- 1Department of Biological Sciences, Indian Institute of Science Education and Research Bhopal, Bhopal, India
- 2School of Biotechnology, Institute of Science, Banaras Hindu University, Varanasi, India
Cyanobacteria are oxygenic photoautotrophs, exhibiting a cosmopolitan distribution in almost all possible environments and are significantly responsible for half of the global net primary productivity. They are well adapted to the diverse environments including harsh conditions by evolving a range of fascinating repertoires of unique biomolecules and secondary metabolites to support their growth and survival. These phototrophs are proved as excellent models for unraveling the mysteries of basic biochemical and physiological processes taking place in higher plants. Several known species of cyanobacteria have tremendous biotechnological applications in diverse fields such as biofuels, biopolymers, secondary metabolites and much more. Due to their potential biotechnological and commercial applications in various fields, there is an imperative need to engineer robust cyanobacteria in such a way that they can tolerate and acclimatize to ever-changing environmental conditions. Adaptations to stress are mainly governed by a precise gene regulation pathways resulting in the expression of novel protein/enzymes and metabolites. Despite the demand, till date few proteins/enzymes have been identified which play a potential role in improving tolerance against abiotic stresses. Therefore, it is utmost important to study environmental stress responses related to post-genomic investigations, including proteomic changes employing advanced proteomics, synthetic and structural biology workflows. In this respect, the study of stress proteomics offers exclusive advantages to scientists working on these aspects. Advancements on these fields could be helpful in dissecting, characterization and manipulation of physiological and metabolic systems of cyanobacteria to understand the stress induced proteomic responses. Till date, it remains ambiguous how cyanobacteria perceive changes in the ambient environment that lead to the stress-induced proteins thus metabolic deregulation. This review briefly describes the current major findings in the fields of proteome research on the cyanobacteria under various abiotic stresses. These findings may improve and advance the information on the role of different class of proteins associated with the mechanism(s) of stress mitigation in cyanobacteria under harsh environmental conditions.
Introduction
Cyanobacteria are the first oxygen evolving organisms which have analogous photosynthesis machinery to higher plants and encompass the ability to fix atmospheric carbon dioxide (CO2) and produce oxygen (O2). The aerobic life on the planet came into existence after the evolution of cyanobacteria. They flourished during the period from 2.8 to 3.5 billion years ago, provided Earth with oxygen and made possible the development of diverse forms of lives (Castenholz, 1996; Fischer, 2008). Cyanobacteria universally inhabit in almost all ecosystems including the Arctic and Antarctic region (Stanier and Bazine, 1977). They also occur in extreme environments, such as high salinity, pH and light irradiances (Castenholz, 1996). Cyanobacteria significantly contribute to the biomass production on the Earth by playing a potential role in the important biogeochemical cycles (e.g., nitrogen, carbon, and oxygen) (Häder et al., 2007). Presence of metalloenzymes known as nitrogenases makes them an excellent natural nitrogen fixer, and therefore they are utilized as biofertilizers in paddy and other nitrogen deficient crop fields (Vaishampayan et al., 2001; Chatterjee et al., 2017). They are massively exploited in biotechnological and pharmaceutical fields because of a unique repository of useful bioactive molecules (Rastogi and Sinha, 2009). They are an excellent source of renewable energy and bio-hydrogen, also genetically engineered for the production of bio-hydrogen and bio-ethanol which can be used as a substitute for conventional sources of energy, as it is clean, safe and environmental friendly (Khetkorn et al., 2017). Many other species are the rich resource of vitamins and minerals therefore used as food and feed supplements (MišurCoVá et al., 2010).
It is believed that the plastids of today’s photosynthetic eukaryotes including plants are the product of an endosymbiotic event between a eukaryotic host and a cyanobacterium (de Vries and Archibald, 2017). This hypothesis proved cyanobacteria as a useful model to study the stress responses, which can be directly or indirectly correlated with higher eukaryotic photosynthetic cells including plant systems (Burnap, 2015). However, there are plant processes such as organ level organization, where cyanobacterial models are not useful. Several molecular approaches are used to investigate the physiology and metabolism of a stressed cell at the DNA, RNA, and protein levels but it is not validated that how transcript and protein expressions link with the specific cellular networks (Gao et al., 2009). Furthermore, it was reported that experimental outcomes of expression levels of mRNA and proteins are contrasting (Zhang et al., 2009). It has been elucidated that the de-regulation in gene expression is not a direct approach to elucidate and understand the precise molecular machinery associated with the different stress-induced responses (Hongsthong et al., 2009; Kurdrid et al., 2011). Proteomics offers advancement to understand the cellular functions because it can crosslink the actual functions of genes, and its translational products and can reflect the cellular protein profile under defined stress conditions (Li et al., 2011). Proteomics can explore the structure and function of proteins by functioning as a connecting link between the transcriptomic and metabolomic profile, thus dissecting the actual physiological and metabolic state of the cells. Furthermore, in a stressful condition, long-term cellular adaptation is governed by the synthesis of “adaptation proteins.” These adaptation proteins are investigated to delineate the mechanism of cellular adaptation under the stress conditions. Identification of deregulated proteins using high throughput proteomics tools (gel based and gel free labeled based approaches) has provided new insights in the field of stress proteomics research. Identification and characterization of deregulated proteins provide necessary information regarding the cellular response of cyanobacteria to a defined stress condition at the level of their functions and therefore advance our understanding toward the cell signaling and stress response pathways that are activated under stress conditions. Several studies on proteomics have been successfully conducted on different cyanobacterial species under various abiotic stresses (Table 1). Literature survey suggests that a huge array of reviews on the physiology of cyanobacteria under stress conditions are available (Castielli et al., 2009; Latifi et al., 2009) but articles specifically dealing with the protein deregulation under abiotic stresses are limited in number. In this review, we made an attempt to describe several stress-responsive proteins differentially expressed in different cyanobacteria under various stress conditions.
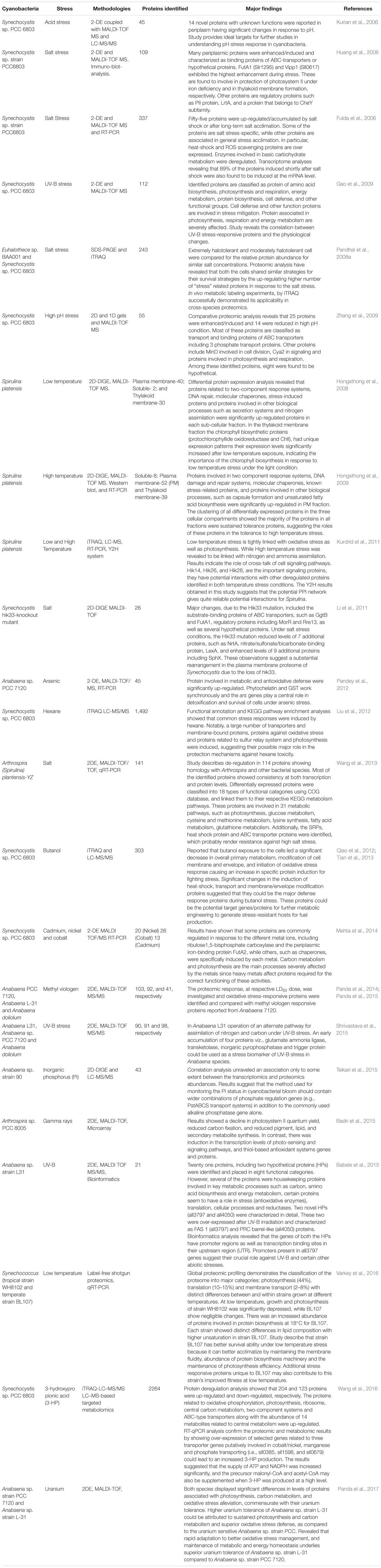
Table 1. A summary of articles published dealing with cyanobacterial proteome analyses in response to various abiotic stresses.
Adaptation Mechanism in Cyanobacteria Under Abiotic Stresses
Stress can be defined as an external factor, which exerts an internal damaging impact on the living organisms. It can also be defined as a significant variation of the most favorable condition of life (Latifi et al., 2009). The stress concept is described according to physiological and ecological requirements of an organism throughout its life-cycle. During the past several years, immense progress in industrialization and anthropogenic activities have resulted in an increase in different types of pollutants which changes the actual environment of the organisms (Bais et al., 2015). At present, abiotic stresses are one of the main concerns imposing a global challenge among us. Cyanobacteria are continuously exposed to various types of abiotic stresses such as solar ultraviolet radiation (UVR), variations in the external environment including light intensity, temperature (high and low), salinity, pH (acidic and basic), heavy metals, drought, and chemical fertilizers in their natural habitat. The growing numbers of cyanobacterial genome sequencing projects of newly identified species from diverse sources have significantly contributed to the modernization of research on cyanobacteria. In addition to conventional biology, advanced molecular biology, computational biology, and system biology methods/techniques have also contributed significantly in co-motivation in the development of high throughput methods to answer these biological questions globally. These technical advancements were made to address various key “-omic” aspects (e.g., genomics, transcriptomics, proteomics, metabolomics, and epigenomics) in accordance with the central dogma of the cellular biology. DNA microarrays are used for transcriptome profiling of organisms under a defined condition, it has been approved as a modern approach to study the deregulation in gene expression at system-level (Hernández-Prieto et al., 2014). Aided with, high throughput proteomic approaches have offered insightful advancements for the study of the function, integration, and regulation of proteome with the recent breakthroughs in biotechnology (Ow and Wright, 2009; Chardonnet et al., 2014). Following this, researchers are utilizing a wide range of high-throughput proteomic techniques to explore one of the most significant loop-hole, i.e., protein redox chemistry and flux analysis. Redox proteomics and fluxomics constitute crucial information about photoautotrophic gene regulation by signaling processes and thus metabolic rearrangements (Ansong et al., 2014; Guo et al., 2014; Diamond et al., 2017).
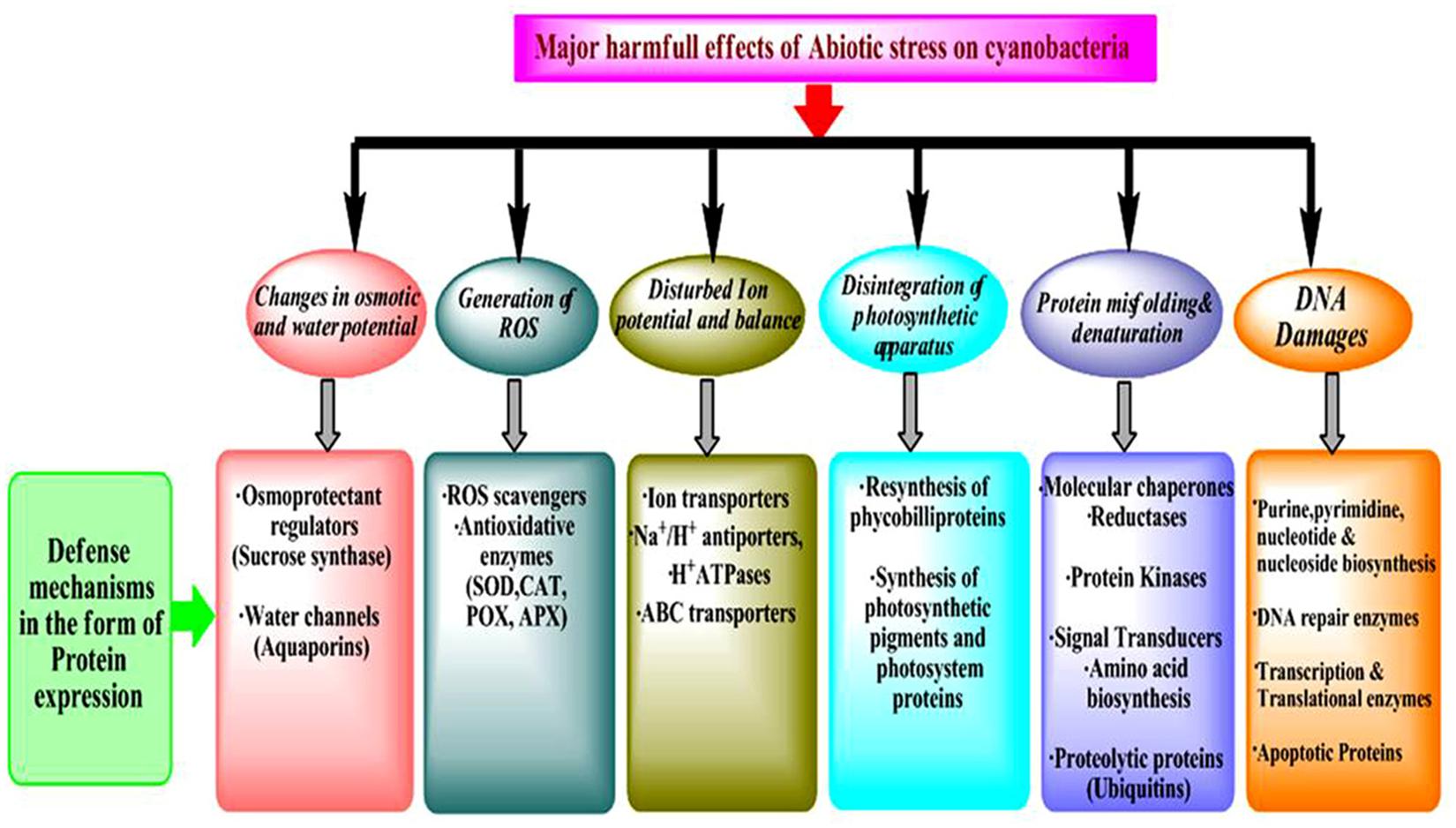
Accuracy and efficacy of protein biosynthesis are very crucial for life since a high degree of dependability of translation of the genetic information is essential to achieve the requirements of the cellular functions and also to preserve the variability developed by evolution. Tolerance to stress is regulated through profound de-regulation in gene expression which leads to changes in the downstream process of the central dogma. Therefore, the analysis of changes in proteome and metabolome is very important since they are direct effectors of stress responses (Burnap et al., 2015). Proteins/enzymes play essential roles inside the cell such as they catalyze various metabolic reactions, function as the components of transcription and translation machinery, and regulate stress response at metabolome levels (Hihara et al., 2001). Furthermore, proteins also have direct functions in the acclimation of stresses leading to changes in the cell physiology and metabolism (Figure 1). As discussed above, a change in protein abundance under stress is an effect of how the cells sense and respond to a particular stressor and most often determines the tolerance to stress. Therefore, investigation of post-translational modifications (PTMs) and identification of novel redox proteins as well as functional sites in the redox-regulated proteins are necessary (Foyer and Noctor, 2009). Elucidation and analysis of the redox regulation mechanisms and its effects on the role of protein along with the distinct pathways in response to environmental stresses will open new avenues in the stress biology of cyanobacteria. These studies considerably contribute to understanding the fundamental physiological mechanisms adopted by cyanobacteria to tolerate the stress responses (Paulsen and Carroll, 2013; Mo et al., 2015).
