- Department of Chemistry and Biochemistry, Brigham Young University, Provo, UT, United States
Use of chlorhexidine in clinical settings has led to concerns that repeated exposure of bacteria to sub-lethal doses of chlorhexidine might result in chlorhexidine resistance and cross resistance with other cationic antimicrobials including colistin, endogenous antimicrobial peptides (AMPs) and their mimics, ceragenins. We have previously shown that colistin-resistant Gram-negative bacteria remain susceptible to AMPs and ceragenins. Here, we investigated the potential for cross resistance between chlorhexidine, colistin, AMPs and ceragenins by serial exposure of standard strains of Gram-negative bacteria to chlorhexidine to generate resistant populations of organisms. Furthermore, we performed a proteomics study on the chlorhexidine-resistant strains and compared them to the wild-type strains to find the pathways by which bacteria develop resistance to chlorhexidine. Serial exposure of Gram-negative bacteria to chlorhexidine resulted in four- to eight-fold increases in minimum inhibitory concentrations (MICs). Chlorhexidine-resistant organisms showed decreased susceptibility to colistin (8- to 32-fold increases in MICs) despite not being exposed to colistin. In contrast, chlorhexidine-resistant organisms had the same MICs as the original strains when tested with representative AMPs (LL-37 and magainin I) and ceragenins (CSA-44 and CSA-131). These results imply that there may be a connection between the emergence of highly colistin-resistant Gram-negative pathogens and the prevalence of chlorhexidine usage. Yet, use of chlorhexidine may not impact innate immune defenses (e.g., AMPs) and their mimics (e.g., ceragenins). Here, we also show that chlorhexidine resistance is associated with upregulation of proteins involved in the assembly of LPS for outer membrane biogenesis and virulence factors in Pseudomonas aeruginosa. Additionally, resistance to chlorhexidine resulted in elevated expression levels of proteins associated with chaperones, efflux pumps, flagella and cell metabolism. This study provides a comprehensive overview of the evolutionary proteomic changes in P. aeruginosa following exposure to chlorhexidine and colistin. These results have important clinical implications considering the continuous application of chlorhexidine in hospitals that could influence the emergence of colistin-resistant strains.
Introduction
Chlorhexidine (Figure 1) is a potent antiseptic agent that has been in use since the 1950s (Davies et al., 1954). It is used extensively in healthcare settings in hand washing, patient bathing, treatment of gingivitis, skin antisepsis, and other antimicrobial applications (Kampf, 2016). Chlorhexidine is sparingly soluble in water, and thereby normally formulated with either acetate or gluconate to form water-soluble salts. Chlorhexidine has been found to have antibacterial, antifungal, and even some anti-viral activities (Lim and Kam, 2008). Although chlorhexidine has proven to be an effective antimicrobial agent in hospital settings, it has limitations. While data show that chlorhexidine is active against planktonic bacterial cells, it is less effective against biofilms. After treatment of Staphylococcus aureus (MRSA) and multi-drug resistant (MDR) Pseudomonas aeruginosa biofilms grown on hospital grade-surfaces, these pathogens were shown to maintain up to 11 and 80% of their cell viability respectively (Smith and Hunter, 2008). Additionally, recent studies found that various clinical isolates of Klebsiella pneumoniae, from before and after the widespread use of chlorhexidine, show a significant difference in susceptibility to the antimicrobial (Bock et al., 2016; Wand et al., 2017). Notably, it has been shown that development of resistance is stable; bacteria do not revert to susceptible forms after being grown for an extended time in antimicrobial-free medium, suggesting that resistance is mutational rather than adaptational (Bock et al., 2016).
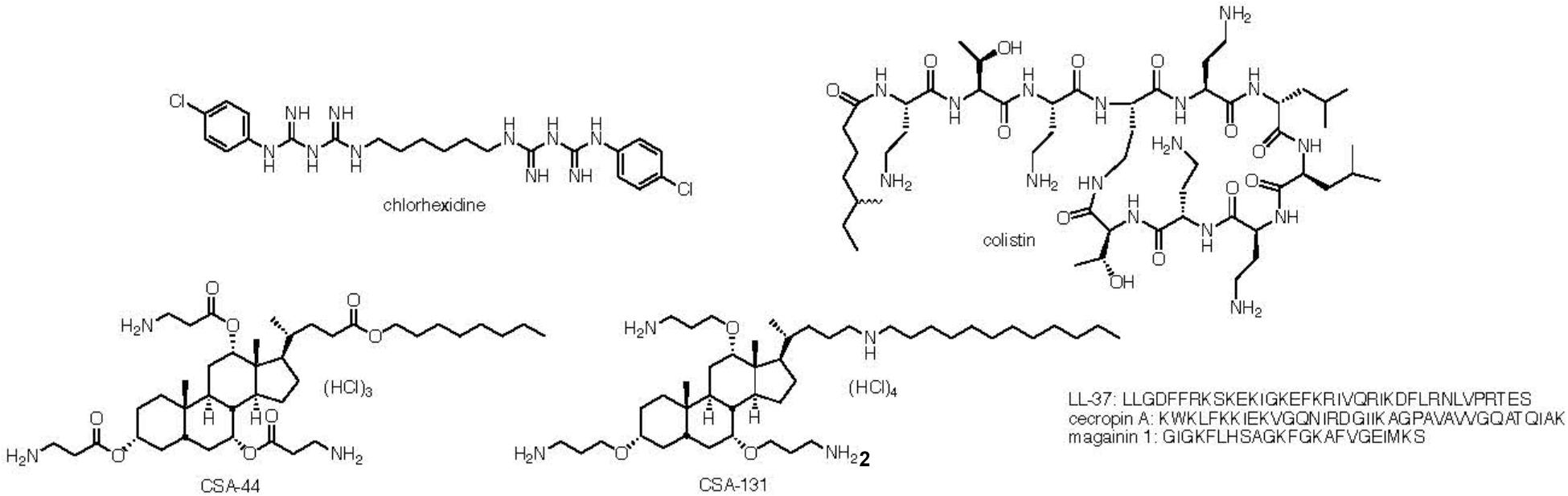
Figure 1. Structure of cholorhexidine, colistin and ceragenins CSA-44 and CSA-131 and sequences of LL-37, cecropin A, and magainin 1.
Due to the prevalent use of chlorhexidine, especially in clinical settings, there has been increasing concern about whether repeated exposures of bacteria to sub-lethal doses of chlorhexidine is a factor in the emergence of MDR bacteria (Denny and Munro, 2017). For example, Palmer and coworkers found that serial exposure of vancomycin-resistant Enterococcus faecium to chlorhexidine resulted in reduced susceptibility to other membrane-targeting antimicrobials such as daptomycin (Bhardwaj et al., 2018). Of particular concern is the role of exposure to chlorhexidine in generation of colistin resistance. The prevalence of Gram-negative, drug-resistant bacteria has resulted in increased use of colistin (Figure 1), which is considered an antibiotic of “last resort.” However, colistin resistant organisms have been increasingly isolated from clinical settings (Wright et al., 2015; Hammerl et al., 2017). Wand et al. (2017) recently reported cross-resistance between chlorhexidine and colistin. They found that in chlorhexidine-adapted strains of K. pneumoniae, the minimum inhibitory concentrations (MICs) for colistin increased by a factor between 16 and 32 with five of the six strains tested. However, when colistin-resistant K. pneumoniae were tested for chlorhexidine susceptibility, Wand et al. (2017) found that MICs of chlorhexidine were the same with colistin-susceptible and colistin-resistant strains. On the other hand, Curiao et al. (2016) reported that at least in Salmonella, there are multiple mutations leading to chlorhexidine resistance that are not necessarily associated with colistin resistance.
Cross resistance of bacteria to chlorhexidine and colistin may be due to common features of these antimicrobials. For example, both are cationic (positively charged) with hydrophobic functionality. Notably, most endogenous antimicrobial peptides (AMPs, representative sequences are shown in Figure 1) also display these features: cationic groups juxtaposed with hydrophobic side chains (Hashemi et al., 2017a, 2018a). We recently reported (Hashemi et al., 2017c) that highly colistin-resistant bacteria are susceptible to both AMPs and non-peptide mimics of AMPs, termed ceragenins (representative structures are shown in Figure 1) (Hashemi et al., 2017b, 2018b,c,d). This finding led to the question of whether generation of chlorhexidine resistance results in cross resistance to ceragenins and AMPs. Reported herein are investigations of the means by which Gram-negative bacteria become resistant to chlorhexidine and how this resistance influences susceptibility to colistin, AMPs and ceragenins. Rather than focus on mutations, changes to the proteome were investigated to obtain an understanding of multiple possible alterations in gene expression that lead to resistance. We found that, as expected, induction of resistance in Gram-negative bacteria to chlorhexidine resulted in decreased susceptibility to colistin; nevertheless, organisms remained susceptible to AMPs and ceragenins. Changes to the proteome of chlorhexidine-resistant organisms, relative to a parent strain, were in the outer membrane proteins as well as proteins associated with chaperones, efflux pumps, flagella and cell metabolism.
