
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 22 March 2019
Sec. Virology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00578
 Xue-yang Wang1,2†
Xue-yang Wang1,2† Zuo-min Shao1†
Zuo-min Shao1† Qian-ying Chen1
Qian-ying Chen1 Jia-ping Xu3
Jia-ping Xu3 Xia Sun1,2
Xia Sun1,2 Zhen-ping Xu1,2
Zhen-ping Xu1,2 Mu-wang Li1,2*
Mu-wang Li1,2* Yang-chun Wu1,2*
Yang-chun Wu1,2*The molecular mechanism of silkworm resistance to Bombyx mori nucleopolyhedrovirus (BmNPV) infection remains unclear. The chaperonin containing t-complex polypeptide 1 (TCP-1) is essential for the folding of tubulin and actin to produce stable and functional competent protein conformation. However, little is known about this protein in silkworm. In the present study, a gene encoding the TCP-1β protein in silkworm was characterized, which has an open reading fragment of 1,611 bp encoding a predicted 536 amino acid residue-protein with a molecular weight of approximately 57.6 kDa containing a Cpn60_TCP1 functional domain. The sequence conservation is 81.52%. The highest level of BmTCP-1β mRNA expression was found in the midgut, while the lowest was in the hemolymph. To further study the function of BmTCP-1β, expression was knocked down with siRNA in vitro, resulting in significant downregulation of the selected cytoskeletal-related genes, actin and tubulin, which was also confirmed by overexpression of BmTCP-1β in BmN cells using the pIZT/V5-His-mCherry insect vector. Moreover, knockdown of BmTCP-1β significantly prolonged the infection process of BmNPV in BmN cells, which was also verified by overexpression of BmTCP-1β in BmN cells. Based on the results of the present study, we concluded that BmTCP-1β plays a vital role in BmNPV infection by regulating the expression of tubulin and actin. Taken together, our work provides valuable data for the clarification of the molecular mechanism of silkworm resistance to BmNPV infection.
The silkworm Bombyx mori L. (Lepidoptera: Bombycidae) has been domesticated for more than 5000 years and still plays an important role in many developing countries. Moreover, B. mori is a good model organism for the study of insect genetics and immunology (Goldsmith et al., 2005; Shao et al., 2012). B. mori nucleopolyhedrovirus (BmNPV) is a primary silkworm pathogen that causes serious economic losses annually. Interestingly, certain silkworm strains exhibit high resistance to BmNPV infection (Bao et al., 2009); however, the molecular underlying mechanism has not yet been fully elucidated.
The chaperonins are key molecular complexes that ensure the correct folding of proteins to produce energetically stable and functionally competent protein conformations (Coghlin et al., 2006). The eukaryotic TRiC (TCP-1 ring complex, also called CCT, chaperonin-containing TCP-1) is composed of two back-to-back stacked rings with eight different related subunits, each composed of eight separate gene products: CCTα, CCTβ, CCTγ, CCTδ, CCT𝜀, CCTζ, CCTη, and CCT𝜃 (Hynes et al., 1995; Liou and Willison, 1997; Gutsche et al., 1999; Bourke et al., 2002). The primary substrates for TCP-1 are cytoskeletal proteins, such as actin (Sternlicht et al., 1993) and tubulin (Yaffe et al., 1992; Sternlicht et al., 1993). The cytoplasmic chaperonin containing TCP-1 is necessary during the process of forming the native conformation of cytoskeletal protein polypeptide chains, in the case of α-tubulin and β-tubulin, and additional protein cofactors (Melki et al., 1997).
The actin and microtubule cytoskeleton is an indispensable compositions of host cells, which plays an important role in the attachment, internalization, endocytosis, nuclear targeting, transcription, replication, assembly and exocytosis of every virus, and the transport of progeny subviral particles (Radtke et al., 2006; Volkman, 2007; Mu et al., 2016). In addition, there exist a plethora of evidence showing that TCP-1 is involved in viral infection. Inoue et al. (2011) reported that chaperonin TRiC/CCT participates in the replication of the hepatitis C viral genome via interaction with the viral NS5B protein. Coghlin et al. (2006) reported that an active TCP-1 is potentially involved in the immune response against viral infection in M. rosenbergii (Arockiaraj et al., 2012). In our previous comparative subcellular proteomics analysis of different resistant silkworm strains following BmNPV infection, we found that BmTCP-1 showed an marked downregulation of expression in the midgut of resistant strains following BmNPV infection (Wang et al., 2017).
To the best of our knowledge, there is no relevant report available to date on TCP-1 in silkworm. To clarify the function of BmTCP-1β, the expression profiles of BmTCP-1β at different developmental stages and in various tissues were analyzed using RT-qPCR. To further define the role of BmTCP-1β in BmNPV infection, the alteration of BmNPV infection in BmN cells and the expression patterns of actin and tubulin were analyzed following knockdown and overexpression of BmTCP-1β using siRNA and the insect pIZT/V5-His-mCherry vector.
The silkworm strain, p50 (‘Dazao’), was maintained in the Key Laboratory of Silkworm and Mulberry Genetic Improvement, Ministry of Agriculture, Sericultural Research Institute, Chinese Academy of Agricultural Sciences, Zhenjiang, China. The first three instar larvae were reared on a fresh artificial diet at 26 ± 1°C, 75 ± 5% relative humidity, and a 12-h day/night cycle. The rearing temperature for the last two instars was reduced to 24 ± 1°C, but the other conditions remained unchanged.
Budded virus containing EGFP-tagged (BV-EGFP) BmNPV were kindly provided by Associate Professor Xudong Tang in School of biotechnology. Polyhedrin promoter was used, the EGFP was inserted between BamHI and XhoI, which is not fused with any protein. The amount of BV-EGFP (pfu/mL) was determined by the standard curve method. Briefly, vp39 of BmNPV was selected to constructed standard plasmid that was used to build a standard curve using RT-qPCR. The equal volume solution containing BV-EGFP (1 × 108 pfu/mL) was added into the culture medium to study the infection process of BmNPV in different cell lines.
The silkworm ovarian cell line, BmN, was cultured in TC-100 medium supplemented with 10% (v/v) FBS, 200 μg/mL penicillin, and 100 μg/mL streptomycin at 28°C. Transfection was performed using NeofectTM DNA transfection reagent (NEOFECT, Chian), according to the manufacturer’s instructions.
Cellular fluorescence images were taken using a Leica inverted research grade microscope DMi3000B camera and processed with the Leica Application Suite V4.6 software.
The cDNA and deduced protein sequence of BmTCP-1β were analyzed using DNAMAN 8.0 (Lynnon Corporation, Quebec, Canada). The conserved motif was predicted on the SMART server1. The signal peptide was predicted by SignalP 4.12. Sequences of orthologs were analyzed using the BLASTP tool3. The amino acid sequences of different species were aligned using the MUSCLE module of the MEGA6 software. A neighbor-joining tree was generated with a bootstrap of 1000 replications using MEGA6, the best DNA/Protein models of LG+G was adopt.
In the present study, STRING4 was adopted to analyze the protein-protein interactions (PPIs) of BmTCP-1β with cytoskeletal proteins. Due to the lack of proteomics information on B. mori in STRING, the PPI network was built using the database of another well-studied insect, Drosophila melanogaster (D. melanogaster).
