
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 24 April 2019
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00691
CYP51 (Erg11) belongs to the cytochrome P450 monooxygenase (CYP) superfamily and mediates a crucial step of the synthesis of ergosterol, which is a fungal-specific sterol. It is also the target of azole drugs in clinical practice. In recent years, researches on fungal CYP51 have stepped into a new stage attributing to the discovery of crystal structures of the homologs in Candida albicans, Cryptococcus neoformans and Aspergillus fumigatus. This review summarizes the functions, structures of fungal CYP51 proteins, and the inhibitors targeting these homologs. In particular, several drug-resistant mechanisms associated with the fungal CYP51s are introduced. The sequences and crystal structures of CYP51 proteins in different fungal species are also compared. These will provide new insights for the advancement of research on antifungal agents.
The incidence of invasive fungal infections has increased during the past three decades, arising more and more concern. For example, statistics show that such incidence ranges from 30 to 40% throughout critical ill patients (Bassetti et al., 2017). Prognosis of invasive fungal infections is serious. Specifically, the mortality rate of systemic C. albicans infection is up to 40% (Gunsalus et al., 2016). Risk factors, such as gastrointestinal surgery, widespread use of broad-spectrum antibiotics and popularization of central venous catheters (CVCs) mainly contribute to the invasive C. albicans infection. At the same time, non-Candida fungal infections should not be neglected, as cryptococcosis is the third prevalent disease in HIV-positive patients, and the mortality of invasive aspergillosis at 30 days in adult ICU patients is estimated to be 33.1% (Lanjewar, 2011; Baddley et al., 2013).
Represented by azole resistance, the continuous emergence of drug-resistant fungal strains has become a serious challenge for public health (Zhang et al., 2017). CYP51 (ERG11) proteins is the target of azoles, which mediates membrane permeability and fluidity by demethylating the 14-α position of lanosterol to form ergosterol (Daum et al., 1998). In attribution to the constant emergence of azole-resistant isolates, a critical understanding of the resistance mechanisms of CYP51 is required for the discovery of novel CYP51 inhibitors.
Sterol synthesis is a very ancient pathway. After the appearance of molecular oxygen in the atmosphere, squalene-2,3-epoxide is formed and then cyclized to steroid precursors, such as lanosterol. Under the oxidative removal of methyl groups by CYP51, these precursors were transformed into ergosterol, which is critical in membrane permeability and fluidity in the fungal kingdom (Rohmer et al., 1979; Daum et al., 1998).
Cytochrome P450s (P450s, CYP) are an abundant hemease superfamily. As the first group of enzymes ranked as “superfamily,” cytochrome P450s play an important role in the primary as well as secondary metabolic pathways (Lamb et al., 2007). Until August 2013, this superfamily contained 10 classes, 267 families and over 21,000 members. These members are important for catalyzing the oxidative process of various organic substrates, and play a critical role during heterogeneous metabolism and steroid conversion in biological kingdoms (Hannemann et al., 2007; Munro et al., 2018).
CYP51 proteins belong to the CYP superfamily and is the most conserved protein in it. Unlike other CYP enzymes, CYP51 has a strong specificity. It only catalyzes the demethylation of a very narrow range of substrates, including lanoserol, obtusifoliol, 24,25-dihydrolanosterol, 24-methylenedihydrolanosterol and 4 β-desmethyllanosterol (Lepesheva and Waterman, 2007). The CYP51-involved catalytic reaction consists of three steps, each of which requires one molecule of oxygen and two molecules of NADPH-sourced reduction equivalent. The first two steps are typical cytochrome P450 monooxygenation processes, during which the 14α methyl is converted to methyl alcohol and further converted to methyl aldehyde. And in the last step, the aldehyde group is transformed into formic acid and detached, accompanied with the synthesis of the Δ-14, 15 double bond (Waterman and Lepesheva, 2005).
The 14α-demethylase is the only invariant P450 present in all sterol biosynthetic pathways, suggesting that all sterol 14α-demethylases share a common prokaryotic ancestor (Lepesheva and Waterman, 2007). CYP51s are widely distributed in the fungal kingdom. However, in different species of fungi, there are still differences in the types and subtypes, as shown in the phylogenetic tree (Režen et al., 2004) (Figure 1). Only one CYP51 gene exists in the pathogenic fungi C. albicans, which belongs to the Ascomycota Saccharomycotina (Hawkins et al., 2014). In contrast, 2 or 3 CYP51 genes are commonly contained in the Ascomycota Pezizomycotina genomes, including CYP51A and CYP51B. CYP51C is exclusive in Fusarium spp. (Becher et al., 2011). Some Aspergillus Spp. such as A. fumigatus carries only one CYP51A and one CYP51B protein, while other Aspergillus species such as A. flavus and A. terreus carry a third paralogous gene, which is a copy of CYP51A or CYP51B. Studies on Aspergillus fumigatus have shown that CYP51B is constitutively expressed, while CYP51A is expressed in an inducible manner. Neither CYP51A nor CYP51B is essential for in vitro growth and virulence, and only the simultaneous inactivation of both genes is lethal (Hu et al., 2007; Hargrove et al., 2015).
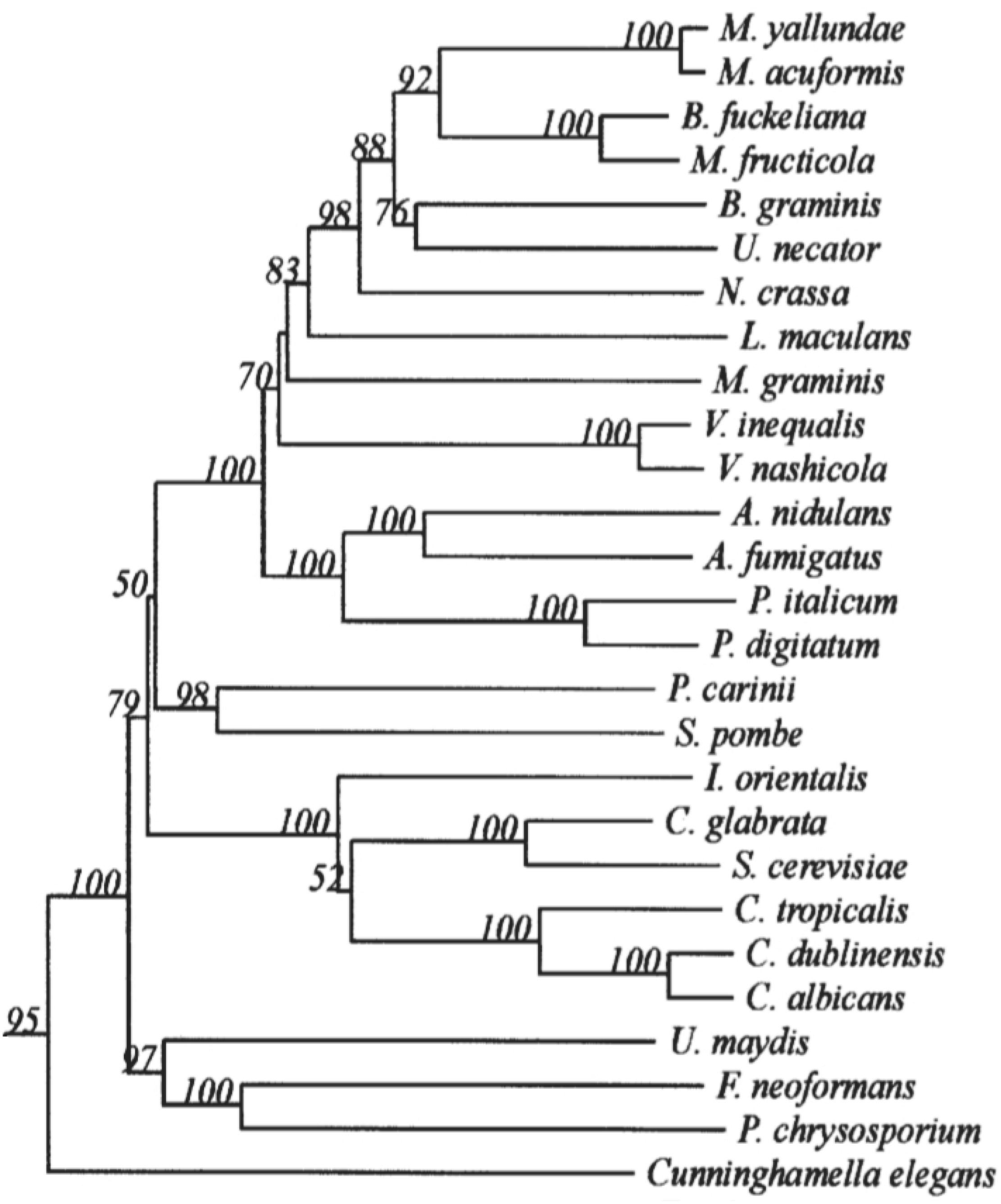
Figure 1. Minor branch of the Fungal CYP51 Phylogenetic Tree. It has been permitted by the copyright holders through RightsLink.
A recent study also showed that CYP51 might have a number of indirect functions. In C. albicans, the deletion of CYP51 (ERG11) reduces mycelial elongation and invasive growth, and causes defects of reactive oxygen elimination, resulting in reduced virulence in vivo. It is worth paying special attention that ERG11-deficient fungi are more susceptible to phagocytosis by macrophages, which indicates that CYP51 may be critical for the immune escape process in fungi (Wu et al., 2018).
Present in all animals, plants, fungi, in some protozoa and bacteria, the CYP51 protein located in the inner face of the endoplasmic reticulum is a membrane monospanning enzyme (Noel, 2012). And its N-terminus includes an amphipathic helix, which links the catalytic subunit to the lipid bilayer (Monk et al., 2014). Besides crystallographic structures of CYP51 proteins from some protozoa and bacteria, and the ligand-free CYP51 proteins from Homo sapiens (3JUV) and complexed with ketoconazole (3LD6) deposited in Protein Data Bank (PDB) (Strushkevich et al., 2010). X-ray structures of CYP51 proteins of some fungi have already been described in literature, including CYP51 proteins from Saccharomyces cerevisiae complexed with the substrate lanosterol (4LXJ) and complexed with itraconazole (5EQB) (Monk et al., 2014), voriconazole (5HS1) and fluconazole (4WMZ) (Sagatova et al., 2015), from C. albicans complexed with posaconazole (5FSA) and the tetrazole-based antifungal drug candidate VT1161 (VT1) (5TZ1) (Hargrove et al., 2017a), and CYP51B structure in complex with the VNI derivative (6CR2) and a tetrazole-based inhibitor VT-1598 (5FRB) from A. fumigates (Hargrove et al., 2017b).
Together with information from the analysis of multiple-sequence alignment of CYP51 proteins from human and fungi including S. cerevisiae, C. albicans, C. glabrata, C. tropicalis, C. krusei, C. dubliniensis, C. parapsilosis, A. fumigatus and Cryptococcus neoformans showing that the identity varied between 36.5 and 93.9% among them (Table 1). The comprehensive comparative analysis of three-dimensional structures uncovered the basic understanding how CYP51 enzymes might maintain their conservation in human and fungi. As shown in Figure 2, the majority of amino-acids residues conserved in the folding of chains might play an essential structural role for their enzymatic function. The residues forming the surface of CYP51 proteins active site were highly conserved, such as Y118, F126, G127, V130 and T311 from the CYP51 proteins helix B’, B” helical turn and Helix I signature regions, respectively (Lepesheva and Waterman, 2011; Hargrove et al., 2017a; Keniya et al., 2018).
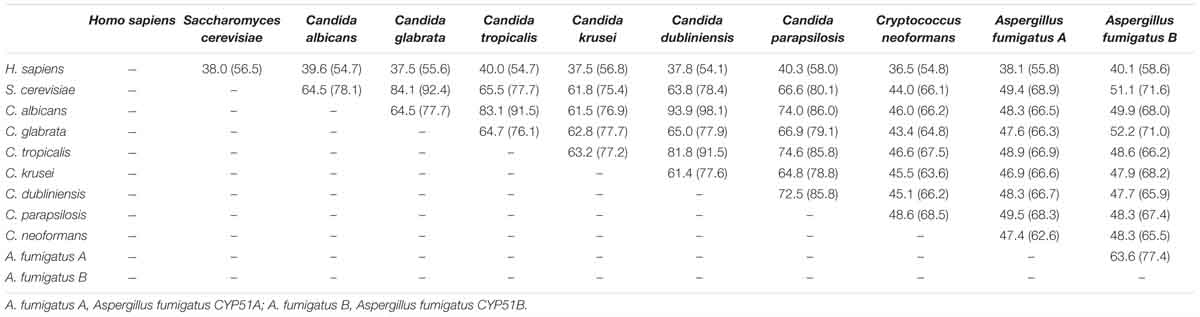
Table 1. Comparison of identity (similarity) % of amino acid residue sequence of CYP51 from different organisms calculated using ClustalW2.
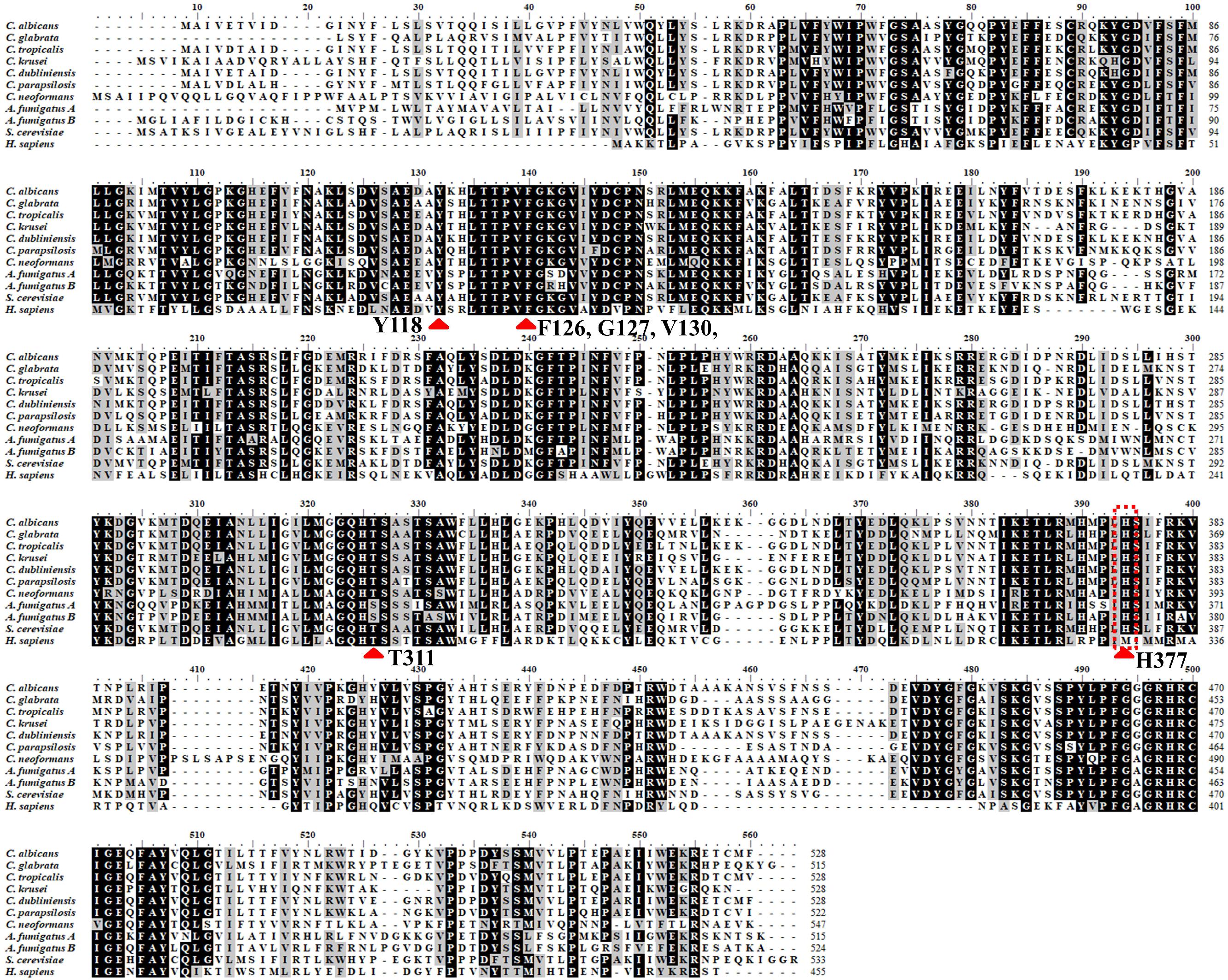
Figure 2. Structure alignment of primary structure of CYP51 from different organisms > 80%. Structure alignment of primary structure of CYP51 from different organisms: H. sapiens, S. cerevisiae, C. albicans, C. glabrata, C. tropicalis, C. krusei, C. dubliniensis, C. parapsilosis, A. fumigatus, and Cryptococcus neoformans. The residues identical in all CYP51s are marked with black, the light gray show the residues conserved in more than 80% sequences. The sequences were from NCBI and CGD database and were aligned using the ClustalW2 program. Red arrow means the most conserved CYP51 active amino acid sites among diffierent species.
