
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 10 April 2019
Sec. Evolutionary and Genomic Microbiology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00741
 Daoyi Xi1,2,3,4†
Daoyi Xi1,2,3,4† Xiaochen Wang1,2,3,4†
Xiaochen Wang1,2,3,4† Kexin Ning1,2,3,4
Kexin Ning1,2,3,4 Qian Liu1,2,3,4
Qian Liu1,2,3,4 Fuyi Jing1,2,3,4
Fuyi Jing1,2,3,4 Xi Guo1,2,3,4
Xi Guo1,2,3,4 Boyang Cao1,2,3,4*
Boyang Cao1,2,3,4*Plesiomonas shigelloides is a Gram-negative, flagellated, rod-shaped, ubiquitous, and facultative anaerobic bacterium. It has been isolated from various sources, such as freshwater, surface water, and many wild and domestic animals. P. shigelloides is associated with diarrheal diseases of acute secretory gastroenteritis, an invasive shigellosis-like disease, and a cholera-like illness in humans. At present, 102 somatic antigens and 51 flagellar antigens of P. shigelloides have been recognized; however, very little is known about variations of O-antigens among P. shigelloides species. In this study, 12 O-antigen gene clusters of P. shigelloides, O2H1a1c (G5877), O10H41 (G5892), O12H35 (G5890), O23H1a1c (G5263), O25H3 (G5879), O26H1a1c (G5889), O32H37 (G5880), O33H38 (G5881), O34H34 (G5882), O66H3 (G5270), O75H34 (G5885), and O76H39 (G5886), were sequenced and analyzed. The genes that control O-antigen synthesis are present as chromosomal gene clusters that maps between rep and aqpZ, and most of the synthesis and translocation of OPS (O-specific polysaccharide) belongs to Wzx/Wzy pathway with the exception of O12, O25, and O66, which use the ATP-binding cassette (ABC) transporter pathway. Phylogenetic analysis of wzx and wzy show that the wzx and wzy genes are specific to individual O-antigens and can be used as targets in molecular typing. Based on the sequence data, an O-antigen specific suspension array that detects 12 distinct OPS’ has been developed. This is the first report to catalog the genetic features of P. shigelloides O-antigen variations and develop a suspension array for the molecular typing. The method has several advantages over traditional bacteriophage and serum agglutination methods and lays the foundation for easier identification and detection of additional O-antigen in the future.
Plesiomonas shigelloides was first isolated from the feces of a patient for whom no clinical history was available. It was reported to be a motile organism with the major antigen of Shigella sonnei phase I, and originally named Paracolon C27 by Ferguson and Henderson (1947), (Hu et al., 2013). Biochemical, morphological, and taxonomical features of the organism were collected by many investigators and the organism was reclassified into the family Enterobacteriaceae, being the only oxidase-positive member of this family (Martinez-Murcia et al., 1992; Huys and Swings, 1999; Garrity et al., 2001). P. shigelloides, the only species of the genus, is a Gram-negative, flagellated, rod-shaped, ubiquitous, and facultative anaerobic bacterium, which has been later isolated from various sources, such as freshwater, surface water, and many wild and domestic animals (Wong and Pang, 2000). P. shigelloides is associated with diarrheal diseases in humans (Brenden et al., 1988), including acute secretory gastroenteritis (Mandal et al., 1982), an invasive shigellosis-like disease (McNeeley et al., 1984), and a cholera-like illness (Tsukamoto et al., 1978). Traveler diarrhea affects millions of tourists each year. P. shigelloides has been ranked third in traveler’s diarrhea in Japan (Schubert and Holz-Bremer, 1999), and third as a cause of diarrhea among a number of military units in China (Bai et al., 2004), as well as among civilians in Hong Kong (Chan et al., 2003).
The O-antigen is a component of lipopolysaccharides (LPS) present in the outer membrane of Gram-negative bacteria and comprised of repeated oligosaccharide units (O-units, OPS). The OPS is the major contribution of the antigenic variability of the cell surface and is subject to intense selection by the host immune system and other environmental factors, such as bacteriophages, which may account for the existence of diverse O-antigen forms within a species (Reeves et al., 1996). The O-antigen gene clusters consist of genes that belong to three main classes: nucleotide sugar biosynthesis pathway genes, glycosyltransferase (GT) genes, and O-unit processing genes. Three distinct pathways participated in the synthesis and translocation of OPS: the Wzx/Wzy pathway, the ATP-binding cassette (ABC) transporter pathway (Hu et al., 2013), and the synthase pathway. O-antigen serotyping, targeting the wzx and wzy genes, is able to detect distinct O-antigen forms, including E. coli, Shigella, Salmonella, Vibrio parahaemolyticus, Acinetobacter, Hafnia, and Proteus (Li et al., 2009, 2010; Guo et al., 2013; Hu et al., 2013; Chen et al., 2015; Duan et al., 2016; Yu et al., 2017). Our previous studies employed wzx and wzy for serotyping E. coli and Shigella by microarray (Li et al., 2006), wzx and wzy for serotyping Pseudomonas aeruginosa (Li et al., 2018) and wzm and wzt for serotyping 15 Legionella pneumophila serogroups by microarray (Cao et al., 2013). In this report, wzx, wzy, and wzm have been screened and used as targets for molecular serotyping of the 12 P. shigelloides strains.
At present, 102 somatic antigens and 51 flagellar antigens of P. shigelloides have been recognized (Aldova and Shimada, 2000). P. shigelloides strain 302-73, isolated in Japan (Shimada and Sakazaki, 1985), is the representative strain for the O1 serogroup, whose complete OPS and the core of LPS has been chemically characterized (Pieretti et al., 2010), and the whole-genome sequence of P. shigelloides 302-73 have been obtained in Pique et al. (2013). P. shigelloides O17 is one of the most frequently encountered serovar, which is involved with protection against shigellosis as it carries a LPS in the cell walls that is also common to S. sonnei (Sayeed et al., 1992).
In this study, 12 O-antigen gene clusters of P. shigelloides, serogroups O2, O10, O12, O23, O25, O26, O32, O33, O34, O66, O75, and O76, were sequenced and analyzed. The genes that control O-antigen synthesis are present as chromosomal gene clusters that maps between rep and aqpZ, and most of the synthesis and translocation of OPS occurs within the Wzx/Wzy pathway, with the exception of O12, O25, and O66, which use ABC transporter pathway. Phylogenetic analysis of wzx and wzy show that the wzx and wzy genes are specific to individual O-antigens and can be used as targets in molecular typing.
In summary, the current study: (1) has determined 12 O-antigen gene clusters of P. shigelloides serogroups O2, O10, O12, O23, O25, O26, O32, O33, O34, O66, O75, and O76; (2) has analyzed the evolutionary relationship of the O-antigen gene clusters in combination with the published O-antigen-specific gene data of P. shigelloides O1; (3) has established a rapid, bead-based suspension array detection technique using the wzx, wzy, or wzm genes of 12 different serogroups of P. shigelloides as the molecular serotyping markers. The suspension array method has been tested for its specificity and sensitivity and found to be suitable for applications in the fields of clinical research, diagnosis, treatment, epidemiology, and prevention of diseases.
The 12 P. shigelloides O-standard reference strains employed in this study are listed in Table 1.
All P. shigelloides strains were cultured in Tryptone Soya Broth (TSB) (QingDao ShuiRi Bio-Technologies Co., Ltd., Qingdao, China) at 37°C overnight with shaking. Genomic DNA was prepared by Bacteria Gen DNA Kit (CWBiotech, Beijing, China).
A library for Illumina Paired-End sequencing was prepared from 5 μg DNA using a Paired-End DNA Sample Prep Kit (Pe-102-1001, Illumina Inc., Cambridge, United Kingdom). DNA was fragmented by nebulization for 6 min at a pressure of 32 psi. For end-repair and phosphorylation, sheared DNA was purified using the QIAquick Nucleotide Removal Kit (Qiagen, Crawley, United Kingdom). The end-repaired DNA was A-tailed and adaptors were ligated according to the manufacturer’s instructions.
