- 1Commonwealth Scientific and Industrial Research Organisation, Health and Biosecurity, Canberra, ACT, Australia
- 2Faculty of Science and Technology, University of Canberra, Canberra, ACT, Australia
- 3Department of Entomology, Pennsylvania State University, University Park, PA, United States
- 4Invasive Animals Cooperative Research Centre, University of Canberra, Canberra, ACT, Australia
The Caliciviridae are viruses with a positive-sense, single-stranded RNA genome that is packaged into an icosahedral, environmentally stable protein capsid. The family contains five genera (Norovirus, Nebovirus, Sapovirus, Lagovirus, and Vesivirus) that infect vertebrates including amphibians, reptiles, birds, and mammals. The RNA-dependent RNA polymerase (RdRp) replicates the genome of RNA viruses and can speed up evolution due to its error-prone nature. Studying calicivirus RdRps in the context of genuine virus replication is often hampered by a lack of suitable model systems. Enteric caliciviruses and RHDV in particular are notoriously difficult to propagate in cell culture; therefore, molecular studies of replication mechanisms are challenging. Nevertheless, research on recombinant proteins has revealed several unexpected characteristics of calicivirus RdRps. For example, the RdRps of RHDV and related lagoviruses possess the ability to expose a hydrophobic motif, to rearrange Golgi membranes, and to copy RNA at unusually high temperatures. This review is focused on the structural dynamics, biochemical properties, kinetics, and putative interaction partners of these RdRps. In addition, we discuss the possible existence of a conserved but as yet undescribed structural element that is shared amongst the RdRps of all caliciviruses.
Introduction
The Caliciviridae family currently consists of five genera (Norovirus, Nebovirus, Sapovirus, Lagovirus, and Vesivirus) (Figure 1). The establishment of three additional genera (i.e., Recovirus, Valovirus, and Balovirus) has been proposed (Farkas et al., 2008; L’Homme et al., 2009; Wang et al., 2017). The exponential increase in metagenomic sequence data is likely to reveal an even higher degree of diversity for this virus family. Not surprisingly, many of the currently known caliciviruses are highly pathogenic (a characteristic that usually leads to discovery), but research using metagenomics is likely to discover more non-pathogenic family members (Mahar et al., 2019). Viruses from the genera Norovirus and Sapovirus are a common cause of gastroenteritis in humans and animals. For example, Norwalk virus and other human noroviruses are responsible for almost half of all gastroenteritis cases globally (Atmar and Estes, 2006). These viruses are easily transmitted either directly from person to person or through contaminated food and water. Infections are especially dangerous for elderly, young, and immunocompromised individuals such as transplant recipients (Schwartz et al., 2011). Despite the high socioeconomic costs associated with human norovirus outbreaks, no approved vaccines or small molecule inhibitors are currently available to prevent or cure infections. Viruses from the genus Vesivirus, such as Feline calicivirus (FCV) and Vesicular exanthema of swine virus (VESV), are highly contagious in animals and can cause persistent infections. FCV causes fever and acute upper respiratory tract and oral cavity disease in all feline species and can lead to a virulent systemic disorder (Hurley and Sykes, 2003). VESV affects pigs and marine mammals, causing fever and epithelial lesions around the mouth, nostrils, and on the feet (Neill et al., 1995). The genus Lagovirus comprises only viruses that infect lagomorphs, especially rabbits and hares. Pathogenicity among lagoviruses can differ dramatically. The Rabbit haemorrhagic disease virus (RHDV) causes acute necrotizing hepatitis and disseminated intravascular coagulation in European rabbits (Oryctolagus cuniculus), which leads to death 48–72 h post-infection, while the Rabbit calicivirus (RCV) causes only mild disease manifestations (Abrantes et al., 2012). Since the mid-1990s, RHDV has been used to control rabbit populations in Australia and New Zealand following the introduction of the European rabbit in the late 1800s (Cooke, 2002; Cooke and Fenner, 2002). Even though RHDV is an important biocontrol agent, it has not yet been studied in great detail; many aspects of viral replication and the function of several proteins remain unknown.
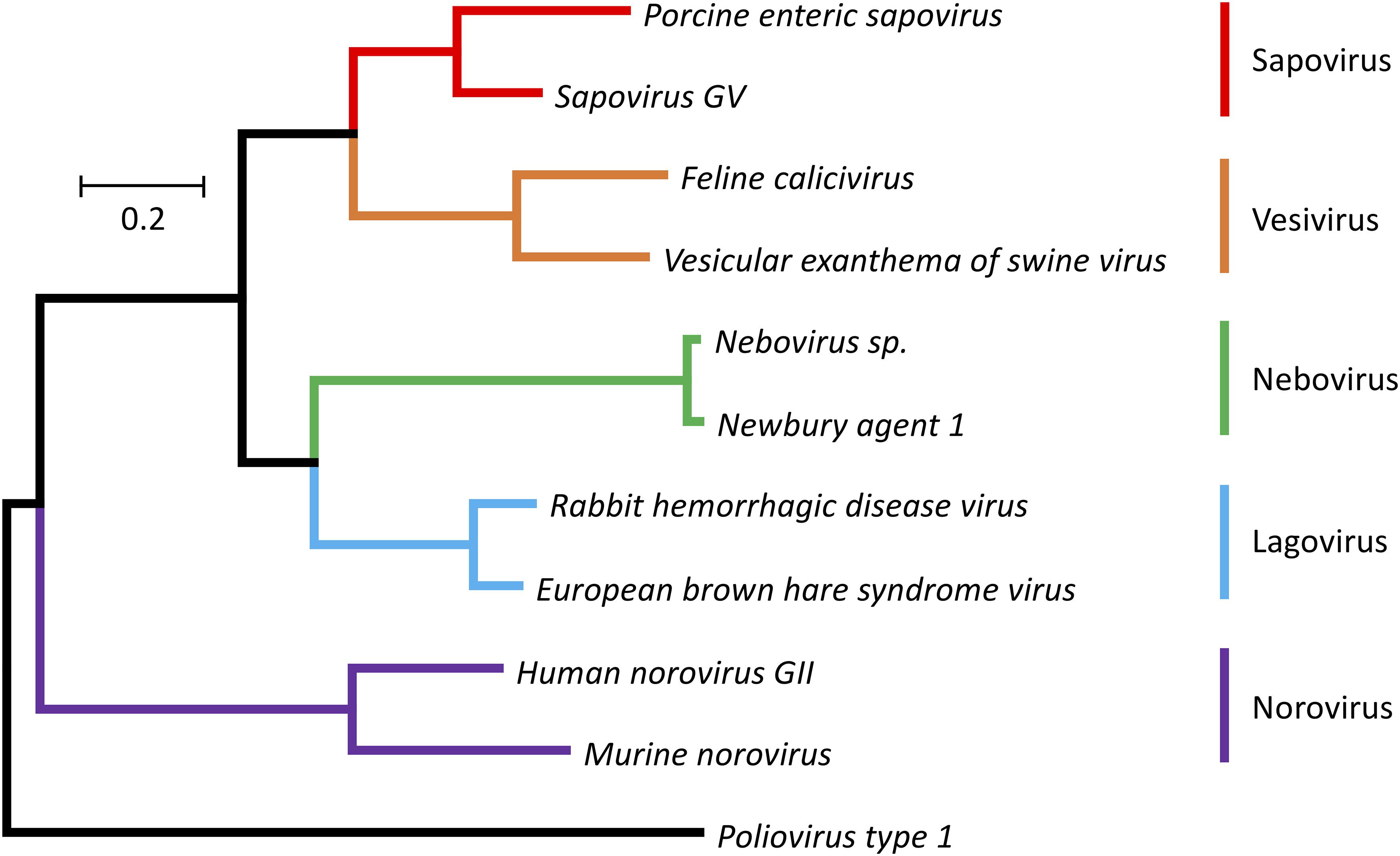
Figure 1. Phylogenetic tree for RdRp protein sequences of the family Caliciviridae and Poliovirus type 1 (Mahoney strain). The evolutionary history was inferred using the Maximum Likelihood method (Jones et al., 1992). The tree is drawn to scale, with branch lengths representing the number of substitutions per site. The analysis involved amino acid sequences from 11 caliciviruses [Porcine enteric sapovirus, A0A348BR93 (UniProt); Sapovirus GV, NP783310 (NCBI Protein); Feline calicivirus, NP786896 (NCBI Protein); Vesicular exanthema of swine virus, AYN44917 (NCBI Protein); Nebovirus sp., YP529897 (NCBI Protein); Newbury agent 1, NP740332 (NCBI Protein); Rabbit haemorrhagic disease virus, NP786902 (NCBI Protein); European brown hare syndrome virus, D0UGI3 (UniProt); Human norovirus GII, AWB14625 (NCBI Protein); Murine norovirus, P03300 (UniProt)] and a poliovirus [Poliovirus type 1, Q6IX02 (UniProt)]. Evolutionary analyses were conducted using the MEGA7 program package (Kumar et al., 2016). Different colors are used for different calicivirus genera.
Viruses of the Caliciviridae family share a number of features. The genome consists of positive-sense, single-stranded RNAs that contain coding sequences in two or more partially overlapping open reading frames (ORFs). The coding sequences are flanked by untranslated regions (UTRs) at both the 5′ and 3′ ends. Genomic RNAs are covalently linked at the 5′ end to a viral protein (VPg, for “virion protein, genome-linked”) and are polyadenylated at the 3′ end. Calicivirus particles contain two types of RNA, a genomic (full-length) RNA of about 7.5 kb and one or more copies of a subgenomic RNA of about 2 kb (Ehresmann and Schaffer, 1977; Meyers et al., 1991a,b). The number of ORFs varies from two to four in full-length genomic RNAs and from two to three in subgenomic RNAs (Wirblich et al., 1996; McFadden et al., 2011; Figure 2). ORF1 is always the largest of the reading frames and encodes a polyprotein that is subsequently cleaved into five non-structural proteins and VPg (genus Norovirus and Vesivirus) or five non-structural proteins, VPg, and the major capsid protein VP1 (genus Lagovirus, Nebovirus, and Sapovirus) (Martín Alonso et al., 1996; Meyers et al., 2000). The second and third ORFs in the genomic RNA of noroviruses encode the structural proteins VP1 and VP2, respectively. In vesiviruses, ORF2 encodes the VP1 precursor protein that is subsequently cleaved into a mature VP1 and a small leader peptide (leader of the capsid protein, LC). The LC protein of FCV is cytopathic and promotes virus spread (Abente et al., 2013). The subgenomic RNAs of all genera are very similar to each other; they contain the 5′ UTR and the VP1 and VP2 coding sequences (Meyers et al., 1991a,b, 2000; Boga et al., 1992). In Murine norovirus (MNV), there is an additional ORF in the VP1 coding region of both genomic and subgenomic RNAs that encodes the viral factor 1 (VF1), an antagonist of the innate antiviral immune response (McFadden et al., 2011).

Figure 2. Schematic representations of typical calicivirus genome organizations. (A–D) Genomic full-length RNAs of about 7.5 kb in size contain either two ORFs (in viruses of the genera Lagovirus, Nebovirus, and Sapovirus) or three ORFs (Norovirus and Vesivirus), except for the genomic RNA of Murine norovirus (MNV; genus Norovirus) that may contain an additional ORF (encoding the VF1 protein). (E) All caliciviruses except MNV and vesiviruses have subgenomic RNAs of about 2.1 kb in size with two ORFs that encode the main structural proteins, VP1 and VP2; the subgenomic RNA of MNV includes three ORFs (similar to the corresponding genomic RNA) and the subgenomic RNA of vesiviruses encodes – apart from proteins VP1 and VP2 – a small leader of the capsid protein (LC). Colored boxes represent coding sequences that are flanked by untranslated leader and trailer sequences (shown as lines). Hexagons represent VPg proteins that are covalently bound to the 5′ end of all genomic and subgenomic RNAs; An represents the poly(A) tail at the 3′ end of all genomic and subgenomic RNAs.
The structural protein VP1 forms an icosahedral, non-enveloped capsid of about 25–40 nm in diameter (Parra and Prieto, 1990; Prasad et al., 1994, 1999). A typical calicivirus capsid contains 90 VP1 dimers. Protruding VP1 (VP60 in RHDV) domains create a surface topography that resembles cup-shaped depressions when viewed using electron microscopy, which inspired the name “calicivirus” (Latin “calyx” = cup). The basic VP2 protein has also been found associated with virus particles (although in much smaller numbers) and plays a role in RNA replication and the maturation of infectious virus particles (Sosnovtsev et al., 2005). In addition, recent studies of FCV suggest a role for VP2 in the formation of a portal-like structure facilitating the delivery of viral RNA into the cytoplasm in the early stages of infection (Conley et al., 2019).
The VPg protein is also found in virus particles and should therefore be categorized as a structural protein, since the components of a mature virus particle are defined as structural proteins. The VPg is covalently linked to the 5′ end of both the full-length genomic and subgenomic RNAs (Black et al., 1978; Burroughs and Brown, 1978; Meyers et al., 1991a). Mass-spectrometry-based assays showed that FCV and MNV VPg proteins are linked to a guanosine diphosphate moiety via tyrosine residues, which is consistent with the presence of a highly conserved 5′ guanosine nucleotide in the genome of all caliciviruses (Olspert et al., 2016). The association between VPg and RNA was recognized for the first time when, following phenol extraction, a significant amount of caliciviral RNA was found in the interphase, along with other viral and cellular proteins. However, when the samples were treated with protease K prior to the extraction, the viral RNA was found in the aqueous phase. Furthermore, when purified RHDV RNA was labeled with 125I, autoradiography revealed two protein bands corresponding to genomic and subgenomic RNAs. The subsequent treatment of the labeled RNAs with RNase produced a single band of about 15 kDa on SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) (Meyers et al., 1991a). The VPg protein also plays a critical role in RNA replication. Following nucleotidylation by the RNA-dependent RNA polymerase (RdRp) or an RdRp precursor, VPg can act as a primer for genome replication (Belliot et al., 2008; Goodfellow, 2011).
The non-structural proteins can be categorized into those with known functions (NTPase, 3C-like protease, and RdRp) and unknown functions (all remaining proteins). The first one or two N-terminal proteins of the full-length genomic RNA (e.g., p16, p23, and p29 in lagoviruses, or p48 and p22 in noroviruses) may have functions similar to the so-called “security proteins” of the Picornaviridae family that counteract host defense mechanisms (Agol and Gmyl, 2010). This hypothesis is based on the fact that the coding sequence for the calicivirus proteins and the picornavirus security proteins have a similar position in the genome of the respective viruses. Although the calicivirus proteins do not share detectable sequence homologies with their picornavirus counterparts, accumulating data from functional studies suggest that these proteins do indeed impede immune responses, e.g., those that depend on cellular secretory pathways. The Norwalk virus protein p48 (when expressed as a recombinant protein in transfected cells) induces Golgi membrane rearrangements (Fernandez-Vega et al., 2004). The p48 protein of both MNV and human noroviruses interacts with the vesicle-associated membrane protein-associated protein A (VAP-A). VAP-A is a soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE)-regulator and is involved in vesicle transport (Weir et al., 1998; Ettayebi and Hardy, 2003). This interaction is likely to disrupt intracellular protein trafficking, as cells that express p48 were unable to expose the vesicular stomatitis G glycoprotein on the cell surface (Ettayebi and Hardy, 2003). Moreover, strand-specific quantitative PCR revealed a delayed accumulation of positive and negative strand MNV RNAs in VAP-A deficient cells (McCune et al., 2017). The p22 protein of Norwalk virus also contributes to Golgi disaggregation and blocks trafficking of vesicles from the ER to the Golgi (Sharp et al., 2010). However, the corresponding proteins in other calicivirus genera have not yet been functionally characterized and, to date, no conserved motifs have been identified that would suggest particular functions. Therefore, their exact role in virus replication and/or pathogenesis remains unknown.
The functions of the remaining non-structural proteins were deduced by comparing calicivirus and picornavirus sequences and by searching for conserved motifs. A 2C-like helicase (named NTPase in Figure 2) was identified after the detection of a nucleotide-binding site that is typical for viral proteins (Neill, 1990). Later, this enzyme was shown to be associated with the replication complex and to destabilize double-stranded RNA in an NTP-independent manner, representing an unexpected RNA chaperone-like activity (Li et al., 2017; Han et al., 2018). Thereafter, the p58 cleavage product of the RHDV polyprotein was found to resemble the 3D polymerase of poliovirus, and its role in RNA replication was subsequently confirmed using functional assays (Wirblich et al., 1996; Vazquez et al., 1998). Similarly, the sequence preceding the RdRp gene was suggested to code for a 3C-like protease (Neill, 1990; Jiang et al., 1993). As with the picornavirus proteases, the calicivirus homologs are responsible for the processing of the polyprotein (on a par with cellular proteases) and for the formation and accumulation of a 3CD-like polymerase precursor (Sosnovtseva et al., 1999; Thumfart and Meyers, 2002; Oka et al., 2005).
RNA-dependent RNA polymerases are the key proteins responsible for viral replication. In all caliciviruses, the RdRp coding sequence follows that of the viral protease at the 3′end of ORF1. Mature RdRps are proteins of about 60 kDa (75 kDa in the precursor form). Remarkably, the calicivirus RdRp precursor protein is also an active polymerase enzyme (Wei et al., 2001). RdRps are usually among the best-characterized proteins of any given virus species; RdRps from several caliciviruses have been crystallized and studied (Table 1).
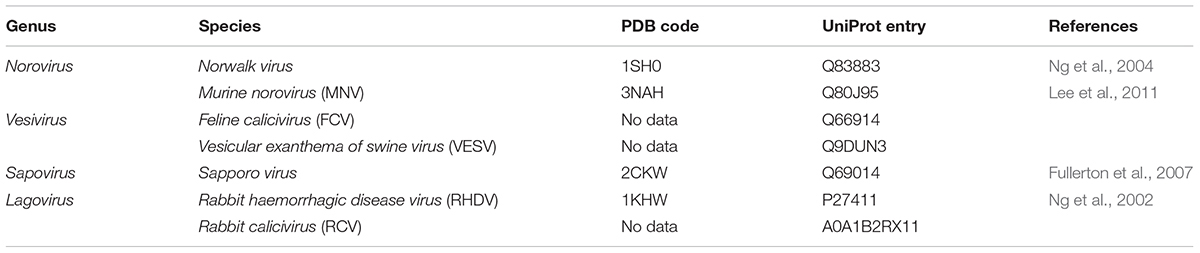
Table 1. Polymerase crystal structures and amino acid sequence information for representative members of the Caliciviridae family.