The conserved amino acid sequences can be classified into three motifs and six putative substrate recognition sites (SRS). Among the three motifs, the most conserved FXXGXXXCXG is a heme binding domain containing a heme axial Cys ligand; and the E–R–R triad, formed by the motifs EXXR and PER, contributes to locking the heme pocket into position and to guarantee stabilization of the core structure (Figure 3). And among the six putative SRSs, the most thoroughly studied SRS1 and SRS4 can be used as landmarks of the fungal CYP51 (Lepesheva and Waterman, 2007) (Figure 4).
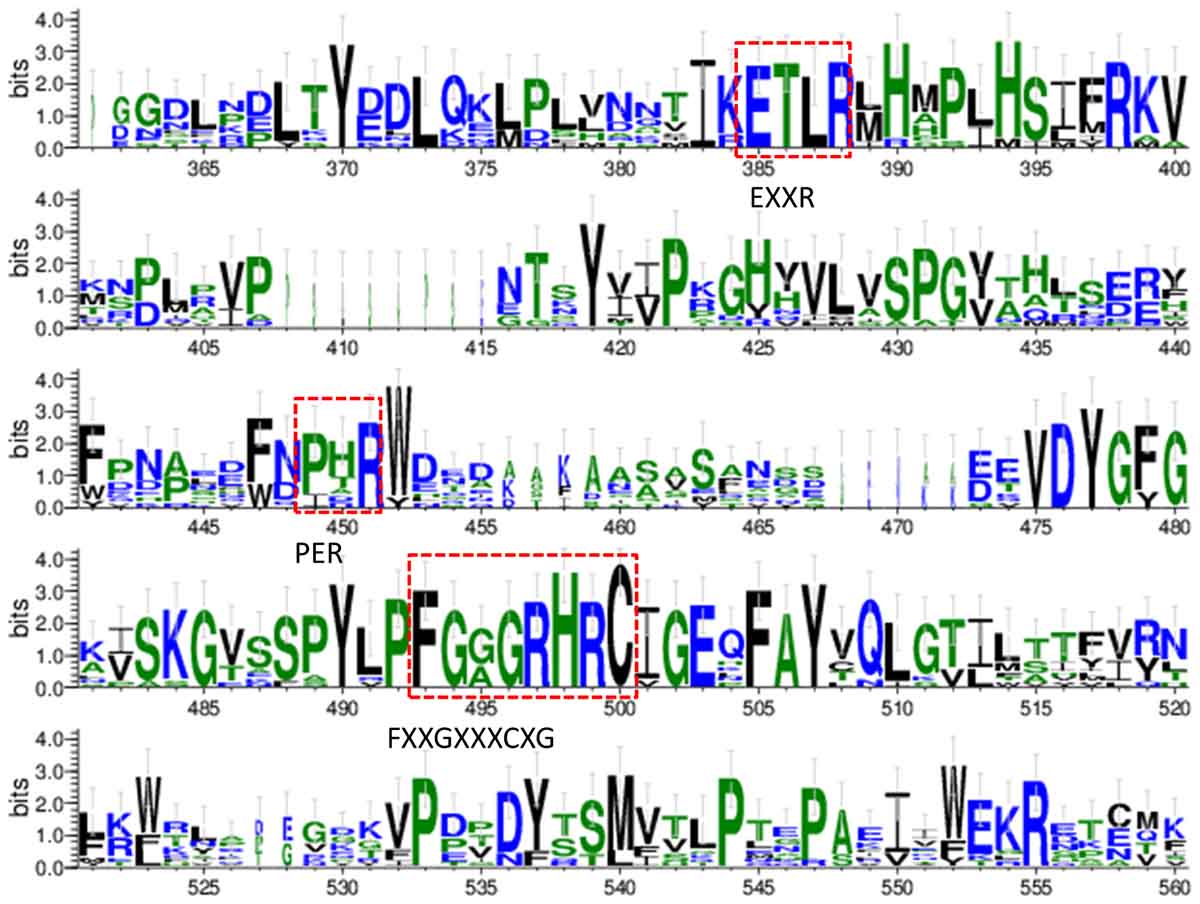
Figure 3. Sequence logos of the conserved CYP motifs from the tested fungi and human’s CYP51. The sequences were from NCBI and CGD database and the consensus logos were generated by WebLogo (http://weblogo.threeplusone.com/create.cgi).
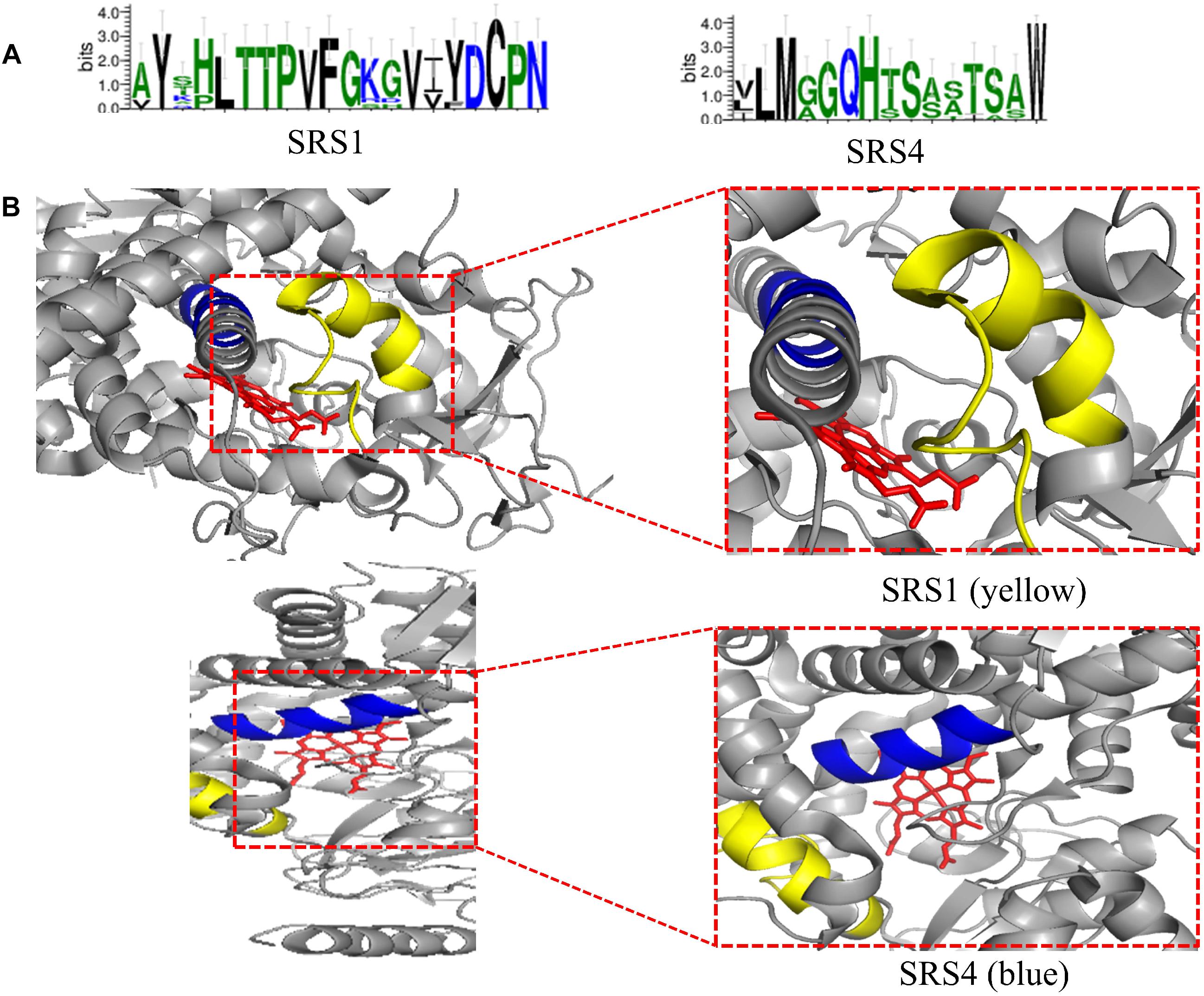
Figure 4. (A) Predicted SRS1 (the positions 117–134) and SRS4 (the positions 305–314) of tested fungal CYP51 (S. cerevisiae, C. albicans, C. glabrata, C. tropicalis, C. krusei, C. dubliniensis, C. parapsilosis, A. fumigatus, and Cryptococcus neoformans). The sequences were from NCBI and CGD database and the consensus logos were generated by WebLogo (http://weblogo.threeplusone.com/create.cgi); (B) Location of SRS1(yellow) and SRS4 (blue) in C. albicans CYP51 (5v5z). HEME is shown in red. They are obtained from the published crystal data file from the PDB database, and then displayed by PyMOL Version 1.5.0.3.
To crystallize C. albicans CYP51 proteins complexes with posaconazole and a tetrazole-based drug candidate VT-1161, Hargrove et al. used E. coli cells to express and purify the truncated C. albicans CYP51 proteins (56 kDa) without the 48-amino-acid-long N-terminal membrane anchor sequence (Hargrove et al., 2017a). The structural analysis of C. albicans CYP51 proteins complexes carried out by Hargrove et al. showed that posaconazole had contacts with a set of 28 residues of C. albicans CYP51 proteins, while VT-1161 interacts with 22 amino acid residues, and forms the H-bond between its trifluoroethoxyphenyl oxygen and the imidazole ring of His377 of C. albicans CYP51 proteins [Table 2 from Hargrove et al. (2017a) with slight modification]. Further, the X-ray structure of A. fumigates CYP51B complex with the tetrazole-based inhibitor VT-1598 (5FRB) determined also by Hargrove et al. (2017b) showed the formation of an optimized hydrogen bond between the phenoxymethyl oxygen of VT-1598 and the imidazole ring nitrogen of His374 of A. fumigates CYP51B. Comparative structural analysis of the CYP51 proteins residue (His377 of C. albicans CYP51 proteins, His374 of A. fumigates CYP51B) among different organisms suggested it was highly conserved across fungal pathogens but not in human, supporting its fungus specificity and the role of H bonding in fungal CYP51/inhibitor complexes (Figures 2, 5).
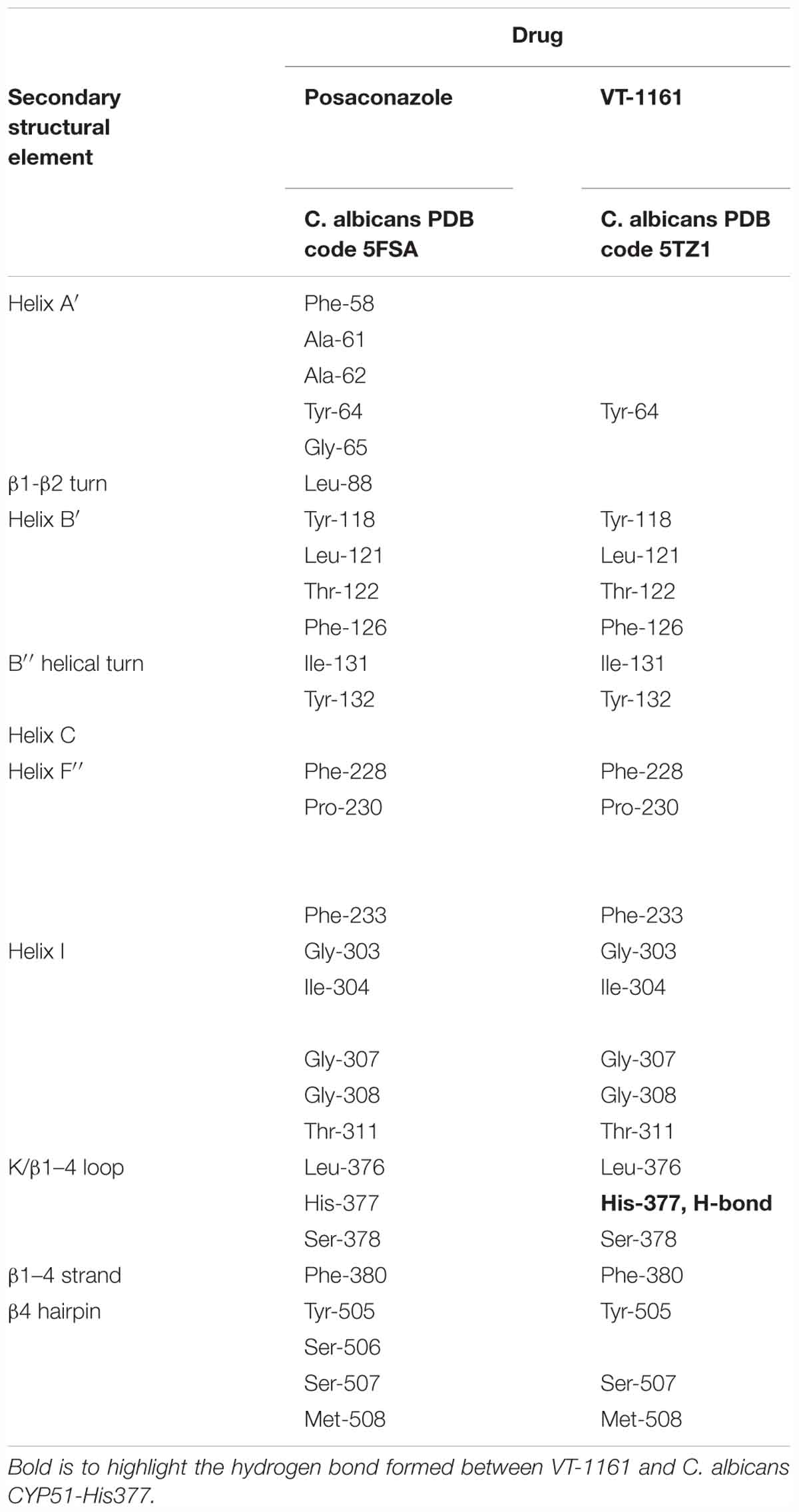
Table 2. Posaconazole and a tetrazole-based drug candidate VT-1161 contacting residues (<4.5 Å) in C. albicans CYP51 structures.