Materials and Methods
Materials
Klebsiella pneumoniae (ATCC 13883), Acinetobacter baumannii (ATCC 19606), and P. aeruginosa (ATCC 27853) were obtained from the American Type Culture Collection (ATCC, Manassas, VA, United States). Ceragenins CSA-131 and CSA-44 were synthesized from a cholic acid scaffolding as previously described (Li et al., 1998). LL-37 (LLGDFFRKSKEKIGKEFKRIVQRIK-DFLRNLVPRTES), cecropin A (KWKLFKKIEKVGQNIRDGIIKAGPAVAVVGQATQIAK), and magainin 1 (GIGKFLHSAGKFGKAFVGEIMKS), all with > 95% purity, were purchased from Anaspec (Fremont, CA, United States) and used without further purification. Chlorhexidine, colistin (polymyxin E), congo red and coomassie brilliant blue dyes were purchased from Sigma-Aldrich (St. Louis, MO, United States).
Susceptibility Testing
Minimum inhibitory concentrations were determined using a broth microdilution method in a 96-well plate according to the Clinical and Laboratory Standards Institute (2006). Briefly, twofold dilutions of antibacterial agents (chlorhexidine, colistin, LL-37, cecropin A, magainin 1, and ceragenins) in tryptic soy broth (TSB) for P. aeruginosa or Mueller Hinton (MH) for K. pneumoniae and A. baumannii were dispensed in separate wells of a 96-well plate. Aliquots (100 μL) of a prepared inoculum (106 cfu/mL) were added, and plates were incubated at 37°C for 18–20 h. Negative and positive controls were included for each set of MIC measurements. The lowest concentrations of antibacterial agents that inhibited visible growth of bacteria were recorded as the MICs.
Serial Passage of Bacteria With Chlorhexidine
Serial passaging of bacteria was performed according to a protocol described previously (Hashemi et al., 2017c). Briefly, MICs (24 h) for the strains were determined, and bacteria growing at the highest concentrations of the antimicrobials were used to inoculate fresh medium. This process was repeated every 18–24 h for 10 days. MICs were recorded every 24 h, and concentrations of the antimicrobials were adjusted to allow for determination of MICs.
Colony Morphology Assay
MHA plates were made in 60 mm × 15 mm Petri dishes with the addition of Congo red (40 mg/L) and Coomassie brilliant blue (20 mg/L) dyes. Bacterial cultures were grown overnight in MH broth at 37°C. An aliquot of bacterial culture (1 μL) was then spotted onto the MHA dyed plates and incubated at 37°C for 24 h (Mukherjee et al., 2017). Images were acquired using a stereo microscope (Stemi 2000, Zeiss).
Proteomic Sample Preparation
Three independent studies were performed for the proteomics analyses and one single colony was used for each independent study. Both wild-type (ATCC 27583) and drug-resistant P. aeruginosa were cultured in TSB (25 mL) to an OD600 nm of 0.5. Cells were pelleted at 3750 × g for 10 min at room temperature followed by three washes with ice-cold PBS. Cell pellets were lysed in 1% sodium dodecyl sulfate (1 mL) in Tris buffer (50 mM, pH 8). Cells were briefly sonicated to further lyse the cells. Protein concentration was determined by bicinchoninic acid (BCA) protein assay (Thermo Fisher Scientific, Waltham, MA, United States). Protein digestion was carried out using filter-assisted digestion. Aliquots of 50 μg of protein were transferred to centrifugal filters (30 kD molecular weight cutoff), denatured with guanidine (100 μL, 6 M guanidinium hydrochloride, 100 mM Tris, pH 8.5), and centrifuged for 15 min at 14000 × g. This step was repeated three times to remove SDS entirely. Cell lysate was reduced by DTT (20 mM in 6 M guanidinium hydrochloride) at 65°C for 5 min. After cooling for 5 min, alkylation was performed with iodoacetamide (50 mM, 1 h incubation at room temperature in the dark). The mixture was then washed two times with ammonium bicarbonate buffer (25 mM, pH 8.5). The washed, alkylated protein sample was re-suspended in ammonium bicarbonate (ABC, 100 μL) followed by overnight digestion at 37°C with trypsin (1 μg, Sigma-Aldrich Proteomics Grade). After trypsin digestion, peptides were isolated by spinning down the mixture (14,000 × g, 30 min) with one rinse of the filter using ABC (100 μL, centrifugation 14,000 × g for 30 min). The combined filtrate was vacuum dried using a speedvac (Sorval). The dried sample was immediately re-dissolved in 50 μL of buffer (0.1% formic acid in Optima-grade water) for mass spectrometry analysis (Wiśniewski et al., 2009).
Mass Spectrometry Data Acquisition
Mass spectrometry data were collected using an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific, Waltham, MA, United States) coupled to an EASY-nLC 1200 liquid chromatography (LC) pump (Thermo Fisher Scientific, Waltham, MA, United States). A capillary RSLC column (EASY-spray column pepMap ® RSLC, C18, 2 μm, 100 Å, 75 μm × 25 cm) was used for separation of peptides. The mobile phase was comprised of buffer A (0.1% formic acid in Optima water) and buffer B (Optima water and 0.1% formic acid in 80% acetonitrile).
Elution of peptides was performed at 300 nL/min with the following gradients over 2 h: 3–25% B for 80 min; 25–35% B for 20 min; 35–45% B for 8 min; 45–85% B for 2 min and 85% for 8 min. Data were acquired using the method described by Peng et al. (2017). Briefly, using the top speed method (3 s cycle) and a resolution of 120,000 at 200 m/z, a full scan MS was obtained in the orbitrap with a target value of 3e5 and a maximum injection time of 60 ms. Detection of fragment ions in the linear ion trap was performed with a target AGC value of 1e4 and a maximum injection time of 250 ms.
Label-free quantification (LFQ) values were obtained using PEAKS (version 8.5) and the Swiss-Prot validated P. aeruginosa proteome (5638 sequences May 2018) with a reverse sequence decoy database concatenated and included during the search. After the removal of contaminants, low scoring peptides and reverse matches, the LFQ values for the top three peptides were summed and transformed by a logarithm of 2. The proteins were then separated into wild-type and mutant groups, and only those with at least three correct LFQ values were used for quantification. Accordingly, two-tailed heteroscedastic t-tests were conducted to compare wild-type versus mutant groups. The acquisition of gene ontology terms and functional clusters, as well as the identification of molecular functions and biological process, was conducted using the Database for Annotation, Visualization and Integrated Discovery (DAVID) and UniProt. Information about protein-protein interaction networks was retrieved from STRING (v10).
Results and Discussion
Susceptibility of Tested Gram-Negative Bacteria to Antimicrobials
Previous work has shown that Gram-negative bacteria become resistant to colistin following exposure to a sublethal concentration of colistin; however, colistin resistance does not correlate with higher MIC levels for AMPs or ceragenins (Hashemi et al., 2017c). To determine whether resistance to chlorhexidine also leads to cross-resistance against other antimicrobials, Gram-negative strains were serially exposed to chlorhexidine for 10 days with MIC determination every 24 h (Figure 2). MICs of AMPs (LL-37, cecropin A, magainin 1), lead ceragenins (CSA-44 and CSA-131) and colistin were measured with the chlorhexidine resistant strains (Table 1). Serial exposure of Gram-negative bacteria to chlorhexidine resulted in a rapid four- to eight-fold increase in MICs of chlorhexidine within 10 days. Notably, the chlorhexidine-resistant organisms showed decreased susceptibility to colistin (8- to 32-fold increases in MICs) in spite of not having been exposed to colistin. In contrast, MICs of representative AMPs were the same with the original strains and the chlorhexidine-resistant organisms. Similarly, the MICs of lead ceragenins (CSA-44 and CSA-131) were unchanged with the original and chlorhexidine resistant bacteria. Although chlorhexidine-resistant Gram-negative bacteria showed decreased susceptibility to colistin, as earlier reported by Wand et al. (2017) and seen in our studies, colistin-resistant Gram-negative bacteria did not show an increased MIC with chlorhexidine (data not shown). Overall, these results support the idea that exposure of Gram-negative bacteria to chlorhexidine can lead to cross-resistance to colistin, while susceptibilities to AMPs and ceragenins remain unchanged.
