
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 21 February 2019
Sec. Microbiotechnology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00334
Enzymatic degumming is an effective approach to produce nutritional, safe, and healthy refined oil. However, the high cost and low efficiency of phospholipase limit the application of enzymatic degumming. In this study, an 879 bp outer membrane phospholipase A (A1) (OM-PLA1) gene encoding 292 amino acid residues was isolated from the genome of Serratia marcescens. The recombinant OM-PLA1 profile of appropriately 33 KDa was expressed by the engineered Pichia pastoris GS115. The OM-PLA1 activity was 21.2 U/mL with the induction of 1 mM methanol for 72 h. The expression efficiencies of OM-PLA1 were 0.29 U/mL/h and 1.06 U/mL/OD600. A complex of magnetic graphene oxide (MGO) and OM-PLA1 (MGO-OM-PLA1) was prepared by immobilizing OM-PLA1 with graphene oxide-based Fe3O4 nanoparticles by cross-linking with glutaraldehyde. The content of phosphorus decreased to 5.1 mg/kg rapeseed oil from 55.6 mg/kg rapeseed oil with 0.02% MGO-OM-PLA1 (w/w) at 50∘C for 4 h. MGO-OM-PLA1 retained 51.7% of the initial activity after 13 times of continuous recycling for the enzymatic degumming of rapeseed oil. This study provided an effective approach for the enzymatic degumming of crude vegetable oil by developing a novel phospholipase and improving the degumming technology.
High-quality edible oils should meet the requirements of good stability, long shelf life, bland odor and taste, good nutritional quality, abundant vitamins, and no contaminants (Nykter et al., 2006; Goyal et al., 2014). Most types of crude vegetable oils extracted using traditional pressing or extraction technology contain colloidal substances, mainly phospholipids (Gofferje et al., 2014). Phospholipids are usually combined with impurities, such as proteins, mucus, and trace metals, in crude vegetable oils (Elena et al., 2016). These colloidal substances directly reduce the quality of oils and affect subsequent refining processes (Elena et al., 2017). Water degumming removes 88.5% and retains 51.1 ppm of phosphorus in oil (Zufarov et al., 2009). The residual phosphorus in hydrated oil could be removed by using acid and enzymatic degumming approaches. Enzymatic oil degumming was industrially proven and well accepted for its advantages of energy saving and environmental protection compared with acid degumming (Sampaio et al., 2015). However, the current high cost of phospholipase increases the production cost of oil and limits the wide application of enzymatic degumming. In addition, the residual phospholipase cannot be recovered and reused and still remain in the oil by free phospholipase degumming. Therefore, the preparation of high-quality phospholipase and its reuse by immobilization determine the application degree of enzymatic degumming (Sampaio et al., 2015).
Outer-membrane phospholipase A (A1) (OM-PLA1) is an enzyme present in the outer membrane of Gram-negative bacteria (Dekker, 2000). OM-PLA1 hydrolyzes the acylester bonds in phospholipids and lysophospholipids. OM-PLA1 possesses a more conserved and specific determinant in the lipid headgroup compared with other phospholipases (Martin et al., 2000). In the glyceryl complexes, OM-PLA1 has high tolerance to the headgroups of triglyceride, phosphatidylglycerol, phosphatidylcholine, and phosphatidylethanolamine (Stanley et al., 2007). In addition, crude vegetable oils widely contain the glyceryl complexes of glycerophosphate, glycerophosphatidyl choline, and glycerophosphatidyl ethanolamine (Sampaio et al., 2015). OM-PLA1 is an effective alternative for the degumming of crude plant oil (Manjula et al., 2010). Therefore, in this study, a novel OM-PLA1 from S. marcescens was applied to crude rapeseed oil degumming.
The protein loading of magnetic graphene oxide (MGO) is much higher than that of granular activated carbon, diatomite and powdered activated carbon (Deng et al., 2013). Graphene oxide is an important derivative of graphene. The surface of graphene oxide has many oxygen-containing functional groups (hydroxyl, epoxy, carboxyl, carbonyl) and can be loaded with metal or metal oxides (Deng et al., 2008). The surface of graphene oxide Fe3O4 nanocomposite is adhering to magnetic nanoiron with superparamagnetism, which makes it have good magnetic separation characteristics and strong adsorption capacity. In addition, graphene oxide Fe3O4 nanocomposite also has advantages of high specific surface area, super strength, high activity recovery, and immobilization efficiency of enzyme compared to other immobilization approaches (Huang et al., 2014; Xie and Huang, 2018).
As an edible vegetable oil, crude rapeseed oil contains substantial phospholipids and causes oil discoloration and low oil quality (Yang et al., 2013; Jiang X. et al., 2014). Engineered P. pastoris can effectively express exogenous protein (Tolner et al., 2006). In this study, the S. marcescens OM-PLA1 expression vector was transformed into the cells of P. pastoris GS115. The gene expression and OM-PLA1 immobilization for crude rapeseed oil degumming were investigated to develop an effective approach for the enzymatic degumming of vegetable oil (Figure 1).
S. marcescens isolated from the intestine of smelly mandarin fish (Siniperca chuatsi) was preserved in College of Food and Biological Engineering, Hefei University of Technology. The smelly mandarin fish is a processed food product sold in market using traditional fermentation technology (Yang et al., 2017). The use of the dead mandarin fish is not unnecessary to obtain the approval from the institutional review board or ethics committee prior to commencing this study. P. pastoris GS115 and plasmid pPIC9K were provided by Dr. Huang from Qingdao Vland Biotech Company. Chemical reagents were from Beijing Transgen Biotech Company. Gene sequencing and primer synthesis were performed by Shanghai Sangon Biotech.
According to the gene sequences of OM-PLA1 in the NCBI database (HG326223.1), primer pair Us-OM-PLA1 and Ds-OM-PLA1 was synthesized to amplify the S. marcescens genome for OM-PLA1 cloning using PCR amplification instrument (TOMOS, United States) (Table 1). A phylogenetic tree was drawn by using Lagergene MegAlign software. The OM-PLA1 gene was inserted into plasmid pEASY-E1. The recombinant plasmid was named pEASY-OM-PLA1 after sequencing confirmation. Then, pEASY-OM-PLA1 was used to amplify OM-PLA1 with upstream primer Us-SnabI-OM-PLA1 and downstream primer Ds-AvrII-OM-PLA1. OM-PLA1 carrying cohesive ends was inserted into the pPIC9K plasmid digested by SnabI and AvrII (Gohel and Singh, 2012; Gamerith et al., 2017). The recombinant plasmid was named pPIC9K-OM-PLA1 by sequencing confirmation. pPIC9K-OM-PLA1 was transformed into P. pastoris GS115 by electroporation (Guo et al., 2015). The electrophoresis device and gel imaging system for DNA test were from Bio-Rad Company (United States).
