
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 30 April 2019
Sec. Food Microbiology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00883
This article is part of the Research Topic New Knowledge of Food Microbiology in Asia View all 48 articles
 Shuai Wei1†
Shuai Wei1† Ramachandran Chelliah2†
Ramachandran Chelliah2† Byung-Jae Park2
Byung-Jae Park2 Se-Hun Kim2
Se-Hun Kim2 Fereidoun Forghani3
Fereidoun Forghani3 Min Seok Cho4
Min Seok Cho4 Dong-Suk Park4
Dong-Suk Park4 Yong-Guo Jin5*
Yong-Guo Jin5* Deog-Hwan Oh2*
Deog-Hwan Oh2*The efficiency of a novel biomarker (the transcriptional regulator, XRE) was tested and evaluated in differentiating Bacillus thuringiensis from Bacillus cereus group species in environmental and spiked samples based on PCR and real-time PCR. Totally 120 strains, representing two bacterial groups, B. cereus group and non-Bacillus sp., were used to evaluate the performance of XRE and crystal protein (cry2, an existing biomarker). Further, three diverse samples (kimbap, lettuce, and spinach) were inoculated with B. thuringiensis and prominent biomarkers XRE and cry2 were used as targets. Direct analysis of the detection results for the pure cultures of B. cereus group wild-types, references and type strains revealed an accuracy rate of 97.5% targeting XRE, and 83.3% targeting cry2. The real-time PCR was constructed with a R2-value of 0.993. For the artificially contaminated samples, a concentration of 103 CFU/g of B. thuringiensis in spiked food samples could be detected using real-time PCR targeting XRE. A good performance was obtained with XRE in discriminating B. thuringiensis from B. cereus groups, as well as detecting B. thuringiensis in spiked food samples with PCR or real-time PCR. Therefore, this real-time PCR targeting XRE can be used as a dependable and promising tool to identify B. thuringiensis in foods.
Bacillus thuringiensis and the other 7 species of spore forming Gram positive bacteria, including B. cereus, B. cytotoxicus, B. anthracis, B. pseudomycoides, B. weihenstephanensis, B. toyonensis, and B. mycoidesare, are primarily detected in soil. Because these bacteria are highly similar in genotype and phenotype, the bacteria are classified as B. cereus group in taxonomy (Park et al., 2007). B. cereus and B. thuringiensis are highly detectable in foods since they are observed in raw materials from agricultural soil during cultivation and distribution. In addition, these two bacteria are usually not discriminated in clinical diagnostics. B. cereus is a second risk priority group of foodborne illness in fresh agricultural products, and its contamination is one of the major problems in vegetables (Felício et al., 2015). In particular, B. thuringiensis was used as a pesticide in the cultivation of certain crops and it is well-known as microbial insecticides that have been used to reduce the amount of chemical pesticides (Azmi et al., 2015).
B. thuringiensis produces insecticidal proteins, which are the main type of Crystalline (Cry) proteins (Kutasi et al., 2016). Conversely, actively growing vegetative cells that lack crystal production lead to non-toxic effects. The δ-endotoxins mainly contains Cytolytic (Cyt) and Cry (Elleuch et al., 2015; Rao et al., 2015). However, Cyt and Cry possess different sequence homologies even though they consist of similar modes of action toward cell lysis, which lead to permanent damage of the insect midgut (Adang et al., 2014).
The structural variation of four δ-endotoxins (Cry1, Cry2, Cry3, and Cyt2Ah) was observed by X-ray crystallography (Dehury et al., 2013). These genes are identified in transgenic cotton and other vegetables, which are considerably effective in controlling pests. Designing a biomarker for the translated product varies with the different categories of cry protein; hence, the detection will be complex. The 16S rRNA gene sequences based on universal primers showed high similarity (>99%) index between B. cereus and B. thuringiensis (Böhm et al., 2015), which cannot be classified using genetic and phenotypic assays (Peng et al., 2015). Further, there has been a discussion since 2000 regarding whether the entire B. cereus group should be treated as a complex species of diverse bacteria (Helgason et al., 2000; Bartoszewicz and Marjańska, 2017). There are also suggestions that phylogeny of these bacteria better fits to their ecological properties (psychrotolerance, virulence) than to taxonomic affiliation (Drewnowska and Swiecicka, 2013; Bartoszewicz and Marjańska, 2017). To resolve this problem, we targeted XRE to detect B. thuringiensis, which controls the major type of crystal protein production.
These two bacteria are highly similar in biochemical results (Böhm et al., 2015), and genetic properties (Osman et al., 2015). Cho et al. (2015) also reported that the genetic and phenotypic properties between these bacteria are barely distinguishable. Furthermore, Pfrunder et al. reported that B. cereus group species cannot be reliably identified using classical biotyping (Pfrunder et al., 2016). Based on these, the biochemical experiments might not be enough for differentiation. Instead, the presence of an insecticidal crystal protein was used as a distinguished characteristic to differentiate these bacteria (Ekino et al., 2014; Wei et al., 2018).
