Corrigendum: A Potential Role for Epigenetic Processes in the Acclimation Response to Elevated pCO2 in the Model Diatom Phaeodactylum tricornutum
- 1State Key Laboratory of Marine Environmental Science,College of Ocean and Earth Sciences, Xiamen University, Xiamen, China
- 2School of Pharmaceutical Sciences, Xiamen University, Xiamen, China
- 3Ecology and Evolutionary Biology Section, Institut de Biologie de l’École Normale Supérieure (IBENS), Département de Biologie, Ecole Normale Supérieure, CNRS UMR8197, Inserm U1024, PSL Research University, Paris, France
- 4Faculté des Sciences et Technologie, Université Paris Est-Créteil, Créteil, France
- 5Faculté des Sciences et Techniques, Université de Nantes, CNRS UMR6286, UFIP, Nantes, France
Understanding of the molecular responses underpinning diatom responses to ocean acidification is fundamental for predicting how important primary producers will be shaped by the continuous rise in atmospheric CO2. In this study, we have analyzed global transcriptomic changes of the model diatom Phaeodactylum tricornutum following growth for 15 generations in elevated pCO2 by strand-specific RNA sequencing (ssRNA-seq). Our results indicate that no significant effects of elevated pCO2 and associated carbonate chemistry changes on the physiological performance of the cells were observed after 15 generations whereas the expression of genes encoding histones and other genes involved in chromatin structure were significantly down-regulated, while the expression of transposable elements (TEs) and genes encoding histone acetylation enzymes were significantly up-regulated. Furthermore, we identified a series of long non-protein coding RNAs (lncRNAs) specifically responsive to elevated pCO2, suggesting putative regulatory roles for these largely uncharacterized genome components. Taken together, our integrative analyses reveal that epigenetic elements such as TEs, histone modifications and lncRNAs may have important roles in the acclimation of diatoms to elevated pCO2 over short time scales and thus may influence longer term adaptive processes in response to progressive ocean acidification.
Introduction
Ocean acidification is caused by the absorption of excessive anthropogenic CO2 emissions by the ocean. It is predicted that the further rise of atmospheric CO2 concentrations under a “business-as-usual” CO2 emission scenario will lead to a further drop of ocean surface pH by 0.4 units by 2100 (Gattuso et al., 2015). Diatoms, responsible for about 20% of global primary production, profoundly influence marine food webs and biogeochemical cycles (Tréguer et al., 2018). Although several studies reporting the effects of ocean acidification on diatoms at the physiological and ecological levels have been reported (Gao and Campbell, 2014), there are only limited studies deciphering the underlying molecular mechanisms of diatoms in response to ocean acidification.
Most studies that have examined the genetic responses of diatoms to ocean acidification have focused on carbon fixation and assimilation genes. It has been reported that the large subunit of ribulose-1, 5-bisphosphate carboxylase/oxygenase (Rubisco), involved in the first committed step of carbon fixation, was down-regulated at the transcriptional and protein levels under elevated pCO2 conditions (Losh et al., 2013; Li et al., 2017). Genes encoding components of carbon concentrating mechanisms (CCMs) including carbonic anhydrases and bicarbonate transporters were also reported to be down-regulated under elevated CO2 conditions (Hopkinson et al., 2011; Li et al., 2014; Hennon et al., 2015). Furthermore, using RNA-seq in the context of a nitrate-limited chemostat experiment, it was additionally reported that elevated CO2 first caused changes in transcriptional regulation and chromosome folding and subsequently metabolic rearrangements in Thalassiosira pseudonana. This study further revealed that genes in one CO2-responsive cluster share a putative cAMP responsive cis-regulatory sequence for a cAMP second messenger system (Hennon et al., 2015). These results imply that in addition to genes encoding central components of carbon fixation and assimilation, the response to elevated CO2 may involve a broader reprogramming of transcription, genome structure, and cell signaling.
Long non-protein coding RNAs (lncRNAs) are transcripts longer than 200 nucleotides which are similar to mRNAs but have low to no apparent protein coding potential. Long considered as transcriptional noise, numerous evidence from animals and plants gathered over recent years indicate that lncRNAs have multiple and important functions, such as histone modification regulation, transcription machinery regulation and post-translational regulation (Lee, 2012; Hon et al., 2017). Furthermore, lncRNAs are often associated with stress responses having fundamental regulatory functions in cellular adaptation and homeostasis (Amaral et al., 2013). LncRNAs may also have important roles in diatoms in response to different environmental stimuli, however, to our knowledge only one study has so far investigated changes in lncRNA levels in response to phosphate fluctuations (Cruz de Carvalho et al., 2016).
Histones play crucial roles in chromosome structure and genome stability. A total of 14 histone genes (H2A, H2B, H3, H4, and H1) are dispersed on 5 of the 34 chromosome scaffolds of the P. tricornutum genome (Bowler et al., 2008). Transposable elements (TEs) are mobile DNA sequences able to move in the genome by generation of new copies. TEs are ancient and active genomic components which have been found in most eukaryotic and prokaryotic genomes (Feschotte and Pritham, 2007). In P. tricornutum and T. pseudonana, 6.4 and 1.9% of the genome are contributed by TEs, respectively (Armbrust et al., 2004; Bowler et al., 2008; Rastogi et al., 2018). TEs are classified into retrotransposons and DNA transposons according to the presence or absence of an RNA transposition. Retrotransposons use a “copy and paste” strategy to proliferate in the genome whereas the majority of DNA transposons use a “cut and paste” strategy to move in the genome. A total of 90% and 58% of TEs are long terminal repeat retrotransposons (LTR-RTs) in the P. tricornutum and T. pseudonana genomes, respectively. Among LTR-RTs, gag pol domains are the most abundant TEs in P. tricornutum (Maumus et al., 2009; Rastogi et al., 2018). Retrotransposons are considered as the major contributors to the expansion of genomes especially in higher eukaryotes and enhance their hosts’ fitness by advantageous genome re-arrangements in a changing environment (Schaack et al., 2010; Casacuberta and González, 2013).
In this study, we have combined physiological performance measurements, with strand-specific RNA sequencing (ssRNA-Seq) to assess global transcriptomic changes in P. tricornutum in response to elevated pCO2 with respect to ambient pCO2 concentrations following growth for 15 generations. Our data provide new insights into how a diatom copes with elevated pCO2 using some potential epigenetic mechanisms and suggest an involvement of TEs and possibly lncRNAs in the acclimation response to elevated pCO2.
Materials and Methods
P. tricornutum Culture Conditions and Physiological Measurements
Axenic cultures of Phaeodactylum tricornutum (Bohlin) strain CCMP 632 were obtained from the Culture Center of Marine Phytoplankton (East Boothbay, ME, United States). P. tricornutum cells were cultured in 2L polycarbonate bottles (Nalgene, United States) containing 1.7–1.8 L artificial seawater enriched with Aquil medium at 20°C under cool white fluorescent lights at 150 mmol m-2 s-1 (12 h:12 h light: dark). Different pCO2 in the medium was achieved by bubbling different concentrations of CO2. The gas streams were pre-filtered through 0.2-μm HEPA filters at a flow rate of 0.6 L min-1. The low CO2 medium was bubbled with ambient air of about 400 ppmv (low CO2, LC) and the high CO2 medium was bubbled with pre-mixed air-CO2 mixtures (1000 ppmv; high CO2, HC) from a plant growth CO2 chamber (HP1000G-D, Ruihua), controlling the CO2 concentration with a less than 3% variation. Triplicates of LC and HC cultures were all started with the same initial cell densities of 2 × 104 cells mL-1. The culture was refreshed with pCO2 and the pH adjusted medium every 48 h to maintain the desired pH and pCO2 values within a variation of less than 0.5%. The carbonate system in the HC cultures differed significantly from the LC culture (Supplementary Table S1). The growth rate was calculated as μ = (ln C1 - ln C0)/(t1 -t0), where C0 and C1 represent the cell density at t0 (initial or just after the dilution) and t1 (before the dilution), respectively. Cell densities were measured by particle count and size analyzer (Z2 Coulter, Beckman). Pigment concentrations were measured as described in (Porra et al., 1989) using spectrophotometry (Beckman DU-800, United States). The maximum quantum yield (Fv/Fm), effective quantum yield (Yield) were measured by a Xenon-Pulse Amplitude Modulated Fluorometer (XE-PAM, Walz, Germany) at the middle of the light period. Particulate organic nitrogen (PON) and carbon (POC) were measured according to (Liu et al., 2017).
RNA Extraction, Library Preparation and ssRNA-seq
Phaeodactylum tricornutum cells from three biological replicates were harvested in the middle of light period by vacuum filtration on 2 mm polycarbonate filters (Millipore, United States) following 15 generations (day 10), flash frozen in liquid N2 and maintained at -80°C until use. Total RNA was extracted from frozen cell pellets using TRIzol® Reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions. RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the 2100 Bioanalyzer (Agilent Technologies, CA, United States) and by RT-qPCR. A total amount of 3 μg RNA per sample was used for construction of RNA sample libraries. Ribosomal RNA was removed by Epicentre Ribo-ZeroTM Gold Kits (Epicentre, United States). The sequencing libraries were generated using NEBNext Ultra Directional RNA Library Prep Kit (NEB, Ipswich, United States) with varied index labeling following the manufacturer’s guidelines. After cluster generation by HiSeq PE Cluster Kit v4-cBot-HS (Illumina), the libraries were sequenced on an Illumina Hiseq 4000 platform and 150 bp paired-end reads were generated. Library preparation and ssRNA-seq were performed offsite at Annoroad Gene Technology Corporation (Beijing, PR China).
ssRNA-seq Data Assembly and Analysis
Raw data from ssRNA-seq were cleaned up by removing adapter-contaminated reads (more than 5 adapter-contaminated bases), poly-N containing (more than 5%) and low quality reads (quality value less than 19, accounting for more than 15% of total bases). All the downstream analyses were based on filtered clean data with high quality. The average clean data was 7.1 Gb for ssRNA-Seq. The reference genome of P. tricornutum was downloaded1. Bowtie2 v2.2.3 was used for building the genome index (Langmead et al., 2009), and clean data was mapped to the reference genome using TopHat v2.0.12 (Trapnell et al., 2009). Read counts for each gene in each sample were counted by HTSeq v0.6.0, and RPKM (Reads Per Kilobase Million Mapped Reads) was then calculated to estimate the expression level of genes in each sample (Wagner et al., 2012).