The inducible expression of OM-PLA1-engineered P. pastoris GS115 was investigated under different methanol concentrations and induction times (Bredell et al., 2018). Fermentation media were prepared by yeast extract 1% (w/v), peptone 2% (w/v), 100 mM pH6 potassium phosphate buffer, YNB 1.34% (w/v), biotin 4 × 10-5 % (w/v), and glycerol 1% (w/v). The engineered P. pastoris colony was cultured in fermentation media at a shaking speed of 200 rpm at 30°C for 36 h. The collected P. pastoris cells were inoculated into the fermentation media added with 1 mM methanol. The final cell concentration at OD600 of 20 was cultured with a shaking speed of 200 rpm at 30°C. A Ni2+-chelating affinity chromatography column (Amersham Pharmacia Biotech, United Kingdom) was pre-equilibrated by a pH 8 buffer prepared by 20 mM imidazole, 50 mM NaH2PO4, and 300 mM NaCl. After the recombinant OM-PLA1 was purified by the column (Wang et al., 2009), OM-PLA1 profile analysis was performed by SDS-PAGE (Bio-Rad, United States) (Bredell et al., 2018).
Magnetic Fe3O4/graphene oxide (MGO) was prepared by mixing 1.5 g of freeze-dried graphene oxide, 0.3 g of FeSO4⋅7H2O, and 0.4 g of FeCl3⋅6H2O in 50 mL of deionized water (Hasanzadeh and Shadjou, 2013; Amirbandeh and Taheri-Kafrani, 2016; Zhuang et al., 2016). OM-PLA1 was immobilized by mixing glutaraldehyde, OM-PLA1, 1 g of MGO, and 50 mL of the immobilization buffer prepared by 0.01 M citric acid and 0.02 M hydrogen phosphate disodium. The parameters of pH, treatment time, and glutaraldehyde concentration were investigated to produce the MGO-OM-PLA1 complex. MGO-OM-PLA1 was separated and recovered by magnets and washed three times by the immobilization buffer (Qiu et al., 2010). Field emission scanning electron microscope (FE-SEM, Hitachi SU8020, Japan) and Fourier transform infrared spectroscope (FTIR, Thermo Nicolet 67, United States) were used to observe the microstructure and the spectral characteristic of MGO-OM-PLA1, respectively (He et al., 2013; Huang et al., 2015).
Enzymatic degumming was performed by adding MGO-OM-PLA1 in the crude rapeseed oil (Jiang X. F. et al., 2014). The phosphorus content of crude rapeseed oil was 55.6 mg/kg. Enzymatic degumming was carried out with the addition of 0.02% MGO-OM-PLA1 (w/w) into a weight of 150 g crude rapeseed oil at 50°C. MGO-OM-PLA1 was recovered by magnets to terminate the degumming reaction. The times of rapeseed oil degumming were determined when the phosphorus concentration was lower than 10 mg/kg rapeseed oil at each time. The recovered MGO-OM-PLA1 particles were reused for the next degumming of rapeseed oil.
Phosphorus content was determined by a spectrophotometric method in accordance with Official Method Ca 12–55 using ultraviolet visible near-infrared spectrophotometer (Agilent CARY5000, United States) (Chen et al., 2014). The activity of OM-PLA1 was measured by the colorimetric assay using a pH indicator (Araújo and Radvanyi, 1987; Pires et al., 2017). Triton–soybean lecithin was prepared by using 10 g of soybean lecithin dissolved in 200 mL of 0.02% Triton X-100 solution (w/v). A 98 mL volume of Triton–soybean lecithin and 2 mL of fermentation solution were mixed. The pH of the mixture was adjusted to 10 by adding 0.01 M NaOH. The activity of OM-PLA1 was determined based on the consumed volume of NaOH. The activity of phospholipase is defined as a unit of phospholipase activity required to hydrolyze phospholipids for 1 min to produce 1 μmol of free fatty acids (Sutto-Ortiz et al., 2017). Data are presented as mean ± standard deviation (SD). Figures were drawn using Software Origin.
The size of the isolated S. marcescens OM-PLA1 is 879 bp after sequencing confirmation (Figure 2), which encodes 292 amino acid residues. The phylogenetic tree of phospholipase sequences from 19 different species was drawn (Figure 3). S. marcescens OM-PLA1 showed the closest relationship to Escherichia coli OM-PLA1 NC_000913.3 among the tested phospholipase-producing microorganisms. A close genetic relationship also existed between Vibrio parahaemolyticus OM-PLA NC_004603.1 and Aliivibrio wodanis PLA NZ_LN554848.1, Ralstonia mannitolilytica PLA NZ_CP010799.1, and Ralstonia pickettii PLA NC_010682.1. However, S. marcescens OM-PLA1 has a far genetic relationship with Sphingomonas sp. OM-PLA NC_008308.1 compared with other phospholipase genes.
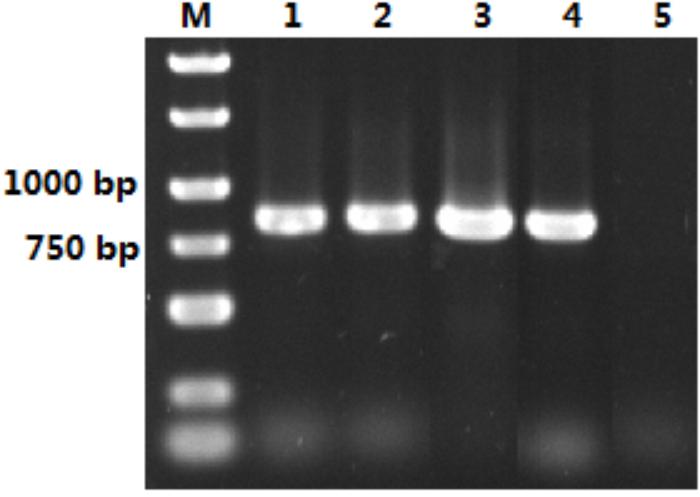
Figure 2. Isolation of OM-PLA1 from the genome of S. marcescens. Note: Lane M represented DNA Marker; lane 1–4 represented PCR amplification product of OM-PLA1; lane 5 represented the control.
