
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 08 May 2019
Sec. Food Microbiology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00981
This article is part of the Research Topic Food Safety and Foodborne Pathogen – A Global Perspective on the Diversity, Combating Multidrug Resistance and Management View all 39 articles
Listeria monocytogenes is an important foodborne pathogen with a significant impact on public health worldwide. A great number of outbreaks caused by L. monocytogenes has been reported, especially in the United States, and European countries. However, listeriosis has not yet been included in notifiable disease in China, and thus information on this infection has been scarce among the Chinese population. In this study, we described a 3-year surveillance of listeriosis in Beijing, China. Fifty-six L. monocytogenes strains isolated from 49 clinical infectious cases (27 pregnancy-associated infections and 22 non-pregnancy-associated infections) were analyzed by serotyping, pulsed field gel electrophoresis (PFGE), multilocus sequence typing (MLST), and antimicrobial susceptibility testing between 2014 and 2016 in Beijing. The predominant serogroups were 1/2a,3a and 1/2b,3b,7 which accounted for 92% of the overall isolates. Four strains were serogroup 4b,4d,4e, isolated from patients with pregnancy-associated infections. Based on PFGE, these isolates were divided into 32 pulsotypes (PTs) and 3 clusters associated with serogroups. Ten PTs were represented by more than one isolate with PT09 containing the most number of isolates. MLST differentiated the isolates into 18 STs, without new ST designated. The three most common STs were ST8 (18.4%), ST5 (16.3%), and ST87 (12.2%), accounting for 46.9% of the isolates. STs prevalent in other parts of the world were also present in China such as ST1, ST2, ST5, ST8, and ST9 which caused maternal fetal infections or outbreaks. However, the STs and serogroup distribution of clinical L. monocytogenes in Beijing, China was different from those in other countries. Strains of ST1 and ST2 were isolated from patients with pregnancy-associated infection, whereas none of ST155 isolates caused pregnancy-associated cases. Surveillance of molecular characterization will provide important information for prevention of listeriosis. This study also enhances our understanding of genetic diversity of clinical L. monocytogenes in China.
Listeria monocytogenes is an important foodborne pathogen with a significant impact on public health worldwide (Schlech, 2000; de Noordhout et al., 2014). It has the potential to cause human diseases ranging from self-limited gastroenteritis and spontaneous abortion in pregnant women to severe invasive infections (sepsis or meningitis) in immuno-compromised patients or older patients (Okutani et al., 2004; Guevara et al., 2009; McCollum et al., 2013; Wang et al., 2013).
Although L. monocytogenes is an uncommon human pathogen, it has a disproportionate share of the food borne disease burden. A previous study reported 1600 human cases of listeriosis annually in the United States, of which 260 resulted in death (Crim et al., 2014). In the European Union, a total of 1763 confirmed human cases of listeriosis (notification rate of 0.44 cases per 100,000 population) were reported with a fatality rate of 15.6% in 2013 (European Food Safety Authority [EFSA], 2015). In China, 253 invasive listeriosis cases were reported in 19 provinces from 2011 to 2016, with a fatality rate of 25.7% (Li et al., 2018). More importantly, the incidence is still increasing worldwide, despite antibiotic treatments (Goulet et al., 2008). A large listeriosis outbreak occurred in Canada in 1981 and provided the first evidence for transmission of listeriosis by foodborne L. monocytogenes (Schlech et al., 1983; Evans et al., 1985). Since then, a series of outbreaks caused by L. monocytogenes have been reported, especially in the United States (Fleming et al., 1985; Linnan et al., 1988; Dalton et al., 1997; Gottlieb et al., 2006; Centers for Disease Control and Prevention, 2008; Chen et al., 2017) and European countries (Bula et al., 1995; Ericsson et al., 1997; Goulet et al., 1998; Aureli et al., 2000; Lyytikainen et al., 2000; Althaus et al., 2017; Amato et al., 2017; Dahl et al., 2017; Kleta et al., 2017). However, listeriosis has not yet been regulated as a notifiable disease in China and therefore information on this infection has been largely scarce among the Chinese population.
Listeria contains multiple species. It has been subtyped using different methods, including serotyping, pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST), multi-virulence-locus sequence typing (MVLST), and restriction fragment length polymorphism (RFLP). L. monocytogenes can be divided into 13 serotypes, of which three serotypes (serotype 1/2a, 1/2b, and 4b) are believed to cause the majority of clinical cases (Gilot et al., 1996). In all of these methods, PFGE provides higher discrimination power than serotyping, making it an important tool in source tracking and outbreak investigation (Graves and Swaminathan, 2001; Centers for Disease Control and Prevention, 2008; Chen et al., 2017; Dahl et al., 2017; Kleta et al., 2017). MLST, based on the analysis of seven housekeeping genes, has been proved as a powerful tool in molecular epidemiological studies and population structure studies of L. monocytogenes (Salcedo et al., 2003; Jadhav et al., 2012). However, only a few studies have approached the molecular characterization of clinical L. monocytogenes in China (Lv et al., 2014; Huang et al., 2015; Wang Y. et al., 2015; Zhang et al., 2016). These studies suggested that listeriosis in China was caused by heterogeneous strains. MLST types (STs) prevalence in other parts of the world were also found in China (Lv et al., 2014; Huang et al., 2015; Wang Y. et al., 2015; Zhang et al., 2016). The objective of the present study was to determine the epidemiological characteristics of listeriosis cases, as well as the characteristics of clinical L. monocytogenes isolates in Beijing, China.
The isolates analyzed in this study were collected from the Survey Project of Human Listeriosis in Beijing. This study was carried out in accordance with the recommendations of Manual of Foodborne Disease Surveillance, China issued by the National Center for Food safety Risk Assessment. The protocol was approved by the ethics committee of Beijing Center for Disease Prevention and Control (Beijing CDC). In this project, 12, 12, and 21 hospitals were covered in 2014, 2015, and 2016, respectively. All the suspected clinical cases of listeriosis were included in the survey. Samples were collected and used to isolate L. monocytogenes. We defined invasive listeriosis as isolation of L. monocytogenes strains from a normally sterile site or from products of conception (Li et al., 2018). All the L. monocytogenes isolates identified by clinical microbiology laboratories were sent to the lab in Beijing CDC.
A total of 56 isolates were isolated from 49 human patients who had a severe illness with serious suspicion of L. monocytogenes infection between 2014 and 2016. All the isolates were firstly identified using VITEK 2-compact system (bioMérieux, Lyons, France) or matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) mass spectrometry (Bruker, Leipzig, Germany); and were further checked by PCR targeting hly fragments specific to L. monocytogenes (Xu et al., 2009).
All the strains were serotyped using multiplex PCR, which was based on the amplification of the following target genes: prs, lmo0737, lmo1118, ORF2110, and ORF2819 described by Doumith et al. (2004).
Antimicrobial susceptibility testing of the L. monocytogenes isolates was performed using broth dilution method. The minimum inhibitory concentrations (MICs) of following 13 antimicrobials were tested: ampicillin (AMP), oxacillin (OXA), vancomycin (VAN), clindamycin (CLI), tetracycline (TET), daptomycin (DAP), erythromycin (ERY), chloramphenicol (CHL), ciprofloxacin (CIP), trimethoprim-sulfamethoxazole (SXT), gentamicin (GEN), penicillin (PEN), and cefoxitin (FOX) (Xingbai, Shanghai, China). Since resistance criteria for AMP, PEN, and SXT of L. monocytogenes exists in the clinical and laboratory standard institute (CLSI) international guidelines, and resistance criteria for ERY exists in European Committee on Antimicrobial Susceptibility Testing (EUCAST) international guidelines, the MICs of AMP, PEN, SXT were interpreted using CLSI international guidelines, and the MIC of ERY was interpreted according to EUCAST international guidelines. No resistance criteria exists for the other 11 antimicrobials of L. monocytogenes, so susceptibility to other tested antimicrobials was interpreted by those recommended for Staphylococcus spp. ATCC29213 was used as the reference strain.
