- 1National Risk Assessment Laboratory for Antimicrobial Resistance of Animal Original Bacteria, College of Veterinary Medicine, South China Agricultural University, Guangzhou, China
- 2Guangdong Key Laboratory for Veterinary Drug Development and Safety Evaluation, College of Veterinary Medicine, South China Agricultural University, Guangzhou, China
- 3Quadram Institute Bioscience, Norwich, United Kingdom
The emergence and increase in prevalence of resistance to cephalosporins amongst isolates of Salmonella from food animals imposes a public health threat. The aim of the present study was to investigate the prevalence and characteristics of CTX-M-producing Salmonella isolates from raw meat and food animals. 27 of 152 (17.76%) Salmonella isolates were ESBL-positive including 21/70 (30%) from food animals and 6/82 (7.32%) from raw meat. CTX-M-55 was the most prevalent ESBL type observed (12/27, 44.44%). 7 of 12 CTX-M-55-positive Salmonella isolates were Salmonella Indiana, 2 were Salmonella Typhimurium, 2 were Salmonella Chester, and the remaining isolate was not typeable. Eight CTX-M-55-positive Salmonella isolates were highly resistant to fluoroquinolones (MICCIP = 64 ug/mL) and co-harbored aac(6’)-Ib-cr and oqxAB. Most of the CTX-M-55 positive isolates (11/12) carried blaCTX-M-55 genes on the chromosome, with the remaining isolate carrying this gene on a transferable 280 kb IncHI2 plasmid. A chromosomal blaCTX-M-55 gene from one isolate transferred onto a 250 kb IncHI2 plasmid which was subsequently conjugated into recipient strain J53. PFGE and MLST profiles showed a wide range of strain types were carrying blaCTX-M-55. Our study demonstrates the emergence and prevalence of foodborne Salmonella harboring a chromosomally located blaCTX-M-55 in China. The co-existence of PMQR genes with blaCTX-M-55 in Salmonella isolates suggests co-selection and dissemination of resistance to both fluoroquinolones and cephalosporins in Salmonella via the food chain in China represents a public health concern.
Introduction
Salmonella species are the second most common bacterial cause of foodborne gastroenteritis worldwide and almost 80.3 million foodborne illness per year are caused by non-typhoid Salmonella infections (Majowicz et al., 2010). Extended-spectrum cephalosporins (ESCs) are effective drugs of choice in children for treatment of non-typhoid salmonellosis, due to the contraindication for use in children of fluoroquinolones (FQs), the classical first-line antibiotics. The emergence of Salmonella isolates resistant to ESC is a worldwide public health concern (Arlet et al., 2006). Resistance to these drugs is mainly mediated by the bacterial production of extended-spectrum β-lactamases (ESBLs) with CTX-M-type enzymes being the most common.
CTX-M genes have successfully disseminated globally and are common in clinical settings, communities, livestock and companion animals. There are many CTX-M variants of which, CTX-M-15 and CTX-M-14 are the most prevalent (Zhao and Hu, 2013; de Jong et al., 2014). However, the epidemiology of CTX-M-type ESBLs is evolving rapidly. A number of minor allelic variants have been described and classified as belonging to one of six groups (CTX-M-1, CTX-M-2, CTX-M-8, CTX-M-9, CTX-M-25 and KLUC, named after the archetypal enzymes of each group) that differ from each other by ≥ 10% amino acid residues (D’Andrea et al., 2013).
CTX-M-55 is a CTX-M-15 variant that contains a substitution of A80V within the β-lactamase possessing enhanced cephalosporin-hydrolyzing activity (He et al., 2015) and has been detected as increasing rapidly in prevalence, especially in Escherichia coli from animals (Zheng et al., 2012; Cunha et al., 2017; Norizuki et al., 2018). Whilst the blaCTX-M-55 gene is less commonly detected in Salmonella from animals or humans, the first report was from human isolates in the United States and China in 2011 (Sjolund-Karlsson et al., 2011; Yu et al., 2011). Since then CTX-M-55 producing Salmonella have been identified from a number of different serotypes from Switzerland (Gallati et al., 2013), Japan (Imoto et al., 2014), China (Wong et al., 2015), Korea (Kim et al., 2017), Denmark (Torpdahl et al., 2017), and Thailand (Luk-In et al., 2018). CTX-M-55 carrying isolates from animals have been isolated from fish, pork and chicken (Nguyen et al., 2016; Nadimpalli et al., 2018). Since CTX-M-55 Salmonella isolates are increasingly detected and show high-level resistance to ESCs and are often cross-resistant to FQs, these Salmonella strains represent a potentially severe clinical and food safety issues and this warrants investigation of the prevalence of blaCTX-M-55-harboring Salmonella.
Carriage of CTX-M genes is mostly associated with a diverse set of transmissible plasmids (Canton et al., 2012). However, a small number of chromosomal CTX-M genes have been identified in several studies in E. coli where transfer into the chromosome was mediated by transposons or insertion sequences (Fabre et al., 2009; Hamamoto and Hirai, 2018).
In the present study, we investigated the prevalence of CTX-M-type ESBL-producing Salmonella isolates from food animals and raw meat in Guangdong province during 2015 and 2017, analyzed the characteristics of these CTX-M-55-positive strains including phenotypes, genotypes, genetic relatedness, and plasmid profiles.
Materials and Methods
Sample Collection and Salmonella Isolation, Identification
A total of 891 specimens were collected from the Guangdong province of China between 2015 and 2017. Of these samples, 453 fecal swabs from free-range food animals (84 from chickens, 249 from ducks, 107 from pigs, and 13 from geese) were obtained from veterinary clinics. 438 raw meat samples (156 from chicken meat, 35 from duck meat, and 247 from pork) were collected from different supermarkets. Fecal swabs and meat samples (cut into pieces) were placed into sterile selenite cysteine broth and incubated for 24 h at 37°C. Aliquots were then streaked on chromogenic medium selective for Salmonella (CHROMagar Microbiology, France) and incubated for another 24 h at 37°C. One purple colony was selected from each plate and then confirmed using the API20E system (bioMérieux, Marcy L’Étoile, France) and identified by MALDI-TOF MS (Axima-Assurance-Shimadzu). All isolates identified as Salmonella were stored at -80°C in Luria-Bertani (LB) broth containing 30% glycerol.
