- 1Floral and Nursery Plants Research Unit, United States National Arboretum, United States Department of Agriculture–Agricultural Research Service, Beltsville, MD, United States
- 2Department of Plant Protection, Faculty of Agriculture, University of Jember, Jember, Indonesia
- 3Department of Plant Pathology, Faculty of Agriculture, Minia University, El-minia, Egypt
The first Ralstonia-infecting bacteriophage from soil of the United States, designated RsoM1USA, was isolated from a tomato field in Florida. Electron microscopy revealed that phage RsoM1USA is member of the genus P2virus in the family Myoviridae with an icosahedral head of about 66 nm in diameter, a contractile tail of about 152 nm in length, and a long “neck.” Phage RsoM1USA infected 12 of the 30 tested R. solanacearum species complex strains collected worldwide in each of the three Ralstonia species: R. solanacearum, R. pseudosolanacearum, and R. syzygii. The phage completed its infection cycle 180 min post infection with a burst size of about 56 particles per cell. Phage RsoM1USA has a genome of 39,309 nucleotides containing 58 open reading frames (ORFs) and is closely related to Ralstonia phage RSA1 of the species Ralstonia virus RSA1. The genomic organization of phage RsoM1USA is also similar to that of phage RSA1, but their integrases share no sequence homology. In addition, we determined that the integration of phage RsoM1USA into its susceptible R. solanacearum strain K60 is mediated by the 3′ 45-base portion of the threonine tRNA (TGT), not arginine tRNA (CCG) as reported for phage RSA1, confirming that the two phages use different mechanism for integration. Our proteomic analysis of the purified virions supported the annotation of the main structural proteins. Infection of a susceptible R. solanacearum strain RUN302 by phage RsoM1USA resulted in significantly reduced growth of the infected bacterium in vitro, but not virulence in tomato plants, as compared to its uninfected RUN302 strain. Due to its differences from phage RSA1, phage RsoM1USA should be considered the type member of a new species with a proposed species name of Ralstonia virus RsoM1USA.
Introduction
Bacterial wilt caused by Ralstonia solanacearum is one of the most destructive plant diseases in the world. The bacterium is known to infect more than 50 families of plants in tropical, subtropical, and warm temperate regions of the world (Hayward, 1991). R. solanacearum is considered a species complex because it contains strains with great genetic variability. It is traditionally classified into five races based on host range and five biovars based on biochemical properties. Using molecular methods, R. solanacearum species complex (referred to as “R. solanacearum” for historic reason in this study) has been divided into four phylotypes and 53 sequevars (Fegan and Prior, 2005), and most recently into three Ralstonia species: R. solanacearum (referred to as “the current R. solanacearum” in this study), R. pseudosolanacearum, and R. syzygii (Safni et al., 2014). R. solanacearum normally enters its plant hosts via openings in roots and colonizes the xylem vessels causing disturbance of water transport and resulting in the appearance of wilt symptoms when its cell densities are over 108 CFU/cm stem (Huang and Allen, 2000). R. solanacearum is very difficult to control because of its wide host range, large genetic diversity and its capability for long-term survival in soil and water. It is, therefore, desirable to explore alternative control strategies such as the use of bacteriophages to combat R. solanacearum. Bacteriophages that have been tested so far for control of R. solanacearum have recently been reviewed by Alvarez and Biosca (2017).
Currently, a wide variety of bacteriophages that specifically infect R. solanacearum have been characterized and belong to the families Inoviridae, Myoviridae, Podoviridae, or Siphoviridae. Interestingly, all Ralstonia-infecting bacteriophages reported so far were isolated from soil collected from Asian countries including Japan (Yamada et al., 2007; Van et al., 2014; Thi et al., 2015; Bhunchoth et al., 2016; Askora et al., 2017), Thailand (Bhunchoth et al., 2015, 2016; Mihara et al., 2016), Korea (Murugaiyan et al., 2010; Park, 2018), China (Su et al., 2017; Liao, 2018), and Indonesia (Addy et al., 2018), except for one from Egypt (Ahmad et al., 2018) and another not from soil but from the culture supernatant of R. solanacearum strain UW551 (Ahmad et al., 2017b). Six Ralstonia phages in the family of Myoviridae have been characterized including RSA1 (Yamada et al., 2007; Fujiwara et al., 2008), RSL1 (Yamada et al., 2007, 2010), RSF1 (Bhunchoth et al., 2016), and RSY1 (Askora et al., 2017) isolated from soil in Japan, as well as RP15 (Mihara et al., 2016) and RSL2 (Bhunchoth et al., 2016) in Thailand. Among the six myoviruses, RSL1, RSL2, and RSF1 are jumbo phages with genome sizes over 220-kb and RP15 has a genome of 168-kb, while RSA1 and RSY1 are relatively small-genome phages of 39- and 40-kb in size, respectively. Although phages RSA1 and RSY1 are similar in genome size and genomic organization, they differ in the orientation of the integrase gene. In addition, RSY1 uses part of serine tRNA (GGA) gene, while RSA1 uses a portion of arginine tRNA (CCG) gene as the att region, resulting in the grouping of phages into RSA1-type and RSY1-type of the P2-like phages of R. solanacearum (Askora et al., 2017). Both the jumbo phage RSL1 and the small-genome phage RSA1 were characterized biologically for their biocontrol potential for R. solanacearum. They both displayed lytic activities against almost all tested R. solanacearum strains (belonging to the current R. pseudosolanacearum strains) isolated from Japan (Yamada et al., 2007) and reduced host bacterial cell density (Fujiwara et al., 2011). Populations of resistant R. solanacearum cells, however, were observed 30 h after phage RSA1 was added to the bacterial culture (Fujiwara et al., 2011), rendering RSA1 not suitable as a biocontrol for R. solanacearum. Subsequent greenhouse experiments with phage RSL1 demonstrated that this phage has a good potential for control of bacterial wilt caused by R. solanacearum (Fujiwara et al., 2011), although its potential against non-Japanese R. solanacearum species complex strains is unknown. These studies suggest that bacteriophages, even in the same virus family and that display similar in vitro lytic activities against large numbers of tested R. solanacearum strains, have different potential in planta as biocontrol agents, and detailed biological characterization of phages are needed before their biocontrol potential can be assessed.
In this study, we purified and characterized the first Ralstonia-infecting bacteriophage from soil samples collected from the United States, isolated from a tomato field in Florida. We also compared the phage to other bacteriophages, determined its integration mechanism, and evaluated its effect in vitro and in planta on its susceptible R. solanacearum host strain under controlled environment conditions.
Materials and Methods
R. solanacearum Strains
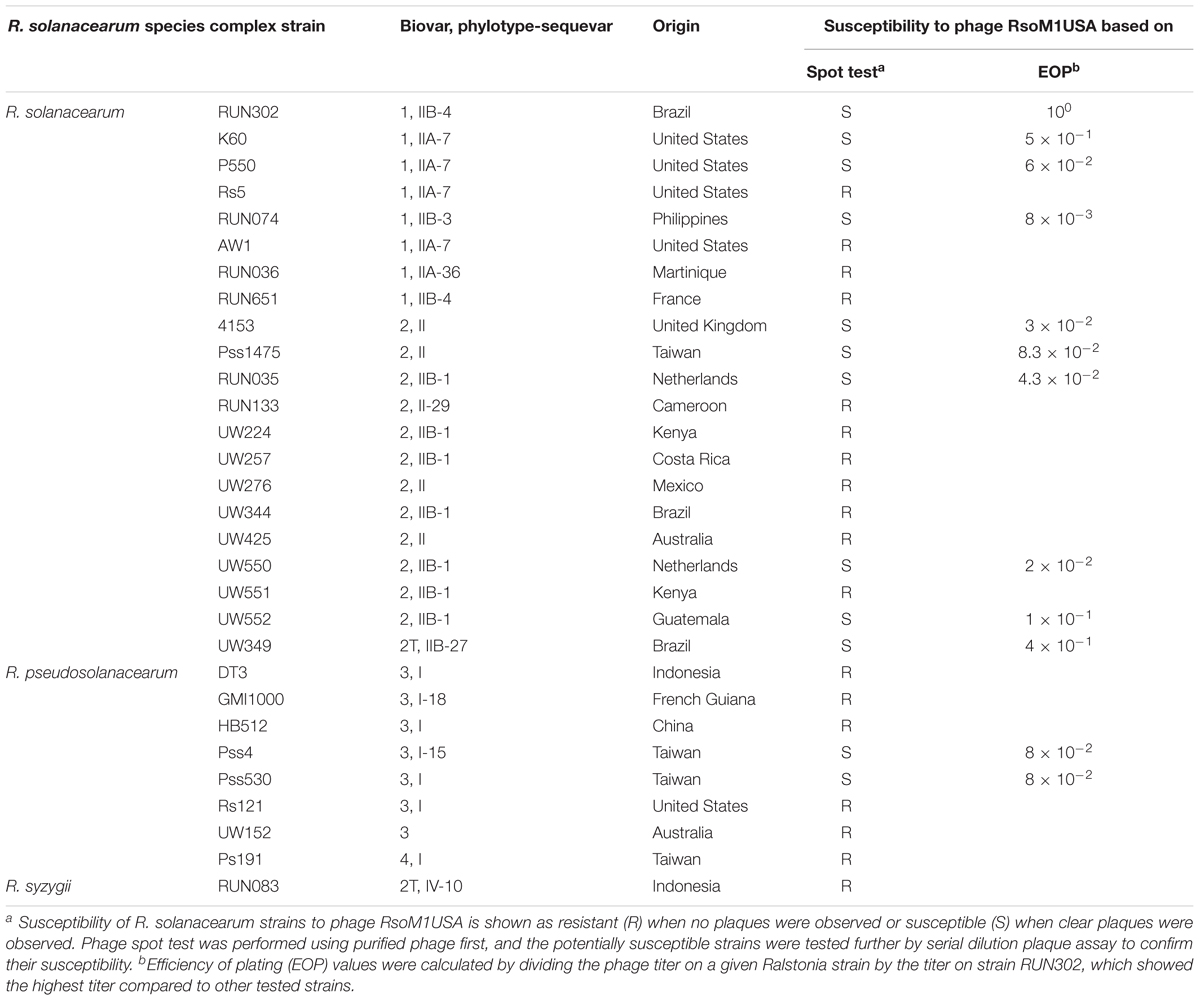
Thirty R. solanacearum species complex strains used in this study are listed in Table 1. They were grown, maintained and their inocula prepared as described before (Stulberg et al., 2015).
