- 1Institute of Postharvest and Food Sciences, Agricultural Research Organization – The Volcani Center, Rishon LeZion, Israel
- 2Dipartimento di Agraria, Università Mediterranea di Reggio Calabria, Calabria, Italy
- 3Department of Ecology, Environment and Plant Sciences, Stockholm University, Stockholm, Sweden
- 4National Residue Control Laboratory, Kimron Veterinary Institute, Beit Dagan, Israel
- 5United States Department of Agriculture, Agricultural Research Service, Kearneysville, WV, United States
While the wheat-associated microbiome is of major agricultural importance, little is known about the alterations in wheat grain microbial community composition during storage. Characterization of the bacterial and fungal communities in stored wheat grains revealed the impact of phosphine fumigation, one of the most effective methods to eliminate insects in stored commodities, on the composition of the wheat grain microbiome. High-throughput amplicon sequencing of the bacterial 16S rRNA gene and fungal internal transcribed spacer (ITS) region was used to analyze the wheat grain microbiome at different times over as 6 months period of storage. Higher bacterial diversity was found across the samples during the first (immediately after harvest) and second (3 months later) time points, with a predominance of Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes and Planctomycetes. A two-fold decrease in the number of bacterial operational taxonomic units (OTUs) was observed in wheat grains at the last time point (6 months later), following phosphine treatment. In contrast to the effect of phosphine on bacteria, it did not affect fungal diversity in stored grains. The majority of fungal sequences were assigned to Ascomycota, followed by Basidiomycota, Glomeromycota, and unidentified fungi, which were evenly distributed throughout the storage period. Alpha and beta diversity analyses were confirmed by examination of the cultured microbial taxa obtained from the stored wheat grains. Mycotoxin analysis of wheat grains collected after phosphine fumigation revealed the presence of Fusarium toxins, primarily deoxynivalenol (DON). Several mycotoxigenic Fusarium spp. were also detected in the same samples. Results of the present study indicate that microbiome of stored, whole wheat grains was strongly affected by phosphine fumigation, which changed the structure of the microbial community leading to shifts in species composition toward mycotoxigenic strains. A better understanding of the complex interactions within the microbial communities of stored grains will assist in the development of novel biocontrol strategies to overcome mycotoxin contamination.
Introduction
Wheat is one of the most important cultivated cereals produced and consumed globally. It provides, on average, about 20% of the daily caloric requirement and about 21% of the daily protein intake in the human diet (Shiferaw et al., 2013). The availability of high-quality wheat grains is essential for maintaining the stability of the world’s food supply and for global food security. Wheat grains are colonized by complex microbial communities that play different roles in grain quality and susceptibility to disease (Links et al., 2014). Some of the bacteria and fungi that are associated with seeds can be harmful to human health and cause plant diseases, while others can have beneficial effects on the host and actually improve nutrient uptake and increase tolerance to biotic and abiotic stresses through multiple mechanisms (Brader et al., 2017). For example, specific species of Fusarium, Aspergillus, Penicillium, and Alternaria can cause spoilage in stored wheat grains and produce mycotoxins that severely decrease crop value and are harmful to animal and human health (Placinta et al., 1999; Magan et al., 2010). Conversely, the plant protective ability of some bacterial epiphytes and endophytes against fungal pathogens has been reported in several crops (Links et al., 2014; Mousa et al., 2016; Gdanetz and Trail, 2017). A better knowledge of the composition and dynamics of wheat-grain-associated microbiota is needed to identify novel beneficial microorganisms that may improve crop health and suppress the growth of potential pathogens in a sustainable manner. Several studies on the composition of the rhizosphere microbiome of wheat and other parts of the wheat plant have been conducted (Ofek et al., 2014; Hartmann et al., 2015; Gdanetz and Trail, 2017; Mahoney et al., 2017). Few studies, however, have characterized the microbiota of wheat grains and the seeds of other crop species (Links et al., 2014; Yuan et al., 2018).
In addition to fungal pathogens that cause seed deterioration during storage, insects can infest and spoil grain that is handled or stored improperly. Thus, a fumigation treatment is often required to prevent insect infestations in stored grains. Phosphine gas is by far the most common fumigation agent used worldwide for the treatment of stored grain against insects, and has recently increased in usage following the prohibition of methyl bromide, an alternative fumigant. Although phosphine is very toxic to insects, it does not leave toxic residues after treatment or impact seed viability (Bell, 2000). While this fumigant is known to have broad biological activity (Berners-Price and Sadler, 1988; Nath et al., 2011), its effect on the microbial community of stored wheat grains is largely unknown. Few studies have been reported on the effect of phosphine on the growth of molds in stored wheat and corn grains (Hocking and Banks, 1991; Castro et al., 2000). These studies reported that phosphine only causes a slight decrease in the population of Aspergillus flavus in stored maize and Eurotium chevalieri, A. flavus, or Aspergillus parasiticus in stored wheat grains.
The objective of the present study was to characterize the composition of the fungal and bacterial communities in stored wheat grains, which produced annually for human and animal consumption, and to determine the impact of phosphine fumigation on the wheat grain microbiome during storage. Amplicon-based high-throughput sequencing (HTS) and chemical analysis of mycotoxins were used to characterize the wheat grain microbiome at different time points over a storage period of 6 months. Significant changes in the composition of the microbial community were identified when phosphine treatment was applied to stored wheat grains. Fusarium-related toxins were also detected in stored wheat grains as a result of shifts in the microbial population profiles which took place following fumigation with phosphine.
Materials and Methods
Wheat Grain Samples
Wheat grain (Triticum aestivum) samples were collected from seven wheat grain storage facilities located in northern and southern districts of Israel. Samples were designated as T1 when obtained immediately after harvest during the first week of storage (June 2016), T2 after storage for 3 months, and T3 after storage for 6 months. Four months after harvest (before collecting T3 samples) the stored grains in all locations were exposed to phosphine (3 g/m3) for a period of 10 days to control insect pests. In each storage facility, three samples (1 kg of grain each), destined for human consumption, were collected at each time point (3 samples × 7 storage sites = a total of 21 samples at each time point) in equal proportions from the front face and the center, at points located 1 m in horizontal depth within the grain mass, and from areas close to the walls. Grain temperature and moisture content were in the ranges of 27–33°C and 10.5–12.9%, respectively, throughout the study. The collected samples were kept in sterile plastic bags during transport to the laboratory and on the same day a 100 g aliquot was taken from each sample, thoroughly mixed, frozen in liquid nitrogen, freeze dried and milled into a fine powder with a grain grinder. The grain grinder was cleaned and disinfected with 70% ethanol solution between sample grinding. Powders were stored at 4°C until further pending analysis (2–4 weeks).
DNA Extraction, Amplification and Sequencing
DNA extraction from wheat grain samples was performed as previously described (Sadhasivam et al., 2017). Total DNA was extracted from 300 mg of wheat powder using a lysis buffer containing hexadecyltrimethylammonium bromide (CTAB). Ten milliliters of DNA extraction buffer (l.0 M Tris–HCl, pH 7.5; 1% (w/v) CTAB; 5 M NaCl; 0.5 M EDTA; 1% (v/v) 2-mercaptoethanol; and proteinase K at 0.3 mg/ml) were added to the powder, mixed gently, and the mixture was incubated at 65°C for 30 min. The extracts were cooled prior to the addition of an equal volume of chloroform, gently mixed and centrifuged at 6000 rpm for 10 min. The aqueous supernatant was recovered and an equal volume of 2-propanol was added. The DNA was precipitated by centrifugation at 4800 ×g for 5 min and resuspended in TE buffer solution (Tris-EDTA, pH 8.0) containing RNase A at 10 μg/ml. DNA was further purified by phenol-chloroform extraction. Finally, the DNA was precipitated with 100% ethanol containing 3 M sodium acetate, rinsed in 70% (v/v) ethanol and resuspended in TE buffer. The purity of the extracted DNA was assayed with a NanoDrop One spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, United States), and the total DNA concentration in each sample was adjusted to 50 ng/μl. The universal primers 515F/926R and 5F/86R were used to amplify the 16S and ITS2 rRNA gene regions of bacteria and fungi, respectively (Supplementary Table S1). The primers were modified to include Illumina adapters1 for subsequent multiplexing. PCR amplification of each sample was performed in triplicate. The PCR mixture (25 μl) contained 12.5 μl 2× DreamTaq green PCR master-mix (Thermo Fisher Scientific, Lithuania), 1 μl of each primer (5 μM), and 1 μl of DNA template. Nuclease-free water (Thermo Fisher Scientific, Lithuania) replaced template DNA in negative controls. All amplicons and amplification mixtures including negative controls were sequenced using Illumina MiSeq V3 (2 × 300 bp) chemistry.
Data Analysis
Illumina adaptors were clipped and low-quality reads removed by Trimmomatic 0.36 (Bolger et al., 2014) using a sliding window trimming, cutting once the average quality within the window of 4 bases falls below a quality threshold of 15. Paired-end reads were merged utilizing PEAR (Zhang et al., 2014) for 16S rRNA gene, and PANDAseq (Masella et al., 2012) for ITS rRNA gene region sequences with default parameters. Chimeric sequences were identified and removed using USEARCH (Edgar, 2010; McDonald et al., 2012) for 16S rRNA gene, and VSEARCH 1.4.0 (Rognes et al., 2016) for ITS rRNA gene region sequences. UCLUST algorithm (Edgar, 2010), as implemented in QIIME 1.9.1 (Caporaso et al., 2010) was used to cluster sequences queried against the Greengenes 13_8_97 database for 16S rRNA gene (DeSantis et al., 2006), and for ITS UNITE dynamic database released on 01.12.2017 (Abarenkov et al., 2010) at a similarity threshold of 97%, respectively. Sequences that failed to cluster against the database were de novo clustered using the same algorithm. After removing singletons, the most abundant sequences in each OTU were selected as representative sequences and used for the taxonomic assignment using the BLAST algorithm (Altschul et al., 1990; McDonald et al., 2012) as implemented in QIIME 1.9.1. The OTU table was normalized by rarefaction to an even sequencing depth in order to remove sample heterogeneity. The rarefied OTU table was used to calculate alpha diversity indices including Observed Species (Sobs), and Shannon metrics. Alpha diversities were compared based on a two-sample t-test using non-parametric (Monte Carlo) methods and 999 Monte Carlo permutations. Results were visualized in boxplots figures.
