
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 19 March 2019
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00544
This article is part of the Research Topic Antimicrobials and Anticancers of Bacterial Origins View all 11 articles
Pseudomonas sp. 11K1, originally isolated from rhizosphere, possesses inhibitory activity against plant pathogenic fungi and bacteria. Herein, the genome of strain 11K1 was sequenced and subjected to in silico, mutational, and functional analyses. The 11K1 genome is 6,704,877 bp in length, and genome mining identified three potential cyclic lipopeptide (CLP) biosynthetic clusters, subsequently named brasmycin, braspeptin, and brasamide. Insertional and deletion mutants displayed impaired brasmycin and braspeptin production, and lost antifungal activity, but retained antibacterial activity against Xanthomonas oryzae. The structures of these two active CLPs were predicted based on adenylation (A) domains. Brasmycin is composed of nine amino acids and belongs to the syringomycin class, while braspeptin is a 22 amino acid cyclic peptide belonging to the tolaasin group. Matrix-Assisted Laser Desorption/Ionization Time-of-Flight (MALDI-TOF) mass spectrometry analysis revealed that brasmycin and braspeptin have different molecular weights compared with known syringomycin and tolaasin members, respectively. Mutation of brasmycin and braspeptin gene clusters affected both biofilm formation and colony morphology. Collectively, these results indicate that Pseudomonas sp. 11K1 produces two novel CLPs that may help bacteria compete for nutrients and niches in the environment.
Pseudomonas spp. are ubiquitous in aquatic and terrestrial habitats, especially plant rhizospheres. Rhizosphere-derived pseudomonads have received much attention in recent decades because many are able to suppress plant diseases (Bender et al., 1999; Raaijmakers et al., 2002; Weller, 2007). Production of antimicrobial metabolites is one of the major mechanisms by which pseudomonads prevent microbial infection. Pseudomonas species exhibit extensive metabolic versatility and produce a remarkable spectrum of antimicrobial metabolites including cyclic lipopeptides (CLPs), 2,4-diacetylphloroglucinol (DAPG), phenazines (PHZs), pyrrolnitrin (PRN), pyoluteorin (PLT), 2,5-dialkylresorcinol, quinolones, gluconic acid, rhamnolipids, siderophores, and hydrogen cyanide (Gross and Loper, 2009; Masschelein et al., 2017).
Comparative genomics studies have revealed substantial diversity among different species of the Pseudomonas genus, and even among strains belonging to the same species (Loper et al., 2012). Many novel antimicrobial compounds have been discovered using genome analysis, including CLPs (de Bruijn et al., 2007; Gross and Loper, 2009; Nikolouli and Mossialos, 2012). CLPs are amphiphilic molecules composed of a cyclic oligopeptide lactone ring coupled to a fatty acid tail (Raaijmakers et al., 2010). CLPs are synthesized by non-ribosomal peptide synthases (NRPSs), large enzymes that are composed of a series of modules. Each module is a building block for the stepwise incorporation of an amino acid in the CLP peptide moiety and consists of an adenylation (A) domain, a thiolation (T) domain, and a condensation (C) domain. The A domain is responsible for amino acid selection and activation, the thiolation (T) domain catalyzes thioesterification of the activated amino acid, and C domain promotes peptide bond formation between two neighboring substrates to elongate the peptide chain. Based on their structural relationships, CLPs produced by Pseudomonas spp. are divided into at least six categories (Raaijmakers et al., 2006; Gross and Loper, 2009). Recently, Geudens and Martins (2018) further classified Pseudomonas spp. CLPs into 14 major groups, according to oligopeptide length and structure. Most CLPs produced by Pseudomonas species exhibit antifungal and antibacterial activity, and some possess antioomycete, antiviral, antiprotozoan, and antitumor activities (Masschelein et al., 2017).
Cyclic lipopeptides exert their antimicrobial activities by targeting either cell wall biosynthesis or cell membrane integrity. Friulimicin B from Actinoplanes friuliensis and the recently discovered CLP malacidin encoded in soil microbiomes inhibit cell wall biosynthesis (Schneider et al., 2009; Hover et al., 2018). Daptomycin produced by Streptomyces roseosporus and tridecaptin A1 produced by Bacillus and Paenibacillus species exert antimicrobial activity by disrupting membrane integrity (Straus and Hancock, 2006; Cochrane et al., 2016). Also Pseudomonas CLPs such as syringopeptins, cormycin, and massetolides exert their antibiosis activity by affecting membrane integrity. Overall, the molecular targets of most CLPs remain unknown (Masschelein et al., 2017).
In addition to antimicrobial activity, many CLPs produced by Pseudomonas spp. are involved in a wide range of other biological functions such as motility, biofilm formation, and virulence. Most CLPs produced by Pseudomonas spp. affect bacterial motility and biofilm formation as biosurfactants (Raaijmakers et al., 2010). Syringomycin and syringpeptin contribute to the virulence of P. syringae pv. syringae in cherry fruits (Scholz-Schroeder et al., 2001), and CLP sessilin hampers orfamide production in Pseudomonas sp. CMR12a (D’Aes et al., 2014).
We previously isolated Pseudomonas sp. 11K1 from Vicia faba rhizosphere in Yunnan province. This strain exhibits strong inhibitory activity against plant pathogens, including the fungal pathogen Botryosphaeria dothidea that causes canker of grapevines, and the bacterial pathogen Xanthomonas oryzae RS105 that causes bacterial blight of rice. In the present work, we sequenced the genome of strain 11K1, performed in silico and genetic analyses of putative secondary metabolite biosynthetic clusters, and identified two CLP clusters contributing to the antifungal activities displayed by this strain.
Bacterial strains and plasmids used in this study are listed in Supplementary Table S1. Pseudomonas sp. 11K1 and its mutants were grown in lysogenic broth (LB), AB medium (ABM), or amended King’s B medium (Proteose peptone No.3, 10 g; K2HPO4, 1.5 g; MgSO4 ⋅ 7H2O, 1.5 g; glucose, 20 g; KBG) at 28°C. Escherichia coli cells were grown at 37°C in LB medium. B. dothidea was cultured in PDA medium, and X. oryzae RS105 cells were grown at 28°C in LB or PDA medium. Where indicated, media was supplemented with ampicillin (50 μg/mL) or kanamycin (50 μg/mL) and/or 5-bromo-4-chloro-3-indolyl-D-galactopyranoside (X-Gal; 40 μg/mL).
Genomic DNA from 11K1 was prepared from a culture grown from a single colony using a DNA purification kit (Promega, Beijing, China) and sequenced on a PacBio RS II platform (Pacific Biosciences, Menlo Park, CA, United States) following the manufacturer’s instructions. De novo assembly of PacBio reads was performed using the Smartanalysis pipeline v2.3.0 in conjunction with the HGAP assembly protocol, and additional assembly was performed with minimus2. Open reading frames were identified and annotated using GeneMarkS, rRNAs were predicted by Barrnap and RNAmmer, and tmRNAs and tRNAs were predicted using ARAGORN and tRNAscan-SE, respectively. Other non-coding RNAs were predicted and classified by infernal conjunction with Rfam. The functions of genes were also annotated using the COG, KEGG, Pfam, TIGRFAMs, and SwissProt databases.
The genome of the 11K1 strain was analyzed using the Antibiotics and Secondary Metabolite Analysis Shell (antiSMASH) pipeline (Blin et al., 2017). ClusterFinder and Use ClusterFinder algorithms were employed for biosynthetic gene cluster (BGC) border prediction and analysis. Complete genome sequences of 11K1, four strains of P. brassicacearum, and 15 well-studied biocontrol agents belonging to other species of the Pseudomonas genus were also analyzed using Extra Features (KnownClusterBlast, SubClusterBlast, and ActiveSiteFinder).
Fragments of genes to be inactivated were PCR-amplified using primer pairs and PCR conditions listed in Supplementary Table S2. PCR products were cloned into the p2P24-Km suicide vector (Yan et al., 2017) to generate insertional mutant constructs, designated as p2P24-GC1, p2P24-GC4, p2P24-GC8, p2P24-GC26, p2P24-GC30, p2P24-GC28-1, and p2P24-28-2 (Supplementary Table S1). These were introduced individually into the E. coli DH5α donor strain then transferred into Pseudomonas sp. 11K1 by triparental mating (Thoma and Schobert, 2009). In brief, cells from 300 μL of overnight LB donor, helper DH5α (λπ) containing plasmid pRK600, and recipient cultures were washed twice with ddH2O and resuspended in 300 μL of ddH2O. The three suspensions were pooled (1:1:1) and cells were harvested by centrifugation. Bacterial pellets were resuspended in 100 μL of ddH2O and spotted onto an LB plate (2 μL per droplet). After incubation for 6 h at 28°C, bacterial cells were removed from the medium, resuspended in 1 mL of ddH2O, and 100 μL aliquots or serial dilutions were plated on ABM plates supplemented with kanamycin.
