
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 15 March 2019
Sec. Aquatic Microbiology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00347
This article is part of the Research TopicHadal Biome Research: Recent Advances and Implication for a Better Understanding of the Deep Ocean Biosphere View all 6 articles
 Logan M. Peoples1
Logan M. Peoples1 Eleanna Grammatopoulou2
Eleanna Grammatopoulou2 Michelle Pombrol1
Michelle Pombrol1 Xiaoxiong Xu1
Xiaoxiong Xu1 Oladayo Osuntokun1
Oladayo Osuntokun1 Jessica Blanton1
Jessica Blanton1 Eric E. Allen1
Eric E. Allen1 Clifton C. Nunnally3
Clifton C. Nunnally3 Jeffrey C. Drazen4
Jeffrey C. Drazen4 Daniel J. Mayor2,5
Daniel J. Mayor2,5 Douglas H. Bartlett1*
Douglas H. Bartlett1*Hadal ocean sediments, found at sites deeper than 6,000 m water depth, are thought to contain microbial communities distinct from those at shallower depths due to high hydrostatic pressures and higher abundances of organic matter. These communities may also differ from one other as a result of geographical isolation. Here we compare microbial community composition in surficial sediments of two hadal environments—the Mariana and Kermadec trenches—to evaluate microbial biogeography at hadal depths. Sediment microbial consortia were distinct between trenches, with higher relative sequence abundances of taxa previously correlated with organic matter degradation present in the Kermadec Trench. In contrast, the Mariana Trench, and deeper sediments in both trenches, were enriched in taxa predicted to break down recalcitrant material and contained other uncharacterized lineages. At the 97% similarity level, sequence-abundant taxa were not trench-specific and were related to those found in other hadal and abyssal habitats, indicating potential connectivity between geographically isolated sediments. Despite the diversity of microorganisms identified using culture-independent techniques, most isolates obtained under in situ pressures were related to previously identified piezophiles. Members related to these same taxa also became dominant community members when native sediments were incubated under static, long-term, unamended high-pressure conditions. Our results support the hypothesis that there is connectivity between sediment microbial populations inhabiting the Mariana and Kermadec trenches while showing that both whole communities and specific microbial lineages vary between trench of collection and sediment horizon depth. This in situ biodiversity is largely missed when incubating samples within pressure vessels and highlights the need for revised protocols for high-pressure incubations.
Ocean sediments make up one of the largest biomes on earth, harboring an estimated 3 × 1029 total microbial cells distributed in 3 × 108 km3 of sediment with 8 × 107 km3 of pore water (Kallmeyer et al., 2012; Amend and LaRowe, 2016). Deep-sea sediment microbial community composition is influenced by organic matter abundance and content (D’Hondt et al., 2009; Bienhold et al., 2012, 2016; Kallmeyer et al., 2012; Jacob et al., 2013), sediment horizon depth (Walsh et al., 2016a,b), water column depth (Jacob et al., 2013), and geographical location (Hamdan et al., 2013; Bienhold et al., 2016). Sediment communities are distinct from those in the water column despite the deposition of sinking taxa from above (Zinger et al., 2011; Hamdan et al., 2013). While surficial sediments include high abundances of Gammaproteobacteria, Deltaproteobacteria, Alphaproteobacteria, and Actinobacteria (Zinger et al., 2011; Ruff et al., 2015), deeper subsurface layers are dominated by the Chloroflexi and Atribacteria (OP9/JS1; Bienhold et al., 2016; Walsh et al., 2016b). These deeper communities may consist of taxa adapted to deep subsurface conditions (Inagaki et al., 2015) or which are found at shallower sediment horizon depths and survive after burial (Starnawski et al., 2017).
Sediment microbial communities at hadal depths remain largely unexplored. These sites are deeper than 6,000 m water depth and are typically affiliated with trenches, steep-walled depressions formed through the subduction of one tectonic plate below another. Sediment oxygen concentrations can drop from 200 μmol l−1 to undetectable levels over the top 10 cm in trenches, indicating high rates of oxygen consumption (Glud et al., 2013; Wenzhöfer et al., 2016). Topographical funneling of organic matter may sustain this activity (Danovaro et al., 2003; Glud et al., 2013; Wenzhöfer et al., 2016) as increases in organic material with water depth have been observed in the Mariana Trench (Luo et al., 2017) and modeled in the Kermadec Trench (Ichino et al., 2015). An alternative source of organic carbon within trenches may be geochemical inputs from below (Li et al., 1999; Fujiwara et al., 2001; Tarn et al., 2016). Until recently, microbial community analyses of hadal sediments have been limited to 16S rRNA gene sequence studies with small sample sizes (Kato et al., 1997; Li et al., 1999; Yanagibayashi et al., 1999; Nunoura et al., 2013; Yoshida et al., 2013; Luo et al., 2015) and cultivation attempts. Culture-independent analyses have identified taxa affiliated with the same lineages as those found within other shallower deep-ocean sediments and suggested the importance of nitrogen cycling within these communities (Nunoura et al., 2013, 2018; Yoshida et al., 2013; Luo et al., 2015; León-Zayas et al., 2017). High-pressure, culture-based analyses have predominantly found copiotrophic members of the Gammaproteobacteria, including Shewanella, Colwellia, Moritella, and Psychromonas (Kato et al., 1998; Nogi et al., 1998, 2002, 2004, 2007).
Two of the deepest locations in the ocean are the Mariana and Kermadec trenches. The Mariana Trench, located in the Northern Hemisphere near the Mariana Islands, extends to 10,984 m at its greatest depth (Gardner et al., 2014). The Kermadec Trench begins off the northeastern coast of New Zealand and reaches a maximum depth of 10,047 m (Angel, 1982). These trenches reside approximately 6,000 km apart within the Pacific Ocean. Deep-sea sediments can show high levels of microbial endemism and significant decay of community similarity over distance (Zinger et al., 2014; Bienhold et al., 2016). Endemism could be especially prevalent in hadal trenches, which are predicted to be rich in endemic taxa due to their extreme depths and geographical isolation (Beliaev, 1989). Furthermore, water mass inputs and annual rates of primary production vary between the two trenches, with primary productivity in the overlying waters of the Kermadec estimated at 87 g C m−2 yr−1 compared to 59 g C m−2 yr−1 in the waters above the Mariana Trench (Longhurst et al., 1995; Jamieson, 2015). Therefore, we hypothesize that geographical isolation and differences in organic matter input lead to distinct community compositions between the two trenches. In this study, we investigated the microbial communities within surficial sediment (0–10 cm) samples collected from 6- to 9-km water depths in the Kermadec Trench and 7- to 8-km depths in the Mariana Trench with both culture-independent high-throughput 16S rRNA gene sequencing and culture-dependent characterization.
Microbial community composition was evaluated within 88 total samples from the Mariana and Kermadec trenches. This included 72 samples belonging to 14 intact, depth-fractioned cores, 3 from the Mariana Trench at depths of 6,844–7,942 m and 11 from the Kermadec Trench ranging from 6,011 to 9,177 m (Supplementary Table 1). Kermadec Trench samples were collected on the R/V Thompson from April to May 2014, and Mariana Trench samples were collected during a cruise on the R/V Falkor from November to December 2014. Bathymetry was obtained from NOAA (Amante and Eakins, 2009) and plotted using the R package marmap (Pante and Simon-Bouhet, 2013). Sediment in the Kermadec Trench was collected using push cores with the HROV Nereus (Fletcher et al., 2009). Samples in the Mariana Trench were recovered using untethered free-falling/ascending landers (Free Vehicle Coring Respirometer (FVCR) and Rock Grabber (Schmidt Ocean Institute, https://schmidtocean.org/technology/elevators-landers/)). Large-diameter (10 cm) megacores were inserted into the seabed by the FVCR using an oil-compensated motor at a steady and slow speed 2 h after landing (Nunally et al., in prep). Megacore tubes were visibly inspected using a task camera on the FVCR to assure sediment integrity was maintained during coring operations. The Rock Grabber lander collected sediments using a Van-Veen Grab. After shipboard recovery, samples were immediately moved to a 4°C cold room to minimize the effects of temperature stress.
Sediment samples were depth fractioned from 0–1, 1–2, 2–5, and 5–10 cm in the Kermadec Trench and down to 10 cm at one-centimeter increments in the Mariana Trench. For samples collected using the Rock Grabber, subsamples were obtained using a sterile syringe inserted up to 10-cm depth, after which subsamples from the top 2–3 cm were extruded into KAPAK bags (Komplete Packaging, Grand Prairie, TX) and homogenized. Samples for DNA extraction were then frozen at −80°C. To determine the effects of long-term pressurization on microbial communities, ~5-g samples were also incubated at in situ pressure in KAPAK bags without amendment for 1 (Mariana) or 1.5 (Kermadec) years. Rock Grabber and other samples not part of intact cores were not included in our analysis of the in situ community as these samples were likely disturbed and mixed with deeper sediment layers and overlying water during ascent. Therefore, they were only used for culturing and experiments under long-term high-hydrostatic pressure conditions.
Sediment (5-g wet-weight) samples from either frozen samples or long-term in situ high-hydrostatic pressure incubations were used for DNA extraction. DNA was extracted using a modified version of Lysis Protocol II described by Lever et al. (2015). 2.5 V of lysis solution (30 mM EDTA, 30 mM Tris–HCl, 800 mM guanidine hydrochloride, 0.5% Triton X-100, final pH 10) and 500 μmol pyrophosphate was added to each sample and the mixture briefly vortexed. Samples were then subjected to two 15-minute freeze–thaw cycles at −80°C, followed by incubation at 50°C with shaking at 150 rpm for 1 h. Samples were centrifuged and the supernatant was treated twice with 1V 24:1 chloroform:isoamyl alcohol. DNA was precipitated using 5 M NaCl and 70% ethanol for 2 h at room temperature and resuspended in nuclease-free water. Extracted DNA was cleaned again using a Quick-gDNA MiniPrep kit (Zymo Research, Irvine, CA). Extraction blanks, consisting of all reagents but no sediment material, were performed in concomitance with every extraction.
