
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 20 February 2019
Sec. Microbial Physiology and Metabolism
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00298
This article is part of the Research TopicMicrobial Sulfur Metabolism - From Symbiosis to Global Nutrient Cycling View all 37 articles
Hydrogen sulfide (H2S) has been proposed to have various physiological functions, and it may function through reactive sulfane sulfur. Since the two sulfur forms often coexist, they are normally considered interchangeable. Here, we characterized the production of H2S and reactive sulfane sulfur in Escherichia coli MG1655 and found that they are not readily interchangeable. They are primarily produced from L-cysteine via different enzymes. L-Cysteine desulfhydrases consumed L-cysteine and directly generated H2S. The produced H2S was mainly lost through evaporation into the gas phase, as E. coli does not have enzymes that easily oxidize H2S to reactive sulfane sulfur. L-Cysteine desulfhydrases were also responsible for the degradation of exogenous L-cysteine, which is toxic at high levels. Conversely, L-cysteine aminotransferase and 3-mercaptopyruvate sulfurtransferase sequentially metabolized endogenous L-cysteine to produce cellular reactive sulfane sulfur; however, it was not a major route of H2S production during normal growth or during the metabolism of exogenous L-cysteine by the resting cells. Noticeably, the 3-mercaptopyruvate sulfurtransferase mutant contained less reactive sulfane sulfur and displayed a greater sensitivity to H2O2 than did the wild type. Thence, reactive sulfane sulfur is likely a common cellular component, involved in protein sulfhydration and protecting cells from oxidative stress.
H2S may protect bacterial cells from oxidative stress (Shatalin et al., 2011) and have physiological roles in cellular signaling through protein sulfhydration (Paul and Snyder, 2012; Giedroc, 2017), in which a sulfur atom is added to a cysteinyl thiol in a target protein to form a hydropersulfide (protein-SSH). Sulfhydration is involved in cellular signaling and in modulating enzyme activity (Toohey, 1986; Wang, 2010; Kimura, 2014). However, the direct involvement of H2S in protein sulfhydration has been questioned, as it does not have the ability to directly react with a thiol group (-SH) (Toohey, 2011; Pan and Carroll, 2013; Yadav et al., 2016). On the other hand, reactive sulfane sulfur (RSSH, RSSnH, n ≥ 1; HSSnH and RS(S)nSR; n ≥ 1), which does not include the insoluble elemental sulfur, can react with protein thiols to generate protein-SSH (Toohey, 2011; Ida et al., 2014). There are appreciable amounts (>100 μM) of reactive sulfane sulfur in the plasma, cells, and tissues of mammals (Ida et al., 2014). Thus, it is unresolved whether H2S or reactive sulfane sulfur is mainly responsible for protein sulfhydration and resistance to oxidative stress in live cells (Toohey, 2011; Ida et al., 2014).
H2S and reactive sulfane sulfur often coexist, making it difficult to precisely distinguish their production and function. H2S can lead to the production of reactive sulfane sulfur via chemically reacting with a disulfide bond and a sulfenic acid (Nagy, 2015) or via oxidation by sulfide:quinone oxidoreductases (SQR) and flavocytochrome c-sulfide dehydrogenases (FCSDs) (Libiad et al., 2014; Shen et al., 2016; Lü et al., 2017). Reactive sulfane sulfur can also be generated from sulfur-containing amino acids by several enzymes. Cystathionine β-synthase (CBS) and cystathionine γ-lyase (CSE) can produce CysSSH from cystine (Cys-SS-Cys) (Ida et al., 2014), but the contribution of these reactions to cellular reactive sulfane sulfur may be limited, as the concentration of reduced thiols is significantly higher than that of disulfides in the reducing cytoplasmic milieu (Yadav et al., 2016). Cysteinyl-tRNA synthetase may directly use L-cysteine to produce Cys-SSH (Akaike et al., 2017). Further, L-cysteine aminotransferase (CAT) can convert L-cysteine to 3-mercaptopyruvate (Nagahara and Sawada, 2006); mercaptopyruvate sulfurtransferase (MST) abstracts the sulfur of 3-mercaptopyruvate to generate a persulfide at its active site (MST-SSH) (Toohey, 2011; Yadav et al., 2013). On the flipside, the various forms of reactive sulfane sulfur can be reduced by cellular thiols such as GSH, thioredoxin, and glutaredoxin to product H2S (Mikami et al., 2011; Yadav et al., 2013; Dóka et al., 2016). However, a systematic investigation on the interchangeability of H2S and sulfane sulfur in live cells has not been investigated.
Escherichia coli MG1655 contains CAT/MST (Nagahara and Sawada, 2006; Shatalin et al., 2011) and at least six enzymes (CysK, CysM, MetC, TnaA, MalY, and YhaM) with L-cysteine desulfhydrase (CD) activity (Awano et al., 2005; Shimada et al., 2016). CDs decompose L-cysteine into pyruvate, ammonium, and H2S. Since E. coli does not have CBS, CSE, SQR, and FCSDs, it is a good system to evaluate the roles of CDs and MST in the production of H2S and reactive sulfane sulfur in live cells. Here, we characterized their roles, evaluated the interchangeability of H2S and reactive sulfane sulfur, and investigated whether H2S or reactive sulfane sulfur is the major antioxidant in E. coli.
All deletions in E. coli MG1655 were performed by using a reported one-step deletion method (Datsenko and Wanner, 2000). For complementation or gene expression, the target genes were amplified by PCR, and the PCR products were cloned into linearized pBBR1MCS2 or pET30-LIC by using the In-Fusion HD cloning kit (Clontech, United States). All bacterial strains, plasmids and recombinant cells are listed in Supplementary Table S1, and all primers are given in Supplementary Table S2. E. coli was cultured in LB medium at 37 or 25°C with shaking (200 rpm). Ampicillin (100 mg/L) or kanamycin (50 mg/L) was added as needed.
Overnight culture of E. coli MG1655 was inoculated in fresh LB medium to OD600 nm of 0.05. The cells were cultured to about OD600 nm of 4.0, harvested, washed and resuspended in 50 mM Tris-HCl buffer, pH 7.6, at OD600 nm of 2.0. Appropriate sulfur-containing compounds were added to a final concentration of 100 μM to the resting cells to initiate the reaction. Glucose was also added to 1% to provide a reducing power. The production of H2S and reactive sulfane sulfur was detected.
Escherichia coli BL21(DE3) cells with the expression plasmid pET-TnaA, pET-MalY, pET-CysK, pET-CysM, pET-YhaM, pET-CAT, or pET-MST were inoculated in LB medium and cultured with shaking to OD600 nm of 0.6 at 25°C, and then isopropyl β-D-thiogalactoside (IPTG) was added to a final concentration of 0.5 mM. The cultures were further cultured for about 10 h. Cells were harvested by centrifugation and disrupted by a pressure cell homogenizer (Stansted Fluid Power LTD, United Kingdom, SPCH-18) at 4°C in buffer I (20 mM Tris-HCl, 0.5 M NaCl, 20 mM imidazole, pH 8.0). The lysate was centrifuged at 12,500 × g for 10 min to remove cell debris 4°C. The target protein was purified by using nickel-nitrilotriacetate agarose (Qiagen, Shanghai, China), according to the supplier’s recommendations. The buffer was exchanged to 20 mM sodium phosphate buffer (pH 7.6) and then 50% glycerol was added to give a final concentration of 10% before storage at -80°C.
The activity of CDs and CAT/MST for the degradation of L-cysteine was assayed. The assay was routinely performed at 25°C. The reaction was carried out in 3 mL of 50 mM Tris-HCl buffer, pH 7.6, in the test tube (1.5 × 10 cm), containing 50 μM pyridoxal phosphate, 1.0 mM L-cysteine, and 1.0 mM α-KG (only for CAT/MST reactive mixture). The purified CD or CAT/MST (both purified CAT and MST) was added at the final concentration of 1 μM to initiate the reaction. After 1 h incubation, the production of H2S were detected by using a reported monobromobimane (mBBr) method (Kolluru et al., 2013) and reactive sulfane sulfur produced in the reaction was detected with SSP4 (Yadav et al., 2016; Bibli et al., 2018).
The H2S generation by E. coli was monitored with both lead acetate [Pb(Ac)2] paper strips (Shatalin et al., 2011) and the mBBr method. Briefly, a [Pb(Ac)2] paper strip with lead acetate was affixed to the inner wall of a glass tube (1.5 × 20 cm) containing 5.0 mL of the culture. H2S was evaporated into the gas phase and reacted with Pb(II) to form dark PbS stains, which were measured via densitometer and evaluated by using a standard curve. To obtain the standard curve, various concentrations NaHS were added to 5 mL of LB medium and after incubation at 37°C with shaking for 12 h, the darkness on the paper strips were measured via densitometer (Chen et al., 2018). Sulfide (H2S, HS-, and S2-), sulfite, and thiosulfate in the liquid phase were detected by using the mBBr method (Kolluru et al., 2013). Briefly, 50 μL of a sample was reacted with 5 μL of 25 mM mBBr at room temperature for 30 min in the dark, and an equal volume of 10% acetic acid in acetonitrile was added. The precipitated proteins were removed by centrifugation at 12,500 × g for 2 min. Then, 20 μL of the supernatant was analyzed by using HPLC (LC-10AT, Shimadzu) with a fluorescence detector (Xin et al., 2017).
