- Division of Biological and Environmental Science and Engineering, Red Sea Research Center, King Abdullah University of Science and Technology, Thuwal, Saudi Arabia
The understanding of host-microbial partnerships has become a hot topic during the last decade as it has been shown that associated microbiota play critical roles in the host physiological functions and susceptibility to diseases. Moreover, the microbiome may contribute to host resilience to environmental stressors. The sea anemone Aiptasia is a good laboratory model system to study corals and their microbial symbiosis. In this regard, studying its bacterial microbiota provides a better understanding of cnidarian metaorganisms as a whole. Here, we investigated the bacterial communities of different Aiptasia host-symbiont combinations under long-term heat stress in laboratory conditions. Following a 16S rRNA gene sequencing approach we were able to detect significant differences in the bacterial composition and structure of Aiptasia reared at different temperatures. A higher number of taxa (i.e., species richness), and consequently increased α-diversity and β-dispersion, were observed in the microbiomes of heat-stressed individuals across all host strains and experimental batches. Our findings are in line with the recently proposed Anna Karenina principle (AKP) for animal microbiomes, which states that dysbiotic or stressed organisms have a more variable and unstable microbiome than healthy ones. Microbial interactions affect the fitness and survival of their hosts, thus exploring the AKP effect on animal microbiomes is important to understand host resilience. Our data contributes to the current knowledge of the Aiptasia holobiont and to the growing field of study of host-associated microbiomes.
Introduction
Corals and their association with different microbes, such as photosynthetic algae from the family Symbiodiniaceae (Lajeunesse et al., 2018) and diverse bacterial communities (Ritchie, 2006; Rosenberg et al., 2007; Ainsworth et al., 2014), is one of the most studied symbioses. The symbiotic microbiota has an important role in the fitness and survival of corals (Bourne et al., 2016), and deviations from a community equilibrium might lead to disease and even death (Bourne et al., 2008; Cárdenas et al., 2012; Closek et al., 2014). Hence, studying the interactions between the host, microbiome and environment is crucial to better comprehend the susceptibility and resilience of coral holobionts to environmental perturbations (Bourne et al., 2008; Kelly et al., 2014; Röthig et al., 2016b; Ziegler et al., 2016; Chen et al., 2017).
Increases in sea surface temperature of just a few degrees can lead to coral bleaching (Hoegh-Guldberg, 1999) and increased susceptibility to disease (Bruno et al., 2007; Harvell et al., 2015; Maynard et al., 2015). Elevated temperatures alter the structure and diversity of the coral-associated microbiota, in many cases resulting in the proliferation of opportunistic bacteria (Ritchie, 2006; Rosenberg et al., 2007; Bourne et al., 2009; Mao-Jones et al., 2010). Even when caused by short-term exposure to high temperatures, thermal stress can cause rapid and long-term shifts in the microbial communities of corals that ultimately favor pathogenic states (Bourne et al., 2009). Similarly, changes in microbiota also occur under acidification, pollution and overfishing stress (Jessen et al., 2013; McDevitt-Irwin et al., 2017). Further, coral responses to heat stress are not ubiquitous; some species can be severely affected (Carpenter et al., 2008; Hoogenboom et al., 2017; Scheufen et al., 2017) while others might be able to persist (Hoogenboom et al., 2017). It is still not fully known what drives resilience but evidence of shifts, or lack thereof, in the microbial composition of heat-stressed corals strongly suggests that microbiota may be a crucial component of coral resilience (Bourne et al., 2008; Lee et al., 2015; Hadaidi et al., 2017).
Corals are, however, notoriously difficult to maintain in aquariums and studying them in situ is not always possible, which makes lab-based molecular and some ecological work challenging (Weis et al., 2008; Voolstra, 2013). Instead, the sea anemone Aiptasia (sensu Exaiptasia pallida, Grajales and Rodríguez, 2014) has been used to study cnidarian-microbial interactions. Among its advantages are that it grows fast, produces large clonal populations, is able to live in a symbiont-free state and that it can be re-infected with different Symbiodiniaceae types under laboratory conditions (Weis et al., 2008; Voolstra, 2013). Furthermore, two distinct populations have been identified based on their endosymbiotic relationship with different Symbiodiniaceae species (Thornhill et al., 2013); H2, a globally distributed lineage (found in Hawaii, Japan, the Mediterranean, and Australia) and CC7, a local lineage (found exclusively in Florida). Recent studies have also investigated their bacterial associations in wild (Brown et al., 2017) and laboratory settings (Röthig et al., 2016a; Herrera et al., 2017). The latter has allowed to culture pure bacterial isolates (Röthig et al., 2016a), thus providing a promising foundation for future functional studies that intend to identify bacteria that are critical for environmental resilience of the holobiont, such as increased thermotolerance.
Here, we assessed changes in the bacterial communities associated with four different Aiptasia host-symbiont combinations across two temperatures (25°C and 32°C) based on a 16S rRNA gene sequencing approach. Moreover, we studied the effect of long-term heat stress on the bacterial diversity and dispersion while taking into account variation resulting from the biological differences between Aiptasia groups and technical replicates (i.e., batch effects).
Materials and Methods
Experimental Set-Up
Symbiotic anemones from three different clonal Aiptasia strains were used in this study; H2 from Hawaii, CC7 from North Carolina and a Red Sea (RS) line collected from the Saudi Arabian coast of the southern RS. These, at the same time, associate with specific Symbiodiniaceae species (Thornhill et al., 2013); H2 occurs in symbiosis with Breviolum minutum (referred to as SSB01, Xiang et al., 2013), whereas CC7 associates with Symbiodinium linucheae (referred to as SSA01, Sunagawa et al., 2009b; Bieri et al., 2016) and RS with Symbiodinium microadriaticum (Cziesielski et al., 2018). Additionally, a new host-symbiont combination was introduced by re-infecting aposymbiotic CC7 individuals with SSB01 Symbiodiniaceae (Baumgarten et al., 2015), herein referred to as CC7-SSB01. Animals were reared in the laboratory as previously described (Röthig et al., 2016a; Herrera et al., 2017); in clear polycarbonate tanks (2 L capacity; Cambro Camwear, Huntington Beach, CA, United States) filled with autoclaved natural seawater collected from the RS (∼39 PSU salinity and pH ∼8) under white-light (∼80 μmol photons m–2 s–1 of photosynthetically active radiation) on a 12 h light: 12 h dark cycle, and fed with freshly hatched Artemia brine shrimp twice per week. Further, anemones were cultured at 25°C (control culture conditions) and 32°C (heat stress treatment). All tanks were maintained under the same conditions, except for temperature, for at least 2 years before performing experiments.
16S rRNA Gene Sequencing
Briefly, a total of 48 samples (3 anemones × 2 replicate tanks × 4 host-symbiont combinations × 2 temperatures, Supplementary Figure S1) were processed according to Röthig et al. (2016a) and Herrera et al. (2017). DNA extractions were performed following the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) manufacturer’s instructions. Regions V5 and V6 of the 16S rRNA gene were amplified using the primers 784F and 1061R (Andersson et al., 2008) with Illumina (San Diego, CA, United States) adapter overhangs. We sequenced the V5-V6 hypervariable regions because these have been shown to have superior phylogenetic resolution for bacterial phyla (Yang et al., 2016) and are more closely aligned with the SILVA database classifier (Zhang et al., 2018). Indeed, many studies target these regions in particular to examine bacterial diversity in corals (Röthig et al., 2016b; Ziegler et al., 2016; Hadaidi et al., 2017) and Aiptasia (Röthig et al., 2016a; Herrera et al., 2017). For each sample, PCRs were done in triplicate using a Qiagen Multiplex PCR kit (Qiagen, Hilden, Germany) with a final primer concentration of 0.6 μM in a 15 μL final reaction volume. Thermocycling conditions were as follows: an initial activation step of 15 min at 95°C, 27 cycles each of 30 s at 95°C, 90 s at 55°C, and 30 s at 72°C, followed by a final extension step of 10 min at 72°C. Triplicate PCRs were then pooled for each sample, cleaned using Agencourt AMPure XP magnetic beads (Beckman Coulter, Indianapolis, IN, United States) and subsequently indexed with Nextera XT barcoded sequencing adapters (Illumina, San Diego, CA, United States). Libraries were quantified using a QuBit dsDNA BR Kit (Thermo Fisher Scientific, Waltham, MA, United States), pooled in equimolar ratios and run on a 2% agarose gel to isolate and purify the final library (MinElute Gel Extraction Kit; Qiagen, Hilden, Germany). Finally, amplicon length and quality were checked on a BioAnalyzer (Agilent Technologies, Santa Clara, CA, United States) before sequencing on the Illumina MiSeq platform (2 × 300 bp, paired-end v3 chemistry) according to the manufacturer’s instructions.
