
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 14 February 2019
Sec. Microbial Physiology and Metabolism
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00160
This article is part of the Research TopicMicrobial Sulfur Metabolism - From Symbiosis to Global Nutrient Cycling View all 37 articles
Soda lakes are saline alkaline lakes characterized by high concentrations of sodium carbonate/bicarbonate which lead to a stable elevated pH (>9), and moderate to extremely high salinity. Despite this combination of extreme conditions, biodiversity in soda lakes is high, and the presence of diverse microbial communities provides a driving force for highly active biogeochemical cycles. The sulfur cycle is one of the most important of these and bacterial sulfur oxidation is dominated by members of the obligately chemolithoautotrophic genus Thioalkalivibrio. Currently, 10 species have been described in this genus, but over one hundred isolates have been obtained from soda lake samples. The genomes of 75 strains were sequenced and annotated previously, and used in this study to provide a comprehensive picture of the diversity and distribution of genes related to dissimilatory sulfur metabolism in Thioalkalivibrio. Initially, all annotated genes in 75 Thioalkalivibrio genomes were placed in ortholog groups and filtered by bi-directional best BLAST analysis. Investigation of the ortholog groups containing genes related to sulfur oxidation showed that flavocytochrome c (fcc), the truncated sox system, and sulfite:quinone oxidoreductase (soe) are present in all strains, whereas dissimilatory sulfite reductase (dsr; which catalyzes the oxidation of elemental sulfur) was found in only six strains. The heterodisulfide reductase system (hdr), which is proposed to oxidize sulfur to sulfite in strains lacking both dsr and soxCD, was detected in 73 genomes. Hierarchical clustering of strains based on sulfur gene repertoire correlated closely with previous phylogenomic analysis. The phylogenetic analysis of several sulfur oxidation genes showed a complex evolutionary history. All in all, this study presents a comprehensive investigation of sulfur metabolism-related genes in cultivated Thioalkalivibrio strains and provides several avenues for future research.
The biogeochemical sulfur cycle is a complex network of biotic and abiotic reactions, involving both organic and inorganic sulfur compounds. This is due to the many possible oxidation states of the sulfur atom, from −II (sulfide) to +VI (sulfate) allowing for multiple energy-generating red-ox transformations in both oxidative and reductive directions. Sulfur cycling is linked to other biogeochemical redox cycles including those of carbon, nitrogen and metals (through interactions with sulfides), and is an important driver of microbial growth and element conversions in a large number of different environments (Sorokin et al., 2011; Bowles et al., 2014; Wasmund et al., 2017). Sulfur-oxidizing bacteria (SOB) derive energy from the oxidation of reduced S compounds, whereas sulfidogens (sulfate-, sulfite-, sulfur- and thiosulfate-reducing, as well as sulfur-disproportionating prokaryotes) utilize oxidized sulfur compounds as electron acceptors in the absence of oxygen. These organisms are the main driving force of the microbial dissimilatory sulfur cycle. Examples of environments with the highest sulfur cycling activity are marine sediments (Wasmund et al., 2017), deep-sea “Black Smoker” hydrothermal vents (Jannasch and Mottl, 1985), acid mine drainage sites (Méndez-García et al., 2015) and alkaline soda lakes (Sorokin et al., 2015). The latter are of particular interest to our understanding of microbial sulfur metabolism, because of some unique features of sulfur chemistry at high pH, such as the reduced toxicity of hydrosulfide (HS−) and the stability of polysulfide (Sn2) (Sorokin et al., 2011).
Soda lakes are double extreme environments characterized by a stable, elevated pH (>pH 9) caused by the presence of high concentrations of sodium carbonate/bicarbonate which can reach up to saturation and result in molar concentrations of soluble alkalinity. They can be found around the globe in arid regions and are formed by evaporation of CO2-rich groundwaters in the absence of divalent cations, such as Ca2+ and Mg2+ (Sorokin et al., 2014). Despite the challenges posed by these saline alkaline conditions, soda lakes support a diverse microbial community, which drives active carbon, nitrogen and sulfur cycles (Sorokin et al., 2014, 2015). Samples taken from south Siberian soda lakes showed high rates of sulfidogenesis (Foti et al., 2007; Sorokin et al., 2010); however, to our knowledge, the rates of sulfide oxidation in soda lakes have not been measured. These rates are likely similarly high, given the fact that sulfide is absent in the top 1–2 cm sediment layer of the soda lakes (Sorokin et al., 2010). Sulfide and other reduced sulfur compounds are oxidized by a community consisting of both phototrophic and chemotrophic SOB (Sorokin et al., 2014). The gammaproteobacterial genus Thioalkalivibrio (Sorokin et al., 2013) is the most abundant group of sulfur-oxidizing bacteria in soda lakes, which was recently confirmed by metagenomic analysis (Vavourakis et al., 2018). Thioalkalivibrio is one of the best-studied groups of bacteria from soda lakes, with nearly a hundred isolates originating from hypersaline soda lakes in seven regions of the world: the Kulunda Steppe, the Buriatia region, the Transbaikal region and North-Eastern Mongolia in Central Asia, Wadi al Natrun (Libyan desert) in Egypt, the East-African Rift Valley in Kenya, and California and Washington State in North-America. Furthermore, Thioalkalivibrio species are the dominant SOB in so-called Thiopaq systems, full-scale bioreactor installations used to remove sulfide from different gas streams at haloalkaline conditions (Sorokin et al., 2008, 2012; Janssen et al., 2009).
Currently, the genus Thioalkalivibrio contains 10 validly described species: Tv. versutus (type species), Tv. nitratis, Tv. denitrificans (Sorokin et al., 2001a), Tv. paradoxus, Tv. thiocyanoxidans (Sorokin et al., 2002b), Tv. jannaschii (Sorokin et al., 2002a), Tv. thiocyanodenitrificans (Sorokin et al., 2004), Tv. nitratireducens (Sorokin, 2003), Tv. halophilus (Banciu et al., 2004), and Tv. sulfidiphilus (Sorokin et al., 2012). These 10 species show a high degree of metabolic flexibility: every isolate from all strains can oxidize sulfide, thiosulfate, elemental sulfur, and polysulfide. Some strains can additionally oxidize tetrathionate, carbon disulfide, and thiocyanate. Although most strains are obligate aerobes, three species are capable of anaerobic growth with either nitrate, nitrite, or nitrous oxide as the electron acceptor (Tv. denitrificans, Tv. thiocyanodenitrificans, and Tv. nitratireducens). Most of the isolates are capable of growth in saturated soda brines, with a total sodium concentration of over 4.3 M Na+ and pH up to 10.5.
Since whole genome sequencing has become widespread, our knowledge of metabolic potential has greatly increased, allowing microbiologists to conduct large-scale studies of the metabolic repertoire of multiple species. In the case of Thioalkalivibrio, the genomes of approximately 70 strains were sequenced as part of the Joint Genome Institute’s Community Science Program around 2012, and several additional type strains were sequenced independently. This created the largest dataset of genomes so far for any group of obligately lithoautotrophic bacteria. Only a small number of those genomes has been annotated and analyzed to date: (i) Tv. sulfidiphilus was selected for its relevance in sulfide-removing bioreactors (Muyzer et al., 2011a), (ii) Thioalkalivibrio sp. K90mix (Muyzer et al., 2011b) was selected for its extreme potassium tolerance, and (iii) Tv. paradoxus, Tv. thiocyanoxidans, and Tv. thiocyanodenitrificans (Berben et al., 2017) were selected for their ability to grow with thiocyanate (NCS−) as the sole electron donor. Additionally, a recent study used the complete set of genomic data to analyze the genomic diversity and phylogeny of the Thioalkalivibrio genus using modern in silico methods—such as average nucleotide identity (ANI), multi-locus sequence analysis (MLSA) and digital DNA–DNA hybridization (dDDH)—compared to classic DNA–DNA hybridization and 16S rRNA phylogeny (Ahn et al., 2017).
Although several studies have reported on individual aspects of sulfur metabolism in members of Thioalkalivibrio, to our knowledge there has been no genus-wide comprehensive analysis of sulfur oxidation-related genes present in the genomes. A complete understanding of the sulfur metabolism pathways in Thioalkalivibrio would provide insights into key microorganisms that carry out an important biogeochemical process in soda lakes and also may help to get insights and possibilities for process optimization of their application in desulfurization of bio- and industrial gas streams.
Here, we present the first comprehensive analysis of the distribution of genes encoding enzymes that catalyze the oxidation of reduced sulfur compounds across the genus Thioalkalivibrio and the diversity of metabolic strategies for dissimilatory sulfur metabolism in these bacteria. We mined the genomes of 75 strains for genes known to be involved in sulfur oxidation using ortholog prediction. These results were further refined with sequence and phylogenetic analyses. Our aim was to provide a comprehensive overview of the potential for the oxidation of a diverse set of sulfur compounds by members of the haloalkaliphilic genus Thioalkalivibrio.
Cultures of 75 Thioalkalivibrio strains were obtained from the personal culture collection of D. Sorokin for genome sequencing. The enrichment and isolation of these strains were described previously (Sorokin et al., 2001a, 2002b, 2004, 2012; Sorokin, 2003; Banciu et al., 2004). A total of 72 of these strains were sequenced by the Joint Genome Institute (JGI) as part of the Community Science Program; cultivation, DNA extraction, sequencing, assembly and annotation procedures have been described previously (Muyzer et al., 2011a). The remaining strains (Tv. versutus AL2T, Tv. denitrificans ALJDT, and Tv. halophilus HL17T) were later sequenced as a separate project (Ahn et al., 2017), because the extracted DNA initially submitted to JGI was of insufficient quality for genome sequencing. Basic genome statistics and accession numbers are included in Supplementary Table S1.
All protein sequences of the 75 Thioalkalivibrio genomes listed in Supplementary Table S1 were subjected to an orthology prediction using OrthoMCL. First we used a locally installed NCBI blastp executable (version 2.7.1) with default parameters to perform all-vs.-all blast of protein sequences annotated in the Thioalkalivibrio genomes. OrthoMCL takes the output of the all-vs.-all blast and uses Markov clustering to calculate clusters of orthologous sequences. We utilized the default Markov inflation factor of 1.5, which balances clustering sensitivity and selectivity (Fischer et al., 2011). The output from OrthoMCL was parsed into a single gene presence/absence matrix using custom Python scripts. The ortholog group table is included as Supplementary Table S2. Ortholog groups representing sulfur oxidation genes were identified by blastp (Altschul et al., 1997), using sequences from well-studied organisms as queries. Sequences from Allochromatium vinosum were used for FccAB (Uniprot accessions: Q06529/Q06530), SoxAXYZB (Q1W3E4, D3RVS6, D3RVA1, D3RVA2, D3RVS5), DsrABC (O33998, D3RSN2, D3RSN6), AprBA (D3RSA0, D3RSA1), Sat (O66036) and SoeABC (NCBI accessions: ADC63403.1, ADC63402.1, ADC63401.1). For SQR a sequence from Acidithiobacillus ferrooxidans (B7JBP8) was used (a type I SQR), for SorAB sequences from Starkeya novella (Q9LA16, Q9LA15) and HdrABC (ADJ22511.1, ADJ22501.1, ADJ22509.1) from Hyphomicrobium denitrificans. For SOR, TcDH and ScnABC hydrolase we used sequences from Thioalkalivibrio paradoxus (WP_006748120.1), Tv. thiocyanoxidans (WP_019623383.1) and Thiobacillus thioparus (O66187, O66187, and O66188), respectively. We used a sequence similarity criterion of >30% and >50% query coverage, as well as organization of genes in operons in case of multi-enzyme systems (for example soeABC).