Total RNA was extracted with TRIzol Reagent (Invitrogen, United States) according to the manufacturer’s instructions. A NanoDrop 2000 spectrophotometer (Thermo Scientific, United States) was used to quantitate the RNA concentration. The purity of all RNA samples was assessed at absorbance ratios of A260/280 and A260/230, and the integrity of the RNA was confirmed by 1% agarose gel electrophoresis. The first strand of cDNA was synthesized using the PrimeScriptTM RT Reagent Kit with gDNA Eraser (TaKaRa, Japan), according to the manufacturer’s instructions.
To analyze the transcriptional levels of BmTCP-1β, actin and tubulin, quantitative reverse transcription PCR (RT-qPCR) was adopted. All primers are listed in Table 1. RT-qPCR reactions were prepared with the SYBR Premix Ex TaqTM Kit (TaKaRa) following the manufacturer’s instructions. Reactions were carried out using the QuantStudioTM Real-Time System (Thermo Fisher Scientific, United States). The thermal cycling profile consisted of an initial denaturation at 95°C for 5 min, 40 cycles at 95°C for 5 s, and 60°C for 31 s. All reactions were performed in triplicate. Relative expression levels were calculated using the 2-ΔΔCt method following the protocol described by Livak and Schmittgen (2001). In the present study, the B. mori glycerol-3-phosphate dehydrogenase-1 (BmGAPDH) gene was used as a reference standard (Guo et al., 2016). Statistical analysis was conducted using the SPSS software (IBM, www.ibm.com). One-way ANOVA with Tukey’s posttest was used to analyze the acquired data. The P-value < 0.05 was believed to be statistically significant.
To knock down BmTCP-1β in vitro, two special targets of this gene’s functional domain were selected. The siRNA oligos were synthesized by Sangon Biotechnology, China (Table 2). The synthesis of siRNA was performed using the in vitro Transcription T7 Kit (for siRNA Synthesis) (TaKaRa, Japan), according to the manufacturer’s instructions. The concentration of siRNA was obtained using a NanoDrop 2000 spectrophotometer. The RNA quality was examined by agarose gel electrophoresis and subsequently stored at -80°C until use.
BmN cells (1 × 106 cells/well) were seeded on to Costar 6-well cell culture clusters and cultured overnight at 28°C. Transfection was performed using NeofectTM DNA transfection reagent (NEOFECT, China), according to the manufacturer’s instructions. Briefly, 4 μg siRNA oligo was added to 0.2 mL serum-free TC-100 media and mixed gently. Subsequently, 4 μL transfection reagent was added to the mixture of TC-100 and siRNA oligo, mixed gently, and incubated for 15–30 min at room temperature (RT). The mixture of siRNA and transfection reagent was then added to the culture medium, mixed gently, and the cells were cultured at 28°C. Each transfection was repeated three times. After a 24-h, 48-h and 72-h incubation with siRNA, BmN cells were harvested to extract the RNA. The RNAi efficiency of BmTCP-1β was assayed by RT-qPCR.
The functional domain of BmTCP-1β was amplified with primers BmTCP-1β FD (Table 1, the underlined portions indicate the Kpn I and EcoRI restriction sites, respectively). The cDNA of p50 midgut was used to amplify the functional domain of BmTCP-1β. Purified PCR products were ligated into pMD-19T for sequencing. The functional domain of BmTCP-1β was obtained by digestion from the recombinant plasmid using KpnI and EcoRI, and subsequent ligation into the pIZT/V5-His-mCherry plasmid to construct the transient expression vector pIZT/V5-His-mCherry-BmTCP-1β. Transfection was performed as described above.
The full-length BmTCP-1β cDNA (GenBank accession number: NM_001046644.1) consists of a 97 bp 5′-untranslated region (5′-UTR), a 928 bp 3′-UTR and a 1,611 bp open reading frame (ORF) encoding a 536 amino acid protein (Supplementary Figure S1). The theoretical MW and pI are 57.59 kDa and 6.32, respectively. BmTCP-1β has one functional domain, Cpn60 TCP-1, which was consisted of several members of HSP60 chaperone family and TCP-1 (T-complex protein) family.
BLASTP search showed that the BmTCP-1β amino acid sequence is most similar to that of Spodoptera litura, Heliothis virescens and Amyelois transitella TCP-1 (94% identity), Helicoverpa armigera, Danaus plexippus, Papilio xuthus, Papilio machaon and Papilio xuthus (93% identity), Pieris rapae, Bicyclus anynana and Papilio polytes (92% identity), followed by Aedes aegypti (78% identity) and Aedes albopictus (77% identity). Therefore, the high conservation of TCP-1β in different species shows that BmTCP-1β may serve as a molecular chaperone during protein folding in silkworm (Supplementary Figure S2).
The entire amino acid sequence of BmTCP-1β and those of other species were used to determine the evolutionary relationships among TCP-1β in different species. A phylogenetic tree consisting of BmTCP-1β and 17 other homologs was constructed. These genes were clearly classified into four groups: Lepidoptera, Hemiptera, Capparidales, and Rodentia. BmTCP-1β and its homologs from 12 other insects including H. armigera, H. virescens, S. litura, D. plexippus, A. transitella, P. xylostella, B. anynana, P. machaon, P. polytes, P. xuthus, and P. rapae were clustered into the Lepidoptera group (Supplementary Figure S3). M. musculus and Arabidopsis sequences shared a lower sequence identity with BmTCP-1β, indicating that the gene in their ancestors may have been diverged before the emergence of these orders.
In living cells, many proteins can interact with each other, and these interacting proteins are expected to be involved in the same biological process or to function in the same subcellular compartment, which is supported by evidence that proteins in the same pathway are more interconnected (Barabasi et al., 2011). STRING contains abundant resources on physical and functional interactions and collects information from numerous sources, including experimental repositories, computational prediction methods, and public text collections (Jensen et al., 2009). To further investigate the relationship between BmTCP-1β and the selected cytoskeletal proteins, Actin and Tubulin, the functional association of these proteins was analyzed using the STRING 9.1 online software. A combined score was assigned to every protein-protein association pair in the software. This score was computed by combining the probabilities from several pieces of evidence and correcting for the probability of randomly observing an interaction. As illustrated in Figure 1, TCP-1β has a close relationship with actin and tubulin, especially actin, which suggests that BmTCP-1β may be involved in BmNPV infection by mediating the expression of cytoskeletal proteins.