Antimicrobial Susceptibility Testing, Detection of ESBL Genes and ESBL Production Verified by Phenotype
The minimum inhibitory concentrations (MICs) of cefotaxime (CTX) and ciprofloxacin (CIP) were determined in triplicate for each bacterial strain using the agar dilution method on Mueller-Hinton agar plates according to the CLSI reference method (CLSI-M100-S26). E. coli ATCC 25922 was used as the quality control strain. Salmonella isolates showing resistance to cefotaxime (with MIC ≥ 4 ug/mL) were screened for the presence of the ESBL-genes blaTEM, blaSHV, blaOXA, blaCTX-M and blaCMY -2 by PCR using the primers and conditions described previously (Jiang et al., 2012). Amplified PCR products were submitted to BGI Life Tech Co., Ltd. (Beijing, China) for DNA sequencing and the identity of specific β-lactamase genes were determined using the protein BLAST algorithm1. Double disk synergy tests were performed to further verify ESBL production by using a central amoxicillin/clavulanic acid (AMC) disk, 15 and 20 mm (center to center) separately away from cefotaxime disks. Synergy was interpreted by a clear-cut enlargement of the inhibition zone of CTX disk near the inhibition zone of AMC disk (Jarlier et al., 1988).
Detection of PMQR Genes, Mutations Within Quinolone Resistance-Determining Region (QRDR) of Target Genes and Serotyping of CTX-M-55-Positive Isolates
The presence of PMQR genes qnrA, qnrB, qnrC, qnrD, qnrS, aac(6’)-Ib-cr, qepA, and oqxAB from blaCTX-M-55-positive isolates was also investigated by PCR using primers and conditions as previously described (Jiang et al., 2012). Mutations in QRDRs of the target genes gyrA, gyrB, parC, parE were confirmed by PCR and sequencing and their DNA sequences were compared with the Salmonella Typhimurium LT2 genome as a reference.
CTX-M-55-producing Salmonella isolates were serotyped using Salmonella specific O and H antigens (Statens Serum Institute, Denmark) by the slide agglutination test according to the Kauffmann-White scheme.
Pulsed-Field Gel Electrophoresis (PFGE) and Multilocus Sequence Typing (MLST)
Genetic relatedness of all blaCTX-M-55-harboring isolates were analyzed by pulsed-field gel electrophoresis (PFGE) of XbaI-digested genomic DNA using a CHEF-MAPPER System (BioRad Laboratories, Hercules, CA, United States) as previously described (Jiang et al., 2014). PFGE patterns were compared using the Dice similarity coefficient with BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium).
MLST was carried out by PCR and DNA sequence analysis of 7 housekeeping genes aroC, dnaN, hemD, hisD, purE, sucA, and thrA to determine the allelic profiles using software available at http://mlst.warwick.ac.uk/mlst/dbs/Senterica.
Conjugation Experiments and Plasmid Analysis
Conjugation experiments of blaCTX-M-55 gene positive Salmonella isolates were conducted by liquid mating in LB broth using sodium azide-resistant E. coli J53 as the recipient strain. Transconjugants were selected on MacConkey agar containing cefotaxime (2 ug/mL) and sodium azide (300 ug/mL). The presence of blaCTX-M-55 in transconjugants was verified by PCR and sequencing as described. PFGE analysis was conducted using S1 nuclease (Takara Biotechnology, Dalian, China) digested genomic DNA as previously described (Barton et al., 1995) to identify the genetic location of blaCTX-M genes. Primers used for blaCTX-M-probes were the same as those used to amplify CTX-M encoding genes. The resulting gels were analyzed by Southern blotting after transfer to Hybond-N+ membranes (GE Healthcare, Little Chalfont, United Kingdom) and probing with a DIG-labeled blaCTX-M gene fragment according to the manufacturer’s instructions (DIG High Prime DNA Labeling and Detection Starter Kit I, Roche Applied Science, Mannheim, Germany). Restriction fragments of agarose-embedded DNA of strain H9812 digested with XbaI(Takara) at 37°C for 4 h was used as DNA size marker during electrophoresis.
Whole Genome Sequencing
To characterize the genetic context of blaCTX-M genes DNA from selected isolates was used to sequence whole genomic content. This was done by MajorBio Co., Shanghai, China. The resulting reads were trimmed and genomes assembled using “SPAdes v 3.11.0” (Nurk et al., 2013), annotated using “Prokka v 1.13” (Seemann, 2014) and mapped reads against reference genomes using “Bowtie2 v 2.3.4.3” (Langmead and Salzberg, 2012). Annotations and alignments were visualized in “Artemis.”
Results
Salmonella Isolation and Antimicrobial Susceptibility Phenotypes
We collected 891 samples for this study and 152 Salmonella were identified by MALDI-TOF MS of which, 46.05% (n = 70) were isolated from animals (13 from chickens, 43 from ducks, 11 from pigs, and 3 from geese). The remaining 82 samples came from raw meat (including 37 chicken meat, 5 duck meat, and 40 pork supermarket samples). The isolation rate of Salmonella strains from raw meat (17.90%) was similar to that from the farm animals (15.45%).
We also examined susceptibility of the 152 Salmonella isolates to cefotaxime and ciprofloxacin. Resistance rates to cefotaxime and ciprofloxacin were 26.97% (n = 41) and 30.26% (n = 46), respectively. In the 70 Salmonella isolates from animals, 30 displayed cefotaxime resistance (42.86%) and 34 ciprofloxacin resistant (48.57%). The 82 Salmonella isolates from raw meat, contained 11 isolates resistant to cefotaxime (13.43%) and 12 resistant to ciprofloxacin (14.63%). The rate of cross-resistant to both antibiotics were significantly higher for the animal isolates (32.86%) than for raw meat (4.88%).
