- 1Department of Biochemistry, Genetics and Microbiology, Forestry and Agricultural Biotechnology Institute, University of Pretoria, Pretoria, South Africa
- 2Agricultural Research Council, Plant Health and Protection Institute, Pretoria, South Africa
- 3South African National Biodiversity Institute, Pretoria National Botanical Garden, Pretoria, South Africa
- 4Department of Botany and Plant Biotechnology, University of Johannesburg, Johannesburg, South Africa
- 5Autonomous Department of Microbial Biology, Faculty of Agriculture and Biology, Warsaw University of Life Sciences – SGGW, Warsaw, Poland
Vachellia karroo (formerly Acacia karroo) is a wide-spread legume species indigenous to southern Africa. Little is known regarding the identity or diversity of rhizobia that associate with this plant in its native range in South Africa. The aims of this study were therefore: (i) to gather a collection of rhizobia associated with V. karroo from a wide range of geographic locations and biomes; (ii) to identify the isolates and infer their evolutionary relationships with known rhizobia; (iii) to confirm their nodulation abilities by using them in inoculation assays to induce nodules under glasshouse conditions. To achieve these aims, soil samples were collected from 28 locations in seven biomes throughout South Africa, which were then used to grow V. karroo seedlings under nitrogen-free conditions. The resulting 88 bacterial isolates were identified to genus-level using 16S rRNA sequence analysis and to putative species-level using recA-based phylogenetic analyses. Our results showed that the rhizobial isolates represented members of several genera of Alphaproteobacteria (Bradyrhizobium, Ensifer, Mesorhizobium, and Rhizobium), as well as Paraburkholderia from the Betaproteobacteria. Our study therefore greatly increases the known number of Paraburkholderia isolates which can associate with this southern African mimosoid host. We also show for the first time that members of this genus can associate with legumes, not only in the Fynbos biome, but also in the Albany Thicket and Succulent Karoo biomes. Twenty-six putative species were delineated among the 88 isolates, many of which appeared to be new to Science with other likely being conspecific or closely related to E. alkalisoli, M. abyssinicae, M. shonense, and P. tropica. We encountered only a single isolate of Bradyrhizobium, which is in contrast to the dominant association of this genus with Australian Acacia. V. karroo also associates with diverse genera in the Grassland biome where it is quite invasive and involved in bush encroachment. Our findings therefore suggest that V. karroo is a promiscuous host capable of forming effective nodules with both alpha- and beta-rhizobia, which could be a driving force behind the ecological success of this tree species.
Introduction
Vachellia karroo (formerly Acacia karroo) (Banfi and Galasso, 2008; Kyalangalilwa et al., 2013) is a legume tree species belonging to the mimosoid clade, which is an informal group nested within the newly recircumscribed Caesalpinioideae subfamily (LPWG, 2017). Vachellia contains 161 species and has a pantropical distribution, although 73 species can be found in Africa and Madagascar (Lewis, 2005; Kyalangalilwa et al., 2013). This is a relatively small genus when compared to the Australian endemic genus Acacia sensu stricto, which contains 1021 species (Kyalangalilwa et al., 2013). V. karroo is commonly referred to as “sweet thorn” and has significant economic importance (Hayward, 2004). It has been shown to have medicinal properties as its leaves and bark can be used to treat diarrhoea, while an exudate from the tree can be used as an emollient for conjunctivitis and parts of the tree also have broad-spectrum anti-microbial activity (van Wyk, 2011; Cock and van Vuuren, 2015; Maroyi, 2017). The leaves, pods and fruits have the potential to be used as fodder for ruminant livestock (Gxasheka et al., 2015; Brown et al., 2016; Dingaan and du Preez, 2018), while its flowers are important for honey production (Dingaan and du Preez, 2018). With regards to human consumption, its roasted seeds can be used as a coffee substitute and its gum in the production of sweets (Cock and van Vuuren, 2015).
Among tree species, V. karroo has the widest distribution in South Africa (Dingaan and du Preez, 2018), extending from the Southwestern Cape northwards into Namibia, Botswana, Zambia, Angola and Zimbabwe (Taylor and Barker, 2012). The species can grow in different soils (ranging from sand to heavy clays), in relatively wet to arid areas (200–1500 mm annual rainfall) and from sea level to an elevation of 1800 m (Maroyi, 2017). As a pioneer, V. karroo inhabits various biomes and has the ability to grow and thrive in a range of climatic and edaphic conditions (Brain et al., 1997; Taylor and Barker, 2012), where it is resistant to factors such as salinity, drought, fire and frost (Brain et al., 1997). This is thought to contribute significantly to its invasiveness, because V. karroo is the foremost woody invader in South African grasslands (O’Connor, 1995; Taylor and Barker, 2012), a phenomenon referred to as “bush encroachment” (Dingaan and du Preez, 2018).
As with most other legumes, V. karroo can form nitrogen-fixing root nodules (Barnes, 2001) in mutualistic symbioses with specific soil bacteria referred to as rhizobia (Poole et al., 2018). These bacteria do not form a monophyletic group and are represented by species in 18 genera of the Alpha- and Betaproteobacteria (Lee and Hirsch, 2006; Poole et al., 2018; de Lajudie et al., 2019). To differentiate between rhizobia from these two classes, they are informally referred to as alpha- and beta-rhizobia (Moulin et al., 2001). Of these two groups, the beta-rhizobia was discovered more recently (Moulin et al., 2001), with one of the first recognised beta-rhizobial species, Paraburkholderia tuberum (formerly Burkholderia tuberum; Sawana et al., 2014), reported as a symbiont of the indigenous South African papilionoid legume, Aspalathus carnosa (Lemaire et al., 2016a), while also being able to nodulate several Cyclopia species and siratro (Macroptilium atropurpureum; Elliott et al., 2007a). Numerous nodulating Paraburkholderia species have since been reported from South Africa, particularly from the Cape Floristic Region in the Western Cape Province (de Meyer et al., 2013a,b, 2014; Steenkamp et al., 2015), and all in association with papilionoid legumes. This is in contrast to other parts of the world, especially South America, where Paraburkholderia is primarily associated with mimosoid legumes (Chen et al., 2005, 2007; Bontemps et al., 2010; Mishra et al., 2012; Bournaud et al., 2013). Various studies thus suggest that South Africa and South America represent distinct centres of diversity for the beta-rhizobial members of this genus (Gyaneshwar et al., 2011; Beukes et al., 2013; Lemaire et al., 2015a; Silva et al., 2018).
Very little information is available regarding the identity and diversity of the rhizobial partners of V. karroo, especially from South Africa and areas where V. karroo occurs naturally (Ngwenya et al., 2016). The only previous study that has focused on V. karroo rhizobia in South Africa showed it is nodulated by diverse symbionts, representing “fast- and slow-growers,” (Ngwenya et al., 2016), which is similar to what has been found in an earlier study from Libya (Mohamed et al., 2000). The terms “fast- and slow-growing rhizobia” typically refer to the alpha-rhizobial genera Rhizobium and Bradyrhizobium, respectively (Leary et al., 2006). The first DNA-based study to attempt identification of the rhizobial symbionts of V. karroo was performed in Kenya with a portion of the 16S ribosomal RNA (rRNA) gene to diagnose an isolate of Rhizobium (McInroy et al., 1999). A more recent study from Algeria employed full-length 16S rRNA gene sequences to identify V. karroo symbionts as members of the alpha-rhizobial genera Rhizobium and Ensifer (Boukhatem et al., 2012). Furthermore, a number of studies on the rhizobia of Acacia sensu lato (excluding V. karroo) in the northern, eastern and western parts of Africa have shown that these isolates represent members of the alpha-rhizobial genera Bradyrhizobium, Ensifer, Mesorhizobium, and Rhizobium (Khbaya et al., 1998; McInroy et al., 1999; Mohamed et al., 2000; Ba et al., 2002; Odee et al., 2002; Wolde-meskel et al., 2004, 2005; Sarr et al., 2005; Romdhane et al., 2006; Diouf et al., 2007; Degefu et al., 2011; Abdi et al., 2012).
The mutualistic interaction between rhizobia and legumes could be an important factor when the plant is introduced into a new environment (Stanton-Geddes and Anderson, 2011; Klock et al., 2015; La Pierre et al., 2017). The symbiosis is especially important to the establishment, growth and survival of the legume when soils are nutrient-poor (Thrall et al., 2000, 2005; Klock et al., 2015). Knowledge regarding the diversity and identity of the rhizobial symbionts of V. karroo would therefore be crucial for understanding their role in the ecological success of this legume. To address this knowledge gap, our overall goal was to determine the identity of the rhizobial symbionts of V. karroo growing in diverse biomes across South Africa. The aims of the study were: (i) to assemble a collection of rhizobial isolates associated with V. karroo from a broad range of geographic locations and biomes, and (ii) to determine the identity of the rhizobia using DNA sequence information from the widely used 16S rRNA and recA genes. How strict or broad the range of possible rhizobial partners of V. karroo is could provide important information on the potential of this legume to find a suitable rhizobial partner in a particular environment.
Materials and Methods
Isolate Collection and Nodulation Confirmation
To collect rhizobial isolates associated with V. karroo from a wide geographic area in South Africa, we used a “trapping” approach. In this approach, soil close to the roots of V. karroo (i.e., rhizosphere soil) was collected and used for planting and growing the germinated seeds of the host plant under nitrogen-free conditions. Such an environment forces the seedlings to interact with the rhizobia in the collected soil and form nodules to obtain nitrogen for growth. To this end, we collected soil from 28 locations across six of South Africa’s nine provinces and included seven biomes [biome definitions sensu Mucina and Rutherford (2006); Table 1 and Figure 1A]. Seedlings of V. karroo were grown from seed that were first scarified with concentrated sulphuric acid for 2 h, followed by five rinses in sterile distilled water (sH2O) and a further three-hour imbibement in sH2O. The swollen seeds (indicative of a damaged seed coat through which water could be absorbed) were plated onto 1.5% water agar (w/v) plates and incubated at 28°C in the dark for 2 days for germination to occur.
