
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 24 April 2019
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00868
Transferable genetic elements conferring macrolide resistance in Streptococcus pneumoniae can encode the efflux pump and ribosomal protection protein, mef(E)/mel, in an operon of the macrolide efflux genetic assembly (Mega) element- or induce ribosomal methylation through a methyltransferase encoded by erm(B). During the past 30 years, strains that contain Mega or erm(B) or both elements on Tn2010 and other Tn916-like composite mobile genetic elements have emerged and expanded globally. In this study, we identify and define pneumococcal isolates with unusually high-level macrolide resistance (MICs > 16 μg/ml) due to the presence of the Mega element [mef(E)/mel] alone. High-level resistance due to mef(E)/mel was associated with at least two specific genomic insertions of the Mega element, designated Mega-2.IVa and Mega-2.IVc. Genome analyses revealed that these strains do not possess erm(B) or known ribosomal mutations. Deletion of mef(E)/mel in these isolates eliminated macrolide resistance. We also found that Mef(E) and Mel of Tn2010-containing pneumococci were functional but the high-level of macrolide resistance was due to Erm(B). Using in vitro competition experiments in the presence of macrolides, high-level macrolide-resistant S. pneumoniae conferred by either Mega-2.IVa or erm(B), had a growth fitness advantage over the lower-level, mef(E)/mel-mediated macrolide-resistant S. pneumoniae phenotypes. These data indicate the ability of S. pneumoniae to generate high-level macrolide resistance by macrolide efflux/ribosomal protection [Mef(E)/Mel] and that high-level resistance regardless of mechanism provides a fitness advantage in the presence of macrolides.
Streptococcus pneumoniae, the pneumococcus, is an obligate commensal of the human nasopharynx and a worldwide opportunistic pathogen. S. pneumoniae causes non-invasive diseases such as acute otitis media, sinusitis, and pneumonia, as well as invasive diseases such as sepsis and meningitis (O’Brien et al., 2009). Antibiotic therapy for community-associated upper respiratory tract bacterial infections where S. pneumoniae is suspected often includes a macrolide (Suda et al., 2014; Hicks et al., 2015). In the United States, macrolides are one of the most prescribed antibiotics with 190 prescriptions per 1000 people in 2011 (Hicks et al., 2015). However, macrolide effectiveness has been compromised by the emergence of macrolide resistance in S. pneumoniae in the early to mid-1990s (Gay et al., 2000; Hyde et al., 2001). The continuing widespread use of macrolides has resulted in a strong selective pressure contributing to the expansion of macrolide-resistant S. pneumoniae (Appelbaum, 2002). While the pneumococcal conjugate vaccines (PCV7, PCV13), introduced in the US in 2000 and 2010, respectively, have significantly reduced the burden of pneumococcal disease as well as the overall incidence of antibiotic resistance in pneumococci, the strong selection pressure for resistance continues in the population.
Macrolide resistance in S. pneumoniae is predominantly due to ribosomal modification or macrolide efflux/ribosomal protection (Roberts et al., 1999; Schroeder and Stephens, 2016; Murina et al., 2018; Su et al., 2018). Macrolides bind to the 23S rRNA (predominantly at residue A2058 for Escherichia coli) of the 50S ribosome to inhibit protein synthesis (Weisblum, 1995b). A ribosomal methyltransferase, encoded by erm(B), prevents binding of macrolides by dimethylating the target site on the ribosome (Weisblum, 1995a). Ribosomal methylation results in high-level macrolide resistance (erythromycin MIC > 256 μg/ml) as well as resistance to lincosamides and streptogramin B (the MLSB phenotype). erm(B) is carried on a group of mobile genetic elements including the erm(B) element and complex elements including the erm(B) element inserted into Tn916 (Tn6002); or erm(B) is carried on Tn917 that can be inserted into Tn916 (Tn3872) (Brenciani et al., 2007; Chancey et al., 2015a).
Macrolide efflux/ribosomal protection is carried on the Macrolide Efflux Genetic Assembly (Mega), a 5.5 kb (Mega-1), or 5.4 kb (Mega-2) genetic element carrying mef(E) and mel, encoding a proton motive force efflux pump and a ribosomal protection protein (Tait-Kamradt et al., 1997; Gay and Stephens, 2001; Schroeder and Stephens, 2016). Mega is found in at least five locations in the pneumococcal genome (most common insertion classes being I, II, III, and IV) and also can be a component of larger genetic elements (e.g., insertion class V) (Gay and Stephens, 2001; Del Grosso et al., 2004; Chancey et al., 2015a). The mef(E)/mel operon confers resistance to 14- and 15-membered macrolides only (the M phenotype) and is inducible by these same macrolide molecules (Ambrose et al., 2005; Chancey et al., 2011). In the United States, most pneumococcal macrolide resistance has been due to mef(E)/mel (Stephens et al., 2005; Rudolph et al., 2013; Schroeder and Stephens, 2016). Mega-mediated macrolide resistance has been generally reported as MICs of erythromycin as 1–16 μg/ml (Ambrose et al., 2005; Chancey et al., 2012), and the clinical significance of Mega-mediated macrolide resistance has been debated (Cilloniz et al., 2015).
S. pneumoniae isolates with both macrolide resistance determinants, erm(B) and Mega [mef(E)/mel], have been identified (Corso et al., 1998; Luna et al., 1999; Nishijima et al., 1999; McGee et al., 2001; Del Grosso et al., 2006; Rudolph et al., 2013). In Atlanta, we documented the emergence and clonal expansion of macrolide-resistant serotype 19A clonal complex 320 isolates that contain Tn2010, containing both the erm(B) element in orf20 and Mega in orf6 of Tn916 (Chancey et al., 2015a; Schroeder et al., 2017).
In this report, new high-level macrolide resistance of S. pneumoniae (MIC ≥ 16–256 μg/ml) due to the Mega element alone was identified and the genetic basis investigated. We also assessed the contribution of erm(B) and mef(E)/mel in the Tn2010 dual element resistance isolates. Further, we found high-level macrolide resistance, regardless of mechanism, provided a competitive growth advantage during exposure to erythromycin.
The characteristics of S. pneumoniae strains used are listed in Tables 1, 2. A panel of 44 epidemiologically distinct, well-characterized Mega containing macrolide-resistant clinical isolates was investigated (Table 1). The S. pneumoniae isolates in this panel were selected from over 13,000 S. pneumoniae isolates identified through active prospective population-based surveillance of invasive pneumococcal disease in Atlanta between 1994 and 2011 (Schuchat et al., 2001; Chancey et al., 2015a). The isolates selected all contained the Mega element [mef(E)/mel] identified by PCR and/or nucleotide sequencing, and were chosen to evaluate resistance profiles based on the different locations of each Mega insertion site (Chancey et al., 2015a). Where possible, five strains for each insertion site were evaluated. Isolates were preferentially selected for which whole genome sequence data was available, and reflected a variety of capsule serotypes, antibiotic resistance phenotypes, and dates of isolation (Chancey et al., 2015a). For experiments, all S. pneumoniae strains were routinely grown on trypticase soy agar II containing 5% sheep’s blood (blood agar) or in Todd-Hewitt broth containing 0.5% yeast extract (THY). Plate cultures were grown at 37°C with 5% CO2 and broth cultures were grown in a 37°C water bath.