Bacteriophage Isolation, Purification, and Characterization
Soil samples from a tomato field infested by R. solanacearum in Florida, United States were used for phage isolation. Ten grams of soil were mixed with water for a total volume of 30 ml in a sterile 50-ml centrifuge tube with gentle shaking overnight at room temperature to release bacteriophages. The tube was then centrifuged at 8,000 × g for 20 min at room temperature, and the supernatant was filtered through a 0.45-μm membrane. A 3 μl aliquot of the filtrate was spotted on a double-layered casamino acid peptone glucose (CPG) plate (Ahmad et al., 2017b) except that R. solanacearum strain RUN302 was used as a bacterial host. The plate was incubated at 28°C for 24 h for the appearance of clear zones caused by phages. A single clear zone was removed, placed into 1 ml of SM buffer (100 mM NaCl, 10 mM MgSO4, 50 mM Tris-HCl at pH 7.5, and 0.01% gelatin) (Sambrook and Russell, 2001), and vortexed vigorously prior to filtering through a 0.45-μm membrane. The filtrate was then serially diluted with the SM buffer. One hundred microliters of the diluted filtrate was added to 400 μl of 2 × 108 cells of R. solanacearum strain RUN302, incubated for 10–20 min at 28°C for phage absorption, and mixed with 3.5 ml of CPG containing 0.45% agar before layering the mixture on top of a CPG plate containing 1.5% of agar for the plaque assay to obtain single plaques. A single plaque was then picked, mixed with SM buffer, vortexed, filtered, serially diluted, and subjected to the plaque assay described above. The process was repeated two more times to complete the triple phage purification process to obtain a pure phage isolate designated RsoM1USA. To make a pure phage stock, a single plaque of phage RsoM1USA was removed and added to a 5 ml liquid CPG culture of 2 × 108 cells of R. solanacearum strain RUN302. It was grown overnight at 28°C with shaking, followed by centrifugation at 8,000 × g for 20 min. The supernatant was filtered through a 0.45 μm membrane, and the filtrate stored in the dark at 4°C. The titer and the optimum multiplicity of infection (MOI) of the phage stock were determined using the plaque assay described above.
The phage particles were propagated, purified, and stored as described (Ahmad et al., 2017b), except that R. solanacearum strain RUN302 was used as the host strain. Briefly, the phage lysate was first cleared by low speed centrifugation (8,000 × g for 15 min), passed through a 0.45 μm membrane filter, and then layered on top of a 30% (wt/vol) sucrose cushion before ultracentrifugation at 50,000 × g for 120 min at 10°C. To obtain a phage titer of about 1010 PFU/ml, the purification process may need to be repeated two or three times using the same volume of lysate each time and by ultracentrifugation through a 30% sucrose cushion in the same tube where the pellet from previous ultracentrifugation was collected. The pellet was then resuspended in either TE or SM buffer.
To characterize the morphology of the phage, purified phage particles were used for negative staining with sodium phosphotungstate (Dykstra, 1993) before observation under a Hitachi HT7700 transmission electron microscope. Phage morphometrics were estimated from more than 10 phage particles using the open source imaging processing program ImageJ 1.50i (National Institutes of Health, United States).
Phage Host Range Test
To determine the host specificity of phage RsoM1USA, the purified phage was subjected to spot testing using 30 R. solanacearum species complex strains as hosts (Table 1). In this test, three microliters of the phage suspension (108 PFU/ml) was spotted on top of the double-layered CPG plate (Ahmad et al., 2017b). The top layer was prepared with 350 μl of each R. solanacearum strain (OD600 of 0.2) mixed with 4.5 ml of CPG. The plate was incubated overnight at 28°C. The formation of a lysis zone indicated that the bacterial strain was susceptible to the phage. Potentially susceptible strains were tested further by serial dilution plaque assay to determine if they were truly susceptible to the phage. In addition, the efficiency of plating (EOP) was determined based on Gill et al. (2011) by calculating the ratio of the phage plaque titer of R. solanacearum strain RUN302 to that of other tested bacterial strains.
One-Step Growth Experiment
The phage infection cycle was characterized with a one-step growth experiment based on Ellis and Delbruck (1939) with modifications. Two hundred microliters of 24-h culture of R. solanacearum strain RUN302 was transferred into 9.8 ml of CPG and grown at 28°C with shaking until the culture reached the OD600 of 0.1 (108 CFU/ml). Phage RsoM1USA was added at a MOI of 0.01 and allowed to adsorb for 15 min at 28°C. The mixture was then centrifuged at 6,000 × g for 5 min to remove any non-absorbed phage particles. The pellet was mixed with 10 ml of CPG, diluted 10,000-fold, and incubated at 28°C without shaking. An aliquot of 500 μl was taken every 30 min for 5 h, filtered through 0.45 μm membrane, diluted, and subjected to the plaque assay described above using RUN302 as a host to estimate phage titers. There were three replicates for each time point, and the experiment was repeated three times.
Thermal Stability Test
To determine the effect of temperature on phage stability, a thermal stability test was conducted at 4°C, and from 10–90°C with 10° intervals. The maximum of 90°C was tested to find out the lethal temperature for the phage, not as a temperature for biocontrol application. Briefly, the purified phage in SM buffer was diluted to 1 × 108 PFU/ml and then 1 ml of the diluted phage suspension was incubated at each of the designated temperatures for 1 h. After incubation, the phage suspension was serially diluted in SM buffer and subjected to plaque assay using R. solanacearum RUN302 as a host to estimate phage numbers. There were three replicates for each temperature and the experiment was repeated three times.
Phage DNA Extraction, Sequencing, and Analysis
To determine the nature of phage RsoM1USA genome, approximately 1 μg of the phage genome was subjected to enzyme digestions with DNase I, RNase A, S1 nuclease, and exonuclease I as described (Ahmad et al., 2017b), as well as with restriction enzymes EcoRV and SmaI using standard molecular biology method (Sambrook and Russell, 2001).
Phage DNA was extracted from purified phage particles (described above) using either a phenol–chloroform method (Sambrook and Russell, 2001) or the Phage DNA Isolation kit (Norgen Biotek Corp., Canada). The phage DNA was sequenced, and the genome sequence assembled commercially by SeqMatic (Fremont, CA, United States). Potential open reading frames (ORFs) larger than 50 amino acids (aa) were identified using the softwares PHASTER (Arndt et al., 2016) and DNASTAR (DNASTAR Inc., United States). The number of aa and the predicted molecular weight for the product of each ORF were calculated using the software ApE version 2.0.51 (University of Utah, United States). To assign possible functions of the ORFs, database searches were performed using BLASTp (Altschul et al., 1990) against NCBI and the Conserved Domain Databases, as well as against motif databases including TIGRFam, Pfam, SMART, PRK, COG and InterPro. The tRNAscan-SE 2.01 was used to search for tRNA genes (Lowe and Eddy, 1997).
Phylogenetic relationships between phage RsoM1USA and 13 phages representing each of the 13-current species in the genus P2virus under the subfamily Peduovirinae of the family Myoviridae were estimated. This was done using the method of Harrison and Langdale (2006) by multiple sequence alignment using Clustal Omega2 (Sievers et al., 2011) to generate nexus-formatted files, followed by execution of the files using downloadable PAUP (Phylogenetic Analysis Using PAUP3) version 4.0a (Swofford, 2003). Phylogenetic analysis based on integrase was performed for 13 phages including RsoM1USA, since no annotation for an integrase has been found for Burkholderia virus phiE122. The accession and ORF numbers of the proteins used for the phylogenetic analysis are listed in Table 2.
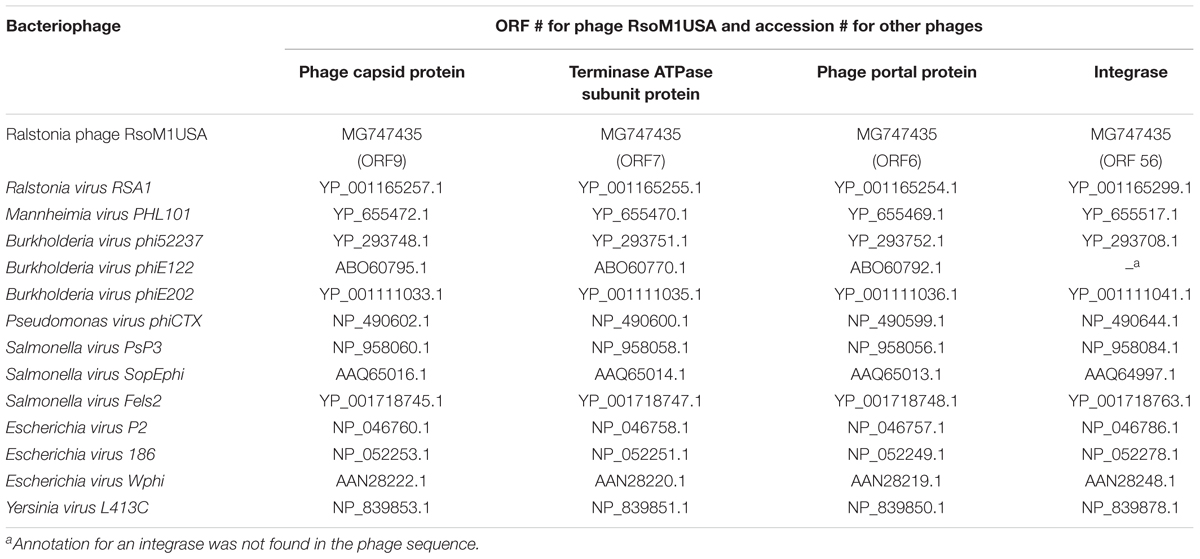
Table 2. List of proteins and their ORFs or accession numbers of bacteriophages used for phylogenetic analysis in this study.
Identification and Proteomic Analysis of RsoM1USA Virion Proteins
Fifty milliliters of purified phage filtrate were treated with 1 μl each of 10 mg/ml RNase A and DNase I for an hour at 37°C prior to adding chloroform at a final concentration of 10%, followed by centrifugation at 7,000 × g for 10 min at 4°C. The top phase containing phage particles was layered on a 20% (w/v) sucrose cushion and centrifuged by ultracentrifugation (Optima MAX-XP Ultracentrifuge, Beckman Coulter, United States) at 50,000 × g for 2 h at 4°C. The phage pellet was suspended in 500 μl of 10 mM Tris-HCl buffer pH 8. Aliquots from purified phage particles was sonicated, mixed with NorexTM tris glycine SDS buffer (2x) (Thermo Fisher Scientific, United States), heated at 85°C for 2 min, and subjected to SDS-PAGE [14% (w/v) polyacrylamide] (Laemmli, 1970). Gels were stained with Coomassie Brilliant Blue R250 stain reagent (Thermo Scientific, United States). The most abundant bands were cut out and sent to Bioproximity, LLC. (Chantilly, VA, United States) for protein identification using liquid chromatography-tandem mass spectrometry (LC-MS/MS).