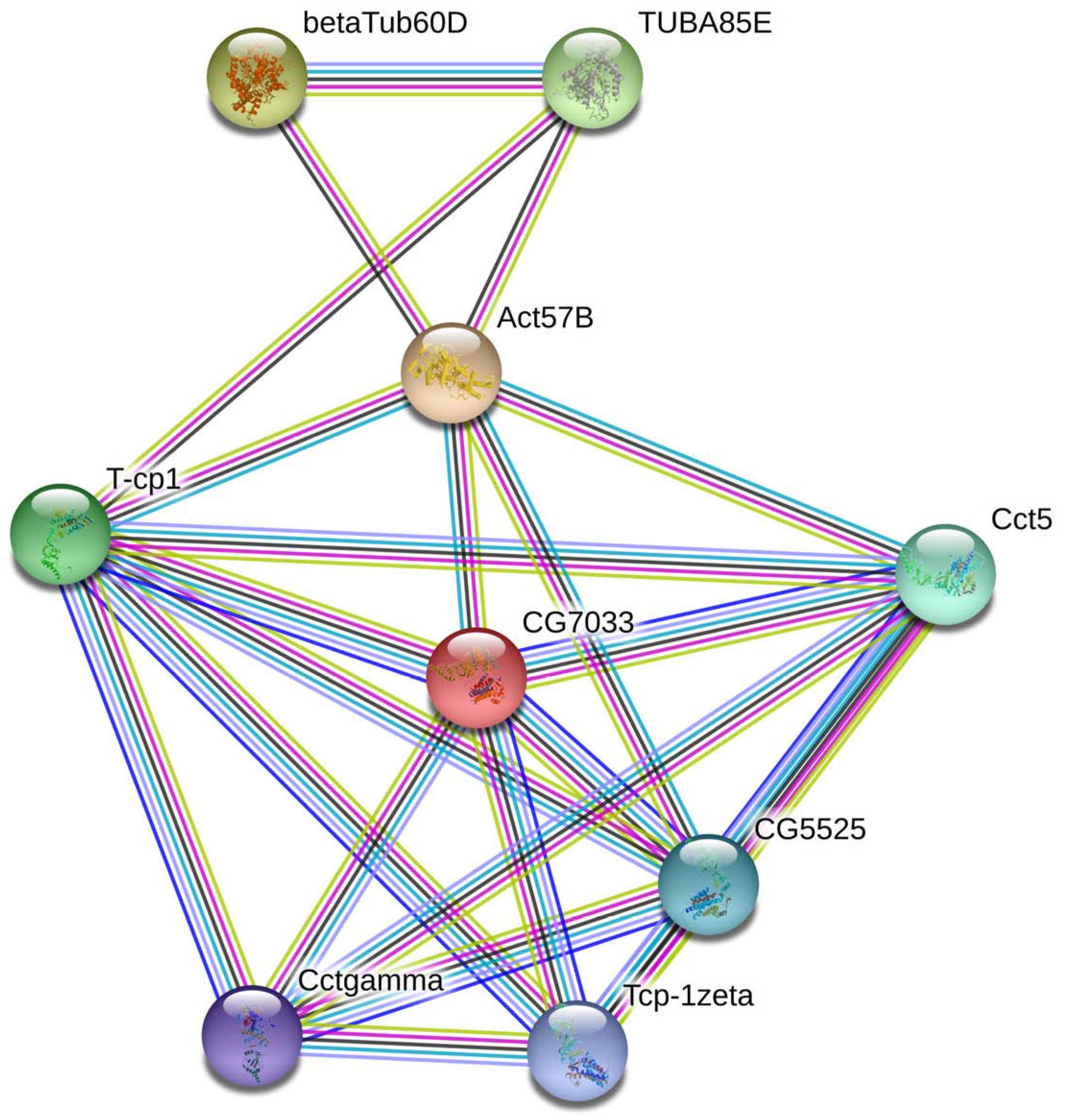
Figure 1. The interaction network of TCP-1β with selected cytoskeletal proteins was constructed based on the STRING website information using the database of another well-studied insect, D. melanogaster. CG7033, TCP-1β; Act57B, actin; β-Tub60D, β-tubulin; TUBAB5E, α-tubulin.
RT-qPCR was used to analyze the transcriptome patterns of BmTCP-1β in different tissues and at various developmental stages. The total RNA of different tissues and developmental stages was extracted from the whole body, and agarose gel electrophoresis was used to assess the integrity of total RNA. The quality of cDNA was assessed using BmGAPDH. We found that the expression pattern of BmTCP-1β was not significantly different among various different developmental stages (Figure 2A). Additionally, BmTCP-1β showed relatively a higher expression level before the molting stage of fourth instar, and reached the highest expression level during the molting stage of fourth instar, while other selected stages showed significantly lower expression levels (Figure 2B). Moreover, BmTCP-1β showed significant specificity in tissue expression, with the lowest expression level observed in the hemolymph and the highest in the midgut (Figure 2C).
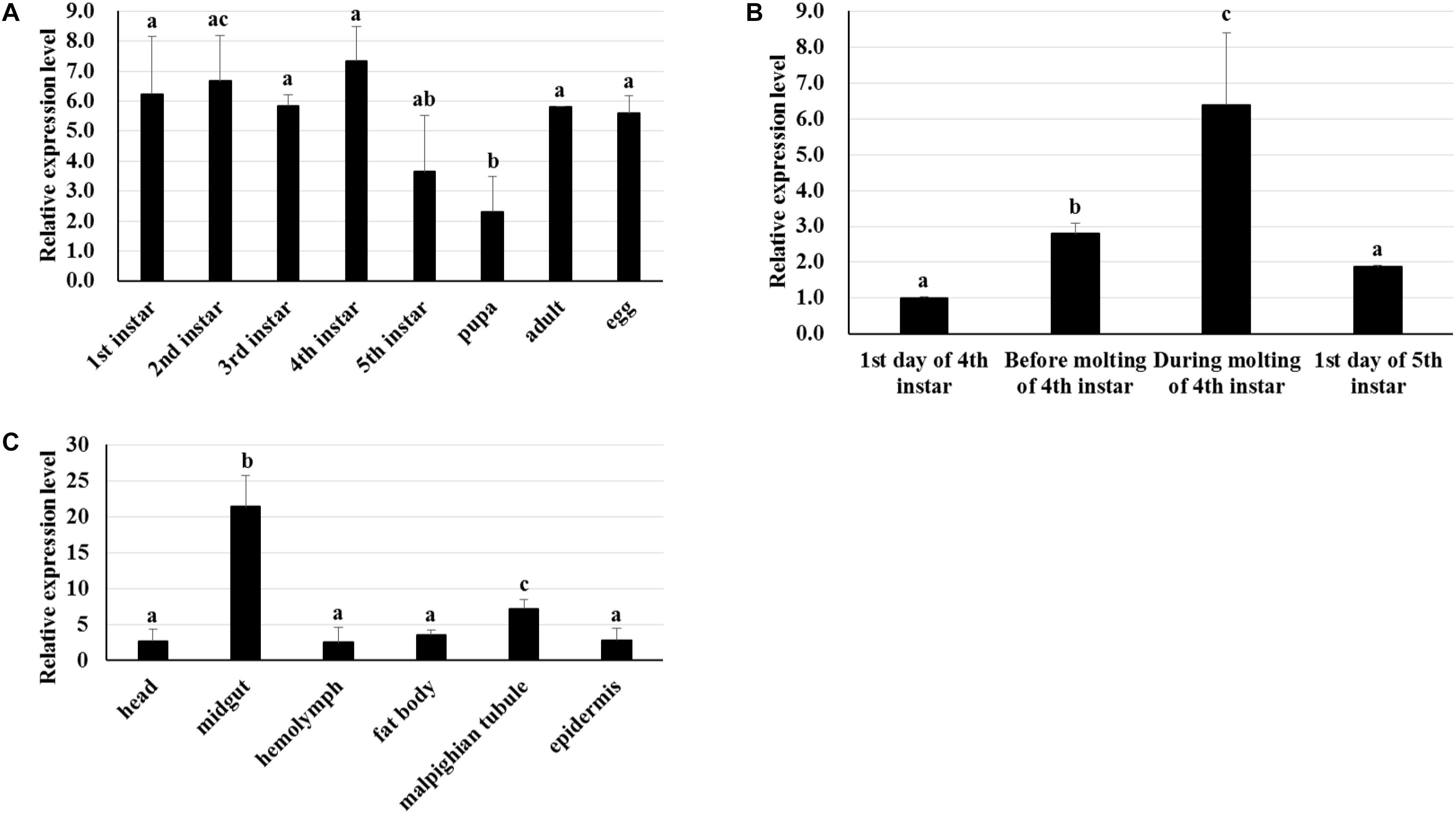
Figure 2. The spatiotemporal expression analysis of BmTCP-1β. (A–C) show the expression patterns of BmTCP-1β at different developmental stages (A), during molting (B), and in different tissues (C). The data were normalized using BmGAPDH and are represented as the mean ± standard error of the mean, from three independent experiments. Relative expression levels were calculated using the 2-ΔΔCt method. Statistical analysis was conducted using the SPSS software. One-way ANOVA with Tukey’s posttest was used. Significant differences are indicated by different letters, e.g., a, b, and c (p < 0.05).