SSP4 was used to detect reactive sulfane sulfur in the enzyme reaction mixtures. 10 mM SSP4 stock solution in acetonitrile was added to a reaction mixture to a final concentration of 10 μM, and the sample was mixed and incubated in the dark at 37°C for 10 min. The fluorescence was detected by using the Synergy H1 microplate reader with excitation of 482 nm and the emission of 515 nm.
Reactive sulfane sulfur produced by E. coli cells in the Tris-HCl buffer during the metabolism of 100 μM sulfur-containing substances was detected by adding 10 μM SSP4 and 0.5 mM CTAB in the reaction mixture. The sample was incubated in the dark at 37°C with shaking for 20 min, and the cells were harvested by centrifugation and washed twice with phosphate buffer saline (PBS). The fluorescence of the resuspended cells (OD600 nm of 0.2) in PBS was detected by using the Synergy H1 microplate reader.
Reactive sulfane sulfur in the supernatant of lysed E. coli cells was also detected. The cells were harvested, washed and resuspended in 50 mM anoxic Tris-HCl buffer, pH 7.6, and disrupted by a pressure cell homogeniser. The lysate was centrifuged at 12,500 × g for 10 min to remove cell debris at 4°C. The sulfane sulfur in the supernatant was measured with SSP4 as above. The fluorescence values were expressed as the intensity per mg of protein.
Escherichia coli MG1655, its mutant E. coli ΔsseA, and its recombinant cells were cultivated in LB medium at 37°C. When OD600 nm reached to 0.6∼0.8, 0.5 mM IPTG was added, and the cells were further cultivated for 12 h at 37°C. The cells were harvested, washed and resuspended at OD600 nm of 2 in 50 mM Tris-HCl buffer, pH 7.6. Ten mL of the cells suspension was transferred to a 50-mL centrifuge tube, and 1% glucose and 0.2 mM MP were added to initiate the reaction. The tube was loosely capped and incubated at 37°C with shaking. The produced sulfide, sulfite, and thiosulfate were analyzed by the mBBr method at various time intervals (Kolluru et al., 2013).
Escherichia coli MG1655 and its mutants were cultured, harvested, washed and resuspended in 50 mM Tris-HCl buffer, pH 7.6, at OD600 nm of 2. Glucose was added to 1%, and L-cysteine and α-KG were added to1 mM to initiate the reaction. Ellman’s reagent (DTNB) was used to detect L-cysteine in the supernatant (Riener et al., 2002). Briefly, 1 mL of a sample was added to 99 μl of the DTNB reagent, mixed, and incubated at 37°C for 10 min. The absorbance at 412 nm was measured to determine L-cysteine contents with a standard curve.
Overnight cultures of E. coli strains were transferred in LB medium at 1% inoculum and grown to OD600 nm of 1 (37°C, 200 rpm). Cells were then treated with 2.5 mM H2O2 at room temperature for 20 min without shaking and diluted to stop the treatment. The diluted cells were spread on LB medium agar plates and incubated at 37°C for 16∼18 h. Cell survival was determined by counting colony forming units (CFU). The averages of three samples and standard deviation were reported.
Bioinformatics analysis was done as reported (Lü et al., 2017; Xia et al., 2017) with minor modification. Briefly, a microbial genomic protein sequence set from NCBI updated until November 11, 2017 was downloaded for MST search. The query sequences of MST were confirmed MSTs (Nagahara and Sawada, 2006; Westrop et al., 2009; Wu et al., 2015) and were used to search the database by using Standalone BLASTP algorithm with conventional criteria (e-value ≤ 1e-10, coveryage ≥ 45%, identity ≥ 35%) to obtain MST candidates from a total of 8671 bacterial genomes. Then the candidates performed multiple sequence alignments to exclude sequences by Clustal Omega1 that are not MST based on conserved cysteine residues at the catalytic site (Yadav et al., 2013). The candidates combined with the seed MSTs for phylogenetic tree analysis by using ClustalW for alignment and MEGA version 7.0 to generate the tree by using neighbor-joining with a pairwise deletion, p-distance distribution, and bootstrap analysis of 1,000 repeats as the parameters (Kumar et al., 2016). The candidates that were in the same clade with the seed MSTs were picked up for further analysis. The identified MST sequences were separately grouped into unique groups by using the CD-HIT program with a threshold of 90% sequence identity or better. Since rhodaneses display clear sequence homology with MST (Nagahara et al., 1995), five published rhodanese sequences were also collected (Cheng et al., 2008) and grouped into unique groups as outgroups. One representative sequence from each unique group was selected and used to build a phylogenetic tree (MEGA Version 7.0).
Escherichia coli MG1655 was grown in Lysogeny broth (LB) medium, harvested, and resuspended in 50 mM Tris buffer (pH 7.6) at OD600 nm of 2.0. The resting cells were then tested to produce H2S and reactive sulfane sulfur from various sulfur-containing compounds. L-Cysteine and L-cystine were the most suitable substrates for the production of H2S (Supplementary Figure S1) and reactive sulfur sulfane compared with other organic (GSH and methionine) and inorganic (Na2SO3 and Na2S2O3) substances (Figure 1A). When GSH was tested, the cells did not generate detectable H2S in 30 min but produced H2S in 1 h of incubation (Supplementary Figures S1, S2A). Methionine did not have a significant effect on the production of H2S and reactive sulfane sulfur even after 12 h incubation (Figure 1 and Supplementary Figure S1A). E. coli released approximately 100 μM H2S after growing in LB medium for 12 h, and the inclusion of methionine up to 10 mM did not affect the H2S production or the cell growth (Supplementary Figure S2B).
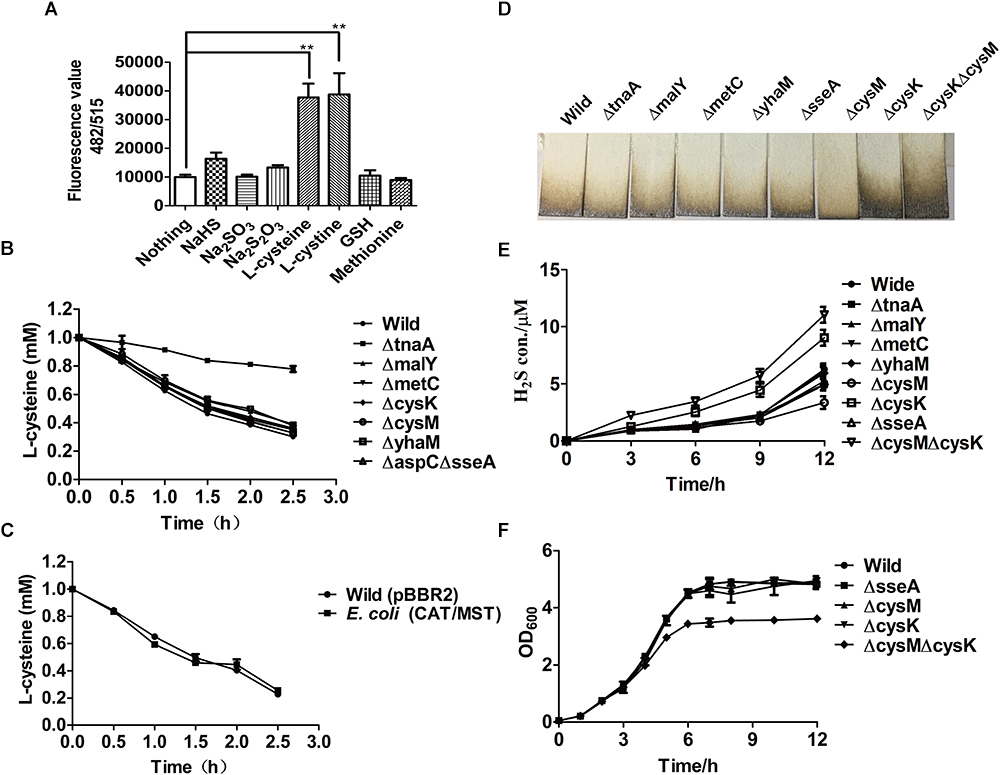
Figure 1. The production of H2S and reactive sulfane sulfur from L-cysteine by the resting cells of E. coli MG1655 and its mutants. (A) Reactive sulfane sulfur as detected with SSP4 within the wild type after 30-min incubation with various substrates. (B) Effects of the deletion of CDs or CAT/MST on L-cysteine degradation. (C) The effect of CAT and MST overexpression on L-cysteine degradation. (D) Pb(Ac)2 paper strips detected the release of H2S into the gas phase by E. coli MG1655 and its mutants grown in LB medium for 12 h. (E) Sulfide was quantified in the supernatant of E. coli MG1655 and mutants grown in LB medium. (F) The growth curve of E. coli MG1655, E. coliΔsseA (ΔsseA), E. coliΔcysM (ΔcysM), E. coliΔcysK (ΔcysK), and E. coliΔcysKΔcysM (ΔcysKΔcysM) in LB medium. Overnight cultures of E. coli MG1655 and mutant strains were inoculated in fresh LB medium to OD600 nm of 0.05, and the growth was monitored via OD600nm at 1-h intervals.