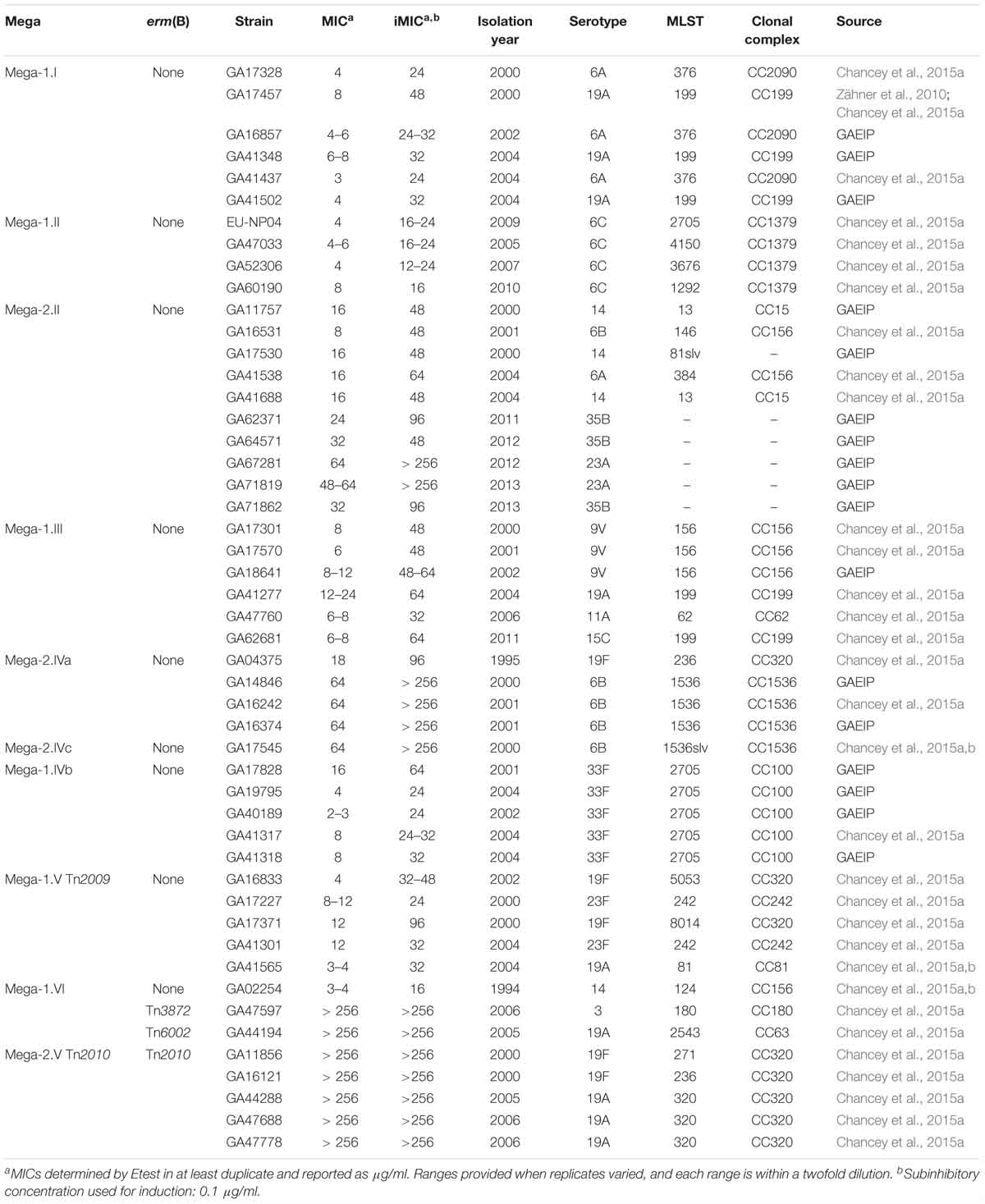
Table 1. S. pneumoniae isolates (including isolation year, serotype, MLST, clonal complex, and source) with Mega (genetic insertion site), erm(B), or both, and minimum inhibitory concentrations (MICs) to erythromycin.
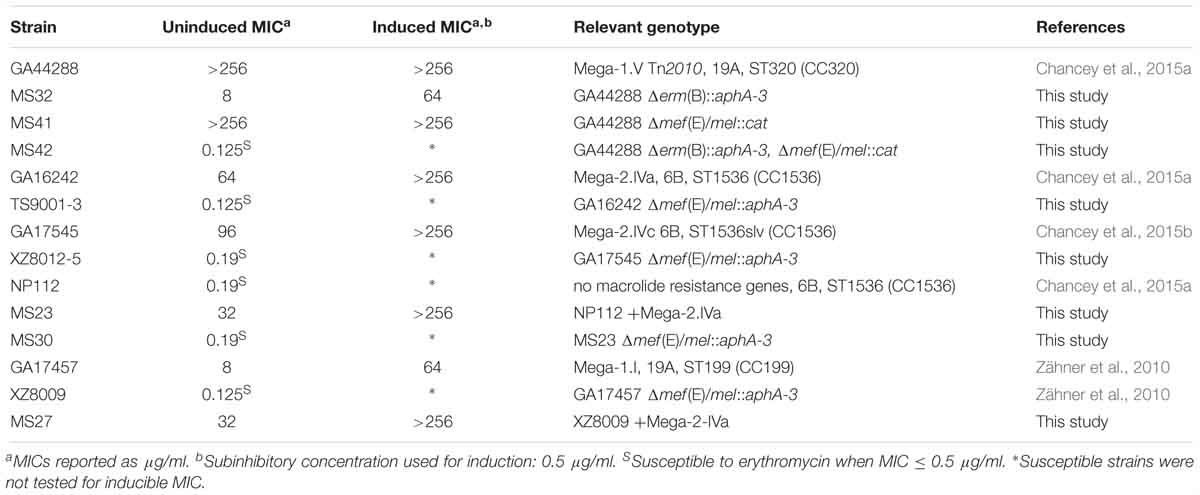
Table 2. Erythromycin minimum inhibitory concentrations (MICs) for S. pneumoniae strains and mutants [Mega insertion, serotype, MLST (clonal complex)] used in this study.
To determine macrolide MICs, bacterial cultures were grown overnight on blood agar and subcultured onto blood agar or blood agar with 0.1 μg/ml erythromycin supplementation to induce resistance expression as a standard protocol in our laboratory (Zähner et al., 2010; Chancey et al., 2011). These overnight cultures were suspended to a density approximately equal to a 0.5 McFarland standard and streaked onto Mueller-Hinton agar containing 5% sheep’s blood. Erythromycin susceptibility tests were performed by applying an erythromycin Etest strip (bioMérieux). After an overnight incubation, erythromycin susceptibility was measured. Uninduced MICs of 1–16 μg/ml were classified as resistant and those >16 μg/ml were classified as high-level resistant.
To evaluate the basis for high level macrolide resistance in S. pneumoniae, a series of mutants were created (Table 2). Primer sequences are listed in Supplemental Table A1. PfuUltra II Fusion DNA polymerase (Agilent Technologies) or Q5 polymerase (New England Biolabs), restriction enzymes (New England Biolabs) and T4 DNA ligase (Invitrogen) were used for mutational cassette construction. Taq DNA polymerase (Applied Biosystems) or One Taq DNA polymerase (New England Biolabs) were used for screening putative mutants.
S. pneumoniae was transformed by a standard method that utilized the competence-stimulating peptide 1 (CSP-1) for induction of competence (Havarstein et al., 1995). CSP-1 was synthesized by the Emory University Microchemical Facility. Transformations were performed using plasmid DNA or PCR products and selected on blood agar containing kanamycin at 400 μg/ml, erythromycin at 1 μg/ml, or chloramphenicol at 3.2 μg/ml as described below. For mutants TS9001-3 and XZ8012-5, competent cells were transformed with a previously created plasmid that replaces mef(E) and mel with an aphA-3 cassette and double crossover mutants were selected on kanamycin and confirmed by PCR and sequencing (Zähner et al., 2010). This method was also used to delete mef(E)/mel from GA16242 to create TS9001-3 and from GA17545 to create XZ8012-5 (Table 2).
To generate mutants MS23 and MS27, a 10.9 kb PCR product containing Mega-2.IVa was amplified using primers SC173 and SC251 and purified using the QIAquick Gel Extraction kit. Purified PCR products were transformed into NP112 to create MS23 and XZ8009 to create MS27 and transformants were selected on erythromycin. Insertions were confirmed by PCR of the left and right junctions of Mega-2 in insertion site IVa with primers SC10 with SC173 and SC70 with SC251 (Table 2). For mutant MS30, MS23 was transformed with BamHI digested mef(E)/mel::aphA-3 plasmid. The desired double crossover was selected on kanamycin. The insertion was confirmed by PCR amplification of a 1521 bp product with primers SC125 and kanA (Table 2).
To generate mutant MS32, GA44288 genomic DNA was amplified upstream (primers MS34 and MS35) and downstream (primers MS36 and MS37), and regions of erm(B) were spliced by overlapping extension (SOE) to create an internal XbaI site using the PCR amplified regions with primers MS34 and MS37. The resulting 1066 bp product was digested with BamHI and PstI and cloned into double-digested pUC19 vector to create pMRS11. The kanamycin resistance cassette, aphA-3 was PCR amplified from pSF151 (Tao et al., 1992) using primers MS53 and MS54 and the product was XbaI digested and cloned into pMRS11 to create pMRS13. The aphA-3 cassette was confirmed to be in the forward direction by PCR with primers MS34 and MS54. Transformation of GA44288 cells with pMRS13 and selection on kanamycin to created strain MS32, which was confirmed by PCR amplification with primers MS27 and kanA as well as MS28 and kanC (Table 2).
Finally, to generate mutants MS41 and MS42, PCR amplification of the mef(E) upstream region by primers MS64 and MS72, the mel downstream region by primers MS63 and MS69, and the chloramphenicol cassette from pEVP3 (Claverys et al., 1995) by primers MS70 and MS71 was performed. A single SOE PCR reaction with the three PCR products and primers MS63 and MS64 created a 2 kb Δmef(E)/mel::cmR cassette. This product was used for transformation and selection on chloramphenicol for GA44288 to create MS41 and MS32 to create MS42, which were confirmed by PCR amplification of a 2 kb product from primers MS63 and MS64 (Table 2).