Features Common to All Calicivirus RdRps
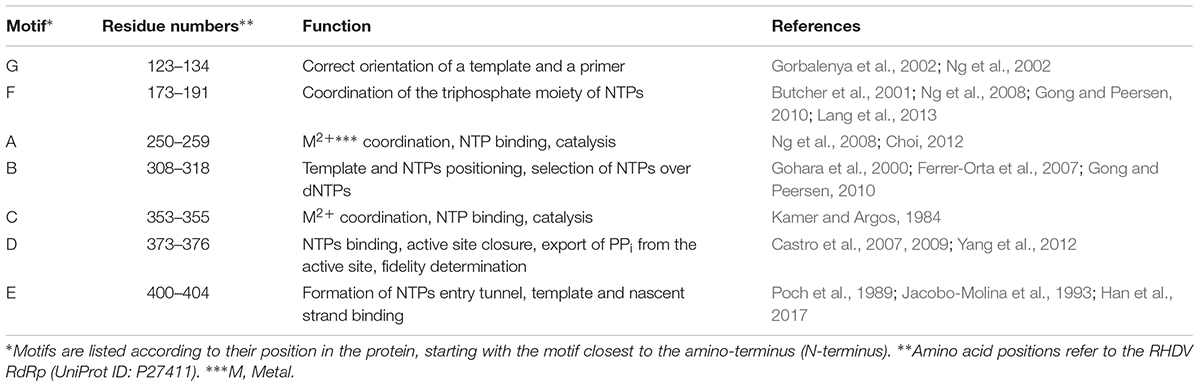
The shape of all RdRps resembles a right hand with fingers, palm, thumb, and an N-terminal domain that links the finger and thumb domains (Figures 3A,B). The active site of the enzyme is located in the palm domain and its architecture is highly conserved. So far, seven highly conserved amino acid sequence motifs have been identified: four motifs in the palm domain (motifs A, B, C, and D), one motif in the thumb domain (motif E), and two motifs in the fingers domain (motifs F and G) (Figures 3A,D; Poch et al., 1989; Koonin, 1991). Whereas these short functional motifs have highly conserved amino acid sequences, the so-called homomorphs encompassing these motifs [except for the newly discovered homomorph H (Černý et al., 2014)] represent protein regions with a conserved structure but no recognizable consensus sequence (Lang et al., 2013; Figure 3C).
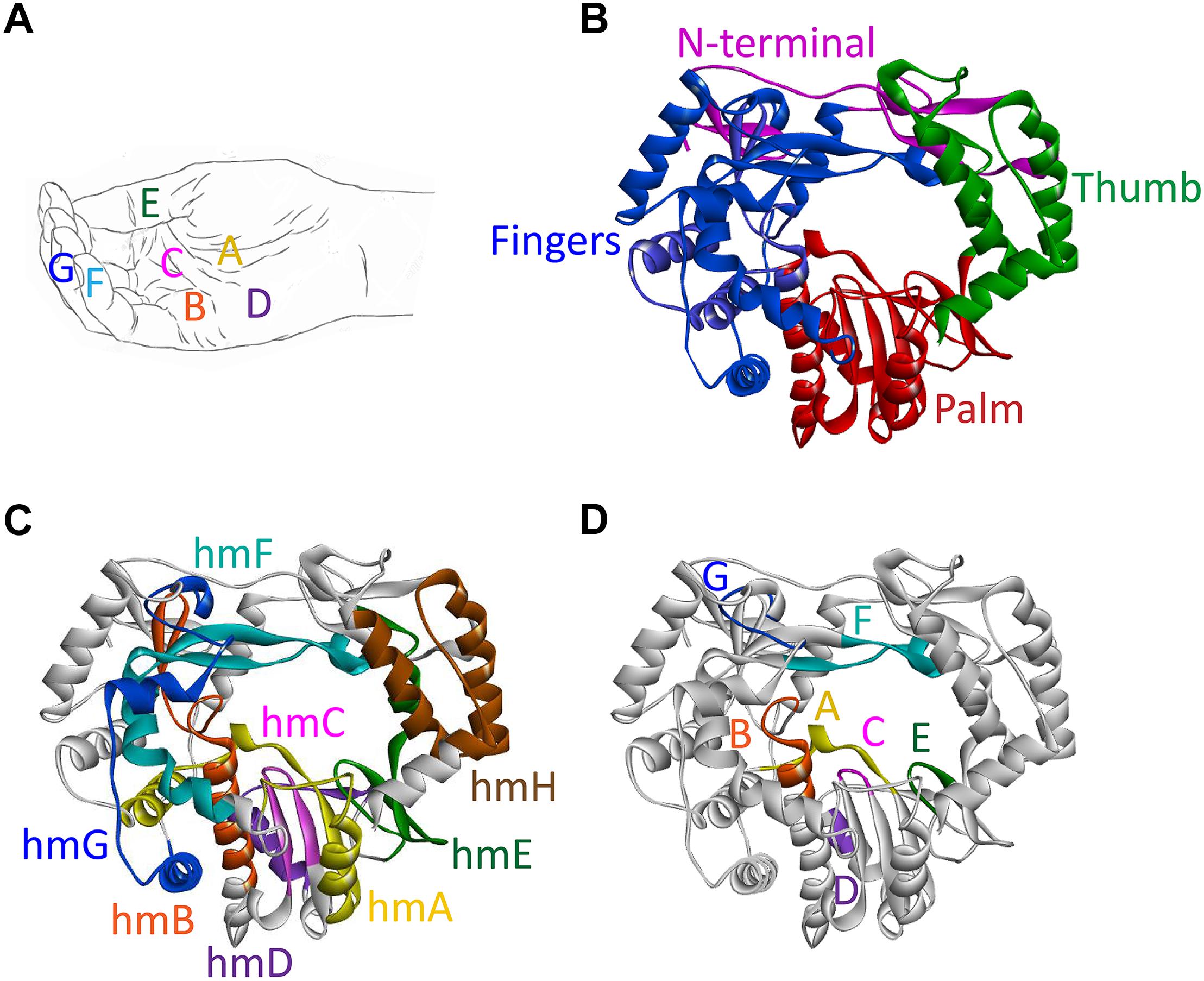
Figure 3. Domains, motifs, and homomorphs of a typical calicivirus RdRp. (A) Representation of a slightly cupped right hand resembling an RdRp with the position of motifs A to G on fingers, palm, and thumb. (B–D) Ribbon diagrams of the RHDV RdRp (PDB ID: 1KHW); (B) fingers, palm, and thumb domains colored blue, red, and green, respectively, and the N-terminal domain colored magenta; (C) structurally conserved homomorphs (hmA to hmH); and (D) functional motifs A to G (the positions of homomorphs and corresponding motifs are indicated by the same color). Ribbon diagrams were generated using Discovery Studio (Dassault Systèmes BIOVIA, Discovery Studio Visualizer v17.2.0, San Diego: Dassault Systèmes, 2016).
Individual motifs cooperate to perform highly specialized functions. Motifs B, D, E, and F are involved in nucleotide recognition and coordination, motifs B and G coordinate template and primer binding, and motifs A and C execute the catalysis of nucleotide binding (Ng et al., 2008; Choi, 2012; Table 2). Motif A comprises two Asp residues separated by up to five amino acids, whereas motif C includes an Asp-Asp dipeptide, forming the highly conserved Gly-Asp-Asp motif (Poch et al., 1989). The Asp residues in motifs A and C coordinate two divalent metal ions that are essential for catalysis, typically Mg2+ or Mn2+. Motif F contains the positively charged residues Arg and Lys that mediate interactions with the triphosphate moieties of incoming nucleoside triphosphates (NTPs) (Butcher et al., 2001; Ng et al., 2008; Gong and Peersen, 2010; Lang et al., 2013). Motif G is located in the template cleft and is involved in protein primer orientation during the initiation of RNA replication (Gorbalenya et al., 2002; Ng et al., 2002).
The thumb domain of calicivirus and picornavirus RdRps is small compared with that of other RdRps and DNA-dependent DNA polymerases. The domain consists of only four helices and forms a relatively large, 15 Å-wide central cleft (also named a channel) that leads to the active site (Ferrer-Orta et al., 2004, 2006). This cleft accommodates both the template and a VPg-linked primer (Choi, 2012).
The main function of RdRps is to copy RNA. This process is based on transferring the α-phosphate moiety of a complementary nucleotide to the 3′-OH end of the nascent strand. This reaction depends on two divalent metal ions (Mn2+ or Mg2+) in the active site. The metal ions are coordinated by the Asp residues of motifs A and C. One of the ions interacts with the 3′-OH group of the primer, which reduces the affinity of this group for the hydrogen and enables a nucleophilic attack of the negatively charged 3′-O- on the α-phosphate residue of the incoming complementary nucleotide (Steitz, 1998). The second metal ion is involved in positioning the incoming NTP and the release of a pyrophosphate (PPi). As a result of the nucleophilic attack, a new phosphoester bond between the 3′-OH terminal group of the protein-linked primer and the α-phosphate of nucleoside monophosphate (NMP) is created and PPi is released (Joyce and Steitz, 1995; Steitz, 1998).
Structural and Functional Characteristics of Norovirus and Lagovirus RdRps
Noroviruses
The overall structure of norovirus RdRps is similar to that of other caliciviruses, but some differences exist (Figures 4A–D). For example, the carboxyl terminus (C-terminus) of the protein is located within the active site cleft close to the two catalytic Asp residues (Ng et al., 2004; Figure 4A). Therefore, the C-terminus is suitably positioned to take part in the initiation of RNA replication. This configuration is similar to that in the RdRps of the Hepatitis C virus (HCV) and the φ6 bacteriophage, in which C-terminal amino acids help to stabilize primers in the active site (Butcher et al., 2001; Laurila et al., 2002; Ranjith-Kumar et al., 2002). This C-terminal addition to the active site would not allow the entry of RNA in the form of a duplex with a long primer, but it does not prevent an interaction of the template with a short dinucleotide primer (Ng et al., 2004). RNA binding to the active site of the norovirus RdRp also causes the rotation of the main helix of the thumb domain (residues 435–449) by 22°, thus forming a suitable groove for a protein-linked primer (Zamyatkin et al., 2008). Sapovirus RdRps share many features with those of noroviruses, e.g., the C-terminus of the sapovirus RdRp is located within the active site cleft (Fullerton et al., 2007; Figure 4D).
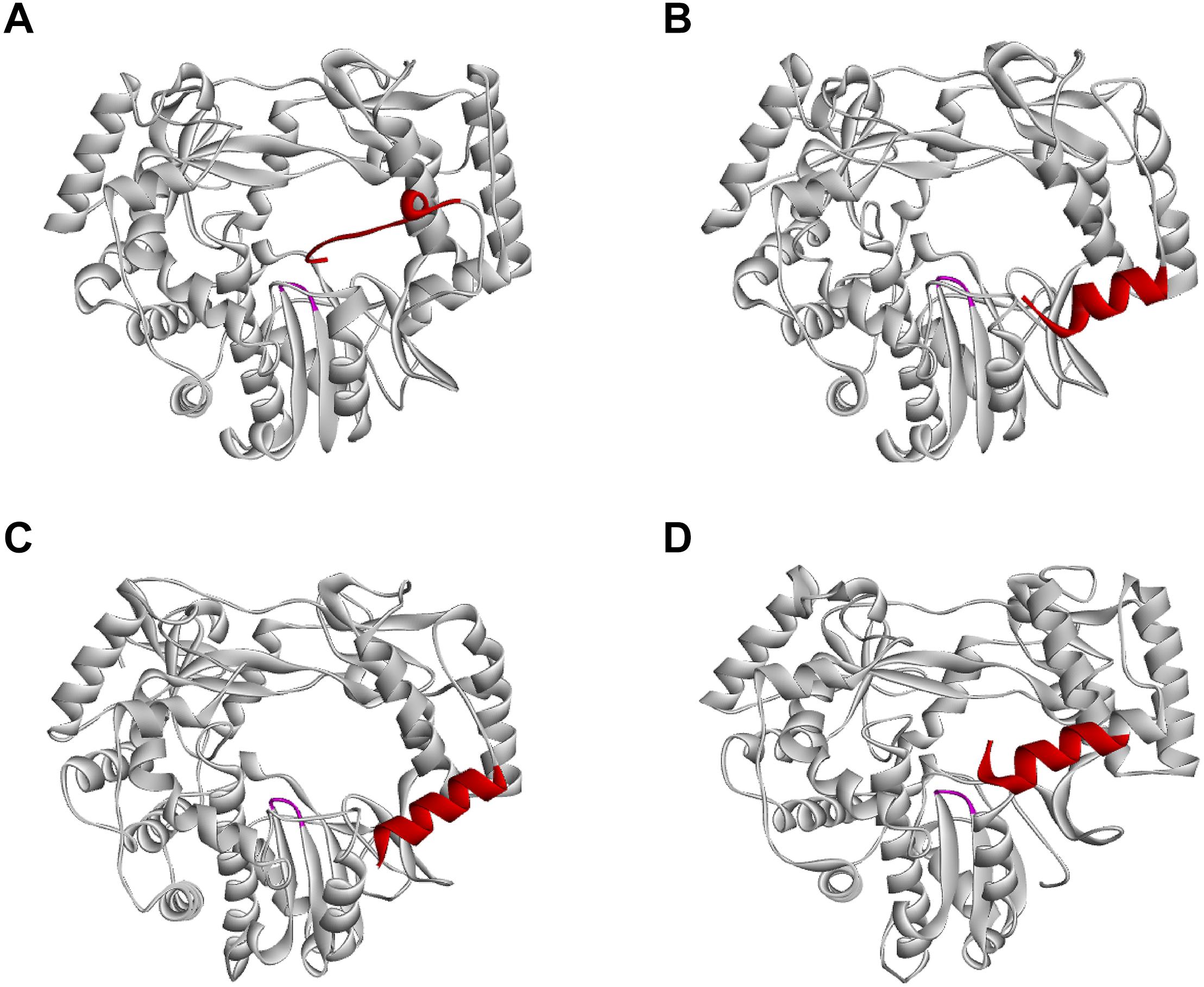
Figure 4. Position of the C-terminus in different calicivirus RdRps. (A) Norwalk virus (PDB ID: 1SH0); (B) MNV (PDB ID: 3NAH); (C) RHDV (PDB ID: 1KHW); (D) Sapporo virus (PDB ID: 1CKW) RdRps, presented as ribbon diagrams. C-terminal amino acids are colored red; the highly conserved motif C is colored magenta. Ribbon diagrams were generated using Discovery Studio (Dassault Systèmes BIOVIA, Discovery Studio Visualizer v17.2.0).
Lagoviruses
Several lines of evidence suggest that functional lagovirus RdRps exist as a 3CD-like precursor protein and a mature protein. Both the in vitro translation of viral RNA with a subsequent precipitation of the products using region-specific antisera, as well as the in vivo analysis of proteins present in RHDV-infected primary hepatocytes revealed a 72 kDa protein corresponding to an uncleaved 15 kDa 3C-protease and 58 kDa polymerase (Martín Alonso et al., 1996; König et al., 1998). Subsequent in vitro studies with recombinant proteins suggest that this 3CD-like precursor possesses both protease and polymerase activities and is able to uridylate VPg (Machín et al., 2009).
Many RNA viruses, including caliciviruses, use cellular membranes to protect and act as a scaffold for their RNA replication machinery (Green et al., 2002). A number of viral proteins recruit intracellular membranes (e.g., p48 of Norwalk virus) but polymerases are usually not involved. One of the most remarkable findings with lagovirus RdRps is their apparent ability to interact with intracellular membranes and to change the architecture of the Golgi apparatus. The expression of recombinant RHDV and RCV RdRps induced a striking rearrangement of cis/medial and medial/trans Golgi membranes (Urakova et al., 2015, 2017a). However, all immunofluorescence studies on the intracellular localization of the recombinant lagovirus RdRps that have been conducted so far have failed to detect a colocalization of RdRps with Golgi (or other) intracellular membranes (Urakova et al., 2015, 2017a). Furthermore, the overexpression of recombinant proteins without viral replication may result in more RdRp proteins being available to change the localization of Golgi membranes (as compared to the situation in virus-infected cells). This may explain why barely detectable amounts of RdRps were observed to be sufficient to induce dramatic changes to the Golgi apparatus (Urakova et al., 2015, 2017b). The enzymatic activity of the RdRp is not required for the RdRp to disaggregate the Golgi apparatus, as active site (motif C) variants with Gly-Asp-Asp to Gly-Asn-Asp and Gly-Asp-Asp to Gly-Ala-Ala substitutions had the same effect on Golgi membranes as proteins with the wild type sequence (Urakova et al., 2017a). The observed Golgi membrane disruption is most likely a consequence of cellular membrane recruitment for the formation of a membranous vesicle network on which virus replication occurs, similar to the membrane recruitment in other caliciviruses and picornaviruses (Schlegel et al., 1996; Green et al., 2002). A hydrophobic motif (residues 189–210) that might be responsible for the interaction with Golgi membranes has been identified (Urakova et al., 2017b; Figure 5A). This motif is located next to the F motif within the F homomorph (this newly identified hydrophobic motif should not be confused with the “classic” conserved motifs A to G). Truncated RHDV RdRp variants without the hydrophobic motif showed a diffuse cytoplasmic localization when expressed in transiently transfected cells. None of these variants accumulated in the distinct cytoplasmic foci that are typical for the intracellular localization of the wild type RdRp (Urakova et al., 2017b). Furthermore, the hydrophobic motif is able to change the localization pattern of other proteins, as it has been demonstrated using the green fluorescent protein (Urakova et al., 2017b). A similar hydrophobic motif was observed in the RdRp of RCV, also in the F homomorph and in the same position as in the RHDV RdRp, but the motif does not exist, or is less obvious in more distantly related caliciviruses (Urakova et al., 2017b). The importance of the hydrophobic amino acids within the motif was demonstrated using variants in which individual Val residues were changed to Ser residues. A variant with two Val to Ser substitutions in the C-terminal part of the motif exhibited a diminished ability to rearrange Golgi membranes, and a variant with four such mutations completely lost this feature (Urakova et al., 2017b).