The matrix of genomes and gene copy numbers was visualized as a heatmap using ggplot2 in the R environment (Wickham, 2009). The dendrogram in Figure 1 was calculated by using Ward hierarchical clustering on the Euclidean distances on a presence/absence matrix of sulfur genes. This matrix contained all genes as separate rows, after clustering identical rows were collapsed for genes that are located in a single operon.
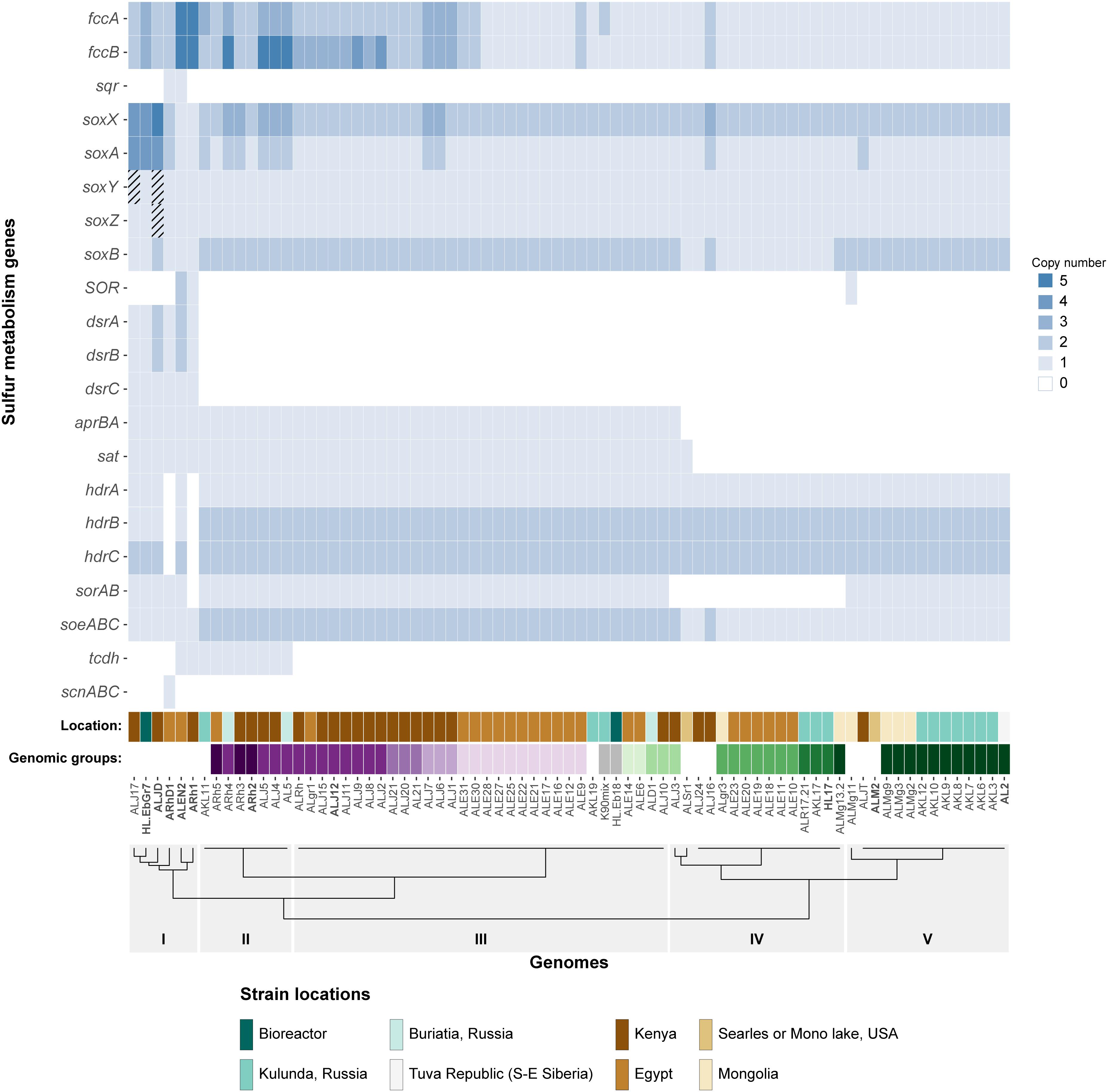
Figure 1. Heatmap showing the copy number of key sulfur oxidation genes in 75 Thioalkalivibrio strains. Cross-hatched cells represent genes which are likely present, but currently unconfirmed. Location denotes the geographical region from which the corresponding strain was isolated. The genomic groups colors correspond to species as defined by Ahn et al. (2017). Genomic groups comprising a single strain are not colored to maintain clarity. The dendrogram on the x-axis shows the Ward clustering of genomes based on presence/absence of the sulfur genes. Please note that SOR is sulfur oxygenase/reductase and sorAB is sulfite:cytochrome c oxidoreductase.
Phylogenetic analysis of selected genes was performed as follows: all available sequences from Thioalkalivibrio (see Supplementary Table S1) were used. Additional sequences from both closely and more distantly related organisms were retrieved from the NCBI protein database, by using protein BLAST. Sequences were aligned using T-COFFEE (Di Tommaso et al., 2011) and the most likely amino acid substitution matrix was determined using prottest 3 (Abascal et al., 2005), which was the LG model (Le and Gascuel, 2008) with gamma-distributed rates and empirical frequencies (model parameter –m PROTGAMMALGF in RAxML) for all alignments. Maximum likelihood trees were calculated using RAxML 8.2.12 (Stamatakis, 2014, 2015), using the rapid bootstrap analysis algorithm. To test the reproducibility of tree branches either 250 or 500 bootstrap replicates were calculated for the final tree. Once generated, the tree was visualized in MEGA7 (Kumar et al., 2016), the root of the tree was placed on the outgroup branch and branches with sequences belonging to a single taxonomic group were collapsed for legibility.
An overview of sulfur oxidation-related genes identified in the 75 Thioalkalivibrio genomes is shown in Table 1. Genes for tetrathionate oxidation (tetrathionate hydrolase, tetrathionate:quinone oxidoreductase) were not detected in any of the Thioalkalivibrio genomes. A putative carbon disulfide hydrolase was only detected in the genome of Tv. nitratireducens ALEN 2T, including the characteristic ‘Phe–Phe’ motif (Smeulders et al., 2011).
Of the 21 genes/operons shown in Figure 1, five genes—flavocytochrome c sulfide dehydrogenase (fccAB), soxAX and soxB—are found in all 75 genomes. Almost half of the strains (37/75) have more than one copy of the flavoprotein subunit fccB, although not all have an equal number of corresponding copies of the cytochrome c subunit fccA. Strains ALEN2T (Tv. nitratireducens) and ARh1T (Tv. paradoxus) carry five copies of both genes, whereas strains AL5, ARh4, ALJ4, and ALJ5 have five copies of fccA, but only three of fccB. Sulfide:quinone oxidoreductase (sqr), which also oxidizes sulfide, but donates electrons to the ubiquinone pool rather than to the cytochrome c pool, is only found in two strains: ARhD1T (Tv. thiocyanodenitrificans) and ALEN2T (Tv. nitratireducens), both of which can grow anaerobically by using nitrate as an electron acceptor.
Genes for soxAX and soxB, two components of the truncated sox system are present in all genomes included in the study: soxAX forms the cytochrome complex that extracts two electrons from the sulfane atom of thiosulfate and binds it covalently to carrier complex soxYZ; thiosulfohydrolase soxB then catalyzes the hydrolysis of the sulfone group, releasing sulfate. The gene for soxY was not found in two genomes of closely related strains Tv. sulfidiphilus ALJ17 and Tv. denitrificans ALJDT (the latter strain also appears to lack the gene for soxZ). The absence of annotated soxY in Thioalkalivibrio sp. ALJ17 and of soxYZ in Tv. denitrificans ALJDT appears to be a result of breaks in the genome assemblies. The soxZ gene is found on the edge of contig 10 in strain ALJ17 and fragments of soxY (annotated as pseudogene features by NCBI) are found on the edges of contigs 5 (locus tag D580_RS0103285) and 54 (D580_RS0114455). Additionally, on contig 54, the partial soxY gene is followed by genes for a sigma 54-dependent Fis family transcriptional regulator (D580_RS0114460), a sensor histidine kinase (D580_RS0114465), a small hypothetical protein (D580_RS16435) and a diaminopimelate decarboxylase (D580_RS0114475), all of which are located downstream from soxY in Tv. sulfidiphilus HL-EbGr7T (Tgr7_0038-0041). In Tv. denitrificans the latter three genes are found on the edge of contig 67 (B1C78_RS11375-385).
The next most abundant set of genes encodes for three subunits of a bacterial heterodisulfide reductase-like system. This is represented in Figure 1 as hdrABC for clarity, but the conserved operon is actually hdrC1B1A-hyp-C2B2. This operon is closely associated with two lipoate-binding proteins LbpA1 and LbpA2 (Cao et al., 2018), which are found adjacent to the hdr-like operon, in all Thioalkalivibrio genomes that contain it, as lbpA1-dsrE-lbpA2. Only two genomes in the dataset appear to not contain these genes: ARh1T and ARhD1T, both of which encode the complete reverse dissimilatory sulfite reductase pathway (dsrABCEFHMKJOPRS). In total, six strains in the genus have the rDsr system, which hdr is speculated to replace (Quatrini et al., 2009; Cao et al., 2018; Koch and Dahl, 2018): ARh1T, ARhD1T, ALEN2T, ALJ17, HL-EbGr17T (Tv. sulfidiphilus) and ALJDT. The latter four genomes contain both dsr and hdr.
There are three known pathways for the oxidation of sulfite to sulfate in SOB: one, the aprBA/sat pathway, is indirect and uses adenosine 5′-phosphosulfate as an intermediate compound (Kappler and Dahl, 2001). This pathway was detected in 47 of the 75 genomes, with one additional strain (ALSr1) containing sat, but not aprBA. The two direct pathways are catalyzed by sulfite:cytochrome c oxidoreductase (sorAB) or sulfite:quinone oxidoreductase (soeABC), although there currently is only genetic evidence for the activity of soeABC (Dahl et al., 2013). Putative soeABC genes are present in all analyzed strains of Thioalkalivibrio and form an operon in all genomes. Genes putatively encoding sorAB are present in 60 of the 75 Thioalkalivibrio strains.