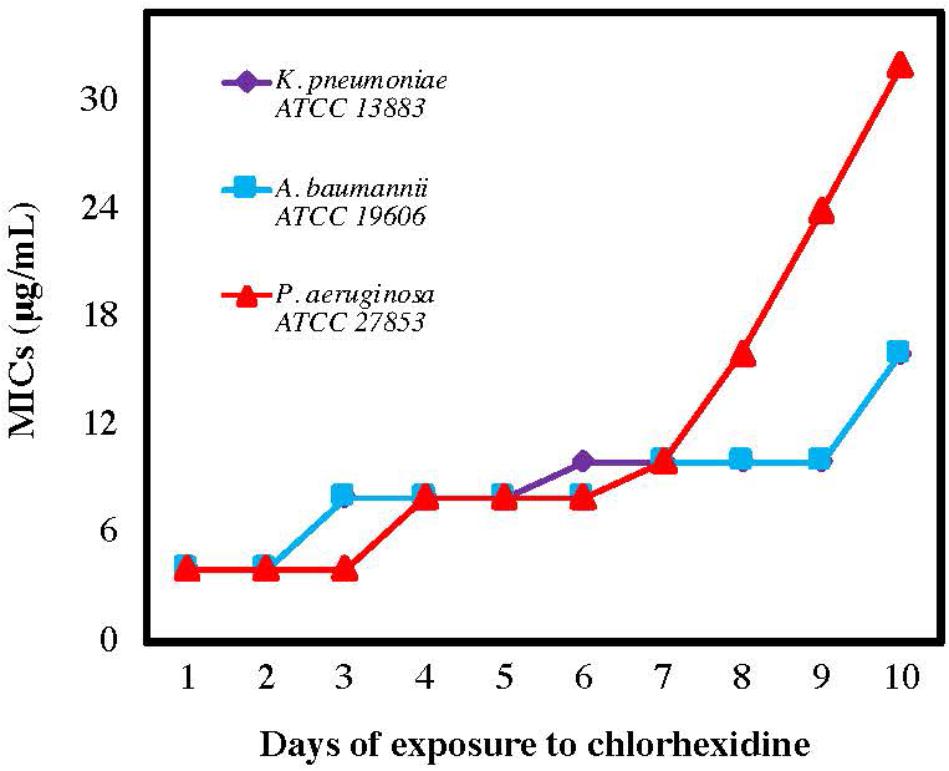
Figure 2. Minimum inhibitory concentrations (MICs) of chlorhexidine against Acinetobacter baumannii, Pseudomonas aeruginosa, and Klebsiella pneumoniae after the number of sequential exposures (24 h) to chlorhexidine.
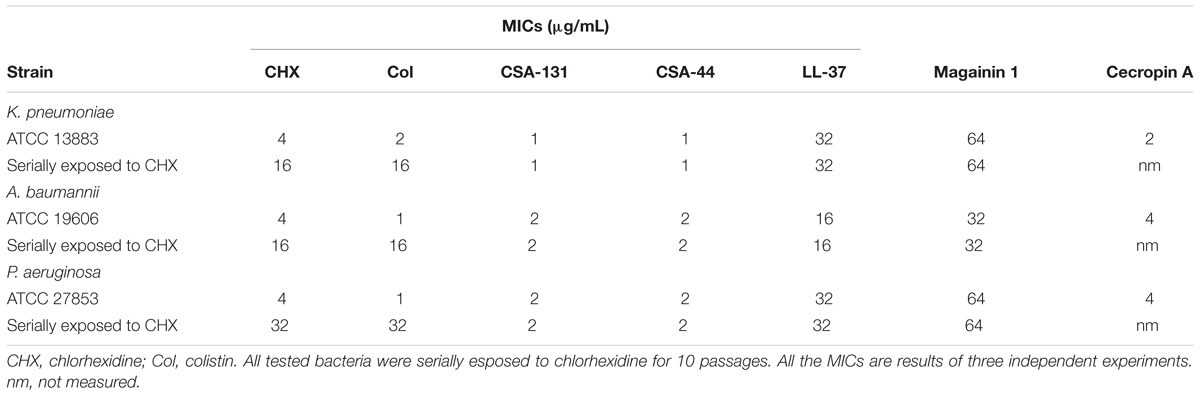
Table 1. Minimum inhibitory concentrations (MICs) of chlorhexidine, colistin, CSA-131, CSA-44, LL-37, magainin 1, and cecropin A with susceptible standard strains of K. pneumoniae, A. baumannii, and P. aeruginosa and with strains serially exposed to chlorhexidine and colistin.
Colony Morphology
Control strains of each type of Gram-negative bacteria produced colonies of circular and smooth morphology (Figures 3A–C). The serially passaged strains of A. baumannii and K. pneumoniae, however, formed colonies of irregular shape and had rough surfaces (Figures 3D–F). Although P. aeruginosa serially exposed to chlorhexidine produced colonies which were circular, they also had slightly rough surfaces and formed undulating margins. All three serially passaged strains grew larger colonies that covered more surface area than their respective controls. These changes in morphology may be a response to LPS modifications. For example, in Pseudomonas stutzeri, it has been shown that chlorhexidine resistance accompanies a loss of low-mass LPS species in SDS-PAGE gels. Because these strains showed cross-resistance, the shift to higher-mass LPS suggests that multidrug resistance is accompanied by LPS production which favors larger polysaccharide components (Tattawasart et al., 2000). And, as demonstrated in Gram-negative Vibrio cholerae, colony morphology can shift from smooth to rough in response to extracellular polysaccharide changes (Limoli et al., 2015). The connection between extracellular polysaccharides and colony morphology is further supported by Salmonella enterica, which shifts from rough to smooth morphology when short O-antigen chain LPS production is up-regulated (Karatzas et al., 2008). Because capsular polysaccharides have been shown to shield K. pneumoniae from polymixins, including colistin (Formosa et al., 2015), the increased mass of the polysaccharides contained within LPS may be the bridge between resistance to the two classes of antimicrobials, either by introducing additional mass or by changing the organization of the capsule. Generally, proposed mechanisms of colistin resistance in Gram-negative bacteria involve LPS modifications (Walsh and Duffy, 2013; Hashemi et al., 2017c; Kim and Ko, 2018). Thus, chlorhexidine resistance potentially brings about colistin resistance through one or more modifications to LPS.
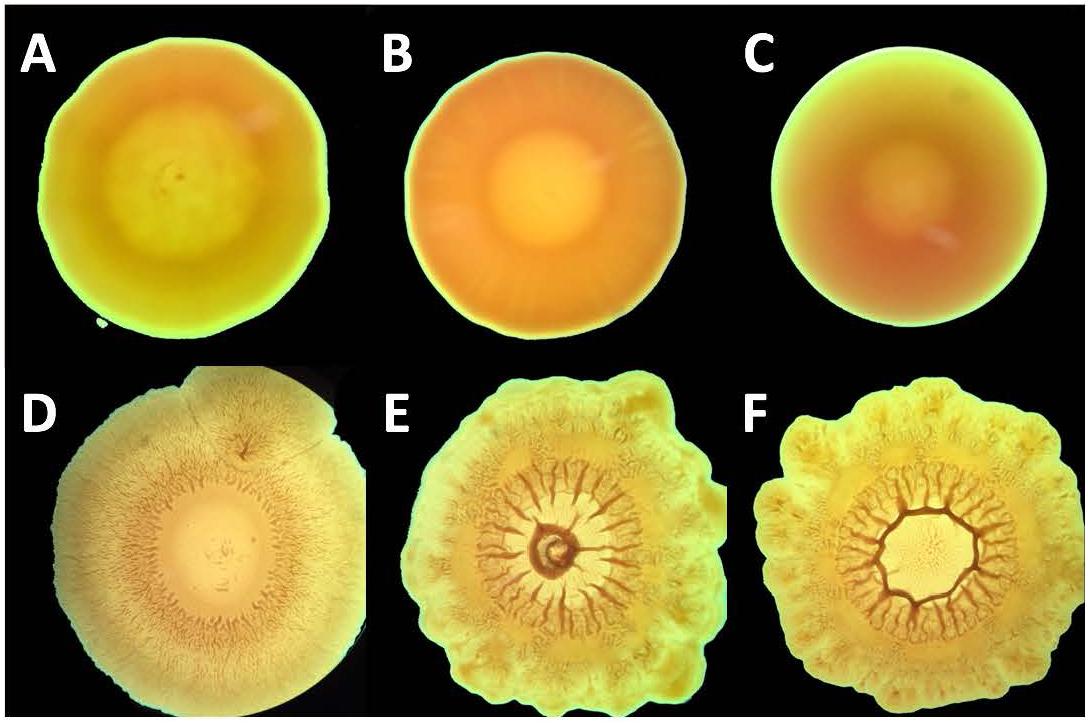
Figure 3. Colony morphology of A. baumannii and, P. aeruginosa, and K. pneumoniae upon expore to chlorhexidine. Bacterial strains P. aeruginosa (A,D), A. baumannii (B,E), and K. pneumoniae (C,F) were serially passaged with chlorhexidine (D–F) and the resulting colony morphology was compared with untreated colonies (A–C).
Proteomics
Proteomics has been established as an effective method of analyzing biological processes and is particularly valuable for examining resistance to antibiotics (Fernández-Reyes et al., 2009). For P. aeruginosa, a recent study has unveiled several differences in protein expression associated with low and high levels of ciprofloxacin resistance (Peng et al., 2017). On the whole, though, proteomic data on chlorhexidine-resistant P. aeruginosa remain scarce, in particular regarding cross-resistance with colistin, for which, to the best of our knowledge, no data on differential expression inherent to resistance are yet available. We have evaluated cross-resistance between colistin, chlorhexidine, AMPs and their mimic, ceragenins, and we further analyzed the proteome of chlorhexidine-resistant P. aeruginosa using reference strain ATCC 27853 for comparison.