Metagenome Seq’s Cumulative Sum Scaling (CSS) (Paulson et al., 2013) was used as a normalization method for other downstream analyses. The CSS normalized OTUs table was analyzed using Bray Curtis metrics (Bray and Curtis, 1957) and utilized to evaluate β-diversity and construct PCoA plots using Emperor (Lozupone and Knight, 2005). Similarity in community composition was tested via ANOSIM in QIIME 1.9.1 using 999 permutations. Differential OTU abundance of the most abundant taxa (≥0.1%) between sample groups were determined using a t-test and the Kruskal-Wallis test (Kruskal and Wallis, 1952). In all tests, significance was determined using 999 Monte Carlo permutations, and the false discovery rate (FDR) was used to adjust the calculated P-values and when the FDR P < 0.05 it was considered significant.
Microbial Isolation and Identification
Grains samples (10 g each) were shaken on an orbital shaker at 150 rpm in 90 ml sterile saline (0.9% NaCl) solution at room temperature for 1 h, then 100 μL of serial 1/10 dilutions were plated on Luria-Bertani (LB) agar plates for isolation of bacteria and Potato Dextrose Agar (PDA) plates supplemented with chloramphenicol (20 μg/ml) for isolation of filamentous fungi and yeasts. LB agar plates were incubated at 37°C for 24–48 h, whereas PDA plates were incubated at 28°C for 48–72 h. Bacterial colonies were randomly selected from the plates and streaked on fresh culture media to obtain pure cultures. Filamentous fungi and yeasts were transferred singly to PDA plates and subcultured twice to obtain a pure culture.
DNA was extracted from each bacterial strain using a lysozyme lysis method described by De et al. (2010). Fungal DNA extraction was performed on lyophilized mycelium/yeasts cells using CTAB-based method as previously described (Sadhasivam et al., 2017). DNA quality and yield were determined using a NanoDrop One spectrophotometer. The 16S rRNA gene in bacteria and ITS rRNA gene region in fungi were amplified with universal primers 27F/1492R and ITS1/ITS4, respectively (Supplementary Table S1). PCR products were purified and sequenced through standard Sanger sequencing; sequences were identified via BLAST matches to the NCBI database2 and deposited as accession number MK229027–MK229144 (Bacterial 16S rRNA gene) and MK226203–MK226306 (Fungal ITS rRNA gene region).
Quantitative Real-Time PCR
qPCR was performed to quantify the presence of mycotoxigenic fungi, such as Fusarium, Aspergillus, and Alternaria spp., in the stored grains at different time points. A broad-spectrum primer pair ITSPF/ITSPR was used to target the fungal ITS region and single genus-specific TaqMan probes were tested for their ability to detect fungal DNA (Supplementary Table S2). The probes were labeled on the 5′ end with the fluorescent reporters FAM, HEX and TEX, and on the 3′ end with TAMRA, BHQ1, and BHQ2. The qPCR reactions were carried out using qPCRBIO Probe Mix (PCR Biosystems, London, United Kingdom), 10 μM of each primer, 10 μM of each probe, and 5 ng of genomic DNA template. Amplification reactions were performed in a total reaction volume of 10 μl and were run on the Eco Real-Time PCR System (Illumina) with the following program: 2 min at 50°C [Uracil-DNA glycosylases (UDG) activation], 2 min at 95°C (denaturation and Taq polymerase activation), an amplification program of 45 cycles at 95°C for 15 s, 55°C for 15 s, and 60°C for 15 s. Real-time qPCR reactions were performed in triplicate for each biological replicate and each sampling date contained three biological replicates. The cycle threshold value (Ct), which refers to the cycle number where the sample’s fluorescence significantly increases above the background level, was calculated automatically by the Eco software. The detection limit of the probes was assessed under optimized PCR conditions with the DNA extracted from pure cultures of Fusarium proliferatum (NRRL 31866), A. flavus (NRRL 3518), and Alternaria infectoria (F11, isolated from stored wheat grains). Serial dilutions were made of DNA from the three fungal species. The real-time PCR data indicated that the Ct values correlated well with known DNA quantities from 8 pg to 5 ng, with R2 values of 0.98 (slope -1.70) for Fusarium, 0.99 (slope -1.48) for Aspergillus, and 0.99 (slope -1.72) for Alternaria (Supplementary Figure S1).
To test the efficacy of the method, sterilized wheat grain samples (10 g) were inoculated with each of A. flavus, F. proliferatum, and A. infectoria cultures at final concentration of 104 spores/g and incubated for 3 days. Non-inoculated sterilized grain samples used as control. DNA extracts isolated from the samples were analyzed using the qPCR assay. The obtained Ct values (23–30) revealed high reproducibility and demonstrated the ability to quantify fungal DNA in artificially contaminated wheat grains. qPCR analysis indicated the average values of DNA content for F. proliferatum, A. flavus, and A. infectoria spp. in seeds at concentrations of 268, 44 and 30 ng/mg, respectively.
Mycotoxin Analysis
Detection, identification, and quantification of mycotoxins in wheat grain samples were performed using high-performance liquid chromatography coupled with tandem mass-spectrometry (LC/MS/MS) as described previously (Sadhasivam et al., 2017). Briefly, individual stock standard solutions (1 mg/ml) of aflatoxin B1, B2, G1, G2 (AFB1, AFB2, AFG1, AFG2), ochratoxin A (OTA), zearalenone (ZEN), deoxynivalenol (DON), fumonisin B1, B2 (FB1, FB2) and T-2 toxin (T-2) (Fermentek, Israel) were prepared in methanol. Multi-toxin working standard solutions with a series of toxin concentrations were prepared by dilution of the stock solutions of the analytes in methanol. For sample preparation, 5 g of the ground seeds were mixed with 20 ml of 25:75 (v/v) water/methanol and placed in an orbital shaker at 200 rpm for 30 min at room temperature. After centrifugation at 8500 ×g for 15 min, 5 ml of the supernatant were transferred to a 15-ml glass tube and evaporated under a stream of nitrogen at 50°C. The dry residue was reconstituted with 0.25 ml of a 95:5 (v/v) water/methanol mixture and centrifuged for 10 min at 17,000 ×g, at 4°C; the supernatant was used directly for the analysis. Three non-contaminated wheat grain samples were spiked with multi-mycotoxin standard solutions at three concentration levels for construction of calibration curves, which were used for mycotoxin quantification. The spiking experiments were performed in triplicate at three different time points. Chromatographic separation was carried out using Nexera X2 UHPLC (Shimadzu, Tokyo, Japan) equipped with 100 × 2.1 mm, 2.6 μm Kinetex C18 column, (Phenomenex, Torrance, CA, United States). The mobile phase consisted of 2.5 mM ammonium acetate acidified with 0.1% acetic acid (A), and methanol (B). The LC system was coupled with API 6500 hybrid triple quadrupole/linear ion trap mass spectrometer (Sciex, Concord, ON, Canada), equipped with a turbo-ion electrospray (ESI) ion source. Validation parameters, such as precision, accuracy, limit of detection (LOD), limit of quantification (LOQ), and specificity, were determined (Supplementary Table S3).
Results
Amplicon Sequencing
After paired-end alignments, quality filtering, and deletion of chimeric, singletons, and plant sequences, 22,338 16S rRNA gene sequences were assigned to 806 bacterial OTUs and 5,383,776 ITS sequences were assigned to 6,064 fungal OTUs. The number of 16S sequences varied between 186 and 5019 (Std. dev. 702) and ITS sequences varied between 23,948 and 359,517 (Std. dev. 42565) reads per sample. The number of OTUs, after rarefaction collapsing biological replicates, varied between 66 and 187 OTUs in the 16S rRNA gene dataset and between 350 and 515 OTUs in the ITS rRNA gene region dataset per category of samples (Supplementary Table S4).
Wheat Grain Microbial Communities
Bacterial OTUs were mainly assigned to Proteobacteria (41.1%), Firmicutes (35.4%), Actinobacteria (11.9%), Bacteroidetes (6.9%), and Acidobacteria (1.9%) (Figure 1A). Proteobacteria was dominated by the presence of Gammaproteobacteria (21.7%), Alphaproteobacteria (8.2%), Betaproteobacteria (7.7%), and Deltaproteobacteria (3.3%).
