
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 13 March 2019
Sec. Microbial Physiology and Metabolism
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00501
More than 200 plants have been suffering from Verticillium wilt caused by Verticillium dahliae (V. dahliae) across the world. The target of rapamycin (TOR) is a lethal gene and controls cell growth and development in various eukaryotes, but little is known about TOR signaling in V. dahliae. Here, we found that V. dahliae strain is hypersensitive to rapamycin in the presence of rapamycin binding protein VdFKBP12 while the deletion mutant △vdfkbp12 is insensitive to rapamycin. Heterologous expressing VdFKBP12 in Arabidopsis conferred rapamycin sensitivity, indicating that VdFKBP12 can bridge the interaction between rapamycin and TOR across species. The key across species of TOR complex 1 (TORC1) and TORC2 have been identified in V. dahliae, suggesting that TOR signaling pathway is evolutionarily conserved in eukaryotic species. Furthermore, the RNA-seq analysis showed that ribosomal biogenesis, RNA polymerase II transcription factors and many metabolic processes were significantly suppressed in rapamycin treated cells of V. dahliae. Importantly, transcript levels of genes associated with cell wall degrading enzymes (CWEDs) were dramatically down-regulated in TOR-inhibited cells. Further infection assay showed that the pathogenicity of V. dahliae and occurrence of Verticillium wilt can be blocked in the presence of rapamycin. These observations suggested that VdTOR is a key target of V. dahliae for controlling and preventing Verticillium wilt in plants.
Verticillium dahliae (V. dahliae) is a soil-borne and hemibiotrophic fungus that causes over 200 plant species wilting including Brassicaceae, Rosaceae, and Solanaceae plants, resulting in tremendous economic losses every year (Pegg and Brady, 2002; Inderbitzin and Subbarao, 2014; Gomez-Lama Cabanas et al., 2015; Chen et al., 2017). V. dahliae is the agent of Verticillium wilt, which is one of the most devastating cotton diseases worldwide. V. dahliae is particularly difficult to manage because it long exists in soil as a dormant structure called microsclerotia. The microsclerotia are primary infectious propagules that can remain alive in soil for more than 20 years (Agrios, 2005; Bui et al., 2018). Hyphopodium differentiates from hypha after conidia germination on the root surface and develops a penetration peg to infect plant roots (Zhao et al., 2016). Hyphal neck from penetration peg partitions the hyphopodium and the invasive hypha and forms a specialized fungus–host interface to deliver secretory proteins into host (Zhou et al., 2017). The plant cell wall is an important interface for the interaction between host and phytopathogenic fungi, which plays a major barrier role in the process of phytopathogenic fungi invading the host. Most fungal pathogens secrete lots of cell wall degrading enzymes (CWDEs) including cellulases, xylanases, and pectinases to depolymerize the host cell wall (Tonukari, 2003; Quoc and Chau, 2017). V. dahliae have been reported to produce CWDEs for degrading plant cell wall (Cooper and Wood, 1980; Tzima et al., 2011; Chen et al., 2016). Endoglucanase-1 (EG-1) is an important enzyme in depolymerization of plant cellulose (Novo et al., 2006; Valaskova and Baldrian, 2006). The EG-1 gene homolog VdEg-1 plays an important role in plant penetration and colonization. The VdEg-1 mutant lost the ability to colonize vascular tissues in inoculated plants (Maruthachalam et al., 2011). Moreover, pectinases play a critical role in pathogenesis and production levels correlated with pathogenicity in different Verticillium strains (Durrands and Cooper, 1988; Fradin and Thomma, 2006; Tzima et al., 2011; Chen et al., 2016).
Target of rapamycin (TOR) is an evolutionarily conserved phosphoinositide-3 kinase-related protein kinase that controls multiple cellular processes in response to various intracellular and extracellular signals (De Virgilio and Loewith, 2006; Shimobayashi and Hall, 2014; Dobrenel et al., 2016; Saxton and Sabatini, 2017). It was originally identified in budding yeast through mutant screens for resistance to the immunosuppressant drug rapamycin (Heitman et al., 1991a). Subsequent identification of TOR in humans and other eukaryotes revealed evolutionary conservation of TOR from the last eukaryotic common ancestor to humans (Soulard et al., 2009; Katz, 2012; Tatebe and Shiozaki, 2017). TOR exists in two functionally and structurally distinct complexes: TOR complex 1 (TORC1) and TORC2. The essential core components of TORC1 are TOR, RAPTOR (regulatory-associated protein of TOR) and LST8 (lethal with SEC thirteen 8), which controls cell growth by regulating translation, transcription and autophagy (Wang and Proud, 2009; Iadevaia et al., 2014; Dobrenel et al., 2016); whereas, those of TORC2 are TOR, RICTOR (rapamycin-insensitive companion of TOR), SIN1 (SAPK-interacting 1) and LST8 (Hara et al., 2002; Jacinto et al., 2004; De Virgilio and Loewith, 2006; Gaubitz et al., 2016). TORC2 responds primarily to growth factors, promoting cell survival, cell cycle and actin cytoskeleton polarization (Jacinto et al., 2004; Oh and Jacinto, 2011; Gaubitz et al., 2016).
Rapamycin (RAP) is a new macrolide immunosuppressant drug produced by Streptomyces hygroscopicus. RAP specifically binds to FKBP12 (FK506 binding protein of 12 kD), which interacts with the FRB domain of TOR kinase to inhibit TORC1 activity (Heitman et al., 1991b; Hara et al., 2002). Crystal structure showed that the ternary complex of RAP-FKBP12-FRB domain of TOR partially occludes substrates to the active site of TOR (Yang et al., 2013; Aylett et al., 2016). However, TORC2 is RAP insensitive (Loewith et al., 2002). The C-terminus of RICTOR prevents the RAP-FKBP12 complex from binding to the FRB domain of TOR protein in TORC2, which makes TORC2 insensitive to RAP (Gaubitz et al., 2015). In addition, the ATP-competitive TOR protein kinase inhibitors including Torin1, Torin2, Ku-0063794 and AZD-8055 can directly bind to the kinase domain of TOR by competing with ATP to inhibit TORC1 and TORC2 activity (Garcia-Martinez et al., 2009; Chresta et al., 2010; Liu et al., 2010, 2011).
The TOR signaling pathway is a central regulator in regulating cell growth, proliferation and metabolism from yeasts to humans (Rexin et al., 2015; Dobrenel et al., 2016; Saxton and Sabatini, 2017). There is no research on the function of TOR signaling pathway in V. dahliae. In this study, we found that the mycelial growth of V. dahliae was retarded by RAP, implying that VdFKBP12 may be functional in mediate RAP and VdTOR. Further functional analysis of △vdfkbp12 and VdFKBP12 overexpression transgenic Arabidopsis suggests that VdFKBP12 can mediate the inhibition of TOR kinase by RAP in V. dahliae. The conserved TOR signaling pathway including TORC1 and TORC2 existing in V. dahliae, indicating that TOR signaling pathway is evolutionarily conserved in eukaryotic species. Additionally, RNA-seq experiments were performed to test the function of VdTOR. A large number of differentially expression genes (DEGs) involving in various cellular processes, such as ribosome biogenesis and CWDEs, were observed in RAP treatment. Importantly, the most of CWDEs are down-regulated in TOR-inhibited cells, implying that TOR involved in the regulation of invasion. Infection assay showed that the pathogenicity of V. dahliae and occurrence of Verticillium wilt can be blocked in the presence of RAP. These independent evidences indicated that RAP inhibits mycelial growth and pathogenicity through reducing VdTOR activity in V. dahliae.
The highly aggressive defoliating isolate Vd991 of V. dahliae was used as the wild-type (WT) strain in this study. The WT strain, deletion mutants and complemented strains were cultured on potato dextrose agar (PDA) at 27°C. For extraction of genomic DNA and conidia production, hyphae were incubated in potato dextrose broth (PDB) at 27°C with shaking at 160 rpm.
The primers for gene deletion and complementation were listed in Supplementary Table 1. Constructs for gene deletion and complementation of V. dahliae were carried out as described previously (Luo et al., 2016). Agrobacterium tumefaciens strain AGL-1 was used to transform the conidia of V. dahliae by using ATMT. The Agrobacterium tumefaciens strain AGL-1 containing gene deletion or complementation vector was mixing with equal volume of conidial suspension of V. dahliae (107 conidia. mL-1). Then, 200 μL of the mixture was placed onto microporous membranes (pore size, 0.45 μm) on cocultivation medium for 48 h. Subsequently, the membranes were transferred to PDA medium containing hygromycin antibiotic (50 mg. mL-1) and cefotaxime (300 mg. mL-1). After 10 days, transformants were transferred to fresh PDA medium with hygromycin (50 mg. mL-1) for further analysis.
Spores of V. dahliae were grown for 5 days in PDB medium at 27°C with shaking at 160 rpm, and then treated with 5 nM RAP and DMSO (as a control) for 24 h, respectively. Total RNA of V. dahliae mycelium was isolated using the Hipure Fungal RNA Kit (Magen, Guangzhou, China). For each treatment, three independent biological replicates were performed. The cDNA library construction was done as described previously (Marioni et al., 2008). An Illumina Hiseq 2000 platform was used to sequence these libraries. The clean reads were mapped to the reference V. dahliae genome database website1 by using TopHat2 software (Kim et al., 2013). Cufflinks and Cuffdiff were used to assemble the mapped reads and identify differentially expressed genes (DEGs), respectively (Trapnell et al., 2010, 2013). Gene ontology (GO) enrichment (corrected P-value < 0.05) of DEGs was performed by using GOseq R package software (Young et al., 2010). The enrichment of DEGs in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (corrected P-value < 0.05) was obtained by using KOBAS software (Kanehisa et al., 2008).
Total RNA of V. dahliae which treated with DMSO and RAP (5 nM) for 24 h in PDB medium was extracted using the Hipure Fungal RNA Kit (Magen, Guangzhou, China). The software Primer premier 5.0 was used to design quantitative real-time primers (Supplementary Table 2). Vd18S rRNA was used as a control. Reaction was performed in a final volume of 25 μL containing 12.5 μL of 2 × SYBR®Premix Ex Taq (Takara, Dalian, China). The relative expression level of each target gene was analyzed with the Bio-Rad CFX Manager software. The data represented the mean ± SD of three independent experiments.
