
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 06 March 2019
Sec. Food Microbiology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00433
This article is part of the Research TopicIndustrial and Health Applications of Lactic Acid Bacteria and Their MetabolitesView all 57 articles
Three Lactobacillus plantarum strains ATG-K2, ATG-K6, and ATG-K8 were isolated from Kimchi, a Korean traditional fermented food, and their probiotic potentials were examined. All three strains were free of antibiotic resistance, hemolysis, and biogenic amine production and therefore assumed to be safe, as supported by whole genome analyses. These strains demonstrated several basic probiotic functions including a wide range of antibacterial activity, bile salt hydrolase activity, hydrogen peroxide production, and heat resistance at 70∘C for 60 s. Further studies of antimicrobial activities against Candida albicans and Gardnerella vaginalis revealed growth inhibitory effects from culture supernatants, coaggregation effects, and killing effects of the three probiotic strains, with better efficacy toward C. albicans. In vitro treatment of bacterial lysates of the probiotic strains to the RAW264.7 murine macrophage cell line resulted in innate immunity enhancement via IL-6 and TNF-α production without lipopolysaccharide (LPS) treatment and anti-inflammatory effects via significantly increased production of IL-10 when co-treated with LPS. However, the degree of probiotic effect was different for each strain as the highest TNF-α and the lowest IL-10 production by the RAW264.7 cell were observed in the K8 lysate treated group compared to the K2 and K6 lysate treated groups, which may be related to genomic differences such as chromosome size (K2: 3,034,884 bp, K6: 3,205,672 bp, K8: 3,221,272 bp), plasmid numbers (K2: 3, K6 and K8: 1), or total gene numbers (K2: 3,114, K6: 3,178, K8: 3,186). Although more correlative inspections to connect genomic information and biological functions are needed, genomic analyses of the three strains revealed distinct genomic compositions of each strain. Also, this finding suggests genome level analysis may be required to accurately identify microorganisms. Nevertheless, L. plantarum ATG-K2, ATG-K6, and ATG-K8 demonstrated their potential as probiotics for mucosal health improvement in both microbial and immunological contexts.
Mucosa or mucosal sites are external tissues of animals covered with mucus, which are exposed to lifespan-ensuring and/or transient microorganisms known as commensal microbiota. The gut is the most studied mucosal site, but other mucosa including air ways, nostril, oral cavity, and vagina also have been thoroughly studied with impacts in the microbiome and immunological fields (Gill et al., 2010). In general, mucosal sites are protected by mucus, secreted immunoglobulin A, antimicrobial peptides, and innate and adaptive immune cells from various non-self-antigens including commensal microbiota and food antigens (Tlaskalová-Hogenová et al., 2004; Brandtzaeg, 2009; McDermott and Huffnagle, 2014). Usually many microbes are tolerated under healthy conditions and are even associated with beneficial effects on immune development, organ development, metabolism, and homeostasis of the host (Sommer and Bäckhed, 2013).
However, some members of commensal microbiota such as Escherichia coli, Staphylococcus spp., Streptococcus spp. in mucosal sites can occasionally be pathogenic in certain cases depending on the type of microorganism and health status of the host, such as invasion of highly virulent microbes, dysbiosis of microbiota by antibiotics, mechanical damage of mucosa, and/or dysregulation of immune system of the host (Honda and Littman, 2012; Khosravi and Mazmanian, 2013). Among commensal microbe-associated mucosal infections related to dysbiosis, candidiasis is one of the more notorious infections, and occurs at most mucosal sites causing inflammatory symptoms and even systemic infection (candidemia) mainly in immunocompromised individuals (Kim and Sudbery, 2011; Moyes and Naglik, 2011). Candidiasis is closely related to the pathogenesis of vulvovaginal candidiasis (Sobel et al., 1998; Kim and Sudbery, 2011), Crohn’s disease (Poulain et al., 2009), oral infections (Ellepola and Samaranayake, 2000) and microbial infections in individuals with acquired immune deficiency syndrome (Kim and Sudbery, 2011). Another typical mucosal disease model caused by dysbiosis of microbiota is bacterial vaginosis (BV), an infectious disease with symptoms of bad odor, excessive discharge, erythema, increment of pH, and inflammation (Morris et al., 2001; Egan and Lipsky, 2002). BV is caused by (1) disruption of vaginal microbiota due to overdosed antibiotics treatment, stress, hormonal changes, and/or physical damage, (2) resultant loss of hydrogen peroxide producing lactobacilli and bifidobacteria, and (3) increased pH in the vagina that allows the growth of other opportunistic pathogens (Sobel, 1997).
To treat such microbial infections in mucosal sites, a probiotic approach is one of the important measures besides chemical or antibiotic treatments that may restore mucosal microbiota to a healthy distribution and/or eliminate certain pathogens in a microbiome environment. Mechanisms of probiotics toward microbial infections include bacteriocin, co-aggregation, production of hydrogen peroxide (H2O2), production of organic acids, reduction of pH, and immunomodulation, which may act singularly or synergistically to benefit the host (Reid et al., 2011).
With mechanistic criteria focused on treating and/or preventing mucosal infection as mentioned above, Lactobacillus plantarum ATG-K2, ATG-K6, and ATG-K8 were isolated in the present study from Kimchi, a Korean traditional fermented vegetable, and examined for their safety and functionalities in biologic and genomic analyses as potential probiotics for improving mucosal health.
Three Lactobacillus plantarum strains, L. plantarum ATG-K2 (K2), L. plantarum ATG-K6 (K6), and L. plantarum ATG-K8 (K8) were isolated from kimchi samples, traditional Korean fermented vegetables, made in the Chungchung region of South Korea. Lactobacillus strains were cultured in de Man Rogosa Sharpe (MRS) medium (Difco Laboratories, United States) at 37°C. Each strain was isolated from different batches of kimchi samples. Lactobacilli were first identified as L. plantarum strains by 16S rRNA sequences using nucleotide BLAST tool of the National Center for Biotechnology Information (NCBI) then processed to whole genome sequencing of section Whole Genome Analysis of L. plantarum Strains of the present study.
For antibacterial effect testing of lactobacilli strains, Staphylococcus aureus KCTC1621 (SA), Escherichia coli KCTC1682 (EC), Pseudomonas aeruginosa KCTC2004 (PA), Listeria monocytogenes KCTC3569 (LM), Cronobacter sakazakii KCTC2949 (CS), Streptococcus mutans KCTC3065 (SM), Streptococcus salivarius ATG-P1 (SS) were used as representative opportunistic pathogens. Opportunistic pathogens were cultured in brain heart infusion medium (BHI, Difco Laboratories, United States) at 37°C.
To determine the impact of L. plantarum strains on infectious mucosal pathogens, Candida albicans KCTC7678 (CA) and Gardnerella vaginalis KCTC5096 (GV) were selected as representative candidiasis and BV pathogens, respectively. CA was cultured in yeast extract maltose medium (YM, Difco Laboratories, United States) at 37°C, and GV was cultured in BHI supplemented with 20% heat-inactivated horse serum (Gibco, United States) under anaerobic conditions using an OxoidTM AnaeroGenTM system (Oxoid, United Kingdom) at 37°C.
Fermentation patterns of L. plantarum strains were tested with an API 50 CHL test (BioMérieux, France) toward 49 selected carbohydrate sources following the manufacturer’s guidelines. Briefly, overnight cultures of lactobacilli were suspended in 10 mL API 50 CHL medium (BioMérieux, France) and each applied to cupels containing different carbohydrates on an API 50 CH test strip. Fermentation patterns were monitored for up to 72 h at 37°C.
To test heat resistance of L. plantarum strains, overnight broth-cultured strains were heated in a water bath at 70°C for 20, 40, 60, and 80 s. Viable bacteria from each time point were measured by 10-fold serial dilution and spread-plate method.
Antibacterial effects of L. plantarum strains were tested by the disc diffusion method. Four Gram-positive bacteria (SA, LM, SM, SS) and three Gram-negative bacteria (EC, PA, CS) were used as target bacteria. Overnight cultures of target bacteria were suspended in 0.9% saline with an optical density (OD) of 0.8 at 600 nm wavelength (OD600). Each bacterial suspension was spread on mixed agar medium of BHI and MRS at a 1:1 ratio in weight. After spreading bacterial suspensions, 8 mm paper discs (Advantec, Japan) were placed on agar plates, and 35 μL of each overnight lactobacillus broth culture was inoculated on the paper discs. After 24–48 h of incubation at 37°C, diameters of inhibition zones were measured. The diameter of the paper disc was subtracted from the total diameter of each measured inhibition zone.
BSH activity of L. plantarum strains was determined using the method described by Dashkevicz and Feighner (1989). Briefly, each strain was streaked on MRS agar medium supplied with 0.5% w/v sodium taurocholic acid (TDCA, Sigma-Aldrich, Germany). Inoculated agar plates were placed in an anaerobic jar and incubated at 37°C for 48–72 h.
H2O2 production ability of L. plantarum strains was determined with 0.25 mg/mL tetramethylbenzidine (TMB, Sigma-Aldrich, Germany) and 0.01 mg/mL horseradish peroxidase (Sigma-Aldrich, Germany) in MRS agar medium as described by McGroarty et al. (1992). Additives were filtered through 0.2 μm size pore filter (Satorious, Germany) and added to autoclaved MRS agar medium when the medium reached approximately 50°C. Test specimens were incubated at 37°C for 48–72 h in an anaerobic jar, and exposed to air at 25°C to monitor color change of colonies.
