
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 04 February 2019
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00076
 Daniela Ceccarelli1*
Daniela Ceccarelli1* Arie Kant1
Arie Kant1 Alieda van Essen-Zandbergen1
Alieda van Essen-Zandbergen1 Cindy Dierikx1†
Cindy Dierikx1† Joost Hordijk1†
Joost Hordijk1† Ben Wit2
Ben Wit2 Dik J. Mevius1,3
Dik J. Mevius1,3 Kees T. Veldman1
Kees T. Veldman1Extended-spectrum β-lactamase (ESBL) and plasmid-mediated AmpC β-lactamase (pAmpC) genes confer resistance to extended spectrum cephalosporin’s. The spread of these genes is mostly facilitated by plasmid-mediated horizontal transfer. National surveillance activities to detect ESBL/pAmpC-producers in commensal bacteria from livestock are in place in the Netherlands since several years. This study aimed at reporting gene and plasmid diversity of commensal ESBL/pAmpC-producing Escherichia coli isolated from healthy animals during surveillance activities between 2007 and 2017. A collection of 2304 extended-spectrum cephalosporin-resistant (ESC-R) E. coli isolated from feces of broilers, dairy cattle, slaughter pigs, turkeys, ducks, and veal calves was investigated and ESBL/pAmpC genes were determined. Gene location of a selection of 473 E. coli isolates was determined and typing of plasmids linked to the ESBL/pAmpC genes was performed. Twenty-two different ESBL/pAmpC genes were identified with blaCTX-M-1 being the most prevalent gene in livestock (43.7%), followed by blaCMY -2 and blaSHV -12, independent of the animal source. Prevalence of typically human associated blaCTX-M-15 was highest in cattle. Less than 10% E. coli isolates owed their ESC-R phenotype to promoter mutations of the chromosomal ampC gene. Majority (92%) of ESBL/pAmpC genes analyzed were plasmid located, with IncI1α being the most represented plasmid family in isolates from all animals, followed by IncF (veal calves, dairy cattle and slaughter pigs), IncK (broilers and laying hens), IncX1 in broilers, and emerging IncX3 in broilers and dairy cattle. Prevalence and molecular diversity of ESC-R E. coli isolated from livestock over an 11-year period revealed a composite scenario of gene-plasmid combinations.
Extended-spectrum β-lactamases (ESBLs) and plasmid-mediated AmpC β-lactamases (pAmpCs) are able to hydrolyse a large variety of β-lactam antibiotics, including cephalosporins and monobactams. The most clinically significant ESBL variants belong to the blaCTX-M, blaTEM, and blaSHV gene families together with pAmpC blaCMY gene family (Bush and Fisher, 2011). The successful spread of ESBL/pAmpC genes is mostly due to their localization on plasmids, resulting in easy transmission between bacteria (Rozwandowicz et al., 2018).
Extended-spectrum cephalosporin-resistant (ESC-R) Enterobacteriaceae have emerged globally in livestock animals during the last decades (Carattoli, 2008), with the consequent concern of animals being a putative source of ESBL/pAmpC-producing bacteria for humans either by direct contact or consumption of contaminated food products, as reviewed by (Ewers et al., 2012). Over the years, measures were implemented to reduce the use of third generation cephalosporins in livestock at national and European level (Speksnijder et al., 2015). Although the impact of transmission from livestock and the food chain on infections in humans is still debated (Madec et al., 2017; Dorado-Garcia et al., 2018), the ESBL/pAmpC reservoir in commensal bacteria from livestock has been increasingly investigated for its potential risk to public health (Michael et al., 2015).
Commensal ESC-R Escherichia coli randomly isolated from livestock feces have been monitored in the Netherlands since 1998, and phenotypic and genotypic results have annually been reported in the Monitoring of Antimicrobial Resistance and Antibiotic Usage in Animals in the Netherlands reports (MARAN Reports). Since 2014, active monitoring through selective culturing and reporting of antimicrobial resistance in several bacteria, including ESC-R E. coli, has become mandatory for member states of the European Union (European Food Safety Authority, 2008, 2013). Results of these activities are yearly published (Maran Reports, 2002/2017) but lack detailed information on plasmid typing and epidemiology.
The aim of this study is to report gene and plasmid diversity observed in ESC-R E. coli isolated from healthy livestock from 2007 to 2017 during surveillance activities in the Netherlands.
All ESC-R E. coli isolates included in this retrospective study originated from fecal samples of livestock collected during different surveillance activities in the Netherlands. Because surveillance activities have changed over the years in terms of sampling and methodologies, full details can be found in the yearly reports (Maran Reports, 2002/2017). Main differences between monitoring activities are briefly described here. Non-selective culturing (2007–2017) was performed by isolation of one randomly selected E. coli colony from a directly inoculated MacConkey agar plate without supplemented antibiotics, each isolate representing one epidemiological unit as prescribed by EFSA guidelines (European Food Safety Authority, 2008). Selective culturing (2014–2017) was performed by overnight incubation of fecal samples in Buffered Peptone Water (BPW) followed by sub-culturing on MacConkey agar plate supplemented with 1 mg/L cefotaxime, according to EURL-AR protocols1. Sampling of ESC-R E. coli via selective isolation was performed on fecal samples from broilers, veal calves, slaughter pigs and dairy cows (European Food Safety Authority, 2013). Outside of mandatory surveillance activities, additional sampling was performed for turkeys in 2011 and 2012 (usually excluded because of low production), laying hens in 2014 and 2016 (typically screened only for Salmonella), and ducks in 2016 (not included in the legislation). Furthermore, ESC-R E. coli isolates obtained during monitoring activities from 2011 to 2013 by the Netherlands Food and Consumer Product Safety Authority (NVWA) with selective culturing (O/N enrichment in BPW followed by selective isolation on MacConkey agar plate with 1 mg/L cefotaxime) of fecal samples from broilers, dairy cattle, slaughter pigs and veal calves were included.
