
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 02 July 2019
Sec. Food Microbiology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.01464
Cronobacter sakazakii is an emerging foodborne pathogen, which is linked to life-threatening infections causing septicemia, meningitis, and necrotizing enterocolitis. These infections have been epidemiologically connected to ingestion of contaminated reconstituted powder infant formula. Even at low water activity C. sakazakii can survive for a long time; it is capable of protective biofilm formation and occasionally shows high virulence and pathogenicity even following stressful environmental conditions. Hence it is a challenging task for the food industry to control contamination of food ingredients and products through the entire production chain, since an increasing number of severe food-related outbreaks of C. sakazakii infections has been observed. The seemingly great capability of C. sakazakii to survive even strict countermeasures combined with its prevalence in many food ingredients requires a greater in depth understanding of its virulence factors to master the food safety issues related to this organism. In this context, we present the whole genome sequence (WGS) of two different C. sakazakii isolated from skimmed milk powder (C7) and ready-to-eat salad mix (C8), respectively. These are compared to other, already sequenced, C. sakazakii genomes. Sequencing of the fusA allele revealed that both isolates were C. sakazakii. We investigated the molecular characteristics of both isolates relevant for genes associated with pathogenesis and virulence factors, resistance to stressful environmental conditions (e.g., osmotic and heat), survival in desiccation as well as conducted a comparative genomic analysis. By using multi-locus sequence typing (MLST), the genetic type of both isolates is assessed and the number of unique genes is determined. DNA of C. sakazakii C8 is shown to hold a novel and unique sequence type; the number of unique genes identified in the genomic sequence of C. sakazakii C7 and C8 were 109 and 188, respectively. Some of the determined unique genes such as the rhs and VgrG genes are linked to the Type VI Secretion System cluster, which is associated with pathogenicity and virulence factors. Moreover, seven genes encoding for multi-drug resistance were found in both isolates. The finding of a number of genes linked to producing capsules and biofilm are likely related to the observed resistance to desiccation.
Cronobacter sakazakii is regarded as an opportunistic foodborne pathogen, which can cause meningitis, bacteremia and necrotizing enterocolitis, particularly in infants (Drudy et al., 2006; Aly et al., 2018). Neonates have suffered from life-threatening infections of C. sakazakii within the first 28 days after birth (Mullane et al., 2007). Also, older infants have suffered fatality rates ranging from 40 to 80% and even immune-compromised adults have suffered fatalities (Nazarowec-White and Farber, 1997; Tsai et al., 2013). Cronobacter is one of the costliest foodborne pathogens due to the debilitation of survivors and loss of life. The cost of each case is estimated to be approximately 1 million USD (Minor et al., 2015). In epidemiological studies, infections in infants and neonates have been connected to ingestion of reconstituted powder infant formula contaminated with C. sakazakii (Hunter et al., 2008). Cronobacter survives under desiccated conditions in powdered infant formula even for extended durations of several months (Bennour Hennekinne et al., 2018). Generally, C. sakazakii can adapt to different adverse environments. It shows an unusual resistance to acid stress (ability to live in pH < 3.9), heat, desiccation (with surviving at a water activity of 0.118) and osmotic stress (continues to grow at 10% NaCl) (Fakruddin et al., 2014). C. sakazakii was shown to produce a capsular form or biofilm layer to protect itself from dry conditions. This helps to explain the high survival capability of C. sakazakii in infant powder milk (Hurrell et al., 2009). In addition, C. sakazakii can resist some antibiotics, thereby raising important issues regarding antibiotic therapies used to combat this organism (Kilonzo-Nthenge et al., 2012; Xu et al., 2015). For example, the resistance gene of colistin (mcr-1) was reported in Cronobacter (Liu et al., 2017). Further investigations of genes encoding for antibiotic resistance in Cronobacter isolates as well as genes responsible for its marvelous ability to survive the extreme environments in the food production chain are therefore of high priority for public health studies.
However, the mechanisms of pathogenicity in C. sakazakii remain largely unknown. It is known that after humans are infected by C. sakazakii, it can infect intestinal epithelial cells; from there, it invades the brain to infect human brain microvascular endothelial cells (HBMEC), confirming a route by which to cause meningitis (Giri et al., 2012). In this context, the outer membrane proteins A (OmpA) and X (OmpX) play important roles to explain how Cronobacter efficiently can adhere to and invade HBMEC, Caco-2, and INT-407 cells (Townsend et al., 2008; Kim et al., 2010; Holý et al., 2019). Strains expressing OmpA also resist killing and multiply in dendritic cells (Mittal et al., 2009; Holý et al., 2019). Moreover, C. sakazakii has been demonstrated not only to survive but also to replicate in macrophages (Townsend et al., 2007). Three putative virulence genes hemolysin (hly), plasminogen activator (cpa), and siderophore interacting protein (sip) were identified in Cronobacter spp. isolates (Cruz et al., 2011). Recently, multi-locus sequence typing (MLST) of 7 housekeeping genes was used to describe the diversity of the genus of Cronobacter from different sources (Joseph and Forsythe, 2012). Alleles known as a sequence type (ST) are determined using MLST schemes that interpret the nucleotide sequence data from a number of conserved housekeeping genes (Joseph and Forsythe, 2012); these enable comparisons and grouping of strains (Joseph and Forsythe, 2012). For example, C. sakazakii ST4 is predominantly connected with meningitis of neonates, while C. sakazakii ST1, 4, 8, and 12 strains are considered pathovars related to human illnesses (Joseph and Forsythe, 2011; Joseph et al., 2012b; Masood et al., 2015; Ogrodzki and Forsythe, 2017). Cronobacter spp. possess ubiquitous presence and have been isolated from cereals, rice, flour, dairy, herbs, medicinal plants, spices, meat, vegetables, and fruits (Cetinkaya et al., 2013; Ye et al., 2014). However, it is of interest to note that ST4 strains usually have been isolated from powdered infant formula and from milk powder processing factories (van Acker et al., 2001; Siqueira Santos et al., 2013; Sonbol et al., 2013; Gicova et al., 2014). Recently, efforts have been made to develop rapid detection schemes for Cronobacter species in food samples (Aly et al., 2018, 2019; Pan et al., 2018). Such technologies would greatly enhance food safety by allowing for continuous monitoring of contamination in the food chain.
Pulsed-field gel electrophoresis (PFGE) has long been considered the gold standard for molecular typing of the pathogens linked to outbreaks (Stranden et al., 2003; Alsonosi et al., 2015). However, PFGE has many limitations, including being incapable of distinguishing strains of highly clonal bacteria that are unrelated, as is the case for Cronobacter species (Alsonosi et al., 2015). Therefore, FoodNet is moving from PFGE to whole genome sequencing (WGS) for epidemiological investigations of foodborne outbreaks (Scharff et al., 2016; Nadon et al., 2017). WGS offers a more detailed resolution of how closely bacterial isolates are related than PFGE does; it additionally provides insights concerning the MLST, pathogenicity genes, as well as a complete molecular characterization of strains (Kozyreva et al., 2016; Forsythe, 2018).
Kozyreva et al. (2016) recently reported that WGS could be used to confirm the Salmonella enterica serovar Poona outbreak in California. Lytsy et al. (2017) examined the applicability, resolution, and reliability of PFGE, MLST, and WGS in three outbreaks in Sweden that occurred 2013–2015 for vancomycin-resistant enterococci. This study led to the recommendation to use WGS-ANI analysis for epidemiological identification of vancomycin-resistant enterococci (Lytsy et al., 2017). Publicly accessible genomic sequence data and the tools utilized to examine these data are now ubiquitous in biological studies, with the food safety research area as no exception (Taboada et al., 2017). Currently available bioinformatics tools for comparative genomic analysis, such as pan-genome analysis, have been employed to characterize the entire gene repertoire of bacterial species (Medini et al., 2005). In molecular biology, the pan-genome is defined as the complete set of non-orthologous genes present in species, consisting of the core and accessory genomes, i.e., sets of genes that are present in all strains and unique to single strains, respectively (Kim et al., 2015). Moreover, this analysis confirms how many new genes can be determined from newly sequenced genomes.
In this work, we use WGS to compare the sequences of C. sakazakii isolated in Austria with other previously published genomes to obtain additional insights into their genetic makeup. We focus our study on parts of the genome that support our understanding of previous observations of pathogenicity and long-term persistence of C. sakazakii. In detail, we investigate and characterize some of the unique genes detected and associated with virulence and pathogenicity mechanisms, resistance to stressful conditions, biofilm formation, and multidrug-resistance.
The two Cronobacter isolates used in this work named C. sakazakii C7 and C8 were isolated from skimmed milk powder and ready-to-eat food (salad mix) samples, respectively. The isolates were obtained in Austria and confirmed as described previously (Aly et al., 2019). They were identified by the rpoB gene (Stoop et al., 2009). Furthermore, fusA sequences were queried in MLST databases1 to confirm their belonging to the Cronobacter species. The bacteria were cultured as described in our earlier work (see text footnote 1; Aly et al., 2019). The genomic DNA (gDNA) of both isolates was extracted and purified using the bacteria genome kit (peqGOLD Bacterial DNA Kit, VWR International GmbH, Germany), according to the guidelines from the manufacturer (Aly et al., 2019). Optical density measurements of the gDNA was performed in a Qubit Fluorometric Spectrophotometer (Life Technologies, Wilmington, DE, United States). Libraries were generated by enzyme fragmentation and constructed using the NEBNext Fast DNA-Library for Ion Torrent sample kit preparation (New England Biolabs, Ipswich, MA, United States) following the manufacturer’s instructions. The fragmented gDNA was end-repaired and ligated to the specific adapters and individual barcodes. The quality control of the gDNA libraries was determined with the Agilent Bioanalyzer 2100. The enriched libraries were amplified using emulsion polymerase chain reaction (ePCR, Ion PITM Hi-QTM OT2 200 kit), following the manufacturer’s instructions (Thermo Fisher Scientific, Inc.). Ion PITM Chip kit v3 chips were employed on the Ion Torrent Proton (Thermo Fisher Scientific, Inc.) platform for the WGS.
The single end sequencing reads generated from the Ion Torrent sequencing were trimmed using the Trimmomatic Tool version 0.36 (Bolger et al., 2014) and filtered by the quality control step of the coupling pipeline in a FASTAQ file format. De novo assembly of high-quality reads is performed with SPAdes (version 3.9.0) assembler (Bankevich et al., 2012). The Quast software was used to evaluate the de novo assembly results. The results were generated on a scaffold based on the number of contigs, GC-depth analysis, coverage analysis of assembly, and comparison and N50 to ensure its quality (Gurevich et al., 2013). Scaffolds greater than 500 bp in sequence length were used for downstream analysis. Next, genome annotation (DNA annotation) was conducted through the “Rapid Annotated using Subsystem Technology” (RAST) with server pipeline (Aziz et al., 2008; Overbeek et al., 2014) in a FASTA file format. RNA gene and protein-coding sequencing (CDS) was used to assign functions and determine the presence of subsystems in the genome. Comparative genome to genome analysis was evaluated in the SEED viewer as previously reported (Overbeek et al., 2014). Homologs of conserved genes in both isolates were identified using the annotation of the NCBI GeneBank of C. sakazakii ATCC BAA-894 (GCA_000017665.1) as the reference genome. The analysis of bacterial pan-genome was performed by the ultra-fast bacterial pan-genome analysis pipeline (BPGA) tool (Chaudhari et al., 2016), and GView Server (Petkau et al., 2010) using the retrieved published complete genomes from the NCBI database including those of, C. sakazakii ES15 (GCF_000263215.1), C. sakazakii BAA-894 (GCA_000017665.1), C. sakazakii NCTC-8155 (GCF_001277275.1), and C. sakazakii Sp291 (GCF_000339015.1). These four genomes were compared to the genomic sequence in our study C. sakazakii C7 and C8. BLASTP search with functional annotation of the unique and core genes was performed by the BLAST2GO analysis pipeline (Götz et al., 2008), applying the default settings for the BLAST search expectation value (E value) of 1.0 × 10–3. Phage-associated gene region clusters in the assembly sequence of C. sakazakii C7 and C8 isolates were identified using the PHASTER server (Arndt et al., 2016). Three scenarios for the wholeness of the identified phage-associated region clusters were assigned according to how many proteins/genes of a known phage the region involved (Dennis et al., 2011): intact (>90%), questionable (70–90%) and incomplete (<70%). Antibiotic resistance genes in the genome assembled for C. sakazakii were detected by a search against a local antibiotic resistance gene sequence from the CARD (Comprehensive Antibiotic Resistance Database) database (Jia et al., 2017).
