- 1State Key Laboratory of Agrobiotechnology, College of Biological Sciences, China Agricultural University, Beijing, China
- 2Guangdong Provincial Key Laboratory of Stomatology, Guanghua School of Stomatology, Hospital of Stomatology, Sun Yat-sen University, Guangzhou, China
Magnetotactic bacteria (MTB) are a large, polyphyletic group of aquatic microorganisms capable of absorbing large amounts of iron and synthesizing intercellular nano-scaled nanoparticles termed magnetosomes. In our previous transcriptomic studies, we discovered that a novel gene (MGMSRv2_2046, termed as mg2046) in Magnetospirillum gryphiswaldense strain MSR-1 was significantly up-regulated during the period of magnetosome synthesis. In the present study, we constructed a MSR-1 mutant strain with deletion of mg2046 (termed Δmg2046) in order to evaluate the role of this gene in cell physiological status and magnetosome formation process. In comparison with wild-type MSR-1, Δmg2046 showed similar cell growth, but much lower cell magnetic response, smaller number and size of magnetosomes, and reduced iron absorption ability. mg2046 deletion evidently disrupted iron uptake, and redox equilibrium, and strongly inhibited transcription of dissimilatory denitrification pathway genes. Our experimental findings, taken together with results of gene homology analysis, indicate that Mg2046 acts as a positive regulator in MSR-1 under microaerobic conditions, responding to hypoxia signals and participating in regulation of oxygen metabolism, in part as a co-regulator of dissimilatory denitrification pathway with oxygen sensor MgFnr (MGMSRv2_2946, termed as Mg2946). Mg2046 is clearly involved in coupled regulation of cellular oxygen, iron and nitrogen metabolism under micro-aerobic or anaerobic conditions. Our findings help explain how MSR-1 cells initiate dissimilatory denitrification pathway and overcome energy deficiency under microaerobic conditions, and have broader implications regarding bacterial survival and energy metabolism strategies under hypoxia.
Introduction
Magnetotactic bacteria (MTB) are a polyphyletic group of prokaryotes found in aquatic and sedimentary environments worldwide (Faivre and Schüler, 2008). A unique characteristic of MTB is the ability to synthesize magnetosomes [nano-sized and single magnetic domain crystals of magnetite (Fe3O4) or greigite (Fe3S4), arranged in chains and enveloped by a lipid bilayer membrane] under oxygen-limited conditions (Lefevre and Bazylinski, 2013). Magnetite formation by MTB is up to 108 kg/year (Lin et al., 2014). MTB play a key role in environmental iron cycling in view of their worldwide distribution and capacity for iron assimilation into magnetosome. They provide a useful model for studies of microbial orientation and navigation, and a source of natural nanomaterials.
Mechanisms of magnetosome formation have been studied for many decades. Clustering of mam and mms (magnetosome membrane protein genes) typically observed in magnetosome island (MAI, part of MTB genome), is genetic determinant of magnetite biomineralization (Komeili, 2012). This sophisticated, stepwise process has been extensively studied based on deletion of single, multi, or full operon of these genes (Murat et al., 2010; Quinlan et al., 2011; Raschdorf et al., 2013; Lohse et al., 2014, 2016). Functions of Mam proteins (e.g., MamA, MamP) in Mam complex assembly and iron mineralization have been examined in vitro using various approaches (Zeytuni et al., 2011; Jones et al., 2015). There has been increasing interest in the roles of genes involved in iron absorption and regulation of magnetosome formation. Genes of feoAB1 operon control transport of ferrous iron (Fe2+) and play an accessory role in magnetosome formation (Rong et al., 2008; Kolinko et al., 2014). Our previous studies demonstrated that the transcriptional regulator protein Fur (Ferric uptake regulator) regulates iron metabolism genes (including feoAB) and affects magnetosome formation in the well-known MTB, Magnetospirillum gryphiswaldense MSR-1 (Qi et al., 2012; Deng et al., 2015). Schüler’s group showed that periplasmic nitrate reductase, terminal oxidase cbb3, and oxygen sensor MgFnr (fumarate and nitrate reduction regulator in M. gryphiswaldense) are also involved in the biomineralization process (Li et al., 2012, 2013, 2014a). We reported recently that a series of redox enzymes and sensors, including OxyR-like (an H2O2 sensor-like protein) play key roles in magnetosome formation (Zhang et al., 2017). Steadily increasing experimental evidence shows clearly that magnetosome synthesis in MTB is controlled by not only mam/mms genes but also various genes related to basic cellular metabolism in MTB.
We applied transcriptome analysis to explore relationships between magnetosomes and cell physiological status (Wang et al., 2016, 2017). Transcription levels of genes involved in certain metabolic pathways, e.g., dissimilatory denitrification, terminal oxidase, and ferrous uptake genes, were affected by changes in dissolved oxygen levels (Wang et al., 2016). Other studies have shown that these pathways are involved in magnetosome formation (Rong et al., 2008; Li et al., 2012, 2014a). Several unknown proteins and their genes showed notably increased or reduced expression under hypoxic (oxygen-deficient) or iron-rich conditions. Among these, gene MGMSRv2_2046 (here termed mg2046) showed significant 5.6-fold upregulation (in association with magnetosome formation), and RPKM (reads per kilobase of transcript, per million mapped reads) values 45.23 and 8.14 (p-value 0.048 < 0.05) under high- and low oxygen conditions, respectively.
In this study, we constructed a MSR-1 mg2046 deletion mutant (termed Δmg2046), and compared wild-type (WT) vs. mutant strains in terms of magnetosome synthesis and cellular physiology. Our findings suggest that Mg2046 is involved indirectly in the early stage of magnetosome synthesis, and directly in the mature stage through regulation of various metabolic pathways – particularly redox reactions driven by terminal oxidases and dissimilatory denitrification. Such pathways involve appropriate redox state, and affect cellular iron absorption and energy production required for magnetosome formation. Mg2046 is an oxygen-sensitive regulator: its synthesis and activity are inhibited by aerobic conditions. Microaerobic conditions are necessary for Mg2046 activity, which indirectly regulates the MSR-1 biomineralization process.
Materials and Methods
Strains and Culture Conditions
Bacterial strains and plasmids used in this study are listed in Table 1.
Magnetospirillum gryphiswaldense MSR-1 (DSM No. 6361) was originally purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen (Brunswick, Germany) and has been cultivated subsequently for >10 years in our lab. MSR-1 cells were cultured in sodium lactate medium (SLM) at 30°C with shaking (100 rpm). SLM contained (per L) 2.60 g sodium lactate solution (55–65%), 1.02 g NaNO3, 0.50 g K2HPO4⋅3H2O, 0.10 g MgSO4, 0.10 g yeast extract, and 5.00 mL trace element mixture (Rong et al., 2008). Sterilized ferric citrate was added (final concentration 60 μM) after autoclaving. For conjugation experiments, MSR-1 were cultured in solid selecting medium, which contained 4.00 g/L sodium glutamate as N source instead of NaNO3 and yeast extract, and 15 g/L agar. MSR-1 strains were cultured in 100-mL serum bottles filled with 50 mL medium. E. coli strain was cultured in Luria broth (LB) at 37°C with shaking (200 rpm). Antibiotics were used at the following concentrations (μg/mL): (for E. coli) ampicillin (Amp) 100, kanamycin (Km) 50, chloramphenicol (Cm) 50, gentamicin (Gm) 20; (for MSR-1) nalidixic acid (Nx) 5, Km 5, Cm 5, Gm 5.
Construction of mg2046 Mutant and Its Complementary Strain
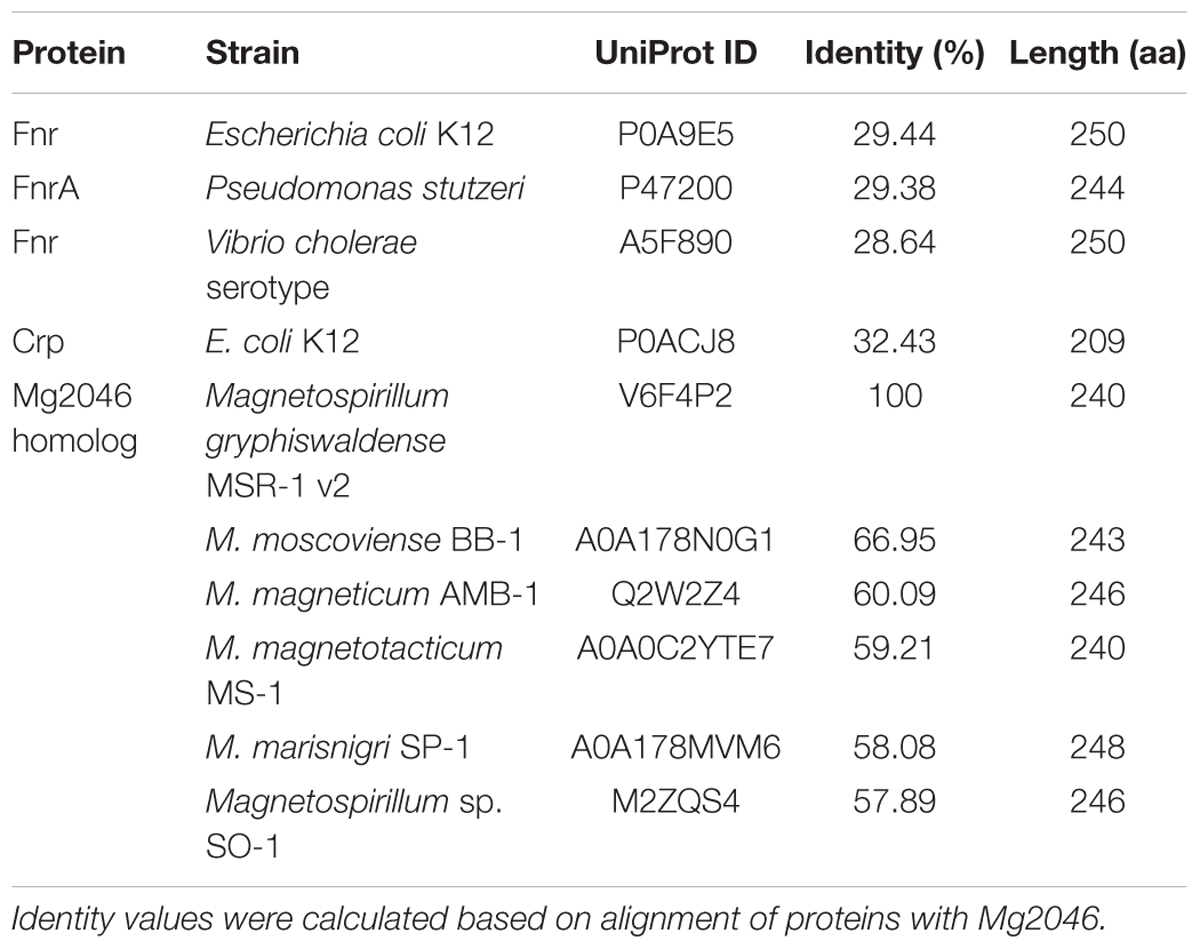
Related genes loci in the genome are illustrated in Figure 1A. Recombinant MSR-1 strain was constructed by biparental conjugation as described previously (Rong et al., 2008). For construction of mg2046-deficient strain, upstream and downstream fragments were amplified and ligated, along with gentamicin resistance cassette from digested pUCGm, into suicide vector pUX19 to form suicide plasmid pUXsuc_mg2046. This plasmid was transformed into MSR-1 by conjugation using E. coli S17-1 as donor strain (Figure 1B). Colonies that showed growth in Gmr/Nxr selecting medium but not in Gmr/Nxr/Kmr selecting medium were considered putative double-crossover strains. Clones were confirmed by PCR. Clones in which mam/mms could be amplified, but mg2046 could not were selected as mg2046 mutants. For construction of complementary strain, mg2046 containing enzyme site amplified by primer were cut and ligated into pBBR1MCS-2 plasmid (Kovach et al., 1995). The recombinant plasmid was transformed into mg2046 mutant by biparental conjugation as above. Clones showing growth in Gmr/Nxr/Kmr selecting medium were confirmed by PCR with mg2046 and mam/mms gene primers (Figure 1C).