To assess the contribution of CAT/MST and CDs in L-cysteine degradation and H2S production by E. coli MG1655 under aerobic conditions when culture in LB medium, a series of mutants were constructed and tested. The resting cells of E. coli strains in the Tris buffer at OD600nm of 2.0 were used to degrade 1.0 mM L-cysteine. After 2.5 h incubation, E. coli MG1655 and its mutants ΔtnaA, ΔmalY, ΔmetC, ΔcysK, ΔcysM, ΔyhaM, and ΔaspCΔsseA degraded 69.9, 22.1, 61.2, 65.0, 63.9, 62.1, 66.9, 67.2% of the added L-cysteine, respectively (Figure 1B). The results indicated that TnaA plays a major role in the degradation of added L-cysteine while others including the CAT/MST pathway have limited contribution to the degradation. The overexpression of CAT/MST [E. coli (CAT/MST)] did not increase the rate of L-cysteine degradation compared with the wild type (Figure 1C).
The effect of the gene deletions on H2S accumulation during growth in LB medium was also tested (Figure 1D). The wild type, ΔtnaA, ΔmalY, ΔmetC, and ΔsseA all released around 100 μM H2S after 12 h of incubation; however, ΔcysK accumulated approximately double H2S and ΔcysM produced only roughly two-thirds H2S compared with wild type (Figure 1D). The double mutant E. coliΔcysKΔcysM accumulated more than twice of H2S compared with wild type (Figure 1D), indicating that the lack of H2S consumption by CysK and CysM contributes to its accumulation. Sulfide in the culture supernatant was further detected with the mBBr method, and the results also showed that ΔtnaA, ΔmalY, ΔmetC, and ΔsseA produced similar amounts of sulfide as the wild type, but ΔcysK and ΔcysKΔcysM produced more sulfide (Figure 1E). The deletion of single genes did not affect the growth of the mutants in LB medium (Figure 1F). The double mutant E. coliΔcysKΔcysM had the same initial growth rate, but had a reduced final yield (Figure 1F). Collectively, the data suggest that E. coli produces H2S during growth in LB medium and uses some of the produced H2S to produce L-cysteine by CysK and CysM.
The purified CDs and CAT/MST all used L-cysteine and released H2S (Supplementary Figure S3 and Figure 2A). However, CysK and CysM had very low activity, suggesting they may not contribute to L-cysteine metabolism in vivo. The production of reactive sulfane sulfur during L-cysteine degradation by these enzymes was also tested with SSP4. CDs did not produce reactive sulfane sulfur, while CAT and MST produced significant amounts of reactive sulfane sulfur during L-cysteine metabolism in the presence of the co-substrate α-KG (Figure 2B). Thus, CDs did not generate reactive sulfane sulfur as an intermediate during L-cysteine degradation, while CAT/MST did.
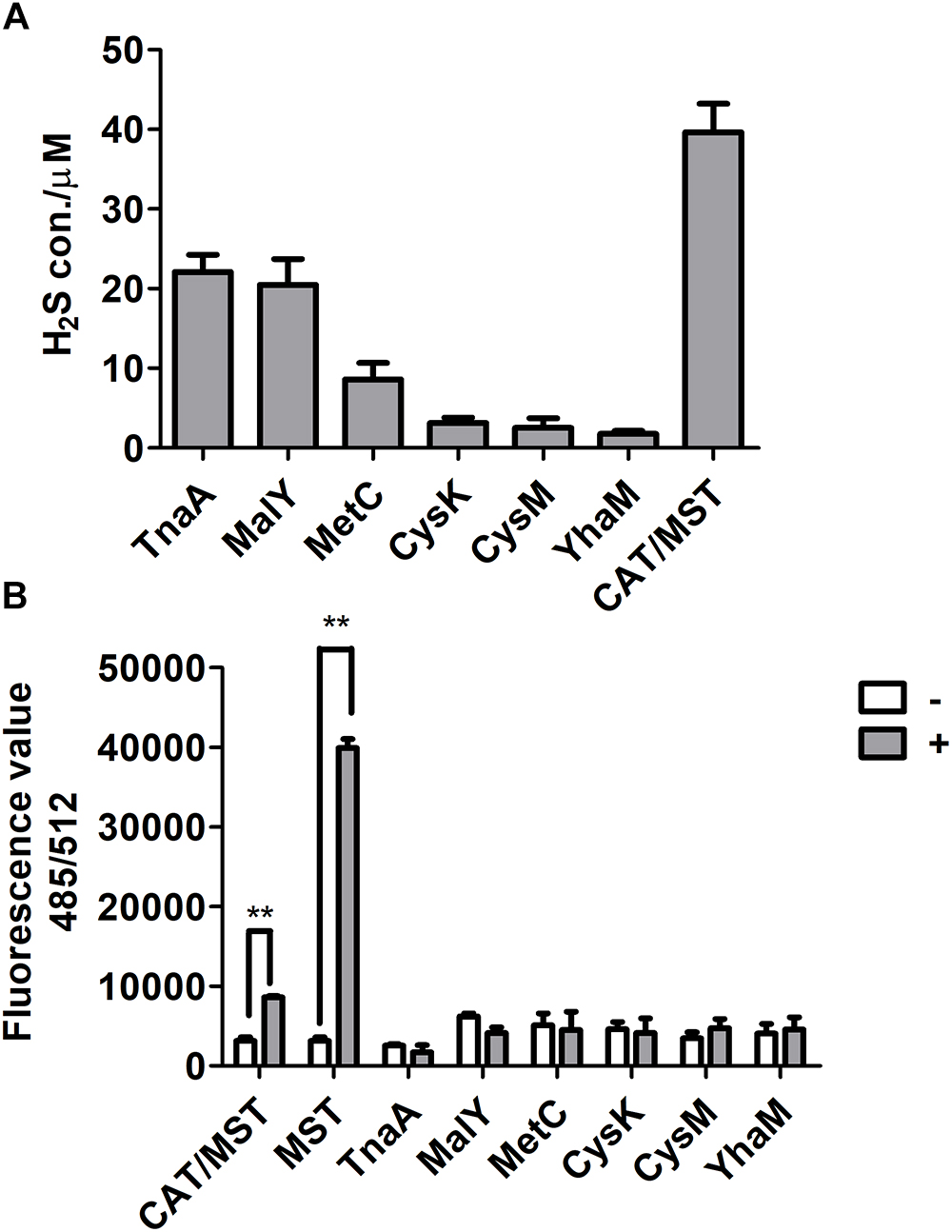
Figure 2. The production of H2S and reactive sulfane sulfur from L-cysteine by purified enzymes. (A) The purified CD or CAT/MST (CAT and MST) at the final concentration of 1 μM was used to metabolism 1 mM L-cysteine. After 1 h, the production of sulfide was quantified with the mBBr method. (B) Simultaneously, the production of reactive sulfane sulfur with (+) or without (-) L-cysteine or MP for MST were monitored by using SSP4. Data were presented as the average ± SD from three independent experiments. T tests were performed to calculate the p-values, and double asterisks indicate statistically significant difference (∗∗p < 0.01).
To clarify that MST consumed MP and produced reactive sulfane sulfur, purified MST was used to metabolize MP in the presence of GSH (Figures 3A–D). With the increase of GSH concentrations in the reaction mixture, GSSH also increased (Figure 3B), but GSSSH decreased (Figure 3C). Sulfide was almost not produced when GSH was less than MP in the reaction mixture (Figure 3C). GSSH and GSSSH reached to the maximum at 10.0 min of the reaction (Figure 3B), while the maximal amount of sulfide was achieved at 30 min (Figure 3D). Thus, MST converted MP mostly to reactive sulfane sulfur (Figures 3B,C), and the latter was reduced by GSH to release sulfide in a delayed reaction (Figure 3D). However, the chemical reaction was concentration-dependent and was slower than the enzymatic reaction.