To identify the best interference time of siRNA in BmTCP-1β expression, BmN cells were transfected with siRNA for three different time points, the control group was treated with transfection reagent without siRNA. The relative expression level of BmTCP-1β showed significant downregulation following siRNA treatment for 24 h, 48 h and 72 h. The significant downregulation of BmTCP-1β was initially 24 h following siRNA transfection; thus, an interference time of 24 h was selected for further analyses (Figure 3A).
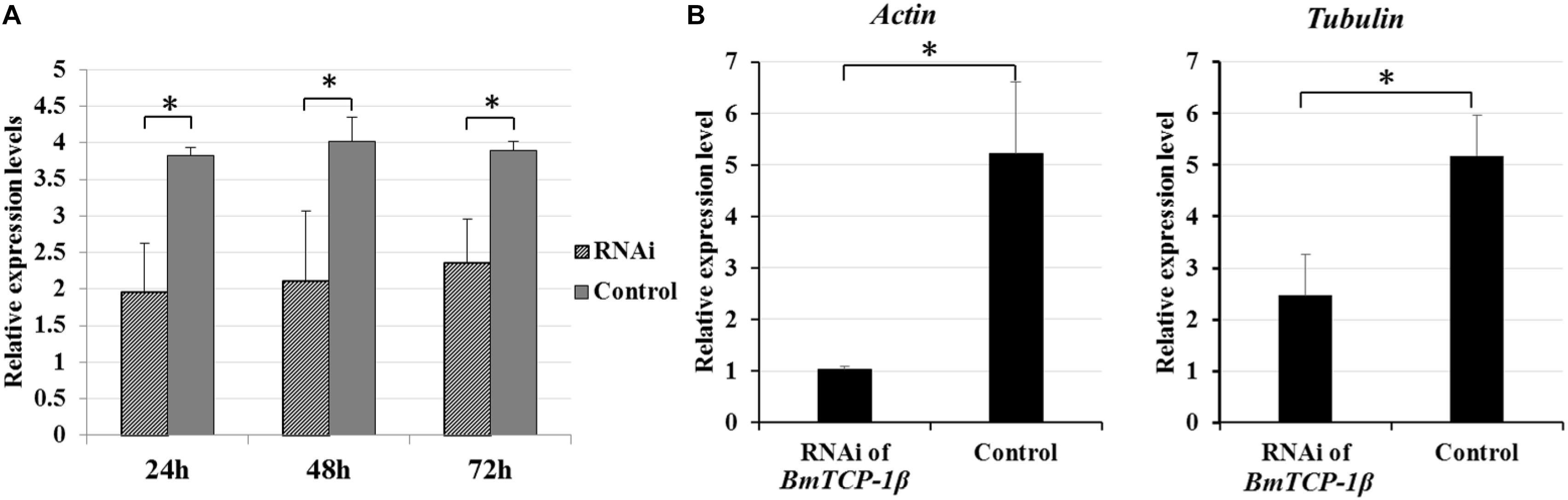
Figure 3. Expression analysis of selected cytoskeletal genes following addition of BmTCP-1β RNAi. (A), expression analysis of BmTCP-1β following siRNA interference using RT-qPCR at different times. (B), expression analysis of selected cytoskeletal genes using RT-qPCR. The data were normalized using BmGAPDH and are represented as the mean ± standard error of the mean, from three independent experiments. Relative expression levels were calculated using the 2-ΔΔCt method. Statistical analysis was conducted using the SPSS software. One-way ANOVA with Tukey’s posttest was used. Significant differences are indicated by asterisks (p < 0.05).
To detect the connection of BmTCP-1β to selected cytoskeletal genes, RT-qPCR was adopted to analyze the expression patterns of actin and tubulin following knockdown of BmTCP-1β. The results show that actin and tubulin were both significantly down-regulated following addition of BmTCP-1β RNAi (Figure 3B), indicating that BmTCP-1β has a close relationship with actin and tubulin.
To identify the function of BmTCP-1β in the process of BmNPV infection, BmN cells infected with BV-EGFP were analyzed at different time points following knockdown of BmTCP-1β by siRNA at 24 h. A significant infection signal of BV-EGFP was found at 48 h post-infection, where the fluorescence signals of BmNPV in the control were significantly higher than those in the BmTCP-1β RNAi group (Figure 4A), indicating that BmTCP-1β promoted the infection of BmNPV. At 72 h post-infection, most BmN cells were dissociated in the control as compared with BmTCP-1β RNAi group (Figure 4B). At 96 h post-infection, there were only few of EGFP signals in the control group, while BmN cells treated with siRNA retained normal cell morphology (Figure 4C), which is consistent with the results obtained at 48 and 72 h, further confirming the vital role of BmTCP-1β in BmNPV infection.
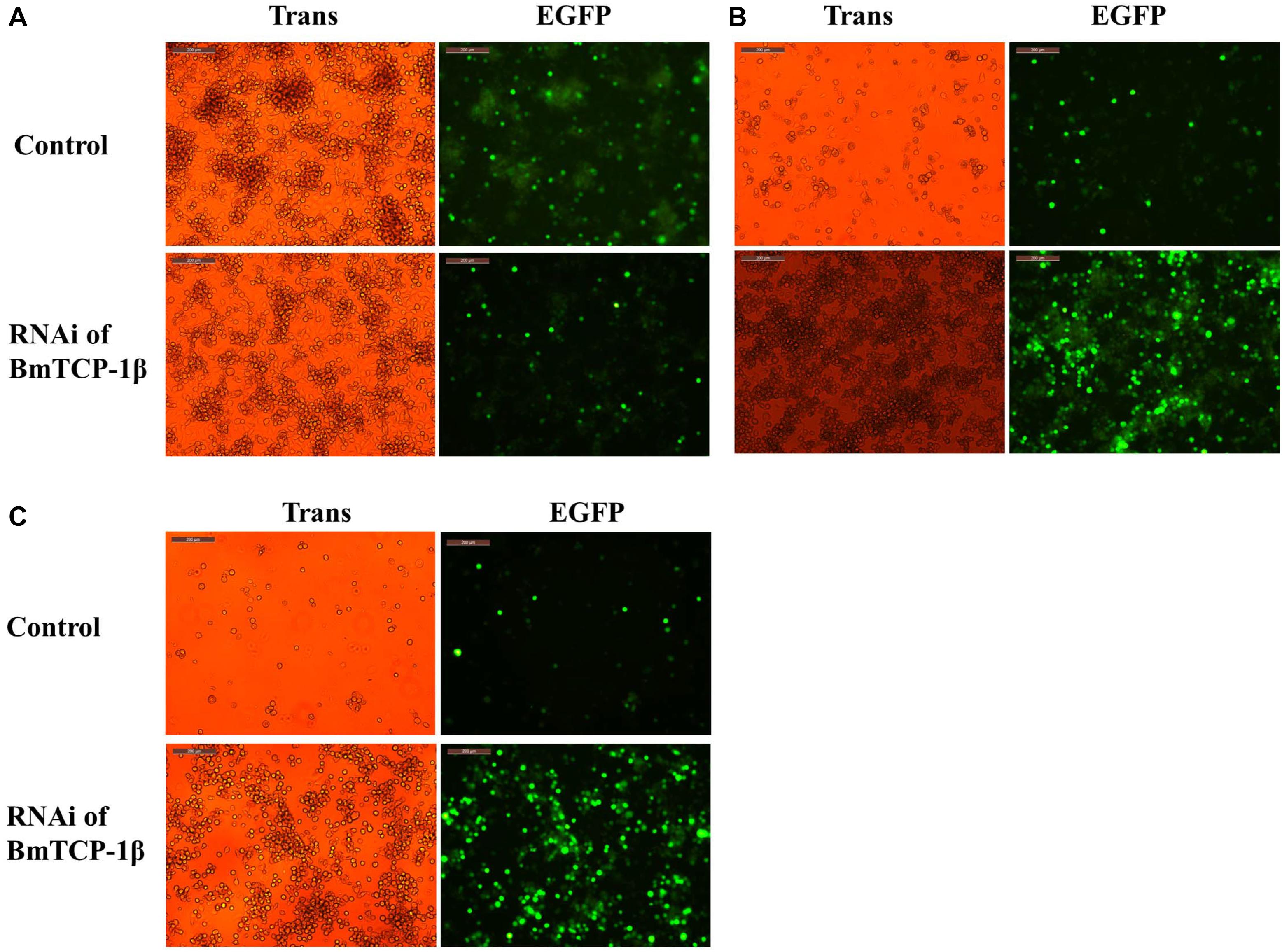
Figure 4. Infection analysis of BmNPV in BmN cells following addition of BmTCP-1β RNAi for different time periods. (A) 48 h after BV-EGFP infection. (B) 72 h after BV-EGFP infection. (C) 96 h after BV-EGFP infection. Scale bar, 200 μm. Trans (white), optical transmission; EGFP (Green), expressed following the replication of BV.
