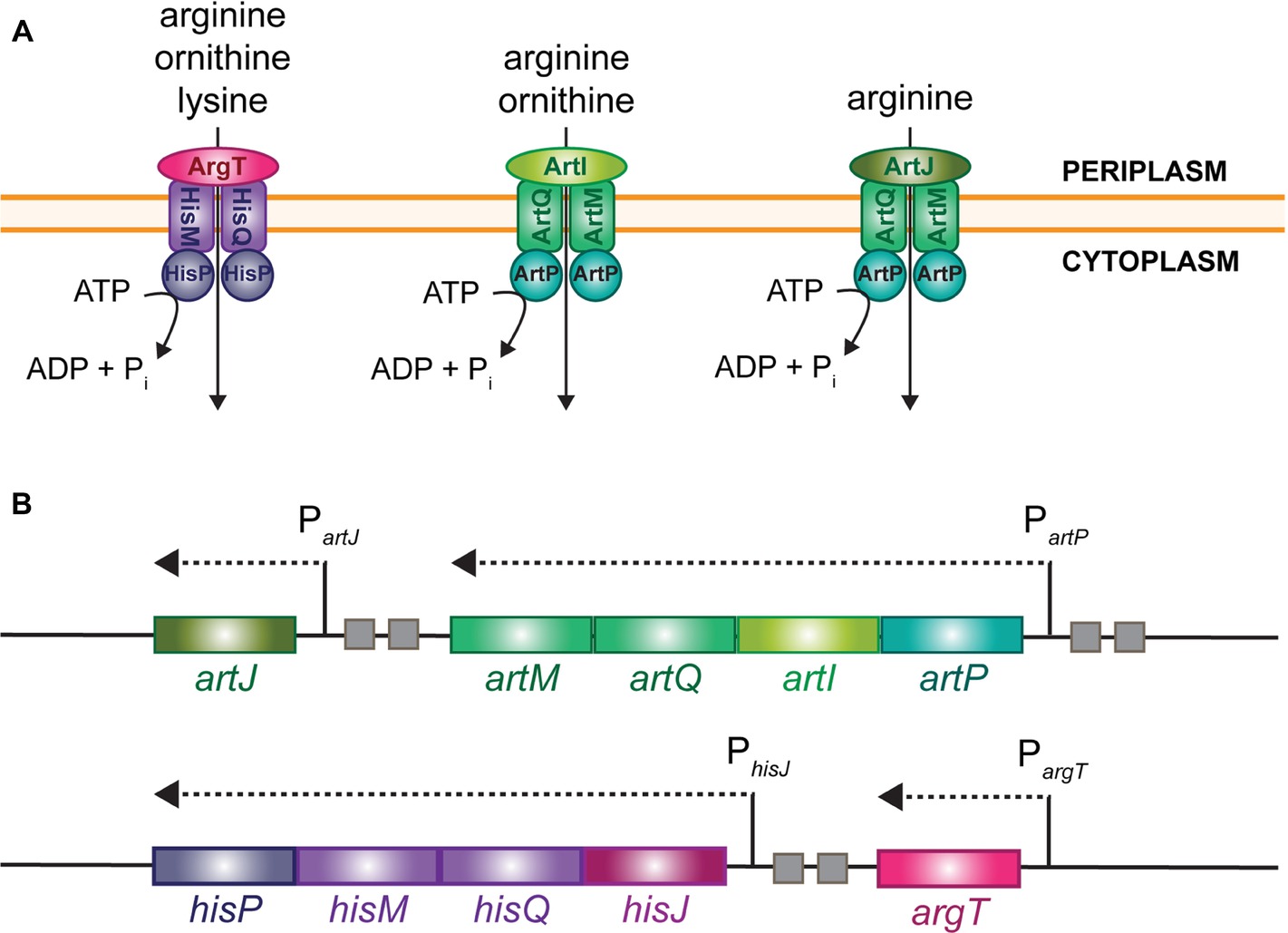
Figure 1. Schematic representation of the different import systems for arginine in E. coli (A) and organization of the cognate gene clusters on the chromosome (B). Small squares represent ARG boxes.
In exponentially growing cells, the artJ, artPIQM, and hisJQMP gene and operons are transcribed from a σ70-dependent promoter and regulated by ArgR (Wissenbach et al., 1995; Caldara et al., 2007), the master regulator of arginine biosynthesis genes and operons (Charlier et al., 1992; Tian et al., 1992; Wang et al., 1998). In contrast, argT is transcribed from a σ54-dependent promoter that is activated by the transcriptional regulator NtrC in conditions of nitrogen limitation, but is apparently not regulated by ArgR (Schmitz et al., 1988; Barrios et al., 1999; Reitzer, 2005; Cho et al., 2011, 2015).
Previously, we have shown that hexameric arginine-bound ArgR binds to two 18 bp semi-palindromic ARG boxes separated by 3 bp, the canonical design of an ArgR binding site in E. coli (Charlier et al., 1992) in the artJ, artP, and hisJ control regions (Caldara et al., 2007). These ArgR binding sites overlap the −35 promoter element of artJ and artP to the same extent, but are located slightly more upstream in the hisJ control region. Furthermore, in vitro binding studies and single-round in vitro transcription assays indicated that arginine-bound ArgR inhibits transcription initiation at PartJ through direct steric exclusion of RNA polymerase binding. This is not the case at PhisJ, where inhibition is less pronounced and repression relies on a different mechanism, likely involving ArgR-induced DNA conformational changes (Caldara et al., 2007).
Genome-wide expression profiling studies, chromatin immunoprecipitation (ChIP-chip), and genomic systematic evolution of ligands by exponential enrichment (SELEX) data indicated that transcription of the artPIQM operon is also down-regulated by the global transcription regulator leucine-responsive regulatory protein (Lrp) in a leucine-sensitive manner (Hung et al., 2002; Cho et al., 2008; Shimada et al., 2015). Furthermore, on the basis of high-throughput sequencing of exonuclease-treated chromatin-immunoprecipitated DNA (ChIP-exo), it was suggested that ArgR binds to the lrp control region and interferes with Lrp-mediated regulation by directly affecting the synthesis of this global regulator (Cho et al., 2011, 2015). In spite of these recent developments, the molecular mechanism of Lrp-mediated control of artP expression and its potential interplay with ArgR in the control of arginine transport remain poorly documented. Furthermore, even though ArgR effectively binds to the long lrp leader region, as also confirmed and analyzed in greater detail in this work, the observed effect on lrp gene expression was small, and the unraveling of the underlying molecular mechanism requires further investigation.
Here, we present an in depth molecular analysis of the effects of the transcriptional regulators ArgR and Lrp on transcription initiation at both PartP and Plrp. To reach this goal, we perform in vivo reporter gene assays in various isogenic backgrounds (WT, ∆argR, lrp::Tn10 and double ∆argR lrp::Tn10 mutants) and in vitro DNA binding assays with purified ArgR and Lrp, alone and in combination. This allowed us to determine the regulatory effects of both transcription factors, to delimit their precise binding sites, to evaluate the effects of ligands on both in vivo regulation and in vitro DNA binding, and to reveal potential interferences in the binding and action of the two transcription factors.
Materials and Methods
E. coli Strains and Plasmid Constructions
DH5α [F− Φ80lacZ∆M15 ∆(lacZYA-argF)U169 hsdR17U (rK− mK+) phoA supE44 recA1 endA1 gyrA96 thi-1 relA1] was used for cloning purposes. Strains CSH100 (F′), FW102 (F−, Smr) and its derivatives FW102 lrp::Tn10 and FW102 ∆argR have been described (Whipple, 1998; Devroede et al., 2004; Peeters et al., 2009). The double mutant FW102 ∆argR lrp::Tn10 was constructed by P1vir-mediated generalized transduction of FW102 ∆argR with a phage lysate prepared on the single lrp::Tn10 derivative of strain FW102 and selection of tetracycline-resistant transductants. Plasmid pFW11-null (Kmr, Cmr) (Whipple, 1998) and its derivative pFW-p/o-hisJ carrying the PhisJ-lacZ fusion (Caldara et al., 2006) and pFW-p/o-artJ carrying the PartJ-lacZ fusion (Caldara et al., 2007) have been described. Plasmids pFW-p/o-artP and pFW-p/o-lrp carrying the PartP-lacZ and Plrp-lacZ fusion, respectively, were constructed by PCR amplification of the control region of artP (222 bp fragment from −195 to +26) and lrp (369 bp fragment from −118 to +250) with genomic DNA of strain FW102 as template and the oligonucleotide pairs DC526f (5′-GGAATTCCGAGAATCGCTAACGACTTG-3′) plus DC392r (5′-CGGGATCCCGTATACTGGCAGTCTGATAGC-3′) and DC1464f (5′-CGGAATTCGCTTTATAAGCCGATTAAATGATG-3′) plus DC1465r (5′-CGGGATCCGTATTCCTTCCCTACTCCTGTC-3′) as primers, and ligation of the EcoRI and BamHI digested amplicon in similarly digested and dephosphorylated pFW11-null plasmid DNA. The 25-bp deletion mutant derivative (∆25) of the Plrp-lacZ construct was obtained by the overlap extension mutagenesis method (Higuchi et al., 1988) with the mutagenic primers DC1562f (5′-TGCGCATAACCATGCATGTAAATACC-3′) and DC1561r (5′-GTATTTACATGCATGGTTATGCGCACTCGAATGTTTTCGCAAAACACCAG-3′). Recombinant plasmids were transformed in strain CSH100 and double cross-over events transferring the promoter-operator-lacZ fusions to the single copy F′-episome were obtained by conjugation with the F− strain FW102 and its single and double ∆argR and lrp::Tn10 derivatives, and selection of Kmr and Smr transconjugants that were further screened for chloramphenicol sensitivity as described (Whipple, 1998). The lrp coding region was amplified with the oligonucleotides DC531f (5′-GGAATTCCATATGGTAGATAGCAAGAAGCGCCCTGG-3′) and DC1463r (5′-CCGCTCGAGTTAGCGCGTCTTAATAACCAGAGC-3′) as primers and genomic DNA of strain FW102 as template. The purified NdeI plus XhoI amplicon was ligated in similarly digested and dephosphorylated plasmid pET28a plasmid DNA and transformed in competent cells of strain DH5α. Plasmid DNA extracted from a correct clone (pET28-lrp) was subsequently transformed in the overexpression strain BL21 (DE3). All constructs were verified by DNA sequencing.