The gnd and galF loci gene clusters are specific for the O-serotype region; they were identified from the BLAST gDNA sequences by the BIGSdb pipeline tools in the PubMLST typing database (see text footnote 1; Jolley and Maiden, 2010; Ogrodzki and Forsythe, 2015).
Briefly, MLST was utilized to query each genome for all known alleles at each locus by homology check, with new alleles recognized and afforded with a unique allele number. MLST of Cronobacter was executed by submitting genome sequences to the PubMLST typing database for Cronobacter (see text footnote 1; Larsen et al., 2012). The 7-loci MLST profiling of Cronobacter being glutaminyl tRNA synthetase (glnS), glutamate synthase large subunit (gltB), ATP synthase b chain (atpD), DNA gyrase subunit B (gyrB), phosphoenolpyruvate synthase A (ppsA), translation initiation factor IF-2 (infB), and elongation factor G (fusA). Speciation of Cronobacter spp. was obtained by a phylogenetic analysis sequence of the fusA allele (Joseph et al., 2012a; Forsythe et al., 2014; Forsythe, 2015).
The whole genomes were submitted to NCBI under the C. sakazakii complete genome bioproject number PRJNA510032 and biosample accession numbers are SAMN10593105 and SAMN10593106 for C. sakazakii C7 and C8, respectively.
Whole-genome sequencing showed a total of 2,193,827,360 bp, and 2,265,893,859 bp with single-end reads from gDNA of C7 (milk powder C. sakazakii isolate) and C8 (ready-to-eat salad C. sakazakii isolate). After quality control, 1409 Mbp of high quality reads were obtained for C7 and 1472 Mbp were obtained for C8. These were kept for de novo assembly, which yielded a total of 64 scaffolds with total scaffold N50 of 344,291 bp for C7 and 61 scaffolds with total scaffold N50 of 436,880 bp for C8. The genomic features of C7 and C8 are summarized in Table 1 and functionally annotated by the RAST server (Figure 1 and Supplementary Table S1). Figure 2 illustrates this phylogenetic analysis of WGS using the MEGA 7 tools. The distances between branches in Figure 2 were calculated using the Maximum Composite Likelihood method (Tamura et al., 2004; Kumar et al., 2016). The phylogeny analysis of the seven species members in genus of Cronobacter strains revealed that the ST of C. sakazakii strains formed their own distinct cluster.
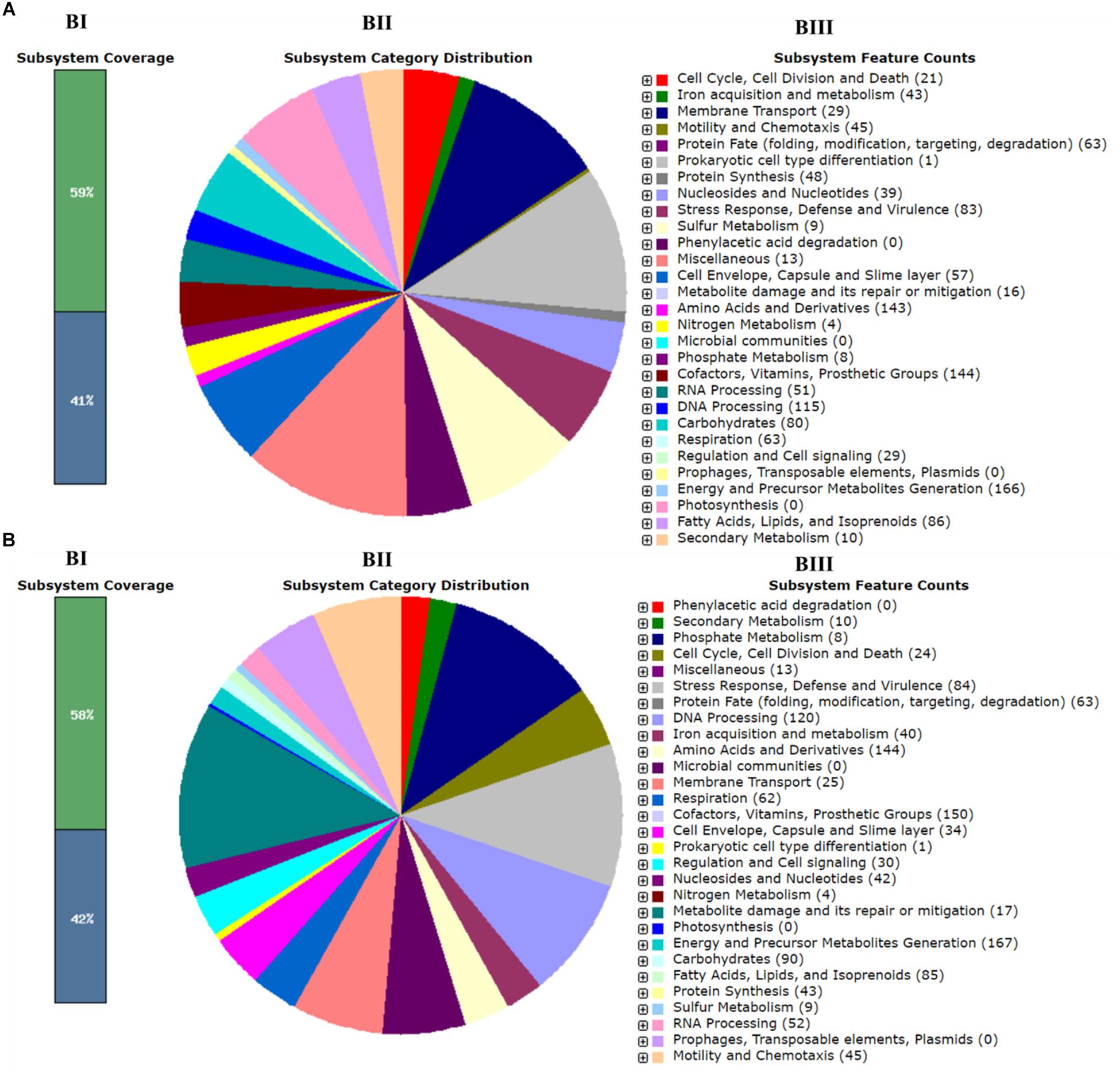
Figure 1. Subsystem category distribution of Cronobacter sakazakii based on the SEED database. (A) C7 and (B) C8. Bar chart (BI) shows the percentage of subsystem coverage (green bar corresponds to the percentage of proteins involved). The pie chart (BII) with legend (BIII) shows the fraction and count (parenthesis in legend) of each subsystem feature.
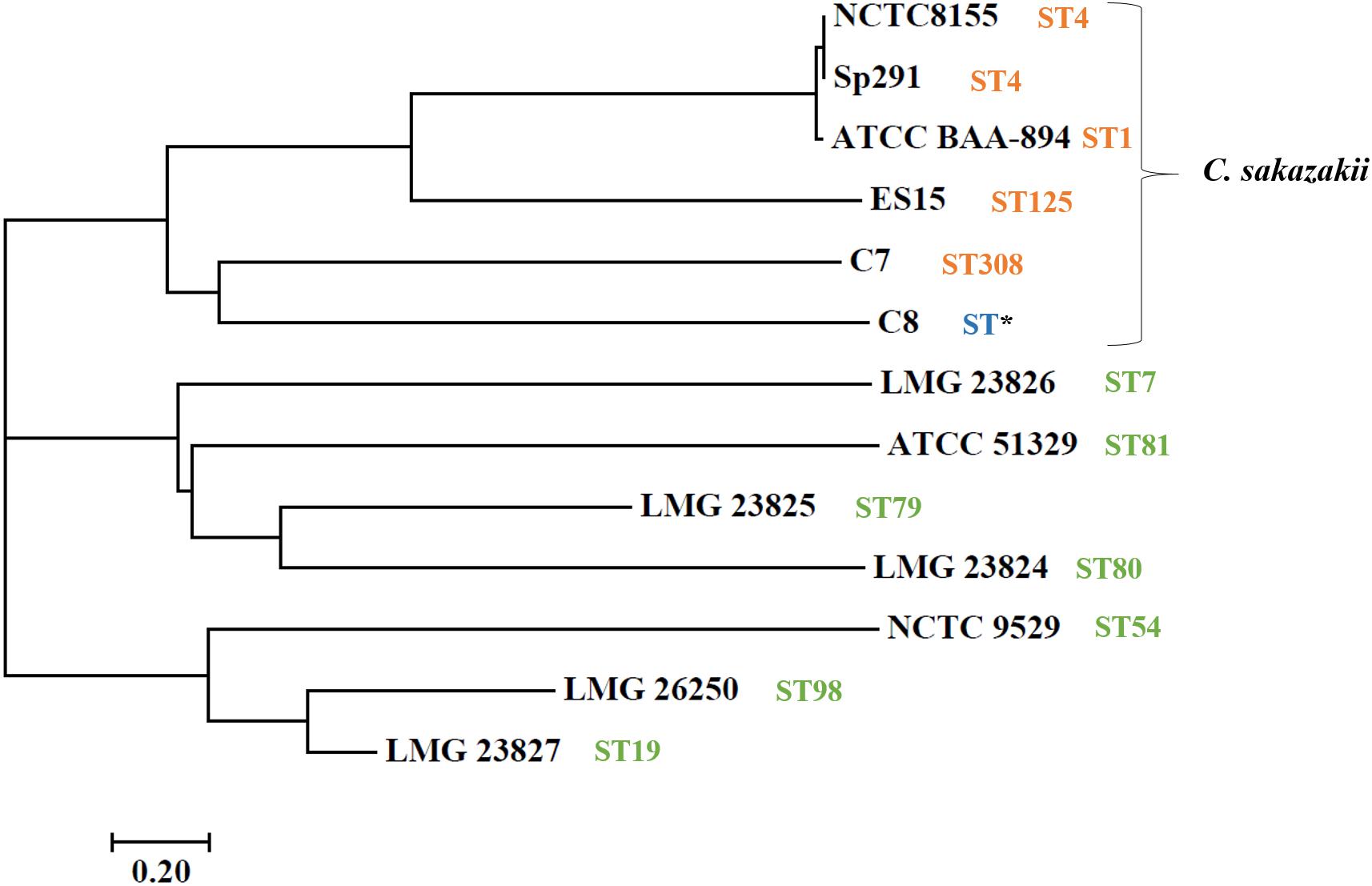
Figure 2. Neighbor-joining tree of C. sakazakii isolated from food, compared with seven representative Cronobacter genus type strains. Accession numbers (NCBI GenBank) of type strains: C. sakazakii ES15 (GCF_000263215.1), C. sakazakii BAA-894 (GCA_000017665.1), C. sakazakii NCTC-8155 (GCF_001277275.1), C. sakazakii Sp291 (GCF_000339015.1), C. universalis NCTC 9529 (NZ_CP012257), C. condimenti LMG 26250 (NZ_CP012264), C. malonaticus LMG 23826T (NZ_CP013940), C. turicensis LMG 23827 (NC_013282), C. dublinensis subsp. lausannensis LMG 23824 (NZ_AJKY00000000), C. dublinensis subsp. lactaridi LMG 23825 (NZ_AJKX00000000), and C. muytjensii ATCC 51329 (NZ_CP012268). The tree was created in MEGA version 7. Sequence type (ST) information was obtained from MLST website (http://pubmlst.org/cronobacter/). Asterisks (*) indicate novel sequence.