Overall, this retrospective study comprises 2304 ESC-R E. coli (Table 1): 330 ESC-R E. coli from non-selective surveillance (2007–2017), 1580 ESC-R E. coli from selective surveillance (2014–2017), and 394 ESC-R E. coli from NVWA selective surveillance activities (2011–2013).
Along the years, different methods to identify ESBL/pAmpC gene families in ESC-R E. coli have been employed, including miniaturized DNA Microarrays (Identibac AMR-ve, Alere Technologies GmbH) (Batchelor et al., 2008), microarray analysis using the Check-MDR CT-101 array platform (Check-Points, Wageningen, Netherlands) or dedicated PCRs (Geurts et al., 2017). DNA was extracted by using the DNeasy Blood and Tissue kit (QIAGEN, Hilden, Germany) according to the manufacturer’s recommendations or DNA lysate preparation (Veldman et al., 2018). Independent of the screening method applied, gene sequences were confirmed by PCR amplification and DNA sequencing (Liakopoulos et al., 2016). Nucleotide and deduced amino acid sequences were compared with sequences in the Lahey clinic database2 and GenBank. Chromosomal mutations of promoters and attenuators of ampC genes were determined by sequencing and compared to GenBank (Mulvey et al., 2005).
A subset of 473 ESC-R E. coli was selected for genomic localization of ESBL/pAmpC genes: 63 from non-selective surveillance and 410 isolates from selective surveillance (Table 2). Over the years, different selection criteria were applied with the aim of including all ESBL/pAmpC genes detected in each animal species, and taking into consideration existing knowledge of gene-plasmid epidemiology. The chosen number of isolates per gene type was dependent on how prevalent the gene was in a given year i.e., for selective surveillance of broilers in 2014, 43% of samples were blaCTXM-1 positive (n = 116 out of 269), and a third of them (n = 42) were analyzed for genomic localization of blaCTXM-1 gene. For non-selective surveillance based on the knowledge that blaCTXM-1 positive E. coli in broilers are usually associated with IncI1 plasmids (Dierikx et al., 2013), majority of these isolates were not typed. Further details can be found in the corresponding results sections. ESC-R E. coli from 2011 to 2012 were not typed because of a temporary change in research priorities. Plasmid typing results for ESC-R E. coli isolated in 2017 via selective surveillance were not available at the time of writing.
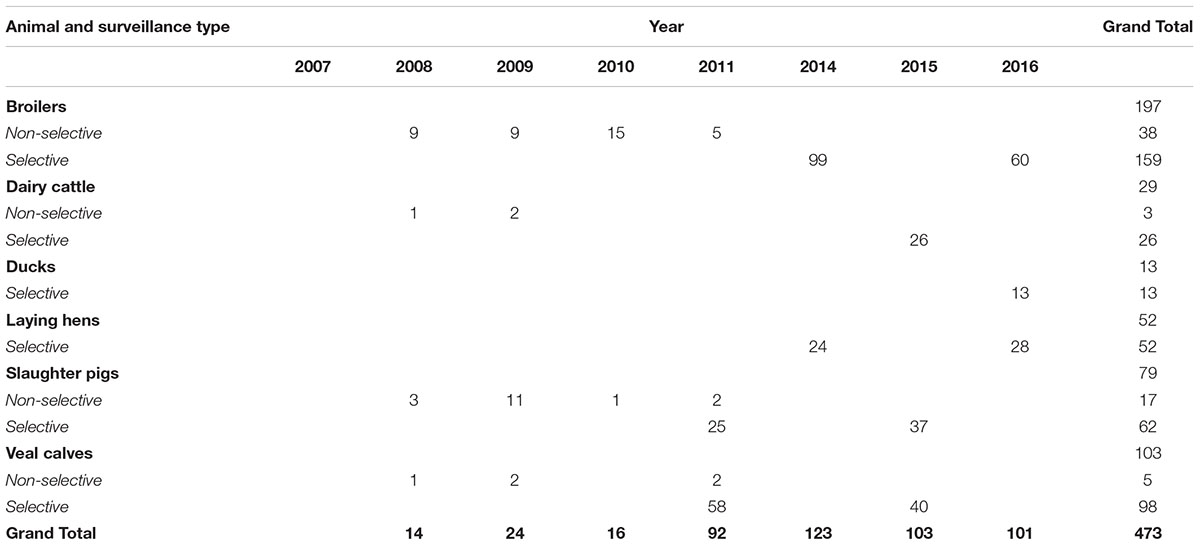
Table 2. Selection of ESC-R E. coli examined to determine the genomic localization of ESBL/pAmpC genes.
Transformation experiments to assess plasmid location of ESBL/pAmpC genes and plasmid classification by PCR-based replicon typing (Carattoli et al., 2005) were performed according to standard procedures, as previously described (Liakopoulos et al., 2016). When transformants could not be retrieved, chromosomal location of ESBL/pAmpC genes was confirmed by I-CeuI Pulsed-Field Gel Electrophoresis (PFGE) of total bacterial DNA, followed by Southern blot hybridization, as previously described (Liu et al., 1993).
In 2087 out of 2304 E. coli isolates (90.5%), the ESC-R phenotype was associated with one or more ESBL/pAmpC genes (Figure 1). In total twenty-two different ESBL/pAmpC genes were detected. blaCTX-M-1 was the most prevalent (43.7%) independent of the animal origin, with the highest frequency observed in isolates from slaughter pigs (49%). Genes blaCMY -2 (14.4%), blaTEM-52 and variants (9.7%), blaSHV -12 (7.6%), and blaCTX-M-15 (5.6%) followed, the latter being the second most prevalent in veal calf (17.5%) after blaCTX-M-1 (44.1%). Eleven E. coli isolates showed co-presence of two ESBL/pAmpC genes: blaCTX-M-1 with blaCTX-M-2 (1 broiler), blaSHV -12 (1 broiler), or blaTEM-52c (1 broiler and 1 veal calf); blaSHV -12 with blaTEM-52c (4 broilers); and blaCMY -2 with blaCTX-M-15 (1 veal calf), blaTEM-32 (1 broiler) or blaTEM-190 (1 dairy cattle). One E. coli isolated from broiler encoded three genes: blaCTX-M-1, blaCMY -2, and blaSHV -12. In general, comparison of ESC-R E. coli derived from selective and non-selective surveillance of the same animal species showed a substantial difference in gene diversity, with selective culturing displaying higher gene variability (Figure 1). This difference is likely dependent on the significantly higher number of isolates available from selective surveillance than from the non-selective one (Table 1).