Post-Translational Modifications (PTMs) in Cyanobacteria
To adapt and survive in a variety of stressful and ever changing conditions, cyanobacteria have evolved intricate signal transduction machinery to sense changing environmental signals. The post-translational modifications (PTMs) systems have vital regulatory role in the signal transduction pathways of cyanobacteria. PTMs are defined as the covalent modifications of proteins. These modifications occur by a number of mechanisms such as protein splicing, and phosphorylation, glutathionylation, and acetylation of specific amino acids, which lead to changes in properties and functions of a protein (Hart and Ball, 2013; Olsen and Mann, 2013; Xiong et al., 2015a). A number of proteins undergo PTMs in response to various abiotic stress signals and are reported in a number of processes in cyanobacteria, such as, photosynthesis (Xu et al., 2001), nitrogen fixation (Gallon et al., 2000), and regulation of circadian rhythms (Axmann et al., 2014). The systematic investigation of PTMs could contribute to the comprehensive description of different types of proteins and to elucidate potential biological roles of each protein in cyanobacteria. Although the proteomic studies of PTMs carried out in cyanobacteria are limited in number, but recently these investigations are gaining much attention. PTMs data have provided clues to elucidate the complex signaling mechanisms that contribute to their evolutionary and ecological success (Xiong et al., 2015a). Among these modifications, phosphorylation of target proteins is well studied mechanism and is reported in cyanobacteria grown under stress conditions (Yang et al., 2013). A TiO2 enrichment and LC-MS/MS based global and site-specific phosphoproteomic analysis of 245 different proteins of Synechococcus sp. PCC 7002 identified 410 phosphorylation sites on 280 phosphopeptides. The characterized phosphoproteins were found to be involved in numerous cellular metabolic processes such as two-component signal transduction pathway and photosynthetic reactions (Yang et al., 2013). Recently, it was studied that Ser/Thr/Tyr kinases and phosphatases also play important role in regulating a number of processes such as photosynthesis and carbon, nitrogen metabolism in cyanobacteria (Spät et al., 2015). The regulatory protein PII (GlnB) involved in nitrogen metabolism, which senses changes in the internal N/C ratio, is the most widely studied and known phosphoprotein in cyanobacteria (Forchhammer, 2008). Mikkat et al. (2014) have utilized 2D gel electrophoresis and a phosphoprotein-specific dye to report the complete protein phosphorylation profile in Synechocystis 6803 (Mikkat et al., 2014). They observed that 32 proteins were phosphoproteins undergoing PTM when cultivated under high- and low-salt concentrations. Another study on direct shotgun membrane phosphoproteome in Synechocystis was studied using gel-based phosphoprotein staining and in-gel digestion leading to identification of 33 phosphoproteins, including 11 membrane bound proteins undergoing PTM (Lee et al., 2015). A global phosphoproteomic profiling of Synechocystis 6803 utilizing TiO2 enrichment of the phosphopeptides, followed by LC-MS/MS, revealed 367 phosphorylation sites on 190 proteins. The characterized proteins were involved in different cellular functions, including photosynthesis-related proteins proposing that phosphorylation of Ser, Thr, and Tyr residues might be involved in the metabolic reactions linked to photosynthesis (Angeleri et al., 2016). These studies show that phosphorylation in cyanobacteria is involved in many metabolic processes such as central carbon, nitrogen metabolism and photosynthesis. In cyanobacteria, stress induced ROS are scavenged by cellular antioxidant defense systems, which involve the redox homeostasis of cellular thiols such as glutathione, present in its reduced form (GSH). Studies have shown that Synechocystis 6803 possesses only one monothiol and two dithiol glutaredoxins (Grxs), which interact with different proteins resistant to toxic metals and other stresses (Sánchez-Riego et al., 2013). Chardonnet et al. (2014) utilized biotinylate doxidized glutathione (BioGSSG) and streptavidin-affinity chromatography in combination with nano LC-MS/MS to accomplish the first detailed study of in vitro glutathionylation in Synechocystis 6803. They identified potential glutathionylation sites on 125 proteins of different metabolic pathways. These studies revealed that glutathionylation is a central mechanism in cyanobacteria to shield themselves from the frequently encountered oxidative stresses caused by high light intensities or presence of toxic metal. PTMs by lysine acetylation are another important mechanism playing an important regulatory function in signal transduction (Kim and Yang, 2011). A recent universal acetylome analysis of 513 acetylated proteins on Synechocystis sp. identified 776 acetylation sites (Mo et al., 2015). These lysine acetylated proteins are functionally classified and most of them involved in cellular metabolism including the subunits of phycobiliproteins (phycocyanin and allophycocyanin) (Mo et al., 2015). Functional significance of lysine malonylation in Synechocystis sp. PCC 6803 was studied using affinity chromatography in combination with tandem mass spectrometry. This global analysis identified 598 lysine malonylation sites on 339 different types of proteins, among them 27 proteins were involved in the reactions of photosynthesis (Ma et al., 2017). Another global proteomic profiling of Synechococcus sp. identified 1653 acetylation sites on 802 acetylproteins involved in a broad range of biological processes including photosynthesis (Chen et al., 2017). These findings on the global acetylation and malonylation of lysine residues on a number of proteins provide new insight into the molecular mechanisms associated with the negative regulation of photosynthetic oxygen evolution in cyanobacteria. In-depth knowledge of these PTMs help in elucidating the physiological role of protein acetylation in regulation of photosynthesis. However, system-wide screening of multiple PTMs is a highly puzzling task, in circumstances where reversible PTMs are caused by a stimulus for a short period of time. Another hindrance in PTM studies is the absence of proficient enrichment strategies for the proteins which are undergoing PTM.
Tools and Techniques Adapted for Proteome Analysis in Cyanobacteria
Cyanobacterial proteomics till date has dealt with many technical challenges. Considerable technical advancements in the field of proteomic research have profoundly contributed to answering the complex biological questions in cyanobacteria. Gel-based proteomics has been very well adapted to analyze the proteomic alterations during growth and development, and for the investigation of proteomic responses to various abiotic and biotic insults. The most extensively used gel-based technique for protein identification is two-dimensional gel electrophoresis (2-DE), in which a large number of cellular proteins appear in the form of spots on the gel matrix and can be clearly visualized after proper protein staining. Thereafter, proteins separated on 2-D gels are preceded for digestion into smaller peptides using proteolytic enzymes and identified by mass spectrometry (MS). Although, the qualities of protein separation by gel-based techniques are debatable, but this separation strategy is extensively used with their own merits and demerits (Jorrín-Novo et al., 2015). Gel based techniques are frequently used because they are simple, reproducible, wide molecular weight coverage, and post-translational modifications detection (Mikkat et al., 2014). However, careful manual editing and optimizations of experimental conditions are required to achieve a high concentration of protein particularly for relative quantitative proteomics. Furthermore, protocols for protein extraction specific to a particular strain are also crucial considering the heterogeneity between species. Although several methods have been developed to enhance the quantity and quality of protein spots in a 2-D gel, these improvements are still not ample to describe the complete proteome profile. For example; isolation and purification of stable and high-quality proteins are one of the crucial parameters for the completion of quantitative proteomic experiments. In the case of cyanobacteria, it is the most challenging task because of their complex and robust nature (Pandhal et al., 2008a). Unlike other microorganisms, cyanobacteria possess a high amount of proteases and oxidative enzymes posing difficulty in extraction of stable proteins. An additional drawback is the abundance of phycobiliproteins and other pigments, which interfere with protein fractionation, thus hinder the effective extraction, purification, and recovery of low abundant protein posing problems in downstream analyses (Ow et al., 2011). Therefore, this technique is not appropriate for comprehensive high-throughput protein functional characterization (Qiao et al., 2012; Guo et al., 2014). Although some of these concerns have been optimized by addition of several steps in protein extraction as well as purification processes, but gel-based approaches are not capable to identify the low abundant proteins expressed exclusively in response to a particular stress (Parimi et al., 2015). Again several proteins undergo one or more post-translational modifications (PTMs), many of which are and could play critical roles in numerous cellular processes cannot be characterized by utilizing gel-based strategies due to its narrow range of pI (isoelectric point) coverage. To tackle these problems several gel-free proteomic techniques have been developed. For identification of proteins, gel-free approaches require protein digestion before chromatographic separation and mass spectrometry (MS). The number of proteins identified and quantified has primarily extended with the advancements of MS. These approaches have revealed to cover a broad range of molecular weight such as identification of proteins present in minute quantity and PTMs (Fournier et al., 2007). The better understanding on the PTMs is utmost important for the biological point of view because it can differentiate between the two identical polypeptides showing the difference in molecular mass due to a single chemical moiety [for, e.g., methyl, acetyl groups (Ghosh and Xu, 2014)]. Gel-free approaches require sophisticated instruments, but we are still facing lack of instruments that enable the assessment of each and every protein deregulated in a system as well as any PTMs at a defined set of conditions (Huang et al., 2006; Mikkat et al., 2014). Recently, gel-free isobaric tags for relative and absolute quantitation (iTRAQ) technique has grown to be more trendy and used frequently due to its better reproducibility as compared to 2-DE (Aggarwal et al., 2006). Global comparison and absolute quantification of complete protein profile due to the emergence of LC-MS based tagging approaches such as isobaric tags for relative and absolute quantitation (iTRAQ) (Stensjö et al., 2007), stable isotope labeling by amino acids in cell culture (SILAC) (Dephoure et al., 2013; Spät et al., 2015), and isotope-coded affinity tags (ICAT) (Sandh et al., 2014) have helped us to discover this field in more detail. The introduction of statistically vigorous label-free quantitative approach is also an important strategy used in quantitative proteomics research to analyze a huge number of samples (Wu et al., 2006). Therefore, the current and continuing developments in the area of mass spectrometry and structural proteomics are likely to provide better outcomes for revealing the biological processes. A typical workflow of proteomic analysis was given in Figure 2. Considerable genome sequences of many cyanobacterial strains can be accessed in the public databases but genome sequences of many important strains are still unavailable and a lot of improvements are required in this particular area. Unavailability of protein databases of unidentified species of cyanobacteria make the proteomic studies difficult but up to some extent, these hurdles are surpassed by comparing the protein sequences from the databases of model cyanobacteria (Pandhal et al., 2008b). Proteomics can successfully lead to the identification and characterization of stress-responsive proteins. Qualitative and quantitative alteration in the abundance could be associated with the physiological and metabolic rearrangements at the genotype level during stress tolerance. Different types of abiotic stresses might have changed proteomic responses. In the following section, we will describe how cyanobacteria respond against abiotic stresses mainly at the protein abundance levels and the de-regulation in proteins will be described as up- or down-regulated.
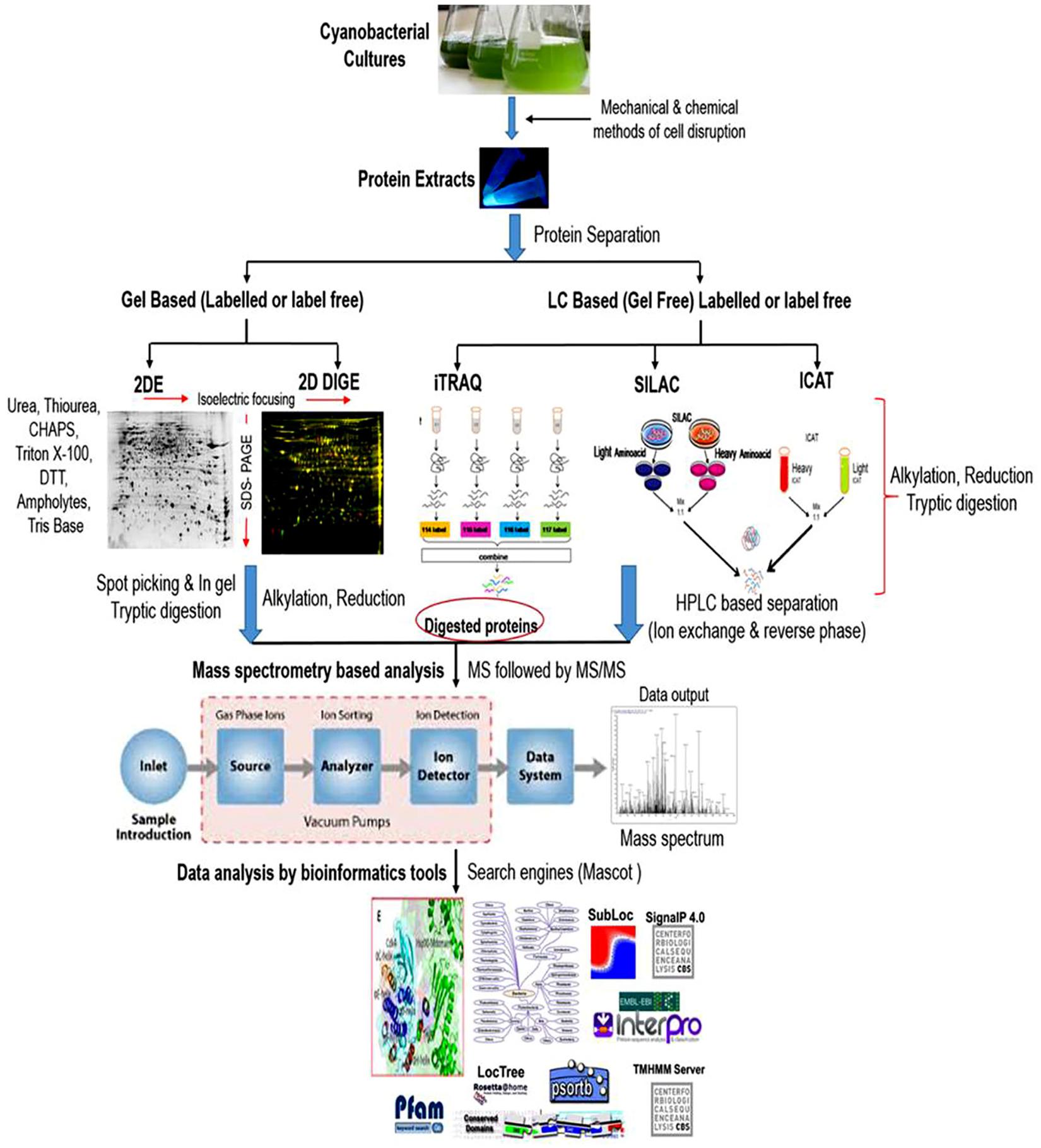
Figure 2. A typical workflow of comparative proteomic analysis in cyanobacteria. Proteins are extracted, cleaned and subjected to separation via gel (2-Dimensional gel electrophoresis) or non-gel (liquid chromatography) based approaches. Reduction, alkylation and trypsin digestion are performed before or after the separation step as per the requirement to convert protein mixtures into peptides. Separated peptides are analyzed through mass spectrometer (MS) followed by tandem MS (MS/MS) for the protein identification. The detected protein list is then used for data analysis using various bioinformatic tools.
Classification of Up-Regulated Proteins in Response to Abiotic Stress
Characterization of proteins that are up-regulated, when cyanobacteria are subjected to different abiotic stress is an essential part of the classification of stress-induced proteins and further includes the identification of the mechanisms involve against these responses. Based on previous studies on cyanobacteria, abiotic stress responsive up-regulated proteins are classified into six groups: (i) DNA repair/protection and transcription regulators (ii) Heat shock proteins (HSPs) and other stress-related proteins, (iii) cellular antioxidative enzymes (iv) proteins of lipid and other cellular metabolisms (v) Two-component system proteins and (vi) hypothetical proteins.
DNA Repair/Protection and Transcription Regulation
DNA repair and transcription regulation mechanisms universally work for all types of living organisms and have been studied comprehensively. Several studies describe the role of many proteins linked with transcriptional regulation and DNA repair mechanisms. Studies conducted on the cyanobacterium Anabaena sp. revealed a higher abundance of proteins having roles in the DNA protection, and transcription like nutrient deficient-induced DNA binding proteins (Babele et al., 2015; Panda et al., 2015; Shrivastava et al., 2015). DNA-binding proteins (Dps) from starved cells have crucial functions in the bacterial defense system against oxidative stress in different cyanobacteria (Moparthi et al., 2016). By binding to DNA, Dps efficiently prevents DNA base modification and strand cleavage while not interfering with normal DNA metabolism (Martinez and Kolter, 1997). These proteins provide the defense to cells during extreme environmental conditions including nutritional deprivation. The structure and function of Dps have been a concern of several studies and reported in several cyanobacteria that possess one or more Dps proteins. Their capability to provide comprehensive defense is mainly due to three integral features of the protein; these are DNA binding, sequestration of iron, and tolerance against reactive oxygen species (ROS). Having these properties, the Dps protein family members are critical in maintaining the tolerance to oxidative stress and iron homeostasis (Shcolnick et al., 2007). During starvation, Dps play important role in gene regulation, thereby rendering the cell more resistant to cytotoxicity by regulating the expression of essential stress resistance genes (Sato et al., 2012).