ESBL Characterization and Production, Serotyping of CTX-M-55-Producing Isolates
A total of 27 CTX-M ESBLs producing isolates were confirmed among the Salmonella isolates. Of these, 21/70 (30%) were from animals and 6/82 (7.32%) were from meat. We found that 12/27 (44.44%) were CTX-M-1 group members and all were confirmed as being blaCTX-M-55. There were also 12/27 that belonged to the CTX-M-9 group and were assigned as blaCTX-M-27 (n = 8), blaCTX-M-14 (n = 2) and blaCTX-M-65 (n = 2). The remaining 3 CTX-M-encoding genes were all identified as the hybrid allele blaCTX-M-64. We also found that 14/41 cefotaxime-resistant isolates were CTX-M negative.
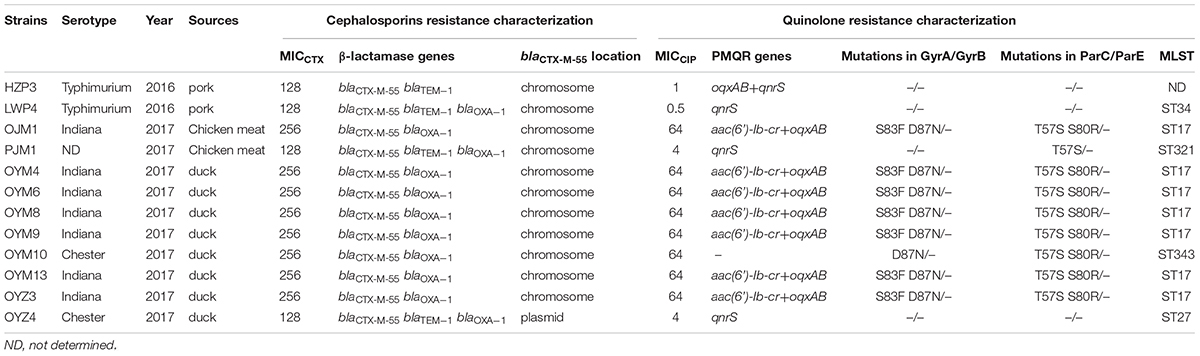
Of the blaCTX-M-55 positive isolates, 10/12 were from animals and the remaining 2 were from meat samples. A clear-cut extension of the edge of the inhibition zone of CTX disk toward the AMC disk was seen from each blaCTX-M-55 carrying Salmonella strain, consistent with ESBL production. These isolates were also serotyped and 7 were S. Indiana, 2 S. Typhimurium, 2 S. Chester and 1 was untypeable (Table 1).
Detection of Ciprofloxacin Resistance Mechanisms in CTX-M-55-Producing Isolates
Multiple QRDR mutations in gyrA and parC were detected in the high-level quinolone resistant isolates that co-harbored aac(6’)-Ib-cr and oqxAB. However, strains with an MICCIP in the 0.5 to 4 ug/mL range did not contain mutations in the QRDRs of target genes, but all contained qnrS (Table 1).
The 12 CTX-M-55 producers were all ciprofloxacin resistant and 8 exhibited high-level resistance (MICCIP = 64 ug/mL), 4 isolates demonstrated lower resistance (MICCIP = 0.5–4 ug/mL). For PMQR determinants, 11 of 12 blaCTX-M-55-positive isolates were found to harbor at least one PMQR gene. The combination of aac(6’) - Ib-cr + oqxAB (n = 7) dominated followed by qnrS (n = 3) and oqxAB + qnrS (n = 1). One isolate completely lacked any PMQR genes (Table 1).
Genetic Relatedness and Molecular Typing Analysis of CTX-M-55-Producing Isolates
The 12 CTX-M-55 isolates produced 11 different profiles that were divided into 9 different PFGE clusters designated 1–9 with 85% genetic similarity. Five MLST profiles were determined including ST17, ST27, ST34, ST321, and ST343. ST17 was most prevalent (n = 7, 58.3%), ST27, ST34, ST321, and ST343 were represented by one isolate each and no ST could be determined for one isolate (Figure 1).
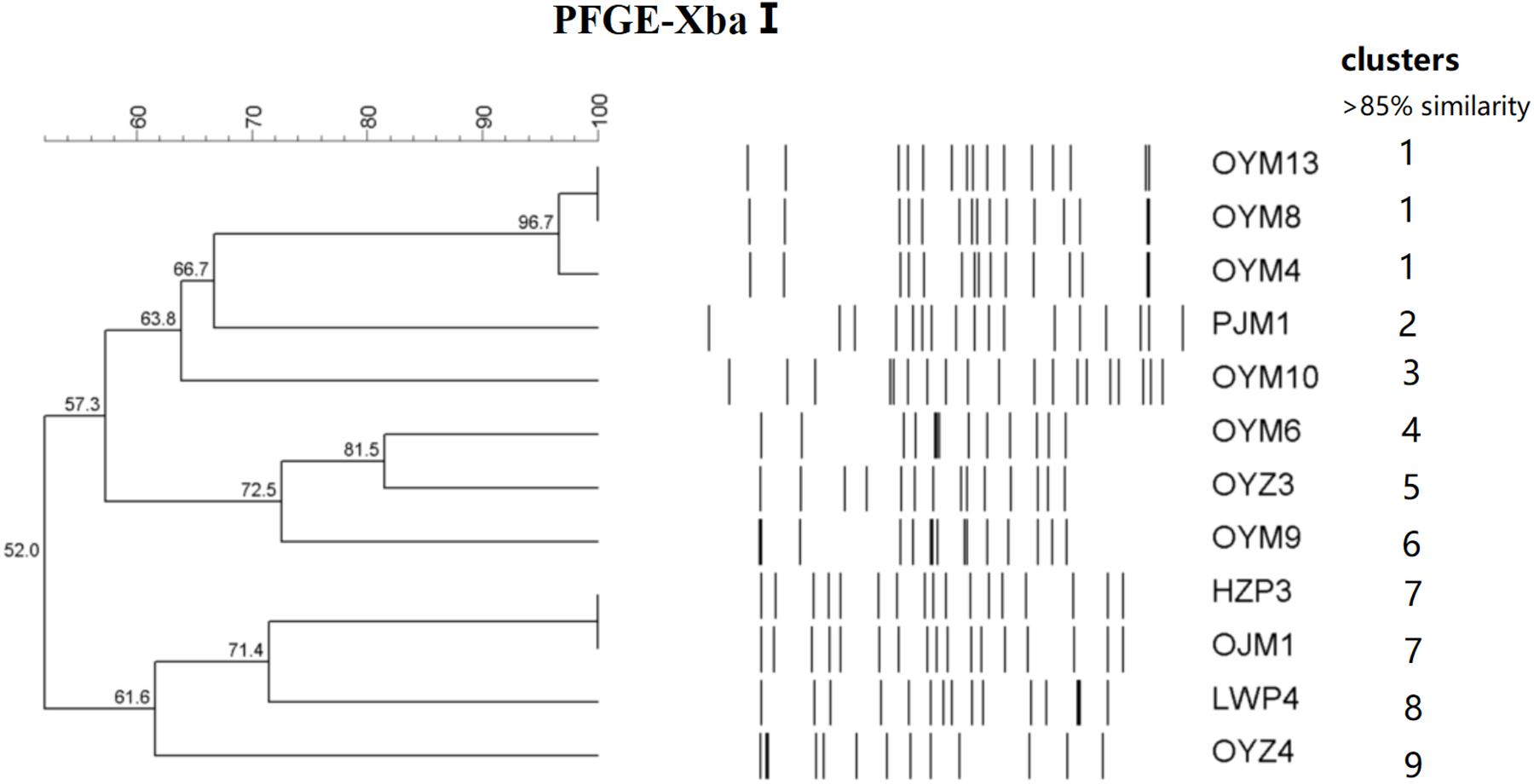
Figure 1. PFGE fingerprinting patterns and other characteristics of Salmonella isolates harboring blaCTX-M-55 genes.
