- 1Department of Ecology and Evolutionary Biology, University of California, Los Angeles, Los Angeles, CA, United States
- 2Centre for Social Evolution, University of Copenhagen, Copenhagen, Denmark
While strict vertical transmission insures the durability of intracellular symbioses, phylogenetic incongruences between hosts and endosymbionts suggest horizontal transmission must also occur. These horizontal acquisitions can have important implications for the biology of the host. Wolbachia is one of the most ecologically successful prokaryotes in arthropods, infecting an estimated 50–70% of all insect species. Much of this success is likely due to the fact that, in arthropods, Wolbachia is notorious for manipulating host reproduction to favor transmission through the female germline. However, its natural potential for horizontal transmission remains poorly understood. Here we evaluate the fundamental prerequisites for successful horizontal transfer, including necessary environmental conditions, genetic potential of bacterial strains, and means of mediating transfers. Furthermore, we revisit the relatedness of Wolbachia strains infecting the Panamanian leaf-cutting ant, Acromyrmex echinatior, and its inquiline social parasite, Acromyrmex insinuator, and compare our results to a study published more than 15 years ago by Van Borm et al. (2003). The results of this pilot study prompt us to reevaluate previous notions that obligate social parasitism reliably facilitates horizontal transfer and suggest that not all Wolbachia strains associated with ants have the same genetic potential for horizontal transmission.
Introduction
Wolbachia pipientis is a maternally inherited α-proteobacterium widely found in arthropods (Werren et al., 2008). Wolbachia exhibits reproductive parasitism in most arthropod species by manipulating the reproductive physiology of hosts and inducing female-biased sex ratios via one of four mechanisms: cytoplasmic incompatibility, feminization of genetic males, male-killing, or enforcing parthenogenesis (for reviews see Stouthamer et al., 1999; Werren et al., 2008). Although transmission within species is strictly vertical, Wolbachia phylogenies rarely correspond to host phylogenies, suggesting horizontal transmission (HT) also occurs (Zhou et al., 1998; Vavre et al., 1999; Raychoudhury et al., 2009; Stahlhut et al., 2010; Ahmed et al., 2013).
Horizontal transmission of intracellular bacterial symbionts require intimate tissue-level interaction between current and future hosts; predator–prey and host–parasitoid relationships have therefore been proposed to explain observed HT events (e.g., Heath et al., 1999; Noda et al., 2001; Yang et al., 2013; Carvalho et al., 2014; Ahmed et al., 2015; Mascarenhas et al., 2016). Ants are the only lineage of social Hymenoptera where permanent social parasites, closely related to their host, commonly invade mature colonies (Boomsma et al., 2014). As ants are perennial, intimate inquiline cohabitation where social parasites live with hosts across generations offers ample opportunities for HT. This idea was first explored in Acromyrmex echinatior colonies, which are considered closed systems for endosymbionts since workers are highly aggressive toward non-nestmates (Larsen et al., 2014). Colonies can, however, be infiltrated by socially parasitic Acromyrmex insinuator queens, which invade and adopt the host colony odor (Lambardi et al., 2007; Nehring et al., 2015). Van Borm et al. (2003) first suggested that HT events occur between Wolbachia endosymbionts of A. echinatior and A. insinuator based on shared bacterial genotypes between cohabiting ant species. Later research suggested the same for Solenopsis daguerrei, a social parasite of S. saevissima (Dedeine et al., 2005; Martins et al., 2012) and for another fungus-growing ant, Sericomyrmex amabilis, and its social parasite Megalomyrmex symmetochus (Adams et al., 2013; Liberti et al., 2015).
In this perspective, we summarize research that has investigated Wolbachia HT events in ants, examine limitations of methods and study systems used, and propose future research. We also partially repeat one of the first field studies (Van Borm et al., 2003) characterizing Wolbachia endosymbionts of two fungus-growing ant species: the leaf-cutting ant, A. echinatior, and its social parasite, A. insinuator. Our pilot results, originating from a single A. echinatior colony parasitized by three A. insinuator queens, only partially confirmed these earlier findings. This highlights the importance of re-evaluating past and current methods and redirecting future efforts to include whole genome sequencing (WGS) data, which could increase the resolution of phylogenetic relationships and reveal pioneering insights into the genes and mechanisms that allow Wolbachia to jump to new hosts.
Methods
A single A. echinatior queen and three parasitic A. insinuator queens cohabiting a mature colony (Ae724; collected in Gamboa, Panama, May 2015) were isolated in separate sterile petri dishes (similar to Stürup et al., 2014). After a 36-h period, ca. 40 eggs were collected from each queen and stored at −20°C. DNA was extracted using the DNeasy Tissue Kit (Qiagen) and a 603 bp region of the Wolbachia surface protein (wsp) was amplified using 81F/691R primers (Braig et al., 1998) and PCR conditions as described in Baldo et al. (2006b). PCR products were purified using the Invitek PCR purification kit, cloned using the TOPO TA cloning kit (Invitrogen, United States), and 24 colonies from each cloning were sent for Sanger sequencing (MWG, Germany). We checked chromatographs and removed primer sequences using Geneious (v. 9.0.4). Trimmed sequences (MG547478-MG547559) were queried against the non-redundant NCBI database to compile the top 100 hits. All sequences were aligned with ClustalW, sites with gaps were removed and sequences that could not align to the entire 426 bp reduced alignment were removed. Maximum Likelihood phylogenetic trees with 1,000 bootstrap iterations and the TVM+G model (jmodeltest v2.1.7) were run in Garli version 2.01.1067 (Zwickl, 2006). A consensus tree was configured in Geneious v9 (Kearse et al., 2012), and one representative wsp sequence from the same host species (>99%) was picked. The tree was further modified in FigTree v1.4.3 (Rambaut, 2016). As described in Baldo et al. (2006b), the strain profiles for each wsp sequence from this study was identified based on four conserved hypervariable regions (HVR) (Supplementary Tables S1, S2). Since eggs were pooled for sampling, coinfecting strains present in each species may not occupy the same individuals.
Results
Van Borm et al. (2003) originally characterized nine Wolbachia infections: two strains in A. echinatior, four in Acromyrmex octospinosus, and three in their social parasite, A. insinuator. Some strains were specific to Acromyrmex ant species (A1 and B2), while others were present across multiple species (B1 and Bcons). Considering this earlier study was conducted >15 years ago, we reconstructed the phylogenetic relationships of previously identified wsp sequences (Van Borm et al., 2003), wsp sequences generated in our pilot study (from one host and three cohabiting parasitic queens), and closely related wsp sequences available on NCBI from other arthropod hosts (Figure 1). Using similar methods as Van Borm et al. (2003) (with the exception of extracting DNA from eggs rather than gynes), we identified three wsp genotypes named HVR1-3 (Supplementary Tables S1, S2). While HVR-1 was the dominant A. echinatior strain in our study (Supplementary Figure S1), it was not identified in the previous study. HVR-2 was identical (>99%) to strain B1 (AF472563; Van Borm study). We identified HVR-2 in both A. insinuator and A. echinatior while Van Borm et al. (2003) found HVR-2 only in A. insinuator and a closely related but not identical strain (Bcons) in A. echinatior. HVR-2 has also been found in A. octospinosus (Van Borm et al., 2003; Andersen et al., 2012). HVR-3 was identical (>99%) to strain B2 (AF472560; Van Borm study) and, as before, was only found in A. insinuator. Two strains identified before (AF472558-9) were not found in the colony we analyzed. The Van Borm study suggested multiple HT events occurred for Acromyrmex ants to acquire their Wolbachia, as evidenced by their findings showing distantly related Wolbachia strains shared by closely related Acromyrmex hosts and the reverse, closely related Wolbachia present in distantly related host species. Our results were consistent with the Van Borm study where wsp sequences from Acromyrmex hosts were most similar to those from distantly related Solenopsis fire ant hosts. Our new phylogeny also revealed additional ant hosts harboring closely related wsp sequences. HVR-2 seemed the most cosmopolitan strain in ant hosts as it is present in at least nine ant genera (Figure 1).
