- 1SAMRC Microbial Water Quality Monitoring Centre, University of Fort Hare, Alice, South Africa
- 2Applied and Environmental Microbiology Research Group, Department of Biochemistry and Microbiology, University of Fort Hare, Alice, South Africa
- 3Department of Biological Sciences, University of Medical Sciences, Ondo City, Nigeria
One of the major challenges of modern medicine includes the failure of conventional protocols to characterize the pathogenicity of emerging pathogens. This is particularly apparent in the case of Plesiomonas shigelloides. Although a number of infections have been linked to this microorganism, experimental evidence of its virulence factors (VFs), obtained by traditional approaches, is somewhat inconclusive. Hence, it remains unclear whether P. shigelloides is a true or opportunistic one. In the current study, four publicly available whole-genome sequences of P. shigelloides (GN7, NCTC10360, 302-73, and LS1) were profiled using bioinformatics platforms to determine the putative candidate VFs to characterize the bacterial pathogenicity. Overall, 134 unique open reading frames (ORFs) were identified that were homologous or orthologous to virulence genes identified in other pathogens. Of these, 52.24% (70/134) were jointly shared by the strains. The numbers of strain-specific virulence traits were 4 in LS1; 7 in NCTC10360; 10 in 302-73; and 15 in GN7. The pathogenicity islands (PAIs) common to all the strains accounted for 24.07% ORFs. The numbers of PAIs exclusive to each strain were 8 in 302-73; 11 in NCTC10360; 14 in GN7; and 18 in LS1. A PAI encoding Vibrio cholerae ToxR-activated gene d protein was specific to 302-73, GN7, and NCTC10360 strains. Out of 33 antibiotic multi-resistance genes identified, 16 (48.48%) genes were intrinsic to all strains. Further, 17 (22.08%) of 77 antibiotic resistance islands were found in all the strains. Out of 23 identified distinct insertion sequences, 13 were only harbored by strain LS1. The number of intact prophages identified in the strains was 1 in GN7; 2 in 302-73; and 2 in NCTC10360. Further, 1 CRISPR element was identified in LS1; 2 in NCTC10360; and 8 in 302-73. Fifteen (78.95%) of 19 secretion systems and secretion effector variants were identified in all the strains. In conclusion, certain P. shigelloides strains might possess VFs associated with gastroenteritis and extraintestinal infections. However, the role of host factors in the onset of infections should not be undermined.
Introduction
Plesiomonas shigelloides (hereafter Plesiomonas) possesses various VFs that result in its emerging pathogen status. E.g., the pathophysiology of Plesiomonas in certain infections has been attributed to possible endotoxin production by this bacterium (Okawa et al., 2004; Kaszowska et al., 2016). Endotoxins have been proposed to be central to septic shock and sepsis associated with plesiomonad infections (Alexander and Rietschel, 2001), and to biofilm formation, attachment, invasion of eukaryotic host cells, complement-resistance in the serum, or blood (Aquilini et al., 2013). Other reported virulence attributes ascribed to plesiomonads include the production of cytotoxic lipopolisaccharide complex (Fisher et al., 1988; Olsvik et al., 1990; Abbott et al., 1991; Ekman, 2003; Falcón et al., 2003; Okawa et al., 2004), β-hemolysin (Aldová et al., 1966; Zajc-Satler et al., 1972; Janda and Abbott, 1993; Santos et al., 1999; Krovacek et al., 2000; Baratéla et al., 2001; Falcón et al., 2003), enterotoxins (Sanyal et al., 1975, 1980; Saraswathi et al., 1983; Matthews et al., 1988; Sears and Kaper, 1996), adherence factors (Schubert and Holz-Bremer, 1999; Theodoropoulos et al., 2001; Ekman, 2003; Tsugawa et al., 2005), invasion factors (Sanyal et al., 1980; Binns et al., 1984; Olsvik et al., 1985; Herrington et al., 1987; Theodoropoulos et al., 2001; Tsugawa et al., 2005), cytolysin (Gurwith and Williams, 1977; Sanyal et al., 1980; Gardner et al., 1987; Olsvik et al., 1990; Abbott et al., 1991; Dumontet et al., 1998; Ekman, 2003), elastin (Ingram et al., 1987; Nolte et al., 1988; Rust et al., 1994; Santos et al., 1999), plasmids (Holmberg and Farmer, 1984; Herrington et al., 1987; Fisher et al., 1988; Nolte et al., 1988; Olsvik et al., 1990; Abbott et al., 1991; Taylor et al., 1993; Marshall et al., 1996; Bravo et al., 1998; Shepherd et al., 2000; Avison et al., 2001; Kubler-Kielb et al., 2008), tetrodotoxin (Wei et al., 1994; Cheng et al., 1995), and histamine (Lopez-Sabater et al., 1996; Dewaal et al., 2006; Bjornsdottir-Butler et al., 2011).
In addition, antibiotic and metal resistance as single or multiple resistance traits (Meyers et al., 1976; Brenden et al., 1988; Fisher et al., 1988; Avison et al., 2000; Stock and Wiedemann, 2001a) or acquired traits (Kain and Kelly, 1989a; Clark et al., 1990; Olsvik et al., 1990; Yeh and Tsai, 1991; Shaw et al., 1993; Bravo-Fariñas et al., 1998; González et al., 1999; Avison et al., 2000; Wong et al., 2000; Stock and Wiedemann, 2001b; Suthienkul et al., 2001; González-Rey et al., 2004; Jun et al., 2011) of plesiomonads have been reported.
Plesiomonas have been credited with causing many infections over the years. These include gastroenteritis and various forms of diarrhea (Tsukamoto et al., 1978; Wong et al., 2000; Chakour et al., 2002; Klontz et al., 2012; Pfeiffer et al., 2012; Novoa-Farías et al., 2016). Of major concern are extraintestinal infections of the central nervous system, such as neonatal meningoencephalitis, meningitis, sepsis, and septic shock (Claesson et al., 1984; Kalotychou et al., 2002; Auxiliadora-Martins et al., 2010; Ozdemir et al., 2010; Xia et al., 2015; Bowman et al., 2016). Other infections include peritonitis (Alcañiz et al., 1995), cellulitis (Gopal and Burns, 1991; Jönsson et al., 1998), wound and foot infections (McCracken and Barkley, 1972; Herve et al., 2007; Pence, 2016), endophthalmitis (Butt et al., 1997; Mahrshmann and Lyons, 1998), and keratitis (Butt et al., 1997; Klatte et al., 2012). Further, pneumonia (Schneider et al., 2009), migratory polyarthritis (Gupta, 1995), cholecystitis (Claesson et al., 1984; Kennedy et al., 1990), pyosalpinx (Roth et al., 2002), pseudo-appendicitis (Fischer et al., 1988), peritonitis (Patel et al., 2016), cholangitis and pancreatitis (Kennedy et al., 1990), and septicemia (Clark et al., 1991) have been attributed to Plesiomonas. However, determination of the associated virulence markers and pathogenic strains has been challenging and frustrating. While the majority of traditional approaches yielded inconclusive data with respect to the bacterium’s pathogenicity, others concluded that the virulence potential of the microorganism is low (Olsvik et al., 1990; Abbott et al., 1991).
Furthermore, suitable models for studying the pathogenesis of Plesiomonas have not yet been established and traditional models proved to be inappropriate. The development of polymerase chain reaction and other rapid low-cost protocols, already used for the analysis of other enteropathogens, for the detection of Plesiomonas VFs is also urgently required. Presently, no specific primer- and molecular-based techniques for the detection of the major, if not all, predicted Plesiomonas VFs exist. Further, strain-specific and rapid systems for the delineation of pathogenic and non-pathogenic Plesiomonas strains are lacking.
Since the emergence of new pathogens (including Plesiomonas) is linked to complex interrelated factors, of which mutation and horizontal gene transfer are the focal driving elements (Che et al., 2014), it is not implausible that several such events occurred in Plesiomonas. In most cases, gene acquisition or loss could impact genome evolution (Che et al., 2014), converting a non-pathogenic strain into a pathogenic one (McDaniel and Kaper, 1997). Although these phenomena may have contributed to the emergence of pathogenicity in Plesiomonas, no studies focusing on this issue have been reported. The study of PAIs, resistance islands, metabolic islands, mobility genes, IS elements, phage-related genes, tRNA genes, and direct repeats will be important for the characterization of the virulence and genome evolution of Plesiomonas.
Consequently, rapid specific culture-independent tools for pathogenicity characterization should be developed for Plesiomonas. A comparative in silico profiling of the available genomes may provide important clues prior to wet-lab studies for the assessment of virulence marker candidates in Plesiomonas and answer other questions about its emerging pathogenicity capabilities. This would not only reveal the as yet unknown virulence traits associated with specific strains, but also inform the development of rapid and inexpensive tools for strain screening and diagnostic characterization of plesiomonads. In the current study, we therefore profiled the publicly available complete genome sequence assemblies of Plesiomonas in silico to identify the associated type III, IV, VI, and VII secretion systems (T3SS, T4SS, T6SS, and T7SS, respectively); integrative and conjugation elements (ICE elements); prophages; VFs; ARs; ARIs; type III, IV, VI, and VII secretion effectors (T3SE, T4SE, T6SE, and T7SE, respectively); class I integrons, IS elements; and CRISPR.
Materials and Methods
Whole-Genome Sequence Assembly Selection
The four publicly available sequence assemblies of Plesiomonas were downloaded from NCBI website via ftp.ncbi.nlm.nih.gov/genomes/ASSEMBLY_BACTERIA (Table 1; last accessed [20 December 2017]).
Detection of Virulence Traits
Presence of PVFs within each genome assembly was predicted using the VRprofile 2.0 (Li et al., 2018). For gene clusters (including genes encoding T3SS, T4SS, T6SS, and T7SS), ICE, and prophage prediction, default parameter settings were used. BLASTP searches were done with a minimum e-value of 1 × 10-4; HMMer search E-value of 1 × 10-4 for ICE and prophage detection; the minimum number of significant hits for each prophage-like region set to 5; and a minimum length of direct repeats of an ICE-like region of 15 bp. Single genes related to VFs, ARs, T3SE, T4SE, T6SE, T7SE, class I integrons, IS elements, GI-like region, PAIs, and ARIs were predicted by setting the BLAST Ha-value to 0.64 (Li et al., 2018). The distribution of PVFs and related genes among the various strains was evaluated using a web-based Venn diagram program1.
In addition, a direct search of the WGS for presence of elements related to already suggested virulence mechanisms in P. shigelloides such as heme iron uptake systems or iron-siderophores (Daskaleros et al., 1991; Santos et al., 1999; Henderson et al., 2001; Gonzalez-Rodriguez et al., 2007; Oldham et al., 2008; Rodríguez-Rodríguez and Santos, 2018) was performed.
Detection of Prophage Sequences
To identify prophages within the Plesiomonas genomes, strain sequences were analyzed using the PHAge Search Tool Enhanced Release (Zhou et al., 2011; Arndt et al., 2016).
Determination of the CRISPR Elements
CRISPR elements in the Plesiomonas genomes were predicted using CRISPR Finder by Random Forest program (Wang and Liang, 2017).
Detection of Plasmids
Plasmid elements in the Plesiomonas genomes were predicted using PlasmidFinder 1.3 (Carattoli et al., 2014) and a direct search for replicon information on NCBI website2.
Results
Figures 1A–D presents the GI, T6SS, and prophage regions harbored by the strains. Strain 302-73 harbors 12 GI regions, 1 T6SS region, and 4 prophage regions, with the GC content of 37.31-54.02% and the size of 8729–14,7461 bp (Figure 1A). Strain LS1 harbors 20 pathogenicity-associated regions comprising 15 GI-like regions, 5 prophages, and 1 T6SS region, with the GC content of 36.75–57.02% and the size of 8203–74,732 bp (Figure 1B). Strain NCTC10360 harbors 12 regions (1 T6SS region, 2 prophage regions, and 9 GI regions) with the GC content of 34.23-51.75% and the size of 9960-81,386 bp (Figure 1C). Finally, strain GN7 harbors 25 regions (18 GI regions, 1 T6SS region, and 6 prophage regions), with the GC content of 36.42-54.15% and the size of 9449-61,142bp (Figure 1D).
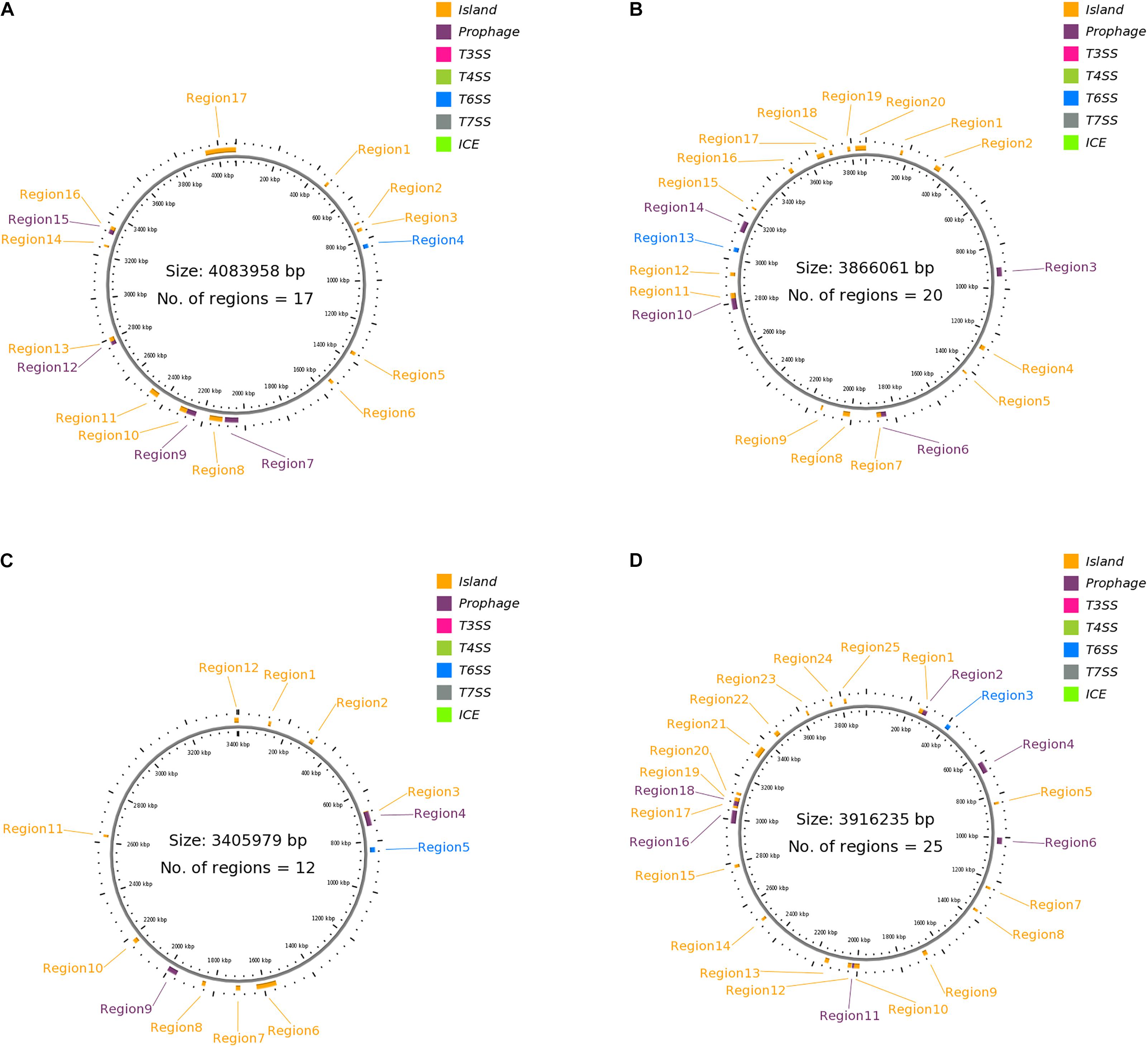
Figure 1. (A) Clustered genes related to pathogenicity traits in P. shigelloides strain 302-73. Detailed attribute of each region is presented as s/n, location, size (bp), GC% and loci tag/ORF coverage as follows: 1:470834..483428, 12595, 44.70, ORF2_383-ORF2_394; 2: 711183..719911, 8729, 40.26, ORF2_576-ORF2_585; 3: 740699..756945, 16247, 46.49, ORF2_607- ORF2_617; 4: 822581..842820, 20240, 54.02, ORF2_668-ORF2_682; 5: 1355512..1371975, 16464, 50.23, ORF2_1092-ORF2_1120; 6: 1528926..1544581, 15656, 39.02, ORF2_1266-ORF2_1280; 7: 2030969..2078415, 47447, 49.39, ORF3_214 - ORF3_276; 8: 2030969..2095574, 64606, 49.46, ORF3_214 - ORF3_291; 9: 2107619..2170457, 62839, 46.35, ORF5_2 - ORF12_27, 10: 2237256..2283735, 46480, 50.17, ORF12_83-ORF12_127; 11: 2284029..2318838, 34810, 44.15, ORF12_128-ORF12_160; 12: 2440089..2484543, 44455, 44.18, ORF12_277-ORF12_312; 13: 2768028..2789058, 21031, 50.25, ORF12_549 - ORF12_565; 14: 2785662..2805932, 20271, 45.82, ORF12_561-ORF12_588; 15: 3247461..3257433, 9973, 51.77, ORF12_977-ORF12_985; 16: 3312901..3342556, 29656, 49.84, ORF12_1032-ORF12_1070; 17: 3333679..3350402, 16724, 46.76, ORF12_1058 - ORF12_1074. (B) Clustered genes related to pathogenicity traits in P. shigelloides strain LS1. Detailed attribute of each region is presented as s/n, location, size (bp), GC% and loci tag/ORF coverage as follows: 1: 159754..171539, 11786, 36.75, ORF1_138 - ORF1_152; 2: 333282..360456, 27175, 38.41, ORF2_2 - ORF2_29, 3: 900943..942310, 41368, 47.42, ORF3_205 - ORF4_6; 4: 924180..938968, 14789, 46.44, ORF3_227 - ORF3_250; 5: 1192553..1222251, 29699, 47.92, ORF4_223 - ORF5_11; 6: 1273566..1293909, 20344, 41.65, ORF5_50 - ORF5_69; 7: 1418262..1426747, 8486, 57.02, ORF5_179 - ORF5_188; 8: 1840332..1883658, 43327, 51.82, ORF8_2 - ORF8_63; 9: 1863820..1884450, 20631, 50.24, ORF8_30 - ORF8_64; 10: 2007292..2039280, 31989, 42.29, ORF8_180 - ORF9_6; 11: 2136454..2144822, 8369, 51.43, ORF9_92 - ORF9_99; 12: 2770842..2845573, 74732, 52.04, ORF13_39 - ORF13_106; 13: 2822483..2847354, 24872, 48.26, ORF13_74 - ORF13_108; 14: 2925054..2941832, 16779, 43.86, ORF14_72 - ORF14_85; 15: 3036530..3056770, 20241, 54.08, ORF15_54 - ORF15_68; 16: 3132048..3183899, 51852, 47.08, ORF16_45 - ORF16_103; 17: 3135832..3161385, 25554, 42.67, ORF16_48 - ORF16_76; 18: 3251054..3259256, 8203, 45.85, ORF17_58 - ORF17_70; 19: 3490616..3508713, 18098, 45.13, ORF20_1 - ORF20_18; 20: 3635149..3669322, 34174, 39.18, ORF22_36 - ORF23_34. (C) Clustered genes related to pathogenicity traits in P. shigelloides strain NCTC10360. Detailed attribute of each region is presented as s/n, location, size (bp), GC% and loci tag/ORF coverage as follows: 1: 121244..132239, 10996, 34.23, ORF1_108-ORF1_117; 2: 303306..319831, 16526, 50.34, ORF1_268-ORF1_296; 3: 675181..700459, 25279, 46.37, ORF1_586-ORF1_617; 4: 677319..735911, 58593, 50.75, ORF1_588-ORF1_640; 5: 823854..844094, 20241, 54.07, ORF1_706 ORF1_720; 6: 1548988..1630373, 81386, 39.03, ORF1_1310-ORF1_1389; 7: 1696284..1714355, 18072, 48.88, ORF1_1445-ORF1_1464; 8:1838362..1852957, 14596, 39.6, ORF1_1583-ORF1_1598; 9: 1961886..2002266, 40381, 50.19, ORF1_1691-ORF1_1748; 10: 1983206..2002266, 19061, 45.73, ORF1_1716-ORF1_1748; 11: 2166969..2184623, 17655, 44.81, ORF1_1884-ORF1_1907; 12:2623511..2633470, 9960, 51.75, ORF1_2292-ORF1_2300. (D) Clustered genes related to pathogenicity traits in P. shigelloides strain GN7. Detailed attribute of each region is presented as s/n, location, size (bp), GC% and loci tag/ORF coverage as follows: 1: 253113...279252, 26140, 46.63, ORF2_172- ORF2_190; 2: 271577..291757, 20181, 45.45, ORF2_185-ORF2_198; 3: 398271..418511, 20241, 54.15, ORF4_86 ORF4_100; 4: 633845..687788, 53944, 51.53, ORF5_172-ORF5_233; 5: 833184..844394, 11211, 46.01, ORF5_354-ORF5_368; 6: 1000747..1030898, 30152, 51.06, ORF7_109-ORF7_149; 7: 1236546..1247816, 11271, 50.27, ORF:ORF9_3-ORF9_26; 8: 1355229..1366836, 11608, 53.68, ORF:ORF13_2-ORF14_8; 9: 1664884..1684947, 20064, 43.91, ORF18_40-ORF18_60; 10: 1990036..2015370, 25335, 40.67, ORF19_207-ORF19_235; 11: 2015357..2025368, 10012, 52.86, ORF19_234-ORF19_244; 12: 2027754..2045137, 17384, 39.50, ORF20_2-ORF20_20; 13: 2135394..2154633, 19240, 39.06, ORF23_1-ORF23_16; 14: 2497318..2509976, 12659, 51.56, ORF23_326-ORF23_338; 15: 2774280..2789036, 14757, 36.42, ORF25_143-ORF25_155; 16: 2981228..3042369, 61142, 47.47, ORF29_1-ORF30_15; 17: 3053731..3064085, 10355, 38.48, ORF30_24-ORF30_35; 18: 3064394..3102497, 38104, 51.14, ORF30_36-ORF30_88; 19: 3085955..3103008, 17054, 47.44, ORF30_61-ORF30_89; 20: 3115857..3128027, 12171, 42.73, ORF31_2-ORF32_8; 21: 3315841..3366201, 50361, 45.74, ORF34_136-ORF37_5; 22: 3448512..3471101, 22590, 46.04, ORF39_2-ORF44_4; 23: 3626872..3636320, 9449, 41.58, ORF58_4-ORF58_14; 24: 3744081..3754030, 9950, 51.86, ORF67_72-ORF67_80.