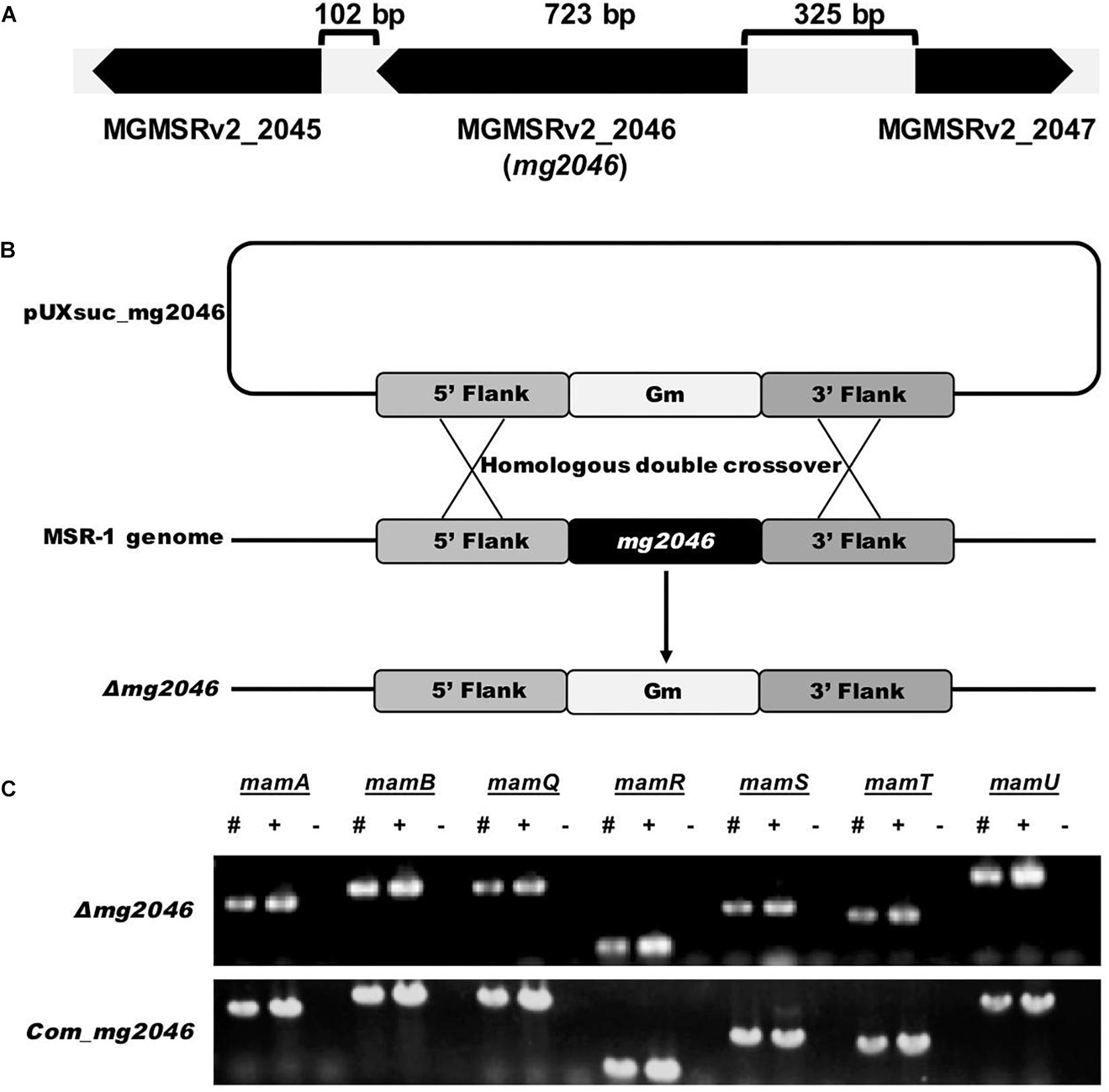
Figure 1. Construction and verification of mg2046 null strain. (A) Schematic representation of mg2046 and flanking genes in MSR-1 genome. Arrows: genes. Brackets: interval gaps between genes. (B) Schematic representation of homologous double-crossover. pUXsuc_mg2046: suicide plasmid. Following double-crossover, mg2046 gene was replaced by gentamicin gene in MSR-1 genome. (C) Detection of mam genes in mutant and complementary strain by PCR. #, Mutant genome; +, complementary strain genome; –, ddH2O used as template. Mutant and complementary strains were successfully constructed without auto-deletion of mam/mms genes.
Cell Growth, Magnetic Response, and TEM Observation
Cell growth and magnetic response were estimated, respectively, as OD565 of culture broth and Cmag value, as described previously (Zhao et al., 2007; Rong et al., 2008). Cells were cultured in SLM for 24 h, washed twice with ddH2O, suspended to a concentration with OD565 ∼1, and dropped onto copper grids. Samples were air-dried and observed directly by TEM (model JEM-1230, JEOL; Tokyo, Japan). Numbers and diameters of magnetosomes were analyzed statistically using ImageJ software program (National Institutes of Health; Bethesda, MD, United States). Lattice structure of magnetosomes was observed by high-resolution TEM (model JEM-2100, JEOL).
Iron Absorption Ability and Intracellular Iron Content
Cells were grown in SLM for 24 h at 30°C, and harvested by centrifugation at 4000×g and 4°C for 20 min. The pellet was washed with equivalent volume of PBS and dried to constant weight. Cells were digested by nitric acid, and total cellular iron content was measured by inductively coupled plasma-optical emission spectrometry (ICP-OES) (Optima 5300 DV system, PerkinElmer; Waltham, MA, United States) (Rong et al., 2008). Residual iron concentration was measured by ferrozine method in broth supernatant aspirated after centrifugation (Dailey and Lascelles, 1977).
Magnetic Properties
Cells were cultured and harvested as in the above section, and room-temperature hysteresis loops and first-order reversal curves (FORCs) were measured using a VSM3900 magnetometer (Princeton Measurements Corp.; Westerville, OH, United States; sensitivity 5 × 10-10 Am2). Saturation magnetization, saturation remanence, and coercivity were determined as described previously (Li et al., 2010), with slight modification of methods.
Real-Time Quantitative PCR (qPCR)
Cells were cultured and harvested as in the preceding sections. Total RNA was isolated using TRIzol reagent (Tiangen Biotech; Beijing, China), and genome DNA in total RNA was digested by DNase I (Takara; Shiga, Japan). DNA remaining in total RNA was detected by PCR using digested product as template. RNA was then reverse-transcribed into cDNA using Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega; Madison, WI, United States). Transcriptional abundance of genes in samples was quantified by qPCR, using housekeeping gene rpoC (encodes RNA polymerase subunit β′) as internal control. qPCR was performed using LightCycler 480 RT-PCR system and LightCycler 480 SYBR green I Master kit (Roche; Mannheim, Germany) as per manufacturer’s instructions. Selected genes and primer sequences are listed in Supplementary Table S1. Relative transcription levels of genes were determined by 2-ΔΔCp method (Zhang et al., 2012; Wang et al., 2015).
Dissimilatory Nitrate Reductase Detection
Cells were cultured and harvested as in the preceding sections. Dissimilatory nitrate reductase enzyme activity of tested bacterial cells was analyzed using bacterial dissimilatory nitrate reductase activity colorimetric assay kit GMS15031.1.3 v.A (Genmed Scientifics; Wilmington, DE, United States) as per manufacturer’s instructions.
