
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 15 March 2019
Sec. Plant Pathogen Interactions
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00514
This article is part of the Research Topic Bacteria Present in Plant Nodules: Diversity, Biogeography and Plant Growth Promotion View all 14 articles
 Vera Safronova1*
Vera Safronova1* Andrey Belimov1,2
Andrey Belimov1,2 Anna Sazanova1
Anna Sazanova1 Elizaveta Chirak1
Elizaveta Chirak1 Irina Kuznetsova1
Irina Kuznetsova1 Evgeny Andronov1
Evgeny Andronov1 Alexander Pinaev1
Alexander Pinaev1 Anna Tsyganova1
Anna Tsyganova1 Elena Seliverstova1,3
Elena Seliverstova1,3 Anna Kitaeva1
Anna Kitaeva1 Viktor Tsyganov1
Viktor Tsyganov1 Igor Tikhonovich1,4
Igor Tikhonovich1,4Two bacterial strains Ach-343 and Opo-235 were isolated, respectively from nodules of Miocene-Pliocene relict legumes Astragalus chorinensis Bunge and Oxytropis popoviana Peschkova originated from Buryatia (Baikal Lake region, Russia). For identification of these strains the sequencing of 16S rRNA (rrs) gene was used. Strain Opo-235 belonged to the species Mesorhizobium japonicum, while the strain Ach-343 was identified as M. kowhaii (100 and 99.9% rrs similarity with the type strains MAFF 303099T and ICMP 19512T, respectively). Symbiotic genes of these strains as well as some genes that promote plant growth (acdS, gibberellin- and auxin-synthesis related genes) were searched throughout the whole genome sequences. The sets of plant growth-promoting genes found were almost identical in both strains, whereas the sets of symbiotic genes were different and complemented each other with several nod, nif, and fix genes. Effects of mono- and co-inoculation of Astragalus sericeocanus, Oxytropis caespitosa, Glycyrrhiza uralensis, Medicago sativa, and Trifolium pratense plants with the strains M. kowhaii Ach-343 and M. japonicum Opo-235 expressing fluorescent proteins mCherry (red) and EGFP (green) were studied in the gnotobiotic plant nodulation assay. It was shown that both strains had a wide range of host specificity, including species of different legume genera from two tribes (Galegeae and Trifolieae). The effects of co-microsymbionts on plants depended on the plant species and varied from decrease, no effect, to increase in the number of nodules, nitrogen-fixing activity and plant biomass. One of the reasons for this phenomenon may be the discovered complementarity in co-microsymbionts of symbiotic genes responsible for the specific modification of Nod-factors and nitrogenase activity. Localization and co-localization of the strains in nodules was confirmed by the confocal microscopy. Analysis of histological and ultrastructural organization of A. chorinensis and O. popoviana root nodules was performed. It can be concluded that the strains M. kowhaii Ach-343 and M. japonicum Opo-235 demonstrate lack of high symbiotic specificity that is characteristic for primitive legume-rhizobia systems. Further study of the root nodule bacteria having complementary sets of symbiotic genes will contribute to clarify the evolutionary paths of legume-rhizobia relationships and the mechanisms of effective integration between partners.
It has been shown that primitive tropical legumes possess relatively low symbiotic specificity, whereas evolutionarily young legumes of temperate climate show higher specificity and can form symbiosis with a single species of root nodule bacteria (Lie et al., 1987; Tikhonovich and Provorov, 2009; Provorov and Vorobyev, 2011; Bakker et al., 2014). The promising models for studying the evolution of such specificity are symbiotic systems of relict legume plants. The species Vavilovia formosa growing in Western Asia and Caucasus mountains as well as Oxytropis triphylla, O. popoviana, O. tragacanthoides, Hedysarum zundukii, Astragalus chorinensis, and Glycyrrhiza uralensis originated from Baikal Lake region are known as Miocene-Pliocene relics (Malyschev, 2006; Sinjushin et al., 2009; Turuta et al., 2015). Information about strains isolated from root nodules of these plant species was recently published (Safronova et al., 2014, 2015a,b, 2017a,b). It was shown that these isolates belonged to different families of rhizobia (Rhizobiaceae, Phyllobacteriaceae, and Bradyrhizobiaceae) and new species Bosea vaviloviae and Phyllobacterium zundukense were described (Safronova et al., 2015a, 2018b).
The impact of natural rhizobial diversity and the resulting multipartite interactions between symbionts on plant growth is currently poorly understood. On the one hand, it was shown that the multi-strain treatments of two Australian wattle species Acacia salicina and A. stenophylla interacting with highly diverse communities of rhizobia had a negative effect on plant growth, probably due to strong competition between strains with different levels of nitrogen-fixing activity and symbiotic effectiveness (Barrett et al., 2015). On the other hand, two rhizobial strains Mesorhizobium japonicum Opo-242 and Bradyrhizobium sp. Opo-243 isolated from the same nodule of a relict legume O. popoviana significantly accelerated the root nodule formation on the host plant after a combined inoculation (Safronova et al., 2018a). The whole genome sequence analysis of this pair of strains showed the presence in Opo-243 additional genes nodPQ, nolK, and noeL involved in the modification of Nod factors (NFs) and affecting the specificity of plant-rhizobia interactions. This report demonstrated that taxonomically different strains forming symbiotic systems with relics can be co-microsymbionts infecting the same nodule and promoting the nodulation process due to complementary sets of symbiotic genes.
The aim of this work was to identify eight new rhizobial strains isolated from nodules of A. chorinensis originated from Buryatia (Baikal Lake region, Russia), to select among the A. chorinensis and previously obtained O. popoviana isolates a pair of strains having the most diverse sets of symbiotic genes, and to study their effects on various legume species (A. sericeocanus, O. caespitosa, G. uralensis, Medicago sativa, and Trifolium pratense) after mono- and co-inoculations. The species A. sericeocanus, O. caespitosa, and G. uralensis were chosen because they have the same distribution area with A. chorinensis and O. popoviana plants. The histological and ultrastructural analyzes of A. chorinensis and O. popoviana root nodules were also conducted to search for specific features of their organization.
Soil samples and seeds of A. chorinensis Bunge were collected in Buryatia (Baikal Lake region, Russia). Seeds were surface sterilized and scarified by treatment with 0.1% HgCl2 for 10 min and then 5% NaOCl for 8 min, rinsed carefully with sterile tap water and germinated on filter paper in Petri dishes at 25°C in the dark for 4 days. Seedlings were transferred to three sterile plastic pots (5 seedlings per pot) containing 250 g of soil. Plants were cultivated for 60 days in the growth chamber with 50% relative humidity and four-level illumination/temperatures mode: night (dark, 18°C, 8 h), morning (200 μmol m-2 s-1, 20°C, 2 h), day (400 μmol m-2 s-1, 23°C, 12 h), evening (200 μmol m-2 s-1, 20°C, 2 h). Illumination was performed by L 36W/77 FLUORA lamps (Osram, Germany). Then roots of individual plants were removed from soil and washed with tap water. Nodules were analyzed on the stereo microscope Stemi 508 (Carl Zeiss, Germany). Strains of nodule bacteria were isolated from the obtained nodules by the standard method described by Novikova and Safronova (1992) using modified yeast extract mannitol agar (YMA, Vincent, 1970) supplemented with 0.5% succinate (YMSA, Safronova et al., 2015a). From one individual plant one strain was isolated. All strains were deposited in the Russian Collection of Agricultural Microorganisms (RCAM, WDCM 966) and stored at -80°C in the automated Tube Store (Liconic Instruments, Lichtenstein) as described previously (Safronova and Tikhonovich, 2012). Isolation of rhizobial strains from nodules of O. popoviana plants originated from the same region as A. chorinensis was carried out in our previous work using similar methods (Safronova et al., 2018a).
For identification of all isolates the following PCR primers were used: fD1 (5′-AGAGTTTGATCCTGGCTCAG -3′) and rD1 (5′-CTTAAGGAGGTGATCCAGCC -3′) for an approximately 1400 bp segment of the 16S rRNA gene (Weisburg et al., 1991). PCR was performed in 25-μL reaction mixtures containing 150 μM dNTPs (Promega, United States), 5 pmol of each primer, 1 U of Taq polymerase (Helicon, Russia) and 50–100 ng of purified template DNA. PCR conditions for amplification of the 16S rRNA gene were following: initial denaturation at 95°C for 3 min 30 s; 35 cycles of denaturation at 94°C for 1 min 10 s, annealing at 56°C for 40 s and extension at 72°C for 2 min 10 s; final extension at 72°C for 6 min 10 s. Electrophoresis was carried out with 1% agarose gel (Invitrogen, United States) in TAE. A 100-bp GeneRulerTM and Lambda DNA/HindIII markers (Fermentas, United States) were used for sizing and approximate quantification of DNA fragments. Purification of the PCR products was usually performed by using PureLinkTM Quick kit (Invitrogen, United States) according to the manufacturer’s guidance. The direct sequencing of PCR products was performed by an ABI PRISM 3500xl genetic analyzer (Applied Biosystems, United States).
The sequences were compared with related sequences of the type strains available in the GenBank database using BLAST analysis (Basic logical alignment search tool) at NCBI. Rrs-dendrogram was constructed using the neighbor-joining method in MEGA 5.0 software package (Tamura et al., 2011). The evolutionary distances were computed using the maximum composite likelihood method. Bootstrap analysis with 1000 replicates was performed to estimate the support of clusters.
All rrs sequences have been deposited to the NCBI GenBank database under accession numbers MH626527, MH628053, MH628054, MH628085, MH628088, MH628090 – MH628092.
Genomic DNA was extracted using Genomic DNA Purification KIT (Thermo Fisher Scientific, Europe) according to recommendation of manufacturer. DNA was fragmented by focused ultrasonicator Covaris S2 (Covaris, United States). Fragment DNA-libraries were prepared with NEBNext DNA Library Kit (NEB, United States), and their quality was estimated with High Sensitivity DNA Kit on Bioanalyzer 2100 (Agilent, United States). DNA amount was estimated with dsDNA High Sensitivity Kit on Qubit 1.0 (Invitrogen, United States). Genome sequencing was performed on a MiSeq genomic sequencer (Illumina, United States) by standard protocol with MiSeq Reagent Kit, 600 Cycles (Illumina, United States) at SB RAS Genomics Core Facility (ICBFM SB RAS). Genome was assembled de novo using the SPAdes 3.5.0 software (Bankevich et al., 2012). Quality control was performed by QUAST 3.0 (Gurevich et al., 2013). Search for genes in the assembled contigs was performed using the RAST annotation service (Overbeek et al., 2014). Search for homologs of the 16S rRNA gene, ITS region, housekeeping genes recA, glnII, and rpoB as well as symbiotic genes in annotated genomes was performed using CLC Genomics Workbench 7.5.1 software using local BLASTn and tBLASTx.
The whole genome sequences have been deposited to the NCBI GenBank database under accession numbers MZXV00000000 for the isolate M. kowhaii Ach-343 and QKOD00000000 for the isolate M. japonicum Opo-235. Genomic features of the complete genomes are given in Supplementary Table S1.