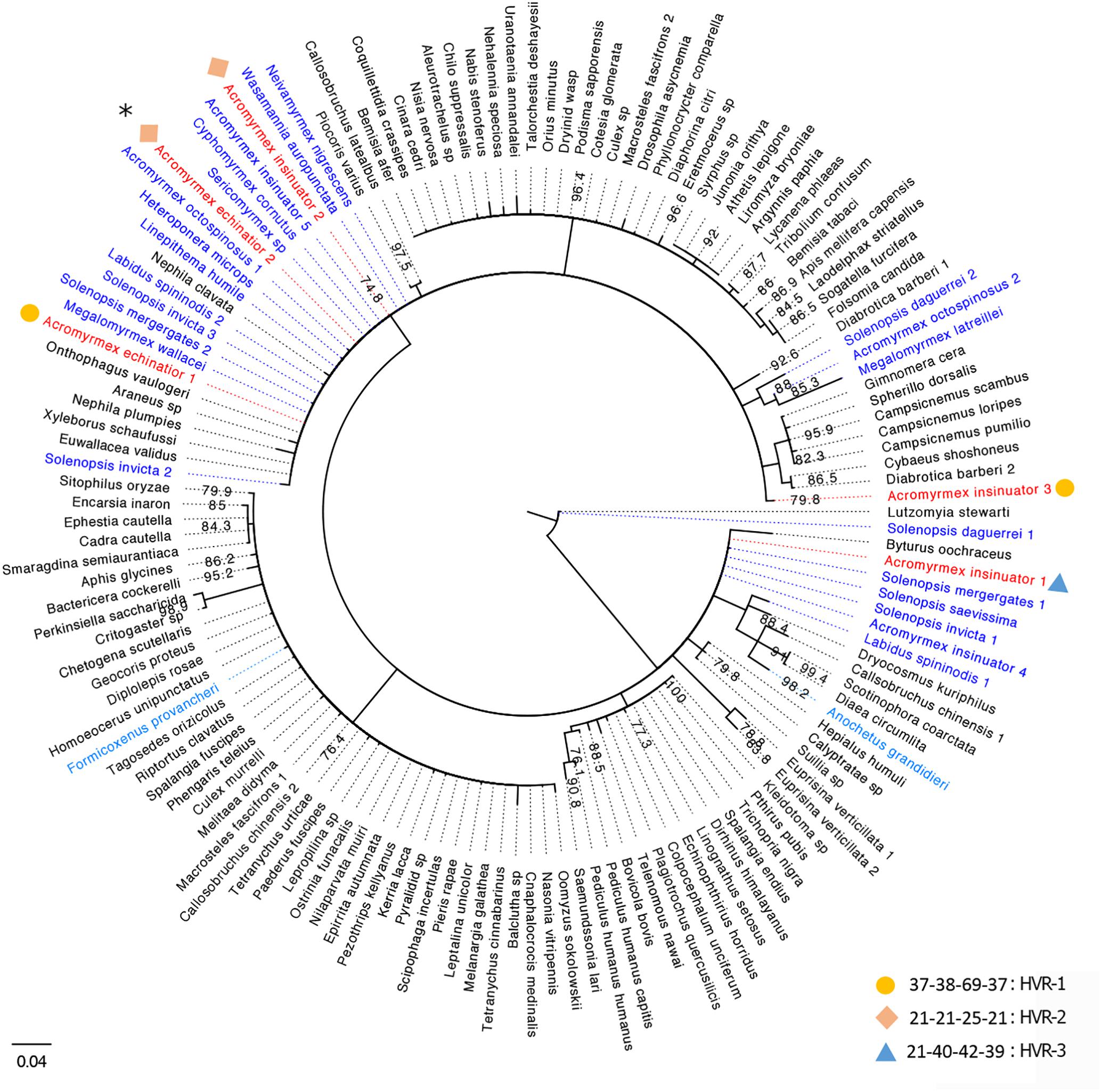
Figure 1. Consensus tree for Wolbachia strains based on the wsp gene. Strains are represented by the infected arthropod host species with which they are associated. Host names based on sequences generated in the current study are colored red and those from previous studies on ant species are colored blue (Neotropical species in dark blue, two others in light blue). Sequences from Wolbachia strains associated with non-ant hosts are presented in black font and bootstrap support is shown at each node. In total, 24 unique Wolbachia sequences from ant hosts and 112 sequences from non-ant arthropod hosts were downloaded from GenBank and used in this phylogenetic analysis. Numbers next to species names represent genetically distinct strains harbored in the same species. Information about the HVR barcoding is given next to each of the Wolbachia strains identified in our study using circles, diamonds, and triangles (legend bottom right), for details see Supplementary Tables S1, S2. Since HVR barcoding was only analyzed for the 83 wsp sequences from the four Acromyrmex queens used in this study, only the representative strains from our study in red font are given a corresponding HVR type. Strains matching the Van Borm et al. (2003) isolates, B2 and B1, are labeled here as Acromyrmex insinuator 4 and 5. The majority of BLAST hits clustering with the A. echinatior and A. insinuator sequences generated in this study are Wolbachia strains from Neotropical New World ant species. The only BLAST hits from ant hosts that are not Neotropical New World ant species were phylogenetically isolated (light blue species; Formicoxenus provancheri, occurring in North America, and Anochetus grandidieri, a species endemic to Madagascar). The asterisk at the top left of the figure marks the HVR-2 strain that is widespread among mainly ants, but also other insect hosts in the Americas.
Discussion
HT events are believed to have largely contributed to the Wolbachia pandemic, where an estimated 50–70% of all insect species are infected (Werren et al., 2008; Saridaki and Bourtzis, 2010; Weinert et al., 2015). High frequency of phylogenetic incongruences between hosts and Wolbachia strains (as seen in Figure 2) suggests HT events are relatively common on an evolutionary time scale despite the fact that they are difficult to predict and observe in nature. The results of our pilot experiment support the hypothesis that HT has occurred between A. echinatior and its social parasite, A. insinuator, originally proposed by Van Borm et al. (2003). As in the Van Borm study, we found distantly related Wolbachia strains occupying the same host (HVR-2 and -3 in A. insinuator) as well as identical strains occupying distantly related hosts (HVR-2; Figures 1, 2). Although social parasitism should provide ample opportunity for HT, our results suggest some strains, like HVR-2, may be better equipped to “jump” between hosts. Although much about HT remains unknown, minimum conditions must be fulfilled for HT to occur: (1) there must be suitable environmental conditions (in the new host as well as the medium/environment the bacteria transitions through), (2) the bacterial strain must have the genetic potential for transfer, and (3) there must be a mechanism that will mediate the HT event.