Overexpression and Purification of E. coli Arginine Repressor and Leucine-Responsive Regulatory Protein
Untagged hexameric ArgR was purified to electrophoretic homogeneity from an IPTG (isopropyl-β-
Enzyme Assays
Specific β-galactosidase activities were determined as described (Nguyen Le Minh et al., 2018) at 28°C in sonicated cell-free extracts of exponentially grown cultures (OD600 0.4), with ortho-nitrophenyl β-
In vitro DNA Binding Experiments
[5′-32P] Single-end labeled DNA fragments used in electrophoretic mobility shift assays (EMSA), and various footprinting and premodification binding interference experiments were prepared and purified as described (Nguyen Le Minh et al., 2018). DNase I, hydroxyl radical (Tullius and Dombroski, 1986) and chemical premodification binding interference experiments (missing contact probing) (Brunelle and Schleif, 1987) were performed as described (Wang et al., 1998). In gel footprinting with the [(OP)2-Cu+] ion (Kuwabara and Sigman, 1987) was performed as described in (Peeters et al., 2004). Binding assays were performed in binding buffer (10 mM Tris-HCl pH 7.4, 5 mM MgCl2, 250 mM KCl, 2.5 mM CaCl2, 0.5 mM DTT and 2.5% glycerol), when indicated supplemented with
Results
Leucine-Responsive Regulatory Protein Represses PartP but Not PartJ and PhisJ Activity
To evaluate the potential effect of Lrp on arginine transport genes in E. coli we assayed gene expression with single-copy F′-borne reporter gene constructs expressing lacZ under the control of the PartP, PartJ, or PhisJ promoter in cells of isogenic wild type and lrp::Tn10 mutant strains grown aerobically on minimal medium and harvested in the exponential growth phase (Figure 2A). The results indicate that Lrp represses PartP activity about three-fold. In contrast, PartJ and PhisJ expression was nearly identical in absence and presence of Lrp. Hence, these two promoters and associated genes are not part of the Lrp regulon. These observations are in full agreement with the genome-wide expression profiling study (Cho et al., 2008) and validate our single copy reporter gene systems.
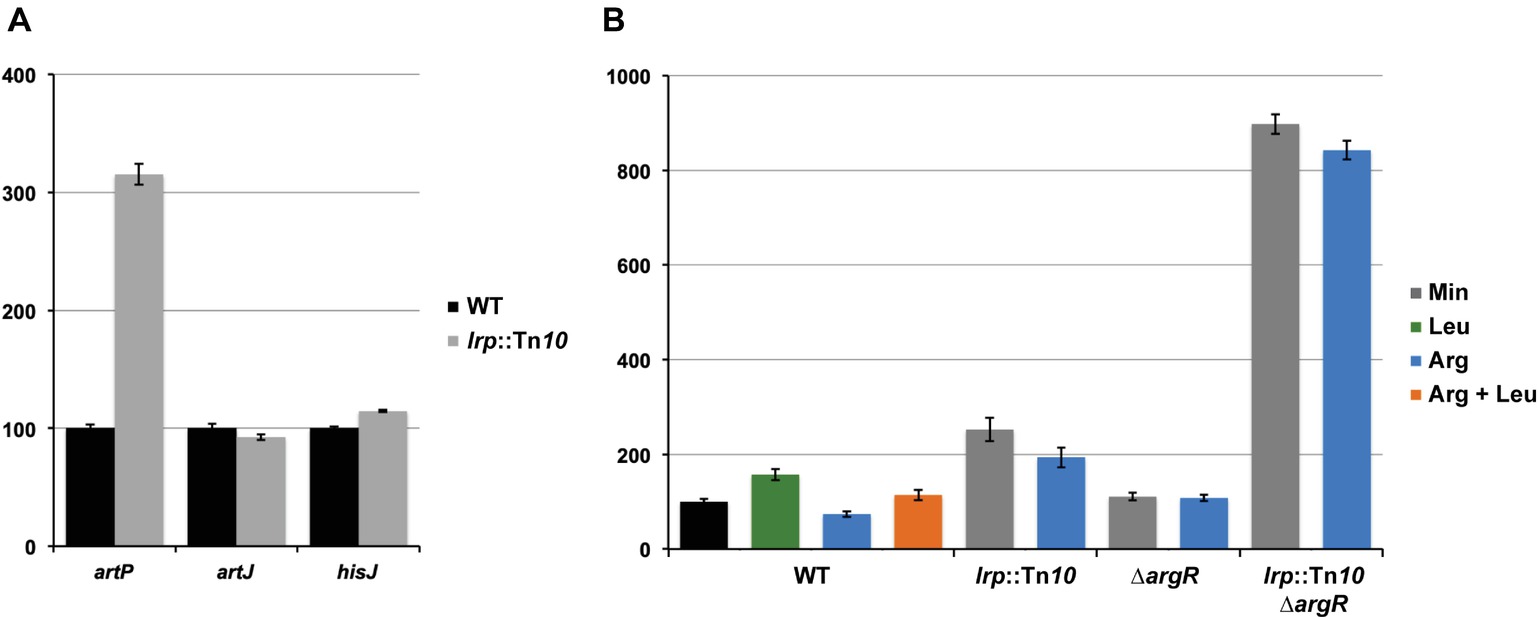
Figure 2. Histogram presentation of β-galactosidase specific activities. Values are the means ± standard deviation of at least three biological replicates. (A) Specific activities measured in cell-free extracts of E. coli strain FW102 (WT) and its isogenic lrp (lrp::Tn10) derivative bearing a single-copy lacZ reporter gene fusion under the control of the artP, artJ, or hisJ promoter/operator grown on minimal medium. 100% corresponds for each fusion construct to the activity measured in the wild type strain. (B) Specific activities of a single-copy lacZ reporter gene under the control of the artP promoter/operator measured in cell-free extracts of FW102 (WT) and its isogenic lrp (lrp::Tn10), argR (∆argR), and double argR lrp (lrp::Tn10/∆argR) mutant derivatives. Cells were grown on minimal medium, supplemented with
Arginine Repressor and Leucine-Responsive Regulatory Protein Act as Competitive Repressors of artP Expression in vivo
Previously, we have shown that arginine-bound ArgR binds to the artP control region and exerts a negative effect on promoter activity (Caldara et al., 2007). To analyze the potential interplay between ArgR and Lrp in the artP control region, we assayed β-galactosidase activities in cell-free extracts of isogenic wild type, ∆argR, lrp::Tn10 and double ∆argR lrp::Tn10 mutants bearing a single-copy F′-borne PartP-lacZ reporter gene construct, grown on minimal medium and minimal medium supplemented with arginine, leucine or both (Figure 2B). The results indicate that repression by Lrp is partially counteracted by leucine supplementation. Furthermore, Lrp has a stronger negative effect than ArgR and both repressors are more potent (approximately three-fold) inhibitors of PartP activity in the absence of the other regulator [compare the ratios double mutant/∆argR (7.8) and lrp::Tn10/WT (2.6) on arginine for repressibility by Lrp in absence and presence of ArgR, respectively, and the ratios double mutant/lrp::Tn10 (4.4) and ∆argR/WT (1.5) on arginine for repressibility by ArgR in absence and presence of Lrp, respectively]. Therefore, we may conclude that Lrp and ArgR act as competitive repressors of PartP.
Interferences of Leucine-Responsive Regulatory Protein and Arginine Repressor Binding to the artP Control Region
To further unravel the molecular basis of ArgR- and Lrp-mediated repression of transcription initiation at PartP, and their interplay, we performed electrophoretic mobility shift assays (EMSAs) with purified Lrp and ArgR, alone and in combination (Figure 3). EMSAs with Lrp binding to the artP control region [apparent overall equilibrium dissociation constant (Kd) of approximately 200 nM] revealed the concentration-dependent formation of four complexes with a different migration velocity: two minor complexes (B1 and B2), and two more abundant complexes (B3 and B4) present at a variable ratio dependent on the Lrp concentration, with higher protein concentrations favoring the formation of the slowest migrating complex B4 at the expense of all other complexes (Figure 3A). Leucine had a negative effect on total complex formation and affected the relative proportions of the different complexes, indicating that leucine lowers the overall affinity and affects the cooperativity in the binding (Figure 3A). Binding of Lrp to its own control region (autoregulation) performed as a control resulted in the formation of one minor and two more abundant complexes (Figure 3A). Again, leucine had a negative effect on binding affinity and affected the relative abundances of the various complexes. This is reminiscent of Lrp binding to its own control region in Salmonella enterica serovar Typhimurium (McFarland and Dorman, 2008) but is in contrast with older observations of E. coli Lrp binding to its own control region, which was claimed to be leucine-insensitive (Lin et al., 1992; Wang et al., 1994). Kd’s for Lrp binding to various targets vary widely but are frequently in the 50–100 nM range, with a very high affinity (8 nM) observed for binding to the six binding sites in the ilvIH control region (Wang and Calvo, 1993) and a more similar Kd (50 nM) for binding to the argO gene for arginine export (Peeters et al., 2009). A large variability in binding affinities for Lrp is not surprising, since the protein exerts various functions, as a structural nucleoid associated protein (NAP) and as a more specific transcription regulator or co-regulator with about 314 binding sites on the E. coli genome identified in the absence of effector molecules (Shimada et al., 2015). Finally, we performed EMSAs with both ArgR and Lrp binding to the artP control region. When a constant amount of ArgR (0.48 nM) was allowed to bind prior to the addition of increasing concentrations of Lrp, the concentration-dependent formation of a new complex (indicated as ArgR-DNA-Lrp) migrating more slowly than the ArgR only complex and in between the Lrp1 and Lrp2 only complexes was observed (Figure 3B). This result indicates that hexameric ArgR and at least one Lrp dimer may bind simultaneously to the artP control region. When inversely, a constant amount of Lrp (300 nM) was allowed to bind to the artP control region prior to the addition of increasing concentrations of ArgR this resulted in the formation of a new complex (Lrp-DNA-ArgR) that migrates slightly below the Lrp2-DNA complex, but more slowly than the ArgR-DNA and Lrp1-DNA complexes (Figure 3C). This result suggests that ArgR is able to displace Lrp from at least one of its binding sites (or at least to bind preferentially after dissociation of Lrp), likely overlapping the ArgR binding site.