Pathogen inoculation was performed by root dip inoculation with conidia of V. dahliae (107 conidia. mL-1) as described previously (Luo et al., 2016). Plants of similar height were selected for each treatment. For the root dip inoculation, 5-week-old roots of cotton plants were immersed in 200 mL of conidia suspension supplemented with or without 50 nM RAP for 10 min. Equal volume DMSO as a solvent control, the final concentration of DMSO was 0.25% (v/v). Then the plants were re-cultivated in soil until the disease symptoms appeared. The roots of cotton plants were immersed in 200 mL sterile water for 10 min and then re-cultured in soil as a control. Cellophane penetration assay was performed as described previously (Prados Rosales and Di Pietro, 2008). The experiments were repeated at least three times.
Cellulose content was measured with the anthrone method previously described (Ververis et al., 2004). Cellulose content of the sample was measured by ultraviolet spectrophotometer of absorbance at 620 nm. Commercial cellulose was used as control for standard curve.
Pectin content was calculated based on the analysis of uronic acid content. The content of uronic acids was measured with the biphenol method as described previously (Blumenkrantz and Asboe-Hansen, 1973). The content of uronic acids was measured by ultraviolet spectrophotometer of absorbance at 525 nm. Commercial galacturonic acid was used as a standard for the calibration curve.
Combination index (CI) values were used to evaluate the interaction between RAP and Torin1. The degree of reagents interaction is based on synergistic effect (CI < 1), additive effect (CI = 1), or antagonism (CI > 1) (Chou, 2006). Spores of V. dahliae were treated with different concentrations RAP and Torin1, or pairwise combination of RAP + Torin1 on PDA medium for 11 days at 27°C. Colony diameter was measured to calculate CI values. Experiments were repeated at least three times. The values of affected fraction (Fa) were calculated according to the CompuSyn sofware program (Chou and Talalay, 1984).
V. dahliae is the main pathogen of Verticillium wilt, which is a devastating plant disease that causes a variety of economic crops wilting, including cotton, tomato, and eggplant (Pegg and Brady, 2002; Inderbitzin and Subbarao, 2014). Rapamycin (RAP) is a broad-spectrum antifungal drug that effectively inhibits pathogenic fungi (Bastidas et al., 2008; Benjamin et al., 2011; Yu et al., 2014). In order to test the antifungal activity of RAP on V. dahliae, RAP was applied to V. dahliae. As expected, with increasing concentrations of RAP, the hyphal growth was subjected to different degrees of inhibition (Figures 1A,C). Meanwhile, Torin1, the second-generation inhibitor of TOR, also inhibits hyphal growth in a dose dependent manner (Figures 1B,D). The IC50 (half-maximal inhibitory concentration) values of RAP and Torin1 were 5 nM and 80 μM, respectively. These results indicated that RAP has a stronger inhibitory effect than Torin1. This may be due to the specific spatial structure of VdTOR, which requires a high concentration of Torin1 to inhibit the kinase activity of VdTOR. Furthermore, the pairwise combination of RAP and Torin1 had enhanced inhibition for hyphal growth compared with RAP or Torin1 alone treatment. The single IC50 value (RAP: 2 nM; Torin1: 5 μM) was significantly reduced when V. dahliae was subjected to RAP and Torin1 combination treatment (Figures 1E,F). The results implied that the potential synergistic effects can be generated by combining RAP and Torin1. Next, Fa-CI curve was generated by using the CompuSyn software (Chou, 2006). Synergistic effect (CI < 1) was observed when hyphae were treated with combination of RAP and Torin1 (Figure 1G), showing that the combination of RAP and Torin1 may have synergistic effect. Additionally, swollen hyphae and shorter septation were observed in RAP, Torin1, and RAP + Torin1 treatment compared with the control (DMSO) (Figure 2). These results implied that RAP and Torin1 might synergistically inhibit hyphal growth in V. dahliae.
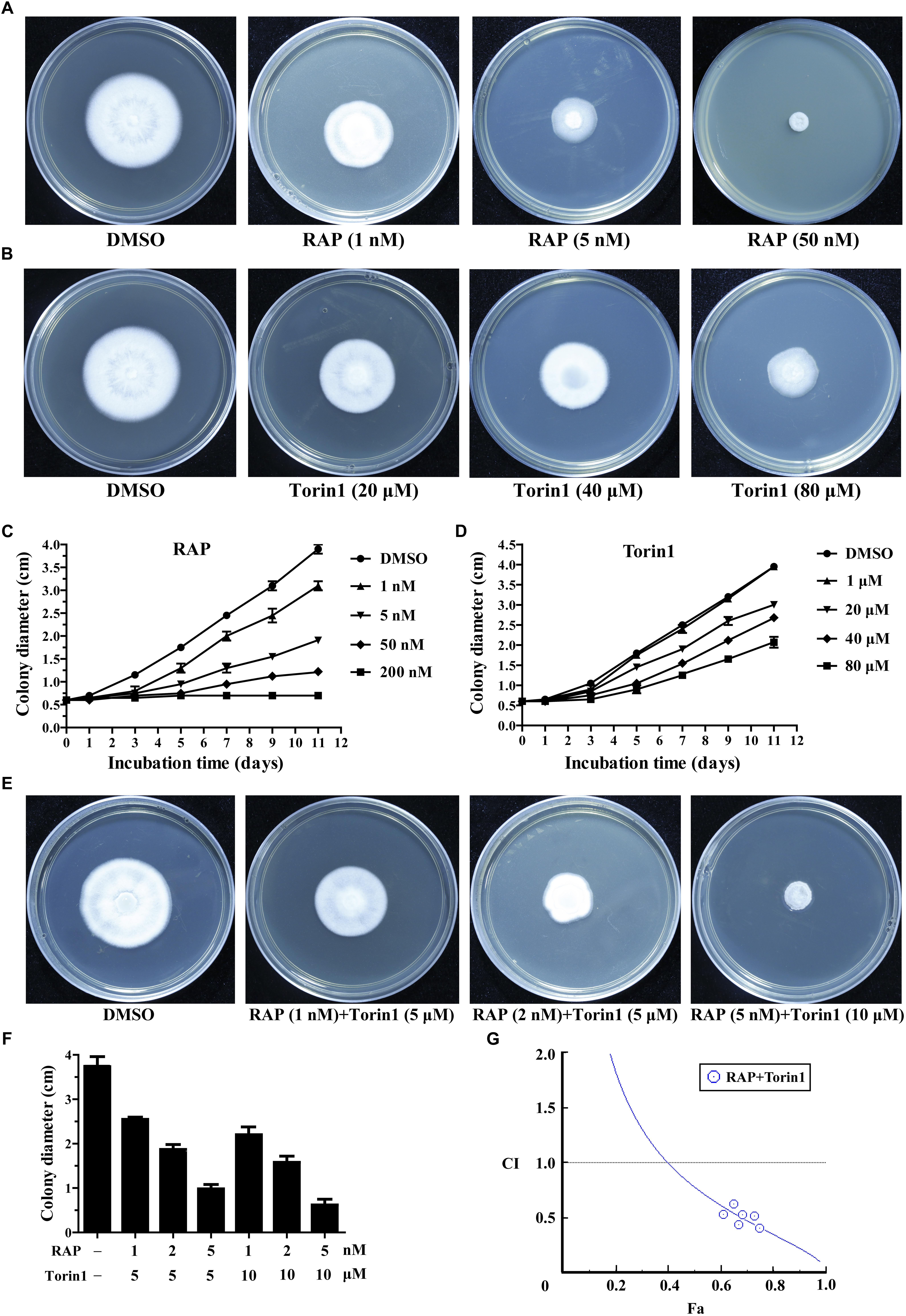
Figure 1. V. dahliae was sensitive to RAP and Torin1 in a dose dependent manner. (A) Spores of V. dahliae were incubated on potato dextrose agar (PDA) including different concentrations RAP for 11 days. (B) Spores of V. dahliae were incubated on PDA medium including different concentrations Torin1 for 11 days. (C) Colony diameter of V. dahliae was incubated on PDA medium including different concentrations RAP from 0 to 11 days. The data represents the mean ± SD of n = 3 independent experiments. (D) Colony diameter of V. dahliae was incubated on PDA medium including different concentrations Torin1 from 0 to 11 days. The data represents the mean ± SD of n = 3 independent experiments. (E) Spores of V. dahliae were incubated on PDA including different concentrations combination of RAP and Torin1 for 11 days. (F) Colony diameter of V. dahliae was incubated on PDA medium including different combination of RAP and Torin1 for 11 days. The data represents the mean ± SD of n = 3 independent experiments. (G) Fa-CI curve shows synergism (CI < 1) between RAP and Torin1.

Figure 2. TOR inhibitors RAP and Torin1 can inhibit the mycelial growth of V. dahliae. Hyphae were incubated in potato dextrose broth (PDB) with RAP (5 nM) and Torin1 (80 μM) for 24 h, and then were photographed by scanning electron microscopy. Bar = 3 μm.
To determine whether RAP and Torin1 inhibit conidial development, we examined germination rate of conidia and spore production. The germination rate of conidia and spore production were obvious reduction by RAP and Torin1 treatment (Supplementary Figure 1A). Meanwhile, the expression levels of genes involved in conidial development including VDAG_JR2_Chr4g03070 (Vdcon10), VDAG_JR2_Chr2g09020 (Vdfluffy), VDAG_JR2_Chr7g08730 (VdPKAC1) and VDAG_JR2_Chr8g02550 (VdSge1) genes (Tzima et al., 2010; Santhanam and Thomma, 2013) were significantly down-regulated in RAP and Torin1 treatment (Supplementary Figure 1B). These data suggested that RAP and Torin1 can inhibit conidial development in V. dahliae. Besides, we examined the expression levels of genes involved in vegetative growth and virulence (Klimes and Dobinson, 2006; Gao et al., 2010; Zhou et al., 2012). The expression of VDAG_JR2_Chr2g02500 (VDH1), VDAG_JR2_Chr3g07080 (GARP1), VDAG_JR2_Chr6g08770 (NLP1), and VDAG_JR2_Chr2g05460 (NLP2) were also significantly down-regulated in RAP and Torin1 treatment (Supplementary Figure 1C), suggesting that TOR inhibition reduced expression levels of vegetative growth and virulence-related genes.