Bioinformatic Analysis
Selected MSR-1 genes were identified by reference to complete genome sequences (M. gryphiswaldense MSR-1 v2) found at MicroScope website1 (note: gene IDs mentioned in this report were taken from this website) or NCBI (GenBank Accession: NC_023065) (Wang et al., 2014).
Results and Discussion
Mg2046 Is a Novel Regulatory Protein and Is Conserved in Genus Magnetospirillum
Gene MGMSRv2_2046 (mg2046) is located from 2160600 to 2161322 nt (723 bp) in the complete M. gryphiswaldense MSR-1 genome (MGMSRv2), and was annotated as putative transcriptional activator FnrA (fumarate and nitrate reduction regulator A). Upstream gene MGMSRv2_2047, mg2046, and downstream gene MGMSRv2_2045 are located at intervals of 325 and 102 bp without overlap (Figure 1A). Transcriptional orientation of each gene is indicated by arrows. mg2046 encodes a protein of 240 amino acids.
Fnr-family proteins (whose members include FnrA and FnrL) belong to the Crp/Fnr superfamily, which contributes to the metabolic versatility of bacteria (Körner et al., 2003). Fnr and Crp (cAMP receptor protein) are the most common members of this superfamily. Searches of genomes of other bacteria showed that Mg2046 has low identity of 29.44% with Fnr from Escherichia coli K12, 29.38% with FnrA from Pseudomonas stutzeri, 28.64% with Fnr from Vibrio cholera, and had similarities to items in the MaGe annotation platform of SwissProt (Table 2). According to NCBI’s Blastp program on NCBI, MGMSRv2_2046 is predicted to have specific hits of cd00038: CRP_ED domain in N-terminus and pfam13545: HTH_Crp_2 domain in C-terminus, suggesting that it is a Crp. However, Mg2046 has low identity (32.43%) with Crp from E. coli K12 (Table 2). Mg2046 can therefore not be identified simply as a Crp or Fnr protein. On the other hand, Mg2046 homologs have identities with proteins from other Magnetospirillum species of 66.95% with MAGMOB_540028 from M. moscoviense BB-1, 60.09% with amb2977 from M. magneticum AMB-1, 59.21% with JXSL01_v1_280147 from M. magnetotacticum MS-1, 58.08% with MAGMAS_370140 from M. marisnigri SP-1, and 57.89% with AONQv1_290020 from Magnetospirillum sp. SO-1 (Table 2). These findings indicate that Mg2046 homologs are highly conserved in genus Magnetospirillum.
Construction of Mutant Strain Δmg2046
To investigate the role of Mg2046 in MSR-1 cell growth and magnetite biomineralization, we constructed null mutant strain Δmg2046 using homologous double-crossover strategy as described previously (Wang et al., 2015; Figure 1B). mam/mms genes were detected by PCR, in consideration of their possible auto-deletion during double-crossover of the target gene (Figure 1C). Upstream and downstream regions (up to 1 kb) of mg2046 in the mutant were sequenced, confirming that there were no other changes. The genes flanking mg2046 were also investigated. The mg2045 transcribes in the same direction with mg2046, while the mg2047 transcribes in the opposite direction to mg2046. The relative transcription levels of mg2045 in the mutant and the wild type strain were analyzed by qPCR, and no significant difference was found.
A complementary strain was constructed, mam/mms genes were again confirmed by PCR, and sequencing of adjacent regions demonstrated that there were no other changes (Figure 1C). We did not observe recovery of magnetic response in the mutant back to WT level. That is, transfer of mg2046 complementary plasmid did not recover the magnetosome formation ability back to WT phenotype. qPCR analysis showed that mg2046 was overtranscribed >60-fold in Δmg2046 relative to WT (Supplementary Figure S1). Expression of complementary gene was too high in the receptor. One possible explanation is that gene copies, promoters, and regulatory processes were distinctive in plasmid vs. genome structure, resulting in different transcription and expression levels. Overexpression of mg2046 disrupted the metabolic balance of cells. On the other hand, magnetosome formation requires proper expression of Mg2046, triggered by low oxygen conditions. Schüler’s group reported a similar phenomenon when MgFnr (regulator of dissimilatory denitrification) was overexpressed (Li et al., 2014b). Therefore, the complementary strain was not used for subsequent experiments.
Δmg2046 Cells Grow Normally but Display Low Magnetic Response
WT and Δmg2046 were cultured under microaerobic conditions, with medium and culture conditions similar to those in our previous studies. As Mg2046 belongs to the Fnr family, and Fnr is one of the major regulatetors of dissimilatory denitrification pathway, ammonium in the medium was replaced by sodium nitrate at equal concentration. Samples were taken every 6 h, and cell growth and magnetic response were estimated, respectively, by OD565 and Cmag. Cell growth curves of Δmg2046 and WT were similar, with maximal OD565 (at 18 h) 0.94 ± 0.05 and 0.89 ± 0.04, respectively (Figure 2A), indicating that mg2046 deletion had no notable effect on cell growth. In contrast, Cmag curves for the two strains were very different. Cmag for WT was 0.93 ± 0.22 at 12 h and 1.31 ± 0.09 at 24 h, whereas Cmag for Δmg2046 was 0.14 ± 0.16 at 18 h and only 0.20 ± 0.12 at 24 h (Figure 2B). The reasons for the strikingly lower Cmag values of Δmg2046 were explored in subsequent experiments, as described in the following sections.
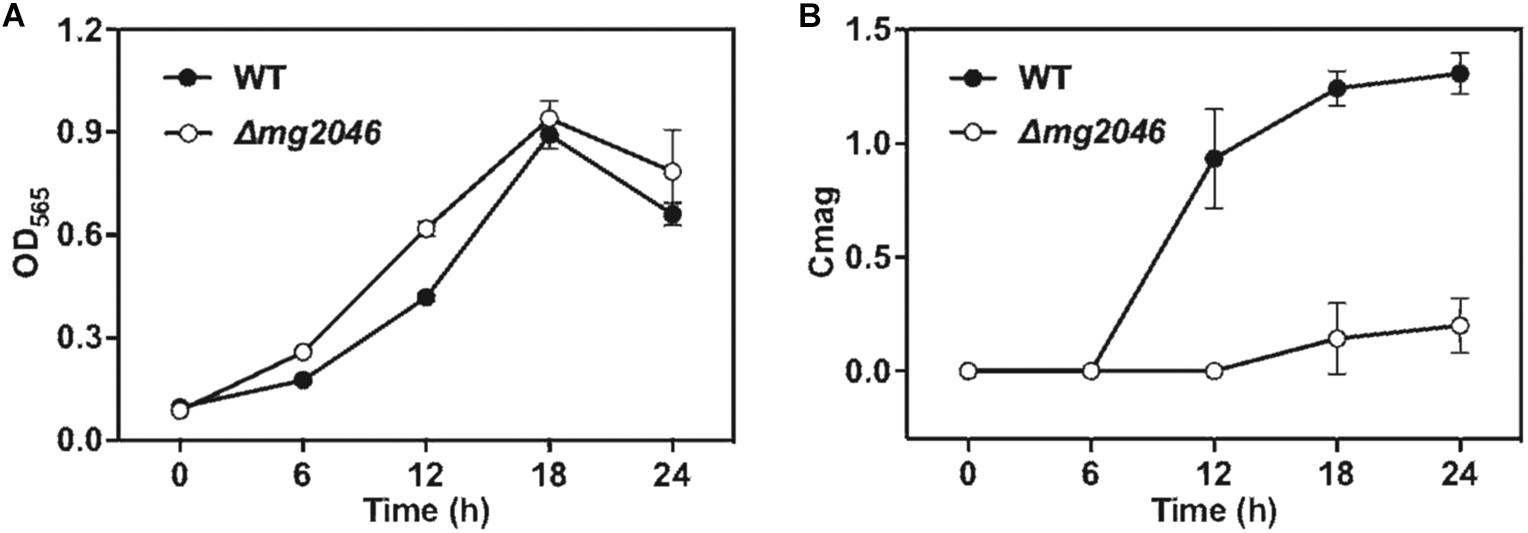
Figure 2. Cell growth and magnetic response of WT and Δmg2046. (A) Cell growth (OD565). (B) Magnetic response (Cmag). Growth was comparable for the two strains, but magnetic response was much lower for Δmg2046.
Synthesis of Abnormal Magnetosome Chains in Δmg2046
Cultured WT and Δmg2046 cells were collected at 24 h and observed by transmission electron microscopy (TEM). Magnetosomes in WT had normal appearance, and formed a regular linear chain along the long axis of the cell, whereas those in Δmg2046 were fewer in number, irregularly shaped, and had large spaces between particles (Figure 3A). High-resolution TEM revealed that Δmg2046 had smaller particles but had typical ferroferric oxide lattice structure similar to that of WT (Figure 3B). Average magnetosome number per cell (8.30 ± 3.02 vs. 18.11 ± 4.89) and magnetosome particle size (16.50 ± 5.26 vs. 36.10 ± 9.17 nm) were much smaller for Δmg2046 than for WT (details in Supplementary Table S2). In box-plot charts, minimum (low bar end), majority (box), and maximum (high bar end) values for these two parameters were also significantly smaller (p < 0.01) for Δmg2046 than for WT (t-test, p < 0.01) (Figures 3C,D).