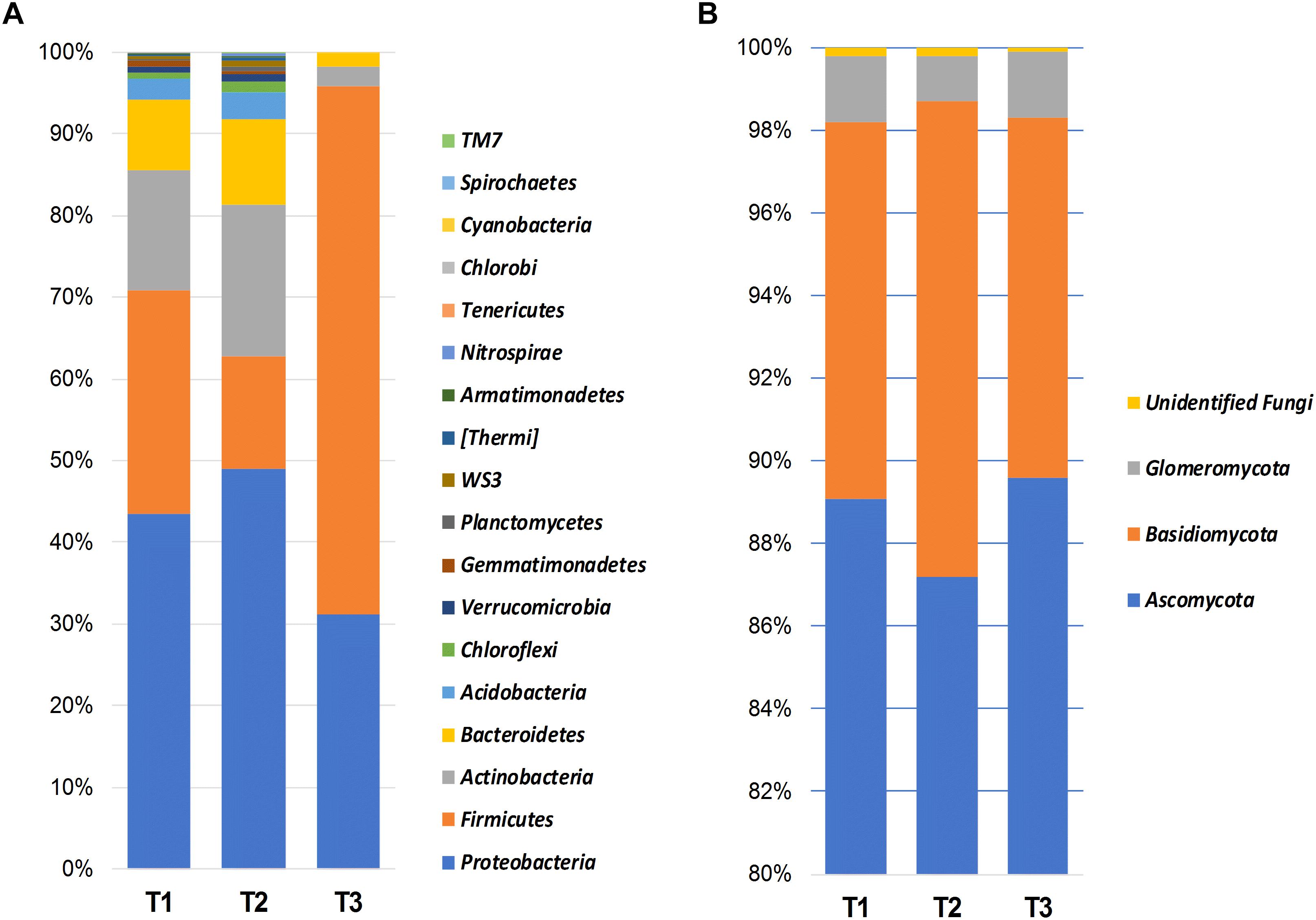
Figure 1. Bar charts showing the relative abundance of most dominant bacterial (A) and fungal (B) phyla detected using high-throughput sequencing technology across the samples collected at three storage time-points (T1, T2, T3).
Fungal members of the Ascomycota were the dominant fungal phylum across all samples, accounting for 88.5% of the total number of detected sequences (Figure 1B). This was followed by Basidiomycota (9.8%), Glomeromycota (1.4%), and unidentified fungi (0.2%). Chytridiomycota, Entorrhizomycota, Mortierellomycota, Mucoromycota, and Olpidiomycota were detected at a very low frequency (<0.1%). Ascomycota were largely identified as members of the classes Dothideomycetes (87.0%), and Sordariomycetes (1.1%). Whereas Basidiomycota were represented by Tremellomycetes (4.4%), Microbotryomycetes (2.8%), and Pucciniomycetes (1.5%), Glomeromycota was almost exclusively dominated by Glomeromycetes (1.4%).
Compositional Differences in the Microbial Community After Different Lengths of Storage
Alpha diversity comparisons based on Shannon index, indicated that bacterial diversity varied significantly (P < 0.003) between T1 and T3, and T2 and T3 in both cases (Table 1 and Figure 2A). In contrast, Shannon index indicated no significant difference (P ≥ 0.1) in fungal community composition between the sampled time-points (Table 1 and Figure 2B). On the other hand, based on the Bray Curtis dissimilarity metric, the three time points (T1, T2, and T3) varied significantly (P < 0.01) in their bacterial and fungal community composition and structure (Table 1). The differences between the time points were also evident in the Principal Coordinate Analysis (PCoA) of the bacterial community, where samples from T3 were positioned far from T1 and T2 samples which formed a separate cluster (Figure 3A). PCoA analysis of the fungal community, however, did not provide any clear clustering based on sampling time (Figure 3B).
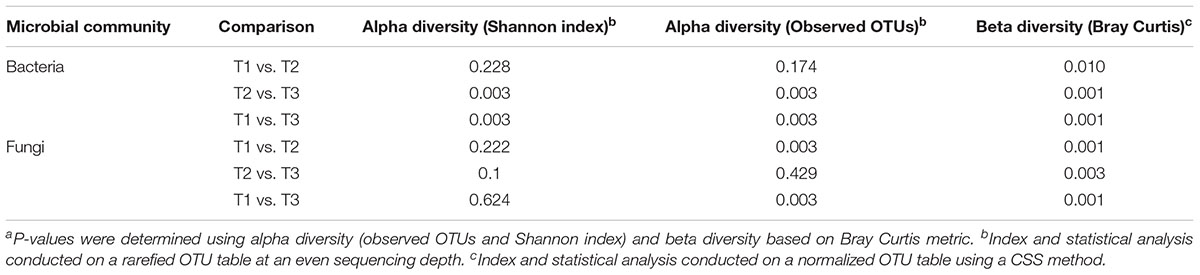
Table 1. P-values of the comparisons between the samples collected at three storage time-points (T1, T2, T3)a.
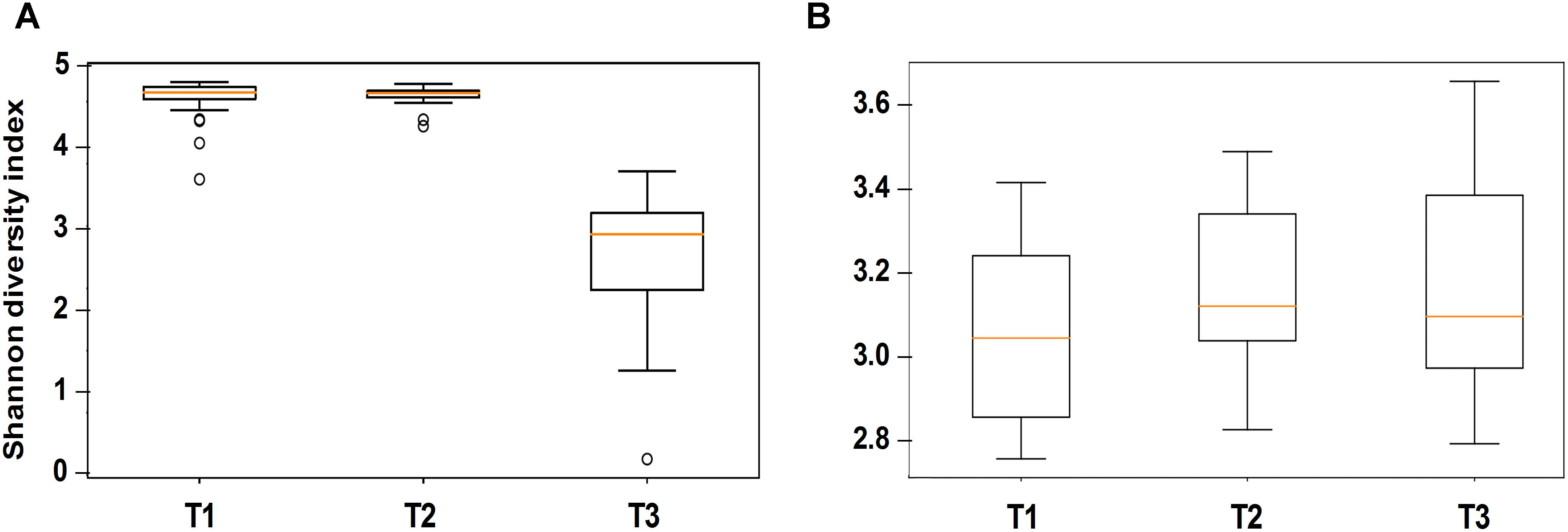
Figure 2. Alpha diversity analysis (based on Shannon diversity index) of the bacterial (A) and fungal (B) communities in stored wheat grain samples. Analyzed samples were collected immediately after harvest during the first week of storage (T1), after 3 months of storage (T2), and after 6 months of storage (T3). The centered square represents the mean, orange line inside the box represents the median, and circles indicate outliers.