To further confirm the antimicrobial activities of lipopeptides, we constructed in-frame deletion mutants of gene clusters encoding brasmycin, braspeptin and brasamide, and combinations thereof (double and triple mutants). In-frame deletions were made using a two-step homologous recombination strategy. Briefly, ∼1 kb upstream and downstream fragments of brasmycin, braspeptin, and brasamide gene clusters were PCR-amplified from 11K1 genomic DNA using primers listed in Supplementary Table S2. PCR products were digested with restriction enzymes and cloned into the suicide vector p2P24-Km to generate plasmids p2P24-Δbam, p2P24-Δbap, and p2P24-Δbaa, which were individually introduced into strain 11K1 by triparental mating. Colonies appeared on LB plates containing kanamycin were picked up and inoculated in liquid LB medium without antibiotics. Overnight cultures were then plated on LB plates supplemented with 20% sucrose to generate the mutants Δbam, Δbap, and Δbaa, in which 38,432, 73,251, and 26,622 bp regions of the biosynthesis gene clusters were removed, respectively (Yan et al., 2017). All deletion mutants were screened and confirmed by PCR amplification (Supplementary Figure S1). The same method was used to generate double gene cluster deletion mutants ΔbamΔbap, ΔbamΔbaa, and ΔbapΔbaa, and the triple gene cluster deletion mutant ΔbamΔbapΔbaa (Supplementary Table S1).
The antiSMASH 4.0 program was used for cluster analysis of the A, T, and C domains of predicted NRPS genes. Sequences from predicted A domains in 11K1 and known A domains from CLPs in other pseudomonads were aligned using ClustalW. Phylogenetic trees were inferred by the neighbor joining method using MEGA5 with 1,000 bootstrap replicates. The peptide moieties of putative CLP signature sequences identified in the A domains were predicted by NRPSpredictor2 as described previously (Röttig et al., 2011).
Overnight liquid cultures of 11K1 in LB were spread onto PDA plates (100 μL). After incubation for 5 days at 28°C, agar was sliced into small pieces and extracted with water overnight at 28°C with shaking at 180 rpm in a 500 mL Erlenmeyer flask. The water extract was separated from agar by cheesecloth filtration and subsequently from cell debris by centrifugation (7000 g, 10 min) (Michelsen et al., 2015a,b). The supernatant pH was adjusted to pH 2 using 6 M HCl, and the sample was incubated overnight at 4°C before centrifugation (7000 g, 10 min). The pellet was collected and dried at 50°C, the crude peptide extract was resuspended in 1 mL of methanol (Biniarz et al., 2016), and analyzed by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS).
MALDI-TOF MS analysis was conducted on a Bruker UltrafleXtreme MALDI-TOF/TOF MS instrument (Ultraflextreme, Bruker, Germany) operated in reflection-positive ion mode with an acceleration voltage of 20 kV. The sample was mixed with matrix in a 1:1 (v/v) ratio. The matrix was α-cyano-4-hydroxycinnamic acid, which was dissolved in 50% acetonitrile containing 0.1% trifluoroacetic acid (TFA) (Monti et al., 2011). The nitrogen laser was set at a threshold adequate for signal generation that minimized fragmentation.
Swarming motility was studied by spotting 5 μL of an overnight cell suspension on KBG medium solidified with 0.6% agar, and evaluating surface swarming motility after incubation at 28°C for 24 h (Olorunleke et al., 2017). Biofilm formation was quantified by incubating 11K1 and its mutants for 3 days at 28°C in glass tubes, and staining with 0.1% crystal violet. Stained biofilms on the inner-surface of glass tubes were subsequently extracted with 95% ethanol and quantified by measuring the absorbance of the solution at 570 nm (Spiers et al., 2003).
The 11K1 complete genome consists of a 6,682,832 bp chromosome and one 22,045 bp plasmid, p11K1. A total of 5,785 CDSs were predicted, of which 5,749 CDSs are chromosomal and 36 are encoded on the plasmid. 84 RNA genes including rRNA, tRNA and tmRNA genes were identified. In addition, 81 miscellaneous RNA genes were identified. The general features of the strain 11K1 genome are summarized in Table 1. Based on 16S rDNA sequence analysis, strain 11K1 was identified as a species in the P. fluorescens group (Supplementary Figure S2; Hesse et al., 2018).
The genome of 11K1 was subjected to an automated search using antiSMASH (version 4.0). When the “Extra Features” settings were applied, 10 gene clusters were identified (Table 2), five of which are NRPS-type gene clusters predicted to be involved in the biosynthesis of lipopeptides, pyoverdine, and cupriachelin, four were predicted to synthesize a bacteriocin, a homoserine lactone (Hserlactone), a aryl polyene, and a lantipeptide, and one designated “Other” accounts for biosynthesis of mangotoxin (Table 2). An “Extended” search found an additional 33 gene clusters (Supplementary Table S3), 24 of which had no predicted products or functions. Other clusters showed only low gene similarities (<40%) with known biosynthetic clusters except for gene cluster 39, which shares 80% gene similarities to the alginate BGC (Supplementary Table S3).
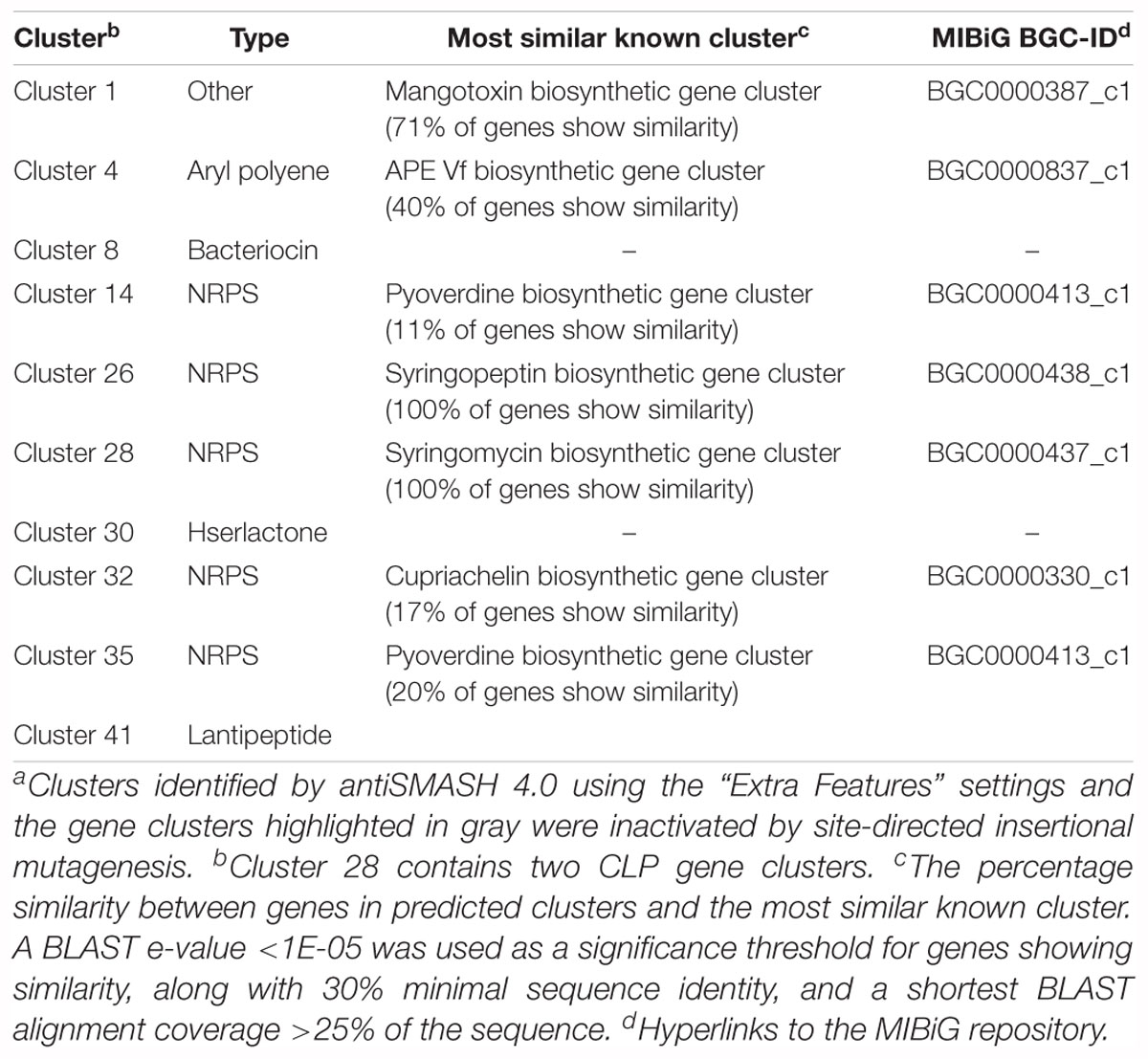
Table 2. Secondary metabolite and antibiotic gene clusters in Pseudomonas sp. 11K1 predicted by antiSMASH 4.0a.