Electroporation of strains was performed in accordance with the previous work (Garg et al., 1999). For this purpose, 90 μl competent cells of strains M. kowhaii Ach-343 and M. japonicum Opo-235 were suspended between two electrodes spaced by 0.1 cm. Electroporation was carried out with an electric pulse of 14 kV/cm and pulse length of ∼7.3 ms (Gene PulserXcell, Bio-Rad, United States). Number of high voltage pulses ranged 1 for each strain. After the pulse was delivered, the cuvettes were kept on ice for 10 min. Then the electroporated cells were suspended in YM broth (Vincent, 1970), incubated for 24 h at 30°C with 200 rpm shaking (Orbital Shaker-Incubator ES20, BioSan, Latvia) and spreaded on the YMA plates. In transformation experiments a derivative of the pHC60 (tetR) plasmid (Cheng and Walker, 1998), in which the GFP coding sequence was replaced by the mCherry coding sequence (J. Fournier, LIPM, Toulouse, France, unpublished results) and pMP4655 (tetR) plasmid harboring the egfp gene were used for electroporation of strains Ach-343 and Opo-235, respectively. Screening of different transformants was performed on YMA medium containing 10 μg/ml tetracycline. PCR was carried out using Thermal Cycler T100 (Bio-Rad, United States) to confirm the transformation using total DNA as template and a pair of primers flanking fluorescent protein gene present on the plasmids pHC60 and pMP4655. The PCR conditions used for the amplification of 1083 and 967 bp fragment (for pHC60 and pMP4655, respectively) included a pre-amplification denaturation at 95°C for 3 min 30 s followed by 35 cycles at 94°C for 1 min 10 s, 54°C for 1 min and 72°C for 2 min 10 s, with a final extension at 72°C for 7 min. The following primers were used: M13F (5′-GTTGTAAAACGACGGCCAGTG-3′) and M13R (5′-AGCGGATAACAATTTCACACAGGA-3′). The PCR products were visualized by electrophoresis on 1.0% agarose gel (Invitrogen, United States) in TAE buffer and purified by using Silica (Helicon, Russia). Sequencing was performed using the ABI PRISM 3500xl (Applied Biosystems, United States) according to the manufacturer’s instructions. Positive clones of each strain were tested for fluorescence in the Axio Imager A1 microscope (Carl Zeiss, Germany).
Seeds of A. chorinensis, A. sericeocanus, O. popoviana, O. caespitosa, G. uralensis, M. sativa, and T. pratense were surface sterilized, scarified and germinated as described above. The uniformly germinated seedlings were transferred to polypropylene pots OS140BOX (Duchefa, Netherlands) containing 20 g of vermiculite (3 seeds per pot, 4 pots per each treatment of A. chorinensis and O. popoviana plants, 10 pots per each treatment of other plant species). Each pot was supplemented with 40 ml of the nutrient solution (g/l): K2HPO4 1.0, KH2PO4 0.25, MgSO4 1.0, Ca3(PO4)2 0.2, FeSO4 0.02, H3BO3 0.005, (NH4)2MoO4 0.005, ZnSO4 × 7 H2O 0.005, MnSO4 0.002 (Novikova and Safronova, 1992). Seedlings were inoculated with unlabeled strains M. kowhaii Ach-343 and M. japonicum Opo-235 or their fluorescent-labeled variants Ach-343(pHC60) and Opo-235(pMP4655) or their combination in the amount of 106 cells per pot. The uninoculated plants were used as negative control. Plants were cultivated for 30 days in the growth chamber as described above. The appearance of nodules on six plants per treatment was recorded after 2 and 3 weeks of cultivation. Nodules were analyzed on the stereo microscope Stemi 508 (Carl Zeiss, Germany) and photos were taken using the microscope color camera AxioCam ERc 5 s (Carl Zeiss, Germany). At the end of experiment the nodules were counted and the fresh biomass of plants (shoots and roots) was determined. The nitrogen fixation of nodules was measured by the acetylene-reduction method (Turner and Gibson, 1980) using gas chromatograph GC-2014 (Shimadzu, Japan). Five nodules from each treatment were used for the confocal microscopy. The data were processed by the standard method of variance analysis using the software STATISTICA version 10 (StatSoft Inc., United States). Fisher’s LSD test was used to evaluate differences between means.
Nodules were molded in 3% agarose gel blocks and prepared in 1/4 MTSB (50 mM PIPES, 5 mM MgSO4 ⋅ 7H2O, 5 mM EGTA, pH 6.9). Nodule sections (50 μm) were prepared using a microtome with a vibrating blade HM650V (Microm, Germany). Sections were analyzed using the laser scanning confocal system LSM 510 META (Carl Zeiss, Germany). EGFP and mCherry were excited at 488 and 543 nm, respectively. Images were acquired with ZEN 2009 software (Carl Zeiss, Germany).
The A. chorinensis and O. popoviana plants inoculated with the strains M. kowhaii Ach-343 and M. japonicum Opo-235, respectively, were grown in the plant nodulation assay as described above. 3-week-old nodules were harvested from roots and placed directly in fixative. The sample preparation and embedding procedure were described previously (Serova et al., 2018). Briefly, the whole nodules were fixed in 2.5% (v/v) glutaraldehyde (Sigma-Aldrich, United States), then were post-fixed in 2% (v/v) osmium tetroxide for 2 h. Samples then were dehydrated in a graded series of increasing ethanol concentrations followed by two changes of 100% acetone. Dehydrated samples were progressively embedded in Epon (Honeywell FlukaTM, Thermo Fisher Scientific, United Kingdom) at room temperature. Embedded samples were transferred to blocks in fresh resin and polymerized at 60°C for 48 h.
For transmission electron microscopy, 90–100-nm-thick ultrathin sections were cut using a diamond knife (Diatome, Switzerland) on a Leica EM UC7 ultramicrotome (Leica Microsystems, Germany) and collected on copper/palladium grids coated with 4% (w/v) pyroxylin and carbon. The grids containing the sections were counterstained with 2% (w/v) aqueous uranyl acetate for 1 h followed by lead citrate for 1 min. Ultrathin sections of the selected area were examined using a Tecnai G2 Spirit electron microscope (FEI, the Netherlands) at 80 kV. Electron micrographs were taken with Mega View G2 CCD Camera (Olympus-SIS, Germany).
Eight fast-growing rhizobial strains (Ach-304, Ach-305, Ach-313, Ach-318, Ach-320, Ach-328, Ach-343 and Ach-347) were isolated from nodules of A. chorinensis. Colonies appeared on YMSA medium at the 5-th day. The identification of these isolates by the sequencing of 16S rRNA gene showed that they belonged to different species of the genus Mesorhizobium (Figure 1 and Supplementary Table S2). The isolates Ach-313, Ach-318, Ach-328 and Ach-347 formed a separate cluster with the type strain M. newzealandense ICMP 19545T (Figure 1) and had 100% rrs similarity with this type strain at 94% query cover (Supplementary Table S2). The isolates Ach-304 and Ach-305 grouped together and were the closest to the type strain M. huakuii IFO 5243T with 99.93% rrs similarity. The isolate Ach-343 formed the high supported cluster with the type strain M. kowhaii ICMP 19512T (99.85% rrs similarity) while the last isolate Ach-320 was the most closely related to the species M. japonicum (99.52% with the type strain MAFF 303099T). Thus the isolates Ach-304 and Ach-305 were assigned to the species M. huakuii and the isolate Ach-343 to M. kowhaii. We preliminarily classified the isolates Ach-313, Ach-318, Ach-328 and Ach-347 as M. newzealandense, and left the isolate Ach-320 without species definition. The species M. newzealandense and M. kowhaii were recently described as microsymbionts of New Zealand endemic species Sophora prostrata and S. microphylla (De Meyer et al., 2016). The host plant range of these rhizobia is not yet studied; however, the genus Sophora includes the extremely promiscuous relict species S. flavescens that forms symbiosis with strains belonging to genera Bradyrhizobium, Sinorhizobium, Mesorhizobium, Rhizobium, and Phyllobacterium (Jiao et al., 2015). On the other hand, the rhizobial species M. huakuii being described long ago (Chen et al., 1991) was up to now known as a specific microsymbiont of the plant species A. sinicus (Jarvis et al., 1997; Fuli et al., 2017).
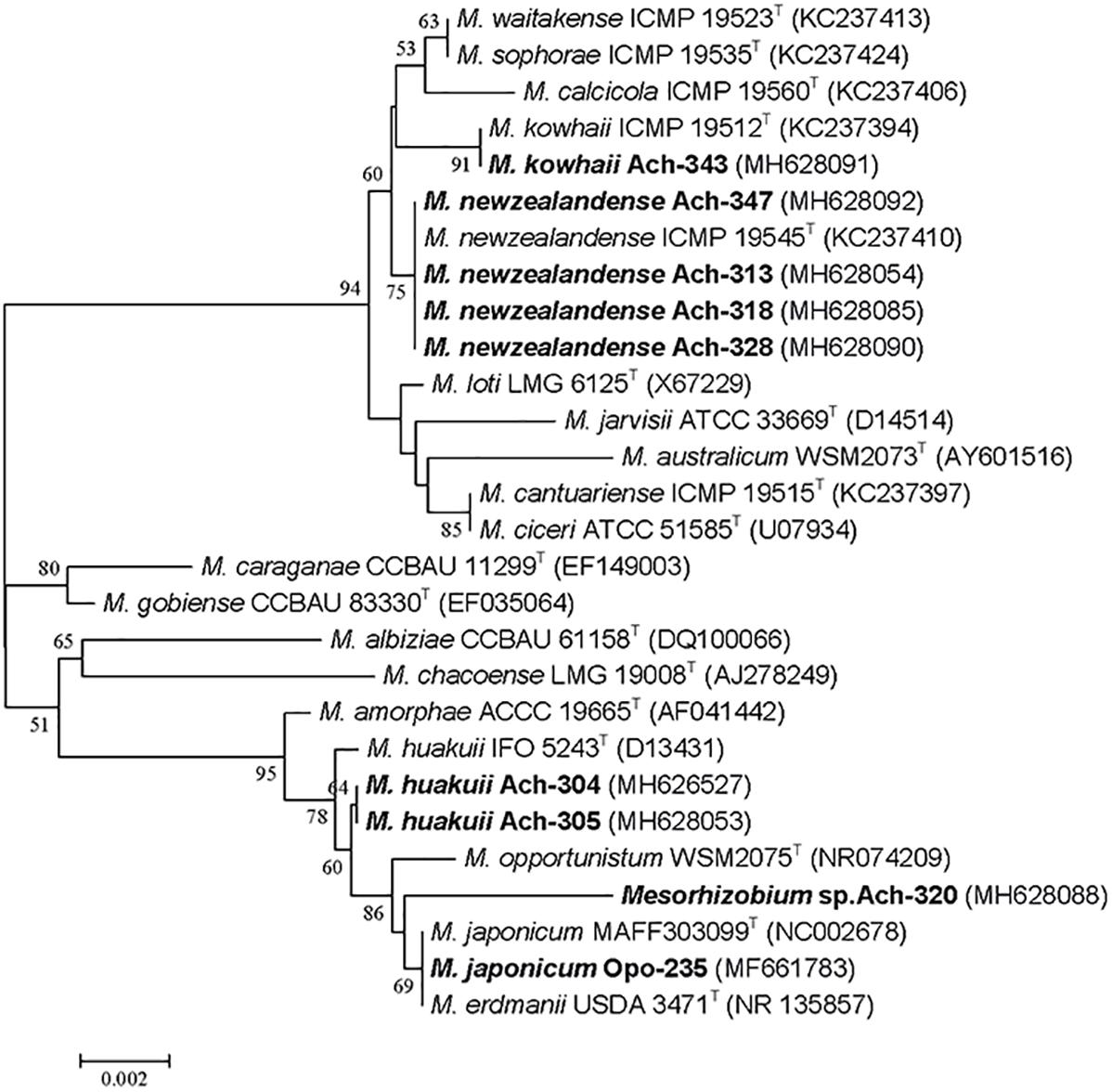
Figure 1. Phylogenetic tree generated by the Neighbor-Joining method using partial 16S rRNA gene sequences (1400 nt) of the Mesorhizobium-related isolates from Astragalus chorinensis and Oxytropis popoviana (in bold) and closely related species. Bootstrap values more than 50% are given. Type strains are indicated by the letter “T.”
Nine other strains belonging to M. japonicum and M. kowhaii were also isolated in our previous work from the nodules of O. popoviana plants (Safronova et al., 2018a). Among them the isolate Opo-235 was chosen as the object of this research and presented in Figure 1, having 100% rrs sequence similarity with the type strain M. japonicum MAFF 303099T isolated from Lotus japonicus (Martínez-Hidalgo et al., 2016).