Some of the factors that differentiate these two bacteria are based on their pathogenicity in samples. B. cereus causes gastrointestinal disorder, while B. thuringiensis has also been involved in epidemics of diarrhea. As reported, the distinguishing characteristic of B. thuringiensis is the presence of insecticidal crystal proteins (δ-endotoxin) encrypted by cry genes (Osman et al., 2015; Albright et al., 2016). Since the identification method using the biomarker for differentiating B. cereus group is complex and time consuming, a highly efficient biomarker is urgently needed to replace the previous ones that gave a lower efficiency and sometimes even showed false results. Based on the two specific genes, the transcriptional regulator and crystal protein genes for B. thuringiensis (Porcar and Juárez-Pérez, 2003; Han et al., 2006), the current study was performed to inspect and compare the efficiency of the designed biomarker (XRE) to that of the existing crystal protein marker (cry2) in identifying B. thuringiensis from B. cereus group strains (Bcg) and non-B. cereus group strains (non-B) in foods. cry2is the most common crystal protein present in B. thuringiensis (Liang et al., 2011; Palma et al., 2014).
All the 120 strains, including 111 of Bcg and 9 non-B, were obtained from bacterial collections in the United States and South Korea (Supplementary Table 1). Among the collections, B. thuringiensis ATCC 10792 (type strain) was used for the optimization of experimental conditions of conventional PCR and real-time PCR. All the strains were thawed at the room temperature (25°C) and streaked on the nutrient agar (Difco, MI, United States). After culturing for 24 h at 35°C in the incubator, a pure colony was picked and inoculated in tryptic soy broth (Difco, MI, United States) and overnight cultures were used for subsequent experiments.
The respective primers targeting XRE for differentiating B. thuringiensis (Table 1) were designed according to the following sequence in Bacillus thuringiensis ATCC 10792 (Gene – 3664610–3665047) using Primer Express® Software (Version 3.0.1) from Applied Biosystems located in CA, United States and synthesized by Bioneer Corporation from Daejeon, Korea. The performance of designed XRE was compared with cry2 (Ben-Dov et al., 1997). After 24 h enrichment in TSB, an aliquot of 1 mL of bacteria was subjected to the genomic DNA extraction using PrepMan® Ultra Sample Preparation Reagent (Applied Biosystems, CA, United States). DNA concentrations were measured by Eppendorf BioSpectrometer® fluorescence (Eppendorf, Hamburg, Germany) and adjusted to 10 ng/μL. DNA samples were stored at -20°C before use.
The conventional PCR were prepared with a C1000 TouchTM Thermal Cycler from Bio-Rad located in Hemel Hempstead, United Kingdom. The reaction tube for the PCR used Accu-Power® Pyro Hot Start Taq PCR Pre-Mix from Bioneer Corporation located in Daejeon, Korea and a volume of 20 μL, including 1 μL of each primer (10 pmoL/μL) and 2 μL of DNA. Amplification conditions of the conventional PCR for transcriptional regulator (XRE) were: initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 49°C for 30 s and elongation at 72°C for 30 s, and a final elongation at 72°C for 5 min. Amplifications conditions for crystal protein (Cry 2) were: initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s and elongation at 72°C for 1 min, and a final elongation at 72°C for 5 min. After DNA amplifications, 1.5% (W/V) agarose gel solution containing RedsafeTM Nucleic Acid Solution 20,000 × (Intron Biotechnology, Inc.) was prepared and gel electrophoresis was performed using Sub-Cell Model 192 from Bio-Rad located in Hemel Hempstead, United Kingdom. The bands were visualized using Smart View Pro UVCI-1100 Imager system from Major Science located in CA, United States. The accuracy (AC) was used to evaluate PCR method using primers XRE and cry2 in detecting B. thuringiensis from Bcg and non-B. Negative agreement (NA) means the same negative result for non-B. thuringiensis using PCR with XRE and cry2. Positive agreement (PA) means the same positive result for B. thuringiensis using PCR with XRE and cry2. The accuracy (AC) is measured using the following equation, (Ma et al., 2014).
Real-time PCR experiments were performed by the StepOneTM real-time PCR System from Applied Biosystems located in CA, United States. A reaction volume of 20 μL consisting of 10 μL of Melt DoctorTM High Resolution Melting (HRM) Master Mix, 2 μL of DNA, 0.5 μL of the F/R primer (10 pmoL/μL) (Table 2). Amplification conditions of the qPCR were: initial denaturation at 95°C for 10 min and 40 cycles at 95°C for 15 s and 60°C for 1 min. Then, in the subsequent melting curve analysis, the temperature was set to raise from 60 to 95°C at 0.3°C/cycle to test the specificity of the primer.