As reported previously (Hu et al., 2013), the whole-genome sequencing was performed using an Illumina/Solexa genome analyzer IIx (Illumina, San Diego, CA, United States). De novo assembly was performed using SOAP de novo (Xie et al., 2014). Gene prediction and annotation were carried out using the NCBI Prokaryotic Genome Annotation Pipeline (Tatusova et al., 2016). The Staden package and the Artemis program were used for sequence assembly and gene annotation, respectively. The BlockMaker program was used to search for conserved motifs. BLAST and PSI-BLAST (Altschul et al., 1997) were used to search sequence databases, including the GenBank database and the Pfam protein motif database (Bateman et al., 2002), to identify potential gene functions. The program TMHMM 2.0 (Krogh et al., 2001) was used to identify potential transmembrane segments. Sequence alignments and comparisons were performed using the ClustalW program (Thompson et al., 1994). For sugar pathway genes, a BLAST search against the UniProt/SwissProt database was used to confirm the allocation of the genes by pathway.
A phylogenetic tree was constructed using the neighbor-joining method and plotted by the Molecular Evolutionary Genetics Analysis (MEGA) 6.0 software package. Bootstrap analysis was carried out based on 1,000 replicates.
The target genes for primer and probe design were wzx genes of P. shigelloides O2, O10, and O26, wzy genes of P. shigelloides O23, O32, O33, O34, O75, and O76, and wzm genes of P. shigelloides O12, O25, and O66 (Table 2). Primers were designed by Primer Premier (Yang et al., 2018). The forward primer was biotinylated at the 5′-end to allow binding to the reporter dye streptavidin-R-phycoerythrin for detection on a Bio-Plex platform. All the primer sequences used for the multiplex PCR are listed in Table 2.
Multiplex PCR was carried out in two groups of six serogroups each; Group A consists of P. shigelloides O12, O25, O26, O33, O75, and O76; Group B consists of P. shigelloides O2, O10, O23, O32, O34, and O66. The multiplex PCR amplification was performed with 50 μL of a reaction mixture that consisted of 100 ng of DNA, 25 μL of HotStar Taq Master Mix (Qiagen), 0.25 μL of the forward primer, and 1.0 μL of the reverse one, with an exception of O25 with double the amount of both primers. Reaction parameters were: 95°C for 5 min; 35 cycles of 94°C for 30 s, 54°C for 45 s, and 72°C for 50 s; and a final extension at 72°C for 5 min. An aliquot of 2 μL of PCR product was run on an agarose gel to examine the amplified DNA. After amplification, the samples were stored at −20°C until used.
All species-specific probes were based on the sequence data from the lab data. Multiple-sequence alignments were carried out using BioEdit version 7.0 software. These species-specific probes were newly designed (Table 2). Probes were synthesized with a 5′-end amino C-12 modification (AuGCT, Beijing, China) and coupled to carboxylated beads (Bio-Rad Laboratories, Hercules, CA, United States) as described by the manufacture’s manual. Briefly, a total of 2.5 × 105 carboxylated beads were suspended in 8.5 μL of 0.1 M MES (pH 4.5) with 2 μL of 0.1 nmol/μL oligonucleotide probes. A 2.5 μL of 10 mg/mL freshly prepared EDC [1-ethyl-3-(3-dimethylaminopropyl) carbodiimide] was added, and vortexed immediately, then incubated at room temperature in the dark for 30 min. After being washed by 0.02% Tween 20 and 0.1% SDS, pellets were centrifuged and resuspended in 20 μL of TE (pH 8.0), and stored at 4°C in the dark until used.
A bead mix set was prepared, consisting of 2,500 beads for each of the 10 probes in 1.5× tetramethylammonium chloride (TMAC) solution (Sigma, St. Louis, MO, United States) that contained 4.5 M TMAC, 0.15% Sarkosyl, 75 mM Tris–HCl (pH 8.0), and 6 mM EDTA (pH 8.0) solution. A total of 17 μL of biotinylated amplicon was added to 33 μl of the bead mix. Amplicon and bead mixture were denatured at 95°C for 5 min and allowed to hybridize at 57°C for 15 min. The mixture was centrifuged at 8,000 rpm and the supernatant carefully discarded, then the beads were resuspended in 75 μL of 1× TMAC solution containing 10 ng/mL streptavidin-R-phycoerythrin (Molecular Probes, Eugene, OR, United States) and incubated for 10 min at 57°C.
The fluorescence intensity of the beads was analyzed in a Bio-Plex 100 suspension array system (Bio-Rad Laboratories). The median fluorescence intensities (MFI) were calculated from 100 replicate measurements with a digital signal processor and BioPlex Manager 4.1 software. A positive signal was defined as MFI is at least >500, and the signal/background ratio (S/B ratio = MFI/Blank) is greater than 3.0.
A serial ten-fold dilution of the genomic DNA of P. shigelloides O26 and O75, ranging from 1.0 fg/uL, 10 fg/uL, 100 fg/uL, 1.0 pg/uL, 10 pg/uL, 100 pg/uL, 1.0 ng/uL, 10 ng/uL, and 100 ng/uL, were used as the template to test the sensitivity of the suspension array assay.
The DNA sequences of the O-antigen gene clusters of 12 O-antigen gene clusters of P. shigelloides serogroups O2, O10, O12, O23, O25, O26, O32, O33, O34, O66, O75, and O76 have been deposited in GenBank under the accession numbers MK551179-MK551190, respectively.
The O-antigen gene cluster of 12 P. shigelloides serogroups were obtained and extracted from the type strains of P. shigelloides serogroups O2, O10, O12, O23, O25, O26, O32, O33, O34, O66, O75, and O76 (Table 1). The clusters located between rep and aqpZ at the chromosome of these 12 isolates had 22,165, 27,407, 16,526, 15,506, 20,682, 29,124, 14,430, 21,362, 25,286, 21,030, 25,342, and 24,898 bp, respectively. Characteristics of the 18, 25, 11, 17, 16, 30, 13, 17, 20, 16, 22, and 18 open reading frames (orfs) of these 12 isolates were identified to each of the serogroups (Figure 1 and Supplementary Tables S1–S12). The functions were assigned based on their similarities to genes of known functions associated with sugar synthesis, sugar transfer, O-antigen processing, and others from available databases (Figures 1, 2).
