- 1Institute for Cell and Molecular Biosciences, Newcastle University, Newcastle upon Tyne, United Kingdom
- 2Department of Biotechnology, Yonsei University, Seoul, South Korea
- 3Department of Molecular Genetics and Microbiology, Duke University Medical Center, Durham, NC, United States
- 4Public Health Research Institute, Rutgers University, Newark, NJ, United States
- 5Department of Molecular Genetics, Biochemistry and Microbiology, Rutgers University, Newark, NJ, United States
More than 1.5 million fungal species are estimated to live in vastly different environmental niches. Despite each unique host environment, fungal cells sense certain fundamentally conserved elements, such as nutrients, pheromones and stress, for adaptation to their niches. Sensing these extracellular signals is critical for pathogens to adapt to the hostile host environment and cause disease. Hence, dissecting the complex extracellular signal-sensing mechanisms that aid in this is pivotal and may facilitate the development of new therapeutic approaches to control fungal infections. In this review, we summarize the current knowledge on how two important pathogenic yeasts, Candida albicans and Cryptococcus neoformans, sense nutrient availability, such as carbon sources, amino acids, and ammonium, and different stress signals to regulate their morphogenesis and pathogenicity in comparison with the non-pathogenic model yeast Saccharomyces cerevisiae. The molecular interactions between extracellular signals and their respective sensory systems are described in detail. The potential implication of analyzing nutrient and stress-sensing systems in antifungal drug development is also discussed.
Carbon Sensing in Fungal Pathogenesis
Fungal pathogens prefer certain carbon sources for rapid uptake and metabolism to provide energy for growth and host colonization. The main carbon sources available in a host during fungal infection are glucose, lactate, and acetate (Ries et al., 2018). In addition, inositol can also be utilized as a carbon source by some fungi, including Cryptococcus neoformans. Multiple carbon sources and fungal sensory systems have been reported in pathogenic yeasts (Figure 1).
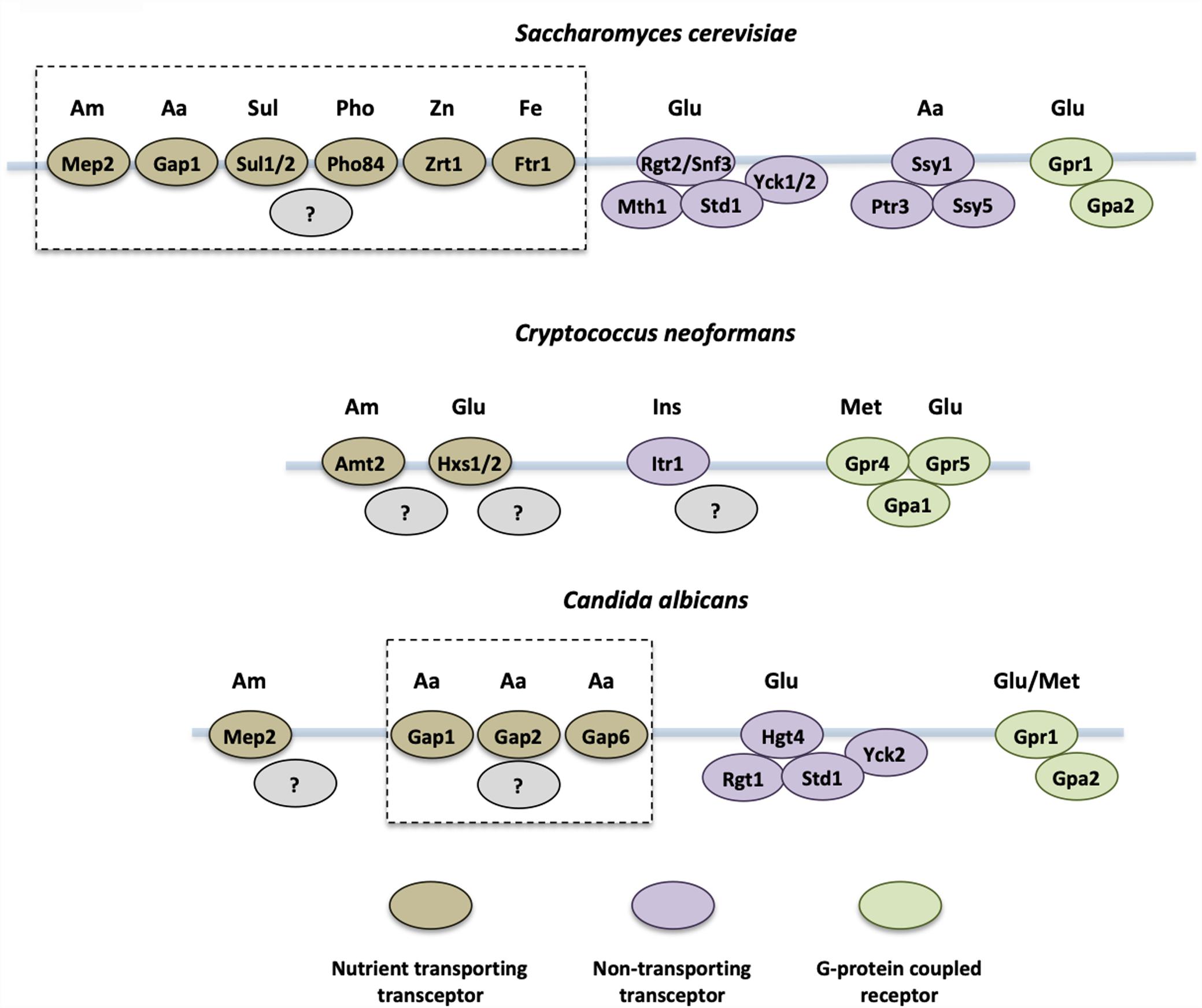
Figure 1. Yeast nutrient receptors that sense external nutrient availability. Nutrient sensing transceptors and G-protein coupled receptors in Saccharomyces cerevisiae, Cryptococcus neoformans, and Candida albicans are shown. Nutrient sensors for glucose (Glu), amino acids (Aa), ammonium (Am), sulfate (Sul), phosphate (Pho), zinc (Zn), iron (Fe), methionine (Met), and inositol (Ins) have been identified. Dashed lines group those transceptors that rapidly activate the PKA pathway by an unknown mechanism. The ammonium transceptors from S. cerevisiae (Mep2), C. albicans (Mep2), and C. neoformans (Amt2) regulate morphology changes in response to ammonium import.
Sugar Transporter Homologs Functioning as Sugar Sensors: Transceptors
Glucose is a preferred carbon and major energy source for most cells. The glucose sensing and signaling networks have been well-characterized in Saccharomyces cerevisiae (Kim and Johnston, 2006; Santangelo, 2006). In S. cerevisiae, two glucose transceptors, Snf3 and Rgt2, sense extracellular glucose levels and regulate the expression of the hexose transporter gene family, which comprises more than 20 genes. Snf3 is a high-affinity glucose sensor and is activated by low glucose levels, while Rgt2 is a low-affinity glucose sensor that is activated by high glucose levels. Both Snf3 and Rgt2 have long C-terminal cytoplasmic tails that interact with casein kinase I (Yck1 and Yck2) and with two transcription regulator proteins (Mth1 and Std1). Although Snf3 and Rgt2 do not transport glucose themselves (Ozcan et al., 1998), it is speculated that glucose binding leads to their conformational changes that activate casein kinase I. In the absence of glucose, Mth1 and Std1 serve as corepressors with the master transcription repressor Rgt1. These three proteins form a complex that binds to the promoter of hexose transporters to repress their expression (Moriya and Johnston, 2004). When glucose is available, Mth1 and Std1 are phosphorylated by casein kinase I and ubiquitinated by the SCF (Skp1-Cullin-F-box protein) E3 ubiquitin ligase Grr1, which leads to their degradation by the 26S proteasome. Depletion of the corepressors dissociates Rgt1 and relieves repression of hexose transporter gene transcription (reviewed in Santangelo, 2006). Hexose transporter genes are the primary regulatory targets of this glucose sensing mechanism in yeast, and this regulation optimizes the expression levels of high- and low-affinity hexose transporters.
The long cytoplasmic tail of Snf3 and Rgt2 plays an important role in glucose sensing by bringing Mth1 and Std1 to Yck1 in the membrane. These cytoplasmic tails can be fused to other sugar transporters and overexpression of these chimeric proteins can derepress expression of hexose transporter genes (Ozcan et al., 1998). Furthermore, overexpression of a membrane-targeted form of the cytoplasmic tail alone leads to constitutive activation of the glucose sensing pathway (Dlugai et al., 2001). However, the tails do not seem to be essential for glucose signaling, because overexpression of an Rgt2 allele lacking the tail also activates hexose transporter (Hxt1) expression (Moriya and Johnston, 2004). It is possible that Rgt2 may stimulate Yck1 through an interaction independent of Yck1 binding to the cytoplasmic tail of Rgt2.
Among over 20 hexose transporter homologs in Candida albicans (Fan et al., 2002), Hgt4 has been identified as a high-affinity glucose transceptor (Brown et al., 2006). Hgt4 shares sequence and structure similarity with other hexose transporters, with the exception of a long C-terminal tail containing 254 amino acids (aa), similar to Snf3 and Rgt2 in S. cerevisiae. Hgt4 is required for glucose induction of other hexose transporter genes, including HGT12, HXT10, and HGT7. Mutagenesis demonstrated that Hgt4 is required for fungal growth on fermentable sugars, such as fructose, mannose, and glucose. Hgt4 is also required for the yeast-to-hyphal morphological switch as well as fungal virulence, demonstrating that C. albicans cells need to sense and regulate sugar levels for filamentous growth during infection. The Hgt4-mediated regulatory mechanism of glucose repression in C. albicans is conserved with its counterpart in S. cerevisiae, and involves the casein kinase Yck2, corepressor Std1, and Rgt1 (Kim and Johnston, 2006; Sabina and Brown, 2009).
Despite the importance of glucose sensing and utilization in fungal development and virulence, glucose sensing is less well-understood in C. neoformans. There are about 50 transporters that share high sequence similarity with known hexose transporters (Liu et al., 2013b). Although some of them may transport sugars other than glucose, it remains elusive how this large family of hexose transporters is regulated. Among these hexose transporter homologs, Hxs1 and Hxs2 share the highest sequence identity with S. cerevisiae Rgt2 and Snf3 glucose transceptors, but neither of them have a long C-terminal tail (Liu et al., 2013b). The expression of Hxs1 is negatively regulated by glucose levels, and mutagenesis analysis showed that Hxs1 is required for efficient glucose uptake and fungal growth under low glucose conditions. Hxs1 is also required for fungal virulence in a murine model of systemic cryptococcosis. However, Hxs1 only modestly regulates the expression of other hexose transporters and it still has glucose uptake activity (Liu et al., 2013b). It is possible that Hxs1 has dual functions as both a glucose sensor and glucose transporter. On the other hand, the function of Hxs2 remains undefined. The downstream regulatory mechanism of glucose repression has not been characterized in detail in C. neoformans, but the function of the two casein kinase I, Cck1 and Cck2, has been studied. While the function of Cck2 is unknown, Cck1 is required for cell integrity and stress response by regulating the phosphorylation of Mpk1 and Hog1 mitogen-activated protein kinases (MAPKs) and also essential for fungal virulence (Wang et al., 2011b).
GPCRs in Glucose Sensing
The role of GPCRs in glucose sensing has been well-characterized in S. cerevisiae. The GPCR receptor Gpr1 encodes a protein containing over 800 aa with a large third cytoplasmic loop and a long C-terminal tail. Gpr1 and its homologs in other fungi share limited sequence homology with the other defined GPCR classes (Attwood and Findlay, 1994; Kolakowski, 1994), and are grouped as a novel evolutionarily distinct GPCR class. Gpr1 senses glucose to activate the G-protein α subunit Gpa2 and regulates yeast cell size and pseudohyphal growth. The binding of glucose with Gpr1 leads to a conformational change that activates Gpa2, which in turn activates adenylyl cyclase to convert ATP into cAMP (Xue et al., 1998; Kraakman et al., 1999; Lorenz et al., 2000b; Rolland et al., 2002; Lemaire et al., 2004). cAMP then binds to the regulatory subunit (Bcy1) of protein kinase A (PKA) and thereby releases the catalytic subunits (Tpk1, 2, 3) of PKA to phosphorylate downstream target proteins. Interestingly, Gpr1 also senses sucrose to activate the Gpa2-cAMP pathway, and the affinity of Gpr1 for sucrose is much higher than for glucose. The half-maximal effective concentration (EC50) for sucrose is around 0.5 mM, compared to approximately 20 to 30 mM for glucose to activate the Gpa2-cAMP pathway. Meanwhile, mannose acts as an antagonist for glucose and sucrose in Gpr1 activation (Versele et al., 2001; Lemaire et al., 2004).
The Gpr1 protein sequence is conserved in C. albicans. Similar to S. cerevisiae, the C. albicans Gpr1 receptor binds to Gpa2 to activate G protein signaling, which in turn activates the cAMP-PKA signaling pathway (Miwa et al., 2004; Maidan et al., 2005a). Gpr1 is important for filamentous growth on solid media, but not in lipid medium (Miwa et al., 2004). However, the role of C. albicans Gpr1 in glucose sensing remains unclear. Some studies showed that Gpr1 and Gpa2 do not have a role in glucose-induced cAMP signaling and may not be involved in glucose sensing (Maidan et al., 2005a). Instead, deletion mutants of Cdc25 or Ras2 lack glucose-induced cAMP signaling, suggesting that the Cdc25-Ras2 branch is instead responsible for glucose sensing in C. albicans. It is possible that amino acids such as methionine are the ligand for Gpr1 in C. albicans, as methionine can trigger Gpr1 internalization and methionine induction of hypha formation on solid media requires a functional Gpr1 (Maidan et al., 2005a). However, the effect of methionine on hypha formation requires the presence of a low level of glucose in the medium. Therefore, it remains possible that Gpr1 may sense methionine, glucose, or both (Maidan et al., 2005b).
In C. neoformans, glucose sensing and utilization is critical for its development and virulence. In addition to being utilized as a preferred energy source for cell growth, glucose is required for capsule production both as a substrate and a signaling molecule. Glucose induces capsule enlargement through the Gpa1-cAMP-PKA signal transduction pathway, which plays a central role in fungal virulence (Alspaugh et al., 1997; Bahn et al., 2007). Although the cAMP-regulated PKA pathway is largely conserved, there is no Gpr1 homolog. Two GPCRs, Gpr4 and Gpr5, share structural similarity with Gpr1. Similar to Gpr1, Gpr4 encodes a large protein containing more than 800 aa with a long third cytoplasmic loop and C-terminal tail, but is not important for glucose sensing because glucose-mediated cAMP signaling activation is independent of Gpr4 function (Xue et al., 2006). Rather, gpr4Δ mutants exhibit defects related to methionine-induced morphogenesis, which is similar to what has been observed in Candida Gpr1. Gpr5 is a smaller protein that shares high sequence identity with Gpr4, and its mutant has shown defects in “Titan” cell production (Okagaki et al., 2011). The gpr4Δ gpr5Δ double mutants have even more pronounced defects in cell size regulation, suggesting these two GPCRs have overlapping functions. It is possible that Gpr5 may be involved in sensing carbon, including glucose. Gpr4 and Gpr5 have been shown to interact with Gpa1 to activate the cAMP-PKA pathway and control cell size and capsule (Okagaki et al., 2011).
Sensing Other Carbon Sources
Inositol is a small carbohydrate molecule that functions as an essential structural and signaling molecule in eukaryotes, including fungi (Fisher et al., 2002; Xue, 2012). The myo-inositol transporter gene family belongs to the sugar transporter superfamily and may also play important roles in myo-inositol sensing in fungi. High sequence similarity within this gene family suggests that these genes likely evolved from a common ancestor. There are two myo-insoitol transporters (ITRs) in S. cerevisiae (Nikawa et al., 1991, 1993) and in C. albicans (Jin and Seyfang, 2003; Chen et al., 2008), but no inositol sensor has been identified in these yeast organisms.