DNA ligases are found to be essential for the DNA repair system. These proteins may be up-regulated during quite harsh DNA damaging conditions due to abiotic stress. When the cyanobacteria cells come under the exposure of DNA damaging agents or replication blockers (e.g., UV irradiation and high temperature), expression of regulatory proteins such as SOS initiates (Domain et al., 2004). These proteins have been known to enhance the survival of the cells by enabling augmented DNA repair capability and also by inhibiting the division of cells (Shinagawa, 1996). Transcription machinery functions as a molecular motor that traverses through genome regularly. It has been recommended that the transcription machinery, for example, DNA-dependent RNA polymerase alpha subunit (all4191) might have a potential role in detecting DNA damage which ultimately leads to the activation of DNA repair and stress-responsive mechanisms (Ljungman, 2007).
A study conducted on Spirulina platensis under cold stress, showed that certain proteins participating in damage, repair and modification of DNA (polymerases, exonucleases, and methylases) were many folds up-regulated in the membrane portions following temperature downshift. It is expected that the induction of SbcC will occur when DNA damage takes place. Therefore, removal of abnormal DNA forms including hairpin loops formed as a result of DNA damage will be performed by SbcC exonuclease (Darmon et al., 2007;Higo et al., 2007). In comparison with cold stress, high-temperature conditions in Spirulina, leads to the drastic induction of exonuclease and endonuclease, thereby explicating an elevated role in the DNA repair pathway (Dwyer et al., 2007). Chromosome segregation ATPase also showed up-regulation and contribute significantly in replication, DNA repair, and genome stability. These outcomes prove the necessity of DNA replication, modification, and repair for the survival of cells exposed to extreme cold and heat stress conditions in Spirulina (Hongsthong et al., 2008, 2009). Proteomic profiling of Synechococcus under high light stress induces the production of several enzymes and transcriptional regulators involved in the process of replication, DNA repair and modification which include ParA, GvrA, and PhrA, NusB, SigD and SYNPCC7002_A2523 as major enzyme and transcriptional regulator (Xiong et al., 2015b). Label-free comparative proteomic profiling of tropical Synechococcus strain WH8102 with temperate strain BL107 suggested that AbrB-like transcriptional regulators were up-regulated in both strains at low temperature condition. Three and two transcriptional regulators were found to show up-regulation in BL107 and WH8102, respectively (Varkey et al., 2016). These regulatory proteins have a significant role in acclimation during low temperature condition. A study on Synechocystis PCC6803 has revealed the up-regulation of AbrB, a transcriptional regulator and it might have a significant role in the uptake of carbon and nitrogen, and may also contribute to the metabolic rearrangements during low temperature stress (Kaniya et al., 2013).
Heat Shock Proteins (HSPs) and Other Stress-Related Proteins
Molecular chaperones play significant roles in the conformational modifications (e.g., folding, misfolding, refolding, aggregation, or degradation), thus maintains the protein homeostasis and provides stress adaptability to cyanobacteria. Molecular chaperones fulfill an essential function in protein biogenesis and protein quality control in the cells. Most of the chaperones are soluble proteins, but some membrane-bound chaperones have also been recognized in cyanobacteria (Rajaram et al., 2014). Studies on Synechococcus sp. reported that DnaK chaperone, present on the thylakoid membrane, which help in protein translocation and translational machinery and play an prominent role in the enhancement of membrane fluidity by modulating membrane lipids under heat stress (Katano et al., 2006). Under salt stress (Fulda et al., 2006), acid stress (Kurian et al., 2006) and UV-B stress (Gao et al., 2009) in Synechocystis sp. molecular chaperones (GroEL, GroEs, 60 kD chaperonin 1, DnaK protein 2, and 60 kD chaperonin 1) are shown to be up-regulated. Study on Anabaena sp., showed up-regulation of chaperones and other stress-related proteins (GrpE, chaperonin GroEL, and DnaK type molecular chaperone) in response to UV-B stress (Shrivastava et al., 2015). Another study on Synechococcus sp. revealed that a number of small heat-shock proteins and many other proteases showed enhanced expression under the stress of low temperature. This observation may suggest that the protein turnover is because of either an increase in protein synthesis or induction in protein misfolding in response to low temperature (Varkey et al., 2016). The enzyme peptidyl-prolyl isomerase (a trigger factor) also known as a cold-shock protein in Synechocystis, acquaintances with the ribosome and assists in the proper folding of newly synthesized polypeptides by increasing the affinity of GroEL with unfolded polypeptides (Prakash et al., 2010). A study conducted on Spirulina sp. in response to heat shock described that glycosyltransferases were found to be up-regulated in all subcellular fractions, moreover, LysR, a membrane helicase, and ferredoxin glutamate synthase were also up-regulated in the fractions of thylakoid and plasma membrane (Hongsthong et al., 2009). Glycosyltransferase has an important role in osmo- and thermo-adaptation (Borges et al., 2004). These studies clearly demonstrate that heat shock proteins and other molecular chaperones support and care for other cellular proteins from their biogenesis, and also significantly contribute in the abolition of polypeptides that are no longer useful and may cause threat to cell viability.
Antioxidative and Cellular Defense Reaction Proteins
Abiotic stress-induced ROS can damage cellular components and act as a stress signaling molecule (Schmitt et al., 2014). ROS generation in cyanobacteria was regulated through complex pathways such as scavenging via antioxidative enzymes for, e.g., superoxide dismutases (SOD), oxidoreductase, catalases, peroxiredoxins, thioredoxins, and AhpC/TSA (Apel and Hirt, 2004; Schmitt et al., 2014). Because of the significant accumulation of H2O2, the H2O2 detoxifying enzymes such as AhpC, catalase, peroxiredoxin showed up-regulation in the proteome map of cyanobacterium, Anabaena (Babele et al., 2015). The overexpression of catalases and homologs of catalase, and oxidoreductase implicit resistance to the cell against oxidative stress which is also reported in case of desiccation stress in Anabaena sp. (Katoh et al., 2004). As evident from the fact that AhpC scavenges intracellular H2O2 at low concentration, whereas, catalase does it at high concentration (Seaver and Imlay, 2004), however, it is also reported that high level of intracellular H2O2 inhibits AhpC resulting in the down-regulation of AhpC on different days. Under these conditions, H2O2 detoxification was governed by the up-regulation of catalase and oxidoreductase when expression of AhpC is very low. Peroxiredoxin (Prx) performs multiple functions during stress conditions; (i) can reduce H2O2 and organic hydroperoxides (Dietz et al., 2006), (ii) acting as a molecular chaperone same as HSPs (Dietz, 2011), and (iii) activators of cell signaling. It is also been reported that the level of Prx was up-regulated in maize (Requejo and Tena, 2006) and Scytosiphon gracilis (Contreras et al., 2010) during the arsenic and copper stress. Therefore, Prx plays an important role in the protection of cyanobacteria from abiotic stress (Hosoya-Matsuda et al., 2005). Another important antioxidative enzyme, i.e., thioredoxin (Trx) known to effectively decrease the intramolecular disulfide bridges in different target proteins (Gelhaye et al., 2005). Trx helps in the maintenance of certain cellular processes like in the reduction of ribonucleotide, suppression of cell death and also provides reducing equivalents to the antioxidant systems (Hishiya et al., 2008; Pandey et al., 2012). A study proves that it regulates hydrophobic amino acids biosynthesis in Chlamydomonas reinhardtii (Lemaire and Miginiac-Maslow, 2004). Thus an up-regulation in Trx may be envisioned to manage oxidative stress in cyanobacteria. These are small ubiquitous proteins having highly conserved Cys–Gly–Pro–Cys active site and are reduced by thioredoxin reductase. They have the potential to reduce the intra- or intermolecular disulfide bridges of polypeptides and associated variety of redox reactions in which electrons are transferred from NADPH to Trx reductase and finally to Trxs. These redox-based reactions strictly control many enzymatic and regulatory activities and fulfill numerous cellular functions involved in cellular responses against oxidative stress. Trxs have been reported in all cyanobacterial genomes to show expression; however, it also expresses in many bacterial species (Zeller and Klug, 2006; Pascual et al., 2010). Trxs also closely associated with other redox partners including peroxiredoxins. They tightly regulate the type II peroxiredoxins, catalase-peroxidase KatG and 1 Cys-Prx in Synechocystis PCC 6803 (Pérez-Pérez et al., 2009). Moreover a number of small ubiquitous proteins known as glutaredoxins (Grx-s) are also present in cyanobacteria. These enzymes maintain the cytoplasmic thiol-redox state but in contrast to thioredoxins, no oxidoreductase reported which can specifically reduce Grx-s. Instead of this, their oxidation is carried out by substrates and non-enzymatic reduction by glutathione, known as the glutathione system (Holmgren, 1989). An evolutionary study on glutathione system suggests that the phototrophic microorganisms including cyanobacteria species show presence of high levels of glutathione during stress condition (Fahey et al., 1987). The primary role of glutathione is the protection of cell during ROS-induced toxicity, this notion was proved in Synechocystis PCC 6803 mutants with glutaredoxin knockdown of genes, these mutants showed enhanced sensitivity against peroxides (Latifi et al., 2009). The direct cellular targets of the glutathione system and the glutaredoxins homology between different cyanobacterial species are mostly unknown and more detailed information is required.
Proteins of Lipid Metabolisms and Transporters
Lipids are storage form of energy, they function as insulators, and are one of the most important structural components of the cell membranes with a significant role in cell signaling. They also play a vital role in stress tolerance against diverse environmental and physiological insults. Tight regulation of membrane fluidity is universal in all the living organisms for the proper functioning of biological membranes. Membrane fluidity is crucial for the mitigation and adaptation against an array of environmental stresses. Unsaturated fatty acids are an essential component in all types of biological membranes and the level of unsaturation in lipids is pertinent for controlling the membrane’s fluidity (Chi et al., 2008). Temperature (high and low) stress modulates membrane fluidity by affecting the saturation and/or unsaturation of fatty acids chains of cell membranes. Heat stress (high temperature) makes membranes more fluid by altering the hydrogen and electrostatic bonding within lipids and between polar groups of proteins, thereby transforming their architecture leading in ion leakage (Los and Murata, 1998; Morgan-Kiss et al., 2006). In contrast, cold stress (low temperature) makes membranes rigid and affect the normal functions of membrane proteins thereby inhibit solute transport of the cells by affecting the activity of H+/ATPase channel (Zhang et al., 2006). Polyunsaturation of membrane fatty acids is critical during the process of stress mitigation and acclimatization and help cyanobacteria to combat cold and heat stress (Singh et al., 2002). Polyunsaturation is catalyzed by the three types of enzymes known as fatty acid desaturases, among them acyl-lipid desaturases are one of important type (Gombos et al., 1992). These enzymes catalyze the incorporation of double bonds into the fatty acid hydrocarbon chains to generate the unsaturated and polyunsaturated fatty acids (Sato et al., 2004; Chi et al., 2008). They enhance the introduction of double bond in fatty acyl chains that have been esterified to a membrane glycerolipid (Sato et al., 2004). These enzymes are found on the surface of the thylakoid membranes of the cyanobacteria. The importance of desaturase enzymes and the expression of respective genes in cold stress acclimation have been well characterized for their significant roles in cyanobacteria (Wada and Murata, 1990; Gombos et al., 1992; Wada et al., 1994; Tasaka et al., 1996). It is believed that the fluidity of thylakoid membranes in cyanobacteria is controlled in such a manner that it maintain the replacement of damaged D1 protein with a newly synthesized protein by providing optimal condition (Inoue et al., 2001). Proteomic profiling of S. platensis culture grown under low temperature stress revealed up-regulation in (3R)-hydroxymyristoyl-[acyl-carrier-protein] -dehydratase or FabZ and an acyl carrier protein (ACP) in soluble and membranous fractions (Jeamton et al., 2008). The up-regulated FabZ protein is the first dehydratase which is a component of fatty acid elongation cycle leading to the biosyntheses of unsaturated fatty acids (Mohan et al., 1994; Heath and Rock, 1996). Furthermore, the substrate of FabZ, (3R)-hydroxymyristoyl acyl carrier protein, is located at a biosynthetic branch point which can also lead to biosynthesis of lipid. Another study on Spirulina plantensis-YZ documented that under salt-stress an important protein of lipid metabolism, i.e., 3-oxoacyl-[acyl-carrier protein] reductase, is up-regulated. This protein might help to promote the process of photosynthesis during salt stress by accelerating the activity of Na+/H+ transporter thus impede the expulsion of Na+ resulting in cell survival by preventing ion poisoning (Wang et al., 2013). Enhanced aggregation of UDP-sulfoquinovose synthase (SQD1), a protein associated with the biosynthesis of sulfolipids is also reported in two Anabaena species upon cadmium toxicity (Singh et al., 2015).
In the natural environment, cyanobacteria face toxicity due to heavy metals, salts, pesticides, and acids. These toxic compounds severely affect the physiology and metabolism by inducing the ion imbalance of cell. Proper ion homeostasis between intra-cellular and intra-thylakoid membrane is universal for growth and survival of a cell. To maintain the ion homeostasis transport proteins or transporters play crucial roles. These membranes bound transporters took part in the transport of ions, small or larger molecules, such as other protein, through the biological membrane. Transporters have been reported for roles against arsenic, copper, salinity, acid, heavy metal and ethanol stresses in different cyanobacteria (Zhang et al., 2015). Among these transporters, ATP binding cassette (ABC) transporters are most studied. They are associated with the transport of different types of substrates actively by using ATP hydrolysis and are located on the inner membrane (Davidson et al., 2008). Whole cell protein profile analysis of acid stress response in Synechocystis sp. PCC 6803 has described the role of many ion transporters. Up-regulation in the bicarbonate transporter, iron(III) dicitrate-binding protein of ABC transporter, probable extracellular solute-binding protein (slr1962), probable sodium/calcium exchanger protein (slr0681), molybdate-binding periplasmic protein were reported in the periplasmic fraction (Kurian et al., 2006). Investigation of the iron uptake protein FutA1 (Slr1295), a major iron-binding protein was found up-regulated in Synechocystis sp. PCC 6803 exposed to either high light or heat shock (Miranda et al., 2013). FutA1, is a part of ABC transporter concerned with the Fe3+ uptake by the cells. FutA1, along with its homologous FutA2 (Slr0513), play an essential role in the protection of PSII following iron depleting conditions. It was suggested that FutA1 is accountable for an effective supply of iron to the D1/D2 reaction center heterodimer and also essential for a rapid PSII repair during intense light exposure. ABC type transporters (phosphate transport) and ATP-binding protein (PstB2) were up-regulated in the three Anabaena species upon cadmium exposure (Singh et al., 2015). Synechococcus strain WH8102 displayed high expression of transport proteins in the periplasmic fraction under low-temperature stress. Increased expression of efflux transporters at low temperature could promote a defense system against accumulated toxic metabolites under stress condition. These proteins may also act as lipid transporters and modulate membrane fluidity by changing the lipid composition the at low temperature (Varkey et al., 2016). ABC transporter and cyanate ABC transporter ATP-binding-component was also significantly up-regulated in the plasma membrane and thylakoid membrane fraction of Spirulina sp. under low-temperature (Hongsthong et al., 2008) and in all the fractions under high temperature (Hongsthong et al., 2009) stress. Proteomic analysis of plasma membrane fraction of Synechocystis sp. under high pH stress describes that more than thirteen ABC type transporters were significantly up-regulated. These proteins are oligopeptide binding protein, ABC transporter subunit ycf24, ABC transporter permease protein, nitrate/nitrite-binding protein (NrtA), phosphate binding protein (PstS1), putative SbtB, phosphate transport ATP-binding protein (PstB1), and iron-binding protein (FutA1) and considered to be present in the periplasmic region of the plasma membrane. On the basis of these, it might be hypothesized that high pH stress cause nutrient deficiency, thus induce the expression of phosphate- and the ATP-binding proteins (Zhang et al., 2009). Proteomic analyses of Anabaena sp. strain 90 following inorganic phosphorus (Pi) stress also describe the role of several ABC transporter proteins. These proteins are significantly up-regulated upon Pi exposure (Teikari et al., 2015).