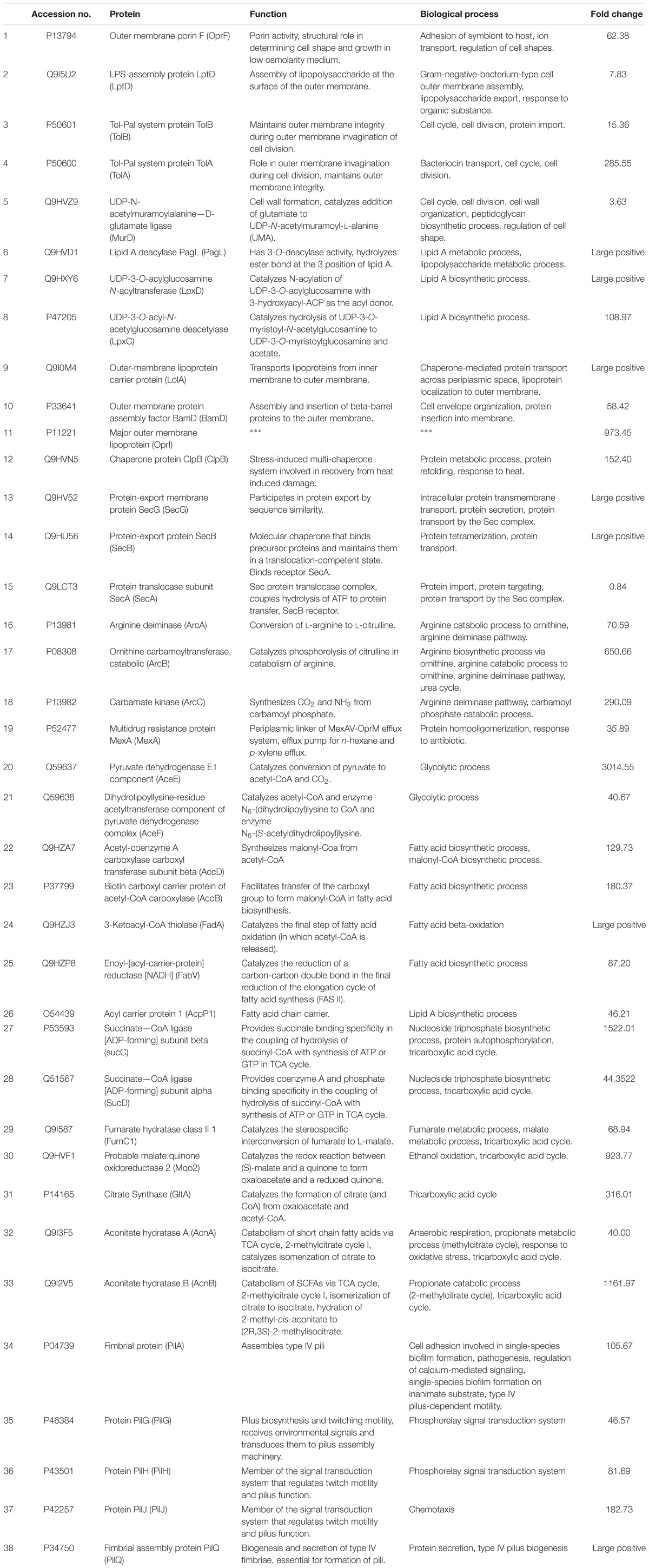
Table 2. Identification, function, and biological process of proteins differentially expressed between chlorhexidine-resistant and wild-type P. aeruginosa.
We used label-free quantitation to compare the relative concentrations of proteins within the proteome of wild-type and chlorhexidine-resistant P. aeruginosa. We observed substantial alterations of the proteome of the resistant strain relative to the wild-type strain, with several highly abundant proteins reduced in relative concentration and increases in a collection of proteins with distinct connections to antibiotic resistance as outlined below. A total of 330 proteins (out of 528 proteins) were differentially expressed at a significant level (twofold-change, p < 0.05), following exposure to chlorhexidine. Complete data analysis is provided as Supplementary Material. Due to the high number of proteins significantly differentially expressed, not all are described or listed in Table 2. Identification, function and biological functions of some proteins differentially expressed between the chlorhexidine-resistant and wild-type P. aeruginosa are listed in Table 2 in the same order that they are mentioned in the text. Large changes in protein expression indicate that the general protein concentration can vary dramatically in response to treatment (Southey-Pillig et al., 2005; Karatzas et al., 2008).
We searched for relationships among the most upregulated (12.5% of the total differentially expressed) and the most downregulated (12.5% of the total differentially expressed) proteins and assigned them to functional clusters. The numbers of differentially expressed proteins for each cluster are shown in Figure 4. Functional clusters containing the largest number of upregulated proteins are involved in microbial metabolism in diverse environments, carbon metabolism, ATP binding, nucleotide binding and translation of proteins. Conversely, functional clusters containing the largest number of downregulated proteins include nucleotide-binding, ATP-binding, cytoplasm and nucleotide phosphate-binding proteins.
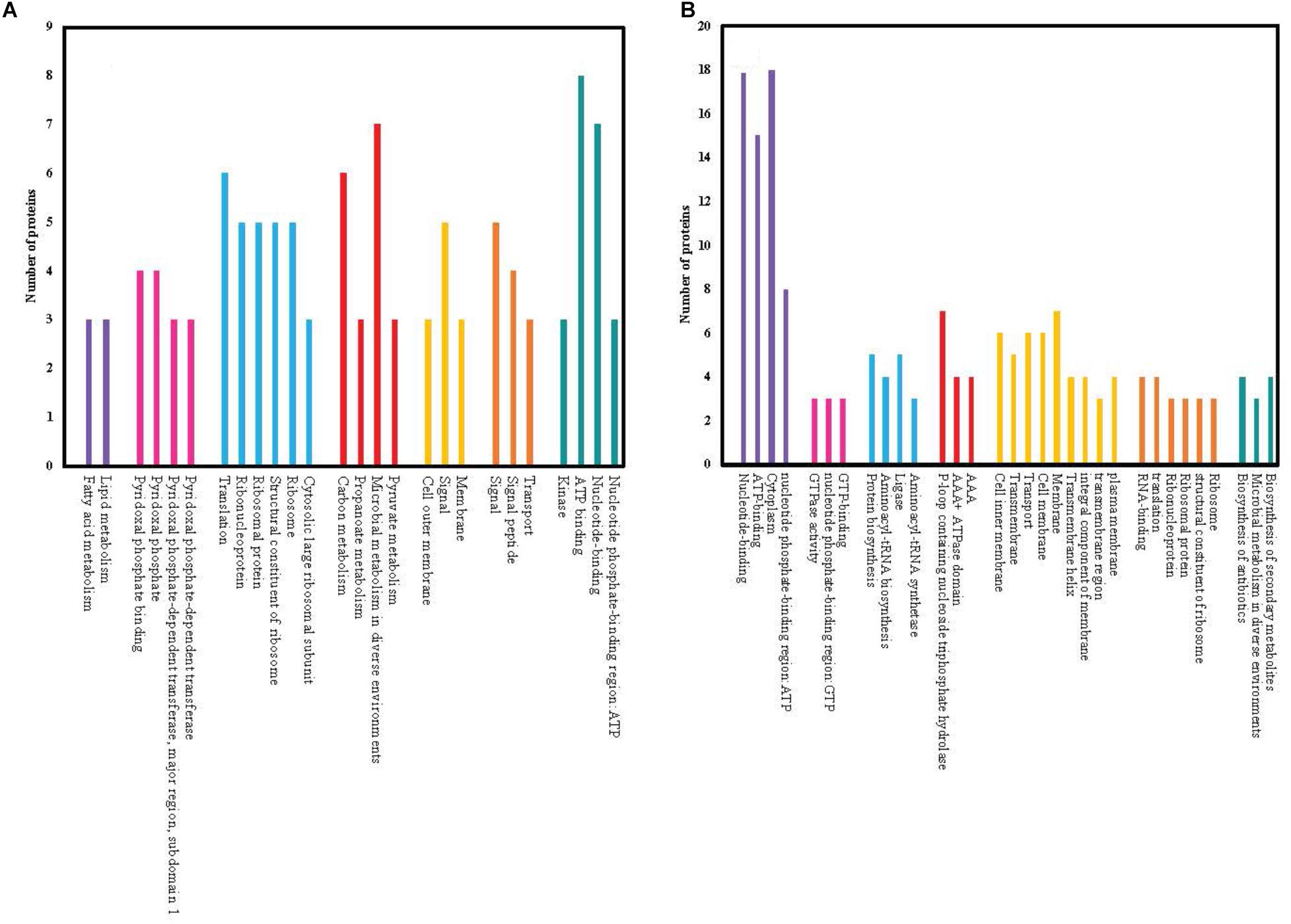
Figure 4. Functionally similar proteins (colored groups) among the most significantly (≥2 fold changes, p < 0.05) upregulated (A) and downregulated (B) proteins of chlorhexidine-resistant P. aeruginosa compared to wild-type. Each bar within the colored group represents a different functional connection between proteins within that group.