Inositol seems to play a significant role in C. neoformans development and pathogenicity. It can be used as a sole carbon source (Healy et al., 1977) and can also stimulate Cryptococcus mating (Xue et al., 2007). As one of the most abundant metabolites in the mammalian brain (Fisher et al., 2002), inositol utilization is required for C. neoformans virulence in murine infection models by promoting brain infection (Xue et al., 2010; Wang et al., 2011a; Liu et al., 2013a, 2014). Therefore, C. neoformans likely utilizes the abundant inositol available inside the mammalian brain for its pathogenicity. Inositol can also stimulate C. neoformans capsule growth, which may contribute to its role in fungal virulence. The cryptococcal genome reflects the evolutionary adaptations associated with the expanded role of inositol in this organism. In particular, C. neoformans contains an unusually large number of ITRs that consists of more than 10 members (Xue et al., 2007, 2010). Functional analysis of the ITR gene family demonstrated that two members (Itr1a and Itr3c) have high inositol uptake activity, while the functions of the other members remain undefined. Of these, Itr1a could be a possible inositol transceptor because it does not show uptake activity in a yeast heterologous system, but it does regulate other ITR gene expression and the itr1aΔ single mutant exhibits defects in mating, hyphal production, and sporulation (Xue et al., 2010; Wang et al., 2011a). Therefore, an inositol transceptor may exist in C. neoformans. Importantly, fungal inositol transporters are proton-dependent symporters, which are pharmacokinetically different from the sodium-dependent human inositol transporters (Jin and Seyfang, 2003). Therefore, fungal inositol transporters may be developed as a valuable antifungal drug target.
Two- and three-carbon (C2 and C3) substrates are another important carbon sources for fungal pathogens. In C. albicans, non-fermentative carbon assimilation by glyoxylate and gluconeogenic pathways, which metabolizes C2 and C3 compounds such as acetate and lactate for glucose production, is critical for its early interaction with host immune cells, in which preferred carbon source such as glucose is limited (Lorenz and Fink, 2001; Barelle et al., 2006). For systemic infection, however, the fermentative glycolysis is the major carbon assimilation pathway for C. albicans (Barelle et al., 2006). In C. neoformans, the gluconeogenic pathway, but not the glyoxylate pathway, is required for the initial establishment of infection in the lungs (Rude et al., 2002; Panepinto et al., 2005; Price et al., 2011). For colonization and proliferation of C. neoformans in the central nervous system, however, glycolysis, but not gluconeogenesis, is critical (Price et al., 2011). Therefore, coordinated regulation of non-fermentative and fermentative carbon assimilation pathways depending on different infection stages appears to be essential for the pathogenicity of the two pathogenic yeasts.
In addition to sugars that are commonly utilized as carbon sources for energy and substrates, fungi also sense other carbon compounds as signaling molecules, mainly alcohol related carbons, to regulate cellular function. Alcohol-related carbon sensing has been reported mostly in S. cerevisiae and C. albicans and is less studied in C. neoformans. S. cerevisiae senses fusel alcohols, such as 1-butanol and isoamyl alcohol, to regulate differentiation of haploid cells. This involves binding of the transcription factor Ste12 to filamentation-specific genes in a Tec1-dependent mechanism (Dickinson, 1996; Lorenz et al., 2000a; Zeitlinger et al., 2003; Chen and Fink, 2006). Butanol also induces pseudohyphal morphology, even in liquid medium, which involves the Swe1-dependent morphogenesis checkpoint and differs from nitrogen-limitation-induced pseudohyphal growth (Martinez-Anaya et al., 2003). Ethanol stimulates hyperfilamentation in diploid cells in a MAPK-dependent manner (Lorenz et al., 2000a; Dickinson, 2008). In addition, aromatic alcohols (such as tryptophol and phenylethanol) secreted by yeast cells may function as quorum sensing molecules and stimulate filamentous growth in response to both cell density and nutritional conditions of the surrounding environment through a Flo11-dependent mechanism in S. cerevisiae (Chen and Fink, 2006). These autoregulatory molecules appear to function in a species-specific manner, because they only trigger the morphological switch in S. cerevisiae, but not in C. albicans (Chen and Fink, 2006).
Other alcohol-related quorum sensing molecules, such as farnesol and tyrosol, have been extensively studied in Candida species (Zhang and Dong, 2004; Hogan, 2006; Nickerson et al., 2006). Farnesol accumulation at the early stationary phase triggers inhibition of both yeast growth and filamentation by blocking GTPase activation, mitosis, and DNA replication in C. albicans (Uppuluri et al., 2007). Farnesol is also a virulence factor and it inhibits macrophage function during fungal-host interactions (Navarathna et al., 2007). In contrast, tyrosol stimulates fungal filamentation at all growth stages. Thus, the quorum-sensing process is under complex positive and negative regulation in response to environmental conditions (Chen et al., 2004). Fungal receptors that sense these quorum sensing molecules are largely unknown, although the Chk1 histidine kinase has been reported to play a role in sensing farnesol in C. albicans (Kruppa et al., 2004).
The C3 compound lactate was also shown to be a signaling molecule in C. albicans. Exposure to L-lactate, but not D-lactate, triggers β-glucan masking in C. albicans by regulating cell wall genes, which allows the pathogen to evade host immune detection (Ballou et al., 2016). Notably, this process is regulated by the Gpr1 GPCR and the Crz1 transcription factor, but is independent of lactate metabolism. Although this lactate-induced β-glucan masking process is conserved in other Candida clade (Ballou et al., 2016), it remains unknown in C. neoformans.
Nitrogen Sensing in Fungal Pathogenicity
Fungal pathogens sense nitrogen levels to control their rate of growth and changes in their morphology, processes that are important for host infection. The extracellular sensing of nitrogen containing compounds occurs via transceptors and GPCRs. Although transceptors are potential antifungal targets due to their cellular localization and their control of major signaling pathways, we currently lack an understanding of the molecular mechanisms involved in transceptor mediated signaling. To explore their therapeutic and economic potential, the molecular mechanisms that underpin fungal transceptor signaling need to be fully characterized.
Transceptors That Control the PKA Pathway
The PKA pathway regulates processes that are associated with cell growth in S. cerevisiae. Under nutrient replete conditions that include a fermentable carbon source, S. cerevisiae grows quickly and exhibits high PKA activity. However, if an essential nutrient is missing from the medium, cells are arrested, enter the stationary phase, and display phenotypes associated with low PKA activity, including high expression of stress-related genes and the production of stored carbohydrates. Under these conditions nutrient replacement results in rapid induction of PKA activity. The signaling pathway that regulates this response is known as the fermentable-growth-medium induced pathway (FGM pathway) (Thevelein, 1994). Activation of the FGM pathway is dependent on a family of transceptors that import amino acids (Gap1), ammonium (Mep1 and Mep2), phosphate (Pho84), sulfate (Sul1 and Sul2), iron (Ftr1), and zinc (Zrt1) (Figure 1) (Donaton et al., 2003; Giots et al., 2003; Van Nuland et al., 2006; Kankipati et al., 2015; Schothorst et al., 2017).
A well-studied model of a PKA regulating transceptor is the general amino acid permease (Gap1) of S. cerevisiae, which is a low affinity permease that imports a broad range of amino acids in cells grown under nitrogen-limiting conditions (Jauniaux and Grenson, 1990; Donaton et al., 2003). Two sites that are important for amino acid binding (Ser388 and Val389) have also been identified in Gap1 (Van Zeebroeck et al., 2009). When ammonium is added to yeast cells growing with proline as the only nitrogen source, Gap1 is rapidly internalized and degraded via a pathway involving the Npi1/Rsp5 ubiquitin ligase (Springael and Andre, 1998). The Ras2/cAMP/PKA pathway may be involved in the ubiquitin-dependent degradation of Gap1 (Garrett, 2008). Gap1 biogenesis is coupled to sphingolipid biosynthesis, which produces a sphingolipid microenvironment essential for the normal conformation, function, and ubiquitination of Gap1 (Lauwers et al., 2007). The activation of the FGM pathway by different nutrient transceptors suggests that they utilize a common signaling mechanism and a number of experimental findings are consistent with nutrient sensing involving an aspect of transporter function rather than changes in internal nutrient metabolism. For example, the FGM pathway is activated in mutants that are unable to metabolize the signaling nutrient or by transceptor-mediated uptake of a non-metabolizable nutrient analog (Donaton et al., 2003; Giots et al., 2003; Van Nuland et al., 2006). Importantly, amino acid substitutions have been identified that separate the transport and signaling functions of transceptors. Mutations of the predicted proton-binding sites within the Pho84 and the Sul1/2 nutrient/proton symporters result in a loss of nutrient transport but do not impact signaling, presumably due to continued substrate binding to the transceptor (Samyn et al., 2012; Kankipati et al., 2015).
A favored hypothesis is that the PKA-regulating transceptors act in a way that is analogous to GPCRs (Holsbeeks et al., 2004; Thevelein and Voordeckers, 2009). This model predicts that conformational changes within the transceptor following substrate binding and/or transport alter the interaction between the transceptor and a signaling partner that regulates the FGM pathway. This potential mechanism is consistent with the hypothesis that transceptors represent intermediates in the evolution of receptors from nutrient transporters and is supported by the existence of the yeast glucose sensors Snf3 and Rgt2 that have homology with nutrient transporters but no transport function (Ozcan et al., 1996; Didion et al., 1998; Thevelein and Voordeckers, 2009). In S. cerevisiae, another non-transporting transceptor plays an important role in amino acid sensing (Didion et al., 1998; Klasson et al., 1999; Forsberg and Ljungdahl, 2001; Wu et al., 2006). In this system, Ssy1 is a transceptor that senses amino acids without transporter activity. Ptr3 and Ssy5 function downstream of Ssy1 and physically interact with Ssy1 to form a signaling complex (Forsberg and Ljungdahl, 2001). Binding of amino acids to Ssy1 activates the Ssy5 protease which, in turn, proteolytically activates the latent transcription factors Stp1 and Stp2 to induce expression of genes encoding amino acid metabolizing enzymes and amino acid permeases (Andreasson et al., 2006). Ssy1 senses extracellular amino acids and activates Ptr3 hyperphosphorylation, which is dependent on SCF (Skp1-Gullin-F-box)-Grr1 E3 ligase complex function, but independent of Ssy5 function. Deletion mutations of Grr1, the F-box protein of this E3 ligase complex, block amino acid-induced Ptr3 hyperphosphorylation (Liu et al., 2008).
Evidence that conformational changes are associated with transceptor function comes from recent X-ray crystal structures of the Mep2 ammonium transceptors from S. cerevisiae and C. albicans (Figure 2) (van den Berg et al., 2016). Mep2 proteins are evolutionarily conserved members of the Amt/Mep/Rh family of ammonium transporters. Several structures of this class of proteins have been determined from bacteria as well as a structure of a human Rhesus protein (Khademi et al., 2004; Zheng et al., 2004; Andrade et al., 2005; Javelle et al., 2006; Gruswitz et al., 2010). They all form stable trimers with each monomer having an extracellular ammonium-binding site, a pair of conserved phenylalanine residues that gate a central narrow hydrophobic pore through which ammonium is conducted. However, there are structural differences that distinguish the fungal Mep2 transceptor from other ammonium transporters, and which may relate to the potential transceptor signaling mechanism (van den Berg et al., 2016). Fungal Mep2 transceptors have a closed conformation with a two-tier block of the central pore whereas all other ammonium transporters with known structures have an open pore. It is likely that the closed state of Mep2 prevents ammonium import, suggesting that the Mep2 transceptor must undergo significant conformational changes if ammonium conductance is to occur. Mep2 is activated via the phosphorylation of a serine residue within the cytoplasmic C-terminal domain (CTD) by the target of rapamycin complex 1 (TORC1) regulated Npr1 kinase, and this is hypothesized to open the ammonium-conducting pore (Boeckstaens et al., 2014; van den Berg et al., 2016). Conversely, the dephosphorylation of this site by the Psr phosphatases blocks ammonium transport (Boeckstaens et al., 2014). The structure of a Mep2 variant with a phosphomimetic mutation of this regulatory serine residue shows a large conformational change within the CTD that includes the formation of a novel 12-residue helix (van den Berg et al., 2016). This structural change is predicted to cause a shift of the CTD toward the main body of the transceptor to open the ammonium channel. It is conceivable that this conformational change also regulates binding of a signaling partner to the Mep2 CTD. While there is evidence that transceptors undergo conformational changes that are linked to signaling, the identification of transceptor signaling partners has not been successful. A split-ubiquitin two-hybrid screen identified proteins that interact with Mep2 and Gap1 and that are involved in different cellular processes (Van Zeebroeck et al., 2011). However, no signaling protein that links Mep2 or Gap1 directly to the FGM pathway was identified.
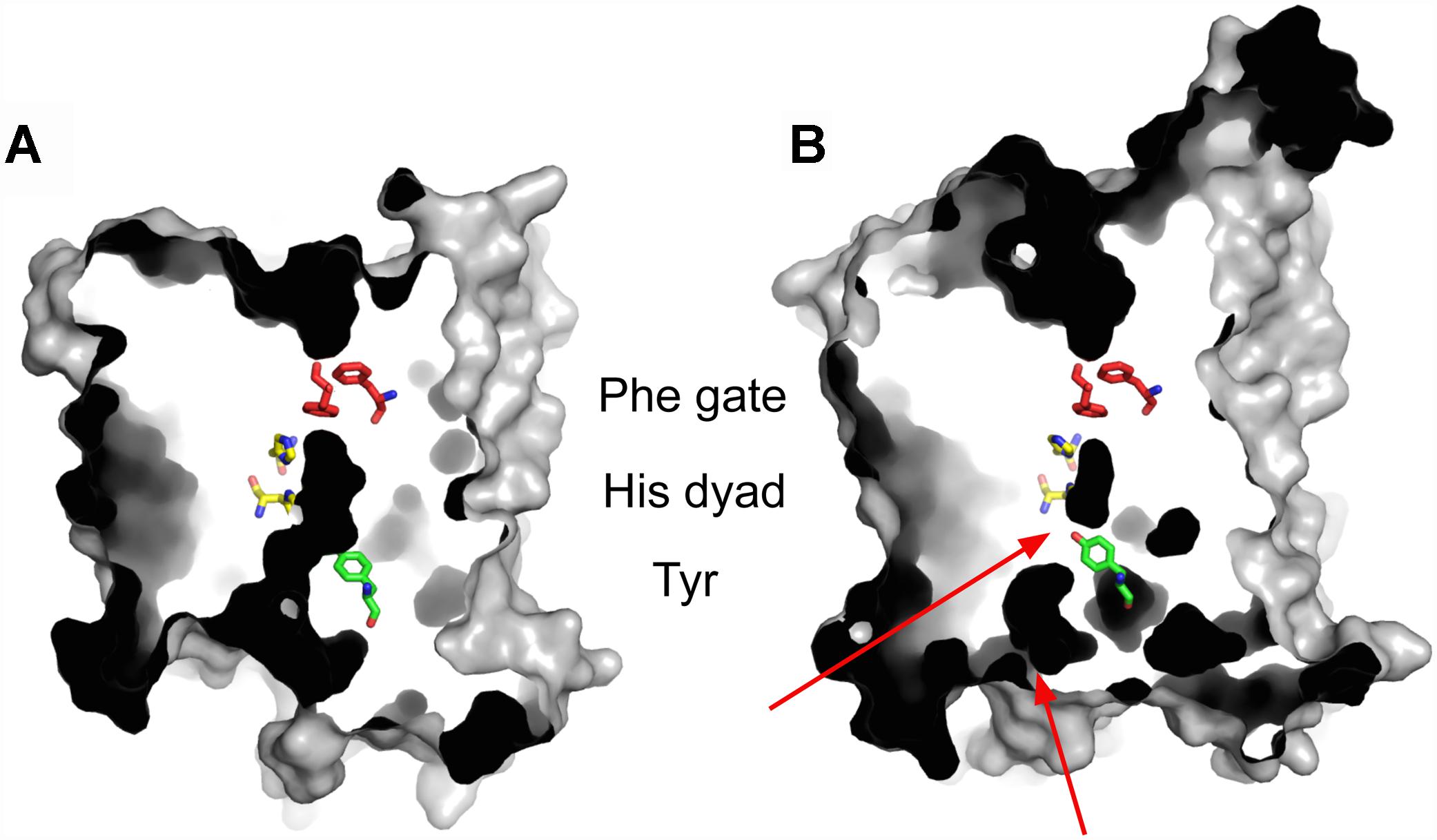
Figure 2. The crystal structure of ammonium transceptor Mep2. Slab views from the membrane plane, showing E. coli AmtB (A) and S. cerevisiae Mep2 (B). The two-tier channel block in Sc Mep2 is indicated by red arrows. In Sc Mep2 Tyr53 makes a strong hydrogen bond with His348, which is one of two histidine residues within the ammonium-conducting channel. The dark areas represent internal cavities and channels.