DESeq (v1.16) was used for differential gene expression analysis. The p-value was assigned to each gene and adjusted by the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with fold change ratio (HC/LC) ≥2 (padj ≤ 0.05) and fold change ratio ≤0.5 (padj ≤ 0.05) were defined as “up-regulated genes” and “down-regulated genes,” respectively. Genes with fold change ratio (HC/LC) between 1.5 and 2 (padj), fold change between 0.5 and 0.67 (padj) were defined as “relatively up-regulated genes” and “relatively down-regulated genes,” respectively.
GO and KEGG Analysis
Gene Ontology analysis2 was conducted to construct meaningful annotation of genes and gene products in different databases and species. GO enrichment analysis was used to assess the significance of GO term enrichment among differentially expressed genes in the domains of biological processes, cellular components and molecular functions. The GO enrichment of differentially expressed genes was implemented by the hypergeometric test, in which p-value was calculated and adjusted as q-value. GO terms with q < 0.05 were considered to be significantly enriched. We further performed KEGG3 pathway analysis to explore the roles of the differentially expressed genes in different metabolic pathways and molecular interactions. The genes related to chromatin, protein DNA, chromosomal part, chromosome, histone modification, glycolysis, tricarboxylic acid cycle (TCA cycle), and photosynthesis (light part), carbon assimilation and N uptake and assimilation were classified manually based on the KEGG and GO gene classification (Supplementary Table S2).
Identification of Novel lncRNAs
LncRNA candidates with length of ≥200 nucleotides, a predicted open reading frame (ORF) of ≤100 amino acids, number of exon ≥ 1 and reads ≥ 3 were selected (Liu et al., 2012; Cruz de Carvalho et al., 2016). Transdecoder was used to remove known mRNA transcripts and candidates with protein coding potential. The novel lncRNAs were classified based on the location of the lncRNA gene region in the reference genome. In order to increase the depth of the reads for the increased detection of non-coding RNA transcripts, the non-coding RNA transcript reads derived from the three replicates were pooled since the Pearson correlation coefficients (PCCs) between the RPKM of three replicates of LC and HCs were high (>0.97). A 1.5-fold/0.67-fold (up-regulated/down regulated) and variance in RPKM and a p-value (Fisher’s exact test) of <0.05 were used as cutoffs to define differentially expressed genes, respectively.
Interaction Network Construction
The P. tricornutum protein–protein interaction network data was downloaded4. The differentially expressed mRNA interaction networks, specifically transcription factor (TF), histone modification genes and mRNAs interaction networks were constructed in Cytoscape 3.4.0 based on the P. tricornutum protein–protein interaction network data database.
Quantitative Real-Time Reverse Transcription PCR
Reverse transcription quantitative PCR (RT-qPCR) was used to verify the transcriptomic data. The primers used for qPCR are listed in Supplementary Table S3. Polyadenylated RNA was converted to cDNA by PrimeScriptTM II Reverse Transcriptase (Takara). The RT product was then used as the template for qPCR. All qPCR reactions were performed on a FTC2000 (Canada) using SYBR Green I mix (Takara) in 96-well plates according to the manufacturer’s recommendations. TBP (TATA box binding protein) and RPS (ribosomal protein small subunit 30S), two housekeeping genes (Siaut et al., 2007), were used as references to calibrate the expression. Three technical and three biological replicates were conducted.
Results and Discussion
Physiological Response of P. tricornutum to Elevated pCO2
No significant differences were detected between cells grown under the HC and LC conditions with respect to pigment contents, Fv/Fm, photosynthetic yield, POC and PON content after growth for 15 generations (Table 1). However, a slight increase in specific growth rate (2.6%) was observed in the HC condition, which is consistent with the previous study (Wu et al., 2010). And the pigment contents, photosynthetic efficiency and growth rate were not significantly different between LC and HC cultures grown for 2 days (∼2.7 generations). The lack of physiological differences between LC and HC suggests an effective and stable acclimated state to the new CO2 saturated environment, revealing a very dynamic plasticity to environmental change in diatoms. The underlying transcriptomic changes may help to maintain the physiological performance in response to elevated CO2.
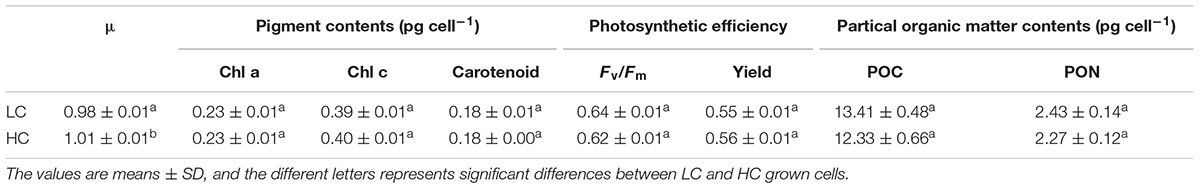
Table 1. The physiological parameters of Phaeodactylum tricornutum after growing for 15 generations.
The Global Transcriptomic Response to Elevated pCO2 After Growing for 15 Generations
ssRNA-seq data generated from the HC and LC physiological states were assembled and mapped. The average mapping rate for ssRNA-seq data was about 76.8%. The average mapping rate on the intergenic regions was 24.8% (Supplementary Table S4). The expression profile of each gene is listed in Supplementary Table S5. The GO enrichment analysis of differentially expressed genes in the domains of biological processes, cellular components and molecular functions is shown in Supplementary Figure S1. Globally, 954 protein coding genes were differentially expressed in response to HC with 179 up-regulated genes, 287 relatively up-regulated genes, 175 down-regulated genes and 313 relatively down-regulated genes compared with the LC condition over 15 generations. In general, the numbers of differentially regulated genes under the HC condition were much lower than induced by other stimuli such as nitrogen starvation and phosphate depletion (Ge et al., 2014; Cruz de Carvalho et al., 2016). This may suggest that elevated pCO2 is not a strong environmental stimulus compared to nitrogen starvation and phosphate depletion for P. tricornutum. A good correlation was found between transcriptomic data and RT-qPCR, thus validating the robustness of the sequencing data (Supplementary Table S6).
The Impact of Elevated pCO2 on Metabolic Genes
Our results showed that some genes involved in CCMs and energy-production/consumption were down-regulated after growing in the HC condition for 15 generations (Figures 1, 5). As expected, some genes involved in CCMs were down-regulated since high CO2 is expected to reduce the need for CCMs. Two genes encoding pyrenoid-localized carbonic anhydrases (Pt CA1: Phatr3_J51305, Pt CA2: Phatr3_J45443) (Tachibana et al., 2011) were down-regulated in the HC condition while other genes encoding carbonic anhydrases were not significantly affected by changing CO2 concentrations. As reported in (Tachibana et al., 2011; Tanaka et al., 2016), Pt CA1 and Pt CA2 was down-regulated under 5% CO2 (50,000 ppmv). Therefore, Pt CA1 and Pt CA2 may be the most responsive carbonic anhydrase gene in face of elevated CO2 and essential in converting from HCO3- to CO2 for carbon fixation by Rubisco. Other CCMs components putative HCO3- transporter (SLC4-5, Phatr3_ J54405), putative CO2 hydration protein (ChpXY, Phatr3_J38227) and malate dehydrogenase (MDH, Phatr3_J42398) were also down-regulated in HC. Furthermore, some genes encoding proteins with roles in energy-producing metabolic pathways such as glycolysis, oxidative phosphorylation and TCA cycle were preferentially down-regulated. This suggested a general reduction in metabolism with an energy saving status in the HC acclimated state. Genes encoding V-type ATPase subunit F (Phatr3_J31133) and mitochondrial F-type ATPase (Phatr3_J39529), which are directly related to energy consumption and production, respectively, showed down-regulation in HC. The down regulation of genes involved in CCMs and energy turnover rate suggest that a general reduction in metabolism may have contributed to the slightly increased growth rate (2.6%) in HC conditions, which is also consistent with previous experimental and modeling results (Wu et al., 2010; Hopkinson et al., 2011).