Sequencing and processing were conducted as previously described (Peoples et al., 2018a). Briefly, the V4-V5 16S rRNA gene region between 515f-926R was amplified (Parada et al., 2015) for 30 cycles and tagged with Illumina barcodes using a secondary PCR procedure. Samples were pooled at equimolar concentrations and sent for sequencing on an Illumina Miseq at the Institute for Genomic Medicine Genomics Center (University of California, San Diego, La Jolla, CA). Paired-end reads were assembled using FLASH (Magoc and Salzberg, 2011) and OTUs picked at 97% similarity using Uclust in QIIME 1.9.1 (Caporaso et al., 2010). Chimeras were identified and removed using VSEARCH (Rognes et al., 2016) and taxonomy assigned against the SILVA 123 release database (Quast et al., 2012). Sequences found within sequenced extraction blanks and which showed similarity to known contaminants were discarded. The exclusion of contaminants was done manually to avoid removing any autochthonous taxa that may have resulted from cross-contamination between samples and controls. Removed genera included Acinetobacter, Stenotrophomonas, Methylobacterium, Achromobacter, Rhizobium, Tetragenococcus, Escherichia-Shigella, Ferribacterium, Massilia, Curvibacter, Ralstonia, Variovorax, Deinococcus, Anoxybacillus, Aeribacillus, Brevundimonas, Enterobacter, Streptococcus, Burkholderia, Staphylococcus, Sphingomonas, Sphingobium, Sphingobacterium, Bradyrhizobium, Paenibacillus, and the family Comamonadaceae. Raw sequence data can be accessed at the Sequence Read Archive under the Biosample Accession numbers SAMN07732241-SAMN07732328.
Sequencing reads were processed with the R package phyloseq (McMurdie and Holmes, 2013). OTUs were removed if they did not have an abundance of at least three reads in at least four samples across the entire dataset. Samples were rarefied to even sampling depth to account for differing sequencing depths between samples. Alpha diversity was calculated using the package vegan (Oksanen et al., 2017) and comparisons between samples were performed using the beta-diversity metric Bray-Curtis in phyloseq. Permutational analysis of variance with adonis was used to determine if trench of collection, sediment horizon depth, and water column depth significantly explained community variation within and between trenches. DESeq2 (Love et al., 2014) was used to identify taxa that were differentially abundant between trench of collection, sediment horizon depth (0–5 or 5–10 cm), water column depth (Kermadec only; 6,000 and 7,000 m vs. 8,000 and 9,000 m), and high-pressure comparison (samples maintained under long-term high hydrostatic pressures and the same samples extracted immediately). DESeq2 analyses were performed on the un-rarefied dataset (McMurdie and Holmes, 2014) with low-abundance (at least <500 total reads per OTU or < 1,000 reads per class or phylum) reads removed. For phylogenetic analysis, representative sequences were aligned with the SINA Aligner (Pruesse et al., 2012; https://www.arb-silva.de/aligner/) and a phylogenetic tree built with FastTree (Price et al., 2010). Trees were visualized with the Interactive Tree of Life (Letunic and Bork, 2007) and GGTREE (Yu et al., 2017). For calculating OTU and sequence percentages shared between trenches, samples were, in order, (1) rarefied to even sequence depth across samples, (2) pooled by trench of collection, and (3) the pooled groups rarefied again to account for different numbers of samples and sequencing depths between trenches. Because of the possibility of losing rare OTUs through pruning and rarefaction (McMurdie and Holmes, 2014), an analysis was also performed where low-abundance OTUs were not removed as described above (at least three reads in at least four samples) but instead only had to be in abundances greater than three reads across the entire dataset. Shared OTU and sequence percentages were then calculated by pooling samples within each trench and rarefying the combined trench communities to even total sequencing depth. This analysis therefore maintained rare OTUs and did not discard large numbers of sequences through rarefaction of each individual sample, but ignored the influence of certain samples having higher sequencing depths than others. For calculating shared OTU percentages between cores, samples were, in order, (1) rarefied to even sampling depth, (2) combined within 0–5 and 5–10 cm groupings within the same core of collection, (3) rarefied to even sampling depth again, and (4) combined by core. This was done to account for different sediment horizon depths, number of samples, and sequencing depth from influencing the abundances of OTUs, although at the cost of losing sequencing depth through rarefaction.
Bacteria were cultured from sediment following plating onto agar or inoculation into pressurizable bulbs (Yayanos, 2001) and incubated at 4°C. Sediment samples from the top 1 cm of sediment, or a homogenized sample from the Rock Grabber, were diluted into 0.1-μm filtered trench water. Samples from the Kermadec Trench were incubated in 2216 Zobell Marine Medium (BD Difco™), A1 Medium (5 g potato starch, 2 g yeast extract, 1 g peptone, 0.5 g NH4Cl, 0.01 g L-methionine, 500 ml 0.2 μm-filtered autoclaved seawater collected from the Ellen Browning Scripps Pier), or a seawater minimal medium (10 mM NH4Cl, 14 mg Na2HPO4, American Type Culture Collection (ATCC) vitamin supplement (ATCC, Manassas, VA), ATCC trace mineral supplement, 1 L 0.2 μm-filtered Scripps Pier seawater), while samples from the Mariana Trench were inoculated in 2216 Marine Medium only. For samples to be incubated at high pressure, the media described above was inoculated, mixed with gelatin (pH 7 in sterile seawater) to a final concentration of 4% (Yayanos, 2001), transferred into polyethylene transfer pipettes (Samco, Thermo Fisher Scientific), heat sealed, and placed inside pressure vessels and incubated at the desired pressure at 4°C. Kermadec Trench samples were incubated at 100 MPa while those from the Mariana Trench were incubated at in situ pressure (40–110 MPa). After approximately 2 months, colony-forming units (CFUs) were calculated and representative isolates selected for 16S rRNA identification. Colonies arising at high pressure in bulbs were picked using a sterile syringe needle, inoculated into liquid medium, and grown until turbid at the original pressure of incubation at 4°C. Selected colonies were boiled and lysate used as template for PCR using the primers 27F and 1492R (Lane, 1991). Partial sequenced reads of at least 500 bp were identified using the SILVA database and compared to known sequences within NCBI.
Hadal trenches are geographically isolated and can have different overlying primary productivity and water mass inputs that may lead to variations in community composition. Therefore, we evaluated the microbial communities within sediments from the Kermadec and Mariana trenches using high-throughput 16S rRNA gene sequencing of 88 samples. Seventy-two of these were from 14 pristine depth-fractioned cores, while 16 were from potentially disturbed sediments and were not included in the comparative analysis of the in situ communities here (Supplementary Figure 1, Supplementary Table 1). Rarefaction resulted in 7,231 sequences per sample and 6,317 total operational taxonomic units (OTUs) at 97% 16S rRNA V4-V5 gene sequence similarity. Phyla with high relative sequence abundances within the sediment samples included the Proteobacteria, Thaumarchaeota, Bacteroidetes, Planctomycetes, and Chloroflexi (Figure 1). While the majority of OTUs were seen in only a few samples each, 27 OTUs were present in all samples in both trenches (Figure 2). These shared OTUs included members related to the Marine Group I (MGI) within the Thaumarchaeota, the JTB255 clade and the genus Marinicella within the Gammaproteobacteria, the Rhodobacterales and Rhodospirillales within the Alphaproteobacteria, the class JTB23 within the Proteobacteria, and BD2–11 within the Gemmatimonadetes (Figure 2, Supplementary Table 2). On average, these OTUs made up 23 ± 4.5% (range 13.7–35.9%) of the total reads in each community and were closely related to sequences from previous studies of abyssal and hadal sediments (Supplementary Figure 2).
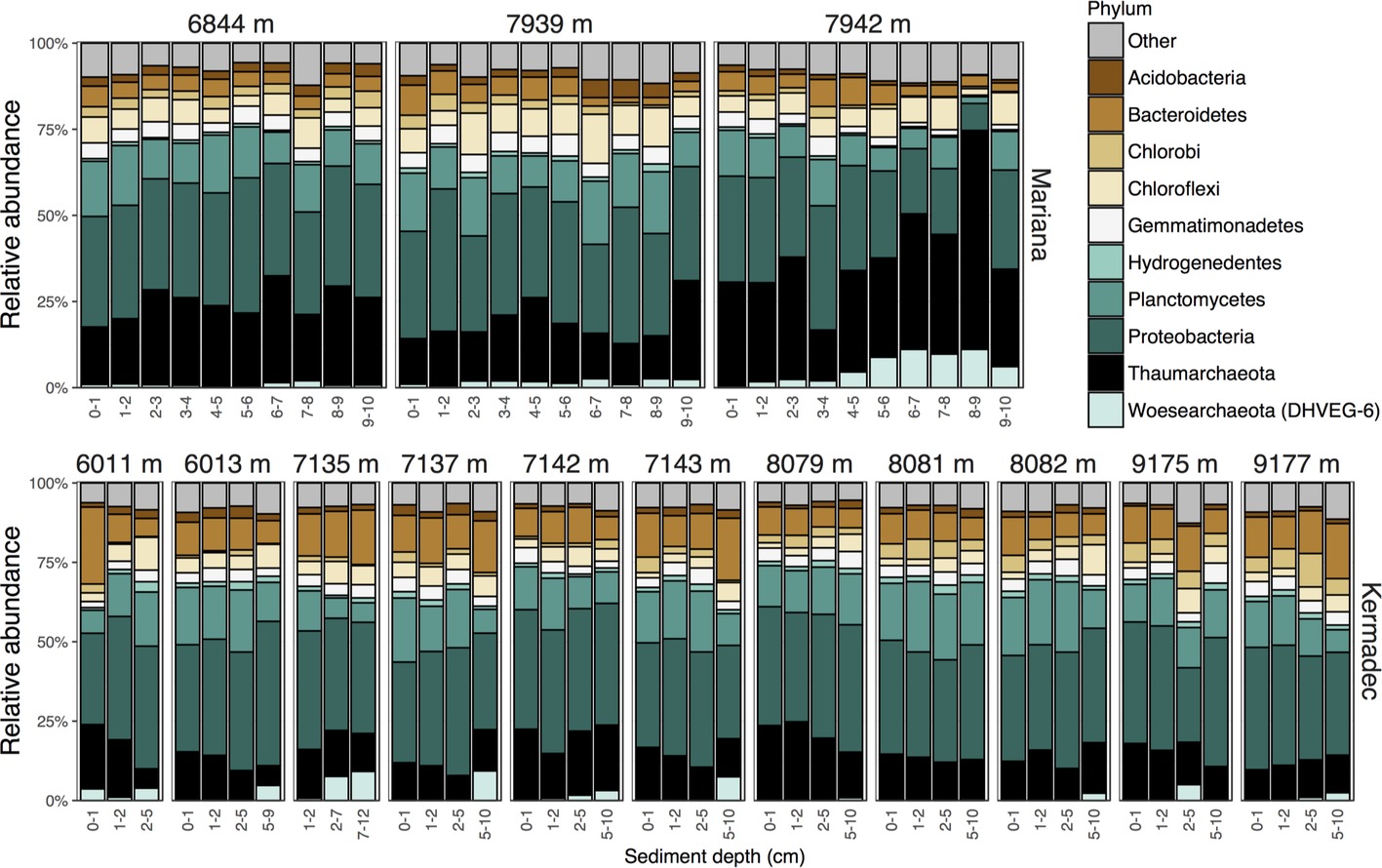
Figure 1. Relative sequence abundances of the 10 most abundant phyla within surficial sediments of the Kermadec and Mariana trenches organized by core (labeled by water depth), trench, and sediment horizon depth.