The insect expression vector pIZT/V5-His-mCherry was used to overexpress BmTCP-1β in vitro. The purified PCR product of BmTCP-1β functional domain was inserted into the pIZT/V5-His-mCherry vector using KpnI and EcoRI restriction enzymes. BmN cells were transfected with the recombinant bacmid to overexpress the BmTCP-1β protein, the stable cell line of pIZT/V5-His-mCherry-BmTCP-1β was screened by final concentration of 200ng/uL zeocin. The red fluorescence protein of mCherry indicates that BmTCP-1β was successfully overexpressed in BmN cells (Figures 5A,B), which was validated by RT-qPCR (Figure 5C). The expression levels of selected cytoskeletal genes in transgenic BmN cells were compared with those in wild-type BmN cells by RT-qPCR. The results show that overexpression of BmTCP-1β could significantly induce up-regulation of actin and tubulin in vitro (Figures 5D,E), which is consistent with the RNAi results.
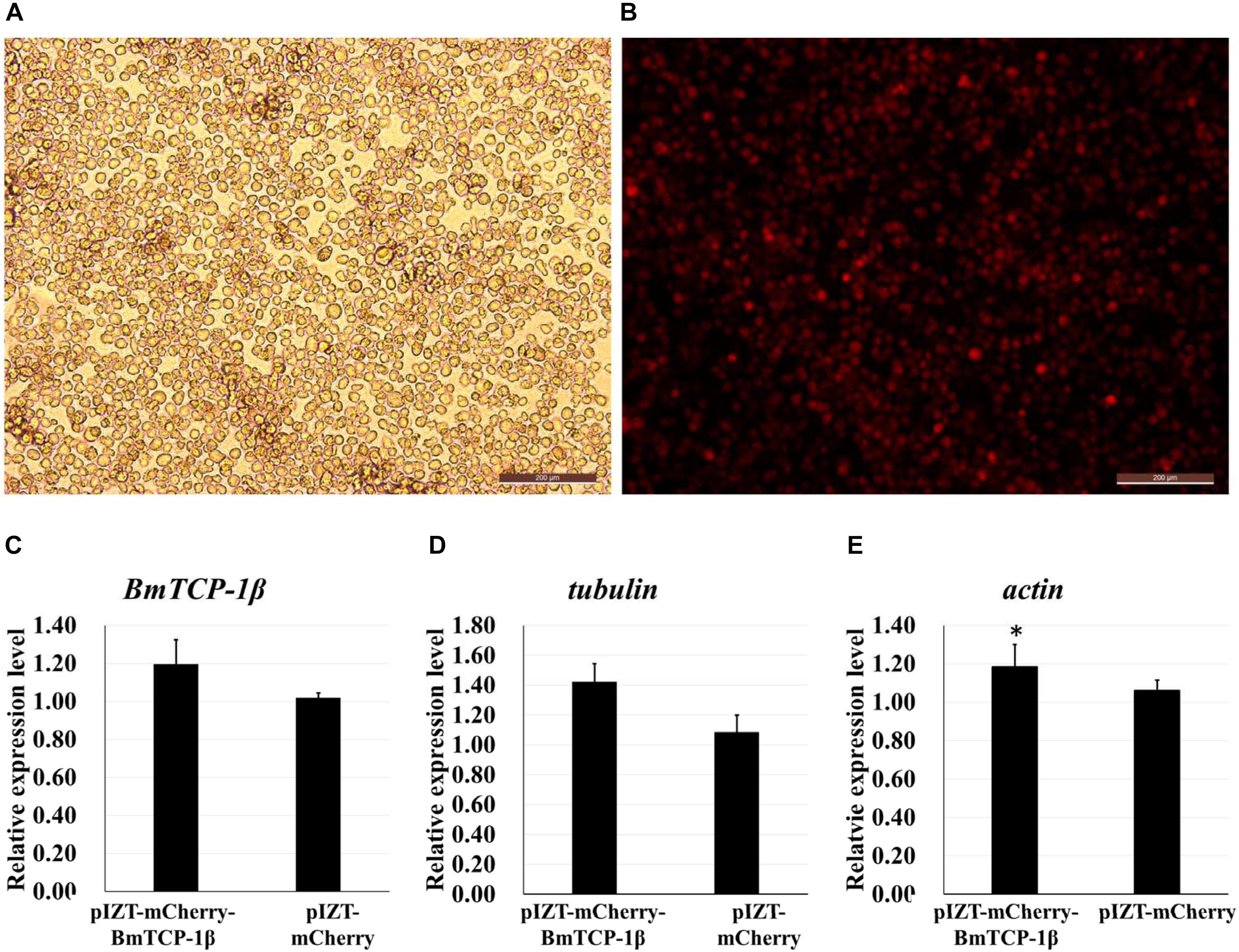
Figure 5. Correlation analysis of BmTCP-1β with selected cytoskeletal genes by overexpression of BmTCP-1β in vitro. (A,B), overexpression of BmTCP-1β in BmN using the pIZT/V5-His-mCherry vector, scale bar = 200 μm. (A) Trans (white), optical transmission. (B) Transfection of BmN cells with the pIZT/V5-His-mCherry-BmTCP-1β vector, mCherry (red). (C–E) expression analysis of BmTCP-1β, actin, and tubulin using RT-qPCR. One-way ANOVA with Tukey’s posttest was used. Data were analyzed as described above. Significant differences are indicated by asterisks (p < 0.05).
