- 1State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, College of Plant Protection, Fujian Agriculture and Forestry University, Fuzhou, China
- 2Key Laboratory of Ministry of Education for Genetics, Breeding and Multiple Utilization of Crops, Plant Immunity Center, Fujian Agriculture and Forestry University, Fuzhou, China
- 3Institute of Oceanography, Minjiang University, Fuzhou, China
- 4Department of Biochemistry and Molecular Biology, University of Oklahoma Health Sciences Center, Oklahoma City, OK, United States
Isovaleryl-CoA dehydrogenase (IVD), a member of the acyl-CoA dehydrogenase (ACAD) family, is a key enzyme catalyzing the conversion of isovaleryl-CoA to β-methylcrotonyl-CoA in the third reaction of the leucine catabolism pathway and simultaneously transfers electrons to the electron-transferring flavoprotein (ETF) for ATP synthesis. We previously identified the ETF ortholog in rice blast fungus Magnaporthe oryzae (MoETF) and showed that MoETF was essential for fungal growth, conidiation and pathogenicity. To further investigate the biological function of electron-transferring proteins and clarify the role of leucine catabolism in growth and pathogenesis, we characterized MoIVD (M. oryzae isovaleryl-CoA dehydrogenase). MoIvd is highly conserved in fungi and its expression was highly induced by leucine. The Δmoivd mutants showed reduced growth, decreased conidiation and compromised pathogenicity, while the conidial germination and appressorial formation appeared normal. Consistent with a block in leucine degradation, the Δmoivd mutants accumulated isovaleric acid, grew more slowly, fully lacked pigmentation and completely failed to produce conidia on leucine-rich medium. These defects were largely rescued by raising the extracellular pH, suggesting that the accumulation of isovaleric acid contributes to the growth and conidiation defects. However, the reduced virulence of the mutants was probably due to their inability to overcome oxidative stress, since a large amount of ROS (reactive oxygen species) accumulated in infected host cell. In addition, MoIvd is localized to mitochondria and interacted with its electron receptor MoEtfb, the β subunit of MoEtf. Taken together, our results suggest that MoIVD functions in leucine catabolism and is required for the vegetative growth, conidiation and full virulence of M. oryzae, providing the first evidence for IVD-mediated leucine catabolism in the development and pathogenesis of plant fungal pathogens.
Introduction
Rice blast caused by Magnaporthe oryzae is a devastating disease in rice growing areas across the world (Valent and Chumley, 1991; Tablot, 2003; Fernandez and Orth, 2018). Each year, this fungus results in an economic loss estimated to be $66 billion, which could feed 60 million people (Pennisi, 2010). The infection begins with three-celled conidia of M. oryzae contacting and germinating on the host surface, and then a dome-shaped infection structure called appressorium is developed from the germ tube end (Hamer et al., 1988). The mature appressoria melanize and accumulate high Turgor pressure (Howard et al., 1991), which assists the penetration peg in mechanically breaching the host cell wall (Kankanala et al., 2007). Once the fungus overcomes host defense response and completes colonization, the infectious hyphae, developed from penetration peg, grow intra- and intercellularly in the host and produce necrotic lesions within 3–5 days (Sakulkoo et al., 2018). Finally, new conidia are formed and released to start a new infection cycle. In addition to rice blast, wheat blast caused by M. oryzae has also recently emerged in South America and Bangladesh (Islam et al., 2016; Sadat and Choi, 2017). Due to its economic importance, genetic tractability and genome sequence availability, M. oryzae has emerged as a model organism to study the fungal pathogenesis and interaction with host plants (Dean et al., 2005; Ebbole, 2007). Understanding important metabolic processes in M. oryzae should help to develop novel and effective strategies to control this disease.
Leucine is one of the three branched amino acids, and its catabolism provides key intermediate metabolites—acetyl-CoA and ATP—for other metabolic processes (Lei et al., 2012; Arany and Neinast, 2018). Isovaleryl-CoA dehydrogenase (IVD) is an enzyme catalyzing the third reaction of the leucine catabolism pathway by conversion of isovaleryl-CoA to β-methylcrotonyl-CoA, and simultaneously electrons transfer to the respiratory chain for ATP production via electron-transferring flavoprotein (ETF) (Mohsen et al., 1998; Zhang et al., 2006; Mohsen and Vockley, 2015). As a flavoprotein with flavin adenine dinucleotide (FAD) as the cofactor, IVD catalyzes α,β-dehydrogenation and removal of one hydrogen as a proton from the branched-chain substrate, isovaleryl-CoA (Tiffany et al., 1997). Structurally, IVD is a member of the acyl-CoA dehydrogenase (ACAD) family and shares high sequence homology and similar mechanism with other family members for the α,β-dehydrogenation of acyl-CoA substrates (Rozen et al., 1994; Mohsen et al., 1998; Ghisla and Thorpe, 2004). According to the chain length and whether it branches, the ACAD family is divided into short/branched-chain acyl-CoA dehydrogenases (SBCAD), short-chain acyl-CoA dehydrogenases (SCAD), medium-chain acyl-CoA dehydrogenases (MCAD), long-chain acyl-CoA dehydrogenases (LCAD), and very long chain acyl-CoA dehydrogenases (VLCAD) (Ghisla and Thorpe, 2004). IVD belongs to the SBCAD subfamily that also includes isobutyryl-CoA dehydrogenase (IBD) and 2-methyl branched-chain acyl-CoA dehydrogenase (MBCAD), which are involved in the catabolism of valine and isoleucine, respectively (Noda et al., 1980; Brosnan and Brosnan, 2006).
Leucine intake and catabolism are important to humans, and the function of human IVD in leucine catabolism has been extensively reported (Vockley and Ensenauer, 2006). Individuals carrying IVD mutations are unable to break down leucine properly and display symptoms including poor feeding, vomiting, seizures, and even stupor (Vockley and Ensenauer, 2006; Kaya et al., 2013). Because isovaleric acid accumulates in the patient’s plasma, this disease is also called isovaleric acidemia (Tanaka, 1990; Pascarella et al., 2011; Erdem et al., 2010). A distinctive odor characterized as “sweaty feet” can be noted in body secretions during acute episodes (Tokatli et al., 1998). Plants, fungi and bacteria can synthesize leucine, unlike humans. However, the function of IVD-mediated leucine catabolism is not as well understood in those species as in humans. In plants, the identification, characterization and purification of pea IVD were reported, and this IVD was localized to the mitochondria and played a regulatory role in development (Däschner et al., 1999; Reinard et al., 2000). Arabidopsis IVD oxidizes catabolic intermediates of both leucine and valine, and also functions as alternative electron donor linking leucine catabolism to the electron transport chain for ATP production (Däschner et al., 2001; Araújo et al., 2010). Likewise, potato IVD is also involved in the catabolism of both leucine and valine (Faivre-Nitschke et al., 2001). In fungi, the only report of an IVD relates to glutamic acid-mediated repression of the promoter activity of Aspergillus oryzae IVD gene (Yamashita et al., 2007). In bacteria, Micrococcus luteus IVD is specific for leucine degradation, and the IVD-deficient mutant cannot grow on leucine (Surger et al., 2018).
The upstream leucine synthesis pathway in M. oryzae is required for conidiation, appressorium Turgor establishment and host penetration as demonstrated by characterizing two acetolactate synthase genes—MoILV2 and MoILV6—and one threonine dehydratase gene—MoILV1—in the pathway (Du et al., 2013, 2014). But the downstream leucine catabolism pathway has not been characterized so far. In our previous study, M. oryzae electron-transferring flavoprotein ETF (MoETF) was functionally characterized and shown to be indispensable for fungal growth, conidiation and pathogenicity (Li et al., 2016). However, the function of its electron donors, acyl-CoA dehydrogenases, remains unknown. Here, we focus on one of the predicted ETF electron donors—MoIVD (M. oryzae Isovaleryl-CoA dehydrogenase gene id MGG_02540)—and demonstrate its biological function in the development and pathogenesis of M. oryzae.
Materials and Methods
Strains and Growth Conditions
The M. oryzae strain Guy11 strain was used as background to construct the Δmoivd mutants (Δmoivd-3 and -6) and complementary strain (Δmoivd-3/MoIVD). The CM (complete medium) and MM (minimal medium) were prepared for testing the fungal growth and conidiation as described previously (Li et al., 2010a). Conidia were harvested from a fungal colony, growing for 12 days in a 9 cm plate, and the production was quantified by a hemacytometer. Conidia suspension was set in a concentration of 5 × 104 conidia ml-1 and placed on a hydrophobic surface in a humid environment at 25°C to induce conidia germination and appressorium formation. Escherichia coli strain DH-5α was used for routine bacterial transformations and maintenance of various plasmid vectors. The culturing medium for DH-5α is liquid and agar LB (Luria-Bertani) medium. By trying a concentration gradient of 0.3 mM, 3 mM, 30 mM, 0.1 M and 1 M, we found that the optimal leucine concentration used in CM and MM to treat the fungus was 0.1 M and 3 mM, respectively. As described previously, 50 mM sodium acetate and sodium carbonate were added to CM to treat the fungus (Li et al., 2016).
Molecular Manipulation
To construct the MoIVD deletion vector, the 1 kb up- and down-stream fragment from the MoIVD coding region were amplified and fused with the up- and down-half of the HPH (hygromycin phosphotransferase) gene, respectively, using overlapping PCR. The up- and down-half fragments of the HPH sequence shared an overlapping region of 100 bp in length. For building MoIVD complementation vectors, the full length MoIVD with a native promoter region (about 1.5-kb) was cloned to the pCB1532 vector. The 1.5 kb GFP sequence harboring a TrpC-terminator was inserted into the C-terminal of MoIVD to construct the MoIvd-GFP strain to explore the protein localization. qRT-PCR was performed by following the kit of “One Step qRT-PCR Kit RNA-direct Realtime PCR Master Mix” (Code Number QRT-101B). To do semi qRT-PCR, the RNA was isolated, and the cDNA was synthesized by using the kit “PrimeScriptTM 1st Strand cDNA Synthesis” (Code Number 6110A) prior to conventional PCR. For doing the Y2H (yeast two-hybrid) assay, MoIVD and MoETFA/B were cloned to pGBKT7 and pGADT7, respectively, and then the resulting plasmids were transformed to the AH109 yeast strain. The yeast clones were grown on -Leu/-Trp/-Ade/-His/SD (synthetical defined) medium to test whether the proteins interact. To do the BIFC (bimolecular fluorescence complementation) assay, MoIVD and MoETFA/B were fused to the N terminal and C terminal of YFP (Yellow Fluorescent Protein), respectively, to construct the MoIvd-NYFP-pKNT and MoEtfa/b-CYFP-pCX62 plasmids, and then the plasmids were transformed to Guy11 to analyze whether the whole YFP signal appears in the conidia of transformants. Primers used in this study are listed in Supplementary Table S1.