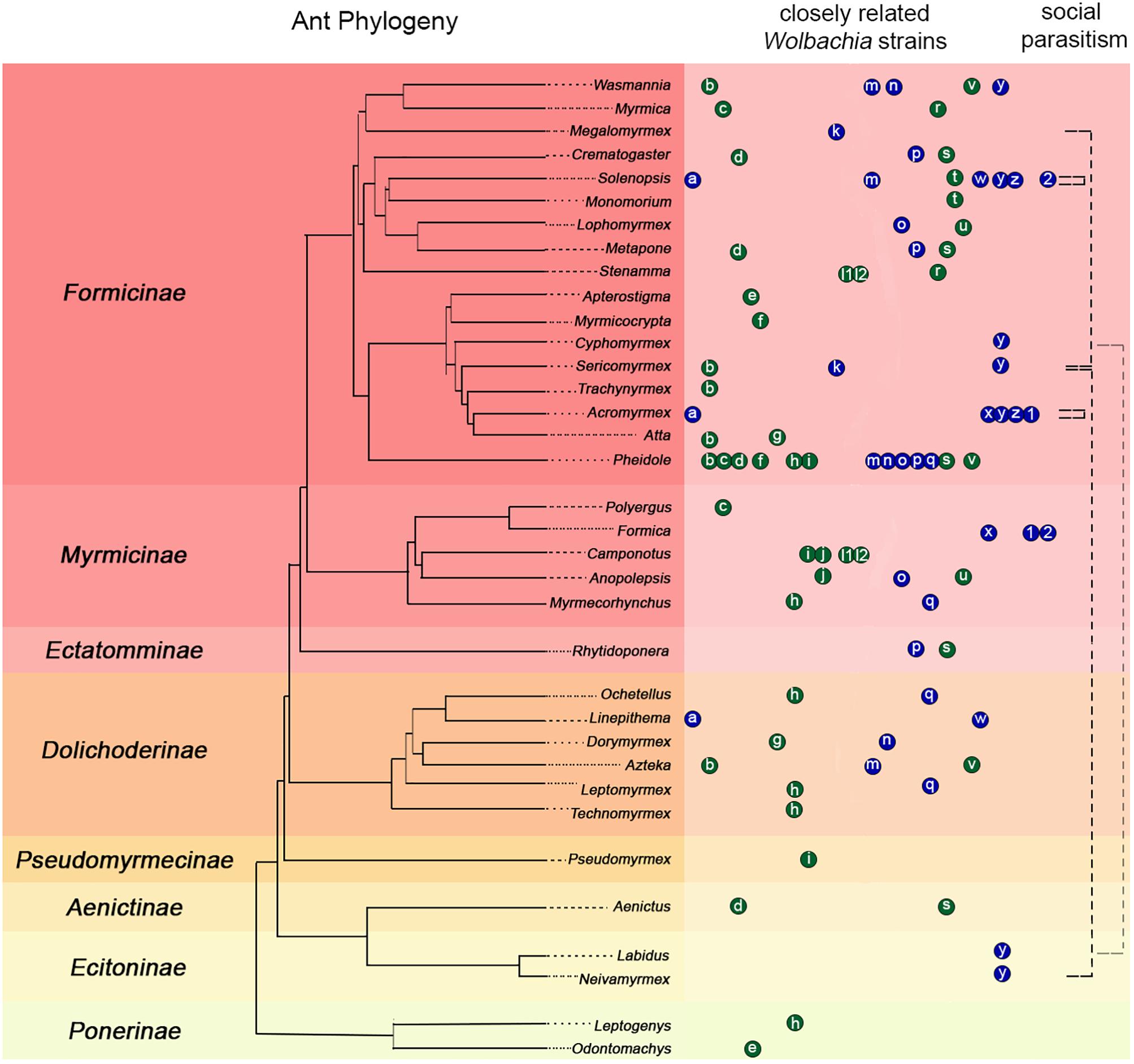
Figure 2. Compilation of previous literature including ant phylogenies, Wolbachia infections, strain typing, and known connections between social parasitism and shared Wolbachia strains. The ant phylogeny on the left was constructed using data from Moreau et al. (2006), Schultz and Brady (2008), and Branstetter et al. (2017). Ant subfamilies are presented on the left of the phylogeny with colored backgrounds separating them. Colored circles with small case letters on the right of the ant phylogeny connect ant hosts suspected to carry similar Wolbachia strains based on previous phylogenies using the wsp gene (blue) or MLST typing (green). Letters in the circles indicate the published source where authors built phylogenies that showed potentially common Wolbachia strains in different host ant species: a: (Dedeine et al., 2005); b–j: (Frost et al., 2010); k: (Liberti et al., 2015); l1: (Ramalho et al., 2017); m–q: (Rey et al., 2013); h, l2, r–v: (Russell et al., 2009); w: (Fernando de Souza et al., 2009); x: (Tsutsui et al., 2003; Reuter et al., 2005); y: (this study); z: (Van Borm et al., 2003); 1, 2: (Viljakainen et al., 2008). Each letter circle occupying the same column represents a set of highly similar Wolbachia strains defined as belonging to the same clade (or a potentially common identical strain) according to the corresponding source publication. Circle order is alphabetical and distances between circles are not indicative of the genetic similarity of strains. Therefore, some heavily sampled genera, such as Pheidole, may have the same strain represented in more than one publication in which different ant genera were analyzed. The black dashed lines in the rightmost column connect known instances of social parasitism between ants based on previous literature (see text for details). Black dashed lines that start and end in the same genus highlight that Solenopsis and Acromyrmex have social parasites within their genera and they share identical Wolbachia strains with them (Van Borm et al., 2003; Dedeine et al., 2005; Martins et al., 2012; our study). The light gray dashed line connecting Labidus and Cyphomyrmex highlight an almost identical shared Wolbachia strain (differing only by 1 bp; Figure 1). However, there is no data suggesting Labidus predates Cyphomyrmex colonies.
Part 1: Wolbachia Genetic Potential
Wolbachia can be artificially transferred across insect genera in the lab (e.g., Zabalou et al., 2004; Hoffmann et al., 2011) and following transfers, adaptations to new hosts may rapidly occur (McMeniman et al., 2008). This ability to invade new hosts is consistent with the identification of genetically similar strains in taxonomically unrelated hosts (e.g., Heath et al., 1999; Raychoudhury et al., 2009). Wolbachia is obligately intracellular yet is capable of surviving extracellularly for several months before reinvading new cells and establishing a stable infection (Rasgon et al., 2006). Although mechanisms of natural HT remain elusive, Wolbachia has demonstrated the ability to successfully “jump” across cells, cross somatic tissues, and reach reproductive organs (Frydman et al., 2006; White et al., 2017). Successful transfers may be attributed to the bacterium’s ability to adapt to new environments. This could be accomplished by recombination, likely mediated by inactive bacteriophages introducing “exotic genes,” resulting in gene gains and diversification of the bacterium’s genome (Wu et al., 2004; Klasson et al., 2009; Vos and Didelot, 2009; Ellegaard et al., 2013). Indeed, the Wolbachia genome has a high number of repetitive elements and ankyrins, mostly introduced by bacteriophages (Ishmael et al., 2009; Kent and Bordenstein, 2010; Leclercq et al., 2011; Siozios et al., 2013). While the function of these gene gains has not been fully deciphered, genomic comparisons with a mutualistic strain infecting nematode hosts, wBm (Foster et al., 2005), suggest they play a role in the bacterium’s ability to induce reproductive phenotypes in arthropods.