Assay to Determine Phage Integration Regions
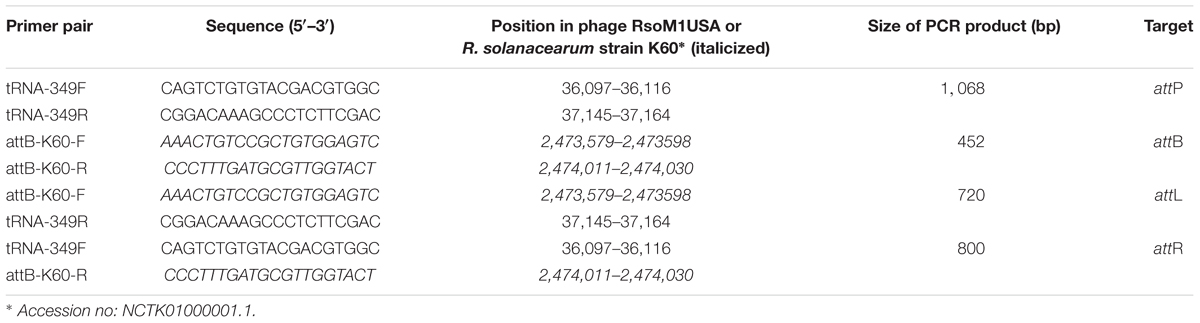
R. solanacearum strain K60 was used to determine where phage RsoM1USA integrated into the bacterial genome, since the complete genome sequence of K60 is publicly available (Hayes et al., 2017). In addition, the bacterial strain did not contain prophage sequence of RsoM1USA in its genome and is susceptible to the phage infection. To infect strain K60, an overnight culture of K60 was adjusted to OD600 of 0.1 in 4.5 ml of liquid CPG. Phage RsoM1USA was then added at a MOI of 0.1 and adsorbed to bacterial cells for 15 min at 28°C, before incubation at 28°C with shaking at 150 rpm for 24 h. The infected bacterial cells were harvested by centrifugation at 6,000 × g for 10 min at 4°C, and the pellet was suspended in sterile water and used for DNA extraction as described by Sambrook and Russell (2001). PCR primers designed and used for this study are listed in Table 3. The primers were designed based on the nucleotide sequences flanking the predicted attP site in phage RsoM1USA and attB site in R. solanacearum strain K60.
PCR was performed in a 20-μl volume containing 1x GoTaq Green Master Mix (Promega, United States), 10 pmol of each primer, and 25 nanograms of DNA. PCR conditions were 1 cycle of 1 min at 94°C, 30 cycles of 30 s at 94°C, 30 s at 60°C, and 1 min at 72°C with a final extension of 10 min at 72°C. The PCR products were purified from agarose gels with the QIAquick gel extraction kit (Qiagen, Inc.) and sequenced by Eurofins Scientific (United States).
Assay for in vitro Growth of Phage-Treated R. solanacearum Strains
To determine the effect of phage infection on the in vitro growth of its susceptible R. solanacearum strain, phage RsoM1USA-treated and untreated R. solanacearum RUN302 strains were grown in CPG in 24-well plates (Corning® Sigma, United States) at 28°C. Briefly, the concentration of the overnight culture of strain RUN302 was adjusted with CPG to approximately 108 CFU/ml, and 1.5 ml of the bacterial suspension was added to each well of the 24-well plate. One hundred and fifty microliters of phage suspension were then added at a MOI of 0.001, 0.01, 0.1, 1.0, and 10, respectively, and the plate was incubated inside a multi-plate reader Epoch2 (BioTek, United States) with slow shaking. SM buffer was used as a phage control (MOI of 0). Bacterial growth indicated by absorbance at 600 nm was measured every 60 min and graphed every 5 h for 35 h. There were three replicates for each MOI treatment and the experiment was repeated three times.
Virulence Assay in Tomato Plants
Tomato plants (Lycopersicon esculentum Mill. cv. ‘Bonnie Best’) were grown, transplanted and inoculated as described previously (Ahmad et al., 2017b), except that 40 ml of bacterial suspension containing a total of 4 × 109 cells of R. solanacearum strain RUN302 was added as a soil drench into each pot. This was followed immediately by pouring 40 ml of phage RsoM1USA suspension (107 PFU/ml) for a MOI of 0.1 into the pot, or water for a control (MOI of 0). Negative control plants were inoculated with 80 ml of water. Inoculated plants were rated daily using a disease index (DI) of 0 to 4 (Roberts et al., 1988). Disease severity was calculated each day using the formula: Disease severity = [Σ(n × DI)/4N] × 100, where n is the number of plants in each DI category and N is the total number of plants in each treatment. There were 12 plants for each treatment and the experiment was repeated three times.
Statistical Analysis
Data for phage thermal stability and in vitro growth of R. solanacearum strains 35 h after phage RsoM1USA treatment at MOIs of 0.001, 0.01, 0.1, 1, and 10 were analyzed by one-way ANOVA using free web-based statistical software4. Tukey’s Honest Significant Difference test included in the software was used to compare means. The mean disease severity between the untreated and phage RsoM1USA-treated R. solanacearum RUN302 strains were analyzed for significant differences using the t-test in Microsoft Excel. The t-test was also performed for in vitro growth of R. solanacearum strain RUN302 between RsoM1USA-untreated (MOI of 0) and treated (MOI of 0.001, 0.01, 0.1, 1 or 10) strains 10, 15, and 35 h after phage treatment. Differences were considered statistically significant if p < 0.05.
Nucleotide Sequence Accession Numbers
The complete genome sequence of phage RsoM1USA has been submitted to GenBank and given accession no. MG747435. The accession numbers for R. solanacearum strain sequences used in this study are: GMI1000, NC_003295; 23-10BR, JQOI01000031.1; Po82, NC_017574.1; and K60, NCTK01000001.1.
Results
Morphology, Host Specificity, and Growth Characteristics of Ralstonia Phage RsoM1USA
A bacteriophage isolated from soil collected from a tomato field in Florida, United States produced clear and round plaques with a diameter of approximately 5 mm on double layered CPG plates using R. solanacearum strain RUN302 as a host. The purified phage had an icosahedral capsid with a size of 62.68 ± 2.17 nm × 66.17 ± 2.61 nm (n = 10) and a long contractile tail with a length of 151.96 ± 4.92 nm and a width of 19.63 ± 0.72 nm (n = 10) (Figure 1A). In addition, the phage has a relatively long “neck” (Figure 1A) compared to other myoviruses such as RP15 (Mihara et al., 2016) and the diameter of its sheath increased approximately 3 nm (n = 12) during contraction, resembling the members of Peduovirinae. The phage was designated Ralstonia phage RsoM1USA using our phage identifier naming system (Ahmad et al., 2018) based on the informal guidelines by Adriaenssens and Brister (2017) and the proposed scheme by Kropinski et al. (2009), since it is the first R. solanacearum-infecting bacteriophage belonging to the family Myoviridae that was isolated from the United States.
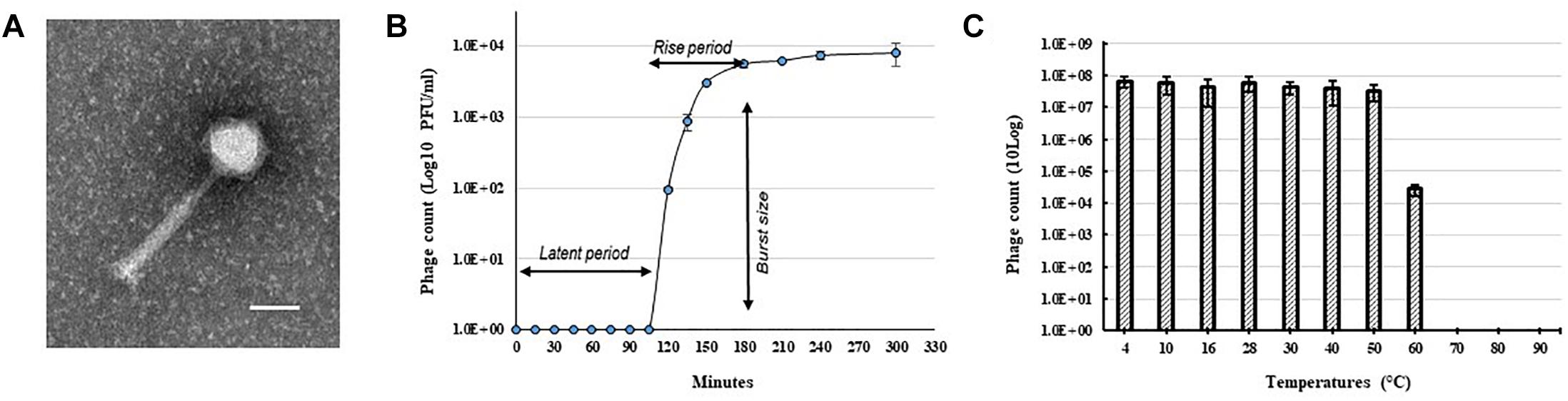
Figure 1. Morphology and growth characteristics of Ralstonia phage RsoM1USA. (A) Transmission electron micrograph of the purified phage virion. The scale represents 50 nm. (B) One-step growth curve of the phage with R. solanacearum strain RUN302 as the host. The phage was added at a MOI of 0.01 and allowed to adsorb for 15 min at 28°C. Phage titers were determined every 30 min using the plaque assay. The latent period is when no release of phage particles was observed. The rise period begins with the end of the latent period and ends when the increase in phage titers ceases. The burst size is the average number of phage particles released per infected cell. (C) Effect of temperature on the stability of phage RsoM1USA. 108 PFU of phage RsoM1USA was incubated at each temperature point and the number of phage was estimated by plaque assay using R. solanacearum RUN302 as a host 1 h after incubation. Means are based on three separate experiments, each containing three replicates. Bars indicate standard errors.
To determine the host specificity of phage RsoM1USA, 30 R. solanacearum strains originally isolated from different geographic locations of the world were tested for their susceptibility to the phage (Table 1). Phage RsoM1USA infected 12 of the 30 tested strains, including members of biovars 1, 2, 2T, and 3 belonging to each of the current R. solanacearum and R. pseudosolanacearum species (Table 1). The efficiency of phage plating was also determined for the 12 susceptible strains, and strain RUN302 was found to have the highest efficiency of phage infection (Table 1). The infection cycle of the phage was determined to be 180 min, including a 105 min of latent period, followed by a 75-min rise period with a burst size of 56 ± 6 particles per infected cell (Figure 1B). Phage RsoM1USA was stable from 4 to 50°C, since its titer at that temperature range remained similar at approximately 108 PFU/ml (Figure 1C). Significant loss in the phage titer, however, was observed at 60°C, and no phage particles were detected after they were incubated at 70, 80, and 90°C for 1 h (Figure 1C).
Genome Characterization of Ralstonia Phage RsoM1USA
The genome of phage RsoM1USA1 was degraded by DNase I, but not by RNase A, Exonuclease I and S1 nuclease (data not shown). The genome was also digested into multiple bands when treated with restriction enzymes EcoRV and SmaI (data not shown). The complete nucleotide sequence of the Ralstonia phage RsoM1USA was determined and submitted to GenBank (accession no. MG747435). The phage genome consists of 39,309 bp with a G+C content of 65.33%. Fifty-eight potential ORFs were identified in phage RsoM1USA (Supplementary Table S1). Among them, 48 started with methionine (ATG), four with leucine (CTG for ORFs 1 and 52 and TTG for 18 and 36), and five with valine (GTG) for ORFs 5, 14, 21, 23, and 37). ORF35, however, started with GTC (valine), an unusual start codon described by Feeney et al. (2017). Forty-two ORFs ended with TGA, nine with TAG (ORFs 2, 18, 22, 23, 26, 29, 52, 57, and 58) and seven with TAA (ORFs 6, 28, 33, 36, 39, 45, and 55). When the whole genome sequence of phage RsoM1USA was used as a query to search GenBank by BLASTn, the phage was found to be most closely related to R. solanacearum phage RSA1 (Yamada et al., 2007; Fujiwara et al., 2008), the member of the species Ralstonia virus RSA1 in the genus P2virus, with 89% nucleotide identity and 79% coverage. Each of the 58 ORFs of phage RsoM1USA was used as a query to search GenBank by BLASTp, and their positions, and predicted functions are summarized in Supplementary Table S1. Each of the ORFs was also compared to ORFs of ϕRSA1 and their similarity levels are indicated in Figure 2.