To determine mef(E)/mel expression, overnight blood agar cultures were first suspended in THY and grown to mid-log phase (OD600 = 0.3–0.5). Each culture was diluted to OD600 = 0.05 in prewarmed THY, grown to mid-log phase, and cultures were divided into tubes with or without erythromycin as indicated and continued to grow until the indicated treatment time was achieved. Culture aliquots were mixed with RNAprotect Bacterial Reagent (Qiagen) and RNA was isolated using the RNeasy Mini Kit (Qiagen). DNA was removed via the TURBO DNA-free (Applied Biosystems) and confirmed to be free of DNA by PCR using primers for genes of interest (Supplemental Table A1). QuantiTect Reverse Transcription Kit (Qiagen) was used to create cDNA from the purified RNA. qRT-PCR was performed using iQ SYBR Green Supermix (BioRad) with an iCycler iQ Real-Time Detection System (BioRad). qRT-PCR primers are listed in Supplemental Table A1. The measured CT values were normalized using 16S rRNA, averaged, and wild type untreated condition was used to calculate the relative expression, ΔΔCT value.
To determine if a competitive advantage resulted from high level macrolide resistance in S. pneumoniae competition assays were performed. Bacterial growth competitions were developed based on methods of Gupta et al. (2013). Overnight blood agar cultures were subcultured onto blood agar with or without supplementation with erythromycin at 0.5 μg/ml. Each strain was suspended in THY broth with or without erythromycin (0.5 μg/ml), grown to OD600 = 0.5–0.7 before dilution to OD600 = 0.050 in fresh media. Diluted cultures were mixed (1:1) for competition assays or grown independently as non-competition controls and grown to OD600 = 0.5–0.7 and diluted 200-fold in fresh media. Cultures were subcultured three times allowing the cultures to grow for approximately 50 generations. Sampling of cultures was performed to monitor growth phase by OD600. At T = 0 and each time the cultures reached late-log/stationary phase, culture aliquots were collected, serially diluted in phosphate-buffered saline, and plated on blood agar without selection (total culture density) and selective blood agar (one of the mutants): kanamycin 400 μg/ml, erythromycin 1 μg/ml, chloramphenicol 3.2 μg/ml, or tetracycline 2 μg/ml. The competitive index (CI) was calculated as CI = (mutant CFUoutput/wildtype CFUoutput)/(mutant CFUinput/wildtype CFUinput) and a CI < 1 indicates the mutant is less fit than then the wildtype.
Unpaired, two-tailed t-tests with 95% confidence intervals were performed using Prism® 5 (GraphPad). For the growth competition experiments, the competitive index values of input were compared to the endpoint of 50 generations of growth.
In a large population-based collection of over 13,000 S. pneumoniae, Mega-containing macrolide resistant pneumococcal isolates were identified (section Materials and Methods). Table 3 shows the incidence of macrolide-resistant invasive pneumococcal disease (MR-IPD) 1999–2016 in Health District-3, Atlanta, GA, by macrolide resistance genotype across all ages before and after PCV7 introduction in 2000 and PCV13 introduction in 2010. Through 2016, 6 years after the introduction of PCV13 in the US, overall invasive pneumococcal disease was 6.94/100,000, macrolide resistance 2.0/100,000, mef(E)/mel resistance 1.11/100,000; and erm(B) resistance 0.67/100,000 (Table 3).
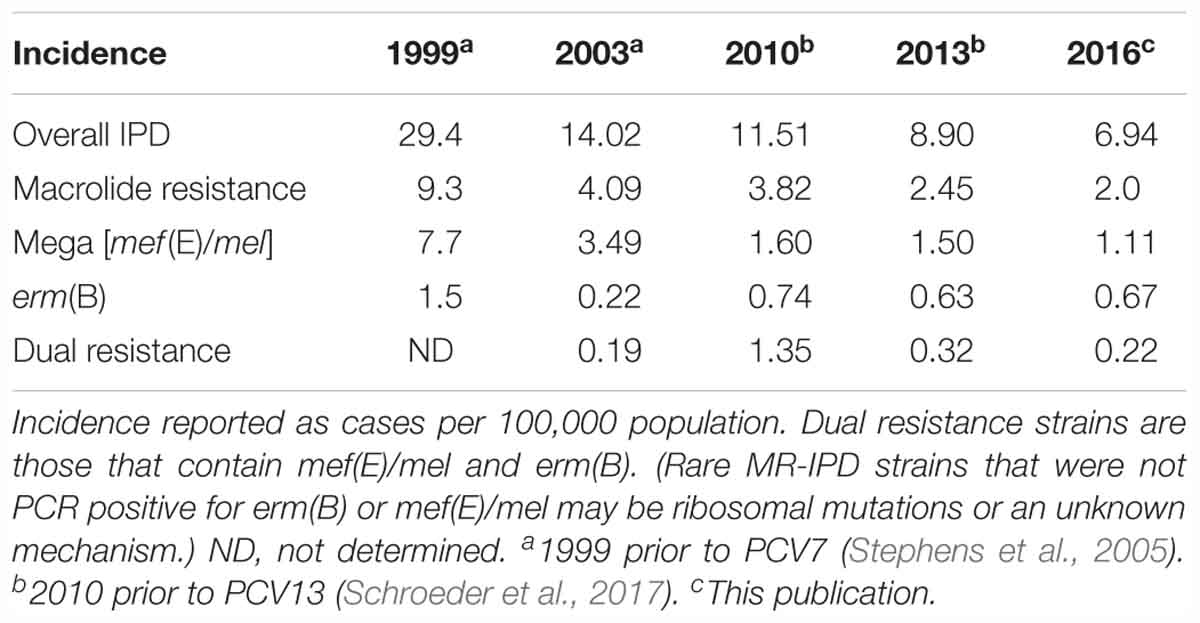
Table 3. Incidence of macrolide-resistant invasive pneumococcal disease (MR-IPD) 1999–2016 in Health District-3, Atlanta, GA, by macrolide resistance genotype across all ages.
Forty-four macrolide resistant S. pneumoniae isolates were chosen for further study, and demonstrated a wide range of resistance to erythromycin with MICs of 2 to > 256 μg/ml (Table 1 and Figure 1). Isolates with erythromycin MICs of 1–16 μg/ml were classified as “resistant” and we defined isolates with MICs > 16 μg/ml as “high-level resistant.” After overnight induction with subinhibitory erythromycin (0.1 μg/ml), the erythromycin MICs for these strains increased to 16 to > 256 μg/ml (Table 1 and Figure 1).
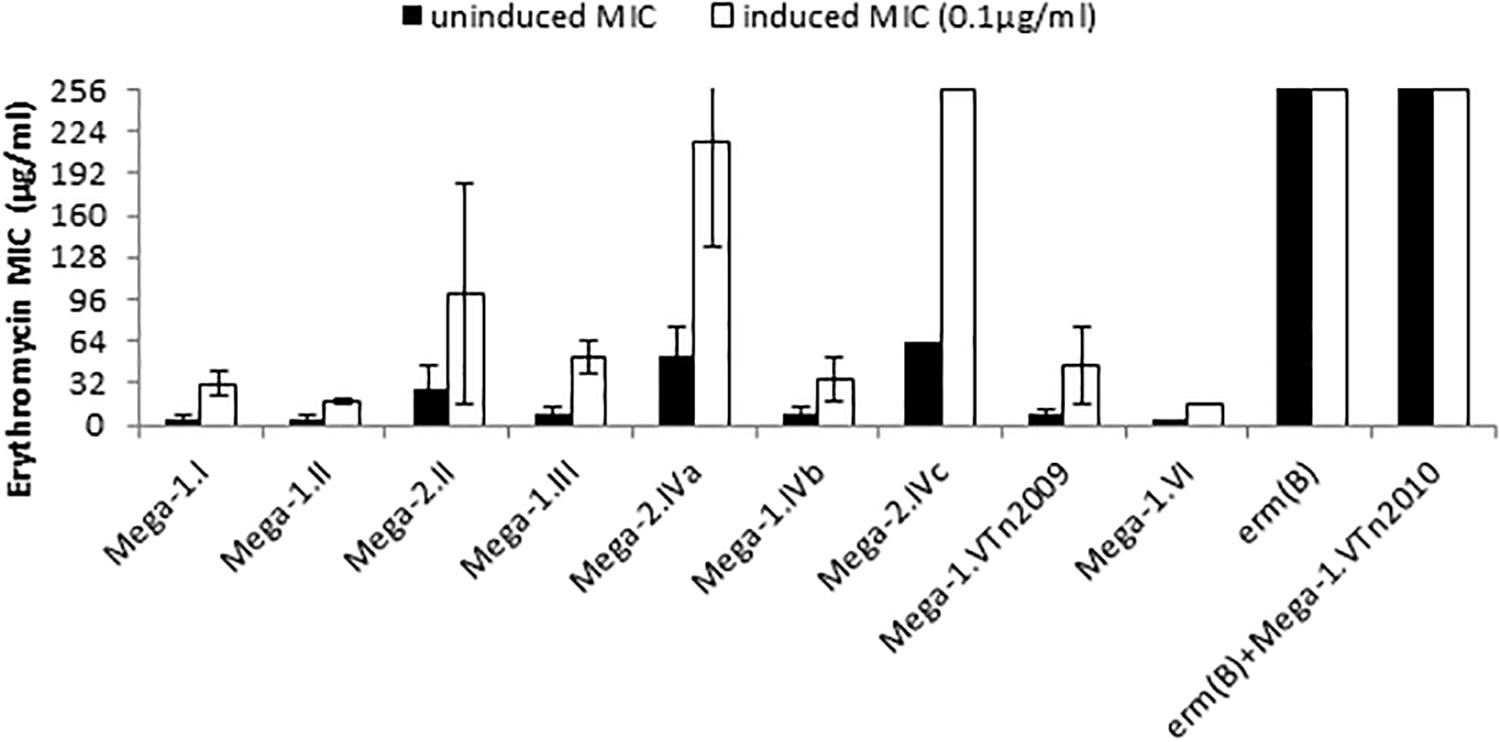
Figure 1. Macrolide resistance phenotypes and genotypes of S. pneumoniae. Erythromycin minimum inhibitory concentrations (MICs) were determined under uninduced conditions (black bars) and cultures induced with 0.1 μg/ml erythromycin (white bars). Each bar is the average MIC for a macrolide resistance genotype and strains in each group are detailed in Table 1.