Considering the significant genomic differences and tissue tropisms between Wolbachia strains, we expect not all strains have the same potential for transmission. For example, while Wolbachia is typically localized in the reproductive tract (e.g., wMel, wSty), there are some B-group strains that colonize somatic (non-reproductive) tissues (e.g., wNo, wMa; Veneti et al., 2004). As expected, not all strains can survive a transfer or induce reproductive phenotypes necessary to facilitate its spread in new host populations (Zabalou et al., 2008; Veneti et al., 2012). Phylogenetic comparisons using wsp sequences (Van Borm et al., 2003; Figure 1) also suggest that one of the strains in Acromyrmex (HVR-2) may have a greater propensity for HT than HVR-1 and HVR-3. HVR-2 is not only common across the Panamanian Acromyrmex species (A. echinatior, A. insinuator, A. octospinosus), where it has been identified as wSinvictaB (Andersen et al., 2012), but also in ant hosts across four subfamilies (Figure 1). In contrast, HVR-1 and HVR-3 appear specific to their respective host species and are far more dominant in those hosts than the shared HVR-2 (Supplementary Figure S1). This distribution suggests that HVR-1 and HVR-3 are better adapted to their respective host species while HVR-2 is a generalist capable of infecting hosts with diverse life histories. Interestingly, HVR-2 (wSinvictaB) appears to be dominant in A. octospinosus (Andersen et al., 2012), but occurs as either a single or double infection with the rare and sparse wSinvictaA (Andersen et al., 2012).
Part 2: Potential Transmission Routes in Ants
Ant sociality offers ample opportunities for Wolbachia transfer across hosts and may be especially favorable for species prone to interspecific social interactions or with less restrictive tissue tropisms. For example, fungus-growing ants are a host where Wolbachia has uncommon tissue tropism; it is present extracellularly in the gut lumen and may reach high titers in the hemolymph (Andersen et al., 2012; Frost et al., 2014; Sapountzis et al., 2015). A common resource, such as a fungal garden, may thus facilitate HT of Wolbachia strains between cohabiting A. echinatior and A. insinuator, as the ants deposit their feces in the fungus, feed on it, and cover their brood with it (which also feeds on the fungus). Similarly, an identical Wolbachia strain has been found between a workerless social parasite, S. daguerrei, and its host ant species S. invicta (Dedeine et al., 2005). However, a shared Wolbachia strain was not found between M. symmetochus social mercenaries and its host, S. amabilis, suggesting cohabitation does not always result in HT (Liberti et al., 2015).
Inquiline mites may also have the capacity to vector Wolbachia between attine species cohabiting the same nest or foraging on the same plants. However, mites in Acromyrmex nests appear to be saprophytic, not parasitic (Peralta and Martínez, 2013), making this alternative transmission route unlikely. Parasitic phorid flies could also serve as a common vector between all three ant species (Brown and Feener, 1998; Fernández-Marín et al., 2006; Pérez-Ortega et al., 2010; Guillade and Folgarait, 2015), however, so far there is no data suggesting they have contributed to HT events (Dedeine et al., 2005).
Independent of being intra- or extra-cellular symbionts, HT may also be mediated by predators such as Neivamyrmex, a genus of army ant known to raid nests of fungus-growing ants and consume their brood (Lapolla et al., 2002; Powell and Clark, 2004). Army ant taxa (subfamilies Aenictinae, Dorylinae, and Ecitoninae) are often infected with Wolbachia and thus offer exciting opportunities for studying potential HT (Figure 2). HVR-2 is distributed across species from the subfamilies Myrmicinae (Acromyrmex and Sericomyrmex) and Ecitoninae (Neivamyrmex; Figures 1, 2). Similarly, an identical Wolbachia strain is shared between Cyphomyrmex and army ants of the genus Labidus (subfamily Ecitoninae; Figure 1), however, there is no known data confirming whether these army ants attack fungus-growing ants (Figure 2).
Part 3: Genomic Data and Sampling Power Limitations
Wolbachia strain typing has relied on several different genes, one of them being the 16S rDNA gene used when performing targeted sequencing (e.g., Kautz et al., 2013; Ramalho et al., 2017). This method is not appropriate to build phylogenies as the 16s gene is highly conserved and cannot distinguish closely related Wolbachia strains (Andersen et al., 2012). The wsp gene has also been used extensively for Wolbachia characterization because its rapid sequence evolution enables differentiation between closely related strains and it contains four HVRs useful in solidifying strain identification (Baldo et al., 2006b). However, the relatively short sequence length (<600 bp), high recombination rate (Baldo et al., 2005) and, in some arthropod hosts, strong positive selection (Jiggins et al., 2002), make wsp suboptimal for constructing phylogenies. Nevertheless, the wsp gene remains a useful “quick and dirty” approach to distinguish phylogenetic relationships of Wolbachia strains and is, in most cases, the only sequence available to build phylogenies. Due to these limitations, multilocus sequence typing (MLST) was introduced, which uses concatenated alignments of five housekeeping genes (Baldo et al., 2006a; Bordenstein et al., 2009). However, due to frequent recombination, WGS is the only accurate method to infer phylogenetic relationships (Bleidorn and Gerth, 2018).
A particular challenge to studying the evolutionary relationships of Wolbachia in arthropods is that hosts are frequently infected with multiple strains (Hiroki et al., 2004; Mouton et al., 2004; Frost et al., 2010; Andersen et al., 2012; Zhao et al., 2013), making even MLST and WGS approaches exceedingly challenging. Acromyrmex ants are one such example as they almost always contain multiple strains (Van Borm et al., 2003; Andersen et al., 2012) and we do not yet have Wolbachia genome data. Wsp typing has confirmed distinct, species-specific Wolbachia strains for A. echinatior (HVR-1) and A. insinuator (HVR-3) as well as a shared strain between the two species and A. octospinosus (HVR-2; Van Borm et al., 2003; Andersen et al., 2012). Differences from this study and Van Borm et al. (2003) could mean strains are transient or that diversity is greater than what is currently known. On the other hand, differences may be related to limited ant colony sampling. Many ant species have wide geographic distributions (e.g., Linepithema, Monomorium, Solenopsis, Atta, and Acromyrmex genera) and show significant differences in infections among colonies and geographic locations (e.g., Reuter et al., 2005; Frost et al., 2010; Martins et al., 2012; Zhukova et al., 2017). Thus, despite previous efforts to illustrate Wolbachia HT events, success has been limited because we have only characterized small subpopulations and because Wolbachia may be evolving and spreading to new hosts faster than we currently study it.