Modifications in Membrane Proteins Following Exposure to Chlorhexidine
Due to the positive charge on chlorhexidine, its initial interactions with Gram-negative bacteria are thought to occur at negatively-charged regions of LPS. Further, reduced susceptibility to chlorhexidine has been previously reported in association with modifications in the cell membrane, including modifications to LPS (Tattawasart et al., 2000; Condell et al., 2014). LPS modifications that have been associated with decreased bacterial susceptibility to cationic agents, such as chlorhexidine, include charge-reducing modifications in LPS and size reduction of the O-antigen polymer (Condell et al., 2014).
In this study, several changes in the expression of proteins associated with the cell membrane were noted following chlorhexidine exposure. As shown in Table 2, OprF was one of the most highly upregulated proteins in our study. OprF is a multifunctional protein that helps maintain cell shape, interacts with peptidoglycan layers, and is associated with forming non-specific channels for passive diffusion of ions, small polar nutrients and antibiotics (Hancock and Brinkman, 2002; Li et al., 2018). It has been shown that inactivated OprF decreases virulence and increases outer membrane vesicle formation in P. aeruginosa (Maccarini et al., 2017). Additional work has been done on the role of OprF in modifying quorum sensing and promoting virulence factor production in P. aeruginosa. Loss of functioning OprF was accompanied by loss of quorum sensing network activity and virulence factor production (Fito-Boncompte et al., 2011).
Protein network analysis (Figure 5) showed that the relative increase in OprF is accompanied by an increase in LptD concentration, a protein involved in the assembly of LPS for OM biogenesis. Mutant, non-functional LptD in P. aeruginosa increases the permeability of the outer membrane and results in compromised barrier function (Balibar and Grabowicz, 2015). Therefore, upregulation of LptD in our data suggests that exposure to chlorhexidine decreases the permeability of the OM in P. aeruginosa.
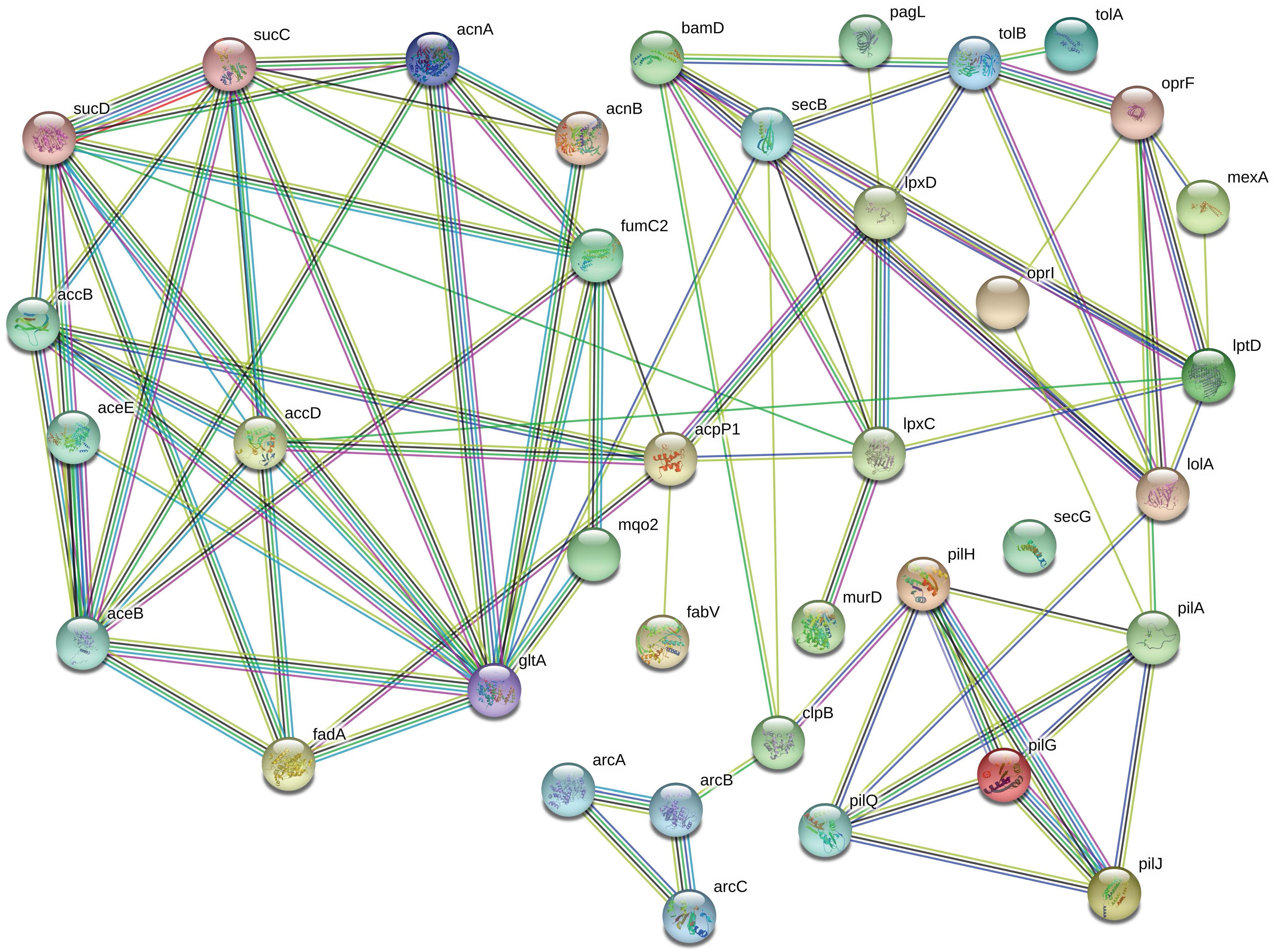
Figure 5. The most significantly upregulated proteins in chlorhexidine-resistant P. aeruginosa are highly interconnected. The connections between OprF to LptD, TolB, OprI, and LolA suggests that LPS production is substantially altered (relevant proteins are in the top right corner).
OprF also is connected to TolB (Figure 5), part of the Tol-Pal system, which contributes to the biogenesis of the OM and plays a key role in maintaining OM integrity. A significant upregulation of TolA, another part of Tol-Pal system, was also observed upon exposure of the cells to chlorhexidine in our proteomics data. Biochemical studies have identified interactions between TolA and outer membrane porins (Derouiche et al., 1995), which are consistent with our results where both TolA and OM porin F, OprF, are upregulated.
Studies have shown that mutant tol genes in E. coli increased susceptibility to antibiotics (Dennis et al., 1996). The absence of TolA in E. coli K-12 has been shown to cause a substantial reduction in the surface expression of O antigen (O7-specific) LPS and less lipid A-core oligosaccharide. Its absence is also correlated with attenuated growth rate and dramatic morphological changes (Gaspar et al., 2000). In tol mutants of V. cholerae, filamentation has also been reported (Heilpern and Waldor, 2000). This observation is consistent with our morphological study in which treatment with chlorhexidine caused dramatic morphological changes and altered flagella protein regulation in the Gram-negative bacteria.
An increase in the expression of a ligase enzyme associated with peptidoglycan biosynthesis (MurD) was also observed. Upregulation of MurD may result in a protective modification by increasing the thickness or cross-linking the peptidoglycan layer, which subsequently reduces the efficacy of chlorhexidine by affecting its transportation. Mur enzymes have been an attractive target for the development of antibacterial inhibitors owing to the fact that these enzymes are highly conserved molecules specific to bacteria with a known structure and function. However, no Mur inhibitors are clinically used as antimicrobials (Nikolaidis et al., 2014).
Of note, a protein which increased with chlorhexidine resistance was PagL, a deacetylase which removes the R-3-hydroxymyristate at position 3 of lipid A. PagL is activated by the phoP/phoQ two-component system, a regulatory system controlling genes for virulence within Salmonella (Olaitan et al., 2014; Park and Groisman, 2014). Though usually inhibited by 4-amino-4-deoxy-L-arabinose and phosphoethanolamine modifications of lipid A, PagL deacetylation of lipid A has been tied to increased polymyxin resistance in Salmonella strains lacking these modifications. In a later study, overexpression of a pagL-specific sRNA, Sr006, was shown to increase pagL mRNA, lipid A deacetylation, and polymyxin B resistance in P. aeruginosa. The same study also demonstrated that a pagL knockout resulted in decreased polymyxin B resistance (Zhang et al., 2017) Therefore, upregulation of PagL in chlorhexidine-resistant P. aeruginosa indicates that the mechanism of action for resistance to chlorhexidine could be partly in common with that of polymyxins.