We compared the secondary compound profile as predicted for strain 11K1 with 19 other Pseudomonas strains, and siderophore production is the only common feature present in all strains (Figure 1). Other antimicrobial compounds (e.g., CLPs, DAPG, PLT, and PRN) are present in conserved genomic locations of some closely related strains (Figure 1). Group III Pseudomonas spp. including strain 11K1 all produce CLPs but not DAPG, PLT, or PRN (Figure 1). Pseudomonas strains from Group II only produce DAPG but not PLT, PRN, or CLPs (Figure 1). Group I, comprising five P. protegens strains, produces all four types of antimicrobial compounds (DAPG, PLT, PRN, and CLPs; Figure 1). These results indicate that different Pseudomonas strains, even those belonging to the same species, may produce different secondary compounds. Closely related strains share similar metabolite profiles, and strain 11K1 may produce multiple CLPs with similar chemical structures to the compounds produced by other members of Group III.
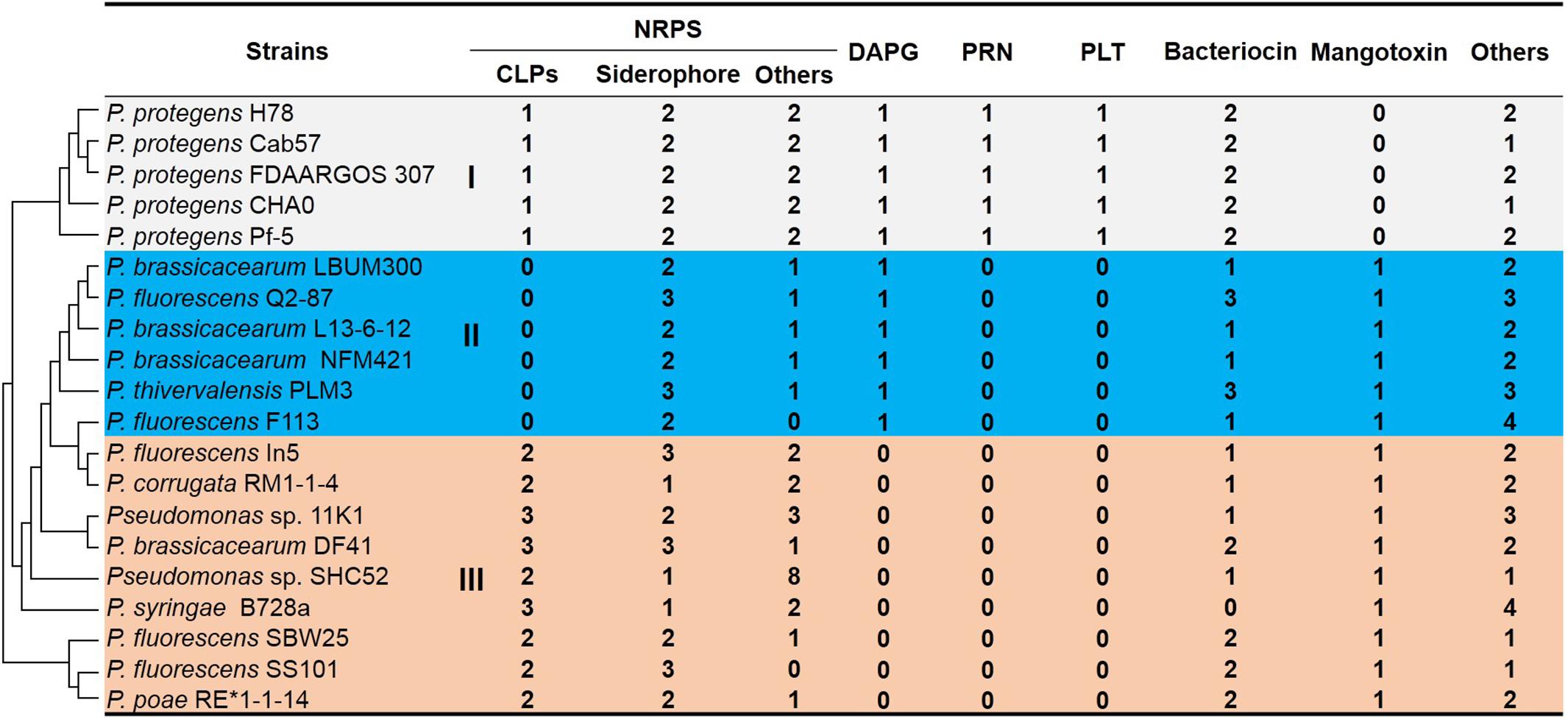
Figure 1. In silico analysis and comparison of secondary metabolite production in Pseudomonas sp. 11K1 and strains of related species. Clusters accounting for biosynthesis of secondary metabolites were predicted by antiSMASH 4.0 using the “Extra Features” settings. Numbers represent the number of gene clusters within a genome. Abbreviations are as follows: CLPs, cyclic lipopeptides; DAPG, 2,4-diacetylphloroglucinol; PRN, pyrrolnitrin; PLT, pyoluteorin; NRPS, non-ribosomal peptide synthase. Pseudomonas strains were divided into groups I (gray), II (blue), and III (orange) based on profiles of secondary metabolites. Group I species produce all four antimicrobial compounds DAPG, PLT, PRN, and CLPs; Pseudomonas strains of Group II produce DAPG but not PLT, PRN, or CLPs; Pseudomonas strains of group III, including strain 11K1, produce CLPs but not DAPG, PLT, or PRN. The phylogenetic tree of 16S rDNA sequences (left) was constructed using the neighbor-joining method in MEGA version 5.0 after multiple sequence alignment by ClustalW.
Based on the results of the in silico analysis, we performed site-directed insertional mutagenesis of seven gene clusters (Table 2), to find out whether one or more of these clusters contribute to the antimicrobial activity of strain 11K1. Inactivated clusters included NRPS-type clusters 26 and 28. The latter cluster is predicted to produce two CLPs. The two CLPs synthetic gene clusters in cluster 28 are located next to each other, but their synthetic genes transcribe in opposite direction (Figure 2). The growth of B. dothidea was not affected by any of the site-directed insertional mutants except mutants 11K1-GC28-1::Km and 11K1-GC28-2::Km. These mutants were both hit in cluster 28. The former mutant displayed a reduced inhibition phenotype whereas the latter completely failed to inhibit B. dothidea. These results indicate that the two CLPs encoded by gene cluster 28 are potential antifungal compounds. The first of the CLP gene clusters in cluster 28 was predicted to encode a 22 amino acid lipopeptide, and the second was predicted to encode a 9 amino acid lipopeptide. Gene cluster 28 is similar to the syringomycin/syringopeptin BGC in P. syringae pv. syringae B301D, and the nunamycin/nunapeptin BGC in P. fluorescens In5 (Supplementary Figure S3A). The third CLP, encoded by gene cluster 26, was predicted to encode an 8 amino acid lipopeptide, and this cluster shares similarity with the syringopeptin gene cluster in P. syringae B301D and the cichopeptin gene cluster in P. cichorii SF1-54 (Scholz-Schroeder et al., 2001; Huang et al., 2015; Supplementary Figure S3B). We designated the predicted 22 amino acid CLP as braspeptin (bap), the 9 amino acid compound as brasmycin (bam), and the 8 amino acid peptide as brasamide (baa; Figure 2). All mutants retained their inhibitory activity against X. oryzae RS105, indicating that other compounds, but not CLPs, were responsible for inhibition of this bacterium.

Figure 2. The CLP gene clusters of Pseudomonas sp. 11K1. CLPs are synthesized by modular non-ribosomal peptide synthases (NRPSs), and each module contains an adenylation (A), condensation (C), and thiolation (T) domain. Red arrows indicate biosynthetic genes, blue arrows indicate transport-related genes, green arrows indicate regulatory genes, the yellow arrow indicates a predicted transposase, and other genes are indicated by black arrows.
To further evaluate the role of CLPs in antibiosis activity, we constructed deletion mutants of single gene clusters Δbam, Δbap, and Δbaa, double gene clusters ΔbamΔbap, ΔbamΔbaa, and ΔbapΔbaa, and the triple gene cluster ΔbamΔbapΔbaa. Mutants Δbam, ΔbamΔbap, ΔbamΔbaa, and ΔbamΔbapΔbaa failed to inhibit B. dothidea growth, whereas mutant Δbaa retained full antifungal activity comparable with that of the WT 11K1 strain. Mutants Δbap and ΔbapΔbaa displayed a reduced inhibition phenotype compared with the Δbaa and WT strains (Figure 3A). These results confirm the results of the insertional mutation analysis, and indicate that brasmycin is the major antifungal compound active against B. dothidea. In addition, all CLP deletion mutants retained antibacterial activity against X. oryzae RS105 (Figure 3B), consistent with the above results obtained using insertional mutants, further confirming that CLPs were not the antibacterial compounds active against X. oryzae RS105.