Thiocyanate dehydrogenase (tcdh) was found in ten strains in two different genomic contexts, as described in detail previously (Berben et al., 2017). Thiocyanate hydrolase (scnABC) was found only in a single strain, ARhD1T, which is capable of oxidizing thiocyanate anaerobically.
Sulfur oxygenase reductase (SOR) was only present in three strains: ARh1T, ALEN2T, and ALMg11, as was recently described by Rühl et al. (2017).
The dendrogram in Figure 1 represents a hierarchical clustering of Thioalkalivibrio genomes based on the presence/absence of sulfur metabolism genes. From left to right, group I, consisting of six strains spanning from ALJ17 (Tv. sulfidiphilus) to ARh1T (Tv. paradoxus) is characterized by the presence of the rDSR pathway. Additionally, the only occurrences of SQR are within this group (ARhD1T and ALEN2T), as well as the only occurrence of thiocyanate hydrolase (ARhD1T). Four of these six genomes contain genes for both dissimilatory sulfite reductase and heterodisulfide reductase. ALEN2T and ARh1T are two of the three strains that have sulfur oxygenase-reductase and fall within this group. Three strains—ALJDT (Tv. denitrificans), HL-EbGr7T (Tv. sulfidiphilus) and ALJ17—have four copies of soxAX, with ALJDT containing a fifth soxX copy. ALEN2T and ARh1T are positive for the thiocyanate dehydrogenase gene and ARh1T is the only strain in this group lacking a putative sorAB. Group II, from ARh5 to AL5, is characterized by the presence of the gene for thiocyanate dehydrogenase, two copies of soeABC and soxB and the presence of aprBA and putative sorAB genes. Copy numbers for fccAB and soxAX are variable within this group. Group III is similar to group II, aside from the lack of TcDH in these strains. Group IV is characterized by the absence of sorAB genes. Genes for aprBA and sat are similarly not found in these genomes, with the exception of strains ALJ3 and ALSr1, the latter of which is the only Thioalkalivibrio genome to contain sat but not aprBA. Genes for fccAB and soeABC are present in single copy, except in strain ALJ16; soxX is present in two copies and soxA in one, except in strain ALJ16 which contains an extra copy of both. Group V is characterized by the presence of sorAB and soeABC and absence of aprBA/sat. ALMg11 is the only strain outside group I that contains sulfur oxygenase/reductase (SOR), placing it on a separate branch within this group.
The genomic groups row in Figure 1 assigns colors to strains based on their assignment to genomic groups by ANIb analysis, as reported by Ahn et al. (2017). Genomic groups that comprise only a single strain were not assigned a color in this figure to maintain clarity. These labels show that strains belonging to the same genomic groups tend to group together closely based on sulfur gene repertoire. The exceptions to this trend are genomic groups 1 (Tv. versutus), as ALMg13-2 is placed in group IV due to its lack of sorAB; genomic group 15, due to the placement of strain ALJ3 in group IV rather than group III for the same reason; and genomic group 11 (Tv. nitratis), in which the presence of thiocyanate dehydrogenase in strains AL5, ALJ4, ALJ5, and ARh4 leads to their assignment to group II, rather than group III.
Figure 2 shows a maximum likelihood phylogenetic tree based on SoxB protein sequences, which is a unique marker of the Sox pathway (Meyer et al., 2007). All Thioalkalivibrio sequences were included, as well as the SoxB sequences of forty other SOB species in a variety of phylogenetic classes. Whereas most classes cluster together, the gammaproteobacterial sequences are found distributed across the tree. One cluster of Gammaproteobacteria is split by the anoxygenic green sulfur bacteria (class Chlorobia) and the Epsilonproteobacteria. A second group of gammaproteobacterial SoxB sequences forms a sister taxa to SoxB sequences of the Betaproteobacteria. The only non-gammaproteobacterial sequence in this cluster belongs to Thermithiobacillus tepidarius. Sequences from Thioalkalivibrio are found in four distinct groups: group I contains a single SoxB sequence from every Thioalkalivibrio strain, with the exception of Tv. nitratireducens (ALEN2T), Tv. paradoxus (ARh1T) and the phylogenetically divergent strains ARhD1 (Tv. thiocyanodenitrificans), HL-EbGr7T and ALJ17 (Tv. sulfidiphilus). Tv. denitrificans (ALJDT) is split from group I by SoxB sequences from Halorhodospira halophilus. Group IIa contains all Thioalkalivibrio strains which possess two copies of the soxB gene, as well as strains ALEN2T and ARh1T. Group IIb contains sequences of strains ARhD1T, HL-EbGr7T, ALJ17, and ALJDT, and is separated from group IIa by a group of sequences belonging to Ectothiorhodospira, Ectothiorhodosinus and one strain of Halorhodospira.
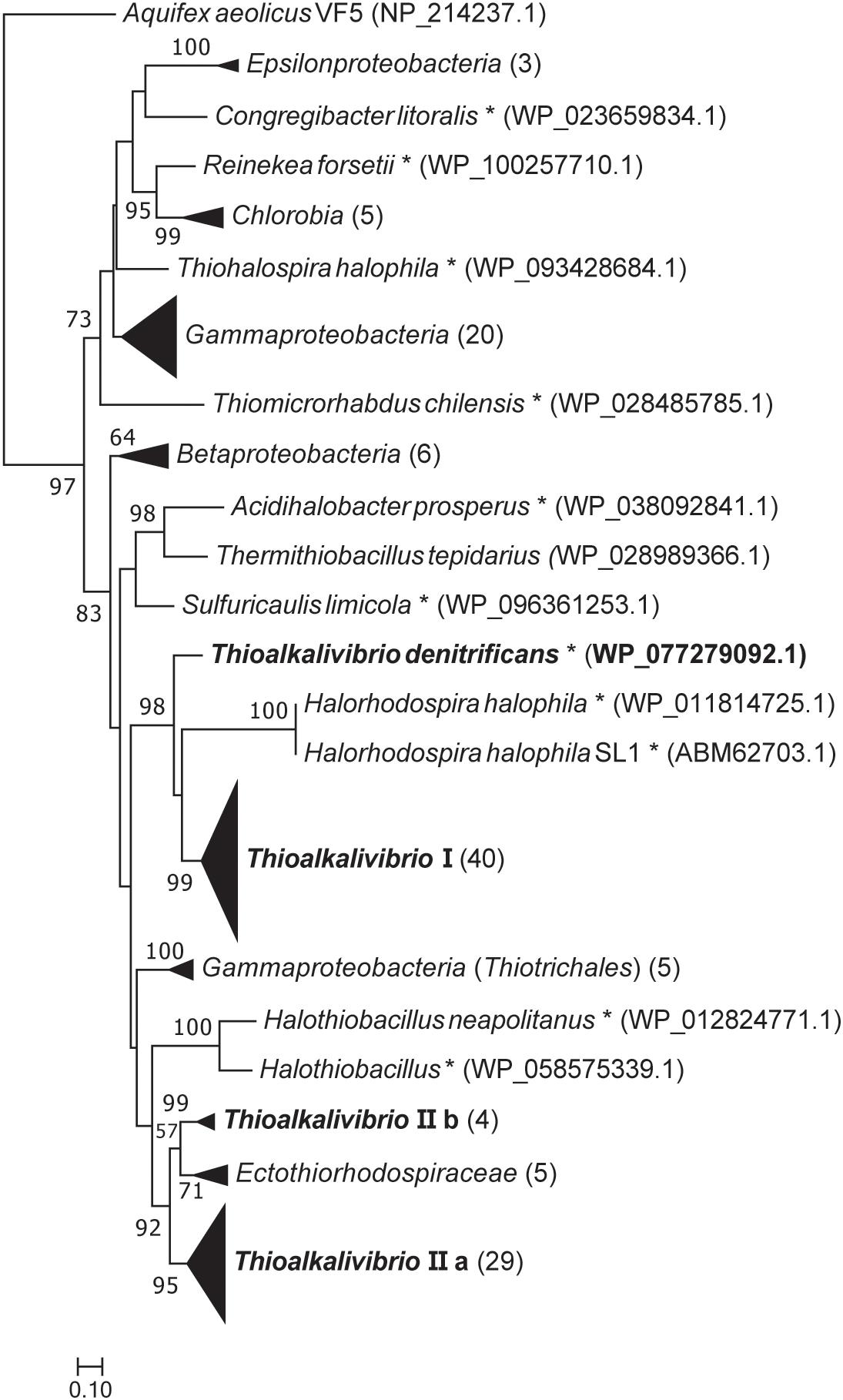
Figure 2. Maximum likelihood tree of SoxB amino acid sequences, based on 250 bootstrap replicates. A group of sequences of 5′-nucleotidase proteins was used as the outgroup, but was pruned from the tree. Sequences of members of the Gammaproteobacteria located on branches that could not be further collapsed are marked with an asterisk. Parenthesized numbers indicate the number of sequences contained in a collapsed branch. The scale bar represents % difference.
Trees based on amino acid sequences for SoxY and SoxZ show similar phylogenetic distributions (see Supplementary Figures S1, S2), however, the bootstrap values in these trees are much lower.
Figure 3 shows the position of putative SQR sequences from Thioalkalivibrio in a previously published phylogenetic tree, where structure information was used to define six types of SQR (Marcia et al., 2010). Candidate SQR sequences from Thioalkalivibrio were assigned to a single ortholog group by the orthoMCL algorithm. Similar sequences from Tv. denitrificans ALJDT, Tv. nitratireducens ALEN2T and Tv. thiocyanodenitrificans ARhD 1T were identified by blastp and added to this group and used to generate a new SQR tree together with the sequences used by Marcia et al. (2010). The figure clearly shows that the putative SQR sequences from ALEN2T and ARhD1T fall within group I, whereas the sequence from ALJDT is a flavocytochrome c sulfide dehydrogenase FccB [which is confirmed by the presence of a gene coding for a diheme cytochrome c subunit (FccA) adjacent to it, locus tag B1C78_RS00715]. The FAD-dependent oxidoreductase sequences contained in ortholog group 1954 form a sister taxa to type II SQR. A recent reclassification of SQR and sulfide dehydrogenases provided by Sousa et al. (2018) maintains consistency with the previously defined type II SQRs (renamed group C), whereas type I SQRs form group D together with types V and VI.