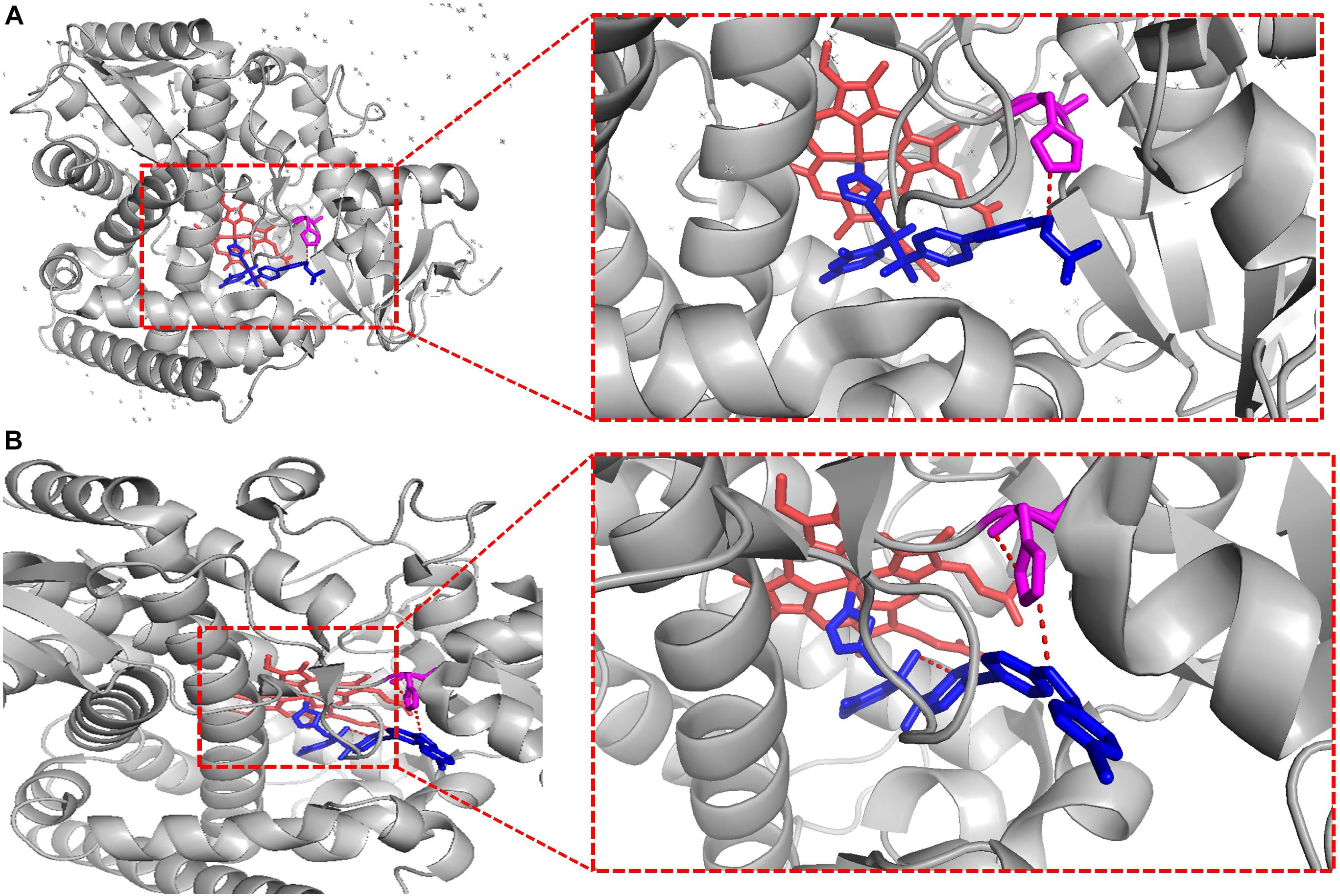
Figure 5. VT-1161 andVT-1598 forming the H bond. VT-1161 and VT-1598 forming the H bond with the His377 of C. albicans CYP51 and the His374 of A. fumigates CYP51B. (A) C. albicans CYP51-His377-VT-1161(5TZ1); (B) A. fumigates CYP51B-His374-VT-1598 (5FRB). They are obtained from the published crystal data file from the PDB database, and then displayed by PyMOL Version 1.5.0.3. The red dot line means H-bonds formed between VT-1161 and C. albicans CYP51-His377 in Figure 5A, and H-bonds formed between VT-1598 and A. fumigates CYP51B-His374 in Figure 5B.
After the first structure of CYP51 proteins with the N-terminal region from S. cerevisiae (5EQB) in complex with itraconazole was elucidated and deposited in PDB, Keniya et al. concluded that the conformations of the three full-length fungal Cyp51 structures binding to itraconazole were close to that of the N-truncated C. albicans Cyp51 with posaconazole. In comparison these had only slight variations in residues conformations, such as C. glabrata Cyp51 I71, T75, I240; C. albicans Cyp51 A62, Q66, I231 and S. cerevisiae Cyp51 V70, M74, I239, all of which were within the ligand binding pocket and may not be significant due to modest electron densities in this region. Besides, the structures showed S. cerevisiae Cyp51-fluconazole or itraconazole had the binding sites in near identical conformations (Sagatova et al., 2015; Keniya et al., 2018). The X-ray crystal structures determined by Keniya et al. (2018) gives us insights into the CYP51 proteins reaction mechanism and emphasizes the identity of ligand-binding sites of fungal CYP51 families, providing a practical basis for the discovery and optimization of novel antifungals targeting at CYP51 families.
In recent years, with the widespread use of CYP51-targeting drugs, drug-resistant Candida, Cryptococcus, and Aspergillus have emerged continuously. As shown in Table 3, in many resistant isolates, the decrease of susceptibility originated from mechanisms associated with CYP51, while other isolates not. Besides, transcription factors Pdr1 and Stb5, or the insertion of the Aft1 transposon into the CYP51 promoter region, can also cause a decrease in the sensitivity of the drugs (Albarrag et al., 2011; Noble et al., 2013; Nishikawa et al., 2016). Transcription factors of non-pathogenic species may also bring some information, such as Set4 in S. cerevisiae, which represses CYP51 expression and reduces drug resistance (Serratore et al., 2018). It is worth noting that drug resistance is often a combination of multiple mechanisms (Berkow and Lockhart, 2017).
The mechanisms associated with CYP51 are discussed below in detail.
Candida spp., represented by C. albicans, is the most prominent pathogenic fungus. The spectrum of disease of invasive candidiasis ranges from minimally symptomatic candidaemia to fulminant sepsis with an associated mortality exceeding 70% (Pappas et al., 2018). The resistance mechanisms of Candida spp. related with CYP51 includes point mutation, genomic plasticity, and upregulation of CYP51 meditated by the transportation factor Upc2.
Point mutation (amino acid non-synonymous substitutions) of CYP51 proteins is a critical origination of reduced drug susceptibility. Three ways were proposed to explain the resistance mechanism: (1) the corresponding amino acids docked with azoles are changed, (2) the structure of the binding cavity is rearranged, leading to changed position of the azole molecule or the heme iron, disturbing the interaction between them, and (3) the access of the drug into the active site is blocked (Becher and Wirsel, 2012).
In C. albicans, mutations of CYP51 proteins amino acid sequences frequently occurs on 105–165, 266–287, and 405–488 (Marichal et al., 1999). Among these mutations, N-terminal and C-terminal are more frequent than central regions, probably due to these terminal substitutions lead to changes in secondary and tertiary structure, especially substrate binding cavities, which are located in α-helix B, B′, C, and in their connection loops (Becher and Wirsel, 2012). And sequential replacement of ERG11 mutant alleles with wild-type alleles contributes to the reduction of resistance (MacCallum et al., 2010). The resistant C. albicans double point mutation Y132F G464S (Y140F G464S by S. cerevisiae numbering) were artificially introduced into S. cerevisiae CYP51, leading to a decrease in sensitivity of the latter (Sagatova et al., 2018). In addition, mutations located at different sites of CYP51 proteins provide different degrees of resistance enhancement. As an instance, K143R is stronger than F449V (Flowers et al., 2015).
The non-albicans Candida (NCAC) species also contains amino acid non-synonymous substitutions, such as Y132F in C. parapsilosis, Y132F and K143R in C. tropicalis (Vandeputte et al., 2005; Xisto et al., 2017; Choi et al., 2018).
The newly emerged C. auris is a multi-drug resistant Candida that causes serious invasive infections, with a mortality rate approximate to 60% (Lee et al., 2011). CYP51 point mutation plays an important role in pan-azole resistance of C. auris. Nine typical amino acid substitutions have been identified, three of which are closely associated with geographic clades: F126T in South Africa, Y132F in Venezuela, and Y132F or K143R in India and Pakistan (Lockhart et al., 2017).
Genomic plasticity, including aneuploidy and loss of heterozygosity (LOH), refers to the abnormal fungal chromosomal behavior induced by heat, oxidative stress or antifungal drugs. It is an important mechanism for fungal adaptation to the environment.
Aneuploidy is a reversible fungal chromosomal adaptive behavior in response to drug stress. If the “selectivity” of the drug disappears, the cells will return to the euploid state. Aneuploidy after azole stress was first discovered in C. glabrata and was detailed studied in C. albicans (Vanden et al., 1992).
The aneuploidy of C. albicans chr1, chr3, chr5, chr6 and chrR has been reported under the stress of azoles treatments (Selmecki et al., 2010; Li et al., 2015). Research on fluconazole-resistant strains has revealed that at least half of them carry aneuploid chromosomes. The duplication of the chr5’s left arm in C. albicans [named i(5L)] results in multiple copies and overexpression of CYP51 gene, contributing to azole resistance (Selmecki et al., 2006). Besides, aneuploidy of chr5 (carrying TAC1), chr3 (MRR1 and CDR1), Chr4 or chr6 (MDR1) also reduce drug susceptibility (Selmecki et al., 2010). Some aneuploid C. albicans chromosomes source from the mitotic defect in the process of quasi-fertility (Forche et al., 2008). There also exists an additional mechanism that, two diploid cells and one tetraploid cell are connected to each other to form a “trimeric,” then the tetraploid cell undergoes defective mitosis and results in two aneuploid progeny cells (Harrison et al., 2014).
An “Evolution Trap” (ET) strategy was proposed to suppress the occurrence of aneuploidy. The aneuploidy of a whole microflora can develop into multiple random directions, but once a specific inducing factor (stress X) is used to strictly limit its development direction, another treatment (treatment Y) can be applied to eliminate this trend and inhibits the generation of resistance. Such strategy has successfully pulled the minimum inhibitory concentration (MIC) against C. albicans carrying aneuploidy-sourced resistance back to normal level (Chen et al., 2015).
Loss of heterozygosity is another branch of genomic plasticity leading to Candida species resistance. It is an irreversible process in diploid fungi, thus resulting the acquired resistance gene mutation (∗) become multiple (e.g., ERG11/ERG11∗→ ERG11∗/ ERG11∗), thus results in overexpression of such gene (White, 1997). LOH contains three mechanisms, (1) local recombination of chromosomes, (2) mitotic recombination between centromeres and related locus, (3) whole-chromosome loss and the remaining chromosome’s duplication (Morschhäuser, 2016).
Transcription factors of CYP51 are tightly related to drug resistance. The Zn2-Cys6 transcription factor Upc2, located in Candida spp., is highly relevant to the increase of azole sensitivity (Vasicek et al., 2014). It has been confirmed that when Upc2 in C. albicans or C. parapsilosis or Upc2A in C. glabrata is deleted, the susceptibility to azoles will be enhanced (Guida et al., 2011; Vasicek et al., 2014; Whaley et al., 2014). The C-terminal domain (CTD) of Upc2 is a novel α-helical fold with a deep hydrophobic pocket. Treatment with azole reduces the membrane ergosterol level, then ergosterol molecules that are previously bound to CTD dissociates from Upc2p. Thereby Upc2p relocates from the cytoplasm to the nucleus to activate CYP51 expression (Yang et al., 2015). It is worth noting that Upc2 only up-regulates the expression of CYP51 under azole stress conditions (Hoot et al., 2011).
The gain-of-function (GOF) mutation of Upc2 also contributes to Candida species increased drug resistance. Typical GOF point mutations inducing overexpression of CYP51 in C. albicans include A643V, G648D, G648S, and Y642F (Dunkel et al., 2008; Flowers et al., 2012). Point mutations in UPC2 can reduce the sensitivity to azoles in combination with amino acid substitution of CYP51. For example, when combined with the CYP51G464S mutation, the MIC value of C. albicans carrying Upc2G648D against fluconazole is increased from 4 μg/ml to 16 μg/ml (Sasse et al., 2012).
Gain-of-function mutations were also found in C. tropicalis Upc2. Nucleotide substitutions T118G and G155A in CtUpc2 promoter increase the expression of this gene, and amino acid substitution G392E in CtUpc2p enhances drug resistance when expressed heterologously in S. cerevisiae (Jiang et al., 2016).
The GOF amino acid substitutions are often localized near the C terminus of Upc2p, where the activation domain of zinc-cluster transcription factors is found. Mutations in this region leading to reduced drug susceptibility include two possible mechanisms: (1) relieve Upc2 from a repressor that would otherwise keep this transcription factor inactive, (2) interfere with the transmembrane region of this protein, leading to Upc2p nuclear localization and initiation of CYP51 transcription (Flowers et al., 2012).
Cryptococcal meningitis (CM) is the most common infection caused by Cryptococcus spp., and is frequently recommended to be treated with fluconazole monotherapy in national guidelines. However, even with such treatment, the mortality of CM can exceed 50%, much of which owing to drug-induced cryptococcal resistance (Rothe et al., 2013; Bongomin et al., 2018). Amino acid point mutation, genomic aneuploidy and Sre1-induced overexpression of CYP51 account for the reduced azole susceptibility.
The amino acid point mutation G344S was found in CYP51 proteins of Cryptococcus neoformans var. grubii, resulting in multi-azole resistance (Kano et al., 2017). Besides, the substitution G484S may confer a change in the orientation of the P450 heme-binding domain, decreasing catalytic activity and azole binding of Cryptococcus neoformans CYP51 proteins (CnCYP51 proteins) (Rodero et al., 2003). And the substitution Y145F affords resistance to voriconazole but attenuates resistance to itraconazole and posaconazole (Sionov et al., 2012). In Cryptococcus gattii CYP51 proteins, amino acid non-synonymous substitution N249D are deduced to result in azole resistance (Gast et al., 2013).
As for genomic aneuploidy, Cryptococcus neoformans Chr1 disomy, which is common in heteroresistant isolates, results in duplication of CnCYP51 and reduced azole sensitivity. A recent report reveals that clinical fluconazole treatment can induce aneuploidy of C. neoformans in CM patients, and relapse of CM is associated with Chr1 disomy (Bongomin et al., 2018; Stone et al., 2019).