blaCTX-M-55 Hybridization and Plasmid Analysis
S1-PFGE and southern hybridization analysis of all 12 blaCTX-M-55-positive Salmonella isolates and 2 transconjugants found that blaCTX-M-55 was chromosomally located in 11 of the isolates and in one isolate was present on a 280 kb plasmid (Figure 2). Interestingly, we successfully obtained one transconjugant from PJM1 whose blaCTX-M-55 gene was chromosomal. This transconjugant carried the blaCTX-M-55 gene on a 250 kb IncHI2 plasmid suggesting a mechanism where transfer from the chromosome onto a plasmid was followed by conjugation of this plasmid, now carrying the resistance gene (Figure 2). Two narrow-spectrum β-lactamase gene, blaTEM-1 and blaOXA-1 and PMQR gene qnrS also co-transferred with blaCTX-M-55 in the transconjugants (Table 2). Analysis of the whole genome assembly of PJM1 identified the blaCTX-M-55 present within an 11 kb contig with 100% identify to plasmid pCSFA1096, previously identified from Salmonella in China. However, mapping of all reads from the total genome sequencing of PJM1 against the pCSFA1096 genome identified alignments only over the elements carrying the resistance genes and the rest of the plasmid backbone was not present. This suggests the blaCTX-M-55 gene originally transferred into the chromosome of PJM1 within a larger mobile element which retains the capacity to be mobilized. This supports our proposed model of transfer of this chromosomal element from PJM1 onto a plasmid and subsequent onward transmission.
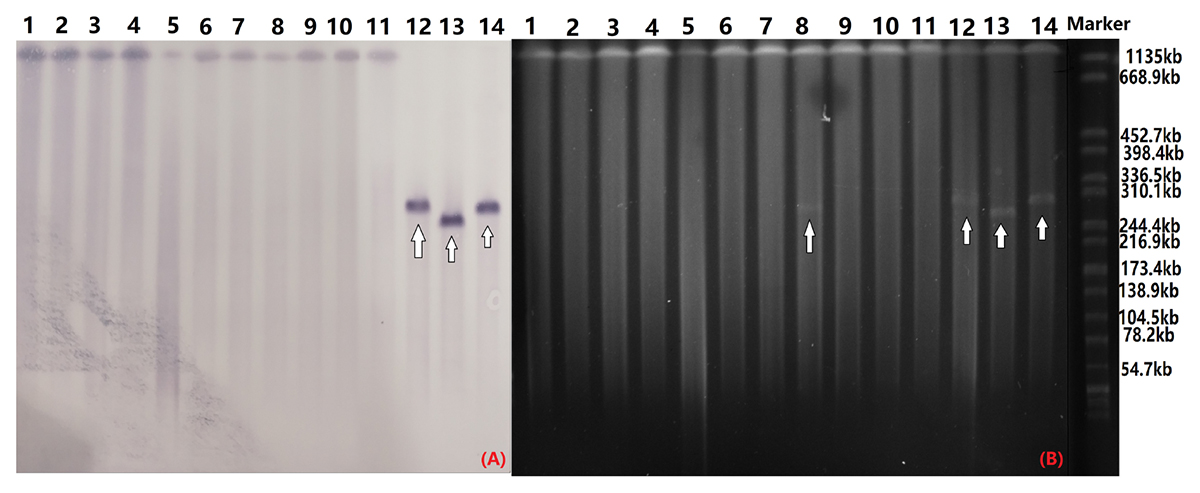
Figure 2. Southern blot hybridization with the blaCTX-M-55 probe (A) and S1-PFGE of S1 nuclease-digested genomic DNA (B). The arrow indicates plasmids in the isolates. OYM4 (lane 1), OYM6 (lane 2), OYM8 (lane 3), OYM9 (lane 4), OYM13 (lane 5), OYZ3 (lane 6), OJM1 (lane 7), HZP3 (lane 8), LWP4 (lane 9), OYM10 (lane 10), PJM1 (lane 11), OYZ4 (lane 12), C-PJM1 (lane 13), C-OYZ4 (lane 14), Marker (H9812).
Discussion
In the present study we found that 27 (65.85%) cefotaxime-resistant Salmonella strains produced CTX-M-type ESBLs. In this group, 21 were from food animals and 6 were from meat. The CTX-M-producing strains included 17 that were ciprofloxacin-resistant and 9 with decreased susceptibility to ciprofloxacin. In addition, at least one PMQR gene was detected in each of the 21 CTX-M isolates from animals and these results were similar to our previous study (Zhang et al., 2016). Together these studies suggest that the co-existence or co-transfer of PMQR genes in CTX-M-producing Salmonella strains increase their probability of survival in the presence of quinolones and/or cephalosporins (Liu et al., 2013).