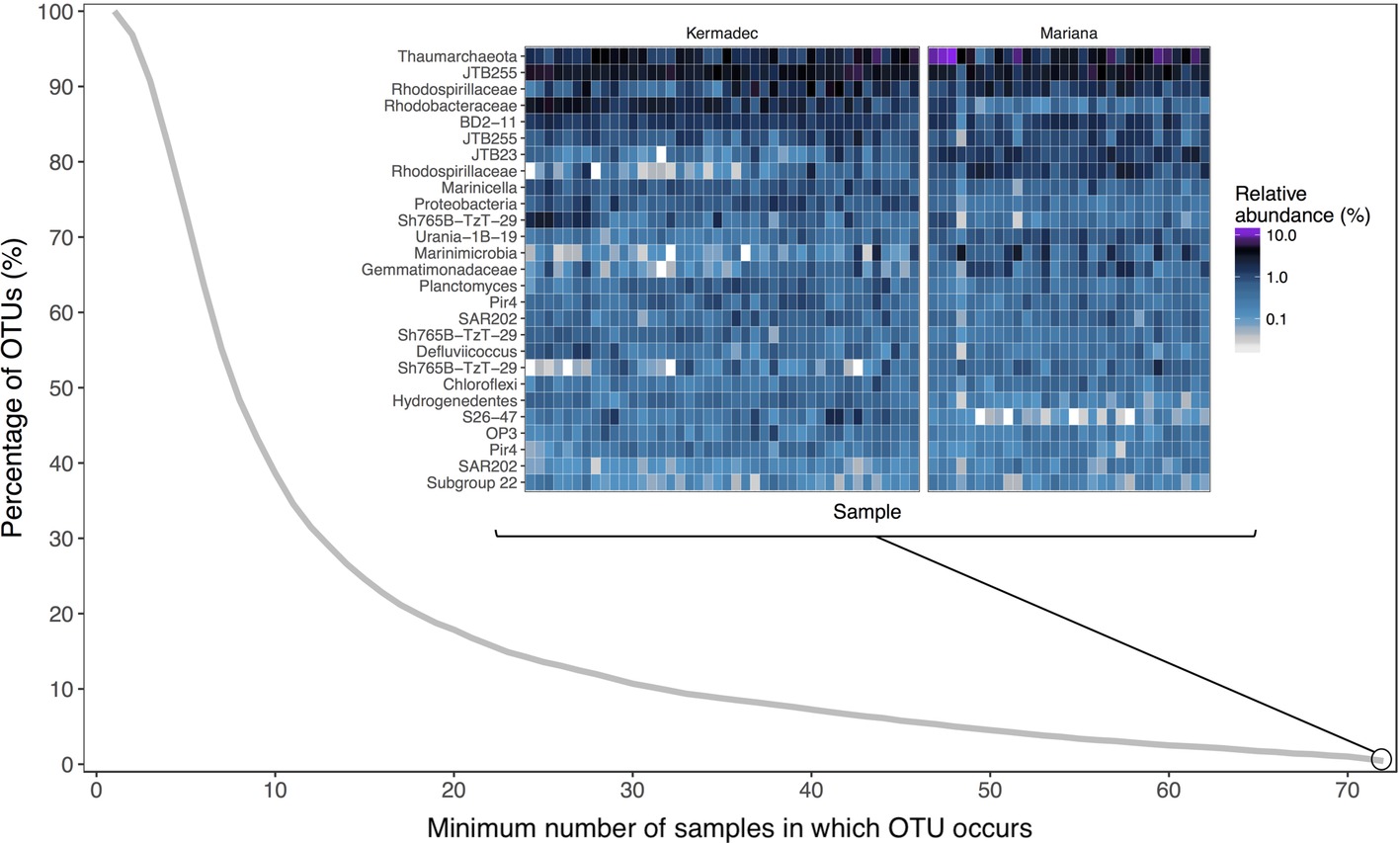
Figure 2. The percentage of the total operational taxonomic units (OTUs) as a function of the minimum number of samples in which they occur. The inset is a relative sequence abundance heat map of OTUs found in all 72 pristine core samples, organized in descending order of average relative sequence abundance across all samples. Labels represent the lowest identifiable taxonomic rank.
Sediment community diversity was structured by trench of collection (Figures 3A,B, Supplementary Figure 4; PERMANOVA, p < 0.001, R2 = 0.23). Fifty-eight percent of OTUs, representing 95% of all sequences, were seen in both trenches (Figure 3C). When the only criterion for maintaining reads was that they had an abundance of at least three across the entire sediment dataset and samples were not rarefied, 40% of the OTUs were shared and still represented greater than 90% of all sequences. More OTUs from any given core could be found within cores from the same trench than cores from the other trench (Figure 3D; minimum found within a comparison core 32%, maximum 58%). Within both trenches assemblages varied by sediment horizon depth (Figure 3B; Kermadec, p < 0.001, R2 = 0.16; Mariana, p < 0.001, R2 = 0.21). Community richness was lower in deeper sediment horizons (Supplementary Figure 3). Community composition also varied by sampling site within each trench (Kermadec, p < 0.001, R2 = 0.25; Mariana, p < 0.001, R2 = 0.18) and water column depth (Kermadec, p < 0.001, R2 = 0.15; Mariana, p < 0.001, R2 = 0.15). Overall these parameters explained half of the variability within each trench.
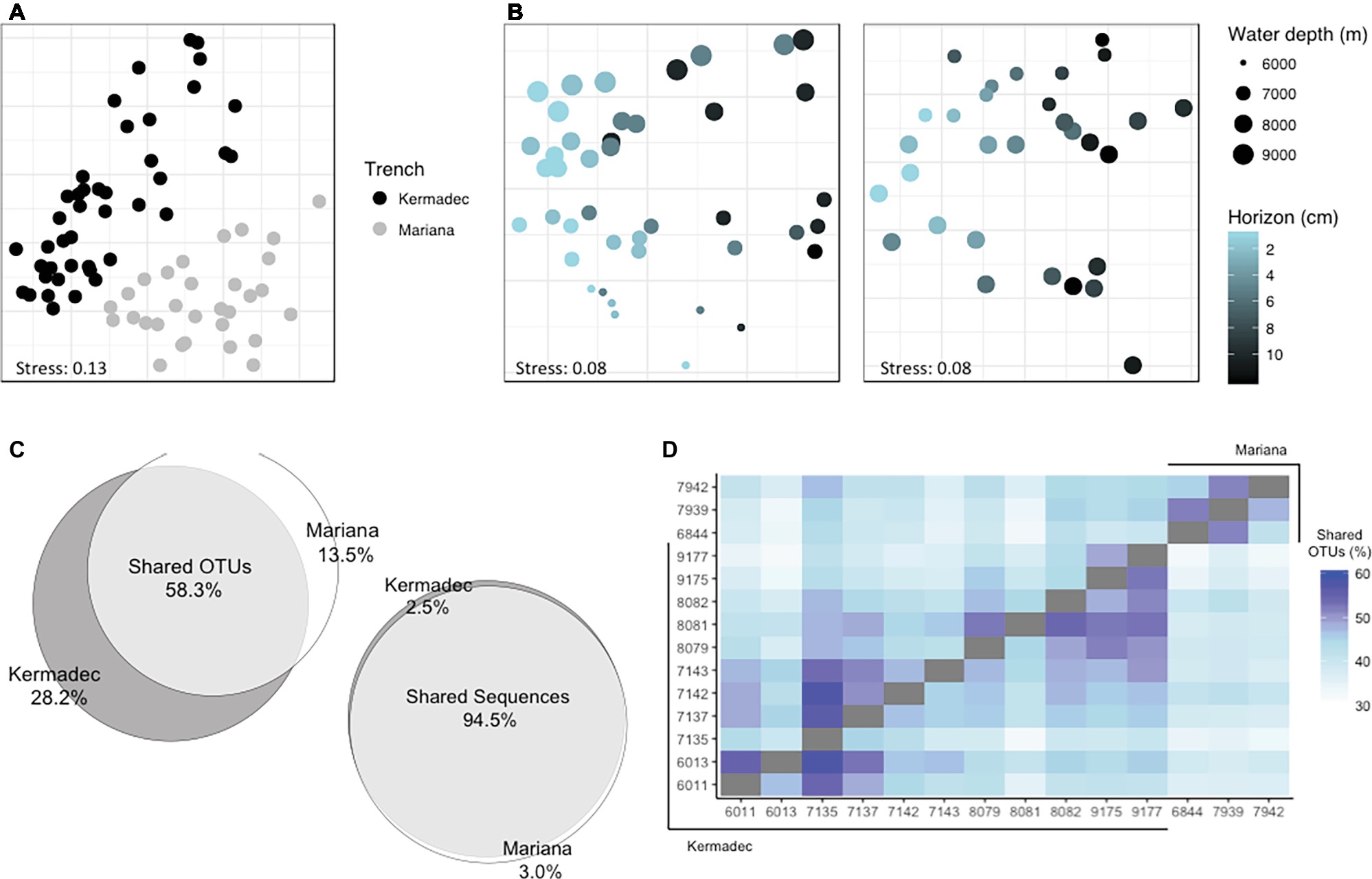
Figure 3. Distances between sediment communities visualized using NMDS ordinations of Bray-Curtis dissimilarities, labeled by trench (A) or separated by trench to examine the importance of sediment and water depth; (B) Left, Kermadec Trench; Right, Mariana Trench. (C) The percentage of OTUs and sequences shared between trenches. (D) The percentage of OTUs from one core found within another core, where the number of shared OTUs between samples on the x and y axes are shown as a percentage of the total OTUs found in the core on the x-axis.