Pulsed-field gel electrophoresis of the strains was processed in accordance with the PulseNet International protocol1. Slices of L. monocytogenes agarose plugs were digested using 50 U of AscI and ApaI (Takara, Dalian, China) per slice for 3 h at 37°C; electrophoresis was performed using a CHEF-DRIII system (Bio-Rad Laboratories, Hercules, CA, United States); electrophoresis was conducted with a switch time of 4–40 s for 19 h; and images were captured using a Gel Doc 2000 system (Bio-Rad) then converted to TIFF files. Salmonella enterica serovar Braenderup strain H9812 restricted with XbaI was used for molecular weight determinations in all PFGE gels. The TIFF files were analyzed using BioNumerics software (version 7.6 Applied Maths, Kortrijk, Belgium). Clustering was performed using the unweighted pair group method with arithmetic mean (UPGMA).
Multilocus sequence typing was performed on all the 56 isolates by amplification and sequencing of internal fragments of seven housekeeping genes (abcZ, bglA, cat, dapE, dat, ldh, and lhkA) (Salcedo et al., 2003). Sequencing was performed on an ABI 3770 automatic sequencer. The alleles and sequences types (STs) were determined by comparison with the allelic profiles for L. monocytogenes in MLST database2.
BioNumerics software was used to create a cluster tree and conduct minimum spanning trees (MST) based on the allelic profiles. In MST, a clonal complex (CC) was formed by STs with six of seven MLST alleles in common and at least two STs; the founder ST was defined as the ST with the highest number of single-locus variants (SLVs); single genotypes that did not correspond to any clone groups were defined as singletons. Types were represented by circles; size of a circle indicated the number of strains of this particular type. Lineages of isolates were assigned as Wiedmann et al. (1997) described.
The origins of the strains were summarized in Table 1. Fifty-six strains were isolated from the 49 cases. Among them, 27 were pregnancy-associated infections, in which all mothers were cured; ten neonates were survived, whereas thirteen fetuses died in the womb or after birth. No data were available for the four fetuses. Among the other 22 non-pregnancy-associated patients, 11 (50%) were males. The median age of patients with non-pregnancy-associated infections was 42 years, five patients were >60 years, and five patients were ≤3 years.
There were five groups of isolates from five patients (Figure 1). The isolates from the same patient had the same serogroup, antimicrobial susceptibilities, PFGE type, and ST. Therefore, only one isolate from each patient was used for the subsequent analysis.
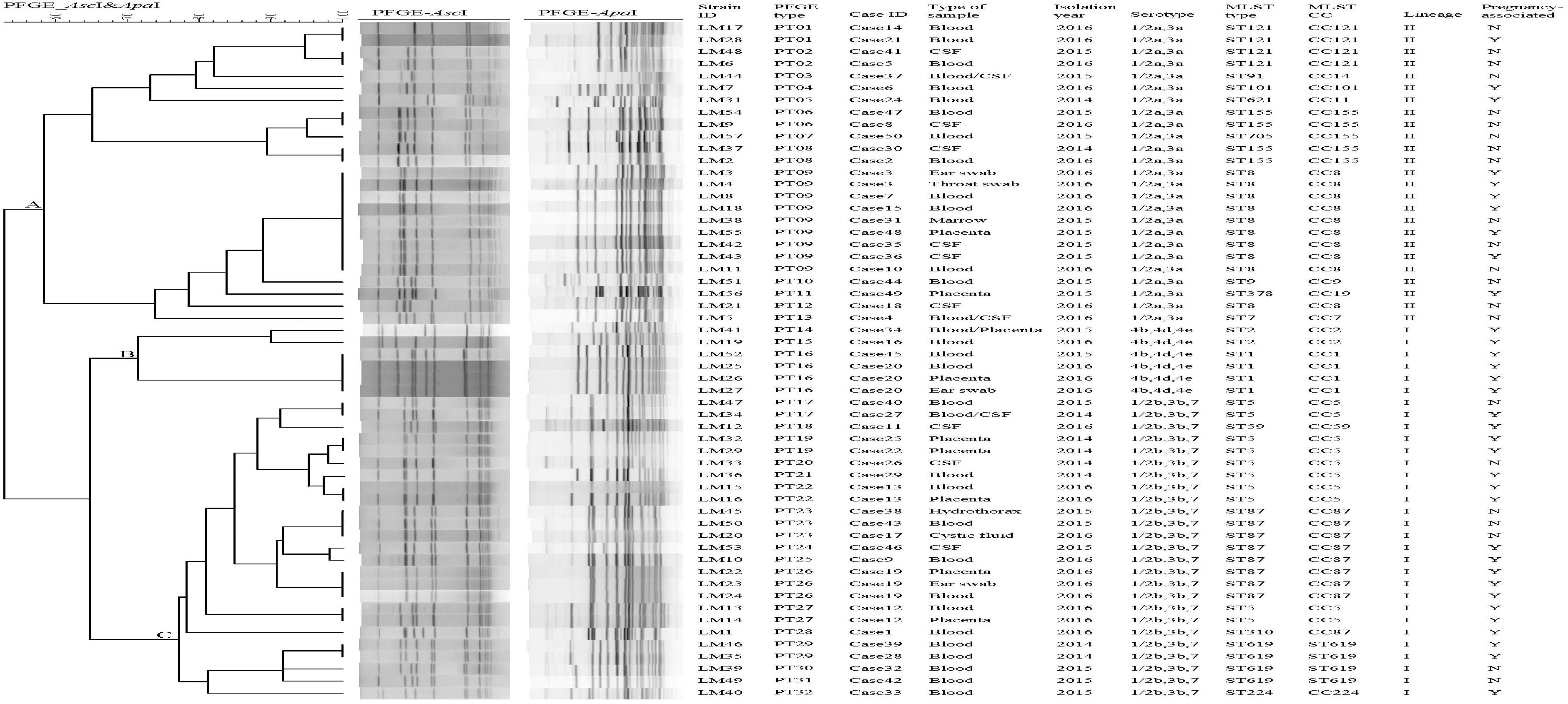
Figure 1. Relationships of the isolates based on PFGE. Forty-nine clinical Listeria monocytogenes strains were analyzed by PFGE using AscI and ApaI. The corresponding data, including the name of the strain (Strain ID), the name of the case (Case ID), PFGE types, the type of sample, isolation date, serotype, MLST type (ST), MLST clonal complexes (CC), lineage, and pregnancy-associated were shown alongside the dendrogram to the right.
Lineage was determined based on the MLST data. We found that 25 strains belonged to lineage I and 24 strains to lineage II. Almost half of the strains belonged to serogroup 1/2a,3a (n = 24,49%) followed by serogroup 1/2b,3b,7 (n = 21,43%), serogroup 4b,4d,4e (n = 4, 8%). Strains of serogroup 1/2b,3b,7 and serogroup 4b,4d,4e belonged to lineage I, whereas strains of serogroup 1/2a,3a belonged to lineage II. All 4 strains of serogroup 4b,4d,4e were isolated from patients with pregnancy-associated infections. More pregnancy-associated cases were caused by lineage I than lineage II strains (72 vs. 37.5%, respectively).
Pulsed-field gel electrophoresis analysis of the comprised AscI and ApaI divided 49 isolates into 32 pulsotypes (PT01-P32) (Figure 1). PT09 accounted for 16.3% (8/49) of isolates, followed by PT23 (3 isolates, 6.12%). Eight PTs (25%) were represented by two isolates. Twenty-two PTs (44.9%) were represented by only one single isolate. A UPGMA dendrogram was constructed for the 32 PTs based on presence or absence of bands. PTs were divided into 3 clusters, respectively (cluster A, B, and C) (Figure 1), corresponding to the three serogroups identified.