Based on the RNAi analysis, knockdown of BmTCP-1β delayed the infection of BmNPV in BmN cells. To further validate the function of BmTCP-1β in BmNPV infection, transcription of BV vp39 gene in control and transgenic BmN cells at different infection times was analyzed following the overexpression of BmTCP-1β using an insect expression system. The results show that a significant infection signal was found 24 h post-infection, which was significantly higher in transgenic BmN cells than in control BmN cells (Figure 6A), consistent with the RNAi results. At 48 h post-infection, significant dissociation of BmN cells was found in the transgenic group as compared with control group (Figure 6B). At 72 h post-infection, most transgenic BmN cells were floating in the culture medium and numerous infected cells were dissociated, while the BmN cells in the control group retained complete cell morphology and were adhered (Figure 6C). Moreover, the number of BmNPV copies was detected using RT-qPCR, and the results show that the replication of BmNPV in the transgenic cells was significantly faster than that of in the control cells at all three selected time points (Figure 6D), which further validated the reasonable suggestion that the early infection signals in the transgenic cells, and BmTCP-1β accelerated BmNPV infection.
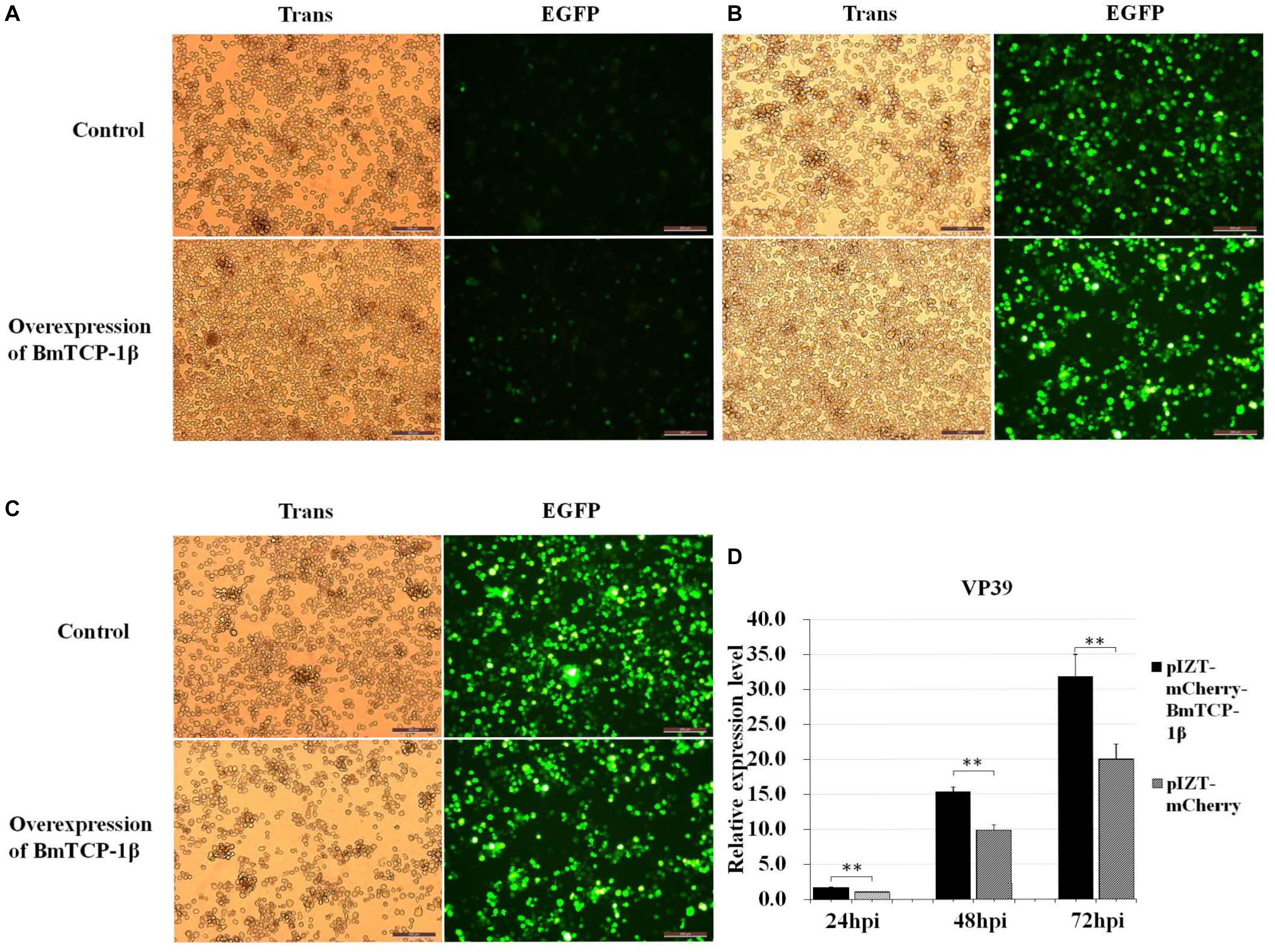
Figure 6. Infection analysis of BmNPV in wild type and transgenic BmN cells. (A) 24 h after BV-EGFP infection. (B) 48 h after BV-EGFP infection. (C) 72 h after BV-EGFP infection. Scale bar = 200 μm. (D) replication analysis of BmNPV following the overexpression of BmTCP-1β. The data was analyzed as described above. One-way ANOVA with Tukey’s posttest was used. Significant differences are indicated by asterisks (p < 0.05).
In our previous comparative subcellular proteomics analysis of different resistant silkworm midguts in response to BmNPV infection, many interesting proteins related to BmNPV infection were identified. Among these, we were interested in BmTCP-1β, which showed a significant downregulation following BmNPV infection, indicating that BmTCP-1β may be involved in BmNPV infection. TCP-1β in response to viral infection has been widely reported in other fields. Inoue et al. (2011) reported that a subunit of CTP1 could ligate to Negri bodies and promote rabies virus transcription and replication (Zhang et al., 2013, 2014), which is in agreement with our previous study reporting notable down-regulation of BmTCP-1β in the resistant strain, BC9, following BmNPV infection, but no differential expression in the susceptible strain, p50. Nevertheless, the underlying mechanism of BmTCP-1β in response to BmNPV infection remains unclear. In the present study, the molecular characterization and functional analysis of BmTCP-1β is described.
The biological information method was used to characterize the sequence of BmTCP-1β. The homologous alignment analysis shows BmTCP-1β shares a high similarity with its homologous genes in other species (Supplementary Figure S2), which was verified by phylogenetic analysis (Supplementary Figure S3), indicating its role in the process of protein folding. Moreover, BmTCP-1β has a close interaction with actin and tubulin on the STRING website using the Drosophila database (Figure 1), which is consistent with previous reports that TCP-1 is vital in protein folding to produce stable and functionally competent conformations (Blitvich et al., 2001; Olshina et al., 2016). The stable expression levels at different developmental stages indicate that BmTCP-1β is essential in silkworm development (Figure 2A). Furthermore, the relatively higher expression levels of BmTCP-1β during the molting stage suggests that the expression of BmTCP-1β may be regulated by molting hormones (Figure 2B). The highest expression level was found in the midgut, indicating its vital role in the process of BmNPV infection of midgut cells (Figure 2C).