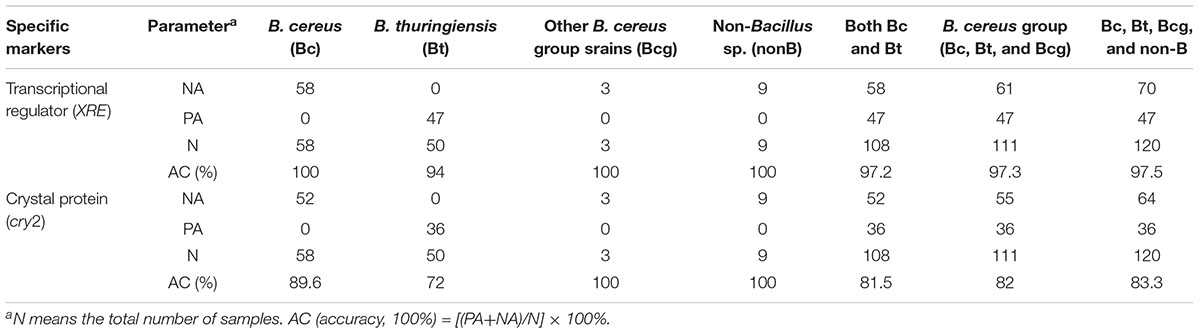
Table 2. Detection of B. cereus group and non-Bacillus sp. targeting biomarkers of XRE and cry2 using PCR.
The performance of XRE and cry2 was evaluated using 50 B. thuringiensis strains, while the exclusivity was tested using 70 non-target bacteria, including 58 B. cereus (Bc), 3 Bcg, and 9 non-B (Chelliah et al., 2017). A standard curve was made using 10-fold dilutions of the DNA extracted from B. thuringiensis ATCC 10792. DNA concentrations were adjusted from 10 ng/μL to 100 fg/μL, corresponding to 2.6 × 106 to 2.6 × 10 CFU, and the amplifications were performed by real-time PCR with triplicates. For negative controls, distilled water was used. Negative results or no amplification curves were considered when the cycle threshold (Ct) values were more than 40.
Lettuce (Lactucasativa), Kimbab, and spinach (Spinaciaoleracea) were bought from a local market in Chuncheon, Korea. The samples were tested for the absence of target bacteria according to ISO 7932 (2004). An aliquot of 50 g of each kind of food was prepared in a sterile stomacher bag from Nasco Whirl-Pak located in WI, United States. Overnight B. thuringiensis cultures were diluted in 0.5% peptone water and used for preparing the artificially contaminated samples with inoculation levels from 103 to 105 CFU/g (Chon et al., 2012; Kim et al., 2013). An aliquot of 1 mL of the suspension of food samples was transferred to a new sterilized tube and used for DNA extraction.
For the 120 strains, amplification products of approximately 700 bp were obtained using the primers cry2 (Table 1). As shown in Table 2 and Figure 1, when using PCR targeting cry2, 6 B. cereus strains and 36 B. thuringiensis strains (72%) were amplified. No other B. cereus group or non-B group was amplified. This showed an accuracy of 82% of cry2 for detecting B. cereus groups. For the 120 strains tested, 100 strains gave NAs or PAs, indicating an overall accuracy percentage of 83.3% (Supplementary Table 1). While Riojas et al. (2015) reported a multiplex PCR of non-ribosomal peptide synthase (NRPS) gene and cry1 to identify B. cereus and B. thuringiensis with a specificity of 24.5% in B. cereus and 15% in B. thuringiensis.

Figure 1. Conventional PCR results using 2 primers (XRE and cry2) targeting 2 genes (transcriptional regulator and crystal protein genes) specific for B. thuringiensis with 120 strains. (A) Is the PCR results of the 120 strains using XRE and (B) is the PCR results using cry-2. Lane M, 100 bp ladder; T1–T16 means the number of 8-strip tube and the details of each lane is shown in the Supplementary Table 1.
It has been reported that the B. thuringiensis strain may synthesize one or more crystal proteins since one or more crystal toxin genes have been found for B. thuringiensis. The main reason for the diversity of toxin genes is the transfer of plasmids in B. thuringiensis (Adang et al., 2014). It is a frequent process for the plasmid transfer by either conjugation or mobilization (Makart et al., 2017). Further, the exchanges of cry genes generate B. cereus with a new binding of cry that leads to the similarity of B. cereus and B. thuringiensis (Fiuza, 2015). In addition, with a different isolation source, the cry genes showed a difference in diversity and distributions. For instance, a novel cry gene can be found in each habitat that shows different insecticidal activity.
Potential coding sequences (CDS) were predicted to be transcriptional regulators. CDS contains a helix-turn-helix (HTH) motif in the XRE-like protein family and MerR family transcriptional regulators, previously the corresponding XRE gene sequences in pBMB28 of B. thuringiensis YBT-020 and pCT281 of B. thuringiensis (GenBank: CP001910). CDS50, which is related to the HTH-type transcriptional regulator, SinR, was identical to the corresponding element pG9842 of B. cereus G9842 (GenBank: CP001187). The HTH proteins, together with sigma factors, participate in a wide range of signaling pathways, for example, as repressors that inhibit sporulation (Murawska et al., 2014), biofilm formation (Colledge et al., 2011), and protease secretion (Pflughoeft et al., 2011). Apart from Cry1Ab21 and δ-endotoxin encoded in the toxigenic plasmid pIS56-63, which are responsible for conjugation and sporulation (Martínez-Núñez et al., 2010; Deng et al., 2014) in addition with 53 different encoded putative proteins.