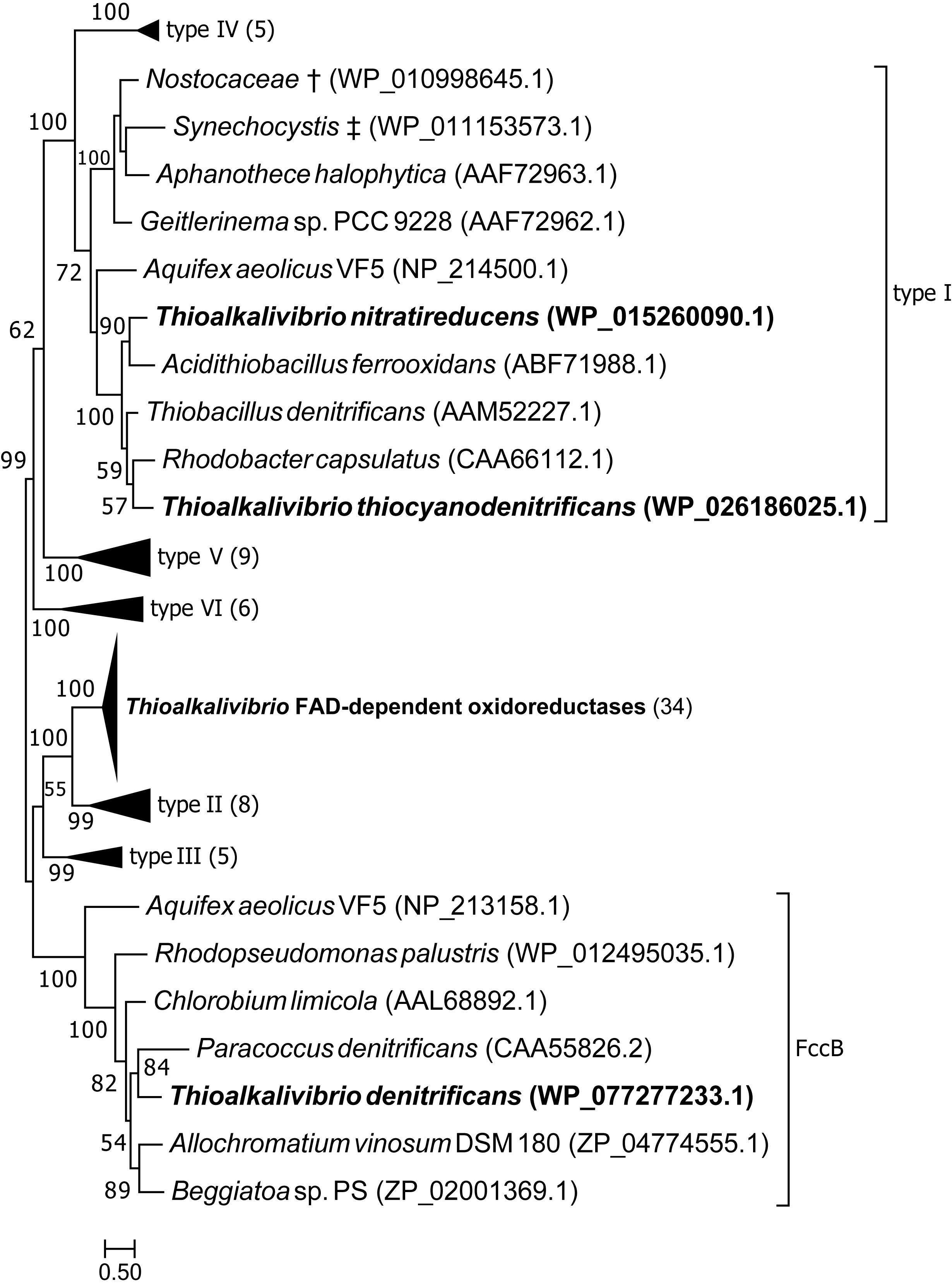
Figure 3. Maximum likelihood tree of SQR amino acid sequences, based on a previously published tree classifying SQR into six groups (Marcia et al., 2010). Sequences from ortholog group 1954 were placed in the tree, as well as sequences similar to SQR by blastp searches of the genomes of strains ALJDT, ARhD1T and ALEN2T. † Multispecies record representing Nostoc sp. PCC 7120 and Trichormus variabilis NIES-23; ‡ multispecies record representing Synechocystis sp. PCC 6803 and Synechocystis sp. IPPAS B-1465. Parenthesized numbers indicate the number of sequences contained in a collapsed branch. The scale bar represents % difference.
Figure 4 is a maximum likelihood tree of amino acid sequences of FccB, the catalytic subunit of sulfide dehydrogenase. In contrast to SoxB, all FccB sequences from Thioalkalivibrio are located in a single cluster. The most closely related sequences are from the other members of the Ectothiorhodospiraceae, which mostly cluster together. The exceptions to this are one sequence of Alkalilimnicola erlichii, which is found as a sister branch to a group of sequences from the Thiotrichales and Betaproteobacteria, and Thiohalomonas denitrificans (Gammaproteobacteria), whose three FccB sequences are located on a branch with mostly other Chromatiales sequences and a single Thiothrix nivea sequence, followed by the Alphaproteobacteria. Sequences from the Betaproteobacteria and gammaproteobacterial orders Chromatiales and Thiotrichales are scattered throughout the tree.

Figure 4. Maximum likelihood tree of FccB (the flavoprotein subunit of flavocytochrome c sulfide dehydrogenase) sequences, based on 500 bootstrap replicates. Members of the family Ectothiorhodospiraceae have been marked with an asterisk. This tree includes all FccB copies shown in Figure 1, identical sequences have been collapsed by NCBI into multispecies records, leaving 59 unique sequences. Parenthesized numbers indicate the number of sequences contained in a collapsed branch. The scale bar represents % difference.
The putative SoeA sequences from Thioalkalivibrio strains form two groups in a tree (Supplementary Figure S3) which includes previously predicted SoeA amino acid sequences from other SOB according to Dahl et al. (2013). Group Thioalkalivibrio I contains a representative sequence from every strain, although the phylogenetically divergent strains ALJDT, ARhD1T, HL-EbGr7T, and ALJ17 are separated from this group with 100% bootstrap support. Thioalkalivibrio group II contains all other SoeA sequences present in the genomes studied here and appears to be more distantly related to the other sequences in the tree, although bootstrap support for this branch is low. The putative SoeA sequences in group II have an average sequence similarity of 55% with those in group I.
To assess the possible function of multiple copies of the same gene present in Thioalkalivibrio genomes, an overview of genomic contexts was generated. Figure 5 shows a schematic representation of this data. Genomic contexts for hdrBC were not included as they are usually present in a single operon (Koch and Dahl, 2018); contexts for dsrAB are not shown as only six genomes contain dsr genes at all, and only two of these contain multiple copies.
Sequences for fccA were assigned to four different ortholog groups by OrthoMCL (Figure 5A). Group OG_435 contains a sequence from every Thioalkalivibrio strain and upstream and downstream contexts are highly similar in all genomes. Strain ALJ16 is an exception to this, as it contains two OG_435 fccA sequences, one of which does not conform to this context. OG_2776 and OG_3025 fccA genes are also always found adjacent to fccB, but show more divergent upstream and/or downstream contexts (see Figure 5A). The fccA sequences that are present in the TcDH operon (Berben et al., 2017) are found in this ortholog group, and only these have the type IIIa context. Only a single fccA sequence was assigned to ortholog group OG_5153, from strain AKL11, and it is not found in an operon with fccB.
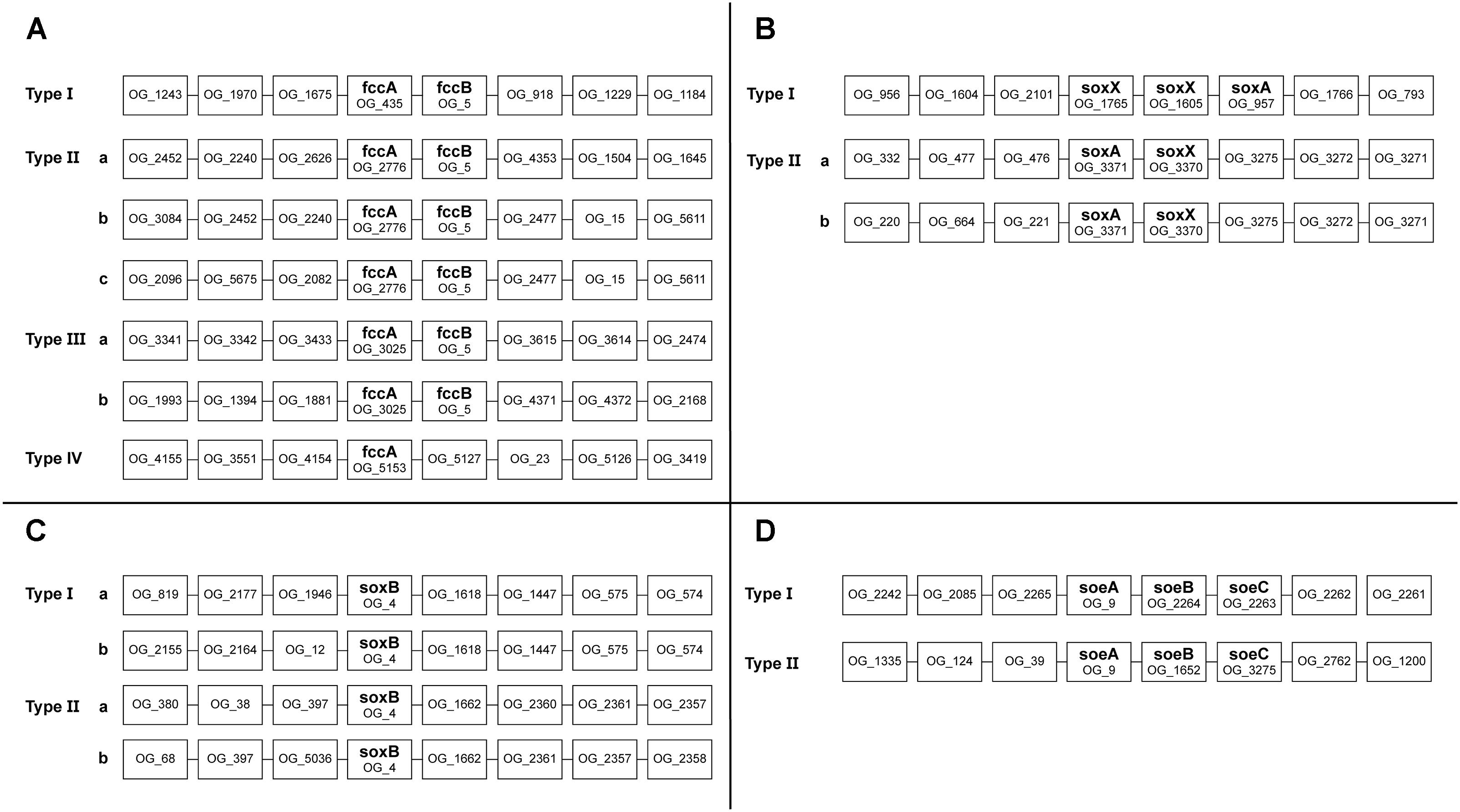
Figure 5. Schematic representation of the genomic contexts of genes present in multiple copies in Thioalkalivibrio genomes. Selected genes were fccA (A); soxX (B); soxB (C), and soeA (D).