Radical scavenging activity of each L. plantarum strain was tested by using a 2, 2′-azino-bis 3-ethylbenzthiazoline-6-sulfonic acid (ABTS, Sigma-Aldrich, Germany) colorimetric assay. The test was conducted with lysates from each strain to avoid growth and to examine both intracellular and extracellular contents of the bacteria. To prepare bacterial lysates, biomass from overnight broth cultured strains were obtained by centrifugation at 4,000 × g for 30 min at 4°C. Bacterial pellets were washed twice with 1× phosphate buffered saline (PBS) and concentrated to 10× by resuspending in PBS. The samples were treated with lysozyme (Sigma-Aldrich, Germany) at 37°C for 2 h, then lysed with sonication. Solid content of resulting lysates was measured with a moisture analyzer (A&D Co., Ltd., Japan), and the final stock solution concentration was adjusted to 50 mg/mL. ABTS was prepared by mixing 14 mM ABTS and 4.9 mM potassium persulfate at a 1:1 ratio and incubating in a darkroom overnight. The OD of the solution was adjusted to 0.7 at 734 nm wavelength to prepare a working solution. Lysate samples were added into the working solution at a 1:10 ratio on microplates and incubated for 10 min in a darkroom. The ABTS-reacted samples were centrifuged at 13,000 rpm for 5 min at 4°C, and the resulting supernatants were measured at 734 nm wavelength with an Epoch microplate spectrophotometer (BioTek Instruments, United States). Percent ABTS radical scavenging activity was calculated using following equation, where ODsample is the measured OD value of ABTS-reacted lysate and ODcontrol is the OD of non-reacted ABTS control:
To determine the safety of L. plantarum strains in the present study, hemolysis activity was first examined with 5% sheep blood supplied in tryptic soybean agar (TSA, Difco Laboratories, United States) by streaking each strain onto the medium followed by overnight incubation at 37°C under anaerobic conditions. SA was also streaked onto a sheep blood agar medium as positive control for the experiment.
Antibiotic susceptibility of lactobacilli was tested with ampicillin, gentamicin, kanamycin, streptomycin, clindamycin, erythromycin, tetracycline, and chloramphenicol E-test strips (BioMérieux, France) to determine minimum inhibitory concentration (MIC) values. Briefly, overnight cultures of lactobacilli strains were suspended in 0.9% saline at an OD600 of 0.8. Bacterial suspensions were spread onto counting agar plates (PCA, Difco Laboratory, United States), and E-test strips were placed on the agar plates. MIC cut-off values were determined using European food safety authority (EFSA) guidelines on antibiotic susceptibility (EFSA, 2012).
Production of the potentially harmful biogenic amines histamine, tyramine, putrescine, and cadaverine was examined using a test medium described by Bover-Cid and Holzapfel (1999). The test medium was made according to the composition of Bover-Cid and Holzapfel and each strain was streaked on the medium. The inocula were incubated at 37°C for 72 h, and colorimetric change of bromocresol purple in the medium from yellow to purple due to pH increase by decarboxylase activity (indicating a positive result) was monitored.
Growth inhibition effects of cell free culture supernatant (CFCS) of L. plantarum strains toward CA and GV were measured. CFCS was prepared from overnight broth cultures of each strain by centrifugation at 5,000 rpm for 30 min. CFCS was filtered through 0.2 μm pore syringe filter for sterilization. CFCS was freeze dried and resuspended in 1× PBS to make a 10× stock solution. No additional process of CFCS was done after stock preparation to preserve all substances in the CFCS. CA was inoculated at 1 × 105 CFU/mL in YM broth supplied with CFCS at 1× final concentration. GV was inoculated at 1 × 106 CFU/mL in BHI broth supplemented with 20% heat inactivated horse serum and CFCS. Samples without CFCS were used as controls. CA was incubated at 37°C for 24 h and GV for 48 h depending on growth properties of each pathogen. Endpoint OD was measured at 600 nm wavelength by a spectrophotometer. Growth inhibition rates were calculated using following equation, where ODcontrol and ODcfcs are the OD of the control and CFCS-treated groups, respectively.
Coaggregation activity to pathogens, another potential antimicrobial ability, was determined for the L. plantarum strains by a method described by Handley et al. (1987). Briefly, overnight broth-cultured microorganisms were collected by centrifugation at 13,000 rpm for 5 min at 4°C, and cell pellets were washed twice with 1× PBS. Each microorganism was resuspended in 1× PBS and adjusted to an OD value of 1.0 at 600 nm wavelength. L. plantarum strain and pathogen suspensions were mixed in equal volumes, vortexed for 5 min and left at room temperature for sedimentation. The upper part of the mixture was carefully removed and its OD was measured at 1, 4, and 8 h time points for each sample. Single suspensions of each microorganism were used as controls. Measured OD values were calculated with following equation to determine coaggregation rate, where ODpatho, ODlp, and ODmix are the ODs of the pathogen groups, L. plantarum strains, and strain and pathogen mixture, respectively.
Coculture experiments of L. plantarum strains and pathogens were conducted to examine direct killing effects. Approximately 2.5 × 106 CFU/mL of CA and of each strain of L. plantarum were inoculated in a 1:1 ratio of mixed YM and MRS broth and incubated at 37°C for 48 h. For the anti-GV test, approximately 1.0 × 108 CFU/mL of GV and of each L. plantarum strain were inoculated in BHI broth supplemented with 20% horse serum, 1% yeast extract, and 0.1% maltose and incubated at 37°C for 48 h in an anaerobic jar. Each test inoculum was sampled and checked for viable pathogens by spread-plate method. For CA growth medium, 100 μg/mL ampicillin supplemented YM agar medium was used to grow CA selectively. For GV growth medium, 5% rabbit blood BHI agar medium was used and GV was selected based on white, slight translucent colony morphology and hemolysis zones formed around GV colonies.
To determine whether L. plantarum strains are capable of immunomodulation, in vitro cell experiments with RAW264.7 murine macrophage cells were performed. Cells were cultured in Dulbecco Modified Eagle Medium (DMEM, Gibco, United States) supplemented with 10% fetal bovine serum (Gibco, United States) and 1% penicillin/streptomycin cocktail (Sigma-Aldrich, Germany) throughout the experiment. RAW264.7 cultured up to 80–90% confluency was collected and 1 × 106 cells were seeded in each well of a 24 well cell culture plate (SPL Life Science, Korea). Seeded cells were stabilized at 37°C in 5% CO2 atmosphere for 24 h. Then, 1 μg/mL of lipopolysaccharide (LPS, Sigma-Aldrich, Germany) was administered was as administered to 100 or 500 μg/mL of lysates from each L. plantarum strain. Non-treated cells were used as a negative control and LPS-treated cell as a positive control. After treating with each substance, cells were incubated at 37°C in 5% CO2 atmosphere for 24 h. Resulting culture supernatants of each treatment group were collected and processed with IL-6 enzyme linked immunosorbent assay (ELISA) with a Mouse IL-6 Quantikine ELISA Kit (R&D Systems, United States) and TNF-α ELISA with a Mouse TNF-alpha Quantikine ELISA Kit (R&D Systems, United States) for quantification of produced cytokines.
Using the same experimental procedure described previously in the present study with some modification, IL-10 production enhancement in macrophages by the lysate of L. plantarum strains was examined, and 1 μg/mL LPS and 100 μg/mL of L. plantarum lysate co-treated groups were added in this experiment. After treatment with each substance, cells were incubated for another 24 h, and culture supernatant samples were collected. IL-10 concentrations of each culture supernatant of the experimental groups were determined with an IL-10 Quantikine ELISA Kit (R&D Systems, United States). The dose of lysate (100 μg/mL) was selected as a representative dose based on the previous experiment of IL-6 and TNF-α quantification.
A single colony from each bacterial strain was inoculated in 3 mL MRS broth in a shaking incubator at 30°C for 6 h. Genomic DNA was extracted from the culture broths after 6 h using a Wizard Genomic DNA Purification Kit (Promega, Madison, WI, United States) according to the manufacturer’s instructions. Genomic DNA concentration was determined using a Qubit 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, United States) and the DNA quality and integrity were checked by electrophoresis on an 0.8% agarose gel. Whole genomes were sequenced on a Pacific Biosciences (Menlo Park, CA, United States) single molecule real-time (SMRT) sequencing platform with P6-C4 chemistry on a PacBio RS II instrument (Eid et al., 2009). The raw reads were generated with SMRT Cells and de novo assembled by the hierarchical genome-assembly process (HGAP) (Chin et al., 2013) protocol RS HGAP Assembly 2 in SMRT analysis version 2.3.0 (Pacific Biosciences)1. De novo assembly parameters were applied as follows: PreAssembler v2: Minimum Seed Read Length: 5000, Celera Assembler v1 Genome Size (Bp): 3,500,000, Target Coverage: 30, Overlapper Error Rate: 0.06, Overlapper Min Length: 40, Overlapper K-mer: 14.
Complete genomes were annotated by NCBI Prokaryotic Genomes Automatic Annotation Pipeline (PGAAP) (Tatusova et al., 2016). PathogenFinder was used to determinate the pathogenic potential (Cosentino et al., 2013). Further, Rapid Annotation using Subsystem Technology (RAST) version 2.0 (Aziz et al., 2008) was applied for genes of interest. The annotated genes were inspected for probiotic potential. The de novo assembled complete genomes were compared using eggnog (evolutionary genealogy of genes: Non-supervised Orthologous Groups)-mapper (Huerta-Cepas et al., 2017) and the associated default settings and Average Nucleotide Identity (ANI) values were obtained using the ANI Calculator tool on EzBiocloud2 (Yoon et al., 2017).
GraphPad Prism 5.0 was used to process data. To compare column to column, unpaired, a two-tailed student’s t-test was used. One-way analysis of variance (ANOVA) with Tukey’s post-test was used for multiple comparison, and two way ANOVA was used with Bonferroni post-test for group analysis.
Fermentation patterns of carbohydrate sources by each L. plantarum strain are noted in Supplementary Table S1. Notable differences in fermentation capability were observed: K6 weakly fermented L-arabinose resulting in a green to blue colorization transition of the API indicator medium instead of yellow, while K2 and K8 were completely negative; K6 fermented raffinose and melizetose while K8 only fermented melizetose, while K2 could not ferment either of the carbohydrates; and turanose was fermented by K6 and K8. Based on the pattern identification through the APIWEB database of BioMérieux, identities (%) of each strain were as follows: K2, 52.0% to L. plantarum group 1; K6, 99.4% to L. plantarum group 1; K8, 99.5% to L. plantarum group 1.