Forty-nine isolates were divided into 18 STs, with no new ST designated. Lineage I included 8 STs of serogroups “1/2b,3b,7” and “4b,4d,4e”; lineage II included 10 STs of serogroups “1/2a,3a” (Figure 2). The most common STs were ST8 (9 isolates, 18.4%, “1/2a,3a”), ST5 (8 isolates, 16.3%, “1/2b,3b,7”), ST87 (6 isolates, 12.2%,“1/2b,3b,7”), followed by ST121 (4 isolates, 8.2%, “1/2a,3a”), ST155 (4 isolates, 8.2%, “1/2a,3a”), and ST619 (4 isolates, 8.2%, “1/2b,3b,7”). Other 12 STs contained one to two isolates, respectively (Figure 2). Among the 18 STs, 17 of them could be assigned clonal complexes and one (ST619) was a singleton based on querying the MLST database (Figure 2). Strains of ST1 and ST2 were isolated from patients with pregnancy-associated infection, whereas none of the ST155 isolates caused pregnancy-associated cases. Five of the nine ST8 strains were isolated from patients with pregnancy-associated infection among which three fetuses died. Six of the eight ST5 strains were isolated from patients with pregnancy-associated infection. Fetuses of pregnancy-associated cases caused by ST5 isolates died while those caused by ST87 isolates survived (except 1 lost to follow-up).
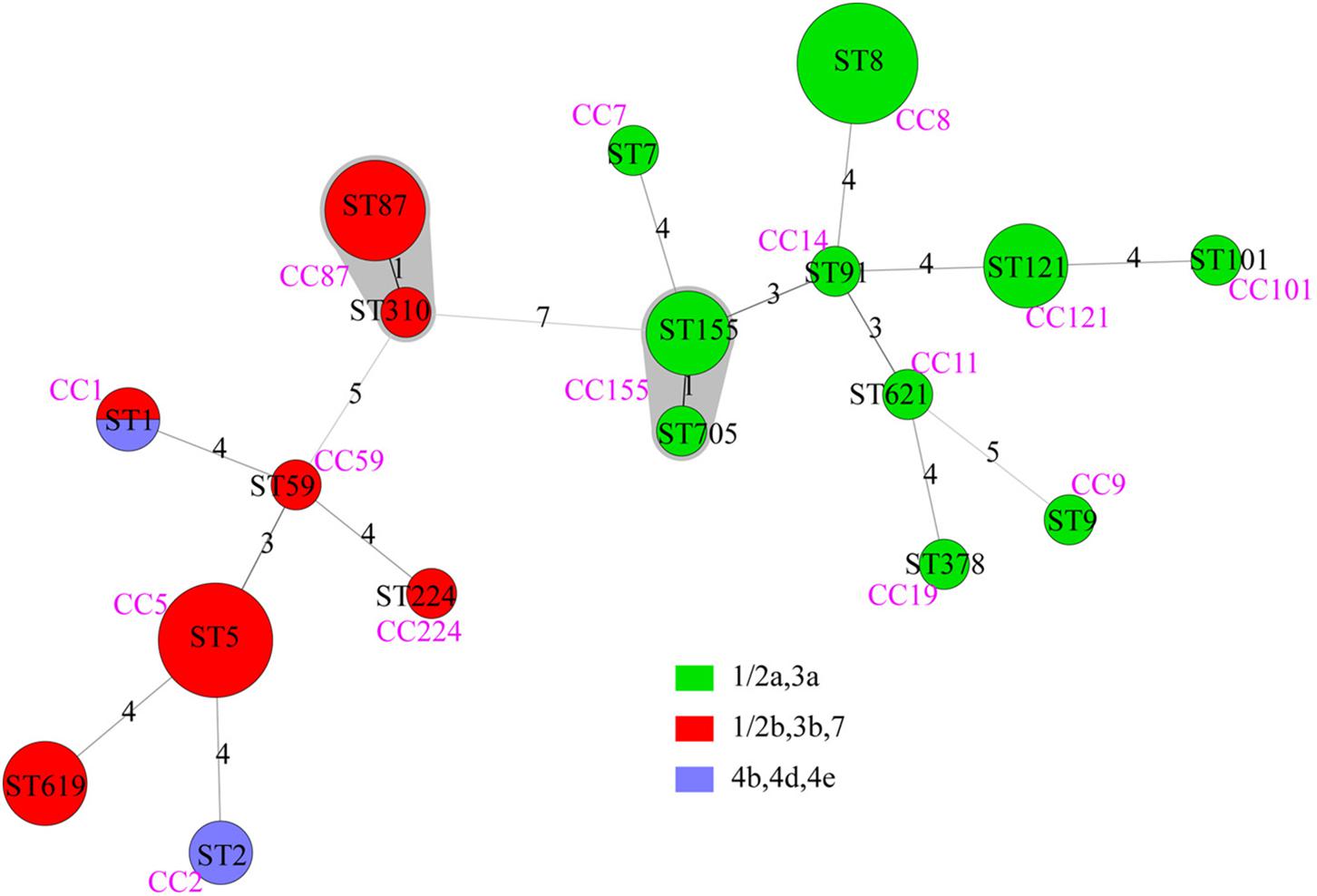
Figure 2. The minimum spanning tree of STs of 49 clinical L. monocytogenes strains isolated from Beijing, China. Each circle represents one sequence type. Gray zones surrounding groups of STs represent CC. The size of the circle is proportional to the number of the isolates and the color within the cycles represents the serotypes of the isolates. Links between circles are represented according to the number of allelic mismatches between STs.
The 49 isolates in this study were compared with 176 other isolates from cases of clinical illness in China reported in previous studies (Lv et al., 2014; Huang et al., 2015; Wang Y. et al., 2015; Zhang et al., 2016). These samples were isolated from Beijing (n = 14), Jiangsu (n = 3), Shandong (n = 4), Shanghai (n = 20), Shanxi (n = 3), and Taiwan (n = 132). Together, these 225 isolates were divided into 28 STs. Among them, 13 ST were found in different regions. There were no obvious regional characteristics of STs (Figure 3).