Currently less is known about the role of transceptors in C. albicans and C. neoformans. Ssy1, Ptr3, and Ssy5 homologs in C. albicans have been reported to function in a pathway that senses amino acids and is important for virulence in the host (Brega et al., 2004; Martinez and Ljungdahl, 2005). There are also six Gap1 homologs in C. albicans, and three (Gap1, Gap2, and Gap6) have transceptor activity to sense amino acids because they can rapidly activate the PKA pathway (Kraidlova et al., 2011). In C. neoformans eight amino acid permease homologs have been identified based on genome information, but their potential role in amino acid sensing has not been analyzed (Fernandes et al., 2015). Also the expression of CFT1 and CFT2, two C. neoformans homologs of iron transceptor Ftr1, are also regulated by PKA activity, and both proteins are required for fungal virulence (Jung et al., 2008). Yet, it is not clear whether either one functions as an iron sensor.
Ammonium Transceptors That Regulate Fungal Morphology
An ammonium transceptor dependent signaling system in dimorphic yeast regulates a switch in morphology in response to limiting nitrogen levels (Figure 1) (Lorenz and Heitman, 1998; Smith et al., 2003; Biswas and Morschhauser, 2005; Rutherford et al., 2008b). In S. cerevisiae, this regulates a switch to a filamentous form of growth known as pseudohyphal growth (Gimeno et al., 1992). Pseudohyphal growth is a Mep2-dependent process and results in S. cerevisiae forming chains of elongated cells that allow the yeast to forage for nutrients when they utilize a poor or limiting nitrogen source (Lorenz and Heitman, 1998; Boeckstaens et al., 2007). A number of signal transduction pathways regulate pseudohyphal growth and include the PKA, MAPK, sucrose non-fermentable, and TORC1 signaling pathways (Cullen and Sprague, 2012). Constitutively active components of the PKA or MAPK pathways restore pseudohyphal growth in a mutant lacking Mep2, suggesting that this transceptor may regulate these pathways during the dimorphic switch (Lorenz and Heitman, 1998; Biswas and Morschhauser, 2005; Rutherford et al., 2008a). The transport and pseudohyphal signaling functions of Mep2 may be linked as a hyperactive transporting and signaling Mep2 variant has been identified and Mep2 variants that do not transport ammonium but are correctly expressed and localized do not induce pseudohyphal growth (Marini et al., 2006; Boeckstaens et al., 2007; Rutherford et al., 2008a). Mep2 separation-of-function variants that transport ammonium but do not induce pseudohyphal growth also establish that ammonium sensing is not a consequence of changes in internal nitrogen metabolism (Van Nuland et al., 2006; Boeckstaens et al., 2008; Rutherford et al., 2008a). Therefore, ammonium sensing and the consequent regulation of pseudohyphal growth are dependent on an aspect of ammonium conductance through Mep2. Mep2 transceptor homologs also regulate morphological changes in C. albicans and C. neoformans (Smith et al., 2003; Biswas and Morschhauser, 2005; Rutherford et al., 2008b). Similar to the transceptor-mediated regulation of the FGM pathway, two models of Mep2 signaling during pseudohyphal growth have been proposed. In the first, Mep2-dependent ammonium transport is predicted to cause changes in cytosolic pH that is then sensed by an internal pH-responsive mechanism (Boeckstaens et al., 2008). This model is based on differences in the optimal pH for ammonium transport between Mep2 and two paralogous non-signaling ammonium transporters, Mep1 and Mep3 (Boeckstaens et al., 2008). Furthermore, a link between pH and polarized growth has been identified in other fungal systems (Bowman et al., 1997; Martinez-Espinoza et al., 2004; Vylkova et al., 2011). The second model of Mep2 signaling involves this transceptor acting analogous to GPCRs by physically interacting with and regulating a signaling partner as has been proposed for regulation of the FGM pathway (Lorenz and Heitman, 1998).
Ammonium metabolism can promote fungal infection as its export can alkalinize the external environment. C. albicans secrets ammonium to raise external pH and induce its filamentous form (Vylkova and Lorenz, 2014). Similarly C. neoformans uses urea as a nitrogen source resulting in the production and secretion of ammonium, which can have diverse impacts on the host. Urease positive C. neoformans promote the induction of a non-protective Type 2 host-immune response (Osterholzer et al., 2009). Furthermore, urease is required for the ability of C. neoformans to cross the host blood brain barrier possibly by disrupting the integrity of the junctions between microvascular endothelial cells in the brain (Shi et al., 2010; Singh et al., 2013). Urease also modulates the pH of macrophage phagolysosomes (Fu et al., 2018). Consistent with these studies urease is required for the virulence of C. neoformans in a mouse model of infection (Cox et al., 2000). In the human host, urea is distributed evenly throughout the body at concentrations in the lower mM range (Liu et al., 2012; Singh et al., 2013). There is therefore a sufficient pool of this metabolite within humans to act as a nutrient source for a fungal pathogen and as a modulator of host pH.
GPCRs and Amino Acid Sensing in Fungi
Amino acid sensing via GPCRs has not been reported in S. cerevisiae. In C. albicans, Gpr1 senses methionine to regulate the yeast-to-hypha transition on solid medium in the presence of carbon sources such as glucose (Maidan et al., 2005a). It is currently unclear whether Gpr1 directly senses extracellular methionine, intracellular amino acids, or both. It is possible that Gpr1 may sense both methionine and glucose.
Similar to Gpr1 in C. albicans, Gpr4 in C. neoformans has also been found to sense amino acids and activate signaling by the Gα protein Gpa1 signaling, which in turn activates cAMP-PKA signaling (Xue et al., 2006). Methionine induces the internalization of a Gpr4-DsRED fusion protein and also induces transient cAMP production in C. neoformans, and both are blocked by gpr4Δ mutations. A low concentration of methionine in the medium stimulates mating hyphae elongation in a Gpr4-dependent manner, but the role of methionine at a molecular level remains to be elucidated. Because Gpr4 contributes to but is not essential for the production of virulence factors controlled by cAMP signaling, and is not important for melanin production or virulence, additional upstream receptors other than Gpr4 may contribute to regulate Gpa1 functions. Activation of cAMP signaling by glucose and amino acids represents a nutrient coincidence detection system conserved in other pathogenic fungi.
The TOR Pathway Is a Globally Conserved Nutrient Sensor
TOR (target of rapamycin) is a serine/threonine kinase of the phosphatidylinositol kinase-related kinase family, which shares conserved motifs (such as HEAT repeats, FAT, and FATC domains), and is structurally and functionally conserved in eukaryotes (Abraham, 2004). The TOR pathway is activated by a variety of environmental signals and acts as a central regulator of cell growth through the phosphorylation of substrates that stimulate anabolic processes and inhibit catabolic process such as autophagy (Wolfson and Sabatini, 2017). The discovery of the TOR signaling pathways began with studies that sought to identify the molecular targets for the novel immunosuppressive, antifungal natural product rapamycin, which was originally isolated from a strain of Streptomyces hygroscopicus, from the beaches of Easter Island (Sehgal et al., 1975). This drug had been discovered in screens for natural products at Wyeth-Ayerst to identify candidate antifungal agents with activity against C. albicans (Sehgal, 2003). Despite having very potent antifungal activity, studies on rapamycin were shelved when it was discovered that the compound caused bone marrow suppression (Martel et al., 1977). When FK506 was discovered as a novel immunosuppressant in a screen at Fujisawa Pharmaceutics for natural products that would inhibit a mixed lymphocyte response assay, rapamycin was appreciated to be structurally related to FK506 and studies began anew to study its immunosuppressive properties and potential (Kino et al., 1987).
The FK506 binding protein FKBP12 was purified from yeast and its mutants were found to be resistant to rapamycin and FK506 (Heitman et al., 1991a,b). Isolation of rapamycin resistant yeast mutants revealed mutations in three genes, FKBP12 and two novel genes (Heitman et al., 1991a), which were named TOR1 and TOR2 for Target of Rapamycin (Kunz et al., 1993; Helliwell et al., 1994), which were later found to form multiprotein complexes known as TORC1 and TORC2 (Loewith et al., 2002; Wedaman et al., 2003). The TORC1 complex is sensitive to rapamycin and involved in a wide range of functions, while the TORC2 complex regulates polarization of the actin cytoskeleton. Several years later studies from multiple groups converged to identify the mammalian ortholog of the yeast TOR proteins (Brown et al., 1994; Chiu et al., 1994; Sabatini et al., 1994; Sabers et al., 1995), now known as mTOR.
Rapamycin inhibits yeast cell growth by inhibiting the ability of yeast cells to appropriately sense and respond to nutrients, particularly nitrogen sources including ammonia and glutamine, for example. Three studies converged to reveal that the TOR pathway functions in nutrient sensing in yeast (Beck and Hall, 1999; Cardenas et al., 1999; Hardwick et al., 1999). These studies revealed two sets of genes to be profoundly disrupted in cells exposed to rapamycin. First, the genes encoding ribosomal proteins, ribosomal RNAs, and many other proteins and enzymes involved in translation were all found to be repressed by rapamycin. Concomitantly, a suite of genes that was induced in cells exposed to rapamycin including many genes encoding transporters for a variety of nitrogen sources, and enzymes and proteins involved in the utilization of alternative nitrogen sources, the so called nitrogen catabolite repression (NCR) response. These later genes are known to be regulated by Ure2 and Gln3, both of which participate in the TOR pathway governing the control of nutrient responsive gene suites. Thus, these studies were paradigmatic in showing that the TOR pathway orchestrates the growth of cells in response to nutrients. Several years later the mTOR pathway was found to similarly control nutrient responsive genes in mammalian cells (Peng et al., 2002). Much more is now known about the intricate pathways that enable TOR and mTOR to sense the availability of several different amino acids and nutrients, and to couple this to appropriate physiological outcomes (reviewed in Jewell et al., 2013; Goberdhan et al., 2016; Gonzalez and Hall, 2017; Wolfson and Sabatini, 2017).
The Tor proteins sense nutrient signals, including amino acids, and regulate a broad range of cell developmental and signaling processes, including ribosome biosynthesis, protein translation, starvation-related transcriptional regulation, and autophagy (Raught et al., 2001; Rohde et al., 2001; Rohde and Cardenas, 2003). Amino acids are sensed by the TORC1 pathway via a variety of molecular mechanisms including the GTP/GDP loading status for the Rag GTPases that are called the Ragulator Rag GTPase in mammals and the EGO (Ego1-Ego2-Ego3) complex in S. cerevisiae. In S. cerevisiae, amino acids activate the SEACAT (Seh1-associated complex subcomplex activating TORC1), which consists of Sec13, Seh1, Sea2, Sea3, and Sea4. The SEACAT binds and negatively regulates SEACIT (Seh1-associated subcomplex inhibiting TORC1), which functions as GTPase-activating proteins (GAPs) for Gtr1, a RAG family small GTPase that binds with EGO complex to tether the TORC1 complex to the vacuole membrane (Binda et al., 2009). The other GTPase Gtr2 is activated by the GAP protein Lst4 in response to amino acids. Amino acid sufficiency promotes the active conformation of the RAG GTPase heterodimer in which Gtr1 is loaded with GTP, while Gtr2 is loaded with GDP (Gong et al., 2011). The active Gtr1GTP-Gtr2GDP heterodimer binds to Kog1 to activate TORC1 (reviewed by Gonzalez and Hall, 2017).
Overall, the core TOR complex is conserved among eukaryotes, while sensors that regulate the TOR pathway are likely more diverse. In mammalian cells, multiple amino acid sensors have been identified, including SLC38A9 as a putative arginine transceptor that positively regulates the mTOR pathway, the Sestrins (Sestrin 1 and 2) as a leucine sensor that functions as a negative regulator of the pathway, and CASTOR1 as a cytosolic arginine sensor for the pathway in a mechanism analogous to that of the Sestrins (reviewed by Saxton et al., 2016). However, similar amino acid sensors have not been identified in S. cerevisiae and most other fungi. It is possible that there are similar sensors in fungi, but with more sequence diversity. In yeast, leucine activates TORC1 via Gtr1. There is no Sestrin homolog and instead the leucyl-tRNA synthase has been shown to act as a cytosolic leucine sensor (Bonfils et al., 2012).
Rapamycin treatment triggers ubiquitination and degradation of some high-affinity amino acid transporters, such as the tryptophan permease Tat2 and the histidine permease Hip1, suggesting that Tor signaling promotes stability of these high-affinity, specialized transporter systems (Schmidt et al., 1998; Beck et al., 1999). On the other hand, Tor signaling also negatively regulates the stability of general amino acid permease systems like Gap1, and thus inversely regulates these two classes of amino acid permeases to balance the nutritional requirements of the cell (Beck et al., 1999). The serine/threonine kinase Npr1 has been recognized to mediate the regulation of these permeases by Tor (Schmidt et al., 1998; Raught et al., 2001).
Glutamine is a preferred nitrogen source and a key intermediate in yeast nitrogen metabolism, and its function is likely regulated by the TOR pathway. Glutamine depletion in yeast triggers nuclear localization and activation of the TOR-inhibited transcription factors Gln3, Rtg1, and Rtg3 (Crespo et al., 2002). Glutamine activation of TORC1 in yeast is independent of Gtr1, and rather requires the vacuolar membrane-associated phosphatidylinositol 3-phosphate binding protein Pib2 (Stracka et al., 2014; Kim and Cunningham, 2015). This nitrogen regulation is also mediated by the PP2A-like phosphatase Sit4. Many of the non-transcriptional effects of Tor, such as the initiation of translation and control of the stability of amino acid permeases, are also regulated via Sit4 (Di Como and Arndt, 1996; Cardenas et al., 1999; Rohde et al., 2004; Jacinto, 2007). Tor-mediated nutrient signaling also triggers nuclear translocation of Gln3 and a role of Golgi-to-endosome vesicular trafficking in TORC1-controlled nuclear translocation has been described (Puria et al., 2008).
In S. cerevisiae, Tor signaling is part of the complex signaling network that controls the yeast-to-filament switch in response to nitrogen limitation (Gimeno et al., 1992; Pan and Heitman, 1999; Cutler et al., 2001). Tor signaling involves the Sit4 protein phosphatase and is independent and parallel to the well-defined MAPK and cAMP-PKA pathways, but it is possible that signaling in response to nutrients involves crosstalk between these pathways (Schmelzle et al., 2004; Zurita-Martinez and Cardenas, 2005; Chen and Powers, 2006). Future studies are needed to understand the precise molecular mechanisms by which Tor regulates nutrient sensing and activation in fungi.