The most resistant strain to 70°C heat treatment for 80 s was K8, followed by K6 and then K2 (Figure 1). As shown in Figure 1, K2 and K6 were unstable and showed low reproducibility in replicate experiments as the error bars indicate. Except for the 80 s time point, all strains showed stable heat resistances up to 60 s. Notably, maximum viable CFU counts of L. plantarum strains when cultured overnight in MRS broth were as follows: K2, 9.15 × 108 CFU/mL; K6, 2.83 × 109 CFU/mL; and K8, 3.57 × 109 CFU/mL.
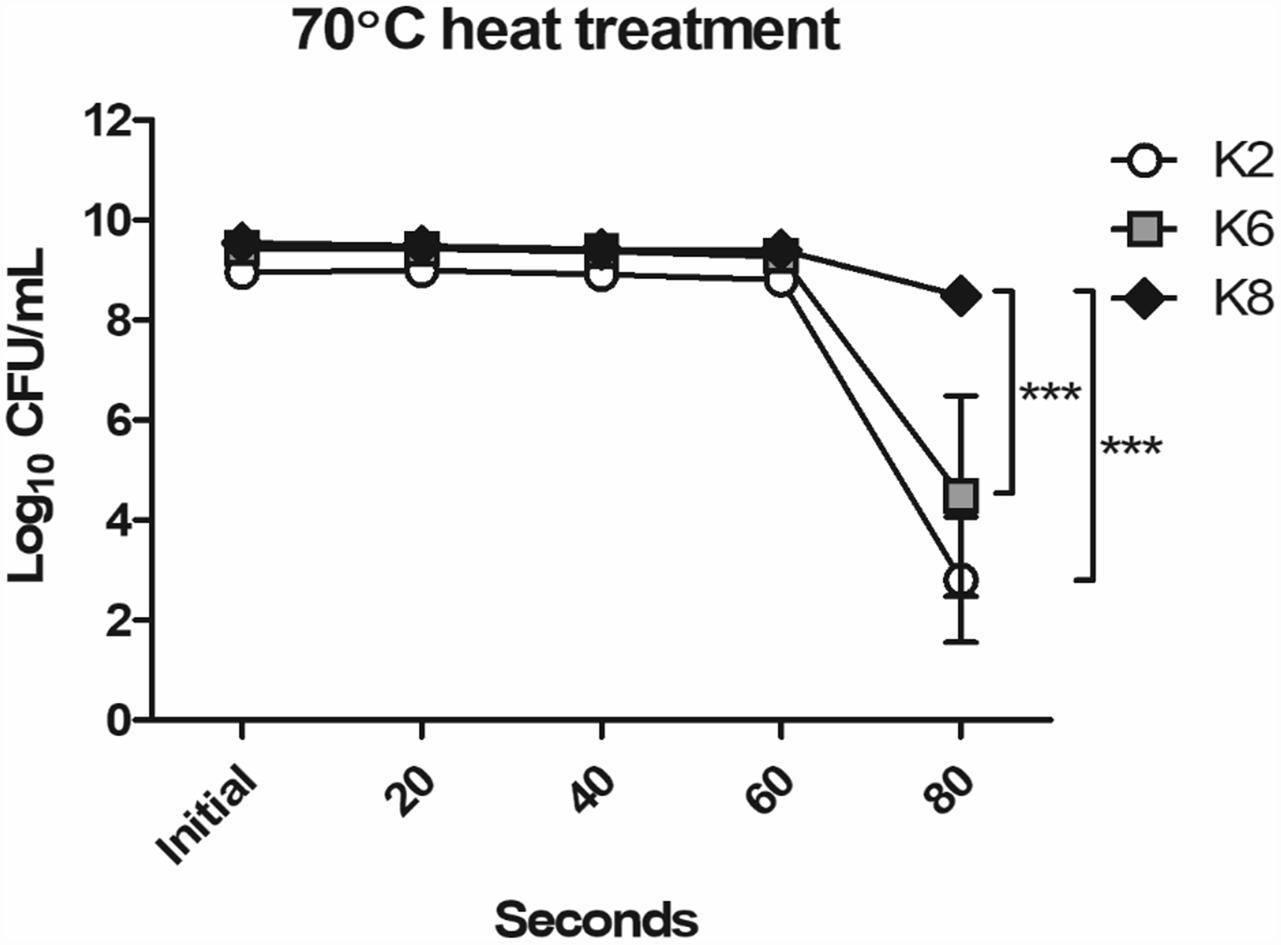
Figure 1. Heat resistance of Lactobacillus plantarum strains ATG-K2, ATG-K6, and ATG-K8. CFU counts of triplicate experiments are expressed as mean ± SEM. Statistical significance between compared groups are indicated as ∗∗∗p< 0.001.
The three L. plantarum strains showed antibacterial effects against all representative opportunistic pathogens in the disc diffusion assay (Table 1). K2 had the largest diameters of inhibition zones except for tests against EC and PA. Tests were conducted with live bacteria suspensions or CFCS, but only live bacteria suspensions showed inhibition activities.

Table 1. Antibacterial effects of Lactobacillus plantarum strains ATG-K2, ATG-K6, and ATG-K8 against Staphylococcus aureus (SA), Listeria monocytogenes (LM), Streptococcus mutans (SM), Streptococcus salivarius (SS), Escherichia coli (EC), Pseudomonas aeruginosa (PA), and Cronobacter sakazakii (CS) determined by disc diffusion assay.
All three strains of L. plantarum showed positive BSH activity which was identified by the formation of convex, shell-like, hardened colonies (Figure 2A). H2O2 production was detected in all three strains of L. plantarum based on the blue pigment of O− from TMB reactions in formed colonies and surroundings (Figure 2B). All lysates from the L. plantarum strains showed radical scavenging activity and K6 had the highest radical scavenging rate, which reached a plateau at 20 mg/mL concentration of the bacterial lysate, followed by K8 and then K2 (Figure 2C).
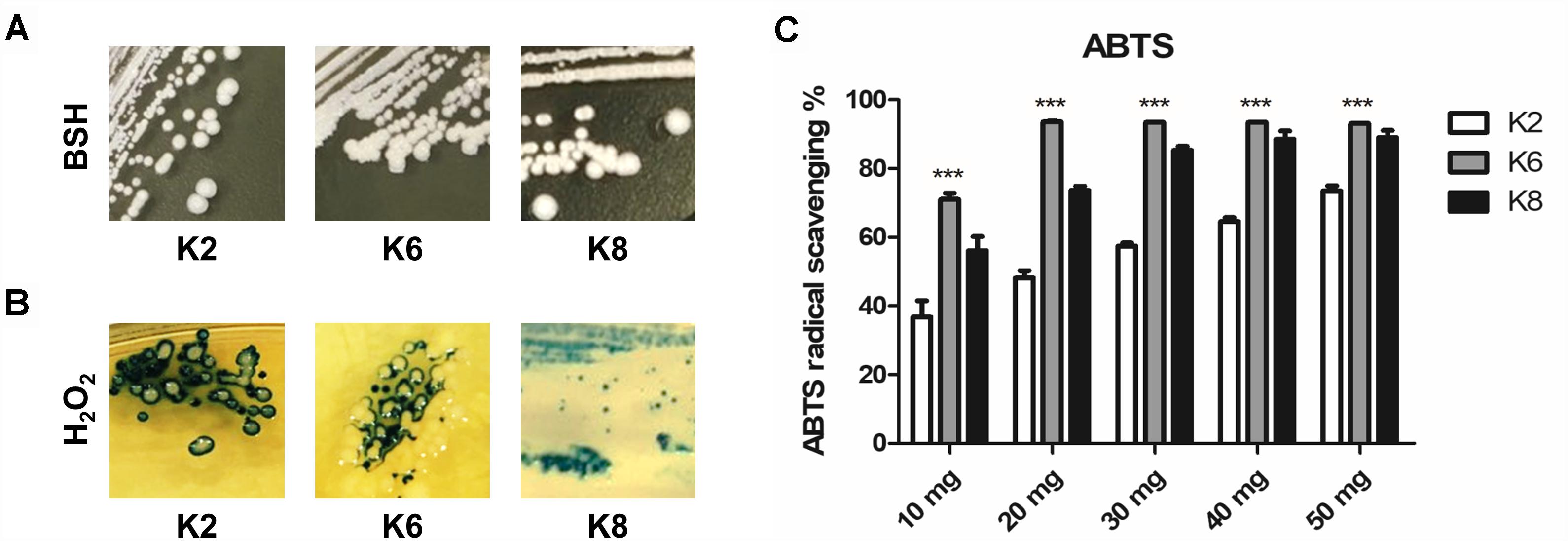
Figure 2. Functional properties of L. plantarum ATG-K2, ATG-K6, and ATG-K8 showing (A) bile salt hydrolase activity, (B) hydrogen peroxide production, and (C) ABTS radical scavenging activities of lysates of each strain. Statistical significance ∗∗∗ of (C) indicates p< 0.001 when K6 compared to K2 and K8.
There was no hemolysis activity detected for all three strains of L. plantarum as is indicated by a lack of clear zone formation (Figure 3A). Also, there was no color change of the test medium in the biogenic amine production test which implies that no histidine, tyramine, putrescine, or cadaverine was produced by any of the three strains of L. plantarum (Figure 3B). Putrescine derived from agmatine was not able to detect due to the composition of the test medium suggested by Bover-Cid and Holzapfel (1999).