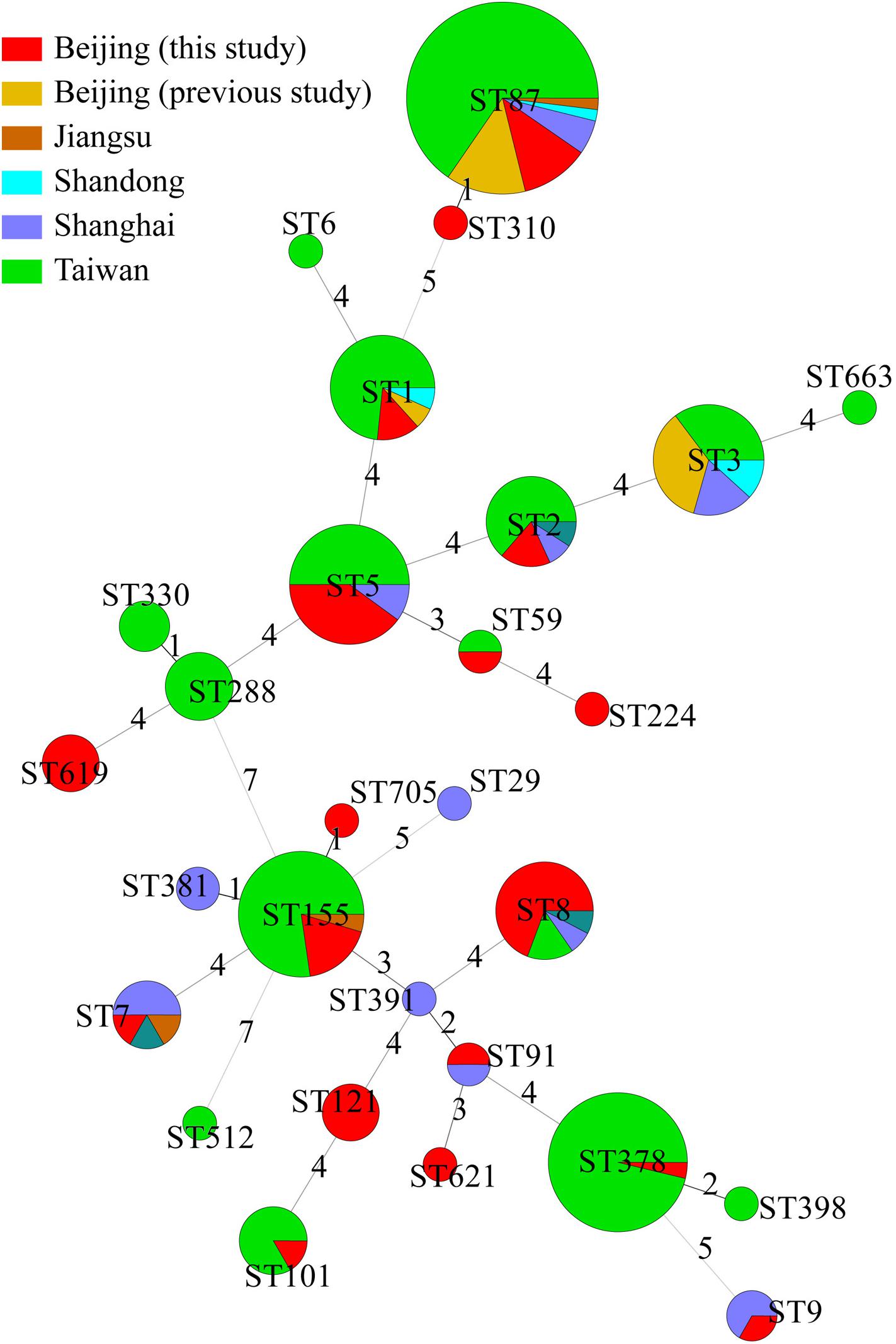
Figure 3. Genetic relationships of the 225 Chinese isolates. A minimum spanning tree was constructed based on STs of 49 isolates from this study and 176 isolates from other studies in China. Each circle represents a sequence type. The size of the circle is proportional to the number of the isolates, and the sources of the isolates were colored as shown in figure. Links between circles are represented according to the number of allelic mismatches between STs.
The 49 isolates were compared with clinical L. monocytogenes isolates from rest of the world (Figure 4). A total of 1094 human L. monocytogenes, screened out from L. monocytogenes MLST database see text footnote 2 in October 08, 2017, were included in the analysis. The 1143 human isolates were divided into 38 CCs and 87 singletons. The most globally prevalent CC in the database were CC1 (224), CC2 (133), CC3 (118), CC9 (65), CC4 (49), CC7 (46), CC8 (45), CC155 (41), and CC101 (32). All isolates obtained in this study, excepting ST619, were co-clustered with foreign isolates. The 15 CCs detected in this study were also found in at least two other countries (Supplementary Table S1).
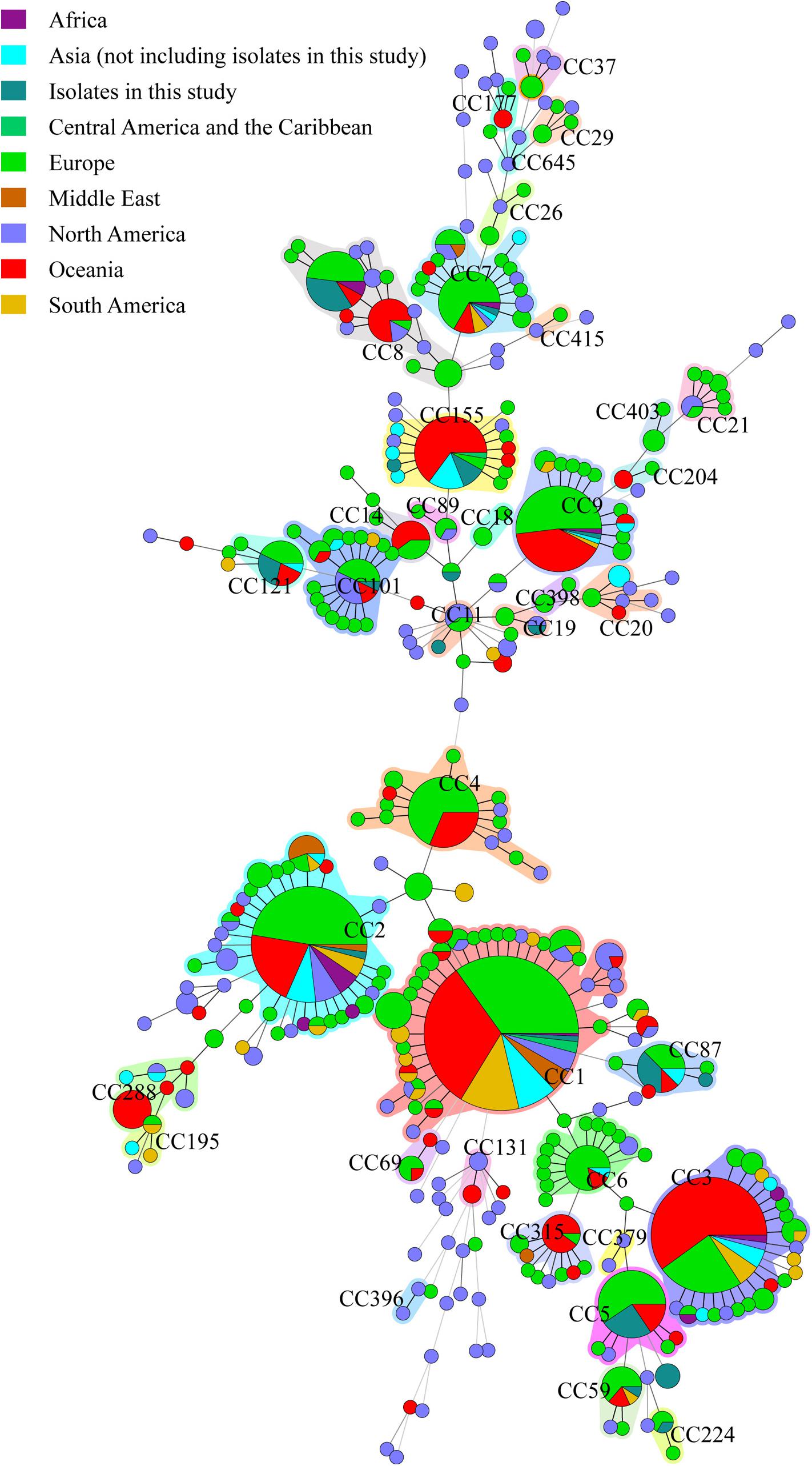
Figure 4. Genetic relationships of the 56 clinical isolates in Beijing and 1094 global isolates from L. monocytogenes MLST database. A minimum spanning tree was constructed based on CCs from this study and MLST database. The size of the circle is proportional to the number of the isolates, and the sources of the isolates were colored as shown in figure. The shadow zones in different color represent different clonal complexes.
Antimicrobial susceptibility testing was conducted for the 49 L.monocytogenes isolates. Details were listed in Table 2. The highest resistance rate was observed for FOX, which reached 100%, followed by DAP (93.9%), OXA (85.7%), and CIP (36.7%). In contrast, three other antimicrobials (TET, 6.1%; PEN, 4.1%; GEN, 2.0%) had resistance rates lower than 10.0%. All the isolates were susceptible to AMP, VAN, CLI, ERY, CHL, and SXT. Among all the 49 isolates, 79.6% (39/49) were co-resistant at least to OXA, DAP, and FOX; 28.6% (14/49) were co-resistant at least to OXA, DAP, CIP, and FOX. Seven isolates (ST5, ST8, ST7, ST155, and ST705) were sensitive to OXA, three isolates (ST5, ST378) were sensitive to DAP, while seventeen isolates (ST1, ST5, ST310, ST619, ST121, ST91, ST8, ST7, and ST155) were resistant to CIP, three isolates (ST155, ST705) were resistant to TET, two isolates (ST101, ST8) were resistant to PEN, one isolates (ST8) was resistant to GEN (Table 2).