Rapamycin is a well-known TOR inhibitor which specifically targets the TOR protein (Benjamin et al., 2011). TOR is a central regulator of cell growth and metabolism in various eukaryotic species from yeasts, plants to humans (Yu et al., 2014; Rexin et al., 2015; Saxton and Sabatini, 2017). In order to identify evolutionary conserved TOR signaling pathway components in V. dahliae. A BLASTp analysis of the V. dahliae genome database (see footnote 1) was performed by using yeast TOR signaling pathway components as reference. A putative homologous gene encoding the key TOR protein (VDAG_JR2_Chr6g10810) locates on chromosome 6 was found in V. dahliae genome database (Table 1). The TOR gene sequence contains 3 introns and 4 exons, which encodes 2442 amino acid residues with molecular mass of 276 kDa (Figure 3A). Alignment of VdTOR with other species TOR proteins showed similar conserved domains including N-term region, FAT, FRB, kinase, and FATC domains as yeast (Figure 3B). Phylogenetic analysis (Figure 3C) and kinase domain alignment with that from other organisms (Figure 3D) indicated that VdTOR was evolutionarily conserved. To further determine VdTOR function, targeted gene replacement was performed in V. dahliae strain. All of hygromycin-resistant transformants of VdTOR deletion mutants were ectopic mutants and failure in creating a null mutant, implying that deletion of VdTOR gene may be lethal. Besides, we also found other homologous genes encoding the key proteins of TORC1 including RAPTOR and LST8. Meanwhile, putative homologs of TORC2 specific proteins including RICTOR and SIN1 were also present in V. dahliae genome (Table 1). These results indicated that exist a conserved and functional TORC1 and TORC2 in V. dahliae.
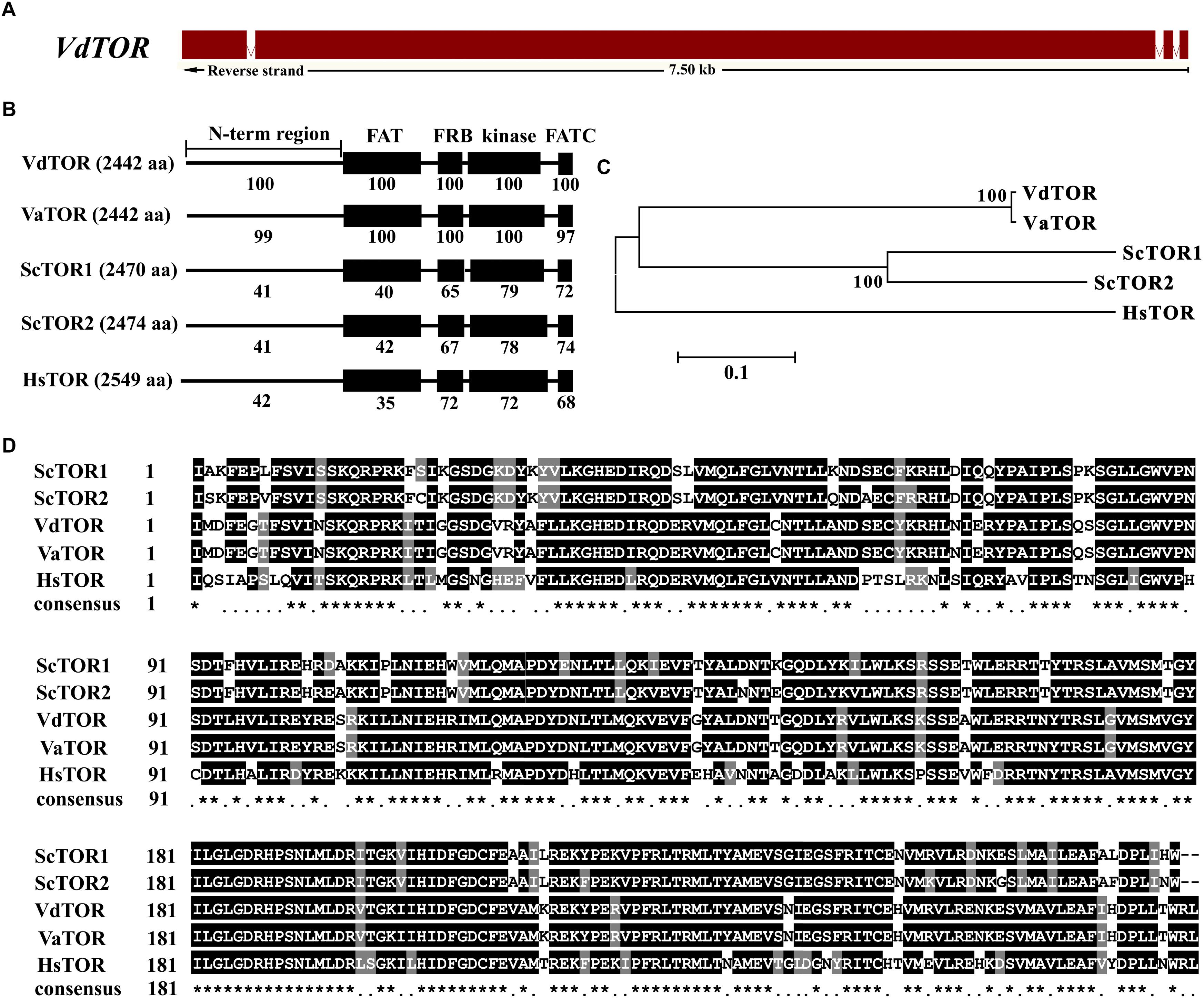
Figure 3. Sequence and structure analysis of VdTOR gene. (A) Sequence of VdTOR gene. Red represents exons and the white rectangles signify introns. (B) Comparison conserved domains of VdTOR proteins with that from other organisms. Each value indicates the percentage of identity with the corresponding domain sequences of VdTOR. The number in brackets represents the number of amino acids. Vd, Verticillium dahliae; Va, Verticillium alfalfa; Hs, Homo sapiens; Sc, Saccharomyces cerevisiae. (C) Phylogenetic relationship between the VdTOR protein and homologs from other organisms. The phylogenetic tree was generated with MEGA4.0 using the neighbor-joining methods. (D) Comparison of amino acid sequences of the kinase domain of VdTOR protein with that from other representative organisms. The ∗ represents identical amino acid residues.
Rapamycin specifically forms non-covalent to link the interaction between FKBP12 and FRB domain of TOR protein. The formation of the ternary complex of RAP-FKBP12-FRB domain of TOR is necessary for RAP response in eukaryotes (Menand et al., 2002; Sormani et al., 2007). V. dahliae owns one FKBP12 ortholog VdFKBP12 (VDAG_JR2_Chr1g18420) locating on chromosome 1, which encodes a protein with 57% similarity to ScFKBP12 (Table 1). The amino acid sequences alignment (Figure 4A) and phylogenetic analysis (Figure 4B) with that from other organisms indicated that VdFKBP12 was evolutionarily conserved. To assess the role of VdFKBP12 gene, knockout transformants (△fkbp12) were generated by using homologous recombination gene deletion strategy (Supplementary Figure 2). No aberrant phenotype was observed under normal condition (Figure 4C). RAP sensitivity test showed that △fkbp12 mutant resistance to RAP, but the sensitivity to RAP was restored in the complementary strain (△fkbp12 + FKBP12) (Figure 4C). Early studies showed that heterologous expressing ScFKBP12 in Arabidopsis can restore the sensitivity to RAP (Sormani et al., 2007; Ren et al., 2012). To further confirm the VdFKBP12 function on bridging the interaction between TOR and RAP, the VdFKBP12 gene was introduced into Arabidopsis and produced VdFKBP12 overexpression transgenic Arabidopsis lines. The transgenic lines displayed the sensitivity to RAP compared with WT Arabidopsis (Figure 4D). Results showed that all transgenic lines displayed shorter primary root length, smaller cotyledon and the decreasing fresh weight compared with WT Arabidopsis (Figures 4D,E), and this observation was consistent with that ScFKBP12 overexpression line (BP12-2) in Arabidopsis (Ren et al., 2012). These results indicated that RAP inhibits the activity of VdTOR protein by VdFKBP12 in V. dahliae.