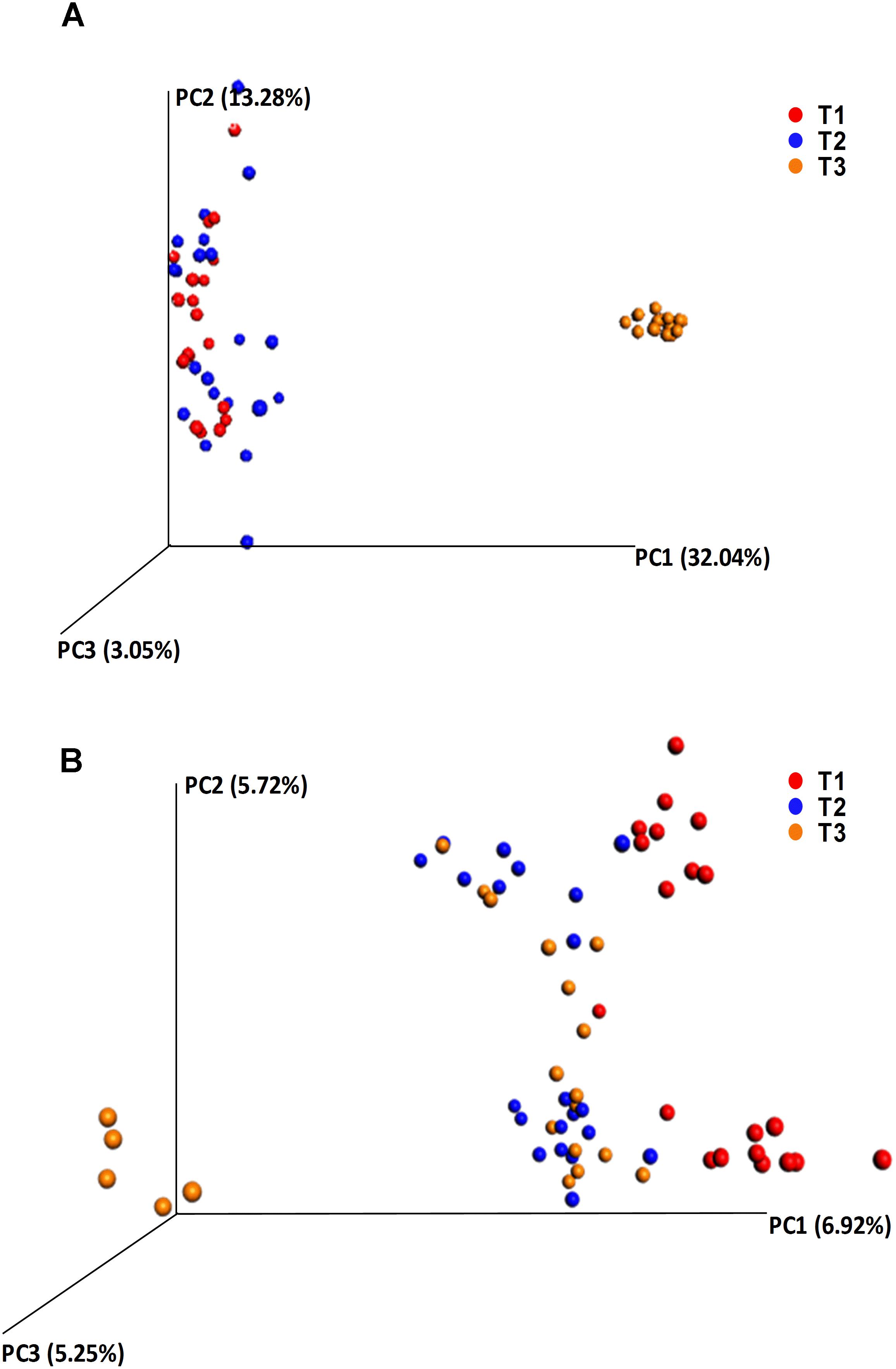
Figure 3. Principle coordinate analysis (PCoA) based on the beta diversity Bray Curtis dissimilarity metrics, showing the distance in the bacterial (A) and fungal (B) communities between the samples collected at T1 (red dots), T2 (blue dots), and T3 (orange dots).
The relative abundance of fungal and bacterial genera detected in wheat grain samples across three sampling time points is shown in Figure 4. Bacterial communities of stored grains mainly consisted of genera of Bacillus (22.5%), followed by Erwinia (9.9%), Pseudomonas (5.9%), unidentified genera that belong to the family Oxalobacteraceae (4.3%), Enterobacteriaceae (3.7%), Streptomyces (3.3%), Curtobacterium (2.2%), and Paenibacillus (1.6%) (Figure 4A). Interestingly, Bacillus and Erwinia were the most abundant bacterial genera found at the last sampling time point (T3), which represented more than 70% of the total bacteria detected after phosphine fumigation (Figure 4A). In addition to the variations in community structure, the relative abundance of several other bacterial taxa, varied significantly between the different time points. Bacterial phylotypes that belongs to Sphingomonas, Frigoribacterium, Ruminococcus, Hymenobacter, Acinetobacter, Chryseobacterium, Methylobacterium, Saccharibacillus, Staphylococcus, Agrobacterium, Pedobacter, and Comamonadaceae were only present at the third sampling time point. In contrast, bacterial taxa such as Ellin6067, Bdellovibrio, Prevotella, Bilophila, Parabacteroides, and an unidentified group of Leuconostocaceae were present at almost equal relative abundance in T1 and T2 and then disappeared entirely in T3 (Supplementary Figure S2). Unlike bacteria, the most abundant fungal genera, such as Alternaria (59.2%), Stemphylium (10%), unidentified Pleosporales (8.6%), Cladosporium (3.4%), Mycosphaerella (3%), Sporobolomyces (2.7%), Filobasidium (2.6%), and Puccinia (1.5%), which accounted for more than 90% of the total fungal community, were equally present in all three of the sampled points (Figure 4B). Only three fungal genera, Cortinarius, Dioszegia, and Monographella, significantly changed in abundance over the three time points (Supplementary Figure S3).
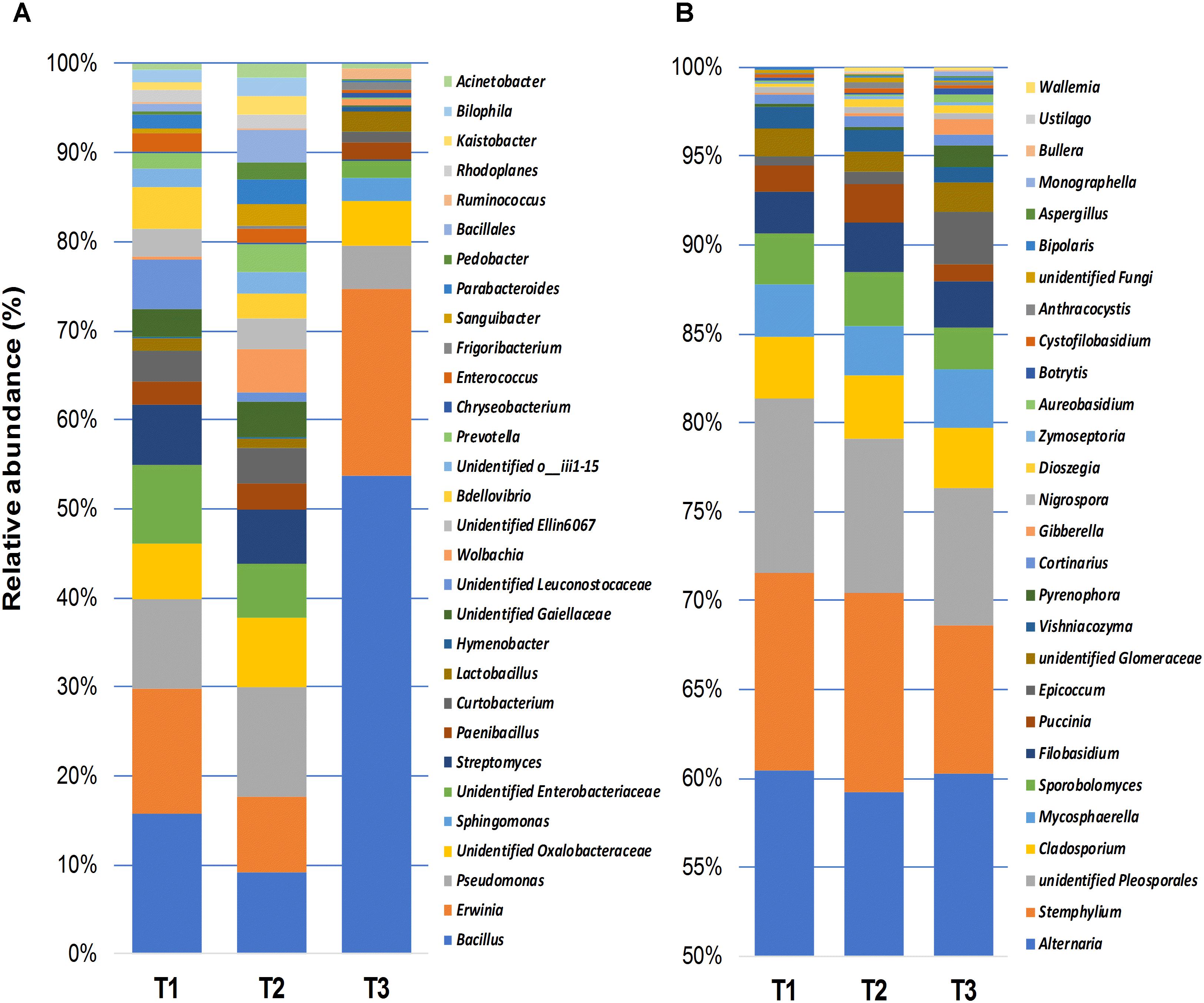
Figure 4. Relative abundance of bacterial (≥0.5%) (A), and fungal (≥0.1%) (B) genera detected using high-throughput sequencing technology across the samples collected at three storage time-points (T1, T2, T3).
Isolation and Identification of Bacteria and Fungi From Wheat Grains
Bacterial and fungal isolates were cultured from wheat grains sampled at the three storage time points to assess the impact of phosphine fumigation on microbial diversity and to evaluate potential interactions between the members of the shared microbiome. Forty bacterial and 44 fungal (24 yeasts and 20 filamentous fungi) isolates were cultured from seeds at the first time point (Figure 5). Bacillus spp. were predominant among cultured bacterial community (55%) followed by Pseudomonas spp. (12.5%) and Pantoea spp. (10%) (Figure 5A). Cryptococcus and Rhodotorula species predominated among the isolated yeasts cultures (39%) based on morphological features (colony color, texture, microscopic observation) and sequencing analysis. The same criteria indicated that 13 out of 44 cultured filamentous fungal isolates were Alternaria spp. (30%), followed by Aspergillus spp. (7%) (Figure 5B).