Fungal Transformation
Fungal protoplast was prepared as described previously (Li et al., 2014). To perform gene deletion transformation, no less than 2 μg MoIVD deletion vector DNA was introduced to Guy11 protoplast, and transformants were selected for hygromycin resistance. Southern blotting was conducted to confirm the deletion events using the digoxigenin (DIG) high prime DNA labeling and detection starter Kit I (11745832910 Roche Germany). A 900 bp upstream fragment of MoIVD locus was selected as a probe for hybridization to digested genomes of all fungal strains. MoIVD complementation transformation was performed by introducing complementation vector DNA into mutant protoplast and screening transformants against chlorimuron-ethyl resistance.
Conidial Induction and Infection Related Assay
To perform conidia induction assay, thin mycelial plugs were excised from colonies and placed on glass slides. After incubating 24 h with constant light, the conidia development state was observed and imaged by microscopy. The conidia suspension was prepared in a concentration of 5 × 104 conidia ml-1 for infection-related assay. To perform the live rice infection assay, an airbrush was used and 5 ml conidial suspension was sprayed on 10 plants of 15-day-old dwarf Indica rice cv. CO-39. Post-spray, inoculated plants were kept in a sealed chamber with 90% relative humidity at 25°C for 24 h. Then, the inoculated plants were removed from the chamber to allow disease symptoms to develop over 4–5 days. The pathogenicity assay on barley was performed by dropping the conidial suspension on excised Golden Promise leaves in 20 μl drops (Li et al., 2010b). The disease spots on barley develop within 5–6 days. The live sheath assay was performed as described previously to evaluate invasive growth (Xu et al., 1998).
Cytorrhysis Assay
The cytorrhysis assay was performed to test appressorium turgor as follows. Conidial suspension was prepared at a concentration of 5 × 104 conidia ml-1 and 10 μl drops were placed on hydrophobic slides for 24 h at 25°C to allow appressoria maturation. Then the covering water was carefully removed and replaced with an equal volume of glycerol in concentrations of 2.0, 3.0, and 4.0 M. After continued incubation for 15 min, the ratio of collapsed appressoria was recorded in each glycerol treatment and to reflect the appressorium turgor difference between different fungal strains.
DAB (3,3′-Diaminobenzidine) Staining
Host ROS accumulation during M. oryzae infection was detected by DAB staining assay as described previously (Chen et al., 2014). The conidial suspension was prepared at a concentration of 5 × 104 conidia ml-1 and sprayed to barley leaves. After 24 h, the leaves were moved to 1 mg/ml DAB solution and kept at room temperature for 8 h for staining. Then, the samples were destained in washing solution (ethanol/acetic acid = 94/4, v/v) for 2–3 h. The ROS in the infected cell are meant to be detected by accumulation of a dark brown precipitate.
Fatty Acids Content Test
The short chain fatty acids content was analyzed as follows. The 1.0 g dry mycelia samples were added to 3 ml water and vortexed with glass beads for 2 min. After centrifugation in the cold, the supernatant was transferred to a new tube and hydrochloric acid (1:10) was added to acidify the fat. Ether was added and the solution was mixed for 10 min for extraction. The ether phase sample was subjected to GC-MS (gas chromatography-mass spectrometer) analysis for short chain fatty acid analysis. For medium and long chain fatty acid testing, the sample was mixed with 2 ml 5% methanol hydrochloride, 3 ml chloroform methanol (1:1), 100 μl of methyl 19 alkylate internal standard and incubated in an 85°C water bath for 1 h. After cooling, 1 ml n-hexane was added for extraction with 2 min of shaking, and then the mixture was left standing for 1 h for stratification prior to GC-MS analysis.
Amino Acids Content Test
Samples of 1.0 g dry mycelia were transferred to 20 ml hydrolysis tube, and 16 ml 6 mol/l HCL solutions was added to resuspend the mycelium. Samples were then degassed for 30 min, sealed by nitrogen and hydrolyzed at 110°C for 22–24 h. After cooling, samples were transferred to a volumetric flask to a final volume of 50 ml with deionized water. A 1 ml sample was transferred to a small bottle, deacidized and dried in a vacuum. The sample was recovered with 1 ml deionized water and dried twice, and 1 ml 0.02 M hydrochloric acid was added and dissolved fully. A 500 μl sample of the hydrolysate was placed in a 5 ml plastic centrifuge tube with 250 μl of 1 M triethylamine acetonitrile and mixed prior to addition of 25 μl of 0.1 M isothiocyanate phenyl acetonitrile. After mixing, the sample was placed at room temperature for 1 h, 2 ml of n-hexane was added, and the mixture was vortexed. After 10 min, the lower phase was collected and filtered through a 0.22-micron filter prior to HPLC (high performance liquid chromatography) analysis.
Results
MoIvd Is Highly Conserved in Fungi
MoIVD is located between +2434073 and +2436238 on chromosome 7 of M. oryzae genome in the 70–15 genome sequence (NCBI assembly GCA_000002495.2). The gene encodes a 470-amino acid (aa) protein including a long ACAD domain located between aa91 and aa464 (Figure 1A). The amino acid sequence of MoIvd was used to search for homologs in the NCBI database by BLAST analysis1. As a result, more than 100 MoIvd homologs with over 70% sequence identity were found from different fungal species (Supplementary Figure S1), suggesting that MoIvd is highly conserved in fungi. With the IVD homologs from several model species, we constructed a phylogenetic tree to analyze their evolutionary relationship. As shown in Figure 1B, MoIvd is more evolutionarily related to fungal homologs of Gaeumannomyces tritici, Neurospora crassa, Thielavia terrestris, Colletotrichum tofieldiae and Sclerotinia sclerotiorum, with 65–70% sequence identity. The plant and animal homologs Arabidopsis thaliana, Oryza sativa, Caenorhabditis elegans, Danio rerio, Homo sapiens and Mus musculus are more distant relatives from MoIvd with only 46–48% sequence identity. The bacterial homolog, Escherichia coli, is on the most distant evolutionary branch, with 30% sequence identity to MoIvd.
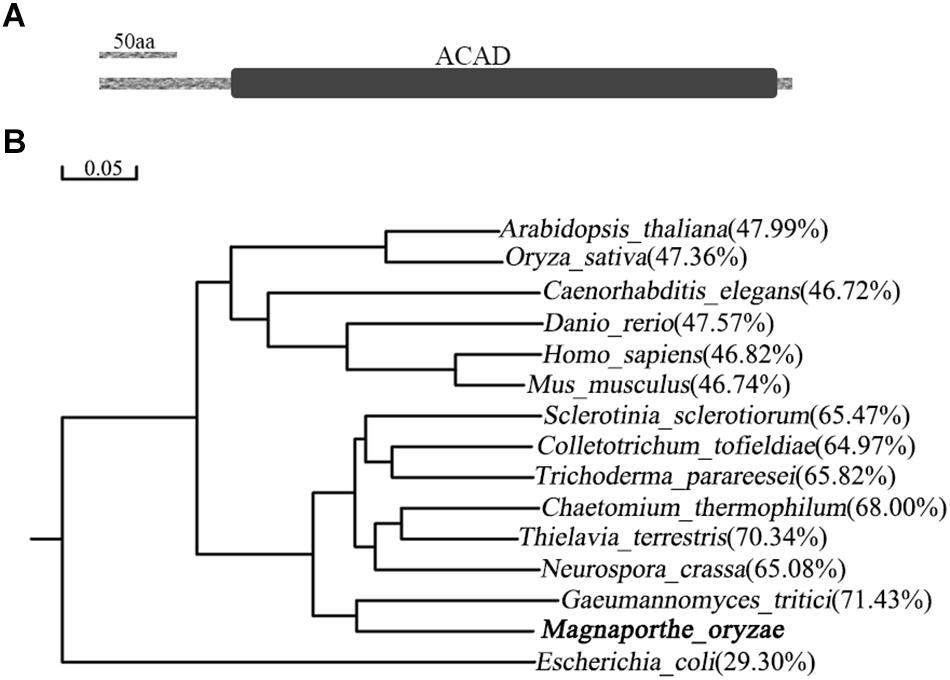
Figure 1. Domain structure of MoIvd and phylogenetic analysis of IVD homologs. (A) MoIvd domain prediction by the PFAM website (http://pfam.xfam.org/). The ACAD domain located between aa91 and aa464. The bar indicates 50 amino acids. (B) IVD phylogenetic tree constructed using the observed divergency method by DNAMAN6.0. The IVD protein sequences of 15 model species were collected from NCBI database. The bar indicates 0.05 distance units. The percentage in brackets indicates sequence identity between MoIvd and corresponding homolog. The sequence accession numbers are listed as follows: Magnaporthe oryzae (XP_003721261.1), Arabidopsis thaliana (NP_190116.1), Oryza sativa (XP_015639342.1), Caenorhabditis elegans (NP_500720.1), Danio rerio (NP_958899.1), Homo sapiens (NP_002216.2), Mus musculus (NP_062800.1), Sclerotinia sclerotiorum (APA11203.1), Colletotrichum tofieldiae (KZL76390.1), Trichoderma parareesei (OTA08745.1), Chaetomium thermophilum (XP_006691847.1), Thielavia terrestris (XP_003651516.1), Neurospora crassa (XP_964284.1), Gaeumannomyces tritici (XP_009227932.1) and Escherichia coli (WP_001189854.1).
MoIVD Expression Is Highly Induced by Exogenous Leucine
To determine if MoIVD is involved in leucine catabolism, we investigated the MoIVD expression level under exogenous leucine stress via the approaches of qRT-RCR and semi qRT-RCR. M. oryzae was cultured in liquid CM containing 0.1 M exogenous leucine or in control CM without leucine or with 0.1 M asparagine, for 48 h at 25°C. In the presence of exogenous leucine, the MoIVD expression was highly induced, and its mRNA level increased by 50-fold and 5-fold, respectively, compared to the CM and the asparagine controls (Figure 2A). The stronger band of the MoIVD cDNA fragment in the leucine treatment also suggested a high mRNA level of MoIVD (Figure 2B).