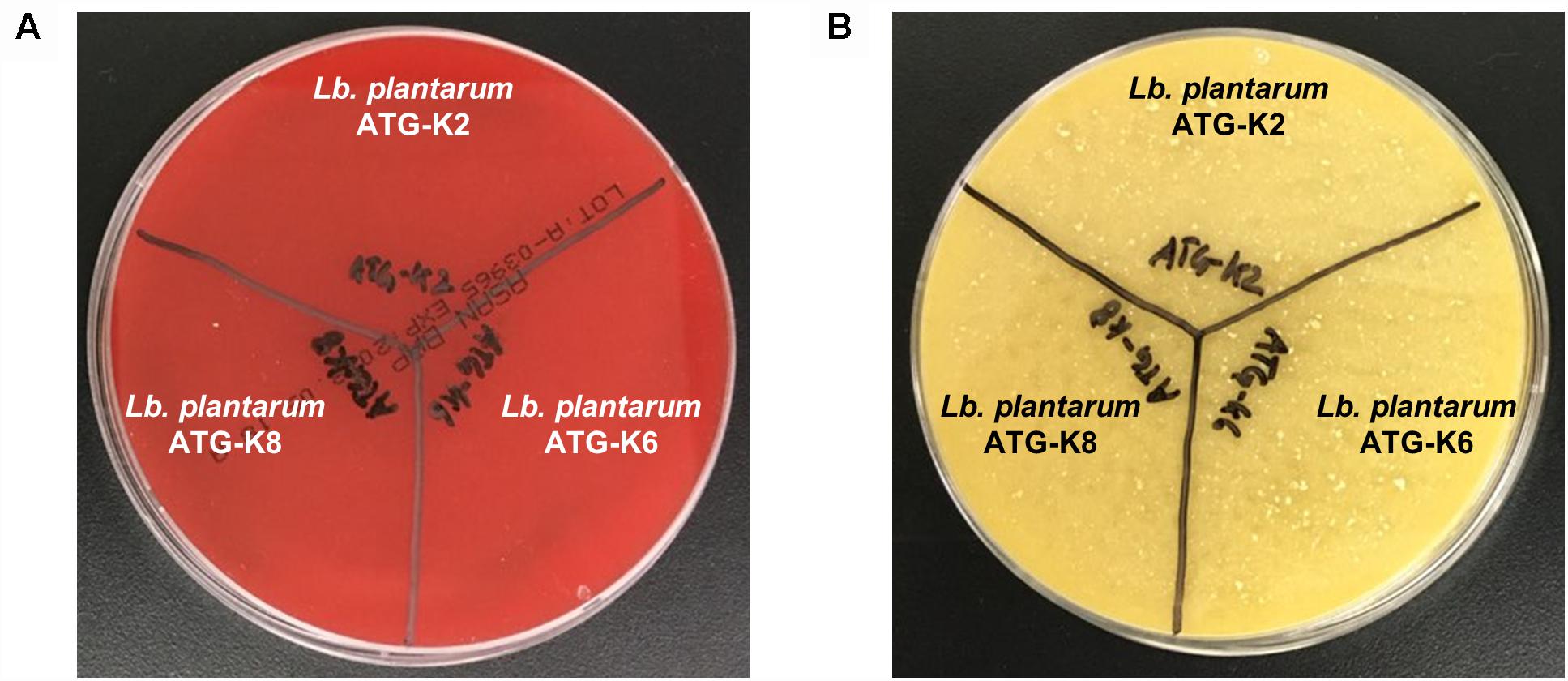
Figure 3. Safety assessment results of L. plantarum ATG-K2, ATG-K6, and ATG-K8 of (A) hemolysis activity (B) biogenic amine production.
All three strains met the requirements of MIC cut-off values suggested by the EFSA guideline on antibiotic susceptibility of lactic acid bacteria (Table 2). Although the EFSA guideline does not require vancomycin and streptomycin susceptibilities, a 94–194 μg/mL MIC for vancomycin and streptomycin was observed from antibiotic susceptibility tests of three strains (data not shown in Table 2).
CFCS of each L. plantarum strain showed growth inhibition toward CA and GV (Figures 4A,B). Against CA at the 24 h mark, K2, K6, and K8 showed 87.05 ± 0.38%, 82.11 ± 0.25%, and 81.34 ± 0.42%, respectively (Figure 4A). After 48 h of incubation, GV growth had inhibition rates by CFCS of K2, K6, and K8 of 37.49 ± 3.17%, 28.25 ± 5.33%, and 44.60 ± 1.51%, respectively (Figure 4B). The coaggregation rate of K2, K6, and K8 were 70.73 ± 3.41%, 58.57 ± 1.32%, and 57.88 ± 2.86%, respectively, when reacted with CA after 8 h (Figure 4C). In comparison to coaggregation with CA, coaggregation rates between GV and each L. plantarum strain were lower than that in the corresponding CA experiment. The coaggregation rates of K2, K6, and K8 were 32.72 ± 0.19%, 30.39 ± 0.13%, 20.45 ± 0.42%, respectively, when reacted with GV after 8 h (Figure 4D).
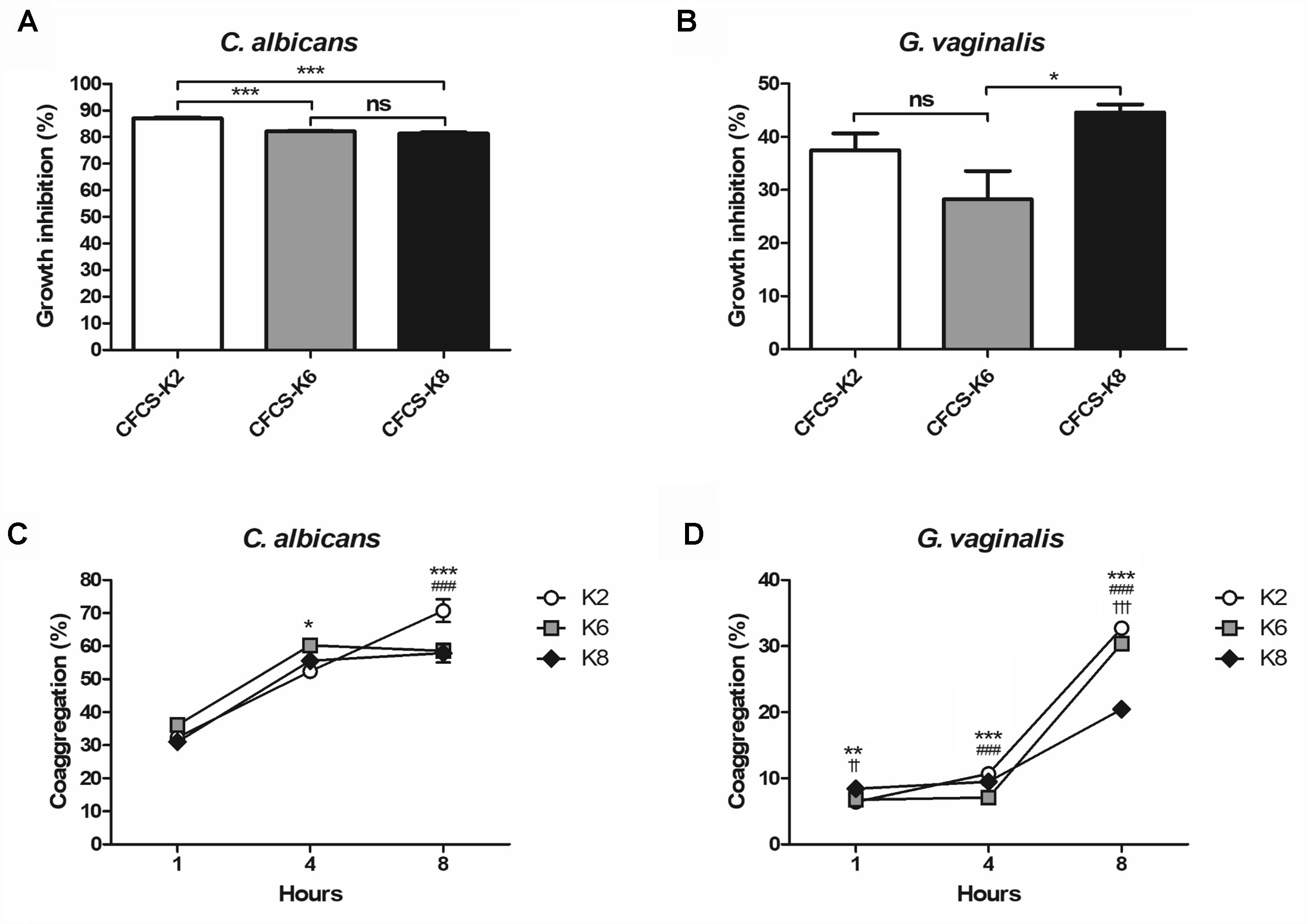
Figure 4. Growth inhibition rates against (A) C. albicans and (B) G. vaginalis by cell free culture supernatant (CFCS) of L. plantarum ATG-K2, ATG-K6, and ATG-K8. Coaggregation of each strain of L. plantarum ATG-K2, ATG-K6, and ATG-K8 reacted with (C) C. albicans and (D) G. vaginalis. Data are expressed as mean ± SEM. Statistical significance in the figures are shown as follows: K2 vs. K6, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; K2 vs. K8, ###p < 0.001; K6 vs. K8, ††p < 0.01, †††p < 0.001.
Complete disappearance of CA after 48 h of coculture was observed for all strains (Figure 5A). At the 24 h mark, the CA reduction rate by K2, K6, and K8 were 99.03 ± 0.06%, 99.49 ± 0.04%, and 99.53 ± 0.02%, respectively (Figure 5B). K6 and K8 showed statistically significant CA reduction rates compared to K2. GV was reduced by K2, K6, and K8, at rates of 87.37 ± 1.03%, 92.50 ± 0.54%, and 93.71 ± 1.10%, respectively, and no significant differences between each test group were found (Figure 5D). However, GV growth was detected at the 48 h mark in a roughly doubled CFU/mL after being reduced to 90% on average by each L. plantarum strain (Figure 5C).