The PCR results with XRE revealed that none of the 58 B. cereus or non-B gave an amplification (Table 2 and Figure 1). In total, out of 50 strains of B. thuringiensis 47 showed positive results (94%). This showed an accuracy of 97.3% of XRE for detecting B. thuringiensis among the tested B. cereus groups. For the 120 strains tested, 117 strains gave NAs or PAs, indicating an overall accuracy percentage of 97.5% (Supplementary Table 1).
The real-time PCR assay was performed using the DNA extracts from the pure cultures of B. thuringiensis type strain ATCC 10792 targeting the XRE gene. The developed real-time PCR assay was efficient for detecting concentrations from 2.6 × 10 to 2.6 × 106 CFU/reaction, which covered 6 orders of magnitude. The equation of log copy number versus the Ct value for B. thuringiensis was y = -3.853x+21.244 with an R-squared value of 0.993, indicating the high linearity (Figure 2).
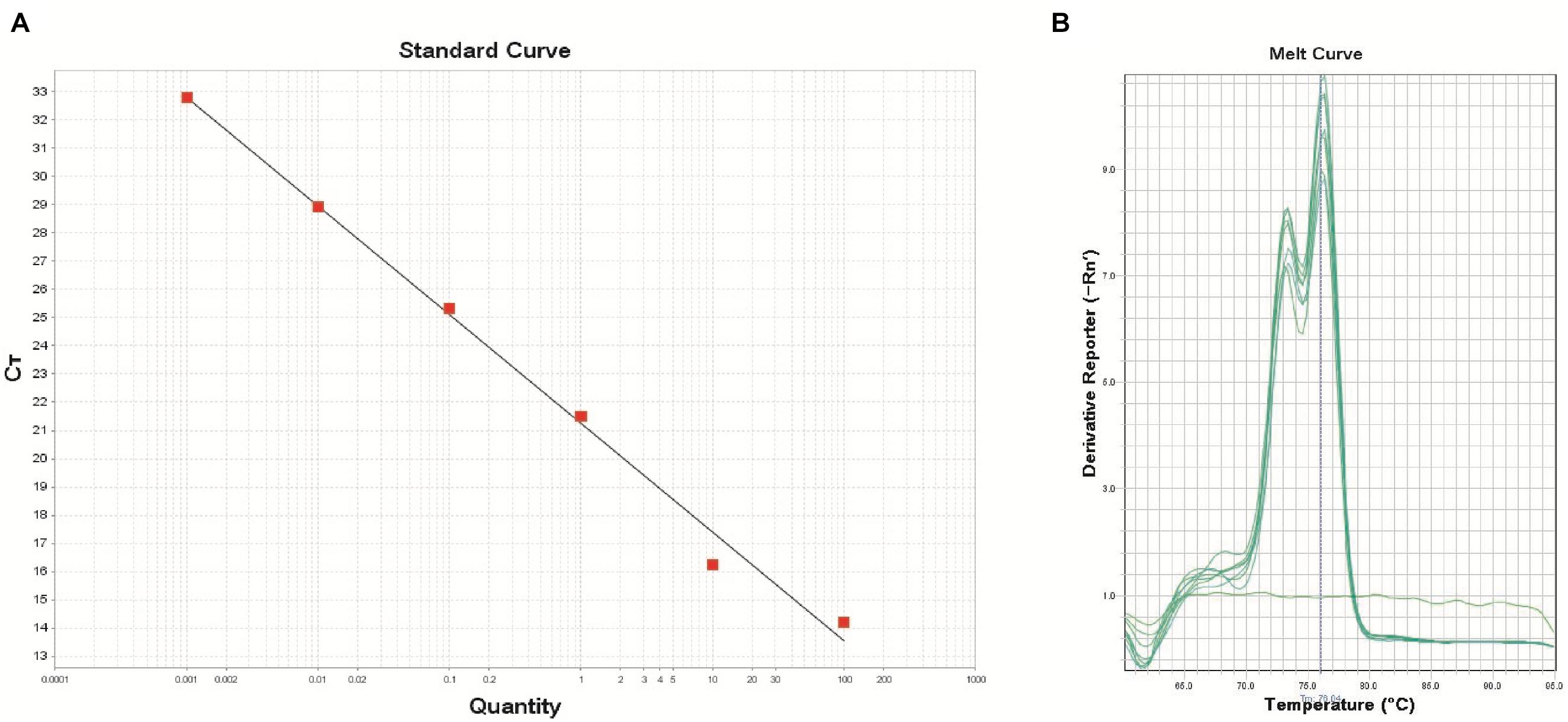
Figure 2. Standard curve (A) using the primer XRE (R2: 0.993, Eff%: 81.8) and melt curve peaks (B) for the detection of the transcriptional regulator and crystal protein genes in the B. thuringiensis strain. The DNA concentrations of B. thuringiensis ATCC 10792 were as follows: 1.0 × 102, 1.0 × 101, 1.0 × 100, 1.0 × 10-1, 1.0 × 10-2, and 1.0 × 10-3ng (from left to right). DNA amounts correspond to 2.6 × 106, 2.6 × 105, 2.6 × 104, 2.6 × 103, 2.6 × 102, and 2.6 × 101 CFU, respectively.