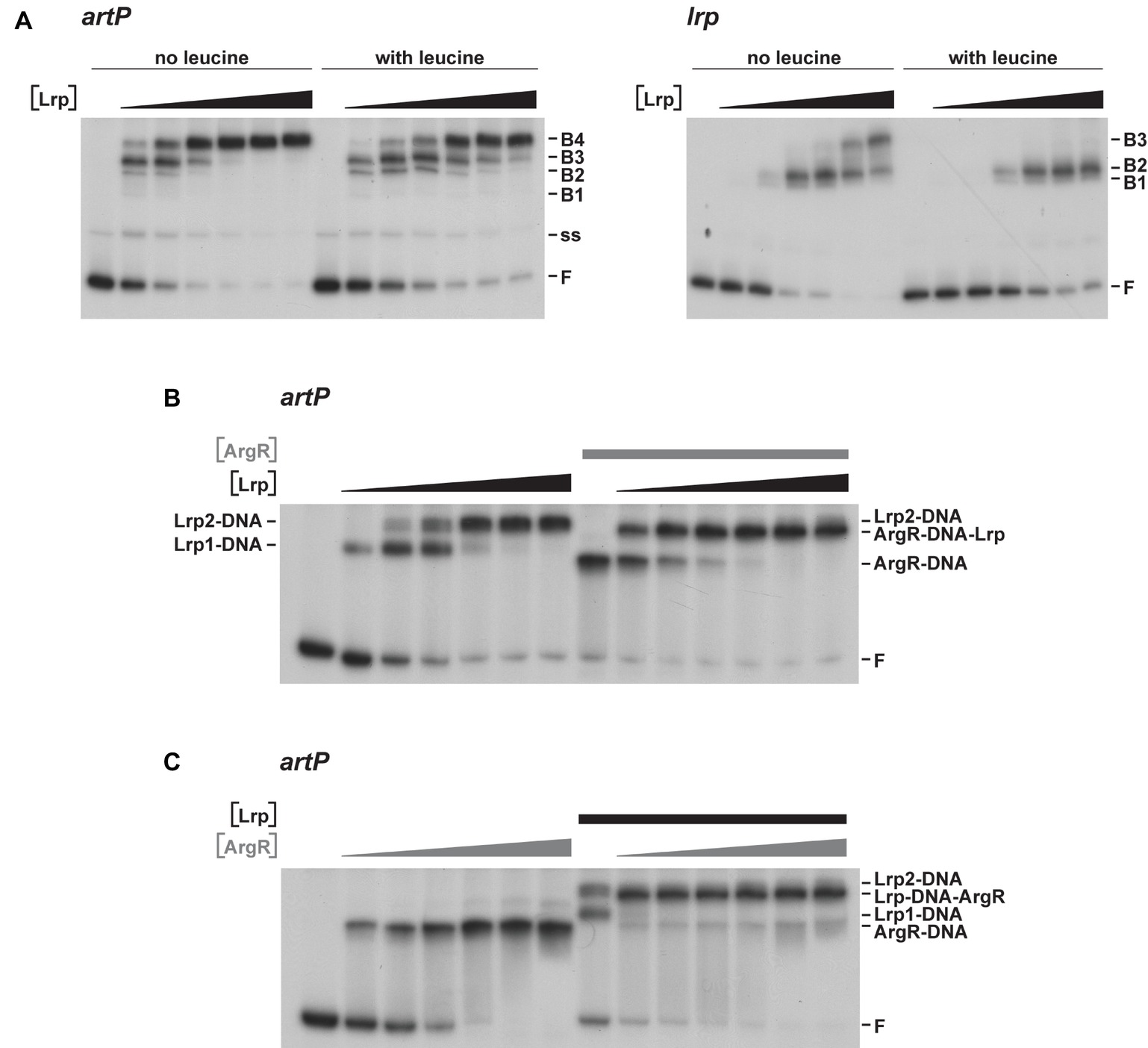
Figure 3. Representative autoradiographs of EMSAs with Lrp and ArgR binding to radiolabeled fragments bearing the artP or lrp control region in the presence of an excess non-labeled non-specific competitor DNA (sonicated harring sperm DNA). (A) Binding of Lrp to the artP and lrp control region in the absence and in the presence of 7.0 mM
As Lrp is known to use several amino acids besides leucine as effector molecules (Hart and Blumenthal, 2011), we also tested the effect of arginine and arginine plus leucine on Lrp binding to the artP and lrp control regions in EMSA (Figure 4). The results indicate that arginine has a small stimulating effect on Lrp binding to its own control region, favoring the formation of the higher order complexes at lower protein concentrations than in the absence of effector (Figures 4A,B). Furthermore, we confirmed that leucine has a negative effect on Lrp binding to both the artP and lrp control regions (Figure 4C) and that in the presence of equimolar concentrations of both amino acids the negative effect of leucine prevails (Figure 4D). To the best of our knowledge, this is the first report of a positive effect of arginine on E. coli Lrp binding, but the Lrp ortholog of Mycobacterium tuberculosis is known to bind arginine and several other amino acids several amino acids (Shrivastava and Ramachandran, 2007).
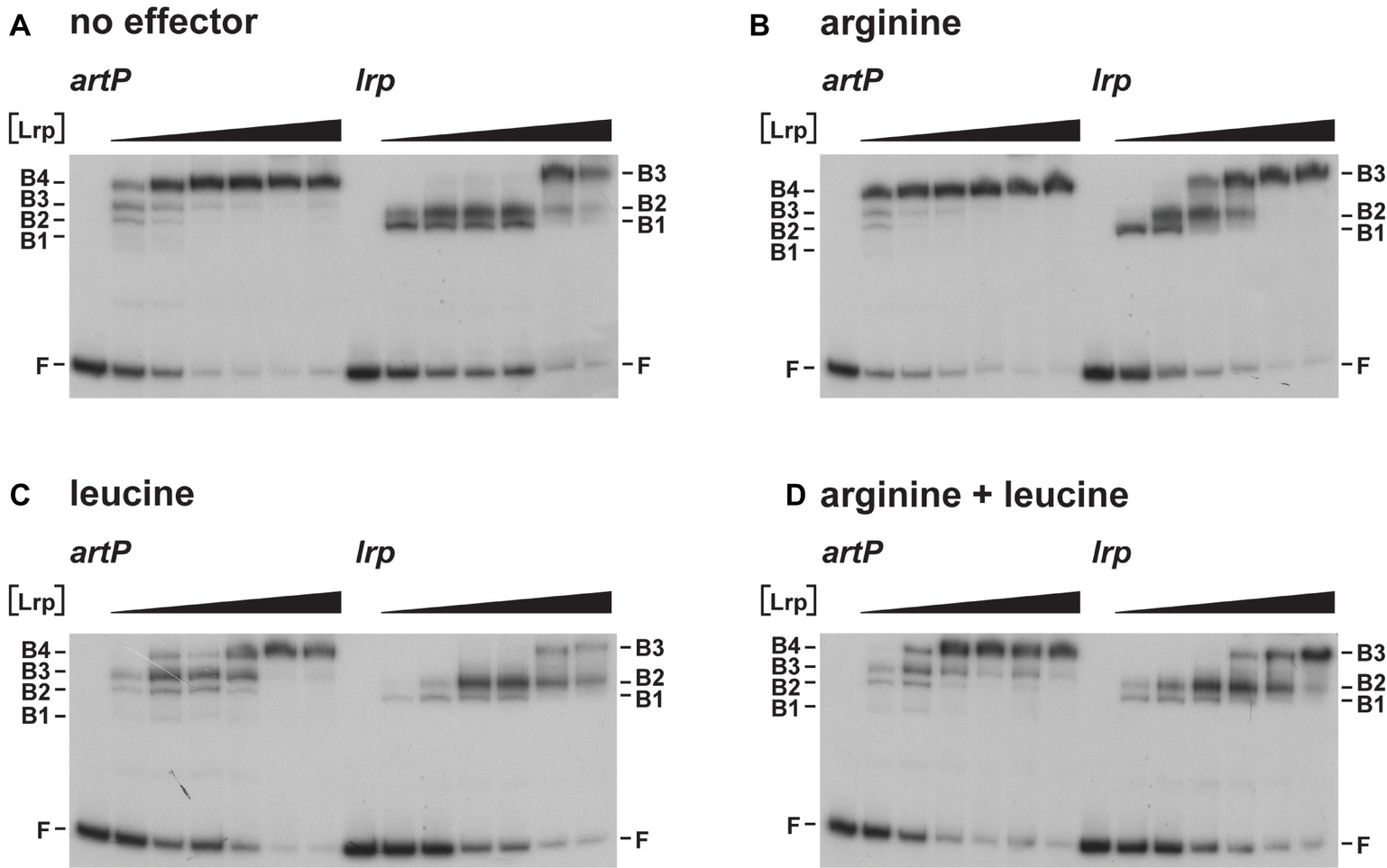
Figure 4. Representative autoradiographs of Lrp binding to the artP and lrp control regions in the absence (A) and in the presence of 10 mM
Identification of Leucine-Responsive Regulatory Protein Binding Sites in the artP Control Region
The Lrp binding sites in the artP control region were identified by DNase I footprinting and in gel footprinting with the [(OP)2-Cu+] ion of the major complexes (B3 and B4) with a different stoichiometry separated by EMSA (Figure 5A). DNase I footprinting indicates a concentration-dependent protection pattern (Figures 5B,C). At the lower Lrp concentrations used, a 27-nt long stretch of the coding strand (from −86 to −112 with respect to the transcription initiation site) was clearly protected against digestion by the nuclease. At higher Lrp concentrations, this zone extended further both upstream and downstream of this nucleation site (from −165 to approximately −10), with concomitant appearance of numerous regularly spaced sites of hyperreactivity, indicative of pronounced DNA deformations resulting in local minor groove widening (Lazarovici et al., 2013). In gel footprinting with the [(OP)2-Cu+] ion on the major complexes (B3, B4) with a different stoichiometry separated by gel electrophoresis (Figure 5A) revealed a 29-nt long zone of protection (from −67 to −95) on the coding strand for the faster migrating complex B3 (Figures 5B,C). This zone partially overlaps the nucleation site observed at low Lrp concentrations in DNase I footprinting. In the slower migrating complex B4, the region of protection is extended on both sides and an additional zone of clear protection is visible between positions −117 and −156. Interestingly, the two zones of clear protection are separated by an approximately 20-bp long G + C rich stretch (−96 to −116, 76% G + C) showing strong hyperreactivity. In complex B4, protection in the downstream direction extends to approximately position −10, which corresponds roughly to the border of protection observed in DNase I footprinting. The extended zones of protection indicate an overlap with the previously identified binding site for ArgR (Figure 5C; Caldara et al., 2007), which is compatible with the binding interference of the two regulators as observed in mixed EMSAs (Figure 3) and the competitive repression detected in vivo (Figure 2B).