Thus, eight new isolates from nodules of Miocene-Pliocene relict legume A. chorinensis originated from the Baikal Lake region (Eastern Siberia) as well as the previously obtained isolates from O. popoviana belonged to mesorhizobial species M. newzealandense, M. huakuii, M. kowhaii, and M. japonicum having East Asian or Polynesian origins. Based on whole genome sequence analysis of twelve strains isolated from O. popoviana, O. triphylla, and A. chorinensis plants (data not presented) two strains M. kowhaii Ach-343 and M. japonicum Opo-235 with the maximum number of symbiotic genes, but with the most different sets of them were selected for further work.
Symbiotic genes of the strains M. kowhaii Ach-343 and M. japonicum Opo-235 isolated, respectively, from A. chorinensis and O. popoviana plants as well as the genes that promote plant growth (acdS, gibberellin- and auxin-biosynthesis related genes) were searched throughout the whole genome sequences. It was shown the presence in both strains the acdS gene encoded anti-stress enzyme 1-aminocyclopropane-1-carboxylate (ACC) deaminase, which is common among root nodule bacteria (Ma et al., 2003) and plays important role in nodulation process (Glick et al., 2007). The following plant growth-promoting genes were also detected in the strains Ach-343 and Opo-235: cpxP, cpxR, cpxU, and ispA related to biosynthesis of phytohormones gibberellins (Nett et al., 2017); the tryptophan synthase (trp) and amine oxidase gene involved in the tryptophan-dependent production of auxins (Ahemad and Kibret, 2014). No difference between both strains in the set of plant growth-promoting genes was found, except for the presence of two copies the acdS gene in the strain Ach-343 and one copy – in the strain Opo-235 (data not presented).
At the same time, the sets of symbiotic genes of the strains Ach-343 and Opo-235 differed and complemented each other with several nod, nif, and fix genes (Table 1). The common nodABC genes necessary for legume nodulation (Wais et al., 2002) were present in both strains. They also contained nif and fix genes required for nitrogen fixation, namely nifHDK and nifENB genes encoding structural and catalytic components of the nitrogenase complex (Dos Santos et al., 2012) as well as fixABCX, fixNOPQ, and fixGHIS genes participating in electron transfer to nitrogenase and symbiotically essential oxidase complex (Fischer, 1994; Edgren and Nordlund, 2004). However, the strain Ach-343 had additional nodG, nodM, nodN, and nifQ genes that were not found in the strain Opo-235, while the nodT, nodW, nifV, and fixJKL genes were observed only in the strain Opo-235. Although the strain Ach-343 contained the protein PZV34926 that has 51% similarity (at 98% coverage) with the NodW protein of the strain Bradyrhizobium diazoefficiens USDA 110 (NODW_BRADU) and can play the same role. The functions of these genes and their possible role in the plant-microbe interactions will be discussed below. Bearing in mind that the strains Ach-343 and Opo-235 belong to the microsymbionts of relict rhizobia-legume systems, we calculated the sequence similarity between their nod, nif, or fix genes and homologs sequences presented in the GenBank database. It was shown that 18 nod genes of our isolates (nodCIJGMPQZ genes of the strain Ach-343 and nodABCDIJEPWZ genes of the strain Opo-235) had a high level of similarity (80 - 99%) simultaneously with representatives of two or three rhizobial families: Rhizobiaceae, Phyllobacteriaceae and Bradyrhizobiaceae (Supplementary Table S3). On the other hand, the nodABDEFLN genes of the strain Ach-343 and the nodFLQT genes of the strain Opo-235 were the closest only to sequences of strains from the family Phyllobacteriaceae (89 – 99% similarity). Phylogenetic trees based on two common nodulation genes nodA and nodC of isolates and representatives of different rhizobial families (Supplementary Figures S1, S2) showed that both genes involved in biosynthesis of the core structure of NFs (Wais et al., 2002) grouped with other Mesorhizobium strains (at 99 and 64% bootstrap support for nodA and nodC, respectively). The strain M. amorphae CCBAU 01583, which is a microsymbiont of Astragalus membranaceus and Caragana intermedia (Yan et al., 2017), was closest to the isolate Opo-235 (99% of nodA and 95% of nodC sequence similarity). The isolate Ach-343 revealed close relatedness to the strain M. septentrionale CCBAU 11244 isolated from Caragana microphylla in China (Chen et al., 2008) with 90% of nodA and 94% of nodC sequence similarity. It should be noted that the nodA gene of the strain Ach-343 was split into two different contigs.
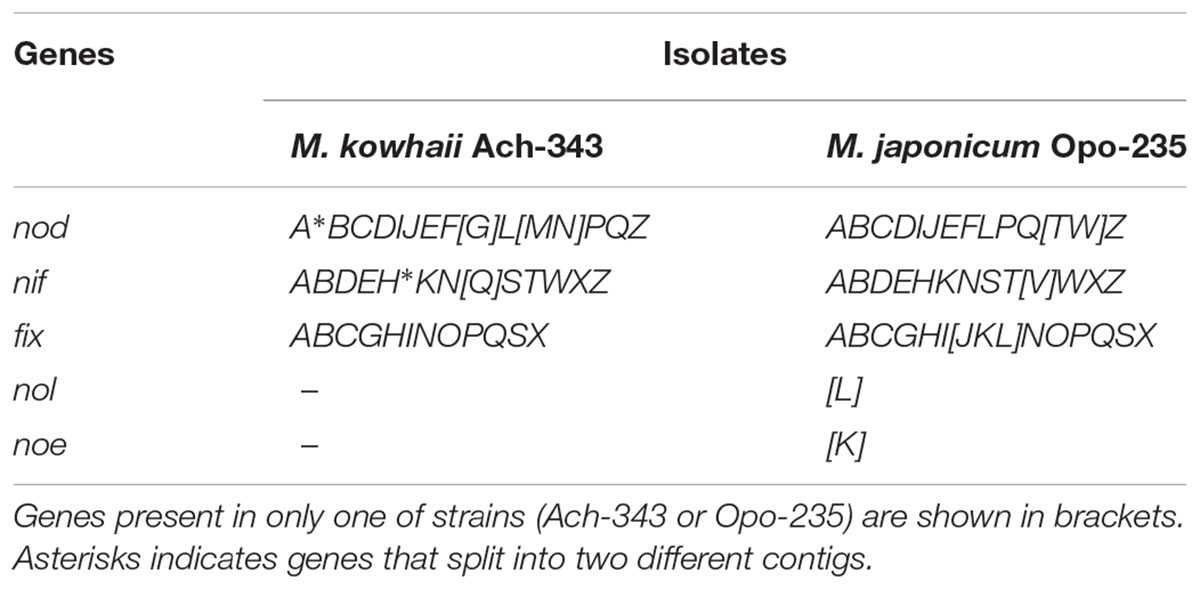
Table 1. Presence of symbiotic genes in the strains Mesorhizobium kowhaii Ach-343 and M. japonicum Opo-235.
Strains M. kowhaii Ach-343 and M. japonicum Opo-235 expressing, respectively, fluorescent proteins mCherry (red) and EGFP (green) were obtained using electroporation method. One positive clone of each strain having the brightest fluorescence was selected for further work.
Symbiotic properties of the unlabeled strains M. kowhaii Ach-343 and M. japonicum Opo-235 as well as their fluorescent-labeled variants Ach-343(pHC60) and Opo-235(pMP4655) were preliminarily studied in gnotobiotic plant nodulation assay with their host plants A. chorinensis and O. popoviana (Supplementary Table S4). It was shown, that all strains nodulated both plant species, but formed low effective symbiosis with O. popoviana plants (no differences with uninoculated control on the plant biomass). However, they were significantly more effective on A. chorinensis plants, although they did not differ in any of the symbiotic parameters. No differences were found between unlabeled strains and their fluorescent-labeled variants (Supplementary Table S4).
Symbiotic phenotype of the fluorescent-labeled strains M. kowhaii Ach-343(pHC60) and M. japonicum Opo-235(pMP4655) was studied in gnotobiotic plant nodulation assay with A. sericeocanus, G. uralensis, O. caespitosa, M. sativa, and T. pratense plants using treatments of mono- and co-inoculation. After 2 weeks of cultivation root nodules were observed only on A. sericeocanus roots (in all variants of inoculation) and G. uralensis (in the co-inoculation treatment), while after 3 weeks of cultivation only uninoculated controls had no nodules (data not shown). The results of plant nodulation assay (Table 2) demonstrated that the strains Ach-343 and Opo-235 had a wide range of host plants including different species and genera of legumes from tribes Galegeae (Astragalus, Oxytropis, and Glycyrrhiza) and Trifolieae (Medicago, Trifolium). However, the number of nodules as well as their acetylene reduction activity was different depending on the plant species and treatments: as a rule the strain Opo-235 induced more nodules and was more active in acetylene reduction than the strain Ach-343 (Table 2). Nitrogen-fixing nodules were not observed on M. sativa and T. pratense plants. The data showed that depending on plant species the effects of a combined inoculation with the strains Ach-343 and Opo-235 on plants varied from the decrease, no effect, to the increase in different symbiotic parameters including plant biomass as an indicator of the symbiotic effectiveness. Figure 2 illustrates this conclusion in the form of a diagram. If the symbiotic effectiveness of the strain Ach-343 is conditionally taken as level 1, the effectiveness of the strain Opo-235 may be taken as levels 1 or 2 (depending on the plant species). With this assumption the symbiotic effectiveness in the variants Ach-343 + Opo-235 can be designated as levels 1, 1.5, or 2, corresponding to the level of one co-microsymbiont (T. pratense and O. caespitosa plants), to an average level of two strains (A. sericeocanus plants) or to the level exceeding both strains (G. uralensis plants).
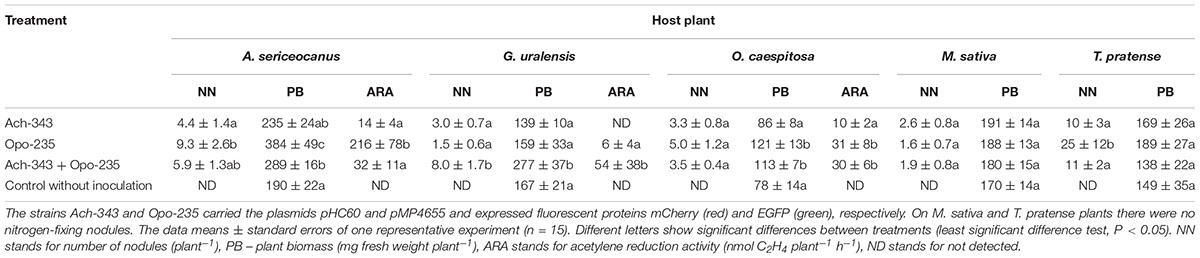
Table 2. Effects of mono- and co-inoculation of Astragalus sericeocanus, Glycyrrhiza uralensis, Oxytropis caespitosa, Medicago sativa and Trifolium pratense plants with the strains M. kowhaii Ach-343 and M. japonicum Opo-235 in the gnotobiotic plant nodulation assay.