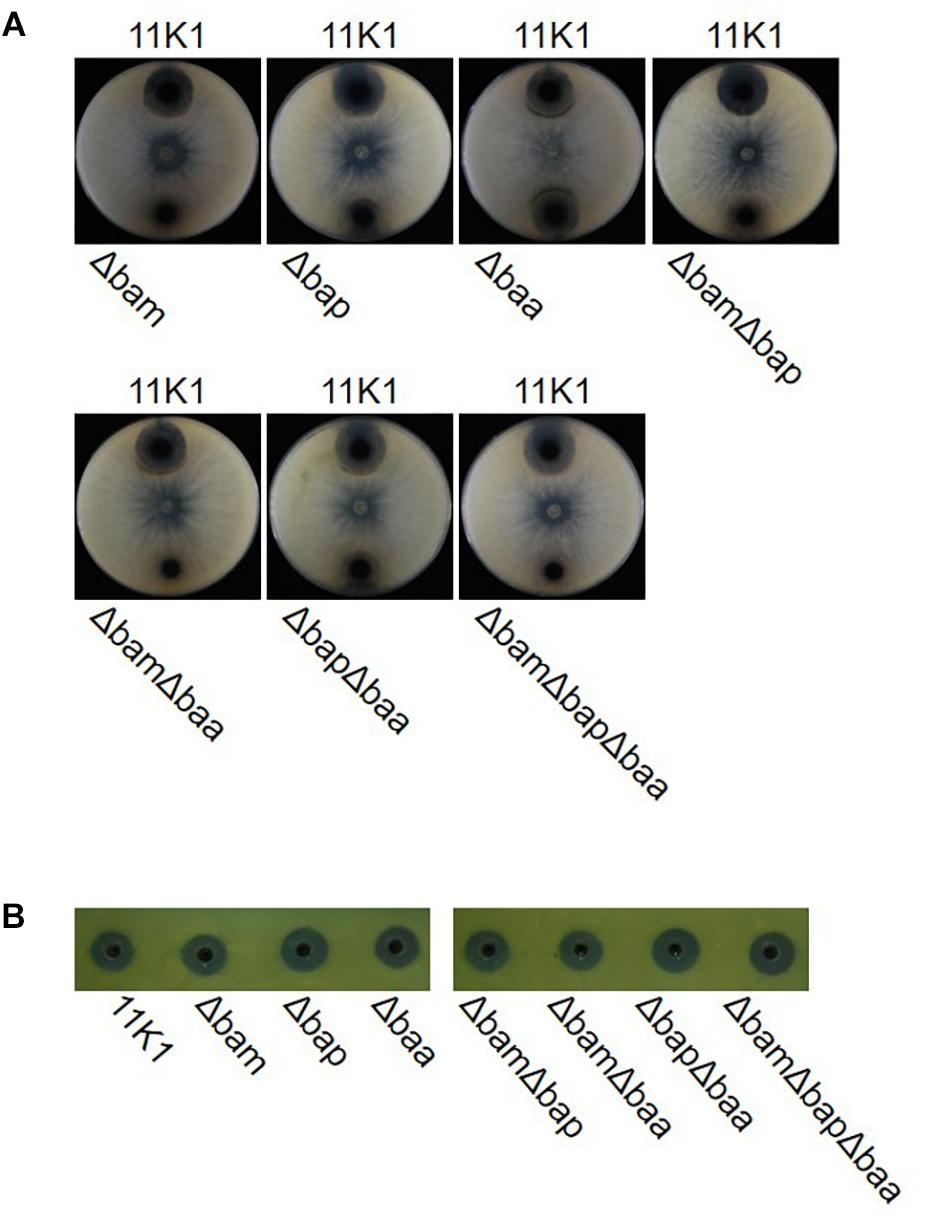
Figure 3. (A) Antimicrobial activity of Pseudomonas sp. 11K1 and its mutants against Botryosphaeria dothidea, and (B) Xanthomonas oryzae RS105. B. dothidea disks (5 mm diameter) were placed in the center of each plate and 5 μL of an overnight culture of wild-type (WT) 11K1 or its mutants were inoculated near the edge of PDA medium plates. For antibacterial assays, 50 μL of sterile fermentation liquor was used to detect bacterial inhibition. Δbam, brasmycin gene cluster deletion mutant; Δbap, braspeptin gene cluster deletion mutant; Δbaa, brasamide gene cluster deletion mutant; ΔbamΔbap, brasmycin and braspeptin double gene cluster deletion mutant; ΔbamΔbaa, brasmycin and brasamide double gene cluster deletion mutant; ΔbapΔbaa, braspeptin and brasamide double gene cluster deletion mutant; ΔbamΔbapΔbaa, brasmycin, braspeptin, and brasamide triple gene cluster deletion mutant.
Bioinformatic analysis revealed that the predicted brasmycin BGC comprises nine modules, each of which consists of an adenylation (A), a thiolation (T), and a condensation (C) domain. Similar to nunamycin and syringomycin BGCs, the last module lacks the A domain, encoded by the first gene (Figure 2; Raaijmakers et al., 2006). The amino acid composition of CLPs could be predicated in silico based on A domain selectivity (Challis et al., 2000), and various A domain specificity features have been empirically determined (Röttig et al., 2011). We predicted the amino acid sequence of brasmycin based on a phylogenetic approach in which all A domains were compared with those of functionally characterized lipopeptides. The predicted brasmycin amino acid sequence was Ser-Orn-Asp-Nrp-His-Thr-Thr-Asp-Thr (Figure 4A and Supplementary Figure S4), which is most similar to thanamycin, although the fourth amino acid was uncertain. The predicted brasmycin sequence differs from nunamycin at amino acid 2 (Orn versus Dab), 3 (Asp versus Gly), and 5 (His versus Dab) and from syringomycin at amino acid 2 (Orn versus Ser), 3 (Asp versus Dab), 5 (His versus Arg), 6 (Thr versus Phe), and 7 (Thr versus Dhb) (Figure 4A), suggesting that brasmycin may represent a new member of the syringomycin group (Gross and Loper, 2009). These results are in agreement with a previous report showing that amino acids 2–7 of syringomycin group members are the most variable (Michelsen et al., 2015b).

Figure 4. Comparison of brasmycin (A) and braspeptin (B) gene clusters and the predicted amino acid sequences with those of similar CLPs. Red arrows indicate biosynthesis genes, while other genes are indicated by black arrows. Amino acid sequences of brasmycin and braspeptin (in red) were predicated based on A domain phylogenetic analysis. Cormycin and corpeptin from P. corrugata CFBP5454 (ATKI00000000), nunamycin and nunapeptin from P. fluorescens In5 (LIRD01000000), thanamycin and thanapeptin from Pseudomonas sp. SHC52 (CBLV000000000), syringomycin and syringopeptin from P. syringae pv. syringae B301D (CP005969). Consensus amino acids are denoted in boxes. Non-standard amino acids are abbreviated as follows: Dab, 2,4-diaminobutyric acid; Dhb, dehydrobutyrine; Dha, dehydroalanine; Orn, ornithine; Hse, homoserine, Nrp, uncertain amino acids.
The predicted amino acid sequence of braspeptin based on A domain phylogenetic analysis suggested that it belonged to the tolaasin group, but with differences from known members (Figure 4B and Supplementary Figure S5). Braspeptin shares conserved sites at amino acid 1 (Dhb), 2 (Pro), 5 (Ala), 13 (Ala), 19 (Ala), and 20 (Dab) with corpeptin, nunapeptin, thanapeptin, and syringopeptin, but differs from corpeptin at amino acid 4 (Ile versus Ala), 7 (Ile versus Val), 11 (Ser versus Ile), 12 (Ser versus Dhp), 15 (Dab versus Ala), and 22 (Val versus Ile); it differs from nunapeptin at amino acid 4 (Ile versus Ala), 7 (Ile versus Ala), 9 (Hse versus Thr), 11 (Ser versus Ile), 12 (Ser versus Dhp), 15 (Dab versus Ala), and 22 (Val versus Ilx); it differs from thanapeptin at amino acid 4 (Ile versus Ala), 7 (Ile versus Val), 11 (Ser versus Ile), 12 (Ser versus Dhb), 15 (Dab versus Ala), and 22 (Val versus Ile); it differs from syringpeptin at amino acid 3 (Ala versus Val), 4 (Ile versus Val), 6 (Val versus Ala), 7 (Ile versus Val), 8 (Dhb versus Val), 9 (Hse versus Dhb), 10 (Val versus Ala), 11 (Ser versus Val), 12 (Ser versus Ala), 14 (Ala versus Dhb), 15 (Dab versus Thr), 16 (Val versus Ser), 18 (Thr versus Dhb), 21 (Ser versus Dab), and 22 (Val versus Tyr). Thus, although amino acid 17 remains uncertain (Figure 4B), braspeptin can be considered a new member of the tolaasin group.