In addition to S. cerevisiae, TOR regulation of nutrient sensing has also been reported in C. albicans. Tor1 has been implicated in the negative regulation of filamentous growth in C. albicans. Inhibition of TORC1 results in the activation of the GATA transcription factor Brg1, which is involved in the regulation of hypha-specific genes and blocking the recruitment of the Nrg1-Tip1 transcriptional repressor complex (Lu et al., 2012; Su et al., 2013). Tor1 also plays a role in the regulation of adhesion gene expression in C. albicans (Bastidas et al., 2009). The Tor pathway in C. albicans has recently also been reported to regulate phosphate sensing that is dependent on the phosphate transporter Pho84 and the function of the TOR-activating small GTPase homologs Gtr1 and Rhb1 in nutrient responses has been characterized (Flanagan et al., 2017; Liu et al., 2017). The mechanisms of nutrient sensing by the TOR pathway in C. neoformans are less studied and remain to be understood.
Stress Sensing in Fungal Pathogenicity
Fungal pathogens are confronted with a series of host-derived stresses during the whole infection process, including temperature shift, fluctuation of oxygen/carbon dioxide levels, osmotic stress, and oxidative/nitrosative stress. To cope with such a variety of environmental stresses, most fungi employs an evolutionarily conserved stress-activated MAPK pathway, commonly known as the high-osmolarity glycerol response (HOG) pathway. Although the HOG pathway was initially discovered as osmosensing system in S. cerevisiae, it was subsequently proved to be a general stress-sensing pathway in most fungi.
Multicomponent Phosphorelay Systems
Fungi utilize phosphorelay systems to sense and respond to a variety of environmental stresses. These phosphorelay systems are widely conserved in prokaryotes and generally consists of two signaling components, sensor histidine kinase (HK), composed of His kinase A and histidine kinase-like ATPase domains serving as an autophosphorylation catalytic core, and its downstream response regulator (RR) (Zschiedrich et al., 2016; Tiwari et al., 2017). In contrast, most fungi contain multicomponent phosphorelay systems, which consist of sensor hybrid histidine kinases (HHKs), histidine-containing phosphotransfer (HPt) protein, and RRs (Figure 3) (Santos and Shiozaki, 2001; Matsushita and Janda, 2002; Laub and Goulian, 2007; Bahn, 2008). The HHKs consist of an HK domain and an aspartate-containing receiver domain (RD). Upon external cues, the HHK undergoes autophosphorylation at the histidine residue in the HK domain and relays its phosphate group to the aspartate residue in the RD. Subsequently, the phosphate group is relayed to the histidine residue in the HPt protein, which is then transferred to the aspartate residue in RRs (Santos and Shiozaki, 2001; Matsushita and Janda, 2002; Laub and Goulian, 2007; Bahn, 2008). Most fungi contain two types of RRs: one RR, similar to a bacterial RR, has a DNA-binding domain and transcriptionally activates effector genes and the other RR activates another signaling cascade, including a Hog1 MAPK module. S. cerevisiae also contains two types of RRs, Ssk1 and Skn7, and the former localizes to the cytoplasm whereas the latter is enriched in the nucleus (Lu et al., 2003). The HPt protein, Ypd1, shuttles between the cytoplasm and nucleus to relay its phosphate group to Ssk1 and Skn7, respectively, depending on the type of eliciting stresses (Lu et al., 2003).
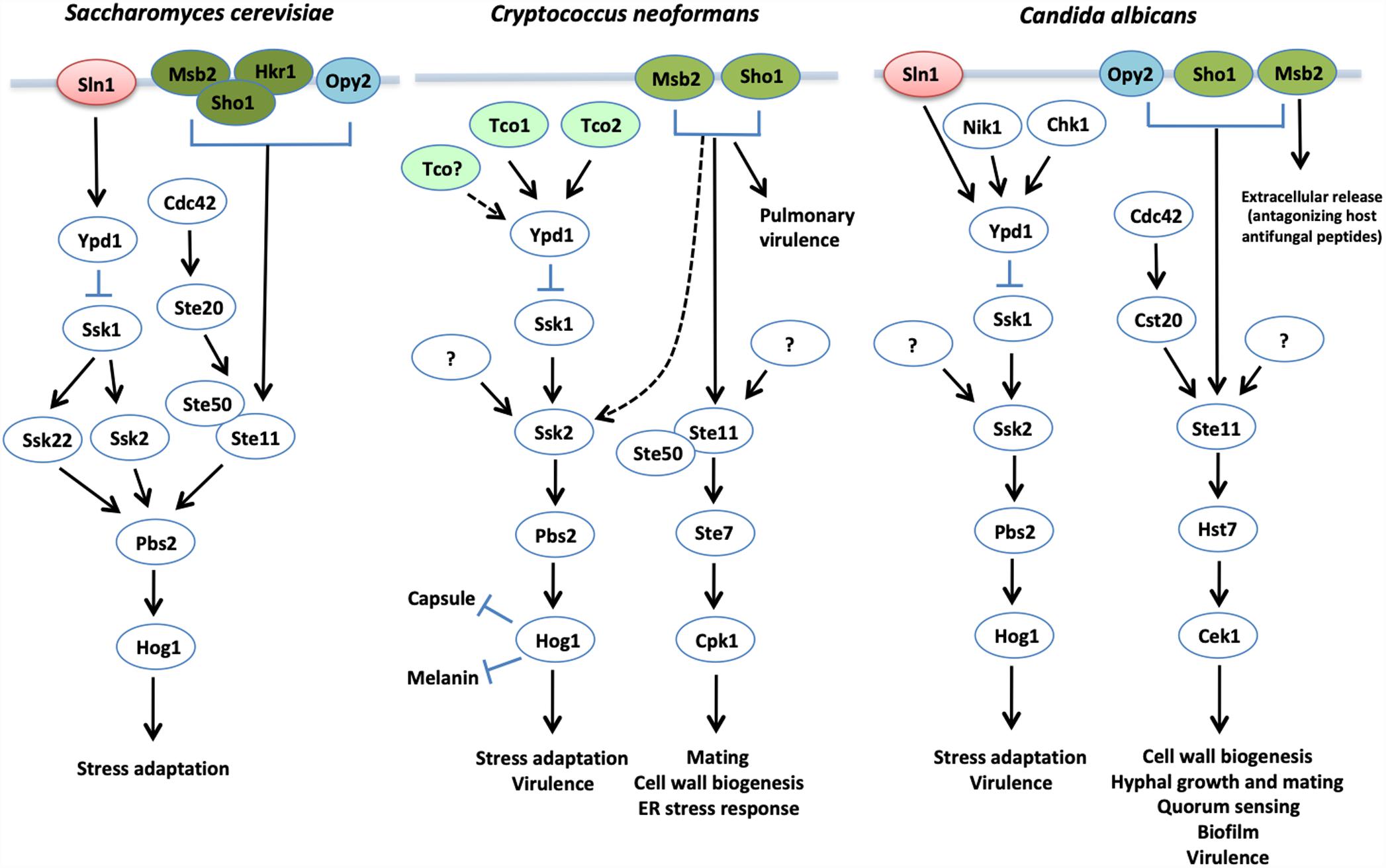
Figure 3. The proposed yeast HOG pathways. In S. cerevisiae, the Ssk2/22-Pbs2-Hog1 MAPK module is modulated by two main upstream branches: the Sln1-Ypd1-Ssk1 phosphorelay system and Msb2/Hkr1/Sho1/Opy2 signaling branches. In contrast, the Ssk2-Pbs2-Hog1 MAPK module is mainly regulated by the multicomponent phosphorelay system in both C. neoformans and C. albicans. In C. neoformans, the Msb2/Sho1 branch pathway plays a minor role in phosphorylating Hog1 in the absence of Ssk1, and functions in phosphorylating Cpk1 for cell wall biogenesis. Although Msb2 and Sho1 are involved in mating process of C. neoformans, they are not directly involved in Cpk1 phosphorylation. In C. albicans, there is no evidence that the Sho1/Msb2/Opy2 branch pathway regulates the Hog1 MAPK module directly.
The most notable feature of the fungal phosphorelay system is a tremendous diversity of HHKs in both number and domain structures (Bahn, 2008; Herivaux et al., 2016). S. cerevisiae contains only a single HHK (Sln1), while some filamentous fungi contains more than 10 HHKs. Although all HHKs commonly contain at least a single HK and one RD, their N-terminal regions contain highly diverse protein domains, which are proposed to play distinct roles for each HHK in sensing different signals and regulating the activity of the HK and RD domains.
Cryptococcus neoformans and Candida albicans contain seven and three HHKs, respectively (Bahn, 2008). This varying number of HHKs may reflect differential host biological niches at which these two fungal pathogenic yeasts reside during infection. C. neoformans HHKs were named Tco1 to Tco7 (Two-component proteins 1–7) (Bahn et al., 2006). Tco1 contains seven HAMP (Histidine kinases-Adenylyl cyclases-Methyl accepting proteins-Phosphatases) domains with an extended N-terminal region (Bahn et al., 2006). Notably, C. neoformans contains two unique dual HHKs, Tco2 and Tco4, which harbor two HK-RD domains in a single polypeptide (Bahn et al., 2006; Bahn, 2008). Since their first discovery in C. neoformans, a recent bioinformatics analysis showed that the dual HHKs are observed only in certain basidiomycetous fungi (Lavin et al., 2014). Tco3 contains GAF (cGMP-specific phosphodiesterases-Adenylyl cyclases-FhlA) and PHY (phytochrome) domains. Tco5 is the only TM domain-containing HHK. Both Tco6 and Tco7 are GAF-containing HHK. In stark contrast to Sln1, which is essential for viability of S. cerevisiae, none of the Tco HHKs are essential for C. neoformans (Bahn et al., 2006; Lee et al., 2016). Among these, Tco1 and Tco2 are two major HHKs in C. neoformans (Bahn et al., 2006). Both HHKs are involved in recognizing fludioxonil, a phenylpyrrole class of fungicide (Bahn et al., 2006). However, Tco1 and Tco2 also have distinct roles. Tco1 is involved in hypoxia sensing, melanin production, the mating process, and virulence, whereas Tco2 is involved in response and adaptation to osmotic and oxidative stress and toxic metabolites (Bahn et al., 2006; Chun et al., 2007).
Candida albicans has three HHKs: Sln1, Nik1/Cos1, and Chk1 (Alex et al., 1998; Nagahashi et al., 1998; Yamada-Okabe et al., 1999). Although C. albicans Sln1 can functionally replace S. cerevisiae Sln1 and is indeed involved in osmosensing, Sln1 is not essential for viability of C. albicans (Nagahashi et al., 1998). The most notable function of these HHKs is the regulation of the morphological transition, which is a crucial virulence factor for C. albicans (Alex et al., 1998; Nagahashi et al., 1998; Yamada-Okabe et al., 1999). Deletion of SLN1, NIK1, or CHK1 causes defects in hyphal development and thereby attenuates the virulence of C. albicans (Yamada-Okabe et al., 1999). However, CHK1 deletion partially restores filamentation and virulence in the sln1Δ and nik1Δ mutants, suggesting that complex cross-talk may occur among these HHKs (Yamada-Okabe et al., 1999).
Regardless of the remarkable diversity of HHKs, most fungi contain only one or two HPt proteins. Therefore, it is still puzzling how different environmental signals sensed by varying HHKs are distinguished by such a small number of HPt proteins. In C. albicans and C. neoformans, a single HPt (Ypd1) has been discovered. In S. cerevisiae, Ypd1 is essential because its absence leads to constitutive dephosphorylation of Ssk1, which overactivates the Ssk2/22-Pbs2-Hog1 MAPK module (Posas et al., 1996). Ypd1 is similarly essential in C. neoformans, but YPD1 deletion is feasible in the hog1Δ mutant background, suggesting that YPD1 deletion also hyperactivates Hog1 in the pathogen (Lee et al., 2011). C. albicans YPD1 can functionally replace S. cerevisiae YPD1. Similar to S. cerevisiae Ypd1, C. albicans Ypd1 also localizes to both the cytoplasm and the nucleus (Mavrianos et al., 2014). Notably, however, Ypd1 is not essential in C. albicans, although its deletion causes growth defects (Mavrianos et al., 2014). YPD1 deletion causes constitutive Hog1 phosphorylation even under unstressed conditions, as expected, and flocculation through enhanced filamentation (Mavrianos et al., 2014).
Cryptococcus neoformans contains two most conserved classes of RRs, Skn7, and Ssk1 (Bahn et al., 2006). Ssk1 plays a major role in relaying the signal from Ypd1 to the Hog1 MAPK module as shown by the fact that the ssk1Δ mutant is phenotypically similar to the hog1Δ mutant (Bahn et al., 2006). However, Hog1 may have another upstream regulator as it can be phosphorylated in the absence of Ssk1 (Bahn et al., 2006). In contrast, Skn7, which contains a heat shock factor-type DNA binding domain at its N-terminus, plays both redundant and distinct roles with Hog1, but the Skn7-mediated signaling is largely independent of the HOG pathway (Bahn et al., 2006). Supporting this, SKN7 deletion does not affect Hog1 phosphorylation and its related phenotypes, such as capsule production and mating efficiency (Bahn et al., 2006). In contrast, C. albicans contains three RRs: Ssk1, Skn7, and Srr1, which are localized to the cytoplasm, the nucleus, and the mitochondria, respectively (Calera et al., 2000; Singh et al., 2004; Mavrianos et al., 2013). Ssk1 is involved in oxidative stress and heat shock response, cell wall biogenesis, adherence, and filamentous growth, whereas Skn7 is involved in the morphological transition and oxidative stress response. Similar to the case in C. neoformans, Skn7 function is largely Hog1-independent in C. albicans (Singh et al., 2004). Srr1 is involved in hyphal development, stress resistance and virulence (Desai et al., 2011).
In S. cerevisiae, the dephosphorylated Ssk1 activates the autophosphorylation activity of the Ssk2/22 MAPK kinase kinase (MAPKKK) by interacting with the autoinhibitory domain of Ssk1 (Posas and Saito, 1998). Activated Ssk2/22 subsequently phosphorylates the MAPK kinase (MAPKK) Pbs2, which then phosphorylates the Hog1 MAPK. In contrast to S. cerevisiae that contains two MAPKKKs, Ssk2 and Ssk22, both C. neoformans and C. albicans contain a single Ssk2 ortholog. Pbs2 and Hog1 are also conserved in the two pathogens (Bahn et al., 2005). Regardless of the conserved Ssk2-Pbs2-Hog1 module, its regulatory mechanism appears to be divergent. Hog1 is not normally phosphorylated under unstressed conditions in both S. cerevisiae and C. albicans, but becomes rapidly phosphorylated in response to environmental stresses (Brewster et al., 1993; Smith et al., 2004). In C. neoformans, however, Hog1 is constitutively phosphorylated under unstressed conditions, but undergoes dephosphorylation in response to environmental stresses, such as osmotic shock (Bahn et al., 2005).