Part 4: Implications for Future Research
Although limited, existing data suggests Wolbachia associated with ants are uniquely shaped by the ant microenvironment and have occasionally taken advantage of opportunities offered by the hosts’ wide range of social interactions to “jump” to other ant species or genera. Comparisons between the widespread HVR-2 and less common strains, HVR-1 and -3, offer an exciting opportunity for future research because these strains (i) have different specificity to ant hosts (frequencies, infection levels), and (ii) have strikingly different distributions across phylogenetically distant ant hosts (although this may be driven by under-sampling). This suggests HVR-2 may have acquired (or lost) a set of genes that have facilitated its “ecological success.” Future genomic comparisons may allow us to answer important questions about Wolbachia evolution and HT including, why strains like HVR-2 have greater ecological success (spread), and what genes and mechanisms are associated with the ability to spread successfully across distantly related host species.
The most reliable Wolbachia phylogenies have been built using WGS data (Klasson et al., 2009; Ellegaard et al., 2013; Gerth et al., 2014; Gerth and Bleidorn, 2016). These phylogenies have resolved important gaps in our knowledge of Wolbachia origin and supergroup diversification as they are typically built using conserved orthologs unaffected by recombination, which would render topologies invalid (Gerth et al., 2014; Gerth and Bleidorn, 2016). Further mapping of Wolbachia diversity on host ant trees and more genomic data, particularly involving ants not hailing from the Americas, will be required to assess biogeography patterns, such as whether there are specialized Wolbachia lineages infecting New World ants (Russell et al., 2009; Frost et al., 2010). The existence of major consortia like the GAGA project1, which aims to sequence and perform comparative bacterial genomics for 200 ant genomes, shows tremendous promise for furthering knowledge of Wolbachia associations with a broader taxonomic host range. Comparative genomics (e.g., identification of selection signatures in genes) can shed light onto genetic prerequisites for HT. Besides advancing phylogenomic and comparative genomic approaches, WGS can provide insight into HT mechanisms for future functional studies (similar to Frydman et al., 2006; White et al., 2017) allowing us to pinpoint specific Wolbachia genes to relevant phenotypes.
Author Contributions
ST performed the experiments and conducted formal analysis of the data with guidance and supervision from PS and PN. ST wrote the original draft of the manuscript. PS and PN reviewed and edited the manuscript.
Funding
Funding was provided by an NSF Graduate Research Fellowship to ST (DGE-1650604).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mariya Zhukova for helping with the collection of eggs and Jacobus Boomsma for comments on an earlier version of the manuscript. The Smithsonian Tropical Research Institute in Panama made facilities available and the Autoridad Nacional del Ambiente (ANAM) of Panama issued collection and export permits.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00296/full#supplementary-material
FIGURE S1 | Relative proportion of HVR types 1–3 across Acromyrmex queen eggs. Pie chart showing the relative proportion of eggs sequenced from queens in this study with each HVR type, as described in Supplementary Table S1. The legend lists the HVR reference number used in this study followed by parentheses including the name of identical strains and the host species harboring those strains as described by Van Borm et al. (2003). Although closely related, the asterisk indicates that the strain Bcons was not a perfect match to HVR-2 from our study, unlike strain B1 (present in A. insinuator and A. octospinosus) which was identical to HVR-2.
TABLE S1 | Top BLAST matches and corresponding HVR type for A. echinatior and A. insinuator wsp sequences generated in this study. Summary of HVR typing results from Supplementary Table S2 and additional wsp characterizations from the A. echinatior host queen (HQ) and the three parasitic A. insinuator (PQ) queens. From left to right: queen ant used to collect egg DNA samples for this study, host species and accession numbers for the top BLAST hits matching respective Wolbachia sequences, the geographic origin of NCBI samples of other ants, the average % match identity in BLAST, the number of sequences that returned these hits, HVR (hypervariable region) typing according to Baldo et al. (2006b) used in this study, and the percentage of sequences from each queen with respective HVR types. HVR typing is based on the four hypervariable regions of the wsp gene which are comprised of relatively conserved amino acid motifs used to identify recombination points, allowing discrimination between closely related Wolbachia strains, comparable to the use of antigens for serotyping pathogenic bacteria (Baldo et al., 2006b). The four HVRs occupy consecutive conserved regions of the wsp gene and the combination of all four HVR haplotypes make up its WSP profile. All A. echinatior and A. insinuator sequences were classified by their four HVRs using the PubMLST database2 (best match to existing sequences in the database), which revealed three distinct wsp genotypes, here referred to as HVR 1-3. Color coding for the HVR types matches those shown in Figure 1, except for one case where sequences were chimeras (type 21-38-69-37/21-21-25-37) of HVR-1 and HVR-2. As identified in other strains, recombination was localized in the HVRs, which suggests the two A. echinatior sequences are true chimeras rather than sequencing errors (Andersen et al., 2012).
TABLE S2 | Individual sequence information and alignment to HVR reference sequences. From left to right: colony of origin, accession numbers for sequences from this study, host species and accession numbers for the top BLAST hits matching respective Wolbachia sequences, % identity to BLAST match, closest match to sequences in pubMLST database (Baldo et al., 2006b) for wsp (nucleotide query), HVR1-4 (amino acid query) with sequence differences listed below. Colors coordinate with HVR types 1–3 used in Figure 1. Asterisks indicate sequences that differed in HVR reference number relative to other strains in the same HVR type (1–3) classified in this study. The three sequences with asterisks only exhibited a 1–3 bp difference from other sequences in their respective HVR characterization (types 1–3) and were thus included in analyses.
Footnotes
References
Adams, R. M. M., Liberti, J., Illum, A. A., Jones, T. H., Nash, D. R., and Boomsma, J. J. (2013). Chemically armed mercenary ants protect fungus-farming societies. Proc. Natl. Acad. Sci. U.S.A. 110, 15752–15757. doi: 10.1073/pnas.1311654110
Ahmed, M. Z., de Barro, P. J., Ren, S.-X., Greeff, J. M., and Qiu, B.-L. (2013). Evidence for horizontal transmission of secondary endosymbionts in the Bemisia tabaci cryptic species complex. PLoS One 8:e53084. doi: 10.1371/journal.pone.0053084
Ahmed, M. Z., Li, S.-J., Xue, X., Yin, X.-J., Ren, S.-X., Jiggins, F. M., et al. (2015). The intracellular bacterium Wolbachia uses parasitoid wasps as phoretic vectors for efficient horizontal transmission. PLoS Pathog. 10:e1004672. doi: 10.1371/journal.ppat.1004672
Andersen, S. B., Boye, M., Nash, D. R., and Boomsma, J. J. (2012). Dynamic Wolbachia prevalence in Acromyrmex leaf-cutting ants: potential for a nutritional symbiosis. J. Evol. Biol. 25, 1340–1350. doi: 10.1111/J.1420-9101.2012.02521.X
Baldo, L., Bordenstein, S., Wernegreen, J. J., and Werren, J. H. (2006a). Widespread recombination throughout Wolbachia genomes. Mol. Biol. Evol. 23, 437–449. doi: 10.1093/molbev/msj049
Baldo, L., Dunning Hotopp, J. C., Jolley, K. A., Bordenstein, S. R., Biber, S. A., Choudhury, R. R., et al. (2006b). Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl. Environ. Microbiol. 72, 7098–7110. doi: 10.1128/AEM.00731-06
Baldo, L., Lo, N., and Werren, J. H. (2005). Mosaic nature of the Wolbachia surface protein. J. Bacteriol. 187, 5406–5418. doi: 10.1128/JB.187.15.5406-5418.2005
Bleidorn, C., and Gerth, M. (2018). A critical re-evaluation of multilocus sequence typing (MLST) efforts in Wolbachia. FEMS Microbiol. Ecol. 94. doi: 10.1093/femsec/fix163
Boomsma, J. J., Huszár, D. B., and Pedersen, J. S. (2014). The evolution of multiqueen breeding in eusocial lineages with permanent physically differentiated castes. Anim. Behav. 92, 241–252. doi: 10.1016/j.anbehav.2014.03.005
Bordenstein, S. R., Paraskevopoulos, C., Dunning Hotopp, J. C., Sapountzis, P., Lo, N., Bandi, C., et al. (2009). Parasitism and mutualism in Wolbachia: what the phylogenomic trees can and cannot say. Mol. Biol. Evol. 26, 231–241. doi: 10.1093/molbev/msn243
Braig, H. R., Zhou, W., Dobson, S. L., and O’Neill, S. L. (1998). Cloning and characterization of a gene encoding the major surface protein of the bacterial endosymbiont Wolbachia pipientis. J. Bacteriol. 180, 2373–2378.