Taxa that showed the largest differences in relative sequence abundances between trenches, sediment horizon depths, and water depths were identified. The Kermadec Trench was enriched in the phyla Bacteroidetes, Hydrogenedentes, Planctomycetes, and Proteobacteria, while the Mariana had higher abundances of the Marinimicrobia, Thaumarchaeota, Woesearchaeota, and Chloroflexi (Figure 4A, Supplementary Figure 5). The OTUs that varied the most between the Mariana and Kermadec trenches belonged to the MGI Thaumarchaeota, with specific OTUs showing enrichment in the Mariana Trench (Supplementary Figure 5). An OTU related to the genus Aquibacter reached abundances up to 11% of certain sediment horizons and was enriched in the Kermadec Trench. This Aquibacter OTU is identical to that which was found to be more abundant in the Kermadec Trench pelagic community than in the Mariana waters (Peoples et al., 2018a) and likely represents a new, hadal-adapted species (Peoples et al., 2018b).
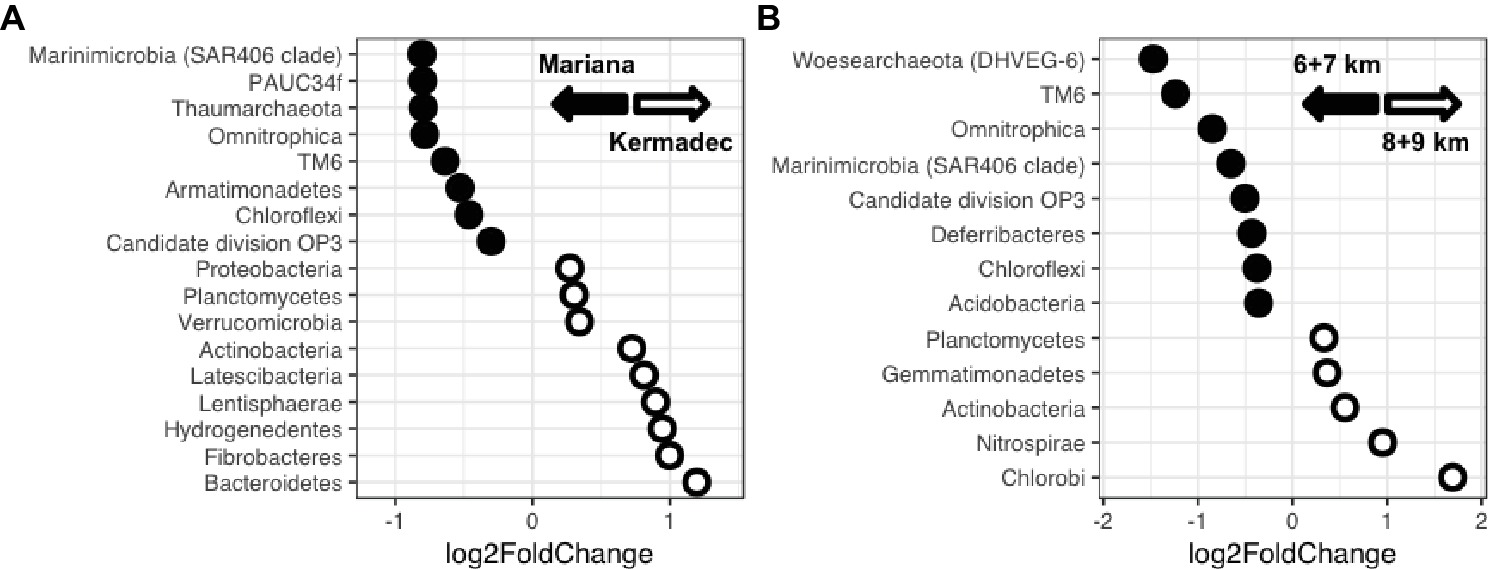
Figure 4. (A) Phyla showing enrichment within either the Kermadec or Mariana Trench. (B) Phyla showing enrichment at either upper hadal (6 + 7 km) or lower hadal (8 + 9 km) water depths in the Kermadec Trench.
Comparisons of surficial (0–5 cm) and deep (5–10 cm) sediments showed that taxa belonging to Woesearchaeota, Marinimicrobia, and Chloroflexi, including members of the clades S085, JG30-KF-CM66, SAR202, and Anaerolineae, were enriched in the deeper sediments (Figure 5A, Supplementary Figure 5, Supplementary Table 3). Phylogenetic analysis of Marinimicrobia OTUs enriched in the deeper sediment horizons showed they are related to sequences predominantly affiliated with deep-ocean sediments (Supplementary Figure 6). In contrast, the shallower sediments were enriched in classes belonging to the Verrucomicrobia, Planctomycetes and Nitrospirae, and OTUs belonging to the Proteobacteria, including Colwellia, and Bacteroidetes. MGI Thaumarchaeota showed strong variation by sediment horizon depth, with certain OTUs reaching up to 45% of some sediment horizon depths. Phylogenetic analysis indicated differentially abundant OTUs belonged to distinct clades, with members of the MGI alpha (α) subgroup more abundant in the upper sediments, while those belonging to the eta (η), upsilon (υ), and zeta (ζ) subgroups were enriched in deeper sediments (Figure 5B).
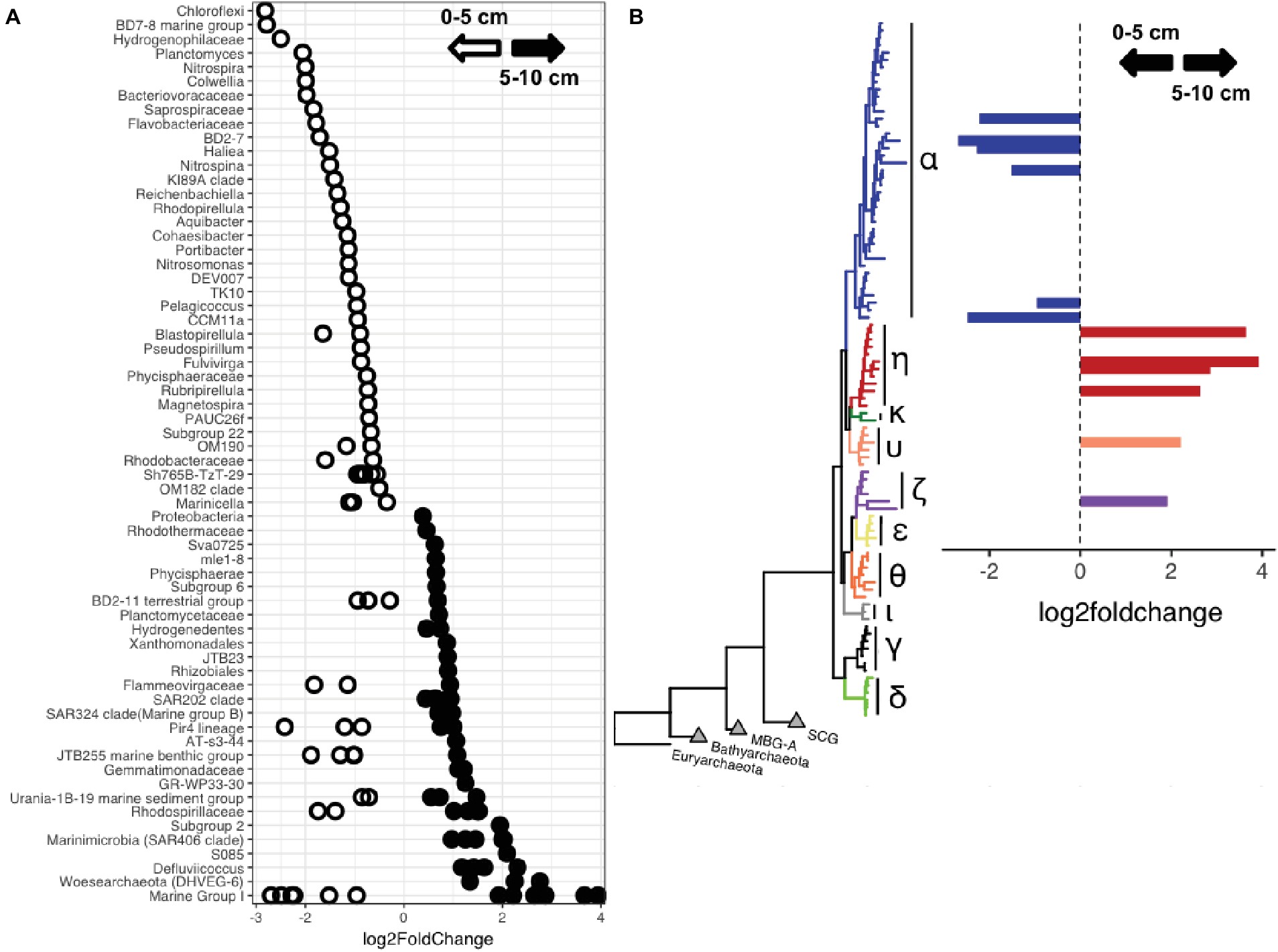
Figure 5. (A) OTUs showing enrichment within shallower (0–5 cm) or deeper (5–10 cm) sediments. The lowest identifiable taxonomic rank is shown. (B) Thaumarchaeota OTUs enriched at different sediment horizon depths, labeled by clade as described in Lauer et al. (2016).
Sediment community composition also varied with water depth. Within the Kermadec Trench, higher percentages of OTUs within a given core could be identified within other cores from similar water depths (Figure 3D). When comparing cores as a function of water depth in the Kermadec Trench, the Chlorobi, Nitrospirae, and Actinobacteria were more abundant in the 8- and 9-km samples, while the Chloroflexi, Acidobacteria, and uncharacterized lineages were enriched at 6 and 7 km (Figure 4B). Cyanobacteria, which could be deposited through topographical funneling, were present, but not found at high abundances regardless of depth. However, an OTU related to the ML635J-21 group of Cyanobacteria was identified at relative sequence abundances of less than 0.1%. This OTU showed highest similarity to taxa from the Japan Trench (99%) and deep-ocean sediments from the Okinawa Trough (99%), with the next closest sequences being only 94% similar.