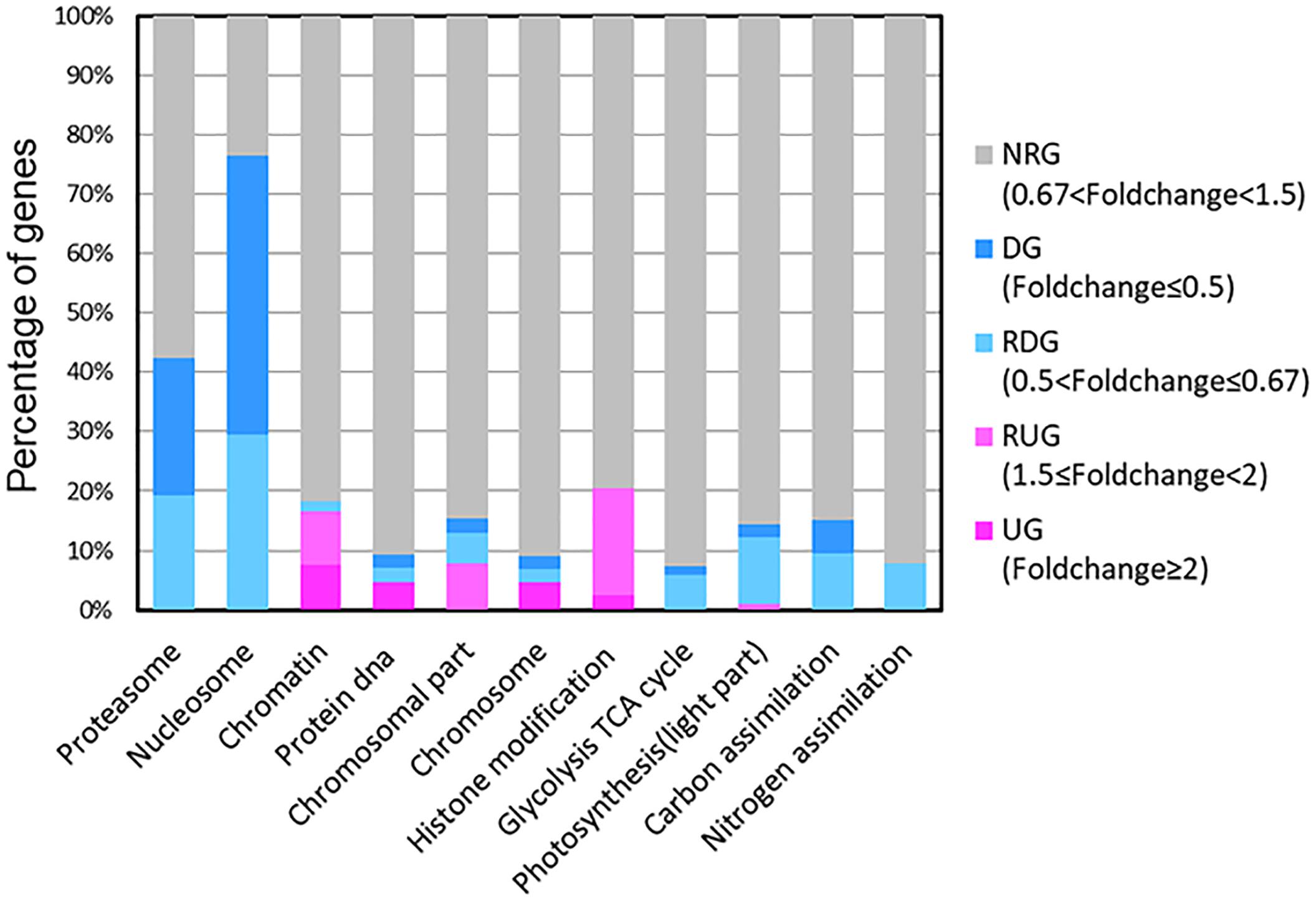
Figure 1. Transcripts abundance (HC/LC) of 11 gene families after Phaeodactylum tricornutum grown for 15 generations. Changes are denoted as the percentage of genes up-regulated with fold change ≥ 2 (dark pink, UGs), relatively up-regulated with 1.5 ≤ fold change < 2 (light pink, RUGs), down regulated with fold change ≤ 0.5(dark blue, DGs), relatively down-regulated with 0.5 < fold change ≤ 0.67 (light blue, RDGs) and non-significantly regulated (gray, NRGs) with 0.67 < fold change < 1.5 within each family. The function description of genes in different gene families and their HC/LC fold change can be found in Supplementary Table S4.
The photochemical performance was not significantly influenced after 15 generations under elevated pCO2; however, some light capture genes such as Fucoxanthin-chlorophyll a-c binding proteins and Fucoxanthin-chlorophyll a/c proteins were down-regulated in the acclimated HC condition (Figure 5). A gene encoding a putative Cytb6f iron-sulfur subunit (Phatr3_J13358), which is essential for electron transport in photosynthesis, was also down-regulated (Figure 5). The down-regulation of genes encoding light capture and electron transport proteins may result from decrease in energy demands of CCMs and maintaining H+ gradient across thylakoid membrane. Although photorespiration was not measured in our study, the putative 2-phosphoglycolate phosphatase gene (Phatr3_Jdraft1186), a key enzyme in the photorespiration pathway, was down-regulated indicating possible decrease in photorespiration rates under the HC condition (Figure 5), which is in line with the previous results in T. pseudonana (Hennon et al., 2015).
A significant proportion of ribosome genes were down-regulated in HC reflecting either a decreased capacity for protein synthesis or a low turnover of ribosomal proteins while a significant proportion of genes encoding proteasome core complex were significantly down-regulated by elevated pCO2 suggesting that the protein degradation system was relatively inactive (Figures 1, 5). All these results indicate a low turnover rate of protein synthesis and degradation, which is somewhat in accordance with the lower energy cost implied by the down-regulation of energy producing genes in HC described above. The down-regulation of ribosome genes in the HC condition was in contrast with the previous study on T. pseudonana acclimated to high CO2 for 10 days (Hennon et al., 2015), probably because the previous study involved low nitrate concentration.
The Impact of Elevated pCO2 on Genes Involved in pH Homeostasis Maintenance
Beside genes directly involved in metabolic pathways being affected by elevated pCO2, some genes related to cellular pH homeostasis maintenance were also affected by elevated pCO2. It is predicted that the bulk seawater H+ concentration (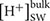 ) will increase by 100–150% by 2100 in response to ocean acidification (Caldeira and Wickett, 2003). It was reported that the increased pH in seawater results in greater diel variation of H+ concentration proximate seawater (
) will increase by 100–150% by 2100 in response to ocean acidification (Caldeira and Wickett, 2003). It was reported that the increased pH in seawater results in greater diel variation of H+ concentration proximate seawater (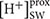 ) of marine organisms by boundary-layer processes (Caldeira and Wickett, 2003). The changes in
) of marine organisms by boundary-layer processes (Caldeira and Wickett, 2003). The changes in 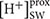 affect the operation of cellular processes involved in pH homeostasis including H+ production/consumption and active/passive H+ transport (Smith and Raven, 1979; Taylor et al., 2011). It has been reported that the down-regulation of CCMs changes intracellular H+ concentration by decreasing HCO3- transport into cytoplasmic matrix and conversion from HCO3- to CO2 (Smith and Raven, 1979). To maintain the neutral and acid pH in cytoplasmic matrix and organelles, the proton pump plays critical roles in H+ transport (Smith and Raven, 1979). In our study, Phatr3_J31133 encoding the subunit F of vacuolar V-type proton pump where H+ is transported from cytoplasmic matrix into vacuoles coupled with ATP hydrolysis was down-regulated in HC; however, Phatr3_J43207 coding inorganic H+ pyrophosphatase was up-regulated stimulating H+ transport from cytoplasmic matrix into vacuoles coupled with pyrophosphate hydrolysis (Figure 5). Similarly, Phatr3_J39529 encoding epsilon chain of mitochondrial F-type proton pump in which H+ is transported from mitochondria into cytoplasmic matrix coupled with ATP synthesis was down-regulated; however, Phatr3_J50171 encoding protein related to mitochondria K+-H+ exchange was up-regulated (Figure 5). These results suggest that the decrease in H+ transport coupled with ATP metabolism may be offset by other H+ transport pathways to maintain the cellular pH homeostasis under high pCO2 conditions in P. tricornutum. More investigations, such as pH measurements in different organelles and the cytoplasm under high pCO2 conditions compared to ambient conditions, are needed for better understanding of pH homeostasis mechanisms operating under conditions reflecting ocean acidification in diatoms.
affect the operation of cellular processes involved in pH homeostasis including H+ production/consumption and active/passive H+ transport (Smith and Raven, 1979; Taylor et al., 2011). It has been reported that the down-regulation of CCMs changes intracellular H+ concentration by decreasing HCO3- transport into cytoplasmic matrix and conversion from HCO3- to CO2 (Smith and Raven, 1979). To maintain the neutral and acid pH in cytoplasmic matrix and organelles, the proton pump plays critical roles in H+ transport (Smith and Raven, 1979). In our study, Phatr3_J31133 encoding the subunit F of vacuolar V-type proton pump where H+ is transported from cytoplasmic matrix into vacuoles coupled with ATP hydrolysis was down-regulated in HC; however, Phatr3_J43207 coding inorganic H+ pyrophosphatase was up-regulated stimulating H+ transport from cytoplasmic matrix into vacuoles coupled with pyrophosphate hydrolysis (Figure 5). Similarly, Phatr3_J39529 encoding epsilon chain of mitochondrial F-type proton pump in which H+ is transported from mitochondria into cytoplasmic matrix coupled with ATP synthesis was down-regulated; however, Phatr3_J50171 encoding protein related to mitochondria K+-H+ exchange was up-regulated (Figure 5). These results suggest that the decrease in H+ transport coupled with ATP metabolism may be offset by other H+ transport pathways to maintain the cellular pH homeostasis under high pCO2 conditions in P. tricornutum. More investigations, such as pH measurements in different organelles and the cytoplasm under high pCO2 conditions compared to ambient conditions, are needed for better understanding of pH homeostasis mechanisms operating under conditions reflecting ocean acidification in diatoms.
Potential Changes in Epigenetic Regulation in Response to Elevated pCO2 Over 15 Generations
After growing in HC and LC conditions for 15 generations, the most striking molecular response was the down-regulation of genes encoding histones and the up-regulation of many TEs. TE and histone gene variations and their changes in expression may have fundamental effects on the adaptive responses to elevated pCO2, as a result stabilizing the physiological performance. 12 out of 14 histone genes (H3, H4, H2B, H3B, and H1) were significantly down-regulated. For example, Phatr3_J54360 encoding H2B 1b isoform showed down-regulation in both RT-qPCR (Supplementary Table S6) and ssRNA-seq data sets in HC. Only Phatr3_J28445 encoding H2A 1 (H2A.Z) and Phatr3_J21239 encoding H3.2 were not significantly affected by elevated pCO2. Phatr3_EG01358 encoding histone H2A isoform 3a, with fold change 0.098 (HC/LC), was the most significantly down-regulated histone gene. On the other hand, genes encoding core domains of TEs, in particular the gag pol genes, were highly expressed in HC compared to LC condition. The LTR-RTs, a group of TEs, are constituted by gag pol genes flanking with long terminal repeat sequences. The gag gene encodes the virus-like particle structural proteins where reverse transcription takes place while the pol genes encode several enzymes, the protease cleaving the POL polyprotein, the reverse transcriptase copying the retrotransposon RNA into cDNA, and the ribonuclease H domain and an integrase integrating the cDNA into the genome. Under HC conditions compared to LC, Phatr3_J48048 and Phatr3_EG00052 encoding reverse transcriptase were up-regulated, and genes encoding integrase core domain (Phatr3_EG00775, Phatr3_J39341, and Phatr3_Jdraft1453 and Phatr3_J33646) also showed up-regulation. In addition, 17 genes encoding putative endonuclease or exonuclease were up-regulated in HC compared to LC. These results demonstrated that TEs were active in response to elevated pCO2.