The cefotaxime-resistant Salmonella strains included 5 CTX-M subtypes and CTX-M-55 was the most prevalent (n = 12). This was inconsistent with our previous studies that CTX-M-27 was the most prevalent ESBL in Salmonella strains isolated in 2009, 2010, and 2014 (Jiang et al., 2014; Zhang et al., 2016). In the current study, all the 12 CTX-M-55-producing strains were simultaneously non-susceptible to ciprofloxacin and almost all harbored PMQR genes. PMQR gene qnrS was only detected in low level ciprofloxacin resistant strains which contained no resistance-associated mutations in the QRDR. Though PMQR determinants only confer low level fluoroquinolone resistance, their existence (especially qnr) provide strains with a selective advantage under fluoroquinolones exposure and can accelerate the development of chromosome-mediated quinolone resistance (Robicsek et al., 2006; Strahilevitz et al., 2009).
Plasmids are key vectors in the global dissemination of antibiotic resistance genes in Gram-negative bacteria. Plasmid families including IncF, IncI1, IncI2, IncX, IncA/C, and IncHI2 play important roles in ESBL gene spread (Wang et al., 2018b). The blaCTX-M-55 genes are the second most abundant ESBL subtype in the Enterobacteriaceae (Zhang et al., 2014; Lupo et al., 2018). This is especially true of E. coli from both humans and animals in Asia and these are usually found on IncF and IncI1 plasmids (Zhang et al., 2014; Lupo et al., 2018; Wang et al., 2018a). Additional data from our laboratory presented evidence that the prevalence of blaCTX-M-55 in E. coli from both livestock and human origin is increasing. The F33:A-:B- and IncI1 plasmids have driven the spread of these genes in China. F33:A-:B- plasmids impart a significant biological advantage to their host and thus contribute to the increasing distribution of blaCTX-M-55 (Wang et al., 2018b).
In the present study, we found chromosomal copies of blaCTX-M-55 in 11/12 CTX-M-55-producing Salmonella strains. We speculate that the cross-species dissemination of blaCTX-M-55 from plasmids in E. coli to Salmonella chromosomes contributes to the spread and stable persistence of this gene in Salmonella (Wong et al., 2015). Following the first isolation from a food animal in 2010 in China (Yan et al., 2010), the detection rate of S. Indiana increase d rapidly, especially from veterinary clinics and food-producing animals. The highly fluoroquinolone and β-lactam-resistant S. Indiana ST17 is the most prevalent sequence type of this serovar in China. This may suggest an increasing disseminating trend of ST17 CTX-M-55-encoding S. Indiana. The highly drug-resistant S. Indiana ST17 is one of the most prevalent antimicrobial-resistant foodborne pathogens in China so that its isolation from animals is a public health concern (Wang et al., 2017; Zhao et al., 2017; Cao et al., 2018). Additionally, the monophasic variant of S. Typhimurium ST34 has already emerged in Europe and Asia (Arnott et al., 2018). A comparison of the genomes of a pork meat and a human isolate revealed only 10 single nucleotide polymorphisms (SNP). This indicated that human bacterium was acquired from pork meat (Arnott et al., 2018).
A major mechanism underpinning the global dissemination of β-lactam resistant bacteria is their possession of resistant plasmids with low fitness cost or stable carriage of ESBL genes in the chromosome although the later are currently uncommon. Before 2013, strains harboring chromosome-located ESBL encoding genes were sporadically detected in Escherichia coli (Garcia et al., 2005; Coque et al., 2008), Salmonella Concord (Fabre et al., 2009), Klebsiella pneumoniae (Coelho et al., 2010), Proteus mirabilis and Morganella morganii (Harada et al., 2012; Mahrouki et al., 2012). Since then, highly prevalent E. coli strains possessing chromosomal CTX-M-14 and CTX-M-15 β-lactamases were identified in 2013 (Hirai et al., 2013), 2016 (Hamamoto et al., 2016) and 2018 (Hamamoto and Hirai, 2018) separately. There may be a chromosomal blaCTX-M transpositional unit responsible for the global dissemination of CTX-M-14 in E. coli. However, the nature and significance of the spread of chromosomally located blaCTX-M genes remains unclear. We found most isolates carrying CTX-M-55 had the gene in a chromosomal context and these were from a diverse set of strain types. Transfer was however, possible from a strain (PJM1) with a chromosomal gene and the transconjugants carried the gene on a 280 kb IncHI2 plasmid. Together these data suggest that CTX-M-55 readily incorporates into the chromosome of Salmonella and that this is associated with PMQR carriage but that the element carrying the CTX-M-55 gene can move onward onto plasmids for further dissemination.
A recent study in Cambodia demonstrated that spread of CTX-M-55-type S. enterica from pork and fish samples was mediated by MDR IncA/C2 and IncHI2 plasmids (Nadimpalli et al., 2018). IncHI2 plasmids are the fifth most widely disseminated plasmid type that mediate transmission of antibiotic resistance genes. These are primarily found in Salmonella, E. coli, Enterobacter cloacae and Klebsiella pneumoniae of human and avian sources (Garcia Fernandez et al., 2007; Li et al., 2013; Haenni et al., 2016; Zhang et al., 2016).
Conclusion
In conclusion, we identified foodborne Salmonella harboring chromosomally located blaCTX-M-55 from China. These strains are simultaneously non-susceptible to fluoroquinolones. The co-existence of PMQR genes and CTX-M ESBL genes indicated co-selection for these determinants which may accelerate the dissemination of multi-drug resistance. Importantly, these strains may promote the development of isolates resistant to both cefotaxime and ciprofloxacin. The determination of the mechanisms and dissemination routes of ESBL-producing Salmonella is critical for animal and human health and understanding the interplay between movement of resistance genes between plasmids and chromosomal locations is important to understand the dynamics and evolutionary consequences of spread of antimicrobial resistance (AMR). This would also provide useful information to effectively control the development of antibiotic resistance to cephalosporins and fluoroquinolones.
Ethics Statement
This study protocol was approved by the South China Agriculture University Animal Ethics Committee. The strains of free-range food animal origin were isolated from fecal swabs of healthy chickens, pigs, ducks, and geese and the owners of the animals gave permission for their animals to be used in this study.