The molecular weight of OM-PLA1 was appropriately 33 KDa by SDS-PAGE (Figure 4). The induction time of methanol affecting the expression of recombinant OM-PLA1 was investigated (Figure 5). The wild-type P. pastoris could not produce OM-PLA1 during the induction processing of methanol. The engineered P. pastoris could effectively produce the recombinant OM-PLA1. The activity of OM-PLA1 was 21.2 U/mL after induction by 1 mM methanol for 72 h. The production efficiencies of treatment time and cell concentration were 0.29 U/mL/h and 1.06 U/mL/OD600, respectively.
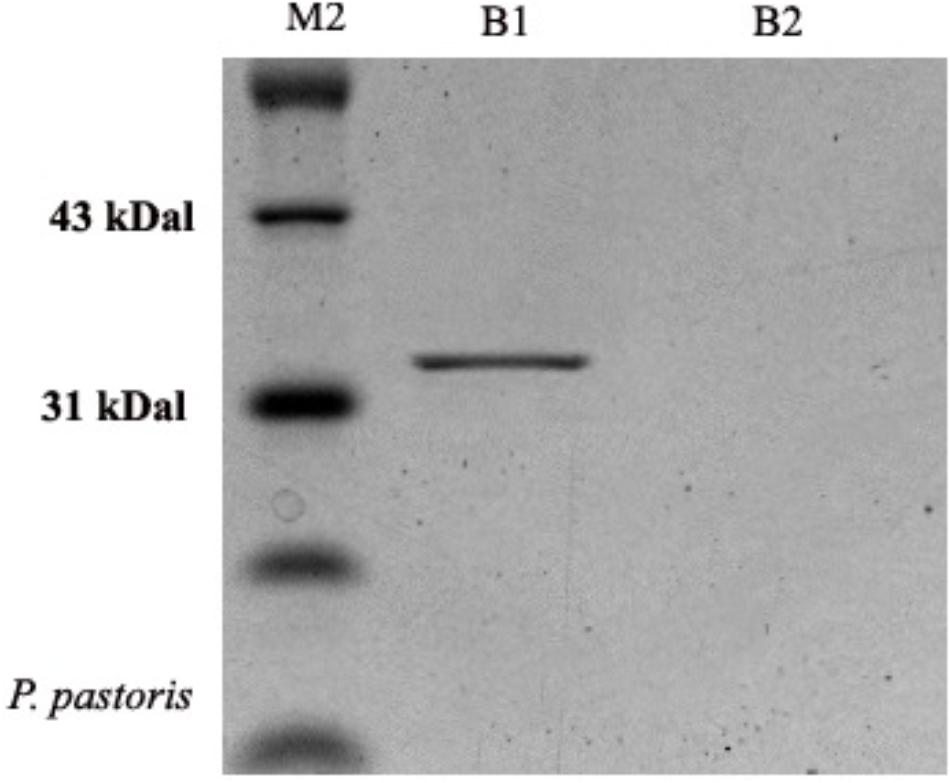
Figure 4. Profile of recombinant OM-PLA1 from P. pastoris by SDS-PAGE approach. Note: Lane M2 represented protein marker; B1 and B2 respectively represented the protein from the engineered and wild type P. pastoris GS 115.
FE-SEM was used to investigate the microcosmic difference between GO and the prepared Fe3O4/GO (Figure 6). The whole structure of GO was complete. The edge outline was clear and neat. The exfoliated lamellae of GO showed a smooth, thin, and large surface. No visible objects attached onto the surface of GO (Figure 6A). Figure 6B shows that the surface of the whole material was rough and convex. The iron oxide nanoparticles were largely anchored on the lamellae of GO in a highly dispersed state.
Very few of the GO were still exposed without attachment of granular objects. Fe3O4 particles showed spherical agglomeration. The thickness of coverage by Fe3O4 particles was inhomogeneous. The Fe3O4 particle size ranged from 10 to 30 nm. A small amount of Fe3O4 particles exceeded 50 nm of size.
FTIR spectroscopy was used to observe the preparation of MGO particles (Figure 7). Multiple oxygen-containing functional groups existed on the surface and edge of GO. The absorption peaks of MGO particles obviously differed from those of graphite oxide. The characteristic absorption peaks at 3412, 1712, 1628, 1197, and 592 cm-1 corresponded to the stretching vibrations of O-H, C = O, C = C, C-O, and Fe-O. In the Fe3O4/GO composite, the absorption peaks at 1712, 1628, and 1197 cm-1 were greatly weakened in comparison with those of GO. The absorption peak of epoxy at 1197 cm-1 almost disappeared in the Fe3O4-GO composite, which reflected that chemical bonding existed between these oxygen-containing functional groups and Fe3O4 particles. In addition, the Fe-O absorption peaks from Fe3O4-GO at 592 cm-1 were significantly strengthened, which further proved that a close combination existed between Fe3O4 and GO.
The effects of pH, glutaraldehyde concentration, and immobilization time on MGO-OM-PLA1 preparation were investigated (Figure 8). At pH 6, the immobilization efficiency of OM-PLA1 reached 61.7% (w/w), which was the highest among the set parameters (Figure 8A). The highest immobilization efficiency was achieved when the glutaraldehyde concentration (v/v) was 7% (Figure 8B). In addition, the efficiency of OM-PLA1 immobilization reached 60.9% (w/w) after 3 h of treatment (Figure 8C), which was the higher than those at other times.
