- 1Key Laboratory of Urban Environment and Health, Institute of Urban Environment, Chinese Academy of Sciences, Xiamen, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3State Key Laboratory of Urban and Regional Ecology, Research Center for Eco-Environmental Sciences, Chinese Academy of Sciences, Beijing, China
Plasmid conjugation is one of the dominant mechanisms of horizontal gene transfer, playing a noticeable role in the rapid spread of antibiotic resistance genes (ARGs). Broad host range plasmids are known to transfer to diverse bacteria in extracted soil bacterial communities when evaluated by filter mating incubation. However, the persistence and dissemination of broad range plasmid in natural soil has not been well studied. In this study, Pseudomonas putida with a conjugative antibiotic resistance plasmid RP4 was inoculated into a soil microcosm, the fate and persistence of P. putida and RP4 were monitored by quantitative PCR. The concentrations of P. putida and RP4 both rapidly decreased within 15-day incubation. P. putida then decayed at a significantly lower rate during subsequent incubation, however, no further decay of RP4 was observed, resulting in an elevated RP4/P. putida ratio (up to 10) after 75-day incubation, which implied potential transfer of RP4 to soil microbiota. We further sorted RP4 recipient bacteria from the soil microcosms by fluorescence-activated cell sorting. Spread of RP4 increased during 75-day microcosm operation and was estimated at around 10-4 transconjugants per recipient at the end of incubation. Analysis of 16S rRNA gene sequences of transconjugants showed that host bacteria of RP4 were affiliated to more than 15 phyla, with increased diversity and shift in the composition of host bacteria. Proteobacteria was the most dominant phylum in the transconjugant pools. Transient transfer of RP4 to some host bacteria was observed. These results emphasize the prolonged persistence of P. putida and RP4 in natural soil microcosms, and highlight the potential risks of increased spread potential of plasmid and broader range of host bacteria in disseminating ARGs in soil.
Introduction
The global increased antimicrobial resistance level in human pathogens has posed significant threat to human health (UNEP, 2017). Other than clinical settings, antibiotic resistance genes (ARGs) have been frequently detected with elevated diversity and abundance in soil (Zhu et al., 2013). Bacteria could become antibiotic resistant via gene mutation, or horizontal gene transfer (HGT) that would facilitate exchange of genetic information and contribute significantly to the evolution of bacteria (Boto, 2010; Soucy et al., 2015). HGT-mediated evolution would confer selective advantages to the host when facing environmental stress and thus promote the adaptation of bacteria under selective pressures (Daubin and Szollosi, 2016).
Plasmid conjugation is one of the most important mechanisms of HGT (Sorensen et al., 2005; Thomas and Nielsen, 2005), contributing to the wide spread of ARGs among environmental bacteria and human pathogens. Conjugative plasmids are often implicated in the rapid propagation and proliferation of ARGs (Dodd, 2012). These plasmids contain genes for the formation of pilus to transfer copies of themselves to other bacteria, enabling horizontal transfer of complete set of genes on the plasmid, including ARGs (Willetts and Wilkins, 1984; Willey et al., 2008). Furthermore, the transfer of diverse non-conjugative resistance plasmids can be facilitated by origin-of-transfer sequences of mobilizable plasmid (O’Brien et al., 2015). The massive use of antibiotics has resulted in high concentration of antibiotic residues in the environments, which may serve as a selection pressure that promote the dissemination of ARGs among environmental bacteria, including pathogenic species (Pruden et al., 2012; Guo et al., 2018). In addition, conjugative plasmids selected with antibiotics have been found to persist among bacterial populations or invade new strains even without antibiotics pressure (Dionisio et al., 2005).
The dissemination of ARGs caused by conjugation highly depends on the conjugation frequency and host range of plasmids. Broad host range plasmids are of high interest due to their transfer potency between phylogenetically distant hosts. Plasmid conjugation has been extensively studied in mating systems using typical pure strains as donors and recipients. Transconjugants can be quantified by colony counting on selective plates and the estimated transfer frequencies differs due to plasmids type, donor and recipient species, density ratio, mating time, and medium (Henschke and Schmidt, 1990; Modrie et al., 2010). However, culture-based screening of transconjugants can only partially determine the transfer frequency and identities of transconjugants in complex bacterial communities due to the limitation in culturing environmental bacteria (Elsas et al., 1989, 1990). Using fluorescent reporter genes to track plasmids made it possible to investigate the spread of resistance plasmids among environmental bacteria. To study the transfer of RP4 in activated sludge, the plasmid was marked with reporter gene gfp and detected by fluorescence microscopy, transfer frequencies were estimated in the range of 4 × 10-6 to 10-5 per recipient over a range of donor to recipient ratios (Geisenberger et al., 1999). Transconjugants obtained plasmids that marked with fluorescent genes can be sorted by fluorescence activated cell sorting (FACS) and identified by 16S rRNA gene sequencing (Musovic et al., 2006; Klumper et al., 2015).
Although these studies have expanded our understanding on plasmid conjugation, these mating experiments were generally conducted in relatively shorter mating period (4–72 h) (Sørensen et al., 2003; Wang et al., 2015b; Lin et al., 2016; Li et al., 2018) and optimized lab conditions that enabling sufficient cell-to-cell contact between donors and recipients (Musovic et al., 2010; Klumper et al., 2015, 2016; Li et al., 2018). Plasmid conjugation in soil could be affected by many factors including soil bacterial community composition, nutrients competition, and selective forces such as antibiotic and metal (Trevors et al., 1989; Elsas et al., 1990; Fox et al., 2008; Musovic et al., 2014; Klumper et al., 2016). However, current knowledge about plasmid conjugation in soil with complex bacterial communities is limited. The concentrations of donor bacteria and plasmid may significantly change after inoculation into soil, and the cell-to-cell contact between donors and recipients in soil is expected to decrease compared with filter mating systems, which could then affect plasmid conjugation in soil. In addition, retransfer of plasmid from transconjugants to the other bacterial populations may occur under extended incubation time.
To address these questions, soil microcosms were set up to investigate plasmid conjugation in soil. The aim of the study is to estimate the spread potential (SP) of plasmid and identify the phylogenetic affiliation of transconjugants in close to natural condition. Pseudomonas putida containing a broad host range plasmid tagged with green fluorescent protein (gfp) gene was used as donors and was inoculated into the soil microcosms. We tracked the abundance of both donor strains and plasmids in soil at intervals by qPCR. Transconjugants were collected with FACS and the 16S rRNA gene of transconjugants were amplified and sequenced.
Materials and Methods
Donor Strain and Plasmid
Pseudomonas putida KT2442 was used as donor, which was chromosomally tagged with dsRed and lacIq and harbored a broad host range IncP conjugative plasmid RP4 (Musovic et al., 2010). Plasmid RP4, containing resistance genes against ampicillin (Amp), kanamycin (Km), and tetracycline (Tc), was marked with gfp and a lacIq repressible promoter upstream of gfp gene, gfp expression was inhibited in donor strains, but when the plasmid transfer to a recipient, gfp expression is possible (both the donor strain and plasmid were kindly provided by Professor Barth F. Smets, Technical University of Denmark). The donor strains were grown aerobically under 180 rpm at 37°C overnight in LB medium supplemented with 100 μg ml-1 Amp, 50 μg ml-1 Km and 20 μg ml-1 Tc. The bacterial cells were harvested by centrifugation at 8500 g for 15 min and the cell pellets were washed twice and re-suspended in 0.9% sterile saline solution, the concentration of resulting donor inoculants was measured by colony-forming unit (CFU) counting.