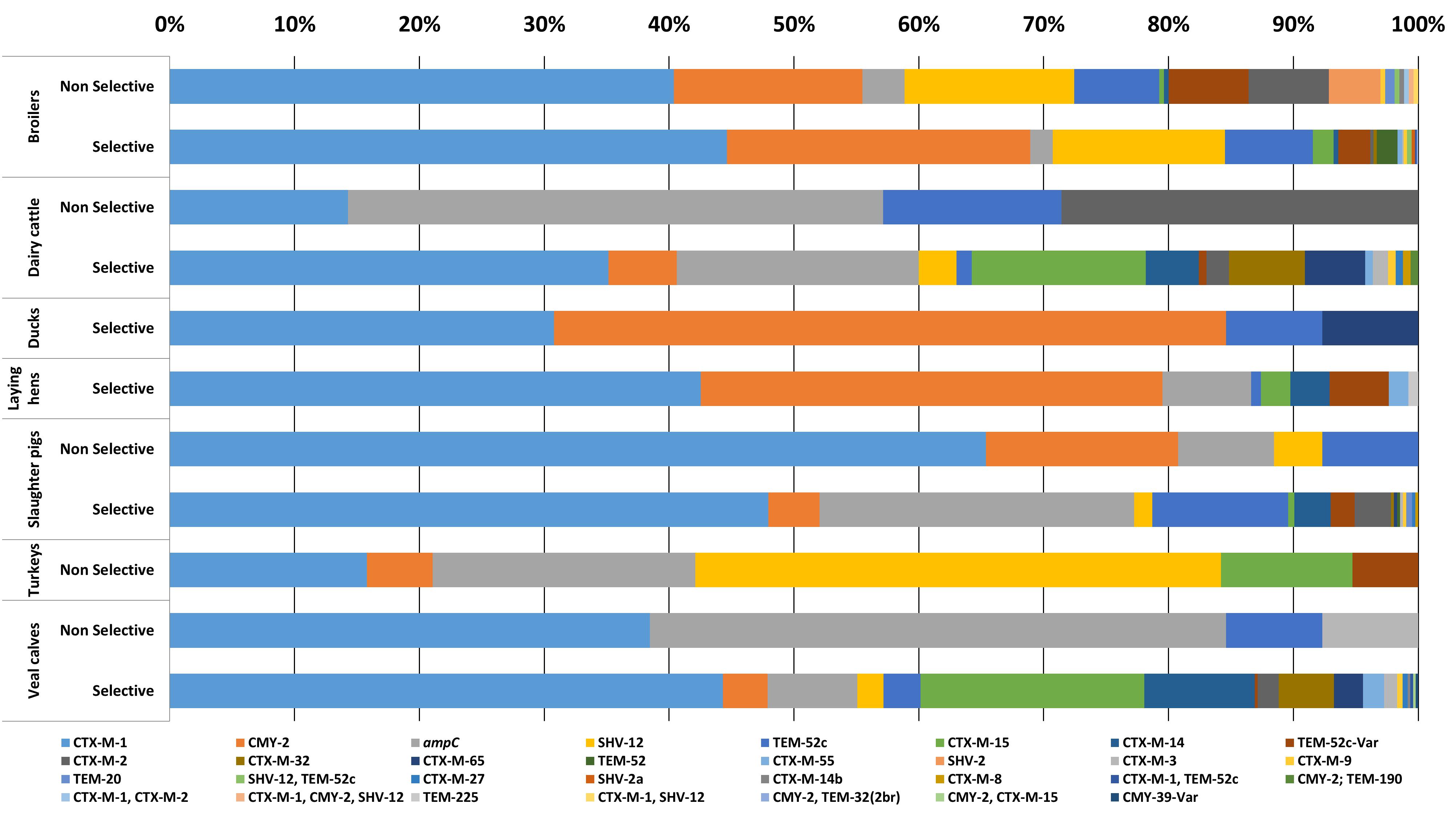
Figure 1. ESC-R E. coli gene distribution from selective and non-selective monitoring (2007–2017, n = 2304). Refer to Table 1 for isolate numbers for each livestock species and to Supplementary Table S1 for % values of each gene variant.
ESC-R E. coli for which no ESBL/pAmpC genes could be detected (n = 217, 9.4%) owed their ESC-R phenotype to promoter mutations of the chromosomally encoded ampC gene, with a peak observed in slaughter pigs (24.1%) (Figure 1). All ampC variants harbored previously described mutations (ampC types 2, 3, 5, 11, 18, 34, 40, and 45) (Mulvey et al., 2005). Majority of chromosomal ampC alterations (88%) created an alternate displaced promoter (type 3) whose mutation at position -42 is thought to have large effect on promoter strength (Caroff et al., 2000).