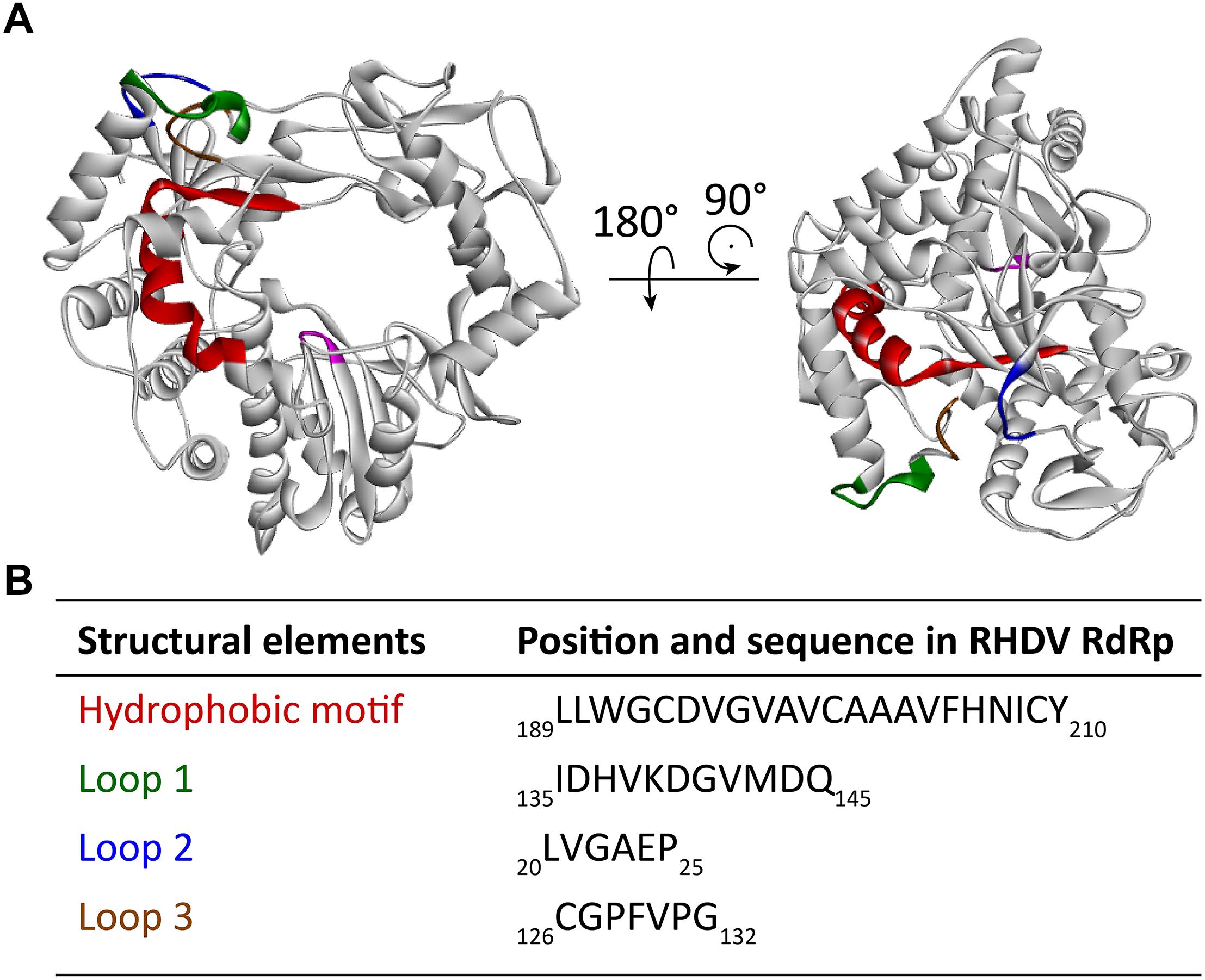
Figure 5. Localization of a partially buried hydrophobic membrane interaction motif in the RHDV RdRp. (A) Ribbon diagrams of the RHDV RdRp (PDB ID: 1KHW). The hydrophobic motif is colored red, loop 1 green, hydrophobic loop 2 blue, and hydrophobic loop 3 brown. The active site (motif C) is highlighted magenta to provide a reference point for the position of the hydrophobic motif in the RdRp. (B) Amino acid positions and sequences of the structural elements highlighted in the diagrams above. Ribbon diagrams were generated using Discovery Studio (Dassault Systèmes BIOVIA, Discovery Studio Visualizer v17.2.0).
Research into the newly identified hydrophobic motif revealed an unexpected structural flexibility of calicivirus RdRps, as the exposure of the partially buried hydrophobic motif requires a series of conformational changes. Molecular dynamics simulations suggest that four regions surrounding the motif possess a high degree of mobility (loops 1–3 in Figure 5B). Two of these loops (loops 1 and 2) flank the “point of access” to the motif, and the third loop covers the motif, much like a “trap door.” The following sequence of movements is thought to bring the RdRp close to an intracellular membrane and allow exposure of the hydrophobic motif (Urakova et al., 2017b): firstly, three collinear, positively charged Lys residues at the edge of a solvent-exposed helix next to the loop 1 interact with the negatively charged surface of the membrane. Next, hydrophobic interactions, including those between the partially hydrophobic loop 2 and the membrane, draw the protein further toward the membrane to a point, at which hydrophobic loop 3 makes contact with the membrane, moves out of the way, and allows the hydrophobic motif to become exposed and to insert itself into the membrane.
Genomic and Subgenomic RNA Replication
Detailed studies on calicivirus replication and pathogenicity often lag behind those in other RNA virus families. For decades, studies on human norovirus and other enteric caliciviruses have been hampered by the lack of a robust cell culture system. Of note, it has been reported that replication competent RHDV RNAs can be generated from plasmids using a T7 promotor and a hepatitis D virus ribozyme (Zhu et al., 2017), but these findings have not yet been independently reproduced. Very recently, however, ground-breaking progress was made in enteroid cultivation methods that show great potential for providing new cell culture systems for noroviruses and lagoviruses (Jones et al., 2015; Ettayebi et al., 2016). The new methods complement and supplant previously developed cell culture models for MNV that relied on bone marrow-derived murine macrophages and dendritic cells. These MNV cell cultures were used as surrogate models to study human noroviruses (Wobus et al., 2004, 2006). However, there is still no general agreement on certain steps of the calicivirus replication process, such as the mechanism of the replication initiation.
Terminal Transferase Activity of RdRps
Terminal transferase activity is the ability to add nucleotides to the 3′ end in a template independent manner. Similar to poliovirus (Arnold et al., 1999) and HCV RdRps (Ranjith-Kumar et al., 2001), human norovirus RdRps possess terminal transferase activity (Rohayem et al., 2006a). The activity is thought to serve as a repair system for 3′ ends that were damaged by cellular exonucleases and, in some cases, it facilitates the initiation of RNA synthesis through the addition of non-templated nucleotides (Wu and Kaper, 1994). For example, the terminal adenylyl transferase activity of the poliovirus 3D polymerase restores the infectivity of poliovirus RNA genomes that lack a poly(A) tail (Neufeld et al., 1994). The terminal transferase activity of calicivirus RdRps generates not only a protective poly(A) tail but may also generate a poly(C) tail that has been suspected to play a critical role in the initiation of genomic and subgenomic RNA synthesis.
Initiation of RNA Synthesis
In calicivirus replication, the initiation of RNA synthesis occurs differently for genomic and antigenomic RNAs (Figures 6A,B,D). The former may occur in a primer independent manner (Rohayem et al., 2006b), and the latter is generally thought to depend on a nucleotidylated VPg primer. However, according to an article by Subba-Reddy et al. (2012), the synthesis of negative-sense RNAs can be initiated de novo. This article was later retracted (Subba-Reddy et al., 2017), although the authors maintain that the overall conclusion in the original article is still valid (“[…] that the norovirus major capsid protein could modulate norovirus RdRp activity has been confirmed in both the Kao and the Goodfellow labs”). Clearly, more research is warranted to better understand the initiation of norovirus RNA synthesis. Interestingly, the VPg of human noroviruses consists of 133 amino acids, which is substantially larger than the 22 amino acids of the homologous protein in picornaviruses. Nevertheless, the larger size does not prevent the norovirus VPg from serving as a protein primer. As shown by Rohayem et al. (2006b) in a series of in vitro experiments, the norovirus RdRp is able to initiate synthesis on subgenomic polyadenylated RNA that carries a VPg protein in the absence of a short poly(U) RNA primer but in the presence of VPg, confirming the notion of a protein-primed initiation. For anti-subgenomic RNA replication to occur, a protein primer is probably not required, as an RNA product is yielded in the absence of exogenous primers (Rohayem et al., 2006b). Interestingly, a poly(C) stretch was detected at the 3′ terminus of newly produced anti-subgenomic norovirus RNA, which may have been the result of a terminal transferase activity of the norovirus RdRp (Figure 6C). Based on these findings, it was proposed that the initiation of subgenomic RNA synthesis occurs de novo on a short 3′-terminal poly(C) stretch of anti-subgenomic RNA, a scenario that is in line with the general observation that the de novo initiation on RNA genomes usually starts on pyrimidines (Kao et al., 2001; Rohayem et al., 2006b). However, other evidence suggests that the synthesis of positive-sense genomic and subgenomic RNAs also depends on a VPg primer, since infectious (positive-sense) RNAs isolated from MNV-infected cells are linked to VPg (Chaudhry et al., 2006). Similar in vitro studies were performed using the 3′-terminal region of genomic norovirus RNA. In these studies, the norovirus RdRp was shown to initiate the synthesis of antigenomic RNA primer-independently (Fukushi et al., 2004). Moreover, a 3′-terminal region without the poly(A) sequence was replicated as effectively as RNA with the original sequence, indicating that a poly(A) sequence is not necessary for the initiation of negative strand RNA synthesis (Fukushi et al., 2004). With sapoviruses, initiation of RNA synthesis was also observed to be different for subgenomic polyadenylated RNA and anti-subgenomic RNA. In the case of subgenomic RNA synthesis, replication initiation is primer independent, whereas the synthesis of anti-subgenomic RNA is strictly primer dependent and occurs only when an oligo(U) primer is added to the reaction (Fullerton et al., 2007). Terminal transferase activity was also shown to be a feature of sapovirus RdRps (Fullerton et al., 2007).
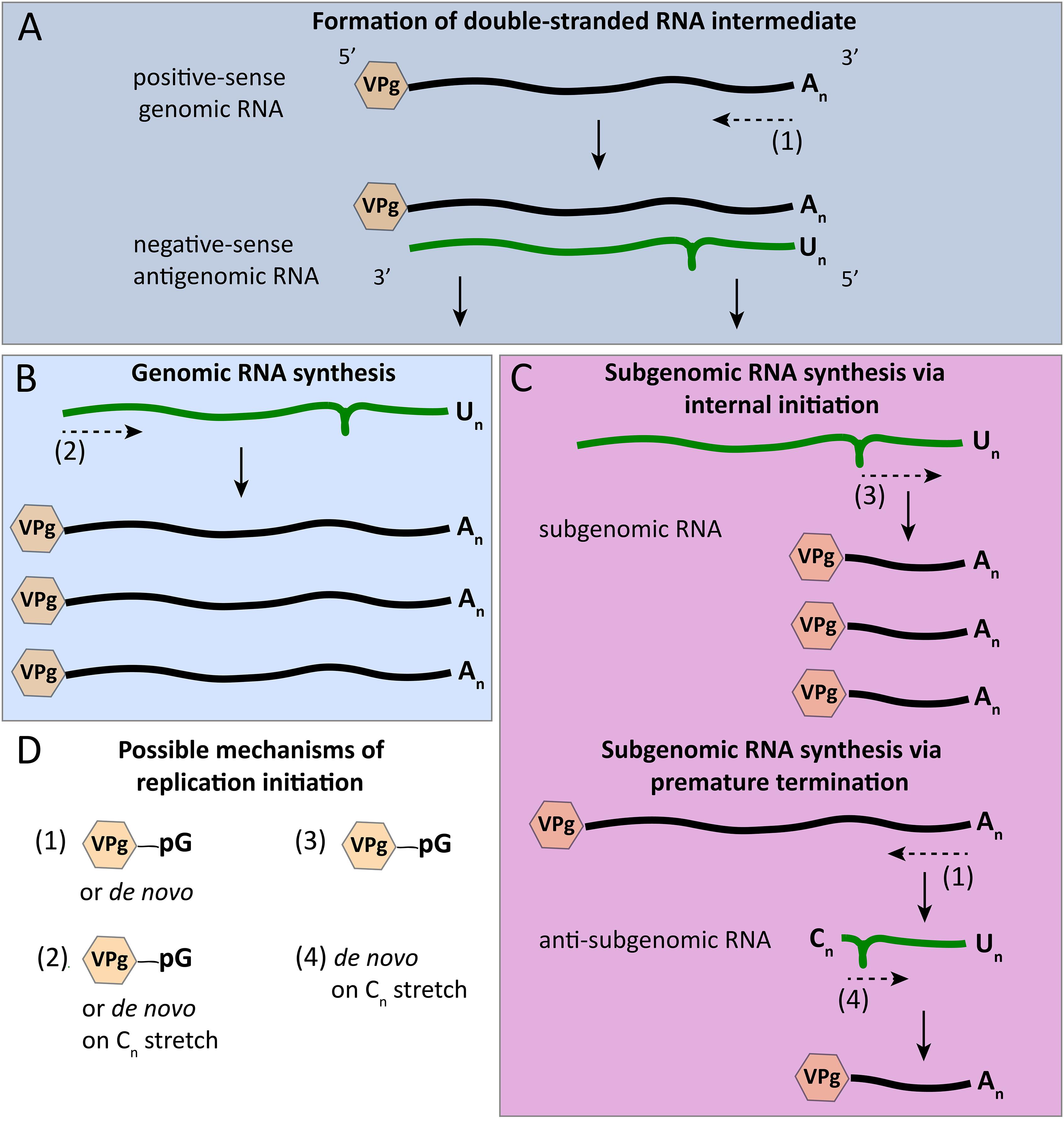
Figure 6. Initiation modes for RNA synthesis during calicivirus replication. (A) The synthesis of antigenomic RNA results in the formation of a double-stranded RNA intermediate; antigenomic RNA synthesis is initiated in a VPg-dependent manner or de novo. (B) The synthesis of new genomic RNA was described to start either de novo or from a poly(C) stretch of nucleotides that were added by the RdRp’s terminal transferase activity. (C) The synthesis of subgenomic RNA may be initialized internally using a stem loop in the negative-sense antigenomic RNA and VPg priming; according to an alternative mechanism, a premature termination of antigenomic RNA synthesis results in anti-subgenomic RNA that is then used as a template for subgenomic RNA synthesis, a process that is suggested to involve a poly(C) stretch similar to the proposed initiation of genomic RNA synthesis. (D) Overview of the various mechanisms that were postulated for the initiation of calicivirus RNA synthesis. Green and black lines symbolize negative- and positive-sense RNAs, respectively; the loop in negative-sense RNAs indicates the position of a stem loop that may act as a subgenomic promoter region; dashed arrows indicate the initiation point and direction of RNA synthesis; hexagons represent VPg proteins that are covalently bound to the 5′ end of all positive-sense RNAs; pG indicates guanylation; An, Un, and Cn represent poly(A), poly(U), and poly(C) sequences, respectively.
Binding of Promoter Regions and Other RdRp-RNA Interactions
The initiation of RHDV subgenomic RNA replication was studied in great detail and these observations may guide a better understanding of calicivirus promoters. There are two possible mechanisms for the synthesis of subgenomic RNA (Figure 6C). It can either be through an internal initiation on a negative strand of genomic RNA, or through a premature termination of genomic negative strand RNA synthesis. The latter would result in negative-sense subgenomic RNA that can be used as a template for positive-sense subgenomic RNA production (Sit et al., 1998; Miller and Koev, 2000). Subgenomic RNA replication in RHDV was shown to be initialized internally on negative strand genomic RNA, and a suitable promoter region was discovered upstream of the subgenomic RNA synthesis start site (Morales et al., 2004). The localization and extent of this subgenomic RNA promoter region was analyzed by constructing deletion mutants with truncated 3′-terminal sequences on the negative strand genomic RNA. At least 50 nucleotide residues preceding the start of the subgenomic RNA were required for subgenomic RNA production (Morales et al., 2004).
Subsequent studies revealed a stable and evolutionarily conserved stem-loop in the negative strand of genomic RNA of all caliciviruses that is located six nucleotides upstream of the start of the subgenomic RNA in the RdRp coding region (Simmonds et al., 2008). The role of this stem loop in subgenomic RNA synthesis was studied by the introduction of nucleotide substitutions in the stem-loop sequence of an MNV replicon that contained the Renilla luciferase gene fused to the foot-and-mouth disease virus (FMDV) 2A protease coding sequence ahead of the VP2-coding region. These reporter replicon variants were used to quantify subgenomic RNA synthesis. Replicons with mutations in the stem-loop produced less luciferase compared with wild type MNV replicons, but similar amounts to a replication-defective replicon. The amount of subgenomic RNA was determined using a primer extension assay, in which a radiolabeled primer complementary to the 5′ region of subgenomic RNA was used to generate a product corresponding to the start of the subgenomic RNA. Subgenomic RNA was detected in cells transfected with the wild type MNV genome but was absent in those transfected with a replicon bearing mutations in the stem-loop region. These results confirm the hypothesis that the stem-loop in the RdRp coding region is essential for the initiation of subgenomic RNA synthesis (Yunus et al., 2015).