Branstetter, M. G., Ješovnik, A., Sosa-Calvo, J., Lloyd, M. W., Faircloth, B. C., Brady, S. G., et al. (2017). Dry habitats were crucibles of domestication in the evolution of agriculture in ants. Proc. Biol. Sci. 284:20170095. doi: 10.1098/rspb.2017.0095
Brown, B. V., and Feener, D. H. (1998). Parasitic phorid flies (Diptera: Phoridae) associated with army ants (Hymenoptera: Formicidae: Ecitoninae, Dorylinae) and their conservation biology. Biotropica 30, 482–487. doi: 10.1111/j.1744-7429.1998.tb00084.x
Carvalho, G. A., Corrêa, A. S., de Oliveira, L. O., and Guedes, R. N. C. (2014). Evidence of horizontal transmission of primary and secondary endosymbionts between maize and rice weevils (Sitophilus zeamais and Sitophilus oryzae) and the parasitoid Theocolax elegans. J. Stored Prod. Res. 59, 61–65. doi: 10.1016/j.jspr.2014.05.004
Dedeine, F., Ahrens, M., Calcaterra, L., and Shoemaker, D. D. (2005). Social parasitism in fire ants (Solenopsis spp.): a potential mechanism for interspecies transfer of Wolbachia. Mol. Ecol. 14, 1543–1548. doi: 10.1111/j.1365-294X.2005.02499.x
Ellegaard, K. M., Klasson, L., Näslund, K., Bourtzis, K., and Andersson, S. G. E. (2013). Comparative genomics of Wolbachia and the bacterial species concept. PLoS Genet. 9:e1003381. doi: 10.1371/journal.pgen.1003381
Fernández-Marín, H., Zimmerman, J. K., and Wcislo, W. T. (2006). Acanthopria and Mimopriella parasitoid wasps (Diapriidae) attack Cyphomyrmex fungus-growing ants (Formicidae, Attini). Naturwissenschaften 93, 17–21. doi: 10.1007/s00114-005-0048-z
Fernando de Souza, R., Daivison Silva Ramalho, J., Santina de Castro Morini, M., Wolff, J. L. C., Araújo, R. C., and Mascara, D. (2009). Identification and characterization of Wolbachia in Solenopsis saevissima fire ants (Hymenoptera: Formicidae) in southeastern Brazil. Curr. Microbiol. 58, 189–194. doi: 10.1007/s00284-008-9301-y
Foster, J., Ganatra, M., Kamal, I., Ware, J., Makarova, K., Ivanova, N., et al. (2005). The Wolbachia Genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 3:e121. doi: 10.1371/journal.pbio.0030121
Frost, C. L., Fernandez-Marin, H., Smith, J. E., and Hughes, W. O. (2010). Multiple gains and losses of Wolbachia symbionts across a tribe of fungus-growing ants. Mol. Ecol. 19, 4077–4085. doi: 10.1111/j.1365-294X.2010.04764.x
Frost, C. L., Pollock, S. W., Smith, J. E., and Hughes, W. O. H. (2014). Wolbachia in the flesh: symbiont intensities in germ-line and somatic tissues challenge the conventional view of Wolbachia transmission routes. PloS One 9:e95122. doi: 10.1371/journal.pone.0095122
Frydman, H. M., Li, J. M., Robson, D. N., and Wieschaus, E. (2006). Somatic stem cell niche tropism in Wolbachia. Nature 441, 509–512. doi: 10.1038/nature04756
Gerth, M., and Bleidorn, C. (2016). Comparative genomics provides a timeframe for Wolbachia evolution and exposes a recent biotin synthesis operon transfer. Nat. Microbiol. 2:16241. doi: 10.1038/nmicrobiol.2016.241
Gerth, M., Gansauge, M.-T., Weigert, A., and Bleidorn, C. (2014). Phylogenomic analyses uncover origin and spread of the Wolbachia pandemic. Nat. Commun. 5:5117. doi: 10.1038/ncomms6117
Guillade, A. C., and Folgarait, P. J. (2015). Effect of phorid fly density on the foraging of Atta vollenweideri leafcutter ants in the field. Entomol. Exp. Appl. 154, 53–61. doi: 10.1111/eea.12255
Heath, B. D., Butcher, R. D., Whitfield, W. G., and Hubbard, S. F. (1999). Horizontal transfer of Wolbachia between phylogenetically distant insect species by a naturally occurring mechanism. Curr. Biol. 9, 313–316. doi: 10.1016/S0960-9822(99)80139-0
Hiroki, M., Tagami, Y., Miura, K., and Kato, Y. (2004). Multiple infection with Wolbachia inducing different reproductive manipulations in the butterfly Eurema hecabe. Proc. Biol. Sci. 271, 1751–1755. doi: 10.1098/rspb.2004.2769
Hoffmann, A. A., Montgomery, B. L., Popovici, J., Iturbe-Ormaetxe, I., Johnson, P. H., Muzzi, F., et al. (2011). Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature 476, 454–457. doi: 10.1038/nature10356
Ishmael, N., Dunning Hotopp, J. C., Ioannidis, P., Biber, S., Sakamoto, J., Siozios, S., et al. (2009). Extensive genomic diversity of closely related Wolbachia strains. Microbiol. Read. Engl. 155, 2211–2222. doi: 10.1099/mic.0.027581-0
Jiggins, F. M., Hurst, G. D. D., and Yang, Z. (2002). Host-symbiont conflicts: positive selection on an outer membrane protein of parasitic but not Mutualistic Rickettsiaceae. Mol. Biol. Evol. 19, 1341–1349. doi: 10.1093/oxfordjournals.molbev.a004195
Kautz, S., Rubin, B. E. R., and Moreau, C. S. (2013). Bacterial Infections across the Ants: frequency and prevalence of Wolbachia, Spiroplasma, and Asaia. Psyche J. Entomol. 2013:e936341. doi: 10.1155/2013/936341
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kent, B. N., and Bordenstein, S. R. (2010). Phage WO of Wolbachia: lambda of the endosymbiont world. Trends Microbiol. 18, 173–181. doi: 10.1016/j.tim.2009.12.011
Klasson, L., Westberg, J., Sapountzis, P., Näslund, K., Lutnaes, Y., Darby, A. C., et al. (2009). The mosaic genome structure of the Wolbachia wRi strain infecting Drosophila simulans. Proc. Natl. Acad. Sci. U.S.A. 106, 5725–5730. doi: 10.1073/pnas.0810753106
Lambardi, D., Dani, F. R., Turillazzi, S., and Boomsma, J. J. (2007). Chemical mimicry in an incipient leaf-cutting ant social parasite. Behav. Ecol. Sociobiol. 61, 843–851. doi: 10.1007/s00265-006-0313-y