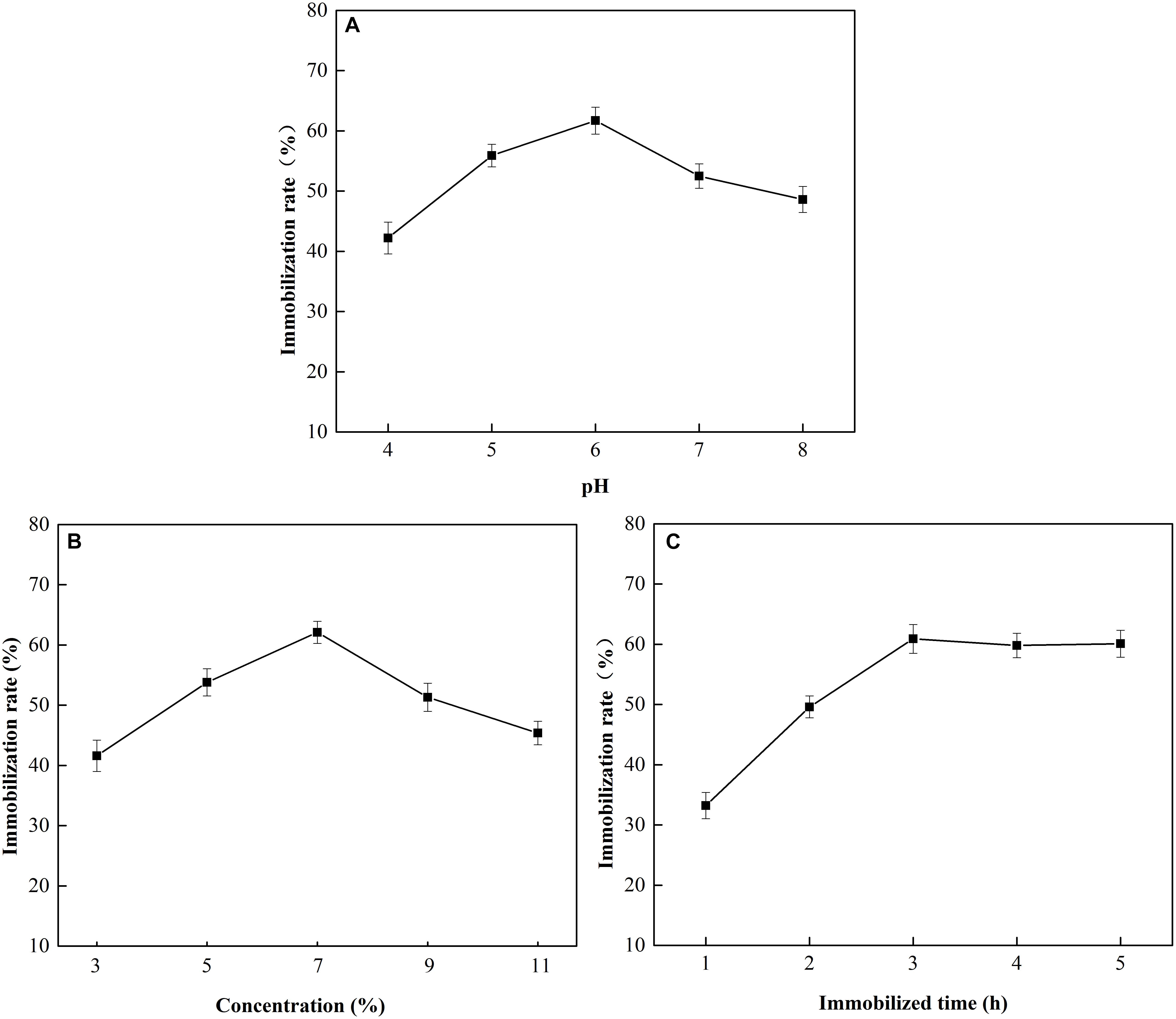
Figure 8. Effect of pH, glutaraldehyde concentration, and immobilization time on MGO-OM-PLA1 preparation. (A–C) Represented pH, glutaraldehyde concentration, and immobilization time, respectively.
The effect of pH and temperature on the activity of MGO-OM-PLA1 was investigated (Figures 9A,B). Under the conditions of pH 7.5 (Figure 9A) and 50°C (Figure 9B), the activity of MGO-OM-PLA1 reached the highest among the set parameters. Therefore, in this study, the parameters of pH 7.5 and 50°C were used to investigate the enzymatic degumming of crude vegetable oils.
MGO-OM-PLA1 was used in the enzymatic degumming of crude rapeseed oil (Figure 10). The contents of phosphorus in crude rapeseed oil gradually decreased with prolonged treatment time. The content of 0.02% MGO-OM-PLA1 (w/w) was used to perform enzymatic degumming with pH 7.5 at 50°C. After degumming for 2.5 h, the phosphorus content was 8.9 mg/kg rapeseed oil (below 10 mg/kg) from the initial concentration of 55.6 mg/kg rapeseed oil. After 4 h of degumming, the phosphorus contents further decreased to 5.1 mg/kg rapeseed oil.
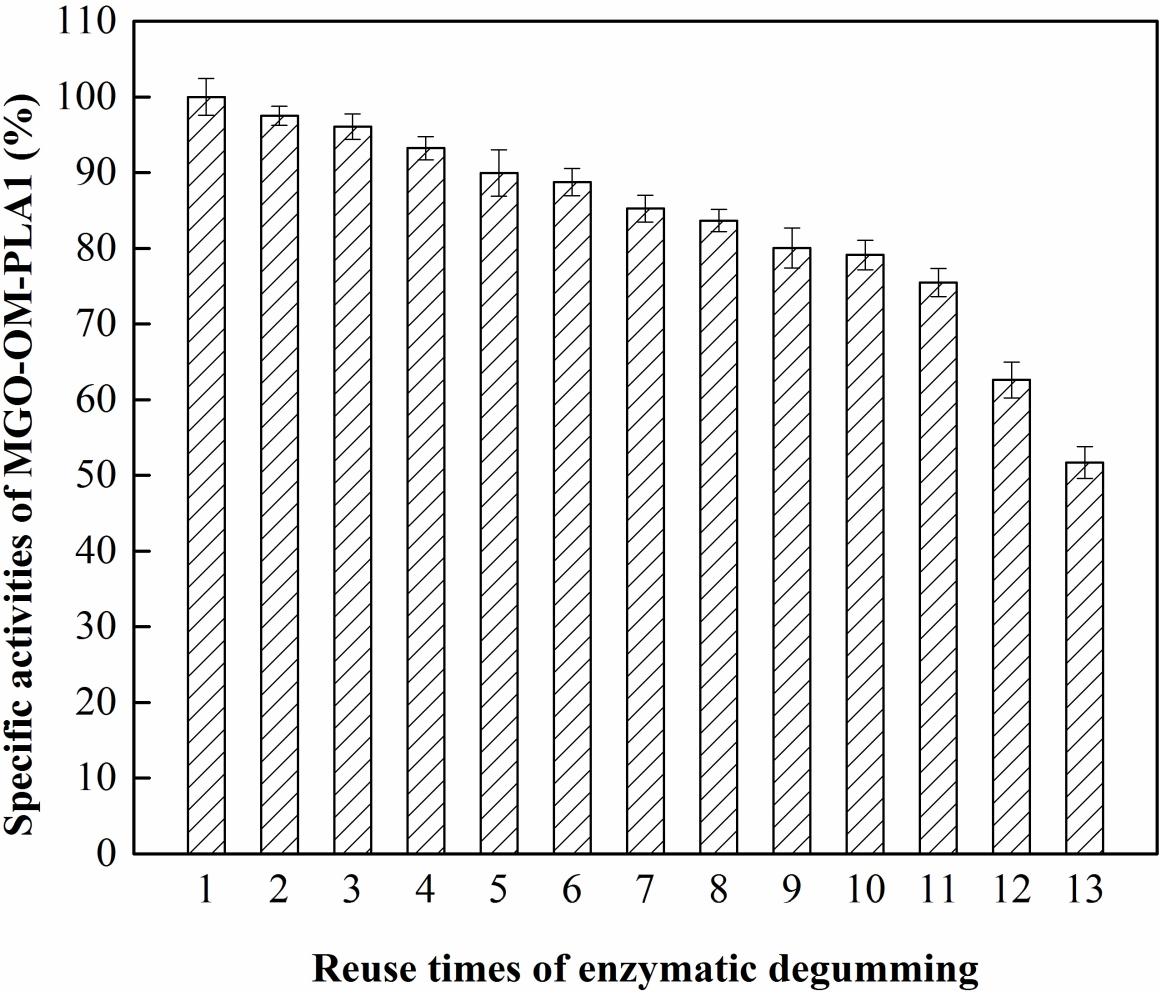
Figure 10. MGO-OM-PLA1 enzymatic degumming of crude rapeseed oil under the conditions of pH 7.5 at 50°C.