Lipopeptides from Pseudomonas sp. 11K1 contributing to its antibiosis activity were extracted, and analyzed using a Bruker UltrafleXtreme MALDI-TOF/TOF MS instrument to determine their molecular masses. In WT 11K1, two peaks were obtained at m/z 1,268.706 and 2,175.493 (Figure 5A), whereas only the m/z 2,175.493 peak was detected for the brasmycin gene cluster deletion mutant Δbam (Figure 5B), and only m/z 1,268.706 was detected in braspeptin gene cluster deletion mutant Δbap (Figure 5C). These results showed that the m/z 1,268.706 ion corresponds to the brasmycin lipopeptide, and the m/z 2,175.493 ion corresponds to the braspeptin lipopeptide. Molecular weights differ between brasmycin (m/z 1,268.706) and known syringomycin members nunamycin (m/z 1,138) (Michelsen et al., 2015a), thanamycin (m/z 1,291) (Van Der Voort et al., 2015), syringomycins (m/z 1,225, m/z 1,253) (Monti et al., 2011), and cormycin A (m/z 1274) (Scaloni et al., 2004). The molecular weight of braspeptin (m/z 2,175.493) is different from nunapeptins (m/z 2,023.22, m/z 2,037.24, and m/z 2,075.22) (Michelsen et al., 2015a), thanamycin (m/z 2,120.9) (Van Der Voort et al., 2015), syringopeptins (m/z 2,400, m/z 2,428) (Monti et al., 2011), corpeptins (m/z 2,095, m/z 2,121.2) (Licciardello et al., 2012; Strano et al., 2015), and sclerosis (m/z 2,095.3, m/z 2,123.3, m/z 2,145.3) (Berry et al., 2012). These results further confirm that brasmycin and braspeptin represent new members of the syringomycin and tolaasin lipopeptide groups, respectively.
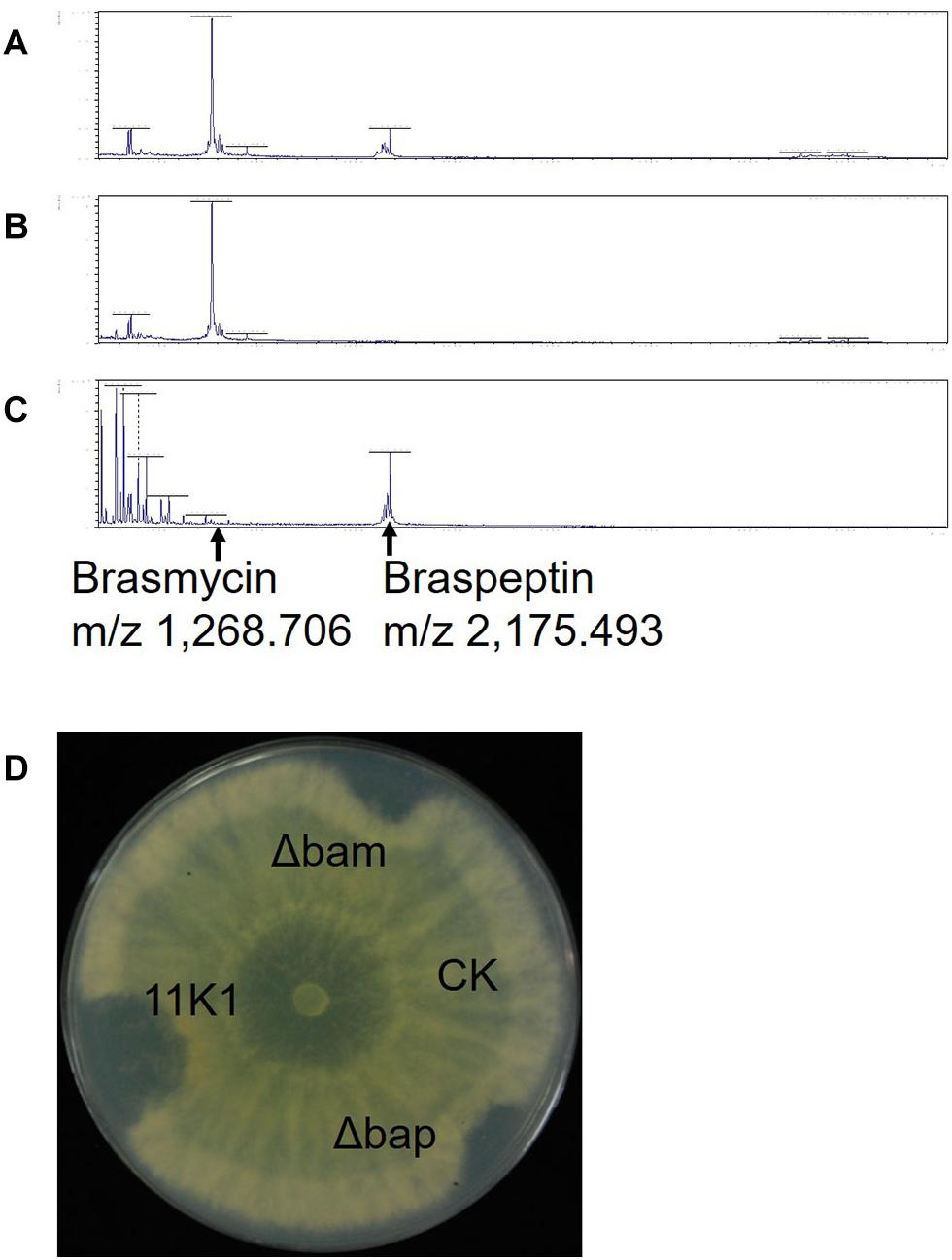
Figure 5. MALDI-TOF MS analysis of CLPs extracted from (A) Pseudomonas sp. 11K1, (B) braspeptin gene cluster mutant Δbap, and (C) brasmycin gene cluster mutant Δbam. (D) Antifungal analysis of crude CLP extracts. Inhibition of B. dothidea mycelial growth by crude peptide extracts derived from Pseudomonas sp. 11K1 and its mutants was assessed. Extracts (20 μL) from 11K1, braspeptin gene cluster deletion mutant Δbap, and brasmycin gene cluster deletion mutant Δbam were spotted on agar plates. Methyl alcohol was applied as a control (CK).
Although it is currently impossible to accurately predict the fatty acid structures of brasmycin and braspeptin, we made a rough prediction based on molecular weights and predicted oligopeptide chains, in which uncertain amino acids were replaced with the most likely amino acids. The brasmycin might have a 12–15 carbon fatty acid tail (Supplementary Figure S6), and the fatty acid tail of braspeptin might contain 10–12 carbon atoms (Supplementary Figure S7).
Corroborating the results from dual-culture experiments (Figure 3A), crude peptides extracted from Pseudomonas sp. 11K1 containing both brasmycin and braspeptin, brasmycin extracted from mutant Δbap, and braspeptin extracted from mutant Δbam all inhibited mycelial growth of B. dothidea (Figure 5D).
Pseudomonas sp. 11K1 was able to swarm on soft KBG plates containing 0.6% agar. All three single lipopeptide gene cluster mutants Δbam, Δbap, and Δbaa exhibited reduced swarming motility to some degree, indicating that all three lipopeptides contribute to swarming motility (Supplementary Figure S8). However, double mutants ΔbamΔbap, ΔbamΔbaa, and ΔbapΔbaa, and triple mutant ΔbamΔbapΔbaa, did not further impair swarming motility (Supplementary Figure S8), indicating that lipopeptides were not the only surfactants contributing to swarming motility; other unknown surfactants were presumably produced under these conditions.
Biofilm formation was tested in glass test tubes containing KGB liquid medium. Strain 11K1 formed a biofilm on the inner wall of tubes at the interface between air and liquid. The brasmycin gene cluster mutant Δbam formed markedly less biofilm, the braspeptin gene cluster mutant Δbap produced more biofilm than the WT 11K1 strain, and biofilm formation by mutant Δbaa was comparable to that of 11K1 (Figure 6A). Double and triple gene cluster mutants ΔbamΔbap, ΔbamΔbaa, and ΔbamΔbapΔbaa formed less biofilm, similar to brasmycin mutant Δbam, indicating that brasmycin was the most important lipopeptide involved in biofilm formation.