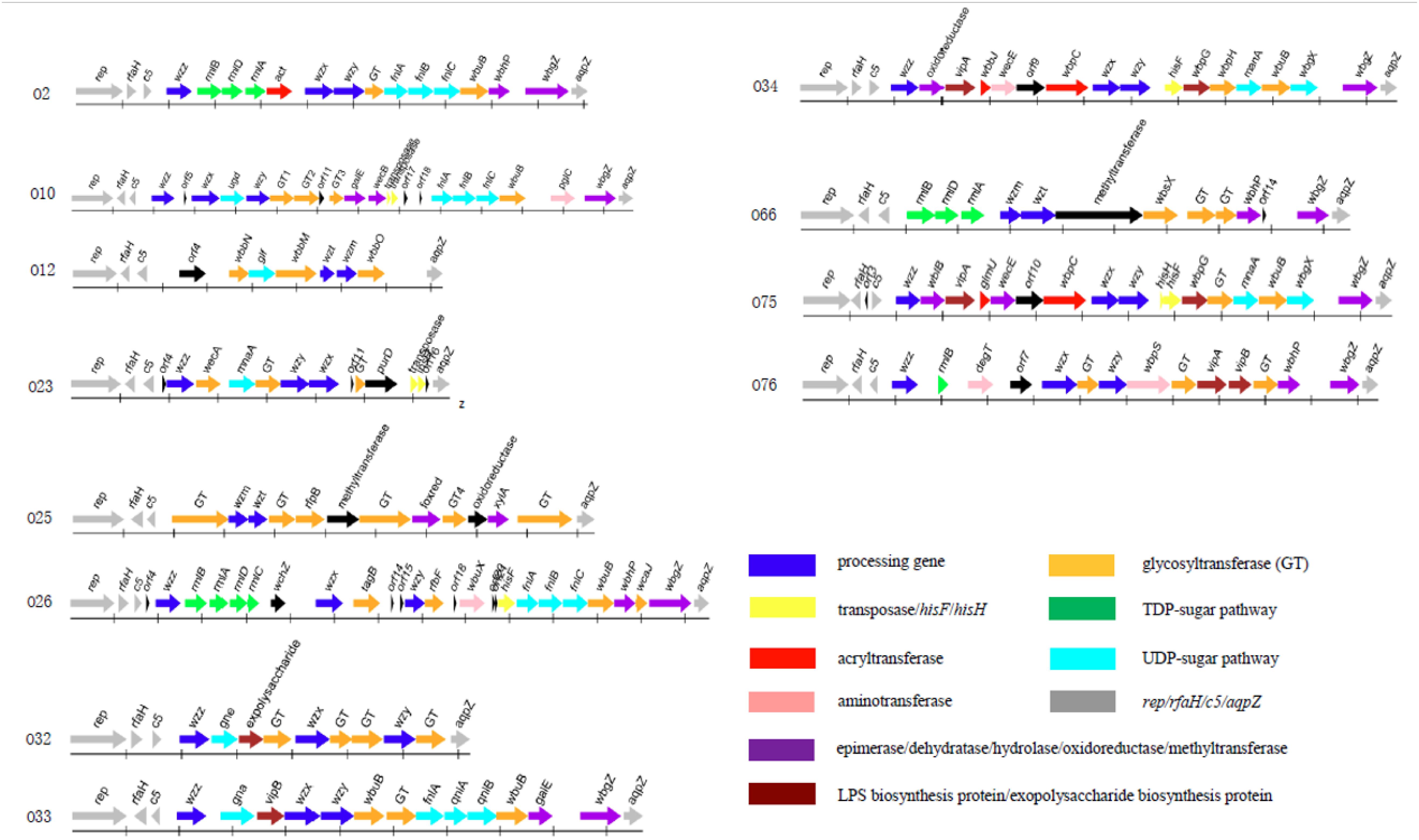
Figure 1. The O-antigen gene clusters of 12 P. shigelloides serogroups O2, O10, O12, O23, O25, O26, O32, O33, O34, O66, O75, and O76.
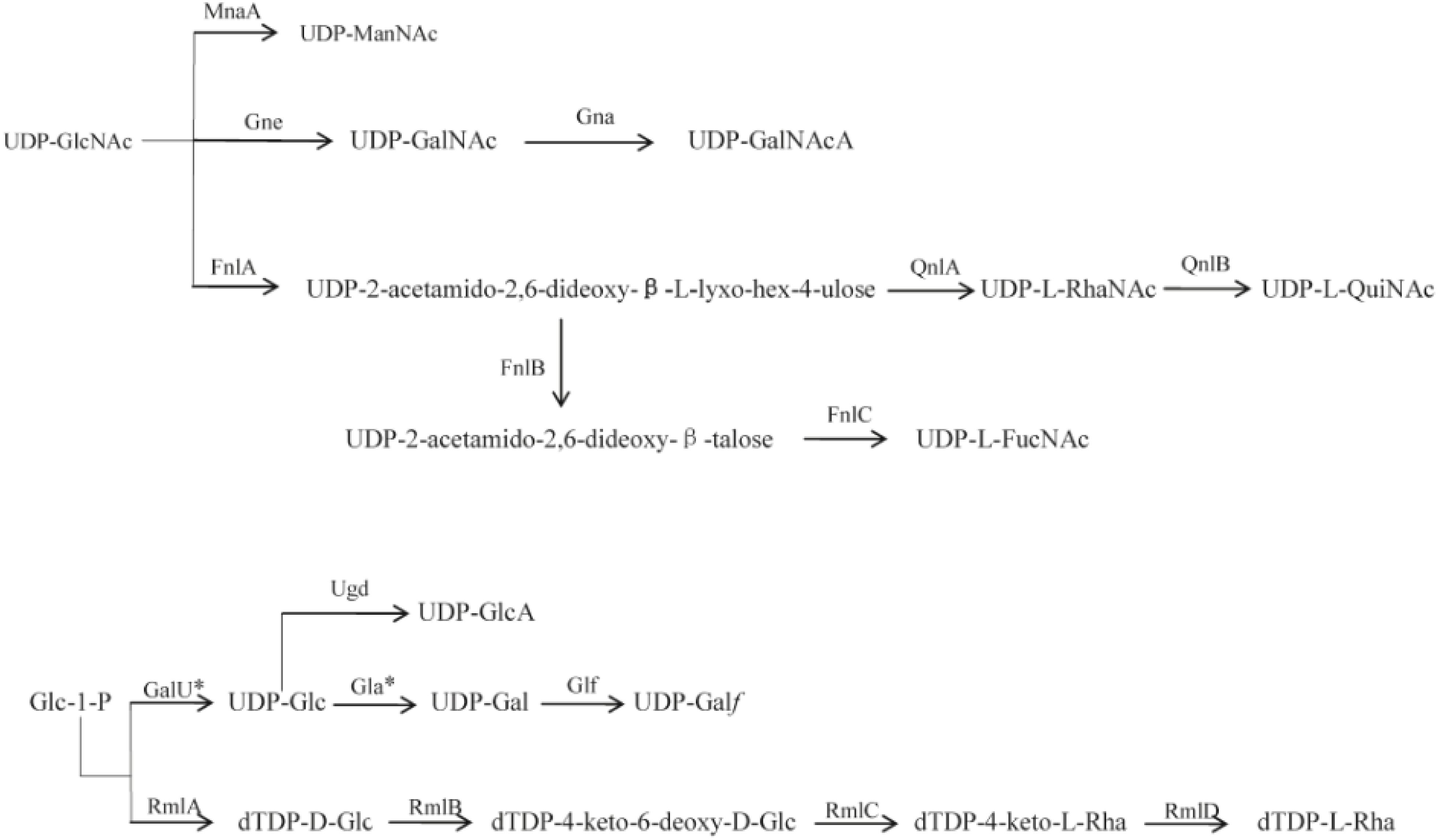
Figure 2. Proposed biosynthesis pathways for sugars in the 12 P. shigelloides serogroups clusters. Putative pathways are denoted by a broken line. MnaA, UDP-N-acetylglucosamine-2-epimerase; Gne, UDP-N-acetylglucosamine-4-epimerase; Gna, UDP-GalNAcA synthetase; FnlA, 4,6-dehydratase, 3- and 5-epimerase; FnlB, reductase; FnlC, C-2 epimerase; QnlA, dTDP-4-dehydrorhamnose reductase; QnlB, C-2 epimerase; GalU∗, UTP-glucose-1-phosphate uridylyltransferase; Ugd, UDP-glucose 6-dehydrogenase; Gla∗, UDP-galacturonase; Glf, UDP-galactopyranose mutase; RmlA, glucose-1-phosphate thymidylyltransferase; RmlB, dTDP-D-glucose 4,6-dehydratase; RmlC, dTDP-4-keto-6-deoxy-D-glucose 3,5-epimerase; RmlD, dTDP-6-deoxy-L-mannose-dehydrogenase. D-GlcNAc, 2-acetamido-2-deoxy-D-glucose; D-ManNAc, 2-acetamido-2-deoxy-D-mannose; D-GalNAc, 2-acetamido-2-deoxy-D-galactose; D-GalNAcA, 2-Acetamido-2-deoxy-D-galacturonic acid; L-RhaNAc, 2-acetamido-2,6-dideoxy-L-mannose (N-acetyl-L-rhamnosamine); L-QuiNAc, 2-Acetamido-2-deoxy-L-quinovose (2-acetamido-2,6-dideoxy-L-glucose); L-FucNAc, 2-Acetamido-2-deoxy-L-fucose; D-GlcA, D-glucuronic acid; D-Glc, D-glucose; D-Gla∗, D-galactose; D-Galf, D-galactofuranose; L-Rha, L-rhamnose (6-deoxy-L-mannose). ∗indicates the genes that were found outside the cluster.