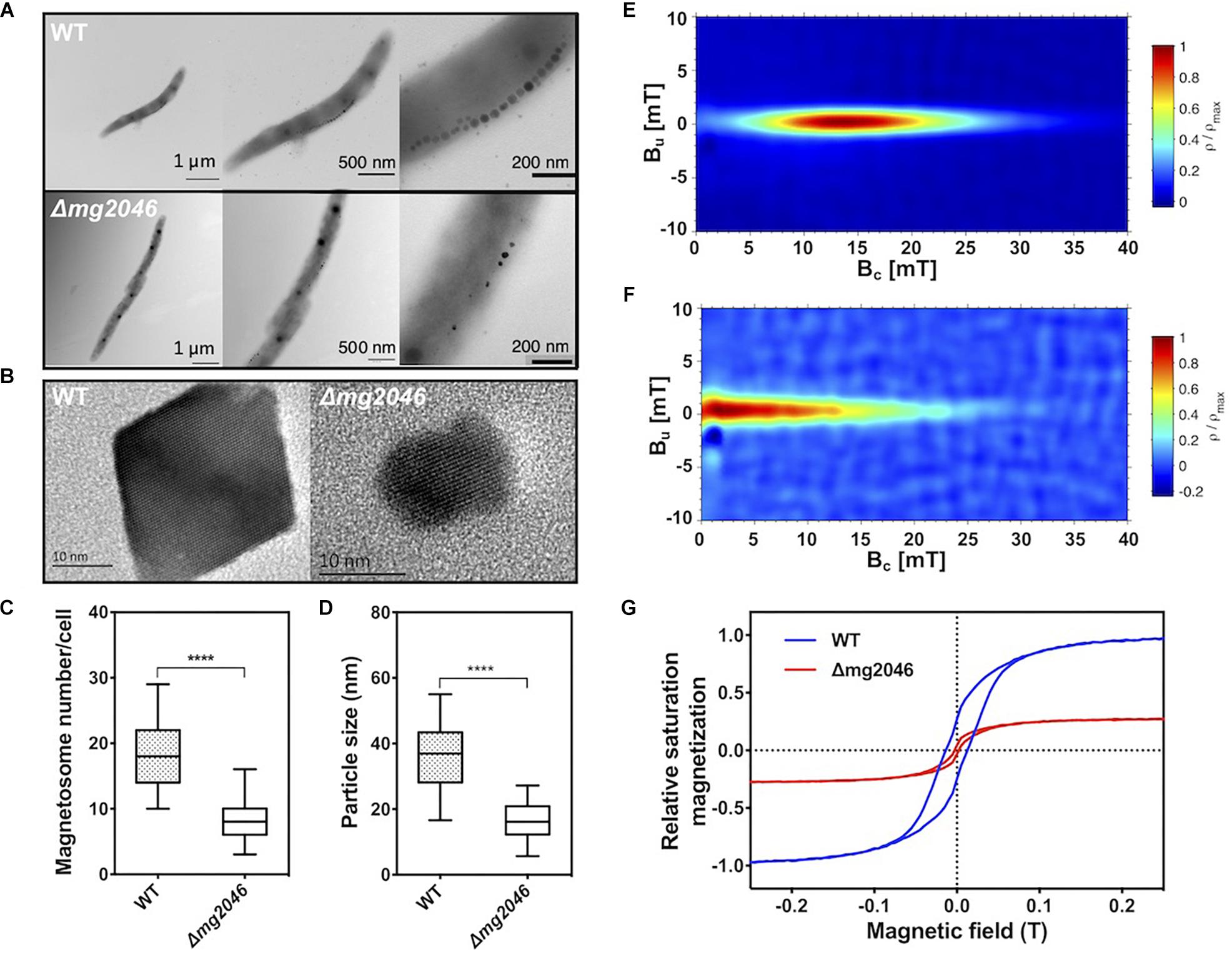
Figure 3. Characteristics of magnetosomes synthesized by WT and Δmg2046. (A) TEM images, with progressive magnification from left to right. Bars: 1 μm, 500 nm, 200 nm. (B) High-resolution TEM image of nanoparticles. Both strains showed typical ferroferric oxide lattice structure. Bar: 10 nm. (C,D) Box-plot charts showing statistical analysis of magnetosome diameter and number. ∗∗∗∗p < 0.05. (E) FORCs diagram of WT. (F) FORCs diagram of Δmg2046. (G) Relative saturation magnetization based on comparison of hysteresis loops of WT (blue) and Δmg2046 (red). Max movement of WT is defined as ±1. Magnetosomes synthesized by Δmg2046 were fewer, smaller, and more weakly magnetic.
To elucidate magnetic properties of magnetosome, we measured room temperature first-order reversal curves (FORCs) and hysteresis loops. FORCs diagram for WT showed closed contours with narrow vertical distribution around coercivity value ∼15 mT (Figure 3E), indicating predominant formation of typical single-domain magnetosomes, whereas FORCs diagram for Δmg2046 showed much lower coercivity value (<5 mT) (Figure 3F). Relative saturation magnetization was compared based on hysteresis loops. Hysteresis loop for WT (Figure 3G; blue line) had “pot-bellied” shape, while that for Δmg2046 (red line) had “thin-waisted” shape.
Comparison of relative saturation magnetization as above indicates that degree of magnetism is less for Δmg2046 than for WT. The difference between the two strains appears to be mainly in their magnetic properties. The above findings, taken together, indicate that mg2046 deletion alters magnetosome synthesis and maturation, resulting in particles that are fewer, smaller, and less magnetic.
Iron Absorption Rate and Intracellular Iron Content
Reduced iron absorption also can reduce magnetosome synthesis in cells. To estimate iron utilization, WT and Δmg2046 were cultured under microaerobic conditions with nitrate supplementation, and sampled at 6-h intervals. OD565 and iron concentration of culture medium were measured. Iron absorption rate [μmol/(OD565∗h)] was calculated as [iron concentration (t h)∗volume – iron concentration (t h + 6 h)∗volume]/{[OD565 (t h + 6 h)∗volume – OD565 (t h)∗volume]∗6 h}. Iron absorption velocities are compared in Figure 4A. Maximal velocities were observed in the initial 0–6 h period: 8.98 ± 0.14 and 4.86 ± 0.22 μmol/(OD565∗h) for WT and Δmg2046, respectively. Values for both strains declined to ∼2 in the 6–12 h and 12–18 h periods. In the 18–24 h period, values increased to 5.76 ± 0.27 and 4.48 ± 0.13, respectively. Because cells were first grown in trace-ferric medium, and then inoculated into rich-ferric medium, it is possible that they absorbed ferric ion quickly and were ready for magnetosome formation within the initial 6-h period. Comparison of the strains over the entire culture process showed that iron absorption rate was lower for Δmg2046 than for WT during the 0–6, 6–12, and 18–24 h periods, and similar during the 12–18 h period. Thus, mg2046 deletion inhibited iron uptake. Intracellular iron contents of the two strains following 24 h were measured. Intracellular iron accounted for 0.34 and 0.26% of cell dry weight in WT and Δmg2046, respectively (Figure 4B). Iron content of WT was 1.31-fold higher than that of Δmg2046. Absence of mg2046 results in partial inhibition of iron absorption and reduction of intracellular iron content, such that cells do not have sufficient iron for synthesis of normal magnetosome chains.
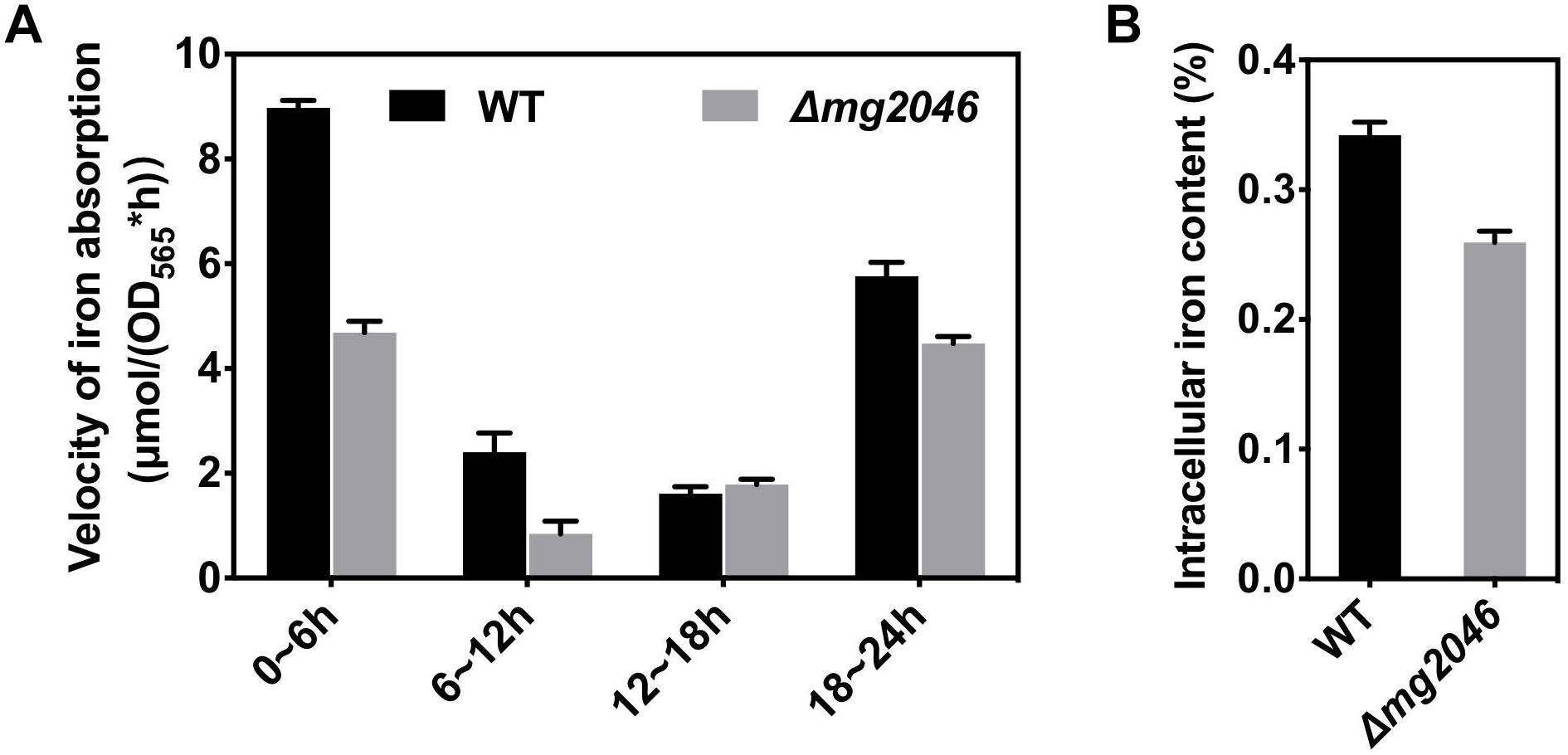
Figure 4. Iron utilization by WT and Δmg2046. (A) Iron absorption rate. (B) Intracellular iron content. Both parameters were lower for Δmg2046 than for WT.