Microbial Data Processing
A total of 6,765,347 reads were demultiplexed and adapter sequences were removed with MiSeq Reporter (v.2.4.60.8; Illumina, San Diego, CA, United States). Data was analyzed using the software MOTHUR (Schloss et al., 2009) following the same procedures described in Röthig et al. (2016a) and Herrera et al. (2017). Reads were assembled into 6,765,347 contigs, trimmed and quality-filtered [i.e., sequences with ambiguous bases, long homopolymers (>5) or insufficient length were removed]. Singletons were removed (1,931,982) and the remaining sequences were aligned against the SILVA database (release 119, Pruesse et al., 2007) and pre-clustered based on 2 bp differences (Huse et al., 2010). Chimeras were removed (113,695) using VSEARCH (Rognes et al., 2016), as were other unwanted sequences (i.e., sequences assigned to chloroplasts, mitochondria, archaea, and eukaryotes). Only sequences that were phylogenetically classified as bacteria (Greengenes release gg_13_8_99; bootstrap = 60, McDonald et al., 2012) were used for further analyses. From the resulting 3,090,432 sequences (average length of 293 bp) further subsampling of 15,028 sequences per sample was done. As shown by a rarefaction analysis (Supplementary Figure S2), this was sufficient to capture the majority of the bacterial diversity. Finally, sequences were clustered into Operational Taxonomic Units (OTUs) using a 97% similarity cutoff. Noteworthy, taxonomic annotation errors have been reported when using the Greengenes database, especially for the orders Vibrionales and Alteromonadales (Edgar, 2018; Lydon and Lipp, 2018). Thus, for those OTUs of interest, representative sequences were BLASTed against NCBI’s GenBank nr to identify previous occurrences of identical or highly similar bacterial sequences (i.e., first best match based on sequence identity and query cover criteria). For the purpose of comparing our results to other studies (Röthig et al., 2016a; Brown et al., 2017; Herrera et al., 2017; Ziegler et al., 2017) both taxonomic annotations were reported, but if different from each other, only the latter was considered in further analyses.
Data Analysis
Methods for analyzing microbiome diversity have been extensively reviewed (Knights et al., 2011; Goodrich et al., 2014); thus standardizing diversity measurements in terms of within (i.e., alpha) and between samples (i.e., beta). First, we examined differences in the bacterial communities within each Aiptasia group across treatments by calculating α-diversity indices (as implemented in MOTHUR). Data was tested for normal distribution using the Shapiro–Wilk test and analyzed via two-way analysis of variance (ANOVA) or two-factorial generalized linear models (GLMs) fitted with gamma distribution and “log” link function (if non-parametric), using host-symbiont combination and temperature as explanatory variables. An overview of the models is provided in the Supplementary Table S2. Significant differences were then identified via post hoc comparisons. Further, permutational multivariant analysis of variance (PERMANOVA, Anderson, 2017) tests (as implemented in the R package vegan, Oksanen et al., 2019) were performed for all Aiptasia groups and temperatures. Differentially abundant OTUs were then determined using the package DESeq2 (Love et al., 2014), which uses a negative binomial distribution model that takes into account sample library size and dispersion for each taxon. Wald test p-values were adjusted with the Benjamini–Hochberg correction method.
β-diversity was also analyzed to better understand the effect of temperature, host-symbiont combination and batch (i.e., technical variation) in shaping the microbiome of Aiptasia. Generally speaking, β-diversity metrics can be quantitative (i.e., using sequence abundance) or qualitative (i.e., considering only presence-absence of sequences), and they can be based on phylogeny or not. While phylogeny-based metrics might outperform other measures in community-level comparisons, the latter can still be useful in clustering analyses. Specific methods used to cluster samples (both distance metrics and clustering algorithms) can affect the outcome and its interpretation. Thus, it is of extreme importance to perform clustering using several different approaches to ensure that sample classification (clustering) is not dependent on a particular parameter (Kuczynski et al., 2010; Goodrich et al., 2014). β-diversity was first analyzed by computing UniFrac dissimilarity matrices (Lozupone et al., 2011) based on the OTU table and phylogenetic tree produced by MOTHUR. Principal coordinates analysis (PCoA) plots were used to visualize distances between samples and thus evaluate the effect of temperature on the bacterial communities of Aiptasia. Changes in the composition and structure of the community were studied using unweighted (i.e., purely based on sequence distances) and weighted UniFrac (i.e., includes both sequence and abundance information) methods, respectively. This was performed first for all the dataset together so that overall patterns could be inspected, and then for each group separately to avoid any confounding effects resulting from the biological variation between different host-symbiont combinations. Dispersion effects (i.e., based on sample distance to the centroid of each group) of the different groups and treatments were quantified by conducting permutation tests of multivariate dispersion (PERMDISP, Anderson, 2008) as implemented in R using the package vegan (Oksanen et al., 2019). Additional clustering analyses were performed using non-phylogenetic metrics such as Bray-Curtis, Pearson and Kendall Tau methods, also widely used in community ecology, to calculate and plot pairwise β-diversity distances (custom Python script available in https://github.com/lyijin/anna_karenina). Ward’s clustering criteria was applied. Different approaches have different merits (Parks and Beiko, 2013; Weiss et al., 2016). Whilst Bray-Curtis measures abundance similarity, the Pearson method evaluates the degree of linear dependence between two variables and Kendall measures how well this relationship can be described using a monotonic function, for example. Therefore, in order to catch all possible correlations, all three methods were simultaneously used.
Results
Here, we investigated the effect of long-term heat stress (>2 years) on the bacterial communities of four different Aiptasia host-symbiont combinations; H2 (associated with its native symbiont B. minutum), CC7 (naturally occurring with S. linucheae), RS (symbiotic with S. microadriaticum) and CC7-SSB01, an experimental combination resulting from re-infecting aposymbiotic CC7 with B. minutum originally from H2. We assessed changes in the microbiota by analyzing within- and between- (Aiptasia group and temperature) diversity, and explored β-diversity patterns using different clustering methods. Moreover, our study offers another level of reproducibility by considering batch effects.
Dysbiotic Microbiota Are More Diverse
The bacterial communities of all host-symbiont combinations and temperatures were examined at the family level (Figure 1). Variations within each Aiptasia group (i.e., batch effects corresponding to the two replicate tanks from which anemones were taken) were detected but not significant (pPERMANOVA > 0.05 for all comparisons). Overall, Aiptasia from 32°C had a higher number of taxa (1088) than those reared at 25°C (1037). Members of the families Alteromonadaceae (10.48%) and Rhodobacteraceae (7.61%) were found in all the Aiptasia examined here, albeit in lower abundances compared to 25°C (24.76 and 12.51%, respectively). Notably, taxa belonging to Rhodospirillaceae and Flammeovirgaceae were also more abundant (100% presence) in anemones cultured at 32°C. Accordingly, OTU richness and diversity were also higher in this treatment (Figure 2 and Supplementary Table S1). All α-diversity estimators were significantly different between temperatures except for the number of observed OTUs and the Inverse Simpson index (Supplementary Table S2). Moreover, from a total of 724 OTUs, the genus Glaciecola sp. (family Alteromonadaceae) was the most dominant at both temperatures. Further analysis of raw abundances showed 17 differentially abundant OTUs, from which 13 were over and 4 under represented at 32°C compared to 25°C, respectively (Figure 3 and Supplementary Table S3). Notably, OTUs belonging to the family Rhodospirillaceae, Planctomycetaceae, and Cytophagaceae were significantly more abundant in heat-stressed anemones compared to Aiptasia reared at 25°C. Whilst, at the same time, OTUs annotated to the orders Actinomycetales and Oceanospirillales were significantly decreased in the bacterial communities of 32°C (Figure 3).