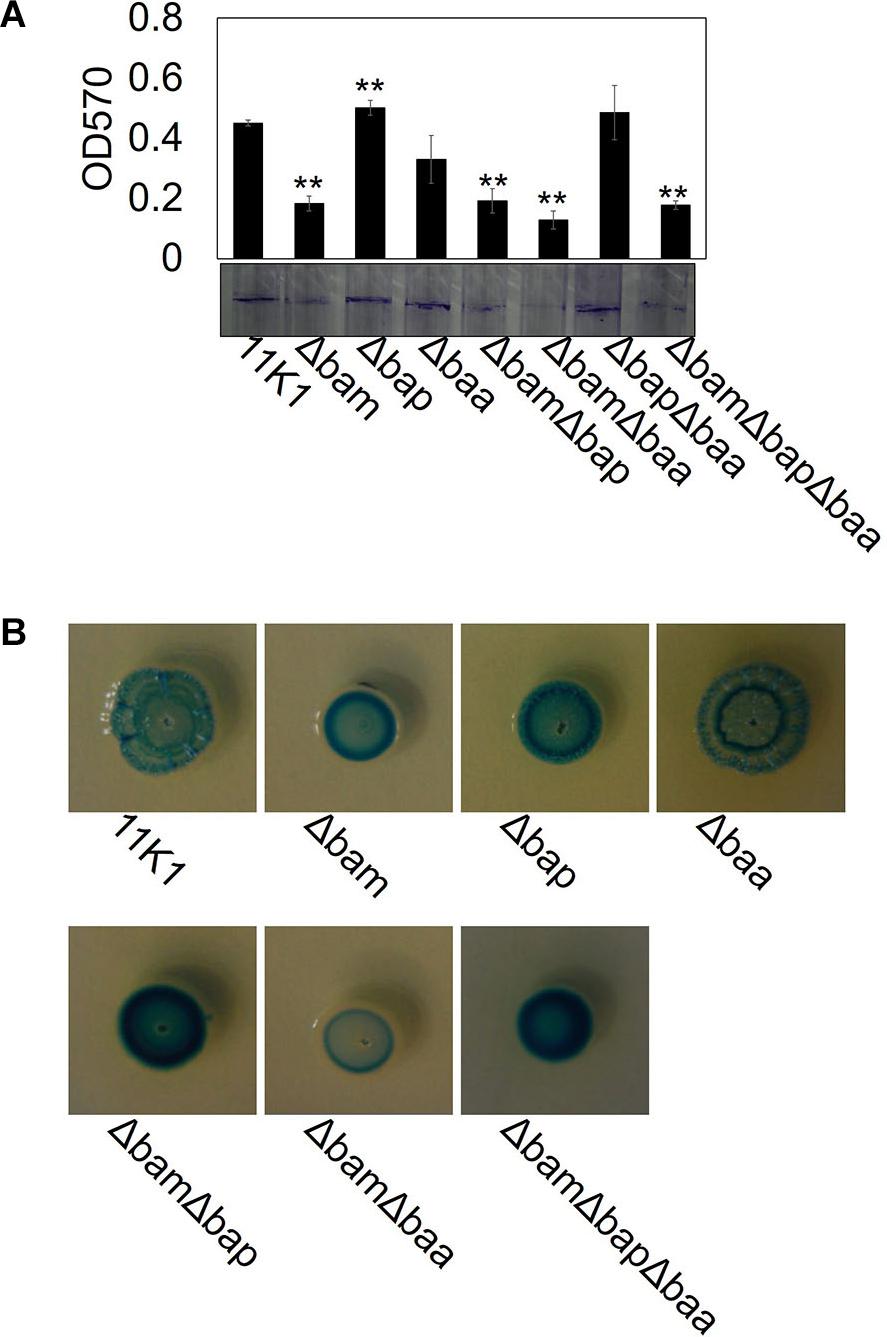
Figure 6. Effects of CLP gene mutations on bacterial biofilm formation (A) and colony morphology (B). (A) Spectrophotometric quantification of biofilms formed by Pseudomonas sp. 11K1 and CLP mutants. Mean values of three replicates are given, and error bars indicate standard error. Visual representation of biofilm formation by Pseudomonas sp. 11K1 and CLP mutants on tubes is shown. Biofilms were stained with Crystal Violet. (B) Colony morphology detected on PDA plates. A 5 μL sample of overnight culture of WT 11K1 or mutant strains was spot-inoculated in the center of a PDA plate and incubated for 48 h at 28°C. Δbam, brasmycin gene cluster deletion mutant; Δbap, braspeptin gene cluster deletion mutant; Δbaa, brasamide gene cluster deletion mutant; ΔbamΔbap, brasmycin and braspeptin double gene cluster deletion mutant; ΔbamΔbaa, brasmycin and brasamide double gene cluster deletion mutant; ΔbapΔbaa, braspeptin and brasamide double gene cluster deletion mutant; ΔbamΔbapΔbaa, brasmycin, braspeptin, and brasamide triple gene cluster deletion mutant.
Mutation of lipopeptide biosynthesis gene clusters caused morphological changes of bacterial colonies on PDA plates. The WT 11K1 strain produced round colonies with a wrinkled surface, whereas all mutants containing brasmycin gene cluster mutations (Δbam, ΔbamΔbap, ΔbamΔbaa, and ΔbamΔbapΔbaa) formed smaller colonies with a smooth surface (Figure 6B). Mutant Δbaa formed typical WT colonies, whereas mutants Δbap and ΔbapΔbaa produced smaller colonies with a less wrinkled surface than WT colonies (Figure 6B).
Genome comparison of twenty Pseudomonas strains highlighted the tremendous diversity of gene sequences encoding secondary metabolites (Figure 1), suggesting strains from various environments encounter selective pressure, resulting in the acquisition or loss of gene clusters during evolution. The 20 Pseudomonas strains were clustered into three distinct groups according to secondary metabolite profiles. Compared to the vertical inheritance that is the process of genetic material from the parent to the offspring, horizontal genes transfer occurs between different species (Eisen, 2000). Group III is composed of different Pseudomonas species including P. fluorescens, P. brassicacearum and strain 11K1, all producing CLPs but not DAPG, PLT, or PRN. In contrast, P. fluorescens and P. brassicacearum in group II do not produce CLPs (Figure 1), indicating the horizontal transfer possibility of CLP biosynthesis gene clusters among Pseudomonas species. In addition, we found one transposase gene flanking the brasmycin and braspeptin synthetic gene cluster, which further indicates CLP gene clusters can be horizontally acquired (Figure 2).
The two active CLPs produced by strain 11K1 (brasmycin and braspeptin) belong to the syringomycin and tolaasin groups, respectively. CLPs in the syringomycin group consist of nine amino acids, including unusual amino acids (Figure 4A). All syringomycin group members possess antifungal ability (Sinden et al., 1971; Michelsen et al., 2015b; Trantas et al., 2015; Van Der Voort et al., 2015), and syringomycin is involved in the full virulence of P. syringae pv. syringae B301D, since syringomycin (syrB1) mutant was significantly reduced in virulence compared with its parental strain (Scholz-Schroeder et al., 2001). Compared with the syringomycin group, CLPs in the tolaasin group are much more diverse, both in the composition of the peptide chain, and in antimicrobial activities (Figure 4B). Syringopeptin and corpeptin display antifungal activity, but not antioomycete activity (Lavermicocca et al., 1997; Strano et al., 2015). By contrast, antifungal activity has not been observed for nunapeptin or thanapeptin, but these compounds inhibit oomycete growth (Michelsen et al., 2015b; Van Der Voort et al., 2015). Braspeptin, produced by 11K1, has antifungal activity, but no antioomycete activity (data not shown). Exactly why CLPs with similar composition exhibit different antimicrobial activities remains unclear. A possible explanation is that small structural changes in the peptide or lipid tail may affect their physicochemical properties and interactions with different types of cellular membranes (Kuiper et al., 2004; Raaijmakers et al., 2010).
Biosurfactants such as CLPs, rhamnolipids, and lipopolysaccharides are essential for bacterial swarming motility (Raaijmakers et al., 2006). Under suitable conditions, bacterial cells differentiate into hyperflagellated swarmers that are more motile on wet and viscous surfaces (Harshey, 2003). Biosurfactants produced by bacteria can change the viscosity of surfaces, thereby influencing cell differentiation and motility (McCarter and Silverman, 1990; Allison et al., 1993). Several studies showed that biosurfactants produced by pseudomonads and other bacteria can promote their surface motility. For example, orfamide and massetolide A are required as surfactants for the swarming ability of Pseudomonas strains CMR12a and SBW25, respectively, because swarming was completely abolished in CLP biosynthesis mutants (de Bruijn et al., 2007; D’Aes et al., 2014). Similarly, swarming of P. aeruginosa 57RP requires alkanoic acids that act as surfactants (Deziel et al., 2003). Salmonella enterica SJW1103 lipopolysaccharide biosynthesis gene mutants lose swarming motility (Toguchi et al., 2000). Our present study shows that the single lipopeptide gene cluster mutants Δbam, Δbap, and Δbaa in 11K1 display partly impaired swarming motility, indicating that all three lipopeptides contribute to swarming motility (Supplementary Figure S8). However, double mutants ΔbamΔbap, ΔbamΔbaa, and ΔbapΔbaa, and triple mutant ΔbamΔbapΔbaa, did not further impair swarming motility (Supplementary Figure S8), indicating that other biosurfactants contribute to the full swarming phenotype (Supplementary Table S3).
Cyclic lipopeptides play an important role in biofilm formation, although this can differ between CLP types. For example, a massetolide A-deficient mutant of P. fluorescens SBW25 forms less biofilm (de Bruijn et al., 2007), whereas a putisolvin-deficient mutant of P. putida PCL1445 displays enhanced biofilm formation (Kuiper et al., 2004). Orfamide has no effect on biofilm formation in P. fluorescens Pf-5 (Gross et al., 2007). In strain 11K1, the brasmycin gene cluster mutant Δbam formed markedly less biofilm, whereas the braspeptin gene cluster mutant Δbap produced more biofilm than the WT 11K1 strain (Figure 6A). The mechanisms by which CLPs affect biofilm formation remain unclear, but because CLPs play an important role in cell surface hydrophobicity, CLPs with different structures might differ in hydrophobicity, and their roles in biofilm formation could be entirely different (de Bruijn et al., 2008; Raaijmakers et al., 2010).