Interestingly, protein network analysis (Figure 5) suggested that PagL is connected to LpxD, a significantly upregulated protein which is involved in lipid A biosynthesis along with LpxC, another upregulated protein. Previous MALDI-TOF analysis of the lipid A extracted from RamA-overexpressing strains of K. pneumoniae revealed that RamA increases colistin/polymyxin resistance levels (De Majumdar et al., 2015). This increase was accomplished through RamA directly binding to lipid A biosynthesis genes such as lpxC causing lipid A structural modifications. However, no observation of RamA overexpression was observed in our study. We also found several outer membrane lipoproteins including LolA, BamD, and OprI upregulated following chlorhexidine exposure. These results suggest that the increased lipoprotein synthesis of BamD and OprI might be required for appropriate sorting of lipoproteins by carrier proteins, like LolA, to the outer membrane under certain stress conditions such as antibiotic exposure (Tao et al., 2012).
Altered Chaperone Protein Regulation Following Exposure to Chlorhexidine
The Clp chaperones and proteases are highly conserved proteins found across the prokaryotes, in the mitochondria of eukaryotes and the chloroplasts of plants. Previous studies showed that ClpB is a chaperone associated with resistance to high temperature in E. coli. Furthermore, inactivation of the clpB gene of Leptospira interrogans resulted in reduced virulence and attenuated the in vitro growth of the cells under stress conditions, suggesting that ClpB plays a major role in bacterial survival under stress conditions (Lourdault et al., 2011). Upregulation of ClpB upon exposure to chlorhexidine was also observed in our study, confirming that ClpB is essential for stress tolerance against inducing factors like antimicrobial agents where proteins tend to unfold and aggregate. Clp protease, a periplasmic chaperone protein, was also reported to be upregulated in biofilm upon chlorhexidine treatment (Rema et al., 2016).
One of the protein translocation systems through the inner membrane involves Sec proteins in a chaperone-based pathway where immature proteins are targeted to a Sec translocase such as SecG, SecE, and SecY using the chaperone SecB for transport across the inner membrane. Our data showed upregulation of SecG and SecB while SecA was downregulated. Previous reports showed that loss of function of SecG leads to a significant increase in membrane depolarization, hydroxyl radical formation and cell death in aminoglycoside-treated cells. The redox-responsive two-component system (Arc) was also shown to have an associated role (Kohanski et al., 2008). Interestingly, our proteomics profile revealed the upregulation of ArcA, ArcB, and ArcC that encode arginine deiminase, ornithine carbamyltransferase, and carbamate kinase, respectively. The Arc two-component system contains ArcB, a quinone-sensitive sensor kinase that controls the redox state of the cellular quinone pool via modulating the phosphorylation state of the transcription factor ArcA (Kohanski et al., 2008; Liu et al., 2008).
Efflux Pump Activity Following Exposure to Chlorhexidine
A previous proteomics study correlated an increase in the expression of MexCD-OprJ efflux systems with a reduction in susceptibility to ciprofloxacin in P. aeruginosa (Peng et al., 2017). Additionally, the transcriptome of P. aeruginosa revealed that MexCD-OprJ efflux pump was significantly upregulated after treatment with chlorhexidine (Nde et al., 2009). Another study showed that increased expression of SmvA, an efflux pump of the major facilitator superfamily (MFS), is associated with increased resistance to chlorhexidine in K. pneumoniae (Wand et al., 2017). However, in our study, exposure to chlorhexidine did not increase the relative expression of SmvA or Mexcd-Oprj efflux systems. Although we did not find upregulation of SmvA or Mexcd-Opri, we found that multidrug resistance protein MexA was significantly upregulated. MexA functions as the periplasmic linker component of the MexAB-OprM efflux system that confers multidrug resistance, possibly indicating that this system plays a role in chlorhexidine response. Conversely, other research has shown that the overexpression of efflux pumps does not impact the antimicrobial efficacy of chlorhexidine, and it has been proposed that this is largely due to the limited solubility of chlorhexidine in the bacterial membrane core (Gilbert and Moore, 2005).
General Cell Metabolism Following Exposure to Chlorhexidine
Proteomic platform analysis showed that many proteins associated with energy metabolism and many enzymes associated with glycolysis, fatty acid biosynthesis, and the TCA cycle were upregulated in the resistant strain. For example, upregulation of AceE and AceF, two enzymes involved in the conversion of pyruvate to acetyl-CoA and CO2, was observed. In addition, proteins associated with acetyl-CoA carboxylase such as AccD and AccB were also upregulated. These proteins are involved in the first step of the subpathway that synthesizes malonyl-CoA from acetyl-CoA. Furthermore, significant upregulation was observed in proteins FadA, FabV and AcpP1, which are all involved in lipid metabolism. FadA catalyzes the final step of fatty acid oxidation, in which acetyl-CoA is released. FabV catalyzes the reduction of a carbon-carbon double bond in an enoyl moiety that is covalently linked to Acp as an acyl carrier protein of the growing fatty acid chain. Consistent with our results, increased expression of several proteins associated with fatty acid synthesis, such as Acp, was also observed in biofilm of Delftia acidovorans following chlorhexidine exposure (Kohanski et al., 2008). Finally, a substantial number of proteins associated with the TCA cycle were upregulated. Succinate-CoA ligase enzymes (SucC and SucD), fumarate hydratase (FumC1), malate:quinone oxidoreductase 2 (MqO2) and citrate synthase (GltA) were the upregulated proteins related to the first step of the cycle. Aconitate hydratase enzymes (AcnA and AcnB) were the upregulated proteins involved in isocitrate synthesis.
The acquisition of antimicrobial resistance may be energetically costly for bacteria, and in a normal environment, high-energy expenditure lowers the general fitness of bacteria. Therefore, bacteria will usually only take on metabolic costs if environmental pressures, such as antibiotics, dictate its necessity (Martínez and Rojo, 2011). The upregulation of metabolic proteins by P. aeruginosa shown in this study could be a result of the intrinsic linkage between bacterial metabolism and antibiotic resistance.
Altered Flagella Protein Regulation in Chlorhexidine Resistant Strain
Of interest in our proteomics data is the upregulation of Pil proteins (PilA, PilG, PilH, PilJ, PilQ) involved in fimbriae formation and motility. Formation of pili, or fimbriae, has been correlated with virulence in pathogens such as P. aeruginosa, E. coli, A. baummanii, V. cholerae, and Streptococcus spp. Presence of pili enables bacteria to bind to a surface or tissue, which initiates interaction with the host and increases biofilm formation (Tomaras et al., 2003; Fernández-Reyes et al., 2009; Martínez and Rojo, 2011). It has been previously reported that overexpression of flagellar genes is associated with increased tolerance to antimicrobial agents, including chlorhexidine, in Salmonella, which may now be extended to chlorhexidine tolerance in Pseudomonas (O’toole and Kolter, 1998; Kim and Surette, 2003; Condell et al., 2014). Twitching motility is a bacterial movement on a solid surface that occurs in a wide variety of pathogens including P. aeruginosa. Twitching motility is mediated by a complex chemosensory pathway including PilG, PilH, PilI, PilJ, and PilK and is also involved in biofilm formation (Whitchurch et al., 2004; Butler et al., 2010). The upregulated proteins identified in our study, including PilG, PilH and PilJ, suggest that exposure to chlorhexidine increased the twitching motility which contributes to virulence and biofilm formation. It is known that type IV pili in P. aeruginosa are assembled from PilA, a single protein subunit, which is transferred out of the cell via outer membrane secretin PilQ to form fimbrial strands. The upregulation of PilA and PilQ suggests that the biosynthesis of pili, which is associated with adhesion and accumulation of P. aeruginosa on surfaces, increased in chlorhexidine-exposed bacteria (Laverty et al., 2014). Overall, the observed alterations in Pil proteins in our comparison of the proteomes of chlorhexidine-susceptible and resistant bacteria are associated with biofilm formation and virulence, suggesting that further investigation of biofilm formation in chlorhexidine-resistant bacteria would be a meaningful line of research.
Conclusion
We have demonstrated that prolonged exposure of Gram-negative bacteria to chlorhexidine results in significantly reduced susceptibility to colistin but not to AMPs and ceragenins. This suggests that there may be a specific mechanism for resistance of Gram-negative bacteria to chlorhexidine that causes even higher resistance to colistin but does not affect susceptibility to AMPs and ceragenins. These observations are consistent with the reported decrease in susceptibility of chlorhexidine-resistant K. pneumoniae to colistin (Wand et al., 2017). The fact that lead ceragenins (CSA-44 and CSA-131) retain bactericidal activity against highly colistin and chlorhexidine resistant bacteria is further support for the development of these compounds as broad-spectrum antibacterial agents to potentially be used in multiple clinical applications.