Recently, B. thuringiensis have been reported to be detected in foods such as milk, fresh fruits and grains that have been cultivated and harvested from soil outside the country. The consumption of raw and minimally processed vegetables is considered natural and healthy but concerns regarding residual chemical pesticides in fresh vegetables have been raised (Chiu et al., 2016). Thus, consumers have turned their interest to organic vegetables (chemical free vegetables), and it is predicted that bio-pesticides, including B. thuringiensis, may account for 20% of the world’s pesticide market by 2020 as a substitute for chemical pesticides (Watts and Williamson, 2015). In this study, the performance of the real-time PCR targeting XRE was evaluated with 3 kinds of artificially contaminated food samples (lettuce, kimbap and spinach). Fresh overnight cultures of B. thuringiensis were inoculated into each food sample. The real-time PCR could successfully detect B. thuringiensis at a concentration of 4.8 × 103, 3.4 × 103, and 1.5 × 103 CFU/g in lettuce, kimbap and spinach, respectively (Figure 3). However, for kimbap samples, the Ct values were higher, since usually it contains more complex materials such as sausage, egg or carrot compared with the vegetables.
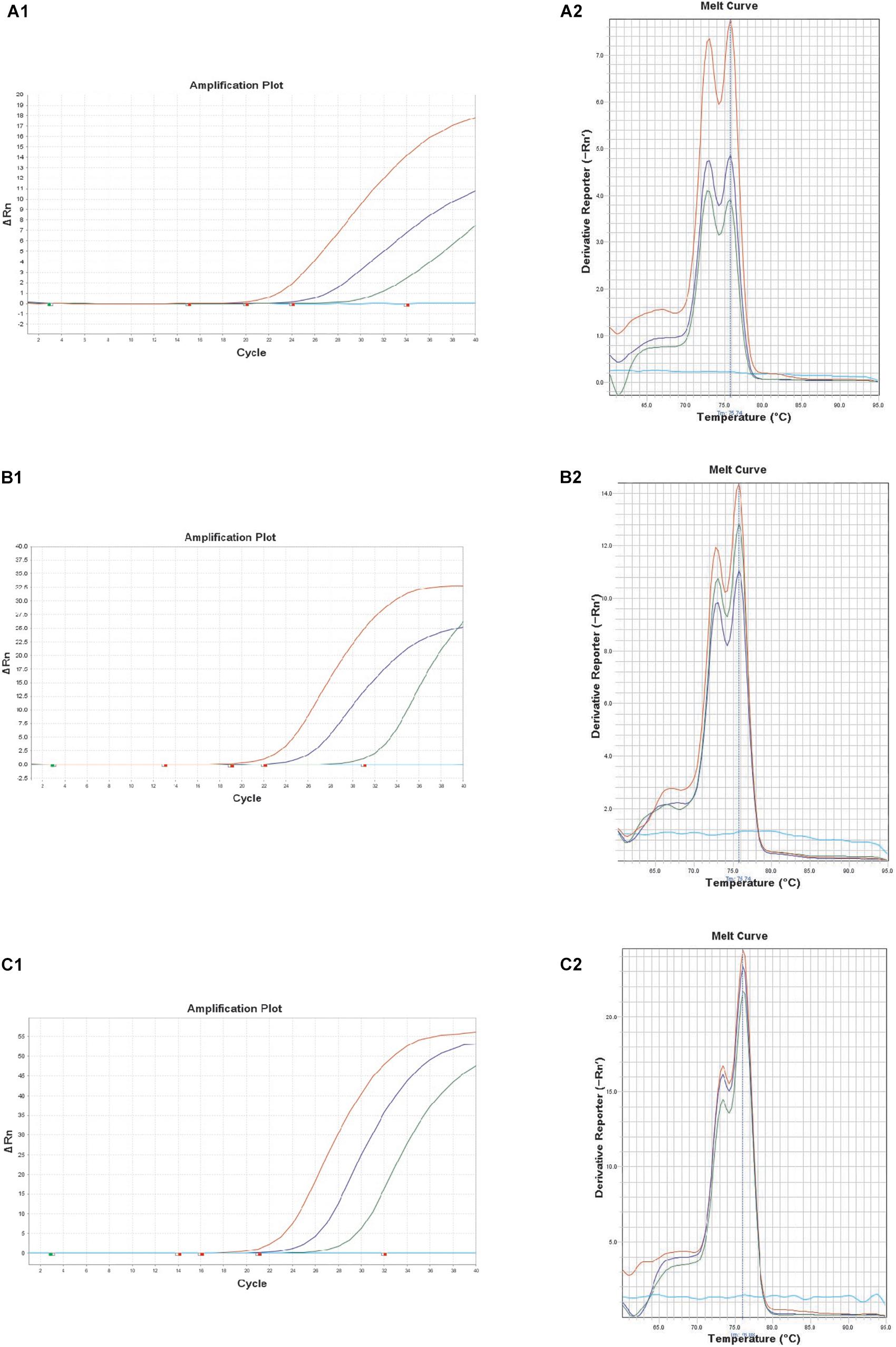
Figure 3. Melt curve peaks and amplification curves of XRE real-time PCR with B. thuringiensis ATCC 10792 in artificially contaminated food samples of lettuce (A1,A2); spinach (B1,B2); and kimbap (C1,C2). Colony forming units of the amplification curves correspond to 8.4 × 102, 8.4 × 101, 8.4 × 100 CFU (lettuce), 9.0 × 102, 9.0 × 101, 9.0 × 100 CFU (spinach), and 2.4 × 102, 2.4 × 101, 2.4 × 100 CFU (kimbap), respectively.