Bacteria were isolated under both low and high hydrostatic pressures to determine the diversity of culturable isolates. Fifty isolates obtained at high pressures under the facultatively anoxic conditions that develop in bulbs included members of the genera Colwellia, Shewanella, Moritella, and Psychromonas (Figure 6A, Supplementary Table 4). Isolates were also obtained from the genus Psychrobium within the Gammaproteobacteria, Arcobacter within the Epsilonproteobacteria, and a member of the Flavobacteriaceae, although none of these isolates survived cryopreservation or repeated subculturing. Qualitatively, the number of isolates obtained per sample was higher from the Kermadec than the Mariana Trench. This is consistent with the pristine sediment community sequence data where OTUs related to previously culturable piezophilic taxa represented ~0.20% of Kermadec samples but less than 0.05% in the Mariana Trench (Supplementary Figure 7; t-test, sediment only, p < 0.115; sediment and water from Peoples et al., 2018a, p < 0.001). One sample from the Kermadec Trench had relative sequence abundances of these taxa in excess of 4.8%. Relative sequence abundances of taxa related to culturable piezophiles were highest in the Kermadec Trench 0–1-cm fraction and lower in deeper sediments (t-test, Kermadec Trench, p < 0.003). No trend was seen among the altogether low relative sequence abundances present in the Mariana samples. Isolates obtained at atmospheric pressure on plates, rather than at high pressure, were related to the genera Pseudoalteromonas, Pseudomonas, Halomonas, and Shewanella. On average in both trenches OTUs related to these genera made up 0.18% relative sequence abundances within each sample.
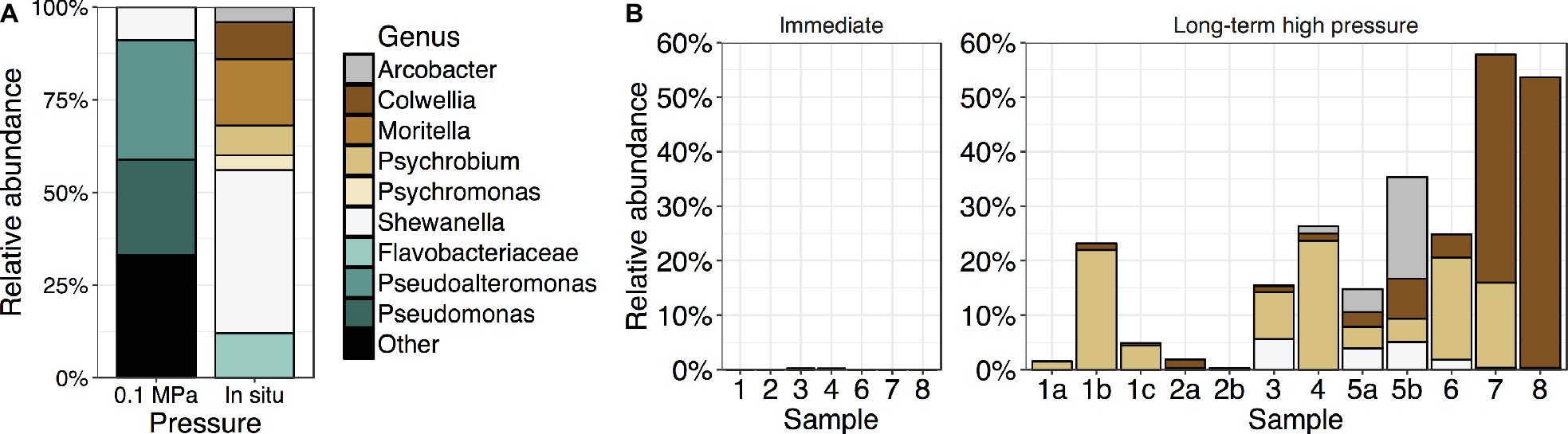
Figure 6. (A) Relative abundances of cultured isolates obtained with nutrient amendment at either 0.1 MPa on plates (n = 119) or high hydrostatic pressure in gelatin bulbs under in situ conditions (n = 50) at 4°C. (B) Relative sequence abundances within whole sediment communities of taxa related to high-pressure isolates (same colors as in A) in either in situ, immediately extracted sediment samples or the same samples after long-term high-pressure static incubations without nutrient amendment.
Sediment samples were maintained at 4°C under static, unamended, in situ pressures for longer than 1 year (hereafter called “long-term” samples) to test the effects of long-term sample incubation on community composition. These analyses were performed with the cores previously described and a number of other samples, including those collected using Van-Veen grabs, that may have been mixed with deeper sediment horizon depths and the overlying water (Supplementary Table 5). The resulting communities were compared to those from the same samples immediately frozen upon collection (“immediate” samples; Supplementary Table 5). DNA gram sediment−1 and alpha diversity were significantly lower in long-term samples relative to immediate samples (Supplementary Figure 8; t-test, DNA gram sediment−1, p < 0.035; Chao1, p < 0.004; Shannon, p < 0.001). OTUs related to the genera Psychrobium, Colwellia, Arcobacter, and Shewanella were enriched after long-term incubation, in some cases representing above 50% of the community despite starting abundances ranging from 0.0 to 0.2% (Figure 6). These OTUs were most similar to cultured piezophiles, including Colwellia marinimaniae (Kusube et al., 2016), Colwellia piezophila, and Shewanella benthica KT99, and to a member of the Psychrobium previously enriched after long-term pressurization (Aoki et al., 2014). Other long-term enriched taxa included members of the Gammaproteobacteria, Rhodobacteraceae, Planctomycetes, and Bacteroidetes (Supplementary Figure 9). In contrast, the long-term pressurized samples showed relative decreases in taxa belonging to uncharacterized and uncultivated lineages, including members of the clades BD2–11, BD7–8, NS72, SAR324, SAR202, Marinimicrobia, and Omnitrophica (OP3), and putative deep-ocean taxa such as Defluviicoccus and Rhodospirillaceae.
In this study, we evaluated the microbial community composition within trench sediments to test the hypothesis that hadal zones have distinct microbial communities from one another. While sediment communities were observed to be distinct between trench of collection and sampling site, there was significant overlap in the abundant taxa found in the Mariana and Kermadec trenches. Shared OTUs within both trenches represented greater than 90% of all reads. Therefore trench endemic OTUs made up small proportions of the communities when evaluated at 97% similarity. Many abundant taxa were closely related to members identified within other abyssal and hadal samples. These taxa belonged to lineages previously identified as having cosmopolitan members at bathyal and abyssal depths (Bienhold et al., 2016; Mußmann et al., 2017), such as sequences related to JTB255, BD2–11, JTB23, and the genus Marinicella. These results indicate that members within these lineages are present even at hadal depths. Similarly, the isolates obtained at high hydrostatic pressures were related to the genera Colwellia, Shewanella, and Moritella, consistent with previous studies. Many of the partial 16S rRNA genes of isolates from the present study were more than 97% similar to those of other piezophilic isolates. New strains were also obtained, including isolates related to the genus Psychrobium within the Gammaproteobacteria, Arcobacter within the Epsilonproteobacteria, and a member of the Flavobacteriaceae. Unfortunately, none of these isolates survived cryopreservation or repeated subculturing. Still, this Psychrobium isolate was related to that previously enriched after long-term pressurization (Aoki et al., 2014). Altogether, our results demonstrate the possible dispersal of OTUs between two widely separated trenches. Deep-ocean currents may lead to the dispersal and deposition of microorganisms in sediments (Müller et al., 2014), such as Antarctic Bottom Water flowing between the Kermadec and Mariana trenches. It is also possible some members do not require dispersal between trenches, but may originate within abyssal sediments, a possibility not yet evaluated. In this scenario, the microbes in question would possess high fitness in both abyssal and trench zones, potentially spreading between the two environments via bottom currents, or perhaps through earthquake-induced mass-wasting deposition down slope (Oguri et al., 2013). If taxa endemic to specific trenches exist in the hadal settings examined, they must exist at the strain rather than the species level, be rare, were lost during sampling, or are present in patchy or sample-specific distributions and were missed by our sampling. Comparisons with microbial communities in more distant trenches, such as the Puerto Rico or Atacama trenches, may show higher sequence abundances of endemic taxa.
Differences in organic matter due to primary production in overlying surface waters and its deposition through topographical funneling may be one of the most important factors structuring communities within trenches (Danovaro et al., 2003; Glud et al., 2013; Ichino et al., 2015; Wenzhöfer et al., 2016). The Kermadec Trench may have higher concentrations of organic matter than the Mariana Trench because of differences in primary productivity at the surface (Longhurst et al., 1995; Jamieson, 2015). While we do not have organic matter concentrations to report here for each core, preliminary data from the 0–1-cm fraction at 8000 m in each trench suggest that percent total organic carbon is higher in the Kermadec Trench (~0.5%) than the Mariana Trench (~0.4%; Grammatopoulou et al., in prep). Therefore, it is reasonable to hypothesize that organic matter is in part responsible for the differences between the Mariana and Kermadec trench communities. Consistent with this, the Kermadec Trench was enriched, relative to the Mariana Trench, in the Bacteroidetes, Actinobacteria, and Proteobacteria, lineages that have been found to correlate with higher concentrations of organic matter (Bienhold et al., 2012; Learman et al., 2016). In contrast, the Mariana Trench had higher proportions of Thaumarchaeota, Chloroflexi, and other uncharacterized lineages. Although Archaea are ubiquitous in marine sediments, organic matter concentrations are thought to negatively correlate with abundances of ammonia-oxidizing Archaea (Luo et al., 2015; Learman et al., 2016), potentially due to differences in electron acceptor availability or Archaea outcompeting Bacteria under energy-starved conditions (Valentine, 2007; Hoehler and Jorgensen, 2013). The Chloroflexi may also be adapted to degrading recalcitrant organic matter (Landry et al., 2017). Furthermore, the number of piezophilic isolates and related sequences in the high-throughput community data were higher within the Kermadec than the Mariana Trench and were enriched in the upper layers of sediment. Their enrichment in the Kermadec Trench pelagic community and on larger size fractions has been suggested to be a result of adaptations to higher concentrations of organic matter, including particulate forms (Peoples et al., 2018a). Intra-trench variability may also be influenced by organic matter due to topographic funneling into the axis of the trench. Within the Kermadec Trench the Chloroflexi and Acidobacteria were enriched at 6- and 7-km depths whereas the Nitrospirae and Actinobacteria are more abundant at 8- and 9-km depths. Enrichments of Nitrospirae within the hadal water column have been attributed to a more eutrophic environment (Nunoura et al., 2015). Altogether, these findings suggest the Kermadec Trench may be enriched in organic matter relative to the Mariana Trench. Ultimately, deep-sea microbial communities are governed by a myriad of variables that may contribute to these inter- and intra-trench differences, such as hydrography (Hamdan et al., 2013), specific location of sample collection, temporal variability and season of sample collection (Lampitt, 1985), sediment lithology (Probandt et al., 2017), or the quality, not just the quantity, of organic matter present. Our findings support the notion that organic matter can contribute to the spatio-temporal variability in deep-sea microbial communities.