Selective surveillance from 2014 onward is based on the use of a harmonized protocol (European Food Safety Authority, 2008), a comparable number of isolates (300–400, depending on the year of sampling) and data are available for a four year period (2014–2017). Therefore, trends in ESC-R E. coli prevalence could be defined (Figure 2). A significant reduction (p < 0.001) from 67.3% (95% CI, 62.4–71.8) to 32.6% (95% CI, 27.3–38.2) was observed in broilers between 2014 and 2017. This trend is in accordance with decreasing prevalence of ESC-R E. coli from non-selective surveillance and in fresh chicken meat (Veldman et al., 2018). Prevalence in dairy cattle and slaughter pigs showed non-significant (p = 0.028, p = 0.2704) fluctuations between 2014 and 2017. A significant increase (p < 0.001) in prevalence was observed in veal calves, from 17.9% (95% CI, 13.8–22.8) in 2014 to 37.8% (95% CI, 32.3–43.5) in 2017. The prevalence of ESC-R in E. coli isolated from veal calves showed an increasing trend already in the years 1997–2010 (Hordijk et al., 2013b); this unexpected increase of ESC-R E. coli in veal calves urges for more research to define possible causes. Interestingly, beside the ubiquitous blaCTX-M-1, prevalence of blaCTX-M-15 and blaCTX-M-14 (ESBLs more associated with human E. coli) was dominant in cattle (veal calves and dairy cattle) (Supplementary Figure S1), a trend already observed since 2005 (Hordijk et al., 2013b) and confirmed in 2017 (Veldman et al., 2018). These findings are in line with recent studies showing that the general human population in the Netherlands has relatively similar ESBL gene profiles to veal calves compared to other reservoirs (Dorado-Garcia et al., 2018).
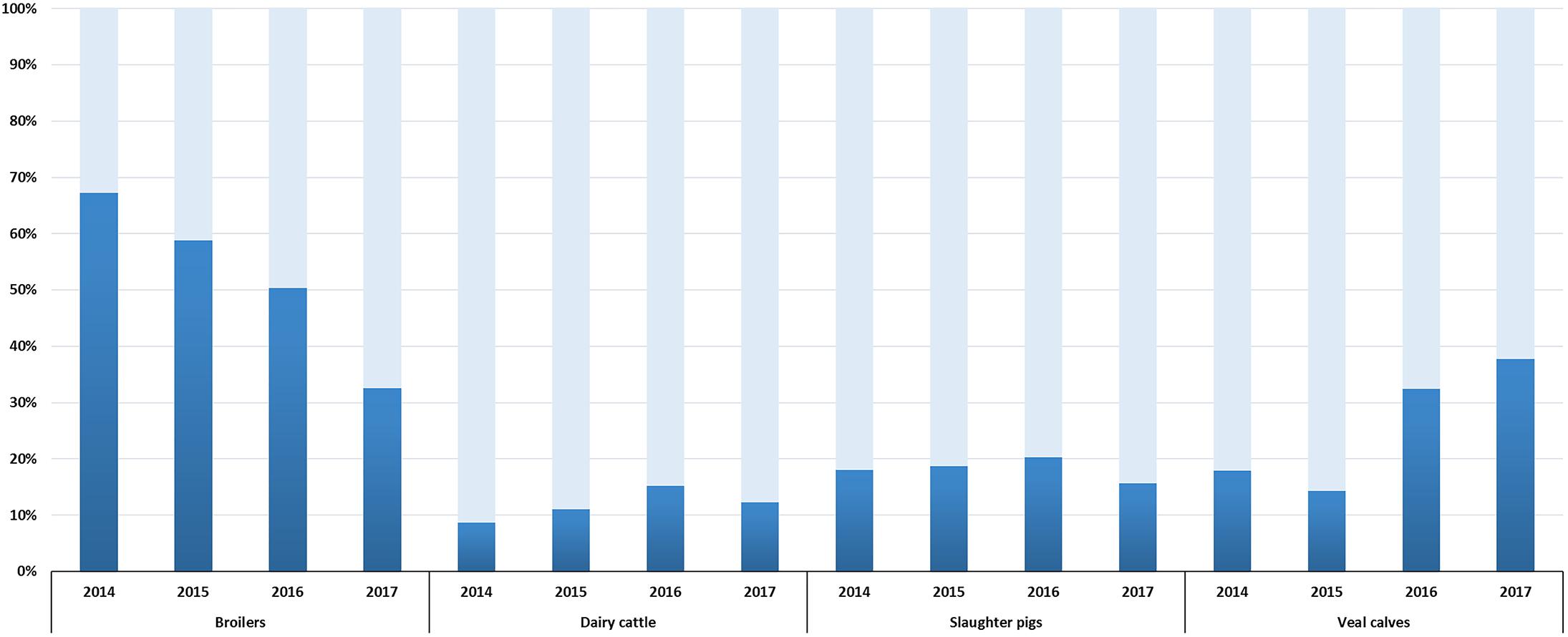
Figure 2. ESC-R E. coli prevalence in broilers, dairy cattle, slaughter pigs and veal calves from selective surveillance (2014–2017). Light gray bars indicate samples negative for the isolation of ESC-R E. coli. Refer to Supplementary Table S2 for isolate numbers and for each livestock species.
Genomic location (plasmid or chromosome) of a subset of ESBL/pAmpC genes was determined in 63 ESC-R E. coli collected over the years with non-selective culturing (Table 2). All ESBL/pAmpC genes were encoded on plasmids with different rep types (B/O, F, I1, K, and X1), in a few cases with multi-replicon plasmids (P/HI2 and P/I1), with distinctive prevalent gene-plasmid combinations per animal species (Supplementary Figure S2). Overall, the most common gene-plasmid combination was IncI1-blaCTX-M-1, detected in 30.1% of the isolates independently on the animal source. In broilers, IncX1-blaTEM-52c-V ar and IncI1-blaSHV -12 were the most prevalent (36.8 and 39.5%, respectively) among the subset of analyzed isolates, excluding blaCTX-M-1 encoding E. coli that were not typed because typically associated with IncI1 plasmids (Dierikx et al., 2013). An E. coli isolate from broiler encoding genes blaCTX-M-1, blaCMY -2, and blaSHV -12 was associated to three IncI1, IncK, and IncX3 plasmids, respectively (Veldman et al., 2012). ESC-R Enterobacteriaceae encoding multiple ESBL/pAmpC genes have been described previously with various genomic settings both on plasmids and/or chromosome in livestock, meat, and clinical isolates (Dhanji et al., 2010; Veldman et al., 2010; Huang et al., 2017), depicting the complex plasmid scenario of cephalosporin-resistance circulation among Enterobacteriaceae. Vast majority of ESC-R E. coli isolates from slaughter pigs were associated to IncI1 plasmids carrying blaCTX-M-1 (76.5%). The most common gene-plasmid combination in veal calves isolates was IncI1-blaCTX-M-1, while ESC-R E. coli isolates from dairy cattle were associated with multi-replicon plasmids IncP/HI2 encoding blaCTX-M-2. All IncI1 plasmid subtyped (89%) were confirmed to be IncI1α (data not shown).