Transcriptome analysis revealed a plethora of Hog1 downstream effector genes in C. albicans and C. neoformans (Enjalbert et al., 2006; Ko et al., 2009). Among these, some of pathogenicity-related effectors are particularly notable. First, the Na+/ATPase efflux pump Ena1 was shown to be strongly induced by salt or osmotic stress in a Hog1-depenent manner and required for pH homeostasis in C. neoformans (Jung et al., 2012; Meyers et al., 2017). Deletion of ENA1 completely abolishes the virulence of C. neoformans (Jung et al., 2012; Meyers et al., 2017). Second, a number of the genes involved in oxidative stress response, including a sulfiredoxin gene (SRX1), are strongly induced by peroxides in a Hog1-dependent manner (Upadhya et al., 2013). Srx1 is required for recycling of peroxiredoxin (Tsa1) and its deletion significantly attenuates the virulence of C. neoformans (Upadhya et al., 2013). Third, several kinases are regulated by the HOG pathway. These include Hrk1 (Hog1-regulated kinase), which is involved in osmoregulation (Kim et al., 2011), and Sch9, which is involved in thermotolerance and oxidative stress response, in C. neoformans (Kim et al., 2009). The C. albicans Hrk1 ortholog Rck2 was also shown to be induced by osmotic stress in a Hog1-dependent manner (Enjalbert et al., 2006). Fourth, Hog1-dependent transcription factors include Mbs1, which is involved in environmental stress response, ergosterol biosynthesis, membrane integrity, and virulence factor production in C. neoformans (Song et al., 2012). In particular, MBS1 deletion attenuates virulence of C. neoformans. In C. albicans, the Sko1 transcription factor is transcriptionally regulated by Hog1 in response to osmotic stress and is also involved in the cell wall damage response (Enjalbert et al., 2006; Rauceo et al., 2008).
As hyperactivation of the HOG pathway is lethal to fungal cells, timely inactivation of the HOG pathway is critical for fungal cell growth. In S. cerevisiae, two tyrosine phosphatases (PTPs; Ptp2 and Ptp3) and three type 2C Ser/Thr phosphatases (PP2C; Ptc1, Ptc2, and Ptc3) are involved in negatively regulation of Hog1 (Maeda et al., 1994; Jacoby et al., 1997; Wurgler-Murphy et al., 1997). In C. neoformans, Ptp1 and Ptp2 were found to be transcriptionally regulated by the HOG pathway as negative feedback regulators (Lee et al., 2014). Particularly, Ptp2 plays a major role in regulating Hog1. Unexpectedly, some of the ptp2Δ mutant phenotypes, including increased susceptibility to osmotic, oxidative, and genotoxic stresses, are similar to those of the hog1Δ mutant, suggesting that coordinated, balanced regulation of the HOG pathway is critical for normal fungal cell physiology. Supporting this, PTP2 deletion also attenuates the virulence of C. neoformans even more strongly than HOG1 deletion (Lee et al., 2014). Notably, PTP1 deletion exacerbates the virulence defect of the ptp2Δ mutant, suggesting that Ptp1 is also involved in the virulence of C. neoformans (Lee et al., 2014). In C. albicans, Ptp2 and Ptp3 are known to repress the basal Hog1 activity in response to Tor1 inhibition, which is required for hyphal maintenance (Su et al., 2013). However, the role of Ptp2 and Ptp3 in virulence of C. albicans has not been addressed. In contrast to PTPs, the role of any PP2Cs in Hog1 regulation remains unknown in C. neoformans and C. albicans.
The Sho1/Msb2/Hkr1-Signaling Pathway
In addition to the multicomponent phosphorelay system, another upstream signaling branch has been reported to regulate the Ssk2-Pbs2-Hog1 MAPK module in S. cerevisiae: the Sho1/Msb2/Hkr1-signaling pathway. Msb2 and Hkr1 are two mucin-like transmembrane proteins and Sho1 is a membrane protein with the SH3 domain (Maeda et al., 1995; O’Rourke and Herskowitz, 2002; Tatebayashi et al., 2007). Msb2 and Hkr1 are bona fide osmosensors, which can physically interact with Sho1 through transmembrane domains and generate intracellular signaling through the cytoplasmic domain of Sho1. Sho1 also serves as an adaptor for recruiting Pbs2 and the Ste11/Ste50 complex (Maeda et al., 1995; Zarrinpar et al., 2004; Tatebayashi et al., 2006). In C. neoformans, Sho1 and Msb2 orthologs have been recently identified and functionally characterized (Kim et al., 2015; So et al., 2018). SHO1 and MSB2 deletions do not markedly affect Hog1 phosphorylation patterns in response to osmotic stress, suggesting that these proteins are not major Hog1 regulators in C. neoformans. Instead, Sho1 and Msb2 play redundant roles in phosphorylating the Cpk1 MAPK for cell wall biogenesis. However, deletion of SHO1 or MSB2 reduces Hog1 phosphorylation in the absence of Ssk1 (So et al., 2018), indicating that the Sho1 and Msb2 pathway may serve as a back-up signaling circuit for Hog1 activation when the major multicomponent phosphorelay system is shut down in C. neoformans. Notably, Msb2 and Sho1 are required for the early acute and later adaptation of C. neoformans, respectively, to the pulmonary environment during mammalian host infection (So et al., 2018).
Surprisingly, however, Ssk1, Sho1, and Msb2 are all dispensable for Hog1 phosphorylation in response to osmotic shock in C. albicans (Roman et al., 2009). In the triple ssk1Δ sho1Δ msb2Δ mutant, Hog1 can still be phosphorylated in response to osmotic shock, like the wild type strain in C. albicans, suggesting that another signaling branch may operate to activate Hog1. In C. albicans, the extracellular domain of Msb2 is cleaved and secreted as a highly glycosylated form during host infection and serves to protect fungal cells from antimicrobial peptides (Szafranski-Schneider et al., 2012). As the extracellular domain is not conserved between C. neoformans and C. albicans Msb2 orthologs, it remains unclear whether the extracellular domain of C. neoformans Msb2 is also cleaved and released during host infection.
Receptors as Potential Drug Targets
Overall, sensing nutrient and stress signals is critical for fungal pathogens to adapt to the host environment to cause infection. As eukaryotic pathogens, fungi and the human host share much similarity in their cellular mechanisms, leading to very few targets for drug development that have both high potency and low toxicity. The cell surface sensors are in general more distinct compared to the intracellular signaling and metabolic pathways, and their accessible location is favorable for drug binding. Therefore, nutrient receptors could represent a potentially rich source of targets for antifungal drug development. Around 40% of all available drugs target GPCRs, making these receptors the most important drug target group. Transceptors are often important for fungal development and virulence, and hence could be an excellent drug target group as well. Indeed, receptors that sense different stress responses have been proposed as a drug targets.
Clinically, the fungal phosphorelay systems have drawn significant attention from a pharmaceutical standpoint because an equivalent system has not been discovered in mammals. In agriculture, some fungicides, for instance phenylpyrrole agents, primarily target the HHKs, which subsequently hyperactivates Hog1 and causes over-accumulation of intracellular glycerols, resulting in growth arrest (Kojima et al., 2006). Due to this reason, Ypd1, which is a central negative regulator of Hog1, has been considered as a prime target for antifungal drug development, particularly because any Ypd1 ortholog is not present in mammals. However, a recent report demonstrated that conditional repression of YPD1 enhances the virulence of C. albicans, perhaps because reduced YPD1 expression enhances stress resistance and filamentation (Day et al., 2017), which raising doubt as to whether Ypd1 is a viable target in C. albicans. In aggregate, increasing our understanding of nutrient sensing in fungal development and pathogenicity is critical and will enhance our ability to develop novel strategies to fight fungal diseases and antifungal drug resistance.
Author Contributions
CX and JH designed the review. JR, Y-SB, BV, JH, and CX contributed to the writing of the manuscript.
Funding
This work was in part supported by the UK Biotechnology and Biological Sciences Research Council to JR, Korea National Research Foundation (NRF) grants (2016R1E1A1A01943365 and 2018R1A5A1025077) and Korea Ministry of Agriculture, Food and Rural Affairs Strategic Initiative for Microbiomes in Agriculture and Food (918012-4) to Y-SB, National Institutes of Health (NIH) R37 AI39115-20 and R01 AI50113-15 to JH, and NIH R01AI123315 to CX.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abraham, R. T. (2004). mTOR as a positive regulator of tumor cell responses to hypoxia. Curr. Top. Microbiol. Immunol. 279, 299–319. doi: 10.1007/978-3-642-18930-2_18
Alex, L. A., Korch, C., Selitrennikoff, C. P., and Simon, M. I. (1998). COS1, a two-component histidine kinase that is involved in hyphal development in the opportunistic pathogen Candida albicans. Proc. Natl. Acad. Sci. U.S.A. 95, 7069–7073. doi: 10.1073/pnas.95.12.7069
Alspaugh, J. A., Perfect, J. R., and Heitman, J. (1997). Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev. 11, 3206–3217. doi: 10.1101/gad.11.23.3206
Andrade, S. L., Dickmanns, A., Ficner, R., and Einsle, O. (2005). Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus. Proc. Natl. Acad. Sci. U.S.A. 102, 14994–14999. doi: 10.1073/pnas.0506254102
Andreasson, C., Heessen, S., and Ljungdahl, P. O. (2006). Regulation of transcription factor latency by receptor-activated proteolysis. Genes Dev. 20, 1563–1568. doi: 10.1101/gad.374206
Attwood, T. K., and Findlay, J. B. (1994). Fingerprinting G-protein-coupled receptors. Protein Eng. 7, 195–203. doi: 10.1093/protein/7.2.195
Bahn, Y. S. (2008). Master and commander in fungal pathogens: the two-component system and the HOG signaling pathway. Eukaryot. Cell 7, 2017–2036. doi: 10.1128/EC.00323-08
Bahn, Y. S., Kojima, K., Cox, G. M., and Heitman, J. (2005). Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol. Biol. Cell 16, 2285–2300. doi: 10.1091/mbc.e04-11-0987
Bahn, Y. S., Kojima, K., Cox, G. M., and Heitman, J. (2006). A unique fungal two-component system regulates stress responses, drug sensitivity, sexual development, and virulence of Cryptococcus neoformans. Mol. Biol. Cell 17, 3122–3135. doi: 10.1091/mbc.e06-02-0113
Bahn, Y. S., Xue, C., Idnurm, A., Rutherford, J. C., Heitman, J., and Cardenas, M. E. (2007). Sensing the environment: lessons from fungi. Nat. Rev. Microbiol. 5, 57–69. doi: 10.1038/nrmicro1578
Ballou, E. R., Avelar, G. M., Childers, D. S., Mackie, J., Bain, J. M., Wagener, J., et al. (2016). Lactate signalling regulates fungal beta-glucan masking and immune evasion. Nat. Microbiol. 2:16238. doi: 10.1038/nmicrobiol.2016.238
Barelle, C. J., Priest, C. L., Maccallum, D. M., Gow, N. A., Odds, F. C., and Brown, A. J. (2006). Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell Microbiol. 8, 961–971. doi: 10.1111/j.1462-5822.2005.00676.x
Bastidas, R. J., Heitman, J., and Cardenas, M. E. (2009). The protein kinase Tor1 regulates adhesin gene expression in Candida albicans. PLoS Pathog. 5:e1000294. doi: 10.1371/journal.ppat.1000294
Beck, T., and Hall, M. N. (1999). The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402, 689–692. doi: 10.1038/45287
Beck, T., Schmidt, A., and Hall, M. N. (1999). Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J. Cell Biol. 146, 1227–1238. doi: 10.1083/jcb.146.6.1227
Binda, M., Peli-Gulli, M. P, Bonfils, G., Panchaud, N., Urban, J., Sturgill, T. W., et al. (2009). The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell 35, 563–573. doi: 10.1016/j.molcel.2009.06.033
Biswas, K., and Morschhauser, J. (2005). The Mep2p ammonium permease controls nitrogen starvation-induced filamentous growth in Candida albicans. Mol. Microbiol. 56, 649–669. doi: 10.1111/j.1365-2958.2005.04576.x
Boeckstaens, M., Andre, B., and Marini, A. M. (2007). The yeast ammonium transport protein Mep2 and its positive regulator, the Npr1 kinase, play an important role in normal and pseudohyphal growth on various nitrogen media through retrieval of excreted ammonium. Mol. Microbiol. 64, 534–546. doi: 10.1111/j.1365-2958.2007.05681.x
Boeckstaens, M., Andre, B., and Marini, A. M. (2008). Distinct transport mechanisms in yeast ammonium transport/sensor proteins of the Mep/Amt/Rh family and impact on filamentation. J. Biol. Chem. 283, 21362–21370. doi: 10.1074/jbc.M801467200
Boeckstaens, M., Llinares, E., Van Vooren P., and Marini, A. M. (2014). The TORC1 effector kinase Npr1 fine tunes the inherent activity of the Mep2 ammonium transport protein. Nat. Commun. 5:3101. doi: 10.1038/ncomms4101
Bonfils, G., Jaquenoud, M., Bontron, S., Ostrowicz, C., Ungermann, C., and De Virgilio C. (2012). Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol. Cell 46, 105–110. doi: 10.1016/j.molcel.2012.02.009
Bowman, E. J., O’Neill F. J, and Bowman, B. J. (1997). Mutations of pma-1, the gene encoding the plasma membrane H+-ATPase of Neurospora crassa, suppress inhibition of growth by concanamycin A, a specific inhibitor of vacuolar ATPases. J. Biol. Chem. 272, 14776–14786. doi: 10.1074/jbc.272.23.14776
Brega, E., Zufferey, R., and Mamoun, C. B. (2004). Candida albicans Csy1p is a nutrient sensor important for activation of amino acid uptake and hyphal morphogenesis. Eukaryot. Cell 3, 135–143. doi: 10.1128/EC.3.1.135-143.2004
Brewster, J. L., de Valoir, T. Dwyer, N. D., Winter, E., and Gustin, M. C. (1993). An osmosensing signal transduction pathway in yeast. Science 259, 1760–1763. doi: 10.1126/science.7681220