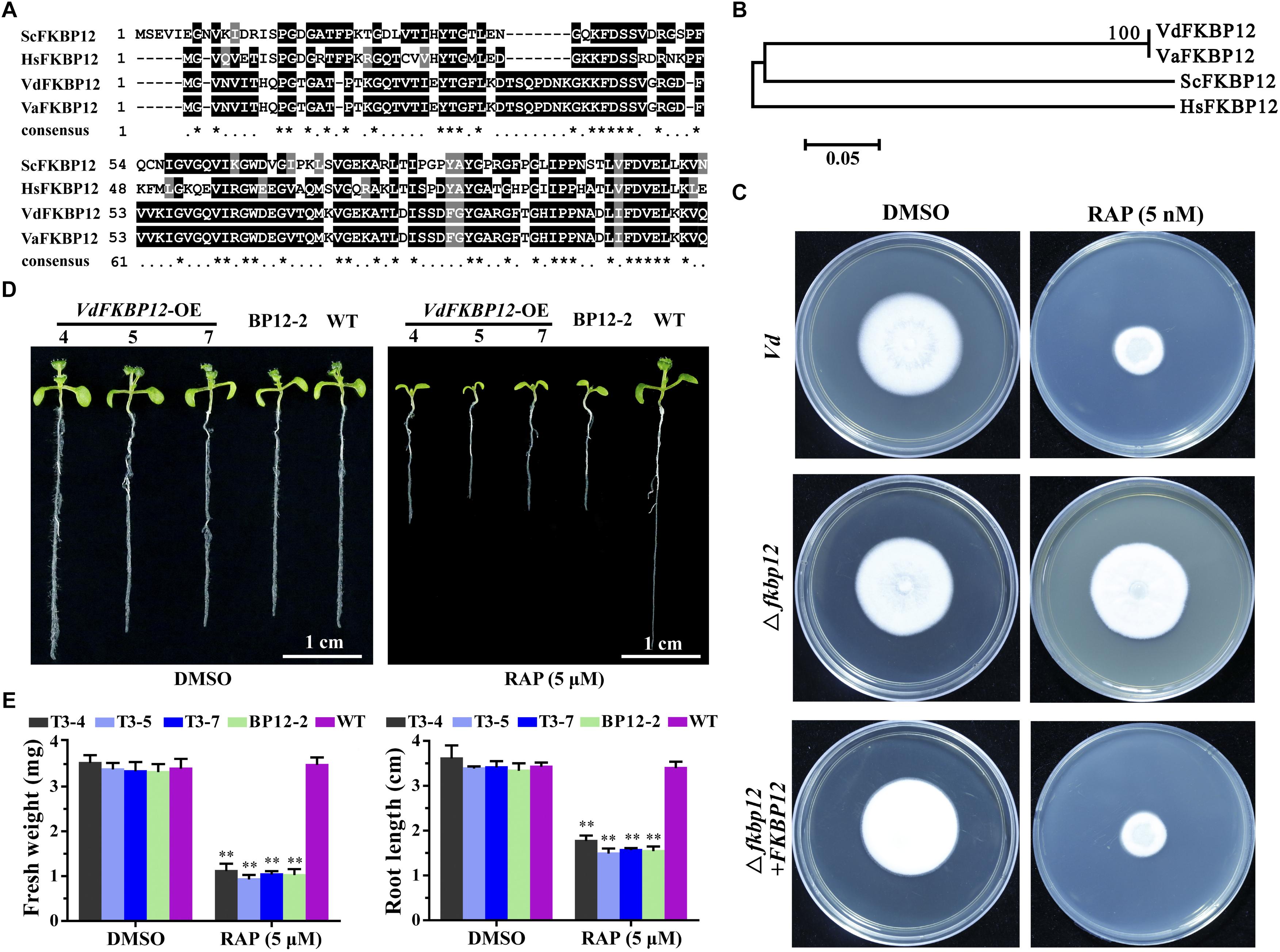
Figure 4. Rapamycin binds to FKBP12 to inhibit TOR activity in V. dahliae. (A) Comparison of amino acid sequences of the FKBP12 from Vd, Va, Hs and Sc. The ∗ represents identical amino acid residues. Vd, Verticillium dahliae; Va, Verticillium alfalfa; Hs, Homo sapiens; Sc, Saccharomyces cerevisiae. (B) Phylogenetic relationship between V. dahliae FKBP12 protein and homologs from other organisms in (A). The phylogenetic tree was generated with MEGA4.0 using the neighbor-joining methods. (C) Deletion of VdFKBP12 (△fkbp12) leads to resistance to RAP in V. dahliae. Spores of Vd, △fkbp12 and the complementary strain (△fkbp12+FKBP12) were incubated on PDA including 5 nM RAP for 11 days. (D) VdFKBP12 overexpression transgenic Arabidopsis lines were sensitive to RAP (5 μM) treatment. Bar = 1 cm. (E) Fresh weight and root length of VdFKBP12 overexpression transgenic Arabidopsis lines treated with RAP (5 μM). The data represents the mean ± SD of n = 3 independent experiments. Asterisks denote Student’s t-test significant difference compared with WT plants (∗∗P < 0.01).
To further elucidate the function of VdTOR signaling pathway on vegetative growth of V. dahliae, gene expression profile analysis was performed in V. dahliae hyphae under the condition of VdTOR inhibition. RNA-seq was conducted in V. dahliae treated with 5 nM RAP and equal volume of DMSO as control, respectively. After stringent quality checking and data cleaning, approximately 70% of the reads can be mapped to the annotated V. dahliae genome (Figure 5A). 5754 differentially expressed genes (DEGs) were found between RAP treatment and DMSO control, of which 2,895 DEGs were up-regulated and 2,859 DEGs were down-regulated (Figures 5B,C). Some DEGs were randomly selected from the RNA-seq data to verify the reliability of RNA-seq data by quantitative real-time PCR. The result displayed the same trends as gained in RNA-seq data (Supplementary Figure 3).

Figure 5. RNA-seq analysis of V. dahliae hyphae treated with DMSO and RAP. (A) Proportions of clean reads of unmapped, mapped to multiple genes and mapped to unique genes, which were plotted by three replicates of RAP and DMSO. (B) The number of down-regulated and up-regulated differentially expressed genes for RAP and DMSO treatment. (C) Cluster analysis of differentially expressed genes for RAP and DMSO treatment. (D) Significantly up-regulated enriched gene ontology for RAP treatment in the RNA-seq database. Gene ontology was ranked by their significance. (E) Significantly down-regulated enriched gene ontology for RAP treatment in the RNA-seq database. Gene ontology was ranked by their significance.
To understand the function of these DEGs, GO assignments and enrichments were analyzed. A total of 214 up-regulated GO terms and 139 down-regulated GO terms were enriched (Supplementary Table 3). In the up-regulated GO pathway category, peptide biosynthetic process (GO: 0043043) was the most significant enrichment (Figure 5D). In the down-regulated group, RNA polymerase II transcription factor activity (GO: 0000981) and cation binding (GO: 0043169) were highly represented (Figure 5E). These data showed that VdTOR regulates multiple cellular processes in V. dahliae.
The process of ribosome biogenesis is conserved from prokaryotes to eukaryotes. TORC1 controls the transcription of genes encoding ribosomal proteins and ribosome biogenesis in response to extracellular and intracellular signals in plants, mammals, and yeasts (Wei and Zheng, 2009; Ren et al., 2011; Chauvin et al., 2014; Kos-Braun and Kos, 2017). Through analysis of the RNA-seq data, we found 47 DEGs associated with ribosome biogenesis genes, including 36 down-regulated genes and 11 up-regulated genes, were assigned to the “ribosome biogenesis in eukaryotes” KEGG pathway (Figure 6 and Supplementary Table 4). Within these DEGs, the genes encoding nucleolar proteins 4 and 58 (NOP4 and NOP58) and U3 small nuclear RNA-associated proteins were down-regulated. These ribosomal core proteins combine with small nucleolar RNAs to form small nucleolar ribonucleoproteins that play an indispensible role in ribosome biogenesis (Sun and Woolford, 1994; Gautier et al., 1997; Qiu et al., 2008). These results indicated that VdTOR involved in the regulation of ribosome biogenesis in V. dahliae.
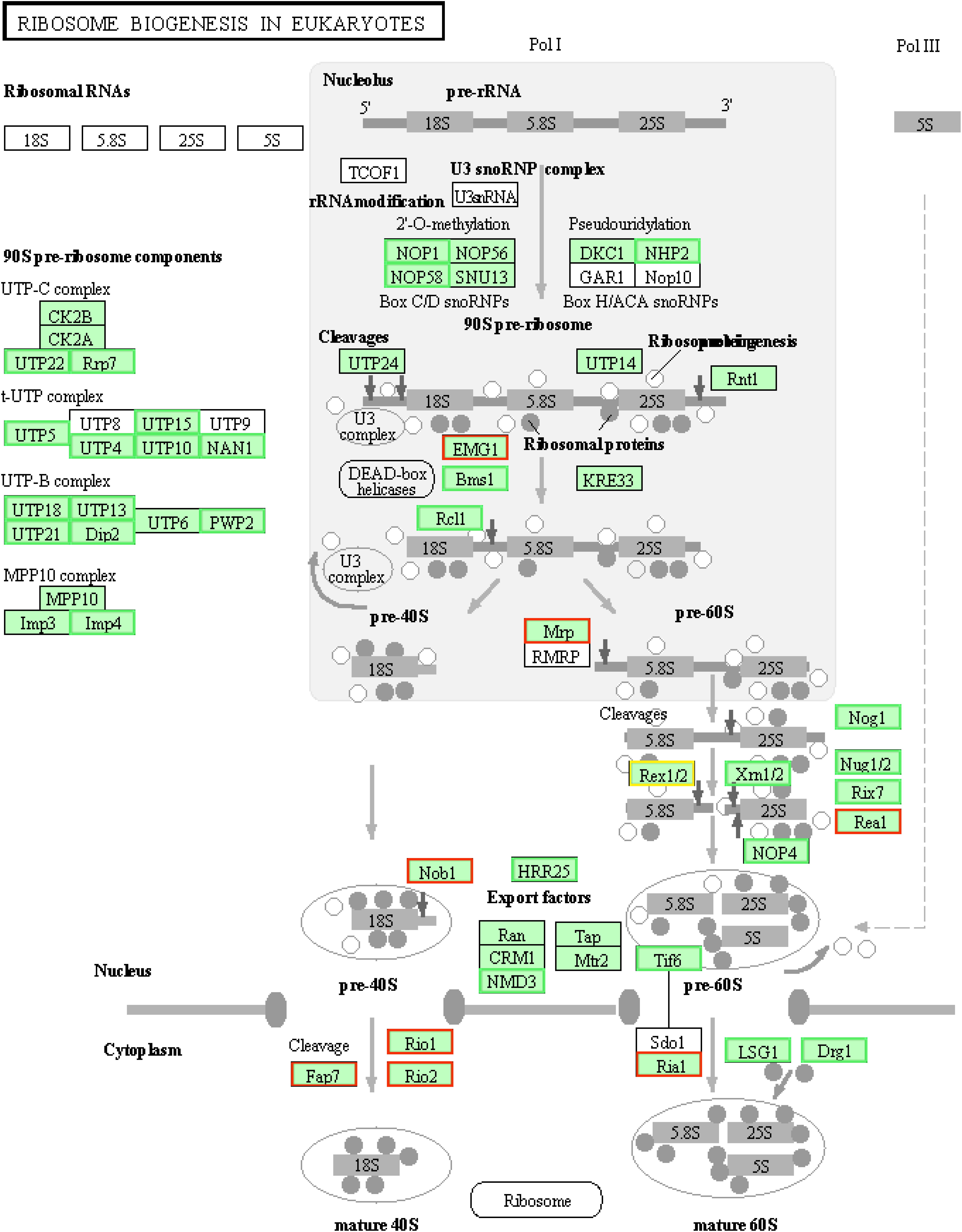
Figure 6. TOR inhibition by RAP induced transcription level changes of ribosome biogenesis-related genes in V. dahliae. Expression changes of genes in the ribosome biogenesis in eukaryotes. Green boxes, down-regulated genes. Red boxes, up-regulated genes.
Carbohydrate metabolism and synthesis of proteins and lipids are important limiting factors for cells growth and proliferation (Ren et al., 2012; Yuan et al., 2013; Saxton and Sabatini, 2017). Carbon metabolism, amino acids metabolism, and fatty acid metabolism were changed in the RNA-seq data (Table 2). Particularly, the genes encoding rate-limiting enzymes of carbohydrate metabolism and biosynthesis of amino acids such as fructose-bisphosphate aldolase and isocitrate dehydrogenase were down-regulated, but expression levels of several genes involved in nitrogen metabolism such as glutamine synthetase, ammonium permease and NAD-specific glutamate dehydrogenase were up-regulated (Table 2). Additionally, the down-regulation of rate-limiting enzyme fatty acid synthase documented that the suppression of fatty acid biosynthesis of V. dahliae by RAP (Table 2). Taken together, the RNA-seq data showed that multiple metabolic processes were affected by RAP, especially carbon metabolism, amino acids and fatty acid metabolism. Metabolic checkpoints of nutrient sensors dictate cell fate in response to metabolic fluctuations (Green et al., 2014), indicating that the disruption of metabolism homeostasis by RAP may contribute to the inhibition of cell growth in V. dahliae.