Cryptococcus neoformans is haploid and aneuploid cells of this species mainly derive from the uncoupling of cell growth and nuclear division (Altamirano et al., 2017). Unlike C. albicans, the aneuploidy of Chr1 in C. neoformans only repeats the entire chromosome without forming segmental isochromosomes (Kwon-Chung and Chang, 2012). A study indicates the detailed mechanism causing aneuploidy: final degradation of the septum is affected by fluconazole during cytokinesis, resulting in Chr1 disomy multinucleated cells, and these cells exhibit an increased potential to proliferate in the presence of fluconazole (Altamirano et al., 2017). However, another study points out that the fluconazole-induced multinucleated cells fail to propagate to form colonies in the presence of fluconazole, and chromosome missegregation of C. neoformans dividing cells has not been detected, suggesting C. neoformans forms aneuploid clones directly from uninucleated cells under fluconazole stress (Chang et al., 2018).
Aneuploidy can be regulated by certain factors in Cryptococcal spp. For example, the decreased expression of AIF1 (apoptosis-inducing factor) conduces to maintain Chr1 aneuploidy, thereby contributes to a stable repeat of CYP51 and preserves resistance to azoles (Semighini et al., 2011).
Sterol regulatory element-binding protein (SREBPs) regulate the CYP51s’ expression in many species of fungal, including C. neoformans (Chang et al., 2009). Under azoles or hypoxic stress, SREBPs regulate the transcription of CYP51 by binding to the sterol regulatory element (SRE) in the promoter
The SREBP in C. neoformans is Sre1. Sre1 is cleaved by Scp1and functions to regulate the expression of CYP51, thus plays a key role in drug resistance (Bien and Espenshade, 2010). Studies have shown that deletion of Sre1 converts the effect of azoles from fungistatic to fungicidal (Bien et al., 2009; Chang et al., 2009).
The ability Aspergillus spp. to adapt to mammal hosts or external environment is a vital fungal characteristic that leads to treatment failure and the emergence of resistant isolates worldwide. Non-synonymous substitution of amino acids, transcription factors SrbA and AtrR, tandem repeats, and Dap proteins constitute the CYP51-related resistance mechanisms of Aspergillus spp.
The lanosterol 14α-demethylase point amino acid mutations of A. fumigatus mainly appear in CYP51A, and G54, L98, G138, M220 and G448 are the hotspots (Denning and Perlin, 2011). G54R/E/V and G138 lead to cross-resistance to itraconazole and posaconazole, and G448S results in voriconazole tolerance (Chowdhary et al., 2017), while M220I/V/T/K can develop resistance to itraconazole, voriconazole, refconazole, and posaconazole (Mellado et al., 2004). When the G138S point-substituted CYP51A in the resistant A. fumigatus was mutated back, the tolerance of the isolate diminished (Umeyama et al., 2018). Moreover, one mutation may have varied effects on resistance for different azoles. For example, in the heterologous expression experiment, G54W significantly reduces the susceptibility to itraconazole and posaconazole, while has almost no effect on voriconazole (Alcazar-Fuoli et al., 2011).
Amino acids substitution also occurs in non-fumigatus Aspergillus species. Four mutations of A. flavus CYP51C (S196F, A324P, N423D and V465M) are correlated with voriconazole resistance (Sharma et al., 2018). For A. clavatus CYP51A, E483K and P486S mutations may narrow the azole transport and therefore confer lower susceptibility (Abastabar et al., 2019). As for A. terreus and related species, M217T and M217V mutations of CYP51Ap was found correlating with posaconazole resistance (Zoran et al., 2018).
Notably, appliance of azole pesticides in agriculture is one of the reasons for the non-synonymous substitution of A. fumigatus CYP51 proteins. Mutations in this type include L98H, Y121F and T289A, which are often accompanied with tandem repeats of the CYP51 promoter (Mellado et al., 2007; Camps et al., 2012; Chowdhary et al., 2013; van der Linden et al., 2013; Isla et al., 2018).
Tandem repeats include 34-base pair (TR34) and 46-base pair (TR46) (Spiess et al., 2014). CBC (CGAAT binding complex) binds to CGAAT of -293 to -289 position in the CYP51 promoter and downregulates CYP51A expression. Tandem repeats reduce the affinity of CBC and the promoter, upregulating CYP51A. Mechanism researches indicate that the presence of eight different nucleotides at the 3′end of TR34 lead to lower CBC affinity (Gsaller et al., 2016).
The combination of CYP51 promoter tandem repeat and CYP51 proteins point mutation contains TR34/L98H/S297T/F495I and TR46/Y121F/T289A, leading to broad-spectrum azole resistance. (Snelders et al., 2015; Chen et al., 2018; Isla et al., 2018; Pinto et al., 2018; Tsitsopoulou et al., 2018; Tsuchido et al., 2019). Studies on TR34/L98H have shown that L98H can cause a flexible change in the BC loop and IH loop of A. fumigatus CYP51A (AfCYP51A), which changes the position of the tyrosine107 and tyrosine 121 side chains. This modifies the ligand access channels in the AfCYP51A and prevents the binding of azoles toward the active heme (Snelders et al., 2011). Moreover, TR34/L98H doesn’t incur a fitness cost or survival disadvantage to A. fumigatus (Beer et al., 2018). On the other hand, studies on TR46/Y121F/T289A indicate that the Y121F substitution seems to disrupt the H-bond between tyrosine and the heme center of AfCYP51A, resulting in the instability of enzyme’s active center (Snelders et al., 2015).
Besides, the insertion of Atf1 in the A. fumigatus CYP51A promoter may also be one of the factors leading to azole resistance (Albarrag et al., 2011). But it requires more researches to confirm whether the effect is direct.
The transportation factor SrbA, as one of A. fumigatus SREBPs, modulates the expression of AfCYP51A. Different from C. neoformans, Scap homologue is absent in A. fumigatus, and SrbA directly binds to the 34 mer of AfCYP51A promoter without cleavage, regulating the synthesis of ergosterol (Blosser and Cramer, 2011; Gsaller et al., 2016). SrbA is directly activated by azole stress and is associated with the intrinsic resistance of A. fumigatus to fluconazole (Song et al., 2017). An azole-resistant strain with TR46/Y121F/T289A can be sensitized to azoles by deletion of srbA (Hagiwara et al., 2016). The intrinsic expression of CYP51 in such strain restores the MIC value to its original level (Willger et al., 2008).
AtrR is a newly discovered fungal-specific Zn2-Cys6 transcription factor in Aspergillus spp. It modulates the expression of CYP51A by directly binding to the promoter of this gene. Deletion of AtrR results in hypersensitivity to azoles and invalidates the CYP51AG54E mutation that would otherwise render azole resistance (Hagiwara et al., 2017).
Damage Resistance Protein A (DapA), which belongs to the cytochrome b5-like heme-binding damage resistance protein (Dap) family, responds to azole treatment in a concentration-dependent manner in A. fumigatus. It co-localizes with CYP51A/B protein in the endoplasmic reticulum (ER), and then binds to the heme group to stabilize these CYP51 proteins. DapA is highly associated with the intrinsic azole resistance of A. fumigatus, and the deletion of DapA leads to hypersensitivity to azoles (Song et al., 2016). Studies show that DapA may indirectly sense the azole stress at the downstream of SrbA (Song et al., 2017).
The study of azole antifungal antifungal agents can be traced back to 1944 (Woolley, 1944). Since then, imidazole (clotrimazole, miconazole, econazole, and ketoconazole) and two generations of triazoles (fluconazole, itraconazole, voriconazole, posaconazole, isavuconazole) successively entered into the clinic (Musiol and Kowalczyk, 2012) (partly shown in Table 4). Besides, there are several other CYP51 inhibitors used in the treatment of topical or superficial fungal infections, such as oxiconazole, sertaconazole, luliconazole, efinaconazole, and ravaconazole.
Both imidazoles and triazoles exits adverse reactions caused by inhibition of human CYP450 (such as CYP3A4 and CYP2C9), due to the strong affinity to heme iron (Hoekstra et al., 2014). Fortunately, replacement by 1-tetrazole could attenuate such affinity. Based on this idea, new compounds VT-1611, VT-1129, and VT-1598 have been developed (Hoekstra et al., 2014) (Figure 6).
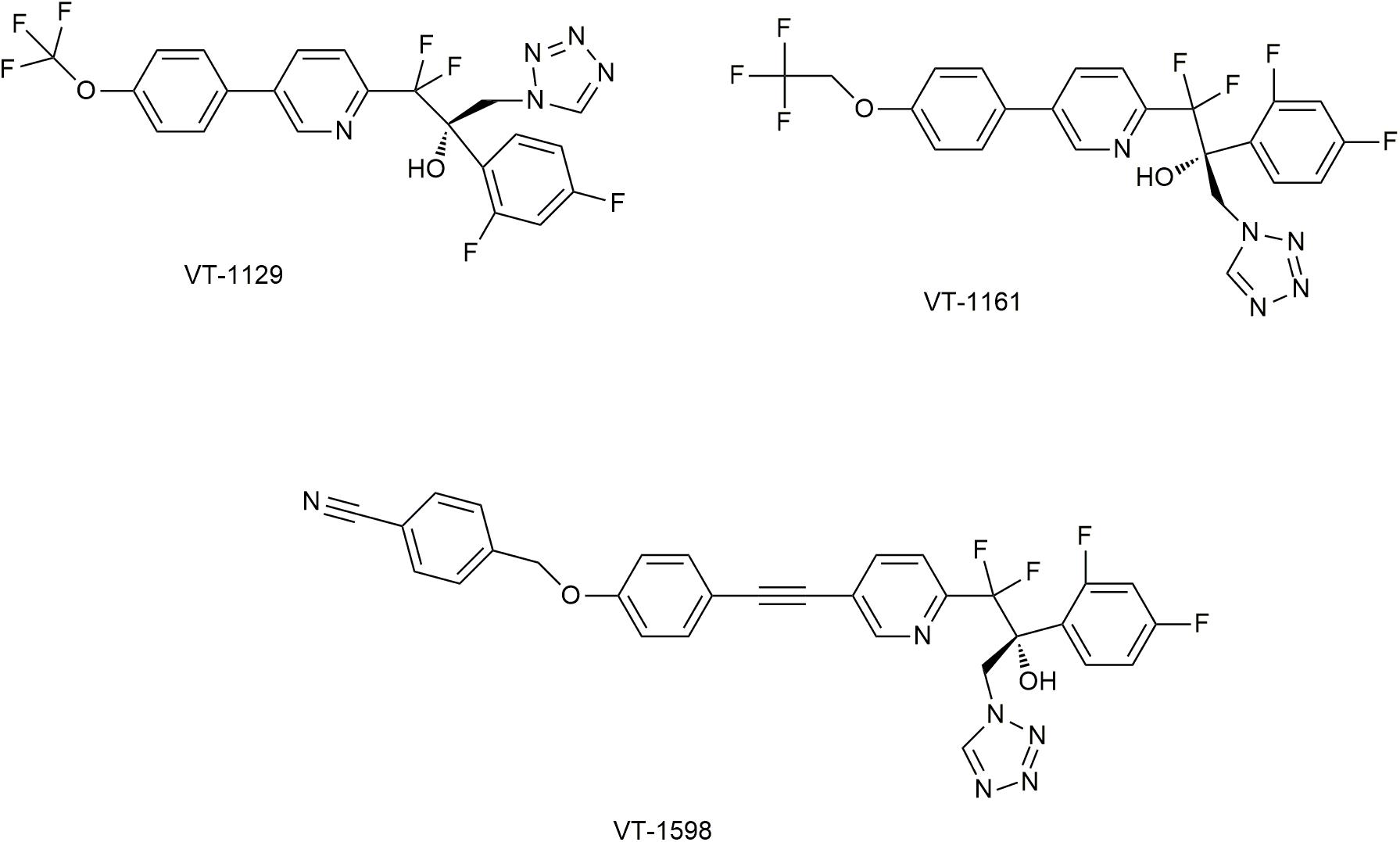
Figure 6. Novel CYP51 inhibitors in studies. It has been permitted by the copyright holders through RightsLink.
VT-1598 has a good affinity for the fungal CYP51 proteins, as the Kd of this compound to A. fumigatus CYP51B is 13 nM (Hargrove et al., 2017b). Its antifungal spectrum is relatively broad. The MIC50 for azole-resistant C. albicans and C. glabrata is 0.124 and 1.19 μg/ml (Wiederhold et al., 2018b). Especially, the inhibition of VT-1598 against the clinical Coccidioides isolates is significantly better than that of fluconazole (McCarthy et al., 2017). The non-selective inhibition of VT-1598 to human CYP450 is weak, as IC50 for human CYP2C9 and CYP3A4 are both more than 200 μM, and the IC50 for CYP2C19 is 138 μM (Yates et al., 2017). When the oral dose (15 mg/kg/day) of VT-1598 is applied to mice, the Cmax is about 13 mg/L, the C24 h level is 6.7 mg/L and the half-life period is 22 h in plasma (Garvey et al., 2018).
VT-1598 has a high binding rate of plasma protein. When co-incubated with plasma at concentration of 1 mg or 5 mg/L in vitro, only less than 1% showed a free state (Garvey et al., 2018). In vivo, when treated with 20 mg/kg of VT-1598 per day, the minimum plasma concentration (Cmin) was 32-fold greater than that of the 25 mg/kg dose of fluconazole (Break et al., 2018a).
In the mouse model, VT-1598 has been used alone or in combination with amphotericin liposomes, and has achieved good efficiency for cryptococcal meningitis caused by C. neoformans or C. gatti infection (Garvey et al., 2018). VT-1598 has also achieved significant preventive effects against mucosal candidiasis induced by sensitive or resistant Candida spp. (Break et al., 2018a). In addition, VT-1598 can be used to treat central nervous system coccidioidomycosis infected by C. posadasii and C. immitis (Wiederhold et al., 2018c). In May 2016, FDA granted orphan drug designation to VT-1598 for the treatment of Valley fever, a disease caused by Coccidioides infection (McCarthy et al., 2017).
VT-1161 can tightly bind to the fungal CYP51 proteins and effectively inhibit the activity of such proteins. For example, the Kd of VT-1161 to C. albicans CYP51 proteins is less than 39 nM and IC50 ranges from 1.4 to 1.6 μM, resulting in the proportion of ergosterol to total sterols drop to only 3% (Warrilow et al., 2014). The MIC50 of VT-1161 against fluconazole-resistant C. albicans is 0.03 μg/ml (Break et al., 2018b). For T. rubrum CYP51 proteins, The Kd and IC50 values are 242 nM and 0.14 μM, respectively (Warrilow et al., 2017). Its non-selective inhibition of human CYP450 is also weak, appears in its IC50 of CYP2C9, CYP2C19 and CYP3A4 are 99, 72, and 65 μM. Pharmacokinetic experiments in mice showed that VT1161 has a wide distribution volume (1.4 L/kg), high oral bioavailability (73%), and long half-life period (>48 h)(Garvey et al., 2015). Phase II clinical trials showed oral VT-1161 plasma exposure for the 150 mg/24 weeks’ or 300 mg/24 weeks’ treatment groups were 3.81 and 8.33 μg/mL (Brand et al., 2018).