Author Contributions
C-ZZ, H-ZD, and H-XJ conceived and designed the experiments. C-ZZ, X-MD, X-LL, R-YS, Y-WL, and R-MC performed the experiments. C-ZZ, MW, H-ZD, and H-XJ analyzed the data. C-ZZ, X-MD, X-LL, Y-WL, and R-MC contributed reagents, materials, and analysis tools. C-ZZ, MW, and H-XJ wrote the manuscript.
Funding
C-ZZ was funded by a China Scholarship Council scholarship. This work was supported in part by the National Natural Science Foundation of China (31772792) and the Guangzhou Science and Technology Project (201607010269).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Arlet, G., Barrett, T. J., Butaye, P., Cloeckaert, A., Mulvey, M. R., and White, D. G. (2006). Salmonella resistant to extended-spectrum cephalosporins: prevalence and epidemiology. Microb. Infect. 8, 1945–1954. doi: 10.1016/j.micinf.2005.12.029
Arnott, A., Wang, Q., Bachmann, N., Sadsad, R., Biswas, C., Sotomayor, C., et al. (2018). Multidrug-resistant Salmonella enterica 4,[5],12:i:- sequence type 34, New South Wales, Australia, 2016-2017. Emerg. Infect. Dis. 24, 751–753. doi: 10.3201/eid2404.171619
Barton, B. M., Harding, G. P., and Zuccarelli, A. J. (1995). A general method for detecting and sizing large plasmids. Anal. Biochem. 226, 235–240. doi: 10.1006/abio.1995.1220
Canton, R., Gonzalez-Alba, J. M., and Galan, J. C. (2012). CTX-M enzymes: origin and diffusion. Front. Microbiol. 3:110. doi: 10.3389/fmicb.2012.00110
Cao, Y., Shen, Y., Cheng, L., Zhang, X., Wang, C., Wang, Y., et al. (2018). Combination of multilocus sequence typing and pulsed-field gel electrophoresis reveals an association of molecular clonality with the emergence of extensive-drug resistance (XDR) in Salmonella. Microbiol. Res. 207, 170–176. doi: 10.1016/j.micres.2017.12.001
Coelho, A., Gonzalez-Lopez, J. J., Miro, E., Alonso-Tarres, C., Mirelis, B., Larrosa, M. N., et al. (2010). Characterisation of the CTX-M-15-encoding gene in Klebsiella pneumoniae strains from the Barcelona metropolitan area: plasmid diversity and chromosomal integration. Int. J. Antimicrob. Agents 36, 73–78. doi: 10.1016/j.ijantimicag.2010.03.005
Coque, T. M., Novais, A., Carattoli, A., Poirel, L., Pitout, J., Peixe, L., et al. (2008). Dissemination of clonally related Escherichia coli strains expressing extended-spectrum β-lactamase CTX-M-15. Emerg. Infect. Dis. 14, 195–200. doi: 10.3201/eid1402.070350
Cunha, M. P., Lincopan, N., Cerdeira, L., Esposito, F., Dropa, M., Franco, L. S., et al. (2017). Coexistence of CTX-M-2, CTX-M-55, CMY-2, FosA3, and QnrB19 in extraintestinal pathogenic Escherichia coli from Poultry in Brazil. Antimicrob. Agents Chemother. 61, e02474-16. doi: 10.1128/AAC.02474-16
D’Andrea, M. M., Arena, F., Pallecchi, L., and Rossolini, G. M. (2013). CTX-M-type β-lactamases: a successful story of antibiotic resistance. Int. J. Med. Microbiol. 303, 305–317. doi: 10.1016/j.ijmm.2013.02.008
de Jong, A., Smet, A., Ludwig, C., Stephan, B., De Graef, E., Vanrobaeys, M., et al. (2014). Antimicrobial susceptibility of Salmonella isolates from healthy pigs and chickens (2008-2011). Vet. Microbiol. 171, 298–306. doi: 10.1016/j.vetmic.2014.01.030
Fabre, L., Delaune, A., Espie, E., Nygard, K., Pardos de la Gandara, M., Polomack, L., et al. (2009). Chromosomal integration of the extended-spectrum β-lactamase gene blaCTX-M-15 in Salmonella enterica serotype Concord isolates from internationally adopted children. Antimicrob. Agents Chemother. 53, 1808–1816. doi: 10.1128/AAC.00451-08
Gallati, C., Stephan, R., Hachler, H., Malorny, B., Schroeter, A., and Nuesch-Inderbinen, M. (2013). Characterization of Salmonella enterica subsp. enterica serovar 4,[5],12:i:- clones isolated from human and other sources in Switzerland between 2007 and 2011. Foodborne Pathog. Dis. 10, 549–554. doi: 10.1089/fpd.2012.1407
Garcia, A., Navarro, F., Miro, E., Mirelis, B., Campoy, S., and Coll, P. (2005). Characterization of the highly variable region surrounding the bla(CTX-M-9) gene in non-related Escherichia coli from Barcelona. J. Antimicrob. Chemother. 56, 819–826. doi: 10.1093/jac/dki345
Garcia Fernandez, A., Cloeckaert, A., Bertini, A., Praud, K., Doublet, B., Weill, F. X., et al. (2007). Comparative analysis of IncHI2 plasmids carrying blaCTX-M-2 or blaCTX-M-9 from Escherichia coli and Salmonella enterica strains isolated from poultry and humans. Antimicrob. Agents Chemother. 51, 4177–4180. doi: 10.1128/AAC.00603-07
Haenni, M., Poirel, L., Kieffer, N., Chatre, P., Saras, E., Metayer, V., et al. (2016). Co-occurrence of extended spectrum β lactamase and MCR-1 encoding genes on plasmids. Lancet Infect. Dis. 16, 281–282. doi: 10.1016/S1473-3099(16)00007-4
Hamamoto, K., and Hirai, I. (2018). Characterization of chromosomally-located blaCTX-M and its surrounding sequence in CTX-M type extended-spectrum β-lactamase-producing Escherichia coli isolates. J. Glob. Antimicrob. Resist. 17, 53–57. doi: 10.1016/j.jgar.2018.11.006