A map showing the BlastN comparison of the plasmids from C. sakazakii C7 and C8 with homologous plasmids from C. sakazakii ATCC BAA-894 plasmid pESA3 is shown in Figure 3. Comparative analysis by RAST pipelines of the WGS assemblies with that of the putative virulence plasmid showed the existence of plasmid genes P1 (126 kbp), pESA3 (131 kbp), and pCTU1 (138 kbp) (see Supplementary Table S2). In silico analysis of plasmid P1, pESA3, and pCTU1 showed that these plasmids possess chromosome (plasmid) partitioning proteins parAB genes immediately upstream of plasmid replication protein repA. The C. sakazakii C7 and C8 strains share some of the alleles present in pESA3 and pCTU1 as described by Franco et al. (2011). This plasmid contains many virulence factors, including iron acquisition systems (iucABCD/iutA), protease VII (Omptin), ABC transporter (iron.B12.siderophore.hemin), aerobactin siderophore and the T6SS gene cluster. The C. sakazakii C7 and C8 strains harbor a plasmidborne T6SS gene cluster such as VgrG (valine-glycine repeat G protein) as described by Franco et al. (2011). These results suggest that the virulence factors cluster in the sequenced genome, as also observed in previous studies (Kucerova et al., 2010; Franco et al., 2011; Yan et al., 2013; Jang et al., 2018; Kadlicekova et al., 2018). A site-specific integrase was detected in the in silico analysis of each plasmid (C7 and pESA3 at position 66 Kbp) and was found to belong to integrases in an operon arrangement with four genes encoding a hypothetical protein. These genes were absent in the C8 plasmid. The same genes were previously observed on the plasmid pESA3 by Franco et al. (2011). A channel-forming transporter/cytolysin activator of the TpsB family gene was found at 80 Kbp. It was clustered in the plasmid sequences of C7 and C8 but was absent in plasmid pESA3. Also, at 130 Kbp a gene encoding a hypothetical protein was found within both the C7 and C8 genomes. In previous studies, C. sakazakii strains have been found to harbor plasmids such as pSP291–3 (Yan et al., 2013), pESA2 (Kucerova et al., 2010), pCTU2, and pCTU3 (Stephan et al., 2011; Yan et al., 2013). However, we did not find these plasmids in the strains that we analyzed.
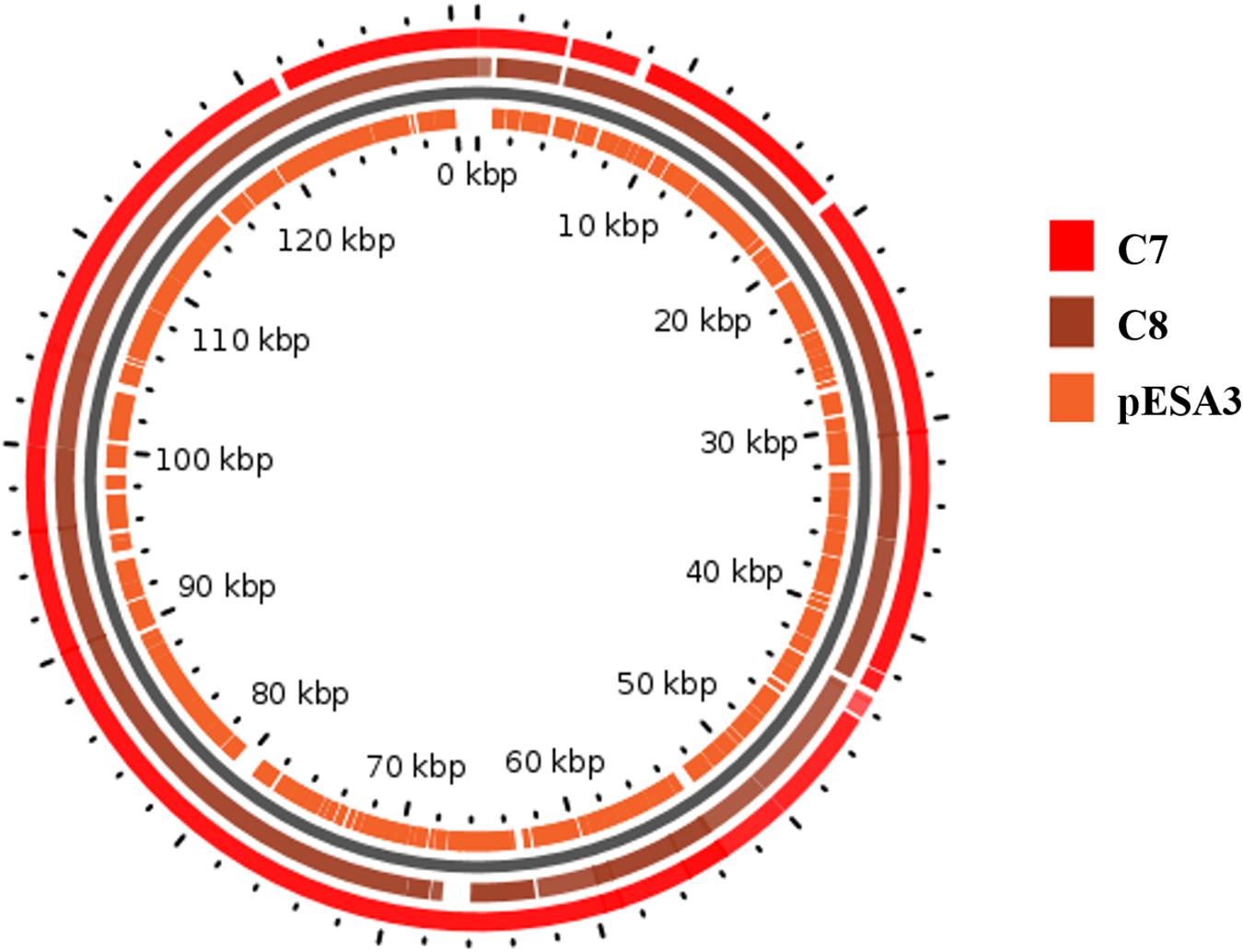
Figure 3. Map of the plasmids from C. sakazakii C7 and C8. The rings show BlastN comparison with homologous plasmids from C. sakazakii ATCC BAA-894 plasmid pESA3. The map was drawn in GView (https://server.gview.ca/).
The prediction of O-serotype was based on the presence of flanking genes gnd and galF on the DNA sequence following the approach of Ogrodzki and Forsythe (2015). In this study, the C. sakazakii C7 isolated from skimmed milk powder was representative of O-antigen serotype (Csak_O2) gene clusters located between the gnd and galF. However, only 17 serotypes, all O-serotypes, have been determined in Cronobacter spp. (Mullane et al., 2008; Jarvis et al., 2011; Sun et al., 2012). The C. sakazakii C8 isolate from ready-to-eat food represents an unknown serotype; there have been no O-antigen genes identified for these flanking loci. This demonstrates that the current standards for molecular serotyping are not adequate and additional protocols for serotyping should be developed to enable serotyping of all C. sakzakii. Additional data on the distribution of serotypes are also required to define whether molecular serotyping is a helpful alternative for epidemiological surveillance of the Cronobacter genus.
Multilocus sequence typing is a method to distinguish strains in the Cronobacter genus, using the seven-loci MLST (Forsythe et al., 2014). It has been reported that the fusA allele sequence is matched with the phylogeny sequence and could be utilized for speciation of the Cronobacter genus (Joseph et al., 2012a). The resting six alleles could then be defined for seven-loci MLST profiling. In the current study, the MLST results for C. sakazakii C7 and C8 isolates are summarized in Table 2. The MLST revealed C7 to be of ST308. Surprisingly, C8 isolated from ready-to-eat food was found to be a new unique ST with novel alleles [gltB, infB, and ppsA]. These are single nucleotide substitution variants when compared with their most homologous alleles. This finding combined with the few investigations to date of the genetic populations of C. sakazakii, indicate that it is essential to recognize more STs to aid in epidemiological investigations and risk assessments of this pathogen.
The complete genomes of four different C. sakazakii strains and their annotation data have been published to date in the GenBank database. As illustrated in Figure 4A, the pan-genome analysis of these 4 complete genomes and those of the 2 new isolates, C7 and C8, presented here were conducted and strain specific-regions were obtained and visualized. An analysis by the BPGA tool was applied to the 6 C. sakazakii strains. First, the core- and pan-genomes and their sizes and trajectories were analyzed using the approach suggested by Knight et al. (2016). Second, the median counts were extrapolated employing two models: exponential regression described in Tettelin et al. (2005) and power-law regression described in Tettelin et al. (2008). The resulting extrapolations from the gene counts were normalized by the mean of the genome sizes of the respective sets to aid in the comparison and visualization of the fits (Figure 4B).
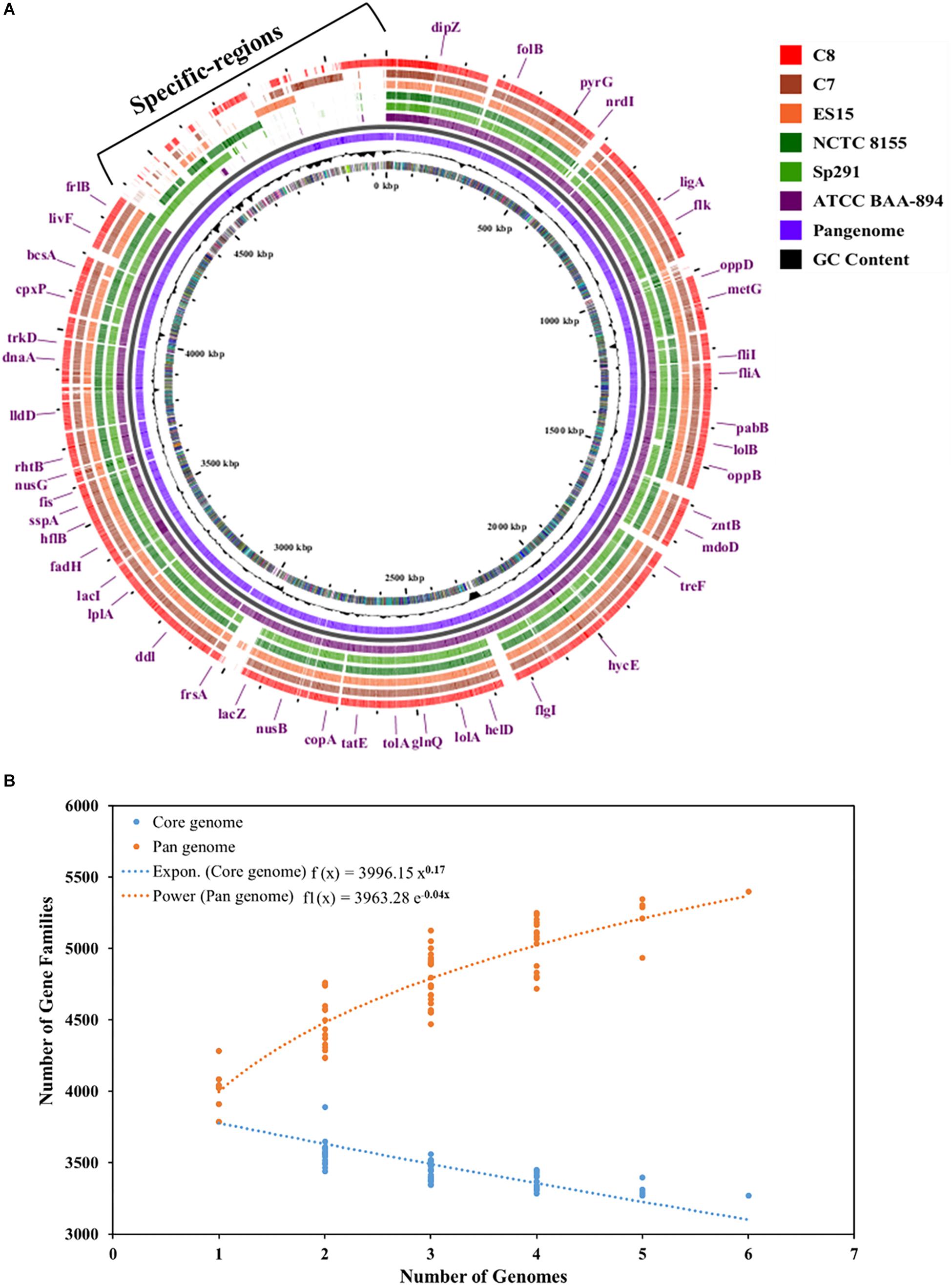
Figure 4. (A) Pan-genome of six C. sakazakii strains (4 database genomes and the 2 isolates) was visualized using the Gview server (Zheng and Huang, 2004; Petkau et al., 2010). The cut-off value was 80% of BlastN homology. The annotation indicates clusters of conserved genes with C. sakazakii (ATCC BAA-894). (B) Plot showing the pan- and core-genome of C. sakazakii. The overall number of genes or pan-genome (Orange) and shared or core-genome (Blue) of 6 C. sakazakii are shown in the plot. The pan-genome consists of 5399 genes, while the core-genomes are made up of 3268 genes.