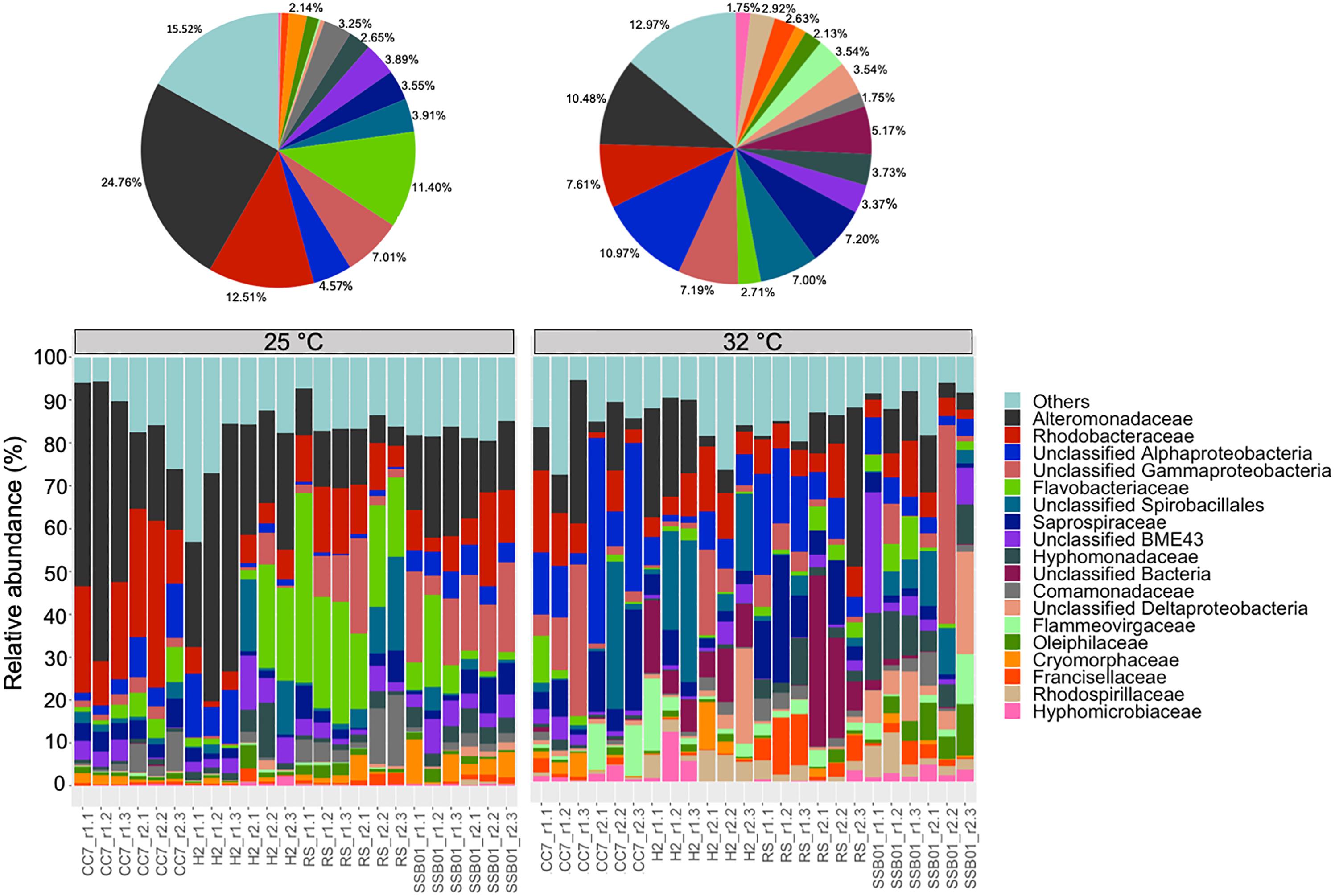
Figure 1. Bacterial community composition of different Aiptasia host-symbiont combinations on the family level (Greengenes database, bootstrap ≥ 60). Each color represents one taxon across all samples. Less abundant taxa <1%), comprising 199 distinct families are grouped under “others.” Pie charts display the average composition of anemones at 25°C (left) and 32°C (right). Sample labels indicate Aiptasia group, batch (i.e., tank) and replicate, respectively (e.g., CC7_r1.1).
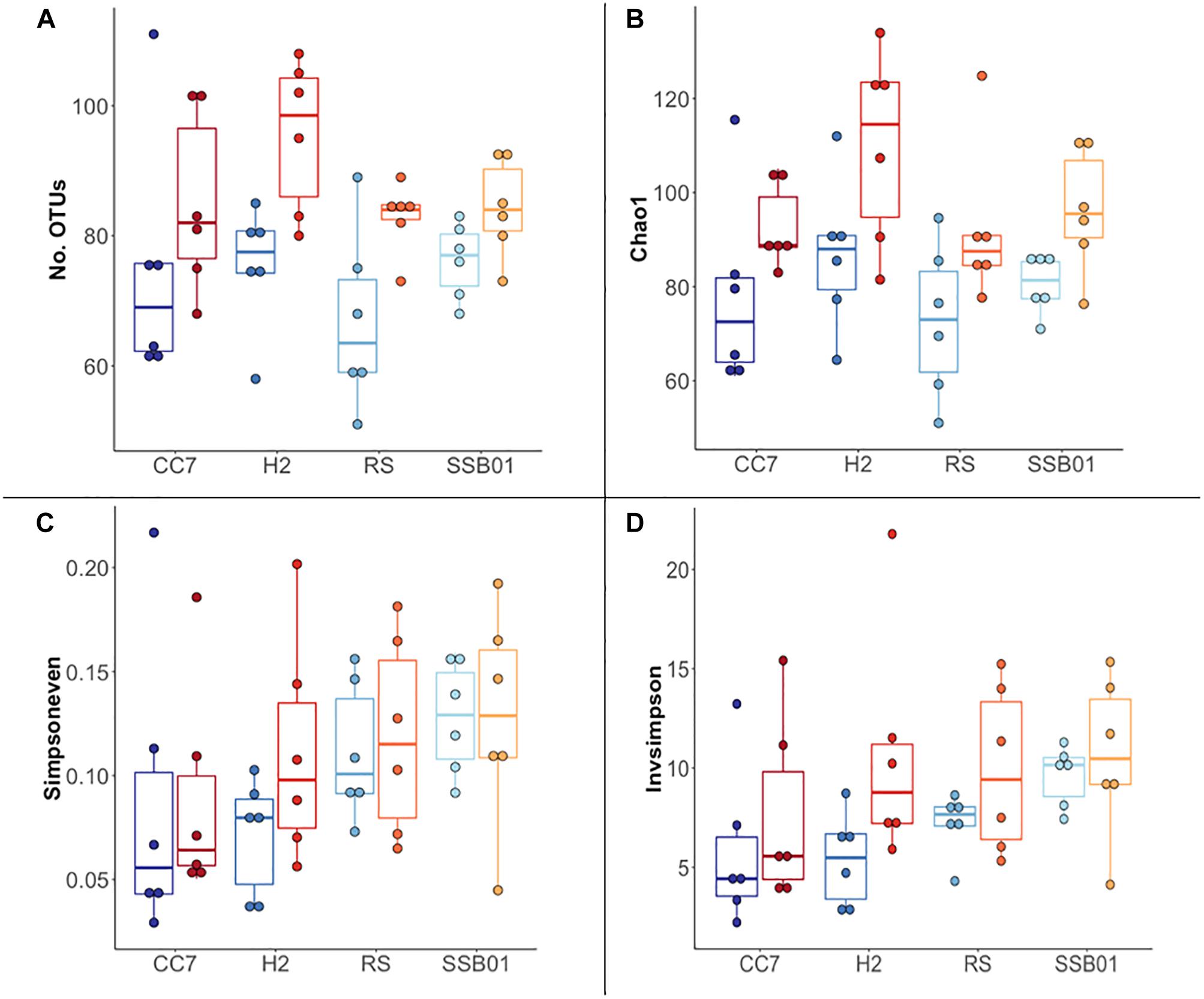
Figure 2. Overview of α-diversity measurements for different Aiptasia host-symbiont combinations. Number of observed OTUs (A), Chao1 richness estimator (B), Inverse Simpson (C), and Simpson’s evenness (D) indexes. Each boxplot shows six data points corresponding to the replicate samples of each host-symbiont combination. Different shades of blue and red represent 25°C and 32°C temperatures, respectively.
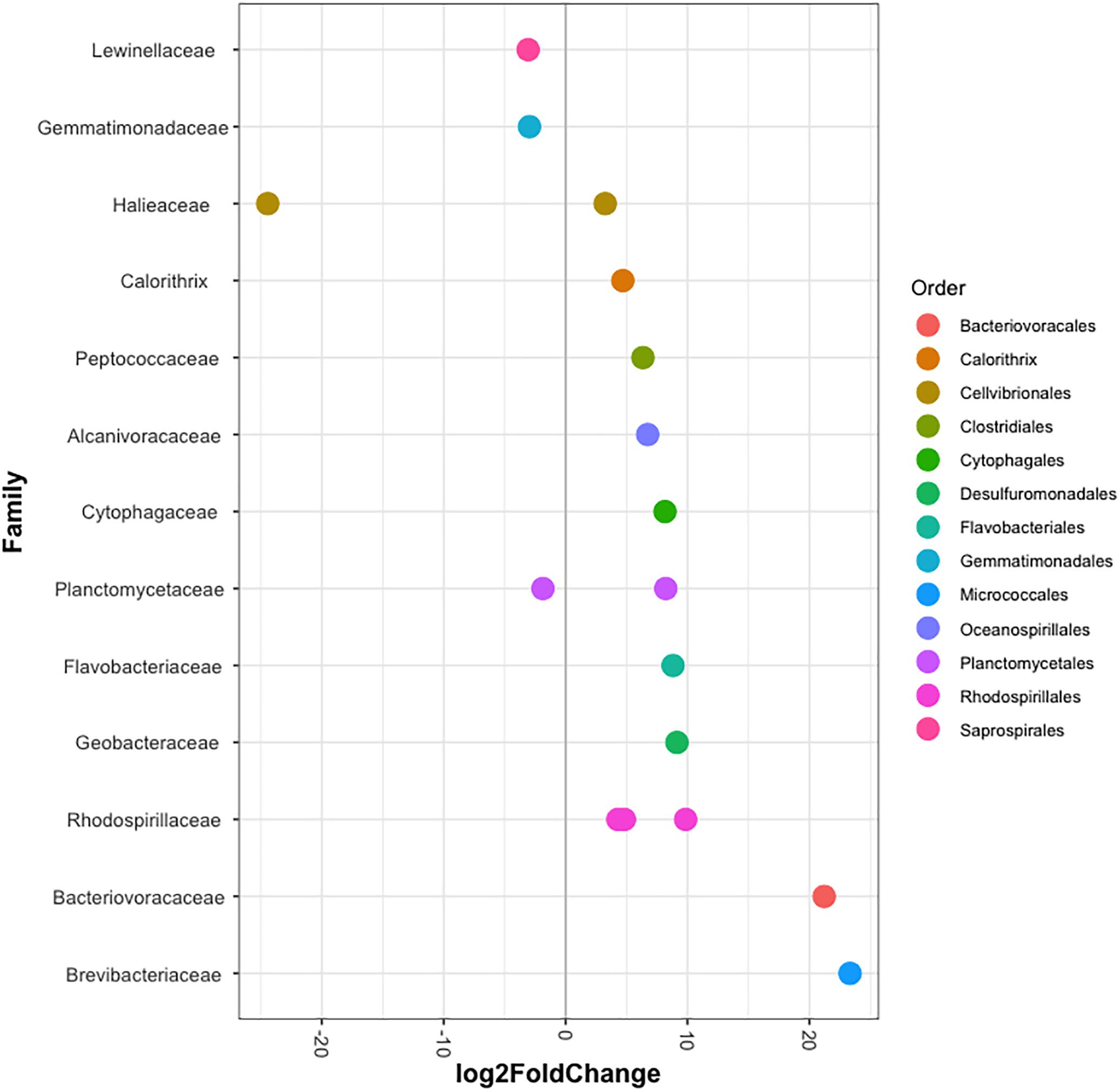
Figure 3. Differentially abundant OTUs (agglomerated by family) in Aiptasia between temperatures. Log2 fold change values indicate OTUs that show significant differential abundance in anemones reared at 32°C compared to those from 25°C (p < 0.05). Taxonomic annotation shown is based on BLASTn closest hit.