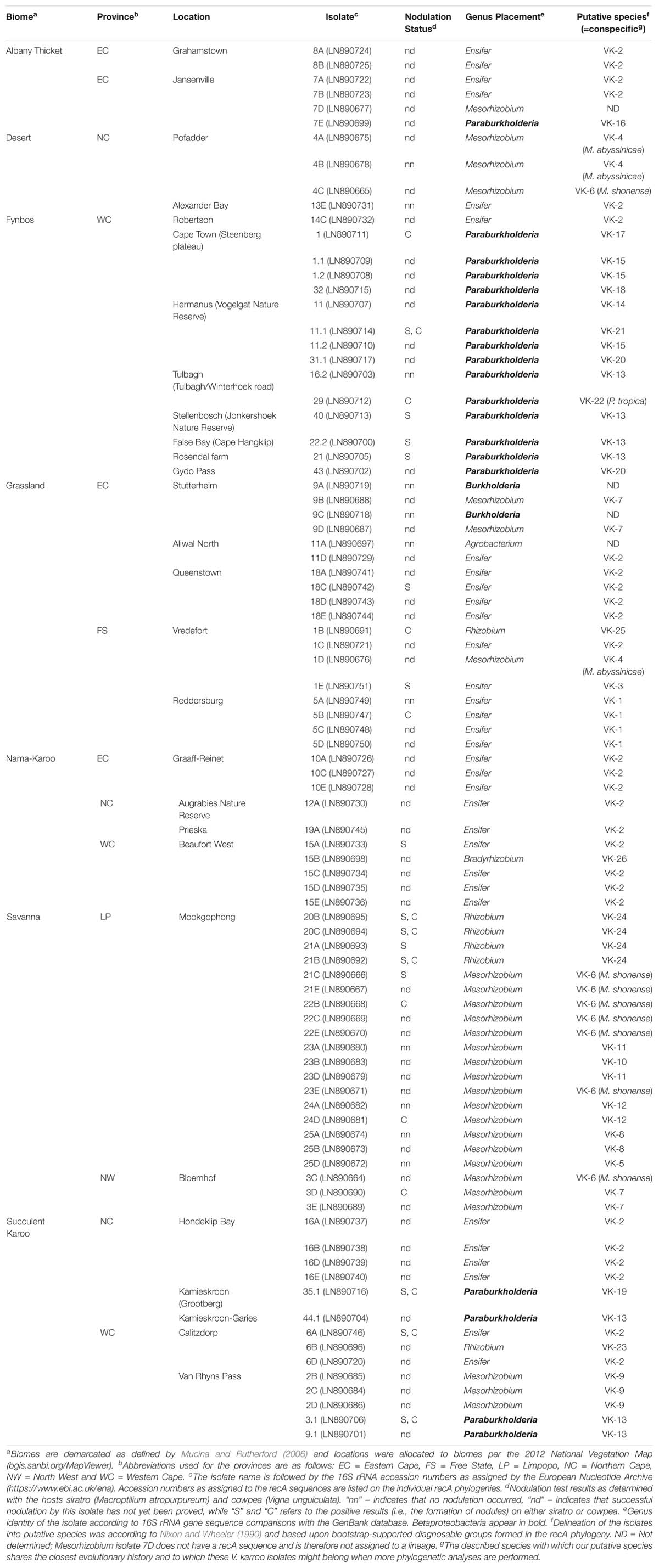
Table 1. Information regarding the rhizobia associated with Vachellia karroo included in this study.
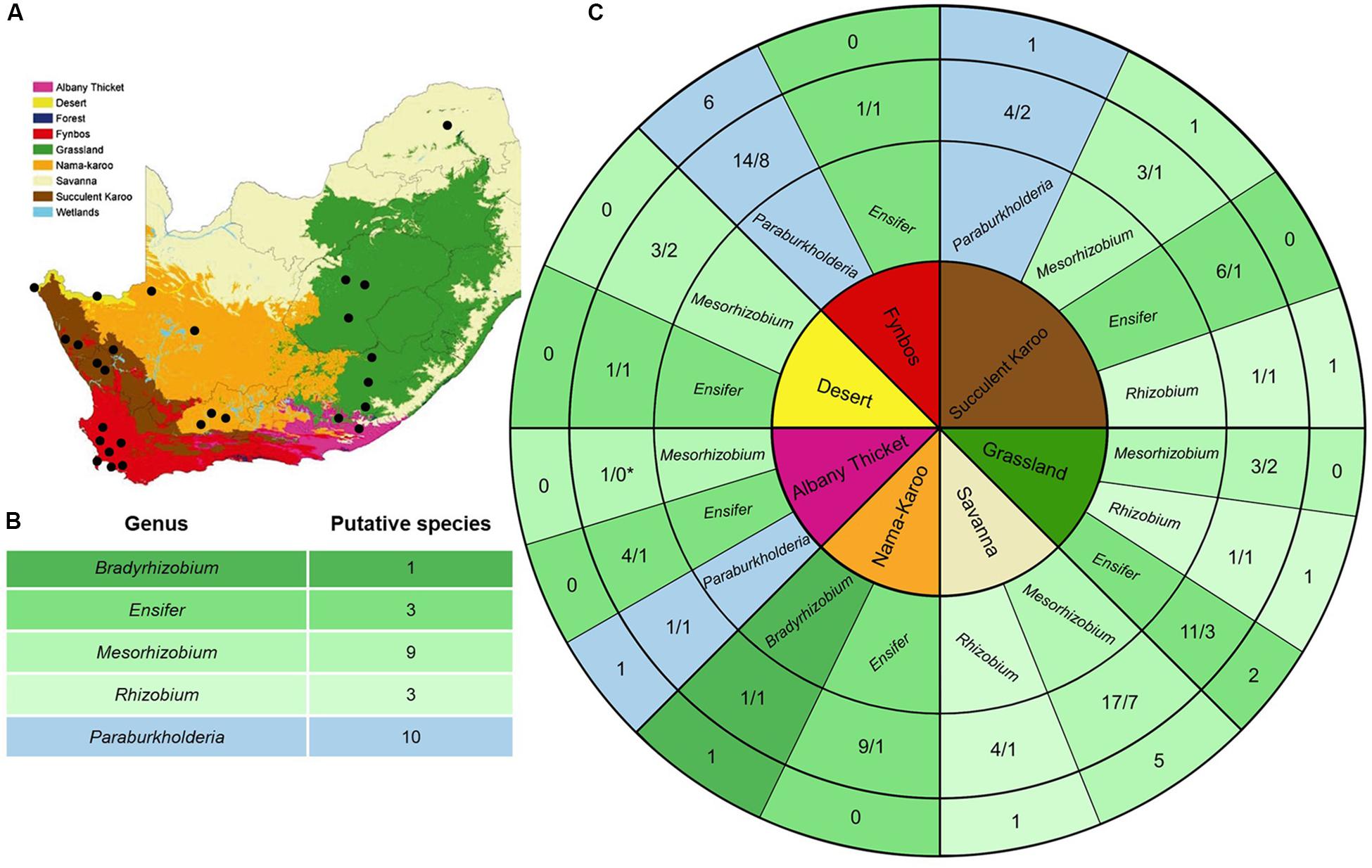
Figure 1. (A) Biome map of South Africa, Lesotho and Swaziland (as in Driver et al., 2005; from the national Spatial Biodiversity Assessment of 2004, courtesy of SANBI). The nine biomes are coloured according to the key and black dots indicate the sampling localities. (B) Summary of the putative species encountered in this study. (C) The core of this diagram lists the seven sampled biomes (colour-coded as in A). The second track indicates the genera isolated from each biome (with alpha-rhizobia appearing in shades of green while beta-rhizobia appear in blue), while the third track shows the number of isolates sampled from each genus for that specific biome relative to the number of recA phylogeny-based putative species they formed part of. The outer track shows the number of unique recA phylogeny-based putative species recovered for a specific biome for each rhizobial genus. We were unable to amplify the recA gene sequence for the Mesorhizobium isolate recovered from the Albany Thicket which is why it is indicated with an asterisk, as it could not be assigned to a species.
The “trapping” experiments were performed in a glasshouse where the seedlings were planted in Leonard jars (Howieson and Dilworth, 2016). Three seedlings were planted in each Leonard jar. The sterile sand in the container was then covered with 20 g of the soil collected from a specific location, thereby filling up the holes in which the seedlings were planted (i.e., seedlings were thus only provided with the nitrogen potentially contained within the 20 g of soil). Five biological replicates were performed for each sampling location and there was also a control jar to which no sampled soil was added. The growth conditions in the glasshouse was set to a day/night cycle of 14 and 10 h with temperatures from 27 to 28 and 15°C, respectively. The plants were checked for nodule formation 4 months after planting.
Vachellia karroo nodules resulting from “trapping” in Western Cape soils were obtained from Dr. M. Maistry (Research leader Dr. S. Chimphango, Department of Biological Sciences, University of Cape Town). The following protocol was used: silica sand was chemically washed with 0.1% (v/v) hydrochloric acid and then rinsed six times with sH2O until a neutral pH was achieved. Scarified V. karroo seeds were then mixed with the treated silica sand and transferred into pots (10 cm in diameter) containing the collected Western Cape soils. Seedlings were watered until they emerged and then treated twice a week with nitrogen-free Hoagland solution (Howieson and Dilworth, 2016) until the plants were harvested after 4 months.
Root nodules that developed on V. karroo plants were surface sterilised (15 min in 3.5% (w/v) sodium hypochlorite), rinsed five times with sH2O and then crushed onto Yeast Mannitol Agar (YMA; Howieson and Dilworth, 2016) to obtain pure single colonies. To prove the nodulation ability of the pure isolates, glasshouse trials were performed with the promiscuous plants (i.e. able to interact with many rhizobial partners) siratro (Macroptilium atropurpureum) and cowpea (Vigna unguiculata). The siratro and cowpea seeds were germinated according to Beukes et al. (2013). Following this procedure, pure cultures of the original “trapped” rhizobia were used as inoculum for the seedlings under nitrogen-free conditions in sterile sand. The resulting nodules were treated as mentioned above and from the pure cultures, 16S rRNA amplification and sequencing were performed (as described below). These sequences were then compared with those of the original “trapping” isolates to confirm their identities. A nodule from each plant (where possible) was also cut open to view the interior, which if pink in colour is due to the presence of leghaemoglobin (an oxygen-carrying protein essential for nitrogen-fixation). The nodules were therefore assumed to be actively fixing nitrogen and were then regarded as effective nodules.
DNA Extraction, PCR and Sequencing
To determine the identity and diversity of the rhizobial symbionts from the different locations, 16S rRNA and recA gene sequences were used. For this purpose, pure cultures of each isolate were grown in 5 ml Yeast Mannitol broth at 28°C with shaking (ca. 130 rpm) for 7 days. Centrifugation for 5 min at 3 824 rcf was used to concentrate the bacterial cells into a pellet. Thereafter DNA was extracted using the protocol of Aljanabi and Martinez (1997).
For amplification of a ca. 1490 bp fragment of the 16S rRNA gene, the primers 16S-F (5′ AGA GTT TGA TCC TGG CTC AG 3′) (Suau et al., 1999) and 16S-R (5′ TAC CTT GTT ACG ACT TCA CCC CA 3′) (Logan et al., 2000) were used. Every 50 μl PCR reaction consisted of 10 μM of each primer, 0.1 U/μL Super-Therm Taq DNA polymerase with reaction buffer (Southern Cross Biotechnology, Cape Town, SA), 25 mM MgCl2, 2.5 mM of each dNTP and 50 to 100 ng genomic DNA. The PCR cycling conditions were: initial denaturation of 94°C for 2 min, followed by 35 cycles of denaturation (94°C for 1 min), annealing (60°C for 1 min) and elongation (72°C for 1 min), with a final elongation step at 72°C for 7 min. The resulting amplicons were cleaned using Centri-SepTM Spin Columns (Life Technologies, CA, United States). The purified amplicons were then subjected to Sanger sequencing, with the same set of primers and the ABI PRISM BigDye Terminator v3.0 Cycle Sequencing Kit on a ABI 3100 Automated Capillary DNA sequencer (Applied Biosystems, CA, United States).
Individual chromatograms were analysed with Chromas Lite v2.01 (Technelysium, Queensland, Australia), before consensus sequences were generated with BioEdit v7.053 (Hall, 1999). By using blastn (Altschul et al., 1990), these nearly complete 16S rRNA sequences were compared to those in GenBank (National Centre for Biotechnology Information http://www.ncbi.nlm.nih.gov/; Benson et al., 2017). The latter allowed identification of the isolates to genus-level.