Putative Virulence Factor
Out of 134 unique PVFs identified in all the strains, strain 302-73 harbored 98 PVFs; strain GN7 harbored 107 PVFs; strain LS1 harbored 97 PVFs; and strain NCTC10360 harbored 90 PVFs. Detail information on the ORFs, amino acid length of predicted encoded proteins, Ha-values, and functional description of the homologous or orthologous predicted genes are provided in the Supplementary Data Sheets S1–S4.
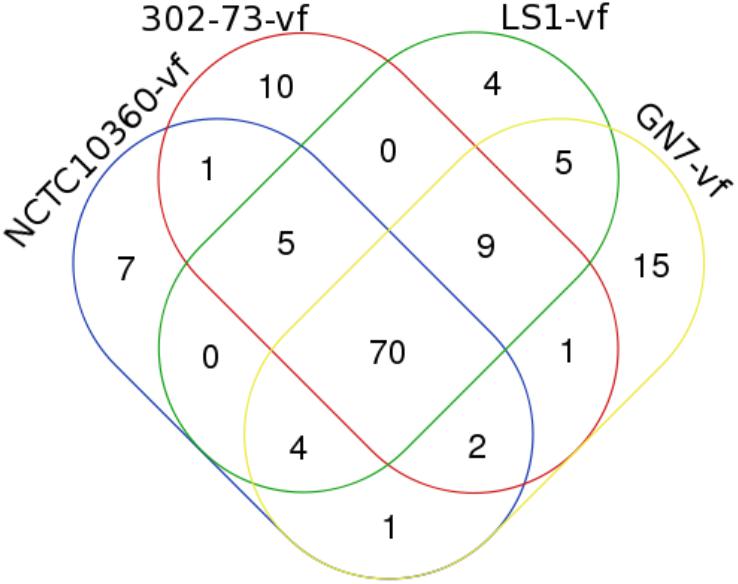
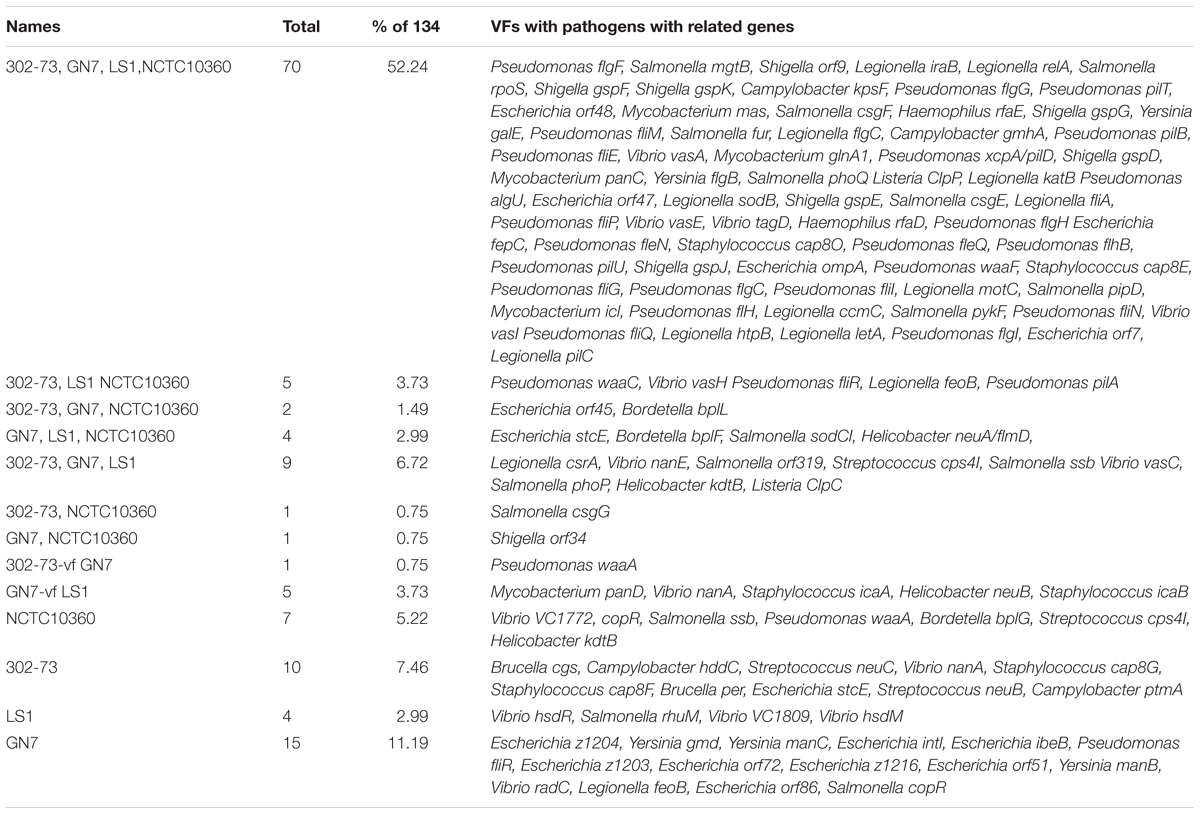
The distribution of VFs among the strains is presented in Figure 2, with the VF summary given in Table 2. Of these, 70 (52.24% of 134) VFs were common to all the strains. The common VFs were mostly flagellum genes (27.14% of 70; flgF, flgG, fliM, flgC, fliE, flgB, fliA, fliP, flgH, fleN, fleQ, flhB, fliG, flgC, fliI, flH, fliN, fliQ, and flgI) related to those found in Pseudomonas, Yersinia, and Legionella. Other VFs common to all the strains included genes homologous or orthologous to Salmonella mgtB, rpoS, csgE, pipD, and phoQ; Shigella orf9, fur, gspD, gspE, gspF, gspG, gspJ, pykF, csgF, and gspK; Legionella iraB, relA, letA, htpB, sodB, katB, ccmC, and motC; Campylobacter kpsF and gmhA; Pseudomonas pilT, algU, pilB, xcpA/pilD, waaF, and pilU; Mycobacterium mas, icl, glnA1, and panC; Haemophilus rfaD and rfaE; Yersinia galE; Vibrio vasA, vasE, tagD, and vasI; Listeria ClpP; Escherichia ompA, orf7, orf47, orf48, and fepC; and Staphylococcus cap8O and cap8E. Five VFs (3.73% of 134) similar to genes found in Pseudomonas (pilA, fliR, and waaC), Vibrio (vasH), and Legionella (feoB) were only found in strains 302-73, LS1, and NCTC10360. Similarly, VF genes homologous or orthologous to Escherichia orf45 and Bordetella bplL were identified in strains 302-73, GN7, and NCTC10360. Further, strains GN7, LS1, and NCTC10360 shared 2.99% (4 of 134) VFs, including Escherichia StcE, Bordetella bplF, Salmonella sodCI, and Helicobacter neuA/flmD homo-orthologs. VF gene subsets (6.72%; 9 of 134) identified only in strains 302-73, GN7 and LS1 were related to Legionella CsrA; Vibrio vasC and nanE; Salmonella orf319, phoP, and ssb; Streptococcus cps4I; Helicobacter kdtB; and Listeria ClpC. Virulence genes that were specifically found only in strains 302-73 and NCTC10360 were homologous to Salmonella csgG; those specific for strains GN7 and NCTC10360 were homologous to Shigella orf34; and those specific for strains 302-73 and GN7 were homologous to Pseudomonas waaA. VF genes homologous to Mycobacterium panD, Vibrio nanA, Staphylococcus icaA, Helicobacter neuB, and Staphylococcus icaB were identified only in strains GN7 and LS1.
The number of strain-specific VFs ranged from 4 to 15 (out of 134) in the four Plesiomonas strains. Specific to strain NCTC10360 were VFs that shared homo/orthology with Vibrio VC1772, copR, Salmonella ssb, Pseudomonas waaA, Bordetella bplG, Streptococcus cps4I, and Helicobacter kdtB. VFs related to Brucella cgs and per, Campylobacter hddC, Streptococcus neuC, Vibrio nanA, Staphylococcus cap8G and cap8F, Escherichia StcE, Streptococcus neuB, and Campylobacter ptmA were specific to strain 302-73. Exclusive to strain LS1 were ORF homologous/orthologous to Vibrio hsdR, VC1809, and hsdM; and Salmonella rhuM. In addition, 15 virulence genes related to Escherichia z1204, intl, ibeB, z1203, orf72, z1216, orf51, and orf86; Pseudomonas fliR; Yersinia gmd, manC, and manB; Vibrio radC; Legionella feoB; and Salmonella copR were only found in strain GN7.
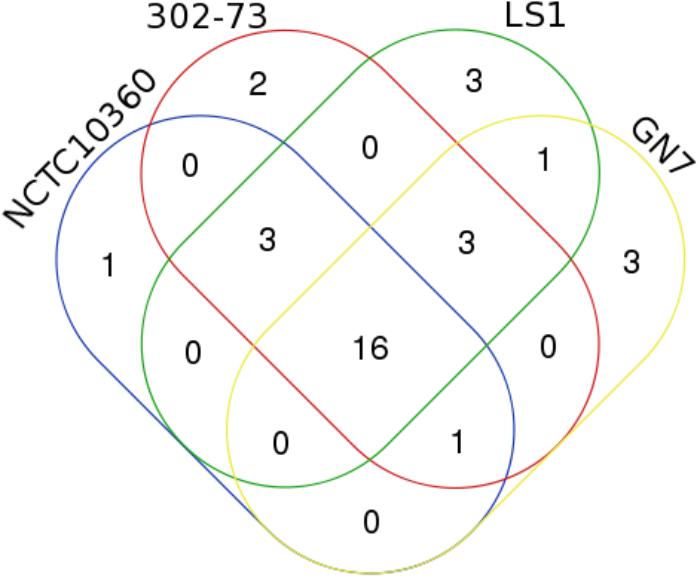
The distribution of IUSRVTs among the strains is presented in Supplementary Figure S1. Of these, 9 IUSRVTs were common to all the strains. The common IUSRVTs were multispecies ubiquinol-cytochrome c reductase iron-sulfur subunit, multispecies iron donor protein CyaY, multispecies iron-sulfur cluster insertion protein ErpA, multispecies succinate dehydrogenase/fumarate reductase iron-sulfur subunit, zinc/iron-chelating domain-containing protein, iron export ABC transporter permease subunit FetB, iron-sulfur cluster repair di-iron protein, ferrous iron transport protein B, multispecies iron-sulfur cluster assembly protein IscA. Two IUSRVTs namely multispecies zinc/iron-chelating domain-containing protein and iron transporter FeoA were only found in strains 302-73, LS1, and NCTC10360. Similarly, the iron ABC transporter permease was identified in strains 302-73, GN7, and NCTC10360. Further, strains 302-73, GN7 and LS1 shared 2 IUSRVTs, including multispecies succinate dehydrogenase iron-sulfur subunit and multispecies iron-sulfur cluster scaffold-like protein. IUSRVTs that were specifically found only in strains 302-73 and LS1 were iron(III) ABC transporter ATP-binding protein. IUSRVTs exclusive to strain NCTC10360, GN7 and 302-73 was succinate dehydrogenase iron-sulfur subunit, multispecies iron transporter FeoA and iron(III) ABC transporter permease respectively.
The distribution of HUSRVTs among the P. shigelloides strains are summarized in Supplementary Figure S2. The various HUSRVTs common to all the strains ranged from heme ABC transporter ATP-binding protein, heme exporter protein CcmB, heme utilization protein HutZ, to heme ABC exporter ATP-binding protein CcmA. Strains GN7, LS1, and NCTC10360 encoded putative heme utilization radical SAM enzyme. Strains 302-73, GN7, and LS1 encoded biliverdin-producing heme oxygenase and multispecies heme exporter protein CcmD. Strains 302-73 and NCTC10360 possessed heme exporter protein CcmC. Specific to strain NCTC10360 were genes encoding heme exporter protein CcmD. Other HUSRVTs were exclusively shared by strains GN7 and LS1 (heme ABC transporter permease and heme peroxidase).
AR Elements
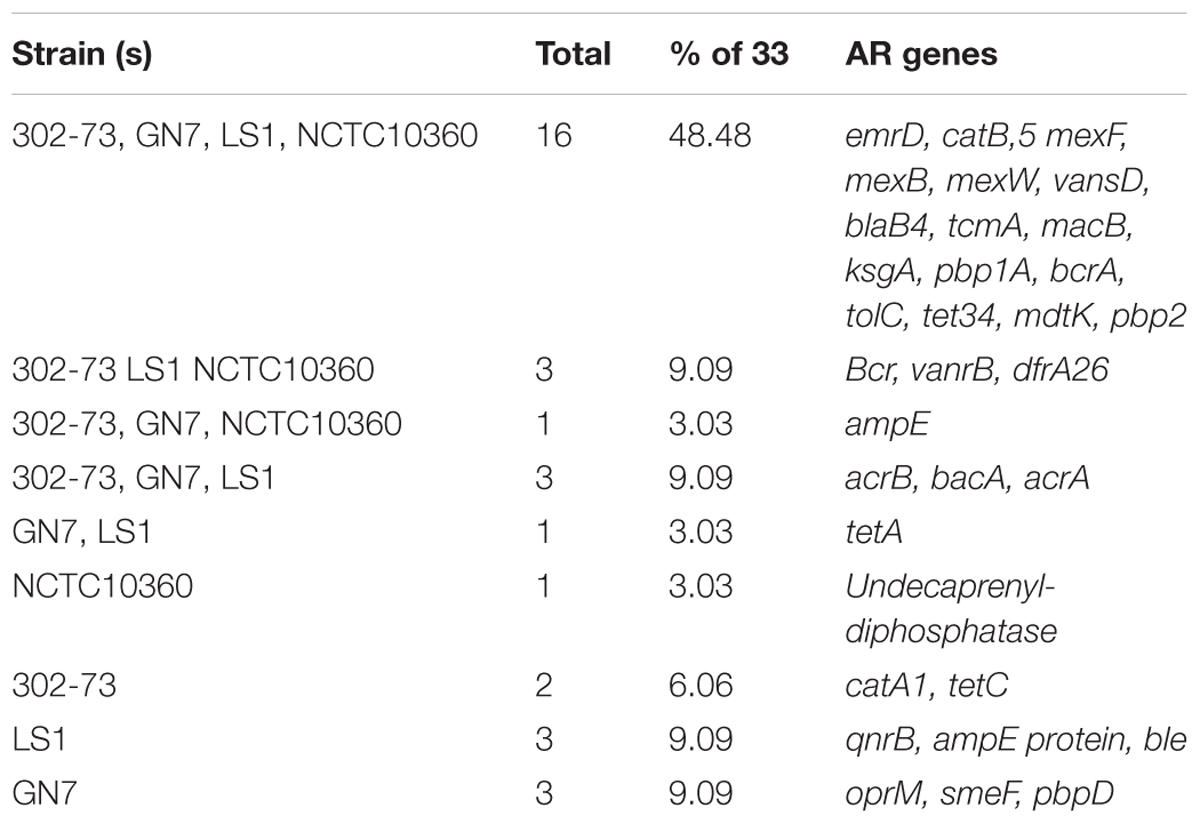
About 20 distinctive AR elements were identified in strains 302-73 (25 genes), GN7 (24 genes), LS1 (26 genes), and NCTC10360 (21 genes) (Figure 3). Overall, 33 unique AR elements were identified in all the strains, as summarized in Table 3. Of the 33 AR elements, 16 (emrD, catB5, mexF, mexB, mexW, vansD, blaB4, tcmA, macB, ksgA, pbp1A, bcrA, tolC, tet34, mdtK, and pbp2) were common to all the strains. Additional three AR elements were found in strains 302-73, LS1, and NCTC10360 (bcr, vanrB, dfra26); three in strains 302-73, GN7, and LS1 (acrB, bacA, and acrA); and one in strains 302-73, GN7, and NCTC10360 (ampE). Undecaprenyl-diphosphatase gene was specifically found in strain NCTC10360, and catA1 and tetC were specifically found in strain 302-73. Other AR elements were exclusively harbored by strain LS1 (qnrB, ampE, and ble) and GN7 (oprM, smeF, and pbpD).