Temperature Effects on β-Dispersion
Significant differences in the overall composition (unweighted UniFrac, Figure 4) and structure (weighted UniFrac, Supplementary Figure S3) of the microbiota were observed. This was further substantiated by PERMANOVA analyses, which showed a clear separation between temperature (p = 0.001) and Aiptasia groups (CC7 p = 0.006, H2 p = 0.004, RS p = 0.002, and SSB01 p = 0.004). Moreover, anemones reared at 32°C also exhibited increased β-diversity dispersion in their bacterial consortia (p = 0.003, PERMDISP based on Bray-Curtis distances). Overall, bacterial communities were more scattered (i.e., higher dispersion, Supplementary Figure S4) in heat-stressed Aiptasia. However, when analyzed separately for each group, only CC7 was significant (p = 0.033).
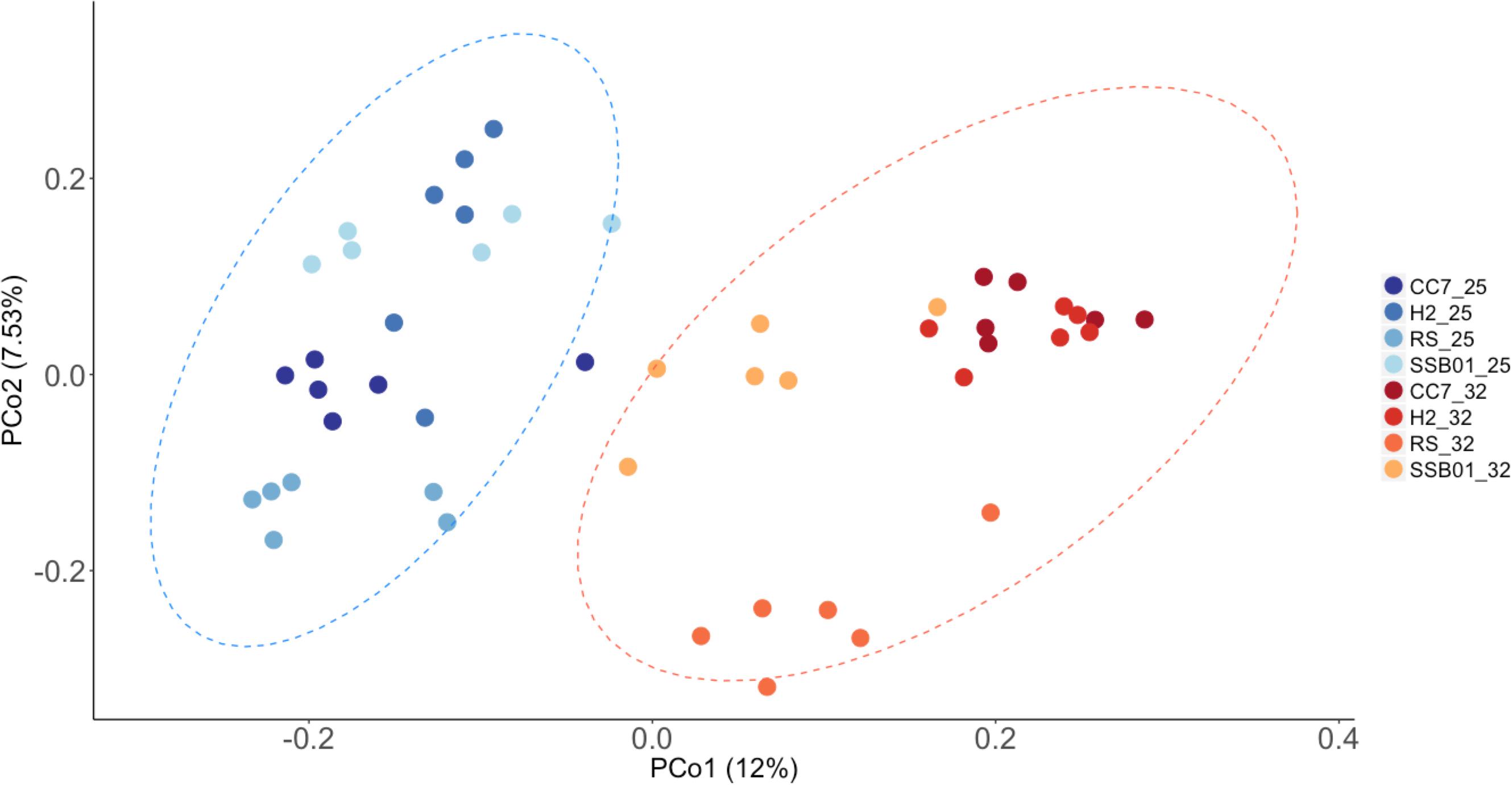
Figure 4. Principal coordinate analysis based on unweigthed Unifrac distances showing changes in the bacterial community composition of Aiptasia across temperatures. Different shades of blue and red show 25°C and 32°C, respectively. Ellipses represent 95% confidence intervals. Percentages on axes indicate the variation explained by the two coordinates.
Temperature Effect on Shaping the Bacterial Composition of Aiptasia
Overall, cluster analyses revealed temperature, and not host-symbiont combination or batch effect, as the main factor driving differences in the bacterial composition of Aiptasia (Supplementary Figure S5). Although there were no clear patterns, differences between treatments were still discernible. Both Bray-Curtis (Supplementary Figure S5A) and Pearson (Supplementary Figure S5B) methods separated the data in two main clusters corresponding to samples from 32°C and 25°C, respectively. Interestingly, no clear distinction between temperature treatments was observed with Kendall’s rank-order correlations (Supplementary Figure S5C). Instead, all RS Aiptasia were clustered together, suggesting this particular host-symbiont combination might have a distinct microbiome composition compared to the other Aiptasia groups. Nonetheless, different methods are based on different assumptions (and constrains), and so data interpretation can vary. Finally, analyses also showed in all cases that individuals first clustered according to the tank they were sampled from, and then by host-symbiont combination and treatment. Thus, highlighting the presence of batch effects (but as mentioned before, not significant).
Discussion
The structure and function of animals’ microbial communities are strongly affected by the environment (Allison and Martiny, 2008; Shade et al., 2012), therefore, having a better understanding of the effects of environment on the associated microbial communities is a key goal in microbial ecology (Green et al., 2008). Many studies have shown that variations in environmental parameters such as temperature (Sunagawa et al., 2015), salinity (Székely et al., 2013), pH (Liu et al., 2015), and organic compounds (Dang and Lovell, 2016) shape the microbial composition and structure of different ecosystems. Stressors in general can change a healthy stable microbiome to a dysbiotic unstable state (Teplitski et al., 2016; McDevitt-Irwin et al., 2017). The mechanisms by which this transition occurs are, however, still poorly understood. Regardless, research has shown that changes in the coral-associated microbial communities can ultimately disrupt holobiont functioning (Thompson et al., 2015; Bourne et al., 2016; Peixoto et al., 2017), thus serving as a diagnostic tool for assessing the health of corals (Glasl et al., 2017).