Brown, E. J., Albers, M. W., Shin, T. B., Ichikawa, K., Keith, C. T., Lane, W. S., et al. (1994). A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 369, 756–758. doi: 10.1038/369756a0
Brown, V., Sexton, J. A., and Johnston, M. (2006). A glucose sensor in Candida albicans. Eukaryot. Cell 5, 1726–1737. doi: 10.1128/EC.00186-06
Calera, J. A., Zhao, X. J., and Calderone, R. (2000). Defective hyphal development and avirulence caused by a deletion of the SSK1 response regulator gene in Candida albicans. Infect. Immun. 68, 518–525. doi: 10.1128/IAI.68.2.518-525.2000
Cardenas, M. E., Cutler, N. S., Lorenz, M. C., Di Como C. J., and Heitman, J. (1999). The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 13, 3271–3279. doi: 10.1101/gad.13.24.3271
Chen, H., and Fink, G. R. (2006). Feedback control of morphogenesis in fungi by aromatic alcohols. Genes Dev. 20, 1150–1161. doi: 10.1101/gad.1411806
Chen, H., Fujita, M., Feng, Q., Clardy, J., and Fink, G. R. (2004). Tyrosol is a quorum-sensing molecule in Candida albicans. Proc. Natl. Acad. Sci. U.S.A. 101, 5048–5052. doi: 10.1073/pnas.0401416101
Chen, J. C., and Powers, T. (2006). Coordinate regulation of multiple and distinct biosynthetic pathways by TOR and PKA kinases in S. cerevisiae. Curr. Genet. 49, 281–293. doi: 10.1007/s00294-005-0055-9
Chen, Y. L., Kauffman, S., and Reynolds, T. B. (2008). Candida albicans uses multiple mechanisms to acquire the essential metabolite inositol during infection. Infect. Immun. 76, 2793–2801. doi: 10.1128/IAI.01514-07
Chiu, M. I., Katz, H., and Berlin, V. (1994). RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc. Natl. Acad. Sci. U.S.A. 91, 12574–12578. doi: 10.1073/pnas.91.26.12574
Chun, C. D., Liu, O. W., and Madhani, H. D. (2007). A link between virulence and homeostatic responses to hypoxia during infection by the human fungal pathogen Cryptococcus neoformans. PLoS Pathog. 3:e22. doi: 10.1371/journal.ppat.0030022
Cox, G. M., Mukherjee, J., Cole, G. T., Casadevall, A., and Perfect, J. R. (2000). Urease as a virulence factor in experimental cryptococcosis. Infect. Immun. 68, 443–448. doi: 10.1128/IAI.68.2.443-448.2000
Crespo, J. L., Powers, T., Fowler, B., and Hall, M. N. (2002). The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc. Natl. Acad. Sci. U.S.A. 99, 6784–6789. doi: 10.1073/pnas.102687599102687599
Cullen, P. J. and Sprague, G. F. Jr. (2012). The regulation of filamentous growth in yeast. Genetics 190, 23–49. doi: 10.1534/genetics.111.127456
Cutler, N. S., Pan, X., Heitman, J., and Cardenas, M. E. (2001). The TOR signal transduction cascade controls cellular differentiation in response to nutrients. Mol. Biol. Cell 12, 4103–4113. doi: 10.1091/mbc.12.12.4103
Day, A. M., Smith, D. A., Ikeh, M. A. C., Haider, M., Herrero-de-Dios, C. M. Brown, A. J. P., et al. (2017). Blocking two-component signalling enhances Candida albicans virulence and reveals adaptive mechanisms that counteract sustained SAPK activation. PLoS Pathog. 13:e1006131. doi: 10.1371/journal.ppat.1006131
Desai, C., Mavrianos, J., and Chauhan, N. (2011). Candida albicans SRR1, a putative two-component response regulator gene, is required for stress adaptation, morphogenesis, and virulence. Eukaryot. Cell 10, 1370–1374. doi: 10.1128/EC.05188-11
Di Como, C. J., and Arndt, K. T. (1996). Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 10, 1904–1916. doi: 10.1101/gad.10.15.1904
Dickinson, J. R. (1996). ‘Fusel’ alcohols induce hyphal-like extensions and pseudohyphal formation in yeasts. Microbiology 142 (Pt. 6), 1391–1397. doi: 10.1099/13500872-142-6-1391
Dickinson, J. R. (2008). Filament formation in Saccharomyces cerevisiae - a review. Folia Microbiol. 53, 3–14. doi: 10.1007/s12223-008-0001-6
Didion, T., Regenberg, B., Jorgensen, M. U., Kielland-Brandt M. C., and Andersen, H. A. (1998). The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol. Microbiol. 27, 643–650. doi: 10.1046/j.1365-2958.1998.00714.x
Dlugai, S., Hippler, S., Wieczorke, R., and Boles, E. (2001). Glucose-dependent and -independent signalling functions of the yeast glucose sensor Snf3. FEBS Lett. 505, 389–392. doi: 10.1016/S0014-5793(01)02854-X
Donaton, M. C., Holsbeeks, I., Lagatie, O., Van Zeebroeck, G. Crauwels, M., Winderickx, J., et al. (2003). The Gap1 general amino acid permease acts as an amino acid sensor for activation of protein kinase A targets in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 50, 911–929. doi: 10.1046/j.1365-2958.2003.03732.x
Enjalbert, B., Smith, D. A., Cornell, M. J., Alam, I., Nicholls, S., Brown, A. J., et al. (2006). Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol. Biol. Cell 17, 1018–1032. doi: 10.1091/mbc.e05-06-0501
Fan, J., Chaturvedi, V., and Shen, S. H. (2002). Identification and phylogenetic analysis of a glucose transporter gene family from the human pathogenic yeast Candida albicans. J. Mol. Evol. 55, 336–346. doi: 10.1007/s00239-002-2330-4
Fernandes, J. D., Martho, K., Tofik, V., Vallim, M. A., and Pascon, R. C. (2015). The role of amino acid permeases and tryptophan biosynthesis in Cryptococcus neoformans survival. PLoS One 10:e0132369. doi: 10.1371/journal.pone.0132369
Fisher, S. K., Novak, J. E., and Agranoff, B. W. (2002). Inositol and higher inositol phosphates in neural tissues: homeostasis, metabolism and functional significance. J. Neurochem. 82, 736–754. doi: 10.1046/j.1471-4159.2002.01041.x
Flanagan, P. R., Liu, N. N., Fitzpatrick, D. J., Hokamp, K., Kohler, J. R., and Moran, G. P. (2017). The Candida albicans TOR-activating GTPases Gtr1 and Rhb1 coregulate starvation responses and biofilm formation. mSphere 2:e00477-17. doi: 10.1128/mSphere.00477-17
Forsberg, H., and Ljungdahl, P. O. (2001). Sensors of extracellular nutrients in Saccharomyces cerevisiae. Curr Genet. 40, 91–109. doi: 10.1007/s002940100244
Fu, M. S., Coelho, C., De Leon-Rodriguez, C. M. Rossi, D. C. P., Camacho, E., Jung, E. H., et al. (2018). Cryptococcus neoformans urease affects the outcome of intracellular pathogenesis by modulating phagolysosomal pH. PLoS Pathog. 14:e1007144. doi: 10.1371/journal.ppat.1007144
Garrett, J. M. (2008). Amino acid transport through the Saccharomyces cerevisiae Gap1 permease is controlled by the Ras/cAMP pathway. Int. J. Biochem. Cell Biol. 40, 496–502. doi: 10.1016/j.biocel.2007.08.012
Gimeno, C. J., Ljungdahl, P. O., Styles, C. A., and Fink, G. R. (1992). Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68, 1077–1090. doi: 10.1016/0092-8674(92)90079-R
Giots, F., Donaton, M. C., and Thevelein, J. M. (2003). Inorganic phosphate is sensed by specific phosphate carriers and acts in concert with glucose as a nutrient signal for activation of the protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 47, 1163–1181. doi: 10.1046/j.1365-2958.2003.03365.x
Goberdhan, D. C., Wilson, C., and Harris, A. L. (2016). Amino acid sensing by mTORC1: intracellular transporters mark the spot. Cell Metab. 23, 580–589. doi: 10.1016/j.cmet.2016.03.013
Gong, R., Li, L., Liu, Y., Wang, P., Yang, H., Wang, L., et al. (2011). Crystal structure of the Gtr1p-Gtr2p complex reveals new insights into the amino acid-induced TORC1 activation. Genes Dev. 25, 1668–1673. doi: 10.1101/gad.16968011
Gonzalez, A., and Hall, M. N. (2017). Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 36, 397–408. doi: 10.15252/embj.201696010
Gruswitz, F., Chaudhary, S., Ho, J. D., Schlessinger, A., Pezeshki, B., Ho, C. M., et al. (2010). Function of human Rh based on structure of RhCG at 2.1 A. Proc. Natl. Acad. Sci. U.S.A. 107, 9638–9643. doi: 10.1073/pnas.1003587107
Hardwick, J. S., Kuruvilla, F. G., Tong, J. K., Shamji, A. F., and Schreiber, S. L. (1999). Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. U.S.A. 96, 14866–14870. doi: 10.1073/pnas.96.26.14866
Healy, M. E., Dillavou, C. L., and Taylor, G. E. (1977). Diagnostic medium containing inositol, urea, and caffeic acid for selective growth of Cryptococcus neoformans. J. Clin. Microbiol. 6, 387–391.
Heitman, J., Movva, N. R., and Hall, M. N. (1991a). Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253, 905–909. doi: 10.1126/science.1715094
Heitman, J., Movva, N. R., Hiestand, P. C., and Hall, M. N. (1991b). FK 506-binding protein proline rotamase is a target for the immunosuppressive agent FK 506 in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 88, 1948–1952. doi: 10.1073/pnas.88.5.1948
Helliwell, S. B., Wagner, P., Kunz, J., Deuter-Reinhard, M. Henriquez, R., and Hall, M. N. (1994). TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol. Biol. Cell 5, 105–118. doi: 10.1091/mbc.5.1.105
Herivaux, A., So, Y. S., Gastebois, A., Latge, J. P., Bouchara, J. P., Bahn, Y. S., et al. (2016). Major sensing proteins in pathogenic fungi: the hybrid histidine kinase family. PLoS Pathog. 12:e1005683. doi: 10.1371/journal.ppat.1005683
Hogan, D. A. (2006). Talking to themselves: autoregulation and quorum sensing in fungi. Eukaryot. Cell 5, 613–619. doi: 10.1128/EC.5.4.613-619.2006
Holsbeeks, I., Lagatie, O., Van Nuland, A. Van de Velde S., and Thevelein, J. M. (2004). The eukaryotic plasma membrane as a nutrient-sensing device. Trends Biochem. Sci. 29, 556–564. doi: 10.1016/j.tibs.2004.08.010
Jacoby, T., Flanagan, H., Faykin, A., Seto, A. G., Mattison, C., and Ota, I. (1997). Two protein-tyrosine phosphatases inactivate the osmotic stress response pathway in yeast by targeting the mitogen-activated protein kinase, Hog1. J. Biol. Chem. 272, 17749–17755. doi: 10.1074/jbc.272.28.17749
Jauniaux, J. C., and Grenson, M. (1990). GAP1, the general amino acid permease gene of Saccharomyces cerevisiae. Nucleotide sequence, protein similarity with the other bakers yeast amino acid permeases, and nitrogen catabolite repression. Eur. J. Biochem. 190, 39–44. doi: 10.1111/j.1432-1033.1990.tb15542.x
Javelle, A., Lupo, D., Zheng, L., Li, X. D., Winkler, F. K., and Merrick, M. (2006). An unusual twin-his arrangement in the pore of ammonia channels is essential for substrate conductance. J. Biol. Chem. 281, 39492–39498. doi: 10.1074/jbc.M608325200
Jewell, J. L., Russell, R. C., and Guan, K. L. (2013). Amino acid signalling upstream of mTOR. Nat. Rev. Mol. Cell Biol. 14, 133–139. doi: 10.1038/nrm3522
Jin, J. H., and Seyfang, A. (2003). High-affinity myo-inositol transport in Candida albicans: substrate specificity and pharmacology. Microbiology 149, 3371–3381. doi: 10.1099/mic.0.26644-0
Jung, K. W., Strain, A. K., Nielsen, K., Jung, K. H., and Bahn, Y. S. (2012). Two cation transporters Ena1 and Nha1 cooperatively modulate ion homeostasis, antifungal drug resistance, and virulence of Cryptococcus neoformans via the HOG pathway. Fungal Genet. Biol. 49, 332–345. doi: 10.1016/j.fgb.2012.02.001
Jung, W. H., Sham, A., Lian, T., Singh, A., Kosman, D. J., and Kronstad, J. W. (2008). Iron source preference and regulation of iron uptake in Cryptococcus neoformans. PLoS Pathog. 4:e45. doi: 10.1371/journal.ppat.0040045
Kankipati, H. N., Rubio-Texeira, M. Castermans, D., Diallinas, G., and Thevelein, J. M. (2015). Sul1 and Sul2 sulfate transceptors signal to protein kinase A upon exit of sulfur starvation. J. Biol. Chem. 290, 10430–10446. doi: 10.1074/jbc.M114.629022
Khademi, S., O’Connell, J. III, Remis, J., Robles-Colmenares, Y. Miercke, L. J., and Stroud, R. M. (2004). Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 A. Science 305, 1587–1594. doi: 10.1126/science.1101952
Kim, A., and Cunningham, K. W. (2015). A LAPF/phafin1-like protein regulates TORC1 and lysosomal membrane permeabilization in response to endoplasmic reticulum membrane stress. Mol. Biol. Cell 26, 4631–4645. doi: 10.1091/mbc.E15-08-0581
Kim, H., Jung, K. W., Maeng, S., Chen, Y. L., Shin, J., Shim, J. E., et al. (2015). Network-assisted genetic dissection of pathogenicity and drug resistance in the opportunistic human pathogenic fungus Cryptococcus neoformans. Sci. Rep. 5:8767. doi: 10.1038/srep08767
Kim, J. H., and Johnston, M. (2006). Two glucose-sensing pathways converge on Rgt1 to regulate expression of glucose transporter genes in Saccharomyces cerevisiae. J. Biol. Chem. 281, 26144–26149. doi: 10.1074/jbc.M603636200
Kim, M. S., Kim, S. Y., Yoon, J. K., Lee, Y. W., and Bahn, Y. S. (2009). An efficient gene-disruption method in Cryptococcus neoformans by double-joint PCR with NAT-split markers. Biochem. Biophys. Res. Commun. 390, 983–988. doi: 10.1016/j.bbrc.2009.10.089
Kim, S. Y., Ko, Y. J., Jung, K. W., Strain, A., Nielsen, K., and Bahn, Y. S. (2011). Hrk1 plays both Hog1-dependent and -independent roles in controlling stress response and antifungal drug resistance in Cryptococcus neoformans. PLoS One 6:e18769. doi: 10.1371/journal.pone.0018769
Kino, T., Hatanaka, H., Miyata, S., Inamura, N., Nishiyama, M., Yajima, T., et al. (1987). FK-506, a novel immunosuppressant isolated from a Streptomyces. II. Immunosuppressive effect of FK-506 in vitro. J. Antibiot. 40, 1256–1265. doi: 10.7164/antibiotics.40.1256
Klasson, H., Fink, G. R., and Ljungdahl, P. O. (1999). Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol. Cell. Biol. 19, 5405–5416. doi: 10.1128/MCB.19.8.5405
Ko, Y. J., Yu, Y. M., Kim, G. B., Lee, G. W., Maeng, P. J., Kim, S., et al. (2009). Remodeling of global transcription patterns of Cryptococcus neoformans genes mediated by the stress-activated HOG signaling pathways. Eukaryot. Cell 8, 1197–1217. doi: 10.1128/EC.00120-09
Kojima, K., Bahn, Y. S., and Heitman, J. (2006). Calcineurin, Mpk1 and Hog1 MAPK pathways independently control fludioxonil antifungal sensitivity in Cryptococcus neoformans. Microbiology 152, 591–604. doi: 10.1099/mic.0.28571-0
Kolakowski, L. F Jr. (1994). GCRDb: a G-protein-coupled receptor database. Receptors Channels 2, 1–7.