Antibiotic Resistance Island
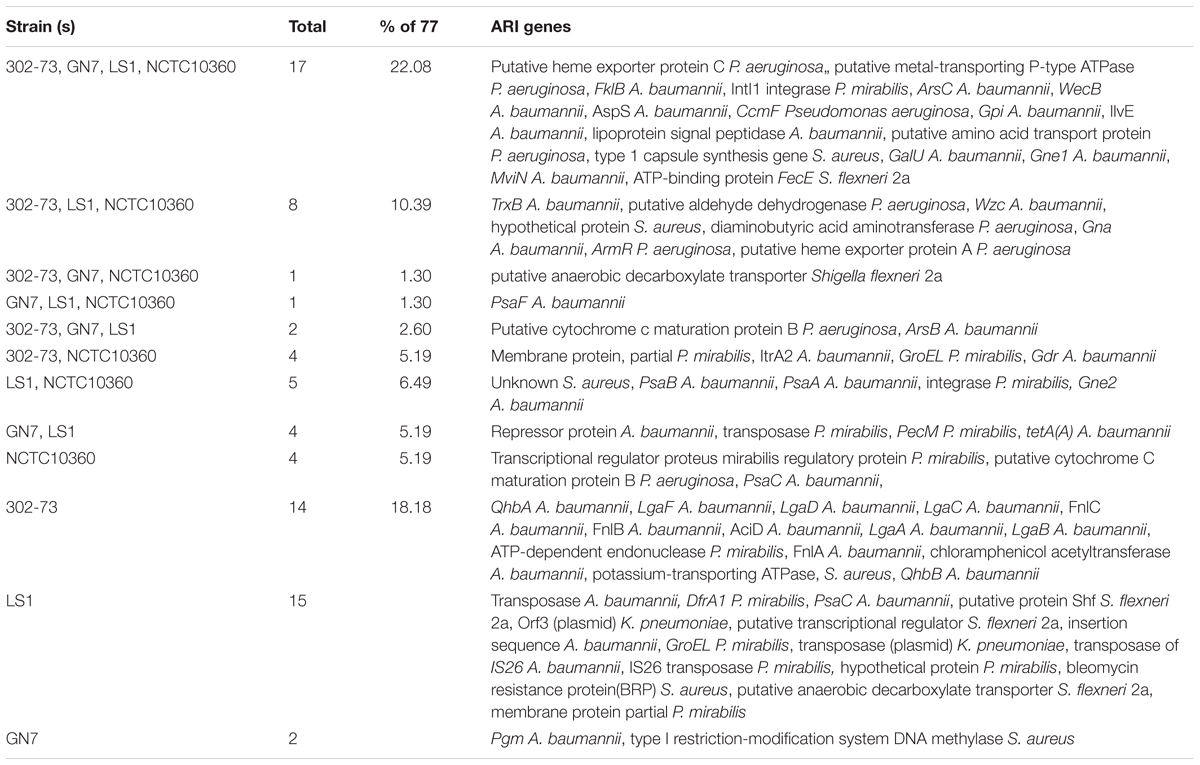
Figure 4 shows the distribution of ARIs among the strains. The various ARIs common to the strains were related to genes from Pseudomonas aeruginosa (4/17), Acinetobacter baumannii (10/17), Pseudomonas mirabilis (1/17), Staphylococcus aureus (1/17), and Shigella flexneri 2a (1/17) (Table 4). These included genes encoding putative heme exporter protein C, putative metal-transporting P-type ATPase, FklB, IntI1 integrase, ArsC, WecB, AspS, CcmF, Gpi, IlvE, lipoprotein signal peptidase, a putative amino acid transport protein, type 1 capsule synthesis protein, GalU, Gne1, MviN, and ATP-binding protein FecE. ARIs specific to strain NCTC10360 encoded a transcriptional regulator, regulatory protein, putative cytochrome C maturation protein B, and PsaC. Most ARIs solely found in strain 302-73 showed homology/orthology with genes encoding QhbA, LgaF, LgaD, LgaC, FnlA, FnlB, FnlC, AciD, LgaA, LgaB, chloramphenicol acetyltransferase, and QhbB from A. baumannii; and ATP-dependent endonuclease and potassium-transporting ATPase from P. mirabilis and S. aureus, respectively. Fifteen ARIs closely related to A. baumannii transposase, IS, transposase of IS26, and PsaC; K. pneumoniae transposase (plasmid) and Orf3 (plasmid); P. mirabilis IS26 transposase, DfrA1 protein, GroEL, hypothetical protein, and membrane partial protein; S. flexneri 2a putative protein Shf, putative transcriptional regulator, and putative anaerobic decarboxylate transporter; and S. aureus bleomycin resistance protein were unique to strain LS1. Sequences similar to ones encoding Pgm and type I restriction-modification system DNA methylase from A. baumannii and S. aureus, respectively, were only identified in strain GN7. Other ARIs shared by either two or three strains included ones encoding TrxB, putative aldehyde dehydrogenase, Wzc, S. aureus hypothetical protein, S. aureus unknown protein, diaminobutyric acid aminotransferase, Gna, ArmR, putative heme exporter protein A, putative anaerobic decarboxylate transporter, PsaF, putative cytochrome C maturation protein B, ArsB, P. mirabilis membrane partial protein, ItrA2, GroEL, Gdr, PsaB, PsaA, integrase, Gne2, A. baumannii repressor protein, P. mirabilis transposase and PecM, TetA(A), P. mirabilis transcriptional regulator and regulatory protein, putative cytochrome C maturation protein B, and PsaC.
Pathogenicity Islands
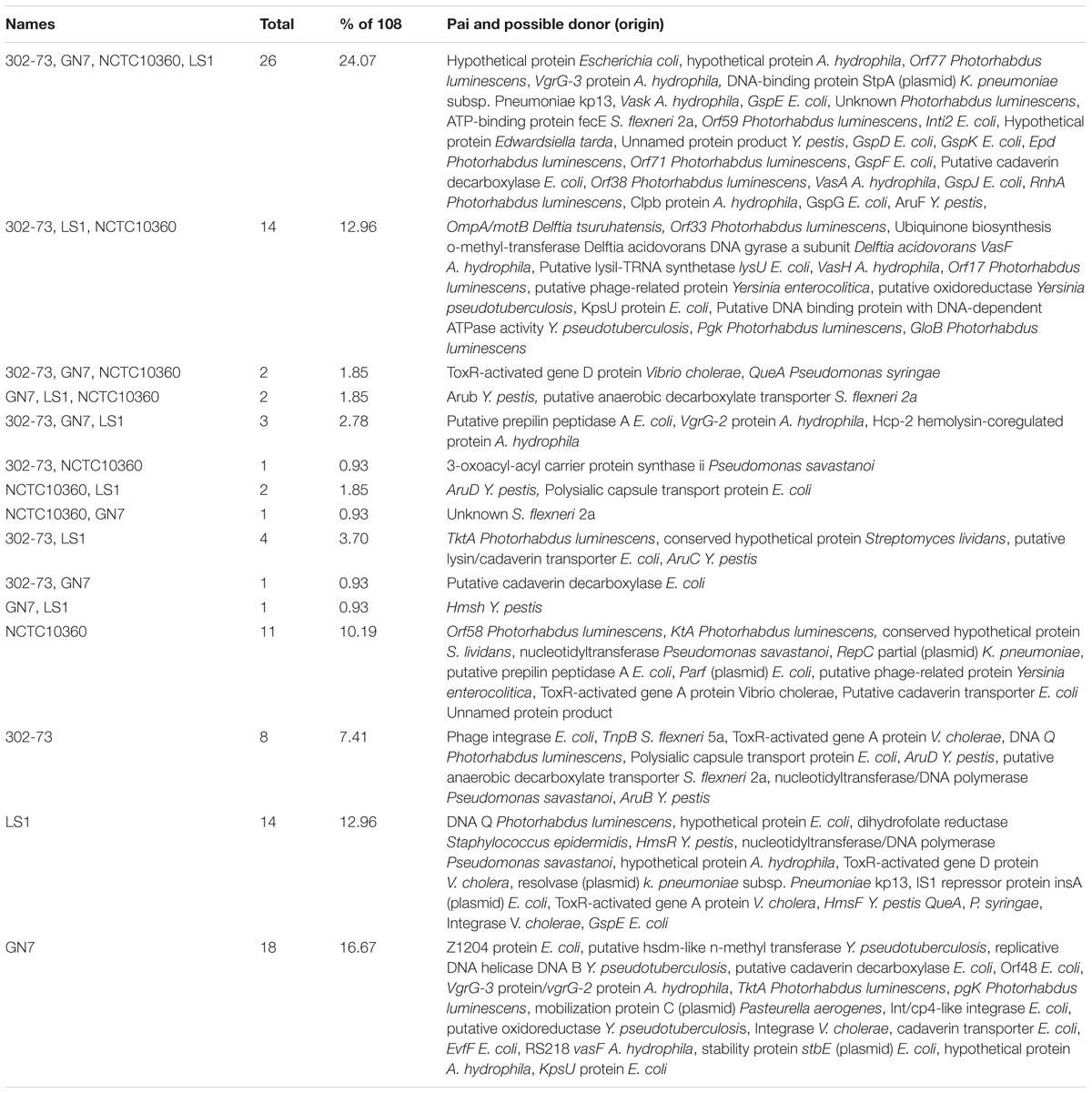
Pathogenicity islands identified in the P. shigelloides strains are summarized in Table 5 and Figure 5 shows the distribution of PAIs among the strains. The various PAIs common to all the strains ranged from unnamed or hypothetical proteins related to Escherichia coli, Aeromonas hydrophila, Edwardsiella tarda, and Yersinia pestis. Examples include PAIs encoding Orf77, Orf59, Epd, RnhA, Orf71, and Orf38 from Photorhabdus luminescens; VasA, Vask, Clpb protein, and VgrG-3 protein from A. hydrophila; DNA-binding protein StpA (plasmid) (Klebsiella pneumoniae subsp. pneumoniae kp13); Inti2, GspD, GspF, putative cadaverin decarboxylase GspE, GspK, GspG, and GspJ (E. coli); ATP-binding protein FecE (S. flexneri 2a), and AruF (Y. pestis).
Strains 302-73, GN7, and NCTC10360 encoded ToxR-activated gene D protein and QueA similar to the respective proteins from V. cholerae and Pseudomonas syringae, respectively. Strains 302-73, GN7, and LS1 encoded proteins similar to putative prepilin peptidase A from E. coli, and VgrG-2 protein and Hcp-2 hemolysin-coregulated protein from A. hydrophila. Specific to strain NCTC10360 were genes encoding Orf58 and KtA related to proteins from Photorhabdus luminescens, conserved hypothetical protein (Streptomyces lividans), nucleotidyltransferase (Pseudomonas savastanoi), RepC partial plasmid (K. pneumoniae), putative prepilin peptidase A (E. coli), ParF plasmid protein (E. coli), putative phage-related protein (Yersinia enterocolitica), and ToxR-activated gene A protein (V. cholerae). PAIs solely harbored by the strain 302-73 encoded phage integrase and polysialic capsule transport protein (E. coli); TnpB (S. flexneri 5a); ToxR-activated gene A protein (V. cholerae); DNA Q protein (Photorhabdus luminescens); putative anaerobic decarboxylate transporter (S. flexneri 2a); nucleotidyltransferase (Pseudomonas savastanoi); and AruB and AruD (Y. pestis). DNA Q protein (Photorhabdus luminescens), hypothetical proteins (E. coli and A. hydrophila), dihydrofolate reductase (Staphylococcus epidermidis), HmsF and HmsR (Y. pestis), DNA polymerase (Pseudomonas savastanoi); integrase, ToxR-activated gene A and D proteins (V. cholerae); resolvase (plasmid) (K. pneumoniae), IS1 repressor protein InsA (plasmid) and GspE (E. coli); and QueA (P. syringae) were putative PAIs specific to strain LS1. Strain GN7 was the only one harboring PAIs encoding Z1204 protein, stability protein StbE (plasmid), EvfF, Int/cp4-like integrase, putative cadaverin decarboxylase, cadaverin transporter, KpsU protein, and Orf48 (E. coli); putative Hsdm-like N-methyl transferase, putative oxidoreductase, and replicative DNA helicase DNA B (Yersinia pseudotuberculosis); RS218, VasF, hypothetical protein, and VgrG-3 protein/VgrG-2 protein (A. hydrophila); Pgk and TktA (Photorhabdus luminescens); mobilization protein C plasmid (Pasteurella aerogenes); and integrase (V. cholerae).
IS Elements
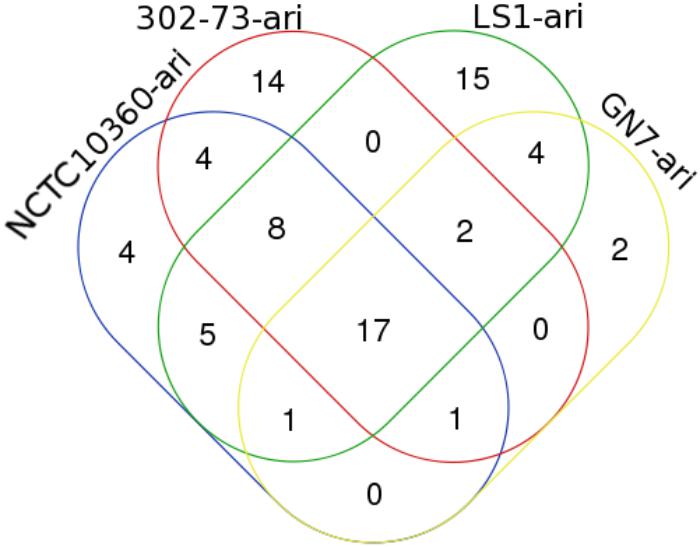
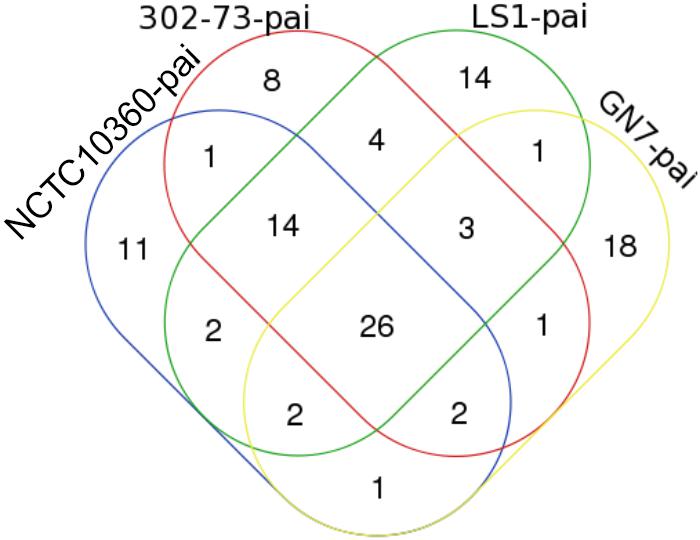
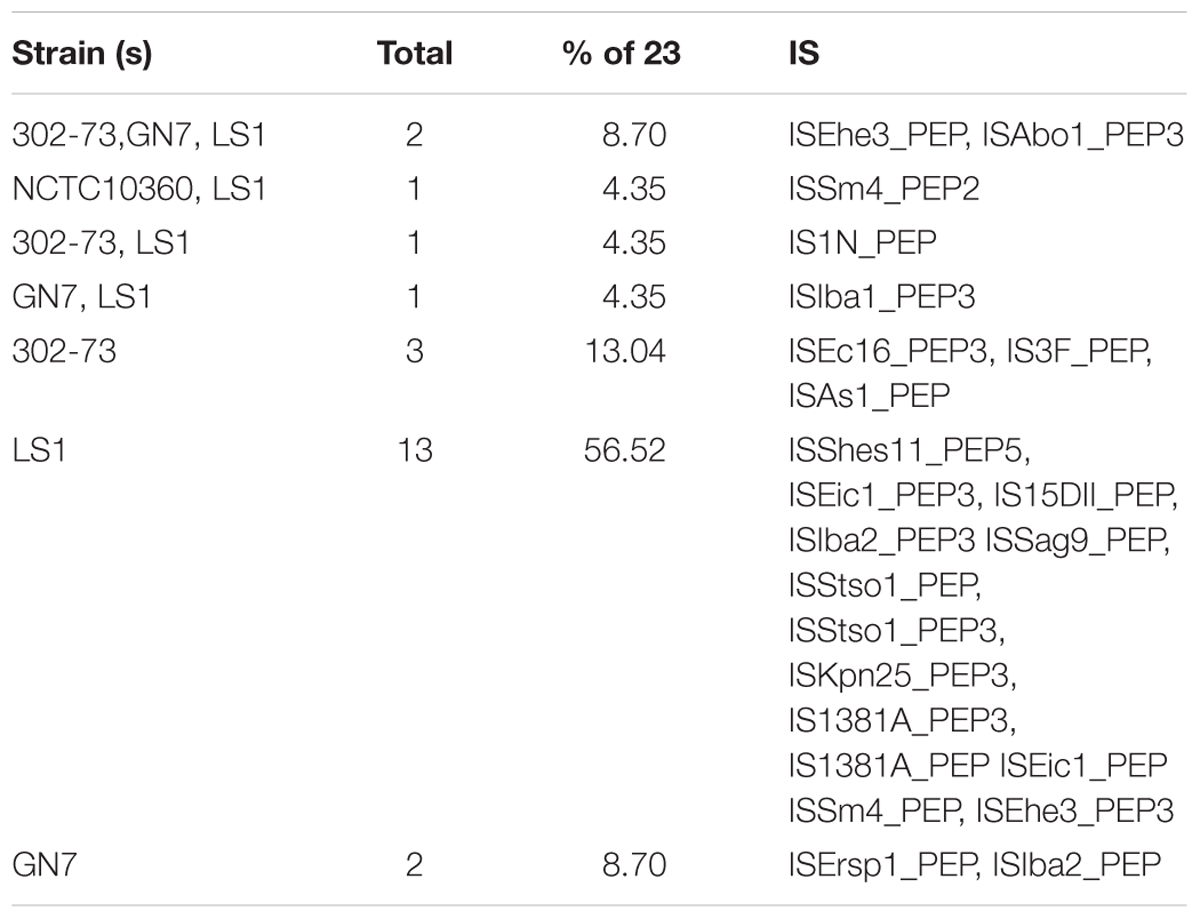
The IS elements identified in the four P. shigelloides strains are presented in Table 6. The IS elements were shared by the strains or specific to a given strain, as follows: ISEhe3_PEP, and ISAbo1_PEP3 were identified in strains 302-73, GN7, and LS1; ISSm4_PEP2 was identified in strains NCTC10360 and LS1; IS1N_PEP was identified in strains 302-73 and LS1; ISIba1_PEP3 was identified in strains GN7 and LS1; ISEc16_PEP3, IS3F_PEP, and ISAs1_PEP were only identified in strain 302-73; ISErsp1_PEP and ISIba2_PEP were only identified in strain GN7; ISShes11_PEP5, ISEic1_PEP3, IS15DII_PEP, ISIba2_PEP3 ISSag9_PEP, ISStso1_PEP, ISStso1_PEP3, ISKpn25_PEP3, IS1381A_PEP3, IS1381A_PEP ISEic1_PEP ISSm4_PEP, and ISEhe3_PEP3 were only identified in strain LS1. The distribution of IS elements among the strains is depicted in Figure 6. LS1 harbored 56.52% (13) of all IS elements identified in the strains.