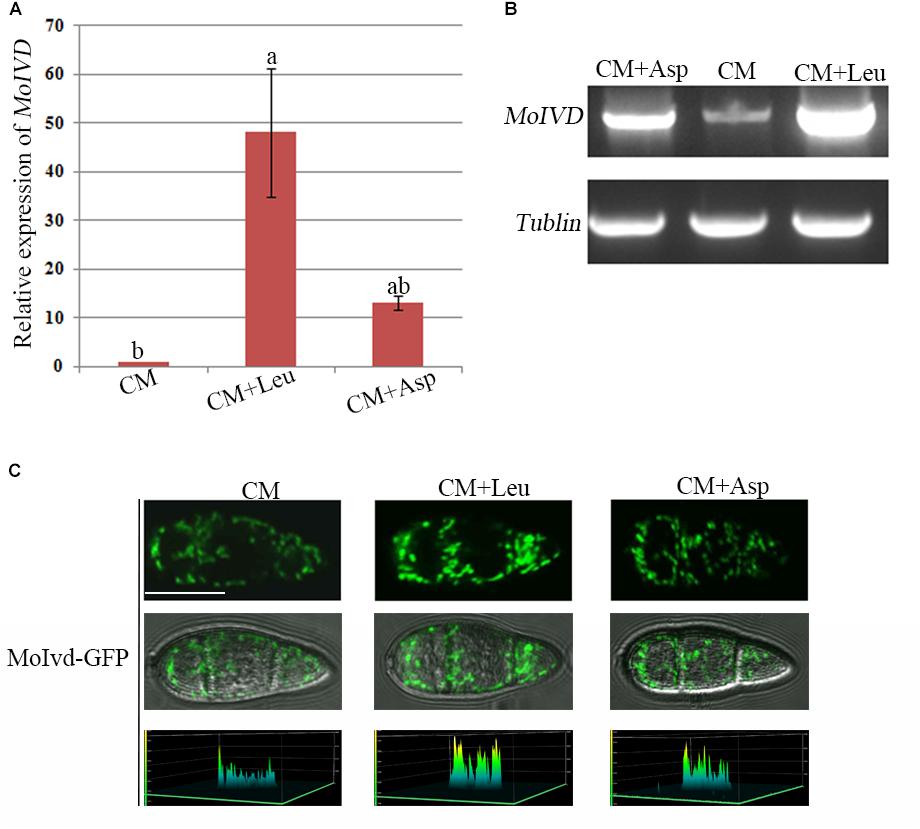
Figure 2. The MoIVD expression was highly induced by exogenous leucine. (A) qRT-PCR test of the relative MoIVD expression in fungus treated with 0.1 M leucine (Leu) and 0.1 M aspartic acid (Asp). (B) Semi qRT-PCR and electrophoresis test of MoIVD expression in fungus treated with 0.1 M Leu and 0.1 M Asp. (C) The fluorescence intensity of MoIvd-GFP in conidia treated with 0.1 M Leu and 0.1 M Asp. The peaks in the 3D chart show the signal intensity. The 3D chart was generated by laser scanning confocal microscopy. Bar = 10 μm.
We also constructed a MoIvd-GFP strain under the control of MoIVD promoter and determined the MoIVD expression level in the cell by confocal fluorescence microscopy. As shown in Figure 2C, MoIvd-GFP conidia displayed much brighter fluorescence in leucine treatment than in controls, suggesting higher expression levels of MoIvd-GFP in the presence of exogenous leucine. These results are consistent with a previous report that the promoter activity of IVD gene in Aspergillus oryzae was highly induced by the hydrophobic amino acid L-leucine, slightly induced by L-asparagine with low hydrophobicity, but not induced by L-glutamic acid sodium salt without hydrophobicity (Yamashita et al., 2007).
MoIVD Is Involved in Vegetative Growth and Conidiation
To investigate the function of MoIVD in M. oryzae, we constructed two Δmoivd mutants (Δmoivd-3 and Δmoivd-6) and one complementation strain Δmoivd-3/MoIVD. All the mutant and complemented strains were confirmed by Southern blot analysis (Supplementary Figure S2). Their growth phenotype was determined by the size and color of fungal colonies on CM medium. Our data indicated that the colony size of the Δmoivd mutants was smaller and the edge of mutant colonies also lost pigmentation in comparison to the controls, Guy11 and Δmoivd-3/MoIVD (Figure 3A). In this regard, the opposites of plates inoculated with the mutant strains completely lost color, while those of Guy11 and Δmoivd-3/MoIVD exhibited the normal black color (Figure 3A). Moreover, when cultured in liquid CM, the mutants produced a very small number of mycelia, and its dry weight was significantly reduced compared to Guy11 and Δmoivd-3/MoIVD (Figure 3B). These results indicated multiple growth defects in the Δmoivd mutants. We also examined conidiation in the mutants. The conidia were harvested from colonies growing on CM plates for 12 days and quantified using a hemacytometer. Our results showed that Δmoivd-3 and Δmoivd-6 produced fewer conidia (1 × 106 spores per plate), which was about 2/3 of the conidia produced by strains Guy11 or Δmoivd-3/MoIVD (Figure 3C). Furthermore, the mutant conidiophores also decreased compared to those of Guy11 and Δmoivd-3/MoIVD (Figure 3D). Taken together, the data suggested that MoIVD is involved in vegetative growth and conidiation in M. oryzae.
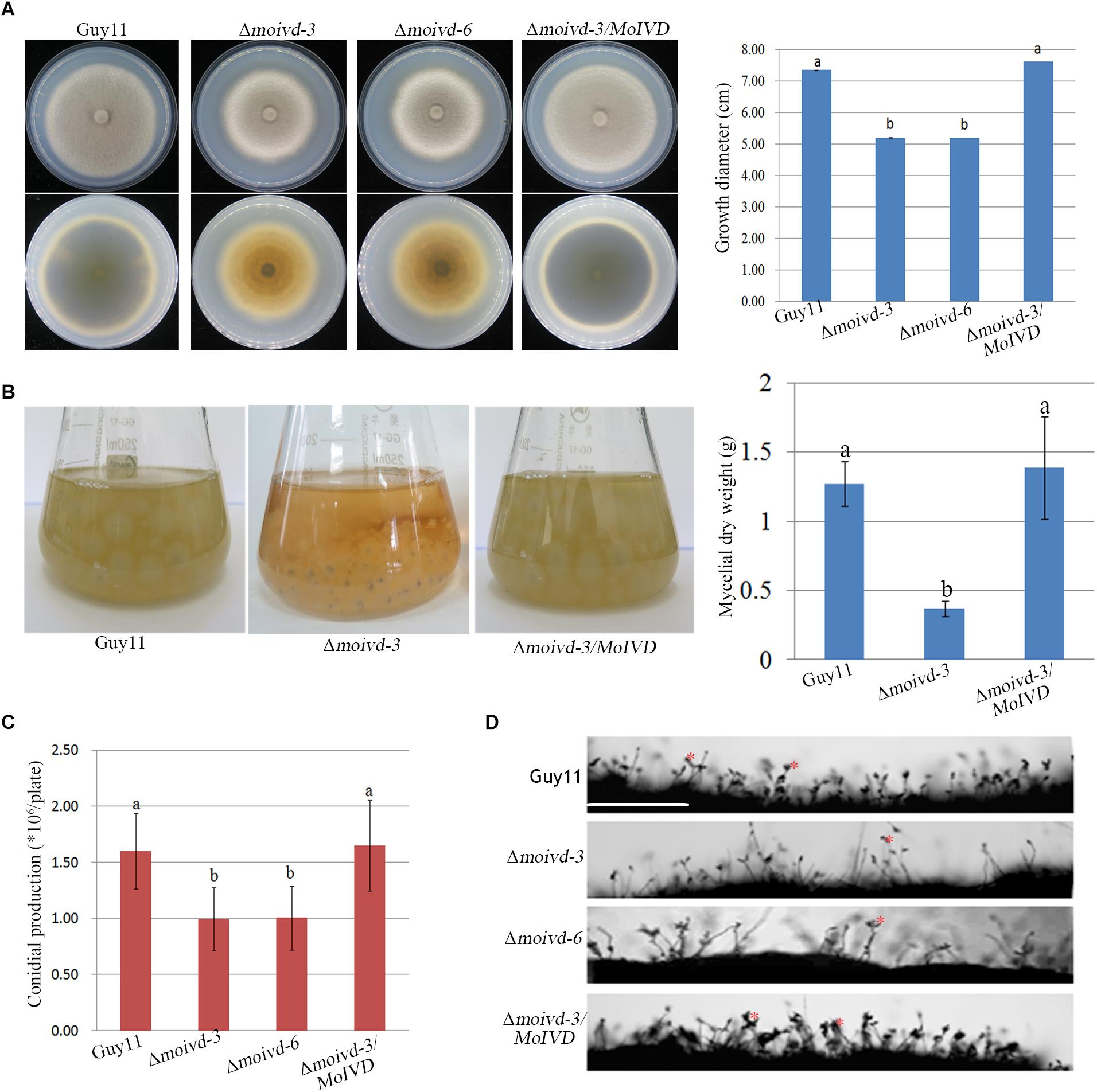
Figure 3. The growth and conidiation phenotype of the Δmoivd mutants. (A) The colonial morphology of the Δmoivd mutants growing on complete medium for 10 days. The upper left panel is the colony outlook. The lower left panel is the opposite side of culture plate. The bar chart in the right shows the growth size of the Δmoivd mutants. (B) The culture morphogenesis of Δmoivd-3 incubated in liquid CM for 48h in flask (left) and bar chart showing the dry weight of Δmoivd-3 culture (right). (C) Bar chart showing the conidial production of the Δmoivd mutants by culture on complete medium for 12 days. (D) Microscopic observation of the sporulating structures of the Δmoivd mutants. ∗ indicates conidia developed from conidiophores. Bar = 50 μm. All the data in this figure were calculated from three independent replicates. Lowercase ‘a’ or ‘b’ on the sample bar indicate no significant differences between samples. The different lowercase letters indicate significant differences (P < 0.05; t test).