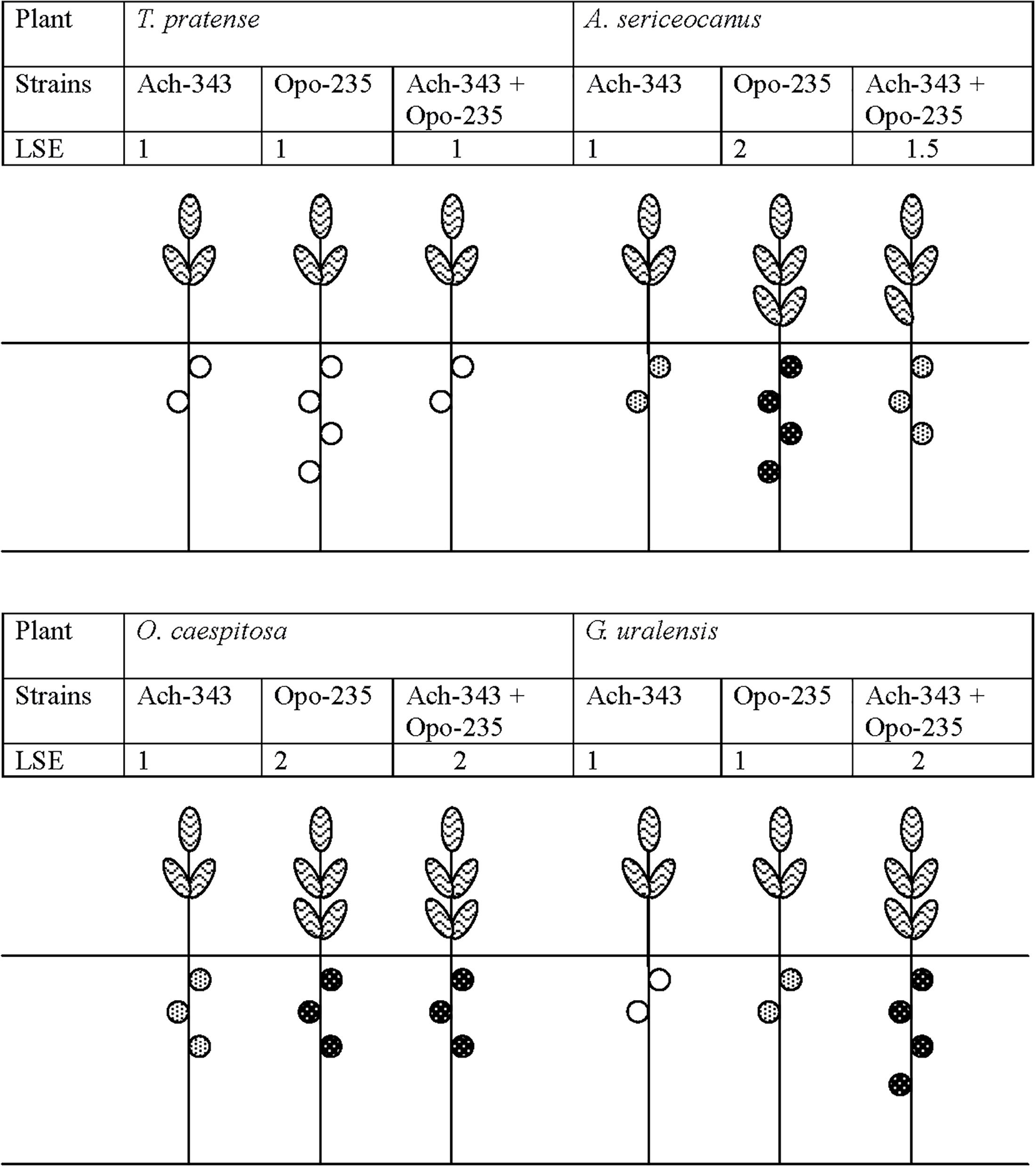
Figure 2. Diagram demonstrating the effects of co-inoculation with strains M. kowhaii Ach-343 and M. japonicum Opo-235 on the effectiveness of symbioses with T. pratense, A. sericeocanus, O. caespitosa, and G. uralensis plants. The strains Ach-343 and Opo-235 carried the plasmids pHC60 and pMP4655 and expressed fluorescent proteins mCherry (red) and EGFP (green), respectively. LSE stands for the conditionally level of symbiotic effectiveness (plant biomass).  Non-active nodules,
Non-active nodules,  lower level of acetylene reduction activity,
lower level of acetylene reduction activity,  higher level of acetylene reduction activity.
higher level of acetylene reduction activity.
Localization and co-localization of the fluorescent-labeled strains M. kowhaii Ach-343(pHC60) and M. japonicum Opo-235(pMP4655) in nodules obtained in the plant nodulation assay was analyzed by the confocal microscopy (Figures 3–5). Figure 3 represents a general view of nodules on the roots of M. sativa and T. pratense plants and confocal images of nodule sections. It can be seen that pseudonodules formed on M. sativa roots in all variants of inoculation did not contain any rhizobia (Figures 3A–C), although bacterial cells of the strain Ach-343 were present on the root surface (Figure 3B). It is known that M. sativa is a highly specific plant that forms a symbiosis only with bacteria Sinorhizobium meliloti and S. medicae, although it has been shown that some other rhizobial strains can also nodulate this plant species (Noreen et al., 2003; Torres Tejerizo et al., 2011). In contrast, in nodules on T. pratense roots inoculated with the strains Opo-235(pMP4655) or Ach-343(pHC60) the corresponding bacteria were present (Figures 3D,E), while in the variant of co-inoculation only nodules with the strain Opo-235(pMP4655) were observed (Figure 3F).
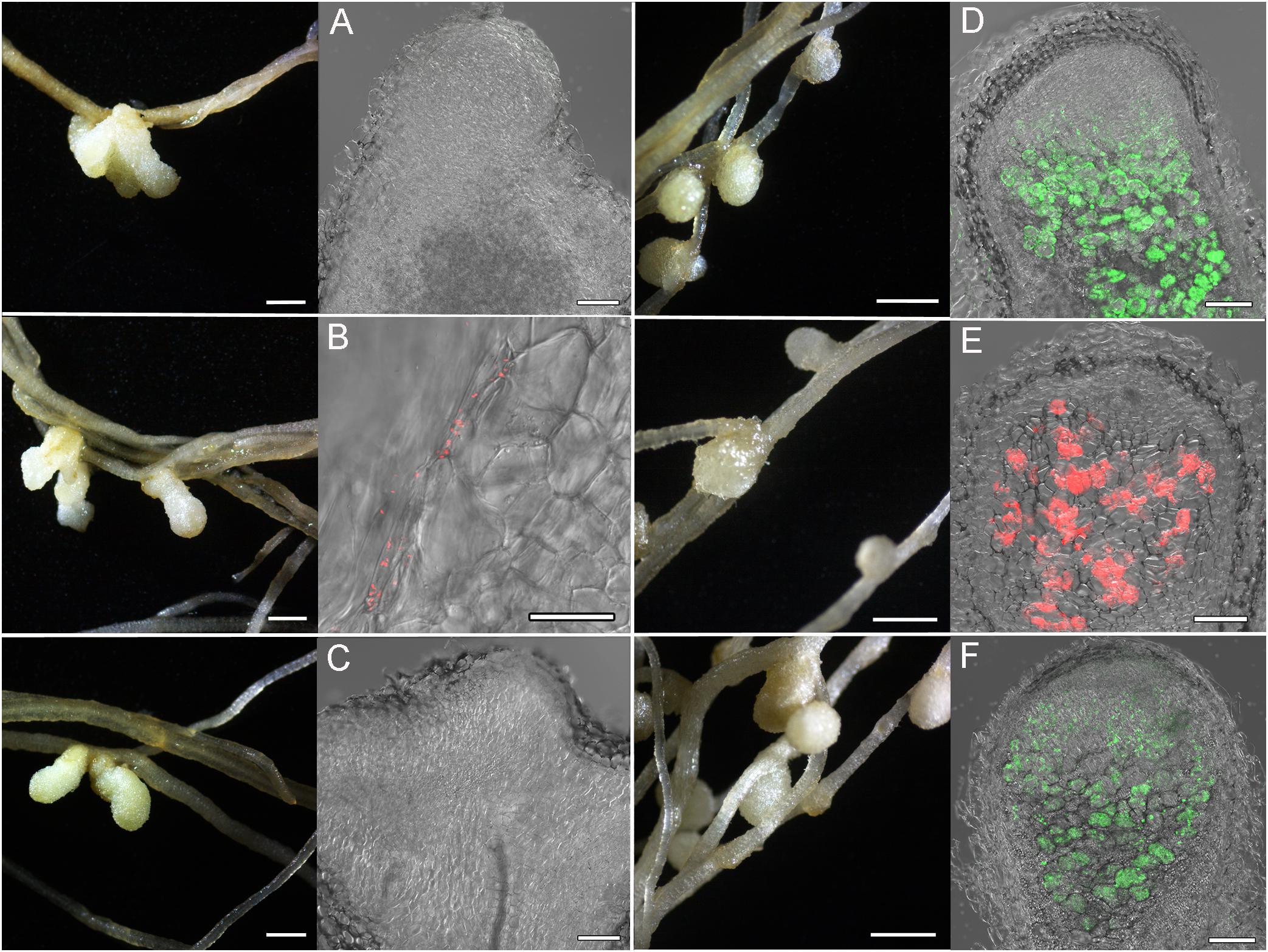
Figure 3. Pictures of root nodules formed by the fluorescent-labeled strains M. kowhaii Ach-343(pHC60) and M. japonicum Opo-235(pMP4655) on the M. sativa and T. pratense plants in gnotobiotic nodulation assay. Plant species: A–C – M. sativa; D–F – T. pratense. Inoculation treatment: A,D – Opo-235, merge of differential interference contrast (DIC) and green channel; B,E – Ach-343, merge of DIC and red channel; C,F – Opo-235 + Ach-343, merge of DIC, green and red channels. Columns 1 and 3 present the general views of nodules and root surface (scale bar = 1 mm), columns 2 and 4 – the confocal microscopy images (scale bar = 100 μm). Opo-235 bacteria in green, Ach-343 bacteria in red.
Figures 4, 5 represent some examples of nodules on the roots of A. sericeocanus, O. caespitosa, and G. uralensis plants. In all variants of mono-inoculation the corresponding strains were detected in nodules. In the co-inoculation treatments all nodules contained the strain Opo-235(pMP4655), however, the strain Ach-343(pHC60) was also observed in some nodules of O. caespitosa (Figure 4C) as well as in all nodules and some infection threads of G. uralensis (Figures 5D–F). The phenomenon of the simultaneous presence of different rhizobial strains in one nodule, which is called the double nodule occupancy, was previously described for common beans (Phaseolus vulgaris) inoculated with the strain Rhizobium tropici CIAT 899 marked with the gus-gene in competition with other isolates from beans (Shamseldin and Werner, 2004). It should be noted that in nodules of O. caespitosa having both strains some plant cells were simultaneously infected by green and red bacteria, but in some plant cells only green or red bacteria were present (Figure 4C). In 3 of the 5 analyzed nodules of G. uralensis both strains were predominantly present in the same plant cells (Figure 5E); however, in two nodules the strains occupied different cells and the cells infected by Ach-343 were significantly smaller than the cells infected by Opo-235 (Figure 5D). The observed small size of cells infected with Ach-343 strain may indicate that they do not undergo typical differentiation, which is accompanied by a significant increase in the infected cell size (Tsyganova et al., 2018). However, further studies are required to confirm this assumption. The form of nodules was different depending on the plant species and rhizobial microsymbiont and varied from irregular in shape to round or elongated with one or several lobes (Figures 3–5).