The relationship of MIC to differences in the mef(E)/mel intergenic region and the genomic sites of Mega insertion was assessed. Mega type 1 (Mega-1) and Mega type 2 (Mega-2) (Gay and Stephens, 2001), differentiated by a 99 bp insertion/deletion of the intergenic region between mef(E) and mel, did not contribute significantly to high-level macrolide resistance. Most Mega-1.II- and Mega-2.II-containing strains were found to have similar erythromycin and induced-erythromycin MICs (Figure 1). Strains with chromosomal insertion sites Mega-1.I, Mega-1.II, Mega-2.II, Mega-1.III, Mega-1.IVb, and Mega-1.VTn2009 typically have uninduced erythromycin MICs 2–16 μg/ml (Table 1 and Figure 1), levels of macrolide resistance previously associated with Mega in S. pneumoniae (Gay and Stephens, 2001; Ambrose et al., 2005; Chancey et al., 2011), but with subinhibitory erythromycin induction MICs increased to 16–64 μg/ml (Table 1 and Figure 1). These isolates were also susceptible to clindamycin and thus have an M-phenotype. The newly described Mega-1.novel insertion (Chancey et al., 2015a), herein named Mega-1.VI, exhibited an MIC of 4 μg/ml and an induced-erythromycin MIC of 16 μg/ml.
S. pneumoniae containing Mega-2.IVa or Mega-2.IVc insertions exhibited intrinsic high-level macrolide resistance, with uninduced erythromycin MICs of >18–256 μg/ml erythromycin and induced MICs of >96 to > 256 μg/ml erythromycin (Table 1 and Figure 1). The Mega-2.IVa and Mega-2.IVc isolates were found to be clindamycin susceptible (M phenotype). No other macrolide resistant determinants were found in these isolates, and mutation of the Mega element resulted in erythromycin sensitivity with MICs of 0.125 μg/ml (Table 2).
As anticipated, Mega-1.V Tn2010 isolates containing both erm(B) and Mega (Del Grosso et al., 2006) had uninduced erythromycin MICs of >256 μg/ml and were clindamycin resistant (MLSB phenotype). To better understand the relative roles of mef(E)/mel and erm(B) in high-level macrolide resistant Tn2010-containing S. pneumoniae, isogenic deletion mutations of erm(B) or mef(E)/mel or both were made in strain GA44288, an invasive pneumococcal disease isolate (Chancey et al., 2015a). The deletion of mef(E)/mel from GA44288, strain MS41 [Tn2010Δmef(E)/mel], had no effect on erythromycin or clindamycin resistance, as MS41 remained highly resistant to erythromycin (MIC of > 256 μg/ml) (Table 2). The deletion of erm(B) in GA44288 generated the mutant MS32 [Tn2010Δerm(B)], which displayed an M phenotype with an erythromycin MIC of 8 μg/ml, an induced erythromycin MIC of 64 μg/ml (Table 2), and susceptibility to clindamycin. These MIC data for MS32 are consistent with the majority of Mega-only containing isolates (Table 1 and Figure 1). The deletion of the dual macrolide resistance determinants [mef(E)/mel and erm(B)] in GA44288 was designated MS42 [Tn2010 Δerm(B)Δmef(E)/mel]. MS42 was susceptible to erythromycin (MIC 0.125 μg/ml) and clindamycin (Table 2). These data confirmed erm(B) and mef(E)/mel as the only macrolide resistance determinants in GA44288 and that the high-level macrolide resistance of Tn2010-containing S. pneumoniae was due to the presence of erm(B).
In S. pneumoniae, the expression of mef(E) and mel is controlled through transcriptional attenuation (Chancey et al., 2015b). Macrolide-induced ribosomal stalling results in deattenuation of mef(E)/mel to produce full-length polycistronic transcripts (Chancey et al., 2015b). To determine if mef(E)/mel was expressed in the presence of erm(B), mef(E) expression was measured by qRT-PCR from GA44288 after a 15 min exposure to erythromycin. The expression of mef(E) was dose-dependent, and 0.5 μg/ml erythromycin was sufficient to induce similar mef(E) expression in both the wild type and the erm(B) deletion mutant (data not shown). Thus, in isolates with both erm(B) and Mega, mef(E)/mel expression was induced by erythromycin and shown by the resistance data to result in a functional efflux pump/ribosomal protection protein.
The molecular basis for Mega-2.IVa and Mega-2.IVc isolates with high-level macrolide resistance but the M phenotype was further assessed. In the high-level macrolide-resistant strain GA16242 with a Mega-2.IVa insertion (uninduced erythromycin MIC of 64 μg/ml) mef(E)/mel was deleted to create TS9001. TS9001 was susceptible to erythromycin at 0.125 μg/ml (Table 2). Similarly, the deletion of mef(E)/mel from the high-level macrolide-resistant Mega-2.IVc strain GA17545 (uninduced erythromycin MIC of 64 μg/ml) resulted in susceptibility to macrolides as the erythromycin MIC of the mutant designated XZ8012-5 was 0.19 μg/ml (Table 2). Thus, mef(E)/mel alone in S. pneumoniae Mega-2.IVa and Mega-2.IVc isolates was responsible for high-level macrolide resistance.
To further confirm that the Mega class IVa and IVc insertions resulted in high-level macrolide resistance and determine whether the high-level macrolide resistance phenotype was transferable, the Mega-2.IVa insertion was transformed into the erythromycin susceptible strain NP112 (MIC 0.19 μg/ml). The resulting NP112 Mega-2.IVa isolate (designated MS23) demonstrated high-level macrolide resistance with an erythromycin MIC of 32 μg/ml and was inducible up to >256 μg/ml (Table 2). The transfer of Mega-2.IVa included the adjacent ISSmi2 element and recreated the pneumococcal pathogenicity island (PPI-1) deletion found in Mega-2.IVa isolates (Chancey et al., 2015a). Deletion of mef(E)/mel from MS23 (designated MS30) restored macrolide susceptibility (Table 2). Mega-mediated high-level macrolide resistance was also transferred to the GA17457 Δmef(E)/mel deletion strain (XZ8009). After transformation of XZ8009 with the Mega-2.IVa insertion (designated MS27), MS27 was found to have an erythromycin MIC of 32 μg/ml inducible up to >256 μg/ml (Table 2).
To determine whether erm(B) and/or mef(E)/mel in S. pneumoniae provided a competitive advantage for growth during exposure to erythromycin, competitive assays using the clinical isolate GA44288 containing erm(B) and mef(E)/mel on Tn2010, and the strains with mutations in these genes (Table 2) were performed. Erythromycin-induced cultures of wild type strain GA44288 [erm(B) and mef(E)/mel on Tn2010] and the isogenic mutants MS32 [Δerm(B), mef(E)/mel] and MS41 [erm(B), Δmef(E)/mel)] (Table 2) were used in an in vitro competitive index (1:1 ratio). The concentration of erythromycin used is known to be achieved in human serum during treatment (0.5 μg/ml) (Metzler et al., 2013). No significant difference in growth was observed between MS41 [erm(B), Δmef(E)/mel] and the wild type strain GA44288 [erm(B), mef(E)/mel] in this assay (Figure 2A, p = 0.5460). This suggested that mef(E)/mel did not provide a growth advantage to an erm(B)-containing strain during exposure to erythromycin. When MS32 [Δerm(B), mef(E)/mel] was competed with GA44288 [wild type, Tn2010 with erm(B), mef(E)mel] the competitive index decreased to approximately 0.01 after 50 generations (Figure 2B). These data indicated a significant growth advantage due to the high-level resistance encoded by erm(B) for GA44288 during exposure to erythromycin (p = 0.0012). Similarly, the competitive index of MS32 [Δerm(B), mef(E)/mel] versus MS41 [erm(B), Δmef(E)/mel] dropped to approximately 0.01 by the endpoint 50 generations (Figure 2C, p < 0.0001), again confirming the importance of erm(B)-induced high-level macrolide resistance during exposure to erythromycin. When these experiments were performed without erythromycin, the competitive indexes were at ∼1 throughout the course of the experiments (Supplemental Figure A1). Thus, in each of the erythromycin-induced competitive index experiments, an active erm(B)-containing strain (GA44288 and MS41) had a competitive advantage over the erm(B) deletion stain (Figure 2). The data suggest that erm(B) provides a growth advantage when S. pneumoniae is exposed to treatment-level concentrations of erythromycin.