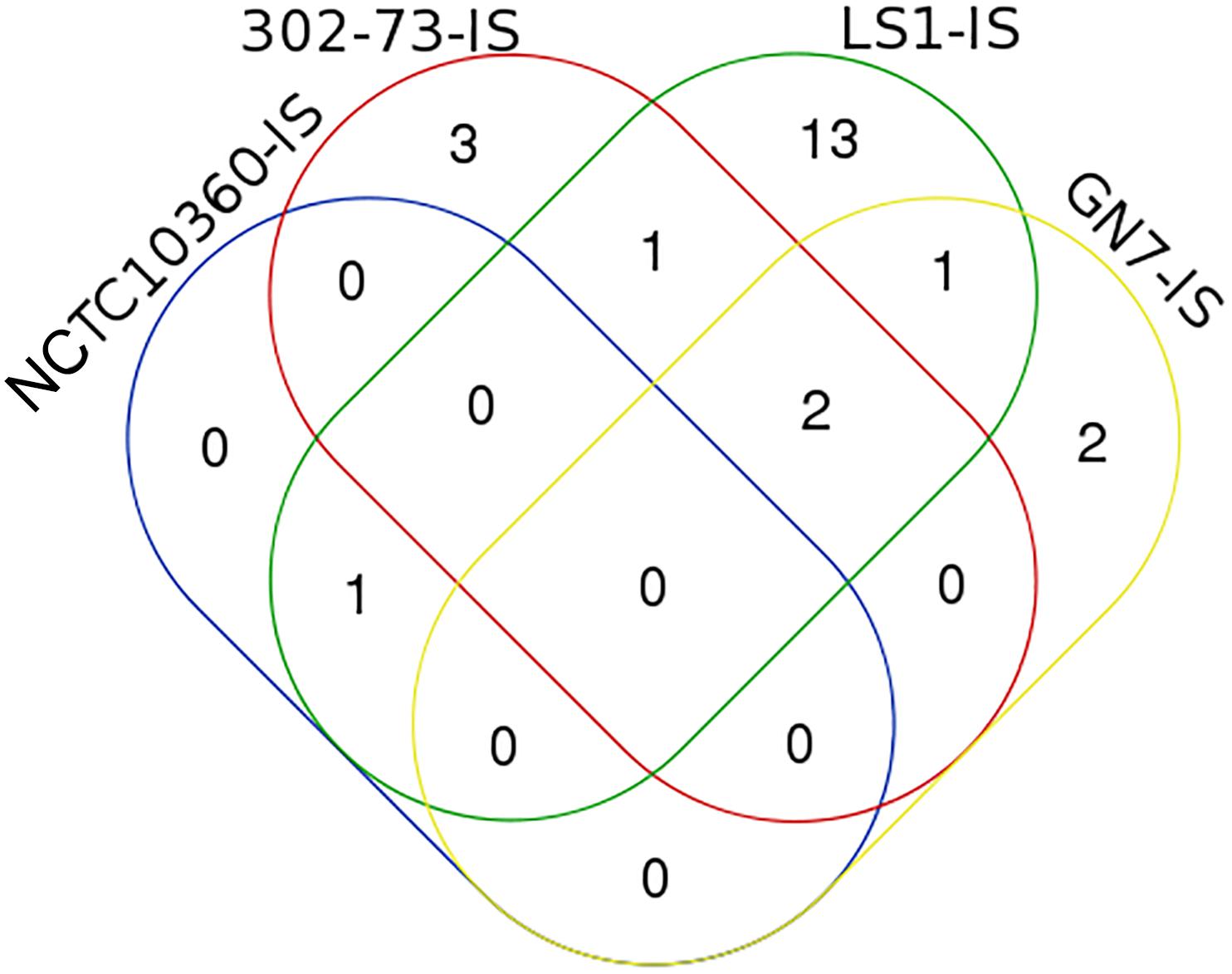
Figure 6. A Venn diagram of ISs in four Plesiomonas shigelloides strains. The identities of elements shared or specific to the sub-region of the Venn diagram is presented against it in parenthesis as follows: 302-73, GN7, and LS1 (ISEhe3_PEP, and ISAbo1_PEP3), NCTC10360 and LS1 (ISSm4_PEP2), 302-73 and LS1 (IS1N_PEP), GN7 and LS1 (ISIba1_PEP3), 302-73 (ISEc16_PEP3, IS3F_PEP, ISAs1_PEP), GN7 (ISErsp1_PEP, ISIba2_PEP), LS1 (ISShes11_PEP5, ISEic1_PEP3, IS15DII_PEP, ISIba2_PEP3 ISSag9_PEP, ISStso1_PEP, ISStso1_PEP3, ISKpn25_PEP3, IS1381A_PEP3, IS1381A_PEP ISEic1_PEP ISSm4_PEP, ISEhe3_PEP3).
SS Elements
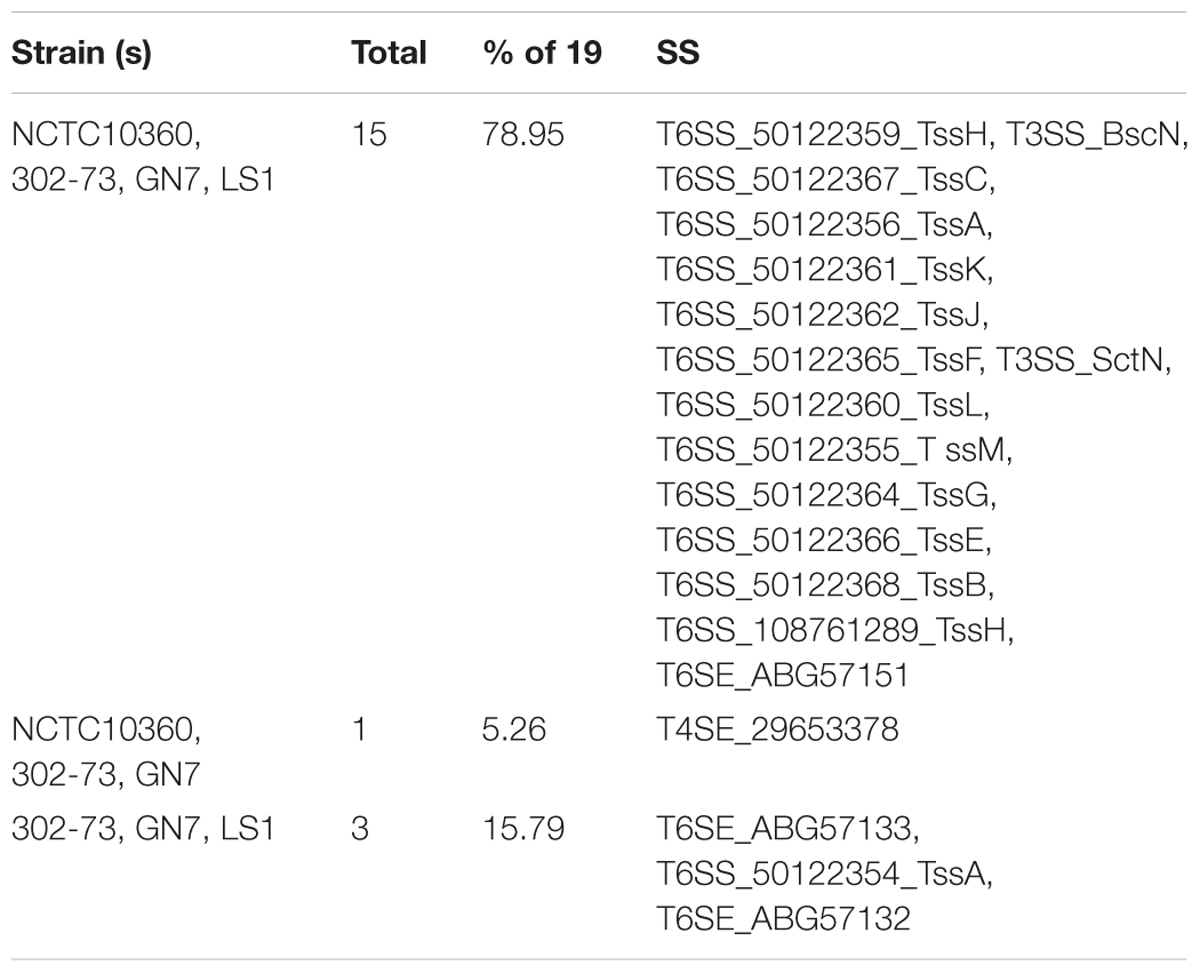
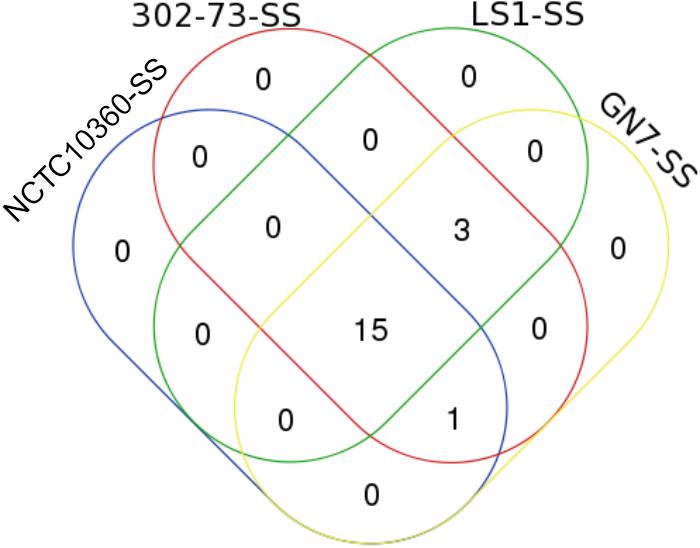
Overall, 19 SS elements were identified in the strains (Table 7). The various SS elements included T6SS (encoding TssA, TssB, TssC, TssH, TssJ, TssK, TssF, TssL, TssM, TssG, and TssE); T3SS (encoding BscN and SctN), T4SE_29653378, and T6SE (encoding ABG57151, ABG57133, and ABG57132). The distribution of SS elements among the strains is shown in Figure 7; 78.95% of all SS elements (15 of 19) were harbored by all the strains, while 15.79% of all SS elements (3) were exclusive to 302-73, GN7, and LS1 strains.
CRISPR Elements
No CRISPR element was identified in strain GN7. Various features of CRISPR elements harbored by the other three Plesiomonas strains are detailed in Table 8. Strain NCTC10360 harbored two CRISPR elements (172 and 185 bp). The 172-bp element harbored three repeated sequences and two spacers, with a repeat length of 25 bp. The 185-bp element harbored four repeated sequences and three spacers, with a repeat length of 20 bp. Strain 302-73 harbored eight CRISPR elements with the total length ranging from 139 to 181 bp. Each of these elements harbored three repeated sequences and two spacers; the repeat length varied from 21 to 28 bp. Finally, in strain LS1, one CRISPR element (142 bp total length) was identified; it harbored three repeats and two spacers, with a repeat length of 22 bp. The structural and compositional details of CRISPRs identified in the P. shigelloides strains are presented in Supplementary Data Sheet S5.
Prophage Entities
The results of comparative phageomics of the four P. shigelloides strains are shown in Table 8. Strain NCTC10360 harbored two intact phages sized 21.9 kb and 53.8 kb, most similar to PHAGE_Entero_mEp235_NC_019708, PHAGE_Klebsi_phiKO2_NC_005857, PHAGE_Yersin_PY54_NC_005069, and PHAGE_Entero_lambda_NC_001416. Strain GN7 harbored two intact and one incomplete phage sized 36.2, 30.3, and 29.5 kb respectively. The GN7 phages were most similar to the following phage species: PHAGE_Flavob_1H_NC_031911, PHAGE_Shigel_Stx_NC_029120, PHAGE_Shigel_POCJ13_NC_025434, PHAGE_Salmon_Fels_2_NC_010463, and PHAGE_Salmon_Fels_2_NC_010463. Three incomplete phages and one questionable phage were identified in strain LS1. The only phage (31.1 kb) identified in strain 302-73 encoded tRNA and was similar to PHAGE_Salmon_RE_2010_NC_019488.
Plasmid Elements
No plasmid element was identified in the four strains. Replicon information of the WGS revealed chromosomes.
Discussion
One major problem of the conventional elucidation of pathogenesis mechanisms of Plesiomonas is the lack of a suitable experimental animal model. In the current study, comparative pathogenomics approach was used to detect virulence traits of the emerging pathogen that, until now, had been unresolved by traditional approaches. As an example, using an adult rabbit model, the ileal loop system, Sanyal et al. (1980) had established the diarrheagenic (enterotoxigenic) potential of Plesiomonas but did not model invasive pathogenesis. Further, pathogenomics studies revealed the presence of several genes associated with adherence and attachment to the villi of enterocytes (including thin aggregative fimbriae genes, such csgE, csgF, csgG, gmhA, pilT, and pilU) (Collinson et al., 1996; Sukupolvi et al., 1997; Römling et al., 1998; Comolli et al., 1999a; Bacon et al., 2001), characteristics necessary for diarrheagenic activity of pathogens. Pathogenomics not only support the findings of Sanyal et al. (1980), provided exhaustive genetic accessories that enable the bacterium with diarrheagenic potential.
Further, invasive models of Plesiomonas involving the eye (guinea pig or rabbit) and HeLa epithelial cells failed either to establish keratoconjunctivitis and internalized by the HeLa epithelial cells, respectively (Sanyal et al., 1980). Failed invasive investigations of Plesiomonas with HeLa cells were also reported by Pitarangsi et al. (1982), Holmberg and Farmer (1984), and Olsvik et al. (1990). Chowdhury et al. (1988) examined enteropathogenicity responses of adult rabbits orograstically dosed with Plesiomonas and reported that the rabbits were neither diarrheic nor deathward, except for soft acute inflammatory effects observed in the ileum mucosa. By contrast, homology shown by various Plesiomonas ORFs to genes linked to the invasiveness of other pathogens suggests its inherent propensity for invasive virulence. For instance, Plesiomonas homologs of invasive genes, such as motC (Heuner and Steinert, 2003), clpP (Gaillot et al., 2000; Gaillot et al., 2001), flgC (Heuner and Steinert, 2003), fliA (Dietrich et al., 2001), flgB (Young et al., 1999; Young et al., 2000), ompA (Prasadarao et al., 2002; Khan et al., 2003; Prasadarao et al., 2003; Sukumaran et al., 2003), LOS (rfaE) (Rao et al., 1999; Swords et al., 2000), waaC and waaF (Rocchetta et al., 1999; Lyczak et al., 2000), and clpC (Rouquette et al., 1996; Rouquette et al., 1998; Nair et al., 2000) have been shown to enhance the adhesion and invasion of different cells by other pathogens. This was further supported by various extraintestinal infections associated with Plesiomonas, including peritonitis (Alcañiz et al., 1995; Patel et al., 2016), cellulitis (Gopal and Burns, 1991; Jönsson et al., 1998), wound and foot infections (McCracken and Barkley, 1972; Herve et al., 2007; Pence, 2016), endophthalmitis (Butt et al., 1997; Mahrshmann and Lyons, 1998), and keratitis (Butt et al., 1997; Klatte et al., 2012). In addition, pneumonia (Schneider et al., 2009), migratory polyarthritis (Gupta, 1995), cholecystitis (Claesson et al., 1984; Kennedy et al., 1990), pyosalpinx (Roth et al., 2002), pseudo-appendicitis (Fischer et al., 1988), cholangitis and pancreatitis (Kennedy et al., 1990) are examples of infections that required invasive factor that have been reported caused by Plesiomonas. Further, Binns et al. (1984) showed that 31.3% of examined strains are invasive toward HeLa cells. Vitovec et al. (2001) showed that Plesiomonas monoinfection of neonatal mice resulted in long-term intestinal colonization and varied patho-histophysiological severity, ranging from an atrophic ileum, and ileal and colonic necrosis to colonic enterocytes. On the other hand, Cryptosporidium parvum co-infection resulted in dense ileal colonization, bacteremia, necrotizing inflammatory responses, diarrheic condition, and death. Of importance in this context is a Plesiomonas ORF homologous to Escherichia ompA identified in the current study. OmpA is associated with several virulence traits, including serum resistance, invasion, binding to C4b-binding protein, evasion of complement-mediated attack, and invasion of the microvascular endothelial cells in the brain via a ligand-receptor interaction (Prasadarao et al., 2002; Khan et al., 2003; Prasadarao et al., 2003; Sukumaran et al., 2003). These observations and comparative pathogenomics of Plesiomonas further attest to the probable invasive pathogenicity potential of this bacterium.
Other core pathogenic traits that remained unidentified or were only weakly supported by conventional experimentation include the cytotoxic and hemolytic potential of Plesiomonas. According to cell culture and animal model studies of Plesiomonas cytotoxicity, some strains produce extracellular cytotoxins with activities against Vero cells (Olsvik et al., 1990). Similarly, vacuolation and cytotoxigenic activities of plesiomonad strains recovered from different environments, including human/animals and aquatic bodies, in Vero cell monolayers were described by Ekman (2003) and Falcón et al. (2003), respectively. Although there is some evidence (as mentioned), other studies yield contradicting data. Vitovec et al. (2001) were unable to demonstrate either cytotoxic or hemolytic activity of Plesiomonas isolates during both, monoinfection and C. parvum co-infection in experimental models. Likewise, Pitarangsi et al. (1982) and Holmberg and Farmer (1984) concluded that the tested Plesiomonas strains are not cytotoxic in the animal model used. Further, the plesiomonad strains studied by Maluping et al. (2004) exhibited no cytotoxicity or hemolytic potential in any of the experimental models explored. Nonetheless, in the current study, we identified some plesiomonad ORFs homologous or orthologous to genes, such as ssb, known to mediate toxin secretion similar to E. coli hemolysin secretion (Wong et al., 1998; Marcus et al., 2000), and xcpA/pilD, known to facilitate the internalization and cytotoxicity in P. aeruginosa (Hahn, 1997; Comolli et al., 1999b). These suggest the cytotoxicity potential of Plesiomonas.
The LOS gene from Haemophilus that showed homology with a Plesiomonas ORF is linked to endotoxin and immunogenic functions, and its phosphoryl choline impacts the invasion via interaction with PAF receptor; and stimulates inflammatory signals, evasion of antigen-specific host defenses, and colonization of diverse microenvironments within the host (Rao et al., 1999; Erwin et al., 2000; Swords et al., 2000). A homologous ORF might provide Plesiomonas with a related pathogenic ability. Other genes related to toxin production and enteropathogenesis with homology to Plesimonas ORFs include waaC and waaF. These genes function in antiphagocytosis and resistance to serum killing (Rocchetta et al., 1999; Lyczak et al., 2000; Schroeder et al., 2002). Further, Plesiomonas possesses ORFs that are homologous to Vibrio PAI genes (tagD, nanE, and nanA). These Vibrio VFs are associated with cholera toxins and a putative protease-modulating VF in toxigenic Vibrio species (Karaolis et al., 1998; Jermyn and Boyd, 2002; Faruque and Mekalanos, 2003; Schmidt and Hensel, 2004). However, Herrington et al. (1987), using early traditional approaches, were unable to establish Plesiomonas enteropathogenesis in an animal model.
Virulence genes required for extraintestinal infections were identified in previous studies. Of these, serum resistance and antiphagocytosis factors, including cap8E and cap8O (Thakker et al., 1998; Cunnion et al., 2001; Luong and Lee, 2002; Cunnion et al., 2003), algU (Pier et al., 2001; Nivens et al., 2001; Song et al., 2003), and bplF (Harvill et al., 2000; Burns et al., 2003; Schaeffer et al., 2004) showed homology to some Plesiomonas ORFs.
Plesiomonad ORF homologs of such genes as Staphylococcus icaA and icaB suggest bacterial ability of intracellular survival within the host. These genes are associated with the synthesis of intercellular adhesin polysaccharide, proteins, and biofilm formation in S. epidermidis (Heilmann et al., 1996; McKenney et al., 1998; Cramton et al., 1999; Götz, 2002). Likewise, the presence of Plesiomonas gene homologous to Salmonella rhuM (Salmonella PAI 3) might suggest that it encodes a similar high-affinity magnesium transport system (mgtCB) required for intramacrophage survival fitness and virulence (Blanc-Potard and Groisman, 1997; Tao et al., 1998; Moncrief and Maguire, 1999). Plesiomonas harbor genes related to katAB, of which katB was shown to encode a catalase-peroxidase (Bandyopadhyay and Steinman, 1998, 2000; Bandyopadhyay et al., 2003), a stress protein essential for intracellular survival and transmission. A Plesiomonas gene homolog of htpB (encoding Hsp60 in Legionella) may function similarly to htpB, i.e., mediate complement-independent attachment to the host cells (Garduño et al., 1998; Swanson and Hammer, 2000). Other notable Plesiomonas VFs include ORFs that show homology to Escherichia enterobactin (fepC), Legionella feoB, and Listeria clpC. While fepC and feoB function in iron uptake in Escherichia (Cao et al., 2000; Sprencel et al., 2000) and Legionella (Robey and Cianciotto, 2002) respectively, ClpC mediates early escape of Listeria from macrophage phagosome, and adhesion and invasion (Rouquette et al., 1996; Rouquette et al., 1998; Nair et al., 2000). Further, Plesiomonas ORF related to Salmonella fur gene might enable iron regulation of gene expression in the bacterium, as reported for Salmonella (Guo et al., 1997; Lucas and Lee, 2000; Lejona et al., 2003).