While trench sediments may become mixed and suspended due to tectonic activity and topographic instability (Kawagucci et al., 2012; Oguri et al., 2013; Nunoura et al., 2016), the sediment communities studied here were stratified and compositionally distinct from those in the water column above them (Peoples et al., 2018a). Because the oxygen penetration depth can be limited to the top 10 cm in trenches (Glud et al., 2013; Wenzhöfer et al., 2016), we compared the microbial communities present in the shallower and deeper sediment horizons. Community richness and diversity varied with sediment horizon depth, consistent with previous comparisons of surficial and deep subsurface environments (Durbin and Teske, 2010; Teske et al., 2011; Shulse et al., 2016; Walsh et al., 2016a,b). Our findings suggest that specific deep-sea lineages are enriched within deeper marine sediments. Within the Thaumarchaeota, MGI-α showed enrichment in the upper sediment while those belonging to the MGI-η subgroup were more abundant in the deeper sediments. Such ecotype differentiation has been previously noted in abyssal sediments (Durbin and Teske, 2010; Tully and Heidelberg, 2013; Lauer et al., 2016) and may be due to changes in organic matter abundance or oxygen availability (Durbin and Teske, 2010). Abundances of alternative electron acceptors, such as nitrate, sulfate, or iron, influence community composition (D’Hondt et al., 2009; Durbin and Teske, 2010, 2012; Nunoura et al., 2013; Jessen et al., 2017) and therefore likely play an important role in hadal sediments (Nunoura et al., 2013, 2018) as the oxygen penetration depth can be shallow (Glud et al., 2013; Wenzhöfer et al., 2016). Relative sequence abundances of the Woesearchaeota increased with increasing sediment horizon depth. This archaeal phylum has members that are specifically enriched in anoxic niches and can have fermentative or symbiotic lifestyles (Castelle et al., 2015; Lazar et al., 2017; Liu et al., 2018). Differences in genomic potential and niche separation of Woesearchaeota suggest that unexplored diversity exists within this phylum (Shcherbakova et al., 2016; Narrowe et al., 2017). OTUs related to the Woesearchaeota within our dataset were remarkably distinct from other previously identified studies, showing highest similarity to sequences from abyssal and hadal sites and low (<95%) similarity to representative sequences from other habitats (Supplementary Figure 10). The Marinimicrobia, which are abundant within the deep-ocean water column (Nunoura et al., 2015; Tarn et al., 2016; Peoples et al., 2018a), also increased with sediment horizon depth. Based on phylogenetic analysis (Supplementary Figure 6) the Marinimicrobia OTUs identified here belong to deep-sea sediment-associated clades that have not been previously identified (Hawley et al., 2017).
Although a large diversity of Bacteria and Archaea were found within the sediments, very few taxa were successfully isolated. Many of those that were belong to the same genera as known piezophiles, consistent with most previous isolation attempts from the deep sea. Interestingly, after unamended, long-term static pressurization of sediment samples, higher relative sequence abundances of taxa related to the cultured piezophiles Psychrobium, Colwellia, Arcobacter, and Shewanella were found. In shallow-ocean communities, Colwellia is one of the first responders to microcosm conditions (Stewart et al., 2012; Mayali et al., 2016) and enrichments of Colwelliaceae, Moritellaceae, Psychromonadaceae, and Shewanellaceae have been observed within enrichments of Arctic bathyal-depth sediment samples under high pressure (Hoffmann et al., 2017). Therefore, regardless of location, depth of collection (surface, bathyal, and in this study hadal), nutrient enrichment, or temperature or pressure incubation conditions, taxa belonging to the same heterotrophic and copiotrophic genera within the Gammaproteobacteria are enriched under mesocosm conditions. These taxa may contain metabolic versatility for colonizing various ecological niches during fluctuating environmental conditions (Stewart et al., 2012). In contrast, microbial populations from immediately recovered samples were enriched in taxa belonging to uncharacterized and uncultivated lineages representative of the in situ diversity. Taken together with the enrichment of culturable taxa, these findings highlight the difficulties in obtaining pure cultures of representative and novel deep-ocean lineages. This study is the first to show that long-term incubations in pressure vessels, regardless of nutrient amendment or collection location, clearly select for the same piezophilic taxa. Decompression during sample retrieval, static incubation conditions in pressure vessels leading to lack of oxygen or other important nutrients, or eukaryotic predation (Tsagaraki et al., 2018) may significantly bias our ability to obtain cultures of piezophiles.
Here, we present the first comparison of microbial communities within hadal sediments of two trenches. Sediment community composition varied by trench of collection, sediment horizon depth, and water column depth, changes predicted to be due in part to variations in organic matter concentrations. Future studies of hadal sediments should couple their analyses of community composition to organic matter concentrations and compositions. While the communities differed between the two trenches, neither appeared to be dominated by endemic microbial communities at the 97% OTU level. Instead, the hadal sediments shared many cosmopolitan taxa similar to those found in other abyssal and hadal sites, suggesting members of these lineages may be ubiquitous at hadal depths. These findings highlight the possibility of microbial dispersal over long distances between hadal zones. Whether these taxa are actually distinct, both between trenches and from those found at abyssal sites, will ultimately require whole genome comparisons and analyses of phenotypic plasticity, and not just partial 16S rRNA gene analyses. Culturing and in situ abundances of known piezophiles showed that these taxa represent a relatively small fraction of environmental samples and were enriched in the Kermadec Trench, perhaps because of affiliation with organic-rich conditions. After long-term batch incubation of sediments at in situ high hydrostatic pressure, these taxa came to dominate the communities at the expense of initially more abundant members. The attempted isolation of piezophiles extends back to the 1940s and yet very few taxa have been isolated, potentially because of the use of pressure vessels. Future work should attempt to more closely mimic in situ conditions using recirculating systems or, perhaps more effectively, attempt to enrich for microorganisms in situ, as current practices involving removing samples from the deep-ocean ultimately select for a few taxa that are not representative of deep-ocean sediment communities at large.
LP and DB were involved in experimental design. LP, EG, CN, JD, DM, and DB collected the samples. LP, EG, MP, XX, and OO performed the research. JB and EA provided methodological and technical advice. LP and DB wrote the manuscript. All authors approved the final version.
We would like to express our appreciation for the financial support provided by the National Science Foundation (0801973, 0827051, 1149552, 1536776), Schmidt Ocean Institute, the National Aeronautics and Space Administration (NNX11AG10G), the Prince Albert II Foundation (Project 1265), and a gift from Earthship Productions. DM received National Capability Funding from the Natural Environment Research Council (NERC).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to the crews of the R/V Falkor, R/V Thompson, and the HADal Ecosystem Studies (HADES) team for help at sea. Casey Machado (WHOI) was instrumental in the building of the FVCR. We would also like to thank NOAA, particularly those from the Mariana Trench Marine National Monument Office, for their assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00347/full#supplementary-material
Amante, C., and Eakins, B. W. (2009). ETOPO1 1 arc-minute global relief model: procedures, data sources and analysis. NOAA Technical Memorandum NESDIS NGDC‐24, 1–19.
Amend, J. P., and LaRowe, D. E. (2016). Ocean Sediments—an enormous underappreciated microbial habitat. Microbe 11, 427–432.
Aoki, M., Ehara, M., Saito, Y., Yoshioka, H., Miyazaki, M., Saito, Y., et al. (2014). A long-term cultivation of an anaerobic methane-oxidizing microbial community from deep-sea methane-seep sediment using a continuous-flow bioreactor. PLoS One 9, 1–14. doi: 10.1371/journal.pone.0105356
Bienhold, C., Boetius, A., and Ramette, A. (2012). The energy-diversity relationship of complex bacterial communities in Arctic deep-sea sediments. ISME J. 6, 724–732. doi: 10.1038/ismej.2011.140
Bienhold, C., Zinger, L., Boetius, A., and Ramette, A. (2016). Diversity and biogeography of bathyal and abyssal seafloor bacteria. PLoS One 11, 1–20. doi: 10.1371/journal.pone.0148016
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Castelle, C. J., Wrighton, K. C., Thomas, B. C., Hug, L. A., Brown, C. T., Wilkins, M. J., et al. (2015). Genomic expansion of domain Archaea highlights roles for organisms from new phyla in anaerobic carbon cycling. Curr. Biol. 25, 690–701. doi: 10.1016/j.cub.2015.01.014
D’Hondt, S., Spivack, A. J., Pockalny, R., Ferdelman, T. G., Fischer, J. P., Kallmeyer, J., et al. (2009). Subseafloor sedimentary life in the South Pacific Gyre. Proc. Natl. Acad. Sci. 106, 11651–11656. doi: 10.1073/pnas.0811793106
Danovaro, R., Della Croce, N., Dell’Anno, A., and Pusceddu, A. (2003). A depocenter of organic matter at 7800 m depth in the SE Pacific Ocean. Deep Sea Res. I 50, 1411–1420. doi: 10.1016/j.dsr.2003.07.001
Durbin, A. M., and Teske, A. (2010). Sediment-associated microdiversity within the Marine Group I Crenarchaeota. Environ. Microbiol. Rep. 2, 693–703. doi: 10.1111/j.1758-2229.2010.00163.x
Durbin, A. M., and Teske, A. (2012). Archaea in organic-lean and organic-rich marine subsurface sediments: an environmental gradient reflected in distinct phylogenetic lineages. Front. Microbiol. 3:168.