According to current guidelines (European Food Safety Authority, 2013), selective surveillance of ESC-R E. coli should be performed following an annual rotation system: broilers and turkeys (years 2014, 2016, 2018, 2020), pigs and bovines (years 2015, 2017, 2019). Although more animal species than the recommended ones are frequently analyzed in the Dutch surveillance program (Maran Reports, 2002/2017), the rotation system was followed to select a subset of ESC-R E. coli (n = 410) for further investigation on the genomic localization of ESBL/pAmpC genes (Table 2). Because poultry ESC-R isolates for 2014 and 2016 were too many to include in the analysis (n = 783), 40–50% of all E. coli from broilers (n = 99 and n = 60, respectively) and laying hens (n = 24 and n = 28, respectively) were screened per year.
Results of this analysis are reported in Figure 3, except for thirteen ESC-R E. coli from ducks (2016) characterized by chromosomal blaCMY -2 (n = 8) or IncI1 plasmids carrying blaCTX-M-1 (n = 5). Transformants for 38 (9.3%) ESC-R E. coli could not be recovered. PFGE and Southern hybridization confirmed the chromosomal location of ESBL/pAmpC genes, mostly belonging to the CTX-M group (Figure 3): blaCTX-M-1, blaCTX-M-14, blaCTX-M-15, blaCTX-M-32, blaCTX-M-55, blaCTX-M-9, and blaCMY -2. Although, the genetic surroundings of these genes were not investigated, it is known that ISEcp1 insertion sequence upstream of ESBL/pAmpC genes are associated with transposition and chromosomal integration of typically plasmid-encoded genes in E. coli, K. pneumoniae, and Shigella flexneri, among others, from animals or humans (Wang et al., 2013; Fang et al., 2015; Huang et al., 2017). Through chromosomal integration, ISEcp1 might contribute to lowering the fitness cost derived from harboring an entire plasmid, while enhancing EBSL/AmpC gene expression under its own promoter (Poirel et al., 2003).
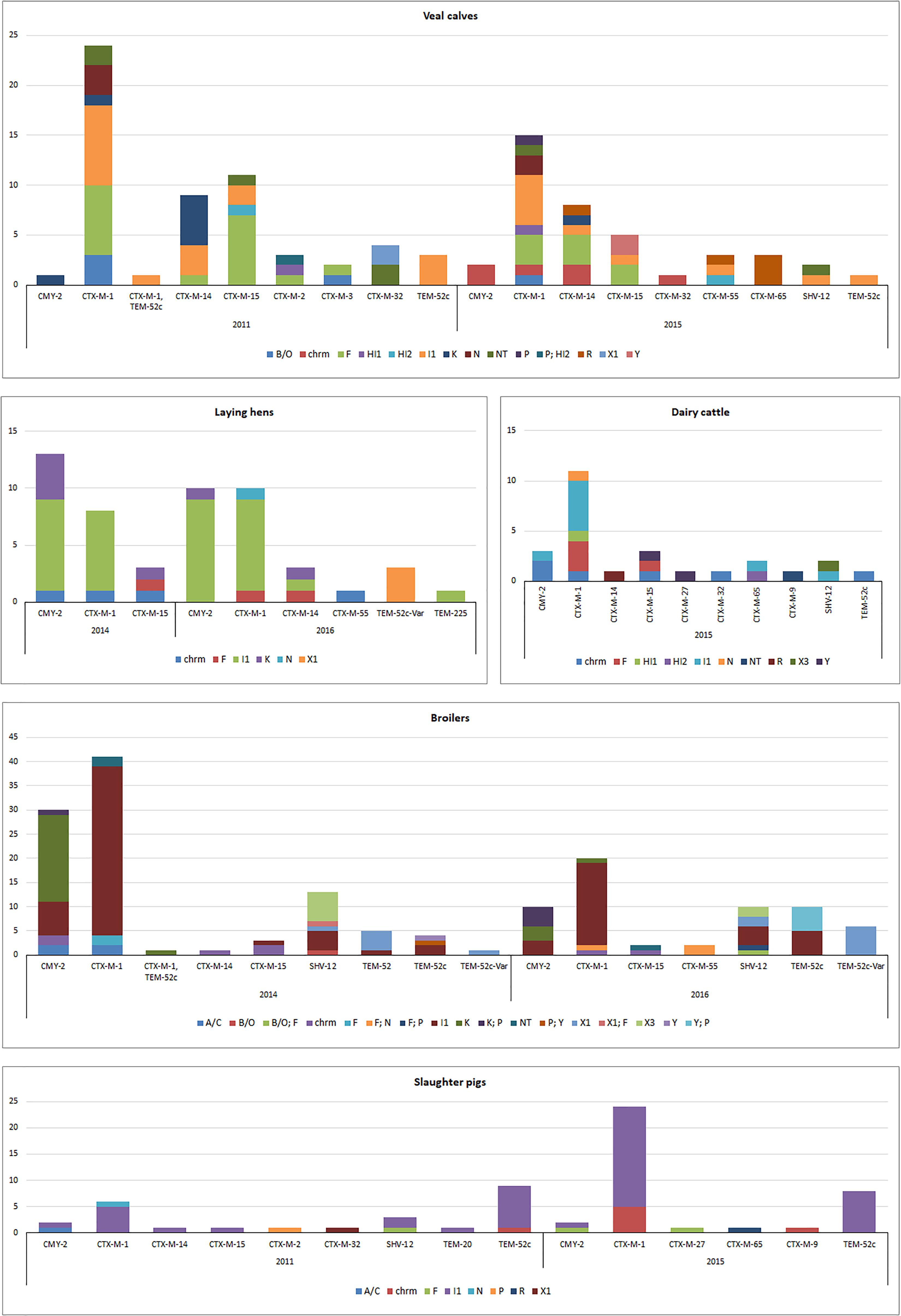
Figure 3. Genomic localization of ESBL/pAmpC genes in ESC-R E. coli from selective monitoring (2011–2016) per animal species. Panels: veal calves (n = 98); broilers (n = 159); laying hens (n = 52); dairy cattle (n = 26); slaughter pigs (n = 62). Refer to Supplementary Table S3 for % values of each gene variant in different livestock species.