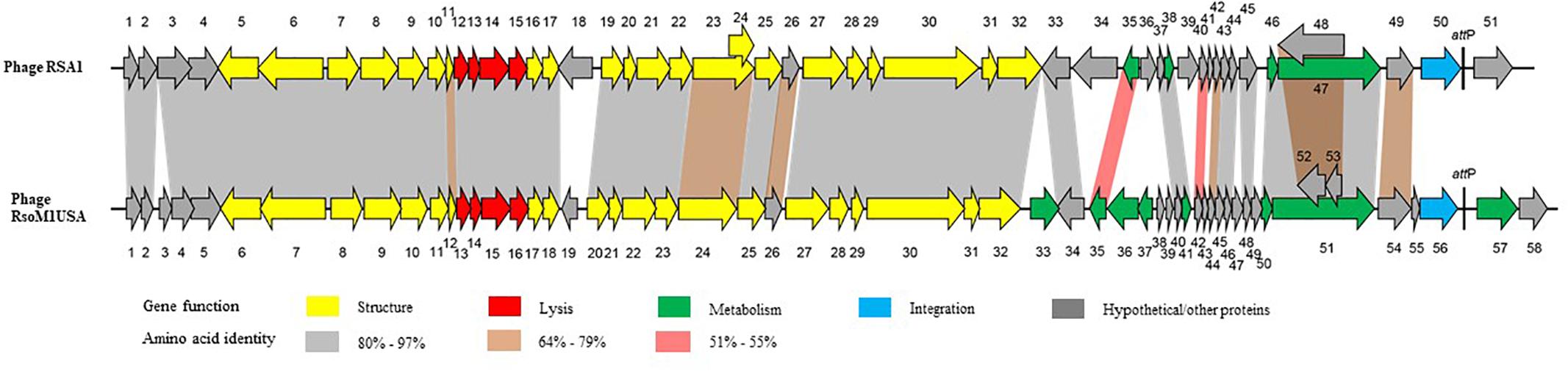
Figure 2. Comparison of genomic organization between phages RsoM1USA and RSA1. Different functional gene groups are represented and different levels of amino acid sequence identity between homologous ORFs are shaded with different colors. The location of the core attachment attP site is indicated in the two phages.
Gene Organization of Phage RsoM1USA and Comparison to Phage RSA1
The predicted ORFs of phage RsoM1USA can be categorized into the following four functional gene groups: metabolism (transcription and regulatory), structure (head and tail), lysis and integration, as well as hypothetical and other proteins.
(i) Metabolism
Eight ORFs were annotated to play a role in the phage’s metabolism, including ORF33 for a site-specific DNA-methyltransferase, ORF35 for a XRE family transcriptional regulator, ORF36 for a transposase, ORF37 for an insertion element repressor, ORF41 for phage transcription activator Org/Delta, a member of the zinc finger recombinase superfamily (pfam04606), ORF50 for a RNA binding protein, and ORF51 for a DNA primase. ORF57 was annotated to encode a ParA family protein, which were identical to its counterpart in R. solanacearum strain 23-10BR but shared no sequence homology to any other Ralstonia phages including RSA1.
(ii) Structure
Twenty-one ORFs were predicted to be involved in morphogenesis of phage RsoM1USA including 6 for capsid- and 15 for tail-related proteins (Supplementary Table S1). ORFs encoding phage capsid-related proteins included ORF 6 to ORF 11 for putative portal protein (ORF6), terminase (ORFs 7 and 10), scaffolding protein (ORF8), major capsid protein (ORF9), and head completion/stabilization protein (ORF11). ORFs encoding phage tail-related proteins included ORFs 12, 17, 18, 23, 29 to 32 for tail, ORFs 20 to 22 for baseplate, ORFs 24 and 25 for tail fiber, and ORFs 27 and 28 for tail sheath proteins. All structural genes of phage RsoM1USA shared aa sequence identity of 70 to 97% with the corresponding ORFs in phage RSA1 (Figure 2 and Supplementary Table S1). The genomic organization of the structural genes in the two phages is also very similar, except that ORF19, located between the structural genes orf18 and orf20 in phage RsoM1USA, was annotated as a hypothetical protein and did not share any aa sequence homology with the similarly located ORF18 in phage RSA1 (Figure 2 and Supplementary Table S1), which was also annotated as a hypothetical protein in phage RSA1 (Fujiwara et al., 2008).
To confirm the identity of the major structural components of phage RsoM1USA, proteomic analysis of the purified phage virions was performed by SDS-PAGE gel. At least 11 proteins ranging from 15 to over 90 kDa were separated in the gel (Figure 3). These bands were cut out and sequenced commercially. Based on the peptides identified using mass spectrometry and their relative abundance, as well as comparison to the deduced amino acid sequences and molecular masses of ORFs in phage RsoM1USA, the 11 bands most likely correspond to phage tail (ORFs 17, 24, 30 and 32), tail sheath (ORF27), portal (ORF 6), capsid (ORF9), baseplate assembly (ORFs 20 and 22), capsid scaffolding (ORF8), major tail tube (ORF28), virion morphogenesis (ORF 18), and bacteriophage P2 tail (ORF 31) proteins (Figure 3).
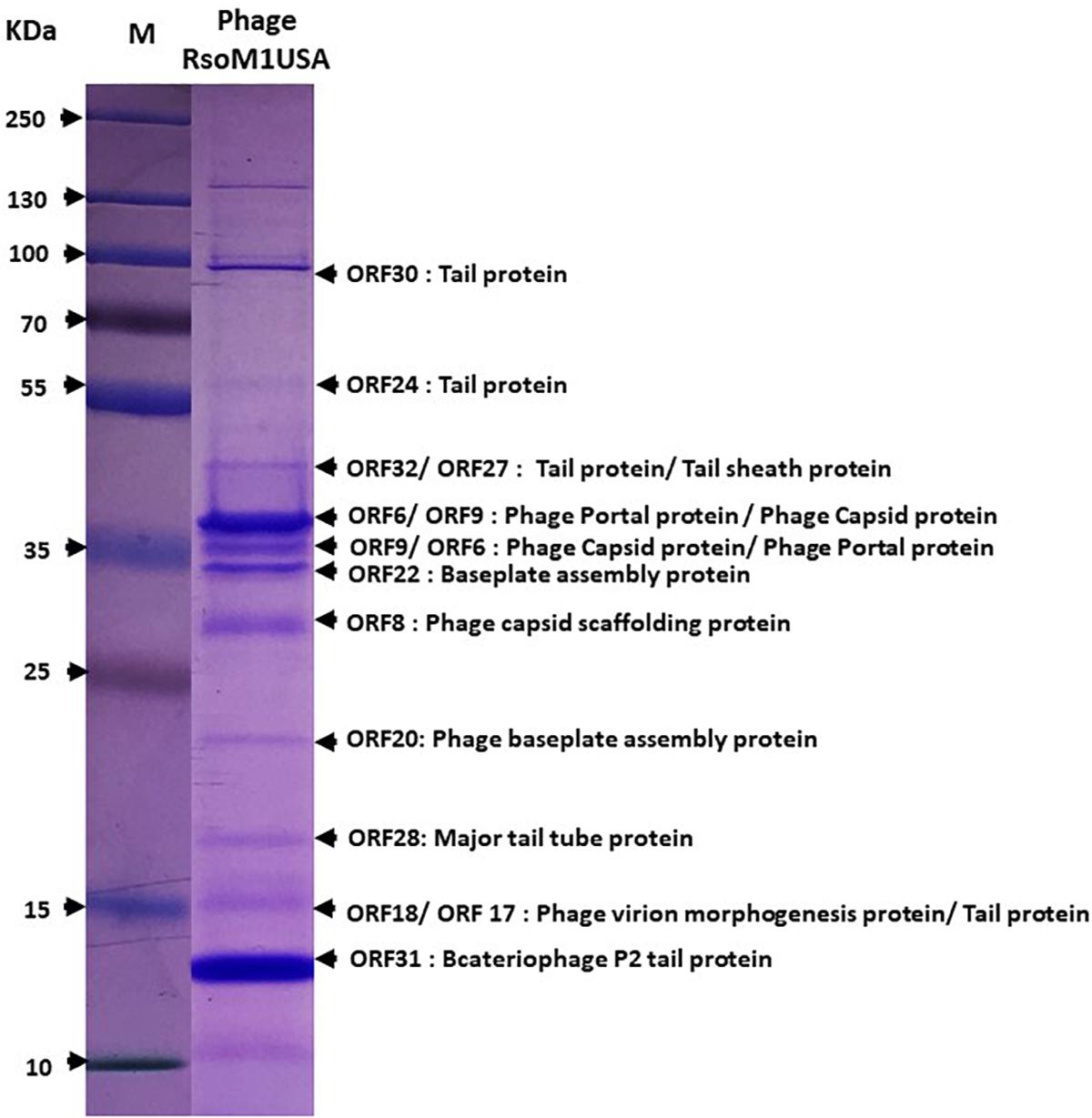
Figure 3. Expression of structural proteins of Ralstonia phage RsoM1USA. Proteins from purified phage virions were separated by SDS-PAGE gel (14%) and stained with Coomassie blue. The functions of the 11 protein bands predicted to correspond to ORFs 6, 8, 9, 17, 18, 20, 22, 24, 28, 30, 31, and 32 were indicated. M: protein ladder, with its molecular weight in kilodaltons (kDa) indicated on the left.
(iii) Lysis and Integration
Four consecutive ORFs (ORFs 13 to 16) were predicted to be involved in lysis of bacterial cells. ORFs 13 and 14 were annotated to encode phage-related transmembrane proteins, sharing aa sequence identity of 100% to the membrane protein and to the phage holin family protein, respectively, of R. solanacearum strain 23-10BR (Supplementary Table S1). ORF15 was predicted to be a peptidoglycan-binding protein (99% identity to R. solanacearum strain 23-10BR sequence) (Supplementary Table S1). ORF16 shared high aa sequence identity to a peptide in R. solanacearum strain 10-23BR (Supplementary Table S1), that may be involved in host lysis through endolysins with signal peptides (Catalao et al., 2013).
ORF56 of phage RsoM1USA was annotated as an integrase, as was ORF50 in phage RSA1. Surprisingly, however, the two integrase sequences did not share any aa sequence identity (Figure 2).