Kraakman, L., Lemaire, K., Ma, P., Teunissen, A. W., Donaton, M. C., and Van Dijck, P. et al. (1999). A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol. Microbiol. 32, 1002–1012. doi: 10.1046/j.1365-2958.1999.01413.x
Kraidlova, L., Van Zeebroeck, G., Van Dijck P., and Sychrova, H. (2011). The Candida albicans GAP gene family encodes permeases involved in general and specific amino acid uptake and sensing. Eukaryot. Cell 10, 1219–1229. doi: 10.1128/EC.05026-11
Kruppa, M., Krom, B. P., Chauhan, N., Bambach, A. V., Cihlar, R. L., and Calderone, R. A. (2004). The two-component signal transduction protein Chk1p regulates quorum sensing in Candida albicans. Eukaryot. Cell 3, 1062–1065. doi: 10.1128/EC.3.4.1062-1065.2004
Kunz, J., Henriquez, R., Schneider, U., Deuter-Reinhard, M., Movva, N. R., and Hall, M. N. (1993). Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 73, 585–596. doi: 10.1016/0092-8674(93)90144-F
Laub, M. T., and Goulian, M. (2007). Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 41, 121–145. doi: 10.1146/annurev.genet.41.042007.170548
Lauwers, E., Grossmann, G., and Andre, B. (2007). Evidence for coupled biogenesis of yeast Gap1 permease and sphingolipids: essential role in transport activity and normal control by ubiquitination. Mol. Biol. Cell 18, 3068–3080. doi: 10.1091/mbc.e07-03-0196
Lavin, J. L., Sarasola-Puente, V., Ramirez, L., Pisabarro, A. G., and Oguiza, J. A. (2014). Dual-histidine kinases in basidiomycete fungi. C. R. Biol. 337, 111–116. doi: 10.1016/j.crvi.2013.12.007
Lee, J. W., Ko, Y. J., Kim, S. Y., and Bahn, Y. S. (2011). Multiple roles of Ypd1 phosphotransfer protein in viability, stress response, and virulence factor regulation in Cryptococcus neoformans. Eukaryot. Cell 10, 998–1002. doi: 10.1128/EC.05124-11
Lee, K. T., Byun, H. J., Jung, K. W., Hong, J., Cheong, E., and Bahn, Y. S. (2014). Distinct and redundant roles of protein tyrosine phosphatases Ptp1 and Ptp2 in governing the differentiation and pathogenicity of Cryptococcus neoformans. Eukaryot. Cell 13, 796–812. doi: 10.1128/Ec.00069-14
Lee, K. T., So, Y. S., Yang, D. H., Jung, K. W., Choi, J., Lee, D. G., et al. (2016). Systematic functional analysis of kinases in the fungal pathogen Cryptococcus neoformans. Nat. Commun. 7:12766. doi: 10.1038/ncomms12766
Lemaire, K., Van de Velde, S., Van Dijck P., and Thevelein, J. M. (2004). Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol. Cell 16, 293–299. doi: 10.1016/j.molcel.2004.10.004
Liu, L., Mo, H., Wei, S., and Raftery, D. (2012). Quantitative analysis of urea in human urine and serum by 1H nuclear magnetic resonance. Analyst 137, 595–600. doi: 10.1039/c2an15780b
Liu, N. N., Flanagan, P. R., Zeng, J., Jani, N. M., Cardenas, M. E., Moran, G. P., et al. (2017). Phosphate is the third nutrient monitored by TOR in Candida albicans and provides a target for fungal-specific indirect TOR inhibition. Proc. Natl. Acad. Sci. U.S.A. 114, 6346–6351. doi: 10.1073/pnas.1617799114
Liu, T. B., Kim, J. C., Wang, Y., Toffaletti, D. L., Eugenin, E., Perfect, J. R., et al. (2013a). Brain inositol is a novel stimulator for promoting Cryptococcus penetration of the blood-brain barrier. PLoS Pathog. 9:e1003247. doi: 10.1371/journal.ppat.1003247
Liu, T. B., Subbian, S., Pan, W., Eugenin, E., Xie, J., and Xue, C. (2014). Cryptococcus inositol utilization modulates the host protective immune response during brain infection. Cell Commun. Signal. 12:51. doi: 10.1186/s12964-014-0051-0
Liu, T. B., Wang, Y., Baker, G. M., Fahmy, H., Jiang, L., and Xue, C. (2013b). The glucose sensor-like protein Hxs1 is a high-affinity glucose transporter and required for virulence in Cryptococcus neoformans. PLoS One 8:e64239. doi: 10.1371/journal.pone.0064239
Liu, Z., Thornton, J., Spirek, M., and Butow, R. A. (2008). Activation of the SPS amino acid-sensing pathway in Saccharomyces cerevisiae correlates with the phosphorylation state of a sensor component, Ptr3. Mol. Cell. Biol. 28, 551–563. doi: 10.1128/MCB.00929-07
Loewith, R., Jacinto, E., Wullschleger, S., Lorberg, A., Crespo, J. L., Bonenfant, D., et al. (2002). Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10, 457–468. doi: 10.1016/S1097-2765(02)00636-6
Lorenz, M. C., Cutler, N. S., and Heitman, J. (2000a). Characterization of alcohol-induced filamentous growth in Saccharomyces cerevisiae. Mol. Biol. Cell 11, 183–199. doi: 10.1091/mbc.11.1.183
Lorenz, M. C., Pan, X., Harashima, T., Cardenas, M. E., Xue, Y., Hirsch, J. P., et al. (2000b). The G protein-coupled receptor gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics 154, 609–622.
Lorenz, M. C., and Fink, G. R. (2001). The glyoxylate cycle is required for fungal virulence. Nature 412, 83–86. doi: 10.1038/3508359435083594
Lorenz, M. C., and Heitman, J. (1998). The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 17, 1236–1247. doi: 10.1093/emboj/17.5.1236
Lu, J. M., Deschenes, R. J., and Fassler, J. S. (2003). Saccharomyces cerevisiae histidine phosphotransferase Ypd1p shuttles between the nucleus and cytoplasm for SLN1-dependent phosphorylation of Ssk1p and Skn7p. Eukaryot. Cell 2, 1304–1314. doi: 10.1128/EC.2.6.1304-1314.2003
Lu, Y., Su, C., and Liu, H. (2012). A GATA transcription factor recruits Hda1 in response to reduced Tor1 signaling to establish a hyphal chromatin state in Candida albicans. PLoS Pathog. 8:e1002663. doi: 10.1371/journal.ppat.1002663
Maeda, T., Takekawa, M., and Saito, H. (1995). Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science 269, 554–558. doi: 10.1126/science.7624781
Maeda, T., Wurgler-Murphy S. M., and Saito, H. (1994). A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369, 242–245. doi: 10.1038/369242a0
Maidan, M. M., De Rop, L., Serneels, J., Exler, S., Rupp, S., Tournu, H., et al. (2005a). The G protein-coupled receptor Gpr1 and the Galpha protein Gpa2 act through the cAMP-Protein Kinase A pathway to induce morphogenesis in Candida albicans. Mol. Biol. Cell 16, 1971–1986. doi: 10.1091/mbc.e04-09-0780
Maidan, M. M., Thevelein, J. M., and Van Dijck P. (2005b). Carbon source induced yeast-to-hypha transition in Candida albicans is dependent on the presence of amino acids and on the G-protein-coupled receptor Gpr1. Biochem. Soc. Trans. 33, 291–293. doi: 10.1042/BST0330291
Marini, A. M., Boeckstaens, M., Benjelloun, F., Cherif-Zahar B., and Andre, B. (2006). Structural involvement in substrate recognition of an essential aspartate residue conserved in Mep/Amt and Rh-type ammonium transporters. Curr. Genet. 49, 364–374. doi: 10.1007/s00294-006-0062-5
Martel, R. R., Klicius, J., and Galet, S. (1977). Inhibition of the immune response by rapamycin, a new antifungal antibiotic. Can. J. Physiol. Pharmacol. 55, 48–51. doi: 10.1139/y77-007
Martinez, P., and Ljungdahl, P. O. (2005). Divergence of Stp1 and Stp2 transcription factors in Candida albicans places virulence factors required for proper nutrient acquisition under amino acid control. Mol. Cell. Biol. 25, 9435–9446. doi: 10.1128/MCB.25.21.9435-9446.2005
Martinez-Anaya, C., Dickinson, J. R., and Sudbery, P. E., (2003). In yeast, the pseudohyphal phenotype induced by isoamyl alcohol results from the operation of the morphogenesis checkpoint. J. Cell Sci. 116, 3423–3431. doi: 10.1242/jcs.00634
Martinez-Espinoza, A. D., Ruiz-Herrera, J., Leon-Ramirez C. G., and Gold, S. E. (2004). MAP kinase and cAMP signaling pathways modulate the pH-induced yeast-to-mycelium dimorphic transition in the corn smut fungus Ustilago maydis. Curr. Microbiol. 49, 274–281. doi: 10.1007/s00284-004-4315-6
Matsushita, M., and Janda, K. D. (2002). Histidine kinases as targets for new antimicrobial agents. Bioorg. Med. Chem. 10, 855–867. doi: 10.1016/S0968-0896(01)00355-8
Mavrianos, J., Berkow, E. L., Desai, C., Pandey, A., Batish, M., Rabadi, M. J., et al. (2013). Mitochondrial two-component signaling systems in Candida albicans. Eukaryot. Cell 12, 913–922. doi: 10.1128/Ec.00048-13
Mavrianos, J., Desai, C., and Chauhan, N. (2014). Two-component histidine phosphotransfer protein Ypd1 is not essential for viability in Candida albicans. Eukaryot. Cell 13, 452–460. doi: 10.1128/Ec.00243-13
Meyers, G. L., Jung, K. W., Bang, S., Kim, J., Kim, S., Hong, J., et al. (2017). The water channel protein aquaporin 1 regulates cellular metabolism and competitive fitness in a global fungal pathogen Cryptococcus neoformans. Environ. Microbiol. Rep. 9, 268–278. doi: 10.1111/1758-2229.12527
Miwa, T., Takagi, Y., Shinozaki, M., Yun, C. W., Schell, W. A., Perfect, J. R., et al. (2004). Gpr1, a putative G-protein-coupled receptor, regulates morphogenesis and hypha formation in the pathogenic fungus Candida albicans. Eukaryot. Cell 3, 919–931. doi: 10.1128/EC.3.4.919-931.2004
Moriya, H., and Johnston, M. (2004). Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase, I. Proc. Natl. Acad. Sci. U.S.A. 101, 1572–1577. doi: 10.1073/pnas.03059011010305901101
Nagahashi, S., Mio, T., Ono, N., Yamada-Okabe, T., Arisawa, M., Bussey, H., et al. (1998). Isolation of CaSLN1 and CaNIK1, the genes for osmosensing histidine kinase homologues, from the pathogenic fungus Candida albicans. Microbiology 144 (Pt. 2), 425–432. doi: 10.1099/00221287-144-2-425
Navarathna, D. H., Nickerson, K. W., Duhamel, G. E., Jerrels, T. R., and Petro, T. M. (2007). Exogenous farnesol interferes with the normal progression of cytokine expression during candidiasis in a mouse model. Infect. Immun. 75, 4006–4011. doi: 10.1128/IAI.00397-07
Nickerson, K. W., Atkin, A. L., and Hornby, J. M. (2006). Quorum sensing in dimorphic fungi: farnesol and beyond. Appl. Environ. Microbiol. 72, 3805–3813. doi: 10.1128/AEM.02765-05
Nikawa, J., Hosaka, K., and Yamashita, S. (1993). Differential regulation of two myo-inositol transporter genes of Saccharomyces cerevisiae. Mol. Microbiol. 10, 955–961. doi: 10.1111/j.1365-2958.1993.tb00967.x
Nikawa, J., Tsukagoshi, Y., and Yamashita, S. (1991). Isolation and characterization of two distinct myo-inositol transporter genes of Saccharomyces cerevisiae. J. Biol. Chem. 266, 11184–11191.
Okagaki, L. H., Wang, Y., Ballou, E. R., O’Meara, T. R., Bahn, Y. S., Alspaugh, J. A., et al. (2011). Cryptococcal titan cell formation is regulated by G-protein signaling in response to multiple stimuli. Eukaryot. Cell 10, 1306–1316. doi: 10.1128/EC.05179-11
O’Rourke, S. M., and Herskowitz, I. (2002). A third osmosensing branch in Saccharomyces cerevisiae requires the Msb2 protein and functions in parallel with the Sho1 branch. Mol. Cell. Biol. 22, 4739–4749. doi: 10.1128/MCB.22.13.4739-4749.2002
Osterholzer, J. J., Surana, R., Milam, J. E., Montano, G. T., Chen, G. H., Sonstein, J., et al. (2009). Cryptococcal urease promotes the accumulation of immature dendritic cells and a non-protective T2 immune response within the lung. Am. J. Pathol. 174, 932–943. doi: 10.2353/ajpath.2009.080673
Ozcan, S., Dover, J., and Johnston, M. (1998). Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 17, 2566–2573. doi: 10.1093/emboj/17.9.2566
Ozcan, S., Dover, J., Rosenwald, A. G., Wolfl, S., and Johnston, M. (1996). Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc. Natl. Acad. Sci. U.S.A. 93, 12428–12432. doi: 10.1073/pnas.93.22.12428
Pan, X., and Heitman, J. (1999). Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 4874–4887. doi: 10.1128/MCB.19.7.4874
Panepinto, J., Liu, L., Ramos, J., Zhu, X., Valyi-Nagy, T., Eksi, S., et al. (2005). The DEAD-box RNA helicase Vad1 regulates multiple virulence-associated genes in Cryptococcus neoformans. J. Clin. Invest. 115, 632–641. doi: 10.1172/JCI23048
Peng, T., Golub, T. R., and Sabatini, D. M. (2002). The immunosuppressant rapamycin mimics a starvation-like signal distinct from amino acid and glucose deprivation. Mol. Cell. Biol. 22, 5575–5584. doi: 10.1128/MCB.22.15.5575-5584.2002
Posas, F., and Saito, H. (1998). Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J. 17, 1385–1394. doi: 10.1093/emboj/17.5.1385
Posas, F., Wurgler-Murphy, S. M., Maeda, T., Witten, E. A., Thai, T. C., and Saito, H. (1996). Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell 86, 865–875. doi: 10.1016/S0092-8674(00)80162-2
Price, M. S., Betancourt-Quiroz, M., Price, J. L., Toffaletti, D. L., Vora, H., Hu, G., et al. (2011). Cryptococcus neoformans requires a functional glycolytic pathway for disease but not persistence in the host. mBio 2:e00103-11. doi: 10.1128/mBio.00103-11
Puria, R., Zurita-Martinez S. A., and Cardenas, M. E. (2008). Nuclear translocation of Gln3 in response to nutrient signals requires Golgi-to-endosome trafficking in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 105, 7194–7199. doi: 10.1073/pnas.0801087105
Rauceo, J. M., Blankenship, J. R., Fanning, S., Hamaker, J. J., Deneault, J. S., Smith, F. J., et al. (2008). Regulation of the Candida albicans cell wall damage response by transcription factor Sko1 and PAS kinase Psk1. Mol. Biol. Cell 19, 2741–2751. doi: 10.1091/mbc.e08-02-0191
Raught, B., Gingras, A. C., and Sonenberg, N. (2001). The target of rapamycin (TOR) proteins. Proc. Natl. Acad. Sci. U.S.A. 98, 7037–7044. doi: 10.1073/pnas.121145898
Ries, L. N. A., Beattie, S., Cramer, R. A., and Goldman, G. H. (2018). Overview of carbon and nitrogen catabolite metabolism in the virulence of human pathogenic fungi. Mol. Microbiol. 107, 277–297. doi: 10.1111/mmi.13887
Rohde, J., Heitman, J., and Cardenas, M. E. (2001). The TOR kinases link nutrient sensing to cell growth. J. Biol. Chem. 276, 9583–9586. doi: 10.1074/jbc.R000034200
Rohde, J. R., Campbell, S., Zurita-Martinez, S. A., Cutler, N. S., Ashe, M., and Cardenas, M. E. (2004). TOR controls transcriptional and translational programs via Sap-Sit4 protein phosphatase signaling effectors. Mol. Cell. Biol. 24, 8332–8341. doi: 10.1128/MCB.24.19.8332-8341.2004
Rohde, J. R., and Cardenas, M. E. (2003). The Tor pathway regulates gene expression by linking nutrient sensing to histone acetylation. Mol. Cell. Biol. 23, 629–635. doi: 10.1128/MCB.23.2.629-635.2003