Questions such as whether the type and magnitude of microbial taxa changes over space and time (Glasl et al., 2018), or how it differs between healthy and diseased ecosystems (Hoisington et al., 2015; Zaneveld et al., 2016; Wang et al., 2017) can be answered by quantifying bacterial community diversity. Richness (i.e., total number of species) and evenness (i.e., proportional abundances of species) are some of the taxon-based measures commonly applied to describe the diversity within a given community (α-diversity). Likewise, the turnover of species between groups (β-diversity) is used to characterize the biological diversity among communities and along environmental gradients (Lozupone and Knight, 2008). One or the other alone cannot describe all aspects of a community’s diversity, especially since taxon-based methods ignore the fact that species are not equally related, thus masking important patterns in the data (Lozupone and Knight, 2008). Instead, divergence-based methods that take into account phylogenetic distance (e.g., Unifrac measures, Lozupone et al., 2011) can provide a better insight into the community structure. These metrics, in conjunction, can then be used to examine variability and stability of the microbiome (Zaneveld et al., 2017).
Distinct Bacterial Microbiomes of Aiptasia Reared at Different Temperatures
Overall, the bacterial composition of all Aiptasia host-symbiont combinations examined here was more diverse in the high temperature (32°C) treatment. Our results showed increased richness (i.e., the total number of observed OTUs) and α-diversity (i.e., the total number of species and their relative abundances) in response to long-term temperature stress. This is not surprising as it has already been shown that corals’ microbial diversity is higher not only in response to increasing seawater temperatures (Tout et al., 2015; Welsh et al., 2015; Lee et al., 2016) but also to reduced pH (Meron et al., 2011), water pollution (Ziegler et al., 2016), and disease (Kimes et al., 2010).
In context, the ability of disturbed corals to regulate or reject incoming bacteria from the environment may be reduced, thus resulting in a higher number of taxa (McDevitt-Irwin et al., 2017). Yet, the opposite can also occur (Meron et al., 2012; Tracy et al., 2015; Morrow et al., 2017). Stress in humans’ gut microbiome, for example, allows for opportunistic bacteria to dominate the microbial community, which in turn leads to a lower α-diversity (Lozupone et al., 2012). Although it is not clear to which degree single strains of bacteria play a role in the tolerance or susceptibility of corals to environmental stressors, meta-analyses of 16S data have suggested that certain taxa in particular are opportunistic and potentially pathogenic thus being highly present in diseased individuals (Teplitski et al., 2016; McDevitt-Irwin et al., 2017). Accordingly, in this study we found that taxa belonging to the families Alteromonadaceae, Flammeovirgaceae, Rhodobacteraceae and Rhodospirillaceae were highly abundant at 32°C compared to 25°C (Figure 1). Three OTUs annotated to Rhodospirillaceae were significantly more abundant in heat-stressed anemones (Figure 4). Similarly, OTUs from the classes Cytophagia and Planctomycetia were also increased at 32°C compared to 25°C. Previous studies in corals have shown that these particular taxonomic groups increase at higher temperatures (McDevitt-Irwin et al., 2017; Ziegler et al., 2017). The family Alteromonadaceae has also been shown to be associated with stressed or diseased corals and has been identified as an age indicator, even though members of this family are also known to reside in healthy corals (Sunagawa et al., 2009a; Glasl et al., 2016). Moreover, Flammeovirgacea has been shown to significantly increase in abundance throughout the coral mucus aging cycle (Sweet and Bulling, 2017). The most dominant taxa in both treatments, although mostly increased at 25°C, was Glaciecola sp. (OTU001), a genus that has already been reported in corals from hot environments (temperatures up to 33°C, Ziegler et al., 2017). This genus is particularly known for having genomic features related to cold adaptation (Qin et al., 2013), and has therefore been linked to thermal tolerance (Ziegler et al., 2017).
Stressed Microbiomes Are More Variable
Only recently, β-diversity dispersion measures have been applied in microbiome studies to show destabilization of the microbiome and bacterial opportunism in stressed and diseased corals (Zaneveld et al., 2016). Here, we observed increased β-diversity and dispersion of Aiptasia bacterial communities in response to temperature stress. Noteworthy, salinity of the seawater used to rear Aiptasia in this study (∼39 PSU from the central Red Sea) is significantly higher than from other geographic regions. High salinity levels have been shown to have a great impact on corals’ microbiome stability (Röthig et al., 2016b). Thus, it is important to take this into account, as salinity is an additional abiotic factor that might confound the effect of temperature we observed in this study. Regardless of not having clear patterns (i.e., cluster analysis) an overall separation between treatments was still observed, supporting the notion that temperature was the main driver of bacterial community differences.
The changes in the bacterial composition observed in this study are in line with the recently described “Anna Karenina principle” for animal microbiomes coined by Zaneveld et al. (2017), which postulates that external stressors often cause a more stochastic (i.e., randomly distributed) community structure due to the host being unable to regulate its microbiome when disturbed. Patterns consistent with this principle have been observed in corals but also in higher organisms like humans (David et al., 2014; Wang et al., 2017). It is interesting, however, that this concept does not hold for sponge microbiomes (Glasl et al., 2018). Moreover, even if this principle has been confirmed in some cases, it still has not been fully established if the observed increase in diversity in response to stress is a common biological phenomenon or rather a result of bias introduced by sampling (i.e., samples taken at different time points might reflect different states of a dynamic microbiome restructuring process). Consequently, comparison of microbiomes sampled at different time points during a dynamic restructuring process would suggest an increase of diversity. This study, however, compared microbiomes of hosts subjected to 2 years of constant heat stress. Hence, it can be assumed that the microbiomes have reached a final stable state and that the observed increase in β-diversity is not due to differences in sampling time. It is then fair to say that Aiptasia falls within this pattern, as increased microbiome β-diversity and dispersion was observed (i.e., greater distances between data points and centroid, Supplementary Figure S4) for individuals reared at higher temperature.
Temperature Shapes the Bacterial Community of Aiptasia
The microbiota of animals is complex, it is shaped through host–specific interactions (e.g., microbial recognition mechanisms and host immune responses, Ding et al., 2016; Neave et al., 2016b), interactions between members of the community (e.g., viruses and fungi, and Symbiodiniaceae in the case of corals (Deines and Bosch, 2016; Duerkop, 2018), geographical location and environmental conditions (Hong et al., 2009; Ceh et al., 2010). Several studies in different coral species have shown how the environment can have a greater impact than the host genetics in driving and shaping the microbiome (Littman et al., 2009; Pantos et al., 2015; Roder et al., 2015; Rothschild et al., 2018). Consistent species-specific associations still occur as it has been suggested for the widespread and highly abundant Endozoicomonas symbionts, found in many corals with contrasting life-history traits and across global scales, for example (Neave et al., 2016a). In Aiptasia, studies showed that the bacterial associations of H2 and CC7 Aiptasia lines cultured under identical laboratory conditions are significantly different, suggesting a species-specific microbiota (Röthig et al., 2016a; Herrera et al., 2017). Although the microbiome of Aiptasia from the Red Sea has still not been described, it is not surprising that these individuals clustered in a separate group (Supplementary Figure S5C). As the Red Sea displays unique physicochemical conditions, so are the microbial communities of corals and epilithic biofilms that thrive in these environments (Roik et al., 2016, 2019; Röthig et al., 2016b; Hadaidi et al., 2017). Thus, studying the bacterial communities from this region may provide a model for understanding the dynamics and functioning of coral reefs under predicted “future ocean” scenarios.
Finally, our study shows that even if host-symbiont combination has a strong effect on the microbiome composition (as shown in the PCoAs and cluster analyses), temperature is the dominating factor structuring the bacterial communities of Aiptasia. Furthermore, we also observed variations within each Aiptasia group, even if all individuals examined here were reared and sampled under the same conditions. This variation most probably stems from batch effects (i.e., replicate tanks), which calls for attention when looking at the reproducibility of microbiome studies. Technical sources of variation are often overlooked even though they are very important to control for statistically, otherwise biased data can lead to potentially erroneous conclusions.
Conclusion
Host-associated microbial partnerships in coral reef ecosystems is a growing field of study, and for this purpose the sea anemone Aiptasia has served as a cnidarian-Symbiodiniaceae model system. We believe that studying the effect of heat stress on the bacterial communities of Aiptasia is a start to better comprehend the microbial dynamics and resilience of this and other holobionts like stony corals. Our data showed that all Aiptasia host-symbiont combinations reared at different temperatures harbored distinct bacterial compositions. Moreover, we found that the bacterial consortia associated with heat-stressed individuals exhibited increased β-diversity and dispersion. The latter led us to explore the recently proposed AKP for disturbed microbiomes, which aims toward a better understanding of host resilience, as the microbiome status can greatly influence the animal fitness and health.
Ethics Statement
This study was exempt due to the use of invertebrate animals.
Author Contributions
MH and MA designed and conceived the experiments. MA contributed with reagents, materials, and analysis tools. HIA and MH generated the data. HIA, MH, YJL, and MA analyzed and interpreted the data. HIA and MH wrote the manuscript with input from all authors. All authors read and approved the final manuscript.
