- Tianjin Key Laboratory of Organic Solar Cells and Photochemical Conversion, College of Chemistry and Chemical Engineering, Tianjin University of Technology, Tianjin, China
Phenanthrene (PHE) is a common pollutant of acidic and non-acidic environments that is recalcitrant to biodegradation. Herein, Burkholderia fungorum FM-2 (GenBank accession no. KM263605) was isolated from oil-contaminated soil in Xinjiang and characterized morphologically, physiologically, and phylogenetically. Environmental parameters including PHE concentration, pH, temperature, and salinity were optimized, and heavy metal tolerance was investigated. The MIC of strain FM-2 tolerant to Pb(II) and Cd(II) was 50 and 400 mg L−1, respectively, while the MIC of Zn(II) was >1,200 mg L−1. Atypically for a B. fungorum strain, FM-2 utilized PHE (300 mg L−1) as a sole carbon source over a wide pH range (between pH 3 and 9). PHE and heavy metal metabolism were assessed using gas chromatography (GC), inductively coupled plasma optical emission spectroscopy (ICP-OES), scanning electron microscopy-energy dispersive X-ray spectroscopy (SEM-EDS), Fourier-transform infrared (FTIR) spectroscopy and ultraviolet (UV) absorption spectrometry. The effects of heavy metals on the bioremediation of PHE in soil were investigated, and the findings suggest that FM-2 has potential for combined bioremediation of soils co-contaminated with PHE and heavy metals.
Introduction
Polycyclic aromatic hydrocarbons (PAHs) fused-ring aromatic compounds consisting of two or more fused benzene and/or pentacyclic rings (Ghosal et al., 2016; Baltrons et al., 2018). PAHs are frequently found alongside heavy metals in contaminated environments including manufacturing plants and refinery sites, and the presence of heavy metals may interfere with PAH biodegradation (Murthy et al., 2014; Quero et al., 2015; Su et al., 2018). Biodegradation is considered an efficient, economic, and facile physicochemical process for the treatment of environments contaminated with PAHs (Mnif et al., 2017; Gong et al., 2018). Novel bacterial strains isolated from soils contaminated with mixtures of pollutants may possess sophisticated catabolic capabilities and the ability to tolerate, extreme conditions, toxic metals, and limited nutrients (Oves et al., 2012; Rashid et al., 2016). Such organisms that can also degrade PAHs may offer an effective and affordable remediation strategy (Kuppusamy et al., 2016).
Members of the Burkholderia genus play significant ecological roles and hold potential for biotechnological applications. Species from the plant-beneficial-environmental (PBE) Burkholderia cluster are able to exploit diverse aromatic compounds as sources of energy and carbon, and some have considerable biotechnological potential due to their ability to degrade chemical pollutants (Suarez-Moreno et al., 2012). Strains of B. xenovorans, B. vietnamiensis, B. fungorum, B. kururiensis, B. unamae, B. sartisoli, and B. phenoliruptrix are employed for bioremediation of polluted environments because they are able to tolerate and metabolize compounds that are recalcitrant to degradation (Caballero-Mellado et al., 2004, 2007; Coenye et al., 2004; O'Sullivan and Mahenthiralingam, 2005; Vanlaere et al., 2008; Felice et al., 2016). However, there are few reports of strains belonging to Burkholderia that can tolerate heavy metals and PAHs (Kuppusamy et al., 2016).
In the present work, the PAH-degrading FM-2 strain was isolated from oil-contaminated soils in an oil field in Xinjiang, and classified as Burkholderia fungorum based on phenotypic and phylogenetic analyses. PHE was selected as a model PAH because it is a widespread pollutant with typical PAH characteristics including a K region and a bent structure. Unusually for a B. fungorum strain, FM-2 can degrade PHE over a wide pH range, including under highly acidic conditions. Herein, we investigated the effectiveness of the PAH-degrading FM-2 for remediating soils co-contaminated with heavy metals and PHE.
Materials and Methods
Sampling, Chemicals, and Culture Media
Samples were collected from oil-contaminated soil in Xinjiang oilfield (Xinjiang, China). Bacterial strains capable of degrading PHE were isolated using the selective enrichment method. PHE (purity ≥97%) and other reagents were purchased from Energy Chemical Technology Co. Ltd.
Minimal medium (pH 7 ± 0.2) consisting of 0.1 g MgSO4, 2.04 g KH2PO4, 12.5 g Na2HPO4·12H2O, and 0.4 g (NH4)2SO4 (per L of distilled water) was used for isolating microorganisms capable of utilizing PHE as a sole source of carbon and energy source.
Enrichment of the strain before screening was performed using mineral salt medium (pH 7 ± 0.2) containing 0.7 g MgSO4, 3.48 g KH2PO4, 1.5 g Na2HPO4·12H2O, 3.96 g (NH4)2SO4, and 0.01 g yeast per L of distilled water.
LMM medium (pH 6.5) contained 0.1 g KH2PO4, 0.1 g Na2HPO4, 0.5 g NH4NO3, 0.5 g (NH4)2SO4, 0.2 g MgSO4, 20 mg CaCl2, 2 mg FeCl2, and 2 mg MnSO4 per L of distilled water (Ramadass et al., 2016).
Enrichment Isolation and Morphological and Biochemical Characterization
Screening of conditions for PHE-degrading strains was performed as described previously (Mnif et al., 2017), and the isolated FM-2 strain was selected for phylogenetic analysis. Typical biochemical and physiological characteristics were systematically evaluated as described in the manual of common bacterial identification (Zhu and Ying, 2001).
PHE Degradation Test
FM-2 was selected using dilution plates based on obvious colony morphology, and transferred to fresh lysogeny broth (LB) agar plates several times to guarantee culture purity. A single colony was picked from the final LB agar plate and inoculated in 30 mL mineral salt medium with 2% glycerol. After culturing for 48 h at 200 rpm and 25°C, a 600 μL aliquot of the fermentation broth was transferred to 30 mL minimal medium containing PHE (300 mg L−1). PHE was dissolved in n-hexane, and then added into 100 ml flask at final concentration of 300–600 mg L−1. The flasks were shaken for 2 h at 200 rpm and 25°C. A thin film of PHE was left at the bottom of the flask after removing n-hexane, and then MM was added. The enrichment culture was sub-cultured aerobically (shaking at 200 rpm) at 25°C to facilitate degradation of PHE. Bacterial cell growth was monitored by measuring the absorbance (Abs) at 600 nm, and degradation of PHE was confirmed by both color change and gas chromatography (GC) analyses. The remaining PHE was extracted from the whole culture (30 mL) in a erlenmeyer flask twice with 30 ml of n-hexane (>98%, benchmark, China). The organic phases were combined and dried with anhydrous Na2SO4. The solvents were removed under reduced pressure and the residues were dissolved in n-hexane. Gas chromatograph equipment (BRUKER 456-GC, USA) was applied to detect the variation of PHE in the n-hexane. Ten microliter of the organic phase was analyzed by using a HP-5 type capillary column (30-m length × 0.32-mm ID, 0.25-μm film thickness). During the analytical process, the column temperature was kept at 80°C for 1 min, then programmed to 290°C at a rate of 10°C min−1 which was maintained for 10 min. Media not inoculated with cells were used as sterile control. Control tests with different PHE concentrations were performed to evaluate the actual PHE recovery efficiency after extraction, which was over 87.2% at 300–600 mg L−1 PHE. The above mentioned methods are based on the approach reported by Tao et al. (2007), similar with the determinating technique of PHE degradation rate (Muangchinda et al., 2013; Oyetibo et al., 2017; Reddy et al., 2017).
C1 - PHE recovery rate in controled experiment after t d, %; D2 - PHE recovery rate in degradation experiment after t d, %.
16S rRNA Gene Sequence Determination and Phylogenetic Analysis
Analysis of the 16S rRNA gene was performed as described in our previous work (Huang et al., 2013), and the nucleotide sequence of the gene in FM-2 has been deposited in GenBank under accession number KM263605.
Determination of PHE Degradation Rate and Optimisation of Biodegradation
The effect of PHE concentration on the growth of FM-2 was assessed in 100 mL Erlenmeyer flasks containing 30 mL minimal medium (2% FM-2, v/v) supplemented with 300, 400, 500, and 600 mg L−1 PHE as a sole carbon source. Flasks were incubated for 3 days at 25°C on rotary shaker (200 rpm), and the initial pH was adjusted to 7.0. A control culture inoculated with boiled FM-2 was also included, and PHE degradation was evaluated by measuring the change in absorbance at 600 nm, and also by GC analysis (Tao et al., 2007).
The pH, salinity, temperature, and cultivation duration were optimized using an appropriate amount of biomass in a 100 mL conical flask containing 30 mL minimal medium and 300 mg L−1 PHE. The pH was tested at intervals of 1.0 between 2.0 and 11.0. NaCl was tested between 0 and 20 g L−1 in minimal medium at pH 7.0, the temperature was assessed between 15 and 40°C, cultivation was performed for 1, 2, and 3 days, and the inoculation volume and sampling operations were as described above.
Effect of Heavy Metal Ions
Heavy metal resistance of the PHE-degrading FM-2 strain was assessed using 20 mg L−1 ZnSO4·7H2O, Pb(NO3)2, and CdCl2·5/2H2O based on minimum inhibitory concentration (MIC) tests, and working test solutions were sterilized filtration trough a 0.45 μm membrane. LMM medium was separately supplemented with each of the heavy metals and 300 mg L−1 PHE, inoculated with 2% (v/v) FM-2, and cultured at 25°C with shaking at 200 rpm for 7 days. Cultures containing LMM, FM-2, and each heavy metal but without PHE were included as control I, and LMM supplemented with FM-2 and 300 mg L−1 PHE but without heavy metals were included as control II. Finally, a sterile control culture without FM-2 was included to assess abiotic loss of PHE.