For all rhizobial isolates, a portion of the recA gene was then sequenced. For all the alpha-rhizobial isolates, a ca. 600 bp region of recA was amplified with the primer pair recA-41F (5′ TTC GGC AAG GGM TCG RTS ATG 3′) and recA-640R (5′ ACA TSA CRC CGA TCT TCA TCG 3′) (Vinuesa et al., 2005), while a ca. 850 bp region for the beta-rhizobia were amplified with the primers BUR1 (5′ GAT CGA RAA GCA GTT CGG CAA 3′) and BUR2 (5′ TTG TCC TTG CCC TGR CCG AT 3′) (Payne et al., 2005). Amplification and sequencing followed the same procedure as for the 16S rRNA gene, except that the annealing temperature of the PCR was 62°C. The resulting recA sequences were analysed with ChromasLite and BioEdit as described above.
Sequence Alignments and Phylogenetic Analyses
After determining the genus to which each isolate belonged, recA nucleotide sequence datasets were constructed by adding the sequence for each described species in the specific genus (as listed on the List of Prokaryotic Names with Standing in Nomenclature (LPSN); www.bacterio.net; Euzéby, 1997; Parte, 2013). We also searched literature and added species which have not yet been included on LPSN (still awaiting validation), as well as any undescribed isolates (which do not yet have species names assigned), originating in Africa or which associates with hosts in Acacia sensu lato (see Supplementary Tables S1–S5 for accession numbers and associated references; also for the accessions of the focal isolates). The multiple sequence datasets were manually aligned in BioEdit by making use of the inferred amino acid sequences.
The aligned datasets were then used to create haplotype files with the software DnaSP v5 (Librado and Rozas, 2009). These files only contained unique taxa (i.e. isolates with identical sequences were collapsed into one haplotype and represented by a single sequence). Parameters for the best-fit evolutionary model of each dataset were estimated with jModelTest v2.1.7 (Guindon and Gascuel, 2003; Darriba et al., 2012). These datasets were subjected to maximum-likelihood phylogenetic analysis using the best-fit model in PhyML v3.1 (Guindon and Gascuel, 2003). Branch support was estimated using the same approach and 1000 pseudoreplicates (Felsenstein, 1985). The initial number of taxa included, the resulting number of haplotypes, the length of the alignment and best-fit evolutionary model for each dataset are summarised in Supplementary Table S6. For each dataset, a group of three isolates from different, but closely related genera, were included for outgroup purposes. The resulting phylogenies were edited in MEGA6 (Tamura et al., 2013), with the final trees showing all the isolates constituting a haplotype, as well as information regarding the country of origin and host for each isolate.
Results
The “trapping” experiments resulted in the isolation of 88 bacterial strains (Table 1). Comparison of their 16S rRNA sequences to those in GenBank revealed that they represented seven genera (Table 1). According to these blastn results, 67 of the isolates represented Alphaproteobacteria and formed part of the genera Ensifer (32 isolates), Mesorhizobium (27 isolates), Rhizobium (six isolates), Bradyrhizobium (one isolate) and Agrobacterium (one isolate). The remaining 21 isolates represented Betaproteobacteria and formed part of the genera Paraburkholderia (19 isolates) and Burkholderia (two isolates).
Isolates identified as belonging to the genera Agrobacterium, Bradyrhizobium and Burkholderia did not form nodules (Table 1), during nodulation tests. Four isolates for Mesorhizobium, eight for Paraburkholderia and five each for Ensifer and Rhizobium did, however, induce effective nodules on both or one of these legumes. Those that induced nodules in both siratro and cowpea were as follows: Paraburkholderia spp. 3.1, 11.1, and 35.1; Ensifer sp. 6A and lastly Rhizobium spp. 20B, 20C, and 21B. Those that nodulated only cowpea were: Ensifer sp. 5B, Mesorhizobium spp. 3D, 22B, 24D; Rhizobium sp. 1B and Paraburkholderia spp. 1 and 29. Those that nodulated only siratro were: Ensifer spp. 1E, 15A, 18C; Paraburkholderia spp. 21, 22.2, 40; Rhizobium sp. 21A and Mesorhizobium sp. 21C. In all these cases, we confirmed that the 16S rRNA sequences generated for the isolates obtained from the sirato and cowpea nodules matched those of the isolates originally used in the individual nodulation tests. Although we could not prove nodulation for all 88 isolates, it could be possible that we have not been able to mimic the conditions required for successful nodulation in the glasshouse. A point also made by Elliott et al. (2007a), when they could not prove nodulation of Aspalathus species by P. tuberum. The hosts cowpea and siratro are also routinely used during nodulation tests as they are considered promiscuous, this is, however, a generalisation and we have encountered situations where it appears as if one of these hosts are less readily nodulated by certain rhizobial isolates (data not shown).
When investigating the recovery of specific rhizobial genera from the soil samples collected in the seven biomes (i.e., Albany Thicket, Desert, Fynbos, Grassland, Nama-Karoo, Savanna and Succulent Karoo), Ensifer isolates were obtained from six of the biomes but were not encountered in soils from the Savanna biome (Table 1 and Figure 1C). In this biome only isolates of Mesorhizobium and Rhizobium were recovered, where Mesorhizobium were commonly isolated (i.e., 17 of the 21 isolates recovered from this biome) (Table 1 and Figure 1C). Mesorhizobium isolates were not found in soils from of the Nama-Karoo or Fynbos biomes. Four of the Rhizobium isolates were recovered from Savanna soil, while one isolate originated from the Succulent Karoo in the Western Cape and one from the Grassland biome (Figure 1C and Table 1). Of the 19 Paraburkholderia isolates, 14 were recovered from soils of the Fynbos biome in the Western Cape, while five were found in soils from the Succulent Karoo and one was recovered from the Albany Thicket biome (Figure 1C and Table 1). The two Burkholderia isolates (9A and 9C) and single Agrobacterium isolate (11A) were all recovered from Grassland biome soil, while the only Bradyrhizobium isolate originated from soil collected in the Nama-Karoo biome. Based on our data, the Grassland biome soils apparently harboured a diverse assemblage of bacteria capable of interacting with V. karroo (either as nodule symbionts or perhaps nodule endophytes) as isolates from five genera were recovered in these samples.
In order to delineate the isolates examined in this study to species level, we utilised the recA sequences. Following Nixon and Wheeler’s (1990) approach, we regarded the smallest diagnosable and bootstrap-supported groups of isolates on our phylogenetic trees as putative species. In this way, a total of 26 putative species (designated VK-1 to VK-26) were identified among 85 isolates (Figure 1B). The two Burkholderia isolates and single Agrobacterium isolate were not included in these analyses, as the focus of our study is potential rhizobia and there is currently very little evidence that these isolates can form functional nodules. As far as the authors are aware there is only one study which has proven that an Agrobacterium isolate (IRBG74) can successfully nodulate Sesbania spp. and does contain a symbiotic plasmid (Cummings et al., 2009). Note also, that the overall species groupings recovered from these phylogenetic trees (Supplementary Figures S1–S5) generally matched those reported in other studies (Martens et al., 2008; Rivas et al., 2009; Degefu et al., 2011, 2013a; Estrada-de los Santos et al., 2013).
Although Ensifer species made up the bulk of the sample set, we only delineated three putative species (VK-1 to VK-3) among the 32 isolates (Supplementary Figure S1). VK-1 contained four of our isolates (i.e., Ensifer spp. 5A-D), as well as isolate AC11d obtained from Vachellia seyal in Ethiopia (Degefu et al., 2012). VK-3 was represented by a large group, containing isolates from a variety of Vachellia hosts in Ethiopia (Degefu et al., 2012), as well as our isolate 1E. The remainder of our Ensifer isolates formed part of VK-2, which also included Ethiopian isolates (from various Vachellia species), as well as an isolate from the root nodules of Lessertia from South Africa (Lemaire et al., 2015a). None of our isolates showed a close phylogenetic relationship with those from Vachellia jacquemontii or Tephrosia spp. from India (Gehlot et al., 2012; Sankhla et al., 2017), nor to isolates of Vachellia tortilis from Morocco (Sakrouhi et al., 2016). No known species appeared to be closely related to VK-3, while the closest phylogenetic neighbour of VK-1 and VK-2 was E. alkalisoli (98% bootstrap support) and E. fredii (87% bootstrap support), respectively.
The Mesorhizobium isolates from this study separated into nine putative species (VK-4 to VK-12) (Supplementary Figure S2). Of these, five were represented by single isolates (VK-5, VK-8, VK-9, VK-10, and VK-12), none of which were closely related to known Mesorhizobium species. Two of our putative species are likely conspecific with M. abyssinicae (i.e., VK-4 with 93% bootstrap support) and M. shonense (i.e., VK-6 with 89% bootstrap support), respectively. Both VK-4 and VK-6 also contained several other Vachellia- or Senegalia-associated isolates from Ethiopia (Degefu et al., 2011). The remaining two putative species (VK-7 and VK-11) included isolates from our study only, and neither received bootstrap-support for their relationships with known species of Mesorhizobium. The phylogenetic analysis included many South African isolates of Mesorhizobium (predominantly from papilionoid hosts) (Supplementary Figure S2), but they did not group closely with any of our isolates from V. karroo.
The 19 Paraburkholderia isolates represented ten putative species (VK-13 to VK-22) (Figure 2; Supplementary Figure S3). Of these, five (VK-14, VK-18, VK-19, VK-21, and VK-22) were represented by single isolates (Figure 2 and Supplementary Figure S3), while VK-15 consisted of three isolates representing one haplotype. The remaining four putative species all associated with South African papilionoid legumes only: VK-13 consisted of isolates from this study only (96% support), VK-16 included ten symbionts of Hypocalyptus sophoroides (Beukes et al., 2013), VK-17 included isolate CS13775 from Indigofera ionii (Lemaire et al., 2016a) and VK-20 included isolate UCT70 from Cyclopia maculata (Kock, 2003; Beukes et al., 2013). The putative species VK-13, VK-14, VK-18, VK-19, and VK-20 did not form supported groupings with known Paraburkholderia species. Of the remaining five, VK-15 was closely related to two species (i.e., P. phenazinium and P. fungorum with 92% bootstrap support), VK-16 was closely related to P. kirstenboschensis (87% bootstrap support), VK-17 grouped closely with P. pallidirosa (99% bootstrap support), VK-19 grouped with P. tuberum (80% bootstrap support), while VK-22 were very closely related and perhaps conspecific with P. tropica (100% bootstrap support) (Figure 2 and Supplementary Figure S3).

Figure 2. A maximum-likelihood phylogeny of the recA locus for the beta-rhizobial genus Paraburkholderia. Isolates from this study are listed in red, while type strains of species appear in bold. The delineated Vachellia karroo-associated lineages (VK-13 to VK-22) appear in blue. In order to minimize the phylogeny some lineages (which do not contain any isolates from this study), have been collapsed. The expanded tree, containing information for the host or source and geographic origin of each isolate, appears as Supplementary Figure S3. GenBank accession numbers for all the isolates included in this phylogeny are listed in Supplementary Table S3. Nodes with ≥50% are indicated, while the scale bar shows the number of nucleotide substitutions per site.