(iv) Hypothetical and Other Proteins
Twenty-four ORFs of phage RsoM1USA were annotated as hypothetical proteins, or proteins with other functions. Nineteen ORFs had homologs in phage RSA1, but ORFs 3, 19, 38, 39, and 49 did not. ORF55 was located before and ORFs 57 and 58 after the integrase (ORF56) and attP region in phage RsoM1USA, while no counterpart for ORF55 was present and only one ORF (ORF51) was found after the integrase (ORF50) and attP region in phage RSA1 (Figure 2). In addition, ORFs 57 and 58 of phage RsoM1USA did not share any aa sequence homology with ORF51 of phage RSA1 (Figure 2).
Phylogenetic Relationships Between Phages RsoM1USA and RSA1
Phylogenetic relationships were determined among phage RsoM1USA and 13 members of the genus P2virus under the subfamily Peduovirinae of the family Myoviridae (Figure 4). A phylogenetic tree generated based on the deduced aa sequences of phage capsid proteins showed that phage RsoM1USA is more closely related to Ralstonia phage RSA1 than to the other 12 members of the P2virus (Figure 4A). Similar trees were also obtained when the deduced aa sequences of terminase ATPase subunit and phage portal proteins were used, respectively (Figures 4B,C). Phage RsoM1USA was shown to be more closely related to Mannheimia virus PHL101 than to phage RSA1, however, when the deduced aa sequences of the integrase protein was used for phylogenetic analysis (Figure 4D).
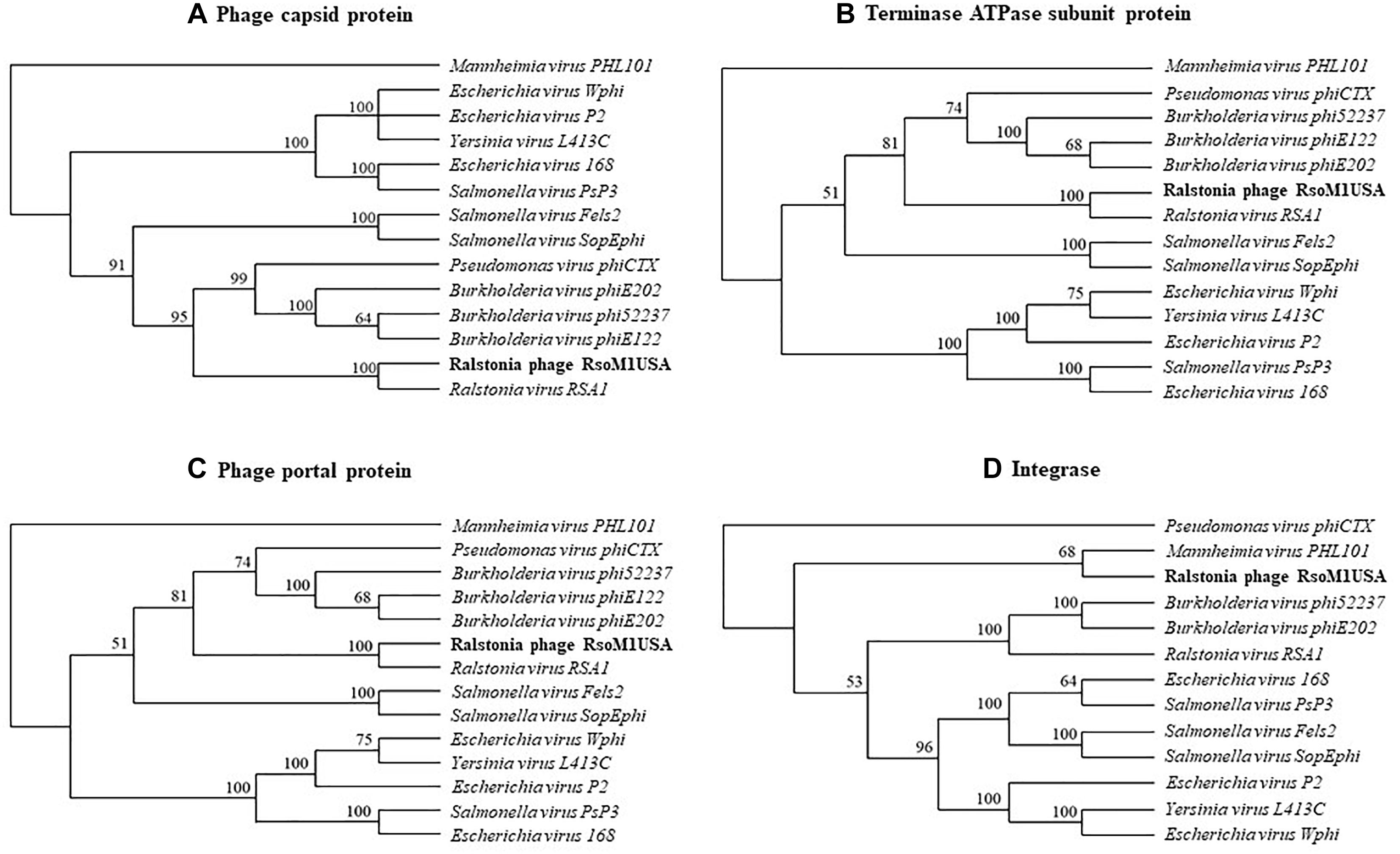
Figure 4. Phylogenetic relationships between Ralstonia phage RsoM1USA and 13 other phages of the genus P2virus (under the subfamily Peduovirinae of the family Myoviridae) including Ralstonia virus RSA1 (phage RSA1), based on deduced amino acid sequences of phage capsid protein (A), terminase ATPase subunit protein (B), and phage portal protein (C), as well as between RsoM1USA and 12 phages based on annotated integrase (D). The phylogenetic trees were generated using the downloadable alpha-test version of PAUP (Phylogenetic Analysis Using PAUP) (http://phylosolutions.com/paup-test). Vertical distances are arbitrary, but the horizontal branches are proportional to genetic distance. Bootstrap values (1,000 replicates) are represented at the nodes of the branches.
Integration and Attachment Site for Phage RsoM1USA
By BLAST search, we identified a tRNA-threonine-like sequence, corresponding to nucleotides 36,657 to 36,732, in phage RsoM1USA (Figure 5A). This sequence matched 100% to the 45 nucleotides at the 3′ end of the 76-bp threonine tRNA sequences in R. solanacearum strains RUN302, Po82, 23-10BR, GMI1000 and K60 (Figure 5A), suggesting that phage RsoM1USA contains a attP site where it recombines with the homologous bacterial attB site for the phage to integrate into the bacterial genome. To confirm that this 45-bp core sequence is involved in the integration of phage RsoM1USA into its susceptible R. solanacearum strain K60, the left and right integration flanking regions, attL and attR, were amplified by PCR using DNA of phage RsoM1USA-infected K60 as a template (Figure 5B) followed by sequencing of the PCR products (Figure 5A). Both the attL and attR fragments contained the 45-bp core att sequence that is in both the bacterial K60 and the phage RsoM1USA genomes (Figure 5A). The attL fragment contained K60 genome sequence upstream and phage RsoM1USA sequence downstream of this core sequence (Figure 5A). The attR site had phage RsoM1USA sequence upstream and K60 sequence downstream of the core sequencing (Figure 5A), the reverse of the orientation found for the attL site, confirming that the 45-bp core sequence is the site of phage integration.
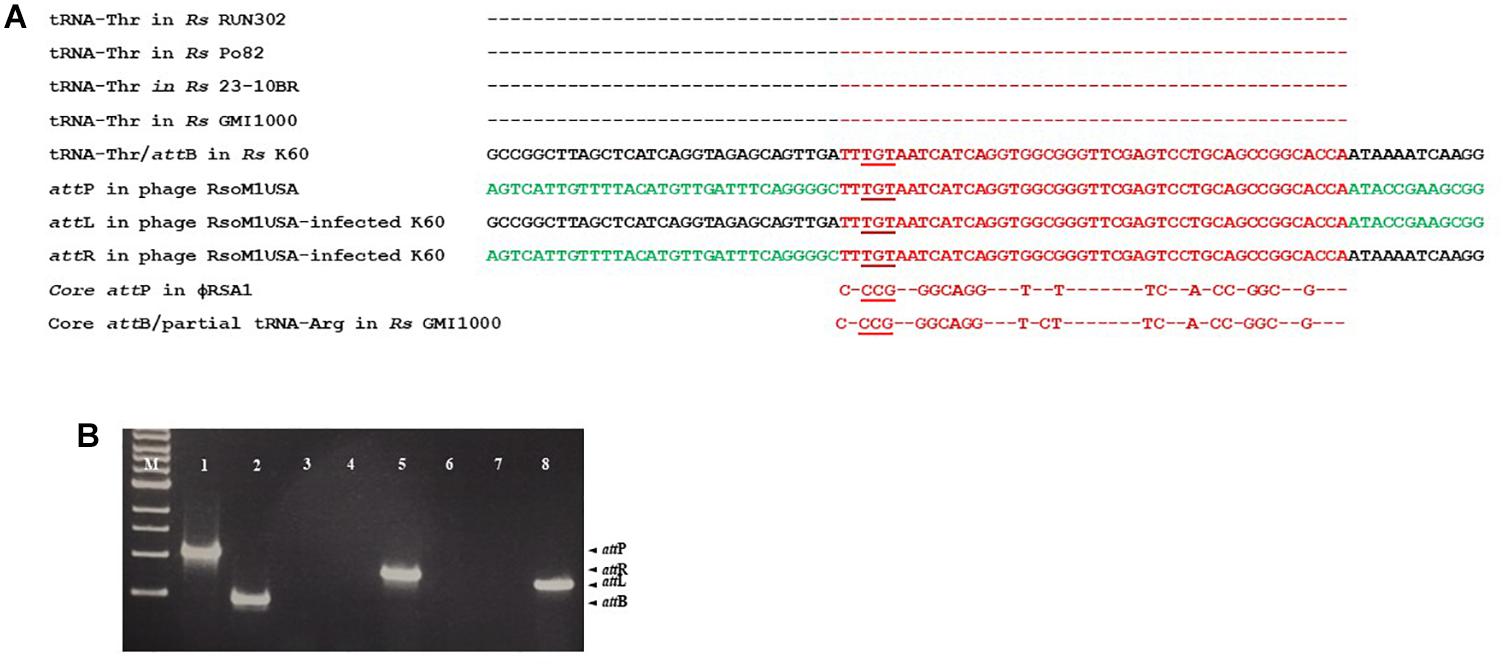
Figure 5. Integration of Ralstonia phage RsoM1USA into the genome of R. solanacearum strain K60. (A) Comparison of the attachment (att) sequences. The attB sequence of R. solanacearum strain K60 (before phage RsoM1USA integration) contains the same 76-bp threonine (Thr) tRNA sequence found in R. solanacearum strains RUN302, P082, 10-23BR, and GMI1000 (the same amino acid is indicated by “–”). The attB sequence in R. solanacearum strain K60 was aligned with the tRNA-threonine-like attP sequence in Ralstonia phage RsoM1USA. The 54-bp core att sequence is shown in red. The sequences after integration of phage RsoM1USA into R. solanacearum strain K60 (attL and attR) are shown. Sequences from the bacterial strain K60 and the phage RsoM1USA are shown in black and green, respectively. The core attP sequence of phage RsoM1USA was also compared to that of phage RSA1 which matched the 3′ arginine (Arg) tRNA in R. solanacearum strain GMI1000 (Fujiwara et al., 2008). Only nucleotides different from phage RsoM1USA’s core attP sequence were shown. The anticodons of threonine (TGT) and arginine (CCG) tRNAs are underlined. Rs: R. solanacearum. (B) Amplification of att regions by PCR and separation by gel electrophoresis. The 1,068-bp attP and 452-bp attB fragments were amplified from DNA of phage RsoM1USA (lane 1) and R. solanacearum strain K60 (lane 2), respectively. The 800-bp attR and 720-bp attL fragments were detected only from DNA of phage RsoM1USA-infected strain K60 (lanes 5 and 8) after integration, but not from phage RsoM1USA (lanes 3 and 6) and strain K60 (lanes 4 and 7). M: 1-kb DNA ladder.