The effect of recycling times on MGO-OM-PLA1 activity was investigated (Figure 11). The activity of MGO-OM-PLA1 gradually decreased with prolonged degumming time. MGO-OM-PLA1 retained 90% of the initial activity after degumming for five times. After 13 times of recycling, the activity of MGO-OM-PLA1 decreased to 51.7% of the initial activity of the enzyme. Therefore, the half life of MGO-OM-PLA1 activity was regarded as 13 times for the enzymatic degumming of rapeseed oil.
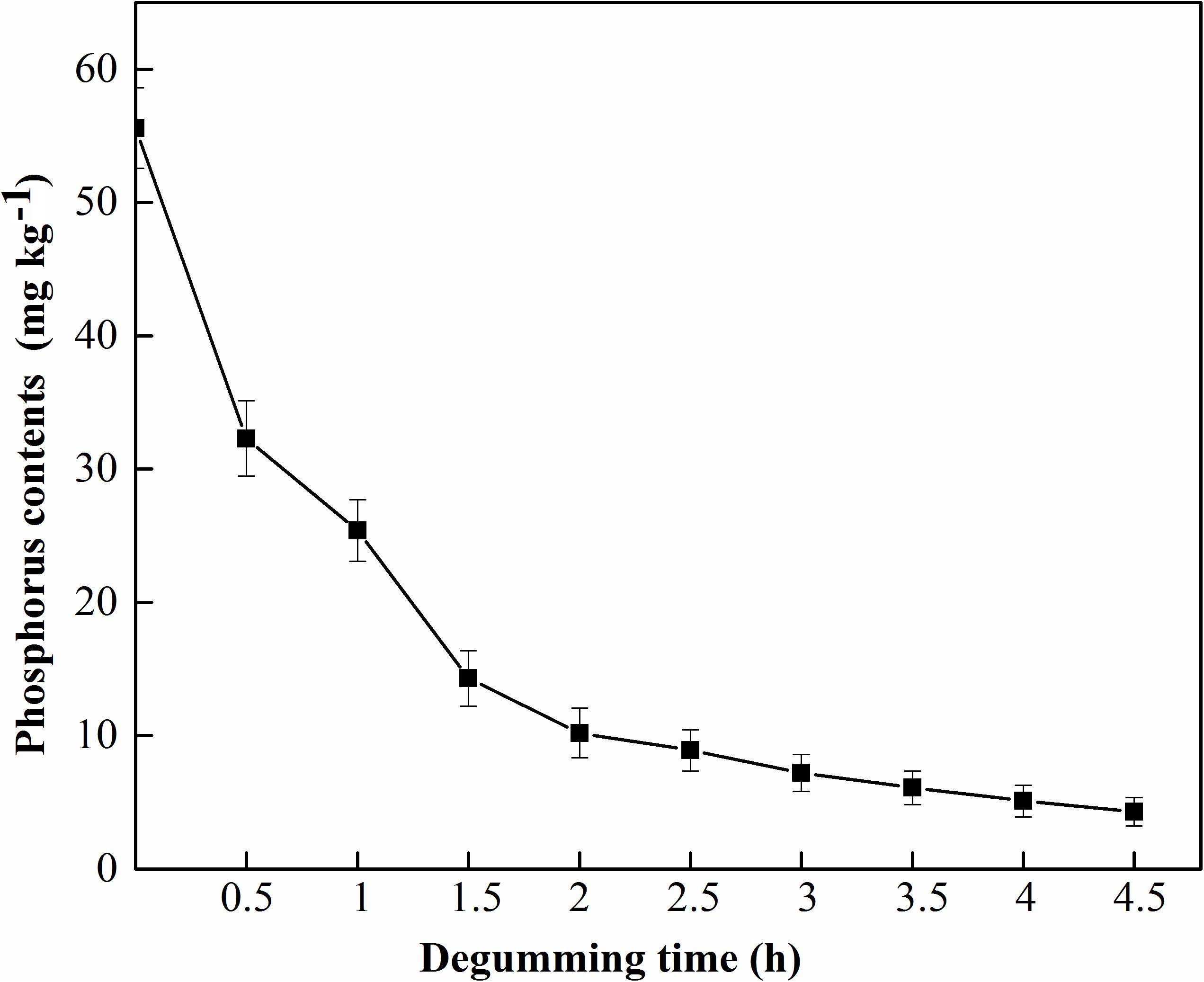
Figure 11. Effect of recycling times on MGO-OM-PLA1 activity under the conditions of pH 7.5 at 50°C.