MoIVD Is Required for M. oryzae Pathogenicity
To investigate the role of MoIVD in M. oryzae pathogenicity, we evaluated the mutant phenotype in infection-related development including conidial germination, appressorium formation and host infection. Although we did not find any significant difference between mutant and wild type strains in conidial germination and appressorium formation (Supplementary Table S2), their virulence on excised barley leaves showed differences, and Δmoivd-3 and Δmoivd-6 produced smaller disease lesions than Guy11 and Δmoivd-3/MoIVD (Figure 4A). Likewise, the mutants also produced much smaller and fewer lesions on rice seedlings than Guy11 and Δmoivd-3/MoIVD (Figures 4B,C). Furthermore, sheath infection assay was used to examine infectious growth in host cells, and Δmoivd-3 and Δmoivd-6 displayed lower penetration frequency and delayed hyphal growth after penetration in comparison to Guy11 and Δmoivd-3/MoIVD at 24 h post-inoculation/infection (Figure 4D). These results suggest MoIVD is required for full pathogenicity of M. oryzae. ROS accumulation is a symbol of host defense against pathogen attack and could be tested by DAB staining assay. Using this method, we found that the barley cells infected by the mutant exhibited a darker brown color than the ones infected by Guy11 and complemented strain Δmoivd-3/MoIVD (Figure 4E), suggesting more ROS accumulation in the mutant-infected cells.
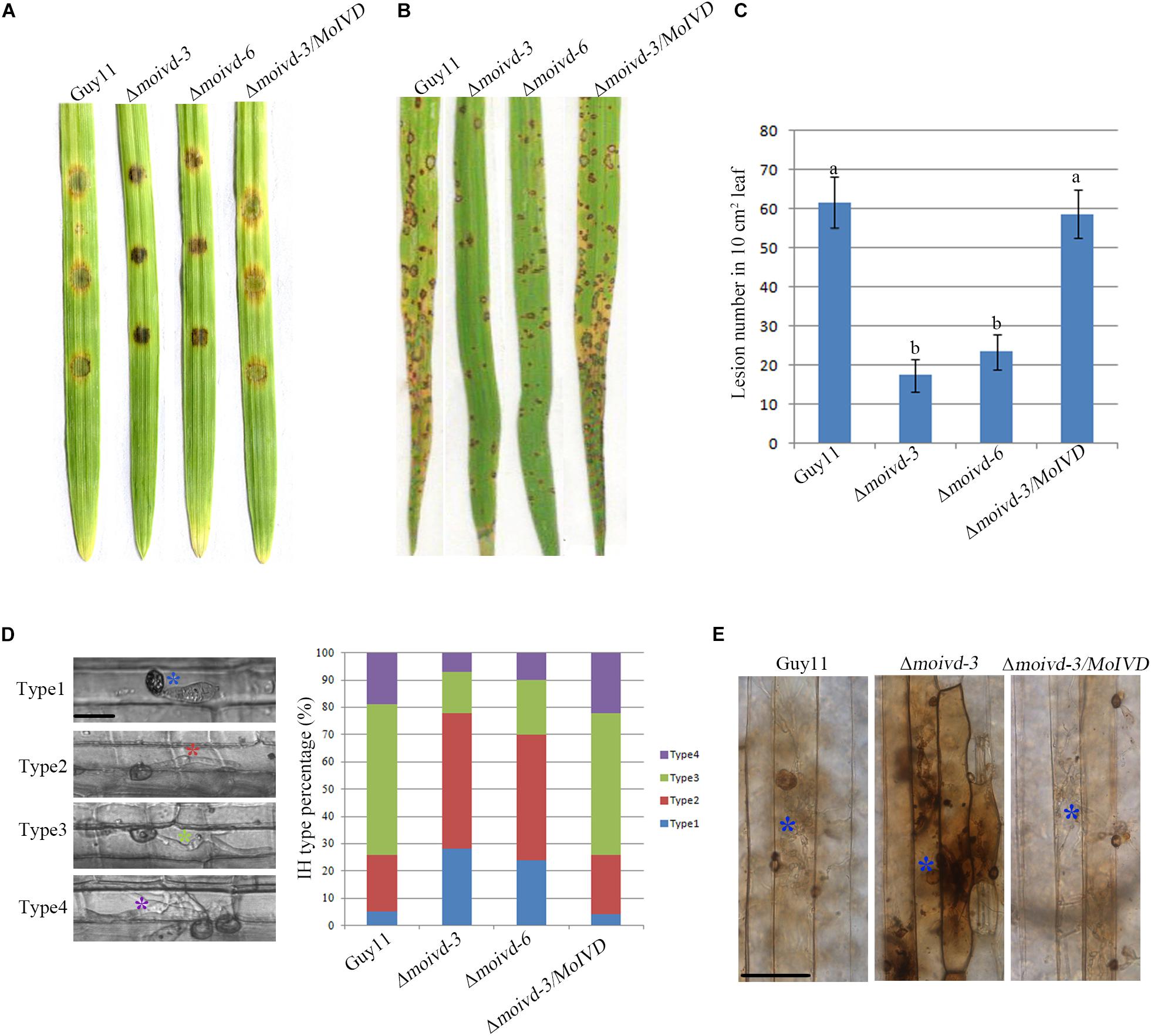
Figure 4. The pathogenicity assay of the Δmoivd mutants. (A) Pathogenicity test of the Δmoivd mutants on excised barley leaves. The conidium was set at a concentration of 1 × 105 spores/ml. (B) Spray inoculation assay of the Δmoivd mutants on rice seedlings. The conidium was set at a concentration of 5 × 104 spores/ml. (C) Quantitative analysis of lesion numbers in 10 cm2 leaf size. The data was calculated from three independent replicates. The same lowercases ‘a’ or ‘b’ on the sample bar indicate no significant differences between samples. The different lowercases indicate significant differences (P < 0.05; t test). (D) Statistic analysis of the invasive hyphae growth rate. The hyphae are divided into four types according to the growth speed. Type 1 indicates no penetration; Type 2 indicates penetration with a small peg; Type 3 represents the invasive hyphae in extension. Type 4 indicates the hyphae crossing the second host cell. Different color asterisk indicates the different hyphae type. Bar = 10 μm. (E) DAB staining of barley leaves infected by Δmoivd-3after inoculation for 24 h. The asterisk indicates the stained infected host cell by DAB (1 mg/ml). Bar = 50 μm.
The Δmoivd Mutants Cannot Utilize Exogenous Leucine Properly
To further investigate whether MoIVD functions in leucine catabolism and whether MoIVD deletion affects the leucine degradation in M. oryzae, we evaluated the ability of utilizing exogenous leucine by the Δmoivd mutants. By adding extra leucine to the CM and MM media and testing the effect on fungal growth, we found that the Δmoivd-3 and Δmoivd-6 grew much slower and their colonies lost pigmentation completely in contrast to those growing on normal CM or MM medium (Figures 5A,B), suggesting a defect in degradation of exogenous leucine. Furthermore, we found that the mutant could not produce any conidia when growing on leucine-containing medium, although Guy11 and Δmoivd-3/MoIVD also reduced conidia production to some extent (Figure 5C). Unlike leucine, two other branched-chain amino acids, isoleucine and valine, had no effect on the growth and conidiation of the mutants (Supplementary Figure S3). These results indicated that MoIVD is essential for leucine degradation and further confirmed MoIVD function in the leucine catabolism pathway.
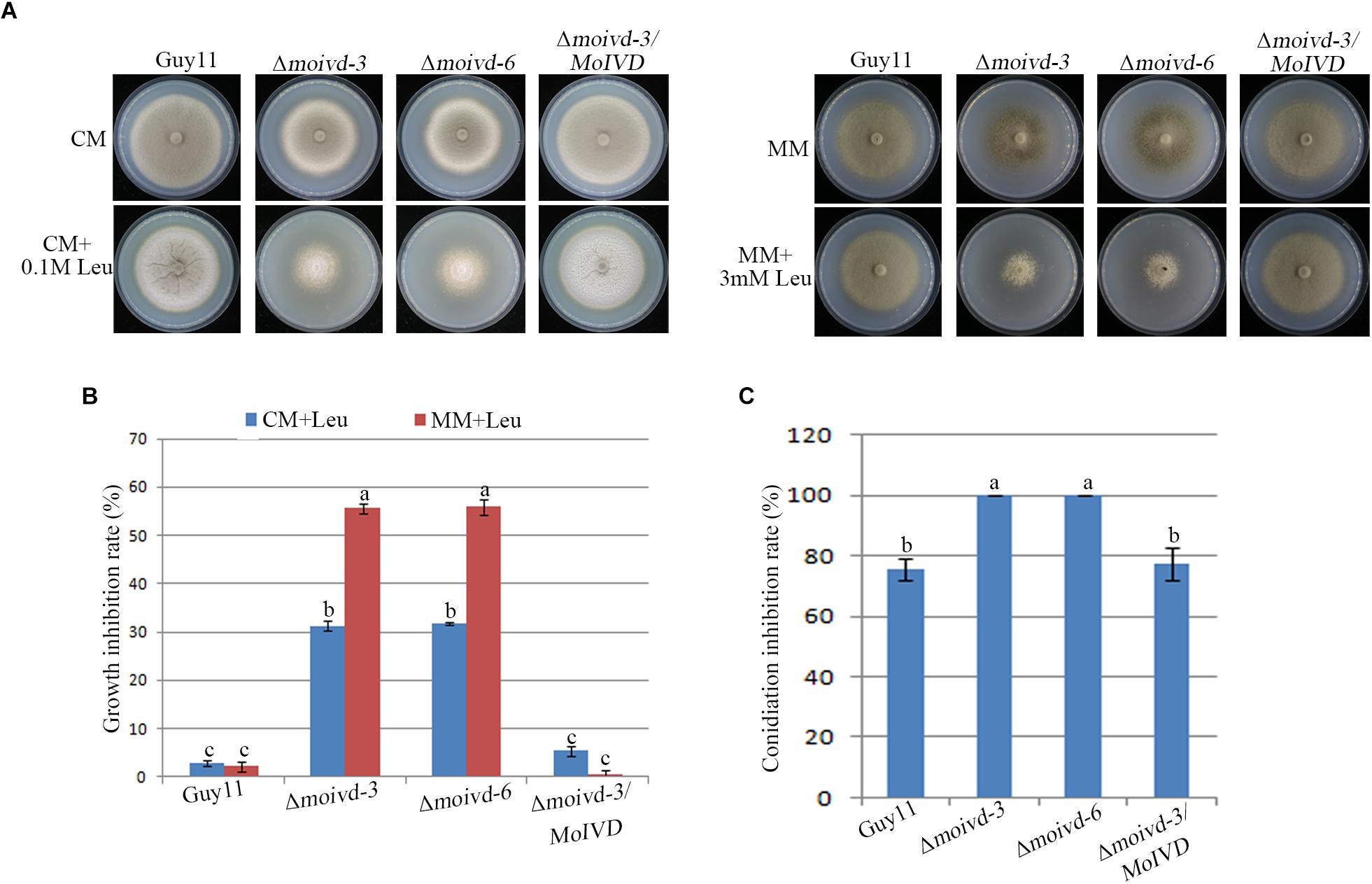
Figure 5. The growth and conidiation phenotype of the Δmoivd mutants under leucine treatment. (A) The growth phenotype of the Δmoivd mutants growing on complete medium (CM) and minimal medium (MM) with adding leucine (Leu). The left panel is the assay performed on CM medium. The right represents MM. The upper panel represents the colony front. The lower panel represents the back of the culture plate. As the mutant growth on MM was very sensitive to high concentrations of leucine, only 3 mM leucine was added in MM. (B) Bar chart showing the growth inhibition rate of the Δmoivd mutants growing on leucine containing medium. (C) Bar chart showing the conidiation inhibition rate of the Δmoivd mutants growing on leucine containing CM medium. All these data were calculated from three independent replicates. The same lowercases ‘a’ or ‘b’ on the sample bar indicate no significant differences between samples. The different lowercases indicate significant differences (P < 0.05; t test).