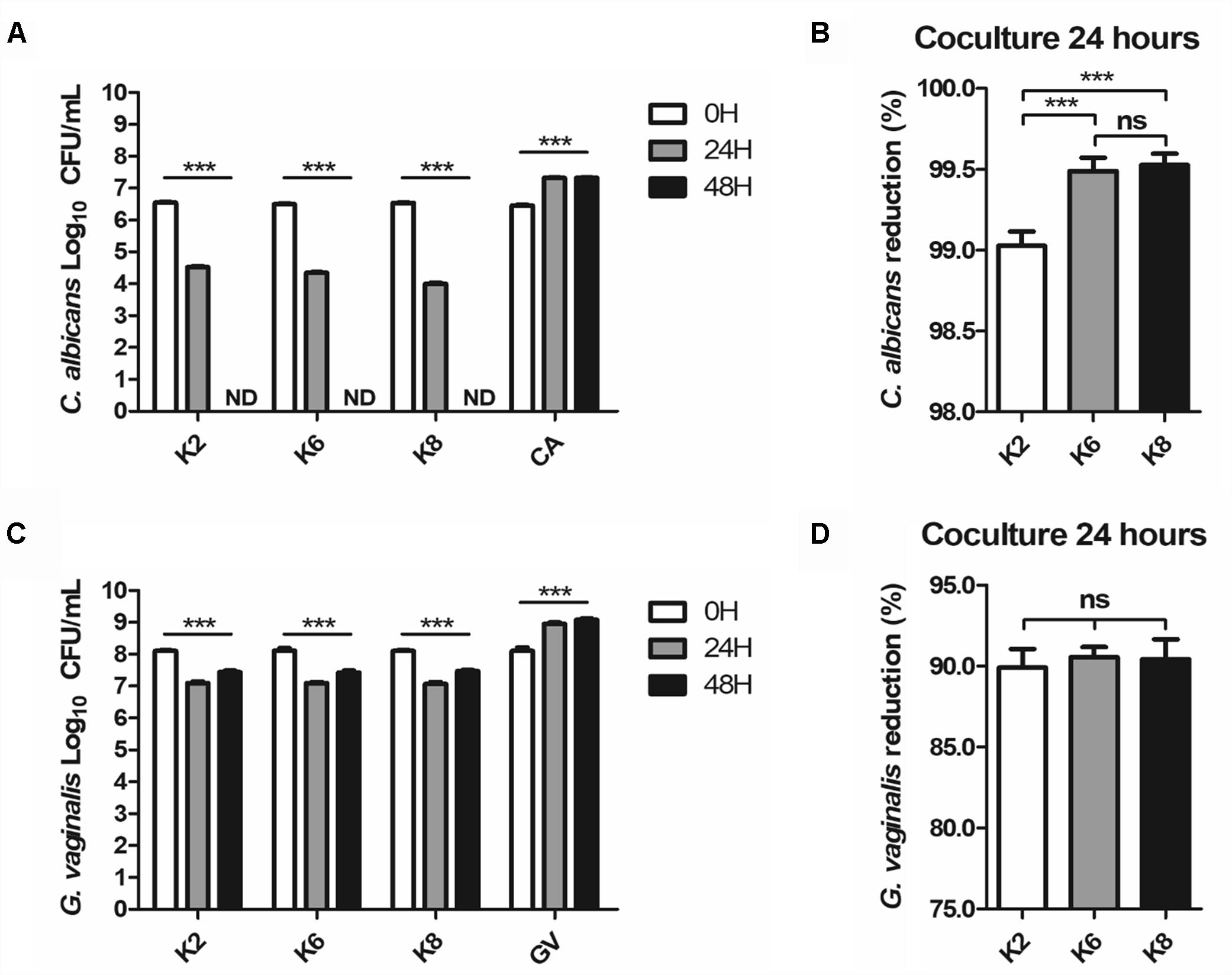
Figure 5. (A,B) C. albicans and (C,D) G. vaginalis killing effects of live L. plantarum ATG-K2, ATG-K6, and ATG-K8 determined by a coculture method. Log10 CFU/mL of each target pathogen measured in 24 h intervals are shown in (A,C). Reduction rates of each target pathogen at the 24 h mark are shown in (B,C). Data are expressed as mean ± SEM for four replicated experiments. Statistical significances are shown as ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
RAW264.7 cells produced significantly increased IL-6 and TNF-α upon treatment with each L. plantarum strain lysate compared to the control and was significantly lower than that of LPS-treated group (Figure 6). Each cytokine concentration was increased to both 100 and 500 μg/mL lysate concentrations. However, there was no significant difference between induced IL-6 concentrations by the same dose of lysates (Figure 6A). For 100 μg/mL lysate treatment, the K8 lysate-treated group (523.47 ± 23.78 pg/mL of TNF-α, p< 0.05) showed significantly higher concentrations of TNF-α compared to that of the K2 lysate (443.25 ± 10.50 pg/mL of TNF-α) and K6 lysate (477.81 ± 7.57 pg/mL of TNF-α)-treated groups (Figure 6B). In the 500 μg/mL lysate dose groups, there were no significant differences between any L. plantarum lysate treated groups for both cytokines.
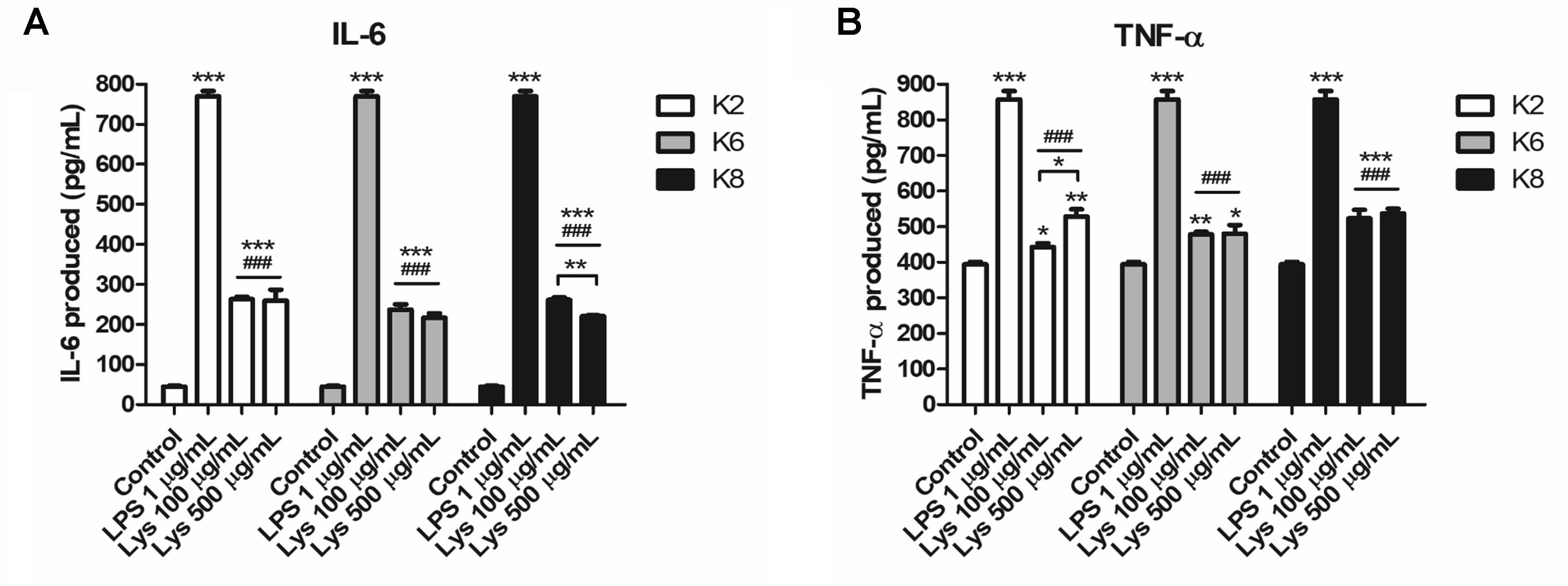
Figure 6. Induction of (A) IL-6 and (B) TNF-α by lysates of L. plantarum ATG-K2, ATG-K6, and ATG-K8 in RAW264.7 murine macrophage cells. The means of triplicated wells are shown from duplicated experiments ± SEM. Statistical significance in the figures are as follows: compared to control, ∗p< 0.05, ∗∗p< 0.01, and ∗∗∗p< 0.001; compared to the LPS group ###p< 0.001. Significant differences between doses in the same group are shown with directional lines as ∗p< 0.05 and ∗∗p< 0.01.
All three lysates of each L. plantarum strain were induced with a significantly higher concentration of IL-10 when treated with LPS compared to the control and LPS-only treated group (Figure 7). Significantly increased IL-10 production was observed in K2 and K6 lysates treated with 1 μg/mL LPS compared to K8 lysate treated with LPS. There was no statistically significant difference between K2 and K6 lysates treated with LPS.