Since B. cereus group are highly similar in biochemical as well as genetic profiles, a new biomarker was developed for identifying and distinguishing B. thuringiensis from the closely related group. The performance of XRE was compared with cry2 gene and artificially contaminated samples were also tested. Compared with cry2, XRE gene was observed to be efficiently accurate in the identification of B. thuringiensis. Further, the developed real-time PCR using XRE successfully identified B. thuringiensis and it could be used to quantify cell numbers with the generated standard curve.
The experiments were conceived and designed by SW, RC, YG-J, and DH-O. SW, BJ-P, FF, MC, and DS-P performed the experiments. BJ-P and SH-K discussed the experiments and results. SW and RC wrote the manuscript.
This research work was supported by the Ministry of Agriculture, Food and Drug Safety (Grant No. 14162MFDS085) of Republic of Korea and Kangwon National University in 2015. SW was grateful for the financial support from the China Scholarship Council (CSC No: 201408260054). The authors would like to thank Hyun-Ah Lee at the Central Laboratory of the Kangwon National University for the technical support. RC is grateful for the financial support from the National Research Foundation of Korea (NRF) 2018007551.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00883/full#supplementary-material
Adang, M. J., Crickmore, N., and Jurat-Fuentes, J. (2014). Diversity of Bacillus thuringiensis crystal toxins and mechanism of action. Adv. Insect. Physiol. 47, 39–70. doi: 10.1016/B978-0-12-800197-4.00002-6
Albright, V. C. III, Hellmich, R. L., and Coats, J. R. (2016). A review of Cry protein detection with enzyme-linked immunosorbent assays. J. Agric. Food Chem. 64, 2175–2189. doi: 10.1021/acs.jafc.5b03766
Azmi, N. U., Ghafar, N. S. A., Yin, C. J., Yakubu, S., Adli, A. A., Aziz, N. A. A., et al. (2015). Toxicity of Bacillus thuringiensis biopesticide produced in shrimp pond sludge as alternative culture medium against Bactrocera dorsalis (Hendel). Acta Biol. Malays. 4, 5–16. doi: 10.7593/abm/4.1.5
Bartoszewicz, M., and Marjańska, P. S. (2017). Milk-originated Bacillus cereus sensu lato strains harbouring Bacillus anthracis-like plasmids are genetically and phenotypically diverse. Food Microbiol. 64, 23–30. doi: 10.1016/j.fm.2017.05.009
Ben-Dov, E., Zaritsky, A., Dahan, E., Barak, Z. E., Sinai, R., Manasherob, R., et al. (1997). Extended screening by PCR for seven cry-group genes from field-collected strains of Bacillus thuringiensis. Appl. Environ. Microbiol. 63, 4883–4890.
Böhm, M.-E., Huptas, C., Krey, V. M., and Scherer, S. (2015). Massive horizontal gene transfer, strictly vertical inheritance and ancient duplications differentially shape the evolution of Bacillus cereus enterotoxin operons hbl, cytK and nhe. BMC Evol. Biol. 15:246. doi: 10.1186/s12862-015-0529-4
Chelliah, R., Wei, S., Park, B. J., Kim, S. H., Park, D. S., Kim, S. H., et al. (2017). Novel motB as a potential predictive tool for identification of B. cereus, B. thuringiensis and differentiation from other Bacillus species by triplex real-time PCR. Microb. Pathog. 111, 22–27. doi: 10.1016/j.micpath.2017.07.050
Chiu, Y.-H., Gaskins, A. J., Williams, P. L., Mendiola, J., Jørgensen, N., Levine, H., et al. (2016). Intake of fruits and vegetables with low-to-moderate pesticide residues is positively associated with semen-quality parameters among young healthy men. J. Nutr. 146, 1084–1092. doi: 10.3945/jn.115.226563
Cho, S. H., Kang, S. H., Lee, Y. E., Kim, S. J., Yoo, Y. B., Bak, Y. S., et al. (2015). Distribution of toxin genes and enterotoxins in Bacillus thuringiensis isolated from microbial insecticide products. J. Microbiol. Biotechnol. 25, 2043–2048. doi: 10.4014/jmb.1506.06025