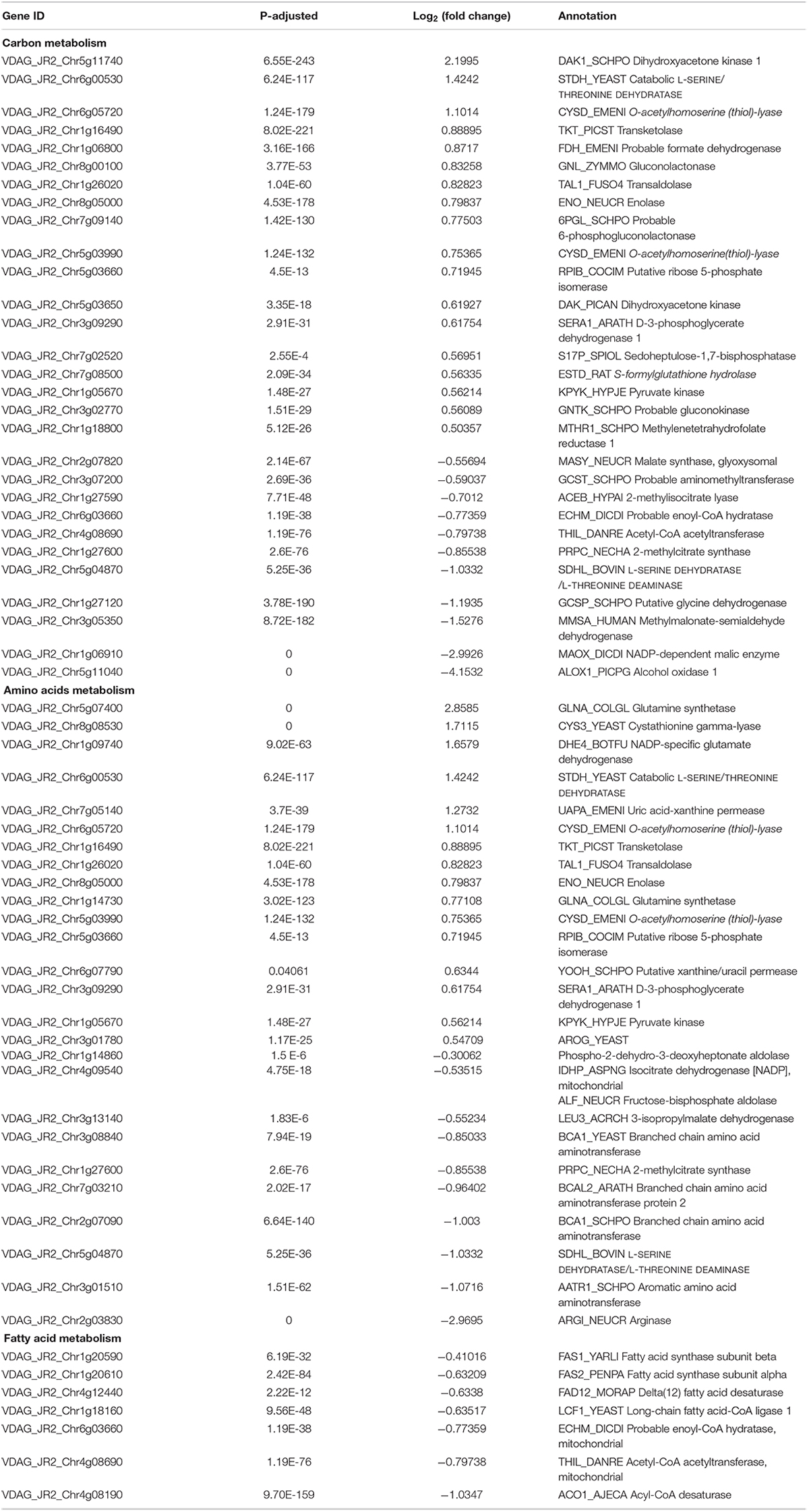
Table 2. DEGs of carbon metabolism, amino acids metabolism, and fatty acid metabolism in RNA-seq data.
Cell wall degrading enzymes produced by phytopathogenic fungi have been proved as virulence factors involving in fungal infection processes (Brito et al., 2006; Mori et al., 2008; Kubicek et al., 2014; Quoc and Chau, 2017), which was also reported in V. dahliae (Cooper and Wood, 1980; Tzima et al., 2011). CWDEs including cellulases, hemicellulases and pectinases were changed under VdTOR inhibition by RAP in the RNA-seq data (Supplementary Table 5). Furthermore, 70.27% differentially expressed CWDE genes were down-regulated (Table 3). Importantly, some pivotal genes of CWDEs such as endoglucanases, exoglucanases (cellobiohydrolases), xyloglucanase, xylanase, polygalacturonase and pectate lyase were significantly down-regulated (Table 3), implying that VdTOR plays a role in the regulation of CWDEs. Besides, VdNLP2 and VdNLP3, which encoded NEP1-like proteins of inducing necrotic lesions and triggering defense responses, were down-regulated 1.93- and 1.76-fold, respectively. The gene VDH1 of encoding a hydrophobin, which played a role in microsclerotia development and the mutant decreased microsclerotia production in V. dahliae (Klimes and Dobinson, 2006), was also down-regulated 2.68-fold in RNA-seq data (Supplementary Table 6). These observations implied that VdTOR has a positive role in the regulation of invasion and virulence in V. dahliae.
To further determine the effect of VdTOR on invasion in V. dahliae, cellophane penetration assay was performed to verify invasive growth of V. dahliae. The V. dahliae wild-type strain was incubated on top of cellophane membrane on PDA medium supplemented with RAP. The hyphae of V. dahliae can efficiently penetrate cellophane membrane on PDA medium, but not PDA medium containing RAP, suggesting an inhibitory effect of V. dahliae on cellophane penetration by RAP treatment (Figure 7A). Importantly, the main component of cellophane is cellulose, which mimics the component of plant cell wall, implying that RAP weakens the activity of CWDEs by inhibiting VdTOR activity. Cotton (Gossypium hirsutum) plants were also used to test the pathogenicity of V. dahliae treated with RAP. After inoculation for 10 days, cotton plants that infection with conidia suspension of V. dahliae occurred wilting of leaves. However, the disease severity of cotton plants that were infected with the conidia suspension containing RAP was reduced compared to that observed with the conidia suspension containing DMSO (Figure 7B). To confirm that the reduction in pathogenicity was caused by reducing the activity of CWDEs, we examined the transcription levels of genes associated with CWDEs. These genes including pectate lyase (VDAG_JR2_Chr1g28940 and VDAG_JR2_Chr2g00430), exoglucanase (VDAG_JR2_Chr1g28900) and endo-1,4-beta-xylanase 1 (VDAG_JR2_Chr3g13470) were significantly down-regulated in TOR-inhibited cells by RAP (Figure 7C). Meanwhile, we also measured the content of cellulose and pectin. The content of cellulose and pectin were significantly increased in cotton roots infected with the conidia suspension-containing RAP compared with the plants that were infected by conidia suspension of V. dahliae (Figure 7D). Taken together, these results suggested that VdTOR positively regulates the pathogenicity of V. dahliae.
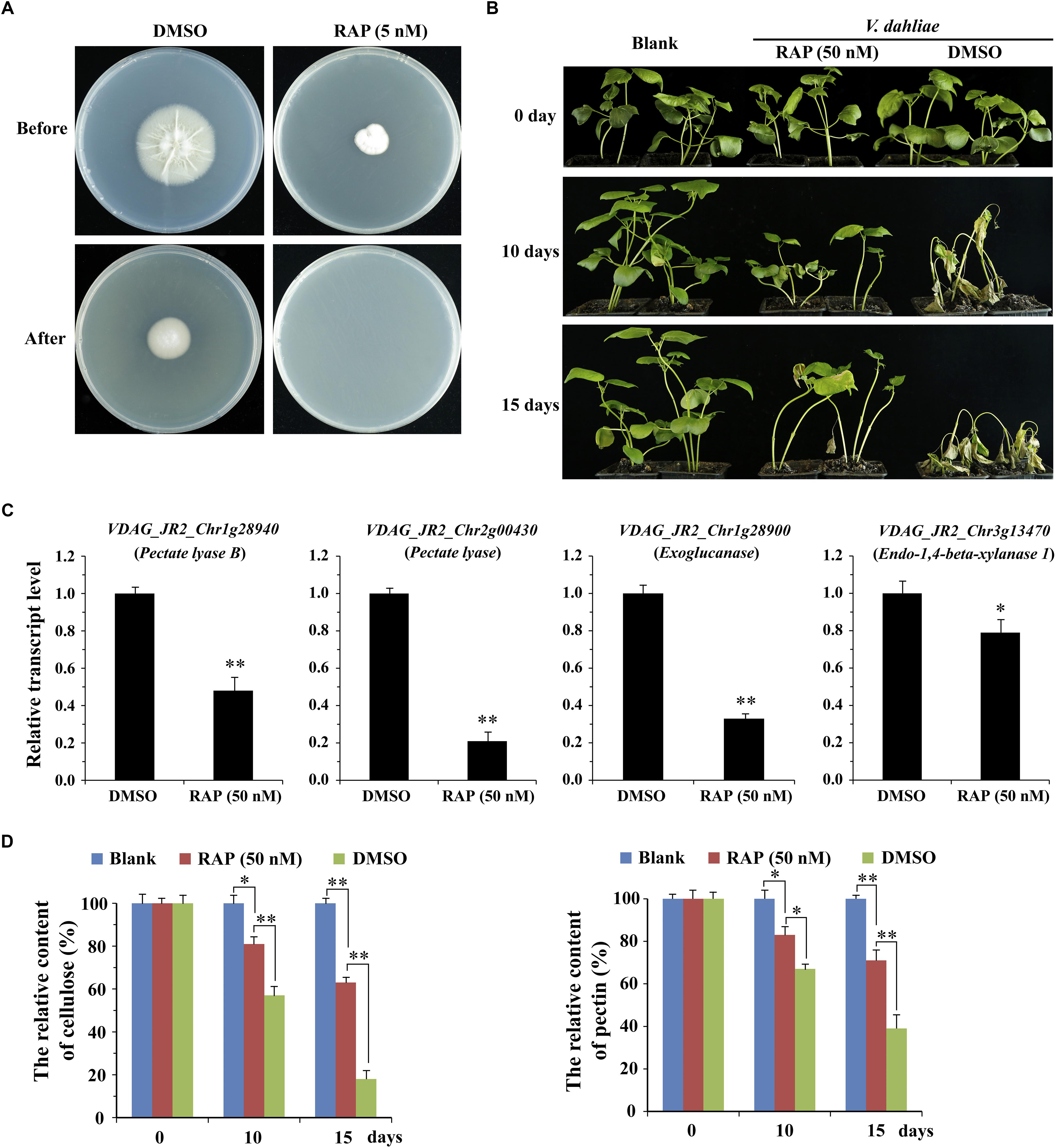
Figure 7. RAP attenuates the invasive ability of V. dahliae. (A) Effect of RAP on penetration of cellophane membranes by V. dahliae. Fungal colonies were grown for 7 days at 27°C on top of cellophane membranes on plates containing RAP (5 nM) (Before). The cellophane with the fungal colony was removed and plates were incubated for 3 days to determine the presence of mycelial growth on the plate (After). (B) Phenotype of cotton plants (Gossypium hirsutum) at 0, 10, and 15 days post-inoculation with conidia suspension supplemented with or without 50 nM RAP. (C) The relative transcript levels of genes associated with CWDEs. The data represents the mean ± SD of n = 3 independent experiments. Asterisks denote Student’s t-test significant difference compared with DMSO (∗P < 0.05; ∗∗P < 0.01). (D) The relative content of cellulose and pectin of cotton roots (Gossypium hirsutum) at 0, 10, and 15 days post-inoculation with conidia suspension supplemented with or without 50 nM RAP. The data represents the mean ± SD of n = 3 independent experiments. Asterisks denote Student’s t-test significant difference compared with RAP (∗P < 0.05; ∗∗P < 0.01).