Analytical Tests
Analysis of the Bioaccumulation of Heavy Metal in FM-2 Cells by Inductively Coupled Plasma Optical Emission Spectroscopy (ICP-OES) and Adsorption Isotherms
Bioaccumulation of heavy metals was evaluated by inoculating B. fungorum FM-2 into LMM medium containing 200 mg L−1 Zn (II), 50 mg L−1 Pb (II), or 200 mg L−1 Cd(II) as well as 300 mg L−1 PHE. Cultures were incubated at 25°C with shaking at 200 rpm for 1, 3, 5, and 7 days. As described previously (Chiboub et al., 2016) with some modifications, cultures were centrifuged (8,000 rpm for 5 min at 4°C) and pellets were washed three times with sterile distilled water to remove free heavy metal ions. Pellets were then treated with 10 mM sterile EDTA at 25°C with agitation at 200 rpm for 30 min. Following centrifugation (8,000 rpm for 10 min at 4°C), the pellet and supernatant (S1) were separated in different 50 mL centrifuge tubes, pellets were washed three times with sterile distilled water, disrupted using an ultrasonic cell disrupter, and the resulting supernatant (S2) was collected by centrifugation (10,000 rpm for 20 min at 4°C). S1 and S2 were analyzed using a Vista MPX ICP-OES instrument (Varian, Palo Alto, CA) to determine the heavy metal content of cell walls (S1) and the intracellular space (S2) (Wei et al., 2009).
Adsorption isotherms was determined by the following experiment: strain FM-2 was inoculated into LMM medium containing 200 mg L−1 Zn (II), 50 mg L−1 Pb (II), or 200 mg L−1 Cd(II) as well as 300 mg L−1 PHE. Cultures were incubated at 25°C with shaking at 200 rpm for 0, 1/2, 1, 2, 3, 4, 5, 6, and 7 days. Specific experiment procedures were carried out according to the literature reported by Vishan et al. (2017b).
Scanning Electron Microscopy-Energy Dispersive X-ray Spectroscopy (SEM/EDS)
PHE-degrading B. fungorum FM-2 was cultured in LMM containing 200 mg L−1 Zn(II), 50 mg L−1 Pb (II), or 200 mg L−1 Cd(II) at 25°C with shaking at 200 rpm for 7 days as described previously (Kang et al., 2014), and a SUPRA 35 VP field emission SEM instrument combined with an X-Max EDS instrument (Oxford Instruments) were used to analyse the surface morphology cells following biosorption of Zn (II), Pb (II), and Cd (II).
Fourier-Transform Infrared Spectroscopy (FTIR)
For FTIR analyses, strain FM-2 was cultured as described in Section Analysis of the Bioaccumulation of Heavy Metal in FM-2 Cells by Inductively Coupled Plasma Optical Emission Spectroscopy (ICP-OES) and Adsorption Isotherms, cell pellets were obtained by centrifugation (8,000 rpm, 10 min, 4°C), washed three times with deionised water, and lyophilized. Bacterial cells grown in LMM without metal ions served as a control. FTIR spectra of KBr pellets of dried cells associated with or without Zn (II), Pb (II), and Cd (II) were obtained using a Vector 33 FTIR instrument (Bruker, Germany).
Bioremediation of Soil Co-contaminated With Heavy Metal Ions and PHE
Soil and Preparation
Topsoil in which PAHs were not detectable was collected from an agricultural field in Xiqing District, Tianjin, China. After transfer to the laboratory, soil was air-dried, and sieved through a 2 mm mesh. Incorperate 10 mg kg−1 zinc sulfate or 6 mg kg−1 cadmium chloride into soils, respectively. All cadmium and zinc amendments were achieved by dissolving 200 mg ZnSO4·7H2O or CdCl2·5/2H2O in 10 mL of sterile deionised water prior to adding to the appropriate soil treatments. Then PHE was mixed with soil at 150 mg kg−1 soil dry weight, and experimental procedures, which lasted 15 days, were performed as previously described (Wong et al., 2005). Four hundred milligram PHE was dissolved in the 10 mL mixture of hexane: acetone (1: 1, v/v) for treating soils. Control soil treatments without heavy metals received 10 mL of sterile deionised water. Abiotic controls were prepared by adding and mixing 10 ml of 20 g L−1 sodium arsenite aqueous solution into soil for measuring the abiotic loss of PHE. Soil samples were placed into a fume hood overnight for the solvent volatilization. For each treatment, the soil was placed in a simulated environment with ambient temperature of 25–30°C and relative humidity of 60–90%. Moisture content of all soil treatments was maintained at relative humidity from 10 to 15% throughout the experiment. All four soil treatments were performed in duplicate, as summarized in Table S1.
PHE Extraction and Analysis
Residual PHE in soil samples (5 g soil, dry-weight equivalent) was prepared based on previous work (Wong et al., 2005) by using the microwave extraction technique (MCR-3, Jiecai Instrument Equipment Co., Ltd, Zhengzhou, China). Extraction parameters were set as follows: mixture of hexane: acetone (1: 1, v/v); at 700 W; 130°C; 30 min. A Hewlett Packard (HP) model 7890A−5975C gas chromatograph-mass spectrometer (GC-MS, HP, USA) was used for PHE analysis. All analysis was carried out using helium as the carrier gas. PHE standards with concentration of 40 mg mL−1 in the mixture of hexane/acetone (1: 1, v/v) were prepared for calibration. GC-MS operating conditions were set as follows: initial oven temperature was held at 40°C for 1 min, followed by an increase to 250°C at 10°C min−1. This temperature was held for 3 min and the total run time was 35 min. PHE analysis was performed on days 0, 1, 3, 5, 7, 9, 11, 13, and 15. These researchers adopt similar methods for the determination of PHE degradation rate (Crampon et al., 2014, 2017; Niepceron et al., 2014).
Results and Discussion
Identification and Classification of a PHE-Degrading Bacterial Strain
Strain FM-2 exhibited Gram-negative (Figure S1a), motile, short rod-like phenotypic characteristics (Figure S1b), without spores. Colonies grown on LB agar were ~1–2 mm in diameter, milky white in color, circular, integrated, convex, and transparent with intact margins (Figure S1c). Analysis of the 16S rRNA gene sequence revealed highest similarity (99%) between strain FM-2 and B. fungorum UFLA04-155 (GenBank accession no. GU144370; Figure S2). Physiological and biochemical characteristics of strain FM-2 are summarized in Table S2, and are shared with Burkholderia fungorum LMG 16225T (Coenye et al., 2001; Andreolli et al., 2011), hence the isolated strain was identified as an B. fungorum strain. Sequence analysis and multilocus sequence typing (MLST) of Burkholderia species has classified them into two main groups; Paraburkholderia (so-called environmental bacteria) and pathogenic species (Oren and Garrity, 2015; Dobritsa and Samadpour, 2016). However, this separation of Burkholderia species into beneficial (good) and detrimental (bad) categories purely based on taxonomy has been questioned (Kaur et al., 2017). Therefore, strain FM-2 was classified as genus and species B. fungorum and named B. fungorum FM-2.
Growth and Degradation of PHE by Strain FM-2
Enrichment and Adaptation of Strain FM-2 During Degradation of PHE
Biodegradation of PAHs is difficult and inefficient (Haritash and Kaushik, 2009). In the present work, strain FM-2 was enriched from MM inoculated with oil-contaminated soil samples from a Xinjiang oilfield, and found to utilize PHE as a sole carbon source when incubated during 2 days under aerobic conditions (shaking at 200 rpm) at 25°C. The culture media containing the enriched FM-2 strain degrading PHE (300 mg L−1) turned from transparent to dark yellow (Figure S3), accompanied by a slight increase in biomass. Following several rounds of sub-culturing, a well-adapted biomass was obtained that could efficiently degrade PHE.
Determination of Cell Growth Curves and PHE Degradation Capability
The growth curve of strain FM-2 revealed that bioactivity was highest after the seed liquid was cultured for 48 h (Figure S4). This was therefore selected as the inoculum, and the density of the seed liquid was 108 colony-forming units (CFU)/mL. PHE degradation by strain FM-2 was investigated using minimal medium supplemented with PHE ranging from 300 to 600 mg L−1. As shown in Figure 1, 300, 400, and 500 mg L−1 PHE was almost completely degraded within 3 days, while the bacterial cell density remained almost unchanged. These results indicate that B. fungorum FM-2 is a more powerful biodegrader of PHE than Sphingobium chlorophenolicum C3R that metabolizes <65% PHE within 3 days at an initial PHE concentration of 300 mg L−1 in a liquid culture comparable to the present work (Colombo et al., 2011). FM-2 is also superior to Pseudomonas stutzeri ZP2 that utilizes 88% of available PHE after 3 days in liquid culture containing 250 mg L−1 PHE (Zhao et al., 2009). A similar result was obtained from Bacillus thuringiensis FQ1 isolated from the soil of steelmaking plant in Heilongjiang Province, China. The degradation rate of PHE was 86.23% after adding PHE at 100–500 mg L−1 for 5 days (Jiang et al., 2015). However, Tao et al. demonstrated that the biodegradation efficiency of Pantoea agglomerans strain at low concentration of PHE (50 mg L−1) was only 78.4% after 15 days of culture, which was isolated from a petroleum-contaminated site at Karamay in Xinjiang in China (Tao et al., 2018). In addition, Janbandhu and Fulekar focused on PHE degradation by bacterial mixed culture, which reported that the microbial consortium (Sphingobacterium sp., Bacillus cereus, and Achromobacter insolitus) could degrade 250 mg L−1 of PHE, and its degradation efficiency was 56.9% after 14 days of culture (Janbandhu and Fulekar, 2011). It can be seen that strain FM-2 has obvious advantages compared with the strains mentioned above for PHE degradation.