The pan-genome was made up of 5399 genes, and the C. sakazakii strains shared 3268 genes as core-genomes (Figure 4B and Supplementary Table S3A). The BGPA power law regression analysis identified the pan-genome of C. sakazakii strains as open (Supplementary Table S3B). The pan-genome is recognized to be “open” as long as new genes significantly increase the total repertoire for each new extra genome and “closed” when newly added genomes do not significantly add to the total repertoire of the genes (Guimarães et al., 2015). Mathematical modeling of the data implies that the pan-genome gene reservoir of unique genes will increase even after hundreds of newly sequenced genomes are added and therefore is considered to be open (Tettelin et al., 2005; He, 2015). The number of unique genes detected in C. sakazakii BAA-894 and C. sakazakii NCTC-8155 were 465 and 53, respectively (Table 3). The C. sakazakii C7 and C. sakazakii C8 have a total of 109 and 188 unique genes conserved in gDNA, respectively. The gene ontology (GO) of the unique genes in gDNA of C. sakazakii C7 revealed that they are rich in proteins associated with molecule binding, heterocyclic, and organic cyclic compound binding, hydrolase activity, drug and ion binding (Supplementary Tables S3C,D).
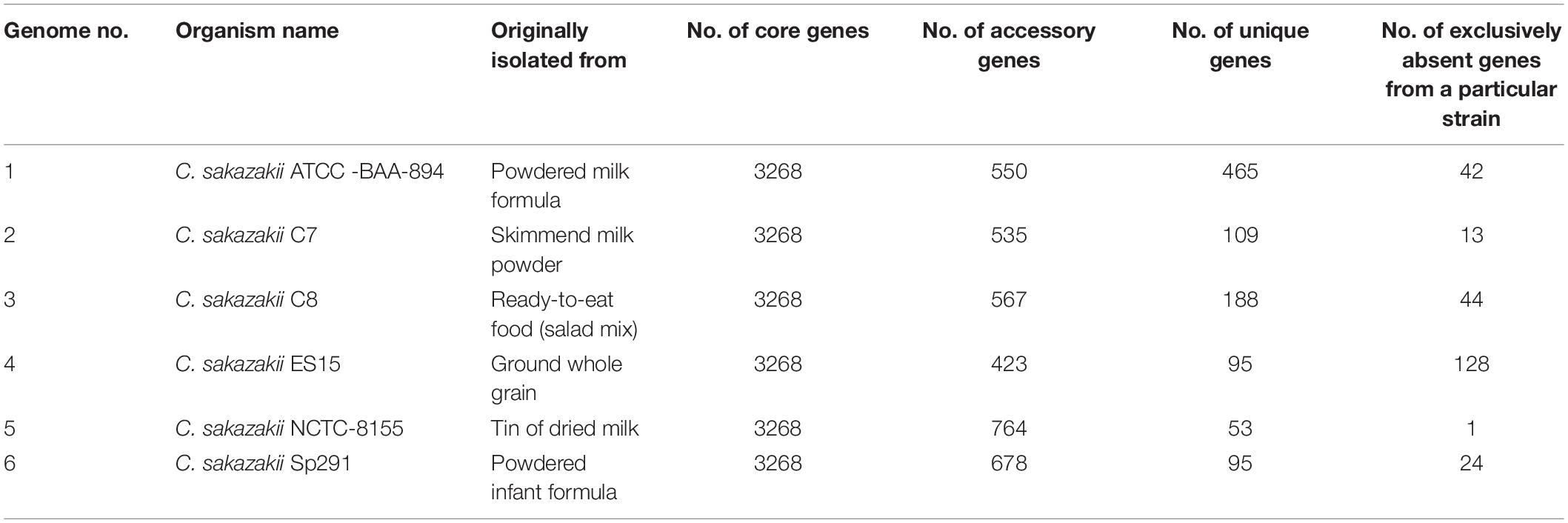
Table 3. Number of core, unique, accessory, and exclusively absent (from a particular strain) genes taken from the pan-genome analysis of 6 C. sakazakii strains.
The presence of multidrug-resistant genes in gDNA that may contribute to antimicrobial resistance was identified by BLAST searching the assembled sequence genome of C. sakazakii isolates against a local copy of CARD sequence data (Supplementary Table S4). As shown in Supplementary Table S4, seven antibiotic resistance genes were found: msbA, emrR, H-NS, emrB, marA, CRP, and PBP3. These provide resistance to multiple antibiotics, such as, beta-lactam antibiotics, tetracycline, a macrolide antibiotic, a fluoroquinolone antibiotic, penams, cephalosporin, and cephamycin. The genomes of both C. sakazakii isolates contained all these genes. Active efflux pumps provide a known mechanism for increased virulence by improving the survival of Enterobacteriaceae in the host’s gastrointestinal tract (Touze et al., 2004). Further, the efflux system enables the invasion of brain microvascular endothelial cells (Franke et al., 2003; Kucerova et al., 2010). Tetracycline-resistance was reported for C. sakazakii isolated from a freshwater Chilean salmon aquaculture farm (Miranda et al., 2003). Previous studies have shown that abuse of antibiotics in such environments and the presence of various antibiotic resistance (mar) operons may enable Cronobacter spp. to develop resistance to numerous different antibiotics (Kim et al., 2008; El-Sharoud et al., 2009; Chon et al., 2012). Hence, the large number of multidrug-resistant genes found here, with an unknown entryway to the C. sakazakii genome, suggests that further studies are required to define the extent of acquired antimicrobial drug resistance and an attempt to map the way it was acquired. Only by such efforts can we prevent the emergence of additional antimicrobial resistance or create early warnings of the emergence of such resistance.
The high survival rate of C. sakazakii, under extreme dessication (e.g., in milk powder, starch, or flour), low pH, heat and osmotically challenging environments, has so far not been examined at the molecular level. We performed WGS also to understand this unusual and challenging property of C. sakazakii. Supplementary Table S1 includes a number of genes associated with resistance to stressful environmental conditions identified in the genomic sequences of the C. sakazakii C7 and C8 isolates. Osmoregulation genes such as osmotically inducible protein (OsmY), transcriptional regulatory protein (YciT), aquaporin Z, hyperosmotic potassium uptake protein (TrkH) potassium uptake protein (TrkA and TrkG), ProP, Betaine aldehyde dehydrogenase (otsA), Trehalose-6-phosphate hydrolase (ostB), and Glutathione-regulated potassium-efflux system protein (KefB, KefC, and KefG) were identified in both genomes of C. sakazakii isolates. In addition, heat/cold shock stress genes were also identified, such as DnaJ and DnaK suppressor proteins and the heat shock proteins (YciM) and (GrpE). It is supposed that when C. sakazakii is exposed to low water activity conditions, it rapidly accumulates electrolytes to increase the internal osmotic pressure; this is the primary fast action by which it protects itself by counteracting the high external osmotic pressure in desiccated environments (Feeney and Sleator, 2011). Jang et al. (2018) also found genes encoding for proteins associated with osmotic response, such as Aquaprotein Z, DnaJ, TreF and ProQ. For example, DnaJ operates actively in the response to hyperosmotic and heat shock by preventing the aggregation of stress-denatured proteins. The physiological mechanisms of survival in desiccated environments are thought to include countermeasures to both primary and secondary desiccation stresses. Glycine betaine, trehalose and other osmoprotectants are used in adaptation strategies by xerotolerant organisms (Santos and Da Costa, 2002; Srikumar et al., 2019). For example, the osmoprotectants found in osmotically stressed E. coli are trehalose, glycine betaine proline, and carnitine (Dinnbier et al., 1988; Lehner et al., 2018; Srikumar et al., 2019). These previous reports describe two genes, otsA and otsB, that are part of the biosynthetic pathway for trehalose, which were transcriptionally highly up-regulated in C. sakazakii cells grown under xerotolerant conditions (Srikumar et al., 2019). The Glutathione-gated potassium-efflux systems (Kef) can also play an important role in desiccation tolerance by preventing extended exposure of bacterial cells to excess potassium ions (Du et al., 2018). The proP gene found in our isolates have been shown to be highly upregulated when the organism has been subjected to a desiccating environment. Deletion of the proP gene in Salmonella Typhimurium led to loss of viability during desiccation for long periods of time (Finn et al., 2013). We also identified the outer membrane protein W (OmpW) gene in the genomes of both C. sakazakii isolates. It was reported that the OmpW gene contributes to survival of C. sakazakii under osmotic and oxidative stress, as well as being associated with increased biofilm formation (Ye et al., 2018; Zhang et al., 2019). Furthermore, epoxide hydrolase was identified as a unique gene in the genome of C. sakazakii C7 (Supplementary Table S3C); it plays a vital role in the degradation of organic compounds and is potentially useful in enantioselective biocatalysis. Being able to survive in dried material is a known characteristic of C. sakazakii and contributing to its virulence. Previous investigations have also described that C. sakazakii is able to survive extreme temperatures, including heat-shock at 47°C and cold-shock at −20°C (Shaker et al., 2008; Chang et al., 2010; Gajdosova et al., 2011). The identified genes provide molecular links to these remarkable properties.