The increase in oil yield and effective application on crude oils determines the development of enzymatic degumming. Enzymatic degumming has been applied in more than 30 crushing-refining plants. In general, a phosphorus content of below 10 mg/kg oil meets the actual requirement (Lamas et al., 2016). Previous reports on crude oil degumming mainly focused on the breeding of strains producing phospholipase A2, B, and C (Table 2). The phospholipases of the recombinant strains possess high application values in enzymatic degumming (Wei et al., 2015; Elena et al., 2017; Su et al., 2017). In this study, S. marcescens OM-PLA1 was effectively expressed in engineered P. pastoris. The activity and expression efficiency of OM-PLA1 were 21.2 U/mL and 0.29 U/mL/h, respectively. The immobilization product MGO-OM-PLA1 possessed excellent degumming capability of oil. The MGO-OM-PLA1 activity still retained 75 and 50% of the initial activity with 11 and 13 recycling times, respectively. In comparison with the reported 70% of the initial activity retained after seven recycles (Zhang et al., 2007), MGO-OM-PLA1 has the advantages of more application frequencies and higher degumming efficiency. Therefore, S. marcescens OM-PLA1 expressed by recombinant P. pastoris exhibited a potential application value in the enzymatic degumming of oil.
The material of immobilization is also a critical factor influencing the degumming of phospholipase. Octyl agarose, gelatin hydrogel, and cellulose triacetate have been applied to immobilize the commercial enzyme Lecitase (R) Ultra for oil degumming (Fernandez-Lorente et al., 2007; Sheelu et al., 2008; da Silva et al., 2017). In addition, sodium alginate and chitosan microparticles were crosslinked with Fe3O4 to immobilize phospholipase (Qu et al., 2016). In this study, graphene oxide was used to immobilize phospholipase OM-PLA1 for its advantage of a large number of functional groups on its surface, such as carboxyl, hydroxyl and epoxy groups, which allow easy reaction with compounds (Li et al., 2015). As a scaffold for enzyme immobilization, graphene oxide-based Fe3O4 nanoparticles possess a magnetic property (Shao et al., 2015), which facilitate easy recovery and reuse.
In this study, graphene oxide-based Fe3O4 nanoparticles were used to immobilize S. marcescens OM-PLA1 for the enzymatic degumming of rapeseed oil. The contents of phosphorus decreased to 8.9 mg/kg rapeseed oil from 55.6 mg/kg rapeseed oil with 2.5 h of degumming. Therefore, this study provided an effective approach for the enzymatic degumming of crude vegetable oil. However, some problems remain to be solved. First, the immobilization efficiencies of OM-PLA1 and graphene oxide-based Fe3O4 nanoparticles can still be increased. Second, the effect of the crosslinking agent glutaraldehyde on the activity of OM-PLA1 need further analysis. Third, the degumming effect of MGO-OM-PLA1 on other crude vegetable oils should be further explored.
In this study, S. marcescens OM-PLA1 gene with a size of 879 bp was isolated and expressed in the recombinant P. pastoris GS 115. The size of OM-PLA1 was appropriately 33 KDa. After 72 h of inducible time, the highest activity of OM-PLA1 was reached (21.2 U/mL). The production efficiencies of treatment time and cell concentration of OM-PLA1 were 0.29 U/mL/h and 1.06 U/mL/OD600, respectively. After 4 h of degumming, the phosphorus contents reached 5.1 mg/kg from 55.6 mg/kg using 0.02% (w/w) MGO-OM-PLA1 prepared by graphene oxide-based Fe3O4 nanoparticles and OM-PLA1. The half life of MGO-OM-PLA1 activity was 13 times for the enzymatic degumming of rapeseed oils with the initial phosphorus content of 55.6 mg/kg. Therefore, S. marcescens OM-PLA1 gene could effectively express in the recombinant P. pastoris. In addition, the cross-linked copolymer MGO-OM-PLA1 possessed good stability and effective degumming of vegetable oils. This study has an important application value in the enzymatic degumming of crude vegetable oil.
PY designed the experiment scheme. SuJ wrote the manuscript. YW performed the experiments. ZH analyzed the data. ZZ drew the figures. LC measured the enzyme activity. MD cloned the OMPLA1 gene. ShJ proposed the idea.
This study was financially supported by the National Key Research and Development Program of China (Project No. 2016YFD0401401-2).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Amirbandeh, M., and Taheri-Kafrani, A. (2016). Immobilization of glucoamylase on triazine-functionalized Fe3O4/graphene oxide nanocomposite: improved stability and reusability. Int. J. Biol. Macromol. 93, 1183–1191. doi: 10.1016/j.ijbiomac.2016.09.092
Araújo, A., and Radvanyi, F. (1987). Determination of phospholipase A2 activity by a colorimetric assay using a pH indicator. Toxicon 25, 1181–1188. doi: 10.1016/0041-0101(87)90136-X
Bredell, H., Smith, J. J., Gorgens, J. F., and van Zyl, W. H. (2018). Expression of unique chimeric human papilloma virus type 16 (HPV-16) L1-L2 proteins in Pichia pastoris and Hansenula polymorpha. Yeast 35, 519–529. doi: 10.1002/yea.3318
Chen, B., Xiao, X., Li, R., Zhao, W. L., Yang, K. D., Chen, G. L., et al. (2014). An improved method for determining the phosphorus content in vegetable oils. Eur. Lipid J. Sci. Technol. 116, 548–552. doi: 10.1002/ejlt.201300378
da Silva, F. B., de Morais, W. G., da Silva, C. V., Vieira, A. T., Batista, A., de Faria, A. M., et al. (2017). Preparation and characterization of cellulose triacetate as support for lecitase ultra immobilization. Molecules 22:E1930. doi: 10.3390/molecules22111930