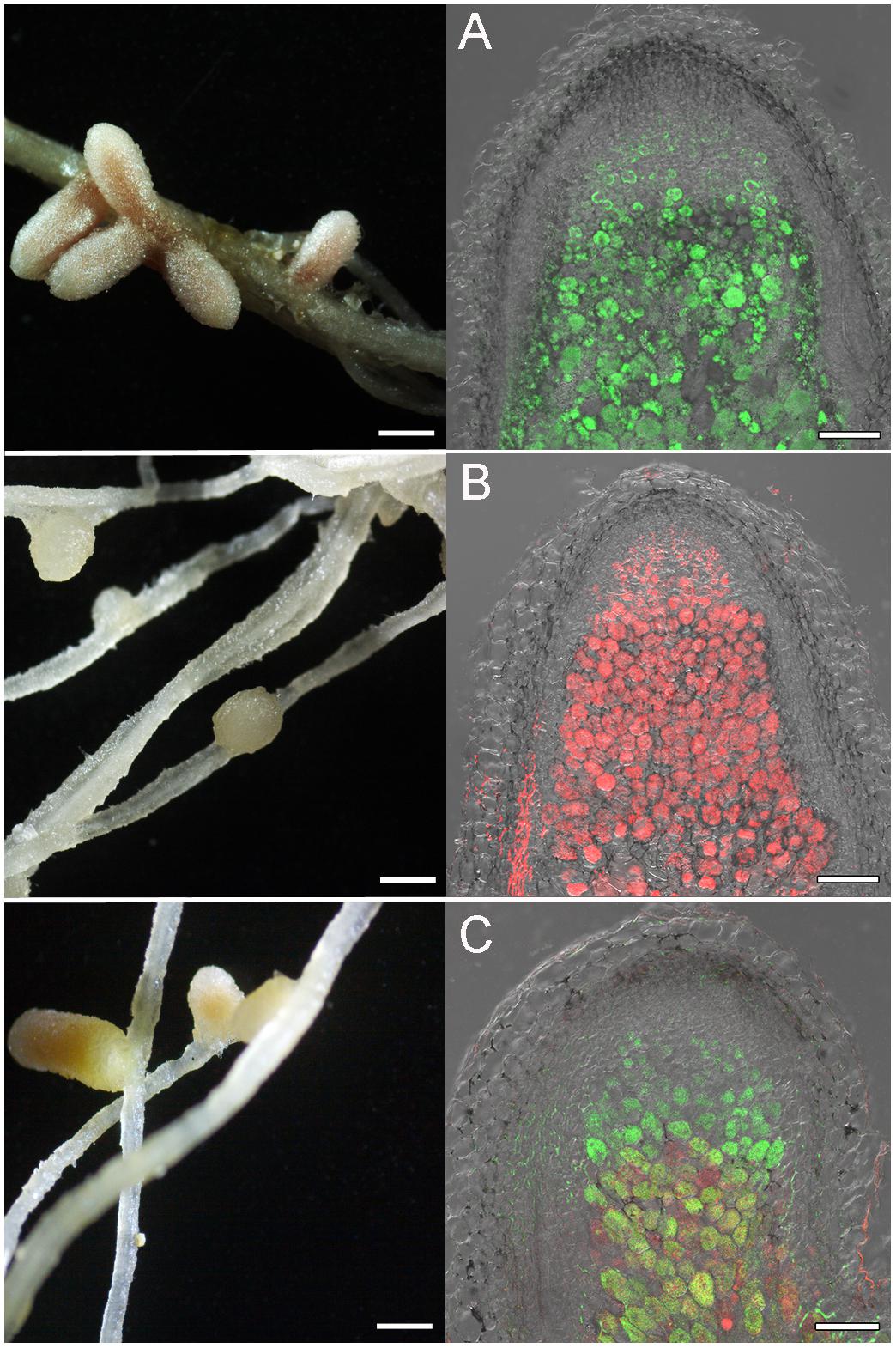
Figure 4. Pictures of root nodules formed by the fluorescent-labeled strains M. kowhaii Ach-343(pHC60) and M. japonicum Opo-235(pMP4655) on the A. sericeocanus and O. caespitosa plants in gnotobiotic nodulation assay. Plant species: A – A. sericeocanus; B,C – O. caespitosa. Inoculation treatment: A – Opo-235 bacteria in green; B – Ach-343 bacteria in red; C – Opo-235 + Ach-343. Column 1 presents the general views of nodules (scale bar = 1 mm), column 2 – the confocal microscopy images (scale bar = 100 μm). A – merge of DIC and green channel; B – merge of DIC and red channel; C – merge of DIC, green and red channels.
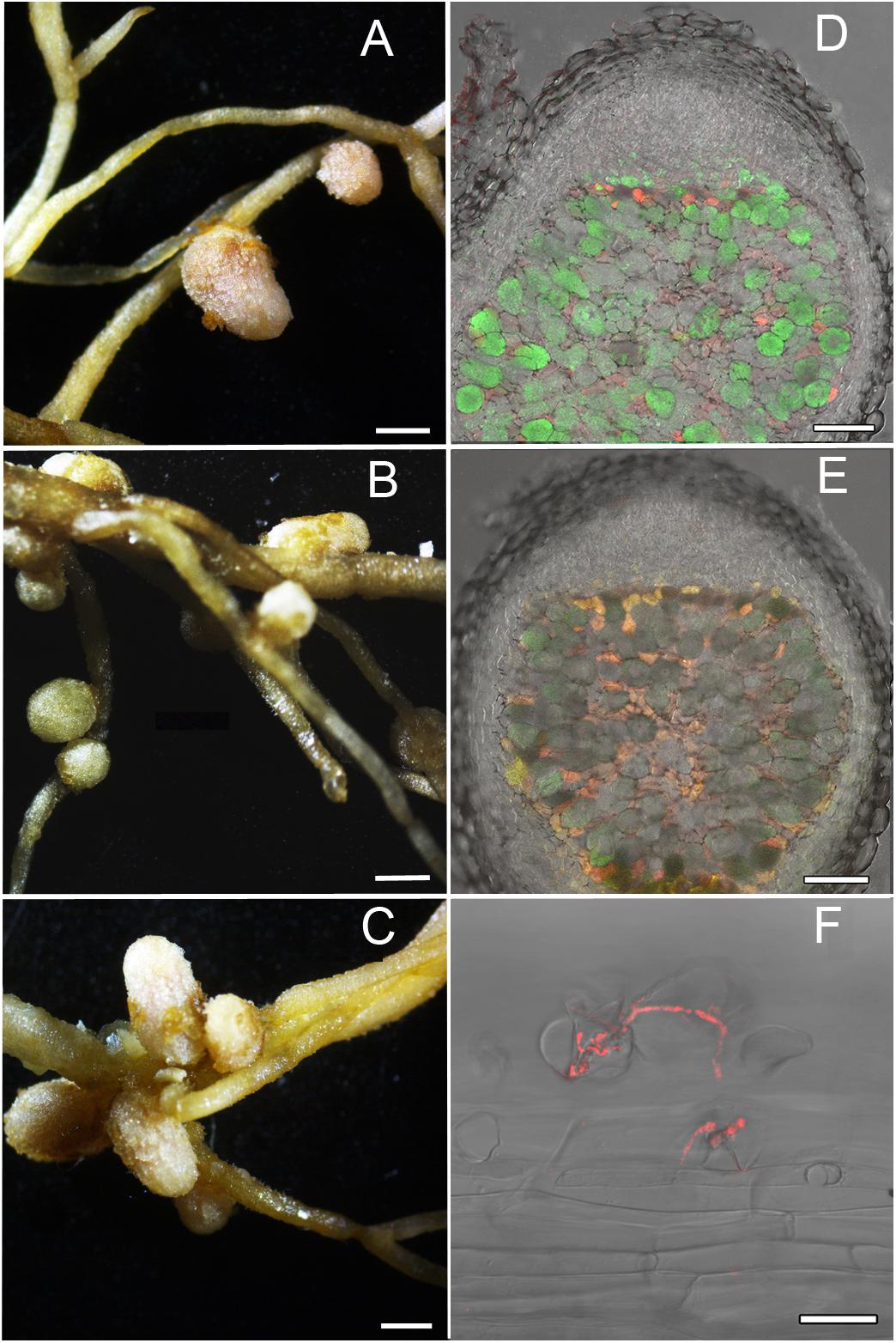
Figure 5. Pictures of root nodules formed by the fluorescent-labeled strains M. kowhaii Ach-343(pHC60) and M. japonicum Opo-235(pMP4655) on the G. uralensis plants in gnotobiotic nodulation assay. Inoculation treatment: A – strain Opo-235; B – strain Ach-343; C–F – Opo-235 + Ach-343; D,E – co-infection of plant cells in nodules; D – nodule in which the strains occupied the different cells; E – nodule in which there are cells with both strains (in yellow-orange); F – infection thread formed by the strain Ach-343. Column 1 presents the general views of nodules (scale bar = 1 mm), column 2 – the confocal microscopy images (scale bar = 100 μm). D,E – merge of DIC, green and red channels; F – merge of DIC and red channel.
Histological organization of A. chorinensis and O. popoviana nodules was typical for indeterminate nodules (Figure 6). Previously, the indeterminate type was described for nodules formed on roots of A. alpinus, O. maydelliana, O. arctobia (Newcomb and Wood, 1986), A. cicer (Wdowiak et al., 2000), A. danicus, A. frigidus, A. glycyphyllos, O. lapponica (Ampomah et al., 2012). The senescence zone occupied the significant part of nodules due to their large age (Figure 6). The infection threads were well developed in the infection zone in nodules of both species (Figures 7A,B). In nodules of A. chorinensis the infected cells were filled by a large number of symbiosomes containing spherical bacteroids individually surrounded with rugose symbiosome membranes (Figure 7C). In O. popoviana nodules in infected cells symbiosomes contained several elongated or elongated-branched bacteroids surrounded by a common symbiosome membrane (Figure 7D). In contrast, in O. maydelliana nodules bacteroids were surrounded by symbiosome membrane individually (Newcomb and Wood, 1986). Previously, it was shown that in nodules of A. canadensis and O. lamberti infected cells are filled with elongated bacteroids (Montiel et al., 2017), in nodules of A. cicer bacteroids are spherical with elongated outgrowths (Wdowiak et al., 2000) and in nodules of O. lapponica bacteroids are elongated-branched (Ampomah et al., 2012). It was suggested that the morphotype of bacteroids correlates with the number of nodule-specific cysteine-rich (NCR) peptides (Montiel et al., 2017). Thus, it is possible that the different morphotypes of bacteroids formed in the nodules of A. canadensis and A. chorinensis are caused by differences in the compositions of NCR peptides produced by these species. In A. chorinensis nodules the numerous mitochondria were present at the periphery of the infected cell (Figure 7E). In nodules of O. popoviana in infected cells the endoplasmic reticulum was powerfully developed (Figure 7F). The extensive endoplasmic reticulum was previously described for A. alpinus, O. maydelliana, O. arctobia as an adaptation for nodule development and functioning in arctic conditions (Newcomb and Wood, 1986).