In the search for the protein region that is involved in RNA recognition and binding, several amino acid residues of the MNV RdRp that potentially interact with genomic RNA were identified: Lys169, Lys183 and 184, Arg185, Lys210, Arg395, and 396, and Lys422. These positively charged amino acid residues are located adjacent to the active site and well conserved across the Caliciviridae family. Using site-directed mutagenesis, seven MNV variants were created, in which positively charged amino acids were substituted with a non-polar Ala (Han et al., 2017). The effect of these substitutions on protein-RNA interactions was examined using electrophoretic mobility shift assays, and the impact of these substitutions on RNA replication was studied in cell culture. The results demonstrate that RdRp variants with Ala substitutions interact with the RNA less efficiently and are either non-viable or replicate with reduced efficiency (Han et al., 2017).
Finally, the FCV polymerase-protease precursor protein was found to interact with the ORF2 region of the viral genome. ORF2 encodes the major structural protein VP1 and the LC protein. This interaction is suspected to be required for the encapsidation of the viral RNA, although this is yet to be proven (Kaiser, 2006).
RdRp-Mediated VPg Nucleotidylation
VPg nucleotidylation is catalyzed much more efficiently by the human norovirus protease-polymerase precursor protein than by the mature enzyme (Medvedev et al., 2017). While nucleotidylation by the precursor protein occurs without a poly(A) template, the mature RdRp requires such a template (Rohayem et al., 2006b; Belliot et al., 2008). Unlike the poliovirus protease-polymerase precursor 3CD that shows only protease activity, the homolog of human noroviruses possesses both protease and polymerase activities, is able to initiate RNA synthesis, and can elongate the nascent RNA (Belliot et al., 2005). The FCV RdRp precursor protein was also suspected to be an active polymerase, because infected cells contain more of the uncleaved precursor than the mature enzyme (Sosnovtseva et al., 1999). Subsequent experiments confirmed that the precursor indeed has polymerase activity. The deletion of 164 amino acids from the amino terminus of the precursor only caused a threefold decrease in polymerase activity, but the deletion of the next amino acid resulted in a 90-fold reduction (Wei et al., 2001). This study defines the boundary of the active form of the FCV RdRp that is located either at Val135 or further toward the amino terminus (Wei et al., 2001).
During FCV replication, a direct interaction of the VPg with the polymerase-protease precursor protein was detected in an ELISA-based assay, in which purified VPg was adsorbed to the wells and the recombinant precursor protein was passed over (Leonard et al., 2000; Kaiser, 2006). The results support the idea of a protein-primed initiation of replication, a concept that was further validated by research into RHDV replication. The RdRp of RHDV also transfers nucleotides to VPg (Machín et al., 2001). Moreover, the RHDV RdRp precursor (p72) catalyzed VPg uridylation more actively than the mature enzyme, although the mature form showed a higher in vitro polymerization activity when a heteropolymeric RNA was used as a template (Machín et al., 2009). Mutational analysis of the FCV VPg further confirmed the hypothesis of a protein-primed initiation of replication: the substitution of Tyr24 to Ala (as well as to Thr, Phe, and Ser) was lethal for the virus (Mitra et al., 2004). A Tyr in this position is believed to be essential for the VPg uridylation in FCV, similar to Tyr21 in the RHDV VPg. The critical amino acid in the RHDV VPg was detected by the deletion of the first 21 N-terminal residues, which completely stopped uridylation. When Tyr21 was substituted either by Phe, Ser, or Thr, the resulting variants were no longer uridylated, due to steric hindrances (in the case of Ser and Thr substitutions), or the lack of a hydroxyl group (in the case of Phe) that is needed as a nucleophile in the uridylation reaction (Machín et al., 2001).
The substrate specificity of RdRps varies. For example, human norovirus RdRps nucleotidylate only human norovirus VPgs, whereas the RdRp of MNV efficiently nucleotidylates the VPgs of both human and murine noroviruses (Min et al., 2012). For the MNV RdRp, this reaction can be enhanced by in vitro-transcribed positive and negative strand subgenomic RNAs. Of all RNAs tested, it was the ORF3 region of the subgenomic negative strand RNA that stimulated nucleotidylation most effectively, indicating that the ORF3 region contains a cis-acting element that stimulates the reaction (Han et al., 2010).
Interactions of RdRps and Other Proteins
Viral Interaction Partners
Many calicivirus protein-protein interactions have been investigated using MNV, because this virus can be propagated in cell culture (Wobus et al., 2004). VPg clearly needs to interact with the calicivirus RdRp. However, this interaction also occurs independent of VPg-priming, because VPg variants that lack the Tyr residue needed for the nucleotidylation process still enhanced the replication process in vitro (Lee et al., 2018).
Further protein-protein interactions were detected using a cell-based assay in which the human norovirus GII.4 RdRp was assessed for its ability to synthesize RNA (Subba-Reddy et al., 2011). The assay uses the ability of various cellular pattern recognition receptors, such as the retinoic acid-inducible gene I (RIG-I) to detect viral RNA to activate and the expression of interferon (IFN)-regulated genes (Patel et al., 2003; Honda et al., 2005). Measuring the luciferase production that is driven by an IFN-β promoter can thus be used to quantify viral RdRp activity, as an increasing virus RNA concentration correlates with an increasing expression of IFN-regulated genes (Subba-Reddy et al., 2011). Using this rather indirect reporter assay, Subba-Reddy and coworkers investigated which viral proteins had stimulatory or inhibitory effects on RNA replication. The researchers reported a stimulatory effect for the human norovirus non-structural protein p48 and the structural protein VP1, and an inhibitory effect for VP2. But when these GII.4 proteins were co-expressed with the RdRps of other viruses, they did not significantly increase virus RNA replication (again quantified via the expression of the IFN-β-dependent reporter), suggesting that these calicivirus protein-protein interactions occur in a species-specific manner (Subba-Reddy et al., 2012, 2017). Further experiments identified the part of the VP1 protein that is responsible for the interaction with RdRp. VP1 consists of two domains, a shell domain and a protruding domain. The co-expression of the RdRp with a series of truncated VP1 proteins revealed that the shell domain was sufficient to modulate the enzyme activity. The findings were confirmed using MNV replicons: the transfection of cells with a replicon defective for VP1 expression showed impaired replication, but when VP1 expression was restored by in trans-complementation, virus replication was rescued. Presumably, this positive feedback (the more positive-sense RNA is synthesized, the more VP1 is translated) slows at the point when VP1 starts to multimerize and assemble into new capsids, which prevents its interaction with RdRp and stops the stimulation of RNA synthesis (Subba-Reddy et al., 2012, 2017).
Cellular Interaction Partners
Only caliciviruses that grow in cell culture, such as FCV and MNV (Wobus et al., 2006; Vashist et al., 2009), allow investigations of RdRp interactions with cellular proteins during genuine virus replication. A redistribution of nucleolin from the nucleoli to the nucleoplasm as well as the perinuclear area was observed in FCV-infected cells. Subsequent studies showed that the FCV RdRp directly interacts and colocalizes with nucleolin, and that this interaction is necessary for efficient virus replication. Given that nucleolin interacts with both RdRp and the 3′ UTR of viral RNAs, it has been suggested that the interaction promotes the formation of replication and/or translation complexes (Cancio-Lonches et al., 2011).
The FCV protease-polymerase precursor inhibits host gene transcription mediated by the cellular RNA polymerase II. The effect was observed using reporter genes under the control of either an endogenous promoter (in this case, the feline IFN-β promoter) or exogenous promoters (simian virus 40, cytomegalovirus, or bacteriophage T7 promoters). Moreover, a domain was identified in the N-terminal region of the protease-polymerase precursor that is responsible for the observed inhibition of the RNA polymerase II (Wu et al., 2016). Similarly, the poliovirus 3C protease shuts off cellular transcription through the cleavage of the TATA-binding protein, which prioritizes the synthesis of viral proteins (Kundu et al., 2005).
Another cellular protein that affects FCV, MNV, and porcine enteric calicivirus (PEC) replication is the lysosomal endopeptidase cathepsin L, a protease that is involved in apoptosis and is primarily located in endosomes. Cathepsin L cleaves the structural protein VP1 of FCV and MNV, and VP2 of PEC. Its inhibition was shown to negatively affect the replication of FCV, MNV, and PEC in cell culture. The effect of cathepsin L inhibition is similar to the inhibition of endosomal acidification (a necessary step during viral entry) and prevents MNV and PEC from endosomal escape. These and possibly other caliciviruses enter host cells via clathrin-mediated endocytosis, thus, it should not come as a surprise that any interference with the endosomal escape of incoming virus particles blocks the initiation of virus replication (Shivanna et al., 2014a,b).
Co- and Post-translational Modifications of Calicivirus RdRps
Co- and post-translational modifications refer to a process in which a protein undergoes enzymatically driven covalent modifications during or following translation. At least some calicivirus RdRps are modified in that manner, e.g., the signaling kinase Akt phosphorylates the norovirus RdRps at residue Thr33 (located at the interface between finger and thumb domains) (Eden et al., 2011). Akt is a serine/threonine protein kinase involved in multiple cellular pathways; it promotes survival through the inhibition of apoptosis and the regulation of the cell cycle (Datta et al., 1999). The consequences of RdRp phosphorylation were studied by comparing the kinetic properties of the wild type enzyme to those of a Thr33 to Glu variant that mimics phosphorylation (Eden et al., 2011). In a de novo GTP incorporation assay that can be used to analyze enzyme kinetics (Bull et al., 2010b), the Thr33 to Glu variant showed a lower maximum enzyme velocity (100 vs. 125 fmol × min-1) and had a lower affinity for the GTP substrate than the wild type, suggesting that phosphorylating Thr33 modulates the activity of the enzyme (Eden et al., 2011).
Oligomerization of RdRps
Norovirus RdRps were shown to form homodimers (Högbom et al., 2009), a phenomenon that had already been described for picornavirus RdRps (Lyle et al., 2002). When different amounts of purified recombinant norovirus RdRp protein were subjected to PAGE in native (non-denaturing) conditions, dimer formation was observed at high protein concentration and subsequently confirmed by a denaturation of the isolated proteins, SDS-PAGE, and Western blotting. The formation of dimers seems to be of biological importance, as norovirus RdRps demonstrate cooperative enzymatic activity. Increasing amounts of RNA can be synthesized in vitro with increasing concentrations of RdRp, until a plateau phase is reached. Högbom and coworkers interpret the data as a shift from active monomeric RdRps to more active dimers. They analyzed the cooperativity between RdRp monomers by calculating the Hill coefficient that, in this case, was determined to have a value greater than one, which is indicative of a positive cooperativity (Högbom et al., 2009).
In MNV, the interaction of RdRp with VPg stimulates RdRp multimerization and formation of large fibril-like structures, a reaction that can be observed by transmission electron microscopy (Lee et al., 2018). Analysis of the crystal structure of the MNV RdRp together with a truncated VPg (consisting of the first 73 amino acids) suggested that two amino acid residues of the RdRp, Asp331, and Leu354, might be involved in the interaction between RdRp and VPg. When Asp331 was changed to Ala and Leu354 to Asp, the resulting RdRp variants were still able to form hexamers in the absence of VPg, but did no longer form higher order protein structures in the presence of VPg. Moreover, the binding affinity of these variants to full length VPg decreased significantly, confirming that Asp331 and Leu354 are critical for the interaction of RdRp with VPg. It has been speculated that the formation of RdRp multimers and tubular fibrils may lead to a better coordination of replication components within larger clusters and thus enhance replication efficiency (Lee et al., 2018).
Enzymatic Properties of Calicivirus RdRps
Polymerase Fidelity, Replication Speed, and Evolutionary Rates
Calicivirus RdRps, as well as the RdRps of other RNA viruses are known to be error-prone enzymes, because they lack the proofreading activities of many DNA polymerases. Approximately one error occurs per replication cycle for RNA viruses compared with one error per 300 cycles for DNA viruses (Drake, 1991, 1993). Comparing studies with different error reporting units is somewhat challenging, but certain trends emerge. The average error rate for HCV (family Flaviviridae) is 3.8 × 10-5, measured as substitutions per nucleotide per cycle of infection (s/n/c) (Sanjuán and Domingo-Calap, 2016; Selisko et al., 2018), and the error frequency of the poliovirus RdRp ranges from 7 × 10-4 to 5.4 × 10-3, as determined by the ratio of non-complementary nucleotides incorporation to the total number of nucleotides (Ward et al., 1988). Similar RdRp error rates were determined for several viruses of the family Caliciviridae, e.g., 6.8 × 10-4 for MNV, 1.6 × 10-4 for sapovirus GI, and 9.0 × 10-4 nucleotide substitutions/site for norovirus GII.4 (Bull et al., 2010b).
RNA-dependent RNA polymerase properties, such as fidelity and replication rate, are important factors that shape virus evolution. For example, RdRps from norovirus GII.4 strains had higher mutation rates (determined using in vitro fidelity assays) compared with those of the closely related but less frequently detected GII.b and GII.7 strains (5.5–9.1 × 10-4 substitutions per site for GII.4 RdRps vs. 1.5 × 10-4 and 2.2 × 10-5 substitutions per site for GII.b and GII.7, respectively). Interestingly, the GII.4 lineage showed an approximately 1.7-fold higher rate of evolution of capsid sequences and a higher frequency of non-synonymous changes compared with non-pandemic norovirus strains (Bull et al., 2010a). Furthermore, Mahar et al. (2013) reported that the acquisition (by recombination) of new GII.3 RdRp variants with higher mutation rates may increase genetic diversity and improve the overall fitness of viral populations under selective pressures. Taken together, a low fidelity rate seems to correlate with a higher evolutionary rate.
The replication rate of a virus is another determinant of viral fitness, since viruses with an increased replication rate can produce more copies of their genome, which would result in more variants even if the RdRp error rate remains the same. For example, the RdRps from the 2006 GII.4 pandemic strains had a higher nucleotide incorporation rate (i.e., they replicated faster) than the recombinant GII.4 RdRps from earlier outbreaks and the US95/96-like pandemic GII.4 strain although the error rates were very similar. The observed increase in the incorporation rate has been associated with the appearance of a mutation outside of the active site, i.e., a Lys291 to Thr substitution in the RdRp finger domain (Bull et al., 2010a). Thus, high mutation and/or replication rate within the GII.4 lineage seem to correlate with the evolution of pandemic strains. However, high replication rates do not always correlate with a high overall fitness of a virus, which suggests that speed needs to be balanced with suitable mutation rates. For example, the GII.7 norovirus lineage, despite having a high replication rate, has a low mutation rate and limited geographic spread (Bull et al., 2010a). It is possible that the speed at which this particular virus replicated was not fast enough to balance its limited ability to produce new variants through the incorporation of mutations.
The contribution of the RdRp to the evolutionary rate of caliciviruses became even more clear with the recent success of recombinant GII.2 and GII.4 viruses that acquired a new polymerase variant. For example, the reemerged recombinant norovirus GII.P16-GII.2 that differs from previous GII.P16-GII.2 strains by 5 amino acids in the RdRp (Ruis et al., 2017), results in high virus loads in feces, possesses a relatively high evolutionary rate (5.5 × 10-3 substitutions/site/year), and has rapidly spread across the world (Ao et al., 2018; Cheung et al., 2019). It has been suggested that the amino acid changes in the new RdRp affect the kinetic properties and the fidelity of the enzyme, but the exact mechanistic details remain unknown.
Genetic recombination events have also been observed between different lagoviruses. RHDV2, originally a virus with moderate virulence and limited geographical range (Le Gall-Reculé et al., 2013), appears to have evolved into a more virulent virus (Capucci et al., 2017), a change that is believed to be at least partially a consequence of recombination with other lagoviruses (Lopes et al., 2015). Some of these recombinant viruses were found to possess the non-structural proteins of benign rabbit calicivirus Australia-1 (RCV-A1)-like viruses (Lopes et al., 2015; Hall et al., 2018). RHDV and RCV-A1 have evolutionary rates of 2.8 × 10-3 and 5.0 × 10-3 substitutions/site/year, respectively (Eden et al., 2015; Mahar et al., 2016). The higher evolutionary rate of RCV-A1 correlates with a higher speed of its RdRp, as determined by in vitro assays (Urakova et al., 2016). It is tempting to speculate that RHDV2 could have acquired a relatively fast polymerase, which may explain its increased virulence and apparent evolutionary success. Within 18 months of its arrival, RHDV2 largely replaced endemic RHDV strains in Australia (Mahar et al., 2017).
The generation of a genetically highly diverse pool of genomes provides an evolutionary advantage, because a diverse virus population can more readily adapt to selective pressures (Domingo, 2002; Lauring and Andino, 2010). If the diversity is the result of a higher error rate, this can also increase the likelihood of acquiring detrimental mutations and it has therefore been suggested that most RNA viruses replicate at the edge of an error threshold that is determined by a complex interplay of several parameters such as genome size, error rates, and replication speed (Duffy et al., 2008). As such, it should not come as a surprise that both increases and decreases in RdRp fidelity can affect viral fitness (Pfeiffer and Kirkegaard, 2005; Xie et al., 2014; Arias et al., 2016; Agol and Gmyl, 2018).