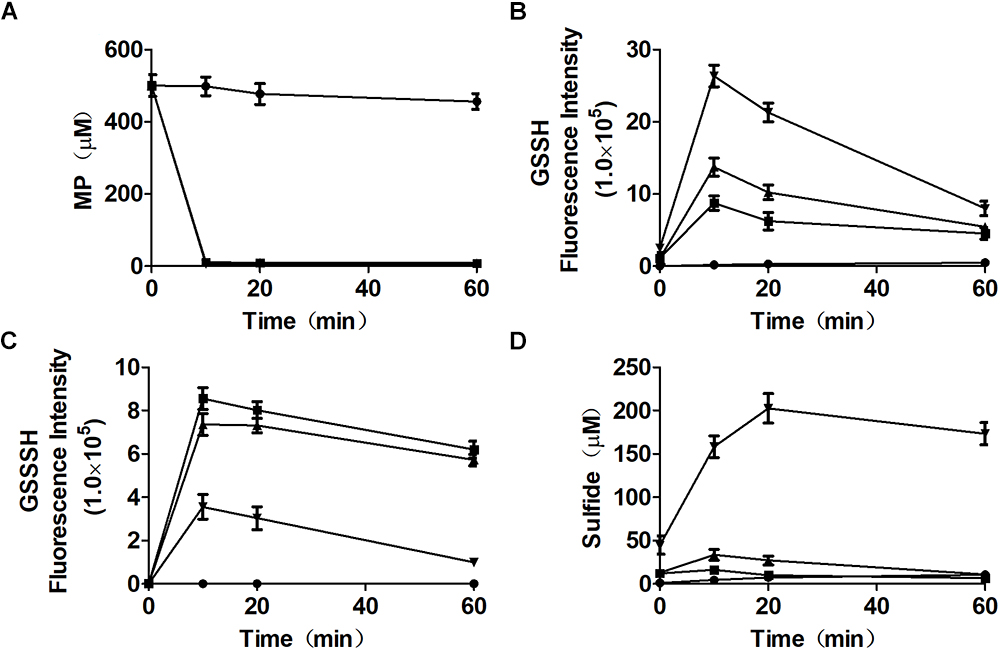
Figure 3. Reactive sulfane sulfur is the direct product of the CAT/MST pathway in vitro. MST catalyzes the reaction of MP with different concentrations of GSH in vitro. MST catalysis of 500 μM MP with 250 μM ( ), 500 μM (
), 500 μM ( ), or 2.0 mM (
), or 2.0 mM ( ) GSH; control without MST (
) GSH; control without MST ( ), containing MP (500 μM) and GSH (500 μM). The substrate (A) MP, or products (B) GSSH, (C), GSSSH, and (D) Sulfide in solution were analyzed via the mBBr method.
), containing MP (500 μM) and GSH (500 μM). The substrate (A) MP, or products (B) GSSH, (C), GSSSH, and (D) Sulfide in solution were analyzed via the mBBr method.
Mutant and recombinant E. coli strains were also used to metabolize MP. MP is unstable in neutral solutions (Cooper et al., 1982), and it rapidly disappeared even in a reaction with the MST mutant E. coli ΔsseA and E. coli ΔsseA (AtBlh) (Figure 4A), but its product was unknown, as sulfide, thiosulfate, and sulfite were not detectable (Figures 4B–D). The wild type with the cloning vector pBBR2 rapidly metabolized 200 μM MP to 105.0 ± 7.5 μM sulfide (Figure 4B) and 7.0 ± 0.86 μM thiosulfate in 5 min, and sulfide in the solution was then rapidly decreased likely due to evaporation with shaking (Figure 4C). To test whether GSSH was generated in vivo, persulfide dioxygenase gene (blh) from Agrobacterium tumefaciens str. C58 was cloned into E. coli MG1655 [E. coli (AtBlh)], and recombinant cells rapidly metabolized 200 μM MP to 38.2 ± 2.1 μM sulfide, 55.9 ± 5.7 μM thiosulfate and 10.7 ± 1.5 μM sulfite (Figures 4B–D). These results suggest that GSH can act as a receptor for reactive sulfane sulfur during the MST reaction to produce GSSH under physiological conditions, and GSSH reacts with small thiols such as GSH to release sulfide. MST did not speed up the reaction of GSSH and GSH (Figure 5), suggesting the reaction occurs non-enzymatically.
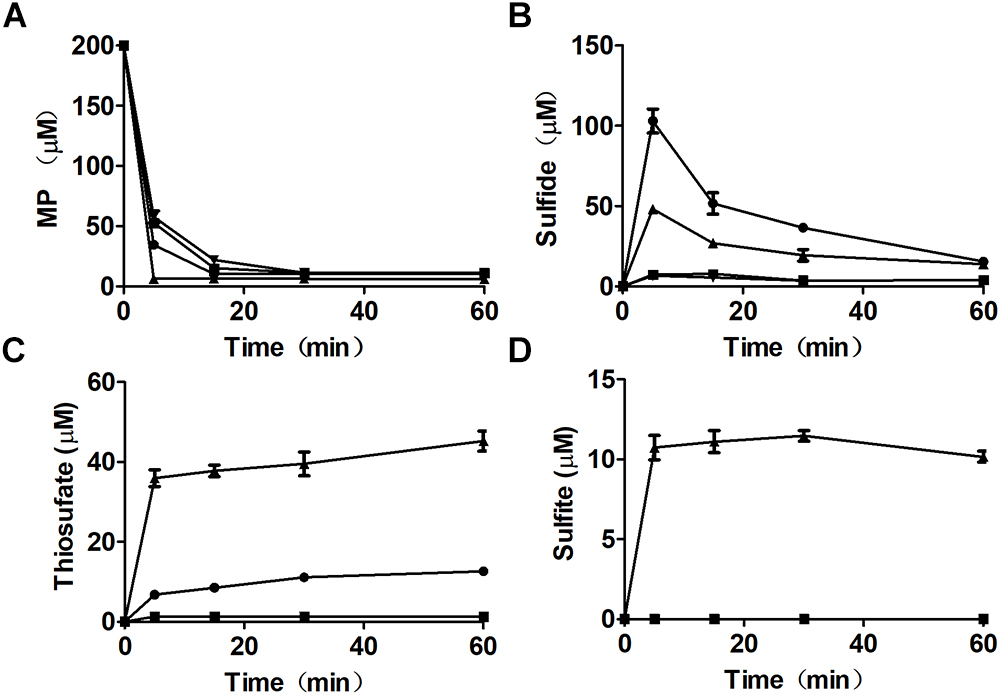
Figure 4. MP metabolism by recombinant E. coli cells. Resting cells of E. coli (pBBR2) ( ), E. coli (AtBlh) (
), E. coli (AtBlh) ( ), E. coli ΔsseA (
), E. coli ΔsseA ( ), and E. coli ΔsseA (AtBlh) (
), and E. coli ΔsseA (AtBlh) ( ) at OD600nm of 2 in 50 mM Tris-HCl buffer, pH 7.6, with 1% glucose was used to degrade 200 μM MP. (A) MP, (B) sulfide, (C) thiosulfate, and (D) sulfite were analyzed.
) at OD600nm of 2 in 50 mM Tris-HCl buffer, pH 7.6, with 1% glucose was used to degrade 200 μM MP. (A) MP, (B) sulfide, (C) thiosulfate, and (D) sulfite were analyzed.
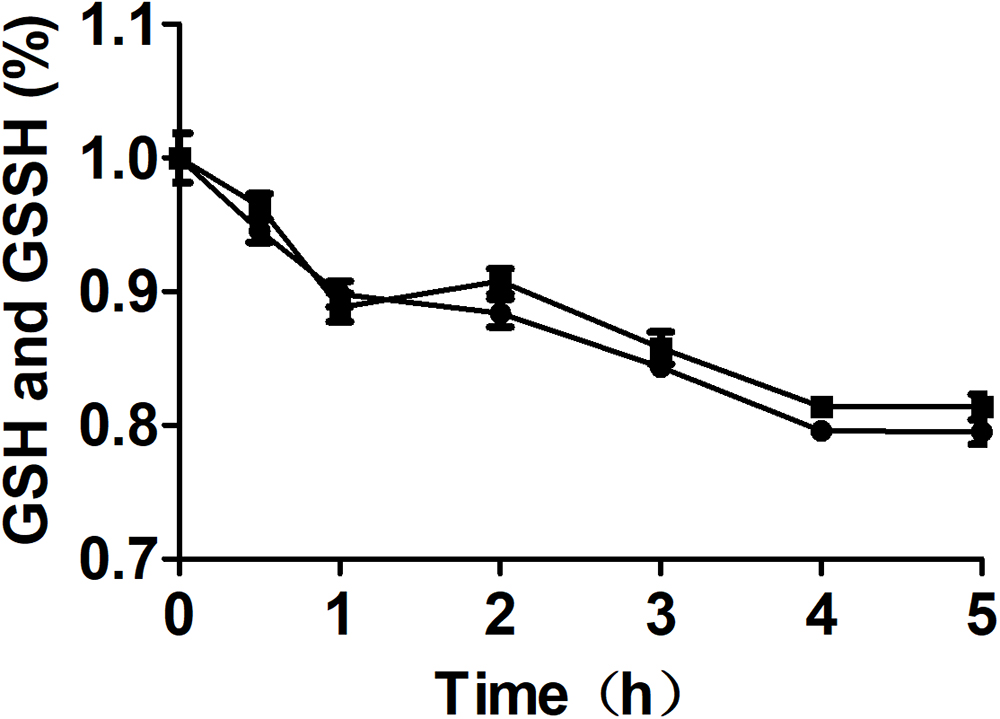
Figure 5. The effect of MST on the stability of GSSH in the presence of GSH. The reaction mixture contained 0.5 mM GSSH, 1.0 mM GSH with ( ) or without (
) or without ( ) 2 μM MST. The decay of total thiol (GSSH and GSH) was analyzed by using Ellman’s reagent.