Soil Microcosms Set Up
Surface soil (sandy loam) was collected from a vegetable field in Xiamen, China, and was passed through a 2 mm sieve. Before inoculation, the original soil microbiota was extracted by Nycodenz density gradient separation (Burmolle et al., 2003; Eichorst et al., 2015) and the concentration of cells was 2.08 × 108 cells g-1 dry soil which was determined by flow cytometry (FlowSight Imaging Flow Cytometer, Amnis Millipore, United States) after SYTO 9 staining. Soil microcosms were prepared in four replicate plastic pots by thoroughly mixing donor bacteria suspension into 500 g soil for each pot, providing a final donor strain density of 1.57 × 108 CFU g-1 dry soil. The microcosms were incubated for 75 days in a greenhouse at room temperature (28°C in the night and 35°C in the daytime on average) and were regularly irrigated twice daily to keep the water content at 60% field water capacity. Control soil without donor inoculation were prepared and incubated at the same condition. Soil samples were collected from each pot on day 0, 1, 2, 3, 4, 5, 10, 15, 20, 30, 60, and 75 and were partitioned into two sub-samples: one stored in 4°C for Nycodenz extraction of bacterial cells and one stored in -20°C for total DNA extraction.
DNA Extraction and qPCR
Soil DNA was extracted from 0.5 g of soil using FastDNA Spin kit for soil (MP Biomedical, Santa Ana, CA, United States). The concentrations of donor strain and plasmid were measured by quantification of dsRed and gfp genes, respectively. All quantifications were performed in triplicate using Roche 480 (Roche Molecular Systems Inc., Branchburg, NJ, United States). Real-Time PCR assays were performed in a total volume of 20 μl with 10 μl 2 × SYBR Premix ExTaq II (TaKaRa, Japan), 10 ng bovine serum albumin, 0.8 μl each primer (10 μM), 1 μl template DNA and 6.4 μl of nuclease-free PCR-grade water. The concentration of donor P. putida KT2442 in each sample was determined by quantifying a 702-bp region of dsRed gene using the primers dsRed-F (5′-ATATAGCATGCGGTCTTCCAAGAATGTTATCAA-3′) and dsRed-R (5′-CTCTCAAGCTTCCCGGGTTAAAGGAACAGATGGTGGCG-3′) (Tolker-Nielsen et al., 2000). Quantitative amplifications were performed with the following thermal cycles: 95°C for 5 min, followed by 40 cycles of 95°C for 50s, 60°C for 50 s, 72°C for 50 s, and a final extension at 72°C for 10 min. The concentration of plasmid RP4 was measured by quantifying a 89-bp region of gfp gene using the primers gfp-F (5′-GAAGATGGAAGCGTTCAA-3′) and gfp-R (5′-AGGTAATGGTTGTCTGGTA-3′) (Hale et al., 2015) with a thermal cycles as follows: 95°C for 5 min, followed by 40 cycles of 95°C for 40 s, 58°C for 30 s, 72°C for 30 s, and a final extension at 72°C for 10 min. Copy numbers of 16S rRNA gene were quantified by qPCR as described previously (Zhu et al., 2017). Details for preparation of standard curves of these genes were provided in Supplemental Method (Supplementary Figure S1). Control soil without inoculation was systematically incubated, sampled and was subjected to DNA extraction and gene quantification in parallel.
Means and standard deviations of qPCR results were calculated with Microsoft Office Excel 2016. Gene copy numbers were normalized by the weight of the dry soil. Concentrations of donor KT2442 and plasmid RP4 in soil microcosms were converted to lg(Ct/C0) (Tolker-Nielsen and Molin, 2000), where Ct corresponds the concentration at time t, C0 corresponds the concentration at time 0 (Al-Jassim et al., 2017). The phrase “spread potential” was selected to indicate the diffusion and proliferation of plasmid RP4 in the soil microbiota during incubation period. SP is considered as the ratio of transconjugants to recipients and was roughly estimated using the formula:
SPqPCR refers to the SP of RP4 estimated by qPCR, C is the concentration of gene determined by qPCR, presented as copies per gram dry soil. Cgfp/x-CdsRed presents the number of transconjugants per gram of dry soil with assumption that every donor strain maintains plasmid. IncP plasmids are typically 1–3 copies per cell, here x refers to the copies of RP4 per cell and 1 or 3 was used for calculation, thus the range of SPqPCR could be estimated. The average number of 16S rRNA-encoding genes per bacterial cell is currently estimated at 4.1 on the basis of the Ribosomal RNA Database (Klappenbach et al., 2001; Chen et al., 2017). C16SrRNA/4 presents the number of total bacteria per gram of dry soil.
Flow Cytometry Sorting of Transconjugants
Bacteria from collected soil samples were extracted by Nycodenz density gradient separation and were re-suspended in PBS. Soil (1.5 g) was homogenized in 5 ml 1× phosphate buffer saline (PBS) supplemented with 0.5% (v/v) Tween-20 in triplicates. The mixtures were shaken for 30 min by a vortex in room temperature then added to the top of Nycodenz (HistoDenz, Sigma, United States) solution at 1.31 g ml-1 (6 g HistoDenz was dissolved in 10 ml sterile ultra-pure water) carefully, followed by centrifugation at 8500 g for 15 min. The upper PBS and bacteria layers were carefully collected, fivefold diluted (v/v) in PBS and centrifuged at 8500 g for 20 min (Musovic et al., 2010). The upper solution was discarded, the resulting pellet was re-suspended and centrifuged again until the bacteria were concentrated to 1 ml suspension.
Bacterial cells including indigenous soil bacteria (no fluorescence), transconjugants (green fluorescence) and donor strains (red fluorescence) were scaled by FlowSight Imaging Flow Cytometer (Amnis Millipore, United States). Transconjugants were sorted using S3e Cell Sorter (Bio-Rad, United States). The flow cytometer was equipped by 488 and 561 nm two-laser systems. GFP fluorescent signal was excited by 488 nm laser and collected in bandpass Filter1 (525/30 nm). DsRed fluorescent was excited by 561 nm and checked in Filter2 (586/25 nm). Escherichia coli K12 was prepared as a fluorescence negative control. E. coli K12 harbored RP4::gfp was used as gfp-positive control. Donor strain P. putida KT2442 (RP4::gfp) was prepared as dsRed-positive and gfp-negative control. According to the fluorescent intensity of control bacteria, the events with high green fluorescence intensity and no red fluorescence intensity were gated, sorted and collected to a 5 ml BD Falcon collection tubes based on the signal of FSC, SSC, Filter1, and Filter 2. Then the enriched gfp-positive events were subjected to an additional sorting under the purity mode to ensure that all collected particles were gfp positive.
To test the accuracy of FACS, green fluorescence of sorted transconjugants were confirmed by confocal laser scanning microscopy immediately. We also conducted a filter mating experiment using several pure bacterial strains as recipients. Mixtures of Mangrovibacter yixingensis, Enterobacter tabaci, Enterobacillus tribolii, Pseudomonas aeruginosa PAO1, Burkholderia sp., Rhodococcus sp., Bacillus sp., and Staphylococcus sp. with equal density were added to KT242/RP4 suspension at 1:1 cell ratio and filtered on 0.22 μm membrane filters (Millipore, United States). The filters with mating mixtures were placed on R2A agar and incubated at 37°C for 10 h. The mating cells were suspended in PBS for further flow cytometric sorting. Each green fluorescence-positive cell was sorted with single mode and collected in 100 μl PCR tubes. The plasmid marker gene gfp was amplified after whole genome amplification from 16 single transconjugants. GFP genes were 100% detected in each single cell and no donor strain or non-mating bacteria were sorted when verified by 16S rRNA gene amplification and sequencing.