A majority of strains have two copies of soxX and a single copy of soxA (Figure 1). SoxX sequences were assigned to three different ortholog groups: OG_1765 and OG_1605 sequences were found directly adjacent to each other, with soxA directly downstream (Figure 5B). SoxX sequences assigned to OG_3370 all have a gene for soxA directly adjacent (Figure 5B). Tv. sulfidiphilus (HL-EbGr7T and ALJ17T) and Tv. denitrificans (ALJDT) contain four repeats of soxXA in a row, with an additional soxX copy in ALJDT. All soxB sequences were assigned to a single ortholog group (OG_4). There are two major genotypes, as shown in Figure 5C. Type I comprises a soxB sequence from every Thioalkalivibrio genome studied, whereas Type II contains a sequence from every strain with two soxB copies. Type I is divided into subtypes by 12 strains which have a divergent genomic context (Figure 5C).
A total of 42 strains have two copies of soeABC (Figure 1), of which all soeA sequences are found in a single ortholog group. These sequences are found in two genomic contexts: all strains have a soeABC cluster in a type I context; additional soeABC copies are found in context type II (Figure 5D).
With the information that is currently available, a complete sulfur oxidation pathway (from sulfide to sulfate) can only be reconstructed for six Thioalkalivibrio strains: ARhD1T (Tv. thiocyanodenitrificans), ALJDT (Thioalkalivibrio denitrificans), HL-EbGr7T (Tv. sulfidiphilus), ALJ17, ARh1T (Tv. paradoxus), and ALEN2T (Tv. nitratireducens). These six strains are capable of zero-valent sulfur oxidation using the reverse dissimilatory sulfite pathway, which was originally characterized in the purple sulfur bacterium Allochromatium vinosum (Frigaard and Dahl, 2009). These strains were previously shown to be phylogenetically divergent from the other Thioalkalivibrio strains, both by 16S rRNA gene analysis and modern phylogenomic methods (Ahn et al., 2017), and may need to be assigned to two different genera in the future. No components of the dsrABEFHCMKLJOPNRS cluster were detected in any of the remaining strains, although aprBA and sat, required for the activation of sulfate in sulfate-reducing bacteria (SRB), were detected in 47 genomes. The oxidation of sulfur in the strains lacking rDsr is more extensively discussed below.
The detection of partial soxYZ genes on contig edges in the genomes of ALJDT and ALJ17, together with the fact that both strains are routinely cultured with thiosulfate as the sole electron donor, support the conclusion that these strains do indeed carry the soxYZ genes. Conclusive proof, however, can only be obtained by either performing additional sequencing runs to close the genome, or by PCR with primers designed using soxYZ sequences of closely related strains. The inclusion of these soxY in ALJ17 and soxYZ in ALJDT does not change the order of the dendrogram in Figure 1, only the branch lengths.
Hierarchical clustering of Thioalkalivibrio strains based on the presence/absence of sulfur oxidation-related genes leads to a dendrogram in which strains that are assigned to individual genomic groups are found on the same branch. Whereas the sulfur gene clustering broadly groups strains within their genomic groups, the order of these clusters does not mirror the phylogenomic distribution of strains. For example, in ANIb analysis, the Kenyan isolate ALJT was shown to be most closely related to another Kenyan strain ALJ16, but based on sulfur gene content it is more similar to strain ALMg11 and the Tv. versutus/Tv. jannaschii group. Similarly, the location in which the strain was initially found seems to be uncorrelated to the distribution of sulfur genes in its genome. Ultimately, the major differences between Thioalkalivibrio strains are the presence or absence of aprBA/sat, sorAB, and the presence of thiocyanate dehydrogenase in ten genomes. The dsr genes, sqr and sulfur oxygenase/reductase are only found in strains that are phylogenetically divergent from the rest of the genus (with the exception of SOR in ALMg11) and thiocyanate hydrolase is only found in a single species, Tv. thiocyanodenitrificans.
The maximum likelihood tree of SoxB amino acid sequences follows a pattern previously seen in the 16S rRNA gene sequence analyses of the type strains of Thioalkalivibrio (Sorokin et al., 2013). Herein, type strains ARhD1T (Tv. thiocyanodenitrificans), ALJDT (Tv. denitrificans) and HL-EbGr7T (Tv. sulfidiphilus) are separated from the remaining type strains by the anoxygenic phototrophic SOB of the Ectothiorhodospiraceae family, such as Ectothiorhodosinus, Ectothiorhodospira, and Thiorhodospira. This holds true for both group I and group II in the SoxB tree (Figure 2), although in group I strain ALJDT is split off by Halorhodospira sequences rather than by Ectothiorhodospira and Ectothiorhodosinus sequences. The division between the ‘core’ soxB gene (groups IIa and IIb combined contain a single representative from all strains) and the ‘accessory’ soxB gene (group I comprises a sequence from all strains that have two copies of soxB, except strain ALJDT) is interesting. A second copy of soxB is present in all strains comprising the clade of species 1–15 in the ANIb tree of Thioalkalivibrio (Ahn et al., 2017; Figure 2). This is possibly the result of a single duplication/horizontal gene transfer event, although it is not possible to infer this definitively from the available data. In this hypothesis, strain ALJDT acquired its second copy of soxB through a separate event. A phylogenetic tree based on AprBA amino acid sequences published previously by Watanabe et al. (2016; Supplementary Figure S3, therein) similarly shows a separation of Tv. thiocyanodenitrificans and Tv. sulfidiphilus from the rest of the Thioalkalivibrio sequences (this tree does not contain sequences from Tv. denitrificans and strain ALJ17), although in this case they are found on a branch with sequences from the family Chromatiaceae.
The phylogenetic tree of FccB (Figure 4), the catalytic FAD-containing subunit of flavocytochrome c, shows all Thioalkalivibrio sequences on a single branch, closely related to other Ectothiorhodospiraceae sequences. On the other hand, sequences from other Chromatiales and Thiotrichales SOB, as well as the betaproteobacterial SOB, are found in multiple clusters spread across the tree, which may be the result of horizontal gene transfer events. However, bootstrap values for most branches are low, making it impossible to infer a strong conclusion from this data.
Of the four ortholog groups that contain fccA sequences, only one contains a representative from all Thioalkalivibrio strains. One of the fccA copies found in strain AKL11 was not located next to an fccB gene and may not function as the electron acceptor to sulfide dehydrogenase. The remaining fccA ortholog groups show a diversity of genomic contexts and we hypothesize that these fccAB clusters were acquired through horizontal gene transfer. The in vivo role of multiple fccAB genes will have to be confirmed with physiological experiments. Likewise, it is not known whether the presence of different genes in the direct genomic context of fccAB has any influence on its expression. A comparison with genome contexts of fccAB in a diverse group of sulfur-oxidizing bacteria using IMG (Markowitz et al., 2014), including Allochromatium vinosum, Chlorobium limicola, Paracoccus denitrificans, and Thiobacillus denitrificans showed no conservation of surrounding genes.
The function of additional copies of soxX is even less clear, as these are in most cases found in soxX-soxX-soxA operons and three of the phylogenetically divergent strains (ALJDT, HL-EbGr7T, and ALJ17) have soxX-soxA-soxX-soxA-soxX-soxA-soxX-soxA operons. In this case, genetic knock-out experiments would likely provide insight into the function of these genes; however, no generically applicable system for genetic manipulation of Thioalkalivibrio currently exists. Additionally, gene expression studies could be used to study the relative expression these multiple copies and show whether all, or just some, are utilized during thiosulfate oxidation.
Genomic contexts for soxB and soeA show a mostly straightforward picture: one genotype which is present in all Thioalkalivibrio strains and one which is present in genomes that have additional copies of these genes. Phylogenetic analysis of soxB showed that the second copies of this gene present in 57 genomes were likely acquired from related Ectothiorhodospiraceae (Figure 2).
A number of sulfur oxidation genes/pathways shown in Figure 1 were detected in each of the 75 strains analyzed in this study. These are: fccAB, which catalyzes the oxidation of sulfide; the truncated sox system, soxAXYZB, which oxidizes thiosulfate to sulfur and sulfate; and soeABC, which oxidizes sulfite to sulfate. A major open question is the pathway involved in the oxidation of sulfur to sulfite. This separates the Thioalkalivibrio strains analyzed in this study in two groups: six strains in which the reverse dissimilatory sulfite pathway was detected and 69 strains which were rDsr-negative. The taxonomic status of four of the six rDsr-positive Thioalkalivibrio strains has been questioned, whereas phylogenomic analysis of Tv. paradoxus ARh1T and Tv. nitratireducens ALEN2T was ultimately inconclusive (Ahn et al., 2017).
A sulfide oxidation mechanism that involves the enzyme persulfide dioxygenase (PDO), which oxidizes the persulfide group of glutathione persulfide (GSSH) to produce GSH and sulfite, was described for heterotrophic sulfur-oxidizing bacteria (Xin et al., 2016; Xia et al., 2017). However, this pathway involves sulfide:quinone reductase, which was only detected in two Thioalkalivibrio strains—both of which are rDsr-positive. Additionally, transcriptomics experiments with persulfide dioxygenase knock-out mutants of Acidithiobacillus caldus did not support a role for PDO in sulfur oxidation in this rDsr and SoxCD negative species (Wu et al., 2017). Another candidate pathway for the oxidation of sulfur is the Hdr-like proteins, which were detected in the genomes of all rDsr-negative strains, as well as in four rDsr-positive ones (ALJDT, HL-EbGr7T, ALJ17, and ALEN2T). Although in vitro biochemical evidence that the Hdr-like pathway catalyzes this reaction has not been published to date, transcriptomics experiments have demonstrated its over expression during growth with sulfur compounds in Acidithiobacillus ferrooxidans (Quatrini et al., 2009) and a recent study involving the dimethyl sulfide-oxidizing bacterium Hyphomicrobium denitrificans showed an increase in Hdr enzyme production during growth on thiosulfate. In the same study it was demonstrated that disrupting the hdr operon abolished thiosulfate oxidation, leaving only a low rate of thiosulfate consumption through a different pathway involving the formation of tetrathionate (Koch and Dahl, 2018). Another recent study highlighted the association of the hdr-like operon with lipoate-binding proteins, which are proposed to function as sulfur carriers presenting the substrate to the catalytic sites of the Hdr-like complex (Cao et al., 2018).
Another question is why many genomes contain multiple putative pathways for the oxidation of sulfite to sulfate. While putative soeABC genes were detected in the genomes of all strains in this study, they are the sole genes implicated in sulfite oxidation in only 12 strains. All other strains contain various combinations of soeABC, aprBA, and sorAB. Additionally, sorAB was previously reported as not widely distributed among Ectothiorhodospiraceae (Ghosh and Dam, 2009). The aprBA genes were shown to not be essential for sulfite oxidation in Allochromatium vinosum, although they did confer a growth advantage under specific conditions (Dahl, 1996; Sánchez et al., 2001). Since complete oxidation of reduced sulfur compounds to sulfate is a common trait among Thioalkalivibrio species, we had expected to find at least one sulfite oxidation system to be ubiquitous among the strains studied here. The only candidate for this currently is soeABC, but phylogenetic analysis shows that not all sequences group together in a tree, which would be expected for a core metabolism gene. The role and importance of soeABC, sorAB, and aprBA in sulfite oxidation in Thioalkalivibrio will have to be studied in more detail using gene expression studies and genetic manipulation.