Majority of ESBL/pAmpC genes (n = 372) were associated with plasmids (Figure 3). All IncI1 plasmid subtyped (86%) were confirmed to be IncI1α (data not shown). Gene-plasmid combinations in broilers did not show major differences between 2014 and 2016. IncI1-blaCTX-M-1 plasmids were the most common, followed by IncI1 encoding blaCMY -2, blaSHV -12 or blaTEM-52c. IncK-blaCMY -2 (or multireplicon IncK/P) plasmids were also commonly detected, suggesting a relatively stable plasmid population in broilers in the Netherlands, as earlier described (Dierikx et al., 2013). Yet, IncX3-blaSHV -12 plasmids, whose emergence in Dutch ESC-R E. coli of animal origin was recently revealed alongside a gradual decrease in the prevalence of IncI1-blaSHV -12 plasmids (Liakopoulos et al., 2018), were detected in both years. Plasmid IncX1 carrying blaSHV -12, blaTEM-52c or blaTEM-52c-V ar followed in prevalence, the latter detected also in ESBL/pAmpC-producing E. coli from laying hens in 2016. Overall, plasmid-gene associations in isolates from laying hens were comparable to broilers, with IncI1-blaCMY -2, IncI1-blaCTX-M-1 and IncK-blaCMY -2 being the most predominant in both 2014 and 2016. The presence of ESC-R E. coli at all levels of the Dutch broiler production pyramid has been demonstrated, as day-old chicks can inherit bacteria from their parents through contaminated egg shells or from the environment (Dierikx et al., 2013).
ESBL/pAmpC-producing E. coli isolated from slaughter pigs from both 2011 and 2015 were dominated by IncI1 plasmids encoding blaCTX-M-1 or blaTEM-52c, recognized as the most prevalent gene-plasmid combinations in Enterobacteriaceae from slaughter pigs worldwide (Geser et al., 2011; Randall et al., 2014; Biasino et al., 2018; Dang et al., 2018). ESC-R E. coli isolates from dairy cattle and veal calves showed a quite variable array of plasmid-gene combinations (Figure 3). Beside predominant IncI1 plasmids, IncF plasmids were detected in both animal reservoirs in association with blaCTX-M-1 and blaCTX-M-14 and blaCTX-M-15 genes. IncR-blaCTX-M-65 and IncR-blaCTX-M-55 were also identified in veal calves in 2015 but no R plasmid was detected in 2011. Gene-plasmid combinations observed in veal calves are coherent with previous studies conducted in the Netherlands and in France (Hordijk et al., 2013a; Haenni et al., 2014) with relatively high prevalence of various blaCTX-M genes located on IncF and IncI1 plasmids. The more variable array of plasmid-gene combinations observed in veal calves compared to other livestock might be a consequence of international trade from different dairy farms to Dutch farms as well as high antimicrobial use and farm management.
In conclusion, the results of this study provide insight in the prevalence and molecular diversity of ESC-R E. coli, revealing a rather composite scenario of plasmid-gene combinations circulating in livestock from the Netherlands over the last decade. Yet, the bias in the selection of isolates for plasmid typing should be kept in mind to avoid risky conclusions on prevalence of ESBL-harboring plasmid types. Nevertheless, the study provides additional information on the occurrence of different plasmid types carrying ESBL/pAmpC-genes in E. coli from livestock in the Netherlands. These findings also demonstrate the added value of selective culturing of ESC-R E. coli and genotyping of genes and plasmids over random isolation for resistance determinants of public health concern.
AK, AE-Z, JH, and BW acquired the data. CD, DC, KV and DM analyzed the data. DC and KV prepared the manuscript. All authors discussed, read, contributed to, and approved the final manuscript.
The Dutch Ministry of Economic Affairs funded surveillance activities over the years (WOT-01-002-003.02). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank: Joop Testerink and Marga Japing (WBVR) for invaluable technical assistance; Michael Brouwer (WBVR) for insightful discussion on plasmid subtyping; Michel Rapallini and everyone at the Netherlands Food and Consumer Product Safety Authority involved in sampling activities.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00076/full#supplementary-material
FIGURE S1 | Gene distribution in ESC-R E. coli isolated from broilers, dairy cattle, slaughter pigs and veal calves during selective surveillance (2014–2017). Refer to Supplementary Table S2 for gene distribution for each livestock species.
FIGURE S2 | Gene-plasmid combinations in ESC-R E. coli from non-selective surveillance per animal species (2008–2011, n = 63). Refer to Supplementary Table S4 for gene-plasmid distribution for each livestock species.
TABLE S1 | % values of ampC and ESBL/pAmpC genes distribution from selective and non-selective monitoring (2007–2017, n = 2304).
TABLE S2 | ESC-R E. coli prevalence in broilers, dairy cattle, slaughter pigs and veal calves from selective surveillance (2014–2017).
TABLE S3 | Genomic localization of ESBL/pAmpC genes in ESC-R E. coli from selective surveillance per animal species (2011–2016, n = 410).
TABLE S4 | Gene-plasmid combinations in ESC-R E. coli from non-selective surveillance per animal species (2008–2011, n = 63).