Funding
This work was supported by KAUST baseline funds to MA.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the two reviewers for their helpful discussions which greatly improved the manuscript. We would also like to thank Ruben Díaz Rúa for his help during library preparation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00975/full#supplementary-material
References
Ainsworth, T. D., Krause, L., Bridge, T., Torda, G., Raina, J.-B., Zakrzewski, M., et al. (2014). The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J. 9, 2261–2274. doi: 10.1038/ismej.2015.39
Allison, S. D., and Martiny, J. B. H. (2008). Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. U.S.A. 105, 11512–11519. doi: 10.1073/pnas.0801925105
Anderson, M. J. (2008). A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x
Anderson, M. J. (2017). Permutational Multivariate Analysis of Variance (PERMANOVA). Wiley StatsRef: Statistics Reference Online. Hoboken, NJ: John Wiley & Sons, Ltd.
Andersson, A. F., Lindberg, M., Jakobsson, H., Bäckhed, F., Nyrén, P., and Engstrand, L. (2008). Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One 3:e0002836. doi: 10.1371/journal.pone.0002836
Baumgarten, S., Simakov, O., Esherick, L. Y., Liew, Y. J., Lehnert, E. M., Michell, C. T., et al. (2015). The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc. Natl. Acad. Sci. U.S.A. 112, 11893–11898. doi: 10.1073/pnas.1513318112
Bieri, T., Onishi, M., Xiang, T., Grossman, A. R., and Pringle, J. R. (2016). Relative contributions of various cellular mechanisms to loss of algae during cnidarian bleaching. PLoS One 11:e0152693. doi: 10.1371/journal.pone.0152693
Bourne, D., Iida, Y., Uthicke, S., and Smith-Keune, C. (2008). Changes in coral-associated microbial communities during a bleaching event. ISME J. 2, 350–363. doi: 10.1038/ismej.2007.112
Bourne, D. G., Garren, M., Work, T. M., Rosenberg, E., Smith, G. W., and Harvell, C. D. (2009). Microbial disease and the coral holobiont. Cell Press Trend Microbiol. 17, 554–562. doi: 10.1016/j.tim.2009.09.004
Bourne, D. G., Morrow, K. M., and Webster, N. S. (2016). Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 70, 317–340. doi: 10.1146/annurev-micro-102215-095440
Brown, T., Otero, C., Grajales, A., Rodriguez, E., and Rodriguez-Lanetty, M. (2017). Worldwide exploration of the microbiome harbored by the cnidarian model, Exaiptasia pallida (Agassiz in Verrill, 1864) indicates a lack of bacterial association specificity at a lower taxonomic rank. PeerJ 5:e3235. doi: 10.7717/peerj.3235
Bruno, J. F., Selig, E. R., Casey, K. S., Page, C. A., Willis, B. L., Harvell, C. D., et al. (2007). Thermal stress and coral cover as drivers of coral disease outbreaks. PLoS Biol. 5:e0050124. doi: 10.1371/journal.pbio.0050124
Cárdenas, A., Rodriguez-R, L. M., Pizarro, V., Cadavid, L. F., and Arévalo-Ferro, C. (2012). Shifts in bacterial communities of two caribbean reef-building coral species affected by white plague disease. ISME J. 6, 502–512. doi: 10.1038/ismej.2011.123
Carpenter, K. E., Abrar, M., Aeby, G., Aronson, R. B., Banks, S., Bruckner, A., et al. (2008). One-third of reef-building corals face elevated extinction risk from climate change and local impacts. Science 321, 560–563. doi: 10.1126/science.1159196
Ceh, J., Keulen, M. V., and Bourne, D. G. (2010). Coral-associated bacterial communities on Ningaloo Reef. FEMS Microbial Ecol. 75, 134–144. doi: 10.1111/j.1574-6941.2010.00986.x
Chen, C. Y., Cheng, P. C., Weng, F. C., Shaw, G. T., and Wang, D. (2017). Habitat and indigenous gut microbes contribute to the plasticity of gut microbiome in oriental river prawn during rapid environmental change. PLoS One 12:e0181427. doi: 10.1371/journal.pone.0181427
Closek, C. J., Sunagawa, S., Desalvo, M. K., Piceno, Y. M., Desantis, T. Z., Brodie, E. L., et al. (2014). Coral transcriptome and bacterial community profiles reveal distinct Yellow Band Disease states in Orbicella faveolata. ISME J. 8, 2411–2422. doi: 10.1038/ismej.2014.85
Cziesielski, M. J., Liew, Y. J., Cui, G., Schmidt-Roach, S., Campana, S., Marondedze, C., et al. (2018). Multi-omics analysis of thermal stress response in a zooxanthellate cnidarian reveals the importance of associating with thermotolerant symbionts. R. Soc. 285:20172654. doi: 10.1098/rspb.2017.2654
Dang, H., and Lovell, C. R. (2016). Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 80, 91–138. doi: 10.1128/MMBR.00037-15
David, L. A., Maurice, C. F., Carmody, R. N., Gootenberg, D. B., Button, J. E., Wolfe, B. E., et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. doi: 10.1038/nature12820
Deines, P., and Bosch, T. C. G. (2016). Transitioning from microbiome composition to microbial community interactions: the potential of the metaorganism hydra as an experimental model. Front. Microbiol. 7:1610. doi: 10.3389/fmicb.2016.01610
Ding, J.-Y., Shiu, J.-H., Chen, W.-M., Chiang, Y.-R., and Tang, S.-L. (2016). Genomic insight into the host–endosymbiont relationship of endozoicomonas montiporae CL-33T with its Coral Host. Front. Microbiol. 7:251. doi: 10.3389/fmicb.2016.00251
Duerkop, B. A. (2018). Bacteriophages shift the focus of the mammalian microbiota. PLoS Pathog. 14:e1007310. doi: 10.1371/journal.ppat.1007310
Edgar, R. (2018). Taxonomy annotation and guide tree errors in 16S rRNA databases. PeerJ 6:e5030. doi: 10.7717/peerj.5030
Glasl, B., Herndl, G. J., and Frade, P. R. (2016). The microbiome of coral surface mucus has a key role in mediating holobiont health and survival upon disturbance. ISME J. 10, 2280–2292. doi: 10.1038/ismej.2016.9
Glasl, B., Smith, C. E., Bourne, D. G., and Webster, N. S. (2018). Exploring the diversity-stability paradigm using sponge microbial communities. Sci. Rep. 8:8425. doi: 10.1038/s41598-018-26641-9
Glasl, B., Webster, N. S., and Bourne, D. G. (2017). Microbial indicators as a diagnostic tool for assessing water quality and climate stress in coral reef ecosystems. Mar. Biol. 164:97. doi: 10.1007/s00227-017-3097-x
Goodrich, J. K., Rienzi, S. C. D., Poole, A. C., Koren, O., Walters, W. A., Caporaso, J. G., et al. (2014). Conducting a microbiome study. Cell 158, 250–262. doi: 10.1016/j.cell.2014.06.037
Grajales, A., and Rodríguez, E. (2014). Morphological revision of the genus Aiptasia and the family Aiptasiidae (Cnidaria, Actiniaria, Metridioidea). Zootaxa 3826, 55–100. doi: 10.11646/zootaxa.3826.1.2
Green, J. L., Bohannan, B. J. M., and Whitaker, R. J. (2008). Microbial biogeography: from taxonomy to traits. Science 320, 1039–1043. doi: 10.1126/science.1153475
Hadaidi, G., RöThig, T., Yum, L. K., Ziegler, M., Arif, C., Roder, C., et al. (2017). Stable mucus-associated bacterial communities in bleached and healthy corals of Porites lobata from the Arabian Seas. Sci. Rep. 7:45362. doi: 10.1038/srep45362
Harvell, D., Jordán-Dahlgren, E., Merkel, S., Rosenberg, E., Raymundo, L., Smith, G., et al. (2015). Coral disease, environmental drivers, and the balance between coral and microbial associates. Oceanography 20, 172–195. doi: 10.5670/oceanog.2007.91
Herrera, M., Ziegler, M., Voolstra, C. R., and Lastra, M. I. A. (2017). Laboratory-cultured strains of the sea anemone Exaiptasia reveal distinct bacterial communities. Front. Mar. Sci. 4:115. doi: 10.3389/fmars.2017.00115
Hoegh-Guldberg, O. (1999). Climate change, coral bleaching and the future of the world’s coral reefs. Mar. Freshw. Res. 50, 839–866. doi: 10.1071/MF99078
Hoisington, A. J., Brenner, L. A., Kinney, K. A., Postolache, T. T., and Lowry, C. A. (2015). The microbiome of the built environment and mental health. BMC Microbiome 3:60. doi: 10.1186/s40168-015-0127-0
Hong, M.-J., Yu, Y.-T., Chen, C. A., Chiang, P.-W., and Tang, S.-L. (2009). Influence of species specificity and other factors on bacteria associated with the coral Stylophora pistillata in Taiwan. Appl. Environ. Microbiol. 75, 7797–7806. doi: 10.1128/AEM.01418-09
Hoogenboom, M. O., Frank, G. E., Chase, T. J., Jurriaans, S., Álvarez-Noriega, M., Peterson, K., et al. (2017). Environmental drivers of variation in bleaching severity of acropora species during an extreme thermal anomaly. Front. Mar. Sci. 4:376. doi: 10.3389/fmars.2017.00376