Two-Component System Proteins
The potential of living organisms to respond against a stress condition mainly depends on its ability to sense and transduce different external and internal signals (Los et al., 2010). The structures of both sensors and regulators are modular, and numerous variations in domain architecture and composition have evolved to adapt according to specific needs in signal perception and signal transduction. In this context, the two-component systems (TCSs) are crucial as these proteins are predominantly associated with signaling pathways and they are transducers of extracellular signals. They strictly regulate the motility, production of secondary metabolites, nutrient and mineral uptake, metabolic rearrangements, and cell division etc. in a variety of bacterial species. These systems also regulate cellular physiological processes in response to environmental stresses and provide adaptation (Mitrophanov and Groisman, 2008). Two-component hybrid sensor and regulatory proteins are involved in different functions such as peptidyl-histidine phosphorylation, phosphorylation, DNA-dependent regulation of transcription, signal transduction, protein histidine kinases activity, signal transducer activity, transferase activity, transfer of phosphorus-containing groups, G-protein coupled photoreceptor activity and ATP binding activity. TCSs family proteins have three main types, these are; two-component hybrid sensor and regulators, two-component sensor histidine kinases and serine/threonine kinases. A typical TCS comprises of a sensor kinase (e.g., histidine kinase; first component) that responds to a second component known as the response regulator. Sensor kinases are usually membrane-bound proteins, having N-terminal ligand-binding domain and a C-terminal kinase domain mainly, however, certain other domains may also present. Kinases function as molecular switches which can exist in either an inactive (‘OFF’) state or an active (‘ON’) state. Upon signals recognition, they can autophosphorylate at conserved histidine residue from ATP and then activate the response regulator by translocating the phosphoryl group to a conserved aspartate residue (Stock et al., 2000). Serine/threonine kinases proteins, along with their associated phosphatases, catalyze reversible phosphorylation of protein having fundamental roles in signal transduction. Kinases generally undergo autophosphorylation and precede to transphosphorylate the downstream substrates in the presence of any external stimuli. Therefore, activity of specific target proteins was controlled by the phosphorylation of specific amino acids in two ways; directly, by inducing the alteration in active sites by modulating their conformation and by indirect regulation via protein-protein interactions (Chao et al., 2010). The activated state of kinases is strongly controlled by different mechanisms, mainly by their subcellular localizations and the binding of allosteric effectors (Huse and Kuriyan, 2002). Activation (phosphorylation) of a response regulator modulates the biochemical properties of its output domain, which can induce the downstream transcription factors and regulate the gene and protein expression.
The endurance of cyanobacteria depends on its capability to promptly respond and adapt against constantly fluctuating harsh environmental conditions. Cyanobacterial chromosomes encode 1,171 potential histidine kinases, hybrid kinases and response regulators. Like most bacteria, cyanobacteria predominantly use TCSs proteins for signal recognition, to sense and respond and for cell–cell communication signals thus regulate physiology and gene expression in response to ever-changing external environment (Grebe and Stock, 1999; Stock et al., 2000; Wolanin et al., 2002; Ashby and Houmard, 2006). Studies conducted on cyanobacterium S. platensis in response to low and high-temperature stress showed differential expression of TCSs protein. Certain proteins present in two-component systems for signal transduction include histidine kinase, Ser/Thr protein kinase and several response regulator, i.e., Period protein–Aryl hydrocarbon receptor nuclear translocator protein-Single-minded protein; PAC, PAS C-terminal (PAC/PAS domains) were shown to be up-regulated in Spirulina during cold stress. Another up-regulated protein in the plasma membrane is forkhead-associated ATP-binding cassette transporter (Hongsthong et al., 2008). This transporter protein contains a forkhead-associated (FHA) domain which can bind to the Ser/Thr kinase and therefore, necessary for phospho-dependent signaling pathways (Curry et al., 2005) and linear signaling pathways (Grundner et al., 2005). Under high-temperature stress, two-component signal transduction proteins, i.e., Ser/Thr protein kinase, histidine kinase, and response regulator domain (Gly-Gly-Asp-Glu-Phe) were also found up-regulated in the both soluble and plasma membrane fraction (Hongsthong et al., 2009). Proteome analysis of Anabaena L31 upon cadmium toxicity also showed up-regulation of signal transduction proteins. It includes two component response regulators such as Ycf27 and Orr-A, ABC transporter family proteins and ATP-binding protein thus signifying a signaling mechanism under cadmium stress (Singh et al., 2015). Another histidine kinase, Hik33 (NblS) was found to be involved in perceiving high temperature and many other environmental insults induced signals in cyanobacteria. This protein shows higher accumulation in a temperate Synechococcus strain BL107 and a response regulator (SrrA) which interacts with Hik33 was induced in tropical strain WH8102 under low-temperature stress (Varkey et al., 2016). It was documented that srrA gene is induced transiently under high light and is involved in regulating the expression of phycobiliproteins in Synechocystis PCC7942 (López-Redondo et al., 2010).
Hypothetical Proteins
Apart from similarities in their amino acids composition whining and between other organisms, the functions of hypothetical proteins are generally not known. A number of hypothetical proteins showed differential abundance during abiotic stresses in different cyanobacteria. Proteome analysis of Synechocystis PCC 6803 showed accumulation of several hypothetical proteins such as sll1863, sll1762, sll0596, slr1485, sll1549, slr2144, slr1535, and slr0711 under salt stress (Fulda et al., 2006). Protein sll0595 has a role in signaling, slr0711 in folate biosynthesis and other protein have domains of unknown functions. Bhargava et al. (2008) have shown the role of alr0803 in Cu homeostasis in Anabaena doliolum. Therefore, alr0803 may act as a sensor of abiotic stress and help in maintaining the homeostasis. It was also observed that hypothetical proteins alr3199, alr0803, all1009, alr4050, all4894, were also over-expressed under desiccation stress thus create desiccation like environment inside the cells. The up-regulation of alr4050 (homolog of akinete marker) protein (Zhou and Wolk, 2002), alr3199 (homolog of HHE domain) and a cation binding protein has also been reported in UV-stressed Nostoc commune (Ehling-Schulz et al., 2002) and in Nostoc sp. strain HK-01 under dehydration like stress (Yoshimura et al., 2006). Expression of a fasciclin superfamily protein (all4894), which play important role in cell–cell adhesion and connection with the extracellular matrix, indicate that this might be involved in aggregation of cyanobacterial filaments to avoid stress exposure (Kim et al., 2000). Induction of all1009 was observed under metal stressed Anabaena cells. This protein has also shown similar induction under drought stress in Anabaena species (Yoshimura et al., 2007). Both hypothetical proteins (all4895 and all1009) are closely related with the outer membrane porin OprB (carbohydrate selective porin) of Pseudomonas aeruginosa and most abundantly present on the outer membrane of heterocysts (Merroun et al., 2005; Moslavac et al., 2007). It is believed that these proteins have a major role in metal sequestration and carbohydrate transport from vegetative cells to heterocyst (Moslavac et al., 2007). A sugar kinase/HSP70/actin superfamily (all5091) protein share a core structural folding this protein coupled with ATP hydrolysis causing conformational changes and thus prevent the protein misfolding by enhancing the affinity of HSP70 toward unfolded protein (Liberek et al., 1991). Another up-regulated protein is alr0893, its accumulation in Anabaena sp. PCC7120 under UV-B stress might be involved in nullifying the damaging effect of UV radiation with its combined ATP-independent repair activity of PfpI protease and ferritin domain (Shrivastava et al., 2015). Proteomic analysis of Anabaena L31 upon cadmium exposure showed up-regulation in a hypothetical protein alr0882, a homolog of universal stress protein (UspA) of A. variabilis ATCC 29413 and in alr0806, a homolog of high light inducible protein (HLIP) from Synechococcus sp. indicate roles of these in photoprotection under cadmium stress. Another up-regulated protein, i.e., alr3199, a homolog of hemerythrin/HHE cation-binding region of A. variabilis, exhibiting DNase activity and might have some role in DNA damage repair mechanism upon cadmium induced genotoxicity in Anabaena species (Singh et al., 2015). Proteomic analysis of Anabaena PCC7120 under salt and UV-B stress describe the up-regulation in alr0882, all5218, all3014, alr3904, and alr3090. Hypothetical proteins alr3904 and all3014 are predicted as glyoxalase II while alr3090, as catalase like, and all5218 as DNA gyrase activities. Hypothetical proteins alr3199 and alr4050 exhibited over accumulation in response to high salt stress in Anabaena PCC7120 (Rai et al., 2013).
Classification of Down-Regulated Protein in Response to Abiotic Stress
Based on several studies on abiotic stresses on cyanobacteria, down-regulated proteins are mainly classified into three families (i) Enzymes of amino acid, fatty acid and cofactor biosynthesis (ii) Proteins of energy metabolism (photosynthesis, respiration, and carbon/nitrogen fixation) and (iii) Enzymes of protein biosynthesis, and transport.
Enzymes of Amino Acid, Fatty Acid, and Cofactor Biosynthesis
Photoautotrophic growth of cyanobacteria requires a highly coordinated and regulated allocation of cellular macromolecules to diverse metabolic processes. These metabolic processes include the de novo synthesis of proteins, lipids, and other cellular macromolecules including cofactors. For unicellular organisms, the optimal allocation of limited resources is a key determinant of evolutionary fitness. Amino acids and fatty acids are the most essential components in all living organisms because they are building blocks of proteins and lipids. Enzymes primarily involved in amino acids, lipid and cofactor biosynthetic pathways are significantly down-regulated upon a variety of abiotic stresses. It was reported that the enzymes cysteine synthase (all2521) and D-3-phosphoglycerate dehydrogenase (alr1890) are drastically down-regulated under UV-B stress, which may negatively influence the amino acid biosynthesis pathway. In our study, we found down-regulation of D-3-phosphoglycerate dehydrogenase in Anabaena L31 under UV-B stress (Babele et al., 2015). Enzymatic activity of D-3-phosphoglycerate dehydrogenase catalyze the conversion of 3-phospho-D-glycerate into 3-phosphohydroxypyruvate which is the committed step in the phosphorylated L-serine biosynthesis pathway. It is also crucial for the biosynthesis of cysteine and glycine amino acids (Grant, 2012). Cysteine is one of the precursor amino acids of glutathione (GSH) biosynthesis, it is a reducing tripeptide thiol, required for maintaining balance in intracellular redox potential and also support in xenobiotic detoxification (Pickering et al., 2000). Proteins involved in amino acid metabolism (3-phosphoshikimate 1-carboxyvinyl-transferase) cofactor biosynthesis (thiamine biosynthesis protein; thiC), fatty acid and phospholipid metabolism (CTP synthetase), and cell envelope proteins (dTDP-glucose 4-6-dehydratase, glucose-1-phosphate thymidylyltransferase) are down-regulated in Synechocystis sp. under sustained UV-B irradiation (Gao et al., 2009). A study conducted on Spirulina platensis under cold shock revealed that macrolide-type polyketide synthase (fragment) and curamycin polyketide synthase acyl carrier protein (ACP) involved in lipid metabolism are many folds down-regulated. Another enzyme of lipid metabolism, i.e., Δ9-desaturase was down-regulated during high-temperature stress in the thylakoid and plasma membrane fraction of Spirulina (Hongsthong et al., 2009). Label-free shotgun proteomic analysis of marine Synechococcuss sp. strain WH8102 showed the down-regulation in the protein involved in coenzyme metabolism (Varkey et al., 2016).
Proteins of Energy Metabolism (Photosynthesis, Respiration, and Carbon/Nitrogen Fixation)
Cyanobacteria are the only photosynthetic prokaryotes, have the capability to convert inorganic carbon into useful carbohydrates at the expense of solar energy. This ability provides them the potentiality to produce bio-energy and other useful biochemical products (Gudmundsson and Nogales, 2015). Photosynthesis is a multistep process of successive redox reactions that comprises a series or proteins/enzymes and accessory pigments. In cyanobacteria, photosynthetic apparatus present in thylakoid membrane usually comprises a chain of membrane-bound multi-subunit protein complexes, consisting of photosystem I (PSI), photosystem II (PSII), cytochrome (Cyt) b6f, and ATP synthase (ATPase) complexes and many small (accessory pigments; plastocyanins, plastoquinones etc.) electron transport molecules (Casella et al., 2017). The thylakoid membranes also harbor the protein machinery of respiratory electron transport chains (ETC) (Mullineaux, 2014). Several studies were performed to describe the harmful effects of abiotic stresses on the photosynthesis and central carbon metabolism (CCM). Different types of oxidative stresses promote over accumulation of ROS thus lead to irreversible damage to photosystems (PS1, PSII) (Latifi et al., 2009). In cyanobacteria, phycobiliproteins are the most abundant accessory photosynthetic pigments of PSII (approximately 40% of total proteins). They are constituted of non-covalently associated subunits of allophycocyanin, phycocyanin, and phycoerythrin and play an important role in light harvesting and photolysis of water (Bryant and Canniffe, 2018). In our study, we found that UV-B stress can severely damage allophycocyanin beta subunit, phycocyanin A subunit, phycoerythrocyanin alpha chain and down-regulate the activity of F0F1 ATP synthase (beta subunit) in Anabaena L31 cells (Babele et al., 2015). A comparative study on three Anabaena species in response to methyl viologen (Panda et al., 2015), UV-B radiation exposure (Shrivastava et al., 2015) and cadmium toxicity (Singh et al., 2015) show reduced levels of most common PSII (ApcA, CpcA, CpcG4, CpcB, and PecC) proteins and ATP synthase alpha subunit. There was many folds down-regulation in phycocyanin α chain, phycoerythrocyanin α chain and phycobilisome rod core linker proteins were observed. Label-free shotgun proteomic analysis of marine Synechococcuss sp. strain WH8102 describes that low temperature has a significant impact on the abundance of photosystems and phycobiliproteins (Varkey et al., 2016). D1 protein (a core subunit of PSII), cytochrome c-550 (PsbV), manganese stabilizing protein of PSII (PsbO), PSI assembly proteins (Ycf3) and ROD linker polypeptides were down-regulated. Significant down-regulation in the proteins of PSI and the levels of phycobiliproteins were observed in Synechococcus sp. exposed to high light stress (Xiong et al., 2015b). Proteomic profiling revealed that under inorganic phosphorus stress, the proteins of PSI and II, ETC and photosynthetic pigments were reduced in Anabaena sp. strain 90 (Teikari et al., 2015). This could be due to the ROS mediated bleaching and over-reduction of PSs under energetic UV-B radiation, high light and low temperature thus affecting the photosynthetic reaction resulting in overall reduction in generation of reduced NADPH and ATP.