Batchelor, M., Hopkins, K. L., Liebana, E., Slickers, P., Ehricht, R., Mafura, M., et al. (2008). Development of a miniaturised microarray-based assay for the rapid identification of antimicrobial resistance genes in Gram-negative bacteria. Int. J. Antimicrob. Agents 31, 440–451. doi: 10.1016/j.ijantimicag.2007.11.017
Biasino, W., De Zutter, L., Garcia-Graells, C., Uyttendaele, M., Botteldoorn, N., Gowda, T., et al. (2018). Quantification, distribution and diversity of ESBL/AmpC-producing Escherichia coli on freshly slaughtered pig carcasses. Int. J. Food Microbiol. 281, 32–35. doi: 10.1016/j.ijfoodmicro.2018.05.021
Bush, K., and Fisher, J. F. (2011). Epidemiological expansion, structural studies, and clinical challenges of new β-lactamases from Gram-negative bacteria. Annu. Rev. Microbiol. 65, 455–478. doi: 10.1146/annurev-micro-090110-102911
Carattoli, A. (2008). Animal reservoirs for extended spectrum β-lactamase producers. Clin. Microbiol. Infect. 14, 117–123. doi: 10.1111/j.1469-0691.2007.01851.x
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., and Threlfall, E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228. doi: 10.1016/j.mimet.2005.03.018
Caroff, N., Espaze, E., Gautreau, D., Richet, H., and Reynaud, A. (2000). Analysis of the effects of -42 and -32 ampC promoter mutations in clinical isolates of Escherichia coli hyperproducing ampC. J. Antimicrob. Chemother. 45, 783–788. doi: 10.1093/jac/45.6.783
Dang, S. T. T., Bortolaia, V., Tran, N. T., Le, H. Q., and Dalsgaard, A. (2018). Cephalosporin-resistant Escherichia coli isolated from farm workers and pigs in northern Vietnam. Trop. Med. Int. Health 23, 415–424. doi: 10.1111/tmi.13054
Dhanji, H., Murphy, N. M., Doumith, M., Durmus, S., Lee, S. S., Hope, R., et al. (2010). Cephalosporin resistance mechanisms in Escherichia coli isolated from raw chicken imported into the UK. J. Antimicrob. Chemother. 65, 2534–2537. doi: 10.1093/jac/dkq376
Dierikx, C., Van Der Goot, J., Fabri, T., Van Essen-Zandbergen, A., Smith, H., and Mevius, D. (2013). Extended-spectrum-β-lactamase- and AmpC-β-lactamase-producing Escherichia coli in dutch broilers and broiler farmers. J. Antimicrob. Chemother. 68, 60–67. doi: 10.1093/jac/dks349
Dierikx, C. M., Van Der Goot, J. A., Smith, H. E., Kant, A., and Mevius, D. J. (2013). Presence of ESBL/AmpC -producing Escherichia coli in the broiler production pyramid: a descriptive study. PLoS One 8:e79005. doi: 10.1371/journal.pone.0079005
Dorado-Garcia, A., Smid, J. H., Van Pelt, W., Bonten, M. J. M., Fluit, A. C., Van Den Bunt, G., et al. (2018). Molecular relatedness of ESBL/AmpC-producing Escherichia coli from humans, animals, food and the environment: a pooled analysis. J. Antimicrob. Chemother. 73, 339–347. doi: 10.1093/jac/dkx397
European Food Safety Authority (2008). Report from the task force on zoonoses data collection including guidance for harmonized monitoring and reporting of antimicrobial resistance in commensal Escherichia coli and Enterococcus spp. from food animals. EFSA J. 6:141r. doi: 10.2903/j.efsa.2008.141r
European Food Safety Authority (2013). Commission implementing decision on the monitoring and reporting of antimicrobial resistance in zoonotic and commensal bacteria, C (2013) 7145. Offic. J. Eur. Union 303, 26–39.
Ewers, C., Bethe, A., Semmler, T., Guenther, S., and Wieler, L. H. (2012). Extended-spectrum beta-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin. Microbiol. Infect. 18, 646–655. doi: 10.1111/j.1469-0691.2012.03850.x
Fang, L. X., Sun, J., Li, L., Deng, H., Huang, T., Yang, Q. E., et al. (2015). Dissemination of the chromosomally encoded CMY-2 cephalosporinase gene in Escherichia coli isolated from animals. Int. J. Antimicrob. Agents 46, 209–213. doi: 10.1016/j.ijantimicag.2015.04.003
Geser, N., Stephan, R., Kuhnert, P., Zbinden, R., Kaeppeli, U., Cernela, N., et al. (2011). Fecal carriage of extended-spectrum beta-lactamase-producing Enterobacteriaceae in swine and cattle at slaughter in Switzerland. J. Food Prot. 74, 446–449. doi: 10.4315/0362-028X.JFP-10-372
Geurts, Y., Brouwer, M., Noorman, K., Kant, A., Ceccarelli, D., Veldman, K., et al. (2017). “Development of sensitive and cost-effective Real-time PCR assays for rapid detection of the beta-lactamase genes CTX-M1, SHV, TEM and AmpC gene CMY2 in Enterobacteriaceae,” in Proceedings of the 27th European Congress of Clinical Microbiology and Infectious Diseases 2017, (Vienna: European Society for Clinical Microbiology and Infectious Diseases).