Isovaleric Acid Accumulation Is Responsible for the Mutant Growth and Conidiation Defects
When growing on CM plates, the Δmoivd mutants released an unpleasant odor, which is consistent with a previous report of smelly odor produced by accumulation of isovaleric acid in blood samples of human patients with IVD deficiency (Tokatli et al., 1998). To this end, we performed GC-MS to test the content of 6 short chain fatty acids in Δmoivd-3. Our results showed a very high concentration of isovaleric acid (54.06 ± 5.55 μg/g) in Δmoivd-3, which was about 450-fold that in Guy11 (0.12 ± 0.03 μg/g) (Figure 6A and Supplementary Figure S4). As a control, we also examined the content of 35 medium and long chain fatty acids by GC-MS and found no significant difference between Δmoivd-3 and Guy11 except several fatty acids with slight changes (Supplementary Figure S5). Thus, MoIVD deletion specifically resulted in isovaleric acid accumulation.
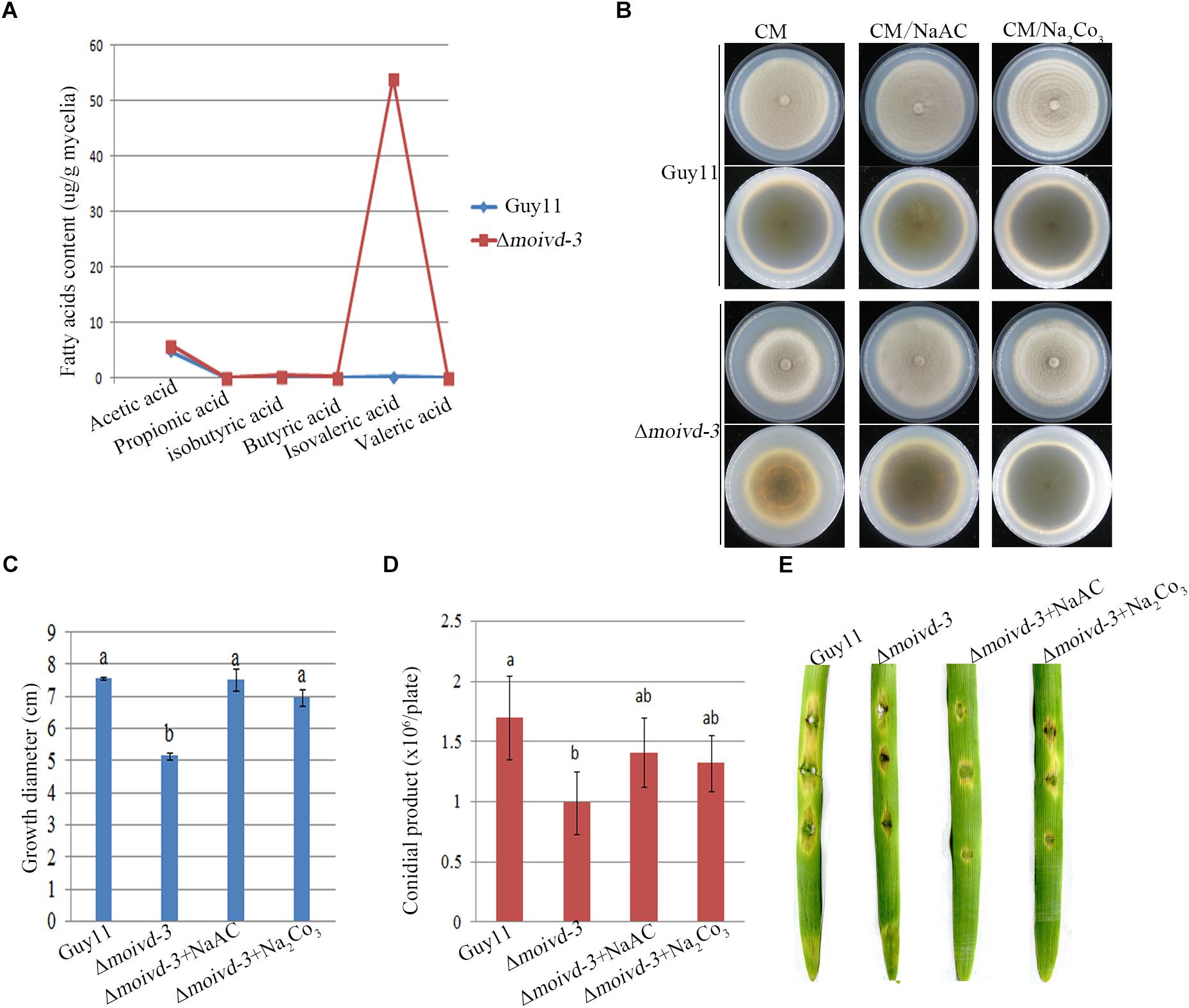
Figure 6. Isovaleric acid accumulation in Δmoivd-3 and the effect of exogenous NaAc and Na2Co3 to mutant phenotype. (A) Line chart showing 6 short chain fatty acid content tested by GCMS. (B) The growth phenotype of Δmoivd-3 growing on NaAc and Na2Co3 treated CM medium. (C) Bar chart showing the growth size of Δmoivd-3 growing on NaAc and Na2Co3 treated CM medium. (D) Bar chart showing the conidial product of Δmoivd-3 growing on NaAc and Na2Co3 treated CM medium. (E) No effect of NaAc or Na2Co3 on the virulence of Δmoivd-3 on excised barley leaves. All the data were calculated from three independent replicates. The same lowercases ‘a’ or ‘b’ on the sample bar indicate no significant differences between samples. The different lowercases indicate significant differences (P < 0.05; t test).
To determine if the accumulation of isovaleric acid contributed to the defects of growth and conidiation in the Δmoivd mutants, we used the weak alkaline of sodium acetate to treat Δmoivd-3 and hoped to neutralize the excessive acid in the mutant. Indeed, Δmoivd-3 almost fully recovered the growth rate and colony color by addition of 50 mM sodium acetate to the medium (Figures 6B,C), and conidia production was also largely restored to the Guy11 level (Figure 6D). Like sodium acetate, sodium carbonate was also able to rescue mutant growth and conidiation (Figures 6B–D). However, both sodium acetate and sodium carbonate were unable to rescue the mutant pathogenicity (Figure 6E).
MoIvd Is Localized in Mitochondria
To further clarify the function of MoIVD, we determined the intracellular localization of MoIvd-GFP in complemented strain Δmoivd-3/MoIVD by confocal microscopy. Our results showed that the green signal of MoIvd-GFP fully co-localized with the red signal of mitochondrial tracker in the conidia, suggesting that MoIvd is localized in the mitochondria (Figure 7A). We further demonstrated the mitochondrial localization of MoIvd-GFP in all developmental stages, including mycelium, conidia, conidial germination, appressoria formation and invasive hyphae (Figures 7B,C). We conclude that MoIvd functions in mitochondria throughout the entire life cycle of M. oryzae.
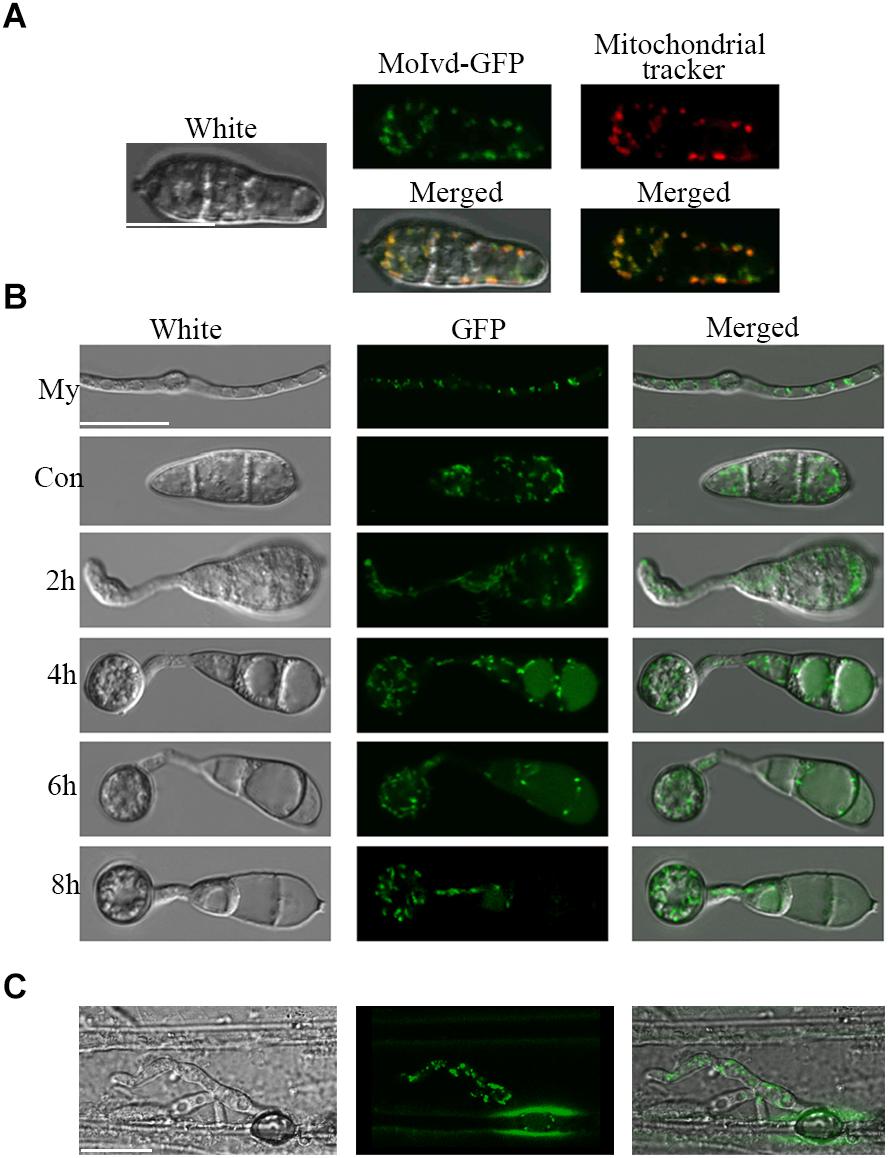
Figure 7. Subcellular localization of MoIvd-GFP. (A) MoIvd-GFP co-localizes with the mitochondrial tracker showing red light analyzed by laser scanning confocal microscopy. The tracker was purchased by Invitrogen with catalog number M7152. Image was taken after the conidia were treated with tracker for 3 min. Bar = 10 μm. (B) MoIvd-GFP stably expressed during mycelia (My) growth, conidia (Con) and appressorium development. Bar = 10 μm. (C) MoIvd-GFP expressed in invasive hyphae by barley infection assay. Bar = 10 μm.