A model of sulfur metabolism in Thioalkalivibrio is shown in Figure 6, similar to earlier models based on a single strain (Ang et al., 2017) or a limited set of thiocyanate-utilizing species (Berben et al., 2017).
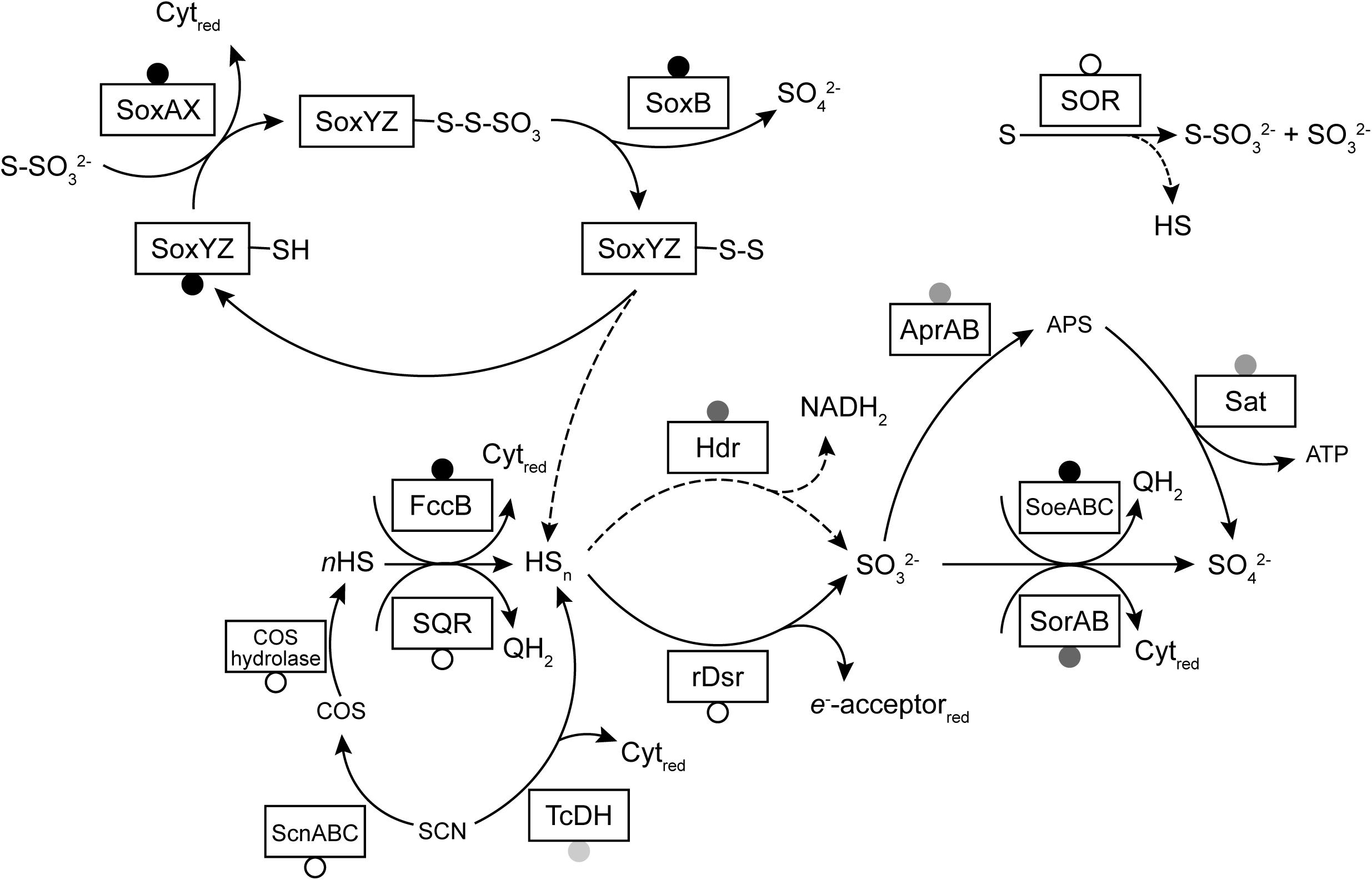
Figure 6. A model of sulfur oxidation pathways in Thioalkalivibrio. Boxes represent enzymes for which genes were detected in the studied genomes. Circles indicate the prevalence of these genes: black, present in all genomes; gray, present in >10 genomes (and <75); unfilled, present in <10 genomes. Dashed lines indicate reactions with unknown mechanism (in the case of sulfane sulfur dissociation from SoxYZ) or low rates (sulfur reduction to sulfide by SOR). The reactions of rDsr and the Hdr-like system have been simplified for clarity.
Apart from the genes of the rDsr pathway, as discussed above, there are four other genes that are uncommon among Thioalkalivibrio. The distribution and genomic context of thiocyanate dehydrogenase (tcdh; found in 10 genomes) and thiocyanate hydrolase (scnABC; found in a single genome) were discussed in detail in a previous study (Berben et al., 2017), where it was shown that the tcdh gene is found in two genomic contexts (one in ARh 1T and ALEN 2T and one in the other cyanate-pathway using strains) that include genes for fccAB, copper-resistance proteins (copCD) and a twin-arginine transporter (tatA). Sulfur oxygenase/reductase (SOR) from Tv. paradoxus was recently biochemically characterized and it was shown that the enzyme has an atypically low reductase activity (Rühl et al., 2017), although the in vivo role of SOR in Thioalkalivibrio has not been determined yet. Genes for tetrathionate degradation were not detected at all in the genomes of Thioalkalivibrio, which appears to contradict reported tetrathionate utilization by some species. However, tetrathionate is unstable in alkaline environments and can spontaneously decompose to form thiosulfate, when the pH is greater than 9 (Varga and Horváth, 2007). Carbon disulfide utilization, at low rates, was only reported for Tv. paradoxus ARh 1 (Sorokin et al., 2002b) and the biochemistry of this reaction remains unknown. A putative homolog of carbon disulfide hydrolase (Smeulders et al., 2011) was not detected in this genome. While oxygen-dependent CS2 consumption was demonstrated in cell-free extracts of Paracoccus denitrificans (Jordan et al., 1997), no further enzymatic characterization was performed. The genome of Tv. nitratireducens ALEN 2T is the only one to contain a gene encoding a putative carbon disulfide hydrolase, but possible growth with CS2 was never tested for this strain.
The phylogenetic tree of putative SQR sequences shown in Figure 3 shows that sequences from Tv. nitratireducens ALEN2T and Tv. thiocyanodenitrificans ARhD1T fall within the type I SQR, as defined by Marcia et al. (2010). The SQR enzymes of this group were shown to have a high affinity for sulfide and are involved in respiratory processes, as well as in photosynthesis in phototrophic sulfide oxidizers (Marcia et al., 2010). In ALEN2T and ARhD1T SQR is likely required for anaerobic growth on sulfide, although this does not explain the fact that SQR was not detected in the genome of Tv. denitrificans ALJDT, which is also capable of anaerobic growth (Sorokin et al., 2001a). The remaining Thioalkalivibrio sequences form a sister clade to type II SQR that is strongly supported by bootstrap analysis. Type II SQR is present in eukaryotes and bacteria. Although SQR activity has been demonstrated in bacterial type II SQR (Han and Perner, 2016; Shen et al., 2016), the affinity for sulfide is low (in the millimolar range) and their physiological roles are unclear (Marcia et al., 2009, 2010). In Thioalkalivibrio, gene expression studies and biochemical characterization of the expressed protein are necessary to determine whether this group is active in sulfide oxidation or has an alternative function.
In this paper we have presented the diversity and distribution of genes related to sulfur oxidation in 75 strains of Thioalkalivibrio, a group of haloalkaliphilic and chemolithoautotrophic SOB from soda lakes. We have shown that flavocytochrome c, the truncated sox system (soxAXYZB) and sulfite:quinone oxidoreductase (soeABC) are present in all strains. The pathway from elemental sulfur to sulfite is currently not resolved for all Thioalkalivibrio, as only six genomes encode the dissimilatory sulfite reductase system. The hdr-like operon is a good candidate for sulfur oxidation, although the release of sulfite from this enzyme system has yet to be demonstrated. It is found in all dsr-negative strains and the genomes of four strains contain both, although the physiological consequences of this are unknown.
Hierarchical clustering showed that the sulfur gene repertoire of individual strains correlates well with genomic groups previously defined by ANIb analysis. Phylogenetic analysis of SoxB, FccB, and SoeA amino acid sequences reaffirms the complex evolutionary history of Thioalkalivibrio that was reported in previous analyses of 16S rRNA and CbbL sequences (Tourova et al., 2006; Ahn et al., 2017). Genomic contexts of genes present in multiple copies show that there is generally one genotype with sequences from all strains and one or more genotypes additional copies that were likely acquired through HGT.
There are a number of major unresolved questions regarding the sulfur metabolism of Thioalkalivibrio, most notably the oxidation of elemental sulfur as mentioned above. The in vivo function of the FAD-dependent oxidoreductases that form a sister group to type II SQR sequences provides another avenue for further research.
TB, LO, DS, and GM contributed to the study design. LO performed the grouping of annotated genes in ortholog groups. TB analyzed the distribution and phylogeny of specific sulfur metabolism-related genes and drafted the manuscript. LO, DS, and GM provided feedback on the analyses and critically reviewed the manuscript. All authors read and approved the final version of the manuscript.
TB, LO, and GM were supported by ERC grant PARASOL (322551); DS was supported by the Russian Ministry of Sciences and Higher Education and by the Russian Foundation for Basic Research (RFBR, Grant 19-04-00401). Genome sequencing was performed at the United States Department of Energy Joint Genome Institute under contract number DE-AC02-05CH11231.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00160/full#supplementary-material
FIGURE S1 | Maximum likelihood tree of SoxY sequences, based on 500 bootstrap replicates. Sequences of desulfoferredoxin-family proteins were used as the outgroup and pruned from the tree. The scale bar represents % difference.
FIGURE S2 | Maximum likelihood tree of SoxZ sequences, based on 500 bootstrap replicates. Sequences of quinoprotein dehydrogenase-associated SoxYZ-like proteins were used as outgroup and later pruned from the tree. The scale bar represents % differences.
FIGURE S3 | Maximum likelihood tree of SoeA sequences, based on 500 bootstrap replicates. Sequences of sulfite:cytochrome c oxidoreductase subunit A (SorA) were used as outgroup and pruned from the tree. The scale bar represents % differences. Bootstrap values below 50% are not shown. Parenthesized numbers indicate the number of sequences included in a collapsed branch.
TABLE S1 | Overview of Thioalkalivibrio strains included in this study, including geographical location from which the strain was isolated, genome size, number of protein-coding genes, genome accession number and the results of the CheckM analysis.
TABLE S2 | Groups of orthologous proteins as predicted by OrthoMCL. Rows represent the different ortholog groups. Accession numbers of protein sequences included in an ortholog group are separated by tabs on the same line.