Dekker, N. (2000). Outer-membrane phospholipase A: known structure, unknown biological function. Mol. Microbiol. 35, 711–717. doi: 10.1046/j.1365-2958.2000.01775.x
Deng, J. H., Zhang, X. R., Zeng, G. M., Gong, J. L., Niu, Q. Y., and Liang, J. (2013). Simultaneous removal of Cd(II) and ionic dyes from aqueous solution using magnetic graphene oxide nanocomposite as an adsorbent. Chem. Eng. J. 226, 189–200. doi: 10.1016/j.cej.2013.04.045
Deng, Y., Qi, D., Deng, C., Zhang, X., and Zhao, D. (2008). Superparamagnetic high-magnetization microspheres with an Fe3O4@SiO2 core and perpendicularly aligned mesoporous SiO2 shell for removal of microcystins. J. Am. Chem. Soc. 130, 28–29. doi: 10.1021/ja0777584
Elena, C., Cerminati, S., Ravasi, P., Rasia, R., Peiru, S., Menzella, H. G., et al. (2017). B-cereus phospholipase C engineering for efficient degumming of vegetable oil. Process Biochem. 54, 67–72. doi: 10.1016/j.procbio.2017.01.011
Elena, C., Ravasi, P., Cerminati, S., Peiru, S., Castelli, M. E., and Menzella, H. G. (2016). Pichia pastoris engineering for the production of a modified phospholipase C. Process Biochem. 51, 1935–1944. doi: 10.1016/j.procbio.2016.08.022
Fernandez-Lorente, G., Palomo, J. M., Guisan, J. M., and Fernandez-Lafuente, R. (2007). Effect of the immobilization protocol in the activity, stability, and enantioslectivity of Lecitase(R) Ultra. J. Mol. Catal. B Enzym. 47, 99–104. doi: 10.1016/j.molcatb.2007.04.008
Gamerith, C., Vastano, M., Ghorbanpour, S. M., Zitzenbacher, S., Ribitsch, D., Zumstein, M. T., et al. (2017). Enzymatic degradation of aromatic and aliphatic polyesters by pastoris, P., expressed cutinase 1 from Thermobifida cellulosilytica. Front. Microbiol. 8:938. doi: 10.3389/fmicb.2017.00938
Gofferje, G., Motulewicz, J., Stabler, A., Herfellner, T., Schweiggert-Weisz, U., and Floter, E. (2014). Enzymatic degumming of crude jatropha oil: evaluation of impact factors on the removal of phospholipids. J. Am. Oil Chem. Soc. 91, 2135–2141. doi: 10.1007/s11746-014-2559-2
Gohel, S. D., and Singh, S. P. (2012). Cloning and expression of alkaline protease genes from two salt-tolerant alkaliphilic actinomycetes in E. coli. Int. J. Biol. Macromol. 50, 664–671. doi: 10.1016/j.ijbiomac.2012.01.039
Goyal, A., Sharma, V., Upadhyay, N., Gill, S., and Sihag, M. (2014). Flax and flaxseed oil: an ancient medicine & modern functional food. J. Food Sci. Technol. 51, 1633–1653. doi: 10.1007/s13197-013-1247-9
Guo, D., Tian, H., Xu, Y., and Zheng, S. (2015). Cloning and expression of β-Glucosidase from Cassava in Pichia pastoris GS115. Adv. Appl. Biotechnol. 333, 11–20. doi: 10.1007/978-3-662-46318-5_2
Hasanzadeh, M., and Shadjou, N. (2013). (Fe3O4)-graphene oxide-SO3H as a new magnetic nanocatalyst for electro-oxidation and determination of selected parabens. J. Nanosci. Nanotechnol. 13, 4909–4916. doi: 10.1166/jnn.2013.7605
He, G. Y., Liu, W. F., Sun, X. Q., Chen, Q., Wang, X., and Chen, H. Q. (2013). Fe3O4@graphene oxide composite: a magnetically separable and efficient catalyst for the reduction of nitroarenes. Mater. Res. Bull. 48, 1885–1890. doi: 10.1016/j.materresbull.2013.01.038
Huang, J., Chang, Q., Ding, Y. B., Han, X. Y., and Tang, H. Q. (2014). Catalytic oxidative removal of 2,4-dichlorophenol by simultaneous use of horseradish peroxidase and graphene oxide/Fe3O4 as catalyst. Chem. Eng. J. 254, 434–442. doi: 10.1016/j.cej.2014.05.136
Huang, Y. H., Wang, Y. Z., Pan, Q., Wang, Y., Ding, X. Q., Xu, K. J., et al. (2015). Magnetic graphene oxide modified with choline chloride-based deep eutectic solvent for the solid-phase extraction of protein. Anal. Chim. Acta 877, 90–99. doi: 10.1016/j.aca.2015.03.048
Jiang, F., Huang, S., Imadad, K., and Li, C. (2012). Cloning and expression of a gene with phospholipase B activity from Pseudomonas fluorescens in Escherichia coli. Bioreour. Technol. 104, 518–522. doi: 10.1016/j.biortech.2011.09.112
Jiang, X., Chang, M., Wang, X., Jin, Q., and Wang, X. (2014). The effect of ultrasound on enzymatic degumming process of rapeseed oil by the use of phospholipase A1. Ultrason. Sonochem. 21, 142–148. doi: 10.1016/j.ultsonch.2013.07.018
Jiang, X. F., Chang, M., Wang, X., Jin, Q., and Wang, X. G. (2014). A comparative study of phospholipase A1 and phospholipase C on soybean oil degumming. J. Am. Oil Chem. Soc. 91, 2125–2134. doi: 10.1007/s11746-014-2555-6
Lamas, D. L., Constenla, D. T., and Raab, D. (2016). Effect of degumming process on physicochemical properties of sunflower oil. Biocatal. Agric. Biotechnol. 6, 138–143. doi: 10.1016/j.bcab.2016.03.007
Li, F., Jiang, X., Zhao, J., and Zhang, S. (2015). Graphene oxide: a promising nanomaterial for energy and environmental applications. Nano Energy 16, 488–515. doi: 10.1016/j.nanoen.2015.07.014
Liu, A. X., Yu, X. W., Sha, C., and Xu, Y. (2015). Streptomyces violaceoruber phospholipase A2: expression in Pichia pastoris, properties, and application in oil degumming. Appl. Biochem. Biotechnol. 175, 3195–3206. doi: 10.1007/s12010-015-1492-7
Manjula, S., Jose, A., Divakar, S., and Subramanian, R. (2010). Degumming rice bran oil using phospholipase-A1. Eur. Lipid J. Sci. Technol. 113, 658–664. doi: 10.1002/ejlt.201000376
Martin, S. F., Follows, B. C., Hergenrother, P. J., and Trotter, B. K. (2000). The choline binding site of phospholipase C (Bacillus cereus): insights into substrate specificity. Biochemistry 39, 3410–3415. doi: 10.1021/bi9919798