The six Rhizobium isolates separated into three putative species (VK-23 to VK-25) (Supplementary Figure S4). Of these, VK-23 was represented by a single isolate and was placed as a sister taxon to a group consisting of R. sophorae and R. sophoriradicis (51% bootstrap). VK-24, including four of our isolates, was closely related to R. calliandrae (57% bootstrap support). VK-25 was also represented by a single isolate (1B) and was closely related to R. endophyticum (93% bootstrap support). The single Bradyrhizobium isolate recovered in this study (15B) represented putative species VK-26, which appeared to be closely related to B. liaoningense (53% bootstrap support; Supplementary Figure S5).
Discussion
This study investigated the root nodule bacteria of V. karroo sampled across a broad geographical area in South Africa, spanning sites in six provinces and seven biomes. Our results revealed that taxonomically diverse bacteria from both the Alpha- and Betaproteobacteria, occupy the root nodules of this legume. Among the 88 isolates collected, 66 belonged to various genera in the Alphaproteobacteria, while 19 were placed in the Betaproteobacteria. These bacteria separated into 26 putative species (VK-1 to VK-26) with representatives from the alpha-rhizobial genera Ensifer, Mesorhizobium, Rhizobium, and Bradyrhizobium and the beta-rhizobial genus Paraburkholderia. Three of the isolates examined were from genera (Agrobacterium and Burkholderia) with no known rhizobia (Mrabet et al., 2006; Vial et al., 2011; Mousavi et al., 2014) and most probably represent endophytes. These three isolates were also not successful in nodulation tests, which is congruent with the fact that members of Burkholderia sensu stricto do not nodulate legumes (Gyaneshwar et al., 2011). Therefore, apart from identifying for the first time the diverse rhizobial symbionts associating with South African V. karroo, this study also extends the known number of beta-rhizobial isolates able to associate with an indigenous South African member of the mimosoid clade.
Although some genera appeared to be common symbionts of V. karroo, they were not necessarily common in all the biomes examined. Ensifer, for instance, was found in all biomes sampled except for the Savanna biome, while Mesorhizobium was particularly common in this biome, but not encountered in the Fynbos or Nama-Karoo biomes. This trend is generally consistent with Mucina and Rutherford’s (2006) view of biomes as “subcontinental biotic super communities.” Accordingly, each community consists of a similar vegetation structure exposed to similar macroclimatic patterns. For example, the Succulent Karoo biome represents a semidesert and the Fynbos biome is a relatively moist winter-rainfall region. The Savanna and Grassland biomes are both summer rainfall regions, but because the Grassland biome is part of the elevated interior of South Africa, it is generally cooler than the Savanna biome (Mucina and Rutherford, 2006). The Albany Thicket biome is characterised by fragmented vegetation types due to the two climate systems present in the area (all-year rainfall in the southwest and summer rainfall in the northeast) (Mucina and Rutherford, 2006). The Desert biome experiences the lowest rainfall and highest temperatures of all South African biomes (Mucina and Rutherford, 2006). Different sets of environmental factors therefore impact upon the ecology of V. karroo in the various biomes. These factors likely also play key roles in shaping, not only the rhizosphere communities associated with this plant, but also its rhizobial symbionts.
As suggested previously in Paraburkholderia, the diversity and distribution of a particular rhizobial group may be determined by environmental factors, as well as life history traits. In the current study, the members of this genus were more prevalent in the Fynbos biome, as was also shown in previous studies (Lemaire et al., 2015b, 2016a,b). This is consistent with the notion that South Africa represents one of the two known centres of diversity for this genus, with the other centre being South America (Gyaneshwar et al., 2011; Beukes et al., 2013; Liu et al., 2014). In Paraburkholderia it has been shown that, instead of legume host range, various environmental factors shape species distribution (Garau et al., 2009; Bontemps et al., 2010; Beukes et al., 2013). These include soil pH (Garau et al., 2009; Bontemps et al., 2010; Gyaneshwar et al., 2011; Mishra et al., 2012; Beukes et al., 2013; de Castro Pires et al., 2018; Silva et al., 2018), and elevation (especially its influence on temperature and precipitation levels) (Bontemps et al., 2010; Beukes et al., 2013; Lemaire et al., 2015a). The tendency of papilionoid legumes in the Cape Floristic Region or Fynbos biome to be nodulated by Paraburkholderia (Elliott et al., 2007a; Beukes et al., 2013; de Meyer et al., 2013a,b; Howieson et al., 2013) may therefore be comparable to that of the Mimosa hosts growing in the Cerrado and Caatinga regions of Brazil (Bontemps et al., 2010; dos Reis et al., 2010), as both of these regions have acidic soils that are low in nutrients (Suárez-Moreno et al., 2012; Beukes et al., 2013; Ndlovu et al., 2013). However, Mimosa hosts in Brazil can associate with Rhizobium species when the soil pH is neutral to alkaline (Baraúna et al., 2016). Dipogon lignosus, an indigenous South African legume from the tribe Phaseoleae (Liu et al., 2014; Lemaire et al., 2016a) is also found as a weed in New Zealand (Liu et al., 2014) where it associates with Paraburkholderia dipogonis (Sheu et al., 2015; Dobritsa and Samadpour, 2016) in soils with an acidic pH but with Bradyrhizobium or Rhizobium in soils with a pH closer to neutral (Liu et al., 2014). These acidic soil conditions are apparently ideally suited to Paraburkholderia species which is why they can outcompete co-existing alpha-rhizobia (Elliott et al., 2009; Suárez-Moreno et al., 2012; Beukes et al., 2013). Similar factors likely also determine the ecological success of the various rhizobial groups in the other six biomes sampled here.
The first report of a rhizobial association between V. karroo and a beta-rhizobium, was a study by Moulin et al. (2014) which showed that Paraburkholderia phymatum STM815T can effectively nodulate this host. The current study contributes another 19 Paraburkholderia isolates with a South African origin to the growing group of beta-rhizobia that can potentially associate with this mimosoid host. In other words, in addition to species of papilionoid genera (e.g., Virgilia, Aspalathus, Amphithalea, Hypocalyptus, Cyclopia, and Indigofera) (Beukes, 2012; Beukes et al., 2013; Lemaire et al., 2015a, 2016a), Paraburkholderia also nodulates V. karroo. This finding is important as symbiotic gene phylogenies have shown that nodulating Paraburkholderia species can be sub-divided into so-called mimosoid and papilionoid nodulating groups (Bontemps et al., 2010; Beukes et al., 2013; Estrada-de los Santos et al., 2018). Only two species, P. tuberum and P. phymatum, are apparently capable of nodulating both papilionoid and mimosoid hosts (Elliott et al., 2007b; Bontemps et al., 2010; Mishra et al., 2012; Suárez-Moreno et al., 2012). In the case of P. tuberum, the biovars mimosae and papilionoideae have been proposed for differentiating among isolates (Mishra et al., 2012). In P. tuberum, this split likely correlates with the distinct origin of the symbiotic loci of the mimosoid and papilionoid-nodulating species (Beukes et al., 2013; de Meyer et al., 2016; Lemaire et al., 2016a). Our future work will seek to obtain sequences for the symbiotic loci of the V. karroo Paraburkholderia isolates and to compare these with sequences from South African papilionoid and South American mimosoid isolates.
Most of the isolates obtained from V. karroo root nodules were members of the alpha-rhizobial genera Ensifer (32 isolates) and Mesorhizobium (27 isolates). Both of these genera include a number of species that have been described from Acacia sensu lato species, and includes Ensifer kostiensis (from Senegalia senegal in Sudan) (Nick et al., 1999), Ensifer terangae (from S. laeta in Senegal) (de Lajudie et al., 1994), Ensifer mexicanus (from Acaciella angustissima in Mexico) (Lloret et al., 2007), Ensifer chiapanecum (from Ac. angustissima in Mexico) (Rincón-Rosales et al., 2009), Ensifer americanus (from Acacia acatlensis in Mexico, now Mariosousa acatlensis) (Toledo et al., 2003; Seigler et al., 2006), M. abyssinicae (from V. abyssinica in Ethiopia) (Degefu et al., 2013b), M. shonense (from V. abyssinicae in Ethiopia) (Degefu et al., 2013b) and M. plurifarium (from S. senegal in Senegal) (de Lajudie et al., 1998). Some of the Mesorhizobium isolates obtained from V. karroo are likely conspecific with these previously identified bacteria, particularly M. abyssinicae and M. shonense. The three putative Ensifer species obtained from V. karroo in the current study are likely conspecific with a variety of rhizobia from V. seyal, V. tortilis and V. abyssinica originating from Ethiopia and that have not yet been formally described (Degefu et al., 2012).
Unlike the genera Ensifer and Mesorhizobium, the genus Rhizobium does not currently contain any named species originating from nodules of legumes in Acacia sensu lato. There are, however, examples where studies have encountered isolates with high similarity to R. leucaenae (McInroy et al., 1999; Romdhane et al., 2006; Ribeiro et al., 2012), R. giardinii (now Pararhizobium giardinii; Mousavi et al., 2015), R. etli and R. leguminosarum (Wolde-meskel et al., 2005). Our study followed a similar trend with none of the identified putative species obtained from V. karroo being conspecific to known species. Nevertheless, none of the Rhizobium isolates from V. karroo showed significant similarity to isolates previously identified from Acacia sensu lato.
The alpha-rhizobial data from this study is comparable to what has been found previously for other African Acacias (i.e., Vachellia and Senegalia species), which are predominantly nodulated by isolates of Mesorhizobium, Rhizobium and Ensifer (Leary et al., 2006). The Senegalia hosts previously examined and for which rhizobial sequence data is available, are S. senegal (Ethiopia, Kenya, Mauritania, Senegal, Sudan) that associates with Mesorhizobium, Rhizobium and Ensifer (de Lajudie et al., 1998; McInroy et al., 1999; Nick et al., 1999; Sarr et al., 2005; Wolde-meskel et al., 2005; Fall et al., 2008; Degefu et al., 2011) and S. laeta (Senegal) that associates with Ensifer (de Lajudie et al., 1994). The Vachellia species that were previously investigated included V. abyssinica (Ethiopia), V. nilotica (Algeria, Mauritania, Senegal, Zimbabwe), V. seyal (Algeria, Ethiopia, Senegal), V. tortilis (Algeria, Ethiopia, Kenya, Mauritania, Morocco, Senegal, Tunisia) that all have Mesorhizobium, Rhizobium, and Ensifer symbionts (de Lajudie et al., 1994, 1998; McInroy et al., 1999; Ba et al., 2002; Sarr et al., 2005; Wolde-meskel et al., 2005; Romdhane et al., 2006; Diouf et al., 2007; Degefu et al., 2011, 2013b; Abdi et al., 2012; Boukhatem et al., 2012), as well as V. horrida (Morocco) that associates with Ensifer (Khbaya et al., 1998). Although V. karroo in Algeria and Kenya have been reported to associate with Rhizobium and Ensifer (McInroy et al., 1999; Boukhatem et al., 2012), our study shows for the first time that this legume is nodulated by Bradyrhizobium, Mesorhizobium and Paraburkholderia in addition to Rhizobium and Ensifer, in its native range in South Africa.