The LTR-RTs are classified into Ty1/copia elements and Ty3/gypsy elements based on the organization of their pol genes and similarities among their encoded reverse transcriptase proteins. Among these active TEs, Copia type retrotransposons were the most active transposons in the HC condition compared to LC based on the order of core domain in the pol region. Based on the TE sequences provided in (Maumus et al., 2009), CoDi 1.1 (EU432476.1, Phatr3_EG00052), CoDi 4.4 (EU432484.1, Phatr3_EG01511), CoDi 6.5 (EU432496.1, Phatr3_EG00775), CoDi 2.6 (EU363804.1, Phatr3_J50428), and CoDi 3.1 (EU432481.1, Phatr3_J33646) were significantly up-regulated in response to the HC condition, as shown in Figure 2. In P. tricornutum, it has been reported that Blackbeard and Surcouf, belonging to the Ty1/copia TEs, were responsive to nitrate starvation and high decadienal condition, respectively (Maumus et al., 2009). In our study, the activation of Copia TEs, especially CoDi 1.1 which was the most active TE, in response to the HC condition after growing for 15 generations was consistent with what has been observed in natural environmental samples that Copia TEs were more active compared to other TEs (Lescot et al., 2016). This suggests that certain TEs specifically respond to given conditions and further emphasize the importance of Copia TEs in response to different environmental stimuli.
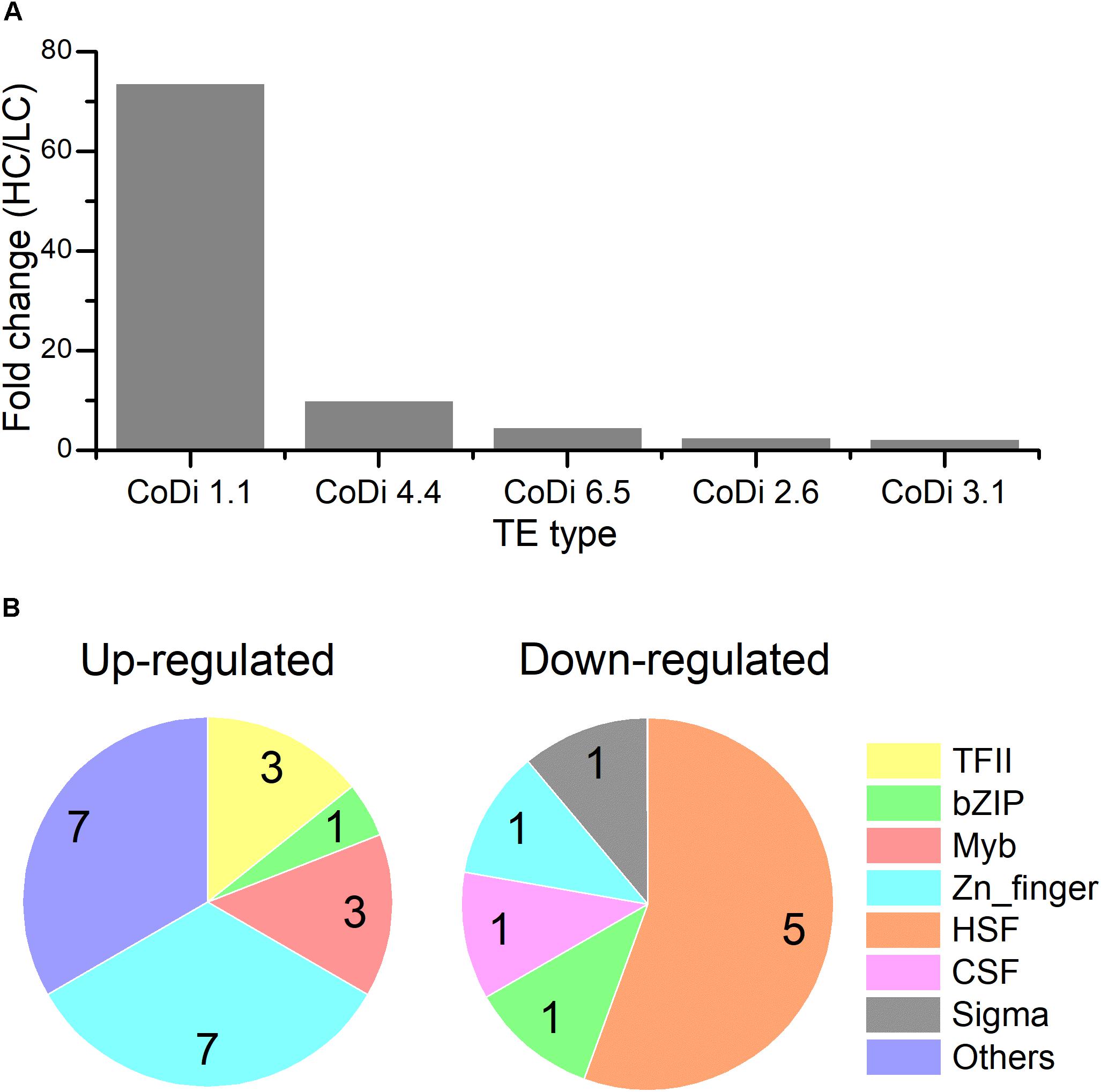
Figure 2. (A) The expression levels of different classes of transposon elements (HC/LC) after P. tricornutum grown for 15 generations. (B) Pie charts of different classes of up-regulated and down-regulated transcription factors (TFs) under the HC condition compared to the LC condition in P. tricornutum after grown for 15 generations (up-regulated: HC/LC fold change ≥ 1.5, down-regulated: HC/LC fold change ≤ 0.67, padj < 0.05).
The mobile nature of TEs can contribute to alterations in genetic regulatory elements, therefore altering levels of gene expression, triggering genome rearrangements and mutations that can accelerate biological diversification and consequently influence genome evolution (Fedoroff, 2012; Lisch, 2013). It is generally considered that the insertion of TEs has detrimental effects, including gene inactivation and alteration of chromosome structure. On the other hand, the beneficial effects of TEs have also been noted. TEs can enhance their hosts’ fitness, induce advantageous rearrangements or enrich the host’s gene pool in the long term (Schaack et al., 2010; Casacuberta and González, 2013). It has been further recognized that TEs as diversifying agents may contribute to adaptive processes occurring over short time scales. These aspects may explain the abundance and ubiquity of TEs in the natural environment (Lisch, 2013; Belyayev, 2014; Schrader et al., 2014).
In our study, the activation of TEs accompanying the down-regulation of histone genes in the HC condition may increase the potential for genome restructuring, and consequently to reshape gene expression patterns by TE insertions in promoters, enhancers and exons, and result in sequence expansion, gene duplication, novel gene formation and expansion, and re-wiring of genetic regulatory networks. More importantly, the effects on the genomes induced by elevated pCO2 on the acclimated state could be transmissible to future generations, which not only increase the fitness and phenotypic variation for acclimation in short time but also tremendously influence the adaptation and evolution of the whole population, such as the whole diatom community, in the long term under the elevated pCO2 conditions (Figure 6). Accumulating data has shown that TEs are not only the most abundant and ubiquitous genes in nature but also are transcriptionally active in marine assemblages and in mutualistic endosymbionts, which further indicate their importance in response to environmental change (Aziz et al., 2010; Kleiner et al., 2013; Lescot et al., 2016).
DNA methylation and histone modifications are two major epigenetic components, which can mediate heritable changes in gene functions beyond DNA sequence changes. In P. tricornutum, cytosine DNA methylation is commonly found in TEs and may be involved in controlling TE mobility in the genome (Veluchamy et al., 2013). As a case in point, the activation of the retrotransposon Blackbeard was accompanied by its hypomethylation under nitrate starvation (Maumus et al., 2009). The “epi-transposon” hypothesis proposes that changing environments can lead to stress-induced breakdown of epigenetic suppression of TEs (such as DNA methylation), which results in extensive transposition, thus providing new material for rapid adaptive shifts in short term acclimation (Zeh et al., 2009). It was reported that some TEs lose DNA methylation in P. tricornutum in response to nitrate starvation (Veluchamy et al., 2015). In our study, besides the down-regulation of histone genes, the burst of TEs was the most significant genetic response in the acclimated HC state. We therefore speculate that the highly expressed TEs may be accompanied by DNA demethylation on TEs in response to elevated pCO2, although there is currently a lack of genome wide DNA methylation data under the HC and LC conditions. It has been shown that reducing the amount of epigenetic variation available to populations can reduce adaptation in the green alga Chlamydomonas reinhardtii, which highlights the importance of epigenetic changes in the adaptation response to environmental changes (Kronholm et al., 2017).
Previous studies have also shown that histone modification may play a role in responses to different environmental stimuli in diatoms. Histone acetylation is usually considered as the positive hallmark for gene expression and H3K9/14 acetylation and H3K4me2 marks were found to be modulated in response to nitrate limitation in P. tricornutum (Veluchamy et al., 2015). It was further reported that genes encoding putative proteins involved in epigenetic modifications were up-regulated under phosphorus depletion in P. tricornutum (Cruz de Carvalho et al., 2016). In this study, the genes encoding proteins responsible for histone acetylation (Phatr3_J45764, Phatr3_J54343, Phatr3_J45703, Phatr3_EG02442, and Phatr3_J45644) were up-regulated in HC. Further studies on whether the up-regulation of histone acetylases leads to acetylation of specific histones residues resulting in up-regulation of histone acetylation marked genes under elevated pCO2 conditions in P. tricornutum will therefore be of interest.