In China, a few studies have reported the prevalence of L. monocytogenes in food (Chen et al., 2009; Wang et al., 2012; Wu et al., 2016). However, the descriptions of clinical L. monocytogenes are very limited. In this study, we described the characteristics of molecular subtyping and antimicrobial susceptibilities of clinical L. monocytogenes strains in Beijing, the capital city of China. The clinical strains analyzed in this study were isolated from a systematic investigation, which provided a unique opportunity to investigate the characterization of this important pathogen in China.
Our findings, for the first time, revealed the predominant serogroups and STs of clinical L. monocytogenes in Beijing, China. The results showed that there were some differences in clinical L. monocytogenes serogroups distribution between Beijing and other countries. Among the 49 isolates in this study, the predominant serogroups were 1/2a,3a (49%) and 1/2b,3b (45%). The serogroup compositions were in agreement with that of previous study, showing 28 L. monocytogenes isolates collected from patients from four cities/provinces in China (Wang Y. et al., 2015). The predominant serogroups were also 1/2a,3a and 1/2b,3b in strains isolated from ready-to-eat foods, raw foods and edible mushrooms in China (Chen et al., 2015, 2018a; Wu et al., 2015, 2016). While, the predominant serogroups of clinical L. monocytogenes strains in our study were different from those from the other countries such as United States, Australia, Brazil, Italy and Portugal (Mammina et al., 2009; Bueno et al., 2010; Centers for Disease Control and Prevention, 2014; Magalhaes et al., 2014; Almeida et al., 2017; Jennison et al., 2017). In United States, serotype 4b (57%) was the most commonly identified serotype of L monocytogenes, followed by serotype 1/2a (26%) and serotype 1/2b (13%) (Centers for Disease Control and Prevention, 2014). In Australia, serogroup 4b,4d,4e was the predominant group, accounting for 56.6% of clinical isolates (Jennison et al., 2017). In Italy, 1/2a and 4b were the predominant serotypes, representing 46.3 and 42.6% of human isolates from sporadic cases (Mammina et al., 2009).
Previous molecular epidemiological studies have detected unreported human outbreaks of listeriosis, for example, Ariza-Miguel et al. (2015) identified an epidemiological connection among strains via analysis of the genomic relationships among isolates through PFGE and MLST subtyping. In our study, ten PFGE clusters possessed strains that were isolated from 2 or more different cases. The isolates with same PFGE patterns had no obvious link as the isolates were obtained from different time and places, with no evidence of epidemiological association. It should be noted that isolates from the case38 and case43 patients, which were identified in the same hospital for 6 days apart, shared an indistinguishable PT, suggesting a possible related source. On the other hand, five groups of isolates from same patients in this study had the same serogroups, molecular subtypes and antimicrobial susceptibility profiles, indicating dissemination of L. monocytogenes in body. In case21, we isolated a L. monocytogenes strain from a beef sandwich in the market, where the pregnant patient always bought foods. This isolate had the same PFGE pattern with the strain LM28, indicating that beef sandwich may be the source of infection (Wang and Chen, 2016). In case13, the PFGE pattern of strains LM15, and LM16 were consistent with a strain isolated from the patient’s home environment swab, indicating possibility of cross contamination (unpublished data).
Most of STs identified in this study were largely distributed across world countries and regions, showing that “everything is everywhere” paradigm of L. monocytogenes clones (Chenal-Francisque et al., 2011). Many STs identified in this study have been associated with outbreaks in other countries, such as ST1 caused outbreaks in France in 1989 and in Sweden in 1995, ST2 caused an outbreak in Italy in 1997, and ST5 strains caused outbreaks in Canada in 1996 as well as in United States in 2011 (Ericsson et al., 1997; Aureli et al., 2000; Lomonaco et al., 2013). In addition, CC3, CC4, CC8, CC88, CC87, CC398, and CC403 were associated with outbreaks in MLST database.
In our study, CC8/ST8 clone was the most common ST, which was distributed globally For instance, in Switzerland, CC8 was the most prevalent clone during 2011–2013 (Althaus et al., 2014); and in Denmark, a CC8/ST8 clone was associated with most of sporadic cases during 2004–2012 (Jensen et al., 2016). Both CC8 clones in Denmark and Canada did not lead to pregnancy-associated infections but were mainly associated with elderly patients (Knabel et al., 2012; Jensen et al., 2016), which was different from our study. In our study, CC8 caused five pregnancy-associated infections. Some studies suggested that the CC8 strains from Canada possessed a strong capacity of biofilm formation, which might support persistence within food production environments, resulting in subsequent contamination of foods (Verghese et al., 2011). In this study, eight CC8/ST8 strains had the indistinguishable pulsotypes; the fact that identical pulsotype had been found in the same period raised the possibility that contamination of food could originate from a common source.
CC5/ST5 clone was the second common ST in our study. Similarly, ST5 was the most predominate ST in ready-to-eat meat product in Nanjing, China (Wang G.Y. et al., 2015). In France, no obvious difference was observed between frequency distribution of CC5 in food samples and clinical samples (Maury et al., 2016). Although CC5 was not considered as a hypervirulent clone in the study of Maury et al. (2016) CC5 caused several outbreaks in the recent years, such as a multistate cantaloupe outbreak in US in 2011 (Lomonaco et al., 2013), a multistate ice cream outbreak in US in 2010–2015 (Centers for Disease Control and Prevention, 2015), and a stone fruit recall in US in 2014 (Jackson et al., 2015). In our study, all fetuses of pregnancy-associated cases caused by ST5 isolates died. Further studies are needed to uncover the virulence of CC5.
Interestingly, ST87 was seldomly linked to human infections in other countries. In 2008, one ST87 isolate from water was reported by Ragon et al. (2008). In 2014, Perez-Trallero et al. (2014) reported two outbreak episodes caused by ST87 strains affecting 15 people in northern Spain. In 2012, ST87, a common MLST type, had been reported from foodborne strains in China (Wang et al., 2012). In 2015, ST87 was reported to be the most frequent ST from clinical strains in Taiwan from 2000 to 2013 (Huang et al., 2015). The ST87 were also found as the most common ST in isolates from rodents in nature environment in China (Wang et al., 2017). Besides, we searched CC87 in MLST database and found that there were a CC87 strain isolated from human in Japan in 1988 see text footnote 2. Our study showed that 12.2% of the human isolates were ST87, demonstrating that ST87 has been already circulating in Beijing. It is necessary to follow the dissemination of this clone to assess its potential emergence. Comparing with ST5, fetuses of pregnancy-associated cases caused by ST87 isolates survived. More studies will be required to further assess the virulent diversity of L. monocytogenes in different STs.
It is widely accepted that food is the source of human L. monocytogenes. The comparison of the population structure of the clinical strains in our study with that from foods, revealed that all STs, except ST621, were also reported in foodborne isolates in China (Wang et al., 2012; Wang Y. et al., 2015; unpublished data). However, there was a notable difference in prevalence of STs between human isolates and foodborne isolates. The most common types of isolates from food sources were ST9, ST8 and ST87 (Wang et al., 2012). ST87 and ST8 were predominant in fresh aquatic products and edible mushroom products in China (Chen et al., 2018a,b). Consistently, ST8 and ST87 were the most predominant STs in human isolates in our study. ST9 (72%) was the most common ST in strains isolated from 356 raw pork samples, 2104 raw pork retail environment swabs and 329 insects in China (Wang et al., 2017). MLST analysis of isolates from France showed a higher prevalence of ST9 and ST121 isolates among food sources when compared with those clinical original (Maury et al., 2016).