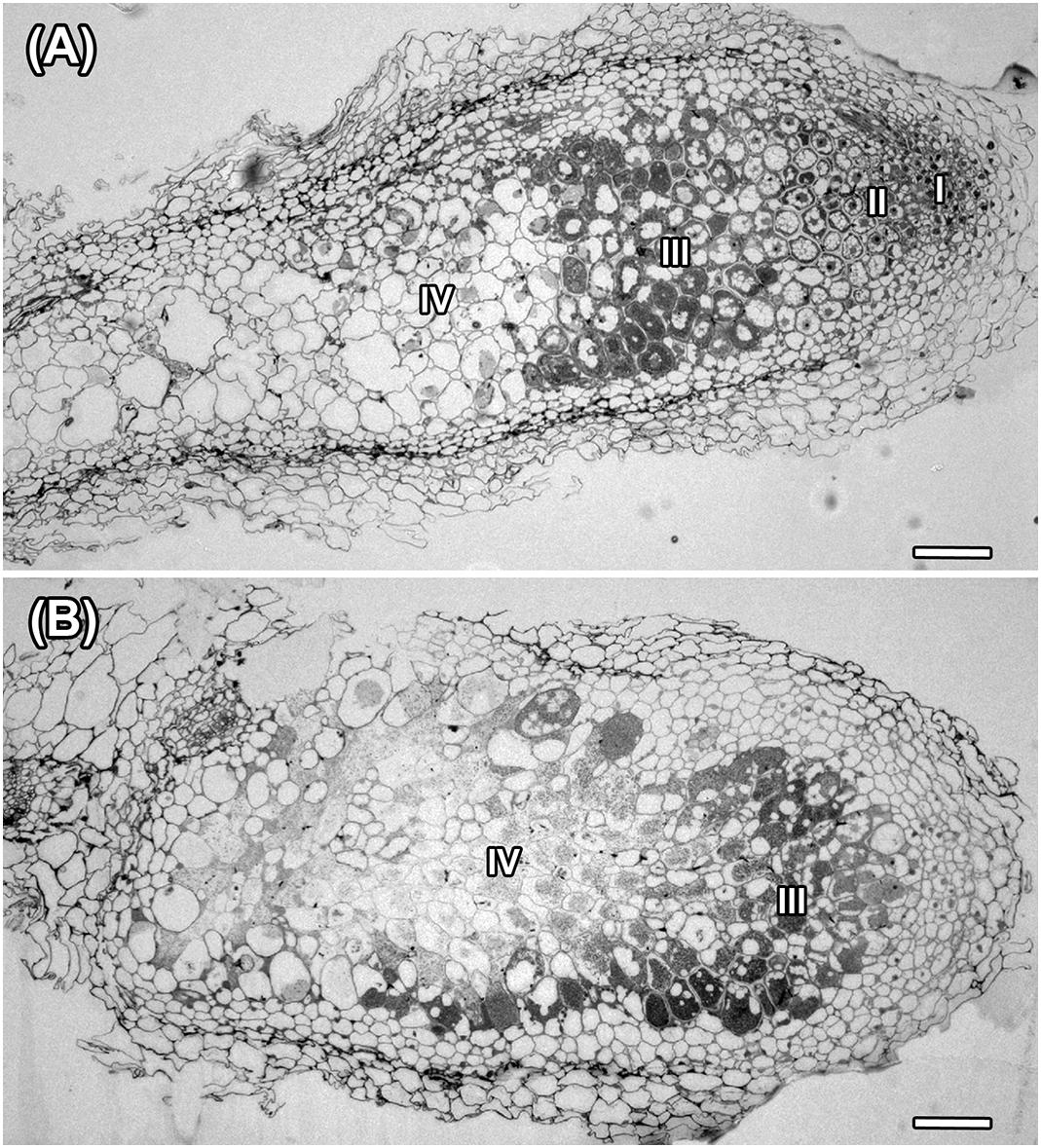
Figure 6. Histological organization in nodules of A. chorinensis and O. popoviana formed by the strains M. kowhaii Ach-343 (A) and M. japonicum Opo-235 (B). Zones in nodule are designated by Roman numerals: I – meristem, II – infection zone, III – nitrogen fixation zone, IV – senescence zone; scale bar = 100 μm.
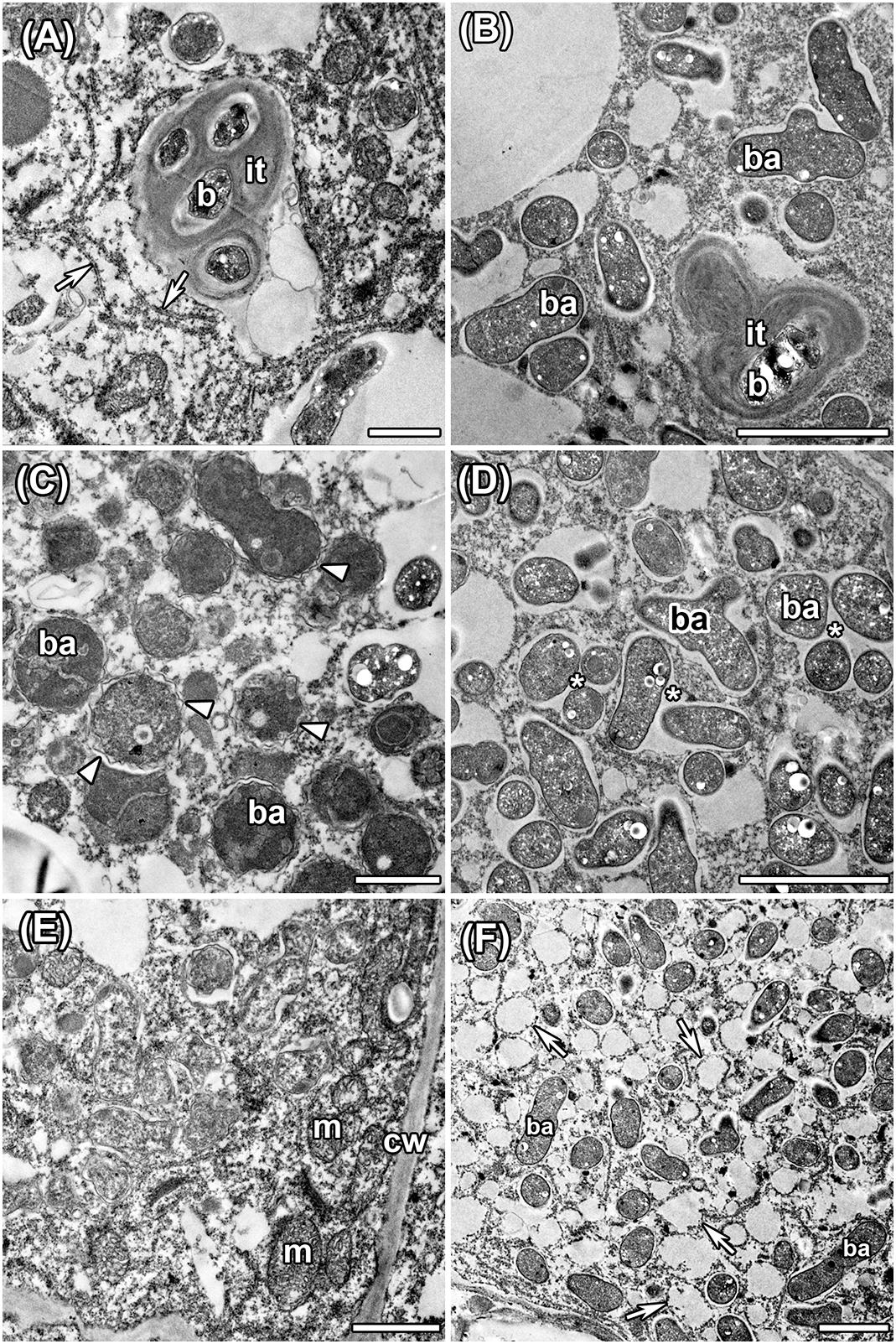
Figure 7. Ultrastructural organization in nodules of A. chorinensis and O. popoviana formed by the strains M. kowhaii Ach-343 (A,C,E) and M. japonicum Opo-235 (B,D,F). it, infection thread; b, bacterium; ba, bacteroid; m, mitochondria; cw, cell wall; arrowheads indicate rugose symbiosome membrane; arrows indicate endoplasmic reticulum, asterisks indicate symbiosomes with several bacteroids. Scale bars: (A,C,E) = 1 μm, (B,D,F) = 2 μm.
Thus, the strains M. kowhaii Ach-343 and M. japonicum Opo-235 isolated from nodules of the relict legumes A. chorinensis and O. popoviana, respectively, had a wide range of host plants, including different genera of legumes (Astragalus, Oxytropis, Glycyrrhiza, Medicago, and Trifolium). The genetic basis for the low specificity of these isolates may be the similarity of their nod genes with representatives of different rhizobial families: Rhizobiaceae, Phyllobacteriaceae and Bradyrhizobiaceae. This data are in agreement with previous reports suggesting that symbiotic systems of relict plants can be formed with different rhizobial strains belonging to various taxonomic groups (Safronova et al., 2014, 2015a,b, 2017b).
The co-inoculation of plants with both strains Ach-343 and Opo-235, which in some cases were localized in the same nodules, led to changes in symbiotic phenotypes (number of nodules, level of nitrogen fixation and plant biomass) compared with the mono-inoculation treatments. Wherein the effectiveness of symbiosis after co-inoculation could both increase and decrease. This phenomenon of rhizobial interaction can be explained by the complementarity of symbiotic genes, which are responsible for the specific modification of NFs, nitrogenase activity and probably other processes related to formation and development of symbiosis. It was shown that the strains Ach-343 and Opo-235 possess quite different sets of nod, nif, fix, nol, and noe genes (Table 1). Among them, nodG, nodM, and nodN genes that were observed only in the strain Ach-343 encode the following enzymes that involved in the biosynthesis and modification of NFs: 3-oxoacyl-acyl carrier protein reductase (Lopez-Lara and Geiger, 2001), glucosamine synthase (Baev et al., 1992; Lohrke et al., 1998) and dehydratase (Baev et al., 1992). The strain Opo-235 had two other genes for NFs modification: noeK gene participates in NFs fucosylation (Lamrabet et al., 1999), nolL gene involved in acetylation of the fucosyl residue (Corvera et al., 1999). The nodT and nodW genes found only in the strain Opo-235 encode an outer membrane lipoprotein playing a role in the secretion of NFs (Rivilla et al., 1995; Pinto et al., 2009) and the nod genes transcription regulator (Perret et al., 2000). It is likely that the differences in the set of nod, nol, and noe genes determine the greater nodule-forming ability of the strain Opo-235 as compared with the strain Ach-343, which was manifested in the plants of A. sericeocanus and T. pratense (Table 2). It can also be assumed that products of different genes responsible for plant nodulation may interact with each other if they are secreted by bacteria into the external environment or localized on the outer membrane of cells. Acceleration of the root nodule formation has been previously observed in the symbiosis between the relict legume O. popoviana and its two co-microsymbionts, one of which could not induce nodules but contained nodPQ, nolK, and noeL genes that could affect the specificity of plant-rhizobia interactions via the sulfation and fucosylation of NFs (Safronova et al., 2018a).
Particular attention should be given to the observed presence of both strains in the same nodules of O. caespitosa and especially G. uralensis, where co-microsymbionts were localized in the same plant cells. Along with an increase in the number of nodules, this could be the reason for significant (nine fold) raising the acetylene reduction activity of G. uralensis nodules (Table 2). The difference in the specific acetylene reduction activity, calculated on the dry nodule weight, in the variant of co-inoculation was not statistically significant compared to the Opo-235 mono-inoculation treatment due to very large standard errors of this parameter (Supplementary Table S5). However, its increase was 56%. It was shown above that the nifQ gene was discovered only in the strain Ach-343, while the nifV and fixJKL genes were found only in the strain Opo-235 (Table 1). The nifV gene is required for a maturation of the nitrogenase MoFe-protein through the homocitrate synthesis (Evans et al., 1991); the fixJKL genes are important for expression of some nif and fix genes (Fischer, 1994). The nifQ gene encodes the molybdenum ion binding protein with reductase activity that is known as a specific molybdenum donor for FeMo cofactor biosynthesis and the incorporation of reduced molybdenum in the MoIV oxidation state into nitrogenase (Hernandez et al., 2009; Black et al., 2012; Boyd et al., 2015). However, the mechanisms of complementation between nif and fix genes localized in different co-microsymbionts inside the same nodule are not clear. Apparently, the alteration of symbiotic activity may occur due to secreted products of these (or some other) genes, extracellular metabolites or any factors released as a result of bacteroid degradation. One the possible way of such complementation having a positive effect on the nitrogen-fixing activity of symbiosis is an increase in the amount of reduced molybdenum required for high nitrogenase activity and provided by the special Mo-binding proteins.