Chon, J. W., Hyeon, J. Y., Park, J. H., Song, K. Y., Kim, J. H., and Seo, K. H. (2012). Improvement of mannitol–yolk–polymyxin B agar by supplementing with trimethoprim for quantitative detection of Bacillus cereus in foods. J. Food Prot. 75, 1342–1345. doi: 10.4315/0362-028X.JFP-11-519
Colledge, V. L., Fogg, M. J., Levdikov, V. M., Leech, A., Dodson, E. J., and Wilkinson, A. J. (2011). Structure and organisation of SinR, the master regulator of biofilm formation in Bacillus subtilis. J. Mol. Biol. 411, 597–613. doi: 10.1016/j.jmb.2011.06.004
Dehury, B., Sahu, M., Sahu, J., Sarma, K., Sen, P., Modi, M. K., et al. (2013). Structural analysis and molecular dynamics simulations of novel δ-endotoxin Cry1Id from Bacillus thuringiensis to pave the way for development of novel fusion proteins against insect pests of crops. J. Mol. Model. 19, 5301–5316. doi: 10.1007/s00894-013-2010-x
Deng, Y., Li, C. Z., Zhu, Y. G., Wang, P. X., Qi, Q. D., Fu, J. J., et al. (2014). ApnI, a transmembrane protein responsible for subtilomycin immunity, unveils a novel model for lantibiotic immunity. Appl. Environ. Microbiol. 80, 6303–6315. doi: 10.1128/AEM.02280-14
Drewnowska, J. M., and Swiecicka, I. (2013). Eco-genetic structure of Bacillus cereus sensu lato populations from different environments in Northeastern Poland. PLoS One 8:e80175. doi: 10.1371/journal.pone.0080175
Ekino, K., Okumura, S., Ishikawa, T., Kitada, S., Saitoh, H., Akao, T., et al. (2014). Cloning and characterization of a unique cytotoxic protein parasporin-5 produced by Bacillus thuringiensis A1100 strain. Toxins 6, 1882–1895. doi: 10.3390/toxins6061882
Elleuch, J., Zghal, R. Z., Fguira, I. B., Lacroix, M. N., Suissi, J., Chandre, F., et al. (2015). Effects of the P20 protein from Bacillus thuringiensis israelensis on insecticidal crystal protein Cry4Ba. Int. J. Biol. Macromol. 79, 174–179. doi: 10.1016/j.ijbiomac.2015.04.035
Felício, M. D. S., Hald, T., Liebana, E., Allende, A., Hugas, M., Nguyen-The, C., et al. (2015). Risk ranking of pathogens in ready-to-eat unprocessed foods of non-animal origin (FoNAO) in the EU: initial evaluation using outbreak data (2007–2011). Int. J. Food Microbiol. 195, 9–19. doi: 10.1016/j.ijfoodmicro.2014.11.005
Fiuza, L. M. (2015). Thuringiensin: a toxin from Bacillus thuringiensis. Bt Res. 6, 1–12. doi: 10.5376/bt.2015.06.0004
Han, C. S., Xie, G., Challacombe, J. F., Altherr, M. R., Bhotika, S. S., Bruce, D., et al. (2006). Pathogenomic sequence analysis of Bacillus cereus and Bacillus thuringiensis isolates closely related to Bacillus anthracis. J. Bacteriol. 188, 3382–3390. doi: 10.1128/JB.188.9.3382-3390.2006
Helgason, E., Økstad, O. A., Caugant, D. A., Johansen, H. A., Fouet, A., Mock, M., et al. (2000). Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensi-one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66, 2627–2630. doi: 10.1128/AEM.66.6.2627-2630.2000
ISO 7932 (2004). Microbiology of Food and Animal Feeding Stuffs – Horizontal Method for the Enumeration of Presumptive Bacillus cereus - Colony-Count Technique at 30 Degrees C 2004. Geneva: FAO.
Kim, D. H., Kim, H., Chon, J. W., Moon, J. S., Song, K. Y., and Seo, K. H. (2013). Development of blood–yolk–polymyxin B–trimethoprim agar for the enumeration of Bacillus cereus in various foods. Int. J. Food Microbiol. 165, 144–147. doi: 10.1016/j.ijfoodmicro.2013.04.015
Kutasi, J., Kovacs, R., Puspan, I., Makk, J., Takacs, K., Erdelyi, B., et al. (2016). Protein patterns and larvicide activity of crystalline inclusions of Bacillus thuringiensis ssp. kumamotoensis DSM 6070. J. Agr. Sci. Tech. 18, 1945–1951.