Some studies reported that reduced susceptibility of L. monocytogenes to many antibiotics in various countries and increases in the frequency of multidrug-resistant strains (Pesavento et al., 2010; Korsak et al., 2012). In our study, the prevalence of some common antibiotic resistance among L. monocytogenes isolated from patients in China was relatively low. PEN alone or with gentamycin is the prioritized antibiotic for treating human listeriosis. Sulfamethoxazole and trimethoprim is used in patients having PEN anaphylaxis. ERY is used in treating pregnant women. Meanwhile, VAN is used in treating L. monocytogenes bacteremia and endocarditis (Janakiraman, 2008). In this study, no resistance to SXT, ERY, VAN, AMP, CLI, and CHL was found, and there was a relatively low resistance rate to PEN (4.1%) and GEN (2.0%). Strains isolated from a Spanish hospital were found to be sensitive to AMP and ERY (Ariza-Miguel et al., 2015), which was consistent with our study. However, most L. monocytogenes isolated from food and food processing environments in China and other countries were resistant to PEN (Obaidat et al., 2015; Lotfollahi et al., 2017; Chen et al., 2018b), which may result from different resistance criteria use. L. monocytogenes is intrinsically resistant to FOX which had a resistance rate of 100% in our study. High resistance to OXA, as reported in previous studies (Khen et al., 2015; Wang et al., 2017), may be intrinsic. In the study, no correlation was found between antimicrobial susceptibility patterns and other characteristics, such as serogroups, PFGE, and MLST types. Further studies are required to survey the antibiotic susceptibility of clinical L. monocytogenes and to explore the potential molecular mechanisms of antibiotic resistance in L. monocytogenes.
In summary, this study observes that more than half of listeria cases were pregnancy-associated infections in Beijing, and as such more attention should be paid to pregnant patients in future studies on this infection. The serogroup and ST distribution of clinical L. monocytogenes in Beijing was different from many other countries. This study enhances our understanding of genetic diversity of clinical L. monocytogenes in China. Continuous surveillance for this pathogen in clinical patients is necessary in China.
YN and XM were involved in the collection of isolates and collected the clinical data. YL, ZL, DW, and XC performed the molecular subtyping and antibiotic susceptibility tests. XZ performed the data analysis. XZ and QC designed the study, drafted, and revised this manuscript.
This work was supported by The National Key Research and Development Program of China (Grant No. 2017YFC1601500), Young Talent Project of Beijing Excellent Talents Funding (Grant No. 2015000021469G186), the Project from the Ministry of Health of the People’s Republic of China (Grant No. 201302005), and Capitals Funds for Health Improvement and Research (Grant No. CFH2011-1013-02).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00981/full#supplementary-material
Almeida, R. M., Barbosa, A. V., Lisboa, R. C., Santos, A., Hofer, E., Vallim, D. C., et al. (2017). Virulence genes and genetic relationship of L. monocytogenes isolated from human and food sources in Brazil. Braz. J. Infect. Dis. 21, 282–289. doi: 10.1016/j.bjid.2017.01.004
Althaus, D., Jermini, M., Giannini, P., Martinetti, G., Reinholz, D., Nuesch-Inderbinen, M., et al. (2017). Local outbreak of Listeria monocytogenes serotype 4b sequence type 6 due to contaminated meat pate. Foodborne Pathog. Dis. 14, 219–222. doi: 10.1089/fpd.2016.2232
Althaus, D., Lehner, A., Brisse, S., Maury, M., Tasara, T., and Stephan, R. (2014). Characterization of Listeria monocytogenes strains isolated during 2011-2013 from human infections in Switzerland. Foodborne Pathog. Dis. 11, 753–758. doi: 10.1089/fpd.2014.1747
Amato, E., Filipello, V., Gori, M., Lomonaco, S., Losio, M. N., Parisi, A., et al. (2017). Identification of a major Listeria monocytogenes outbreak clone linked to soft cheese in Northern Italy - 2009-2011. BMC Infect. Dis. 17:342. doi: 10.1186/s12879-017-2441-6
Ariza-Miguel, J., Fernandez-Natal, M. I., Soriano, F., Hernandez, M., Stessl, B., and Rodriguez-Lazaro, D. (2015). Molecular epidemiology of invasive listeriosis due to Listeria monocytogenes in a spanish hospital over a nine-year study period, 2006-2014. BioMed Res. Int. 2015:191409. doi: 10.1155/2015/191409
Aureli, P., Fiorucci, G. C., Caroli, D., Marchiaro, G., Novara, O., Leone, L., et al. (2000). An outbreak of febrile gastroenteritis associated with corn contaminated by Listeria monocytogenes. N. Engl. J. Med. 342, 1236–1241. doi: 10.1056/nejm200004273421702
Bueno, V. F., Banerjee, P., Banada, P. P., Jose de Mesquita, A., Lemes-Marques, E. G., and Bhunia, A. K. (2010). Characterization of Listeria monocytogenes isolates of food and human origins from Brazil using molecular typing procedures and in vitro cell culture assays. Int. J. Environ. Health Res. 20, 43–59. doi: 10.1080/09603120903281283
Bula, C. J., Bille, J., and Glauser, M. P. (1995). An epidemic of food-borne listeriosis in western Switzerland: description of 57 cases involving adults. Clin. Infect. Dis. 20, 66–72. doi: 10.1093/clinids/20.1.66
Centers for Disease Control and Prevention. (2008). Outbreak of Listeria monocytogenes infections associated with pasteurized milk from a local dairy–Massachusetts, 2007. MMWR Morb. Mortal. Wkly. Rep. 57, 1097–1100.
Centers for Disease Control and Prevention. (2014). Data From: National Enteric Disease Surveillance: Listeria Annual Summary. Washington, D.C.: Centers for Disease Control and Prevention.
Centers for Disease Control and Prevention. (2015). Data from: Multistate Outbreak of Listeriosis Linked to Blue Bell Creameries Products (Final Update). Washington, D.C.: Centers for Disease Control and Prevention.
Chen, J., Zhang, X., Mei, L., Jiang, L., and Fang, W. (2009). Prevalence of listeria in chinese food products from 13 provinces between 2000 and 2007 and virulence characterization of Listeria monocytogenes isolates. Foodborne Pathog. Dis. 6, 7–14. doi: 10.1089/fpd.2008.0139
Chen, M., Cheng, J., Wu, Q., Zhang, J., Chen, Y., Xue, L., et al. (2018a). Occurrence, antibiotic resistance, and population diversity of Listeria monocytogenes isolated from fresh aquatic products in china. Front. Microbiol. 9:2215. doi: 10.3389/fmicb.2018.02215
Chen, M., Cheng, J., Wu, Q., Zhang, J., Chen, Y., Zeng, H., et al. (2018b). Prevalence, potential virulence, and genetic diversity of listeria monocytogenes isolates from edible mushrooms in chinese markets. Front. Microbiol. 9:1711. doi: 10.3389/fmicb.2018.01711
Chen, M., Wu, Q., Zhang, J., Wu, S., and Guo, W. (2015). Prevalence, enumeration, and pheno- and genotypic characteristics of Listeria monocytogenes isolated from raw foods in south china. Front. Microbiol. 6:1026. doi: 10.3389/fmicb.2015.01026
Chen, Y., Luo, Y., Carleton, H., Timme, R., Melka, D., Muruvanda, T., et al. (2017). Whole genome and core genome multilocus sequence typing and single nucleotide polymorphism analyses of Listeria monocytogenes associated with an outbreak linked to cheese, united states, 2013. Appl. Environ. Microbiol. 83:e00633-17. doi: 10.1128/AEM.00633-17
Chenal-Francisque, V., Lopez, J., Cantinelli, T., Caro, V., Tran, C., Leclercq, A., et al. (2011). Worldwide distribution of major clones of Listeria monocytogenes. Emerg. Infect. Dis. 17, 1110–1112. doi: 10.3201/eid1706.101778
Crim, S. M., Iwamoto, M., Huang, J. Y., Griffin, P. M., Gilliss, D., Cronquist, A. B., et al. (2014). Incidence and trends of infection with pathogens transmitted commonly through food-foodborne diseases active surveillance network, 10 U.S.sites, 2006–2013. MMWR Morb. Mortal. Wkly. Rep. 63, 328–332.