Huse, S. M., Welch, D. M., Morrison, H. G., and Sogin, M. L. (2010). Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol. 12, 1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x
Jessen, C., Lizcano, J. F. V., Bayer, T., Roder, C., Aranda, M., Wild, C., et al. (2013). In-situ effects of eutrophication and overfishing on physiology and bacterial diversity of the red sea coral Acropora hemprichii. PLoS One 8:e0062091. doi: 10.1371/journal.pone.0062091
Kelly, L. W., Williamsb, G. J., Barotta, K. L., Carlson, C. A., Dinsdalea, E. A., Edwardsd, R. A., et al. (2014). Local genomic adaptation of coral reef-associated microbiomes to gradients of natural variability and anthropogenic stressors. Proc. Natl. Acad. Sci. U.S.A. 111, 10227–10232. doi: 10.1073/pnas.1403319111
Kimes, N. E., Nostrand, J. D. V., Weil, E., Zhou, J., and Morris, P. J. (2010). Microbial functional structure of Montastraea faveolata, an important Caribbean reef-building coral, differs between healthy and yellow-band diseased colonies. Environ. Microbiol. 12, 541–556. doi: 10.1111/j.1462-2920.2009.02113.x
Knights, D., Costello, E. K., and Knight, R. (2011). Supervised classification of human microbiota. FEMS Microbiol. Rev. 35, 343–359. doi: 10.1111/j.1574-6976.2010.00251.x
Kuczynski, J., Liu, Z., Lozupone, C., Mcdonald, D., Fierer, N., and Knight, R. (2010). Microbial community resemblance methods differ in their ability to detect biologically relevant patterns. Nat. Methods 7, 813–819. doi: 10.1038/nMeth.1499
Lajeunesse, T. C., Parkinson, J. E. W., Gabrielson, P., Jeong, H. J., Davisreimer, J. R., Voolstra, C., et al. (2018). Systematic revision of symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 28, 2570–2580. doi: 10.1016/j.cub.2018.07.008
Lee, S. T. M., Davy, S. K., Tang, S.-L., Fan, T.-Y., and Kench, P. S. (2015). Successive shifts in the microbial community of the surface mucus layer and tissues of the coral Acropora muricata under thermal stress. FEMS Microbial Ecol. 91:fiv142. doi: 10.1093/femsec/fiv142
Lee, S. T. M., Davy, S. K., Tang, S.-L., and Kench, P. S. (2016). Mucus sugar content shapes the bacterial community structure in thermally stressed acropora muricata. Front. Microbiol. 7:371. doi: 10.3389/fmicb.2016.00371
Littman, R. A., Willis, B. L., Pfeffer, C., and Bourne, D. G. (2009). Diversities of coral-associated bacteria differ with location, but not species, for three acroporid corals on the Great Barrier Reef. FEMS Microbial Ecol. 68, 152–163. doi: 10.1111/j.1574-6941.2009.00666.x
Liu, S., Ren, H., Shen, L., Lou, L., Tian, G., Zheng, P., et al. (2015). pH levels drive bacterial community structure in sediments of the Qiantang River as determined by 454 pyrosequencing. Front. Microbiol. 6:285. doi: 10.3389/fmicb.2015.00285
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. BMC Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Lozupone, C., Lladser, M. E., Knights, D., Stombaugh, J., and Knight, R. (2011). UniFrac: an effective distance metric for microbial community comparison. ISME J. 5, 169–172. doi: 10.1038/ismej.2010.133
Lozupone, C. A., and Knight, R. (2008). Species divergence and the measurement of microbial diversity. FEMS Microbiol. Rev. 32, 557–578. doi: 10.1111/j.1574-6976.2008.00111.x
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K., and Knight, R. (2012). Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230. doi: 10.1038/nature11550
Lydon, K. A., and Lipp, E. K. (2018). Taxonomic annotation errors incorrectly assign the family Pseudoalteromonadaceae to the order Vibrionales in Greengenes: implications for microbial community assessments. PeerJ 6:e5248. doi: 10.7717/peerj.5248
Mao-Jones, J., Ritchie, K. B., Jones, L. E., and Ellner, S. P. (2010). How microbial community composition regulates coral disease development. PLoS Biol. 8:e1000345. doi: 10.1371/journal.pbio.1000345
Maynard, J., Hooidonk, R. V., Eakin, C. M., Puotinen, M., Garren, M., Williams, G., et al. (2015). Projections of climate conditions that increase coral disease susceptibility and pathogen abundance and virulence. Nat. Clim. Chang. 5, 688–694. doi: 10.1038/nclimate2625
McDevitt-Irwin, J. M., Baum, J. K., Garren, M., and Thurber, R. L. V. (2017). Responses of coral-associated bacterial communities to local and global stressors. Front. Mar. Sci. 4:262. doi: 10.3389/fmars.2017.00262
McDonald, D., Price, M. N., Goodrich, J., Nawrocki, E. P., Desantis, T. Z., Probst, A., et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618. doi: 10.1038/ismej.2011.139
Meron, D., Atias, E., Kruh, L. I., Elifantz, H., Minz, D., Fine, M., et al. (2011). The impact of reduced pH on the microbial community of the coral Acropora eurystoma. ISME J. 5, 51–60. doi: 10.1038/ismej.2010.102
Meron, D., Rodolfo-Metalpa, R., Cunning, R., Baker, A. C., Fine, M., and Banin, E. (2012). Changes in coral microbial communities in response to a natural pH gradient. ISME J. 6, 1775–1785. doi: 10.1038/ismej.2012.19
Morrow, K. M., Bromhall, K., Motti, C. A., Munn, C. B., and Bourne, D. G. (2017). Allelochemicals produced by brown macroalgae of the lobophora genus are active against coral larvae and associated bacteria, supporting pathogenic shifts to vibrio dominance. Appl. Environ. Microbiol. 83:AEM.2391–AEM.2316. doi: 10.1128/AEM.02391-16
Neave, M. J., Apprill, A., Ferrier-Pagès, C., and Voolstra, C. R. (2016a). Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas. Appl. Microbiol. Biotechnol. 100, 8315–8324. doi: 10.1007/s00253-016-7777-0
Neave, M. J., Rachmawati, R., Xun, L., Michell, C. T., Bourne, D. G., Apprill, A., et al. (2016b). Differential specificity between closely related corals and abundant Endozoicomonas endosymbionts across global scales. ISME J. 11, 186–200. doi: 10.1038/ismej.2016.95
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., Mcglinn, D., et al. (2019). Package ‘Vegan’. Available at: https://cran.r-project.org/web/packages/vegan/vegan.pdf (accessed February 4, 2019).
Pantos, O., Bongaerts, P., Dennis, P. G., Tyson, G. W., and Hoegh-Guldberg, O. (2015). Habitat-specific environmental conditions primarily control the microbiomes of the coral Seriatopora hystrix. ISME J. 9, 1916–1927. doi: 10.1038/ismej.2015.3
Parks, D. H., and Beiko, R. G. (2013). Measures of phylogenetic differentiation provide robust and complementary insights into microbial communities. ISME J. 7, 173–183. doi: 10.1038/ismej.2012.88
Peixoto, R. S., Rosado, P. M., Leite, D. C. D. A., Rosado, A. S., and David, G. B. (2017). Beneficial Microorganisms for Corals (BMC): proposed mechanisms for coral health and resilience. Front. Microbiol. 8:341. doi: 10.3389/fmicb.2017.00341
Pruesse, E., Quast, C., Knittel, K., Fuchs, B. M., Ludwig, W., Peplies, J., et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196. doi: 10.1093/nar/gkm864
Qin, Q. L., Xie, B. B., Yu, Y., Shu, Y. L., Rong, J. C., Zhang, Y. J., et al. (2013). Comparative genomics of the marine bacterial genus Glaciecola reveals the high degree of genomic diversity and genomic characteristic for cold adaptation. Environ. Microbiol. 16, 1642–1653. doi: 10.1111/1462-2920.12318
Ritchie, K. B. (2006). Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Progr. Ser. 322, 1–14. doi: 10.3354/meps322001
Roder, C., Bayer, T., Aranda, M., Kruse, M., and Voolstra, C. R. (2015). Microbiome structure of the fungid coral Ctenactis echinata aligns with environmental differences. Mol. Ecol. 24, 3501–3511. doi: 10.1111/mec.13251
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
Roik, A., Röthig, T., Roder, C., Ziegler, M., Kremb, S. G., and Voolstra, C. R. (2016). Year-long monitoring of physico-chemical and biological variables provide a comparative baseline of coral reef functioning in the central red sea. PLoS One 11:e0163939. doi: 10.1371/journal.pone.0163939
Roik, A., Ziegler, M., and Voolstra, C. R. (2019). “Physicochemical dynamics, microbial community patterns, and reef growth in coral reefs of the central red sea,” in Oceanographic and Biological Aspects of the Red Sea, eds N. Rasul and I. Stewart (Cham: Springer).