To understand the changes to Gram-negative bacteria that lead to chlorhexidine and colistin resistance but do not impact AMPs and ceragenins, an in-depth quantitative proteomic study was performed with a parent strain of P. aeruginosa and its chlorhexidine-resistant progeny. It is likely that proteins involved in resistance to chlorhexidine function similarly in other Gram-negative bacteria. Our study suggests that increased expression of membrane proteins such as OprF, LptD, and the Tol-Pal system is responsible for chlorhexidine resistance, but this is not the only pathway contributing to resistance. For example, chlorhexidine resistance is also associated with upregulation of PagL, which is affected by the two-component regulatory system phoP/phoQ. Additionally, upregulation of flagella proteins, chaperones and proteins associated with energy metabolism were also observed in the chlorhexidine-resistance pathway. Upregulation of some of these proteins also correlates with colistin resistance. Co-resistance with colistin suggests that LPS modification along with changes in expression of associated membrane proteins could be involved in the primary mechanism of chlorhexidine resistance. The fact that exposure of Gram-negative bacteria to chlorhexidine may lead to colistin resistance and possible drug resistance among Gram-positive organisms suggests that wide-spread use of chlorhexidine should be reconsidered. In order to decrease the speculated effect of chlorhexidine on the development of colistin-resistant strains, application of other antimicrobials such as ceragenins could be a safe choice considering its lower risk of cross-resistance.
Author Contributions
MH and PS designed the experiments and wrote the manuscript. BSH, JC, MT, SW, BH, AZ, CM, RC, and SD performed the experiments. MH, JA, and JP performed data analysis.
Funding
This work was carried out with funding from Brigham Young University, N8 Medical, and CSA Biotech. Each provided funding for research supplies and salaries for researchers.
Conflict of Interest Statement
PS is a paid consultant for N8 Medical and CSA Biotech.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge National Institutes of Health funding to JA (R15CA202619) and Fritz B. Burns Foundation to JP.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00210/full#supplementary-material
TABLE S1 | Proteomic data analysis: peptides and proteins.
References
Balibar, C. J., and Grabowicz, M. (2015). Mutant alleles of lptD increase the permeability of Pseudomonas aeruginosa and define determinants of intrinsic resistance to antibiotics. Antimicrob. Agents Chemother. 2, 845–854. doi: 10.1128/AAC.01747-15
Bhardwaj, P., Hans, A., Ruikar, K., Guan, Z., and Palmer, K. L. (2018). Reduced chlorhexidine and daptomycin susceptibility in vancomycin-resistant Enterococcus faecium after serial chlorhexidine exposure. Antimicrob. Agents Chemother. 62:e01235-17. doi: 10.1128/AAC.01235-17
Bock, L., Wand, M., and Sutton, J. (2016). Varying activity of chlorhexidine-based disinfectants against Klebsiella pneumoniae clinical isolates and adapted strains. J. Hosp. Infect. 93, 42–48. doi: 10.1016/j.jhin.2015.12.019
Butler, M. T., Wang, Q., and Harshey, R. M. (2010). Cell density and mobility protect swarming bacteria against antibiotics. Proc. Natl. Acad. Sci. U.S.A. 107, 3776–3781. doi: 10.1073/pnas.0910934107
Clinical and Laboratory Standards Institute (2006). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically: Approved Standard, 7th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Condell, O., Power, K. A., Händler, K., Finn, S., Sheridan, A., Sergeant, K., et al. (2014). Comparative analysis of Salmonella susceptibility and tolerance to the biocide chlorhexidine identifies a complex cellular defense network. Front. Microbiol. 5:373. doi: 10.3389/fmicb.2014.00373
Curiao, T., Marchi, E., Grandgirard, D., León-Sampedro, R., and Viti, C. (2016). Multiple adaptive routes of Salmonella enterica Typhimurium to biocide and antibiotic exposure. BMC Genomics 17:491. doi: 10.1186/s12864-016-2778-z
Davies, G. E., Francis, J., Martin, A. R., Rose, F. L., and Swain, G. (1954). 1:6-di-4′-chlorophenyldiguanidohexane (“hibitane”). Laboratory investigation of a new antibacterial agent of high potency. Br. J. Pharmacol. Chemother. 9, 192–196.
De Majumdar, S., Yu, J., Fookes, M., McAteer, S. P., Llobet, E., Finn, S., et al. (2015). Elucidation of the RamA regulon in Klebsiella pneumoniae reveals a role in LPS regulation. PLoS Pathog. 11:e1004627. doi: 10.1371/journal.ppat.1004627
Dennis, J. J., Lafontaine, E. R., and Sokol, P. A. (1996). Identification and characterization of the tolQRA genes of Pseudomonas aeruginosa. J. Bacteriol. 178, 7059–7068.
Denny, J., and Munro, C. L. (2017). Chlorhexidine bathing effects on health-care-associated infections. Biol. Res. Nurs. 2, 123–136. doi: 10.1177/1099800416654013
Derouiche, R., Benedetti, H., Lazzaroni, J.-C., Lazdunski, C., and Lloubes, R. (1995). Protein complex within Escherichia coli inner membrane. TolA N-terminal domain interacts with TolQ and TolR proteins. J. Biol. Chem. 270, 11078–11084.
Fernández-Reyes, M., Rodríguez-Falcón, M., Chiva, C., Pachón, J., Andreu, D., and Rivas, L. (2009). The cost of resistance to colistin in Acinetobacter baumannii: a proteomic perspective. Proteomics 9, 1632–1645. doi: 10.1002/pmic.200800434
Fito-Boncompte, L., Chapalain, A., Bouffartigues, E., Chaker, H., Lesouhaitier, O., Gicquel, G., et al. (2011). Full virulence of Pseudomonas aeruginosa requires OprF. Infect. Immun. 79, 1176–1186. doi: 10.1128/IAI.00850-10
Formosa, C., Herold, M., Vidaillac, C., Duval, R., and Dague, E. (2015). Unravelling of a mechanism of resistance to colistin in Klebsiella pneumoniae using atomic force microscopy. J. Antimicrob. Chemother. 70, 2261–2270. doi: 10.1093/jac/dkv118
Gaspar, J. A., Thomas, J. A., Marolda, C. L., and Valvano, M. A. (2000). Surface expression of O-specific lipopolysaccharide in Escherichia coli requires the function of the TolA protein. Mol. Microbiol. 38, 262–275.
Gilbert, P., and Moore, L. (2005). Cationic antiseptics: diversity of action under a common epithet. J. Appl. Microbiol. 99, 703–715.
Hammerl, J. A., Grobbel, M., Tenhagen, B.-A., and Kaesbohrer, A. (2017). Impact of mcr-1 harbouring bacteria in clinical settings and the public health sector: how can we act against this novel threat? J. Public Health Emerg. 1:51. doi: 10.21037/jphe.2017.05.08
Hancock, R. E., and Brinkman, F. S. (2002). Function of Pseudomonas porins in uptake and efflux. Annu. Rev. Microbiol. 56, 17–38.
Hashemi, M. M., Holden, B. S., Durnaś, B., Bucki, R., and Savage, P. B. (2017a). Ceragenins as mimics of endogenous antimicrobial peptides. J. Antimicrob. Agents 3:1000141. doi: 10.4172/2472-1212.1000141
Hashemi, M. M., Rovig, J., Bateman, J., Holden, B. S., Modelzelewski, T., Gueorguieva, I., et al. (2017b). Preclinical testing of a broad-spectrum antimicrobial endotracheal tube coated with an innate immune synthetic mimic. J. Antimicrob. Chemother. 73, 143–150. doi: 10.1093/jac/dkx347
Hashemi, M. M., Rovig, J., Weber, S., Hilton, B., Forouzan, M. M., and Savage, P. B. (2017c). Susceptibility of colistin-resistant, Gram-negative bacteria to antimicrobial peptides and ceragenins. Antimicrob. Agents Chemother 61:e00292-17. doi: 10.1128/AAC.00292-17
Hashemi, M. M., Holden, B. S., and Savage, P. B. (2018a). “Ceragenins as non-peptide mimics of endogenous antimicrobial peptides,” in Fighting Antimicrobial Resistance, ed. A. Budimir (Zagreb: IAPC Publishing), 139–169.
Hashemi, M. M., Holden, B. S., Taylor, M. F., Wilson, J., Coburn, J., Hilton, B., et al. (2018b). Antibacterial and antifungal activities of poloxamer micelles containing ceragenin CSA-131 on ciliated tissues. Molecules 23, 596. doi: 10.3390/molecules23030596
Hashemi, M. M., Mmuoegbulam, A., Holden, B., Coburn, J., Wilson, J., Taylor, M., et al. (2018c). Susceptibility of multidrug-resistant bacteria. isolated from water and plants in Nigeria, to Ceragenins. Int. J. Environ. Res. Public Health 15:2758. doi: 10.3390/ijerph15122758
Hashemi, M. M., Rovig, J., Holden, B. S., Taylor, M. F., Weber, S., Wilson, J., et al. (2018d). Ceragenins are active against drug-resistant Candida auris clinical isolates in planktonic and biofilm forms. J. Antimicrob. Chemother. 73, 1537–1545. doi: 10.1093/jac/dky085
Heilpern, A. J., and Waldor, M. K. (2000). CTXφ infection of Vibrio cholerae requires the tolQRA gene products. J. Bacteriol. 182, 1739–1747.