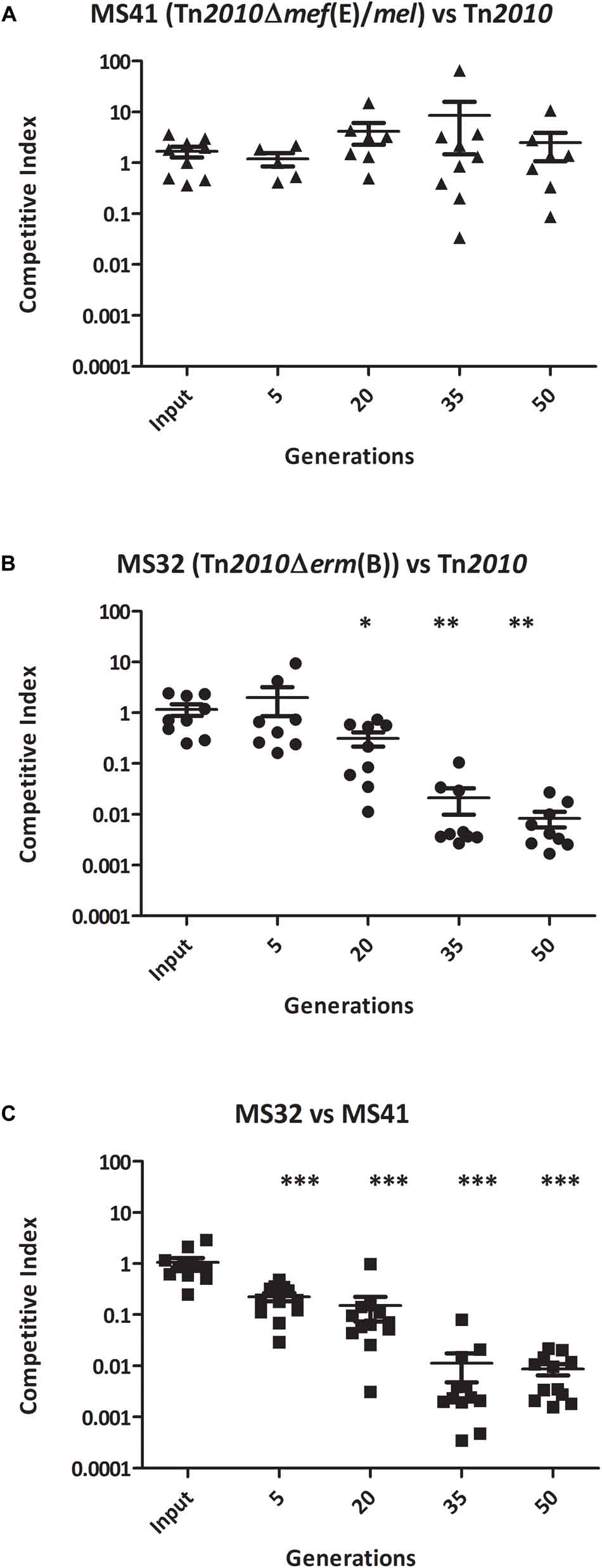
Figure 2. The competitive index of isogenic GA44288 Tn2010 mutants grown in vitro with erythromycin (0.5 μg/ml) for approximately 50 generations: (A) MS41 [Tn2010Δmef(E)/mel] versus Tn2010, (B) MS32 [Tn2010Δerm(B)] versus Tn2010, and (C) MS32 [Tn2010Δerm(B)] versus MS41 [Tn2010Δmef(E)/mel]. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
To determine whether the competitive advantage for growth of erm(B) during erythromycin exposure was due Erm(B)-mediated ribosomal methylation or to the high-level macrolide resistance of the erm(B)-containing strains, the competitive index assay was performed with the GA44288 isogenic strains in competition with GA16242, the Mega-2.IVa strain that produced high-level macrolide resistance due only to the presence of mef(E)/mel (Figure 1 and Table 2). The competitive index for GA44288 versus GA16242 remained ∼1 throughout the course of the experiments (Figure 3A, p = 0.3088). The competitive index for MS41 [erm(B), Δmef(E)/mel] versus GA16242 [mel(E)/mef, Mega-2.IVa] also did not change throughout the experiments (Figure 3B, p = 0.4397). These data suggest that high-level macrolide resistance and not Erm(B)-mediated ribosomal methylation was responsible for the growth advantage in erythromycin. Finally, we assayed MS32 [GA44288 Δerm(B), mel(E)/mef] with GA16242 and found the competitive index decreased below 0.1 after 50 generations of growth (Figure 3C, p = 0.0316). Thus, both high-level macrolide resistance strains (GA44288 and GA16242), albeit generated by different mechanisms, provided the growth competitive advantage during exposure to erythromycin.
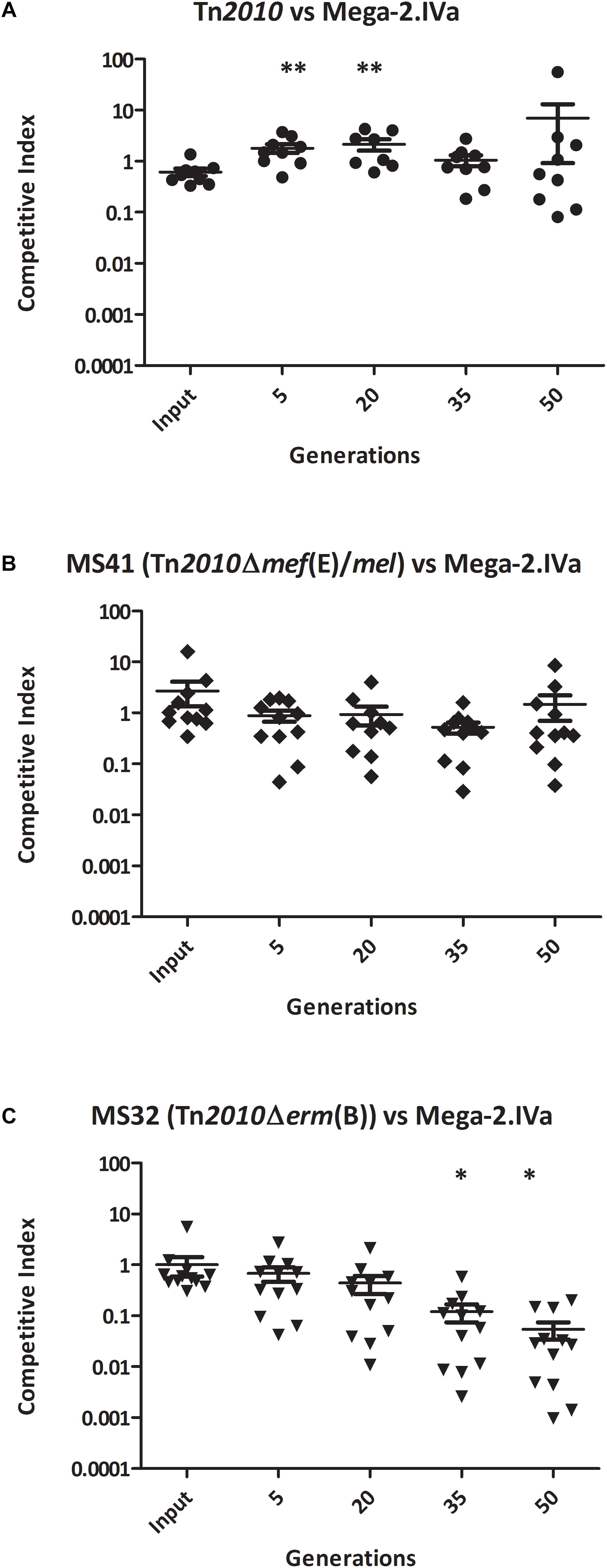
Figure 3. The competitive index of high-level macrolide resistance strains with distinct mechanisms [erm(B) and Mega-2.Iva] in vitro with erythromycin (0.5 μg/ml) grown for approximately 50 generations: (A) GA44288 [erm(B)-and mef(E)/mel-containing] versus GA16242 (Mega-2.IVa-containing), (B) MS41 [GA44288 Δmef(E)/mel, erm(B)-containing] versus GA16242 (Mega-2.IVa-containing), and (C) MS32 [GA44288 Δerm(B)] versus GA16242 (Mega-2.IVa-containing). ∗p < 0.05, ∗∗p < 0.01.