Rolland, F., Winderickx, J., and Thevelein, J. M. (2002). Glucose-sensing and -signalling mechanisms in yeast. FEMS Yeast Res. 2, 183–201. doi: 10.1111/j.1567-1364.2002.tb00084.x
Roman, E., Cottier, F., Ernst, J. F., and Pla, J. (2009). Msb2 signaling mucin controls activation of Cek1 mitogen-activated protein kinase in Candida albicans. Eukaryot. Cell 8, 1235–1249. doi: 10.1128/EC.00081-09
Rude, T. H., Toffaletti, D. L., Cox, G. M., and Perfect, J. R. (2002). Relationship of the glyoxylate pathway to the pathogenesis of Cryptococcus neoformans. Infect. Immun. 70, 5684–5694. doi: 10.1128/IAI.70.10.5684-5694.2002
Rutherford, J. C., Chua, G., Hughes, T., Cardenas, M. E., and Heitman, J. (2008a). A Mep2-dependent transcriptional profile links permease function to gene expression during pseudohyphal growth in Saccharomyces cerevisiae. Mol. Biol. Cell 19, 3028–3039. doi: 10.1091/mbc.E08-01-0033
Rutherford, J. C., Lin, X., Nielsen, K., and Heitman, J. (2008b). Amt2 permease is required to induce ammonium-responsive invasive growth and mating in Cryptococcus neoformans. Eukaryot. Cell 7, 237–246. doi: 10.1128/EC.00079-07
Sabatini, D. M., Erdjument-Bromage, H., Lui, M., Tempst, P., and Snyder, S. H. (1994). RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 78, 35–43. doi: 10.1016/0092-8674(94)90570-3
Sabers, C. J., Martin, M. M., Brunn, G. J., Williams, J. M., Dumont, F. J., Wiederrecht, G., et al. (1995). Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J. Biol. Chem. 270, 815–822. doi: 10.1074/jbc.270.2.815
Sabina, J., and Brown, V. (2009). Glucose sensing network in Candida albicans: a sweet spot for fungal morphogenesis. Eukaryot. Cell 8, 1314–1320. doi: 10.1128/EC.00138-09
Samyn, D. R., Ruiz-Pavon, L., Andersson, M. R., Popova, Y., Thevelein, J. M., and Persson, B. L. (2012). Mutational analysis of putative phosphate- and proton-binding sites in the Saccharomyces cerevisiae Pho84 phosphate:H+ transceptor and its effect on signalling to the PKA and PHO pathways. Biochem. J. 445, 413–422. doi: 10.1042/BJ20112086
Santangelo, G. M. (2006). Glucose signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70, 253–282. doi: 10.1128/MMBR.70.1.253-282.2006
Saxton, R. A., Chantranupong, L., Knockenhauer, K. E., Schwartz, T. U., and Sabatini, D. M. (2016). Mechanism of arginine sensing by CASTOR1 upstream of mTORC1. Nature 536, 229–233. doi: 10.1038/nature19079
Schmelzle, T., Beck, T., Martin, D. E., and Hall, M. N. (2004). Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol. Cell. Biol. 24, 338–351. doi: 10.1128/MCB.24.1.338-351.2004
Schmidt, A., Beck, T., Koller, A., Kunz, J., and Hall, M. N. (1998). The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 17, 6924–6931. doi: 10.1093/emboj/17.23.6924
Schothorst, J., Zeebroeck, G. V., and Thevelein, J. M. (2017). Identification of Ftr1 and Zrt1 as iron and zinc micronutrient transceptors for activation of the PKA pathway in Saccharomyces cerevisiae. Microb. Cell 4, 74–89. doi: 10.15698/mic2017.03.561
Sehgal, S. N. (2003). Sirolimus: its discovery, biological properties, and mechanism of action. Transplant Proc. 35, 7S–14S. doi: 10.1016/S0041-1345(03)00211-2
Sehgal, S. N., Baker, H., and Vezina, C. (1975). Rapamycin (AY-22,989), a new antifungal antibiotic. II Fermentation., isolation and characterization. J. Antibiot. 28, 727–732. doi: 10.7164/antibiotics.28.727
Shi, M., Li, S. S., Zheng, C., Jones, G. J., Kim, K. S., Zhou, H., et al. (2010). Real-time imaging of trapping and urease-dependent transmigration of Cryptococcus neoformans in mouse brain. J. Clin. Invest. 120, 1683–1693. doi: 10.1172/JCI41963
Singh, A., Panting, R. J., Varma, A., Saijo, T., Waldron, K. J., Jong, A., et al. (2013). Factors required for activation of urease as a virulence determinant in Cryptococcus neoformans. mBio 4:e00220-13. doi: 10.1128/mBio.00220-13
Singh, P., Chauhan, N., Ghosh, A., Dixon, F., and Calderone, R. (2004). SKN7 of Candida albicans: mutant construction and phenotype analysis. Infect. Immun. 72, 2390–2394. doi: 10.1128/IAI.72.4.2390-2394.2004
Smith, D. A., Nicholls, S., Morgan, B. A., Brown, A. J., and Quinn, J. (2004). A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol. Biol. Cell 15, 4179–4190. doi: 10.1091/mbc.e04-03-0181
Smith, D. G., Garcia-Pedrajas, M. D., Gold, S. E., and Perlin, M. H. (2003). Isolation and characterization from pathogenic fungi of genes encoding ammonium permeases and their roles in dimorphism. Mol. Microbiol. 50, 259–275. doi: 10.1046/j.1365-2958.2003.03680.x
So, Y. S., Jang, J., Park, G., Xu, J., Olszewski, M. A., and Bahn, Y. S. (2018). Sho1 and Msb2 play complementary but distinct roles in stress responses, sexual differentiation, and pathogenicity of Cryptococcus neoformans. Front. Microbiol. 9:2958. doi: 10.3389/fmicb.2018.02958.
Song, M. H., Lee, J. W., Kim, M. S., Yoon, J. K., White, T. C., Floyd, A., et al. (2012). A flucytosine-responsive Mbp1/Swi4-like protein, Mbs1, plays pleiotropic roles in antifungal drug resistance, stress response, and virulence of Cryptococcus neoformans. Eukaryot. Cell 11, 53–67. doi: 10.1128/EC.05236-11
Springael, J. Y., and Andre, B. (1998). Nitrogen-regulated ubiquitination of the Gap1 permease of Saccharomyces cerevisiae. Mol. Biol. Cell 9, 1253–1263. doi: 10.1091/mbc.9.6.1253
Stracka, D., Jozefczuk, S., Rudroff, F., Sauer, U., and Hall, M. N. (2014). Nitrogen source activates TOR (target of rapamycin) complex 1 via glutamine and independently of Gtr/Rag proteins. J. Biol. Chem. 289, 25010–25020. doi: 10.1074/jbc.M114.574335
Su, C., Lu, Y., and Liu, H. (2013). Reduced TOR signaling sustains hyphal development in Candida albicans by lowering Hog1 basal activity. Mol. Biol. Cell 24, 385–397. doi: 10.1091/mbc.E12-06-0477
Szafranski-Schneider, E., Swidergall, M., Cottier, F., Tielker, D., Roman, E., Pla, J., et al. (2012). Msb2 shedding protects Candida albicans against antimicrobial peptides. PLoS Pathog. 8:e1002501. doi: 10.1371/journal.ppat.1002501
Tatebayashi, K., Tanaka, K., Yang, H. Y., Yamamoto, K., Matsushita, Y., Tomida, T., et al. (2007). Transmembrane mucins Hkr1 and Msb2 are putative osmosensors in the SHO1 branch of yeast HOG pathway. EMBO J. 26, 3521–3533. doi: 10.1038/sj.emboj.7601796
Tatebayashi, K., Yamamoto, K., Tanaka, K., Tomida, T., Maruoka, T., Kasukawa, E., et al. (2006). Adaptor functions of Cdc42, Ste50, and Sho1 in the yeast osmoregulatory HOG MAPK pathway. EMBO J. 25, 3033–3044. doi: 10.1038/sj.emboj.7601192
Thevelein, J. M. (1994). Signal transduction in yeast. Yeast 10, 1753–1790. doi: 10.1002/yea.320101308
Thevelein, J. M., and Voordeckers, K. (2009). Functioning and evolutionary significance of nutrient transceptors. Mol. Biol. Evol. 26, 2407–2414. doi: 10.1093/molbev/msp168
Tiwari, S., Jamal, S. B., Hassan, S. S., Carvalho, P., Almeida, S., Barh, D., et al. (2017). Two-component signal transduction systems of pathogenic bacteria as targets for antimicrobial therapy: an overview. Front. Microbiol. 8:1878. doi: 10.3389/fmicb.2017.01878
Upadhya, R., Kim, H., Jung, K. W., Park, G., Lam, W., Lodge, J. K., et al. (2013). Sulphiredoxin plays peroxiredoxin-dependent and -independent roles via the HOG signalling pathway in Cryptococcus neoformans and contributes to fungal virulence. Mol. Microbiol. 90, 630–648. doi: 10.1111/mmi.12388
Uppuluri, P., Mekala, S., and Chaffin, W. L. (2007). Farnesol-mediated inhibition of Candida albicans yeast growth and rescue by a diacylglycerol analogue. Yeast 24, 681–693. doi: 10.1002/yea.1501
van den Berg, B., Chembath, A., Jefferies, D., Basle, A., Khalid, S., and Rutherford, J. C. (2016). Structural basis for Mep2 ammonium transceptor activation by phosphorylation. Nat. Commun. 7:11337. doi: 10.1038/ncomms11337
Van Nuland, A., Vandormael, P., Donaton, M., Alenquer, M., Lourenco, A., Quintino, E., et al. (2006). Ammonium permease-based sensing mechanism for rapid ammonium activation of the protein kinase A pathway in yeast. Mol. Microbiol. 59, 1485–1505. doi: 10.1111/j.1365-2958.2005.05043.x
Van Zeebroeck, G., Bonini, B. M., Versele, M., and Thevelein, J. M. (2009). Transport and signaling via the amino acid binding site of the yeast Gap1 amino acid transceptor. Nat. Chem. Biol. 5, 45–52. doi: 10.1038/nchembio.132
Van Zeebroeck, G., Kimpe, M., Vandormael, P., and Thevelein, J. M. (2011). A split-ubiquitin two-hybrid screen for proteins physically interacting with the yeast amino acid transceptor Gap1 and ammonium transceptor Mep2. PLoS One 6:e24275. doi: 10.1371/journal.pone.0024275
Versele, M., Lemaire, K., and Thevelein, J. M. (2001). Sex and sugar in yeast: two distinct GPCR systems. EMBO Rep. 2, 574–579. doi: 10.1093/embo-reports/kve132
Vylkova, S., Carman, A. J., Danhof, H. A., Collette, J. R., Zhou, H., and Lorenz, M. C. (2011). The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. mBio 2:e00055-11. doi: 10.1128/mBio.00055-11
Vylkova, S., and Lorenz, M. C. (2014). Modulation of phagosomal pH by Candida albicans promotes hyphal morphogenesis and requires Stp2p, a regulator of amino acid transport. PLoS Pathog. 10:e1003995. doi: 10.1371/journal.ppat.1003995
Wang, L. I., Lin, Y. S., Liu, K. H., Jong, A. Y., and Shen, W. C. (2011a). Cryptococcus neoformans mediator protein Ssn8 negatively regulates diverse physiological processes and is required for virulence. PLoS One 6:e19162. doi: 10.1371/journal.pone.0019162
Wang, Y., Liu, T. B., Patel, S., Jiang, L., and Xue, C. (2011b). The casein kinase I protein Cck1 regulates multiple signaling pathways and is essential for cell integrity and fungal virulence in Cryptococcus neoformans. Eukaryot. Cell 10, 1455–1464. doi: 10.1128/EC.05207-11
Wedaman, K. P., Reinke, A., Anderson, S., Yates, J. III, McCaffery, J. M., and Powers, T. (2003). Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 1204–1220. doi: 10.1091/mbc.e02-09-0609
Wolfson, R. L., and Sabatini, D. M. (2017). The dawn of the age of amino acid sensors for the mTORC1 pathway. Cell Metab. 26, 301–309. doi: 10.1016/j.cmet.2017.07.001
Wu, B., Ottow, K., Poulsen, P., Gaber, R. F., Albers, E., and Kielland-Brandt, M. C. (2006). Competitive intra- and extracellular nutrient sensing by the transporter homologue Ssy1p. J. Cell Biol. 173, 327–331. doi: 10.1083/jcb.200602089
Wurgler-Murphy, S. M., Maeda, T., Witten, E. A., and Saito, H. (1997). Regulation of the Saccharomyces cerevisiae HOG1 mitogen-activated protein kinase by the PTP2 and PTP3 protein tyrosine phosphatases. Mol. Cell. Biol. 17, 1289–1297. doi: 10.1128/MCB.17.3.1289
Xue, C. (2012). Cryptococcus and beyond–inositol utilization and its implications for the emergence of fungal virulence. PLoS Pathog. 8:e1002869. doi: 10.1371/journal.ppat.1002869PPATHOGENS-D-12-00981
Xue, C., Bahn, Y. S., Cox, G. M., and Heitman, J. (2006). G protein-coupled receptor Gpr4 senses amino acids and activates the cAMP-PKA pathway in Cryptococcus neoformans. Mol. Biol. Cell 17, 667–679. doi: 10.1091/mbc.E05-07-0699
Xue, C., Liu, T., Chen, L., Li, W., Liu, I., Kronstad, J. W., et al. (2010). Role of an expanded inositol transporter repertoire in Cryptococcus neoformans sexual reproduction and virulence. mBio 1:e00084-10. doi: 10.1128/mBio.00084-10
Xue, C., Tada, Y., Dong, X., and Heitman, J. (2007). The human fungal pathogen Cryptococcus can complete its sexual cycle during a pathogenic association with plants. Cell Host Microbe 1, 263–273. doi: 10.1016/j.chom.2007.05.005
Xue, Y., Batlle, M., and Hirsch, J. P. (1998). GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Gα subunit and functions in a Ras-independent pathway. EMBO J. 17, 1996–2007. doi: 10.1093/emboj/17.7.1996
Yamada-Okabe, T., Mio, T., Ono, N., Kashima, Y., Matsui, M., Arisawa, M., et al. (1999). Roles of three histidine kinase genes in hyphal development and virulence of the pathogenic fungus Candida albicans. J. Bacteriol. 181, 7243–7247.
Zarrinpar, A., Bhattacharyya, R. P., Nittler, M. P., and Lim, W. A. (2004). Sho1 and Pbs2 act as coscaffolds linking components in the yeast high osmolarity MAP kinase pathway. Mol. Cell 14, 825–832. doi: 10.1016/j.molcel.2004.06.011
Zeitlinger, J., Simon, I., Harbison, C. T., Hannett, N. M., Volkert, T. L., Fink, G. R., et al. (2003). Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell 113, 395–404. doi: 10.1016/S0092-8674(03)00301-5
Zhang, L. H., and Dong, Y. H. (2004). Quorum sensing and signal interference: diverse implications. Mol. Microbiol. 53, 1563–1571. doi: 10.1111/j.1365-2958.2004.04234.x
Zheng, L., Kostrewa, D., Berneche, S., Winkler, F. K., and Li, X. D. (2004). The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 101, 17090–17095. doi: 10.1073/pnas.0406475101
Zschiedrich, C. P., Keidel, V., and Szurmant, H. (2016). Molecular mechanisms of two-component signal transduction. J. Mol. Biol. 428, 3752–3775. doi: 10.1016/j.jmb.2016.08.003
Keywords: yeast, nutrient sensing, transceptor, G protein-coupled receptor, Mep2, fungal pathogen, stress response, Tor
Citation: Rutherford JC, Bahn Y-S, van den Berg B, Heitman J and Xue C (2019) Nutrient and Stress Sensing in Pathogenic Yeasts. Front. Microbiol. 10:442. doi: 10.3389/fmicb.2019.00442
Received: 09 January 2019; Accepted: 20 February 2019;
Published: 08 March 2019.
Edited by:
Gustavo Henrique Goldman, University of São Paulo, BrazilReviewed by:
Slavena Vylkova, Friedrich-Schiller-Universität Jena, GermanyCarol Munro, University of Aberdeen, United Kingdom
Copyright © 2019 Rutherford, Bahn, van den Berg, Heitman and Xue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joseph Heitman, aGVpdG0wMDFAZHVrZS5lZHU= Chaoyang Xue, eHVlY2hAcnV0Z2Vycy5lZHU=
†These authors have contributed equally to this work
 Julian C. Rutherford1†
Julian C. Rutherford1† Yong-Sun Bahn
Yong-Sun Bahn Joseph Heitman
Joseph Heitman Chaoyang Xue
Chaoyang Xue