For the synthesis of UDP-L-FucNAc, FnlABC is reportedly involved in synthesizing O-polysaccharides in several species, e.g., in Salmonella O47, O48, and O61 (Liu et al., 2014). The set of orf12, orf13, and orf14 in the cluster of P. shigelloides O2 shares 93% identity with FnlA of P. shigelloides WP_010862887, 65 and 84% with FnlBC of E. coli, indicating the existence of a rare sugar, UDP-L-FucNAc. Rhamnose is widely present as a bacterial surface polysaccharide, and its biosynthesis pathway is well known to require four enzymes that are encoded by rmlABCD in the polysaccharide gene clusters of E. coli, Shigella, and Salmonella (Liu et al., 2008, 2014). For P. shigelloides O2, the set of orf5, orf6, and orf7 of P. shigelloides O2 shows around 90% identity to RmlBDA in E. coli, indicating the existence of another rare sugar, dTDP-L-Rha. The orf9 shows 39% identity to Wzx in Escherichia coli and the orf10 shows 40% identity to Wzy in Pseudomonas mucidolens. The set of wzx and wzy genes suggests the presence of Wzx/Wzy pathway related O-antigen processing. There are two GTs encoded by the orf11 and orf15, which share 37% identity with GT family 2 protein of Pseudomonas sp. NFR02, and 58% identity with GT/WbuB of Burkholderia territorii.
The set of FnlABC in the cluster of P. shigelloides O10 shares 98% identity with FnlA of P. shigelloides, 63% identity with FnlB of E. coli, and 99% identity with FnlC of P. shigelloides, indicating the existence of the rare sugar, UDP-L-FucNAc. The function of Ugd is to convert UDP-Glc to UDP-GlcA. The orf7 shares more than 94% identity with Ugd in Shewanella algae. The orf6 shows 79% identity to Wzx in Shewanella decolorationis and the orf8 shows 32% identity to Wzy in Marinobacter sp. EN3. The set of wzx and wzy genes suggests the presence of Wzx/Wzy pathway related O-antigen processing. There are three GTs encoded by the orf9, orf10, and orf12 which share 40% identity with GT family 2 protein of Prosthecochloris sp. CIB 2401, 35% identity with GT family 1 protein of P. shigelloides, and 88% identity with GT of S. algae, respectively.
There is no rare sugar in the cluster of P. shigelloides O12. The function of Glf is to convert UDP-Gal to UDP-Galf. The orf6 in the cluster shares 79% identity with the corresponding gene of Klebsiella pneumoniae. The set of wzm and wzt genes suggests the presence of ABC transporter pathway related O-antigen processing. There are three GTs encoded by the orf5, orf7, and orf10 which share 50% identity with GT/WbbN of Serratia liquefaciens, 67% identity with GT/WbbM of K. pneumoniae subsp. rhinoscleromatis SB3432, and 57% identity with GT/WbbO of Serratia sp. Leaf50, respectively.
The function of MnaA is to convert UDP-GlcNAc to UDP-ManNAc. MnaA in P. shigelloides O23 share more than 81% identity with the one in E. coli, indicating there is a rare sugar UDP-ManNAc in the structure. The orf10 shows 27% identity to Wzx in E. coli S88 and the orf9 shows 29% identity to Wzy in Salmonella enterica. The set of wzx and wzy genes suggests the presence of Wzx/Wzy pathway related O-antigen processing. WecA in P. shigelloides O23 shares more than 93% identity with the corresponding gene of P. shigelloides. There is one GT encoded by the orf8 which shares 42% identity with GT family 4 protein of Vibrio cholerae. There are two transposes encoded by the orf14 and orf15. As Duan et al. (2016) reported in most E. coli and Shigella strains, the first sugar residue in the O-unit is GlcNAc or GalNAc, and the IT (Initial transferase) encoded by wecA, which is responsible for initiating the synthesis of GlcNAc- and GalNAc- initiated OPS. Usually, the wecA gene is located outside the cluster and acts as a UDP-GlcNAc:undecaprenyl phosphate GlcNAc-1-phosphate transferase, however, it presents in the O23 gene cluster.
There is no rare sugar in the cluster of P. shigelloides O25. The set of wzm and wzt genes suggests the presence of the ABC transporter pathway related O-antigen processing. Interestingly, there are six GTs encoded by the orf4, orf7 orf8, orf10, orf12, and orf15 which share 54% identity with GT family 1 protein of Enterobacteriaceae, 56% identity with GT family 1 protein of Citrobacter europaeus, 67% identity with GT/RfbP of C. europaeus, 67% identity with GT family 1 protein of C. europaeus, 65% identity with GT family 2 protein of Serratia sp. DD3, and 77% identity with GT family 2 protein of Duganella sp. Root336D2, respectively.
The orf6 and orf7 of P. shigelloides O26 share 89 and 95% identity with RmlB and RmlA of V. cholerae, and the orf8 and orf9 share 85 and 83% identity with RmlD and RmlC of Vibrio anguillarum, indicating the presence of a rare sugar, dTDP-L-Rha. The set of orf23, orf24, and orf25 in the cluster shares 93% identity with FnlA of P. shigelloides WP_010862887, 65 and 84% with FnlBC of E. coli, indicating there is a rare sugar UDP-L-FucNAc. The orf12 shows 36% identity to Wzx in E. coli and the orf16 shows 36% identity to Wzy in Flavobacterium sp. GSP27. The presence of wzx and wzy genes suggests the presence of Wzx/Wzy pathway related O-antigen processing in O26. Except the two GTs of orf13 and orf28 encoding TagD for teichoic acid biosynthesis and WcaJ for colanic acid biosynthesis. There are other two GTs encoded by the orf17 and orf26 which share 38% identity with GT/RfbF of Shigella flexneri, and 89% identity with GT/WbuB of V. cholerae, respectively.
There is no rare sugar in the cluster of P. shigelloides O32. The orf8 shows 45% identity to Wzx in E. coli and the orf11 shows 33% identity to Wzy in S. enterica. The set of wzx, wzy, and wzz genes suggests the presence of Wzx/Wzy pathway related O-antigen processing. The orf5 in the cluster of P. shigelloides O32 shares 64% identity with Gne of Yersinia pseudotuberculosis, indicating there is a sugar UDP- GalNAc. There are four GTs encoded by the orf7, orf9, orf10, and orf12, which share 57% identity with GT of S. enterica, 57% identity with GT family 2 protein of Proteus mirabilis, 58% identity with GT family 4 protein of Edwardsiella tarda, and 55% identity with GT of Shewanella sp. SACH, respectively.
The orf5 in the cluster of P. shigelloides O33 shares 83% identity with Gna of P. shigelloides, indicating the presence of a sugar, UDP- GalNAcA. The set of orf11, orf12, and orf13 in the cluster of P. shigelloides O33 shares more than 93, 69, and 88% identity with FnlA, QnlA, and QnlB of E. coli, indicating there is a rare sugar, UDP-L-QuiNAc. The orf7 shows 38% identity to Wzx in E. coli and the orf8 shows 46% identity to Wzy in Moritella viscosa. There are wzx and wzy genes in the cluster, indicating the presence of Wzx/Wzy pathway related O-antigen processing. There are three GTs encoded by orf9, orf10, and orf14 which share 23% identity with GT/WbuB of Pseudomonas fluorescens, 76% identity with GT family 1 protein of Aeromonas fluvialis, and 74% identity with GT/WbuB of Shewanella xiamenensis, respectively.