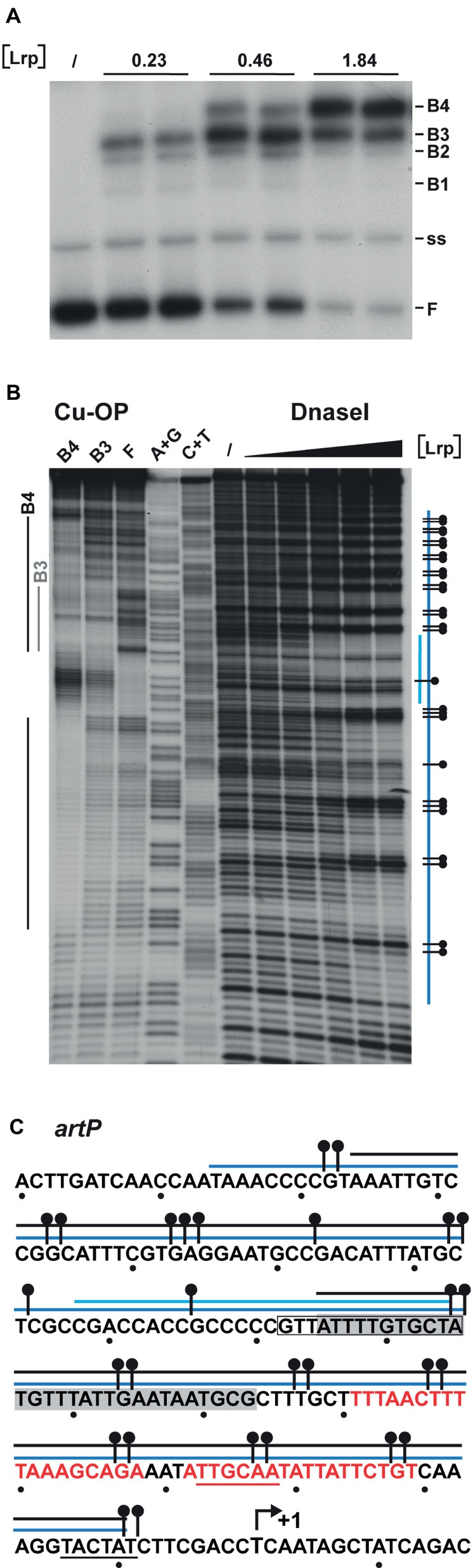
Figure 5. DNAse I and in gel copper-phenanthroline (OP-Cu) footprinting of Lrp binding to the artP control region (top or coding strand revealed). (A) EMSA with Lrp binding to the artP control region used for the in gel footprinting with the [(OP)2-Cu+] ion. Lrp concentrations (in μM) are expressed in monomer equivalents. The position of free DNA (F), single stranded DNA (ss) and the various complexes with a different migration velocity (B1 to B4) are indicated. (B) Autoradiograph of in gel footprinting of free DNA (F) and the major Lrp-DNA complexes B3 and B4 with a different migration velocity extracted from the gel presented in panel (A) after treatment with the [(OP)2-Cu+] ion and separation of the reaction products by gel electrophoresis in denaturing conditions. Regions of protection in the different complexes are indicated with gray and black colored lines for the complexes B3 and B4, respectively. A + G and C + T represent the Maxam-Gilbert sequencing ladders. In DNase I footprinting the black triangle on top of the autoradiograph represents increasing Lrp concentrations corresponding to 0, 0.46, 0.92, 1.86, 3.68, and 7.36 μM. Regions protected against DNase I cleavage are indicated with a blue colored line. The short lightblue colored line represents the region that is already protected at the lower Lrp concentrations (1.86 μM, nucleation site), the long blue colored line represents the extended zone of protection observed at higher Lrp concentrations. A short horizontal bar with a dot represents a site that becomes hyperreactive to DNase I cleavage upon Lrp binding. (C) Sequence of the artP control region (coding strand) with indication of regions of protection against enzymatic and chemical cleavage. The region of protection in complex B3 is gray shaded. Other symbols are as in panel B. The −10 and − 35 promoter elements are underlined. A short arrow represents the start of transcription (+1). The in silico predicted Lrp binding site (−85 to −98) (Cho et al., 2008) is boxed. Red colored letters represent ARG box sequences.
Identification of Arginine Repressor-DNA Contacts in the lrp Control Region
Previously it was shown that ArgR binds to the lrp control region in vivo (Cho et al., 2011, 2015). Here, we perform a detailed in vitro interaction study of purified ArgR binding to the lrp control region by a combination of enzymatic and chemical footprinting techniques (DNase I, hydroxyl radical, “in-gel” footprinting with the [(OP)2-Cu+] ion), and establish a high-resolution base-specific contact map based on premodification binding interference assays (Figures 6, 7). DNase I revealed the protection of a 37-nt long stretch on both stands (Figures 6A,D) extending from position +29 to +65 downstream of the transcription initiation site. This zone of protection comprises two 18-bp imperfect palindromes separated by 3 bp, a canonical configuration for an E. coli ArgR binding site associated with arginine biosynthetic and transport genes (Figure 6D; Charlier et al., 1992; Tian et al., 1992; Caldara et al., 2007). Hyperreactivity for DNase I interrupting the zone of protection and in the adjacent flanking regions is indicative of DNA bending associated with minor groove widening. In gel footprinting of the single ArgR-DNA complex separated from free DNA by gel electrophoresis with the [(OP)2-Cu+] ion confirmed the position of this zone of interaction (Figures 6C,D) that also corresponds to the in vivo protected target (Cho et al., 2015). Chemical footprinting with the very small and highly reactive hydroxyl radical (Tullius and Dombroski, 1986), allowed to break up this global region of protection into five small subzones on both strands, each one being 2 to 4 nt long, with the centers of protection separated by approximately one full helical turn and slightly shifted toward the 3′-end on complementary strands (Figures 6B,D). Such a pattern is indicative of ArgR binding to one face of the DNA helix (Oackley and Dervan, 1990), as observed previously with ArgR binding to the ARG boxes of arginine biosynthetic genes (Charlier et al., 1992; Wang et al., 1998). The position of the ArgR binding site in the lrp control region, downstream of the transcription start, is unusual for strong ArgR-repressed promoters since in arginine biosynthetic operators the ARG boxes are located upstream of the transcription start and partially overlap the −10 and/or − 35 promoter elements (Charlier et al., 1992; Tian et al., 1992; Caldara et al., 2007).
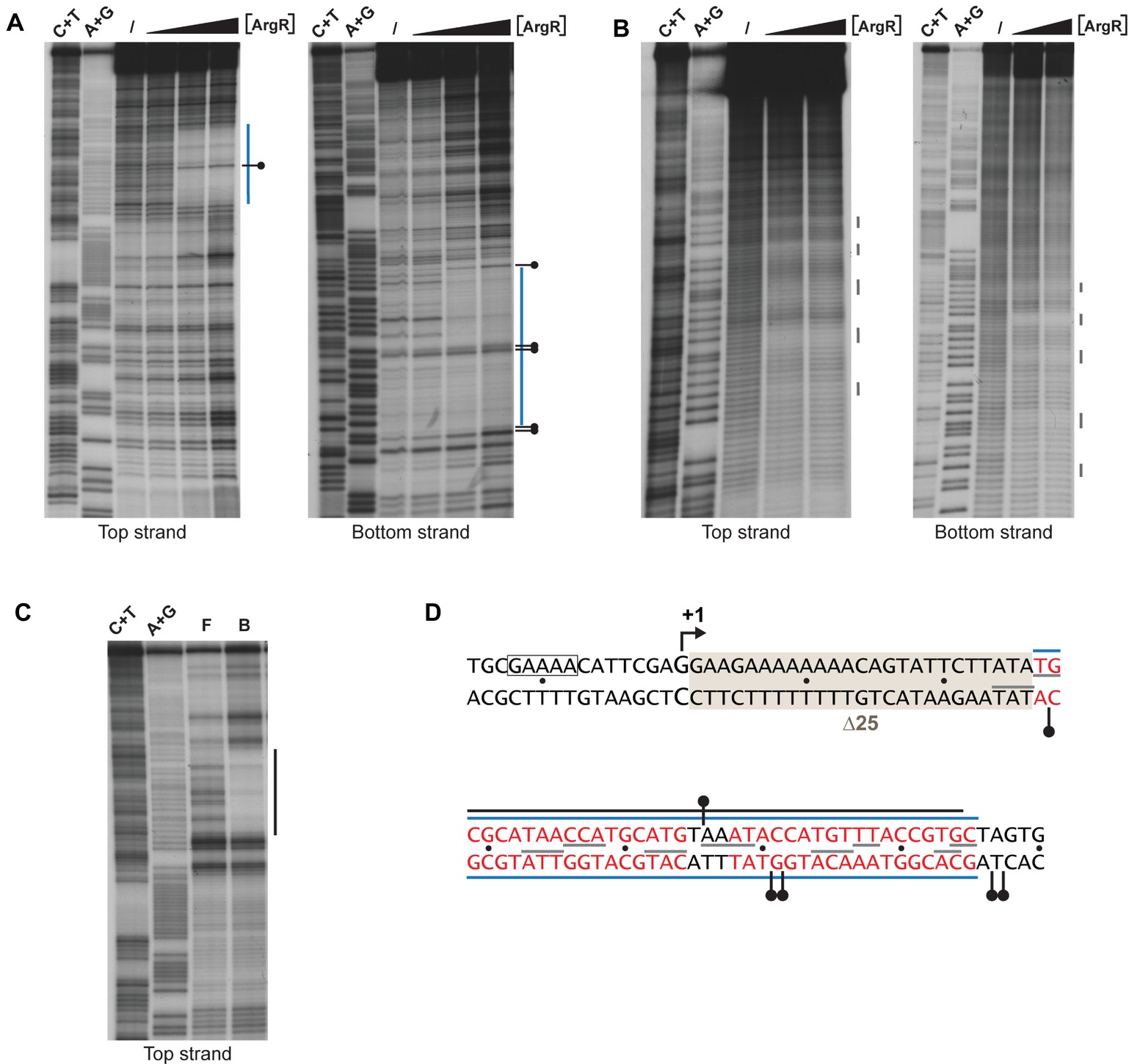
Figure 6. Enzymatic and chemical footprinting of ArgR binding to the lrp control region. (A) DNase I footprinting. A blue bar represents the zone of protection. A short horizontal bar with a dot represents a site that becomes hyperreactive to DNase I cleavage upon ArgR binding. ArgR concentrations used are 0, 0.39, 3.91, and 39.1 nM. C + T and A + G correspond to the Maxam-Gilbert sequencing reactions. (B) Hydroxyl radical footprinting. ArgR concentrations used are 0, 39.1 and 78.3 nM. Short gray colored vertical lines represent sites of protection. (C) In gel footprinting with the [(OP)2-Cu+] ion. F and B correspond to free and ArgR-bound DNA recovered from gel after treatment with the [(OP)2-Cu+] ion. (D) Nucleotide sequence with indication of zones of protection. Symbols are as in panels A, B and C. Red colored letters represent the 18 bp ARG boxes. The −10 promoter element is boxed. The 25-bp deletion of mutant ∆25 used for reporter gene assays (Figure 8) is gray shaded. The Lrp binding site as defined by DNase I footprinting (Wang et al., 1994) extends from position −106 to −20 and is not included in this figure.