Hamamoto, K., Ueda, S., Toyosato, T., Yamamoto, Y., and Hirai, I. (2016). High prevalence of chromosomal blaCTX-M-14 in Escherichia coli isolates possessing blaCTX-M-14. Antimicrob. Agents Chemother. 60, 2582–2584. doi: 10.1128/AAC.00108-16
Harada, S., Ishii, Y., Saga, T., Kouyama, Y., Tateda, K., and Yamaguchi, K. (2012). Chromosomal integration and location on IncT plasmids of the blaCTX-M-2 gene in Proteus mirabilis clinical isolates. Antimicrob. Agents Chemother. 56, 1093–1096. doi: 10.1128/AAC.00258-11
He, D., Chiou, J., Zeng, Z., Liu, L., Chen, X., Zeng, L., et al. (2015). Residues distal to the active site contribute to enhanced catalytic activity of variant and hybrid β-Lactamases derived from CTX-M-14 and CTX-M-15. Antimicrob. Agents Chemother. 59, 5976–5983. doi: 10.1128/AAC.04920-14
Hirai, I., Fukui, N., Taguchi, M., Yamauchi, K., Nakamura, T., Okano, S., et al. (2013). Detection of chromosomal blaCTX-M-15 in Escherichia coli O25b-B2-ST131 isolates from the Kinki region of Japan. Int. J. Antimicrob. Agents 42, 500–506. doi: 10.1016/j.ijantimicag.2013.08.005
Imoto, A., Ooi, Y., Edogawa, S., Ogura, T., Masuda, D., Mohamed, M., et al. (2014). Liver abscess caused by CTX-M-55-type extended-spectrum β-lactamase (ESBL)-producing Salmonella enteritidis. Int. Med. 53, 1699–1703. doi: 10.2169/internalmedicine.53.2407
Jarlier, V., Nicolas, M. H., Fournier, G., and Philippon, A. (1988). Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10, 867–878. doi: 10.1093/clinids/10.4.867
Jiang, H. X., Song, L., Liu, J., Zhang, X. H., Ren, Y. N., Zhang, W. H., et al. (2014). Multiple transmissible genes encoding fluoroquinolone and third-generation cephalosporin resistance co-located in non-typhoidal Salmonella isolated from food-producing animals in China. Int. J. Antimicrob. Agents 43, 242–247. doi: 10.1016/j.ijantimicag.2013.12.005
Jiang, H. X., Tang, D., Liu, Y. H., Zhang, X. H., Zeng, Z. L., Xu, L., et al. (2012). Prevalence and characteristics of β-lactamase and plasmid-mediated quinolone resistance genes in Escherichia coli isolated from farmed fish in China. J. Antimicrob. Chemother. 67, 2350–2353. doi: 10.1093/jac/dks250
Kim, J. S., Kim, S., Park, J., Shin, E., Yun, Y. S., Lee, D. Y., et al. (2017). Plasmid-mediated transfer of CTX-M-55 extended-spectrum β-lactamase among different strains of Salmonella and Shigella spp. in the Republic of Korea. Diagn. Microbiol. Infect. Dis. 89, 86–88. doi: 10.1016/j.diagmicrobio.2017.03.014
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, L., Liao, X., Yang, Y., Sun, J., Li, L., Liu, B., et al. (2013). Spread of oqxAB in Salmonella enterica serotype Typhimurium predominantly by IncHI2 plasmids. J. Antimicrob. Chemother. 68, 2263–2268. doi: 10.1093/jac/dkt209
Liu, B. T., Yang, Q. E., Li, L., Sun, J., Liao, X. P., Fang, L. X., et al. (2013). Dissemination and characterization of plasmids carrying oqxAB-blaCTX-M genes in Escherichia coli isolates from food-producing animals. PLoS One 8:e73947. doi: 10.1371/journal.pone.0073947
Luk-In, S., Chatsuwan, T., Pulsrikarn, C., Bangtrakulnonth, A., Rirerm, U., and Kulwichit, W. (2018). High prevalence of ceftriaxone resistance among invasive Salmonella enterica serotype Choleraesuis isolates in Thailand: the emergence and increase of CTX-M-55 in ciprofloxacin-resistant S. Choleraesuis isolates. Int. J. Med. Microbiol. 308, 447–453. doi: 10.1016/j.ijmm.2018.03.008
Lupo, A., Saras, E., Madec, J. Y., and Haenni, M. (2018). Emergence of blaCTX-M-55 associated with fosA, rmtB and mcr gene variants in Escherichia coli from various animal species in France. J. Antimicrob. Chemother. 73, 867–872. doi: 10.1093/jac/dkx489
Mahrouki, S., Belhadj, O., Chihi, H., Mohamed, B. M., Celenza, G., Amicosante, G., et al. (2012). Chromosomal blaCTX-M-(1)(5) associated with ISEcp1 in Proteus mirabilis and Morganella morganii isolated at the Military Hospital of Tunis, Tunisia. J. Med. Microbiol. 61(Pt 9), 1286–1289. doi: 10.1099/jmm.0.039487-0
Majowicz, S. E., Musto, J., Scallan, E., Angulo, F. J., Kirk, M., O’Brien, S. J., et al. (2010). The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 50, 882–889. doi: 10.1086/650733
Nadimpalli, M., Fabre, L., Yith, V., Sem, N., Gouali, M., Delarocque-Astagneau, E., et al. (2018). CTX-M-55-type ESBL-producing Salmonella enterica are emerging among retail meats in Phnom Penh, Cambodia. J. Antimicrob. Chemother. 74, 342–348. doi: 10.1093/jac/dky451
Nguyen, D. T., Kanki, M., Nguyen, P. D., Le, H. T., Ngo, P. T., Tran, D. N., et al. (2016). Prevalence, antibiotic resistance, and extended-spectrum and AmpC β-lactamase productivity of Salmonella isolates from raw meat and seafood samples in Ho Chi Minh City, Vietnam. Int. J. Food Microbiol. 236, 115–122. doi: 10.1016/j.ijfoodmicro.2016.07.017