The SP of RP4 estimated by flow cytometric sorting (SPFCM) was roughly calculated using the formula:
The plasmid-carrying donor strain was inoculated in the soil microcosms, which were incubated for 75 days. Plasmids transfer to recipients at different time and green fluorescent proteins in those transconjugant cells express with various levels. We only sorted out the events with higher fluorescent intensity to avoid the negative cells without RP4 being sorted.
Amplification and Analysis of 16S rRNA Gene of Transconjugants
Sorted cells of each sample were concentrated in 100 μl PCR tubes and were subjected to three thermal cycles including 15 s in 100°C water and 45 s on ice. The V4–V5 region of 16S rRNA gene was amplified using primer 515F (5′-GTGCCAGCMGCCGCGG-3′) and 907R (5′-CCGTCAATCMTTTRAGTTT-3′) (Zhou et al., 2011). Each 2 × 50 μl PCR reactions contained 25 μl ExTaq (TaKaRa, Japan), 1 μl BSA of 20 μg ml-1, 1 μl each primer of 10 μM, 5 μl cell lysate as template and 17.5 μl nuclease-free PCR-grade water. The thermal program consisted of 95°C for 5 min, followed by 25 cycles of 95°C for 30 s, 58°C for 30 s, 72°C for 30 s, and a final extension at 72°C for 10 min. Non-template controls were included for detecting any contamination during PCR preparation (Chen et al., 2018). The amplicons were purified, quantified and sequenced on an Illumina Hiseq 2500 platform (Novogene, China) (Zhu et al., 2017). Raw pair-end reads were filtered to remove reads with low average quality scores and short reads (<300 nt). Sequences were processed with the Quantitative Insights Into Microbial Ecology (QIIME 1.9.1) pipeline (Caporaso et al., 2010; Edgar, 2010) as described previously (Zhu et al., 2017) for Operational Taxonomic Units (OTUs) clustering and taxonomic affiliation. Shannon index was generated to compare the level of bacterial diversity. Phylogenetic trees were constructed using MEGA 7 and iTOL1 (Letunic and Bork, 2016).
To determine the potential pathogen species in the transconjugant pools, all the high-quality 16S rRNA gene sequences from each sample were blasted against a bacterial pathogen database with an E-value < 1 × 10-10 and a sequence identity >99%. The bacterial pathogen database including 557 pathogenic species was constructed in our previous study (Chen et al., 2016).
Results
Fate of Donor Strain and Plasmid RP4
We quantified the marker genes gfp and dsRed to determine the concentrations of the plasmid RP4 and the donor strain KT2442, respectively. The fate of RP4 and KT2442 showed a similar decreased trend at the early stage of the microcosm operation, but differed at the late phase, resulting in a significantly increased RP4/KT2442 ratio during the incubation. The copy numbers of 16S rRNA gene in soil with KT2442/RP4 inoculation and control soil were at similar levels (3.3–4.9 × 109 copies g-1 dry soil), and they both remained relatively stable during the incubation.
Both the fate of KT2442 and RP4 showed biphasic behaviors during the 75-days incubation. KT2442 was inoculated into the soil at an initial concentration of about 108 CFU g-1 dry soil. They rapidly decreased 2.1 and 1.6 log to 106 copies g-1 dry soil within 15 days incubation, respectively (Figure 1). After 15-days of incubation, the fate of KT2442 and RP4 followed different trends. KT2442 decayed at a significantly lower rate during subsequent incubation and persisted at a concentration around 105 copies g-1 dry soil on the 60th and 75th day. However, no further decay of RP4 was observed, the concentration of RP4 persisted at a level of 106 copies g-1 dry soil since the 10th day. Consequently, RP4/ KT2442 ratio significantly increased to around 10, indicating the occurrence of conjugation of RP4 to soil microbiota.
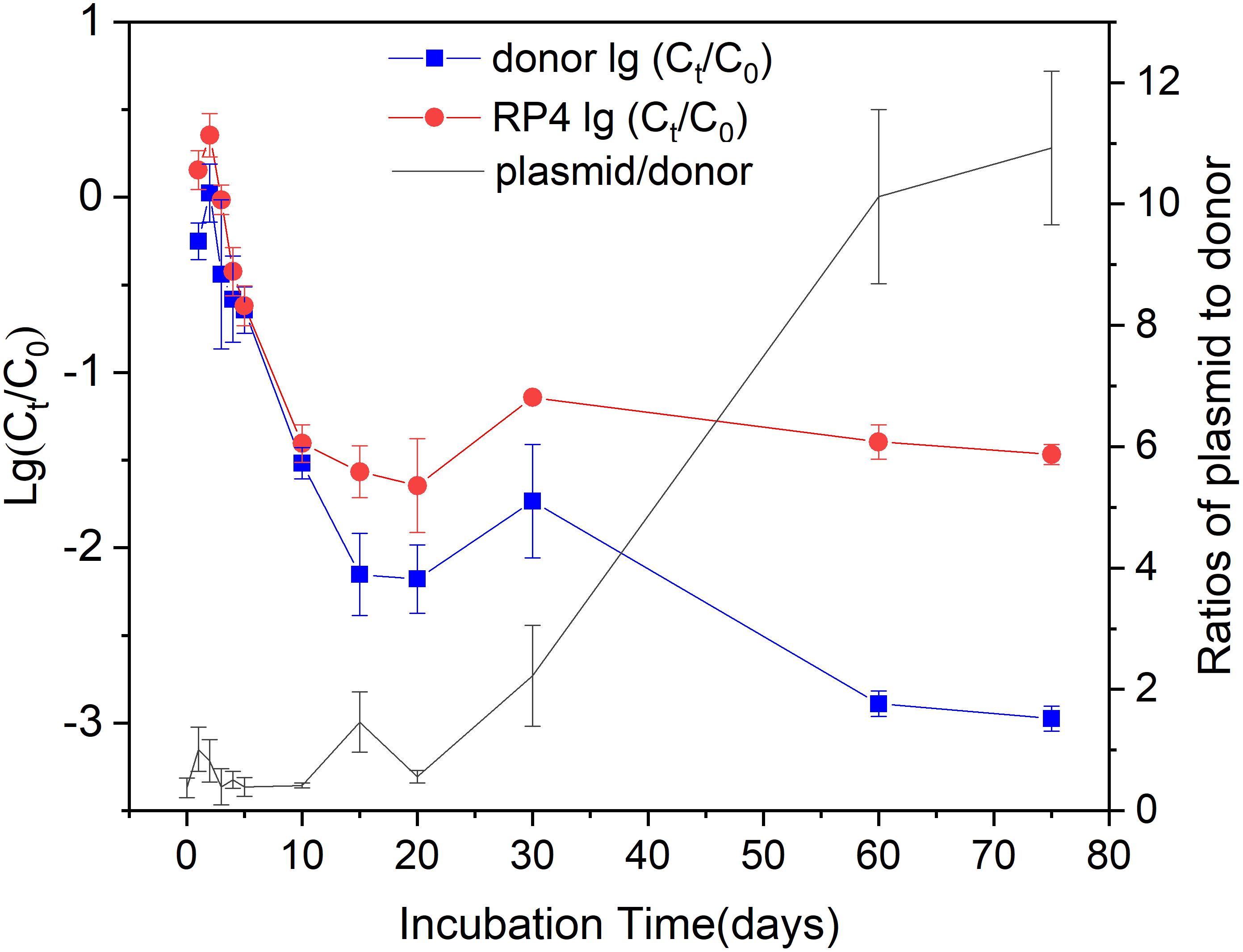
Figure 1. The decay of the donor and plasmid during incubation. Log values of the ratios of donor (plasmid) concentration at time t to the initial concentration were shown on the left Y-axis. The donor and plasmid concentrations were represented by the copy numbers of dsRed and gfp gene, respectively. The right Y-axis showed the ratios of plasmid RP4 to donor. Error bars represent standard error (SE) of sampling replicates on each time point (n = 3 or 4).