In order to further analyze the potential roles of histone modification in response to HC, the network of differentially expressed putative histone modification genes and their associated genes was constructed in Cytoscape (Figure 3). All putative histone modification genes, including putative histone deacetylase Phatr3_J4423, putative histone acetyltransferases Phatr3_J45703, Phatr3_J54343, and Phatr3_J45764 and putative H3K4 methyltransferase Phatr3_J44935 were up-regulated in the constructed network in HC. The putative H3K4 methyltransferase (Phatr3_J44935), histone deacetylase genes (Phatr3_J4423) and histone acetyltransferases (Phatr3_J45703 and Phatr3_J45764) are highly connected with other genes, which implies that H3K4 methylation and histone deacetylation or/and acetylation may be involved in regulating genes in response to elevated pCO2, such as genes involved in TCA cycle and carbon assimilation (Phatr3_J53935, Phatr3_J22122).
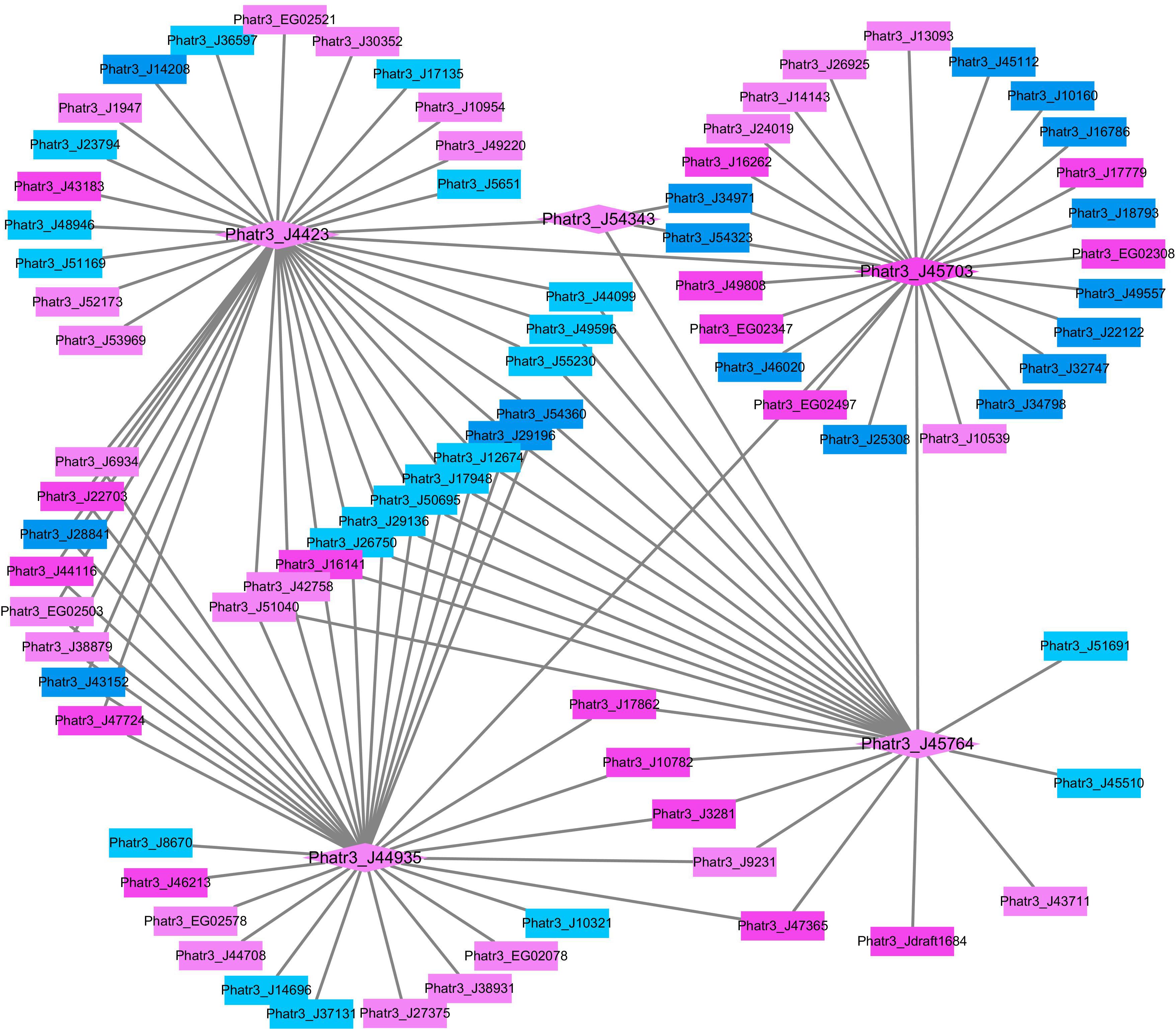
Figure 3. The network of differentially expressed histone modification genes and their correlated differentially expressed mRNAs under the HC relative to the LC conditions after growing for 15 generations. The diamonds illustrate histone modification genes while the rectangles illustrate other mRNAs. The up-regulated genes are filled with pink color and the down-regulated genes are filled with blue color (UGs: dark pink, DGs: dark blue, RUGs:light pink, RDGs:light blue).
The Response of Transcription Factors to Elevated pCO2
The TFs of diatoms are able to change their activity and ultimately control the diatom transcriptomic response to different signals. We investigated the expression profile of 289 genes encoding putative TFs (Rayko et al., 2010; Cruz de Carvalho et al., 2016). Among them, 30 TFs (21 up-regulated TFs with HC/LC fold change ≥ 1.5 and 9 down-regulated TFs with HC/LC fold change ≤ 0.67) were differentially expressed after growing for 15 generations in HC compared to LC (Supplementary Table S7 and Figure 2). Phatr3_J37556 encoding transcription initiation factor TFIIB was the most up-regulated TF while Phatr3_J45112 encoding a heat shock factor was the most down-regulated TF. A total of seven up-regulated TFs belonged to Zn-finger and more than 50% up-regulated TFs were C2H2-type. In addition, three up-regulated TFs belonged to “Myeloblastosis” family, which may mediate signal transduction pathway in response to abiotic drivers (Chen et al., 2006). Meanwhile, 50% of the down-regulated TFs belong to heat shock factors (HSFs) which can bind to the conserved heat shock elements found in the promoters of target genes, including heat shock proteins (HSPs) (Åkerfelt et al., 2010). The down-regulation of HSFs correlated with the decreased expression of HSPs, an important group of molecular chaperones involved in protein assembly and folding, suggesting an acquired acclimated state under elevated pCO2.
Long Non-coding Transcripts Specifically Induced by Elevated pCO2
We analyzed the P. tricornutum non-coding transcriptome and identified 189 lncRNAs which are all intergenic lncRNAs (lincRNAs). The majority of the lincRNAs identified are between 600 and 700 nt in length and are significantly shorter than mRNAs, which range from 100 nt to more than 3000 nt. The majority of the detected P. tricornutum lincRNAs were also intronless, with one exon of similar size to the exons found in mRNAs previously described (Cruz de Carvalho et al., 2016) (Supplementary Figure S2). This is a common feature of lincRNAs and has been suggested to be related to the nuclear localization of these transcripts (Niazi and Valadkhan, 2012). The expression levels of lincRNAs were lower than mRNA in P. tricornutum as shown in Supplementary Figure S3 which is also a common feature of this class of transcripts. Among them, 54 lincRNAs were up-regulated in HC while 59 lincRNAs were down-regulated. 28 up-regulated lincRNAs and 35 down-regulated lincRNAs are correlated with lincRNAs identified in the phosphate fluctuation study (Cruz de Carvalho et al., 2016) (Figure 4). The most significantly up-regulated lincRNA was lnc_117 while the most down regulated lincRNA was lnc_145 (Supplementary Table S8).
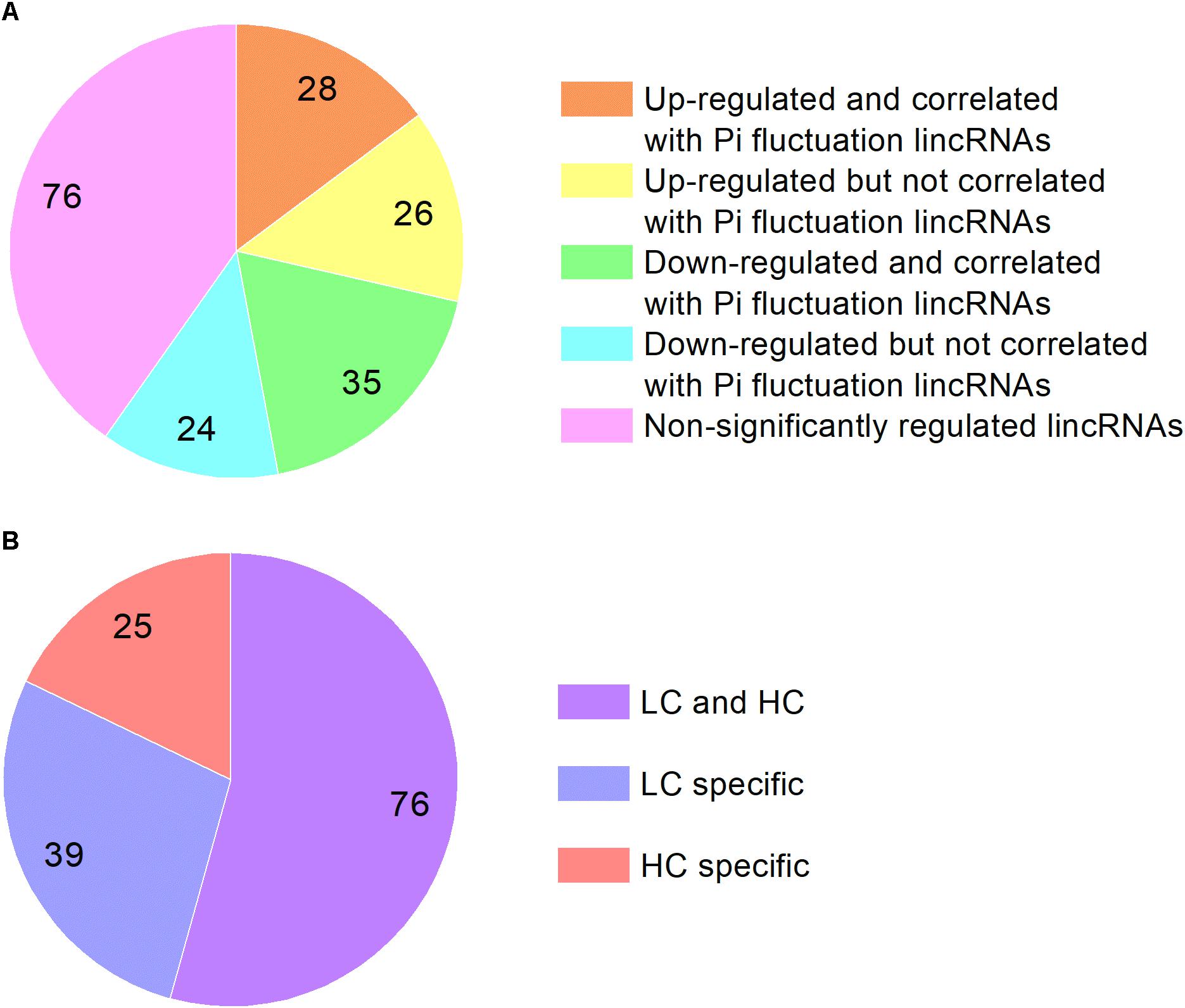
Figure 4. (A) The classification of lincRNAs identified in this study based on the correlation with lincRNAs identified in phosphate fluctuation study and expression. (B) The classification of lincRNAs identified in phosphate fluctuation study based on the correlation with elevated CO2 responsive lincRNAs.