Base-specific contacts of ArgR with purine and pyrimidine bases in the lrp control region were identified with chemical premodification binding interference experiments (Figure 7; Brunelle and Schleif, 1987). Sparingly modified 5′-single-end-labeled DNA (treated with either citrate at pH 4.0 and 80°C or hydrazine at 25°C for purine and pyrimidine missing contact probing, respectively) was incubated with various concentrations of ArgR and separated on basis of affinity for the regulator in a native EMSA. Free and ArgR-bound DNA molecules were subsequently extracted from the gel, cleaved at the position of modification by piperidine treatment, and the reaction products analyzed by gel electrophoresis in denaturing conditions. In such an experiment, molecules modified at a position important for complex formation, and thus exhibiting a reduced affinity for the repressor, are expected to be overrepresented in the free DNA and underrepresented in the bound form, whereas molecules with a modification at a position that does not significantly contribute to complex formation are expected to be equally distributed over free and bound forms. The results indicate that all strong and moderate negative effects of base removal on ArgR binding are located within the ARG boxes as defined above (Figure 7). In the promoter-proximal ARG box, the strongest negative effects were observed upon removal of A6, T7, A8, A9, A12, and T13 from the top strand (coding strand) and T3’, G4’, A6’, G9’, T10’, T11’, A12’, and T13’ of the bottom strand. In the promoter-distal box, only removal of T11 occasioned a strong negative effect on ArgR binding. This is in full agreement with the more degenerate character of this ARG box compared to the consensus sequence (Figure 7C; Charlier et al., 1992). Of the strong interfering positions, A6, T7, A12, T13 and the symmetrical counterparts are among the most conserved positions in ARG box sequences previously described, and their importance in complex formation and gene regulation has been demonstrated for various genes and operons of the ArgR regulon (Charlier et al., 1992; Wang et al., 1998; Caldara et al., 2007). In various instances (C4, C5, C13, and C14 of the top strand and T16’ of the bottom strand), base removal in the downstream ARG box of the lrp control region resulted in enhanced ArgR binding. Some of these inverse signals are generated upon removal of bases occupying highly conserved positions among ARG boxes (especially positions 4 and 13) but do not correspond to the consensus sequence (G4 and T13, respectively) in the lrp operator. These results indicate that the presence of a non-consensus base, showing a different distribution of base-specific groups as potential donors and acceptors in hydrogen bond formation in the major groove segment contacted by ArgR, at these positions in the lrp control region does not contribute to ArgR binding but instead generates a steric hindrance for the establishment of another, nearby contact (see “Discussion”).
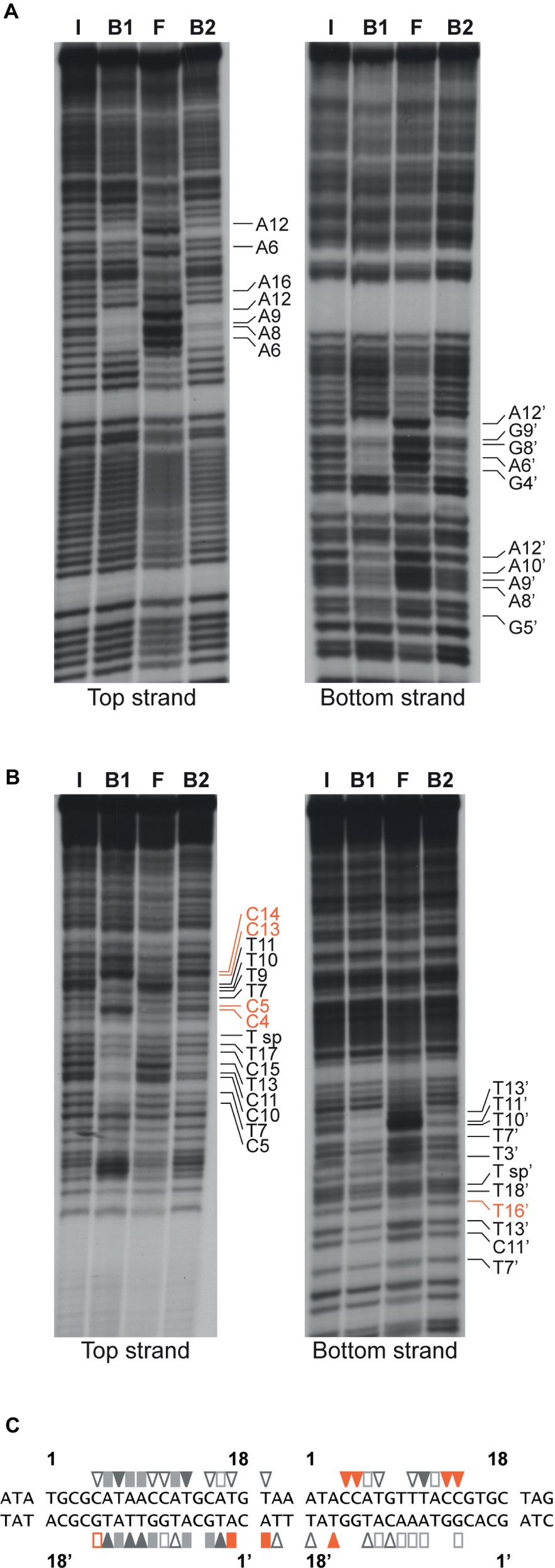
Figure 7. Missing contact probing with ArgR binding to the lrp control region. (A) Depurination (citrate) and (B) depyrimidination (hydrazine). Sparingly modified DNA molecules incubated in the absence (I = input) and in the presence of various concentrations of ArgR, resulting in approximately 50% binding for F1 and B1, and > 90% for F2 and B2, were separated on basis of their affinity for the protein by gel electrophoresis in native conditions, extracted from gel, cleaved at the positions of modification with piperidine and equal amounts of the reaction products separated by gel electrophoresis in denaturing conditions. Positions that upon base removal interfere strongly or moderately are indicated. Positions that upon removal result in better binding are indicated with orange colored letters (C) Nucleotide sequence of the ArgR binding site in the lrp control region with indication of positions that upon modification interfere strongly (filled symbols) or moderately (open symbols) with ArgR binding are indicated. Orange colored symbols represent inverse effects. ARG box sequences separated by a 3 bp spacer are numbered 1 to 18 for the top (coding) strand and 1′ to 18′ for the bottom strand in the 5′-3′ direction.
Effects of Arginine Repressor and Leucine-Responsive Regulatory Protein on in vivo lrp Gene Expression
The effect of the transcriptional regulators ArgR and Lrp on lrp gene expression was determined with a single-copy F′-borne reporter gene construct expressing lacZ under the control of the Plrp promoter in isogenic wild type, ∆argR, lrp::Tn10 and double ∆argR lrp::Tn10 mutant strains grown on minimal medium and minimal medium supplemented with arginine, leucine or both. The results (Figure 8A) indicate negative autoregulation (about four-fold, compare WT and lrp::Tn10 on minimal medium) that is partially counteracted by leucine as effector molecule (about two-fold, compare WT minimal and leucine). This reciprocal mode of autoregulation has also been proposed for lrp in Salmonella typhimurium (McFarland and Dorman, 2008), but is in contradiction with previous work indicating leucine-independent negative regulation of E. coli lrp (Lin et al., 1992; Wang et al., 1994). Besides autoregulation, we observed little or no negative effect of ArgR and arginine on lrp promoter activity. Instead, slightly lower values (about 1.3-fold) were measured in the ∆argR mutant compared to the WT. This reduction might be due to a stronger repression exerted by Lrp in the absence of any interfering ArgR binding [5.6-fold (ratio double mutant/∆argR) compared to 3.7-fold in the presence of liganded ArgR (ratio lrp::Tn10/WT grown on arginine)]. As shown above, the ARG boxes in the lrp control region are located downstream of the transcription start site. In this context, it is worth noticing that ArgR binding to the ARG boxes overlapping the E. coli carP2 promoter strongly represses transcription initiation from this promoter but does not hinder transcription initiated 67 nt more upstream at the carP1 promoter (Charlier et al., 1988, 2018). Hence, it appears that ArgR is unable to function as an efficient roadblock for an elongating RNA polymerase. To further verify this hypothesis, we constructed a 25 bp deletion mutant (∆25) of the lrp operator-lacZ fusion construct in which the ARG boxes are positioned immediately downstream of the transcription initiation site (Figure 6D). Reporter gene assays performed with this construct indicate that the deletion has a negative effect on intrinsic promoter strength [about 1.7-fold, compare WT and mutant operator in the absence of regulation (double ∆argR lrp::Tn10 background)] (Figures 8A,B). In the WT background arginine has a slight negative (1.7-fold) and leucine a slight positive effect (1.7-fold) on the ∆25 mutant promoter activity. This arginine-dependent reduction is indicative of a weak ArgR-mediated repression, which is, however, not reflected in the activities measured in the ∆argR background. This might again be due to a stronger repression exerted by Lrp in the absence of interfering ArgR binding. Indeed, repression by Lrp is about 1.5-fold stronger in the absence of ArgR than in its presence [compare the 6.6 to 7.0-fold repression in the absence (ratio double mutant/∆argR on minimal or arginine supplemented medium, respectively) with the 4.4-fold repression in the presence of liganded ArgR (ratio lrp::Tn10/WT on arginine supplemented medium)]. In agreement with this observation repression by liganded ArgR is also stronger in the absence of Lrp (2.5-fold, ratio double mutant/lrp::Tn10 grown both on arginine) than in its presence.