Effects of Temperature, pH, and Salt Conditions on RdRp Performance
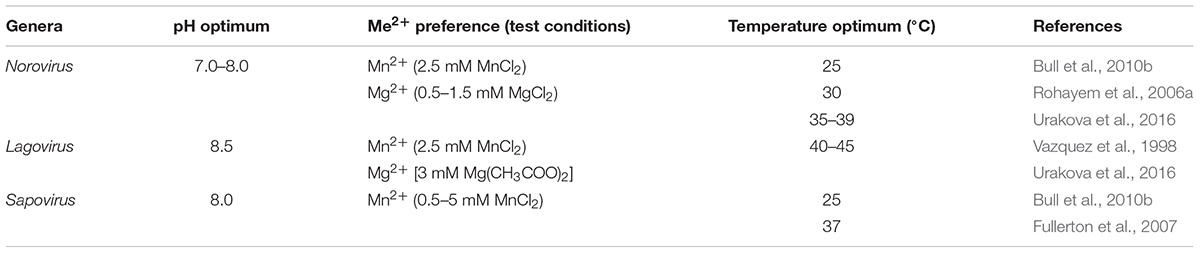
The conditions for an optimal performance of calicivirus RdRps were determined for viruses from the genera Norovirus, Sapovirus, and Lagovirus (Table 3). The activity of viral RdRps is temperature dependent, although the optimal temperature is not necessarily that of the host’s body. In early studies, the highest sapovirus RdRp activity was detected at 37°C (Fullerton et al., 2007). However, more recent studies indicate that many calicivirus RdRps work in an environment that does not allow for maximal performance. For example, a human norovirus RdRp demonstrated a higher activity at 30 than at 37°C according to in vitro assays (Rohayem et al., 2006a). Moreover, when a broader temperature range was studied (i.e., 5, 25, 37, 55, 65, and 75°C) with human norovirus and sapovirus RdRps, the activity was highest at 25°C, and only about 50% of the optimal enzymatic activity was exhibited at 37°C (Bull et al., 2010b). Furthermore, the norovirus and sapovirus RdRps displayed only approximately 20% of their optimal activity at 5°C and only about 1% at 55°C. No activity was detected at 65 or 75°C for any of the RdRps except sapovirus RdRp, which still exhibited 13% of the optimal activity at 65°C (Bull et al., 2010b). Interestingly, the optimal temperature for some if not all lagoviruses is higher than that of human noroviruses and sapoviruses. Using recombinant proteins, it was found that the RdRps of the non-pathogenic RCV and the highly pathogenic RHDV performed best between 40 and 45°C (Urakova et al., 2016), a feature that can be explained as an adaptation of rabbit caliciviruses to their hosts, as the body temperature of healthy rabbits ranges from 38.3 to 39.4°C. Moreover, the fever associated with rabbit haemorrhagic disease often raises the body temperature to 42°C (Strive et al., 2010), but this temperature is not high enough to slow down the activity of the RHDV RdRp (Urakova et al., 2016). The reason why caliciviruses other than lagoviruses seem to possess a temperature optimum that is different from the core body temperature of the host is presently unknown and further research is required to answer this question.
The optimal pH for rabbit calicivirus RdRps was found to be 8.5, which is higher than that of norovirus RdRps (7.0–8.0) (Bull et al., 2010b; Urakova et al., 2016). For optimal catalytic function, the norovirus and lagovirus RdRps can utilize either Mn2+ or Mg2+, but not Fe2+ (Vazquez et al., 1998; Rohayem et al., 2006a; Urakova et al., 2016). Sapovirus RdRp demonstrated a higher activity with Mn2+, but it was also active when Mg2+ was added as a cofactor to the reaction, indicating some flexibility in the use of cofactors (Fullerton et al., 2007).
A Putative Undescribed Conserved Motif in Calicivirus RdRps
Our own sequence comparison of calicivirus RdRps revealed a conserved motif that had not previously been described. This short motif in the RHDV RdRp is located in the thumb domain and consists of four amino acids: 46Pro-Ala-Asn-Leu49 (Figures 7D,E). The flanking amino acids Pro and Leu are significantly conserved, whereas the internal Ala and Asn are not (Figures 7A–C). This motif is present in all calicivirus and picornavirus RdRps, but does not extend beyond the order Picornavirales. We propose to name the new motif “I motif” in accordance with the established nomenclature for previously described motifs and homomorphs. A literature search revealed that several FMDV variants with amino acid substitutions in the region of the I motif have been investigated. Pro36 to Lys, Ala37 to Val, and Leu39 to Phe were all non-viable, supporting the hypothesis that this RdRp region is critical for the enzymatic function of the protein (Xie et al., 2014). Interestingly, an Ala38 to Val substitution changed the fidelity of the FMDV RdRp (Zeng et al., 2014). This variant was selected as ribavirin-resistant during exposure to ribavirin and demonstrated a 1.65-fold increase in fidelity compared with the wild type FMDV (Zeng et al., 2014), a finding that is in line with similar reports on other polymerases (Mansky and Cunningham, 2000; Pfeiffer and Kirkegaard, 2003). While no specific function has as yet been assigned to the I motif, its high level of conservation warrants further investigation. Future studies should be directed at its possible involvement in regulating polymerase fidelity.
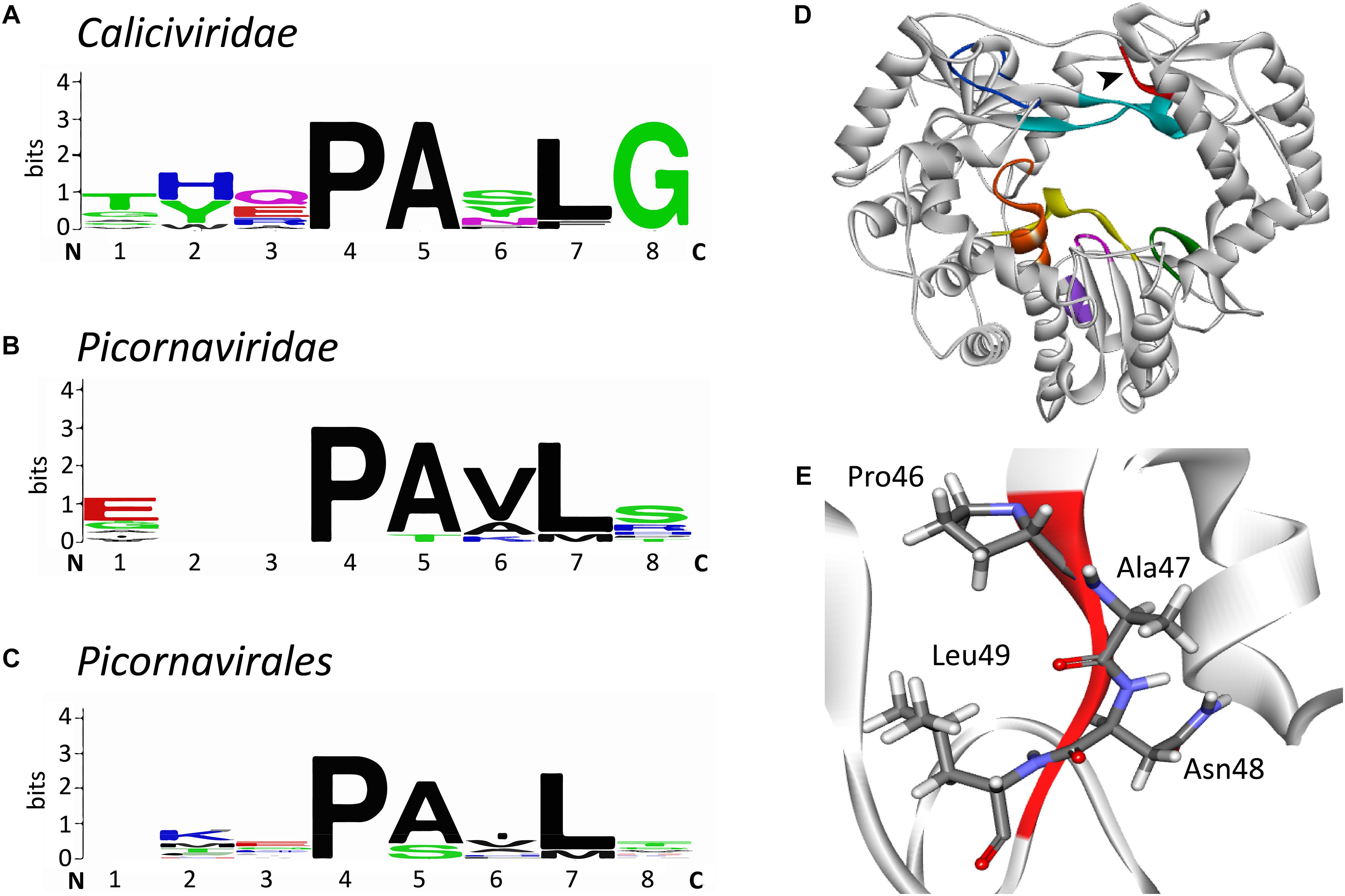
Figure 7. Sequence alignment logos of a putative new conserved motif (“motif I”) and the localization of the motif in the RHDV RdRp. (A) Sequence logo alignment for the putative motif of the following viruses in the family Caliciviridae: European brown hare syndrome virus and Rabbit haemorrhagic disease virus (both genus Lagovirus); Norwalk virus, Lordsdale virus, Murine norovirus (genus Norovirus); Sapporo virus (genus Sapovirus); Feline calicivirus, Vesicular exanthema of swine virus, and San Miguel sea lion virus (genus Vesivirus); Newbury 1 virus (genus Nebovirus). (B) Sequence logo alignment for the putative motif of the following viruses in the family Picornaviridae: Poliovirus, Bovine enterovirus, Coxsackievirus B3, Human rhinovirus A, and Echovirus (genus Enterovirus); Foot and mouth disease virus (genus Aphtovirus); Hepatitis A virus (genus Hepatovirus); Human parechovirus (genus Parechovirus); Theiler’s murine encephalomyelitis virus and Encephalomyocarditis virus (genus Cardiovirus); Avian encephalomyelitis virus (genus Tremovirus). (C) Sequence logo alignment for the putative motif of the following viruses in the order Picornavirales: Poliovirus, Foot and mouth disease virus, Hepatitis A virus, and Human parechovirus (family Picornaviridae); Cricket paralysis virus and Drosophila C virus (family Dicistroviridae); Parsnip yellow fleck virus, Broad bean wilt virus, Cowpea mosaic virus, and Beet ringspot virus (family Secoviridae). N and C indicate N- and C-terminal directions, respectively. Sequence conservation is measured in bits and is indicated by the height of each letter’s stack. Amino acids are colored according to their chemical properties: polar amino acids (Gly, Ser, Thr, Tyr, Cys, Gln, Asn), green; basic (Lys, Arg, His), blue; acidic (Asp, Glu), red; and hydrophobic (Ala, Val, Leu, Ile, Pro, Trp, Phe, Met), black (Crooks et al., 2004). (D) Ribbon diagram of Rabbit haemorrhagic disease virus RdRp (PDB ID: 1KHW). The black arrow head points at the new motif I that is colored red, other conserved motifs are colored as in Figure 3D. (E) Structure of the motif I. Sequence alignments were performed with the multiple sequence alignment tool MUSCLE (Edgar, 2004); sequence logo pictures were created with Weblogo (Crooks et al., 2004). The ribbon diagram was generated using Discovery Studio (Dassault Systèmes BIOVIA, Discovery Studio Visualizer v17.2.0).
Calicivirus RdRp Inhibitors
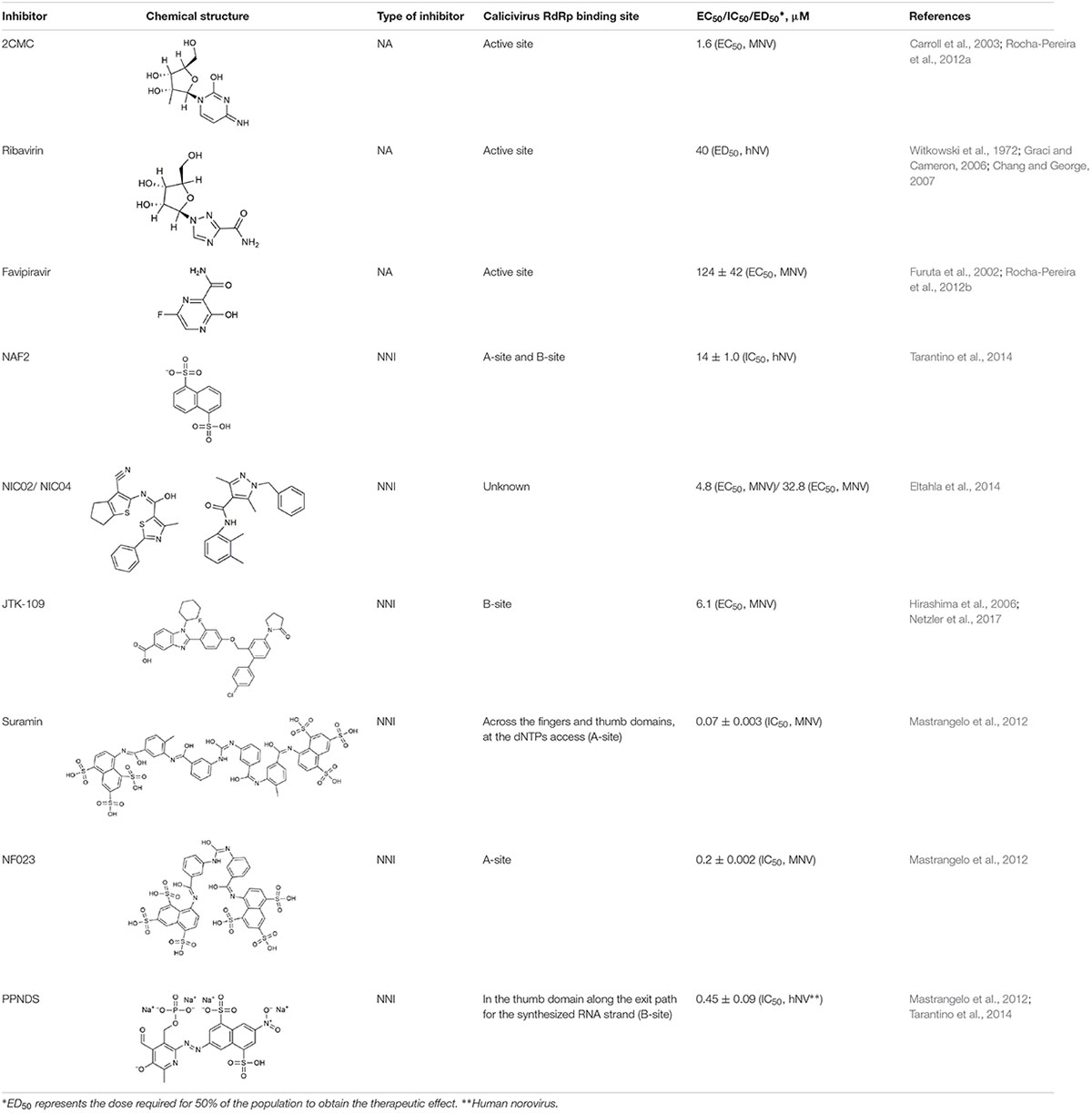
RNA-dependent RNA polymerases are attractive targets for antiviral intervention, because these enzymes are indispensable for virus replication and are very different from any of the host polymerases, which greatly reduces off target effects. RdRp inhibitors can be classified into two major groups: nucleoside analogs (NAs) and non-nucleoside inhibitors (NNIs) (Table 4). NAs are treated by an RdRp as “normal” nucleotides (once an NA is phosphorylated and is in its active form). When they are incorporated into a nascent RNA strand, they can cause a termination of the RNA synthesis or lethal mutagenesis (Galmarini et al., 2001; Costantini et al., 2012). NNIs are aimed to bind an RdRp allosterically, i.e., they bind outside of the active center (Caillet-Saguy et al., 2011; Netzler et al., 2017).
Nucleoside Analogs
2CMC
The active 5′-triphosphate form of 2′-C-methylcytidine (2CMC) is an HCV polymerase inhibitor that competes with the nucleotide cytidine triphosphate (CTP) for binding to the active site of RdRps. Incorporation of 2CMC into a nascent RNA strand leads to the termination of RNA synthesis. In cell culture, this compound is also active against Dengue virus, Yellow fever virus, and West Nile virus (Pierra et al., 2006). 2CMC also inhibits calicivirus replication in cell culture: as demonstrated by time-of-drug-addition assays, 2CMC inhibited MNV replication and plaque formation (Rocha-Pereira et al., 2012a). Furthermore, 2CMC was able to “cure” cultured cells from Norwalk virus replicons (Rocha-Pereira et al., 2013).
Ribavirin
Ribavirin (1-β-D-ribofuranosyl-1,2,4-triazole-3-carboxamide) mimics the guanosine nucleotide and inhibits the replication of a broad range of DNA and RNA viruses (Kanda et al., 2004; Leyssen et al., 2005; Graci and Cameron, 2006). In cell culture experiments, ribavirin significantly reduced norovirus replicon RNA production (Chang and George, 2007). Various mechanisms of the ribavirin-mediated inhibitory effect on virus replication have been proposed, including indirect mechanisms such as guanosine triphosphate (GTP) depletion via the downregulation of inosine monophosphate dehydrogenase, an enzyme that catalyzes GTP synthesis. More direct mechanisms include the ribavirin incorporation into the nascent RNA strand, which may increase mutation frequencies and lead to an “error catastrophe” (Graci and Cameron, 2006).
Favipiravir (T-705)
Originally, T-705 (6-fluoro-3-hydroxy-2-pyrazinecarboxamide), a purine nucleoside analog, was developed as an influenza virus inhibitor. T-705 is a prodrug which is turned into its active form (favipiravir-ribofuranosyl-5′-triphosphate) by cellular enzymes (Furuta et al., 2002, 2013). This compound proved also to be a potent inhibitor of bunyaviruses, arenaviruses, and flaviviruses (Gowen et al., 2007; Morrey et al., 2008). Furthermore, it inhibits MNV replication in cell culture, although at a relatively high EC50 (half maximal effective concentration) (Rocha-Pereira et al., 2012b). The mechanism through which favipiravir inhibits virus multiplication is most probably lethal mutagenesis, because this NA in its active form is incorporated opposite C and U by “susceptible” RdRps (Jin et al., 2013). This hypothesis was confirmed when increased mutation frequencies were observed in MNV-infected mice after the treatment with favipiravir (Arias et al., 2014). Moreover, after the treatment of a human norovirus-infected patient with favipiravir, a distinct viral variant was observed that differed greatly from all variants that were detected before the treatment commenced and contained 118 non-synonymous substitutions (Ruis et al., 2018).