In the United States, the rapid rise of macrolide resistance (includes resistance to erythromycin, azithromycin, clarithromycin and other 14- and 15-membered macrolides) in S. pneumoniae throughout the 1990s was due to the horizontal transfer and clonal expansion of stains containing the mef(E)/mel encoded efflux pump and ribosomal protection protein. The mef(E)/mel operon is found in the pneumococcal genome on the 5.4–5.5 kb Mega element (Stephens et al., 2005; Rudolph et al., 2013). Mega-mediated pneumococcal macrolide resistance has generally been associated with lower levels of macrolide MICs (1–16 μg/ml to erythromycin) compared to high levels of macrolide resistance observed for erm(B) (usually > 256 μg/ml to erythromycin) (Rudolph et al., 2013). The use of in vitro assays to query MIC values may be a limitation, however, macrolide resistance in the pneumococcus, caused by either erm(B) or mef(E)/mel-containing isolates, has been linked to treatment failures for lower respiratory tract infections and bacteremia (Klugman, 2002; Lonks et al., 2002; Schentag et al., 2007; Zähner et al., 2010). In one study by Gonzalez et al. (2004) 10/11 isolates from azithromycin treatment failures, who did not have underlying medical conditions, had the erm(B) (two patients) or Mega (eight patients) phenotype and three of the six available Mega-containing isolates had MICs ≥ 16 μg/ml.
We previously defined the genetic basis for the resistance mediated by the mef(E)/mel-containing Mega element in S. pneumoniae, demonstrated that mef(E) and mel are inducible by most 14- and 15-membered macrolides (including erythromycin, clarithromycin, and azithromycin) and by antimicrobial peptides, defined the Mega element that contains mef(E)/mel, identified the mechanism of macrolide induction of mef(E)/mel, demonstrated Mega’s relationship to conjugative transposons, and mapped the locations of Mega in the pneumococcal genome (Gay and Stephens, 2001; Zähner et al., 2010; Chancey et al., 2011, 2015a,b). Mega elements are either type 1 or 2, distinguished by a 99 bp insertion/deletion (Mega-1 at 5.5 kb and Mega-2 at 5.4 kb) (Gay and Stephens, 2001). Mega is found in the pneumococcal genome in six distinct sites, termed Mega classes (Chancey et al., 2015a). Insertion sites numbered I-IV were originally described (Gay and Stephens, 2001). When inserted into orf6 of Tn916-like elements, Mega is classified as insertion site V (Del Grosso et al., 2004, 2006, 2007). Recently, we reported a new chromosomal insertion site, VI (Chancey et al., 2015a). Mega class IV has been further classified into three subclasses: Mega-2.IVa, Mega-1.IVb, and Mega-2.IVc all of which are upstream of the Pneumococcal Pathogenicity Island-1 (PPI-1) (Chancey et al., 2015a).
High-level macrolide resistance (MIC > 16 μg/ml) due to Mega has been observed in clinical isolates (Gonzalez et al., 2004; Rudolph et al., 2013) but has not been not characterized at a genomic level. We found high-level macrolide resistance in S. pneumoniae due to Mega related to specific genomic insertions Mega-2.IVa and Mega-2.IVc. We also have recent evidence of M-phenotype high-level macrolide resistance in Mega-2.II strains (unpublished data). Most Mega-2.IVa and Mega-2.IVc have high-level macrolide resistance while the Mega-1.IVb insertions have MICs similar to the other Mega insertion sites (Chancey et al., 2015a). The molecular mechanism of high-level macrolide resistance due to efflux and ribosomal protection in Mega-2.IVa/c and the newly discovered Mega-2.II isolates is not understood and is a limitation of this study, but is currently under investigation. One possibility is that high level macrolide resistance is related to genomic “trans” factors favoring higher expression the mef/mel operon, and this is associated with specific clonal complexes such as ST156 or ST15. Baseline MICs and mef/mel operon expression are elevated in strains with high level resistance. The 99-bp insertion in the intergenic region between mef(E) and mel, the distinguishing characteristic between Mega-1 from Mega-2, does not appear to significantly contribute to high-level macrolide resistance.
S. pneumoniae containing both macrolide resistance genetic elements, mef(E)/mel and erm(B), were first noted in the late-1990s from the US, Japan, and South Africa (Corso et al., 1998; Luna et al., 1999; Nishijima et al., 1999; McGee et al., 2001). Many of these strains were initially noted to belong to a single 19F multidrug resistant clone and subsequent work identified this clone as lineage Taiwan19F-14, or PMEN14 (McGee et al., 2001; Croucher et al., 2014). Dual element macrolide-resistant serotypes, especially 19A S. pneumoniae belonging to CC320 (formerly CC271) a PMEN14 lineage (Del Grosso et al., 2007; Bowers et al., 2012; Chancey et al., 2015a), have increased steadily in the US and worldwide (Moore et al., 2008; Mahjoub-Messai et al., 2009; Schroeder et al., 2017). For example, clonal expansion of dual macrolide-resistant 19A S. pneumoniae expanded dramatically in the US after the introduction of the pneumococcal conjugate vaccine, PCV7 (Moore et al., 2008). We documented the decline, 2010–2013 of serotype 19A (CC 320) with dual element macrolide resistance mechanisms following the introduction of PCV13 in 2010 (Schroeder et al., 2017). As part of this study, we analyzed our 2013–2016 population data set and isolate collection to further understand the impact of PCV13 on macrolide resistance in the population and the continued importance of the dual element resistance strains.
Through 2016, 6 years after the introduction of PCV13, the incidence of invasive pneumococcal disease has declined but the percent of these isolates that are macrolide resistance has remained at ∼30%. The erm(B)/mef(E)/mel dual element resistance strains continue to circulate with an incidence of 0.22/100,000 in the population (Table 3). These dual element resistance strains remain primarily 19A (CC320). While the samples for this study were collected from a defined geographic area, this potential limitation is moderated by the persistence of dual element resistant strains which were also observed throughout the US in 2016.
S. pneumoniae isolates with the dual element macrolide resistance genotype are carried on the mobile Tn2010 and are multidrug resistant (McGee et al., 2001; Bowers et al., 2012; Chancey et al., 2015a). Tn2010 is a large 26⋅4-kb element with Mega [mef(E)/mel] and Tn917 [erm(B)] inserted at two distinct sites into a Tn916-like conjugative transposon (Del Grosso et al., 2007). A possible origin of Tn2010 is via homologous recombination of a Tn2009 (Mega-containing) strain acquiring a Tn6002 fragment with erm(B) flanked by Tn916 orf20, or a Tn6002 [erm(B)-containing] strain acquiring a Tn2009 fragment with Mega flanked by Tn916 orf6 (Chancey et al., 2015a; Schroeder and Stephens, 2016). This dual element macrolide resistance genotype results in an MLSB phenotype with high-level macrolide resistance. These elements are not only able to transmit between co-resident serotypes of S. pneumoniae within the nasopharyngeal niche, but also have the possibility of transmitting to other naturally competent bacteria present in the nasopharynx, such as the clinically relevant Haemophilus influenzae (Lattar et al., 2018).
In this study, we found both mef(E)/mel and erm(B) are expressed and functional in the Tn2010 background. This indicates the co-expression of macrolide determinants in these strains. While high-level macrolide resistance of these S. pneumoniae isolates with the dual element macrolide resistance genotype was dominated by erm(B), which also confers resistance to lincosamides and streptogramin B, mef(E)/mel may provide other selective advantages. The presence of mef(E)/mel has been found to enhance resistance to the human antimicrobial peptide LL-37 (Zähner et al., 2010). Thus, selective pressure for the acquisition and maintenance of both mef(E)/mel and erm(B) in S. pneumoniae may be encountered in human colonization and disease.
Antibiotic resistance determinants are often inducible and provide a selective advantage over non-resistant organisms in an antibiotic-containing environment. However, the expression of these determinants can be associated with a fitness cost (Andersson and Hughes, 2010). In the pneumococcus, erm(B) commonly is inducible and tightly regulated through translational attenuation (Montanari et al., 2001; Min et al., 2008). The mef(E)/mel operon is also tightly regulated but through transcriptional attenuation (Chancey et al., 2015b). While the expression of erm(B) in S. pneumoniae by a partial attenuator deletion did not cause a growth defect when the strain was grown in vitro as a pure culture (Wolter et al., 2008), in Staphylococcus aureus deregulation of erm(C), a homolog of erm(B), results in increased expression of a subset of the proteome that causes a 10-fold fitness defect in vitro (Gupta et al., 2013).
We found that erm(B) in S. pneumoniae did not cause a growth defect and provided a competitive advantage up to 100-fold over a lower-level resistant Mega-containing strain during growth in a macrolide containing environment. Insertions of mef(E)/mel causing high-level macrolide resistance when used in the competitive growth assays also provided a substantial growth fitness increase over the lower-level resistant mef(E)/mel phenotype when exposed to erythromycin. With a low fitness burden for maintenance of mef(E)/mel and/or erm(B), these determinants are unlikely to be lost from the pneumococcal population even with reduced use of antibiotics in clinical settings (Andersson and Hughes, 2010). Active efflux of the macrolide into the environment by Mega high-level resistant strains growing at high density may provide a further fitness advantage over low-level resistant strains.