) 2 μM MST. The decay of total thiol (GSSH and GSH) was analyzed by using Ellman’s reagent.
Escherichia coli and it mutants were grown in LB medium to OD600 nm of 1. The wild type, ΔcysM, ΔcysK, and ΔsseA produced and released 0.94, 0.9.6, 1.25, and 0.98 μM sulfide into the culture supernatants, respectively (Figure 1E). At OD600 nm of 1, they were also challenged with H2O2. E. coli ΔsseA exhibited a much greater sensitivity to H2O2 than did the wild type, while E. coliΔcysK and E. coli ΔcysM showed similar sensitivity to H2O2 (Figure 6A). Compared with the wild type, the intracellular content of reactive sulfane sulfur per mg of protein in the supernatant of E. coli ΔsseA lysate was clearly reduced (Figure 6B). The complemented mutant E. coli ΔsseA (MST) recovered the content of reactive sulfane sulfur and the survival rate when treated with H2O2 (Figures 6C,D). Thus, MST contributes to the production of cellular reactive sulfane sulfur that confers resistance to H2O2.
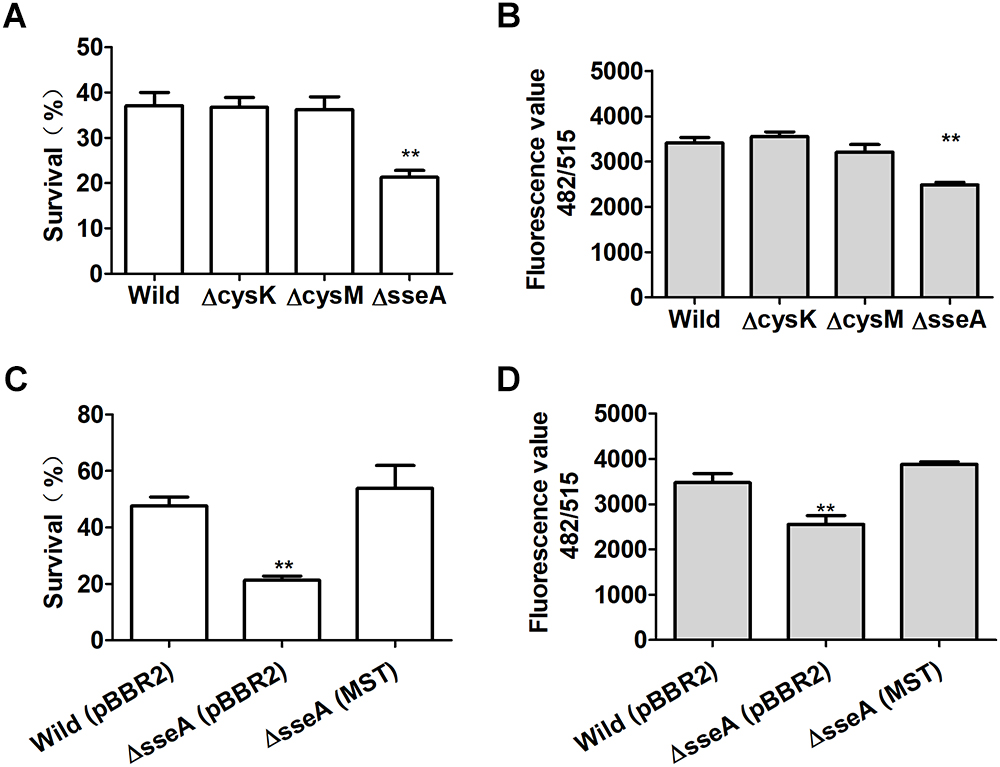
Figure 6. Reactive sulfane sulfur produced by MST protects E. coli from H2O2. (A) The survival of H2S underproducing (ΔcysM), overproducing (ΔcysK) or normal (ΔsseA and the wild type) strains under H2O2 challenge. (B) The intracellular reactive sulfane sulfur content as measured with SSP4 per mg of protein in the supernatant of the lysed E. coli strains. (C) The survival of the complemented E. coli ΔsseA (MST) under H2O2 challenge. (D) The intracellular reactive sulfane sulfur content in the supernatant of the lysed E. coli ΔsseA (MST). Data were presented as average ± SD of three independent experiments. T-tests were performed to calculate the p-values, and double asterisks indicate statistically significant difference (∗∗p < 0.01).
To assess the importance of MST, we evaluated the conservation and distribution of MST in bacteria. Five confirmed MSTs (Spallarossa et al., 2004; Nagahara and Sawada, 2006; Mikami et al., 2011) were used as seed sequences to BLAST a microbial genomic protein sequence set of 8671 bacterial genomes (NCBI updated until November 11, 2017). Our result showed that 48.2% the sequenced genomes contained MST, 97% of which had only one MST; most of MSTs (89.2%) were distributed in six classes: Gammaproteobacteria (45.4%), Betaproteobacteria (18.3%), Alphaproteobacteria (12.3%), Bacilli (7.3%), Corynebacteriales (6.0%), and Streptomycetales (1.9%) (Supplementary Table S3). We also searched the conserved domain (CDD) of the MSTs at the NCBI website and found that all MSTs contained the CDD COG2897 belonging to the cl25399 family. While the five rhodaneses that were used as the outgroup references of the phylogenetic tree contained the CDD PRK00162 belonging to the cl00125 family (Figure 7). The phylogenetic tree showed that MSTs were not grouped into distinct branches, corresponding to their wide distribution.
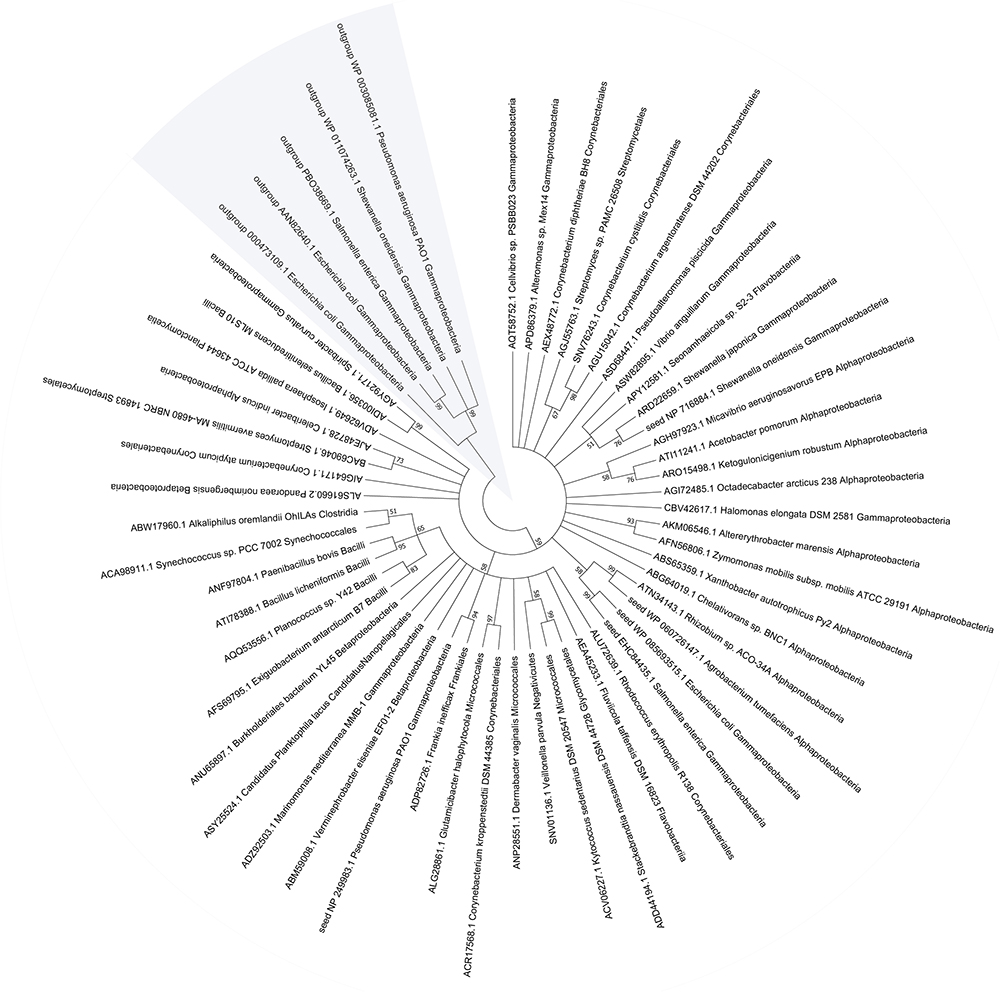
Figure 7. The phylogenetic tree and distribution of MST in bacteria. The representative MST sequences were used for phylogenetic tree construction with reported seed sequences.