Spread Potential of Plasmid RP4
We estimated the SP based on qPCR and flow cytometric sorting data, respectively. Copy numbers of gfp were consistently lower than that of dsRed until the 60th day, so it was not possible to estimate the SP in early soil microcosm samples using data of qPCR. SPqPCR in samples of 60th day and 75th day was calculated using the formula described before, the results were 4.65 × 10-4–1.79 × 10-3 transconjugants per recipient (TC/R) and 3.63 × 10-4–1.42 × 10-3 TC/R, respectively.
Soil microbiota extracted from collected samples during incubation was analyzed and sorted by fluorescence activated cell sorter (FACS), including the early samples. The sorting results showed that RP4 had transferred to soil microbiota in the early stage of incubation. SPFCM of samples on the 5th day was 7.5 × 10-5 TC/R, while it increased to 2.5 × 10-4 TC/R on the 75th day. Only cells with strong green fluorescence signal were collected by flow cytometry, so the transconjugant numbers could be underestimated.
Phylogenetic Analysis of Transconjugant Pools
Transconjugants were sorted and subjected to taxonomic identification based on 16S rRNA gene sequences. The results indicated that transconjugants covered most of the major bacterial phyla, and the diversity of transconjugants differed at different time points. More than 300 transconjugant OTUs over 15 phyla were acquired from D5 samples. Proteobacteria was the most dominant phylum, contributing 85.7% of the transconjugant pools, followed by Bacteroidetes (3.17%), gram-positive Firmicutes (2.53%), and Actinobacteria (1.64%) (Figure 2A). The diversity of transconjugants significantly increased in D75 samples, in which three times more transconjugant OTUs were identified and affiliated to 20 phyla. Shannon index of D5 transconjugants was 2.38, while it was 4.41 of D75 transconjugant pools. Proteobacteria, Bacteroidetes, Firmicutes, and Actinobacteria were still the dominant phyla, contributing 95.6% of total transconjugants (Figure 2A). The relative abundance of Actinobacteria increased to 4.32% including 30 genera, while there were only 21 genera with lower abundance in D5 samples. The proportion of Gammaproteobacteria increased from 36.8 to 60.7% and Alphaproteobacteria decreased from 21.4 to 4.58%.
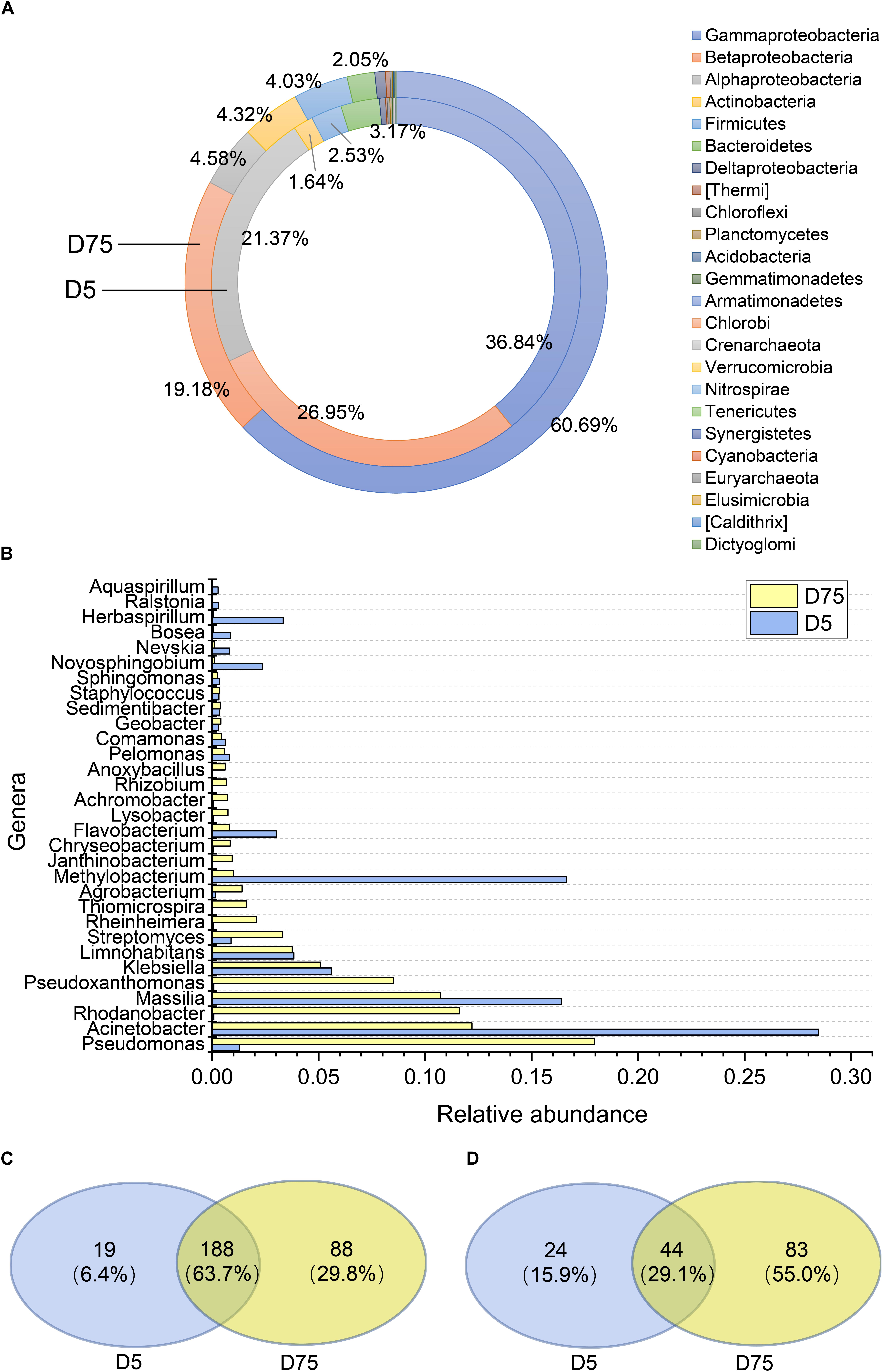
Figure 2. Composition of sorted transconjugant pools on day 5 and day 75. (A) Phylum distribution. (B) The relative abundance of the top 20 genera on day 5 or day 75. (C) Venn diagram showing the number of shared and unique genera between D5 and D75. (D) Venn diagram showing the number of shared and unique core OTUs that contributed 95% sequences among D5 and D75 samples.
Nine of the top 20 genera of D75 were shared with D5 (Figure 2B), they were Acinetobacter, Methylobacterium, Massilia, Klebsiella, Limnohabitans, Flavobacterium, Pseudomonas, Streptomyces, and Pelomonas, among which 7 genera were affiliated to Proteobacteria. The relative abundance of shared top genera changed over time. Acinetobacter and Massilia occupied high proportions in both transconjugant pools. Methylobacterium was the second dominant genus of D5 transconjugants, with a relative proportion of 16.6%, while it decreased to 1.01% in D75. On the contrary, Pseudomonas was the most dominant genus of D75, contributed to 18.0% of transconjugant pool when excluding out the donor strain species P. putida. While it was 1.28% in D5. Pseudomonas, Acinetobacter, and Staphylococcus were detected with relative high frequency. Opportunistic pathogenic species such as Acinetobacter baumannii, Aeromonas veronii, Enterobacter cloacae, Pseudomonas mendocina, and Staphylococcus aureus were identified in the further sequence analysis (Supplementary Table S1). Transconjugant pools between D5 and D75 shared 63.7% genera (Figure 2C), indicating that plasmid transferred into most genera of the recipients at the early invasion stage. However, a total of 88 unique genera were detected in D75 samples, indicating significantly increased diversity of transconjugants. In addition, only 19 unique genera were detected in D5 samples, implying that these recipients may lose RP4 in subsequent incubation. Analysis of core OTUs of transconjugants accounting for 95% of the sequences indicated 127 OTUs in D75 and 66 OTUs of D5 (Figure 2D), among which, only 44 OTUs were shared between D5 and D75 samples.