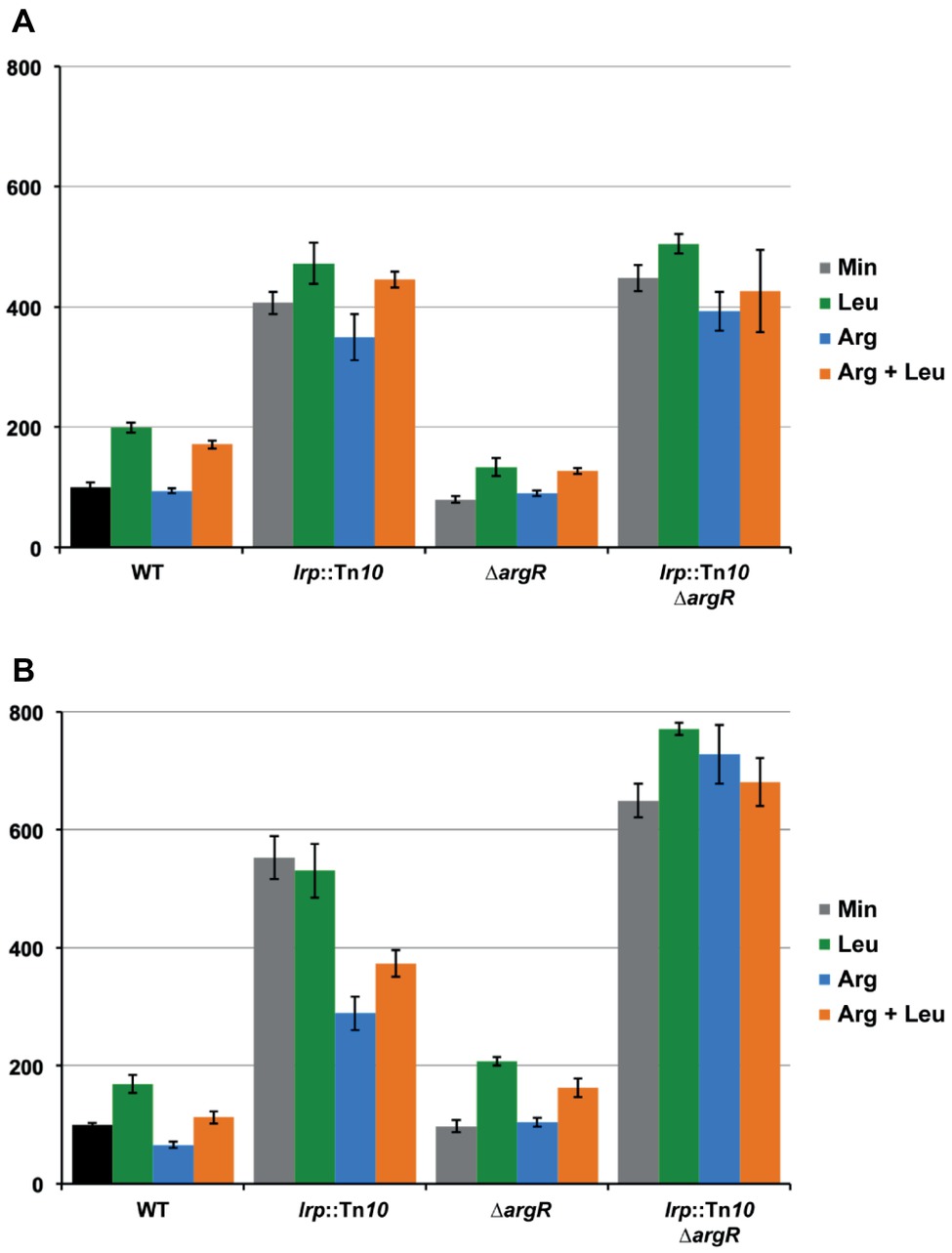
Figure 8. Histogram presentation of β-galactosidase specific activities measured in cell-free extracts of E. coli strain FW102 (WT) and its isogenic lrp (lrp::Tn10), argR (∆argR) and double lrp argR (lrp::Tn10 ∆argR) mutants bearing the wild type lrp promoter/operator-lacZ (A) or mutant lrp∆25 promoter/operator-lacZ fusion (B) on a single-copy F’ episome. Cells were grown in minimal medium, when indicated supplemented with
Discussion
Independent studies using different approaches have shown that transcription of the artPIQM operon is negatively regulated by ArgR (Caldara et al., 2006, 2007; Cho et al., 2015) and Lrp (Cho et al., 2008; Shimada et al., 2015). Arginine as an effector molecule of ArgR is known to enhance its negative effect (corepressor) on PartP, whereas leucine partially counteracts the negative effect of Lrp (reciprocal regulation mode) and thus acts as an inducer. Many synthesis, degradation and transport systems for amino acids and amino acid sensing transcription factors are subjected to transcriptional regulation by Lrp (Cho et al., 2011; Shimada et al., 2015). Several amino acid uptake and export systems are either negatively or positively regulated by Lrp and leucine in a reciprocal manner, whereby leucine attenuates the effect of Lrp. This is for instance the case for serine (sdaC), alanine (cycA), proline (proY), leucine (leuE), and also arginine export (argO) (Kutukova et al., 2005; Cho et al., 2008; Peeters et al., 2009). In contrast, Lrp-mediated regulation of several transporters for aromatic amino acids occurs according to the concerted mode, whereby leucine potentiates the negative effect of the regulator (Cho et al., 2008). As a consequence of the widespread effect of Lrp on amino acid metabolism, amino acid pools in isogenic WT and lrp mutant strains may vary to some extent (Shimada et al., 2015), which may in turn affect the action of amino acid sensing transcription factors, especially upon growth in minimal or leucine supplemented medium.
Many bacterial promoters are regulated by multiple transcription factors for the integration of distinct environmental signals (Paul et al., 2007; Ogasawara et al., 2010). Their positive or negative interplay in the control of one or more promoters illustrates just one aspect of the complexity, diversity and versatility of bacterial gene regulation mechanisms, which are frequently multi-layered and operate at different levels and stages of gene expression (for an overview, see Bervoets and Charlier, 2019). Here, we analyzed the interplay between ArgR and Lrp in the control of PartP activity in vivo and in vitro and demonstrate that each repressor is a more potent inhibitor of PartP activity in the absence of the other. Hence, they act as competitive repressors. This observation is in agreement with, and may be explained by, the existence of partially overlapping binding sites for ArgR and Lrp in the artP control region. Binding interferences indicate that even though ArgR and Lrp may bind simultaneously to the artP control region, the number of Lrp dimers that can bind in the presence of ArgR is reduced, and thus the formation of higher order Lrp-DNA complexes is inhibited. This may be explained by the observation that the nucleation site for Lrp binding does not overlap the ARG boxes, whereas the more extended zone of interaction identified at higher Lrp concentrations does overlap (Figure 5C). The concentration-dependent formation of four complexes and the effect of leucine on binding affinity and cooperativity is reminiscent of Lrp binding to several other Lrp-regulated genes and artificial operator constructs (Azam and Ishihama, 1999), among which the E. coli argO gene for arginine export, that is activated in a competitive manner by Lrp and ArgP (a LysR-type transcriptional regulator) (Peeters et al., 2009; Nguyen Le Minh et al., 2018). Our findings are also fully compatible with the results of a detailed analysis of cooperative Lrp binding to various targets, indicating that leucine decreases the intrinsic affinity of Lrp for a single binding site but enhances the cooperativity in the binding to multiple sites (in a spacer length dependent manner) (Chen et al., 2005).
At low concentrations (nM range) Lrp exists primarily as a homodimer in solution (Chen et al., 2001; Chen and Calvo, 2002), but the protein can form higher oligomeric structures at higher concentrations in vitro, upon DNA binding, and in vivo (where it is present at μM concentrations), with leucine favoring the dissociation of hexadecameric Lrp into two leucine-bound octamers (Chen et al., 2005). In view of this effect of leucine on the self-association of Lrp, it has been postulated that the hexadecameric form would be the active one for promoters that are either activated or repressed by Lrp and leucine in the reciprocal mode, whereas the leucine-bound octamer would be the active form for promoters that are regulated in the concerted manner by the regulator and its effector (Chen et al., 2001). Our results suggest the cooperative binding of at least four Lrp dimers to the artP control region, although we cannot exclude the existence of some higher oligomeric forms of the protein in solution. The consensus Lrp binding site [CAG-N9(A + T rich)-CTG] proposed on basis of ChIP-chip experiments (Cho et al., 2008) is rather degenerated and various slightly different consensus sites have been proposed in the past (Cui et al., 1996; Shultzaberger and Schneider, 1999; Peterson et al., 2007). This is not surprising, since Lrp, as a nucleoid associated protein (NAP), also plays an important structural role. The nucleation site identified by different footprinting methods in the artP control region bears the motif G-N2–3-TTT, which is also found in the six well-characterized Lrp binding sites in the E. coli pap control region as part of the consensus (Nou et al., 1995). Several stretches showing imperfect conservation of this motif can also be found in the regions flanking the nucleation site that are protected in the higher oligomeric Lrp-DNA complexes (in gel footprinting) and at higher Lrp concentrations (DNase I) (Figure 5). Furthermore, a unique potential Lrp binding site predicted on basis of in silico analyses (Cho et al., 2008) is comprised within the nucleation site (Figure 5C). The extended and regularly spaced hyperreactivity for DNase I cleavage observed in complexes made at higher Lrp concentrations are indicative of pronounced DNA deformations (Figure 5B), an observation that is compatible with DNA wrapping around octameric Lrp as in the Lrp-DNA cocrystal structure (de los Rios and Perona, 2007). It is therefore likely that individual DNA-bound Lrp dimers interact to form a higher oligomeric assembly in which the artP operator DNA is highly deformed.
The interference of Lrp binding with the ArgR-specific control of PartP activity may also explain to a large extent the major differences observed in the in vivo repressibility of artJ and artPIQM transcription (Caldara et al., 2006, 2007) and the higher degree of in vivo occupancy by ArgR of the artJ operator compared to artP (Cho et al., 2011). Indeed, PartJ is about nine-fold more repressible by liganded ArgR than PartP even though in vitro both control regions exhibit a very similar affinity for purified ArgR and an identical degree of overlap of the ARG boxes with the promoter elements (Caldara et al., 2007). In this context, it is worth noticing that in different growth conditions, and besides Lrp, additional regulatory molecules may influence artP transcription and its ArgR-specific regulation in vivo. In a genomic SELEX experiment the artP control region was also identified as a target site for AllR (Hasegawa et al., 2008), a member of the large IclR family of transcription factors that controls a set of genes for degradation and reutilization of purines. It senses both allantoin and glyoxylate (Walker et al., 2006). However, potential binding site(s) for AllR in the artP operator have not been identified and an effect of AllR on PartP has not been demonstrated. Furthermore, artP mRNA levels were found to be about two-fold upregulated under stressful conditions, including the presence of the superoxide generating compound paraquat, and the weak acid salt sodium salicylate (Pomposiello et al., 2001). Furthermore, upregulation of the artP mRNA level was also observed at the onset of stationary phase growth, likely by initiation from additional EσS-specific promoters located upstream of the major PartP studied here (Lacour and Landini, 2004). This all appears now to be linked to the effect of the small regulatory RNA (sRNA) SdsR that is transcribed by EσS and accumulates in stationary phase (Fröhlich et al., 2012). SdsR is highly conserved among enterobacteriaceae and was shown to directly affect artPIQM expression in Salmonella in a Hfq-dependent manner (Fröhlich et al., 2016).
ArgR binds to two 18 bp ARG boxes located downstream of the transcription initiation site (Figure 6), an unusual position for an ArgR binding site, at least in the control region of highly repressible genes and operons, where they overlap the promoter (Charlier et al., 1992; Tian et al., 1992; Caldara et al., 2007). High-resolution contact probing revealed base specific details of ArgR-DNA contacts, and indicates that the promoter-proximal ARG box contributes more to the energy of complex formation than the promoter distal box (Figure 7). Inverse effects (higher affinity binding) upon removal of particular bases in this promoter distal ARG box was observed as well and was particularly striking for the cytosine residues in position 4, 5, 13 and 14 of the top strand (Figures 7B,C). The removal of these cytosine residues appears to eliminate a hindrance for the establishment of nearby contacts. Position 4 and 13 are highly conserved and generally consist of a G-C and T-A pair, respectively, and are located in operator segments where the major groove is facing the repressor (Wang et al., 1998). Since the removal of the complementary guanine residues of the bottom strand did not show such an inverse effect, we may hypothesize that the negative effect on ArgR binding of C-G base pairs at positions 4 and 13 is due to the presence of the exocyclic amino group of the cytosine ring pointing into the major groove. Such a group, which may serve as a donor in the formation of a hydrogen bond, is neither present on the G4 and T13 residues found at these positions on the top strand of the consensus sequence. Instead, G and T carry a carbonyl group pointing in the major groove, which may function as an acceptor in the formation of a hydrogen bond.