Fletcher, B., Bowen, A., Yoerger, D. R., and Whitcomb, L. L. (2009). Journey to the Challenger Deep: 50 years later with the Nereus hybrid remotely operated vehicle. Mar. Technol. Soc. J. 43, 65–76. doi: 10.4031/MTSJ.43.5.26
Fujiwara, Y., Kato, C., Masui, N., Fujikura, K., and Kojima, S. (2001). Dual symbiosis in the cold-seep thyasirid clam Maorithyas hadalis from the hadal zone in the Japan Trench, western Pacific. Mar. Ecol. Prog. Ser. 214, 151–159. doi: 10.3354/meps214151
Gardner, J. V., Armstrong, A. A., Calder, B. R., and Beaudoin, J. (2014). So, how deep is the Mariana Trench? Mar. Geod. 37, 1–13. doi: 10.1080/01490419.2013.837849
Glud, R. N., Wenzhöfer, F., Middelboe, M., Oguri, K., Turnewitsch, R., Canfield, D. E., et al. (2013). High rates of microbial carbon turnover in sediments in the deepest oceanic trench on Earth. Nat. Geosci. 6, 284–288. doi: 10.1038/ngeo1773
Hamdan, L. J., Coffin, R. B., Sikaroodi, M., Greinert, J., Treude, T., and Gillevet, P. M. (2013). Ocean currents shape the microbiome of Arctic marine sediments. ISME J. 7, 685–696. doi: 10.1038/ismej.2012.143
Hawley, A. K., Nobu, M. K., Wright, J. J., Durno, W. E., Morgan-Lang, C., Sage, B., et al. (2017). Diverse Marinimicrobia bacteria may mediate coupled biogeochemical cycles along eco-thermodynamic gradients. Nat. Commun. 8:1507. doi: 10.1038/s41467-017-01376-9
Hoehler, T. M., and Jorgensen, B. B. (2013). Microbial life under extreme energy limitation. Nat. Rev. Microbiol. 11, 83–94. doi: 10.1038/nrmicro2939
Hoffmann, K., Hassenrück, C., Salman-Carvalho, V., Holtappels, M., and Bienhold, C. (2017). Response of bacterial communities to different detritus compositions in Arctic deep-sea sediments. Front. Microbiol. 8, 1–18. doi: 10.3389/fmicb.2017.00266
Ichino, M. C., Clark, M. R., Drazen, J. C., Jamieson, A., Jones, D. O. B., Martin, A. P., et al. (2015). The distribution of benthic biomass in hadal trenches: a modeling approach to investigate the effect of vertical and lateral organic matter transport to the seafloor. Deep-Sea Res. I 100, 21–33. doi: 10.1016/j.dsr.2015.01.010
Inagaki, F., Hinrichs, K. -U., Kubo, Y., Bowles, M. W., Heuer, V. B., Hong, W. -L., et al. (2015). Exploring deep microbial life in coal-bearing sediment down to ~2.5 km below the ocean floor. Science 349, 420–424. doi: 10.1126/science.aaa6882
Jacob, M., Soltwedel, T., Boetius, A., and Ramette, A. (2013). Biogeography of deep-sea benthic bacteria at regional scale (LTER HAUSGARTEN, Fram Strait, Arctic). PLoS One 8, 1–10. doi: 10.1371/journal.pone.0072779
Jamieson, A. J. (2015). The hadal zone: life in the deepest oceans. London: Cambridge University Press.
Jessen, G. L., Lichtschlag, A., Ramette, A., Pantoja, S., Rossel, P. E., Schubert, C. J., et al. (2017). Hypoxia causes preservation of labile organic matter and changes seafloor microbial community composition (Black Sea). Sci. Adv. 3:e16017897. doi: 10.1126/sciadv.1601897
Kallmeyer, J., Pockalny, R., Adhikari, R. R., Smith, D. C., and D’Hondt, S. (2012). Global distribution of microbial abundance and biomass in subseafloor sediment. Proc. Natl. Acad. Sci. 109, 16213–16216. doi: 10.1073/pnas.1203849109
Kato, C., Li, L., Tamaoka, J., and Horikoshi, K. (1997). Molecular analyses of the sediment of the 11000-m deep Mariana Trench. Extremophiles 1, 117–123. doi: 10.1007/s007920050024
Kato, C., Li, L., Nogi, Y., Nakamura, Y., Tamaoka, J., and Horikoshi, K. (1998). Extremely barophilic bacteria isolated from the Mariana Trench, Challenger Deep, at a depth of 11,000 meters. Appl. Environ. Microbiol. 64, 1510–1513.
Kawagucci, S., Yoshida, Y. T., Noguchi, T., Honda, M. C., Uchida, H., Ishibashi, H., et al. (2012). Disturbance of deep-sea environments induced by the M9.0 Tohoku Earthquake. Sci. Rep. 2, 1–7. doi: 10.1038/srep00270
Kusube, M., Kyaw, T. S., Tanikawa, K., Chastain, R. A., Hardy, K. M., Cameron, J., et al. (2016). Colwellia marinimaniae sp. nov., a hyperpiezophilic species isolated from an amphipod within the Challenger Deep, Mariana Trench. Int. J. Syst. Evol. Microbiol. 67, 824–831. doi: 10.1099/ijsem.0.001671
Lampitt, R. S. (1985). Evidence for seasonal deposition of detritus to the deep-sea floor and its subsequent resuspension. Deep-Sea Res. 32, 885–897. doi: 10.1016/0198-0149(85)90034-2
Landry, Z., Swan, B. K., Herndl, G. J., Stepanauskas, R., and Giovannoni, S. J. (2017). SAR202 genomes from the dark ocean predict pathways for the oxidation of recalcitrant dissolved organic matter. MBio 8, 1–19. doi: 10.1128/mBio.00413-17
Lane, D. J. (1991). “16S/23S rRNA sequencing” in Nucleic acid techniques in bacterial systematics. eds. E. Stackebrandt and M. Goodfellow (New York: John Wiley and Sons), 115–175.
Lauer, A., Sorensen, K. B., and Teske, A. (2016). Phylogenetic characterization of marine benthic archaea in organic-poor sediments of the eastern equatorial Pacific Ocean (ODP Site 1225). Microorganisms 4, 1–14. doi: 10.3390/microorganisms4030032
Lazar, C. S., Baker, B. J., Seitz, K. W., and Teske, A. P. (2017). Genomic reconstruction of multiple lineages of uncultured benthic archaea suggests distinct biogeochemical roles and ecological niches. ISME J. 11, 1118–1129. doi: 10.3390/microorganisms4030032
Learman, D. R., Henson, M. W., Thrash, J. C., Temperton, B., Brannock, P. M., Santos, S. R., et al. (2016). Biogeochemical and microbial variation across 5500 km of Antarctic surface sediment implicates organic matter as a driver of benthic community structure. Front. Microbiol. 7, 1–11. doi: 10.3389/fmicb.2016.00284
León-Zayas, R., Peoples, L., Biddle, J. F., Podell, S., Novotny, M., Cameron, J., et al. (2017). The metabolic potential of the single cell genomes obtained from the Challenger Deep, Mariana Trench within the candidate superphylum Parcubacteria (OD1). Environ. Microbiol. 19, 2769–2784. doi: 10.1111/1462-2920.13789
Letunic, I., and Bork, P. (2007). Interactive Tree of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128. doi: 10.1093/bioinformatics/btl529
Lever, M. A., Torti, A., Eickenbusch, P., Michaud, A. B., Santl-Temkiv, T., and Jorgensen, B. B. (2015). A modular method for the extraction of DNA and RNA, and the separation of DNA pools from diverse environmental sample types. Front. Microbiol. 6, 1–25. doi: 10.3389/fmicb.2015.00476
Li, L., Kato, C., and Horikoshi, K. (1999). Microbial diversity in sediments collected from the deepest cold-seep area, the Japan Trench. Mar. Biotechnol. 1, 391–400.
Liu, X., Li, M., Castelle, C. J., Probst, A. J., Zhou, Z., Pan, J., et al. (2018). Insights into the ecology, evolution, and metabolism of the widespread Woesearchaeotal lineages. Microbiome 6:102. doi: 10.1186/s40168-018-0488-2
Longhurst, A., Sathyendranath, S., Platt, T., and Caverhill, C. (1995). An estimate of global primary production in the ocean from satellite radiometer data. J. Plankton Res. 17, 1245–1271. doi: 10.1093/plankt/17.6.1245
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Luo, M., Gieskes, J., Chen, L., Shi, X., and Chen, D. (2017). Provenances, distribution, and accumulation of organic matter in the southern Mariana Trench rim and slope: implication for carbon cycle and burial in hadal trenches. Mar. Geol. 386, 98–106. doi: 10.1016/j.margeo.2017.02.012
Luo, Z., Xu, W., Li, M., Gu, J., and Zhong, T. (2015). Spatial distribution and abundance of ammonia-oxidizing microorganisms in deep-sea sediments of the Pacific Ocean. Van Leeuw J. Microb. 108, 329–342. doi: 10.1007/s10482-015-0485-4
Magoc, T., and Salzberg, S. L. (2011). FlASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Mayali, X., Stewart, B., Mabery, S., and Weber, P. K. (2016). Temporal succession in carbon incorporation from macromolecules by particle-attached bacteria in marine microcosms. Environ. Microbiol. Rep. 8, 68–75. doi: 10.1111/1758-2229.12352
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
McMurdie, P. J., and Holmes, S. (2014). Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 10:e1003531. doi: 10.1371/journal.pcbi.1003531
Mußmann, M., Pjevac, P., Krüger, K., and Dyksma, S. (2017). Genomic repertoire of the Woeseiaceae/JTB255, cosmopolitan and abundant core members of microbial communities in marine sediments. ISME J. 11, 1276–1281. doi: 10.1038/ismej.2016.185
Müller, A. L., Rosa de Rezende, J., Hubert, C. R. J., Kjeldsen, K. U., Lagkouvardos, I., Berry, D., et al. (2014). Endospores of thermophilic bacteria as tracers of microbial dispersal by ocean currents. ISME J. 8, 1153–1165. doi: 10.1038/ismej.2013.225
Narrowe, A. B., Angle, J. C., Daly, R. A., Stefanik, K. C., Wrighton, K. C., and Miller, C. S. (2017). High-resolution sequencing reveals unexplored archaeal diversity in freshwater wetland soils. Environ. Microbiol. 19, 2192–2209. doi: 10.1111/1462-2920.13703
Nogi, Y., Kato, C., and Horikoshi, K. (1998). Taxonomic studies of deep-sea barophilic Shewanella strains and description of Shewanella violacea sp. nov. Arch. Microbiol. 170, 331–338. doi: 10.1007/s002030050650
Nogi, Y., Kato, C., and Horikoshi, K. (2002). Psychromonas kaikoae sp. nov., a novel piezophilic bacterium from the deepest cold-seep sediments in the Japan Trench. Int. J. Syst. Evol. Microbiol. 52, 1527–1532. doi: 10.1099/00207713-52-5-1527
Nogi, Y., Hosoya, S., Kato, C., and Horikoshi, K. (2004). Colwellia piezophila sp. nov., a novel piezophilic species from deep-sea sediments of the Japan Trench. Int. J. Syst. Evol. Microbiol. 54, 1627–1631. doi: 10.1099/ijs.0.03049-0
Nogi, Y., Hosoya, S., Kato, C., and Horikoshi, K. (2007). Psychromonas hadalis sp. nov., a novel piezophilic bacterium isolated from the bottom of the Japan Trench. Int. J. Syst. Evol. Microbiol. 57, 1360–1364. doi: 10.1099/ijs.0.64933-0
Nunoura, T., Nishizawa, M., Kikuchi, T., Tsubouchi, T., Hirai, M., Koide, O., et al. (2013). Molecular biological and isotopic biogeochemical prognoses of the nitrification-driven dynamic microbial nitrogen cycle in hadopelagic sediments. Environ. Microbiol. 15, 3087–3107. doi: 10.1111/1462-2920.12152
Nunoura, T., Takaki, Y., Hirai, M., Shimamura, S., Makabe, A., Koide, O., et al. (2015). Hadal biosphere: insight into the microbial ecosystem in the deepest ocean on Earth. Proc. Natl. Acad. Sci. 112, E1230–E1236. doi: 10.1073/pnas.1421816112
Nunoura, T., Hirai, M., Yoshida-Takashima, Y., Nishizawa, M., Kawagucci, S., Yokokawa, T., et al. (2016). Distribution and niche separation of planktonic microbial communities in the water columns from the surface to the hadal waters of the Japan Trench under eutrophic ocean. Front. Microbiol. 7, 1–12. doi: 10.3389/fmicb.2016.01261
Nunoura, T., Nishizawa, M., Hirai, M., Shimamura, S., Harnvoravongchai, P., Koide, O., et al. (2018). Microbial diversity in sediments from the bottom of the Challenger Deep, the Mariana Trench. Microbes Environ. 33, 186–194. doi: 10.1264/jsme2.ME17194
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2017). vegan: Community Ecology Package. https://cran.r-project.org/web/packages/vegan/vegan.pdf.