The basic requirements for absorption by MTB of iron source rapidly and in large quantity are low-oxygen (microaerobic) and high-iron conditions. The reduction of iron absorption by mg2046 knockout may conceivably be due to loss of oxygen -regulating ability, insufficient energy, and/or iron supply. To clarify the reasons, we evaluated and compared transcription of selected target genes in WT and Δmg2046. The genes and their qPCR primers of selected target genes are listed in Supplementary Table S1.
Effects of mg2046 Deletion on Expression of Genes in Various Key Metabolic Pathways
WT and Δmg2046 cultured for 8 and 16 h, and gene transcription were measured by qPCR. Target genes included mam/mms, iron transport, terminal oxidase, and dissimilatory denitrification pathway genes.
mam/mms Genes
Relative transcription levels were measured for mamY, mamH, mamF, mms6, and the first genes of mamXY, mamAB, mamCD, and mms operon (Figures 5A,B). Differences (fold changes in detail) between samples were calculated and summarized in Figure 5E. Values for WT at 8 h were defined as 1. Fold changes of transcription levels ranged from 0.70 to 1.56 (changes <twofold, Figure 5E), except that mms6 level in mg2046 was 0.43-fold that in WT at 8 h (Figure 5B). For comparisons of other mam/mms genes (mamE, -A, -B, -P, -Q, -S, -T, belonging to mamAB cluster, and mmsF fold changes ranged from 0.58 to 1.87 (changes <twofold, Figure 5E), except for mamE fold change 2.23 for Δmg2046-16 h vs. WT-16 h, and mmsF fold change 0.42 for Δmg2046-8 h vs. WT-8 h (Figures 5B,E). Thus, various mam/mms genes were stably transcribed at 8 and 16 h, transcription levels differed slightly between WT and Δmg2046, and most mam/mms genes appeared to be expressed in constitutive form. These findings are consistent with our previous transcriptomic analysis (Wang et al., 2016). mg2046 deletion had minimal effect on transcription of mam genes, but reduced mms6 and mmsF expression in early-stage (0–8 h) magnetosome synthesis, the period when cells absorb large quantities of iron. Functions of mam/mms genes have been suggested previously to be related to membrane invagination, vesicle formation, ion transport, or control of magnetosome arrangement. Mms6 protein expression is closely related to crystal nucleus formation in magnetosomes. Mms6 was proposed to bind iron ions to control magnetic particle size (Arakaki et al., 2003; Tanaka et al., 2011; Wang et al., 2012). Recent Mms6 structure prediction suggests that its C-terminus is hydrophilic and enriched in acidic amino acid residues, such that Mms6 can aggregate spontaneously on the surface of solution or solid phase to form polymers of varying sizes, which determine magnetic particle size (Arakaki et al., 2016; Staniland and Rawlings, 2016). Other studies suggest that self-assembled MmsF controls magnetite nanoparticle formation in vitro (Murat et al., 2011), and is a key regulator of magnetite biomineralization (Rawlings et al., 2014). Our finding that mg2046 deletion reduces transcription of mms6 and mmsF suggests that Mg2046 protein is involved in regulation of certain pathways and affects the biomineralization process during early stages of cell growth.
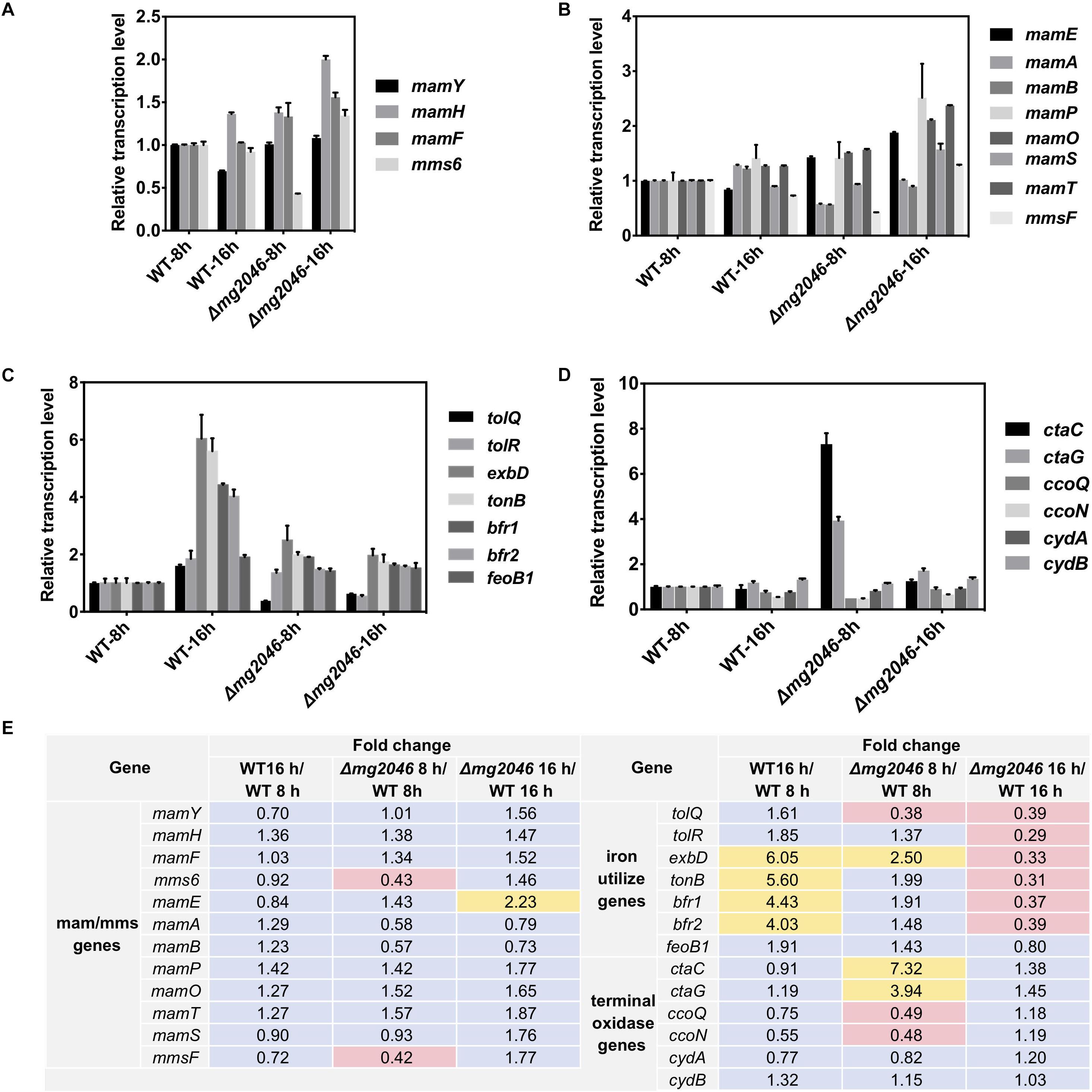
Figure 5. Relative transcription levels of key pathway genes in WT and Δmg2046. (A) First genes of mam/mms operons (see text). (B) Important mam/mms genes. (C) Iron metabolism related genes. (D) Terminal oxidase genes. Transcription levels of genes in WT 8 h sample were used as reference (defined as 1) for expression of fold changes in other samples. (E) Fold changes between samples. Yellow: fold change >2. Pink: fold change <0.5. Δmg2046 showed disrupted transcription of mms, terminal oxidase, iron uptake, and storage related genes.
Iron Metabolism Related Genes
We examined transcription in WT and Δmg2046, at 8 and 16 h, of ion transport Tol system genes tolQ and tolR (Goemaere et al., 2007), iron affinity uptake system TonB-ExbBD component genes tonB and exbB (Wang et al., 2014; Klebba, 2016), ferrous uptake system gene feoB1 (Rong et al., 2008), and iron storage protein bacterioferritin bfr1 and bfr2 genes (Wang et al., 2016). In WT, transcription levels of these genes were relatively consistent and stable up to 8 h culture. Transcription of tonB, exbB, bfr1, and bfr2 was upregulated >fourfold in WT-16 h compared to WT-8 h. In Δmg2046, transcription levels of all genes were unstable during the initial stage (0–8 h), when activity of two iron transport systems is relatively low. All genes except feoB1 were downregulated (fold change 0.31–0.39) in Δmg2046-16 h relative to WT-16 h (Figures 5C,E). Loss of Mg2046 reduced expression of genes related to iron transport. Iron affinity and absorption of Δmg2046 showed a slight increase at 16h, but this increase was limited and unevenly expressed in comparison with WT.
The TonB-ExbBD system in bacteria is generally a high ferric affinity transport system, and Bfr stores or releases iron to maintain iron balance in cells. Studies of iron utilization and regulation in MSR-1 to date have focused mainly on the ferrous uptake (Feo) system and ferrous uptake regulator (Fur). In the present study, transcription of tonB, exbD, bfrs, and feoB1 were much higher in WT 16 h than in WT 8h. Such increase was not observed in Δmg2046 16 h relative to Δmg2046-8 h.