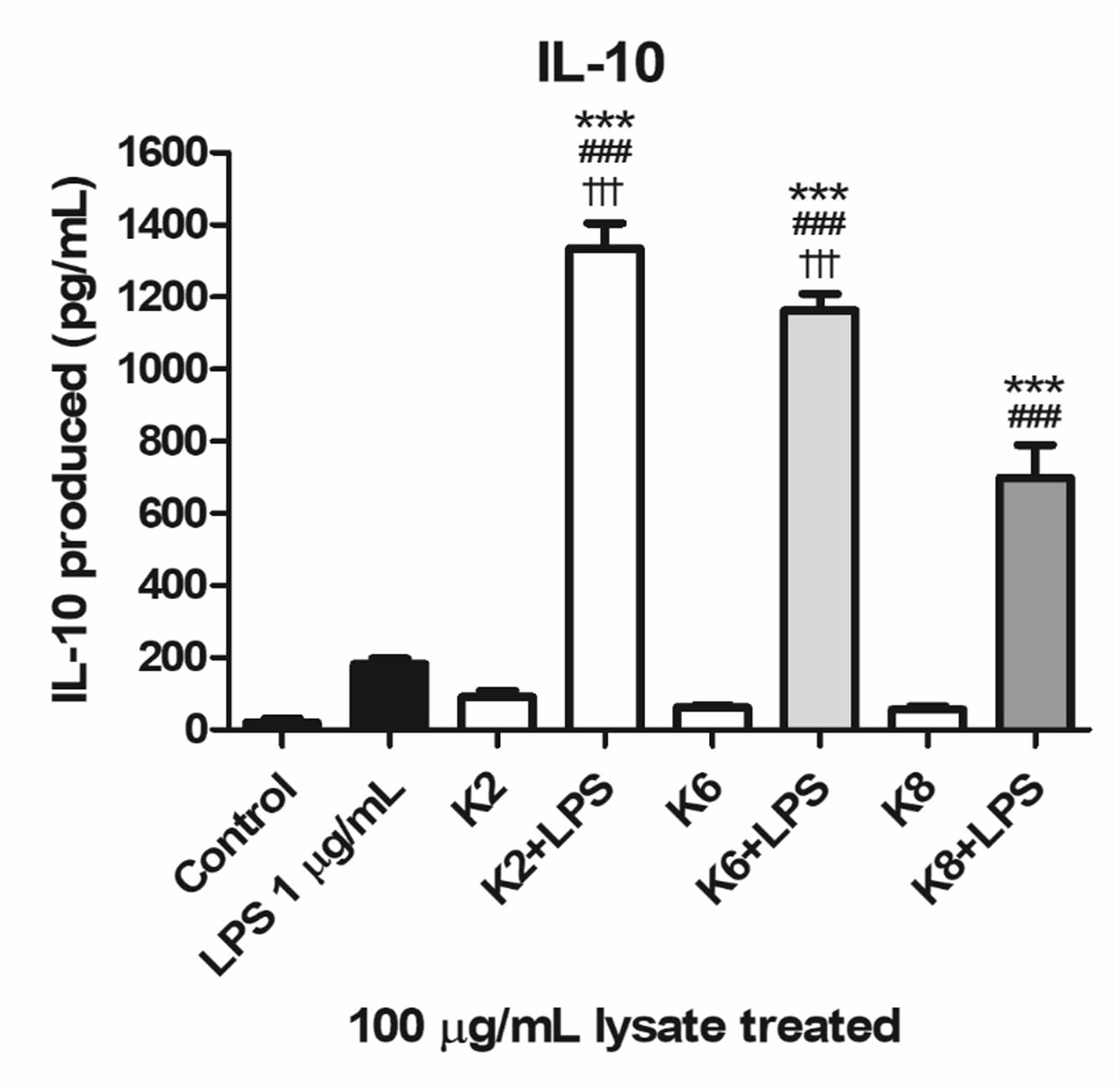
Figure 7. Induction of IL-10 lysates of L. plantarum ATG-K2, ATG-K6, and ATG-K8 in RAW264.7 murine macrophage cells upon LPS challenge. LPS (1 μg/mL) challenged groups are marked as +LPS in X axis labels. The means of triplicated wells are shown as a representative of duplicated experiments ± SEM. Statistical significance in the figures are as follows: compared to control, ∗p< 0.05, ∗∗p< 0.01, and ∗∗∗p< 0.001; to the LPS group, ##p< 0.01, ###p< 0.001; to the K8+LPS group †††p< 0.001.
The genomes of L. plantarum strains were sequenced with the PacBio SMRT platform with resulting 330× coverage for K2, 384× for K6, and 432× for K8. The genome of K2 was assembled into one circular chromosome and three plasmids, while the genomes of K6 and K8 were assembled into one circular chromosome and one plasmid (Figure 8). The total genome size of K8 (3,275,764 bp) is slightly larger than those of K2 (3,175,098 bp) and K6 (3,262,505 bp). The protein coding sequences were predicted using NCBI PGAAP and deposited in GenBank with accession numbers GCA_003597635.1 (K2), GCA_003597595.1 (K6), and GCA_003597615.1 (K8). A total of 3,114 open reading frames (ORFs) were predicted from the genome of the strain K2; 2,857 are protein coding genes and 85 are RNA genes. A total of 3,178 ORFs were predicted from the genome of the strain K6; 2,999 are protein coding genes and 87 are RNA genes. Finally, a total of 3,186 ORFs were predicted from the genome of the strain K6; 2,999 are protein coding genes and 87 are RNA genes (Table 3).
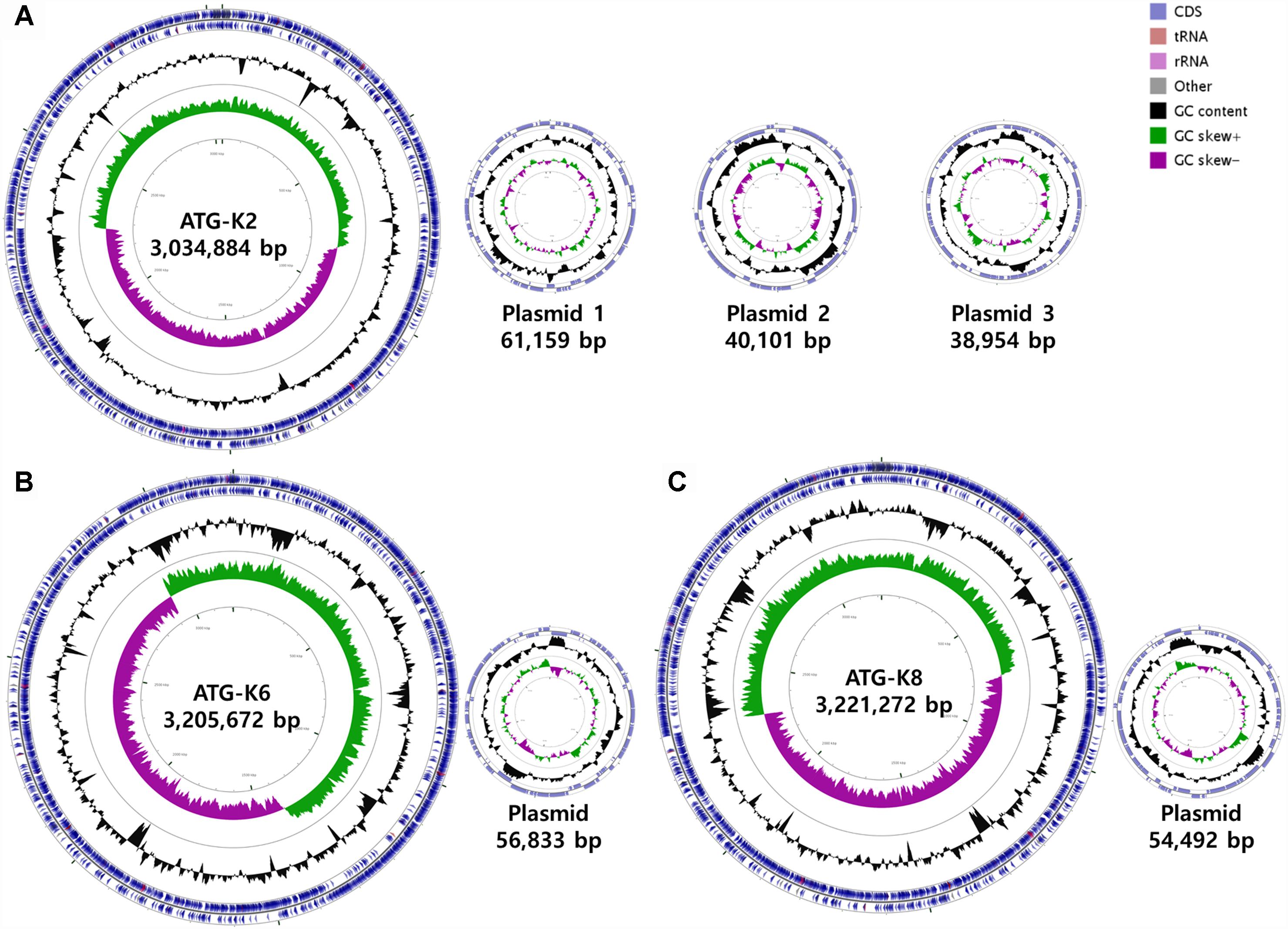
Figure 8. Circular maps of (A) Lactobacillus plantarum ATG-K2, (B) L. plantarum ATG-K6, and (C) L. plantarum ATG-K8 complete genomes.
Based on ANI analyses, K2 showed 96.30 and 96.12% sequence identity with K6 and K8, respectively. Strain K6 and K8 showed 99.92% sequence identity. All three strains were predicted to be non-human pathogens with PathogenFinder. Coding sequences were identified and grouped in COG classes (Figure 9). The coding sequences were categorized as by their involvement in carbohydrate transport and metabolism (7.9–8.8%); transcription (8.0–8.5%); amino acid transport and metabolism (6.9–7.2%); translation, ribosomal structure and biogenesis (4.9–5.2%); cell wall, membrane, envelope biogenesis (5.6–5.9%); and replication, recombination and repair of nucleic acids (5.0–6.2%). K2 (counted number: 223) showed a lower gene number for carbohydrate transport and metabolism compared with K6 (counted number: 261) and K8 (counted number: 263), while K2 (counted number: 175) showed a higher gene number for replication, recombination and repair compared with K6 (counted number: 149) and K8 (counted number: 154).