Lapolla, J. S., Mueller, U. G., Seid, M. A., and Cover, S. P. (2002). Predation by the army ant Neivamyrmex rugulosus on the fungus-growing ant Trachymyrmex arizonensis - semantic scholar. Insect. Soc. 49, 251–256. doi: 10.1007/s00040-002-8310-2
Larsen, J., Fouks, B., Bos, N., d’Ettorre, P., and Nehring, V. (2014). Variation in nestmate recognition ability among polymorphic leaf-cutting ant workers. J. Insect Physiol. 70, 59–66. doi: 10.1016/j.jinsphys.2014.09.002
Leclercq, S., Giraud, I., and Cordaux, R. (2011). Remarkable abundance and evolution of mobile group II introns in Wolbachia bacterial endosymbionts. Mol. Biol. Evol. 28, 685–697. doi: 10.1093/molbev/msq238
Liberti, J., Sapountzis, P., Hansen, L. H., Sørensen, S. J., Adams, R. M. M., and Boomsma, J. J. (2015). Bacterial symbiont sharing in Megalomyrmex social parasites and their fungus-growing ant hosts. Mol. Ecol. 24, 3151–3169. doi: 10.1111/mec.13216
Martins, C., Souza, R. F., and Bueno, O. C. (2012). Presence and distribution of the endosymbiont Wolbachia among Solenopsis spp. (Hymenoptera: Formicidae) from Brazil and its evolutionary history. J. Invertebr. Pathol. 109, 287–296. doi: 10.1016/j.jip.2012.01.001
Mascarenhas, R. O., Prezotto, L. F., Perondini, A. L. P., Marino, C. L., and Selivon, D. (2016). Wolbachia in guilds of Anastrepha fruit flies (Tephritidae) and parasitoid wasps (Braconidae). Genet. Mol. Biol. 39, 600–610. doi: 10.1590/1678-4685-GMB-2016-0075
McMeniman, C. J., Lane, A. M., Fong, A. W. C., Voronin, D. A., Iturbe-Ormaetxe, I., Yamada, R., et al. (2008). Host adaptation of a Wolbachia strain after long-term serial passage in mosquito cell lines. Appl. Environ. Microbiol. 74, 6963–6969. doi: 10.1128/AEM.01038-08
Moreau, C. S., Bell, C. D., Vila, R., Archibald, S. B., and Pierce, N. E. (2006). Phylogeny of the ants: diversification in the age of angiosperms. Science 312, 101–104. doi: 10.1126/science.1124891
Mouton, L., Dedeine, F., Henri, H., Boulétreau, M., Profizi, N., and Vavre, F. (2004). Virulence, multiple infections and regulation of symbiotic population in the Wolbachia-Asobara tabida symbiosis. Genetics 168, 181–189. doi: 10.1534/genetics.104.026716
Nehring, V., Dani, F. R., Turillazzi, S., Boomsma, J. J., and d’Ettorre, P. (2015). Integration strategies of a leaf-cutting ant social parasite. Anim. Behav. 108, 55–65. doi: 10.1016/j.anbehav.2015.07.009
Noda, H., Miyoshi, T., Zhang, Q., Watanabe, K., Deng, K., and Hoshizaki, S. (2001). Wolbachia infection shared among planthoppers (Homoptera: Delphacidae) and their endoparasite (Strepsiptera: Elenchidae): a probable case of interspecies transmission. Mol. Ecol. 10, 2101–2106. doi: 10.1046/j.0962-1083.2001.01334.x
Peralta, L., and Martínez, P. A. (2013). Ensambles de ácaros oribátidos en hormigueros de Acromyrmex spp. (Hymenoptera, Formicidae). Ecol. Austral 23, 209–217.
Pérez-Ortega, B., Fernández-Marín, H., Loiácono, M., Galgani, P., and Wcislo, W. T. (2010). Biological notes on a fungus-growing ant, Trachymyrmex cf. zeteki (Hymenoptera, Formicidae, Attini) attacked by a diverse community of parasitoid wasps (Hymenoptera, Diapriidae). Insectes Sociaux 57, 317–322. doi: 10.1007/s00040-010-0086-1
Powell, S., and Clark, E. (2004). Combat Between Large Derived Societies: A Subterranean Army Ant Established As a Predator of Mature Leaf-Cutting Ant Colonies. Available at: http://repository.si.edu//handle/10088/3728 [Accessed December 7, 2017].
Ramalho, M. O., Martins, C., Silva, L. M. R., Martins, V. G., and Bueno, O. C. (2017). Intracellular symbiotic bacteria of camponotus textor, Forel (Hymenoptera, Formicidae). Curr. Microbiol. 74, 589–597. doi: 10.1007/s00284-017-1201-6
Rambaut, A. (2016). FigTree Version 1.4.3 [Computer Program]. Available at: http://tree.bio.ed.ac.uk/software/figtree/
Rasgon, J. L., Gamston, C. E., and Ren, X. (2006). Survival of Wolbachia pipientis in cell-free medium. Appl. Environ. Microbiol. 72, 6934–6937. doi: 10.1128/AEM.01673-06
Raychoudhury, R., Baldo, L., Oliveira, D. C., and Werren, J. H. (2009). Modes of acquisition of Wolbachia: horizontal transfer, hybrid introgression, and codivergence in the Nasonia species complex. Evol. Int. J. Org. Evol. 63, 165–183. doi: 10.1111/j.1558-5646.2008.00533.x
Reuter, M., Pedersen, J. S., and Keller, L. (2005). Loss of Wolbachia infection during colonisation in the invasive argentine ant linepithema humile. Heredity 94, 364–369. doi: 10.1038/sj.hdy.6800601
Rey, O., Estoup, A., Facon, B., Loiseau, A., Aebi, A., Duron, O., et al. (2013). Distribution of endosymbiotic reproductive manipulators reflects invasion process and not reproductive system polymorphism in the little fire ant Wasmannia auropunctata. PloS One 8:e58467. doi: 10.1371/journal.pone.0058467
Russell, J. A., Goldman-Huertas, B., Moreau, C. S., Baldo, L., Stahlhut, J. K., Werren, J. H., et al. (2009). Specialization and geographic isolation among Wolbachia symbionts from ants and lycaenid butterflies. Evol. Int. J. Org. Evol. 63, 624–640. doi: 10.1111/j.1558-5646.2008.00579.x
Sapountzis, P., Zhukova, M., Hansen, L. H., Sørensen, S. J., Schiøtt, M., and Boomsma, J. J. (2015). Acromyrmex leaf-cutting ants have simple gut microbiota with nitrogen-fixing potential. Appl. Environ. Microbiol. 81, 5527–5537. doi: 10.1128/AEM.00961-15
Saridaki, A., and Bourtzis, K. (2010). Wolbachia: more than just a bug in insects genitals. Curr. Opin. Microbiol. 13, 67–72. doi: 10.1016/j.mib.2009.11.005
Schultz, T. R., and Brady, S. G. (2008). Major evolutionary transitions in ant agriculture. Proc. Natl. Acad. Sci. U.S.A. 105, 5435–5440. doi: 10.1073/pnas.0711024105