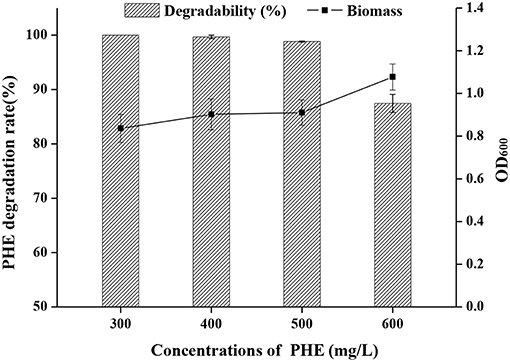
Figure 1. Effect of initial concentration of PHE (300–600 mg L−1) on a degradation rate and OD600 by strain FM-2.
Optimal Cultivation Duration, Temperature, Salinity and pH for PHE Degradation
As shown in Figure 2, both FM-2 cell growth and PHE degradation were increased after incubation for 2 days. Strain FM-2 degraded 96.31, 98.45, and 99.67% of PHE within 48, 60, and 72 h, respectively. Therefore, a culture duration of 2 days was chosen for subsequent experiments.
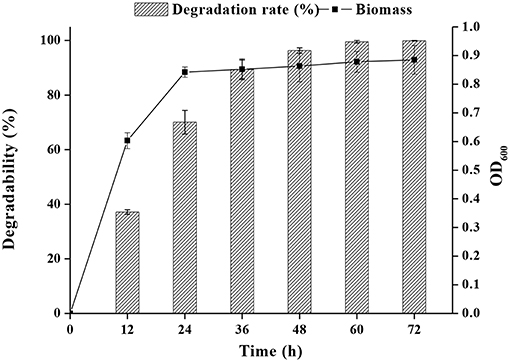
Figure 2. Effect of incubation time on PHE degradation at 25°C under shaking conditions (200 rpm) within 72 h.
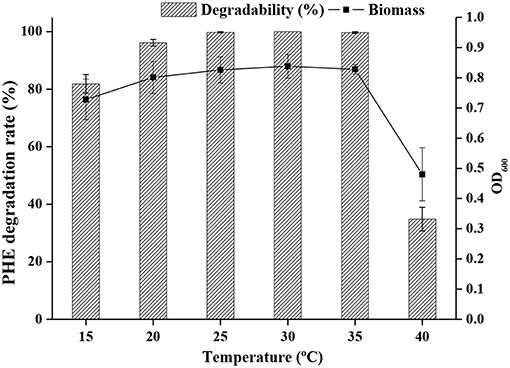
The effect of temperature on PHE degradation was investigated, and maximum PHE degradation was observed at 25°C, followed by 30 and 35°C (Figure 3).
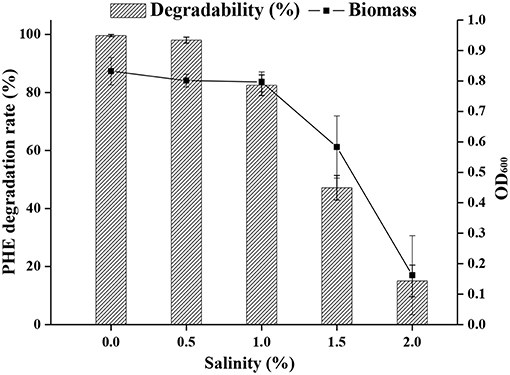
The optimal salinity for cell growth and PHE removal was investigated, and 0.5 % (w/v) NaCl was found to be optimal (Figure 4). A relatively high cell density was achieved in media containing up to 1% NaCl, but NaCl higher than 1% was detrimental to cell growth.
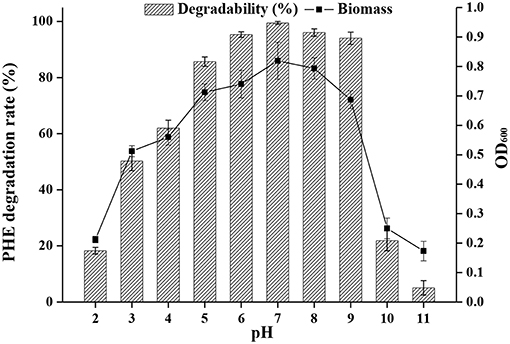
The pH of effluents from industrial processes can fluctuate widely due to their complex composition, and this represents a challenge for biodegradation by microorganisms (Alva and Peyton, 2003). The effects of pH on PAH bioremediation have been investigated (Birolli et al., 2017), and a pH range of 3.7 to 7.7 generally favors bacterial growth. Bacterial strains are remarkably sensitive to a pH <5 and >9, and removal of pollutants is generally decreased under such extreme acidic or basic conditions (Al-Thukair and Malik, 2016). However, biodegradation of PHE has been shown to occur even at pH 10 (Lin et al., 2014). Indeed, as shown in Figure 5, strain FM-2 could degrade over 94% of PHE at an initial concentration of 300 mg L−1 over a pH range from 6.0 to 9.0. The highest degradation rate occurred at pH 7.0, and degradation was slightly lower at pH 8.0. Furthermore, the bacterial cell density (OD600) was comparable at pH 7.0 and 8.0, in accordance with the PHE degradation rate, but the cell density was decreased markedly at pH 2.0, 10.0, and 11.0, indicating some limitations for cell growth. However, when the pH of the culture medium was three, the OD600 value of strain FM-2 was 0.512, and the PHE-degrading rate was 50.23%, demonstrating that the strain is strongly resistant to acidic conditions. This finding is important because only very few PAH-degrading bacteria such as Mycobacterium (Grosser et al., 1991) and Sphingomonas (Kästner et al., 1998) are capable of oxidizing PAHs at acidic pH. B. fungorum T3A13001 is reportedly active over a wide pH range, and can biodegrade 59–62% of available pyrene in liquid culture at pH 5.0 and 9.0 (Al-Thukair and Malik, 2016). This study only surveyed the degradation of pyrene at pH 5.0 and 9.0, while our current research found that PHE degradation by strain FM-2 was influenced over a broader pH range. Burkholderia species such as the identified FM-2 are renowned for their acid tolerance as well as their PHE catabolic potential.
Heavy Metal Tolerance of the PHE-Degrading FM-2 Strain
Although the impact of heavy metals on microbes has received much attention (Teitzel and Parsek, 2003; Tang et al., 2006a; Abou-Shanab et al., 2007; Choudhary and Sar, 2009; Pepi et al., 2009), the impact of heavy metals on the degradation of PHE by microorganisms is poorly understood. In the present study, the free Cd ion concentration in the culture medium was 39% according to Cd-selective electrode measurements. Similarly, ion-selective electrodes in previous studies have measured free ion concentrations 20 and 48% for Pb and Zn, respectively (Ramadass et al., 2016). Given that the free metal ion concentration in the culture medium is generally less than these values, subsequent experiments were carried out with LMM medium (Ramadass et al., 2016).
As shown in Table S3, strain FM-2 exhibited an MIC of 400 mg L−1 for Cd (II) and 50 mg L−1 for Pb (II). However, an MIC value for Zn (II) was not obtained because strain FM-2 still displayed potent degradation of PHE at a Zn concentration of 1,200 mg L−1 (data not shown) after 7 days. Addition of Zn at 1,200 mg L−1 did not inhibit bacterial growth, possibly due to complexation of Zn (II) by bacterial biosurfactants. As shown in Figure 6A, PHE biodegradation by strain FM-2 was almost unaffected by addition of Zn (II).
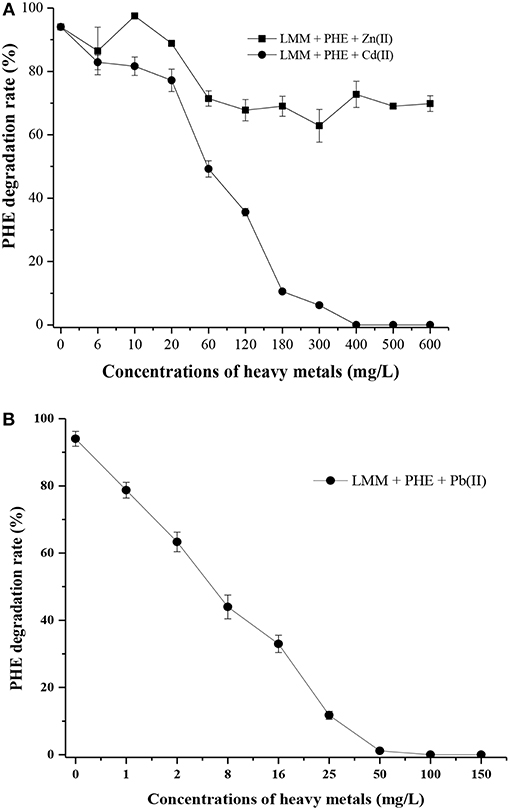
Figure 6. Effect of heavy metals on PHE degradation within 7 d (A), Zn (II) and Cd (II); (B), Pb (II).