Vachellia karroo and Australian Acacia (Acacia sensu stricto) appear to differ markedly in their symbiotic preference. Only one Bradyrhizobium isolate was recovered here, which is very low when compared to Acacia s. s. species which predominantly nodulates with this alpha-rhizobial genus (Marsudi et al., 1999; Lafay and Burdon, 2001; Rodríguez-Echeverría et al., 2011; Crisóstomo et al., 2013). Leary et al. (2006) attributed these differences in symbiont associations in Africa and Australia to an apparent correlation between host phylogeny and the rhizobial groups nodulating them. They reported that the majority of Australian acacias nodulated with Bradyrhizobium while African and American acacias nodulated with Ensifer and Mesorhizobium species. However, improved resolution of legume taxonomy and wider sampling of symbionts suggest that the distribution of these associations is more complex. For example, certain species in the tribe Ingeae, a close relative of Acacia s. s. (including the Australian Acacia) (LPWG, 2013) are also nodulated by Bradyrhizobium species (Robinson and Harris, 2000; Miller and Bayer, 2001, 2003; Leary et al., 2006), but several Calliandra species (tribe Ingeae) associate with Paraburkholderia (Silva et al., 2018). Additionally, African acacias (represented by Vachellia and Senegalia) are mostly nodulated by Mesorhizobium and Ensifer species (Zhang et al., 1991; Ba et al., 2002; Leary et al., 2006), but their close relatives in the Piptadenia group (including the genus Mimosa; LPWG, 2013) associate predominantly with Paraburkholderia and other beta-rhizobia (Suárez-Moreno et al., 2012; Taulé et al., 2012; Bournaud et al., 2013).
It is more likely that the observed legume-rhizobium distribution patterns reflect the symbiotic compatibilities between the partners. This is because the symbiosis rely on successful recognition of the rhizobial Nod factor by the host nodulin genes (Ferro et al., 2000). Ferro et al. (2000) demonstrated that the Nod factors of rhizobia in the genera Rhizobium, Mesorhizobium and Ensifer, which associate with Vachellia or Senegalia hosts, does not contain a glycosyl group and are instead predominantly sulfated (Ferro et al., 2000). By contrast, Bradyrhizobium isolates associating with Faidherbia albida (previously Acacia albida; phylogenetically closely related to Acacia sensu stricto) produce Nod factors that are sulfated at a different postion and that are also fucosylated (Ferro et al., 2000).
The single Bradyrhizobium V. karroo-associated isolate (15B) obtained in this study grouped with B. liaoningense (originally isolated from Glycine max nodules in China) (Xu et al., 1995). Although an isolate obtained from Vachellia sieberiana var. woodii (formerly Acacia sieberiana var. woodii; Kyalangalilwa et al., 2013) in a previous study was designated B. liaoningense, that identification should be viewed with caution as it was based on partial 16S rRNA gene sequence information (Jaftha, 2000) which is known to have limited resolution in Bradyrhizobium at the species level (Willems et al., 2001; Vinuesa et al., 2008). Also, B. liaoningense is believed to be scarce in southern African soils (Steenkamp et al., 2008). Increasing the sample size in follow-up studies would facilitate a better geographical representation and analysis of whether the low numbers and diversity of Bradyrhizobium strains associated with V. karroo in South Africa are indeed a true reflection of what is occurring in these environments.
Taken together, our findings confirmed that the symbionts of V. karroo were indeed members of the alpha-rhizobial genera Mesorhizobium, Ensifer and Rhizobium. We were also able to isolate 19 South African Paraburkholderia isolates from the root nodules of this mimosoid host. With regard to the distribution of the V. karroo-symbionts across the various biomes, our study is also the first to report the legume-Paraburkholderia association in regions outside the Fynbos biome (i.e., one and three putative species were recovered as symbionts of V. karroo in one and four locations in the Albany Thicket and Succulent Karoo biomes, respectively). We also showed that a high diversity of rhizobial symbionts associate with V. karroo in the various regions, even in the Grassland biome where we identified putative species from three genera. The latter is important because V. karroo contributes significantly to bush encroachment (i.e., the change of grasslands to ecosystems containing more shrubs and trees), which is particularly evident in the Grassland biome in the Eastern Cape, North West and Free State provinces of South Africa (Dingaan and du Preez, 2018). The promiscuity of V. karroo in these and other settings therefore suggest that finding a suitable rhizobial partner is not a hurdle to the establishment or spread of this legume and likely represents an important driver or facilitator of its ecological success.
Author Contributions
CB and FB planned the study, obtained the samples and cultures, tested for nodulation ability, analysed datasets, submitted sequence data, interpreted the results, and wrote the manuscript. FP and AH assisted with testing the nodulation ability, proofread the manuscript. MlR and TS proofread the manuscript. SV and ES planned the study, interpreted the results, wrote and proofread the manuscript.
Funding
This research was made possible by financial support from the National Research Foundation (NRF) of South Africa. Opinions expressed and conclusions arrived at, are those of the author and are not necessarily to be attributed to the NRF.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Drs. Maistry and Chimphango for the nodules resulting from Western Cape soils.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01195/full#supplementary-material
References
Abdi, D. T., Wolde-meskel, E., and Frostegård, Å. (2012). Phylogenetic multilocus sequence analysis identifies seven novel Ensifer genospecies isolated from a less-well-explored biogeographical region in East Africa. Int. J. Syst. Evol. Microbiol. 62, 2286–2295. doi: 10.1099/ijs.0.039230-0
Aljanabi, S. M., and Martinez, I. (1997). Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 25, 4692–4693. doi: 10.1093/nar/25.22.4692
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local allignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Ba, S., Willems, A., de Lajudie, P., Roche, P., Jeder, H., Quatrini, P., et al. (2002). Symbiotic and taxonomic diversity of rhizobia isolated from Acacia tortilis subsp. raddiana in Africa. Syst. Appl. Microbiol. 25, 130–145. doi: 10.1078/0723-2020-00091
Banfi, E., and Galasso, G. (2008). “New combinations in Vachellia Wight & Arn., formerly Acacia Mill. ss (Fabaceae),” in Proceedings of the Atti della Società Italiana di Scienze Naturali e del Museo Civico di Storia Naturale di Milano, Vol. 149, (Milan), 149–150.
Baraúna, A. C., Rouws, L. F. M., Simoes-Araujo, J. L., dos Reis Junior, F. B., Iannetta, P. P. M., Maluk, M., et al. (2016). Rhizobium altiplani sp. nov., isolated from effective nodules on Mimosa pudica growing in untypically alkaline soil in central Brazil. Int. J. Syst. Evol. Microbiol. 66, 4118–4124. doi: 10.1099/ijsem.0.001322
Barnes, R. (2001). The African acacias-a thorny subject. South. Afr. For. J. 190, 9–18. doi: 10.1080/20702620.2001.10434110
Benson, D. A., Cavanaugh, M., Clark, K., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., et al. (2017). GenBank. Nucleic Acids Res. 45, D37–D42. doi: 10.1093/nar/gkx1094
Beukes, C. W. (2012). Characterisation of Bacteria Associated with the Root Nodules of Hypocalyptus and Related Genera. master’s dissertation. Pretoria: University of Pretoria.
Beukes, C. W., Venter, S. N., Law, I. J., Phalane, F. L., and Steenkamp, E. T. (2013). South African papilionoid legumes are nodulated by diverse Burkholderia with unique nodulation and nitrogen-fixation loci. PloS One 8:e68406. doi: 10.1371/journal.pone.0068406
Bontemps, C., Elliott, G. N., Simon, M. F., Dos Reis Junior, F. B., Gross, E., Lawton, R. C., et al. (2010). Burkholderia species are ancient symbionts of legumes. Mol. Ecol. 19, 44–52. doi: 10.1111/j.1365-294X.2009.04458.x
Boukhatem, Z. F., Domergue, O., Bekki, A., Merabet, C., Sekkour, S., Bouazza, F., et al. (2012). Symbiotic characterization and diversity of rhizobia associated with native and introduced acacias in arid and semi-arid regions in Algeria. FEMS Microbiol. Ecol. 80, 534–547. doi: 10.1111/j.1574-6941.2012.01315.x
Bournaud, C., de Faria, S. M., dos Santos, J. M. F., Tisseyre, P., Silva, M., Chaintreuil, C., et al. (2013). Burkholderia species are the most common and preferred nodulating symbionts of the piptadenia group (Tribe Mimoseae). PLoS One 8:e63478. doi: 10.1371/journal.pone.0063478
Brain, P., Harris, S., and Barnes, R. (1997). Leaf peroxidase types in Acacia karroo Hayne (Acacieae, Leguminosae): a range-wide study. Silvae Genet. 46, 88–93.
Brown, D., N’gambi, J. W., and Norris, D. (2016). Feed potential of Acacia karroo leaf meal for communal goat production in southern Africa: a review. J. Anim. Plant Sci. 26, 1178–1186.