Abascal, F., Zardoya, R., and Posada, D. (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105. doi: 10.1093/bioinformatics/bti263
Ahn, A.-C., Meier-Kolthoff, J. P., Overmars, L., Richter, M., Woyke, T., Sorokin, D. Y., et al. (2017). Genomic diversity within the haloalkaliphilic genus Thioalkalivibrio. PLoS One 12:e0173517. doi: 10.1371/journal.pone.0173517
Algueró, M., Dahl, C., and Trüper, H. G. (1988). Partial purification and characterization of ADP sulfurylase from the purple sulfur bacterium Thiocapsa roseopersicina. Microbiologia 4, 149–160.
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Ang, W. K., Mahbob, M., Dhouib, R., and Kappler, U. (2017). Sulfur compound oxidation and carbon co-assimilation in the haloalkaliphilic sulfur oxidizers Thioalkalivibrio versutus and Thioalkalimicrobium aerophilum. Res. Microbiol. 168, 255–265. doi: 10.1016/j.resmic.2016.12.004
Arakawa, T., Kawano, Y., Kataoka, S., Katayama, Y., Kamiya, N., Yohda, M., et al. (2007). Structure of thiocyanate hydrolase: a new nitrile hydratase family protein with a novel five-coordinate cobalt(III) center. J. Mol. Biol. 366, 1497–1509. doi: 10.1016/j.jmb.2006.12.011
Banciu, H., Sorokin, D. Y., Galinski, E. A., Muyzer, G., Kleerebezem, R., and Kuenen, J. G. (2004). Thialkalivibrio halophilus sp. nov., a novel obligately chemolithoautotrophic, facultatively alkaliphilic, and extremely salt-tolerant, sulfur-oxidizing bacterium from a hypersaline alkaline lake. Extremophiles 8, 325–334. doi: 10.1007/s00792-004-0391-6
Berben, T., Overmars, L., Sorokin, D. Y., and Muyzer, G. (2017). Comparative genome analysis of three thiocyanate oxidizing Thioalkalivibrio species isolated from soda lakes. Front. Microbiol. 8:254. doi: 10.3389/fmicb.2017.00254
Boughanemi, S., Lyonnet, J., Infossi, P., Bauzan, M., Kosta, A., Lignon, S., et al. (2016). Microbial oxidative sulfur metabolism: biochemical evidence of the membrane-bound heterodisulfide reductase-like complex of the bacterium Aquifex aeolicus. FEMS Microbiol. Lett. 363:fnw156. doi: 10.1093/femsle/fnw156
Bowles, M. W., Mogollon, J. M., Kasten, S., Zabel, M., and Hinrichs, K.-U. (2014). Global rates of marine sulfate reduction and implications for sub-sea-floor metabolic activities. Science 344, 889–891. doi: 10.1126/science.1249213
Brüser, T., Selmer, T., and Dahl, C. (2000). “ADP sulfurylase” from Thiobacillus denitrificans is an adenylylsulfate:phosphate adenylyltransferase and belongs to a new family of nucleotidyltransferases. J. Biol. Chem. 275, 1691–1698. doi: 10.1074/jbc.275.3.1691
Cao, X., Koch, T., Steffens, L., Finkensieper, J., Zigann, R., Cronan, J. E., et al. (2018). Lipoate-binding proteins and specific lipoate-protein ligases in microbial sulfur oxidation reveal an atpyical role for an old cofactor. eLife 7:e37439. doi: 10.7554/eLife.37439
Chen, Z. W., Koh, M., Van Driessche, G., Van Beeumen, J. J., Bartsch, R. G., Meyer, T. E., et al. (1994). The structure of flavocytochrome c sulfide dehydrogenase from a purple phototrophic bacterium. Science 266, 430–432. doi: 10.1126/science.7939681
Dahl, C. (1996). Insertional gene inactivation in a phototrophic sulphur bacterium: APS-reductase-deficient mutants of Chromatium vinosum. Microbiology 142, 3363–3372. doi: 10.1099/13500872-142-12-3363
Dahl, C. (2015). Cytoplasmic sulfur trafficking in sulfur-oxidizing prokaryotes. IUBMB Life 67, 268–274. doi: 10.1002/iub.1371
Dahl, C., Engels, S., Pott-sperling, A. S., Schulte, A., Sander, J., Lübbe, Y., et al. (2005). Novel genes of the dsr gene cluster and evidence for close interaction of Dsr proteins during sulfur oxidation in the phototrophic sulfur bacterium Allochromatium vinosum. PLoS One 12:e0183668. doi: 10.1128/JB.187.4.1392-1404.2005
Dahl, C., Franz, B., Hensen, D., Kesselheim, A., and Zigann, R. (2013). Sulfite oxidation in the purple sulfur bacterium Allochromatium vinosum: Identification of SoeABC as a major player and relevance of SoxYZ in the process. Microbiology 159, 2626–2638. doi: 10.1099/mic.0.071019-0
Di Tommaso, P., Moretti, S., Xenarios, I., Orobitg, M., Montanyola, A., Chang, J.-M., et al. (2011). T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39, W13–W17. doi: 10.1093/nar/gkr245
Fischer, S., Brunk, B. P., Chen, F., Gao, X., Omar, S., Iodice, J. B., et al. (2011). Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr. Protoc. Bioinformatics 6, 12.1–12.19. doi: 10.1002/0471250953.bi0612s35
Foti, M., Sorokin, D. Y., Lomans, B., Mussman, M., Zacharova, E. E., Pimenov, N. V., et al. (2007). Diversity, activity, and abundance of sulfate-reducing bacteria in saline and hypersaline soda lakes. Appl. Environ. Microbiol. 73, 2093–2100. doi: 10.1128/AEM.02622-06
Friedrich, C. G., Quentmeier, A., Bardischewsky, F., Rother, D., Kraft, R., Kostka, S., et al. (2000). Novel genes coding for lithotrophic sulfur oxidation of Paracoccus pantotrophus GB17. J. Bacteriol. 182, 4677–4687. doi: 10.1128/JB.182.17.4677-4687.2000
Frigaard, N.-U. U., and Dahl, C. (2009). Sulfur metabolism in phototrophic sulfur bacteria. Adv. Microb. Physiol. 54, 103–200. doi: 10.1016/S0065-2911(08)00002-7
Fritz, G., Büchert, T., Huber, H., Stetter, K. O., and Kroneck, P. M. (2000). Adenylylsulfate reductases from archaea and bacteria are 1:1 alphabeta-heterodimeric iron-sulfur flavoenzymes–high similarity of molecular properties emphasizes their central role in sulfur metabolism. FEBS Lett. 473, 63–66. doi: 10.1016/S0014-5793(00)01500-3
Ghosh, W., and Dam, B. (2009). Biochemistry and molecular biology of lithotrophic sulfur oxidation by taxonomically and ecologically diverse bacteria and archaea. FEMS Microbiol. Rev. 33, 999–1043. doi: 10.1111/j.1574-6976.2009.00187.x
Han, Y., and Perner, M. (2016). Sulfide consumption in Sulfurimonas denitrificans and heterologous expression of its three sulfide-quinone reductase homologs. J. Bacteriol. 198, 1260–1267. doi: 10.1128/JB.01021-15
Jannasch, H. W., and Mottl, M. J. (1985). Geomicrobiology of deep-sea hydrothermal vents. Science 229, 717–725. doi: 10.1126/science.229.4715.717
Janssen, A. J. H., Lens, P. N. L., Stams, A. J. M., Plugge, C. M., Sorokin, D. Y., Muyzer, G., et al. (2009). Application of bacteria involved in the biological sulfur cycle for paper mill effluent purification. Sci. Total Environ. 407, 1333–1343. doi: 10.1016/j.scitotenv.2008.09.054
Jordan, S. L., McDonald, I. R., Kraczkiewicz-Dowjat, A. J., Kelly, D. P., Rainey, F. A., Murrell, J. C., et al. (1997). Autotrophic growth on carbon disulfide is a property of novel strains of Paracoccus denitrificans. Arch. Microbiol. 168, 225–236. doi: 10.1007/s002030050492
Kappler, U., Bennett, B., Rethmeier, J., Schwarz, G., Deutzmann, R., McEwan, A. G., et al. (2000). Sulfite:Cytochrome c oxidoreductase from Thiobacillus novellus. Purification, characterization, and molecular biology of a heterodimeric member of the sulfite oxidase family. J. Biol. Chem. 275, 13202–13212. doi: 10.1074/jbc.275.18.13202
Kappler, U., and Dahl, C. (2001). Enzymology and molecular biology of prokaryotic sulfite oxidation. FEMS Microbiol. Lett. 203, 1–9. doi: 10.1111/j.1574-6968.2001.tb10813.x
Katayama, Y., Matsushita, Y., Kaneko, M., Kondo, M., Mizuno, T., and Nyunoya, H. (1998). Cloning of genes coding for the three subunits of thiocyanate hydrolase of Thiobacillus thioparus THI 115 and their evolutionary relationships to nitrile hydratase. J. Bacteriol. 180, 2583–2589.