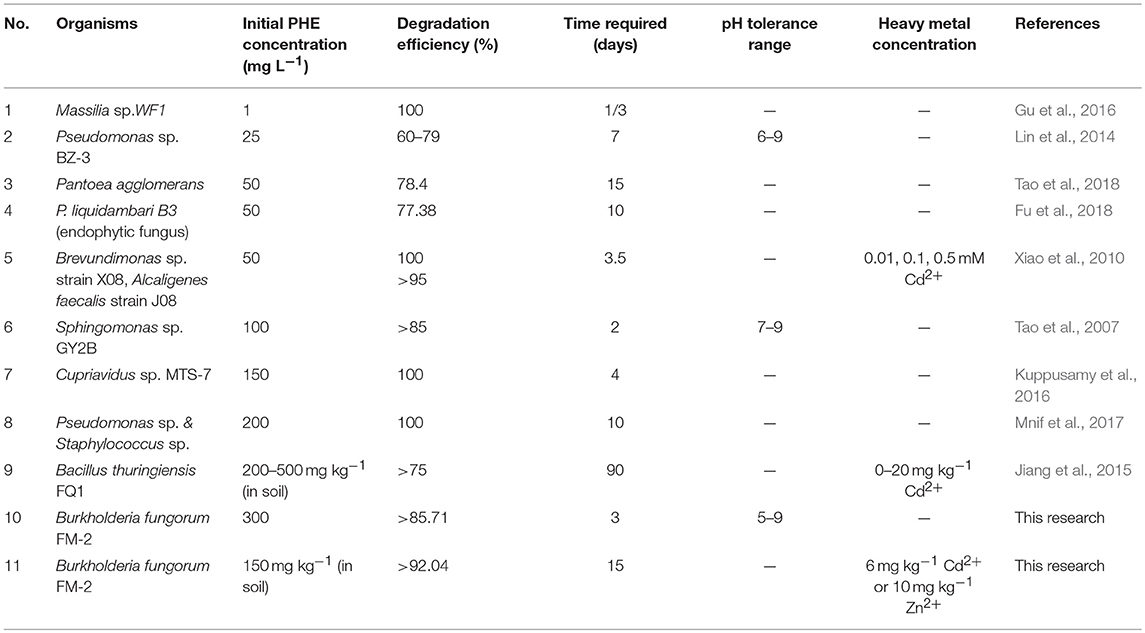
When Cd (II) was added to LMM medium at low concentrations (0–20 mg L−1), it had little effect on PHE degradation or FM-2 cell growth. However, when the Cd (II) concentration was increased to 60 mg L−1, cell growth was significantly diminished, and PHE degradation was also decreased to 49.25%. As shown in Figure 6B, low concentrations (1–25 mg L−1) of Pb (II) in LMM medium lowered the PHE degradation rate after 7 days. Our results indicate that Pb (II) was the most toxic heavy metal and potent inhibitor of PHE degradation by strain FM-2, whereas low concentrations of Zn (II) promoted cell growth. Therefore, resistance to heavy metals was ordered Zn (II) >Cd (II) >Pb (II). This could be due to different responses to heavy metals in PHE degraders (Thavamani et al., 2015), and strain FM-2 clearly displayed maximum resistance against Zn. As Table 1 showed, strain FM-2 exhibited higher PHE degradation efficiency compared with strain species reported previously under different PHE concentrations or pH or supplement of heavy metal ions.
Analysis of Heavy Metal Bioaccumulation in FM-2
ICP-OES Analysis and Isotherm Study
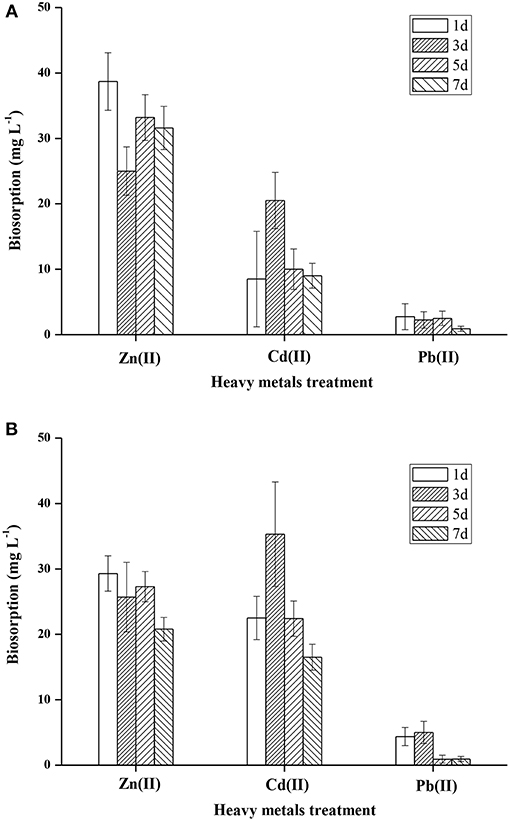
It is well-documented that microorganisms have a high affinity for metals, and can accumulate heavy metals by a variety of mechanisms (Irawati et al., 2017). Heavy metal accumulation in the cell wall of FM-2 was investigated, and the mean Zn (II) content was 38.7, 25, 33.2, and 31.6 mg L−1 after 1, 3, 5, and 7 days, respectively (Figure 7A). Thus, strain FM-2 exhibited effective bioaccumulation of Zn (II) after only 1 day of incubation at 25°C. By contrast, Cd (II) bioaccumulation peaked at 3 days at 35.3 mg L−1, then declined by 37 and 53% after 5 and 7 days, respectively (Figure 7B). Accumulation of Pb (II) was lowest after 5 days. Moderate amounts of Zn (II) accumulated in the intracellular space, and overall, strain FM-2 achieved high bioaccumulation of Zn (II) and Cd (II). Similarly, Durve et al. (2013) reported a slower accumulation of Pb over time in comparison with cadmium (Durve et al., 2013), and whole-cell accumulation of Cd (II) was highest in the most Cd-resistant Pseudomonas sp. (strain I10). Furthermore, intracellular Cd (II) bioaccumulation was found to be greater than that in the cell wall, demonstrating that Cd (II) bioaccumulation was related to cell growth and/or increased biomass (Chiboub et al., 2016). The Cd (II) bioaccumulation by strain FM-2 demonstrated in the present work is therefore consistent with previous reports. However, in FM-2, Zn (II) accumulation was greater in the cell wall than in the intracellular space, suggesting extrusion as a potential mechanism underpinning heavy metal resistance in FM-2 (Wei et al., 2009).
As shown in Figures S5, S6, the Langmuir and Freundlich isotherm models were used to predict the adsorption behaviors of strain FM-2 on Zn(II), Pb(II), or Cd(II). After different time of adsorption, the results led to better accordance to Langmuir model than the Freundlich model for the adsorption of Zn(II) or Pb(II), owing to higher R2 value in Langmuir model. Compared with Zn(II) or Pb(II), the result of Cd(II) fitted better in Freundlich adsorption model. The Langmuir isotherm suggests that the biosorption of metal ions can be regareded as a single-layer adsorption with no interaction between metal ions (Prasad et al., 2013).
SEM-EDS Analysis
Figure S7 shows SEM images of dried FM-2 biomass after Zn (II), Pb (II), and Cd (II) biosorption at a magnification of 20,000×. In the absence of metal ions, cells were short rods with smooth surfaces in a loosely-bound form (Figure S1b). However, after biosorption of Zn(II), Pb(II), and Cd(II), biomass was much reduced, and cell surfaces became rough, wrinkled, and porous. This might be due to deposition of heavy metal ions, precipitation of organic functional groups, or metal sequestration (Vishan et al., 2017a), but irrespective of the mechanism, these results suggest that heavy metal ions altered the cell morphology (Chatterjee et al., 2011) and caused significant toxicity, as shown previously for other Burkholderia species (Wei-hua et al., 2009). SEM images were confirmed by EDS spectra, which revealed the presence of Zn, Pb, and Cd on the cell surface.
FTIR Spectroscopy Analysis
FTIR spectroscopy was conducted in the range of 400–4,000 cm−1 to identify functional groups involved in Zn (II), Pb (II), and Cd (II) biosorption (Figures S8a–d). FTIR spectra of FM-2 biomass prepared without metal ions displayed typical absorption peaks consistent with the complex nature of bacterial biomass. Strain FM-2 exhibited a broad absorption band corresponding to hydroxyl groups between 3,500 and 3,300 cm−1 due to complexation of -OH groups with Zn (II), Pb (II), and Cd (II) ions. Similar results have been reported for Bacillus cereus after treatment with As (III) (Bahari et al., 2013; Giri et al., 2013), chromium-treated Pseudomonas aeruginosa (Chatterjee et al., 2011), and Escherichia coli after treatment of cells with Cd (II), Cr (VI), Fe (III), and Ni (II) (Abhay et al., 2016). A peak at ~2,922 cm−1 was assigned to stretching vibrations of C–H bonds of methylene groups (Figure S8c), in accordance with previous studies (Zhang and Min, 2009; Vishan et al., 2017b). According to reports by Chandra et al. (2014) and Abhay et al. (2016), the strong stretching vibration band at 2,361 cm−1 representing CO2 hydrates is shifted to a higher frequency. A peak was observed at ~1,634 cm−1, which is in 1,640–1,550 cm−1 region that corresponds to the bending vibrations of amines (–NH), consistent with primary and secondary amide groups of amide and protein peptide bonds. This peak was shifted to a higher frequency (~1,636 cm−1) following complexation of amide groups with Zn (II), Pb (II), and Cd (II) ions. The observed absorption band around 1,398 cm−1 could be assigned to the C = O stretching of carboxyl groups (Kazy et al., 2006), and peaks at 1,175 and 1,049 cm−1 may be attributed to C–N stretching vibrations of amine groups, P–O–C links of organic phosphate groups, or P–O vibrations of the (C–) moiety (Jiang et al., 2004). Additional small peaks were also observed at 1,003 cm−1 that may be attributed to substituted ethylenic CH–CH groups or glycoside bonds in the polysaccharide structure (Pagnanelli et al., 2000). The peak observed at 669 cm−1 in the fingerprint region was shifted to 613 cm−1 (in Figure S8c), suggesting the involvement of aromatic amino acids during the biosorption of heavy metals (Prasad et al., 2011). Binding of these groups to metal may form intense δ (M–O) and δ (O–M–O) bonds with stretching vibrations at 800–400 cm−1 (Kazy et al., 2006). These FTIR spectroscopy results strongly indicate that carboxyl, amide and phosphate groups are the dominant functional groups involved in metal ions interactions in FM-2.