Dahl, V., Sundqvist, L., Hedenstrom, I., Lofdahl, M., Alm, E., Ringberg, H., et al. (2017). A nationwide outbreak of listeriosis associated with cold-cuts, Sweden 2013-2014. Infect. Ecol. Epidemiol. 7:1324232. doi: 10.1080/20008686.2017.1324232
Dalton, C. B., Austin, C. C., Sobel, J., Hayes, P. S., Bibb, W. F., Graves, L. M., et al. (1997). An outbreak of gastroenteritis and fever due to Listeria monocytogenes in milk. N. Engl. J. Med. 336, 100–105.
de Noordhout, C. M., Devleesschauwer, B., Angulo, F. J., Verbeke, G., Haagsma, J., Kirk, M., et al. (2014). The global burden of listeriosis: a systematic review and meta-analysis. Lancet Infect. Dis. 14, 1073–1082. doi: 10.1016/S1473-3099(14)70870-9
Doumith, M., Buchrieser, C., Glaser, P., Jacquet, C., and Martin, P. (2004). Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 42, 3819–3822. doi: 10.1128/jcm.42.8.3819-3822.2004
Ericsson, H., Eklow, A., Danielsson-Tham, M. L., Loncarevic, S., Mentzing, L. O., Persson, I., et al. (1997). An outbreak of listeriosis suspected to have been caused by rainbow trout. J. Clin. Microbiol. 35, 2904–2907.
European Food Safety Authority [EFSA] (2015). The European union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013. EFSA J. 13:3991. doi: 10.2903/j.efsa.2015.3991
Evans, J. R., Allen, A. C., Stinson, D. A., Bortolussi, R., and Peddle, L. J. (1985). Perinatal listeriosis: report of an outbreak. Pediatr. Infect. Dis. 4, 237–241. doi: 10.1097/00006454-198505000-00005
Fleming, D. W., Cochi, S. L., MacDonald, K. L., Brondum, J., Hayes, P. S., Plikaytis, B. D., et al. (1985). Pasteurized milk as a vehicle of infection in an outbreak of listeriosis. N. Engl. J. Med. 312, 404–407. doi: 10.1056/nejm198502143120704
Gilot, P., Genicot, A., and Andre, P. (1996). Serotyping and esterase typing for analysis of Listeria monocytogenes populations recovered from foodstuffs and from human patients with listeriosis in Belgium. J. Clin. Microbiol. 34, 1007–1010.
Gottlieb, S. L., Newbern, E. C., Griffin, P. M., Graves, L. M., Hoekstra, R. M., Baker, N. L., et al. (2006). Multistate outbreak of listeriosis linked to turkey deli meat and subsequent changes in US regulatory policy. Clin. Infect. Dis. 42, 29–36. doi: 10.1086/498113
Goulet, V., Hedberg, C., Le Monnier, A., and de Valk, H. (2008). Increasing incidence of listeriosis in france and other european countries. Emerg. Infect. Dis. 14, 734–740. doi: 10.3201/eid1405.071395
Goulet, V., Rocourt, J., Rebiere, I., Jacquet, C., Moyse, C., Dehaumont, P., et al. (1998). Listeriosis outbreak associated with the consumption of rillettes in France in 1993. J. Infect. Dis. 177, 155–160. doi: 10.1086/513814
Graves, L. M., and Swaminathan, B. (2001). PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int. J. Food Microbiol. 65, 55–62. doi: 10.1016/s0168-1605(00)00501-8
Guevara, R. E., Mascola, L., and Sorvillo, F. (2009). Risk factors for mortality among patients with nonperinatal listeriosis in los angeles county, 1992-2004. Clin. Infect. Dis. 48, 1507–1515. doi: 10.1086/598935
Huang, Y. T., Ko, W. C., Chan, Y. J., Lu, J. J., Tsai, H. Y., Liao, C. H., et al. (2015). Disease burden of invasive listeriosis and molecular characterization of clinical isolates in taiwan, 2000-2013. PloS One. 10:e0141241. doi: 10.1371/journal.pone.0141241
Jackson, B. R., Salter, M., Tarr, C., Conrad, A., Harvey, E., Steinbock, L., et al. (2015). Notes from the field: listeriosis associated with stone fruit-United States, 2014. MMWR Morb. Mortal. Wkly. Rep. 64, 282–283.
Jadhav, S., Bhave, M., and Palombo, E. A. (2012). Methods used for the detection and subtyping of Listeria monocytogenes. J. Microbiol. Methods 88, 327–341. doi: 10.1016/j.mimet.2012.01.002
Janakiraman, V. (2008). Listeriosis in pregnancy: diagnosis, treatment, and prevention. Rev. Obstet. Gynecol. 1, 179–185.
Jennison, A. V., Masson, J. J., Fang, N., Graham, R. M., Bradbury, M. I., Fegan, N., et al. (2017). Analysis of the Listeria Monocytogenes population structure among isolates from 1931 to 2015 in Australia. Front. Microbiol. 8:603. doi: 10.3389/fmicb.2017.00603
Jensen, A. K., Bjorkman, J. T., Ethelberg, S., Kiil, K., Kemp, M., and Nielsen, E. M. (2016). Molecular typing and epidemiology of human listeriosis cases, Denmark, 2002-2012. Emerg. Infect. Dis. 22, 625–633. doi: 10.3201/eid2204.150998
Khen, B. K., Lynch, O. A., Carroll, J., McDowell, D. A., and Duffy, G. (2015). Occurrence, antibiotic resistance and molecular characterization of Listeria monocytogenes in the beef chain in the republic of Ireland. Zoonoses Public Health 62, 11–17. doi: 10.1111/zph.12106
Kleta, S., Hammerl, J. A., Dieckmann, R., Malorny, B., Borowiak, M., Halbedel, S., et al. (2017). Molecular tracing to find source of protracted invasive listeriosis outbreak, southern Germany, 2012-2016. Emerg. Infect. Dis. 23, 1680–1683. doi: 10.3201/eid2310.161623
Knabel, S. J., Reimer, A., Verghese, B., Lok, M., Ziegler, J., Farber, J., et al. (2012). Sequence typing confirms that a predominant Listeria monocytogenes clone caused human listeriosis cases and outbreaks in Canada from 1988 to 2010. J. Clin. Microbiol. 50, 1748–1751. doi: 10.1128/JCM.06185-11
Korsak, D., Borek, A., Daniluk, S., Grabowska, A., and Pappelbaum, K. (2012). Antimicrobial susceptibilities of Listeria monocytogenes strains isolated from food and food processing environment in Poland. Int. J. Food Microbiol. 158, 203–208. doi: 10.1016/j.ijfoodmicro.2012.07.016
Li, W., Bai, L., Fu, P., Han, H., Liu, J., and Guo, Y. (2018). The epidemiology of Listeria. monocytogenes in China. Foodborne Pathog. Dis. 15, 459–466. doi: 10.1089/fpd.2017.2409
Linnan, M. J., Mascola, L., Lou, X. D., Goulet, V., May, S., Salminen, C., et al. (1988). Epidemic listeriosis associated with mexican-style cheese. N. Engl. J. Med. 319, 823–828. doi: 10.1056/nejm198809293191303
Lomonaco, S., Verghese, B., Gerner-Smidt, P., Tarr, C., Gladney, L., Joseph, L., et al. (2013). Novel epidemic clones of Listeria monocytogenes, United States, 2011. Emerg. Infect. Dis. 19, 147–150. doi: 10.3201/eid1901.121167
Lotfollahi, L., Chaharbalesh, A., Ahangarzadeh Rezaee, M., and Hasani, A. (2017). Prevalence, antimicrobial susceptibility and multiplex PCR-serotyping of Listeria monocytogenes isolated from humans, foods and livestock in Iran. Microb. Pathog. 107, 425–429. doi: 10.1016/j.micpath.2017.04.029
Lv, J., Qin, Z., Xu, Y., and Xie, Q. (2014). Listeria infection in chinese pregnant women and neonates from shandong. Int. J. Clin. Exp. Med. 7, 2730–2734.