It has been demonstrated that C. sakazakii can produce biofilms on stainless steel, glass, polyvinyl chloride, polystyrene, silicone, and latex surfaces (Kim et al., 2006; Hurrell et al., 2009; Aly et al., 2019); these biofilms could not always be eliminated even by thorough washing procedures using disinfectants and water. This capability of biofilm formation by C. sakazakii makes it a more capable pathogen (Shi et al., 2017). We found genes linked to the ability to form biofilms in the genomes of both C. sakazakii isolates (Supplementary Table S1). These include genes for biosynthesis, such as colanic acid biosynthesis process, flagellar assembly protein (FliH), flagellar protein (FlgA-K) and FliCJLPQSTZ, FlhA-E genes linked to the biosynthesis of flagella. Bacterial flagella have been shown to have multiple important roles in biofilm development (Wood, 2013; Kang et al., 2015). They provide cell motility, which is important during formation of biofilms as well as for dispersal bacteria from biofilms, and they play an important role in sensing and colonization of surfaces (Cammarota et al., 2012; Friedlander et al., 2015; Ratthawongjirakul et al., 2016). The flgJ gene is associated with the biosynthesis of a flagellum and it has been described that the presence of the flgJ gene led to increased ability to form biofilms (Ye et al., 2015; Bao et al., 2017a). A unique sequence annotated as filamentous hemagglutinin protein (FHA) was identified in the genome of isolate C7 (Supplementary Tables S2, S3C). In Cronobacter spp. this gene is present on the pCTU1 plasmid and it encodes a filamentous hemagglutinin (Franco et al., 2011). This gene is suggested to play a role in promoting bacterial aggregation and adhesion. In Bordetella, the filamentous hemagglutinin leads to adhesion both in a secreted and surface-associated form (Lambert-Buisine et al., 1998; Jacob-Dubuisson et al., 2000; Franco et al., 2011). In Xanthomonas axonopodis, genes encoding for filamentous hemagglutinin protein is needed for surface attachment, tissue colonization and linked to biofilm formation (Gottig et al., 2009). The grxA family glutaredoxin was identified as a unique gene in the genome of C. sakazakii C8 (Supplementary Table S3D); it is also implicated to play a vital role in biofilm formation. It was recently demonstrated in E. coli that grxA is associated with biofilm formation and with prolonging attachment periods following cell attachment to the substrate (Hancock and Klemm, 2007; Bhomkar et al., 2010). E. coli strains that were functionally deleted for grxA formed biofilms that had lower survival rates after exposure to metal ions (Harrison et al., 2007). Recently, the link between cellulose (a component of the biofilm matrix) production and biofilm formation ability of C. sakazakii was experimentally demonstrated (Hu et al., 2015; Aly et al., 2019).
Cronobacter sakazakii have been shown to have the gene cluster of nanAKTR to utilize sialic acid as a carbon source for bacterial growth (Joseph et al., 2013). We found that C. sakazakii C7 and C8 have the gene clusters nanT, nanR, and for the first time found nanC, for sialic acid metabolism. This unique capability to utilize sialic acid for biosynthesis during biofilm formation could be due to an adaptation to the milk powder environment, because milk contains sialic acid (Wang et al., 2001).
The genes associated with desiccation resistance were identified in both genomes; they include Capsular polysaccharide ABC transporter, Colanic acid capsular biosynthesis activation proteins rcsA and rcsB, Putative capsular polysaccharide transport protein (YegH) and Capsular polysaccharide genes cluster (KpsCDES). The ability to produce a heteropolysaccharide capsule by both C. sakazakii isolates can enhance their ability to form biofilms, which also contributes to their desiccation resistance. Recently, (Ogrodzki and Forsythe, 2015) reported that capsule-associated genes in a C. sakazakii clinical strain is a potentially important virulence trait linked to severe neonatal infection. Furthermore, colanic acid is known as a component of exopolysaccharides in the genus of Cronobacter, which promotes adhesion to different abiotic surfaces with increased resistance for environmental stress factors such as heat, acid, antibiotics and desiccation (Jung et al., 2013). Our observations here agree well with the biofilm formation ability under different conditions of both C. sakazakii C7 and C8 described in our earlier study (Aly et al., 2019). We emphasize that the two unique sequences annotated with filamentous hemagglutinin protein (C7) and grxA gene (C8) associated with the biofilm-forming ability have previously not been observed in C. sakazakii.
To define the conserved genes associated with pathogenicity, we identified the virulence factors of C. sakazakii C7 and C8 isolates (Supplementary Tables S1, S3). The genes of C7 and C8 associated with virulence can be classified into 5 categories. These are: adherence, invasion, secretion system, iron uptake, and toxins.
Starting with the adherence category, we found outer membrane protein A (OmpA), FliR, fliC, and FlhA adhesion genes in the genomes of both the C7 and C8 isolates. It has been reported that the OmpA gene led to improving cell adhesion of C. sakazakii and invasion of HBMEC (Nair et al., 2009), suggesting both isolates may invade HBMEC. The FliR gene is mainly responsible for the structural components of the flagellum. Flagella are implicated in many mechanisms of adhesion to host cells and pathogenicity; the FliR gene is therefore instrumental in both adherence and invasion (Du et al., 2016). The fliC gene has also been strongly associated with adherence and virulence traits of the pathogens (Dingle et al., 2011; Aldubyan et al., 2017; Holý et al., 2019). The FlhA gene is implicated as required for export of flagellin and therefore assembly of flagella; it therefore has been described to play a role in invasion into and adhesion to epithelial cells by various bacteria, such as Pseudomonas aeruginosa, Bacillus thuringiensis and B. cereus and C. sakazakii (Fleiszig et al., 2001; Ramarao and Lereclus, 2006; Du et al., 2016).
Additionally, identified genes that potentially can aid invasion and contribute to pathogenicity and virulence were annotated as efflux system component, zinc metalloprotease YfgC precursor, putative S-ribosyl homocysteine lyase encoded by the luxS gene and protein hemolysin III homologs. The silver and copper cation efflux system has also been implicated in facilitated invasion of HBMEC (Kucerova et al., 2010). Furthermore, the superoxide dismutase gene was present in the genome; it has been implicated in improved survival of bacteria in macrophages (Townsend et al., 2007; Bao et al., 2017b).
In the secretion system category, based on sequence analysis, we identified several putative genes of Type VI Secretion System cluster in the genomic sequence of both isolates. In addition, we identified unique sequences linked to the Type VI Secretion, such as the rhs gene in the genome of C7 and the VgrG gene cluster in the genome of C8. The Type VI Secretion System (T6SS) is a versatile protein secretion machinery able to immediately deliver protein toxins into eukaryotic cells (Durand et al., 2014; Wang et al., 2018). Its functions are linked to virulence factors and it delivers bacteriolytic effectors to target cells. In P. aeruginosa, it has been reported that the Type VI Secretion System can secrete three types of exported effectors (Tse1-3) that act to destroy cell membranes, peptidoglycans and cytoplasmic components in infected organisms (Russell et al., 2011, 2013). The VgrG genes are part of the Type VI Secretion System machinery, also acting as effectors; VgrG1 possesses a C-terminal extension, defined as an actin cross-linking domain of the Vibrio cholerae Type VI Secretion System (Ma et al., 2009).
In the iron uptake category, a number of genes responsible for production of metal binding proteins were identified in both genomic sequences; these include the iron ABC transporter system and the iron-sulfur cluster binding protein. Both are associated with bacterial virulence. The iron ABC transporter permease was also identified as unique gene in the genome sequence of C7. Iron is an important element for survival and colonization by bacteria since it plays a considerable role in the electron transport chain to produce energy (Mietzner and Morse, 1994). Successful competition for iron uptake is therefore crucial for pathogenicity. The privileged iron acquisition system, containing siderophore biosynthesis (iucABD/iutA operon) and EfeO systems for the acquisition of ferrous iron were identified in C. sakazakii C7 and C8. This ability may lead to the survival of C. sakazakii in blood and thereby its ability to invade the central nervous system (Singh et al., 2015) by crossing the blood-brain barrier. The latter is a characteristic responsible for the worst diseases observed to be caused by C. sakazakii.
In the toxin category, many genes associated with toxins such as a HigB toxin protein, RelE/StbE replicon stabilization toxin and HigA protein (antitoxin to HigB) were identified in the genomic sequence of both C7 and C8. Furthermore, C. sakazakii C7 exhibited one unique toxin RelE gene associated with cellular processes that include persistence. RelE bacterial toxin is structurally like microbial endoribonucleases (Griffin et al., 2013); RelE has been suggested to “represent a shift in the RNase general acid-base catalytic paradigm and promote catalysis predominantly by leaving-group protonation” and charge stabilization (Griffin et al., 2013). In summary, several unique genes associated with pathogenicity and virulence were identified in the genomic sequences of C7 (milk powder) and C8 (ready-to-eat mixed salad) isolates.
Prophages are mobile genetic elements able to deliver virulence factors (O’Brien et al., 1984) or antimicrobial-resistance genes (Colomer-Lluch et al., 2011) to bacterial hosts and increase the diversity of the host genome (Ventura et al., 2006). We found three regions associated with phages (A1–A3) in the genome of C7. We also identified three regions associated with phages (S1–S3) in the genome of C8 (Supplementary Table S5). Each isolate contains two intact phages. Further, we found one questionable prophage in the S1 region (Entero-Tyrion phage) of the C8 genome, whereas for C7 one incomplete prophage was identified.
In this study, we present the WGS results of C. sakazakii strains isolated from ready-to-eat mixed salad and skimmed milk powder. Many identified genes harbored by both isolates are associated with multidrug-resistance, pathogenesis, virulence, and biofilm formation ability. Several identified genes associated with producing capsules and biofilms point to this as an important mechanism for the C. sakazakii defense against desiccation and its ability to survive in, e.g., milk powder for a long time. Analyzing all these traits gives a molecular basis to understand their ability to survive the extremely stressful environments met in food production and storage, including the highly desiccated and heat-treated environment of dried milk powder. The genome of C. sakazakii isolated in Austria from ready-to-eat mixed salad held a unique ST not previously observed, but also most of the genes implicating C. sakazakii as a highly virulent pathogen. The presence of such C. sakazakii in ready-to-eat food indicates another potential route for infection and pathogenicity by C. sakazakii, besides the established route of powdered milk formulas. Especially disturbing is the multidrug antibiotics resistance found in both isolates. Our study provides new data to better understand the pathogenicity mechanism and virulence of C. sakazakii in food ingredients, and to improve monitoring and tracking of the source of food contamination. It highlights the use of WGS for traceability and detection of C. sakazakii strains. Based on our findings we call for further research to clarify the potential reservoirs of this emerging pathogen as well as to how and where it has acquired its multidrug antibiotics resistance.
The datasets generated in this manuscript can be found on NCBI under the C. sakazakii complete genome bioproject number PRJNA510032 and biosample accession numbers are SAMN10593105 and SAMN10593106 for C. sakazakii C7 and C8, respectively.
MA designed and carried out the work. ER and KD supervised the work. ER and MA wrote the manuscript. All authors reviewed the manuscript.
We acknowledge support for this work by the Egypt Ministry of Higher Education (Ph.D. grant program from the Egyptian Cultural Affairs Sector and Missions) under the Fifth Mission Plan for the year 2010–2011 (Announcement No. 2), as well as the Austrian Science Fund (FWF) grant number I 3064.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01464/full#supplementary-material
Aldubyan, M. A., Almami, I. S., Benslimane, F. M., Alsonosi, A. M., and Forsythe, S. J. (2017). Comparative outer membrane protein analysis of high and low-invasive strains of Cronobacter malonaticus. Front. Microbiol. 8:2268. doi: 10.3389/fmicb.2017.02268
Alsonosi, A., Hariri, S., Kajsik, M., Orieskova, M., Hanulik, V., Roderova, M., et al. (2015). The speciation and genotyping of Cronobacter isolates from hospitalised patients. Eur. J. Clin. Microbiol. Infect. Dis. 34, 1979–1988. doi: 10.1007/s10096-015-2440-8
Aly, M., Domig, K., Kneifel, W., and Reimhult, E. (2018). Immunogold nanoparticles for rapid plasmonic detection of C. sakazakii. Sensors 18:2028. doi: 10.3390/s18072028
Aly, M. A., Reimhult, E., Kneifel, W., and Domig, K. J. (2019). Characterization of biofilm formation by cronobacter spp. isolates of different food origin under model conditions. J. Food Prot. 82, 65–77. doi: 10.4315/0362-028x.jfp-18-036
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bao, X., Jia, X., Chen, L., Peters, B. M., Lin, C.-W., Chen, D., et al. (2017a). Effect of polymyxin resistance (pmr) on biofilm formation of Cronobacter sakazakii. Microb. Pathog. 106, 16–19. doi: 10.1016/j.micpath.2016.12.012
Bao, X., Yang, L., Chen, L., Li, B., Li, L., Li, Y., et al. (2017b). Analysis on pathogenic and virulent characteristics of the Cronobacter sakazakii strain BAA-894 by whole genome sequencing and its demonstration in basic biology science. Microb. Pathog. 109, 280–286. doi: 10.1016/j.micpath.2017.05.030
Bennour Hennekinne, R., Guillier, L., Fazeuilh, L., Ells, T., Forsythe, S., Jackson, E., et al. (2018). Survival of Cronobacter in powdered infant formula and their variation in biofilm formation. Lett. Appl. Microbiol. 66, 496–505. doi: 10.1111/lam.12879
Bhomkar, P., Materi, W., Semenchenko, V., and Wishart, D. S. (2010). Transcriptional response of E. coli upon FimH-mediated fimbrial adhesion. Gene Regul. Syst. Biol. 4, 1–17.