Non-nucleoside RdRp Inhibitors
Suramin, NF023, and PPNDS
Suramin, NF023, and PPNDS are naphthylurea derivatives. Originally, Suramin was developed as a medication for African sleeping sickness and river blindness (Voogd et al., 1993). However, Suramin and NF023 also inhibit a broad range of viruses, including human norovirus and MNV. The calicivirus RdRps were inhibited in a dose-dependent manner indicating a binding of the drug to the free enzyme or enzyme-substrate complex (Mastrangelo et al., 2012). In norovirus RdRps, there are two defined binding sites for naphthylurea derivatives: one site (retrospectively named “A-site”) is located across the fingers and thumb domains where NTPs access the active site (Mastrangelo et al., 2012), the other one is named the “B-site” and is situated in the thumb domain (Tarantino et al., 2014). The A-site was identified by studying the crystal structures of the MNV RdRp in a complex with Suramin and NF023 (Mastrangelo et al., 2012). The B-site is located along the exit path for the synthesized RNA strand and was identified by analyzing a crystal structure of the RdRp in complex with NAF2 (naphthalene-1,5-disulphonic acid). NAF2 is a fragment derived from the aforementioned compounds (Suramin and NF023) and is likely to represent the most active inhibitory part of larger naphthylurea derivatives. The naphthalene sulfonate-based compound pyridoxal-50-phosphate-6-(20-naphthylazo-60-nitro-40,80-disulfonate) tetrasodium salt (PPNDS) was selected in a docking assay (a method widely used to identify drug binding sites) for further characterization of the B-site. Subsequent studies proved that PPNDS inhibits the RdRp at a much lower IC50 (half maximal inhibitory concentration) value compared with NAF2 (Tarantino et al., 2014).
NIC02 and NIC04
Phenylthiazole carboxamide (NIC02) and pyrazole acetamide (NIC04) are novel compounds that inhibit GII.4 norovirus RdRps at relatively low IC50 values. The mode of inhibition was determined by examining the kinetics of GTP incorporation in the presence of increasing concentrations of these inhibitors. With higher inhibitor concentrations, the reaction velocity and the substrate affinity decreased, suggesting that these compounds can bind both free enzyme and the enzyme-substrate complex (Eltahla et al., 2014). The inhibitory potential of both compounds was also tested using calicivirus RdRps from sapovirus GII, MNV, and different human norovirus strains. NIC02 has a broad inhibitory range (as far as calicivirus RdRps are concerned), which suggests that it binds within a conserved but as yet unidentified motif (Eltahla et al., 2014).
JTK-109
JTK-109 is a benzimidazole derivative that is known as an allosteric inhibitor of the HCV RdRp (Hirashima et al., 2006). JTK-109 also possesses inhibitory activity against a variety of caliciviruses (including members of the genera Norovirus, Sapovirus, and Lagovirus), as measured in a quantitative fluorescent de novo RdRp activity assay (Eltahla et al., 2013; Netzler et al., 2017). In cell culture experiments, this compound inhibited MNV plaque formation and virus growth (Netzler et al., 2017). Using molecular docking, (Netzler et al., 2017) showed that JTK-109 targets calicivirus RdRps by binding to the B-site of the thumb domain.
Outlook
Caliciviruses, like almost all other RNA viruses, depend on their RdRps for genome replication. All virus RdRps possess a conserved core structure that is different from cellular polymerases. This dependency on a unique enzyme class provides a soft target for antiviral intervention. However, little information is currently available on how exactly calicivirus RdRps initiate, elongate, and terminate RNA synthesis, how they contribute to the establishment of viral replication factories, and how they escape host cell innate immune responses. Research into the replication of human caliciviruses is especially lacking compared with other small RNA viruses, such as picornaviruses. Norovirus research has long relied on surrogate models or cell-free in vitro studies, but recent progress with organoid culture techniques has provided new models that will soon allow the study of RdRp functions in the context of genuine virus replication and host cell countermeasures. As RdRps are often the principal targets in drug-based antiviral therapies, a better understanding of their enzymatic activities and interactions with viral and cellular partners will likely aid in the development of a new generation of highly effective and more specific antivirals.
Author Contributions
TS and MF developed the conceptual outline. ES, NU, and MF wrote the manuscript (ES wrote the first draft). All authors contributed to editing and revising the manuscript. All authors read and approved the final manuscript.
Funding
ES was supported by a University of Canberra Higher Degree by Research Stipend Scholarship and a CSIRO Postgraduate Studentship.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Robyn Hall and Ina Smith for helpful discussions, Kerry Mills, Andrew Warden, Alexander G. Litov, and Dmitrii Y. Travin for their critical reading of the manuscript.
References
Abente, E. J., Sosnovtsev, S. V., Sandoval-Jaime, C., Parra, G. I., Bok, K., and Green, K. Y. (2013). The feline calicivirus leader of the capsid protein is associated with cytopathic effect. J. Virol. 87, 3003–3017. doi: 10.1128/JVI.02480-12
Abrantes, J., Van Der Loo, W., Le Pendu, J., and Esteves, P. J. (2012). Rabbit haemorrhagic disease (RHD) and rabbit haemorrhagic disease virus (RHDV): a review. Vet. Res. 43:12. doi: 10.1186/1297-9716-43-12
Agol, V. I., and Gmyl, A. P. (2010). Viral security proteins: counteracting host defences. Nat. Rev. Microbiol. 8, 867–878. doi: 10.1038/nrmicro2452
Agol, V. I., and Gmyl, A. P. (2018). Emergency services of viral RNAs: repair and remodeling. Microbiol. Mol. Biol. Rev. 82:e00067-17. doi: 10.1128/MMBR.00067-17
Ao, Y., Cong, X., Jin, M., Sun, X., Wei, X., Wang, J., et al. (2018). Genetic analysis of reemerging GII.P16-GII.2 noroviruses in 2016-2017 in China. J. Infect. Dis. 218, 133–143. doi: 10.1093/infdis/jiy182
Arias, A., Thorne, L., Ghurburrun, E., Bailey, D., and Goodfellow, I. (2016). Norovirus polymerase fidelity contributes to viral transmission in vivo. mSphere 1:e00279-16. doi: 10.1128/mSphere.00279-16
Arias, A., Thorne, L., and Goodfellow, I. (2014). Favipiravir elicits antiviral mutagenesis during virus replication in vivo. eLife 3:3679. doi: 10.7554/elife.03679
Arnold, J. J., Ghosh, S. K. B., and Cameron, C. E. (1999). Poliovirus RNA-dependent RNA polymerase (3D(pol)). Divalent cation modulation of primer, template, and nucleotide selection. J. Biol. Chem. 274, 37060–37069. doi: 10.1074/jbc.274.52.37060
Atmar, R. L., and Estes, M. K. (2006). The epidemiologic and clinical importance of norovirus infection. Gastroenterol. Clin. North Am. 35, 275–290. doi: 10.1016/j.gtc.2006.03.001
Belliot, G., Sosnovtsev, S. V., Chang, K.-O., Babu, V., Uche, U., Arnold, J. J., et al. (2005). Norovirus proteinase-polymerase and polymerase are both active forms of RNA-dependent RNA polymerase. J. Virol. 79, 2393–2403. doi: 10.1128/JVI.79.4.2393-2403.2005
Belliot, G., Sosnovtsev, S. V., Chang, K. O., McPhie, P., and Green, K. Y. (2008). Nucleotidylylation of the VPg protein of a human norovirus by its proteinase-polymerase precursor protein. Virology 374, 33–49. doi: 10.1016/j.virol.2007.12.028
Black, D. N., Burroughs, J. N., Harris, T. J. R., and Brown, F. (1978). The structure and replication of calicivirus RNA. Nature 274, 614–615. doi: 10.1038/274614a0
Boga, J., Marín, M. S., Casais, R., Prieto, M., and Parra, F. (1992). In vitro translation of a subgenomic mRNA from purified virions of the Spanish field isolate AST/89 of rabbit hemorrhagic disease virus (RHDV). Virus Res. 26, 33–40. doi: 10.1016/0168-1702(92)90144-X
Bull, R. A., Eden, J. S., Rawlinson, W. D., and White, P. A. (2010a). Rapid evolution of pandemic noroviruses of the GII.4 lineage. PLoS Pathog. 6:1000831. doi: 10.1371/journal.ppat.1000831
Bull, R. A., Hyde, J., MacKenzie, J. M., Hansman, G. S., Oka, T., Takeda, N., et al. (2010b). Comparison of the replication properties of murine and human calicivirus RNA-dependent RNA polymerases. Virus Genes 42, 16–27. doi: 10.1007/s11262-010-0535-y
Burroughs, J. N., and Brown, F. (1978). Presence of a covalently linked protein on calicivirus RNA. J. Gen. Virol. 41, 443–446. doi: 10.1099/0022-1317-41-2-443
Butcher, S. J., Grimes, J. M., Makeyev, E. V., Bamford, D. H., and Stuart, D. I. (2001). A mechanism for initiating RNA-dependent RNA polymerization. Nature 410, 235–240. doi: 10.1038/35065653
Caillet-Saguy, C., Simister, P. C., and Bressanelli, S. (2011). An objective assessment of conformational variability in complexes of hepatitis C virus polymerase with non-nucleoside inhibitors. J. Mol. Biol. 414, 370–384. doi: 10.1016/j.jmb.2011.10.001
Cancio-Lonches, C., Yocupicio-Monroy, M., Sandoval-Jaime, C., Galvan-Mendoza, I., Ureña, L., Vashist, S., et al. (2011). Nucleolin interacts with the Feline calicivirus 3’ untranslated region and the protease-polymerase NS6 and NS7 proteins, playing a role in virus veplication. J. Virol. 85, 8056–8068. doi: 10.1128/JVI.01878-10
Capucci, L., Cavadini, P., Schiavitto, M., Lombardi, G., and Lavazza, A. (2017). Increased pathogenicity in rabbit haemorrhagic disease virus type 2 (RHDV2). Vet. Rec. 180:426. doi: 10.1136/vr.104132
Carroll, S. S., Tomassini, J. E., Bosserman, M., Getty, K., Stahlhut, M. W., Eldrup, A. B., et al. (2003). Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J. Biol. Chem. 278, 11979–11984. doi: 10.1074/jbc.M210914200
Castro, C., Smidansky, E., Maksimchuk, K. R., Arnold, J. J., Korneeva, V. S., Gotte, M., et al. (2007). Two proton transfers in the transition state for nucleotidyl transfer catalyzed by RNA- and DNA-dependent RNA and DNA polymerases. Proc. Natl. Acad. Sci. U.S.A. 104, 4267–4272. doi: 10.1073/pnas.0608952104
Castro, C., Smidansky, E. D., Arnold, J. J., Maksimchuk, K. R., Moustafa, I., Uchida, A., et al. (2009). Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat. Struct. Mol. Biol. 16, 212–218. doi: 10.1038/nsmb.1540
Černý, J., Bolfíková, B. Č, Valdés, J. J., Grubhoffer, L., and Růžek, D. (2014). Evolution of tertiary structure of viral RNA dependent polymerases. PLoS One 9:96070. doi: 10.1371/journal.pone.0096070
Chang, K.-O., and George, D. W. (2007). Interferons and ribavirin effectively inhibit Norwalk virus replication in replicon-bearing cells. J. Virol. 81, 12111–12118. doi: 10.1128/jvi.00560-07
Chaudhry, Y., Nayak, A., Bordeleau, M. E., Tanaka, J., Pelletier, J., Belsham, G. J., et al. (2006). Caliciviruses differ in their functional requirements for eIF4F components. J. Biol. Chem. 281, 25315–25325. doi: 10.1074/jbc.M602230200
Cheung, S. K. C., Kwok, K., Zhang, L. Y., Mohammad, K. N., Lui, G. C. Y., Lee, N., et al. (2019). Higher viral load of emerging norovirus GII.P16-GII.2 than pandemic GII.4 and epidemic GII.17, Hong Kong, China. Emerg. Infect. Dis. 25, 119–122. doi: 10.3201/eid2501.180395
Choi, K. H. (2012). Viral polymerases. Adv. Exp. Med. Biol. 726, 267–304. doi: 10.1007/978-1-4614-0980-9_12
Conley, M. J., McElwee, M., Azmi, L., Gabrielsen, M., Byron, O., Goodfellow, I. G., et al. (2019). Calicivirus VP2 forms a portal-like assembly following receptor engagement. Nature 565, 377–381. doi: 10.1038/s41586-018-0852-1
Cooke, B. D. (2002). Rabbit haemorrhagic disease: field epidemiology and the management of wild rabbit populations. Rev. Sci. Tech. 21, 347–358. doi: 10.20506/rst.21.2.1337
Cooke, B. D., and Fenner, F. (2002). Rabbit haemorrhagic disease and the biological control of wild rabbits, Oryctolagus cuniculus, in Australia and New Zealand. Wildl. Res. 29, 689–706. doi: 10.1071/WR02010
Costantini, V. P., Whitaker, T., Barclay, L., Lee, D., McBrayer, T. R., Schinazi, R. F., et al. (2012). Antiviral activity of nucleoside analogues against norovirus. Antivir. Ther. 17, 981–991. doi: 10.3851/IMP2229
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Datta, S. R., Brunet, A., and Greenberg, M. E. (1999). Cellular survival: a play in three akts. Genes Dev. 13, 2905–2927. doi: 10.1101/gad.13.22.2905
Domingo, E. (2002). Quasispecies theory in virology. J. Virol. 76, 463–465. doi: 10.1128/JVI.76.1.463-465.2002
Drake, J. W. (1991). A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl. Acad. Sci. U.S.A. 88, 7160–7164.
Drake, J. W. (1993). Rates of spontaneous mutation among RNA viruses. Proc. Natl. Acad. Sci. U.S.A. 90, 4171–4175.
Duffy, S., Shackelton, L. A., and Holmes, E. C. (2008). Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 9, 267–276. doi: 10.1038/nrg2323
Eden, J.-S., Read, A. J., Duckworth, J. A., Strive, T., and Holmes, E. C. (2015). Resolving the origin of rabbit hemorrhagic disease virus: insights from an investigation of the viral stocks released in Australia. J. Virol. 89, 12217–12220. doi: 10.1128/JVI.01937-15
Eden, J.-S., Sharpe, L. J., White, P. A., and Brown, A. J. (2011). Norovirus RNA-dependent RNA polymerase is phosphorylated by an important survival kinase, Akt. J. Virol. 85, 10894–10898. doi: 10.1128/JVI.05562-11
Edgar, R. C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Ehresmann, D. W., and Schaffer, F. L. (1977). RNA synthesized in calicivirus infected cells is atypical of picornaviruses. J. Virol. 22, 572–576.
Eltahla, A. A., Lackovic, K., Marquis, C., Eden, J. S., and White, P. A. (2013). A fluorescence-based high-throughput screen to identify small compound inhibitors of the genotype 3a hepatitis c virus RNA polymerase. J. Biomol. Screen. 18, 1027–1034. doi: 10.1177/1087057113489883
Eltahla, A. A., Lim, K. L., Eden, J.-S., Kelly, A. G., Mackenzie, J. M., and White, P. A. (2014). Nonnucleoside inhibitors of norovirus RNA polymerase: scaffolds for rational drug design. Antimicrob. Agents Chemother. 58, 3115–3123. doi: 10.1128/aac.02799-13
Ettayebi, K., Crawford, S. E., Murakami, K., Broughman, J. R., Karandikar, U., Tenge, V. R., et al. (2016). Replication of human noroviruses in stem cell-derived human enteroids. Science 353, 1387–1393. doi: 10.1126/science.aaf5211
Ettayebi, K., and Hardy, M. E. (2003). Norwalk virus nonstructural protein p48 forms a complex with the SNARE regulator VAP-A and prevents cell surface expression of vesicular stomatitis virus G protein. J. Virol. 77, 11790–11797. doi: 10.1128/JVI.77.21.11790
Farkas, T., Sestak, K., Wei, C., and Jiang, X. (2008). Characterization of a rhesus monkey calicivirus representing a new genus of caliciviridae. J. Virol. 82, 5408–5416. doi: 10.1128/JVI.00070-08
Fernandez-Vega, V., Sosnovtsev, S. V., Belliot, G., King, A. D., Mitra, T., Gorbalenya, A., et al. (2004). Norwalk virus N-terminal nonstructural protein is associated with disassembly of the Golgi complex in transfected cells. J.Virol. 78, 4827–4837. doi: 10.1128/JVI.78.9.4827
Ferrer-Orta, C., Arias, A., Escarmís, C., Verdaguer, N., Van Duyne, G. D., and Yang, W. (2006). A comparison of viral RNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 16, 27–34. doi: 10.1016/j.sbi.2005.12.002
Ferrer-Orta, C., Arias, A., Perez-Luque, R., Escarmís, C., Domingo, E., and Verdaguer, N. (2004). Structure of foot-and-mouth disease virus RNA-dependent RNA polymerase and its complex with a template-primer RNA. J. Biol. Chem. 279, 47212–47221. doi: 10.1074/jbc.M405465200
Ferrer-Orta, C., Arias, A., Perez-Luque, R., Escarmis, C., Domingo, E., and Verdaguer, N. (2007). Sequential structures provide insights into the fidelity of RNA replication. Proc. Natl. Acad. Sci. U.S.A. 104, 9463–9468. doi: 10.1073/pnas.0700518104
Fukushi, S., Kojima, S., Takai, R., Hoshino, F. B., Oka, T., Takeda, N., et al. (2004). Poly(A)- and primer-independent RNA polymerase of Norovirus. J.Virol. 78, 3889–3896. doi: 10.1128/JVI.78.8.3889
Fullerton, S. W. B., Blaschke, M., Coutard, B., Gebhardt, J., Gorbalenya, A., Canard, B., et al. (2007). Structural and functional characterization of sapovirus RNA-dependent RNA polymerase. J. Virol. 81, 1858–1871. doi: 10.1109/BIOCAS.2017.8325071
Furuta, Y., Gowen, B. B., Takahashi, K., Shiraki, K., Smee, D. F., and Barnard, D. L. (2013). Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antiviral Res. 100, 446–454. doi: 10.1016/j.antiviral.2013.09.015
Furuta, Y., Takahashi, K., Fukuda, Y., Kuno, M., Kamiyama, T., Kozaki, K., et al. (2002). In vitro and in vivo activities of anti-influenza virus compound T-705. Antimicrob. Agents Chemother. 46, 977–981. doi: 10.1128/AAC.46.4.977-981.2002
Galmarini, C. M., Mackey, J. R., and Dumontet, C. (2001). Nucleoside analogues: mechanisms of drug resistance and reversal strategies. Leukemia 15, 875–890.