It should be noted that some other factors besides symbiotic may be involved in the processes of integration between symbiotic partners: plant hormones (auxin, ethylene, gibberellin, cytokinin, abscisic acid), exo- and lipopolysaccharides, cellulases, rhizopines, components of secretion systems (T3SS, T6SS) encoding broad classes of effectors proteins (Russell et al., 2014; Notti and Stebbins, 2016; Okazaki et al., 2016). It was previously shown that the strains M. kowhaii Ach-343 and M. japonicum Opo-235 did not differ statistically in the activity of the ACC-deaminase reducing the level of the plant stress ethylene and the ability to produce auxins: indole-3-acetic acid (IAA), indole-3-carboxylic acid (ICA), and indole-3-lactic acid (ILA), which are one of the main factors affecting the nodulation process (data not published). The auxin producing activities for the strains Ach-343 and Opo-235 were at the following levels, respectively: 199 and 175 ng/ml for IAA, 111 and 103 ng/ml for ICA, 12 and 15 ng/ml for ILA production. The ACC-deaminase activities of free-living bacteria determined by monitoring the amount of a-ketobutyrate (αKB) generated after hydrolysis of ACC (Saleh and Glick, 2001) by modified method in suspensions of disrupted cells (Belimov et al., 2015) were 14.5 and 12.3 μM αKB mg-1 h-1 for the strains Ach-343 and Opo-235, respectively. However, it should be taken into account that the levels of ACC deaminase activity of mesorhizobia in free-living cells and inside nodules can be different because within this genus the expression of acdS genes are usually regulated by NifA protein (Nascimento et al., 2012).
The presence of some components of the T6SS (icmF, tssABCEGJKL, tagFH, vgrG, vasA, hcp, and clpV genes) was detected in both strains, with the gene sets being identical and the gene similarity between strains was 76 – 81% (data not presented). The genes found are known to encode key components of the bacterial pathogenicity (penetrating system; secretion of cytotoxins, lysozymes, homologs to phage tail proteins, lipoproteins, factors of adherence to epithelial cells). Based on the analysis of secretion systems it remains unclear why the strain Ach-343 demonstrates lower virulence compared with the strain Opo-235 (forms statistically fewer nodules on some plants and is usually not localized inside nodules in the co-inoculation treatments). Further comparative analysis of the factors related to formation and functioning of symbiosis in taxonomic different co-microsymbionts will contribute to the study of the phenomenon of rhizobial interaction, based on genetic complementarity, as well as to the disclosure of the evolutionary paths of legume-rhizobia relationships and the mechanisms of efficient integration between partners.
VS and IK participated in microbiology work. AB and AP participated in obtaining of fluorescent-labeled strains. AS and EC participated in molecular work. EA participated in photography of nodules. AT, ES, and VT participated in ultrastructural analysis of root nodules. AK participated in confocal microscopy. IT participated in bio-informatics work.
This work was supported by the Russian Science Foundation (Grant 16-16-00080 for microbiology and molecular work; Grant 17-76-30016 for photography of nodules and bio-informatics work; Grant 16-16-10035 for ultrastructural analysis of root nodules, and Grant 17-14-01363 for obtaining of fluorescent-labeled strains). Deposition of strains in the RCAM collection was supported by the Program for the Development and Inventory of Bioresource Collections. The research was performed using equipment of the Core Centrum “Genomic Technologies, Proteomics and Cell Biology” in ARRIAM.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank Jan Puhalsky and Tatiana Aksenova (ARRIAM, Saint Petersburg, Russia) for technical assistance and Dr. Kirill Demchenko (Komarov Botanical Institute, Saint Petersburg, Russia) for fruitful discussion.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00514/full#supplementary-material
Ahemad, M., and Kibret, M. (2014). Mechanisms and applications of plant growth promoting rhizobacteria: current perspective. J. King Saud. Univ. Sci. 26, 1–20. doi: 10.1016/j.jksus.2013.05.001
Ampomah, O. Y., James, E. K., Iannetta, P. P., Kenicer, G., Sprent, J. I., and Huss-Danell, K. (2012). Nodulation and ecological significance of indigenous legumes in Scotland and Sweden. Symbiosis 57, 133–148. doi: 10.1007/s13199-012-0188-9
Baev, N., Schultze, M., Barlier, I., Ha, D. C., Virelizier, H., Kondorosi, E., et al. (1992). Rhizobium nodM and nodN genes are common nod genes: nodM encodes functions for efficiency of nod signal production and bacteroid maturation. J. Bacteriol. 174, 7555–7565. doi: 10.1128/jb.174.23.7555-7565.1992
Bakker, M. G., Schlatter, D. C., Otto-Hanson, L., and Kinkel, L. L. (2014). Diffuse symbioses: roles of plant-plant, plant-microbe and microbe-microbe interactions in structuring the soil microbiome. Mol. Ecol. 23, 1571–1583. doi: 10.1111/mec.12571
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Barrett, L. G., Bever, J. D., Bissett, A., and Thrall, P. H. (2015). Partner diversity and identity impacts on plant productivity in Acacia-rhizobial interactions. J. Ecol. 103, 130–142. doi: 10.1111/1365-2745.12336
Belimov, A. A., Dodd, I. C., Safronova, V. I., Shaposhnikov, A. I., Azarova, T. S., Makarova, N. M., et al. (2015). Rhizobacteria that produce auxins and contain 1-amino-cyclopropane-1-carboxylic acid deaminase decrease amino acid concentrations in the rhizosphere and improve growth and yield of well-watered and water-limited potato (Solanum tuberosum). Ann. Appl. Biol. 167, 11–25. doi: 10.1111/aab.12203
Black, M., Moolhuizen, P., Chapman, B., Barrero, R., Howieson, J., Hungria, M., et al. (2012). The genetics of symbiotic nitrogen fixation: comparative genomics of 14 Rhizobia strains by resolution of protein clusters. Genes 3, 138–166. doi: 10.3390/genes3010138
Boyd, E. S., Costas, A. M. G., Hamilton, T. L., Mus, F., and Peters, J. W. (2015). Evolution of molybdenum nitrogenase during the transition from anaerobic to aerobic metabolism. J. Bacteriol. 197, 1690–1699. doi: 10.1128/JB.02611-14
Chen, W. F., Guan, S. H., Zhao, C. T., Yan, X. R., Man, C. X., Wang, E. T., et al. (2008). Different Mesorhizobium species associated with Caragana symbiotic genes and have common host ranges. FEMS Microbiol. Lett. 283, 203–209. doi: 10.1111/j.1574-6968.2008.01167.x
Chen, W. X., Li, G. S., Qi, Y. L., Wang, E. T., Yuan, H. L., and Li, J. L. (1991). Rhizobium huakuii sp. nov. isolated from the root nodules of Astragalus sinicus. Int. J. Syst. Bacteriol. 41, 275–280. doi: 10.1099/00207713-41-2-275
Cheng, H. P., and Walker, G. C. (1998). Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J. Bacteriol. 180, 5183–5191.
Corvera, A., Promé, D., Promé, J.-C., Martínez-Romero, E., and Romero, D. (1999). The nolL gene from Rhizobium etli determines nodulation efficiency by mediating the acetylation of the fucosyl residue in the nodulation factor. Mol. Plant Microbe Interact. 12, 236–246. doi: 10.1094/MPMI.1999.12.3.236
De Meyer, S. E., Tan, H. W., Andrews, M., Heenan, P. B., and Willems, A. (2016). Mesorhizobium calcicola sp. nov., Mesorhizobium waitakense sp. nov., Mesorhizobium sophorae sp. nov., Mesorhizobium newzealandense sp. nov. and Mesorhizobium kowhaii sp. nov. isolated from Sophora root nodules. Int. J. Syst. Evol. Microbiol. 66, 786–795. doi: 10.1099/ijsem.0.000796
Dos Santos, P. C., Fang, Z., Mason, S. W., Setubal, J. C., and Dixon, R. (2012). Distribution of nitrogen fixation and nitrogenase-like sequences amongst microbial genomes. BMC Genomics 13:162. doi: 10.1186/1471-2164-13-162
Edgren, T., and Nordlund, S. (2004). The fixABCX genes in Rhodospirillum rubrum encode a putative membrane complex participating in electron transfer to nitrogenase. J. Bacteriol. 186, 2052–2060. doi: 10.1128/JB.186.7.2052-2060.2004
Evans, D. J., Jones, R., Woodley, P. R., Wilborn, J. R., and Robson, R. L. (1991). Nucleotide sequence and genetic analysis of the Azotobacter chroococcum nifUSVWZM gene cluster, including a new gene (nifP) which encodes a serine acetyltransferase. J. Bacteriol. 173, 5457–5469. doi: 10.1128/jb.173.17.5457-5469.1991
Fischer, H.-M. (1994). Genetic regulation of nitrogen fixation in Rhizobia. Microbiol. Rev. 58, 352–386.
Fuli, X., Wenlong, Z., Xiao, W., Jing, Z., Baohai, H., Zhengzheng, Z., et al. (2017). A genome-wide prediction and identification of intergenic small RNAs by comparative analysis in Mesorhizobium huakuii 7653R. Front. Microbiol. 8:1730. doi: 10.3389/fmicb.2017.01730
Garg, B., Dogra, R. C., and Sharma, P. K. (1999). High-efficiency transformation of Rhizobium leguminosarum by electroporation. Appl. Environ. Microbiol. 65, 2802–2804.