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Cammarota, G., Sanguinetti, M., Gallo, A., and Posteraro, B. (2012). Review article: biofilm formation by Helicobacter pylori as a target for eradication of resistant infection. Aliment. Pharmacol Ther. 36, 222–230. doi: 10.1111/j.1365-2036.2012.05165.x
Cetinkaya, E., Joseph, S., Ayhan, K., and Forsythe, S. J. (2013). Comparison of methods for the microbiological identification and profiling of Cronobacter species from ingredients used in the preparation of infant formula. Mol. Cell. Probes 27, 60–64. doi: 10.1016/j.mcp.2012.10.003
Chang, C. H., Chiang, M. L., and Chou, C. C. (2010). The effect of heat shock on the response of Cronobacter sakazakii to subsequent lethal stresses. Foodborne Pathog. Dis. 7, 71–76. doi: 10.1089/fpd.2009.0345
Chaudhari, N. M., Gupta, V. K., and Dutta, C. (2016). BPGA- an ultra-fast pan-genome analysis pipeline. Sci. Rep. 6:24373. doi: 10.1038/srep24373
Chon, J.-W., Song, K.-Y., Kim, S.-Y., Hyeon, J.-Y., and Seo, K.-H. (2012). Isolation and characterization of Cronobacter from desiccated foods in Korea. J. Food Sci. 77, M354–M358. doi: 10.1111/j.1750-3841.2012.02750.x
Colomer-Lluch, M., Imamovic, L., Jofre, J., and Muniesa, M. (2011). Bacteriophages carrying antibiotic resistance genes in fecal waste from cattle, pigs, and poultry. Antimicrob. Agents Chemother. 55, 4908–4911. doi: 10.1128/aac.00535-11
Cruz, A., Xicohtencatl-Cortes, J., González-Pedrajo, B., Bobadilla, M., Eslava, C., and Rosas, I. (2011). Virulence traits in Cronobacter species isolated from different sources. Can. J. Microbiol. 57, 735–744. doi: 10.1139/w11-063
Dennis, J. J., Lynch, K. H., Liang, Y., Zhou, Y., and Wishart, D. S. (2011). PHAST: a fast phage search tool. Nucleic Acids Res. 39, W347–W352. doi: 10.1093/nar/gkr485
Dingle, T. C., Mulvey, G. L., and Armstrong, G. D. (2011). Mutagenic analysis of the Clostridium difficile Flagellar proteins, FliC and FliD, and their contribution to virulence in Hamsters. Infect. Immun. 79, 4061–4067. doi: 10.1128/iai.05305-11
Dinnbier, U., Limpinsel, E., Schmid, R., and Bakker, E. P. (1988). Transient accumulation of potassium glutamate and its replacement by trehalose during adaptation of growing cells of Escherichia coli K-12 to elevated sodium chloride concentrations. Arch. Microbiol. 150, 348–357. doi: 10.1007/bf00408306
Drudy, D., Mullane, N. R., Quinn, T., Wall, P. G., and Fanning, S. (2006). Enterobacter sakazakii: an emerging pathogen in powdered infant formula. Clin. Infect. Dis. 42, 996–1002. doi: 10.1086/501019
Du, X.-J., Wang, X.-Y., Dong, X., Li, P., and Wang, S. (2018). Characterization of the desiccation tolerance of Cronobacter sakazakii Strains. Front. Microbiol. 9:2867. doi: 10.3389/fmicb.2018.02867
Du, X. J., Zhang, X., Li, P., Xue, R., and Wang, S. (2016). Screening of genes involved in interactions with intestinal epithelial cells in Cronobacter sakazakii. AMB Express 6:74. doi: 10.1186/s13568-016-0246-4
Durand, E., Cambillau, C., Cascales, E., and Journet, L. (2014). VgrG, Tae, Tle, and beyond: the versatile arsenal of Type VI secretion effectors. Trend. Microbiol. 22, 498–507. doi: 10.1016/j.tim.2014.06.004
El-Sharoud, W. M., O’Brien, S., Negredo, C., Iversen, C., Fanning, S., and Healy, B. (2009). Characterization of Cronobacter recovered from dried milk and related products. BMC Microbiol. 9:9. doi: 10.1186/1471-2180-9-24
Fakruddin, M., Rahaman, M., Ahmed, M. M., and Hoque, M. M. (2014). Stress tolerant virulent strains of Cronobacter sakazakii from food. Biol. Res. 47:63. doi: 10.1186/0717-6287-47-63
Feeney, A., and Sleator, R. D. (2011). An in silico analysis of osmotolerance in the emerging gastrointestinal pathogen Cronobacter sakazakii. Bioeng Bugs 2, 260–270. doi: 10.4161/bbug.2.5.17238
Finn, S., Handler, K., Condell, O., Colgan, A., Cooney, S., McClure, P., et al. (2013). ProP is required for the survival of desiccated Salmonella enterica serovar Typhimurium cells on a stainless steel surface. Appl. Environ. Microbiol. 79, 4376–4384. doi: 10.1128/aem.00515-13
Fleiszig, S. M. J., Arora, S. K., Van, R., and Ramphal, R. (2001). FlhA, a component of the flagellum assembly apparatus of Pseudomonas aeruginosa, plays a role in internalization by corneal epithelial cells. Infect. Immun. 69, 4931–4937. doi: 10.1128/iai.69.8.4931-4937.2001
Forsythe, S. (2018). Updates on the Cronobacter genus. Annu. Rev. Food Sci. Technol. 9, 23–44. doi: 10.1146/annurev-food-030117-012246
Forsythe, S. J. (2015). “New insights into the emergent bacterial pathogen Cronobacter,” in Food Safety Emerging Issues, Technologies and Systems, eds S. C. Ricke, J. R. Donaldson, and C. A. Phillips (San Diego: Academic Press), 265–308. doi: 10.1016/b978-0-12-800245-2.00013-7
Forsythe, S. J., Dickins, B., and Jolley, K. A. (2014). Cronobacter, the emergent bacterial pathogen Enterobacter sakazakii comes of age; MLST and whole genome sequence analysis. BMC Genomics 15:1121. doi: 10.1186/1471-2164-15-1121
Franco, A. A., Hu, L., Grim, C. J., Gopinath, G., Sathyamoorthy, V., Jarvis, K. G., et al. (2011). Characterization of putative virulence genes on the related RepFIB plasmids harbored by Cronobacter spp. Appl. Environ. Microbiol. 77, 3255–3267. doi: 10.1128/aem.03023-10
Franke, S., Grass, G., Rensing, C., and Nies, D. H. (2003). Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 185, 3804–3812. doi: 10.1128/jb.185.13.3804-3812.2003
Friedlander, R. S., Vogel, N., and Aizenberg, J. (2015). Role of flagella in adhesion of Escherichia coli to abiotic surfaces. Langmuir 31, 6137–6144. doi: 10.1021/acs.langmuir.5b00815
Gajdosova, J., Benedikovicova, K., Kamodyova, N., Tothova, L., Kaclikova, E., Stuchlik, S., et al. (2011). Analysis of the DNA region mediating increased thermotolerance at 58 degrees C in Cronobacter sp. and other enterobacterial strains. Antonie Van Leeuwenhoek 100, 279–289. doi: 10.1007/s10482-011-9585-y
Gicova, A., Orieskova, M., Oslanecova, L., Drahovska, H., and Kaclikova, E. (2014). Identification and characterization of Cronobacter strains isolated from powdered infant foods. Lett. Appl. Microbiol. 58, 242–247. doi: 10.1111/lam.12179
Giri, C. P., Shima, K., Tall, B. D., Curtis, S., Sathyamoorthy, V., Hanisch, B., et al. (2012). Cronobacter spp. (previously Enterobacter sakazakii) invade and translocate across both cultured human intestinal epithelial cells and human brain microvascular endothelial cells. Microb. Pathog. 52, 140–147. doi: 10.1016/j.micpath.2011.10.003
Gottig, N., Garavaglia, B. S., Garofalo, C. G., Orellano, E. G., and Ottado, J. (2009). A filamentous hemagglutinin-like protein of Xanthomonas axonopodis pv. citri, the phytopathogen responsible for citrus canker, is involved in bacterial virulence. PLoS One 4:e4358. doi: 10.1371/journal.pone.0004358
Götz, S., García-Gómez, J. M., Terol, J., Williams, T. D., Nagaraj, S. H., Nueda, M. J., et al. (2008). High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435. doi: 10.1093/nar/gkn176
Griffin, M. A., Davis, J. H., and Strobel, S. A. (2013). Bacterial toxin RelE: a highly efficient nuclease with exquisite substrate specificity using atypical catalytic residues. Biochemistry 52, 8633–8642. doi: 10.1021/bi401325c
Guimarães, L. C., Florczak-Wyspianska, J., de Jesus, L. B., Viana, M. V. C., Silva, A., Ramos, R. T. J., et al. (2015). Inside the pan-genome - methods and software overview. Curr. Genomics 16, 245–252. doi: 10.2174/1389202916666150423002311
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Hancock, V., and Klemm, P. (2007). Global gene expression profiling of asymptomatic bacteriuria Escherichia coli during biofilm growth in human urine. Infect. Immun. 75, 966–976. doi: 10.1128/iai.01748-06
Harrison, J. J., Ceri, H., and Turner, R. J. (2007). Multimetal resistance and tolerance in microbial biofilms. Nat. Rev. Microbiol. 5, 928–938. doi: 10.1038/nrmicro1774
He, Y. (2015). “Chapter 20 - bacterial whole-genome determination and applications,” in Molecular Medical Microbiology (Second Edition), eds Y.-W. Tang, M. Sussman, D. Liu, I. Poxton, and J. Schwartzman (Boston: Academic Press), 357–368. doi: 10.1016/b978-0-12-397169-2.00020-2
Holý, O., Cruz-Córdova, A., Xicohtencatl-Cortes, J., Hochel, I., Parra-Flores, J., Petrželová, J., et al. (2019). Occurrence of virulence factors in Cronobacter sakazakii and Cronobacter malonaticus originated from clinical samples. Microbial Pathog. 127, 250–256. doi: 10.1016/j.micpath.2018.12.011
Hu, L., Grim, C. J., Franco, A. A., Jarvis, K. G., Sathyamoorthy, V., Kothary, M. H., et al. (2015). Analysis of the cellulose synthase operon genes, bcsA, bcsB, and bcsC in Cronobacter species: prevalence among species and their roles in biofilm formation and cell-cell aggregation. Food Microbiol. 52, 97–105. doi: 10.1016/j.fm.2015.07.007
Hunter, C. J., Petrosyan, M., Ford, H. R., and Prasadarao, N. V. (2008). Enterobacter sakazakii: an emerging pathogen in infants and neonates. Surg. Infect. 9, 533–539. doi: 10.1089/sur.2008.006