Effects of Phage RsoM1USA on R. solanacearum Strain RUN302 in vitro and in planta
To study the effect of phage RsoM1USA on the in vitro growth and virulence of its susceptible R. solanacearum strain RUN302, we compared the in vitro growth and virulence of the wild type RUN302 to those of the phage-treated RUN302 of R. solanacearum. Strain RUN302 was chosen for the study because it showed the highest efficiency of phage infection (Table 1) and did not contain any intact prophages like prophage RsoM1USA in its genome before phage treatment. The growth of phage-treated R. solanacearum strain RUN302 was significantly reduced 15 h after phage treatment at MOI of 0.1, 1, and 10 (p < 0.05), and 35 h at all tested MOIs (p < 0.05) (Figure 6A). Since MOIs of 10, 1 and 0.1 resulted in similarly reduced bacterial growth 35 h after the phage treatment (p < 0.05) (Figure 6A), the lowest MOI of 0.1 was chosen for virulence assays in tomato plants for practical purposes. Results from our virulence assays revealed that both the untreated and the phage-treated RUN302 strains started to wilt tomato plants 6 days after soil drenching inoculation, and reached disease severity of over 80% by day 8 (Figures 6B,C). The overall disease severity caused by untreated RUN302 was not statistically different from that caused by the phage-treated RUN302 (p = 0.423 by the t-test at day 9) (Figure 6B). Tomato plants inoculated with water displayed no disease symptoms (Figures 6B,C).
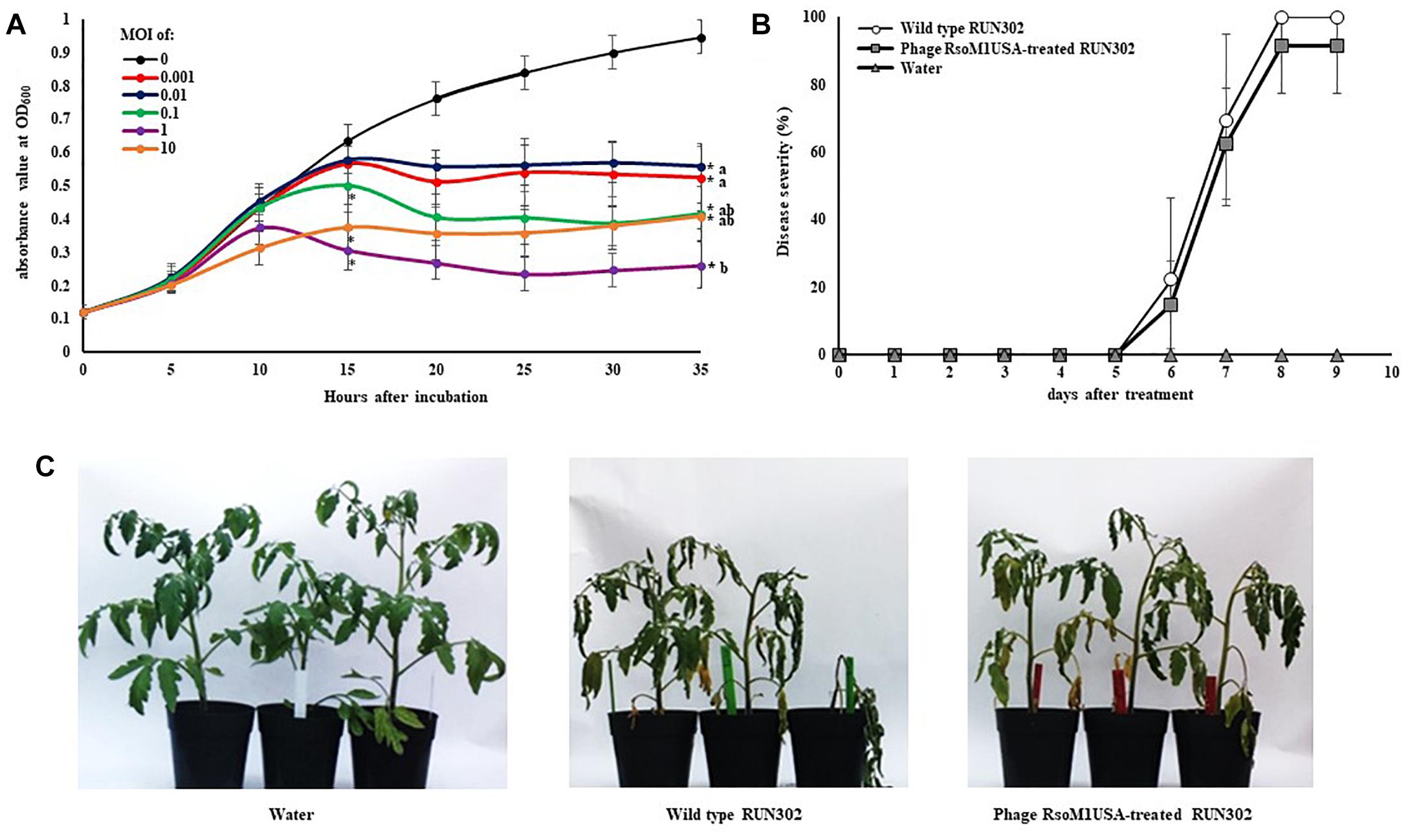
Figure 6. Effects of Ralstonia phage RsoM1USA on R. solanacearum strain RUN302. (A) In vitro growth of R. solanacearum strain RUN302 inoculated with phage RsoM1USA at a multiplicity of infection (MOI) of 0 (black), 0.001 (red), 0.01 (blue), 0.1 (green), 1 (pink) and 10 (orange), respectively. Bacterial growth indicated by absorbance at 600 nm was measured every 60 min and graphed every 5 h for 35 h. Points shown are the means of three separate experiments, each containing three replicates for each MOI treatment. Bars indicate standard errors. ∗ indicates significant difference (p < 0.05) at hours 15 and 35 between MOI of 0 and each of the other MOIs by the t-test. Means with different letters at hour 35 are significantly different based on the Tukey’s Honest Significant Difference test (p < 0.05). (B) Virulence of R. solanacearum strain RUN302 alone (open circle) and co-inoculated with phage RsoM1USA at the MOI of 0.1 (closed square). Disease severity was calculated based on a 0 (healthy) to 4 (75 to 100% leaves wilted) disease index of each plant. Points shown are means of three separate experiments, each containing 12 plants per treatment. Bars indicate standard errors. Significance of means was measured using the t-test (p < 0.05). (C) Appearance of the inoculated plants 8 days after soil drenching inoculation with water (left), RUN302 (middle), or RUN302 followed by phage RsoM1USA at the MOI of 0.1 (right).
Discussion
RsoM1USA is the first Ralstonia-infecting phage isolated from soil of the United States from a tomato field in Florida. We purified the phage and determined the phage to be a member of the genus P2virus in the family Myoviridae based on its morphology. The phage was designated RsoM1USA using the systematic phage identifier naming approach we proposed previously to make the phage name not only unique but also meaningful regarding the phage’s host species, morphology and origin (Ahmad et al., 2018).
Phage RsoM1USA contains all genes responsible for the morphogenesis of the phage head or capsid assembly as described by Fujiwara et al. (2008) and Fokine and Rossmann (2014), and those genes have high sequence homology (70–97%) with those of phage RSA1. It is, therefore, not surprising that virions of phage RsoM1USA resemble those of phage RSA1 (Yamada et al., 2007), displaying an icosahedral capsid and a long contractile tail with a tail sheath. Like phage RSA1, phage RsoM1USA encodes putative lysis genes commonly found in P2-like phages (Christie and Calendar, 2016) such as holin, suggesting that RsoM1USA may lyse host cells using holin and endolysin. Holin plays a key role in host lysis by forming large pores that are utilized as transport channels for endolysin to access and degrade the peptidoglycan layer resulting in host lysis (Shin et al., 2014). It is worth noting that both phages contain ORF1, encoding a protein similar to the PAAR (proline-alanine-alanine-arginine)-domain containing protein. This protein has been found mostly in beta- and gamma-proteobacteria such as Serratia marcescens, Vibrio cholerae, or Acinetobacter baylyi (Shneider et al., 2013; Cianfanelli et al., 2016). It is known to be a component of the spike complex of the bacterial type VI secretion system (T6SS) and has been reported to play a role in bacterial virulence (Shneider et al., 2013; Cianfanelli et al., 2016). In addition to the similarity in gene organization, the two phages are also similar in size (39,309 vs. 38,760-bp), and are closely related to each other (Figures 4A–C). Since phages RsoM1USA and RSA1 were isolated from soil in the United States and Japan, respectively, it raises an interesting question how the phages obtained from two different countries across continents share morphological and genomic similarities. It is likely that the two phages share common ancestry, as Hendrix et al. (1999) proposed for “all of the dsDNA tailed phages.” Many ORFs of phage RsoM1USA also have high sequence identity with prophage sequences in R. solanacearum strain 23-10BR (Supplementary Table S1), which is a sequevar 27 strain isolated from Brazil (Clarke et al., 2015), suggesting evolutionary relationships between the phage and bacterial strain.