The limited or diminished supply of ATP and NADPH2 may also inhibit the CO2-fixing ability. CO2 fixation by CCM is the most important pathway for the synthesis of these bio-products. CCM include the Calvin-Benson cycle that is closely associated with photorespiratory 2-phosphoglycolate (2PG) metabolism, glycolysis, oxidative pentose phosphate (OxPP) and TCA cycle. Excess organic carbon (i.e., glucose) that is not instantly used for growth and metabolism can be stored as glycogen (storage form of organic carbon). Abiotic stresses severely affect these CCM eventually halt the growth and metabolism of cyanobacteria. Several proteins associated with CCM and thus involved in fulfilling the energy demand of the cells are the primary target of the abiotic stress. We found that the expression of phosphoribulokinase was down-regulated under UV-B stress (Babele et al., 2015). This enzyme catalyzes the conversion of ribulose-5-phosphate into ribulose-1, 5-phosphate during the process of CO2 assimilation. Many energy metabolic enzymes, i.e., aminomethyltransferase, carbamoyl-phosphate synthase large subunit, GDP-mannose pyrophosphorylase, phosphoglycerate mutase, and OxPP Cycle gene were down-regulated upon prolonged UV-B exposure to Synechocystis sp. (Gao et al., 2009). Glucokinase and glyceraldehydes 3-phosphate dehydrogenase-2 showed reduced levels in methyl viologen treated Anabaena PCC7120 cells (Panda et al., 2014). Enzymes of Calvin cycle [ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCO) large subunit, and phosphoribulokinase], pentose phosphate pathway (6-phosphogluconate dehydrogenase, transketolase and fructose bisphosphate aldolase) and glycolysis (glucose-6-phosphate isomerase, glyceraldehyde 3-phosphate dehydrogenase, fructose-1,6-bisphosphate aldolase, and phosphoglycerate kinase) were depressed in three Anabaena species exposed to cadmium (Singh et al., 2015). It has been demonstrated that RuBisCO might show structural modifications this could be the results of photodegradation, fragmentation and denaturation, active site modifications and solubility of membrane proteins (Feller et al., 2008). Down-regulation in these enzymes suggest noticeable inhibition of photosynthetic dark reaction in Anabaena species and their inability to generate ATP, which is used for maintaining metabolic processes under stressful condition. Furthermore, inhibition in the activity of Kreb’s cycle enzymes in all three studies Anabaena species not only lowers the generation of ATP and NADPH but also reduce the production of many other metabolic precursors of different metabolic pathways. It was also reported that under the stress of inorganic phosphorus there was a reduction in the protein level of large and small subunit of RuBisCO in Anabaena sp. strain 90 (Teikari et al., 2015). Transketolase connects the pentose phosphate pathway to glycolytic intermediates (Schenk et al., 1998; Jung et al., 2004). Its down-regulation might be due to its sensitivity to accumulated ROS in the cells and competitive inhibition of TK catalyzed reactions (Kruger and von Schaewen, 2003; Singh et al., 2015). In the Calvin cycle FBA II reversibly catalyze the conversion of fructose 1, 6-bis phosphate (FBP) into glyceraldehydes 3-phosphate (GAP) and dihydroxy acetone 3-phosphate (DHAP) (Nakahara et al., 2003). During stress, GAP and FBP might be converted into glucose 6-phosphate for re-entry into the PPP for NADPH synthesis and FBA also causes RuBP regeneration (Fridlyand and Scheibe, 1999). Another Calvin cycle enzyme, i.e., fructose 1,6-bisphosphatase (FBPase) increases the production of fructose 6-phosphate and glucose by catalyzing the conversion of fructose-1,6-bisphosphate (Yan and Xu, 2008). Thus down-regulation in the activity of FBPase indicates the decline in carbon fixation, NADPH/NADH production and therefore the loss in cell growth (Gao et al., 2009). The loss in the activity of all these enzymes were reported in different Anabaena sp. under temperature (Mishra et al., 2009) and UV radiation stress (Shrivastava et al., 2015). Subcellular proteomic profiling of Spirulina sp. under low-temperature stress indicate that the expression levels of phosphoenolpyruvate synthase and phosphoenolpyruvate carboxylase were repressed (Hongsthong et al., 2008). The enzyme phosphoenolpyruvate synthase catalyzes the essential step in Calvin cycle and converts pyruvate into phosphoenolpyruvate (PEP). Gluconeogenesis occurs in the cells when pyruvate or lactate is used as a carbon source. PEP can also be utilized as an energy source for the phosphotransferase system of sugar uptake. Phosphoenolpyruvate carboxylase plays an antipleurotic role in bacteria; it supplies oxaloacetate to the TCA cycle, which requires an uninterrupted supply of C4 molecules, so as to fulfill the demand of intermediate molecules which are consumed in amino acid biosynthesis.
Defects in the proteins/enzymes of carbon metabolism also negatively affect the protein machinery of nitrogen metabolism. Comparative proteomic analyses of three Anabaena species under cadmium toxicity indicate defects in nitrogen metabolism. A key enzyme, i.e., glutamine synthetase (GlnA) of nitrogen assimilation which catalyzes the production of glutamine from glutamate and ammonia is severely down-regulated across three Anabaena species. Another protein involved in reassembly of intracellular membranes during heterocyst differentiation and nitrogen fixation, i.e., putative heterocyst to vegetative cell connection (FraH) showed a decline in all three Anabaena species upon cadmium exposure (Singh et al., 2015). Enzymes of nitrogen metabolism such as nitrogenase, nitrate, and nitrite reductases are severely affected by UV radiations and other abiotic stresses. Nitrogenase has been found to be extremely sensitive to UV-B radiation (Kumar et al., 2003).
Enzymes of RNA and Protein Biosynthesis and Transport
One of the important classes of down-regulated proteins is involved in the transcription and translation process. Defects in the CCM leads to drastic down-regulation of many proteins/enzymes of transcription and translation due to the limited source of energy. Many transcriptional proteins such as thymidine kinase, oligoribonuclease, exodeoxyribonuclease VII small subunit, and phosphoribosylformylglycinamidine synthase I (FGAM synthase) and translational protein, i.e., ATP-dependent Clp protease proteolytic subunit 2 down-regulated many folds in S. platensis upon cold stress (Jeamton et al., 2008). A transcriptional regulator (abrb family) protein is also down-regulated in S. plantensis-YZ under salt-stress conditions. Proteomic analysis of Synechocystis sp. following prolonged UV-B irradiation showed down-regulation in transcriptional (ribonuclease D) and translational (methionine sulfoxide reductase A) proteins. Proteome analysis of three Anabaena species describe that ribosomal proteins and other translational proteins such as EF-Tu, EF-Ts, EF-G, RPS-1, and Pmb-A were significantly down-regulated upon cadmium exposure in all the Anabaena species. These proteins are associated in proper ribosome assembly, post-transcriptional processes and translation indicating cadmium mediated obstruction in protein biosynthesis by inducing degradation and alteration in the tertiary structure of proteins (Singh et al., 2015). A translational protein slr1463, an elongation factor EF-G is down-regulated in Synechocystis PCC 6803 under high pH stress (Zhang et al., 2009). Label-free shotgun proteomic analysis of marine Synechococcus sp. strain WH8102 describes the down-regulation in several translational, ribosomal biogenesis and transcriptional proteins under low-temperature stress (Varkey et al., 2016). Among these proteins, many ribosomal proteins, RNA binding proteins, translation initiation factors, tRNA synthetases and GTP binding proteins are many folds down-regulated. Proteins such as transcription elongation factor NusA, DNA directed RNA polymerase beta subunit and RNA polymerase sigma factor (type 1) were also down-regulated under low temperature stress. Some of the protein with similar names and functions are also found to be down-regulated in Synechocystis sp. under nickel, cobalt and cadmium toxicity (Mehta et al., 2014). Under the long-term UV-B stress in Synechocystis sp., the enzymes, i.e., three isoforms of Msr-A, aminopeptidase P, and 30S-RP that are associated in translation were down-regulated (Gao et al., 2009). Msr has two important functions, first, it repairs denatured proteins and maintains the proper functioning of important housekeeping proteins (Ezraty et al., 2004). Second, it helps in the reduction of methionine residues rich proteins thus facilitates these proteins to act as ROS quenchers or sinks by a progressive redox cycle on the surface of methionines which is very important under stressful condition (Stadtman et al., 2003).
Concluding Remarks and Future Perspectives
The present review deals with the effect of different abiotic stresses on the cyanobacterial proteome. Efficient protein extraction protocols and advanced proteomic tools/techniques for identification of novel/hypothetical proteins have been standardized in different strains of cyanobacteria for better knowledge on the molecular mechanism(s) of abiotic stress sensing and intracellular signal transduction in response to abiotic stresses. Interestingly, 2-DE/MALDI-TOF MS and gel free-LC-MS/MS systems have been exploited for the identification of novel proteins expressed under stress conditions. One of the exciting areas will be investigating the roles of hypothetical proteins up-/down-regulated during stress situations. Their functional characterization will illuminate new mechanisms involved in complex mitigation process. Till date, it is not well known that how cyanobacteria perceive changes in stress that lead to the stress-induced expression of various proteins. Another important question is to how the expression of the genes for stress sensors and transducers occurs (Murata and Suzuki, 2006). Research has mainly focussed on photosynthetic ETS chain as the major source of ROS production, but not much is known about the contribution of the respiratory chain in ROS production. Inside a photosynthetic cell, many types of ROS are produced simultaneously, therefore it is complicated to determine the specific targets of ROS. In future, research should be focussed toward the identification and characterization of the physiological/molecular responses they provoke and specific targets of each ROS. Research should also focus on the characterization of truly specific sensory genes and proteins up and/or down-regulated upon a particular type of abiotic stress. In order to identify and characterize possible sensor protein and their regulatory molecules, systematic analysis of gene’s promoter structure and its potential sigma and transcriptional factors proteins and the role of small, non-coding RNAs (microRNAs) could be a good point to initiate.
Redox changes taking place inside the photosynthetic organism can affect the effectiveness of biofuel production. Despite the utmost importance of cyanobacteria for future industrial setups, the knowledge of site-specific redox changes are insufficient, especially under light conditions, therefore, critical analysis of whole proteome is urgently required at quantitative and site-specific levels. These understanding will enlighten the specific growth conditions necessary for cyanobacteria to thrive in industrial settings. Cyanobacteria are termed as prominent microbial biofactories for producing biochemicals and biofuels harnessing solar energy and carbon dioxide. Little if any, is known about the specific cysteine sites in the organism, which act as switches, turning redox on and off, especially since light changes from day to night (Guo et al., 2014). With the help of cyanobacterial genomics, and different ‘-omics’ technologies, such as transcriptomics, proteomics, and metabolomics, have been used in cyanobacteria to study acclimation toward different environmental stresses. In this respect, a critical evaluation of the transcriptomic and proteomic data by advanced bioinformatic tools is utmost necessary. These technologies, along with the site-directed mutagenesis will generate the knockouts of the desired genes and allow us to screen genes and their coding proteins as potential sensory polypeptides. These advancements in the field of cyanobacterial research make them feasible to study different types of stress responses. More recently, cyanobacteria are receiving a lot of attention due to their immense application in the production of biofuel and bioremediation. Advanced knowledge of stress mitigation will be beneficial in these areas. It is apparent that in the near future environment will be more polluted and hence harsher than present environment. Therefore, the use of stress tolerant cyanobacteria will be beneficial industrially. We feel that the present review would provide new insights into the cyanobacterial stress responses (Figure 3), which are required for the development of stress-tolerant GEMS for the benefit of humankind. It will also help in studying de-regulation in protein abundance within the cyanobacteria cell in response to abiotic stress.
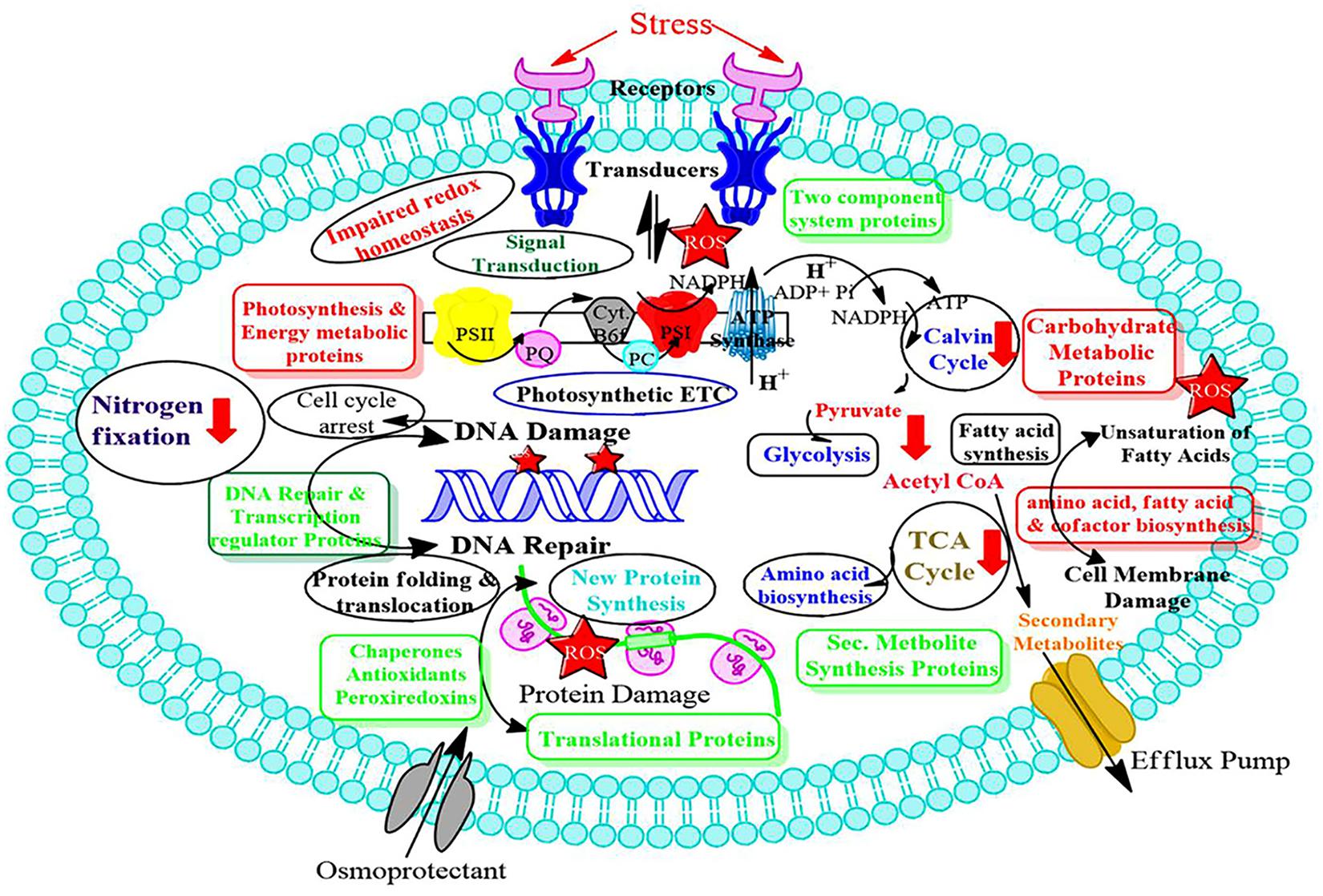
Figure 3. Scheme of abiotic stress responsive protein networks involved in cyanobacteria. Proteins shown in red boxes are down regulated while proteins shown in green boxes are upregulated. Metabolic processes having red arrows are affected by abiotic stresses.