Our reporter gene assays point to an indirect effect of ArgR on lrp expression through interference with negative autoregulation (Figure 8). In this context, it is worth noticing that the intracellular concentration of free arginine in E. coli cells grown on minimal medium (0.14 mM; Caldara et al., 2008) is already responsible for significant in vivo repression (Charlier and Glansdorff, 2004; Caldara et al., 2006) and ArgR binding (Cho et al., 2015), and that the ARG boxes of the lrp control region are located at an unusual position downstream of the transcription initiation site (Figure 7). A stronger effect of arginine supplementation on Plrp activity was measured in a 25 bp deletion mutant in which the ARG boxes are positioned immediately downstream of the start of transcription. Previously we have shown that ArgR binding in overlap with the carP2 promoter elements strongly inhibits transcription initiation from this promoter but does not interfere with transcription initiated 67 nt upstream, at the carP1 promoter (Charlier et al., 1988). Combined, these observations suggest that ArgR is unable to function as an efficient roadblock, as some other repressors including PurR do at specific promoters (He and Zalkin, 1992). Interestingly, an indirect effect of ArgR on gene expression exerted from a far upstream binding site has been demonstrated for the gltBDF operon encoding one of the two major pathways for ammonia assimilation in E. coli (Paul et al., 2007). gltB promoter activity is regulated by Lrp, IHF, CRP, and ArgR, and the latter mainly interferes with the activation by Lrp (Paul et al., 2007). Hence, here ArgR acts as an anti-activator rather than as a repressor. ArgR also binds upstream (17 bp) of the −35 element of the hisJ promoter and exerts a weak repression. However, this inhibitory effect of ArgR on promoter activity is direct as it was also observed in a pure in vitro transcription assay (Caldara et al., 2007). The recent genome-wide in vivo ArgR binding study of Cho et al. (2015) has revealed numerous additional ArgR binding sites in intergenic regions of the E. coli chromosome. However, how ArgR exerts a potential direct or indirect regulatory effect on adjacent promoters and how it may interfere negatively or positively with the action of other transcription factors, including NAPs, remains to be elucidated.
Data Availability
All datasets generated for this study are included in the manuscript and/or the supplementary files.
Author Contributions
DC, OT, and IB conceived and designed the experiments. OT, EP, and DC performed the experiments. OT, IB, and DC analyzed and interpreted the data. DC wrote the first draft of the paper. All authors contributed to manuscript revisions, read, and approved the submitted version.
Funding
This work was supported by the Fonds voor Wetenschappelijk Onderzoek Vlaanderen (G.0321.13N to DC) and the Research Council of the Vrije Universiteit Brussel.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge the expert technical assistance of Karl Jonckheere with protein purification.
References
Ames, G. F. L., Nikaido, K., Wang, I. X., Liu, P. Q., Liu, C. E., and Hu, C. (2001). Purification and characterization of the membrane-bound complex of an ABC transporter, the histidine permease. J. Bioenerg. Biomembr. 33, 79–92. doi: 10.1023/A:1010797029183
Azam, T. A., and Ishihama, A. (1999). Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J. Mol. Biol. 274, 33105–33113.
Barrios, H., Valderrama, B., and Morett, E. (1999). Compilation and analysis of σ54-dependent promoter sequences. Nucleic Acids Res. 27, 4305–4313.
Bervoets, I., and Charlier, D. (2019). Diversity, versatility and complexity of bacterial gene regulation mechanisms: opportunities and drawbacks for applications in synthetic biology. FEMS Microbiol. Rev. 43, 304–339. doi: 10.1093/femsre/fuz001
Biemans-Oldehinkel, E., Doeven, M. K., and Poolman, B. (2006). ABC transporter architecture and regulatory roles of accessory domains. FEBS Lett. 580, 1023–1035. doi: 10.1016/j.febslet.2005.11.079
Brunelle, A., and Schleif, R. F. (1987). Missing contact-probing of DNA-protein interactions. Proc. Natl. Acad. Sci. USA 84, 6673–6676.
Burkovski, A., and Krämer, R. (2002). Bacterial amino acid transport proteins: occurrence, functions, and significance for biotechnological applications. Appl. Microbiol. Biotechnol. 58, 265–274. doi: 10.1007/s00253-001-0869-4
Caldara, M., Charlier, D., and Cunin, R. (2006). The arginine regulon of Escherichia coli: whole-system transcriptome analysis discovers new genes and provides an integrated view on arginine regulation. Microbiology 152, 3343–3354. doi: 10.1099/mic.0.29088-0
Caldara, M., Dupont, G., Leroy, F., Goldbeter, A., De Vuyst, L., and Cunin, R. (2008). Arginine biosynthesis in Escherichia coli: experimental perturbation and mathematical modeling. J. Biol. Chem. 283, 6347–6358. doi: 10.1074/jbc.M705884200
Caldara, M., Nguyen Le Minh, P., Bostoen, S., Massant, J., and Charlier, D. (2007). ArgR-dependent repression of arginine and histidine transport genes in Escherichia coli K-12. J. Mol. Biol. 373, 251–267. doi: 10.1016/j.jmb.2007.08.013
Celis, R. T. (1981). Chain-terminating mutants affecting a periplasmic binding protein involved in the active transport of arginine and ornithine in Escherichia coli. J. Biol. Chem. 256, 773–779.
Celis, R. T., Rosenfeld, H. J., and Maas, W. K. (1973). Mutant of Escherichia coli defective in transport of basic amino acids. J. Bacteriol. 116, 619–626.
Charlier, D., and Glansdorff, N. (2004). Biosynthesis of arginine and polyamines. EcoSal Plus 1. doi: 10.1128/ecosalplus.3.6.1.10
Charlier, D., Nguyen Le Minh, P., and Roovers, M. (2018). Regulation of carbamoylphosphate synthesis in Escherichia coli: an amazing metabolite at the crossroad of arginine and pyrimidine biosynthesis. Amino Acids 50, 1647–1661. doi: 10.1007/s00726-018-2654-z
Charlier, D., Roovers, M., Van Vliet, F., Boyen, A., Cunin, R., Nakamura, Y., et al. (1992). Arginine regulon of Escherichia coli K-12. A study of repressor-operator interactions and of in vitro binding affinities versus in vivo repression. J. Mol. Biol. 226, 367–386.
Charlier, D., Weyens, G., Roovers, M., Piette, J., Bocquet, C., Piérard, A., et al. (1988). Molecular interactions in the control region of the carAB operon encoding Escherichia coli carbamoylphosphate synthetase. J. Mol. Biol. 204, 867–877. doi: 10.1016/0022-2836(88)90047-2
Chen, S., and Calvo, J. M. (2002). Leucine-induced dissociation of Escherichia coli Lrp hexadecamers to octamers. J. Mol. Biol. 318, 1031–1042. doi: 10.1016/S0022-2836(02)00187-0
Chen, S., Iannolo, M., and Calvo, J. M. (2005). Cooperative binding of the leucine-responsive regulatory protein (Lrp) to DNA. J. Mol. Biol. 345, 251–264. doi: 10.1016/j.jmb.2004.10.047
Chen, S., Rosner, M. H., and Calvo, J. M. (2001). Leucine-regulated self association of leucine-responsive regulatory protein (Lrp) from Escherichia coli. J. Mol. Biol. 312, 625–635. doi: 10.1006/jmbi.2001.4955
Cho, B., Barrett, C. L., Knight, E. M., Park, Y. S., and Palsson, B. Ø. (2008). Genome-scale reconstruction of the Lrp regulatory network in Escherichia coli. Proc. Natl. Acad. Sci. USA 105, 19462–19467. doi: 10.1073/pnas.0807227105
Cho, S., Cho, Y. B., Kang, T. J., Kim, S. C., Palsson, B., and Cho, B. K. (2015). The architecture of ArgR-DNA complexes at the genome-scale in Escherichia coli. Nucleic Acids Res. 43, 3079–3088. doi: 10.1093/nar/gkv150
Cho, B. K., Federowicz, S., Park, Y. S., Zengler, K., and Palsson, B. Ø. (2011). Deciphering the transcriptional regulatory logic of amino acid metabolism. Nat. Chem. Biol. 8, 65–71. doi: 10.1038/nchembio.710
Cui, Y., Midkiff, M. A., Wang, Q., and Calvo, J. M. (1996). The leucine-responsive regulatory protein (Lrp) from Escherichia coli. J. Biol. Chem. 271, 6611–6617.
Davidson, A. L., and Chen, J. (2004). ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 73, 241–268. doi: 10.1146/annurev.biochem.73.011303.073626
de los Rios, S., and Perona, J. J. (2007). Structure of the Escherichia coli leucine-responsive regulatory protein lrp reveals a novel octameric assembly. J. Mol. Biol. 366, 1589–1602. doi: 10.1016/j.jmb.2006.12.032
Devroede, N., Thia-Toong, T. L., Gigot, D., Maes, D., and Charlier, D. (2004). Purine and pyrimidine-specific repression of the Escherichia coli carAB operon are functionally and structurally coupled. J. Mol. Biol. 336, 25–42. doi: 10.1016/j.jmb.2003.12.024
Fröhlich, K. S., Haneke, K., Papenfort, K., and Vogel, J. (2016). The target spectrum of SdsR small RNA in salmonella. Nucleic Acids Res. 44, 10406–10422. doi: 10.1093/nar/gkw632
Fröhlich, K. S., Papenfort, K., Berger, A. A., and Vogel, J. (2012). A conserved RpoS-dependent small RNA controls the synthesis of major porin OmpD. Nucleic Acids Res. 40, 3623–3640. doi: 10.1093/nar/gkr1156
Hart, B. R., and Blumenthal, R. M. (2011). Unexpected coregulator range for the global regulator Lrp of Escherichia coli and Proteus mirabilis. J. Bacteriol. 193, 1054–1064. doi: 10.1128/JB.01183-10
Hasegawa, A., Ogasawara, H., Kori, A., Teramoto, J., and Ishihama, A. (2008). The transcriptional regulator AllR senses both allantoin and glyoxylate and controls a set of genes for degradation and reutilization of purines. Microbiology 154, 3366–3378. doi: 10.1099/mic.0.2008/020016-0
He, B., and Zalkin, H. (1992). Repression of Escherichia coli purB is by a transcriptional roadblock mechanism. J. Bacteriol. 174, 7121–7127. doi: 10.1128/jb.174.22.7121-7127.1992
Higuchi, R., Krummel, B., and Saiki, R. K. (1988). A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16, 7351–7367. doi: 10.1093/nar/16.15.7351
Hosie, A., and Poole, P. (2001). Bacterial ABC transporters of amino acids. Res. Microbiol. 152, 259–270. doi: 10.1016/S0923-2508(01)01197-4
Hung, S. P., Baldi, P., and Hatfield, G. W. (2002). Global gene expression profiling in Escherichia coli K12. The effects of leucine-responsive regulatory protein. J. Biol. Chem. 277, 40309–40323. doi: 10.1074/jbc.M204044200
Kerppola, R. E., Shyamala, V. K., Klebba, P., and Ames, G. F. L. (1991). The membrane-bound proteins of periplasmic permeases form a complex. Identification of the histidine permease HisQMP complex. J. Biol. Chem. 266, 9857–9865.