Kelly, D. P., Shergill, J. K., Lu, W. P., and Wood, A. P. (1997). Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie Van Leeuwenhoek 71, 95–107. doi: 10.1023/A:1000135707181
Koch, T., and Dahl, C. (2018). A novel bacterial sulfur oxidation pathway provides a new link between the cycles of organic and inorganic sulfur compounds. ISME J. 12, 2479–2491. doi: 10.1038/s41396-018-0209-7
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Le, S. Q., and Gascuel, O. (2008). An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320. doi: 10.1093/molbev/msn067
Marcia, M., Ermler, U., Peng, G., and Michel, H. (2009). The structure of Aquifex aeolicus sulfide:quinone oxidoreductase, a basis to understand sulfide detoxification and respiration. Proc. Natl. Acad. Sci. U.S.A. 106, 9625–9630. doi: 10.1073/pnas.0904165106
Marcia, M., Ermler, U., Peng, G., and Michel, H. (2010). A new structure-based classification of sulfide:quinone oxidoreductases. Proteins Struct. Funct. Bioinforma. 78, 1073–1083. doi: 10.1002/prot.22665
Markowitz, V. M., Chen, I.-M. A., Palaniappan, K., Chu, K., Szeto, E., Pillay, M., et al. (2014). IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 42, D560–D567. doi: 10.1093/nar/gkt963
Méndez-García, C., Peláez, A. I., Mesa, V., Sánchez, J., Golyshina, O. V., and Ferrer, M. (2015). Microbial diversity and metabolic networks in acid mine drainage habitats. Front. Microbiol. 6:475. doi: 10.3389/fmicb.2015.00475
Meyer, B., Imhoff, J. F., and Kuever, J. (2007). Molecular analysis of the distribution and phylogeny of the soxB gene among sulfur-oxidizing bacteria - evolution of the Sox sulfur oxidation enzyme system. Environ. Microbiol. 9, 2957–2977. doi: 10.1111/j.1462-2920.2007.01407.x
Muyzer, G., Sorokin, D. Y., Mavromatis, K., Lapidus, A., Clum, A., Ivanova, N., et al. (2011a). Complete genome sequence of “Thioalkalivibrio sulfidophilus” HL-EbGr7. Stand. Genomic Sci. 4, 23–35. doi: 10.4056/sigs.1483693
Muyzer, G., Sorokin, D. Y., Mavromatis, K., Lapidus, A., Foster, B., Sun, H., et al. (2011b). Complete genome sequence of Thioalkalivibrio sp. K90mix. Stand. Genomic Sci. 5, 341–355. doi: 10.4056/sigs.2315092
Ogawa, T., Noguchi, K., Saito, M., Nagahata, Y., Kato, H., Ohtaki, A., et al. (2013). Carbonyl sulfide hydrolase from Thiobacillus thioparus strain thi115 is one of the β-carbonic anhydrase family enzymes. J. Am. Chem. Soc. 135, 3818–3825. doi: 10.1021/ja307735e
Pott, A. S., and Dahl, C. (1998). Sirohaem sulfite reductase and other proteins encoded by genes at the dsr locus of Chromatium vinosum are involved in the oxidation of intracellular sulfur. Microbiology 144, 1881–1894. doi: 10.1099/00221287-144-7-1881
Quatrini, R., Appia-Ayme, C., Denis, Y., Jedlicki, E., Holmes, D. S., and Bonnefoy, V. (2009). Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans. BMC Genomics 10:394. doi: 10.1186/1471-2164-10-394
Rühl, P., Pöll, U., Braun, J., Klingl, A., and Kletzin, A. (2017). A sulfur oxygenase from the haloalkaliphilic bacterium Thioalkalivibrio paradoxus with Atypically low reductase activity. J. Bacteriol. 199:e00675-16. doi: 10.1128/JB.00675-16
Sánchez, O., Ferrera, I., Dahl, C., and Mas, J. (2001). In vivo role of adenosine-5’-phosphosulfate reductase in the purple sulfur bacterium Allochromatium vinosum. Arch. Microbiol. 176, 301–305. doi: 10.1007/s002030100327
Shen, J., Peng, H., Zhang, Y., Trinidad, J. C., and Giedroc, D. P. (2016). Staphylococcus aureus sqr encodes a type ii sulfide:quinone oxidoreductase and impacts reactive sulfur speciation in cells. Biochemistry 55, 6524–6534. doi: 10.1021/acs.biochem.6b00714
Smeulders, M. J., Barends, T. R. M., Pol, A., Scherer, A., Zandvoort, M. H., Udvarhelyi, A., et al. (2011). Evolution of a new enzyme for carbon disulphide conversion by an acidothermophilic archaeon. Nature 478, 412–416. doi: 10.1038/nature10464
Sorokin, D. Y. (2003). Thialkalivibrio nitratireducens sp. nov., a nitrate-reducing member of an autotrophic denitrifying consortium from a soda lake. Int. J. Syst. Evol. Microbiol. 53, 1779–1783. doi: 10.1099/ijs.0.02615-0
Sorokin, D. Y., Banciu, H., Robertson, L. A., Kuenen, J. G., Muntyan, M. S., Muyzer, G., et al. (2013). “Halophilic and haloalkaliphilic sulfur-oxidizing bacteria,” in The Prokaryotes, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer), 529–554.
Sorokin, D. Y., Banciu, H. L., and Muyzer, G. (2015). Functional microbiology of soda lakes. Curr. Opin. Microbiol. 25, 88–96. doi: 10.1016/j.mib.2015.05.004
Sorokin, D. Y., Berben, T., Melton, E. D., Overmars, L., Vavourakis, C. D., and Muyzer, G. (2014). Microbial diversity and biogeochemical cycling in soda lakes. Extremophiles 18, 791–809. doi: 10.1007/s00792-014-0670-9
Sorokin, D. Y., Gorlenko, V. M., Tourova, T. P. T. P., Tsapin, A. I., Nealson, K. H., Kuenen, G. J., et al. (2002a). Thioalkalimicrobium cyclicum sp. nov. and Thioalkalivibrio jannaschii sp. nov., novel species of haloalkaliphilic, obligately chemolithoautotrophic sulfur-oxidizing bacteria from hypersaline alkaline Mono Lake (California). Int. J. Syst. Evol. Microbiol. 52, 913–920.
Sorokin, D. Y., Tourova, T. P., Lysenko, A. M., Mityushina, L. L., and Kuenen, J. G. (2002b). Thioalkalivibrio thiocyanoxidans sp. nov. and Thioalkalivibrio paradoxus sp. nov., novel alkaliphilic, obligately autotrophic, sulfur-oxidizing bacteria capable of growth on thiocyanate, from soda lakes. Int. J. Syst. Evol. Microbiol. 52, 657–664.
Sorokin, D. Y., Kuenen, J. G., and Muyzer, G. (2011). The microbial sulfur cycle at extremely haloalkaline conditions of soda lakes. Front. Microbiol. 2:44. doi: 10.3389/fmicb.2011.00044
Sorokin, D. Y., Muntyan, M. S., Panteleeva, A. N., and Muyzer, G. (2012). Thioalkalivibrio sulfidiphilus sp. nov., a haloalkaliphilic, sulfur-oxidizing gammaproteobacterium from alkaline habitats. Int. J. Syst. Evol. Microbiol. 62, 1884–1889. doi: 10.1099/ijs.0.034504-0
Sorokin, D. Y., Rusanov, I. I., Pimenov, N. V., Tourova, T. P., Abbas, B., and Muyzer, G. (2010). Sulfidogenesis under extremely haloalkaline conditions in soda lakes of Kulunda Steppe (Altai, Russia). FEMS Microbiol. Ecol. 73, 278–290. doi: 10.1111/j.1574-6941.2010.00901.x
Sorokin, D. Y., Tourova, T. P., Antipov, A. N., Muyzer, G., and Kuenen, J. G. (2004). Anaerobic growth of the haloalkaliphilic denitrifying sulfur-oxidizing bacterium Thialkalivibrio thiocyanodenitrificans sp. nov. with thiocyanate. Microbiology 150, 2435–2442. doi: 10.1099/mic.0.27015-0
Sorokin, D. Y., Lysenko, A. M., Mityushina, L. L., Tourova, T. P., Jones, B. E., Rainey, F. A., et al. (2001a). Thioalkalimicrobium aerophilum gen. nov., sp. nov. and Thioalkalimicrobium sibericum sp. nov., and Thioalkalivibrio versutus gen. nov., sp. nov., Thioalkalivibrio nitratis sp.nov., novel and Thioalkalivibrio denitrificancs sp. nov., novel obligately alkal. Int. J. Syst. Evol. Microbiol. 51, 565–580.
Sorokin, D. Y., Tourova, T. P., Lysenko, A. M., and Kuenen, J. G. (2001b). Microbial thiocyanate utilization under highly alkaline conditions. Appl. Environ. Microbiol. 67, 528–538.
Sorokin, D. Y., van den Bosch, P. L. F., Abbas, B., Janssen, A. J. H., and Muyzer, G. (2008). Microbiological analysis of the population of extremely haloalkaliphilic sulfur-oxidizing bacteria dominating in lab-scale sulfide-removing bioreactors. Appl. Microbiol. Biotechnol 80, 965–975. doi: 10.1007/s00253-008-1598-8
Sousa, F. M., Pereira, J. G., Marreiros, B. C., and Pereira, M. M. (2018). Taxonomic distribution, structure/function relationship and metabolic context of the two families of sulfide dehydrogenases: SQR and FCSD. Biochim. Biophys. Acta Bioenerg. 1859, 742–753. doi: 10.1016/j.bbabio.2018.04.004
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stamatakis, A. (2015). Using RAxML to infer phylogenies. Curr. Protoc. Bioinforma 51, 6.14.1–6.14.14. doi: 10.1002/0471250953.bi0614s51
Tourova, T. P., Spiridonova, E. M., Berg, I. A., Kuznetsov, B. B., and Sorokin, D. Y. (2006). Occurrence, phylogeny and evolution of ribulose-1,5-bisphosphate carboxylase/oxygenase genes in obligately chemolithoautotrophic sulfur-oxidizing bacteria of the genera Thiomicrospira and Thioalkalimicrobium. Microbiology 152, 2159–2169. doi: 10.1099/mic.0.28699-0
Varga, D., and Horváth, A. K. (2007). Kinetics and mechanism of the decomposition of tetrathionate ion in alkaline medium. Inorg. Chem. 46, 7654–7661. doi: 10.1021/ic700992u
Vavourakis, C. D., Andrei, A.-S., Mehrshad, M., Ghai, R., Sorokin, D. Y., and Muyzer, G. (2018). A metagenomics roadmap to the uncultured genome diversity in hypersaline soda lake sediments. Microbiome 6:168. doi: 10.1186/s40168-018-0548-7
Venceslau, S. S., Stockdreher, Y., Dahl, C., and Pereira, I. A. C. (2014). The “bacterial heterodisulfide” DsrC is a key protein in dissimilatory sulfur metabolism. Biochim. Biophys. Acta 1837, 1148–1164. doi: 10.1016/j.bbabio.2014.03.007
Wasmund, K., Mußmann, M., and Loy, A. (2017). The life sulfuric: microbial ecology of sulfur cycling in marine sediments. Environ. Microbiol. Rep. 9, 323–344. doi: 10.1111/1758-2229.12538
Watanabe, T., Kojima, H., and Fukui, M. (2016). Identity of major sulfur-cycle prokaryotes in freshwater lake ecosystems revealed by a comprehensive phylogenetic study of the dissimilatory adenylylsulfate reductase. Sci. Rep. 6:36262. doi: 10.1038/srep36262
Wu, W., Pang, X., Lin, J., Liu, X., Wang, R., Lin, J., et al. (2017). Discovery of a new subgroup of sulfur dioxygenases and characterization of sulfur dioxygenases in the sulfur metabolic network of Acidithiobacillus caldus. PLoS One 12:e0183668. doi: 10.1371/journal.pone.0183668
Xia, Y., Lü, C., Hou, N., Xin, Y., Liu, J., Liu, H., et al. (2017). Sulfide production and oxidation by heterotrophic bacteria under aerobic conditions. ISME J. 11, 2754–2766. doi: 10.1038/ismej.2017.125
Keywords: Thioalkalivibrio, soda lakes, chemolithoautotrophs, sulfur oxidation, thiocyanate oxidation, denitrification
Citation: Berben T, Overmars L, Sorokin DY and Muyzer G (2019) Diversity and Distribution of Sulfur Oxidation-Related Genes in Thioalkalivibrio, a Genus of Chemolithoautotrophic and Haloalkaliphilic Sulfur-Oxidizing Bacteria. Front. Microbiol. 10:160. doi: 10.3389/fmicb.2019.00160
Received: 05 November 2018; Accepted: 22 January 2019;
Published: 14 February 2019.
Edited by:
Christiane Dahl, Universität Bonn, GermanyReviewed by:
Marianne Guiral, Centre National de la Recherche Scientifique (CNRS), FranceCopyright © 2019 Berben, Overmars, Sorokin and Muyzer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gerard Muyzer, Zy5tdWlqemVyQHV2YS5ubA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.