The four P. shigelloides strains were replete with heme iron utilization systems (IUSRVTs/ HUSRVTs) early proposed as virulence mechanisms (Daskaleros et al., 1991; Santos et al., 1999; Henderson et al., 2001; Gonzalez-Rodriguez et al., 2007; Oldham et al., 2008; Rodríguez-Rodríguez and Santos, 2018). However, the roles of these iron acquisition systems in P. shigelloides pathogenesis require further transcriptomic validation in suitable models. While many authors found an association of iron uptake systems with virulence in some pathogens (Fisher et al., 2009; Watson et al., 2010), others found no direct relationship between them (Lindgren et al., 2011; Fletcher et al., 2018). Most importantly, the iron uptake systems (Feo Systems) are the essential cellular component for survival in pathogens, non-pathogens and commensal bacteria (Cartron et al., 2006; Lau et al., 2015). Some pathogens whose virulence depend on iron uptake systems include Neisseria meningitidis (Stojijkovic et al., 1995), Salmonella (Velayudhan et al., 2007; Nagy et al., 2014), Campylobacter (Naikare et al., 2006), Helicobacter (Velayudhan et al., 2000), Legionella (Robey and Cianciotto, 2002), Haemophilus influenzae (Morton et al., 2009), and others (Aranda et al., 2009; Cassat and Skaar, 2013).
The presence of ARIs and clinically relevant antibiotic (multi)resistance genes might limit the treatment and management options for the infections caused by Plesiomonas. This could have potentially serious consequences since Plesiomonas infections are not routinely diagnosed in the medical setting (Chen et al., 2013). Similar to many cases of antimicrobial resistance shown by various microorganisms, infections involving resistant Plesiomonas might be very difficult to manage, leading to extended hospitalization, high morbidity, and, ultimately, elevated mortality rates (Kain and Kelly, 1989b). Multi-resistance of zoonotic Plesiomonas could pose increased threat to human and veterinary health. This might further raise the associated possibility of dissemination and contact between human and animals, especially pets and livestock. Plesiomonas are by nature, β-lactamase producers (resistant to β-lactam drugs) (Meyers et al., 1976; Brenden et al., 1988; Fisher et al., 1988; Avison et al., 2000; Stock and Wiedemann, 2001a). Plesiomonad tetracycline resistance has been reported by Kain and Kelly (1989a); González et al. (1999), Wong et al. (2000), and Suthienkul et al. (2001). Co-resistance of plesiomonad strains to antibiotics, including ampicillin, amoxicillin/clavulanic acid, trimethoprim/sulfamethoxazole, chloramphenicol, streptomycin, tetracycline, erythromycin, sulphametoxazole, and neomycin has also been documented (Olsvik et al., 1990; González et al., 1999; Wong et al., 2000; Jun et al., 2011). Chloramphenicol-, co-trimoxazole-, erythromycin-, and rifampin-resistant Plesiomonas strains are frequently reported (Wong et al., 2000; Stock and Wiedemann, 2001b; González-Rey et al., 2004). Tobramycin- and gentamicin-resistant strains have also been documented (Clark et al., 1990). Tetracycline-resistant strains of Plesiomonas veterinary isolates are not uncommon. González-Rey et al. (2004) documented a multi-drug resistant Plesiomonas and tetracycline-resistant strain recovered from a Cuban dog and an environmental sample, respectively. Widespread tetracycline-resistant strains have also been noted among plesiomonad isolates from catfish ponds (DePaola et al., 1993). Further, aminoglycoside-resistant and penicillin-resistant Plesiomonas isolates were reported (Clark et al., 1990; Bravo-Fariñas et al., 1998; Avison et al., 2000; Stock and Wiedemann, 2001b). Yeh and Tsai (1991) have documented vancomycin-resistant strains of Plesiomonas. Furthermore, Jun et al. (2011) reported Plesiomonas resistance to amikacin, cefotaxime, cefepime, gentamicin, and ciprofloxacin. Plesiomonas inactivation of aminoglycoside antibiotics has been attributed to the production of specific enzymes, such as aminoglycoside-modifying enzymes (Shaw et al., 1993). Homology of Plesiomonas ORFs to heavy-metal resistance genes closely related to arsenic resistance in Acinetobacter spp. suggests the bacterium’s increasing ability to evade potential antiseptic treatment. Previously, Chao et al. (2016) reported cadmium-resistance in Plesiomonas, with a maximum tolerance concentration as high as 150 mg/L. Although Chao et al. (2016) concluded that this trait could be applicable in wastewater treatment, employed as an indicator of heavy metal pollution for eco-monitoring and environmental impact assessment, heavy metal resistance in Plesiomonas also suggests possible adverse implications for infectious cases.
The presence of Plesiomonas ORFs homologous to genes encoding different secretory and effector systems (T6SS, T3SS, T4SE, and T6SE) found in other pathogens might be indicative of a similar virulence function in the microorganism. For example, a Yersinia and Salmonella effector molecule that depolymerizes actin plays a similar role in tissue culture cells (Rosqvist et al., 1995). Secretory systems directly transport effector molecules from the cytoplasmic compartment to the cell surface to interact with and modify mammalian host cell proteins (Michiels et al., 1990). Other functional consequences of secretory systems in enteric and many other pathogens, such as Yersinia spp., Shigella spp., and Salmonella spp., include abolition of the immune cell function and engulfment ability, and macrophage protein modification (Rosqvist et al., 1990; Bliska et al., 1991; Cornelis, 1992); and enhanced pathogen entry into non-phagocytic cells (Galán, 1996; Ménard et al., 1996). Similar virulence traits are probably linked to the extraintestinal infections caused by Plesiomonas. Many genes harbored on or related to TSS elements, such as gspD–J, play roles in protecting pathogens against human complement activity and serum (Aiello et al., 2010; Johnson et al., 2016; Cianciotto and White, 2017; Waack et al., 2017). TSS elements are also involved in the mediation of cascades of secreted virulence effectors in gram-negative pathogens (Aiello et al., 2010; Johnson et al., 2016; Cianciotto and White, 2017).
Almost all Plesiomonas strains analyzed in the current study possessed PAIs homologous to those found in most enteropathogens. One of the PAIs, for instance, contained cadavarine decarboxylase gene (cadA). Probable alteration of the Plesiomonas cadA gene could be responsible for the virulence of some strains. Deletion of the cadA gene in Shigella reportedly promotes virulence to evade host cell protection against Shigella enterotoxin that relies on the inhibition of cadaverine synthesis (Maurelli et al., 1998; Brosch et al., 2001). Virulence plasticity is attributed principally to insertions and deletions in a pathogen genome (Brosch et al., 2001). Plesiomonas harbors IS elements that share homology with those found in pathogenic bacteria. The identified Plesiomonas IS elements could have selected for the pathogenic potential of this microorganism in many instances. IS elements are known mutagenic systems of the bacterial genome that modify genes by deletion, disruption, or up-regulation of neighboring genes (Mahillon and Chandler, 1998; Gyles and Boerlin, 2014). The identified IS elements might have impacted the evolutionary advantage of virulence in Plesiomonas, with both diarrheagenic and extraintestinal pathogenesis of this microorganism increasingly emerging.
The prophages detected in Plesiomonas strain genomes contribute to their genetic identity and possible differences of virulence traits. Prophage entities are known to impact bacterial pathogen fitness, from increasing pathogenicity potential, to chromosomal rearrangement and prophage-encoded VFs (Canchaya et al., 2004; Bondy-Denomy and Davidson, 2014).
The observed numerical and structural variation in Plesiomonas CRISPR elements might be invaluable for strain typing. CRISPR variability is employed as a gold standard for strain-typing purposes in epidemiological studies of Mycobacterium tuberculosis (Kamerbeek et al., 1997; Comas et al., 2009; Abadia et al., 2010; Gomgnimbou et al., 2012), Campylobacter jejuni (van Belkum et al., 2001; Schouls et al., 2003; Louwen et al., 2013), Erwinia amylovora (Rezzonico et al., 2011), Salmonella (Liu et al., 2011), Propionibacterium acnes (Bruggemann et al., 2012), and Corynebacterium diphtheria (Mokrousov et al., 2007, 2009). Further, the discriminatory power of CRISPR typing techniques is enhanced by combining with multilocus sequence typing and amplified-fragment length polymorphism analyses (Schouls et al., 2003), CRISPR-MVLST (Shariat et al., 2013), or spoligotyping (Mokrousov et al., 2009; Ginevra et al., 2012). In addition, CRISPR-Cas systems have been adopted for bacterial virulence profiling and resistance studies of enterococci (Frimodt-Moller, 1993; Bourgogne et al., 2008; Palmer and Gilmore, 2010; van Schaik et al., 2010; Yosef et al., 2012; Tremblay et al., 2013), Salmonella (Sheikh et al., 2011), S. aureus (Kinnevey et al., 2013; Otto, 2013), enterohemorrhagic E. coli (Delannoy et al., 2012), C. jejuni (Louwen et al., 2013), Streptococcus pyogenes (McShan et al., 2008; Nozawa et al., 2011), and Mycoplasma gallisepticum (Delaney et al., 2012). Generally, studies revealed that type II CRISPR-Cas–harboring species, including the genera Campylobacter, Neisseria, and Streptococcus, exert tremendous pressure on the food industry and healthcare system (Louwen et al., 2014). Therefore, in-depth exploration of CRISPR systems might enable strain delineation and pathotyping in Plesiomonas.
The absence of plasmids in the four strains revealed that most virulence traits were chromosomally borne. Nevertheless, presence of plasmids and plasmid-borne traits have been reported in some P. shigelloides isolates (Herrington et al., 1987; Fischer et al., 1988; Olsvik et al., 1990; Kelly and Kain, 1991; Avison et al., 2001; Al-Gelawi and Al-Jeboory, 2011; Abdelhamed et al., 2018).
Conclusion
As demonstrated in the current study, comparative genomics of Plesiomonas strains revealed the presence of probable virulence traits that could not be unambiguously determined using traditional approaches. The remarkable homology or orthology of Plesiomonas ORFs with VFs and antibiotic resistance genes from species commonly identified in the healthcare and food industries are non-accidental and further attest to the pathogenic capacity of plesiomonads. We hypothesize that differential expression and regulation of virulence traits in the presence of different host factors or under different conditions might be responsible for the lack of reproducibility (i.e., dissimilar results) of data on Plesiomonas pathogenicity obtained during early investigations that relied on conventional experimental models. That probably stems from the facts that the conditions that warrant the (over)expression of a Plesiomonas virulence gene in the host might not be replicable under experimental conditions, compromising the efficiency of conventional protocols for the assessment of the bacterial virulence potential. Expression of such virulence genes or validation of our hypothesis might be of interest to future transcriptomic investigations. Further, the current study provides clues on an alternative exploratory approach that could be adapted for strain typing and delineation of pathogenic and non-pathogenic variants of the bacterium. In-depth exploration of numerical and structural variation of CRISPR systems in Plesiomonas in combination with other specific high-throughput techniques could provide low-cost but sufficient discriminatory power invaluable for strain typing and virulence profiling. Thus, the quest for developing rapid and inexpensive tools for strain and diagnostic characterization of Plesiomonas becomes imperative. We conclude that P. shigelloides possesses some VFs associated with gastroenteritis and extraintestinal infections; however, the role of host factors in the onset of infections cannot be undermined.
Data Availability Statement
All datasets generated for this study are included in the manuscript and the Supplementary Files.
Author Contributions
TE conceptualized idea for the study, responsible for data collection, analysis, interpretation, and wrote the first draft of the manuscript. AO participated in result interpretation, proofread the first draft, and supervised the study. All authors read and approved the final manuscript.
Funding
We are grateful to the National Research Foundation of South Africa, the South Africa Medical Research Council, and the World Academy of Science for financial support (Grant No: 99796).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to acknowledge Govan Mbeki Research Development Centre (GMRDC), Applied and Environmental Microbiology Research Group (AEMREG), members of the University of Fort Hare.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.03077/full#supplementary-material
FIGURE S1 | A Venn diagram showing distribution of IUSRVTs in four Plesiomonas shigelloides strains. 302-73, GN7, LS1, and NCTC10360 (9): multispecies ubiquinol-cytochrome c reductase iron-sulfur subunit, multispecies iron donor protein CyaY, multispecies iron-sulfur cluster insertion protein ErpA, multispecies succinate dehydrogenase/fumarate reductase iron-sulfur subunit, zinc/iron-chelating domain-containing protein, iron export ABC transporter permease subunit FetB, iron-sulfur cluster repair di-iron protein, ferrous iron transport protein B, multispecies iron-sulfur cluster assembly protein IscA; GN7, LS1, NCTC10360 (1): iron ABC transporter permease; 302-73, LS1 and NCTC10360 (2): multispecies zinc/iron-chelating domain-containing protein and iron transporter FeoA; 302-73, GN7 and LS1 (2): multispecies succinate dehydrogenase iron-sulfur subunit and multispecies iron-sulfur cluster scaffold-like protein; 302-73 and LS1 (1): iron(III) ABC transporter ATP-binding protein; NCTC10360 (2): succinate dehydrogenase iron-sulfur subunit; GN7 (1): multispecies iron transporter FeoA; 302-73 (1): iron(III) ABC transporter permease.
FIGURE S2 | A Venn diagram showing distribution of HUSRVTs in four Plesiomonas shigelloides strains. 302-73, GN7, LS1 and NCTC10360 (4): heme ABC transporter ATP-binding protein, heme exporter protein CcmB, heme utilization protein HutZ, heme ABC exporter ATP-binding protein CcmA; GN7, LS1 and NCTC10360 (1): putative heme utilization radical SAM enzyme HutW; 302-73, GN7 and LS1 (2): biliverdin-producing heme oxygenase and multispecies: heme exporter protein CcmD; 302-73 and NCTC10360 (1): heme exporter protein CcmC; GN7 and LS1 (2): heme ABC transporter permease and heme peroxidase; NCTC10360 (2): heme exporter protein CcmD.
DATA SHEETS S1–S4 | Detailed VRprofile query outputs for the four P. shigelloides genomes.
DATA SHEET S5 | Structural and compositional details of CRISPRs obtained in P. shigelloides strain.
Abbreviations
AR, acquired antibiotic resistance determinant; ARI, antibiotic resistance island; CRISPR, clustered regularly interspaced palindromic repeats; GI, genomic island; HUSRVT, heme uptake system related virulence trait; ICE, integrative and conjugation element; IS, insertion sequence; IUSRVT, iron uptake system related virulence trait; ORF, open reading frame; PAI, pathogenicity island; PVF, putative virulence factor; TSE, type secretion effector; TSS, type secretion system; VF, virulence factor.
Footnotes
- ^http://bioinformatics.psb.ugent.be/webtools/Venn/
- ^https://www.ncbi.nlm.nih.gov/genome/genomes/17449?
References
Abadia, E., Zhang, J., dos Vultos, T., Ritacco, V., Kremer, K., Aktas, E., et al. (2010). Resolving lineage assignation on Mycobacterium tuberculosis clinical isolates classified by spoligotyping with a new high-throughput 3R SNPs based method. Infect. Genet. Evol. 10, 1066–1074. doi: 10.1016/j.meegid.2010.07.006
Abbott, S. L., Kokka, R. P., and Janda, J. M. (1991). Laboratory investigations on the low pathogenic potential of Plesiomonas shigelloides. J. Clin. Microbiol. 29, 148–153.
Abdelhamed, H., Ozdemir, O., Tekedar, H. C., Arick, M. A., Hsu, C. Y., Karsi, A., et al. (2018). Complete genome sequence of multidrug-resistant Plesiomonas shigelloides strain MS-17-188. Genome Announcem. 6:e387-18. doi: 10.1128/genomeA.00387-18
Aiello, D., Williams, J. D., Majgier-Baranowska, H., Patel, I., Peet, N. P., Huang, J., et al. (2010). Discovery and characterization of inhibitors of Pseudomonas aeruginosa type III secretion. Antimicrob. Agents Chemother. 54, 1988–1999. doi: 10.1128/AAC.01598-09
Alcañiz, J. P., de Cuenca Morón, B., Gómez Rubio, M., Martínez Albares, J. L., and Garcia Alvarez, J. (1995). Spontaneous bacterial peritonitis due to Plesiomonas shigelloides. Am. J. Gastroenterol. 90, 1529–1530.
Aldová, E., Rakovsky, J., and Chovancova, A. (1966). The microbiological diagnostics of Aeromonas shigelloides. J. Hyg. Epidem. (Praha) 10, 470–482.
Alexander, C., and Rietschel, E. T. (2001). Bacterial lipopolysaccharides and innate immunity. J. Endotoxin Res. 7, 167–202. doi: 10.1179/096805101101532675
Alexander, S., Fazal, M. A., Burnett, E., Deheer-Graham, A., Oliver, K., Holroyd, N., et al. (2016). Complete genome sequence of Plesiomonas shigelloides type strain NCTC10360. Genome Announcem. 4:e1031-16. doi: 10.1128/genomeA.01031-16
Al-Gelawi, M. H., and Al-Jeboory, M. R. (2011). The role of plasmids in pathogenicity of locally isolated Plesiomonas shigelloides. J. Biotechnol. Res. Center 5, 46–54.
Aquilini, E., Merino, S., and Tomás, J. M. (2013). The Plesiomonas shigelloides wb O1 gene cluster and the role of O1-antigen LPS in pathogenicity. Microb. Pathog. 63, 1–7. doi: 10.1016/j.micpath.2013.05.010
Aranda, J., Cortés, P., Garrido, M. E., Fittipaldi, N., Llagostera, M., Gottschalk, M., et al. (2009). Contribution of the FeoB transporter to Streptococcus suis virulence. Int. Microbiol. 12:137.
Arndt, D., Grant, J., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016:3. doi: 10.1093/nar/gkw387
Auxiliadora-Martins, M., Bellissimo-Rodrigues, F., Viana, J. M., Teixeira, G. C., Nicolini, E. A., Cordeiro, K. S., et al. (2010). Septic shock caused by Plesiomonas shigelloides in a patient with sickle beta-zero thalassemia. Hear. Lung J. Acute Crit. Care 39, 335–339. doi: 10.1016/j.hrtlng.2009.06.015
Avison, M. B., Bennett, P. M., and Walsh, T. R. (2000). β-Lactamase expression in Plesiomonas shigelloides. J. Antimicrob. Chemother. 45, 877–880. doi: 10.1093/jac/45.6.877
Avison, M. B., Walsh, T. R., and Bennett, P. M. (2001). PUB6060: a broad-host-range, DNA polymerase-I-independent ColE2-like plasmid. Plasmid 45, 88–100. doi: 10.1006/plas.2000.1511
Bacon, D. J., Alm, R. A., Hu, L., Hickey, T. E., Ewing, C. P., Batchelor, R. A., et al. (2002). DNA sequence and mutational analysis of the pVir plasmid of Campylobacter jejuni 81-176. Infect. Immun. 70, 6242–6250. doi: 10.1128/IAI.70.11.6242-6250.2002
Bacon, D. J., Szymanski, C. M., Burr, D. H., Silver, R. P., Alm, R. A., and Guerry, P. (2001). A phase-variable capsule is involved in virulence of Campylobacter jejuni 81-176. Mol. Microbiol. 40, 769–777. doi: 10.1046/j.1365-2958.2001.02431.x
Bandyopadhyay, P., Byrne, B., Chan, Y., Swanson, M. S., and Steinman, H. M. (2003). Legionella pneumophila catalase-peroxidases are required for proper trafficking and growth in primary macrophages. Infect. Immun. 71, 4526–4535. doi: 10.1128/IAI.71.8.4526-4535.2003
Bandyopadhyay, P., and Steinman, H. M. (1998). Legionella pneumophila catalase-peroxidases: cloning of the katB gene and studies of KatB function. J. Bacteriol. 180, 5369–5374.