An effective measure to date in combating antibiotic-resistant pneumococcal infections has been the introduction of pneumococcal conjugate vaccines (Stephens et al., 2005). These vaccines provide individual protection for vaccinated individuals and reduce transmission of vaccine serotypes leading to herd protection for unvaccinated individuals in the same population (Rodgers and Klugman, 2011). The targeted vaccine serotypes have often had the highest rates of antibiotic resistance determinants (Stephens et al., 2005). However, as shown in the recent surveillance data macrolide resistance in genetic elements capable of horizontal transfer to new serotypes continues to persist in invasive S. pneumoniae.
In summary, likely driven by macrolide use in the population, high-level macrolide resistance in S. pneumoniae due to the Mega element alone has emerged, specifically in the context of the Mega-2.IVa or Mega-2.IVc and Mega-2.II genomic insertions. In addition, Tn2010-containing S. pneumoniae isolates that carry both erm(B) and mef(E)/mel have rapidly expanded with both resistant determinants functional in these strains. High-level macrolide resistance regardless of mechanism provides a competitive advantage for growth during exposure to macrolides.
MS, SC, and DS designed the study. MS and SL collected the data. MS, SL, SC, and DS performed the analysis. MS, SL, and DS wrote the manuscript.
The work was supported by a VA Merit Award grant, an NIH grant (RO1 AI07829) to DS, CDC’s Emerging Infections Programs funding (CDC-RFA-ck17-1701), with federal funds from the HHS NIH National Institute of Allergy and Infectious Diseases under contract number HHSN272200900009C. Funding agencies had no role in study design, data collection or interpretation, or the decision to submit the work for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Tammy Schwalm Tollison, Xiaoliu Zhou, and Aditya Gudlavalleti for assisting in laboratory work and creation of mutants. We would also like to thank the Georgia Emerging Infections Program for support and to members Sarah Satola, Stephanie Thomas, and Monica Farley. Strains were provided by the Georgia Emerging Infections Program and strain characterizations were aided by the CDC Streptococcus Laboratory and CDC Respiratory Disease Branch.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00868/full#supplementary-material
Ambrose, K. D., Nisbet, R., and Stephens, D. S. (2005). Macrolide efflux in Streptococcus pneumoniae is mediated by a dual efflux pump (mel and mef) and is erythromycin inducible. Antimicrob. Agents Chemother. 49, 4203–4209. doi: 10.1128/aac.49.10.4203-4209.2005
Andersson, D. I., and Hughes, D. (2010). Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 8, 260–271. doi: 10.1038/nrmicro2319
Appelbaum, P. C. (2002). Resistance among Streptococcus pneumoniae: implications for drug selection. Clin. Infect. Dis. 34, 1613–1620. doi: 10.1086/340400
Bowers, J. R., Driebe, E. M., Nibecker, J. L., Wojack, B. R., Sarovich, D. S., Wong, A. H., et al. (2012). Dominance of multidrug resistant CC271 clones in macrolide-resistant Streptococcus pneumoniae in Arizona. BMC Microbiol. 12:12. doi: 10.1186/1471-2180-12-12
Brenciani, A., Bacciaglia, A., Vecchi, M., Vitali, L. A., Varaldo, P. E., and Giovanetti, E. (2007). Genetic elements carrying erm(B) in Streptococcus pyogenes and association with tet(M) tetracycline resistance gene. Antimicrob. Agents Chemother. 51, 1209–1216. doi: 10.1128/AAC.01484-06
Chancey, S. T., Agrawal, S., Schroeder, M. R., Farley, M. M., Tettelin, H., and Stephens, D. S. (2015a). Composite mobile genetic elements disseminating macrolide resistance in Streptococcus pneumoniae. Front. Microbiol. 6:26. doi: 10.3389/fmicb.2015.00026
Chancey, S. T., Bai, X., Kumar, N., Drabek, E. F., Daugherty, S. C., Colon, T., et al. (2015b). Transcriptional attenuation controls macrolide inducible efflux and resistance in Streptococcus pneumoniae and in other Gram-positive bacteria containing mef/mel(msr(D) elements. PLoS One 10:e0116254. doi: 10.1371/journal.pone.0116254
Chancey, S. T., Zähner, D., and Stephens, D. S. (2012). Acquired inducible antimicrobial resistance in Gram-positive bacteria. Future Microbiol. 7, 959–978. doi: 10.2217/fmb.12.63
Chancey, S. T., Zhou, X., Zahner, D., and Stephens, D. S. (2011). Induction of efflux-mediated macrolide resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 55, 3413–3422. doi: 10.1128/AAC.00060-11
Cilloniz, C., Albert, R. K., Liapikou, A., Gabarrus, A., Rangel, E., Bello, S., et al. (2015). The effect of macrolide resistance on the presentation and outcome of patients hospitalized for Streptococcus pneumoniae pneumonia. Am. J. Respir. Crit. Care Med. 191, 1265–1272. doi: 10.1164/rccm.201502-0212OC
Claverys, J. P., Dintilhac, A., Pestova, E. V., Martin, B., and Morrison, D. A. (1995). Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene 164, 123–128. doi: 10.1016/0378-1119(95)00485-O
Corso, A., Severina, E. P., Petruk, V. F., Mauriz, Y. R., and Tomasz, A. (1998). Molecular characterization of penicillin-resistant Streptococcus pneumoniae isolates causing respiratory disease in the United States. Microb. Drug Resist. 4, 325–337. doi: 10.1089/mdr.1998.4.325
Croucher, N. J., Chewapreecha, C., Hanage, W. P., Harris, S. R., McGee, L., van der Linden, M., et al. (2014). Evidence for soft selective sweeps in the evolution of pneumococcal multidrug resistance and vaccine escape. Genome Biol. Evol. 6, 1589–1602. doi: 10.1093/gbe/evu120
Del Grosso, M., Camilli, R., Iannelli, F., Pozzi, G., and Pantosti, A. (2006). The mef(E)-carrying genetic element (mega) of Streptococcus pneumoniae: insertion sites and association with other genetic elements. Antimicrob. Agents Chemother. 50, 3361–3366. doi: 10.1128/AAC.00277-06
Del Grosso, M., Northwood, J. G., Farrell, D. J., and Pantosti, A. (2007). The macrolide resistance genes erm(B) and mef(E) are carried by Tn2010 in dual-gene Streptococcus pneumoniae isolates belonging to clonal complex CC271. Antimicrob. Agents Chemother. 51, 4184–4186. doi: 10.1128/AAC.00598-07
Del Grosso, M., Scotto d’Abusco, A., Iannelli, F., Pozzi, G., and Pantosti, A. (2004). Tn2009, a Tn916-like element containing mef(E) in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 48, 2037–2042. doi: 10.1128/aac.48.6.2037-2042.2004
Gay, K., Baughman, W., Miller, Y., Jackson, D., Whitney, C. G., Schuchat, A., et al. (2000). The emergence of Streptococcus pneumoniae resistant to macrolide antimicrobial agents: a 6-year population-based assessment. J. Infect. Dis. 182, 1417–1424. doi: 10.1086/315853
Gay, K., and Stephens, D. S. (2001). Structure and dissemination of a chromosomal insertion element encoding macrolide efflux in Streptococcus pneumoniae. J. Infect. Dis. 184, 56–65. doi: 10.1086/321001
Gonzalez, B. E., Martinez-Aguilar, G., Mason, E. O. Jr., and Kaplan, S. L. (2004). Azithromycin compared with beta-lactam antibiotic treatment failures in pneumococcal infections of children. Pediatr. Infect. Dis. J. 23, 399–405. doi: 10.1097/01.inf.0000122605.34902.49
Gupta, P., Sothiselvam, S., Vazquez-Laslop, N., and Mankin, A. S. (2013). Deregulation of translation due to post-transcriptional modification of rRNA explains why erm genes are inducible. Nat. Commun. 4:1984. doi: 10.1038/ncomms2984
Havarstein, L. S., Coomaraswamy, G., and Morrison, D. A. (1995). An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U.S.A. 92, 11140–11144. doi: 10.1073/pnas.92.24.11140
Hicks, L. A., Bartoces, M. G., Roberts, R. M., Suda, K. J., Hunkler, R. J., Taylor, T. H., et al. (2015). US outpatient antibiotic prescribing variation according to geography, patient population, and provider specialty in 2011. Clin. Infect. Dis. 60, 1308–1316. doi: 10.1093/cid/civ076