Glick, B. R., Biljana, T., Czarny, J., Cheng, Z., Duan, J., and McConkey, B. (2007). Promotion of plant growth by bacterial ACC deaminase. Crit. Rev. Plant Sci. 26, 227–242. doi: 10.1080/07352680701572966
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Hernandez, J. A., George, S. J., and Rubio, L. M. (2009). Molybdenum trafficking for nitrogen fixation. Biochemistry 48, 9711–9721. doi: 10.1021/bi901217p
Jarvis, B. D. W., Van Berkum, P., Chen, W. X., Nour, S. M., Fernandez, M. P., Cleyet-Mare, J.-C., et al. (1997). Transfer of Rhizobium loti, Rhizobium huakuii, Rhizobium ciceri, Rhizobium mediterraneum, and Rhizobium tianshanense to Mesorhizobium gen. nov. Int. J. Syst. Bacteriol. 47, 895–898. doi: 10.1099/00207713-47-3-895
Jiao, Y. S., Liu, Y. H., Yan, H., Wang, E. T., Tian, C. F., Chen, W. X., et al. (2015). Rhizobial diversity and nodulation characteristics of the extremely promiscuous legume Sophora flavescens. Mol. Plant Microbe Interact. 28, 1338–1352. doi: 10.1094/MPMI-06-15-0141-R
Lamrabet, Y., Bellogín, R. A., Cubo, T., Espuny, R., Gil, A., Krishnan, H. B., et al. (1999). Mutation in GDP-fucose synthesis genes of Sinorhizobium fredii alters Nod factors and significantly decreases competitiveness to nodulate soybeans. Mol. Plant Microbe Interact. 12, 207–217. doi: 10.1094/MPMI.1999.12.3.207
Lie, T. A., Göktan, D., Engin, M., Pijnenborg, J., and Anlarsal, E. (1987). Co-evolution of the legume-Rhizobium association. Plant Soil 100, 171–181. doi: 10.1007/BF02370940
Lohrke, S. M., Day, B., Kolli, V. S., Hancock, R., Yuen, J. P., de Souza, M. L., et al. (1998). The Bradyrhizobium japonicum noeD gene: a negatively acting, genotype-specific nodulation gene for soybean. Mol. Plant Microbe Interact. 11, 476–488. doi: 10.1094/MPMI.1998.11.6.476
Lopez-Lara, I. M., and Geiger, O. (2001). The nodulation protein NodG shows the enzymatic activity of an 3-oxoacyl-acyl carrier protein reductase. Mol. Plant Microbe Interact. 14, 349–357. doi: 10.1094/MPMI.2001.14.3.349
Ma, W., Sebestianova, S. B., Sebestian, J., Burd, G. I., Guinel, F. C., and Glick, B. R. (2003). Prevalence of 1-aminocyclopropane-1-1carboxylate deaminase in Rhizobium spp. Anthony van Leeuwenhoek 83, 285–291. doi: 10.1023/A:1023360919140
Malyschev, L. I. (2006). Flora of Siberia, Fabaceae (Leguminosae), Vol. 9. Boca Raton, FL: CRC Press. doi: 10.1201/b10743
Martínez-Hidalgo, P., Ramírez-Bahena, M. H., Flores-Félix, J. D., Igual, J. M., Sanjuán, J., León-Barrios, M., et al. (2016). Reclassification of strains MAFF 303099T and R7A into Mesorhizobium japonicum sp. nov. Int. J. Syst. Evol. Microbiol. 66, 4936–4941. doi: 10.1099/ijsem.0.001448
Montiel, J., Downie, J. A., Farkas, A., Bihari, P., Herczeg, R., Bálint, B., et al. (2017). Morphotype of bacteroids in different legumes correlates with the number and type of symbiotic NCR peptides. Proc. Natl. Acad. Sci. U.S.A. 114, 5041–5046. doi: 10.1073/pnas.1704217114
Nascimento, F. X., Brígido, C., Glick, B. R., and Oliveira, S. (2012). ACC deaminase genes are conserved among Mesorhizobium species able to nodulate the same host plant. FEMS Microbiol. Lett. 336, 26–37. doi: 10.1111/j.1574-6968.2012.02648.x
Nett, R. S., Montanares, M., Marcassa, A., Lu, X., Nagel, R., Charles, T. C., et al. (2017). Elucidation of gibberellin biosynthesis in bacteria reveals convergent evolution. Nat. Chem. Biol. 13, 69–74. doi: 10.1038/nchembio.2232
Newcomb, W., and Wood, S. M. (1986). Fine structure of nitrogen-fixing leguminous root nodules from the Canadian Arctic. Nord. J. Bot. 6, 609–626. doi: 10.1111/j.1756-1051.1986.tb00461.x
Noreen, S., Schlaman, W. R. M., Bellogin, R. A., Buendia-Claveria, A. M., Espuny, M. R., Harteveld, M., et al. (2003). Alfalfa nodulation by Sinorhizobium fredii does not require sulfated Nod-factors. Funct. Plant Biol. 30, 1219–1232. doi: 10.1071/FP03093
Notti, R. Q., and Stebbins, C. E. (2016). The structure and function of type III secretion systems. Microbiol. Spectr. 4, 1–18. doi: 10.1128/microbiolspec.VMBF-0004-2015
Novikova, N., and Safronova, V. (1992). Transconjugants of Agrobacterium radiobacter harbouring sym genes of Rhizobium galegae can form an effective symbiosis with Medicago sativa. FEMS Microbiol. Lett. 93, 261–268. doi: 10.1111/j.1574-6968.1992.tb05107.x
Okazaki, S., Tittabutr, P., Teulet, A., Thouin, J., Fardoux, J., Chaintreuil, C., et al. (2016). Rhizobium-legume symbiosis in the absence of Nod factors: two possible scenarios with or without the T3SS. ISME J. 10, 64–74. doi: 10.1038/ismej.2015.103
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Perret, X., Staehelin, C., and Broughton, W. J. (2000). Molecular basis of symbiotic promiscuity. Microbiol. Mol. Biol. Rev. 64, 180–201. doi: 10.1128/MMBR.64.1.180-201.2000
Pinto, F. G., Chueire, L. M., Vasconcelos, A. T., Nicolás, M. F., Almeida, L. G., Souza, R. C., et al. (2009). Novel genes related to nodulation, secretion systems, and surface structures revealed by a genome draft of Rhizobium tropici strain PRF 81. Funct. Integr. Genomics 9, 263–270. doi: 10.1007/s10142-009-0109-z
Provorov, N. A., and Vorobyev, N. I. (2011). Evolution of legume-rhizobium symbiosis for an improved ecological efficiency and genotypic specificity of partner interactions. Genetika 47, 417–424.
Rivilla, R., Sutton, J. M., and Downie, J. A. (1995). Rhizobium leguminosarum NodT is related to a family of outer-membrane transport proteins that includes TolC, PrtF, CyaE and AprF. Gene 161, 27–31. doi: 10.1016/0378-1119(95)00235-X
Russell, A. B., Peterson, S. B., and Mougous, J. D. (2014). Type VI secretion system effectors: poisons with a purpose. Nat. Rev. Microbiol. 51, 584–593. doi: 10.1038/nrmicro3185
Safronova, V., Belimov, A., Andronov, E., Popova, J., Tikhomirova, N., Orlova, O., et al. (2017a). Method for obtaining root nodules of the Baikal relict legumes in laboratory pot experiments. Int. J. Environ. Stud. 74, 700–705. doi: 10.1080/00207233.2017.1283948
Safronova, V., Belimov, A., Sazanova, A., Kuznetsova, I., Popova, J., Andronov, E., et al. (2017b). Does the miocene-pliocene relict legume Oxytropis triphylla form nitrogen-fixing nodules with a combination of bacterial strains? Int. J. Environ. Stud. 74, 706–714. doi: 10.1080/00207233.2017.1283947
Safronova, V., Belimov, A., Sazanova, A., Chirak, E., Verkhozina, A., Kuznetsova, I., et al. (2018a). Taxonomically different co-microsymbionts of a relict legume Oxytropis popoviana have complementary sets of symbiotic genes and together increase the efficiency of plant nodulation. Mol. Plant Microbe Interact. 31, 833–841. doi: 10.1094/MPMI-01-18-0011-R
Safronova, V. I., Sazanova, A. L., Kuznetsova, I. G., Belimov, A. A., Andronov, E. E., Chirak, E. R., et al. (2018b). Phyllobacterium zundukense sp. nov., a novel species of rhizobia isolated from root nodules of the legume species Oxytropis triphylla (Pall.) Pers. Int. J. Syst. Evol. Microbiol. 68, 1644–1651. doi: 10.1099/ijsem.0.002722
Safronova, V., and Tikhonovich, I. (2012). “Automated cryobank of microorganisms: unique possibilities for long-term authorized depositing of commercial microbial strains,” in Microbes in Applied Research: Current Advances and Challenges, ed. A. Mendez-Vilas (Hackensack, NJ: World Scientific Publishing Co), 331–334.
Safronova, V. I., Kimeklis, A. K., Chizhevskaya, E. P., Belimov, A. A., Andronov, E. E., Pinaev, A. G., et al. (2014). Genetic diversity of rhizobia isolated from nodules of the relict species Vavilovia formosa (Stev.) Fed. Antonie van Leeuwenhoek 105, 389–399. doi: 10.1007/s10482-013-0089-9
Safronova, V. I., Kuznetsova, I. G., Sazanova, A. L., Kimeklis, A. K., Belimov, A. A., Andronov, E. E., et al. (2015a). Bosea vaviloviae sp. nov. a new species of slow-growing rhizobia isolated from nodules of the relict species Vavilovia formosa (Stev.) Fed. Antonie van Leeuwenhoek 107, 911–920. doi: 10.1007/s10482-015-0383-9
Safronova, V. I., Kuznetsova, I. G., Sazanova, A. L., Kimeklis, A. K., Belimov, A. A., Andronov, E. E., et al. (2015b). Extra slow-growing Tardiphaga strains isolated from nodules of Vavilovia formosa (Stev.) Fed. Arch. Microbiol. 197, 889–898. doi: 10.1007/s00203-015-1122-3
Saleh, S. S., and Glick, B. R. (2001). Involvement of gacS and rpoS in enhancement of the plant growth-promoting capabilities of Enterobacter cloacae CAL2 and UW4. Can. J. Microbiol. 47, 698–705. doi: 10.1139/w01-072
Serova, T. A., Tsyganova, A. V., and Tsyganov, V. E. (2018). Early nodule senescence is activated in symbiotic mutants of pea (Pisum sativum L.) forming ineffective nodules blocked at different nodule developmental stages. Protoplasma 255, 1443–1459. doi: 10.1007/s00709-018-1246-9
Shamseldin, A., and Werner, D. (2004). Selection of competitive strains of Rhizobium nodulating Phaseolus vulgaris and adapted to environmental conditions in Egypt, using the gus-reporter gene technique. World J. Microbiol. Biotechnol. 20, 377–382. doi: 10.1023/B:WIBI.0000033060.27180.8c
Sinjushin, A. A., Demidenko, N. V., and Gostimskii, S. A. (2009). Preliminary report on taxonomical position of Vavilovia formosa (Stev.) Fed. evidenced from morphological and molecular data. Pisum Genet. 41, 15–20.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parismony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tikhonovich, I. A., and Provorov, N. A. (2009). From plant-microbe interactions to symbiogenetics: a universal paradigm for the inter-species genetic integration. Ann. Appl. Biol. 154, 341–350. doi: 10.1111/j.1744-7348.2008.00306.x
Torres Tejerizo, G., Del Papa, M. F., Soria-Diaz, M. E., Draghi, W., Lozano, M., Giusti Mde, L., et al. (2011). The nodulation of alfalfa by the acid-tolerant Rhizobium sp. strain LPU83 does not require sulfated forms of lipochitooligosaccharide nodulation signals. J. Bacteriol. 193, 30–39. doi: 10.1128/JB.01009-10
Tsyganova, A. V., Kitaeva, A. B., and Tsyganov, V. E. (2018). Cell differentiation in nitrogen-fixing nodules hosting symbiosomes. Funct. Plant Biol. 45, 47–57. doi: 10.1071/FP16377
Turner, G. L., and Gibson, A. H. (1980). “Measurement of nitrogen fixation by indirect means,” in Methods for Evaluating Biological Nitrogen Fixation, ed. F. J. Bergensen (Toronto: Wiley), 111–138.
Turuta, O., Ryabtsev, V., Novitskaya, N., and Vakarenko, L. (2015). Protected Microhabitats as a Part of Baikal Regional Ecological Network. Plant Microreserves the Site Based Plant Conservation And Monitoring Network. Available at: http://cretaplant.biol.uoa.gr/docs/Turuta2001.pdf
Vincent, J. M. (1970). A Manual for the Practical Study of Root Nodule Bacteria in IBP Handbook. Oxford: Blackwell Scientific Publications, 73–97.
Wais, R. J., Keating, D. H., and Long, S. R. (2002). Structure-function analysis of Nod factor-induced root hair calcium spiking in Rhizobium-legume symbiosis. Plant Physiol. 129, 211–224. doi: 10.1104/pp.010690
Wdowiak, S., Małek, W., Sajnaga, E., Łotocka, B., Stepkowski, T., and Legocki, A. (2000). Symbiosis of Astragalus cicer with its microsymbionts: partial nodC gene sequence, host plant specificity, and root nodule structure. Antonie Van Leeuwenhoek 78, 63–71. doi: 10.1023/A:1002738609114
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. doi: 10.1128/jb.173.2.697-703.1991
Keywords: plant-microbe interaction, symbiosis, nodulation, complementarity of genes, Astragalus chorinensis, Oxytropis popoviana
Citation: Safronova V, Belimov A, Sazanova A, Chirak E, Kuznetsova I, Andronov E, Pinaev A, Tsyganova A, Seliverstova E, Kitaeva A, Tsyganov V and Tikhonovich I (2019) Two Broad Host Range Rhizobial Strains Isolated From Relict Legumes Have Various Complementary Effects on Symbiotic Parameters of Co-inoculated Plants. Front. Microbiol. 10:514. doi: 10.3389/fmicb.2019.00514
Received: 06 October 2018; Accepted: 28 February 2019;
Published: 15 March 2019.
Edited by:
Lorena Carro, Newcastle University, United KingdomReviewed by:
Andrea Squartini, University of Padova, ItalyCopyright © 2019 Safronova, Belimov, Sazanova, Chirak, Kuznetsova, Andronov, Pinaev, Tsyganova, Seliverstova, Kitaeva, Tsyganov and Tikhonovich. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vera Safronova, di5zYWZyb25vdmFAcmFtYmxlci5ydQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.