A tight correlation between wrinkled colony morphology and increased biofilm formation has been reported for various bacteria including P. aeruginosa PA14, Burkholderia cenocepacia H111, and P. fluorescens SBW25 (Spiers et al., 2003; Friedman and Kolter, 2004; Fazli et al., 2013), but there are exceptions. B. subtilis NCIB 3610 produces robust wrinkled colonies, but displays poor biofilm formation on glass surfaces, whereas B. subtilis JH642 readily forms biofilms on glass surfaces, but does not produce robust wrinkled colonies (Vlamakis et al., 2013). In the present work, both Δbam and Δbap mutants produce smooth surface colonies, but only Δbam forms less biofilm than the WT 11K1 strain, which produces wrinkled colonies (Figure 6B). Both biofilm formation and wrinkled colony morphology require exopolysaccharides, but the exopolysaccharides involved are not always the same (Fazli et al., 2014). Although acetylation of cellulose is essential for wrinkled colonies, it is not required for biofilm formation on tubes by P. fluorescens SBW25 (Spiers et al., 2003). Thus, the molecular relationship between biofilm formation and wrinkled colony morphology remains unclear.
The gene cluster of brasamide shares high similarity with some predicted CLP gene clusters in strains of the P. fluorescens group (Supplementary Figure S9), but the potential CLPs they encode have not yet been investigated. The predicted brasamide and its homologs contain eight amino acids with identical sequence, which are distinctly different from two known eight amino acid CLPs bananamide and pseudofactin (Supplementary Figure S9B; Janek et al., 2010; Nguyen et al., 2016). We cannot assign brasamide to any existing group because its chemical structure has not been experimentally resolved, but the distinctly different composition of predicted amino acids suggests that brasamide may belong to a new group of lipopeptides. Except for its contribution to bacterial swarming (Supplementary Figure S6), few information is available about its biological function. However, conserved distribution of brasamide-like CLPs in closely related pseudomonad strains strongly suggest that these CLPs fulfill an ecological function, which will be subject of future study.
This complete genome project has been deposited at GenBank under the accession numbers CP035088 (chromosome sequence) and CP035089 (plasmid sequence).
HZ and L-QZ designed the research. L-QZ supervised the study. Y-PL contributed to the MALDI-TOF MS. HZ performed the other experiments, analyzed the data, and wrote the manuscript together with L-QZ. All authors revised the manuscript and approved the final version for submission.
This work was supported by grants from Special Fund for Agro-scientific Research in the Public Interest (201503109), National Key Research and Development Program (2017YFD0201106), National Technology System for Grape Industry (CARS-29), and the National Natural Science Foundation of China (31572045 and 31872020).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00544/full#supplementary-material
Allison, C., Lai, H. C., Gygi, D., and Hughes, C. (1993). Cell differentiation of Proteus mirabilis is initiated by glutamine, a specific chemoattractant for swarming cells. Mol. Microbiol. 8, 53–60. doi: 10.1111/j.1365-2958.1993.tb01202.x
Bender, C. L., Rangaswamy, V., and Loper, J. (1999). Polyketide production by plant-associated Pseudomonads. Annu. Rev. Phytopathol. 37, 175–196. doi: 10.1146/annurev.phyto.37.1.175
Berry, C. L., Brassinga, A. K. C., Donald, L. J., Fernando, W. G. D., Loewen, P. C., and de Kievi, T. R. (2012). Chemical and biological characterization of sclerosin, an antifungal lipopeptide. Can. J. Microbiol. 58, 1027–1034. doi: 10.1139/w2012-079
Biniarz, P., Łukaszewicz, M., and Janek, T. (2016). Screening concepts, characterization and structural analysis of microbial-derived bioactive lipopeptides: a review. Crit. Rev. Biotechnol. 37, 393–410. doi: 10.3109/07388551.2016.1163324
Blin, K., Wolf, T., Chevrette, M. G., Lu, X., Schwalen, C. J., and Kautsar, S. A. (2017). AntiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 45, 36–41. doi: 10.1093/nar/gkx319
Challis, G. L., Ravel, J., and Townsend, C. A. (2000). Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7, 211–224. doi: 10.1016/S1074-5521(00)00091-0
Cochrane, S. A., Findlay, B., Bakhtiary, A., Acedo, J., Rodriguez-Lopez, E. M., Mercier, P., et al. (2016). Antimicrobial lipopeptide tridecaptin A1 selectively binds to Gram-negative lipid II. Proc. Natl Acad. Sci. 113, 11561–11566. doi: 10.1073/pnas.1608623113
D’Aes, J., Kieu, N. P., Léclère, V., Tokarski, C., Olorunleke, F. E., De Maeyer, K., et al. (2014). To settle or to move? The interplay between two classes of cyclic lipopeptides in the biocontrol strain Pseudomonas CMR12a. Environ. Microbiol. 16, 2282–2300. doi: 10.1111/1462-2920.12462
de Bruijn, I., de Kock, M., Yang, M., de Waard, P., van Beek, A., and Raaijmakers, J. M. (2007). Genome-based discovery, structure prediction and functional analysis of cyclic lipopeptide antibiotics in Pseudomonas species. Mol. Microbiol. 63, 417–428. doi: 10.1111/j.1365-2958.2006.05525.x
de Bruijn, I., de Kock, M. J., Waard, P., van Beek, T. A., and Raaijmakers, J. M. (2008). Massetolide A biosynthesis in Pseudomonas fluorescens. J. Bacteriol. 190, 2777–2789. doi: 10.1128/JB.01563-07
Deziel, E., Lepine, F., Milot, S., and Villemur, R. (2003). rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy) alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 149, 2005–2013. doi: 10.1099/mic.0.26154-0
Eisen, J. A. (2000). Horizontal gene transfer among microbial genomes: new insights from complete genome analysis. Curr. Opin. Gent. Dev. 10, 606–611. doi: 10.1016/S0959-437X(00)00143-X
Fazli, M., Almblad, H., Rybtke, M. L., Givskov, M., Eberl, L., and Tolker-Nielsen, T. (2014). Regulation of biofilm formation in Pseudomonas and Burkholderia species. Environ. Microbiol. 16, 1961–1981. doi: 10.1111/1462-2920.12448
Fazli, M., McCarthy, Y., Givskov, M., Ryan, R., and Tolker-Nielsen, T. (2013). The exopolysaccharide gene cluster Bcam1330–Bcam1341 is involved in Burkholderia cenocepacia biofilm formation, and its expression is regulated by c-di-GMP and Bcam1349. Microbiologyopen 2, 105–122. doi: 10.1002/mbo3.61
Friedman, L., and Kolter, R. (2004). Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51, 675–690. doi: 10.1046/j.1365-2958.2003.03877.x
Geudens, N., and Martins, J. C. (2018). Cyclic lipodepsipeptides from Pseudomonas spp. - biological swiss-army knives. Front. Microbiol. 9:1867. doi: 10.3389/fmicb.2018.01867
Gross, H., and Loper, J. E. (2009). Genomics of secondary metabolite production by Pseudomonas spp. Nat. Prod. Rep. 26, 1408–1446. doi: 10.1039/b817075b
Gross, H., Stockwell, V. O., Henkels, M. D., Nowak-Thompson, B., Loper, J. E., and Gerwick, W. H. (2007). The genomisotopic approach: a systematic method to isolate products of orphan biosynthetic gene clusters. Chem. Biol. 14, 53–63. doi: 10.1016/j.chembiol.2006.11.007
Harshey, R. M. (2003). Bacterial motility on a surface: many ways to a common goal. Annu. Rev. Microbiol. 57, 249–273. doi: 10.1146/annurev.micro.57.030502.091014
Hesse, C., Schulz, F., Bull, C. T., Elbourne, L. D. H., Yan, Q., Shapiro, N., et al. (2018). Genome-based evolutionary history of Pseudomonas spp. Environ microbial. 6, 2142–2159. doi: 10.1111/1462-2920.14130
Hover, B., Kim, S. H., Katz, M., Charlop-Powers, Z., Owen, J., Melinda, A., et al. (2018). Culture-independent discovery of the malacidins as calcium-dependent antibiotics with activity against multidrug-resistant Gram-positive pathogens. Nat. Microbiol. 3, 415–422. doi: 10.1038/s41564-018-0110-1
Huang, C., Pauwelyn, E., Ongena, M., Debois, D., Leclere, V., Jacques, P., et al. (2015). Characterization of Cichopeptins, new phytotoxic cyclic lipodepsipeptides produced by Pseudomonas cichorii SF1-54, and their role in bacterial midrib rot disease of lettuce. Mol. Plant. Microbe Interact. 28, 1109–1022. doi: 10.1094/MPMI-03-15-0061-R