Nykter, M., Kymäläinen, H. R., Gates, F., and Sjöberg, A. M. (2006). Quality characteristics of edible linseed oil. Agric. Food Sci. 15, 402–413. doi: 10.15171/apb.2018.013
Pires, L. N., Brandão, G. C., and Teixeira, L. S. (2017). Determination of phospholipids in soybean lecithin samples via the phosphorus monoxide molecule by high-resolution continuum source graphite furnace molecular absorption spectrometry. Food Chem. 225, 162–166. doi: 10.1016/j.foodchem.2017.01.019
Qiu, J. D., Peng, H. P., Liang, R. P., and Xia, H. (2010). Facile preparation of magnetic core-shell Fe3O4@Au nanoparticle/myoglobin biofilm for direct electrochemistry. Biosens. Bioelectron. 25, 1447–1453. doi: 10.1016/j.bios.2009.10.043
Qu, Y. F., Sun, L. X., Li, X., Zhou, S., Zhang, Q., Sun, L. B., et al. (2016). Enzymatic degumming of soybean oil with magnetic immobilized phospholipase A(2). LWT Food Sci. Technol. 73, 290–295. doi: 10.1016/j.lwt.2016.06.026
Ravasi, P., Braia, M., Eberhardt, F., Elena, C., Cerminati, S., Peiru, S., et al. (2015). High-level production of Bacillus cereus phospholipase C in Corynebacterium glutamicum. J. Biotechnol. 216, 142–148. doi: 10.1016/j.jbiotec.2015.10.018
Sampaio, K. A., Zyaykina, N., Wozniak, B., Tsukamoto, J., De Greyt, W., and Stevens, C. V. (2015). Enzymatic degumming: degumming efficiency versus yield increase. Eur. Lipid J. Sci. Technol. 117, 81–86. doi: 10.1002/ejlt.201400218
Shao, Y., Jing, T., Tian, J., and Zheng, Y. (2015). Graphene oxide-based Fe3O4 nanoparticles as a novel scaffold for the immobilization of porcine pancreatic lipase. RSC Adv. 5, 103943–103955. doi: 10.1039/C5RA19276E
Sheelu, G., Kavitha, G., and Fadnavis, N. W. (2008). Efficient immobilization of lecitase in gelatin hydrogel and degumming of rice bran oil using a spinning basket reactor. J. Am. Oil Chem. Soc. 85, 739–748. doi: 10.1007/s11746-008-1261-7
Stanley, A. M., Treubrodt, A. M., Chuawong, P., Hendrickson, T. L., and Fleming, K. G. (2007). Lipid chain selectivity by outer membrane phospholipase A. Mol. J. Biol. 366, 461–468. doi: 10.1016/j.jmb.2006.10.055
Su, L. Q., Ji, D. N., Tao, X. M., Yu, L. G., Wu, J., and Xia, Y. M. (2017). Recombinant expression, characterization, and application of a phospholipase B from Fusarium oxysporum. J. Biotechnol. 242, 92–100. doi: 10.1016/j.jbiotec.2016.12.009
Sutto-Ortiz, P., Camacho-Ruiz, M. A., Kirchmayr, M. R., Camacho-Ruiz, R. M., Mateos-Díaz, J., Noiriel, A., et al. (2017). Screening of phospholipase A activity and its production by new actinomycete strains cultivated by solid-state fermentation. PeerJ 5:e3524. doi: 10.7717/peerj.3524
Tolner, B., Smith, L., Begent, R., and Chester, K. (2006). Production of recombinant protein in Pichia pastoris by fermentation. Nat. Protoc. 1, 1006–1021. doi: 10.1038/nprot.2006.126
Wang, Y., Xuan, Y. J., Zhang, P., Jiang, X., Ni, Z. H., Tong, L. J., et al. (2009). Targeting expression of the catalytic domain of the kinase insert domain receptor (KDR) in the peroxisomes of Pichia pastoris. FEMS Yeast Res. 9, 732–743. doi: 10.1111/j.1567-1364.2009.00521.x
Wei, T., Xu, C. P., Yu, X., Jia, W. W., Yang, K. P., Jia, C. X., et al. (2015). Characterization of a novel thermophilic phospholipase B from Thermotoga lettingae TMO: applicability in enzymatic degumming of vegetable oils. J. Ind. Microbiol. Biotechnol. 42, 515–522. doi: 10.1007/s10295-014-1580-7
Xie, W. L., and Huang, M. Y. (2018). Immobilization of Candida rugosa lipase onto graphene oxide Fe3O4 nanocomposite: characterization and application for biodiesel production. Energy Convers. Manag. 159, 42–53. doi: 10.1016/j.enconman.2018.01.021
Yang, M., Zheng, C., Zhou, Q., Huang, F. H., Liu, C. S., and Wang, H. (2013). Minor components and oxidative stability of cold-pressed oil from rapeseed cultivars in China. J. Food Compost. Anal. 29, 1–9. doi: 10.1016/j.jfca.2012.08.009
Yang, P., Zhu, X., Cao, L., Cheng, J., Zheng, Z., and Jiang, S. (2017). Safety evaluation of Bacillus cereus isolated from smelly mandarin fish. J. Food Meas. Charact. 11, 726–735. doi: 10.1007/s11694-016-9442-9
Zhang, L., Hellgren, L., and Xu, X. B. (2007). Immobilization of phospholipase C for the production of ceramide from sphingomyelin hydrolysis. J. Am. Oil Chem. Soc. 84, 237–247. doi: 10.1007/s11746-006-1028-y
Zhuang, W., He, J., Zhu, J., Zheng, J. W., Liu, X. J., Dong, Y. H., et al. (2016). Efficient nanobiocatalytic systems of nuclease P1 immobilized on PEG-NH2 modified graphene oxide: effects of interface property heterogeneity. Colloids Surf. B Biointerfaces 145, 785–794. doi: 10.1016/j.colsurfb.2016.05.074
Keywords: phospholipase, enzymatic degumming, immobilization, graphene oxide, magnetic nanoparticle, Pichia pastoris
Citation: Yang P, Jiang S, Wu Y, Hou Z, Zheng Z, Cao L, Du M and Jiang S (2019) Recombinant Expression of Serratia marcescens Outer Membrane Phospholipase A (A1) in Pichia pastoris and Immobilization With Graphene Oxide-Based Fe3O4 Nanoparticles for Rapeseed Oil Degumming. Front. Microbiol. 10:334. doi: 10.3389/fmicb.2019.00334
Received: 19 November 2018; Accepted: 08 February 2019;
Published: 21 February 2019.
Edited by:
Qiang Wang, Institute of Hydrobiology (CAS), ChinaReviewed by:
Xiaoli Qin, Southwest University, ChinaCopyright © 2019 Yang, Jiang, Wu, Hou, Zheng, Cao, Du and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peizhou Yang, eWFuZ3BlaXpob3VAMTYzLmNvbQ== Shaotong Jiang, amlhbmdzaGFvdG9uZ0AxNjMuY29t
†They are co-first authors
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.