Hurrell, E., Kucerova, E., Loughlin, M., Caubilla-Barron, J., and Forsythe, S. J. (2009). Biofilm formation on enteral feeding tubes by Cronobacter sakazakii, Salmonella serovars and other Enterobacteriaceae. Int. J. Food Microbiol. 136, 227–231. doi: 10.1016/j.ijfoodmicro.2009.08.007
Jacob-Dubuisson, F., Kehoe, B., Willery, E., Reveneau, N., Locht, C., and Relman, D. A. (2000). Molecular characterization of Bordetella bronchiseptica filamentous haemagglutinin and its secretion machinery. Microbiology 146(Pt 5), 1211–1221. doi: 10.1099/00221287-146-5-1211
Jang, H., Woo, J., Lee, Y., Negrete, F., Finkelstein, S., Chase, H. R., et al. (2018). Draft genomes of Cronobacter sakazakii strains isolated from dried spices bring unique insights into the diversity of plant-associated strains. Stand Genomic Sci. 13:35. doi: 10.1186/s40793-018-0339-6
Jarvis, K. G., Grim, C. J., Franco, A. A., Gopinath, G., Sathyamoorthy, V., Hu, L., et al. (2011). Molecular characterization of Cronobacter lipopolysaccharide O-antigen gene clusters and development of serotype-specific PCR assays. Appl. Environ. Microbiol. 77, 4017–4026. doi: 10.1128/aem.00162-11
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Jolley, K. A., and Maiden, M. C. J. (2010). BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinform. 11:595. doi: 10.1186/1471-2105-11-595
Joseph, S., Desai, P., Ji, Y., Cummings, C. A., Shih, R., and Degoricija, L. (2012a). Comparative analysis of genome sequences covering the seven Cronobacter species. PLoS One 7:e49455. doi: 10.1371/journal.pone.0049455
Joseph, S., Sonbol, H., Hariri, S., Desai, P., McClelland, M., and Forsythe, S. J. (2012b). Diversity of the Cronobacter genus as revealed by multilocus sequence typing. J. Clin. Microbiol. 50, 3031–3039. doi: 10.1128/jcm.00905-12
Joseph, S., and Forsythe, S. J. (2011). Predominance of Cronobacter sakazakii sequence type 4 in neonatal infections. Emerging. Infect. Dis. 17, 1713–1715. doi: 10.3201/eid1709.110260
Joseph, S., and Forsythe, S. J. (2012). Insights into the emergent bacterial pathogen cronobacter spp., generated by multilocus sequence typing and analysis. Front. Microbiol. 3:397. doi: 10.3389/fmicb.2012.00397
Joseph, S., Hariri, S., Masood, N., and Forsythe, S. (2013). Sialic acid utilization by Cronobacter sakazakii. Microb. Inform. Exp. 3:3. doi: 10.1186/2042-5783-3-3
Jung, J.-H., Choi, N. Y., and Lee, S. Y. (2013). Biofilm formation and exopolysaccharide (EPS) production by Cronobacter sakazakii depending on environmental conditions. Food. Microbiol. 34, 70–80. doi: 10.1016/j.fm.2012.11.008
Kadlicekova, V., Kajsik, M., Soltys, K., Szemes, T., Slobodnikova, L., Janosikova, L., et al. (2018). Characterisation of Cronobacter strains isolated from hospitalised adult patients. Antonie van Leeuwenhoek 111, 1073–1085. doi: 10.1007/s10482-017-1008-2
Kang, X. M., Wang, F. F., Zhang, H., Zhang, Q., and Qiana, W. (2015). Genome-wide identification of genes necessary for biofilm formation by nosocomial pathogen Stenotrophomonas maltophilia reveals that orphan response regulator FsnR is a critical modulator. Appl. Environ. Microbiol. 81, 1200–1209. doi: 10.1128/aem.03408-14
Kilonzo-Nthenge, A., Rotich, E., Godwin, S., Nahashon, S., and Chen, F. (2012). Prevalence and antimicrobial resistance of Cronobacter sakazakii isolated from domestic kitchens in middle Tennessee, United States. J. Food Prot. 75, 1512–1517. doi: 10.4315/0362-028x.jfp-11-442
Kim, H., Ryu, J. H., and Beuchat, L. R. (2006). Attachment of and biofilm formation by Enterobacter sakazakii on stainless steel and enteral feeding tubes. Appl. Environ. Microbiol. 72, 5846–5856. doi: 10.1128/Aem.00654-06
Kim, J. N., Kim, Y., Jeong, Y., Roe, J. H., Kim, B. G., and Cho, B. K. (2015). Comparative genomics reveals the core and accessory genomes of Streptomyces species. J. Microbiol. Biotechnol. 25, 1599–1605. doi: 10.4014/jmb.1504.04008
Kim, K., Jang, S. S., Kim, S. K., Park, J. H., Heu, S., and Ryu, S. (2008). Prevalence and genetic diversity of Enterobacter sakazakii in ingredients of infant foods. Int. J. Food Microbiol. 122, 196–203. doi: 10.1016/j.ijfoodmicro.2007.11.072
Kim, K., Kim, K. P., Choi, J., Lim, J. A., Lee, J., Hwang, S., et al. (2010). Outer membrane proteins A (OmpA) and X (OmpX) are essential for basolateral invasion of Cronobacter sakazakii. Appl. Environ. Microbiol. 76, 5188–5198. doi: 10.1128/aem.02498-09
Knight, D. R., Squire, M. M., Collins, D. A., and Riley, T. V. (2016). Genome analysis of Clostridium difficile PCR Ribotype 014 lineage in australian pigs and humans reveals a diverse genetic repertoire and signatures of long-range interspecies transmission. Front. Microbiol. 7:2138. doi: 10.3389/fmicb.2016.02138
Kozyreva, V. K., Crandall, J., Sabol, A., Poe, A., Zhang, P., Concepcion-Acevedo, J., et al. (2016). Laboratory investigation of Salmonella enterica serovar poona outbreak in California: comparison of pulsed-field gel electrophoresis (PFGE) and whole genome sequencing (WGS) results. PLoS Curr. 8:ecurrents.outbreaks.1bb3e36e74bd5779bc43ac3a8dae52e6. doi: 10.1371/currents.outbreaks.1bb3e36e74bd5779bc43ac3a8dae52e6
Kucerova, E., Clifton, S. W., Xia, X. Q., Long, F., Porwollik, S., Fulton, L., et al. (2010). Genome sequence of Cronobacter sakazakii BAA-894 and comparative genomic hybridization analysis with other Cronobacter species. PLoS One 5:e9556. doi: 10.1371/journal.pone.0009556
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lambert-Buisine, C., Willery, E., Locht, C., and Jacob-Dubuisson, F. (1998). N-terminal characterization of the Bordetella pertussis filamentous haemagglutinin. Mol. Microbiol. 28, 1283–1293. doi: 10.1046/j.1365-2958.1998.00892.x
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/jcm.06094-11
Lehner, A., Tall, B. D., Fanning, S., and Srikumar, S. (2018). Cronobacter spp.—opportunistic foodborne pathogens: an update on evolution, osmotic adaptation and pathogenesis. Curr. Clin. Microb. Rep. 5, 97–105. doi: 10.1007/s40588-018-0089-7
Liu, B.-T., Song, F.-J., Zou, M., Hao, Z.-H., and Shan, H. (2017). Emergence of colistin resistance gene mcr-1 in Cronobacter sakazakii producing NDM-9 and in Escherichia coli from the same animal. Antimicrob. Agents Chemother. 61:e1444-16. doi: 10.1128/aac.01444-16
Lytsy, B., Engstrand, L., Gustafsson, A., and Kaden, R. (2017). Time to review the gold standard for genotyping vancomycin-resistant enterococci in epidemiology: comparing whole-genome sequencing with PFGE and MLST in three suspected outbreaks in Sweden during 2013-2015. Infect. Genet. Evol. 54, 74–80. doi: 10.1016/j.meegid.2017.06.010
Ma, A. T., McAuley, S., Pukatzki, S., and Mekalanos, J. J. (2009). Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe 5, 234–243. doi: 10.1016/j.chom.2009.02.005
Masood, N., Moore, K., Farbos, A., Paszkiewicz, K., Dickins, B., McNally, A., et al. (2015). Genomic dissection of the 1994 Cronobacter sakazakii outbreak in a French neonatal intensive care unit. BMC Genomics 16:750. doi: 10.1186/s12864-015-1961-y
Medini, D., Donati, C., Tettelin, H., Masignani, V., and Rappuoli, R. (2005). The microbial pan-genome. Curr. Opin. Genet. Dev. 15, 589–594. doi: 10.1016/j.gde.2005.09.006
Mietzner, T. A., and Morse, S. A. (1994). The role of iron-binding proteins in the survival of pathogenic bacteria. Annu. Rev. Nutr. 14, 471–493. doi: 10.1146/annurev.nu.14.070194.002351
Minor, T., Lasher, A., Klontz, K., Brown, B., Nardinelli, C., and Zorn, D. (2015). The per case and total annual costs of foodborne illness in the United States. Risk Anal. 35, 1125–1139. doi: 10.1111/risa.12316
Miranda, C. D., Kehrenberg, C., Ulep, C., Schwarz, S., and Roberts, M. C. (2003). Diversity of tetracycline resistance genes in bacteria from chilean salmon farms. Antimicrob. Agents Chemother. 47, 883–888. doi: 10.1128/aac.47.3.883-888.2003
Mittal, R., Bulgheresi, S., Emami, C., and Prasadarao, N. V. (2009). Enterobacter sakazakii targets DC-SIGN to induce immunosuppressive responses in dendritic cells by modulating MAPKs. J. Immunol. 183, 6588–6599. doi: 10.4049/jimmunol.0902029
Mullane, N., O’Gaora, P., Nally, J. E., Iversen, C., Whyte, P., Wall, P. G., et al. (2008). Molecular analysis of the Enterobacter sakazakii O-antigen gene locus. Appl. Environ. Microbiol. 74, 3783–3794. doi: 10.1128/aem.02302-07
Mullane, N. R., Iversen, C., Healy, B., Walsh, C., Whyte, P., Wall, P. G., et al. (2007). Enterobacter sakazakii an emerging bacterial pathogen with implications for infant health. Minerva Pediatr. 59, 137–148.
Nadon, C., Van Walle, I., Gerner-Smidt, P., Campos, J., Chinen, I., Concepcion-Acevedo, J., et al. (2017). PulseNet International: vision for the implementation of whole genome sequencing (WGS) for global food-borne disease surveillance. Euro. Surveill. 22:30544. doi: 10.2807/1560-7917.es.2017.22.23.30544
Nair, M. K. M., Venkitanarayanan, K., Silbart, L. K., and Kim, K. S. (2009). Outer membrane protein A (OmpA) of Cronobacter sakazakii Binds Fibronectin and Contributes to Invasion of Human Brain Microvascular Endothelial Cells. Foodborne Pathog. Dis. 6, 495–501. doi: 10.1089/fpd.2008.0228
Nazarowec-White, M., and Farber, J. M. (1997). Enterobacter sakazakii: a review. Int. J. Food Microbiol. 34, 103–113.