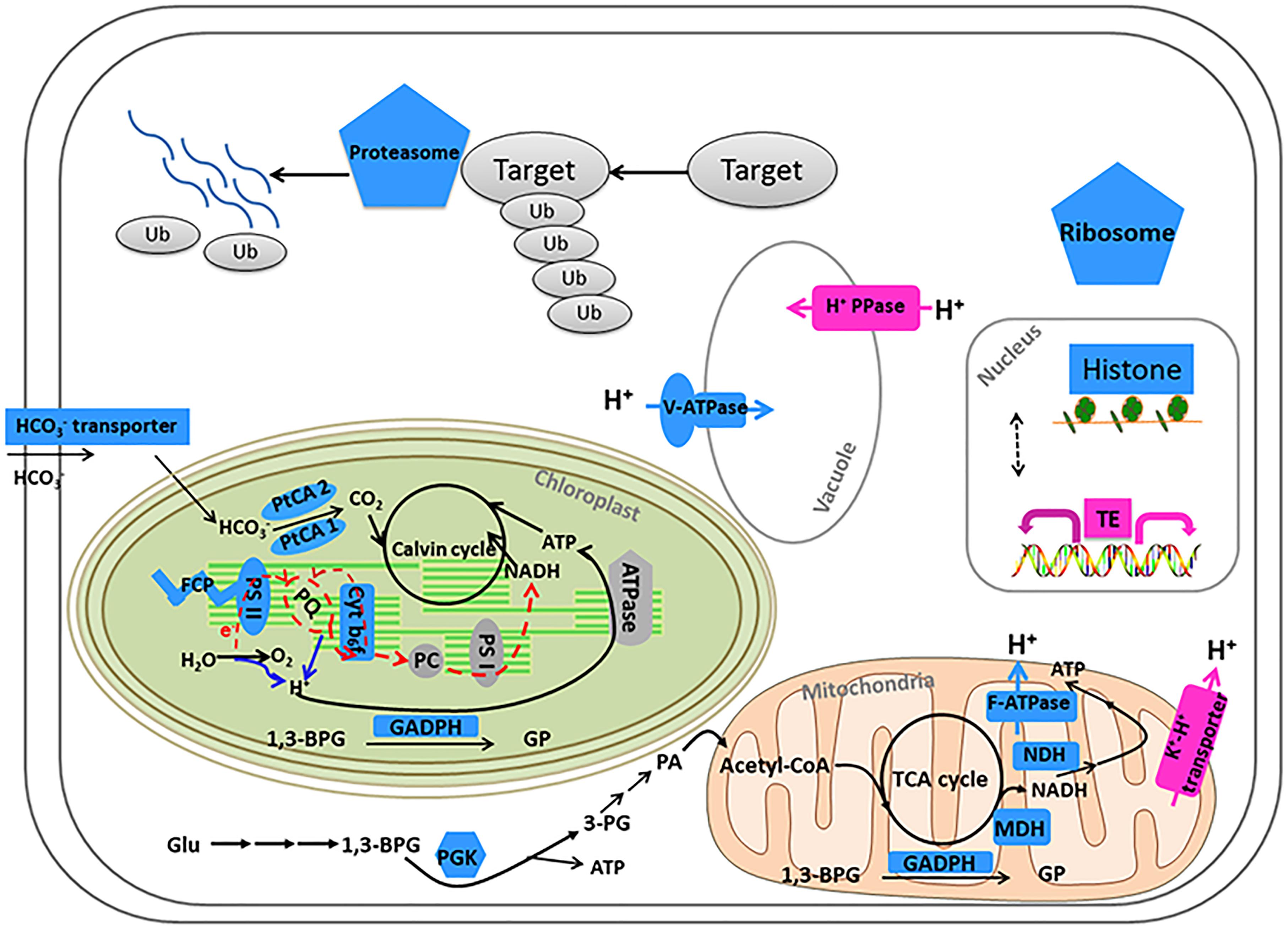
Figure 5. Model of metabolic and signaling pathways in P. tricornutum after acclimation to HC for 15 generations. Gene expression changes (HC/LC) are indicated by different colors: pink (up-regulation) and blue (down-regulation). Solid arrows indicate reactions and dashed arrows indicate regulatory relationships. HCO3- transporter: Phatr3_J54405; Fucoxanthin chlorophyll a-c binding protein or Fucoxanthin chlorophyll a/c protein (FCP): Phatr3_J24119, Phatr3_J25893, Phatr3_J18049, Phatr3_J27278, Phatr3_J54027; Photosystem II subunit (PS II): Phar3_J55057, Phatr3_J44899; Carbonic anhydrase (PtCA 2): Phar3_J45443; Carbonic anhydrase (PtCA 1): Phar3_J51305; Cytb6f iron-sulfur subunit: Phatr3_J13358; NADH dehydrogenase (NDH): Phatr3_EG00870, Phatr3_EG01423; Malate dehydrogenase, mitochondrial (MDH): Phar3_J42398; Mitochondrial ATPase: Phar3_J39529; F-ATPase: Phatr3_J31133; 1,3-BPG: 1,3-bisphosphoglycerate; 3-PG: 3-phosphoglycerate; GP: Glyceraldehyde 3-phosphate; Glyceraldehyde 3-phosphate dehydrogenase (GADPH): Phar3_J25308 (mitochondria), Phar3_J22122 (chloroplast), Phar3_J32747; Glu: Glucose; PA: pyroracemic acid; Phosphoglycerate kinase (PGK): Phar3_J48983; Histones: Phatr3_J34971, Phatr3_J54360, Phatr3_J46020, Phatr3_J11841, Phatr3_J26896, Phatr3_J26802, Phatr3_J34798, Phatr3_J50872, Phatr3_J11823, Phatr3_EG02092, Phatr3_EG01358, Phatr3_J50695; Ribosome: Phatr3_J17519, Phatr3_Jdraft477, Phatr3_J28562, Phatr3_J10196, Phatr3_J47804, Phatr3_J36226, Phatr3_J51066; Proteasome: Phatr3_Jdraft611, Phatr3_Jdraft866, Phatr3_EG00973, Phatr3_J30003, Phatr3_J5685, Phatr3_EG02026, Phatr3_J27508, Phatr3_J49897, Phatr3_J51691, Phatr3_J20007, Phatr3_EG02638, Phatr3_J24474; V-ATPase: Phatr3_J31133; Inorganic H+ pyrophosphatase (H+ PPase): Phatr3_J43207; K+-H+ transporter: Phatr3_J50171.
We compared the lincRNAs identified in our study with the lincRNAs associated with a phosphate fluctuation response (Cruz de Carvalho et al., 2016). The results showed that 140 lincRNAs associated with phosphate fluctuations are also responsive to CO2 concentrations. Among these 140 lincRNAs, 25 lincRNAs are associated HC conditions, 39 lincRNAs are associated with LC conditions, and 76 lincRNAs are associated with both HC and LC conditions as shown in Supplementary Table S9. Most of the lincRNAs identified in this study are also involved in phosphate stress responses suggesting that linRNAs may play central regulatory roles in response to different environmental conditions. LincRNAs have been described previously to be involved in a multitude of regulatory processes at the transcriptional, post-transcriptional and epigenetic levels (Niazi and Valadkhan, 2012; Amaral et al., 2013; Hon et al., 2017). LincRNAs have been namely reported to associate with several regulatory proteins such as TF and chromatin modifying complexes and have central roles in cellular homeostasis and stress response adaptations (Amaral et al., 2013). Although the regulatory roles of lincRNAs in P. tricornutum in response to environmental stimuli remain to be determined, their expression patterns under HC and phosphate fluctuations indicate a tight regulation, which strongly suggests function.