Lyytikainen, O., Autio, T., Maijala, R., Ruutu, P., Honkanen-Buzalski, T., Miettinen, M., et al. (2000). An outbreak of Listeria monocytogenes serotype 3a infections from butter in Finland. J. Infect. Dis. 181, 1838–1841. doi: 10.1086/315453
Magalhaes, R., Ferreira, V., Santos, I., Almeida, G., Teixeira, P., and Research, T. (2014). Genetic and phenotypic characterization of Listeria monocytogenes from human clinical cases that occurred in Portugal between 2008 and 2012. Foodborne Pathog. Dis. 11, 907–916. doi: 10.1089/fpd.2014.1806
Mammina, C., Aleo, A., Romani, C., Pellissier, N., Nicoletti, P., Pecile, P., et al. (2009). Characterization of Listeria monocytogenes isolates from human listeriosis cases in Italy. J. Clin. Microbiol. 47, 2925–2930. doi: 10.1128/JCM.00102-09
Maury, M. M., Tsai, Y. H., Charlier, C., Touchon, M., Chenal-Francisque, V., Leclercq, A., et al. (2016). Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 48, 308–313. doi: 10.1038/ng.3501
McCollum, J. T., Cronquist, A. B., Silk, B. J., Jackson, K. A., O’Connor, K. A., Cosgrove, S., et al. (2013). Multistate outbreak of listeriosis associated with cantaloupe. N. Engl. J. Med. 369, 944–953. doi: 10.1056/NEJMoa1215837
Obaidat, M. M., Bani Salman, A. E., Lafi, S. Q., and Al-Abboodi, A. R. (2015). Characterization of Listeria monocytogenes from three countries and antibiotic resistance differences among countries and Listeria monocytogenes serogroups. Lett. Appl. Microbiol. 60, 609–614. doi: 10.1111/lam.12420
Okutani, A., Okada, Y., Yamamoto, S., and Igimi, S. (2004). Nationwide survey of human Listeria monocytogenes infection in Japan. Epidemiol. Infect. 132, 769–772. doi: 10.1017/s0950268804001967
Perez-Trallero, E., Zigorraga, C., Artieda, J., Alkorta, M., and Marimon, J. M. (2014). Two outbreaks of Listeria monocytogenes infection, northern Spain. Emerg. Infect. Dis. 20, 2155–2157. doi: 10.3201/eid2012.140993
Pesavento, G., Ducci, B., Nieri, D., Comodo, N., and Lo Nostro, A. (2010). Prevalence and antibiotic susceptibility of Listeria spp. isolated from raw meat and retail foods. Food Control 21, 708–713. doi: 10.1089/fpd.2012.1361
Ragon, M., Wirth, T., Hollandt, F., Lavenir, R., Lecuit, M., Le Monnier, A., et al. (2008). A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 4:e1000146. doi: 10.1371/journal.ppat.1000146
Salcedo, C., Arreaza, L., Alcala, B., de la Fuente, L., and Vazquez, J. A. (2003). Development of a multilocus sequence typing method for analysis of Listeria monocytogenes clones. J. Clin. Microbiol. 41, 757–762. doi: 10.1128/jcm.41.2.757-762.2003
Schlech, W. F. III. (2000). Foodborne listeriosis. Clin. Infect. Dis. 31, 770–775. doi: 10.1086/314008
Schlech, W. F. III., Lavigne, P. M., Bortolussi, R. A., Allen, A. C., Haldane, E. V., Wort, A. J., et al. (1983). Epidemic listeriosis–evidence for transmission by food. N. Engl. J. Med. 308, 203–206. doi: 10.1056/nejm198301273080407
Verghese, B., Lok, M., Wen, J., Alessandria, V., Chen, Y., Kathariou, S., et al. (2011). Comk prophage junction fragments as markers for Listeria monocytogenes genotypes unique to individual meat and poultry processing plants and a model for rapid niche-specific adaptation, biofilm formation, and persistence. Appl. Environ. Microbiol. 77, 3279–3292. doi: 10.1128/AEM.00546-11
Wang, G. Y., Qian, W. J., Zhang, X. X., Wang, H. H., Ye, K. P., Bai, Y., et al. (2015). Prevalence, genetic diversity and antimicrobial resistance of Listeria monocytogenes isolated from ready-to-eat meat products in nanjing China. Food Control 50, 202–208. doi: 10.1016/j.foodcont.2014.07.057
Wang, Y., Jiao, Y., Lan, R., Xu, X., Liu, G., Wang, X., et al. (2015). Characterization of Listeria monocytogenes isolated from human listeriosis cases in China. Emerg. Microbes Infect. 4:e50. doi: 10.1038/emi.2015.50
Wang, H. L., Ghanem, K. G., Wang, P., Yang, S., and Li, T. S. (2013). Listeriosis at a tertiary care hospital in beijing, china: high prevalence of nonclustered healthcare-associated cases among adult patients. Clin. Infect. Dis. 56, 666–676. doi: 10.1093/cid/cis943
Wang, L., and Chen, Q. (2016). An etiological analysis and molecular characteristics studies of Listeria monocytogenes isolated from a pregnant woman (in Chinese). Capital J. Public Health 10, 103–106.
Wang, Y., Lu, L., Lan, R., Salazar, J. K., Liu, J., Xu, J., et al. (2017). Isolation and characterization of Listeria species from rodents in natural environments in China. Emerg. Microbes Infect. 6:e44. doi: 10.1038/emi.2017.28
Wang, Y., Zhao, A., Zhu, R., Lan, R., Jin, D., Cui, Z., et al. (2012). Genetic diversity and molecular typing of Listeria monocytogenes in China. BMC Microbiol. 12:119. doi: 10.1186/1471-2180-12-119
Wiedmann, M., Bruce, J. L., Keating, C., Johnson, A. E., McDonough, P. L., and Batt, C. A. (1997). Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65, 2707–2716.
Wu, S., Wu, Q., Zhang, J., Chen, M., and Guo, W. (2016). Analysis of multilocus sequence typing and virulence characterization of Listeria monocytogenes isolates from chinese retail ready-to-eat food. Front. Microbiol. 7:168. doi: 10.3389/fmicb.2016.00168
Wu, S., Wu, Q., Zhang, J., Chen, M., Yan, Z. A., and Hu, H. (2015). Listeria monocytogenes prevalence and characteristics in retail raw foods in China. PloS One. 10:e0136682. doi: 10.1371/journal.pone.0136682
Xu, X. K., Wu, Q. P., Zhang, J. M., Deng, M. Q., and Zhou, Y. H. (2009). Studies on specific detection of Listeria monocytogenesin foods by duplex PCR(in Chinese). J. Health Lab. Technol. 19, 1199–1221.
Keywords: Listeria monocytogenes, human, MLST, PFGE, antimicrobial susceptibility, China
Citation: Zhang X, Niu Y, Liu Y, Lu Z, Wang D, Cui X, Chen Q and Ma X (2019) Isolation and Characterization of Clinical Listeria monocytogenes in Beijing, China, 2014–2016. Front. Microbiol. 10:981. doi: 10.3389/fmicb.2019.00981
Received: 23 September 2018; Accepted: 18 April 2019;
Published: 08 May 2019.
Edited by:
Learn-Han Lee, Monash University, MalaysiaReviewed by:
Edward M. Fox, Northumbria University, United KingdomCopyright © 2019 Zhang, Niu, Liu, Lu, Wang, Cui, Chen and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qian Chen, Y2NoZW5xaWFuQDI2My5uZXQ= Xiaochen Ma, eGlhb2NoLW1hQDEyNi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.