Our previous proteomic data showed that BmTCP-1β may be involved in BmNPV infection. By combining the transcriptome data (Wang et al., 2016), we deduced that BmTCP-1β is involved in BmNPV infection by regulating the expression of cytoskeletal proteins. To validate this point, here, the expression of BmTCP-1β was knocked down using siRNA in BmN cells. The result showed that BmTCP-1β not only down-regulated the expression of selected cytoskeletal genes, including actin and tubulin (Figure 3), but also delayed the infection of BmNPV (Figure 4), indicating that our inference is reasonable. Moreover, to further validate the results of the BmTCP-1β RNAi described above, overexpression of BmTCP-1β was also carried out using the insect pIZT/V5-His-mCherry vector. The functional domain of BmTCP-1β was ligated into pIZT/V5-His-mCherry to overexpress BmTCP-1β in BmN cells (Figure 5). The results show that the expression levels of cytoskeletal genes were up-regulated in the transgenic cell lines and the infection time of BmNPV was also shorter than that in the control group (Figure 6). Based on the above analysis, it is reasonable to suggest that BmTCP-1β plays a vital role in the response to BmNPV infection by regulating the expression of cytoskeletal genes. Taken together, our work provides valuable data for the clarification of the underlying molecular mechanism of silkworm resistance to BmNPV infection.
Based on the above analysis in combination with a previous report about the role of the cytoskeleton in the process of baculoviral infection (Radtke et al., 2006), we hypothesize that BmTCP-1β plays a vital role in the process of BmNPV infection of host cells by regulating cytoskeletal proteins. The actin-mediated endocytic process is triggered by BV-envelope contact with the plasma membrane (Ascough, 2004) and surfing via filopodia toward an area with high endocytic activity (Pollard and Borisy, 2003), where it is internalized by endocytosis. Alternatively, BmNPV nucleocapsids can fuse with the plasma membrane. After traversing the actin cortex, either inside endocytic vesicles or by itself, free nucleocapsids or BV-envelopes inside vesicles are transported along tubulin toward the microtubule-organizing center (MTOC). From the MTOC, nucleocapsids are transported toward the nucleus, and upon binding to the nuclear pore, the nucleocapsids release their genome for replication (Radtke et al., 2006; Figure 7). The actin and tubulin involved in this process acquire their native confirmation in the presence of BmTCP-1β, and in the case of α-tubulin and β-tubulin, additional protein cofactors, which may explain the lower expression level of BmTCP-1β in the resistant strain, BC9, as compared with the susceptible strain, p50.
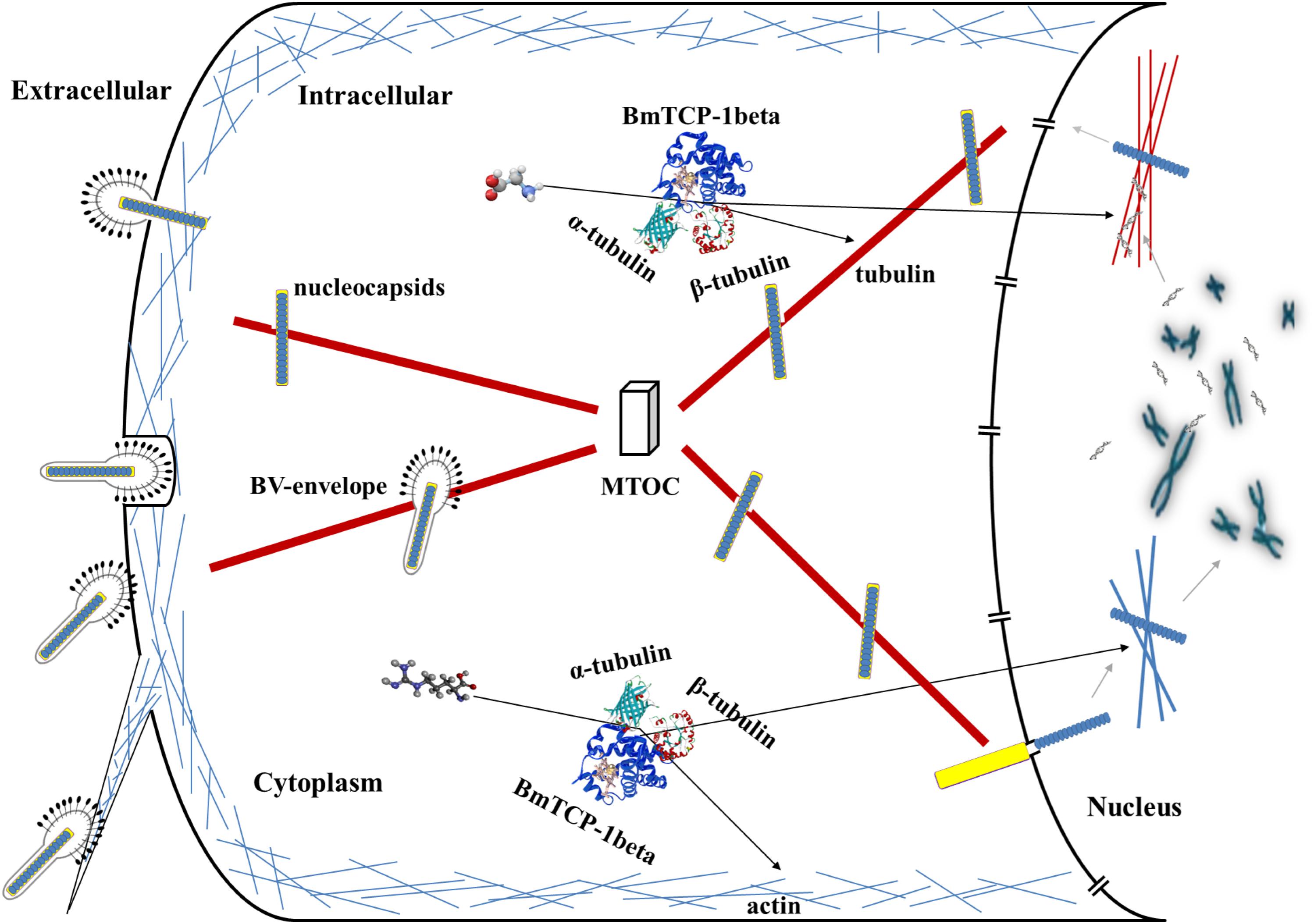
Figure 7. Hypothesized roles of BmTCP-1β in the response to BmNPV infection. MTOC, microtubule-organizing center.
X-yW conceived and designed the experiments. X-yW, Z-mS, Q-yC, and XS performed the experiments. X-yW, Z-mS, and Q-yC analyzed the data. M-wL, Z-pX, and Y-cW contributed to reagents, materials, and analysis tools. X-yW, Z-mS, and J-pX wrote the manuscript.