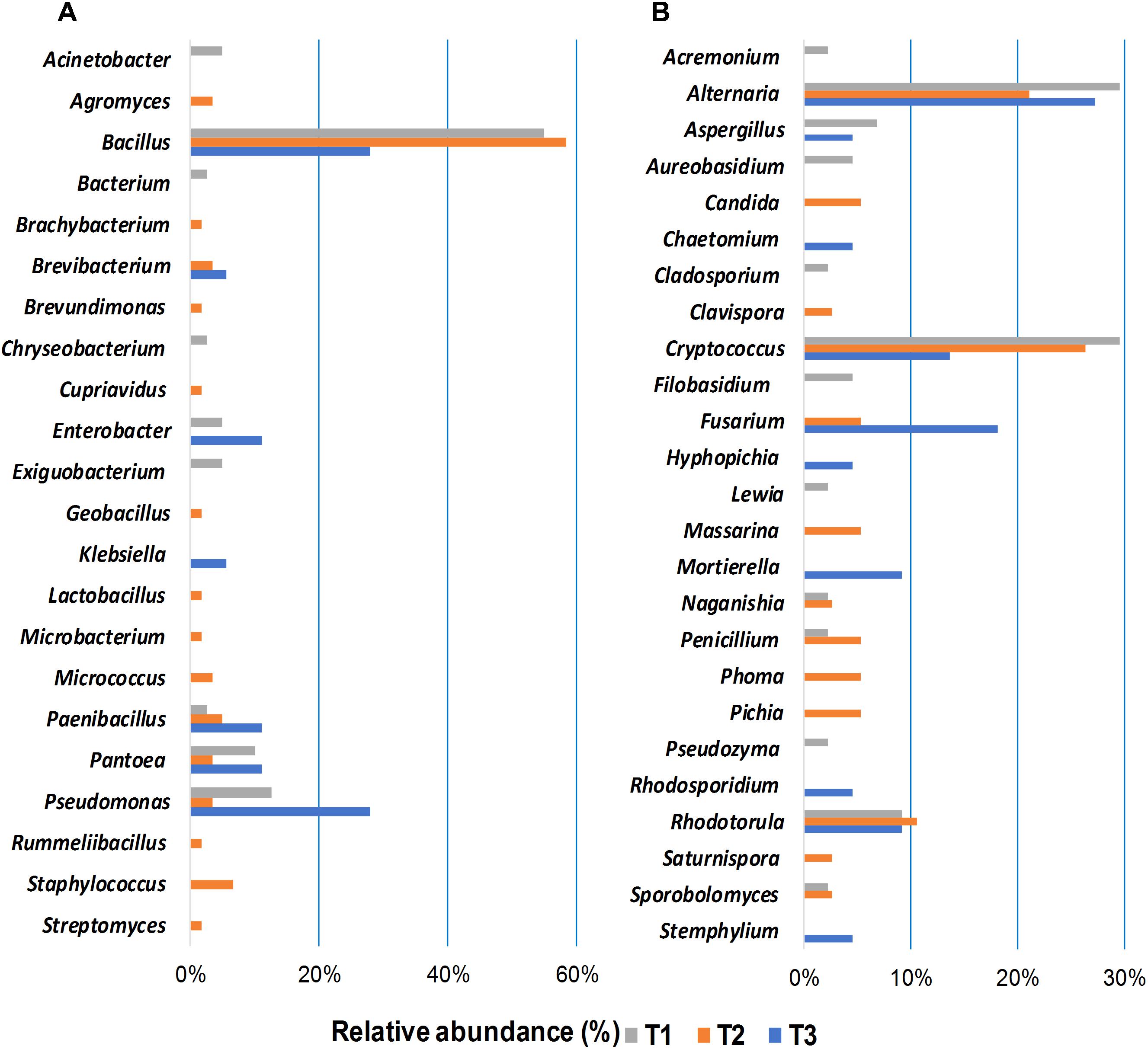
Figure 5. Relative abundance of bacterial (A), and fungal (B) genera isolated by plating from grain samples collected at three storage time-points (T1, T2, T3). The isolates were identified by sequencing the 16S rRNA gene in bacteria and ITS region in fungi; the sequences were determined via BLAST matches to the NCBI database.
A more diverse bacterial community was identified in the bacterial isolates cultured at the second time point (60 isolates), where Bacillus spp. were most common (58%) (Figure 5A). The 38 fungal isolates cultured from wheat grains at the second time point were identified as Alternaria spp. (21%), Fusarium spp., Massarina spp., Penicillium spp., and Phoma spp. (5% for each genus). Cryptococcus and Rhodotorula spp. were still the most abundant (36%) yeast taxa (Figure 5B). A dramatic decrease was observed in the number of bacterial strains isolated from wheat grains sampled at the third time point (18 isolates). Bacillus and Pseudomonas spp. were the predominant isolates cultured from T3 samples (28% for each genus) (Figure 5A). The number of yeast isolates cultured from T3 samples was also significantly reduced compared to T1 and T2 (Figure 5B). Fungal abundance and diversity in T3 samples did not change, however, relative to T1 and T2. Fifteen filamentous fungal isolates, out of a total 22 fungal microorganisms, were cultured from stored seeds collected at the T3 time point. Mycotoxigenic fungi, such as Alternaria (27%) and Fusarium (18%) species, were predominant. The changes in microbial community composition identified in the culturing assay were in good agreement with results obtained in the alpha- and beta-diversity analyses (Figures 2, 3).
Analysis of Mycotoxigenic Fungi in Stored Grains by Real-Time PCR
Since cultured fungal diversity was not influenced by phosphine fumigation and was generally consistent across the three sampled time points, real-time PCR method was utilized to quantify fungal DNA in stored grain samples. The broad-spectrum primers and specific fluorescent probes were used to amplify the targeted genomic DNA and to detect and quantify the major mycotoxigenic fungal genera, Aspergillus, Fusarium, and Alternaria.
The wheat grain samples collected throughout the entire study did not exhibit any evidence of spoilage. Nevertheless, the qPCR analysis indicated the presence of all three pathogenic genera in majority of the samples, which corroborates the results obtained in the culture assay of wheat grains. Fusarium DNA was detected in 11, 12, and 15 out of 21 wheat grain samples at T1, T2, and T3 time points, respectively, with an increased DNA content at T3 (Figure 6A). The obtained Ct values ranging between 27.62 and 32.78 indicated the presence of relatively low concentrations of Fusarium DNA in the samples (0.5 – 8 ng/mg; Figure 6A), suggesting that qPCR is more sensitive in detecting mycotoxigenic fungi compared to culture-based approaches. Aspergillus DNA was detected in only 10 and 3 out of 21 wheat grain samples collected at the T1 and T3 time points, respectively, with no detection in the T2 samples (Figure 6B). In contrast, 9 to 11 wheat grain samples in all three time points were found to be positive for Alternaria spp. by qPCR, and with a relatively higher fungal DNA content relative to the other tested mycotoxigenic species (Figure 6C).
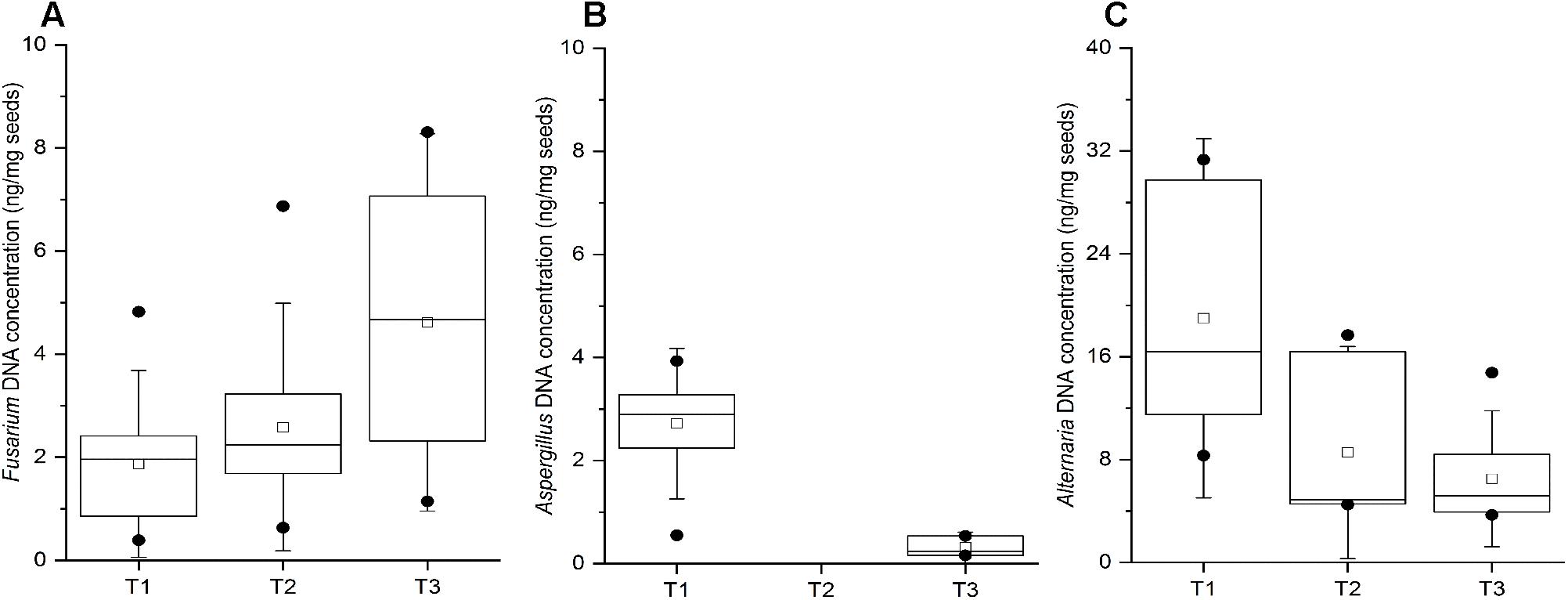
Figure 6. Quantification by qPCR of Fusarium spp. (A) Aspergillus spp. (B), and Alternaria spp. (C) in stored wheat grain at three storage time-points (T1, T2, T3). qPCR results are from at least two biological replicates (DNA extractions) and three technical replicates per sample. The lower and upper edges of each box correspond to the 25th and 75th percentiles. The black line inside the box represents the median, and centered square represents the mean. The whiskers represent 1.5× interquartile range. Dots indicate outliers.