Siozios, S., Ioannidis, P., Klasson, L., Andersson, S. G. E., Braig, H. R., and Bourtzis, K. (2013). The diversity and evolution of Wolbachia ankyrin repeat domain genes. PLoS One 8:e55390. doi: 10.1371/journal.pone.0055390
Stahlhut, J. K., Desjardins, C. A., Clark, M. E., Baldo, L., Russell, J. A., Werren, J. H., et al. (2010). The mushroom habitat as an ecological arena for global exchange of Wolbachia. Mol. Ecol. 19, 1940–1952. doi: 10.1111/j.1365-294X.2010.04572.x
Stouthamer, R., Breeuwer, J. A., and Hurst, G. D. (1999). Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu. Rev. Microbiol. 53, 71–102. doi: 10.1146/annurev.micro.53.1.71
Stürup, M., Nash, D. R., Hughes, W. O. H., and Boomsma, J. J. (2014). Sperm mixing in the polyandrous leaf-cutting ant Acromyrmex echinatior. Ecol. Evol. 4, 3571–3582. doi: 10.1002/ece3.1176
Tsutsui, N. D., Kauppinen, S. N., Oyafuso, A. F., and Grosberg, R. K. (2003). The distribution and evolutionary history of Wolbachia infection in native and introduced populations of the invasive argentine ant (Linepithema humile). Mol. Ecol. 12, 3057–3068. doi: 10.1046/j.1365-294X.2003.01979.x
Van Borm, S., Wenseleers, T., Billen, J., and Boomsma, J. J. (2003). Cloning and sequencing of wsp encoding gene fragments reveals a diversity of co-infecting Wolbachia strains in Acromyrmex leafcutter ants. Mol. Phylogenet. Evol. 26, 102–109. doi: 10.1016/S1055-7903(02)00298-1
Vavre, F., Fleury, F., Lepetit, D., Fouillet, P., and Boulétreau, M. (1999). Phylogenetic evidence for horizontal transmission of Wolbachia in host-parasitoid associations. Mol. Biol. Evol. 16, 1711–1723. doi: 10.1093/oxfordjournals.molbev.a026084
Veneti, Z., Clark, M. E., Karr, T. L., Savakis, C., and Bourtzis, K. (2004). Heads or tails: host-parasite interactions in the Drosophila-Wolbachia system. Appl. Environ. Microbiol. 70, 5366–5372. doi: 10.1128/AEM.70.9.5366-5372.2004
Veneti, Z., Zabalou, S., Papafotiou, G., Paraskevopoulos, C., Pattas, S., Livadaras, I., et al. (2012). Loss of reproductive parasitism following transfer of male-killing Wolbachia to Drosophila melanogaster and Drosophila simulans. Heredity 109, 306–312. doi: 10.1038/hdy.2012.43
Viljakainen, L., Reuter, M., and Pamilo, P. (2008). Wolbachia transmission dynamics in formica wood ants. BMC Evol. Biol. 8:55. doi: 10.1186/1471-2148-8-55
Vos, M., and Didelot, X. (2009). A comparison of homologous recombination rates in bacteria and archaea. ISME J. 3, 199–208. doi: 10.1038/ismej.2008.93
Weinert, L. A., Araujo-Jnr, E. V., Ahmed, M. Z., and Welch, J. J. (2015). The incidence of bacterial endosymbionts in terrestrial arthropods. Proc. Biol. Sci. 282:20150249. doi: 10.1098/rspb.2015.0249
Werren, J. H., Baldo, L., and Clark, M. E. (2008). Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6, 741–751. doi: 10.1038/nrmicro1969
White, P. M., Pietri, J. E., Debec, A., Russell, S., Patel, B., and Sullivan, W. (2017). Mechanisms of horizontal cell-to-cell transfer of Wolbachia spp. in Drosophila melanogaster. Appl. Environ. Microbiol. 83, e03425-e16. doi: 10.1128/AEM.03425-16
Wu, M., Sun, L. V., Vamathevan, J., Riegler, M., Deboy, R., Brownlie, J. C., et al. (2004). Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2:E69. doi: 10.1371/journal.pbio.0020069
Yang, X.-H., Zhu, D.-H., Liu, Z., Zhao, L., and Su, C.-Y. (2013). High levels of multiple infections, recombination and horizontal transmission of Wolbachia in the Andricus mukaigawae (Hymenoptera; Cynipidae) communities. PloS One 8:e78970. doi: 10.1371/journal.pone.0078970
Zabalou, S., Apostolaki, A., Pattas, S., Veneti, Z., Paraskevopoulos, C., Livadaras, I., et al. (2008). Multiple rescue factors within a Wolbachia strain. Genetics 178, 2145–2160. doi: 10.1534/genetics.107.086488
Zabalou, S., Riegler, M., Theodorakopoulou, M., Stauffer, C., Savakis, C., and Bourtzis, K. (2004). Wolbachia-induced cytoplasmic incompatibility as a means for insect pest population control. Proc. Natl. Acad. Sci. U.S.A. 101, 15042–15045. doi: 10.1073/pnas.0403853101
Zhao, D.-X., Chen, D.-S., Ge, C., Gotoh, T., and Hong, X.-Y. (2013). Multiple infections with Cardinium and two strains of Wolbachia in the spider mite Tetranychus phaselus Ehara: revealing new forces driving the spread of Wolbachia. PLoS One 8:e54964. doi: 10.1371/journal.pone.0054964
Zhou, W., Rousset, F., and O’Neil, S. (1998). Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc. Biol. Sci. 265, 509–515. doi: 10.1098/rspb.1998.0324
Zhukova, M., Sapountzis, P., Schiøtt, M., and Boomsma, J. J. (2017). Diversity and transmission of gut bacteria in Atta and Acromyrmex leaf-cutting ants during development. Front. Microbiol. 8:1942. doi: 10.3389/fmicb.2017.01942
Keywords: symbiosis, fungus-growing ants, horizontal transmission, social interactions, Wolbachia
Citation: Tolley SJA, Nonacs P and Sapountzis P (2019) Wolbachia Horizontal Transmission Events in Ants: What Do We Know and What Can We Learn? Front. Microbiol. 10:296. doi: 10.3389/fmicb.2019.00296
Received: 09 October 2018; Accepted: 04 February 2019;
Published: 06 March 2019.
Edited by:
George Tsiamis, University of Patras, GreeceReviewed by:
Sandra Breum Andersen, NYU Langone Health, United StatesAurélien Vigneron, Yale University, United States
Ana Ješovnik, Smithsonian Institution, United States
Copyright © 2019 Tolley, Nonacs and Sapountzis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sarah J. A. Tolley, c3RvbGxleUB1Y2xhLmVkdQ==
 Sarah J. A. Tolley
Sarah J. A. Tolley Peter Nonacs
Peter Nonacs Panagiotis Sapountzis
Panagiotis Sapountzis