In view of the fact that iron utilization and balance in bacteria are co-regulated by Fur family proteins and intracellular ferrous/ferric concentration, the inhibitory effect of TonB, ExbD, and Bfr is presumably exerted in indirect manner, perhaps through disruption of cellular redox balance. MSR-1 has certain iron absorption and regulatory genes (including Tol and TonB-ExbBD system genes) that are ubiquitous in non-magnetic bacteria, but has greater affinity for Fe3+ than for Fe2+, and does not secrete siderophores (Schüler and Baeuerlein, 1996; Wang et al., 2014); therefore, it may have yet-unknown genes involved in Fe3+ absorption (Wang et al., 2016). Coupling of regulation of iron and oxygen metabolism has been clearly demonstrated in MTB. Many aspects of this complex regulatory network remain to be clarified. The present findings indicate that loss of mg2046 greatly reduces transcription of ferric transport and iron storage related genes during the magnetosome formation process.
Terminal Oxidase Operon Genes
The aerobic respiration pathway in MSR-1 has three branches, which allow adaptation and energy production to changing dissolved oxygen levels. When cellular oxygen level decreases to hypoxic conditions, bacterial heme is expressed in large quantities, combines with oxygen with high affinity, and facilitates oxygen transportation and utilization. We examined transcription levels in WT and Δmg2046, at 8 and 16 h, of three terminal oxidase operons: ctaC and ctaG (ctaCDGE operon), ccoQ and ccoN (ccoNOQP operon), and cydA and cydB (cydAB operon) (Wang et al., 2016). In WT, transcription of these genes was relatively stable, with slight differences among genes at 16 h, with fold changes in the range 0.55–1.32 (changes <twofold) relative to WT 8 h (Figures 5D,E). In Δmg2046 8 h, ctaC and ctaG were upregulated 7.32–and 3.94-fold, and ccoQ and ccoN were downregulated 0.48-fold relative to WT 8 h. These findings suggest that in early-stage cell growth intracellular oxygen level fluctuates because of the absence of mg2046, and increased amounts of ctaC and ctaG expression products are required to maintain oxygen balance. In Δmg2046 16 h, levels of these genes were more stable, and were only 1.18- to 1.45-fold higher than in WT 16 h (Figures 5D,E). Transcription levels of cydA and cydB ranged from 0.77 to 1.32 for all comparison sets, indicating more consistent expression in both WT or Δmg2046 during culture. In summary, mg2046 deletion had opposite effects on transcription of cta and cco genes; the former were upregulated and the latter downregulated during early stage (0–8 h) when cells experienced microaerobic culture conditions.
Terminal oxidases are components of the final-step reaction in respiratory chain. Three types of terminal oxidase complex (Cyd, Cta, Cco) are found in Magnetospirillum, and Cta and Cco are conserved in strains MSR-1, AMB-1, and SO-1 (Wang et al., 2016). Cco terminal oxidases are involved in maintenance of the proper redox state required for magnetosome formation (Li et al., 2014a). In the present study, mg2046 deletion resulted in low cco expression. Terminal oxidases are regulated by sensing/control system RegBA/PrrBA (the name varies depending on strain) activated by redox state under low-oxygen condition (Bueno et al., 2012). In microarray analysis of Pseudomonas aeruginosa genome, RoxSR (component of RegBA/PrrBA) upregulated transcription of ccoNOQP operon and downregulated cox gene (Cox and Cta are both aa3 terminal oxidases) under low-oxygen conditions (Kawakami et al., 2010; Bueno et al., 2012). mg2046 deletion thus appears to cause striking changes during early-stage cell growth. Upregulation of cta transcription failed to restore normal oxygen balance or iron absorption in cells, and was therefore inadequate to support normal biomineralization process.
Faivre’s group used X-ray absorption spectroscopy and TEM to show that magnetite particles were formed by a reductive process from Fh (Fe2O3⋅n H2O) (Baumgartner et al., 2013). We demonstrated that reducing power (NADH/NAD+ ratio) in MSR-1 increased rapidly when a large number of magnetosomes were synthesized (Yang et al., 2013). Redox status clearly plays an important role in magnetosome formation. Dissimilatory denitrification and terminal oxidases (particularly Cco) are closely associated with redox status and reducing power. Kralova et al. (1992) reported that at low redox level dissimilatory denitrification products (dinitrogen or nitrous oxide) were generated, and dinitrogen transformation (the final step of dissimilatory denitrification) was enhanced. In Rhodobacter sphaeroides, Cco functions as a redox sensor under aerobic conditions, but enhances reducing power under anaerobic conditions (Oh and Kaplan, 2002). In contrast to bacteria (e.g., E. coli) that display only partial dissimilatory denitrification and lack Cta and Cco terminal oxidases, Magnetospirillum strains have complete dissimilatory denitrification pathway and multiple types of terminal oxidases (Wang et al., 2016). Thus, redox sensing/regulation in Magnetospirillum differs from that in E. coli. In Magnetospirillum, Mg2046 acts as a redox regulator that promotes cco transcription and iron absorption.
Dissimilatory Denitrification Pathway Genes
This pathway has four operons (nap, nir, nor, nos; the first genes are, respectively, napF, nirT, norC, nosZ) and is responsible for efficient transformation of nitrate → nitrite → nitric oxide → nitrous oxide → nitrogen. Several previous studies indicate that dissimilatory denitrification genes are strongly upregulated under microaerobic conditions and play essential roles in magnetosome formation (Li et al., 2012, 2013; Wang et al., 2016). In the present study, transcription levels of the above four genes in Δmg2046 vs. WT were similar during the 0–8 h period, but differed significantly during the 8–16 h period (Figure 6A; details in Supplementary Table S3). The levels were 10- to 103-fold higher in WT 16 h than in WT 8 h. In Δmg2046 16 h, transcription levels of napF, nirT, norC, and nosZ were much lower (respectively, 0.28, 1.13 × 10-3, 1.42 × 10-2, and 2.16 × 10-2 fold) than those in WT 16 h. These findings indicate that in middle- and late-stage cell growth (magnetosome maturation stage), the dissimilatory denitrification pathway is strongly activated, and that mg2046 deletion blocks such activation, particularly in the last three steps. We further investigated activity of nitrate reductase, the key catalytic enzyme in first step. Samples were taken from the two strains at 3-h intervals after 6 h culture. In WT, enzyme activity [nmol nitrate/(mg∗min)] was ∼3.0 at 6 and 9 h, 16.1 at 12 h, and 30.0 at 18 h (Figure 6B). In Δmg2046, enzyme activity values were also ∼3.0 at 6 and 9 h, but did not reach any higher than 7.0 at subsequent sampling times. It appears that enzyme activity of WT and Δmg2046 was similar from 6 to 9 h, but dissolved oxygen level in culture medium subsequently decreased, and nitrate reductase activity in WT strongly increased. Enzyme activity of WT, relative to Δmg2046, was threefold higher at 12 and 15 h, and almost 10-fold higher at 18 h.
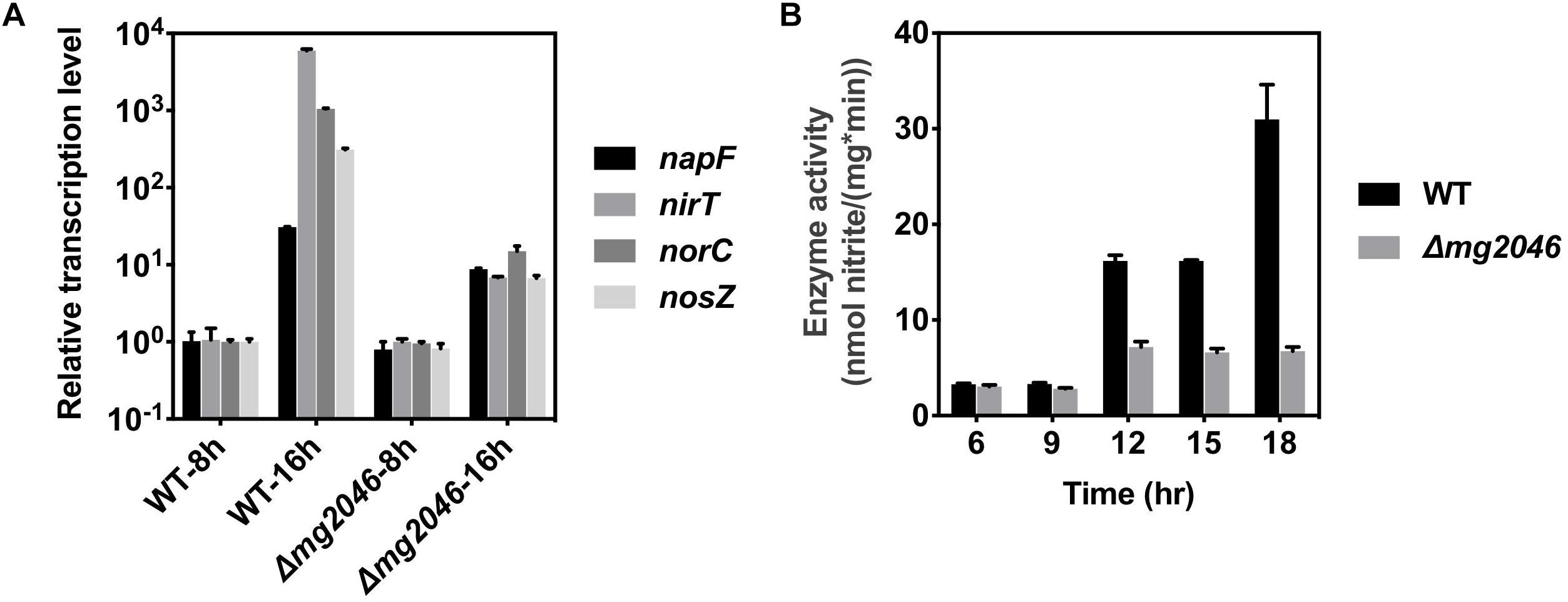
Figure 6. Transcription levels of nap, nir, nor, nos genes, and enzyme activity of nitrate reductase, in WT and Δmg2046. (A) Relative transcription levels of first genes (nap, nir, nor, nos) of four operons of dissimilatory denitrification pathway (see text) in WT 8 h, WT 16 h, Δmg2046 8 h, and Δmg2046 16 h. (B) Nitrate reductase activity at times 6, 9, 12, 15, and 18 h.