V. dahliae is a phytopathogenic fungus that causes serious wilt disease in various plants including many economically important crops, especially for cotton (Agrios, 2005; Chen et al., 2016, 2017). TOR signaling pathway controls a wide variety of cellular processes in response to nutrients, growth factors, stresses and other environmental signals (Yuan et al., 2013; Rexin et al., 2015; Dobrenel et al., 2016; Saxton and Sabatini, 2017). Analyze the functions of VdTOR signaling pathway, which is important to gain insight into molecular processes involved in the cell growth and pathogenicity of V. dahliae. In this study, we provided some insights into how VdTOR modulates cell growth and pathogenicity through controls multiple cellular processes. We identified the putative components of VdTOR signaling pathway including TORC1 and TORC2 based on released genome database of V. dahliae (Table 1). The high similarity of the kinase domain of TOR protein was obtained among V. dahliae and other organisms (Figure 3), suggesting TOR is a structurally conserved protein in various species.
Rapamycin is an allosteric inhibitor of TOR and was approved as an immunosuppressant (Zaragoza et al., 1998). Since the function mutation of plants FKBP12 results in plants insensitivity to RAP (Sormani et al., 2007; Ren et al., 2012), RAP can be used as a potential fungicide for plant fungal diseases. For example, in comparison with chemical fungicides tebuconazole and carbendazim, RAP showed a stronger inhibitory effect on hyphal growth of Fusarium graminearum (Yu et al., 2014). Owing to its specificity, RAP has proven to be an invaluable drug in the discovery of TOR and as a pharmacological tool to dissect TOR’s cellular function (Benjamin et al., 2011; Ren et al., 2012). RAP was applied to elucidate the function of VdTOR in V. dahliae. As expected, RAP can effectively inhibit mycelial growth and conidial development of V. dahliae in a dose dependent manner (Figure 1). To further confirm whether RAP mediates the inhibition of VdTOR protein by VdFKBP12, △vdfkbp12 mutant and VdFKBP12 overexpression transgenic Arabidopsis lines were generated. RAP sensitivity test showed that △vdfkbp12 mutant resistance to RAP, but VdFKBP12 overexpression transgenic Arabidopsis lines were sensitive to RAP (Figure 4), suggesting that the ternary complex of RAP-VdFKBP12-FRB domain of VdTOR is necessary for TOR inhibition.
Due to the high specificity and minimal off-target effects of RAP, it was employed to further elucidate the function of VdTOR signaling pathway by RNA sequencing. The RNA-seq analysis showed that VdTOR inhibition resulted in changes in many metabolic processes (Figures 5, 6 and Table 2). Importantly, the disruption of carbon metabolism, biosynthesis of proteins and fatty acid metabolism destroyed metabolic homeostasis. Analysis of the RNA-seq data suggested that the inhibitory effect of RAP on cell growth of V. dahliae is most likely due to disruption the homeostasis of some important metabolic processes. The TOR kinase is a central regulator of growth and metabolism in all eukaryotic species including animals, plants and fungi (Yang et al., 2013; Rexin et al., 2015; Saxton and Sabatini, 2017). Metabolism changes from anabolism to catabolism leads to a massive accumulation of starch, triacylglycerols and amino acids after TOR inhibition (Imamura et al., 2015; Juppner et al., 2018). Interestingly, TOR inhibition increased nitrogen uptake and activities of glutamine synthetase and glutamine oxoglutarate aminotransferase in Chlamydomonas (Mubeen and Juppner, 2018). We observed that VdTOR inhibition led to transcriptional up-regulation of genes involved in nitrogen metabolism such as glutamine synthetase, the main nitrogen assimilating enzymes. This result is, at least in part, in agreement with previous study showing that RAP activates expression of nitrogen metabolism related genes in Chlamydomonas reinhardtii and Fusarium fujikuroi (Teichert et al., 2006; Mubeen and Juppner, 2018). Furthermore, RAP also altered the expression of important genes associated with CWDEs and virulence (Figure 7, Table 3, and Supplementary Table 5). In the process of recognition of plant pathogenic fungi and the host, CWDEs secreted by phytopathogenic fungi can degrade the cell wall of the host plant, which is conducive to the invasion and colonization of pathogenic fungi (Quoc and Chau, 2017; Liu et al., 2018). Transcriptional regulation of genes encoding CWDEs was controlled by transcription factors. The zinc finger transcription factor XlnR is a major activator of CWDEs in pathogenic fungi. Deletion of XlnR gene lacked transcriptional activation of xylanase and cellulase genes which resulted in failure in xylan and cellulose degradation (Calero-Nieto et al., 2007; Battaglia et al., 2011; Klaubauf et al., 2014). Besides, other transcription factors such as ACEII, PacC and CRE were documented to be involved in regulating the expression of pectinases, cellulases and xylanases encoding genes (Aro et al., 2001, 2005; Quoc and Chau, 2017). TOR and the bZIP protein MeaB control vegetative hyphal invasion and root adhesion in plant pathogenic fungi (Lopez-Berges et al., 2010). These observations implied that transcription factors play important roles in various intracellular processes regulated by TOR signaling pathway. Whether TOR regulates the activity of CWDEs through some transcription factors such as zinc finger proteins and bZIP proteins still needs further study.
In conclusion, TOR specific inhibitor RAP can inhibit the mycelial growth of V. dahliae in a dose dependent manner, suggesting that VdTOR plays an essential role in hyphal growth and development. These observations indicated direct inhibitory effects of RAP on the hyphal growth of V. dahliae and provided some insights into the interaction between RAP and plant pathogens. RNA-seq analysis indicated that VdTOR inhibition resulted in changes in various metabolic processes. Importantly, many genes of CWDEs were down-regulated during VdTOR inhibition by RAP, suggesting that VdTOR positively involved in the regulation of CWDEs. Further infection assay showed that the pathogenicity of V. dahliae and occurrence of Verticillium wilt can be blocked by RAP, indicating that RAP can be used as a potential bio-fungicide instead of chemical fungicides to prevent the occurrence of Verticillium wilt.
All datasets analyzed for this study are included in the manuscript and the Supplementary Files.
MR, FL, and LL designed the experiments. LL, TZ, YS, XL, LF, and FZ performed the experiments. LL, TZ, and YS analyzed the data. MR and LL wrote the manuscript.
This work was supported by grants from the National Natural Science Foundation of China (Grant Nos. U1804231, 31672206, and 31801911), National Key R&D Program of China (Grant Nos. 2018YFD0200808, 2016YFD0100306, and 2017YFE0115500), and the Fundamental and Frontier Research Project of Chongqing (Grant Nos. cstc2018jcyjA3432 and cstc2016jcyjA0822).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00501/full#supplementary-material
Aro, N., Pakula, T., and Penttila, M. (2005). Transcriptional regulation of plant cell wall degradation by filamentous fungi. FEMS Microbiol. Rev. 29, 719–739. doi: 10.1016/j.femsre.2004.11.006
Aro, N., Saloheimo, A., Ilmén, M., and Penttilä, M. (2001). ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J. Biol. Chem. 276:24309. doi: 10.1074/jbc.M003624200
Aylett, C. H., Sauer, E., Imseng, S., Boehringer, D., Hall, M. N., Ban, N., et al. (2016). Architecture of human mTOR complex 1. Science 351, 48–52. doi: 10.1126/science.aaa3870
Bastidas, R. J., Reedy, J. L., Morales-Johansson, H., Heitman, J., and Cardenas, M. E. (2008). Signaling cascades as drug targets in model and pathogenic fungi. Curr. Opin. Investig. Drugs 9, 856–864.