Animal model studies have shown that VT1161 can be used to prevent or treat mucormycosis caused by Rhizopus arrhizus, and can also be used for treatment of mouse modeling infection or canine naturally occurring coccidioidomycosis (Shubitz et al., 2015, 2017; Gebremariam et al., 2017). In addition, in a mouse model, VT1611 can treat oropharynx or vaginal Candidiasis caused by fluconazole sensitive or resistant C. albicans (Break et al., 2018b). Phase II clinical trials indicate that oral VT1161 has a good effect on treating human vulvar candidiasis, with a satisfactory tolerance and a low incidence of adverse reactions (Brand et al., 2018). Currently, oral VT-1161 has completed Phase II clinical trials for moderate – severe interdigital tinea pedis, vaginal candidiasis, and onychomycosis of the toenail. And phase III clinical trial is underway for Vaginal Candidiasis.
VT-1129 has a similar skeleton to VT-1161, and shows a remarkable inhibitory activity against Cryptococcus CYP51 protein. For example, the Kd for C. neoformans, C. gattii, and C. grubii CYP51 proteins are about 11, 24, and 25 nM, respectively, and the corresponding IC50 are 0.16, 0.15, and 0.18 μM. VT-1129 can reduce the proportion of ergosterol to total sterols in C. neoformans to 11.5%, and only 0.12 μg/ml can totally inhibit the growth of C. neoformans (Warrilow et al., 2016; Wiederhold et al., 2018a). Its non-specific inhibition to human CYP450 is lower than that of the previous azole antifungals, appears in the IC50 to CYP2C9 and CYP2C19 are 87 and 110 μM, respectively, and for CYP3A4 is also higher than 79 μM (Warrilow et al., 2016). VT-1129 also inhibits the growth of azole or echinocandin resistant C. glabrata and C. krusei (Schell et al., 2017).
The pharmacokinetic experiments in mice model showed that VT-1129’s half-life period is long (>6 days). The plasma and brain concentrations were still above the MIC values even after 20 and 32 days stopping oral treatment of VT-1129. The non-linear pharmacokinetic model has been approved to describe the correlation between concentrations of VT-1129 in plasma and in brain (Wiederhold et al., 2018a,d).
At present, the research on VT-1129 in animal models is mainly focused on cryptococcal meningitis. VT-1129 significantly reduces fungal burden and improve survival rates during treatments. When treated with a dose of ≥ 3 mg/kg/day, the fungal burden was undetectable in most mice even 20 days after dosing was stopped. And treating at a dose of 20 mg/kg once daily reached a maximal survival benefit (100%). Because VT-1129 plasma and brain concentrations are related with fungal burden reductions, the loading dose-maintenance dose (LD-MD) strategy to treat cryptococcal meningitis seems feasible (Wiederhold et al., 2018a,d).
CYP51 plays a crucial role in fungal invasive growth, hyphae formation and virulence, and inhibitors targeting CYP51 have always been an important component of antifungal agents. Further researches on fungal CYP51s might set about from the following aspects: First, while the detailed crystal structures of several susceptible pathogenic fungi CYP51 have been elaborated, those of drug-resistant variants are still in hypothesis. If those structures were elucidated, targeting at common drug-resistant CYP51 protein variants could probably be promising. Second, it deserves more in-depth researches to find out why some kind of amino acid point mutations (such as G54W in A. fumigatus CYP51A) could have varied effects on resistance for different azoles. Such researches may provide important ideas to minimize drug resistance. Third, the study of model organisms may also bring some inspiration. Such as Set4, which targets to ergosterol gene promoters with a Hap1-dependent manner under hypoxic conditions in S. cerevisiae, could downregulates the expression of CYP51. Whether homologous proteins and similar mechanisms exist in pathogenic fungi requires further researches. As long as they exist, new remedies may be put forward to activate Set4 to inhibit azole-induced CYP51 overexpression. Forth, it might be achievable to target other proteins as well as CYP51 simultaneously to maximize the therapeutic effect. There already exists some preliminary works focusing on this, and it is probably requisite to find out more accompanying targets. Fifth, the ”Evolution Trap”(ET) strategy, which successfully restored the sensitivity of C. albicans in vitro, might be practiced in vivo to investigate whether similar effects exist in animal models and further in humans. Aside from these, it is also hopeful for pharmaceutics amelioration to improve treatment efficacy. Novel drug delivery systems, such as liposomes, may confer better oral bioavailability on azoles.
JZ contributed to the section of CYP51’s functions, inhibitors and drug resistance. LL contributed the structural analysis part of CYP51. QL, LY, YW, and YJ provided the revisions for this article.
LY was supported by Natural Science Foundation of Shanghai (17ZR1437700). YJ was supported by Natural Science Foundation of China (81330083 and 81573473).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All the authors acknowledge and thank their respective Institutes and Universities.
Abastabar, M., Hosseini, T., Valadan, R., Lagzian, M., Haghani, I., and Aslani, N. (2019). Novel point mutations in cyp51a and cyp51b genes associated with itraconazole and posaconazole resistance in Aspergillus clavatus isolates. Microb. Drug Resist. doi: 10.1089/mdr.2018.0300
Abuhelwa, A. Y., Foster, D. J. R., Mudge, S., Hayes, D., and Upton, R. N. (2015). Population pharmacokinetic modeling of itracsonazole and hydroxyitraconazole for oral SUBA-itraconazole and sporanox capsule formulations in healthy subjects in fed and fasted states. Antimicrob. Agents Chemother. 59, 5681–5696. doi: 10.1128/AAC.00973-15
Albarrag, A. M., Anderson, M. J., Howard, S. J., Robson, G. D., Warn, P. A., and Sanglard, D. (2011). Interrogation of related clinical pan-azole-resistant Aspergillus fumigatus strains: G138C, Y431C, and G434C single nucleotide polymorphisms in cyp51A, upregulation of cyp51A, and integration and activation of transposon Atf1 in the cyp51A promoter. Antimicrob. Agents Chemother. 55, 5113–5121. doi: 10.1128/AAC.00517-11
Alcazar-Fuoli, L., Mellado, E., Cuenca-Estrella, M., and Sanglard, D. (2011). Probing the role of point mutations in the CYP51A gene from Aspergillus fumigatus in the model yeast Saccharomyces cerevisiae. Med. Mycol. 49, 276–284. doi: 10.3109/13693786.2010.512926
Altamirano, S., Fang, D., Simmons, C., Sridhar, S., Wu, P., and Sanyal, K. (2017). Fluconazole-induced ploidy change in Cryptococcus neoformans results from the uncoupling of cell growth and nuclear division. mSphere 2, e00205-17. doi: 10.1128/mSphere.00205-17
Amichai, B., and Grunwald, M. H. (1998). Adverse drug reactions of the new oral antifungal agents–terbinafine, fluconazole, and itraconazole. Int. J. Dermatol. 37, 410–415. doi: 10.1046/j.1365-4362.1998.00496.x
Baddley, J. W., Stephens, J. M., Ji, X., Gao, X., Schlamm, H. T., and Tarallo, M. (2013). Aspergillosis in intensive care unit (ICU) patients: epidemiology and economic outcomes. BMC Infect. Dis. 13:29. doi: 10.1186/1471-2334-13-29
Bassetti, M., Garnacho-Montero, J., Calandra, T., Kullberg, B., Dimopoulos, G., and Azoulay, E. (2017). Intensive care medicine research agenda on invasive fungal infection in critically ill patients. Intens. Care Med. 43, 1225–1238. doi: 10.1007/s00134-017-4731-2
Becher, R., Weihmann, F., Deising, H. B., and Wirsel, S. G. (2011). Development of a novel multiplex DNA microarray for Fusarium graminearum and analysis of azole fungicide responses. BMC Genomics 12:52. doi: 10.1186/1471-2164-12-52
Becher, R., and Wirsel, S. G. R. (2012). Fungal cytochrome P450 sterol 14α-demethylase (CYP51) and azole resistance in plant and human pathogens. Appl. Microbiol. Biot. 95, 825–840. doi: 10.1007/s00253-012-4195-9
Beer, K. D., Farnon, E. C., Jain, S., Jamerson, C., Lineberger, S., and Miller, J. (2018). Multidrug-resistant Aspergillus fumigatus carrying mutations linked to environmental fungicide exposure - three states, 2010-2017. MMWR 67, 1604–1607. doi: 10.15585/mmwr.mm6738a5
Behtash, A., Nafisi, S., and Maibach, H. I. (2017). New generation of fluconazole: a review on existing researches and technologies. Curr. Drug Deliv. 14, 2–15. doi: 10.2174/1567201813666160502125620
Berkow, E., and Lockhart, S. (2017). Fluconazole resistance in candida species: a current perspective. Infect. Drug Resist. 10, 237–245. doi: 10.2147/IDR.S118892
Bernhardt, A., Meyer, W., Rickerts, V., Aebischer, T., and Tintelnot, K. (2018). Identification of 14-alpha-lanosterol demethylase (CYP51) in Scedosporium species. Antimicrob. Agents Chemother. 62:e02599-17. doi: 10.1128/AAC.02599-17
Bien, C. M., Chang, Y. C., Nes, W. D., Kwon-Chung, K. J., and Espenshade, P. J. (2009). Cryptococcus neoformans Site-2 protease is required for virulence and survival in the presence of azole drugs. Mol. Microbiol. 74, 672–690. doi: 10.1111/j.1365-2958.2009.06895.x
Bien, C. M., and Espenshade, P. J. (2010). Sterol regulatory element binding proteins in fungi: hypoxic transcription factors linked to pathogenesis. Eukaryotic Cell 9, 352–359. doi: 10.1128/EC.00358-09
Blankenship, J. R., Steinbach, W. J., Perfect, J. R., and Heitman, J. (2003). Teaching old drugs new tricks: reincarnating immunosuppressants as antifungal drugs. Curr. Opin. Investig. Drugs 4, 192–199.
Blosser, S. J., and Cramer, R. A. (2011). SREBP-dependent triazole susceptibility in Aspergillus fumigatus is mediated through direct transcriptional regulation of erg11A (cyp51A). Antimicrob. Agents Chemother. 56, 248–257. doi: 10.1128/AAC.05027-11
Bongomin, F., Oladele, R. O., Gago, S., Moore, C. B., and Richardson, M. D. (2018). A systematic review of fluconazole resistance in clinical isolates of Cryptococcus species. Mycoses 61, 290–297. doi: 10.1111/myc.12747
Branco, J., Ola, M., Silva, R. M., Fonseca, E., Gomes, N. C., and Martins-Cruz, C. (2017). Impact of ERG3 mutations and expression of ergosterol genes controlled by UPC2 and NDT80 in Candida parapsilosis azole resistance. Clin. Microbiol. Infect. 23, 575.e1–575.e8. doi: 10.1016/j.cmi.2017.02.002
Brand, S. R., Degenhardt, T. P., Person, K., Sobel, J. D., Nyirjesy, P., and Schotzinger, R. J. (2018). A phase 2, randomized, double-blind, placebo-controlled, dose-ranging study to evaluate the efficacy and safety of orally administered VT-1161 in the treatment of recurrent vulvovaginal candidiasis. Am. J. Obstet. Gynecol. 218, 624.e1–624.e9. doi: 10.1016/j.ajog.2018.03.001
Break, T. J., Desai, J. V., Healey, K. R., Natarajan, M., Ferre, E., and Henderson, C. (2018a). VT-1598 inhibits the in vitro growth of mucosal Candida strains and protects against fluconazole-susceptible and -resistant oral candidiasis in IL-17 signalling-deficient mice. J. Antimicrob. Chemother. 73, 2089–2094. doi: 10.1093/jac/dky170
Break, T. J., Desai, J. V., Natarajan, M., Ferre, E., Henderson, C., and Zelazny, A. M. (2018b). VT-1161 protects mice against oropharyngeal candidiasis caused by fluconazole-susceptible and -resistant Candida albicans. J. Antimicrob. Chemother. 73, 151–155. doi: 10.1093/jac/dkx352
Camps, S. M., Rijs, A. J., Klaassen, C. H., Meis, J. F., O’Gorman, C. M., and Dyer, P. S. (2012). Molecular epidemiology of Aspergillus fumigatus isolates harboring the TR34/L98H azole resistance mechanism. J. Clin. Microbiol. 50, 2674–2680. doi: 10.1128/JCM.00335-12
Cannon, R. D., Lamping, E., Holmes, A. R., Niimi, K., Baret, P. V., and Keniya, M. V. (2009). Efflux-mediated antifungal drug resistance. Clin. Microbiol. Rev. 22, 291–321. doi: 10.1128/CMR.00051-08
Chang, Y. C., Ingavale, S. S., Bien, C., Espenshade, P., and Kwon-Chung, K. J. (2009). Conservation of the sterol regulatory element-binding protein pathway and its pathobiological importance incryptococcus neoformans. Eukaryotic Cell 8, 1770–1779. doi: 10.1128/EC.00207-09
Chang, Y. C., Khanal, L. A., and Kwon-Chung, K. J. (2018). Cryptococcus neoformans, unlike candida albicans, forms aneuploid clones directly from uninucleated cells under fluconazole stress. mBio 9, e01290–18. doi: 10.1128/mBio.01290-18
Chen, G., Mulla, W. A., Kucharavy, A., Tsai, H., Rubinstein, B., and Conkright, J. (2015). Targeting the adaptability of heterogeneous aneuploids. Cell 160, 771–784. doi: 10.1016/j.cell.2015.01.026
Chen, Y., Li, Z., Han, X., Tian, S., Zhao, J., and Chen, F. (2018). Elevated MIC values of imidazole drugs against Aspergillus fumigatus isolates with TR34/L98H/S297T/F495I mutation. Antimicrob. Agents Chemother. 62, e01549-17. doi: 10.1128/AAC.01549-17
Chien, R. N., Yang, L. J., Lin, P. Y., and Liaw, Y. F. (1997). Hepatic injury during ketoconazole therapy in patients with onychomycosis: a controlled cohort study. Hepatology 25, 103–107. doi: 10.1002/hep.510250119
Choi, Y. J., Kim, Y. J., Yong, D., Byun, J. H., Kim, T. S., and Chang, Y. S. (2018). Fluconazole-resistant candida parapsilosis bloodstream isolates with y132f mutation in erg11 gene, south korea. Emerg. Infect. Dis. 24, 1768–1770. doi: 10.3201/eid2409.180625
Chowdhary, A., Kathuria, S., Xu, J., and Meis, J. F. (2013). Emergence of azole-resistant Aspergillus fumigatus strains due to agricultural azole use creates an increasing threat to human health. PLoS Pathog. 9:e1003633. doi: 10.1371/journal.ppat.1003633
Chowdhary, A., Sharma, C., and Meis, J. F. (2017). Azole-resistant Aspergillosis: epidemiology, molecular mechanisms, and treatment. J. Infect. Dis. 216, S436–S444. doi: 10.1093/infdis/jix210
Clark, N. M., Grim, S. A., and Lynch, J. R. (2015). Posaconazole: use in the prophylaxis and treatment of fungal infections. Semin. Respir. Crit. Care Med. 36, 767–785. doi: 10.1055/s-0035-1562902
Cormican, S., Adams, N., O’Connell, P., McErlean, A., and de Freitas, D. (2018). Voriconazole-induced periostitis deformans: serial imaging in a patient with ANCA vasculitis. Skeletal Radiol. 47, 191–194. doi: 10.1007/s00256-017-2764-9
Cowen, L. E., Singh, S. D., Kohler, J. R., Collins, C., Zaas, A. K., and Schell, W. A. (2009). Harnessing Hsp90 function as a powerful, broadly effective therapeutic strategy for fungal infectious disease. Pro. Natl. Acad. Sci. U.S.A. 106, 2818–2823. doi: 10.1073/pnas.0813394106
Cruz, M. C., Goldstein, A. L., Blankenship, J., Del, P. M., Perfect, J. R., and McCusker, J. H. (2001). Rapamycin and less immunosuppressive analogs are toxic to Candida albicans and Cryptococcus neoformans via FKBP12-dependent inhibition of TOR. Antimicrob. Agents Chemother. 45, 3162–3170. doi: 10.1128/AAC.45.11.3162-3170.2001
Daneshmend, T. K., and Warnock, D. W. (1988). Clinical pharmacokinetics of ketoconazole. Clin. Pharmacokinet. 14, 13–34. doi: 10.2165/00003088-198814010-00002
Daum, G., Lees, N. D., Bard, M., and Dickson, R. (1998). Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast 14, 1471–1510.