Author Contributions
PB wrote the review manuscript. JK and VC corrected the manuscript. All authors approved and revised the final version of the manuscript.
Funding
This work was supported by a project grant sanctioned to PB in the form of National Postdoctoral fellowship by SERB-DST, New Delhi, India (File No. PDF/000200).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
JK is very much thankful to UGC, New Delhi, India for providing financial support in the form of junior and senior research fellowship. VC acknowledges his gratitude to UGC, New Delhi, India in the form of research fellowship.
References
Aggarwal, K., Choe, L. H., and Lee, K. H. (2006). Shotgun proteomics using the iTRAQ isobaric tags. Brief. Funct. Genomics Proteomics 5, 112–120. doi: 10.1093/bfgp/ell018
Angeleri, M., Muth-Pawlak, D., Aro, E.-M., and Battchikova, N. (2016). Study of O-phosphorylation sites in proteins involved in photosynthesis-related processes in Synechocystis sp. strain PCC 6803: application of the SRM approach. J. Proteome Res. 15, 4638–4652. doi: 10.1021/acs.jproteome.6b00732
Ansong, C., Sadler, N. C., Hill, E. A., Lewis, M. P., Zink, E. M., Smith, R. D., et al. (2014). Characterization of protein redox dynamics induced during light-to-dark transitions and nutrient limitation in Cyanobacteria. Front. Microbiol. 5:325. doi: 10.3389/fmicb.2014.00325
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Ashby, M. K., and Houmard, J. (2006). Cyanobacterial two-component proteins: structure, diversity, distribution, and evolution. Microbiol. Mol. Biol. Rev. 70, 472–509. doi: 10.1128/mmbr.00046-05
Axmann, I. M., Hertel, S., Wiegard, A., Dörrich, A. K., and Wilde, A. (2014). Diversity of KaiC-based timing systems in marine Cyanobacteria. Mar. Genomics 14, 3–16. doi: 10.1016/j.margen.2013.12.006
Babele, P. K., Singh, G., Kumar, A., and Tyagi, M. B. (2015). Induction and differential expression of certain novel proteins in Anabaena L31 under UV-B radiation stress. Front. Microbiol. 6:133.
Badri, H., Monsieurs, P., Coninx, I., Wattiez, R., and Leys, N. (2015). Molecular investigation of the radiation resistance of edible cyanobacterium Arthrospira sp. PCC 8005. Microbiologyopen 4, 187–207. doi: 10.1002/mbo3.229
Bais, A., McKenzie, R., Bernhard, G., Aucamp, P., Ilyas, M., Madronich, S., et al. (2015). Ozone depletion and climate change: impacts on UV radiation. Photochem. Photobiol. Sci. 14, 19–52.
Bhargava, P., Mishra, Y., Srivastava, A. K., Narayan, O. P., and Rai, L. C. (2008). Excess copper induces anoxygenic photosynthesis in Anabaena doliolum: a homology based proteomic assessment of its survival strategy. Photosynth. Res. 96, 61–74. doi: 10.1007/s11120-007-9285-7
Borges, N., Marugg, J. D., Empadinhas, N., da Costa, M. S., and Santos, H. (2004). Specialized roles of the two pathways for the synthesis of mannosylglycerate in osmoadaptation and thermoadaptation of Rhodothermus marinus. J. Biol. Chem. 279, 9892–9898. doi: 10.1074/jbc.m312186200
Bryant, D. A., and Canniffe, D. P. (2018). How nature designs light-harvesting antenna systems: design principles and functional realization in chlorophototrophic prokaryotes. J. Phys. B At. Mol. Opt. Phys. 51:033001. doi: 10.1088/1361-6455/aa9c3c
Burnap, R. L. (2015). Systems and photosystems: cellular limits of autotrophic productivity in cyanobacteria. Front. Bioeng. Biotechnol. 3:1.
Burnap, R. L., Hagemann, M., and Kaplan, A. (2015). Regulation of CO2 concentrating mechanism in cyanobacteria. Life 5, 348–371. doi: 10.3390/life5010348
Casella, S., Huang, F., Mason, D., Zhao, G.-Y., Johnson, G. N., Mullineaux, C. W., et al. (2017). Dissecting the native architecture and dynamics of cyanobacterial photosynthetic machinery. Mol. Plant 10, 1434–1448. doi: 10.1016/j.molp.2017.09.019
Castenholz, R. W. (1996). Endemism and biodiversity of thermophilic cyanobacteria. Nova Hedwig. Beih. 112, 33–48.
Castielli, O., De la Cerda, B., Navarro, J. A., Hervás, M., and De la Rosa, M. A. (2009). Proteomic analyses of the response of cyanobacteria to different stress conditions. FEBS Lett. 583, 1753–1758. doi: 10.1016/j.febslet.2009.03.069
Chao, J. D., Papavinasasundaram, K. G., Zheng, X., Chávez-Steenbock, A., Wang, X., Lee, G. Q., et al. (2010). Convergence of Ser/Thr and two-component signaling to coordinate expression of the dormancy regulon in Mycobacterium tuberculosis. J. Biol. Chem. 285, 29239–29246. doi: 10.1074/jbc.M110.132894
Chardonnet, S., Sakr, S., Cassier-Chauvat, C., Le Mareìchal, P., Chauvat, F., Lemaire, S. P. D., et al. (2014). First proteomic study of S-glutathionylation in cyanobacteria. J. Proteome Res. 14, 59–71. doi: 10.1021/pr500625a
Chatterjee, A., Singh, S., Agrawal, C., Yadav, S., Rai, R., and Rai, L., (eds). (2017). “Role of algae as a biofertilizer,” in Algal Green Chemistry (Amsterdam: Elsevier), 189–200. doi: 10.1016/b978-0-444-63784-0.00010-2
Chen, Z., Zhang, G., Yang, M., Li, T., Ge, F., and Zhao, J. (2017). Lysine acetylome analysis reveals photosystem II manganese-stabilizing protein acetylation is involved in negative regulation of oxygen evolution in model Cyanobacterium Synechococcus sp. PCC 7002. Mol. Cell. Proteomics 16, 1297–1311. doi: 10.1074/mcp.M117.067835
Chi, X., Yang, Q., Zhao, F., Qin, S., Yang, Y., Shen, J., et al. (2008). Comparative analysis of fatty acid desaturases in cyanobacterial genomes. Comp. Funct. Genomics 2008:284508.
Contreras, L., Moenne, A., Gaillard, F., Potin, P., and Correa, J. A. (2010). Proteomic analysis and identification of copper stress-regulated proteins in the marine alga Scytosiphon gracilis (Phaeophyceae). Aquat. Toxicol. 96, 85–89. doi: 10.1016/j.aquatox.2009.10.007
Curry, J. M., Whalan, R., Hunt, D. M., Gohil, K., Strom, M., Rickman, L., et al. (2005). An ABC transporter containing a forkhead-associated domain interacts with a serine-threonine protein kinase and is required for growth of Mycobacterium tuberculosis in mice. Infect. Immun. 73, 4471–4477. doi: 10.1128/iai.73.8.4471-4477.2005
Darmon, E., Lopez-Vernaza, M. A., Helness, A. C., Borking, A., Wilson, E., Thacker, Z., et al. (2007). SbcCD regulation and localization in Escherichia coli. J. Bacteriol. 189, 6686–6694. doi: 10.1128/jb.00489-07
Davidson, A. L., Dassa, E., Orelle, C., and Chen, J. (2008). Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72, 317–364. doi: 10.1128/MMBR.00031-07
de Vries, J., and Archibald, J. M. (2017). Endosymbiosis: did plastids evolve from a freshwater cyanobacterium? Curr. Biol. 27, R103–R105. doi: 10.1016/j.cub.2016.12.006
Dephoure, N., Gould, K. L., Gygi, S. P., and Kellogg, D. R. (2013). Mapping and analysis of phosphorylation sites: a quick guide for cell biologists. Mol. Biol. Cell 24, 535–542. doi: 10.1091/mbc.E12-09-0677
Diamond, S., Rubin, B. E., Shultzaberger, R. K., Chen, Y., Barber, C. D., and Golden, S. S. (2017). Redox crisis underlies conditional light-dark lethality in cyanobacterial mutants that lack the circadian regulator, RpaA. Proc. Natl. Acad. Sci. U.S.A. 114, E580–E589. doi: 10.1073/pnas.1613078114
Dietz, K.-J. (2011). Peroxiredoxins in plants and cyanobacteria. Antioxid. Redox Signal. 15, 1129–1159. doi: 10.1089/ars.2010.3657
Dietz, K.-J., Jacob, S., Oelze, M.-L., Laxa, M., Tognetti, V., de Miranda, S. M. N., et al. (2006). The function of peroxiredoxins in plant organelle redox metabolism. J. Exp. Bot. 57, 1697–1709. doi: 10.1093/jxb/erj160
Domain, F., Houot, L., Chauvat, F., and Cassier-Chauvat, C. (2004). Function and regulation of the cyanobacterial genes lexA, recA and ruvB: LexA is critical to the survival of cells facing inorganic carbon starvation. Mol. Microbiol. 53, 65–80. doi: 10.1111/j.1365-2958.2004.04100.x
Dwyer, D. J., Kohanski, M. A., Hayete, B., and Collins, J. J. (2007). Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol. Syst. Biol. 3:91.
Ehling-Schulz, M., Schulz, S., Wait, R., Görg, A., and Scherer, S. (2002). The UV-B stimulon of the terrestrial cyanobacterium Nostoc commune comprises early shock proteins and late acclimation proteins. Mol. Microbiol. 46, 827–843. doi: 10.1046/j.1365-2958.2002.03209.x
Ezraty, B., Grimaud, R., El Hassouni, M., Moinier, D., and Barras, F. (2004). Methionine sulfoxide reductases protect Ffh from oxidative damages in Escherichia coli. EMBO J. 23, 1868–1877. doi: 10.1038/sj.emboj.7600172
Fahey, R. C., Buschbacher, R. M., and Newton, G. L. (1987). The evolution of glutathione metabolism in phototrophic microorganisms. J. Mol. Evol. 25, 81–88. doi: 10.1007/bf02100044
Feller, U., Anders, I., and Demirevska, K. (2008). Degradation of rubisco and other chloroplast proteins under abiotic stress. Gen. Appl. Plant Physiol. 34, 5–18.
Fischer, W. W. (2008). Biogeochemistry: life before the rise of oxygen. Nature 455, 1051–1052. doi: 10.1038/4551051a
Forchhammer, K. (2008). PII signal transducers: novel functional and structural insights. Trends Microbiol. 16, 65–72. doi: 10.1016/j.tim.2007.11.004
Fournier, M. L., Gilmore, J. M., Martin-Brown, S. A., and Washburn, M. P. (2007). Multidimensional separations-based shotgun proteomics. Chem. Rev. 107, 3654–3686. doi: 10.1021/cr068279a
Foyer, C. H., and Noctor, G. (2009). Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antioxid. Redox Signal. 11, 861–905. doi: 10.1089/ars.2008.2177
Fridlyand, L. E., and Scheibe, R. (1999). Regulation of the Calvin cycle for CO 2 fixation as an example for general control mechanisms in metabolic cycles. Biosystems 51, 79–93. doi: 10.1016/s0303-2647(99)00017-9
Fulda, S., Mikkat, S., Huang, F., Huckauf, J., Marin, K., Norling, B., et al. (2006). Proteome analysis of salt stress response in the cyanobacterium Synechocystis sp. strain PCC 6803. Proteomics 6, 2733–2745. doi: 10.1002/pmic.200500538
Gallon, J. R., Cheng, J., Dougherty, L. J., Gallon, V. A., Hilz, H., Pederson, D. M., et al. (2000). A novel covalent modification of nitrogenase in a cyanobacterium. FEBS Lett. 468, 231–233. doi: 10.1016/s0014-5793(00)01229-1
Gao, Y., Xiong, W., Li, X.-B., Gao, C.-F., Zhang, Y.-L., Li, H., et al. (2009). Identification of the proteomic changes in Synechocystis sp. PCC 6803 following prolonged UV-B irradiation. J. Exp. Bot. 60, 1141–1154. doi: 10.1093/jxb/ern356
Gelhaye, E., Rouhier, N., Navrot, N., and Jacquot, J. (2005). The plant thioredoxin system. Cell. Mol. Life Sci. 62, 24–35.
Ghosh, D., and Xu, J. (2014). Abiotic stress responses in plant roots: a proteomics perspective. Front. Plant Sci. 5:6.