Janek, T., Lukaszewicz, M., Rezanka, T., and Krasowska, A. (2010). Isolation and characterization of two new lipopeptide biosurfactants produced by Pseudomonas fluorescens BD5 isolated from water from the arctic archipelago of svalbard. Bioresour. Technol. 101, 6118–6123. doi: 10.1016/j.biortech.2010.02.109
Kuiper, I., Lagendijk, E. L., Pickford, R., Derrick, J., Lamers, G., Thomas-Oates, J., et al. (2004). Characterization of two Pseudomonas putida lipopeptide biosurfactants, putisolvin I and II, which inhibit biofilm formation and break down existing biofilms. Mol. Microbiol. 51, 97–113. doi: 10.1046/j.1365-2958.2003.03751.x
Lavermicocca, P., Iacobellis, N. S., and Simmaco, M. (1997). Biological properties and spectrum of activity of Pseudomonas syringae pv. syringae toxins. Physiol. Mol. Plant 50, 129–140. doi: 10.1006/pmpp.1996.0078
Licciardello, G., Strano, C. P., Bertani, I., Bella, P., Fiore, A., Fogliano, V., et al. (2012). N-acyl-homoserine-lactone quorum sensing in tomato phytopathogenic Pseudomonas spp. is involved in the regulation of lipodepsipeptide production. J. Biotechnol. 159, 274–282. doi: 10.1016/j.jbiotec.2011.07.036
Loper, J. E., Hassan, K. A., Mavrodi, D. V., Davis, E. W., Lim, C. K., Shaffer, B. T., et al. (2012). Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 8:e1002784. doi: 10.1371/journal.pgen.1002784
Masschelein, J., Jenner, M., and Challis, G. L. (2017). Antibiotics from Gram-negative bacteria: a comprehensive overview and selected biosynthetic highlights. Nat. Pro. Rep. 34, 712–783. doi: 10.1039/c7np00010c
McCarter, L., and Silverman, M. (1990). Surface-induced swarmer cell differentiation of Vibrio parahaemolyticus. Mol. Microbiol. 4, 1057–1062. doi: 10.1111/j.1365-2958.1990.tb00678.x
Michelsen, C. F., Jensen, H., Venditto, V. J., Hennessy, R. C., and Stougaard, P. (2015a). Bioactivities by a crude extract from the Greenlandic Pseudomonas sp. In5 involves the nonribosomal peptides, nunamycin and nunapeptin. PeerJ 3:e1476. doi: 10.7717/peerj.1476
Michelsen, C. F., Watrous, J., Glaring, M. A., Kersten, R., Koyamae, N., and Stougaarda, P. (2015b). Nonribosomal peptides, key biocontrol components for Pseudomonas fluorescens In5, isolated from a Greenlandic suppressive soil. mBio 6:e00079. doi: 10.1128/mBio.00079-15
Monti, S. M., Gallo, M., Ferracane, R., Borrelli, R. C., Ritieni, A., Greco, M. L., et al. (2011). Analysis of bacterial lipodepsipeptides by matrix-assisted laser desorption/ionisation time-of-flight and high-performance liquid chromatography with electrospray mass spectrometry. Rapid Commun. Mass Spectrom. 15, 623–628. doi: 10.1002/rcm.277
Nguyen, D. D., Melnik, A. V., Koyama, N., Lu, X. W., Schorn, M., Fang, J. S., et al. (2016). Indexing the Pseudomonas specialized metabolome enabled the discovery of poaeamide B and the bananamides. Nat. Microbiol. 2:16197. doi: 10.1038/NMICROBIOL.2016.197
Nikolouli, K., and Mossialos, D. (2012). Bioactive compounds synthesized by non-ribosomal peptide synthetases and type-I polyketide synthases discovered through genome-mining and metagenomics. Biotechnol. lett. 34, 1393–1403. doi: 10.1007/s10529-012-0919-2
Olorunleke, F. E., Kieu, N. P., De Waele, E., Timmerman, M., Ongena, M., and Höfte, M. (2017). Coregulation of the cyclic lipopeptides orfamide and sessilin in the biocontrol strain Pseudomonas sp. CMR12a. Microbiologyopen 6:e00499. doi: 10.1002/mbo3.499
Raaijmakers, J. M., de Bruijn, I., and de Kock, M. J. (2006). Cyclic lipopeptide production by plant-associated Pseudomonas spp.: diversity, activity, biosynthesis, and regulation. Mol. Plant. Microbe. Interact. 19, 699–710. doi: 10.1094/MPMI-19-0699
Raaijmakers, J. M., de Bruijn, I., Nybroe, O., and Ongena, M. (2010). Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol. Rev. 34, 1037–1062. doi: 10.1111/j.1574-6976.2010.00221.x
Raaijmakers, J. M., Vlami, M., and De Souza, J. T. (2002). Antibiotic production by bacterial biocontrol agents. Antonie Van Leeuwenhoek 81, 537–547. doi: 10.1023/A:1020501420831
Röttig, M., Medema, M. H., Blin, K., Weber, T., Rausch, C., and Kohlbacher, O. (2011). NRPSpredictor2-a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 39, 362–367. doi: 10.1093/nar/gkr323
Scaloni, A., Dalla Serra, M., Amodeo, P., Mannina, L., Vitale, R. M., Segre, A. L., et al. (2004). Structure, conformation and biological activity of a novel lipodepsipeptide from Pseudomonas corrugata: cormycin A1. Biochem. J. 384, 25–36. doi: 10.1042/BJ20040422
Schneider, T., Gries, K., Josten, M., Wiedemann, I., Pelzer, S., Labischinski, H., et al. (2009). The lipopeptide antibiotic friulimicin B inhibits cell wall biosynthesis through complex formation with bactoprenol phosphate. Antimicrob. Agents Chemother. 53, 1610–1618. doi: 10.1128/AAC.01040-08
Scholz-Schroeder, B. K., Hutchison, M. L., Grgurina, I., and Gross, D. (2001). The contribution of syringopeptin and syringomycin to virulence of Pseudomonas syringae pv. syringae strain B301D on the basis of sypA and syrB1 biosynthesis mutant analysis. Mol. Plant Microbe. Interact. 14, 336–348. doi: 10.1094/MPMI.2001.14.3.336
Sinden, S. L., De Vay, J. E., and Backman, P. A. (1971). Properties of syringomycin, a wide spectrum antibiotic and phytotoxin produced by Pseudomonas syringae, and its role in the bacterial canker disease of peach trees. Physiol. Plant Pathol. 1, 199–200. doi: 10.1016/0048-4059(71)90029-4
Spiers, A. J., Bohannon, J., Gehrig, S. M., and Rainey, P. (2003). Biofilm formation at the air-liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol. Microbiol. 50, 15–27. doi: 10.1046/j.1365-2958.2003.03670.x
Strano, C. P., Bella, P., Licciardello, G., Lo Piero, A. R., Fogliano, V., Venturi, V., et al. (2015). Pseudomonas corrugata crpCDE is part of the cyclic lipopeptide corpeptin biosynthetic gene cluster and is involved in bacterial virulence in tomato and in hypersensitive response in Nicotiana benthamiana. Mol. Plant Pathol. 16, 495–506. doi: 10.1111/mpp.12207
Straus, S. K., and Hancock, R. E. W. (2006). Mode of action of the new antibiotic for Gram-positive pathogens daptomycin: comparison with cationic antimicrobial peptides and lipopeptides. Biochim. Biophys. Acta 1758, 1215–1223. doi: 10.1016/j.bbamem.2006.02.009
Thoma, S., and Schobert, M. (2009). An improved Escherichia coli donor strain for diparental mating. FEMS Microbiol. Lett. 294, 127–132. doi: 10.1111/j.1574-6968.2009.01556.x
Toguchi, A., Siano, M., Burkkart, M., and Harshey, R. (2000). Genetics of swarming motility in Salmonella enterica serovar typhimurium: critical role for lipopolysaccharide. J. Bacteriol. 182, 6308–6321. doi: 10.1128/JB.182.22.6308-6321.2000
Trantas, E. A., Licciardello, G., Nalvo, F., Almeida, N., Witek, K., Strano, C., et al. (2015). Comparative genomic analysis of multiple strains of two unusual plant pathogens: Pseudomonas corrugata and Pseudomonas mediterranea. Front. Microbiol. 6:811. doi: 10.3389/fmicb.2015.00811
Van Der Voort, M., Meijer, H., Schmidt, Y., Watrous, G., Dekkers, E., Mendes, E., et al. (2015). Genome mining and metabolic profiling of the rhizosphere bacterium Pseudomonas sp. SH-C52 for antimicrobial compounds. Front. Microbiol. 6:693. doi: 10.3389/fmicb.2015.00693
Vlamakis, H., Chai, Y., Beauregard, P., Losick, R., and Kolter, R. (2013). Sticking together: building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 11, 157–168. doi: 10.1038/nrmicro2960
Weller, D. M. (2007). Pseudomonas biocontrol agents of soilborne pathogens: looking back over 30 years. Phytopathology 97, 250–256. doi: 10.1094/phyto-97-2-0250
Keywords: Pseudomonas, genome mining, secondary metabolites, cyclic lipopeptides, NRPS, rhizosphere, antifungal activity, biofilm formation
Citation: Zhao H, Liu Y-P and Zhang L-Q (2019) In silico and Genetic Analyses of Cyclic Lipopeptide Synthetic Gene Clusters in Pseudomonas sp. 11K1. Front. Microbiol. 10:544. doi: 10.3389/fmicb.2019.00544
Received: 06 December 2018; Accepted: 01 March 2019;
Published: 19 March 2019.
Edited by:
Ana R. Freitas, Universidade do Porto, PortugalReviewed by:
Maarten Ghequire, KU Leuven, BelgiumCopyright © 2019 Zhao, Liu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li-Qun Zhang, emhhbmdscUBjYXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.