Denis, J., Ledoux, M. P., Nivoix, Y., and Herbrecht, R. (2018). Isavuconazole: a new broad-spectrum azole. part 1: in vitro activity. J. Mycol. Méd. 28, 8–14. doi: 10.1016/j.mycmed.2018.02.005
Denning, D. W., and Perlin, D. S. (2011). Azole resistance in Aspergillus: a growing public health menace. Future Microbiol. 6, 1229–1232. doi: 10.2217/fmb.11.118
Desai, J. V., Mitchell, A. P., and Andes, D. R. (2014). Fungal biofilms, drug resistance, and recurrent infection. Cold Spring Harb. Perspect. Med. 4:a019729. doi: 10.1101/cshperspect.a019729
Dunkel, N., Liu, T. T., Barker, K. S., Homayouni, R., Morschhauser, J., and Rogers, P. D. (2008). A gain-of-function mutation in the transcription factor Upc2p causes upregulation of ergosterol biosynthesis genes and increased fluconazole resistance in a clinical Candida albicans isolate. Eukaryot. Cell 7, 1180–1190. doi: 10.1128/EC.00103-08
Eddouzi, J., Parker, J. E., Vale-Silva, L. A., Coste, A., Ischer, F., and Kelly, S. (2013). Molecular mechanisms of drug resistance in clinical Candida species isolated from tunisian hospitals. Antimicrob. Agents Chemother. 57, 3182–3193. doi: 10.1128/AAC.00555-13
Espinel-Ingroff, A., Aller, A. I., Canton, E., Castanon-Olivares, L. R., Chowdhary, A., and Cordoba, S. (2012). Cryptococcus neoformans-Cryptococcus gattii species complex: an international study of wild-type susceptibility endpoint distributions and epidemiological cutoff values for fluconazole, itraconazole, posaconazole, and voriconazole. Antimicrob. Agents Chemother. 56, 5898–5906. doi: 10.1128/AAC.01115-12
Fischer, J., Ganellin, C. R., Ganesan, A., and Proudfoot, J. (2010). Analogue-Based Drug Discovery. Hoboken, NJ: Wiley-VCH.
Flowers, S. A., Barker, K. S., Berkow, E. L., Toner, G., Chadwick, S. G., and Gygax, S. E. (2012). Gain-of-function mutations in UPC2 are a frequent cause of ERG11 upregulation in azole-resistant clinical isolates of Candida albicans. Eukaryot. Cell 11, 1289–1299. doi: 10.1128/EC.00215-12
Flowers, S. A., Colon, B., Whaley, S. G., Schuler, M. A., and Rogers, P. D. (2015). Contribution of clinically derived mutations in ERG11 to azole resistance in Candida albicans. Antimicrob. Agents Chemother. 59, 450–460. doi: 10.1128/AAC.03470-14
Forche, A., Alby, K., Schaefer, D., Johnson, A. D., Berman, J., and Bennett, R. J. (2008). The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol. 6:e110. doi: 10.1371/journal.pbio.0060110
Garvey, E. P., Hoekstra, W. J., Schotzinger, R. J., Sobel, J. D., Lilly, E. A., and Fidel, P. J. (2015). Efficacy of the clinical agent VT-1161 against fluconazole-sensitive and -resistant Candida albicans in a murine model of vaginal candidiasis. Antimicrob. Agents Chemother. 59, 5567–5573. doi: 10.1128/AAC.00185-15
Garvey, E. P., Sharp, A. D., Warn, P. A., Yates, C. M., and Schotzinger, R. J. (2018). The novel fungal CYP51 inhibitor VT-1598 is efficacious alone and in combination with liposomal amphotericin B in a murine model of cryptococcal meningitis. J. Antimicrob. Chemother. 73, 2815–2822. doi: 10.1093/jac/dky242
Gast, C. E., Basso, L. J., Bruzual, I., and Wong, B. (2013). Azole resistance in Cryptococcus gattii from the Pacific Northwest: Investigation of the role of ERG11. Antimicrob. Agents Chemother. 57, 5478–5485. doi: 10.1128/AAC.02287-12
Gebremariam, T., Alkhazraji, S., Lin, L., Wiederhold, N. P., Garvey, E. P., and Hoekstra, W. J. (2017). Prophylactic treatment with VT-1161 protects immunosuppressed mice from Rhizopus arrhizus var. arrhizus infection. Antimicrob. Agents Chemother. 61, e00390-17. doi: 10.1128/AAC.00390-17
Greenblatt, D. J., Zhao, Y., Venkatakrishnan, K., Duan, S. X., Harmatz, J. S., and Parent, S. J. (2011). Mechanism of cytochrome P450-3A inhibition by ketoconazole. J. Pharm. Pharmacol. 63, 214–221. doi: 10.1111/j.2042-7158.2010.01202.x
Greenblatt, H. K., and Greenblatt, D. J. (2014). Liver injury associated with ketoconazole: review of the published evidence. J. Clin. Pharmacol. 54, 1321–1329. doi: 10.1002/jcph.400
Groll, A. H., and Walsh, T. J. (2014). Posaconazole: clinical pharmacology and potential for management of fungal infections. Expert Rev. Anti Infe. 3, 467–487. doi: 10.1586/14787210.3.4.467
Gsaller, F., Hortschansky, P., Furukawa, T., Carr, P. D., Rash, B., and Capilla, J. (2016). Sterol biosynthesis and azole tolerance is governed by the opposing actions of SrbA and the CCAAT binding complex. PLoS Pathog. 12:e1005775. doi: 10.1371/journal.ppat.1005775
Guida, A., Lindstadt, C., Maguire, S. L., Ding, C., Higgins, D. G., and Corton, N. J. (2011). Using RNA-seq to determine the transcriptional landscape and the hypoxic response of the pathogenic yeast Candida parapsilosis. BMC Genomics 12:628. doi: 10.1186/1471-2164-12-628
Gunsalus, K. T., Tornberg-Belanger, S. N., Matthan, N. R., Lichtenstein, A. H., and Kumamoto, C. A. (2016). Manipulation of host diet to reduce gastrointestinal colonization by the opportunistic pathogen Candida albicans. mSphere 1, e00020-15. doi: 10.1128/mSphere.00020-15
Gupta, A. K., Daigle, D., and Foley, K. A. (2014). Drug safety assessment of oral formulations of ketoconazole. Expert Opin. Drug Saf. 14, 325–334. doi: 10.1517/14740338.2015.983071
Gupta, A. K., and Lyons, D. C. A. (2015). The Rise and fall of oral ketoconazole. J. Cutan. Med. Surg. 19, 352–357. doi: 10.1177/1203475415574970
Hagiwara, D., Miura, D., Shimizu, K., Paul, S., Ohba, A., and Gonoi, T. (2017). A novel Zn2-Cys6 transcription factor AtrR plays a key role in an azole resistance mechanism of Aspergillus fumigatus by co-regulating cyp51A and cdr1B Expressions. PLoS Pathog. 13:e1006096. doi: 10.1371/journal.ppat.1006096
Hagiwara, D., Watanabe, A., and Kamei, K. (2016). Sensitisation of an azole-resistant aspergillus fumigatus strain containing the Cyp51A-related mutation by deleting the SrbA gene. Sci. Rep. 6:38833. doi: 10.1038/srep38833
Hannemann, F., Bichet, A., Ewen, K. M., and Bernhardt, R. (2007). Cytochrome P450 systems–biological variations of electron transport chains. Biochim. Biophys. Acta 1770, 330–344. doi: 10.1016/j.bbagen.2006.07.017
Hargrove, T. Y., Friggeri, L., Wawrzak, Z., Qi, A., Hoekstra, W. J., and Schotzinger, R. J. (2017a). Structural analyses of Candida albicans sterol 14alpha-demethylase complexed with azole drugs address the molecular basis of azole-mediated inhibition of fungal sterol biosynthesis. J. Biol. Chem. 292, 6728–6743. doi: 10.1074/jbc.M117.778308
Hargrove, T. Y., Garvey, E. P., Hoekstra, W. J., Yates, C. M., Wawrzak, Z., and Rachakonda, G. (2017b). Crystal structure of the new investigational drug candidate VT-1598 in complex with Aspergillus fumigatus sterol 14alpha-demethylase provides insights into its broad-spectrum antifungal activity. Antimicrob. Agents Chemother. 61, e00570-17. doi: 10.1128/AAC.00570-17
Hargrove, T. Y., Wawrzak, Z., Lamb, D. C., Guengerich, F. P., and Lepesheva, G. I. (2015). Structure-functional characterization of cytochrome P450 Sterol 14α-Demethylase (CYP51B) from Aspergillus fumigatus and molecular basis for the development of antifungal drugs. J. Biol. Chem. 290, 23916–23934. doi: 10.1074/jbc.M115.677310
Harrison, B. D., Hashemi, J., Bibi, M., Pulver, R., Bavli, D., and Nahmias, Y. (2014). A tetraploid intermediate precedes aneuploid formation in yeasts exposed to fluconazole. PLoS Biol. 12:e1001815. doi: 10.1371/journal.pbio.1001815
Hawkins, N. J., Cools, H. J., Sierotzki, H., Shaw, M. W., Knogge, W., and Kelly, S. L. (2014). Paralog re-emergence: a novel, historically contingent mechanism in the evolution of antimicrobial resistance. Mol. Biol. Evol. 31, 1793–1802. doi: 10.1093/molbev/msu134
Heel, R. C., Brogden, R. N., Carmine, A., Morley, P. A., Speight, T. M., and Avery, G. S. (1982). Ketoconazole: a review of its therapeutic efficacy in superficial and systemic fungal infections. Drugs 23, 1–36.
Hoekstra, W. J., Garvey, E. P., Moore, W. R., Rafferty, S. W., Yates, C. M., and Schotzinger, R. J. (2014). Design and optimization of highly-selective fungal CYP51 inhibitors. Bioorg. Med. Chem. Lett. 24, 3455–3458. doi: 10.1016/j.bmcl.2014.05.068
Hoot, S. J., Smith, A. R., Brown, R. P., and White, T. C. (2011). An A643V amino acid substitution in Upc2p contributes to azole resistance in well-characterized clinical isolates of Candida albicans. Antimicrob. Agents Chemother. 55, 940–942. doi: 10.1128/AAC.00995-10
Hu, W., Sillaots, S., Lemieux, S., Davison, J., Kauffman, S., and Breton, A. (2007). Essential gene identification and drug target prioritization in Aspergillus fumigatus. PLoS Pathog. 3:e24. doi: 10.1371/journal.ppat.0030024
Hyland, R., Jones, B. C., and Smith, D. A. (2003). Identification of the cytochrome P450 enzymes involved in the N-oxidation of voriconazole. Drug Metab. Dispos. 31, 540–547.
Isla, G., Leonardelli, F., Tiraboschi, I. N., Refojo, N., Hevia, A., and Vivot, W. (2018). First clinical isolation of an azole-resistant Aspergillus fumigatus isolate harboring a TR46 Y121F T289A mutation in south america. Antimicrob. Agents Chemother. 62, e00872-18. doi: 10.1128/AAC.00872-18
Isoherranen, N. (2004). Role of itraconazole metabolites in CYP3A4 inhibition. Drug Metab. Dispos. 32, 1121–1131. doi: 10.1124/dmd.104.000315
Jiang, C., Ni, Q., Dong, D., Zhang, L., Li, Z., and Tian, Y. (2016). The role of UPC2 gene in azole-resistant Candida tropicalis. Mycopathologia 181, 833–838. doi: 10.1007/s11046-016-0050-3
Job, K. M., Olson, J., Stockmann, C., Constance, J. E., Enioutina, E. Y., and Rower, J. E. (2016). Pharmacodynamic studies of voriconazole: informing the clinical management of invasive fungal infections. Expert Rev. Anti. Infect. Ther. 14, 731–746. doi: 10.1080/14787210.2016.1207526
Kano, R., Okubo, M., Hasegawa, A., and Kamata, H. (2017). Multi-azole-resistant strains of Cryptococcus neoformans var. grubii isolated from a FLZ-resistant strain by culturing in medium containing voriconazole. Med. Mycol. 55, 877–882. doi: 10.1093/mmy/myw101
Keniya, M. V., Sabherwal, M., Wilson, R. K., Woods, M. A., Sagatova, A. A., and Tyndall, J. (2018). Crystal structures of full-length lanosterol 14alpha-demethylases of prominent fungal pathogens Candida albicans and Candida glabrata provide tools for antifungal discovery. Antimicrob. Agents Chemother. 62, e01134-18. doi: 10.1128/AAC.01134-18
Kim, H., Kumari, P., Laughlin, M., Hilbert, M. J., Indelicato, S. R., and Lim, J. (2003). Use of high-performance liquid chromatographic and microbiological analyses for evaluating the presence or absence of active metabolites of the antifungal posaconazole in human plasma. J. Chromatogr. A 987, 243–248.