Phylogenetic trees were constructed using the sequences of OTUs with relative abundance more than 0.01% in the transconjugant pools. The phylogenetic distance between donor strain P. putida and the transconjugants was shown, as well as the relative abundance of OTUs (Figure 3). RP4 transferred from P. putida to a broad range of recipients including phylogenetic distant group. However, the advantage of Proteobacteria was outstanding, with more than half of branches were identified to be Proteobacteria. Remarkably, much more branches identified to be Pseudomonas appeared in tree of D75, indicating the advantage of intra-genus transfer.
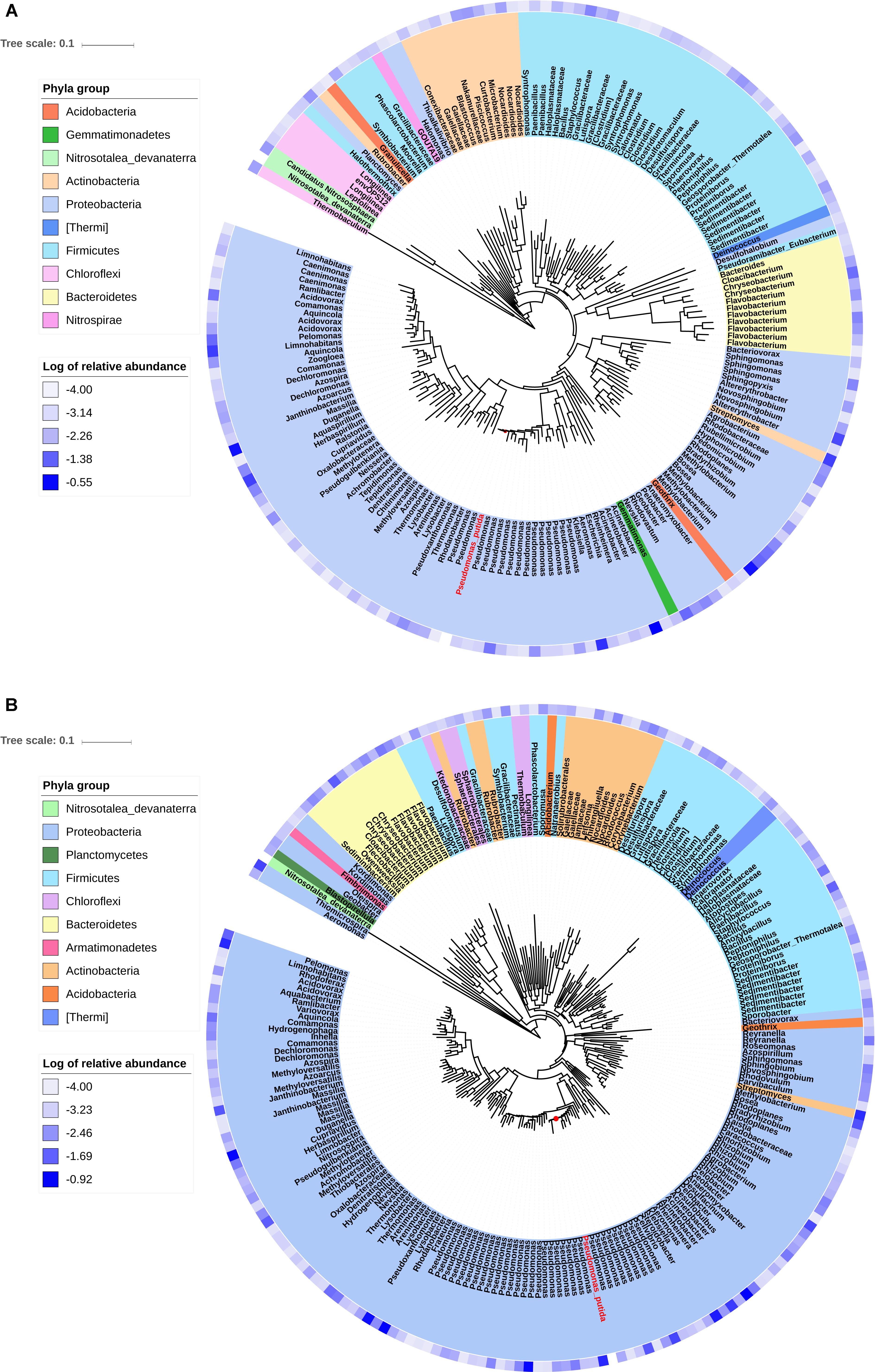
Figure 3. Phylogenetic tree showing the identified OTUs that contributed more than 0.01% in the transconjugant pools. (A) Showing transconjugants on D5. (B) Showing transconjugants on D75. The 16S rRNA gene sequence of Pseudomonas putida (donor strain) was shown in red letters and Nitrosotalea devanaterra (distant relative species to most of the transconjugants) was imported as the reference. The blue gradient circle at the periphery of the tree represents log of relative abundance of the OTU in the transconjugant pools.
Discussion
Human activities, including land application of manure, sewage sludge and wastewater, are providing direct routes for disseminating antibiotic resistant bacteria and genes to soil (Zhu et al., 2018). The spread of ARGs in soil is closely associated with HGT, however, studies on the HGT in soil are challenging due to the complexity of HGT process in soil (Williams-Nguyen et al., 2016). In this study, we use quantitative PCR and an established fluorescent bioreporter system to estimate the SP and delineate the taxonomic affiliation of recipient populations of a plasmid carrying multiple ARGs in soil. Occurrence of HGT need physical contact between donor and recipient populations, thus the population density of donor strain in soil is an important parameter for HGT (Martinez et al., 2015).
Using P. putida KT2442 harboring RP4 as a model strain, we thus investigated the dynamics of donor in a vegetable soil microcosm. We observed a biphasic decay behavior of both P. putida KT2442 and RP4. This biphasic decay had been well documented in P. putida, E. coli and pathogens (Brouwer et al., 2017; Mantilla-Calderon and Hong, 2017). The concentrations of dsRed and gfp were average 1.3 × 108 and 4.3 × 107 copies g-1 dry soil after initial spiking event, and they slightly increased at first 2 days after inoculation, which might be due to the division of donor strains and was in accordance with previous studies (Elsas et al., 1989; Henschke and Schmidt, 1990). Concentration of P. putida KT2442 rapidly decreased but a significantly reduced decay rate was observed after 15 days, and the copy number of dsRed maintained at 105 copies g-1 dry soil at the late stage of incubation. P. putida is frequently detected in natural soil and this result suggested that P. putida KT2442 survived and established in soil for a relatively longer period, which may highly affect the persistence and proliferation of plasmid, as well as the transfer of plasmid (Turner et al., 1998).