Chen, W.-M., de Faria, S. M., James, E. K., Elliott, G. N., Lin, K.-Y., Chou, J.-H., et al. (2007). Burkholderia nodosa sp. nov., isolated from root nodules of the woody Brazilian legumes Mimosa bimucronata and Mimosa scabrella. Int. J. Syst. Evol. Microbiol. 57, 1055–1059. doi: 10.1099/ijs.0.64873-0
Chen, W.-M., de Faria, S. M., Straliotto, R., Pitard, R. M., Simões-Araùjo, J. L., Chou, J.-H., et al. (2005). Proof that Burkholderia strains form effective symbioses with legumes: a study of novel Mimosa-nodulating strains from South America. Appl. Environ. Microbiol. 71, 7461–7471. doi: 10.1128/AEM.71.11.7471-7471.2005
Cock, I. E., and van Vuuren, S. F. (2015). South African food and medicinal plant extracts as potential antimicrobial food agents. J. Food Sci. Technol. 52, 6879–6899. doi: 10.1007/s13197-015-1806-3
Crisóstomo, J. A., Rodríguez-Echeverría, S., and Freitas, H. (2013). Co-introduction of exotic rhizobia to the rhizosphere of the invasive legume Acacia saligna, an intercontinental study. Appl. Soil Ecol. 64, 118–126. doi: 10.1016/j.apsoil.2012.10.005
Cummings, S. P., Gyaneshwar, P., Vinuesa, P., Farruggia, F. T., Andrews, M., Humphry, D., et al. (2009). Nodulation of Sesbania species by Rhizobium (Agrobacterium) strain IRGB74 and other rhizobia. Environ. Microbiol. 11, 2510–2525. doi: 10.1111/j.1462-2920.2009.01975.x
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
de Castro Pires, R., dos Reis Junior, F. B., Zilli, J. E., Fischer, D., Hofmann, A., James, E. K., et al. (2018). Soil characteristics determine the rhizobia in association with different species of Mimosa in central Brazil. Plant Soil 423, 411–428. doi: 10.1007/s11104-017-3521-5
de Lajudie, P., Willems, A., Nick, G., Moreira, F., Molouba, F., Hoste, B., et al. (1998). Characterization of tropical tree rhizobia and description of Mesorhizobium plurifarium sp. nov. Int. J. Syst. Bacteriol. 48, 369–382. doi: 10.1099/00207713-48-2-369
de Lajudie, P., Willems, A., Pot, B., Dewettinck, D., Maestrojuan, G., Neyra, M., et al. (1994). Polyphasic taxonomy of rhizobia: emendation of the genus Sinorhizobium and description of Sinorhizobium meliloti comb. nov., Sinorhizobium saheli sp. nov., and Sinorhizobium teranga sp. nov. Int. J. Syst. Bacteriol. 44, 715–733. doi: 10.1099/00207713-44-4-715
de Lajudie, P. M., Andrews, M., Ardley, J., Eardly, B., Jumas-Bilak, E., Kuzmanovic, N., et al. (2019). Minimal standards for the description of new genera and species of rhizobia and agrobacteria. Int. J. Syst. Evol. Microbiol. doi: 10.1099/ijsem.0.003426
de Meyer, S. E., Briscoe, L., Martínez-Hidalgo, P., Agapakis, C. M., Estrada de-los Santos, P., Seshadri, R., et al. (2016). Symbiotic Burkholderia species show diverse arrangements of nif/fix and nod genes and lack typical high-affinity cytochrome ccb3 oxidase genes. Mol. Plant Microbe Interact. 29, 609–619. doi: 10.1094/MPMI-05-16-0091-R
de Meyer, S. E., Cnockaert, M., Ardley, J. K., Maker, G., Yates, R., Howieson, J. G., et al. (2013a). Burkholderia sprentiae sp. nov. isolated from Lebeckia ambigua root nodules from South Africa. Int. J. Syst. Evol. Microbiol. 63, 3950–3957. doi: 10.1099/ijs.0.048777-0
de Meyer, S. E., Cnockaert, M., Ardley, J. K., Trengove, R. D., Garau, G., Howieson, J. G., et al. (2013b). Burkholderia rhynchosiae sp. nov. isolated from Rhynchosia ferulifolia root nodules from South Africa. Int. J. Syst. Evol. Microbiol. 63, 3344–3949. doi: 10.1099/ijs.0.048751-0
de Meyer, S. E., Cnockaert, M., Ardley, J. K., van Wyk, B.-E., Vandamme, P. A., and Howieson, J. G. (2014). Burkholderia dilworthii sp. nov., isolated from Lebeckia ambigua root nodules. Int. J. Syst. Evol. Microbiol. 64, 1090–1095. doi: 10.1099/ijs.0.058602-0
Degefu, T., Wolde-meskel, E., and Frostegård, A. (2012). Phylogenetic multilocus sequence analysis identifies seven novel Ensifer genospecies isolated from a less-well-explored biogeographical region in East Africa. Int. J. Syst. Evol. Microbiol. 62, 2286–2295. doi: 10.1099/ijs.0.039230-0
Degefu, T., Wolde-meskel, E., and Frostegård, Å (2011). Multilocus sequence analyses reveal several unnamed Mesorhizobium genospecies nodulating Acacia species and Sesbania sesban trees in Southern regions of Ethiopia. Syst. Appl. Microbiol. 34, 216–226. doi: 10.1016/j.syapm.2010.09.006
Degefu, T., Wolde-meskel, E., and Frostegård, Å (2013a). Phylogenetic diversity of Rhizobium strains nodulating diverse legume species growing in Ethiopia. Syst. Appl. Microbiol. 36, 272–280. doi: 10.1016/j.syapm.2013.03.004
Degefu, T., Wolde-meskel, E., Liu, B., Cleenwerck, I., Willems, A., and Frostegård, Å (2013b). Mesorhizobium shonense sp. nov., Mesorhizobium hawassense sp. nov. and Mesorhizobium abyssinicae sp. nov. isolated from root nodules of different agroforestry legume trees growing in southern Ethiopia. Int. J. Syst. Evol. Microbiol. 63, 1746–1753. doi: 10.1099/ijs.0.044032-0
Dingaan, M., and du Preez, P. J. (2018). “Chapter 6. Vachellia (Acacia) karroo communities in South Africa: an overview”. Available at: http://www.intechopen.com/books/pure-and-applied-biogeography (accessed February 7, 2018).
Diouf, D., Samba-Mbaye, R., Lesueur, D., Ba, A. T., Dreyfus, B., de Lajudie, P., et al. (2007). Genetic diversity of Acacia seyal Del. rhizobial populations indigenous to Senegalese soils in relation to salinity and pH of the sampling sites. Microb. Ecol. 54, 553–566. doi: 10.1007/s00248-007-9243-0
Dobritsa, A. P., and Samadpour, M. (2016). Transfer of eleven species of the genus Burkholderia to the genus Paraburkholderia and proposal of Caballeronia gen. nov. to accommodate twelve species of the genera Burkholderia and Paraburkholderia. Int. J. Syst. Evol. Microbiol. 66, 2836–2846. doi: 10.1099/ijsem.0.001065
dos Reis, F. B. Jr., Simon, M. F., Gross, E., Boddey, R. M., Elliott, G. N., Neto, N. E., et al. (2010). Nodulation and nitrogen fixation by Mimosa spp. in the Cerrado and Caatinga biomes of Brazil. New Phytol. 186, 934–946. doi: 10.1111/j.1469-8137.2010.03267.x
Driver, A., Maze, K., Rouget, M., Lombard, A. T., Nel, J., Turpie, J. K., et al. (2005). National Spatial Biodiversity Assessment 2004: Priorities for Biodiversity Conservation in South Africa, Vol. 17. Pretoria: South African National Biodiversity Institute.
Elliott, G. N., Chen, W.-M., Bontemps, C., Chou, J.-H., Young, J. P. W., Sprent, J. I., et al. (2007a). Nodulation of Cyclopia spp.(Leguminosae, Papilionoideae) by Burkholderia tuberum. Ann. Bot. 100, 1403–1411. doi: 10.1093/aob/mcm227
Elliott, G. N., Chen, W.-M., Chou, J.-H., Wang, H.-C., Sheu, S.-Y., Perin, L., et al. (2007b). Burkholderia phymatum is a highly effective nitrogen-fixing symbiont of Mimosa spp. and fixes nitrogen ex planta. New Phytol. 173, 168–180. doi: 10.1111/j.1469-8137.2006.01894.x
Elliott, G. N., Chou, J.-H., Chen, W.-M., Bloemberg, G. V., Bontemps, C., Martínez-Romero, E., et al. (2009). Burkholderia spp. are the most competitive symbionts of Mimosa, particularly under N-limited conditions. Environ. Microbiol. 11, 762–778. doi: 10.1111/j.1462-2920.2008.01799.x
Estrada-de los Santos, P., Palmer, M., Chávez-Ramírez, B., Beukes, C., Steenkamp, E. T., Briscoe, L., et al. (2018). Whole genome analyses suggests that Burkholderia sensu lato contains two additional novel genera (Mycetohabitans gen. nov., and Trinickia gen. nov.): implications for the evolution of diazotrophy and nodulation in the Burkholderiaceae. Genes 9:E389. doi: 10.3390/genes9080389
Estrada-de los Santos, P., Vinuesa, P., Martínez-Aguilar, L., Hirsch, A. M., and Caballero-Mellado, J. (2013). Phylogenetic analysis of Burkholderia species by multilocus sequence analysis. Curr. Microbiol. 67, 51–60. doi: 10.1007/s00284-013-0330-9
Euzéby, J. P. (1997). List of bacterial names with standing in nomenclature: a folder available on the internet. Int. J. Syst. Bacteriol. 47, 590–592. doi: 10.1099/00207713-47-2-590
Fall, D., Diouf, D., Ourarhi, M., Faye, A., Abdelmounen, H., Neyra, M., et al. (2008). Phenotypic and genotypic characteristics of Acacia senegal (L.) Willd. root-nodulating bacteria isolated from soils in the dryland part of Senegal. Lett. Appl. Microbiol. 47, 85–97. doi: 10.1111/j.1472-765X.2008.02389.x
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Ferro, M., Lorquin, J., Ba, S., Sanon, K., Promé, J.-C., and Boivin, C. (2000). Bradyrhizobium sp. strains that nodulate the leguminous tree Acacia albida produce fucosylated and partially sulfated Nod factors. Appl. Environ. Microbiol. 66, 5078–5082. doi: 10.1128/AEM.66.11.5078-5082.2000
Garau, G., Yates, R. J., Deiana, P., and Howieson, J. G. (2009). Novel strains of nodulating Burkholderia have a role in nitrogen fixation with papilionoid herbaceous legumes adapted to acid, infertile soils. Soil Biol. Biochem. 41, 125–134. doi: 10.1016/j.soilbio.2008.10.011
Gehlot, H. S., Panwar, D., Tak, N., Tak, A., Sankhla, I. S., Poonar, N., et al. (2012). Nodulation of legumes from the Thar desert of India and molecular characterization of their rhizobia. Plant Soil 357, 227–243. doi: 10.1007/s11104-012-1143-5
Guindon, S., and Gascuel, O. (2003). A simple, fast and accurate algorithm to estimate large phylogenies by maximum-likelihood. Syst. Biol. 52, 696–704. doi: 10.1080/10635150390235520
Gxasheka, M., Tyasi, T. L., Qin, N., and Lyu, Z.-C. (2015). An overview of tannins rich plants as alternative supplementation on ruminant animals: a review. Int. J. Agric. Res. Rev. 3, 343–349.
Gyaneshwar, P., Hirsch, A. M., Moulin, L., Chen, W.-M., Elliott, G. N., Bontemps, C., et al. (2011). Legume-nodulating betaproteobacteria: diversity, host range, and future prospects. Mol. Plant Microbe Interact. 24, 1276–1288. doi: 10.1094/MPMI-06-11-0172
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hayward, R. (2004). The Acacia Tree: a Sustainable Resource for Africa. Swindon: Natural Environmental Research Council.
Howieson, J. G., de Meyer, S. E., Vivas-Marfisi, A., Ratnayake, S., Ardley, J. K., and Yates, R. J. (2013). Novel Burkholderia bacteria isolated from Lebeckia ambigua – a perennial suffrutescent legume of the fynbos. Soil Biol. Biochem. 60, 55–64. doi: 10.1016/j.soilbio.2013.01.009
Howieson, J. G., and Dilworth, M. J. (2016). Working with Rhizobia. Canberra, ACT: Australian Centre for International Agricutlural Research.
Jaftha, J. B. (2000). Polyphasic Taxonomy of Rhizobia Associated with Legumes Occuring in South Africa. PhD thesis. Pretoria: University of Pretoria.
Khbaya, B., Neyra, M., Normand, P., Zerhari, K., and Filali-Maltouf, A. (1998). Genetic diversity and phylogeny of rhizobia that nodulate Acacia spp. in Morocco assessed by analysis of rRNA genes. Appl. Environ. Microbiol. 64, 4912–4917.
Klock, M. M., Barrett, L. G., Thrall, P. H., and Harms, K. E. (2015). Host promiscuity in symbiont associations can influence exotic legume establishment and colonization of novel ranges. Divers. Distrib. 21, 1193–1203. doi: 10.1111/ddi.12363
Kock, M. M. (2003). Diversity of Root Nodulating Bacteria Associated with Cyclopia Species. PhD thesis. Pretoria: University of Pretoria.