Kushima, H., Tokimatsu, I., Ishii, H., Kawano, R., Watanabe, K., and Kadota, J. I. (2017). A new amino acid substitution at G150S in Lanosterol 14-alpha demethylase (Erg11 protein) in Multi-azole-resistant Trichosporon asahii. Med. Mycol. J. 58, E23–E28. doi: 10.3314/mmj.16-00027
Kwon-Chung, K. J., and Chang, Y. C. (2012). Aneuploidy and drug resistance in pathogenic fungi. PLoS Pathog. 8:e1003022. doi: 10.1371/journal.ppat.1003022
Lamb, D. C., Waterman, M. R., Kelly, S. L., and Guengerich, F. P. (2007). Cytochromes P450 and drug discovery. Curr. Opin. Biotechnol. 18, 504–512. doi: 10.1016/j.copbio.2007.09.010
Lamoth, F., Alexander, B. D., Juvvadi, P. R., and Steinbach, W. J. (2015). Antifungal activity of compounds targeting the Hsp90-calcineurin pathway against various mould species. J. Antimicrob. Chemother. 70, 1408–1411. doi: 10.1093/jac/dku549
Lamoureux, F., Duflot, T., Woillard, J. B., Metsu, D., Pereira, T., and Compagnon, P. (2016). Impact of CYP2C19 genetic polymorphisms on voriconazole dosing and exposure in adult patients with invasive fungal infections. Int. J. Antimicrob. Agents 47, 124–131. doi: 10.1016/j.ijantimicag.2015.12.003
Lanjewar, D. N. (2011). The spectrum of clinical and pathological manifestations of AIDS in a consecutive series of 236 autopsied cases in mumbai, India. Patholog Res. Int. 2011:547618. doi: 10.4061/2011/547618
Ledoux, M. P., Denis, J., Nivoix, Y., and Herbrecht, R. (2018). Isavuconazole: a new broad-spectrum azole. Part 2: pharmacokinetics and clinical activity. J. Mycol. Méd. 28, 15–22. doi: 10.1016/j.mycmed.2018.02.002
Lee, W. G., Shin, J. H., Uh, Y., Kang, M. G., Kim, S. H., and Park, K. H. (2011). First three reported cases of nosocomial fungemia caused by Candida auris. J. Clin. Microbiol. 49, 3139–3142. doi: 10.1128/JCM.00319-11
Lepesheva, G. I., and Waterman, M. R. (2007). Sterol 14alpha-demethylase cytochrome P450 (CYP51), a P450 in all biological kingdoms. Biochim. Biophys. Acta 1770, 467–477. doi: 10.1016/j.bbagen.2006.07.018
Lepesheva, G. I., and Waterman, M. R. (2011). Structural basis for conservation in the CYP51 family. Biochim Biophys Acta 1814, 88–93. doi: 10.1016/j.bbapap.2010.06.006
Lestner, J., and Hope, W. W. (2013). Itraconazole: an update on pharmacology and clinical use for treatment of invasive and allergic fungal infections. Expert Opin. Drug Met. 9, 911–926. doi: 10.1517/17425255.2013.794785
Levine, M. T., and Chandrasekar, P. H. (2016). Adverse effects of voriconazole: over a decade of use. Clin. Transplant. 30, 1377–1386. doi: 10.1111/ctr.12834
Li, T. Y., Liu, W., Chen, K., Liang, S. Y., and Liu, F. (2017). The influence of combination use of CYP450 inducers on the pharmacokinetics of voriconazole: a systematic review. J. Clin. Pharm. Ther. 42, 135–146. doi: 10.1111/jcpt.12493
Li, X., Yang, F., Li, D., Zhou, M., Wang, X., and Xu, Q. (2015). Trisomy of chromosome R confers resistance to triazoles in Candida albicans. Med. Mycol. 53, 302–309. doi: 10.1093/mmy/myv002
Listed, N. A. (1981). Ketoconazole approved for systemic fungal infections. FDA Drug Bull 11, 18–19.
Liu, S., Hou, Y., Chen, X., Gao, Y., Li, H., and Sun, S. (2014). Combination of fluconazole with non-antifungal agents: A promising approach to cope with resistant Candida albicans infections and insight into new antifungal agent discovery. Int. J. Antimicrob. Ag. 43, 395–402. doi: 10.1016/j.ijantimicag.2013.12.009
Lockhart, S. R., Etienne, K. A., Vallabhaneni, S., Farooqi, J., Chowdhary, A., and Govender, N. P. (2017). Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin. Infect. Dis. 64, 134–140. doi: 10.1093/cid/ciw691
MacCallum, D. M., Coste, A., Ischer, F., Jacobsen, M. D., Odds, F. C., and Sanglard, D. (2010). Genetic dissection of azole resistance mechanisms in Candida albicans and their validation in a mouse model of disseminated infection. Antimicrob. Agents Chemother. 54, 1476–1483. doi: 10.1128/AAC.01645-09
Maertens, J. A., Raad, I. I., Marr, K. A., Patterson, T. F., Kontoyiannis, D. P., and Cornely, O. A. (2016). Isavuconazole versus voriconazole for primary treatment of invasive mould disease caused by Aspergillus and other filamentous fungi (SECURE): a phase 3, randomised-controlled, non-inferiority trial. Lancet 387, 760–769. doi: 10.1016/S0140-6736(15)01159-9
Malani, A. N., Kerr, L. E., and Kauffman, C. A. (2015). Voriconazole: how to use this antifungal agent and what to expect. Semin. Respir. Crit. Care Med. 36, 786–795. doi: 10.1055/s-0035-1562903
Marichal, P., Koymans, L., Willemsens, S., Bellens, D., Verhasselt, P., and Luyten, W. (1999). Contribution of mutations in the cytochrome P450 14alpha-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology 145(Pt 10), 2701–2713. doi: 10.1099/00221287-145-10-2701
Martel, C. M., Parker, J. E., Bader, O., Weig, M., Gross, U., and Warrilow, A. G. (2010). Identification and characterization of four azole-resistant erg3 mutants of Candida albicans. Antimicrob. Agents Chemother. 54, 4527–4533. doi: 10.1128/AAC.00348-10
McCarthy, M. W., Kontoyiannis, D. P., Cornely, O. A., Perfect, J. R., and Walsh, T. J. (2017). Novel agents and drug targets to meet the challenges of resistant fungi. J. Infect. Dis. 216, S474–S483. doi: 10.1093/infdis/jix130
McCormack, P. L. (2015). Isavuconazonium: first global approval. Drugs 75, 817–822. doi: 10.1007/s40265-015-0398-6
Mellado, E., Garcia-Effron, G., Alcazar-Fuoli, L., Cuenca-Estrella, M., and Rodriguez-Tudela, J. L. (2004). Substitutions at methionine 220 in the 14alpha-sterol demethylase (Cyp51A) of Aspergillus fumigatus are responsible for resistance in vitro to azole antifungal drugs. Antimicrob. Agents Chemother. 48, 2747–2750. doi: 10.1128/AAC.48.7.2747-2750.2004
Mellado, E., Garcia-Effron, G., Alcazar-Fuoli, L., Melchers, W. J., Verweij, P. E., and Cuenca-Estrella, M. (2007). A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob. Agents Chemother. 51, 1897–1904. doi: 10.1128/AAC.01092-06
Monk, B. C., Tomasiak, T. M., Keniya, M. V., Huschmann, F. U., Tyndall, J. D., and O’Connell, J. R. (2014). Architecture of a single membrane spanning cytochrome P450 suggests constraints that orient the catalytic domain relative to a bilayer. Proc. Natl. Acad. Sci. U.S.A. 111, 3865–3870. doi: 10.1073/pnas.1324245111
Moore, J. N., Healy, J. R., and Kraft, W. K. (2015). Pharmacologic and clinical evaluation of posaconazole. Expert Rev. Clin. Pharmacol. 8, 321–334. doi: 10.1586/17512433.2015.1034689
Morio, F., Jensen, R. H., Le Pape, P., and Arendrup, M. C. (2017). Molecular basis of antifungal drug resistance in yeasts. Int. J. Antimicrob. Ag. 50, 599–606. doi: 10.1016/j.ijantimicag.2017.05.012
Morita, K., Konishi, H., and Shimakawa, H. (1992). Fluconazole: a potent inhibitor of cytochrome P-450-dependent drug-metabolism in mice and humans in vivo. comparative study with ketoconazole. Chem. Pharm. Bull. 40, 1247–1251.
Morschhäuser, J. (2016). The development of fluconazole resistance in Candida albicans – an example of microevolution of a fungal pathogen. J. Microbiol. 54, 192–201. doi: 10.1007/s12275-016-5628-4
Mounier, A., Agard, E., Douma, I., Chehab, H. E., Vie, A. L., and Dot, J. M. (2018). Macular toxicity and blind spot enlargement during a treatment by voriconazole: a case report. Eur. J. Ophthalmol. 28, N11–N14. doi: 10.1177/1120672117750051
Munro, A. W., McLean, K. J., Grant, J. L., and Makris, T. M. (2018). Structure and function of the cytochrome P450 peroxygenase enzymes. Biochem. Soc. T. 46, 183–196. doi: 10.1042/BST20170218
Musiol, R., and Kowalczyk, W. (2012). Azole antimycotics–a highway to new drugs or a dead end? Curr. Med. Chem. 19:1378. doi: 10.2174/092986712799462621
Nishikawa, J. L., Boeszoermenyi, A., Vale-Silva, L. A., Torelli, R., Posteraro, B., and Sohn, Y. (2016). Inhibiting fungal multidrug resistance by disrupting an activator–Mediator interaction. Nature 530, 485–489. doi: 10.1038/nature16963
Noble, J. A., Tsai, H. F., Suffis, S. D., Su, Q., Myers, T. G., and Bennett, J. E. (2013). STB5 is a negative regulator of azole resistance in Candida glabrata. Antimicrob. Agents Chemother. 57, 959–967. doi: 10.1128/AAC.01278-12
Noel, T. (2012). The cellular and molecular defense mechanisms of the Candida yeasts against azole antifungal drugs. J. Mycol. Med. 22, 173–178. doi: 10.1016/j.mycmed.2012.04.004
Owusu, O. A., Egelund, E. F., Alsultan, A., Peloquin, C. A., and Johnson, J. A. (2014). CYP2C19 polymorphisms and therapeutic drug monitoring of voriconazole: are we ready for clinical implementation of pharmacogenomics? Pharmacotherapy 34, 703–718. doi: 10.1002/phar.1400
Pais, P., Costa, C., Cavalheiro, M., Romão, D., and Teixeira, M. C. (2016). Transcriptional control of drug resistance, virulence and immune system evasion in pathogenic fungi: a cross-species comparison. Front. Cell. Infect. Microbiol. 6:131. doi: 10.3389/fcimb.2016.00131
Pappas, P. G., Lionakis, M. S., Arendrup, M. C., Ostrosky-Zeichner, L., and Kullberg, B. J. (2018). Invasive candidiasis. Nat. Rev. Dis. Primers 4:18026. doi: 10.1038/nrdp.2018.26
Parker, J. E., Warrilow, A. G. S., Price, C. L., Mullins, J. G. L., Kelly, D. E., and Kelly, S. L. (2014). Resistance to antifungals that target CYP51. J. Chem. Biol. 7, 143–161. doi: 10.1007/s12154-014-0121-1
Patterson, T. F., Thompson, G. R., Denning, D. W., Fishman, J. A., Hadley, S., and Herbrecht, R. (2016). Practice guidelines for the diagnosis and management of Aspergillosis: 2016 update by the infectious diseases society of america. Clin. Infect. Dis. 63, e1–e60. doi: 10.1093/cid/ciw326
Pettit, N. N., and Carver, P. L. (2015). Isavuconazole: a new option for the management of invasive fungal infections. Ann. Pharmacother. 49, 825–842. doi: 10.1177/1060028015581679
Pinto, E., Monteiro, C., Maia, M., Faria, M. A., Lopes, V., and Lameiras, C. (2018). Aspergillus species and antifungals susceptibility in clinical setting in the north of portugal: cryptic species and emerging azoles resistance in a. fumigatus. Front. Microbiol. 9:1656. doi: 10.3389/fmicb.2018.01656
Pont, A., Goldman, E. S., Sugar, A. M., Siiteri, P. K., and Stevens, D. A. (1985). Ketoconazole-induced increase in estradiol-testosterone ratio. probable explanation for gynecomastia. Arch. Intern. Med. 145, 1429–1431.
Pont, A., Williams, P. L., Loose, D. S., Feldman, D., Reitz, R. E., and Bochra, C. (1982). Ketoconazole blocks adrenal steroid synthesis. Ann. Intern. Med. 97, 370–372.
Režen, T., Debeljak, N., Kordiš, D., and Rozman, D. (2004). New Aspects on lanosterol 14α-demethylase and cytochrome p450 evolution: lanosterol/cycloartenol diversification and lateral transfer. J. Mol. Evol. 59, 51–58. doi: 10.1007/s00239-004-2603-1
Rodero, L., Mellado, E., Rodriguez, A. C., Salve, A., Guelfand, L., and Cahn, P. (2003). G484S amino acid substitution in lanosterol 14-alpha demethylase (ERG11) is related to fluconazole resistance in a recurrent Cryptococcus neoformans clinical isolate. Antimicrob. Agents Chemother. 47, 3653–3656. doi: 10.1128/AAC.47.11.3653-3656.2003
Rodriguez, R. J., and Acosta, D. J. (1997). N-deacetyl ketoconazole-induced hepatotoxicity in a primary culture system of rat hepatocytes. Toxicology 117, 123–131.
Rodriguez, R. J., Proteau, P. J., Marquez, B. L., Hetherington, C. L., Buckholz, C. J., and O’Connell, K. L. (1999). Flavin-containing monooxygenase-mediated metabolism of N-deacetyl ketoconazole by rat hepatic microsomes. Drug Metab. Dispos. 27, 880–886.