Enzyme activities of the Δmg2046 mutant strain were slightly higher at 12 and 15 h than at 6 or 9 h, perhaps because of regulation of MgFnr (Mg2946); however, energy produced by dissimilatory denitrification pathway was limited because cells were entering the late stage of growth. The observed patterns of enzyme activity in the two strains were consistent with those of gene transcription levels. Inhibition of magnetosome synthesis in Δmg2046 resulted from not only iron deficiency, but also lack of normal energy supply. It is reasonable to conclude that Mg2046 strongly activates transcription of dissimilatory denitrification pathway genes, and that this pathway is essential for magnetosome maturation.
In our previous study, MSR-1 cells showed comparable growth under aerobic and microaerobic conditions, but synthesized magnetosomes only under microaerobic conditions (Wang et al., 2016). High vs. low oxygen supply results in striking differences of MSR-1 metabolism. Understanding the mechanism of biomineralization in MTB requires clarifying the links between cell physiological status and magnetosome formation process. Specific regulators control metabolic pathways of bacteria cultured under differing oxygen concentrations. Two of the best-studied regulators are oxygen- and redox-sensitive proteins Fnr and OxyR. Homologs (or similar proteins) of Fnr and OxyR have been reported in MSR-1. Deletion of MgFnr (Fnr homolog in MSR-1) slightly affected magnetosome formation, mainly by inhibiting nitrous oxide reductase activity under microaerobic conditions (Li et al., 2014b). Deletion of OxyR-like resulted in abnormally shaped magnetosomes, but apparently not through disruption of carbon metabolism (Zhang et al., 2017). Mg2046, the focus of the present study, is clearly involved in basic metabolism and magnetosome formation in MSR-1. In its role as a regulatory protein, its transcription is significantly inhibited by hyperoxia, whereas its function is activated under appropriate microaerobic conditions. Mg2046 function is induced by hypoxic signaling, resulting in activated transcription of dissimilatory denitrification pathway genes, sufficient energy for cell growth and high iron absorption under microaerobic conditions, and consequent magnetosome synthesis.
Conclusion
This study was focused on the role of novel protein Mg2046 in MSR-1 cell growth and magnetosome synthesis. Major conclusions are as follow. (1) Mg2046 positively regulates magnetosome synthesis in MSR-1, in indirect manner. Absence of Mg2046 disrupts oxygen and iron metabolism, resulting in a series of associated negative effects: reduced iron absorption ability, reduced transcription levels of dissimilatory denitrification pathway genes, inadequate energy supply, and inability to synthesize normal magnetosomes. (2) Mg2046 is a hypoxia-dependent redox regulator involved in maintenance of proper redox state required for magnetosome formation. It appears to exert its regulatory function during early-stage cell growth, and may cooperate with Mg2946 (MgFnr) in regulation of dissimilatory denitrifying pathway, but the two proteins are activated by different oxygen levels. These findings help explain how MSR-1 cells initiate dissimilatory denitrification pathway and overcome energy deficiency under microaerobic conditions, and have broader implications regarding bacterial survival and energy metabolism strategies under hypoxia. Interestingly, Mg2046 is unique to the genus Magnetospirillum according to the protein sequence, while the Mg2946 (MgFnr) is widely distributed in almost all bacteria. Mg2046 may thus specially involve in magnetosome formation and dissimilatory denitrification pathway in magnetotactic spiral bacteria.
Our ongoing studies will extend these findings by examining the relationship between Mg2046 and Mg2946, and their roles in regulation of dissimilatory denitrification pathway.
Author Contributions
YL, XW, and JT conceived the project and designed the experiments. XW, QW, HZ, and YW conducted the experiments and data analysis. JL, JT, YW, and JS provided useful discussion. XW and YL wrote the manuscript. All authors read and approved the finalized manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (Grants 31270093 and 31570037) and the Project for Extramural Scientists of State Key Laboratory of Agrobiotechnology (2019SKLAB6-9).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Tongwei Zhang, Dr. Yinzhao Wang, and Prof. Yongxin Pan (Institute of Geology and Geophysics, Chinese Academy of Sciences) for technical assistance in measurement of cellular magnetic properties and to Dr. S. Anderson for language editing of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01478/full#supplementary-material
FIGURE S1 | Relative transcription level of mg2046 gene in Δmg2046 and complementary strain (Com2046), with level in WT as reference (defined as 1). Level in Com2046 was ∼60-fold higher than in WT.
TABLE S1 | qPCR primers used in this study. F: forward primers. R: reverse primers.
TABLE S2 | Average magnetosome number and particle size for WT and Δmg2046.
TABLE S3 | Relative transcription levels of napF, nirT, norC, and nosZ genes in WT vs. Δmg2046.
Footnotes
References
Arakaki, A., Kikuchi, D., Tanaka, M., Yamagishi, A., Yoda, T., and Matsunaga, T. (2016). Comparative subcellular localization analysis of magnetosome proteins reveals a unique localization behavior of Mms6 protein onto magnetite crystals. J. Bacteriol. 198, 2794–2802. doi: 10.1128/JB.00280-16
Arakaki, A., Webb, J., and Matsunaga, T. (2003). A novel protein tightly bound to bacterial magnetic particles in Magnetospirillum magneticum strain AMB-1. J. Biol. Chem. 278, 8745–8750. doi: 10.1074/jbc.m211729200
Baumgartner, J., Morin, G., Menguy, N., Perez Gonzalez, T., Widdrat, M., Cosmidis, J., et al. (2013). Magnetotactic bacteria form magnetite from a phosphate-rich ferric hydroxide via nanometric ferric (oxyhydr) oxide intermediates. Proc. Natl. Acad. Sci. U.S.A. 110, 14883–14888. doi: 10.1073/pnas.1307119110
Bueno, E., Mesa, S., Bedmar, E. J., Richardson, D. J., and Delgado, M. J. (2012). Bacterial adaptation of respiration from oxic to microoxic and anoxic conditions: redox control. Antioxid. Redox Sign. 16, 819–852. doi: 10.1089/ars.2011.4051
Dailey, H. A., and Lascelles, J. (1977). Reduction of iron and synthesis of protoheme by Spirillum itersonii and other organisms. J. Bacteriol. 129, 815–820.
Deng, Z., Wang, Q., Liu, Z., Zhang, M., Machado, A. C., Chiu, T. P., et al. (2015). Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator. Nat. Commun. 6:7642. doi: 10.1038/ncomms8642
Faivre, D., and Schüler, D. (2008). Magnetotactic bacteria and magnetosomes. Chem. Rev. 108, 4875–4898.
Goemaere, E. L., Devert, A., Lloubès, R., and Cascales, E. (2007). Movements of the TolR C-terminal domain depend on TolQR ionizable key residues and regulate activity of the Tol complex. J. Biol. Chem. 282, 17749–17757. doi: 10.1074/jbc.m701002200
Jones, S. R., Wilson, T. D., Brown, M. E., Rahnlee, L., Yu, Y., Fredriksen, L. L., et al. (2015). Genetic and biochemical investigations of the role of MamP in redox control of iron biomineralization in Magnetospirillum magneticum. Proc. Natl. Acad. Sci. U.S.A. 112, 3904–3909. doi: 10.1073/pnas.1417614112
Kawakami, T., Kuroki, M., Ishii, M., Igarashi, Y., and Arai, H. (2010). Differential expression of multiple terminal oxidases for aerobic respiration in Pseudomonas aeruginosa. Environ. Microbiol. 12, 1399–1412. doi: 10.1111/j.1462-2920.2009.02109.x
Klebba, P. E. (2016). ROSET model of TonB action in gram-negative bacterial iron acquisition. J. Bacteriol. 198:1013. doi: 10.1128/JB.00823-15
Kolinko, I., Lohse, A., Borg, S., Raschdorf, O., Jogler, C., Tu, Q. Y., et al. (2014). Biosynthesis of magnetic nanostructures in a foreign organism by transfer of bacterial magnetosome gene clusters. Nat. Nanotechnol. 9, 193–197. doi: 10.1038/nnano.2014.13
Komeili, A. (2012). Molecular mechanisms of compartmentalization and biomineralization in magnetotactic bacteria. FEMS Microbiol. Rev. 36, 232–255. doi: 10.1111/j.1574-6976.2011.00315.x
Körner, H., Sofia, H. J., and Zumft, W. G. (2003). Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 7, 559–592. doi: 10.1016/s0168-6445(03)00066-4
Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., and Roop, R. M. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. doi: 10.1016/0378-1119(95)00584-1
Kralova, M., Masscheleyn, P. H., Lindau, C. W., and Patrick, W. H. (1992). Production of dinitrogen and nitrous oxide in soil suspensions as affected by redox potential. Water Air Soil Pollut. 61, 37–45. doi: 10.1007/bf00478364
Lefevre, C. T., and Bazylinski, D. A. (2013). Ecology, diversity, and evolution of magnetotactic bacteria. Microbiol. Mol. Biol. Rev. 77, 497–526. doi: 10.1128/MMBR.00021-13
Li, J., Pan, Y., Chen, G., Liu, Q., Tian, L., and Lin, W. (2010). Magnetite magnetosome and fragmental chain formation of Magnetospirillum magneticum AMB-1: transmission electron microscopy and magnetic observations. Geophys. J. Int. 177, 33–42. doi: 10.1111/j.1365-246x.2009.04043.x
Li, Y., Bali, S., Borg, S., Katzmann, E., Ferguson, S. J., and Schüler, D. (2013). Cytochrome cd1 nitrite reductase NirS is involved in anaerobic magnetite biomineralization in Magnetospirillum gryphiswaldense and requires NirN for proper d1 heme assembly. J. Bacteriol. 195, 4297–4309. doi: 10.1128/JB.00686-13
Li, Y., Katzmann, E., Borg, S., and Schüler, D. (2012). The periplasmic nitrate reductase nap is required for anaerobic growth and involved in redox control of magnetite biomineralization in Magnetospirillum gryphiswaldense. J. Bacteriol. 194, 4847–4856. doi: 10.1128/JB.00903-12
Li, Y., Raschdorf, O., Silva, K. T., and Schüler, D. (2014a). The terminal oxidase cbb3 functions in redox control of magnetite biomineralization in Magnetospirillum gryphiswaldense. J. Bacteriol. 196, 2552–2562. doi: 10.1128/JB.01652-14
Li, Y., Sabaty, M., Borg, S., Silva, K. T., Pignol, D., and Schüler, D. (2014b). The oxygen sensor MgFnr controls magnetite biomineralization by regulation of denitrification in Magnetospirillum gryphiswaldense. BMC Microbiol. 14:153. doi: 10.1186/1471-2180-14-153
Lin, W., Bazylinski, D. A., Xiao, T., Wu, L., and Pan, Y. (2014). Life with compass: diversity and biogeography of magnetotactic bacteria. Environ. Microbiol. 16, 2646–2658. doi: 10.1111/1462-2920.12313
Lohse, A., Borg, S., Raschdorf, O., Kolinko, I., Tompa, E., Posfai, M., et al. (2014). Genetic dissection of the mamAB and mms6 operons reveals a gene set essential for magnetosome biogenesis in Magnetospirillum gryphiswaldense. J. Bacteriol. 196, 2658–2669. doi: 10.1128/JB.01716-14
Lohse, A., Kolinko, I., Raschdorf, O., Uebe, R., Borg, S., Brachmann, A., et al. (2016). Overproduction of magnetosomes by genomic amplification of biosynthesis-related gene clusters in a magnetotactic bacterium. Appl. Environ. Microbiol. 82, 3032–3041.