This work was supported by the National Natural Science Foundation of China (31802137) and the funding gave by Jiangsu University of Science and Technology (1732931704).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00578/full#supplementary-material
Arockiaraj, J., Vanaraja, P., Easwvaran, S., Singh, A., Othman, R. Y., and Bhassu, S. (2012). Molecular functions of chaperonin gene, containing tailless complex polypeptide 1 from Macrobrachium rosenbergii. Gene 508, 241–249. doi: 10.1016/j.gene.2012.07.050
Ascough, K. R. (2004). Endocytosis: actin in the driving seat. Curr. Biol. 14, R124–R126. doi: 10.1016/j.cub.2004.01.021
Bao, Y. Y., Tang, X. D., Lv, Z. Y., Wang, X. Y., Tian, C. H., Xu, Y. P., et al. (2009). Gene expression profiling of resistant and susceptible Bombyx mori strains reveals nucleopolyhedrovirus-associated variations in host gene transcript levels. Genomics 94, 138–145. doi: 10.1016/j.ygeno.2009.04.003
Barabasi, A. L., Gulbahce, N., and Loscalzo, J. (2011). Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 12, 56–68. doi: 10.1038/nrg2918
Blitvich, B. J., Rayms-Keller, A., Blair, C. D., and Beaty, B. J. (2001). Complete cDNA and deduced amino acid sequence of the chaperonin containing T-complex polypeptide 1 (CCT) delta subunit from Aedes triseriatus mosquitoes. DNA Seq. 12, 203–208. doi: 10.3109/10425170109080776
Bourke, G. J., El, A. W., Wilson, S. J., Yuan, A., Roobol, A., and Carden, M. J. (2002). Slow axonal transport of the cytosolic chaperonin CCT with Hsc73 and actin in motor neurons. J. Neurosci. Res. 68, 29–35. doi: 10.1002/jnr.10186
Coghlin, C., Carpenter, B., Dundas, S. R., Lawrie, L. C., Telfer, C., and Murray, G. I. (2006). Characterization and over-expression of chaperonin t-complex proteins in colorectal cancer. J. Pathol. 210, 351–357. doi: 10.1002/path.2056
Goldsmith, M. R., Shimada, T., and Abe, H. (2005). The genetics and genomics of the silkworm, Bombyx mori. Annu. Rev. Entomol. 50, 71–100. doi: 10.1146/annurev.ento.50.071803.130456-3
Guo, H., Jiang, L., and Xia, Q. (2016). Selection of reference genes for analysis of stress-responsive genes after challenge with viruses and temperature changes in the silkworm Bombyx mori. Mol. Genet. Genomics 291, 999–1004. doi: 10.1007/s00438-015-1125-4
Gutsche, I., Essen, L. O., and Baumeister, W. (1999). Group II chaperonins: new TRiC(k)s and turns of a protein folding machine. J. Mol. Biol. 293, 295–312. doi: 10.1006/jmbi.1999.3008
Hynes, G., Kubota, H., and Willison, K. R. (1995). Antibody characterisation of two distinct conformations of the chaperonin-containing TCP-1 from mouse testis. FEBS Lett. 358, 129–132. doi: 10.1016/0014-5793(94)01408-S
Inoue, Y., Aizaki, H., Hara, H., Matsuda, M., Ando, T., Shimoji, T., et al. (2011). Chaperonin TRiC/CCT participates in replication of hepatitis C virus genome via interaction with the viral NS5B protein. Virology 410, 38–47. doi: 10.1016/j.virol.2010.10.026
Jensen, L. J., Kuhn, M., Stark, M., Chaffron, S., Creevey, C., Muller, J., et al. (2009). STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 37, D412–D416. doi: 10.1093/nar/gkn760
Liou, A. K. F., and Willison, K. R. (1997). Elucidation of the subunit orientation in CCT (chaperonin containing TCP1) from the subunit composition of CCT micro-complexes. EMBO J. 16, 4311–4316. doi: 10.1093/emboj/16.14.4311
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Melki, R., Batelier, G., and Stéphanie Soulié, A. (1997). RCW. Cytoplasmic chaperonin containing TCP-1:‡ structural and functional characterization†. Biochemistry 36, 5817–5826. doi: 10.1021/bi962830o
Mu, J., Zhang, Y., Hu, Y., Xue, H., Yuan, Z., He, Z., et al. (2016). Autographa californica multiple nucleopolyhedrovirus Ac34 protein retains cellular actin-related protein 2/3 complex in the nucleus by subversion of CRM1-dependent nuclear export. PLoS Pathog. 12:e1005994. doi: 10.1371/journal.ppat.1005994
Olshina, M. A., Baumann, H., Willison, K. R., and Baum, J. (2016). Plasmodiumactin is incompletely folded by heterologous protein-folding machinery and likely requires the native Plasmodium chaperonin complex to enter a mature functional state. FASEB J. 30, 405–416. doi: 10.1096/fj.15-276618
Pollard, T. D., and Borisy, G. G. (2003). Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–465. doi: 10.1016/S0092-8674(03)00120-X
Radtke, K., Döhner, K., and Sodeik, B. (2006). Viral interactions with the cytoskeleton: a hitchhiker’s guide to the cell. Cell. Microbiol. 8, 387–400. doi: 10.1111/j.1462-5822.2005.00679.x
Shao, Q. M., Yang, B., Xu, Q. Y., Li, X. Q., Lu, Z. Q., Wang, C. S., et al. (2012). Hindgut innate immunity and regulation of fecal microbiota through melanization in insects. J. Biol. Chem. 287, 14270–14279. doi: 10.1074/jbc.M112.354548
Sternlicht, H., Farr, G. W., Sternlicht, M. L., Driscoll, J. K., Willison, K., and Yaffe, M. B. (1993). The t-complex polypeptide 1 complex is a chaperonin for tubulin and actin in vivo. Proc. Natl. Acad. Sci. U.S.A. 90, 9422–9426. doi: 10.2307/2363166
Volkman, L. E. (2007). Baculovirus infectivity and the actin cytoskeleton. Curr. Drug Targets 8, 1075–1083. doi: 10.2174/138945007782151379
Wang, X. Y., Yu, H. Z., Geng, L., Xu, J. P., Yu, D., Zhang, S. Z., et al. (2016). Comparative transcriptome analysis of Bombyx mori (Lepidoptera) larval midgut response to BmNPV in susceptible and near-isogenic resistant strains. PLoS One 11:e0155341. doi: 10.1371/journal.pone.0155341
Wang, X. Y., Yu, H. Z., Xu, J. P., Zhang, S. Z., Yu, D., Liu, M. H., et al. (2017). Comparative subcellular proteomics analysis of susceptible and near-isogenic resistant Bombyx mori (Lepidoptera) larval midgut response to BmNPV infection. Sci. Rep. 7:45690. doi: 10.1038/srep45690
Yaffe, M. B., Farr, G. W., Miklos, D., Horwich, A. L., Sternlicht, M. L., and Sternlicht, H. (1992). TCP1 complex is a molecular chaperone in tubulin biogenesis. Nature 358, 245–248. doi: 10.1038/358245a0
Zhang, J. Y., Wu, X. P., Zan, J., Wu, Y. P., Ye, C. J., Ruan, X. Z., et al. (2013). Cellular chaperonin CCT gamma contributes to rabies virus replication during infection. J. Virol. 87, 7608–7621. doi: 10.1128/Jvi.03186-12
Keywords: Bombyx mori, BmNPV, chaperonin containing t-complex polypeptide 1β, protein folding, response mechanism
Citation: Wang X-y, Shao Z-m, Chen Q-y, Xu J-p, Sun X, Xu Z-p, Li M-w and Wu Y-c (2019) Knockdown of BmTCP-1β Delays BmNPV Infection in vitro. Front. Microbiol. 10:578. doi: 10.3389/fmicb.2019.00578
Received: 23 January 2019; Accepted: 06 March 2019;
Published: 22 March 2019.
Edited by:
Akio Adachi, Kansai Medical University, JapanReviewed by:
Zhaojun Wei, Hefei University of Technology, ChinaCopyright © 2019 Wang, Shao, Chen, Xu, Sun, Xu, Li and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mu-wang Li, bXdsaUBqdXN0LmVkdS5jbg== Yang-chun Wu, amtkd3ljQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.