Despite the similarities between phages RsoM1USA and RSA1, the two phages are different in the following ways: (1) The genes for metabolism are highly variable between the two phages. Among the eight ORFs annotated to have metabolic functions in phage RsoM1USA, one (ORF 35) shared low sequence homology with and four (ORFs 33, 36, 37, and 57) have no counterpart ORFs in phage RSA1 (Figure 2). Instead, they share some sequence homology with R. solanacearum strains including 23-10BR and CQPS-1 (Supplementary Table S1). (2) Seven ORFs in phage RsoM1USA annotated as hypothetical/other proteins have no corresponding ORFs in phage RSA1. (3) Myoviruses are known to integrate into bacterial genomes mediated by a portion of bacterial tRNA, an att site, as in the cases of phages RSA1 and RSY 1 of R. solanacearum (Fujiwara et al., 2008; Askora et al., 2017), phage 16-3 of Rhizobium meliloti (Blaha et al., 2004), and phage vB_RleM_PPF1 on Rhizobium leguminosarum (Halmillawewa et al., 2016). We confirmed experimentally that phage RsoM1USA uses the 3′ 45-base portion of the threonine tRNA (TGT) gene as its att site (Figure 5), which is different from RSA1 which uses the 3′ 45-base of the arginine tRNA (CCG) gene (Fujiwara et al., 2008). This is also different from the Ralstonia-infecting myovirus RSY1 which uses a 3′ 15-base portion of the serine tRNA (GGA) gene for integration and contains an integrase gene in opposite orientation (Askora et al., 2017). In addition, putative integrases encoded by ORF56 of phage RsoM1USA and ORF 50 of phage RSA1 have no sequence identity. The phylogenetic trees derived from the deduced amino acid sequences of capsid, terminase ATPase subunit and portal proteins all suggest that phage RsoM1USA is more closely related to the Ralstonia phage RSA1 than to other myoviruses in the genus P2virus (Figures 4A–C). A phylogenetic tree derived from deduced amino acid sequences of integrase, however, revealed that RsoM1USA is more closely related to Mannheimia virus PHL101 than to phage RSA1 (Figure 4D). These results suggest that integrases in the two phages may be evolved from different sources and they use different mechanism for integration. In view of these differences, phage RsoM1USA is either a variant of phage RSA1 or represents a third group, the RsoM1USA-type, in the p2-like lineages of R. solanacearum phages. At the species level, phages RsoM1USA and RSA1 are different based on their 89% nucleotide sequence identity, which is less than the 95% identity in DNA sequence conventionally used as a criterion to be considered the same species (Addy et al., 2018). As a result, the Ralstonia phage RsoM1USA should be considered a member of a new species with a proposed species name of Ralstonia virus RsoM1USA under the genus P2virus in the family Myoviridae.
Phage RsoM1USA is capable of infecting strains belonging to different Ralstonia species and originated from different geographic locations. Out of the 30 tested R. solanacearum species complex strains collected worldwide, 12 were susceptible to phage RsoM1USA including biovars 1, 2, 2T and 3 strains in two of the recently classified Ralstonia species: the current R. solanacearum (phylotypes II and III) and R. pseudosolanacearum (phylotypes I) originally isolated from the United States, Brazil, United Kingdom, Netherlands, Guatemala, and Taiwan (Table 1). It is worth noting that previous host range studies for Ralstonia phages used strains isolated only from the testing country (Yamada et al., 2007; Murugaiyan et al., 2010). In addition, those tested bacterial strains belong only to the current R. pseudosolanacearum species, making it harder to assess the biocontrol potential of the Ralstonia phages on Ralstonia strains beyond that country and species.
Compared to the untreated R. solanacearum strain RUN302, the in vitro growth of phage RsoM1USA-treated R. solanacearum strain RUN302 was significantly reduced when the phage was added to the bacterial cells at MOIs of 0.001 to 10. We used MOI of 0.1, not a higher one like 1 or 10, in our virulence assay is because by the Tukey’s Host Significant Difference test for the five MOIs used (0.001, 0.01, 0.1, 1, and 10), no significant difference in in vitro growth of R. solanacearum strain RUN302 treated by phage RsoM1USA was found at a MIO of 0.1, 1 or 10. We therefore chose the lowest MOI for practical purposes. The disease severity caused by the phage-treated RUN302 strain, however, was not affected in tomato plants. This suggests that the activity of a phage in vitro on a synthetic medium is not always correlated with its activity in planta under artificial or natural environment (soil or potting mix). The latter is chemically and physically more complex and may affect phage-bacterial host interaction and phage infectivity, particularly during phage absorption stage (Marsh and Wellington, 1994). Kimura et al. (2008) found that phage particles can be absorbed to soil with the degree of absorption over 90%, therefore decreasing the number of free phage particles available to infect bacterial hosts.
By BLAST search, the 45-bp attP site was found with 100% identity to the 45 nucleotides at the 3′ end of the 76-bp threonine tRNA sequences corresponding to nucleotide 1,494,395 to 1,494,470 in R. solanacearum strain RUN302 [(with an alternative name of IBSBF 1503 (Wicker et al., 2007)] (Figure 5), suggesting that phage RsoM1USA is capable of integrating into the genome of RUN302, causing no significant effect on bacterial virulence. This is similar to what was found for Ralstonia phage RSA1, the most closely related phage to RsoM1USA. Phage RSA1 uses the 3′ 45-base arginine gene for integration and results in no significant changes in virulence with RSA1 lysogenic cells in planta (Fujiwara et al., 2008). This, however, would be different from previously characterized lysogenic bacteriophages, which have been demonstrated under greenhouse conditions to either significantly enhance virulence (e.g., Ralstonia phage RSS1 in Addy et al., 2012b) or reduce virulence (e.g., Ralstonia phage Rs551 in Ahmad et al., 2017b and Xanthomonas phage XacF1 in Ahmad et al., 2014). Ralstonia phage RSM3 even caused loss of virulence of phage-infected R. solanacearum strain MAFF106611 (Addy et al., 2012a), but further studies are needed to develop this phage into a biocontrol agent. Whether these are the reasons why phage RsoM1USA did not prevent plant disease caused by R. solanacearum is worthy of future research. Future research is also needed to determine whether phage RsoM1USA plays an important ecological role in “regulating the virulence of and offering competitive fitness to its carrier bacterial strain for persistence of the bacterium in the environment,” similar to what Ralstonia phage Rs551 does (Ahmad et al., 2017a). These studies are important for a better understanding of the relationships among phages, bacterial hosts and their environments for effective control of R. solanacearum.
Conclusion
A Ralstonia-infecting bacteriophage designated RsoM1USA was isolated from soil of the United States. It is a member of Myoviridae containing 39,309 nucleotides with 58 ORFs. It is closely related to Ralstonia phage RSA1 but is different from RSA1 mainly in integrase and in utilizing the threonine tRNA as the att region for integration. Phage RsoM1USA should be considered a member of a new species with a proposed species name of Ralstonia virus RsoM1USA. Although phage RsoM1USA significantly reduced the in vitro growth of R. solanacearum strain RUN302 at MOIs between 0.001 and 10, it had no significant effect in reducing disease symptoms of RUN302 in tomato plants at MOI of 0.1 when compared to the untreated controls. The role of phage RsoM1USA, if any, in providing competitive fitness to its carrier bacterial strains remains to be determined.
Author Contributions
HA, AA, and QH conceived and designed the experiments and analyzed the data. HA and AA performed the experiments. QH contributed to the reagents, materials, and analysis tools. HA and QH wrote the manuscript.
Funding
This research was financially supported by the United States Department of Agriculture, Agricultural Research Service.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Joseph Mowery and the Beltsville Electron and Confocal Microscope Unit for technical assistance, and Dr. John Hartung at USDA/ARS for critical review of the manuscript. We also thank Dr. Mathews Paret and his students at University of Florida for providing soil samples.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00267/full#supplementary-material
Footnotes
- ^http://lowelab.ucsc.edu/tRNAscan-SE/
- ^https://www.ebi.ac.uk/Tools/msa/clustalo
- ^http://phylosolutions.com/paup-test
- ^http://vassarstats.net/anova1u.html
References
Addy, H. S., Askora, A., Kawasaki, T., Fujie, M., and Yamada, T. (2012a). Loss of virulence of the phytopathogen Ralstonia solanacearum through infection by ϕRSM filamentous phages. Phytopathology 102, 469–477. doi: 10.1094/PHYTO-11-11-0319-R
Addy, H. S., Askora, A., Kawasaki, T., Fujie, M., and Yamada, T. (2012b). The filamentous phage ϕRSS1 enhances virulence of phytopathogenic Ralstonia solanacearum on tomato. Phytopathology 102, 244–251. doi: 10.1094/PHYTO-10-11-0277
Addy, H. S., Farid, M. M., Ahmad, A. A., and Huang, Q. (2018). Host range and molecular characterization of a lytic Pradovirus-like Ralstonia phage RsoP1IDN isolated from Indonesia. Arch. Virol. 163, 3409–3414. doi: 10.1007/s00705-018-4033-1
Adriaenssens, E. M., and Brister, J. R. (2017). How to name and classify your phage: an informal guide. Viruses 9:E70. doi: 10.3390/v9040070
Ahmad, A. A., Askora, A., Kawasaki, T., Fujie, M., and Yamada, T. (2014). The filamentous phage XacF1 causes loss of virulence in Xanthomonas axonopodis pv. citri, the causative agent of citrus canker disease. Front. Microbiol. 5:321. doi: 10.3389/fmicb.2014.00321
Ahmad, A. A., Elhalag, K. M., Addy, H., Hussien, A. S., Nasr-Eldin, M. A., and Huang, Q. (2018). Sequencing, genome analysis and host range of a novel Ralstonia phage RsoP1EGY isolated from Egypt. Arch. Virol. 163, 2271–2274. doi: 10.1007/s00705-018-3844-4
Ahmad, A. A., Stulberg, M. J., and Huang, Q. (2017a). Prophage Rs551 and its repressor gene orf14 reduce virulence and increase competitive fitness of its Ralstonia solanacearum carrier strain UW551. Front. Microbiol. 8:2480. doi: 10.3389/fmicb.2017.02480
Ahmad, A. A., Stulberg, M. J., Mershon, J. P., Mollov, D. S., and Huang, Q. (2017b). Molecular and biological characterization of ϕRs551, a filamentous bacteriophage isolated from a race 3 biovar 2 strain of Ralstonia solanacearum. PLoS One 12:e0185034. doi: 10.1371/journal.pone.0185034
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Alvarez, B., and Biosca, E. G. (2017). Bacteriophage-based bacterial wilt biocontrol for an environmentally sustainable agriculture. Front. Plant Sci. 8:1218. doi: 10.3389/fpls.2017.01218
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Askora, A., Kawasaki, T., Fujie, M., and Yamada, T. (2017). Lysogenic conversion of the phytopathogen Ralstonia solanacearum by the P2virus ϕRSY1. Front. Microbiol. 8:2212. doi: 10.3389/fmicb.2017.02212
Bhunchoth, A., Blanc-Mathieu, R., Mihara, T., Nishimura, Y., Askora, A., Phironrit, N., et al. (2016). Two jumbo phages, ϕRSL2 and ϕRSF1, infect Ralstonia solanacearum and show common features of ϕKZ-related phages. Virology 494, 56–66. doi: 10.1016/j.virol.2016.03.028
Bhunchoth, A., Phironrit, N., Leksomboon, C., Chatchawankanphanich, O., Kotera, S., Narulita, E., et al. (2015). Isolation of Ralstonia solanacearum-infecting bacteriophages from tomato fields in Chiang Mai, Thailand, and their experimental use as biocontrol agents. J. Appl. Microbiol. 118, 1023–1033. doi: 10.1111/jam.12763
Blaha, B., Semsey, S., Ferenczi, S., Csiszovszki, Z., Papp, P. P., and Orosz, L. (2004). A proline tRNA(CGG) gene encompassing the attachment site of temperate phage 16-3 is functional and convertible to suppressor tRNA. Mol. Microbiol. 54, 742–754. doi: 10.1111/j.1365-2958.2004.04300.x
Catalao, M. J., Gil, F., Moniz-Pereira, J., Sao-Jose, C., and Pimentel, M. (2013). Diversity in bacterial lysis systems: bacteriophages show the way. FEMS Microbiol. Rev. 37, 554–571. doi: 10.1111/1574-6976.12006
Christie, G. E., and Calendar, R. (2016). Bacteriophage P2. Bacteriophage 6:e1145782. doi: 10.1080/21597081.2016.1145782
Cianfanelli, F. R., Alcoforado, D. J., Guo, M., De Cesare, V., Trost, M., Coulthurst, S. J., et al. (2016). VgrG and PAAR Proteins define distinct versions of a functional type VI secretion system. PLoS Pathog. 12:e1005735. doi: 10.1371/journal.ppat.1005735
Clarke, C. R., Studholme, D. J., Hayes, B., Runde, B., Weisberg, A., Cai, R., et al. (2015). Genome-enabled phylogeographic investigation of the quarantine pathogen Ralstonia solanacearum race 3 biovar 2 and screening for sources of resistance against its core effectors. Phytopathology 105, 597–607. doi: 10.1094/PHYTO-12-14-0373-R
Dykstra, M. J. (1993). A Manual of Applied Technique for Biological Electron Microscopy. New York, NY: Plenum Press. doi: 10.1007/978-1-4684-0010-6
Ellis, E. L., and Delbruck, M. (1939). The growth of bacteriophage. J. Gen. Physiol. 22, 365–384. doi: 10.1085/jgp.22.3.365
Feeney, M. A., Chandra, G., Findlay, K. C., Paget, M. S. B., and Buttner, M. J. (2017). Translational control of the SigR-directed oxidative stress response in Streptomyces via IF3-mediated repression of a noncanonical GTC start codon. mBio 8:e00815-17. doi: 10.1128/mBio.00815-17
Fegan, M., and Prior, P. (2005). “How complex is the Ralstonia solanacearum species complex?” in Bacterial Wilt Disease and the Ralstonia solanacearum Species Complex, eds C. Allen, P. Prior, and A. C. Hayward (Saint Paul, MN: APS Press), 449–461.