Kyalangalilwa, B., Boatwright, J. S., Daru, B. H., Maurin, O., and van der Bank, M. (2013). Phylogenetic position and revised classification of Acacia sl (Fabaceae: Mimosoideae) in Africa, including new combinations in Vachellia and Senegalia. Bot. J. Linn. Soc. 172, 500–523. doi: 10.1111/boj.12047
La Pierre, K. J., Simms, E. L., Tariq, M., Zafar, M., and Porter, S. S. (2017). Invasive legumes can associate with many mutualists of native legumes, but usually do not. Ecol. Evol. 7, 8599–8611. doi: 10.1002/ece3.3310
Lafay, B., and Burdon, J. J. (2001). Small-Subunit rRNA genotyping of rhizobia nodulating Australian Acacia spp. Appl. Environ. Microbiol. 67, 396–402. doi: 10.1128/AEM.67.1.396-402.2001
Leary, J. K., Singleton, P. W., Scowcroft, P. G., and Borthakur, D. (2006). Symbiotic diversity in the cosmopolitan genus Acacia. Symbiosis 41, 107–117.
Lee, A., and Hirsch, A. M. (2006). Signals and responses. Choreographing the complex interaction between legumes and α- and β-rhizobia. Plant Signal. Behav. 1, 161–168. doi: 10.4161/psb.1.4.3143
Lemaire, B., Chimphango, S. B. M., Stirton, C., Rafudeen, S., Honnay, O., Smets, E., et al. (2016a). Biogeographical patterns of legume-nodulating Burkholderia spp.: from African Fynbos to continental scales. Appl. Environ. Microbiol. 82, 5099–5115. doi: 10.1128/AEM.00591-16
Lemaire, B., Van Cauwenberghe, J., Verstraete, B., Chimphango, S., Stirton, C., Honnay, O., et al. (2016b). Characterization of the papiloinoid-Burkholderia interactions in the Fynbos biome: the diversity and distribution of beta-rhizobia nodulating Podalyria calyptrata (Fabaceae, Podalyrieae). Syst. Appl. Microbiol. 39, 41–48. doi: 10.1016/j.syapm.2015.09.006
Lemaire, B., Dlodlo, O., Chimphango, S., Stirton, C., Schrire, B., Boatwright, J. S., et al. (2015a). Symbiotic diversity, specificity and distribution of rhizobia in native legumes of the core Cape Subregion (South Africa). FEMS Microbiol. Ecol. 91, 1–17. doi: 10.1093/femsec/fiu024
Lemaire, B., Van Cauwenberghe, J., Chimphango, S., Stirton, C., Honnay, O., Smets, E., et al. (2015b). Recombination and horizontal transfer of nodulation and ACC deaminase (acdS) genes within Alpha- and Betaproteobacteria nodulating legumes of the Cape Fynbos biome. FEMS Microbiol. Ecol. 91:fiv118. doi: 10.1093/femsec/fiv118
Lewis, G. P. (2005). “Tribe Acacieae,” in Legumes of the World, eds G. Lewis, B. Schrire, B. Mackinder, and M. Lock (Richmond: The Royal Botanic Gardens Kew).
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Liu, W. Y. Y., Ridgway, H. J., James, T. K., James, E. K., Chen, W.-M., Sprent, J. I., et al. (2014). Burkholderia sp. induces functional nodules on the South African invasive legume Dipogon lignosus (Phaseoleae) in New Zealand Soils. Microb. Ecol. 68, 542–555. doi: 10.1007/s00248-014-0427-0
Lloret, L., Ormeño-Orrillo, E., Rincón, R., Martínez-Romero, J., Rogel-Hernández, M. A., and Martínez-Romero, E. (2007). Ensifer mexicanus sp. nov. a new species nodulating Acacia angustissima (Mill.) Kuntze in Mexico. Syst. Appl. Microbiol. 30, 280–290. doi: 10.1061/j.syapm.2006.12.002
Logan, N. A., Lebbe, L., Hoste, B., Goris, J., Forsyth, G., Heyndrickx, M., et al. (2000). Aerobic endospore-forming bacteria from geothermal environments in northern Victoria Land, Antarctica, and Candlemas Island, South Sandwich archipelago, with the proposal of Bacillus fumarioli sp. nov. Int. J. Syst. Evol. Microbiol. 50, 1741–1753. doi: 10.1099/00207713-50-5-1741
LPWG (2013). Legume phylogeny and classification in the 21st century: progress, prospects and lessons for other species-rich clades. Taxon 62, 217–248. doi: 10.5167/uzh-78167
LPWG (2017). A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny. Taxon 66, 44–77. doi: 10.12705/661.3
Maroyi, A. (2017). Acacia karroo Hayne: ethnomedical uses, phytochemistry and pharmacology of an important medicinal plant in southern Africa. Asian Pac. J. Trop. Med. 10, 351–360. doi: 10.1016/j.apjtm.2017.03.021
Marsudi, N. D. S., Glenn, A. R., and Dilworth, M. J. (1999). Identification and characterization of fast- and slow-growing root nodule bacteria from South-Western Australian soils able to nodulate Acacia saligna. Soil Biol. Biochem. 31, 1229–1238. doi: 10.1016/S0038-0717(99)00032-2
Martens, M., Dawyndt, P., Coopman, R., Gillis, M., De Vos, P., and Willems, A. (2008). Advantages of multilocus sequence analysis for taxonomic studies: a case study using 10 housekeeping genes in the genus Ensifer (including former Sinorhizobium). Int. J. Syst. Evol. Microbiol. 58, 200–214. doi: 10.1099/ijs.0.65392
McInroy, S. G., Campbell, C. D., Haukka, K. E., Odee, D. W., Sprent, J. I., Wang, W. J., et al. (1999). Characterisation of rhizobia from African acacias and other tropical woody legumes using BiologTM and partial 16S rRNA sequencing. FEMS Microbiol. Lett. 170, 111–117. doi: 10.1111/j.1574-6968.1999.tb13362.x
Miller, J. T., and Bayer, R. J. (2001). Molecular phylogenetics of Acacia (Fabaceae: Mimosoideae) based on the chloroplast matK coding sequence and flanking trnK intron spacer regions. Am. J. Bot. 88, 697–705. doi: 10.2307/2657071
Miller, J. T., and Bayer, R. J. (2003). Molecular phylogenetics of Acacia subgenera Acacia and Aculeiferum (Fabaceae: Mimosoideae), based on the chloroplast matK coding sequence and flanking trnK intron spacer regions. Aust. Syst. Bot. 16, 27–33. doi: 10.1071/SB01035
Mishra, R. P., Tisseyre, P., Melkonian, R., Chaintreuil, C., Miché, L., Klonowska, A., et al. (2012). Genetic diversity of Mimosa pudica rhizobial symbionts in soils of French Guiana: investigating the origin and diversity of Burkholderia phymatum and other beta-rhizobia. FEMS Microbiol. Ecol. 79, 487–503. doi: 10.1111/j.1574-6941.2011.01235.x
Mohamed, S. H., Smouni, A., Neyra, M., Kharchaf, D., and Filali-Maltouf, A. (2000). Phenotypic characteristics of root-nodulating bacteria isolated from Acacia spp. grown in Libya. Plant Soil 224, 171–183. doi: 10.1023/A:1004838218642
Moulin, L., Klonowska, A., Bournaud, C., Booth, K., Vriezen, J. A. C., Melkonian, R., et al. (2014). Complete genome sequence of Burkholderia phymatum STM815T, a broad host range and efficient nitrogen-fixing symbiont of Mimosa species. Stand. Genomic Sci. 9, 763–774. doi: 10.4056/sigs.4861021
Moulin, L., Munive, A., Dreyfus, B., and Boivin-Masson, C. (2001). Nodulation of legumes by members of the β-subclass of Proteobacteria. Nature 411, 948–950. doi: 10.1038/35082070
Mousavi, S. A., Österman, J., Wahlberg, N., Nesme, X., Lavire, C., Vial, L., et al. (2014). Phylogeny of the Rhizobium-Allorhizobium-Agrobacterium calde supports the delineation of Neorhizobium gen. nov. Syst. Appl. Microbiol. 37, 208–215. doi: 10.1016/j.syapm.2013.12.007
Mousavi, S. A., Willems, A., Nesme, X., de Lajudie, P., and Lindström, K. (2015). Revised phylogeny of Rhizobiaceae: proposal of the delineation of Pararhizobium gen. nov., and 13 new species combinations. Syst. Appl. Microbiol. 38, 84–90. doi: 10.1016/j.syapm.2014.12.003
Mrabet, M., Mnasri, B., Romdhane, S. B., Laguerre, G., Aouani, M. E., and Mhamdi, R. (2006). Agrobacterium strains isolated from root nodules of common bean specifically reduce nodulation by Rhizobium gallicum. FEMS Microbiol. Ecol. 56, 304–309. doi: 10.1111/j.1574-6941.2006.00069.x
Mucina, L., and Rutherford, M. C. (2006). The Vegetation of South Africa, Lesotho and Swaziland. Pretoria: South African National Biodiversity Institute.
Ndlovu, J., Richardson, D. M., Wilson, J. R., and Le Roux, J. J. (2013). Co-invasion of South African ecosystems by an Australian legume and its rhizobial symbionts. J. Biogeogr. 40, 1240–1251. doi: 10.1111/jbi.12091
Ngwenya, N., Zharare, G. E., Basson, A. K., and Mthembu, M. S. (2016). Phenotypic characterisation of rhizobia nodulating different morphotypes of Vachellia karroo. Legume Res. 39, 722–728.