Kampf, G. (2016). Acquired resistance to chlorhexidine – is it time to establish an ‘antiseptic stewardship’ initiative? J. Hosp. Infect. 94, 213–227. doi: 10.1016/j.jhin.2016.08.018
Karatzas, K. A., Randall, L. P., Webber, M., Piddock, L. J., Humphrey, T. J., Woodward, M. J., et al. (2008). Phenotypic and proteomic characterization of multiply antibiotic-resistant variants of Salmonella enterica serovar Typhimurium selected following exposure to disinfectants. Appl. Environ. Microbiol. 74, 1508–1516. doi: 10.1128/AEM.01931-37
Kim, S. J., and Ko, K. S. (2018). Diverse genetic alterations responsible for post-exposure colistin resistance in populations of the same strain of Klebsiella pneumoniae. Int. J. Antimicrob. Agents 52, 425–429. doi: 10.1016/j.ijantimicag.2018.06.010
Kim, W., and Surette, M. G. (2003). Swarming populations of Salmonella represent a unique physiological state coupled to multiple mechanisms of antibiotic resistance. Biol. Proced. Online 5, 189–196.
Kohanski, M. A., Dwyer, D. J., Wierzbowski, J., Cottarel, G., and Collins, J. J. (2008). Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell 135, 679–690. doi: 10.1016/j.cell.2008.09.038
Laverty, G., Gorman, S. P., and Gilmore, B. F. (2014). Biomolecular mechanisms of Pseudomonas aeruginosa and Escherichia coli biofilm formation. Pathogens 3, 596–632. doi: 10.3390/pathogens3030596
Li, C., Peters, A. S., Meredith, E. L., Allman, G. W., and Savage, P. B. (1998). Design and synthesis of potent sensitizers of Gram-negative bacteria based on a cholic acid scaffolding. J. Am. Chem. Soc. 120, 2961–2962. doi: 10.1021/ja973881r
Li, X., Gu, G.-Q., Chen, W., Gao, L.-J., Wu, X.-H., and Zhang, L. Q. (2018). The outer membrane protein OprF and the sigma factor SigX regulate antibiotic production in Pseudomonas fluorescens 2P24. Microbiol. Res. 206, 159–167. doi: 10.1016/j.micres.2017.10.006
Lim, K., and Kam, P. (2008). Chlorhexidine-pharmacology and clinical applications. Anaesth. Intensive Care 36, 502–512.
Limoli, D. H., Jones, C. J., and Wozniak, D. J. (2015). Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol. Spectr. 3, 1–19. doi: 10.1128/microbiolspec.MB-0011-2014
Liu, Y., Dong, Y., Chen, Y.-Y. M., and Burne, R. A. (2008). Environmental and growth phase regulation of the Streptococcus gordonii arginine deiminase genes. Appl. Environ. Microbiol. 74, 5023–5030. doi: 10.1128/AEM.00556-08
Lourdault, K., Cerqueira, G. M., Wunder, E. A., and Picardeau, M. (2011). Inactivation of clpB in the pathogen Leptospira interrogans reduces virulence and resistance to stress conditions. Infect. Immun. 79, 3711–3717. doi: 10.1128/IAI.05168-11
Maccarini, M., Gayet, L., Alcaraz, J.-P., Liguori, L., Stidder, B., Watkins, E. B., et al. (2017). Functional characterization of cell-free expressed OprF Porin from Pseudomonas aeruginosa stably incorporated in tethered lipid Bilayers. Langmuir 33, 9988–9996. doi: 10.1021/acs.langmuir.7b01731
Martínez, J. L., and Rojo, F. (2011). Metabolic regulation of antibiotic resistance. FEMS Microbiol. Rev. 35, 768–789. doi: 10.1111/j.1574-6976.2011.00282.x
Mukherjee, S., Moustafa, D., Smith, C. D., Goldberg, J. B., and Bassler, B. L. (2017). The RhlR quorum-sensing receptor controls Pseudomonas aeruginosa pathogenesis and biofilm development independently of its canonical homoserine lactone autoinducer. PLoS Pathog. 13:e1006504. doi: 10.1371/journal.ppat.1006504
Nde, C. W., Jang, H. J., Toghrol, F., and Bentley, W. E. (2009). Global transcriptomic response of Pseudomonas aeruginosa to chlorhexidine diacetate. Environ. Sci. Technol. 43, 8406–8415. doi: 10.1021/es9015475
Nikolaidis, I., Favini-Stabile, S., and Dessen, A. (2014). Resistance to antibiotics targeted to the bacterial cell wall. Protein Sci. 23, 243–259. doi: 10.1002/pro.2414
Olaitan, A. O., Morand, S., and Rolain, J.-M. (2014). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643. doi: 10.3389/fmicb.2014.00643
O’toole, G. A., and Kolter, R. (1998). Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30, 295–304.
Park, S. Y., and Groisman, E. A. (2014). Signal-specific temporal response by the Salmonella PhoP/PhoQ regulatory system. Mol. Microbiol. 91, 135–144. doi: 10.1111/mmi.12449
Peng, J., Cao, J., Ng, F. M., and Hill, J. (2017). Pseudomonas aeruginosa develops Ciprofloxacin resistance from low to high level with distinctive proteome changes. J. Proteomics 152, 75–87. doi: 10.1016/j.jprot.2016.10.005
Rema, T., Medihala, P., Lawrence, J. R., Vidovic, S., Leppard, G. G., Reid, M., et al. (2016). ). Proteomic analyses of chlorhexidine tolerance mechanisms in delftia acidovorans biofilms. mSphere 1:e0017-15. doi: 10.1128/mSphere.00017-15
Smith, K., and Hunter, I. S. (2008). Efficacy of common hospital biocides with biofilms of multi-drug resistant clinical isolates. J. Med. Microbiol. 57, 966–973. doi: 10.1099/jmm.0.47668-0
Southey-Pillig, C. J., Davies, D. G., and Sauer, K. (2005). Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J. Bacteriol. 187, 8114–8126. doi: 10.1128/JB.187.23.8114-26.2005
Tao, K., Narita, S.-I., and Tokuda, H. (2012). Defective lipoprotein sorting induces lolA expression through the Rcs stress response phosphorelay system. J. Bacteriol. 194, 3643–3650. doi: 10.1128/JB.00553-12
Tattawasart, U., Maillard, J.-Y., Furr, J., and Russell, A. (2000). Outer membrane changes in Pseudomonas stutzeri resistant to Chlorhexidine diacetate and Cetylpyridinium chloride. Int. J. Antimicrob. Agents 16, 233–238.
Tomaras, A. P., Dorsey, C. W., Edelmann, R. E., and Actis, L. A. (2003). Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149, 3473–3484.
Walsh, F., and Duffy, B. (2013). The culturable soil antibiotic resistome: a community of multi-drug resistant bacteria. PLoS One 8:e65567.
Wand, M. E., Bock, L. J., Bonney, L. C., and Sutton, J. M. (2017). Mechanisms of increased resistance to chlorhexidine and cross-resistance to colistin following exposure of Klebsiella pneumoniae clinical isolates to chlorhexidine. Antimicrob. Agents Chemother. 61:e1162-16. doi: 10.1128/AAC.01162-16
Whitchurch, C. B., Leech, A. J., Young, M. D., Kennedy, D., Sargent, J. L., Bertrand, J. J., et al. (2004). Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 52, 873–893.
Wiśniewski, J. R., Zougman, A., Nagaraj, N., and Mann, M. (2009). Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362. doi: 10.1038/nmeth.1322
Wright, M. S., Suzuki, Y., Jones, M. B., Marshall, S. H., Rudin, S. D., van Duin, D., et al. (2015). Genomic and transcriptomic analyses of colistin-resistant clinical isolates of Klebsiella pneumoniae reveal multiple pathways of resistance. Antimicrob. Agents Chemother. 59, 536–543. doi: 10.1128/AAC.04037-14
Keywords: chlorhexidine, ceragenins, colistine, proteomic, cross-resistance, Pseudomonas aeruginosa, Gram-negative bacteria
Citation: Hashemi MM, Holden BS, Coburn J, Taylor MF, Weber S, Hilton B, Zaugg AL, McEwan C, Carson R, Andersen JL, Price JC, Deng S and Savage PB (2019) Proteomic Analysis of Resistance of Gram-Negative Bacteria to Chlorhexidine and Impacts on Susceptibility to Colistin, Antimicrobial Peptides, and Ceragenins. Front. Microbiol. 10:210. doi: 10.3389/fmicb.2019.00210
Received: 16 October 2018; Accepted: 24 January 2019;
Published: 18 February 2019.
Edited by:
Jose L. Martinez, Centro Nacional de Biotecnología (CSIC), SpainReviewed by:
Karl Hassan, University of Newcastle, AustraliaFernando Baquero, Ramón y Cajal Institute for Health Research, Spain
Copyright © 2019 Hashemi, Holden, Coburn, Taylor, Weber, Hilton, Zaugg, McEwan, Carson, Andersen, Price, Deng and Savage. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paul B. Savage, cGF1bF9zYXZhZ2VAYnl1LmVkdQ==; cGJzQGJ5dS5lZHU=
 Marjan M. Hashemi
Marjan M. Hashemi Brett S. Holden
Brett S. Holden Jordan Coburn
Jordan Coburn Scott Weber
Scott Weber Paul B. Savage
Paul B. Savage