Kustu, S. G., and Ames, G. F. L. (1973). The HisP protein, a known histidine transport component in Salmonella typhimurium, is also an arginine transport component. J. Bacteriol. 116, 107–113.
Kutukova, E. A., Livshits, V. A., Altman, I. A., Ptitsyn, L. R., Zyiatdinov, M. H., Tokmakova, I. L., et al. (2005). The yeaS (leuE) gene of Escherichia coli encodes an exporter of leucine, and the Lrp protein regulates its expression. FEBS Lett. 579, 4629–4634. doi: 10.1016/j.febslet.2005.07.031
Kuwabara, M., and Sigman, D. (1987). Footprinting DNA-protein complexes in situ following gel retardation assay using 1, 10-phenanthroline copper ion: Escherichia coli RNA polymerase-lac promoter complexes. Biochemistry 26, 7234–7238.
Lacour, S., and Landini, P. (2004). σS-dependent gene expression at the onset of stationary phase in Escherichia coli: function of σS-dependent genes and identification of their promoter sequences. J. Bacteriol. 186, 3139–3146. doi: 10.1128/JB.186.21.7186-7195.2004
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of the bacteriophage T4. Nature 227, 680–685.
Lazarovici, A., Zhou, T., Shafer, A., Dantas Machado, A. C., Riley, T. R., Sandstrom, R., et al. (2013). Probing DNA shape and methylation state on a genomic scale with DNase I. Proc. Natl. Acad. Sci. USA 110, 6376–6381. doi: 10.1073/pnas.1216822110
Leroy, F., and Charlier, D. (2017). “Arginine deiminase in microorganisms” in The handbook of microbial metabolism of amino acids. ed. J. P. F. D’Mello (Wallingford: CAB International Nosworthy Way).
Lim, D., Oppenheim, J. D., Eckhardt, T., and Maas, W. K. (1987). Nucleotide sequence of the argR gene of Escherichia coli K-12 and isolation of its product, the arginine repressor. Proc. Natl. Acad. Sci. USA 84, 6697–6701.
Lin, R., D’Ari, R., and Newman, E. B. (1992). Lambda placMu insertions in genes of the leucine regulon: extension of the regulon to genes not regulated by leucine. J. Bacteriol. 174, 1948–1955.
McFarland, K. A., and Dorman, C. J. (2008). Autoregulated expression of the gene coding for the leucine-responsive protein, Lrp, a global regulator in Salmonella enterica serovar Typhimurium. Microbiology 154, 2008–2016. doi: 10.1099/mic.0.2008/018358-0
Nguyen Le Minh, P., Velázquez Ruiz, C., Vandermeeren, S., Abwoyo, P., and Charlier, D. (2018). Differential protein-DNA contacts for activation and repression by ArgP, a LysR-type (LTTR) transcriptional regulator in Escherichia coli. Microbiol. Res. 206, 141–158. doi: 10.1016/j.micres.2017.10.009
Nou, X., Braaten, B., Kaltenbach, L., and Low, D. A. (1995). Differential binding of Lrp to two sets of pap DNA binding sites mediated by Pap I regulates Pap phase variation in Escherichia coli. EMBO J. 14, 5785–5797. doi: 10.1002/j.1460-2075.1995.tb00267.x
Oackley, M. G., and Dervan, P. (1990). Structural motif of the GCN4-DNA binding domain characterized by affinity cleaving. Science 248, 847–850. doi: 10.1126/science.2111578
Ogasawara, H., Yamada, K., Kori, A., Yamamoto, K., and Ishihama, A. (2010). Regulation of the Escherichia coli csgD promoter: interplay between five transcription factors. Microbiology 156, 2470–2483. doi: 10.1099/mic.0.039131-0
Paul, L., Mishra, P. K., Blumenthal, R. M., and Matthews, R. G. (2007). Integration of regulatory signals through involvement of multiple global regulators: control of the Escherichia coli gltBDF operon by Lrp, IHF, CRP and ArgR. BMC Microbiol. 7:2. doi: 10.1186/1471-2180-7-2
Peeters, E., Nguyen Le Minh, P., Foulquié-Moreno, M., and Charlier, D. (2009). Competitive activation of the Escherichia coli argO gene encoding for an arginine exporter by the transcriptional regulators Lrp and ArgP. Mol. Microbiol. 74, 1513–1526. doi: 10.1111/j.1365-2958.2009.06950.x
Peeters, E., Thia-Toong, T. L., Gigot, D., Maes, D., and Charlier, D. (2004). Ss-LrpB, a novel Lrp-like regulator of Sulfolobus solfataricus P2, binds cooperatively to three conserved targets in its own control region. Mol. Microbiol. 54, 321–336. doi: 10.1111/j.1365-2958.2004.04274.x
Peterson, S. N., Dahlquist, F. W., and Reich, N. O. (2007). The role of high affinity non-specific DNA binding by Lrp in transcriptional regulation and DNA organization. J. Mol. Biol. 369, 1307–1317. doi: 10.1016/j.jmb.2007.04.023
Pomposiello, P. J., Bennik, M. H. J., and Demple, B. (2001). Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183, 3890–3902. doi: 10.1128/JB.183.13.3890-3902.2001
Reitzer, L. (2005). Catabolism of amino acids and related compounds. EcoSal Plus 1. doi: 10.1128/ecosalplus.3.4.7
Rosen, B. P. (1973). Basic amino acid transport in Escherichia coli. II. Purification and properties of arginine-binding protein. J. Biol. Chem. 248, 1211–1218.
Saier, H. J. (2000). Families of transmembrane transporters selective for amino acids and their derivatives. Microbiology 146, 1775–1795. doi: 10.1099/00221287-146-8-1775
Schmitz, G., Nikaido, K., and Ames, G. F. L. (1988). Regulation of transport operon promoter in Salmonella typhimurium: identification of sites essential for nitrogen assimilation. Mol. Gen. Genet. 215, 107–117. doi: 10.1007/BF00331311
Shimada, T., Saito, N., Maeda, M., Tanaka, K., and Ishihama, A. (2015). Expanded roles of leucine-responsive regulatory protein in transcription regulation of the Escherichia coli genome: genomic SELEX screening of the regulation targets. Microb. Genom. 1:e00001. doi: 10.1099/mgen.0.000001
Shrivastava, T., and Ramachandran, R. (2007). Mechanistic insights from the crystal structures of a feast/famine regulatory protein from Mycobacterium tuberculosis H37Rv. Nucleic Acids Res. 35, 7324–7335. doi: 10.1093/nar/gkm850
Shultzaberger, R. K., and Schneider, T. D. (1999). Using sequence logos and information analysis of Lrp binding sites to investigate discrepancies between natural selection and SELEX. Nucleic Acids Res. 27, 882–887. doi: 10.1093/nar/27.3.882
Tian, G., Lim, D., Carey, J., and Maas, W. K. (1992). Binding of the arginine repressor of Escherichia coli to its operator sites. J. Mol. Biol. 226, 387–397. doi: 10.1016/0022-2836(92)90954-I
Tullius, T. D., and Dombroski, B. A. (1986). Hydroxyl radical footprinting: high resolution information about DNA-protein contacts and application to λ repressor and Cro protein. Proc. Natl. Acad. Sci. USA 83, 5469–5473.
Walker, J. R., Altamentova, S., Ezersky, A., Lorca, G., Skarina, T., Kudritska, M., et al. (2006). Structural and biochemical study of effector molecule recognition by the E. coli glyoxylate and allantoin utilization regulatory protein AllR. J. Mol. Biol. 358, 810–828. doi: 10.1016/j.jmb.2006.02.034
Wang, Q., and Calvo, J. M. (1993). Lrp, a global regulatory protein of E. coli, binds cooperatively to multiple sites and activates transcription of ilvIH. J. Mol. Biol. 229, 306–318. doi: 10.1006/jmbi.1993.1036
Wang, H., Glansdorff, N., and Charlier, D. (1998). The arginine repressor of Escherichia coli K-12 makes direct contacts to minor and major groove determinants of the operators. J. Mol. Biol. 277, 805–824. doi: 10.1006/jmbi.1998.1632
Wang, Q., Wu, J., Friedberg, D., Platko, J., and Calvo, J. M. (1994). Regulation of the Escherichia coli lrp gene. J. Bacteriol. 176, 1831–1839. doi: 10.1128/jb.176.7.1831-1839.1994
Whipple, F. W. (1998). Genetic analysis of prokaryotic and eukaryotic DNA-binding proteins in Escherichia coli. Nucleic Acids Res. 26, 3700–3706.
Wissenbach, U., Keck, B., and Unden, G. (1993). Physical map location of the new artPIQMJ genes of Escherichia coli, encoding a periplasmic arginine transport system. J. Bacteriol. 175, 3687–3688. doi: 10.1128/jb.175.11.3687-3688.1993
Keywords: ArgR, Lrp, arginine transport, ornithine, Escherichia coli, reporter gene assays, protein-DNA interactions, transcription factors
Citation: Torres Montaguth OE, Bervoets I, Peeters E and Charlier D (2019) Competitive Repression of the artPIQM Operon for Arginine and Ornithine Transport by Arginine Repressor and Leucine-Responsive Regulatory Protein in Escherichia coli. Front. Microbiol. 10:1563. doi: 10.3389/fmicb.2019.01563
Edited by:
Harold J. Schreier, University of Maryland, Baltimore County, United StatesReviewed by:
Boris Belitsky, Tufts University School of Medicine, United StatesAkira Ishihama, Hosei University, Japan
Copyright © 2019 Torres Montaguth, Bervoets, Peeters and Charlier. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel Charlier, ZGNoYXJsaWVAdnViLmJl
†Present address: Oscar E. Torres Montaguth, DNA-Protein Interactions Unit, Faculty of Sciences, School of Biochemistry, University of Bristol, Bristol, United Kingdom
 Oscar E. Torres Montaguth
Oscar E. Torres Montaguth Indra Bervoets
Indra Bervoets Eveline Peeters
Eveline Peeters Daniel Charlier
Daniel Charlier