Battaglia, E., Hansen, S. F., Leendertse, A., Madrid, S., Mulder, H., Nikolaev, I., et al. (2011). Regulation of pentose utilisation by AraR, but not XlnR, differs in Aspergillus nidulans and Aspergillus niger. Appl. Microbiol. Biotechnol. 91, 387–397. doi: 10.1007/s00253-011-3242-2
Benjamin, D., Colombi, M., Moroni, C., and Hall, M. N. (2011). Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat. Rev. Drug Discov. 10, 868–880. doi: 10.1038/nrd3531
Blumenkrantz, N., and Asboe-Hansen, G. (1973). New method for quantitative determination of uronic acids. Anal. Biochem. 54:484. doi: 10.1016/0003-2697(73)90377-1
Brito, N., Espino, J. J., and González, C. (2006). The endo-beta-1,4-xylanase xyn11A is required for virulence in Botrytis cinerea. Mol. Plant Microbe Interact. 19, 25–32. doi: 10.1094/MPMI-19-0025
Bui, T. T., Harting, R., Braus-Stromeyer, S. A., Tran, V. T., Leonard, M., Hofer, A., et al. (2018). Verticillium dahliae transcription factors Som1 and Vta3 control microsclerotia formation and sequential steps of plant root penetration and colonisation to induce disease. New Phytol. 221, 2138–2159. doi: 10.1111/nph.15514
Calero-Nieto, F., Di Pietro, A., Roncero, M. I., and Hera, C. (2007). Role of the transcriptional activator xlnR of Fusarium oxysporum in regulation of xylanase genes and virulence. Mol. Plant Microbe Interact. 20, 977–985. doi: 10.1094/MPMI-20-8-0977
Chauvin, C., Koka, V., Nouschi, A., Mieulet, V., Hoareau-Aveilla, C., Dreazen, A., et al. (2014). Ribosomal protein S6 kinase activity controls the ribosome biogenesis transcriptional program. Oncogene 33, 474–483. doi: 10.1038/onc.2012.606
Chen, J. Y., Liu, C., Gui, Y. J., Si, K. W., Zhang, D. D., Wang, J., et al. (2017). Comparative genomics reveals cotton-specific virulence factors in flexible genomic regions in Verticillium dahliae and evidence of horizontal gene transfer from Fusarium. New Phytol. 217, 756–770. doi: 10.1111/nph.14861
Chen, J. Y., Xiao, H. L., Gui, Y. J., Zhang, D. D., Li, L., Bao, Y. M., et al. (2016). Characterization of the Verticillium dahliae exoproteome involves in pathogenicity from cotton-containing medium. Front. Microbiol. 7:1709. doi: 10.3389/fmicb.2016.01709
Chou, T. C. (2006). Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 58, 621–681. doi: 10.1124/pr.58.3.10
Chou, T. C., and Talalay, P. (1984). Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 22, 27–55. doi: 10.1016/0065-2571(84)90007-4
Chresta, C. M., Davies, B. R., Hickson, I., Harding, T., Cosulich, S., Critchlow, S. E., et al. (2010). AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res. 70, 288–298. doi: 10.1158/0008-5472.CAN-09-1751
Cooper, R. M., and Wood, R. K. S. (1980). Cell wall degrading enzymes of vascular wilt fungi. III. Possible involvement of endo-pectin lyase in Verticillium wilt of tomato. Physiol. Plant Pathol. 16, 285–300. doi: 10.1016/0048-4059(80)90043-0
De Virgilio, C., and Loewith, R. (2006). Cell growth control: little eukaryotes make big contributions. Oncogene 25, 6392–6415. doi: 10.1038/sj.onc.1209884
Dobrenel, T., Caldana, C., Hanson, J., Robaglia, C., Vincentz, M., Veit, B., et al. (2016). TOR signaling and nutrient sensing. Plant Biol. 67, 261–285. doi: 10.1146/annurev-arplant-043014-114648
Durrands, P. K., and Cooper, R. M. (1988). Selection and characterization of pectinase-deficient mutants of the vascular wilt pathogen Verticillium albo-atrum. Physiol. Mol. Plant Pathol. 32, 343–362. doi: 10.1016/S0885-5765(88)80029-8
Fradin, E. F., and Thomma, B. P. (2006). Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol. 7, 71–86. doi: 10.1111/j.1364-3703.2006.00323.x
Gao, F., Zhou, B. J., Li, G. Y., Jia, P. S., Li, H., Zhao, Y. L., et al. (2010). A glutamic acid-rich protein identified in Verticillium dahliae from an insertional mutagenesis affects microsclerotial formation and pathogenicity. PLoS One 5:e15319. doi: 10.1371/journal.pone.0015319
Garcia-Martinez, J. M., Moran, J., Clarke, R. G., Gray, A., Cosulich, S. C., Chresta, C. M., et al. (2009). Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR). Biochem. J. 421, 29–42. doi: 10.1042/BJ20090489
Gaubitz, C., Oliveira, T. M., Prouteau, M., Leitner, A., Karuppasamy, M., Konstantinidou, G., et al. (2015). Molecular basis of the rapamycin insensitivity of target of rapamycin complex 2. Mol. Cell 58, 977–988. doi: 10.1016/j.molcel.2015.04.031
Gaubitz, C., Prouteau, M., Kusmider, B., and Loewith, R. (2016). TORC2 Structure and function. Trends Biochem. Sci. 41, 532–545. doi: 10.1016/j.tibs.2016.04.001
Gautier, T., Berges, T., Tollervey, D., and Hurt, E. (1997). Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell Biol. 17, 7088–7098. doi: 10.1128/MCB.17.12.7088
Gomez-Lama Cabanas, C., Schiliro, E., Valverde-Corredor, A., and Mercado-Blanco, J. (2015). Systemic responses in a tolerant olive (Olea europaea L.) cultivar upon root colonization by the vascular pathogen Verticillium dahliae. Front. Microbiol. 6:928. doi: 10.3389/fmicb.2015.00928
Green, D. R., Galluzzi, L., and Kroemer, G. (2014). Cell biology. Metabolic control of cell death. Science 345:1250256. doi: 10.1126/science.1250256
Hara, K., Maruki, Y., Long, X., Yoshino, K., Oshiro, N., Hidayat, S., et al. (2002). Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110, 177–189. doi: 10.1016/S0092-8674(02)00833-4
Heitman, J., Movva, N. R., and Hall, M. N. (1991a). Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253, 905–909. doi: 10.1126/science.1715094
Heitman, J., Movva, N. R., Hiestand, P. C., and Hall, M. N. (1991b). FK-506-binding protein proline rotamase is a target for the immunosuppressive agENT FK-506 in saccharomyces-cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 88, 1948–1952. doi: 10.1073/pnas.88.5.1948
Iadevaia, V., Liu, R., and Proud, C. G. (2014). mTORC1 signaling controls multiple steps in ribosome biogenesis. Semin. Cell Dev. Biol. 36, 113–120. doi: 10.1016/j.semcdb.2014.08.004
Imamura, S., Kawase, Y., Kobayashi, I., Sone, T., Era, A., Miyagishima, S. Y., et al. (2015). Target of rapamycin (TOR) plays a critical role in triacylglycerol accumulation in microalgae. Plant Mol. Biol. 89, 309–318. doi: 10.1007/s11103-015-0370-6
Inderbitzin, P., and Subbarao, K. V. (2014). Verticillium systematics and evolution: how confusion impedes Verticillium wilt management and how to resolve it. Phytopathology 104, 564–574. doi: 10.1094/PHYTO-11-13-0315-IA
Jacinto, E., Loewith, R., Schmidt, A., Lin, S., Ruegg, M. A., Hall, A., et al. (2004). Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 6, 1122–1128. doi: 10.1038/ncb1183
Juppner, J., Mubeen, U., Leisse, A., Caldana, C., Wiszniewski, A., Steinhauser, D., et al. (2018). The target of rapamycin kinase affects biomass accumulation and cell cycle progression by altering carbon/nitrogen balance in synchronized Chlamydomonas reinhardtii cells. Plant J. 93, 355–376. doi: 10.1111/tpj.13787
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–484. doi: 10.1093/nar/gkm882
Katz, L. A. (2012). Origin and diversification of eukaryotes. Ann. Rev. Microbiol. 66, 411–427. doi: 10.1146/annurev-micro-090110-102808
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., and Salzberg, S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. doi: 10.1186/gb-2013-14-4-r36
Klaubauf, S., Narang, H. M., Post, H., Zhou, M., Brunner, K., Mach-Aigner, A. R., et al. (2014). Similar is not the same: differences in the function of the (hemi-)cellulolytic regulator XlnR (Xlr1/Xyr1) in filamentous fungi. Fungal Genet. Biol. 72, 73–81. doi: 10.1016/j.fgb.2014.07.007
Klimes, A., and Dobinson, K. F. (2006). A hydrophobin gene, VDH1, is involved in microsclerotial development and spore viability in the plant pathogen Verticillium dahliae. Fungal Genet. Biol. 43, 283–294. doi: 10.1016/j.fgb.2005.12.006
Kos-Braun, I. C., and Kos, M. (2017). Post-transcriptional regulation of ribosome biogenesis in yeast. Microb. Cell 4, 179–181. doi: 10.15698/mic2017.05.575
Kubicek, C. P., Starr, T. L., and Glass, N. L. (2014). Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi. Ann. Rev. Phytopathol. 52, 427–451. doi: 10.1146/annurev-phyto-102313-045831
Liu, N., Sun, Y., Pei, Y., Zhang, X., Wang, P., Li, X., et al. (2018). A pectin methylesterase inhibitor enhances resistance to Verticillium wilt. 176, 2202–2220.