L-Cysteine is the major source of H2S and reactive sulfane sulfur for E. coli, while methionine is not (Figure 1A and Supplementary Figure S1). Although thiosulfate can be used directly by CysM for the production of L-cysteine (Zhao et al., 2006), the resting E. coli cell did not use it to contribute to H2S production in the presence of glucose. It did not use the assimilatory sulfite reduction pathway to generate extra H2S, either, as sulfite did not stimulate H2S production (Supplementary Figure S1). E. coli mainly used CDs and CAT/MST to metabolize L-cysteine (Figures 2–4).
CDs convert L-cysteine to H2S, while the CAT/MST pathway produces reactive sulfane sulfur, due to different catalytic mechanisms. CDs catalyze L-cysteine metabolism through a mechanism of α,β-elimination, β-replacement, or α-hydrogen exchange with the formation of a double-bond intermediate, leading to co-elimination of H2S and NH3 (Watanabe and Snell, 1977). In the CDs catalyzed reaction, the sulfur with a valence of -2 does not undergo oxidoreduction. In the CAT/MST pathway, MST metabolizes MP to pyruvate and produces sulfane sulfur, which is initially in the form of a persulfide at the active site (Toohey, 2011; Yadav et al., 2013); MST can pass the sulfane sulfur to GSH producing GSSH (Figures 3, 4). Although the produced sulfane sulfur can be reduced to H2S by thioredoxin and glutaredoxin inside cells (Dóka et al., 2016), the CAT/MST pathway did not clearly contribute to H2S production during normal growth (Figures 1D,E) or during the metabolism of exogenous L-cysteine (Figure 1B).
GSSH produced by MST is relatively stable. GSSH was the direct product of MP oxidation by MST in the presence of GSH, and H2S was the subsequent reaction product of GSSH with excessive GSH via a relatively slow process (Figures 3, 4). This finding is in agreement with the kinetic analysis of MST for the release of H2S from MP in the presence of various small thiols and thioredoxin (Mikami et al., 2011; Yadav et al., 2013). Further, E. coli (AtBlh) metabolized MP and produced significant amounts of sulfite and thiosulfate (Figure 3), suggesting that GSSH is an intermediate and is oxidized by the recombinant persulfide dioxygenase AtBlh to sulfite. Sulfite spontaneously reacts with reactive sulfane sulfur to generate thiosulfate (Xin et al., 2017). The results are consistent with the sulfur part of L-cysteine is metabolized by CAT/MST/PDO to thiosulfate in the plant Arabidopsis thaliana (Höfler et al., 2016). However, in vivo release of H2S by E. coli cells metabolizing MP was relatively rapid (Figure 4). This is likely because the elevated levels of reactive sulfane sulfur inside E. coli cells during MP degradation. The high concentration promotes its reduction by cellular thiols, including GSH, thioredoxin, and glutaredoxin (Fahey et al., 1978; Mclaggan et al., 1990; Westrop et al., 2009; Mikami et al., 2011; Dóka et al., 2016).
CDs are multifunctional enzymes (Han et al., 2001), and L-cysteine metabolism is usually not their main physiological function. For example, TnaA is better known as tryptophanase that degrades L-tryptophan to indole, and it is also important in degrading excessive L-cysteine, as the mutant is inhibited by L-cysteine in the medium (Awano et al., 2005). Our data support the role of TnaA in degrading exogenous L-cysteine (Figure 1C). However, its deletion did not affect H2S production during the growth in LB medium (Figures 1D,E). This is expected since TnaA has an apparent Km for L-cysteine of 11 mM(Snell, 1975; Oguri et al., 2012) and L-cysteine is normally around 200 μM in E coli (Park and Imlay, 2003). Collectively, the physiological role of CDs is likely to detoxify excess L-cysteine, as previously suggest (Awano et al., 2005). Although CysK and CysM have CD activity (Awano et al., 2005), they are better known as cysteine synthases that consume H2S (Kredich and Tomkins, 1966; Zhao et al., 2006). Thus, it is expected to see H2S accumulation in E. coliΔcysK and E. coliΔcysKΔcysM (Figures 1D,E). The same phenomenon has been observed with Shewanella oneidensis and its ΔcysK mutant produces more H2S (Wu et al., 2015). The double mutant E. coliΔcysKΔcysM showed a reduced biomass (Figure 1F), likely due to the short supply of L-cysteine. Surprisingly, E. coliΔcysM accumulated slightly less H2S than the wild type. Perhaps, CysM also uses thiosulfate in LB medium to produce L-cysteine (Zhao et al., 2006), and the extra L-cysteine may contribute to H2S production in the wild type.
Nagahara and Sawada report that the CAT/MST pathway dose not contribute much to L-cysteine degradation (Nagahara and Sawada, 2006), which is in agreement with our data that the mutant has a limited impact on L-cysteine degradation and H2S production (Figures 1D,E). This is not a surprise, since CAT is also a multifunctional enzyme that also catalyzes the metabolism of aspartate, phenylalanine and tyrosine via transamination (Gelfand and Steinberg, 1977; Han et al., 2001). The ability of the CAT to catalyze the transamination for L-cysteine is much weaker than for aspartate (Sung et al., 1990), implying that L-cysteine is not a preferred substrate for CAT under the physiological conditions. This is also consistent with our observation that E. coli ΔsseA released similar amount of H2S as the wild type when growing in LB medium; the finding is different from a previous observation (Shatalin et al., 2011) that the sseA mutant produced less amount of H2S. However, our results are consistent with the sseA mutant of S. oneidensis that produced the same amount of H2S as the wild type(Wu et al., 2015).
The deletion of sseA has been reported to make the mutant sensitive to H2O2 (Shatalin et al., 2011) which is in agreement with our results with E. coli ΔsseA, but they hypothesized that H2S was responsible for the resistance. They also provided additional evidence to support the function of H2S and proposed H2S to sequestrate Fe2+ inside the cell to minimize the Fenton’s reaction as a mechanism (Mironov et al., 2017). Since we only observed a reduction in reactive sulfane sulfur, we attributed the antioxidant activity to the latter, which is in line with its chemical and physical properties (Greiner et al., 2013; Ida et al., 2014; Toohey and Cooper, 2014). Reactive sulfane sulfur GSSH and CysSSH are approximately 10∼100 times more reactive toward H2O2 than GSH and L-cysteine (Saund et al., 2015; Kasamatsu et al., 2016), and GSSH is approximately 50 times more reactive than H2S toward H2O2 (Ida et al., 2014). E. coli is known to use different approaches to resist H2O2, such as the Oxy regulon (Zheng et al., 2001) and the L-cysteine/L-cystine shuttle system (Iwao et al., 2010). Here we presented evidence to support that reactive sulfane sulfur is an additional mechanism.
Bioinformatics data showed that MST is widely distributed in bacteria. About 48.2% sequenced bacterial genomes contain MST, which is significantly more than the 20.6% for SQR (Xia et al., 2017). The wide distribution implies for its important roles such as protecting cells from oxidative stress. MSTs are also present in plants and animals (Höfler et al., 2016; Kuo et al., 2016). The phylogenetic tree of MSTs suggests that they are highly heterogeneous, but clearly separated from rhodaneses that are also highly heterogeneous (Cipollone et al., 2010). Cysteinyl-tRNA synthetase is universal in all organisms, and it can produce reactive sulfane sulfur (Akaike et al., 2017). However, cysteinyl-tRNA synthetase is relatively inefficient in producing Cys-SSH, and only about 4 μM Cys-SSH is generated after it metabolizes 100 μM L-cysteine in 60 min (Akaike et al., 2017). In comparison, MST is a specific enzyme for the production of reactive sulfane sulfur and quickly generating elevated reactive sulfane sulfur (Figure 3A). Thus, cysteinyl-tRNA synthetase may play a significant role in generating low levels of reactive sulfane sulfur, and MST has the ability to produce additional reactive sulfane sulfur, coffering E. coli with resistance to oxidative stress.
L-cysteine is the major substrate for the production of H2S and reactive sulfane sulfur in E. coli (Figure 8). CDs produce H2S, and the CAT/MST pathway generates reactive sulfane sulfur. The two forms are not readily interchangeable in E. coli, as it cannot easily oxidize H2S to reactive sulfane sulfur and the reduction of reactive sulfane sulfur by cellular thiols is not likely a major route for H2S production, as E. coli ΔsseA generated similar amount of H2S but contained less sulfane sulfur (Figures 6B,D). The produced H2S is mainly lost through evaporation into the gas phase. Therefore, H2S is not accumulated inside the cell and its concentration is likely not high enough to be responsible for protein sulfhydration under normal growth conditions. Reactive sulfane sulfur is relatively stable and is constantly present inside cells, and it is likely involved in protein sulfhydration and confers the cell with resistance to oxidative stress. Our findings provide a basis for further investigation of the cellular functions of reactive sulfane sulfur.