MoIvd Interacts With MoEtfb, the Electron-Transferring Flavoprotein β Subunit in M. oryzae
Our previous study identified the MoEtf, which consists of two subunits—MoEtfa and MoEtfb—as a key regulator of fungal growth, conidiation and pathogenictiy in M. oryzae (Li et al., 2016), and IVD was predicated as one of the ETF electron donors in human (Watmough and Frerman, 2010). To clarify whether there is any interaction between the two proteins in M. oryzae, we conducted Y2H and BIFC assays. As shown in Figure 8A, MoIvd was able to interact with MoEtfb but not MoEtfa in the Y2H assay. In the BIFC assay, MoIvd and MoEtf were fused to the N- and C- terminus of YFP, respectively, and then co-transformed into M. oryzae. Our results showed the interaction fluorescent signal in the conidia co-expressing MoIvd and MoEtfb, but not in the negative controls, and also showed the conidia expressing MoIvd and MoEtfa (Figure 8B). Furthermore, we demonstrated co-localization of MoIvd-GFP and MoEtfb-RFP in the mitochondria of conidia (Figure 8C). These results suggested that MoIvd and MoEtf may function in the same complex for transferring electrons in the mitochondria of M. oryzae.
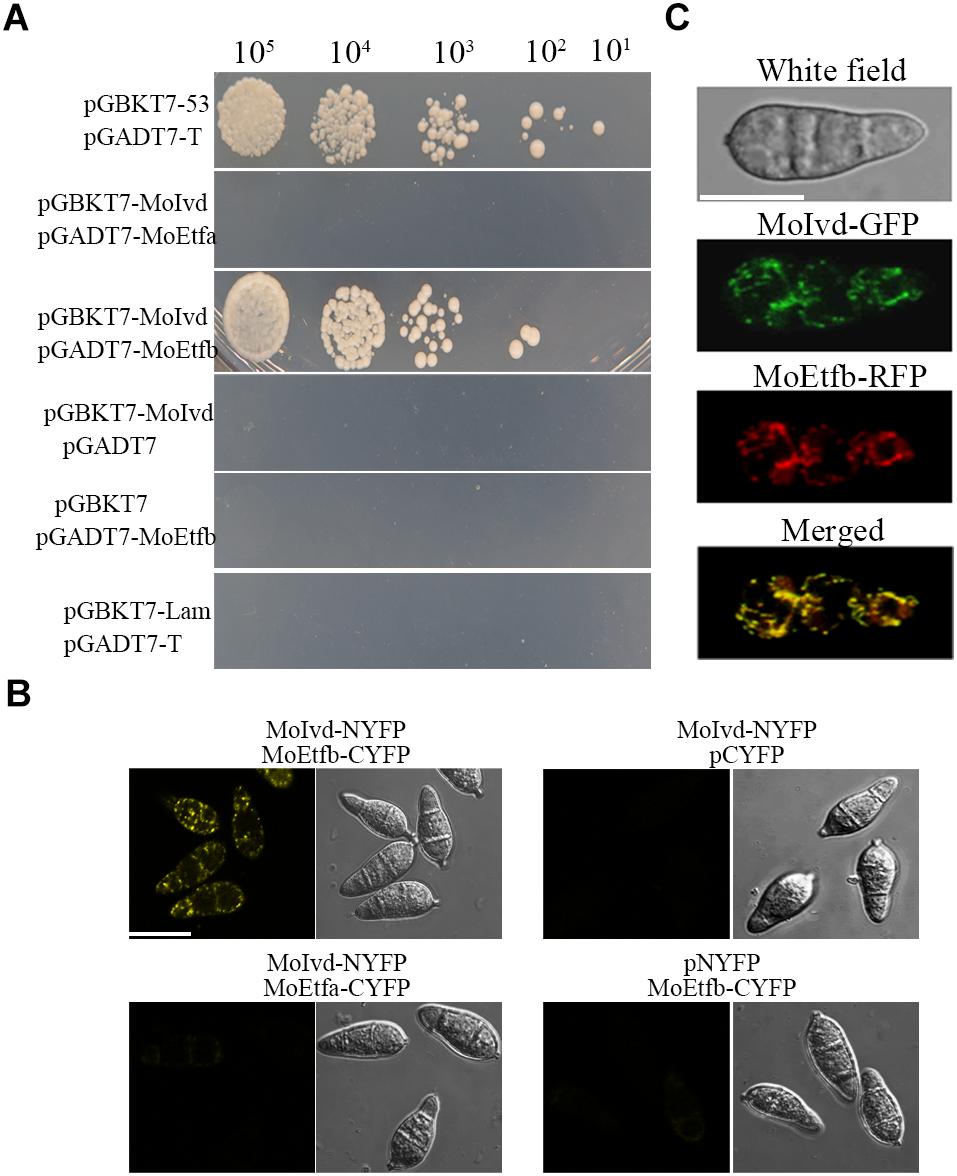
Figure 8. MoIvd interacted and co-localized with MoEtfb. (A) Yeast two hybrids showing that MoIvd interacted with MoEtfb, but not MoEtfa. The yeast transformants were grown on -His/-Ade/-Trp/-Leu/SD medium with 2 mM 3AT to test the interaction. (B) BIFC showing that MoIvd interacted with MoEtfb by displaying whole yellow signal in conidia transformed with MoIvd-NYFP and MoEtfb-CYFP. Bar = 10 μm. (C) MoIvd-GFP was co-localized with MoEtfb-RFP by showing overlapping GFP and RFP fluorescence in conidia transformed with MoIvd-GFP and MoEtfb-RFP. Bar = 10 μm.
Discussion
In this study, we show that MoIVD deletion in M. oryzae resulted in reduced growth, decreased conidiation and compromised pathogenicity of MoIVD-defective strains. The mutant accumulated with a high level of isovaleric acid, and exogenous sodium acetate or sodium carbonate could rescue the mutant growth and conidiation defects (Figures 6B–D). To our knowledge, this is the first report that IVD-mediated leucine catabolism is essential to the development and pathogenesis of plant pathogens. As reported, the human IVD mutation resulted in the isovaleryl-CoA not being degraded normally in the body, where the by product isovaleric acid accumulated and yielded a sweaty foot smell (Tokatli et al., 1998; Erdem et al., 2010; Pascarella et al., 2011). In our study, the Δmoivd mutants also displayed a similar odor and excessive accumulation of isovaleric acid was detected. However, unlike the human case (Fries et al., 1996), the administration of glycine and L-carnitine in M. oryzae could not rescue the defects of Δmoivd-3 (Supplementary Figures S6A,B). It is unclear if the alkalinization of the environment that we used to suppress the mutant defect could be applied to human IVD mutation.
Electron-transferring flavoprotein is predicated as the electron receptor of IVD. Our previous study functionally characterized the MoETF but did not investigate its association with MoIVD (Li et al., 2016). In this study, by a series of protein interaction assays (Figures 8A–C), we confirmed that MoIvd could constitute a complex with MoEtf by interacting with the β subunit of MoEtf (MoEtfb) for possible electron transferring. By further comparing the mutant phenotype, we found both mutants showed defects in vegetative growth, conidiation and virulence, but those in the Δmoetf mutant were more severe than those in the Δmoivd mutant strain. For instance, the Δmoetf mutant fully unpigemented, failed to produce conidia and completely lost virulence (Li et al., 2016). We suggest this is because MoETF functions as electron receptor of 8 predicted acyl-CoA dehydrogenases (gene id MGG_16316, MGG_03418, MGG_05949, MGG_15041, MGG_08690, MGG_08661, MGG_06561, and MGG_02540) in M. oryzae, and MoETF may play a much more extensive and crucial role than a single partner, acyl-CoA dehydrogenase gene (MoIVD). The function of other 7 acyl-CoA dehydrogenases in M. oryzae will be of interest in this regard. Another similar phenotype trait was that the Δmoetf mutant also released an unpleasant odor, probably from accumulated butyrate, and the mutant growth and conidiation, but not pathogenicity, could also be largely recovered by exogenous sodium acetate or carbonate (Li et al., 2016). These data indicate that, as electron transferring proteins, both MoIVD and MoETF are involved in vegetative growth, conidiation and pathogenicity, and the accumulated acid metabolites in the mutants specifically contribute to the growth and conidiation defects.
One of the important products of leucine metabolism is acetyl-CoA, but no report has shown that an IVD mutation in leucine metabolism could cause a shortage of acetyl-CoA and thereby lead to defects. This is likely because leucine catabolism is not the only pathway to produce acetyl-CoA (Lei et al., 2012) and other metabolic pathways would also have a compensation mechanism. Therefore, the byproducts caused by IVD mutation, such as isovaleric acid and hydroxyisovalerate (Erdem et al., 2010; Pascarella et al., 2011), would play more important roles in leading to mutant defects. Also confirmed by our study in M. oryzae, supplementation of acetyl-CoA could not rescue Δmoivd-3 defects (Supplementary Figure S6C). This finding further suggests that the major reason responsible for the Δmoivd mutants’ defects is the excess isovaleric acid. However, the mutant defects in virulence were not caused by isovaleric acid, as they were not responsive to sodium acetate or carbonate (Figure 6E). Considering that the mutant could produce normal appressorium with enough turgor pressure for penetration (Supplementary Figure S7), we speculated that the reduced virulence of the mutant was more likely the result of not coping with oxidative stress during initial colonization. In our study, we confirmed this hypothesis by the result that ROS production was enhanced significantly during mutant-mediated infection (Figure 5E). To analyze whether the ROS production was induced by isovaleric acid, we treated barley leaves with 0.1, 1 and 10% isovaleric acid in vitro and then stained the leaves with DAB. Similar to the untreated leaves, no ROS was stained. This suggests that it is not isovaleric acid that induced high ROS production in the mutant infected host cells (Supplementary Figure S8). Given that MoIVD is involved in electron transferring and electron transferring dysfunction probably caused an imbalance of redox state (Lämmer et al., 2011), we speculate that the mutant defect in managing oxidative stress possibly results from imbalance of redox state, although more evidence is needed.