Rohmer, M., Bouvier, P., and Ourisson, G. (1979). Molecular evolution of biomembranes: structural equivalents and phylogenetic precursors of sterols. Proc. Natl. Acad. Sci. U.S.A. 76, 847–851. doi: 10.1073/pnas.76.2.847
Rothe, C., Sloan, D. J., Goodson, P., Chikafa, J., Mukaka, M., and Denis, B. (2013). A prospective longitudinal study of the clinical outcomes from cryptococcal meningitis following treatment induction with 800 mg oral fluconazole in Blantyre. Malawi. PLoS One 8:e67311. doi: 10.1371/journal.pone.0067311
Sagatova, A. A., Keniya, M. V., Tyndall, J., and Monk, B. C. (2018). Impact of homologous resistance mutations from pathogenic yeast on Saccharomyces cerevisiae lanosterol 14alpha-demethylase. Antimicrob. Agents Chemother. 62, e02242-17. doi: 10.1128/AAC.02242-17
Sagatova, A. A., Keniya, M. V., Wilson, R. K., Monk, B. C., and Tyndall, J. D. (2015). Structural Insights into Binding of the antifungal drug fluconazole to Saccharomyces cerevisiae Lanosterol 14alpha-demethylase. Antimicrob. Agents Chemother. 59, 4982–4989. doi: 10.1128/AAC.00925-15
Sansone-Parsons, A., Krishna, G., Martinho, M., Kantesaria, B., Gelone, S., and Mant, T. G. (2007). Effect of oral posaconazole on the pharmacokinetics of cyclosporine and tacrolimus. Pharmacotherapy 27, 825–834. doi: 10.1592/phco.27.6.825
Sasse, C., Dunkel, N., Schafer, T., Schneider, S., Dierolf, F., and Ohlsen, K. (2012). The stepwise acquisition of fluconazole resistance mutations causes a gradual loss of fitness in Candida albicans. Mol. Microbiol. 86, 539–556. doi: 10.1111/j.1365-2958.2012.08210.x
Schell, W. A., Jones, A. M., Garvey, E. P., Hoekstra, W. J., Schotzinger, R. J., and Alexander, B. D. (2017). Fungal CYP51 Inhibitors VT-1161 and VT-1129 Exhibit Strong In Vitro Activity against Candida glabrata and C. krusei isolates clinically resistant to azole and echinocandin antifungal compounds. Antimicrob. Agents Chemother. 61, e1817–e1816. doi: 10.1128/AAC.01817-16
Schmitt-Hoffmann, A., Roos, B., Heep, M., Schleimer, M., Weidekamm, E., and Brown, T. (2006). Single-ascending-dose pharmacokinetics and safety of the novel broad-spectrum antifungal triazole BAL4815 after intravenous infusions (50, 100, and 200 milligrams) and oral administrations (100, 200, and 400 milligrams) of its prodrug, BAL8557, in healthy volunteers. Antimicrob. Agents Chemother. 50, 279–285. doi: 10.1128/AAC.50.1.279-285.2006
Selmecki, A., Forche, A., and Berman, J. (2006). Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313, 367–370. doi: 10.1126/science.1128242
Selmecki, A., Forche, A., and Berman, J. (2010). Genomic plasticity of the human fungal pathogen Candida albicans. Eukaryot Cell 9, 991–1008. doi: 10.1128/EC.00060-10
Semighini, C. P., Averette, A. F., Perfect, J. R., and Heitman, J. (2011). Deletion of Cryptococcus neoformans AIF ortholog promotes chromosome aneuploidy and fluconazole-resistance in a metacaspase-independent manner. PLoS Pathog. 7:e1002364. doi: 10.1371/journal.ppat.1002364
Serratore, N. D., Baker, K. M., Macadlo, L. A., Gress, A. R., Powers, B. L., and Atallah, N. (2018). A novel sterol-signaling pathway governs azole antifungal drug resistance and hypoxic gene repression in Saccharomyces cerevisiae. Genetics 208, 1037–1055. doi: 10.1534/genetics.117.300554
Sharma, C., Kumar, R., Kumar, N., Masih, A., Gupta, D., and Chowdhary, A. (2018). Investigation of multiple resistance mechanisms in voriconazole-resistant Aspergillus flavus clinical isolates from a chest hospital surveillance in delhi, india. Antimicrob. Agents Chemother. 62, e01928-17. doi: 10.1128/AAC.01928-17
Shubitz, L. F., Roy, M. E., Trinh, H. T., Hoekstra, W. J., Schotzinger, R. J., and Garvey, E. P. (2017). Efficacy of the investigational antifungal VT-1161 in treating naturally occurring coccidioidomycosis in dogs. Antimicrob. Agents Chemother. 61, e00111-17. doi: 10.1128/AAC.00111-17
Shubitz, L. F., Trinh, H. T., Galgiani, J. N., Lewis, M. L., Fothergill, A. W., and Wiederhold, N. P. (2015). Evaluation of VT-1161 for treatment of coccidioidomycosis in murine infection models. Antimicrob. Agents Chemother. 59, 7249–7254. doi: 10.1128/AAC.00593-15
Silva, D. B. D. S., Rodrigues, L. M. C., Almeida, A. A. D., Oliveira, K. M. P. D., and Grisolia, A. B. (2016). Novel point mutations in the ERG11 gene in clinical isolates of azole resistant Candida species. Mem. Inst. Oswaldo Cruz 111, 192–199. doi: 10.1590/0074-02760150400
Singh-Babak, S. D., Babak, T., Diezmann, S., Hill, J. A., Xie, J. L., and Chen, Y. L. (2012). Global analysis of the evolution and mechanism of echinocandin resistance in Candida glabrata. PLoS Pathog. 8:e1002718. doi: 10.1371/journal.ppat.1002718
Sionov, E., Chang, Y. C., Garraffo, H. M., Dolan, M. A., Ghannoum, M. A., and Kwon-Chung, K. J. (2012). Identification of a Cryptococcus neoformans cytochrome P450 lanosterol 14alpha-demethylase (Erg11) residue critical for differential susceptibility between fluconazole/voriconazole and itraconazole/posaconazole. Antimicrob. Agents Chemother. 56, 1162–1169. doi: 10.1128/AAC.05502-11
Snelders, E., Camps, S. M., Karawajczyk, A., Rijs, A. J., Zoll, J., and Verweij, P. E. (2015). Genotype-phenotype complexity of the TR46/Y121F/T289A cyp51A azole resistance mechanism in Aspergillus fumigatus. Fungal Genet. Biol. 82, 129–135. doi: 10.1016/j.fgb.2015.06.001
Snelders, E., Karawajczyk, A., Verhoeven, R. J., Venselaar, H., Schaftenaar, G., and Verweij, P. E. (2011). The structure-function relationship of the Aspergillus fumigatus cyp51A L98H conversion by site-directed mutagenesis: the mechanism of L98H azole resistance. Fungal Genet. Biol. 48, 1062–1070. doi: 10.1016/j.fgb.2011.08.002
Song, J., Zhai, P., and Lu, L. (2017). Damage resistance protein (Dap) contributes to azole resistance in a sterol-regulatory-element-binding protein SrbA-dependent way. Appl. Microbiol. Biotechnol. 101, 3729–3741. doi: 10.1007/s00253-016-8072-9
Song, J., Zhai, P., Zhang, Y., Zhang, C., Sang, H., and Han, G. (2016). The Aspergillus fumigatus damage resistance protein family coordinately regulates ergosterol biosynthesis and azole susceptibility. mBio 7, e01919-15. doi: 10.1128/mBio.01919-15
Spiess, B., Postina, P., Reinwald, M., Cornely, O. A., Hamprecht, A., and Hoenigl, M. (2014). Incidence of Cyp51 A key mutations in Aspergillus fumigatus-a study on primary clinical samples of immunocompromised patients in the period of 1995-2013. PLoS One 9:e103113. doi: 10.1371/journal.pone.0103113
Stone, N. R., Rhodes, J., Fisher, M. C., Mfinanga, S., Kivuyo, S., and Rugemalila, J. (2019). Dynamic ploidy changes drive fluconazole resistance in human cryptococcal meningitis. J. Clin. Invest. 129, 999–1014. doi: 10.1172/JCI124516
Strushkevich, N., Usanov, S. A., and Park, H. W. (2010). Structural basis of human CYP51 inhibition by antifungal azoles. J. Mol. Biol. 397, 1067–1078. doi: 10.1016/j.jmb.2010.01.075
Traynor, K. (2015). Isavuconazonium sulfate approved for invasive fungal infections. Am. J. Health Syst. Pharm. 72:592. doi: 10.2146/news150027
Tsitsopoulou, A., Posso, R., Vale, L., Bebb, S., Johnson, E., and White, P. L. (2018). Determination of the prevalence of triazole resistance in environmental Aspergillus fumigatus strains isolated in south wales. U.K. Front. Microbiol. 9:1395. doi: 10.3389/fmicb.2018.01395
Tsuchido, Y., Tanaka, M., Nakano, S., Yamamoto, M., Matsumura, Y., and Nagao, M. (2019). Prospective multicenter surveillance of clinically isolated Aspergillus species revealed azole-resistant Aspergillus fumigatus isolates with TR34/L98H mutation in the Kyoto and Shiga regions of Japan. Med. Mycol. doi: 10.1093/mmy/myz003
Umeyama, T., Hayashi, Y., Shimosaka, H., Inukai, T., Yamagoe, S., and Takatsuka, S. (2018). CRISPR/Cas9 genome editing to demonstrate the contribution of Cyp51A Gly138Ser to azole resistance in Aspergillus fumigatus. Antimicrob Agents Chemother 62, e00894-18. doi: 10.1128/AAC.00894-18
van der Linden, J. W., Camps, S. M., Kampinga, G. A., Arends, J. P., Debets-Ossenkopp, Y. J., and Haas, P. J. (2013). Aspergillosis due to voriconazole highly resistant Aspergillus fumigatus and recovery of genetically related resistant isolates from domiciles. Clin. Infect. Dis. 57, 513–520. doi: 10.1093/cid/cit320
Vanden, B. H., Marichal, P., Odds, F. C., Le Jeune, L., and Coene, M. C. (1992). Characterization of an azole-resistant Candida glabrata isolate. Antimicrob Agents Chemother 36, 2602–2610. doi: 10.1128/AAC.36.12.2602
Vandeputte, P., Larcher, G., Berges, T., Renier, G., Chabasse, D., and Bouchara, J. P. (2005). Mechanisms of azole resistance in a clinical isolate of Candida tropicalis. Antimicrob. Agents Chemother. 49, 4608–4615. doi: 10.1128/AAC.49.11.4608-4615.2005
Vasicek, E. M., Berkow, E. L., Flowers, S. A., Barker, K. S., and Rogers, P. D. (2014). UPC2 is universally essential for azole antifungal resistance in Candida albicans. Eukaryot. Cell 13, 933–946. doi: 10.1128/EC.00221-13
Warrilow, A. G., Parker, J. E., Price, C. L., Nes, W. D., Garvey, E. P., and Hoekstra, W. J. (2016). The investigational drug VT-1129 is a highly potent inhibitor of Cryptococcus species CYP51 but only weakly inhibits the human enzyme. Antimicrob. Agents Chemother. 60, 4530–4538. doi: 10.1128/AAC.00349-16
Warrilow, A. G. S., Hull, C. M., Parker, J. E., Garvey, E. P., Hoekstra, W. J., and Moore, W. R. (2014). The clinical candidate VT-1161 Is a highly potent inhibitor of Candida albicans CYP51 but fails to bind the human enzyme. Antimicrob. Agents Chemother. 58, 7121–7127. doi: 10.1128/AAC.03707-14
Warrilow, A. G. S., Parker, J. E., Price, C. L., Garvey, E. P., Hoekstra, W. J., and Schotzinger, R. J. (2017). The tetrazole VT-1161 is a potent inhibitor of Trichophyton rubrum through Its Inhibition of T. rubrum CYP51. Antimicrob. Agents Chemother. 61, e00333-17. doi: 10.1128/AAC.00333-17
Washton, H. (1989). Review of fluconazole: a new triazole antifungal agent. Diagn. Micr. Infec. Dis. 12(4 Suppl.), 229S–233S.
Waterman, M. R., and Lepesheva, G. I. (2005). Sterol 14 alpha-demethylase, an abundant and essential mixed-function oxidase. Biochem. Biophys. Res. Commun. 338, 418–422. doi: 10.1016/j.bbrc.2005.08.118
Whaley, S. G., Caudle, K. E., Vermitsky, J. P., Chadwick, S. G., Toner, G., and Barker, K. S. (2014). UPC2A is required for high-level azole antifungal resistance in Candida glabrata. Antimicrob. Agents Chemother. 58, 4543–4554. doi: 10.1128/AAC.02217-13
White, T. C. (1997). The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14alpha demethylase in Candida albicans. Antimicrob. Agents Chemother. 41, 1488–1494. doi: 10.1128/AAC.41.7.1488
Wiederhold, N. P., Najvar, L. K., Garvey, E. P., Brand, S. R., Xu, X., and Ottinger, E. A. (2018a). The Fungal Cyp51 Inhibitor VT-1129 Is Efficacious in an Experimental Model of Cryptococcal Meningitis. Antimicrob. Agents Chemother. 62, e01071-18. doi: 10.1128/AAC.01071-18
Wiederhold, N. P., Patterson, H. P., Tran, B. H., Yates, C. M., Schotzinger, R. J., and Garvey, E. P. (2018b). Fungal-specific Cyp51 inhibitor VT-1598 demonstrates in vitro activity against Candida and Cryptococcus species, endemic fungi, including Coccidioides species, Aspergillus species and Rhizopus arrhizus. J. Antimicrob. Chemother. 73, 404–408. doi: 10.1093/jac/dkx410
Wiederhold, N. P., Shubitz, L. F., Najvar, L. K., Jaramillo, R., Olivo, M., and Catano, G. (2018c). The novel fungal cyp51 inhibitor vt-1598 is efficacious in experimental models of central nervous system coccidioidomycosis caused by Coccidioides posadasii and Coccidioides immitis. Antimicrob. Agents Chemother. 62, e02258-17. doi: 10.1128/AAC.02258-17
Wiederhold, N. P., Xu, X., Wang, A., Najvar, L. K., Garvey, E. P., and Ottinger, E. A. (2018d). In vivo efficacy of vt-1129 against experimental cryptococcal meningitis with the use of a loading dose-maintenance dose administration strategy. Antimicrob. Agents Chemother. 62, e01315-18. doi: 10.1128/AAC.01315-18
Willger, S. D., Puttikamonkul, S., Kim, K. H., Burritt, J. B., Grahl, N., and Metzler, L. J. (2008). A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog. 4:e1000200. doi: 10.1371/journal.ppat.1000200
Wong, G. K., Griffith, S., Kojima, I., and Demain, A. L. (1998). Antifungal activities of rapamycin and its derivatives, prolylrapamycin, 32-desmethylrapamycin, and 32-desmethoxyrapamycin. J Antibiot. 51, 487–491.
Wood, N., Tan, K., Purkins, L., Layton, G., Hamlin, J., and Kleinermans, D. (2003). Effect of omeprazole on the steady-state pharmacokinetics of voriconazole. Br. J. Clin. Pharmacol. 56(Suppl. 1), 56–61.
Woolley, D. W. (1944). Some biological effects produced by benzimidazole and their reversal by purines. J. Biol. Chem. 152, 225–232.
Wu, Y., Wu, M., Wang, Y., Chen, Y., Gao, J., and Ying, C. (2018). ERG11 couples oxidative stress adaptation, hyphal elongation, and virulence in Candida albicans. FEMS Yeast Res. 18:foy057. doi: 10.1093/femsyr/foy057
Xing, Y., Chen, L., Feng, Y., Zhou, Y., Zhai, Y., and Lu, J. (2017). Meta-analysis of the safety of voriconazole in definitive, empirical, and prophylactic therapies for invasive fungal infections. BMC Infect. Dis. 17:798. doi: 10.1186/s12879-017-2913-8
Xisto, M. I., Caramalho, R. D., Rocha, D. A., Ferreira-Pereira, A., Sartori, B., and Barreto-Bergter, E. (2017). Pan-azole-resistant Candida tropicalis carrying homozygous erg11 mutations at position K143R: a new emerging superbug? J. Antimicrob. Chemother. 72, 988–992. doi: 10.1093/jac/dkw558
Yan, J. Y., Nie, X. L., Tao, Q. M., Zhan, S. Y., and Zhang, Y. D. (2013). Ketoconazole associated hepatotoxicity: a systematic review and meta- analysis. Biomed. Environ. Sci. 26, 605–610. doi: 10.3967/0895-3988.2013.07.013
Yang, H., Tong, J., Lee, C. W., Ha, S., Eom, S. H., and Im, Y. J. (2015). Structural mechanism of ergosterol regulation by fungal sterol transcription factor Upc2. Nat. Commun. 6:6129. doi: 10.1038/ncomms7129
Yates, C. M., Garvey, E. P., Shaver, S. R., Schotzinger, R. J., and Hoekstra, W. J. (2017). Design and optimization of highly-selective, broad spectrum fungal CYP51 inhibitors. Bioorg. Med. Chem. Lett. 27, 3243–3248. doi: 10.1016/j.bmcl.2017.06.037
Zhang, H. Z., Gan, L. L., Wang, H., and Zhou, C. H. (2017). New progress in azole compounds as antimicrobial agents. Mini. Rev. Med. Chem. 17, 122–166. doi: 10.2174/1389557516666160630120725
Keywords: CYP51, antifungal, crystal structure, azoles, resistance
Citation: Zhang J, Li L, Lv Q, Yan L, Wang Y and Jiang Y (2019) The Fungal CYP51s: Their Functions, Structures, Related Drug Resistance, and Inhibitors. Front. Microbiol. 10:691. doi: 10.3389/fmicb.2019.00691
Received: 10 November 2018; Accepted: 19 March 2019;
Published: 24 April 2019.
Edited by:
Santi M. Mandal, Indian Institute of Technology Kharagpur, IndiaReviewed by:
Praveen Rao Juvvadi, Duke University, United StatesCopyright © 2019 Zhang, Li, Lv, Yan, Wang and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lan Yan, eWxhbnNtbXVAc2luYS5jb20= Yan Wang, d2FuZ3lhbnNtbXVAMTI2LmNvbQ== Yuanying Jiang, MTM3NjE1NzE1NzhAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.