The orf16 in the cluster of P. shigelloides O34 shares more than 89% identity with MnaA of V. cholerae, indicating the presence of UDP-ManNAc. The orf11 shows 36% identity to Wzx in Janthinobacterium sp. CG23_2 and the orf12 shows 35% identity to Wzy in Pseudomonas sp. BAY1663. The set of wzx, wzy, and wzz genes suggest the presence of Wzx/Wzy pathway related O-antigen processing. There are two GTs encoded by the orf15 and orf17 which share 57% identity with GT/WbpH of Pseudomonas sp. CFII68, and 55% identity with GT of Photorhabdus luminescens.
The set of orf4, orf5 and orf6 in the cluster of P. shigelloides O66 share 91, 94, and 98% identity with RmlBDA of Aeromonas sp. indicating there is a rare sugar dTDP-L-Rha. The galE is the gene that is responsible for the synthesis of UDP-Gal in the gene cluster. The set of wzm and wzt genes suggest the presence of the ABC transporter pathway related O-antigen processing. There are three GTs encoded by the orf10, orf11, and orf12 which share 63% identity with GT/WbsX of Nitrosomonas europae, 81% identity with GT of Aeromonas hydrophila, and 75% identity with GT family 2 protein of Shigella boydii, respectively.
The MnaA in the cluster of P. shigelloides O75 shares more than 89% identity with the one of V. cholerae, indicating there is a rare sugar, UDP-ManNAc, in the structure. The orf12 shows 42% identity to Wzx and the orf13 shows 34% identity to Wzy in Pseudomonas sp. BAY1663. The set of wzx, wzy, and wzz genes suggests the presence of Wzx/Wzy pathway related O-antigen processing. There are two GTs encoded by the orf17 and orf19, which share 87% identity with GT of Vibrio ordalii, and 55% identity of GT/WbuB of P. luminescens.
There is no rare sugar in the cluster of P. shigelloides O76. The orf8 shows 51% identity to Wzx in Acinetobacter baumannii and the orf10 shows 45% identity to Wzy in E. coli. The set of wzx, wzy, and wzz genes suggests the presence of Wzx/Wzy pathway related O-antigen processing. There are three GTs encoded by the orf9, orf12, and orf15, which share 81% identity with GT family 2 protein of Pseudomonas oleovorans, 91% identity with GT of V. cholerae, and 91% identity with GT/WbuB of V. cholerae, respectively.
Previous studies suggest that O-antigen processing genes, wzx and wzy for Wzx/Wzy pathway or wzm and wzt for ABC transporter pathway, are highly specific to individual O-antigens (Greenfield and Whitfield, 2012; Islam and Lam, 2014). We therefore selected them as targets for PCR-based typing methods. Out of the 12 P. shigelloides O-antigens, three of them contain both wzm and wzt, the remaining nine contain both wzx and wzy. We have constructed the maximum likelihood phylogenetic trees using the wzx and wzy genes, and the results show the high levels of diversity of these two genes among different strains (Figure 3). The diversity of the wzx and wzy genes provided us a basis to develop molecular techniques to detect and identify different P. shigelloides O serogroups. Accordingly, the specific primer pairs were designed based on the wzx genes of O2, O10, and O26; the wzy genes of O23, O32, O33, O34, O75, and O76; and the wzm genes of O12, O25, and O66.
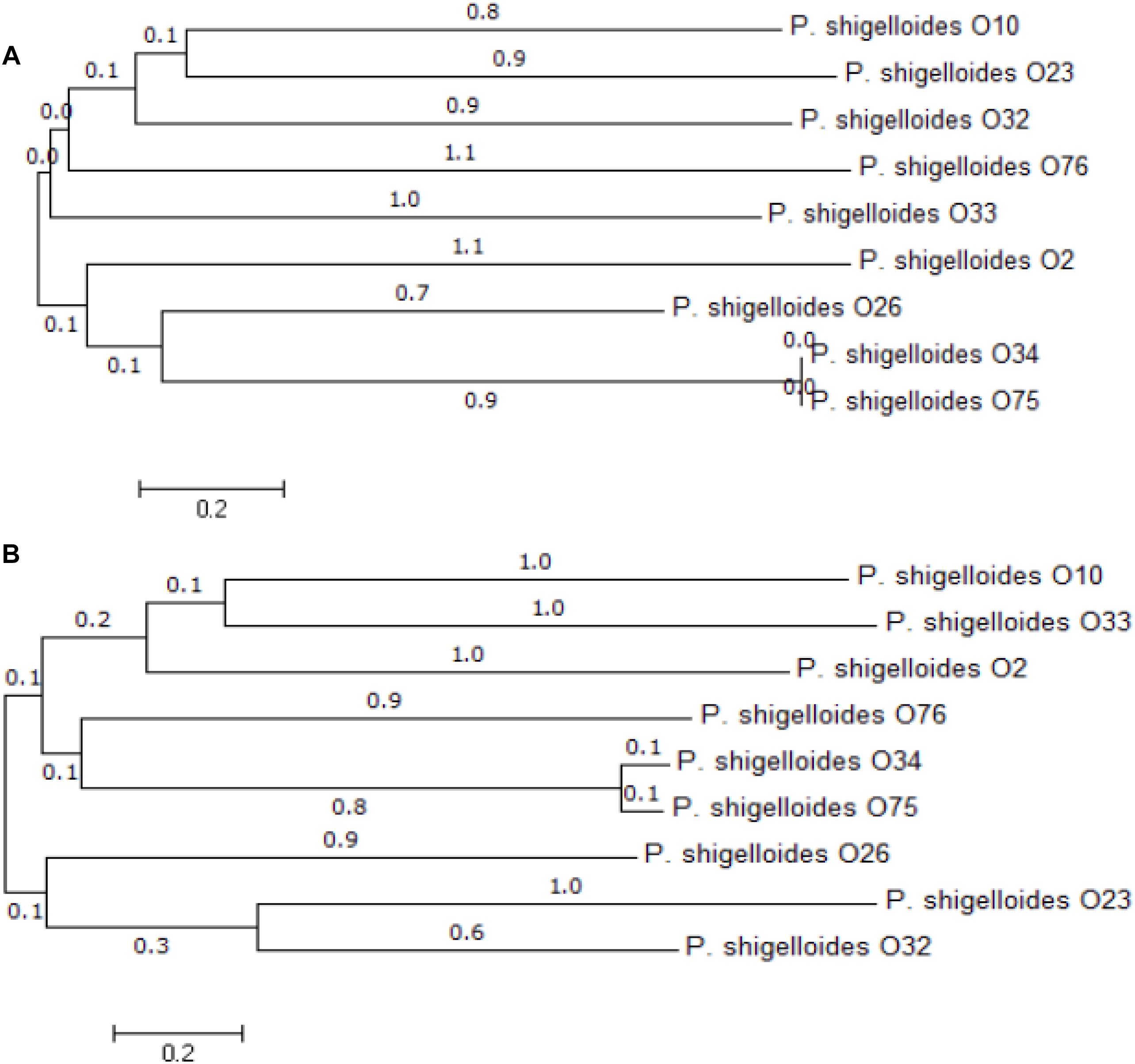
Figure 3. Phylogenetic analysis. An unrooted phylogenetic tree constructed by the neighbor joining method based on the wzx (A) and wzy (B) genes is shown. Bootstrap values were based on 1000 replications and only values greater than 50% are shown.
Multiplex PCR was used to amplify and generate the single strands of individual amplicons. Under our optimized conditions, the amplicons of the 12 O-antigens were amplified simultaneously with the PCR products ranging from 82 to 351 bp in length for clear hybridization signals. For all of the 12 strains tested, the S/B ratio for each probe tested against its homologous DNA was significantly greater than the ratio that was obtained when the probes tested against non-homologous DNA. The S/B ratios of the positive samples are set as greater than 3.0. No cross reactivity was observed for any probe that was tested against non-homologous DNA (Figure 4).