Rosenberg, E., Koren, O., Reshef, L., Efrony, R., and Zilber-Rosenberg, L. (2007). The role of microorganisms in coral health. Dis. Evol. Nat. Rev. Microbiol. 5, 355–362. doi: 10.1038/nrmicro1635
Röthig, T., Costa, R. B. M., Simona, F., Baumgarten, S., Torres, A. F., Radhakrishnan, A., et al. (2016a). Distinct bacterial communities associated with the coral model aiptasia in aposymbiotic and symbiotic states with symbiodinium. Front. Mar. Sci. 3:234. doi: 10.3389/fmars.2016.00234
Röthig, T., Ochsenkühn, M. A., Roik, A., Merwe, R. V. D., and Voolstra, C. R. (2016b). Long-term salinity tolerance is accompanied by major restructuring of the coral bacterial microbiome. Mol. Ecol. 25, 1308–1323. doi: 10.1111/mec.13567
Rothschild, D., Weissbrod, O., Barkan, E., Kurilshikov, A., Korem, T., Zeevi, D., et al. (2018). Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215. doi: 10.1038/nature25973
Scheufen, T., Krämer, W. E., Iglesias-Prieto, R., and Enríquez, S. (2017). Seasonal variation modulates coral sensibility to heat-stress and explains annual changes in coral productivity. Sci. Rep. 7:4937. doi: 10.1038/s41598-017-04927-8
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Shade, A., Read, J. S., Youngblut, N. D., Fierer, N., Knight, R., Kratz, T. K., et al. (2012). Lake microbial communities are resilient after a whole-ecosystem disturbance. ISME J. 6, 2153–2167. doi: 10.1038/ismej.2012.56
Sunagawa, S., Coelho, L. P., Chaffron, S., Kultima, J. R., Labadie, K., Salazar, G., et al. (2015). Structure and function of the global ocean microbiome. Science 348:1261356. doi: 10.1126/science.1261359
Sunagawa, S., Desantis, T. Z., Piceno, Y. M., Brodie, E. L., Desalvo, M. K., Voolstra, C. R., et al. (2009a). Bacterial diversity and white plague disease-associated community changes in the Caribbean coral Montastraea faveolata. ISME J. 3, 512–521. doi: 10.1038/ismej.2008.131
Sunagawa, S., Wilson, E. C., Thaler, M., Smith, M. L., Caruso, C., Pringle, J. R., et al. (2009b). Generation and analysis of transcriptomic resources for a model system on the rise: the sea anemone Aiptasia pallida and its dinoflagellate endosymbiont. BMC Genomics 10:258. doi: 10.1186/1471-2164-10-258
Sweet, M. J., and Bulling, M. T. (2017). On the importance of the microbiome and pathobiome in coral health and disease. Front. Mar. Sci. 4:9. doi: 10.3389/fmars.2017.00009
Székely, A. J., Berga, M., and Langenheder, S. (2013). Mechanisms determining the fate of dispersed bacterial communities in new environments. ISME J. 7, 61–71. doi: 10.1038/ismej.2012.80
Teplitski, M., Krediet, C. J., Meyer, J. L., Ritchie, K. B. (2016). “Microbial interactions on coral surfaces and within the coral holobiont,” in The Cnidaria, Past, Present and Future, eds Z. Dubinsky and S. Goffredo (Cham: Springer), 331–364.
Thompson, J. R., Rivera, H. E., Closek, C. J., and Medina, M. (2015). Microbes in the coral holobiont: partners through evolution, development, and ecological interactions. Front. Cell. Infect. Microbiol. 4:176. doi: 10.3389/fcimb.2014.00176
Thornhill, D. J., Xiang, Y., Pettay, D. T., Zhong, M., and Santos, S. R. (2013). Population genetic data of a model symbiotic cnidarian system reveal remarkable symbiotic specificity and vectored introductions across ocean basins. Mol. Ecol. 22, 4499–4515. doi: 10.1111/mec.12416
Tout, J., Siboni, N., Messer, L. F., Garren, M., Stocker, R., Webster, N. S., et al. (2015). Increased seawater temperature increases the abundance and alters the structure of natural Vibrio populations associated with the coral Pocillopora damicornis. Front. Microbiol. 6:432. doi: 10.3389/fmicb.2015.00432
Tracy, A. M., Koren, O., Douglas, N., Weil, E., and Harvell, C. D. (2015). Persistent shifts in Caribbean coral microbiota are linked to the 2010 warm thermal anomaly. Environ. Microbiol. Rep. 7, 471–479. doi: 10.1111/1758-2229.12274
Voolstra, C. R. (2013). A journey into the wild of the cnidarian model system Aiptasia and its symbionts. Mol. Ecol. 22, 4366–4368. doi: 10.1111/mec.12464
Wang, B., Yao, M., Lv, L., Ling, Z., and Li, L. (2017). The human microbiota in health and disease. Engineering 3, 71–82. doi: 10.1016/J.ENG.2017.01.008
Weis, V. M., Davy, S. K., Guldberg, O. H., Lanetty, M. R., and Pringle, J. R. (2008). Cell biology in model systems as the key to understanding corals. Trends Ecol. Evol. 23, 369–376. doi: 10.1016/j.tree.2008.03.004
Weiss, S., Treuren, W. V., Lozupone, C., Faust, K., Friedman, J., Deng, Y., et al. (2016). Correlation detection strategies in microbial data sets vary widely in sensitivity and precision. ISME J. 10, 1669–1681. doi: 10.1038/ismej.2015.235
Welsh, R. M., Rosales, S. M., Zaneveld, J. R. R., Payet, J. P., Mcminds, R., Hubbs, S. L., et al. (2015). Alien vs. Predator: pathogens open niche space for opportunists, unless controlled by predator. PeerJ 3:e1537v1. doi: 10.7287/peerj.preprints.1537v1
Xiang, T., Hambleton, E. A., Denofrio, J. C., Pringle, J. R., and Grossman, A. R. (2013). Isolation of clonal axenic strains of the symbiotic dinoflagellate Symbiodinium and their growth and host specificity. Phycol. Soc. America 49, 447–458. doi: 10.1111/jpy.12055
Yang, B., Wang, Y., and Qian, P.-Y. (2016). Sensitivity and correlation of hypervariable regions in 16S rRNA genes in phylogenetic analysis. BMC Genomics 17:135. doi: 10.1186/s12859-016-0992-y
Zaneveld, J. R., Burkepile, D. E., Shantz, A. A., Pritchard, C. E., Mcminds, R., Payet, J. P., et al. (2016). Overfishing and nutrient pollution interact with temperature to disrupt coral reefs down to microbial scales. Nat. Commun. 7:11833. doi: 10.1038/ncomms11833
Zaneveld, J. R., Mcminds, R., and Thurber, R. V. (2017). Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat. Microbiol. 2:17121. doi: 10.1038/nmicrobiol.2017.121
Zhang, J., Ding, X., Guan, R., Zhu, C., Xu, C., Zhu, B., et al. (2018). Evaluation of different 16S rRNA gene V regions for exploring bacterial diversity in a eutrophic freshwater lake. Sci. Total Environ. 618, 1254–1267. doi: 10.1016/j.scitotenv.2017.09.228
Ziegler, M., Roik, A., Porter, A., Zubier, K., Mudarris, M. S., Ormond, R., et al. (2016). Coral microbial community dynamics in response to anthropogenic impacts near a major city in the central Red Sea. Mar. Pollut. Bull. 105, 629–640. doi: 10.1016/j.marpolbul.2015.12.045
Keywords: β-diversity, dispersion, microbiome, 16S rRNA gene, temperature
Citation: Ahmed HI, Herrera M, Liew YJ and Aranda M (2019) Long-Term Temperature Stress in the Coral Model Aiptasia Supports the “Anna Karenina Principle” for Bacterial Microbiomes. Front. Microbiol. 10:975. doi: 10.3389/fmicb.2019.00975
Received: 17 December 2018; Accepted: 18 April 2019;
Published: 08 May 2019.
Edited by:
George Tsiamis, University of Patras, GreeceReviewed by:
Julie L. Meyer, University of Florida, United StatesMauricio Rodriguez-Lanetty, Florida International University, United States
Copyright © 2019 Ahmed, Herrera, Liew and Aranda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manuel Aranda, bWFudWVsLmFyYW5kYUBrYXVzdC5lZHUuc2E=
†These authors have contributed equally to this work
‡Present address: Yi Jin Liew, CSIRO Health and Biosecurity, North Ryde, NSW, Australia
 Hanin Ibrahim Ahmed
Hanin Ibrahim Ahmed Marcela Herrera
Marcela Herrera Yi Jin Liew‡
Yi Jin Liew‡ Manuel Aranda
Manuel Aranda