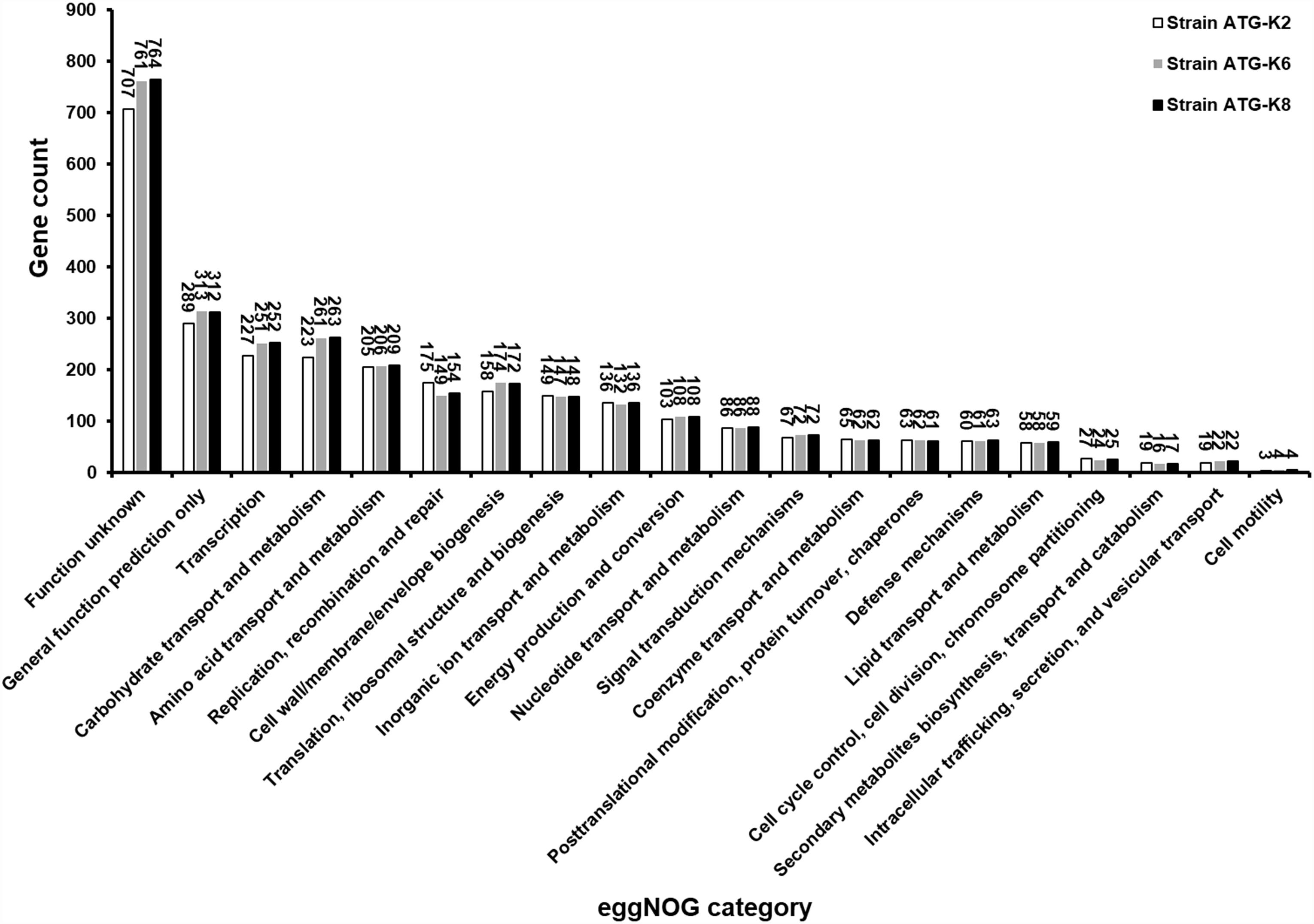
Figure 9. Numbers of proteins encoded by Lactobacillus plantarum ATG-K2, L. plantarum ATG-K6, and L. plantarum ATG-K8 annotated with the corresponding eggNOG functional category.
Three strains of L. plantarum in the present study, namely K2, K6, and K8, demonstrated BSH activity, H2O2 production, immunomodulatory effects, and antimicrobial functions especially against CA which revealed potential probiotic effects of these strains. Also, safety assessment results support that K2, K6, and K8 are safe for practical use with along with the safety of the L. plantarum species in general described by De Vries et al. (2006).
From a microbial perspective, BSH activity is important for secondary bile salt metabolism which may contribute to bile resistance of bacteria and benefit cholesterol metabolism of the host by lowering the host’s bile salt concentration, but excessive deconjugated bile salt may interfere digestive activity or intestinal health of the host, deconjugated bile salt production of BSH positive strains should be further investigated in quantitative manner (Begley et al., 2006). H2O2 produced by L. plantarum strains of the present study may be involved in killing opportunistic pathogens (De Vuyst and Vandamme, 1994). In addition to H2O2 production, radical scavenging activity from produced H2O2 might be an important factor for their own survival (Condon, 1987) as well as removing excessive radicals produced in local sites of the host through their oxidative enzymes (Lin and Yen, 1999). Among antimicrobial activities against several pathogens, anti-CA effects can benefit prevention of candidiasis in mucosal sites which is the first line of defense in the host. Particularly, coaggregation ability of L. plantarum strains with CA can be synergic with growth inhibition, H2O2, and killing effects depending on proximity. Although relatively weaker antimicrobial activities against GV were observed compared to anti-CA effects, it may still contribute to a prevention of BV taken together with antibacterial effects toward other opportunistic pathogens. Also, anti-CA effects also may benefit vaginal health since CA is also a pathogen that causes vaginitis (Sobel et al., 1998). In short, microbial properties of L. plantarum strains in the present study suggest the potential of the strains as probiotics for mucosal health.
From an immunology perspective, induction of IL-6 and TNF-α in macrophages by lysates of K2, K6, and K8 implicates that innate immunity is enhanced. IL-6 is a cytokine which is involved in activation of macrophages, recruitment and chemotaxis of neutrophils, activation of inflammatory responses and regulation of the immune response (Fielding et al., 2008; Scheller et al., 2011). TNF-α is another important cytokines that has roles in anti-tumor and proinflammatory responses, pyrexia, inhibition of viral replication, and activation of monocytes (Vujanovic, 2011). However, these proinflammatory cytokines must be tightly regulated to not result in pathological excessive inflammation. For regulation of such excessive inflammatory responses, IL-10 needs to be produced. IL-10 is an anti-inflammatory and regulatory cytokine that is produced by dendritic cells, macrophages, and regulatory T cells, and thus elevated concentration of IL-10 will suppress excessive activation of proinflammatory responses (Fiorentino et al., 1991; Saraiva and O’Garra, 2010). As shown in the co-treatment of LPS and lysate of L. plantarum strains in the present study, IL-10 production is enhanced when macrophages are activated by LPS. In addition, the presence of IL-6 and TNF-α is important for enhanced production of IL-10 which as demonstrated in in vitro human cell experiments (Daftarian et al., 1996). Interestingly, enhanced IL-10 production was lower in the K8 lysate-treated group compared to that of the K2 and K6 lysate treated groups, which it may be related to the formation of differential immune states or “immune tones” at a strain-specific level (Beck et al., 2016). However, this needs to be examined in a more complex animal model system to dissect strain-specific immunomodulatory effects in further studies. As macrophages have essential roles in mucosal immunity (Mowat and Bain, 2011; Ginhoux and Jung, 2014), the results of macrophage experiments supports that the enhanced host immunity and probiotic effects of L. plantarum strains in the present study might be synergistic.
From a genomic perspective, although all three strains were of the same species, K2 showed smaller genome size and different compositions compared to the other two strains, which implies the importance of genomic studies to distinguish and explore characteristics of microorganisms at a strain level. For example, lower numbers of carbohydrate transport and metabolism, and transcription-related genes in the K2 strain compared to other strains may influence maximum growth (CFU/mL), which was approximately 30% of that of K6 and K8 as noted in the results of heat resistance examination. An absence of potential virulence factors reconfirms the safety of L. plantarum strains. On the other hand, antimicrobial activities of three strains were positive but to different degrees which may relate to differential composition of the plantaricin biosynthesis gene cluster (Li et al., 2016). Varying degrees of immunological responses of RAW264.7 cells to the each strain may be due to differential activation of pattern recognition receptors due to differences in cell membrane components, or production of different metabolites (Hu et al., 2011; Xue et al., 2014). In short, genomic analyses of three strains strongly support the uniqueness of each strain. These factors should be considered in future genome-wide studies of these three strains.
In conclusion, L. plantarum strains ATG-K2, ATG-K6, and ATG-K8 in the present study demonstrated their potentials for mucosal health improvement in both microbiological and immunological contexts which may result in synergistic effects by influencing both microbial and host factors.
BB contributed to the experimental designs, experimented on the microbiological assays and the in vitro assays with cells, the writing and editing of the manuscript, and agreed to be listed as a corresponding author. G-SP contributed to the genome sequencing and analyses, and the writing of the manuscript. YL experimented on the in vitro assays with cells. SI oversaw the culture and management of L. plantarum strains. DJ experimented on the microbiological assays. JK contributed to the experimental designs, the editing of the manuscript editing, and agreed to be listed as a corresponding author.
There was no third-party funding, and AtoGen Co., Ltd. supported the present study with non-financial supports.
All authors were employed by AtoGen Co., Ltd.
We would like to thank Na-Rae Lee for providing Kimchi samples and lactic acid bacteria.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00433/full#supplementary-material
Aziz, R. K., Bartels, D., Best, A. A., Dejongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Beck, B. R., Song, J. H., Park, B. S., Kim, D., Kwak, J. H., Do, H. K., et al. (2016). Distinct immune tones are established by Lactococcus lactis BFE920 and Lactobacillus plantarum FGL0001 in the gut of olive flounder (Paralichthys olivaceus). Fish Shellfish Immunol. 55, 434–443. doi: 10.1016/j.fsi.2016.06.022
Begley, M., Hill, C., and Gahan, C. G. (2006). Bile salt hydrolase activity in probiotics. Appl. Environ. Microbiol. 72, 1729–1738. doi: 10.1128/AEM.72.3.1729-1738.2006
Bover-Cid, S., and Holzapfel, W. H. (1999). Improved screening procedure for biogenic amine production by lactic acid bacteria. Int. J. Food Microbiol. 53, 33–41. doi: 10.1016/S0168-1605(99)00152-X
Brandtzaeg, P. (2009). Mucosal immunity: induction, dissemination, and effector functions. Scand. J. Immunol. 70, 505–515. doi: 10.1111/j.1365-3083.2009.02319.x
Chin, C. S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569. doi: 10.1038/nmeth.2474
Condon, S. (1987). Responses of lactic acid bacteria to oxygen. FEMS Microbiol. Lett. 46, 269–280. doi: 10.1111/j.1574-6968.1987.tb02465.x
Cosentino, S., Larsen, M. V., Aarestrup, F. M., and Lund, O. (2013). PathogenFinder-distinguishing friend from foe using bacterial whole genome sequence data. PLoS One 8:e77302. doi: 10.1371/journal.pone.0077302
Daftarian, P. M., Kumar, A., Kryworuchko, M., and Diaz-Mitoma, F. (1996). IL-10 production is enhanced in human T cells by IL-12 and IL-6 and in monocytes by tumor necrosis factor-alpha. J. Immunol. 157, 12–20.