Bioremediation of Soil Co-contaminated With PHE and Heavy Metals
In a previous time-course metagenomic study (Kato et al., 2015), a soil sample artificially contaminated with four aromatic compounds indicated syntrophic degradation of PHE by Mycobacterium and Burkholderia (Ohtsubo et al., 2016). Illumina sequencing was performed to analyze changes in bacterial communities, and Burkholderia was the dominant genus in all treated soils (Guo et al., 2017; Li et al., 2017). In another study, Alcaligenes sp. strain J08 was found to biodegrade PHE effectively, even under 0.5 mM Cd stress conditions (Xiao et al., 2010). Moreover, addition of Bacillus thuringiensis FQ1 into the soils containing PHE of 300–500 mg kg−1 with Cd treatments, increased PHE biodegradation efficiencies that were more than 75% for 3 months (Jiang et al., 2015). Similarly, biodegradation of PHE was boosted in soils co-contaminated with 140 mg kg−1 zinc (Wong et al., 2005). Song et al. have reported that saponin (a plant-derived biosurfactant) could desorb PHE from contaminated soils, and it also had the potential to remove heavy metals and PAHs from heavily contaminated soils (Song et al., 2008). Given that strain FM-2 displayed a low tolerance of Pb (II), as described in section Heavy metal tolerance of the PHE-degrading FM-2 strain above, soil experiments were carried out with the optimum PHE-degrading concentrations, which were 10 mg L−1 for Zn (II) and 6 mg L−1 for Cd (II), as summarized in Table S1. Figure 8 shows that PHE was removed effectively in treatments 2, 3, and 4 after culturing for only 1 day, and the removal efficiency for treatment 2 was 90.81% after 9 days, with treatment 4 showing similar efficacy. The PHE degradation rate of treatment 4 was higher than that of treatment 2 after 3 days, which indicates that Zn (II) significantly enhanced he ability of strain FM-2 to degrade PHE, consistent with a previous report on PHE degradation in Zn-containing soils (Wong et al., 2005). However, the degradation rate was slower after 3 days in treatment 3, and reached 82.91% after 9 days. At this point, the PHE degradation rate was <7.9% that of treatment 2, and Cd (II) strongly inhibited the ability of strain FM-2 to degrade PHE. The residual rates of PHE degradation was 89% after 15 days in treatment 1 not inoculated with strain FM-2 due to natural attenuation. PHE was almost completely degraded within 11 days in treatment 4. Figure 8 shows that addition of strain FM-2 significantly increased the degradation rate of PHE in soil. Although the addition of 6 mg L−1 Cd (II) clearly inhibited degradation of PHE in soil, strain FM-2 still enhanced PHE degradation under these conditions, suggesting that strain FM-2 is a candidate strain for bioremediation of PHE and heavy metals. The capacity of strain FM-2 to tolerate Cd (II) and Zn (II) while degrading PHE demonstrates its feasibility for the bioremediation of soils and sediments co-contaminated with PAHs and heavy metals.

Figure 8. PHE residual rate (%) in co-contaminated soil (all soils had a concentration of PHE 150 mg kg−1 dry soil on day zero) (Treatment 1, sterilized soil + PHE; Treatment 2, sterilized soil + PHE + FM-2; Treatment 3, sterilized soil + PHE + Cd (II) + FM-2; Treatment 4, sterilized soil + PHE + Zn (II) + FM-2).
Up to now, many researches focus on genes related to PHE degradation and heavy metal removal (Beard et al., 1997; Spain and Alm, 2003; Singleton et al., 2009; Izmalkova et al., 2013; Olaniran et al., 2013; Gran-Scheuch et al., 2017). As Figure S9 shown, there are one amino acid sequence and two gene sequences of strain FM-2. Based on previous reports, we designed the primers and cloned PAHs dioxygenase gene of strain FM-2. The BLAST analysis showed that the PAHs dioxygenase gene of B. fungorum FM-2 had a 99% identity with PAHs dioxygenase subunit alpha of Burkholderia sp. (WP_030101499.1), which was initially determined the gene as PAHs dioxygenase gene. We also obtained the Protocatechuate 3,4-dioxygenase gene related to PHE degradation, which had 100% homology of the pcaH gene (Protocatechuate 3,4-dioxygenase beta subunit) sequence from Paraburkholderia fungorum strain ATCC BAA-463 (CP010027.1). The heavy metal removal genes of strain FM-2 were detected, and it has an identical function with the TonB dependent receptor family protein of Paraburkholderia xenovorans LB400 (CP008760.1), but their homology is only about 64%. TonB gene has been reported broardly, which generally involves in the transportation of many important substances such as iron, heme, vitamin B12, carbohydrates, and various metal elements transition (Wexler et al., 2002; Schauer et al., 2008; Lim, 2010; Porcheron et al., 2013; Anugraha, 2015). Due to the low similarity of the BLAST results, we assume that detected gene may be a novel one in the TonB family. In order to explore more functions of genes related to heavy metal removal, it will be studied by carring out whole genome sequencing of strain FM-2. We will modify partial genome sequence of strain FM-2 by gene editing technology to increase the removal efficiencies of heavy metals.
Potential environmental issues have been considered into microbial bioremediation process. Minerals in the environment, including Kaolinite and quartz, were found to be useful for PAHs degradation (Duval et al., 2002; Gong et al., 2016). A recent study also conducted: Sphingomonas sp. GY2B was attached on different minerals (kaolinite and quartz) that enhanced PHE degradation, which revealed the effects of different minerals on the microbial degradation for PAHs and the underlying mechanism (Gong et al., 2018). In biodegradation process, researchers also placed a high value on aerobic and anaerobic environmental conditions for microorganisms (Tang et al., 2006a; Kong et al., 2017; Himmelberg et al., 2018; Ni et al., 2018). Tang et al. performed experiments on PHE degradation under both microaerobic and anaerobic conditions in polluted marine sediments [surface layer (0–10 cm)] and measured the PHE degradation rate (Tang et al., 2006b). Combined kinetic analysis method, the study revealed two important factors of microbial biodegradation: bacterial adaptation state and presence of a adsorption phase (Tang et al., 2005). In addition, mini-bioreactor was used to simulate the actual environment conditions including pH, temperature and dissolved oxygen, which had strong influence on heavy metals removal (Tang et al., 2006a). In this study, strain FM-2 can degrade PHE (300 mg L−1) under both sufficient oxygen and microaerobic conditions. After 3 days, the degradation efficiencies of PHE were 100 and 69.7%, respectively. However, strain FM-2 cannot grow under anaerobic conditions and did not degrade PHE and adsorb heavy metals. We would further explore the removal efficiencies of strain FM-2 on heavy metals under different culture conditions (pH, temperature, dissolved oxygen, etc.). We also would use mini-bioreactor to simulate different external environments, and determine the optimal growth conditions of strain FM-2 for removing heavy metals, which aims to improve its efficiencies for practical applications.
Conclusions
In the present study, strain FM-2 was enriched from oil-contaminated soil collected from an oilfield in Xinjiang, and found to degrades PHE as a sole energy and carbon source when supplied at 300 mg L−1 in minimal medium. Low concentrations of Zn (II) had a positive effect on PHE degradation, while low concentrations of Cd (II) had the opposite effect. However, the presence of Pb (II) strongly inhibited the PHE-degrading capability of strain FM-2. ICP-OES, SEM-EDS, and FTIR spectroscopy analyses following metal accumulation further confirmed that metal ions were associated with the cell wall and intracellular space of strain FM-2. Importantly, strain FM-2 displayed versatile catabolic activity and the ability to tolerate high concentrations of toxic metals while degrading PHE. Additionally, FM-2 exhibited high tolerance to acidic pH and very few PAH-degrading bacteria capable of oxidizing PAHs at acidic pH have been reported to date. These findings suggest that the multifunctional B. fungorum FM-2 strain may offer a cost-effective bioremediation strategy for soils and sediments contaminated with multiple pollutants, most notably in acidic environments suffering long-term pollution.
Author Contributions
LH made substantial contributions to the experimental design and revised the article critically for important intellectual content. XL made substantial contributions to the acquisition of data, and the analysis and interpretation of data as well as drafting the article. XH conducted the Fourier-transform infrared spectroscopy test. YC tested the soil. WP was responsible for scanning the electron microscopy-energy and undertaking the dispersive X-ray spectroscopy test. JH was responsible for inductively coupled plasma optical emission spectroscopy test. PG conducted the phenanthrene biodegradation test.
Funding
This work was supported by National Natural Science Foundation of China (Grant No. 21777113), Natural Science Foundation of Tianjin City (Grant No. 15JCQNJC08800), National Undergraduate Training Programs for Innovation and Entrepreneurship Nos. (201610060037 and 201710060027) and Training Project of Innovation Team of Colleges and Universities in Tianjin.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00408/full#supplementary-material
References
Abhay, P. C., Rawat, P., and Singh, D. P. (2016). Isolation of alkaliphilic bacterium Citricoccus alkalitolerans CSB1: an efficient biosorbent for bioremediation of tannery waste water. Cell. Mol. Biol. 62:3. doi: 10.4172/1165-158X.1000135
Abou-Shanab, R. A., van Berkum, P., and Angle, J. S. (2007). Heavy metal resistance and genotypic analysis of metal resistance genes in gram-positive and gram-negative bacteria present in Ni-rich serpentine soil and in the rhizosphere of Alyssum murale. Chemosphere. 68, 360–367. doi: 10.1016/j.chemosphere.2006.12.051
Al-Thukair, A. A., and Malik, K. (2016). Pyrene metabolism by the novel bacterial strains Burkholderia fungorum (T3A13001) and Caulobacter sp (T2A12002) isolated from an oil-polluted site in the Arabian Gulf. Int. Biodeterior. Biodegradation. 110, 32–37. doi: 10.1016/j.ibiod.2016.02.005
Alva, V. A., and Peyton, B. M. (2003). Phenol and catechol biodegradation by the haloalkaliphile Halomonas campisalis: influence of pH and salinity. Environ. Sci. Technol. 37, 4397–4402. doi: 10.1021/es0341844
Andreolli, M., Lampis, S., Zenaro, E., Salkinoja-Salonen, M., and Vallini, G. (2011). Burkholderia fungorum DBT1: a promising bacterial strain for bioremediation of PAHs-contaminated soils. FEMS Microbiol. Lett. 319, 11–18. doi: 10.1111/j.1574-6968.2011.02259.x
Anugraha, M. (2015). Iron Transport Strategies of the Genus Burkholderia. Zurich: Faculty of Science.