Bandyopadhyay, P., and Steinman, H. M. (2000). Catalase-peroxidases of Legionella pneumophila: cloning of the katA gene and studies of KatA function. J. Bacteriol. 182, 6679–6686. doi: 10.1128/JB.182.23.6679-6686.2000
Baratéla, K. C., Saridakis, H. O., Gaziri, L. C. J., and Pelayo, J. S. (2001). Effects of medium composition, calcium, iron and oxygen on haemolysin production by Plesiomonas shigelloides isolated from water. J. Appl. Microbiol. 90, 482–487. doi: 10.1046/j.1365-2672.2001.01270.x
Binns, M. M., Vaughan, S., Sanyal, S. C., and Timmis, K. N. (1984). Invasive ability of Plesiomonas shigelloides. Zentralbl. Bakteriol. Hyg. A 257, 343–347.
Bjornsdottir-Butler, K., Jones, J. L., Benner, R., and Burkhardt, W. (2011). Development of a real-time PCR assay with an internal amplification control for detection of gram-negative histamine-producing bacteria in fish. Food Microbiol. 28, 356–363. doi: 10.1016/j.fm.2010.06.013
Bjornsdottir-Butler, K., Jones, J. L., Benner, R., and Burkhardt, W. III. (2011). Development of a real-time PCR assay with an internal amplification control for detection of Gram-negative histamine-producing bacteria in fish. Food Microbiol. 28, 356–363. doi: 10.1016/j.fm.2010.06.013
Blanc-Potard, A. B., and Groisman, E. A. (1997). The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J. 16, 5376–5385. doi: 10.1093/emboj/16.17.5376
Bliska, J. B., Guan, K., Dixon, J. E., and Falkow, S. (1991). Tyrosine phosphatase hydrolysis of host proteins by an essential Yersinia virulence determinant. Proc. Natl. Acad. Sci. U.S.A. 61, 3914–3921.
Bondy-Denomy, J., and Davidson, A. R. (2014). When a virus is not a parasite: the beneficial effects of prophages on bacterial fitness. J. Microbiol. 52, 235–242. doi: 10.1007/s12275-014-4083-3
Bourgogne, A., Garsin, D. A., Qin, X., Singh, K. V., Sillanpaa, J., Yerrapragada, S., et al. (2008). Large scalevariation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 9:R110. doi: 10.1186/gb-2008-9-7-r110
Bowman, J. K., Zhang, X. C., and Hack, J. B. (2016). Plesiomonas shigelloides meningitis in an adult in the ED. Am. J. Emerg. Med. 34:1329.e1-2. doi: 10.1016/j.ajem.2015.12.046
Bravo, L., Monté, R., Rámirez, M., Garcia, B., Urbášková, P., and Aldová, E. (1998). Characterization of Plesiomonas shigelloides from diarrheic children. Cent. Eur. J. Public Health 6, 67–70.
Bravo-Fariñas, L., De Paula-Almeida, O. S., Maestre-Mesa, J., Ramírez-Àlvarez, M., and García-Rodríguez, B. (1998). The antimicrobial sensitivity and plasmid isolation in Plesiomonas shigelloides. Rev. Cubana Med. Trop. 50, 203–206.
Brenden, R. A., Miller, M. A., and Janda, J. M. (1988). Clinical disease spectrum and pathogenic factors associated with Plesiomonas shigelloides infections in humans. Rev. Infect. Dis. 10:16. doi: 10.1093/clinids/10.2.303
Brosch, R., Pym, A. S., Gordon, S. V., and Cole, S. T. (2001). The evolution of mycobacterial pathogenicity: clues from comparative genomics. Trends Microbiol. 9, 452–458. doi: 10.1016/S0966-842X(01)02131-X
Bruggemann, H., Lomholt, H. B., Tettelin, H., and Kilian, M. (2012). CRISPR-casloci of type II Propionibacterium acnes confer immunity against acquisition of mobile elements present in type I P. acnes. PLoS One 7:e34171. doi: 10.1371/journal.pone.0034171
Burns, V. C., Pishko, E. J., Preston, A., Maskell, D. J., and Harvill, E. T. (2003). Role of bordetella O antigen in respiratory tract infection. Infect. Immun. 71, 86–94. doi: 10.1128/IAI.71.1.86-94.2003
Butt, A. A., Figueroa, J., and Martin, D. H. (1997). Ocular infection caused by three unusual marine organisms. Clin. Infect. Dis. 24, 740. doi: 10.1093/clind/24.4.740
Canchaya, C., Fournous, G., and Brüssow, H. (2004). The impact of prophages on bacterial chromosomes. Mol. Microbiol. 53, 9–18. doi: 10.1111/j.1365-2958.2004.04113.x
Cao, Z., Qi, Z., Sprencel, C., Newton, S., and Klebba, P. E. (2000). Aromatic components of two ferric enterobactin binding sites in Escherichia coli FepA. Mol. Microbiol. 37, 1306–1317. doi: 10.1046/j.1365-2958.2000.02093.x
Carattoli, A., Zankari, E., García-Fernández, A., Larsen, M. V., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Cartron, M. L., Maddocks, S., Gillingham, P., Craven, C. J., and Andrews, S. C. (2006). Feo–transport of ferrous iron into bacteria. Biometals 19, 143–157. doi: 10.1007/s10534-006-0003-2
Cassat, J. E., and Skaar, E. P. (2013). Iron in infection and immunity. Cell Host Microbe 13:5. doi: 10.1016/j.chom.2013.04.010
Chakour, M., Koeck, J. L., Boulland, P., Trueba, F., Ruttimann, M., and Teyssou, R. (2002). Severe choleriform diarrhea from Plesiomonas shigelloides in an expatriate from Djibouti. Méd. Trop. 62:205.
Chao, X., Peishi, Q., Mengsha, L., and Yunzhi, L. (2016). Characterization and sorptivity of the Plesiomonas shigelloides strain and its potential use to remove Cd2+from wastewater. Water 8, 1–12. doi: 10.3390/w8060241
Che, D., Hasan, M. S., and Chen, B. (2014). Identifying pathogenicity islands in bacterial pathogenomics using computational approaches. Pathogens 3, 36–56. doi: 10.3390/pathogens3010036
Chen, X., Chen, Y., Yang, Q., Kong, H., Yu, F., Han, D., et al. (2013). Plesiomonas shigelloides infection in Southeast China. PLoS One 8:e77877. doi: 10.1371/journal.pone.0077877
Cheng, C. A., Hwang, D. F., Tsai, Y. H., Chen, H. C., Jeng, S. S., Noguchi, T., et al. (1995). Microflora and tetrodotoxin-producing bacteria in a gastropod, Niotha clathrata. Food Chem. Toxicol. 33, 929–934. doi: 10.1016/0278-6915(95)00061-6
Chowdhury, K., Sack, D. A., Rahman, A., and Rahim, Z. (1988). Enteropathogenicity of Plesiomonas shigelloides by oral inoculation in adult conditioned rabbits. J. Diarrhoeal Dis. Res. 6, 221–227.
Cianciotto, N. P., and White, R. C. (2017). Expanding role of type II secretion in bacterial pathogenesis and beyond. Infect. Immun. 85:e00014-17. doi: 10.1128/IAI.00014-17
Claesson, B. E. B., Holmlund, D. E. W., Lindhagen, C. A., and Matzch, T. W. (1984). Plesiomonas shigelloides in acute cholecystitis: a case report. J. Clin. Microbiol. 20, 985–987.
Clark, R. B., Lister, P. D., Arneson-Rotert, L., and Janda, J. M. (1990). In vitro susceptibilities of Plesiomonas shigelloides to 24 antibiotics and antibiotic-beta-lactamase-inhibitor combinations. Antimicrob. Agents Chemother. 34, 159–160. doi: 10.1128/AAC.34.1.159
Clark, R. B., Westby, G. R., Spector, H., Soricelli, R. R., and Young, C. L. (1991). Fatal Plesiomonas shigelloides septicaemia in a splenectomised patient. J. Infect. 23, 89–92. doi: 10.1016/0163-4453(91)94217-8
Collinson, S. K., Clouthier, S. C., Doran, J. L., Banser, P. A., and Kay, W. W. (1996). Salmonella enteritidis agfBAC operon encoding thin, aggregative fimbriae. J. Bacteriol. 178, 662–667. doi: 10.1128/jb.178.3.662-667.1996
Comas, I., Homolka, S., Niemann, S., and Gagneux, S. (2009). Genotyping of genetically monomorphic bacteria: DNA sequencing in Mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS One 4:e7815. doi: 10.1371/journal.pone.0007815
Comolli, J. C., Hauser, A. R., Waite, L., Whitchurch, C. B., Mattick, J. S., and Engel, J. N. (1999a). Pseudomonas aeruginosa gene products PilT and PilU are required for cytotoxicity in vitro and virulence in a mouse model of acute pneumonia. Infect. Immun. 67, 3625–3630.
Comolli, J. C., Waite, L. L., Mostov, K. E., and Engel, J. N. (1999b). Pili binding to asialo-GM1 on epithelial cells can mediate cytotoxicity or bacterial internalization by Pseudomonas aeruginosa. Infect. Immun. 67, 3207–3214.
Cornelis, G. R. (1992). “Yersinia pathogenicity factors,” in Molecular Biology of Bacterial Infection: Current Status and Future Perspectives, eds C. E. Hormaeche, C. W. Penn, C. J. Smyth, C. E. Hormaeche, C. W. Penn, and C. J. Smyth (Cambridge: Cambridge University Press).
Cramton, S. E., Gerke, C., Schnell, N. F., Nichols, W. W., and Götz, F. (1999). The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67, 5427–5433.
Cunnion, K. M., Lee, J. C., and Frank, M. M. (2001). Capsule production and growth phase influence binding of complement to Staphylococcus aureus. Infect. Immun. 69, 6796–6803. doi: 10.1128/IAI.69.11.6796-6803.2001
Cunnion, K. M., Zhang, H. M., and Frank, M. M. (2003). Availability of complement bound to Staphylococcus aureus to interact with membrane complement receptors influences efficiency of phagocytosis. Infect. Immun. 71, 656–662. doi: 10.1128/IAI.71.2.656-662.2003
Daskaleros, P. A., Stoebner, J. A., and Payne, S. M. (1991). Iron uptake in Plesiomonas shigelloides: cloning of the genes for the heme-iron uptake system. Infect. Immun. 59, 2706–2711.
Delaney, N. F., Balenger, S., Bonneaud, C., Marx, C. J., Hill, G. E., FergusonNoel, N., et al. (2012). Ultrafast evolution and loss of CRISPRs following a host shift in a novel wildlife pathogen, Mycoplasma gallisepticum. PLoS Genet. 8:e1002511. doi: 10.1371/journal.pgen.1002511
Delannoy, S., Beutin, L., and Fach, P. (2012). Use of clustered regularly interspaced short palindromic repeat sequence polymorphisms for specific detection of enterohemorrhagic Escherichia coli strains of serotypes O26: H11, O45:H2, O103:H2, O111:H8, O121:H19, O145:H28, and O157:H7 by real-time PCR. J. Clin. Microbiol. 50, 4035–4040. doi: 10.1128/JCM.02097-12
DePaola, A., Hill, W. E., and Harrell, F. M. (1993). Oligonucleotide probe determination of tetracycline-resistant bacteria isolated from catfish ponds. Mol. Cell. Probes 7, 345–348. doi: 10.1006/mcpr.1993.1051
Dewaal, C. S., Hicks, G., Barlow, K., Alderton, L., and Vegosen, L. (2006). Foods associated with foodborne illness outbreaks from 1990 through 2003. Food Protect. Trends, 26, 466–473.
Dietrich, C., Heuner, K., Brand, B. C., Hacker, J., and Steinert, M. (2001). Flagellum of Legionella pneumophila positively affects the early phase of infection of eukaryotic host cells. Infect. Immun. 69, 2116–2122. doi: 10.1128/IAI.69.4.2116-2122.2001
Dumontet, S., Aliberti, F., Pasquale, V., Gonzalez-Rey, C., Svenson, S.B., Feinstein, R., et al. (1998). “Enteropathogenicity of Plesiomonas shigelloides isolated from fresh water,” in Atti dell’XI Congresso Internazionale. Autocontrollo ed Eco Audit: Concrete Opportunita Professionali per il Biologo, eds S. Dumontet, E. Landi, and F. Pastoni (Napoli: Ordine Nazionale dei Biologi), 237–245.
Ekman, H. (2003). Bakteriella Virulencefaktorer-Studie Kring Adhesiva Egenskaper Hos Plesiomonas shigelloides. Examensarbete 2003:9. Uppsala: Institutionen för Veterinärmedicisnk Mikrobiologi, avdelningen för Bakteriologi, Veterinärmedicisnka fakulteten, SLU.
Erwin, A. L., Brewah, Y. A., Couchenour, D. A., Barren, P. R., Burke, S. J., Choi, G. H., et al. (2000). Role of lipopolysaccharide phase variation in susceptibility of Haemophilus influenzae to bactericidal immunoglobulin M antibodies in rabbit sera. Infect. Immun. 68, 2804–2807. doi: 10.1128/IAI.68.5.2804-2807.2000
Falcón, R., Carbonell, G. V., Figueredo, P. M. S., Butiao, F., Saridakis, H. O., Pelayo, J. S., et al. (2003). Intracellular vacuolation induced by culture filtrates of Plesiomonas shigelloides isolated from environmental sources. J. Appl. Microbiol. 95, 273–278. doi: 10.1046/j.1365-2672.2003.01983.x
Faruque, S. M., and Mekalanos, J. J. (2003). Pathogenicity islands and phages in Vibrio cholerae evolution. Trends Microbiol. 11, 505–510. doi: 10.1016/j.tim.2003.09.003
Fischer, K., Chakraborty, T., Hof, H., Kirchner, T., and Wamsler, O. (1988). Pseudoappendicitis caused by Plesiomonas shigelloides. J. Clin. Microbiol. 26, 2675–2677.
Fisher, C. R., Davies, N. M., Wyckoff, E. E., Feng, Z., Oaks, E. V., and Payne, S. M. (2009). Genetics and virulence association of the Shigella flexneri sit iron transport system. Infect. Immun. 77, 1992–1999. doi: 10.1128/IAI.00064-09
Fisher, K., Chakraborty, T., Hof, H., Kirchner, T., and Wamsler, O. (1988). Pseudoappendicitis caused by Plesiomonas shigelloides. J. Clin. Microbiol. 26, 2675–2677.
Fletcher, J. R., Crane, D. D., Wehrly, T. D., Martens, C. A., Bosio, C. M., and Jones, B. D. (2018). The ability to acquire iron is inversely related to virulence and the protective efficacy of francisella tularensis live vaccine strain. Front. Microbiol. 9:607. doi: 10.3389/fmicb.2018.00607
Frimodt-Moller, N. (1993). The mouse peritonitis model: present and future use. J. Antimicrob. Chemother. 31(Suppl D), 55–60. doi: 10.1093/jac/31.suppl_D.55
Gaillot, O., Bregenholt, S., Jaubert, F., Di Santo, J. P., and Berche, P. (2001). Stress-induced ClpP serine protease of listeria monocytogenes is essential for induction of listeriolysin O-dependent protective immunity. Infect. Immun. 69, 4938–4943. doi: 10.1128/IAI.69.8.4938-4943.2001
Gaillot, O., Pellegrini, E., Bregenholt, S., Nair, S., and Berche, P. (2000). The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol. Microbiol. 35, 1286–1294. doi: 10.1046/j.1365-2958.2000.01773.x
Galán, J. E. (1996). Molecular genetic bases of Salmonella entry into host cells. Mol. Microbiol. 20, 263–271. doi: 10.1111/j.1365-2958.1996.tb02615.x
Gardner, S. E., Fowlston, S. E., and George. W. L. (1987). In vitro production of a cholera-like activity by Plesiomonas shigelloides. J. Infect. Dis. 156, 720–722. doi: 10.1093/infdis/156.5.720
Garduño, R. A., Garduño, E., and Hoffman, P. S. (1998). Surface-associated hsp60 chaperonin of Legionella pneumophila mediates invasion in a HeLa cell model. Infect. Immun. 66, 4602–4610.
Ginevra, C., Jacotin, N., Diancourt, L., Guigon, G., Arquilliere, R., Meugnier, H., et al. (2012). Legionella pneumophila sequence type 1/Paris pulsotype subtyping by spoligotyping. J. Clin. Microbiol. 50, 696–701. doi: 10.1128/JCM.06180-11
Gomgnimbou, M. K., Abadia, E., Zhang, J., Refregier, G., Panaiotov, S., Bachiyska, E., et al. (2012). “Spoligoriftyping,” a dual-primingoligonucleotide-based direct-hybridization assay for tuberculosis control with a multianalyte microbead-based hybridization system. J. Clin. Microbiol. 50, 3172–3179. doi: 10.1128/JCM.00976-12
González, C. J., López-Díaz, T. M., García-López, M. L., Prieto, M., and Otero, A. (1999). Bacterial microflora of wild brown trout (Salmo trutta), wild pike (Esox lucius), andaquacultured rainbow trout (Oncorhynchus mykiss). J. Food Prot. 62, 1270–1277. doi: 10.4315/0362-028X-62.11.1270
González-Rey, C., Svenson, S. B., Bravo, L., Siitonen, A., Pasquale, V., Dumontet, S., et al. (2004). Serotypes and anti-microbial susceptibility of Plesiomonas shigelloides isolates from humans, animals and aquatic environments in different countries. Comp. Immunol. Microbiol. Infect. Dis 27, 129–139. doi: 10.1016/j.cimid.2003.08.001
Gonzalez-Rodriguez, N., Santos, J. A., Otero, A., and Garcia-Lopez, M. L. (2007). Cell-associated hemolytic activity in environmental strains of Plesiomonas shigelloides expressing cell-free, iron-influenced extracellular hemolysin. J. Food Prot. 70, 885–890. doi: 10.4315/0362-028X-70.4.885
Gopal, V., and Burns, F. E. (1991). Cellulitis and compartment syndrome due to Plesiomonas shigelloides: a case report. Mil. Med. 156:43. doi: 10.1093/milmed/156.1.43
Götz, F. (2002). Staphylococcus and biofilms. Mol. Microbiol. 43, 1367–1378. doi: 10.1046/j.1365-2958.2002.02827.x
Guo, L., Lim, K. B., Gunn, J. S., Bainbridge, B., Darveau, R. P., Hackett, M., et al. (1997). Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science 276, 250–253. doi: 10.1126/science.276.5310.250
Gupta, S. (1995). Migratory polyarthritis associated with Plesiomonas shigelloides infection. Scand. J. Rheumatol. 24, 323–325. doi: 10.3109/03009749509095174
Gurwith, M. J., and Williams, T. W. (1977). Gastroenteritis in children: a two-year review in Manitoba. J. Infect. Dis. 136, 239–247. doi: 10.1093/infdis/136.2.239
Gyles, C., and Boerlin, P. (2014). Horizontally transferred genetic elements and their role in pathogenesis of bacterial disease. Vet. Pathol. 51, 328–340. doi: 10.1177/0300985813511131
Hahn, H. P. (1997). The type-4 pilus is the major virulence-associated adhesin of Pseudomonas aeruginosa–a review. Gene 192, 99–108. doi: 10.1016/S0378-1119(97)00116-9
Harvill, E. T., Preston, A., Cotter, P. A., Allen, A. G., Maskell, D. J., and Miller, J. F. (2000). Multiple roles for bordetella lipopolysaccharide molecules during respiratory tract infection. Infect. Immun. 68, 6720–6728. doi: 10.1128/IAI.68.12.6720-6728.2000
Heilmann, C., Schweitzer, O., Gerke, C., Vanittanakom, N., Mack, D., and Götz, F. (1996). Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20, 1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x
Henderson, D. P., Wyckoff, E. E., Rashidi, C. E., Verlei, H., and Oldham, A. L. (2001). Characterization of the Plesiomonas shigelloides genes encoding the heme iron utilization system. J. Bacteriol. 183, 2715–2723. doi: 10.1128/JB.183.9.2715-2723.2001
Herrington, D. A., Tzipori, S., Robins-Browne, R. M., Tall, B. D., and Levine, M. M. (1987). In vitro and in vivo pathogenicity of Plesiomonas shigelloides. Infect. Immun. 55, 979–985.