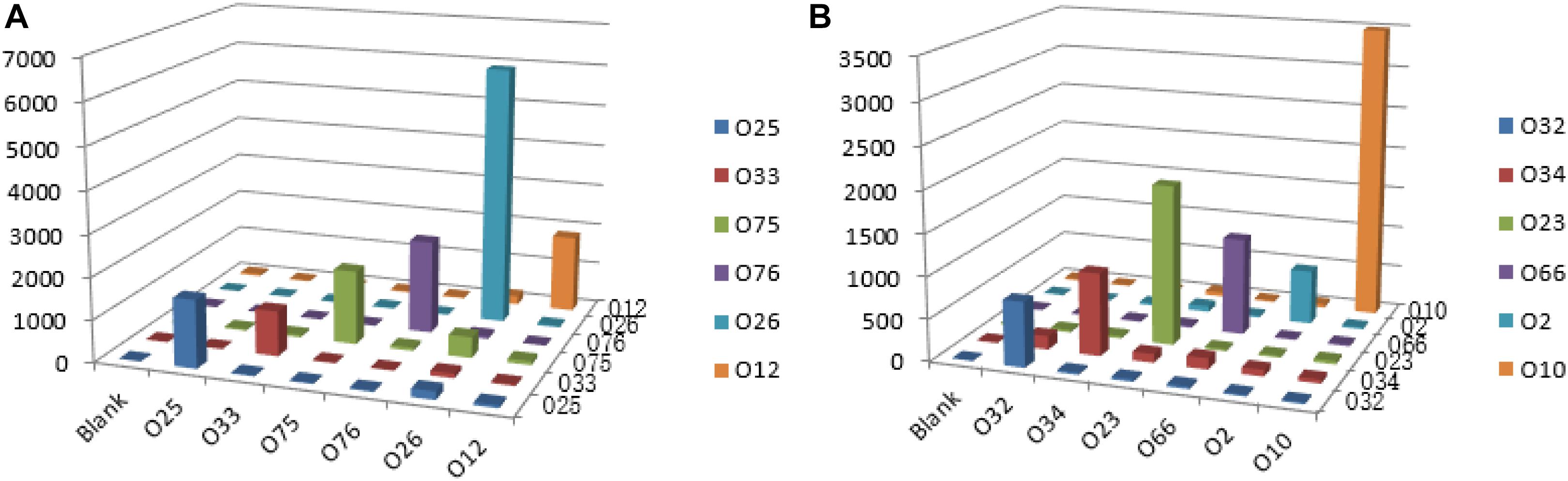
Figure 4. Specific detection by the xMAP bead-based multiplex assay on the 12 P. shigelloides O-antigens. MFI, median fluorescent intensity; the X-axis represents P. shigelloides strains of the 12 O-antigens. (A) Group A; (B) Group B.
The lowest amount of DNA that produces positive signals were: 100 pg DNA for P. shigelloides O26, and 10 pg DNA for P. shigelloides O75 (Figure 5). And the sensitivity of the assay with genomic DNA was set at 1.0 ng of DNA.
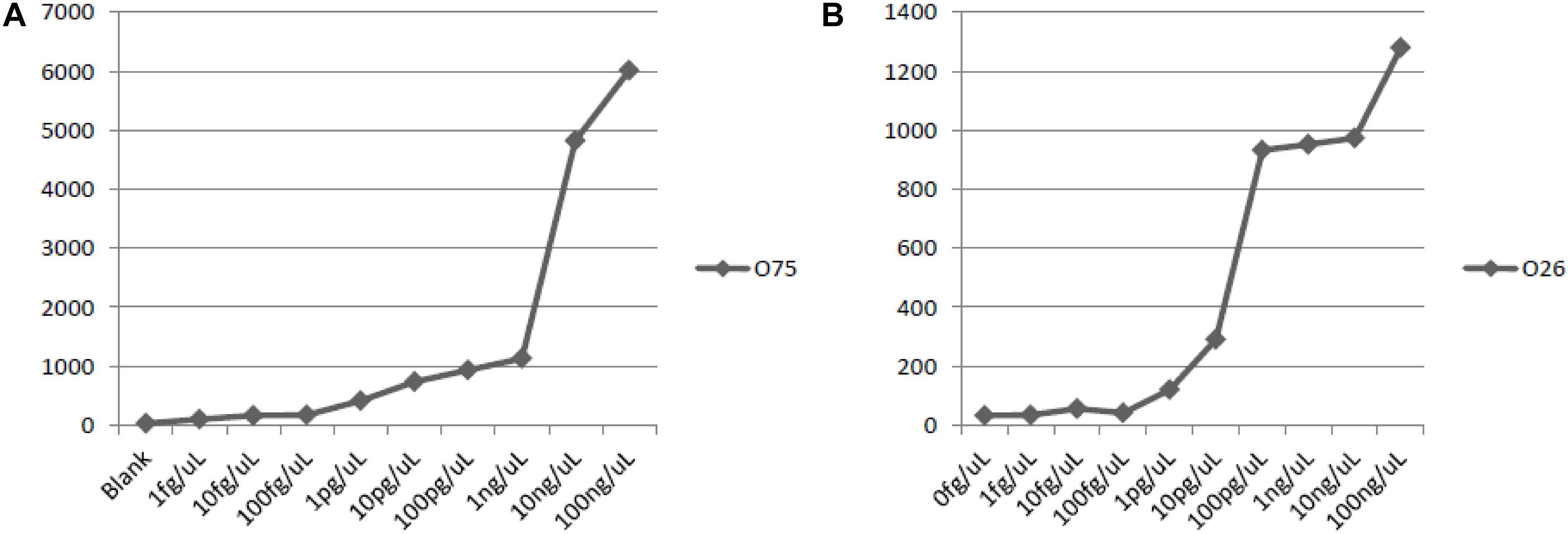
Figure 5. Sensitivity of detection with genomic DNA of O75 and O26. (A) sensitivity of O75 genomic DNA; (B) sensitivity of O26 genomic DNA.
L-Rha is widely distributed in O-antigens of bacteria, and the four rml genes related to the biosynthesis are usually grouped together and easily identified in a range of species. In E. coli and Salmonella, the genes are generally in the order of rmlBDAC at the 5′ end of the O-antigen gene cluster downstream of wzz (Liu et al., 2014). Out of the 12 P. shigelloides O-antigens studied in this report, L-Rha is present in three of them, O2, O26, and O66. rmlC is present in O26 cluster, and the set of four rml genes is in the order of rmlBADC at the 5′ end of the O-antigen gene cluster downstream of wzz, rmlC is absent in O2 and O66 clusters, and the rest of the three genes of the two O-antigens are in the order of rmlBDA at the 5′ end of the O-antigen gene cluster downstream of wzz. The two missing rmlC of O2 and O66 outside the O-antigen clusters are likely located somewhere in the chromosome. It is in line with the fact that rmlB, rmlD, and rmlA genes of E. coli are conserved and have characteristics of housekeeping genes (Li and Reeves, 2000), while the rmlC gene is more varied and O-antigen specific.