Nick, G., de Lajudie, P., Eardly, B. D., Suomalainen, S., Paulin, L., Zhang, X., et al. (1999). Sinorhizobium arboris sp. nov. and Sinorhizobium kostiense sp. nov., isolated from leguminous trees in Sudan and Kenya. Int. J. Syst. Evol. Microbiol. 49, 1359–1368. doi: 10.1099/00207713-49-4-1359
Nixon, K. C., and Wheeler, Q. D. (1990). An amplification of the phylogenetic species concept. Cladistics 6, 211–223. doi: 10.1111/j.1096-0031.1990.tb00541.x
O’Connor, T. G. (1995). Acacia karroo invasion of grassland: environmental and biotic effects influencing seedling emergence and establishment. Oecologia 103, 214–223. doi: 10.1007/BF00329083
Odee, D., Haukka, K., McInroy, S. G., Sprent, J. I., Sutherland, J. M., and Young, J. P. W. (2002). Genetic and symbiotic characterization of rhizobia isolated from tree and herbaceous legumes grown in soils from ecologically diverse sites in Kenya. Soil Biol. Biochem. 34, 801–811. doi: 10.1016/S0038-0717(02)00009-3
Parte, A. C. (2013). LPSN - list of prokaryotic names with standing in nomenclature. Nucleic Acids Res. 42, D613–D616. doi: 10.1093/nar/gkt1111
Payne, G. W., Vandamme, P., Morgan, S. H., LiPuma, J. J., Coenye, T., Weightman, A. J., et al. (2005). Development of a recA gene-based identification approach for the entire Burkholderia genus. Appl. Environ. Microbiol. 71, 3917–3927. doi: 10.1128/AEM.71.7.3917-3927.2005
Poole, P., Ramachandran, V., and Terpolilli, J. (2018). Rhizobia: from saprophytes to endosymbionts. Nat. Rev. Microbiol. 16, 291–303. doi: 10.1038/nrmicro.2017.171
Ribeiro, R. A., Rogel, M. A., López- López, A., Ormeño-Orrillo, E., Barcellos, F. G., Martínez, J., et al. (2012). Reclassification of Rhizobium tropici type A strains as Rhizobium leucaenae sp. nov. Int. J. Syst. Evol. Microbiol. 62, 1179–1184. doi: 10.1099/ijs.0.032912-0
Rincón-Rosales, R., Lloret, L., Ponce, E., and Martínez-Romero, E. (2009). Rhizobia with different symbiotic efficiences nodulate Acaciella angustissima in Mexico, including Sinorhizobium chiapanecum sp. nov. which has common symbiotic genes with Sinorhizobium mexicanum. FEMS Microbiol. Ecol. 6, 103–117. doi: 10.1111/j.1574-6941.2008.00590.x
Rivas, R., Martens, M., de Lajudie, P., and Willems, A. (2009). Multilocus sequence analysis of the genus Bradyrhizobium. Syst. Appl. Microbiol. 32, 101–110. doi: 10.1016/j.syapm.2008.12.005
Robinson, J., and Harris, S. (2000). A plastid DNA phylogeny of the genus Acacia Miller (Acacieae, Leguminosae). Bot. J. Linn. Soc. 132, 195–222. doi: 10.1111/j.1095-8339.2000.tb01527.x
Rodríguez-Echeverría, S., Le Roux, J. J., Crisóstomo, J. A., and Ndlovu, J. (2011). Jack-of-all-trades and master of many? How does associated rhizobial diversity influence the colonization success of Australian Acacia species? Divers. Distrib. 17, 946–957. doi: 10.1111/j.1472-4642.2011.00787.x
Romdhane, S. B., Nasr, H., Samba-Mbaye, R., Neyra, M., Ghorbal, M. H., and De Lajudie, P. (2006). Genetic diversity of Acacia tortilis ssp. raddiana rhizobia in Tunisia assessed by 16S and 16S-23S rDNA genes analysis. J. Appl. Microbiol. 100, 436–445. doi: 10.1111/j.1365-2672.2005.02765.x
Sakrouhi, I., Belfquih, M., Sbabou, L., Moulin, P., Bena, G., Filali-Maltouf, A., et al. (2016). Recovery of symbiotic nitrogen fixing acacia rhizobia from Merzouga Desert sand dunes in South East Morocco - identification of a probable new species of Ensifer adapted to stressed environments. Syst. Appl. Microbiol. 39, 122–131. doi: 10.1016/j.syapm.2016.01.001
Sankhla, I. S., Tak, N., Meghwal, R. R., Choudhary, S., Tak, A., Rathi, S., et al. (2017). Molecular characterization of nitrogen-fixing microsymbionts from root nodules of Vachellia (Acacia) jacquemontii, a native legume from the Thar Desert of India. Plant Soil 410, 21–40. doi: 10.1007/s11104-016-2838-9
Sarr, A., Neyra, M., Houeibib, M. A. O., Ndoye, I., Oihabi, A., and Lesueur, D. (2005). Rhizobial populations in soils from natural Acacia senegal and Acacia nilotica forests in Mauritania and the Senegal River Valley. Microb. Ecol. 50, 152–162. doi: 10.1007/s00248-004-0077-8
Sawana, A., Adeolu, M., and Gupta, R. S. (2014). Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front. Genet. 5:429. doi: 10.3389/fgene.2014.00429
Seigler, D. S., Ebinger, J. E., and Miller, J. T. (2006). Mariosousa, a new segregate genus from Acacia s.l. (Fabaceae, Mimosoideae) from Central and North America. Novon 16, 413–420. doi: 10.3417/1055-3177(2006)16
Sheu, S.-Y., Chen, M.-H., Liu, W. Y. Y., Andrews, M., James, E. K., Ardley, J. K., et al. (2015). Burkholderia dipogonis sp. nov., isolated from root nodules of Dipogon lignosus in New Zealand and Western Australia. Int. J. Syst. Evol. Microbiol. 65, 4716–4723. doi: 10.1099/ijsem.0.000639
Silva, V. C., Alves, P. A. C., Rhem, M. F. K., dos Santos, J. M. F., James, E. K., and Gross, E. (2018). Brazilian species of Calliandra Benth. (tribe Ingeae) are nodulated by diverse strains of Paraburkholderia. Syst. Appl. Microbiol. 41, 241–250. doi: 10.1016/j.syapm.2017.12.003
Stanton-Geddes, J., and Anderson, C. G. (2011). Does a facultative mutualism limit species range expansion? Oecologia 167, 149–155. doi: 10.1007/s00442-011-1958-4
Steenkamp, E. T., Stȩpkowski, T., Przymusiak, A., Botha, W. J., and Law, I. J. (2008). Cowpea and peanut in southern Africa are nodulated by diverse Bradyrhizobium strains harboring nodulation genes that belong to the large pantropical clade common in Africa. Mol. Phylogenet. Evol. 48, 1131–1144. doi: 10.1016/j.ympev.2008.04.032
Steenkamp, E. T., van Zyl, E., Beukes, C. W., Avontuur, J. R., Chan, W. Y., Palmer, M., et al. (2015). Burkholderia kirstenboschensis sp. nov. nodulates papilionoid legumes indigenous to South Africa. Syst. Appl. Microbiol. 38, 545–554. doi: 10.1016/j.syapm.2015.09.003
Suárez-Moreno, Z. R., Caballero-Mellado, J., Coutinho, B. G., Mendonça-Previato, L., James, E. K., and Venturi, V. (2012). Common features of environmental and potentially beneficial plant-associated Burkholderia. Microb. Ecol. 63, 249–266. doi: 10.1007/s00248-011-9929-1
Suau, A., Bonnet, R., Sutren, M., Godon, J.-J., Gibson, G. R., Collins, M. D., et al. (1999). Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65, 4799–4807.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Taulé, C., Zabaleta, M., Mareque, C., Platero, R., Sanjurjo, L., Sicardi, M., et al. (2012). New betaproteobacterial rhizobium strains able to efficiently nodulate Parapiptadenia rigida (Benth.) Brenan. Appl. Environ. Microb. 78, 1692–1700. doi: 10.1128/AEM.06215-11
Taylor, C. L., and Barker, N. P. (2012). Species limits in Vachellia (Acacia) karroo (Mimosoideae: Leguminoseae): evidence from automated ISSR DNA “fingerprinting”. South. Afr. J. Bot. 83, 36–43. doi: 10.1016/j.sajb.2012.07.014
Thrall, P. H., Burdon, J. J., and Woods, M. J. (2000). Variation in the effectiveness of symbiotic associations between native rhizobia and temperate Australian legumes: interactions within and between genera. J. Appl. Ecol. 37, 52–65. doi: 10.1046/j.1365-2664.2000.00470.x
Thrall, P. H., Millsom, D. A., Jeavons, A. C., Waayers, M., Harvey, G. R., Bagnall, D. J., et al. (2005). Seed inoculation with effective root-nodule bacteria enhances revegetation success. J. Appl. Ecol. 42, 740–751. doi: 10.1111/j.1365-2664.2005.01058.x
Toledo, I., Lloret, L., and Martínez-Romero, E. (2003). Sinorhizobium americanus sp. nov., a new Sinorhizobium species nodulating native Acacia spp. in Mexico. Syst. Appl. Microbiol. 26, 54–64. doi: 10.1078/072320203322337317
van Wyk, B.-E. (2011). The potential of South African plants in the development of new medicinal products. S. Afr. J. Bot. 77, 812–829. doi: 10.1016/j.sajb.2011.08.011
Vial, L., Chapalain, A., Groleau, M.-C., and Déziel, E. (2011). The various lifestyles of the Burkholderia cepacia complex species: a tribute to adaptation. Environ. Microbiol. 13, 1–12. doi: 10.1111/j.1462-2920.2010.02343.x
Vinuesa, P., Rojas-Jiménez, K., Contreras-Moreira, B., Mahna, S. K., Prasad, B. N., Moe, H., et al. (2008). Multilocus sequence analysis for assessment of the biogeography and evolutionary genetics of four Bradyrhizobium species that nodulate soybeans on the Asiatic Continent. Appl. Environ. Microbiol. 74, 6987–6996. doi: 10.1128/AEM.00875-08
Vinuesa, P., Silva, C., Werner, D., and Martínez-Romero, E. (2005). Population genetics and phylogenetic inference in bacterial molecular systematics: the roles of migration and recombination in Bradyrhizobium species cohesion and delineation. Mol. Phylogent. Evol. 34, 29–54. doi: 10.1016/j.ympev.2004.08.020
Willems, A., Coopman, R., and Gillis, M. (2001). Phylogenetic and DNA-DNA hybridization analyses of Bradyrhizobium species. Int. J. Syst. Evol. Microbiol. 51, 111–117. doi: 10.1099/00207713-51-1-111
Wolde-meskel, E., Terefework, Z., Frostegård, Å, and Lindström, K. (2005). Genetic diversity and phylogeny of rhizobia isolated from agroforestry legume species in southern Ethiopia. Int. J. Syst. Evol. Microbiol. 55, 1439–1452. doi: 10.1099/ijs.0.63534-0
Wolde-meskel, E., Terefework, Z., Lindström, K., and Frostegård, Å (2004). Rhizobia nodulating African Acacia spp. and Sesbania sesban trees in southern Ethiopian soils are metabolically and genomically diverse. Soil Biol. Biochem. 36, 2013–2025. doi: 10.1016/j.soilbio.2004.05.020
Xu, L. M., Ge, C., Cui, Z., Li, J., and Fan, H. (1995). Bradyrhizobium liaoningense sp. nov., isolated from the root nodules of soybeans. Int. J. Syst. Bacteriol. 45, 706–711. doi: 10.1099/00207713-45-4-706
Keywords: alpha-rhizobia, beta-rhizobia, Acacia karroo, Vachellia karroo, Paraburkholderia, Bradyrhizobium, South Africa
Citation: Beukes CW, Boshoff FS, Phalane FL, Hassen AI, le Roux MM, Stȩpkowski T, Venter SN and Steenkamp ET (2019) Both Alpha- and Beta-Rhizobia Occupy the Root Nodules of Vachellia karroo in South Africa. Front. Microbiol. 10:1195. doi: 10.3389/fmicb.2019.01195
Received: 13 August 2018; Accepted: 13 May 2019;
Published: 04 June 2019.
Edited by:
Encarna Velázquez, University of Salamanca, SpainReviewed by:
Euan James, The James Hutton Institute, United KingdomErnesto Ormeño-Orrillo, National Agrarian University, Peru
Rajaa Chahboune, Faculty of Medicine and Pharmacy of Tangier, Morocco
Copyright © 2019 Beukes, Boshoff, Phalane, Hassen, le Roux, Stȩpkowski, Venter and Steenkamp. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stephanus N. Venter, ZmFudXMudmVudGVyQHVwLmFjLnph
†These authors have contributed equally to this work
 Chrizelle W. Beukes
Chrizelle W. Beukes Francois S. Boshoff1†
Francois S. Boshoff1† Francina L. Phalane
Francina L. Phalane Ahmed I. Hassen
Ahmed I. Hassen Tomasz Stȩpkowski
Tomasz Stȩpkowski Stephanus N. Venter
Stephanus N. Venter