Oguri, K., Kawamura, K., Sakaguchi, A., Toyofuku, T., Kasaya, T., Murayama, M., et al. (2013). Hadal disturbance in the Japan Trench induced by the 2011 Tohoku-Oki Earthquake. Sci. Rep. 3, 1–6. doi: 10.1038/srep01915
Pante, E., and Simon-Bouhet, B. (2013). marmap: a package for importing, plotting, and analyzing bathymetric and topographic data in R. PLoS One 8:e73051. doi: 10.1371/journal.pone.0073051
Parada, A. E., Needham, D. M., and Fuhrman, J. A. (2015). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Peoples, L. M., Donaldson, S., Osuntokun, O., Xia, Q., Nelson, A., Blanton, J., et al. (2018a). Vertically distinct microbial communities in the Mariana and Kermadec trenches. PLoS One 13:e0195102. doi: 10.1371/journal.pone.0195102
Peoples, L. M., Norenberg, M., Price, D., McGoldrick, M., Novotny, M., Bochdansky, A., et al. (2018b). A full-ocean-depth rated modular lander and pressure-retaining sampler capable of collecting hadal-endemic microbes under in situ conditions. Deep Search Res. 143, 50–57. doi: 10.1016/j.dsr.2018.11.010
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2 – Approximately maximum-likelihood trees for large alignments. PLoS One 5, 1–10. doi: 10.1371/journal.pone.0009490
Probandt, D., Knittel, K., Tegetmeyer, H. E., Ahmerkamp, S., Holtappels, M., and Amann, R. (2017). Permeability shapes bacterial communities in sublittoral surface sediments. Environ. Microbiol. 19, 1584–1599. doi: 10.1111/1462-2920.13676
Pruesse, E., Peplies, J., and Glockner, F. O. (2012). SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28, 1823–1829. doi: 10.1093/bioinformatics/bts252
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Research 41, D590–D596. doi: 10.1093/nar/gks1219
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahe, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
Ruff, S. E., Biddle, J. F., Teske, A. P., Knittel, K., Boetius, A., and Ramette, A. (2015). Global dispersion and local diversification of the methane seep microbiome. Proc. Natl. Acad. Sci. 112, 4015–4020. doi: 10.1073/pnas.1421865112
Shulse, C. N., Maillot, B., Smith, C. R., and Church, M. J. (2016). Polymetallic nodules, sediments, and deep waters in the equatorial North Pacific exhibit highly diverse and distinct bacterial, archaeal, and microeukaryotic communities. Microbiol. Open 6, 1–16. doi: 10.1002/mbo3.428
Shcherbakova, V., Yoshimura, Y., Ryzhmanova, Y., Taguchi, Y., Segawa, T., Oshurkova, V., et al. (2016). Archaeal communities of Arctic methane-containing permafrost. FEMS Microbiol. Ecol. 92, 1–11. doi: 10.1093/femsec/fiw135
Starnawski, P., Bataillon, T., Ettema, T. J. G., Jochum, L. M., Schreiber, L., Chen, X., et al. (2017). Microbial community assembly and evolution in subseafloor sediment. Proc. Natl. Acad. Sci. 114, 2940–2945. doi: 10.1073/pnas.1614190114
Stewart, F. J., Dalsgaard, T., Young, C. R., Thamdrup, B., Revsbech, N. P., Ulloa, O., et al. (2012). Experimental incubations elicit profound changes in community transcription in OMZ bacterioplankton. PLoS One 7, 1–12. doi: 10.1371/journal.pone.0037118
Tarn, J., Peoples, L. M., Hardy, K., Cameron, J., and Bartlett, D. H. (2016). Identification of free-living and particle-associated microbial communities present in hadal regions of the Mariana Trench. Front. Microbiol. 7:665. doi: 10.3389/fmicb.2016.00665
Teske, A., Durbin, A., Ziervogel, K., Cox, C., and Arnosti, C. (2011). Microbial community composition and function in permanently cold seawater and sediments from an Arctic fjord of Svalbard. Appl. Environ. Microbiol. 77, 2008–2018. doi: 10.1128/AEM.01507-10
Tsagaraki, T. M., Pree, B., Leiknes, Ø., Larsen, A., Bratbak, G., Øvreås, L., et al. (2018). Bacterial community composition responds to changes in copepod abundance and alters ecosystem function in an Arctic mesocosm study. ISME J. 12, 2694–2705. doi: 10.1038/s41396-018-0217-7
Tully, B. J., and Heidelberg, J. F. (2013). Microbial communities associated with ferromanganese nodules and the surrounding sediments. Front. Microbiol. 4, 1–10. doi: 10.3389/fmicb.2013.00161
Valentine, D. L. (2007). Adaptations to energy stress dictate the ecology and evolution of the archaea. Nat. Rev. Microbiol. 5, 316–323. doi: 10.1038/nrmicro1619
Walsh, E. A., Kirkpatrick, J. B., Pockalny, R., Sauvage, J., Spivack, A. J., Murray, R. W., et al. (2016a). Relationship of bacterial richness to organic degradation rate and sediment age in subseafloor sediment. Appl. Environ. Microbiol. 82, 4994–4999. doi: 10.1128/AEM.00809-16
Walsh, E. A., Kirkpatrick, J. B., Rutherford, S. D., Smith, D. C., Sogin, M., and D’Hondt, S. (2016b). Bacterial diversity and community composition from sea surface to subseafloor. ISME J. 10, 979–989. doi: 10.1038/ismej.2015.175
Wenzhöfer, F., Oguri, K., Middelboe, M., Turnewitsch, R., Toyofuku, T., Kitazato, H., et al. (2016). Benthic carbon remineralization in hadal trenches: assessment by in situ O2 microprofile measurements. Deep-Search Res. 116, 276–286. doi: 10.1016/j.dsr.2016.08.013
Yanagibayashi, M., Nogi, Y., Li, L., and Kato, C. (1999). Changes in the microbial community in the Japan Trench sediment from a depth of 6292 m during cultivation without decompression. FEMS Microbiol. Lett. 170, 271–279. doi: 10.1111/j.1574-6968.1999.tb13384.x
Yayanos, A. A. (2001). Deep-sea piezophilic bacteria. Methods Microbiol. 30, 615–637. doi: 10.1016/S0580-9517(01)30065-X
Yoshida, M., Takaki, Y., Eitoku, M., Nunoura, T., and Takai, K. (2013). Metagenomic analysis of viral communities in (hado)pelagic sediments. PLoS One 8, 1–14. doi: 10.1371/journal.pone.0057271
Yu, G., Smith, D. K., Zhu, H., Guan, Y., and Lam, T. T. Y. (2017). GGTREE: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36. doi: 10.1111/2041-210X.12628
Zinger, L., Amaral-Zettler, L. A., Furhman, J. A., Claire Horner-Devine, M., Huse, S. M., Mark Welch, D. B., et al. (2011). Global patterns of bacterial beta-diversity in seafloor and seawater ecosystems. PLoS One 6, 1–11. doi: 10.1371/journal.pone.0024570
Keywords: hadal, trench, sediment, pressure, piezophile
Citation: Peoples LM, Grammatopoulou E, Pombrol M, Xu X, Osuntokun O, Blanton J, Allen EE, Nunnally CC, Drazen JC, Mayor DJ and Bartlett DH (2019) Microbial Community Diversity Within Sediments from Two Geographically Separated Hadal Trenches. Front. Microbiol. 10:347. doi: 10.3389/fmicb.2019.00347
Received: 06 September 2018; Accepted: 11 February 2019;
Published: 15 March 2019.
Edited by:
Ian Hewson, Cornell University, United StatesReviewed by:
Rulong Liu, Shanghai Ocean University, ChinaCopyright © 2019 Peoples, Grammatopoulou, Pombrol, Xu, Osuntokun, Blanton, Allen, Nunnally, Drazen, Mayor and Bartlett. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Douglas H. Bartlett, ZGJhcnRsZXR0QHVjc2QuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.