The O-antigen gene cluster of strain 302-73 (serotype O1) and the chemical structure of the O-specific chains have been reported (Aquilini et al., 2013; Pique et al., 2013) with the genes rep and aqpZ located at the two ends of the O-antigen cluster. The fnlABC, pglB, and pglD in the cluster indicate the presence of UDP-L-FucNAc, PneNAc, and QuiNAcHb in the structure. And SDS-PAGE showed that both pglD and fnlA mutants lack the O-antigen LPS by their gel profile (Aquilini et al., 2013). The O-antigen gene cluster and chemical structure of O-specific chain of PCM2231 (serotype O17) have also been studied (Chida et al., 2000; Maciejewska et al., 2013). The genes rep and aqpZ were found in the O-antigen cluster anchors. The presence of wbgVX and wbgZ indicate there is a UDP-D-FucNAc4N and UDP-L-AltNAcA. All of the 12 P. shigelloides strains used in this report are reference strains for their corresponding serogroups, and are in line with the O1 and O17. Out of these 12 O-antigen clusters, several rare sugars can be deduced based on the highly homologous sugar synthetic genes, for example, dTDP-L-Rha in O2, O26, and O66; UDP-L-FucNAc in O2, O10, and O26; UDP-L-QuiNAc in O33; and UDP-ManNAc in O23, O34, and O75. In addition, there are two rare sugars of dTDP-L-Rha and UDP-L-FucNAc in both O2 and O26. However, the existence of these rare sugars need to be confirmed by chemical structure determination, such as, NMR spectrum (Perepelov et al., 2016).
In most E. coli and Shigella strains WecA, which is encoded by a gene in the enterobacterial common antigen (ECA) gene cluster, is responsible for initiating the synthesis of GlcNAc- and GalNAc-initiated O-antigens by transferring GlcNAc-1-phosphate to the UndP carrier (Liu et al., 2008). The existence of wecA in O23 cluster indicating the existence of either GlcNAc or GalNAc in its chemical structure. We used O23 WecA to blast the genomes of the other 11 P. shigelloides and found that all the other 11 wecAs are located in the chromosomes outside the O-antigen gene clusters with similarity of 37% and e-value of 1.00E-66.
In the 12 P. shigelloides serogroups O-antigens, ABC transporter pathways encoded by wzm/wzt are employed for O-antigen processing of O12, O25, and O66; and Wzx/Wzy pathways encoded by wzx/wzy are employed for O-antigen processing of the other nine ones, O2, O10, O23, O26, O32, O33, O34, O75, and O76.
We took efforts to improve the reproducibility and sensitivity of the suspension array. We have compared three methods of amplification, (1) in the two-step PCR method, at the first step, the same concentration of forward and reverse primers were used to amplify the target genes at the first step; followed by the labeling of single–strand DNA using the reverse primers only; (2) the one-step symmetry PCR method, which uses the same concentration of both forward and reverse primers for PCR reactions; and (3) the one-step asymmetry PCR method, the different concentration of forward and reverse primers were used for PCR reactions. We found the one-step asymmetry PCR method achieved the best results. Also, in order to obtain a better hybridization signal, the PCR amplification fragments were kept shorter than 350 bp. The sensitivity of the method reaches 100 pg DNA for P. shigelloides O26, and 10 ng DNA for P. shigelloides O75.
Shepherd et al. (2000) reported that S. sonnei gained its current plasmid-borne O-antigen genes from P. shigelloides O17 in a recent event. As described in this report, we established the method to distinguish them by targeting wzy and wbgV. We expected that the suspension array is able to detect and dissect new serogroups of recombinant P. shigelloides that may arise from time to time.
However, the array has its limitations as the design probes requires prior knowledge of the sequence. In summary, the results in the study demonstrated that the probes and primers worked well on target strains and generated the specific signals and the suspension array is able to provide an easy, rapid, and accurate detection for P. shigelloides to facilitate better monitor the disease.
BC conceived the project. DX and XG purchased the strains. DX prepared the sample DNA, performed the sequence analyses, and developed the molecular serotyping system. XW and QL conducted the bioinformatics analyses. DX, KN, and FJ developed the multiplexed Luminex – based array. BC and DX prepared the manuscript. All authors read and approved the final manuscript.
This work was supported by the National Special Project on Research and Development of Key Bio-safety Technologies (Grant No. 2016YFC1200100) and the National Key Programs for Infectious Diseases of China (Grant Nos. 2017ZX10303405-001 and 2017ZX10104002-001-006).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00741/full#supplementary-material
Aldova, E., and Shimada, T. (2000). New O and H antigens of the international antigenic scheme for Plesiomonas shigelloides. Folia Microbiol. 45, 301–304. doi: 10.1007/BF02817550
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Aquilini, E., Merino, S., and Tomas, J. M. (2013). The Plesiomonas shigelloides wb(O1) gene cluster and the role of O1-antigen LPS in pathogenicity. Microb. Pathog. 63, 1–7. doi: 10.1016/j.micpath.2013.05.010
Bai, Y., Dai, Y. C., Li, J. D., Nie, J., Chen, Q., Wang, H., et al. (2004). Acute diarrhea during army field exercise in southern China. World J. Gastroenterol. 10, 127–131. doi: 10.3748/wjg.v10.i1.127
Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S. R., et al. (2002). The pfam protein families database. Nucleic Acids Res. 30, 276–280. doi: 10.1093/nar/30.1.276
Brenden, R. A., Miller, M. A., and Janda, J. M. (1988). Clinical disease spectrum and pathogenic factors associated with Plesiomonas shigelloides infections in humans. Rev. Infect. Dis. 10, 303–316. doi: 10.1093/clinids/10.2.303
Cao, B., Yao, F., Liu, X., Feng, L., and Wang, L. (2013). Development of a DNA microarray method for detection and identification of all 15 distinct O-antigen forms of Legionella pneumophila. Appl. Environ. Microbiol. 79, 6647–6654. doi: 10.1128/aem.01957-13
Chan, S. S., Ng, K. C., Lyon, D. J., Cheung, W. L., Cheng, A. F., and Rainer, T. H. (2003). Acute bacterial gastroenteritis: a study of adult patients with positive stool cultures treated in the emergency department. Emerg. Med. J. 20, 335–338. doi: 10.1136/emj.20.4.335
Chen, M., Shpirt, A. M., Guo, X., Shashkov, A. S., Zhuang, Y., Wang, L., et al. (2015). Identification serologically, chemically and genetically of two Escherichia coli strains as candidates for new O serogroups. Microbiology 161, 1790–1796. doi: 10.1099/mic.0.000136
Chida, T., Okamura, N., Ohtani, K., Yoshida, Y., Arakawa, E., and Watanabe, H. (2000). The complete DNA sequence of the O antigen gene region of Plesiomonas shigelloides serotype O17 which is identical to Shigella sonnei form I antigen. Microbiol. Immunol. 44, 161–172. doi: 10.1111/j.1348-0421.2000.tb02478.x
Duan, Z., Niedziela, T., Lugowski, C., Cao, B., Wang, T., Xu, L., et al. (2016). Genetic diversity of O-Antigens in Hafnia alvei and the development of a suspension array for serotype detection. PLoS One 11:e0155115. doi: 10.1371/journal.pone.0155115
Ferguson, W. W., and Henderson, N. D. (1947). Description of strain C27: a motile organism with the major antigen of Shigella sonnei Phase I. J. Bacteriol. 54, 179–181.
Garrity, G., Winters, M., and Searles, D. B. (2001). “Taxonomicoutlineof Tatherosclerotic”in Be Bergen Mas EmiannuaLnd EdnEdensLeeds. New York, NY: Springer-Verlag, 13.