Bahari, M. Z., Altowayti, W. A., Ibrahim, Z., Jaafar, J., and Shahir, S. (2013). Biosorption of As (III) by non-living biomass of an arsenic-hypertolerant Bacillus cereus strain SZ2 isolated from a gold mining environment: equilibrium and kinetic study. Appl. Biochem. Biotechnol. 171, 2247–2261. doi: 10.1007/s12010-013-0490-x
Baltrons, O., Lopez-Mesas, M., Vilaseca, M., Gutierrez-Bouzan, C., Le Derf, F., Portet-Koltalo, F., et al. (2018). Influence of a mixture of metals on PAHs biodegradation processes in soils. Sci. Total Environ. 628–629, 150–158. doi: 10.1016/j.scitotenv.2018.02.013
Beard, S. J., Hashim, R., Hernandez, J. M., Hughes, M. N., and Poole, R. K. (1997). Zinc(II) tolerancein Escherichia coli K-12 : evidence that the zntA gene (o732) encodes a cation transport ATPase. Mol. Microbiol. 25, 883–891. doi: 10.1111/j.1365-2958.1997.mmi518.x
Birolli, W. G., Santos, D. A., Alvarenga, N., Garcia, A., Romao, L. P., and Porto, A. L. (2017). Biodegradation of anthracene and several PAHs by the marine-derived fungus Cladosporium sp. CBMAI 1237. Mar. Pollut. Bull. 129, 525–533. doi: 10.1016/j.marpolbul.2017.10.023
Caballero-Mellado, J., Martinez-Aguilar, L., Paredes-Valdez, G., and Santos, P. E. (2004). Burkholderia unamae sp. nov., an N2-fixing rhizospheric and endophytic species. Int. J. Syst. Evol. Microbiol. 54, 1165–1172. doi: 10.1099/ijs.0.02951-0
Caballero-Mellado, J., Onofre-Lemus, J., Estrada-de Los Santos, P., and Martinez-Aguilar, L. (2007). The tomato rhizosphere, an environment rich in nitrogen-fixing Burkholderia species with capabilities of interest for agriculture and bioremediation. Appl. Environ. Microbiol. 73, 5308–5319. doi: 10.1128/AEM.00324-07
Chandra, P., Barsainya, M., and Singh, D. P. (2014). A fourier transform infrared (FTIR) spectroscopic study on cellular changes in the Marinococcus Luteus sslb 1 under different salinity regime. Int. J. Pharm. Bio. Sci. 5, 848–854.
Chatterjee, S., Ghosh, I., and Mukherjea, K. K. (2011). Uptake and removal of toxic Cr(VI) by Pseudomonas aeruginosa: physicochemical and biological evaluation. Curr. Sci. 101, 645–652.
Chiboub, M., Saadani, O., Fatnassi, I. C., Abdelkrim, S., Abid, G., Jebara, M., et al. (2016). Characterization of efficient plant-growth-promoting bacteria isolated from Sulla coronaria resistant to cadmium and to other heavy metals. C. R. Biol. 339, 391–398. doi: 10.1016/j.crvi.2016.04.015
Choudhary, S., and Sar, P. (2009). Characterization of a metal resistant Pseudomonas sp. isolated from uranium mine for its potential in heavy metal (Ni2+, Co2+, Cu2+, and Cd2+) sequestration. Bioresour. Technol. 100, 2482–2492. doi: 10.1016/j.biortech.2008.12.015
Coenye, T., Henry, D., Speert, D. P., and Vandamme, P. (2004). Burkholderia phenoliruptrix sp. nov., to accommodate the 2,4,5-trichlorophenoxyacetic acid and halophenol-degrading strain AC1100. Syst. Appl. Microbiol. 27, 623–627. doi: 10.1078/0723202042369992
Coenye, T., Laevens, S., Willems, A., Ohlén, M., Hannant, W., Govan, J. R. W., et al. (2001). Burkholderia fungorum sp. nov. and Burkholderia caledonica sp. nov., two new species isolated from the environment, animals and human clinical samples. Int. J. Syst. Evol. Microbiol. 51, 1099–1107. doi: 10.1099/00207713-51-3-1099
Colombo, M., Cavalca, L., Bernasconi, S., and Andreoni, V. (2011). Bioremediation of polyaromatic hydrocarbon contaminated soils by native microflora and bioaugmentation with Sphingobium chlorophenolicum strain C3R: a feasibility study in solid- and slurry-phase microcosms. Int. Biodeterior. Biodegradation. 65, 191–197. doi: 10.1016/j.ibiod.2010.11.002
Crampon, M., Bureau, F., Akpa-Vinceslas, M., Bodilis, J., Machour, N., Le Derf, F., et al. (2014). Correlations between PAH bioavailability, degrading bacteria, and soil characteristics during PAH biodegradation in five diffusely contaminated dissimilar soils. Environ. Sci. Pollut. Res. Int. 21, 8133–8145. doi: 10.1007/s11356-014-2799-6
Crampon, M., Cebron, A., Portet-Koltalo, F., Uroz, S., Le Derf, F., and Bodilis, J. (2017). Low effect of phenanthrene bioaccessibility on its biodegradation in diffusely contaminated soil. Environ. Pollut. 225, 663–673. doi: 10.1016/j.envpol.2017.03.053
Dobritsa, A. P., and Samadpour, M. (2016). Transfer of eleven species of the genus Burkholderia to the genus Paraburkholderia and proposal of Caballeronia gen. nov. to accommodate twelve species of the genera Burkholderia and Paraburkholderia. Int. J. Syst. Evol. Microbiol. 66, 2836–2846. doi: 10.1099/ijsem.0.001065
Durve, A., Naphade, S., Bhot, M., Varghese, J., and Chandra, N. (2013). Quantitative evaluation of heavy metal bioaccumulation by microbes. J. Microbiol. Biotech. Res. 6, 21–32.
Duval, Y., Mielczarski, J. A., Pokrovsky, O. S., Mielczarski, E., and Ehrhardt, J. J. (2002). Evidence of the existence of three types of species at the quartz-aqueous solution interface at pH 0-10_ XPS surface group quantification and surface complexation modeling. J. Phys. Chem. B. 106, 2937–2945. doi: 10.1021/jp012818s
Felice, A. D., Lorenzo, F. D., Scherlach, K., Ross, C., Silipo, A., and Hertweck, C. (2016). Structural investigation of the lipopolysaccharide O-chain isolated from Burkholderia fungorum strain DSM 17061. Carbohydr. Res. 433, 31–35. doi: 10.1016/j.carres.2016.07.008
Fu, W., Xu, M., Sun, K., Hu, L., Cao, W., Dai, C., et al. (2018). Biodegradation of phenanthrene by endophytic fungus Phomopsis liquidambari in vitro and in vivo. Chemosphere. 203, 160–169. doi: 10.1016/j.chemosphere.2018.03.164
Ghosal, D., Ghosh, S., Dutta, T. K., and Ahn, Y. (2016). Current state of knowledge in microbial degradation of polycyclic aromatic hydrocarbons (PAHs): a review. Front. Microbiol. 7:1369. doi: 10.3389/fmicb.2016.01369
Giri, A. K., Patel, R. K., Mahapatra, S. S., and Mishra, P. C. (2013). Biosorption of arsenic (III) from aqueous solution by living cells of Bacillus cereus. Environ. Sci. Pollut. Res. Int. 20, 1281–1291. doi: 10.1007/s11356-012-1249-6
Gong, B., Wu, P., Huang, Z., Li, Y., Dang, Z., Ruan, B., et al. (2016). Enhanced degradation of phenol by Sphingomonas sp. GY2B with resistance towards suboptimal environment through adsorption on kaolinite. Chemosphere 148, 388–394. doi: 10.1016/j.chemosphere.2016.01.003
Gong, B., Wu, P., Ruan, B., Zhang, Y., Lai, X., Yu, L., et al. (2018). Differential regulation of phenanthrene biodegradation process by kaolinite and quartz and the underlying mechanism. J. Hazard. Mater. 349, 51–59. doi: 10.1016/j.jhazmat.2018.01.046
Gran-Scheuch, A., Fuentes, E., Bravo, D. M., Jimenez, J. C., and Perez-Donoso, J. M. (2017). Isolation and characterization of phenanthrene degrading bacteria from diesel fuel-contaminated antarctic soils. Front. Microbiol. 8:1634. doi: 10.3389/fmicb.2017.01634
Grosser, R., Warshawsky, D., and Vestal, J. R. (1991). Indigenous and enhancedmineralization of pyrene benzo[a]pyrene, and carbazole in soils. Appl. Environ. Microbiol. 57, 3462–3469.