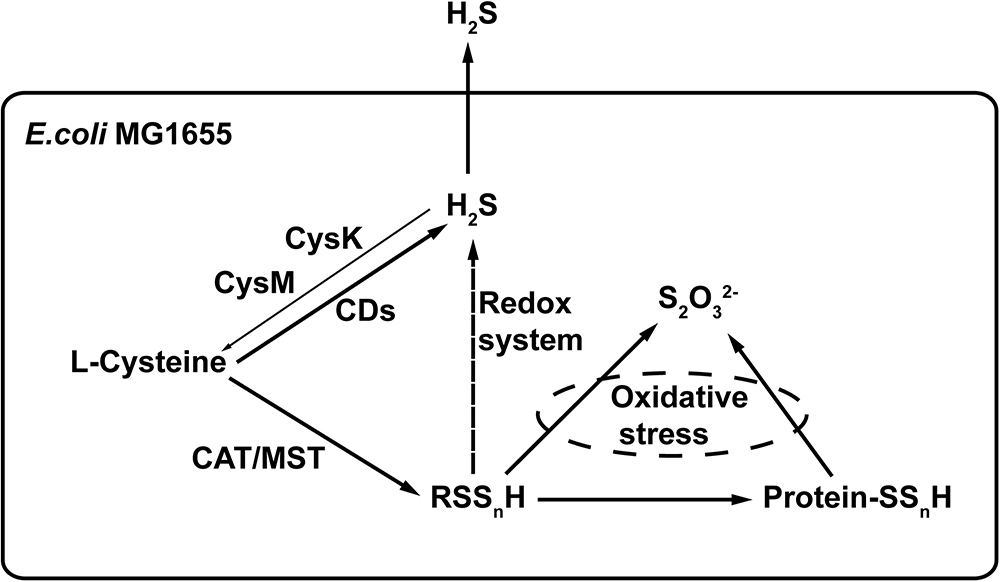
Figure 8. The proposed model of H2S and reactive sulfane sulfur production in E. coli. CDs convert L-cysteine to H2S. CAT/MST metabolize L-cysteine to generate reactive sulfane sulfur that can render protein sulfhydration. Excessive reactive sulfane sulfur is reduced to H2S by cellular thiols and thioredoxin/glutaredoxin, but this is not an active route of H2S production during normal growth. Although CysK and CysM use H2S to produce L-cysteine, most of the produced H2S is lost via evaporation. H2O2 can oxidize reactive sulfane sulfur to thiosulfate (Nagahara et al., 2012).
KL acquired and analyzed the data. YFX and GX contributed to the gene deletion and plasmid construction. RZ helped with the bioinformatics. HL and YZX supervised the research. KL, YZX, and LX designed the study and wrote the manuscript.
The work was financially supported by grants from the National Natural Science Foundation of China (91751207 and 21477062).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00298/full#supplementary-material
Akaike, T., Ida, T., Wei, F. Y., Nishida, M., Kumagai, Y., Alam, M. M., et al. (2017). Cysteinyl-tRNA synthetase governs cysteine polysulfidation and mitochondrial bioenergetics. Nat. Commun. 8:1177. doi: 10.1038/s41467-017-01311-y
Awano, N., Wada, M., Mori, H., Nakamori, S., and Takagi, H. (2005). Identification and functional analysis of Escherichia coli cysteine desulfhydrases. Appl. Environ. Microbiol. 71, 4149–4152. doi: 10.1128/AEM.71.7.4149-4152.2005
Bibli, S. I., Luck, B., Zukunft, S., Wittig, J., Chen, W., Xian, M., et al. (2018). A selective and sensitive method for quantification of endogenous polysulfide production in biological samples. Redox Biol. 18, 295–304. doi: 10.1016/j.redox.2018.07.016
Chen, Z., Zhang, X., Li, H., Liu, H., Xia, Y., and Xun, L. (2018). The complete pathway of thiosulfate utilization in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 84:e1241-18. doi: 10.1128/AEM.01241-18
Cheng, H., Donahue, J. L., Battle, S. E., Ray, W. K., and Larson, T. J. (2008). Biochemical and genetic characterization of PspE and GlpE, two single-domain sulfurtransferases of Escherichia coli. Open Microbiol. J. 2, 18–28. doi: 10.2174/1874285800802010018
Cipollone, R., Ascenzi, P., and Visca, P. (2010). Common themes and variations in the rhodanese superfamily. IUBMB Life 59, 51–59. doi: 10.1080/15216540701206859
Cooper, A. J., Haber, M. T., and Meister, A. (1982). On the chemistry and biochemistry of 3-mercaptopyruvic acid, the alpha-keto acid analog of cysteine. J. Biol. Chem. 257, 816–826.
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Dóka,É, Pader, I., Bíró, A., Johansson, K., Cheng, Q., Ballagó, K., et al. (2016). A novel persulfide detection method reveals protein persulfide and polysulfide-reducing functions of thioredoxin and glutathione systems. Sci. Adv. 2:e1500968. doi: 10.1126/sciadv.1500968
Fahey, R. C., Brown, W. C., Adams, W. B., and Worsham, M. B. (1978). Occurrence of glutathione in bacteria. J. Bacteriol. 133, 1126–1129.
Gelfand, D. H., and Steinberg, R. A. (1977). Escherichia coli mutants deficient in the aspartate and aromatic amino acid aminotransferases. J. Bacteriol. 130, 429–440.
Giedroc, D. P. (2017). A new player in bacterial sulfide-inducible transcriptional regulation. Mol. Microbiol. 105, 347–352. doi: 10.1111/mmi.13726
Greiner, R., Zoltán, P., Katrin, B., Dörte, B., Haike, A., Péter, N., et al. (2013). Polysulfides link H2S to protein thiol oxidation. Antioxid. Redox Signal. 19, 1749–1765. doi: 10.1089/ars.2012.5041
Han, Q., Fang, J., and Li, J. (2001). Kynurenine aminotransferase and glutamine transaminase K of Escherichia coli: identity with aspartate aminotransferase. Biochem. J. 360, 617–623. doi: 10.1042/bj3600617
Höfler, S., Lorenz, C., Busch, T., Brinkkötter, M., Tohge, T., Fernie, A. R., et al. (2016). Dealing with the sulfur part of cysteine: four enzymatic steps degrade l-cysteine to pyruvate and thiosulfate in Arabidopsis mitochondria. Physiol. Plant. 157, 352–366. doi: 10.1111/ppl.12454
Ida, T., Sawa, T., Ihara, H., Tsuchiya, Y., Watanabe, Y., Kumagai, Y., et al. (2014). Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proc. Natl. Acad. Sci. U.S.A. 111, 7606–7611. doi: 10.1073/pnas.1321232111
Iwao, O., Natthawut, W., Susumu, M., Takeshi, N., Hiroshi, K., and Hiroshi, T. (2010). The L-cysteine/L-cystine shuttle system provides reducing equivalents to the periplasm in Escherichia coli. J. Biol. Chem. 285, 17479–17487. doi: 10.1074/jbc.M109.081356
Kasamatsu, S., Nishimura, A., Morita, M., Matsunaga, T., Abdul, H. H., and Akaike, T. (2016). Redox signaling regulated by cysteine persulfide and protein polysulfidation. Molecules 21, e1721–e1731. doi: 10.3390/molecules21121721
Kimura, H. (2014). Production and physiological effects of hydrogen sulfide. Antioxid. Redox Signal. 20, 783–793. doi: 10.1089/ars.2013.5309
Kolluru, G. K., Shen, X., Bir, S. C., and Kevil, C. G. (2013). Hydrogen sulfide chemical biology: pathophysiological roles and detection. Nitric Oxide 35, 5–20. doi: 10.1016/j.niox.2013.07.002
Kredich, N. M., and Tomkins, G. M. (1966). The enzymic synthesis of L-cysteine in Escherichia coli and Salmonella typhimurium. J. Biol. Chem. 241, 4955–4965.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kuo, M. M., Kim, D. H., Jandu, S., Bergman, Y., Tan, S., Wang, H., et al. (2016). MPST but not CSE is the primary regulator of hydrogen sulfide production and function in the coronary artery. Am. J. Physiol. Heart Circ. Physiol. 310, H71–H79. doi: 10.1152/ajpheart.00574.2014
Libiad, M., Yadav, P. K., Vitvitsky, V., Martinov, M., and Banerjee, R. (2014). Organization of the human mitochondrial hydrogen sulfide oxidation pathway. J. Biol. Chem. 289, 30901–30910. doi: 10.1074/jbc.M114.602664
Lü, C., Xia, Y., Liu, D., Zhao, R., Gao, R., Liu, H., et al. (2017). Cupriavidus necator H16 uses flavocytochrome c-sulfide dehydrogenase to oxidize self-produced and spiked sulfide. Appl. Environ. Microbiol. 83:e01610-17. doi: 10.1128/AEM.01610-17
Mclaggan, D., Logan, T. M., Lynn, D. G., and Epstein, W. (1990). Involvement of gamma-glutamyl peptides in osmoadaptation of Escherichia coli. J. Bacteriol. 172, 3631–3636. doi: 10.1128/jb.172.7.3631-3636.1990
Mikami, Y., Shibuya, N., Kimura, Y., Nagahara, N., Ogasawara, Y., and Kimura, H. (2011). Thioredoxin and dihydrolipoic acid are required for 3-mercaptopyruvate sulfurtransferase to produce hydrogen sulfide. Biochem. J. 439, 479–485. doi: 10.1042/BJ20110841
Mironov, A., Seregina, T., Nagornykh, M., Luhachack, L. G., Korolkova, N., Lopes, L. E., et al. (2017). Mechanism of H2S mediated protection against oxidative stress in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 114, 6022–6027. doi: 10.1073/pnas.1703576114