Plasmid RP4 showed a similar decay at the early stage of incubation, however, no further decay was observed after 15 day. The copy numbers of gfp plateaued at average 106 copies g-1 dry soil during the following incubation period, resulting an escalated plasmid/donor ratio that peaked at 10 on day 60. The plasmid decay kinetics may mainly be affected by three factors, the lysis of host bacteria and subsequent release of plasmid, the stability of plasmid in host bacteria, and plasmid transfer between host and recipient populations. Previous reports have shown that IncP plasmid is stable in Pseudomonas (Shintani et al., 2006; Yano et al., 2007), thus cellular decay of P. putida KT2442 may drive the kinetic of RP4 at the early stage of incubation. Though the copies of RP4 decreased rapidly and were lower to the concentrations of KT2442 at the early stage, plasmid transfer potentially occurred during the period, which was confirmed by flow cytometric sorting and analysis of transconjugants. It was reported that transconjugants appeared immediately after introduction of E. coli C600(RP4) or P. putida BH(RP4) into soil microcosms (Inoue et al., 2009). The donor bacteria were added with a cell ratio at 1:1 to indigenous soil bacteria, which was a relative high level although this ratio had been reported in previous studies (Elsas et al., 1990; Kozdrój, 2008; Lin et al., 2016; Stalder and Top, 2016). We conducted the experiment with a cell ratio at 1:1 for two reasons. Firstly, a higher conjugation frequency was observed when the mating densities of donor and recipients were equal according to a early study (Qiu et al., 2012). Secondly, added bacteria would decay rapidly when inoculated into microcosms (Elsas et al., 1989; Inoue et al., 2009; Bonot and Merlin, 2010), a high initial concentration of donors may be conducive to maintaining the donor cells and the subsequent analysis of plasmid transfer in this long-term experiment. Further experiments should be conducted with a gradient of donor cells for better understanding the transfer of plasmid in soil.
An established fluorescent bioreporter approach was adopted to estimate the SP of RP4 in soil, which was 7.5 × 10-5 TC/R on day 5 and 2.5 × 10-4 TC/R on day 75. The SP significantly increased during incubation, the longer incubation period with donor in soil microcosms may partially contribute to the spread of plasmid in this study, which provide sufficient time for plasmid transfer between donors and recipients, or even between transconjugants and recipients. The composition of recipient bacterial community is another factor that could significantly affect the SP of plasmid (Lopatkin et al., 2016). For example, the transfer frequency that estimated via a 48-h filter mating experiment using the same donor strain and plasmid with sewage sludge microbiota ranged from 3 to 50 conjugation events per 100000 cells (Li et al., 2018). Other factors, including antibiotics (Subbiah et al., 2011), nanomaterials (Qiu et al., 2012), metals (Klumper et al., 2016), UV irradiation (Lin et al., 2016), or ionic liquid (Wang et al., 2015a) may also affect the plasmid transfer in soil. Further studies should be conducted to investigate the effect of multiple factors on plasmid transfer and persistence in soil. The overall conjugation dynamics were determined by conjugation efficiency and propagation of transconjugants (Lopatkin et al., 2016). Discriminating the contribution of these two processes is challenging in soil. Nevertheless, our results provided an overall estimation of the proportion of host bacteria of plasmid in soil microbiota, which is essential for evaluating the potential role of plasmid transfer in mediating the dissemination of ARGs in soil after long-term exposure to antibiotic resistant bacteria.
Phylogenetic analysis of sorted transconjugants indicated that RP4 disseminated to a broad range of hosts, which had been documented in previous reports, with Proteobacteria being the predominant phylum (Klumper et al., 2015, 2016; Li et al., 2018), followed by Bacteroidetes, Firmicutes, and Actinobacteria. Gammaproteobacteria, known as the dominant hosts for the broad host range plasmid (Suzuki et al., 2010; Norberg et al., 2011), occupied 41.75% of total transconjugants in day 75. The most dominant genera in transconjugant pool of D75 including Pseudomonas, Acinetobacter, and Rhodanobacter were all from this class. However, it was observed that the phylogenetic diversity increased during the incubation with 88 unique genera were detected in day 75, implying that the plasmid may transfer to a broader range of taxa upon extended exposure time in soil than we expected. As the concentrations of P. putida KT2442 and RP4 were at roughly the same level from day 15 to 30, the increased diversity of transconjugant pools could be attributed, at least in part, to the plasmid transfer between transconjugants and recipient populations. A significant shift of transconjugant pool composition was also observed, for examples, 11 of the top 20 genera in day 75 were different from those in day 5 samples (Figure 3B). Rhodovastum (0.15%)/Sphingopyxis (0.13%) (Alphaproteobacteria), and Planctomyces (0.17%, Planctomycetes) were detected on day 5, but were not recovered on day 75. These data indicated that RP4 transiently transferred to members of these genera but they lost the plasmid during the following incubation, probably due to the fitness cost of the plasmid or the plasmid cannot replicate in these recipients. Nevertheless, the existing of transient host bacteria could aid in the survival and persistence of plasmid (Yano et al., 2013) in soil, and consequently contribute to the dissemination of ARGs in soil.
Author Contributions
J-QS and Y-GZ designed the experiments. X-TF and HL performed the microcosm experiments and qPCR. X-TF performed the flow cytometric sorting. Q-LC, Y-SZ, and JY conducted the phylogenetic analysis. All authors contributed to data analysis. X-TF and J-QS wrote the manuscript with comments from all authors.
Funding
This study was financially supported by the National Key Research and Development Program from Ministry of Science and Technology (2016YFD0800205), National Natural Science Foundation of China (31722004), K.C. Wong Education Foundation and Youth Innovation Promotion Association, CAS.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Barth F. Smets (Technical University of Denmark) for kindly providing P. putida KT2442::dsRed and RP4::gfp, and Tian-Lei Qiu and Xu-Ming Wang from Beijing Academy of Agriculture and Forestry Sciences for support in cell sorting by flow cytometry.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00194/full#supplementary-material
Footnotes
References
Al-Jassim, N., Mantilla-Calderon, D., Wang, T., and Hong, P. Y. (2017). Inactivation and gene expression of a virulent wastewater Escherichia coli strain and the nonvirulent commensal Escherichia coli DSM1103 strain upon solar irradiation. Environ. Sci. Technol. 51, 3649–3659. doi: 10.1021/acs.est.6b05377