Haenni, M., Chatre, P., Metayer, V., Bour, M., Signol, E., Madec, J. Y., et al. (2014). Comparative prevalence and characterization of ESBL-producing Enterobacteriaceae in dominant versus subdominant enteric flora in veal calves at slaughterhouse, France. Vet. Microbiol. 171, 321–327. doi: 10.1016/j.vetmic.2014.02.023
Hordijk, J., Wagenaar, J. A., Kant, A., Van Essen-Zandbergen, A., Dierikx, C., Veldman, K., et al. (2013a). Cross-sectional study on prevalence and molecular characteristics of plasmid mediated ESBL/AmpC-producing Escherichia coli isolated from veal calves at slaughter. PLoS One 8:e65681. doi: 10.1371/journal.pone.0065681
Hordijk, J., Wagenaar, J. A., Van De Giessen, A., Dierikx, C., Van Essen-Zandbergen, A., Veldman, K., et al. (2013b). Increasing prevalence and diversity of ESBL/AmpC-type beta-lactamase genes in Escherichia coli isolated from veal calves from 1997 to 2010. J. Antimicrob. Chemother. 68, 1970–1973. doi: 10.1093/jac/dkt132
Huang, W., Wang, G., Sebra, R., Zhuge, J., Yin, C., Aguero-Rosenfeld, M. E., et al. (2017). Emergence and evolution of multidrug-resistant Klebsiella pneumoniae with both blaKPC and blaCTX-M integrated in the chromosome. Antimicrob. Agents Chemother. 61:e0076-17. doi: 10.1128/AAC.00076-17
Liakopoulos, A., Geurts, Y., Dierikx, C. M., Brouwer, M. S., Kant, A., Wit, B., et al. (2016). Extended-spectrum cephalosporin-resistant Salmonella enterica serovar Heidelberg strains, the Netherlands. Emerg. Infect. Dis. 22, 1257–1261. doi: 10.3201/eid2207.151377
Liakopoulos, A., Van Der Goot, J., Bossers, A., Betts, J., Brouwer, M. S. M., Kant, A., et al. (2018). Genomic and functional characterisation of IncX3 plasmids encoding blaSHV-12 in Escherichia coli from human and animal origin. Sci. Rep. 8:7674. doi: 10.1038/s41598-018-26073-5
Liu, S. L., Hessel, A., and Sanderson, K. E. (1993). Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. U.S.A. 90, 6874–6878. doi: 10.1073/pnas.90.14.6874
Madec, J. Y., Haenni, M., Nordmann, P., and Poirel, L. (2017). Extended-spectrum β-lactamase/AmpC- and carbapenemase-producing Enterobacteriaceae in animals: a threat for humans? Clin. Microbiol. Infect. 23, 826–833. doi: 10.1016/j.cmi.2017.01.013
Maran Reports (2002/2017). Monitoring of Antimicrobial Resistance and Antibiotic Usage in Animals in the Netherlands. Available at: https://www.wur.nl/nl/Onderzoek-Resultaten/Onderzoeksinstituten/Bioveterinary-Research/Publicaties/MARAN-Rapporten.htm
Michael, G. B., Freitag, C., Wendlandt, S., Eidam, C., Fessler, A. T., Lopes, G. V., et al. (2015). Emerging issues in antimicrobial resistance of bacteria from food-producing animals. Fut. Microbiol. 10, 427–443. doi: 10.2217/fmb.14.93
Mulvey, M. R., Bryce, E., Boyd, D. A., Ofner-Agostini, M., Land, A. M., Simor, A. E., et al. (2005). Molecular characterization of cefoxitin-resistant Escherichia coli from Canadian hospitals. Antimicrob. Agents Chemother. 49, 358–365. doi: 10.1128/AAC.49.1.358-365.2005
Poirel, L., Decousser, J.-W., and Nordmann, P. (2003). Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob. Agents Chemother. 47, 2938–2945. doi: 10.1128/AAC.47.9.2938-2945.2003
Randall, L. P., Lemma, F., Rogers, J. P., Cheney, T. E., Powell, L. F., and Teale, C. J. (2014). Prevalence of extended-spectrum-beta-lactamase-producing Escherichia coli from pigs at slaughter in the UK in 2013. J. Antimicrob. Chemother. 69, 2947–2950. doi: 10.1093/jac/dku258
Rozwandowicz, M., Brouwer, M. S. M., Fischer, J., Wagenaar, J. A., Gonzalez-Zorn, B., Guerra, B., et al. (2018). Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 73, 1121–1137. doi: 10.1093/jac/dkx488
Speksnijder, D. C., Mevius, D. J., Bruschke, C. J., and Wagenaar, J. A. (2015). Reduction of veterinary antimicrobial use in the Netherlands. The dutch success model. Zoonoses Public Health. 62(Suppl. 1), 79–87. doi: 10.1111/zph.12167
Veldman, K., Dierikx, C., Van Essen-Zandbergen, A., Van Pelt, W., and Mevius, D. (2010). Characterization of multidrug-resistant, qnrB2-positive and extended-spectrum-beta-lactamase-producing Salmonella concord and Salmonella senftenberg isolates. J. Antimicrob. Chemother. 65, 872–875. doi: 10.1093/jac/dkq049
Veldman, K., Swanenburg, M., Ceccarelli, D., and Mevius, D. (2018). Monitoring of Antimicrobial Resistance and Antibiotic Usage in Animals (MARAN) in the Netherlands in 2017. Lelystad: Wageningen Bioveterinary Research.
Veldman, K., Van Essen-Zandbergen, A., Kant, A., and Mevius, D. (2012). Characterization of qnr-positive Escherichia coli isolates from food-producing animals in the Netherlands. J. Antimicrob. Chemother. 67, 239–240. doi: 10.1093/jac/dkr424
Keywords: ESBL, pAmpC, Escherichia coli, plasmid, livestock
Citation: Ceccarelli D, Kant A, van Essen-Zandbergen A, Dierikx C, Hordijk J, Wit B, Mevius DJ and Veldman KT (2019) Diversity of Plasmids and Genes Encoding Resistance to Extended Spectrum Cephalosporins in Commensal Escherichia coli From Dutch Livestock in 2007–2017. Front. Microbiol. 10:76. doi: 10.3389/fmicb.2019.00076
Received: 25 September 2018; Accepted: 15 January 2019;
Published: 04 February 2019.
Edited by:
Sebastian Guenther, University of Greifswald, GermanyReviewed by:
María de Toro, Centro de Investigación Biomédica de La Rioja, SpainCopyright © 2019 Ceccarelli, Kant, van Essen-Zandbergen, Dierikx, Hordijk, Wit, Mevius and Veldman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniela Ceccarelli, ZGFuaWVsYS5jZWNjYXJlbGxpQHd1ci5ubA==
†Present address:Cindy Dierikx, National Institute for Public Health and the Environment (RIVM), Centre for Infectious Disease Control, Bilthoven, Netherlands Joost Hordijk, Faculty of Veterinary Medicine, Utrecht University, Utrecht, Netherlands
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.