O’Brien, A. D., Newland, J. W., Miller, S. F., Holmes, R. K., Smith, H. W., and Formal, S. B. (1984). Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226, 694–696. doi: 10.1126/science.6387911
Ogrodzki, P., and Forsythe, S. (2015). Capsular profiling of the Cronobacter genus and the association of specific Cronobacter sakazakii and C. malonaticus capsule types with neonatal meningitis and necrotizing enterocolitis. BMC Genomics 16:758. doi: 10.1186/s12864-015-1960-z
Ogrodzki, P., and Forsythe, S. J. (2017). DNA-sequence based typing of the Cronobacter genus using MLST, CRISPR-cas array and capsular profiling. Front. Microbiol. 8:1875. doi: 10.3389/fmicb.2017.01875
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Pan, R., Jiang, Y., Sun, L., Wang, R., Zhuang, K., Zhao, Y., et al. (2018). Gold nanoparticle-based enhanced lateral flow immunoassay for detection of Cronobacter sakazakii in powdered infant formula. J. Dairy Sci. 101, 3835–3843. doi: 10.3168/jds.2017-5
Petkau, A., Stuart-Edwards, M., Stothard, P., and Van Domselaar, G. (2010). Interactive microbial genome visualization with GView. Bioinformatics 26, 3125–3126. doi: 10.1093/bioinformatics/btq588
Ramarao, N., and Lereclus, D. (2006). Adhesion and cytotoxicity of Bacillus cereus and Bacillus thuringiensis to epithelial cells are FlhA and PlcR dependent, respectively. Microbes infect. 8, 1483–1491. doi: 10.1016/j.micinf.2006.01.005
Ratthawongjirakul, P., Thongkerd, V., and Chaicumpa, W. (2016). The impacts of a fliD mutation on the biofilm formation of Helicobacter pylori. Asian Pac. J. Trop. Biomed. 6, 1008–1014. doi: 10.1016/j.apjtb.2016.10.005
Russell, A. B., Hood, R. D., Bui, N. K., LeRoux, M., Vollmer, W., and Mougous, J. D. (2011). Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–347. doi: 10.1038/nature10244
Russell, A. B., LeRoux, M., Hathazi, K., Agnello, D. M., Ishikawa, T., Wiggins, P. A., et al. (2013). Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 496, 508–512. doi: 10.1038/nature12074
Santos, H., and Da Costa, M. S. (2002). Compatible solutes of organisms that live in hot saline environments. Environ. Microbiol. 4, 501–509. doi: 10.1046/j.1462-2920.2002.00335.x
Scharff, R. L., Besser, J., Sharp, D. J., Jones, T. F., Peter, G. S., and Hedberg, C. W. (2016). An economic evaluation of PulseNet: a network for foodborne disease surveillance. Am. J. Prev. Med. 50(5 Suppl. 1), S66–S73. doi: 10.1016/j.amepre.2015.09.018
Shaker, R. R., Osaili, T. M., Abu Al-Hasan, A. S., Ayyash, M. M., and Forsythe, S. J. (2008). Effect of desiccation, starvation, heat, and cold stresses on the thermal resistance of Enterobacter sakazakii in rehydrated infant milk formula. J. Food Sci. 73, M354–M359. doi: 10.1111/j.1750-3841.2008.00880.x
Shi, C., Sun, Y., Liu, Z., Guo, D., Sun, H., Sun, Z., et al. (2017). Inhibition of Cronobacter sakazakii virulence factors by citral. Sci. Rep. 7:43243. doi: 10.1038/srep43243
Singh, N., Goel, G., and Raghav, M. (2015). Insights into virulence factors determining the pathogenicity of Cronobacter sakazakii. Virulence 6, 433–440. doi: 10.1080/21505594.2015.1036217
Siqueira Santos, R. F., da Silva, N., Amstalden Junqueira, V. C., Kajsik, M., Forsythe, S., and Pereira, J. L. (2013). Screening for Cronobacter species in powdered and reconstituted infant formulas and from equipment used in formula preparation in maternity hospitals. Ann. Nutr. Metabol. 63, 62–68. doi: 10.1159/000353137
Sonbol, H., Joseph, S., McAuley, C. M., Craven, H. M., and Forsythe, S. J. (2013). Multilocus sequence typing of Cronobacter spp. from powdered infant formula and milk powder production factories. Int. Dairy J. 30, 1–7. doi: 10.1016/j.idairyj.2012.11.004
Srikumar, S., Cao, Y., Yan, Q., Van Hoorde, K., Nguyen, S., Cooney, S., et al. (2019). RNA sequencing-based transcriptional overview of xerotolerance in Cronobacter sakazakii SP291. Appl. Environ. Microbiol. 85:e1993-18. doi: 10.1128/aem.01993-18
Stephan, R., Lehner, A., Tischler, P., and Rattei, T. (2011). Complete genome sequence of Cronobacter turicensis LMG 23827, a food-borne pathogen causing deaths in Neonates. J. Bacteriol. 193, 309–310. doi: 10.1128/jb.01162-10
Stoop, B., Lehner, A., Iversen, C., and Fanning, S. (2009). Development and evaluation of rpoB based PCR systems to differentiate the six proposed species within the genus Cronobacter. Int. J. Food Microbiol. 136, 165–168. doi: 10.1016/j.ijfoodmicro.2009.04.023
Stranden, A., Frei, R., and Widmer, A. F. (2003). Molecular typing of methicillin-resistant Staphylococcus aureus: can PCR replace pulsed-field gel electrophoresis? J. Clin. Microbiol. 41, 3181–3186. doi: 10.1128/jcm.41.7.3181-3186.2003
Sun, Y., Wang, M., Wang, Q., Cao, B., He, X., Li, K., et al. (2012). Genetic analysis of the Cronobacter sakazakii O4 to O7 O-antigen gene clusters and development of a PCR assay for identification of all C. sakazakii O serotypes. Appl. Environ. Microbiol. 78, 3966–3974. doi: 10.1128/aem.07825-11
Taboada, E. N., Graham, M. R., Carriço, J. A., and Van Domselaar, G. (2017). Food safety in the age of next generation sequencing, bioinformatics, and open data access. Front. Microbiol. 8:909. doi: 10.3389/fmicb.2017.00909
Tamura, K., Nei, M., and Kumar, S. (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U.S.A. 101, 11030–11035. doi: 10.1073/pnas.0404206101
Tettelin, H., Masignani, V., Cieslewicz, M. J., Donati, C., Medini, D., Ward, N. L., et al. (2005). Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. U.S.A. 102, 13950–13955. doi: 10.1073/pnas.0506758102
Tettelin, H., Riley, D., Cattuto, C., and Medini, D. (2008). Comparative genomics: the bacterial pan-genome. Curr. Opin. Microbiol. 11, 472–477. doi: 10.1016/j.mib.2008.09.006
Touze, T., Eswaran, J., Bokma, E., Koronakis, E., Hughes, C., and Koronakis, V. (2004). Interactions underlying assembly of the Escherichia coli AcrAB-TolC multidrug efflux system. Mol. Microbiol. 53, 697–706. doi: 10.1111/j.1365-2958.2004.04158.x
Townsend, S., Hurrell, E., and Forsythe, S. (2008). Virulence studies of Enterobacter sakazakii isolates associated with a neonatal intensive care unit outbreak. BMC Microbiol. 8:64. doi: 10.1186/1471-2180-8-64
Townsend, S. M., Hurrell, E., Gonzalez-Gomez, I., Lowe, J., Frye, J. G., Forsythe, S., et al. (2007). Enterobacter sakazakii invades brain capillary endothelial cells, persists in human macrophages influencing cytokine secretion and induces severe brain pathology in the neonatal rat. Microbiology 153(Pt 10), 3538–3547. doi: 10.1099/mic.0.2007/009316-0
Tsai, H. Y., Liao, C. H., Huang, Y. T., Lee, P. I., and Hsueh, P. R. (2013). Cronobacter infections not from infant formula. Taiwan. Emerg. Infect. Dis. 19, 167–169. doi: 10.3201/eid1901.120774
van Acker, J., de Smet, F., Muyldermans, G., Bougatef, A., Naessens, A., and Lauwers, S. (2001). Outbreak of necrotizing enterocolitis associated with Enterobacter sakazakii in powdered milk formula. J. Clin. Microbiol. 39, 293–297. doi: 10.1128/jcm.39.1.293-297.2001
Ventura, M., Canchaya, C., Bernini, V., Altermann, E., Barrangou, R., McGrath, S., et al. (2006). Comparative genomics and transcriptional analysis of prophages identified in the genomes of Lactobacillus gasseri, Lactobacillus salivarius, and Lactobacillus casei. Appl. Environ. Microbiol. 72, 3130–3146. doi: 10.1128/AEM.72.5.3130-3146.2006
Wang, B., Brand-Miller, J., McVeagh, P., and Petocz, P. (2001). Concentration and distribution of sialic acid in human milk and infant formulas. Am. J. Clin. Nutr. 74, 510–515. doi: 10.1093/ajcn/74.4.510
Wang, M., Cao, H., Wang, Q., Xu, T., Guo, X., and Liu, B. (2018). The roles of two type VI secretion systems in Cronobacter sakazakii ATCC 12868. Front. Microbiol. 9:2499. doi: 10.3389/fmicb.2018.02499
Wood, T. K. (2013). Precedence for the structural role of flagella in biofilms. mBio 4:e225-13. doi: 10.1128/mBio.00225-13
Xu, X., Li, C., Wu, Q., Zhang, J., Huang, J., and Yang, G. (2015). Prevalence, molecular characterization, and antibiotic susceptibility of Cronobacter spp. in Chinese ready-to-eat foods. Int. J. Food Microbiol. 204, 17–23. doi: 10.1016/j.ijfoodmicro.2015.03.003
Yan, Q., Power, K. A., Cooney, S., Fox, E., Gopinath, G. R., Grim, C. J., et al. (2013). Complete genome sequence and phenotype microarray analysis of Cronobacter sakazakii SP291: a persistent isolate cultured from a powdered infant formula production facility. Front. Microbiol. 4:256. doi: 10.3389/fmicb.2013.00256
Ye, Y., Li, H., Wu, Q., Chen, M., Lu, Y., and Yan, C. (2014). Isolation and phenotypic characterization of Cronobacter from dried edible macrofungi samples. J. Food Sci. 79, M1382–M1386. doi: 10.1111/1750-3841.12513
Ye, Y., Ling, N., Gao, J., Zhang, X., Zhang, M., Tong, L., et al. (2018). Roles of outer membrane protein W (OmpW) on survival, morphology, and biofilm formation under NaCl stresses in Cronobacter sakazakii. J. Dairy Sci. 101, 3844–3850. doi: 10.3168/jds.2017-13791
Ye, Y. W., Ling, N., Jiao, R., Wu, Q. G., Han, Y. J., and Gao, J. N. (2015). Effects of culture conditions on the biofilm formation of Cronobacter sakazakii strains and distribution of genes involved in biofilm formation. LWT-Food Sci. Technol. 62, 1–6. doi: 10.1016/j.lwt.2015.01.035
Zhang, X., Gao, J., Ling, N., Zeng, H., Tong, L., Zhang, M., et al. (2019). Short communication: roles of outer membrane protein W on survival, cellular morphology, and biofilm formation of Cronobacter sakazakii in response to oxidative stress. J. Dairy Sci. 102, 2017–2021. doi: 10.3168/jds.2018-14643
Keywords: Cronobacter sakazakii, WGS, pathogenicity, biofilm formation, desiccation, multidrug-resistant
Citation: Aly MA, Domig KJ, Kneifel W and Reimhult E (2019) Whole Genome Sequencing-Based Comparison of Food Isolates of Cronobacter sakazakii. Front. Microbiol. 10:1464. doi: 10.3389/fmicb.2019.01464
Received: 28 February 2019; Accepted: 11 June 2019;
Published: 02 July 2019.
Edited by:
Julio Parra-Flores, University of the Bío Bío, ChileReviewed by:
Ben Davies Tall, United States Food and Drug Administration, United StatesCopyright © 2019 Aly, Domig, Kneifel and Reimhult. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohamed A. Aly, bW9oYW1lZC5hbHlAYm9rdS5hYy5hdA==; bW9oYW1lZC5hZGVsQGFnci5hc3UuZWR1LmVn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.