Nagahara, N., Nirasawa, T., Yoshii, T., and Niimura, Y. (2012). Is novel signal transducer sulfur oxide involved in the redox cycle of persulfide at the catalytic site cysteine in a stable reaction intermediate of mercaptopyruvate sulfurtransferase? Antioxid. Redox Signal. 16, 747–753. doi: 10.1089/ars.2011.4468
Nagahara, N., Okazaki, T., and Nishino, T. (1995). Cytosolic mercaptopyruvate sulfurtransferase is evolutionarily related to mitochondrial rhodanese. J. Biol. Chem. 270, 16230–16235. doi: 10.1074/jbc.270.27.16230
Nagahara, N., and Sawada, N. (2006). The mercaptopyruvate pathway in cysteine catabolism: a physiologic role and related disease of the multifunctional 3-mercaptopyruvate sulfurtransferase. Curr. Med. Chem. 13, 1219–1230. doi: 10.2174/092986706776360914
Nagy, P. (2015). Mechanistic chemical perspective of hydrogen sulfide signaling. Nitric Oxide 47, S5–S6. doi: 10.1016/bs.mie.2014.11.036
Oguri, T., Schneider, B., and Reitzer, L. (2012). Cysteine catabolism and cysteine desulfhydrase (CdsH/STM0458) in Salmonella enterica serovar typhimurium. J. Bacteriol. 194, 4366–4376. doi: 10.1128/JB.00729-12
Pan, J., and Carroll, K. S. (2013). Persulfide reactivity in the detection of protein s-sulfhydration. ACS Chem. Biol. 8, 1110–1116. doi: 10.1021/cb4001052
Park, S., and Imlay, J. A. (2003). High levels of intracellular cysteine promote oxidative DNA damage by driving the fenton reaction. J. Bacteriol. 185, 1942–1950. doi: 10.1128/JB.185.6.1942-1950.2003
Paul, B. D., and Snyder, S. H. (2012). H2S signalling through protein sulfhydration and beyond. Nat. Rev. Mol. Cell Biol. 13, 499–507. doi: 10.1038/nrm3391
Riener, C. K., Kada, G., and Gruber, H. J. (2002). Quick measurement of protein sulfhydryls with ellman’s reagent and with 4,4′-dithiodipyridine. Anal. Bioanal. Chem. 373, 266–276. doi: 10.1007/s00216-002-1347-2
Saund, S. S., Sosa, V., Henriquez, S., Nguyen, Q. N., Bianco, C. L., Soeda, S., et al. (2015). The chemical biology of hydropersulfides (RSSH): chemical stability, reactivity and redox roles. Arch. Biochem. Biophys. 588, 15–24. doi: 10.1016/j.abb.2015.10.016
Shatalin, K., Shatalina, E., Mironov, A., and Nudler, E. (2011). H2S: a universal defense against antibiotics in bacteria. Science 334, 986–990. doi: 10.1126/science.1209855
Shen, J., Hui, P., Zhang, Y., Trinidad, J. C., and Giedroc, D. P. (2016). Staphylococcus aureus sqr encodes a type II sulfide:quinone oxidoreductase and impacts reactive sulfur speciation in cells. Biochemistry 55, 6524–6534. doi: 10.1021/acs.biochem.6b00714
Shimada, T., Tanaka, K., and Ishihama, A. (2016). Transcription factor DecR (YbaO) controls detoxification of L-cysteine in Escherichia coli. Microbiology 162, 1698–1707. doi: 10.1099/mic.0.000337
Snell, E. E. (1975). Tryptophanase: structure, catalytic activities, and mechanism of action. Adv. Enzymol. Relat. Areas Mol. Biol. 42, 287–333. doi: 10.1002/9780470122877.ch6
Spallarossa, A., Forlani, F., Carpen, A., Armirotti, A., Pagani, S., Bolognesi, M., et al. (2004). The “rhodanese” fold and catalytic mechanism of 3-mercaptopyruvate sulfurtransferases: crystal structure of SseA from Escherichia coli. J. Mol. Biol. 335, 583–593. doi: 10.1016/j.jmb.2003.10.072
Sung, M. H., Tanizawa, K., Tanaka, H., Kuramitsu, S., Kagamiyama, H., and Soda, K. (1990). Purification and characterization of thermostable aspartate aminotransferase from a thermophilic Bacillus species. J. Bacteriol. 172, 1345–1351. doi: 10.1128/jb.172.3.1345-1351.1990
Toohey, J. I. (1986). Persulfide sulfur is a growth factor for cells defective in sulfur metabolism. Biochem. Cell Biol. 64, 758–765. doi: 10.1139/o86-103
Toohey, J. I. (2011). Sulfur signaling: is the agent sulfide or sulfane? Anal. Biochem. 413, 1–7. doi: 10.1016/j.ab.2011.01.044
Toohey, J. I., and Cooper, A. J. L. (2014). Thiosulfoxide (sulfane) sulfur: new chemistry and new regulatory roles in biology. Molecules 19, 12789–12813. doi: 10.3390/molecules190812789
Wang, R. (2010). Hydrogen sulfide: the third gasotransmitter in biology and medicine. Antioxid. Redox Signal. 12, 1061–1064. doi: 10.1089/ars.2009.2938
Watanabe, T., and Snell, E. E. (1977). The interaction of Escherichia coli tryptophanase with various amino and their analogs. Active site mapping. J. Biochem. 82, 733–745. doi: 10.1093/oxfordjournals.jbchem.a131750
Westrop, G. D., Georg, I., and Coombs, G. H. (2009). The mercaptopyruvate sulfurtransferase of Trichomonas vaginalis links cysteine catabolism to the production of thioredoxin persulfide. J. Biol. Chem. 284, 33485–33494. doi: 10.1074/jbc.M109.054320
Wu, G., Li, N., Mao, Y., Zhou, G., and Gao, H. (2015). Endogenous generation of hydrogen sulfide and its regulation in Shewanella oneidensis. Front. Microbiol. 6:374. doi: 10.3389/fmicb.2015.00374
Xia, Y., Lü, C., Hou, N., Xin, Y., Liu, J., Liu, H., et al. (2017). Sulfide production and oxidation by heterotrophic bacteria under aerobic conditions. ISME J. 11, 2754–2766. doi: 10.1038/ismej.2017.125
Xin, Y., Liu, H., Cui, F., Liu, H., and Xun, L. (2017). Recombinant Escherichia coli with sulfide:quinone oxidoreductase and persulfide dioxygenase rapidly oxidises sulfide to sulfite and thiosulfate via a new pathway. Environ. Microbiol. 18, 5123–5136. doi: 10.1111/1462-2920.13511
Yadav, P. K., Martinov, M., Vitvitsky, V., Seravalli, J., Wedmann, R., Filipovic, M. R., et al. (2016). Biosynthesis and reactivity of cysteine persulfides in signaling. J. Am. Chem. Soc. 138, 289–299. doi: 10.1021/jacs.5b10494
Yadav, P. K., Yamada, K., Chiku, T., Koutmos, M., and Banerjee, R. (2013). Structure and kinetic analysis of H2S production by human mercaptopyruvate sulfurtransferase. J. Biol. Chem. 288, 20002–20013. doi: 10.1074/jbc.M113.466177
Zhao, C., Kumada, Y., Imanaka, H., Imamura, K., and Nakanishi, K. (2006). Cloning, overexpression, purification, and characterization of O -acetylserine sulfhydrylase-B from Escherichia coli. Protein Expr. Purif. 47, 607–613. doi: 10.1016/j.pep.2006.01.002
Keywords: L-cysteine, hydrogen sulfide, reactive sulfane sulfur, antioxidant, 3-mercaptopyruvate sulfurtransferase
Citation: Li K, Xin Y, Xuan G, Zhao R, Liu H, Xia Y and Xun L (2019) Escherichia coli Uses Separate Enzymes to Produce H2S and Reactive Sulfane Sulfur From L-cysteine. Front. Microbiol. 10:298. doi: 10.3389/fmicb.2019.00298
Received: 26 October 2018; Accepted: 04 February 2019;
Published: 20 February 2019.
Edited by:
Christiane Dahl, Universität Bonn, GermanyReviewed by:
Evgeny Nudler, New York University, United StatesCopyright © 2019 Li, Xin, Xuan, Zhao, Liu, Xia and Xun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongzhen Xia, eGlheW9uZ3poZW4yMDAyQHNkdS5lZHUuY24= Luying Xun, bHV5aW5nX3h1bkB3c3UuZWR1; bHV5aW5nX3h1bkB2ZXRtZWQud3N1LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.