Herve, V., Bigaillon, C., Duhamel, P., Petit, M. P., and Soler, C. (2007). Cutaneous abscess due to Plesiomonas shigelloïdes consecutive to a trauma in fresh water. Méd. Malad. Infect. 37:840. doi: 10.1016/j.medmal.2007.03.003
Heuner, K., and Steinert, M. (2003). The flagellum of Legionella pneumophila and its link to the expression of the virulent phenotype. Int. J. Med. Microbiol. 293, 133–143. doi: 10.1078/1438-4221-00259
Holmberg, S. D., and Farmer, J. J. (1984). Aeromonas hydrophila and Plesiomonas shigelloides as causes of intestinal infections. Rev. Infect. Dis. 6, 633–639. doi: 10.1093/clinids/6.5.633
Ingram, C. W., Morrison, A. J. Jr., and Levitz, R. E. (1987). Gastroenteritis, sepsis, and osteomyelitis caused by Plesiomonas shigelloides in an immunocompetent host: case report and review of the literature. J. Clin. Microbiol. 25, 1791–1793.
Janda, J. M., and Abbott, S. L. (1993). Expression of hemolytic activity by Plesiomonas shigelloides. J. Clin. Microbiol. 31, 1206–1208.
Jermyn, W. S., and Boyd, E. F. (2002). Characterization of a novel Vibrio pathogenicity island (VPI-2) encoding neuraminidase (nanH) among toxigenic Vibrio cholerae isolates. Microbiology 148, 3681–3693. doi: 10.1099/00221287-148-11-3681
Johnson, T. L., Waack, U., Smith, S., Mobley, H., and Sandkvist, M. (2016). Acinetobacter baumannii is dependent on the type II secretion system and its substrate LipA for lipid utilization and in vivo fitness. J. Bacteriol. 198, 711–719. doi: 10.1128/JB.00622-15
Jönsson, I., Monsen, T., and Wiström, J. (1998). A case report of Plesiomonas shigelloides cellulitis and bacteraemia from Northern Europe. Scand. J. Infect Dis. 29, 631–632. doi: 10.3109/00365549709035909
Jun, J. W., Kim, J. H., Choresca, C. H. Jr., Shin, S. P., Han, J. E., Jeong, D. S., et al. (2011). Isolation and molecular detection of Plesiomonas shigelloides containing tetA gene from asian arowana (Scleropages formosus) in a Korean aquarium. Afr. J. Microbiol. Res. 5, 5019–5021.
Kain, K. C., and Kelly, M. T. (1989a). Antimicrobial susceptibility of Plesiomonas shigelloides from patients with diarrhea. Antimicrob. Agents Chemother. 33, 1609–1610. doi: 10.1128/AAC.33.9.1609
Kain, K. C., and Kelly, M. T. (1989b). Clinical features, epidemiology, and treatment of Plesiomonas shigelloides diarrhea. J. Clin. Microbiol. 27, 998–1001.
Kalotychou, V., Rombos, Y., Tzanetea, R., Konstantopoulos, K., Spiliopoulou, C., Xanthaki, A., et al. (2002). Plesiomonas shigelloides sepsis in a thalassemia intermedia patient. Scand. J. Infect. Dis. 34, 687–689. doi: 10.1080/00365540210147877
Kamerbeek, J., Schouls, L., Kolk, A., van Agterveld, M., van Soolingen, D., Kuijper, S., et al. (1997). Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35, 907–914.
Karaolis, D. K., Johnson, J. A., Bailey, C. C., Boedeker, E. C., Kaper, J. B., and Reeves, P. R. (1998). A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc. Natl. Acad. Sci. U.S.A. 95, 3134–3139. doi: 10.1073/pnas.95.6.3134
Kaszowska, M., Stojkovic, K., Niedziela, T., and Lugowski, C. (2016). The O-antigen of Plesiomonas shigelloides serotype O36 containing pseudaminic acid. Carbohydr. Res. 434, 1–5. doi: 10.1016/j.carres.2016.07.007
Kelly, M. T., and Kain, K. C. (1991). Biochemical characteristics and plasmids of clinical and environmental Plesiomonas shigelloides. Experientia 47, 439–441.
Kennedy, C. A., Goetz, M. B., and Mathisen, G. E. (1990). Postoperative pancreatic abscess due to Plesiomonas shigelloides. Rev. Infect. Dis. 12, 813–816. doi: 10.1093/clinids/12.5.813
Khan, N. A., Shin, S., Chung, J. W., Kim, K. J., Elliott, S., Wang, Y., et al. (2003). Outer membrane protein A and cytotoxic necrotizing factor-1 use diverse signaling mechanisms for Escherichia coli K1 invasion of human brain microvascular endothelial cells. Microb. Pathog. 35, 35–42. doi: 10.1016/S0882-4010(03)00090-1
Kinnevey, P. M., Shore, A. C., Brennan, G. I., Sullivan, D. J., Ehricht, R., and Monecke, S., (2013). Emergence of sequence type 779 methicillin-resistant Staphylococcus aureus harboring a novel pseudo staphylococcal cassette chromosome mec (SCCmec)-SCC-SCCCRISPR composite element in Irish hospitals. Antimicrob. Agents Chemother. 57, 524–531. doi: 10.1128/AAC.01689-12
Klatte, J. M., Dastjerdi, M. H., Clark, K., Harrison, C. J., Grigorian, F., and Stahl, E. D. (2012). Hyperacute infectious keratitis with Plesiomonas shigelloides following traumatic lamellar corneal laceration. Pediatr. Infect. Dis. J. 31, 1200–1201. doi: 10.1097/INF.0b013e318266b61f
Klontz, E. H., Faruque, A. S., Das, S. K., Malek, M. A., Islam, Z., Luby, S. P., et al. (2012). Clinical and epidemiologic features of diarrheal disease due to Aeromonas hydrophila and Plesiomonas shigelloides infections compared with those due to Vibrio cholerae Non-O1 and Vibrio parahaemolyticus in Bangladesh. ISRN Microbiol. 2012:6. doi: 10.5402/2012/654819
Krovacek, K., Eriksson, L. M., González-Rey, C., Rosinsky, J., and Ciznar, I. (2000). Isolation, biochemical and serological characterisation of Plesiomonas shigelloides from freshwater in Northern Europe. Comp. Immunol. Microbiol. Infect. Dis. 23, 45–51. doi: 10.1016/S0147-9571(99)00058-2
Kubler-Kielb, J., Schneerson, R., Mocca, C., and Vinogradov, E. (2008). The elucidation of the structure of the core part of the LPS from Plesiomonas shigelloides serotype O17 expressing O-polysaccharide chain identical to the Shigella sonnei O-chain. Carbohydr. Res. 343, 3123–3127. doi: 10.1016/j.carres.2008.09.017
Lau, C. K., Krewulak, K. D., and Vogel, H. J. (2015). Bacterial ferrous iron transport: the Feo system. FEMS Microbiol. Rev. 40, 273–298. doi: 10.1093/femsre/fuv049
Lei, P., Shuiyi L., Xuwei, C., Yiting T., Tao Y., Peipei L., et al. (2017). Isolation, Identification and genomic analysis of Plesiomonas shigelloides isolated from diseased percocypris pingi (Tchang, 1930). Am. J. Biochem. Biotechnol. 13, 226–232. doi: 10.3844/ajbbsp.2017.226.232
Lejona, S., Aguirre, A., Cabeza, M. L., Véscovi, E. G., and Soncini, F. C. (2003). Molecular characterization of the Mg2+-responsive PhoP-PhoQ regulon in Salmonella enterica. J. Bacteriol. 185, 6287–6294. doi: 10.1128/JB.185.21.6287-6294.2003
Li, J., Tai, C., Deng, Z., Zhong, W., He, Y., Ou, H. Y. (2018). VRprofile: gene-cluster-detection-based profiling of virulence and antibiotic resistance traits encoded within genome sequences of pathogenic bacteria. Brief. Bioinform. 19, 566–574. doi: 10.1093/bib/bbw141
Lindgren, H., Honn, M., Salomonsson, E., Kuoppa, K., Forsberg,Å., and Sjöstedt, A. (2011). Iron content differs between Francisella tularensis subspecies tularensis and subspecies holarctica strains and correlates to their susceptibility to H2O2-induced killing. Infect. Immun. 79, 1218–1224. doi: 10.1128/IAI.01116-10
Liu, F., Kariyawasam, S., Jayarao, B. M., Barrangou, R., Gerner-Smidt, P., Ribot, E. M., et al. (2011). Subtyping Salmonella enterica serovar enteritidis isolates from different sources by using sequence typing based on virulence genes and clustered regularly interspaced short palindromic repeats (CRISPRs). Appl. Environ. Microbiol. 77, 4520–4526. doi: 10.1128/AEM.00468-11
Lopez-Sabater, E. I., Rodriguez-Jerez, J. J., Hernandez-Herrero, M., and Mora-Ventura, M. T. (1996). Incidence of histamine-forming bacteria and histamine content in scombroid fish species from retail markets in the Barcelona area. Int. J. Food Microbiol. 28, 411–418. doi: 10.1016/0168-1605(94)00007-7
Louwen, R., Horst-Kreft, D., de Boer, A. G., van der Graaf, L., de Knegt, G., Hamersma, M., et al. (2013). A novel link between Campylobacter jejuni bacteriophage defence, virulence and Guillain-Barré syndrome. Eur. J. Clin. Microbiol. Infect. Dis. 32, 207–226. doi: 10.1007/s10096-012-1733-4
Louwen, R., Staals, R. H., Endtz, H. P., van Baarlen, P., and van der Oost, J. (2014). The Role of CRISPR-Cas systems in virulence of pathogenic bacteria. Microbiol. Mol. Biol. Rev. 78, 74–88. doi: 10.1128/MMBR.00039-13
Lucas, R. L., and Lee, C. A. (2000). Unravelling the mysteries of virulence gene regulation in Salmonella typhimurium. Mol. Microbiol. 36, 1024–1033. doi: 10.1046/j.1365-2958.2000.01961.x
Luong, T. T., and Lee, C. Y. (2002). Overproduction of type 8 capsular polysaccharide augments Staphylococcus aureus virulence. Infect. Immun. 70, 3389–3395. doi: 10.1128/IAI.70.7.3389-3395.2002
Lyczak, J. B., Cannon, C. L., and Pier, G. B. (2000). Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist1. Microbes Infect. 2, 1051–1060. doi: 10.1016/S1286-4579(00)01259-4
Mahrshmann, W. E., and Lyons, C. J. (1998). Congenital endophthalmitis following maternal shellfish ingestion. Aust. N. Z. Ophthalmol. 26, 161–163. doi: 10.1111/j.1442-9071.1998.tb01534.x
Maluping, R. P., Lavilla-Pitogo, C. R., DePaola, A., Janda, J. M., and Krovacek, K. (2004). Occurrence, characterisation and detection of potential virulence determinants of emerging aquatic bacterial pathogens from the Philippines and Thailand. New Microbiol. 27, 381–389.
Marcus, S. L., Brumell, J. H., Pfeifer, C. G., and Finlay, B. B. (2000). Salmonella pathogenicity islands: big virulence in small packages. Microbes Infect. 2, 145–156. doi: 10.1016/S1286-4579(00)00273-2
Marshall, D. L., Kim, J. J., and Donnelly, S. P. (1996). Antimicrobial susceptibility and plasmid-mediated streptomycin resistance of Plesiomonas shigelloides isolated from blue crab. J. Appl. Bacteriol. 81, 195–200. doi: 10.1111/j.1365-2672.1996.tb04500.x
Matthews, B. G., Douglas, H., and Guiney, D. G. (1988). Production of a heat stable enterotoxin by Plesiomonas shigelloides. Microb. Pathog. 5, 207–213. doi: 10.1016/0882-4010(88)90023-X
Maurelli, A. T., Fernández, R. E., Bloch, C. A., Rode, C. K., and Fasano, A. (1998). ‘Black holes’ and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella s and enteroinvasive Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 95, 3943–3948. doi: 10.1073/pnas.95.7.3943
McCracken, A. W., and Barkley, R. (1972). Isolation of Aeromonas species from clinical sources. J. Clin. Pathol. 25, 970–975. doi: 10.1136/jcp.25.11.970
McDaniel, T. K., and Kaper, J. B. (1997). A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23, 399–407. doi: 10.1046/j.1365-2958.1997.2311591.x
McKenney, D., Hübner, J., Muller, E., Wang, Y., Goldmann, D. A., and Pier, G. B. (1998). The ica locus of Staphylococcus epidermidis encodes production of the capsular polysaccharide/adhesin. Infect. Immun. 66, 4711–4720.
McShan, W. M., Ferretti, J. J., Karasawa, T., Suvorov, A. N., Lin, S., Qin, B., et al. (2008). Genome sequence of a nephritogenic and highly transformable M49 strain of Streptococcus pyogenes. J. Bacteriol. 190, 7773–7785. doi: 10.1128/JB.00672-08
Ménard, R., Dehio, C., and Sansonetti, P. J. (1996). Bacterial entry into epithelial cells: the paradigm of Shigella. Trends Microbiol. 4, 220–226. doi: 10.1016/0966-842X(96)10039-1
Meyers, J. A., Sanchez, D., Elwell, L. P., and Falkow, S. (1976). Simple agarose gel electrophoretic method for the identification and characterization of plasmid deoxyribonucleic acid. J. Bacteriol. 127, 1529–1537.
Michiels, T., Wattiau, P., Brasseur, R., Ruysschaert, J.-M., and Cornelis, G. R. (1990). Secretion of yop proteins by yersiniae. Infect. Immun. 58, 2840–2849.
Mokrousov, I., Limeschenko, E., Vyazovaya, A., and Narvskaya, O. (2007). Corynebacterium diphtheriae spoligotyping based on combined use of two CRISPR loci. Biotechnol. J. 2, 901–906. doi: 10.1002/biot.200700035
Mokrousov, I., Vyazovaya, A., Kolodkina, V., Limeschenko, E., Titov, L., and Narvskaya, O. (2009). Novel macroarray-based method of Corynebacterium diphtheriae genotyping: evaluation in a field study in Belarus. Eur. J. Clin. Microbiol. Infect. Dis. 28, 701–703. doi: 10.1007/s10096-008-0674-4
Moncrief, M. B. C., and Maguire, M. E. (1999). Magnesium transport in prokaryotes. JBIC J. Biol. Inorgan. Chem. 4, 523–527. doi: 10.1007/s007750050374
Morton, D. J., Seale, T. W., Bakaletz, L. O., Jurcisek, J. A., Smith, A., VanWagoner, T. M., et al. (2009). The heme-binding protein (HbpA) of Haemophilus influenzae as a virulence determinant. Int. J. Med. Microbiol. 299, 479–488. doi: 10.1016/j.ijmm.2009.03.004
Nagy, T. A., Moreland, S. M., and Detweiler, C. S. (2014). Salmonella acquires ferrous iron from haemophagocytic macrophages. Mol. Microbiol. 93, 1314–1326.
Naikare, H., Palyada, K., Panciera, R., Marlow, D., and Stintzi, A. (2006). Major role for FeoB in Campylobacter jejuni ferrous iron acquisition, gut colonization, and intracellular survival. Infect. Immun. 74, 5433–5444. doi: 10.1128/IAI.00052-06
Nair, S., Milohanic, E., and Berche, P. (2000). ClpC ATPase is required for cell adhesion and invasion of Listeria monocytogenes. Infect. Immun. 68, 7061–7068. doi: 10.1128/IAI.68.12.7061-7068.2000
Nivens, D. E., Ohman, D. E., Williams, J., and Franklin, M. J. (2001). Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J. Bacteriol. 183, 1047–1057. doi: 10.1128/JB.183.3.1047-1057.2001
Nolte, F. S., Poole, R. M., Murphy, G. W., Clark, C., and Panner, B. J. (1988). Proctitis and fatal septicemia by Plesiomonas shigelloides in a bisexual man. J. Clin. Microbiol. 26, 388–391.
Novoa-Farías, O., Frati-Munari, A. C., Peredo, M. A., Flores-Juárez, S., Novoa-García, O., Galicia-Tapia, J., et al. (2016). Susceptibilidad de las bacterias aisladas de infecciones gastrointestinales agudas a la rifaximina y otros agentes antimicrobianos en México. Rev. Gastroenterol. México 81, 3–10. doi: 10.1016/j.rgmx.2015.07.003
Nozawa, T., Furukawa, N., Aikawa, C., Watanabe, T., Haobam, B., Kurokawa, K., et al. (2011). CRISPR inhibition of prophage acquisition in Streptococcus pyogenes. PLoS One 6:e19543. doi: 10.1371/journal.pone.0019543
Okawa, Y., Ohtomo, Y., Tsugawa, H., Matsuda, Y., Kobayashi, H., and Tsukamoto, T. (2004). Isolation and characterization of a cytotoxin produced by Plesiomonas shigelloides P-1 strain. FEMS Microbiol. Lett. 239, 125–130. doi: 10.1016/j.femsle.2004.08.026
Oldham, A. L., Wood, T. A., and Henderson, D. P. (2008). Plesiomonas shigelloides hugZ encodes an iron-regulated heme binding protein required for heme iron utilization. Can. J. Microbiol. 54, 97–102. doi: 10.1139/W07-122
Olsvik, O., Wachsmuth, K., Kay, B., Birkness, K. A., Yi, A., and Sack, B. (1990). Laboratory observations on Plesiomonas shigelloides strains isolated from children with diarrhea in Peru. J. Clin. Microbiol. 28, 886–889.
Olsvik, O., Wachsmuth, K., Kay, B. A., Hale, T. L., and Sack, R. B. (1985). “Pathogenicity studies of clinical isolates of Plesiomonas shigelloides [abstract no. B168],” in Proceedings of the Abstracts of the 85th annual meeting of the American Society for Microbiology (Washington, DC: American Society for Microbiology), 46.
Otto, M. (2013). Coagulase-negative staphylococci as reservoirs of genes facilitating MRSA infection: staphylococcal commensal species such as Staphylococcus epidermidis are being recognized as important sources of genes promoting MRSA colonization and virulence. Bioessays 35, 4–11. doi: 10.1002/bies.201200112
Ozdemir, O., Sari, S., Terzioglu, S., and Zenciroglu, A. (2010). Plesiomonas shigelloides sepsis and meningoencephalitis in a surviving neonate. J. Microbiol. Immuno. Infect. 43, 344–346. doi: 10.1016/S1684-1182(10)60053-9
Palmer, K. L., and Gilmore, M. S. (2010). Multidrug-resistant enterococci lack CRISPR-Cas. mBio 1:e00227-10. doi: 10.1128/mBio.00227-10
Patel, S., Gandhi, D., Mehta, V., Bhatia, K., and Epelbaum, O. (2016). Plesiomonas shigelloides: an extremely rare cause of spontaneous bacterial peritonitis. Acta Gastroenterol. Belg. 79, 52–53.
Pence, M. A. (2016). The brief case: wound infection with Plesiomonas shigelloides following a freshwater injury. J. Clin. Microbiol. 54, 1180–1182. doi: 10.1128/JCM.02651-15
Pfeiffer, M. L., DuPont, H. L., and Ochoa, T. J. (2012). The patient presenting with acute dysentery–a systematic review. J. Infect. 64, 374–386. doi: 10.1016/j.jinf.2012.01.006
Pier, G. B., Coleman, F., Grout, M., Franklin, M., and Ohman, D. E. (2001). Role of alginate O acetylation in resistance of mucoid Pseudomonas aeruginosa to opsonic phagocytosis. Infect. Immun. 69, 1895–1901. doi: 10.1128/IAI.69.3.1895-1901.2001
Piqué, N., Aquilini, E., Alioto, T., Miñana-Galbis, D., and Tomás, J. M. (2013). Genome sequence of Plesiomonas shigelloide Piqué, N., Aquilini, E., Alioto, T., Miñana-Galbis, D. and Tomás, J.M., 2013. Genome sequence of Plesiomonas shigelloides strain 302-73 (serotype O1). Genome Announcem. 1:e00404-13.