Mycotoxin Detection in Stored Wheat Grains
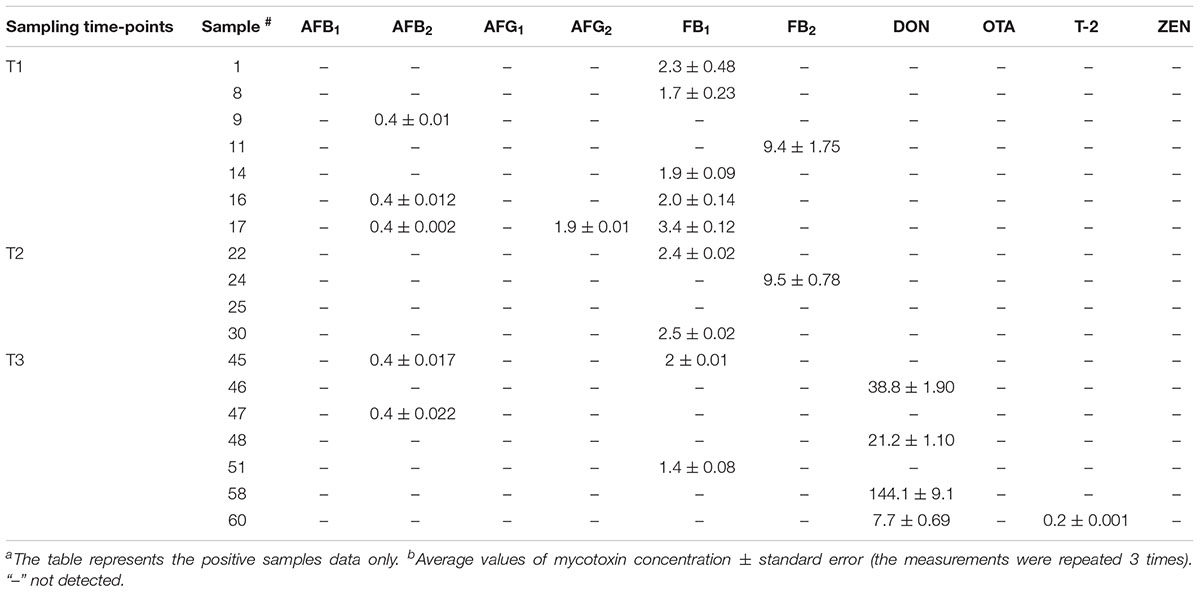
The LC/MS/MS multi-toxin method was optimized for the simultaneous detection and quantification of 10 mycotoxins in wheat grain samples to investigate potential changes in mycotoxin levels in grains over the storage period of 6 months, including after phosphine fumigation for insect control. Results indicated that 17 wheat samples were positive for mycotoxin contamination and were above their limit of detection (LOD) but under the maximum EU regulatory limits (Table 2). AFB2 was detected at 0.4 ppb in three wheat samples collected at T1 and the co-occurrence of AFG2 was only observed in one of the samples. Two grain samples from T3 were also found to be contaminated with AFB2 (Table 2). These findings are consistent with data obtained in the culturing assay and by qPCR, where Aspergillus spp. were also detected in T1 and T3 samples. The LC/MS/MS analysis also indicated the presence of fumonisins at values below the regulatory guidelines in the samples analyzed at all three time points. Notably, a strong link was observed between increased abundance of Fusarium spp. in T3 wheat grain samples, and the presence of DON toxin only in T3 samples (Table 2). These results indicate that phosphine fumigation significantly altered the structure of microbial community of stored wheat grains and induced shift in the abundance of mycotoxin-producing Fusarium taxa.
Discussion
An objective of the present study was to characterize the composition of microbial communities associated with stored wheat grains and to identify any changes in the diversity and composition of the wheat microbiome during storage and in response to phosphine fumigation to control insect pests. Characterizing the microbiota of wheat grains after harvest is an essential step to understanding the interactions that potentially occur between different members of the community in relation to the colonization of the grains with common mycotoxigenic fungi (Aspergillus, Fusarium, Penicillium, and Alternaria species). Results of the HTS analysis indicated that fumigation of the grains with phosphine had a significant effect on the diversity and abundance of various components of the wheat-grain microbiome. It is known that many other factors could affect the stored seed microbial community such as temperature, humidity, water activity, grain moisture (Schmidt et al., 2018). Nevertheless, in the current study, the impact of such environmental factors was minor due to the optimal and stable weather conditions during a 6-months storage period. In general, the composition of the bacterial and fungal microbiota of wheat grains found in the current study is in agreement with previous studies (Karlsson et al., 2014, 2017; Comby et al., 2016; Díaz Herrera et al., 2016; Gdanetz and Trail, 2017). Genera, including Bacillus, Erwinia, Pseudomonas, and Paenibacillus were dominant bacterial taxa identified, while Alternaria, Stemphylium, Cladosporium, Sporobolomyces, Mycosphaerella, and Filobasidium were the most prevalent fungi and represent the most common members of the wheat grain mycobiome. The majority of the detected bacteria taxa, with the exception of Erwinia, are known to have a beneficial impact on plants. For instance, Bacillus species are known to have a growth promotive effect on wheat plants and are generally isolated from both grains and the rhizosphere (Tao et al., 2014; Pan et al., 2015; Cherif-Silini et al., 2016). Paenibacillus is recognized as a predominantly endophytic bacterium in wheat plants and seeds, also with growth promotive effects (Díaz Herrera et al., 2016; Hao and Chen, 2017). Similarly, Pseudomonas contains several species that promote plant growth by suppressing pathogenic microorganisms, and synthesizing growth-stimulating plant hormones (Preston, 2004). Notably, however, other species of Pseudomonas may be phytopathogenic. In the present study, the results obtained with the culturing assay of stored wheat grains microbes were in agreement with the results obtained by HTS sequencing data. Bacillus, Pseudomonas, and Pantoea were found to be most abundant genera obtained in the cultured bacterial taxa isolated from stored wheat grains throughout the study, and the same was true in the HTS analysis.
The data indicated that phosphine fumigation significantly affected the microbial community associated with wheat grains. Phosphine or hydrogen phosphide (PH3) is a low molecular weight compound that diffuses rapidly and penetrates deeply into materials, such as the bulk storage of grain (Bond, 1989). It is the dominant fumigant used to control insect pests in stored grain and many other stored commodities. Since phosphine is toxic to aerobically respiring organisms, it also has effects on the survival and growth of some aerobic bacteria and fungi (Hocking and Banks, 1991; Castro et al., 2000). In the present study, bacterial population exhibited a drastic reduction in their level of diversity and the number of observed species after fumigation. Highly abundant bacteria, including Streptomyces, Ellin606, Bdellovibrio, Prevotella, Bilophila, and unidentified groups of Parabacteroides, and Leuconostocaceae that collectively accounted for approximately 45% of the total bacterial OTUs in the first two time points (T1 and T2), disappeared entirely in T3 after the phosphine treatment. This change corresponded with a significant increase in Bacillus from an average of 2.2% in T1 and T2 to more than 50% in T3 (Supplementary Figure S2). A similar trend was reported by Yuan et al. (2018), who reported that the abundance of Bacillus spp. in wheat grains increased from 2 to 36% after 9 months of storage and up to 48% after 12 months of storage at different position within a storage silo. Interestingly, in that study, the authors indicated that stored wheat grain was treated with ozone to kill insects.
Small quantities of stable but harmless breakdown products of phosphine, which become incorporated into normal cellular metabolism as phosphates and phosphites, may remain in fumigated materials (Nath et al., 2011). Bacillus and Pseudomonas spp. are capable of utilizing phosphite and belong to the group of phosphate solubilizing bacteria, which play an important role in plant growth promotion (Foster et al., 1978; Metcalf and Wolfe, 1998; Igual et al., 2001; Park et al., 2009; Kang et al., 2014). Bacillus and Pseudomonas may take advantage of these unique abilities after phosphine fumigation to enhance their survival and proliferation. The exclusive increase in the abundance of Sphingomonas species, which are aerobic bacteria utilized for environmental remediation due to their ability to degrade aromatic compounds (Story et al., 2004), after phosphine fumigation, may indicate their involvement in the degradation of this toxic substance. Since one of the main modes of action of phosphine is the inhibition of cytochrome c oxidase (Price, 1980), apparently some oxidase-negative bacteria (i.e., those that do not rely on cytochrome c oxidase in respiration), such as Acinetobacter, Lactobacillus, Ruminococcus, Methylobacterium, Saccharibacillus, and Frigoribacterium species, may survive and proliferate following phosphine treatment.
Results indicated that neither the diversity nor abundance of fungi were affected by phosphine fumigation. Only a few studies have described the influence of phosphine on the survival and growth of molds in stored grains. Phosphine was reported to have little effect on populations of fungi that were unable to grow in stored grains under conditions of limiting water activity (aw 0.8–0.86) (Raghunathan et al., 1969; Sinha et al., 1967; Hocking and Banks, 1991). Natarajan and Bagyaraj (1984) noted some reduction in fungal growth on legumes when exposed to very high phosphine levels (100 g/m3), particularly at limiting aw conditions (0.8). It seems that the effect of phosphine on fungal growth, however, may vary between fungal species and the type of stored grain or seed. Phosphine was shown to inhibit the growth of A. flavus and/or A. parasiticus and aflatoxin production on peanuts in both laboratory and warehouse experiments (Castro et al., 1992, 1995, 1996). A reduction of A. flavus growth and aflatoxin production was also observed in response to phosphine treatment of maize kernels stored at different moisture levels (Castro et al., 2000). In the same study, however, the authors stated that Penicillium species and F. verticillioides, which are commonly found in freshly harvested grains, were tolerant to phosphine (Castro et al., 2000). Those findings are consistent with the results obtained in the current study of wheat-grain-associated microflora, in which a greater number of Fusarium species, such as F. culmorum and F. proliferatum, were detected at the last sampling time point (T3), probably due to the changes that occurred in microbial community composition following phosphine fumigation. Notably, the presence of mycotoxigenic Fusarium isolates in the stored wheat grains after fumigation was strongly associated with the occurrence of DON in T3 samples, suggesting that phosphine induced a shift in the microbial composition toward more toxigenic strains. Shifts in Fusarium species composition on cereal grain due to changes in climate and cultivation practices have been previously reported (Garrett et al., 2011). Magan et al. (2011) investigated the effect of climate change on mycotoxin-producing fungi and indicated that shifts toward Fusarium pathogens and DON contamination in cereals may occur due to changes in management practices. For example, the effect of fungicides on Fusarium infections could be different depending on the antifungal compound, the time of treatment, and the composition of the microbial flora in cereals (Pirgozliev et al., 2003). Henriksen and Elen (2005) reported that different fungicides (such as propiconazole and cyprodinil) significantly increased the infection level of DON-producing F. avenaceum, F. tricinctum, and F. culmorum in grains.