Liu, Q., Chang, J. W., Wang, J., Kang, S. A., Thoreen, C. C., Markhard, A., et al. (2010). Discovery of 1-(4-(4-propionylpiperazin-1-yl)-3-(trifluoro-methyl)phenyl)-9-(quinolin-3-yl)benz o[h][1,6]naphthyridin-2(1H)-one as a highly potent, selective mammalian target of rapamycin (mTOR) inhibitor for the treatment of cancer. J. Med. Chem. 53, 7146–7155. doi: 10.1021/jm101144f
Liu, Q., Wang, J., Kang, S. A., Thoreen, C. C., Hur, W., Ahmed, T., et al. (2011). Discovery of 9-(6-aminopyridin-3-yl)-1-(3-(trifluoro-methyl)phenyl)benzo[h][1,6]naphthyridin-2(1H)-one (Torin2) as a potent, selective, and orally available mammalian target of rapamycin (mTOR) inhibitor for treatment of cancer. J. Med. Chem. 54, 1473–1480. doi: 10.1021/jm101520v
Loewith, R., Jacinto, E., Wullschleger, S., Lorberg, A., Crespo, J. L., Bonenfant, D., et al. (2002). Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10, 457–468. doi: 10.1016/S1097-2765(02)00636-6
Lopez-Berges, M. S., Rispail, N., Prados-Rosales, R. C., and Di Pietro, A. (2010). A nitrogen response pathway regulates virulence functions in Fusarium oxysporum via the protein kinase TOR and the bZIP protein MeaB. Plant Cell 22, 2459–2475. doi: 10.1105/tpc.110.075937
Luo, X., Mao, H., Wei, Y., Cai, J., Xie, C., Sui, A., et al. (2016). The fungal-specific transcription factor Vdpf influences conidia production, melanized microsclerotia formation, and pathogenicity in Verticillium dahliae. Mol. Plant Pathol. 17, 1364–1381. doi: 10.1111/mpp.12367
Marioni, J. C., Mason, C. E., Mane, S. M., Stephens, M., and Gilad, Y. (2008). RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 18:1509. doi: 10.1101/gr.079558.108
Maruthachalam, K., Klosterman, S. J., Kang, S., Hayes, R. J., and Subbarao, K. V. (2011). Identification of pathogenicity-related genes in the vascular wilt fungus Verticillium dahliae by Agrobacterium tumefaciens-mediated T-DNA insertional mutagenesis. Mol. Biotechnol. 49, 209–221. doi: 10.1007/s12033-011-9392-8
Menand, B., Desnos, T., Nussaume, L., Berger, F., Bouchez, D., Meyer, C., et al. (2002). Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc. Natl. Acad. Sci. U.S.A. 99, 6422–6427. doi: 10.1073/pnas.092141899
Mori, T., Jung, H. Y., Maejima, K., Hirata, H., Himeno, M., Hamamoto, H., et al. (2008). Magnaporthe oryzae endopolygalacturonase homolog correlates with density-dependent conidial germination. FEMS Microbiol. Lett. 280, 182–188. doi: 10.1111/j.1574-6968.2008.01062.x
Mubeen, U., and Juppner, J. (2018). Target of rapamycin inhibition in Chlamydomonas reinhardtii triggers de novo amino acid synthesis by enhancing nitrogen assimilation. 30, 2240–2254. doi: 10.1105/tpc.18.00159
Novo, M., Pomar, F., Gayoso, C., and Merino, F. (2006). Cellulase activity in isolates of V. dahliae differing in aggressiveness. Plant Dis. 90, 155–160. doi: 10.1094/PD-90-0155
Oh, W. J., and Jacinto, E. (2011). mTOR complex 2 signaling and functions. Cell Cycle 10, 2305–2316. doi: 10.4161/cc.10.14.16586
Pegg, G. F., and Brady, B. L. (2002). Verticillium wilts. Wallingford: CABI Publishing. doi: 10.1079/9780851995298.0000
Prados Rosales, R. C., and Di Pietro, A. (2008). Vegetative hyphal fusion is not essential for plant infection by Fusarium oxysporum. Eukaryot. Cell 7:162. doi: 10.1128/EC.00258-07
Qiu, H., Eifert, J., Wacheul, L., Thiry, M., Berger, A. C., Jakovljevic, J., et al. (2008). Identification of genes that function in the biogenesis and localization of small nucleolar RNAs in Saccharomyces cerevisiae. Mol. Cell Biol. 28, 3686–3699. doi: 10.1128/MCB.01115-07
Quoc, N. B., and Chau, N. N. B. (2017). The role of cell wall degrading enzymes in pathogenesis of Magnaporthe oryzae. Curr. Protein Pept. Sci. 18, 1019–1034. doi: 10.2174/1389203717666160813164955
Ren, M., Qiu, S., Venglat, P., Xiang, D., Feng, L., Selvaraj, G., et al. (2011). Target of rapamycin regulates development and ribosomal RNA expression through kinase domain in Arabidopsis. Plant Physiol. 155, 1367–1382. doi: 10.1104/pp.110.169045
Ren, M., Venglat, P., Qiu, S., Feng, L., Cao, Y., Wang, E., et al. (2012). Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis. Plant Cell 24, 4850–4874. doi: 10.1105/tpc.112.107144
Rexin, D., Meyer, C., Robaglia, C., and Veit, B. (2015). TOR signalling in plants. Biochem. J. 470, 1–14. doi: 10.1042/BJ20150505
Santhanam, P., and Thomma, B. P. (2013). Verticillium dahliae Sge1 differentially regulates expression of candidate effector genes. Mol. Plant Microbe Interact. 26, 249–256. doi: 10.1094/MPMI-08-12-0198-R
Saxton, R. A., and Sabatini, D. M. (2017). mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976. doi: 10.1016/j.cell.2017.02.004
Shimobayashi, M., and Hall, M. N. (2014). Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat. Rev. Mol. Cell Biol. 15, 155–162. doi: 10.1038/nrm3757
Sormani, R., Yao, L., Menand, B., Ennar, N., Lecampion, C., Meyer, C., et al. (2007). Saccharomyces cerevisiae FKBP12 binds Arabidopsis thaliana TOR and its expression in plants leads to rapamycin susceptibility. BMC Plant Biol. 7:8. doi: 10.1186/1471-2229-7-26
Soulard, A., Cohen, A., and Hall, M. N. (2009). TOR signaling in invertebrates. Curr. Opin. Cell Biol. 21, 825–836. doi: 10.1016/j.ceb.2009.08.007
Sun, C., and Woolford, J. L. Jr. (1994). The yeast NOP4 gene product is an essential nucleolar protein required for pre-rRNA processing and accumulation of 60S ribosomal subunits. EMBO J. 13, 3127–3135. doi: 10.1002/j.1460-2075.1994.tb06611.x
Tatebe, H., and Shiozaki, K. (2017). Evolutionary conservation of the components in the TOR signaling pathways. Biomolecules. 7:E77. doi: 10.3390/biom7040077
Teichert, S., Wottawa, M., Schonig, B., and Tudzynski, B. (2006). Role of the Fusarium fujikuroi TOR kinase in nitrogen regulation and secondary metabolism. Eukaryot. Cell 5, 1807–1819. doi: 10.1128/EC.00039-06
Tonukari, N. J. (2003). Enzymes and fungal virulence. J. Appl. Sci. Environ. Manag. 7, 5–8. doi: 10.4314/jasem.v7i1.17158
Trapnell, C., Hendrickson, D. G., Sauvageau, M., Goff, L., Rinn, J. L., and Pachter, L. (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53. doi: 10.1038/nbt.2450
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., Van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Tzima, A., Paplomatas, E. J., Rauyaree, P., and Kang, S. (2010). Roles of the catalytic subunit of cAMP-dependent protein kinase A in virulence and development of the soilborne plant pathogen Verticillium dahliae. Fungal Genet Biol. 47, 406–415. doi: 10.1016/j.fgb.2010.01.007
Tzima, A. K., Paplomatas, E. J., Rauyaree, P., Ospina-Giraldo, M. D., and Kang, S. (2011). VdSNF1, the sucrose nonfermenting protein kinase gene of Verticillium dahliae, is required for virulence and expression of genes involved in cell-wall degradation. Mol. Plant Microbe Interact. 24, 129–142. doi: 10.1094/MPMI-09-09-0217
Valaskova, V., and Baldrian, P. (2006). Degradation of cellulose and hemicelluloses by the brown rot fungus Piptoporus betulinus–production of extracellular enzymes and characterization of the major cellulases. Microbiology 152, 3613–3622. doi: 10.1099/mic.0.29149-0
Ververis, C., Georghiou, K., Christodoulakis, N., Santas, P., and Santas, R. (2004). Fiber dimensions, lignin and cellulose content of various plant materials and their suitability for paper production. Ind. Crops Prod. 19, 245–254. doi: 10.1016/j.indcrop.2003.10.006
Wang, X., and Proud, C. G. (2009). Nutrient control of TORC1, a cell-cycle regulator. Trends Cell Biol. 19, 260–267. doi: 10.1016/j.tcb.2009.03.005
Wei, Y., and Zheng, X. F. (2009). Sch9 partially mediates TORC1 signaling to control ribosomal RNA synthesis. Cell Cycle 8, 4085–4090. doi: 10.4161/cc.8.24.10170
Yang, H., Rudge, D. G., Koos, J. D., Vaidialingam, B., Yang, H. J., and Pavletich, N. P. (2013). mTOR kinase structure, mechanism and regulation. Nature 497, 217–223. doi: 10.1038/nature12122
Young, M. D., Wakefield, M. J., Smyth, G. K., and Alicia, O. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, 1–12. doi: 10.1186/gb-2010-11-2-r14
Yu, F., Gu, Q., Yun, Y., Yin, Y., Xu, J. R., Shim, W. B., et al. (2014). The TOR signaling pathway regulates vegetative development and virulence in Fusarium graminearum. New Phytol. 203, 219–232. doi: 10.1111/nph.12776
Yuan, H. X., Xiong, Y., and Guan, K. L. (2013). Nutrient sensing, metabolism, and cell growth control. Mol. Cell 49, 379–387. doi: 10.1016/j.molcel.2013.01.019
Zaragoza, D., Ghavidel, A., Heitman, J., and Schultz, M. C. (1998). Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol. Cell Biol. 18, 4463–4470. doi: 10.1128/MCB.18.8.4463
Zhao, Y. L., Zhou, T. T., and Guo, H. S. (2016). Hyphopodium-specific VdNoxB/VdPls1-dependent ROS-Ca2+ signaling is required for plant infection by Verticillium dahliae. PLoS Pathog. 12:e1005793. doi: 10.1371/journal.ppat.1005793
Zhou, B. J., Jia, P. S., Gao, F., and Guo, H. S. (2012). Molecular characterization and functional analysis of a necrosis- and ethylene-inducing, protein-encoding gene family from Verticillium dahliae. Mol. Plant Microbe Interact. 25, 964–975. doi: 10.1094/MPMI-12-11-0319
Keywords: rapamycin, target of rapamycin, pathogenicity, Verticillium dahliae, Verticillium wilt
Citation: Li L, Zhu T, Song Y, Luo X, Feng L, Zhuo F, Li F and Ren M (2019) Functional Characterization of Target of Rapamycin Signaling in Verticillium dahliae. Front. Microbiol. 10:501. doi: 10.3389/fmicb.2019.00501
Received: 13 December 2018; Accepted: 27 February 2019;
Published: 13 March 2019.
Edited by:
Hari S. Misra, Bhabha Atomic Research Centre, IndiaReviewed by:
Ken Yokawa, Kitami Institute of Technology, JapanCopyright © 2019 Li, Zhu, Song, Luo, Feng, Zhuo, Li and Ren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maozhi Ren, cmVubWFvemhpQGNxdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.