Gohara, D. W., Crotty, S., Arnold, J. J., Yoder, J. D., Andino, R., and Cameron, C. E. (2000). Poliovirus RNA-dependent RNA polymerase (3D(pol)): Structural, biochemical, and biological analysis of conserved structural motifs A and B. J. Biol. Chem. 275, 25523–25532. doi: 10.1074/jbc.M002671200
Gong, P., and Peersen, O. B. (2010). Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 107, 22505–22510. doi: 10.1073/pnas.1007626107
Goodfellow, I. (2011). The genome-linked protein VPg of vertebrate viruses – a multifaceted protein. Curr. Opin. Virol. 1, 355–362. doi: 10.1016/j.coviro.2011.09.003
Gorbalenya, A. E., Pringle, F. M., Zeddam, J.-L., Luke, B. T., Cameron, C. E., Kalmakoff, J., et al. (2002). The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J. Mol. Biol. 324, 47–62. doi: 10.1016/S0022-2836(02)01033-1
Gowen, B. B., Wong, M.-H., Jung, K.-H., Sanders, A. B., Mendenhall, M., Bailey, K. W., et al. (2007). In vitro and in vivo activities of T-705 against arenavirus and bunyavirus infections. Antimicrob. Agents Chemother. 51, 3168–3176. doi: 10.1128/AAC.00356-07
Graci, J. D., and Cameron, C. E. (2006). Mechanisms of action of ribavirin against distinct viruses. Rev. Med. Virol. 16, 37–48. doi: 10.1002/rmv.483
Green, K. Y., Mory, A., Fogg, M. H., Weisberg, A., Belliot, G., Wagner, M., et al. (2002). Isolation of enzymatically active replication complexes from feline calicivirus-infected cells. J. Virol. 76, 8582–8595. doi: 10.1128/JVI.76.17.8582-8595.2002
Hall, R. N., Mahar, J. E., Read, A. J., Mourant, R., Piper, M., Huang, N., et al. (2018). A strain-specific multiplex RT-PCR for Australian rabbit haemorrhagic disease viruses uncovers a new recombinant virus variant in rabbits and hares. Transbound. Emerg. Dis. 65, e444–e456. doi: 10.1111/tbed.12779
Han, K. R., Alhatlani, B. Y., Cho, S., Lee, J. H., Hosmillo, M., Goodfellow, I. G., et al. (2017). Identification of amino acids within norovirus polymerase involved in RNA binding and viral replication. J. Gen. Virol. 98, 1311–1315. doi: 10.1099/jgv.0.000826
Han, K. R., Choi, Y., Min, B. S., Jeong, H., Cheon, D., Kim, J., et al. (2010). Murine norovirus-1 3Dpol exhibits RNA-dependent RNA polymerase activity and nucleotidylylates on Tyr of the VPg. J. Gen. Virol. 91, 1713–1722. doi: 10.1099/vir.0.020461-0
Han, K. R., Lee, J.-H., Kotiguda, G. G., Jung, K. H., Chung, M. S., Kang, S., et al. (2018). Nucleotide triphosphatase and RNA chaperone activities of murine norovirus NS3. J. Gen. Virol. 99, 1482–1493. doi: 10.1099/jgv.0.001151
Hirashima, S., Suzuki, T., Ishida, T., Noji, S., Yata, S., Ando, I., et al. (2006). Benzimidazole derivatives bearing substituted biphenyls as hepatitis C virus NS5B RNA-dependent RNA polymerase inhibitors: structure-activity relationship studies and identification of a potent and highly selective inhibitor JTK-109. J. Med. Chem. 49, 4721–4736. doi: 10.1021/jm060269e
Högbom, M., Jäger, K., Robel, I., Unge, T., and Rohayem, J. (2009). The active form of the norovirus RNA-dependent RNA polymerase is a homodimer with cooperative activity. J. Gen. Virol. 90, 281–291. doi: 10.1099/vir.0.005629-0
Honda, K., Yanai, H., Negishi, H., Asagiri, M., Sato, M., Mizutani, T., et al. (2005). IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434, 772–777. doi: 10.1038/nature03464
Hurley, K. F., and Sykes, J. E. (2003). Update on feline calicivirus: new trends. Vet. Clin. North Am. Small Anim. Pract. 33, 759–772. doi: 10.1016/S0195-5616(03)00025-1
Jacobo-Molina, A., Ding, J., Nanni, R. G., Clark, A. D., Lu, X., Tantillo, C., et al. (1993). Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc. Natl. Acad. Sci. U.S.A. 90, 6320–6324. doi: 10.1073/pnas.90.13.6320
Jiang, X., Wang, M., Wang, K., and Estes, M. K. (1993). Sequence and genomic organization of Norwalk virus. Virology 195, 51–61. doi: 10.1006/viro.1993.1345
Jin, Z., Smith, L. K., Rajwanshi, V. K., Kim, B., and Deval, J. (2013). The ambiguous base-pairing and high substrate efficiency of T-705 (Favipiravir) ribofuranosyl 5′-triphosphate towards influenza a virus polymerase. PLoS One 8:68347. doi: 10.1371/journal.pone.0068347
Jones, D. T., Taylor, W. R., and Thornton, J. M. (1992). The rapid generation of mutation data matrices. Bioinformatics 8, 209–213.
Jones, M. K., Grau, K. R., Costantini, V., Kolawole, A. O., De Graaf, M., Freiden, P., et al. (2015). Human norovirus culture in B cells HHS Public Access Author manuscript. Nat. Protoc. 10, 1939–1947. doi: 10.1038/nprot.2015.121
Joyce, C. M., and Steitz, T. A. (1995). Polymerase structures and function: variations on a theme? J. Bacteriol. 177, 6321–6329. doi: 10.1128/jb.177.22.6321-6329.1995
Kaiser, W. J. (2006). Analysis of protein-protein interactions in the feline calicivirus replication complex. J. Gen. Virol. 87, 363–368. doi: 10.1099/vir.0.81456-0
Kamer, G., and Argos, P. (1984). Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 12, 7269–7282. doi: 10.1093/nar/12.18.7269
Kanda, T., Yokosuka, O., Imazeki, F., Tanaka, M., Shino, Y., Shimada, H., et al. (2004). Inhibition of subgenomic hepatitis C virus RNA in Huh-7 cells: ribavirin induces mutagenesis in HCV RNA. J. Viral Hepat. 11, 479–487. doi: 10.1111/j.1365-2893.2004.00531.x
Kao, C. C., Singh, P., and Ecker, D. J. (2001). De novo initiation of viral RNA-dependent RNA synthesis. Virology 287, 251–260. doi: 10.1006/viro.2001.1039
König, M., Thiel, H.-J. J., and Meyers, G. (1998). Detection of viral proteins after infection of cultured hepatocytes with rabbit hemorrhagic disease virus. J. Virol. 72, 4492–4497. doi: 10.3732/ajb.90.5.796
Koonin, E. V. (1991). The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 72, 2197–2206. doi: 10.1099/0022-1317-72-9-2197
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kundu, P., Raychaudhuri, S., Tsai, W., and Dasgupta, A. (2005). Shutoff of RNA polymerase II transcription by poliovirus involves 3C protease-mediated cleavage of the TATA-binding protein at an alternative site: incomplete shutoff of transcription interferes with efficient viral replication. J. Virol. 79, 9702–9713. doi: 10.1128/JVI.79.15.9702-9713.2005
Lang, D. M., Zemla, A. T., and Ecale Zhou, C. L. (2013). Highly similar structural frames link the template tunnel and NTP entry tunnel to the exterior surface in RNA-dependent RNA polymerases. Nucleic Acids Res. 41, 1464–1482. doi: 10.1093/nar/gks1251
Laurila, M. R. L., Makeyev, E. V., and Bamford, D. H. (2002). Bacteriophage φ6 RNA-dependent RNA polymerase. Molecular details of initiating nucleic acid synthesis without primer. J. Biol. Chem. 277, 17117–17124. doi: 10.1074/jbc.M111220200
Lauring, A. S., and Andino, R. (2010). Quasispecies theory and the behavior of RNA viruses. PLoS Pathog. 6:e1001005. doi: 10.1371/journal.ppat.1001005
Le Gall-Reculé, G., Lavazza, A., Marchandeau, S., Bertagnoli, S., Zwingelstein, F., Cavadini, P., et al. (2013). Emergence of a new lagovirus related to Rabbit Haemorrhagic Disease Virus. Vet. Res. 44:81. doi: 10.1186/1297-9716-44-81
Lee, J. H., Alam, I., Han, K. R., Cho, S., Shin, S., Kang, S., et al. (2011). Crystal structures of murine norovirus-1 RNA-dependent RNA polymerase. J. Gen. Virol. 92, 1607–1616. doi: 10.1099/vir.0.031104-0
Lee, J. H., Park, B. S., Han, K. R., Biering, S. B., Kim, S. J., Choi, J., et al. (2018). Insight into the interaction between RNA polymerase and VPg for murine norovirus replication. Front. Microbiol. 9:1466. doi: 10.3389/fmicb.2018.01466
Leonard, S., Plante, D., Wittmann, S., Daigneault, N., Fortin, M. G., and Laliberte, J.-F. (2000). Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlates with virus infectivity. J. Virol. 74, 7730–7737. doi: 10.1128/JVI.74.17.7730-7737.2000
Leyssen, P., Balzarini, J., De Clercq, E., and Neyts, J. (2005). The predominant mechanism by which ribavirin exerts its antiviral activity in vitro against flaviviruses and paramyxoviruses is mediated by inhibition of IMP dehydrogenase. J. Virol. 79, 1943–1947. doi: 10.1128/jvi.79.3.1943-1947.2005
L’Homme, Y., Sansregret, R., Plante-Fortier,É, Lamontagne, A. M., Ouardani, M., Lacroix, G., et al. (2009). Genomic characterization of swine caliciviruses representing a new genus of caliciviridae. Virus Genes 39, 66–75. doi: 10.1007/s11262-009-0360-3
Li, T.-F., Hosmillo, M., Schwanke, H., Shu, T., Wang, Z., Yin, L., et al. (2017). Human norovirus NS3 has RNA helicase and chaperoning activities. J. Virol. 92:e01606-17. doi: 10.1128/jvi.01606-17
Lopes, A. M., Dalton, K. P., Magalhães, M. J., Parra, F., Esteves, P. J., Holmes, E. C., et al. (2015). Full genomic analysis of new variant rabbit hemorrhagic disease virus revealed multiple recombination events. J. Gen. Virol. 96, 1309–1319. doi: 10.1099/vir.0.000070
Lyle, J. M., Bullitt, E., Bienz, K., and Kirkegaard, K. (2002). Visualization and functional analysis of RNA-dependent RNA polymerase lattices. Science 296, 2218–2222. doi: 10.1126/science.1070585
Machín,Á, Martín Alonso, J. M., Dalton, K. P., and Parra, F. (2009). Functional differences between precursor and mature forms of the RNA-dependent RNA polymerase from rabbit hemorrhagic disease virus. J. Gen. Virol. 90, 2114–2118. doi: 10.1099/vir.0.011296-0
Machín,Á, Martín Alonso, J. M., and Parra, F. (2001). Identification of the amino acid residue involved in rabbit hemorrhagic disease virus VPg uridylylation. J. Biol. Chem. 276, 27787–27792. doi: 10.1074/jbc.M100707200
Mahar, J. E., Bok, K., Green, K. Y., and Kirkwood, C. D. (2013). The importance of intergenic recombination in norovirus GII.3 evolution. J. Virol. 87, 3687–3698. doi: 10.1128/JVI.03056-12
Mahar, J. E., Hall, R. N., Shi, M., Mourant, R., Huang, N., Strive, T., et al. (2019). The discovery of three new hare lagoviruses reveals unexplored viral diversity in this genus. Virus Evol. 5. doi: 10.1093/ve/vez005.
Mahar, J. E., Hall, R. N., Peacock, D., Kovaliski, J., Piper, M., Mourant, R., et al. (2017). Rabbit hemorrhagic disease virus 2 (RHDV2; GI.2) is replacing endemic strains of RHDV in the Australian landscape within 18 months of its arrival. J. Virol. 92, 1374–1391. doi: 10.1128/jvi.01374-17
Mahar, J. E., Nicholson, L., Eden, J.-S., Duchêne, S., Kerr, P. J., Duckworth, J., et al. (2016). Benign rabbit caliciviruses exhibit evolutionary dynamics similar to those of their virulent relatives. J. Virol. 90, 9317–9329. doi: 10.1128/JVI.01212-16
Mansky, L. M., and Cunningham, K. S. (2000). Virus mutators and antimutators. Trends Genet. 16, 512–517. doi: 10.1016/S0168-9525(00)02125-9
Martín Alonso, J. M., Casais, R., Boga, J. A., and Parra, F. (1996). Processing of rabbit hemorrhagic disease virus polyprotein. J. Virol. 70,1261–1265.
Mastrangelo, E., Pezzullo, M., Kramer, D., Germani, F., Petazzi, R., Tarantino, D., et al. (2012). Structure-based inhibition of norovirus RNA-dependent RNA polymerases. J. Mol. Biol. 419, 198–210. doi: 10.1016/j.jmb.2012.03.008
McCune, B. T., Tang, W., Lu, J., Eaglesham, J. B., Thorne, L., Mayer, A. E., et al. (2017). Noroviruses co-opt the function of host proteins VAPA and VAPB for replication via a phenylalanine–phenylalanine- acidic-tract-motif mimic in nonstructural viral protein NS1/2. mBio 8, 668–685. doi: 10.1128/mBio.00668-17
McFadden, N., Bailey, D., Carrara, G., Benson, A., Chaudhry, Y., Shortland, A., et al. (2011). Norovirus regulation of the innate immune response and apoptosis occurs via the product of the alternative open reading frame 4. PLoS Pathog. 7:e1002413. doi: 10.1371/journal.ppat.1002413
Medvedev, A., Viswanathan, P., May, J., and Korba, B. (2017). Regulation of human norovirus VPg nucleotidylylation by ProPol and nucleoside triphosphate binding by its amino terminal sequence in vitro. Virology 503, 37–45. doi: 10.1016/j.virol.2017.01.003
Meyers, G., Wirblich, C., and Thiel, H. J. (1991a). Genomic and subgenomic RNAs of rabbit hemorrhagic disease virus are both protein-linked and packaged into particles. Virology 184, 677–686. doi: 10.1016/0042-6822(91)90437-G
Meyers, G., Wirblich, C., and Thiel, H. J. (1991b). Rabbit hemorrhagic disease virus-molecular cloning and nucleotide sequencing of a calicivirus genome. Virology 184, 664–676. doi: 10.1016/0042-6822(91)90436-F
Meyers, G., Wirblich, C., Thiel, H. J., and Thumfart, J. O. (2000). Rabbit hemorrhagic disease virus: Genome organization and polyprotein processing of a calicivirus studied after transient expression of cDNA constructs. Virology 276, 349–363. doi: 10.1006/viro.2000.0545
Miller, W. A., and Koev, G. (2000). Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology 273, 1–8. doi: 10.1006/viro.2000.0421
Min, B. S., Han, K. R., Lee, J. I., and Yang, J. M. (2012). cDNA cloning of Korean human norovirus and nucleotidylylation of VPg by norovirus RNA-dependent RNA polymerase. J. Microbiol. 50, 625–630. doi: 10.1007/s12275-012-2087-4
Mitra, T., Sosnovtsev, S. V., and Green, K. Y. (2004). Mutagenesis of tyrosine 24 in the VPg protein is lethal for feline calicivirus. J. Virol. 78, 4931–4935. doi: 10.1128/JVI.78.9.4931-4935.2004
Morales, M., Bárcena, J., Ramírez, M. A., Boga, J. A., Parra, F., and Torres, J. M. (2004). Synthesis in vitro of rabbit hemorrhagic disease virus subgenomic RNA by internal initiation on (-)sense genomic RNA: mapping of a subgenomic promoter. J. Biol. Chem. 279, 17013–17018. doi: 10.1074/jbc.M313674200
Morrey, J. D., Taro, B. S., Siddharthan, V., Wang, H., Smee, D. F., Christensen, A. J., et al. (2008). Efficacy of orally administered T-705 pyrazine analog on lethal West Nile virus infection in rodents. Antiviral Res. 80, 377–379. doi: 10.1016/j.antiviral.2008.07.009
Neill, J. D. (1990). Nucleotide sequence of a region of the feline calicivirus genome which encodes picornavirus-like RNA-dependent RNA polymerase, cysteine protease and 2C polypeptides. Virus Res. 17, 145–160. doi: 10.1016/0168-1702(90)90061-F
Neill, J. D., Meyer, R. F., and Seal, B. S. (1995). Genetic relatedness of the caliciviruses: San Miguel sea lion and vesicular exanthema of swine viruses constitute a single genotype within the Caliciviridae. J. Virol. 69, 4484–4488.
Netzler, N. E., Enosi Tuipulotu, D., Eltahla, A. A., Lun, J. H., Ferla, S., Brancale, A., et al. (2017). Broad-spectrum non-nucleoside inhibitors for caliciviruses. Antivir. Res. 146, 65–75. doi: 10.1016/j.antiviral.2017.07.014
Neufeld, K. L., Galarza, J. M., Richards, O. C., Summers, D. F., and Ehrenfeld, E. (1994). Identification of terminal adenylyl transferase activity of the poliovirus polymerase 3D pol. J. Virol. 68, 5811–5818.