Conclusion
In summary, our study demonstrated that phosphine fumigation had a significant impact on microbial community composition in stored wheat grain. The shifts in bacterial and yeast populations, coincident with the fumigant application may have also led to changes in the functional diversity of those communities. Fumigation did not change filamentous fungal abundance in stored grains at T3; however, phosphine treatment apparently altered the community composition of molds, relative to T1 and T2. Previous studies have extensively investigated the effect of different fumigants on soil bacterial communities (Engelen et al., 1998; Ibekwe et al., 2001; van Agtmaal et al., 2015; Zhang et al., 2017). To our knowledge, however, there are no published studies describing the impact of pesticide treatment on the microbiome of stored wheat grains using combined HTS analysis and culture assays. The development of next generation sequencing (NGS) technologies have provided a deeper understanding of the microbiome on plant material, as well human and animal subjects, and a host of different physical environments. Since many of the microorganisms identified by NGS technology are not culturable, it would be very difficult to detect alterations in microbial community diversity and abundance after phosphine treatment using only classical culturing and molecular identification approaches. Findings in the current study are consistent with previous studies, where NGS technology was able to identify changes in the microbiome that were not detectable using classical methodologies (Sylla et al., 2013; Schmidt et al., 2014). A better understanding of the interactions that occur among the different members of the microbial community of stored wheat grains at specific time points (e.g., before and/or after fumigant application) can assist in the prediction of fungal disease incidence and mycotoxin production, as well as in the development of novel approaches for controlling mycotoxin contamination in grains and managing crop diseases in general.
Data Availability
The raw sequence files supporting the findings of this article are available in the NCBI Sequence Read Archive (SRA) under the BioProject ID PRJNA503713.
Author Contributions
MKS, AA, SD, and ES conceived and designed the experiments, analyzed the data, and wrote the manuscript. MKS, AA, MB, VZ, and MW performed the experiments. MW provided critical comments on the study and edited the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by a grant (No. 20–06–0055) from the Chief Scientist of the Israeli Ministry of Agriculture and Rural Development.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Moshe Kostyukovsky for his assistance during the wheat grains samples collection from storage facilities.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01098/full#supplementary-material
Footnotes
References
Abarenkov, K., Henrik Nilsson, R., Larsson, K.-H., Alexander, I. J., Eberhardt, U., Erland, S., et al. (2010). The UNITE database for molecular identification of fungi - recent updates and future perspectives. New Phytol. 186, 281–285. doi: 10.1111/j.1469-8137.2009.03160.x
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Bell, C. (2000). Fumigation in the 21st century. Crop Prot. 19, 563–569. doi: 10.1016/S0261-2194(00)00073-9
Berners-Price, S. J., and Sadler, P. J. (1988). Phosphines and metal phosphine complexes: relationship of chemistry to anticancer and other biological activity. Bioinorg. Chem. 70, 27–102. doi: 10.1007/3-540-50130-4_2
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bond, E. J. (1989). Manual of Fumigation for Insect Control. FAO Plant Protection Paper no. 54. Rome: Food and Agriculture Organization of the United Nations, 432.
Brader, G., Compant, S., Vescio, K., Mitter, B., Trognitz, F., Ma, L.-J., et al. (2017). Ecology and genomic insights into plant-pathogenic and plant-nonpathogenic endophytes. Annu. Rev. Phytopathol. 55, 61–83. doi: 10.1146/annurev-phyto-080516-0-35641
Bray, J. R., and Curtis, J. T. (1957). An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27, 325–349. doi: 10.2307/1942268
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Castro, M. F. P. M., De Pacheco, I. A., and Taniwaki, M. H. (1992). Effects of phosphine on aflatoxin production in peanuts stored with a high content. Methyl Bromide Technical Options Workshop, Washington.
Castro, M. F. P. P. M., de Pacheco, I. A., Soares, L. M. V., Furlani, R. P. Z., Paula, D. C., de, et al. (1996). Warehouse control of Aspergillus flavus Link and A. Parasiticus speare on peanuts (Arachis hypogaea) by phosphine fumigation and its effect on aflatoxin production. J. Food Prot. 59, 407–411. doi: 10.4315/0362-028X-59.4.407
Castro, P. P. M. F. M., Pacheco, I. A., Soares, L. M. V., and Furlani, R. P. (1995). Phosphine fumigation of stored peanuts for Aspergillus flavus Link/ Aspergillus parasiticus speare and aflatoxins control. Rev. Microbiol. 26,296–301.
Castro, M. F. P. M., De Leitao, M. F. F., Do Vale, J. O., Bragnolo, N., Anichiareo, E. S., and Mills, K. A. (2000). “Effects of phosphine in the development of Aspergillus flavus aflatoxin production in maize grains stored at different moisture contents,” in Proceedings of the International Working Conference on Controlled Atmosphere and Fumigation in Stored Products, Vol. 29, eds E. J. Donahaye, S. Navarro, and J. G. Leesch (Fresno, CA: Executive Printing Services), 179–191.
Cherif-Silini, H., Silini, A., Yahiaoui, B., Ouzari, I., and Boudabous, A. (2016). Phylogenetic and plant-growth-promoting characteristics of Bacillus isolated from the wheat rhizosphere. Ann. Microbiol. 66, 1087–1097. doi: 10.1007/s13213-016-1194-6
Comby, M., Lacoste, S., Baillieul, F., Profizi, C., and Dupont, J. (2016). Spatial and temporal variation of cultivable communities of co-occurring endophytes and pathogens in wheat. Front. Microbiol. 7:403. doi: 10.3389/fmicb.2016.00403
De, S., Kaur, G., Roy, A., Dogra, G., Kaushik, R., Yadav, P., et al. (2010). A simple method for the efficient isolation of genomic DNA from Lactobacilli isolated from traditional indian fermented milk (dahi). Indian J. Microbiol. 50, 412–418. doi: 10.1007/s12088-011-0079-4
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi: 10.1128/AEM.03006-05
Díaz Herrera, S., Grossi, C., Zawoznik, M., and Groppa, M. D. (2016). Wheat seeds harbour bacterial endophytes with potential as plant growth promoters and biocontrol agents of Fusarium graminearum. Microbiol. Res. 186, 37–43. doi: 10.1016/J.MICRES.2016.03.002
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Engelen, B., Meinken, K., von Wintzingerode, F., Heuer, H., Malkomes, H. P., and Backhaus, H. (1998). Monitoring impact of a pesticide treatment on bacterial soil communities by metabolic and genetic fingerprinting in addition to conventional testing procedures. Appl. Environ. Microbiol. 64, 2814–2821.
Foster, T. L., Winans, L., Helms, S. J., and Helms, S. J. (1978). Anaerobic utilization of phosphite and hypophosphite by bacillus sp. Appl. Environ. Microbiol. 35, 937–944.