Gombos, Z., Wada, H., and Murata, N. (1992). Unsaturation of fatty acids in membrane lipids enhances tolerance of the cyanobacterium Synechocystis PCC6803 to low-temperature photoinhibition. Proc. Natl. Acad. Sci. U.S.A. 89, 9959–9963. doi: 10.1073/pnas.89.20.9959
Grant, G. A. (2012). Contrasting catalytic and allosteric mechanisms for phosphoglycerate dehydrogenases. Arch. Biochem. Biophys. 519, 175–185. doi: 10.1016/j.abb.2011.10.005
Grebe, T. W., and Stock, J. B. (1999). The histidine protein kinase superfamily. Adv. Microb. Physiol. 41, 139–227. doi: 10.1016/s0065-2911(08)60167-8
Grundner, C., Gay, L. M., and Alber, T. (2005). Mycobacterium tuberculosis serine/threonine kinases PknB, PknD, PknE, and PknF phosphorylate multiple FHA domains. Protein Sci. 14, 1918–1921. doi: 10.1110/ps.051413405
Gudmundsson, S., and Nogales, J. (2015). Cyanobacteria as photosynthetic biocatalysts: a systems biology perspective. Mol. Biosyst. 11, 60–70. doi: 10.1039/c4mb00335g
Guo, J., Nguyen, A. Y., Dai, Z., Su, D., Gaffrey, M. J., Moore, R. J., et al. (2014). Proteome-wide light/dark modulation of thiol oxidation in cyanobacteria revealed by quantitative site-specific redox proteomics. Mol. Cell. Proteomics 13, 3270–3285. doi: 10.1074/mcp.M114.041160
Häder, D.-P., Kumar, H., Smith, R., and Worrest, R. (2007). Effects of solar UV radiation on aquatic ecosystems and interactions with climate change. Photochem. Photobiol. Sci. 6, 267–285. doi: 10.1039/b700020k
Hart, G. W., and Ball, L. E. (2013). Post-translational modifications: a major focus for the future of proteomics. Mol. Cell. Proteomics 12:3443. doi: 10.1074/mcp.e113.036491
Heath, R. J., and Rock, C. O. (1996). Roles of the FabA and FabZ β-hydroxyacyl-acyl carrier protein dehydratases in Escherichia coli fatty acid biosynthesis. J. Biol. Chem. 271, 27795–27801. doi: 10.1074/jbc.271.44.27795
Hernández-Prieto, M. A., Semeniuk, T. A., and Futschik, M. E. (2014). Toward a systems-level understanding of gene regulatory, protein interaction, and metabolic networks in cyanobacteria. Front. Genet. 5:191. doi: 10.3389/fgene.2014.00191
Higo, A., Suzuki, T., Ikeuchi, M., and Ohmori, M. (2007). Dynamic transcriptional changes in response to rehydration in Anabaena sp. PCC 7120. Microbiology 153, 3685–3694. doi: 10.1099/mic.0.2007/009233-0
Hihara, Y., Kamei, A., Kanehisa, M., Kaplan, A., and Ikeuchi, M. (2001). DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 13, 793–806. doi: 10.1105/tpc.13.4.793
Hishiya, S., Hatakeyama, W., Mizota, Y., Hosoya-Matsuda, N., Motohashi, K., Ikeuchi, M., et al. (2008). Binary reducing equivalent pathways using NADPH-thioredoxin reductase and ferredoxin-thioredoxin reductase in the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Cell Physiol. 49, 11–18. doi: 10.1093/pcp/pcm158
Hongsthong, A., Sirijuntarut, M., Prommeenate, P., Lertladaluck, K., Porkaew, K., Cheevadhanarak, S., et al. (2008). Proteome analysis at the subcellular level of the cyanobacterium Spirulina platensis in response to low-temperature stress conditions. FEMS Microbiol. Lett. 288, 92–101. doi: 10.1111/j.1574-6968.2008.01330.x
Hongsthong, A., Sirijuntarut, M., Yutthanasirikul, R., Senachak, J., Kurdrid, P., Cheevadhanarak, S., et al. (2009). Subcellular proteomic characterization of the high-temperature stress response of the cyanobacterium Spirulina platensis. Proteome Sci. 7, 1–19. doi: 10.1186/1477-5956-7-33
Hosoya-Matsuda, N., Motohashi, K., Yoshimura, H., Nozaki, A., Inoue, K., Ohmori, M., et al. (2005). Anti-oxidative stress system in cyanobacteria SIGNIFICANCE OF TYPE II PEROXIREDOXIN AND THE ROLE OF 1-Cys PEROXIREDOXIN IN SYNECHOCYSTIS SP. STRAIN PCC 6803. J. Biol. Chem. 280, 840–846. doi: 10.1074/jbc.m411493200
Huang, F., Fulda, S., Hagemann, M., and Norling, B. (2006). Proteomic screening of salt-stress-induced changes in plasma membranes of Synechocystis sp. strain PCC 6803. Proteomics 6, 910–920. doi: 10.1002/pmic.200500114
Huse, M., and Kuriyan, J. (2002). The conformational plasticity of protein kinases. Cell 109, 275–282. doi: 10.1016/s0092-8674(02)00741-9
Inoue, N., Taira, Y., Emi, T., Yamane, Y., Kashino, Y., Koike, H., et al. (2001). Acclimation to the growth temperature and the high-temperature effects on photosystem II and plasma membranes in a mesophilic cyanobacterium, Synechocystis sp. PCC6803. Plant Cell Physiol. 42, 1140–1148. doi: 10.1093/pcp/pce147
Jeamton, W., Mungpakdee, S., Sirijuntarut, M., Prommeenate, P., Cheevadhanarak, S., Tanticharoen, M., et al. (2008). A combined stress response analysis of Spirulina platensis in terms of global differentially expressed proteins, and mRNA levels and stability of fatty acid biosynthesis genes. FEMS Microbiol. Lett. 281, 121–131. doi: 10.1111/j.1574-6968.2008.01100.x
Jorrín-Novo, J. V., Pascual, J., Sánchez-Lucas, R., Romero-Rodríguez, M. C., Rodríguez-Ortega, M. J., Lenz, C., et al. (2015). Fourteen years of plant proteomics reflected in Proteomics: moving from model species and 2DE-based approaches to orphan species and gel-free platforms. Proteomics 15, 1089–1112. doi: 10.1002/pmic.201400349
Jung, Y.-M., Lee, J.-N., Shin, H.-D., and Lee, Y.-H. (2004). Role of tktA gene in pentose phosphate pathway on odd-ball biosynthesis of poly-β-hydroxybutyrate in transformant Escherichia coli harboring phbCAB operon. J. Biosci. Bioeng. 98, 224–227. doi: 10.1016/s1389-1723(04)00272-5
Kaniya, Y., Kizawa, A., Miyagi, A., Kawai-Yamada, M., Uchimiya, H., Kaneko, Y., et al. (2013). Deletion of the transcriptional regulator cyAbrB2 deregulates primary carbon metabolism in Synechocystis sp. PCC 6803. Plant Physiol. 162, 1153–1163. doi: 10.1104/pp.113.218784
Katano, Y., Nimura-Matsune, K., and Yoshikawa, H. (2006). Involvement of DnaK3, one of the three DnaK proteins of cyanobacterium Synechococcus sp. PCC7942, in translational process on the surface of the thylakoid membrane. Biosci. Biotechnol. Biochem. 70, 1592–1598. doi: 10.1271/bbb.50631
Katoh, H., Asthana, R., and Ohmori, M. (2004). Gene expression in the cyanobacterium Anabaena sp. PCC7120 under desiccation. Microb. Ecol. 47, 164–174. doi: 10.1007/s00248-003-1043-6
Khetkorn, W., Rastogi, R. P., Incharoensakdi, A., Lindblad, P., Madamwar, D., Pandey, A., et al. (2017). Microalgal hydrogen production–A review. Bioresour. Technol. 243, 1194–1206. doi: 10.1016/j.biortech.2017.07.085
Kim, G.-W., and Yang, X.-J. (2011). Comprehensive lysine acetylomes emerging from bacteria to humans. Trends Biochem. Sci. 36, 211–220. doi: 10.1016/j.tibs.2010.10.001
Kim, J.-E., Kim, S.-J., Lee, B.-H., Park, R.-W., Kim, K.-S., and Kim, I.-S. (2000). Identification of motifs for cell adhesion within the repeated domains of transforming growth factor-β-induced gene, βig-h3. J. Biol. Chem. 275, 30907–30915. doi: 10.1074/jbc.m002752200
Kruger, N. J., and von Schaewen, A. (2003). The oxidative pentose phosphate pathway: structure and organisation. Curr. Opin. Plant Biol. 6, 236–246. doi: 10.1016/s1369-5266(03)00039-6
Kumar, A., Tyagi, M. B., Jha, P. N., Srinivas, G., and Singh, A. (2003). Inactivation of cyanobacterial nitrogenase after exposure to ultraviolet-B radiation. Curr. Microbiol. 46, 380–384. doi: 10.1007/s00284-001-3894-8
Kurdrid, P., Senachak, J., Sirijuntarut, M., Yutthanasirikul, R., Phuengcharoen, P., Jeamton, W., et al. (2011). Comparative analysis of the Spirulina platensis subcellular proteome in response to low-and high-temperature stresses: uncovering cross-talk of signaling components. Proteome Sci. 9:39. doi: 10.1186/1477-5956-9-39
Kurian, D., Phadwal, K., and Mäenpää, P. (2006). Proteomic characterization of acid stress response in Synechocystis sp. PCC 6803. Proteomics 6, 3614–3624. doi: 10.1002/pmic.200600033
Latifi, A., Ruiz, M., and Zhang, C.-C. (2009). Oxidative stress in cyanobacteria. FEMS Microbiol. Rev. 33, 258–278. doi: 10.1111/j.1574-6976.2008.00134.x
Lee, D.-G., Kwon, J., Eom, C.-Y., Kang, Y.-M., Roh, S. W., Lee, K.-B., et al. (2015). Directed analysis of cyanobacterial membrane phosphoproteome using stained phosphoproteins and titanium-enriched phosphopeptides. J. Microbiol. 53, 279–287. doi: 10.1007/s12275-015-5021-8
Lemaire, S. D., and Miginiac-Maslow, M. (2004). The thioredoxin superfamily in Chlamydomonas reinhardtii. Photosynth. Res. 82, 203–220. doi: 10.1007/s11120-004-1091-x
Li, T., Yang, H.-M., Cui, S.-X., Suzuki, I., Zhang, L.-F., Li, L., et al. (2011). Proteomic study of the impact of Hik33 mutation in Synechocystis sp. PCC 6803 under normal and salt stress conditions. J. Proteome Res. 11, 502–514. doi: 10.1021/pr200811s
Liberek, K., Skowyra, D., Zylicz, M., Johnson, C., and Georgopoulos, C. (1991). The Escherichia coli DnaK chaperone, the 70-kDa heat shock protein eukaryotic equivalent, changes conformation upon ATP hydrolysis, thus triggering its dissociation from a bound target protein. J. Biol. Chem. 266, 14491–14496.
Liu, J., Chen, L., Wang, J., Qiao, J., and Zhang, W. (2012). Proteomic analysis reveals resistance mechanism against biofuel hexane in Synechocystis sp. PCC 6803. Biotechnol. Biofuels 5:68. doi: 10.1186/1754-6834-5-68
Ljungman, M. (2007). The transcription stress response. Cell Cycle 6, 2252–2257. doi: 10.4161/cc.6.18.4751
López-Redondo, M. L., Moronta, F., Salinas, P., Espinosa, J., Cantos, R., Dixon, R., et al. (2010). Environmental control of phosphorylation pathways in a branched two-component system. Mol. Microbiol. 78, 475–489. doi: 10.1111/j.1365-2958.2010.07348.x
Los, D. A., and Murata, N. (1998). Structure and expression of fatty acid desaturases. Biochim. Biophys. Acta 1394, 3–15. doi: 10.1016/s0005-2760(98)00091-5
Los, D. A., Zorina, A., Sinetova, M., Kryazhov, S., Mironov, K., and Zinchenko, V. V. (2010). Stress sensors and signal transducers in cyanobacteria. Sensors 10, 2386–2415. doi: 10.3390/s100302386
Ma, Y., Yang, M., Lin, X., Liu, X., Huang, H., and Ge, F. (2017). Malonylome analysis reveals the involvement of lysine malonylation in metabolism and photosynthesis in cyanobacteria. J. Proteome Res. 16, 2030–2043. doi: 10.1021/acs.jproteome.7b00017
Martinez, A., and Kolter, R. (1997). Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J. Bacteriol. 179, 5188–5194. doi: 10.1128/jb.179.16.5188-5194.1997
Mehta, A., López-Maury, L., and Florencio, F. J. (2014). Proteomic pattern alterations of the cyanobacterium Synechocystis sp. PCC 6803 in response to cadmium, nickel and cobalt. J. Proteomics 102, 98–112. doi: 10.1016/j.jprot.2014.03.002
Merroun, M. L., Raff, J., Rossberg, A., Hennig, C., Reich, T., and Selenska-Pobell, S. (2005). Complexation of uranium by cells and S-layer sheets of Bacillus sphaericus JG-A12. Appl. Environ. Microbiol. 71, 5532–5543. doi: 10.1128/aem.71.9.5532-5543.2005
Mikkat, S., Fulda, S., and Hagemann, M. (2014). A 2D gel electrophoresis-based snapshot of the phosphoproteome in the cyanobacterium Synechocystis sp. strain PCC 6803. Microbiology 160(Pt 2), 296–306. doi: 10.1099/mic.0.074443-0
Miranda, H., Cheregi, O., Netotea, S., Hvidsten, T. R., Moritz, T., and Funk, C. (2013). Co-expression analysis, proteomic and metabolomic study on the impact of a Deg/HtrA protease triple mutant in Synechocystis sp. PCC 6803 exposed to temperature and high light stress. J. Proteomics 78, 294–311. doi: 10.1016/j.jprot.2012.09.036
Mishra, Y., Bhargava, P., Chaurasia, N., and Rai, L. C. (2009). Proteomic evaluation of the non-survival of Anabaena doliolum (Cyanophyta) at elevated temperatures. Eur. J. Phycol. 44, 551–565. doi: 10.1080/09670260902947001
MišurCoVá, L., KráčMar, S., Klejdus, B., and Vacek, J. (2010). Nitrogen content, dietary fiber, and digestibility in algal food products. Czech J. Food Sci. 28, 27–35. doi: 10.17221/111/2009-cjfs
Mitrophanov, A. Y., and Groisman, E. A. (2008). Signal integration in bacterial two-component regulatory systems. Genes Dev. 22, 2601–2611. doi: 10.1101/gad.1700308
Mo, R., Yang, M., Chen, Z., Cheng, Z., Yi, X., Li, C., et al. (2015). Acetylome analysis reveals the involvement of lysine acetylation in photosynthesis and carbon metabolism in the model cyanobacterium Synechocystis sp. PCC 6803. J. Proteome Res. 14, 1275–1286. doi: 10.1021/pr501275a
Mohan, S., Kelly, T. M., Eveland, S. S., Raetz, C., and Anderson, M. S. (1994). An Escherichia coli gene (FabZ) encoding (3R)-hydroxymyristoyl acyl carrier protein dehydrase. Relation to fabA and suppression of mutations in lipid A biosynthesis. J. Biol. Chem. 269, 32896–32903.
Moparthi, V. K., Li, X., Vavitsas, K., Dzhygyr, I., Sandh, G., Magnuson, A., et al. (2016). The two Dps proteins, NpDps2 and NpDps5, are involved in light-induced oxidative stress tolerance in the N 2-fixing cyanobacterium Nostoc punctiforme. Biochim. Biophys. Acta 1857, 1766–1776. doi: 10.1016/j.bbabio.2016.08.003
Morgan-Kiss, R. M., Priscu, J. C., Pocock, T., Gudynaite-Savitch, L., and Huner, N. P. (2006). Adaptation and acclimation of photosynthetic microorganisms to permanently cold environments. Microbiol. Mol. Biol. Rev. 70, 222–252. doi: 10.1128/mmbr.70.1.222-252.2006
Moslavac, S., Reisinger, V., Berg, M., Mirus, O., Vosyka, O., Plöscher, M., et al. (2007). The proteome of the heterocyst cell wall in Anabaena sp. PCC 7120. Biol. Chem. 388, 823–829.
Mullineaux, C. W. (2014). Co-existence of photosynthetic and respiratory activities in cyanobacterial thylakoid membranes. Biochim. Biophys. Acta 1837, 503–511. doi: 10.1016/j.bbabio.2013.11.017
Murata, N., and Suzuki, I. (2006). Exploitation of genomic sequences in a systematic analysis to access how cyanobacteria sense environmental stress. J. Exp. Bot. 57, 235–247. doi: 10.1093/jxb/erj005
Nakahara, K., Yamamoto, H., Miyake, C., and Yokota, A. (2003). Purification and characterization of class-I and class-II fructose-1, 6-bisphosphate aldolases from the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 44, 326–333. doi: 10.1093/pcp/pcg044
Olsen, J. V., and Mann, M. (2013). Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol. Cell. Proteomics 12, 3444–3452. doi: 10.1074/mcp.o113.034181
Ow, S. Y., Salim, M., Noirel, J., Evans, C., and Wright, P. (2011). Minimising iTRAQ ratio compression through understanding LC-MS elution dependence and high-resolution HILIC fractionation. Proteomics 11, 2341–2346. doi: 10.1002/pmic.201000752
Ow, S. Y., and Wright, P. C. (2009). Current trends in high throughput proteomics in cyanobacteria. FEBS Lett. 583, 1744–1752. doi: 10.1016/j.febslet.2009.03.062
Panda, B., Basu, B., Acharya, C., Rajaram, H., and Apte, S. K. (2017). Proteomic analysis reveals contrasting stress response to uranium in two nitrogen-fixing Anabaena strains, differentially tolerant to uranium. Aquat. Toxicol. 182, 205–213. doi: 10.1016/j.aquatox.2016.12.002
Panda, B., Basu, B., Rajaram, H., and Apte, S. K. (2015). Comparative proteomics of oxidative stress response in three cyanobacterial strains native to Indian paddy fields. J. Proteomics 127(Pt A), 152–160.