Conclusion
Our results suggest protectthat chromatin-based processes may be important for the response of P. tricornutum to ocean acidification, not only in short term acclimation but also in long term adaptation (Figure 6). The striking activation of TEs combined with down regulation of histone genes indicates the potential epigenetic component in the response to ocean acidification in P. tricornutum. Some studies have shown that the dynamic of histone modifications are accompanied with the changes of histone genes expression (Busslinger et al., 1985; Rao et al., 2007; Zager and Johnson, 2009). This indicates that ocean acidification may profoundly influence the adaptive processes of diatoms, important primary producers in the oceanic environment with important impacts in marine ecosystems. It was proposed that when organisms are challenged by new conditions, they can improve their fitness by a multitude of processes involving physiological acclimation, epigenetic changes, structural re-arrangements of the genome, and changes in DNA sequence, described as the adaptation spectrum. Different adaptations are characterized by the time needed for organisms to attain them and by their duration (Yona et al., 2015). Physiological adaptation is what most studies focus on, but while they may confer selective advantages, they are not actively amplified, memorized, or propagated over many generations. On the next level are epigenetic adaptations and DNA copy-number adaptations, which constitute a molecular “memory” of relatively labile genetic changes, although they do not involve changes in the actual nucleotide sequence of the genome. The observed transcriptional activation of TEs and the down-regulation of histone genes may induce further changes that influence the long term adaptation of P. tricornutum, such as after growing 1000 or longer generations under elevated pCO2 (Figure 6). In contrast with physiological adjustments, this level of adaptation could be transmitted to subsequent generations and thus have more profound effects on the whole community. New genes and new regulatory elements may appear in P. tricornutum due to potential TE insertions and chromosomal arrangement, which may lead to new metabolic state in response to elevated pCO2 as reported in Li et al. (2017). On the other hand, TE insertions and chromosomal arrangement may also lead to deleterious effects in P. tricornutum. The importance of epigenetic regulation in response to environmental changes in P. tricornutum may be extrapolated to the whole diatom community.
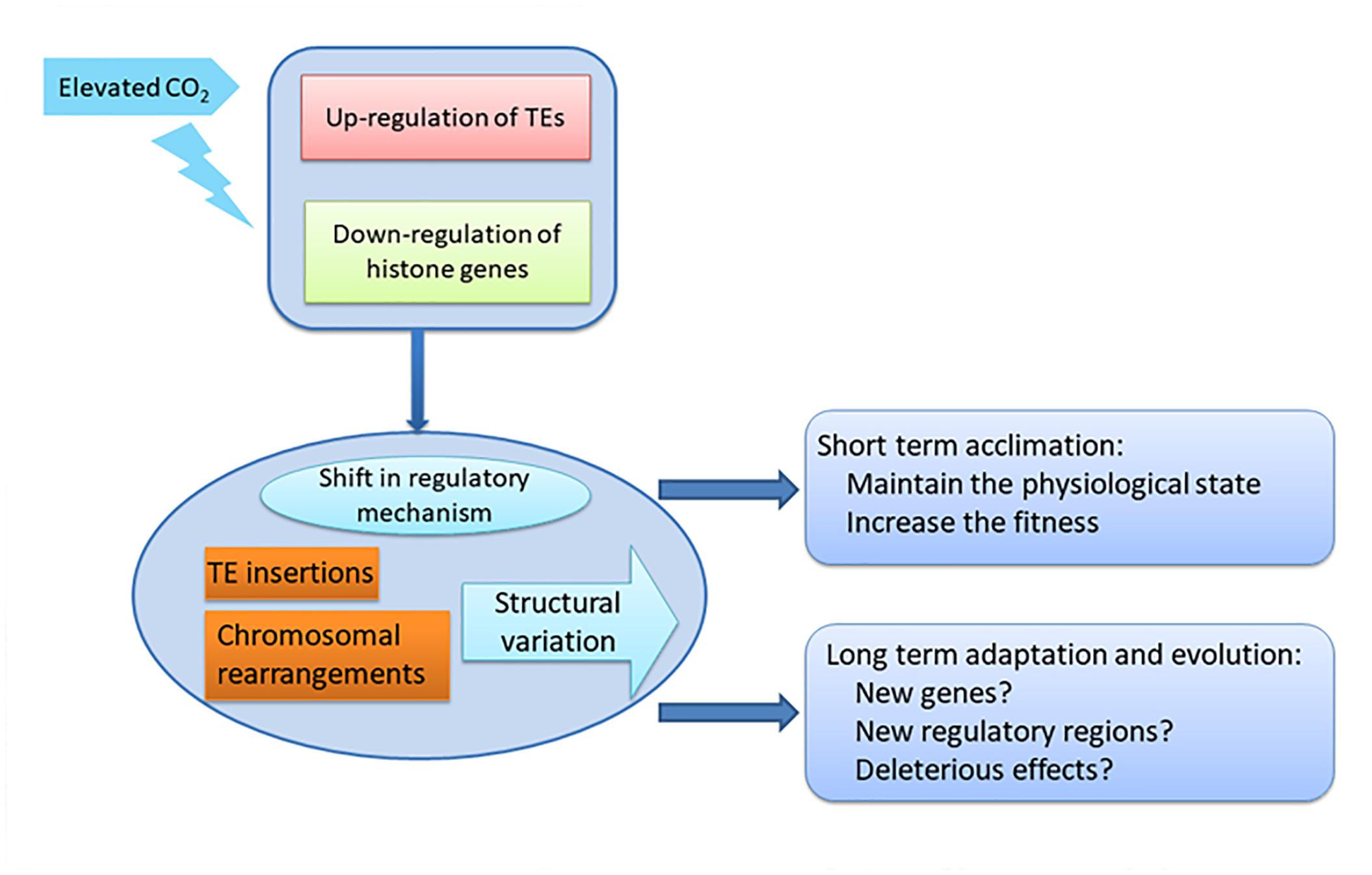
Figure 6. Schematic of the impact of down-regulation of histone genes and up-regulation of TEs on acclimation and adaptation of diatoms under elevated CO2.
In order to better understand the effects of ocean acidification, in addition to the ecological responses, the evolutionary responses and the underlying molecular mechanisms should be taken into account. It will be very interesting and worthwhile to investigate how epigenetic regulation and variation contribute to the acclimation in short term and adaptation in long term in response to elevated pCO2 using epigenetic approaches, such as genome wide methylome analysis and histone modification analysis (such as histone acetylation) by ChIP-seq. Furthermore, the expression profiles of several lincRNAs also suggests their involvement in the acclimation to elevated pCO2 responses which is worth to be further investigated in the future.
Data Availability Statement
The sequencing data using in this study have been deposited in the NCBI (PRJNA484278).
Author Contributions
XL, RH, and KG planned and designed the research. RH performed the experiments. RH, JD, and XL analyzed the data. XL, RH, MCC, CB, and LT wrote the manuscript.
Funding
This study was supported by the National Key Research and Development Program of China (Grant No. 2016YFA0601302) and National Natural Science Foundation of China (Grant Nos. 41306096 and 41430967).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Professor Senjie Lin for revising the manuscript. We also thank bioRxiv for posting this manuscript as a pre-print (Huang et al., 2018).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.03342/full#supplementary-material
Footnotes
- ^http://protists.ensembl.org/Phaeodactylum_tricornutum/Info/Index
- ^http://www.geneontology.org
- ^http://www.kegg.jp/
- ^http://string-db.org/
References
Åkerfelt, M., Morimoto, R. I., and Sistonen, L. (2010). Heat shock factors: integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 11, 545–555. doi: 10.1038/nrm2938
Amaral, P. P., Dinger, M. E., and Mattick, J. S. (2013). Non-coding RNAs in homeostasis, disease and stress responses: an evolutionary perspective. Brief. Funct. Genomics 12, 254–278. doi: 10.1093/bfgp/elt016
Armbrust, E. V., Berges, J. A., Bowler, C., Green, B. R., Martinez, D., Putnam, N. H., et al. (2004). The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79–86. doi: 10.1126/science.1101156
Aziz, R. K., Breitbart, M., and Edwards, R. A. (2010). Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res. 38, 4207–4217. doi: 10.1093/nar/gkq140
Belyayev, A. (2014). Bursts of transposable elements as an evolutionary driving force. J. Evol. Biol. 27, 2573–2584. doi: 10.1111/jeb.12513
Bowler, C., Allen, A. E., Badger, J. H., Grimwood, J., Jabbari, K., Kuo, A., et al. (2008). The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244. doi: 10.1038/nature07410
Busslinger, M., Schümperli, D., and Birnstiel, M. L. (1985). Regulation of histone gene expression. Cold Spring Harb. Syrup. Quant. Biol. 50, 665–670. doi: 10.1101/SQB.1985.050.01.081
Caldeira, K., and Wickett, M. E. (2003). Oceanography: anthropogenic carbon and ocean pH. Nature 425:365. doi: 10.1038/425365a
Casacuberta, E., and González, J. (2013). The impact of transposable elements in environmental adaptation. Mol. Ecol. 22, 1503–1517. doi: 10.1111/mec.12170
Chen, Y., Yang, X., He, K., Liu, M., Li, J., Gao, Z., et al. (2006). The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 60, 107–124. doi: 10.1007/s11103-005-2910-y
Cruz de Carvalho, M. H., Sun, H. X., Bowler, C., and Chua, N. H. (2016). Noncoding and coding transcriptome responses of a marine diatom to phosphate fluctuations. New Phytol. 210, 497–510. doi: 10.1111/nph.13787
Fedoroff, N. V. (2012). Transposable elements, epigenetics, and genome evolution. Science 338, 758–767. doi: 10.1126/science.338.6108.758
Feschotte, C., and Pritham, E. J. (2007). DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 41, 331–368. doi: 10.1146/annurev.genet.40.110405.090448
Gao, K., and Campbell, D. A. (2014). Photophysiological responses of marine diatoms to elevated CO2 and decreased pH: a review. Funct. Plant Biol. 41, 449–459. doi: 10.1071/FP13247
Gattuso, J.-P., Magnan, A., Billé, R., Cheung, W. W. L., Howes, E. L., Joos, F., et al. (2015). Contrasting futures for ocean and society from different anthropogenic CO2 emissions scenarios. Science 349:aac4722. doi: 10.1126/science.aac4722
Ge, F., Huang, W., Chen, Z., Zhang, C., Xiong, Q., Bowler, C., et al. (2014). Methylcrotonyl-CoA carboxylase regulates triacylglycerol accumulation in the model diatom Phaeodactylum tricornutum. Plant Cell 26, 1681–1697. doi: 10.1105/tpc.114.124982
Hennon, G. M. M., Ashworth, J., Groussman, R. D., Berthiaume, C., Morales, R. L., Baliga, N. S., et al. (2015). Diatom acclimation to elevated CO2 via cAMP signalling and coordinated gene expression. Nat. Clim. Chang. 5, 761–765. doi: 10.1038/nclimate2683
Hon, C. C., Ramilowski, J. A., Harshbarger, J., Bertin, N., Rackham, O. J. L., Gough, J., et al. (2017). An atlas of human long non-coding RNAs with accurate 5’ ends. Nature 543, 199–204. doi: 10.1038/nature21374