Bonot, S., and Merlin, C. (2010). Monitoring the dissemination of the broad-host-range plasmid pB10 in sediment microcosms by quantitative PCR. Appl. Environ. Microbiol. 76, 378–382. doi: 10.1128/AEM.01125-09
Boto, L. (2010). Horizontal gene transfer in evolution: facts and challenges. Proc. Biol. Sci. 277, 819–827. doi: 10.1098/rspb.2009.1679
Brouwer, A. F., Eisenberg, M. C., Remais, J. V., Collender, P. A., Meza, R., and Eisenberg, J. N. (2017). Modeling biphasic environmental decay of pathogens and implications for risk analysis. Environ. Sci. Technol. 51, 2186–2196. doi: 10.1021/acs.est.6b04030
Burmolle, M., Hansen, L. H., Oregaard, G., and Sorensen, S. J. (2003). Presence of N-acyl homoserine lactones in soil detected by a whole-cell biosensor and flow cytometry. Microb. Ecol. 45, 226–236. doi: 10.1007/s00248-002-2028-6
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335. doi: 10.1038/nmeth.f.303
Chen, Q., An, X., Li, H., Su, J., Ma, Y., and Zhu, Y. G. (2016). Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil. Environ. Int. 9, 1–10. doi: 10.1016/j.envint.2016.03.026
Chen, Q.-L., Fan, X.-T., Zhu, D., An, X.-L., Su, J.-Q., and Cui, L. (2018). Effect of biochar amendment on the alleviation of antibiotic resistance in soil and phyllosphere of Brassica chinensis L. Soil Biol. Biochem. 119, 74–82. doi: 10.1016/j.soilbio.2018.01.015
Chen, Q. L., Li, H., Zhou, X. Y., Zhao, Y., Su, J. Q., Zhang, X., et al. (2017). An underappreciated hotspot of antibiotic resistance: the groundwater near the municipal solid waste landfill. Sci. Total Environ. 609, 966–973. doi: 10.1016/j.scitotenv.2017.07.164
Daubin, V., and Szollosi, G. J. (2016). Horizontal gene transfer and the history of life. Cold Spring Harb. Perspect. Biol. 8:a018036. doi: 10.1101/cshperspect.a018036
Dionisio, F., Conceicao, I. C., Marques, A. C., Fernandes, L., and Gordo, I. (2005). The evolution of a conjugative plasmid and its ability to increase bacterial fitness. Biol. Lett. 1, 250–252. doi: 10.1098/rsbl.2004.0275
Dodd, M. C. (2012). Potential impacts of disinfection processes on elimination and deactivation of antibiotic resistance genes during water and wastewater treatment. J. Environ. Monit. 14, 1754–1771. doi: 10.1039/c2em00006g
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Eichorst, S. A., Strasser, F., Woyke, T., Schintlmeister, A., Wagner, M., and Woebken, D. (2015). Advancements in the application of NanoSIMS and Raman microspectroscopy to investigate the activity of microbial cells in soils. FEMS Microbiol. Ecol. 91:fiv106. doi: 10.1093/femsec/fiv106
Elsas, J. D. V., Trevors, J. T., Overbeek, L. S. V., and Starodub, M. E. (1989). Survival of Pseudomonas fluorescens containing plasmids RP4 or pRK2501. Can. J. Microbiol. 35, 951–959. doi: 10.1139/m89-157
Elsas, J. D. V., Trevors, J. T., Starodub, M. E., and Overbeek, L. S. V. (1990). Transfer of plasmid RP4 between pseudomonads after introduction into soil; influence of spatial and temporal aspects of inoculation. FEMS Microbiol. Lett. 73, 1–11. doi: 10.1016/0378-1097(90)90718-6
Fox, R. E., Zhong, X., Krone, S. M., and Top, E. M. (2008). Spatial structure and nutrients promote invasion of IncP-1 plasmids in bacterial populations. ISME J. 2, 1024–1039. doi: 10.1038/ismej.2008.53
Geisenberger, O., Ammendola, A., Christensen, B. B., Molin, S., Schleifer, K.-H., and Eberl, L. (1999). Monitoring the conjugal transfer of plasmid RP4 in activated sludge and in situ identification of the transconjugants. FEMS Microbiol. Lett. 174, 9–17. doi: 10.1111/j.1574-6968.1999.tb13543.x
Guo, X., Yan, Z., Zhang, Y., Xu, W., Kong, D., Shan, Z., et al. (2018). Behavior of antibiotic resistance genes under extremely high-level antibiotic selection pressures in pharmaceutical wastewater treatment plants. Sci. Total Environ. 612, 119–128. doi: 10.1016/j.scitotenv.2017.08.229
Hale, L., Luth, M., and Crowley, D. (2015). Biochar characteristics relate to its utility as an alternative soil inoculum carrier to peat and vermiculite. Soil Biol. Biochem. 81, 228–235. doi: 10.1016/j.soilbio.2014.11.023
Henschke, R. B., and Schmidt, F. R. J. (1990). Plasmid mobilization from genetically engineered bacteria to members of the indigenous soil microflora in situ. Curr. Microbiol. 20, 105–110. doi: 10.1007/bf02092881
Inoue, D., Soda, S., Tsutsui, H., Yamazaki, Y., Murashige, K., Sei, K., et al. (2009). Occurrence and persistence of indigenous transconjugants carrying conjugative plasmids in soil. J. Biosci. Bioeng. 108, 231–234. doi: 10.1016/j.jbiosc.2009.03.014
Klappenbach, J. A., Saxman, P. R., Cole, J. R., and Schmidt, T. M. (2001). rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 29, 181–184. doi: 10.1093/nar/29.1.181
Klumper, U., Dechesne, A., Riber, L., Brandt, K. K., Gulay, A., Sorensen, S. J., et al. (2016). Metal stressors consistently modulate bacterial conjugal plasmid uptake potential in a phylogenetically conserved manner. ISME J. 11, 152–165. doi: 10.1038/ismej.2016.98
Klumper, U., Riber, L., Dechesne, A., Sannazzarro, A., Hansen, L. H., Sorensen, S. J., et al. (2015). Broad host range plasmids can invade an unexpectedly diverse fraction of a soil bacterial community. ISME J. 9, 934–945. doi: 10.1038/ismej.2014.191
Kozdrój, J. (2008). Survival, plasmid transfer and impact of Pseudomonas fluorescens introduced into soil. J. Environ. Sci. Health A Environ. Sci. Eng. Toxicol. 32, 1139–1157. doi: 10.1080/10934529709376601
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Li, L., Dechesne, A., He, Z., Madsen, J. S., Nesme, J., Sørensen, S. J., et al. (2018). Estimating the transfer range of plasmids encoding antimicrobial resistance in a wastewater treatment plant microbial community. Environ. Sci. Technol. Lett. 5, 260–265. doi: 10.1021/acs.estlett.8b00105
Lin, W., Li, S., Zhang, S., and Yu, X. (2016). Reduction in horizontal transfer of conjugative plasmid by UV irradiation and low-level chlorination. Water Res. 91, 331–338. doi: 10.1016/j.watres.2016.01.020
Lopatkin, A. J., Huang, S., Smith, R. P., Srimani, J. K., Sysoeva, T. A., Bewick, S., et al. (2016). Antibiotics as a selective driver for conjugation dynamics. Nat. Microbiol. 1:16044. doi: 10.1038/nmicrobiol.2016.44
Mantilla-Calderon, D., and Hong, P.-Y. (2017). Fate and persistence of a pathogenic NDM-1-positive Escherichia coli strain in anaerobic and aerobic sludge microcosms. Appl. Environ. Microbiol. 83:e00640-17. doi: 10.1128/aem.00640-17
Martinez, J. L., Coque, T. M., and Baquero, F. (2015). Prioritizing risks of antibiotic resistance genes in all metagenomes. Nat. Rev. Microbiol. 13:396. doi: 10.1038/nrmicro3399-c2
Modrie, P., Beuls, E., and Mahillon, J. (2010). Differential transfer dynamics of pAW63 plasmid among members of the Bacillus cereus group in food microcosms. J. Appl. Microbiol. 108, 888–897. doi: 10.1111/j.1365-2672.2009.04488.x
Musovic, S., Dechesne, A., Sorensen, J., and Smets, B. F. (2010). Novel assay to assess permissiveness of a soil microbial community toward receipt of mobile genetic elements. Appl. Environ. Microbiol. 76, 4813–4818. doi: 10.1128/AEM.02713-09