Panda, B., Basu, B., Rajaram, H., and Kumar Apte, S. (2014). Methyl viologen responsive proteome dynamics of Anabaena sp. strain PCC7120. Proteomics 14, 1895–1904. doi: 10.1002/pmic.201300522
Pandey, S., Rai, R., and Rai, L. C. (2012). Proteomics combines morphological, physiological and biochemical attributes to unravel the survival strategy of Anabaena sp. PCC7120 under arsenic stress. J. Proteomics 75, 921–937. doi: 10.1016/j.jprot.2011.10.011
Pandhal, J., Ow, S. Y., Wright, P. C., and Biggs, C. A. (2008a). Comparative proteomics study of salt tolerance between a nonsequenced extremely halotolerant cyanobacterium and its mildly halotolerant relative using in vivo metabolic labeling and in vitro isobaric labeling. J. Proteome Res. 8, 818–828. doi: 10.1021/pr800283q
Pandhal, J., Wright, P. C., and Biggs, C. A. (2008b). Proteomics with a pinch of salt: a cyanobacterial perspective. Saline Systems 4:1. doi: 10.1186/1746-1448-4-1
Parimi, N. S., Singh, M., Kastner, J. R., Das, K. C., Forsberg, L. S., and Azadi, P. (2015). Optimization of protein extraction from Spirulina platensis to generate a potential co-product and a biofuel feedstock with reduced nitrogen content. Front. Energy Res. 3:30. doi: 10.3389/fenrg.2015.00030
Pascual, M. B., Mata-Cabana, A., Florencio, F. J., Lindahl, M., and Cejudo, F. J. (2010). Overoxidation of 2-Cys peroxiredoxin in prokaryotes cyanobacterial 2-Cys peroxiredoxins sensitive to oxidative stress. J. Biol. Chem. 285, 34485–34492. doi: 10.1074/jbc.M110.160465
Paulsen, C. E., and Carroll, K. S. (2013). Cysteine-mediated redox signaling: chemistry, biology, and tools for discovery. Chem. Rev. 113, 4633–4679. doi: 10.1021/cr300163e
Pérez-Pérez, M. E., Martín-Figueroa, E., and Florencio, F. J. (2009). Photosynthetic regulation of the cyanobacterium Synechocystis sp. PCC 6803 thioredoxin system and functional analysis of TrxB (Trx x) and TrxQ (Trx y) thioredoxins. Mol. Plant 2, 270–283. doi: 10.1093/mp/ssn070
Pickering, I. J., Prince, R. C., George, M. J., Smith, R. D., George, G. N., and Salt, D. E. (2000). Reduction and coordination of arsenic in Indian mustard. Plant Physiol. 122, 1171–1178. doi: 10.1104/pp.122.4.1171
Prakash, J. S., Krishna, P. S., Sirisha, K., Kanesaki, Y., Suzuki, I., Shivaji, S., et al. (2010). An RNA helicase, CrhR, regulates the low-temperature-inducible expression of heat-shock genes groES, groEL1 and groEL2 in Synechocystis sp. PCC 6803. Microbiology 156, 442–451. doi: 10.1099/mic.0.031823-0
Qiao, J., Wang, J., Chen, L., Tian, X., Huang, S., Ren, X., et al. (2012). Quantitative iTRAQ LC–MS/MS proteomics reveals metabolic responses to biofuel ethanol in cyanobacterial Synechocystis sp. PCC 6803. J. Proteome Res. 11, 5286–5300. doi: 10.1021/pr300504w
Rai, S., Singh, S., Shrivastava, A. K., and Rai, L. (2013). Salt and UV-B induced changes in Anabaena PCC 7120: physiological, proteomic and bioinformatic perspectives. Photosynth. Res. 118, 105–114. doi: 10.1007/s11120-013-9931-1
Rajaram, H., Chaurasia, A. K., and Apte, S. K. (2014). Cyanobacterial heat-shock response: role and regulation of molecular chaperones. Microbiology 160(Pt 4), 647–658. doi: 10.1099/mic.0.073478-0
Rastogi, R. P., and Sinha, R. P. (2009). Biotechnological and industrial significance of cyanobacterial secondary metabolites. Biotechnol. Adv. 27, 521–539. doi: 10.1016/j.biotechadv.2009.04.009
Requejo, R., and Tena, M. (2006). Maize response to acute arsenic toxicity as revealed by proteome analysis of plant shoots. Proteomics 6, S156–S162.
Sánchez-Riego, A. M., López-Maury, L., and Florencio, F. J. (2013). Glutaredoxins are essential for stress adaptation in the cyanobacterium Synechocystis sp. PCC 6803. Front. Plant Sci. 4:428. doi: 10.3389/fpls.2013.00428
Sandh, G., Ramström, M., and Stensjö, K. (2014). Analysis of the early heterocyst Cys-proteome in the multicellular cyanobacterium Nostoc punctiforme reveals novel insights into the division of labor within diazotrophic filaments. BMC Genomics 15:1064. doi: 10.1186/1471-2164-15-1064
Sato, N., Moriyama, T., Toyoshima, M., Mizusawa, M., and Tajima, N. (2012). The all0458/lti46. 2 gene encodes a low temperature-induced Dps protein homologue in the cyanobacteria Anabaena sp. PCC 7120 and Anabaena variabilis M3. Microbiology 158, 2527–2536. doi: 10.1099/mic.0.060657-0
Sato, N., Ohmori, M., Ikeuchi, M., Tashiro, K., Wolk, C. P., Kaneko, T., et al. (2004). Use of segment-based microarray in the analysis of global gene expression in response to various environmental stresses in the cyanobacterium Anabaena sp. PCC 7120. J. Gen. Appl. Microbiol. 50, 1–8. doi: 10.2323/jgam.50.1
Schenk, G., Duggleby, R. G., and Nixon, P. F. (1998). Properties and functions of the thiamin diphosphate dependent enzyme transketolase. Int. J. Biochem. Cell Biol. 30, 1297–1318. doi: 10.1016/s1357-2725(98)00095-8
Schmitt, F.-J., Renger, G., Friedrich, T., Kreslavski, V. D., Zharmukhamedov, S. K., Los, D. A., et al. (2014). Reactive oxygen species: re-evaluation of generation, monitoring and role in stress-signaling in phototrophic organisms. Biochim. Biophys. Acta 1837, 835–848. doi: 10.1016/j.bbabio.2014.02.005
Seaver, L. C., and Imlay, J. A. (2004). Are respiratory enzymes the primary sources of intracellular hydrogen peroxide? J. Biol. Chem. 279, 48742–48750. doi: 10.1074/jbc.m408754200
Shcolnick, S., Shaked, Y., and Keren, N. (2007). A role for mrgA, a DPS family protein, in the internal transport of Fe in the cyanobacterium Synechocystis sp. PCC6803. Biochim. Biophys. Acta 1767, 814–819. doi: 10.1016/j.bbabio.2006.11.015
Shinagawa, H. (1996). “SOS response as an adaptive response to DNA damage in prokaryotes,” in Stress-Inducible Cellular Responses, eds U. Feige, R. I. Morimoto., and B. Polla (Berlin: Springer), 221–235. doi: 10.1007/978-3-0348-9088-5_14
Shrivastava, A. K., Chatterjee, A., Yadav, S., Singh, P. K., Singh, S., and Rai, L. (2015). UV-B stress induced metabolic rearrangements explored with comparative proteomics in three Anabaena species. J. Proteomics 127(Pt A), 122–133.
Singh, P. K., Shrivastava, A. K., Chatterjee, A., Pandey, S., Rai, S., Singh, S., et al. (2015). Cadmium toxicity in diazotrophic Anabaena spp. adjudged by hasty up-accumulation of transporter and signaling and severe down-accumulation of nitrogen metabolism proteins. J. Proteomics 127, 134–146. doi: 10.1016/j.jprot.2015.05.019
Singh, S. C., Sinha, R. P., and Hader, D.-P. (2002). Role of lipids and fatty acids in stress tolerance in cyanobacteria. Acta Protozool. 41, 297–308.
Spät, P., Maèek, B., and Forchhammer, K. (2015). Phosphoproteome of the cyanobacterium Synechocystis sp. PCC 6803 and its dynamics during nitrogen starvation. Front. Microbiol. 6:248.
Stadtman, E. R., Moskovitz, J., and Levine, R. L. (2003). Oxidation of methionine residues of proteins: biological consequences. Antioxid. Redox Signal. 5, 577–582. doi: 10.1089/152308603770310239
Stanier, R., and Bazine, G. (1977). Phototrophic prokaryotes: the cyanobacteria. Annu. Rev. Microbiol. 31, 225–274.
Stensjö, K., Ow, S. Y., Barrios-Llerena, M. E., Lindblad, P., and Wright, P. C. (2007). An iTRAQ-based quantitative analysis to elaborate the proteomic response of Nostoc sp. PCC 7120 under N2 fixing conditions. J. Proteome Res. 6, 621–635. doi: 10.1021/pr060517v
Stock, A. M., Robinson, V. L., and Goudreau, P. N. (2000). Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215.
Tasaka, Y., Gombos, Z., Nishiyama, Y., Mohanty, P., Ohba, T., Ohki, K., et al. (1996). Targeted mutagenesis of acyl-lipid desaturases in Synechocystis: evidence for the important roles of polyunsaturated membrane lipids in growth, respiration and photosynthesis. EMBO J. 15, 6416–6425. doi: 10.1002/j.1460-2075.1996.tb01033.x
Teikari, J., Österholm, J., Kopf, M., Battchikova, N., Wahlsten, M., Aro, E.-M., et al. (2015). Transcriptomics and proteomics profiling of Anabaena sp. strain 90 under inorganic phosphorus stress. Appl. Environ. Microbiol. 81, 5212–5222. doi: 10.1128/aem.01062-15
Tian, X., Chen, L., Wang, J., Qiao, J., and Zhang, W. (2013). Quantitative proteomics reveals dynamic responses of Synechocystis sp. PCC 6803 to next-generation biofuel butanol. J. Proteomics 78, 326–345. doi: 10.1016/j.jprot.2012.10.002
Vaishampayan, A., Sinha, R., Hader, D.-P., Dey, T., Gupta, A., Bhan, U., et al. (2001). Cyanobacterial biofertilizers in rice agriculture. Bot. Rev. 67, 453–516. doi: 10.1007/bf02857893
Varkey, D., Mazard, S., Ostrowski, M., Tetu, S. G., Haynes, P., and Paulsen, I. T. (2016). Effects of low temperature on tropical and temperate isolates of marine Synechococcus. ISME J. 10, 1252–1263. doi: 10.1038/ismej.2015.179
Wada, H., Gombos, Z., and Murata, N. (1994). Contribution of membrane lipids to the ability of the photosynthetic machinery to tolerate temperature stress. Proc. Natl. Acad. Sci. U.S.A. 91, 4273–4277. doi: 10.1073/pnas.91.10.4273
Wada, H., and Murata, N. (1990). Temperature-induced changes in the fatty acid composition of the cyanobacterium, Synechocystis PCC6803. Plant Physiol. 92, 1062–1069. doi: 10.1104/pp.92.4.1062
Wang, H., Yang, Y., Chen, W., Ding, L., Li, P., Zhao, X., et al. (2013). Identification of differentially expressed proteins of Arthrospira (Spirulina) plantensis-YZ under salt-stress conditions by proteomics and qRT-PCR analysis. Proteome Sci. 11:6. doi: 10.1186/1477-5956-11-6
Wang, Y., Chen, L., and Zhang, W. (2016). Proteomic and metabolomic analyses reveal metabolic responses to 3-hydroxypropionic acid synthesized internally in cyanobacterium Synechocystis sp. PCC 6803. Biotechnol. Biofuels 9:209.
Wolanin, P. M., Thomason, P. A., and Stock, J. B. (2002). Histidine protein kinases: key signal transducers outside the animal kingdom. Genome Biol. 3, 3013.1–3013.8.
Wu, W. W., Wang, G., Baek, S. J., and Shen, R.-F. (2006). Comparative study of three proteomic quantitative methods, DIGE, cICAT, and iTRAQ, using 2D gel-or LC-MALDI TOF/TOF. J. Proteome Res. 5, 651–658. doi: 10.1021/pr050405o
Xiong, Q., Chen, Z., and Ge, F. (2015a). Proteomic analysis of post translational modifications in cyanobacteria. J. Proteomics 134, 57–64.
Xiong, Q., Feng, J., Li, S.-T., Zhang, G.-Y., Qiao, Z.-X., Chen, Z., et al. (2015b). Integrated transcriptomic and proteomic analysis of the global response of Synechococcus sp. PCC 7002 to high light stress. Mol. Cell. Proteomics, 14, 1038–1053. doi: 10.1074/mcp.M114.046003
Xu, W., Tang, H., Wang, Y., and Chitnis, P. R. (2001). Proteins of the cyanobacterial photosystem I. Biochim. Biophys. Acta 1507, 32–40.
Yan, C., and Xu, X. (2008). Bifunctional enzyme FBPase/SBPase is essential for photoautotrophic growth in cyanobacterium Synechocystis sp. PCC 6803. Prog. Nat. Sci. 18, 149–153. doi: 10.1016/j.pnsc.2007.09.003
Yang, M.-K., Qiao, Z.-X., Zhang, W.-Y., Xiong, Q., Zhang, J., Li, T., et al. (2013). Global phosphoproteomic analysis reveals diverse functions of serine/threonine/tyrosine phosphorylation in the model cyanobacterium Synechococcus sp. strain PCC 7002. J. Proteome Res. 12, 1909–1923. doi: 10.1021/pr4000043
Yoshimura, H., Ikeuchi, M., and Ohmori, M. (2006). Up-regulated gene expression during dehydration in a terrestrial cyanobacterium, Nostoc sp. strain HK-01. Microbes Environ. 21, 129–133. doi: 10.1264/jsme2.21.129
Yoshimura, H., Okamoto, S., Tsumuraya, Y., and Ohmori, M. (2007). Group 3 sigma factor gene, sigJ, a key regulator of desiccation tolerance, regulates the synthesis of extracellular polysaccharide in cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 14, 13–24. doi: 10.1093/dnares/dsm003
Zeller, T., and Klug, G. (2006). Thioredoxins in bacteria: functions in oxidative stress response and regulation of thioredoxin genes. Naturwissenschaften 93, 259–266. doi: 10.1007/s00114-006-0106-1
Zhang, J.-H., Liu, Y.-P., Pan, Q.-H., Zhan, J.-C., Wang, X.-Q., and Huang, W.-D. (2006). Changes in membrane-associated H+-ATPase activities and amounts in young grape plants during the cross adaptation to temperature stresses. Plant Sci. 170, 768–777. doi: 10.1016/j.plantsci.2005.11.009
Zhang, L.-F., Yang, H.-M., Cui, S.-X., Hu, J., Wang, J., Kuang, T.-Y., et al. (2009). Proteomic analysis of plasma membranes of cyanobacterium Synechocystis sp. strain PCC 6803 in response to high pH stress. J. Proteome Res. 8, 2892–2902. doi: 10.1021/pr900024w
Zhang, Y., Niu, X., Shi, M., Pei, G., Zhang, X., Chen, L., et al. (2015). Identification of a transporter Slr0982 involved in ethanol tolerance in cyanobacterium Synechocystis sp. PCC 6803. Front. Microbiol. 6:487. doi: 10.3389/fmicb.2015.00487
Keywords: cyanobacteria, abiotic stress, proteomics, PTMs, up-regulated protein, down-regulated protein
Citation: Babele PK, Kumar J and Chaturvedi V (2019) Proteomic De-Regulation in Cyanobacteria in Response to Abiotic Stresses. Front. Microbiol. 10:1315. doi: 10.3389/fmicb.2019.01315
Received: 08 October 2018; Accepted: 27 May 2019;
Published: 12 June 2019.
Edited by:
Ivan Mijakovic, Chalmers University of Technology, SwedenReviewed by:
Feng Ge, Chinese Academy of Sciences, ChinaRajesh P. Rastogi, Ministry of Environment, Forest and Climate Change, India
Copyright © 2019 Babele, Kumar and Chaturvedi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Piyoosh Kumar Babele, cGl5b29zaDIwMDZAeWFob28uY28uaW4=
 Piyoosh Kumar Babele
Piyoosh Kumar Babele Jay Kumar
Jay Kumar Venkatesh Chaturvedi
Venkatesh Chaturvedi