Fokine, A., and Rossmann, M. G. (2014). Molecular architecture of tailed double-stranded DNA phages. Bacteriophage 4:e28281. doi: 10.4161/bact.28281
Fujiwara, A., Fujisawa, M., Hamasaki, R., Kawasaki, T., Fujie, M., and Yamada, T. (2011). Biocontrol of Ralstonia solanacearum by treatment with lytic bacteriophages. Appl. Environ. Microbiol. 77, 4155–4162. doi: 10.1128/AEM.02847-10
Fujiwara, A., Kawasaki, T., Usami, S., Fujie, M., and Yamada, T. (2008). Genomic characterization of Ralstonia solanacearum phage ϕRSA1 and its related prophage (ϕRSX) in strain GMI1000. J. Bacteriol. 190, 143–156. doi: 10.1128/JB.01158-07
Gill, J. J., Summer, E. J., Russell, W. K., Cologna, S. M., Carlile, T. M., Fuller, A. C., et al. (2011). Genomes and characterization of phages Bcep22 and BcepIL02, founders of a novel phage type in Burkholderia cenocepacia. J. Bacteriol. 193, 5300–5313. doi: 10.1128/JB.05287-11
Halmillawewa, A. P., Restrepo-Córdoba, M., Perry, B. J., Yost, C. K., and Hynes, M. F. (2016). Characterization of the temperate phage vB_RleM_PPF1 and its site-specific integration into the Rhizobium leguminosarum F1 genome. Mol. Genet. Genomics 291, 349–362. doi: 10.1007/s00438-015-1113-8
Harrison, C. J., and Langdale, J. A. (2006). A step by step guide to phylogeny reconstruction. Plant J. 45, 561–572. doi: 10.1111/j.1365-313X.2005.02611.x
Hayes, M. M., MacIntyre, A. M., and Allen, C. (2017). Complete genome sequences of the plant pathogens Ralstonia solanacearum type strain K60 and R. solanacearum race 3 biovar 2 strain UW551. Genome Announc. 5:e01088-17. doi: 10.1128/genomeA.01088-17
Hayward, A. C. (1991). Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 29, 65–87. doi: 10.1146/annurev.py.29.090191.000433
Hendrix, R. W., Smith, M. C. M., Burns, R. N., Ford, M. E., and Hatfull, G. F. (1999). Evolutionary relationships among diverse bacteriophages and prophages: all the world’s a phage. Proc. Natl. Acad. Sci. U.S.A. 96, 2192–2197. doi: 10.1073/pnas.96.5.2192
Huang, Q., and Allen, C. (2000). Polygalacturonases are required for rapid colonization and full virulence of Ralstonia solanacearum on tomato plants. Physiol. Mol. Plant Pathol. 57, 77–83. doi: 10.1006/pmpp.2000.0283
Kimura, M., Jia, Z. J., Nakayama, N., and Asakawa, S. (2008). Ecology of viruses in soils: past, present and future perspective. Soil Sci. Plant Nutr. 54, 1–32. doi: 10.1111/j.1747-0765.2007.00197.x
Kropinski, A. M., Prangishvili, D., and Lavigne, R. (2009). Position paper: the creation of a rational scheme for the nomenclature of viruses of bacteria and archaea. Environ. Microbiol. 11, 2775–2777. doi: 10.1111/j.1462-2920.2009.01970.x
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. doi: 10.1038/227680a0
Liao, M. (2018). Genomic characterization of the novel Ralstonia phage RPSC1. Arch. Virol. 163, 1969–1971. doi: 10.1007/s00705-018-3713-1
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.955
Marsh, P., and Wellington, E. M. H. (1994). Phage-host interaction in soil. FEMS Microbiol. Ecol. 15, 99–108. doi: 10.1111/j.1574-6941.1994.tb00234.x
Mihara, T., Nasr-Eldin, M. A., Chatchawankanphanich, O., Bhunchoth, A., Phironrit, N., Kawasaki, T., et al. (2016). A phage ϕRP15 is closely related to and encodes 19 tRNA-related sequences. Virol. Rep. 6, 61–73. doi: 10.1016/j.virep.2016.07.001
Murugaiyan, S., Bae, J. Y., Wu, J., Lee, S. D., Um, H. Y., Choi, H. K., et al. (2010). Characterization of filamentous bacteriophage PE226 infecting Ralstonia solanacearum strains. J. Appl. Microbiol. 110, 296–303. doi: 10.1111/j.1365-2672.2010.04882.x
Park, T.-H. (2018). Complete genome sequence of DU_RP_II, a novel Ralstonia solanacearum phage of the family Podoviridae. Arch. Virol. 163, 269–271. doi: 10.1007/s00705-017-3577-9
Roberts, D. P., Denny, T. P., and Schell, M. (1988). Cloning of the egl gene of Pseudomonas solanacearum and analysis of its role in pathogenicity. J. Bacteriol. 170, 1445–1451. doi: 10.1128/jb.170.4.1445-1451.1988
Safni, I., Cleenwerck, I., De Vos, P., Fegan, M., Sly, L., and Kappler, U. (2014). Polyphasic taxonomic revision of the Ralstonia solanacearum species complex: proposal to emend the descriptions of Ralstonia solanacearum and Ralstonia syzygii and reclassify current R. syzygii strains as Ralstonia syzygii subsp. syzygii subsp. nov., R. solanacearum phylotype IV strains as Ralstonia syzygii subsp. indonesiensis subsp. nov., banana blood disease bacterium strains as Ralstonia syzygii subsp. celebesensis subsp. nov. and R. solanacearum phylotype I and III strains as Ralstonia pseudosolanacearum sp. nov. Int. J. Syst. Evol. Microbiol. 64, 3087–3103. doi: 10.1099/ijs.0.066712-0
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning: A Laboratory Manual, 3rd Edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Shin, H., Lee, J. H., Yoon, H., Kang, D. H., and Ryu, S. (2014). Genomic investigation of lysogen formation and host lysis systems of the Salmonella temperate bacteriophage SPN9CC. Appl. Environ. Microbiol. 80, 374–384. doi: 10.1128/AEM.02279-13
Shneider, M. M., Buth, S. A., Ho, B. T., Basler, M., Mekalanos, J. J., Leiman, P. G., et al. (2013). PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500, 350–353. doi: 10.1038/nature12453
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Stulberg, M. J., Shao, J., and Huang, Q. (2015). A multiplex PCR assay to detect and differentiate select agent strains of Ralstonia solanacearum. Plant Dis. 99, 333–341. doi: 10.1094/PDIS-05-14-0483-RE
Su, J., Sun, H., Liu, J., Guo, Z., Fan, G., Gu, G., et al. (2017). Complete genome sequence of a novel lytic bacteriophage isolated from Ralstonia solanacearum. Arch. Virol. 162, 3919–3923. doi: 10.1007/s00705-017-3555-2
Swofford, D. L. (2003). PAUP∗. Phylogenetic Analysis Using Parsimony (∗and Other Methods). Version 4. Sunderland, MA: Sinauer Associates.
Thi, B. V. T., Khanh, N. H. P., Namikawa, R., Miki, K., Kondo, A., Thi, P. T. D., et al. (2015). Genomic characterization of Ralstonia solanacearum phage RS138 of the family Siphoviridae. Arch. Virol. 161, 483–486. doi: 10.1007/s00705-0152654-1
Van, T. T. B., Yoshida, S., Miki, K., Kondo, A., and Kamei, K. (2014). Genomic characterization of ϕRS603, a filamentous bacteriophage that is infectious to the phytopathogen Ralstonia solanacearum. Microbiol. Immunol. 58, 697–700. doi: 10.1111/1348-0421.12203
Wicker, E., Grassart, L., Coranson-Beaudu, R., Mian, D., Guilbaud, C., Fegan, M., et al. (2007). Ralstonia solanacearum strains from Martinique (French west indies) exhibiting a new pathogenic potential. Appl. Environ. Microbiol. 73, 6790–6801. doi: 10.1128/AEM.00841-07
Yamada, T., Kawasaki, T., Nagata, S., Fujiwara, A., Usami, S., and Fujie, M. (2007). New bacteriophages that infect the phytopathogen Ralstonia solanacearum. Microbiology 153, 2630–2639. doi: 10.1099/mic.0.2006/001453-0
Keywords: Ralstonia solanacearum, Ralstonia phage, isolation, characterization, P2virus, Myoviridae
Citation: Addy HS, Ahmad AA and Huang Q (2019) Molecular and Biological Characterization of Ralstonia Phage RsoM1USA, a New Species of P2virus, Isolated in the United States. Front. Microbiol. 10:267. doi: 10.3389/fmicb.2019.00267
Received: 05 October 2018; Accepted: 01 February 2019;
Published: 19 February 2019.
Edited by:
Ramy Karam Aziz, Cairo University, EgyptReviewed by:
Jessica Sacher, University of Alberta, CanadaHany Anany, Agriculture and Agri-Food Canada (AAFC), Canada
Copyright © 2019 Addy, Ahmad and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qi Huang, cWkuaHVhbmdAYXJzLnVzZGEuZ292
 Hardian Susilo Addy
Hardian Susilo Addy Abdelmonim Ali Ahmad
Abdelmonim Ali Ahmad Qi Huang
Qi Huang