Greenfield, L. K., and Whitfield, C. (2012). Synthesis of lipopolysaccharide O-antigens by ABC transporter-dependent pathways. Carbohydr. Res. 356, 12–24. doi: 10.1016/j.carres.2012.02.027
Guo, D., Liu, B., Liu, F., Cao, B., Chen, M., Hao, X., et al. (2013). Development of a DNA microarray for molecular identification of all 46 Salmonella O serogroups. Appl. Environ. Microbiol. 79, 3392–3399. doi: 10.1128/aem.00225-13
Hu, D., Liu, B., Dijkshoorn, L., Wang, L., and Reeves, P. R. (2013). Diversity in the major polysaccharide antigen of Acinetobacter baumannii assessed by DNA sequencing, and development of a molecular serotyping scheme. PLoS One 8:e70329. doi: 10.1371/journal.pone.0070329
Huys, G., and Swings, J. (1999). Evaluation of a fluorescent amplified fragment length polymorphism (FAFLP) methodology for the genotypic discrimination of Aeromonas taxa. FEMS Microbiol. Lett. 177, 83–92. doi: 10.1111/j.1574-6968.1999.tb13717.x
Islam, S. T., and Lam, J. S. (2014). Synthesis of bacterial polysaccharides via the Wzx/Wzy-dependent pathway. Can. J. Microbiol. 60, 697–716. doi: 10.1139/cjm-2014-0595
Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Li, D., Liu, B., Chen, M., Guo, D., Guo, X., Liu, F., et al. (2010). A multiplex PCR method to detect 14 Escherichia coli serogroups associated with urinary tract infections. J. Microbiol. Methods 82, 71–77. doi: 10.1016/j.mimet.2010.04.008
Li, H., Du, Y., Qian, C., Li, L., Jiang, L., Jiang, X., et al. (2018). Establishment of a suspension array for Pseudomonas aeruginosa O-antigen serotyping. J. Microbiol. Methods 155, 59–64. doi: 10.1016/j.mimet.2018.11.006
Li, Q., and Reeves, P. R. (2000). Genetic variation of dTDP-L-rhamnose pathway genes in Salmonella enterica. Microbiology 146(Pt 9), 2291–2307. doi: 10.1099/00221287-146-9-2291
Li, Y., Cao, B., Liu, B., Liu, D., Gao, Q., Peng, X., et al. (2009). Molecular detection of all 34 distinct O-antigen forms of Shigella. J. Med. Microbiol. 58(Pt 1), 69–81. doi: 10.1099/jmm.0.000794-0
Li, Y., Liu, D., Cao, B., Han, W., Liu, Y., Liu, F., et al. (2006). Development of a serotype-specific DNA microarray for identification of some Shigella and pathogenic Escherichia coli strains ∇. J. Clin. Microbiol. 44, 4376–4383. doi: 10.1128/jcm.01389-06
Liu, B., Knirel, Y. A., Feng, L., Perepelov, A. V., Senchenkova, S. N., Reeves, P. R., et al. (2014). Structural diversity in Salmonella O antigens and its genetic basis. FEMS Microbiol. Rev. 38, 56–89. doi: 10.1111/1574-6976.12034
Liu, B., Knirel, Y. A., Feng, L., Perepelov, A. V., Senchenkova, S. N., Wang, Q., et al. (2008). Structure and genetics of Shigella O antigens. FEMS Microbiol. Rev. 32, 627–653. doi: 10.1111/j.1574-6976.2008.00114.x
Maciejewska, A., Lukasiewicz, J., Kaszowska, M., Man-Kupisinska, A., Jachymek, W., and Lugowski, C. (2013). Core oligosaccharide of Plesiomonas shigelloides PCM 2231 (Serotype O17) lipopolysaccharide–structural and serological analysis. Mar. Drugs 11, 440–454. doi: 10.3390/md11020440
Mandal, B. K., Schofield, P. F., and Morson, B. C. (1982). A clinicopathological study of acute colitis: the dilemma of transient colitis syndrome. Scand. J. Gastroenterol. 17, 865–869. doi: 10.3109/00365528209181107
Martinez-Murcia, A. J., Benlloch, S., and Collins, M. D. (1992). Phylogenetic interrelationships of members of the genera Aeromonas and Plesiomonas as determined by 16S ribosomal DNA sequencing: lack of congruence with results of DNA-DNA hybridizations. Int. J. Syst. Bacteriol. 42, 412–421. doi: 10.1099/00207713-42-3-412
McNeeley, D., Ivy, P., Craft, J. C., and Cohen, I. (1984). Plesiomonas: biology of the organism and diseases in children. Pediatr. Infect. Dis. 3, 176–181. doi: 10.1097/00006454-198403000-00023
Perepelov, A. V., Guo, X., Senchenkova, S. N., Shashkov, A. S., Liu, B., and Knirel, Y. A. (2016). Structure elucidation and analysis of biosynthesis genes of the O-antigen of Escherichia coli O131 containing N-acetylneuraminic acid. Carbohydr. Res. 436, 41–44. doi: 10.1016/j.carres.2016.11.003
Pieretti, G., Carillo, S., Lindner, B., Lanzetta, R., Parrilli, M., Jimenez, N., et al. (2010). The complete structure of the core of the LPS from Plesiomonas shigelloides 302-73 and the identification of its O-antigen biological repeating unit. Carbohydr. Res. 345, 2523–2528. doi: 10.1016/j.carres.2010.09.007
Pique, N., Aquilini, E., Alioto, T., Minana-Galbis, D., and Tomas, J. M. (2013). Genome sequence of Plesiomonas shigelloides strain 302-73 (Serotype O1). Genome Announc. 1, e404–e413. doi: 10.1128/genomeA.00404-13
Reeves, P. R., Hobbs, M., Valvano, M. A., Skurnik, M., Whitfield, C., Coplin, D., et al. (1996). Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 4, 495–503. doi: 10.1016/S0966-842X(97)82912-5
Sayeed, S., Sack, D. A., and Qadri, F. (1992). Protection from Shigella sonnei infection by immunisation of rabbits with Plesiomonas shigelloides (SVC 01). J. Med. Microbiol. 37, 382–384. doi: 10.1099/00222615-37-6-382
Schubert, R. H., and Holz-Bremer, A. (1999). Cell adhesion of Plesiomonas shigelloides. Zentralbl Hyg Umweltmed 202, 383–388. doi: 10.1016/S0934-8859(99)80003-0
Shepherd, J. G., Wang, L., and Reeves, P. R. (2000). Comparison of O-antigen gene clusters of Escherichia coli (Shigella) sonnei and Plesiomonas shigelloides O17: sonnei gained its current plasmid-borne O-antigen genes from P. shigelloides in a recent event. Infect. Immun. 68, 6056–6061. doi: 10.1128/IAI.68.10.6056-6061.2000
Shimada, T., and Sakazaki, R. (1985). New O and H antigens and additional serovars of Plesiomonas shigelloides. Jpn. J. Med. Sci. Biol. 38, 73–76. doi: 10.7883/yoken1952.38.73
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Tsukamoto, T., Kinoshita, Y., Shimada, T., and Sakazaki, R. (1978). Two epidemics of diarrhoeal disease possibly caused by Plesiomonas shigelloides. J. Hyg. 80, 275–280. doi: 10.1017/S0022172400053638
Wong, T. K., and Pang, S. M. (2000). Holism and caring: nursing in the Chinese health care culture. Holist. Nurs. Pract. 15, 12–21. doi: 10.1097/00004650-200010000-00004
Xie, Y., Wu, G., Tang, J., Luo, R., Patterson, J., Liu, S., et al. (2014). SOAPdenovo-Trans: de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics 30, 1660–1666. doi: 10.1093/bioinformatics/btu077
Yang, S., Xi, D., Jing, F., Kong, D., Wu, J., Feng, L., et al. (2018). Genetic diversity of K-antigen gene clusters of Escherichia coli and their molecular typing using a suspension array. Can. J. Microbiol. 64, 231–241. doi: 10.1139/cjm-2017-0620
Keywords: Plesiomonas shigelloides, O-antigen gene clusters, serogroups, PCR, luminex-based array
Citation: Xi D, Wang X, Ning K, Liu Q, Jing F, Guo X and Cao B (2019) O-Antigen Gene Clusters of Plesiomonas shigelloides Serogroups and Its Application in Development of a Molecular Serotyping Scheme. Front. Microbiol. 10:741. doi: 10.3389/fmicb.2019.00741
Received: 12 December 2018; Accepted: 25 March 2019;
Published: 10 April 2019.
Edited by:
Baolei Jia, Chung-Ang University, South KoreaReviewed by:
Susana Merino, University of Barcelona, SpainCopyright © 2019 Xi, Wang, Ning, Liu, Jing, Guo and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Boyang Cao, Ym95YW5nY2FvQG5hbmthaS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.