Norizuki, C., Kawamura, K., Wachino, J. I., Suzuki, M., Nagano, N., Kondo, T., et al. (2018). Detection of Escherichia coli producing CTX-M-1-group extended-spectrum β-lactamases from pigs in aichi prefecture, Japan, between 2015 and 2016. Jpn. J. Infect. Dis. 71, 33–38. doi: 10.7883/yoken.JJID.2017.206
Nurk, S., Bankevich, A., Antipov, D., Gurevich, A. A., Korobeynikov, A., Lapidus, A., et al. (2013). Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 20, 714–737. doi: 10.1089/cmb.2013.0084
Robicsek, A., Jacoby, G. A., and Hooper, D. C. (2006). The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 6, 629–640. doi: 10.1016/S1473-3099(06)70599-0
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Sjolund-Karlsson, M., Howie, R., Krueger, A., Rickert, R., Pecic, G., Lupoli, K., et al. (2011). CTX-M-producing non-Typhi Salmonella spp. isolated from humans, United States. Emerg. Infect. Dis. 17, 97–99. doi: 10.3201/eid1701.100511
Strahilevitz, J., Jacoby, G. A., Hooper, D. C., and Robicsek, A. (2009). Plasmid-mediated quinolone resistance: a multifaceted threat. Clin. Microbiol. Rev. 22, 664–689. doi: 10.1128/CMR.00016-09
Torpdahl, M., Hasman, H., Litrup, E., Skov, R. L., Nielsen, E. M., and Hammerum, A. M. (2017). Detection of mcr-1-encoding plasmid-mediated colistin-resistant Salmonella isolates from human infection in Denmark. Int. J. Antimicrob. Agents 49, 261–262. doi: 10.1016/j.ijantimicag.2016.11.010
Wang, J., Yao, X., Luo, J., Lv, L., Zeng, Z., and Liu, J. H. (2018a). Emergence of Escherichia coli co-producing NDM-1 and KPC-2 carbapenemases from a retail vegetable. China J. Antimicrob. Chemother. 73, 252–254. doi: 10.1093/jac/dkx335
Wang, J., Zeng, Z. L., Huang, X. Y., Ma, Z. B., Guo, Z. W., Lv, L. C., et al. (2018b). Evolution and Comparative Genomics of F33:A-:B- Plasmids Carrying blaCTX-M-55 or blaCTX-M-65 in Escherichia coli and Klebsiella pneumoniae Isolated from animals, food products, and humans in China. mSphere 3, e00137-18. doi: 10.1128/mSphere.00137-18
Wang, Y., Zhang, A., Yang, Y., Lei, C., Jiang, W., Liu, B., et al. (2017). Emergence of Salmonella enterica serovar Indiana and California isolates with concurrent resistance to cefotaxime, amikacin and ciprofloxacin from chickens in China. Int. J. Food Microbiol. 262, 23–30. doi: 10.1016/j.ijfoodmicro.2017.09.012
Wong, M. H., Liu, L., Yan, M., Chan, E. W., and Chen, S. (2015). Dissemination of IncI2 plasmids that harbor the blaCTX-M element among clinical Salmonella isolates. Antimicrob. Agents Chemother. 59, 5026–5028. doi: 10.1128/AAC.00775-15
Yan, H., Li, L., Alam, M. J., Shinoda, S., Miyoshi, S., and Shi, L. (2010). Prevalence and antimicrobial resistance of Salmonella in retail foods in northern China. Int J Food Microbiol 143, 230–234. doi: 10.1016/j.ijfoodmicro.2010.07.034
Yu, F., Chen, Q., Yu, X., Li, Q., Ding, B., Yang, L., et al. (2011). High prevalence of extended-spectrum β lactamases among Salmonella enterica Typhimurium isolates from pediatric patients with diarrhea in China. PLoS One 6:e16801. doi: 10.1371/journal.pone.0016801
Zhang, J., Zheng, B., Zhao, L., Wei, Z., Ji, J., Li, L., et al. (2014). Nationwide high prevalence of CTX-M and an increase of CTX-M-55 in Escherichia coli isolated from patients with community-onset infections in Chinese county hospitals. BMC Infect. Dis. 14:659. doi: 10.1186/s12879-014-0659-0
Zhang, W. H., Lin, X. Y., Xu, L., Gu, X. X., Yang, L., Li, W., et al. (2016). CTX-M-27 Producing Salmonella enterica Serotypes Typhimurium and indiana are prevalent among food-producing animals in China. Front. Microbiol. 7:436. doi: 10.3389/fmicb.2016.00436
Zhao, W. H., and Hu, Z. Q. (2013). Epidemiology and genetics of CTX-M extended-spectrum β-lactamases in Gram-negative bacteria. Crit. Rev. Microbiol. 39, 79–101. doi: 10.3109/1040841X.2012.691460
Zhao, X., Ye, C., Chang, W., and Sun, S. (2017). Serotype distribution, antimicrobial resistance, and class 1 integrons profiles of Salmonella from Animals in Slaughterhouses in Shandong Province, China. Front. Microbiol. 8:1049. doi: 10.3389/fmicb.2017.01049
Keywords: chromosomal, blaCTX-M-55, Salmonella, Indiana, transfer
Citation: Zhang C-Z, Ding X-M, Lin X-L, Sun R-Y, Lu Y-W, Cai R-M, Webber MA, Ding H-Z and Jiang H-X (2019) The Emergence of Chromosomally Located blaTX-M-55 in Salmonella From Foodborne Animals in China. Front. Microbiol. 10:1268. doi: 10.3389/fmicb.2019.01268
Received: 03 December 2018; Accepted: 22 May 2019;
Published: 05 June 2019.
Edited by:
Shaolin Wang, China Agricultural University, ChinaReviewed by:
Liang Li, Los Angeles Biomedical Research Institute, United StatesKristina Kadlec, Friedrich Loeffler Institute, Germany
Copyright © 2019 Zhang, Ding, Lin, Sun, Lu, Cai, Webber, Ding and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huan-Zhong Ding, aHpkaW5nQHNjYXUuZWR1LmNu; Hong-Xia Jiang, aHhqaWFuZ0BzY2F1LmVkdS5jbg==
†These authors have contributed equally to this work
 Chuan-Zhen Zhang1,2†
Chuan-Zhen Zhang1,2† Yue-Wei Lu
Yue-Wei Lu Huan-Zhong Ding
Huan-Zhong Ding Hong-Xia Jiang
Hong-Xia Jiang