Murat, D., Falahati, V., Bertinetti, L., Csencsits, R., Kornig, A., Downing, K., et al. (2011). The magnetosome membrane protein, MmsF, is a major regulator of magnetite biomineralization in Magnetospirillum magneticum AMB-1. Mol. Microbiol. 85, 684–699. doi: 10.1111/j.1365-2958.2012.08132.x
Murat, D., Quinlan, A., Vali, H., and Komeili, A. (2010). Comprehensive genetic dissection of the magnetosome gene island reveals the step-wise assembly of a prokaryotic organelle. Proc. Natl. Acad. Sci. U.S.A. 107, 5593–5598. doi: 10.1073/pnas.0914439107
Oh, J. I., and Kaplan, S. (2002). Oxygen adaptation: the role of the CcoQ subunit of the cbb3 cytochrome c oxidase of Rhodobacter sphaeroides 2.4.1. J. Biol. Chem. 277, 16220–16228.
Qi, L., Li, J., Zhang, W., Liu, J., Rong, C., Li, Y., et al. (2012). Fur in Magnetospirillum gryphiswaldense influences magnetosomes formation and directly regulates the genes involved in iron and oxygen metabolism. PLoS One 7:e29572. doi: 10.1371/journal.pone.0029572
Quinlan, A., Murat, D., Vali, H., and Komeili, A. (2011). The HtrA/DegP family protease MamE is a bifunctional protein with roles in magnetosome protein localization and magnetite biomineralization. Mol. Microbiol. 80, 1075–1087. doi: 10.1111/j.1365-2958.2011.07631.x
Raschdorf, O., Muller, F. D., Posfai, M., Plitzko, J. M., and Schüler, D. (2013). The magnetosome proteins MamX, MamZ and MamH are involved in redox control of magnetite biomineralization in Magnetospirillum gryphiswaldense. Mol. Microbiol. 89, 872–886. doi: 10.1111/mmi.12317
Rawlings, A. E., Bramble, J. P., Walker, R., Bain, J., Galloway, J. M., and Staniland, S. S. (2014). Self-assembled MmsF proteinosomes control magnetite nanoparticle formation in vitro. Proc. Natl. Acad. Sci. U.S.A. 111, 16094–16099. doi: 10.1073/pnas.1409256111
Rong, C., Huang, Y., Zhang, W., Jiang, W., Li, Y., and Li, J. (2008). Ferrous iron transport protein B gene (feoB1) plays an accessory role in magnetosome formation in Magnetospirillum gryphiswaldense strain MSR-1. Res. Microbiol. 159, 530–536. doi: 10.1016/j.resmic.2008.06.005
Schüler, D., and Baeuerlein, E. (1996). Iron-limited growth and kinetics of iron uptake in Magnetospirillum gryphiswaldense. Arch. Microbiol. 166, 301–307. doi: 10.1007/s002030050387
Staniland, S. S., and Rawlings, A. E. (2016). Crystallizing the function of the magnetosome membrane mineralization protein Mms6. Biochem. Soc. Trans. 44, 883–890. doi: 10.1042/BST20160057
Tanaka, M., Mazuyama, E., Arakaki, A., and Matsunaga, T. (2011). MMS6 protein regulates crystal morphology during nano-sized magnetite biomineralization in vivo. J. Biol. Chem. 286, 6386–6392. doi: 10.1074/jbc.M110.183434
Wang, L., Prozorov, T., Palo, P. E., Liu, X., Vaknin, D., Prozorov, R., et al. (2012). Self-assembly and biphasic iron-binding characteristics of Mms6, a bacterial protein that promotes the formation of superparamagnetic magnetite nanoparticles of uniform size and shape. Biomacromolecules 13, 98–105. doi: 10.1021/bm201278u
Wang, Q., Wang, M., Wang, X., Guan, G., Li, Y., Peng, Y., et al. (2015). Iron response regulator protein IrrB in Magnetospirillum gryphiswaldense MSR-1 helps control the iron/oxygen balance, oxidative stress tolerance, and magnetosome formation. Appl. Environ. Microbiol. 81, 8044–8053. doi: 10.1128/AEM.02585-15
Wang, Q., Wang, X., Zhang, W., Li, X., Zhou, Y., Li, D., et al. (2017). Physiological characteristics of Magnetospirillum gryphiswaldense MSR-1 that control cell growth under high-iron and low-oxygen conditions. Sci. Rep. 7:2800. doi: 10.1038/s41598-017-03012-4
Wang, X., Wang, Q., Zhang, W., Wang, Y., Li, L., Wen, T., et al. (2014). Complete genome sequence of Magnetospirillum gryphiswaldense MSR-1. Genome Announc. 2:e00171-14. doi: 10.1128/genomeA.00171-14
Wang, X., Wang, Q., Zhang, Y., Wang, Y., Zhou, Y., Zhang, W., et al. (2016). Transcriptome analysis reveals physiological characteristics required for magnetosome formation in Magnetospirillum gryphiswaldense MSR-1. Environ. Microbiol. Rep. 8, 371–381. doi: 10.1111/1758-2229.12395
Yang, J., Li, S., Huang, X., Tang, T., Jiang, W., Zhang, T., et al. (2013). A key time point for cell growth and magnetosome synthesis of Magnetospirillum gryphiswaldense based on real-time analysis of physiological factors. Front. Microbiol. 4:210. doi: 10.3389/fmicb.2013.00210
Zeytuni, N., Ozyamak, E., Benharush, K., Davidov, G., Levin, M., Gat, Y., et al. (2011). Self-recognition mechanism of MamA, a magnetosome-associated TPR-containing protein, promotes complex assembly. Proc. Natl. Acad. Sci. U.S.A. 108, 13369–13370. doi: 10.1073/pnas.1103367108
Zhang, W. J., Santini, C. L., Bernadac, A., Ruan, J., Zhang, S. D., Kato, T., et al. (2012). Complex spatial organization and flagellin composition of flagellar propeller from marine magnetotactic ovoid strain MO-1. J. Mol. Biol. 416, 558–570. doi: 10.1016/j.jmb.2011.12.065
Zhang, Y., Wen, T., Guo, F., Geng, Y., Liu, J., Peng, T., et al. (2017). The disruption of an OxyR-Like protein impairs intracellular magnetite biomineralization in Magnetospirillum gryphiswaldense MSR-1. Front. Microbiol. 8:208. doi: 10.3389/fmicb.2017.00208
Keywords: Magnetospirillum gryphiswaldense, Mg2046, magnetosome, regulator, redox status, dissimilatory denitrification pathway
Citation: Wang X, Zheng H, Wang Q, Jiang W, Wen Y, Tian J, Sun J, Li Y and Li J (2019) Novel Protein Mg2046 Regulates Magnetosome Synthesis in Magnetospirillum gryphiswaldense MSR-1 by Modulating a Proper Redox Status. Front. Microbiol. 10:1478. doi: 10.3389/fmicb.2019.01478
Received: 16 November 2018; Accepted: 13 June 2019;
Published: 26 June 2019.
Edited by:
Rich Boden, University of Plymouth, United KingdomReviewed by:
Islam S. M. Khalil, University of Twente, NetherlandsDenis Trubitsyn, Longwood University, United States
Copyright © 2019 Wang, Zheng, Wang, Jiang, Wen, Tian, Sun, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiesheng Tian, MjAxNzk2ODUyNUBxcS5jb20=; Ying Li, eWluZ2xpNTI4QHZpcC5zaW5hLmNvbQ==
 Xu Wang
Xu Wang Haolan Zheng
Haolan Zheng Qing Wang
Qing Wang Wei Jiang
Wei Jiang Ying Wen
Ying Wen Jiesheng Tian
Jiesheng Tian Jianbo Sun
Jianbo Sun Ying Li
Ying Li Jilun Li1
Jilun Li1