Gu, H., Lou, J., Wang, H., Yang, Y., Wu, L., Wu, J., et al. (2016). Biodegradation, biosorption of phenanthrene and its trans-membrane transport by Massilia sp. WF1 and phanerochaete chrysosporium. Front. Microbiol. 7:38. doi: 10.3389/fmicb.2016.00038
Guo, G., Tian, F., Ding, K., Wang, L., Liu, T., and Yang, F. (2017). Effect of a bacterial consortium on the degradation of polycyclic aromatic hydrocarbons and bacterial community composition in Chinese soils. Int. Biodeterior. Biodegradation. 123, 56–62. doi: 10.1016/j.ibiod.2017.04.022
Haritash, A. K., and Kaushik, C. P. (2009). Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): a review. J. Hazard. Mater. 169, 1–15. doi: 10.1016/j.jhazmat.2009.03.137
Himmelberg, A. M., Bruls, T., Farmani, Z., Weyrauch, P., Barthel, G., Schrader, W., et al. (2018). Anaerobic degradation of phenanthrene by a sulfate-reducing enrichment culture. Environ. Microbiol. 20, 3589–3600. doi: 10.1111/1462-2920.14335
Huang, L., Xie, J., Lv, B. Y., Shi, X. F., Li, G. Q., Liang, F. L., et al. (2013). Optimization of nutrient component for diesel oil degradation by Acinetobacter beijerinckii ZRS. Mar. Pollut. Bull. 76, 325–332. doi: 10.1016/j.marpolbul.2013.03.037
Irawati, W., Parhusip, A. J. N., Christlan, S., and Yuwono, T. (2017). The potential capability of bacteria and yeast strains isolated from Rungkut Industrial Sewage in Indonesia as a bioaccumulators and biosorbents of copper. Biodiversitas 18, 971–977. doi: 10.13057/biodiv/d180315
Izmalkova, T. Y., Sazonova, O. I., Kosheleva, I. A., and Boronin, A. M. (2013). Phylogenetic analysis of the genes for naphthalene and phenanthrene degradation in Burkholderia sp. strains. Russ. J. Genet. 49, 609–616. doi: 10.1134/S1022795413060033
Janbandhu, A., and Fulekar, M. H. (2011). Biodegradation of phenanthrene using adapted microbial consortium isolated from petrochemical contaminated environment. J. Hazard. Mater. 187, 333–340. doi: 10.1016/j.jhazmat.2011.01.034
Jiang, J., Liu, H., Li, Q., Gao, N., Yao, Y., and Xu, H. (2015). Combined remediation of Cd-phenanthrene co-contaminated soil by Pleurotus cornucopiae and Bacillus thuringiensis FQ1 and the antioxidant responses in Pleurotus cornucopiae. Ecotoxicol. Environ. Saf. 120, 386–393. doi: 10.1016/j.ecoenv.2015.06.028
Jiang, W., Saxena, A., Song, B., Ward, B. B., Beveridge, T. J., and Myneni, S. C. B. (2004). Elucidation of functional groups in gram-positive and gram-negative bacterial surfaces using infrared spectroscopy. Langmuir 20, 11433–11442. doi: 10.1021/la049043+
Kang, C., Wu, P., Li, Y., Ruan, B., Zhu, N., and Dang, Z. (2014). Estimates of heavy metal tolerance and chromium(VI) reducing ability of Pseudomonas aeruginosa CCTCC AB93066: chromium(VI) toxicity and environmental parameters optimization. World J. Microbiol. Biotechnol. 30, 2733–2746. doi: 10.1007/s11274-014-1697-x
Kästner, M., Breuer-Jammali, M., and Mahro, B. (1998). Impact of inoculation protocols, salinity, and pH on the degradation of polycyclic aromatic hydrocarbons (PAHs) and survival of PAH-degrading bacteria introduced into soil. Appl. Environ. Microbiol. 64, 359–362.
Kato, H., Mori, H., Maruyama, F., Toyoda, A., Oshima, K., Endo, R., et al. (2015). Time-series metagenomic analysis reveals robustness of soil microbiome against chemical disturbance. DNA Res. 22, 413–424. doi: 10.1093/dnares/dsv023
Kaur, C., Selvakumar, G., and Ganeshamurthy, A. N. (2017). “Burkholderia to Paraburkholderia: the journey of a plant-beneficial-environmental bacterium,” Recent Advances in Applied Microbiology, ed P. Shukla (Singapore: Springer), 213–228.
Kazy, S. K., Das, S. K., and Sar, P. (2006). Lanthanum biosorption by a Pseudomonas sp. equilibrium studies and chemical characterization. J. Ind. Microbiol. Biotechnol. 33, 773–783. doi: 10.1007/s10295-006-0108-1
Kong, J., Wang, H., Liang, L., Li, L., Xiong, G., and Hu, Z. (2017). Phenanthrene degradation by the bacterium Pseudomonas stutzeri JP1 under low oxygen condition. Int. Biodeterior. Biodegradation 123, 121–126. doi: 10.1016/j.ibiod.2017.06.001
Kuppusamy, S., Thavamani, P., Megharaj, M., Lee, Y. B., and Naidu, R. (2016). Polyaromatic hydrocarbon (PAH) degradation potential of a new acid tolerant, diazotrophic P-solubilizing and heavy metal resistant bacterium Cupriavidus sp. MTS-7 isolated from long-term mixed contaminated soil. Chemosphere. 162, 31–39. doi: 10.1016/j.chemosphere.2016.07.052
Li, J., Zhang, D., Song, M., Jiang, L., Wang, Y., Luo, C., et al. (2017). Novel bacteria capable of degrading phenanthrene in activated sludge revealed by stable-isotope probing coupled with high-throughput sequencing. Biodegradation 28, 423–436. doi: 10.1007/s10532-017-9806-9
Lim, B. L. (2010). TonB-dependent receptors in nitrogen-fixing nodulating bacteria. Microbes Environ. 25, 67–74. doi: 10.1264/jsme2.ME10102
Lin, M., Hu, X., Chen, W., Wang, H., and Wang, C. (2014). Biodegradation of phenanthrene by Pseudomonas sp. BZ-3, isolated from crude oil contaminated soil. Int. Biodeterior. Biodegradation. 94, 176–181. doi: 10.1016/j.ibiod.2014.07.011
Mnif, S., Chebbi, A., Mhiri, N., Sayadi, S., and Chamkha, M. (2017). Biodegradation of phenanthrene by a bacterial consortium enriched from Sercina oilfield. Process. Saf. Environ. 107, 44–53. doi: 10.1016/j.psep.2017.01.023
Muangchinda, C., Pansri, R., Wongwongsee, W., and Pinyakong, O. (2013). Assessment of polycyclic aromatic hydrocarbon biodegradation potential in mangrove sediment from Don Hoi Lot, Samut Songkram Province, Thailand. J. Appl. Microbiol. 114, 1311–1324. doi: 10.1111/jam.12128
Murthy, S., Bali, G., and Sarangi, S. K. (2014). Effect of lead on growth, protein and biosorption capacity of Bacillus cereus isolated from industrial effluent. J. Environ. Biol. 35, 407–411.
Ni, N., Shi, R., Liu, Z., Bian, Y., Wang, F., Song, Y., et al. (2018). Effects of biochars on the bioaccessibility of phenanthrene/pyrene/zinc/lead and microbial community structure in a soil under aerobic and anaerobic conditions. J. Environ. Sci. 63, 296–306. doi: 10.1016/j.jes.2017.05.023
Niepceron, M., Beguet, J., Koltalo, F. P., Laurent, F. M., Quillet, L., and Bodilis, J. (2014). Low impact of phenanthrene dissipation on the bacterial community in grassland soil. Environ. Sci. Pollut. Res. 21, 2977–2987. doi: 10.1007/s11356-013-2258-9
Ohtsubo, Y., Nonoyama, S., Ogawa, N., Kato, H., Nagata, Y., and Tsuda, M. (2016). Complete genome sequence of Burkholderia caribensis Bcrs1W (NBRC110739), a strain co-residing with phenanthrene degrader Mycobacterium sp. EPa45. J. Biotechnol. 228, 67–68. doi: 10.1016/j.jbiotec.2016.04.042
Olaniran, A. O., Balgobind, A., and Pillay, B. (2013). Bioavailability of heavy metals in soil: impact on microbial biodegradation of organic compounds and possible improvement strategies. Int. J. Mol. Sci. 14, 10197–10228. doi: 10.3390/ijms140510197
Oren, A., and Garrity, G. M. (2015). List of new names and new combinations previously effectively, but not validly, published. Int. J. Syst. Evol. Microbiol. 65, 2017–2025. doi: 10.1099/ijs.0.000317
O'Sullivan, L. A., and Mahenthiralingam, E. (2005). Biotechnological potential within the genus Burkholderia. Lett. Appl. Microbiol. 41, 8–11. doi: 10.1111/j.1472-765X.2005.01758.x
Oves, M., Khan, M. S., Zaidi, A., and Ahmad, E. (2012). “Soil contamination, nutritive value, and human health risk assessment of heavy metals: an overview,” in Toxicity of Heavy Metals to Legumes and Bioremediation, eds A. Zaidi, P. A. Wani, and M. S. Khan (Vienna: Springer), 1–27. doi: 10.1007/978-3-7091-0730-0_1
Oyetibo, G. O., Chien, M.-F., Ikeda-Ohtsubo, W., Suzuki, H., Obayori, O. S., Adebusoye, S. A., et al. (2017). Biodegradation of crude oil and phenanthrene by heavy metal resistant Bacillus subtilis isolated from a multi-polluted industrial wastewater creek. Int. Biodeterior. Biodegradation. 120, 143–151. doi: 10.1016/j.ibiod.2017.02.021
Pagnanelli, F., Papini, M. P., Toro, L., and Trifoni, M. (2000). Biosorption of metal ions on Arthrobacter sp.: biomass characterization and biosorption modeling. Environ. Sci. Technol. 34, 2773–2778. doi: 10.1021/es991271g
Pepi, M., Lobianco, A., Renzi, M., Perra, G., Bernardini, E., Marvasi, M., et al. (2009). Two naphthalene degrading bacteria belonging to the genera Paenibacillus and Pseudomonas isolated from a highly polluted lagoon perform different sensitivities to the organic and heavy metal contaminants. Extremophiles 13, 839–848. doi: 10.1007/s00792-009-0271-1
Porcheron, G., Garenaux, A., Proulx, J., Sabri, M., and Dozois, C. M. (2013). Iron, copper, zinc, and manganese transport and regulation in pathogenic Enterobacteria: correlations between strains, site of infection and the relative importance of the different metal transport systems for virulence. Front. Cell. Infect. Microbiol. 3:90. doi: 10.3389/fcimb.2013.00090
Prasad, K. S., Ramanathan, A. L., Paul, J., Subramanian, V., and Prasad, R. (2013). Biosorption of arsenite (As(+3)) and arsenate (As(+5)) from aqueous solution by Arthrobacter sp. biomass. Environ. Technol. 34, 2701–2708. doi: 10.1080/09593330.2013.786137
Prasad, K. S., Srivastava, P., Subramanian, V., and Paul, J. (2011). Biosorption of As(III) Ion on Rhodococcus sp. WB-12: biomass characterization and kinetic studies. Sep. Sci. Technol. 46, 2517–2525. doi: 10.1080/01496395.2011.597040
Quero, G. M., Cassin, D., Botter, M., Perini, L., and Luna, G. M. (2015). Patterns of benthic bacterial diversity in coastal areas contaminated by heavy metals, polycyclic aromatic hydrocarbons (PAHs) and polychlorinated biphenyls (PCBs). Front. Microbiol. 6:1053. doi: 10.3389/fmicb.2015.01053
Ramadass, K., Megharaj, M., Venkateswarlu, K., and Naidu, R. (2016). Soil bacterial strains with heavy metal resistance and high potential in degrading diesel oil and n-alkanes. Int. J. Environ. Sci. Technol. 13, 2863–2874. doi: 10.1007/s13762-016-1113-1
Rashid, M. I., Mujawar, L. H., Shahzad, T., Almeelbi, T., Ismail, I. M., and Oves, M. (2016). Bacteria and fungi can contribute to nutrients bioavailability and aggregate formation in degraded soils. Microbiol. Res. 183, 26–41. doi: 10.1016/j.micres.2015.11.007
Reddy, P. V., Karegoudar, T. B., and Nayak, A. S. (2017). Mineralization of phenanthrene by Paenibacillus sp. PRNK-6_ effect of synthetic surfactants on its mineralization. J. Microbiol. Virol. 7, 1–7.