Liang, H., Liu, Y., Zhu, J., Guan, P., Li, S., Wang, S., et al. (2011). Characterization of cry2-type genes of Bacillus thuringiensis strains from soil-isolated of Sichuan basin. China. Braz. J. Microbiol. 42, 140–146. doi: 10.1590/S1517-83822011000100018
Ma, K., Deng, Y., Bai, Y., Xu, D., Chen, E., Wu, H., et al. (2014). Rapid and simultaneous detection of Salmonella, Shigella, and Staphylococcus aureus in fresh pork using a multiplex real-time PCR assay based on immunomagnetic separation. Food Control. 42, 87–93. doi: 10.1016/j.foodcont.2014.01.042
Makart, L., Commans, F., Gillis, A., and Mahillon, J. (2017). Horizontal transfer of chromosomal markers mediated by the large conjugative plasmid pXO16 from Bacillus thuringiensis serovar israelensis. Plasmid 91, 76–81. doi: 10.1016/j.plasmid.2017.04.001
Martínez-Núñez, M. A., Perez-Rueda, E., Gutiérrez-Ríos, R. M., and Merino, E. (2010). New insights into the regulatory networks of paralogous genes in bacteria. Microbiology 156, 14–22. doi: 10.1099/mic.0.033266-0
Murawska, E., Fiedoruk, K., and Swiecicka, I. (2014). Modular genetic architecture of the toxigenic plasmid pIS56–63 harboring cry1Ab21 in Bacillus thuringiensis subsp. thuringiensis strain IS5056. Pol. J. Microbiol. 63, 147–156.
Osman, G., Already, R., Assaeedi, A., Organji, S., El-Ghareeb, D., Abulreesh, H., et al. (2015). Bioinsecticide Bacillus thuringiensis a comprehensive review. Egyptian J. Biol. Pest Control. 25, 271–288.
Palma, L., Muñoz, D., Berry, C., Murillo, J., and Caballero, P. (2014). Bacillus thuringiensis toxins: an overview of their biocidal activity. Toxins 6, 3296–3325. doi: 10.3390/toxins6123296
Park, S., Kim, H., Kim, J., Kim, T., and Kim, H. (2007). Simultaneous detection and identification of Bacillus cereus group bacteria using multiplex PCR. J. Microbiol. Biotechnol. 17, 1177–1182.
Peng, D. H., Pang, C. Y., Wu, H., Huang, Q., Zheng, J. S., and Sun, M. (2015). The expression and crystallization of Cry65Aa require two C-termini, revealing a novel evolutionary strategy of Bacillus thuringiensis Cry proteins. Sci. Rep. 5:8291. doi: 10.1038/srep08291
Pflughoeft, K. J., Sumby, P., and Koehler, T. M. (2011). Bacillus anthracis sin locus and regulation of secreted proteases. J. Bacteriol. 193, 631–639. doi: 10.1128/JB.01083-10
Pfrunder, S., Grossmann, J., Hunziker, P., Brunisholz, R., Gekenidis, M. T., and Drissner, D. (2016). Bacillus cereus group-type strain-specific diagnostic peptides. J. Proteome Res. 15, 3098–3107. doi: 10.1021/acs.jproteome.6b00216
Porcar, M., and Juárez-Pérez, V. (2003). PCR-based identification of Bacillus thuringiensis pesticidal crystal genes. FEMS Microbiol. Rev. 26, 419–432. doi: 10.1111/j.1574-6976.2003.tb00624.x
Rao, F., Short, F. L., Voss, J. E., Blower, T. R., Orme, A. L., Whittaker, T. E., et al. (2015). Co-evolution of quaternary organization and novel RNA tertiary interactions revealed in the crystal structure of a bacterial protein–RNA toxin–antitoxin system. Nucleic Acids Res. 43, 9529–9540. doi: 10.1093/nar/gkv868
Riojas, M. A., Kiss, K., McKee, M. L., and Hazbón, M. H. (2015). Multiplex PCR for Species-Level Identification of Bacillus anthracis and Detection of pXO1, pXO2, and Related Plasmids. Health Sec. 13, 122–129. doi: 10.1089/hs.2014.0056
Watts, M., and Williamson, S. (2015). Replacing Chemicals with Biology: Phasing Out Highly Hazardous Pesticides with Agroecology. Taiwan: PAN International, 1–210.
Wei, S., Chelliah, R., Park, B. J., Park, J. H., Forghani, F., Park, Y. S., et al. (2018). Molecular discrimination of Bacillus cereus group species in foods (lettuce, spinach, and kimbap) using quantitative real-time PCR targeting groEL and gyrB. Microb. Pathog. 115, 312–320. doi: 10.1016/j.micpath.2017.12.079
Keywords: B. thuringiensis, transcriptional regulator, crystal protein, kimbap, spinach, lettuce
Citation: Wei S, Chelliah R, Park B-J, Kim S-H, Forghani F, Cho MS, Park D-S, Jin Y-G and Oh D-H (2019) Differentiation of Bacillus thuringiensis From Bacillus cereus Group Using a Unique Marker Based on Real-Time PCR. Front. Microbiol. 10:883. doi: 10.3389/fmicb.2019.00883
Received: 10 September 2018; Accepted: 05 April 2019;
Published: 30 April 2019.
Edited by:
Qingli Dong, University of Shanghai for Science and Technology, ChinaReviewed by:
Govindarajan Prasanna, Shanghai Jiao Tong University, ChinaCopyright © 2019 Wei, Chelliah, Park, Kim, Forghani, Cho, Park, Jin and Oh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong-Guo Jin, amlueW9uZ2d1b0BtYWlsLmh6YXUuZWR1LmNu Deog-Hwan Oh, ZGVvZ2h3YUBrYW5nd29uLmFjLmty
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.