Musovic, S., Klumper, U., Dechesne, A., Magid, J., and Smets, B. F. (2014). Long-term manure exposure increases soil bacterial community potential for plasmid uptake. Environ. Microbiol. Rep. 6, 125–130. doi: 10.1111/1758-2229.12138
Musovic, S., Oregaard, G., Kroer, N., and Sorensen, S. J. (2006). Cultivation-independent examination of horizontal transfer and host range of an IncP-1 plasmid among gram-positive and gram-negative bacteria indigenous to the barley rhizosphere. Appl. Environ. Microbiol. 72, 6687–6692. doi: 10.1128/AEM.00013-06
Norberg, P., Bergstrom, M., Jethava, V., Dubhashi, D., and Hermansson, M. (2011). The IncP-1 plasmid backbone adapts to different host bacterial species and evolves through homologous recombination. Nat. Commun. 2:268. doi: 10.1038/ncomms1267
O’Brien, F. G., Yui Eto, K., Murphy, R. J., Fairhurst, H. M., Coombs, G. W., Grubb, W. B., et al. (2015). Origin-of-transfer sequences facilitate mobilisation of non-conjugative antimicrobial-resistance plasmids in Staphylococcus aureus. Nucleic Acids Res. 43, 7971–7983. doi: 10.1093/nar/gkv755
Pruden, A., Arabi, M., and Storteboom, H. N. (2012). Correlation between upstream human activities and riverine antibiotic resistance genes. Environ. Sci. Technol. 46, 11541–11549. doi: 10.1021/es302657r
Qiu, Z., Yu, Y., Chen, Z., Jin, M., Yang, D., Zhao, Z., et al. (2012). Nanoalumina promotes the horizontal transfer of multiresistance genes mediated by plasmids across genera. Proc. Natl. Acad. Sci. U.S.A. 109, 4944–4949. doi: 10.1073/pnas.1107254109
Shintani, M., Yano, H., Habe, H., Omori, T., Yamane, H., Tsuda, M., et al. (2006). Characterization of the replication, maintenance, and transfer features of the IncP-7 plasmid pCAR1, which carries genes involved in carbazole and dioxin degradation. Appl. Environ. Microbiol. 72, 3206–3216. doi: 10.1128/AEM.72.5.3206-3216.2006
Sorensen, S. J., Bailey, M., Hansen, L. H., Kroer, N., and Wuertz, S. (2005). Studying plasmid horizontal transfer in situ: a critical review. Nat. Rev. Microbiol. 3, 700–710. doi: 10.1038/nrmicro1232
Sørensen, S. J., Sørensen, A. H., Hansen, L. H., Oregaard, G., and Veal, D. (2003). Direct detection and quantification of horizontal gene transfer by using flow cytometry and gfp as a reporter gene. Curr. Microbiol. 47, 129–133. doi: 10.1007/s00284-002-3978-0
Soucy, S. M., Huang, J., and Gogarten, J. P. (2015). Horizontal gene transfer: building the web of life. Nat. Rev. Genet. 16, 472–482. doi: 10.1038/nrg3962
Stalder, T., and Top, E. (2016). Plasmid transfer in biofilms: a perspective on limitations and opportunities. NPJ Biofilms Microb. 2:16022. doi: 10.1038/npjbiofilms.2016.22
Subbiah, M., Top, E. M., Shah, D. H., and Call, D. R. (2011). Selection pressure required for long-term persistence of blaCMY-2-positive IncA/C plasmids. Appl. Environ. Microbiol. 77, 4486–4493. doi: 10.1128/AEM.02788-10
Suzuki, H., Yano, H., Brown, C. J., and Top, E. M. (2010). Predicting plasmid promiscuity based on genomic signature. J. Bacteriol. 192, 6045–6055. doi: 10.1128/JB.00277-10
Thomas, C. M., and Nielsen, K. M. (2005). Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3, 711–721. doi: 10.1038/nrmicro1234
Tolker-Nielsen, T., Brinch, U. C., Ragas, P. C., Andersen, J. B., Jacobsen, C. S., and Molin, S. (2000). Development and dynamics of Pseudomonas sp. Biofilms J. Bacteriol. 182, 6482–6489. doi: 10.1128/jb.182.22.6482-6489.2000
Tolker-Nielsen, T., and Molin, S. (2000). Spatial organization of microbial biofilm communities. Microb. Ecol. 40, 75–84.
Trevors, J. T., Elsas, J. D. V., Starodub, M. E., and Overbeek, L. S. V. (1989). Survival of and plasmid stability in Pseudomonas and Klebsiella spp. introduced into agricultural drainage water. Can. J. Microbiol. 35, 675–680. doi: 10.1139/m89-110
Turner, P. E., Cooper, V. S., and Lenski, R. E. (1998). Tradeoff between horizontal and vertical modes of transmission in bacterial plasmids. Evolution 52, 315–329. doi: 10.1111/j.1558-5646.1998.tb01634.x
UNEP (2017). Frontiers 2017 Emerging Issues of Environmental Concern. Nairobi: United Nations Environment Programme.
Wang, Q., Lu, Q., Mao, D., Cui, Y., and Luo, Y. (2015a). The horizontal transfer of antibiotic resistance genes is enhanced by ionic liquid with different structure of varying alkyl chain length. Front. Microbiol. 6:864. doi: 10.3389/fmicb.2015.00864
Wang, Q., Mao, D., and Luo, Y. (2015b). Ionic liquid facilitates the conjugative transfer of antibiotic resistance genes mediated by plasmid RP4. Environ. Sci. Technol. 49, 8731–8740. doi: 10.1021/acs.est.5b01129
Willetts, N., and Wilkins, B. (1984). Processing of plasmid DNA during bacterial conjugation. Microbiol. Rev. 48, 24–41.
Willey, J., Sherwood, L. M., and Woolverton, C. (2008). Prescott/Harley/Klein’s Microbiology, Seventh Edn. Cardiff: Colin Wheatley/Janice Roerig-Blong.
Williams-Nguyen, J., Sallach, J. B., Bartelt-Hunt, S., Boxall, A. B., Durso, L. M., McLain, J. E., et al. (2016). Antibiotics and antibiotic resistance in agroecosystems: state of the science. J. Environ. Qual. 45, 394–406. doi: 10.2134/jeq2015.07.0336
Yano, H., Garruto, C. E., Sota, M., Ohtsubo, Y., Nagata, Y., Zylstra, G. J., et al. (2007). Complete sequence determination combined with analysis of transposition/site-specific recombination events to explain genetic organization of IncP-7 TOL Plasmid pWW53 and related mobile genetic elements. J. Mol. Biol. 369, 11–26. doi: 10.1016/j.jmb.2007.02.098
Yano, H., Rogers, L. M., Knox, M. G., Heuer, H., Smalla, K., Brown, C. J., et al. (2013). Host range diversification within the IncP-1 plasmid group. Microbiology 159(Pt 11), 2303–2315. doi: 10.1099/mic.0.068387-0
Zhou, J., Wu, L., Deng, Y., Zhi, X., Jiang, Y. H., Tu, Q., et al. (2011). Reproducibility and quantitation of amplicon sequencing-based detection. ISME J. 5, 1303–1313. doi: 10.1038/ismej.2011.11
Zhu, Y. G., Gillings, M., Simonet, P., Stekel, D., Banwart, S., and Penuelas, J. (2018). Human dissemination of genes and microorganisms in Earth’s critical zone. Glob. Change Biol. 24, 1488–1499. doi: 10.1111/gcb.14003
Zhu, Y. G., Johnson, T. A., Su, J. Q., Qiao, M., Guo, G. X., Stedtfeld, R. D., et al. (2013). Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. U.S.A. 110, 3435–3440. doi: 10.1073/pnas.1222743110
Keywords: antibiotic resistance, horizontal gene transfer, conjugation, quantitative PCR, spread potential, flow cytometric sorting, phylogenetic analysis
Citation: Fan X-T, Li H, Chen Q-L, Zhang Y-S, Ye J, Zhu Y-G and Su J-Q (2019) Fate of Antibiotic Resistant Pseudomonas putida and Broad Host Range Plasmid in Natural Soil Microcosms. Front. Microbiol. 10:194. doi: 10.3389/fmicb.2019.00194
Received: 30 September 2018; Accepted: 23 January 2019;
Published: 01 March 2019.
Edited by:
Dongchang Sun, Zhejiang University of Technology, ChinaReviewed by:
Peter Mullany, University College London, United KingdomChristopher Morton Thomas, University of Birmingham, United Kingdom
Barth F. Smets, Technical University of Denmark, Denmark
Copyright © 2019 Fan, Li, Chen, Zhang, Ye, Zhu and Su. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-Qiang Su, anFzdUBpdWUuYWMuY24=
 Xiao-Ting Fan1,2
Xiao-Ting Fan1,2 Jun Ye
Jun Ye Jian-Qiang Su
Jian-Qiang Su