The IVD mutation destroys leucine catabolism at the third step, involving the conversion of isovaleryl-CoA to β-methylcrotonyl-CoA. An accumulation of leucine has not been observed in IVD mutants (Mohsen et al., 1998; Mohsen and Vockley, 2015) and we also did not find an obvious high level of leucine in Δmoivd-3 (Supplementary Figure S9), but rather a high level of isovaleric acid was present (Figure 6A and Supplementary Figure S4). These findings suggest that the IVD mutation led to a block of leucine catabolism at the third step, and that accumulated isovaleryl-CoA is converted into isovaleric acid that is not further degraded. This explains why the Δmoivd mutants displayed a more severe defect in growth and conidiation when leucine was added to the culture medium (Figure 5). Addition of two other branched amino acids, valine and isoleucine, to the growth medium caused no obvious change in the growth and conidiation characteristics displayed by the mutant strain (Supplementary Figure S3). This result further suggested that MoIVD is specific for leucine catabolism in M. oryzae, which is not similar to the potato IVD, which has an additional role in degrading valine (Faivre-Nitschke et al., 2001).
In summary, we functionally characterized MoIvd-mediated leucine catabolism in M. oryzae and proposed a working model in which the leucine-inducible MoIvd functions specifically in the third reaction of leucine catabolism in mitochondria (Figure 9). Upon MoIVD deletion, the isovaleryl-CoA could not be degraded and is converted to isovaleric acid, which accumulates in the mutant, possibly within mitochondria. The isovaleric acid accumulation at least partially contributed to the mutant growth and conidiation defects due to intracellular acidification, as those phenotypes were largely recovered by alkalinization of the medium. As the electron donor of MoETF, MoIVD deletion would block the MoIvd-MoEtf-mediated electron transfer reaction and probably result in an imbalance of redox state, which explains why the mutant could not overcome oxidative stress during infection and subsequently displayed a reduced virulence.
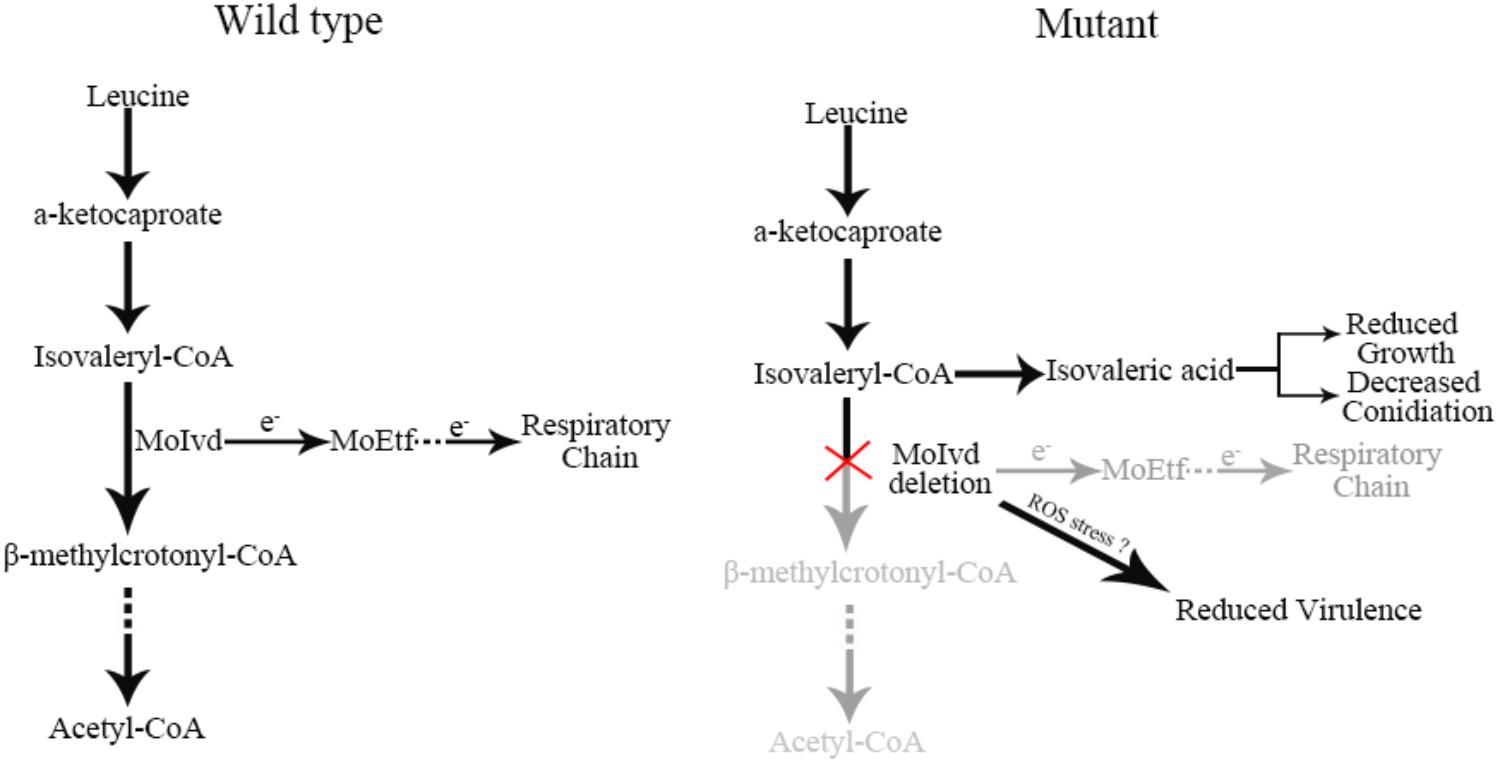
Figure 9. Proposed working model of MoIVD in M. oryzae. In wild-type strain, leucine is normally degraded by the leucine catabolism pathway and electrons are transferred to the respiratory chain from MoIvd, while in the Δmoivd mutants, the pathway was blocked at the third step where isovaleryl-CoA was converted into isovaleric acid. The isovaleric acid accumulation in mutant resulted in the reduced growth and decreased conidiation. The reduced virulence probably resulted from the mutant’s inability to overcome host ROS defense. “e-” indicates electron. “?” means a probable virulence reduction mechanism of mutant not coping with ROS stress during infection.
Author Contributions
YL and GDL designed the experiments. YL and XZ analyzed the mutant phenotypes, processed the data and wrote the manuscript. MC, MZ, YHZ, WW, XC, and SZ constructed the gene deletion and complementation and collected the mutant phenotype data. FH, JL, MP, and YZ completed the protein localization experiments. JZ did the DAB staining assay. GPL, MW, and ZW revised the manuscript. All the authors approved the manuscript.
Funding
This research was financially supported by the National Science Foundation of China (31500120), Special Fund of Fujian Agriculture and Forestry University for Science and Technology Innovation (KFA17384A), and the Natural Science Foundation of Fujian Province (2018J01615).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Daniel J. Ebbole from Texas A&M University for making a thorough grammar and writing revision to this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00444/full#supplementary-material
Footnotes
References
Arany, Z., and Neinast, M. (2018). Branched chain amino acids in metabolic disease. Curr. Diab. Rep. 18:76. doi: 10.1007/s11892-018-1048-7
Araújo, W. L., Ishizaki, K., Nunes-Nesi, A., Larson, T. R., Tohge, T., Krahnert, I., et al. (2010). Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22, 1549–1563. doi: 10.1105/tpc.110.075630
Brosnan, J. T., and Brosnan, M. E. (2006). Branched-chain amino acids: enzyme and substrate regulation. J. Nutr. 136, 207–211. doi: 10.1093/jn/136.1.207S
Chen, X. L., Shi, T., Yang, J., Shi, W., Gao, X., Chen, D., et al. (2014). N-glycosylation of effector proteins by an α-1,3-mannosyltransferase is required for the rice blast fungus to evade host innate immunity. Plant Cell 26, 1360–1376. doi: 10.1105/tpc.114.123588
Däschner, K., Couée, I., and Binder, S. (2001). The mitochondrial isovaleryl-coenzyme a dehydrogenase of arabidopsis oxidizes intermediates of leucine and valine catabolism. Plant Physiol. 126, 601–612. doi: 10.1104/pp.126.2.601
Däschner, K., Thalheim, C., Guha, C., Brennicke, A., and Binder, S. (1999). In plants a putative isovaleryl-CoA-dehydrogenase is located in mitochondria. Plant Mol. Biol. 39, 1275–1282.