Hopkinson, B. M., Dupont, C. L., Allen, A. E., and Morel, F. M. M. (2011). Efficiency of the CO2-concentrating mechanism of diatoms. Proc. Natl. Acad. Sci. U.S.A. 108, 3830–3837. doi: 10.1073/pnas.1018062108
Huang, R., Ding, J., Gao, K. S., Cruz de Carvalho, M. H., Bowler, C., Tirichine, L., et al. (2018). Apotential role for epigenetic processes in the acclimation response to elevated pCO2 in the model diatom Phaeodactylum tricornutum. bioRxiv [Preprint]. doi: 10.1101/364604
Kleiner, M., Young, J. C., Shah, M., Verberkmoes, N. C., and Dubilier, N. (2013). Metaproteomics reveals abundant transposase expression in mutualistic endosymbionts. mBio 4:e00223-13. doi: 10.1128/mBio.00223-13
Kronholm, I., Bassett, A., Baulcombe, D., and Collins, S. (2017). Epigenetic and genetic contributions to adaptation in Chlamydomonas. Mol. Biol. Evol. 34, 2285–2306. doi: 10.1093/molbev/msx166
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. doi: 10.1186/gb-2009-10-3-r25
Lee, J. T. (2012). Epigenetic regulation by long noncoding RNAs. Science 338, 1435–1439. doi: 10.1126/science.1231776
Lescot, M., Hingamp, P., Kojima, K. K., Villar, E., Romac, S., Veluchamy, A., et al. (2016). Reverse transcriptase genes are highly abundant and transcriptionally active in marine plankton assemblages. ISME J. 10, 1134–1146. doi: 10.1038/ismej.2015.192
Li, Y., Xu, J., and Gao, K. (2014). Light-modulated responses of growth and photosynthetic performance to ocean acidification in the model diatom Phaeodactylum tricornutum. PLoS One 9:e96173. doi: 10.1371/journal.pone.0096173
Li, Y., Zhuang, S., Wu, Y., Ren, H., Chen, F., Lin, X., et al. (2017). Ocean acidification modulates expression of genes and physiological performance of a marine diatom. PLoS One 12:e0170970. doi: 10.1371/journal.pone.0170970
Lisch, D. (2013). How important are transposons for plant evolution? Nat. Rev. Genet. 14, 49–61. doi: 10.1038/nrg3374
Liu, J., Jung, C., Xu, J., Wang, H., Deng, S., Bernad, L., et al. (2012). Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24, 4333–4345. doi: 10.1105/tpc.112.102855
Liu, N., Tong, S., Yi, X., Li, Y., Li, Z., Miao, H., et al. (2017). Carbon assimilation and losses during an ocean acidification mesocosm experiment, with special reference to algal blooms. Mar. Environ. Res. 129, 229–235. doi: 10.1016/j.marenvres.2017.05.003
Losh, J. L., Young, J. N., and Morel, F. M. M. (2013). Rubisco is a small fraction of total protein in marine phytoplankton. New Phytol. 198, 52–58. doi: 10.1111/nph.12143
Maumus, F., Allen, A. E., Mhiri, C., Hu, H., Jabbari, K., Vardi, A., et al. (2009). Potential impact of stress activated retrotransposons on genome evolution in a marine diatom. BMC Genomics 10:624. doi: 10.1186/1471-2164-10-624
Niazi, F., and Valadkhan, S. (2012). Computational analysis of functional long noncoding RNAs reveals lack of peptide-coding capacity and parallels with 3’ UTRs. RNA 18, 825–843. doi: 10.1261/rna.029520.111
Porra, R. J., Thompson, W. A., and Kriedemann, P. E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta Bioenerg. 975, 384–394. doi: 10.1016/S0005-2728(89)80347-0
Rao, J., Bhattacharya, D., Banerjee, B., Sarin, A., and Shivashankar, G. V. (2007). Trichostatin-A induces differential changes in histone protein dynamics and expression in HeLa cells. Biochem. Biophys. Res. Commun. 363, 263–268. doi: 10.1016/j.bbrc.2007.08.120
Rastogi, A., Maheswari, U., Dorrell, R. G., Vieira, F. R. J., Maumus, F., Kustka, A., et al. (2018). Integrative analysis of large scale transcriptome data draws a comprehensive landscape of Phaeodactylum tricornutum genome and evolutionary origin of diatoms. Sci. Rep. 8:4838. doi: 10.1038/s41598-018-23106-x
Rayko, E., Maumus, F., Maheswari, U., Jabbari, K., and Bowler, C. (2010). Transcription factor families inferred from genome sequences of photosynthetic stramenopiles. New Phytol. 188, 52–66. doi: 10.1111/j.1469-8137.2010.03371.x
Schaack, S., Gilbert, C., and Feschotte, C. (2010). Promiscuous DNA: horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol. Evol. 25, 537–546. doi: 10.1016/j.tree.2010.06.001
Schrader, L., Kim, J. W., Ence, D., Zimin, A., Klein, A., Wyschetzki, K., et al. (2014). Transposable element islands facilitate adaptation to novel environments in an invasive species. Nat. Commun. 5:5495. doi: 10.1038/ncomms6495
Siaut, M., Heijde, M., Mangogna, M., Montsant, A., Coesel, S., Allen, A., et al. (2007). Molecular toolbox for studying diatom biology in Phaeodactylum tricornutum. Gene 406, 23–35. doi: 10.1016/j.gene.2007.05.022
Smith, F. A., and Raven, J. A. (1979). Intracellular pH and its regulation. Annu. Rev. Plant Physiol. 30, 289–311. doi: 10.1146/annurev.pp.30.060179.001445
Tachibana, M., Allen, A. E., Kikutani, S., Endo, Y., Bowler, C., and Matsuda, Y. (2011). Localization of putative carbonic anhydrases in two marine diatoms, Phaeodactylum tricornutum and Thalassiosira pseudonana. Photosynth. Res. 109, 205–221. doi: 10.1007/s11120-011-9634-4
Tanaka, A., Ohno, N., Nakajima, K., and Matsuda, Y. (2016). Light and CO2 /cAMP signal cross talk on the promoter elements of chloroplastic β -carbonic anhydrase genes in the marine diatom Phaeodactylum tricornutum. Plant Physiol. 170, 1105–1116. doi: 10.1104/pp.15.01738
Taylor, A. R., Chrachri, A., Wheeler, G., Goddard, H., and Brownlee, C. (2011). A voltage-gated H+ channel underlying pH homeostasis in calcifying coccolithophores. PLoS Biol. 9:e1001085. doi: 10.1371/journal.pbio.1001085
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Tréguer, P., Bowler, C., Moriceau, B., Dutkiewicz, S., Gehlen, M., Aumont, O., et al. (2018). Influence of diatom diversity on the ocean biological carbon pump. Nat. Geosci. 11, 27–37. doi: 10.1038/s41561-017-0028-x
Veluchamy, A., Lin, X., Maumus, F., Rivarola, M., Bhavsar, J., Creasy, T., et al. (2013). Insights into the role of DNA methylation in diatoms by genome-wide profiling in Phaeodactylum tricornutum. Nat. Commun. 4:2091. doi: 10.1038/ncomms3091
Veluchamy, A., Rastogi, A., Lin, X., Lombard, B., Murik, O., Thomas, Y., et al. (2015). An integrative analysis of post-translational histone modifications in the marine diatom Phaeodactylum tricornutum. Genome Biol. 16:102. doi: 10.1186/s13059-015-0671-8
Wagner, G. P., Kin, K., and Lynch, V. J. (2012). Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 131, 281–285. doi: 10.1007/s12064-012-0162-3
Wu, Y., Gao, K., and Riebesell, U. (2010). CO2-induced seawater acidification affects physiological performance of the marine diatom Phaeodactylum tricornutum. Biogeosciences 7, 2915–2923. doi: 10.5194/bg-7-2915-2010
Yona, A. H., Frumkin, I., and Pilpel, Y. (2015). A relay race on the evolutionary adaptation spectrum. Cell 163, 549–559. doi: 10.1016/j.cell.2015.10.005
Zager, R. A., and Johnson, A. C. (2009). Renal ischemia-reperfusion injury upregulates histone-modifying enzyme systems and alters histone expression at proinflammatory/profibrotic genes. Am. J. Physiol. 296, F1032–F1041. doi: 10.1152/ajprenal.00061.2009
Keywords: ocean acidification, climate change, diatom, transposable element, histone, long non-coding RNA
Citation: Huang R, Ding J, Gao K, Cruz de Carvalho MH, Tirichine L, Bowler C and Lin X (2019) A Potential Role for Epigenetic Processes in the Acclimation Response to Elevated pCO2 in the Model Diatom Phaeodactylum tricornutum. Front. Microbiol. 9:3342. doi: 10.3389/fmicb.2018.03342
Received: 28 August 2018; Accepted: 27 December 2018;
Published: 14 January 2019.
Edited by:
Susana Agusti, King Abdullah University of Science and Technology, Saudi ArabiaReviewed by:
Giovanna Benvenuto, Stazione Zoologica Anton Dohrn, ItalyRamiro Logares, Instituto de Ciencias del Mar (ICM), Spain
Copyright © 2019 Huang, Ding, Gao, Cruz de Carvalho, Tirichine, Bowler and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Lin, eGlubGludWxtQHhtdS5lZHUuY24=
 Ruiping Huang1
Ruiping Huang1 Kunshan Gao
Kunshan Gao Leila Tirichine
Leila Tirichine Chris Bowler
Chris Bowler Xin Lin
Xin Lin