Garrett, K. A., Forbes, G. A., Savary, S., Skelsey, P., Sparks, A. H., Valdivia, C., et al. (2011). Complexity in climate-change impacts: an analytical framework for effects mediated by plant disease. Plant Pathol. 60, 15–30. doi: 10.1111/j.1365-3059.2010.02409.x
Gdanetz, K., and Trail, F. (2017). The wheat microbiome under four management strategies, and potential for endophytes in disease protection. Phytobiomes 1, 158–168. doi: 10.1094/PBIOMES-05-17-0023-R
Hao, T., and Chen, S. (2017). Colonization of wheat, maize and cucumber by Paenibacillus polymyxa WLY78. PLoS One 12:e0169980. doi: 10.1371/journal.pone.0169980
Hartmann, M., Frey, B., Mayer, J., Mäder, P., and Widmer, F. (2015). Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 9, 1177–1194. doi: 10.1038/ismej.2014.210
Henriksen, B., and Elen, O. (2005). Natural Fusarium grain infection level in wheat, barley and oat after early application of fungicides and herbicides. J. Phytopathol. 153, 214–220. doi: 10.1111/j.1439-0434.2005.00955.x
Hocking, A. D., and Banks, H. J. (1991). Effects of phosphine fumigation on survival and growth of storage fungi in wheat. J. Stored Prod. Res. 27, 115–120. doi: 10.1016/0022-474X(91)90021-4
Ibekwe, A. M., Papiernik, S. K., Gan, J., Yates, S. R., Yang, C. H., and Crowley, D. E. (2001). Impact of fumigants on soil microbial communities. Appl. Environ. Microbiol. 67, 3245–3257. doi: 10.1128/AEM.67.7.3245-3257.2001
Igual, J., Valverde, A., Cervantes, E., and Velazquez, E. (2001). Phosphate-solubilizing bacteria as inoculants 979 for agriculture: use updated molecular techniques in their study. Agronomie 21, 561–568. doi: 10.1051/agro:2001145
Kang, S.-M., Radhakrishnan, R., You, Y.-H., Joo, G.-J., Lee, I.-J., Lee, K.-E., et al. (2014). Phosphate solubilizing Bacillus megaterium mj1212 regulates endogenous plant carbohydrates and amino acids contents to promote mustard plant growth. Indian J. Microbiol. 54, 427–433. doi: 10.1007/s12088-014-0476-6
Karlsson, I., Friberg, H., Kolseth, A.-K., Steinberg, C., and Persson, P. (2017). Organic farming increases richness of fungal taxa in the wheat phyllosphere. Mol. Ecol. 26, 3424–3436. doi: 10.1111/mec.14132
Karlsson, I., Friberg, H., Steinberg, C., and Persson, P. (2014). Fungicide effects on fungal community composition in the wheat phyllosphere. PLoS One 9:e111786. doi: 10.1371/journal.pone.0111786
Kruskal, W. H., and Wallis, W. A. (1952). Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 47, 583–621. doi: 10.1080/01621459.1952.10483441
Links, M. G., Demeke, T., Gräfenhan, T., Hill, J. E., Hemmingsen, S. M., and Dumonceaux, T. J. (2014). Simultaneous profiling of seed-associated bacteria and fungi reveals antagonistic interactions between microorganisms within a shared epiphytic microbiome on Triticum and Brassica seeds. New Phytol. 202, 542–553. doi: 10.1111/nph.12693
Lozupone, C., and Knight, R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005
Magan, N., Aldred, D., Hope, R., and Mitchell, D. (2010). Grain and Grapes: effects on growth, deoxynivalenol and ochratoxin production by Fusarium culmorum and Aspergillus carbonarius. Toxins 2, 353–366. doi: 10.3390/toxins2030353
Magan, N., Medina, A., and Aldred, D. (2011). Possible climate-change effects on mycotoxin contamination of food crops pre- and postharvest. Plant Pathol. 60, 150–163. doi: 10.1111/j.1365-3059.2010.02412.x
Mahoney, A. K., Yin, C., and Hulbert, S. H. (2017). Community structure, species variation, and potential functions of rhizosphere-associated bacteria of different winter wheat (Triticum aestivum) cultivars. Front. Plant Sci. 8:132. doi: 10.3389/fpls.2017.00132
Masella, A. P., Bartram, A. K., Truszkowski, J. M., Brown, D. G., and Neufeld, J. D. (2012). PANDAseq: paired-end assembler for Illumina sequences. BMC Bioinformatics 13:31. doi: 10.1186/1471-2105-13-31
McDonald, D., Price, M. N., Goodrich, J., Nawrocki, E. P., DeSantis, T. Z., Probst, A., et al. (2012). An improved greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618. doi: 10.1038/ismej.2011.139
Metcalf, W. W., and Wolfe, R. S. (1998). Molecular genetic analysis of phosphite and hypophosphite oxidation by Pseudomonas stutzeri WM88. J. Bacteriol. 180, 5547–5558.
Mousa, W. K., Shearer, C., Limay-Rios, V., Ettinger, C. L., Eisen, J. A., and Raizada, M. N. (2016). Root-hair endophyte stacking in finger millet creates a physicochemical barrier to trap the fungal pathogen Fusarium graminearum. Nat. Microbiol. 1:16167. doi: 10.1038/nmicrobiol.2016.167
Natarajan, T., and Bagyaraj, D. J. (1984). Fumigation effect on micro flora and viability of black gram and field bean seeds. Pesticides 18, 40–42.
Nath, N. S., Bhattacharya, I., Tuck, A. G., Schlipalius, D. I., and Ebert, P. R. (2011). Mechanisms of phosphine toxicity. J. Toxicol. 2011:494168. doi: 10.1155/2011/494168
Ofek, M., Voronov-Goldman, M., Hadar, Y., and Minz, D. (2014). Host signature effect on plant root-associated microbiomes revealed through analyses of resident vs. Active communities. Environ. Microbiol. 16, 2157–2167. doi: 10.1111/1462-2920.12228
Pan, D., Mionetto, A., Tiscornia, S., and Bettucci, L. (2015). Endophytic bacteria from wheat grain as biocontrol agents of Fusarium graminearum and deoxynivalenol production in wheat. Mycotoxin Res. 31, 137–143. doi: 10.1007/s12550-015-0224-8
Park, K.-H., Lee, C.-Y., and Son, H.-J. (2009). Mechanism of insoluble phosphate solubilization by Pseudomonas fluorescens RAF15 isolated from ginseng rhizosphere and its plant growth-promoting activities. Lett. Appl. Microbiol. 49, 222–228. doi: 10.1111/j.1472-765X.2009.02642.x
Paulson, J. N., Stine, O. C., Bravo, H. C., and Pop, M. (2013). Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 10, 1200–1202. doi: 10.1038/nmeth.2658
Pirgozliev, S. R., Edwards, S. G., Hare, M. C., and Jenkinson, P. (2003). Strategies for the control of Fusarium head blight in cereals. Eur. J. Plant Pathol. 109, 731–742. doi: 10.1023/A:1026034509247
Placinta, C. M., D’Mello, C. P. F., and MacDonald, A. M. C. (1999). A review of worldwide contamination of cereal grains and animal feed with Fusarium mycotoxins. Anim. Feed Sci. Technol. 78, 21–37. doi: 10.1016/s0377-8401(98)00278-8
Preston, G. M. (2004). Plant perceptions of plant growth-promoting Pseudomonas. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 359, 907–918. doi: 10.1098/rstb.2003.1384
Price, N. R. (1980). Some aspects of the inhibition of cytochrome c oxidase by phosphine in susceptible and resistant strains of Rhyzopertha dominicia. Insect Biochem. 10, 147–150. doi: 10.1016/0020-1790(80)90065-7
Raghunathan, A. N., Muthu, M., and Majumder, S. K. (1969). Control of internal fungi of sorghum by fumigation. J. Stored Prod. Res. 5, 389–392. doi: 10.1016/0022-474X(69)90012-5
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
Sadhasivam, S., Britzi, M., Zakin, V., Kostyukovsky, M., Trostanetsky, A., Quinn, E., et al. (2017). Rapid detection and identification of mycotoxigenic fungi and mycotoxins in stored wheat grain. Toxins 9:302. doi: 10.3390/toxins9100302
Schmidt, M., Zannini, E., and Arendt, E. (2018). Recent advances in physical post-harvest treatments for shelf-life extension of cereal crops. Foods 7:45. doi: 10.3390/foods7040045
Schmidt, R., Köberl, M., Mostafa, A., Ramadan, E. M., Monschein, M., Jensen, K. B., et al. (2014). Effects of bacterial inoculants on the indigenous microbiome and secondary metabolites of chamomile plants. Front. Microbiol. 5:64. doi: 10.3389/fmicb.2014.00064
Shiferaw, B., Smale, M., Braun, H.-J., Duveiller, E., Reynolds, M., and Muricho, G. (2013). Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Secur. 5, 291–317. doi: 10.1007/s12571-013-0263-y
Sinha, R. N., Berck, B., and Wallace, H. A. H. (1967). Effect of phosphine on mites, insects, and microorganisms. J. Econ. Entomol. 60, 125–132. doi: 10.1093/jee/60.1.125
Story, S. P., Kline, E. L., Hughes, T. A., Riley, M. B., and Hayasaka, S. S. (2004). Degradation of aromatic hydrocarbons by Sphingomonas paucimobilis Strain EPA505. Arch. Environ. Contam. Toxicol. 47, 168–176. doi: 10.1007/s00244-004-3069-2
Sylla, J., Alsanius, B. W., Krüger, E., Reineke, A., Strohmeier, S., and Wohanka, W. (2013). Leaf microbiota of strawberries as affected by biological control agents. Phytopathology 103, 1001–1011. doi: 10.1094/PHYTO-01-13-0014-R
Tao, A., Pang, F., Huang, S., Yu, G., Li, B., and Wang, T. (2014). Characterisation of endophytic Bacillus thuringiensis strains isolated from wheat plants as biocontrol agents against wheat flag smut. Biocontrol Sci. Technol. 24, 901–924. doi: 10.1080/09583157.2014.904502
van Agtmaal, M., van Os, G. J., Hol, W. H. G., Hundscheid, M. P. J., Runia, W. T., Hordijk, C. A., et al. (2015). Legacy effects of anaerobic soil disinfestation on soil bacterial community composition and production of pathogen-suppressing volatiles. Front. Microbiol. 6:701. doi: 10.3389/fmicb.2015.00701
Yuan, Q.-S., Yang, P., Wu, A.-B., Zuo, D.-Y., He, W.-J., Guo, M.-W., et al. (2018). Variation in the microbiome, trichothecenes, and aflatoxins in stored wheat grains in Wuhan, China. Toxins 10:171. doi: 10.3390/toxins10050171
Zhang, J., Kobert, K., Flouri, T., and Stamatakis, A. (2014). PEAR: a fast and accurate illumina paired-end reAd mergeR. Bioinformatics 30, 614–620. doi: 10.1093/bioinformatics/btt593
Keywords: stored wheat grain, microbiome analysis, phosphine fumigation, mycotoxigenic fungi, mycotoxins
Citation: Solanki MK, Abdelfattah A, Britzi M, Zakin V, Wisniewski M, Droby S and Sionov E (2019) Shifts in the Composition of the Microbiota of Stored Wheat Grains in Response to Fumigation. Front. Microbiol. 10:1098. doi: 10.3389/fmicb.2019.01098
Received: 12 March 2019; Accepted: 30 April 2019;
Published: 17 May 2019.
Edited by:
Jia Liu, Chongqing University of Arts and Sciences, ChinaReviewed by:
Simona Marianna Sanzani, University of Bari Aldo Moro, ItalyXuehong Wu, China Agricultural University, China
Copyright © 2019 Solanki, Abdelfattah, Britzi, Zakin, Wisniewski, Droby and Sionov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Samir Droby, c2FtaXJkQHZvbGNhbmkuYWdyaS5nb3YuaWw= Edward Sionov, ZWR3YXJkc2lvQHZvbGNhbmkuYWdyaS5nb3YuaWw=
†These authors have contributed equally to this work
 Manoj Kumar Solanki
Manoj Kumar Solanki Ahmed Abdelfattah
Ahmed Abdelfattah Malka Britzi4
Malka Britzi4 Michael Wisniewski
Michael Wisniewski Samir Droby
Samir Droby Edward Sionov
Edward Sionov