Ng, K. K. S., Arnold, J. J., and Cameron, C. E. (2008). Structure-function relationships among RNA-dependent RNA polymerases. Curr. Top. Microbiol. Immunol. 320, 137–156. doi: 10.1007/978-3-540-75157-1_7
Ng, K. K. S., Cherney, M. M., Vázquez, A. L., Machín,Á, Martín Alonso, J. M., Parra, F., et al. (2002). Crystal structures of active and inactive conformations of a caliciviral RNA-dependent RNA polymerase. J. Biol. Chem. 277, 1381–1387. doi: 10.1074/jbc.M109261200
Ng, K. K. S., Pendás-Franco, N., Rojo, J., Boga, J. A., Machín,Á, Martín Alonso, J. M., et al. (2004). Crystal structure of Norwalk virus polymerase reveals the carboxyl terminus in the active site cleft. J. Biol. Chem. 279, 16638–16645. doi: 10.1074/jbc.M400584200
Oka, T., Katayama, K., Ogawa, S., Hansman, G. S., Kageyama, T., Ushijima, H., et al. (2005). Proteolytic processing of sapovirus ORF1 polyprotein. J. Virol. 79, 7283–7290. doi: 10.1128/JVI.79.12.7283-7290.2005
Olspert, A., Truve, E., Chaudhry, Y., Hosmillo, M., Peil, L., and Goodfellow, I. (2016). Protein-RNA linkage and posttranslational modifications of feline calicivirus and murine norovirus VPg proteins. PeerJ 4:e2134. doi: 10.7717/peerj.2134
Parra, F., and Prieto, M. (1990). Purification and characterization of a Calicivirus as the causative agent of a lethal hemorrhagic disease in rabbits. J. Virol. 64, 4013–4015.
Patel, K., Bukh, J., Shi, S. T., Romano, P. R., Barber, G. N., Lai, M. M. C., et al. (2003). Triggering the interferon antiviral response through an IKK-related pathway. Science 300, 1148–1151.
Pfeiffer, J. K., and Kirkegaard, K. (2003). A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. PNAS 100, 7289–7294. doi: 10.1073/pnas.1232294100
Pfeiffer, J. K., and Kirkegaard, K. (2005). Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice. PLoS Pathog. 1:e11. doi: 10.1371/journal.ppat.0010011
Pierra, C., Amador, A., Benzaria, S., Cretton-Scott, E., D’Amours, M., Mao, J., et al. (2006). Synthesis and pharmacokinetics of valopicitabine (NM283), an efficient prodrug of the potent anti-HCV agent 2′-C-methylcytidine. J. Med. Chem. 49, 6614–6620. doi: 10.1021/jm0603623
Poch, O., Sauvaget, I., Delarue, M., and Tordo, N. (1989). Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 8, 3867–3874. doi: 10.1093/emboj/16.6.1248
Prasad, B. V. V., Hardy, M. E., Dokland, T., Bella, J., Rossmann, M. G., and Estes, M. K. (1999). X-ray crystallographic structure of the Norwalk virus capsid. Science 286, 287–290. doi: 10.1126/science.286.5438.287
Prasad, B. V. V., Matson, D. O., and Smith, A. W. (1994). Three-dimensional structure of calicivirus. J. Mol. Biol. 240, 256–264. doi: 10.1006/jmbi.1994.1439
Ranjith-Kumar, C. T., Gajewski, J., Gutshall, L., Maley, D., Sarisky, R. T., and Cheng Kao, C. (2001). Terminal nucleotidyl transferase activity of recombinant flaviviridae RNA-dependent RNA polymerases: implication for viral RNA synthesis. J. Virol. 75, 8615–8623. doi: 10.1128/JVI.75.18.8615-8623.2001
Ranjith-Kumar, C. T., Kim, Y.-C., Gutshall, L., Silverman, C., Khandekar, S., Sarisky, R. T., et al. (2002). Mechanism of de novo initiation by the hepatitis C virus RNA-dependent RNA polymerase: role of divalent metals. J. Virol. 76, 12513–12525. doi: 10.1128/JVI.76.24.12513-12525.2002
Rocha-Pereira, J., Jochmans, D., Dallmeier, K., Leyssen, P., Cunha, R., Costa, I., et al. (2012a). Inhibition of norovirus replication by the nucleoside analogue 2’-C-methylcytidine. Biochem. Biophys. Res. Commun. 427, 796–800. doi: 10.1016/j.bbrc.2012.10.003
Rocha-Pereira, J., Jochmans, D., Dallmeier, K., Leyssen, P., Nascimento, M. S. J., and Neyts, J. (2012b). Favipiravir (T-705) inhibits in vitro norovirus replication. Biochem. Biophys. Res. Commun. 424, 777–780. doi: 10.1016/j.bbrc.2012.07.034
Rocha-Pereira, J., Verbeken, E., Jochmans, D., Nascimento, M. S. J., Debing, Y., and Neyts, J. (2013). The viral polymerase inhibitor 2’-C-methylcytidine inhibits norwalk virus replication and protects against norovirus-induced diarrhea and mortality in a mouse model. J. Virol. 87, 11798–11805. doi: 10.1128/jvi.02064-13
Rohayem, J., Jäger, K., Robel, I., Scheffler, U., Temme, A., and Rudolph, W. (2006a). Characterization of norovirus 3Dpol RNA-dependent RNA polymerase activity and initiation of RNA synthesis. J. Gen. Virol. 87, 2621–2630. doi: 10.1099/vir.0.81802-0
Rohayem, J., Robel, I., Jager, K., Scheffler, U., and Rudolph, W. (2006b). Protein-primed and de novo initiation of RNA synthesis by norovirus 3Dpol. J. Virol. 80, 7060–7069. doi: 10.1128/JVI.02195-05
Ruis, C., Brown, L.-A. K., Roy, S., Atkinson, C., Williams, R., Burns, S., et al. (2018). Mutagenesis in norovirus in response to favipiravir treatment. N. Engl. J. Med. 379, 2173–2176. doi: 10.1056/NEJMc1806941
Ruis, C., Roy, S., Brown, J. R., Allen, D. J., Goldstein, R. A., and Breuer, J. (2017). The emerging GII.P16-GII.4 Sydney 2012 norovirus lineage is circulating worldwide, arose by late-2014 and contains polymerase changes that may increase virus transmission. PLoS One 12:e0179572. doi: 10.1371/journal.pone.0179572
Sanjuán, R., and Domingo-Calap, P. (2016). Mechanisms of viral mutation. Cell. Mol. Life Sci. 73, 4433–4448. doi: 10.1007/s00018-016-2299-6
Schlegel, A., Giddings, T. H., Ladinsky, M. S., and Kirkegaard, K. (1996). Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 70, 6576–6588.
Schwartz, S., Vergoulidou, M., Schreier, E., Loddenkemper, C., Reinwald, M., Schmidt-Hieber, M., et al. (2011). Norovirus gastroenteritis causes severe and lethal complications after chemotherapy and hematopoietic stem cell transplantation. Blood 117, 5850–5856. doi: 10.1182/blood-2010-12-325886
Selisko, B., Papageorgiou, N., Ferron, F., and Canard, B. (2018). Structural and functional basis of the fidelity of nucleotide selection by flavivirus RNA-dependent RNA polymerases. Viruses 10:E59. doi: 10.3390/v10020059
Sharp, T. M., Guix, S., Katayama, K., Crawford, S. E., and Estes, M. K. (2010). Inhibition of cellular protein secretion by norwalk virus nonstructural protein p22 requires a mimic of an endoplasmic reticulum export signal. PLoS One 5:13130. doi: 10.1371/journal.pone.0013130
Shivanna, V., Kim, Y., and Chang, K. O. (2014a). Endosomal acidification and cathepsin L activity is required for calicivirus replication. Virology 46, 287–295. doi: 10.1016/j.virol.2014.07.025
Shivanna, V., Kim, Y., and Chang, K. O. (2014b). The crucial role of bile acids in the entry of porcine enteric calicivirus. Virology 45, 268–278. doi: 10.1016/j.virol.2014.04.002
Simmonds, P., Karakasiliotis, I., Bailey, D., Chaudhry, Y., Evans, D. J., and Goodfellow, I. G. (2008). Bioinformatic and functional analysis of RNA secondary structure elements among different genera of human and animal caliciviruses. Nucleic Acids Res. 36, 2530–2546. doi: 10.1093/nar/gkn096
Sit, T. L., Vaewhongs, A. A., and Lommel, S. A. (1998). RNA-mediated trans-activation of transcription from a viral RNA. Science 281, 829–832. doi: 10.1126/science.281.5378.829
Sosnovtsev, S. V., Belliot, G., Chang, K.-O., Onwudiwe, O., and Green, K. Y. (2005). Feline calicivirus VP2 is essential for the production of infectious virions. J. Virol. 79, 4012–4024. doi: 10.1128/JVI.79.7.4012-4024.2005
Sosnovtseva, S. A., Sosnovtsev, S. V., and Green, K. Y. (1999). Mapping of the feline calicivirus proteinase responsible for autocatalytic processing of the nonstructural polyprotein and identification of a stable proteinase-polymerase precursor protein. J. Virol. 73, 6626–6633.
Strive, T., Wright, J., Kovaliski, J., Botti, G., and Capucci, L. (2010). The non-pathogenic Australian lagovirus RCV-A1 causes a prolonged infection and elicits partial cross-protection to rabbit haemorrhagic disease virus. Virology 398, 125–134. doi: 10.1016/j.virol.2009.11.045
Subba-Reddy, C. V., Goodfellow, I., and Cheng Kao, C. (2011). VPg-primed RNA synthesis of norovirus RNA-dependent RNA polymerases by using a novel cell-based assay. J. Virol. 85, 13027–13037. doi: 10.1128/JVI.06191-11
Subba-Reddy, C. V., Yunus, M. A., Goodfellow, I. G., and Kao, C. C. (2012). Norovirus RNA synthesis is modulated by an interaction between the viral RNA-dependent RNA polymerase and the major capsid protein. VP1. J. Virol. 86, 10138–10149. doi: 10.1128/JVI.01208-12
Subba-Reddy, C. V., Yunus, M. A., Goodfellow, I. G., and Kao, C. C. (2017). Retraction for Subba-Reddy et al., “norovirus RNA synthesis is modulated by an interaction between the viral RNA-dependent RNA polymerase and the major capsid protein, VP1”. J. Virol. 91:e01708-17. doi: 10.1128/JVI.01708-17
Tarantino, D., Pezzullo, M., Mastrangelo, E., Croci, R., Rohayem, J., Robel, I., et al. (2014). Naphthalene-sulfonate inhibitors of human norovirus RNA-dependent RNA-polymerase. Antivir. Res. 102, 23–28. doi: 10.1016/j.antiviral.2013.11.016
Thumfart, J. O., and Meyers, G. (2002). Rabbit hemorrhagic disease virus: identification of a cleavage site in the viral polyprotein that is not processed by the known calicivirus protease. Virology 304, 352–363. doi: 10.1006/viro.2002.1660
Urakova, N., Frese, M., Hall, R. N., Liu, J., Matthaei, M., and Strive, T. (2015). Expression and partial characterisation of rabbit haemorrhagic disease virus non-structural proteins. Virology 484, 69–79. doi: 10.1016/j.virol.2015.05.004
Urakova, N., Netzler, N., Kelly, A. G., Frese, M., White, P. A., and Strive, T. (2016). Purification and biochemical characterisation of rabbit calicivirus RNA-dependent RNA polymerases and identification of non-nucleoside inhibitors. Viruses 8:100. doi: 10.3390/v8040100
Urakova, N., Strive, T., and Frese, M. (2017a). RNA-dependent RNA polymerases of both virulent and benign Rabbit caliciviruses induce striking rearrangement of Golgi membranes. PLoS One 12:e0169913. doi: 10.1371/journal.pone.0169913
Urakova, N., Warden, A. C., White, P. A., Strive, T., and Frese, M. (2017b). A motif in the F homomorph of rabbit haemorrhagic disease virus polymerase is important for the subcellular localisation of the protein and its ability to induce redistribution of Golgi membranes. Viruses 9:202. doi: 10.3390/v9080202
Vashist, S., Bailey, D., Putics, A., and Goodfellow, I. (2009). Model systems for the study of human norovirus biology. Future Virol. 4, 353–367. doi: 10.2217/fvl.09.18
Vazquez, A. L., Alonso, J. M. M., Casais, R., Boga, J. A., and Parra, F. (1998). Expression of enzymatically active rabbit hemorrhagic disease virus RNA-dependent RNA polymerase in Escherichia coli. J. Virol. 72, 2999–3004. doi: 10.7551/mitpress/9780262033589.003.0011
Voogd, T. E., Vansterkenburg, E. L. M., Wilting, J., and Janssen, L. H. M. (1993). Recent research on the biological activity of Suramin. Pharmacol. Rev. 45, 177–203.
Wang, F., Wang, M., Dong, Y., Zhang, B., and Zhang, D. (2017). Genetic characterization of a novel calicivirus from a goose. Arch. Virol. 162, 2115–2118. doi: 10.1007/s00705-017-3302-8
Ward, C. D., Stokes, M. A., and Flanegan, J. B. (1988). Direct measurement of the poliovirus RNA polymerase error frequency in vitro. J. Virol. 62, 558–562.
Wei, L., Huhn, J. S., Mory, A., Pathak, H. B., Sosnovtsev, S. V., Green, K. Y., et al. (2001). Proteinase-polymerase precursor as the active form of feline calicivirus RNA-dependent RNA polymerase. J. Virol. 75, 1211–1219. doi: 10.1128/JVI.75.3.1211-1219.2001
Weir, M. L., Klip, A., and Trimble, W. S. (1998). Identification of a human homologue of the vesicle-associated membrane protein (VAMP)-associated protein of 33 kDa (VAP-33): a broadly expressed protein that binds to VAMP. Biochem. J. 333, 247–251. doi: 10.1042/bj3330247
Wirblich, C., Thiel, H. J., and Meyers, G. (1996). Genetic map of the calicivirus rabbit hemorrhagic disease virus as deduced from in vitro translation studies. J. Virol. 70, 7974–7983. doi: 10.1006/viro.2000.0579
Witkowski, J. T., Robins, R. K., Sidwell, R. W., and Simon, L. N. (1972). Design, synthesis, and broad spectrum antiviral activity of 1-β-D-ribofuranosyl-1,2,4-triazole-3-carboxamide and related nucleosides. J. Med. Chem. 15, 1150–1154.
Wobus, C. E., Karst, S. M., Thackray, L. B., Chang, K. O., Sosnovtsev, S. V., Belliot, G., et al. (2004). Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2:e432. doi: 10.1371/journal.pbio.0020432
Wobus, C. E., Thackray, L. B., and Virgin, H. W. (2006). Murine norovirus: a model system to study norovirus biology and pathogenesis. J. Virol. 80, 5104–5112. doi: 10.1128/JVI.02346-05
Wu, G., and Kaper, J. M. (1994). Requirement of 3’-terminal guanosine in(-)-stranded RNA for in vitro replication of cucumber mosaic virus satellite RNA by viral RNA-dependent RNA polymerase. J. Mol. Biol. 238, 655–657. doi: 10.1006/jmbi.1994.1326
Wu, H., Zu, S., Sun, X., Liu, Y., Tian, J., and Qu, L. (2016). N-terminal domain of feline calicivirus (FCV) proteinase-polymerase contributes to the inhibition of host cell transcription. Viruses 8:199. doi: 10.3390/v8070199
Xie, X., Wang, H., Zeng, J., Li, C., Zhou, G., Yang, D., et al. (2014). Foot-and-mouth disease virus low-fidelity polymerase mutants are attenuated. Arch. Virol. 159, 2641–2650. doi: 10.1007/s00705-014-2126-z
Yang, X., Smidansky, E. D., Maksimchuk, K. R., Lum, D., Welch, J. L., Arnold, J. J., et al. (2012). Motif D of viral RNA-dependent RNA polymerases determines efficiency and fidelity of nucleotide addition. Structure 20, 1519–1527. doi: 10.1016/j.str.2012.06.012
Yunus, M. A., Lin, X., Bailey, D., Karakasiliotis, I., Chaudhry, Y., Vashist, S., et al. (2015). The Murine norovirus core subgenomic RNA promoter consists of a stable stem-loop that can direct accurate initiation of RNA synthesis. J. Virol. 89, 1218–1229. doi: 10.1128/JVI.02432-14
Zamyatkin, D. F., Parra, F., Martín Alonso, J. M., Harki, D. A., Peterson, B. R., Grochulski, P., et al. (2008). Structural insights into mechanisms of catalysis and inhibition in Norwalk virus polymerase. J. Biol. Chem. 283, 7705–7712. doi: 10.1074/jbc.M709563200
Zeng, J., Wang, H., Xie, X., Li, C., Zhou, G., Yang, D., et al. (2014). Ribavirin-resistant variants of foot-and-mouth disease virus: the effect of restricted quasispecies diversity on viral virulence. J. Virol. 88, 4008–4020. doi: 10.1128/JVI.03594-13
Keywords: polymerase, RdRp, replication, motif, RNA virus, Caliciviridae, Lagovirus, RHDV
Citation: Smertina E, Urakova N, Strive T and Frese M (2019) Calicivirus RNA-Dependent RNA Polymerases: Evolution, Structure, Protein Dynamics, and Function. Front. Microbiol. 10:1280. doi: 10.3389/fmicb.2019.01280
Received: 29 January 2019; Accepted: 22 May 2019;
Published: 06 June 2019.
Edited by:
Mathilde Richard, Erasmus University Rotterdam, NetherlandsReviewed by:
Kyung Hyun Kim, Korea University, South KoreaHirokazu Kimura, Gunma Paz University, Japan
Copyright © 2019 Smertina, Urakova, Strive and Frese. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael Frese, bWljaGFlbC5mcmVzZUBjYW5iZXJyYS5lZHUuYXU=
 Elena Smertina
Elena Smertina Nadya Urakova
Nadya Urakova Tanja Strive
Tanja Strive Michael Frese
Michael Frese