Schauer, K., Rodionov, D. A., and de Reuse, H. (2008). New substrates for TonB-dependent transport: do we only see the ‘tip of the iceberg’? Trends. Biochem. Sci. 33, 330–338. doi: 10.1016/j.tibs.2008.04.012
Singleton, D. R., Ramirez, L. G., and Aitken, M. D. (2009). Characterization of a polycyclic aromatic hydrocarbon degradation gene cluster in a phenanthrene-degrading Acidovorax strain. Appl. Environ. Microbiol. 75, 2613–2620. doi: 10.1128/AEM.01955-08
Song, S., Zhu, L., and Zhou, W. (2008). Simultaneous removal of phenanthrene and cadmium from contaminated soils by saponin, a plant-derived biosurfactant. Environ. Pollut. 156, 1368–1370. doi: 10.1016/j.envpol.2008.06.018
Spain, A., and Alm, E. (2003). Implications of microbial heavy metal tolerance in the environment. Rev. Undergrad. Res. 2, 1–6.
Su, X. M., Bamba, A. M., Zhang, S., Zhang, Y. G., Hashmi, M. Z., Lin, H. J., et al. (2018). Revealing potential functions of VBNC bacteria in polycyclic aromatic hydrocarbons biodegradation. Lett. Appl. Microbiol. 66, 277–283. doi: 10.1111/lam.12853
Suarez-Moreno, Z. R., Caballero-Mellado, J., Coutinho, B. G., Mendonca-Previato, L., James, E. K., and Venturi, V. (2012). Common features of environmental and potentially beneficial plant-associated Burkholderia. Microb. Ecol. 63, 249–266. doi: 10.1007/s00248-011-9929-1
Tang, Y. J., Carpenter, S. D., Deming, J. W., and Krieger-Brockett, B. (2006b). Depth-related influences on biodegradation rates of phenanthrene in polluted marine sediments of Puget Sound, W. A. Mar. Pollut. Bull. 52, 1431–1440. doi: 10.1016/j.marpolbul.2006.04.009
Tang, Y. J., Laidlaw, D., Gani, K., and Keasling, J. D. (2006a). Evaluation of the effects of various culture conditions on Cr(VI) reduction by Shewanella oneidensis MR-1 in a novel high-throughput mini-bioreactor. Biotechnol. Bioeng. 95, 176–184. doi: 10.1002/bit.21002
Tang, Y. J., Qi, L., and Krieger-Brockett, B. (2005). Evaluating factors that influence microbial phenanthrene biodegradation rates by regression with categorical variables. Chemosphere 59, 729–741. doi: 10.1016/j.chemosphere.2004.10.037
Tao, K., Zhao, S., Gao, P., Wang, L., and Jia, H. (2018). Impacts of Pantoea agglomerans strain and cation-modified clay minerals on the adsorption and biodegradation of phenanthrene. Ecotoxicol. Environ. Saf. 161, 237–244. doi: 10.1016/j.ecoenv.2018.05.091
Tao, X.-Q., Lu, G.-N., Dang, Z., Yang, C., and Yi, X.-Y. (2007). A phenanthrene-degrading strain Sphingomonas sp. GY2B isolated from contaminated soils. Process. Biochem. 42, 401–408. doi: 10.1016/j.procbio.2006.09.018
Teitzel, G. M., and Parsek, M. R. (2003). Heavy metal resistance of biofilm and planktonic Pseudomonas aeruginosa. Appl. Environ. Microbiol. 69, 2313–2320. doi: 10.1128/AEM.69.4.2313-2320.2003
Thavamani, P., Megharaj, M., and Naidu, R. (2015). Metal-tolerant PAH-degrading bacteria: development of suitable test medium and effect of cadmium and its availability on PAH biodegradation. Environ. Sci. Pollut. Res. Int. 22, 8957–8968. doi: 10.1007/s11356-013-1850-3
Vanlaere, E., Sergeant, K., Dawyndt, P., Kallow, W., Erhard, M., Sutton, H., et al. (2008). Matrix-assisted laser desorption ionisation-time-of of-flight mass spectrometry of intact cells allows rapid identification of Burkholderia cepacia complex. J. Microbiol. Methods 75, 279–286. doi: 10.1016/j.mimet.2008.06.016
Vishan, I., Laha, A., and Kalamdhad, A. (2017b). Biosorption of Pb(II) by Bacillus badius AK strain originating from rotary drum compost of water hyacinth. Water Sci. Technol. 75, 1071–1083. doi: 10.2166/wst.2016.590
Vishan, I., Sivaprakasam, S., and Kalamdhad, A. (2017a). Biosorption of lead using Bacillus badius AK strain isolated from compost of green waste (water hyacinth). Environ. Technol. 38, 1812–1822. doi: 10.1080/09593330.2017.1298674
Wei, G., Fan, L., Zhu, W., Fu, Y., Yu, J., and Tang, M. (2009). Isolation and characterization of the heavy metal resistant bacteria CCNWRS33-2 isolated from root nodule of Lespedeza cuneata in gold mine tailings in China. J. Hazard. Mater. 162, 50–56. doi: 10.1016/j.jhazmat.2008.05.040
Wei-hua, X., Yun-guo, L., Guang-ming, Z., Xin, L., Hua-xiao, S., and Qing-qing, P. (2009). Characterization of Cr(VI) resistance and reduction by Pseudomonas aeruginosa. T. Nonferr. Metal. Soc. 19, 1336–1341. doi: 10.1016/S1003-6326(08)60446-X
Wexler, M., Yeoman, K. H., Stevens, J. B., Luca, N. G., Sawers, G., and Johnston, A. W. (2002). The Rhizobium leguminosarum tonB gene is required for the uptake of siderophore and haem as sources of iron. Mol. Microbiol. 41, 801–816. doi: 10.1046/j.1365-2958.2001.02556.x
Wong, K. W., Toh, B. A., Ting, Y. P., and Obbard, J. P. (2005). Biodegradation of phenanthrene by the indigenous microbial biomass in a zinc amended soil. Lett. Appl. Microbiol. 40, 50–55. doi: 10.1111/j.1472-765X.2004.01623.x
Xiao, J., Guo, L., Wang, S., and Lu, Y. (2010). Comparative impact of cadmium on two phenanthrene-degrading bacteria isolated from cadmium and phenanthrene co-contaminated soil in China. J. Hazard. Mater. 174, 818–823. doi: 10.1016/j.jhazmat.2009.09.126
Zhang, J., and Min, H. (2009). Characterization of a multimetal resistant Burkholderia fungorum isolated from an e-waste recycling area for its potential in Cd sequestration. World J. Microbiol. Biotechnol. 26, 371–374. doi: 10.1007/s11274-009-0163-7
Keywords: phenanthrene biodegradation, Burkholderia fungorum, heavy metal resistance, acid-tolerant bacteria, bioremediation
Citation: Liu X, Hu X, Cao Y, Pang W, Huang J, Guo P and Huang L (2019) Biodegradation of Phenanthrene and Heavy Metal Removal by Acid-Tolerant Burkholderia fungorum FM-2. Front. Microbiol. 10:408. doi: 10.3389/fmicb.2019.00408
Received: 30 May 2018; Accepted: 18 February 2019;
Published: 14 March 2019.
Edited by:
Zofia Piotrowska-Seget, University of Silesia of Katowice, PolandReviewed by:
Yinjie Tang, Washington University in St. Louis, United StatesM. Oves, King Abdulaziz University, Saudi Arabia
Copyright © 2019 Liu, Hu, Cao, Pang, Huang, Guo and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Huang, aHVhbmdsZWlAdGp1dC5lZHUuY24=
 Xin-xin Liu
Xin-xin Liu Lei Huang
Lei Huang