Dashkevicz, M. P., and Feighner, S. D. (1989). Development of a differential medium for bile salt hydrolase-active Lactobacillus spp. Appl Environ Microbiol 55, 11–16.
De Vries, M. C., Vaughan, E. E., Kleerebezem, M., and De Vos, W. M. (2006). Lactobacillus plantarum—survival, functional and potential probiotic properties in the human intestinal tract. Int. Dairy J. 16, 1018–1028. doi: 10.1016/j.idairyj.2005.09.003
De Vuyst, L., and Vandamme, E. J. (1994). “Antimicrobial potential of lactic acid bacteria,” in Bacteriocins of Lactic Acid Bacteria: Microbiology, Genetics and Applications, eds L. De Vuyst and Vandamme (United Kingdom: Chapman & Hall), 91–142. doi: 10.1007/978-1-4615-2668-1_3
EFSA (2012). Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J. 10:2740. doi: 10.2903/j.efsa.2012.2740
Egan, M., and Lipsky, M. S. (2002). Vaginitis: case reports and brief review. AIDS Patient Care STDS 16, 367–373. doi: 10.1089/10872910260196396
Eid, J., Fehr, A., Gray, J., Luong, K., Lyle, J., Otto, G., et al. (2009). Real-time DNA sequencing from single polymerase molecules. Science 323, 133–138. doi: 10.1126/science.1162986
Ellepola, A., and Samaranayake, L. (2000). Oral candidal infections and antimycotics. Criti. Rev. Oral Biol. Med. 11, 172–198. doi: 10.1177/10454411000110020301
Fielding, C. A., Mcloughlin, R. M., Mcleod, L., Colmont, C. S., Najdovska, M., Grail, D., et al. (2008). IL-6 regulates neutrophil trafficking during acute inflammation via STAT3. J. Immunol. 181, 2189–2195. doi: 10.4049/jimmunol.181.3.2189
Fiorentino, D. F., Zlotnik, A., Mosmann, T. R., Howard, M., and O’garra, A. (1991). IL-10 inhibits cytokine production by activated macrophages. J. Immunol. 147, 3815–3822.
Gill, N., Wlodarska, M., and Finlay, B. B. (2010). The future of mucosal immunology: studying an integrated system-wide organ. Nat. Immunol. 11:558. doi: 10.1038/ni0710-558
Ginhoux, F., and Jung, S. (2014). Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat. Rev. Immunol. 14, 392–404. doi: 10.1038/nri3671
Handley, P. S., Harty, D. W., Wyatt, J. E., Brown, C. R., Doran, J. P., and Gibbs, A. C. (1987). A comparison of the adhesion, coaggregation and cell-surface hydrophobicity properties of fibrillar and fimbriate strains of Streptococcus salivarius. J. Gen. Microbiol. 133, 3207–3217. doi: 10.1099/00221287-133-11-3207
Honda, K., and Littman, D. R. (2012). The microbiome in infectious disease and inflammation. Ann. Rev. Immunol. 30, 759–795. doi: 10.1146/annurev-immunol-020711-074937
Hu, S., Dong, T. S., Dalal, S. R., Wu, F., Bissonnette, M., Kwon, J. H., et al. (2011). The microbe-derived short chain fatty acid butyrate targets miRNA-dependent p21 gene expression in human colon cancer. PLoS One 6:e16221. doi: 10.1371/journal.pone.0016221
Huerta-Cepas, J., Forslund, K., Coelho, L. P., Szklarczyk, D., Jensen, L. J., Von Mering, C., et al. (2017). Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 34, 2115–2122. doi: 10.1093/molbev/msx148
Khosravi, A., and Mazmanian, S. K. (2013). Disruption of the gut microbiome as a risk factor for microbial infections. Curr. Opin. Microbiol. 16, 221–227. doi: 10.1016/j.mib.2013.03.009
Kim, J., and Sudbery, P. (2011). Candida albicans, a major human fungal pathogen. J. Microbiol. 49, 171–177. doi: 10.1007/s12275-011-1064-7
Li, P., Gu, Q., and Zhou, Q. (2016). Complete genome sequence of Lactobacillus plantarum LZ206, a potential probiotic strain with antimicrobial activity against food-borne pathogenic microorganisms. J. Biotechnol. 238, 52–55. doi: 10.1016/j.jbiotec.2016.09.012
Lin, M. Y., and Yen, C. L. (1999). Antioxidative ability of lactic acid bacteria. J. Agric. Food Chem. 47, 1460–1466. doi: 10.1021/jf981149l
McDermott, A. J., and Huffnagle, G. B. (2014). The microbiome and regulation of mucosal immunity. Immunology 142, 24–31. doi: 10.1111/imm.12231
McGroarty, J. A., Tomeczek, L., Pond, D. G., Reid, G., and Bruce, A. W. (1992). Hydrogen peroxide production by Lactobacillus species: correlation with susceptibility to the spermicidal compound nonoxynol-9. J. Infect. Dis. 165, 1142–1144. doi: 10.1093/infdis/165.6.1142
Morris, M., Nicoll, A., Simms, I., Wilson, J., and Catchpole, M. (2001). Bacterial vaginosis: a public health review. BJOG 108, 439–450. doi: 10.1111/j.1471-0528.2001.00124.x
Mowat, A. M., and Bain, C. C. (2011). Mucosal macrophages in intestinal homeostasis and inflammation. J. Innate Immun. 3, 550–564. doi: 10.1159/000329099
Moyes, D. L., and Naglik, J. R. (2011). Mucosal immunity and candida albicans infection. Clin. Dev. Immunol. 2011:346307. doi: 10.1155/2011/346307
Poulain, D., Sendid, B., Standaert-Vitse, A., Fradin, C., Jouault, T., Jawhara, S., et al. (2009). Yeasts: neglected pathogens. Dig. Dis. 27(Suppl. 1), 104–110. doi: 10.1159/000268129
Reid, G., Younes, J. A., Van Der Mei, H. C., Gloor, G. B., Knight, R., and Busscher, H. J. (2011). Microbiota restoration: natural and supplemented recovery of human microbial communities. Nat. Rev. Microbiol. 9, 27–38. doi: 10.1038/nrmicro2473
Saraiva, M., and O’Garra, A. (2010). The regulation of IL-10 production by immune cells. Nat. Rev. Immunol. 10, 170–181. doi: 10.1038/nri2711
Scheller, J., Chalaris, A., Schmidt-Arras, D., and Rose-John, S. (2011). The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim. Biophys. Acta 1813, 878–888. doi: 10.1016/j.bbamcr.2011.01.034
Sobel, J. D., Faro, S., Force, R. W., Foxman, B., Ledger, W. J., Nyirjesy, P. R., et al. (1998). Vulvovaginal candidiasis: epidemiologic, diagnostic, and therapeutic considerations. Am. J. Obstet. Gynecol. 178, 203–211. doi: 10.1016/S0002-9378(98)80001-X
Sommer, F., and Bäckhed, F. (2013). The gut microbiota—masters of host development and physiology. Nat. Rev. Microbiol. 11, 227–238. doi: 10.1038/nrmicro2974
Tatusova, T., Dicuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Tlaskalová-Hogenová, H., Štěpánková, R., Hudcovic, T., Tučková, L., Cukrowska, B., Lodinová-Žádnıková, R., et al. (2004). Commensal bacteria (normal microflora), mucosal immunity and chronic inflammatory and autoimmune diseases. Immunol. Lett. 93, 97–108. doi: 10.1016/j.imlet.2004.02.005
Vujanovic, N. L. (2011). Role of TNF superfamily ligands in innate immunity. Immunol. Res. 50, 159–174. doi: 10.1007/s12026-011-8228-8
Xue, X., Cao, A. T., Cao, X., Yao, S., Carlsen, E. D., Soong, L., et al. (2014). Downregulation of micro RNA-107 in intestinal CD 11c+ myeloid cells in response to microbiota and proinflammatory cytokines increases IL-23p19 expression. Eur. J. Immunol. 44, 673–682. doi: 10.1002/eji.201343717
Keywords: Lactobacillus plantarum, probiotics, Candida albicans, candidiasis, Gardnerella vaginalis, bacterial vaginosis, macrophage, whole genome sequencing
Citation: Beck BR, Park G-S, Lee YH, Im S, Jeong DY and Kang J (2019) Whole Genome Analysis of Lactobacillus plantarum Strains Isolated From Kimchi and Determination of Probiotic Properties to Treat Mucosal Infections by Candida albicans and Gardnerella vaginalis. Front. Microbiol. 10:433. doi: 10.3389/fmicb.2019.00433
Received: 20 November 2018; Accepted: 19 February 2019;
Published: 06 March 2019.
Edited by:
Paloma López, Centro de Investigaciones Biológicas (CSIC), SpainReviewed by:
Giuseppe Spano, University of Foggia, ItalyCopyright © 2019 Beck, Park, Lee, Im, Jeong and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Ram Beck, YnJicjc3N0BoYW5tYWlsLm5ldA== Jihee Kang, amhrYW5nQGF0b2dlbi5jby5rcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.