Dean, R. A., Talbot, N. J., Ebbole, D. J., Farman, M. L., Mitchell, T. K., Orbach, M. J., et al. (2005). The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434, 980–986. doi: 10.1038/nature03449
Du, Y., Hong, L., Tang, W., Li, L., Wang, X., Ma, H., et al. (2014). Threonine deaminase MoIlv1 is important for conidiogenesis and pathogenesis in the rice blast fungus Magnaporthe oryzae. Fungal Genet. Biol. 73, 53–60. doi: 10.1016/j.fgb.2014.10.001
Du, Y., Zhang, H., Hong, L., Wang, J., Zheng, X., and Zhang, Z. (2013). Acetolactate synthases MoIlv2 and MoIlv6 are required for infection-related morphogenesis in Magnaporthe oryzae. Mol. Plant Pathol. 14, 870–884. doi: 10.1111/mpp.12053
Ebbole, D. J. (2007). Magnaporthe as a model for understanding host pathogen interactions. Annu. Rev. Phytopathol. 45, 437–456. doi: 10.1146/annurev.phyto.45.062806.094346
Erdem, E., Cayonu, N., Uysalol, E., and Yildirmak, Z. Y. (2010). Chronic intermittent form of isovaleric acidemia mimicking diabetic ketoacidosis. J. Pediatr. Endocrinol. Metab. 23, 503–505. doi: 10.1515/jpem.2010.082
Faivre-Nitschke, S. E., Couée, I., Vermel, M., Grienenberger, J. M., and Gualberto, J. M. (2001). Purification, characterization and cloning of isovaleryl-CoA dehydrogenase from higher plant mitochondria. Eur. J. Biochem. 268, 1332–1339. doi: 10.1046/j.1432-1327.2001.01999.x
Fernandez, J., and Orth, K. (2018). Rise of a cereal killer: the biology of Magnaporthe oryzae biotrophic growth. Trends Microbiol. 26, 582–597. doi: 10.1016/j.tim.2017.12.007
Fries, M. H., Rinaldo, P., Schmidt-Sommerfeld, E., Jurecki, E., and Packman, S. (1996). Isovaleric acidemia: response to a leucine load after three weeks of supplementation with glycine, L-carnitine, and combined glycine-carnitine therapy. J. Pediatr. 129, 449–452. doi: 10.1016/S0022-3476(96)70081-1
Ghisla, S., and Thorpe, C. (2004). Acyl-CoA dehydrogenases. A mechanistic overview. Eur. J. Biochem. 271, 494–508. doi: 10.1046/j.1432-1033.2003.03946.x
Hamer, J. E., Howard, R. J., Chumley, F. G., and Valent, B. (1988). A mechanism for surface attachment in spores of a plant pathogenic fungus. Science 239, 288–290. doi: 10.1126/science.239.4837.288
Howard, R. J., Ferrari, M. A., Roach, D. H., and Money, N. P. (1991). Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc. Natl. Acad. Sci. U.S.A. 88, 11281–11284. doi: 10.1073/pnas.88.24.11281
Islam, M. T., Croll, D., Gladieux, P., Soanes, D. M., Persoons, A., Bhattacharjee, P., et al. (2016). Emergence of wheat blast in Bangladesh was caused by a South American lineage of Magnaporthe oryzae. BMC Biol. 14:84. doi: 10.1186/s12915-016-0309-7
Kankanala, P., Czymmek, K., and Valent, B. (2007). Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19, 706–724. doi: 10.1105/tpc.106.046300
Kaya, N., Colak, D., Al-Bakheet, A., Al-Younes, B., Tulbah, S., Daghestani, M., et al. (2013). Identification of a novel IVD mutation in a consanguineous family with isovaleric acidemia. Gene 513, 297–300. doi: 10.1016/j.gene.2012.09.097
Lämmer, A. B., Rolinski, B., Ahting, U., and Heuss, D. (2011). Multiple acyl-CoA-dehydrogenase deficiency (MADD)–a novel mutation of electron-transferring-flavoprotein dehydrogenase ETFDH. J. Neurol. Sci. 307, 166–167. doi: 10.1016/j.jns.2011.05.001
Lei, J., Feng, D., Zhang, Y., Dahanayaka, S., Li, X., Yao, K., et al. (2012). Regulation of leucine catabolism by metabolic fuels in mammary epithelial cells. Amino Acids 45, 2179–2189. doi: 10.1007/s00726-012-1302-2
Li, Y., Liang, S., Yan, X., Wang, H., Li, D., Soanes, D. M., et al. (2010a). Characterization of MoLDB1 required for vegetative growth, infection-related morphogenesis, and pathogenicity in the rice blast fungus Magnaporthe oryzae. Mol. Plant Microbe Interact. 23, 1260–1274. doi: 10.1094/MPMI-03-10-0052
Li, Y., Yan, X., Wang, H., Liang, S., Ma, W. B., Fang, M. Y., et al. (2010b). MoRic8 is a novel component of G-protein signaling during plant infection by the rice blast fungus Magnaporthe oryzae. Mol. Plant Microbe Interact. 23, 317–331. doi: 10.1094/MPMI-23-3-0317
Li, Y., Yue, X., Que, Y., Yan, X., Ma, Z., Talbot, N. J., et al. (2014). Characterisation of four LIM protein-encoding genes involved in infection-related development and pathogenicity by the rice blast fungus Magnaporthe oryzae. PLoS One 9:e88246. doi: 10.1371/journal.pone.0088246
Li, Y., Zhu, J., Hu, J., Meng, X., Zhang, Q., Zhu, K., et al. (2016). Functional characterization of electron-transferring flavoprotein and its dehydrogenase required for fungal development and plant infection by the rice blast fungus. Sci. Rep. 6:24911. doi: 10.1038/srep24911
Mohsen, A. W., Anderson, B. D., Volchenboum, S. L., Battaile, K. P., Tiffany, K., Roberts, D., et al. (1998). Characterization of molecular defects in isovaleryl-CoA dehydrogenase in patients with isovaleric acidemia. Biochemistry 37, 10325–10335. doi: 10.1021/bi973096r
Mohsen, A. W., and Vockley, J. (2015). Kinetic and spectral properties of isovaleryl-CoA dehydrogenase and interaction with ligands. Biochimie 108, 108–119. doi: 10.1016/j.biochi.2014.11.007
Noda, C., Rhead, W. J., and Tanaka, K. (1980). Isovaleryl-CoA dehydrogenase: demonstration in rat liver mitochondria by ion exchange chromatography and isoelectric focusing. Proc. Natl. Acad. Sci. U.S.A. 77, 2646–2650. doi: 10.1073/pnas.77.5.2646
Pascarella, A., Rosa, M., della Casa, R., Andria, G., and Parenti, G. (2011). Isovaleric academia. J. Pediatr. Endocrinol. Metab. 24:399. doi: 10.1515/jpem.2011.046
Reinard, T., Janke, V., Willard, J., Buck, F., Jacobsen, H. J., and Vockley, J. (2000). Cloning of a gene for an acyl-CoA dehydrogenase from Pisum sativum L. and purification and characterization of its product as an isovaleryl-CoA dehydrogenase. J. Biol. Chem. 275, 33738–33743. doi: 10.1074/jbc.M004178200
Rozen, R., Vockley, J., Zhou, L., Milos, R., Willard, J., Fu, K., et al. (1994). Isolation and expression of a cDNA encoding the precursor for a novel member (ACADSB) of the acyl-CoA dehydrogenase gene family. Genomics 24, 280–287. doi: 10.1006/geno.1994.1617
Sadat, M. A., and Choi, J. (2017). Wheat blast: a new fungal inhabitant to bangladesh threatening world wheat production. Plant Pathol. J. 33, 103–108. doi: 10.5423/PPJ.RW.09.2016.0179
Sakulkoo, W., Osés-Ruiz, M., Garcia, E. O., Soanes, D. M., Littlejohn, G. R., Hacker, C., et al. (2018). A single fungal MAP kinase controls plant cell-to-cell invasion by the rice blast fungus. Science 359, 1399–1403. doi: 10.1126/science.aaq0892
Surger, M. J., Angelov, A., Stier, P., Übelacker, M., and Liebl, W. (2018). Impact of branched-chain amino acid catabolism on fatty acid and alkene biosynthesis in Micrococcus luteus. Front. Microbiol. 9:374. doi: 10.3389/fmicb.2018.00374
Tablot, N. J. (2003). On the trail of a cereal killer: exploring the biology of Magnaporthe grisea. Annu. Rev. Microbiol. 57, 177–202. doi: 10.1146/annurev.micro.57.030502.090957
Tanaka, K. (1990). Isovaleric acidemia: personal history, clinical survey and study of the molecular basis. Prog. Clin. Biol. Res. 321, 273–290.
Tiffany, K. A., Roberts, D. L., Wang, M., Paschke, R., Mohsen, A. W., Vockley, J., et al. (1997). Structure of human isovaleryl-CoA dehydrogenase at 2.6 A resolution: structural basis for substrate specificity. Biochemistry 36, 8455–8464. doi: 10.1021/bi970422u
Tokatli, A., Coşkun, T., and Ozalp, I. (1998). Isovaleric acidemia. Clinical presentation of 6 cases. Turk. J. Pediatr. 40, 111–119.
Valent, B., and Chumley, F. G. (1991). Molecular genetic analysis of the rice blast fungus, Magnaporthe grisea. Annu. Rev. Phytopathol. 29, 443–467. doi: 10.1146/annurev.py.29.090191.002303
Vockley, J., and Ensenauer, R. (2006). Isovaleric acidemia: new aspects of genetic and phenotypic heterogeneity. Am. J. Med. Genet. C Semin. Med. Genet. 142C, 95–103. doi: 10.1002/ajmg.c.30089
Watmough, N. J., and Frerman, F. E. (2010). The electron transfer flavoprotein: ubiquinone oxidoreductases. Biochim. Biophys. Acta 1797, 1910–1916. doi: 10.1016/j.bbabio.2010.10.007
Xu, J. R., Staiger, C. J., and Hamer, J. E. (1998). Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc. Natl. Acad. Sci. U.S.A. 95, 12713–12718. doi: 10.1073/pnas.95.21.12713
Yamashita, N., Sakamoto, K., Yamada, O., Akita, O., and Nishimura, A. (2007). The promoter activity of isovaleryl-CoA dehydrogenase-encoding gene (IvdA) from Aspergillus oryzae is strictly repressed by glutamic acid. Biosci. Biotechnol. Biochem. 71, 1561–1563. doi: 10.1271/bbb.60712
Keywords: Magnaporthe oryzae, isovaleryl-CoA dehydrogenase, leucine catabolism, biological function, fungal development and pathogenesis
Citation: Li Y, Zheng X, Zhu M, Chen M, Zhang S, He F, Chen X, Lv J, Pei M, Zhang Y, Zhang Y, Wang W, Zhang J, Wang M, Wang Z, Li G and Lu G (2019) MoIVD-Mediated Leucine Catabolism Is Required for Vegetative Growth, Conidiation and Full Virulence of the Rice Blast Fungus Magnaporthe oryzae. Front. Microbiol. 10:444. doi: 10.3389/fmicb.2019.00444
Received: 06 November 2018; Accepted: 20 February 2019;
Published: 14 March 2019.
Edited by:
Gustavo Henrique Goldman, University of São Paulo, BrazilReviewed by:
Qing-Ming Qin, Jilin University, ChinaChaoyang Xue, Rutgers, The State University of New Jersey, United States
Copyright © 2019 Li, Zheng, Zhu, Chen, Zhang, He, Chen, Lv, Pei, Zhang, Zhang, Wang, Zhang, Wang, Wang, Li and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ya Li, bGl5YS04MUAxNjMuY29t Guodong Lu, Z2RsdWZhZnVAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Ya Li
Ya Li Xiuxia Zheng1†
Xiuxia Zheng1† Yunhui Zhang
Yunhui Zhang Mo Wang
Mo Wang Zonghua Wang
Zonghua Wang Guangpu Li
Guangpu Li Guodong Lu
Guodong Lu