Hyde, T. B., Gay, K., Stephens, D. S., Vugia, D. J., Pass, M., Johnson, S., et al. (2001). Macrolide resistance among invasive Streptococcus pneumoniae isolates. JAMA 286, 1857–1862. doi: 10.1001/jama.286.15.1857
Klugman, K. P. (2002). Bacteriological evidence of antibiotic failure in pneumococcal lower respiratory tract infections. Eur. Respir. J. Suppl. 36, 3s–8s. doi: 10.1183/09031936.02.00400402
Lattar, S. M., Wu, X., Brophy, J., Sakai, F., Klugman, K. P., and Vidal, J. E. (2018). A mechanism of unidirectional transformation, leading to antibiotic resistance, occurs within nasopharyngeal pneumococcal biofilm consortia. mBio 9:e00561-18. doi: 10.1128/mBio.00561-18
Lonks, J. R., Garau, J., Gomez, L., Xercavins, M., Ochoa de Echaguen, A., Gareen, I. F., et al. (2002). Failure of macrolide antibiotic treatment in patients with bacteremia due to erythromycin-resistant Streptococcus pneumoniae. Clin. Infect. Dis. 35, 556–564. doi: 10.1086/341978
Luna, V. A., Coates, P., Eady, E. A., Cove, J. H., Nguyen, T. T., and Roberts, M. C. (1999). A variety of gram-positive bacteria carry mobile mef genes. J. Antimicrob. Chemother. 44, 19–25. doi: 10.1093/jac/44.1.19
Mahjoub-Messai, F., Doit, C., Koeck, J. L., Billard, T., Evrard, B., Bidet, P., et al. (2009). Population snapshot of Streptococcus pneumoniae serotype 19A isolates before and after introduction of seven-valent pneumococcal vaccination for French children. J. Clin. Microbiol. 47, 837–840. doi: 10.1128/JCM.01547-08
McGee, L., Klugman, K. P., Wasas, A., Capper, T., and Brink, A. (2001). Serotype 19f multiresistant pneumococcal clone harboring two erythromycin resistance determinants (erm(B) and mef(A)) in South Africa. Antimicrob. Agents Chemother. 45, 1595–1598. doi: 10.1128/aac.45.5.1595-1598.2001
Metzler, K., Drlica, K., and Blondeau, J. M. (2013). Minimal inhibitory and mutant prevention concentrations of azithromycin, clarithromycin and erythromycin for clinical isolates of Streptococcus pneumoniae. J. Antimicrob. Chemother. 68, 631–635. doi: 10.1093/jac/dks461
Min, Y. H., Kwon, A. R., Yoon, E. J., Shim, M. J., and Choi, E. C. (2008). Translational attenuation and mRNA stabilization as mechanisms of erm(B) induction by erythromycin. Antimicrob. Agents Chemother. 52, 1782–1789. doi: 10.1128/AAC.01376-07
Montanari, M. P., Mingoia, M., Giovanetti, E., and Varaldo, P. E. (2001). Differentiation of resistance phenotypes among erythromycin-resistant pneumococci. J. Clin. Microbiol. 39, 1311–1315. doi: 10.1128/jcm.39.4.1311-1315.2001
Moore, M. R., Gertz, R. E. Jr., Woodbury, R. L., Barkocy-Gallagher, G. A., Schaffner, W., Lexau, C., et al. (2008). Population snapshot of emergent Streptococcus pneumoniae serotype 19A in the United States, 2005. J. Infect. Dis. 197, 1016–1027. doi: 10.1086/528996
Murina, V., Kasari, M., Hauryliuk, V., and Atkinson, G. C. (2018). Antibiotic resistance ABCF proteins reset the peptidyl transferase centre of the ribosome to counter translational arrest. Nucleic Acids Res. 46, 3753–3763. doi: 10.1093/nar/gky050
Nishijima, T., Saito, Y., Aoki, A., Toriya, M., Toyonaga, Y., and Fujii, R. (1999). Distribution of mefE and ermB genes in macrolide-resistant strains of Streptococcus pneumoniae and their variable susceptibility to various antibiotics. J. Antimicrob. Chemother. 43, 637–643. doi: 10.1093/jac/43.5.637
O’Brien, K. L., Wolfson, L. J., Watt, J. P., Henkle, E., Deloria-Knoll, M., McCall, N., et al. (2009). Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374, 893–902. doi: 10.1016/S0140-6736(09)61204-6
Roberts, M. C., Sutcliffe, J., Courvalin, P., Jensen, L. B., Rood, J., and Seppala, H. (1999). Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43, 2823–2830. doi: 10.1128/AAC.43.12.2823
Rodgers, G. L., and Klugman, K. P. (2011). The future of pneumococcal disease prevention. Vaccine 29S, C43–C48. doi: 10.1016/j.vaccine.2011.07.047
Rudolph, K., Bulkow, L., Bruce, M., Zulz, T., Reasonover, A., Harker-Jones, M., et al. (2013). Molecular resistance mechanisms of macrolide-resistant invasive Streptococcus pneumoniae isolates from Alaska, 1986 to 2010. Antimicrob. Agents Chemother. 57, 5415–5422. doi: 10.1128/AAC.00319-13
Schentag, J. J., Klugman, K. P., Yu, V. L., Adelman, M. H., Wilton, G. J., Chiou, C. C., et al. (2007). Streptococcus pneumoniae bacteraemia: pharmacodynamic correlations with outcome and macrolide resistance–a controlled study. Int. J. Antimicrob. Agents 30, 264–269. doi: 10.1016/j.ijantimicag.2007.04.013
Schroeder, M. R., Chancey, S. T., Thomas, S., Kuo, W. H., Satola, S. W., Farley, M. M., et al. (2017). A population-based assessment of the impact of 7- and 13-valent pneumococcal conjugate vaccines on macrolide-resistant invasive pneumococcal disease: emergence and decline of Streptococcus pneumoniae serotype 19A (CC320) with dual macrolide resistance mechanisms. Clin. Infect. Dis. 65, 990–998. doi: 10.1093/cid/cix446
Schroeder, M. R., and Stephens, D. S. (2016). Macrolide resistance in Streptococcus pneumoniae. Front. Cell. Infect. Microbiol. 6:98. doi: 10.3389/fcimb.2016.00098
Schuchat, A., Hilger, T., Zell, E., Farley, M. M., Reingold, A., Harrison, L., et al. (2001). Active bacterial core surveillance of the emerging infections program network. Emerg Infect. Dis. 7, 92–99. doi: 10.3201/eid0701.700092
Stephens, D. S., Zughaier, S. M., Whitney, C. G., Baughman, W. S., Barker, L., Gay, K., et al. (2005). Incidence of macrolide resistance in Streptococcus pneumoniae after introduction of the pneumococcal conjugate vaccine: population-based assessment. Lancet 365, 855–863. doi: 10.1016/S0140-6736(05)71043-6
Su, W., Kumar, V., Ding, Y., Ero, R., Serra, A., Lee, B. S. T., et al. (2018). Ribosome protection by antibiotic resistance ATP-binding cassette protein. Proc. Natl. Acad. Sci. U.S.A. 115, 5157–5162. doi: 10.1073/pnas.1803313115
Suda, K. J., Hicks, L. A., Roberts, R. M., Hunkler, R. J., and Taylor, T. H. (2014). Trends and seasonal variation in outpatient antibiotic prescription rates in the United States, 2006 to 2010. Antimicrob. Agents Chemother. 58, 2763–2766. doi: 10.1128/AAC.02239-13
Tait-Kamradt, A., Clancy, J., Cronan, M., Dib-Hajj, F., Wondrack, L., Yuan, W., et al. (1997). mefE is necessary for the erythromycin-resistant M phenotype in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41, 2251–2255. doi: 10.1128/AAC.41.10.2251
Tao, L., LeBlanc, D. J., and Ferretti, J. J. (1992). Novel streptococcal-integration shuttle vectors for gene cloning and inactivation. Gene 120, 105–110. doi: 10.1016/0378-1119(92)90016-I
Weisblum, B. (1995a). Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39, 577–585. doi: 10.1128/AAC.39.3.577
Weisblum, B. (1995b). Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob. Agents Chemother. 39, 797–805. doi: 10.1128/AAC.39.4.797
Wolter, N., Smith, A. M., Farrell, D. J., Northwood, J. B., Douthwaite, S., and Klugman, K. P. (2008). Telithromycin resistance in Streptococcus pneumoniae is conferred by a deletion in the leader sequence of erm(B) that increases rRNA methylation. Antimicrob. Agents Chemother. 52, 435–440. doi: 10.1128/AAC.01074-07
Keywords: Streptococcus pneumoniae, pneumococcus, macrolide resistance, Mega, mef(E)/mel, erm(B), Tn2010
Citation: Schroeder MR, Lohsen S, Chancey ST and Stephens DS (2019) High-Level Macrolide Resistance Due to the Mega Element [mef(E)/mel] in Streptococcus pneumoniae. Front. Microbiol. 10:868. doi: 10.3389/fmicb.2019.00868
Received: 10 December 2018; Accepted: 04 April 2019;
Published: 24 April 2019.
Edited by:
Henrietta Venter, University of South Australia, AustraliaReviewed by:
Annalisa Pantosti, Istituto Superiore di Sanità (ISS), ItalyCopyright © 2019 Schroeder, Lohsen, Chancey and Stephens. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David S. Stephens, ZHN0ZXAwMUBlbW9yeS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.