Pitarangsi, C., Echeverria, P., Whitmire, R., Tirapat, C., Formal, S., Dammin, G. J., et al. (1982). Enteropathogenicity of Aeromonas hydrophila and Plesiomonas shigelloides: prevalence among individuals with and without diarrhea in Thailand. Infect. Immun. 35, 666–673.
Prasadarao, N. V., Blom, A. M., Villoutreix, B. O., and Linsangan, L. C. (2002). A novel interaction of outer membrane protein A with C4b binding protein mediates serum resistance of Escherichia coli K1. J. Immunol. 169, 6352–6360. doi: 10.4049/jimmunol.169.11.6352
Prasadarao, N. V., Srivastava, P. K., Rudrabhatla, R. S., Kim, K. S., Huang, S. H., and Sukumaran, S. K. (2003). Cloning and expression of the Escherichia coli K1 outer membrane protein A receptor, a gp96 homologue. Infect. Immun. 71, 1680–1688. doi: 10.1128/IAI.71.4.1680-1688.2003
Rao, V. K., Krasan, G. P., Hendrixson, D. R., Dawid, S., and St. Geme, J. W. III. (1999). Molecular determinants of the pathogenesis of disease due to non-typable Haemophilus influenzae. FEMS Microbiol. Rev. 23, 99–129. doi: 10.1111/j.1574-6976.1999.tb00393.x
Rezzonico, F., Smits, T. H., and Duffy, B. (2011). Diversity, evolution, and functionality of clustered regularly interspaced short palindromic repeat (CRISPR) regions in the fire blight pathogen Erwinia amylovora. Appl. Environ. Microbiol. 77, 3819–3829. doi: 10.1128/AEM.00177-11
Robey, M., and Cianciotto, N. P. (2002). Legionella pneumophila feoAB promotes ferrous iron uptake and intracellular infection. Infect. Immun. 70, 5659–5669. doi: 10.1128/IAI.70.10.5659-5669.2002
Rocchetta, H. L., Burrows, L. L., and Lam, J. S. (1999). Genetics of O-antigen biosynthesis in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 63, 523–553.
Rodríguez-Rodríguez, S., and Santos, J. A. (2018). Detection and characterization of the ferric uptake regulator (fur) gene in Plesiomonas shigelloides. Lett. Appl. Microbiol. 66, 347–351. doi: 10.1111/lam.12858
Römling, U., Bian, Z., Hammar, M., Sierralta, W. D., and Normark, S. (1998). Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 180, 722–731.
Rosqvist, R., Forsberg,Å., Rimpiläinen, M., Bergman, T., and Wolf-Watz, H. (1990). The cytotoxic proteinYopE of Yersinia obstructs the primary host defense. Mol. Microbiol. 4, 657–667. doi: 10.1111/j.1365-2958.1990.tb00635.x
Rosqvist, R., Håkansson, S., Forsberg,Å., and WolfWatz, H. (1995). Functional conservation of the secretion and translocation machinery for virulence proteins of Yersiniae, Salmonellae and Shigellae. EMBO J. 14, 4187–4195. doi: 10.1002/j.1460-2075.1995.tb00092.x
Roth, T., Hentsch, C., Erard, P., and Tschantz, P. (2002). Pyosalpinx: not always a sexual transmitted disease? Pyosalpinx caused by Plesiomonas shigelloides in an immunocompetent host. Clin. Microbiol. Infect. 8, 803–805. doi: 10.1046/j.1469-0691.2002.00443.x
Rouquette, C., De Chastellier, C., Nair, S., and Berche, P. (1998). The ClpC ATPase of Listeria monocytogenes is a general stress protein required for virulence and promoting early bacterial escape from the phagosome of macrophages. Mol. Microbiol. 27, 1235–1245. doi: 10.1046/j.1365-2958.1998.00775.x
Rouquette, C., Ripio, M. T., Pellegrini, E., Bolla, J. M., Tascon, R. I., Vázquez-Boland, J. A., et al. (1996). Identification of a ClpC ATPase required for stress tolerance and in vivo survival of Listeria monocytogenes. Mol. Microbiol. 21, 977–987. doi: 10.1046/j.1365-2958.1996.641432.x
Rust, L., Messing, C. R., Iglewski, B. H. (1994). “Elastase assays,” in Methods in Enzymolgy: Bacterial Pathogenesis. Part A: Identification and Regulation of Virulence Factors, Vol. 235, eds V. L. Clark, P. M. Bavoil (London: Academic Press), 554–562. doi: 10.1016/0076-6879(94)35170-8
Santos, J. A., Gonzalez, C. J., Lopez, T. M., Otero, A., and Garcia-Lopez, M. L. (1999). Hemolytic and elastolytic activities influenced by iron in Plesiomonas shigelloides. J. Food Prot. 62, 1475–1477. doi: 10.4315/0362-028X-62.12.1475
Sanyal, S. C., Saraswathi, B., and Sharm, P. (1980). Enteropathogenicity of Plesiomonas shigelloides. J. Med Microb. 13, 41–409. doi: 10.1099/00222615-13-3-401
Sanyal, S. C., Singh, S. J., and Sen, P. C. (1975). Enterotoxigenicity of Aeromonas hydrophila and Plesiomonas shigelloides. J. Med. Microbiol. 195–198. doi: 10.1099/00222615-8-1-195
Saraswathi, B., Agarwal, R. K., and Sanyal, S. C. (1983). Further studies on enteropathogenicity of Plesiomonas shigelloides. Indian J. Med. Res. 78, 12–18.
Schaeffer, L. M., McCormack, F. X., Wu, H., and Weiss, A. A. (2004). Bordetella pertussis lipopolysaccharide resists the bactericidal effects of pulmonary surfactant protein A. J. Immunol. 173, 1959–1965. doi: 10.4049/jimmunol.173.3.1959
Schmidt, H., and Hensel, M. (2004). Pathogenicity islands in bacterial pathogenesis. Clin. Microbiol. Rev. 17, 14–56. doi: 10.1128/CMR.17.1.14-56.2004
Schneider, F., Lang, N., Reibke, R., Michaely, H. J., Hiddemann, W., and Ostermann, H. (2009). Plesiomonas shigelloides pneumonia. Méd. Malad. Infect. 39, 397–400. doi: 10.1016/j.medmal.2008.11.010
Schouls, L. M., Reulen, S., Duim, B., Wagenaar, J. A., Willems, R. J., Dingle, K. E., et al. (2003). Comparative genotyping of Campylobacter jejuni by amplified fragment length polymorphism, multilocus sequence typing, and short repeat sequencing: strain diversity, host range, and recombination. J. Clin. Microbiol. 41, 15–26. doi: 10.1128/JCM.41.1.15-26.2003
Schroeder, T. H., Lee, M. M., Yacono, P. W., Cannon, C. L., Gerçeker, A. A., Golan, D. E., et al. (2002). CFTR is a pattern recognition molecule that extracts Pseudomonas aeruginosa LPS from the outer membrane into epithelial cells and activates NF-κB translocation. Proc. Natl. Acad. Sci. U.S.A. 99, 6907–6912. doi: 10.1073/pnas.092160899
Schubert, R. H. W., and Holz-Bremer, A. (1999). Cell adhesion of Plesiomonas shigelloides. Zentralbl. Hyg. Umweltmed. 202, 383–388. doi: 10.1016/S0934-8859(99)80003-0
Sears, C. L., and Kaper, J. B. (1996). Enteric bacterial toxins; mechanisms of action and linkage to intestinal secretion. Microbiol. Rev. 60, 167–215.
Shariat, N., Kirchner, M. K., Sandt, C. H., Trees, E., Barrangou, R., and Dudley, E. G. (2013). Subtyping of Salmonella enterica serovar Newport outbreak isolates by CRISPR MVLST and determination of the relationship between CRISPR-MVLST and PFGE results. J. Clin. Microbiol. 51, 2328–2336. doi: 10.1128/JCM.00608-13
Shaw, K. J., Rather, P. N., Hare, R. S., and Miller, G. H. (1993). Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57, 138–163. Available at: http://mmbr.asm.org/content/57/1/138.full.pdf+html
Sheikh, A., Charles, R. C., Sharmeen, N., Rollins, S. M., Harris, J. B., Bhuiyan, M. S., et al. (2011). In vivo expression of Salmonella enterica serotype Typhi genes in the blood of patients with typhoid fever in Bangladesh. PLoS Negl. Trop. Dis. 5:e1419. doi: 10.1371/journal.pntd.0001419
Shepherd, J. G., Wang, L., and Reeves, P. R. (2000). Comparison of O-antigen gene clusters of Escherichia coli (Shigella) Sonnei and Plesiomonas shigelloides O17: sonnei gained its current plasmid-borne o-antigen genes from P. shigelloides in a recent event. Infect. Immun. 68, 6056–6061. doi: 10.1128/IAI.68.10.6056-6061.2000
Song, Z., Wu, H., Ciofu, O., Kong, K. F., Høiby, N., Rygaard, J., et al. (2003). Pseudomonas aeruginosa alginate is refractory to Th1 immune response and impedes host immune clearance in a mouse model of acute lung infection. J. Med. Microbiol. 52, 731–740. doi: 10.1099/jmm.0.05122-0
Sprencel, C., Cao, Z., Qi, Z., Scott, D. C., Montague, M. A., Ivanoff, N., et al. (2000). Binding of ferric enterobactin by the Escherichia coli periplasmic protein FepB. J. Bacteriol. 182, 5359–5364. doi: 10.1128/JB.182.19.5359-5364.2000
Stock, I., and Wiedemann, B. (2001a). β-Lactam-susceptibility patterns of Plesiomonas shigelloides strains: importance of inoculum and medium. Scand. J. Infect. Dis. 33, 692–696. doi: 10.1080/00365540110026872
Stock, I., and Wiedemann, B. (2001b). Natural antimicrobial susceptibilities of Plesiomonas shigelloides strains. J. Antimicrob. Chemother. 48, 803–811. doi: 10.1093/jac/48.6.803
Stojijkovic, I., Hwa, V., de Saint Martin, L., O’Gaora, P., Nassif, X., Heffron, F., et al. (1995). The Neisseria meningitidis haemoglobin receptor: its role in iron utilization and virulence. Mol. Microbiol. 15, 531–541. doi: 10.1111/j.1365-2958.1995.tb02266.x
Sukumaran, S. K., Shimada, H., and Prasadarao, N. V. (2003). Entry and intracellular replication of Escherichia coli K1 in macrophages require expression of outer membrane protein A. Infect. Immun. 71, 5951–5961. doi: 10.1128/IAI.71.10.5951-5961.2003
Sukupolvi, S., Lorenz, R. G., Gordon, J. I., Bian, Z., Pfeifer, J. D., Normark, S. J., et al. (1997). Expression of thin aggregative fimbriae promotes interaction of Salmonella typhimurium SR-11 with mouse small intestinal epithelial cells. Infect. Immun. 65, 5320–5325.
Suthienkul, O., Aiumlaor, P., Siripanichgon, K., Eampokalap, B., Likhanonsakul, S., Utrarachkij, F., et al. (2001). Bacterial causes of AIDSassociated diarrhea in Thailand. Southeast Asian J. Trop. Med. Public Health 32, 158–170.
Swanson, M. S., and Hammer, B. K. (2000). Legionella pneumophila pathogenesis: a fateful journey from amoebae to macrophages. Ann. Rev. Microbiol. 54, 567–613. doi: 10.1146/annurev.micro.54.1.567
Swords, W. E., Buscher, B. A., Ver Steeg Ii, K., Preston, A., Nichols, W. A., Weiser, J. N., et al. (2000). Non-typeable Haemophilus influenzae adhere to and invade human bronchial epithelial cells via an interaction of lipooligosaccharide with the PAF receptor. Mol. Microbiol. 37, 13–27. doi: 10.1046/j.1365-2958.2000.01952.x
Tao, T., Grulich, P. F., Kucharski, L. M., Smith, R. L., and Maguire, M. E. (1998). Magnesium transport in Salmonella typhimurium: biphasic magnesium and time dependence of the transcription of the mgtA and mgtCB loci. Microbiology 144, 655–664. doi: 10.1099/00221287-144-3-655
Taylor, D. N., Trofa, A. C., Sadoff, J., Chu, C., Bryla, D., Shiloach, J., et al. (1993). Synthesis, characterization, and clinical evaluation of conjugate vaccines composed of the O-specific polysaccharides of Shigella dysenteriae type 1, Shigella flexneri type 2a, and Shigella sonnei (Plesiomonas shigelloides) bound to bacterial toxoids. Infect. Immun. 61, 3678–3687.
Thakker, M., Park, J. S., Carey, V., and Lee, J. C. (1998). Staphylococcus aureus serotype 5 capsular polysaccharide is antiphagocytic and enhances bacterial virulence in a murine bacteremia model. Infect. Immun. 66, 5183–5189.
Theodoropoulos, C., Wong, T. H., O’Brien, M., and Stenzel, D. (2001). Plesiomonas shigelloides enters polarized human intestinal Caco-2 cells in an in vitro model system. Infect. Immun. 69, 2260–2269. doi: 10.1128/IAI.69.4.2260-2269.2001
Tremblay, C. L., Charlebois, A., Masson, L., and Archambault, M. (2013). Characterization of hospital-associated lineages of ampicillin resistant Enterococcus faecium from clinical cases in dogs and humans. Front. Microbiol. 4:245. doi: 10.3389/fmicb.2013.00245
Tsugawa, H., Ono, T., Murakami, H., and Okawa, Y. (2005). Invasive phenotype and apoptosis induction of Plesiomonas shigelloides P-1 strain to Caco-2 cells. J. Appl. Microbiol. 99, 1435–1443. doi: 10.1111/j.1365-2672.2005.02721.x
Tsukamoto, T., Kinoshita, Y., Shimada, T., and Sakazaki, R. (1978). Two epidemics of diarrhoeal disease possibly caused by Plesiomonas shigelloides. J. Hygiene 80, 275–280. doi: 10.1017/S0022172400053638
van Belkum, A., van den Braak, N., Godschalk, P., Ang, W., Jacobs, B., Gilbert, M., et al. (2001). A Campylobacter jejuni gene associated with immune-mediated neuropathy. Nat. Med. 7, 752–753. doi: 10.1038/89831
van Schaik, W., Top, J., Riley, D. R., Boekhorst, J., Vrijenhoek, J. E., Schapendonk, C. M., et al. (2010). Pyrosequencing-based comparative genome analysis of the nosocomial pathogen Enterococcus faecium and identification of a large transferable pathogenicity island. BMC Genomics 11:239. doi: 10.1186/1471-2164-11-239
Velayudhan, J., Castor, M., Richardson, A., Main-Hester, K. L., and Fang, F. C. (2007). The role of ferritins in the physiology of Salmonella enterica sv. Typhimurium: a unique role for ferritin B in iron-sulphur cluster repair and virulence. Mol. Microbiol. 63, 1495–1507. doi: 10.1111/j.1365-2958.2007.05600.x
Velayudhan, J., Hughes, N. J., McColm, A. A., Bagshaw, J., Clayton, C. L., Andrews, S. C., et al. (2000). Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter. Mol. Microbiol. 37, 274–286. doi: 10.1046/j.1365-2958.2000.01987.x
Vitovec, J., Aldova, E., Vladik, P., and Krovacek, K. (2001). Enteropathogenicity of Plesiomonas shigelloides and Aeromonas s in experimental mono-and coinfection with Cryptosporidium parvum in the intestine of neonatal BALB/c mice. Compar. Immunol. Microbiol. Infect. Dis. 24, 39–55. doi: 10.1016/S0147-9571(00)00012-6
Waack, U., Johnson, T. L., Chedid, K., Xi, C., Simmons, L. A., Mobley, H. L., et al. (2017). Targeting the type II secretion system: development, optimization, and validation of a high-throughput screen for the identification of small molecule inhibitors. Front. Cell. Infect. Microbiol. 7:380. doi: 10.3389/fcimb.2017.00380
Wang, K., and Liang, C. (2017), CRF: detection of CRISPR arrays using random forest. PeerJ 5:e3219. doi: 10.7717/peerj.3219
Watson, R. J., Millichap, P., Joyce, S. A., Reynolds, S., and Clarke, D. J. (2010). Research article The role of iron uptake in pathogenicity and symbiosis in Photorhabdus luminescens TT01. BMC Microbiol. 10:177. doi: 10.1186/1471-2180-10-177
Wei, H. F., Wu, P. H., Chas, T. Y., Chen, K. T., Hwang, D. F., and Horng, C. B. (1994). A snail poisoning outbreak in Fangliao, Pintung County. Epidemiol. Bull. 10, 115–122.
Wong, K. K., McClelland, M., Stillwell, L. C., Sisk, E. C., Thurston, S. J., and Saffer, J. D. (1998). Identification and sequence analysis of a 27-kilobase chromosomal fragment containing a Salmonella pathogenicity island located at 92 minutes on the chromosome map of Salmonella enterica serovar Typhimurium LT2. Infect. Immun. 66, 3365–3371.
Wong, T. Y., Tsui, H. Y., So, M. K., Lai, J. Y., Lai, S. T., Tse, C. W., et al. (2000). Plesiomonas shigelloides infection in Hong Kong: retrospective study of 167 laboratory-confirmed cases. Hong Kong Med. J. 6, 375–380.
Xia, F. Q., Liu, P. N., and Zhou, Y. H. (2015). Meningoencephalitis caused by Plesiomonas shigelloides in a Chinese neonate: case report and literature review. Ital. J. Pediatr. 41:1. doi: 10.1186/s13052-014-0107-1
Yeh, T. J., and Tsai, W. C. (1991). Plesiomonas shigelloides–associated diarrhea. Zhonghua Chin. Med. J. 47, 362–368.
Yosef, I., Goren, M. G., and Qimron, U. (2012). Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 40, 5569–5576. doi: 10.1093/nar/gks216
Young, G. M., Badger, J. L., and Miller, V. L. (2000). Motility is required to initiate host cell invasion by Yersinia enterocolitica. Infect. Immun. 68, 4323–4326. doi: 10.1128/IAI.68.7.4323-4326.2000
Young, G. M., Schmiel, D. H., and Miller, V. L. (1999). A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc. Natl. Acad. Sci. U.S.A. 96, 6456–6461. doi: 10.1073/pnas.96.11.6456
Zajc-Satler, J., Dragas, A. Z., and Kumelj, M. (1972). Morphological and biochemical studies of 6 strains of Plesiomonas shigelloides isolated from clinical studies. Zentralbl. Bakteriol. Hyg. [Orig.] 219, 514–521.
Keywords: Plesiomonas shigelloides, virulence factor, antibiotic resistance, bioinformatics, pathogenicity island
Citation: Ekundayo TC and Okoh AI (2018) Pathogenomics of Virulence Traits of Plesiomonas shigelloides That Were Deemed Inconclusive by Traditional Experimental Approaches. Front. Microbiol. 9:3077. doi: 10.3389/fmicb.2018.03077
Received: 08 May 2018; Accepted: 29 November 2018;
Published: 21 December 2018.
Edited by:
George Tsiamis, University of Patras, GreeceReviewed by:
Jesús Santos, Universidad de León, SpainNikolaos Remmas, Democritus University of Thrace, Greece
Copyright © 2018 Ekundayo and Okoh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Temitope C. Ekundayo, cyruscyrusthem@gmail.com
 Temitope C. Ekundayo
Temitope C. Ekundayo Anthony I. Okoh
Anthony I. Okoh